Abstract
Mild to moderate equine asthma syndrome (mEAS) affects horses of all ages and breeds. To date, the etiology and pathophysiology of mEAS are still active areas of research, and it remains incompletely understood whether mEAS horses with different immune cell ‘signatures’ on BAL cytology represent different phenotypes, distinct pathobiological mechanisms (endotypes), varied environmental conditions, disease severity, genetic predispositions, or all of the above. In this descriptive study, we compared gene expression data from BAL cells isolated from horses with normal BALF cytology (n=5), to those isolated from horses with mild/moderate neutrophilic inflammation (n=5), or mild/moderate mastocytic inflammation (n=5). BAL cell protein lysates were analyzed using Multiplex Bead Immunoassay to evaluate cytokine/chemokine levels, and using immunoblot to evaluate levels of select proteins, on 11 out of 15 horses used for RNA-seq. The transcriptome, determined by RNA-seq and analyzed with DEseq2, contained 20, 63, and 102 significantly differentially expressed genes in horses with normal vs. neutrophilic, normal vs. mastocytic, and neutrophilic vs. mastocytic BALF cytology, respectively. Pathway analyses revealed that BAL-isolated cells from horses with neutrophilic vs. normal cytology showed enrichment in inflammation pathways, and horses with mastocytic vs. normal cytology showed enrichment in pathways involved in fibrosis and allergic reaction. BAL cells from horses with mastocytic mEAS, compared to neutrophilic mEAS, showed enrichment in pathways involved in alteration of tissue structures. Cytokine analysis determined that IL-1β was significantly different in the lysates from horses with neutrophilic inflammation compared to those with normal or mastocytic BAL cytology. Immunoblot revealed significant difference in the relative level of MMP2 in horses with neutrophilic vs. mastocytic mEAS. Upregulation of mRNA transcripts involved in the IL-1 family cytokine signaling axis (IL1a, IL1b, and IL1R2) in neutrophilic mEAS, as well as KIT mRNA in mastocytic mEAS, are novel, potentially clinically relevant, findings of this study. These findings further inform our understanding of inflammatory cell subtypes in mEAS.
Keywords: Equine asthma syndrome, heaves, inflammatory airway disease, recurrent airway obstruction, asthma phenotype, gene expression, RNA-seq, BAL
1. Introduction
Mild/moderate equine asthma (mEAS), previously referred to as inflammatory airway disease (IAD), is an extremely common noninfectious respiratory disease that can affect horses of any age or breed (Couëtil et al., 2016). Clinical signs of mEAS are often subtle and/or variable and can include exercise intolerance, decreased racing performance and/or chronic, occasional coughing with no increase in respiratory rate or effort at rest (Bond et al., 2018; Couëtil et al., 2016; Lavoie et al., 2011). The lower airway inflammation of EAS can be diagnosed by bronchoalveolar lavage fluid (BALF) cytology. Advanced diagnostics such as pulmonary function testing are also used, although they are typically limited to veterinary referral or research facilities. Therefore, in the majority of cases, BALF cytology is the sole means to determine the inflammatory status of the lower airways in horses.
In both equine and human medicine, asthma is increasingly recognized as a complex and heterogenous disease in which the etiology and pathophysiology remain incompletely understood. In human asthma, phenotypic terms such as ‘atopic’, ‘non-atopic’, and ‘late-onset’ are used to describe various clinical presentations, while endotypes, such as ‘Th2-high’ and ‘Th2-low’, refer to the varied underlying pathobiological mechanisms of asthma (Braido et al., 2018). Recently, it has been proposed that the inflammatory cell signature of BALF cytology in horses with mEAS could be due to differences in asthma pathophysiology, and that understanding these differences could contribute to the development of specific therapeutic targets for horses with different phenotypes and/or endotypes of mEAS (Klier et al., 2015; RS., 2017). There are several subtypes/phenotypes of mEAS that have been proposed based on the inflammatory cell populations present in the BALF (Bedenice et al., 2008; Couëtil et al., 2016; Lavoie et al., 2011). Neutrophilic or mastocytic inflammation are most commonly observed in the BALF of horses with mEAS (Davis and Sheats, 2019). An eosinophilic phenotype is less common overall, but is described in horses less than 5 years of age (Ivester et al., 2014; Lavoie et al., 2011). Argument for these inflammatory cell subtypes of EAS is supported by studies that show cytokine profiles in BAL fluid supernatant tend to differ coincident with the immune cell “signature” of the BALF cytology (Beekman et al., 2012; Lavoie et al., 2011; Lavoie et al., 2001). Additional research is needed to investigate potential similarities and differences in asthma immunopathology in horses with different inflammatory cell profiles on BAL cytology, with the ultimate goal that a better understanding of disease mechanism could help identify targets for future therapies (Bond et al., 2018).
The objective of this study was to determine whether the BAL cells from horses with different inflammatory cell profiles of lower airway inflammation had differentially expressed genes and cell signaling pathways compared to BAL cells from horses with normal BAL cytology and no evidence of respiratory disease. We also sought to identify genes that were differentially expressed (DE) in BAL cells from horses with neutrophilic vs. mastocytic lower airway inflammation. Limited protein analysis of BAL cell lysates by Multiplex Bead Immunoassay and immunoblot adds additional insight to several of our findings.
2. Materials and Methods
2.1. Horses
The North Carolina State University Institutional Animal Care and Use Committee (IACUC #16–074-O) approved all procedures performed for the purposes of this study. Horses included in this study were university owned teaching animals that lived on pasture in the southeastern United States, received the same feed, and received no medications for the duration of this study. Each horse received a physical exam, clinical score, and a BAL. The clinical score rubric (Table S1) was previously described and included respiratory rate, cough score, nostril flare score, abdominal lift score, nasal discharge score, and lung auscultation score (Davis and Sheats, 2019). Fifteen horses were included in the RNA sequencing and DE analysis of this study. Samples from seven of these horses (4 normal, 1 neutrophilic, and 2 mastocytic) were chosen at random for Multiplex Bead Immunoassay; which also utilized samples from an additional eight horses (Table S2).
2.2. BALF Collection and Sample Processing
BALF collection was performed as described previously (Davis and Sheats, 2019). Briefly, horses were placed under standing sedation with detomidine (0.005–0.01 mg/kg IV) and butorphanol (0.02–0.04 mg/kg IV), and 300 total mL of warmed sterile saline solution was infused and re-aspirated, 150mL at a time, through a cuffed catheter. Procedure time took an average of 10 minutes and horses were monitored until they recovered from sedation (approximately 45 minutes). The pooled sample was submitted for 300 cell differential count by blinded clinical pathologists (see Supplemental Methods for explanation of additional validation of differential count). BALF cytology was used to assign horses to one of three groups: normal, mild/moderate neutrophilic inflammation or mild/moderate mastocytic inflammation. Differential count was performed on a cytospin preparation slide. A previously published criterion was utilized for BALF cytology classification (Davis and Sheats, 2019). Horses in the normal group had ≤ 6% neutrophils, ≤ 2% mast cells and ≤ 1% eosinophils, horses in mild/moderate neutrophilic group had 7–19% neutrophils but ≤ 2% mast cells and horses in mild/moderate mastocytic group had ≥ 3% mast cells but ≤ 6% neutrophils in the BALF (Bedenice et al., 2008; Lavoie et al., 2011).
2.3. Statistical Analysis
Physical examination results (except for sex) and BALF differential cell counts, multiplex immunoassay, and immunoblot results were analyzed by One-way ANOVA with Tukey’s post-hoc test. Chi-square test was performed to determine whether there was a significant difference in the sex distribution among groups. P<0.05 was considered statistically significant. All analyses were performed using GraphPad Prism (Version 8.0, GraphPad Software, La Jolla, CA).
2.4. RNA Isolation
RNA isolation and DNAse materials were obtained from QIAgen (Valencia, CA, USA). All samples were processed within an hour of collection. Following isolation, BAL cells were briefly centrifuged and supernatant was removed. RNA was then isolated from the BAL cells using an RNeasy Mini Kit with QIA shredder column homogenization per manufacturer’s protocol. An RNase-free DNAse set was used to perform DNase digestion on-column per manufacturer’s protocol. Following DNase digestion, RNA cleanup was performed using the RNeasy Mini Kit. The isolated RNA samples were kept frozen at −80°C for 8 to 18 months.
2.5. mRNA Sequences and Data Analysis
Messenger RNA sequencing via polyA selection was carried out (Illumina, San Diego, CA) by an external laboratory (Genewiz, Inc; South Plainfield, NJ). Quality and concentration of RNA samples were determined with Bioanalyzer RNA nanochip (Agilent 2100) (Table S3). Samples were expanded, with individual sample reads ranging from 23–42 M at 2 X150 bp, and sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. For read quality, a score of Q30 ≧ 80% was considered acceptable. The trimmed reads were mapped to the Equus caballus EquCab3.0 reference genome available on ENSEMBL using the STAR aligner v.2.5.2b. For mapping quality, % Total Mapped Reads ≧ 90% and % Unique Mapped Reads ≧ 80% were considered acceptable. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Only unique reads that fell within exon regions were counted. Using DESeq2, the raw counts were normalized using variance-stabilizing transformation and a comparison of gene expression between the defined groups of samples was performed (Love et al., 2014). The Wald test was used to generate p-values and log2 fold changes. After Benjamini-Hochberg false discovery correction, genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 were identified as differentially expressed genes for each comparison.
2.6. Quantitative RT-PCR
Quantitative RT-PCR was performed to further evaluate differential gene expression of three select genes: IL1b, MMP2 and KIT. For these assays we used RNA isolated from BAL cells of 2 horses with normal BALF cytology, 2 horses with mild/moderate neutrophilic inflammation and 2 horses with mild/moderate mastocytic inflammation (population 1), selected at random. Primers and probes were obtained from Invitrogen’s proprietary equine-specific gene expression assay database and have been validated by the company (Invitrogen, Thermo Fisher Scientific, Grand Island, NY, USA). All samples were assayed in triplicate and the mean was used for analysis. Fold change in mRNA level was determined using the ΔΔCt data analysis method with SDHA as an endogenous control (Livak and Schmittgen, 2001). For each gene, the directionality of normalized mRNA fold change, as determined by RNA-seq or qRT-PCR, was compared. See supplemental methods for additional information on the qRT-PCR protocol.
2.7. Pathway Analysis
Functional annotation was performed using Ingenuity Pathway Analysis (IPA, www.ingenuity.com) using predefined pathways and functional categories of the Ingenuity Knowledge Base. Fisher’s exact test was applied to identify significantly enriched differentially expressed genes as members of pathways and functional categories. Relevant gene regulatory networks were identified using the Ingenuity Knowledge Base.
2.8. Multiplex Bead Immunoassay sample collection and analysis
Protein lysates prepared from BAL cells were utilized for cytokine protein analysis. Cell supernatants were collected and stored at −80°C. Protein analysis was carried out using an equine-specific Milliplex® Map Magnetic Bead Panel (EMD Millipore, Billerica, MA, USA) per manufacturer’s protocol (see Supplemental Methods for additional information). Mean fluorescence intensity data using five-parameter logistic standard curve was used to calculate analyte concentration, and the results were normalized to total cell counts. Levels of 23 different cytokines and chemokines were assayed. Cytokines and chemokines included in the analysis were as follows: IL-1α (interleukin-1α), IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12 (p70), IL-13, IL-17A, IL-18, IFNγ (interferon γ), TNFα (tumor necrosis factor α), FGF-2 (fibroblast growth factor-2), Eotaxin (CCL11), G-CSF (granulocyte-stimulating factor), GM-CSF (granulocyte monocyte-stimulating factor), GRO (growth-regulated protein or CXCL1), MCP-1 (monocyte chemotactic protein-1 or CCL2), fractalkine (CX3CL1), RANTES (CCL5) and IP-10 (IFNγ-induced protein 10).
2.9. Immunoblot
Samples were resolved by gel electrophoresis using MES running buffer (Thermo Scientific™) and transferred to Immobilon-P PVDF membrane (Millipore, Billerica, MA, USA) with NuPAGE™ Transfer Buffer (Thermo Scientific™). Following blocking with 5% nonfat dry milk in TBS/T, membranes were incubated overnight with anti-MMP2, anti-CD117/c-KIT, or anti-beta actin antibodies at 4°C. Following incubation with the appropriate HRP-conjugated secondary antibody for 1 hour at room temperature, the membranes were developed using enhanced chemiluminescence (ChemiDoc™ XRS+ System, Bio-Rad, Hercules, CA, USA) for 5 minutes at room temperature and imaged using the ChemiDoc MP imager (Bio-Rad). The Band Analysis tool of ImageLab software version 6.0.1 (Bio-Rad) was used to determine the background-subtracted density of the bands. Relative expression levels of proteins of interest were determined by dividing the relative density of the protein band of interest by the relative density of the corresponding beta-actin loading control band, and expressed as arbitrary units. See supplemental methods for the complete immunoblot protocol.
3. Results
3.1. Physical Examination and BALF Cytology
The horses included in this study ranged in age from 6 to 20 years and were of mixed breed and gender (Table 1, Table S2). No horses had abnormal respiratory signs at rest. Five horses had normal BALF cytology, five horses had mild/moderate neutrophilic inflammation and five horses had mild/moderate mastocytic inflammation (Table 1, Table S2). One-way ANOVA with Tukey’s post-hoc test revealed significant difference in the % neutrophils in mild/moderate neutrophilic group compared to normal and mild/moderate mastocytic groups (p<0.0001 for both), and significant difference in the % mast cells in mastocytic group compared to normal and mild/moderate neutrophilic group (p=0.002 and p=0.0012, respectively). Along with other measured parameters, BAL cellularity (total cell count/uL) was not significantly different among the groups.
Table 1.
(a) Clinical characteristics and BALF cytology results of horses included in the RNA-seq experiment. P values on the tables represent ANOVA p values. (b) Clinical characteristics and BALF cytology results of horses included in the Multiplex Bead Immunoassay.
| (a) | Normal | Mild/moderate neutrophilic | Mild/moderate mastocytic | p value |
|---|---|---|---|---|
| Number of horses | 5 | 5 | 5 | |
| Mean (range) age, years | 14 (9–20) | 10.8 (6–19) | 11.8 (7–18) | 0.5752 |
| Sex, mare/gelding | 5/1 | 4/1 | 2/3 | 0.092 |
| Mean (range) resting respiratory | ||||
| rate | 16.8 (12–20) | 16 (12–24) | 14.4 (12–20) | 0.638 |
| Mean (range) clinical score | 1.2 (0–3) | 1.08 (0–1) | 0.8 (0–1) | 0.2814 |
| Mean (range) total cell count, /uL | 419.2 (310–588) | 439.8 (150–663) | 286 (208–346) | 0.2186 |
| Neutrophils, mean (range) % | 3.68 (2.7–5) | 11.62 (9–16.3)a,b | 2.72 (1–5) | <0.0001 |
| Mast cells, mean (range) % | 1.44 (0–2.3) | 1.2 (0–2)a,b | 5.06 (3.3–8) | 0.0007 |
| Eosinophils, mean (range) % | 0.52 (0–1) | 0.44 (0–1.7) | 0.06 (0–0.3) | 0.3406 |
| Macrophages, mean (range) % | 49.82 (42.3–57.3) | 41.54 (37–45) | 48.48 (39–60.3) | 0.1632 |
| Lymphocytes, mean (range) % | 44.54 (38.7–49.7) | 45.2 (40–53) | 43.46 (35.3–54) | 0.8942 |
| (b) | Normal | Mild/moderate neutrophilic | Mild/moderate mastocytic | p value |
| Number of horses | 6 | 4 | 5 | |
| Mean (range) age, years | 14 (9–20) | 12.25 (9–21) | 11.2 (6–18) | 0.6548 |
| Sex, mare/gelding | 6/0 | 3/1 | 2/3 | 0.0809 |
| Mean (range) resting respiratory rate | 18.67 (12–28) | 17 (12–24) | 16 (12–20) | 0.6696 |
| Mean (range) clinical score | 1.33 (1–3) | 0.5 (0–2) | 0.4 (0–1) | 0.1445 |
| Mean (range) total cell count, /uL | 351 (240–588) | 334.5 (235–443) | 196.6 (118–346) | 0.0744 |
| Neutrophils, mean (range) % | 3.9 (1.7–6.3) | 12.53 (7–17.3)a,b | 2.74 (1.7–5) | 0.0003 |
| Mast cells, mean (range) % | 1.63 (0.3–2.3) | 1.53 (0.7–2) | 6.18 (3.6–9)a, c | 0.0003 |
| Eosinophils, mean (range) % | 0.38 (0–1) | 0.38 (0–1) | 0.26 (0–7) | 0.854 |
| Macrophages, mean (range) % | 46.4 (37–57.3) | 51.4 (34.3–66) | 50.16 (39–58.4) | 0.689 |
| Lymphocytes, mean (range) % | 47.67 (38.7–56.3) | 34.18 (16–47.7) | 40.66 (30.3–47.7) | 0.0961 |
different from normal
different from mastocytic
different from neutrophilic. All p-values were calculated with One-way ANOVA test except for sex, which was calculated with Chi-square test. The bold values are p values were there was a statistically significant difference.
3.2. mRNA Sequencing and Differential Gene Expression
All RNA samples met minimum quality control criteria established by the sequencing laboratory (Table S3). All samples yielded data with ≧ 80% Q30 quality scores, % Total Mapped Reads ≧ 90% and % Unique Mapped Reads ≧ 80%. On average, 29,618,357 total reads were analyzed per sample. The raw read data sets for each sample are available in the NCBI sequence Read Archive (SRA accession: PRJNA589665).
Gene expression comparison between horses with normal BAL cytology vs. horses with mild/moderate neutrophilic inflammation identified 14,328 unique genes. Of those, 20 genes were significantly differentially expressed; 18 were upregulated genes, while 2 were downregulated (Table 2, Table 3, Figure S1).
Table 2.
Number of differentially expressed genes in three comparison groups (P adjusted ≤ 0.05).
| Normal vs. Neutrophilic | Normal vs. Mastocytic | Neutrophilic vs. Mastocytic | |
|---|---|---|---|
| Number of upregulated genes | 18 | 60 | 80 |
| Number of downregulated genes | 2 | 3 | 22 |
| Total number of differentially expressed genes | 20 | 63 | 102 |
Table 3.
Top 20 differentially expressed genes in the comparison between BAL cells from samples with no vs. neutrophilic mild/moderate lower airway inflammation as ranked by adjusted p value.
| Gene Name | Normalized Mean Expression Value | log2FoldChange | P adjusted |
|---|---|---|---|
| IL1A• | 601.7532302 | 1.466654334 | 7.14E-26 |
| IL1B | 1531.549148 | 1.680370875 | 1.08E-12 |
| SOCS3 | 928.6570283 | 1.03059484 | 2.00E-11 |
| MEFV | 93.60427726 | 1.826504204 | 8.62E-06 |
| CA12‡ | 231.4487655 | 1.463863307 | 0.000109 |
| HRH1 | 106.4905373 | 1.665822377 | 0.000255 |
| CXCL2 | 1110.789335 | 1.105310608 | 0.00283 |
| LOC111768766 | 184.8342195 | 2.221595825 | 0.003553 |
| KCNJ2 | 1012.908394 | 1.544264791 | 0.003918 |
| KCNJ15 | 437.9658698 | 1.058194198 | 0.004821 |
| PLA2G2D | 543.1491738 | 1.509280014 | 0.005019 |
| CRISPLD2‡ | 134.3474915 | 1.472082729 | 0.006033 |
| TGM3 | 149.8147891 | 1.285760594 | 0.007896 |
| LOC100058291‡ | 313.0407757 | 1.057526648 | 0.007896 |
| IL1R2 | 37.2824749 | 1.426952822 | 0.013795 |
| LOC111768069 | 17.7114675 | −3.585449383 | 0.013795 |
| LOC102149298 | 66.80223464 | 1.103784651 | 0.015772 |
| A2ML1 | 50.36311547 | 2.120189457 | 0.016433 |
| LOC111773283‡ | 72.98146484 | −2.253909193 | 0.019715 |
| LOC100061252 | 109.8851836 | 6.11209782 | 0.034616 |
indicates transcript is also differentially expressed in normal vs. mastocytic
indicates transcript is also differentially expressed in neutrophilic vs. mastocytic.
Gene expression comparison between horses with normal BAL cytology vs. horses with mild/moderate mastocytic inflammation identified 14,331 unique genes. Of those, 63 genes were significantly differentially expressed; 60 were upregulated genes, while 3 were downregulated (Table 2, Table 4, Supplemental Table S4, Figure S1).
Table 4.
Top 20 differentially expressed genes in the comparison between BAL cells from samples with no vs. mastocytic mild/moderate lower airway inflammation as ranked by adjusted p value.
| Gene Name | Normalized Mean Expression Value | log2FoldChange | P adjusted |
|---|---|---|---|
| LOC100069290‡ | 476.8198255 | 1.441352317 | 1.35E-09 |
| HDC‡ | 196.0091624 | 1.755953953 | 1.37E-06 |
| FBLN2‡ | 86.68946323 | 1.926733234 | 1.17E-05 |
| ATP8B1 | 43.66733507 | 2.376097341 | 1.22E-05 |
| MS4A2 | 758.3826438 | 2.03602185 | 1.22E-05 |
| BDKRB2 | 33.04048547 | 1.822029257 | 1.22E-05 |
| FSTL1 | 32.91000323 | 1.679590784 | 1.22E-05 |
| KIT‡ | 797.552028 | 1.906031326 | 2.21E-05 |
| DSCAML1‡ | 47.67390766 | 2.048198364 | 0.000143 |
| PTPRM‡ | 51.81351726 | 2.209788787 | 0.000159 |
| LOC102149826‡ | 29.61753678 | 2.616200332 | 0.00017 |
| PODXL | 64.76425103 | 1.94288855 | 0.00022 |
| SOX13‡ | 95.100782 | 1.762004081 | 0.00022 |
| LOC100060011 | 38.36159103 | 1.48841687 | 0.00022 |
| MAST4‡ | 33.69569004 | 2.461504003 | 0.00027 |
| STX3‡ | 53.69883963 | 1.690978348 | 0.000296 |
| PTGS1‡ | 138.7606042 | 1.692464108 | 0.000407 |
| PLCB4‡ | 80.77665148 | 1.835501008 | 0.000488 |
| COL23A1‡ | 35.56291541 | 2.861169681 | 0.000556 |
| TAL1‡ | 59.7161359 | 1.866790726 | 0.000649 |
indicates transcript is also differentially expressed in normal vs. neutrophilic
indicates transcript is also differentially expressed in neutrophilic vs. mastocytic. A complete list of DEG is in Table S4.
Gene expression comparison between horses with mild/moderate neutrophilic inflammation vs. mild/moderate mastocytic inflammation identified 14,401 unique genes. Of those, 102 genes were significantly differentially expressed; 80 were upregulated, while 22 were downregulated (Table 2, Table 5, Supplemental Table S5, Figure S1).
Table 5.
Top 20 differentially expressed genes in the comparison between BAL cells from samples with neutrophilic vs. mastocytic mild/moderate lower airway inflammation as ranked by adjusted p value.
| Gene Name | Normalized Mean Expression Value | log2FoldChange | P adjusted |
|---|---|---|---|
| LOC100069290• | 505.9593753 | 1.4608234 | |
| SCN3B• | 120.1143648 | −2.6518194 | 4.01E-13 |
| LOC100629802 | 321.9237115 | −8.1917093 | 1.35E-12 |
| HDC• | 226.9187589 | 1.2973479 | 4.28E-08 |
| RGS13 | 78.25494936 | 1.38684148 | 1.55E-05 |
| CBLN3 | 133.0952636 | −3.2494852 | 3.37E-05 |
| COL23A1• | 37.9219877 | 2.87296169 | 3.75E-05 |
| FBLN2• | 96.50730284 | 1.65514937 | 6.64E-05 |
| LOC100063525 | 38.54942578 | 5.66089589 | 0.000132 |
| TMEM35B | 188.207381 | −1.5496595 | 0.000207 |
| LTC4S | 192.4608192 | 1.57097899 | 0.000222 |
| C1H10orf128 | 98.8223954 | 1.621126 | 0.00023 |
| TRPC6 | 35.47032587 | 1.63006794 | 0.00026 |
| RET | 1211.0403 | 1.03017536 | 0.000283 |
| CD1A6 | 37.39546429 | 1.94542597 | 0.000349 |
| PTGS1• | 152.9107785 | 1.50526771 | 0.00042 |
| LOC102149751• | 210.6348848 | 1.38781634 | 0.00042 |
| CHRM4• | 79.64067396 | 2.26087112 | 0.000494 |
| LOC100057298 | 165.627884 | 1.57776757 | 0.000517 |
| STEAP4 | 169.5686494 | −1.4026596 | 0.000537 |
indicates also transcript is also differentially expressed in normal vs. mastocytic
indicates transcript is also differentially expressed in normal vs. neutrophilic. A complete list of DEG is in Table S5.
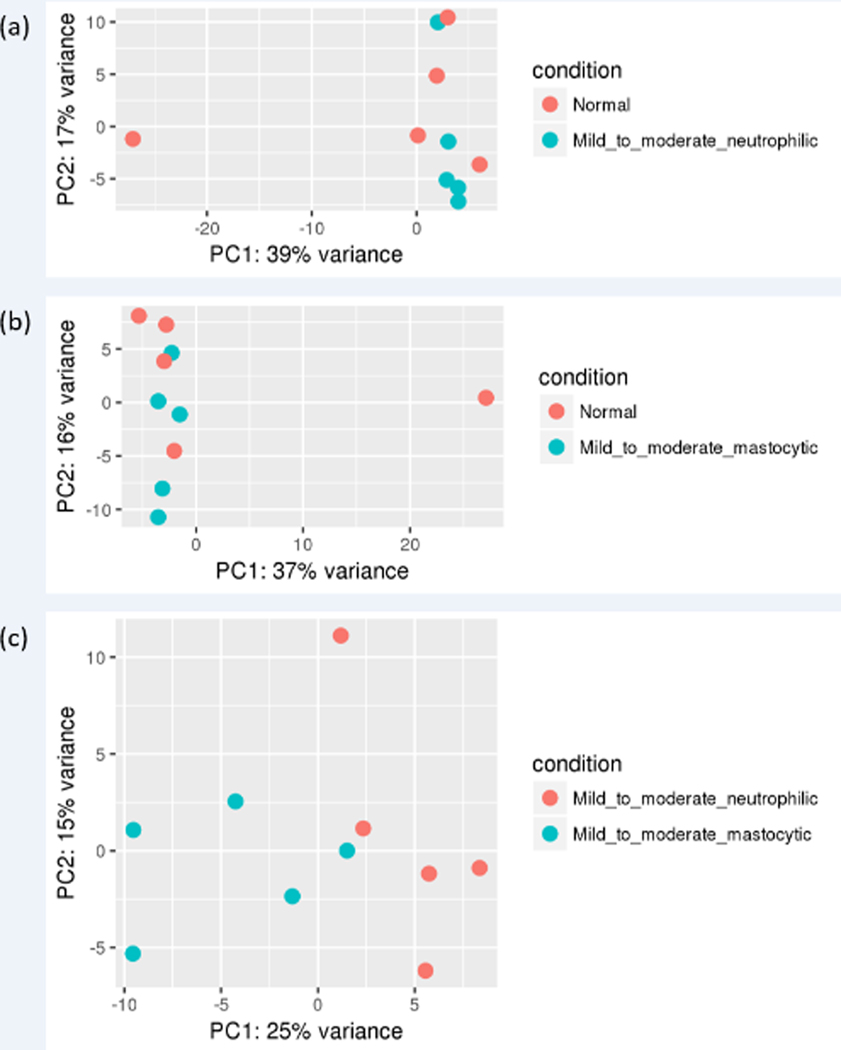
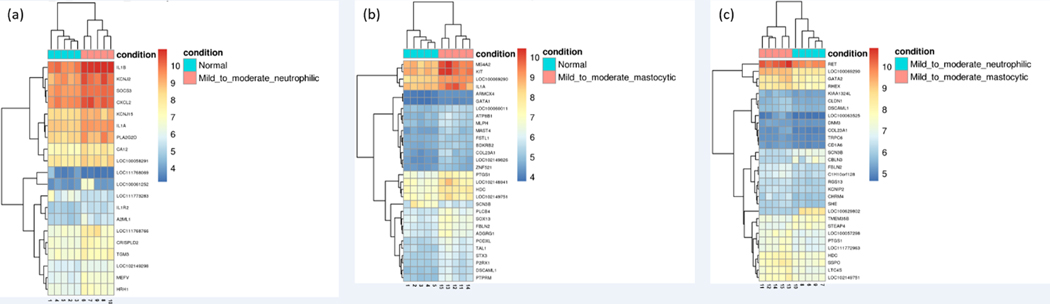
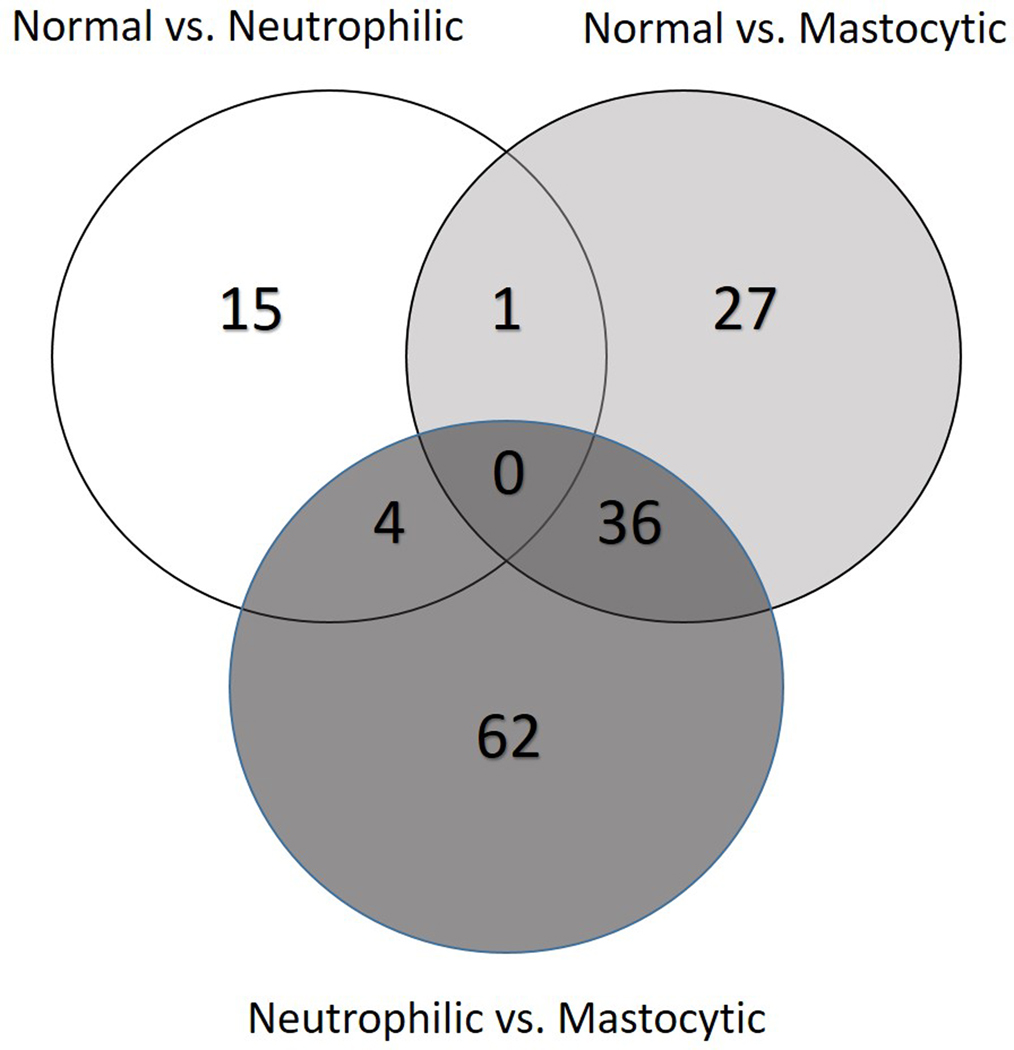
PCA plot of differential gene expression shows some overlap of individuals with different inflammatory cell subtypes of mEAS with normal horses, as well as a potential outlier in the normal group (Figure 1). Overall, gene expression profiles for the top 30 DE genes in BAL cells from horses with the same inflammatory cell profile were similar (Figure 2). A comparison of unique vs. overlapping DEG in the 3 groups identified 36 DEG in common between normal vs. mastocytic and neutrophilic vs. mastocytic results, 1 DEG in common between normal vs. mastocytic and normal vs. neutrophilic, and 4 DEG in common between normal vs. neutrophilic and neutrophilic vs. mastocytic comparison (Figure 3). There were no DEG in common for all three comparison groups (Figure 3).
Figure 1.
Principal component analysis plots of RNA-seq data from BAL cells isolated from horses with normal vs. neutrophilic (a), normal vs. mastocytic (b), and neutrophilic vs. mastocytic (c), BAL cytology. These plots show the characteristics of samples according to gene expression (FPKM) levels. FPKM, fragments per kilobase of transcript per million mapped reads.
Figure 2.
Bi-clustering heat map of top statistically significant differentially expressed genes between normal vs. neutrophilic (a), normal vs. mastocytic (b), and neutrophilic vs. mastocytic (c). Color intensity was normalized to log2 (fragments per kilobase of transcript per million mapped reads +1), with red colors representing more highly expressed genes and blue colors representing less highly expressed genes. Hierarchial clustering is based on the Manhattan distance among samples and is intended to highlight relative differences in expression between groups.
Figure 3.
Venn diagram of the three sets of differentially expressed genes between the three comparisons: ‘Normal vs. Neutrophilic’, ‘Normal vs. Mastocytic’, and ‘Neutrophilic vs. Mastocytic’. The three overlapping sets shared between two contrasts varied from 1 to 36 genes (Table 2, S4, S5).
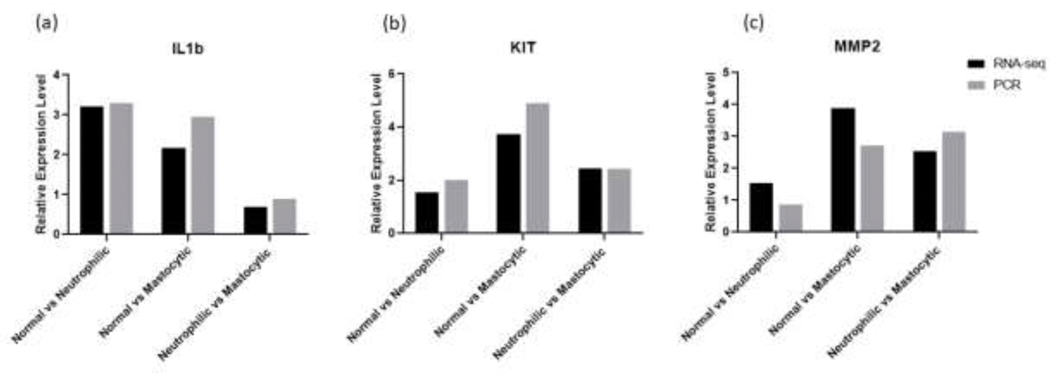
3.3. Quantitative RT-PCR
Three gene transcripts identified as differentially expressed by RNAseq, IL1β, MMP2, and KIT, were also examined by qRT-PCR. Results from horses with normal (n=2), neutrophilic (n=2), or mastocytic (n=2) BAL cytology yielded similar fold change for the three examined transcripts (Figure 4). Figure S2 shows the normalized expression of these genes for each individual in pairwise comparisons. These data can also be found in Table S6(a-c). Figure S3 shows the ΔCt values of the 3 genes for each individual horse. The qRT-PCR data show a 3.3, and 3.0, fold increase in IL1β mRNA expression in mild/moderate neutrophilic and mild/moderate mastocytic groups, compared to normal, respectively. Compared to normal, there was a 2.0, and 4.9, fold increase in KIT mRNA expression in mild/moderate neutrophilic and mastocytic groups, respectively. There was a 0.9, and 2.7, fold increase in MMP2 mRNA expression in mild/moderate neutrophilic and mastocytic groups compared to normal, respectively. Across the 6 samples, the endogenous control SDHA had a mean, standard deviation (SD) and coefficient of variation (CV) of threshold cycles of 22.9373112 (mean), 0.52724221 (SD), and 2.29862255 (CV) (Table S7). Log 2 scale of the relative expression levels of the three target genes compared to the reference gene are shown in Table S8.
Figure 4.
Fold change of IL1b (a), KIT (b) and MMP2 (c) mRNA, determined via RNA-seq differential gene expression analysis (black bars, n=5 per group) and qRT-PCR (white bars, n=2 per group) performed on BAL cell RNA samples. For RNA-seq, Log2fold change was calculated as Log2 (Group 2 mean normalized counts/Group 1 mean normalized counts). For the qRT-PCR assays, relative quantification was calculated as dCT with equine succinate dehydrogenase complex, subunit A, flavoprotein (SDHA) used as an endogenous control, and ΔΔCT was used to calculate fold change.
3.4. Pathway Analysis
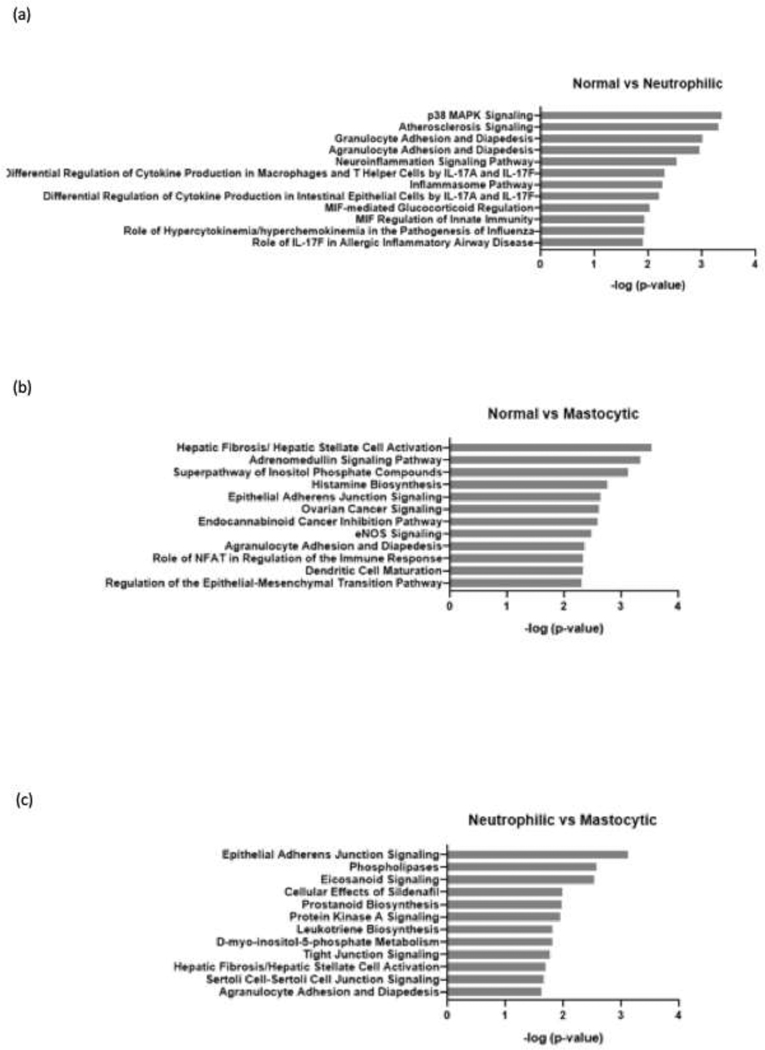
Use of the Ingenuity Knowledge Base to analyze differentially expressed genes revealed that compared with normal horses, horses with mild/moderate neutrophilic inflammation had enrichment in inflammation pathways such as p38 MAPK signaling, cell adhesion and diapedesis, and cytokine production. Compared to normal horses, horses with mild/moderate mastocytic inflammation showed enrichment in pathways that are involved in fibrosis and allergic reaction. Additionally, when compared to horses with mild/moderate neutrophilic inflammation, horses with mild/moderate mastocytic inflammation showed enrichment in pathways that are involved in tissue remodeling such as epithelial adherens junction signaling, tight junction signaling, and fibrosis (Figure 4).
3.5. Protein Analysis
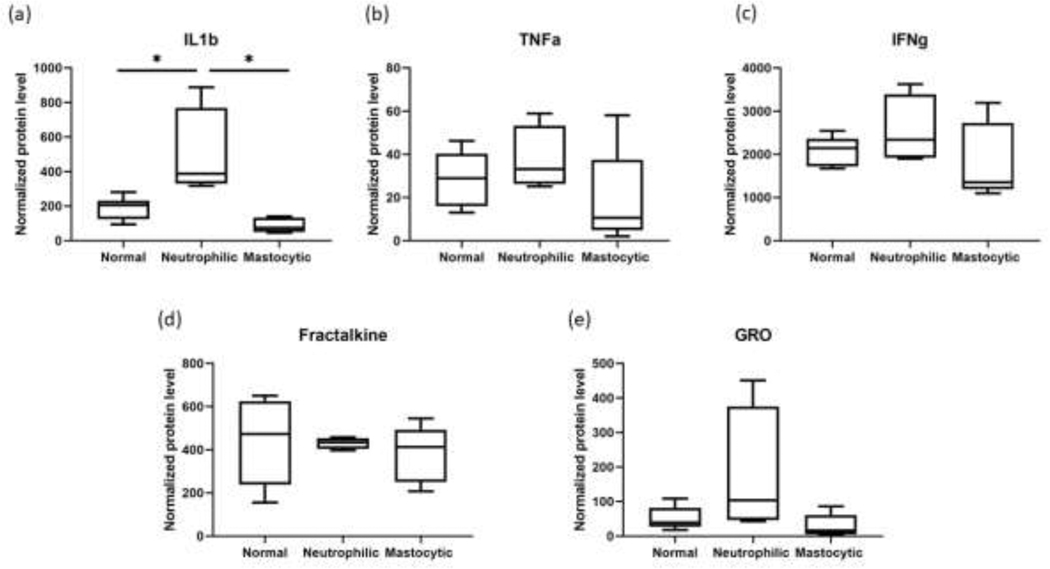
Samples from 15 horses were used for Multiplex Bead Immunoassay analysis of BAL protein lysate. Of those, 6 horses had normal BALF cytology, 4 horses had mild/moderate neutrophilic inflammation, and 5 horses had mild/moderate mastocytic inflammation. Most proteins in the assay kit were below the lower limit of detection. Those that were detected included IL-1β, TNFα, IFNγ, Fractalkine, and GRO/CXCL1. One horse in the mastocytic group had TNFα concentration that was below the lower limit of detection (2.7 pg/mL). Another horse in the mastocytic group had GRO concentration that was below the detection limit (4.42 pg/mL). For those samples with results below the detection limit, a value less than the detection limit (2.0 pg/mL for TNFα and 4.0 pg/mL for GRO) was assigned for statistical analysis. There was a statistically significant difference in the IL-1β level in the lysates from horses with mild/moderate neutrophilic inflammation compared to mild/moderate mastocytic horses. There was a trend towards increased TNFα, IFNγ and GRO levels in mild/moderate neutrophilic group compared to normal and mild/moderate mastocytic groups (Figure 5).
Figure 5.
Summary of pathway analysis for differentially expressed genes. The differentially expressed genes were compared between normal vs. neutrophilic phenotype (a), normal vs. masotytic phenotype (b), and neutrophilic vs. mastocytic phenotype (c). Functional annotation was performed using Ingenuity Pathway Analysis using predefined pathways and functional categories of the Ingenuity Knowledge Base. Fisher’s exact test was applied to identify significantly enriched differentially expressed genes as members of pathways and functional categories. Relevant gene regulatory networks were identified using the Ingenuity Knowledge Base.
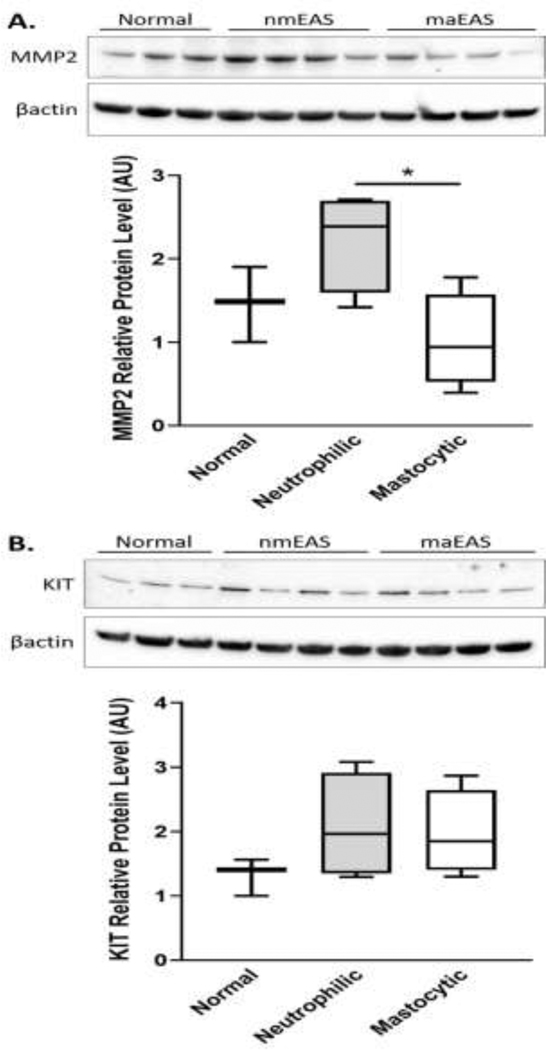
BAL cell protein lysates from 11 of the 15 horses used for RNA-seq were also analyzed by immunoblot for relative levels of MMP2 and KIT proteins. There was a significant difference in the relative level of MMP2 protein detected in BAL cell lysates from horses with neutrophilic mEAS vs. mastocytic mEAS inflammation (Figure 6).
Figure 6.
IL-1β(a), TNFα(𝑏), IFNγ (c), Fractalkine (d) and GRO (e) levels in BAL cell lysates determined by Multiplex Bead Immunoassay. 6 normal horses, 4 mild/moderate neutrophilic horses and 4 mild/moderate mastocytic horses were sampled. Protein concentrations normalized to total protein concentrations were averaged and compared between groups shown on the x-axis.
4. Discussion
In horses, several categories of mEAS inflammation have been identified based on the immune cells featured in the BALF cytology, including neutrophilic, mastocytic, eosinophilic, and mixed inflammation (Bond et al., 2018). However, it remains unclear whether the inflammatory cell signature on BAL cytology informs mEAS phenotype, endotype, or both. In this study, our goal was to determine which genes and molecular pathways were differentially expressed in horses with different inflammatory cell ‘signatures’ on BAL cytology, as this could inform our understanding of both unique and shared disease immunopathology and/or therapeutic targets.
We performed DE analysis on three different pairwise comparisons of horses that were grouped as ‘normal’, ‘neutrophilic mEAS’, or ‘mastocytic mEAS’ based on BALF cytology. All horses were members of a teaching animal herd housed on adjacent pastures. Despite being housed on pasture, the majority of these horses had BAL cytology consistent with mEAS of varying inflammatory cell subtypes. Potential triggers in the environment of these horses include year-round exposure to round-bale hay, environmental pollution from a pasture-adjacent roadway, and allergens associated with pasture. DE analyses identified 20, 63, and 102 DE genes (Table 2) between horses with no vs. neutrophilic (Table 3), no vs. mastocytic (Table 4, Table S4), and neutrophilic vs. mastocytic inflammation (Table 5, Table S5), respectively.
Of the 20 significant DEG in the normal vs. neutrophilic comparison, only 1 was also DE in the normal vs. mastocytic comparison (Figure 3, Table 2, 3, Table S4). While this result suggests a consistent role for IL1A in mEAS lower airway inflammation regardless of cell signature, it also suggests that the top DEG in mEAS differ with the type of inflammation present. The normal vs. neutrophilic and neutrophilic vs. mastocytic comparisons shared 4 DEG, while the normal vs. mastocytic and neutrophilic vs. mastocytic comparisons shared 36 DEG (Figure 3, Table 2, S4, S5). Interestingly, the neutrophilic vs. mastocytic comparison identified 62 DEG that were not identified in the other comparisons (Figure 3, Table S5). In addition to the normal vs. cell specific comparisons, these results may help to identify cell signaling and cellular targets unique to the inflammatory cell subtypes of mEAS. Further work is needed to determine whether these genes could provide insight into differences in the pathophysiology of neutrophilic vs. mastocytic mEAS.
Previous findings suggest that Th1 and Th2 immune responses, as well as IL-17, are implicated in mild/moderate EAS (Bond et al., 2018). Transcripts for four of the classic T helper cytokines, IL-4, IL-9, IL-13, and IL-2, as well as IL-17, were not identified at all in our RNAseq results, presumably due to lack of mRNA transcript in the sample. Th2 cytokines reported included IL-5, and Th1 cytokines reported included IFNγ and TNFα. While expression of these cytokine transcripts was not significantly different between groups, there was a trend towards increased TNFα in both mastocytic and neutrophilic samples compared to normal (consistent with Multiplex results), and a trend towards increased IL-5 in neutrophilic vs. normal. Possible reasons for the lack of significant findings include small sample size, mild clinical phenotype, and differences in cytokine mRNA vs. protein levels. Future studies to address cytokine profiles in mEAS inflammatory cell subtypes would benefit from simultaneous analysis of cytokine mRNA in both bronchial epithelial and BAL isolated cells, as well as cytokine protein levels in BALF supernatant.
In the comparison between normal vs. neutrophilic BALs, several transcripts of interest were upregulated, including CXCL2, IL1β, IL1α, IL1R2, and LOC100058291 mRNA. CXCL2 protein, also known as macrophage inflammatory protein 2-alpha (MIP2-α) or Growth-regulated protein beta (Gro-β), is a small cytokine that is primarily produced by macrophages, but also neutrophils and mast cells, and is chemotactic for neutrophils. Previous findings by Deluca et al. reported increased CXCL2 expression in BAL cells from horses challenged with stabling and hay feeding (DeLuca et al., 2008). LOC100058291 mRNA, which is a predicted transcript variant for Equus caballus CXC chemokine receptor type 2, is a receptor for both CXCL2 and IL8. IL8 has already been identified as playing a significant role in the pathophysiology of severe EAS (Tessier et al., 2017). In a mouse model, CXCR2 signaling was identified as playing a key role in IL-17A/IL33-induced neutrophilic inflammation and airway hyperresponsiveness, and inhibition of CXCR2 with anti-CXCR2 mAb attenuated early-phase airway obstruction and subsequent Th2 cytokine responses (Mizutani et al., 2014). Interleukin-1, comprised of subtypes IL1α and IL1β, is a proinflammatory cytokine with diverse effects on both innate and adaptive immune responses. While IL1α and IL1β are encoded by different genes, they have similar biological activities and activate the same cell surface receptors (Dinarello, 2018). BAL cell IL1R2 (Interleukin-1 receptor type 2) mRNA was also significantly upregulated in horses with neutrophilic mEAS compared to normal horses, in our study. This is consistent with a recent report by Hansen et al. (Hansen et al., 2020). IL1R2, also known as IL1RA, is a non-signaling receptor that acts as an anti-inflammatory member of the IL-1 family. This receptor competes with IL1R1 (Interleukin 1 receptor type 1) for IL-1α/β binding, and because it is a “dummy” receptor, it reduces the proinflammatory activity of IL1 cytokines (Dinarello, 2018). IL1β has been repeatedly linked to lower airway inflammation in horses with and without clinical signs (Beekman et al., 2012; Hughes et al., 2011; Joubert et al., 2011; Lavoie et al., 2011; Padoan et al., 2013). Upregulation and expression of anti-inflammatory components of the IL-1 family could have a role in balancing IL-1α/β mediated inflammation in the lower airway. Research into both CXCR2 and IL-1 signaling as potential therapeutic targets in inflammatory diseases such as asthma is ongoing (Al-Alwan et al., 2013; Osei et al., 2019).
Our results show that BAL cells from horses with mastocytic inflammation have increased expression of MMP2, and KIT mRNA, compared to horses with no inflammation. Several previous studies evaluated matrix metalloproteinases (MMPs) in equine BALF samples. Similar to our results, Koivunen et al. report increased concentrations of MMP2 in tracheal aspirates from horses with sEAS compared to those from healthy controls (Koivunen et al., 1997). A positive correlation between BALF MMP2 concentration and stable dust concentrations in horses has also been reported (Nevalainen et al., 2002; Simonen-Jokinen et al., 2005). Unlike other differentially expressed genes highlighted in our study, our finding that KIT is upregulated in BAL cells from horses with mastocytic mEAS is novel. KIT is a proto-oncogene which encodes a receptor tyrosine kinase, located at the cell surface of certain cell types, including mast cells (Da Silva et al., 2006). KIT plays an essential role in mast cell homeostasis, and both experimental animal models and human clinical trial data support KIT/c-kit as a potential therapeutic target for asthma. Using a mouse asthma model, Wu et al. report that intranasal delivery of siRNA nanoparticles targeting c-kit reduced airway mucus secretion and the infiltration of eosinophils in bronchoalveolar lavage fluid (Wu et al., 2014). Cahill et al. report that treatment of patients with severe, poorly controlled asthma with imatinib, a KIT-receptor inhibitor, decreased airway hyperresponsiveness, mast-cell counts, and tryptase release, compared with placebo (Cahill et al., 2017). Further investigations are warranted to determine whether KIT/c-kit may be a potential novel therapeutic target of mastocytic equine asthma.
Compared to horses with neutrophilic inflammation, horses with mastocytic inflammation had increased transcript levels of RET and LOC100069290 mRNA, and decreased transcript levels CRISPLD2 mRNA. “Rearranged duration translation” (RET) is a proto-oncogene that encodes a transmembrane receptor tyrosine kinase RET. This receptor is expressed in lung tissues in immune and epithelial cells and is preferentially activated by neurotrophic factor neurturin (NTN) (Mauffray et al., 2015). NTN signaling through RET was recently shown to decrease IL-6 and TNF-α by immune and epithelial cells (Mauffray et al., 2015). Authors of this research suggest that NTN may be useful as a novel anti-inflammatory treatment for asthma. LOC100069290 is a predicted transcript variant for Equus caballus caspase-12 (XM_023644724.1, NCBI), analogous to the human caspase 12 gene/pseudogene, but the predicted amino acid sequence shares homology with human caspase-1 (NP_001244047.1, NCBI). Caspase 1 is known to be involved in cytokine maturation, and caspase 12 has been linked with inflammatory/innate immune response and sepsis risk in African Americans (Saleh et al., 2004). CRISPLD2 mRNA encodes for a cysteine-rich secretory protein known as Cysteine-rich secretory protein LCCL domain-containing 2. CRISPLD2 mRNA expression and protein levels are increased in primary human airway smooth muscle (ASM) cells following dexamethasone treatment and IL1β stimulation. Further, siRNA knockdown of CRISPLD2 in ASM cells increased IL1β, IL6, and IL8 (Himes et al., 2014). Due to these effects on inflammatory cytokines, CRISPLD2 is being investigated as an asthma pharmacogenetics candidate gene.
In addition to RNA-seq, we performed two different methods of protein analysis of BAL cell lysates from horses with normal, mild/moderate neutrophilic, and mild/moderate mastocytic lower airway inflammation. Multiplex Bead Immunoassay revealed a significant difference in the IL-1β protein level in the BAL cell lysates from horses with mild/moderate neutrophilic inflammation compared to normal horses and horses with mild/moderate mastocytic inflammation. These findings are consistent with the RNA-seq data, which showed a significant difference in the IL1β gene expression level in the mild/moderate neutrophilic group compared to normal group. Interestingly, the multiplex bead immunoassay also showed a significant difference in the IL-1β protein level in the mild/moderate neutrophilic group vs. the mild/moderate mastocytic group, although there was no significant difference in the IL1β gene expression level between these groups. IL-1β is initially translated as the inactive, pre-cursor protein pro-IL-1β, that must be cleaved into a mature form (Dinarello, 2018). Intracellularly, caspase-1 mediates IL-1β cleavage and activation. Alternatively, extracellular proteases produced by both neutrophils (i.e. elastase and cathepsin G) and mast cells (i.e. chymase) are also capable of cleaving pro-IL-1β into a biologically active form (Afonina et al., 2015). Increased levels of secreted IL-1β in BALF supernatant, without increased transcription of IL1β mRNA, could be explained by increased activity of proteases in horses with increased airway neutrophils. Multiplex also showed a trend towards increased GRO/CXCL1 in the neutrophilic group. CXCL1, a chemokine produced by both macrophages and TNF-stimulated endothelial cells, works in concert with CXCL2 to promote neutrophil diapedesis (Girbl et al., 2018). Although increases in CXCL1 in BALF supernatant did not achieve significance, this trend is consistent with the upregulation of CXCL2/GRO mRNA identified in horses with neutrophilic mEAS.
Based on immunoblot, the mean relative level of MMP2 protein was significantly different in BAL cell lysates from horses with neutrophilic vs. mastocytic mEAS. This observed increase in MMP2 in horses with neutrophilic inflammation is consistent with previous reports of increased MMP2 and MMP9 protein expression and gelatinolytic activity in BALF supernatant in horses with both mild and severe neutrophilic EAS (Barton et al., 2015). Interestingly, our DE analysis and immunoblot findings offer conflicting results regarding MMP2 in BAL cells from horses with mastocytic inflammation. While analysis of RNA-seq indicated an increase in MMP2 mRNA expression in BAL cells from mastocytic vs. normal horses, immunoblot results show a trend towards decreased mean protein levels of MMP2 in mastocytic vs. normal horses. MMPs are zinc-dependent proteolytic enzymes that contribute to airway inflammation by degrading extracellular matrix. Because these proteins are secreted by white blood cells, the discrepancy between the mRNA and protein expression data could be explained by MMP2 protein levels in the BALF supernatant, which were not examined in this study. Alternatively, enhanced mast cell accumulation and activation could lead to increased secretion of MMP2, causing a decreased amount of protein within BAL cells. Mast cell tryptase and chymase have been implicated in the regulation of MMP-2 and −9 (Iddamalgoda et al., 2008; Tchougounova et al., 2005). It is therefore possible that horses with increases in airway mast cells have increased tryptase and/or chymase activity, which could lead to activation and subsequent release of MMPs from other immune cells (i.e. lymphocytes and macrophages). Simultaneous evaluation of BAL cell lysate and supernatant proteins would further inform our understanding of the different immunopathologies that could be contributing to mEAS in horses with different inflammatory cell phenotypes.
Similar to MMP2, conclusions from our KIT protein immunoblot data are also limited. Despite a trend towards increasing, mean KIT protein levels in BAL cell lysates from horses with neutrophilic and mastocytic inflammation compared to normal were not significantly different; while mRNA differential expression analysis showed a 4.9 fold change in the KIT mRNA expression in mild/moderate mastocytic horses compared to normal horses. It is possible that our ability to measure KIT by immunoblot was limited by protein stability. The signaling of the KIT cell surface receptor is modulated in vivo both by protein phosphatases, and by rapid internalization and degradation of the receptor. Even though protease and phosphatase inhibitors were added to our BAL samples as soon as they were collected, KIT protein in our samples could have degraded, particularly in the samples containing mast cells, which are prone to degranulate when manipulated.
In addition to the DE analysis, our results using Ingenuity Pathway Analysis software also highlighted enrichment in distinct pathways associated with different BALF cytology profiles. BAL cells from horses with neutrophilic inflammation showed enrichment in pathways involved in inflammatory reactions, such as p38 MAPK signaling, granulocyte adhesion and diapedesis, and cytokine signaling pathways. BAL cell from horses with mastocytic inflammation showed enrichment in pathways involved in tissue structure alteration or remodeling, such as fibrosis, epithelial adherens junction signaling, tight junction signaling and epithelial-mesenchymal transition pathway (Figure 4). These IPA results are consistent with previous reports documenting the significant role of mast cell-derived mediators in the pathogenesis of airway remodeling. Mediators such as tryptase and chymase are known to induce proliferation of fibroblasts, airway smooth muscle cells, endothelial cells, and airway epithelial cells, and degradation of the basement membrane components (Okayama et al., 2007). Our IPA results are also consistent with reports that neutrophils both respond to and perpetuate a cycle of airway inflammation during asthma. In neutrophilic asthma, inflammatory cytokines (i.e. CXCR2) and bacterial derived triggers (i.e. fMLP) activate neutrophil p38 MAPK, which regulates neutrophil chemotaxis by controlling surface expression of receptors including CXCR1, leading to increased airway neutrophils (Kim and Haynes, 2013). It is important to point out that IPA analyses of our data are limited by the fact that several of the top DE genes we identified had no associated HGNC symbol. Additional investigation is needed to further characterize these potentially relevant genes.
One of the limitations of this study was that BAL cytology reference ranges used to group horses into inflammatory cell subtypes of mEAS are not a definitive diagnosis. While the reference ranges used for normal and abnormal cytology have been previously published, debate remains regarding which cytology values should be used to determine normal vs. abnormal. Therefore, we suggest that our findings regarding the molecular mechanisms associated with inflammatory cell subtypes in mild/moderate Equine Asthma Syndrome should be viewed as initial, rather than definitive, findings. In futures studies, additional diagnostics, including airway endoscopy and/or pulmonary function testing, could help to further define subtypes/phenotypes, and potentially even endotypes, of EAS (Kuruvilla et al., 2019).
Another limitation of the data presented here is the small sample size. Differential expression analysis based on a small sample size can be greatly impacted by each individual’s RNAseq data. RNAseq data is in turn affected by factors including RNA integrity at the sample and individual transcript level, methods of normalization, as well as inclusion of samples that do not truly represent the condition and treatment (Soneson and Delorenzi, 2013). Methods to avoid potential sampling errors in analysis of RNAseq data have been described (Son et al., 2018). Because the RNA integrity of our samples was high (i.e. RIN>8.0), bias in gene expression levels due to poor transcript integrity is unlikely (Gallego Romero et al., 2014). Gene expression principal component analysis of our RNAseq data did identify an outlier in our normal group of horses. While it is unclear whether this individual had a significant impact on results of our DE analysis, this individual met the definitions of our study criteria for “normal BAL cytology” and therefore had no reason to be excluded.
Finally, because horses with mEAS do not have clinical signs at rest, mEAS is arguably a disease of performance horses, and our study used samples from non-performance horses. However, it is reasonable to think that non-performance horses can experience the same underlying immunopathology of mEAS as performance horses. While the sedentary lifestyle of horses in our study precluded our ability to know whether they were performance limited, they were determined to have normal or abnormal BAL cytology per previously published guidelines (Couëtil et al., 2016). Mild to moderate changes in BAL cytology have repeatedly been associated with airway hyperreactivity and performance impacts (Ivester et al., 2018; Rossi et al., 2018; Secombe et al., 2019). Due to lack of pulmonary function testing equipment, we were unable to investigate possible alterations in airway function in horses in our study. However, the similarities between our findings and previously identified equine asthma mediators, such as CXCL2, MMP2, IL1R1, and IL1β mRNA, and MMP2, and TNFα protein, support the relevance of our study population for investigating mEAS immunopathology, and lend credibility to our more novel findings. An important follow up to the current study will be to determine whether novel transcripts and proteins, which we identified as differentially expressed in neutrophilic and mastocytic airway inflammation, are also altered in performance horses with mEAS.
5. Conclusion
In this study, horses with the same cellular subtypes of mEAS (neutrophilic or mastocytic) had similar expression patterns of top DEG. Additionally, the top DEG in the neutrophilic subtype showed very little overlap with the top DEG in the mastocytic subtype. Many of the genes identified as differentially expressed in this population of horses with inflammatory BAL cytology have been previously implicated in asthma pathophysiology in human and/or mouse studies and warrant further investigation. Upregulation of mRNA transcripts involved in the IL-1 family cytokine signaling axis (IL1α, IL1β, and IL1R2) in BAL cells from horses with neutrophilic mEAS, as well as KIT mRNA in BAL cells from horses with mastocytic mEAS, may offer novel, inflammatory-cell specific targets for future investigation. Whether these findings are consistent or relevant in other populations of horses with mEAS remains to be determined.
Supplementary Material
Figure 7.
MMP2 (a) and KIT (b) immunoblot results from horses with normal BALF cytology (n=3), neutrophilic inflammation (n=4), and mastocytic inflammation (n=4). Box-and-whisker plot (arbitrary units) for the relative protein levels of MMP2 and KIT in BAL cells, respectively. *p<0.05.
6. Acknowledgments
We thank Ms. Jenna Schirmer for assistant with the BAL procedures, and Mr. Brian Williams for assistant with the qRT-PCR procedures.
Footnotes
Conflict of interest
The authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonina IS, Müller C, Martin SJ, Beyaert R, 2015. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity 42, 991–1004. [DOI] [PubMed] [Google Scholar]
- Al-Alwan LA, Chang Y, Mogas A, Halayko AJ, Baglole CJ, Martin JG, Rousseau S, Eidelman DH, Hamid Q, 2013. Differential roles of CXCL2 and CXCL3 and their receptors in regulating normal and asthmatic airway smooth muscle cell migration. J Immunol 191, 2731–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AK, Shety T, Bondzio A, Einspanier R, Gehlen H, 2015. Metalloproteinases and Their Tissue Inhibitors in Comparison between Different Chronic Pneumopathies in the Horse. Mediators Inflamm 2015, 569512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedenice D, Mazan MR, Hoffman AM, 2008. Association between cough and cytology of bronchoalveolar lavage fluid and pulmonary function in horses diagnosed with inflammatory airway disease. J Vet Intern Med 22, 1022–1028. [DOI] [PubMed] [Google Scholar]
- Beekman L, Tohver T, Dardari R, Léguillette R, 2011. Evaluation of suitable reference genes for gene expression studies in bronchoalveolar lavage cells from horses with inflammatory airway disease. BMC Mol Biol 12, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman L, Tohver T, Léguillette R, 2012. Comparison of cytokine mRNA expression in the bronchoalveolar lavage fluid of horses with inflammatory airway disease and bronchoalveolar lavage mastocytosis or neutrophilia using REST software analysis. J Vet Intern Med 26, 153–161. [DOI] [PubMed] [Google Scholar]
- Bond S, Léguillette R, Richard EA, Couetil L, Lavoie JP, Martin JG, Pirie RS, 2018. Equine asthma: Integrative biologic relevance of a recently proposed nomenclature. J Vet Intern Med 32, 2088–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braido F, Tiotiu A, Kowal K, Mihaicuta S, Novakova P, Oguzulgen IK, 2018. Phenotypes/endotypes-driven treatment in asthma. Curr Opin Allergy Clin Immunol 18, 184–189. [DOI] [PubMed] [Google Scholar]
- Cahill KN, Katz HR, Cui J, Lai J, Kazani S, Crosby-Thompson A, Garofalo D, Castro M, Jarjour N, DiMango E, Erzurum S, Trevor JL, Shenoy K, Chinchilli VM, Wechsler ME, Laidlaw TM, Boyce JA, Israel E, 2017. KIT Inhibition by Imatinib in Patients with Severe Refractory Asthma. N Engl J Med 376, 1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Waldenström J, 2015. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS One 10, e0141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couëtil LL, Cardwell JM, Gerber V, Lavoie JP, Léguillette R, Richard EA, 2016. Inflammatory Airway Disease of Horses--Revised Consensus Statement. J Vet Intern Med 30, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva CA, Reber L, Frossard N, 2006. Stem cell factor expression, mast cells and inflammation in asthma. Fundam Clin Pharmacol 20, 21–39. [DOI] [PubMed] [Google Scholar]
- Davis KU, Sheats MK, 2019. Bronchoalveolar Lavage Cytology Characteristics and Seasonal Changes in a Herd of Pastured Teaching Horses. Front Vet Sci 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca L, Erb HN, Young JC, Perkins GA, Ainsworth DM, 2008. The effect of adding oral dexamethasone to feed alterations on the airway cell inflammatory gene expression in stabled horses affected with recurrent airway obstruction. J Vet Intern Med 22, 427–435. [DOI] [PubMed] [Google Scholar]
- Dinarello CA, 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev 281, 8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulkerson PC, 2017. Transcription Factors in Eosinophil Development and As Therapeutic Targets. Front Med (Lausanne) 4, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pai AA, Tung J, Gilad Y, 2014. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbl T, Lenn T, Perez L, Rolas L, Barkaway A, Thiriot A, Del Fresno C, Lynam E, Hub E, Thelen M, Graham G, Alon R, Sancho D, von Andrian UH, Voisin MB, Rot A, Nourshargh S, 2018. Distinct Compartmentalization of the Chemokines CXCL1 and CXCL2 and the Atypical Receptor ACKR1 Determine Discrete Stages of Neutrophil Diapedesis. Immunity 49, 1062–1076.e1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S, Otten ND, Birch K, Skovgaard K, Hopster-Iversen C, Fjeldborg J, 2020. Bronchoalveolar lavage fluid cytokine, cytology and IgE allergen in horses with equine asthma. Vet Immunol Immunopathol 220, 109976. [DOI] [PubMed] [Google Scholar]
- Himes BE, Jiang X, Wagner P, Hu R, Wang Q, Klanderman B, Whitaker RM, Duan Q, Lasky-Su J, Nikolos C, Jester W, Johnson M, Panettieri RA, Tantisira KG, Weiss ST, Lu Q, 2014. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One 9, e99625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KJ, Nicolson L, Da Costa N, Franklin SH, Allen KJ, Dunham SP, 2011. Evaluation of cytokine mRNA expression in bronchoalveolar lavage cells from horses with inflammatory airway disease. Vet Immunol Immunopathol 140, 82–89. [DOI] [PubMed] [Google Scholar]
- Iddamalgoda A, Le QT, Ito K, Tanaka K, Kojima H, Kido H, 2008. Mast cell tryptase and photoaging: possible involvement in the degradation of extra cellular matrix and basement membrane proteins. Arch Dermatol Res 300 Suppl 1, S69–76. [DOI] [PubMed] [Google Scholar]
- Ivester KM, Couëtil LL, Moore GE, 2018. An observational study of environmental exposures, airway cytology, and performance in racing thoroughbreds. J Vet Intern Med 32, 1754–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivester KM, Couëtil LL, Moore GE, Zimmerman NJ, Raskin RE, 2014. Environmental exposures and airway inflammation in young thoroughbred horses. J Vet Intern Med 28, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert P, Cordeau ME, Lavoie JP, 2011. Cytokine mRNA expression of pulmonary macrophages varies with challenge but not with disease state in horses with heaves or in controls. Vet Immunol Immunopathol 142, 236–242. [DOI] [PubMed] [Google Scholar]
- Kim D, Haynes CL, 2013. The role of p38 MAPK in neutrophil functions: single cell chemotaxis and surface marker expression. Analyst 138, 6826–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klier J, Lehmann B, Fuchs S, Reese S, Hirschmann A, Coester C, Winter G, Gehlen H, 2015. Nanoparticulate CpG immunotherapy in RAO-affected horses: phase I and IIa study. J Vet Intern Med 29, 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen AL, Maisi P, Konttinen YT, Sandholm M, 1997. Gelatinolytic activity in tracheal aspirates of horses with chronic obstructive pulmonary disease. Acta Vet Scand 38, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla ME, Lee FE, Lee GB, 2019. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin Rev Allergy Immunol 56, 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie JP, Cesarini C, Lavoie-Lamoureux A, Moran K, Lutz S, Picandet V, Jean D, Marcoux M, 2011. Bronchoalveolar lavage fluid cytology and cytokine messenger ribonucleic Acid expression of racehorses with exercise intolerance and lower airway inflammation. J Vet Intern Med 25, 322–329. [DOI] [PubMed] [Google Scholar]
- Lavoie JP, Maghni K, Desnoyers M, Taha R, Martin JG, Hamid QA, 2001. Neutrophilic airway inflammation in horses with heaves is characterized by a Th2-type cytokine profile. Am J Respir Crit Care Med 164, 1410–1413. [DOI] [PubMed] [Google Scholar]
- Leroy P, Tham A, Wong H, Tenney R, Chen C, Stiner R, Balmes JR, Paquet AC, Arjomandi M, 2015. Inflammatory and repair pathways induced in human bronchoalveolar lavage cells with ozone inhalation. PLoS One 10, e0127283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CC, Aronow B, Hutton J, Santeliz J, Dienger K, Herman N, Finkelman FD, Wills-Karp M, 2009. Unique and overlapping gene expression patterns driven by IL-4 and IL-13 in the mouse lung. J Allergy Clin Immunol 123, 795–804.e798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall J, Akdis CA, Bacharier LB, Bjermer L, Casale TB, Custovic A, Lemanske RF, Wardlaw AJ, Wenzel SE, Greenberger PA, 2011. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol 127, 355–360. [DOI] [PubMed] [Google Scholar]
- Mauffray M, Domingues O, Hentges F, Zimmer J, Hanau D, Michel T, 2015. Neurturin influences inflammatory responses and airway remodeling in different mouse asthma models. J Immunol 194, 1423–1433. [DOI] [PubMed] [Google Scholar]
- Miki Y, Kidoguchi Y, Sato M, Taketomi Y, Taya C, Muramatsu K, Gelb MH, Yamamoto K, Murakami M, 2016. Dual Roles of Group IID Phospholipase A2 in Inflammation and Cancer. J Biol Chem 291, 15588–15601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani N, Nabe T, Yoshino S, 2014. IL-17A promotes the exacerbation of IL-33-induced airway hyperresponsiveness by enhancing neutrophilic inflammation via CXCR2 signaling in mice. J Immunol 192, 1372–1384. [DOI] [PubMed] [Google Scholar]
- Nevalainen M, Raulo SM, Brazil TJ, Pirie RS, Sorsa T, McGorum BC, Maisi P, 2002. Inhalation of organic dusts and lipopolysaccharide increases gelatinolytic matrix metalloproteinases (MMPs) in the lungs of heaves horses. Equine Vet J 34, 150–155. [DOI] [PubMed] [Google Scholar]
- Nigo YI, Yamashita M, Hirahara K, Shinnakasu R, Inami M, Kimura M, Hasegawa A, Kohno Y, Nakayama T, 2006. Regulation of allergic airway inflammation through Toll-like receptor 4-mediated modification of mast cell function. Proc Natl Acad Sci U S A 103, 2286–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama Y, Ra C, Saito H, 2007. Role of mast cells in airway remodeling. Curr Opin Immunol 19, 687–693. [DOI] [PubMed] [Google Scholar]
- Osei ET, Brandsma CA, Timens W, Heijink IH, Hackett TL, 2019. Current perspectives on the role of Interleukin-1 signaling in the pathogenesis of Asthma and COPD. Eur Respir J. [DOI] [PubMed] [Google Scholar]
- Padoan E, Ferraresso S, Pegolo S, Castagnaro M, Barnini C, Bargelloni L, 2013. Real time RT-PCR analysis of inflammatory mediator expression in recurrent airway obstruction-affected horses. Vet Immunol Immunopathol 156, 190–199. [DOI] [PubMed] [Google Scholar]
- Rossi H, Virtala AM, Raekallio M, Rahkonen E, Rajamäki MM, Mykkänen A, 2018. Comparison of Tracheal Wash and Bronchoalveolar Lavage Cytology in 154 Horses With and Without Respiratory Signs in a Referral Hospital Over 2009–2015. Front Vet Sci 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RS P, 2017. RAO/SPRAOD - severe equine asthma? World Equine Airway Symposium. Copenhagen, Denmark: University of Copenhagen. [Google Scholar]
- Saleh M, Vaillancourt JP, Graham RK, Huyck M, Srinivasula SM, Alnemri ES, Steinberg MH, Nolan V, Baldwin CT, Hotchkiss RS, Buchman TG, Zehnbauer BA, Hayden MR, Farrer LA, Roy S, Nicholson DW, 2004. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature 429, 75–79. [DOI] [PubMed] [Google Scholar]
- Secombe CJ, van Eps AW, Bruce M, Lester GD, 2019. The relationship between bronchoalveolar lavage fluid cytology and airway hyper-reactivity in a population of Australian horses presented for poor performance. Aust Vet J 97, 343–350. [DOI] [PubMed] [Google Scholar]
- Seki Y, Inoue H, Nagata N, Hayashi K, Fukuyama S, Matsumoto K, Komine O, Hamano S, Himeno K, Inagaki-Ohara K, Cacalano N, O’Garra A, Oshida T, Saito H, Johnston JA, Yoshimura A, Kubo M, 2003. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nat Med 9, 1047–1054. [DOI] [PubMed] [Google Scholar]
- Sharma S, Ghosh B, 2009. Promoter polymorphism in the MS4A2 gene and asthma in the Indian population. Int Arch Allergy Immunol 149, 208–218. [DOI] [PubMed] [Google Scholar]
- Simonen-Jokinen T, Pirie RS, McGorum BC, Maisi P, 2005. Effect of composition and different fractions of hay dust suspension on inflammation in lungs of heaves-affected horses: MMP-9 and MMP-2 as indicators of tissue destruction. Equine Vet J 37, 412–417. [DOI] [PubMed] [Google Scholar]
- Son K, Yu S, Shin W, Han K, Kang K, 2018. A Simple Guideline to Assess the Characteristics of RNA-Seq Data. Biomed Res Int 2018, 2906292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Delorenzi M, 2013. A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchougounova E, Lundequist A, Fajardo I, Winberg JO, Abrink M, Pejler G, 2005. A key role for mast cell chymase in the activation of pro-matrix metalloprotease-9 and pro-matrix metalloprotease-2. J Biol Chem 280, 9291–9296. [DOI] [PubMed] [Google Scholar]
- Tessier L, Côté O, Clark ME, Viel L, Diaz-Méndez A, Anders S, Bienzle D, 2017. Impaired response of the bronchial epithelium to inflammation characterizes severe equine asthma. BMC Genomics 18, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal CA, Riffel AK, Dai H, Rosenwasser LJ, Jones BL, 2013. Detecting gene expression in buccal mucosa in subjects with asthma versus subjects without asthma. Pediatr Allergy Immunol 24, 138–143. [DOI] [PubMed] [Google Scholar]
- Weathington N, O’Brien ME, Radder J, Whisenant TC, Bleecker ER, Busse WW, Erzurum SC, Gaston B, Hastie AT, Jarjour NN, Meyers DA, Milosevic J, Moore WC, Tedrow JR, Trudeau JB, Wong HP, Wu W, Kaminski N, Wenzel SE, Modena BD, 2019. BAL Cell Gene Expression in Severe Asthma Reveals Mechanisms of Severe Disease and Influences of Medications. Am J Respir Crit Care Med 200, 837–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Chen H, Li YM, Wang SY, Diao X, Liu KG, 2014. Intranasal sirna targeting c-kit reduces airway inflammation in experimental allergic asthma. Int J Clin Exp Pathol 7, 5505–5514. [PMC free article] [PubMed] [Google Scholar]
- Zafra MP, Mazzeo C, Gámez C, Rodriguez Marco A, de Zulueta A, Sanz V, Bilbao I, Ruiz-Cabello J, Zubeldia JM, del Pozo V, 2014. Gene silencing of SOCS3 by siRNA intranasal delivery inhibits asthma phenotype in mice. PLoS One 9, e91996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.