Abstract
Decellularized extracellular matrix (ECM) scaffolds are complex biomaterials derived from tissues and organs used in clinical and research applications. A number of decellularization protocols have been described for ECM biomaterials derivation, each adapted to a particular tissue and use, restricting comparisons among materials. One of the major sources of variability in ECM products comes from the tissue source and animal age. Although this variability could be minimized by using established tissue sources, other sources arise from the decellularization process itself. Overall, current protocols require manual work and are poorly standardized with regard to the choice of reagents, the order by which they are added, and exposure times. The combination of these factors add variability affecting the uniformity of the final product between batches. Furthermore, each protocol needs to be optimized for each tissue and tissue source making tissue-to-tissue comparisons difficult. Automation and standardization of ECM scaffold development constitute a significant improvement to current biomanufacturing techniques but remains poorly explored. This study aims to develop a biofabrication method for fast and automated derivation of raw material for ECM hydrogel production while preserving ECM composition and controlling lot-to-lot variability. The main result is a closed semi-batch bioreactor system with automated dosing of decellularization reagents capable of deriving ECM material from pre-treated soft tissues. The ECM was further processed into hydrogels to demonstrate gelation and cytocompatibility. This work presents a versatile, scalable, and automated platform for the rapid production of ECM scaffolds.
Keywords: Automation, Decellularization, Extracellular Matrix, Hydrogel, Biomaterial, Biofabrication
Graphical Abstract
1. INTRODUCTION
Limited availability of donor tissues and/or organs for allotransplantation combined with an increase in donor site morbidity risk associated with autografting, led to high demand for off-the-shelf tissue replacements.1 Extracellular matrix (ECM) biomaterials are complex, three-dimensional, heterogeneous networks of structural and functional proteins (e.g., collagens, elastins, fibronectins, laminins, etc.) that promote tissue-specific remodeling, repair, and regeneration at the site of implantation.2–3 ECM-based biomaterials are commonly derived from decellularized organs or tissues from various species (e.g., human, porcine, bovine, murine, etc.) derived following a process to remove cellular content while maintaining the integrity of the main features of the extracellular matrix.4 The ultimate goal is to produce a bioactive material to promote tissue remodeling while eliminating adverse immune reactions.5–6 ECM scaffolds can be derived from a variety of tissues such as hearts, vocal folds, lungs, skeletal muscle, pancreas, and dermis, resulting in scaffolds with unique compositions and structures.4,7 Porcine tissues are readily available given their ubiquitous use for human consumption and represent an abundant source of tissue for the derivation of ECM biomaterials such as hydrogels.8 ECM hydrogels can be used as both in vitro cell culture platforms as well as in vivo therapies (e.g., coatings, hybrids, embedded with cells and/or growth factors etc.).9
Moreover, ECM scaffolds are currently used for a variety of applications such as implantable or injectable materials, ECM-based bio-inks, and substrates for cell growth.8,10 Depending on the application, ECM scaffolds can be processed into various formulations such as single sheets, multi-laminated sheets, powders, and hydrogels.9 Many materials are in preclinical stages or are currently commercially available.11 Patches or sheets derived from skin, small intestine submucosa (SIS), and urinary bladder matrix (UBM) are commonly used in clinical settings that include burn wounds, diabetic ulcers, tendon repair, hernias, etc.12–13 However, ECM sheets require surgical access when implanted at the wound site and cannot be delivered via minimally invasive techniques.14 These limitations make injectable ECM hydrogels an attractive alternative for clinical use.
Existing decellularization protocols require prolonged exposure times for each chemical wash that could result in potential loss of important ECM components, ultimately leading to decreased bioactivity.15 Chemical agents (e.g. acids and bases, non-ionic and ionic detergents, etc.) and biological agents (e.g. enzymes and chelating agents, etc.) disrupt ECM ultrastructure and damage important ECM components such as collagen, glycosaminoglycans (GAGs), elastin, and laminin if utilized for extended periods of time.16 For example, sodium dodecyl sulfate (SDS), a commonly used decellularization reagent, can effectively remove cellular and nuclear content. However, SDS can also affect the overall ultrastructure of the ECM and damage collagens, GAGs, and growth factors.4 Another example includes sodium deoxycholate, which can disrupt the ECM ultrastructure and remove GAGs.17 These reagents are typically combined with tissue sheets and subjected to agitation and manual liquid changes. This approach can result in nonconformities in the homogeneity of the product due to the inherent variability of a manual decellularization process and high variability in the end product.
Currently, no standard decellularization protocols are available for all tissue types and each protocol requires optimization for the specific tissue source and configuration (sheet vs. whole organ decellularization).10,18 Depending on parameters such as pre- and post-decellularization processing steps, biofabrication method, tissue source, and tissue thickness, the final ECM product may have inconsistent physical and biochemical properties.19 These variables can affect standardization and reproducibility of the final product and represent challenges for the scale-up, commercialization, and the translation of ECM-based biomaterials to the clinic.2 Deficiencies in decellularization methods provide an opportunity for significant improvement to harness the full potential of ECM-based biomaterials. By optimizing factors such as manufacturing method, production costs, production time, and complexity of the manufacturing process, it is likely to decrease the time necessary to deliver standardized, high quality, and reproducible ECM materials to the clinic in a cost-effective manner.10, 20
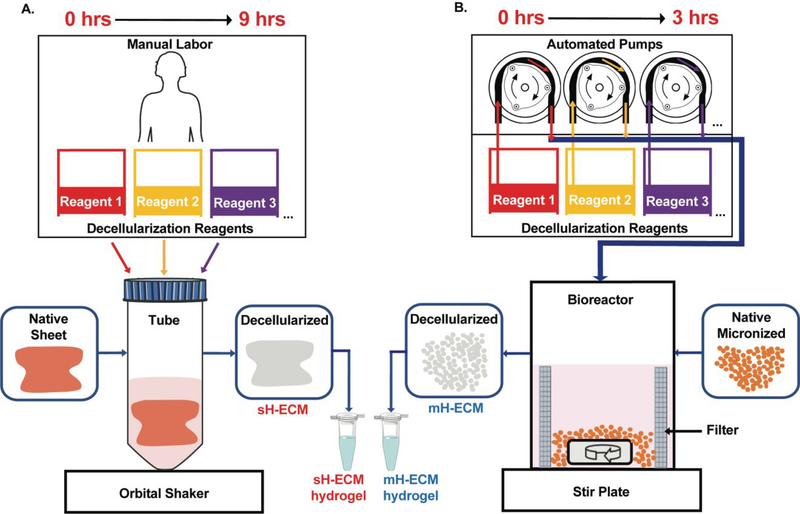
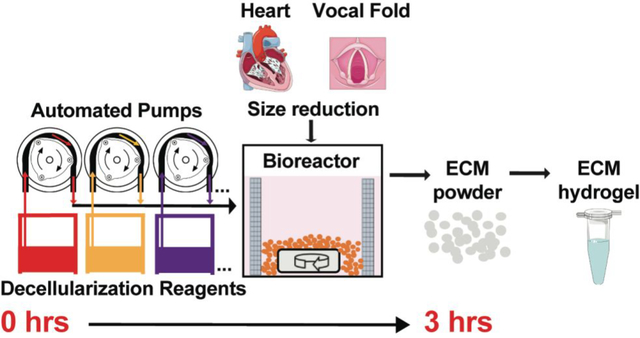
Our goal was to automate and optimize the decellularization process of representative soft tissues for ECM-based hydrogel production. Hydrogel and injectable forms of ECM scaffolds from different sources (e.g., human, porcine) are attractive as clinical materials given their ability to elicit constructive remodeling while being amenable to minimally invasive delivery methods.14 For example, ECM hydrogels have been extensively researched for the treatment of myocardial infarction, one of the most common causes of heart disease. As an example, a porcine derived myocardial ECM hydrogel completed phase I clinical trials and will soon proceed to phase II trials.11 Other target tissues for injectable ECM hydrogels include, but are not limited to, musculoskeletal tissues, neural tissues, liver, lung, kidney, and vocal fold (VF). The lamina propria (LP) layer of the vocal folds, involved in sound production, has a unique and rich ECM protein composition. Currently, no injectable biomaterials regenerate the vocal folds following injury. However, our group previously showed that a porcine derived injectable VFLP-ECM hydrogel can modulate the expression of ECM proteins associated with scar formation.21 As shown in Figure 1A, current methods are heavily dependent on user intervention, adding significant time and variability to the process. The present study describes a fast, controllable, and automated semi-batch system for the derivation of tissue specific ECM-based biomaterials in a micronized form for hydrogel production (Figure 1B). The automated decellularization process described significantly reduces the exposure time to reagents, minimizes lot-to-lot variability, and preserves the native ECM-protein composition. In this study we focus on the optimization and comparison between an automated and manual method for two different ECM scaffolds that are commonly targeted as injectable materials: heart and vocal fold derived hydrogels.
Figure 1. Overview.
A.) Currently available method for the decellularization of porcine heart sheets (sH-ECM).22 B.) Automated decellularization method with custom-made bioreactor connected to automated pumps for the decellularization of porcine micronized heart (mH-ECM).
2. EXPERIMENTAL SECTION
2.1. Tissue Decellularization
2.1.1. Tissue Dissection and Decellularization (Control)
Heart decellularization
Three porcine heart dissections and independent decellularizations were performed as previously described.22 Briefly, porcine hearts (Nahunta Pork Outlet, Raleigh, NC) were procured from market weight pigs, thoroughly cleaned of excess connective tissue, blood, and debris, and frozen at −80°C for at least 24 hours. The frozen heart was sliced into thin (2–3mm) sheets (sH) using a commercially available meat slicer. Random slices from throughout the entire porcine heart were selected for the manual decellularization method. The remaining portions of the heart were randomized, micronized, and collected in order to obtain a more uniform sample to be decellularized as described in section 2.1.2. The sheets were placed in 50mL tubes and treated with the following solutions under constant agitation on an orbital shaker: deionized (DI) water for 5 minutes, 2X DPBS (GeneClone, Raleigh, NC) for 15 minutes, 0.02% trypsin (Life Technologies, Carlsbad, CA) for 2 hours, DI water for 5 minutes, 2X DPBS for 15 minutes, 3% Tween-20 (Sigma-Aldrich, St. Louis, MO) for 2 hours, DI water for 5 minutes, 2X DPBS for 15 minutes, 4% w/v sodium deoxycholate (Sigma-Aldrich) for 2 hours, DI water for 5 minutes, 2X DPBS for 15 minutes, 0.1% v/v peracetic acid (Sigma-Aldrich) in 4% v/v ethanol solution for 1 hour, 1X DPBS for 5 minutes, DI water for 5 minutes, and 1X DPBS for 15 minutes. The decellularized heart sheets (sH-ECM) were stored in 1X DPBS with 1% penicillin/streptomycin (Life Technologies) at 4°C. Three independent organs were decellularized at least three independent times (n=9).
Vocal Fold Lamina Propria (VFLP) Decellularization
Porcine VFLP dissection and decellularization were performed as previously described.23 Porcine larynges (Nahunta Pork Outlet, Raleigh, NC) were dissected and the VFLP of each of the true vocal folds was cleaned from surrounding connective tissue and frozen at −80°C for at least 24 hours. The VFLPs were treated with the following solutions under constant agitation on an orbital shaker: three times with 1X DPBS for 15 minutes each, 4% sodium deoxycholate w/v (Sigma-Aldrich, St. Louis, MO) for 2 hours, 1X DPBS for 15 minutes, deoxyribonuclease (DNase) I (Sigma-Aldrich) at 273Kunitz/mL in PBS pH 7.4 supplemented with 2.5mM Mg and 0.1mM Ca for 2 hours, 1X DPBS for 15 minutes, 0.1% v/v peracetic acid (Sigma-Aldrich) in 4% v/v ethanol solution for 30 minutes, 1X DPBS for 15 minutes. The decellularized VFLPs (sVFLP-ECM) were stored in 1X DPBS with 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA) at 4°C. A pool of 40 VFLPs were combined from 20 different animals. Half of the batch was used for the manual decellularization at three independent times (n=3) of 7, 7, and 6 VFLP tissues respectively.
2.1.2. Automated 3-hour Tissue Decellularization Protocol
The remaining heart pieces from the control method were immediately cut into thick slices and micronized using a meat grinder (Altra Model AZ-MG090, Foshan, Guangdong, China). Three independent hearts were decellularized at least three independent times (n=9). Meanwhile, half of the dissected pool of the VFLP batch (20 VFLPs) were ground after freezing overnight using a Ninja Blender® (Amazon, Seattle, WA). At least three independent decellularizations were performed (n=3). The micronized tissues were then placed in a custom-made bioreactor (described in section 2.2) and treated with the solutions listed in Table 1 (heart) or Table 2 (VFLP) under constant stirring. Decellularized micronized heart-ECM (mH-ECM) or micronized VFLP-ECM (mVFLP-ECM) scaffolds were lyophilized overnight and stored at room temperature.
Table 1.
Steps in the 3-hour automated decellularization of the porcine heart including reagent and exposure time.
| # | Reagent | Time (minutes) |
|---|---|---|
| 1 | DI Water | 5 |
| 2 | 2X DPBS | 5 |
| 3 | 0.02% Trypsin | 30 |
| 4 | DI Water | 5 |
| 5 | 2X DPBS | 5 |
| 6 | 3% Tween-20 | 30 |
| 7 | DI Water | 5 |
| 8 | 2X DPBS | 5 |
| 9 | 4% Sodium Deoxycholate | 30 |
| 10 | DI Water | 5 |
| 11 | 2X DPBS | 5 |
| 12 | 0.1% Peracetic Acid | 30 |
| 13 | 1X DPBS | 15 |
| 14 | 1X DPBS | 15 |
Table 2.
Steps in the 2.5-hours automated decellularization of porcine VFLP including reagent and exposure time.
| # | Reagent | Time (minutes) |
|---|---|---|
| 1 | DI Water | 5 |
| 2 | 2X DPBS | 5 |
| 3 | 4% Sodium Deoxycholate | 30 |
| 4 | DI Water | 5 |
| 5 | 2X DPBS | 5 |
| 6 | DNAse (273Kunitz/mL) | 30 |
| 7 | DI Water | 5 |
| 8 | 2X DPBS | 5 |
| 9 | 0.1% Peracetic Acid | 30 |
| 10 | 1X DPBS | 15 |
| 11 | 1X DPBS | 15 |
2.2. Bioreactor Setup
The bioreactor consisted of modular components to allow for continuous decellularization. An inline mesh filter (Bouncer, Wilmington, NC) and screen (Bouncer) were used to form the body of the bioreactor. To ensure consistent interactions between tissue samples and reagents, mechanical stirring of the system was accomplished using a magnetic stir bar (Fisher Scientific) placed at the center of the bioreactor. The bioreactor was placed on top of a magnetic stirrer. Programmable auto dosing pumps (Jebao, Guangdong, China) were used to load the reagents into the bioreactor input by connecting 8mm silicone tubing (Uxcell, Hong Kong, China) and T shaped three-way valves (Uxcell) to the main 8mm input tube. The number of active auto dosing pumps was adapted to the decellularization protocol used. Finally, waste was removed by feeding an 8mm silicone tube (Uxcell) through the outlet to the bottom of the system. The waste was pumped to an external waste tank. A more detailed description is found in section 3.2.
2.3. Histological Analysis
Tissue samples of native and decellularized sH-ECM and mH-ECM were fixed in 4% formaldehyde (Sigma-Aldrich) overnight and stored in 70% ethanol. The samples were trimmed and sectioned at a thickness of 5μm and subjected to Hematoxylin and Eosin (H&E) staining. The histological staining was performed at the Histology Laboratory in the College of Veterinary Medicine at North Carolina State University.
2.4. mH-ECM vs. sH-ECM Proteomics
Discovery proteomic strategies were used to characterize and compare the overall protein composition of acellular sH-ECM and mH-ECM scaffolds.
2.4.1. Sample Preparation
Three porcine hearts were used to produce three independent decellularized mH-ECM or sH-ECM scaffolds. To generate a biological representative sample, 10mg each of mH-ECM or sH-ECM were pooled into a final 30mg sample per condition. Samples were suspended in 1mL of 50mM ammonium bicarbonate (pH=8.0) with 5% Sodium Deoxycholate (SDC) for digestion and homogenization. To determine protein concentration, a bicinchoninic acid assay (BCA assay) was performed. Samples were prepared prior to injection into the mass spectrometry (MS) system by using a filter-aided sample preparation (FASP) approach.24
2.4.2. Liquid chromatography (LC)
All samples were processed according to a discovery proteomics workflow using an Easy Nano-LC 1200 complexed to a Thermo Scientific Q-Exactive HFX with an EASY-Spray source for acquisition. A ThermoFisher Scientific Acclaim™ PepMap™ 100 trap column (C18 LC Columns, 3μm particle size, 75μm ID, 20mm length (164946)) was utilized in line with an EASY-Spray™ analytical column (2μm particle size, 75μm ID, 250 mm length (ES802A)) at 35 °C.
2.4.3. Data analysis
Raw data were loaded into Proteome Discoverer 2.4.0.305 (ThermoFisher Scientific) for analysis. A label-free quantitation (LFQ) workflow was used. For peptide searching, protein FASTA database was downloaded via Proteome Discoverer from SwissProt (fully annotated) and TrEMBL (unreviewed proteins) databases for Sus Scrofa (taxonomy ID = 9823). A maximum of 8 equal mods and a maximum of 3 total dynamic mods were used per peptide. The following post translational modifications (PTMs) were accommodated in the search algorithm (modified amino acids in parentheses): oxidation (K, M, P), deamidation (N, Q), galactosyl (K), Glucosylgalactosyl (K), and Lys → Allysine (K).
2.5. Hydrogel Preparation
ECM scaffolds were frozen in liquid nitrogen, powdered using a mortar and pestle, and lyophilized overnight. The lyophilized ECM was digested at 10mg/mL in a ratio of 10:0.6:1 of H-ECM, pepsin (3200–4500units/mg; Sigma-Aldrich), and 0.1M HCl (Sigma-Aldrich) on a magnetic stir plate at room temperature for 48 hours. The resulting ECM digestion was aliquoted and stored at −20°C until use. ECM hydrogels (mH-ECM, sH-ECM, mVFLP-ECM, sVFLP-ECM; 6 mg/mL) were prepared by thawing the ECM digestion, adjusting its pH to 7.3 ± 0.2 using 0.1M NaOH (Sigma-Aldrich), and balancing the salt content using 10X DPBS and 1X DPBS. FibriCol I, Collagen Type I >97% (Advanced Biomatrix, Carlsbad, CA) was used as a collagen control (Col). Next, 250μL of the ECM hydrogels or Col control were pipetted into each well of a 24-well plate (Corning, NY). Crosslinking into a gel was achieved by placing the plate in the incubator at 37°C for 30–45 minutes. Gelation kinetics of ECM and control hydrogels were measured as described in Method S1.
2.6. Cell Culture Conditions
Primary human dermal fibroblast normal cells (HDFn) were purchased from ATCC® (Manassas, VA). HDFn were cultured in tissue culture plastic flasks (VWR, Radnor, PA) using Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum heat-inactivated (Genesee Scientific, San Diego, CA) and 1% penicillin/streptomycin (ThermoFisher Sciesntific). Media was changed every 3 days. HDFn cell passages 10–18 were used for this study. HDFn were passed when reaching 80–90% confluency by incubation with 0.05% trypsin-EDTA (Life Technologies) for 5 minutes and seeded onto tissue culture plastic flasks.
2.7. LIVE/DEAD Viability Assay
HDFn cells were seeded on top of Col, sH-ECM, mH-ECM, sVFLP-ECM, and mVFLP-ECM hydrogels (40,000 cells/condition). The hydrogels seeded with HDFn were stained at 24 and 48 hours using the LIVE/DEAD Viability/Cytotoxicity Kit (Life Technologies) according to the manufacturer’s instructions. The samples were imaged using bright field and fluorescence microscopy (Revolve microscope, Echo, San Diego, CA).
2.8. Double stranded DNA (dsDNA) Quantification
Native heart tissue and decellularized ECM scaffolds were lyophilized overnight. Approximately 3mg per sample was then digested in 20μL (at 20mg/mL) Proteinase K Solution (Qiagen) and 180μL Buffer ATL (Qiagen) overnight at 60°C. For cells cultured on the ECM hydrogels, three hydrogel replicates of 100μL/each were collected and digested using the same ratio of Proteinase K/ ATL buffer.
dsDNA quantification was performed using the QuantiFluor® dsDNA System (Promega, Madison, WI) according to the manufacturer’s instructions. The digested samples were diluted and mixed thoroughly using 800μL of TE pH 7.4 buffer (ThermoFisher Scientific). A second dilution (1:50) was prepared using the same buffer. Further dilutions were required for native samples to reach a signal in the same threshold as the standards supplied by the kit. The samples were read using an Infinite M200 Pro plate reader (Tecan, Männedorf, Switzerland).
2.9. Statistical Analysis
GraphPad PRISM 8.0 software was used to perform statistical analyses. All experiments were performed at least three independent times, unless otherwise noted. Proteomic discovery analysis was performed using ThermoFisher Proteome Discoverer 2.4 with an ANOVA hypothesis test (individual proteins). Student unpaired t-test with Welch’s correction was performed for dsDNA quantification analysis. A value of P<0.05 was considered significant unless otherwise noted.
3. RESULTS
3.1. Fast automated decellularization approach
Lot-to-lot variation represents one challenge to the development of scalable and continuous manufacturing of decellularized ECM-biomaterials. External processing factors, such as the variety of decellularization protocols and the manual work required (i.e., manual working time > 8 hours and often by different people), hinder the standardization of the process and affect optimization. Figure 1 provides a schematic of the overall experimental approach.
As shown in Figure 1A, one current method for decellularization of porcine heart sheets requires ~9 hours to obtain the ECM for hydrogel production. In this case, each reagent is fed in a batch reaction configuration.22 The method starts with a thin sheet of heart (sH) placed inside a container and exposed to a combination of decellularization reagents. Homogeneity and thickness (2–3mm) of the tissue impact the success of decellularization by affecting the penetration and effectiveness of the decellularization reagents. Additionally, as this method is dependent on manual labor, variations in the end product are frequent and often unavoidable.
The automated system described in this study is illustrated in Figure 1B and involves pre-treatment of the native soft-tissue (i.e., micronization) and a custom-adapted inline filtration bioreactor connected to automated dosing pumps allowing a semi-batch reactor setting. The system can produce decellularized porcine heart ECM for hydrogel production in approximately 3 hours. The automated approach starts with a pre-treatment of the tissue via micronization to increase the surface area of the tissue and reduce exposure times required for each decellularization reagent. The settings used can be adjusted to further optimize the decellularization protocol as this design allows for changes in the exposure time, the type of reagent, and the order in which the tissue is exposed to a reagent. In addition, the platform can be tailored to decellularize other soft tissues.
3.2. Platform design
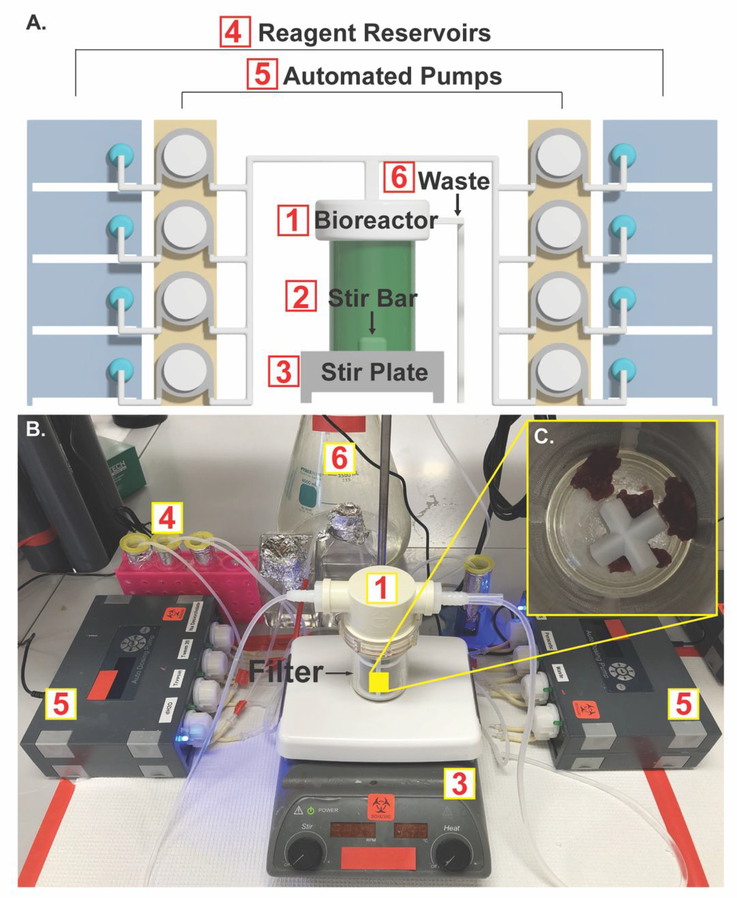
Figure 2A shows a computer-generated schematic of the final prototype of the automated decellularization system. We identified that vertical filtration resulted in clogging of the system due to the nature of soft tissues. Therefore, a lateral inline filtration system was utilized. The stirring was a requirement to ensure a homogenous decellularization environment. The semi-batch system can be connected to various dosing pumps in order to automate the reagent addition and waste removal.
Figure 2. Bioreactor design.
A.) Computer-generated schematic of the automated decellularization system. B.) Photograph showing the system assembled on the benchtop. C.) Micronized native heart tissue placed inside the bioreactor containing a 178μm mesh filter and a cross stir bar.
Figure 2B shows the benchtop system. The micronized native tissue was resuspended in DI water and transferred by pipetting into the bioreactor (Figure 2A1) through the top inlet. Figure 2C shows the bioreactor with a 178μm mesh filter placed inside. Different filter sizes can be used depending on the size of the pretreated native tissue. The filter was used to retain the decellularized tissue within the bioreactor while allowing the removal of waste during the process. Waste was removed from the bottom of the system through a tube that was placed outside of the filter and connected through the output of the bioreactor lid. Stir bars (Figure 2A2) were placed inside the bioreactor to provide mechanical stirring. Individual peristaltic pump (Figure 2A5) outputs were connected in parallel by tubes joined through connectors that converge on the input of the bioreactor lid. The pump inputs were then connected to individual reagent reservoirs (Figure 2A4). An additional peristaltic pump was used to pump waste from the bioreactor to the waste tank (Figure 2A6). The pump outlets were connected in the manifold to the bioreactor input and arranged to prevent unwanted interactions between reagents. In addition, the pumps responsible for DPBS and DI water washes were located at the distal end of the manifold to allow for clearing of any residual decellularization reagents in the tubing. The pumps were programmed to deliver 30mL of each reagent and remove waste at designated time points. Finally, the bioreactor was placed on a magnetic stir plate (Figure 2A3) and set to 500RPM. The final decellularized tissue is shown in Figure S1. Following decellularization, samples were characterized to evaluate the efficacy of the process.
3.3. Assessment of decellularization (sH-ECM vs. mH-ECM)
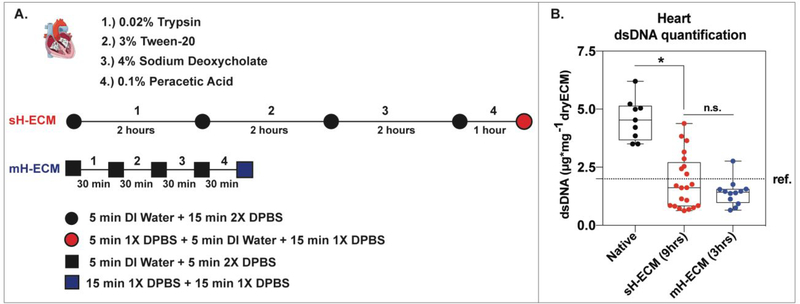
Timelines of the decellularization processes are shown in Figure 3A. The method used to decellularize sH-ECM required approximately 9 hours to complete, substantively longer than the 3 hours using the automated method developed in this study.
Figure 3. Heart tissue decellularization.
A.) Schematic comparing the two methods (sH-ECM vs. mH-ECM) used to decellularize porcine heart tissue. B.) Box and Whisker plot showing double stranded DNA (dsDNA) quantification per mg of dry tissue for native and decellularized sH-ECM and mH-ECM. Three independent organs were decellularized at least three independent times (n=9). The error bars represent the standard error of the mean (SEM). * = P<0.05. n.s. = not significant (P>0.05). ref. ~2μg/mg (for Urinary Bladder Matrix–UBM).25
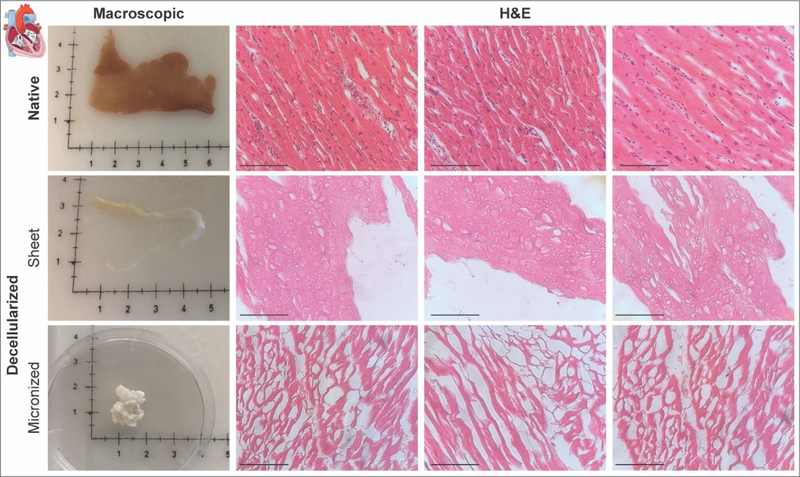
The same decellularization reagents as well as washing steps were used. The steps include hypotonic conditions via DI water followed by isotonic or hypertonic conditions using DPBS between every stage for both methods. However, in the case of mH-ECM (Table 1 and Figure 3A), the exposure time was reduced by 1.5 hours for 0.02% trypsin, 3% Tween, 4% sodium deoxycholate, and by 30 minutes for 0.1% peracetic acid. The removal of nuclear content was evaluated via double-stranded DNA (dsDNA) quantification (Figure 3B). Native tissue yielded a dsDNA content per dry tissue weight of 4.564±0.299μg/mg. Significantly decreased dsDNA content was observed for both sH-ECM (1.872±0.253μg/mg) and mH-ECM (1.399±0.158μg/mg). Furthermore, no statistically significant differences were observed between sH-ECM and mH-ECM (P>0.05). Additionally, values for sH-ECM and mH-ECM were similar to the dsDNA reported for a commercially available scaffold UBM-ECM used as a reference.25 H&E was employed to further evaluate decellularization quality (Figure 4). The decellularization of both sH-ECM and mH-ECM resulted in a significant reduction in nuclei and dsDNA content.
Figure 4. Histological analysis.
Macroscopic images and Hematoxylin and Eosin (H&E) staining for native and decellularized sH-ECM (sheet) and mH-ECM (micronized). (Scale bar = 100μm).
3.4. Discovery proteomics comparing sH-ECM and mH-ECM
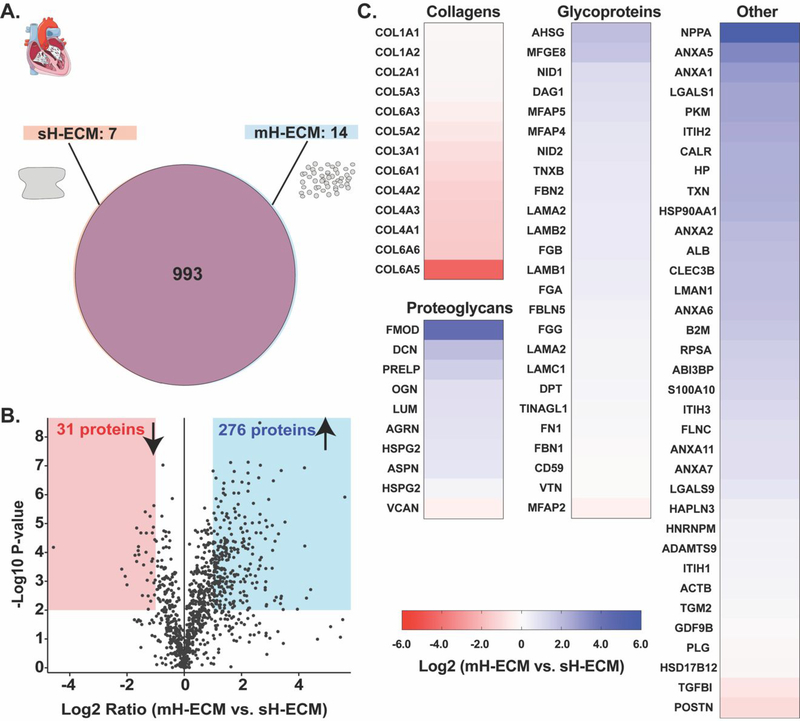
To further characterize the decellularized scaffolds, we utilized discovery proteomics strategies to evaluate the main difference in protein composition between automated mH-ECM and the standard sH-ECM (Figure 5). The assay identified approximately 1000 proteins for both acellular scaffolds, as depicted in Figure 5A. The Venn diagram shows no major differences in the total number of identified proteins between the two protocols tested. The protocol for sH-ECM yielded a scaffold with seven proteins not identified in mH-ECM (Figure 5A) and mH-ECM yielded 14 proteins not identified in sH-ECM (Figure 5A). The volcano plot shown in Figure 5B presents the overall changes in protein abundance. In general, the plot displays a large number of identified proteins with a positive Log2 ratio, suggesting that the mH-ECM scaffold derived using the automated method preserves larger proteins quantities. This trend was also seen when identifying low or high abundance proteins by selecting proteins with P-values equal or lower than 0.01 (i.e., Log10 P-values > 2) and at least +/− 2X ratios (i.e., Log2 ratio > 1). A total of 31 proteins were identified as less abundant in the mH-ECM (red area), which is significantly lower than the 276 proteins identified with a higher abundance in the mH-ECM (blue area).
Figure 5. Discovery proteomics.
A.) Venn diagram representing the overall number of proteins identified per condition (n=3). B.) Volcano plot representing in the x-axis the Log2 ratio for the identified proteins according to the label-free quantification analysis and the y-axis the −Log10 P-value according to the statistical analysis considering a false discovery rate of 0.01. The red shadow area corresponds to the statistically significant area for less abundant proteins identified (i.e., Log2 ratio<1 & P-value<0.01). The blue area corresponds to the statistically significant area for the identified proteins with higher abundance (i.e., Log2 ratio>1 & P-value>0.01). C.) Heatmaps showing the abundance ratio as fold change between automated mH-ECM and manual sH-ECM for collagens, proteoglycans, glycoproteins, and other ECM-related proteins.
In Figure 5C, proteins are listed and categorized as extracellular matrix proteins by using cellular component gene ontology analysis. The proteins are categorized into collagens, proteoglycans, glycoproteins and other ECM-related proteins. Analysis revealed that structural Collagens type 3, 4, and 6 were less abundant in the mH-ECM than in the sH-ECM. Conversely, ECM proteins with highest abundance were in a range of 4–40 times than in the mH-ECM.
3.5. mH-ECM optimization (mH-ECM-O) and cytocompatibility
Although no standard parameters are available for decellularized ECM, Crapo et al. proposed the following criteria: <50ng dsDNA per mg of dry weight ECM, <200 base pairs DNA fragment length, and no visible nuclei in 4′,6-diamidino-2-phenylindole (DAPI) and hematoxylin and eosin (H&E) staining.4 However, these criteria have been reconsidered; Cramer et al. suggested these criteria were too strict for certain tissues and applications.2 However, dsDNA removal is accepted as a valuable indicator of decellularization level and a good parameter for monitoring the optimization of the process. The system described in this study can be used as a platform to further optimize the decellularization process for heart tissue as well as adapt it to other soft tissues such as VFLP.
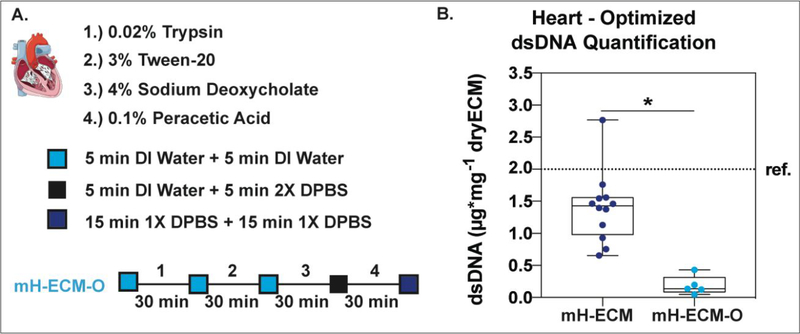
The decellularization protocol was adjusted by focusing on increasing dsDNA removal. Figure 6A shows a schematic of the protocol used to further optimize the dsDNA removal of porcine heart tissue. Fabrication time and decellularization reagents were the same as those used for the mH-ECM protocol described in Figure 3A.
Figure 6. Optimized heart tissue decellularization.
A.) Schematic of the decellularization protocol used to further optimize the DNA removal of porcine heart tissue. B.) Box and Whisker plot comparing dsDNA quantification per mg of dry tissue for decellularized mH-ECM (From Figure 3B) and optimized mH-ECM (mH-ECM-O). Three independent organs were decellularized at least three independent times (n=9). * = P<0.05.). ref. ~2μg/mg (for Urinary Bladder Matrix–UBM).
The difference between the protocols in Figure 6A vs. Figure 3A is the first three wash steps. The 5 minutes DI water followed by 5 minutes 2XDPBS steps were replaced with two consecutive DI water washes of 5 minutes each to facilitate nucleic acid solubility and removal from the material.
Additionally, mechanical stirring was used. The outcome of the adjusted system is presented in Figure 6B; optimized mH-ECM (mH-ECM-O) yielded a DNA content per dry tissue weight of 0.187±0.065μg/mg which is significantly lower (P<0.05) than mH-ECM.
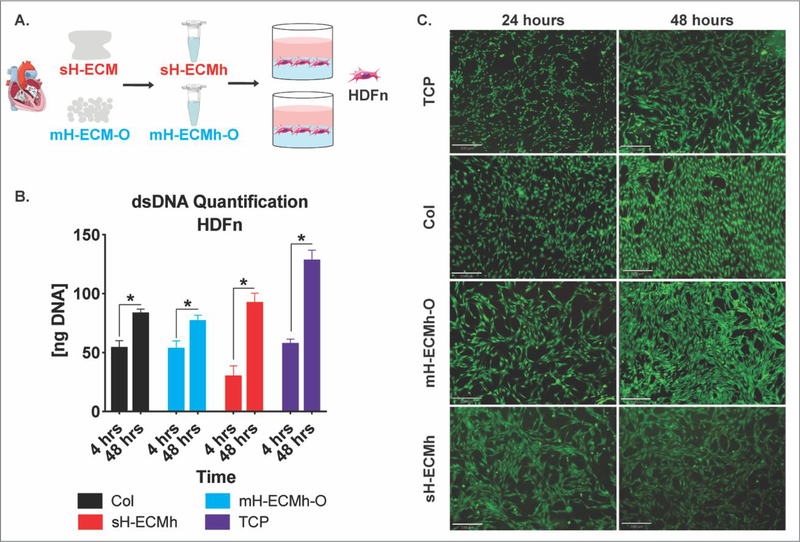
The mH-ECM-O was collected and solubilized into an ECM-hydrogel (mH-ECMh-O) to study its cytocompatibility evaluated via dsDNA quantification on cells cultured on the hydrogel and by imaging using the LIVE/DEAD Cell Viability assay (Figure 7). The hydrogel without cells was used as a blank to subtract any potential background from the scaffold in the dsDNA measurement. In addition, tissue culture plastic (TCP), Collagen Type I (Col), and sH-ECMh were used as controls. HDFn cells were cultured and the dsDNA was measured after 4 hours of cell attachment and at the end point of the experiment after 48 hours. Figure 7B shows dsDNA values; a significant (P≤0.05) increase in dsDNA content was observed over time across all conditions. The dsDNA measurement was corroborated with LIVE/DEAD imaging (Figure 7C). Live HDFn cells cultured on TCP, mH-ECMh-O, sH-ECMh, and Col were stained green (Calcein-AM) and dead cells were stained red (Ethidium Homodimer-1) after 24 and 48 hours in culture. The majority of the cells were alive in all conditions.
Figure 7. Cytocompatibility studies.
A.) ECM scaffolds were enzymatically digested and self-assembled into ECM hydrogels; human dermal fibroblasts (HDFn) were seeded on top and cytocompatibility determined. B.) dsDNA quantification of HDFn cultured on Col control, mH-ECMh-O, sH-ECMh, and tissue culture plastic (TCP). * = P≤0.05. Bar graphs represent mean ± SEM. C.) LIVE/DEAD imaging of HDFn cells cultured on TCP, Col control, mH-ECMh-O, and sH-ECMh. Calcein-AM staining (Green) shows live cells while Ethidium Homodimer-1 staining (Red) shows dead cells (Scale bar = 330μm).
3.6. Automated decellularization of VFLP and cytocompatibility
Vocal fold lamina propria (VFLP) was selected to evaluate the robustness of the decellularization platform. A biological pool of 40 VFLPs was prepared from 20 different animals and decellularization was performed three independent times (n=3). The decellularization approach was similar to micronized heart tissue and was developed to reduce the total time required for decellularization. Figure 8A shows a schematic of the automated method used to decellularize porcine VFLP in approximately 2.5 hours. The automated method was 2.5X faster in comparison with the method used to decellularize porcine VFLP sheets (sVFLP-ECM), which takes approximately 6 hours.23
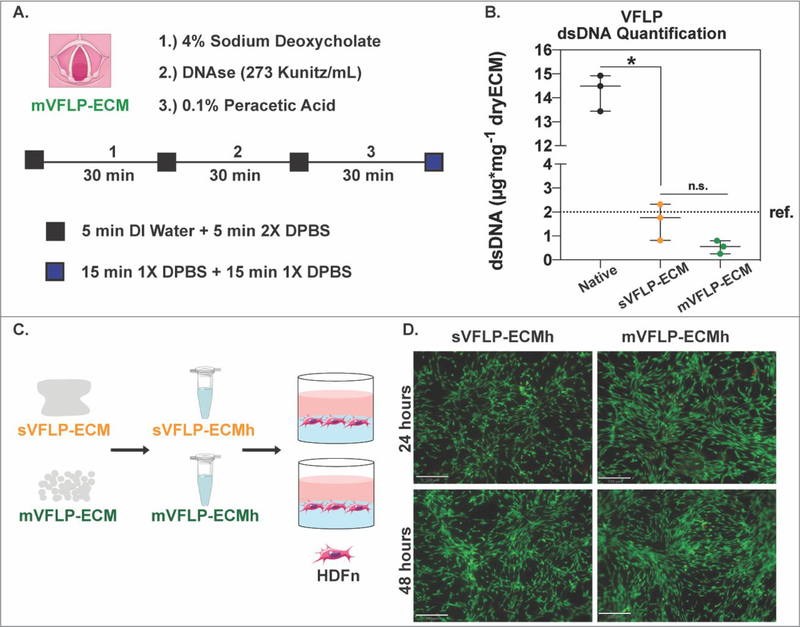
Figure 8. Vocal fold lamina propria decellularization.
A.) Schematic showing the protocol for the automated decellularization of porcine vocal fold lamina propria (mVFLP-ECM). B.) Box and Whisker plot showing dsDNA quantification per mg of dry tissue for native and decellularized sVFLP-ECM and mVFLP-ECM. Approx. 40 VFLPs were combined from 20 animals and the decellularization was performed three independent times (n=3). * = P<0.05. n.s. = not significant (P>0.05). ref. ~2μg/mg (for Urinary Bladder Matrix–UBM). C.) ECM scaffolds were enzymatically digested, self-assembled into ECM hydrogels, and seeded with HDFn for cytocompatibility studies. D.) LIVE/DEAD imaging of HDFn cells cultured on sVFLP-ECM, and mVFLP-ECM. Calcein-AM staining (Green) shows live cells while Ethidium Homodimer-1 staining (Red) shows dead cells (Scale bar = 330μm).
The same decellularization reagents were used for both methods. However, in the case of mVFLP-ECM, exposure time was reduced by 1.5 hours for 4% sodium deoxycholate and by 1.5 hours for the DNAse treatment in comparison to sVFLP-ECM. The first three washes with 1X DPBS for 15 minutes each were replaced with 5 minutes DI water followed by 5 minutes 2X DPBS washes. As with mH-ECM, mechanical stirring was used. The dsDNA content (Figure 8B) for both sVFLP-ECM (1.630±0.439μg/mg) and mVFLP-ECM (0.535±0.158μg/mg) was significantly lower (P<0.05) than native VFLP (14.285±0.442μg/mg). Although the dsDNA content of the mVFLP-ECM was lower and the lot-to-lot variability was minimized, no statistically significant differences were observed between sVFLP-ECM and mVFLP-ECM (P>0.05). Both sVFLP-ECM and mVFLP-ECM were similar to dsDNA reported for the commercially available UBM-ECM.
Finally, mVFLP-ECM was solubilized into a hydrogel (mVFLP-ECMh) to study its cytocompatibility by using the LIVE/DEAD Cell Viability assay after culturing HDFn cells. Figure 8D shows cell survival after 24 and 48 hours of seeding on top of mVFLP-ECMh and sVFLP-ECMh.
4. DISCUSSION
Decellularized ECM-based products have been on the market for over 20 years. More and more acellular ECM scaffolds have gathered enough evidence to transition to preclinical trials and most recently an injectable heart ECM hydrogel finished phase I trials and will soon continue to phase II trials.11 New biofabrication technologies such as 3-D printers that use ECM based bio-inks and injectable therapeutic materials will increase the demand for ECM scaffolds. These applications require biomaterial fabrication processes with controlled lot-to-lot variation and the capacity for customization. Even though decellularization of various tissues from different sources has been widely explored, automation and the manufacturing process to standardize derivation of injectable ECM scaffolds remain poorly studied. Factors such as the inherent variability of tissues and organs, harvesting conditions, methods and reagents used, and the labor-intensive requirements prevent standardization and optimization of decellularization processes.22–23, 26–34
In order to achieve decellularization and significantly reduce production time at each step, our experimental approach (Figure 1) involved a closed semi-batch bioreactor and a specific pre-treatment of the native tissue. The baseline method selected for comparison was the production of a heart ECM scaffold (sH-ECM-Figure 1A) derived from a batch decellularization protocol in which the user manually switches each of the decellularization reagents in a stepwise process. It is important to note that current decellularization processes involve manual labor (often by multiple users) and require user input at every step of the batch process.19,35 This personnel requirement represents a limiting step to reduced production time and can introduce substantial variability in the final product.
A previous study described a platform that reduced the labor-demanding decellularization process of porcine abdominal aortas using an automated and modular device.28 The method described by Pellegata et al. involved approximately 1–2 days of tissue preparation: drying, storing at −80°C, defrosting for 12 hours at −20°C and overnight at 4°C. After tissue preparation, it took four (4) cycles of decellularization adding to approximately 87 hours to produce a successful ECM scaffold comparable to their control (i.e., manual aortic decellularization). Several groups described automated platforms for whole organ decellularization for heart, lungs, and kidneys.32–34 These whole organs have been decellularized for the purpose of recellularization with the ultimate goal of obtaining autologous organs for transplantation. However, the decellularization time for these protocols is often longer than methods for tissue sheets (over 10 hours) due to the size of whole organs. Even though the production yield is increased, this approach likely remains limited for the production of ECM hydrogels as the organs would require further processing (e.g., powdering/micronization) for digestion and will require larger quantities of reagents. Additionally, the platforms do not allow for in situ hydrogel production and further modifications would be needed to allow for successful digestion of the scaffolds. In the method described in this study, which focuses on ECM hydrogel production, micronization and the semi-batch reactor settings led to significantly reduced decellularization times compared to flat or cylindrical sheets and whole organs. Reduced size of native tissue required the adaptation of a filtration method and proper mechanical stirring of the micronized soft tissue. The system described in the present study allowed for automated decellularization for a variety of soft tissues (e.g., heart, VFLP) in a shorter time, facilitating future downstream production of ECM hydrogels and other solubilized forms of the ECM.
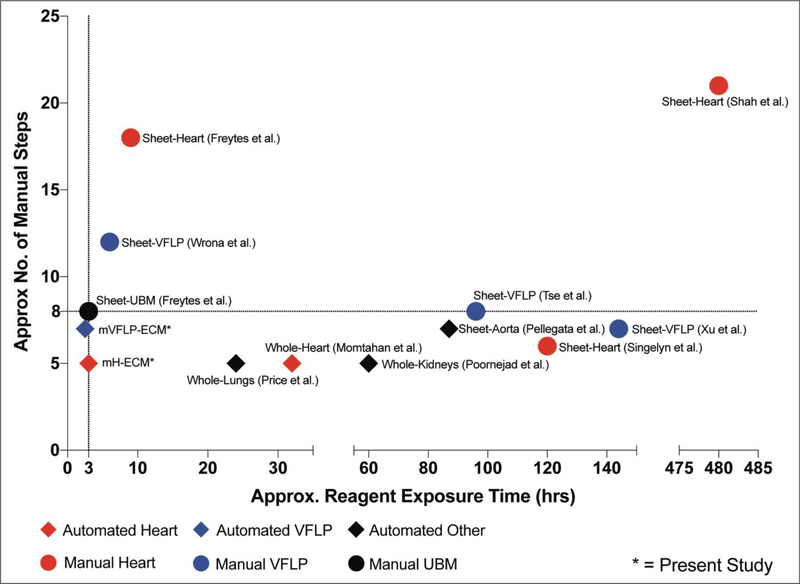
The efficiency for heart tissue decellularization was compared against the sH-ECM (reference protocol selected) that, if run continuously, should take approximately 9 hours.18 It is important to note that the selected protocol is shorter than some of the other protocols available for heart tissues requiring 48–72 hours or even weeks depending on the reagents used (Figure 9). As an example, the method used by Singelyn et al. to produce decellularized myocardial matrix (currently used in clinical trials), may take approximately 96–144 hours.11,29 Similarly, the method by Shah et al. requires approximately 2.5 weeks to obtain thin slices of decellularized porcine myocardium.27 These data suggest that the 3-hour automated semi-batch process demonstrated similar dsDNA removal to the 9-hour method using the same sequence of decellularization reagents. The similarity in the decellularization sequence between both methods (3- and 9-hour) allows for improved comparisons regarding cytocompatibility elicited by the scaffold as well as compositional comparisons. The significant number of lab-specific decellularization protocols, as well as the lot-to-lot variation, represent a challenge for studying tissue specificity and limit experimental reproducibility. Typically, each laboratory group selects or develops their own decellularization protocol depending on the particular tissue of interest, site-specific limitations, and scientific approaches used by the lab.36 For example, as shown in Figure 9, at least 3 different protocols are commonly used to decellularize heart sheets. Even though the ECM derived from these studies had encouraging biological responses, it remains a challenge to compare them due to the different decellularization methods used and our inability to distinguish if the effect is a result of the final product, the decellularization protocol used or the ECM itself. Nevertheless, Figure 9 highlights the significant improvements in both time and required steps to decellularize two soft tissues when using the automated system described in this study. Ultimately, we seek to develop a “universal” decellularization method for any tissue or organ in order to remove the decellularization protocol as a variable impeding biological comparisons.
Figure 9. Automated vs. manual methods.
Graph showing the approximate reagent exposure time vs. approximate number of manual steps for the automated method (mH-ECM and mVFLP-ECM) and currently available methods for the decellularization of heart, aorta, VFLP, and UBM. The approximate number of manual steps was calculated as shown in Table S1. The approximate reagent exposure time was calculated without taking into account tissue processing steps. Overnight steps were assumed to equal 8 hours.22–23, 26–34
The method described in this study involves automated dosing pumps connected to an inline filtration bioreactor, allowing for the production of ECM scaffolds. Pre-processing the native tissue via micronization allowed for significantly reduced decellularization time as shown in Figure 3A. When using the same 3-hour protocol to decellularize heart sheets (Figure S2 for sH-ECM (3 hrs W)), dsDNA removal was above acceptable levels when compared to sH-ECM or the values for UBM-ECM. The data show high variability in the standard method and all the measured values were higher than the references selected (sH-ECM and UBM-ECM). The size reduction increases available surface area and facilitates diffusion between the tissue and the reagents. By reducing decellularization time, this method can significantly reduce production costs. Additionally, due to reduced exposure time to reagents, ECM native protein composition may be better preserved compared to other methods.
We compared the effects of starting the decellularization using a dry (D) native tissue and its effect on batch-to-batch variability and dsDNA removal. DNA quantification was highly variable and insufficient with regard to DNA removal for sH-ECM (9 hrs D), sH-ECM (3 hrs D), and mH-ECM (3 hrs D) as shown in Figure S2. These results were supported by histological examination via H&E which showed intact nuclei after decellularization for freeze-dried tissues (Figure S2). Therefore, hydration of the initial micronized material could play a role in the overall efficiency and variability of the process. Future studies will address the use of lyophilized raw material given its potential to help with raw material storage and handling during the scale up process.
The semi-batch geometry was designed to facilitate waste removal to avoid potential interactions between the released dsDNA and the acellular ECM scaffold. Even though the semi-batch system was designed as a closed environment to maintain sterility of the product during the decellularization process, future studies will address additional pre-treatments such as bioburden reduction. However, Hernandez et al. tested this variable on ECM decellularization and concluded that there was no major effect on ECM scaffolds.10 The process described in this study opens up the possibility of monitoring parameters of interest in real time during the decellularization process to ensure optimal ECM scaffold production.
In order to evaluate if the semi-batch method affects global protein content, discovery proteomic analysis was performed comparing the manual heart protocol (sH-ECM) with the automated heart protocol (mH-ECM). The overall number of proteins present after decellularization between the automated (3 hrs) method and the standard protocol (9 hrs) was not affected. However, the relative abundances of a few structural collagens (type 3, 4, 5, and 6) were significantly reduced in the automated method. This finding could be explained by the increase in surface area and potential elution of these proteins from the final product during waste removal. However, a larger number of proteoglycans, glycoproteins, and other ECM-related proteins remained in the scaffold obtained with the automated method. Natriuretic peptides precursor A and Fibromodulin (NPPA, FMOD) were the most abundant proteins identified in the 3-hour acellular ECM (about 30X higher compared to the 9-hour protocol). NPPA belongs to the family of natriuretic peptides which role has been associated with cardiovascular physiology and host metabolism.37 Also, previous studies have reported the potential benefits of increased concentration of these peptides in patients.38–39 FMOD is a proteoglycan that has been reported as an important ECM protein involved in angiogenesis, the regulation of scar formation in skin, and in cardiac remodeling. Another abundant proteoglycan in the scaffold obtained with the automated method was Decorin (DCN), which has been associated with regulating extracellular matrix integrity, angiogenesis, and fibrosis. Previous studies suggest DCN interacts with various growth factors such as transforming growth factor beta (TGFβ). An in vitro study with human cardiac fibroblasts showed that DCN downregulated collagen production following TGFβ stimulation.40 In spite of the lack of universal standard quality control methods to evaluate decellularization efficiency or the removal of cellular components,40 we evaluated the abundance of five cellular proteins in order to further compare the decellularization efficiency of both automated (mH-ECM) and manual (sH-ECM) methods as shown in Figure S5.41–43 From the analysis, THY-1/CD90, smooth muscle alpha-2 actin (ACTA2), and myelin protein zero (MPZ) yielded similar abundances for both processes. Interestingly, vimentin (VIM) presented a Log2 abundance of around 1.75. However, this abundance ratio provides a comparison between methods and cannot quantify the actual amount in each scaffold. Overall, the relative abundance of these proteins in the ECM and their contribution to decellularization efficiency warrant further investigation. As described by Hill et al., the specific type and amount of cellular proteins required to elicit an immune response is not known. The study reported the retention of approximately 2% of cellular proteins in the decellularized scaffolds, suggesting that the complete removal of cellular components is highly unlikely.44 However, the current clinical success and lack of meaningful immune rejection of ECM scaffolds such as UBM and SIS suggest current decellularization methods sufficiently remove cellular components. The automated platform presented in this study proposes an efficient tool for screening multiple protocols aiming at the retention or removal of particular proteins and could be used in future studies to further study the effects of cellular remnants on the host tissue response.
Further optimization can be achieved with the semi-batch system by focusing on dsDNA removal. Although some commercial acellular ECMs may contain a higher amount of dsDNA, a recent report stated that this does not limit clinical applications.2 Furthermore, the mH-ECM scaffold and the mH-ECM-O described in this study had lower dsDNA concentration than UBM-ECM.25 Therefore, the platform was used to optimize the mH-ECM by improving mechanical stirring of the tissue and by adding a 5-minute wash/rinse using DI water immediately after exposure to reagents. The data suggest this condition increased the removal of the dsDNA with a final ECM scaffold with dsDNA values in the nanogram range per mg of dry tissue. In addition to dsDNA removal, future studies will target other parameters such as preservation of specific ECM proteins, reduced variability, and inline monitoring of the decellularization process. The semi-batch system developed is a versatile platform that produces ECM scaffolds ready for transformation into hydrogel forms in an efficient manner. In addition, the platform can be adjusted for the in situ production of other solubilized scaffolds or biomaterials such as Matrix-Bound vesicles (MBVs) production using downstream solubilization methods.25, 45–47
To illustrate adaptability of the automated system, the semi-batch bioreactor system was tested using vocal fold lamina propria (VFLP). The VFLP protocol selected for comparison took approximately 6 hours to complete in a labor dependent manner using sodium deoxycholate, DNAse I, and peracetic acid.23 Another available protocol for the decellularization of VFs involves a 4-day treatment with SDS.30 SDS treatment for an extended period of time is known to be cytotoxic if not properly removed, disrupt the ECM ultrastructure, remove GAGs and growth factors, and damage collagens.17 As the VFLP is rich in ECM proteins, the method employed did not include either trypsin or Tween in order to increase ECM preservation. To reduce reagent exposure times, the tissue was frozen and grounded, and the pumps were set to run a 2–3 hours protocol following the timeline presented in Figure 8A. The dsDNA removal met the target of less than 2μg/mg of dry tissue. Similar retention of ECM proteins is expected for VFLP-ECM and future studies will compare the proteomic content of both methods. These results show the versatility of this new system and its potential use with a variety of soft tissues.
ECM scaffolds can be used for various clinical applications in sheet form typically sutured into the wound or as a hydrogel injected into the site of injury using minimally invasive methods. It is important to note that different authors reported in vitro and clinical studies regarding the preservation of particular molecular cues from the ECM in its solubilized form or its re-assembled hydrogel version.11,48–50 Therefore, cytocompatibility of heart ECM hydrogels derived with this automated method was tested using HDFn. The hydrogel was produced outside of the bioreactor using a previously described method by Freytes et al.15 Viability of HDFns was determined via dsDNA quantification at 24 and 48 hours of culture on sH-ECMh and mH-ECMh-O. HDFns grew and proliferated in the micronized ECM hydrogels (mH-ECMh-O) as shown in Figure 7B. The LIVE/DEAD (green/red) staining confirmed these results and showed the cells were alive and with an elongated morphology typically associated with healthy fibroblasts.51 As shown in Figure 7C, the green stain was heavily represented throughout the samples and the red stain was barely detected suggesting a high survival rate over 24 and 48 hours. Similar results were obtained for HDFns cultured on mVFLP-ECMh hydrogels derived with the automated system as shown in Figure 8D. To investigate self-assembly, we measured gelation kinetics of ECM hydrogels derived from each method. Our data (Figure S4) shows that sH-ECMh, mH-ECMh, sVFLP-ECMh, mVFLP-ECMh hydrogels exhibited sigmoidal curves similar to collagen type I confirming hydrogel formation. These results suggest that the automated system described in this study can produce high quality ECM as a foundation for hydrogels from multiple soft tissues.
We acknowledge that the automated decellularization system described in this study has three major limitations. First, the method described in this study is limited to ECM hydrogels and other solubilized forms of the ECM and cannot be readily translated to whole organs or sheets. Second, the automated system is not currently set up to allow for in situ production of ECM hydrogels. Therefore, sterility issues may arise when transferring the ECM scaffold from the bioreactor to the lyophilizer and finally to the digestion chamber. Lastly, ECM yield is decreased compared to the manual sheet method or whole organ methods.
Future studies will focus on using this approach for other soft tissues (e.g., skeletal muscle, tendon, dermis, lung, etc.) and ultimately developing a standard “universal” decellularization method applicable to any soft tissue. Additionally, we will focus on the production of ECM hydrogels and MBVs inside the bioreactor to maintain sterility and the automated scale-up the automated decellularization process. The semi-batch process described in this study can be adjusted for further downstream fine-tuning of material properties (e.g., inclusion of crosslinkers and initiators for bio-ink manufacturing).18,48 The decellularization protocol in this study was performed in 50mL bioreactors, however, future work will incorporate the transition to industrial scale bioreactor systems (0.5 to 5L) to increase ECM yields and allow for higher production rates. By incorporating inline monitoring of the process, the automated system may be able to self-adjust in the future providing a novel platform for rapid tissue decellularization.
5. CONCLUSIONS
The combination of pre-treating soft tissues via size reduction, bioreactor design, inline filtration, the addition of hypotonic or hypertonic short washes, and mechanical dissociation of the tissue via continuous stirring has enabled us to develop an automated semi-batch bioreactor system for the production of ECM hydrogels. The system significantly reduced the production time and consistency of ECM scaffolds from soft tissues (heart and vocal fold lamina propria) while preserving ECM content and maintaining cytocompatibility.
Supplementary Material
List of abbreviations S1
Method S1 Gelation kinetics
Figure S1 Final decellularized product inside the bioreactor
Figure S2 Hematoxylin and Eosin stain and dsDNA quantification for native and decellularized dry (D) and wet (W) sH-ECM and mH-ECM after 3 and/or 9 hours
Figure S3 Gene ontology
Figure S4 Gelation kinetics curves
Figure S5 Differences in selected cellular proteins between automated (mH-ECM) vs. manual (sH-ECM)
Table S1 Approx. total number of manual steps for different decellularization protocols
ACKNOWLEDGEMENTS
We thank our funding sources: National Institutes of Health/National Institute on Deafness and Other Communication Disorders (R01DC017139, R01DC017743), and The American Heart Association Predoctoral Fellowship (19PRE34380006) to Emily W. Ozpinar. We also acknowledge the Comparative Medicine Institute (CMI) at North Carolina State University. We thank the Histology Laboratory in the College of Veterinary Medicine at North Carolina State University for their contributions. This work was performed in part by the Molecular Education, Technology and Research Innovation Center (METRIC) at North Carolina State University, which is supported by the state of North Carolina. The figures in this work made use of Servier Medical ART (SMART).
REFERENCES
- 1.Zhu M; Li W; Dong X; Yuan X; Midgley AC; Chang H; Wang Y; Wang H; Wang K; Ma PX; Wang H; Kong D, In vivo engineered extracellular matrix scaffolds with instructive niches for oriented tissue regeneration. Nat Commun 2019, 10 (1), 4620. DOI: 10.1038/s41467-019-12545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer MC; Badylak SF, Extracellular Matrix-Based Biomaterials and Their Influence Upon Cell Behavior. Ann Biomed Eng 2019. DOI: 10.1007/s10439-019-02408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade RJ; Burdick JA, Engineering ECM signals into biomaterials. Mater Today 2012, 15 (10), 454–459. DOI: Doi 10.1016/S1369-7021(12)70197-9. [DOI] [Google Scholar]
- 4.Crapo PM; Gilbert TW; Badylak SF, An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32 (12), 3233–3243. DOI: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown BN; Badylak SF, Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl Res 2014, 163 (4), 268–285. DOI: 10.1016/j.trsl.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez-Gastelum GR; Aguilar-Medina EM; Soto-Sainz E; Ramos-Payan R; Silva-Benitez EL, Antimicrobial Properties of Extracellular Matrix Scaffolds for Tissue Engineering. Biomed Res Int 2019, 2019, 9641456. DOI: 10.1155/2019/9641456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sackett SD; Tremmel DM; Ma F; Feeney AK; Maguire RM; Brown ME; Zhou Y; Li X; O’Brien C; Li L; Burlingham WJ; Odorico JS, Extracellular matrix scaffold and hydrogel derived from decellularized and delipidized human pancreas. Sci Rep 2018, 8 (1), 10452. DOI: 10.1038/s41598-018-28857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury D; Tun HW; Wang TY; Naing MW, Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol 2018, 36 (8), 787–805. DOI: 10.1016/j.tibtech.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Badylak SF; Freytes DO; Gilbert TW, Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater 2009, 5 (1), 1–13. DOI: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez MJ; Yakutis GE; Zelus EI; Hill RC; Dzieciatkowska M; Hansen KC; Christman KL, Manufacturing considerations for producing and assessing decellularized extracellular matrix hydrogels. Methods 2019. DOI: 10.1016/j.ymeth.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Traverse JH; Henry TD; Dib N; Patel AN; Pepine C; Schaer GL; DeQuach JA; Kinsey AM; Chamberlin P; Christman KL, First-in-Man Study of a Cardiac Extracellular Matrix Hydrogel in Early and Late Myocardial Infarction Patients. JACC: Basic to Translational Science 2019, 357. DOI: 10.1016/j.jacbts.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussey GS; Cramer MC; Badylak SF, Extracellular Matrix Bioscaffolds for Building Gastrointestinal Tissue. Cell Mol Gastroenter 2018, 5 (1), 1–13. DOI: 10.1016/j.jcmgh.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussey GS; Dziki JL; Badylak SF, Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater 2018, 3 (7), 159–173. DOI: 10.1038/s41578-018-0023-x. [DOI] [Google Scholar]
- 14.Spang MT; Christman KL, Extracellular matrix hydrogel therapies: In vivo applications and development. Acta Biomater 2018, 68, 1–14. DOI: 10.1016/j.actbio.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saldin LT; Cramer MC; Velankar SS; White LJ; Badylak SF, Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomater 2017, 49, 1–15. DOI: 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Efraim Y; Schoen B; Zahran S; Davidov T; Vasilyev G; Baruch L; Zussman E; Machluf M, 3D Structure and Processing Methods Direct the Biological Attributes of ECM-Based Cardiac Scaffolds. Sci Rep-Uk 2019, 9. DOI: ARTN 5578 10.1038/s41598-019-41831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Perez J; Ahearne M, Author Correction: The impact of decellularization methods on extracellular matrix derived hydrogels. Sci Rep 2019, 9 (1), 19818. DOI: 10.1038/s41598-019-56283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawan KC; Hong Y; Zhang G, Cardiac tissue-derived extracellular matrix scaffolds for myocardial repair: advantages and challenges. Regen Biomater 2019, 6 (4), 185–199. DOI: 10.1093/rb/rbz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simsa R; Padma AM; Heher P; Hellstrom M; Teuschl A; Jenndahl L; Bergh N; Fogelstrand P, Systematic in vitro comparison of decellularization protocols for blood vessels. Plos One 2018, 13 (12). DOI: ARTN e0209269 10.1371/journal.pone.0209269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzobo K; Motaung K; Adesida A, Recent Trends in Decellularized Extracellular Matrix Bioinks for 3D Printing: An Updated Review. Int J Mol Sci 2019, 20 (18). DOI: 10.3390/ijms20184628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L; Stiadle JM; Lau HK; Zerdoum AB; Jia X; Thibeault SL; Kiick KL, Tissue engineering-based therapeutic strategies for vocal fold repair and regeneration. Biomaterials 2016, 108, 91–110. DOI: 10.1016/j.biomaterials.2016.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freytes DO; O’Neill JD; Duan-Arnold Y; Wrona EA; Vunjak-Novakovic G, Natural Cardiac Extracellular Matrix Hydrogels for Cultivation of Human Stem Cell-Derived Cardiomyocytes. Radisic M; Black Iii LD, Eds. Springer New York: New York, NY, 2014; Vol. 1181, pp 69–81. DOI: 10.1007/978-1-4939-1047-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrona EA; Peng R; Born H; Amin MR; Branski RC; Freytes DO, Derivation and characterization of porcine vocal fold extracellular matrix scaffold. Laryngoscope 2016, 126 (4), 928–35. DOI: 10.1002/lary.25640. [DOI] [PubMed] [Google Scholar]
- 24.Wisniewski JR; Zougman A; Nagaraj N; Mann M, Universal sample preparation method for proteome analysis. Nature Methods 2009, 6 (5), 359–U60. DOI: 10.1038/Nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 25.Huleihel L; Hussey GS; Naranjo JD; Zhang L; Dziki JL; Turner NJ; Stolz DB; Badylak SF, Matrix-bound nanovesicles within ECM bioscaffolds. Sci Adv 2016, 2 (6). DOI: UNSP e1600502 10.1126/sciadv.1600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freytes DO; Martin J; Velankar SS; Lee AS; Badylak SF, Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix. Biomaterials 2008, 29 (11), 1630–7. DOI: 10.1016/j.biomaterials.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Shah M; Pawan KC; Copeland KM; Liao J; Zhang G, A Thin Layer of Decellularized Porcine Myocardium for Cell Delivery. Sci Rep-Uk 2018, 8. DOI: ARTN 16206 10.1038/s41598-018-33946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellegata AF; Asnaghi MA; Zonta S; Zerbini G; Mantero S, A novel device for the automatic decellularization of biological tissues. Int J Artif Organs 2012, 35 (3), 191–198. DOI: 10.5301/ijao.5000079. [DOI] [PubMed] [Google Scholar]
- 29.Singelyn JM; DeQuach JA; Seif-Naraghi SB; Littlefield RB; Schup-Magoffin PJ; Christman KL, Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials 2009, 30 (29), 5409–5416. DOI: 10.1016/j.biomaterials.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse JR; Long JL, Microstructure Characterization of a Decellularized Vocal Fold Scaffold for Laryngeal Tissue Engineering. Laryngoscope 2014, 124 (8), E326–E331. DOI: 10.1002/lary.24605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu CC; Chan RW; Tirunagari N, A biodegradable, acellular xenogeneic scaffold for regeneration of the vocal fold lamina propria. Tissue Eng 2007, 13 (3), 551–566. DOI: 10.1089/ten.2006.0169. [DOI] [PubMed] [Google Scholar]
- 32.Poornejad N; Momtahan N; Salehi ASM; Scott DR; Fronk CA; Roeder BL; Reynolds PR; Bundy BC; Cook AD, Efficient decellularization of whole porcine kidneys improves reseeded cell behavior. Biomed Mater 2016, 11 (2), 025003–025003. DOI: 10.1088/1748-6041/11/2/025003. [DOI] [PubMed] [Google Scholar]
- 33.Momtahan N; Poornejad N; Struk JA; Castleton AA; Herrod BJ; Vance BR; Eatough JP; Roeder BL; Reynolds PR; Cook AD, Automation of Pressure Control Improves Whole Porcine Heart Decellularization. Tissue Eng Part C Methods 2015, 21 (11), 1148–61. DOI: 10.1089/ten.TEC.2014.0709. [DOI] [PubMed] [Google Scholar]
- 34.Price AP; Godin LM; Domek A; Cotter T; D’Cunha J; Taylor DA; Panoskaltsis-Mortari A, Automated decellularization of intact, human-sized lungs for tissue engineering. Tissue Eng Part C Methods 2015, 21 (1), 94–103. DOI: 10.1089/ten.TEC.2013.0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kusuma GD; Yang MC; Brennecke SP; O’Connor AJ; Kalionis B; Heath DE, Transferable Matrixes Produced from Decellularized Extracellular Matrix Promote Proliferation and Osteogenic Differentiation of Mesenchymal Stem Cells and Facilitate Scale-Up. Acs Biomater Sci Eng 2018, 4 (5), 1760–1769. DOI: 10.1021/acsbiomaterials.7b00747. [DOI] [PubMed] [Google Scholar]
- 36.Gilpin A; Yang Y, Decellularization Strategies for Regenerative Medicine: From Processing Techniques to Applications. Biomed Research International 2017. DOI: Artn 9831534 10.1155/2017/9831534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song W; Wang H; Wu QY, Atrial natriuretic peptide in cardiovascular biology and disease (NPPA). Gene 2015, 569 (1), 1–6. DOI: 10.1016/j.gene.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunner-La Rocca HP; Kiowski W; Ramsay D; Sutsch G, Therapeutic benefits of increasing natriuretic peptide levels. Cardiovasc Res 2001, 51 (3), 510–520. [DOI] [PubMed] [Google Scholar]
- 39.Palmer BF; Clegg DJ, An Emerging Role of Natriuretic Peptides: Igniting the Fat Furnace to Fuel and Warm the Heart. Mayo Clinic Proceedings 2015, 90 (12), 1666–1678. DOI: 10.1016/j.mayocp.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Christensen G; Herum KM; Lunde IG, Sweet, yet underappreciated: Proteoglycans and extracellular matrix remodeling in heart disease. Matrix Biol 2019, 75–76, 286–299. DOI: 10.1016/j.matbio.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Ivey MJ; Tallquist MD, Defining the Cardiac Fibroblast. Circ J 2016, 80 (11), 2269–2276. DOI: 10.1253/circj.CJ-16-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philips C; Cornelissen M; Carriel V, Evaluation methods as quality control in the generation of decellularized peripheral nerve allografts. J Neural Eng 2018, 15 (2), 021003. DOI: 10.1088/1741-2552/aaa21a. [DOI] [PubMed] [Google Scholar]
- 43.Zhou P; Pu WT, Recounting Cardiac Cellular Composition. Circ Res 2016, 118 (3), 368–70. DOI: 10.1161/CIRCRESAHA.116.308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill RC; Calle EA; Dzieciatkowska M; Niklason LE; Hansen KC, Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics 2015, 14 (4), 961–73. DOI: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huleihel L; Bartolacci JG; Dziki JL; Vorobyov T; Arnold B; Scarritt ME; Molina CP; LoPresti ST; Brown BN; Naranjo JD; Badylak SF, Matrix-Bound Nanovesicles Recapitulate Extracellular Matrix Effects on Macrophage Phenotype. Tissue Eng Pt A 2017, 23 (21–22), 1283–1294. DOI: 10.1089/ten.tea.2017.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hussey GS; Dziki JL; Lee YC; Bartolacci JG; Behun M; Turnquist HR; Badylak SF, Matrix bound nanovesicle-associated IL-33 activates a pro-remodeling macrophage phenotype via a non-canonical, ST2-independent pathway. Journal of Immunology and Regenerative Medicine 2019, 3, 26–35. DOI: 10.1016/j.regen.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mora-Navarro C; Badileanu A; Gracioso Martins AM; Ozpinar EW; Gaffney L; Huntress I; Harrell E; Enders JR; Peng X; Branski RC; Freytes DO, Porcine Vocal Fold Lamina Propria-Derived Biomaterials Modulate TGF-β1-Mediated Fibroblast Activation in Vitro. Acs Biomater Sci Eng 2020. DOI: 10.1021/acsbiomaterials.9b01837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeffords ME; Wu JL; Shah M; Hong Y; Zhang G, Tailoring Material Properties of Cardiac Matrix Hydrogels To Induce Endothelial Differentiation of Human Mesenchymal Stem Cells. Acs Appl Mater Inter 2015, 7 (20), 11053–11061. DOI: 10.1021/acsami.5b03195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Neill JD; Freytes DO; Anandappa AJ; Oliver JA; Vunjak-Novakovic GV, The regulation of growth and metabolism of kidney stem cells with regional specificity using extracellular matrix derived from kidney. Biomaterials 2013, 34 (38), 9830–41. DOI: 10.1016/j.biomaterials.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reing JE; Zhang L; Myers-Irvin J; Cordero KE; Freytes DO; Heber-Katz E; Bedelbaeva K; McIntosh D; Dewilde A; Braunhut SJ; Badylak SF, Degradation products of extracellular matrix affect cell migration and proliferation. Tissue Eng Part A 2009, 15 (3), 605–14. DOI: 10.1089/ten.tea.2007.0425. [DOI] [PubMed] [Google Scholar]
- 51.Sarker B; Singh R; Silva R; Roether JA; Kaschta J; Detsch R; Schubert DW; Cicha I; Boccaccini AR, Evaluation of Fibroblasts Adhesion and Proliferation on Alginate-Gelatin Crosslinked Hydrogel. Plos One 2014, 9 (9). DOI: ARTN e107952 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of abbreviations S1
Method S1 Gelation kinetics
Figure S1 Final decellularized product inside the bioreactor
Figure S2 Hematoxylin and Eosin stain and dsDNA quantification for native and decellularized dry (D) and wet (W) sH-ECM and mH-ECM after 3 and/or 9 hours
Figure S3 Gene ontology
Figure S4 Gelation kinetics curves
Figure S5 Differences in selected cellular proteins between automated (mH-ECM) vs. manual (sH-ECM)
Table S1 Approx. total number of manual steps for different decellularization protocols