Abstract
In legumes, symbiotic nitrogen (N) fixation (SNF) occurs in specialized organs called nodules after successful interactions between legume hosts and rhizobia. In a nodule, N-fixing rhizobia are surrounded by symbiosome membranes, through which the exchange of nutrients and ammonium occurs between bacteria and the host legume. Phosphorus (P) is an essential macronutrient, and N2-fixing legumes have a higher requirement for P than legumes grown on mineral N. As in the previous studies, in P deficiency, barrel medic (Medicago truncatula) plants had impaired SNF activity, reduced growth, and accumulated less phosphate in leaves, roots, and nodules compared with the plants grown in P sufficient conditions. Membrane lipids in M. truncatula tissues were assessed using electrospray ionization–mass spectrometry. Galactolipids were found to increase in P deficiency, with declines in phospholipids (PL), especially in leaves. Lower PL losses were found in roots and nodules. Subsequently, matrix-assisted laser desorption/ionization–mass spectrometry imaging was used to spatially map the distribution of the positively charged phosphatidylcholine (PC) species in nodules in both P-replete and P-deficient conditions. Our results reveal heterogeneous distribution of several PC species in nodules, with homogeneous distribution of other PC classes. In P poor conditions, some PC species distributions were observed to change. The results suggest that specific PC species may be differentially important in diverse nodule zones and cell types, and that membrane lipid remodeling during P stress is not uniform across the nodule.
ESI–MS and matrix-assisted laser desorption ionization–mass spectrometry imaging reveal alterations in Medicago truncatula nodules membrane lipid composition and spatial distribution in phosphorus deficiency.
Introduction
Legumes have the distinctive ability to interact with symbiotic nitrogen (N)-fixing soil bacteria called rhizobia. A mature interaction culminates in the formation of nodules, specialized plant structures on legume roots. With nodules, symbiotic N fixation (SNF) takes place in the internalized rhizobia, which biosynthesize ammonia from atmospheric N. Rhizobia exchange the ammonia for photosynthate-derived carbon from the plant in complex processes (Fuchs et al., 2010; Udvardi and Poole, 2013; Flores-Tinoco et al., 2020). In N deficiency, a molecular signal exchange between the host plant and appropriate rhizobial species initiates nodule development (Oldroyd, 2013). Plant roots secrete flavonoids and rhizobia responds by producing Nod factors. Nod factor perception leads to curling of the epidermal root hair cells in the plant. Plant cell division in the root cortical cells initiates the formation of the nodule primordia, leading to the development of a nodule meristem. Concomitantly, rhizobia enter root hairs enclosed in plant-derived infection threads (ITs) that bring them into the underlying dividing root cells. There they are released into symbiosomes (Syms), organelle-like structures containing the rhizobia, now called bacteroids, enclosed in a plant-derived Sym membrane (SymM), also called the peribacteroid membrane (Verma and Hong, 1996). Bacteroids and plant cells then start a differentiation process that culminates in induction of SNF genes in the rhizobia and a mature functional leghemoglobin-containing pink nodule.
Tropical legumes like soybean (Glycine max), and birdsfoot trefoil (Lotus japonicus) forms determinate nodules that contain one symbiotic state at a time (Hirsch, 1992). In contrast, temperate legumes like barrel medic (Medicago truncatula), and pea (Pisum sativum), form indeterminate nodules. Indeterminate nodules are elongated because of a persistent apical meristem that leads to the formation of different developmental stages or zones (Vasse et al., 1990; Roux et al., 2014). Zone I (ZI) is the rhizobia-free meristematic region, containing mostly non-differentiated dividing cells. Zone II (ZII) corresponds to the infection zone, comprising the pre-infection zone with non-infected differentiating cells and cells releasing rhizobia from ITs into Syms with rhizobia enclosed within the SymM; the proximal ZII includes the area where plant and bacterial cells differentiate and enlarge through endoreduplication (Cebolla et al., 1999). Zone III (ZIII) comprise large cells containing N-fixing, Sym-enclosed bacteroids interspersed with uninfected plant cells. Between ZII and III is Interzone II–III (IZII–III), a region several cell layers wide. The senescent zone, Zone IV (ZIV), gradually develops in the proximal part of the nodule and becomes apparent after several weeks (Perez Guerra et al., 2010). These cells are contained within the epidermal, outer cortical, vascular, and inner cortical cells of the nodule.
Most agricultural fields lack sufficient phosphorus (P) for maximum crop yields. Legumes are especially affected because they are typically grown symbiotically, and it has been shown that legumes reliant on SNF require higher P compared to legumes grown with N fertilizer. Studies in diverse legumes have shown growth parameters and symbiotic responses are improved by P fertilization (Israel, 1987; Itoh, 1987; Pereira and Bliss, 1989; Al-Niemi et al., 1997; Vance, 2001; Vance et al., 2003; Schulze and Drevon, 2005; Hernandez et al., 2009; Liu et al., 2018). Legume nodules act as a P sink in P deprivation, with nodule P concentration increased up to three times compared to that of leaves and roots (Sa and Israel, 1991; Schulze et al., 2006). Nodulation is accompanied by reprogramming of both plant and bacterial metabolism including membrane biosynthesis required for division and differentiation of both rhizobial and plant cells, organelle function, and formation and development of SymMs. These membrane lipids are among the P sinks in nodules and are typically rich in phospholipids (PLs) primarily followed by galactolipids (GLs; Hernández and Cooke, 1996; Whitehead and Day, 1997; Gaude et al., 2004). The major PLs present in nodules are phosphatidylcholine (PC) and phosphatidylethanolamine (PE), while phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidylserine (PS), and phosphatidic acid (PA) are low in abundance (Gaude et al., 2004). The major GLs reported in nodules are monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) where they are found in the SymM and likely other plant membranes (Gaude et al., 2004).
Because nodules are symbiotic organs, bacteroid membrane lipids are also present (de Rudder et al., 1999; Lopez-Lara et al., 2003, 2005). Rhiziobial membrane lipid composition has been extensively studied in Bradyrhizobium and Sinorhizobium meliloti spp. in cell cultures (Miller et al., 1990; de Rudder et al., 1997) and has been found to contain PG, cardiolipin and PE as major membrane lipids, followed by PC in substantial amounts. Under P limiting cell culture conditions, rhizobial membrane PLs serve as a pool of metabolizable phosphate (Pi) for the synthesis of P-free lipids such as sulfoquinovosyl diacylglycerol, an ornithine containing lipid and diacylglyceryl trimethylhomoserine (Zavaleta-Pastor et al., 2010). The composition and changes in bacteroid membrane lipids in absence of P have been studied in the large determinate nodules formed by G. max–Bradyrhizobium japonicum (Gaude et al., 2004).
The conventional way of analyzing lipids from plant tissues or seeds is through organic extraction followed by analysis by mass spectrometry (Narasimhan et al., 2013). The extracted lipids are typically introduced in the mass spectrometer by direct infusion using an electrospray ionization (ESI) source (Brügger et al., 1997). Precursor and neutral loss scans are performed for specific lipid detection and quantification, with standards (Shiva et al., 2013). The biggest problem with this type of analyses is the information on the spatial distribution of where these lipid metabolites originated is lost during their extraction processes (Sturtevant et al., 2016, 2017). Imaging techniques using genetically encoded biosensors have been developed and used to reveal tissue and subcellular spatial differences in specific membrane lipid classes; however, these biosensors cannot discriminate degree of lipid unsaturation and can be difficult to use quantitatively (Colin and Jaillais, 2020). Semi-quantitative matrix-assisted laser desorption ionization–mass spectrometry (MALDI–MS) imaging has emerged as a promising alternative to analyze the spatial distribution of plant membrane lipids (Horn et al., 2012; Dueñas et al., 2017; Woodfield et al., 2017). MALDI–MS imaging (MALDI–MSI) has the advantages of high resolution leading to accurate mass measurements of the metabolites which are critical for imaging studies (Gemperline et al., 2015). Among membrane lipids, PC species are amenable for MALDI–MSI because of their abundance and positively charged quaternary ammonia group. In addition, MALDI–MSI allows determination of PC lipid unsaturation.
Inspired by reports showing spatial differences in PC species distribution in seed tissues (Sturtevant et al. 2017; Woodfield et al., 2017), we wondered whether PC distribution would be evenly across nodule zones that have distinct physiological functions in indeterminate nodules. Further, we were curious about whether PC species distribution would change in P deprivation. In this study, we investigated membrane lipid composition in nodulated M. truncatula plants that were P deprived compared to controls. We analyzed the spatial distribution of PC in nodules from both P starved and control plants. Our findings showed expected changes in membrane lipid composition during P deprivation and present insights of PC distribution and remodeling in nodule development, in SNF, and those processes during P deprivation.
Results
P deficiency has differing effects on M. truncatula membrane lipid composition in leaves, roots, and nodules
To induce P deprivation, we grew plants in aeroponics chambers to control the growth medium conditions. R108 genotype plants were grown for 5 d in N and P replete media, starved for N for 5 d while receiving full P, and subsequently starved for P at the same time as they were inoculated with Sinorhizobium melliloti (−P). In these conditions, nodules started to develop at the onset of P starvation. Control plants were grown side-by-side in replete P conditions (+P). Media was assessed daily for P concentrations (Supplemental Table S1). After 4 weeks, −P plants were assessed for P deprivation characteristics (Li et al., 2001; Williamson et al., 2001; Schulze et al., 2006; Hernandez et al., 2007; Morcuende et al., 2007; Hernandez et al., 2009; Sulieman et al., 2013a, 2013b; Ding et al., 2020). Medicago truncatula −P plants showed impaired growth compared to those in +P, with lower fresh and dry weights (DWs) and lower inorganic Pi and total P in leaves, roots, and nodules (Supplemental Figure S1, A–E). The nodulated root systems from −P plants were found to have higher numbers of lateral roots and fewer and smaller nodules than those from +P plants (Supplemental Figure S2, A–H). Plants had reduced nitrogenase activity, as revealed by the acetylene reduction assay (ARA; Supplemental Figure S2I). This confirmed that our −P conditions were effective in inducing P deprivation.
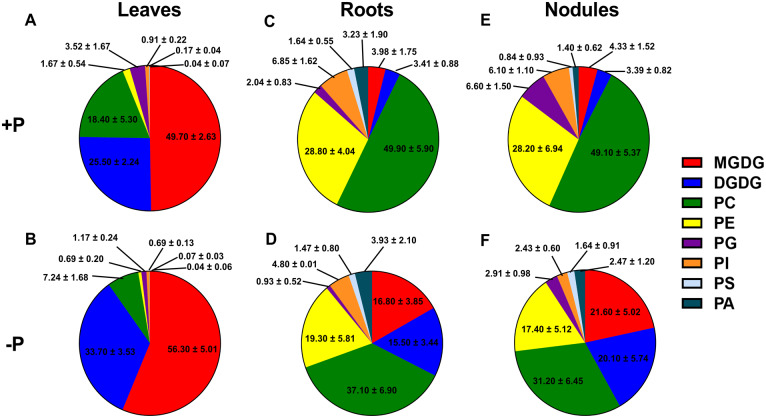
To gain insight into membrane lipid composition changes under P deprivation versus sufficiency, lipid profiling using ESI–MS was carried out to determine the polar membrane glycerolipid composition of leaves, roots, and nodules, including PLs and GLs. In leaves, in +P conditions, the GLs MGDG and DGDG were found to be highly abundant, comprising almost 75% of total glycerolipids measured (Figure 1A). In −P leaves, the composition of GLs increased to 90% of total membrane lipids (Figure 1B). In both +P and −P, MGDG species were found to be more abundant than DGDG species, with increases in DGDG species higher than MGDG in −P as compared to +P (Figure 1, A and B). MGDG and DGDG species of 36:6 fatty acid composition predominated in both +P and −P leaves (Figure 1, A and B). In −P, DGDG 36:3 species were found to increase markedly compared to +P, although their levels were found to be ∼10-fold lower than those of DGDG 36:6 species (Supplemental Figure S3A). MGDG and DGDG 36:6 and DGDG 36:3 species accumulated close to two-fold higher in −P leaves (Supplemental Figure S3, A and B), accounting for most of the GL increases observed (Figure 1, A and B). In +P leaves, PLs were found to comprise only ∼25% of total membrane glycerolipids measured, with PC and PG species encompassing 75% and 14% of the total PLs, respectively (Figure 1A). Smaller contributions to total PL were made by PE (1.67%), PI (0.91%), PS (0.17%), and PA (0.04%; Figure 1A). In −P leaves, PLs comprised only 10% of the total glycerolipids, with a significant decrease observed in quantities of PC (7.24%) and PG (1.17%; Figure 1, A and B). PC species with fatty acid compositions 34:3, 34:2, 36:6, 36:5, 36:4, and 36:3 were markedly reduced in −P (Supplemental Figure S3C). Levels of PE species with fatty acids compositions 34:3, 34:2, 36:5, and 36:4 were lower in −P leaves (Supplemental Figure S3D). PG 34:4 significantly contributed to decrease in the PG species (Supplemental Figure S3E) as it was reduced almost by two-fold in −P leaves. No significant difference in PA and PI species was observed between +P and −P leaves (Supplemental Figure S3, F and H). Two PS species with fatty acid composition 40:3 and 42:3 were reduced in −P leaves (Supplemental Figure S3G).
Figure 1.
Total lipid composition for leaves, roots, and nodules in mol%. A and B, represent total lipid composition in leaves. A, +P; B −P. C and D, represent total lipid composition in roots. C, +P; D −P. E and F, represent total lipid composition in nodules. E, +P; F −P. The values are mean ± standard deviation (sd).
In roots, contrasting differences in the membrane composition were observed in +P versus −P compared to leaves. PL species contributed 92% to the total glycerolipids in +P roots and showed a decline to 68% in −P (Figure 1, C and D). PC species were the most abundant membrane lipids encompassing 50% of the total lipids in roots in +P which decreased to 37% in −P roots, respectively (Figure 1, C and D). Relatively abundant PC species with fatty acid compositions of 34:3, 34:2, 36:5, 36:6, 36:4, 36:3, and 36:2 showed a significant reduction in −P (Supplemental Figure S4A). In +P roots, lower contributions to total lipid composition were made by PE (28.80%), PG (2.04%), PI (6.85%), PS (1.64%), and PA (3.23%; Figure 1C). P deprivation resulted in lower concentrations of root PE (19.30%), PG (0.93%), and PI (4.80%), while PS (1.47%) and PA did not significantly change (3.93%; Figure 1D). Like PC, 34- and 36-carbon containing PE species (34:3, 34:2, 36:6, 36:5, 36:3, and 36:2) were reduced in −P roots (Supplemental Figure S4B). PG 32:0, 34:2, and 34:3 and PI 34:3 and 34:2 were also found to be decreased in −P roots (Supplemental Figure S4, C and D). Most of the PS and PA species showed no significant difference between +P and −P roots, except PS 40:2 and 42:2 which decreased in –P (Supplemental Figure S4, E and F). In +P roots, the GLs MGDG and DGDG comprised only 7.39% of the total glycerolipid composition, with 3.98% and 3.41%, respectively (Figure 1C). In –P roots, the GL content increased to 32.30% of total glycerolipids, with MGDG comprising 16.80% and DGDG 15.50% of the total (Figure 1D). Similar to leaves, the 36:6 fatty acid moieties of both MGDG and DGDG were found to predominate in +P and −P roots (Supplemental Figure S4, G and H). MGDG 36:6 was relatively more abundant compared to DGDG 36:6 in roots in −P conditions (Supplemental Figure S4, G and H). Levels of most 34- and 36-carbon species of DGDG and 36-carbon species of MGDG (including 34:3) were increased in −P roots in (Supplemental Figure S4, G and H).
We found that nodules had a similar membrane glycerolipid profile to roots, with PLs most abundant. In +P nodules, PC (49.10%) species comprised the most abundant class of membrane lipids, with PE (28.20%) second in abundance of the total glycerolipids (Figure 1E). In −P nodules, PC species remained the most abundant class of membrane glycerolipids (31.20%). Major PC species that showed decreases in −P nodules included those of 34:3, 34:2 34:1, 36:6, 36:5, 36:4, 36:3, and 36:2 compositions (Supplemental Figure S5A). PE species were reduced to 17.40% in −P nodules (Figure 1F), with the 34- and 36-carbon fatty acid-containing species showing the most significant changes (Supplemental Figure S5B). Other PL species in nodules that showed changes in their levels between the two different growth conditions were PG (6.60% in +P to 2.91% in −P) and PI (6.10% in +P to 2.43% in −P; Figure 1, E and F). Similar to PC and PE, the 34- and 36-carbon containing fatty acid species of PG and PI showed the largest changes in −P nodules (Supplemental Figure S5, C and D). No significant differences were observed in PS and PA species in −P nodules compared to +P nodules (Figure 1, E and F; Supplemental Figure S5, E and F). In +P nodules, the GLs MGDG (4.33%) and DGDG (3.39%) comprised 7.72% of the total glycerolipid composition (Figure 1E), similar to that of roots. In −P nodules, MGDG (21.60%) and DGDG (20.10%) increased to 41.70% of the total glycerolipids in nodules, noticeably (8%) higher than that observed in −P roots (Figure 1, D and F). The 36:6 fatty acid species were the most abundant species of GLs in nodules regardless of the growth conditions (Supplemental Figure S5, G and H). DGDG 36:6 accumulated approximately four-fold in −P nodules and MGDG 36:6 showed a three-fold increase (Supplemental Figure S5, G and H). Most 34- and 36-carbon species of DGDG and 36-carbon species of MGDG (including 34:3) showed increased levels in nodules in P deficiency (Supplemental Figure S5, G and H).
MALDI–MSI shows PC species are non-uniformly distributed in M. truncatula nodules
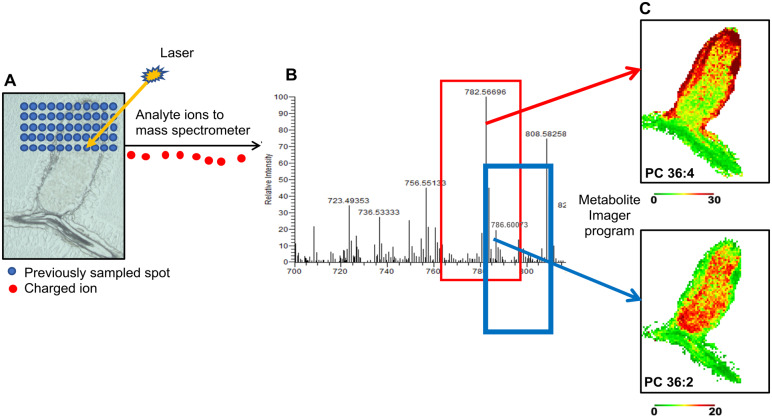
MALDI–MSI is a semi-quantitative technique that permits mapping of metabolites in biological sections, yielding a spatial metabolic context (Chen et al., 2009; Fuchs et al., 2010; Gemperline et al., 2015; Sturtevant et al., 2016; Lu et al., 2018). With this technique, matrix-coated nodule sections are subjected to laser rastering in a series of points, collected as x–y coordinates. The laser-excited metabolite ions enter a mass spectrometer where their m/z values are measured. Subsequently, metabolites are identified and used to show individual metabolite intensity at each x–y coordinate via false-color images (Figure 2). Among the membrane glycerolipids, PC species are abundant and contain a quaternary ammonia group with a permanent positive charge and are thus detectable in tissue sections. MALDI–MSI has been used to investigate PC species distribution in animal tissues (Jackson et al., 2007; Chen et al., 2009; Fuchs et al., 2010). Among plants, barley germinating seeds and Brassica napus developing seeds have been subjected to MALDI–MSI for PC distribution (Gorzolka et al., 2016; Woodfield et al., 2017; Lu et al., 2018; Colin and Jaillais, 2020). We sought to bring MALDI–MSI to indeterminate M. truncatula nodules to examine PC species distribution, the most abundant membrane glycerolipid type found there (Figure 1, E and F). The instrument we used had 40-µm resolution, capable of analyses at the tissue scale.
Figure 2.
Schematic showing the MALDI–MSI method. A, A laser is rastered over a DHB-coated nodule cross-section in a series of spot points with a given x–y coordinate. The example shown is a nodule cross-section of M. truncatula R108 (25 µm). B, The analyte ions produced are directed to a mass spectrometer, where their m/z values are measured. C, Metabolites are identified, and images are generated using Metabolite Imager. For example, PC 36:4 and PC 36:2 as shown.
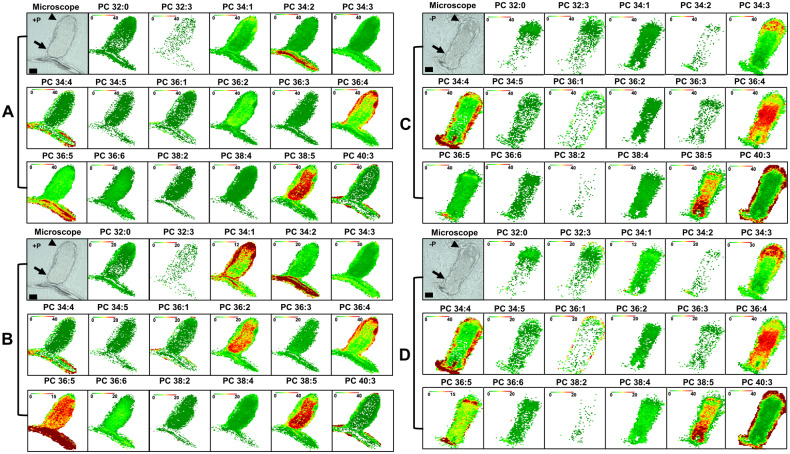
To examine nodule PC species distribution, we first evaluated the PC distribution in +P nodules (Figure 3, A and B; Supplemental Figure S6, A and B). Figure 3A and Supplemental Figure S6A show false-color image maps where all the species are shown at the same intensity scale (40 mol%), revealing the absolute abundance of each PC, allowing comparison of PC species abundance to one another. For example, this analysis shows that PC species 34:1, 36:2, 36:5, and 38:5 are more abundant in nodules, and for 36:4 in the surrounding root tissue, in comparison with other species (Figure 3A;Supplemental Figure S6A). Overall, it can be seen that the PC species abundance determined by semi-quantitative MALDI–MSI agrees reasonably well with results obtained via the more quantitative ESI–MS (Supplemental Figure S5A) where PC species 34:1, 36:2, 36:4, and 36:5 were abundantly present in nodules, as has been similarly shown in seed studies where PC species have been imaged by MALDI–MS (Woodfield et al., 2017). The disadvantage of fixed value false-color MALDI–MS images is that the spatial distribution of lipid molecular species is difficult to see for species with high or low amounts in tissues: either overexposed in brown/dark red for high levels or all green for low levels. To surmount this, we adjusted the scale for in each MALDI–MS image and created a relative distribution profile, so that the maximum ion intensity (in mol%) was brown/dark red to show the distribution of particular lipid molecular species in each nodule section (Figure 3B;Supplemental Figure S6B).
Figure 3.
MALDI–MSI of PC metabolites in M. truncatula nodules. A and B, Bright-field images of nodule cross-section (25 µm) before coating with DHB matrix grown under +P. C and D, Bright-field images of nodules cross-section (25 µm) before coating with DHB matrix grown under −P is shown on the left. A and C, Distributions of selected PC molecular species are shown with fixed mol% to show absolute distribution profiles. B and D, Adjusted mol % to show relative distribution profiles. Bars = 500 µm. Proximal zone is indicated by arrowhead and distal zone by arrow. MALDI scale shows mol% with green and red representing low and high levels, respectively. Representative images of N = 5 each for +P and −P.
The adjusted image maps reveal dramatic differences in distributions of PC species in +P nodules (Figure 3B;Supplemental Figure S6B). For example, PC 34:2 and 36:5 have high distribution in the root tissue attached to the nodule, while PC 36:5 shows fairly high distribution in the central parts of the nodule (Figure 3B). In contrast, PC 36:2 and 38:5 have a similar distribution in nodules but are not as abundant in root tissue. PC 36:2 and 38:5 species are not found as distally in nodules as 36:5. Other PC species, for example, PC 34:1 and 36:4, show higher abundance in the nodule epidermal, parenchymal, and vascular regions, with lower accumulations in the nodule’s central region (Figure 3B). Other species, for example, PC 36:3 and 36:6 show a relatively uniform distribution across the nodule (Figure 3B;Supplemental Figure S6B). Thus, these data show that in M. truncatula +P nodules, PC species differentially accumulate in different regions of nodules.
Different nodule PC species show differential spatial changes in P stress
We sought to determine changes in PC distribution in −P nodules as compared to +P nodules using MALDI–MSI (Figure 3, C and D; Supplemental Figure S6, C and D). Maps created at the same intensity scale showed that most PC species declined in abundance in −P nodules, in agreement with the quantitative results determined by the ESI–MS analysis (Figure 3C;Supplemental Figure S5A). To compare whether there were changes in distribution of particular PC species in −P nodules compared with +P nodules, we used image maps for −P nodules that were scaled to the same mol% values used in +P nodules (compare Figure 3, B and D; Supplemental Figure S6, B and D). These data revealed that many PC species changed distribution in response to P stress. For instance, PC 34:2, 36:2, 36:5, and 38:5 species were found in lower abundance across the nodule in −P conditions. These PCs were also found to be consistently lower in the ESI–MS lipidomic analysis (Supplemental Figure S5A). Curiously, PC species 34:3 had higher distributions in the distal part of nodule zone II in −P conditions. PC species 34:1 and 36:4 showed relatively similar localization pattern with higher abundance in the peripheral tissues of +P nodules, PC 34:1 was less abundant in −P nodules and PC 36:4 was found at higher abundance in the central part of −P nodules (Figure 3, B and D). In a similar vein, PC 36:2 and 38:5 species, which were both abundant in the central parts of +P nodules, were both observed to have lower distribution in −P nodules, but PC 38:5 had fairly high abundance in the proximal parts of the −P nodule (Figure 3, B and D). PC 36:5 also had lower abundance in −P nodules compared to +P (Figure 3, B and D). Some PC species did not show major changes in spatial distribution in −P nodules compared to +P nodules; their image maps are shown in Supplemental Figure S6. Unfortunately, we were not able to image the non-P containing GLs (MGDG and DGDG) in nodule sections. This is likely because of ion suppression caused by PC (Supplemental Figure S7; Supplemental Dataset 1). The phenomenon of ion suppression caused by PC has been observed previously (Petkovic et al., 2001; Fuchs et al., 2009, 2010; Emerson et al., 2010; Horn et al., 2012; Woodfield et al., 2017; Stopka et al., 2018; Colin and Jaillais, 2020).
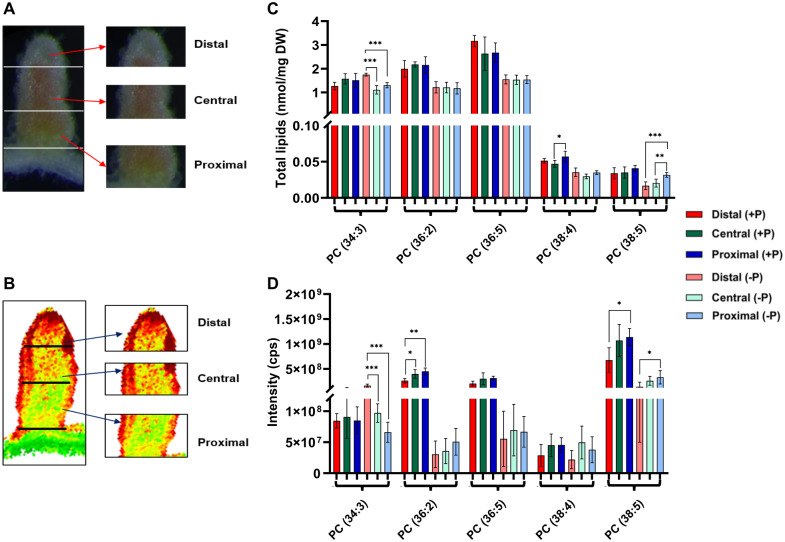
Validation of the differences in PC distribution in dissected nodules
There are known limitations of the MALDI–MSI technique, which include ion suppression, lack of absolute quantitation (Horn et al., 2012; Woodfield et al., 2017), and the observation that a longitudinal section does not represent the entire nodule in three dimensions. To address this, we used a second approach to compare and validate the differences in spatial distributions observed by MALDI–MSI. Twenty-eight days post-inoculation (dpi) nodules from plants grown in +P and in −P conditions were dissected into distal (approximately ZI to ZII), central (IZII/III and ZIII), and proximal (proximal zone III–IV) sections (Figure 4A). We extracted lipids from the separated nodule sections and subjected them to ESI–MS to determine the PC species composition of each. The resulting data were compared to those obtained from nodule MALDI–MS images that were “virtually” sectioned (Figure 4B), followed by a computational determination of the intensity of PC species in each virtual section (Figure 4B). Comparison of the ESI–MS to MALDI–MS results showed reasonable agreement with each other. Our results showed that PC 34:3 was similarly abundant in all nodule zones in +P nodules. In −P nodules statistically significant differences were found similarly when comparing distal to central and distal to proximal areas in both ESI–MS and MALDI analyses (Figure 4, C and D). Differences between central and proximal regions in −P were not significantly different. For PC 36:2 ESI–MS analysis showed relatively uniform concentrations of this PL in all three nodule areas. In contrast, MALDI results showed statistically significant differences in +P conditions when comparing distal to central and distal to proximal areas of the nodule (Figure 4, C and D). PC 36:5 and PC 38:4 were found at similar abundance in +P and −P nodules with one exception, PC 38:4 was found to be significantly lower in the central area of the nodule compared to the proximal area of the nodule in ESI results; in comparison MALDI showed a similar distribution of PC 38:4 in these zones (Figure 4, C and D). PC 38:5 was found to be similarly distributed in −P nodules by each analysis with the highest abundance in the proximal part (Figure 4, C and D). However, it was significantly lower in distal compared to the proximal part of +P nodules by MALDI–MSI. Similar differences in trend between ESI–MS and MALDI–MS were also observed for PG 34:4 in a study on maize leaf (Dueñas et al., 2017).
Figure 4.
Comparison of ESI–MS and MALDI–MSI results for PC species in different sections of dissected nodules. A, Bright-field image of a nodule dissected into distal, central, and proximal sections used for ESI–MS analysis. B, Virtually dissected nodule into distal, central, and proximal sections used for MALDI–MSI analysis. C, Quantitative data of representative PC species obtained from distal, central, and proximal nodules sections using ESI–MS. Values are expressed as total lipids (nmol/mg DW of tissue) after normalizing with the amount of respective standards. D, Representative PC species obtained from distal, central, and proximal nodules sections using MALDI–MSI are expressed in intensities count per second; values are mean ± sd (N = 5). Error bars indicate sd. Asterisks indicate a significant difference between the control plants (+P) and test plants (−P), as determined using one-way ANOVA analysis; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Discussion
P deficiency is one of the limiting factors adversely affecting worldwide legume growth and productivity (Tesfaye et al., 2007). Legumes that are nodulated and fixing N2 are known to have a higher demand for P than those grown on mineral N (Graham, 1981; Vance et al., 2003; Sulieman and Tran, 2015). Processes that contribute to N2-fixing legumes’ high P requirement include the high energy required for nitrogenase to function at a high level, signal transduction/enzyme regulation particularly those pathways regulated by protein phosphorylation, nucleic acid biosynthesis, and membrane lipid biosynthesis and homeostasis (Graham and Vance, 2000).
This study focused on membrane lipid homeostasis in M. truncatula plants undergoing severe P deficiency. We confirmed that M. truncatula growth was impaired in P deprivation compared to control P replete (+P) plants in our aeroponic growth system (Supplemental Figures S1 and S2), showing that M. truncatula plants were stressed similarly to those in previous studies on P deprivation in M. truncatula (Sulieman et al., 2013a, 2013b), M. sativa (Sulieman et al., 2013a, 2013b), rice (Li et al., 2001), A. thaliana (Williamson et al., 2001; Morcuende et al., 2007), common bean (Hernandez et al., 2007; Hernandez et al., 2009), white lupin (Schulze et al., 2006), and switchgrass (Ding et al., 2020).
The glycerolipid composition of Sinorhizobium-nodulated M. truncatula leaves and roots (Figure 1, A and C; Supplemental Figures S3 and S4) was found to be similar to that observed in Bradyrhizobium-nodulated soybean leaves and roots (Gaude et al., 2004; Narasimhan et al., 2013) with plastidial DGDG and MGDG species found in highest abundance in leaves, and PC and PE species in highest amounts in roots. M. truncatula nodule glycerolipid content was also found to be similar to that of soybean (Gaude et al., 2004), with a similar profile as roots, but with more PG found in nodules as compared to roots (Figure 1E). As a symbiotic organ, a fraction of the glycerolipid content in nodules comes from the internalized bacteroids, which in the larger, determinate Bradyrhizobium-soybean nodules contain predominantly PE, PC, PG, and PI in their membranes (Gaude et al., 2004). To our knowledge, the polar lipid composition of the bacteroids from S. meliloti-M. truncatula nodules has not yet been assessed, but since PG is a prominent lipid found in free-living S. meliloti (Lopez-Lara et al., 2005), the increased PG found in nodules compared to roots may come from the bacteroid component of nodules. The lipid profiling results reported here show higher content of PC than PE in roots and nodules (Figure 1, C and E). This differs from lipid compositions found in soybean (Gaude et al., 2004) and in a previous M. truncatula study in genotype A17 (Si et al., 2019), who observed more PE than PC in roots and nodules. The differences might be attributable to the different growth conditions employed in the different studies and the different species and M. truncatula genotypes studied.
In P deprived M. truncatula, we observed changes in glycerolipid composition that were overall similar to those observed previously in soybean (Gaude et al., 2004) and M. truncatula (Si et al., 2019). Leaves, roots and nodules showed increases in GLs MGDG and DGDG, as anticipated (Figure 1, B, D, and F), although PC still comprised the largest single class of glycerolipid in roots and nodules (Figure 1, D and F). The increases in the non-P-containing GLs during P stress in the roots and nodules instead of PLs is likely necessary to conserve P for other essential biological processes like biosynthesis of nucleic acids, phosphorylated sugars, and signaling by phosphorylated proteins. In −P nodules, similar to +P nodules, PL species detected in our study could originate from bacteroid membranes. We note that S. meliloti possesses three Pi transport systems each of which is sufficient for SNF in alfalfa, M. truncatula’s close relative, in normal +P conditions (Yuan et al., 2006). It is not yet known how these are regulated in internalized bacteroids of nodulated plants when P is depleted from the environment.
One of the advantages of studying the effect of P deprivation on membrane lipids in M. truncatula is that its indeterminate nodules make it possible to examine whether specific PLs change abundance at different stages of symbiotic development using the developmental gradient found in the continually differentiating nodules. MALDI–MSI, a semi-quantitative technique, allows visualization of metabolites at the tissue level with fairly high resolution (Gemperline et al., 2015). For membrane lipids, MALDI–MSI is currently limited to abundant and well-ionizable PLs and has the advantage of being able to discriminate their degree of unsaturation. PC is the most abundant membrane lipid in nodules and possesses a quaternary ammonia group containing a permanent positive charge (Fuchs et al., 2010; Horn et al., 2012; Woodfield et al., 2017). Therefore, detection of PC species is sensitively detected in complex lipid mixtures or tissue sections than other less abundant or poorly ionizable molecular species (Petkovic et al., 2001; Emerson et al., 2010; Fuchs et al., 2010; Horn et al., 2012; Woodfield et al., 2017; Stopka et al., 2018; Colin and Jaillais, 2020). To address the question of PC distribution in nodules, we used this technique. Our results (Figure 3, A and B) showed spatial heterogeneity of many PC species in nodules that are obscured when using regular analytical extraction-based techniques. The differences in spatial distribution of PC species observed here open the possibility that there may be cell- and tissue-specific regulation of membrane lipid metabolism during nodule development. We then used MALDI–MSI to address PC distribution in −P nodules, with our results showing a markedly different spatial heterogeneity for many PCs from patterns observed in +P nodules (Figure 3). Traditional organic extractions of membrane lipids of dissected nodule sections followed by with ESI–MS analysis of PC content for selected PC species showed strong associations with virtual MALDI–MSI sections (Figure 4), validating the comparison. For example, the MALDI–MSI results show that PC 38:5 abundant in +P inner nodule tissues changes localization to several cell layers at the distal end of −P nodules (compare Figure 3, B with D). In contrast PC 36:4, different from PC 36:5 by one desaturation step each, is found in peripheral nodule cells in +P conditions, while in −P nodules was found as an abundant species in the central nodule zones. These data suggest that other membrane lipid species in addition to PC, particular fatty acid biosynthetic enzymes, and specific desaturases may be differentially regulated in nodule cells at different stages of nodule cell maturation and in +P versus −P environmental conditions. Another possibility is that different PC species may be better substrates for MGDG and DGDG synthetases (Si et al., 2019), which may themselves be heterogeneously distributed spatially in nodules.
To fully understand the spectrum of membrane lipid patterning in P deprived nodules compared to those from legumes grown in P replete conditions will require new methods to quantitate and image PL and GL accumulation and turnover at tissue and cellular resolution (Stopka et al., 2017, 2018; Agtuca et al., 2020; Colin and Jaillais, 2020). GLs will be of interest to localize in nodules because they are found in nodules and increase during P deprivation (Gaude et al 2004, Si et al. 2019; this study). However, the ion suppression caused by PC in nodules interferes with GL detection in the same tissue using MALDI–MSI (Supplemental Figure S7; Supplemental Dataset S1). Ion suppression by PC of other lipids has been previously observed and discussed (Petkovic et al., 2001; Fuchs et al., 2009; Emerson et al., 2010; Fuchs et al., 2010; Horn et al., 2012; Woodfield et al., 2017; Stopka et al., 2018; Colin and Jaillais, 2020). Thus, technological advances are needed to localize GLs and less ionizable PLs found in cell membranes (Agtuca et al., 2020; Colin and Jaillais, 2020).
PC species are abundant membrane lipids that are mostly thought of as structural lipids as compared to less abundant PLs, like PI species, that have additional well-established signaling roles (Boss and Im, 2012; Noack and Jaillais, 2020). When one considers PC in structural roles, our results suggest the possibility that modifying plants to engineer higher abundance of PC species to favor those species found in higher amounts in the N-fixing zones of nodules could result in altered, perhaps increased, SNF efficiency. The distinction between structural and signaling roles for PCs are becoming blurred, with the finding that flowering time in Arabidopsis is influenced by PC content and that florigen FLOWERING LOCUS T binds preferentially to PC species with 18:3 fatty acids in vitro (Nakamura et al., 2014). Plants that are engineered to contain higher 18:3 fatty acid have been shown to flower later than controls (Arondel et al., 1992). Together these data suggest that specific PC species or their metabolites could have signaling roles (Colin and Jaillais, 2020). Thus, we speculate that in nodules, different PC species may have specific roles, signaling as well as structural, in different zones of M. truncatula nodules and that these roles may change in P deprivation. We further suggest that other membrane lipids, such as GLs and other PL species are likely to demonstrate non-homogeneous spatial distribution in nodules and this is likely to be of functional importance. Because of the technical challenges in localizing these molecules spatially, especially at the subcellular level, it is not possible to speculate about function. It is known that altering the GL composition by modifying DGDG abundance in nodules alters nodule function (Si et al. 2019), but the mechanism by which nodules are functionally altered, or which intracellular membranes are affected, is not yet clear. Further work to investigate whether manipulating the membrane lipid composition in nodules could alter nodule function and/or P deprivation effects on N fixation will address issues about particular membrane lipids and function.
Materials and methods
Plant growth conditions
Seed collection, surface sterilization, and germination
Medicago truncatula R108 wild type (WT) pods were crushed to release their seeds. Seeds were scarified with concentrated H2SO4 for 5 min, rinsed in sterile water, surface sterilized with 6% (v/v) sodium hypochlorite for 1.5 min, and rinsed with sterile water. The seeds were then imbibed in sterile water for 3 d at 4°C in the dark. Seeds were subsequently placed on 1% water agar petri dishes for germination. The plates were inverted and kept in the dark at room temperature (RT) for 2 d to allow seeds to germinate.
Plant growth in aeroponic systems
Germinated seedlings were grown using aeroponic chamber systems prepared similarly as those described in the Medicago handbook (Barker et al., 2006) and previously (Veereshlingam et al., 2004). Briefly, our aeroponic chamber is a 20 gal. trashcan (Rubbermaid, Atlanta, GA, USA) modified with a black Plexiglas lid containing 4 mm holes to accommodate seedlings. It is powered by a misting humidifier (Defensor 505; Texas Air Systems, Irving, TX, USA) sealed with silicone sealant. Plants were grown in two aeroponics chambers side by side. Lullien’s medium was used for growth (Lullien et al., 1987). Lullien’s medium contains 5 mM NH4NO3, 0.52 mM K2SO4, 0.25 mM MgSO4, 1 mM CaCl2, 50 µM Na2EDTA, 30 µM H3BO3, 10 µM MnSO4, 0.7 µM ZnSO4, 0.2 µM CuSO4, 0.1 µM Na2MoO4, 0.04 µM CoCl2, 23.7 µM FeSO4, 33.2 µM FeCl2, and 5.5 mM potassium Pi buffer at pH 6.9. Seedlings were grown in full Lullien’s medium for 5 d to encourage robust growth, then in Lullien’s medium without 5 mM NH4NO3 for 5 d to induce N starvation. Then media was changed to fresh Lullien’s medium without 5 mM NH4NO3 for +P plants. For the −P plants, fresh Lullien’s medium without 5 mM NH4NO3 and without potassium Pi buffer was used. 1 M KOH was added as a potassium source and to adjust the pH to 6.9. Plants were inoculated with S. meliloti Rm41 and grown for 4 weeks.
Plant phenotypes
Plants were phenotyped at 28 dpi. Plant fresh and DW were measured. Plants and nodules were photographed by camera (Canon model PC1562; Canon USA, Huntington NY, USA) and microscopically (Leica dissection microscope, MSV269; Buffalo Grove, IL, USA). Nodule length and width were measured, and lateral roots were enumerated. The ARA was used to measure nitrogenase activity (Dilworth, 1966). Glass bottles (38 mL) were preloaded with 1 mL of Lullien’s medium with Pi and without Pi in order to maintain the consistency of the growth conditions for +P and −P plants, respectively. The bottles were sealed with gas-tight rubber cap. Immediately, 3.7 mL gas was withdrawn from the bottle with a gas-tight syringe and 3.7 mL of acetylene was introduced back in the bottle. Plants in acetylene-contained bottles were maintained at RT and after 2 h 300 µL of gas was removed from the bottle and injected into a gas chromatograph –flame ionization detector (Shimadzu GC-14A Gas Chromatograph, Dallas, TX, USA) to quantitate the amount of acetylene reduced to ethylene. Haysep T80/100 column (*6' x 1/8" stainless steel) from Alltech (Maynooth, Ireland) was used. Helium was used as the carrier gas. The GC conditions were as follows: injection temperature 120°C, constant isothermal column temperature at 55°C, and detector temperature 120°C. Fifteen plants were randomly sampled from +P and −P treatment conditions for determining plant fresh weight, DW, lateral root number, and nodule number. Fifteen individual nodules were arbitrarily picked from plant subjected or not to P stress for estimating nodule length, nodule width, nodule fresh weight, and nodule DW. For ARA five replicates from each growth conditions were used. One-way analysis of variance (ANOVA) was used to determine the statistical significance. Data from at least two independent experiments were used for this and all following experiments.
Total P and inorganic Pi quantitation
Fifteen to 30 mg of tissues were collected, frozen, and ground to a fine powder in liquid N. The ground tissues were suspended in 1% glacial acetic acid and vortexed thoroughly. Cellular debris was separated by a brief centrifugation and aliquots of the solution were assayed for Pi using a phosphomolybdate colorimetric assay as described previously (Chiou et al., 2006). For total P quantification, tissues were ashed, hydrolyzed, and subjected to colorimetric assay (Chiou et al., 2006). One milliliter of media was collected daily for 28 d from +P and −P growth conditions to assay amount of Pi in the growth media using colorimetric assay described in (Chiou et al., 2006). Six replicates of each growth conditions for each tissue type (leaves, roots, and nodules) were processed and analyzed. One-way ANOVA test was used to determine the statistical significance. Data from two independent experiments were used for inorganic Pi and total P experiments.
Lipid extraction and profiling
Leaves, roots, nodules, and nodules sections sliced with razor blades from 28 dpi M. truncatula plants were lyophilized at −80°C. Lipids were extracted from the tissues as described in (Shiva et al., 2013). Briefly, lyophilized tissues were homogenized in hot (75°C) isopropanol with 0.01% butylated hydroxytoluene and incubated at 75°C for 15 min to inactivate endogenous phospholipases. After incubation, a stream of N was used to evaporate the extract. The dry extract was dissolved in 1 mL CHCl3 and stored at −20°C. Lipid samples were analyzed using an ESI triple quadrupole mass spectrometer (ABI 3000; Applied Biosystems, Foster City, CA, USA). Internal standards were used for quantification of PL and GL species. The internal standards for GLs were 1.665 nmol 16:0–18:0-MGDG, 1.405 nmol di18:0-MGDG, 0.44 nmol 16:0–18:0-DGDG, and 1.48 nmol di18:0-DGDG, purchased from the Kansas Lipidomics Research Center (Manhattan, KS, USA). The internal standards for PLs were 0.6 nmol di14:0-PC, 0.6 nmol di24:1-PC, 0.3 nmol di14:0-PE, 0.3 nmol di24:1-PE, 0.3 nmol di14:0-PG, 0.3 nmol di24:1- PG, 0.3 nmol di14:0-PA, 0.3 nmol di20:0 (phytanoyl)-PA, 0.2 nmol di14:0-PS, 0.2 nmol di20:0 (phytanoyl)-PS, 0.287 nmol 16:0–18:0-PI, and 0.105 nmol di18:0-PI. We used the previously determined limit of detection (0.002 nmol) to filter lipids for analysis (Devaiah et al., 2006). The quantity of each lipid was determined after normalizing with the amount of the internal standard added and later expressed in per milligram DW of tissue. Five replicates of each treatment for each tissue type (leaves, roots, and nodules) were processed and analyzed. One-way ANOVA test was used to determine the statistical significance. Data from two independent experiments were used for lipidomic analysis.
MALDI–MSI
Twenty eight dpi nodules were fixed under vacuum with 4% paraformaldehyde in 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)-NaOH, pH 7.2 buffer for 15–45 min at RT. Nodules were rinsed 3 times with 50 mM PIPES-NaOH pH 7.2 for 10 min each rinse, replacing the buffer after each wash. A 10% (w/v) porcine gelatin solution in deionized water was prepared and equilibrated in a 37°C water bath with shaking for 2 h. Fixed nodules were embedded in the gelatin solution, flash frozen at −80°C at least overnight, and then were transferred to −20°C for 48 h prior to sectioning. Embedded nodules were sectioned at 25-μm tissue thickness using a cryo-microtome (Leica CM1950, Leica Microsystems, Buffalo Grove, IL, USA). Sections were collected using forceps and then placed on Superfrost microscope slides (Fisherbrand, 12-544-7). Slides with the thaw-mounted sections were lyophilized for 3 h, then used immediately for MALDI–MSI. All MALDI–MSI occurred within 12 h of cryo-sectioning. Bright-field images were taken of all sections used for MALDI–MSI using Leica microscope MSV269.
For MALDI–MSI, the matrix 2,5 dihydroxybenzoic acid (DHB) was used for analysis of PLs and GLs (MGDG and DGDG). DHB was applied by sublimation using an adapted method developed from (Hankin et al., 2007). MALDI–MSI data were collected on a hybrid MALDI-LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Waltham, MA, USA). The instrument laser was equipped with an N laser with a spot size of ∼40 μm. MALDI–MSI data acquisition conditions were as follows: laser energy was 16-µJ/pulse, a raster step size of 40 μm, 10 laser shots per raster step with zero sweepshot. Data were acquired using the Orbitrap mass analyzer with a resolution of 60,000, between an m/z scan range of 700–1,200. Raw mass spectra were processed into MALDI–MS images using ImageQuest software (Thermo Scientific). Images were generated using Metabolite Imager software (Horn and Chapman, 2014) and plotted as mol% on a color scale of green (low mol%) to red (high mol%). Imaging of PC molecular species are sums of intensities of [M + H]+ adducts. Each nodule image was carefully dissected virtually using the Metabolite Imager filter into distal, central, and proximal zones (Figure 4B) and in order to determine the amount of PC content for the zonal profiling study. Five individual nodules from different plants grown in +P and −P conditions were used for MALDI–MSI. One-way ANOVA was used to calculate the statistical differences. One independent experiment was done for MALDI–MSI.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characteristics of M. truncatula in P depleted conditions.
Supplemental Figure S2. Characteristics of nodulated root system of M. truncatula R108 in P depleted conditions.
Supplemental Figure S3. Total polar glycerolipids from leaves of M. truncatula R108 plants.
Supplemental Figure S4. Total polar glycerolipids from roots of M. truncatula R108 plants.
Supplemental Figure S5. Total polar glycerolipids from nodules of M. truncatula R108 plants.
Supplemental Figure S6. MALDI–MSI of PC metabolites in M. truncatula nodules.
Supplemental Figure S7. Suppression of GLs by PC in MALDI.
Supplemental Table S1. Pi levels in media during M. truncatula growth.
Supplemental Dataset S1. Suppression study of PC on GL detection in MALDI.
Supplementary Material
Acknowledgments
We thank Dr Guido Verbeck and the UNT Laboratory of Imaging Mass Spectroscopy for access to the ESI–MS and MALDI–MSI equipment. We also thank Kansas Lipidomics Research Center for providing PL and GL standards. We thank Dr Ruth Welti and Dr Kent Chapman for their expert guidance on our ESI–MS analysis, and Dr Kent Chapman for advice on comparing MALDI–MS and ESI–MS.
Funding
Research supported by US National Science Foundation grant NSF IOS-1733470 and University of North Texas Research funding to R.D.
Conflict of interest statement. The authors declare no conflict of interest.
R.D. conceived the project; R.D., A.P.A., and D.D. designed experiments; D.D., T.R., and D.K. carried out experiments; R.D., D.D., A.P.A., and T.R. analyzed the results; D.D., R.D., and A.P.A. wrote the manuscript; all authors edited the manuscript.
References
- Agtuca BJ, Stopka SA, Evans S, Samarah L, Liu Y, Xu D, Stacey MG, Koppenaal DW, Pasa-Tolic L, Anderton CR, et al. (2020) Metabolomic profiling of wild-type and mutant soybean root nodules using laser-ablation electrospray ionization mass spectrometry reveals altered metabolism. Plant J 103: 1937–1958 [DOI] [PubMed] [Google Scholar]
- AI-Niemi TS, Kahn ML, McDermott TR (1997) P Metabolism in the Bean-Rhizobium tropici symbiosis. Plant Physiol 113: 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arondel V, Lemieux B, Hwang I, Gibson S, Goodman HM, Somerville CR (1992) Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258: 1353–1355 [DOI] [PubMed] [Google Scholar]
- Barker DG, TP DM, EG SR, Lepetit M SW, FM RMN, Journet EP (2006) Growing M. truncatula: choice of substrates and growth conditions. In The Medicago truncatula Handbook. Mathesius U, Journet EP, Sumner LW (eds). ISBN 0-9754303-1-9 http://www.noble.org/MedicagoHandbook/
- Boss WF, Im YJ (2012) Phosphoinositide signaling. Annu Rev Plant Biol 63: 409–429 [DOI] [PubMed] [Google Scholar]
- Brügger B, , Erben G, , Sandhoff R, , Wieland FT, , Lehmann WD ( 1997) Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Natl Acad Sci USA 94: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, , Vinardell JM, , Kiss E, , Olah B, , Roudier F, , Kondorosi A, , Kondorosi E ( 1999) The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18: 4476–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hui L, Sturm RM, Li L (2009) Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectrom 20: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin LA, Jaillais Y (2020) Phospholipids across scales: lipid patterns and plant development. Curr Opin Plant Biol 53: 1–9 [DOI] [PubMed] [Google Scholar]
- de Rudder KE, , Thomas-Oates JE, , Geiger O ( 1997) Rhizobium meliloti mutants deficient in phospholipid N-methyltransferase still contain phosphatidylcholine. J Bacteriol 179: 6921–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rudder KE, Sohlenkamp C, Geiger O (1999) Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J Biol Chem 274: 20011–20016 [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X (2006) Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dilworth MJ (1966) Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim Biophys Acta 127: 285–294 [DOI] [PubMed] [Google Scholar]
- Ding N, Huertas R, Torres-Jerez I, Liu W, Watson B, Scheible WR, Udvardi M (2020) Transcriptional, metabolic, physiological and developmental responses of switchgrass to phosphorus limitation. Plant Cell Environ 44: 186–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueñas ME, Klein AT, Alexander LE, Yandeau-Nelson MD, Nikolau BJ, Lee YJ (2017) High spatial resolution mass spectrometry imaging reveals the genetically programmed, developmental modification of the distribution of thylakoid membrane lipids among individual cells of maize leaf. Plant J 89: 825–838 [DOI] [PubMed] [Google Scholar]
- Emerson B, Gidden J, Lay JO Jr., Durham B (2010) A rapid separation technique for overcoming suppression of triacylglycerols by phosphatidylcholine using MALDI-TOF MS. J Lipid Res 51: 2428–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Tinoco CE, Tschan F, Fuhrer T, Margot C, Sauer U, Christen M, Christen B (2020) Co-catabolism of arginine and succinate drives symbiotic nitrogen fixation. Mol Syst Biol 16: e9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B, Bischoff A, Suss R, Teuber K, Schurenberg M, Suckau D, Schiller J (2009) Phosphatidylcholines and -ethanolamines can be easily mistaken in phospholipid mixtures: a negative ion MALDI-TOF MS study with 9-aminoacridine as matrix and egg yolk as selected example. Anal Bioanal Chem 395: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Fuchs B, Suss R, Schiller J (2010) An update of MALDI-TOF mass spectrometry in lipid research. Prog Lipid Res 49: 450–475 [DOI] [PubMed] [Google Scholar]
- Gaude N, Tippmann H, Flemetakis E, Katinakis P, Udvardi M, Dormann P (2004) The galactolipid digalactosyldiacylglycerol accumulates in the peribacteroid membrane of nitrogen-fixing nodules of soybean and Lotus. J Biol Chem 279: 34624–34630 [DOI] [PubMed] [Google Scholar]
- Gemperline E, Jayaraman D, Maeda J, Ane JM, Li L (2015) Multifaceted investigation of metabolites during nitrogen fixation in Medicago via high resolution MALDI-MS imaging and ESI-MS. J Am Soc Mass Spectrom 26: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzolka K, Kolling J, Nattkemper TW, Niehaus K (2016) Spatio-temporal metabolite profiling of the barley germination process by MALDI MS imaging. PLoS One 11: e0150208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH (1981) Some problems of nodulation and symbiotic nitrogen fixation in Phaseolus vulgaris L.: a review. Field Crops Res 4: 93–112 [Google Scholar]
- Graham PH, Vance CP (2000) Nitrogen fixation in perspective: an overview of research and extension needs. Field Crops Res 65: 93–106 [Google Scholar]
- Hankin JA, Barkley RM, Murphy RC (2007) Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom 18: 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Ramirez M, Valdes-Lopez O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al. (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez G, Valdes-Lopez O, Ramirez M, Goffard N, Weiller G, Aparicio-Fabre R, Fuentes SI, Erban A, Kopka J, Udvardi MK, et al. (2009) Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol 151: 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández LE, Cooke DT (1996) Lipid composition of symbiosomes from pea root nodules. Phytochemistry 42: 341–346 [Google Scholar]
- Hirsch AM (1992) Developmental biology of legume nodulation. New Phytol 122: 211–237 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Chapman KD (2014) Metabolite Imager: customized spatial analysis of metabolite distributions in mass spectrometry imaging. Metabolomics 10: 337–348 [Google Scholar]
- Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee YJ, Chapman KD (2012) Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24: 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DW (1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S (1987) Characteristics of phosphorus uptake of chickpea in comparison with pigeonpea, soybean, and maize. Soil Sci Plant Nutr 33: 417–422 [Google Scholar]
- Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J, Woods AS (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom 42: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xia M, Wu P (2001) Effect of phosphorus deficiency stress on rice lateral root growth and nutrient absorption. Acta Bot Sin 43: 1154–1160 [Google Scholar]
- Liu A, Contador CA, Fan K, Lam HM (2018) Interaction and regulation of carbon, nitrogen, and phosphorus metabolisms in root nodules of legumes. Front Plant Sci 9: 1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lara IM, Gao JL, Soto MJ, Solares-Perez A, Weissenmayer B, Sohlenkamp C, Verroios GP, Thomas-Oates J, Geiger O (2005) Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol Plant Microbe Interact 18: 973–982 [DOI] [PubMed] [Google Scholar]
- Lopez-Lara IM, Sohlenkamp C, Geiger O (2003) Membrane lipids in plant-associated bacteria: their biosyntheses and possible functions. Mol Plant Microbe Interact 16: 567–579 [DOI] [PubMed] [Google Scholar]
- Lu S, Sturtevant D, Aziz M, Jin C, Li Q, Chapman KD, Guo L (2018) Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J 94: 915–932 [DOI] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469–478 [DOI] [PubMed] [Google Scholar]
- Miller KJ, , Shon BC, , Gore RS, , Hunt WP ( 1990) The phospholipid composition of Bradyrhizobium spp. Current Microbiology 21: 205–210 [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Blasing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Andres F, Kanehara K, Liu YC, Dormann P, Coupland G (2014) Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat Commun 5: 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan R, Wang G, Li M, Roth M, Welti R, Wang X (2013) Differential changes in galactolipid and phospholipid species in soybean leaves and roots under nitrogen deficiency and after nodulation. Phytochemistry 96: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack LC, Jaillais Y (2020) Functions of anionic lipids in plants. Annu Rev Plant Biol 71: 71–102 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Pereira PAA, Bliss FA (1989) Selection of common bean (Phaseolus vulgaris L.) for N2 fixation at different levels of available phosphorus under field and environmentally-controlled conditions. Plant Soil 115: 75–82 [Google Scholar]
- Perez Guerra JC, Coussens G, De Keyser A, De Rycke R, De Bodt S, Van De Velde W, Goormachtig S, Holsters M (2010) Comparison of developmental and stress-induced nodule senescence in Medicago truncatula. Plant Physiol 152: 1574–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic M, Schiller J, Muller M, Benard S, Reichl S, Arnold K, Arnhold J (2001) Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal Biochem 289: 202–216 [DOI] [PubMed] [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrere S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Sa TM, Israel DW (1991) Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol 97: 928–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J, Drevon JJ (2005) P-deficiency increases the O2 uptake per N2 reduced in alfalfa. J Exp Bot 56: 1779–1784 [DOI] [PubMed] [Google Scholar]
- Schulze J, Temple G, Temple SJ, Beschow H, Vance CP (2006) Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot 98: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiva S, Vu HS, Roth MR, Zhou Z, Marepally SR, Nune DS, Lushington GH, Visvanathan M, Welti R (2013) Lipidomic analysis of plant membrane lipids by direct infusion tandem mass spectrometry. Methods Mol Biol 1009: 79–91 [DOI] [PubMed] [Google Scholar]
- Si Z, Yang Q, Liang R, Chen L, Chen D, Li Y (2019) Digalactosyldiacylglycerol synthase gene MtDGD1 plays an essential role in nodule development and nitrogen fixation. Mol Plant Microbe Interact 32: 1196–1209 [DOI] [PubMed] [Google Scholar]
- Stopka SA, Agtuca BJ, Koppenaal DW, Pasa-Tolic L, Stacey G, Vertes A, Anderton CR (2017) Laser-ablation electrospray ionization mass spectrometry with ion mobility separation reveals metabolites in the symbiotic interactions of soybean roots and rhizobia. Plant J 91: 340–354 [DOI] [PubMed] [Google Scholar]
- Stopka SA, Khattar R, Agtuca BJ, Anderton CR, Pasa-Tolic L, Stacey G, Vertes A (2018) Metabolic noise and distinct subpopulations observed by single cell LAESI mass spectrometry of plant cells in situ. Front Plant Sci 9: 1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant D, Duenas ME, Lee YJ, Chapman KD (2017) Three-dimensional visualization of membrane phospholipid distributions in Arabidopsis thaliana seeds: a spatial perspective of molecular heterogeneity. Biochim Biophys Acta Mol Cell Biol Lipids 1862: 268–281 [DOI] [PubMed] [Google Scholar]
- Sturtevant D, Lee YJ, Chapman KD (2016) Matrix assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) for direct visualization of plant metabolites in situ. Curr Opin Biotechnol 37: 53–60 [DOI] [PubMed] [Google Scholar]
- Sulieman S, Ha CV, Schulze J, Tran LS (2013a) Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J Exp Bot 64: 2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Schulze J, Tran LS (2013b) Comparative analysis of the symbiotic efficiency of Medicago truncatula and Medicago sativa under phosphorus deficiency. Int J Mol Sci 14: 5198–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Tran LS (2015) Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci 239: 36–43 [DOI] [PubMed] [Google Scholar]
- Tesfaye M, Liu J, Allan DL, Vance CP (2007) Genomic and genetic control of phosphate stress in legumes. Plant Physiol 144: 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Poole PS (2013) Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol 64: 781–805 [DOI] [PubMed] [Google Scholar]
- Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127: 390–397 [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172: 4295–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H, Haynes JG, Penmetsa RV, Cook DR, Sherrier DJ, Dickstein R (2004) nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol 136: 3692–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DP, Hong Z (1996) Biogenesis of the peribacteroid membrane in root nodules. Trends Microbiol 4: 364–368 [DOI] [PubMed] [Google Scholar]
- Whitehead LF, Day DA (1997) The peribacteroid membrane. Physiol Plant 100: 30–44 [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfield HK, Sturtevant D, Borisjuk L, Munz E, Guschina IA, Chapman K, Harwood JL (2017) Spatial and temporal mapping of key lipid species in Brassica napus seeds. Plant Physiol 173: 1998–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZC, Zaheer R, Finan TM (2006) Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J Bacteriol 188: 1089–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaleta-Pastor M, Sohlenkamp C, Gao JL, Guan Z, Zaheer R, Finan TM, Raetz CR, Lopez-Lara IM, Geiger O (2010) Sinorhizobium meliloti phospholipase C required for lipid remodeling during phosphorus limitation. Proc Natl Acad Sci USA 107: 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.