Abstract
Activation of plasma membrane (PM) H+-ATPase activity is crucial in guard cells to promote light-stimulated stomatal opening, and in growing organs to promote cell expansion. In growing organs, SMALL AUXIN UP RNA (SAUR) proteins inhibit the PP2C.D2, PP2C.D5, and PP2C.D6 (PP2C.D2/5/6) phosphatases, thereby preventing dephosphorylation of the penultimate phosphothreonine of PM H+-ATPases and trapping them in the activated state to promote cell expansion. To elucidate whether SAUR–PP2C.D regulatory modules also affect reversible cell expansion, we examined stomatal apertures and conductances of Arabidopsis thaliana plants with altered SAUR or PP2C.D activity. Here, we report that the pp2c.d2/5/6 triple knockout mutant plants and plant lines overexpressing SAUR fusion proteins exhibit enhanced stomatal apertures and conductances. Reciprocally, saur56 saur60 double mutants, lacking two SAUR genes normally expressed in guard cells, displayed reduced apertures and conductances, as did plants overexpressing PP2C.D5. Although altered PM H+-ATPase activity contributes to these stomatal phenotypes, voltage clamp analysis showed significant changes also in K+ channel gating in lines with altered SAUR and PP2C.D function. Together, our findings demonstrate that SAUR and PP2C.D proteins act antagonistically to facilitate stomatal movements through a concerted targeting of both ATP-dependent H+ pumping and channel-mediated K+ transport.
SMALL AUXIN UP RNA (SAUR) proteins and PP2C.D phosphatases antagonistically regulate stomatal aperture in Arabidopsis by modulating the activities of plasma membrane H+-ATPases and K+ channels.
Introduction
The stomatal pores on the epidermis of plant leaves are flanked by pairs of guard cells that swell and shrink to cause stomatal opening and closing, respectively, thereby regulating gaseous exchange between the leaf interior and the atmosphere to support photosynthesis while minimizing water loss through transpiration (Inoue and Kinoshita, 2017; Jezek and Blatt, 2017). Plasma membrane (PM) H+-ATPases play an important role in stomatal aperture regulation. These H+-ATPases are activated in response to red and blue light, energizing the guard cell PM by transporting protons into the apoplast (Kinoshita and Shimazaki, 1999; Inoue and Kinoshita, 2017; Ando and Kinoshita, 2018) to generate a H+ electrochemical gradient across the PM and negative membrane voltage. Membrane energization powers ion flux, notably through inward-rectifying K+ and Cl− channels to drive solute and water into guard cells, generating turgor pressure and an increase in guard cell volume to open the stomatal pore (Chen et al., 2012; Hills et al., 2012; Minguet-Parramona et al., 2016).
During stomatal opening, guard cells accumulate solutes, mainly K+, Cl−, malate, and sugar (Jezek and Blatt, 2017). The negative membrane voltage activates inward-rectifying K+ channels, primarily K+ CHANNEL IN ARABIDOPSIS THALIANA (KAT1), to promote K+ influx (Nakamura et al., 1995; Pilot et al., 2001, 2003; Lebaudy et al., 2008). Proton-coupled K+ transport via HIGH AFFINITY K+-type transporters also contributes to K+ accumulation (Blatt and Slayman, 1987; Blatt and Clint, 1989; Maathuis and Sanders, 1994; Osakabe et al., 2013; Véry et al., 2014). During stomatal closing, H+-ATPase activity is suppressed and Cl− channels are activated to promote membrane depolarization, thereby suppressing K+ uptake through inward-rectifying K+ channels and promoting K+ efflux through outward-rectifying K+ channels, including the gated outwardly-rectifying K+ (GORK) channel of Arabidopsis (Jezek and Blatt, 2017). Such changes in ion flux are evident also in response to abscisic acid (ABA; Blatt, 1990; Linder and Raschke, 1992). ABA promotes an increase in cytosolic free calcium ([Ca2+]i) by triggering Ca2+ influx and its release via endomembrane Ca2+ channels (Grabov and Blatt, 1997, 1998, 1999; Hamilton et al., 2000; Garcia-Mata et al., 2003). The increase of [Ca2+]i, together with ABA-mediated activation of SLAC (S-type) and ALMT (R-type) anion channels, leads to anion efflux and reduced PM H+-ATPase activity, resulting in membrane depolarization (Schmidt et al., 1995; Grabov and Blatt, 1997; Merlot et al., 2007; Geiger et al., 2009; Chen et al., 2010; Meyer et al., 2010). The elevated [Ca2+]i also prevents K+ uptake by suppressing inward-rectifying K+ channels (KAT1; Grabov and Blatt, 1997 , 1999) and GORK channel activity is further enhanced by increased cytosolic pH (pHi; Blatt, 1990; Blatt and Armstrong, 1993; Grabov and Blatt, 1997; Suhita et al., 2004).
In our prior work on auxin-mediated cell expansion, we found that members of the PP2C.D family of type 2C protein phosphatases repress cell expansion by dephosphorylating PM H+-ATPases to inhibit H+ pumping activity (Spartz et al., 2014; Ren et al., 2018; Du et al., 2020). Several SMALL AUXIN UP RNA (SAUR) proteins antagonize PP2C.D activity by binding these phosphatases to inhibit their enzymatic activity, thereby promoting cell expansion (Spartz et al., 2014, 2017; Ren et al., 2018). Given that PM H+-ATPases are critical regulators of both cell expansion and stomatal aperture control, we examined whether similar SAUR–PP2C.D regulatory modules might also govern stomatal movements. Our prior findings hinted at this possibility, as both Arabidopsis and tomato (Solanum lycopersicum) plants ectopically expressing SAUR19 fusion proteins driven by the Cauliflower mosaic virus 35S promoter exhibited reduced drought tolerance (Spartz et al., 2014, 2017). Although SAUR19 is not normally expressed in guard cells and is therefore unlikely to regulate stomatal movements, several other SAUR and PP2C.D phosphatase genes are expressed in guard cells (Bauer et al., 2013; Adrian et al., 2015), supporting the possibility that SAUR–PP2C.D modules may play a role in stomatal function.
Here we report that SAUR proteins and PP2C.D phosphatases regulate stomatal aperture in Arabidopsis. Similar to their roles in controlling cell expansion during organ growth, SAUR and PP2C.D proteins function antagonistically in guard cells, with SAURs inhibiting PP2C.D activity to promote stomatal opening and conductance. Although these findings are consistent with SAUR–PP2C.D modules controlling stomatal aperture via the PM H+-ATPases, voltage clamp analysis in plants with perturbed SAUR or PP2C.D function uncovered substantial changes in the activities of both inward- and outward-rectifying K+ channels, including a dramatic effect on voltage-dependence of gating of the outward-rectifying K+ channels of guard cells. Thus, our findings indicate that SAUR–PP2C.D control modules target multiple transport processes to govern stomatal movements.
Results
Gain-of-function SAUR fusion proteins increase stomatal apertures
We previously noticed that plants expressing 35S:GFP-SAUR19 or 35S:StrepII-SAUR19 transgenes exhibited larger stomatal apertures and accelerated water loss compared to the wild-type (Spartz et al., 2014, 2017). To determine if these phenotypes were a general property of SAUR overexpression, we conducted water loss assays on plants overexpressing a SAUR63-YFP-HA fusion protein. SAUR63 and SAUR19 represent distinct clades of the 79-member Arabidopsis SAUR gene family. Although distantly related to one another (Ren and Gray, 2015), they share the highly conserved 60–70 amino acid core SAUR domain and both are stabilized as xFP-fusion proteins to cause a gain of function (Chae et al., 2012; Spartz et al., 2012). As previously observed with SAUR19 overexpression lines (Spartz et al., 2014, 2017), detached leaves of SAUR63-YFP-HA overexpression plants lost water faster than did leaves of wild-type controls (Figure 1A). The water loss phenotype is similar to the dominant open stomata2 (ost2-2) mutant, which harbors two-point mutations (L169F and G867S) in AUTOINHIBITED H+ ATPASE (AHA1) that confer constitutive proton pump activity (Merlot et al., 2007).
Figure 1.
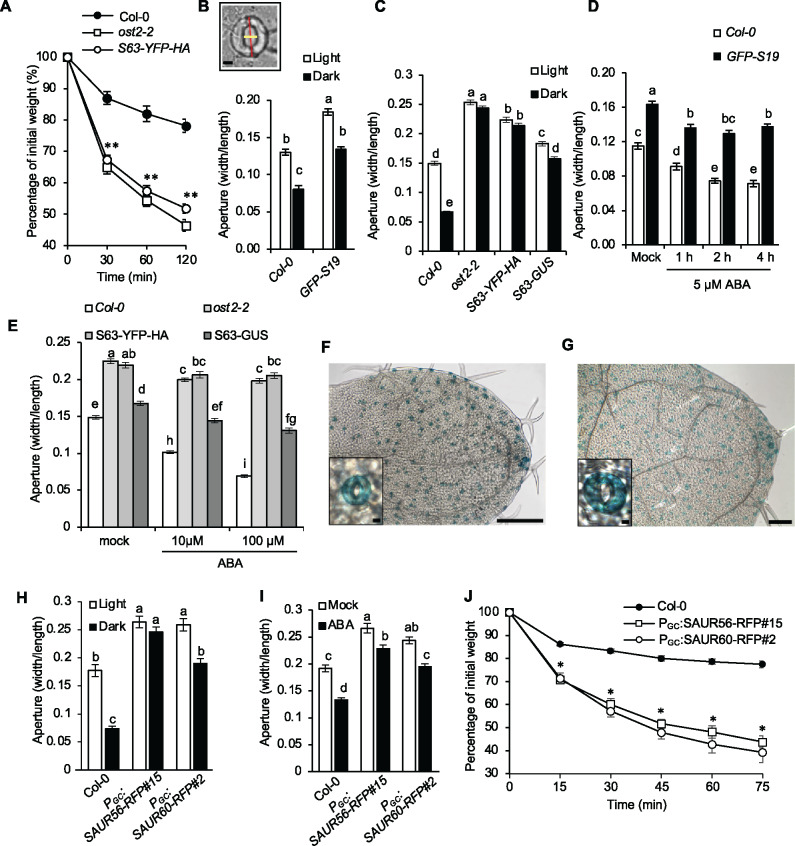
SAUR overexpression confers larger stomatal aperture. (A) Water loss in leaf detachment assays of wild-type Col-0, ost2-2, and 35S:SAUR63-YFP-HA plants. Data are means ± se (n = 15). (B) Stomatal aperture measurements of wild-type Col-0 and 35S:GFP-SAUR19 plants during light and dark conditions for 2 h. Data are means ± se (n = 60). Inset depicts aperture width/length stomate measurements. Scale bar = 5 µm. (C) Stomatal aperture measurements of wild-type Col-0, ost2-2, 35S:SAUR63-YFP-HA, and 35S:SAUR63-GUS plants during light and dark conditions for 2 h. Data are means ± se (n ≥ 106). (D) Stomatal aperture measurement of wild-type Col-0 and 35S:GFP-SAUR19 plants following treatment with 5 µM ABA. Data are means ± se (n = 60). (E) Stomatal aperture measurements of wild-type, ost2-2, 35S:SAUR63-YFP-HA, and 35S:SAUR63-GUS after 1h treatments with 10 or 100 µM ABA. Data are means ± se (n ≥ 113). X-Gluc staining of young leaves from PSAUR56:SAUR56-GUS (F) and PSAUR60:GUS (G) seedlings. Scale bars = 250 µm (whole leaf), 5 µm (inset). Stomatal aperture measurements of wild-type, PGC:SAUR56-RFP, and PGC:SAUR60-RFP plants during light and dark conditions (n ≥ 48) (H) or following a 1 h treatment with 100 µM ABA (n ≥ 65) (I). (J) Water loss in leaf detachment assays of wild-type, PGC:SAUR56-RFP and PGC:SAUR60-RFP plants. Data are means ± se (n = 6). All experiments were repeated at least twice with similar results. Different letters above error bars indicate significant difference (P < 0.05) by one-way analysis of variance analysis with Tukey’s HSD test. (A and J) Asterisks indicate significant difference from the wild-type (P < 0.05, Student’s t test).
We examined stomatal responses to light and dark treatments after pre-incubating epidermal peels to induce stomatal opening. Protocols for these experiments differed slightly between our laboratories (see Materials and methods section), but yielded similar results, confirming the robustness of these phenotypes. Plants harboring 35S:GFP-SAUR19 or 35S:StrepII-SAUR19 overexpression constructs exhibited larger stomatal apertures than did wild-type plants under both light and dark conditions (Figure 1B; Supplemental Figure S1A). Likewise, plants overexpressing SAUR63-YFP-HA or -GUS fusion proteins also displayed increased aperture diameter (Figure 1C). Similarly, compared to the wild-type control, SAUR19 and SAUR63 overexpression lines displayed reduced aperture closing in response to ABA (Figure 1, D and E; Supplemental Figure S1B).
Guard cells in the SAUR19 and SAUR63 overexpression lines were also larger than Col-0 guard cells (Supplemental Figure S1, C and D), reflecting roles for these proteins in promoting cell expansion during growth (Chae et al., 2012; Spartz et al., 2012, 2014). To confirm that the aperture phenotypes exhibited by SAUR overexpressors were due to altered guard cell responsiveness, we measured the apertures of estradiol-inducible SAUR63-CeruleanFP-HA (PEST:SAUR63-CerFP-HA) leaves subjected to a 1 h, 5 µM ABA treatment. Apertures of uninduced PEST:SAUR63-CerFP-HA stomata were comparable to Col-0 controls and closed in response to ABA (Supplemental Figure S1E). In contrast, induction of SAUR63-CerFP-HA expression with 10 µM estradiol for 1 h increased the aperture diameter of PEST:SAUR63-CerFP-HA stomata, and largely prevented closure in response to ABA (Supplemental Figure S1E). These findings indicate a significant effect on aperture regulation, and are supported by kinetic analysis of stomatal conductances (below).
We used reverse transcription–PCR (RT–PCR) to examine the expression of SAUR genes in guard cells, comparing data from isolated guard cell protoplasts (GCPs) to that from whole Arabidopsis leaves. This analysis revealed that although neither SAUR19 nor SAUR63 was expressed in guard cells, expression of several other SAUR family members was observed (Supplemental Figure S2A). These findings are consistent with prior transcriptomic studies (Yang et al., 2008; Wang et al., 2011; Bauer et al., 2013). SAUR56 and SAUR60 comprise a subclade in the phylogeny of Arabidopsis SAUR proteins (Ren and Gray, 2015), and expression of both genes is repressed by ABA, suggesting a possible role in stomatal aperture control (Bauer et al., 2013). To examine the expression of these SAUR genes, PSAUR56:SAUR56-GUS and PSAUR60:GUS reporter constructs were transformed into wild-type Arabidopsis plants. Both reporters were expressed specifically in the guard cells of cotyledons and leaves, and could not be detected in other shoot cells examined (Figure 1, F and G; Supplemental Figure S2, B and C).
To test whether SAUR proteins that are normally expressed in guard cells could also promote stomatal opening, we made PGC:SAUR56-RFP and PGC:SAUR60-RFP plants, expressing putative gain-of-function fusion proteins under the control of an ABA-insensitive form of the guard cell-specific pMYB60 promoter (PGC; Cominelli et al., 2005, 2011). PGC:SAUR56-RFP and PGC:SAUR60-RFP lines each had more open stomata than did the wild-type in the light, after shift to darkness, and after treatment with ABA (Figure 1, H and I; Supplemental Figure S2, D and E), similarly to the SAUR19 and SAUR63 overexpression lines. Detached leaves of these plants also lost water faster than did wild-type leaves (Figure 1J). Thus, both the native SAUR proteins and ectopically expressed SAURs promote an increase in stomatal aperture under a variety of conditions.
Several PP2C.D proteins repress stomatal opening
We hypothesized that the more open stomatal aperture phenotypes of the SAUR overexpression lines might be due to inhibition of PP2C.D activity. To explore this possibility, we first examined the expression of PP2C.D phosphatase genes in guard cells. RT–PCR assays were carried out as before to compare the expression from GCPs and from intact leaves. With the exception of PP2C.D1, substantial expression of all PP2C.D family members was detected in the guard cells (Supplemental Figure S3A). Largely consistent with this finding, β-glucuronidase (GUS) staining of leaf epidermal peels of Arabidopsis plants expressing native PP2C.D promoter-driven GUS reporter constructs (Ren et al., 2018) was evident for all PP2C.D family members except PP2C.D1 and D7 (Figure 2, A;Supplemental Figure S3B).
Figure 2.
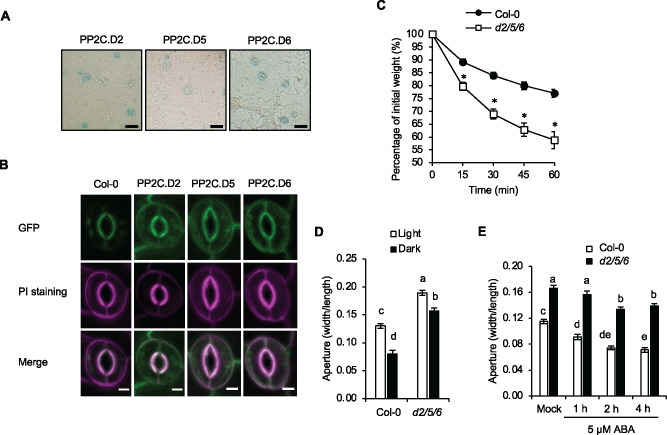
The pp2c.d2/5/6 triple mutant exhibits increased stomatal aperture diameter. (A) X-Gluc staining of native promoter:PP2C.D-GUS translational fusion proteins in leaf epidermal peels. Scale bar = 25 µm. (B) Subcellular localization of native promoter:PP2C.D-GFP fusion proteins (green) in guard cells of 7-d-old cotyledons. Propidium iodide (PI) (magenta) was used to mark the cell periphery. Scale bar = 5 µm. (C) Water loss in leaf detachment assays of wild-type Col-0 and pp2c.d2/5/6 triple mutants. Data are means ± se (n = 8). Asterisks indicate significant difference from the wild-type (P < 0.05, Student’s t test). (D) Stomatal aperture measurements of wild-type Col-0 and pp2c.d2/5/6 triple mutants during light and dark conditions for 2 h. Data are means ± se (n = 60). (E) Stomatal aperture measurements of the wild-type and pp2c.d2/5/6 triple mutants following treatment with 5 µM ABA. Data are means ± se (n = 60). Different letters above error bars indicate significant difference (P < 0.05) by one-way analysis of variance analysis with Tukey’s HSD test.
Of the nine Arabidopsis PP2C.D phosphatases, the PM-localized PP2C.D2, PP2C.D5, and PP2C.D6 subset (hereafter denoted as PP2C.D2/5/6) inhibit PM H+-ATPase activity to repress organ growth (Ren et al., 2018). Among these family members, PP2C.D6 was strongly expressed in guard cells, with PP2C.D2 and D5 expressed to a lesser extent (Figure 2A). Confocal imaging of PP2C.D2/5/6-GFP fusion proteins driven by their native promoters confirmed their localization at the periphery of the guard cells (Figure 2B), much as reported in other cell types (Tovar-Mendez et al., 2014; Ren et al., 2018).
To test whether PP2C.D2/5/6 normally regulate stomatal aperture, we assayed water loss from detached leaves from single-, double-, and triple-mutant combinations. None of the single mutants exhibited an obvious difference from the wild-type, whereas the pp2c.d2/6 and d5/6 double mutants displayed a slight increase in the rate of water loss (Supplemental Figure S4A). The pp2c.d2/5/6 triple mutant exhibited an even greater increase in water loss, similar to those of 35S:GFP-SAUR19, 35S:SAUR63-YFP-HA, PGC:SAUR56-RFP, PGC:SAUR60-RFP and ost2-2 plants (Figures 1, A, J and 2, C; Supplemental Figure S4A).
We next examined stomatal apertures in the pp2c.d2/5/6 triple mutants. Under both light and dark conditions, the triple mutant exhibited larger apertures than the wild-type (Figure 2D). When treated with 5 µM ABA to induce closing, stomata of the pp2c.2/5/6 mutant displayed both slower kinetics and a reduction in closure compared to the wild-type control (Figure 2E). The aperture phenotype of pp2c.d2/5/6 mutants was complemented by PP2C.D5-GFP, PP2C.D6-GFP, and to lesser extent, PP2C.D2-GFP transgenes (Supplemental Figure S4, B–D). Thus, we conclude that PP2C.D2, PP2C.D5, and PP2C.D6 act redundantly to inhibit stomatal opening, as they also do in inhibiting cell growth (Ren et al., 2018).
SAUR overexpression and pp2c.d2/5/6 mutant lines exhibit abnormal stomatal kinetics
We examined stomatal kinetics of SAUR19/63 overexpressors and pp2c.d2/5/6 mutants during dark/light/dark transitions using infrared gas analysis to determine the stomatal conductance (gs; Merlot et al., 2007; Wang et al., 2012, 2017). Plants were pre-adapted to the dark and gs was determined on stepping to 400 µmol m−2 s−1 light and on return to the dark (Figure 3A). The kinetics of stomatal opening was fitted to an exponential function of the following form:
| (1) |
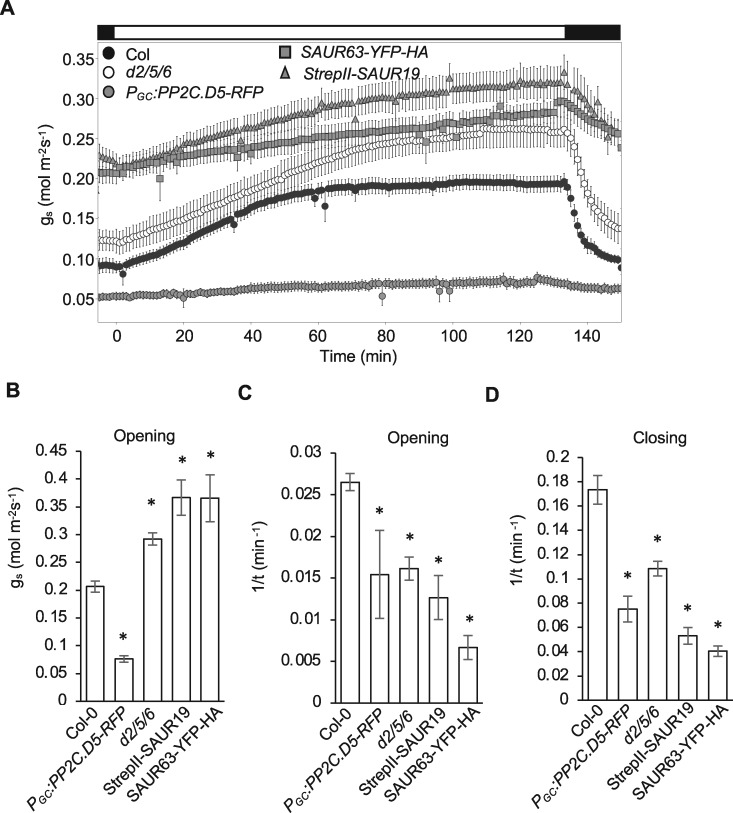
where y0 is the value of conductance at time 0, a is the range of conductance, b is the apparent rate constant of the relaxation, and t is the time from the start of the light period. The steady-state value of gs for the open stomata is defined by the value (y0+ a) and for the closed stomata by the value y0. We found that the pp2c.d2/5/6 mutant and SAUR19/63 overexpressors exhibited increased gs at all times (Figure 3, A and B). The opening kinetics were also slowed in comparison to the wild-type (Figure 3C).
Figure 3.
Gas exchange analyses of pp2c.d2/5/6 mutant and SAUR19/63 overexpression plants. (A) Stomatal conductance (gs) response from dark-adapted leaves exposed to light and subsequent transfer back to darkness. Data are means ± se (n ≥ 5). Bar above figure indicates dark/light/dark transitions. Bar graphs represent (B) maximum stomatal opening, (C) b value of opening, and (D) b value of closing. Data are means ± se (n ≥ 4). Asterisks indicate statistically significant differences relative to wild-type Col-0 determined by one-way analysis of variance Holm test (P < 0.05).
For the measurement of stomatal closure kinetics, plants were returned to dark conditions, and conductance curves were fitted with an exponential decay function of the following form:
| (2) |
where the parameters have the same meanings as before. Once again, the pp2c.d2/5/6 mutant and SAUR overexpressor lines exhibited greater baseline values for gs compared with the wild-type (Figure 3A) and, similarly, the triple mutant and SAUR overexpressors displayed slower closure kinetics (Figure 3D). Thus, we concluded that the pp2c.d2/5/6 mutant and SAUR19/63 overexpression lines were affected in the baseline gs in both light and darkness, and were impaired in the dynamic range and kinetics for stomatal movements between the two conditions.
PP2C.D5 overexpression in guard cells closes stomata constitutively
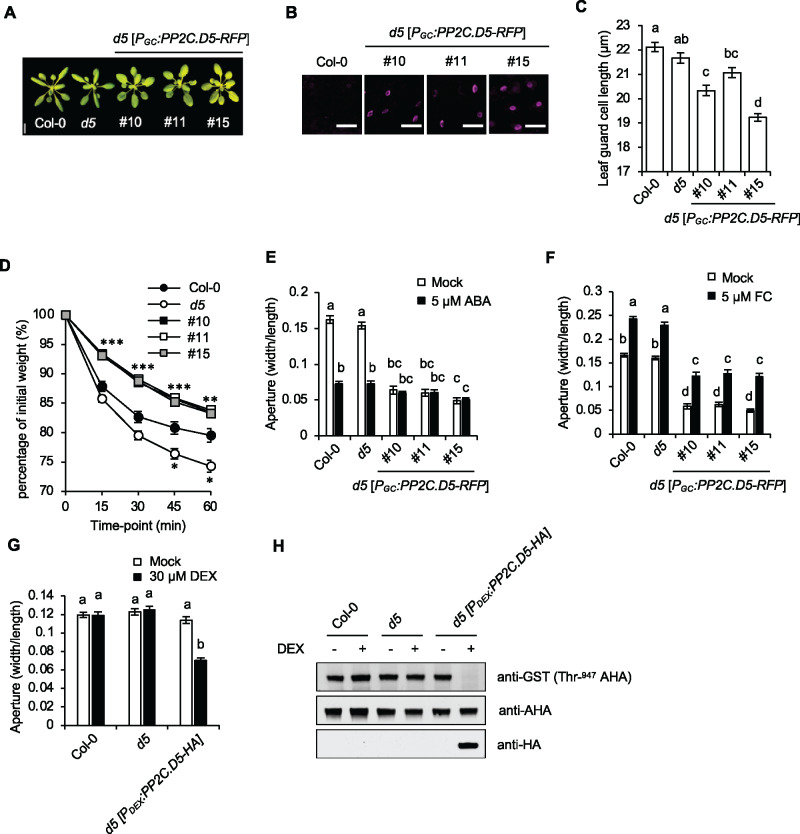
To explore further the mechanisms by which PP2C.D2/5/6 promote stomatal closure, we overexpressed PP2C.D2/5/6-RFP fusion genes under the control of the guard cell-specific PGC promoter (Cominelli et al., 2005; Cominelli et al., 2011). Three independent lines of each PP2C.D2/6-RFP construct were generated in the wild-type background, and the PP2C.D5-RFP construct was introduced into the pp2c.d5 single mutant background. Each line was examined by confocal microscopy to confirm guard cell-specific expression (Figure 4, B andSupplemental Figure S5A). Although overexpression of PP2C.D5 from the 35S promoter or even its native promoter confers severe reductions in plant stature due to its role in regulating cell expansion (Ren et al., 2018), the PGC:PP2C.D5-RFP transgenic plants displayed no obvious growth defects (Figure 4A). Examination of stomata in these lines, however, showed a modest reduction in guard cell size (Figure 4C). Additionally, using detached leaves we found that all PGC:PP2C.D2/5/6-RFP transgenic lines exhibited a reduced rate of water loss compared to the wild-type (Figure 4, D andSupplemental Figure S5B).
Figure 4.
Overexpression of PP2C.D5 in guard cell confers constitutively closed stomata. (A) 24-d-old plants. Three independent PGC:PP2C.D5-RFP lines are shown. Scale bar = 1 cm. (B) Guard cell-specific expression of PGC:PP2C.D5-RFP (magenta) in 14-d-old leaves with wild-type Col-0 as the negative control. Scale bar = 50 µm. (C) Stomatal length of leaf guard cells of wild-type Col-0, pp2c.d5 and pp2c.d5 [PGC:PP2C.D5-RFP] plants. Data are means ± se (n = 120). (D) Water loss in leaf detachment assays of wild-type Col-0, the pp2c.d5 single mutant, and PGC:PP2C.D5-RFP lines. Data are means ± se (n = 11). Asterisks indicate significant difference from the wild-type (P < 0.05, Student’s t test). The number of asterisks indicate the number of PGC:PP2C.D5-RFP lines that differed significantly from wild-type at each time point. (E) Stomatal aperture measurements of wild-type Col-0, the pp2c.d5 single mutant, and PGC:PP2C.D5-RFP lines following 2 h mock or 5 µM ABA treatments. Data are means ± se (n = 60). (F) Stomatal aperture measurements of wild-type Col-0, the pp2c.d5 single mutant, and PGC:PP2C.D5-RFP lines following 2 h mock or 5 µM FC treatments. Data are means ± se (n = 60). (G) Stomatal aperture measurements of wild-type Col-0, the pp2c.d5 single mutant, and PDEX:PP2C.D5-HA lines in the absence or presence of 30 µM DEX treatment for 5 h. Data are means ± se (n = 60). (H) Examination of AHA Thr-947 phosphorylation in 3-week-old rosette leaves by GST-14-3-3 far-western gel assay. PP2C.D5-HA protein expression levels are shown in the bottom panel. For (C, E–G), different letters above error bars indicate significant difference (P < 0.05) by one-way analysis of variance analysis with Tukey’s HSD test.
To establish how stomatal behavior is affected by PP2C.D2/5/6 overexpression, we measured stomatal apertures in epidermal peels of PGC:PP2C.D2/5/6-RFP leaves under mock and 5 µM ABA treatments. All PGC:PP2C.D2/5/6-RFP lines exhibited constitutively closed stomatal apertures in the light, both in the presence and absence of ABA (Figure 4, E; Supplemental Figure S5C). In agreement with our aperture measurements, gas exchange analysis of PGC:PP2C.D5-RFP plants indicated a dramatic reduction in gs, with only minor opening or closing responses observed in response to light or darkness, respectively (Figure 3, A–D).
To test whether the constitutively closed aperture phenotype of PGC:PP2C.D2/5/6-RFP plants was related to guard cell size, we examined the apertures of plants expressing dexamethasone (DEX)-inducible PP2C.D5 fused with an HA tag (PDex:PP2C.D5-HA). Uninduced PDex:PP2C.D5-HA plants displayed apertures comparable to wild-type controls. However, following a 5-h induction with DEX, PDex:PP2C.D5-HA stomata closed (Figure 4G), confirming that PP2C.D5 promotes stomatal closure.
Overexpression of PP2C.D5 represses H+-ATPase activity
In expanding cells, PP2C.D2/5/6 inhibit PM H+-ATPase activity by catalyzing dephosphorylation of the penultimate Thr residue in the autoinhibitory domain (corresponding to Thr-947 of AHA2; Ren et al., 2018; Wong et al., 2019). Indeed, penultimate Thr phosphorylation of PM H+-ATPases was dramatically diminished in induced PDex:PP2C.D5-HA plants (Figure 4H) as shown by GST-14-3-3 far-western blotting assays (Fuglsang et al., 1999; Kinoshita and Shimazaki, 1999; Hayashi et al., 2010). Unfortunately, such biochemical assays of PM H+-ATPase phosphorylation status specifically in guard cells are not possible due to high background signal from other cells. We therefore tested whether the closed aperture phenotype of PGC:PP2C.D5-RFP plants could be rescued by the fungal toxin fusicoccin (FC). FC activates PM H+-ATPases by promoting irreversible 14-3-3 protein binding to the C-terminal autoinhibitory domain, preventing dephosphorylation of the penultimate Thr residue (Olsson et al., 1998; Svennelid et al., 1999). FC treatment partially rescued the closed stomatal phenotype (Figure 4F), demonstrating that PGC:PP2C.D5-RFP stomata can in fact open, and confirming that the constitutively closed phenotype is not due to the effect of PP2C.D5 overexpression on guard cell size. Importantly, these results suggest that low H+-ATPase activity impacts stomatal opening in these plants.
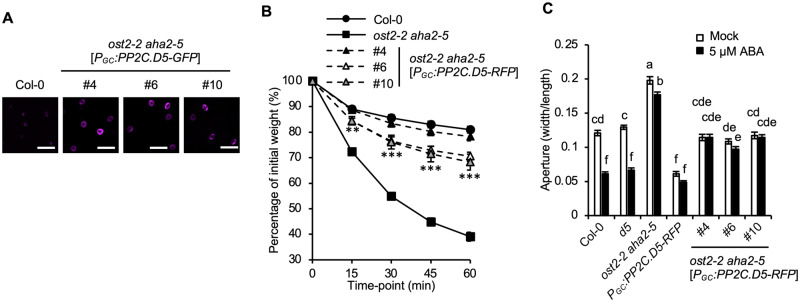
AHA1 encodes the primary PM H+-ATPase responsible for driving stomatal opening, with AHA2 playing a minor supporting role (Merlot et al., 2007; Wang et al., 2014; Yamauchi et al., 2016). To test genetically whether increased H+-ATPase activity could rescue the closed stomata of PGC:PP2C.D5-RFP plants, we transformed the PGC:PP2C.D5-RFP construct into the ost2-2 aha2-5 genetic background, in which the gain-of-function ost2-2 allele increases AHA1 activity. Three independent lines displaying guard cell-specific expression of PP2C.D5-RFP were analyzed (Figure 5A). ost2-2 aha2-5[PGC:PP2C.D5-RFP] plants exhibited phenotypes intermediate between those of PGC:PP2C.D5-RFP and ost2-2 aha2-5 plants in both water loss (Figure 5B) and stomatal aperture assays (Figure 5C). Again, these results suggest that H+-ATPase activity limits stomatal opening in the PGC:PP2C.D5-RFP plants. However, as neither FC treatment nor the ost2-2 mutation could entirely suppress the PGC:PP2C.D5-RFP stomatal phenotypes, the data raise the possibility that PP2C.D5 affects other targets in addition to the H+-ATPase.
Figure 5.
The ost2-2 mutation partially suppresses the PGC:PP2C.D5-RFP closed stomata phenotype. (A) Guard cell-specific expression of PP2C.D5-RFP (magenta) in 14-d-old leaves of ost2-2 aha2-5 mutants with wild-type Col-0 as a negative control. Scale bar = 50 µm. (B) Water loss in leaf detachment assays of wild-type Col-0, the ost2-2 aha2-5 double mutant, and ost2-2 aha2-5[PGC:PP2C.D5-RFP] plants. Data are means ± SE (n = 12). Asterisks indicate significant difference from the ost2-2 aha2-5 parental line (P < 0.05, Student’s t test). The number of asterisks indicate the number of PGC:PP2C.D5-RFP lines that differed significantly from ost2-2 aha2-5 at each time point. (C) Stomatal aperture measurements of wild-type Col-0, pp2c.d5 single mutant, ost2-2 aha2-5 double mutant, and PGC:PP2C.D5-RFP in either the pp2c.d5 or ost2-2 aha2-5 genetic backgrounds following 2 h mock or 5 µM ABA treatments. Data are means ± se (n = 60). Different letters above error bars indicate significant difference (P < 0.05) by one-way analysis of variance analysis with Tukey’s HSD test.
Guard cell SAURs interact with PP2C.D proteins to control stomatal aperture
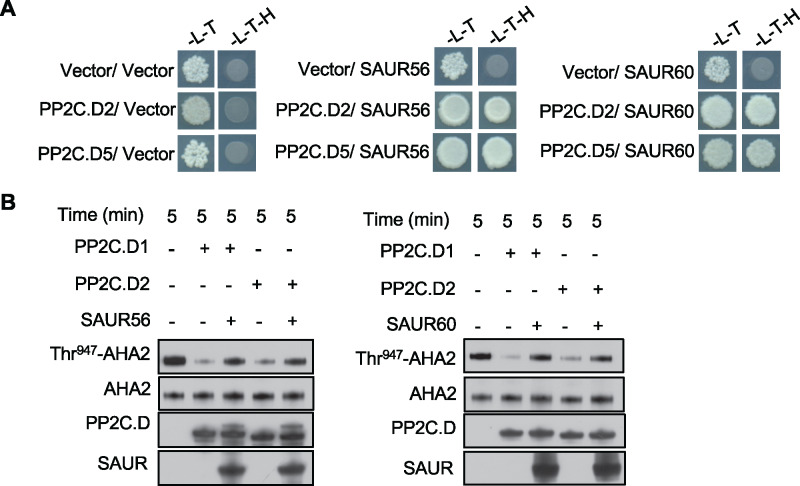
As described above, SAUR56 and SAUR60 expression are highly enriched in guard cells. Furthermore, overexpression of these SAURs in guard cells confers a constitutively open stomatal phenotype (Figure 1, H–J) similar to the pp2c.d2/5/6 triple mutant, suggesting that these SAURs may antagonize PP2C.D function as has been described for SAUR19 during organ growth (Spartz et al., 2014, 2017). To test this possibility, we examined the ability of SAUR56 and SAUR60 to interact with PP2C.D phosphatases. In both yeast two-hybrid and bimolecular fluorescence complementation assays, both SAUR proteins interacted with PP2C.D proteins (Figure 6, A; Supplemental Figure S6). Moreover, in in vitro phosphatase assays, recombinant SAUR56 or SAUR60 proteins inhibited PP2C.D-mediated dephosphorylation of Thr-947 of AHA2 (Figure 6B). Together, these findings suggest that SAUR56 and SAUR60 may inhibit PP2C.D phosphatases to promote PM H+-ATPase activity specifically in guard cells.
Figure 6.
SAUR56 and SAUR60 repress PP2C.C phosphatase activity. (A) Interaction between SAUR56/SAUR60 and PP2C.D phosphatases in yeast two-hybrid assays. Diluted yeast cultures were spotted onto Leu, Trp (-L-T) or Leu, Trp, His (-L-T-H) dropout plates and incubated for 3–4 d. (B) In vitro phosphatase assays employing yeast-expressed AHA2. Recombinant SAUR56 (left) and SAUR60 (right) inhibit PP2C.D-mediated dephosphorylation of Thr-947 of AHA2. Thr-947 phosphorylation status was assessed by GST-14-3-3 far-western blotting, and PP2C.D1/2 and SAUR56/60 abundance was monitored by detection of the S-tag.
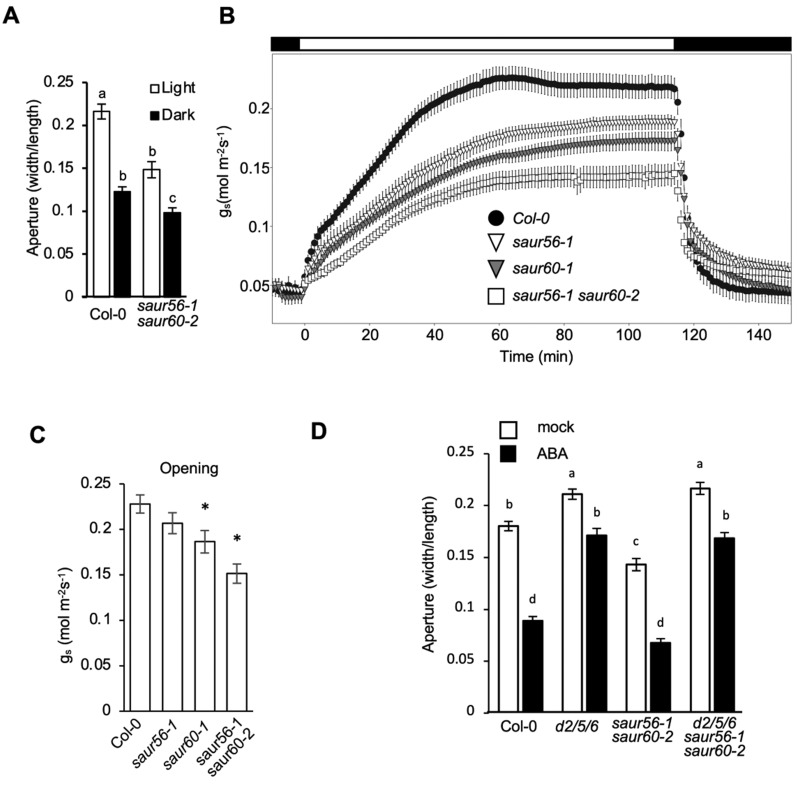
To explore further the roles of SAUR56 and SAUR60 in stomatal physiology, we used CRISPR/Cas9-mediated genome editing to make frameshift mutations in the genomic coding sequences of both genes (Supplemental Figure S7A), and crossed these lines to obtain double mutants among the progeny. Two lines containing independent mutations in both SAUR genes were obtained (saur56-1 saur60-2 and saur56-2 saur60-1). In stomatal aperture assays, these double mutants displayed decreased apertures compared to the wild-type in the light (Figure 7, A; Supplemental Figure S7B). Consistent with this finding, saur56-1 saur60-2 plants exhibited reduced gs in the light, as did the corresponding single mutants, albeit to a lesser extent (Figure 7, B and C). Moreover, a genomic SAUR56 transgene rescued the reduced stomatal aperture phenotype of saur56-1 saur60-2 double mutant plants partially or fully, depending on the transformant (Supplemental Figure S7D). Taken together, the results indicate that SAUR56 and SAUR60 act partially redundantly to open stomata in the light.
Figure 7.
SAUR56 and SAUR60 regulate stomatal aperture. (A) Stomatal aperture measurements of wild-type and saur56-1 saur60-2 mutant plants under light and dark conditions Data are means ± se (n ≥ 80). (B) Stomatal conductance (gs) response from dark-adapted plants exposed to light and subsequent transfer back to darkness. Data are means ± se (n ≥ 7). Bar above figure indicates dark/light/dark transitions. (C) Maximum stomatal opening obtained in gs assays. Data are means ± se (n ≥ 7). Asterisks indicate statistically significant differences relative to wild-type Col-0 determined by one-way analysis of variance Holm test (P < 0.05). (D) Stomatal aperture measurements following a 2 h treatment with 5 µM ABA or solvent control. Data are means ± se (n ≥ 65). For (A and D), different letters above error bars indicate significant difference (P < 0.05) by one-way analysis of variance analysis with Tukey’s HSD test.
Given that SAUR56 and SAUR60 can inhibit PP2C.D phosphatase activity, we hypothesized that the saur56/60 mutant phenotypes would be dependent upon PP2C.D function. To examine genetic interactions between these SAUR and PP2C.D proteins, we measured stomatal apertures of pp2c.d2/5/6 saur56/60 quintuple mutants. The pp2c.d2/5/6 mutations were largely epistatic to saur56/60, with the quintuple mutant stomata displaying a nearly identical increased aperture phenotype as seen with pp2c.d2/5/6 stomata in both the absence and presence of ABA (Figure 7D). Excised leaves of pp2c.d2/5/6 saur56/60 quintuple mutant plants also lost water at a rate comparable to the pp2c.d2/5/6 triple mutant (Supplemental Figure S7C). These results suggest that SAUR56 and SAUR60 require the PP2C.D proteins for their action, consistent with the ability of these SAUR proteins to inhibit PP2C.D activity.
SAURs and PP2C.D2/5/6 regulate K+ channel activities in guard cells
Our findings that FC treatment or the ost2-2 mutation each only partially suppressed stomatal closure in the PGC:PP2C.D5-RFP plants suggest that PP2C.D phosphatases may have additional targets beside H+-ATPases in the regulation of stomatal movements. To explore this question in detail, we carried out voltage clamp analyses to quantify the dominant K+ channel activities in the guard cells. After instantaneous background current subtraction, steady-state currents were fitted to a Boltzmann function of the following form:
| (3) |
where V1/2 is the voltage giving half-maximal activation of the current IK, δ is the voltage sensitivity coefficient, gmax is the maximum conductance, EK is the K+-equilibrium voltage, and R and T have their usual meanings.
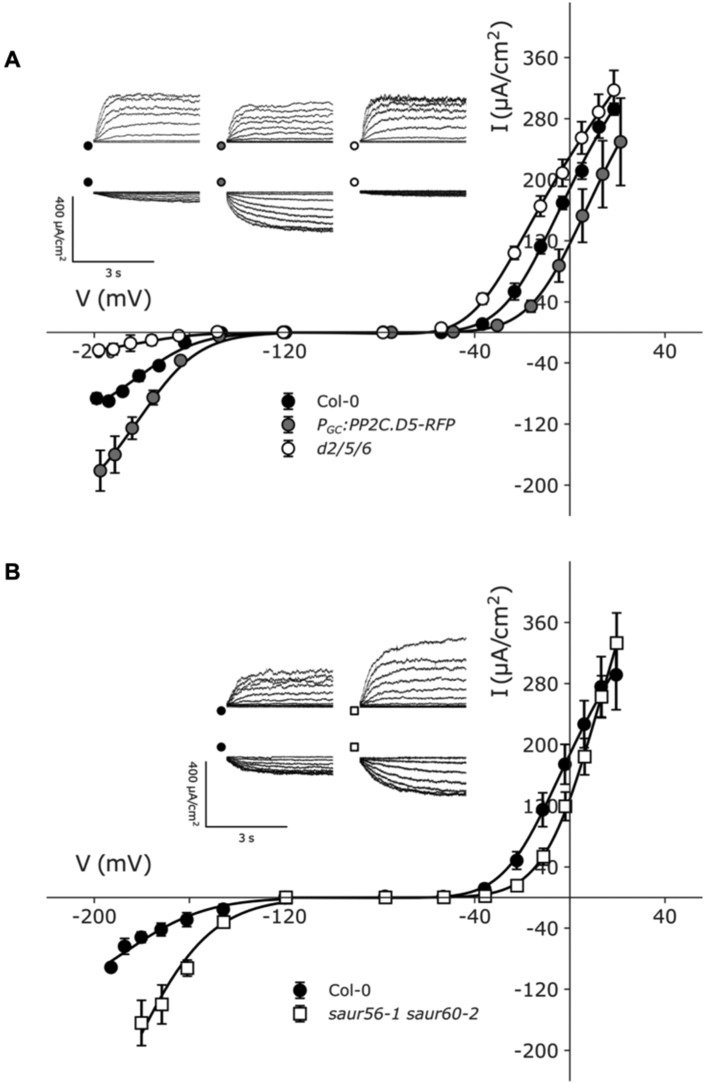
We first examined the inward- (IK,in) and outward-rectifying (IK,out) K+ channel activities in the pp2c.d2/5/6 mutant and gain-of-function PGC:PP2C.D5-RFP lines. Fittings for each current yielded a common set of voltage-sensitivity coefficients (δ; Figure 8, A andTable 1, top), consistent with the previous analyses of the channels (Hosy et al., 2003; Wang et al., 2012, 2017; Jezek and Blatt, 2017). Additionally, IK,in data were fitted with a common V1/2, much as reported in the past (Wang et al., 2012; Jezek and Blatt, 2017; Lefoulon et al., 2018).
Figure 8.
Characteristics of K+-out and K+-in channels in guard cells of plants with altered SAUR–PP2C.D function. Voltage clamp measurements of guard cells of wild-type Col-0, pp2c.d2/5/6 triple mutant, PGC:PP2C.D5-RFP and saur56-1 saur60-2 double mutant. Voltage stepped from a holding potential of −120 to +20 mV for IK,out and from −120 to −210 mV for IK,in. Data are means ± se (n ≥ 4) for each dataset. Fittings were performed jointly for each current. Scale: 400 μA vertical, 3 s horizontal. (A) PGC:PP2C.D5-RFP K+ channel data are plotted together with pp2c.d2/5/6 mutant and wild-type Col-0. Fittings for IK,out yielded common voltage-sensitivity coefficients (δ) of −2.12 ± 0.08. The data for IK,in were fitted with common voltage-sensitivity coefficients (δ) −1.47 ± 0.21 and the common midpoint voltage (V1/2) for IK,in −176 ± 2 mV. (B) saur56-1 saur60-2 data are plotted together with wild-type Col-0. Fittings for IK,out yielded common voltage-sensitivity coefficients (δ) of 2.12 ± 0.15. The data for IK,in were fitted with common voltage-sensitivity coefficients (δ) −1.40 ± 0.24 and the common midpoint voltage (V1/2) for IK,in −179 ± 8 mV.
Table 1.
Steady-state currents of wild-type, pp2c.d2/5/6 triple mutant, PP2C.D5 and SAUR19/63 overexpression lines and saur56-1 saur60-2 double mutant fitted to a Boltzmann function
| Genotype | gmax (mS cm−2) | V 1/2 (mV) | δ |
|---|---|---|---|
| I K, out | |||
| Col 0 | 3.93 ± 0.05 (A) | –16 ± 1 (X) | 2.12 ± 0.08 |
| pp2c.d2/5/6 | 4.07 ± 0.03 (A) | –33 ± 1 (Y) | |
| PGC:PP2C.D5 | 3.49 ± 0.0(B) | –3 ± 1 (Z) | |
| StrepII-SAUR19 | 1.81 ± 0.03 (C) | –31 ± 2 (Y) | |
| SAUR63-YFP-HA | 1.98 ± 0.02 (C) | –42 ± 3 (Y) | |
| I K,in | |||
| Col-0 | 0.79 ± 0.06 (a) | –176 ± 2 | –1.47 ± 0.21 |
| pp2c.d2/5/6 | 0.19 ± 0.03 (b) | ||
| PGC:PP2C.D5 | 1.55 ± 0.10 (c) | ||
| StrepII-SAUR19 | 0.14 ± 0.03 (b) | ||
| SAUR63-YFP-HA | 0.00 ± 0.03 (d) | ||
|
I K, out | |||
| Col-0 | 3.98 ± 0.10 (I) | –15 ± 2 (x) | 2.12 ± 0.15 |
| saur56-1 saur60-2 | 5.33 ± 0.33 (J) | 5 ± 2 (y) | |
| I K,in | |||
| Col-0 | 0.91 ± 0.19 (i) | –179 ± 8 | –1.40 ± 0.24 |
| saur56-1 saur60-2 | 2.86 ± 0.62 (j) | ||
After instantaneous background current subtraction, steady-state currents were fitted to a Boltzmann function of the form IK = gmax* (V–EK)/(1+e δF(V–V1/2)/RT), where V1/2 is the voltage giving half-maximal activation of the current IK, δ is the voltage sensitivity coefficient, gmax is the maximum conductance, EK is the K+ equilibrium voltage, and R and T have their usual meanings. The data were adjusted to ensure the equivalents between Col-0 currents. Fittings for IK,out yielded common voltage-sensitivity coefficients (δ), and IK,in were fitted with common voltage-sensitivity coeficients (δ) and common midpoint voltage (V1/2). Different letters within each dataset indicate statistically significant differences determined by one-way analysis of variance Scheffe test (P < 0.05).
Analysis of the pp2c.d2/5/6 mutant showed, for IK,out, a small increase in gmax and a highly significant shift in V1/2 to more negative voltages (Figure 8, A and Table 1, top), whereas the opposite was observed in the PGC:PP2C.D5-RFP transgenic guard cells, which exhibited reduced gmax and a significant shift in V1/2 to more positive voltages relative to the wild-type (Figure 8, A and Table 1, top). A complementary pattern in response was observed for IK,in. In this case, the current in the pp2c.d2/5/6 mutant was greatly reduced, whereas in the PGC:PP2C.D5-RFP guard cells it was enhanced when compared with wild-type guard cells (Figure 8, A andTable 1, top). Thus, loss and gain of PP2C.D2/5/6 function in the guard cells led to reciprocal changes in the K+ channel activities, the effect being opposite between the two currents. Significantly, the effects on IK,out were not restricted to scalar changes in the ensemble conductance gmax, but also affected the mid-point voltage, V1/2, for gating. Such an effect cannot be explained on the basis of a change in the population of channels and implies a direct effect on the gating process itself (Jezek and Blatt, 2017). Thus, our findings suggest that these phosphatases positively regulate the inward-rectifying channels and affect the voltage-dependence of gating of the outward-rectifying K+ channels.
We also analyzed the K+ channel currents recorded from SAUR19/63 overexpression lines and saur56/60 mutants. Recordings of IK,out in both SAUR overexpression lines showed characteristics similar to those of the pp2c.d2/5/6 triple mutant, with a negative shift in V1/2 compared with wild-type guard cells (Supplemental Figure S8 and Table 1, top). However, gmax values for IK,out in the SAUR19/63 overexpressing lines were only half that of the pp2c.d2/5/6 mutant and the wild-type. In contrast, the saur56 saur60 double mutant exhibited increased IK,out conductance in comparison with wild-type guard cells and a highly significant positive shift in V1/2 relative to the wild-type (Figure 8, B and Table 1, bottom). In both SAUR overexpression lines, values for gmax of IK,in were much lower than in the wild-type and slightly lower than the pp2c.d2/5/6 knockout mutants (Supplemental Figure S8 and Table 1, top). Conversely, for the saur56 saur60 guard cells, gmax of this current was enhanced by more than three-fold in comparison with the wild-type (Figure 8, B and Table 1, bottom). Thus, we conclude that, in general, the SAURs reduce IK,in ensemble activity, plausibly through a reduction in the population of activatable channels. Together, these findings suggest that SAUR and PP2C.D proteins have largely opposing effects on inward-rectifying and outward-rectifying K+ channel conductances. Their effects on the outward-rectifying K+ channel are more complex and include significant effects on gating as indicated by the effects on V1/2.
Discussion
Regulation of stomatal aperture by SAUR and PP2C.D proteins
Our results using both gain- and loss-of-function plants indicate that SAUR and PP2C.D proteins act antagonistically to regulate stomatal aperture. In aperture assays, we demonstrated that overexpression of SAUR19, SAUR63, SAUR56, or SAUR60 fusion proteins all resulted in increased apertures under multiple conditions. Of these, SAUR56 and SAUR60 are expressed in guard cells and normally regulate stomatal aperture, whereas SAUR19 and SAUR63 are not expressed in guard cells and instead normally regulate cell expansion in growing tissues (Chae et al., 2012; Spartz et al., 2012). Likewise, the SAUR-regulated PP2C.D2, PP2C.D5, and PP2C.D.6 phosphatases are also expressed in guard cells, and loss-of-function pp2c.d2/5/6 mutants exhibit increased stomatal apertures. Reciprocally, gain-of-function PP2C.D5 lines and loss-of-function saur56/60 mutants display reductions in stomatal aperture. These findings are further supported by results obtained in stomatal conductance and water loss assays and are consistent with SAUR proteins inhibiting PP2C.D phosphatase function to control stomatal aperture. Moreover, the SAUR56 and SAUR60 proteins, which are expressed in guard cells, can interact with the PP2C.D2/5/6 phosphatases and inhibit their enzymatic activity. Together, these findings implicate SAUR–PP2C.D regulatory modules in stomatal aperture control, with SAURs inhibiting PP2C.D2/5/6 activity to promote stomatal opening.
During organ growth, auxin-induced SAUR proteins inhibit PP2C.D2/5/6-mediated dephosphorylation of the penultimate Thr residue within the autoinhibitory domain of PM H+-ATPases, resulting in cell expansion via an acid growth mechanism (Spartz et al., 2014; Ren et al., 2018; Wong et al., 2019; Du et al., 2020). Given the well-established role of H+ ATPases in driving stomatal opening (Kinoshita and Shimazaki, 1999; Inoue and Kinoshita, 2017; Ando and Kinoshita, 2018), it seems likely that guard cell SAURs, including SAUR56 and SAUR60, may similarly activate H+-ATPases by inhibiting PP2C.D function to regulate stomatal aperture. The similar aperture behaviors of SAUR overexpressors, pp2c.d2/5/6 mutants and the ost2-2 gain-of-function allele of the AHA1 PM H+-ATPase support this hypothesis. Furthermore, the finding that the pp2c.d2/5/6 open stomate phenotype is epistatic to the reduced aperture phenotype of saur56/60 mutants strongly suggests that these SAURs work through the PP2C.D2/5/6 phosphatases. Consistent with this notion, we demonstrated that SAUR56 and SAUR60 inhibit PP2C.D-mediated dephosphorylation of the penultimate Thr residue of H+-ATPases in vitro. Direct measurements of H+-ATPase activity and phosphorylation specifically in guard cells would be needed to confirm this possibility. However, our data also clearly demonstrate that the SAUR–PP2C.D actions are more complex than this. In short, the effects on the H+-ATPase are not sufficient to fully explain the physiological consequences for the guard cell.
Our findings suggest that SAUR56/60-PP2C.D2/5/6 regulatory modules function in guard cells to control stomatal aperture. Notably however, the stomatal opening defect of saur56/60 double mutants was considerably weaker than that observed for PGC:PP2C.D5-RFP plants. Likewise, pp2c.d2/5/6 gas exchange phenotypes were noticeably weaker than those elicited by SAUR overexpression lines. These differences are likely due, at least in part, to additional functional redundancy within the SAUR and PP2C.D gene families. Several other SAUR genes are expressed in guard cells (Supplemental Figure S2A; Bauer et al., 2013; Adrian et al., 2015) and may act together with SAUR56 and SAUR60 to inhibit PP2C.D proteins and promote stomatal opening. Similarly, several additional PP2C.D family members are expressed in guard cells (Supplemental Figure S3), have been previously reported to bind SAUR proteins (Sun et al., 2016; Ren et al., 2018), and therefore may contribute to stomatal aperture control.
Arabidopsis has nine PP2C.D genes, at least three of which, PP2C.D2, PP2C.D5, and PP2C.D6, plays important roles both in regulating cell expansion during organ growth and in stomatal aperture control. In contrast, Arabidopsis has 79 SAUR genes. Many of these are induced by growth hormones and expressed in growing portions of the plant (Ren and Gray, 2015), whereas a distinct set of SAUR genes, including SAUR56 and SAUR60 are expressed in guard cells (Yang et al., 2008; Wang et al., 2011; Bauer et al., 2013). It thus appears that over evolutionary time, SAUR genes have diversified to acquire cell type-specific functions, at least in part by engaging the PP2C.D-H+-ATPase cellular machinery that is present in nearly all plant cells. It seems plausible that, in parallel to SAUR genes acquiring cell type-specific gene expression, SAUR proteins may also have diversified in their quantitative biochemical or regulatory properties.
Effects on K+ currents
PM H+-ATPase activation by either FC or the ost2-2 allele was able to suppress the closed stomata phenotype of PGC:PP2C.D5-RFP plants only partially. This raises the possibility that SAUR–PP2C.D modules may regulate additional targets that contribute to guard cell dynamics. The demonstrable effects of SAUR and PP2C.D expression we identify for K+ channels strongly support this possibility. Indeed, our detailed knowledge of guard cell membrane transport shows that the concerted action on several transporters during stomatal movements is the norm rather than the exception (Jezek and Blatt, 2017).
The pp2c.d2/5/6 mutant, as well as SAUR19/63 overexpressors, each exhibited dramatic reductions in inward-rectifying K+ channel activity. These results are similar to previous findings with the ost2-2 mutant, which also exhibited reduced IK,in currents, reportedly as a consequence of elevated [Ca2+]i and pHi (Wang et al., 2017). Conversely, the PGC:PP2C.D5-RFP and saur56 saur60 guard cells showed increased activity of the inward-rectifying K+ current. In each case, SAUR and PP2C.D proteins appear to affect this K+ current antagonistically and in a manner consistent with their predicted effects on H+-ATPase activity (Wang et al., 2017). The effects on stomatal aperture and kinetics, however, defy naive expectations with these changes in current. Such counterintuitive associations are typical for a system of transporters in which the activities of several system components interact in nonlinear fashion across a common membrane. That an increase in the inward-rectifying K+ current accompanied a decrease in stomatal opening, thus highlighting the mechanistic complexity of solute transport that drives stomatal physiology.
Changes in SAUR and PP2C.D activity also affected the outward-rectifying K+ channels, but in this case, not in a manner that could be explained through a simple SAUR–PP2C.D antagonism. Although both ost2-2 (Wang et al., 2017) and pp2c.d2/5/6 mutants exhibited increased activity of the outward-rectifying K+ channels, as evident in the gmax, unlike ost2-2, the pp2c.d2/5/6 mutants also showed a shift in V1/2 to the left along the voltage axis. Consistent with these findings, PP2C.D5 overexpression conferred reciprocal effects on gmax and V1/2. In contrast, the SAUR19 and SAUR63 overexpression lines displayed reductions in gmax for IK,out, and displaced V1/2 to the left along the voltage axis, whereas the saur56/60 plants showed an increase in gmax, and displaced V1/2 to the right along the voltage axis. Thus, the effect on V1/2 of the pp2c.d2/5/6 mutant was similar to that of the SAUR19 and SAUR63 overexpression lines, but the two sets of data differed in the consequences for gmax. To our knowledge, such disparate effects of SAUR overexpression and pp2c.d2/5/6 mutation have not been observed before, and the finding points to SAUR regulation of the outward-rectifying K+ channels that is independent of the PP2C.D2/5/6 phosphatases. Curiously, while SAUR overexpression and pp2c.d2/5/6 mutation had opposing effects on outward-rectifying K+ channel activity, both perturbations similarly altered voltage-dependent gating, shifting V1/2 toward more negative voltages than observed in wild-type guard cells. Conversely, the saur56 saur60 mutant and PGC:PP2C.D5-RFP guard cells had a more positive V1/2 threshold. These findings may suggest that SAUR proteins regulate multiple aspects of outward-rectifying K+ channel activity, some of which are dependent upon PP2C.D2/5/6 (i.e. voltage gating) and others that are not. Such regulation could occur via other PP2C.D family members or as yet to-be-discovered SAUR targets. Importantly, the effects of SAUR and PP2C.D perturbation on the outward-rectifying K+ channels do not match any effects previously associated with H+-ATPase superactivity, as reflected in the ost2 mutants (Wang et al., 2017), and therefore cannot be ascribed solely to an effect of elevated H+-ATPase activity, such as a rise in pHi. One intriguing possibility is that SAUR–PP2C.D regulatory modules control the phosphorylation status of K+ channels or regulators thereof to alter channel regulatory properties.
Conclusion
Our findings provide evidence that SAUR–PP2C.D regulatory modules control stomatal movements, demonstrating that a common regulatory mechanism controls cell expansion during organ growth and reversible guard cell movements. We also demonstrate that SAUR and PP2C.D proteins affect not just the PM H+-ATPase but also the activities of two different guard cell K+ channels. The identification of additional SAUR/PP2C.D regulatory targets and studies examining mechanisms of effects on K+ channels are promising areas for future research aimed at elucidating the regulation of stomatal opening and closing dynamics.
Materials and methods
Plant materials and growth conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this article were in the Col-0 ecotype. Seeds were sterilized using 30% (v/v) bleach and 0.04% (v/v) Triton X-100 for 15 min, and then washed three times with sterile water. The sterilized seeds were grown on ATS media at 22°C after 3 d of stratification. Generation of transgenic plants was done by the floral dip method using Agrobacterium tumefaciens strain GV3101 (helper plasmid pMP90) harboring the binary vector with desired target genes (Clough and Bent, 1998). The GFP-SAUR19 and StrepII-SAUR19 transgenic lines have been described previously (Spartz et al., 2012).
Molecular cloning
Primers used for all cloning steps and genotyping assays are listed in Supplemental Table S1. For the guard cell promoter-driven PP2C.D (PGC:PP2C.D-RFP) and SAUR constructs, a full-length PP2C.D2 cDNA lacking a stop codon, or genomic fragments of PP2C.D5 and PP2C.D6 flanking the start codon and last amino acid, were cloned into pENTR/D-TOPO entry vector (Life Technologies). SAUR56 and SAUR60 open reading frames were similarly cloned into pENTR/D-TOPO. The entry clones harboring respective genes were recombined into V400/W008 destination vector containing an ABA-insensitive derivative of the guard cell-specific MYB60 promoter using Gateway LR clonase II enzyme mix (Life Technologies; Karimi et al., 2002).
For the 35S:SAUR63-YFP-HA construct, the full-length SAUR63 coding sequence without a stop codon was cloned into pENTR/D-TOPO entry clone. The entry clone harboring SAUR63 cDNA was recombined into destination vector pEarleyGate101 containing YFP and HA tags, pMDC140 containing GUS (Curtis and Grossniklaus, 2003), or a pMDC7 derivative vector containing an estradiol induction system and C-terminal CerFP-HA fusion protein using Gateway LR clonase II enzyme (Earley et al., 2006; Akimoto-Tomiyama et al., 2012).
For the PSAUR56:SAUR56-GUS construct, a fragment incorporating ∼1 kB directly upstream of the start codon and the SAUR56 coding sequence lacking a stop codon was amplified by PCR. For PSAUR60:GUS, only the upstream 1 kB region was amplified. These PCR products were cloned into pENTR/D-TOPO entry vector (Life Technologies), verified by sequencing, and recombined into destination vector pGWB3 containing a GUS reporter tag using Gateway LR clonase II enzyme (Life Technologies; Nakagawa et al., 2007).
For the PSAUR56:SAUR56 complementation construct, a fragment incorporating ∼1 kB directly upstream of the start codon, the SAUR56 coding sequence, and ∼500 bp downstream from the stop codon was amplified by PCR. This product was cloned into pENTR/D-TOPO entry vector (Life Technologies), verified by sequencing, and recombined into destination vector pGWB1 using Gateway LR clonase II enzyme (Life Technologies; Nakagawa et al., 2007). This construct was introduced into saur56-1 saur60-2 double knockout plants via floral dip for isolation of transgenic lines.
For CRISPR/Cas9 targeting of SAUR56 and SAUR60, single guide RNAs targeting SAUR56 or SAUR60 were cloned by SLIM tailed PCR (Chiu et al., 2004) into pDONR207 vector (Life Technologies), subcloned into destination binary vector pCUT6 (Peterson et al., 2016) by LR clonase (Invitrogen), and introduced into Arabidopsis ecotype Columbia plants by floral dip (Clough and Bent, 1998) using A. tumefaciens strain GV3101 pMP90. Basta-resistant T1 transformed plants were selected on soil by spraying with 0.01% (v/v) Finale (Bayer Environmental Science, RTP, NC, USA). Mutations were identified in the T2 generation by detecting size differences on 15% (w/v) polyacrylamide gels of PCR products spanning the target sites, and by sequencing PCR products spanning the targeted sites.
GCPs isolation
Arabidopsis GCPs were isolated enzymatically from leaves of 3- to 4-week-old plants according to previously reported procedures with some modifications (Ueno et al., 2005). Blended epidermal tissues were collected on 80 µm nylon mesh, rinsed with cold sterile water, and incubated with first-step enzymatic digestion medium containing 0.5% (w/v) cellulase (Onozuka R-10; RPI Research Products International), 0.05% (w/v) macerozyme R-10 (PhytoTechnology Laboratories), 0.1% (w/v) polyvinylpyrrolidone, 0.2% (w/v) bovine serum albumin (BSA), 0.25 M mannitol, 1 mM CaCl2, and 10 mM MES-KOH, pH 5.4 at 22°C for 1 h with 70 r.p.m. shaking. For the subsequent second-step enzymatic digestion, the epidermal tissues were incubated with 1.5% (w/v) cellulase RS (Onozuka RS; RPI Research Products International), 0.5% (w/v) macerozyme R-10 (PhytoTechnology Laboratories), 0.2% (w/v) BSA, 1 mM CaCl2, and 0.4 M mannitol, pH 5.4 at 22°C for 1 h with 50 r.p.m. shaking. The digestion contents were passed through 80 µm nylon mesh, and the filtrate was further passed through two layers of 10 µm nylon mesh. The released GCPs were collected by centrifugation at 510 g for 12 min. The pellet was washed two times with 0.4 M mannitol with 1 mM CaCl2 and finally suspended in the same solution. The suspended GCPs were kept in the dark on ice until further use the following day.
Phosphatase assays
AHA2 dephosphorylation assays were conducted as described by Spartz et al. (2014, 2017) using recombinant 6xHis-TRX-SAUR and -PP2C.D proteins and PM fractions prepared from AHA2-expressing yeast cells.
RT–PCR
Primers used for semi-quantitative PCR of PP2C.D genes have been reported previously (Ren et al., 2018), except a different primer pair was used for PP2C.D3 in this study as listed in Supplemental Table S1. Total RNA was prepared from rosette leaves or GCPs from 4-week-old plants using the Nucleospin RNA Plant (Macherey-Nagel) kit according to the manufacturer’s instruction. On-column DNase treatment was performed to eliminate genomic DNA in RNA samples before cDNA synthesis using M-MLV reverse transcriptase (Promega). Semi-quantitative PCR was performed on the cDNAs using GoTaq DNA Polymerase (Promega) on a Techne TC-3000G PCR Thermal Cycler (GMI Inc.).
Water loss assays and stomatal aperture measurement
For water loss assays, plants were acclimated to room humidity for 1 h prior to the experiment, or were assayed in the same walk-in growth room where they were growing. Age-matched rosette leaves were excised from 4-week-old plants and weighed using an analytical scale. Relative water loss was calculated based on the initial leaf weight at the 0 min time point. For stomatal aperture measurements, leaf epidermal peels were prepared from young expanded rosette leaves of 3-week-old plants, adhered onto a coverslip with silicone adhesive (Factor II, Inc. or Hollister Adapt 7730), and apertures examined as previously described with minor modifications (Azoulay-Shemer et al., 2015). Briefly, adhered epidermal peels were incubated in opening buffer (either 30 mM KCl, 10 mM MES, pH 6.1 or 10 mM KCl, 5 mM MES, 0.1 mM CaCl2, pH 6.1) under white light for 3 h. For closing assays, peels were subsequently transferred to fresh opening buffer or to closing buffer (0.1 mM KCl, 8 mM CaCl2, 10 mM MES titrated to pH 6.1 with Ca(OH)2) and transferred to darkness; or transferred to 10 mM KCl, 5 mM MES titrated to pH 6.1 with Ca(OH)2 and treated with ABA or solvent control. In an alternative protocol, full leaves or leaf halves with midveins removed were immersed in 30 mM KCl 10 mM MES opening buffer for 2 h in the light, and then treated with or without ABA in the light, or transferred to light or darkness, for 1 h before epidermal peels were made and imaged immediately. Both protocols gave similar results. Images of at least 45 stomata for each data point were captured using a Leica DM5000B or Zeiss Axio Imager A1 microscope. The stomatal aperture was calculated as the aperture width divided by the longitudinal length the same stomate. ImageJ software (http://rsb.info.nih.gov/ij) was used for the stomatal aperture measurement (Abramoff et al., 2004).
Membrane fraction isolation, western, and far-western blot assays
Microsomal proteins of 7-d-old Arabidopsis seedlings grown on ATS plates containing 1% (w/v) sucrose were prepared as previously reported (Ito and Gray, 2006). Protein samples were separated on NuPage 4–12% acrylamide gels (Life Technologies) and blotted to nitrocellulose membranes. Blots were blocked with 5% (w/v) milk in TBS containing 0.1% (v/v) Tween-20 for 1 h and then incubated with antibodies or 0.1 µM purified GST-14-3-3 (GF14ø) protein overnight at 4°C. GST-14-3-3 binding was subsequently detected using an α-GST-HRP conjugate antibody (GE Healthcare) and chemiluminescence detection reagents (GE Healthcare) as previously described (Spartz et al., 2014).
Bimolecular fluorescence complementation (BiFC)
BiFC assays were performed as previously described (Ren et al., 2018; Wong et al., 2019). Briefly, pSPYCE and pSPYNE expression constructs were infiltrated into approximately 5-week-old Nicotiana benthamiana leaves, and YFP fluorescence signals were observed 3-d post-infiltration using a Nikon Ti2 A1si Confocal system (Nikon USA) with a 488 nm excitation wavelength and 545/40 nm emission filter.
Gas exchange
Plants were grown in short-day conditions (8 h light/16 h dark cycle, 22°C/18°C, 70% relative humidity). Daylight regimes with steady 150 μmol m−2 s−1 light intensity were used. Gas exchange measurements were carried out using the LI-COR 6400 XT Infrared Gas Analyzer (LICOR Biosciences) and a whole-plant Arabidopsis chamber (LI-COR 6400-17). Measurements were carried out at 22°C, 55% relative humidity, and at 400 p.p.m. CO2. Gas exchange responses were measured using an external light source (LI-COR 6400-18) after the leaves were dark-adapted for 3 h. Measurements were carried out over the same period of the diurnal cycle and normalized to rosette area calculated using ImageJ (http://rsb.info.nih.gov/ij). The conductance curves for stomatal opening were fitted using the Marquardt-Levenberg algorithm of SigmaPlot v.11 (Systat, Cambridge USA) with Equations (1)–(3).
Electrophysiology
Voltages clamp data were recorded from guard cells in epidermal peels using Henry’s EP Software Suite (http://www.psrg.org.uk). Double-barreled microelectrodes (tip resistances >100 MΩ) were filled with 200 mM K+ acetate, pH 7.5 after equilibration against resin-bound BAPTA [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid] (Invitrogen) to prevent Ca2+ loading of the cytosol from the microelectrode as previously described (Blatt and Slayman, 1983; Blatt, 1987; Wang et al., 2013). Epidermal peels were pre-incubated in opening buffer (5 mM Ca2+-MES, pH 6.1, with 60 mM KCl) for 1 h. Measurements were carried out in 5 mM Ca2+-MES, pH 6.1, with 10 mM KCl. Surface areas of impaled guard cells were calculated assuming a spheroid geometry and voltages were analyzed using Henry’s EP Software Suite (Blatt and Slayman, 1983; Wang et al., 2013). Where necessary for comparisons of lines between sets of experiments, data were referenced to the amplitudes of concurrent measurements from wild-type guard cells.
Statistical analyses
All statistical analyses were done using JMP Pro 13.1 software suite (SAS Institute) to perform analysis of variance or otherwise mentioned in the text. The results were grouped under letters, with different letters showing significant differences (P < 0.05) based on Tukey’s HSD (honestly significant difference) test.
Accession numbers
All sequence data can be found in the GenBank/EMBL data libraries under the following accession numbers: PP2C.D1 (At5g02760), PP2C.D2 (At3g17090), PP2C.D3 (At3g12620), PP2C.D4 (At3g55050), PP2C.D5 (At4g38520), PP2C.D6 (At3g51370), PP2C.D7 (At5g66080), PP2C.D8 (At4g33920), PP2C.D9 (At5g06750), SAUR19 (At5g18010), SAUR56 (At1g76190), SAUR60 (At1g20470), SAUR63 (At1g29440), ACTIN7 (At5g09810).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 Stomatal aperture measurements of SAUR19 and SAUR63 gain-of-function lines.
Supplemental Figure S2 SAUR56 and SAUR60 are expressed in guard cells and regulate stomatal aperture.
Supplemental Figure S3 Expression of PP2C.D genes in guard cells.
Supplemental Figure S4 Water loss assay of pp2c.d mutants and complementation lines.
Supplemental Figure S5 Guard cell overexpression of PP2C.D2-RFP and PP2C.D6-RFP.
Supplemental Figure S6 SAUR56 and SAUR60 interact with PP2C.D proteins in BiFC assays.
Supplemental Figure S7 Loss-of-function alleles of SAUR56 and SAUR60.
Supplemental Figure S8 Characteristics of and channels in guard cells of SAUR overexpression plants.
Supplemental Table S1 Oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank Dr Masaki Okumura for his helpful advice on the isolation of Arabidopsis GCPs and Dr Nathalie Leonhardt for providing the ABA-insensitive MYB60 promoter derivative used to construct the V400/W008 expression vector. We also thank the UM College of Biological Sciences Imaging Center for assistance with confocal microscopy.
Funding
This work was supported by the National Science Foundation (MCB-1613809 to W.M.G., MCB-1615557 to J.W.R.); the Biotechnology and Biological Sciences Research Council (BB/P011586/1 to M.R.B.); the National Institutes of Health (GM067203 to W.M.G.); and a Summer Undergraduate Research Fellowship from the UNC Office of Undergraduate Research to B.W.
W.M.G., M.R.B., and J.W.R. designed the research; J.H.W., M.K., S.A.S., P.N., S.D., M.P., G.B., B.W., E.N.K., B.T., and W.M.G. performed the experiments; W.M.G., M.R.B., and J.W.R. supervised the experiments; J.H.W., J.W.R., M.K., M.R.B., and W.M.G. wrote the article with contributions from all authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: William M. Gray (grayx051@umn.edu).
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11: 36–42 [Google Scholar]
- Adrian J, Chang J, Ballenger CE, Bargmann BO, Alassimone J, Davies KA, Lau OS, Matos JL, Hachez C, Lanctot A, et al. (2015) Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev Cell 33: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto-Tomiyama C, Furutani A, Tsuge S, Washington EJ, Nishizawa Y, Minami E, Ochiai H (2012) XopR, a type III effector secreted by Xanthomonas oryzae pv. oryzae, suppresses microbe-associated molecular pattern-triggered immunity in Arabidopsis thaliana. Mol Plant Microbe Interact 25: 505–514 [DOI] [PubMed] [Google Scholar]
- Ando E, Kinoshita T (2018) Red light-induced phosphorylation of plasma membrane H. Plant Physiol 178: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay-Shemer T, Palomares A,, Bagheri A, Israelsson-Nordstrom M, Engineer CB, Bargmann BO, Stephan AB, Schroeder JI (2015) Guard cell photosynthesis is critical for stomatal turgor production, yet does not directly mediate CO2- and ABA-induced stomatal closing. Plant J 83: 567–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Blatt MR (1987) Electrical characteristics of stomatal guard cells: the ionic basis of the membrane potential and the consequence of potassium chlorides leakage from microelectrodes. Planta 170: 272–287 [DOI] [PubMed] [Google Scholar]
- Blatt MR (1990) Potassium channel currents in intact stomatal guard cells: rapid enhancement by abscisic acid. Planta 180: 445–455 [DOI] [PubMed] [Google Scholar]
- Blatt MR, Armstrong F (1993) K+ channels of stomatal guard cells: abscisic acid-evoked control of the outward rectifier mediated by cytoplasmic pH. Planta 191: 330–341 [Google Scholar]
- Blatt MR, Clint GM (1989) Mechanisms of fusicoccin action: kinetic modification and inactivation of K(+) channels in guard cells. Planta 178: 509–523 [DOI] [PubMed] [Google Scholar]
- Blatt MR, Slayman CL (1983) KCl leakage from microelectrodes and its impact on the membrane parameters of a nonexcitable cell. J Membr Biol 72: 223–234 [DOI] [PubMed] [Google Scholar]
- Blatt MR, Slayman CL (1987) Role of “active” potassium transport in the regulation of cytoplasmic pH by nonanimal cells. Proc Natl Acad Sci U S A 84: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Bätz U, Amtmann A, Lew VL, Blatt MR (2012) Systems dynamic modeling of the stomatal guard cell predicts emergent behaviors in transport, signaling, and volume control. Plant Physiol 159: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Lim CK, Blatt MR (2010) Dynamic regulation of guard cell anion channels by cytosolic free Ca2+ concentration and protein phosphorylation. Plant J 61: 816–825 [DOI] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D (2004) Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res 32: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G, Tonelli C (2011) DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol 11: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15: 1196–1200 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Spalding EP, Gray WM (2020) Rapid auxin-mediated cell expansion. Annu Rev Plant Biol 71: 379–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG (1999) Binding of 14-3-3 protein to the plasma membrane H(+)-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J Biol Chem 274: 36774–36780 [DOI] [PubMed] [Google Scholar]
- Garcia-Mata C, Gay R, Sokolovski S, Hills A, Lamattina L, Blatt MR (2003) Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc Natl Acad Sci U S A 100: 11116–11121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Blatt MR (1997) Parallel control of the inward-rectifier K+ channel by cytosolic-free Ca2+ and pH in Vicia guard cells. Planta 201: 84–95 [Google Scholar]
- Grabov A, Blatt MR (1998) Membrane voltage initiates Ca2+ waves and potentiates Ca2+ increases with abscisic acid in stomatal guard cells. Proc Natl Acad Sci USA 95: 4778–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Blatt MR (1999) A steep dependence of inward-rectifying potassium channels on cytosolic free calcium concentration increase evoked by hyperpolarization in guard cells. Plant Physiol 119: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DW, Hills A, Kohler B, Blatt MR (2000) Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc Natl Acad Sci USA 97: 4967–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Nakamura S, Takemiya A, Takahashi Y, Shimazaki K, Kinoshita T (2010) Biochemical characterization of in vitro phosphorylation and dephosphorylation of the plasma membrane H+-ATPase. Plant Cell Physiol 51: 1186–1196 [DOI] [PubMed] [Google Scholar]
- Hills A, Chen ZH, Amtmann A, Blatt MR, Lew VL (2012) OnGuard, a computational platform for quantitative kinetic modeling of guard cell physiology. Plant Physiol 159: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Poree F, Boucherez J, Lebaudy A, Bouchez D, Very AA, et al. (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci USA 100: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SI, Kinoshita T (2017) Blue light regulation of stomatal opening and the plasma membrane H. Plant Physiol 174: 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gray WM (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142: 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezek M, Blatt MR (2017) The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiol 174: 487–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H(+)-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaudy A, Vavasseur A, Hosy E, Dreyer I, Leonhardt N, Thibaud JB, Véry AA, Simonneau T, Sentenac H (2008) Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci USA 105: 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefoulon C, Waghmare S, Karnik R, Blatt MR (2018) Gating control and K(+) uptake by the KAT1 K(+) channel leaveraged through membrane anchoring of the trafficking protein SYP121. Plant Cell Environ 41: 2668–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Raschke K (1992) A slow anion channel in guard cells, activating at large hyperpolarization, may be principal for stomatal closing. FEBS Lett 313: 27–30 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D (1994) Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc Natl Acad Sci USA 91: 9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Leonhardt N, Fenzi F, Valon C, Costa M, Piette L, Vavasseur A, Genty B, Boivin K, Müller A, Giraudat J, Leung J (2007) Constitutive activation of a plasma membrane H(+)-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J 26: 3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Mumm P, Imes D, Endler A, Weder B, Al-Rasheid KA, Geiger D, Marten I, Martinoia E, Hedrich R (2010) AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J 63: 1054–1062 [DOI] [PubMed] [Google Scholar]
- Minguet-Parramona C, Wang Y, Hills A, Vialet-Chabrand S, Griffiths H, Rogers S, Lawson T, Lew VL, Blatt MR (2016) An optimal frequency in Ca2+ oscillations for stomatal closure is an emergent property of ion transport in guard cells. Plant Physiol 170: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR (1995) Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiol 109: 371–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Arinaga N, Umezawa T, Katsura S, Nagamachi K, Tanaka H, Ohiraki H, Yamada K, Seo SU, Abo M, et al. (2013) Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BA, Haak DC, Nishimura MT, Teixeira PJ, James SR, Dangl JL, Nimchuk ZL (2016) Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One 11: e0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Gaymard F, Mouline K, Chérel I, Sentenac H (2003) Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol Biol 51: 773–787 [DOI] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Cherel I, Boucherez J, Thibaud JB, Sentenac H (2001) Guard cell inward K+ channel activity in arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem 276: 3215–3221 [DOI] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Park MY, Spartz AK, Wong JH, Gray WM (2018) A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet 14: e1007455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Schelle I, Liao YJ, Schroeder JI (1995) Strong regulation of slow anion channels and abscisic acid signaling in guard cells by phosphorylation and dephosphorylation events. Proc Natl Acad Sci USA 92: 9535–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lor VS, Ren H, Olszewski NE, Miller ND, Wu G, Spalding EP, Gray WM (2017) Constitutive expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiol 173: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A (2004) Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennelid F, Olsson A, Piotrowski M, Rosenquist M, Ottman C, Larsson C, Oecking C, Sommarin M (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H(+)-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell 11: 2379–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Mendez A, Miernyk JA, Hoyos E, Randall DD (2014) A functional genomic analysis of Arabidopsis thaliana PP2C clade D. Protoplasma 251: 265–271 [DOI] [PubMed] [Google Scholar]
- Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K (2005) Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol 46: 955–963 [DOI] [PubMed] [Google Scholar]
- Véry AA, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H (2014) Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol 171: 748–769 [DOI] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM (2011) Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen ZH, Zhang B, Hills A, Blatt MR (2013) PYR/PYL/RCAR abscisic acid receptors regulate K+ and Cl- channels through reactive oxygen species-mediated activation of Ca2+ channels at the plasma membrane of intact Arabidopsis guard cells. Plant Physiol 163: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hills A, Vialet-Chabrand S, Papanatsiou M, Griffiths H, Rogers S, Lawson T, Lew VL, Blatt MR (2017) Unexpected connections between humidity and ion transport discovered using a model to bridge guard cell-to-leaf scales. Plant Cell 29: 2921–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Noguchi K, Ono N, Inoue S, Terashima I, Kinoshita T (2014) Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proc Natl Acad Sci USA 111: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Papanatsiou M, Eisenach C, Karnik R, Williams M, Hills A, Lew VL, Blatt MR (2012) Systems dynamic modeling of a guard cell Cl- channel mutant uncovers an emergent homeostatic network regulating stomatal transpiration. Plant Physiol 160: 1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Spartz AK, Park MY, Du M, Gray WM (2019) Mutation of a conserved motif of PP2C.D phosphatases confers SAUR immunity and constitutive activity. Plant Physiol 181: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi S, Takemiya A, Sakamoto T, Kurata T, Tsutsumi T, Kinoshita T, Shimazaki K (2016) The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiol 171: 2731–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI (2008) Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.