The available information on the plant-specific plasma membrane-bound family of REMORIN proteins points to their potential role as scaffolding proteins in a plethora of cellular processes.
Abstract
REMORINs (REMs) are a plant-specific protein family, proposed regulators of membrane-associated molecular assemblies and well-established markers of plasma membrane nanodomains. REMs play a diverse set of functions in plant interactions with pathogens and symbionts, responses to abiotic stresses, hormone signaling and cell-to-cell communication. In this review, we highlight the established and more putative roles of REMs throughout the literature. We discuss the physiological functions of REMs, the mechanisms underlying their nanodomain-organization and their putative role as regulators of nanodomain-associated molecular assemblies. Furthermore, we discuss how REM phosphorylation may regulate their functional versatility. Overall, through data-mining and comparative analysis of the literature, we suggest how to further study the molecular mechanisms underpinning the functions of REMs.
REMORIN discovery
REMORINs (REMs) were first identified in tomato (Solanum lycopersicum) and potato (Solanum tuberosum) by Edward Farmer, Gregory Pearce, and Clarence Ryan in 1989, in a will of identifying molecular actors involved in the perception of polygalacturonides (PGAs) by plant cells (Farmer et al., 1989). PGAs are pectic polysaccharides present in the plant cell wall released during wounding and pathogen attack to sensitize plant defense (Voxeur and Höfte, 2016). By in vitro phosphorylation assays on isolated tomato and potato plasma membranes (PMs), Farmer et al. (1989) found that PGA treatment caused the phosphorylation of a protein of 34 kDa, named Phosphorylated Protein of 34 kilodaltons pp34. Pp34 was then used as a marker of the plant response to wounding (Farmer et al., 1991). Unintuitively, the purification (Jacinto et al., 1993) and cloning of pp34 (Reymond et al., 1996) revealed that its sequence turned out to be one of a hydrophilic protein even though it is tightly bound to the PM. Pp34 was then renamed REMORIN (later named StREM1.3 for S. tuberosum REMORIN of Group 1 isoform 3) in reference to remora or “suckerfish” depicted as attaching itself to vessels and larger fish described by J.L. Borges in “Book of Imaginary Beings” (El libro de los seres imaginarios, J.L. Borges, 1969), and reflecting REM’s ability to bind to the PM whilst displaying an overall hydrophilic residue profile. Concurrently to the experiments of Farmer et al., Alliote et al. isolated a similar protein in Arabidopsis (Arabidopsis thaliana) termed DNA-binding protein. This gene would be later known as AtREM1.3 (At2g45820) and was first characterized as a DNA-binding protein due to its highly hydrophilic nature permitting electrostatic interactions with DNA and its amino-acid composition similar to Histone 1 proteins (Alliotte et al., 1989).
Genome-wide analyses have shown that the REM family is specific to the land-plant lineage (Raffaele et al., 2007). REM proteins present a highly conserved C-terminal domain and a divergent N-terminal domain, which has been the basis for their phylogenetic classification into six separate groups (Raffaele et al., 2007). For example, we count 19 OsREMs in rice (Oryza sativa) and 16 AtREMs in Arabidopsis.
Since their discovery in the late 1980s, REMs have been the subject of ever-increasing attention. Be they directly the subject of studies or found in different screens of “omics” approaches, REMs are consistently found in a large variety of academic inquiries, providing grist to the mill to the idea that they could play a central role in plant development and adaptation. The first REM protein was identified in potato, which would be later named StREM1.3 for S. tuberosum REMORIN of Group 1 isoform 3. REM’s polyanion binding capacity, originally linked to binding cell wall compounds (Reymond et al., 1996), would be later mitigated by the discovery of REM’s presence in the inner-leaflet of the PM.
In this review, we address the different characteristics that have been described for REMs as well as an overview of the physiological roles in which REMs may participate. We will discuss how REMs are anchored to the PM and cluster into PM nanodomains, how they are phosphorylated and the subsequent role of these post-translational modifications. Finally, we provide a number of perspectives on how REMs should be further studied to better understand the many physiological conditions involving REMs.
Advances box
REMs are a plant-specific membrane-bound protein family involved in response to biotic (bacteria, viruses, fungi, oomycetes, mycorrhizae) and abiotic stresses (cold, mannitol, salt…), as well as developmental cues.
REMs are strongly embedded in the inner-leaflet of the plasma membrane by an unconventional mechanism involving anionic lipids and sterols.
REMs are proposed as nanodomain-organizing proteins.
REMs are highly phosphorylated proteins containing putative intrinsically disordered regions, which likely play a role in scaffolding protein complexes.
REMs regulate cell-to-cell connectivity through PD.
REMs localize in diverse and coexisting nanodomains
REMs predominantly associate with the PM (Raffaele et al., 2009a; Marín et al., 2012; Jarsch et al., 2014; Konrad et al., 2014; Perraki et al., 2014). Moreover, isoforms from Group 1 and Group 6 REMs are partially associated with the plasmodesmata (PD)–PM in rice and in Solanaceae (Raffaele et al., 2009a; Fernandez-Calvino et al., 2011; Gui et al., 2014; Perraki et al., 2018). Nonetheless, translocation into the nucleus upon interaction with α-importins (Marín et al., 2012) and re-localization to intracellular foci upon perception of an immunogenic epitope of bacterial flagellum (Albers et al., 2019) have been reported for Group 1 and Group 4 REMs, respectively. However, the endoplasmic reticulum ER–PM contact sites observed by bimolecular fluorescence complementation (BiFC) for StREM1.3 are likely artifactual and highlight the risk of using the BiFC to study membrane protein interactions in plants (Tao et al., 2019).
Concerning their PM localization, the identification of Group 1 REMs in tobacco (Nicotiana tabacum) Detergent-resistant membrane fractions (DRM; a biochemical counterpart of membrane sub-compartmentalization) first suggested a lateral organization of REMs into clusters at the PM (Mongrand et al., 2004). REMs’ co-purification with DRM appeared to be dependent on the presence of phytosterols as this co-purification was not reported in the sterol methyl-transferase mutant smt1 albeit still present in the DRM fraction in the sterol glycosylation mutant ugt80A2;B1 (Zauber et al., 2014).
In relation with the above biochemical data, electron microscopy immunolocalization, stimulated-emission depletion microscopy and photoactivated localization microscopy studies have all showed that Group 1 REMs organize into nanodomains around about 70–90 nm in diameter in Arabidopsis and Solanaceae species (Raffaele et al., 2009a; Demir and Horntrich, 2013; Gronnier et al., 2017). Members of other groups in Arabidopsis and Medicago truncatula are organized in PM domains that can be distinguished in density, size, and shape (Jarsch et al., 2014). Interestingly, evolutionary distant REMs (belonging to distinct groups e.g. Groups 1 and 6) localize in separate nanodomains suggesting that distinct REMs cluster into separate domains to play independent functions (Jarsch et al., 2014; Bücherl et al., 2017). It must also be noted that not all REMs are found in DRM-fractions of different plant tissues (Stanislas et al., 2009; Keinath et al., 2010; Srivastava et al., 2013; Takahashi et al., 2013) hinting at a diversity of REM-associated PM domains.
Mechanisms of REMs association to PM nanodomains
A number of works have been performed to understand how highly hydrophilic REMs could be tightly anchored to the PM and the molecular interactions leading to the clustering of REMs into nanodomains. REMs are anchored to the PM inner-leaflet via the unconventional lipid-binding motif called REMORIN C-terminal Anchor (REM-CA; Raffaele et al., 2009a; Perraki et al., 2012; Konrad et al., 2014; Gronnier et al., 2017). In the past few years, several molecular determinants regulating REMs’ nanodomain organization have emerged mostly for Group 1 and 2 REMs and need to be further studied to fully understand the diversity of REMs’ clustering (Jarsch et al., 2014). There are at least four determinants: the lipid-binding properties of REM–CA domain, REM–CA S-acylation, REM–REM oligomerization, and REM–cytoskeleton interactions. Here, we review the knowledge obtained to explain the molecular mechanisms at play.
The unconventional membrane-anchoring REMORIN C-terminal anchor domain (REM-CA) and S-acylation
REMs do not contain target-peptides or transmembrane domains (Raffaele et al., 2007) and attach to the PM independently of the conventional secretory pathway (Gui et al., 2015; Gronnier et al., 2017). StREM1.3, SYMREM1 (also termed MtREM2.2) as well as AtREM1.2, AtREM1.3, AtREM4.2, AtREM6.1, and AtREM6.4 have been described to be strictly PM localized thanks to their REM-CA moieties (Raffaele et al., 2009a; Lefebvre et al., 2010; Perraki et al., 2012; Raffaele, 2013; Jarsch et al., 2014; Konrad et al., 2014; Gronnier et al., 2017). In the case of StREM1.3, the last 28 amino acids (i.e. REM-CA) partially folds into an alpha helix in the presence of phosphatidylinositol-4-phosphate (PI4P) and sitosterol and also inserts itself into the hydrophobic core of the bilayer (Gronnier et al., 2017). This is driven by protein–lipid interactions between the REM-CA and the PM’s inner-leaflet lipids: negatively charged phosphoinositides (PIPs), notably PI4P, and sterols, notably sitosterol (Raffaele et al., 2009a; Legrand et al., 2019), (Figure 1). REM-CA binds to PI4P with a yet unknown stoichiometry through its Lysine/Arginine residues via electrostatic interactions. Association of StREM1.3’s REM-CA with negatively charged phospholipids regulates its nanodomain organization (Gronnier et al., 2017).
Figure 1.
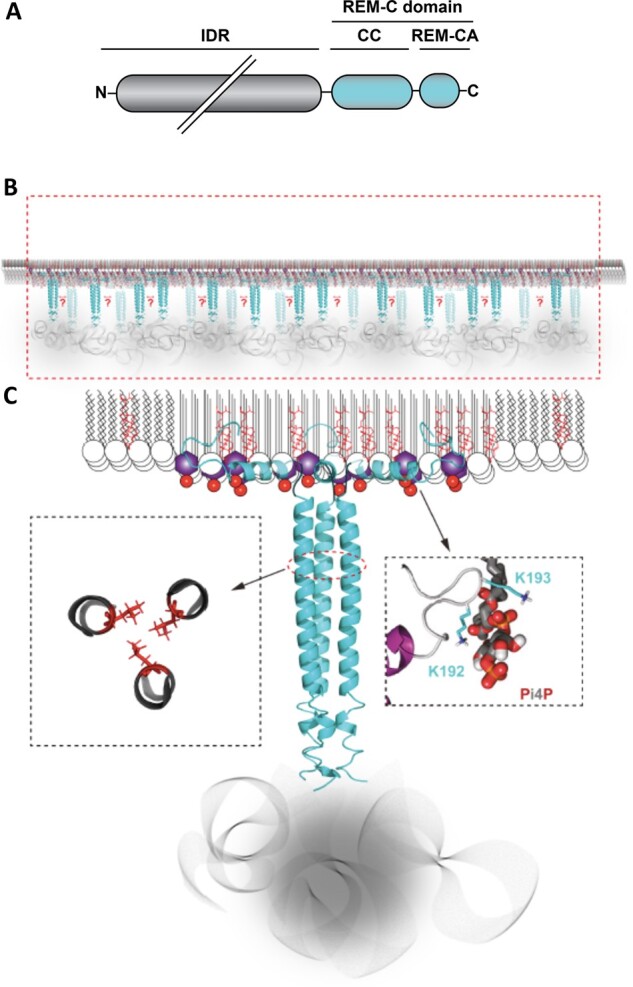
StREM1.3-enriched PM nanodomains based on structural analysis. A, Sequence features of all REMs with IDR domain at the N-terminal highly variable in length, and a REM-C domain composed of a coiled-coil region (CC) and the REM-C-terminal anchor (REM-CA). B, StREM1.3 (blue) clusters at the PM’s inner leaflet into nanodomains (red rectangle) enriched in phytosterols and phosphoinositides PIPs, the most common being sitosterol and PI4P (Palta et al., 1993; Furt et al., 2010, 2011; Gronnier et al., 2017; Legrand et al., 2019). The overall REM orientation is likely to be perpendicular to the membrane plane to minimize steric hindrance, though the precise angle distribution is unknown. Lipids herein are more ordered than the bulk of the PM and the membrane therefore is slightly thicker even though the PM’s complex composition may greatly attenuate this tendency (Gronnier et al., 2018, Legrand et al., 2019). C, StREM1.3 forms homo-trimers through a CC (Martinez et al., 2018) bundling together three REM-CAs (Gronnier et al., 2017) and three IDRs bearing phosphorylation sites (Perraki et al., 2018). REM-CA binds to PI4P through electrostatic interactions involving, notably, positively charged amino-acids K192 and K193, as proposed in the right panel by molecular dynamics (Gronnier et al., 2017). The most C-terminal part (region 2) of REM-CA is embedded inside the inner leaflet, as supported by solid-state NMR and molecular dynamics (Gronnier et al., 2017). Nanodomain-associated PIPs enriched in saturated acyl chains and sitosterol (Furt et al., 2010). The left panel shows hydrophobic contacts between the three L155 residues (red), which are critical to the CC structure, supporting membrane-association (Martinez et al., 2018). Membrane binding is accompanied by an enrichment in PI4P and sitosterol in the vicinity of the binding site (Legrand et al., 2019). A relative impoverishment in such lipids in the surrounding bulk membrane would therefore be a logical consequence. Note here that only REM molecules are presented, and the other PM proteins are omitted.
In mammals, PIPs have been reported to possess the ability to cluster in vivo to form nanodomains (van den Bogaart et al., 2011). In vitro, cholesterols are found in the vicinity of saturated phospholipids rather than unsaturated ones (Engberg et al., 2016). This mechanism creates a so-called liquid-ordered domain in the lipid bilayer. In plants, the fact that PIPs bear mostly saturated acyl-chains suggests that phytosterols may also actively participate in the formation of nanodomains (Furt et al., 2010). REMs could be either targeted to pre-existing lipid-mediated nanodomains or participate in the organization of their own lipid environments. Both the latter and former possibilities for REMs’ association to PM nanodomains still remain open questions.
The presence of cysteine residues in the REM-CA of many REMs raises the possibility of membrane association due to S-acylation. AtREM1.2 was the first REM found to be S-acylated (Hemsley et al., 2013). AtREM1.2 and AtREM6.4 lost strict-PM localization when their REM-CA cysteine residues were substituted by alanine residues (Konrad et al., 2014). Altered localization when S-acylation is abolished via 2-bromopalmitate is also observed with the rice REM OsREM6.6 (Gui et al., 2015). This is reminiscent of the relocalization observed for the S-acylation site mutant NbREM1.1C206A to RFP-ATG8e-tagged vesicles (Fu et al., 2018). It is important to note that NbREM1.1C206A is no longer organized in nanodomains, nor co-purifies with the DRM biochemical fraction (Fu et al., 2018). Yet for SYMREM1, the cysteine substitution does not reduce its PM localization and does not change its segregation pattern in the PM, hinting to a more complex PM association mechanism. Altogether, all of the above-mentioned data highlight the importance of the REM-CA as a determinant of REM’s general membrane affinity as well as REM’s organization within these membranes. Interestingly, many REMs do not have predicted S-acylation sites in their REM-CA (Konrad et al., 2014; Gronnier et al., 2017), suggesting a degree of diversity in the mechanisms regulating REMs’ PM targeting and nanodomain organization.
REM oligomerization
REM oligomerization is highly important for its targeting and function. REMs were first proposed to form oligomeric structures. The first evidence of oligomerization was reported in Bariola et al. (2004) where the group 1 REM coiled-coil domain was described to participate in the formation of REM oligomers. Electron microscopy and glutaraldehyde crosslinking assays evidenced REM multimerization via the C-terminal region of two Group 1 REMs i.e. StREM1.3 and SlREM1.2 (Bariola et al., 2004). Bariola et al. (2004) proposed via analytical ultracentrifugation of recombinant proteins and cross-linking experiments on isolated PM that Group 1 REMs associate into dimers, tetramers, or higher order oligomeric structures via their coiled-coil domains. Alternatively, Perraki et al. (2012) developed the hypothesis that StREM1.3 would trimerize either before or after the anchoring event at the PM. This hypothesis was upheld by gel filtration assays with the Escherichia coli purified 6His-tagged full-length StREM1.3 protein (Perraki et al., 2012). The exact order of oligomerization of REMs in planta is still lacking (Jaillais and Ott, 2020).
In our current view, StREM1.3 is homo-trimeric in solution bundling three REM-CA domains together (Bariola et al., 2004; Martinez et al., 2018). This trimeric hypothesis was further developed in Martinez et al. (2018) upon in silico modeling confirmed by solid-state nuclear magnetic resonance (NMR) in conjunction with cryo-electron microscopy (cryo-EM) and in vivo observations of mutations in the coiled-coil domain that impair PM association (Figure 1). These in vivo observations seem to corroborate the hypothesis that in a minimal state, the trimeric form is necessary for PM anchoring. Cryo-EM further revealed a possible lateral association of the trimeric REMs in the presence of an N-terminal 6His-tag (Martinez et al., 2018), yet the presence of a 6His-tag may also alter the oligomeric behavior of proteins (Amor-Mahjoub et al., 2006). This casts a doubt on the in vivo relevance of the trimer of StREM1.3 observed by gel filtration (Perraki et al., 2012). In any case, the existence in vivo of homo-oligomers and higher order oligomers has not been invalidated by the latest data. The formation of higher order oligomers, i.e. oligomers of trimers, as a means to drive nanoclustering cannot be excluded but has yet to be tested. Interactions between neighboring REM trimers are likely to further stabilize nanodomain clustering.
Group 1 AtREMs have been found to form oligomers in a large-scale study of PM protein oligomerization in leaves (McBride et al., 2017). AtREM1.2, AtREM1.3, and AtREM1.4 were predicted to form oligomeric complexes according to their ratio of the calculated mass to the apparent mass (Rapp) score, which calculates a ratio between the predicted molecular mass of the monomer and the measured molecular mass of the complex in which the protein is found. AtREM1.2, AtREM1.3, and AtREM1.4 were found in different complexes to have a ratio of the calculated mass to the apparent mass scores (Rapp) ranging on average from 35 to 60 for AtREM1.2, 25 for AtREM1.3, and 30 to 95 for AtREM1.4 (McBride et al., 2017). These data may suggest that in addition to being homo-oligomerized, Group 1 REMs are also forming multiprotein complexes. This idea has already been suggested in the literature, notably in Lefebvre et al. (2010) where REMs are compared to caveolins considering the common properties they share: small, oligomeric, lipid raft-associated scaffold proteins that can form filamentous structures.
REM association with the cytoskeleton
In the seminal work of Jarsch et al. (2014), filamentous exclusion zones observed at the surface of the PM have suggested that REMs could be associated with the cortical cytoskeleton. A localization dependent on microtubule polymerization has been reported: the microtubule depolymerizing drug oryzalin lead to a smoother localization, i.e. REMs was not clustered in nanodomains (Szymanski et al., 2015). Moreover, AtREM6.6, which localizes at fibrillar structures and to PM domains, has been shown to be affected in its fibrillar localization by oryzalin treatment (Jarsch et al., 2014; Konrad et al., 2014).
This interplay between the cortical cytoskeleton and PM nanodomains fits with the “fences and pickets” paradigm (Kusumi et al., 2012), where REMs could play a role as a putative scaffold protein. This interplay with actin has been further demonstrated for other REMs, such as SYMREM1. SYMREM1’s association to PM nanodomains is destabilized by the actin depolymerizing drug, latrunculin B, yet not by the microtubule depolymerizing drug, oryzalin. SYMREM1’s presence at the PM is also essential for the establishment of FLOTILIN4-LYK3 domains that are dependent on the contact between FLOTILIN4 and the actin cytoskeleton (Liang et al., 2018; see Figure 2B). The rice Group 6 REM, OsREM6.6 (or GSD1 for grain setting defect 1, see below), associates with the actin cytoskeleton. Treatment with cytochalasin D (a potent inhibitor of actin polymerization) completely disrupted the punctate localization pattern of AtREM1.2 and AtREM1.3 to a more uniform distribution with smaller punctate patterns of lower intensity (Szymanski et al., 2015). Additionally, AtREM1.2 associates with the actin cytoskeleton under viral infection (Cheng et al., 2020). These data reveal the strong relationship between REMs’ anchoring to the PM and the actin cytoskeleton, although the functional mechanism remains to be tackled.
Figure 2.
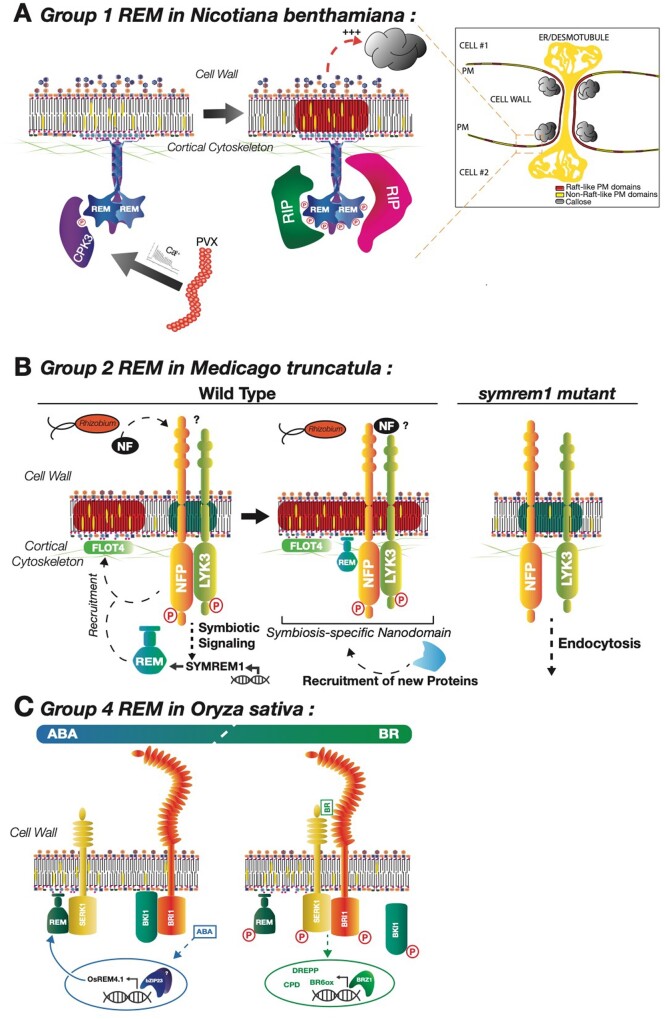
Proposed model for REM-mediated signaling. A, Group 1 REM in N. benthamiana. Proposed model for the molecular mechanisms involved in the StREM1.3-dependent limiting of PVX cell-to-cell movement according to the data from (Raffaele et al., 2009; Perraki et al., 2012, 2018; Gronnier et al., 2017). Perception of PVX leads to the production of a calcium burst that activates a group 2 calcium-dependent protein kinase (here CPK3), which in turn phosphorylates group 1 REMs. Group 1 REMs’ phosphorylated state increases their PM mobility, and they interact with phospho-REM interacting proteins (RIP). These interactions cause an increase in callose deposition at the PD. B, Group 2 REM in M. truncatula. Constitutively-expressed FLOT4 forms a primary PM-scaffold that is unable to recruit LYK3 in the absence of SYMREM1 (MtREM2.2). Nod factor (NF) perception by NFP and LYK3 PM-receptors triggers the activation of a symbiosis-specific signaling cascade that leads to the expression of SYMREM1. Due to its ability to directly bind LYK3, SYMREM1 actively recruits the receptor into the FLOT4 domain. In symrem1 mutants, LYK3 is destabilized and endocytosed upon rhizobial inoculation. Illustrations adapted from Liang et al. (2018). C, Group 4 REM in Oryza sativa. OsREM4.1 is PM-localized in association with the OsBRI1 (brassinosteroid insensitive)–OsSERK1 somatic embryogenesis receptor kinase 1 complex. Upon elevated ABA levels, the ABA-responsive transcription factor OsbZIP23 basic leucine zipper is activated and upregulates the OsREM4.1 expression. OsREM4.1 interacts with OsSERK1 and interferes with OsBRI1–OsSERK1 active complex formation, therefore repressing the BR signaling initiation. Increased BR levels cause the binding of BR to the extracellular domain of OsBRI1 and the activation of Os BKI1 BRI1 kinase to phosphorylate OsREM4.1. The phosphorylated OsREM4.1 has lower binding affinity to OsSERK1, and therefore the OsREM4.1–OsSERK1 complex is dissociated. Therefore, OsSERK1 is able to interact with OsBRI1 to form the OsBRI1–OsSERK1 receptor kinase complex and activate BR signaling. OsREM4.1 function is similar to BKI1, which upon BR binding to BRI1, is phosphorylated and released from the PM, allowing the formation of the BRI1/SERK1 complex. Illustrations are adapted from Gui et al. (2016).
The biological functions of REMs
Established functions of REM proteins
Plant–microbe interactions
Numerous studies have reported the implication of REMs in plant interactions with microorganisms.
Viruses
The first REM implicated in the context of viral infection is StREM1.3. Its overexpression limits the cell-to-cell movement of Potato Virus X (PVX), and its underexpression (RNAi lines) accelerates PVX movement (Raffaele et al., 2009; Perraki et al., 2018). A general effect on the REM-dependent gating of PD has also been shown in the presence of viral movement proteins, such as 30K from the Tobacco Mosaic Virus (TMV) or Hc-Pro from the Potato Virus Y (PVY; Perraki et al., 2014). In addition to limiting PVX cell-to-cell movement, StREM1.3 also limits TMV cell-to-cell propagation (Perraki et al., 2018). The overexpression of StREM1.3 does not impair the silencing suppressor activity of the PVX-encoded TRIPLE-GENE BLOCK protein 1 (TGBp1; Perraki et al., 2012), but rather induces the increase in callose accumulation at PD pit-fields (Perraki et al., 2018). Furthermore, StREM1.3’s capacity in hindering PVX cell-to-cell movement is abolished when REM-CA is mutated, thereby modifying its targeting to the PM (Gronnier et al., 2017). Additionally, StREM1.3 physically interacts with the PVX movement protein TGBp1 with or without an impaired REM-CA domain (Perraki et al., 2012).
Perraki et al., (2018) showed that PVX-activated kinases are responsible for StREM1.3’s phosphorylation potentially on residues S74/T86/S91 and that expression of the phosphomimetic, but not the phosphoablative mutant of StREM1.3 hampers virus cell-to-cell propagation to similar levels with the wild type. Importantly, the calcium-dependent protein kinase AtCPK3, which could phosphorylate StREM1.3 in vitro, could also restrict PVX cell-to-cell movement in a REM-dependent manner. In vitro, the StREM1.3 phosphomutants are not phosphorylated by AtCPK3 (Perraki et al., 2018). This suggests that an AtCPK3 ortholog in potato could potentially regulate StREM1.3 in vivo for anti-viral defense. Moreover, the phosphorylation mutants impact the localization of REM at PD and on callose deposition, associating phosphorylation status, protein mobility, PD permeability and cell-to-cell viral propagation (Perraki et al., 2018; Figure 2A).
Solanaceae Group 1 REMs have also been studied in the context of Tenuivirus infection. Nicotiana benthamiana NbREM1.1 and NbREM1.2 degrade during Rice Stripe Virus (RSV) infection in consequence to the interference of the RSV-encoded protein, NSvc4. The degradation of NbREM1.1 and NbREM1.2 via the autophagy pathway leads to RSV circumventing NbREM1-associated resistance (Fu et al., 2018). In contrast, a recent study has underlined the effect of the tobacco REM NtREM1.2 on the cell-to-cell movement of a Tobamovirus, the Tomato Mosaic Virus (ToMV; Sasaki et al., 2018). ToMV infection and the overexpression of ToMV movement proteins separately affect NtREM1.2’s localization by inducing its aggregation at the PM. Dissimilarly to StREM1.3, which is known to localize in a patchy pattern at the PM and is present in PM nanodomains, NtREM1.2 localized in a uniform fashion throughout the PM regardless of N-terminal or C-terminal fusion to fluorescent tags. Considering the high sequence conservation of the C-terminal region, this suggests that the N-terminal region is involved in NtREM1.2’s characteristic localization. Interestingly, NtREM1.2 interacts and colocalizes with ToMV’s 30-kDa movement protein at PD. Agrobacterium tumefaciens-mediated transient expression of NtREM1.2 slightly increases ToMV infection foci size (Sasaki et al., 2018). Moreover, in ToMV infected cells, NtREM1.2 aggregates occur close to tubular ER structures and are associated with ToMV’s 30-kDa movement protein bodies that appeared to be linked to the ER-Actin network (Sasaki et al., 2018). Finally, Arabidopsis AtREM1.2 has been proposed to play a role in Turnip Mosaic Virus (TuMV) cell-to-cell movement by competing with the TuMV protein VPg in the actin-dependent regulation of PD aperture (Cheng et al., 2020).
Group 4 REMs have been reported as positive regulators of viral infection. AtREM4.1 and AtREM4.2 are susceptibility factors during Beet Curly Top Virus (BCTV) and Beet Severe curly Top Virus (BSCTV) infection. This was shown using single- and double-knock-out (KO) and overexpressing lines for AtREM4.1 and AtREM4.2. This effect of Group 4 REMs on geminiviral infection could be linked to SnRK1, an important positive regulator of plant stress involved in antiviral defense, which could phosphorylate AtREM4.1 in vitro (Son et al., 2014).
Bacterial symbiosis
The Group 2 REMs merely constituted of a C_domain (c.f. Figure 1A; Raffaele et al., 2007) have been characterized by their role in the establishment of infection threads (IT) during the symbiosis of M. truncatula and Lotus japonicus with Rhizobiaceae family bacteria by participating in the formation of root nodules (Lefebvre et al., 2010; Tóth et al., 2012). SYMREM1 is specifically expressed in M. truncatula roots and nodules with expression strongly induced by Nod factor treatment (Lefebvre et al., 2010), which are lipo-chitooligosaccharides produced by symbiotic bacteria to initiate symbiosis (Oldroyd, 2013). Furthermore, SYMREM1 knock-down lines strongly reduce nodule establishment and growth (Lefebvre et al., 2010). Analogously, the overexpression of Group 2 REM LjSYMREM1 significantly increases the number of nodules per root in L. japonicus, establishing these orthologs as key players for symbiosis in legumes (Tóth et al., 2012). In this context, SYMREM1 has been further characterized at the molecular level. It has been described as a scaffold protein due to its interaction with three receptor-like kinases (RLKs) essential for nodule establishment and maintenance i.e. NFP, DMI2, and LYK3 (Lefebvre et al., 2010). This could be a recruiting process of RLKs to specific nanodomains to initiate IT along with FLOT4 (Liang et al., 2018). SYMREM1 is believed to stabilize the interactions between FLOT4 and both Nod factor perception NFP and LysM-receptor like kinase LYK3, which would prevent the RLKs’ endocytosis and ensure nodulation-promoting signaling (Liang et al., 2018; see Figure 2B).
Fungi and oomycetes
REMs have been implicated in interactions with filamentous pathogens/symbionts. StREM1.3 has long been established as a marker of Phytophthora infestans extrahaustorial membrane (EHM; Lu et al., 2012; Bozkurt et al., 2014). StREM1.3 labels haustoria approximately 50% of the time and specifically labels noncallosic haustoria (Bozkurt et al., 2014). Moreover, the overexpression of StREM1.3 in N. benthamiana and tomato increases susceptibility to P. infestans infection (Bozkurt et al., 2014). The precise role of StREM1.3 during P. infestans infection has yet to be precisely determined. Interestingly, recent studies show the specific recruitment of PIPs to the plant–pathogen interfacial membrane during fungal infection (Qin et al., 2020). Different PIPs are enriched at the EHM. Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) is dynamically upregulated at powdery mildew infection sites and recruited to the EHM, whereas PI4P is absent in the EHM. Furthermore, the depletion of PI(4,5)P2 in the pip5k1 pip5k2 mutant lines inhibits fungal pathogen development and causes disease resistance, independent of cell death-associated defenses, involving impaired host susceptibility. The fact that REMs both bind to PIPs and are involved in immune responses to fungi incites a detailed examination of the link between these two observations in future work.
The first report of a possible implication of REMs in plant–fungal interactions was a maize (Zea mays) ZmREM6.3 that was found in a quantitative trait loci analysis for resistance to northern leaf blight caused by the fungal pathogen Setosphaeria turcica (Jamann et al., 2016). A recent study further highlighted the importance of REMs in the resistance to fungal pathogens by identifying ZmREM1.3 in a large-scale proteomic analysis (Wang et al., 2019). The comparison between southern corn rust (Puccinia polysora) sensitive- and resistant-maize inbred lines revealed that ZmREM1.3 protein amount was increased in the resistant line, whereas it was decreased in the sensitive line. Genetic approaches (overexpression and KO) confirmed that ZmREM1.3 mediated maize resistance to P. polysora through salicylic acid (SA)/jasmonic acid signaling and defense gene upregulation (Wang et al., 2019).
A recent study shows that the overexpression of tomato (S. lycopersicum) SlREM1 increases susceptibility to the necrotrophic fungus Botrytis cinerea (Cai et al., 2020). Heterologous expression of SlREM1 increased reactive oxygen species accumulation and triggered other cell-death regulators, suggesting a positive regulatory role of SlREM1 in programmed cell-death (PCD) and providing clues for understanding the PCD molecular regulatory networks in plants.
Hormone signaling and abiotic stress
Rice OsREM4.1 is upregulated by abscisic acid (ABA) treatment via the bZIP transcription factor OsZIP23, which can bind the OsREM4.1 promoter. The OsREM4.1 overexpression inhibits brassinosteroid (BR) signaling by inhibiting OsSERK1 and OsBRI1 complex formation. This REM effect is undermined by the phosphorylation of REM. Under high BR concentrations, BR stabilizes the OsSERK1/OsBRI1 complex, which activates the phosphorylation of OsREM4.1 by OsBRI1, thereby reducing OsREM4.1’s affinity for OsSERK1 and enabling the BR signaling cascade. This signaling system maintains a dynamic equilibrium between ABA and BR signaling (Gui et al., 2016; Figure 2C). In Arabidopsis, AtREM1.2 and AtREM1.3 organize lipid raft nanodomains in a SA-dependent fashion. Indeed, by interaction with GRF10, both REMs participate in PD closure after SA treatment (Huang et al., 2019). Moreover, AtREM1.2 is crucial for SA-induced asymmetric auxin flux during root gravitropic response as well as for the regulation of clathrin-mediated endocytosis (Ke et al., 2020).
The tomato SlREM1.2 has recently been shown to be involved in fruit ripening by interacting with ethylene biosynthesis proteins. The overexpression of SlREM1.2 upregulates key genes involved in ethylene biosynthesis, lycopene biosynthesis and ripening regulators, resulting in an effective increase in the ethylene and lycopene content of fruit (Cai et al., 2018).
The functional characterization of a mulberry (Morus indica) Group 1 isoform MiREM1 evidenced its transcriptional upregulation under a number of abiotic stresses and hormone treatments. The gain of function via the heterologous overexpression of MiREM1 in Arabidopsis conferred resistance to salt stress and drought (Checker and Khurana, 2013).
The Group 6 REM SiREM6 (Setaria italica, fox-tail millet) is transcriptionally up regulated under salt, cold, ABA, and osmotic stress. Yet, this upregulation is weakly observed under drought stress, and the SiREM6 overexpression does not impact drought tolerance. The SiREM6 overexpression in Arabidopsis increased germination rate and seedling biomass as well as seedling survival under salt stress. These characteristics are linked to an increase in proline content and a reduction in electrolyte leakage. Additionally, the overexpression of SiREM6 in Arabidopsis enhances sensitivity to ABA treatment (Yue et al., 2014). In rice, OsREM6.5 is also highly upregulated during drought stress and ABA treatment (Wu et al., 2006)
In Populus euphratica, a salt-resistant poplar, PeREM6.5 is induced by NaCl stress. Interestingly, PeREM6.5 recombinant protein significantly increases the H+-ATPase hydrolytic activity and proton transport activity in P. euphratica PM vesicles. Yeast two-hybrid assays show that PeREM6.5 interacts with RPM1-interacting protein 4 (PeRIN4). Notably, the increase of H+-ATPase activity induced by PeREM6.5 is further enhanced by PeRIN4 recombinant protein. Finally, the overexpression of PeREM6.5 in Arabidopsis improves salt tolerance. PeREM6.5, by regulating H+-ATPase activity in the PM, may therefore enhance the plant capacity to maintain ionic homeostasis under salinity (Zhang et al., 2020).
Cell-to-cell connectivity via PD
The first direct evidence of the implication of REMs in cell-to-cell connectivity via PD was revealed by the ability of StREM1.3 to hinder PVX cell-to-cell movement as well as GFP diffusion in nonvirally-infected plants in N. benthamiana leaf epidermal cells (Raffaele et al., 2009; Perraki et al., 2012, 2014). REM transcript and protein levels increased in mature, aging, and senescing tissues, as well as in “source parts” of tobacco leaves (i.e. the tip of the leaf), where a majority of mature- and branched-PD are present (Raffaele et al., 2009b). These results suggest that tobacco Group 1 REMs predominantly associate with mature-branched-PD.
The Group 6 REM OsREM6.6 regulates PD permeability in rice, leading to the filling of the seed. The mutant termed GSD-1 (grain setting defect 1) shows an altered and ubiquitous expression of OsREM6.6, which is natively only expressed in phloem companion cells, thereby inducing an accumulation of starch and free-sugars in the leaf blades and consequently a reduction of starch accumulation in grains (Gui et al., 2014).
Data mining toward the putative functions of REMs
Throughout the relatively extensive literature that links REMs to biological functions (from transcriptomic, proteomic and phosphoproteomic data), several patterns may be found. Considering the overlapping nature of these types of events, we summarize those data in Table 1 by grouping REMs by phylum and by putative function. These links remain to be further experimentally confirmed but deserve to be pointed out to pave the way in understanding the biological functions of REMs.
Table 1.
Data mining toward the putative functions of REMORINs
| Plant species | Group of REMs | Methods / Stimuli | Biological effect(s) | Putative associated function(s) | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | AtREM1.2 | Protein–protein interaction (in vivo co-purification) | Interaction with RESISTANCE TO P. SYRINGAE PV MACULICOLA 1 RPM1- INTERACTING PROTEIN 4 (RIN4) | Bacterial immunity | (Mackey et al., 2002; Liu et al., 2009; Lee et al., 2015) |
| AtREM1.2 | Proteomics and phosphoproteomics / AvrRPM1 | Increased protein amount and phosphorylation | Bacterial immunity | (Widjaja et al., 2009) | |
|
AtREM1.2 AtREM1.3 |
Protein–protein interaction (in vivo proximity-dependent biotin identification BioID) | Interaction with HopF2bPtODC3000 | Bacterial immunity | (Khan et al., 2018) | |
| AtREM1.2 AtREM1.3 | Proteomics | Presence in extracellular vesicles | Immunity | (Rutter and Innes, 2017) | |
| AtREM1.3 | Phosphoproteomics / flagellin (flg22) | Increased phosphorylation | Bacterial immunity | (Benschop et al., 2007) | |
| AtREM1.3 | Transcriptomics / water deficit, cold, mannitol, salt stress | Transcript upregulation | Abiotic stress | (Reymond et al., 2000; Bray, 2002; Kreps et al., 2002) | |
| AtREM1.3 | Transcriptomics / auxin | Transcript upregulation | Development | (Alliotte et al., 1989) | |
| AtREM1.3 | Protein–protein interaction (yeast two-hybrid) | Interaction with Arabidopsis Response-Regulator 4 (ARR4) | Cytokinin response | (Yamada et al., 1998) | |
| AtREM1.3 | Affinity-based chromatography enrichment with immobilized ergosterol/ ergosterol | Interaction with ergosterol a fungal Microbe Associated Molecular Patterns MAMP | Fungal immunity | (Khoza et al., 2019) | |
| AtREM4.1 AtREM4.2 | Transcriptomics / drought, osmotic, salt stress and ABA treatment |
Transcript upregulation |
Abiotic stress | (Reymond et al., 2000; Bray, 2002; Son et al., 2014). | |
| AtREM6.7 | Transcriptomics / overexpression of DAYSLEEPER |
Transcript upregulation |
Flowering, PD connectivity in the meristematic zones | (Bundock and Hooykaas, 2005) | |
| Curcumis sativus | Group 1 and 4 CsREMs | Quantitative trait loci and transcriptomics / Podosphaera fusca |
Transcript upregulation associated with increased fungal resistance |
Fungal immunity | (Xu et al., 2017) |
| Glycine max | GmREM4 | Transcriptomics / drought treatment |
Transcript upregulation |
Abiotic stress, circadian rhythm | (Marcolino-Gomes et al., 2014) |
|
Hordeum vulgare |
Group 1 REM | Transcriptomics / gibberellic acid (GA) and ABA treatments |
Transcript downregulation |
Hormone response | (Chen and An, 2006) |
| Lotus japonicus | Group 1 REMs | Transcriptomics / Glomus intraradices |
Transcript upregulation |
Fungal symbiosis | (Kistner et al., 2005) |
| Oryza sativa | OsREM1.5 | Transcriptomics / ABA, BR treatments |
Transcript upregulation |
Hormone response | (Lin et al., 2003) |
| OsREM5.3 (Long Panicle 1 LP1) | Quantitative trait loci | Strongly expressed in young panicle, associated with longer panicle | Panicle size | (Liu et al., 2016) | |
| OsREM4.1 | Transcriptomics / overexpression of Deschampsia antartica C-repeat binding factor /dehydration-responsive element binding protein (CBF/DREB), DaCBF7 |
Transcript upregulation |
Stress adaptation, cold recovery | (Byun et al., 2015) | |
| OsREM6.5 | Drought stress and ABA treatment |
Transcript upregulation |
Stress adaptation, | (Wu et al., 2006) | |
| Quercus robur | QrREM4.1 | Transcriptomics / Piloderma croceum |
Transcript upregulation |
Ectomycorrhizal symbiosis | (Tarkka et al., 2013) |
| Solanum tuberosum | StREMa4 | Transcriptomics / Ralstonia solanacearum |
Transcript upregulation |
Bacterial immunity | (Kong et al., 2016) |
| StREMa4 | Transcriptomics/ ABA, SA, methyl jasmonate (MeJa) |
Transcript upregulation |
Hormone response | (Kong et al., 2016) | |
| Solanum lycopersicum | SlREM1.2 | Tomato plants overexpressing PHY-interacting factor CaPIF1 |
Transcript upregulation |
Enhanced resistance to cold stress | (Seong et al., 2007). |
| SlREM1.2 | Proteomics / Verticillium dahlia | Increased protein amount during incompatible interaction | Fungal immunity | (Hu et al., 2019) | |
| Triticum aestivum | 12 members of TaREM | Transcriptomics / cold acclimation | Transcript upregulation during early (7 TaREMs) and late (3 TaREMs) cold response, transcript downregulation (2 TaREM) | Cold adaptation | (Badawi et al., 2019) |
| Zea mays | ZmREM4.1 | Transcriptomics / cold | Transcript downregulation | Cold sensitivity | (Bilska-Kos et al., 2016). |
| ZmREM4.1 | Transcripts localized near PD linking cells from the Kranz mesophyll and the bundle sheath | Transcript localization | PD connectivity | (Bilska-Kos et al., 2016) |
Structural and biological implications of REM phosphorylation
Since the first identification of REM as an in vitro phosphorylated protein, pp34 (Farmer et al., 1989), members of all REM groups have been detected in phosphoproteomes from diverse tissues and biological contexts, suggesting phosphorylation as a major in vivo regulatory mechanism of REMs (see Supplemental Table 1 for a compilation of REM phosphoproteomic data). A recent study established the phosphorylation pattern of 14 AtREMs in 30 different plant tissues (Mergner et al., 2020). Apart from this broad-scale analysis, other studies have shown that, for some residues, the phosphorylation of REMs is modulated by stress conditions such as flg22 and PGA treatment, nitrogen deprivation, ABA, H2O2, cold, osmotic, and salt stresses (Benschop et al., 2007; Kohorn et al., 2016; Menz et al., 2016; Nikonorova et al., 2018; Wang et al., 2020; see Supplemental Table 1), suggesting functional roles of phosphorylation that remain to be explored.
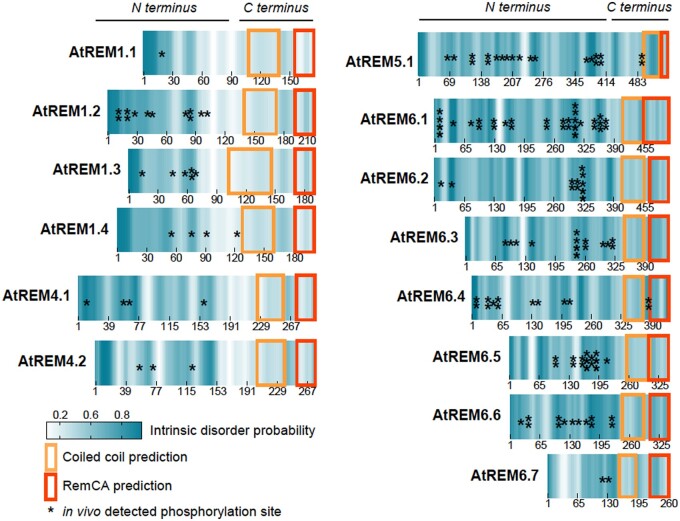
REMs are phosphorylated in vivo mainly at the putatively intrinsically disordered region (IDR) located in the N-terminal domain (Figure 3; Marín and Ott, 2012). IDR regions are flexible and extended protein segments that provide dynamic structural remodeling and protein–protein interaction plasticity (Iakoucheva et al., 2004; Dyson and Wright, 2005; Uversky, 2013). They are often regulated via phosphorylation that may act as a regulatory switch leading to structural changes such as folding and the sequestration of binding sites (Bah et al., 2015). Because they facilitate protein recognition and binding by acting as adaptable interaction surfaces, IDRs are often found in hub proteins that link interaction networks and integrate signals (Kim et al., 2008). REMs’ basal and signal-induced phosphorylation regions could putatively regulate interactions with its protein partners upon stimulus (Figure 3). This, and the fact that REMs participate in different plant signaling networks, ground the idea that they act as PM-bound-complex signaling “hubs” able to interact with many different proteins at specific times and locations and in response to specific stimuli. However, little is known about how REMs integrate diverse signaling cues in plants.
Figure 3.
Phosphorylation of Arabidopsis REMs occurs mostly at putative intrinsically disordered regions at its N-terminus. Graphs show the intrinsic disorder probability on the Arabidopsis REM proteins that were found to be phosphorylated to date by in vivo studies. In vivo phosphorylation site positions on REM sequences are indicated by an asterisk. Intrinsic disorder predictions were calculated using the protein disorder prediction (PrDOS) server (http://prdos.hgc.jp/cgi-bin/top.cgi). Coiled-coil predictions were calculated using the COILS program (https://embnet.vital-it.ch/software/COILS_form.html). REM-CA prediction was estimated based on homology with the REM-CA protein from StREM1.3.
Only a few studies have drawn links between REM phosphorylation and intracellular signaling. Marín and Ott (2012) showed that AtREM1.3 phosphorylation at S66 located in the IDR led to a reduced affinity for importins in yeast two-hybrid assays. However, the impact of this phosphorylation on the structure and subcellular localization of AtREM1.3 remains to be investigated. Furthermore, Gui et al. (2016) demonstrated the role of OsREM4.1 phosphorylation by OsBRI1 in activation of the BR signaling pathway (Gui et al., 2016; Figure 2C). While the phosphosites remain to be identified, this model connects REM phosphorylation, hormone-signaling balance and rice development, giving an important insight on one of the functional roles of REM phosphorylation in rice. In Perraki et al. (2018), the phosphorylation of StREM1.3 on residues S74, T86, S91 was essential for its anti-viral function and dynamic nanodomain association (Perraki et al., 2018; Figure 2A). Yet, the in vivo phosphorylation at those sites in response to PVX infection still remains to be determined.
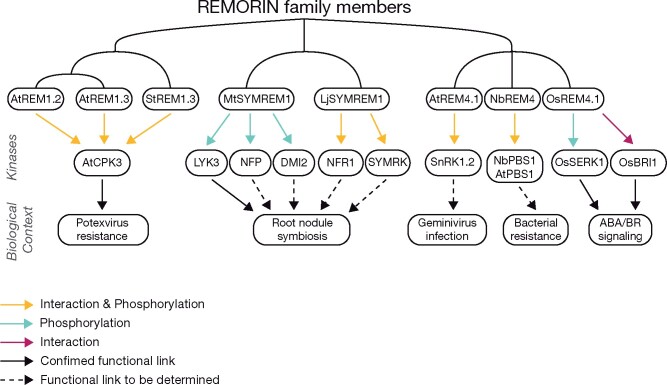
Although multiple REM phosphosites have been described in phosphoproteomes or with directed approaches, only few REM-associated kinases have been identified (see Supplemental Table 1). A recent large-scale phosphoproteomic study identified 8 Arabidopsis kinases (calcium protein kinase CPK11, mitogen-activated protein kinase MPK6, open stomata kinase OST1, sucrose nonfermenting-related kinase SnRK2.4, salt overly sensitive SOS2, oxidative signal-inducible OXI1, casein kinase-like CKL2, CT10 regulator of kinase-like CRKL2) that could phosphorylate, in vitro, AtREM phosphopeptides generated in vivo (Wang et al., 2020). Additional kinases responsible for REM phosphorylation have been described in rice, N. benthamiana, M. truncatula, L. japonicus, and Arabidopsis (SnRK1.2, AtCPK3, and various RLKs; Mehlmer et al., 2010; Tóth et al., 2012; Son et al., 2014; Gui et al., 2016; Perraki et al., 2018; Albers et al., 2019; see a summary in Figure 4). However, all these data have been obtained in vitro and will require further validation. Notably, in vivo interactions between REMs and protein kinases had already been reported for some of the kinases identified in Wang et al. (2020), Tóth et al. (2012), Perraki et al. (2018), and Albers et al. (2019). These kinases have been reported to phosphorylate a unique REM or several of them (Mehlmer et al., 2010; Tóth et al., 2012; Son et al., 2014; Gui et al., 2016; Perraki et al., 2018; Albers et al., 2019; Wang et al., 2020; Supplemental Table 1). Interestingly, most of those kinases are associated with DRMs, (Majeran et al., 2018) but some of them are putatively soluble, suggesting a PM recruitment.
Figure 4.
REM proteins are involved in independent signaling pathways and are phosphorylated by different kinases. Schematic representation of all the known interactions between REM proteins from groups 1, 2, 4, and protein kinases that were described in plant immunity, symbiosis, and hormonal signaling. Dotted lined arrow (i.e. functional link to be determined) signifies that the given REM and kinases were identified as being involved in the given biological context yet direct experimental data has yet to be established. See text for details.
This protein–protein interaction conferred by regions with high scores of intrinsic disorder (Raffaele et al., 2007; Marín and Ott, 2012) and phosphorylation (Reymond et al., 1996; Marín and Ott, 2012; Gui et al., 2016; Kohorn et al., 2016; Perraki et al., 2018; see Figure 3) are characteristics reminiscent of scaffold proteins, such as the 14-3-3 family of proteins. This analogy holds on the cases where REMs’ interaction capacity was reported to differ upon phosphorylation of either the interaction partner(s) or the REM isoform itself (Gui et al., 2016; Liang et al., 2018). GRF10 (General regulatory factor 14-3-3 epsilon) was recently shown to interact with and mediate AtREM1.2 and AtREM1.3 interaction (Huang et al., 2019). The 14-3-3 family proteins are involved in phospho-relay events, preeminently in facilitating and hindering protein–protein interactions by binding to the phosphorylated residues of interaction partners (Lozano-Durán and Robatzek, 2015).
Perspectives
The amount of data that has been accumulated since the cloning of the first REM in 1989 on the characteristics and the cellular and physiological role of REMs is very heterogeneous. Although many articles have shown REM expression as being related with a number of physiological processes, a clear role for the REM family has not yet arose. Nevertheless, many trails have emerged from the 30 years of research on REMs that show great promise in the search for REMs’ functionalities. One of these functionalities stands out, considering the multiple instances where REMs are linked to intercellular connectivity. These instances reinforce the idea that the establishment of REMs as a genetic family could have been important for the adaptation of photosynthetic algae during the land plant invasion that occurred approximately 475 million years ago (Steemans et al., 2009). Indeed, the control of water and solute exchanges between cells and tissues was a major necessary adaptation to handle water fluxes in dry land. Complex PD along with a complex vasculature necessary for the establishment of root systems and hormone signaling were the fundamental developments that enabled adaptation and establishment of embryophytes (Rensing et al., 2008). Moreover, the fact REMs are involved in biotic interactions, notably with symbiotic fungi and bacteria, is also a clue to their potential involvement in the conquest of land by plants, considering the hypothetical importance of symbiotic microorganisms in helping the first rootless land plants to secure nutrients (for review see Raffaele et al., 2009a; Rensing, 2018). The recent discovery of Group 1 REMs in a proteomic analysis of Arabidopsis extracellular vesicles may potentially change our view of REM’s strictly-PM function (Rutter and Innes, 2017, 2018).
Genetics
A major weakness in the functional study of REMs is the lack of genetic tools. Overall, REM single mutants have not yet displayed any striking or strong phenotypes, although AtREM1.2 alone has recently been shown to be involved in SA-dependent gravitropism response (Ke et al., 2020). To study REMs, the most notable problem is currently the lack of multiple KO-lines for each isoform, although the generation of these lines is currently ongoing with the recently published CRISPR-Cas9 KO-lines for two Group 1 REMs in N. benthamiana (Fu et al., 2018; Huang et al., 2019) and the quadruple CRISPR-Cas9 KO-line for all four Group 1 REMs in Arabidopsis that will soon be available (T. Ott, personal communication). KO lines for each REM Group will be essential for our understanding of the role played by each REM Group that could entail not only different possible cellular functions but also different tissue specificities (e.g. Gui et al., 2014). Nevertheless, we cannot exclude that REMs from different sub-groups are at least partially redundant or can functionally substitute for other REMs, even if in the wild-type situation they fulfill a different function.
Several REM Groups, such as Groups 5 and 3, could consequently become less illusive as to their function. The generation of KO-lines should be combined with gain-of-function assays as those undertaken in the characterization of OsREM6.6 (or GSD1; Gui et al., 2014) and OsREM4.1 (Gui et al., 2016) to develop true functional approaches in deciphering the cellular and physiological purposes of REMs.
Protein–protein interactions
Phosphorylation of REMs is suspected to be involved in protein–protein interactions (Raffaele et al., 2007; Marín et al., 2012; Figure 2;Supplemental Table 1) reminiscent of scaffold proteins, notably of 14-3-3 family proteins. Recently, GRF10 was shown to interact with AtREM1.2 and to be crucial for its partitioning at the PM (Huang et al., 2019). REMs and 14-3-3 family proteins could indeed co-depend on each other considering REMs’ interaction capacity varies upon phosphorylation of either the interaction partner(s) or the REM isoform itself (Gui et al., 2016; Liang et al., 2018;). In both cases, the REM isoforms i.e. OsREM4.1 or SYMREM1, interact with RLKs OsSERK1 (Gui et al., 2016) and LYK3 and NFP (Liang et al., 2018), respectively (Figure 2B). The interplay between REMs and RLKs deserves to be further investigated, particularly in view of the recent emerging role of RLK signaling in PD physiology (Grison et al., 2019; Cheval et al., 2020)
Considering that phosphorylation is at the center of the interaction events recorded for REMs, it seems crucial to develop phospho-dependent interactomic screens for REMs to understand their function. This type of screen should importantly be performed under different stress conditions and in different tissues as REMs’ interactions with its cognate partners appear to be highly determined by plant development and stress.
The PM’s complexity
Important advancements have recently been established in the deciphering of the PM-anchoring properties of REMs (Perraki et al., 2012; Gronnier et al., 2017; Martinez et al., 2018; Legrand et al., 2019). In as much as REMs are dependent of their C-terminal region or REM-CA for their anchoring to the PM, more must be developed in the understanding of the exact biophysical properties that govern this anchoring. Critically, decrypting the determinants of the PM-nanodomain association of REMs is a top priority, as it has already been shown to be a determining factor of REM function (Gronnier et al., 2017; Fu et al., 2018; Ke et al., 2020). In particular, affiliating the presence of Group 1 REMs in PM nanodomains with their observed effect on the accumulation of callose at PD pit-fields (Gronnier et al., 2017; Perraki et al., 2018) raises a critical question: how does a protein, present in the PM inner-leaflet, influence enzymatic activities (i.e. callose synthesis/degradation) occurring in the cell wall or, at best in the PM outer-leaflet?
Several hypotheses may be explored to answer this question: (1) Can Group 1 REMs or their interaction partners regulate callose synthases or other partners of the callose synthesis complex present at PD pit-fields to stimulate callose deposition? (2) Can Group 1 REMs or their interaction partners regulate the activity or localization of ß-1,3-glucanases? This latter question can be a particularly interesting hypothesis to investigate considering the different subcellular and extracellular localizations of ß-1,3-glucanases (for review see Zavaliev et al., 2011). Group 1 REMs could potentially regulate the extracellular secretion of ß-1,3-glucanases as it was reported in Zavaliev et al. 2013 in response to SA treatments. Yet, some ß-1,3-glucanases have been reported to be glycosylphosphatidylinositol (GPI)-anchored proteins within the PM outer-leaflet associated with PD (Levy et al., 2007). The enrichment of GPI-anchored proteins in the DRM biochemical fraction (for review on plant GPI-anchored proteins see Yeats et al., 2018) licenses the hypothesis that there could be trans-bilayer coupling between the inner-leaflet PM domains containing REMs and the outer-leaflet PM domains containing GPI-anchored ß-1,3-glucanases. In this context, the concept of trans-bilayer coupling refers to the presence of each component in a common PM domain that spans from one PM-leaflet to the other. There could therefore be interaction in trans between inner- and outer-leaflet proteins via the interdigitation of saturated and/or very-long-chain acyl chains of the lipids that are present in these PM domains. This functional mechanism has been demonstrated in animal cells by showing the necessity of long-chain phosphatidylserine species with at least one long saturated chain to register a GPI-anchored protein to a lipid–protein complex that virtually anchored the GPI-anchored protein to the actin cytoskeleton (Raghupathy et al., 2015; Skotland and Sandvig, 2019). The registration hypothesis is supported by the altered StREM1.3 PM-dynamics under a phosphorylated state (Perraki et al., 2018), which could be due to the regulation of StREM1.3’s PM-lateral-segregation via the cortical cytoskeleton. This change in REM PM-patterning could affect the PM-dynamics of PM-outer-leaflet-associated proteins e.g. GPI-anchored ß-1,3-glucanases. Ultimately, elucidating not only the protein–protein but also the protein–lipid interactions necessary for REMs’ PM-nanodomain-association constitutes a great challenge that will participate in the unfurling of the complex and dynamic molecular mechanisms that govern REMs particular role(s) at the PM.
Outstanding questions
How can REM functional versatility be explained?
What are the consequences of the phosphorylation(s) of REM’s putative intrinsically disordered regions in terms of 3D structure, protein–protein interaction and nanodomain organization?
How do REMs regulate cell-to-cell connectivity through PD?
New methodologies must be developed to study the PM-based events and interactions. These methodologies should include: (1) cutting-edge proteomic and lipidomic techniques (e.g. identifying lipids in close interaction with proteins), including phosphoproteomics and the study of lipidated proteins (myristoylation, palmitoylation, isoprenylation, and GPI-anchoring; reviewed in Ray et al., 2017); (2) biophysical tools to study lipid–protein interactions via surface plasmon resonance (e.g. Lenarčič et al., 2017), liposome binding assays and lipid blotting assays (e.g. Perraki et al., 2012), solid-state NMR, cryo-electron microscopy, atomic force microscopy, computational modeling, Langmuir monolayer tensiometry, and Fourier transform infrared spectrometry (e.g. Gronnier et al., 2017; methods reviewed in Zhou and Hancock 2018); (3) tools to obtain the structure of these lipid–protein complexes, for example, by solid-state NMR; (4) generation of mutants impaired in PM-domain-association and the use of super-resolution imaging to visualize the in vivo segregation and dynamics of PM-components, which should be a focus to better understand the molecular interplay occurring at the protein and lipid clustering sites that are PM-domains. These important tools can considerably be assisted by in silico modeling such as molecular dynamics simulations (for review see Ulmschneider and Ulmschneider, 2018). Overall, these techniques and methods have been used and described in numerous research articles and reviews that focus on understanding the function of PM-based proteins (ZHou et al., 2015, 2017; Gronnier et al., 2018; Maxwell et al., 2018).
Accession numbers
Accession numbers are indicated in Supplemental Table S1.
Supplemental data
Supplemental Table S1. Compilation of phosphoproteomic datasets on REMs of different plant species.
Supplementary Material
Acknowledgments
This work was sustained by CNRS (Centre National de la Recherche Scientifique) and the University of Bordeaux. This work has benefited from the facilities of the Bordeaux Metabolome/lipidome Facility-MetaboHUB (ANR–11–INBS–0010).
Funding
A.F.D., A.L., J.G., and P.G. were supported by grants from the French Ministère de l’Enseignement Supérieur et de la Recherche (MESR). J.G. was supported by EMBO LTF 438-2018 and ERC-2017-COG IMMUNO-PEPTALK ID: 773153. S.M., B.H., M.B., and V.G. are supported by ANR project Phospho-REM domain (ANR 2019). S.G., S.M., V.G. are supported by ANR project Potymove The IPS2 (MB) benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). B.H. was supported by the CNRS Momentum.
Conflict of interest statement. None declared.
P.G. and J.G. developed the rationale of the review and wrote the text with the help of all the co-authors. A.L. and B.H. made Figure 1 and wrote the corresponding text. P.G. made Figure 2. A.P. made Figures 3 and 4. M.D.J. and M.B. gathered the data for the Supplementary Table S1 and wrote the corresponding text. S.M. and V.G. supervised the writing and figures. All authors read and corrected the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is: Sébastien Mongrand (sebastien.mongrand@u-bordeaux.fr).
References
- Albers P, Üstün S, Witzel K, Kraner M, Börnke F (2019) A Remorin from Nicotiana benthamiana interacts with the pseudomonas type-III effector protein HopZ1a and is phosphorylated by the immune-related kinase PBS1. Mol Plant-Microbe Interact 32:1229–1242 [DOI] [PubMed] [Google Scholar]
- Alliotte T, Tiré C, Engler G, Peleman J, Caplan a, Van Montagu M, Inzé D (1989) An auxin-regulated gene of Arabidopsis thaliana encodes a DNA-binding protein. Plant Physiol 89:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor-Mahjoub M, Suppini JP, Gomez-Vrielyunck N, Ladjimi M (2006) The effect of the hexahistidine-tag in the oligomerization of HSC70 constructs. J Chromatogr B Anal Technol Biomed Life Sci 844:328–334 [DOI] [PubMed] [Google Scholar]
- Badawi MA, Agharbaoui Z, Zayed M, Li Q, Byrns B, Zou J, Fowler DB, Danyluk J, Sarhan F (2019) Genome-wide identification and characterization of the wheat Remorin (Ta REM) family during cold acclimation. Plant Genome 12:180040. [DOI] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD (2015) Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519:106–109 [DOI] [PubMed] [Google Scholar]
- Bariola PA, Retelska D, Stasiak A, Kammerer RA, Fleming A, Hijri M, Frank S, Farmer EE (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol Biol 55:579–594 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJR, Slijper M, Menke FLH (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6:1198–1214 [DOI] [PubMed] [Google Scholar]
- Bilska-Kos A, Grzybowski M, Jończyk M, Sowiński P (2016) In situ localization and changes in the expression level of transcripts related to intercellular transport and phloem loading in leaves of maize (Zea mays L.) treated with low temperature. Acta Physiol Plant. DOI: 10.1007/s11738-016-2151-5 [Google Scholar]
- van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmüller H, Diederichsen U, et al. (2011) Membrane protein sequestering by ionic protein–lipid interactions. Nature 479:552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Richardson A, Dagdas YF, Mongrand S, Kamoun S, Raffaele S (2014) The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol 165:1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EA (2002) Classification of genes differentially expressed during water-deficit stress in Arabidopsis thaliana: an analysis using microarray and differential expression data. Ann Bot 89:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücherl CA, Jarsch IK, Schudoma C, Segonzac C, Mbengue M, Robatze S, MacLean D, Ott T, Zipfe C (2017) Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. eLife 6: 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P (2005) An Arabidopsis hAT-like transposase is essential for plant development. Nature 436: 282–284 [DOI] [PubMed] [Google Scholar]
- Byun MY, Lee J, Cui LH, Kang Y, Oh TK, Park H, Lee H, Kim WT (2015) Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci 236:61–74 [DOI] [PubMed] [Google Scholar]
- Cai J, Chen T, Wang Y, Qin G, Tian S (2020) SlREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiol 183:717–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Qin G, Chen T, Tian S (2018) The mode of action of remorin1 in regulating fruit ripening at transcriptional and post-transcriptional levels. New Phytol 219:1406–1420 [DOI] [PubMed] [Google Scholar]
- Checker VG, Khurana P (2013) Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep 32:1729–1741 [DOI] [PubMed] [Google Scholar]
- Chen K, An YQC (2006) Transcriptional responses to gibberellin and abscisic acid in Barley aleurone. J Integr Plant Biol 48:591–612 [Google Scholar]
- Cheng G, Yang Z, Zhang H, Zhang J, Xu J (2020) Remorin interacting with PCaP1 impairs Turnip mosaic virus intercellular movement but is antagonised by VPg. New Phytol 225:2122–2139 [DOI] [PubMed] [Google Scholar]
- Cheval C, Samwald S, Johnston MG, de Keijzer J, Breakspear A, Liu X, Bellandi A, Kadota Y, Zipfel C, Faulkner C (2020) Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc Natl Acad Sci USA 117:9621–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F, Horntrich C (2013) Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc Natl Acad Sci USA 2013; 110:8296–8301 [DOI] [PMC free article] [PubMed]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208 [DOI] [PubMed] [Google Scholar]
- Engberg O, Hautala V, Yasuda T, Dehio H, Murata M, Slotte JP, Nyholm TKM (2016) The affinity of cholesterol for different phospholipids affects lateral segregation in bilayers. Biophys J 111:546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E, Pearce G, Ryan C (1989) In vitro phosphorylation of plant plasma membrane proteins in response to the proteinase inhibitor inducing factor. Proc Natl Acad Sci USA 86:1539–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Moloshok TD, Saxtonj MJ (1991) Oligosaccharide signalling in plants: specificity of oligouronide-enhanced plasma membrane protein phosphorylation. J Biol Chem 266:3140–3145. [PubMed]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A (2011) Arabidopsis plasmodesmal proteome. PLoS One. DOI: 10.1371/journal.pone.0018880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Xu Y, Li C, Li Y, Wu J, Zhou X (2018) Rice stripe virus interferes with S-acylation of Remorin and induces its autophagic degradation to facilitate virus infection. Mol Plant 11: 269–287 [DOI] [PubMed] [Google Scholar]
- Furt F, König S, Bessoule JJ, Sargueil F, Zallot R, Stanislas T, Noirot E, Lherminier J, Simon-Plas F, Heilmann I, et al. (2010) Polyphosphoinositides are enriched in plant membrane rafts and form microdomains in the plasma membrane. Plant Physiol 152:2173–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F, Mongrand S, Simon-Plas F (2011) The plant plasma membrane. Plant Plasma Membr. DOI: 10.1007/978-3-642-13431-9 [Google Scholar]
- Grison MS, Kirk P, Brault ML, Wu XN, Schulze WX, Benitez-Alfonso Y, Immel F, Bayer EM (2019) Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol 181:142–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Crowet JM, Habenstein B, Nasir MN, Bayle V, Hosy E, Platre MP, Gouguet P, Raffaele S, Martinez D, et al. (2017) Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife 6:26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronnier J, Gerbeau-Pissot P, Germain V, Mongrand S, Simon-Plas F (2018) Divide and rule: plant plasma membrane organization. Trends Plant Sci 23: 899–917 [DOI] [PubMed] [Google Scholar]
- Gui J, Liu C, Shen J, Li L (2014) Grain setting defect1, encoding a Remorin protein, affects the grain setting in rice through regulating plasmodesmatal conductance. Plant Physiol 166:1463–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Zheng S, Liu C, Shen J, Li J, Li L (2016) OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev Cell 38:201–213 [DOI] [PubMed] [Google Scholar]
- Gui J, Zheng S, Shen J, Li L (2015) Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front Plant Sci 6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 197:805–814 [DOI] [PubMed] [Google Scholar]
- Hou Y, Qiu J, Tong X, Wei X, Nallamilli BR, Wu W, Huang S, Zhang J (2015) A comprehensive quantitative phosphoproteome analysis of rice in response to bacterial blight. BMC Plant Biol 15:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Puri KD, Gurung S, Klosterman SJ, Wallis CM, Britton M, Durbin-Johnson B, Phinney B, Salemi M, Short DPG, et al. (2019) Proteome and metabolome analyses reveal differential responses in tomato - Verticillium dahliae-interactions. J Proteomics 207:103449. [DOI] [PubMed] [Google Scholar]
- Huang D, Sun Y, Ma Z, Ke M, Cui Y, Chen Z, Chen C, Ji C, Tran TM, Yang L, et al. (2019) Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc Natl Acad Sci USA 116:21274–21284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK (2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32:1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto T, Farmer E, Ryan C (1993) Purification of potato leaf plasma membrane protein pp34, a protein phosphorylated in response to oligogalacturonide signals for defense and development. Plant Physiol 103: 1393–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Ott T (2020) The nanoscale organization of the plasma membrane and its importance in signaling: a proteolipid perspective. Plant Physiol 182:1682–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamann TM, Luo X, Morales L, Kolkman JM, Chung CL, Nelson RJ (2016) A remorin gene is implicated in quantitative disease resistance in maize. Theor Appl Genet 129:591–602 [DOI] [PubMed] [Google Scholar]
- Jarsch IK, Konrad SSA, Stratil TF, Urbanus SL, Szymanski W, Braun P, Braun K-HK-H, Ott T (2014) Plasma membranes are subcompartmentalized into a plethora of coexisting and diverse microdomains in Arabidopsis and Nicotiana benthamiana. Plant Cell 26:1698–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke M, Ma Z, Wang D, Sun Y, Wen C, Huang D, Chen Z, Yang L, Tan S, Li R, et al. (2020) Salicylic acid regulates PIN2 auxin transporter hyper‐clustering and root gravitropic growth via Remorin‐dependent lipid nanodomain organization in Arabidopsis thaliana. New Phytol. DOI: 10.1111/nph.16915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (Pathogen-associated Molecular Pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285:39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Youn J-Y, Gingras A-C, Subramaniam R, Desveaux D (2018) In planta proximity dependent biotin identification (BioID). Sci Rep 8:9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoza TG, Dubery IA, Piater LA (2019) Identification of candidate ergosterol-responsive proteins associated with the plasma membrane of Arabidopsis thaliana. Int J Mol Sci 20:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Sboner A, Xia Y, Gerstein M (2008) The role of disorder in interaction networks: a structural analysis. Mol Syst Biol 4:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stoudgard J, Webb KJ, et al. (2005) Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17:2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Hoon D, Minkoff BB, Sussman MR, Kohorn SL (2016) Rapid oligo-galacturonide induced changes in protein phosphorylation in Arabidopsis. Mol Cell Proteomics 15:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong CY, Luo YP, Duan TT, Xue Z, Gao XD, Zhao X, Gao G (2016) Potato remorin gene StREMa4 cloning and its spatiotemporal expression pattern under Ralstonia solanacearum and plant hormones treatment. Phytoparasitica 44: 575–584 [Google Scholar]
- Konrad SSA, Popp C, Stratil TF, Jarsch IK, Thallmair V, Folgmann J, Marín M, Ott T (2014) S-acylation anchors Remorin proteins to the plasma membrane but does not primarily determine their localization in membrane microdomains. New Phytol 203:758–769 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang H, Zhu T, Wang X, Harper JF, Mesa T, Row M, Diego S, California JAK, et al. (2002) Transcriptome Changes for Arabidopsis in Response to Salt, Osmotic, and Cold Stress. Plant Physiol 130:2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KGN (2012) Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol 28:215–250 [DOI] [PubMed] [Google Scholar]
- Lee D, Bourdais G, Yu G, Robatzek S, Coaker G (2015) Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H+-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell 27:2042–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Timmers T, Mbengue M, Moreau S, Hervé C, Tóth K, Bittencourt-Silvestre J, Klaus D, Deslandes L, Godiard L, et al. (2010) A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc Natl Acad Sci USA 107:2343–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand A, Martinez D, Grélard A, Berbon M, Morvan E, Tawani A, Loquet A, Mongrand S, Habenstein B (2019) Nanodomain clustering of the plant protein Remorin by solid-state NMR. Front Mol Biosci 6:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarčič T, Albert I, Böhm H, Hodnik V, Pirc K, Zavec AB, Podobnik M, Pahovnik D, Žagar E, Pruitt R, et al. (2017) Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358:1431–1434 [DOI] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL (2007) A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J 49:669–682 [DOI] [PubMed] [Google Scholar]
- Liang P, Stratil TF, Popp C, Marín M, Folgmann J, Mysore KS, Wen J, Ott T (2018) Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc Natl Acad Sci 115: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Xu SL, Ni WM, Chu ZQ, Xu ZH, Xue HW (2003) Identification of ABA-responsive genes in rice shoots via cDNA macroarray. Cell Res 13:59–68 [DOI] [PubMed] [Google Scholar]
- Liu E, Liu Y, Wu G, Zeng S, Tran Thi TG, Liang L, Liang Y, Dong Z, She D, Wang H, et al. (2016) Identification of a candidate gene for panicle length in rice (Oryza sativa L.) via association and linkage analysis. Front Plant Sci 7: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Coaker G (2009) Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant Signal Behav 4:1107–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Robatzek S (2015) 14-3-3 proteins in plant-pathogen interactions. Mol Plant-Microbe Interact 28:511–518 [DOI] [PubMed] [Google Scholar]
- Lu YJ, Schornack S, Spallek T, Geldner N, Chory J, Schellmann S, Schumacher K, Kamoun S, Robatzek S (2012) Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol 14:682–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Iii BFH, Wiig A, Dangl JL, Hill C, Carolina N (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108:743–754 [DOI] [PubMed] [Google Scholar]
- Majeran W, Le Caer JP, Ponnala L, Meinnel T, Giglione C (2018) Targeted profiling of Arabidopsis thaliana subproteomes illuminates co- and posttranslationally N-terminal myristoylated proteins. Plant Cell 30:543–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, Bendix C, Nakayama TJ, Celaya B, Molinari HBC, De Oliveira MCN, Harmon FG, Nepomuceno A (2014) Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One. DOI: 10.1371/journal.pone.0086402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Ott T (2012) Phosphorylation of intrinsically disordered regions in remorin proteins. Front Plant Sci 3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Thallmair V, Ott T (2012) The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein-protein interactions. J Biol Chem 287:39982–39991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Legrand A, Gronnier J, Decossas M, Gouguet P, Lambert O, Berbon M, Verron L, Grélard A, Germain V, et al. (2018) Coiled-coil oligomerization controls localization of the plasma membrane REMORINs. J Struct Biol 206:12–19 [DOI] [PubMed] [Google Scholar]
- Maxwell KN, Zhou Y, Hancock JF (2018) Clustering of Rac1: selective lipid sorting drives signaling. Trends Biochem Sci 43:75–77 [DOI] [PubMed] [Google Scholar]
- McBride Z, Chen D, Reick C, Xie J, Szymanski DB (2017) Global analysis of membrane-associated protein oligomerization using protein correlation profiling. Mol Cell Proteomics 16:1972–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, Bayer R, Teige M (2010) The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J 63:484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz J, Li Z, Schulze WX, Ludewig U (2016) Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J 88:717–734 [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, Papacek M, Chen X, Chaudhary A, Samaras P, Richter S, Shikata H, Messerer M, et al. (2020) Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579:409–414 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde J-P, Hartmann M-A, Bonneu M, Simon-Plas F, Lessire R, Bessoule J-J (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279:36277–36286 [DOI] [PubMed] [Google Scholar]
- Nikonorova N, Van Den Broeck L, Zhu S, Van De Cotte B, Dubois M, Gevaert K, Inzé D, De Smet I (2018) Early mannitol-triggered changes in the Arabidopsis leaf (phospho)proteome reveal growth regulators. J Exp Bot 69:4591–4607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252–263 [DOI] [PubMed] [Google Scholar]
- Palta JP, Whitaker BD, Weiss LS (1993) Plasma membrane lipids associated with genetic variability in freezing tolerance and cold acclimation of Solanum species. Plant Physiol 103:793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Binaghi M, Mecchia M a, Gronnier J, German-Retana S, Mongrand S, Bayer E, Zelada AM, Germain V (2014) StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett 588:1699–1705 [DOI] [PubMed] [Google Scholar]
- Perraki A, Cacas J-L, Crowet J-M, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol 160:624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Gronnier J, Gouguet P, Boudsocq M, Deroubaix F, Simon V, Legrand A, German-Retana S, Zipfel C, Bayer EMF, et al. (2018) REM1.3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog 14: e1007378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Zhou Z, Li Q, Zhai C, Liu L, Quilichini TD, Gao P, Kessler SA, Jaillais Y, Datla R, et al. (2020) Specific recruitment of phosphoinositide species to the plant-pathogen interfacial membrane underlies arabidopsis susceptibility to fungal infection. Plant Cell 32: tpc.00970.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S (2013) The Remorin C-terminal Anchor was shaped by convergent evolution among membrane binding domains. Plant Signal Behav 8: e23207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde J-P, Lherminier J, Noirot E, et al. (2009a) Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21:1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Mongrand S (2009b) Upregulation of the plant protein remorin correlates with dehiscence and cell maturation: a link with the maturation of plasmodesmata? Plant Signal Behav 4:915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T (2007) Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol 145:593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy R, Anilkumar AA, Polley A, Singh PP, Yadav M, Johnson C, Suryawanshi S, Saikam V, Sawant SD, Panda A, et al. (2015) Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell 161:581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Jatana N, Thukral L (2017) Lipidated proteins: spotlight on protein-membrane binding interfaces. Prog Biophys Mol Biol 128:74–84 [DOI] [PubMed] [Google Scholar]
- Rensing SA (2018) Great moments in evolution: the conquest of land by plants. Curr Opin Plant Biol 42:49–54 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD (2008) Conquest of land by plants. Science 319:64–69 [DOI] [PubMed] [Google Scholar]
- Reymond P, Kunz B, Paul-Pletzer K, Grimm R, Eckerskorn C, Farmer EE (1996) Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8:2265–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12:707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter BD, Innes RW (2018) Extracellular vesicles as key mediators of plant–microbe interactions. Curr Opin Plant Biol 44:16–22 [DOI] [PubMed] [Google Scholar]
- Rutter BD, Innes RW (2017) Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 173:728–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N, Takashima E, Nyunoya H (2018) Altered subcellular localization of a tobacco membrane raft-associated remorin protein by tobamovirus infection and transient expression of viral replication and movement proteins. Front Plant Sci 9: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong ES, Baek KH, Oh SK, Jo SH, Yi SY, Park JM, Joung YH, Lee S, Cho HS, Choi D (2007) Induction of enhanced tolerance to cold stress and disease by overexpression of the pepper CaPIF1 gene in tomato. Physiol Plant 129:555–566 [Google Scholar]
- Skotland T, Sandvig K (2019) The role of PS 18:0/18:1 in membrane function. Nat Commun 10:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Oh C, An C (2014) Arabidopsis thaliana remorins interact with SnRK1 and play a role in susceptibility to beet curly top virus and beet severe curly top virus. Plant Pathol J 30:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Malm E, Sundqvist G, Bulone V (2013) Quantitative proteomics reveals that plasma membrane microdomains from poplar cell suspension cultures are enriched in markers of signal transduction, molecular transport, and callose biosynthesis. Mol Cell Proteomics 12:3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T, Bouyssie D, Rossignol M, Vesa S, Fromentin J, Morel J, Pichereaux C, Monsarrat B, Simon-Plas F (2009) Quantitative proteomics reveals a dynamic association of proteins to detergent-resistant membranes upon elicitor signaling in tobacco. Mol Cell Proteomics 8:2186–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steemans P, Hérissé A Le, Melvin J, Miller MA, Paris F, Verniers J, Wellman CH (2009) Origin and radiation of the earliest vascular land plants. Science 324:353. [DOI] [PubMed] [Google Scholar]
- Szymanski WG, Zauber H, Erban A, Wu XN, Schulze WX (2015) Cytoskeletal components define protein location to membrane microdomains. Mol Cell Proteomics 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D, Kawamura Y, Uemura M (2013) Detergent-resistant plasma membrane proteome to elucidate microdomain functions in plant cells. Front Plant Sci 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Waletich JR, Arredondo F, Tyler BM (2019) Manipulating endoplasmic reticulum-plasma membrane tethering in plants through fluorescent protein complementation. Front Plant Sci 10:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkka MT, Herrmann S, Wubet T, Feldhahn L, Recht S, Kurth F, Mailänder S, Bönn M, Neef M, Angay O, et al. (2013) OakContigDF159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol 199:529–540 [DOI] [PubMed] [Google Scholar]
- Tóth K, Stratil TF, Madsen EB, Ye J, Popp C, Antolín-Llovera M, Grossmann C, Jensen ON, Schüßler A, Parniske M, et al. (2012) Functional domain analysis of the remorin protein LjSYMREM1 in lotus japonicus. PLoS One. DOI: 10.1371/journal.pone.0030817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider JP, Ulmschneider MB (2018) Molecular dynamics simulations are redefining our view of peptides interacting with biological membranes. ACC Chem Res 51:1106–1116 [DOI] [PubMed] [Google Scholar]
- Uversky VN (2013) A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci 22:693–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Höfte H (2016) Cell wall integrity signaling in plants: “To grow or not to grow that’s the question.” Glycobiology 26:950–960 [DOI] [PubMed] [Google Scholar]
- Wang P, Hsu CC, Du Y, Zhu P, Zhao C, Fu X, Zhang C, Paez JS, Macho AP, Andy Tao W, et al. (2020) Mapping proteome-wide targets of protein kinases in plant stress responses. Proc Natl Acad Sci USA 117:3270–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen Z, Tian L, Ding Y, Zhang J, Zhou J, Liu P, Chen Y, Wu L (2019) Comparative proteomics combined with analyses of transgenic plants reveal ZmREM1.3 mediates maize resistance to southern corn rust. Plant Biotechnol J 17:2153–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widjaja I, Naumann K, Roth U, Wolf N, Mackey D, Dangl JL, Scheel D, Lee J (2009) Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9:138–147 [DOI] [PubMed] [Google Scholar]
- Wu C-Q, Hu H-H, Zeng Y, Liang D-C, Xie K-B, Zhang J-W, Chu Z-H, Xiong L-Z (2006) Identification of novel stress-responsive transcription factor genes in rice by cDNA array analysis. J Integr Plant Biol 48:1216–1224 [Google Scholar]
- Xu Q, Xu X, Shi Y, Qi X, Chen X (2017) Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genomics 18:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Hanaki N, Imamura A, Ueguchi C, Mizuno T (1998) An Arabidopsis protein that interacts with the cytokinin-inducible response regulator, ARR4, implicated in the His-Asp phosphorylay signal transduction. FEBS Lett 436:76–80 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Bacic A, Johnson KL (2018) Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J Integr Plant Biol 60:649–669 [DOI] [PubMed] [Google Scholar]
- Yue J, Li C, Liu Y, Yu J (2014) A remorin gene SiREM6, the target gene of SiARDP, from foxtail millet (Setaria italica) promotes high salt tolerance in transgenic Arabidopsis. PLoS One 9:e100772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber H, Burgos A, Garapati P, Schulze WX (2014) Plasma membrane lipid-protein interactions affect signaling processes in sterol-biosynthesis mutants in Arabidopsis thaliana. Front Plant Sci 5:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R, Levy A, Gera A, Epel BL (2013) Subcellular dynamics and role of Arabidopsis β-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol Plant Microbe Interact 26:1016–1030 [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V (2011) Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248:117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deng C, Wu X, Yao J, Zhang Y, Zhang Y, Deng S, Zhao N, Zhao R, Zhou X, et al. (2020) Populus euphratica remorin 6.5 activates plasma membrane H+-ATPases to mediate salt tolerance. Tree Physiol 40: 731–745 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hancock JF (2018) Deciphering lipid codes: K-Ras as a paradigm. Traffic 19:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Prakash P, Liang H, Cho K-J, Gorfe AA, Hancock JF (2017) Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wong CO, Cho KJ, Van Der Hoeven D, Liang H, Thakur DP, Luo J, Babic M, Zinsmaier KE, Zhu MX, et al. (2015) Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 349:873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.