Abstract
Optimal plant growth performance requires that the presence and action of growth signals, such as gibberellins (GAs), are coordinated with the availability of photo-assimilates. Here, we studied the links between GA biosynthesis and carbon availability, and the subsequent effects on growth. We established that carbon availability, light and dark cues, and the circadian clock ensure the timing and magnitude of GA biosynthesis and that disruption of these factors results in reduced GA levels and expression of downstream genes. Carbon-dependent nighttime induction of gibberellin 3-beta-dioxygenase 1 (GA3ox1) was severely hampered when preceded by reduced daytime light availability, leading specifically to reduced bioactive GA4 levels, and coinciding with a decline in leaf expansion rate during the night. We attributed this decline in leaf expansion mostly to reduced photo-assimilates. However, plants in which GA limitation was alleviated had significantly improved leaf expansion, demonstrating the relevance of GAs in growth control under varying carbon availability. Carbon-dependent expression of upstream GA biosynthesis genes (Kaurene synthase and gibberellin 20 oxidase 1, GA20ox1) was not translated into metabolite changes within this short timeframe. We propose a model in which the extent of nighttime biosynthesis of bioactive GA4 by GA3ox1 is determined by nighttime consumption of starch reserves, thus providing day-to-day adjustments of GA responses.
GA-sugar matching occurs specifically at night and determines day to day adjustment of GA levels and subsequent growth.
Introduction
Growth in plants is controlled by many signals, consisting of both environmental and internal cues. Growth is a complex parameter often interpreted as the increase in the number and size of organs (Nelissen et al., 2016). Others consider growth as the gain of biomass, sometimes more specifically as the incorporation of carbon into structural carbohydrates (Sulpice et al., 2014; Mengin et al., 2017). Growth parameters like protein and cell wall synthesis directly follow carbon availability (Pal et al., 2013; Ikakov et al., 2017). Carbon storage and utilization are organized such that carbohydrates are roughly evenly incorporated into structural biomass throughout the day (Sulpice et al., 2014; Mengin et al., 2017). However, size increases of plants do not necessarily follow carbon availability. Hypocotyl and leaf expansion rates have been shown to vary throughout the day, and these expansion patterns have been attributed to the circadian clock and light signaling (Farré, 2012; Dornbusch et al., 2014; Apelt et al., 2017). Interestingly, expansion and metabolic biosynthesis can be distinctly regulated. In hypocotyls, cell wall biosynthesis is exclusively dependent on metabolic signals and cell expansion on the circadian clock (Ikakov et al., 2017). For optimal plant performance, size increase must match biomass integration. The ability to adjust leaf expansion rates based on resource availability would be especially important when conditions vary from day to day.
In conjunction with the clock and light signals, a variety of growth-stimulating signaling networks exists within the plant, including a set that rely on phytohormones (Nelissen et al., 2016). Pivotal among these are gibberellins (GAs), which positively regulate cell expansion and cell division (Olszewski et al., 2002; Achard et al., 2009). Plants deficient in GAs develop slowly and are typically very small in stature (Hedden and Thomas, 2012), whereas overexpression of biosynthetic routes or GA signaling leads to enhanced plant size (Gonzalez et al., 2010; VanHaeren et al. 2014; Nam et al., 2017; Pullen, 2019). GA biosynthesis occurs via a sequence of enzymatic steps, starting in the plastids and subsequently the endoplasmic reticulum, which convert geranylgeranyl diphosphate to GA12. GA12 is considered the common precursor to all GAs in plants and is further processed into a variety of forms by gibberellin 20-oxidases (GA20ox) and a gibberellin 13-oxidase (GA13ox). The final step to bioactive GA is catalyzed by gibberellin 3-beta-dioxygenase (GA3ox; Hedden and Thomas, 2012; He et al., 2019, 2020). In angiosperms, GA4, GA1, and GA3 have been confirmed to bind the gibberellin insenstive dwarf1 (GID1) receptor (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Yoshida et al., 2018). These bioactive GAs can also be made inactive by the enzyme GA2ox (Hedden and Thomas, 2012).
Established aspects of the regulation of GA biosynthesis concern localized suppression in the shoot apical meristem to ensure stem cell maintenance, and feedback regulation that is considered important to maintain GA homeostasis (Yamaguchi, 2008; Middleton et al., 2012). Feedback regulation is structured such that it aims to keep GA levels constant (Middleton et al., 2012) and is therefore not adequate to explain growth adjustments to varying conditions. To identify the role of GAs in vegetative growth and adjustment to adverse conditions, we studied the regulation of key GA biosynthetic genes, the subsequent variation in GA abundance, and ultimately their role in growth in Arabidopsis thaliana. This led to the finding that nighttime conversion of precursors to bioactive GA4 by starvation-sensitive GA3ox1 expression plays a role in adjusting growth to adverse conditions.
Results
Light, the circadian clock, and carbon availability regulate the timing and transcriptional induction of GA biosynthetic enzymes
Several enzymatic steps of GA biosynthesis are encoded by multigene families. To identify the key family members with relatively high expression during vegetative growth, two public microarray datasets that followed transcript abundance over 48 h in adult Arabidopsis rosettes were investigated (Smith et al., 2004; Bläsing et al., 2005). Further indication of a role in adjusting growth control was based on oscillatory behavior over the day–night cycle, indicating a responsiveness to changes in carbon supply, light signals, or the clock. These data (Supplemental Figure 1) pointed to Kaurene synthase (KS), GA20ox1, and GA3ox1 as main candidates (Figure 1).
Figure 1.
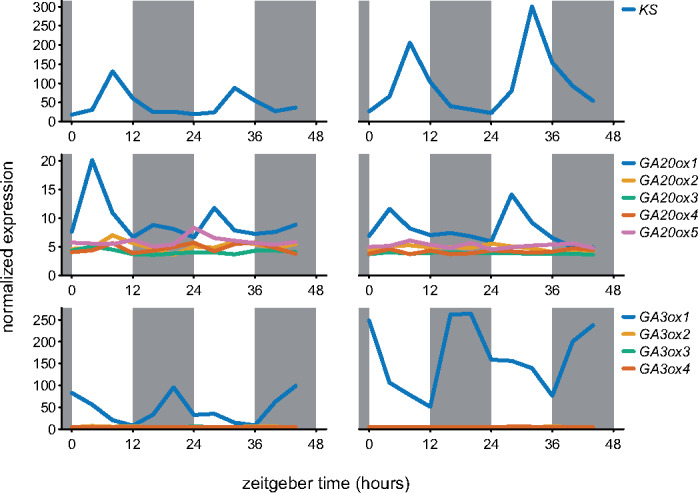
mRNA abundance over the day–night cycle of the gene family members of rhythmic GA biosynthetic genes. Left and right column based on data from 35- and 29-d-old, respectively, soil-grown Col-0 rosettes (Smith et al. 2004; Bläsing et al., 2005). Gray boxes represent the night period. All family members are shown.
Clock components and clock-regulated genes retain their oscillations after a transfer to continuous light conditions (Supplemental Figure 2A). Of the daytime expressed candidates, oscillations were retained under continuous light for KS, but not for GA20ox1. However, their peak in expression during normal day/night cycles, start of the day (GA20ox1), and afternoon (KS), matched the publicly available datasets (Figures 1, 2A). GA20ox1 oscillations seemed independent of the clock (Figure 2A), and a role for light in inducing this gene was investigated by exposing plants to light 3 h prior to expected dawn. An earlier start of the day resulted in a concomitant earlier peak of GA20ox1 (Figure 2B). Since an early day also led to an earlier increase in sugars (Figure 2B and Supplemental Figure 2B), a role for photosynthates to induce the earlier peak could not be excluded. After inhibiting photosynthesis with DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea), GA20ox1 still peaked upon earlier light exposure. However, DCMU did reduce the extent of induction and abolished GA20ox1 induction at later time points (Figure 2B).
Figure 2.
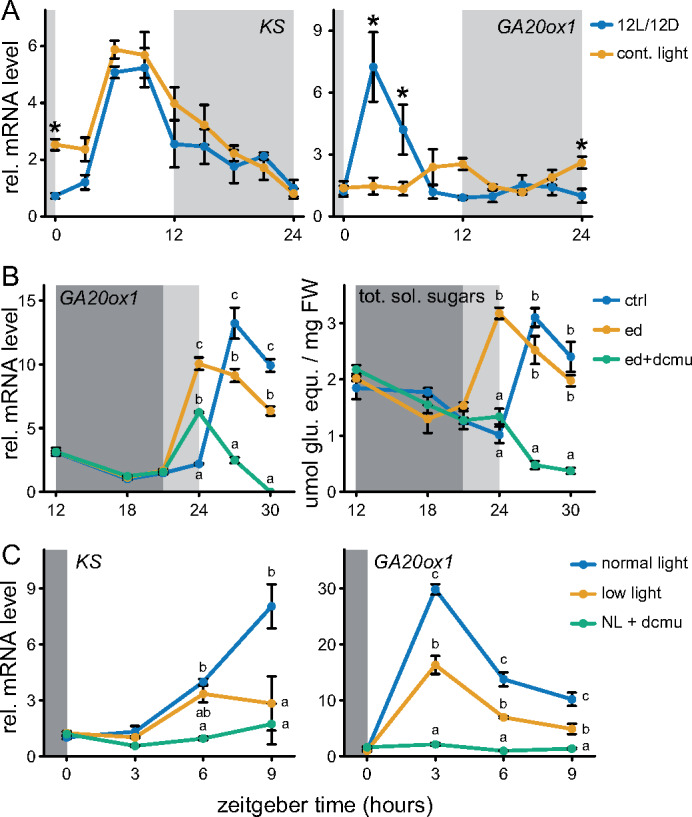
Transcriptional regulation of the daytime peaking GA biosynthetic genes, KS and GA20ox1. (A) The role of the circadian clock in mediating rhythmic expression. Light gray boxes represent subjective night. Sampling started the second day after beginning continuous light conditions. (B) Effect of a 3-h early start of day. Dark and light gray represent night and the subjective night, respectively. DCMU and mock solution (100 µM) was applied at zeitgeber time 12. Ctrl: 12L/12D, ed: early start of day. (C) The effect of reduced light availability or DCMU (NL + dcmu) on KS and GA20ox1 transcript abundance. DCMU was applied at the start of the preceding night. All data are from ∼10 leaf stage soil grown Col-0 rosettes. Means ± SE are shown, n = 4. Asterisks (planned comparisons) and letters (Tukey honestly significant difference (HSD)) represent statistically significant difference at specific time points, P < 0.05).
DCMU is a potent inhibitor of photosystem II and can cause harsh carbon starvation. The classical starvation marker genes DIN6 and TPS8 (Usadel et al., 2008) were strongly induced following DCMU treatment (Supplemental Figure 2B). Therefore, plants were also exposed to low-light (LL) conditions that were sufficient to maintain a positive carbon balance but did reduce photosynthesis and also showed minor induction of the starvation markers compared to the DCMU treatment (Supplemental Figure 2, C–E). Both KS and GA20ox1 showed severely reduced daytime expression after DCMU treatment, whereas the LL treatment led, as expected, to an intermediate reduction in their expression levels (Figure 2C).
Regarding the nighttime-expressed candidate, GA3ox1 expression was induced upon transition to darkness, in line with public data (Figures 1, 3A). Oscillations of GA3ox1 were not retained under continuous light, indicating that a shift to darkness is essential for GA3ox1 induction (Figure 3A). However, starting the night period earlier or later did not lead to a concomitant shift in GA3ox1 induction (Figure 3, B and C). This implies that GA3ox1 responds to darkness, but only when it occurs simultaneously with the expected onset of the night. The effect of carbon starvation on nighttime GA3ox1 induction was investigated by DCMU and LL treatments, as done for daytime-expressed KS and GA20ox1. Although LL treatments led to an intermediate starvation response compared to DCMU (Supplemental Figure 2E), it was sufficient to completely abolish the nighttime induction of GA3ox1, identical to the effect of DCMU (Figure 3D). These observations suggest that GA3ox1 is more sensitive to carbon starvation.
Figure 3.
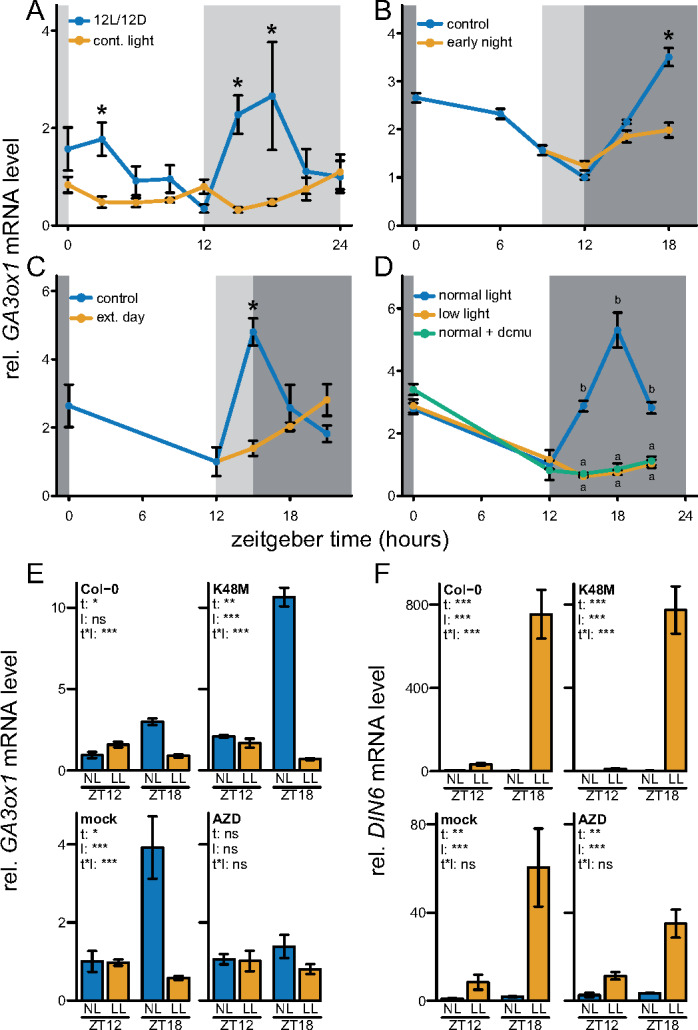
Transcriptional regulation of the nighttime peaking GA biosynthetic gene, GA3ox1. (A) The role of the circadian clock in mediating rhythmic expression. Light gray boxes represent subjective night. Sampling started second day after starting continuous light conditions. (B) Effect of a 3-h early start of the night. Dark and light gray represent night and the subjective night, respectively. (C) Effect of a 3-h extension of the day. Dark and light gray represent night and the subjective night, respectively. (D) The effect of reduced light availability or DCMU on nighttime GA3ox1 transcript abundance. DCMU was applied at the start of the preceding night. All data are from ∼10 leaf-stage, soil-grown Col-0 rosettes. Means ± SE are shown, n = 4. Asterisks (planned comparisons) and letters (Tukey HSD) represent statistically significant difference at specific time points, P < 0.05). (E, F) Manipulation of energy signaling pathways and the mRNA abundance of GA3ox1 and DIN6 at the end of the day (ZT12) and during the subsequent night (ZT18), both after control NL and LL levels. K48M: overexpresses the dominant inactive form of SnRK1.1/KIN10, where lysine residue 48 is mutated (Baena-Gonzalez et al., 2007). Blocking of TOR kinase activity by AZD8055 (AZD, 30 µM). Mock treatment (DMSO 0.01%) or AZD was applied to Col-0 at 6 a.m., 2 h prior the start of the day (ZT0, 8 a.m.). Means ± SE are shown (N = 5). Asterisks indicate the significance of effects in a two-way analysis of variance (ANOVA). *P < 0.05; **P < 0.01; ***P < 0.001. t – effect of time (ZT12, ZT18), l – effect of NL, LL, t * l, interaction of time and light.
The relevance of energy and carbon signaling for GA3ox1 regulation was further investigated by transgenic-, mutant-, and pharmaceutical inhibitor-based analysis of players in the three main energy signaling pathways, namely target of rapamycin (TOR) kinase, SNF1-related protein kinase 1 (SnRK1.1/KIN10), and glucose insensitive2, the latter of which functions in glucose signaling (Moore et al., 2003; Baena-González et al., 2007; Deprost et al., 2007). Overexpression of the wild-type alleles of either KIN10 or TOR, or the glucose insensitive2-1 null allele, did not affect the behavior of GA3ox1 during the night (Supplemental Figure 3). However, removal of a starvation signal by an active site-specific mutation (K48M) in KIN10 led to enhanced GA3ox1 expression (Figure 3E and F). Pharmaceutical inhibition of TOR kinase activity, an energy abundance signal, abolished nighttime GA3ox1 induction (Figure 3, E, and F). The direction of change in GA3ox1 upon manipulation of KIN10 or TOR kinase activity corresponded with DCMU or LL suppression of GA3ox1 (Figure 3D). The starvation reporter DIN6 is considered a target of KIN10 via bZIP transcription factors (Baena-González et al., 2007). However, DIN6 was not affected by manipulation of any of the energy signaling pathways (Figure 3F and Supplemental Figure 3, B, D, and F) indicating some redundancy in DIN6 starvation-dependent regulation. Overall, these results show that GA3ox1 is induced at night through a clock-gated darkness signal, where the magnitude of induction depends on carbon or energy signaling.
Bioactive GAs accumulate during the night and are severely reduced in the morning after a day of LL levels
Patterns of transcriptional regulation suggest that GA precursors are made during the day via the upstream enzyme KS and pre-final enzyme GA20ox1. These accumulated precursors would then be converted to bioactive GA by GA3ox1, which peaks during the night, leading to high bioactive GA at the end of the night. Moreover, reduced light availability affected all three transcripts, especially GA3ox1 (Figures 2, C and 3, D). Additionally, we confronted plants with an extended day to reduce GA3ox1 induction while maintaining carbon availability (Figure 3C). To see how such GA3ox1 regulation changed metabolite levels, we investigated the substrates of GA3ox1, their products, and inactivated forms of GA. GA20ox metabolism includes many intermediates, though a best estimate of a flux through a pathway is obtained from the end products (Fernie et al., 2005), which in this case would accumulate or deplete depending on the extent of GA20ox versus GA3ox activity. The enzymatic reaction from GA19 to GA20 was reported to be very slow (He et al., 2019) therefore, also GA19 was investigated.
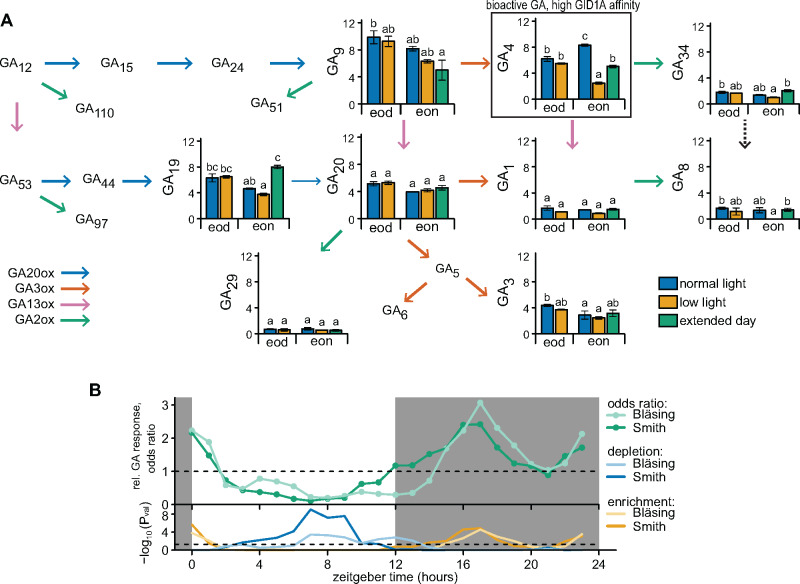
For all three measured precursors (GA9, GA19, and GA20), the differences between the treatments and time of day was very minor. Though levels tended to be higher at the end of the day, and reduced after the night of a LL day, no significant differences were observed (Figure 4A). An extended day, which reduces nighttime GA3ox1 expression, did result in elevated GA19 levels (Figure 4A). The bioactive GAs differ in their affinity for the receptor GID1, with GA3 and GA1 having a weak affinity compared to strong binding affinity of GA4 (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Yoshida et al., 2018). The low-affinity bioactive GAs (GA1 and GA3) had no clear differences between treatments (Figure 4A). The high-affinity GA, GA4, was the most abundant bioactive GA, and its levels were highest at the end of the night (Figure 4A). Reducing nighttime GA3ox1 induction by an extended day (Figure 3C) led to a subsequent drop in GA4 the following morning (Figure 4A). LL levels led to an even stronger drop in GA4 the following morning, whereas GA4 levels remained unchanged at the end of the actual LL day (Figure 4A). These results suggest an important role of nighttime GA3ox1 expression in determining bioactive GA levels in response to LL, whereas the drop of KS and GA20ox1 was not translated into reduced GA biosynthesis within the time frame studied here. GA inactivating enzymes could also play a role in determining the dynamics of bioactive GA. However, no consistent changes of inactivated GAs (GA8, GA29, and GA34) were found (Figure 4A).
Figure 4.
GA abundance and responses over time and after LL conditions. (A) Abundance of GA metabolites, directly following a control (NL) or LL day (end of day, ZT12) and after the following subsequent night (end of night, ZT24). Extended day (under control light conditions) represents a 3-h delay in the start of the night as in Figure 2C. Data are means ± SE and are shown as ng g−1 fresh weight (FW). Letters indicate significant differences (P < 0.05, Tukey HSD, N = 3). The narrow arrow from GA19 to GA20 represents the slow reaction rate and the dashed line represents a predicted, but unconfirmed, reaction in planta (He et al., 2019, 2020). (B) The extent of association or dissociation between a GA signature and time of day-specific expression. The odds ratio is defined as the odds of a gene being GA responsive among a set of time of day-specific genes divided by the odds of not being GA responsive among the genes that are not specific to that time of the day. Therefore, relative enrichment (odd ratio >1) or depletion (odd ratio <1) for each time point indicates more or less GA-responsive genes, respectively. Gene sets for each time of day were obtained from previous studies (Smith et al., 2004; Bläsing et al., 2005; Mockler et al., 2007). GA-responsive genes were based on GA-treated rosettes (Ribeiro et al., 2012a, 2012b). Below is the statistical significance of depletion or enrichment of GA-responsive genes (Hypergeometric distribution). Dashed line indicates P = 0.05 (Fisher’s exact test).
Lower and higher levels of GA4 at the end of the day and night, respectively, suggest temporal dynamics in GA abundance. These direct GA measurements are supported by in silico investigation of genes responsive to GA in Arabidopsis rosettes (Ribeiro et al., 2012a, 2012b). For two independent datasets (Smith et al., 2004; Bläsing et al., 2005), the GA-responsive genes were enriched among gene sets that peaked in expression during the night, but underrepresented (depleted) among daytime peaking genes (Figure 4B and Supplemental Figure 4 and Supplemental Table S1). These dynamics of GA-responsive genes could also be ascribed to changes in GA sensitivity, as also observed in seedling hypocotyls (Arana et al., 2011). Indeed, transcript levels of GID1A seem to be highest during the evening and start of the night in rosettes (Supplemental Figure 1). The starchless mutant phosphoglucomutase (pgm) typically suffers from severe starvation at night (Caspar et al., 1985; Bläsing et al., 2005). Subsequently, the nighttime induction of GA3ox1 was abolished, whereas the daytime GA biosynthetic genes (KS and GA20ox1) remained mostly unaffected (Supplemental Figure 5A). Indeed, pgm and other starch mutants are susceptible to altered GA metabolism and reduced GA levels (Paparelli et al., 2013). Subsequently, we found that rhythmic expression of GA-responsive genes was lost in pgm, despite retaining rhythmicity in GID1A (Supplemental Figure 5B).
The relevance of GAs in time of day-specific leaf expansion was further investigated in the gibberellic acid insensitive (gai) and della pentuple (pent) mutants, which cannot respond to varying GA levels (Pullen et al., 2019). gai plants had smaller rosettes and reduced rates of lamina length increase. Pent plants had larger rosette sizes, but no difference in lamina size increase rate could be detected (Figure 5, A and B). The relative contribution to leaf expansion measured as lamina length increase, of the morning, afternoon, and night was determined (Figure 5C). Leaf expansion of gai and pent was decreased at night (Figure 5C) when the expression of GA3ox1 determines the bioactive GA levels (Figures 3 and 4B). The gai plants had the strongest drop in the nighttime expansion, and also had elevated expansion rates in the morning (Figure 5C), which may represent a GA-independent behavior that might utilize untapped nighttime growth potential. Likewise, a GA3ox1-knockout mutant, ga3ox1-3 (Mitchum et al., 2006), also has reduced nighttime expansion and elevated morning expansion (Figure 5C). Another measure of changes in rhythmicity, the largest differences in expansion found between the three time periods, was lower in the GA mutants (Figure 5D). Overall, these results suggest that an ability to respond to changes in GA levels and GA biosynthesis contributes to time of day-specific expansion.
Figure 5.
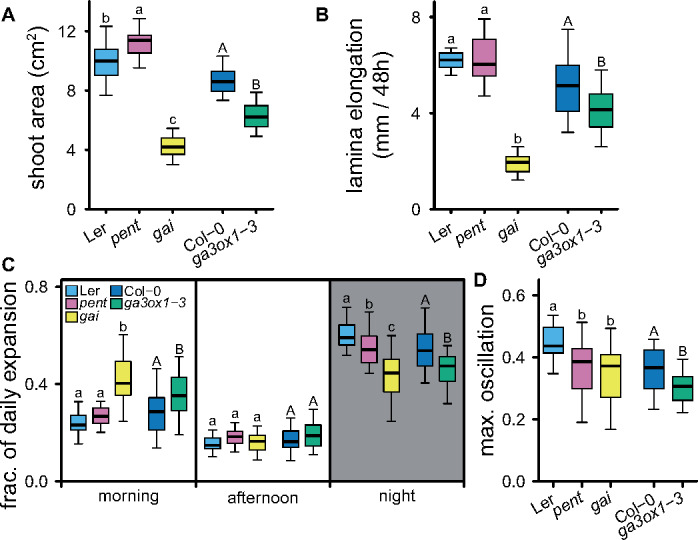
Daily changes in expansion. (A) Projected shoot area 3 weeks after transplanting. (B) Leaf expansion (lamina length increase) of leaf 7 in a 10-leaf rosette over 48 h. (C) Portion of the daily expansion occurring during the morning (ZT0–ZT6), afternoon (ZT6–ZT12), and night (ZT12–ZT24). The average of 2 d is shown. (D) The extent of growth rhythmicity expressed as the maximum difference in fraction of daily expansion between the three time periods investigated (morning, afternoon, and night). Letters indicate significant differences (P < 0.05, Tukey HSD, N = 25), lower and uppercase distinguish comparisons for the two different genetic backgrounds used. Pent = della pentuple mutant.
Reduced light availability leads to reduced leaf expansion rates only during the following night, which is partially mediated by GA3ox1
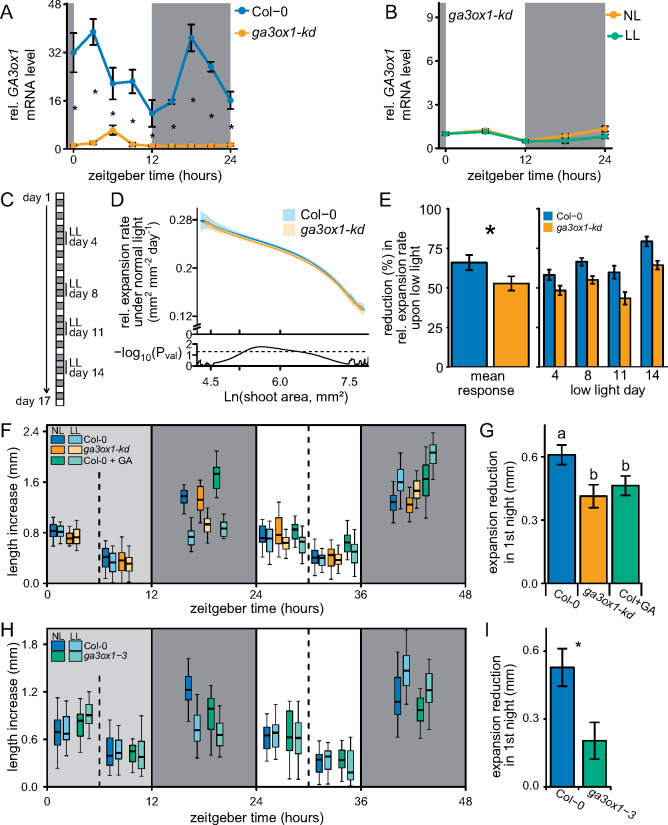
The next step was to investigate whether the carbon-dependent regulation of GA3ox1 transcription and subsequent GA4 levels are important to adjust growth to periodic LL levels. To effectively investigate growth reductions, it is important that a mutant only has minor growth effects under control conditions (termed normal light, NL); therefore, a ga3ox1 knock down (SALK_025076) with a T-DNA insertion in the intron was used. This ga3ox1-kd had more than a 10-fold drop in GA3ox1-kd mRNA abundance and less than half the GA4 levels (Figure 6A and Supplemental Figure 6, A–C). Furthermore, the low GA3ox1-kd mRNA abundance of the knock down was not significantly further reduced by a LL treatment (Figure 6B). The specific leaf area, an important growth parameter, was unaffected. However, ga3ox1-kd did flower slightly later and had reduced bolt length (Supplemental Figure 6D).
Figure 6.
Carbon and GA dependent growth under constant and variable conditions. (A) Transcript levels of GA3ox1 in Col-0 and the ga3ox1-kd line. Data are means ± SE (N = 4). (B) The behavior of GA3ox1 during LL and NL conditions in ga3ox1-kd. Data are as means ± SE (N = 5). (C) A brief schematic of the measuring and treatment regime. (D) Expansion rate during constant high-light conditions for a given projected shoot area. The shaded areas indicate the 95% confidence interval of the mean. The statistical difference between genotypes (Student’s t test) is given below (N = 40). Dashed line indicates P = 0.05. Based on control plants (NL) not confronted by LL days. (E) The reduction in relative expansion rate during a LL day and its subsequent night compared to the expansion rate of the previous days. The reduction for each individual LL day, and the mean LL response over all 4 d are shown. Asterisks indicate the significant difference (ANOVA). Data are means ± SE (N = 40). (F) Lamina length increase (leaf 7) over measured time intervals (6 or 12 h), during and following a LL day. Dark and light gray boxes indicate the night and the LL treatment, respectively. GA4 + 7/mock was applied at ZT12. N = 25, 10-leaf stage plants. (G) Reduction in expansion compared to NL plants specifically at the night following a LL treatment (ZT12–ZT24). Data are means ± SE (N = 25). LL Col+GA growth reduction is compared to Col-0 NL + mock. Letters indicate significant differences (P < 0.05, Tukey HSD). (H, I) Experiment identical to F and G, but with Col-0 and the full knock out ga3ox1-3. Means ± SE (N = 25) are shown, asterisk indicates statistical significance (P < 0.05, Tukey HSD). NL: control normal-light conditions, LL: low-light conditions.
With the automated phenotyping platform Weighing, Imaging, and Watering Automated Machine (WIWAM) XY to precisely control soil water content and to image individual plants over time (Clauw et al., 2015; Dubois et al., 2017), we followed the expansion rates of Col-0 and ga3ox1-kd plants under constant day/night cycles and when confronted with four interspersed LL days (Figure 6C). To obtain expansion rates under NL and to determine LL-mediated reductions in expansion we fitted growth curves to individual plants (Supplemental Figure 7). Under control conditions, expansion rates were close to identical between Col-0 and ga3ox1-kd (Figure 6D). We concluded that GA3ox1-dependent fluctuations in GA levels could not be the main driver of growth in ga3ox1 under NL conditions. Exposure to the four LL days led to strongly reduce final rosette area in both genotypes (Supplemental Figure 7), implying that a few LL days can have dramatic effects. Quantification of the reduction in expansion during the LL day and subsequent night showed that ga3ox1-kd had a smaller reduction in expansion rate than Col-0 (Figure 6E). This suggests that expansion in ga3ox1-kd, in which the nighttime expression and regulation of GA3ox1 is negligible (Figure 6, A and B), does not suffer as strongly from LL as the wild-type, where LL severely dampens the expression of GA3ox1 (Figure 3D) and GA4 levels (Figure 4B).
To investigate the exact time of the day at which the growth reduction takes place and to further explore the role of GA3ox1 in such reductions, leaf expansion rates of the fastest growing leaf (seventh leaf; Supplemental Figure 8) were followed over 6- or 12-h intervals. During the LL treatment, leaf expansion rates remained unchanged, whereas in the subsequent night, expansion was severely reduced (Figure 6F). Preventing a drop in GA4 after LL by either GA application or the ga3ox1-kd, mitigated the reduction in expansion to a similar degree (Figure 6G). However, neither manipulation of GAs could completely abolish the growth penalty (Figure 6G). Similarly, for the full knock out of GA3ox1, ga3ox1-3, LL-induced reduction of nighttime leaf expansion was mitigated (Figure 6, H and I). LL levels also led to altered carbon dynamics in the subsequent day. The following day, less carbon is allocated to structural biomass and subsequently soluble carbohydrates (including starch) accumulate to higher levels (Moraes et al., 2019), and this accumulation of the subsequent day was independent of GAs (Supplemental Figure 9, A and B). The higher carbon availability coincided with enhanced growth during the second night after the LL treatment, especially in Col-0 where the LL-induced expansion was of a similar magnitude as the expansion induced solely by exogenous GAs. ga3ox1-kd seemed not able to benefit from this growth increase to the same extent (Supplemental Figure 9C).
Discussion
A capacity to adjust hormonal profiles to prevailing environmental conditions is essential for optimal plant performance. For this reason, we aimed to identify how GA metabolism was affected by carbon availability and the subsequent effects on growth. Nighttime production of GA4 by GA3ox1 was found to be highly sensitive to LL levels, which led to a reduction of GA4 levels only in the subsequent night and not the actual day. Similarly, effects on leaf expansion upon LL were only noticeable during the night and were mitigated in plants whose growth was not driven by nighttime expression of GA3ox1. The results presented here provide new insights into the regulation of GA metabolism and add perspective on the mechanisms of growth control.
We found that reducing GA3ox1 mRNA levels, either through an extended day, LL availability, or a genetic knockdown, consistently led to reduced GA4 abundance, but did not affect the levels of the low-affinity bioactive GAs (GA3 and GA1; Figure 4). Indeed, in vitro activity of GA3ox1 was shown to have a strong preference for converting precursors to GA4 (Williams et al., 1998). Full knockouts of GA3ox1 were also reported to have reduced GA4 levels (Mitchum et al., 2006; Hu et al., 2008). GA3 and GA1 are clearly not affected by the environmental perturbations and represent a set of background GAs. Their affinity for the GID1A receptor is low compared to GA4 (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006; Yoshida et al., 2018) and their constitutive presence could provide a weak baseline GA response. GA3 has rarely been reported in Arabidopsis and could possibly be subject to a developmental gradient (Derkx et al., 1994; Eriksson et al., 2006; Fambrini et al., 2015). Furthermore, inactivation pathways for GA3 are so far unknown, which suggest it could linger in the plant for long periods after biosynthesis.
In contrast to GA3ox1, clearly reduced mRNA levels of upstream biosynthesis genes (KS and GA20ox1; Figure 2) did not lead to a corresponding reduction of precursor end-products (Figure 4). GA20ox is generally considered the rate-limiting step, since constitutive overexpression leads to higher GA4 levels, in contrast to overexpression of other biosynthesis genes (Yamaguchi, 2008). Longevity of prior produced enzymes could explain a lack of responses at the metabolite level observed after the short transcriptional perturbation of this study. Likely, successive days of reduced light availability would eventually also lead to an effect on precursor metabolism. However, GA4 levels were quick to respond to changes in GA3ox1, suggesting that GA3ox1 proteins need to be resynthesized daily. This implies that GA metabolism is reset every day via GA3ox1. The results presented here suggest that the abundance of GA3ox1 and GA20ox1 vary throughout the day–night cycle, and subsequently also the flux through each of these pathways.
It is tempting to attribute the low afternoon-growth and high night- and morning-growth rates (Figures 5, C and 6, F) with the corresponding changes in GA4 levels and behavior of GA4-responsive genes (Figure 4). GA sensitivity was shown to be regulated by the circadian clock in hypocotyls (Arana et al., 2011), here we show that also the actual levels of GA4 vary throughout the day and probably coincide with high GA sensitivity (Figure 4 and Supplemental Figure 1). Varying expansion rates of leaves throughout the day–night cycle are also regulated by carbon availability, water availability, light cues, and the circadian clock (Nozue and Maloof, 2006; Pantin et al., 2011; Farré, 2012; Dornbusch et al., 2014; Apelt et al., 2017; Dubois et al., 2017). It is not straight forward to piece apart the contributions of these growth cues and discern how they act in concert. Nevertheless, the results presented here show that light and dark cues, and the clock ensure timing of GA biosynthesis and that disruption of biosynthesis patterns in the starchless pgm abolishes the rhythmic nature of downstream GA responses. Moreover, removing the ability to respond to varying GA levels or disrupting biosynthesis impacts growth dynamics corresponding to the rhythmicity in GA biosynthesis.
The nighttime induction of GA3ox1 is highly dependent on carbon availability (Figure 3D). In plants without the capacity of GA3ox1 regulation, the consistently reduced nighttime growth penalty upon LL, either for whole rosettes or a single leaf, demonstrates the relevance of this gene and subsequent GAs in adapting growth to the prevailing conditions (Figure 6). Although the difference in the LL-induced growth penalty between Col-0 and ga3ox1-kd is seemingly small, around 20%–30%, it is of a similar magnitude as a rescue by GAs (Figure 6, E and G). A variety of growth-stimulating gene regulatory networks has been identified in Arabidopsis and many of these acts independent of one another (Gonzalez et al., 2010; Vanhaeren et al., 2014). Likewise, the different plant hormones have surprisingly little overlap in downstream genes and appear to operate in distinct fashions (Nemhauser et al., 2006). Alternative mechanisms that connect carbon and growth networks likely exist in plants (Ljung et al., 2015). One is the starvation-induced autophagy of brassinosteroid signaling components (Zhang et al., 2016; Nolan et al., 2017). A collection of numerous such growth-regulating mechanisms will buffer against the loss of the single mechanism identified here, which could explain the small effect size. Similarly, boosting carbon assimilation by elevated CO2 can still enhance growth in paclobutrazol-treated plants (Ribeiro et al., 2012a, 2012b). In this study, plants were confronted by fewer photo-assimilates, which places physical constraints on growth regardless of the manipulation of growth signaling mechanisms.
GAs are crucial hormones for plants, which stimulate growth and development. This study reveals how GA metabolism is balanced to match the physiology of the plant. We present a model (Figure 7) of GA biosynthesis that is coordinated throughout the day–night cycle by light cues, dark cues, and the circadian clock. Here, the nighttime GA4 biosynthesis rate is reset daily and strongly depends on carbon availability. Starch, as the nighttime carbon source (Graf et al., 2010), plays a pivotal role in this model and acts as a robust integrator of daytime performance, unlike the highly variable nature of photosynthesis and light availability. Using starch, rather than photosynthesis directly, could prevent excessive starting and stopping of growth. Though the exact mechanism is unclear, plants can precisely estimate starch levels to ensure proper nighttime utilization rates (Graf et al., 2010; Scialdone et al., 2013). Subsequently, starch utilization rates can be accurately used to pace GA metabolism and aid in adjusting GA responses, such as growth, to the prevailing conditions when plants grow in variable environments.
Figure 7.
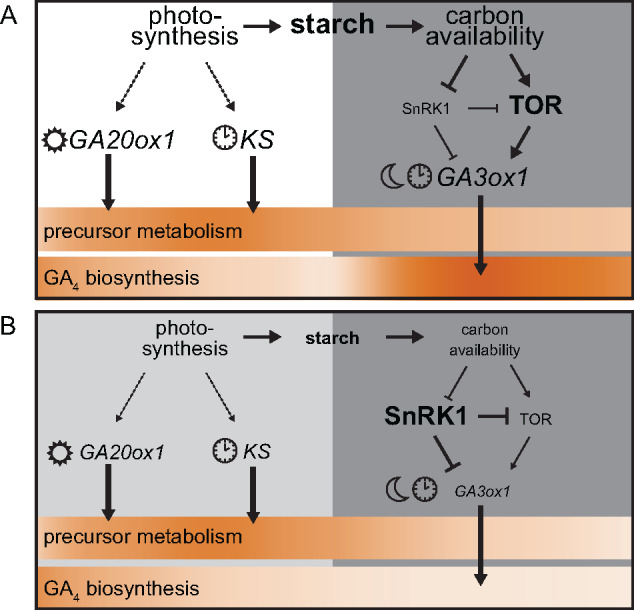
Model describing GA and associated leaf expansion dynamics. A schematic representation of the day and subsequent night under normal conditions (A) and when confronted with reduced photosynthetic capacity (B). White, light gray, and dark gray boxes represent the day, reduced‐light day, and night, respectively. Size difference between A and B indicates dominant processes in either condition. The heatbar indicates the strength (dark) of the process over the 24‐h cycle. Under normal conditions, a portion of photosynthesis is stored as starch to be used as a carbon source during the night. Photosynthesis is required for full daytime induction of GA20ox1 and KS. These are also regulated by a transition to light or the clock (Figure 2). Despite strong rhythmic expression, this is only translated into slight temporal effects of precursor metabolism (Figure 4A). GA3ox1 is induced at night through a transition to dark and gating by the clock. Moreover, induction is highly sensitive to starvation mediated through SnRK1 and TOR kinase activity (Figure 3). As a result of temporal and carbohydrate regulation of GA3ox1, GA4 biosynthesis occurs predominately at night (Figure 4). This corresponds with the behavior of downstream GA signaling genes and changes in leaf expansion dynamics of GA mutants (Figures 4B, 5). When confronted with LL, KS and GA20ox1 expressions are reduced. However, precursor metabolites remain unaffected (Figure 4A). The night following a LL day leads to a strong starvation response as little starch was accumulated. This leads to a shift from TOR kinase activity to SnRK1 activity, suppressing GA3ox1 induction (Figure 3). Subsequently, GA4 biosynthesis is rapidly and dramatically reduced (Figure 4A). Penalties on nighttime leaf expansion by a preceding LL day are thus reduced in genotypes that do not have a peak (normal) and drop (low light) in nighttime GA3ox1 (Figure 6).
Materials and methods
Plant material, growth, and treatments
Col-0, ga3ox1-kd (NASC: N670439, SALK_025076), and ga3ox1-3 (NASC: N6943, SALK_004521) were grown on a perlite soil mixture. Light levels used for the experiments ranged from 100 to 130 µmol m−2 s−1 photosynthetic active radiation (PAR) and 50%–70% relative humidity and a temperature of 20°C. LL treatments were 30–45 µmol m−2 s−1 PAR. DCMU (100 µM) and GA4 + 7 (100 µM) were applied by spraying on the plants. Experiments were started once the plants had 11 visible leaves.
Gene expression and carbohydrate analyses
For gene expression, two entire rosettes were pooled per replicate. RNA isolation was performed according to Kiefer method (Kiefer et al., 2000). DNA contaminant was removed with RQ1 RNase-Free DNase™ (Promega) and cDNA was synthesized using Maxima First Strand cDNA Synthesis Kit™ (Thermofisher). The cDNA was used as template for real-time PCR (primers in Supplemental Table 2) using CFV384 Touch™ Real-Time PCR Detection System (Bio-Rad). Housekeeping genes (AT4G34270, AT1G13320) were selected based on Czechowski et al. (2005), with an extra focus on constant expression levels during the day–night cycle. Sugars and starch were determined through coupled enzymatic reactions and NADH absorbance at 340 nm as described previously (Paparelli et al., 2013).
GA metabolites
GA levels were determined as previously described in Mariotti et al. (2011) with some modifications. In short, 2–4 g of pooled shoot was homogenized in cold 80% (v/v) methanol (1:5, w/v) using a mortar and pestle. Fifty nanograms of deuterated GAs ([17,17-2H2]-GA9, [17,17-2H2]-GA4, [17,17-2H2]-GA34, [17,17-2H2]-GA19, [17,17-2H2]-GA20, [17,17-2H2]-GA29, [17,17-2H2]-GA1, [17,17-2H2]-GA8, [17,17-2H2]-GA3) were added as internal standards to account for purification losses. Methanol was evaporated under a vacuum at 35°C, and the aqueous phase was partitioned against ethyl acetate, after adjusting the pH to 2.8. The extracts were dried and suspended in 0.3–0.5 mL of distilled water with 0.01% acetic acid [w/v] and 10% methanol [v/v]. High-performance liquid chromatography (HPLC) analysis was performed with a Kontron instrument (Munich, Germany) equipped with a UV absorbance detector operating at 214 nm. The samples were applied before to a 150 × 4.6-mm ID column packed with ODS Hypersil (Thermo Fischer Scientific, Milan Italy), particle size 5 μm, eluted at a flow rate of 1 mL min−1. The column was held constant at 10% methanol for 4 min, followed by a double gradient elution from 10% to 30% and 30% to 100% [v/v] over 40 min. The GAs were collected in four fractions: fraction n°1: GA8 and GA29; fraction n°2: GA1 and GA3; fraction n°3: GA20, GA19, GA34; fraction n°4: GA9 and GA4. Subsequently, the fractions were applied to 250 × 4.6-mm ID, Nucleosil 100-5 N(CH3)2 column (Macherey-Nagel GmbH & Co, Düren, Germany) and eluted isocratically with 100% methanol [v/v] containing 0.01% acetic acid [w/v] at a flow rate of 1 ml min−1. The fractions corresponding to the elution volumes of standard GAs were collected, dried and silylated with N,O bis (trimethylsilyl) trifluoroacetamide containing 1% trimethylchlorosilane [w/v] (Pierce, Rockford, IL, USA) at 70°C for 1 h. The gas chromatography–tandem mass spectrometry (GC-MS) analysis was performed on a Saturn 2200 quadruple ion trap mass spectrometer coupled to a CP-3800 gas chromatograph (Varian Analytical Instruments) equipped with a MEGA (http://www.mega.mi.it) 1MS capillary column (30 m X 0.25-mm i.d. and 0.25-µm film thickness). The carrier gas was helium, which was dried and air free, with a linear speed of 60 cm s−1. The oven temperature for fraction n°1 was maintained at 120°C for 2 min and increased to 280°C at a rate of 20°C min−1, and then increased to 300°C at a rate of 5°C min−1. The oven temperature for fraction n°2 was maintained at 120°C for 2 min and increased to 255°C at a rate of 30°C min−1, and then increased to 260°C at a rate of 1°C min−1 and finally arrived at 300°C at a rate of 30°C min−1. The oven temperature for fraction n° 3 and 4 was maintained at 120°C for 2 min and increased to 240°C at a rate of 30°C in−1, and increased to 250°C at a rate of 1°C min−1 and finally arrived at 300°C at a rate of 30°C min−1. Injector and transfer line were set at 250°C and the ion source temperature at 200°C. Full scan mass spectra were obtained in EI+ mode with an emission current of 10 µA and an axial modulation of 4 V. Data acquisition was carried out in SIM (Selected Ion Monitoring) mode from 150 to 650 Da at a speed of 1.4 scan s−1. GAs were identified by comparing the full mass spectra with those of the authentic compounds (Supplemental Table S3) and those reported by Gaskin and MacMillan (1991), and quantification with reference to standard plots of concentration ratios versus ion ratios that were obtained by analyzing known mixtures of unlabeled and labeled GAs.
Growth measurements
The WIWAM (www.wiwam.com) to precisely control soil water content and image plants was used to follow size increase of individuals over time. The PSB (Plant Systems Biology) Interface for Plant Phenotype Analysis (https://pippa.psb.ugent.be) was used for analysis, visualization, and management of phenotypic datasets and images. A four-parameter logistic model (constant conditions) or a series of exponential models (LL treatment) were fitted to individual plants. This allowed us to track growth rates over time and rosette area. Contributions of distinct leaves to total size increase were determined by two destructive harvests, separated by 24 h. Leaves were dissected and arranged on 0.59% agarose-filled plates, leaf incisions ensured proper flattening of the leaves. Areas were determined with ImageJ software on pictures obtained by scanning the plates. Growth of individual leaves over time was determined, nondestructively, by manual length measurements with a digital calliper.
In silico analysis and statistics
Data analysis and statistics were done using R software. Data were transformed if needed to satisfy the assumptions of statistical tests. Calling of rhythmic genes in pgm was done by the HAYSTACK tool from the DIURNAL project (Mockler et al., 2007).
Accession Numbers
IDs of the genes studied here can be found in Supplemental Table 2 and Supplemental Figure 1.
Supplemental Data
Supplemental Figure S1. mRNA abundance over the day–night cycle of GA-biosynthetic and -signaling genes.
Supplemental Figure S2. The effect of treatments on the clock, sugar dynamics, and starvation responses.
Supplemental Figure S3. The relevance of sugar/energy signaling pathways on GA3ox1 expression.
Supplemental Figure S4. Rhythmic expression of gene sets tested for GA enrichment.
Supplemental Figure S5. GA biosynthesis gene behavior and GA-responsive genes in pgm.
Supplemental Figure S6. Characterization of ga3ox1-kd.
Supplemental Figure S7. Carbon- and GA-dependent growth under constant and variable conditions.
Supplemental Figure S8. Contribution of different leaves to total rosette growth.
Supplemental Figure S9. Non-structural carbohydrates during and after a LL day.
Supplemental Table S1. Number of GA-induced genes in and out of the phase of rhythmically regulated genes.
Supplemental Table S2. Primers used for RT-qPCR.
Supplemental Table S3. GAs identified by full-scan GC-MS of their TMS ester TMS ether or TMS ether derivatives in extracts from A. thaliana.
Funding
This work was supported by the Scuola Superiore Sant’ Anna (SSSA); the Plantecophysiology group (Utrecht University); Agrobiodiversity (SSSA); CISUP at University of Pisa; Research Foundation Flanders (FWO); the Dutch scientific organization (grant no. ALWOP.419); Horizon2020 Programme of the EU (EPPN2020 Grant Agreement 731013).
Conflict of interest statement. None declared.
Supplementary Material
P.Pe and H.v.V conceived the project; P.Pr, S.D., D.I. P.Pe, and H.v.V designed experiments; P.Pr, L.M., M.C.V., G.N., and H.v.V performed experiments; P.Pr, S.D., and H.v.V analysed the data; H.v.V. and P.Pe wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys) is Pierdomenico Perata (p.perata@santannapisa.it) and Hans van Veen (h.vanveen@uu.nl).
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GTS, Genschik P (2009) Gibberellin signaling controls cell proliferation rate in arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Apelt F, Breuer D, Olas JJ, Annunziata MG, Flis A, Nikoloski Z, Kragler F, Stitt M (2017) Circadian, carbon, and light control of expansion growth and leaf movement. Plant Physiol 174: 1949–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville C (1985) Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana (L.) deficient in chloroplast phosphoglucomutase activity. Plant Physiol 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw P, Coppens F, De Beuf K, Dhondt S, Van Daele T, Maleux K, Storme V, Clement L, Gonzalez N, Inzé D (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol 167: 800–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation EMBO Reports 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkx MP, Vermeer E, Karssen CM (1994) Gibberellins in seeds of Arabidopsis thaliana: biological activities, identification and effects of light and chilling on endogenous levels. Plant Growth Regul 15: 223–234 [Google Scholar]
- Dornbusch T, Michaud O, Xenarios I, Fankhauser C (2014) Differentially phased leaf growth and movements in Arabidopsis depend on coordinated circadian and light regulation. Plant Cell 26: 3911–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Claeys H, Van den Broeck L, Inzé D (2017) Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant, Cell Environ 40: 180–189 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M, Mariotti L, Parlanti S, Salvini M, Pugliesi C (2015) A GRAS‐like gene of sunflower (Helianthus annuus L.) alters the gibberellin content and axillary meristem outgrowth in transgenic Arabidopsis plants. Plant Biol 17: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Farré EM (2012) The regulation of plant growth by the circadian clock. Plant Biol 14: 401–410 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Geigenberger P, Stitt M (2005) Flux an important, but neglected, component of functional genomics. Curr Opin Pant Biol 8: 174–182. [DOI] [PubMed] [Google Scholar]
- Gaskin P, MacMillan J (1991) GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra, Cantock’s Enterprises, Bristol. [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: Different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM (2010) Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. PNAS 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Chen Q, Xin P, Yuan J, Ma Y, Wang X, Xu M, Chu J, Peters RJ, Wang G (2019) CYP72A enzymes catalyse 13-hydrolyzation of gibberellins. Nat Plants 5: 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Xin P, Ma X, Chu J, Wang G (2020) Gibberellin metabolism in flowering plants: An update and perspectives. Front Plant Sci 11: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai WC, Hanada A, Alonso JM, Ecker JR, et al. (2008) Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20: 320–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikakov A, Flis A, Apelt F, Fünfgeld M, Scherer U, Stitt M, Kragler F, Vissenberg K, Persson S, Suslov D (2017) Cellulose synthesis and cell expansion are regulated by different mechanisms in growing Arabidopsis hypocotyls. Plant Cell 29: 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer E, Heller W, Ernst D (2000) A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol Biol Rep 18: 33–39 [Google Scholar]
- Ljung K, Nemhauser JL, Perata P (2015) New mechanistic links between sugar and hormone signalling networks. Curr Opin Plant Biol 25: 130–137 [DOI] [PubMed] [Google Scholar]
- Mariotti L, Picciarelli P, Lombardi L, Ceccarelli N (2011) Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. J Plant Growth Regul 30: 405–415 [Google Scholar]
- Mengin V, Pyl E-T, Moraes TA, Sulpice R, Krohn N, Encke B, Stitt M (2017) Photosynthate partitioning to starch in Arabidopsis thaliana is insensitive to light intensity but sensitive to photoperiod due to a restriction on growth in the light in short photoperiods. Plant Cell Environ 40: 2608–2627 [DOI] [PubMed] [Google Scholar]
- Middleton AM, Úbeda-Tomás S, Griffiths J, Holman T, Hedden P, Thomas SG, Phillips AL, Holdsworth MJ, Bennett MJ, King JR, et al. (2012) Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. PNAS 109: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3‐oxidases in Arabidopsis development. Plant J 45: 804–818 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harbor Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Moraes TA, Mengin V, Annunziata MG, Encke B, Krohn N, Hoehne M, Stitt M (2019) Response of the circadian clock and diel starch turnover to one day of low light or low CO2. Plant Physiol 179: 1457–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi‐Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M,. et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Nam YJ, Herman D, Blomme J, Chae E, Kojima M, Coppens F, Storme V, Van Daele T, Dhondt S, Sakakibara H, et al. (2017) Natural variation of molecular and morphological gibberellin responses. Plant Physiol 173: 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Gonzalez N, Inzé D (2016) Leaf growth in dicots and monocots: so different yet so alike. Curr Opin Plant Biol 33: 72–76 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Fangxin H, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y (2017) Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell 41: 33–46.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Maloof JN (2006) Diurnal regulation of plant growth. Plant Cell Environ 9: 396–408 [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14: S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MCM, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al. (2013) Diurnal changes of polysome loading track sucrose content in the rosette of wild-type arabidopsis and the starchless pgm Mutant Plant Physiol 162: 1246–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantin F, Simonneau T, Rolland G, Dauzat M, Muller B (2011) Control of leaf expansion: a developmental switch from metabolics to hydraulics. Plant Physiol 156: 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparelli E, Parlanti S, Gonzali S, Novi G, Mariotti L, Ceccarelli N, van Dongen JT, Kölling K, Zeeman SC, Perata P (2013) Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25: 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen N, Zhang N, Alonso AD, Penfield S (2019) Growth rate regulation is associated with developmental modification of source efficiency. Nat Plants 5: 148–152 [DOI] [PubMed] [Google Scholar]
- Ribeiro DM, Araújo WL, Fernie AR, Schippers JHM, Mueller-Roeber B (2012a) Translatome and metabolome effects triggered by gibberellins during rosette growth in Arabidopsis. J Exp Bot 63: 2769–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro DM, Araújo WL, Fernie AR, Schippers JHM, Mueller-Roeber B (2012b) Action of gibberellins on growth and metabolism of arabidopsis plants associated with high concentration of carbon dioxide. Plant Physiol 160: 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Mugford ST, Feike D, Skeffington A, Borrill P, Graf A, Smith AM, Howard M (2013) Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife 2: e00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Alexander AF, Ivakov A, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, Stitt M (2014) Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant 7: 137–155 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YIC, Kitano H, Yamaguchi II, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaeren H, Gonzalez N, Coppens F, De Milde L, Van Daele T, Vermeersch M, Eloy NB, Storme V, Inzé D (2014) Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. eLife 3: e02252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Phillips AL, Gaskin P, Hedden P (1998) Function and substrate specificity of the gibberellin 3β-hydroxylase encoded by the arabidopsis GA4 gene. Plant Physiol 117: 559–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Tanimoto E, Hirai T, Miyanoiri Y, Mitani R, Kawamura M, Takeda M, Takehara S, Hirano K, Kainosho M, et al. (2018) Evolution and diversification of the plant gibberellin receptor GID1. Proc Natl Acad Sci USA 115: E7844–E7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu J-Y, Roh J, Marchive C, Kim S-K, Meyer C, Sun Y, Wang W, Wang Z-Y (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in arabidopsis. Current Biol 26: 1854–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.