Cardiac-derived exosomes have received intense interest for their roles in paracrine communications and regenerative therapies. However, current understanding of how exosomes mediate cellular signalling is incomplete, in part because the contents of exosomes from different cardiac cell types are poorly defined. To learn what signals cardiac cells release, we examined the microRNA (miRNA) compositions secreted in exosomes from human induced pluripotent stem cells (iPSCs) and 3 major iPSC-derived cardiac cell types.
With approval by the Stanford University Institutional Review committee and informed consent, human iPSC lines from two healthy donors were reprogrammed in the Stanford Cardiovascular Institute Biorepository, and differentiated into cardiomyocytes (iPSC-CMs), endothelial cells (iPSC-ECs), and cardiac fibroblasts (iPSC-CFs) using established protocols1. Low yield has been a bottleneck in elucidating the composition and function of extracellular exosomes. To boost isolation yields, we applied a mechanical-sorting device (ExoTIC) that isolates exosomes from small volumes of culture medium2, 3. Briefly, the device is an engineered fluidic system that delivers culture medium through nanoporous membranes to selectively capture vesicles at 50–200 nm in diameter. We validated the extracted exosomes using nanoparticle tracking analysis and immunoblots, and confirmed superior yield over conventional precipitation methods (Fig. 1A). We then isolated total RNA (≥18 nt) from exosomes of 2 biological replicate lines for library generation and small RNA sequencing on an Illumina NextSeq platform. Sequencing reads were mapped to GRCh38 human reference genome after adaptor clipping and annotated against miRBase v.22.1 coordinates using conventional pipelines.
Figure 1. Distinct exosomal miRNAs secreted by iPSCs and differentiated cardiac cell types.
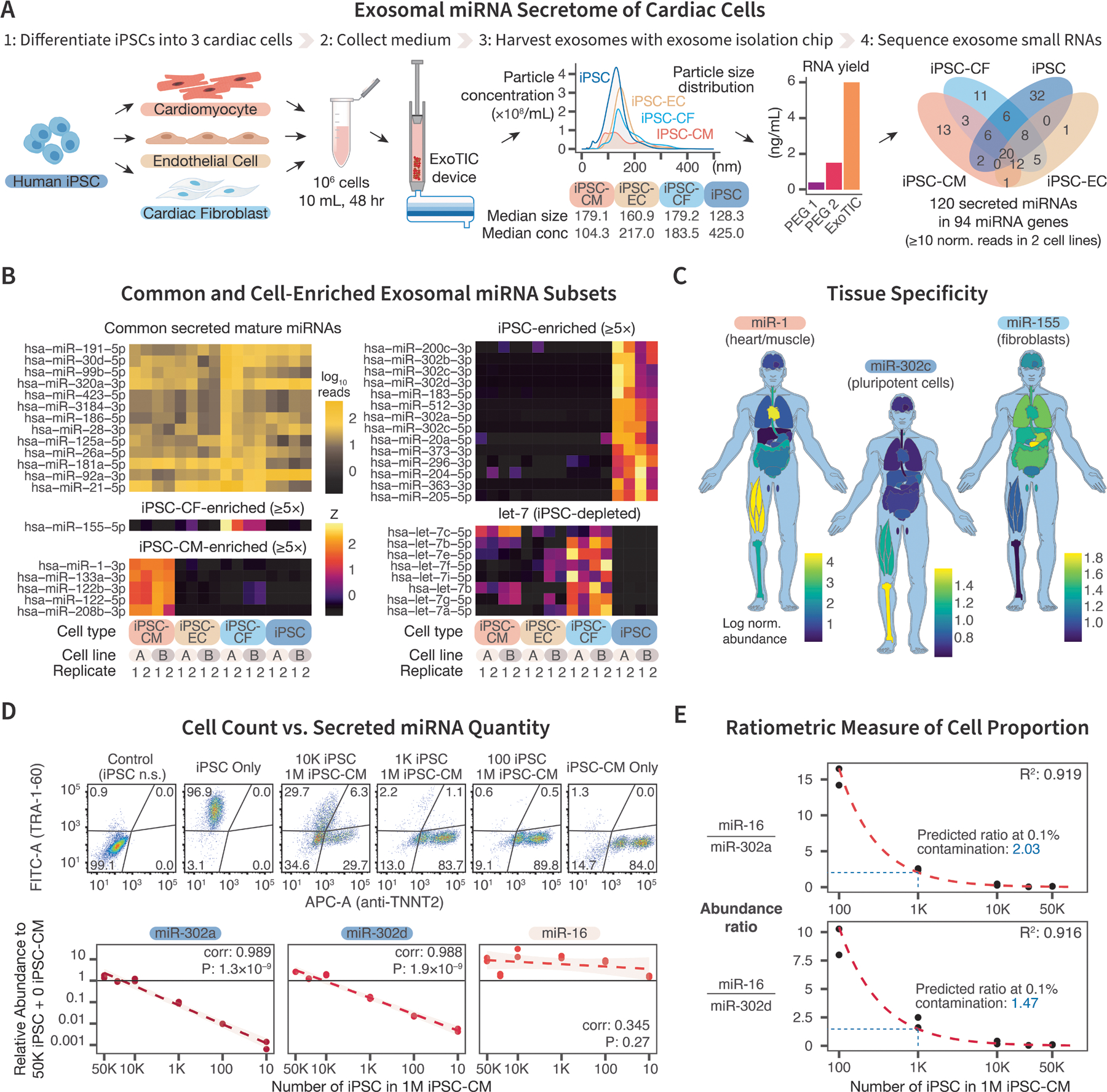
(A) Experimental schema. Human iPSC lines from 2 healthy donors were differentiated into cardiomyocytes (iPSC-CMs), endothelial cells (iPSC-ECs), and fibroblasts (iPSC-CFs), from which 10 mL of culture medium per ~1M cells were collected after 48 hr and processed for small RNA sequencing. An engineered device (ExoTIC) specifically captured extracellular vesicles 50–200 nm in diameter and improved RNA yield by ≥3-fold over conventional polyethylene glycol (PEG) precipitation. Line chart: distribution of particle sizes isolated from the culture medium of each cell type using nanoparticle tracking. Bar chart: RNA yield in ng/mL. (B) Small RNA sequencing uncovered 120 miRNAs secreted from the cultured cells, including miRNAs enriched from specific cell types. Heat map represents quantile-normalized read counts of commonly secreted miRNAs or standardized counts of iPSC-CF-, iPSC-CM-, or iPSC-enriched mature miRNAs (≥10 counts for both biological replicates, ≥5-fold enrichment over other tested cell types for enriched miRNAs; n=2 biological × 2 technical replicates). Let-7 family precursors, known to be miRNA markers of differentiated (i.e., non-pluripotent) cells, were depleted in iPSCs. (C) We selected a subset of enriched, secreted, mature miRNAs and compared their total tissue expression in 17 different tissues in a published human miRNA atlas and graphed the abundance of miR-1, miR-155, and miR-203c in the body. Data on other miRNAs are accessible on our web-app (https://bit.ly/heartsecretome). (D) Upper: Co-culture setup between iPSC-CMs and various numbers of iPSCs for 48 hours. Plating was validated using CM-specific (TNNT2) and iPSC-specific (TRA-1–60) antibodies in flow cytometry. Numbers: cell percentage in gated regions; n.s.: no staining. Lower: Secretion of miR-302a and miR-302d in the culture medium scaled with decreasing numbers of plated iPSCs (from 50,000 down to 10) co-cultured with iPSC-CMs over ≥2 orders of magnitude. Y-axis: relative abundance of miRNA compared to 5×105 iPSCs plated without iPSC-CMs. Secretion of miR-16 remained constant in these co-culture samples. (E) The ratiometric quantity between miR-16 and miR-302a or miR302d inversely reflects the ratio of iPSCs in iPSC-CMs co-culture (Top: miR-16/miR-302a, bottom: miR-16/miR-302d). R2: goodness-of-fit of linear model between log(abundance) vs. log(iPSC counts).
From the data, we identified 120 miRNAs (mature or stem loop) from 94 miRNA genes to be secreted at in the analyzed cell types (normalized read counts ≥10 in both lines). The exosomal miRNA profiles revealed that (i) let-7 miRNA precursors, which are suppressed by Lin28, are depleted in iPSC secretomes as expected; (ii) only a subset of total cellular miRNAs are secreted (e.g., both miR-155 and miR-143 are preferentially found in intracellular over exosomal pools in iPSCs); (iii) a common core of miRNAs is secreted by all three cardiac cell types (e.g., miR-320); and (iv) importantly, we found distinct enrichment or depletion of different mature miRNAs in each exosome type (Fig. 1B). For instance, we found miR-1, critical for cardiac development and pathology, is only secreted by iPSC-CMs in our data. Using a published human miRNA atlas4, we next compared total tissue expression of miR-1 in 17 different tissues, which showed miR-1 to be specific to striated muscles (Fig. 1C). Similarly, miR-302c, known to be specific to pluripotent cells, is enriched in iPSC-derived vesicles5 and also appears to be enriched in the bone, likely reflecting bone marrow hematopoietic stem cells4. Finally, miR-155 is primarily secreted in iPSC-CFs in our data and is diffusely expressed in the body in the miRNA atlas4.
Taken together, these data suggest that exosomal miRNAs reflect the biology of their cell type of origin, yet are also distinct from the intracellular total miRNA pools. Additionally, we also found several secreted cardiac miRNAs with still unclear roles in the cardiovascular system, including miR-423 and miR-125a, suggesting they may have function in intercellular communication. To facilitate data sharing, we created an interactive web-app that empowers users to perform exploratory analysis and download the relative abundance of all exosomal miRNAs discovered in our experiment, freely accessible at https://bit.ly/heartsecretome. Sequencing data are on GEO (GSE149290).
To explore the utility of this secretome map, we asked whether the detected miRNAs scale quantitatively with cell counts, and, if so, whether they can be utilized to assess the compositions of heterogeneous cell mixtures. To test this, we co-cultured decreasing numbers of undifferentiated iPSCs (102–105) in a mixture of iPSC-CMs (total 106 cells) over 48 hours. After verifying cell plating with flow cytometry, we found that the secretion of miR-302a and miR-302d into the culture medium decreased quantitatively and proportionally with diminishing numbers of plated iPSCs ≥2 orders of magnitude, with a detection limit between 10–100 cells (Fig. 1D). As the total cell counts in the co-cultures were comparable, we showed that a non-iPSC-enriched miRNA (miR-16) remained invariant to iPSC proportions. We next derived a ratiometric quantity between a constitutively expressed miRNA vs. iPSC-enriched miRNA (e.g., miR-16/miR-302a or miR-302d) and showed that this unitless measurement scaled inversely to the proportion of iPSCs within the assessed dynamic range (Fig. 1E). Hence, we propose that this exosomal miRNA ratio may provide a new, facile, and non-destructive readout of iPSC contaminant following differentiation, which would be useful for regenerative medicine, disease modelling, and drug screening.
In summary, we present an atlas of exosomal miRNAs from human iPSCs and three differentiated cardiac cell types. We found distinct secretory profiles from each cell type, supporting the importance of direct analysis of isolated exosomes. This resource may avail understanding of exosome biology and presents a potential method to monitor the purity of cardiac cell preparations.
Acknowledgements
We thank Dr. Aleksandra Leligdowicz for analysis of flow cytometry data.
Funding Sources
This project was funded in part by Stanford Chemistry, Engineering & Medicine for Human Health (ChEM-H) Seed Grant (M.C. & M.O.O.); Canary Center at Stanford for Cancer Early Detection Seed Grant (U.D.); American Heart Association 19CDA34680002 (J.-W.R) and 19CDA34760019 (C.L.); NIH Research Supplements to Promote Diversity in Health-Related Research R01 HL113006 (D.R.W) and R01 HL123968 (J.M.); NIH F32 HL139045 and K99 HL144829 (E.L.); and R01HL139664 (U.D.), R01 HL141371, R01 HL123968, R01 HL145676, and P01 HL141084 (J.C.W.).
Footnotes
Disclosures
J.C.W. is a co-founder of Khloris Biosciences. U.D. is a co-founder of DxNow Inc, Koek Biotech, Levitas, and Hillel. The work presented here is independent and managed in accordance with conflict of interest policies.
Bibliography
- 1.Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmström A, Matsa E, Zhang Y, Kumar A, Fan AC, del Álamo JC, Wu SM, Moslehi JJ, Mercola M and Wu JC. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Science Translational Medicine 2017;9:eaaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS and Demirci U. The Exosome Total Isolation Chip. ACS Nano 2017;11:10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan K, Shamskhou EA, Orcholski ME, Nathan A, Reddy S, Honda H, Mani V, Zeng Y, Ozen MO, Wang L, Demirci U, Tian W, Nicolls MR and de Jesus Perez VA. Loss of endothelium-derived Wnt5a is associated with reduced pericyte recruitment and small vessel loss in pulmonary arterial hypertension. Circulation 2019;139:1710–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E and Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016;44:3865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parr CJ, Katayama S, Miki K, Kuang Y, Yoshida Y, Morizane A, Takahashi J, Yamanaka S and Saito H. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep 2016;6:32532. [DOI] [PMC free article] [PubMed] [Google Scholar]


