Abstract
Regulation of translation via stop codon readthrough (SC-RT) expands not only tissue-specific but also viral proteomes in humans and, therefore, represents an important subject of study. Understanding this mechanism and all involved players is critical also from a point of view of prospective medical therapies of hereditary diseases caused by a premature termination codon. tRNAs were considered for a long time to be just passive players delivering amino acid residues according to the genetic code to ribosomes without any active regulatory roles. In contrast, our recent yeast work identified several endogenous tRNAs implicated in the regulation of SC-RT. Swiftly emerging studies of human tRNA-ome also advocate that tRNAs have unprecedented regulatory potential. Here, we developed a universal U6 promotor-based system expressing various human endogenous tRNA iso-decoders to study consequences of their increased dosage on SC-RT employing various reporter systems in vivo. This system combined with siRNA-mediated downregulations of selected aminoacyl-tRNA synthetases demonstrated that changing levels of human tryptophan and tyrosine tRNAs do modulate efficiency of SC-RT. Overall, our results suggest that tissue-to-tissue specific levels of selected near-cognate tRNAs may have a vital potential to fine-tune the final landscape of the human proteome, as well as that of its viral pathogens.
INTRODUCTION
Protein synthesis is a central, essential biological process during which the genetic information imprinted on mRNAs is decoded into the necessary protein context required for cell's viability by tRNAs carrying cognate amino acids onto ribosomes. Regulation of translation rates is cell- or tissue-specific and defines cell's life cycle, providing the means of cross-talk for virtually every known biochemical pathway to maintain cellular and tissue homeostasis (1,2). tRNA molecules, generated by the RNA polymerase III (PolIII), were until recently considered ‘housekeeping’ RNAs and passive carriers of amino acids, representing adaptor molecules between the RNA and protein worlds (3). However, surprisingly high number of tRNA-encoding genes (∼600 in humans) and advances in quantitative tRNA sequencing revealing unexpected variations in tRNA copy numbers in different cells/tissues strongly suggest additional roles for these ancient molecules (4–6). Indeed, several recent studies demonstrated that cells can respond to various environmental signals by changing their tRNA expression patterns, tRNA modifications and tRNA usage, and thus promote adaptive translation in a very dynamic way (7). Undoubtedly, the gene expression world is dumbfounded by the profound complexity of the human tRNA-ome (including various functional tRNA-derived fragments) affecting cell cycle, development, and numerous pathologies, such as cancer and neurodegenerative and metabolic diseases (8,9). For example, it has been shown that activation of the oncogenic signaling pathways, including AKT-mTOR, RAS-MAPK and MYC, or loss of the tumor suppressor TP53 can regulate RNA polymerase III expression, thus leading to altered tRNA expression (10–12). As a result, increased levels of tRNAs may contribute to tumor progression by supplying the high demand codons of oncogenic pathways (13).
Codon degeneracy for the 20 amino acids requires up to 5 tRNAs with distinct anticodons (tRNA iso-acceptors) to read codons for each amino acid. There are 21 iso-acceptor families, one for each amino acid and one for seleno-cysteine. An iso-acceptor family may consist of one tRNA member, e.g. tRNATrp, or of up to five tRNA members, e.g. tRNALeu. Each family contains numerous iso-decoders; i.e. tRNA genes having the same anticodon but different sequences elsewhere in the tRNA body (14). For example, nine genes code for iso-decoder tRNATrp with the CCA anticodon in the human genome GtRNAdb (5,15). The tRNA abundance has been initially correlated with iso-decoder tRNA gene copy number, however, recent studies suggest that tRNA expression regulation is far more complex than previously anticipated (16). Variations in the expression levels of iso-decoder tRNAs logically modulate levels of iso-acceptor tRNAs, whose net changes are nowadays seen as either natural condition-stimulated or pathology-forced adaptations to the transcriptome codon usage alterations in healthy or transformed cells, respectively (17–19).
The only step of the translational cycle where there should be no place for regulatory tRNAs is, by definition, translation termination. Indeed, to capture protein synthesis, occurrence of one of the three stop codons UAA, UAG and UGA in the ribosomal A-site is recognized by the complex of the release factors eRF1 and eRF3. Following stop codon recognition, the nascent peptide is hydrolyzed by eRF1 - releasing the mature protein product - and ribosomes are subsequently removed from the mRNA by recycling (reviewed in (20,21)). However, it has been shown that competition between eRF1 and certain elongator tRNAs can infrequently result in accommodation of a so-called near-cognate tRNA in the A-site, resulting in translational readthrough of the stop codon and thus production of C-terminally extended protein isoforms (22,23). While natural stop codon readthrough (SC-RT) is quite rare (lower than 0.1%), its so-called programmed form with frequencies up to 20% does play important biological roles in cells (24,25). tRNAs, therefore, hold a control of every single translational phase.
High rates of SC-RT have been documented in diverse organisms including viruses (26), yeast (27), flies (28) and mammals (25). Viruses rely on the host cell translation machinery for efficient synthesis of their own proteins. Programmed SC-RT in viruses like Sindbis virus (SINV) and Colorado thick fever virus (CTFV) enables the production of a diverse array of structural and enzymatic proteins at appropriate stoichiometric levels required for a proper completion of a viral life cycle (29,30). Eukaryotic cells, on the other hand, take advantage of programmed SC-RT to produce C-terminally extended protein isoforms with distinct functions (24,31,32). Intriguingly, the extreme case of SC-RT is programmed reassignment of all three stop-codons as sense codons in some organisms, such as ciliates or some trypanosomatids (33,34). Not surprisingly, SC-RT is also linked to human disease. More than 15% of all human genetic diseases can be attributed to the presence of a premature stop codon (PTC) in the coding region of an essential protein (35,36). Importantly, for mRNAs containing PTC, the frequency of readthrough defines the efficiency of functional protein synthesis (37).
Over the years, the efficiency of SC-RT was shown to rely on the presence of stimulatory elements downstream of the stop codon (38–41), specific protein factors such as for example eIF3 (42), the identity of the +1 nucleotide immediately following the stop codon (the +4 base) that together define a so-called stop codon tetranucleotide (43–49), and other features. Indeed, much effort was also invested into deciphering the reading of the genetic code by near-cognate tRNAs (50–52). With respect to the latter, we recently demonstrated that it is the identity of the stop codon tetranucleotide that largely determines preferences for specific near-cognate tRNAs in such a way that new decoding rules could have been proposed, at least for the yeast genome (42,47–49). In particular, four highly readthrough-efficient near-cognate tRNAs (tRNATrp, tRNACys, tRNAGln, tRNATyr) were identified with a defined stop codon tetranucleotide-specificity that we designated as readthrough-inducing tRNAs (rti-tRNAs) (47,48). Whether the same stop codon tetranucleotide decoding rules apply also on mammals remains to be determined.
Therefore, here we wished to expand our analysis of SC-RT into human cells. The first goal was to create a tool for tRNA overexpression in cellulo that we fulfilled with the development of our universal U6 promotor-based overexpressing system. Since SC-RT in human cells impacts regulation of expression of three main groups of genes: (i) endogenous genes with C-terminal extensions; (ii) genes with PTCs and (iii) viral genes of human pathogenes, our second goal was to uncover specific human tRNA iso-decoders that are able to regulate these three events in vivo. With this approach, we demonstrate that modified levels of tRNATrp and tRNATyr specifically impact SC-RT on UGA or UAG and UAA stop codons in human cells, respectively. Please note that only tRNA iso-decoders with high probability tRNA scores, as specified in the GtRNAdb database (5,15), are functional in vivo. Finally, using various reporters, we also show that overexpression of tryptophan tRNA: 1) boosts SC-RT of reporters carrying termination sequences of selected cellular and viral genes that are known to be a subject of this type of regulation, and 2) enhances restoration of a functional p53 protein production from mutant PTC-containing mRNA.
MATERIALS AND METHODS
Plasmids
The lists and descriptions of plasmids, primers, and GeneArt™ Strings™ DNA Fragments used throughout this study (summarized in Tables S1–S3) can be found in the Supplementary Information. The tRNA sequences were obtained by tRNAscan-SE Analysis of Homo sapiens (hg19 – NCBI Build 37.1 February 2009) (53).
Cell lines manipulations
Hek293T cells (human embryonic kidney cells obtained from Petr Svoboda, IMG of Czech Academy of Sciences) were cultured in DMEM (Sigma, cat # D6429) supplemented with 10% fetal bovine serum FBS (Sigma, cat # F7524). The H1299 cells were cultured in RPMI plus GlutaMAX (Gibco by Life Technologies) supplemented with 10% fetal bovine serum (FBS, Gibco by Life Technologies) and 100 U/ml of penicillin/streptomycin. Cells were kept in a humidified atmosphere containing 5.5% CO2 at 37°C. H1299 is p53-null cell line generated from a human lung carcinoma cells (provided by ATCC to the O. Namy lab).
Stop codon readthrough assays
For one reaction, ∼40 000 cells were plated into a single well of a 24-well plate and 24 h later transfected with 200 ng of DNA (for UAG, UGA and sense control reporters) or 500 ng of DNA (for UAA) using Turbofect (Invitrogen). No cell toxicity was observed for these DNA doses. Please note that when using a six-well plate, five times more of each component must be taken into each reaction. A properly mixed reaction (100 μl of DMEM (Sigma) with DNA and Turbofect (1 μl per 500 ng of DNA)) was incubated at the room temperature for 20 min and then applied onto the cells. SC-RT was assayed 24 h after the transfection as follows. The media mixture was removed from each well and the cells were lysed with a room tempered 1× Glo buffer (Promega) for 5 min at 25°C while shaking at 550 rpm. Equal amounts of the lysate were transferred into two Microtiter plates (one for Renilla and the second for Bright Glo (Promega)). Substrates for both luciferases were added in a one-to-one ratio and the reactions were incubated for 5 min (BrightGlo) or 15 min (Renilla) at 25°C while shaking at 225 rpm. The luminiscence was measured by Tecan Infinite 200Pro. The readthrough efficiency was estimated by calculating the ratio of Firefly luciferase to Renilla luciferase activity obtained with the test constructs and normalizing it to the ratio obtained with an in-frame control construct. For each construct with two or three reactions at the time; at least five independent transfection experiments were performed.
Factor downregulation
Hek293T cells were grown at 37°C and 5% CO2 in six-well plates in DMEM (Sigma, cat # D6429) supplemented with 10% FBS (Sigma, cat # F7524). Twenty-four hours after seeding 200 000 cells per well, cells were transfected with ON-TARGETplus siRNA cocktail system from Dharmacon with the 5 nM final concentration. The catalog numbers of all siRNAs used are: NT (D-001810-03), eRF1 (L-019840-00-0005), YARS (L-011498-00-0005) and WARS (L-008322-00-0005). Targeted sequences were for eRF1 (GAGCUACGUUGGAAAUUGU; UAACUAUGUUCGGAAAGUA; AAACUGAACUAAGUCAAUC; AACAUUAAGUCACGAGUAA); for YARS (GAACGGCAACCACGGGCAA; UGAAGGAGCGGGAACUUAA; AAGAGAAACUGCACCUUAU; ACGCAUACCUGGAUAACAU) and for WARS (GAGCUCAGGUUUCUACAAA; UGACGGAUGACGAGAAGUA; GAGCUAAUAAACCGAAUAG; GGUCACCUCAUUCCAUUUA). INTERFERin (Polyplus, cat # 409) was used as a transfection reagent and transfection was carried out according to the vendor‘s instructions. Twenty-four hours after the transfection the cells were plated for individual experiments.
RNA isolation, reverse transcription and qPCR
Total RNA was isolated using the 1ml RNA Blue reagent (Top Bio, cat #R013) 48 h post-seeding (200 000 cells per well in a six-well plate) according to the manufacturer's instructions. Glycoblue (Thermofisher, cat #AM9516) was used as a co-precipitant. The resulting RNA pellets were resuspended in RNase-free water and the concentration was quantified by NanoDropOne (Thermo Scientific) followed by the TurboDNAse digestion (Ambion, cat # AM2238). Subsequently, 500 ng of DNA-free RNA samples with identical amounts of ‘spike RNA’ (the yeast RPL41 mRNA produced in vitro using MAXIscript SP6/T7 transcription kit Thermofisher, cat #AM1320) were used for reverse transcription in a 20 μl reaction with the High Capacity cDNA Reverse Transcription kit (Thermofisher, cat # 4368814). qPCR was carried out according to vendor's instructions (Solis BioDyne, cat #08-25-00020) using Bio-Rad CFX384 Real-Time PCR System. qPCR reactions were prepared by mixing 5× HOTFIREPol EvaGreen qPCR Mix Plus (no ROX) with 0.3 M primers and 5 μl of five times (or 500 times in case of 18S rRNA) diluted cDNA, and run using the following program: 95°C for 15′ followed by 44 cycles of 95°C for 15″, 60°C for 22″, and 72°C for 20″. Melting curves were analyzed between 65 and 95°C. Levels of eRF1, YARS, WARS, 18S rRNA, ALAS and spike RNA were measured with at least three 10-fold serial dilutions as described before (54,55), together with no reverse transcription (NRT) and no template (NTC) controls. Results were analyzed using Bio-Rad CFX Manager. For each downregulation experiment, the obtained data within the primer pair were normalized to samples treated with non-targeted siRNA and expressed as a cycle difference (dCq; note that 0 value means no change in RNA levels; value 1 indicates ∼50% reduction in RNA levels, etc.). All qPCR primers are listed in Supplementary Table S2.
p53 activity assay and p53 visualization
H1299 cells were seeded (200 000 cells per well) into a six-well plate (Falcon) and 24 h later co-transfected—in the presence of the Jet Pei reagent (Ozyme), as recommended by the manufacturer—with pCMVp53WT or its mutant derivatives (3 μg), variants of p53BS-luc (2 μg) and pCMLacZ (0.6 μg). Media was exchanged six hours after transfection. The antibiotic G418 (200 μg/ml) was added with the fresh media 6 h post-transfection. Protein extracts were prepared 24 h after transfection and enzymatic activities were measured as previously described (46). For each experimental set-up, at least five independent transfection experiments were performed. For visualization of short and long forms of the p53 protein, protein extracts were prepared using 200 μl of a room tempered 1× Glo buffer; incubation for 5 min at 25°C while shaking at 550 rpm. Approximately 0.5 μg of protein extracts were subjected to the ‘Western-like’ analyses using the Jess Protein Simple device; the total protein was measured by Bradford. The p53 antibody (DO-1; the N-terminal epitope mapping between amino acid residues 11–25 of p53; Santa Cruz Biotechnology) was used at its saturating conditions 1:100. The equal loading was monitored by the total protein staining (Protein Simple).
RESULTS and DISCUSSION
Plasmid born in cellulo production of human tRNAs from the U6 promoter
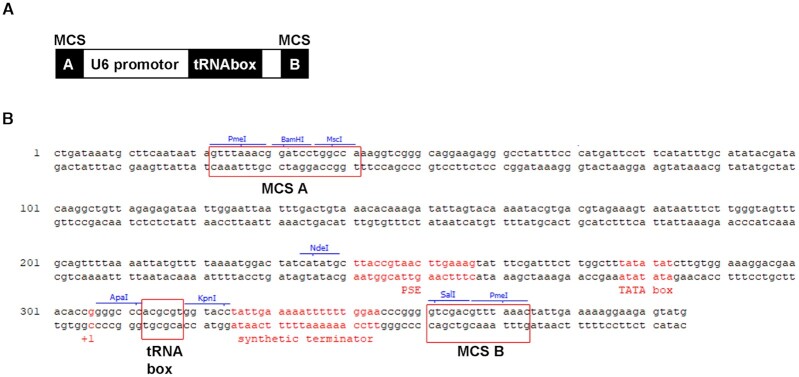
To create a tool for tRNA overexpression in human cells, we first had to select an optimal promoter. The fact is that the strength and functionality of tRNA promoters cannot be easily predicted bioinformatically. To overcome this problem, we took advantage of the fact that the endogenous U6 promoter was successfully used for in vivo expression of a suppressor tRNA in the past (56). Therefore, we designed our own U6 promoter with its natural PSE (proximal sequence element), TATA box and +1 position for production of all tRNAs of our interest. In particular, we created a U6-tRNA-Box cassette including a self-designed synthetic terminator with poly(T) flanked by multicloning sites (MCS) A and B that can be easily subcloned into any reporter used throughout the study (Figure 1). A DNA sequence of any mature tRNA of interest was used to replace the indicated ‘tRNA-Box’ backbone sequence between the ApaI and KpnI sites (Figure 1B) to create individual U6-X-tRNA overexpression cassettes. Indeed, immature tRNA-encoding DNA sequences containing introns can be used too. Please note that even though our goal was to drive a tRNA expression solely from the U6 promoter, we are aware of the fact that all tRNA inserts contain their own residual internal promoters that could—in co-operation with the artificial U6-TATA box—contribute to the overall expression.
Figure 1.
Plasmid born in cellulo production of human tRNAs from the U6 promoter. (A) Schematic of the U6-tRNAbox cassette used for overexpression of human tRNAs. (B) The sequence of U6-tRNAbox cassette including U6 promotor features. Individual U6-tRNAbox cassettes with various tRNA sequences subcloned between ApaI and KpnI sites of the ‘tRNAbox’ were always cut at the specific restriction sites within the multicloning sites (MCS) A and B and inserted into the reporter of interest cleaved with the corresponding restriction sites. The PSE, TATAbox, the +1 position of the human U6 promoter, and a synthetic terminator are indicated in red.
Overexpression of tryptophan and tyrosine tRNAs increase stop codon readthrough in human cells
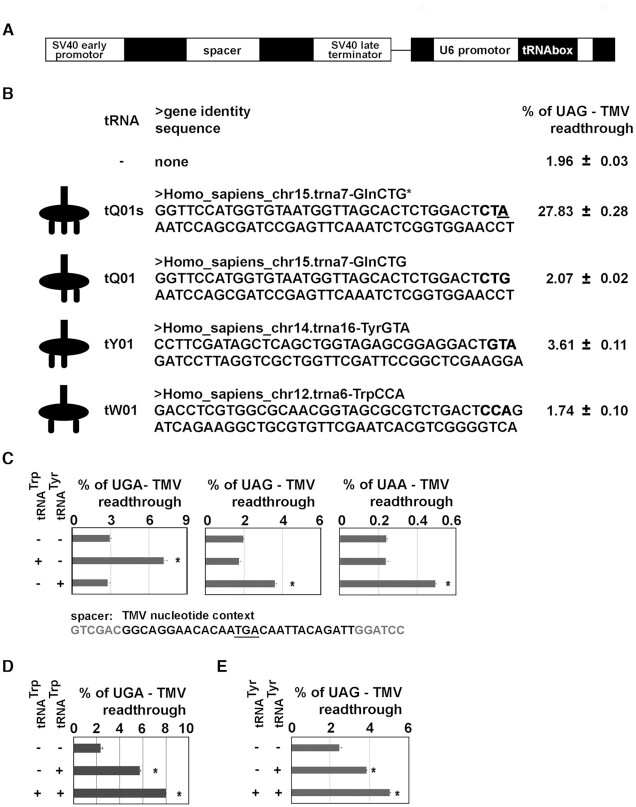
We have recently published a comprehensive analysis of all natural tRNAs that are near-cognate with some of three stop codons in yeast in order to identify all those that are capable of efficient decoding of stop codons (47,48). This analysis uncovered altogether four so-called readthrough-inducing (rti) tRNAs - Trp, Cys, Gln and Tyr. To determine whether the same tRNAs work also in human cells, we decided to take advantage of the pre-existing human readthrough reporter system p2luci (57), as well as of its new StopGo generation pSGDluc (58). In both variants the in-frame renilla and firefly luciferases, under the control of an early SV40 promoter, are separated by a spacer containing either a stop codon of choice or a sense codon, as a control sample. We further modified the reporter plasmids by inserting the aforementioned U6-tRNAbox; either empty or bearing a human tRNA of our interest (Figure 2A). This arrangement ensures that the measured readthrough values originated only from cells that did contain an ectopically-expressed tRNA and thus reflects its capacity to boost stop codon recoding.
Figure 2.
Overexpression of tryptophan and tyrosine tRNAs increase stop codon readthrough in human cells. (A) Schematic of the readthrough reporter containing the U6-tRNAbox cassette. For more details please the main text. (B) Readthrough measurements using the UAG—TMV reporter (see panel C) with or without selected human tRNAs in the HEK293T cells. The schematics of base-pairing between individual tRNAs and the UAG stop codon is shown to the right of the name of each construct, followed by the gene identity with the tRNA sequence and the relative percentage of the UAG readthrough. The tRNA sequences were individually placed under the U6 promoter of the PBB168 reporter or its sense control PBB172 reporter (see Figure 1). The tQ01s (marked by an asterisk) is the mutated variant of tQ01; a single-point mutation in the anticodon is underlined. (C) Readthrough measurements of all three stop codon reporters in Hek293T cells overexpressing Trp tRNA (tW01) or Tyr tRNA (tY01). The corresponding sequences of these two tRNAs shown in panel 2B were placed under the U6 promoter of PBB171 (for the UGA reporter); PBB168 (UAG); PBB170 (UAA), or its sense control PBB172. As an example, the DNA spacer sequence containing the UGA stop codon followed by six nucleotides representing the TMV stop codon nucleotide context is shown below. Changes in readthrough levels to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P<0.05). (D) The UGA – Trp tRNA (tW01) readthrough measurements as in panel C only two copies of the U6-tRNATrp cassettes was added (plasmid PBB288). Changes in readthrough levels to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). (E) The UAG—Tyr tRNA (tY01) readthrough measurements as in panel D (plasmid PBB292). Changes in readthrough levels to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05).
Naturally, our newly developed tRNA overexpression system had to be validated for its functionality. Therefore, to demonstrate that an ectopically expressed tRNA is fully engaged with the process of translation, we exploited a specific form of human glutamine tRNA where the CUG* anticodon is mutated to CUA (56). This single point mutation renders this tRNA fully cognate for the UAG stop codon (Figure 2B, left column) and, therefore, it is expected to very efficiently suppress translation termination in vivo. Hence, we inserted the mutated form of this tRNA (tQ01s) into our expression vector and measured UAG readthrough in HEK293T cells; please note that UAG is followed by the very potent Tobacco Mosaic Virus (TMV) nucleotide context (59) (Figure 2B). As expected, mutated Gln tRNA expressed in our system suppressed very robustly translation termination of our dual luciferase reporter by showing ∼27.83% of UAG readthrough compared to ∼1.93% measured with a no tRNA control vector (Figure 2B).
In contrast, the wild-type form of the CUG* isoform of Gln tRNA (tQ01) base-pairing with last two nucleotides of the UAG stop codon of our reporter (Figure 2B, left column), which also features as one of the rti-tRNAs for the UAG stop codon in the budding yeast (48), produced only ∼2.07% of readthrough, practically matching the value obtained with no tRNA control (Figure 2B). Naturally, our system by itself does not distinguish between no effect versus improper folding and/or acetylation of a tested tRNA, and we have no means to examine it at present. Similarly, we are unable to quantify exact amounts of an ectopically expressed tRNA because we perform only transient transfections, the efficiency of which varies and we have no means to determine the number of non-transfected cells that would need to be accounted for. Therefore, we are entitled to make solid claims only about tRNAs showing statistically significant increase of readthrough. Actually, this technical limitation prohibited us from carrying out any omics studies so far. Nonetheless, taking into account the robust readthrough potential of mutated tQ01s differing from the tQ01 sequence only by a single nucleotide, we assume that also the wild-type form of this particular glutamine tRNA is most probably properly processed and, therefore, does not induce readthrough in humans in contrast to yeast.
Next, we decided to examine another tRNA whose counterpart was identified as rti-tRNA for UAG and UAA stop codons in yeast, human Tyr tRNA (iso-decoder tY01). Expressing this tRNA - base-pairing with the first two nucleotides of the UAG stop codon of our reporter (Figure 2B, left column) - revealed that the selected tY01 iso-decoder significantly increased readthrough (from ∼1.93% of no tRNA to 3.61%; Figure 2B). This suggests that Tyr tRNA promotes readthrough also in humans.
The last human tRNA that we employed in this experimental set-up with the UAG reporter was Trp tRNA (iso-decoder tW01), whose yeast counterpart was identified as rti-tRNA for the UGA stop codon. However, in this particular case the main purpose was obviously not to examine its rti-tRNA character (this we show below), but to complete the validation of our system by testing all three possible combinations of a two bases-based base-pairing (the Trp tRNA anticodon is complementary to the first and third nucleotides of UAG; Figure 2B, left column). As expected, despite the full functionality of this tRNA (documented below), we observed no increase above the no tRNA control (Figure 2B), demonstrating the specificity of our system. Please also note that increased cellular levels of all these tRNAs do not affect Renilla to Firefly ratio for a reporter bearing the sense codon (CAA) (Supplementary Figure S1). To conclude, we created a versatile overexpressing reporter system to monitor effects of any tRNA iso-decoder of interest on a relevant molecular mechanism such as SC-RT. In principle, any other small RNA under the control of PolIII, like miRNAs and shRNAs (60,61), can be investigated using our expression system.
Next, we wished to examine the rti-tRNA character of human Trp and Tyr tRNAs in detail. Towards this end, we individually overexpressed them in combination with all three stop codon reporters with the TMV nucleotide context (59) in the p2luci vector and revealed that, as in yeast, human Trp tRNA specifically and significantly increased readthrough only with the UGA reporter, whereas Tyr tRNA did so with both UAG and UAA reporters (Figure 2C). These findings clearly demonstrate that both these tRNAs do get overexpressed in cells but act only where expected. In agreement, doxycycline inducible TetU6-tW01 expression in HEK-Trex cells displayed the anticipated Dox-specific effect on the efficiency of SC-RT in our system (Supplementary Figure S2). In addition, increasing the number of Trp or Tyr tRNA cassettes in the UGA or UAG reporter, respectively, from one to two further increased the efficiency of readthrough (Figure 2D and E). Taken together, our results clearly illustrate that both Trp and Tyr tRNAs promote readthrough also in humans when overexpressed and, in addition, demonstrate the tRNA dosage dependence of our system.
The last of the four rti-tRNAs identified in yeast to be tested in human cell lines is Cys tRNA. Four structurally distinct cysteine tRNA ACG iso-decoders were commercially synthetized by Genestring and cloned under the U6 promoter to be tested in our system. Unexpectedly, individual expression of all of them had a negative impact on cell proliferation resulting in a translational shut down (data not shown). This effect could be explained by spontaneous cleavage of Cys tRNAs generating the 5′ Cys tRNA fragment forming G-quadruplexes that were shown to act as particularly potent translation repressors (62). In any case, this ‘technical’ problem prevented us from including iso-decoders of this particular near cognate human Cys tRNA into our analysis. As expected, no increase was seen with the ACA iso-acceptor Cys tRNA possessing an intron and a low tRNA score (data not shown).
To summarize this part of our work, we conclude that yeast (47,48) and humans share at least two rti-tRNAs (Trp and Tyr) as dosage-dependent enhancers of readthrough in living cells. More work is needed to examine all functional iso-decoders of Cys and Gln tRNAs for their readthrough potential before they can be included or definitely excluded from the rti-tRNA class of tRNAs in humans.
Detailed analysis of the readthrough potential of all predicted human tRNATyr and tRNATrp iso-decoders
The Tyr tRNA tY01 was initially selected from the GtRNAdb list (5,15) of tRNAs with the GUA anticodon based on high reads in the publicly available RNAseq data. Interestingly, this list for example contained five identical sequences of mature Tyr tRNA encoded by unique human tRNA genes differing only in their UTR and intron sequences. Hence, we became curious to examine whether any variations anywhere in the sequence of Tyr tRNA genes would affect the readthrough inducing potential of a mature tRNA. Therefore, all Tyr tRNA-encoding iso-decoders with the GTA anticodon deprived of computationally predicted introns (Supplementary Figures S3 and S4) were subcloned into our reporters and tested for the readthrough efficiency. Based on the outcome, we divided them into two groups: (i) featuring those iso-decoders displaying increased readthrough (Supplementary Figure S3) and (ii) those with no change compared to our no tRNA control (Supplementary Figure S4). The A group encompasses Tyr tRNAs varying by one to three nucleotides including tY01 (Supplementary Figure S3A and B). All increased readthrough but some to somewhat reduced level compared to our reference tY01 iso-decoder (Supplementary Figure S3C). No obvious patterns with respect to the individual sequence variations were identified. Nonetheless, these findings indicate that the observed nucleotide polymorphism determines the degree of the readthrough-inducing potential of individual tRNAs by a yet-to-be explored mechanism. As could have been expected, the B group with no effect is represented by tyrosine tRNA genes with a low ‘tRNA score’ (tRNA score defines a probability with what a given sequence encodes a real tRNA), strongly suggesting that these genes do not encode functional tRNAs.
To capture our effort with this particular tRNA, we also analyzed the singleton human tyrosine iso-acceptor tRNA with the ATA anticodon that we retrieved from the GtRNAdb (5,15) (Supplementary Figure S5). Based on its tRNA score (∼55.93), it can be also considered as a low score tRNA, nonetheless, we were curious to examine the potential effect of a different anticodon that retains complementarity to the first two nucleotides of the UAG stop codon. However, no effect was observed compared to the no tRNA control (Supplementary Figure S5).
Finally, we subjected all discernible Trp tRNAs iso-decoders to analogous analysis, using the pSGDluc reporters this time, and obtained analogous results with similar conclusions. The Trp tRNA iso-decoders with a high tRNA score increased readthrough by ∼2–2.5-fold compared to the no tRNA control (Supplementary Figure S6), in contrast to those with a low score showing no increase (Supplementary Figure S7).
Based on these analysis we propose that the ‘tRNA score’ (5,15) is a reliable predictor of a functionality of a given tRNA in translation. Our assay thus may serve as a rapid tester of functional consequences of spontaneous mutations - like SNPs - on high tRNA score iso-decoders.
Depletion of tryptophan or tyrosine aminoacyl-tRNA synthetases decreases stop codon readthrough
As a complementary approach to the tRNA overexpression strategy we attempted to decrease levels of amino acylated tRNAs under study via siRNA-mediated downregulation of their corresponding aminoacyl-tRNA synthetases; the cytoplasmic tryptophanyl-tRNA synthetase (WARS) for Trp and tyrosyl-tRNA synthetase (YARS) for Tyr. As a specific control we also downregulated eukaryotic release factor 1 (eRF1). As expected, upon eRF1 downregulation (eRF1K.D.) the SC-RT efficiency increased by ∼3-fold irrespective of the nature of a stop codon in our reporter (Figure 3A). Please note that we achieved ∼50% reduction in the eRF1-encoding mRNA level and ∼60% of the eRF1 protein level (Figure 3B). Upon successful completion of this proof of principle experiment, we knock down YARS (YARSK.D.; ∼50% reduction on the mRNA level, no working antibodies available—Figure 3B) and WARS (WARSK.D.; ∼50% reduction on the mRNA level and ∼40% on the protein level—Figure 3B). As shown in Figure 3A, we observed that YARSK.D. specifically and significantly (by ∼50%) reduced readthrough with UAG but not UGA reporters, and conversely WARSK.D. reduced readthrough with UGA but not UAG reporters, though to a smaller, yet still statistically significant degree. These results not only support our conclusion that Trp tRNA increases readthrough for UGA and Tyr tRNA for the two remaining stop codons, they also suggest that varying cellular levels of rti-tRNAs influence the efficiency of SC-RT in both directions.
Figure 3.
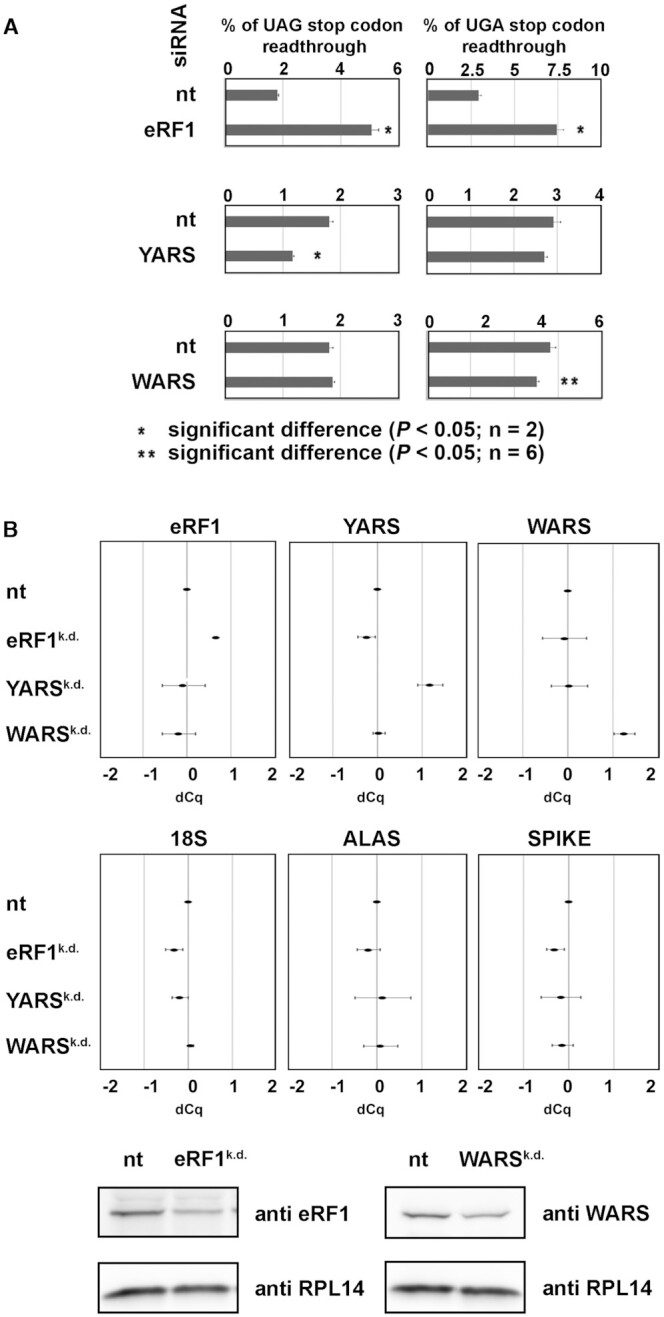
Depletion of tryptophan and tyrosine aminoacyl-tRNA synthetases decreases stop codon readthrough. (A) The UAG and UGA readthrough measurements in HEK293T cells after siRNA-mediated downregulation of eRF1 (upper panel), YARS (middle panel), and WARS (lower panel). For details please see Materials and Methods. Changes in readthrough levels to non-targeted siRNA control were analyzed by the Student's t-test (mean + SD; n = 2 or n = 6) and shown to be statistically significant for those values marked (P< 0.05). (B) mRNA and protein levels of selected factors in individual knock-downs determined at the time of the readthrough measurement; same seeding as in panel A was used. The mRNA levels were established by RT-qPCR and quantified as described in Materials and Methods.
Tryptophan tRNA overexpression increases SC-RT on reporters bearing stop codon contexts of mRNAs encoding human proteins known to be natural readthrough substrates
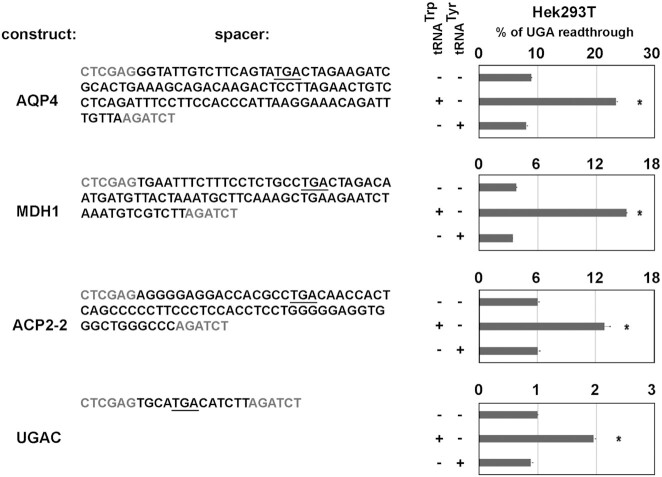
Functional translation readthrough in humans was reviewed recently with the focus put on three human genes (VEGFA, LDHB and MDH1) by (23), where the authors described distinct cellular functions of parent and extended forms of these proteins. Even though functions of many of the best known, highly conserved mammalian mRNAs that are subjects of SC-RT are still unclear (25), the list of readthrough-governed proteins with ascribed cellular roles continues to grow steadily. The most recent additions are AQP4 (25,63) and a vitamin D receptor (31). In case of the former, translational readthrough was shown to generate new astrocyte AQP4 isoforms that modulate supramolecular clustering, glial endfeet localization, and water transport. In case of the later, a C-terminally extended variant of the human vitamin D receptor generated by SC-RT displayed a reduced transcriptional response to calcitriol. Expectedly, the list of tissue-specific, readthrough-regulated mRNAs will be in reality much bigger, as can be deduced from high throughput studies such as this (28), with only a handful of examples where functions of both variants have been characterized experimentally. To demonstrate that overexpression of readthrough-inducing Trp tRNA may influence the efficiency of SC-RT of human mRNAs known to be regulated this way, we first prepared a set of reporters where the spacer bears sequences encompassing the end of the coding region including a stop codon of a gene of interest followed by its well-described the C-terminal extension, the so called stop codon context (SCC) (Figure 4). Next, we measured readthrough with or without overexpressed Trp or Tyr tRNAs in HEK cells. All of these sequences – including a non-genomic control – naturally contain the UGA stop codon, which is in case of AQP4 and MDH1 insertions followed by the well-known readthrough-promoting CUAG SCC (24,25). In addition, the ACP2-2 insertion also contains a sequence following the stop codon, which supposedly forms a specific stem loop structure representing an important readthrough driving force (58). Importantly, functionality of insertions of all selected gene derivatives (namely AQP4, MDH1, ACP2-2 and the UGAC negative control) in the very pSGDluc reporter system was verified in the past by others (58). As can be seen in Figure 4, overexpressing Trp but not Tyr tRNA did significantly increase readthrough with all these constructs. These findings lead us to propose that tissue-specific and/or stress-induced changes in cellular levels of readthrough-inducing tRNAs, such as Trp tRNA, may represent one of the important regulatory means of readthrough with a direct impact on the ratio between the parent versus extended form protein production.
Figure 4.
Tryptophan tRNA overexpression increases SC-RT on reporters bearing stop codon contexts of mRNAs encoding human proteins known to be natural readthrough substrates. The UGA readthrough measurements in Hek293T cells overexpressing tRNATrp (tW01) or tRNATyr (tY01) transfected with the pSGDluc-based plasmids bearing genomic sequences surrounding stop codons of three genes of interest as spacers, or UGAC as a basal readthrough control. Their respective sense (UGG) controls were used in parallel. Changes in readthrough levels to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). For details please see Materials and Methods.
As an example, ∼4% of malate dehydrogenase (MDH1) is physiologically extended by translational readthrough and the actual ratio of MDH1x (extended protein) to regular MDH1 is dependent on the cell type. All vertebrate MDH1x is directed to peroxisomes via a hidden peroxisomal targeting signal in the readthrough extension, where it is critically required for their proper functioning (64). In the light of our results, it is tempting to speculate that the cell type-specificity in the amount of MDH1x that gets delivered to peroxisomes is determined by the cell type-specificity of Trp tRNA levels. On a global scale, we propose that tissue-to-tissue varying expression of tRNA iso-decoder genes could be genetically programmed to contribute to the phenotypical variability of cells.
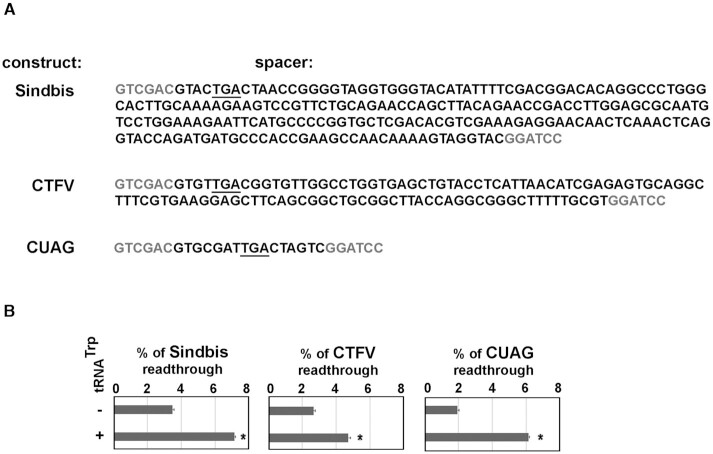
Tryptophan tRNA overexpression increases SC-RT on reporters bearing SCCs of mRNAs encoding viral proteins from two human pathogenes
Viruses rely on the host cell translation machinery for efficient synthesis of their own proteins. It is known that they use translational readthrough and other recoding mechanisms to maximize the coding capacity of their often small genomes (30). Emerging evidence also shows that RNA viruses exploit distinct aspects of the host tRNA biology in the process of viral infection for their overall benefit (26). Here, we wished to examine whether the capacity of human Trp tRNA to induce readthrough may represent one of these aspects. As examples of viral translational readthrough we chose well-described Sindbis virus (SINV) (41) and a functionally uncharacterized 65-kDa product VP9′ of Colorado thick fever virus (CTFV) (29). Although these two human pathogens represent species of a distinctive genus, Alphavirus for SINV and Coltivirus for CTFV, they possess a similar readthrough-promoting feature; i.e. the UGA stop codon followed by a distinct stem loop structure in their 3′ UTR (Supplementary Figure S8) (29,41). To examine if the readthrough-mediated expression of these two viral mRNAs is responsive to tryptophan tRNA overexpression, we prepared reporters where the spacer bears sequences encompassing the end of their coding regions including UGA followed by well-described the C-terminal extensions (Figure 5A), and measured readthrough with or without our Trp tRNA overexpression cassette in HEK cells. Overexpressing tRNATrp significantly increased readthrough for both Sindbis (from ∼3.5% to ∼7%) and CTFV (from ∼2.5% to ∼4.5%) constructs (Figure 5B). Hence, as in the previous case, we propose that alterations in cellular levels of readthrough-inducing tRNAs may impact the proteome composition of readthrough-exploiting viruses.
Figure 5.
Tryptophan tRNA overexpression increases SC-RT on reporters bearing SCCs of mRNAs encoding viral proteins from two human pathogenes - Alphavirus and Coltivirus. (A) Spacer sequences of the p2luci reporters containing the UGA stop codon genomic sequences of Sindbis virus, CTFV virus, or the UGA-CUAG human readthrough permissive nucleotide context. (B) The UGA readthrough measurements in Hek293T cells overexpressing tRNATrp (tW01) transfected with the p2luci reporters described in panel A; their respective sense (UGG) controls were used in parallel. Changes in readthrough levels to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). For details, see Materials and Methods.
Since the balance between the regular versus extended forms of viral proteins is critical for the life cycle of relevant viruses, such as for example Moloney murine leukemia virus (MuLV) (65,66), it is conceivable that even a small, transient change in cellular levels of corresponding tRNAs could stop the pathogenic viral propagation. For example, MuLV has gag and pol in the same reading frame, separated by a UAG stop codon, and termination codon readthrough is required for expression of the viral Gag-Pol fusion protein. It was demonstrated that reducing the MuLV readthrough efficiency by ∼10-fold essentially abolished viral replication (65). Hence, antiviral therapies targeting readthrough via directed manipulations of cellular levels of corresponding rti-tRNAs may represent a future avenue of our defense against these pathogens.
Tryptophan tRNA overexpression enhances restoration of a functional p53 protein production from a mutant PTC-containing mRNA
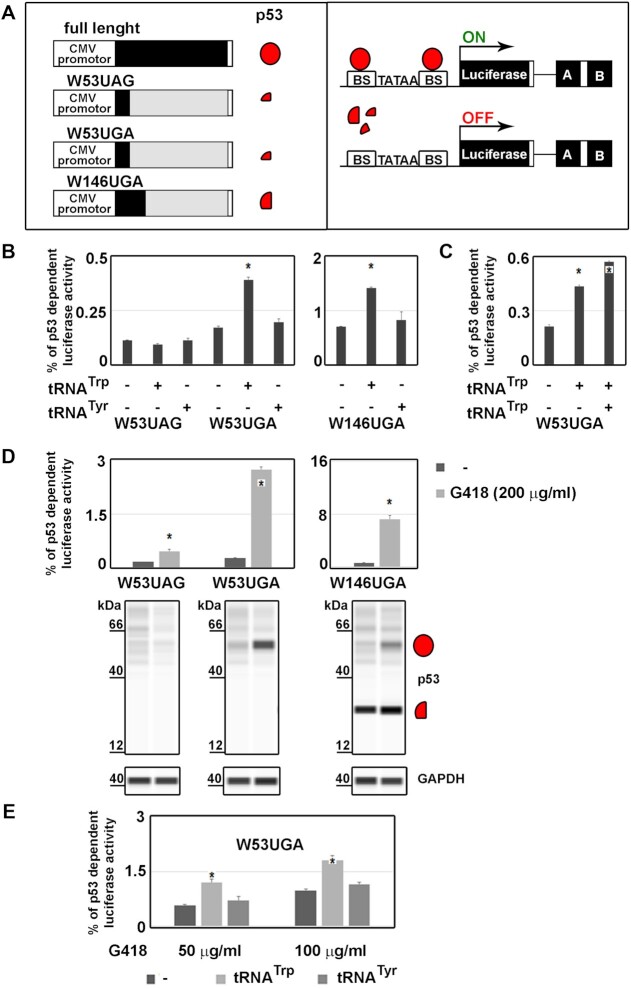
Large number of human diseases and cancer syndromes are caused or accompanied by premature termination codons (PTCs) occurring in the coding sequence of numerous genes, resulting in a production of a truncated protein and mRNA degradation by non-sense mediated mRNA (NMD) decay (35–37,67). To investigate a potential effect of the readthrough-inducing tRNA overexpression on the PTC-associated pathogenesis, we adopted an experimental set-up developed by others (46). It enables monitoring of the restored production of the fully functional tumor suppressor p53 protein from its PTC-containing mRNA in cellulo (Figure 6A). Briefly, we selected several mRNA variants of p53 under the CMV promoter carrying various naturally occurring PTCs. Their presence results in synthesis of various non-functional p53 truncated isoforms that are incapable of switching on transcription of the Luciferase gene under control of the p53 transcription factor. A factor X-driven restoration of the fully functional, full-length p53 production can be quantitatively measured (Figure 6A). The utmost relevance of this assay is demonstrated by the fact that many cancers are linked to the presence of a SNP in a tumor suppressor gene and that in more than 50% of these cases, it is the p53 tumor suppressor gene that is mutated (68). Moreover, ∼8% of all cancer-related p53 mutations are non-sense, PTC mutations.
Figure 6.
Tryptophan tRNA overexpression enhances restoration of a functional p53 protein production from a mutant PTC-containing mRNA. (A) The schematics of the p53-based functional assay for determination of production of functional p53 protein from PTC-containing mRNAs. The full-length p53 as well as its three PTC-terminated forms are co-expressed from the CMV promoter together with the luciferase under the control of the p53-dependent promoter (from Bcl-G gene) containing p53-binding sites (BS) surrounding its TATA box. In addition, the luciferase-based reporter also contains and expresses the Trp or Tyr U6-tRNAbox cassettes described in Figure 1B. (B) The p53-dependent luciferase activity from PTC-containing p53 mRNAs normalized to the luciferase activity obtained with the full-length p53 protein. p53 null cells H1299 were co-transfected with p53 expression plasmids and luciferase/tRNA reporters described in panel A, as well as with a plasmid bearing β-Galactosidase for internal normalization and subjected to measurement analyses as described previously (46). Changes in levels of p53 dependent luciferase activity to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). (C) Restoration of the p53 functional protein expression from the PTC-containing mRNA (W53UGA-U) responds to the Trp tRNA (tW01) overexpression. Same as in panel B, except that luciferase reporters PBB321, PBB301 and PBB330 bearing none, one or two tRNATrp cassettes were used. Changes in levels of p53 dependent luciferase activity to no tRNA control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). (D) Overnight G418 treatment (200 μg/ml) boosts restoration of the p53 functional protein expression from the PTC-containing mRNAs (W53UGA-U and W146UGA-G). Changes in levels of p53 dependent luciferase activity to no drug control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P< 0.05). Production of the full length p53 protein was monitored by the ‘Western-like’ analysis using the Jess system from Protein Simple; for details please see Materials and Methods. (E) Combining G418 treatment with overexpression of Trp-tRNA displays an additive effect on restoration of the p53 functional protein expression from the PTC-containing mRNA (W53UGA). Changes in levels of p53 dependent luciferase activity to no tRNA no drug control were analyzed by the Student's t-test (mean + SD; n = 2) and shown to be statistically significant for those values marked with the asterisk (P<0.05).
In analogy with our analysis described above, we focused on the Trp tRNA and thus tryptophan codon (UGG) naturally occurring PTC mutations. In particular, we picked W53UAG with the estimated mutation frequency about 0.6% (46), W53UGA (0.81%), and W146UGA (4.71%) (Figure 6A). These frequencies were given relative to total non-sense mutations listed for p53 gene in IARC TP53 Database. The percentage of the p53-dependent luciferase activity produced in the presence or absence of overexpressed Trp or Tyr tRNAs was estimated for all three mutations relative to the luciferase activity produced in cells transfected with a wild-type p53 mRNA, which was set to 100%. Whereas no relative induction was observed for W53UAG when Trp or Tyr tRNAs were overexpressed (Trp tRNA does not incorporate at UAG (42) and Tyr tRNA would incorporate tyrosine instead of tryptophan rendering—most probably—the full-length protein non-functional), Trp tRNA overexpression significantly and specifically induced luciferase activity for both W53UGA and W146UGA (Figure 6B). By definition, this implies that overexpression of the UGA-specific rti-tRNA boosted restoration of the full-length p53 protein production from the latter two PTC-containing mRNAs. In support, increasing the copy number of a Trp tRNA cassette led to a further activity induction (Figure 6C).
Next we compared individual effects of overexpression of Trp tRNA with the aminoglycoside G418-dependent activation reported earlier (46); G418 is one of the extensively studied readthrough-inducing drugs (RTIDs). Administration of 200 μg/ml of G418 for 18 hours prior to measurements induced the luciferase activity for all three PTCs, however, only W53UGA (∼3% induction) and mainly W146UGA (∼8%) displayed potentially medically-relevant increases (Figure 6D). Thanks to these robust results, we could employ the Jess Protein Simple analyzer to visualize both truncated (obtainable only for W146UGA), as well as full-length forms of p53 protein (Figure 6D, bottom panels). A marked, drug-dependent increase in the expression levels of mature p53 is seen for both W53UGA and W146UGA. Combining the tRNA overexpression and RTID approaches displayed an additive effect with the high rti-tRNA specificity for W53UGA (Figure 6E).
Taken together, we demonstrate here that overexpression of human tryptophan tRNA enhances restoration of a functional p53 protein production from a mutant PTC-containing mRNA in a PTC-specific manner, however, to a smaller degree than that observed for G418. Obviously, to achieve the Trp tRNA-mediated restoration of the full-length protein production that would be comparable in its effect to the drug-driven approach, a more robust tRNA overexpression strategy must be engineered. Only then this approach could be considered as medically interesting, which is a future direction of our research. Perhaps, combining both approaches—RTIDs and rti-tRNAs—could be beneficial in the sense that lower, non-deleterious concentrations of often very toxic RTIDs could be used to maximize targeted readthrough while minimizing off-target miscoding.
CONCLUDING REMARKS
Here, the universal U6 promotor-based system overexpressing various human endogenous tRNA iso-decoders was developed. Using several experimental set-ups we demonstrated the following. i) Yeast and humans share at least two rti-tRNAs (Trp and Tyr) as dosage-dependent enhancers of readthrough on UGA or UAG and UAA stop codons, respectively. (ii) The ‘tRNA score’ is a reliable predictor of a functionality of a given tRNA iso-decoder in translation. (iii) Overexpressing tryptophan tRNA: (a) boosts SC-RT on reporters bearing SCCs of mRNAs encoding cellular and viral genes known to be SC-RT subjects, and (b) enhances restoration of a functional p53 protein production from its mRNA containing mutations converting codons cognate to tRNATrp to nonsense PTCs. Therefore, we propose that tissue-to-tissue specific levels of selected readthrough-inducing tRNAs might have a significant impact on the balanced expression of the parent versus extended forms of cellular, as well as viral proteins. As such, we envisage that a cell-type specific modulation of the rti-tRNA levels could help to combat human pathogens whose life cycle relies on SC-RT and, perhaps, also support the developing RTID-driven treatments of PTC-caused diseases.
DATA AVAILABILITY
All data and reagents are freely available. Further information and requests for reagents should be direct to L.S.V. (valasekl@biomed.cas.cz).
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Zuzana Pavlíková for critical reading of the manuscript, Olga Krýdová and Kristína Koudelová for technical and administrative assistance, and all lab members for fruitful discussions. We are indebted to Dr Tomáš Mašek for his gift of the HEK293Trex FlipIN cells.
Contributor Information
Petra Beznosková, Laboratory of Regulation of Gene Expression, Institute of Microbiology ASCR, Videnska 1083, 142 20 Prague, the Czech Republic.
Laure Bidou, Sorbonne Universités, Paris, France; Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France.
Olivier Namy, Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91198 Gif-sur-Yvette, France.
Leoš Shivaya Valášek, Laboratory of Regulation of Gene Expression, Institute of Microbiology ASCR, Videnska 1083, 142 20 Prague, the Czech Republic.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Czech Science Foundation [18-02014S, 20-00579S to L.S.V.]; Programme for research and mobility of starting researchers from the Czech Academy of Sciences (to P.B.); ANR grant Actimeth [19-CE12-0004-02]; French foundation ARC grant [PJA20131200234 to O.N.]. Funding for open access charge: Czech Science Foundation [20-00579S].
Conflict of interest statement. None declared.
REFERENCES
- 1. Roux P.P., Topisirovic I.. Signaling pathways involved in the regulation of mRNA translation. Mol. Cell. Biol. 2018; 38:e00070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buszczak M., Signer R.A., Morrison S.J.. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014; 159:242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018; 19:45–58. [DOI] [PubMed] [Google Scholar]
- 4. Parisien M., Wang X., Pan T.. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013; 10:1853–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Orioli A. tRNA biology in the omics era: stress signalling dynamics and cancer progression. Bioessays. 2017; 39:1600158. [DOI] [PubMed] [Google Scholar]
- 7. Kirchner S., Ignatova Z.. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015; 16:98–112. [DOI] [PubMed] [Google Scholar]
- 8. Huang S.Q., Sun B., Xiong Z.P., Shu Y., Zhou H.H., Zhang W., Xiong J., Li Q.. The dysregulation of tRNAs and tRNA derivatives in cancer. J. Exp. Clin. Cancer Res. 2018; 37:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santos M., Fidalgo A., Varanda A.S., Oliveira C., Santos M.A.S.. tRNA deregulation and its consequences in cancer. Trends Mol. Med. 2019; 25:853–865. [DOI] [PubMed] [Google Scholar]
- 10. Gomez-Roman N., Grandori C., Eisenman R.N., White R.J.. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003; 421:290–294. [DOI] [PubMed] [Google Scholar]
- 11. Felton-Edkins Z.A., Fairley J.A., Graham E.L., Johnston I.M., White R.J., Scott P.H.. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 2003; 22:2422–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei Y., Tsang C.K., Zheng X.F.. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009; 28:2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Truitt M.L., Ruggero D.. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016; 16:288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodenbour J.M., Pan T.. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006; 34:6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mahlab S., Tuller T., Linial M.. Conservation of the relative tRNA composition in healthy and cancerous tissues. RNA. 2012; 18:640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M.R., Pan T.. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009; 37:7268–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sorensen K.D.et al.. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014; 158:1281–1292. [DOI] [PubMed] [Google Scholar]
- 19. Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F.. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016; 165:1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson R.J., Hellen C.U., Pestova T.V.. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 2012; 86:45–93. [DOI] [PubMed] [Google Scholar]
- 21. Valasek L.S., Zeman J., Wagner S., Beznoskova P., Pavlikova Z., Mohammad M.P., Hronova V., Herrmannova A., Hashem Y., Gunisova S.. Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res. 2017; 45:10948–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E.. Translational readthrough potential of natural termination codons in eucaryotes–The impact of RNA sequence. RNA Biol. 2015; 12:950–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schueren F., Thoms S.. Functional translational readthrough: a systems biology perspective. PLos Genet. 2016; 12:e1006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gartner J., Thoms S.. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014; 3:e03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F.. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014; 42:8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nunes A., Ribeiro D.R., Marques M., Santos M.A.S., Ribeiro D., Soares A.R.. Emerging roles of tRNAs in RNA virus infections. Trends Biochem. Sci. 2020; 45:794–805. [DOI] [PubMed] [Google Scholar]
- 27. Namy O., Duchateau-Nguyen G., Hatin I., Denmat Hermann-Le, Termier S., Rousset J.P.. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2003; 31:2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Napthine S., Yek C., Powell M.L., Brown T.D., Brierley I.. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA. 2012; 18:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Firth A.E., Brierley I.. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012; 93:1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Loughran G., Jungreis I., Tzani I., Power M., Dmitriev R.I., Ivanov I.P., Kellis M., Atkins J.F.. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018; 293:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yamaguchi Y., Hayashi A., Campagnoni C.W., Kimura A., Inuzuka T., Baba H.. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J. Biol. Chem. 2012; 287:17765–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zahonova K., Kostygov A.Y., Sevcikova T., Yurchenko V., Elias M.. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr. Biol. 2016; 26:2364–2369. [DOI] [PubMed] [Google Scholar]
- 34. Heaphy S.M., Mariotti M., Gladyshev V.N., Atkins J.F., Baranov P.V.. Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in Condylostoma magnum. Mol. Biol. Evol. 2016; 33:2885–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kurosaki T., Maquat L.E.. Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci. 2016; 129:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Linde L., Kerem B.. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008; 24:552–563. [DOI] [PubMed] [Google Scholar]
- 37. Keeling K.M., Xue X., Gunn G., Bedwell D.M.. Therapeutics based on stop codon readthrough. Annu. Rev. Genomics Hum. Genet. 2014; 15:371–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Namy O., Hatin I., Rousset J.P.. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001; 2:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skuzeski J.M., Nichols L.M., Gesteland R.F., Atkins J.F.. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991; 218:365–373. [DOI] [PubMed] [Google Scholar]
- 40. Harrell L., Melcher U., Atkins J.F.. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res. 2002; 30:2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Firth A.E., Wills N.M., Gesteland R.F., Atkins J.F.. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011; 39:6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beznoskova P., Wagner S., Jansen M.E., Haar T., Valasek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bonetti B., Fu L., Moon J., Bedwell D.M.. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995; 251:334–345. [DOI] [PubMed] [Google Scholar]
- 44. McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P.. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cassan M., Rousset J.P.. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol. 2001; 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Floquet C., Hatin I., Rousset J.P., Bidou L.. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet. 2012; 8:e1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beznoskova P., Gunisova S., Valasek L.S.. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016; 22:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Beznoskova P., Pavlikova Z., Zeman J., Echeverria Aitken C., Valasek L.S.. Yeast applied readthrough inducing system (YARIS): an invivo assay for the comprehensive study of translational readthrough. Nucleic Acids Res. 2019; 47:6339–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gunisova S., Beznoskova P., Mohammad M.P., Vlckova V., Valasek L.S.. In-depth analysis of cis-determinants that either promote or inhibit reinitiation on GCN4 mRNA after translation of its four short uORFs. RNA. 2016; 22:542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blanchet S., Cornu D., Argentini M., Namy O.. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blanchet S., Cornu D., Hatin I., Grosjean H., Bertin P., Namy O.. Deciphering the reading of the genetic code by near-cognate tRNA. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:3018–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roy B., Leszyk J.D., Mangus D.A., Jacobson A.. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lowe T.M., Eddy S.R.. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chiu W.-L., Wagner S., Herrmannová A., Burela L., Zhang F., Saini A.K., Valášek L., Hinnebusch A.G.. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 2010; 30:4415–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khoshnevis S., Gunišová S., Vlčková V., Kouba T., Neumann P., Beznosková P., Ficner R., Valášek L.S.. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res. 2014; 42:4123–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Koukuntla R., Ramsey W.J., Young W.B., Link C.J.. U6 promoter-enhanced GlnUAG suppressor tRNA has higher suppression efficacy and can be stably expressed in 293 cells. J. Gene Med. 2013; 15:93–101. [DOI] [PubMed] [Google Scholar]
- 57. Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F.. A dual-luciferase reporter system for studying recoding signals. RNA. 1998; 4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 58. Loughran G., Howard M.T., Firth A.E., Atkins J.F.. Avoidance of reporter assay distortions from fused dual reporters. RNA. 2017; 23:1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pelham H.R. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978; 272:469–471. [DOI] [PubMed] [Google Scholar]
- 60. Borchert G.M., Lanier W., Davidson B.L.. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006; 13:1097–1101. [DOI] [PubMed] [Google Scholar]
- 61. Wang Z., Rao D.D., Senzer N., Nemunaitis J.. RNA interference and cancer therapy. Pharm. Res. 2011; 28:2983–2995. [DOI] [PubMed] [Google Scholar]
- 62. Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P.. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011; 43:613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. De Bellis M., Pisani F., Mola M.G., Rosito S., Simone L., Buccoliero C., Trojano M., Nicchia G.P., Svelto M., Frigeri A.. Translational readthrough generates new astrocyte AQP4 isoforms that modulate supramolecular clustering, glial endfeet localization, and water transport. Glia. 2017; 65:790–803. [DOI] [PubMed] [Google Scholar]
- 64. Hofhuis J., Schueren F., Notzel C., Lingner T., Gartner J., Jahn O., Thoms S.. The functional readthrough extension of malate dehydrogenase reveals a modification of the genetic code. Open Biol. 2016; 6:160246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Csibra E., Brierley I., Irigoyen N.. Modulation of stop codon read-through efficiency and its effect on the replication of murine leukemia virus. J. Virol. 2014; 88:10364–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Irigoyen N., Dinan A.M., Brierley I., Firth A.E.. Ribosome profiling of the retrovirus murine leukemia virus. Retrovirology. 2018; 15:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bidou L., Allamand V., Rousset J.P., Namy O.. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 2012; 18:679–688. [DOI] [PubMed] [Google Scholar]
- 68. Perri F., Pisconti S., Della Vittoria Scarpati G.. P53 mutations and cancer: a tight linkage. Ann. Transl. Med. 2016; 4:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and reagents are freely available. Further information and requests for reagents should be direct to L.S.V. (valasekl@biomed.cas.cz).