Abstract
Communication between cellular compartments is vital for development and environmental adaptation. Signals emanating from organelles, so-called retrograde signals, coordinate nuclear gene expression with the developmental stage and/or the functional status of the organelle. Plastids (best known in their green photosynthesizing differentiated form, the chloroplasts) are the primary energy-producing compartment of plant cells, and the site for the biosynthesis of many metabolites, including fatty acids, amino acids, nucleotides, isoprenoids, tetrapyrroles, vitamins, and phytohormone precursors. Signals derived from plastids regulate the accumulation of a large set of nucleus-encoded proteins, many of which localize to plastids. A set of mutants defective in retrograde signaling (genomes uncoupled, or gun) was isolated over 25 years ago. While most GUN genes act in tetrapyrrole biosynthesis, resolving the molecular function of GUN1, the proposed integrator of multiple retrograde signals, has turned out to be particularly challenging. Based on its amino acid sequence, GUN1 was initially predicted to be a plastid-localized nucleic acid-binding protein. Only recently, mechanistic information on the function of GUN1 has been obtained, pointing to a role in plastid protein homeostasis. This review article summarizes our current understanding of GUN-related retrograde signaling and provides a critical appraisal of the various proposed roles for GUNs and their respective pathways.
This review summarizes new insights in GUN-mediated retrograde signaling, and highlights outstanding questions and challenges that should be addressed in future research.
Introduction
During photosynthesis, sunlight is converted to chemical energy and ultimately stored as biomass. In photosynthetic eukaryotes, the biophysical processes involved in energy conversion as well as in the biochemical reactions involved in carbon fixation take place in the chloroplast (plastid). In addition to being the site of photosynthesis, plastids also harbor various pathways of primary and secondary metabolism, including the biosynthesis of fatty acids, many amino acids, pigments, and plant hormones, and also have key roles in nitrogen and sulfur assimilation (Neuhaus and Emes, 2000; Lopez-Juez and Pyke, 2005). Plastids originated from a free-living cyanobacterium that, in a phenomenon known as endosymbiosis, was engulfed by a non-photosynthetic eukaryotic cell (Archibald, 2009; Criscuolo and Gribaldo, 2011; Keeling, 2013). Most genes of the cyanobacterial endosymbiont were subsequently lost or transferred to the nuclear genome of the host cell (Bock and Timmis, 2008; Bock, 2017). Present-day plastids still contain a small genome of 50–200 protein-encoding genes depending on the species (Martin and Herrmann, 1998; Martin et al., 1998; Ponce-Toledo et al., 2019). Many protein complexes in plastids (e.g. ribosomes and photosystems) comprise subunits encoded by the plastid genome and others by the nuclear genome. This dual encoding of multi-subunit protein complexes therefore necessitates a high degree of synchronization in gene expression between the nucleus and the plastid to ensure the required subunit stoichiometry.
The coordinated expression of the two genomes is ensured by two types of signals: signals from the nucleus that are sent to the chloroplast and control plastid gene expression (PGE; so-called anterograde signals), and signals emanating from plastids that regulate nuclear gene expression (referred to as retrograde signals; Nott et al., 2006; Woodson and Chory, 2008; Chan et al., 2016; de Souza et al., 2017). The retrograde signals identified so far can be grouped into two classes: (i) “biogenic control” signals that act predominantly during early chloroplast biogenesis (i.e. chloroplast differentiation from proplastids or etioplasts), germination and/or seedling development to inform the nucleus about the developmental status of the plastid and the resulting demands for nucleus-encoded proteins, and (ii) “operational control” signals that are mainly generated by mature chloroplasts in response to environmental stimuli that alter the demand for nuclear gene products by plastids (Pogson et al., 2008; Barajas-Lopez et al., 2013; Hernández-Verdeja and Strand, 2018).
Important knowledge about biogenic plastid-to-nucleus signaling has come from the isolation and characterization of a set of mutants, called genomes uncoupled (abbreviated as gun mutants) that disrupt this type of retrograde signaling (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Koussevitzky et al., 2007; Woodson et al., 2011). This review will mainly focus on recent progress in our understanding of biogenic control and the roles that the GENOMES UNCOUPLED (GUN) genes play in retrograde signaling.
Retrograde signaling in biogenic control and the GUN genes
Chloroplasts can develop directly from proplastids, which are the undifferentiated progenitor organelles of all type of plastids, or from etioplasts upon illumination. One of the most delicate tasks during chloroplast biogenesis is the assembly of the photosynthetic complexes necessary for the electron transfer reactions of photosynthesis (Nelson and Ben-Shem, 2004; Woodson and Chory, 2008; Pogson and Albrecht, 2011; Jarvis and López-Juez, 2013). This task is achieved by the coordinated expression and assembly of plastid- and nucleus-encoded protein subunits, and the simultaneous incorporation of photosynthetic pigments (chlorophylls and carotenoids) and redox-active co-factors (e.g. hemes and iron–sulfur clusters). Inhibition of photosynthetic pigment biosynthesis and perturbation of PGE were demonstrated to affect retrograde control of the expression of photosynthesis-associated nuclear genes (PhANGs; Barajas-Lopez et al., 2013; Chan et al., 2016; Hernández-Verdeja and Strand, 2018). For example, the exogenous application of the carotenoid biosynthesis inhibitor norflurazon (NF; Oelmüller, 1989) or plastid translation inhibitors such as lincomycin (Lin; Mulo et al., 2003) trigger strong responses in nuclear gene expression, and studies with these inhibitors provided a deeper understanding of biogenic retrograde signaling pathways. Most importantly, inhibitor experiments revealed a large set of nuclear genes that are strongly regulated by retrograde signals from developmentally impaired plastids, including many PhANGs, and also various genes involved in other biological processes. Prominent PhANGs responsive to retrograde signals are genes for light-harvesting complex proteins (LHCB) and for the small subunit of the central carbon-fixing enzyme Rubisco (RBCS). The expression of these genes is repressed when chloroplast biogenesis is disturbed by inhibited pigment biosynthesis and/or impaired PGE (Nott et al., 2006; Chan et al., 2016).
Elegant genetic screens were conducted in Arabidopsis (Arabidopsis thaliana) to isolate mutants that display derepressed expression of LHCB genes upon NF treatment (Susek et al., 1993; Woodson et al., 2011). The underlying assumption was that such mutant screens would reveal genes that encode components of retrograde communication pathways. A number of mutants with deregulated LHCB (and RBCS) gene expression were isolated and dubbed gun mutants. The corresponding genes mapped to six loci in the Arabidopsis genome (Supplemental Table S1; Mochizuki et al., 2001; Larkin et al., 2003; Koussevitzky et al., 2007; Woodson et al., 2011). Notably, with a single exception, all identified GUN genes (GUN2-GUN6) are directly involved in the tetrapyrrole biosynthesis (TPB) pathway (Figure 1), by encoding either enzymes or regulator proteins of the pathway. These findings strongly suggested an important role of TPB-derived metabolite(s) as retrograde signals in chloroplast biogenesis (Strand et al., 2003; Zhang et al., 2015b). The signaling function of TPB intermediates has also been demonstrated in the unicellular green alga Chlamydomonas reinhardtii, suggesting evolutionary conservation (von Gromoff et al., 2008; Voss et al., 2011).
Figure 1.
Retrograde signals derived from the TPB pathway. The debate about positive versus negative signals from the TPB pathway is not yet resolved. A proposed negative model posits that Mg-ProtoIX and its monomethylester (Mg-ProtoIX-ME) act as possible signaling molecules by leaving the plastid via an unknown transporter (shown in brown at the plastid membrane) and then binding to HSP90 to inhibit its activity. Reduced HSP90 inhibition results in a gun phenotype, because HSP90 activity is positively correlated with PhANG expression (Strand et al., 2003; Kindgren et al., 2011; Kindgren et al., 2012a; Wu et al., 2019b). Under the positive model, heme synthesized specifically by FC1, but not FC2, is exported out of plastids by an unknown transporter (shown in red at the plastid membrane) and acts as a putative positive signal that actives PhANG expression (Woodson et al., 2011). In the nucleus, HY5, GLKs, and likely other currently unknown positive or negative transcriptional regulators participate in the control of PhANG expression. GluTR, glutamyl-tRNA reductase; POR, pchlide oxidoreductase.
TPB-derived retrograde signals
A large body of research has suggested that intermediates of the TPB pathway serve as plastid retrograde signals (Figure 1; Kropat et al., 1997; Vasileuskaya et al., 2004). In seed plants, initial evidence for the involvement of tetrapyrroles in plastid-to-nucleus signaling emerged from a genetic screen for Arabidopsis gun mutants in the presence of the carotenoid biosynthesis inhibitor NF and the subsequent identification of the mutated genes (Susek and Chory, 1992; Susek et al., 1993; Mochizuki et al., 2001; Surpin et al., 2002). In NF-treated gun mutants, LHCB gene expression was significantly higher than in NF-treated wild-type plants (but still lower than in untreated plants). Interestingly, the expression of LHCB was negatively correlated with the levels of magnesium protoporphyrin IX (Mg-ProtoIX), an intermediate of chlorophyll biosynthesis (Figure 1), hinting that Mg-ProtoIX may be the signaling molecule. For example, the gun5-1 mutant harbors a point mutation (A990V) in the gene for the H subunit of magnesium chelatase, reducing the activity of the enzyme (Mochizuki et al., 2001). Upon NF treatment, the gun5-1 mutant accumulates less Mg-ProtoIX than the wild type (Strand et al., 2003). The TPB inhibitor α,α′-dipyridyl (DP), an iron chelator, blocks protoporphyrin flux to heme and also inhibits the conversion of Mg-ProtoIX methyl ester (Mg-ProtoIX-ME) toward chlorophyll (Duggan and Gassman, 1974). Treatment of the gun5 and gun2 mutants (GUN2 encodes heme oxygenase, the enzyme conducting the committed step of phytochromobilin biosynthesis by catalyzing the oxidative cleavage of heme) with DP increases Mg-ProtoIX levels and abolishes the gun phenotype of both mutants (Strand et al., 2003), thus suggesting a negative regulatory role of Mg-ProtoIX in plastid retrograde signaling. Additional studies with seed plants and algae, including the analysis of mutants in the TPB pathway, the use of TPB inhibitors, and feeding with δ-aminolevulinic acid (ALA, an early precursor of all tetrapyrroles), also supported the correlation between Mg-ProtoIX (and Mg-ProtoIX-ME) and the repression of LHCB and RBCS gene expression (Johanningmeier and Howell, 1984; Oster et al., 1996; Vinti et al., 2000; La Rocca et al., 2001; Nott et al., 2006; Pontier et al., 2007).
However, several observations argue against a direct role of Mg-ProtoIX (or Mg-ProtoIX-ME) as a plastid retrograde signal. First, although gun5 and gun4 (encoding an activator of Mg-chelatase; Larkin et al., 2003; Peter and Grimm, 2009) mutants display a gun phenotype, mutants lacking the activity for another subunit of this enzyme (the I subunit, CHLI; cs and ch42-1 mutants), although also producing less Mg-ProtoIX, do not develop a gun phenotype, in that they do not show derepression of LHCB upon NF treatment (Mochizuki et al., 2001). Second, LHCB gene expression in barley is greatly reduced in NF-treated plants, but with no detectable accumulation of Mg-ProtoIX (or Mg-ProtoIX-ME; Gadjieva et al., 2005). Third, although cytoplasmic accumulation of Mg-ProtoIX can be observed by confocal laser-scanning microscopy, it can only be seen in seedlings fed with ALA prior to NF treatment (Ankele et al., 2007). Fourth, detailed quantification of tetrapyrroles in Arabidopsis seedlings grown in the presence of NF does not support a correlation of Mg-ProtoIX levels with the molecular phenotypes of several gun mutants (Mochizuki et al., 2008; Moulin et al., 2008), although it was argued later that the accumulation of Mg-ProtoIX may be transient and thus may still be negatively linked to the gun phenotype (Kindgren et al., 2011, 2012a; Zhang et al., 2011).
To resolve the conflicting information obtained from the different studies addressing a possible negative role of Mg-ProtoIX in plastid retrograde communication, a new gain-of-function screen was carried out to identify regulator molecules in this signaling pathway (Woodson et al., 2011). This screen led to the isolation of the dominant gun6-1D mutant allele that results in the overexpression of plastid ferrochelatase 1 (FC1), thereby increasing FC1 enzyme activity. Overexpression of FC1 is expected to enhance the flux of tetrapyrroles into the heme branch of TPB. The study positively correlated PhANG expression with the size of the heme pool. This relationship appears to be specific to FC1, in that overexpression of FC2 does not produce a gun phenotype, even though both ferrochelatase isoforms localize to plastids (Woodson et al., 2011). Interestingly, heme was also shown to be emitted as a positive retrograde signal from mitochondria in yeast (Kwast et al., 1998; Zhang et al., 1998; Zhang and Hach, 1999), and plastids in green algae (von Gromoff et al., 2008). Based on the knowledge obtained from the gun6-1D mutant, a model for retrograde signaling was proposed, in which healthy plastids emit a positive signal (e.g. heme) to support the expression of PhANGs (Figure 1). PhANG expression is lost upon disruption of key plastid functions (e.g. by NF treatment) that are required to generate this positive signal (Woodson et al., 2011; Terry and Smith, 2013). However, the existence of negative signals cannot be ultimately excluded. ALA feeding experiments and DP suppression of the gun phenotype of the gun2 and gun5 mutants still support a negative correlation of PhANG expression with Mg-ProtoIX levels (Strand et al., 2003). There is also evidence from transcriptome analysis of the pap7 mutant (defective in a protein associated with the plastid-encoded RNA polymerase, PEP, thus causing impaired plastid transcription) for the additional involvement of negative signal(s) in retrograde communication (Grubler et al., 2017).
PGE-dependent retrograde signals
The organellar and nuclear gene expression systems are tightly coordinated. When PGE is disturbed, the resulting imbalance triggers a response in nuclear gene expression and, for example, leads to repression of many PhANGs. Experimentally, this can be shown by pharmacological inhibition of PGE, as with tagetitoxin, an inhibitor of plastid transcription (Rapp and Mullet, 1991) or Lin, a specific inhibitor of plastid translation (Mulo et al., 2003). Mutations in genes encoding components of the PGE machinery (e.g. transcription, RNA processing, protein biosynthesis, and protein import; Sullivan and Gray, 1999; Hricova et al., 2006; Kakizaki et al., 2009, 2012; Kindgren et al., 2012b; Woodson et al., 2013; Díaz et al., 2018; Paieri et al., 2018) have also been extensively used to demonstrate that proper expression of plastid genome-encoded genes is required to sustain PhANG expression in the nucleus (Figure 2, A). Interestingly, the suppressive effects of PGE inhibitor treatments on PhANG expression are only seen in seedlings, that is within the first few days after seed germination, indicating that PGE-derived retrograde signals function mainly during early chloroplast biogenesis (Oelmüller et al., 1986).
Figure 2.
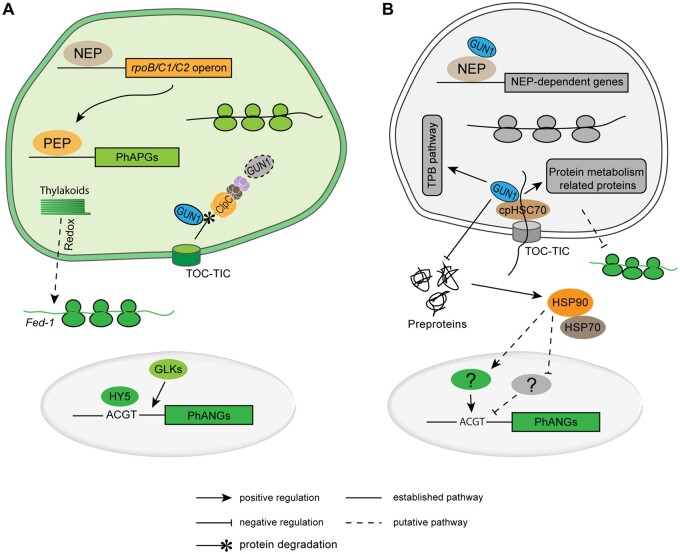
PGE-dependent retrograde control of nuclear gene expression. (A) In green tissues where chloroplast biogenesis is completed, PEP is responsible for the transcription of most plastid-encoded genes (including all PhAPGs: photosynthesis-associated plastid genes), while NEP mainly transcribes the rpoB/C1/C2 operon and a few genes containing only NEP promoters (e.g. accD, the only plastid-encoded gene involved in fatty acid metabolism). Positive regulators such as HY5 and GLKs activate PhANG transcription, the resulting mRNAs are translated in the cytosol and the protein products imported into chloroplasts through the TOC-TIC apparatus. GUN1 has no function at this stage and is rapidly degraded by the Clp protease upon import through the action of the chaperone ClpC1 (Wu et al., 2018). Chloroplast-derived redox signals related to PET activity regulate cytosolic mRNA stability by promoting the translation of specific mRNAs (e.g. Fed-1), thus preventing their ribonucleolytic degradation (Petracek et al., 1998). The light green ribosomes denote translation of PhAPGs in the chloroplast, the dark green ribosomes represent the translation of PhANGs in the cytosol. (B) When PGE is impaired (e.g. upon inhibition of translation with Lin), GUN1 interacts with cpHSC70-1, a motor protein of the TIC complex, and facilitates import of plastid-targeted proteins to maintain proteostasis of the plastid under stressful conditions (and preventing aberrant accumulation of preproteins in the cytosol). GUN1 clients (i.e. proteins whose import is GUN1-dependent) include proteins involved in various steps of plastid protein metabolism, and several enzymes of the TPB pathway (Wu et al., 2019a, 2019b). In the absence of GUN1, unimported preproteins accumulate in the cytosol, where their presence causes folding stress and triggers the accumulation of the cytosolic HSP90/HSP70 chaperone complex, which, in turn, derepresses PhANG expression. Thus, HSP90 activity is positively correlated with PhANG expression (Wu et al., 2019b), and may act by either activating positive or inhibiting negative transcriptional regulators in the nucleus. PhANGs and ribosomal proteins were shown to be regulated translationally (e.g. RBCS and LHCA4 are translationally repressed upon Lin treatment) and/or post-translationally either in a GUN1-dependent or independent manner (Wu et al., 2019a). GUN1 was also reported to be able to directly bind NEP to promote the expression of NEP-dependent genes (Tadini et al., 2020), although this possible function is likely independent of the gun phenotype of gun1 mutants. The gray ribosomes denote the translation of house-keeping genes in the plastid, the dark green ribosomes represent the translation of PhANGs in the cytosol.
Five non-allelic GUN loci were identified in the original screen for gun mutants (Susek et al., 1993). Unlike gun2–gun5 which affect enzymes (or regulator proteins) of the TPB pathway, GUN1 encodes a pentatricopeptide repeat (PPR) protein with a C-terminal small MutS-related (SMR) domain, which was suggestive of nucleic acid-binding activity (Koussevitzky et al., 2007). In contrast to all other gun mutants, which only show a gun phenotype upon NF treatment, gun1 mutations derepress PhANG expression upon Lin inhibition of PGE as well (Susek et al., 1993; Koussevitzky et al., 2007). Thus, GUN1 appears to participate in multiple plastid retrograde signaling pathways and its loss of function suppresses the reduced PhANG expression seen in mutants with impaired plastid transcription (e.g. the sigma factor mutants sig2 and sig6; Woodson et al., 2013), translation (e.g. prors1; Tadini et al., 2016), protein import (e.g. ppi2; Kakizaki et al., 2009), or defects in metabolism and chloroplast development (e.g. slow green1, sugar inducible cotyledon yellow‐192; Hu et al., 2014; Maruta et al., 2015). GUN1 was also implicated in operational control by plastid retrograde signaling (Koussevitzky et al., 2007), based on the findings that the high light repression of LHCB expression is released in gun1 mutants, and that the double mutant gun1 prin2 (plastid redox insensitive2, encoding a PEP-associated protein that appears to participate in high light-mediated retrograde signaling) alleviates the repressed PhANG expression seen in the prin2 single mutant (Kindgren et al., 2012b). Together, these findings were interpreted as evidence for GUN1 functioning as a central node that integrates signals from multiple retrograde signaling pathways (Figure 2, B; Koussevitzky et al., 2007; Wu et al., 2019b).
The role of GUN1 in plastid protein homeostasis and retrograde signaling
Because of its unique features and, especially, its response to both Lin and NF treatments, the gun1 mutant attracted tremendous interest among plant biologists. Yet, the molecular function of GUN1 has remained elusive for more than 25 years, and has only recently started to unfold (Jia et al., 2019). A major obstacle to all efforts toward elucidating the function of GUN1 has been its extremely low protein levels in most tissues and developmental stages. In fact, the GUN1 protein had never been seen in any proteomic study (Plant Proteome DataBase; http://ppdb.tc.cornell.edu/).
This unfortunate situation changed with a recent study that discovered a developmental window in which GUN1 protein accumulation occurs, by systematically investigating developmental stages, tissues, and environmental conditions (Wu et al., 2018). The protein was shown to accumulate only when active chloroplast biogenesis takes place, for example, in cotyledons and shoot apical meristems very early (36–72 h) after germination. GUN1 accumulation initiates prior to cotyledon greening, when the cotyledons are still largely enclosed by the seed coat, and peaks 72 h after germination (when the cotyledons rapidly transition from pale green to green), suggesting a connection to active chloroplast biogenesis (Wu et al., 2018). Consistent with this interpretation, the GUN1 protein is also highly expressed in the basal part of newly emerging true leaves, where expansion growth occurs and active chloroplast biogenesis takes place, and upon interference with plastid retrograde signaling (e.g. by Lin or NF treatment). Interestingly, at the transcript level, GUN1 is highly expressed in various tissues and developmental stages, indicating that post-transcriptional mechanisms control the accumulation of the GUN1 protein. This post-transcriptional regulation is achieved through the rapid degradation of GUN1 by the stromal Clp protease, with strong involvement of the chaperone ClpC1 (Figure 2, A; Wu et al., 2018; Pesaresi and Kim, 2019).
GUN1 contains PPR tracts and a C-terminal SMR domain. PPR proteins form a large protein family whose members are involved in various steps of organellar gene expression, including transcript processing, mRNA editing, RNA stability, and translational regulation (Lurin et al., 2004; Kotera et al., 2005; Schmitz-Linneweber et al., 2005, 2006; Schmitz-Linneweber and Small, 2008; Zoschke et al., 2013). By contrast, the SMR domain is a DNA-binding motif that is present in proteins participating in DNA repair and recombination, but also in a few organellar proteins related to RNA metabolism (Moreira and Philippe, 1999; Wu et al., 2016; Zhou et al., 2017). When the GUN1 gene was identified, it was therefore suggested that GUN1 may act as a nucleic acid-binding protein that may regulate PGE, plastid DNA metabolism, or DNA repair (Koussevitzky et al., 2007). This assumption misguided many subsequent studies on GUN1, and thus the realization that GUN1 interacts with proteins rather than nucleic acids was an important finding that changed the direction of investigations (Tadini et al., 2016; Jia et al., 2019; Wu et al., 2019b). However, a new complication arose from the large number of putative physical GUN1-interacting partners (Table 1), which may indicate that GUN1 is either a rather promiscuous or an unusually sticky protein. Putative GUN1-interacting proteins include enzymes of the TPB pathway and several proteins that participate in PGE and protein homeostasis (proteostasis) such as plastid chaperones (cpHSC70, ClpC1, CPN60) and ribosomal proteins (Tadini et al., 2016). The identity of these putative interactors raised the intriguing possibility that they provide a direct mechanistic link between GUN1 and the TPB and PGE pathways of retrograde signaling.
Table 1.
Proposed functions of GUN1 and current controversies (see text for details)
| Proposed function | Molecular interactions | Suggested mechanism | Possible problems | References |
|---|---|---|---|---|
| Nucleic acids binding | Plastid DNA | Regulation of PGE |
|
(Koussevitzky et al., 2007; Sun et al., 2011; Tadini et al., 2016; Page et al., 2017; Wu et al., 2019b; Kacprzak et al., 2019) |
| Protein binding | Plastid translation machinery (PRPS1, FUG1) | Regulation of plastid proteostasis |
|
(Tadini et al., 2016; Marino et al., 2019) |
| Plastid chaperones (ClpC1, cpHSC70-1) | Interaction with ClpC1 for rapid turnover of GUN1, interaction with cpHSC70-1 to regulate protein import and maintain plastid proteostasis |
|
(Tadini et al., 2016; Wu et al., 2018, 2019a, 2019b) | |
| NEP | Activation of NEP activity upon depletion of PEP |
|
(Tadini et al., 2020) | |
| MORF2 | Regulation of plastid RNA editing |
|
(Zhao et al., 2019b) | |
| Tetrapyrroles and TPB enzymes | Tetrapyrroles and related enzymes (CHLD, PBGD, UROD2, FC1) |
|
|
(Tadini et al., 2016; Shimizu et al., 2019) |
RS, retrograde signaling; OE, overexpression; TF, transcription factor; CHLD, D subunit of Mg-chelatase; PBGD, porphobilinogen deaminase; UROD2, uroporphyrinogen III decarboxylase.
Although evidence has accumulated for GUN1 engaging in protein–protein interactions, the mechanism of action and the origin of the gun phenotype in gun1 mutants has remained elusive. A new entry point into the elucidation of the molecular function of GUN1 was provided by a recent study that implicates GUN1 in the control of plastid protein import under conditions that interfere with retrograde signaling (Wu et al., 2019b). Indeed, when GUN1 function is dispensable (e.g. in mature, unstressed leaves), the GUN1 protein is rapidly degraded after its entry into the chloroplast. Degradation is mediated by the Clp protease and triggered by direct interaction between GUN1 and the chaperone ClpC1 (Figure 2, A). By contrast, GUN1 interacts with cpHSC70-1, a motor protein of the chloroplast protein import apparatus (Shi and Theg, 2010; Su and Li, 2010; Liu et al., 2014), and facilitates efficient protein import when the demand for nucleus-encoded proteins in the plastid is particularly high. This is the case, for example, when active chloroplast biogenesis occurs, or under stress conditions that lead to disturbed plastid proteostasis (Figure 2, B; Wu et al., 2018, 2019b). The precise mechanism behind how GUN1 stimulates protein import, and the physiological role of its interaction with cpHSC70, is currently unknown. Besides acting as a motor protein in protein import, cpHSC70 functions as a canonical chaperone in plastids. cpHSC70 may therefore interact with GUN1 and stabilize it, possibly in competition with ClpC1.
Consistent with the proposed function of GUN1 in plastid protein import, unimported precursor proteins (preproteins) accumulate in the cytosol of gun1 mutants upon Lin or NF treatment. This accumulation of preproteins cannot be explained by the derepressed expression of PhANGs, since the expression of the genes encoding these unimported preproteins does not differ substantially between the wild type and the gun1 mutant (Wu et al., 2019b). The accumulation of unimported precursors does not affect all plastid-targeted proteins equally, in that the preproteins encoded by several PhANGs (e.g. LHCB, RBCS) remain undetectable also in the gun1 mutant (Wu et al., 2019a). Whether this is due to the lower stability of their precursors in the cytosol or, alternatively, some substrate selectivity of the import apparatus, remains to be investigated. In any case, the currently available data suggest that the physiological role of GUN1 is to promote protein import under stressful conditions, thus maintaining plastid protein homeostasis upon rising demands for nucleus-encoded gene products.
Loss of GUN1 function impairs the capacity of the protein import to adjust to stressful conditions, thus causing imbalances in plastid proteostasis and, at the same time, disturbing cytosolic proteostasis by the accumulation of plastid preproteins. In response to this perturbation, cytosolic HSP90 (and, to a lesser extent, HSP70) chaperones highly accumulate in gun1 mutant plants under these conditions. The activity of HSP90 is positively correlated with PhANG expression, and was proposed to be directly involved in the development of the gun phenotype seen in gun1 mutants (Wu et al., 2019b). Notably, HSP90 can interact with Mg-ProtoIX in an in vitro pull-down assay (Kindgren et al., 2011). Further studies revealed that the binding of Mg-ProtoIX to HSP90 can inhibit the ATPase activity of the HSP90 chaperone. In addition, knock-down of HSP90 by RNA interference partially suppresses the gun phenotype of the gun5 mutant (Kindgren et al., 2012a). The model derived from these findings suggests that Mg-ProtoIX acts as a negative signal that exits from stressed plastids (Ankele et al., 2007) and binds HSP90 to inhibit its activity (Figure 1). In the gun5 mutant, the reduced contents of Mg-ProtoIX (and Mg-ProtoIX-ME) result in less inhibition of HSP90 activity and elevated expression of PhANGs, since HSP90 activity is positively correlated with the expression of PhANGs (Wu et al., 2019b).
The cytosolic chaperone HSP90 participates in both the TPB and PGE pathways of retrograde signaling (Kindgren et al., 2011, 2012a; Wu et al., 2019b), and also in various other signaling pathways in yeast, animals, and plants (Picard et al., 1990; Cutforth and Rubin, 1994; Kimura et al., 1995; Zhang et al., 2015a; Wang et al., 2016). Thus, HSP90 may represent a central cytosolic transducer of plastid retrograde signals (Wu et al., 2019b). Intriguingly, the efficient import of several key enzymes of the TPB pathway is dependent on GUN1 function, providing a potential mechanistic explanation for the role of GUN1 in retrograde regulation by TPB-derived signals (Figure 2, B; Wu et al., 2019b). Inhibition of the PGE pathway induces expression of chloroplast chaperones (e.g. ClpB3 and cpHSC70-2), presumably to increase protein folding activity in the plastid. This, in turn, improves the solubility of deoxyxylulose 5-phosphate synthase (DXS), the enzyme that catalyzes the initial (and rate-limiting) step of the methylerythritol 4-phosphate (MEP) pathway of isoprenoid biosynthesis, and increases the resistance of seedlings to treatment with inhibitors of the MEP pathway (Llamas et al., 2017). Interestingly, Lin treatment leads to increased resistance to MEP pathway inhibitors in wild-type Arabidopsis seedlings, but not in the gun1 mutant, despite similar transcriptional induction of the chloroplast chaperones in the two genotypes (Llamas et al., 2017). This finding is consistent with the hypothesis that GUN1 acts as a pivotal component of global plastid proteostasis. Taken together, the protein import model of GUN1 function suggests straightforward explanations for (i) how GUN1 regulates plastid proteostasis (Llamas et al., 2017), by facilitating the import of key chaperones, and (ii) the hypersensitivity of gun1 mutants to Lin and NF, and many other stress treatments (Cottage et al., 2010; Llamas et al., 2017; Song et al., 2018; Zhao et al., 2018; Wu et al., 2019b).
The finding that GUN1 regulates plastid proteostasis gained further support from the proteomic comparison of the wild type and the gun1 mutant upon Lin treatment (Wu et al., 2019a). Although the wild-type and gun1 mutant proteomes are relatively similar under unstressed conditions, they show dramatic differences when the seedlings are grown in the presence of Lin. Most of the proteins displaying lower abundance in gun1 are nucleus-encoded plastid proteins, including many proteins implicated in diverse processes in protein metabolism like translation, folding, post-translational modification, translocation, and degradation (Wu et al., 2019a). These proteins represent possible clients whose import is regulated by GUN1.
A recent study has shown that the gun1 mutation aggravates the phenotype of the fug1 mutant (defective in a nucleus-encoded plastid translation initiation factor) under normal growth conditions and under cold stress (Marino et al., 2019). However, plastid translation activity is similar in the gun1 fug1 double mutant and the fug1 single mutant. One possible explanation may call upon inefficient protein import in the gun1 fug1 double mutant, which would exacerbate the imbalance in plastid proteostasis already seen in the fug1 single mutant. Interestingly, GUN1 regulates protein import only upon interference with retrograde signaling (Wu et al., 2019b), which is consistent with the idea that GUN1 does not represent a constitutive component of the protein import apparatus, but rather functions as a conditional modulator of import and, consequently, proteostasis. In fact, GUN1 has no role under harmonious conditions (when it is rapidly degraded by the Clp protease; Wu et al., 2018) and the gun1 mutant is undistinguishable from the wild type, but functions as an important regulator when plastid proteostasis is disturbed. Such disturbances may result from genetic lesions, including mutations in genes for Clp subunits (clpc1, clpr1), chHSC70-1, components of the Translocon at the Inner (or Outer) Chloroplast membrane (tic40, toc33 [also called plastid protein import 1, ppi1] and toc159 [also called ppi2]; Kakizaki et al., 2009; Wu et al., 2019b; Tadini et al., 2020), plastid ribosomal proteins (prpl11, prpl24, and prps17; Tadini et al., 2016; Paieri et al., 2018), plastid translation initiation factors (fug1; Marino et al., 2019), FtsH proteases (var1 and var2; Marino et al., 2019; Tadini et al., 2020), the nucleus-encoded plastid RNA polymerase (NEP; sca3-1; Supplemental Table S1; Tadini et al., 2020) as well as stress resulting from Lin and NF treatment or environmental cues such as cold stress (Marino et al., 2019; Wu et al., 2019b). The protein import-related function of GUN1 was recently confirmed by an independent study (Tadini et al., 2020).
Open questions related to the gun phenotype in gun1 and the import model of GUN1 function
An unresolved issue in the import model of retrograde signaling control by GUN1 relates to the, at first glance, puzzling observation that mutants for core subunits of the chloroplast protein import apparatus do not show a gun phenotype upon Lin or NF treatment. Mutants that have been tested include ppi1 (defective in the TOC component TOC33), toc75-III-3, and tic40-4 (Wu et al., 2019b). All three mutants show a pale green phenotype (Jarvis et al., 1998; Kovacheva et al., 2005; Huang et al., 2011). TOC33 was shown to be particularly important for the import of photosynthesis-related proteins. Considering that tic and toc mutants do not show a classic gun phenotype, whether preprotein accumulation (and induction of HSP90) in these mutants occurs under conditions that interfere with retrograde signaling (e.g. Lin or NF treatment) should be investigated. In particular, preproteins that are known to accumulate in gun1 upon Lin and NF treatment (e.g. preClpD) should be analyzed to see if they also accumulate in these (and potentially other) tic and toc mutants.
Proteomic analysis of the ppi2 mutant (lacking TOC159 function) revealed that several plastid preproteins accumulate in the cytosol of ppi2. In addition, HSP90.1 was significantly upregulated at the protein level in the ppi2 mutant (Bischof et al., 2011), somewhat resembling gun1 mutants upon Lin or NF treatment. Inhibition of plastid protein import in the ppi2 mutant was shown to repress PhANG expression (Kakizaki et al., 2009), possibly because the loss of TOC159 disrupts general chloroplast functionality, and may consequently prevent the emission of the (positive) retrograde signal. Interestingly, introduction of the gun1 mutation into the ppi2 background partially restores PhANG expression (Kakizaki et al., 2009). It will be worth examining whether the HSP90 and HSP70 chaperones accumulate to higher levels in the gun1 ppi2 double mutant relative to the ppi2 single mutant.
Reduced PhANG expression in the ppi2 mutant is not mediated by the nucleus-encoded transcription factor ABISCISIC ACID INSENSITIVE 4 (ABI4, which, according to a recent study, is unlikely to have a role in plastid retrograde signaling; Kacprzak et al., 2019), but rather by the GOLDEN2-LIKE1 (GLK1) transcription factor. GLK transcriptional activators positively regulate the expression of a large number of PhANGs in plants (Yasumura et al., 2005; Waters et al., 2009). The expression of GLK1 is upregulated in the gun1 ppi2 double mutant compared with the ppi2 single mutant, possibly suggesting that GLK1 represents a master regulator of plastid retrograde signaling to control PhANG expression in the nucleus (Table 2).
Table 2.
Nuclear transcriptional regulators with proposed roles in plastid retrograde signaling
| TF | Proposed function and key evidence | Possible problems | Reference |
|---|---|---|---|
| ABI4 | Binds CCAC motifs and inhibits the binding of light-responsive factors to the G-box cis-element. | The function of ABI4 in RS has since been questioned. | (Koussevitzky et al., 2007; Kacprzak et al., 2019) |
| PTM | The C-terminal part of PTM travels to the nucleus and activates ABI4. | The ptm mutant does not show a gun phenotype in independent studies. | (Sun et al., 2011; Page et al., 2017) |
| HY5 | Binds to the promoter of PhANGs and activates their expression. The hy5 mutant does not respond to Mg-ProtoIX feeding to inhibit PhANG expression. | – | (Kindgren et al., 2012a) |
| GLK1 | Binds to the promoter of PhANGs and activates their expression. OE of GLK1 produces a gun phenotype. | – | (Kakizaki et al., 2009) |
TF, transcription factor; RS, retrograde signaling; OE, overexpression.
Somewhat counterintuitively, the phenotype of the gun1 ppi2 double mutant is aggravated in comparison to the ppi2 single mutant, and this despite restored PhANG expression in the gun1 ppi2 double mutant (Kakizaki et al., 2009). This observation may indicate that, in this genetic background, the functional significance of GUN1 is not related to the repression of PhANG expression, but rather to the maintenance of proteostasis by, for example, promotion of the import of relevant nucleus-encoded proteins. Since PhANG expression is positively correlated with HSP90 activity, it seems possible that the increased expression of PhANGs (i.e. the gun phenotype) in the gun1 mutant merely represents a secondary consequence of impaired protein import and the resulting overaccumulation of preproteins and HSP90. If this scenario can be bolstered by additional experimental data, it would question the proposed role of GUN1 as central integrator of plastid retrograde signaling. Instead, GUN1 would solely act as a regulator of protein import and plastid proteostasis whose loss of function will cause protein folding stress in the cytosol and trigger the accumulation of HSP90.
Other proposed functions for GUN1
In addition to the cpHSC70-1 chaperone, many other putative GUN1 interacting partners have been suggested (Table 1 and Supplemental Table S1). These include the plastid ribosomal protein S1 (PRPS1) and several TPB enzymes (Tadini et al., 2016), the auxiliary subunit of plastid RNA editing complexes MULTIPLE ORGANELLAR RNA EDITING FACTOR2 (MORF2; Zhao et al., 2019a, b), the RNA polymerase NEP (Tadini et al., 2020), and even various tetrapyrroles (Shimizu et al., 2019). These interactions would lend support to the previously raised hypothesis that GUN1 may act as a scaffold protein that promotes protein complex formation in various biological contexts (Colombo et al., 2016). However, it should be noted that some of the proposed macromolecular interactions are not supported by strong experimental evidence, and the presented protein interaction assays often lack essential controls (for accepted community standards, see, e.g. Kudla and Bock, 2016; Table 1). Below, we explore some of the proposed GUN1 interactors and speculate as to how they may help explain the gun phenotype of gun1 mutants.
Is RNA editing in gun1 mutants linked to retrograde signaling?
It was recently proposed that GUN1 interacts with MORF2 and regulates plastid RNA editing (Zhao et al., 2019b). The possible link to RNA editing was postulated based on the observation that the editing efficiency at multiple sites is altered in the gun1 mutant upon NF or Lin treatment. Whether these quantitative effects on RNA editing are primary or secondary consequences of the gun1 mutation is currently unclear. RNA editing efficiency is known to be sensitive to various stresses, physiological disturbances and, especially, to inhibition of plastid protein biosynthesis (e.g. Karcher and Bock, 1998; Nakajima and Mulligan, 2001; Karcher and Bock, 2002a, 2002b). The stress sensitivity of the gun1 mutant (Cottage et al., 2010) may therefore explain the observed quantitative changes in editing efficiency upon treatment with NF and Lin.
Interestingly, MORF9, another plastid MORF protein that, similar to MORF2, affects almost all RNA editing sites in plastids (Takenaka et al., 2012) was not identified in co-immunoprecipitation experiments with GUN1 and MORF2 (Zhao et al., 2019b). This is surprising, because MORF9 interacts with MORF2, and the two proteins are believed to be jointly present in RNA editing complexes (dubbed editosomes; Takenaka et al., 2012; Zehrmann et al., 2015; Sun et al., 2016). In this context, it would be interesting to determine if morf9 mutants or MORF9-overexpression lines display a gun phenotype. RNA editing efficiency at multiple but distinct sites have been shown to be up or down in gun1 mutants upon NF or Lin treatment, which makes it somewhat difficult to propose a consistent model of how these editing events may correlate with retrograde signaling. Moreover, the differences in RNA editing between gun1 and the wild type are very different upon Lin versus NF treatment (with only four editing sites showing similar trends). Compared with gun1 mutants, MORF2-overexpression lines show dramatically impaired RNA editing in plastids, but develop only a relatively mild gun phenotype (Zhao et al., 2019b), possibly indicating that the editing changes are not directly related to retrograde signaling. More importantly, the gun phenotype of MORF2-overexpression lines can only be seen when they are treated with NF, but not with Lin (Zhao et al., 2019b), which is difficult to reconcile with the proposed central function of GUN1 in multiple retrograde signaling pathways (Koussevitzky et al., 2007; Hernández-Verdeja and Strand, 2018). In summary, the effects of the gun1 mutation on plastid RNA editing represent an interesting observation, but the underlying mechanistic basis and functional significance remain to be elucidated.
Can interaction between GUN1 and NEP explain the gun phenotype?
Interestingly, mRNAs transcribed by NEP are induced in the wild type treated with Lin, a response that is compromised in the gun1 mutant (Wu et al., 2019b; Tadini et al., 2020). GUN1 was found to physically interact with NEP and is postulated to promote NEP activity (Tadini et al., 2020). This proposed function would provide a reasonable explanation for the observed differential expression of NEP-transcribed genes. However, the induction of NEP-dependent transcripts and the difference between the wild type and the gun1 mutant are seen only upon Lin treatment, but not upon NF treatment (Wu et al., 2019b). Furthermore, a mutation inactivating the NEP-encoding gene RpoTp (in the sca3-1 mutant) does not lead to a gun phenotype (Tadini et al., 2020). These findings suggest that neither reduced NEP activity nor reduced expression of NEP-dependent genes are the cause of the gun phenotype of the gun1 mutant.
Does GUN1 specifically bind tetrapyrroles?
A recent study suggests that GUN1 may bind various intermediates in TPB (heme, ProtoIX, and Mg-ProtoIX) and activates the enzyme FC1 (Shimizu et al., 2019). The seemingly appealing aspect of this hypothesis is that it potentially links GUN1 function to the TPB pathway and, as such, would connect it to the functions of all other classical GUN factors (GUN2-GUN6). The effects of GUN1 on the flux through the tetrapyrrole pathway were determined in dark-grown seedlings (Shimizu et al., 2019). The use of dark-grown material was justified by the claim that GUN1 is rapidly degraded in white light (Shimizu et al., 2019), which represents a misunderstanding of the study (Wu et al., 2018) cited in support of this claim. Indeed, the cited work demonstrated the dependence of GUN1 turnover on the developmental status of the plastid, and not on light quality or quantity. Moreover, a significant accumulation of protochlorophyllide (Pchlide) in gun1 mutants can only be seen upon exogenous ALA feeding. By contrast, in the absence of exogenous ALA application, a difference in Pchlide levels relative to the wild type is detectable only in the weak allele gun1-1, but not in a knock-out allele (gun1-103 T-DNA insertion mutant). Hence, Pchlide contents do not correlate with GUN1 activity, as the weak gun1-1 allele accumulates more Pchlide than the strong gun1-103 allele (Shimizu et al., 2019). Moreover, reduced Pchlide accumulation was observed only in one of the two GUN1 overexpression lines analyzed, although GUN1 transcript levels were similar in both lines.
GUN1 was further proposed to stimulate FC1 activity by enhancing substrate affinity (Shimizu et al., 2019). However, contradictory to the suggested positive effect of GUN1 on FC1 activity, the heme content in the gun1 mutant increases, while it decreases in GUN1 overexpression lines. It was further suggested that GUN1 may enhance FC1 activity via a similar mechanism to how GUN4 stimulates Mg-chelatase activity, that is by binding heme, the product of the reaction. However, GUN1 was proposed to not only bind heme as the reaction product, but also the substrate (ProtoIX) and the reaction product of the chlorophyll branch (Mg-ProtoIX; synthesized by Mg-chelatase). Furthermore, GUN1 binding to tetrapyrroles is more than 10-fold weaker than that of GUN4 to Mg-ProtoIX, thus raising questions about the possibility that GUN1 regulates FC1 activity in a similar manner.
Finally, heme-binding assays and FC1 activity assays need to be interpreted with caution, given that even bovine serum albumin (BSA) binds heme in vitro (Hargrove et al., 1996). Future attempts to verify tetrapyrrole binding to GUN1 should include stringent negative controls, including a related PPR protein and an inactive GUN1 variant. The removal of the N-terminal domain abolishes the ability of GUN1 to complement the gun phenotype of the gun1 mutant (Wu et al., 2018). This finding raises the possibility that the (N-terminally truncated) GUN1 protein variant used for the in vitro heme-binding and FC1 activity assays (Shimizu et al., 2019) was, in fact, non-functional. Thus, more work will be needed to substantiate the proposed link between GUN1 function and metabolites of the TPB pathway and, most importantly, test the intuitively appealing hypothesis that tetrapyrroles mediate altered retrograde signaling in all identified gun mutants. As discussed above, while metabolites from the chlorophyll branch of the TPB pathway were initially proposed as negative retrograde signals, later research has cast considerable doubt on this mode of regulation. Instead, heme was suggested more recently to act as positive signal based on the study of the gun6-1D mutant (Woodson et al., 2011). Overall, additional evidence from well-designed studies on the effects of TPB metabolites on the function of GUN proteins is needed to ultimately clarify the role(s) of protein–metabolite interactions in retrograde signaling.
Post-transcriptional regulation by retrograde signaling
The majority of published studies on retrograde signaling focused on the transcriptional control of nuclear genes by plastid-derived signals. Recently, post-transcriptional regulation by retrograde signals has started to draw more attention. Ferredoxin I (Fed-I) transcript levels are positively correlated with their translation by cytosolic ribosomes in tobacco (Nicotiana tabacum), presumably because their association with ribosomes in the light protects the mRNAs from ribonucleolytic degradation, an effect that has been attributed to the activity of photosynthetic electron transport (PET) in the chloroplast (Elliott et al., 1989; Dickey et al., 1998; Petracek et al., 1998). Translational responses in retrograde signaling were also studied in Arabidopsis plants shifted from low to high light, a treatment that triggers operational retrograde signaling (Oelze et al., 2014). Accumulation of GLK1, the transcription factor mentioned earlier that controls the expression of many PhANGs and participates in GUN1-dependent retrograde signaling (Kakizaki et al., 2009; Waters et al., 2009), is regulated post-translationally by protein degradation via the ubiquitin proteasome system (Tokumaru et al., 2017). Moreover, accumulation of the (nucleus-encoded) plastid ribosomal protein PRPS1 is regulated by GUN1 in a manner that is independent of PRPS1 transcript accumulation (Tadini et al., 2016).
Recently, a systematic correlation analysis between transcripts and their protein products was carried out for wild-type Arabidopsis plants and gun1 mutant plants to explore the extent of post-transcriptional regulation in plastid retrograde signaling (Wu et al., 2019a). Remarkably, while the gun1 mutant accumulates high transcript levels for many PhANGs, the corresponding PhANG proteins did not overly accumulate. Many proteins related to various steps in protein metabolism (translation, folding, modification, translocation, and degradation) accumulate to lower levels in the gun1 mutant upon Lin treatment, supporting a pivotal function for GUN1 in maintenance of plastid proteostasis under these conditions (Wu et al., 2019a). Highly translationally or post-translationally regulated (HiToP) genes, whose regulation at the protein level is much stronger than at the transcript level, were also identified in the wild type. These findings indicate that post-transcriptional regulation likely represents a general mechanism employed by plants during retrograde communication between plastids and the nucleus. Interestingly, HiToP PhANGs are regulated translationally in a manner dependent on the action of GUN1 (Figure 2, B), whereas HiToP-encoded ribosomal proteins are regulated post-translationally and likely independently of GUN1 function (Wu et al., 2019a).
Other biogenic retrograde signals and their interaction with GUN proteins
The identification of the genes GUN2 to GUN6 pointed to a close link between photosynthetic pigment biosynthesis and PhANG expression in the nucleus. The herbicide NF used in the initial genetic screens for gun mutants inhibits phytoene desaturase, an enzyme involved in carotenoid biosynthesis (Oelmüller, 1989). Remarkably, several compounds derived from carotenoid metabolism (including carotenoid cleavage products, so-called apocarotenoids) have been demonstrated to act as signaling molecules that regulate chloroplast biogenesis and leaf development (Avendano-Vazquez et al., 2014). For example, in the ζ-carotene desaturase/chloroplast biogenesis5 (zds/clb5) mutant, defective carotenoid biosynthesis results in the repression of PhANGs and several plastid-encoded genes (rpo genes, clpP1), thus blocking chloroplast biogenesis. Mutant characterization provided evidence for a signaling role of a phytofluene and/or ζ-carotene-derived apocarotenoid in the control of chloroplast biogenesis (Avendano-Vazquez et al., 2014). A recent study with the carotenoid biosynthesis mutant carotenoid chloroplast regulation2 (ccr2) suggests that apocarotenoid(s) derived from cis-carotenes regulate both etioplast development and chloroplast biogenesis (Cazzonelli et al., 2020). Whether there is crosstalk between apocarotenoid signals and tetrapyrrole signals, or any other interaction of carotenoid-derived signals with the GUN-dependent pathways of retrograde communication, is currently unknown.
The redox state of the PET chain actively participates in the regulation of nuclear gene expression in response to altered chloroplast homeostasis, as triggered by environmental cues such as excess light (operational control). The prin2 mutant, identified through a forward genetic approach, shows misregulated PhANG expression under high light or when treated with PET inhibitors, suggesting that PRIN2 participates in retrograde control via redox signaling (Kindgren et al., 2012b). The PRIN2 gene encodes a protein associated with PEP, and itself is a redox-regulated protein. In the prin2 mutant, the expression of plastid-encoded genes that are transcribed by PEP is disturbed and chloroplast biogenesis is impaired (Díaz et al., 2018), thus revealing a potential link between operational control and biogenic control. Interestingly, PhANG expression in the prin2 mutant is rescued by the introduction of the gun1 mutation, although PEP-dependent PGE is not restored (Kindgren et al., 2012b), confirming that GUN1 is not involved in the regulation of (PEP-dependent) PGE.
Biogenic control and retrograde signaling in non-green plastids
In contrast to chloroplast development, the biogenesis of non-green plastids (e.g. chromoplasts in flowers and fruits, amyloplasts in tubers and roots) is much less studied. Also, very little is known about the role of biogenic retrograde signals during the differentiation of non-photosynthetic plastid types. The differentiation of carotenoid-storing chromoplasts is regulated by multiple environmental factors and developmental cues (SadaLi et al., 2019). Studies on chromoplast development during fruit ripening in tomato (Solanum lycopersicum) have revealed massive reprogramming of the nuclear transcriptome during chloroplast-to-chromoplast conversion (Vrebalov et al., 2002; Klee and Giovannoni, 2011; Matas et al., 2011; Nguyen et al., 2014; Shinozaki et al., 2018).
The tomato plastid genome encodes 114 genes, comprising 61 genetic system genes (encoding components of the transcriptional apparatus and the translation machinery), 47 photosynthesis-related genes, and a few other genes and conserved open-reading frames (Kahlau et al., 2006). Chromoplast differentiation is accompanied by the loss of photosynthetic activity and the stimulation of carotenoid and lipid biosynthesis (Pesaresi et al., 2014). During chromoplast development, plastid photosynthesis-related genes are largely repressed. By contrast, the expression of accD, the only plastid gene involved in lipid biosynthesis (encoding a subunit of acetyl-CoA carboxylase, the rate-limiting enzyme of fatty acid biosynthesis), is upregulated (Kahlau and Bock, 2008). Whether this distinct gene expression profile serves as a source of a retrograde signal that contributes to the adjustment of nuclear gene expression to the needs of chromoplast formation is not known, but given the availability of plastid genome engineering in tomato (Ruf et al., 2001), this question is addressable by conditional knock-down of accD expression during fruit ripening (Verhounig et al., 2010).
It was recently shown that overexpression of a bacterial phytoene synthase (crtB), the enzyme that catalyzes the committed step of carotenoid biosynthesis, can induce the transition from chloroplasts to chromoplasts in various tissues of different plant species (Llorente et al., 2020). This is likely due to the reduction of chloroplast photosynthetic competence as caused by excessive metabolic flux into carotenoid biosynthesis. It seems conceivable that a retrograde signal emitted from carotenoid-accumulating plastids may reprogram nuclear gene expression to support chromoplast formation. β-Cyclocitral, an apocarotenoid produced upon carotenoid degradation, was identified as an operational retrograde signal (D’Alessandro et al., 2018) in response to excess light. β-Cyclocitral was also shown to participate in developmental processes such as root branching and lateral root development (Van Norman et al., 2014; Dickinson et al., 2019). However, whether carotenoids and/or their degradation products also act in the biogenic control of plastid differentiation remains to be established.
Interaction of plastid retrograde signals with light signals and developmental signals
One of the most challenging tasks for plants is to manage the transition from skotomorphogenesis (development in the dark, before etiolated seedlings reach the soil surface) to photomorphogenesis (light-mediated development). Exposure to light requires the rapid generation of photosynthetically active chloroplasts to facilitate autotrophic growth while, at the same time, avoiding photooxidative damage from reactive oxygen species that are produced by an incompletely assembled photosynthetic machinery. It has long been established that plastid-derived biogenic retrograde signals cross-talk with light signals during photomorphogenesis (Pogson et al., 2015). Of the five GUN loci identified through the original genetic screen (Susek et al., 1993), GUN2 (encoding heme oxygenase 1) and GUN3 (encoding phytochromobilin synthase) are also known as ELONGATED HYPOCOTYL 1 (HY1) and HY2, respectively (Figure 1). Because the defects in the gun2 and gun3 mutants affect the biosynthesis of phytochromobilin, the chromophore of the red and far-red light photoreceptors phytochromes (Bae and Choi, 2008; Li et al., 2011), both hy1 and hy2 were initially identified as key components of light signaling pathways (Chory et al., 1989; Parks and Quail, 1991). Their later identification as gun mutants indicated a close link between light and retrograde signaling (Ruckle et al., 2012; Larkin, 2014).
It was suggested that the nucleus-encoded transcription factor ABI4 responds to plastid retrograde signals and mediates downstream transcriptional responses. ABI4 binds to the cis-element CCAC, which is often found in close proximity to a G-box (ACGT), the cis-element that acts as binding site for transcription factors conferring light-regulated gene expression (e.g. HY5; Lee et al., 2007). It was proposed that ABI4 directly competes with the transcription factors mediating light regulation, thereby suppressing PhANG expression (Koussevitzky et al., 2007). Although seemingly appealing, this model has been called into question by the finding that the abi4 mutant does not show a gun phenotype (Kacprzak et al., 2019; Mhamdi and Gommers, 2019). In a second genetic screen for additional gun mutants (using the more sensitive reporter gene luciferase), mutations in CRYPTOCHROME 1 (CRY1), the blue-light photoreceptor, were identified as gun mutants. Loss of cryptochrome function was suggested to confer repression of PhANG expression by converting HY5 from a positive to a negative regulator of PhANG transcription (Figure 3; Ruckle et al., 2007; Ruckle and Larkin, 2009).
Figure 3.
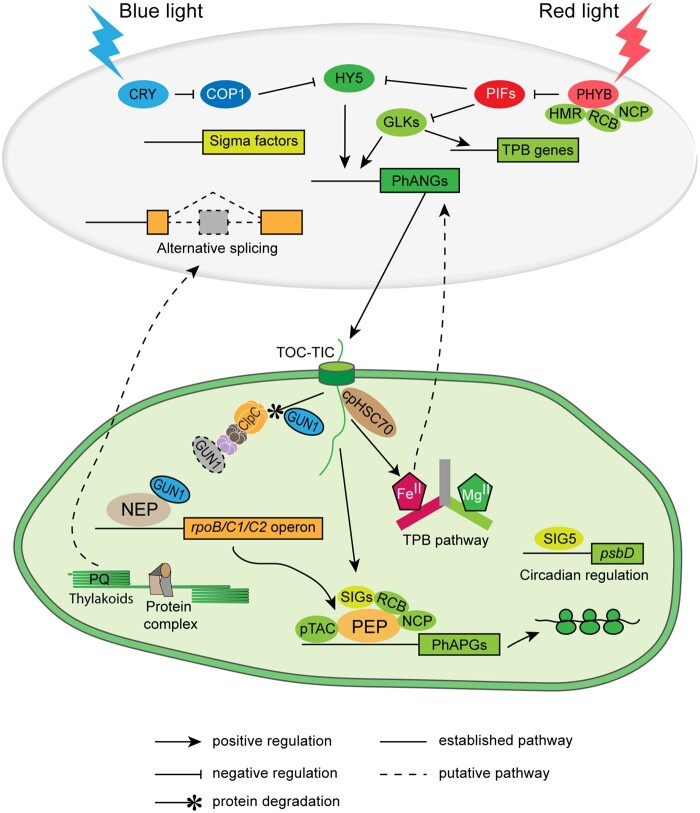
Interaction of light and plastid signals during chloroplast biogenesis. In the dark, active plastid-encoded genes are mainly transcribed by NEP. GUN1 was reported to directly bind NEP and promote the expression of NEP-dependent genes upon inhibited plastid translation (Tadini et al., 2020). Upon illumination, the activated form of blue-light and red-light photoreceptors (CRY and phy) degrade negative regulators (COP1 and PIFs, respectively) of photomorphogenesis, thus activating the expression of positive transcriptional regulators (e.g. HY5 and GLKs), thereby promoting the expression of PhANGs and TPB genes. The PhANG-encoded proteins and TPB enzymes are imported into plastids (proplastids, etioplasts, and etiochloroplasts) through the TOC-TIC apparatus. GUN1 facilitates protein import during chloroplast biogenesis, and is degraded by the Clp protease when chloroplast biogenesis is complete. PEP is the major RNA polymerase in chloroplasts, and its activity requires the plastid-encoded core components RpoA, RpoB, RpoC1, and RpoC2, and nucleus-encoded accessory factors, including bacterial-type Sigma factors (SIGs) for promoter recognition. Other nucleus-encoded factors support complex assembly (e.g. pTACs [PAP8, HMR], REGULATOR OF CHLOROPLAST BIOGENESIS [RCB], and NUCLEAR CONTROL OF PEP ACTIVITY [NCP]) and regulate PEP activity, for example, PLASTID REDOX INSENSITIVE2 (PRIN2) that is involved in redox regulation of PEP activity (Díaz et al., 2018). The nucleus-encoded Sigma factor SIG5 communicates circadian information from the nucleus to the chloroplast and controls expression of at least 12% of all chloroplast-encoded genes (Noordally et al., 2013). Chloroplast-derived signals promote PhANG expression through positive signaling molecules such as heme, regulate alternative splicing of transcripts from nuclear genes (through the redox status of the plastoquinone pool, PQ; Petrillo et al., 2014), and facilitate nuclear body-localization of the red-light photoreceptor phyB and the degradation of negative regulators (PIFs; Chen et al., 2010; Galvao et al., 2012; Yang et al., 2019; Yoo et al., 2019).
The finding that gun1 mutants display delayed greening when etiolated seedlings are illuminated (Mochizuki et al., 1996; Wu et al., 2019a) suggests an interaction between light signals and plastid signals during de-etiolation. A mechanistic model was proposed in which a retrograde signal transduced by GUN1 in concert with PHD-TYPE TRANSCRIPTION FACTOR WITH TRANSMEMBRANE DOMAINS (PTM) and ABI4 modulates the light signal transduced by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and HY5 (Xu et al., 2016). However, the GUN1-PTM-ABI4 model has been questioned by two independent studies that show that neither ptm nor abi4 mutants have a gun phenotype (Page et al., 2017; Kacprzak et al., 2019; Table 2). The delayed greening of gun1 mutant seedlings during de-etiolation may be instead explained by a slower import of nucleus-encoded plastid proteins (Wu et al., 2019b).
Retrograde signaling is also mechanistically connected to red and far-red light perception. The localization of phytochrome B (phyB), the major red light photoreceptor in Arabidopsis, to nuclear bodies, is regulated by HEMERA (HMR, also named PLASTID TRANSCRIPTIONALLY ACTIVE CHROMOSOME PROTEIN 12, pTAC12; Chen et al., 2010). HMR appears to be dually localized to plastids and the nucleus (Figure 3; Nevarez et al., 2017). The dual localization of HMR, and possibly other proteins that participate in PEP complex assembly (including REGULATOR OF CHLOROPLAST BIOGENESIS [RCB], NUCLEAR CONTROL OF PEP ACTIVITY [NCP], and PAP8; Figure 3 and Supplemental Table S1; Yang et al., 2019; Yoo et al., 2019; Liebers et al., 2020), suggests a link between the PGE pathway of retrograde signaling and phyB-mediated light signaling. Recently, the ALBOSTRIANS gene in barley (Hordeum vulgare) was found to encode the transcription factor CONSTANS, CO-LIKE, and TOC1 (CCT) MOTIF FAMILY 7 (HvCMF7; Li et al., 2019). HvCMF7 and its Arabidopsis ortholog CHLOROPLAST IMPORT APPARATUS 2 (CIA2; Gawronski et al., 2020) localize to both chloroplasts and the nucleus and affect chloroplast biogenesis, possibly by regulating plastid ribosome assembly. The growing number of transcriptional regulator proteins identified as being dually localized may well turn out to provide a list of important players in intracellular communication.
The plastid-derived retrograde signaling metabolite methylerythritol cyclodiphosphate (MEcPP) was proposed to participate in operational control of nuclear gene expression (Xiao et al., 2012). MEcPP influences hypocotyl length under red light by regulating phyB protein levels in a calcium-dependent manner, via CALMODULIN-BINDING TRANSCRIPTIONAL ACTIVATOR 3 (CAMTA3; Jiang et al., 2019, 2020). GLK1, a key transcriptional regulator of photomorphogenesis (Fitter et al., 2002; Waters et al., 2008, 2009), is also a major nuclear regulator of plastid retrograde signals (Kakizaki et al., 2009). The GLK1 locus is also a direct target of PHYTOCHROME-INTERACTING FACTOR4 (PIF4; Oh et al., 2012), one of the main repressors of photomorphogenesis regulated by phytochrome (Bae and Choi, 2008; Jeong and Choi, 2013). It was proposed that, during photomorphogenesis, phytochrome-mediated degradation of PIFs releases the repression of GLK1 expression, thus promoting the expression of photosynthesis-related genes (Martin et al., 2016). Under stress conditions and when plastids are dysfunctional, GLK1 expression will be repressed in a GUN1-dependent manner (a negative signal was proposed by the authors), antagonizing the phytochrome signal and attenuating photomorphogenesis (Martin et al., 2016).
In addition to cross-talk with photoreceptor-mediated light signals, chloroplast signals are also involved in rhythmic gene expression. For example, the plastid genome-encoded psbD transcripts are regulated by the circadian clock via the sigma factor SIG5, a nucleus-encoded component of the PEP complex (Noordally et al., 2013; Figure 3), and the diurnal variation in thermotolerance and alternative splicing of nucleus-encoded genes was shown to be dependent on chloroplast-derived signals (presumably the redox status of the plastoquinone pool; Petrillo et al., 2014; Dickinson et al., 2018).
Outlook
The major challenge in the field of plastid retrograde signaling will be to obtain a complete picture of individual signaling pathways. At present, our understanding of retrograde signaling is rather fragmented. It is clear that several pathways exist and a few of their constituents have been identified. What is lacking in all cases is a complete logic chain connecting the signal generator in the chloroplast to the signal receiver in the nucleus. Of key importance will be the identification of the transducer molecules that travel between compartments or relay the signal across the double membrane of the plastid. Previous studies have yielded conflicting evidence with respect to both the TPB intermediates that may act as signals and the nature of the signal (positive or negative). An FC1-specific heme pool was proposed as a positive signal (Woodson et al., 2011). A future challenge will be to identify the parts of the signaling chain that link heme with PhANG expression in various environmental conditions and developmental states.
The recent determination of the accumulation pattern of GUN1 protein and the identification of the cytosolic transducer HSP90 (Wu et al., 2018, 2019b) have provided fresh entry points into the study of GUN1 molecular function(s) and its role in retrograde signaling. Future studies should be directed toward the elucidation of the precise mechanism by which GUN1 affects plastid protein import and proteostasis in response to environmental stimuli and developmental cues. Finally, the exact mechanism that underlies HSP90 regulation of PhANG expression in cooperation with the gene expression machinery in the nucleus remains to be elucidated.
Supplemental data
Supplemental Table S1. Accession numbers of the major genes mentioned in this article.
Funding
Research on retrograde signaling in the authors’ laboratories has been financed by grants from the National Natural Science Foundation of China (NSFC; 32070299), the Shanghai Pujiang Program (20PJ1405600), the Bio-X Interdisciplinary Fund of Shanghai Jiao Tong University (20CX-04 to G.-Z.W), and the Max Planck Society and grants from the Deutsche Forschungsgemeinschaft (FOR 804, BO1482/15-2, SFB-TR 175 to R.B.).
Conflict of interest statement. None declared.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are: Guo-Zhang Wu (gzwu@sjtu.edu.cn) and Ralph Bock (rbock@mpimp-golm.mpg.de).
References
- Ankele E, Kindgren P, Pesquet E, Strand Å (2007) In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19: 1964–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM (2009) The puzzle of plastid evolution. Curr Biol 19: R81–R88 [DOI] [PubMed] [Google Scholar]
- Avendano-Vazquez AO, Cordoba E, Llamas E, San Roman C, Nisar N, De la Torre S, Ramos-Vega M, Gutierrez-Nava MD, Cazzonelli CI, Pogson BJ, et al. (2014) An uncharacterized apocarotenoid-derived signal generated in zeta-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26: 2524–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Barajas-Lopez JdD, Blanco NE, Strand Å (2013) Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833: 425–437 [DOI] [PubMed] [Google Scholar]
- Bischof S, Baerenfaller K, Wildhaber T, Troesch R, Vidi PA, Roschitzki B, Hirsch-Hoffmann M, Hennig L, Kessler F, Gruissem W, et al. (2011) Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins. Plant Cell 23: 3911–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R (2017) Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu Rev Genet 51: 1–22 [DOI] [PubMed] [Google Scholar]
- Bock R, Timmis JN (2008) Reconstructing evolution: gene transfer from plastids to the nucleus. Bioessays 30: 556–566 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Hou X, Alagoz Y, Rivers J, Dhami N, Lee J, Marri S, Pogson BJ (2020) A cis-carotene derived apocarotenoid regulates etioplast and chloroplast development. eLife 9: e45310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chen M, Galvao RM, Li M, Burger B, Bugea J, Bolado J, Chory J (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P (2016) GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front Plant Sci 7: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC (2010) The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J Exp Bot 61: 3773–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S (2011) Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol Biol Evol 28: 3019–3032 [DOI] [PubMed] [Google Scholar]
- Cutforth T, Rubin GM (1994) Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell 77: 1027–1036 [DOI] [PubMed] [Google Scholar]
- D’Alessandro S, Ksas B, Havaux M (2018) Decoding beta-cyclocitral-mediated retrograde signaling reveals the role of a detoxification response in plant tolerance to photooxidative stress. Plant Cell 30: 2495–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza A, Wang J-Z, Dehesh K (2017) Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol 68: 85–108 [DOI] [PubMed] [Google Scholar]
- Díaz MG, Hernández-Verdeja T, Kremnev D, Crawford T, Dubreuil C, Strand Å (2018) Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat Commun 9: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey LF, Petracek ME, Nguyen TT, Hansen ER, Thompson WF (1998) Light regulation of Fed-1 mRNA requires an element in the 5′-untranslated region and correlates with differential polyribosome association. Plant Cell 10: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson AJ, Lehner K, Mi J, Jia KP, Mijar M, Dinneny J, Al-Babili S, Benfey PN (2019) β-Cyclocitral is a conserved root growth regulator. Proc Natl Acad Sci U S A 116: 10563–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson PJ, Kumar M, Martinho C, Yoo SJ, Lan H, Artavanis G, Charoensawan V, Schottler MA, Bock R, Jaeger KE, et al. (2018) Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Rep 22: 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan J, Gassman M (1974) Induction of porphyrin synthesis in etiolated bean leaves by chelators of iron. Plant Physiol 53: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Dickey LF, White MJ, Thompson WF (1989) Cis-acting elements for light regulation of Pea Ferredoxin I gene expression are located within transcribed sequences. Plant Cell 1: 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Gadjieva R, Axelsson E, Olsson U, Hansson M (2005) Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol Biochem 43: 901–908 [DOI] [PubMed] [Google Scholar]
- Galvao RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M (2012) Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev 26: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski P, Burdiak P, Scharff LB, Mielecki J, Gorecka M, Zaborowska M, Leister D, Waszczak C, Karpinski S (2020) CIA2 and CIA2-LIKE are required for optimal photosynthesis and stress responses in Arabidopsis thaliana. Plant J (doi: 10.1111/tpj.15058) [DOI] [PubMed] [Google Scholar]
- Grubler B, Merendino L, Twardziok SO, Mininno M, Allorent G, Chevalier F, Liebers M, Blanvillain R, Mayer KFX, Lerbs-Mache S, et al. (2017) Light and plastid signals regulate different sets of genes in the albino mutant Pap7-1. Plant Physiol 175: 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove MS, Barrick D, Olson JS (1996) The association rate constant for heme binding to globin is independent of protein structure. Biochemistry 35: 11293–11299 [DOI] [PubMed] [Google Scholar]
- Hernández-Verdeja T, Strand Å (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricova A, Quesada V, Micol JL (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Xu F, Guan L, Qian P, Liu Y, Zhang H, Huang Y, Hou S (2014) The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in Arabidopsis. J Exp Bot 65: 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Ling Q, Bédard J, Lilley K, Jarvis P (2011) In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol 157: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Chen LJ, Li H, Peto CA, Fankhauser C, Chory J (1998) An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Jeong J, Choi G (2013) Phytochrome-interacting factors have both shared and distinct biological roles. Mol Cells 35: 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Tian H, Zhang S, Ding Z, Ma C (2019) GUN1-interacting proteins open the door for retrograde signaling. Trends Plant Sci 24: 884–887 [DOI] [PubMed] [Google Scholar]
- Jiang J, Xiao Y, Chen H, Hu W, Zeng L, Ke H, Ditengou FA, Devisetty U, Palme K, Maloof J, et al. (2020) Retrograde induction of phyB orchestrates ethylene–auxin hierarchy to regulate growth. Plant Physiol 183: 1268–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zeng L, Ke H, De La Cruz B, Dehesh K (2019) Orthogonal regulation of phytochrome B abundance by stress-specific plastidial retrograde signaling metabolite. Nat Commun 10: 2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanningmeier U, Howell S (1984) Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. Possible involvement of chlorophyll synthesis precursors. J Biol Chem 259: 13541–13549 [PubMed] [Google Scholar]
- Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ (2019) Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol 179: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Aspinall S, Gray JC, Bock R (2006) Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J Mol Evol 63: 194–207 [DOI] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol 151: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizaki T, Yazu F, Nakayama K, Ito-Inaba Y, Inaba T (2012) Plastid signalling under multiple conditions is accompanied by a common defect in RNA editing in plastids. J Exp Bot 63: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Bock R (1998) Site-selective inhibition of plastid RNA editing by heat shock and antibiotics: a role for plastid translation in RNA editing. Nucleic Acids Res 26: 1185–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Bock R (2002a) Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr Genet 41: 48–52 [DOI] [PubMed] [Google Scholar]
- Karcher D, Bock R (2002b) The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J Biol Chem 277: 5570–5574 [DOI] [PubMed] [Google Scholar]
- Keeling PJ (2013) The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64: 583–607 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yahara I, Lindquist S (1995) Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Eriksson MJ, Benedict C, Mohapatra A, Gough SP, Hansson M, Kieselbach T, Strand Å (2011) A novel proteomic approach reveals a role for Mg-protoporphyrin IX in response to oxidative stress. Physiol Plant 141: 310–320 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Kremnev D, Blanco NE, de Dios Barajas Lopez J, Fernandez AP, Tellgren-Roth C, Kleine T, Small I, Strand Å (2012b) The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J 70: 279–291 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Noren L, Lopez Jde D, Shaikhali J, Strand Å (2012a) Interplay between heat shock protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol Plant 5: 901–913 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim IJ, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Kovacheva S, Bedard J, Patel R, Dudley P, Twell D, Rios G, Koncz C, Jarvis P (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rudiger W, Beck CF (1997) Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci U S A 94: 14168–14172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Bock R (2016) Lighting the way to protein–protein interactions: recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell 28: 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO (1998) Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol 201: 1177–1195 [DOI] [PubMed] [Google Scholar]
- La Rocca N, Rascio N, Oster U, Rüdiger W (2001) Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213: 101–108 [DOI] [PubMed] [Google Scholar]
- Larkin RM (2014) Influence of plastids on light signalling and development. Phil Trans R Soc Lond B Biol Sci 369: 20130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM, Alonso JM, Ecker JR, Chory J (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Deng XW (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hensel G, Mascher M, Melzer M, Budhagatapalli N, Rutten T, Himmelbach A, Beier S, Korzun V, Kumlehn J, et al. (2019) Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 31: 1430–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebers M, Gillet FX, Israel A, Pounot K, Chambon L, Chieb M, Chevalier F, Ruedas R, Favier A, Gans P, et al. (2020) Nucleo-plastidic PAP8/pTAC6 couples chloroplast formation with photomorphogenesis. EMBO J 39: e104941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, McNeilage RT, Shi LX, Theg SM (2014) ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas E, Pulido P, Rodriguez-Concepcion M (2017) Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet 13: e1007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Torres-Montilla S, Morelli L, Florez-Sarasa I, Matus JT, Ezquerro M, D’Andrea L, Houhou F, Majer E, Pico B, et al. (2020) Synthetic conversion of leaf chloroplasts into carotenoid-rich plastids reveals mechanistic basis of natural chromoplast development. Proc Natl Acad Sci U S A 117: 21796–21803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Naranjo B, Wang J, Penzler JF, Kleine T, Leister D (2019) Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J 99: 521–535 [DOI] [PubMed] [Google Scholar]
- Martin G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Herrmann RG (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol 118: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165 [DOI] [PubMed] [Google Scholar]
- Maruta T, Miyazaki N, Nosaka R, Tanaka H, Padilla-Chacon D, Otori K, Kimura A, Tanabe N, Yoshimura K, Tamoi M, et al. (2015) A gain-of-function mutation of plastidic invertase alters nuclear gene expression with sucrose treatment partially via GENOMES UNCOUPLED1-mediated signaling. New Phytol 206: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Gommers CMM (2019) Another gun dismantled: ABSCISIC ACID INSENSITIVE4 is not a target of retrograde signaling. Plant Physiol 179: 13–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci U S A 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Susek R, Chory J (1996) An intracellular signal transduction pathway between the chloroplast and nucleus is involved in de-etiolation. Plant Physiol 112: 1465–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci U S A 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Philippe H (1999) Smr: a bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem Sci 24: 298–300 [DOI] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG (2008) Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc Natl Acad Sci U S A 105: 15178–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulo P, Pursiheimo S, Hou CX, Tyystjarvi T, Aro EM (2003) Multiple effects of antibiotics on chloroplast and nuclear gene expression. Funct Plant Biol 30: 1097–1103 [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Mulligan RM (2001) Heat stress results in incomplete C-to-U editing of maize chloroplast mRNAs and correlates with changes in chloroplast transcription rate. Curr Genet 40: 209–213 [DOI] [PubMed] [Google Scholar]
- Nelson N, Ben-Shem A (2004) The complex architecture of oxygenic photosynthesis. Nat Rev Mol Cell Biol 5: 971–982 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Nevarez PA, Qiu Y, Inoue H, Yoo CY, Benfey PN, Schnell DJ, Chen M (2017) Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiol 173: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CV, Vrebalov JT, Gapper NE, Zheng Y, Zhong S, Fei Z, Giovannoni JJ (2014) Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26: 585–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally ZB, Ishii K, Atkins KA, Wetherill SJ, Kusakina J, Walton EJ, Kato M, Azuma M, Tanaka K, Hanaoka M, et al. (2013) Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339: 1316–1319 [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Oelmüller R (1989) Photooxidative destruction of chloroplasts and its effect on nuclear gene-expression and extraplastidic enzyme levels. Photochem Photobiol 49: 229–239 [Google Scholar]
- Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H (1986) Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168: 482–492 [DOI] [PubMed] [Google Scholar]
- Oelze ML, Muthuramalingam M, Vogel MO, Dietz KJ (2014) The link between transcript regulation and de novo protein synthesis in the retrograde high light acclimation response of Arabidopsis thaliana. BMC Genomics 15: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster U, Brunner H, Rüdiger W (1996) The greening process in cress seedlings. V. Possible interference of chlorophyll precursors, accumulated after thujaplicin treatment, with light-regulated expression of Lhc genes. J Photochem Photobiol B Biol 36: 255–261 [Google Scholar]
- Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paieri F, Tadini L, Manavski N, Kleine T, Ferrari R, Morandini P, Pesaresi P, Meurer J, Leister D (2018) The DEAD-box RNA helicase RH50 is a 23S–4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol 176: 634–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH (1991) Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi P, Kim C (2019) Current understanding of GUN1: a key mediator involved in biogenic retrograde signaling. Plant Cell Rep 38: 819–823 [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Mizzotti C, Colombo M, Masiero S (2014) Genetic regulation and structural changes during tomato fruit development and ripening. Front Plant Sci 5: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter E, Grimm B (2009) GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol Plant 2: 1198–1210 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Gatz C, Sowinski DA, Allen GC, Thompson WF (1998) Ferredoxin-1 mRNA is destabilized by changes in photosynthetic electron transport. Proc Natl Acad Sci U S A 95: 9009–9013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JW, Barta A, Kalyna M, et al. (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR (1990) Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature 348: 166–168 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Ganguly D, Albrecht-Borth V (2015) Insights into chloroplast biogenesis and development. Biochim Biophys Acta 1847: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Forster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Ponce-Toledo RI, Lpez-Garca P, Moreira D (2019) Horizontal and endosymbiotic gene transfer in early plastid evolution. New Phytol 224: 618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Albrieux C, Joyard J, Lagrange T, Block MA (2007) Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. J Biol Chem 282: 2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JC, Mullet JE (1991) Chloroplast transcription is required to express the nuclear genes rbcS and cab. Plastid DNA copy number is regulated independently. Plant Mol Biol 17: 813–823 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, Larkin RM (2009) Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol 182: 367–379 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM (2012) Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol 159: 366–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R (2001) Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol 19: 870–875 [DOI] [PubMed] [Google Scholar]
- Sadali NM, Sowden RG, Ling Q, Jarvis RP (2019) Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep 38: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kacprzak SM, Mochizuki N, Nagatani A, Watanabe S, Shimada T, Tanaka K, Hayashi Y, Arai M, Leister D, et al. (2019) The retrograde signaling protein GUN1 regulates tetrapyrrole biosynthesis. Proc Natl Acad Sci U S A 116: 24900–24906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Nicolas P, Fernandez-Pozo N, Ma Q, Evanich DJ, Shi Y, Xu Y, Zheng Y, Snyder SI, Martin LBB, et al. (2018) High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat Commun 9: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Chen Z, Larkin RM (2018) The genomes uncoupled mutants are more sensitive to norflurazon than wild type. Plant Physiol 178: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand Å, Asami T, Alonso J, Ecker JR, Chory J (2003) Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83 [DOI] [PubMed] [Google Scholar]
- Su P-H, Li H-M (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Gray JC (1999) Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR (2016) The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci 21: 962–973 [DOI] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nature Commun 2: 477. [DOI] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J (2002) Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl): S327–S338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek R, Chory J (1992) A tale of two genomes: role of a chloroplast signal in coordinating nuclear and plastid genome expression. Aust J Plant Physiol 19: 387–399 [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tadini L, Peracchio C, Trotta A, Colombo M, Mancini I, Jeran N, Costa A, Faoro F, Marsoni M, Vannini C, et al. (2020) GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J 101: 1198–1220 [DOI] [PubMed] [Google Scholar]
- Tadini L, Pesaresi P, Kleine T, Rossi F, Guljamow A, Sommer F, Mühlhaus T, Schroda M, Masiero S, Pribil M, et al. (2016) GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol 170: 1817–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Hartel B, Brennicke A (2012) Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci U S A 109: 5104–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MJ, Smith AG (2013) A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front Plant Sci 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru M, Adachi F, Toda M, Ito-Inaba Y, Yazu F, Hirosawa Y, Sakakibara Y, Suiko M, Kakizaki T, Inaba T (2017) Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol 173: 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TD, Chan KX, Thompson AJ, Benfey PN (2014) Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc Natl Acad Sci U S A 111: E1300–E1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF (2004) Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth Res 82: 289–299 [DOI] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci U S A 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinti G, Hills A, Campbell S, Bowyer J, Mochizuki N, Chory J, Lόpez-Juez E (2000) Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J 24: 883–894 [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF (2008) Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20: 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss B, Meinecke L, Kurz T, Al-Babili S, Beck CF, Hess WR (2011) Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol 155: 892–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M (2016) HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nature Commun 7: 10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J (2008) Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 9: 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J (2011) Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol 21: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J 73: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Chalvin C, Hoelscher MP, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176: 2472–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Richter AS, Schuster M, Ling Q, Schottler MA, Walther D, Zoschke R, Grimm B, Jarvis RP, et al. (2019b) Control of retrograde signalling by protein import and cytosolic folding stress. Nat Plants 5: 525–538 [DOI] [PubMed] [Google Scholar]
- Wu GZ, Meyer EH, Wu S, Bock R (2019a) Extensive post-transcriptional regulation of nuclear gene expression by plastid retrograde signals. Plant Physiol 180: 2034–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, Howell KA, Huang J (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J 85: 607–621 [DOI] [PubMed] [Google Scholar]
- Xiao YM, Savchenko T, Baidoo EEK, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Xu X, Chi W, Sun X, Feng P, Guo H, Li J, Lin R, Lu C, Wang H, Leister D, et al. (2016) Convergence of light and chloroplast signals for de-etiolation through ABI4-HY5 and COP1. Nat Plants 2: 16066. [DOI] [PubMed] [Google Scholar]
- Yang EJ, Yoo CY, Liu J, Wang H, Cao J, Li FW, Pryer KM, Sun TP, Weigel D, Zhou P, Chen M (2019) NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat Commun 10: 2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA (2005) A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17: 1894–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, Weigel D, Chen M (2019) Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat Commun 10: 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehrmann A, Hartel B, Glass F, Bayer-Csaszar E, Obata T, Meyer E, Brennicke A, Takenaka M (2015) Selective homo- and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J Biol Chem 290: 6445–6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hach A (1999) Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell Mol Life Sci 56: 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hach A, Wang C (1998) Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol 18: 3819–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Millet YA, Cheng Z, Bush J, Ausubel FM (2015a) Jasmonate signalling in Arabidopsis involves SGT1b-HSP70-HSP90 chaperone complexes. Nat Plants 1: 15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Yuan S, Feng H, Xu F, Cheng J, Shang J, Zhang DW, Lin HH (2011) Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs—new evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J Plant Physiol 168: 714–721 [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Zhang GC, Zhu F, Zhang DW, Yuan S (2015b) The roles of tetrapyrroles in plastid retrograde signaling and tolerance to environmental stresses. Planta 242: 1263–1276 [DOI] [PubMed] [Google Scholar]
- Zhao X, Huang J, Chory J (2018) Genome uncoupled 1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol 178: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Huang J, Chory J (2019a) Unraveling the linkage between retrograde signaling and RNA metabolism in plants. Trends Plant Sci 25: 141–147 [DOI] [PubMed] [Google Scholar]
- Zhao X, Huang J, Chory J (2019b) GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc Natl Acad Sci U S A 116: 10162–10167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lu Q, Li Q, Wang L, Ding S, Zhang A, Wen X, Zhang L, Lu C (2017) PPR-SMR protein SOT1 has RNA endonuclease activity. Proc Natl Acad Sci U S A 114: E1554–E1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Qu Y, Zubo YO, Börner T, Schmitz-Linneweber C (2013) Mutation of the pentatricopeptide repeat-SMR protein SVR7 impairs accumulation and translation of chloroplast ATP synthase subunits in Arabidopsis thaliana. J Plant Res 126: 403–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.