Abstract
Phosphate is a vital macronutrient for plant growth, and its availability in soil is critical for agricultural sustainability and productivity. A substantial amount of cellular phosphate is used to synthesize phospholipids for cell membranes. Here, we identify a key enzyme, nonspecific phospholipase C4 (NPC4) that is involved in phosphosphingolipid hydrolysis and remodeling in Arabidopsis during phosphate starvation. The level of glycosylinositolphosphorylceramide (GIPC), the most abundant sphingolipid in Arabidopsis thaliana, decreased upon phosphate starvation. NPC4 was highly induced by phosphate deficiency, and NPC4 knockouts in Arabidopsis decreased the loss of GIPC and impeded root growth during phosphate starvation. Enzymatic analysis showed that NPC4 hydrolyzed GIPC and displayed a higher activity toward GIPC as a substrate than toward the common glycerophospholipid phosphatidylcholine. NPC4 was associated with the plasma membrane lipid rafts in which GIPC is highly enriched. These results indicate that NPC4 uses GIPC as a substrate in planta and the NPC4-mediated sphingolipid remodeling plays a positive role in root growth in Arabidopsis response to phosphate deficiency.
Nonspecific phospholipase C4 (NPC4), which is induced by phosphate deficiency, hydrolyzes the common phosphosphingolipid glycosylinositolphosphorylceramide and mediates sphingolipid remodeling that supports root growth in Arabidopsis response to phosphate deficiency.
Introduction
Phosphate is an essential nutrient for plant growth and productivity, but plants frequently suffer from phosphate deficiency due to the limited availability of phosphate in many types of soils (Raghothama, 1999, 2000). A better understanding of the mechanism by which plants respond to phosphate deficiency can aid the development of plants with improved phosphate-use efficiency. Approximately one-third of organic phosphate in plants is in the form of phospholipids (Gaude et al., 2008). There are two major types of phosphorus-containing membrane lipids: (1) phosphoglycerolipids, which have the three-carbon glycerol backbone and (2) phosphosphingolipids, which have the long-chain base (LCB) sphingosine-like backbone. Upon phosphate deficiency, one major metabolic change is membrane lipid remodeling, by which a significant portion of phosphoglycerolipids such as phosphatidylcholine (PC), the most abundant glycerophospholipid in plants, is replaced by nonphosphorus glycerolipids, such as digalactosyldiacylglycerol (DGDG; Hartel et al., 2000; Andersson et al., 2003, 2005; Jouhet et al., 2004; Su et al., 2018). Impairment of PC hydrolysis impedes plant growth during phosphate-deficient condition (Cruz-Ramirez et al., 2006; Li et al., 2006).
The most abundant phosphosphingolipid in plants is glycosylinositolphosphorylceramide (GIPC), which has a sphingosine-like backbone, with an amide-linked fatty acid and a phosphoinositol head group. GIPC constitutes ∼25%–50% of lipids in the plasma and tonoplast membranes (Sperling et al., 2005; Markham et al., 2006, 2013). GIPC is abundant in plants but not in mammals, whereas sphingomyelin is specifically found in mammalian tissues. GIPC plays important roles in many essential cellular and physiological processes, such as pathogen defense (Wang et al., 2008; Mortimer et al., 2013), symbiosis (Borner et al., 2005), formation of lipid rafts (Borner et al., 2005), and pollen function (Rennie et al., 2014). Studies indicated a decrease in the level of GIPC during phosphate starvation (Andersson et al., 2005; Okazaki et al., 2013). However, the functional significance and enzymes mediating the decrease in GIPC remain unknown.
During phosphate deprivation, phospholipase Dζ2 (PLDζ2) and nonspecific phospholipase C4 (NPC4), are the two most highly induced lipid-hydrolyzing enzymes in Arabidopsis (Cruz-Ramirez et al., 2006; Gaude et al., 2008). PLDζ2 hydrolyzes PC to generate phosphatidic acid (PA), which can be hydrolyzed by phosphatases to release phosphate and generate diacylglycerol (DAG). DAG is a substrate for synthesis of monogalactosyldiacylglycerol (MGDG) by MGDG synthases and subsequently the synthesis of DGDG by DGDG synthases (Awai et al., 2001; Kelly and Dormann, 2002; Su et al., 2018). On the other hand, NPC4 could potentially hydrolyze PC directly to generate DAG, and NPC4 was shown to hydrolyze PC to DAG in vitro (Nakamura et al., 2005; Peters et al., 2010). Disruption of NPC4 shortens root length during phosphate deficiency (Su et al., 2018). However, the gene knockout (KO) of NPC4 did not result in significant changes in the level of PC or DGDG in Arabidopsis rosettes during phosphate deficiency when compared with a wild-type (WT; Nakamura et al., 2005).
To determine whether NPC4 is involved in glycerolipid remodeling in a tissue and/or time-specific manner, we analyzed the effect of NPC4-KO, PLDζ2-KO, and NPC4 PLDζ2 double KO Arabidopsis on lipid changes in seedlings after different durations of phosphate starvation. PLDζ2 has a predominant effect on decreasing glycerophospholipids (Cruz-Ramirez et al., 2006; Li et al., 2006), whereas the single KO of NPC4 had no impact on the glycerophospholipid decrease in either leaves or roots (Su et al., 2018). Therefore, the metabolic function of the highly induced NPC4 during phosphate deficiency is unclear. Here, we report the change of sphingolipid metabolism in phosphate-starved plants and we identify NPC4 as an enzyme that hydrolyzes the most abundant sphingolipid GIPC in plants. The disruption of NPC4 diminished GIPC hydrolysis and impeded root growth during phosphate deprivation.
Results
The level of phosphosphingolipids decreases in phosphate-deprived Arabidopsis
To determine the effect of phosphate deprivation on phosphosphingolipids in Arabidopsis, 3-day-old seedlings grown on half-strength MS (Murashige and Skoog) vertical plates were transferred to similar plates with or without phosphate. After growing for an additional 10 days, rosettes and roots were harvested separately for sphingolipid analysis. The most abundant phosphorus-containing sphingolipid in Arabidopsis GIPC was quantified by mass spectrometry. The level of total GIPC decreased by 61% in rosettes and by 59% in roots after the phosphate-starvation treatment (Figure 1A). Different molecular species of GIPC displayed similar trends of decrease in rosettes and roots, with 38 out of 40 GIPC species showing decreased levels during phosphate deficiency (Figure 1B).
Figure 1.
Change of GIPC and hCer Levels in Arabidopsis rosettes and roots in response to phosphate starvation. A, The level of GIPC in rosettes and roots with and without phosphate. Levels of GIPC are shown relative to the level of GIPC in rosettes during sufficient phosphate (set at 100%). B, Heat map showing the level of GIPC species in rosettes and roots with and without phosphate. Each horizontal colored bar represents the log2 of the concentration (nmol g−1 dry weight) as shown in the color key. C, Levels of hCer in rosettes and roots with and without phosphate. The levels of hCer are shown relative to the level of hCer in rosettes during sufficient phosphate (100%). D, Heat map showing the levels of hCer species in rosettes and roots with and without phosphate. Each horizontal colored bar represents the log2 of the concentration [(nmol g−1 dry weight) × 10] as shown in the color key. Three-day-old seedlings were transferred to a phosphate-sufficient or -deficient half-strength MS medium for 10 days. In (B) and (D), five replicates of rosette and root samples were harvested for lipid analysis by mass spectrometry and the average data of each species were normalized by a log2 calculation method (n = 5; Szymanski et al., 2014). The heat map was drawn using Heatmap Illustrator (version of HemI 1.0.3.3). *Significant at P < 0.05; **Significant at P < 0.01 compared with the phosphate-sufficient condition in the same tissue, based on Student’s t test (Supplemental Data Set 1). The phosphate treatments, lipid extraction and analysis in (A) and (C) were repeated three times with consistent results, and the results of one representative lipid analysis are shown.
Because the GIPC level decreased in plant tissues during phosphate starvation, we thought it is likely that the phosphorus-containing head group of GIPC was cleaved for phosphate recycling and that hydroxyceramide (hCer) and glucosylceramide (GlcCer) might be generated. Thus, we quantified hCer and GlcCer in rosettes and roots with and without phosphate starvation. The total hCer level was comparable between phosphate-sufficient and deficient conditions in rosettes and roots (Figure 1C). However, during phosphate deficiency, 18 hCer species increased significantly in roots, and in rosettes 2 hCer species increased, whereas 4 hCer species displayed a decrease compared to those during sufficient phosphate (Figure 1D). The level of total GlcCer increased by 110% in roots and 20 GlcCer species displayed a similar trend of increase in roots during phosphate deficiency (Supplemental Figure 1, A and B). By comparison, in rosettes during phosphate deficiency, only three GlcCer species increased, whereas seven species decreased and the level of total GlcCer decreased by 30% relative to that during sufficient phosphate (Supplemental Figure 1, A and B). These results showed that the level of the phosphorus sphingolipid, GIPC, drastically decreased in both rosettes and roots and that GlcCer increased in roots upon phosphate starvation.
Disruption of NPC4 impedes the GIPC decrease during phosphate deficiency
To investigate the molecular basis for the observed GIPC decrease, we hypothesized that the ester linkage between the hCer backbone and phosphorus-containing group in GIPC could be hydrolyzed by a phospholipase C-like enzyme. The fungal enzyme inositol phosphosphingolipid phospholipase C1 is capable of hydrolyzing mannosyl inositol phosphoceramide (MIPC; Sawai et al., 2000). In addition, neutral sphingomyelinase 2 in mammals was reported to hydrolyze sphingomyelin (Hofmann, 2000). MIPC, sphingomyelin, and GIPC are all phosphosphingolipids. MIPC is found in fungi but not in mammals and plants, and sphingomyelin is specifically found in mammal tissues. GIPC is abundant in plants but not found in mammals. Two distinctively different families of phospholipase C have been reported in Arabidopsis, phosphatidylinositol-specific phospholipase C and the NPC, encoded by nine and six genes, respectively (Wang, 2012). Among those, the transcript of NPC4 was the most highly induced during phosphate deficiency (Nakamura et al., 2005; Gaude et al., 2008). These results prompted us to determine the effect of NPC4 on GIPC hydrolysis during phosphate starvation.
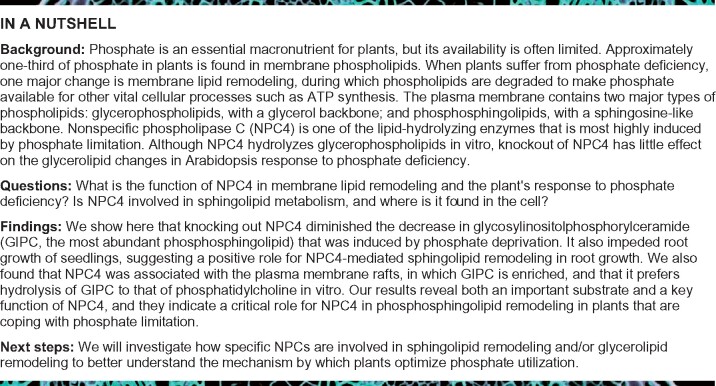
We isolated three T-DNA KO mutants of NPC4 (Supplemental Figure 2A). The presence of T-DNA inserts and the loss of NPC4 expression were verified by PCR using genomic DNA and cDNA as templates, respectively (Supplemental Figure 2, B–D). To test the effect of NPC4 disruption on plant growth during phosphate starvation, 3-day-old seedlings grown on a phosphate-sufficient medium were transferred to either phosphate-sufficient or -deficient medium for 5 days and then the primary root length was measured. The primary root length showed no significant difference among WT and NPC4 mutants during the phosphate-sufficient condition. However, the primary root length of NPC4 mutants was about 25% shorter compared to WT grown during the phosphate-deficient condition (Figure 2, A and B). The cell length and width of primary roots were similar between the mutants and WT (Supplemental Figure 3), indicating that reduced cell proliferation contributed to the shorter roots. All three NPC4 mutants were impeded in root growth during phosphate deficiency. These results indicate that NPC4 sustains root growth during phosphate deficiency.
Figure 2.
Effect of NPC4–KO on plant growth and GIPC levels during phosphate-sufficient and -deficient conditions. A, Representative images of WT and npc4 mutant seedlings growth during phosphate-sufficient or -deficient conditions. Bars = 1 cm. B, Primary root length of WT and npc4 mutants during Pi-sufficient and Pi-deficient conditions. Values are means ± sd (n = 5). C, GIPC levels in Arabidopsis roots. The relative level of GIPC was calculated by comparison to the level of GIPC in roots during sufficient phosphate. Values are means ± sd (n = 5). D, GIPC levels in Arabidopsis rosettes. The relative level of GIPC was calculated by comparison to the level of GIPC in rosettes during sufficient phosphate. Values are means ± sd (n = 5). E, The level of GIPC species in Arabidopsis roots. The average data of GIPC content of each species were normalized by a log2 calculation method (n = 5; Szymanski et al., 2014). The heatmap was generated using Heatmap Illustrator (version of HemI 1.0.3.3). *Significant at P < 0.05; **Significant at P < 0.01 compared with WT during the same condition, based on Student’s t test (Supplemental Data Set 1). Each horizontal-colored bar represents the log2 of the concentration (nmol g−1 dry weight) as shown in the color key. The phosphate treatments, lipid extractions, and analyses for (A) to (E) were repeated three times with similar results. The results of one representative experiment are shown.
The mutant seedlings were subjected to phosphate starvation along with WT plants. Three-days-old seedlings grown on a phosphate-sufficient medium were transferred to phosphate-sufficient and -deficient media for additional 10 days. During the phosphate-sufficient condition, there were no differences in the level of GIPC in rosettes or roots among WT and NPC4 mutant plants. During phosphate deficiency, compared to WT, the GIPC level in roots was almost two-fold higher in all three NPC4 mutants, with most GIPC species displaying significant increases (Figure 2, C and E). In rosettes, the GIPC level in NPC4-KO mutants tended to be higher than WT during phosphate deficiency (Figure 2D), with many GIPC species displaying significant increases relative to those in WT rosettes (Supplemental Figure 4). However, the magnitude of phosphate deficiency-induced difference in GIPC levels was much greater between NPC4-KO mutants and WT in roots than leaves, suggesting that NPC4 affects GIPC hydrolysis more in roots than in leaves.
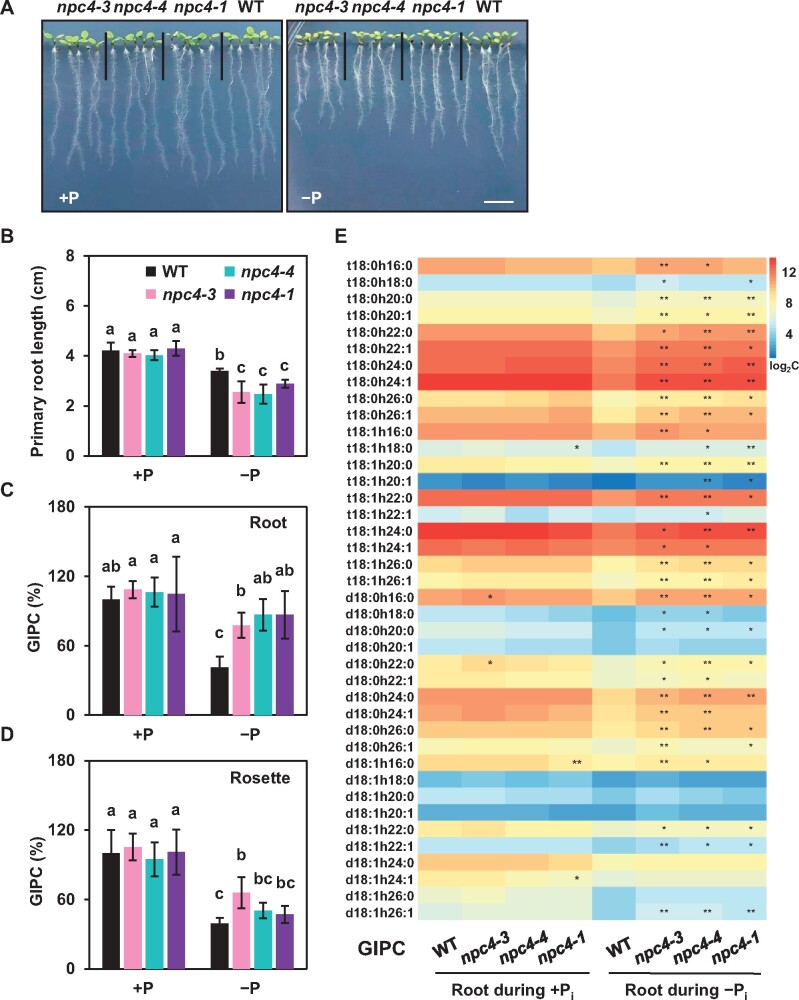
The level of the nonphosphorus sphingolipid hCer in both rosettes and roots was comparable between WT and NPC4-KO mutants grown during phosphate-sufficient and -deficient conditions (Supplemental Figure 5, A–D). The GlcCer level in roots was significantly increased both in WT and NPC4 mutants during phosphate deficiency, but the GlcCer increase in the two NPC4-KO mutants was smaller than that in WT roots (Figure 3A). The level of several GlcCer species in phosphate-starved roots tended to be lower in the two NPC4-KO mutants than WT (Figure 3B). However, the GlcCer level in rosettes was similar in WT and NPC4-KO mutants (Supplemental Figure 6, A and B). We measured the transcript levels of glucosylceramide synthase (GCS), which converts hCer to GlcCer (Msanne et al., 2015). The levels of NPC4 and GCS transcripts were significantly increased during phosphate deficiency, and the magnitude of increase of NPC4 and GCS expression was greater in roots than that in rosettes (Figure 3C). In addition, we analyzed the level of LCB and long-chain base phosphate (LCBP). Relative to that during sufficient phosphate, the LCB level during phosphate deficiency increased in rosettes but decreased in roots. Whereas it was comparable in WT and NPC4-KO rosettes, the LCB level in two NPC4-KO roots was lower than that in WT roots during phosphate deficiency (Supplemental Figure 7, A and C). In comparison, during phosphate deficiency, the LCBP level increased in both roots and rosettes and was similar between WT and NPC4-KO roots and rosettes and it increased leaves (Supplemental Figure 7B).
Figure 3.
Effect of NPC4–KO on GlcCer levels in Arabidopsis roots. A, Total GlcCer level in Arabidopsis roots during phosphate-sufficient and -deficient conditions. Three-day-old seedlings were transferred to phosphate-sufficient or -deficient half-strength MS media for an additional 10 days. Lipids were extracted and analyzed by mass spectrometry. The levels of GlcCer are shown relative to the level of GlcCer in roots during phosphate-sufficient condition (100%). Values are means ± sd (n = 5 biological repeats) and different letters indicate differences at P < 0.05 using two-way ANOVA (Supplemental Data Set 1). B, Level of GlcCer species in Arabidopsis roots. Average data of GlcCer content of each species were normalized by a log2 calculation method (n = 5) (Szymanski et al., 2014). The heatmap was drawn using Heatmap Illustrator (version of HemI 1.0.3.3). Each horizontal colored bar represents the log2 of concentration [(nmol g−1 dry weight) × 10] as shown in the color key. *Significant at P < 0.05; **Significant at P < 0.01 compared with WT during the same condition, based on Student’s t test (Supplemental Data Set 1). C, Relative level of NPC4 and GCS expression during phosphate-sufficient or -deficient conditions. Total RNA was extracted from Arabidopsis rosettes and roots during both phosphate-sufficient and -deficient conditions. UBQ10 was used as an internal standard control. Values are means ± sd (n = 3 biological repeats) and different letters indicate difference at P < 0.05 using two-way ANOVA (Supplemental Data Set 1).
NPC4 prefers GIPC to PC as a substrate
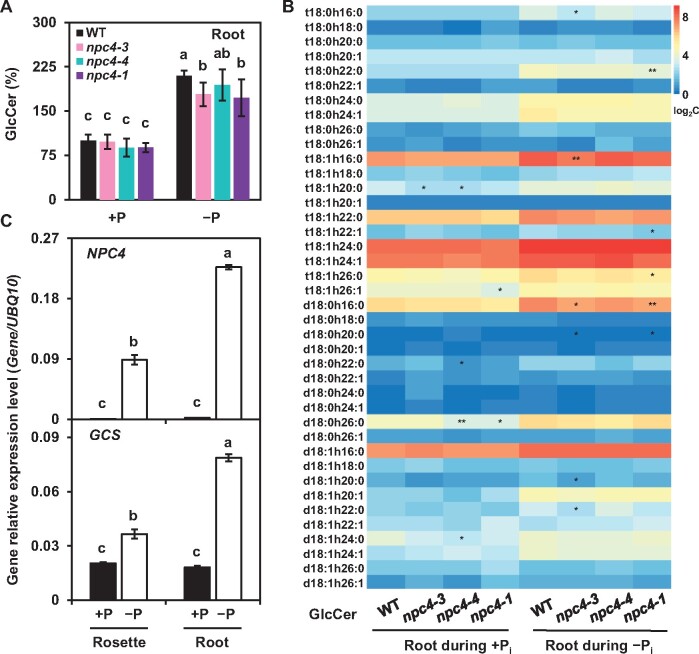
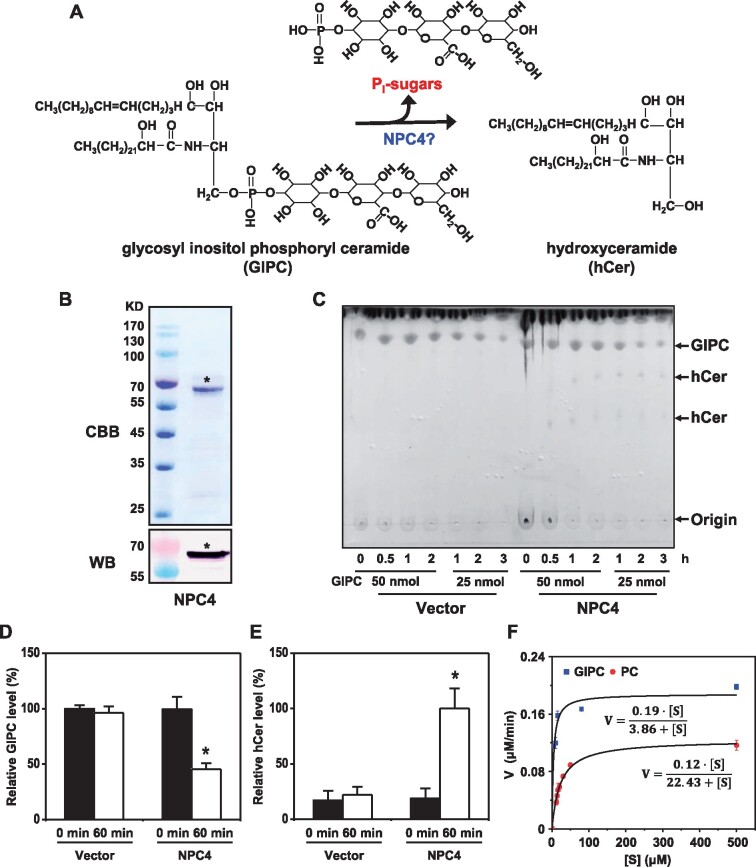
To determine whether NPC4 hydrolyzes GIPC (Figure 4A), recombinant protein NPC4-6xHis was purified from Escherichia coli and different amounts of NPC4-6xHis were incubated with different amounts of GIPC for different durations (Figure 4, B and C; Supplemental Figure 8A). With increasing time of incubation, the GIPC level decreased in NPC4-containing reactions, but that of GIPC remained the same in a protein preparation from the empty vector control (Figure 4C). Meanwhile, the production of hCer was increased when NPC4 was present (Figure 4C; Supplemental Figure 8A). After incubation for 60 min, the level of GIPC had decreased by 50% in the presence of NPC4-6xHis protein, while there was no change in GIPC for samples from the vector control (Figure 4D). The resulting reactions were also analyzed by mass spectrometry for potential products. hCer was generated in the reactions with the presence of NPC4 while no production of hCer occurred in the vector control sample (Figure 4E). In addition, we measured the soluble phosphate released from GIPC hydrolysis and the level of phosphate was significantly higher in the NPC4 reaction than that of the vector control (Supplemental Figure 8B). These results indicate that NPC4 hydrolyzes GIPC to produce hCer.
Figure 4.
NPC4 Hydrolyzes GIPC to Generate hCer in vitro. A, Diagram showing NPC4-mediated hydrolysis of GIPC to hCer. B, Coomassie Brilliant Blue staining (upper) and immunoblotting (lower) of purified recombinant NPC4 expressed in E. coli. C, Changes in GIPC levels after incubation of GIPC with purified NPC4. GIPCs were chromatographed in CHCl3/CH3OH/4M NH4OH (9:7:2, by vol) with 0.2 M ammonium acetate. Values are means ± sd (n = 3). *Significant difference compared with vector control at P < 0.05. D, Changes in GIPC levels after incubation of GIPC with purified NPC4. Relative GIPC level (%) refers to GIPC level relative to that of GIPC added to the assay. Values are means ± sd (n = 3). *Significant difference compared with vector control at P < 0.05. E, Production of hCer after incubation of GIPC with purified 10 µg NPC4. Relative hCer level (%) refers to the level of hCer produced by NPC4 after a 60-min reaction. Vector refers to a negative control using an equal volume of eluents from an identically processed sample containing empty vector alone. F, Kinetic parameters of NPC4 hydrolysis of GIPC and PC. GIPC was isolated from Arabidopsis leaves; commercial PC from soybean seeds (Avanti Polar Lipids Inc. 840054P) was used. For GIPC, Km = 3.86 μM; for PC, Km = 22.43 μM. Values are means ± sd (n = 3). Origin 8.0 was used to plot the data and calculate the kinetic parameters.
Previous studies reported that NPC4 was able to hydrolyze glycerophospholipids such as PC (Nakamura et al., 2005; Peters et al., 2010). To compare the activity of NPC4 toward GIPC and PC, we measured NPC4 hydrolysis of PC and GIPC under the same assay conditions. For the GIPC substrate, NPC4 had a Km at 3.86 µM and a Vmax of 0.19 µM min−1, whereas for PC, NPC4 had a Km at 22.43 µM and a Vmax of 0.12 µM min−1 (Figure 4F). The kinetic data indicated that NPC4 had nearly six-fold higher affinity toward GIPC than toward PC and nearly a 1.5-fold greater maximal velocity for GIPC than PC (Figure 4F), suggesting that NPC4 prefers GIPC to PC.
NPC4 is mostly associated with membrane rafts
The in planta use of GIPC requires NPC4 to have access to its lipid substrate in the cell. NPC4 is associated with the plasma membrane (PM; Peters et al., 2010), whereas GIPC is a major PM lipid, and it is particularly enriched in PM lipid rafts (Lefebvre et al., 2007; Cacas et al., 2012). We first verified the PM association of NPC4 by colocalizing NPC4-GFP with the PM marker calcineurin B-like calcium sensor protein tagged with red fluorescence protein (CBL1-RFP; Figure 5A). When the microsomal membranes were solubilized with 1% Triton X-100, a majority of NPC4 was present in the detergent-insoluble fraction (Figure 5B), a hallmark for membrane raft-associated protein. To isolate membrane rafts, we isolated the PM fraction from tobacco leaves using two-phase partitioning and then treated the PM fraction with chilled Triton X-100, followed by discontinuous sucrose gradient centrifugation. An opaque band was visible near the 30%–35% interface, and this was shown to contain detergent-insoluble membranes (Mongrand et al., 2004; Lefebvre et al., 2007). The potential detergent-insoluble membranes were fractionated mostly in fractions 4 and 5. Immunoblotting of the sucrose-gradient fractions indicated that most NPC4 were present in fractions 4, 5, and 6 (Figure 5C). Lipid analysis of those fractions revealed that fractions 4 and 5 contained the highest level of GIPC (Figure 5D). Similarly, hCer and GlcCer mainly were found in fractions 3–5 (Supplemental Figures S9 and S10). These results support the idea that NPC4 and GIPC colocalize in PM lipid rafts.
Figure 5.
NPC4 is Mostly Associated with PM Rafts. A, Colocalization of NPC4-GFP with the PM marker CBL1-RFP in tobacco leaves. Bars = 50 μm. B, Immunoblotting of NPC4-GFP in Triton X-100-insoluble and -soluble microsomal membrane fractions of tobacco (Nicotiana benthamiana) leaves NPC4-GFP after centrifugation at 100,000 g. P, Triton X-100-insoluble microsomal precipitate; S, soluble proteins after Triton X-100 treatment; T, total protein extract from tobacco leaves. C, Immunoblotting of NPC4-GFP in the discontinuous sucrose gradient fractions of the PM. PM from tobacco leaves isolated by two-phase partitioning was treated with Triton X-100 and then subjected to sucrose gradient flotation. The gradient was separated into nine 1-mL fractions from top (1) to bottom (9). Fractions 4 and 5 were near the 30%–35% interface. Proteins from an equal volume of each fraction were used for 10% SDS–PAGE, followed by immunoblotting with an anti-GFP antibody for NPC4-GFP. D, Detection of GIPC in discontinuous sucrose gradient fractions. Lipids extracted from the equal volume of each fraction was analyzed by LC–MS/MS and GPLC levels were expressed as mass signal per the same volume from each fraction.
Discussion
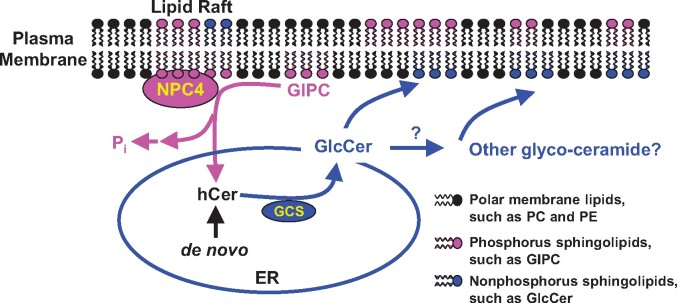
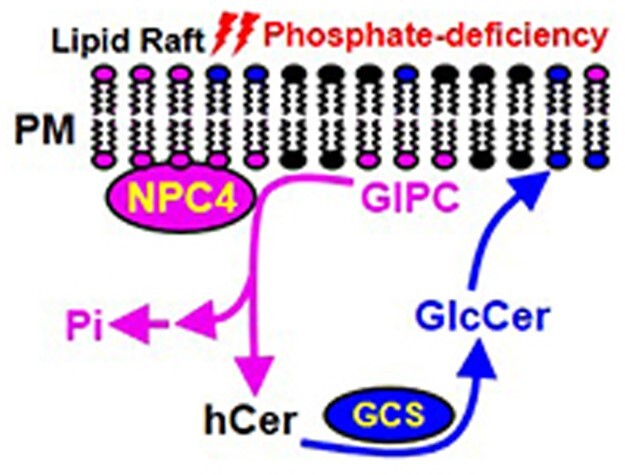
The ability to change membrane glycerolipid composition is an adaptive mechanism by which plants cope with phosphate deficiency (Awai et al., 2001; Cruz-Ramirez et al., 2006; Li et al., 2006; Nakamura et al., 2009; Okazaki et al., 2013). In glycerolipid remodeling, the decrease in PC cannot be completely compensated functionally by an increase in nonphosphorus-containing glycerolipids because plant growth is compromised during Pi deficiency (Gaude et al., 2008; Su et al., 2018). Sphingolipids are another important class of membrane lipids, particularly abundant in plasma and tonoplast membranes (Sperling et al., 2005; Markham et al., 2006; Markham et al., 2013). GIPC is the most abundant phosphosphingolipid in plants (Sperling et al., 2005; Markham et al., 2006, 2013) and its level decreases substantially in leaves and roots in response to phosphate deficiency, as shown in this study. Moreover, this work showed that NPC4 hydrolyzes GIPC, revealing a new function for NPC. Although the importance of sphingolipid metabolism in plant growth, development, and stress response has begun to be appreciated (Markham et al., 2013; Luttgeharm et al., 2016), little is known about the specific enzymes involved in sphingolipid hydrolysis. The present study provides evidence that NPC4 hydrolyzes GIPC, thus identifying a major metabolic step in plant sphingolipid metabolism (Figure 6).
Figure 6.
Proposed model of sphingolipid hydrolysis by NPC4 in Arabidopsis roots in response to phosphate-deficiency. When phosphate is limiting, the expression of NPC4 increases and resulting NPC4 is associated mostly with the PM rafts in which GIPC is enriched. NPC4 hydrolyzes GIPC to release phosphate for other essential cellular processes. The hydrolysis of GIPC produces hCer, which serves as substrate for the synthesis of GlcCer that can be further glycosylated to produce other glycosyleramides. ER, endoplasmic reticulum; GCS, glucosylceramide synthase; GlcCer, glucosylceramide; GIPC, glycosyl inositol phosphoryl ceramide; hCer, hydroxyceramide; PE, phosphatidylethanolamine; Pi, inorganic phosphate; PM, plasma membrane.
Previous studies showed that NPC4 also hydrolyzed glycerophospholipids such as PC when assayed in vitro (Nakamura et al., 2005; Peters et al., 2010). This raises an important, yet challenging question: What lipids does NPC4 hydrolyze in plants? It is known that in vitro a lipolytic enzyme may hydrolyze various lipids when an individual lipid is present (Kirk Pappan, 1997). But, the seemly broad substrate use in vitro may not represent the lipid substrate of the enzyme in vivo. Various factors, including enzyme kinetics and intracellular distribution of the substrate lipids and enzymes, affect its substrate access and use in plant tissues.
Our comparative kinetic analyses indicated that NPC4 had about a six-fold higher affinity for GIPC than for PC. In addition, we showed that NPC4 was associated with the PM and was enriched in the membrane rafts, whereas GIPC is known to be enriched in PM lipid rafts. GIPC constitutes about 65 mol% of the lipids in isolated PM rafts from tobacco, which represents a 2.5-fold enrichment of GIPC compared to the rest of the nonraft PM regions (Lefebvre et al., 2007; Cacas et al., 2012). This PM raft association of NPC4 places NPC4 in the same sub-membrane locale as GIPC. On the other hand, GIPC is thought to be localized primarily in the outer leaflet of PM in plants, and this raises the question of how NPC4 at the cytoplasmic side accesses GIPC at the other side. The experimental evidence for the transverse PM membrane distribution of sphingolipids is scarce in plants. In oat roots during phosphate sufficiency, the distribution of GlcCer between the cytosolic:outer leaflets of the PM was 30:70 (mol:mol). However, during phosphate deficiency, the transversal distribution of GlcCer in PM changed to 65:35 (cytosolic:outer leaflets) in oat roots (Tjellstrom et al., 2010). This result suggests that sphingolipids including GIPC are localized in the cytosolic leaflet and accessible by NPC4.
It is worth-noting that in vitro, NPCs other than NPC4 may hydrolyze GIPC, as they have highly conserved catalytic domains. However, a clear difference between NPC4 and other NPCs is its unique association with the PM in Arabidopsis (Pokotylo et al., 2013; Pejchar et al., 2015). NPCs2 and 6 are found in the plastid and have predicted signal peptides at the N terminus (Song et al., 2017; Ngo et al., 2018). NPC3 is associated with the tonoplast (Pokotylo et al., 2013), whereas NPC5 is cytosolic (Gaude et al., 2008; Peters et al., 2014). The different subcellular locations underlie a basis for diverse functions of different NPCs, ranging from responses to various stress conditions to seed oil production (Cai et al., 2020). Among six NPCs in Arabidopsis, NPC4 and NPC5 are most similar in sequences, but NPC5 is not membrane rafts associated, which may limit its access to GIPC in vivo. NPC5 is involved in glycerophospholipid remodeling in leaves undergoing phosphate deficiency (Gaude et al., 2008), whereas NPC4 affects sphingolipid changes more in roots than leaves as shown in the present study. The demonstration of GIPC hydrolysis in vitro, together with the comparative lipid analysis between WT and NPC4-KO mutants supports the conclusion that NPC4 hydrolyzes GIPC in plants during phosphate deficiency in Arabidopsis. Compared to the decrease in WT roots, the loss of NPC4 in three NPC-KO mutants impeded ∼80% of GIPC decrease during phosphate deficiency, Thus, we have identified NPC4 as a major enzyme mediating a decrease of GIPC in roots during phosphate deficiency, and that its PM association could be critical to its access to and hydrolysis of GIPC in plants. However, the loss of NPC4 did not impede completely phosphate deficiency-induced decrease in GIPC, particularly in rosettes. This could mean that GIPC is degraded by other enzymatic activities, such as GIPC-PLD activities (Tanaka et al., 2013; Kida et al., 2017) and potentially other NPCs that may be more active in rosettes.
Although the level of GIPC was dramatically decreased in roots and rosettes during phosphate deficiency, the increase in hCer was limited to specific species. These results suggest that hCer produced from NPC4 activity is further metabolized to nonphosphorus-containing glycosylceramides (Figure 6). Indeed, during phosphate deficiency, GlcCer showed a significant increase in roots, and the increase was attenuated in roots deficient in NPC4. The gene transcript of GCS that catalyzes the synthesis of GlcCer using hCer also increased significantly in roots. These results are consistent with the proposition that hCer released from NPC4-mediated GIPC hydrolysis is further metabolized to GlcCer by GCS in roots during phosphate-deficiency. However, no difference in GlcCer levels was observed between WT and NPC4-KO rosettes during phosphate deficiency. Those results are consistent with the attenuated decrease in GIPC in rosettes relative to that in roots. In addition, NPC4-KO roots exhibited an attenuated decrease in the LCB level compared to WT, but no such difference occurred in rosettes. All the results indicate that NPC4 has a more profound effect on sphingolipid changes in roots than in leaves.
The functional significance of NPC4 in sphingolipid hydrolysis is shown by changes in root growth in NPC4-KO plants. The impairment of NPC4-mediated hydrolysis of GIPC resulted in reduced root hair elongation during phosphate deficiency (Su et al., 2018). Forward genetic screening of defects in root hair elongation identified a mutant per2 (Pi deficiency root hair defective 2; Chandrika et al., 2013). PER2 encodes Alfin-like 6, a protein that belongs to a small family of nuclear localized plant homeodomain-containing putative transcription factors. Interestingly, transcriptomic analysis of per2 for altered gene expression upon phosphate deficiency revealed downregulation of NPC4 (Chandrika et al., 2013). In addition to root hairs, our data show that disruption of NPC4 decreased overall root growth. Collectively, these results indicate that NPC4 sustains root growth during phosphate deficiency. In addition, NPC4 has a much higher expression level in old leaves than that in young leaves (Peters et al., 2010). It is likely that NPC4 plays a role in recycling of phosphate by hydrolysis of GIPC not only during phosphate deficiency but also in phosphate recycling from GIPC in old tissues to young tissues. Furthermore, NPC4 is involved in response to other stress conditions, such as high salinity (Peters et al., 2010; Kocourkova et al., 2011) and nitrogen deficiency (Mei et al., 2016). GIPC was reported to play a key role in sensing salt stress to trigger Ca2+ influx in plants (Kocourkova et al., 2011; Jiang et al., 2019). It will be of great interest to investigate whether the NPC4-catalyzed GIPC hydrolysis plays a role in plants response to other stressors.
In summary, we have identified a sphingolipid metabolic process in which GIPC, the most abundant sphingolipid in the PM, is hydrolyzed in response to phosphate deficiency in plants and that NPC4 is primarily responsible for the GIPC hydrolysis and the decreased plant response to phosphate deficiency (Figure 6). Disruption of NPC4 function impairs the GIPC decline and root growth during phosphate deprivation. The genetic, physiological, biochemical and metabolic evidence presented support the importance of the NPC4-mediated hydrolysis of GIPC and sphingolipid remodeling in plants coping with phosphate limitation (Figure 6).
Materials and methods
Confirmation of NPC4 KO plants
The isolation of the npc4-1 (Salk_046713)KO was described previously (Peters et al., 2010). T-DNA insertional mutants for npc4-3 (CS349184) and npc4-4 (CS345155) were identified from the Salk Arabidopsis T-DNA KO collection obtained from the Ohio State University ABRC. The homozygous T-DNA insertional NPC4 (At3g03530) mutant was verified by PCR-based screening using a T-DNA left border primer and gene-specific primers. Expression of NPC4 in WT, npc4-1, npc4-3, and npc4-4 was analyzed by RT-PCR. Total RNA was extracted from rosette using a cetyl-trimethylammonium bromide method (Li et al., 2006). RNase-free DNase was used to remove the DNA contamination in RNA samples. cDNA was synthesized using the iScript kit (Bio-Rad) from isolated RNA template through reverse transcription. PCR was cycled as following: one cycle of 95°C for 1 min; 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and final extension of DNA at 72°C for 10 min. Primers used for the PCR verification of npc4-1, npc4-3, and npc4-4 are listed in Supplemental Table S1.
Plant growth, sphingolipid extraction, and sphingolipid analysis by mass spectrometry
Plants were grown in a growth room with a 12-h light/12-h dark cycle at 23/21°C, 50% humidity, and 200 µmol m−2 s−1 of light intensity. Seedlings were grown on a phosphate-sufficient medium (0.5 mM KH2PO4, half MS agar plate, pH 5.8) or phosphate-deficient medium (0 mM KH2PO4, half MS agar plate, pH 5.8; Li et al., 2006; Gaude et al., 2008). Sphingolipids were extracted as described previously (Markham and Jaworski, 2007; Bure et al., 2011). Briefly, 10–30 mg of freeze-dried Arabidopsis tissues were homogenized and 3-mL extraction solvent (isopropanol/heptane/water 55:20:25) was added followed by incubation at 60°C for 15 min. The supernatant was transferred to a new Teflon-lined screw cap glass tube after centrifugation at 500g for 10 min. The extraction was repeated 2–3 times and the supernatants were pooled. The supernatant was then dried under a stream of nitrogen in a heating block at 60°C. Samples were heated in 1 mL of tetrahydrofuran (THF)/methanol/water (2:1:2 v/v/v) containing 0.1% formic acid and dissolved by ultrasound sonication, then centrifuged at 500g for 10 min to remove insoluble material, and samples were then stored at −80°C prior to further analysis.
Sphingolipids were analyzed using an Exion UPLC system coupled with a triple quadrupole/ion trap mass spectrometer (6500 Plus QTRAP; SCIEX) according to a previous method with modifications (Markham and Jaworski, 2007). Multiple reaction monitoring transitions were set up for analysis of sphingolipids including GIPC, hCer, and GlcCer as described previously (Markham and Jaworski, 2007). Standards for quantification of sphingolipids were purchased from Avanti Polar Lipids Inc. Ganglioside GM1 (Ovine brain), Cer (d18:1c14:0), GlcCer (d18:2c16:0), LCB (d17:0), and LCBP (d17:1) were used to quantify GIPC, hCer, GlcCer, LCB, and LCBP abundance. Lipids were eluted from a column (SUPELCOSIL ABZ+Plus, 150 × 3 mm, 5-µm particle size) at 1 mL min−1 with a binary-gradient system consisting of solvent A, THF/methanol/5 mM ammonium formate (3:2:5 v/v/v) + 0.1% formic acid, and solvent B, THF/methanol/5 mM ammonium formate (7:2:1 v/v/v) + 0.1% formic acid.
RNA extraction and real-time PCR
Total RNA was extracted from 2-week old Arabidopsis rosettes and roots using the RNAprep pure plant kit (DP432, http://www.tiangen.com/). Isolated RNA was used as template for cDNA synthesis through reverse transcription using an iScript kit (Bio-Rad). PCR products were quantitatively monitored by SYBR green fluorescent labeling of double-stranded DNA using MyiQ (Bio-Rad). The expression level was normalized to that of UBQ10 (At4g05320). PCR reactions were as follows: one cycle of 95°C for 1 min; 50 cycles at 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s; and final extension of DNA at 72°C for 5 min. The real-time PCR primers for NPC4 (At3g03530) and GCS (At2G19880) are listed in Supplemental Table S1.
Protein expression, purification, and immunoblotting
The cDNA encoding NPC4 was cloned into pET28a vector (Novagen) before the 6xHis coding sequence. The construct was sequenced and introduced into E. coli strain Rosetta (DE3, Amersham Biosciences). Isopropyl β-d-1-thiogalactopyranoside (0.1 mM) was added to the bacterial culture at OD600 of 0.7 to induce NPC4 protein production for 16 h at 16°C. The NPC4-6xHis fusion protein was purified as described previously (Peters et al., 2010; Li et al., 2011). The bacterial cells were harvested by centrifugation and resuspended in Tris-buffered saline (TBS) buffer containing 1 mg mL−1 lysozyme (50 mM Tris–HCl, pH 7.3, 50 mM NaCl, 5% glycerol, 1 mM DTT, and 0.5 mM PMSF). The samples were kept on ice for 30 min to digest cell walls, and then 5 mM dithiothreitol and 1.5% (w/v) N-laurylsarcosine (Sarkosyl) were added. The sample mix was vortexed and sonicated on ice for 5 min. The supernatant was obtained by centrifugation at 10,000g for 20 min and transferred to a new tube. Triton X-100 was added to the supernatant to a final concentration of 1% (v/v) and Ni-NTA-agarose beads were added (10%, w/v). The sample mix was gently rotated at 4°C for 1 h. Twenty volumes of wash buffer (50 mM Tris–HCl, pH 7.3, 50 mM imidazole, 5% glycerol, 1 mM DTT, and 0.5 mM PMSF) were used to wash the agarose beads. Triple volumes of elution buffer (50 mM Tris–HCl, pH 7.3, 250 mM imidazole, 5% glycerol, 1 mM DTT, and 0.5 mM PMSF) were applied to elute the fusion protein bound to agarose beads. The concentration of purified protein was measured with a protein assay kit (Bio-Rad). The purified proteins were separated by 10% SDS–PAGE, stained with Coomassie Brilliant Blue and immunoblotted using His-tag antibodies (1:3,000 dilution; A-5588, Sigma).
NPC activity assays
GIPC was purified from Arabidopsis leaves according to a method described previously (Bure et al., 2011). Briefly, ∼25 g of well-expanded Arabidopsis leaves were ground with 400 mL cold 0.1 N aqueous acetic acid. The sample was filtered through eight layers of acid-washed Miracloth and the filtrate was discarded. This step was repeated one more time and the residue was then extracted with hot acidic 70% ethanol containing 0.1 N HCl (70°C). The extract was filtered through Miracloth and washed with hot acidic 70% ethanol. The process of hot acidic ethanol extraction was repeated twice. The combined filtrates were chilled immediately and put at −20°C overnight. The sample was then centrifuged at 2,000g at 4°C for 15 min to obtain the GIPC-containing pellet. The pellet was washed with cold acetone several times until it became colorless. Then, the pellet was washed with cold diethyl ether to obtain a whitish precipitate, which was then dissolved in THF/methanol/water (4:4:1, v/v/v) containing 0.1% formic acid. The sample was heated at 60°C and sonicated gently to be dissolved well. After centrifugation at 2,000g at 4°C for 5 min, the supernatant containing GIPC was retained and dried under a stream of N2. Then, 1-mL butanol/water (1:1, v/v) was added for phase partitioning. While the debris of protein and cell walls were in the lower aqueous phase, the upper butanolic phase contained GIPC. The GIPC extract was dried and the residue was dissolved in THF/methanol/water (4:4:1, v/v/v) containing 0.1% formic acid. The concentration of dissolved GIPC was quantified using LC–MS (6500 Plus QTRAP; SCIEX) with GM1 as standard. Soy PC from soybean seeds was purchased from Avanti Polar Lipids Inc. (840054P) and used for activity assays.
GIPC and PC were suspended in reaction buffer (25 mM HEPES, pH 7.5, 10 mM CaCl2, and 10 mM MgCl2) by sonication on ice for 5 min. Ten micrograms of purified NPC4 were added to the reaction mixture in a final volume of 200 µL For empty vector control, an equal volume of eluents from an identically processed sample containing empty vector alone was added to the assay mixture. The reaction was incubated at 30°C for 30, 60, 90, and 120 min, and stopped by adding of 200 µL of butanol followed by vigorously vortexing. The sample was centrifuged at 12,000g and 200 µL of the lower phase was used to measure the phosphate released from the head group, as determined by the molybdenum blue method (Peters et al., 2010). Then, 200-µL water was added to the remaining mixture followed by vortexing and centrifugation and. the lower phase was discarded. GIPC in the samples was mixed with solvent H (isopropanol:hexane:water (55:20:25) with the upper phase removed) containing 30 mM ammonia acetic acid and analyzed by mass spectrometry as described below. For TLC, the solvent from the organic, upper phase of the 200 µL of butanol extract of the reaction was evaporated under a stream of N2, and the residue was dissolved in 20 µL of chloroform and separated on a TLC plate. GIPC was chromatographed in CHCl3:CH3OH:4M NH4OH (9:7:2, by vol.) with 0.2 M ammonium acetate (Voxeur and Fry, 2014) and visualized by copper sulfate as described (Kim et al., 2009).
Subcellular localization of NPC4
The constructs of 35Spro:NPC4-GFP and PM marker RFP (CBL1, Calcineurin B-like Calcium Sensor Proteins) were delivered into the epidermis of tobacco leaves by infiltration with Agrobacterium tumefaciens (EHA105) harboring the plasmid constructs. Five days after infection, the fluorescence images were observed using a Lecia TCS SP2 confocal microscope. For subcellular fractionation, infected tobacco leaves were homogenized in a chilled buffer A containing 1% Triton X-100 (Gaude et al., 2008; Li et al., 2011; 50 mM Tris–HCl, pH 8.0, 1mM ethylenediaminetetraacetic acid, 10 mM KCl, 2 mM DTT, 0.5 mM PMSF, and 0.5 M sucrose). The homogenate was centrifuged at 6,000g for 10 min, and resulting supernatants were centrifuged at 100,000g for 60 min to obtain cytosolic plus Triton X-100-soluble (supernatant) and Triton X-100-insoluble microsomal (pellet) fractions.
Lipid rafts were prepared as described previously (Mongrand et al., 2004; Lefebvre et al., 2007). Briefly, 100 g tobacco leaves after 5 days of infiltration with the 35Spro:NPC4-GFP construct were homogenized in 30 mL of buffer B containing 50 mM Tris–HCl (pH 7.4), 50 mM NaCl, 250 mM sucrose, 1 mM DTT, 0.5 mM sodium orthovanadate, 1 mM PMSF, 10 μg mL−1 aprotinin, 1 μg mL−1 pepstatin A, and 1 μg mL−1 leupeptin. Homogenates were centrifuged at 1,000g for 10 min at 4°C, and resulting supernatants were centrifuged at 100,000g for 60 min to obtain microsomal fractions. This microsomal pellet was resuspended in 330 mM sorbitol, 5 mM KCl, and 5 mM K2HPO4 pH 7.8. PMs were purified twice in an aqueous polymer two-phase system with 23 g of phase mixture (6.2% PEG3350 [w/w], 6.2% DextranT-500 [w/w] in 330 mM sorbitol, 5 mM KCl, 5 mM K2HPO4 pH 7.8) including 6 g of microsomal fraction. The final upper phase was diluted with three volumes in 4 mL of TBS buffer (140 mM NaCl, 3 mM KCl, 25 mM Tris–HCl, pH 7.5) with 1 mM PMSF to remove residual polyethylene glycol-Dextran. After a 60-min centrifugation at 100,000g, the pellet was washed and then resuspended in 0.5 mL of TBS buffer. Triton X-100 was added to 1% final concentration, and the membranes were solubilized at 4°C for 30 min, then brought to a final concentration of 48% sucrose (w/w), overlaid with successive 2 mL layers of 40%, 35%, and 30% sucrose in TBS buffer (w/w), and then centrifuged for 16 h at 200,000g at 4°C and 9 portions (1 mL each) were collected from the top. Detergent insoluble membrane fraction could be recovered near the 30%–35% interface as an opaque band. Proteins from the equal volume of each fraction were precipitated with 10% TCA acetone, followed by 10% SDS–PAGE and were then transferred onto a polyvinylidene difluoride membrane for immunoblotting. Membrane was blocked with phosphate-buffered saline containing 3% bovine serum albumin, followed by incubation with anti-GFP antibody (1:3,000 dilution; sc-9996, Santa Cruz) conjugated with horse radish peroxidase (HRP). Protein bands were detected by chemi-luminescent immunodetection of HRP activity. Meanwhile, Lipids in the fraction were extracted from the equal volume of each fraction with chloroform and butanol (1:1 v/v) and GIPC was analyzed by LC–MS/MS as described above and expressed as mass signal per the same volume in each fraction.
Statistical analysis
Values are means ± sd. Different lower letters indicate differences at P < 0.05 among genotypes during phosphate-sufficient and -deficient conditions using two-way ANOVA. *Significant at P < 0.05; **Significant at P < 0.01 compared with the control based on Student’s t test. ANOVA and t-test results are provided in Supplemental Data Set 1.
Accession numbers
Sequences of A. thaliana (At-) genes used in this study can be found in GenBank under accession numbers At-NPC4 (At3G03530), At-NPC5 (At3G03540), At-GCS (At2G19880), At-CBL1 (At4G17615), At-UBQ10 (At4g05320).
Supplemental data
Supplemental Figure 1. Changes of GlcCer Level in Arabidopsis Rosettes and Roots in Response to Pi Starvation.
Supplemental Figure 2. Confirmation of T-DNA Mutants of NPC4.
Supplemental Figure 3. Root Cell Length and Width of WT and npc4 Seedlings.
Supplemental Figure 4. Effect of NPC4–KO on GIPC Level in Arabidopsis Rosettes during Pi-Sufficient and Pi-Deficient Conditions.
Supplemental Figure 5. Effect of NPC4–KO on hCer Levels in Arabidopsis Rosettes and Roots.
Supplemental Figure 6. Effect of NPC4–KO on GlcCer Levels in Arabidopsis Rosettes.
Supplemental Figure 7. Effect of NPC4–KO on LCB and LCBP Levels in Arabidopsis Rosettes and Roots.
Supplemental Figure 8. NPC4 Hydrolyzes GIPC to Generate hCer in vitro.
Supplemental Figure 9. The Level of GIPC Species in Discontinuous Sucrose Flotation Gradient.
Supplemental Figure 10. The Level of hCer and GlcCer in Discontinuous Sucrose Flotation Gradient.
Supplemental Table 1. Primers for Gene Cloning, PCR Confirmation and Real-Time PCR.
Supplemental Data Set 1. Statistical Analysis Results.
Supplementary Material
Acknowledgments
The authors thank Dr David Osborn at the Center for Nanoscience at the University of Missouri-St Louis for maintenance of mass spectrometry.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31570808), and by Agriculture and Food Research Initiative (AFRI) award no. (2020-67013-30908/project accession number 1022148) from the USDA National Institute of Food and Agriculture and the National Science Foundation (MCB-1412901).
Conflict of interest statement. None declared.
L.G., M.L., and X.W. designed and supervised the study. M.L., B.Y., A.P., L.L., U.A., and Q.L. performed the experiments. M.L. and B.Y. analyzed the data. M.L. and B.Y. wrote the manuscript. L.G., X.W., S.L., and Y.H. revised the manuscript. All authors read and approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell/pages/General-Instructions) are: Liang Guo (guoliang@mail.hzau.edu.cn) and Xuemin Wang (swang@danforthcenter.org).
References
- Andersson MX, Larsson KE, Tjellstrom H, Liljenberg C, Sandelius AS (2005) Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J Biol Chem 280: 27578–27586 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 [DOI] [PubMed] [Google Scholar]
- Awai K, Marechal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J (2001) Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc Natl Acad Sci U S A 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bure C, Cacas JL, Wang F, Gaudin K, Domergue F, Mongrand S, Schmitter JM (2011) Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun Mass Spectrom 25: 3131–3145 [DOI] [PubMed] [Google Scholar]
- Cacas J-L, Furt F, Le Guédard M, Schmitter J-M, Buré C, Gerbeau-Pissot P, Moreau P, Bessoule J-J, Simon-Plas F, Mongrand S (2012) Lipids of plant membrane rafts. Progress in Lipid Research 51: 272–299 [DOI] [PubMed] [Google Scholar]
- Cai G, , Fan C, , Liu S, , Yang Q, , Liu D, , Wu J, , Li J, , Zhou Y, , Guo L, , Wang X ( 2020) Nonspecific phospholipaseC6 increases seed oil production in oilseed Brassicaceae plants. New Phytol 226: 1055–1073 [DOI] [PubMed] [Google Scholar]
- Carter HE, Koob JL (1969) Sphingolipids in bean leaves (Phaseolus vulgaris). J Lipid Res 10: 363–369 [PubMed] [Google Scholar]
- Chandrika NNP, Sundaravelpandian K, Yu S-M, Schmidt W (2013) ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198: 709–720 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramirez A, Oropeza-Aburto A, Razo-Hernandez F, Ramirez-Chavez E, Herrera-Estrella L (2006) Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci U S A 103: 6765–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dormann P (2008) Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Hartel H, Dormann P, Benning C (2000) DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc Natl Acad Sci U S A 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Tomiuk S, Wolff G, Stoffel W (2000) Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc Natl Acad Sci U S A 97: 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Zhou X, Tao M, Yuan F, Liu L, Wu F, Wu X, Xiang Y, Niu Y, Liu F, et al. (2019) Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572: 341–346 [DOI] [PubMed] [Google Scholar]
- Jouhet J, Marechal E, Baldan B, Bligny R, Joyard J, Block MA (2004) Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J Cell Biol 167: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, Dormann P (2002) DGD2, an arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J Biol Chem 277: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Kida T, Itoh A, Kimura A, Matsuoka H, Imai H, Kogure K, Tokumura A, Tanaka T (2017) Distribution of glycosylinositol phosphoceramide-specific phospholipase D activity in plants. J Biochem 161: 187–195 [DOI] [PubMed] [Google Scholar]
- Kim K, Zhang O, Wilson MC, Xu W, Hsu F-F, Turk J, Kuhlmann FM, Wang Y, Soong L, Key P, et al. (2009) Degradation of Host Sphingomyelin Is Essential for Leishmania Virulence. PLoS Pathogens 5: e1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk Pappan SA-B, Chapman KD, Wang X (1997) Substrate Selectivities and Lipid Modulation of Plant Phospholipase Da, -β, and -γ. Arch Biochem Biophys 353: 131–140 [DOI] [PubMed] [Google Scholar]
- Kocourkova D, Krckova Z, Pejchar P, Veselkova S, Valentova O, Wimalasekera R, Scherer GF, Martinec J (2011) The phosphatidylcholine-hydrolysing phospholipase C NPC4 plays a role in response of Arabidopsis roots to salt stress. J Exp Bot 62: 3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B, Furt F, Hartmann MA, Michaelson LV, Carde JP, Sargueil-Boiron F, Rossignol M, Napier JA, Cullimore J, Bessoule JJ, et al. (2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol 144: 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, Wang X (2011) Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23: 1107–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Welti R, Wang X (2006) Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D zeta1 and D zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol 142: 750–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttgeharm KD, Cahoon EB, Markham JE (2016) Substrate specificity, kinetic properties and inhibition by fumonisin B1 of ceramide synthase isoforms from Arabidopsis. Biochem J 473: 593–603 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG (2007) Rapid measurement of sphingolipids fromArabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG (2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281: 22684–22694 [DOI] [PubMed] [Google Scholar]
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB (2013) Plant sphingolipids: function follows form. Curr Opin Plant Biol 16: 350–357 [DOI] [PubMed] [Google Scholar]
- Mei CE, Cussac M, Haslam RP, Beaudoin F, Wong YS, Marechal E, Rebeille F (2016) C1 Metabolism inhibition and nitrogen deprivation trigger triacylglycerol accumulation in Arabidopsis thaliana cell cultures and highlight a role of NPC in phosphatidylcholine-to-triacylglycerol pathway. Front Plant Sci 7: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ (2004) Lipid rafts in higher plant cells: purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Yu X, Albrecht S, Sicilia F, Huichalaf M, Ampuero D, Michaelson LV, Murphy AM, Matsunaga T, Kurz S, et al. (2013) Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25: 1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msanne J, Chen M, Luttgeharm KD, Bradley AM, Mays ES, Paper JM, Boyle DL, Cahoon RE, Schrick K, Cahoon EB (2015) Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J 84: 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H (2005) A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J Biol Chem 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H (2009) Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci U S A 106: 20978–20983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo AH, Lin Y-C, Liu Y-C, Gutbrod K, Peisker H, Dörmann P, Nakamura Y (2018) A pair of nonspecific phospholipases C, NPC2 and NPC6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytol 219: 163–175 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K (2013) A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun 4: 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchar P, Potocky M, Krckova Z, Brouzdova J, Danek M, Martinec J (2015) Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana. Front Plant Sci 6: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perotto S, Donovan N, Drobak BK, Brewin NJ (1995) Differential expression of a glycosyl inositol phospholipid antigen on the peribacteroid membrane during pea nodule development. Mol Plant Microbe Interact 8: 560–568 [Google Scholar]
- Peters C, Kim SC, Devaiah S, Li M, Wang X (2014) Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ 37: 2002–2013 [DOI] [PubMed] [Google Scholar]
- Peters C, Li M, Narasimhan R, Roth M, Welti R, Wang X (2010) Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22: 2642–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokotylo I, Pejchar P, Potocký M, Kocourková D, Krčková Z, Ruelland E, Kravets V, Martinec J (2013) The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog Lipid Res 52: 62–79 [DOI] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raghothama KG (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Rennie EA, Ebert B, Miles GP, Cahoon RE, Christiansen KM, Stonebloom S, Khatab H, Twell D, Petzold CJ, Adams PD, et al. (2014) Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 26: 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA (2000) Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem 275: 39793–39798 [DOI] [PubMed] [Google Scholar]
- Song J, Zhou Y, Zhang J, Zhang K (2017) Structural, expression and evolutionary analysis of the non-specific phospholipase C gene family in Gossypium hirsutum. BMC Genomics 18: 979–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling P, Franke S, Luthje S, Heinz E (2005) Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol Biochem 43: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Su Y, Li M, Guo L, Wang X (2018) Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J 94: 315–326 [DOI] [PubMed] [Google Scholar]
- Szymanski J, Brotman Y, Willmitzer L, Cuadros-Inostroza A (2014) Linking gene expression and membrane lipid composition of Arabidopsis. Plant Cell 26: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kida T, Imai H, Morishige J, Yamashita R, Matsuoka H, Uozumi S, Satouchi K, Nagano M, Tokumura A (2013) Identification of a sphingolipid-specific phospholipase D activity associated with the generation of phytoceramide-1-phosphate in cabbage leaves. FEBS J 280: 3797–3809 [DOI] [PubMed] [Google Scholar]
- Tjellstrom H, Hellgren LI, Wieslander A, Sandelius AS (2010) Lipid asymmetry in plant plasma membranes: phosphate deficiency-induced phospholipid replacement is restricted to the cytosolic leaflet. FASEB J 24: 1128–1138 [DOI] [PubMed] [Google Scholar]
- Voxeur A, Fry SC (2014) Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J 79: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ryu S, Wang X (2012) Plant phospholipases: an overview. Methods Mol Biol 861: 123–137 [DOI] [PubMed] [Google Scholar]
- Wang W, Yang X, Tangchaiburana S, Ndeh R, Markham JE, Tsegaye Y, Dunn TM, Wang GL, Bellizzi M, Parsons JF, et al. (2008) An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.