Abstract
In angiosperms, endosperm development comprises a series of developmental transitions controlled by genetic and epigenetic mechanisms that are initiated after double fertilization. Polycomb repressive complex 2 (PRC2) is a key component of these mechanisms that mediate histone H3 lysine 27 trimethylation (H3K27me3); the action of PRC2 is well described in Arabidopsis thaliana but remains uncertain in cereals. In this study, we demonstrate that mutation of the rice (Oryza sativa) gene EMBRYONIC FLOWER2a (OsEMF2a), encoding a zinc-finger containing component of PRC2, causes an autonomous endosperm phenotype involving proliferation of the central cell nuclei with separate cytoplasmic domains, even in the absence of fertilization. Detailed cytological and transcriptomic analyses revealed that the autonomous endosperm can produce storage compounds, starch granules, and protein bodies specific to the endosperm. These events have not been reported in Arabidopsis. After fertilization, we observed an abnormally delayed developmental transition in the endosperm. Transcriptome and H3K27me3 ChIP-seq analyses using endosperm from the emf2a mutant identified downstream targets of PRC2. These included >100 transcription factor genes such as type-I MADS-box genes, which are likely required for endosperm development. Our results demonstrate that OsEMF2a-containing PRC2 controls endosperm developmental programs before and after fertilization.
Polycomb Repressive Complex 2 containing EMBRYONIC FLOWER2a controls rice endosperm development before and after fertilization by repressing target genes via epigenetic H3K27me3 modification.
Introduction
In angiosperms, formation of the embryo and endosperm results from separate fertilization events by two sperm cells. The role of the endosperm is to provide nourishment to the embryo and to support seedling growth after seed germination (Lopes and Larkins, 1993). In rice (Oryza sativa), the endosperm stores starches and storage proteins; it has been exploited as a major source of calories in the human diet along with those of similar cereals, such as maize (Zea mays), barley, and wheat. In the present study, we classified rice endosperm development into three stages: coenocyte (previously inappropriately termed “syncytium”), cell division, and maturation. These categories are defined by cytological differences during endosperm development (Ishikawa et al., 2011; Sekine et al., 2013; Wu et al., 2016). In the coenocytic stage, the endosperm nuclei divide without cytokinesis and form a single large cell with multiple cytoplasmic nuclear domains within the ovule (Olsen, 2004). After a number of synchronous nuclear divisions (the actual number varies among species), the endosperm exits from the coenocytic stage and initiates cytokinesis accompanied by phragmoplast and cell wall formation in a process termed cellularization (Otegui and Staehelin, 2000; Olsen, 2004). The cells further divide in a centripetal direction and begin to deposit storage compounds during the maturation stage.
The transition from the coenocytic to cellularization stage is a developmental landmark and a useful cytological marker for characterizing early endosperm development in rice and Arabidopsis thaliana (Arabidopsis). This transition is thought to be necessary for embryonic growth and the biosynthesis of storage products in Arabidopsis; it has been proposed that stored hexoses are converted to sucrose to be utilized for growth of the globular stage embryo and thereafter (Hehenberger et al., 2012; Lafon-Placette and Köhler, 2014). Cellularization is controlled by the agamous-like MADS-box protein AGL62 together with the paternally expressed imprinted gene PHERES1 (PHE1; de Folter et al., 2005; Kang et al., 2008). In rice, the timing of cellularization is altered by parental genome balance in interspecific or interploidy crosses (Ishikawa et al., 2011; Sekine et al., 2013; Zhang et al., 2016; Tonosaki et al., 2018) and may be mediated by type-I MADS-box transcription factors (Ishikawa et al., 2011; Chen et al., 2016).
Endosperm development is also controlled by genomic imprinting (Haig and Westoby, 1991; Haig, 2013). Genomic imprinting refers to the monoallelic expression of a gene depending on its parent-of-origin. Unequal gene expression patterns affecting maternally and paternally derived genomes during endosperm development can confer opposite functions (Feil and Berger, 2007; Kinoshita et al., 2008). Genome-wide analyses have shown limited overlap of imprinted genes in Arabidopsis and cereals (Waters et al., 2013; Xin et al., 2013; Yuan et al., 2017; Chen et al., 2018; Yang et al., 2018; Wyder et al., 2019), likely reflecting differences in the ephemeral versus persistent nature of endosperm in eudicots and monocot species. Recent studies have shown that many imprinted genes in rice are associated with quantitative trait loci for grain size (Yuan et al., 2017). One of the most striking characteristics of imprinting in many plant species is that some components of the polycomb repressive complex 2 (PRC2), which catalyzes histone H3 lysine 27 methylation (H3K27me3) to repress gene expression, are maternally expressed and are involved in the control of endosperm development (Tonosaki and Kinoshita, 2015). Therefore, there may be an evolutionary constraint on the machinery that allocates nutrition to the embryo, which is consistent with the proposed theory (Haig, 2013).
In Arabidopsis, different forms of PRC2 govern distinct developmental transitions during the plant lifecycle (Butenko and Ohad, 2011; Mozgova and Hennig, 2015). For example, FERTILIZATION-INDEPENDENT SEED2 (FIS2)-containing PRC2 (FIS-class PRC2) specifically controls both endosperm proliferation and the repression of autonomous endosperm development without fertilization (Kiyosue et al., 1999; Luo et al., 2000; Zhang et al., 2018). In Arabidopsis, endosperm-specific FIS-class PRC2 is composed of four core component proteins: FIS2, MEDEA (MEA), FERTILIZATION-INDEPENDENT ENDOSPERM, and MULTICOPY SUPPRESSOR OF IRA1 (Hennig and Derkacheva, 2009; Bemer and Grossniklaus, 2012). PRC2 also contributes to the silencing of the maternal alleles of paternally expressed imprinted genes (PEGs) (Batista and Kohler, 2020); one of the components of MEA controls its own imprinting (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006). Thus, FIS-class PRC2 controls endosperm development in Arabidopsis through imprinted gene expression.
The identities of the component proteins in the counterpart complex in rice are currently unclear (Tonosaki and Kinoshita, 2015). Although OsFIE1 and OsFIE2 are well documented in many reports (Nallamilli et al., 2013; Folsom et al., 2014; Li et al., 2014; Huang et al., 2016; Liu et al., 2016; Zhong et al., 2018), mutations in their respective genes do not result in comparably severe phenotypes to those reported in Arabidopsis. This may be due to genetic redundancy with genes with overlapping expression patterns in rice endosperm, unlike in Arabidopsis. OsFIE1 is a maternally expressed imprinted gene (MEG) that is specifically expressed in the endosperm; in contrast, OsFIE2 is not imprinted and is expressed in various organs including endosperm (Luo et al., 2009). Thus, OsFIE1- and OsFIE2-containing PRC2 might redundantly contribute to endosperm development. Accordingly, while the insertion of a T-DNA sequence into OsFIE1 did not cause an abnormal phenotype (Luo et al., 2009), over-expression of OsFIE1 caused precocious cellularization and reduced seed size (Folsom et al., 2014).
Two homologs of EMF2, Oryza sativa gene EMBRYONIC FLOWER2a (OsEMF2a), and OsEMF2b, have been identified in the rice genome; these homologs encode zinc-finger containing components of PRC2 (Luo et al., 2009). OsEMF2a has been shown by a whole-genome survey to be a maternally expressed imprinted gene (Chen et al., 2018); however, exploration of the role of OsEMF2a has only recently been undertaken (Cheng et al., 2020a). In contrast to OsEMF2a, OsEMF2b has been shown to control flowering time and to determine floral identity (Luo et al., 2009; Yang et al., 2013; Conrad et al., 2014; Xie et al., 2015).
In the current study, we focused on the imprinted gene OsEMF2a and found that PRC2 represses central cell nuclear division before fertilization and the endosperm developmental transition after fertilization. These characteristic phenotypes are conserved in the eudicot species Arabidopsis and the monocot species rice. However, we also found striking phenotypes that were characteristic of rice PRC2. The autonomous endosperm showed increased expression of genes encoding storage compounds, which are normally expressed during the later maturation stage. Consistently, we found starch granules and protein storage vacuole-like structures in the autonomous endosperm of plants heterozygous for a mutation of OsEMF2a. Transcriptomic and H3K27me3 ChIP-seq analyses revealed that OsEMF2a controls endosperm proliferation and the developmental transition after fertilization by repressing specific target genes, including type-I MADS-box transcription factor genes. Thus, analyses of emf2a mutations allowed us to identify both conserved activities and a role of PRC2 not identified in Arabidopsis, namely, repression of the production of storage nutrients before fertilization.
Results
Proliferation of nuclear cytoplasmic domains and accumulation of starch granules in emf2a autonomous endosperm without fertilization
Since duplicated genes tend to be epigenetically neo-functionalized in the endosperm (Dickinson et al., 2012; Yoshida et al., 2018), we focused on the imprinted gene OsEMF2a to explore the role of PRC2 in rice endosperm. We first confirmed maternal-specific expression of OsEMF2a in our materials and cultivation conditions (Supplemental Figure 1, A and B). To generate a loss-of-function mutation of OsEMF2a, we designed a CRISPR/Cas9 vector targeting the zinc-finger domain of OsEMF2a, which is located in the 10th exon, and introduced it into rice calli. Three independent emf2a alleles were generated: a di-nucleotide deletion (emf2a-3), a di-nucleotide insertion (emf2a-4), and a single nucleotide deletion (emf2a-5). All three alleles were frame-shift mutations (Supplemental Figure 1, C). We also selected lines that were null segregants of the CRISPR/Cas9 construct for these mutants. In the emf2a/+ heterozygotes, we did not detect clear abnormal phenotypes from seedling growth to the flowering stages (Supplemental Figure 1, D). All three mutations allowed viable seeds to be produced; however, no seedlings homozygous for an emf2a mutation were obtained. Therefore, we carried out a detailed investigation of seed development in emf2a/+ plants.
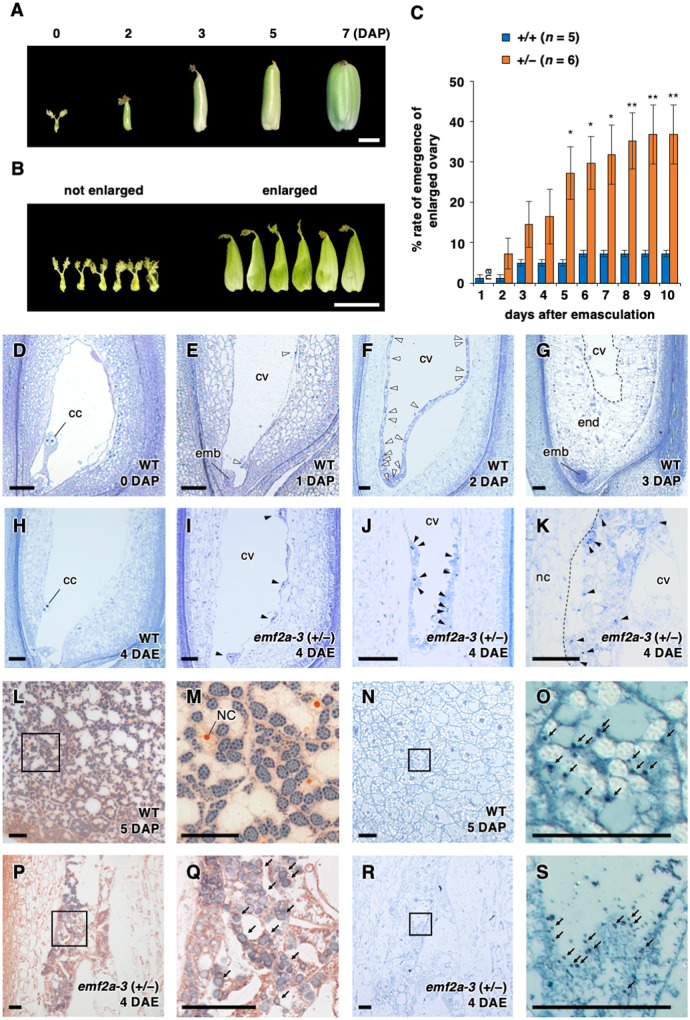
Many studies have attempted to characterize autonomous endosperm development in rice, however, the results were unclear or showed very low penetrance (Luo et al., 2009; Nallamilli et al., 2013; Li et al., 2014; Cheng et al., 2020a, 2020b), thereby precluding functional characterization. In the present study, we noticed a high-frequency emergence of enlarged ovaries after emasculation of emf2a-4/+ plants (Figure 1, A and B) during the course of genetic cross experiments. Therefore, we carefully investigated the rate of emergence of enlarged ovaries using emf2a-4/+ plants and found that 36.8 ± 7.3% of florets had enlarged ovaries at 10 days after emasculation (DAE; Figure 1, C). In contrast, the rate was much lower in wild-type plants under the same conditions (7.3% ± 6.1%; Figure 1, C). This ovary enlargement without fertilization was almost fully penetrant and reproducible in both emf2a-3/+ and emf2a-4/+ (Supplemental Figure 2, A and B).
Figure 1.
Phenotypes of emf2a/+ ovaries without fertilization. (A) Developing seeds of wild-type plants up to 7 days after pollination (DAP). Scale bar = 2 mm. (B) Two ovary classes, not enlarged (left) and enlarged (right), in emf2a-4/+ plants at 10 days after emasculation (DAE). Scale bar = 5 mm. (C) Rate of emergence of an enlarged ovary at 0–10 DAE in wild-type and emf2a-4/+ plants. Data are means ± se of 5–6 plants at each time point. Asterisks indicate significant differences between different emf2a genotypes (Student’s t-test, *P < 0.05, **P < 0.01). (D–K) Toluidine blue stained thin sections. Fertilized seeds of wild-type at 0–3 DAP (D–G) and enlarged ovaries in wild-type at 4 DAE (H) and in emf2a-3/+ plants at 4 DAE (I–K). Empty and solid arrowheads indicate dividing nuclear cytoplasmic domains in the presence or absence of fertilization, respectively. emb, embryo; end, endosperm; cc, central cell nuclei; cv, central vacuole; nc, nucellus. Scale bars = 50 µm. (L–S) Accumulation of starch grains and protein storage vacuoles in fertilized seeds of wild-type at 5 DAP (L–O) and emf2a-3/+ plants at 4 DAE (P–S). Sections doubly stained with iodine and safranin to detect starch grains and cell walls, respectively (L–M) and (P–Q). Differential interference contrast images of CBB stained thin sections (N–O) and (R–S). The boxed regions in (L), (N), (P), and (R) are enlarged as (M), (O), (Q), and (S), respectively. NC, endosperm nuclei. Arrows indicate stained starch grains or protein storage vacuoles. Scale bars = 50 µm.
We investigated the developmental processes in these enlarged ovaries via light microscopy of plastic-embedded thin sections (Figure 1, D–K). Although the polar nuclei did not proliferate in the cavity of the wild-type central cell before fertilization (Figure 1, D), multiple nuclear cytoplasmic domains were observed at 1–2 days after pollination (DAP; Figure 1, E and F). These were already cellularized and started growing in the centripetal direction by 3 DAP (Figure 1, G). Without fertilization, enlarged ovaries of wild-type plants were retained the polar nuclei at 4 DAE (Figure 1, H); however, multiple nuclear cytoplasmic domains were present in the enlarged cavity of the embryo sac in emf2a-3/+ plants at 4 DAE (Figure 1, I–K) and at 3 and 5 DAE in emf2a-4/+ plants (Supplemental Figure 2, C and D). We did not observe any sign of egg cell division in the same series of thin sections or in the images obtained using a confocal microscope (Supplemental Figure 2, E–G), suggesting that the autonomous nuclear divisions in emf2a/+ plants were central cell specific.
The enlarged ovaries of wild-type plants always contained solutes but no other visible substances; in contrast, an iodine-positive whitish mass was visible in the enlarged ovaries of both emf2a-3/+ and emf2a-4/+ plants (Supplemental Figure 3, A–C). This result is consistent with the finding of iodine positive starch granules in the autonomous endosperm and higher starch content in the enlarged ovaries of emf2a-3/+ (Figure 1, P and Q; Supplemental Figure 3, D). Similarly, we found Coomassie Brilliant Blue (CBB)-positive granular structures in the cytosol in endosperm of wild-type plants at 5 DAP (Figure 1, N and O) and in the autonomous endosperm of emf2a-3/+ plants at 5 DAE (Figure 1R and 1S). These structures closely resembled protein storage vacuoles. Thus, OsEMF2a plays a role in repressing nuclear divisions and the acquisition of endosperm characteristics in the central cell before fertilization.
Gene expression profile of emf2a autonomous endosperm without fertilization
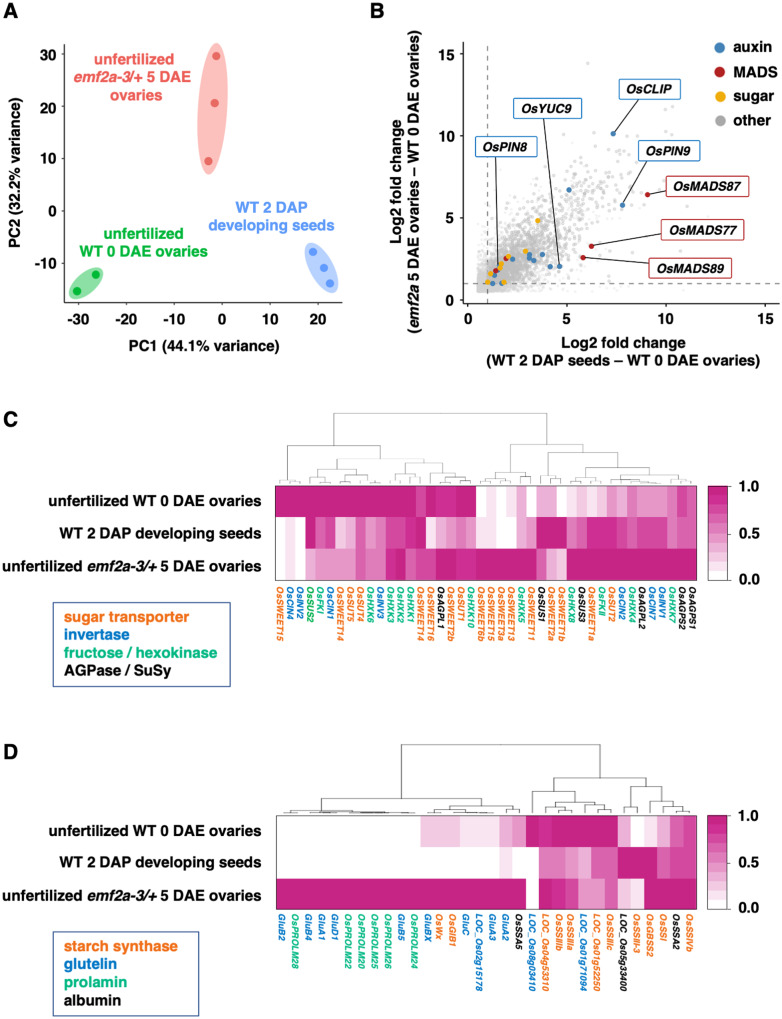
To determine the gene expression profiles in the enlarged ovaries that developed in endosperm of emf2a-3/+ without fertilization, we performed RNA-seq analyses of wild-type unfertilized ovaries at 0 DAE, enlarged emf2a-3/+ ovaries at 5 DAE, and wild-type developing ovaries without the embryo-containing region at 2 DAP. We selected 2 DAP fertilized developing ovaries as a wild-type control because the wild-type endosperm is generally at the coenocytic stage, which is similar to emf2a autonomous endosperm (Figure 1, F). We carried out principal component analysis (PCA) of transcripts and found that each biological replicate clustered in the same proximate areas (Figure 2, A). The transcriptomes of the emf2a-3/+ ovaries, which contained autonomous endosperm, shifted to the region of the fertilized seeds on PC1 without the paternal genome contribution (Figure 2, A). We identified 5,293 differentially expressed genes (DEGs), including 3,298 upregulated and 1,995 downregulated genes, between the control and the unfertilized ovaries (Supplemental Data Sets 1 and 2); 3,889 DEGs, including 2,775 upregulated and 1,114 downregulated genes, were present in the emf2a-3/+ ovaries compared with the unfertilized ovaries (Supplemental Data Sets 3 and 4). In total, 1,494 (32.6%) of the upregulated genes and 683 (28.2%) of the downregulated genes in these comparisons overlapped in Venn diagrams (Supplemental Figure 4).
Figure 2.
Transcriptome analysis of autonomous endosperm development in emf2a-3/+. (A) PCA of variance in normalized read counts for whole transcriptomes in three different libraries prepared from unfertilized wild-type 0 DAE ovaries (green), fertilized wild-type 2 DAP developing seeds without embryos (blue), and unfertilized emf2a-3/+ 5 DAE enlarged ovaries (red), with two or three replicates. The two axes represent the first two principal components (PCs) and the percentages indicate the variance contribution. (B) Scatter plot of commonly upregulated genes in two comparisons: fertilized wild-type 2 DAP developing seeds versus unfertilized wild-type 0 DAE ovaries (X-axis); unfertilized emf2a-3/+ 5 DAE ovaries versus unfertilized wild-type 0 DAE ovaries (Y-axis). Gray points are upregulated genes, and colored points indicate related pathways of gene families for auxin, MADS-box, sugar, and others (detailed information is given in Supplemental Figure 5). The two stippled axes show log2 fold changes in expression of upregulated genes in the two comparisons. (C) and (D) Heatmaps of hierarchical clustering of DEGs that show a higher expression state in unfertilized emf2a-3/+ 5 DAE ovaries with respect to the sugar metabolism pathway (C) and for storage compounds (D). For each gene, the average transcripts per million value normalized by the maximum value of the gene is shown. The different colors of the gene names indicate encoded proteins of each gene.
Autonomous emf2a endosperm may have acquired a genetic program for endosperm development without fertilization, which is likely repressed by the OsEMF2a-containing PRC2 complex in the central cell prior to fertilization. Therefore, we focused on upregulated genes that overlapped in the Venn diagram (Supplemental Figure 4). These genes were representative of those expressed after fertilization, such as auxin biosynthesis genes, type-I MADS box genes, and sugar metabolism genes (Figure 2, B; Supplemental Figure 5). Auxin production and signaling in the primary endosperm and the integuments are characteristic events after fertilization by a sperm cell (Chen et al., 2014; Figueiredo et al., 2015; Figueiredo and Köhler, 2018). In our transcriptome data, genes involved in auxin biosynthesis (OsYUC9) and auxin transport (OsPIN8 and OsPILS1) were upregulated in both fertilized developing seeds and unfertilized emf2a ovaries (Figure 2, B; Supplemental Figure 5, A). Auxin-responsive genes such as auxin response factor genes (OsARF7, OsARF13, and OsARF22), auxin/indole-3-acetic acid family gene (OsIAA10 and OsIAA15), and small auxin upregulated RNA genes (OsSAUR28 and OsSAUR57) were also upregulated in these samples (Supplemental Data Sets 1 and 3). This pattern of expression is consistent with the occurrence of autonomous endosperm development in emf2a-3/+ and emf2a-4/+ plants.
In Arabidopsis, the Mα-type MADS-box gene AGL62 (AGAMOUS-LIKE 62) is required for autonomous endosperm development (Figueiredo et al., 2015). Although the rice orthologues of AGL62 have not been identified, a Mα-type MADS-box gene, OsMADS77, was activated in emf2a-3/+ ovaries (Figure 2, B; Supplemental Figure 5, B). In addition to OsMADS77, the Mγ-type MADS-box gene OsMADS87 was also activated in emf2a-3/+ ovaries (Figure 2, B; Supplemental Figure 5, B); OsMADS87 expression is correlated with coenocytic endosperm (Ishikawa et al., 2011; Tonosaki et al., 2018).
In Arabidopsis, hexoses are converted from sucrose and stored in the central vacuole during the coenocytic endosperm stage; these hexoses are required for turgor pressure and rapid seed growth (Hehenberger et al., 2012; Beauzamy et al., 2016). We found that genes related to hexose synthesis and sucrose transport, namely, fructokinase (OsFKII), hexokinase (OsHXK8), sucrose synthase (OsSUS1 and OsSUS3), sucrose transporters (OsSUT2, OsSWEET1a, and OsSWEET11), and cell wall invertase (OsCIN2), were commonly expressed in emf2a-3/+ ovaries and developing seeds (Supplemental Figure 5C). We also analyzed the expression patterns of genes related to the sugar metabolism pathway and maturation stage-specific storage products (Figure 2, C and D), as we observed structures similar to starch granules and protein storage vacuoles in the autonomous endosperm. Waxy (OsWx) was upregulated in these samples, along with genes related to storage proteins, which are specific to the protein storage organelles in the seed, even in the absence of fertilization (Figure 2, D). Taken together, these results indicate that the gene expression profile of unfertilized emf2a-3/+ ovaries acquires, at least in part, similar characteristics to those observed during rice endosperm development.
Maternal OsEMF2a is involved in seed development after fertilization
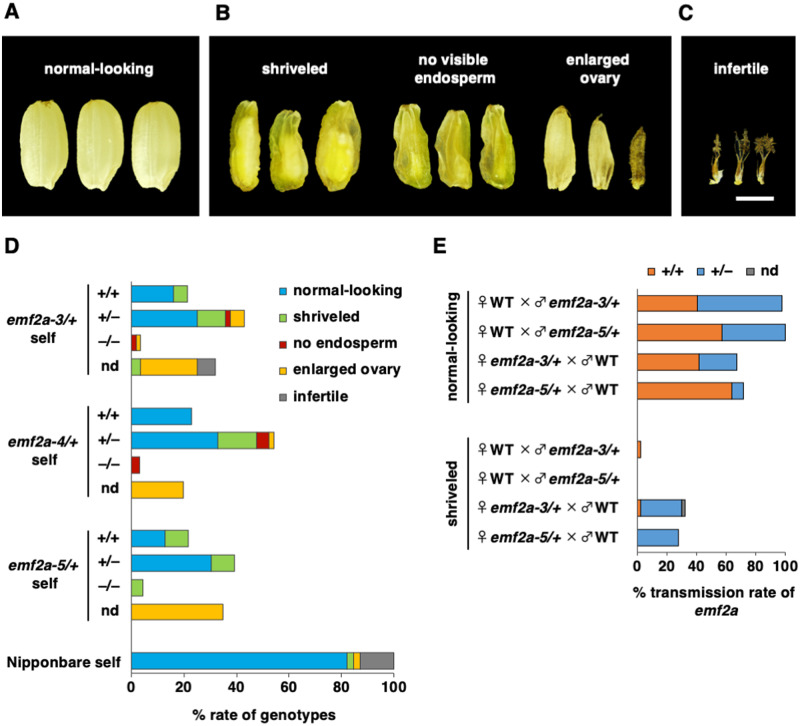
Next, we investigated seed phenotypes of the emf2a mutants to characterize the role of OsEMF2a in seed development. The results from self-pollinated seeds from the three emf2a/+ alleles were consistent. Approximately half of all seeds had a wild-type-like appearance (normal-looking), while the remainder displayed a range of abnormal phenotypes: shriveled seeds with some endosperm and a viable embryo (shriveled seed); flat seeds with little or no endosperm (no visible endosperm); enlarged ovary without a visible embryo or endosperm (enlarged ovary); and nongrowing ovary (infertile) (Table 1; Figure 3, A–C). The segregation ratios of normal-looking seeds to abnormal seeds for each emf2a mutant did not conform to a Mendelian 3:1 inheritance pattern (Table 1). All of the normal-looking seeds that were tested and some of the shriveled seeds germinated, but no germination occurred from the other abnormal categories, namely, seeds with no visible endosperm and seeds with an enlarged ovary (Table 1).
Table 1.
Seed phenotypes of F1 generations derived from self-pollinated emf2a/+ plants and reciprocal crosses
| Plant materials (generation) |
Total seeds | Normal seeds (n: germinated, seedling)a |
Abnormal seeds (n: germinated, seedling) |
Infertile (n) | Chi-square test (3:1) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maternal | Paternal | Normal looking | Shriveled | No visible endosperm | Enlarged ovary | X2 | P-value | |||||||
| Self-pollinationb | ||||||||||||||
| Wild-type | Wild-type | 39 | 82.05% | (32) | 2.56% | (1) | 0.00% | (0) | 2.56% | (1) | 12.82% | (5) | − | |
| emf2a-3/+ (T2) | emf2a-3/+ (T2) | 56 | 41.07% | (23: 23, 21) | 19.64% | (11: 5, 1) | 3.57% | (2: 0, 0) | 28.57% | (16: 0, 0) | 7.14% | (4) | 26.26 | 3.00E-07 |
| emf2a-4/+ (T4) | emf2a-4/+ (T4) | 61 | 55.74% | (34: 34, 34) | 14.75% | (9: 4, 1) | 8.20% | (5: 0, 0) | 21.31% | (13: 0, 0) | 0% | (0) | 12.07 | 5.10E-04 |
| emf2a-5/+ (T4) | emf2a-5/+ (T4) | 23 | 43.48% | (10: 10, 10) | 21.74% | (5: 1, 1) | 0.00% | (0) | 34.78% | (8: 0, 0) | 0% | (0) | 12.19 | 4.80E-04 |
| Reciprocal crossesc | ||||||||||||||
| Wild-type | emf2a-3/+ (T4) | 44 | 97.73% | (43: 43, 43) | 2.27% | (1: 1, 0) | – | – | – | – | ||||
| emf2a-3/+ (T4) | Wild-type | 48 | 62.50% | (30: 29, 29) | 37.50% | (18: 14, 10) | – | – | – | – | ||||
| Wild-type | emf2a-5/+ (T6) | 14 | 100.00% | (14: 14, 14) | 0.00% | (0) | – | – | – | – | ||||
| emf2a-5/+ (T6) | Wild-type | 28 | 64.29% | (18: 18, 18) | 35.71% | (10: 7, 2) | – | – | – | – | ||||
n: number of seeds, germinated; number of germinated seeds, seedling; number of germinated seeds that survived as seedlings.
All flowers on a panicle were investigated.
Plump and shriveled seeds were investigated.
Figure 3.
Parent-of-origin effect of the emf2a mutation during seed development. (A–C) Seed phenotypes at the maturation stage from self-pollinated emf2a-3/+ plants. Phenotypes were categorized as normal-looking (A), three abnormal types (B), and infertile (C). Scale bar = 2 mm. (D) Distribution of each seed phenotype depending on genotype from self-pollination of wild-type plants and plants carrying an emf2a mutation. Y-axis indicates the mutant allele and genotype of self-pollinated seeds. (E) Transmission rate of the emf2a allele in the F1 progeny of reciprocal crosses between wild-type plants and plants carrying either emf2a-3 or emf2a-5. nd, not determined.
We determined the genotypes of seedlings from seeds that successfully germinated and of small samples of tissues dissected from inviable seeds. We were unable to extract DNA from enlarged ovaries due to the lack of internal tissue. Wild-type segregants produced seeds with a normal-looking phenotype and seeds with a shriveled appearance in some population; heterozygotes produced normal-looking seeds and a variety of abnormal seeds (Figure 3, D). All three emf2a mutations fell into the inviable seed categories when homozygous (Figure 3, D); thus, viable homozygous emf2a plants were never obtained.
If the paternal OsEMF2a allele is completely silenced by genomic imprinting and there is no genetic redundancy with OsEMF2b, then half of the seeds would appear normal-looking and the other half would be aborted in a cross between a maternal emf2a heterozygote and a wild-type male. In contrast, only normal-looking seeds would be expected in the reciprocal cross; these would consist of a 1:1 ratio of wild-type to heterozygous seeds. To address this question, we analyzed the parental transmission rate of emf2a in reciprocal crosses between emf2a-3 or emf2a-5 heterozygotes and wild-type plants by genotyping germinated viable seeds (Figure 3, E).
In crosses in which emf2a-3/+ or emf2a-5/+ plants were used as the pollen donor, almost all germinated seeds were normal looking regardless of their genotypes (Figure 3, E; Table 1). By contrast, a number of shriveled seeds were observed when emf2a-3/+ or emf2a-5/+ plants were used as the maternal parent (Figure 3, E; Table 1). Thus, shriveled seeds appeared after the maternal transmission but not paternal transmission of emf2a. Importantly, the maternally transmitted emf2a-3 or emf2a-5 alleles did not always cause a shriveled phenotype, suggesting that there was incomplete silencing of the paternal allele or genetic redundancy, possibly with OsEMF2b. Consistent with these observations, we observed a relatively high amount of paternally-derived read counts from OsEMF2a compared with the imprinted OsFIE1 control (Supplemental Figure 1A and 1B); we also observed the expression of OsEMF2b during endosperm development (Supplemental Figure 6). Overall, these results suggest that the paternal transmission of emf2a had little effect on seed development, whereas maternal transmission was of considerable importance to the development of severe defects.
OsEMF2a controls the timing of cellularization
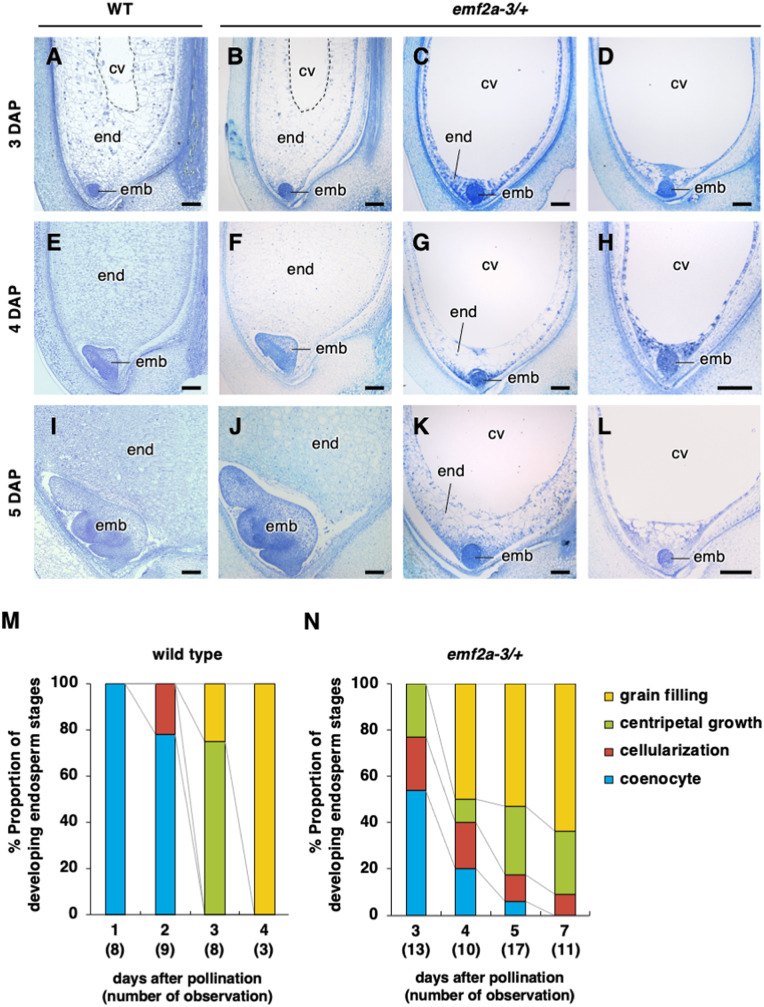
To clarify the effect of emf2a on embryo and endosperm development after fertilization, we analyzed tissue sections of developing seeds derived from emf2a-3/+ plants. Since the genotypes were difficult to determine from plastic-embedded tissue sections, and because emf2a-bearing seeds showed a broad spectrum of phenotypes (described above), we analyzed multiple seed samples. Under our cultivation conditions using a biotron chamber, seed development was stable and highly reproducible (Ohnishi et al., 2011). In seeds of wild-type plants, the globular embryo and the coenocytic endosperm formed by 2 DAP, and cellularization and cell divisions continued in the centripetal direction at ∼3 DAP. At the maturation stage, starch granules were observed in the amyloplasts in the endosperm (grain filling stage) (Figure 4, A, E, I, and M).
In contrast, in seeds of self-pollinated emf2a-3/+ plants at 3 DAP, approximately half of the endosperm had passed the cellularization stage and divided in the centripetal direction (Figure 4, B and 4); however, the other half had not initiated cellularization and remained at the coenocytic stage (Figure 4, C, D. and N). In addition to this delayed development, the coenocytic endosperm displayed unusual vacuolated structures and centripetal proliferation without cellularization, especially at the embryo periphery (Figure 4, C and D). Similarly, in some seeds at 4 DAP, the endosperm had reached the grain filling stage (Figure 4, F and N), whereas other seeds showed delayed developmental transition with unusual cellular masses in the endosperm (Figure 4, G, H, and N). At 5 and 7 DAP, we still observed the coenocytic, cellularized, and centripetal growth endosperm stages in tissue sections, similar to those in seeds at earlier stages of development. However, the proportion of seeds at the grain filling stage had increased to over 50% (Figure 4, N), which is consistent with the finding that some normal-looking seeds were generated after maternal transmission of the emf2a-3 or emf2a-5 alleles. We also observed delayed or arrested embryo development accompanied by unusual endosperm (Figure 4, L). The perceived stage of embryo development was likely affected by the degree of endosperm abnormality; thus, the accumulation of starch granules correlated roughly with the stage of embryo development (Supplemental Figure 7). These observations indicate that OsEMF2a play a primary role in endosperm development.
Figure 4.
Embryo and endosperm development in seeds from self-pollinated wild-type and emf2a-3/+ plants. (A–L) Representative images of seeds of wild-type (A), (E), and (I) and emf2a-3/+ plants (B–D), (F–H), and (J–L) at 3 (A–D), 4 (E–H), and 5 DAP (I–L). Each panel of emf2a-3/+ plants displays wild-type appearance, centripetal growth stage (B) and grain filling stage (F) and (J), and unusual phenotypes, cellularization stage (C), (G), and (K) and coenocytic stage (D), (H), and (L). end, endosperm; emb, embryo; cv, central vacuole. Scale bars = 100 µm. (M) and (N) Proportions of different endosperm developmental stages from self-pollinated wild-type at 1–4 DAP (M) and emf2a-3/+ plants at 3–5, and 7 DAP (N).
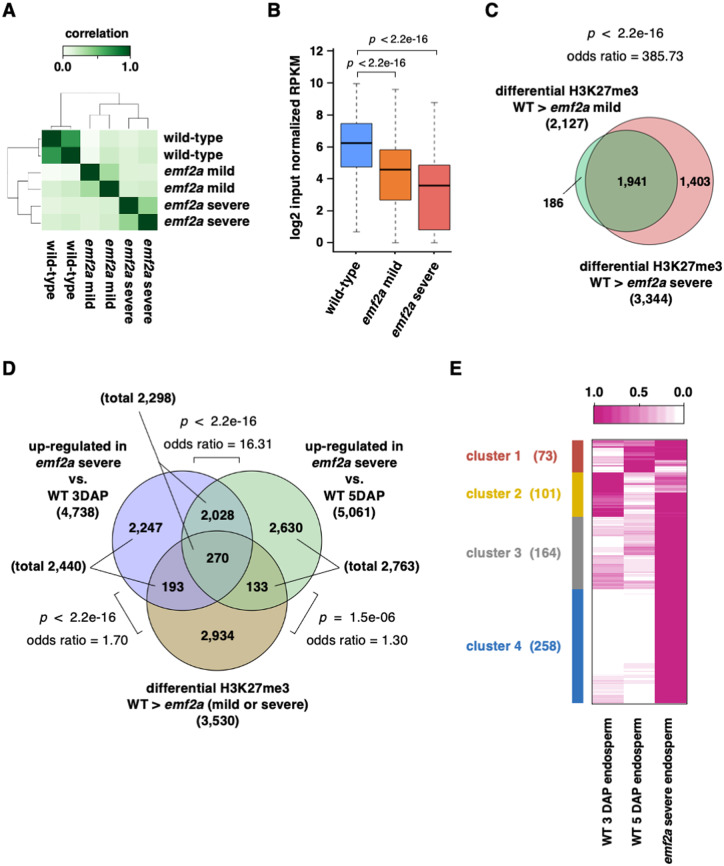
Identification of OsEMF2a-containing PRC2 targets in developing endosperm by H3K27me3 ChIP-seq analysis
Our phenotypic analyses of emf2a endosperm identified the roles of OsEMF2a-containing PRC2 in the repression of autonomous endosperm development before fertilization and the developmental transition of endosperm after fertilization. We reasoned that OsEMF2a-containing PRC2 might repress the key regulatory genes for these events via deposition of H3K27me3. Therefore, we sought to identify potential downstream direct target genes of OsEMF2a-containing PRC2 using a combination of RNA-seq and ChIP-seq analyses. For the RNA-seq analysis, we harvested wild-type endosperm at 3 and 5 DAP; these sampling times correspond to the centripetal growth and grain-filling stages of seed development, respectively (Supplemental Figure 8, A and B). We also harvested endosperm samples from self-pollinated emf2a-3/+ plants at 5 DAP. We selected relatively severe phenotypes showing liquid uncellularized endosperm (Supplemental Figure 8, C) and confirmed that the majority of read counts in the RNA-seq analysis arose from the mutant allele (Supplemental Figure 8, D). Consistent with delayed development, severely abnormal emf2a endosperm at 5 DAP was relatively close to the wild-type endosperm at 3 DAP on PC1 (Supplemental Figure 8E).
For the H3K27me3 ChIP-seq analysis, we collected wild-type endosperm at 5 DAP to estimate total H3K27me3 marked regions as a reference. In addition, we selected two types of emf2a endosperm phenotypes, partially cellularized and liquid uncellularized, which we refer to as mildly and severely abnormal endosperm, respectively. We sought to determine whether these phenotypic variations could be explained by their respective levels of H3K27me3 enrichment. The ChIP-seq signals were highly correlated in each biological replicate (Supplemental Figure 9, A). To estimate genotype proportions in the mild and severe emf2a endosperm phenotypes, we aligned control reads derived from input DNA materials before immunoprecipitation with an H3K27me3 antibody to the reference genome.
In the mild emf2a endosperm fractions, the proportion of mutant to wild-type alleles was close to a 2:1 ratio (note that the ratio is two maternal genomes to one paternal genome in the endosperm; Supplemental Figure 9, B). In contrast, the ratio of mutant to wild-type sequences deviated from a 2:1 ratio in the severe emf2a endosperm fractions (Supplemental Figure 9, B), suggesting that some homozygotes were present in this fraction.
We then performed peak calling to identify H3K27me3-enriched regions. In wild-type endosperm, we identified 19,514 H3K27me3 peaks in the whole genome, 10,308 of which overlapped with the gene bodies (Supplemental Figure 9, C; Supplemental Data Set 5). In mild and severe emf2a endosperms, we also found 24,104 and 11,548 peaks in the whole genome and 12,369 and 5,415 peaks that overlapped with gene bodies, respectively (Supplemental Figure 9, C; Supplemental Data Set 5). A relatively large number of H3K27me3 peaks were identified in the mild emf2a endosperm compared to wild type. This is likely due to residual shorter peaks in the mild emf2a endosperm; the peak widths and the levels of enrichment were significantly lower in this tissue (Supplemental Figure 9, D and E). We found that 10,265, 11,248, and 5,144 genes were associated with at least one of the H3K27me3 peaks in the wild-type, mild, and severe emf2a endosperms, respectively (Supplemental Data Set 5). Since the developmental stages of the wild-type, mild, and severe endosperms varied as described above, the genes associated with H3K27me3 did not fully overlap (Supplemental Figure 9, F). Nevertheless, we developed a list of H3K27me3-marked genes in the endosperm for further analyses (Supplemental Data Set 5).
Next, we performed a comparative analysis of the read counts on all peak regions using three samples. The normalized read counts were concordant between replicates (Figure 5, A). In mild and severe emf2a endosperms, we identified 4,755 and 5,806 differentially enriched regions in pairwise comparisons to wild type, respectively. The levels of H3K27me3 enrichment gradually decreased in mild and severe endosperms (Figure 5, B). In these emf2a endosperms, 2,627 and 4,544 peaks had significantly reduced H3K27me3 levels, and these peaks were associated with 2,127 and 3,344 genes, respectively, most of which overlapped (Figure 5, C; Supplemental Data Set 6). Thus, we identified 3,530 genes as OsEMF2a-containing PRC2 target genes and analyzed their H3K27me3 levels (Figure 5, C).
Figure 5.
Analysis of the OsEMF2a target genes in emf2a endosperm based on transcriptome analysis and H3K27me3 ChIP-seq data. (A) Correlation heatmaps of H3K27me3 enrichment on peak locations detected in three different libraries: wild-type endosperm, emf2a mild endosperm, and emf2a severe endosperm at 5 DAP. (B) Boxplots of H3K27me3 enrichment on significant enrichment peaks for three different samples, showing the median and 25th and 75th percentiles and error bars show the 10th and 90th percentiles. P-values were calculated with Wilcoxon rank-sum test. (C) Venn diagram showing genes with significantly decreased H3K27me3 enrichment in mild or severe emf2a endosperm compared with wild-type, and the overlap between each set of genes. P-value and odds ratio were calculated with Fisher’s exact test. (D) Venn diagram showing overlapping upregulated genes in emf2a endosperm in two comparisons: emf2a 5 DAP versus wild-type 3 DAP endosperm and emf2a 5 DAP versus wild-type 5 DAP endosperm; and overlapping genes with decreased H3K27me3 enrichment in emf2a endosperm compared with wild-type 5 DAP endosperm. P-value and odds ratios were calculated with Fisher’s exact test. (E) Heatmap of k-means clustering of OsEMF2a targeted genes upregulated by emf2a mutation. For each gene, the average transcripts per million value normalized by the maximum value of the gene is shown. The different colored boxes on the left indicate different clusters of genes with similar expression patterns.
Gene ontology (GO) analysis of the 3,530 target genes showed they were particularly enriched for sequence-specific DNA binding genes, including 29 MADS-box genes, 31 WRKY genes, and 27 homeobox genes (Supplemental Table 1 and Supplemental Data Set 6), indicating that OsEMF2a represses over 100 transcription factor genes. In our analysis, the listed 3,530 target genes overlapped with known imprinted genes (Luo et al., 2011; Yuan et al., 2017; Chen et al., 2018). While only 14 out of 300 MEGs were targeted, almost two-thirds of PEGs (190 out of 288) were targeted by OsEMF2a-containing PRC2 (Supplemental Figure 10, A and B, Supplemental Data Set 7). Thus, rice PRC2 is an important regulator that represses the maternal alleles of PEGs to establish genomic imprinting.
In an Arabidopsis PRC2 mutant, only some PRC2 target genes are up-regulated (Weinhofer et al., 2010). Therefore, we searched emf2a endosperm for genes with reduced levels of H3K27me3 and increased levels of expression. Among the 3,530 putative targets, 596 genes (16.9%) were upregulated in emf2a endosperm compared to 3 or 5 DAP wild-type endosperm (Figure 5D; Supplemental Data Set 6). The de-regulated genes were assigned to four groups by k-means clustering based on their expression patterns (Figure 5, E). Some of the de-regulated genes in emf2a endosperm were also preferentially expressed in 3 or 5 DAP wild-type endosperm (clusters 1 and 2). However, many of the de-regulated genes showing higher expression in emf2a endosperm were not expressed or had low levels of expression in wild-type endosperm; these genes fell into cluster 3 and 4 (Figure 5, E, see also the GO terms in Supplemental Table 2).
Consistent with a recent report (Cheng et al., 2020a), many up-regulated genes in emf2a endosperm, in addition to putative direct targets of OsEMF2-containing PRC2, were not marked with H3K27me3 or did not show changes in the level of H3K27me3 between the mutant and wild type (Supplemental Figure 10, C, Supplemental Data Set 6). This group of genes is likely to include indirect targets of OsEMF2a-containing PRC2. Interestingly, we observed upregulation of many cell cycle-related genes, which is consistent with a recent report (Cheng et al., 2020a); many of these genes fell into this grouping, with the exception of Orysa;CYCD2;3 (LOC_Os03g27420) and Orysa;DEL1 (LOC_Os06g13670) (Supplemental Data Set 6).
OsEMF2a targets MADS-box transcription factor genes
Our analysis identified more than 100 transcription factor genes among OsEMF2a-containing PRC2 target genes. In Arabidopsis, the type-I MADS-box transcription factors AGL62 and PHE1 control the transition from the coenocytic to cellular stages of endosperm development and are known to be targets of FIS-class PRC2 (Hehenberger et al., 2012; Batista et al., 2019). In rice, we observed a delayed cellularization phenotype in emf2a endosperm (Figure 4). We examined the expression levels of OsEMF2a-containing PRC2 target genes to identify which rice type-I MADS-box genes displayed expression that was correlated with the timing of cellularization, i.e., that showed higher expression in the early stages of endosperm development, as has been found for AGL62 and PHE1 in Arabidopsis.
In our analysis, 29 type-I MADS-box transcription factor genes were targeted by OsEMF2a (Supplemental Data Set 8). Approximately half of the annotated MIKCc-type MADS-box genes were targeted by OsEMF2a and highly expressed in wild-type endosperm at 3 and 5 DAP (Supplemental Data Sets 8 and 9), although they were not upregulated in emf2a endosperm (Figure 6A). In contrast, one Mα-type and five Mγ-type MADS-box genes were highly expressed in 3 DAP wild-type endosperm and upregulated in emf2a endosperm (Figure 6A). Interestingly, the Mγ-type MADS-box gene OsMADS87 (Figure 6, A and B) and three other Mγ-type genes were not direct targets of OsEMF2a, whereas Mα-type OsMADS77 and the Mγ-type OsMADS89 were OsEMF2a targets and were upregulated in emf2a endosperm (Figure 6, A, C, and D). Thus, we succeeded in narrowing down the many candidate type-I MADS box genes to two genes: OsMADS77 and OsMADS89; these genes are directly controlled by OsEMF2a-containing PRC2 and therefore might control the timing of cellularization in rice endosperm in a similar manner to AGL62 and PHE1 in Arabidopsis.
Figure 6.
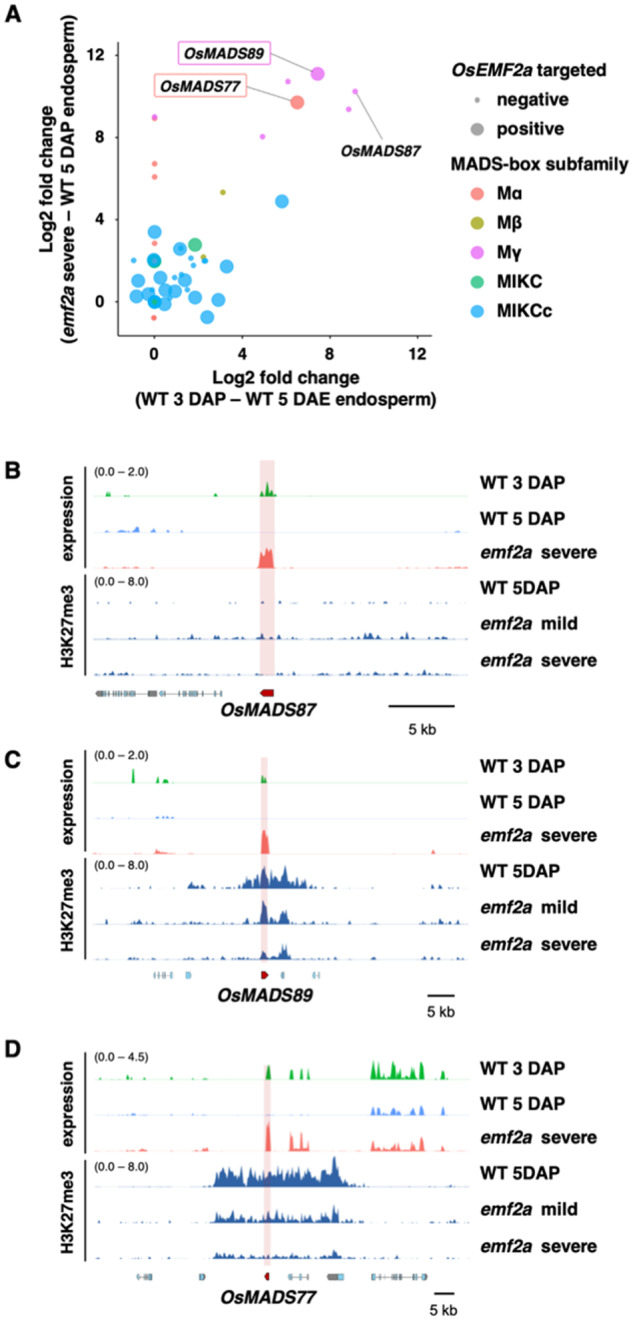
Expression and H3K27me3 profiles of MADS-box genes in wild-type and emf2a-3/+ endosperm. (A) Upregulated type-I MADS-box genes in two comparisons (emf2a 5 DAP versus wild-type 5 DAP endosperm and wild-type 3 DAP versus wild-type 5 DAP endosperm). Each MADS-box subfamily is shown by a colored circle. The two axes show log2-fold changes in expression of upregulated genes. OsEMF2a targeted genes are indicated by the size of the closed circle and their gene names are shown in the open boxes. (B–D) Transcription and H3K27me3 enrichment in the vicinity of OsMADS87 (B), OsMADS89 (C), and OsMADS77 (D). The upper three panels indicate the expression levels of log2 bins per kilobase million values of wild-type 3 DAP and 5 DAP endosperm and emf2a 5 DAP endosperm. The lower three panels show H3K27me3 enrichment of wild-type 5 DAP endosperm and emf2a 5 DAP mild and severe endosperm.
Discussion
PRC2 is an indispensable, conserved epigenetic component that catalyzes H3K27me3 in plants and mammals (Guitton and Berger, 2005; Grossniklaus and Paro, 2014). In plants, the role of PRC2 is well characterized, such as the repression of key transcription factor genes and genes related to many aspects of plant development (Hennig and Derkacheva, 2009; Tonosaki and Kinoshita, 2015). In Arabidopsis, three different PRC2 complexes, including FIS-class PRC2, control distinct developmental transitions. Mutants in each of the FIS-class components, including fis2, mea, and fie, exhibit similar phenotypic defects with respect to the repression of autonomous endosperm before fertilization and cellularization after fertilization (Guitton and Berger, 2005; Hands et al., 2016). This effect might be simply explained by low genetic redundancy of the PRC2 complexes in Arabidopsis. However, in rice, gene duplication can be seen with OsEMF2a and OsEMF2b and with OsFIE1 and OsFIE2 (Furihata et al., 2016; Cheng et al., 2020a, 2020b). These genes are redundantly expressed in the endosperm; therefore, until recently, it has been difficult to demonstrate the role of PRC2 in rice endosperm. Our analysis and recent reports have shown the occurrence of autonomous endosperm and cellularization defects in CRISPR/Cas9-based mutants of OsEMF2a (Cheng et al., 2020a) and the redundant genes OsFIE1 and OsFIE2 (Cheng et al., 2020b)
In this study, we exploited heterozygous emf2a mutants to demonstrate repression of autonomous endosperm and delayed cellularization. The occurrence of an autonomous endosperm phenotype in rice was a controversial topic: one study reported no autonomous development in an OsFIE2 RNAi line (Nallamilli et al., 2013), whereas another study provided evidence for cellularized autonomous endosperm using a different OsFIE2 RNAi line (Li et al., 2014). Recent studies have reported that homozygous mutations in OsEMF2a or OsFIE2 cause nuclear divisions in the unfertilized central cell, albeit with low penetrance (Cheng et al., 2020a, 2020b). Since heterozygosity for our mutant emf2a-3 and emf2a-4 alleles resulted in almost full penetrance for nuclear divisions without fertilization, we further characterized the autonomous endosperm phenotype in rice. We observed structures similar to starch granules and protein bodies under the microscope. Consistent with these observations, we also detected upregulation of genes related to starch synthesis and storage proteins specific to the endosperm. The different levels of penetrance of the mutant alleles between the previous reports and the current one may be due to the use of homozygous and heterozygous emf2a plants. Homozygous mutants exhibit severe defects in general; however, sporophytic and gametophytic effects cannot be separated during sexual reproduction. Since the PRC2 plays a role in seed coat development in Arabidopsis (Figueiredo and Köhler, 2018), heterozygous mutants have an advantage in that they can be used to investigate gametophytic function and monoallelically expressed genes in the endosperm. In any cases, these results provide strong evidence that rice PRC2 represses central cell proliferation and endosperm formation before fertilization.
Our analyses provided mechanistic insights into imprinting of the OsEMF2a locus. This gene locus did not show deposition of H3K27me3 in either wild-type or emf2a mutant endosperm. Instead, the promoter region of the gene was highly DNA methylated in the embryos, while the level of methylation was reduced in the endosperm (Chen et al., 2018). These observations suggest that maternal hypomethylation of this locus in rice endosperm was likely due to genome-wide DNA demethylation in rice endosperm (Rodrigues et al., 2013). Although DNA methylation may also be involved in silencing of the paternal OsEMF2a, imprinting of this gene may not be strictly controlled. We observed substantial expression of the paternal OsEMF2a allele at relatively high levels compared with OsFIE1. In addition, variations in OsEMF2a expression patterns have been described by various studies. Luo et al. reported that OsEMF2a was not imprinted, whereas Chen et al. provided evidence for imprinting at this locus (Luo et al., 2009; Chen et al., 2018). Expression of the paternal allele seems to depend on developmental timing and subspecies combination (Kuang et al., 2019; Cheng et al., 2020a). These issues may explain the phenotypic variation of emf2a mutants in the endosperm in our analysis. Further investigations will be required to clarify the control of OsEMF2a imprinting together with the other possibility of genetic redundancy with OsEMF2b.
The timing of cellularization is an important aspect of endosperm development in rice; precocious and delayed timing are associated with abnormal development of the endosperm in interspecific and interploidy crosses of rice (Ishikawa et al., 2011; Sekine et al., 2013; Zhang et al., 2016; Tonosaki et al., 2018). In our analyses, we showed that OsMADS77 and OsMADS89 were predominantly expressed during earlier developmental stages and that repression of expression after cellularization was associated with H3K27me3 deposition; the timing of expression of these genes was altered in emf2a endosperm. Although we did not detect significant enrichment of H3K27me3 at the OsMADS87 locus, the role of this gene during endosperm development is still of interest, as it is upregulated in emf2a plants. In addition, OsMADS87 is a maternally expressed imprinted gene whose expression pattern correlates with the timing of cellularization (Ishikawa et al., 2011; Tonosaki et al., 2018). OsMADS87 and OsMADS77 are also highly expressed in autonomous endosperm. These genes may play a role in early rice endosperm development similar to that of Arabidopsis AGL62 (Figueiredo et al., 2016) and are of interest for future investigations together with OsMADS89, which is a direct target of OsEMF2a. Interestingly, a recent study succeeded in constructing the quintuple mutation combination osemf2a osmads77 osmads78 osmads79 osmads87, which showed no complementation of the phenotypic defect in the homozygous emf2a mutant caryopsis (Cheng et al., 2020a). This observation exemplifies the need for further analyses using mutations of OsMADS89.
The evolutionary origin of endosperm is unclear (Friedman, 2001; Baroux et al., 2002). Studies from several basal angiosperms led to the proposal that a supernumerary embryo may have evolved into nutritional tissue similar to endosperm (Williams and Friedman, 2002). An alternative hypothesis is that endosperm is derived from an embryo-nourishing female gametophyte (Williams and Friedman, 2002). The validity of these hypotheses has not been determined, and recent molecular evidence still supports both possibilities (Yuan et al., 2018). In our analysis, we showed that the rice central cell has the ability to proliferate nuclear cytoplasmic domains without fertilization and, furthermore, to accumulate iodine-positive starch granules and Coomassie Brilliant Blue (CBB)-positive protein storage vacuoles. Our cytological findings are consistent with the upregulation of genes encoding the storage proteins albumin, globulin, and prolamin in emf2a seeds (Supplemental Figure 11). Accumulation of storage proteins in the protein storage vacuoles is a specific characteristic of the endosperm at maturity (Shimada et al., 2018). Therefore, it is likely that OsEMF2a-containing PRC2 represses the genetic program in the central cell toward endosperm, such as proliferation activity and storage of nutrients required for embryonic growth, until fertilization with the paternal sperm cell. This hidden genetic program by PRC2 in the rice central cell is similar to the developmental process observed in the embryo-nourishing female gametophyte in gymnosperms (Brown and Lemmon, 2008), where haploid endosperm cells already proliferate before fertilization to store nutrition for embryonic growth after fertilization similar to emf2a mutants. This is consistent with the possibility of an evolutionary transition from maternal gametophytic to biparental control. Such a transition may have been selected to better balance parental inputs into the seed (Dilkes and Comai, 2004).
It is also worth noting that the genetic programs identified by analysis of the emf2a mutations are different before and after fertilization. The most prominent difference is seen for gene expression patterns related to storage compounds (Figure 2 and Supplemental Figures 7 and 11). Genes related to storage proteins are expressed in the autonomous endosperm, whereas their expression seems to be delayed in the fertilized emf2a endosperm. Mechanistic insights into this phenomenon are not yet available. However, as the maternal epigenetic states in the two processes should be the same before fertilization, the key difference could be attributed to the paternally derived genome. It is tempting to consider that the paternal genome has a promotive effect on endosperm development in many interspecies and interploidy crosses, and, conversely, that the OsEMF2a-containing PRC2 derived from the maternal genome has a repressive effect during rice endosperm development. In the present study, we identified potential candidates for these processes: ∼200 PEGs were directly controlled by OsEMF2a-containing PRC2 (Supplemental Figure 10).
Methods
Construction of CRISPR/Cas9 lines
The rice (Oryza sativa ssp. japonica cv. Nipponbare) plants were used to create the CRISPR/Cas9 lines as they grow under short-day conditions as described previously (Ohnishi et al., 2011). A CRISPR/Cas9 guide RNA sequence (5′-CCGTTTTGCTTGGTACGGTG-3′) was chosen by CRISPR-P (http://CRISPR.hzau.edu.cn/CRISPR/); this sequence corresponds to a region of the 10th exon of OsEMF2a that encodes a zinc-finger domain. The 20-nt guide sequence was cloned into the BbsI site of pU6gRNA-oligo (Mikami et al., 2015). sgRNA expression constructs consisting of OsU6pro:sgRNA:polyT were transferred into the binary vector pZH_OsU3gYSA_MMCas9 (Mikami et al., 2015) harboring SpCas9 and hygromycin phosphotransferase gene (HPT) expression constructs using AscI and PacI sites. Rice transformation with the CRISPR/Cas9 construct was performed using Agrobacterium tumefaciens strain EHA105 as described previously (Mikami et al., 2015). Transformed calli were identified by hygromycin screening, and selected calli were used to regenerate transgenic plants. Mutations of OsEMF2a were identified by CAPS analysis using a pair of gene-specific primers (F: 5′-CTGCTGTTGGCCAATAATGA-3′, R: 5′-AGCCAACCAGTGAAATGAGC-3′) and the restriction enzyme HpyCH4III (NEB, https://www.neb.com). Sanger sequencing was used to confirm the OsEMF2a mutations. For detailed analysis of transgenic plants, the vector construct was removed by multiple backcrosses to wild-type plants.
Emasculation and induction of enlarged ovaries
Emasculation was performed by immersing pre-anthesis panicles in hot water for 7 min at 42°C. After the hot water treatment (Ohnishi et al., 2011), the stamens were removed from open florets; unopened florets were excised from the panicles. The plants were placed under short-day conditions (see above) for 3–10 days to allow development of enlarged ovaries. The number of enlarged ovaries was counted every 24 h after emasculation.
Histological analysis
Ovaries and seeds were fixed under a vacuum in FAA solution (5% v/v formaldehyde, 5% v/v acetic acid, 63% v/v ethanol). The samples were then dehydrated through a graded ethanol series and embedded in Technovit 7100 resin (Kulzer, https://www.kulzer-technik.de), according to the manufacturer’s instructions. Sections (2-µm thick) were cut using an RM2245 microtome (Leica, http://www.leica-microsystems.com) and stained with 0.02% toluidine blue. To visualize cell walls and starch granules, the sections were stained with 1% w/v safranin in 50% ethanol and counterstained with 1% w/v iodine and 1% w/v potassium iodide. Protein bodies were identified by staining with 0.01% w/v Coomassie Brilliant Blue R-250. Images of stained sections were captured using an Axioimager M1 (Zeiss, http://www.zeiss.com). Propidium iodide staining was performed as previously described (Sekine et al., 2013). A series of optical images was obtained under a TCS-SP8 MP confocal microscope (Leica).
RNA isolation and RT-qPCR analysis
Leaf, root, and spikelet tissues were homogenized in liquid nitrogen and the powdered tissue resuspended in RNA extraction buffer. Endosperm tissues were isolated from developing seeds and homogenized in RNA extraction buffer. Total RNA was extracted using an RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/) with RNase-free DNase (Qiagen). Total RNA was synthesized to cDNAs using a PrimeScript II first strand cDNA Synthesis Kit (TaKaRa, http://www.takara-bio.co.jp/) according to the manufacturer’s instructions. For allele-specific expression analysis, DNA sequences, including an Single Nucleotide Polymorphism (SNP) site from the Nipponbare and Kitaake cultivars, were amplified by PCR using gene-specific primers (Supplemental Table 3). Quantitative RT-PCR was performed using SYBR Premix EX Taq (TaKaRa) and gene-specific primers (Supplemental Table 3) on a Thermal Cycler DICE Real-time System TP800 (TaKaRa; cycling conditions were 98°C for 1 min, followed by 35 repeats of 98°C for 10 s, 60°C for 30 s and 72°C for 20 s). Three independent samples were analyzed as biological replicates.
Analysis of parent-of-origin expression
Endosperm RNA was extracted from an F1 seed produced by crossing cv. Nipponbare and cv. Kitaake; extraction was performed at 7 DAP. cDNA was synthesized from each RNA using a PrimeScript II first-strand cDNA Synthesis Kit (TaKaRa). Parent-of-origin expression patterns were confirmed by Sanger sequencing of PCR products amplified from cDNAs using gene-specific primers (Supplemental Table 3). The proportions of maternal and paternal gene products were estimated by amplicon sequence analysis of PCR products. Amplification was performed by PCR using gene-specific primers (Supplemental Table 3) with linker sequences; a multiplex library was then prepared as described by the 16S Metagenomic Sequencing Library Preparation Illumina protocol (Part #15044223 Rev. A, Illumina, http://www.illumina.com). Sequencing was performed on the Illumina MiSeq platform in the 300 bp paired-end mode. Adapter sequences were removed and low-quality reads were filtered from the raw reads using cutadapt version 1.17 (Martin, 2011) and Trimmomatic version 0.36 (Bolger et al., 2014), respectively. Clean reads were aligned to the coding sequence of each gene using bwa version 0.7.17 (Li and Durbin, 2009). The SNP information was extracted from the alignment files using igvtools version 2.3.98 (Robinson et al., 2011). The proportion of maternal and paternal expression was calculated based on the frequency of the SNPs.
RNA-seq library preparation and transcriptome data analyses
Total RNA was extracted from endosperm tissue that had been manually dissected under a binocular microscope. A PicoPure RNA Isolation Kit (ThermoFisher, https://www.thermofisher.com) was used for 3 DAP endosperm from wild-type plants, and an RNeasy Plant Mini Kit (Qiagen) was used for 5 DAP endosperm from wild-type and emf2/+ plants. Extracted RNAs were treated with RNase-free DNase (Qiagen). RNA-seq libraries were produced using TruSeq Stranded Total RNA with a Ribo-Zero Plant Kit (Illumina, http://www.illumina.com) according to the manufacturer’s instructions. Each library was sequenced using the Illumina HiSeq X Ten platform the 150-bp paired-end mode (Supplemental Data Set 10). The raw RNA reads were processed for trimming and quality control. Adapter and poly-A tail sequences were removed using cutadapt version 1.17 (Martin, 2011) and the Perl script prinseq-lite.pl version 0.20.4 (Schmieder and Edwards, 2011), respectively. Quality trimming and filtering were performed with Trimmomatic version 0.36 (Bolger et al., 2014).
All clean reads were aligned to the reference genome sequence of O. sativa ssp. japonica cv. Nipponbare (IRGSP-1.0) (Kawahara et al., 2013) using HISAT version 2.1.0 (Kim et al., 2015). Uniquely aligned read counts were obtained from alignment files with featureCounts version 1.4.6 (Liao et al., 2014). Differentially expressed genes were detected by Fisher’s exact test using the R package edgeR with 5% False Discovery Rate (FDR) and log2 fold change >1 (Robinson et al., 2010). The R package clusterProfiler (Yu et al., 2012) and gene sets from the PANTHER database were used to test for overrepresented Gene Ontology terms with P < 0.01 and enrichment of over five genes. Expression data were visualized using ggplot2 (Wickham, 2016), function heatmap.2 in the gplots package (Warnes et al., 2020). Venn diagrams were generated by VennDiagram (Chen, 2018).
ChIP-seq library preparation
Endosperm was homogenized in liquid nitrogen using a multibeads shocker (YASUI KIKAI, http://www.yasuikikai.co.jp). The samples were resuspended in 10-ml extraction buffer 1 (10 mM Tris–HCl, 0.4 M sucrose, 0.035% 2-ME, 1-mM PMSF, 1× Roche complete protease inhibitor ethylenediaminetetraacetic acid (EDTA)-free, pH 7.5) and filtered through 70- and 40-µm cell strainers. The samples were crosslinked by 1% formaldehyde for 10 min, after which the formaldehyde was quenched with glycine. Crude nuclei were pelleted at 1,000g for 10 min and washed twice with 10 mL extraction buffer 1. The nuclei were resuspended in 1 mL extraction buffer 2 (10-mM Tris–HCl, 0.25-M sucrose, 1% Triton X-100, 10-mM MgCl2, 0.035% 2-ME, 1-mM Phenylmethylsulfonyl fluoride (PMSF), 1× Roche complete protease inhibitor EDTA-free, pH 7.5) and pelleted at 12,000g for 10 min at 4°C. The nuclei were resuspended in 500-µL extraction buffer 3 (10-mM Tris–HCl, 1.7 M sucrose, 0.15% Triton X-100, 2-mM MgCl2, 0.035% 2-ME, 1-mM PMSF, 1× Roche complete protease inhibitor EDTA-free, pH 7.5) and layered on 500-µL extraction buffer 3. The nuclei were pelleted at 16,000g for 60 min at 4°C.
The isolated nuclei were resuspended in 1-mL nuclei lysis buffer (50-mM Tris–HCl, 0.5% sodium lauroyl sarcosinate, 100-mM NaCl, 2-mM EDTA, 1-mM PMSF, 1× Roche complete protease inhibitor EDTA-free, pH 7.5). Chromatin was sheared with a Covaris M220 focused-ultrasonicator (https://covaris.com/; duty cycle, 10%, intensity peak incident power, 75 W; cycles per burst, 200; bath temperature, 7°C; 25 min). Following centrifugation at 16,000g for 10 min at 4°C, the supernatant was collected and diluted with 4-mL dilution buffer (50-mM Tris–HCl, 1.25% Triton X-100, 100-mM NaCl, 2-mM EDTA, 1-mM PMSF, 1× Roche complete protease inhibitor EDTA-free, pH 7.5). H3K27me3 was immunoprecipitated using rabbit polyclonal antibodies (Millipore 07-449, http://www.merckmillipore.com) and washed with low-salt wash buffer (50-mM Tris–HCl, 150-mM NaCl, 2-mM EDTA, 0.5% Triton X-100), high-salt wash buffer (50-mM Tris–HCl, 500-mM NaCl, 2-mM EDTA, 0.5% Triton X-100) and final wash buffer (50-mM Tris–HCl, 50-mM NaCl, 2-mM EDTA). Immunoprecipitated chromatin was eluted with 200-µL elution buffer (50-mM Tris–HCl, 10-mM EDTA, 1% SDS, pH 8.0) at 65°C overnight and de-crosslinked with proteinase K. DNA was purified with 1.5× AMPure XP beads (Beckman, https://ls.beckmancoulter.co.jp), and Illumina-compatible libraries were prepared with a NEBNext Ultra II library preparation kit (NEB) according to the manufacturer’s protocol, except that KAPA HiFi HotStart ReadyMix (KAPA Biosystems, https://www.kapabiosystems.com/) was used for amplification.
ChIP-seq data analysis
Each library was sequenced using an Illumina HiSeq X Ten in paired-end mode (Supplemental Data Set 10). Adapter sequence and low-quality sequences were removed from the raw reads using cutadapt version 1.17 (Martin, 2011) and Trimmomatic version 0.36 (Bolger et al., 2014). All clean reads were aligned to the reference genome sequence of O. sativa ssp. japonica cv. Nipponbare (IRGSP-1.0; Kawahara et al. 2013) using bowtie2 version 2.3.4.1 (Langmead and Salzberg, 2012). Multi-mapped reads were discarded using samtools versions 1.9 (Li et al., 2009) and subsequently filtered for PCR duplicated reads using Picard version 2.18.3 (Broad Institute, 2019). A heatmap of Pearson’s correlations was created from filtered alignment files using deeptools version 3.4.3 utility multiBamSummary (Ramirez et al., 2014). Bigwig coverage files subtracted from input were generated using deeptools version 3.4.3 utility bamCoverage and bigwigCompare (Ramirez et al., 2014) and visualized with pyGenomeTracks (Lopez-Delisle et al., 2020). Significant H3K27me3 peaks were called by the callpeak function of MACS2 version 2.2.5 with options [-broad --broad-cutoff 0.1-g 3.7e8] (Zhang et al., 2008). Differential enrichment of peaks was identified using the R package DiffBind version 2.16.0 with 5% FDR (Stark and Brown, 2011).
Accession numbers
Sequence data from this article can be found in the rice annotation project database (MSU) under the following accession numbers: OsEMF2a (LOC_Os04g08034), OsEMF2b (LOC_Os09g13630), OsFIE1 (LOC_Os08g04290), OsFIE2 (LOC_Os08g04270), OsMADS77 (LOC_Os09g02780), OsMADS87 (LOC_Os03g38610), OsMADS89 (LOC_Os01g18440), OsYUC9 (LOC_Os01g16714), OsPIN8 (LOC_Os01g51780), OsPILS1 (LOC_Os09g31478), OsARF7 (LOC_Os02g35140), OsARF13 (LOC_Os04g59430), OsARF22 (LOC_Os10g33940), OsIAA10 (LOC_Os02g57250), OsIAA15 (LOC_Os05g08570), OsSAUR28 (LOC_Os06g48860), OsSAUR57 (LOC_Os12g41600), OsFKII (LOC_Os08g02120), OsHXK8 (LOC_Os05g09500), OsSUS1 (LOC_Os03g28330), OsSUS3 (LOC_Os07g42490), OsSUT2 (LOC_Os12g44380), OsSWEET1a (LOC_Os01g65880), OsSWEET11 (LOC_Os08g42350), OsCIN2 (LOC_Os04g33740), OsWx (LOC_Os06g04200). All RNA-seq and ChIP-seq data generated from this study have been deposited into the DDBJ Sequence Read Archive (DRA, https://www.ddbj.nig.ac.jp/dra/index-e.html). Accession numbers for RNA-seq and ChIP-seq are DRA009458 and DRA010700, respectively.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. OsEMF2a maternal expression and the CRISPR/Cas9 mutant lines of OsEMF2a.
Supplemental Figure S2. Phenotypes of the enlarged ovary in wild-type and emf2a/+ plants.
Supplemental Figure S3. Starch accumulation in the enlarged ovary in wild-type and emf2a/+ plants.
Supplemental Figure S4. Significantly up- and downregulated genes in fertilized wild-type seeds and unfertilized emf2a-3/+ ovaries.
Supplemental Figure S5. Scatter plots of commonly upregulated genes in fertilized wild-type seeds and unfertilized emf2a-3/+ ovaries.
Supplemental Figure S6. Expression and H3K27me3 enrichment of OsEMF2a and OsEMF2b.
Supplemental Figure S7. Sections of the developing embryo and endosperm in self-pollinated emf2a-3/+ plants.
Supplemental Figure S8. Endosperm samples used for RNA-seq analysis.
Supplemental Figure S9. H3K27me3 ChIP-seq analysis of wild-type and emf2a-3/+ endosperm.
Supplemental Figure S10. Comparative analyses of OsEMF2a target genes with reported data sets, imprinted genes, and upregulated genes in emf2a caryopses.
Supplemental Figure S11. Heatmaps of hierarchical clustering of DEGs with higher expression levels in fertilized emf2a-3 endosperm at 5 DAP.
Supplemental Table S1. Gene ontology analysis of OsEMF2a-PRC2 target genes.
Supplemental Table S2. Gene ontology analysis of de-regulated genes in emf2a endosperm.
Supplemental Table S3. Primer sequences used in this study.
Supplemental Data Set 1. Upregulated genes in wild-type developing seeds at 2 DAP compared with wild-type unfertilized ovaries at 0 DAE.
Supplemental Data Set 2. Downregulated genes in wild-type developing seeds at 2 DAP compared with wild-type unfertilized ovaries at 0 DAE.
Supplemental Data Set 3. Upregulated genes in emf2a-3/+ ovaries at 5 DAE compared with wild-type unfertilized ovaries at 0 DAE.
Supplemental Data Set 4. Downregulated genes in emf2a-3/+ ovaries at 5 DAE compared with wild-type unfertilized ovaries at 0 DAE.
Supplemental Data Set 5. Genes marked with H3K27me3 peaks in wild-type 5 DAP endosperm and in mild and severe emf2a endosperm.
Supplemental Data Set 6. OsEMF2a-containing PRC2 target genes and their expression profiles.
Supplemental Data Set 7. Change in expression of imprinted genes that are targets of OsEMF2a-containing PRC2.
Supplemental Data Set 8. Upregulated genes in emf2a-3/+ endosperm at 5 DAP compared with wild-type 3 DAP endosperm.
Supplemental Data Set 9. Upregulated genes in emf2a-3/+ endosperm at 5 DAP compared with wild-type 5 DAP endosperm.
Supplemental Data Set 10. Summary of sequencing data.
Supplemental File 1. Statistic tables.
Supplementary Material
Acknowledgments
We thank H. Maruoka for providing some images of histological sections.
Funding
This work was partly supported by Grant-in-Aid for Scientific Research on Innovative Area from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant nos. 16H06464, 16H06471, and 16H21727 to T.Ki., 19H04873 to T.Ka.); Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Fellows (grant no.16J02580 to K.T.) and Young Scientists (B; grant no. 17K15210 to K.T.) from JSPS; Japan Science and Technology Agency (JST) Advanced Low Carbon Technology Research and Development Program Grant number JPMJAL1510 (to M.O.-T. and T.Ki.). Computations were partially performed on the NIG supercomputer at Research Organization of Information and Systems (ROIS) National Institute of Genetics, Japan.
Conflict of interest statement. None declared.
K.T., A.O., and T.Ki. conceived research plans; T.Ki. supervised the experiments; L.C. advised on experiments; K.T., A.O., M.K., M.N., H.N., S.S., S.K., and T.Ka., performed the experiments; K.T. analyzed the data; H.F., K.N., Y.S., M.O-.T., M.E., and K.H. provided research materials; K.T. and T.Ki. wrote the paper. L.C. edited the paper.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Tetsu Kinoshita (tkinoshi@yokohama-cu.ac.jp) and Kaoru Tonosaki (tonosaki@iwate-u.ac.jp).
References
- Baroux C, Spillane C, Grossniklaus U (2002) Evolutionary origins of the endosperm in flowering plants. Genome Biol 3: reviews1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U (2006) Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista RA, Kohler C (2020) Genomic imprinting in plants-revisiting existing models. Genes Dev 34: 24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista RA, Figueiredo DD, Santos-Gonzalez J, Köhler C (2019) Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev 33: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauzamy L, Fourquin C, Dubrulle N, Boursiac Y, Boudaoud A, Ingram G (2016) Endosperm turgor pressure decreases during early Arabidopsis seed development. Development 143: 3295–3299 [DOI] [PubMed] [Google Scholar]
- Bemer M, Grossniklaus U (2012) Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol 15: 523–529 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad Institute (2019) Picard Toolkit. GitHub repository http://broadinstitute.github.io/picard/ (December 22, 2020)
- Brown RC, Lemmon BE (2008) Microtubules in early development of the megagametophyte of Ginkgo biloba. J Plant Res 121: 397–406 [DOI] [PubMed] [Google Scholar]
- Butenko Y, Ohad N (2011) Polycomb-group mediated epigenetic mechanisms through plant evolution. Biochim Biophys Acta 1809: 395–406 [DOI] [PubMed] [Google Scholar]
- Chen C, Begcy K, Liu K, Folsom JJ, Wang Z, Zhang C, Walia H (2016) Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol 171: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li T, Zhu S, Liu Z, Shi Z, Zheng X, Chen R, Huang J, Shen Y, Luo S, et al. (2018) Characterization of imprinted genes in rice reveals conservation of regulation and imprinting with other plant species. Plant Physiol 177: 1754–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H (2018) VennDiagram: Generate High-Resolution Venn and Euler Plots. https://CRAN.R-project.org/package=VennDiagram (December 22, 2020)
- Chen J, Lausser A, Dresselhaus T (2014) Hormonal responses during early embryogenesis in maize. Biochem Soc Trans 42: 325–331 [DOI] [PubMed] [Google Scholar]
- Cheng X, Pan M,, Zhou Y, Niu B, Chen C (2020a) The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice. Plant Comm 1: 100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Pan M, Zhou Y, Niu B, Chen C (2020b) Functional divergence of two duplicated Fertilization Independent Endosperm genes in rice with respect to seed development. Plant J 104: 124–137 [DOI] [PubMed] [Google Scholar]
- Conrad LJ, Khanday I, Johnson C, Guiderdoni E, An G, Vijayraghvan U, Sundaresan V (2014) The polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J 80: 883–894 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson H, Costa L, Gutierrez-Marcos J (2012) Epigenetic neofunctionalisation and regulatory gene evolution in grasses. Trends Plant Sci 17: 389–394 [DOI] [PubMed] [Google Scholar]
- Dilkes BP, Comai L (2004) A differential dosage hypothesis for parental effects in seed development. Plant Cell 16: 3174–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Berger F (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23: 192–199 [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Köhler C (2018) Auxin: a molecular trigger of seed development. Genes Dev 32: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C (2015) Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C (2016) Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 5: e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom JJ, Begcy K, Hao X, Wang D, Walia H (2014) Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol 165: 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman WE (2001) Developmental and evolutionary hypotheses for the origin of double fertilization and endosperm. C R Acad Sci III 324: 559–567 [DOI] [PubMed] [Google Scholar]
- Furihata HY, Suenaga K, Kawanabe T, Yoshida T, Kawabe A (2016) Gene duplication, silencing and expression alteration govern the molecular evolution of PRC2 genes in plants. Genes Genet Syst 91: 85–95 [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Paro R (2014) Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton AE, Berger F (2005) Control of reproduction by Polycomb Group complexes in animals and plants. Int J Dev Biol 49: 707–716 [DOI] [PubMed] [Google Scholar]
- Haig D (2013) Kin conflict in seed development: an interdependent but fractious collective. Annu Rev Cell Dev Biol 29: 189–211 [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M (1991) Genomic imprinting in endosperm: its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Phil Trans R Soc Lond B 333: 1–14 [Google Scholar]
- Hands P, Rabiger DS, Koltunow A (2016) Mechanisms of endosperm initiation. Plant Reprod 29: 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Kohler C (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Hennig L, Derkacheva M (2009) Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet 25: 414–423 [DOI] [PubMed] [Google Scholar]
- Huang X, Lu Z, Wang X, Ouyang Y, Chen W, Xie K, Wang D, Luo M, Luo J, Yao J (2016) Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J 87: 305–317 [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T (2011) Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F (2006) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Ikeda Y, Ishikawa R (2008) Genomic imprinting: a balance between antagonistic roles of parental chromosomes. Semin Cell Dev Biol 19: 574–579 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Mardossian L, Harada JJ, Goldberg RB, et al. (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Q, Wang Y, Li S (2019) Detailed observation on expression dynamics of Polycomb group genes during rice early endosperm development in subspecies hybridization reveals their characteristics of parent-of-origin genes. Rice 12: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C (2014) Embryo and endosperm, partners in seed development. Curr Opin Plant Biol 17: 64–69 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhou B, Peng X, Kuang Q, Huang X, Yao J, Du B, Sun MX (2014) OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol 201: 66–79 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Liu X, Wei X, Sheng Z, Jiao G, Tang S, Luo J, Hu P (2016) Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS One 11: e0164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MA, Larkins BA (1993) Endosperm origin, development, and function. Plant Cell 5: 1383–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Delisle L, Rabbani L, Wolff J, Bhardwaj V, Backofen R, Gruning B, Ramirez F, Manke T (2020) pyGenomeTracks: reproducible plots for multivariate genomic data sets. Bioinformatics 3: btaa692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2: 711–723 [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88: 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Hennig L (2015) The polycomb group protein regulatory network. Annu Rev Plant Biol 66: 269–296 [DOI] [PubMed] [Google Scholar]
- Nallamilli BR, Zhang J, Mujahid H, Malone BM, Bridges SM, Peng Z (2013) Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet 9: e1003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yoshino M, Yamakawa H, Kinoshita T (2011) The biotron breeding system: a rapid and reliable procedure for genetic studies and breeding in rice. Plant Cell Physiol 52: 1249–1257 [DOI] [PubMed] [Google Scholar]
- Olsen OA (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 (Suppl): S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA (2000) Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T (2014) deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D (2013) Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci U S A 110: 7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, Kinoshita T (2013) Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 76: 792–799 [DOI] [PubMed] [Google Scholar]
- Shimada T, Takagi J, Ichino T, Shirakawa M, Hara-Nishimura I (2018) Plant Vacuoles. Annu Rev Plant Biol 69: 123–145 [DOI] [PubMed] [Google Scholar]
- Stark R, Brown G (2011) DiffBind: differential binding analysis of ChIP-Seq peak data. Bioconductor. http://bioconductor.org/packages/release/bioc/html/DiffBind.html (December 22, 2020)
- Tonosaki K, Kinoshita T (2015) Possible roles for polycomb repressive complex 2 in cereal endosperm. Front Plant Sci 6: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonosaki K, Sekine D, Ohnishi T, Ono A, Furuumi H, Kurata N, Kinoshita T (2018) Overcoming the species hybridization barrier by ploidy manipulation in the genus Oryza. Plant J 93: 534–544 [DOI] [PubMed] [Google Scholar]
- Warnes GR Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. (2020) gplots: Various R Programming Tools for Plotting Data. https://github.com/talgalili/gplots (December 22, 2020)
- Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM (2013) Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc Natl Acad Sci U S A 110: 19639–19644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Kohler C (2010) H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer, New York [Google Scholar]
- Williams JH, Friedman WE (2002) Identification of diploid endosperm in an early angiosperm lineage. Nature 415: 522–526 [DOI] [PubMed] [Google Scholar]
- Wu X, Liu J, Li D, Liu CM (2016) Rice caryopsis development II: Dynamic changes in the endosperm. J Integr Plant Biol 58: 786–798 [DOI] [PubMed] [Google Scholar]
- Wyder S, Raissig MT, Grossniklaus U (2019) Consistent reanalysis of genome-wide imprinting studies in plants using generalized linear models increases concordance across datasets. Sci Rep 9: 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Chen M, Pei R, Ouyang Y, Yao J (2015) OsEMF2b acts as a regulator of flowering transition and floral organ identity by mediating H3K27me3 deposition at OsLFL1 and OsMADS4 in rice. Plant Mol Biol Rep 33: 121–132 [Google Scholar]
- Xin M, Yang R, Li G, Chen H, Laurie J, Ma C, Wang D, Yao Y, Larkins BA, Sun Q, et al. (2013) Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25: 3212–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Liu Z, Gao L, Yu K, Feng M, Yao Y, Peng H, Hu Z, Sun Q, Ni Z, et al. (2018) Genomic imprinting was evolutionarily conserved during wheat polyploidization. Plant Cell 30: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee S, Hang R, Kim SR, Lee YS, Cao X, Amasino R, An G (2013) OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J 73: 566–578 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Kawanabe T, Bo Y, Fujimoto R, Kawabe A (2018) Genome-wide analysis of parent-of-origin allelic expression in endosperms of Brassicaceae species, Brassica rapa. Plant Cell Physiol 59: 2590–2601 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen S, Jiao W, Wang L, Wang L, Ye W, Lu J, Hong D, You S, Cheng Z, et al. (2017) Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol 216: 373–387 [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu Z, Song X, Jernstedt J, Sundaresan V (2018) The gymnosperm ortholog of the angiosperm central cell-specification gene CKI1 provides an essential clue to endosperm origin. New Phytol 218: 1685–1696 [DOI] [PubMed] [Google Scholar]
- Zhang HY, Luo M, Johnson SD, Zhu XW, Liu L, Huang F, Liu YT, Xu PZ, Wu XJ (2016) Parental genome imbalance causes post-zygotic seed lethality and deregulates imprinting in rice. Rice 9: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang D, Zhang H, Skaggs MI, Lloyd A, Ran D, An L, Schumaker KS, Drews GN, Yadegari R (2018) FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 plays a dual role in regulating type I MADS-box genes in early endosperm development. Plant Physiol 177: 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Peng Z, Peng Q, Cai Q, Peng W, Chen M, Yao J (2018) Regulation of plant height in rice by the Polycomb group genes OsEMF2b, OsFIE2 and OsCLF. Plant Sci 267: 157–167 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.