Abstract
A polyphasic study was designed to determine the taxonomic provenance of a strain, isolate PRKS01-29T, recovered from an Indonesian sand dune and provisionally assigned to the Streptomyces violaceusniger clade. Genomic, genotypic and phenotypic data confirmed this classification. The isolate formed an extensively branched substrate mycelium which carried aerial hyphae that differentiated into spiral chains of rugose ornamented spores, contained LL-as the wall diaminopimelic acid, MK-9 (H6, H8) as predominant isoprenologues, phosphatidylethanolamine as the diagnostic phospholipid and major proportions of saturated, iso- and anteiso- fatty acids. Whole-genome sequences generated for the isolate and Streptomyces albiflaviniger DSM 41598T and Streptomyces javensis DSM 41764T were compared with phylogenetically closely related strains, the isolate formed a branch within the S. violaceusniger clade in the resultant phylogenomic tree. Whole-genome sequences data showed that isolate PRKS01-29T was most closely related to the S. albiflaviniger strain but was distinguished from the latter and from other members of the clade using combinations of phenotypic properties and average nucleotide identity and digital DNA:DNA hybridization scores. Consequently, it is proposed that isolate PRKS01-29T (= CCMM B1303T = ICEBB-02T = NCIMB 15210T) should be classified in the genus Streptomyces as Streptomyces sabulosicollis sp. nov. It is also clear that streptomycetes which produce spiral chains of rugose ornamented spores form a well-defined monophyletic clade in the Streptomyces phylogenomic tree., the taxonomic status of which requires further study. The genome of the type strain of S. sabulosicollis contains biosynthetic gene clusters predicted to produce new natural products.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10482-021-01564-0.
Keywords: Streptomyces sabulosicollis, Polyphasic taxonomy, Streptomyces violaceusniger clade, Genomics, Genome mining
Introduction
The classification of Streptomyces species is especially challenging given the high number of validly published species (https://www.bacterio.net.streptomyces.html), the limited resolution of 16S rRNA gene sequences in their delineation (Labeda et al. 2012, 2017) and evidence that the genus is underspeciated (Yamac et al. 2011; Hamm et al. 2017). However, multi-locus sequence analyses (MLSA) of concatenated protein-coding house-keeping genes (Ayed et al. 2018; Kusuma et al. 2020; Li et al. 2020; Martinet et al. 2020) and comparative surveys of whole-genome sequences (Nouioui et al. 2018) provide invaluable data for the circumscription of novel Streptomyces species. MLSA analyses have revealed a correlation between the delineation of phylogenetic clades and associated phenotypic properties (Rong and Huang 2014; Labeda et al. 2014), as exemplified by the assignment of streptomycetes with spiral chains of rugose ornamented spores to a well supported taxon (Labeda et al. 2017), known as the Streptomyces violaceusniger clade (Sembiring et al. 2000; Kumar and Goodfellow 2008, 2010). Representatives of this clade show the same pattern of HPLC-detected metabolites (Ward and Goodfellow 2004; Goodfellow et al. 2007), give a characteristic amplification product with taxon-specific primers (Kumar et al. 2007) and form a characteristic grey aerial spore mass and a greyish yellow substrate mycelium on oatmeal agar (International Streptomyces Project medium 3 [ISP 3]., Shirling and Gottlieb 1966) (Sembiring et al. 2000; Kumar and Goodfellow 2008, 2010; Goodfellow et al. 2007)
Improvements in the classification of the S. violaceusniger clade (Rong and Huang 2012; Komaki et al. 2017; Labeda et al. 2017; Zhou et al. 2017) led to the recognition of 16 species which include Streptomyces albiflaviniger (Goodfellow et al. 2007, Euzéby 2008), Streptomyces himastatinicus (Kumar and Goodfellow 2008), Streptomyces hygroscopicus (Jensen 1931) Waksman and Henrici 1948, Streptomyces iranensis (Hamedi et al. 2010), Streptomyces javensis (Sembiring et al. 2000, 2001), Streptomyces malaysiensis (Al-Tai et al. 1999), Streptomyces melanosporofaciens (Arcamone et al. 1959), Streptomyces rapamycinicus (Kumar and Goodfellow 2008), Streptomyces rhizosphaericus (Sembiring et al. 2000, 2001), Streptomyces solisilvae (Zhou et al. 2017) and Streptomyces violaceusniger corrig (Waksman and Curtis 1916) Pridham et al. 1958, as emended by Labeda and Lyons (1991), the earliest validly published species in the taxon. An additional species, “Streptomyces ruani” (Kumar and Goodfellow 2008) was shown to be invalid by Tindall (2014). Strains assigned to the clade have been detected in diverse habitats (Kumar et al. 2007) but are usually associated with rhizosphere and non-rhizosphere soil (Sembiring et al. 2000; Sahin et al. 2010).
Strains classified in the S. violaceusniger clade have an impressive track record as a source of new antibiotics (DeBoer et al. 1970; Chen et al. 2003; Cheng et al. 2010; Xie et al. 2019), antiparasitic metabolites (Sun et al. 2002), antitumour compounds (Lam et al. 1990; Wang et al. 2013), enzymes (Rabe et al. 2017) and immunosuppressants (Vezina et al. 1975) and biocontrol agents (Clermont et al. 2010; Palaniyandi et al. 2016; Sarwar et al. 2019) hence the continued interest in them for genome mining and natural product discovery. Members of this taxon are gifted in the sense of Baltz (2017) as they have large genomes (> 8 Mbp) rich in biosynthetic gene clusters (BGCs) predicted to encode for specialised metabolites (Baranasic et al. 2013; Horn et al. 2014; Komaki et al. 2018). Prospecting for Streptomyces diversity also shows that sampling strains from unexplored, including extreme habitats, raises the probability of finding new compounds (Nicault et al. 2020) and that streptomycete genomes are a prolific source of novel BGCs (Vicente et al. 2018; Martinet et al. 2020).
The present study was designed to classify a putative new member of the S. violaceusniger clade based on genomic, genotypic and phenotypic data and to gain an insight into its potential as a source of new specialised metabolites. The resultant datasets showed that the isolate represents a novel species, named Streptomyces sabulisicollis sp. nov. Associated phylogenomic data clarified the internal taxonomic structure of the S. violaceusniger clade and relationships to its closest phylogenetic neighbours.
Materials and methods
Isolation, maintenance and cultivation
Isolate PRKS01-29T was isolated from an arid, non-saline soil sample (pH 5.8., organic matter content 0.06%) collected just below the surface of a sand dune in the Parangkusumo Region (8° 1′7 513″ S/ 110° 19′ 11.04″ E) of Yogyakarta Province, Java, Indonesia following incubation on Actinomycete Isolation Agar (HiMedia, Einhausen, Germany), pH 7.3, supplemented with cycloheximide (50 µg/mL), nalidixic acid (25 µg/mL) and nystatin (25 µg/mL) and incubated for 7 days at 45 °C, as described previously (Kusuma et al. 2020). The isolate and S. albiflaviniger DSM 41598T, S. iranensis DSM 41954T, S. javensis DSM 41764T, S. malaysiensis NBRC 13472T, S. rapamycinicus NRRL 5491T and S. rhizosphaericus NRRL B-24304T and S. violaceusniger DSM 40583T were maintained on yeast extract-malt extract agar (International Streptomyces Project medium 2 [ISP 2]., Shirling and Gottlieb 1966) and as mixtures of hyphal fragments and spores in 20%, v/v glycerol at −20 °C and −80 °C. The type strains of S. albiflaviniger and S. iranensis were obtained from the Leibniz Institute DSMZ German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany and the remaining reference strains were from the personal collection of Professor Michael Goodfellow, Newcastle University, Newcastle-upon-Tyne, United Kingdom. Biomass for the chemotaxonomic studies carried out on the isolate was prepared in 1L Erlenmeyer flasks containing 250 mL of sterile ISP 2 broth (Shirling and Gottlieb 1966), the flasks were shaken at 180 rpm for 14 days at 28 °C and the resultant biomass harvested by centrifugation at 4000 rpm for 10 min, washed twice in sterile distilled water and freeze dried for 3 days.
Acquisition of chemotaxonomic, cultural and morphological properties
The isolate was examined for chemotaxonomic, cultural and morphological properties of value in Streptomyces systematics (Kämpfer 2012; van der Aart et al. 2019). Gram-stain (Hucher’s modification, Society for American Bacteriology 1957) and micromorphological features were recorded following growth on ISP 3 agar for 7 days at 28 °C. Growth from the ISP 2 preparation was examined for spore-chain arrangement and spore-surface ornamentation using a scanning electron microscope (Tescan Vega 3, LMU instrument) and the procedure described by O’Donnell et al. (1993). The ability of the test and associated marker strains to grow at different temperatures, pH regimes and in the presence of various concentrations of sodium chloride was carried out in triplicate, as mentioned by Kusuma et al. (2020). Standard chromatographic methods were used to detect the isomers of diaminopimelic acid (A2pm) (Staneck and Roberts 1974), whole-organism sugars (Lechevalier and Lechevalier 1970) and for menaquinones and polar lipids by applying the integrated procedure of Minnikin et al. (1984), using appropriate controls. Cellular fatty acids were extracted from freeze dried cells of the isolate and fatty acid methyl esters (FAMES) prepared following saponification and methylation using the procedure described by Miller (1982), as modified by Kuykendall et al. (1988). The FAMES were separated by gas chromatography (Agilent 68,908 instrument), the resulted peaks automatically integrated and the fatty acid names and properties determined using the standard Microbial Identification (MIDI) system, version 4.5 and the ACTIN 6 database (Sasser 1990). The growth and cultural characteristics of the isolate and reference strains were determined on tryptone yeast extract, yeast extract-malt extract, oatmeal, inorganic salts-starch, glycerol-asparagine, peptone-yeast extract-iron and tyrosine agar plates (ISP media 1–7; Shirling and Gottlieb 1966) for 21 days at 28 °C., aerial spore mass and substrate mycelial colours and those of diffusible pigments were recorded using colour charts (Kelly 1958).
Whole genome sequencing
Genomic DNA was extracted from wet biomass of single colonies of the isolate, S. albiflaviniger DSM 41598T and S. javensis DSM 41764T, grown on ISP 2 agar for 7 days at 28 °C, following the protocol provided by MicrobesNG (Birmingham, UK) (http://www.microbesng.uk) and sequenced on an Miseq instrument (Illumina, San Diego, USA). The quality of the extracted DNA preparations and the sequencing of genomic DNA libraries was achieved, as described by Kusuma et al. (2020). The libraries were sequenced following the 2 × 250-bp paired-end protocol (MicrobesNG, Birmingham, UK). Reads under 200 bp were discarded and contigs assembled using SPAdes software version 3.1.1 (Bankevich et al. 2012). The draft genome assemblies of the strains were annotated using the RAST-SEED web server (Aziz et al. 2008; Overbeek et al. 2014) with default options and are available from GenBank database.
Phylogeny
An almost complete 16S rRNA gene sequence (1454 nucleotides [nt]) (GenBank accession number MK503616) was taken directly from the draft genome of the isolate using the ContEst16S tool from the EZBioCloud webserver (https://www.ezbiocloud.net/tools/contest16s) (Lee et al. 2017); this had been compared with the associated 16S rRNA gene sequence generated using Sanger method. The gene sequence was aligned with corresponding sequences of the most closely related type strains of Streptomyces species retrieved from the EzBiocloud webserver (Yoon et al. 2017) using MUSCLE software (Edgar 2004). Pairwise sequence similarities were determined using the single-gene tree option from the Genome-to-Genome Distance Calculator (GGDC) webserver (Meier-Kolthoff et al. 2013a, b). Phylogenetic trees were inferred using the maximum-likelihood (ML., Felsenstein 1981), maximum-parsimony (MP., Fitch 1971) and neighbour-joining (NJ., Saitou and Nei 1987) algorithms. A ML tree was inferred from alignments with RAxML (Stamatakis 2014) using rapid bootstrapping with the auto Maximum-Relative-Error (MRE) criterion (Pattengale et al. 2010) and a MP tree was constructed from the alignments with the Tree Analysis New Technology (TNT) program (Goloboff et al. 2008) using 1000 bootstraps together with tree-bisection-and-reconnection branch swapping and ten random sequence replicates. The sequences were checked for computational bias using the X2 test from PAUP*(Phylogenetic Analysis Using Parsimony) (Swofford 2002).The trees were evaluated using bootstrap analyses based on 1000 replicates (Felsenstein 1985) from the MEGA X software package (Kumar et al. 2018) and the two-parameter model of Jukes and Cantor (1969) then rooted with the 16S rRNA gene sequence from Streptomyces albus subsp. albus NRRL B-1811 T (GenBank accession number JX486031.1), the type strain of the type species of the genus Streptomyces.
Comparison of genomes
The draft genome sequences generated for isolate PRKS01-29T, S. albiflaviniger DSM 41598T and S. javensis DSM 41764T were compared with corresponding sequences of type strains of species classified in the S. violaceusniger 16S rRNA gene clade. The ML phylogenomic tree inferred using the codon tree option in the PATRIC webserver (Wattam et al. 2017), which was based on aligned amino acids and nucleotides derived from 453 single copy genes in the genome dataset matched against the PATRIC PGFams database (http://www.patricbrc.org), was generated using the RAxML algorithm (Stamatakis 2006). The genome sequences of isolate PRKS01-29T and the S. albiflaviniger and S. javensis strains were compared with one another and with those of S. antimycoticus NRRL B-24289T, S. himastatinicus ATCC 53653T, S. hygroscopicus subsp. hygroscopicus NBRC 16556T, S. iranensis DSM 41954T, S. malaysiensis DSM 4137T, S. melanosporofaciens DSM 40318T, S. milbemycinicus NRRL 5739T, S. rapamycinicus NRRL 5491T, S. rhizosphaericus NRRL-24304T, S. sparsogenes DSM 40356T and S. violaceusniger DSM 40503T. Average nucleotide identity (orthoANI., Lee et al. 2016) and digital DNA-DNA hybridisation (dDDH., Meier-Kolthoff et al. 2013a) values were determined between the isolate and members of the S. violaceusniger clade using the ANI calculator from the EzBioCloud (https://www.ezbiocloud.net/tools/ani) and the GGDC webserver (http://ggdc.dsmz.de/ggdc), respectively. The presence of natural product-BGCs in the genome of the strains were detected using the antiSMASH 5.0 platform (Blin et al. 2019) with default option available at https://antismash.secondarymetabolites.org.
Phenotypic tests
Isolate PRKS01-29T and the type strains of its closest phylogenomic neighbours were examined for phenotypic properties that distinguish between species classified in the S. violaceusniger 16S rRNA gene clade (Sembiring et al. 2000; Goodfellow et al. 2007; Kumar and Goodfellow 2008, 2010; Hamedi et al. 2010; Zhou et al. 2017). Biochemical, degradation and physiological properties were acquired using media and methods described by Williams et al. (1983) and enzyme profiles with API-ZYM strips (BioMériux, France). All of the tests were carried out in duplicate using a standard inoculum equivalent to 5.0 on the McFarland scale (Murray et al. 1999).
Screening for bioactivity
The isolate was screened for antimicrobial activity against a panel of wild type microorganisms (primary screens) and Bacillus subtilis reporter strains (secondary screens) using a standard plug assay (Fiedler 2004). Plugs of isolate PRKS01-29T were taken from ISP 2 and ISP 3 agar (Shirling and Gottlieb 1966) and from MMM and from 410 agar (Goodfellow and Fiedler2010) plates incubated for 14 days at 28 °C and added to cultures of wild type strains of Bacillus subtilis, Candida albicans, Escherichia coli, Micrococcus luteus, Pseudomonas aeruginosa and Staphylococcus aureus., all of the strains were obtained from Public Health Laboratory Service, Freeman Hospital, Newcastle-upon-Tyne, United Kingdom. The wild type strains were prepared by inoculating 500 μL of overnight cultures grown at 37 °C in 25 mL Luria Bertani (LB) broth (Sigma Aldrich, UK) to an optical density (OD) of 0.6 and the resultant preparations diluted to give an OD value of 0.0125 by mixing 100 mL of the LB media with the same proportion of nutrient agar (Sigma Aldrich, UK)., each of the resultant preparations was carefully mixed, poured into the square Petri dishes containing the agar plugs of the isolate and the plate incubated overnight at 37 °C. The incubated plates were observed for the presence and sizes (in millimetres) of inhibition zones around the agar plugs. In the secondary assays, agar plugs were added to overnight cultures of six B. subtilis reporter strains grown as described above., the reporter strains were designed to detect modes of action of antimicrobial compound(s) produced by the isolate, as shown in Table S1. Overnight cultures of the strains were grown at 37 °C in Luria Bertani broth then mixed with a similar volume of nutrient agar (Sigma-Aldrich, UK) to give an optical density reading of 0.0125. The resultant preparations were examined for the presence of blue halos around the circumference of inhibition zones, the latter are formed when bioactive compound(s) produced by the isolate cleave X-gal in the agar media to 5-bromo-4-chloro-3-hydroxy indole (blue compound) and galactose.
Results and discussion
The chemotaxonomic, colonial and morphological properties of the isolate showed that it was a bona fide member of the S. violaceusniger clade (Sembiring et al. 2000; Goodfellow et al. 2007; Kumar and Goodfellow 2008, 2010; Hamedi et al. 2010; Nguyen and Kim 2015; Zhou et al. 2017). The organism was found to be aerobic, Gram-stain positive, formed an extensively branched substrate mycelium and aerial hyphae that differentiated into spiral chains of rugose ornamented spores (Fig S1), produced a dark grey to black aerial spore mass and a grey yellow substrate mycelium on oatmeal agar (Fig S2), contained LL-A2pm as the diamino acid of the peptidoglycan, MK-9 (H6) (58.4%) and MK-9 (H8) (41.6%) as the predominant isoprenologues, galactose, glucose, mannose and ribose as whole cell sugars and gave a polar lipid profile consisting of diphosphatidylglycerol, two phosphatidylglycerols, phosphatidylinositol, two phosphatidylinositol mannosides and two unknown phospholipids (Fig S3).
The major fatty acids (> 10%) of the isolate were iso-C15:0 (14.4%), anteiso-C15:0 (13.8%) and iso-C16:0 (27.2%) with lower proportions of iso-C14:0 (4.9%), C14:0 (1.0%), iso-H-C16:1 (1.2%), C16:0 (9.3%), anteiso-ω9c-C17:1 (1.8%), iso-C17:0 (6.5%), anteiso-C17:0 (9.9%), cyclo C17:0 (1.9%), C17:0 (1.1%), C16:1-ω7c/ C16:1-ω6c (1.2%) and iso-C17:1 ω9c/10-methyl C16:0 (2.8%)., trace components made up the balance of the profile. Complex mixtures of saturated straight chain and iso- and anteiso- fatty acids have been reported for the type strains of S. fabae (Nguyen and Kim 2015), S. iranensis (Hamedi et al. 2010), S. malaysiensis (Al-Tai et al. 1999) and S. solisilvae (Zhou et al. 2017).
The genomic features of the isolate, S. albiflaviniger DSM 41598T and S. javensis DSM 41764T are shown in Table 1. It is interesting that these strains have draft genomes over 8 Mbp in size and hence can be considered to be gifted after Baltz (2017). Available whole genome sequences of type strains of species classified in the S. violaceusniger 16S rRNA gene clade have larger genome sizes, as exemplified by S. iranensis HM 35T (12.1 Mb; Horn et al. 2014) and S. rapamycinicus (12.7 Mb; Baranasic et al. 2013), the genome of the latter contains 48 BGCs including the biocluster expressing for rapamycin biosynthesis.
Table 1.
Genomic features of the isolate and type strains of S. albiflaviniger and S. javensis
| Genomic features | Isolate PRKS01-29T | S. albiflaviniger DSM 11483T | S. javensis DSM 41764T |
|---|---|---|---|
| Genome size (Mbp) | 10.2 | 10.3 | 11.1 |
| Mean coverage | 56.92 | 9.93 | 35.33 |
| Number of contigs | 1104 | 3530 | 1486 |
| Number of rRNA operons | 8 | 8 | 8 |
| Number of tRNA operons | 64 | 59 | 71 |
| G+C (mol%) | 71.66 | 70.90 | 71.23 |
| GenBank accessions | JAEEAP000000000.1 | JAEEAR000000000.1 | JAEEAQ000000000.1 |
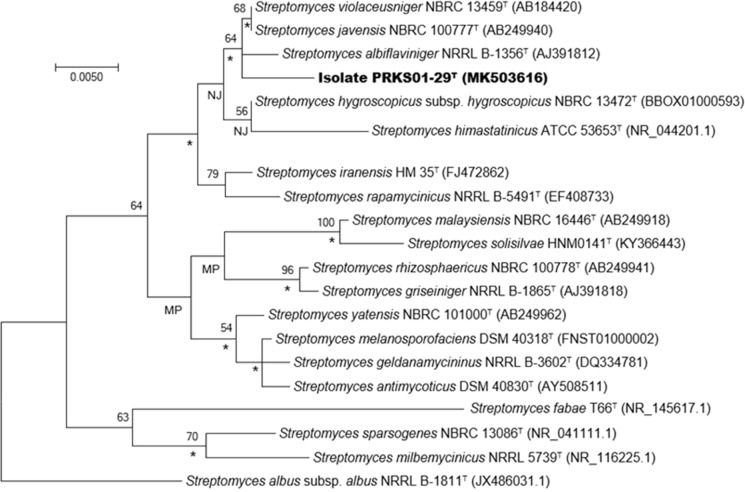
The phylogenetic tree (Fig. 1) based on 16S rRNA gene sequences shows that the isolate forms a clade in the Streptomyces gene tree together with the type strains of S. albiflaviniger, S. javensis and S. violaceusniger. It is most closely related to S. javensis NBRC 100777T and S. violaceusniger NBRC 13459T sharing a similarity with these strains of 99.4%, a value which corresponds to 9 nucleotide (nt) differences., the corresponding values with S. albiflaviniger NRRL B-1356T are 99.3% (10 nt differences in 1414 sites). The 16S rRNA gene similarities between the isolate and the remaining representatives of the S. violaceusniger clade were within the range 96.8% to 99.2%. In general, these results are in agreement with those reported by Labeda et al. (2012) who found that streptomycetes producing spores with rugose or rough surfaces belonged to six highly related clades.
Fig. 1.
Maximum-likelihood tree based on 16S rRNA gene sequences showing relationships between isolate PRKS01-29T and closely related type strains of Streptomyces species classified in the Streptomyces violaceusniger clade. Asterisks indicate branches of the tree that were found using the neighbour-joining and maximum-parsimony algorithms. NJ and MP denote nodes recovered using the neighbour-joining and maximum-parsimony tree-making algorithms, respectively. Numbers at the nodes show bootstrap values, only those above 50% are shown. The root of the tree was established using Streptomyces albus subsp. albus NRRL B-1811T. Bar indicates 0.005 substitutions per nucleotide position
The phylogenomic tree (Fig. 2) shows that the isolate forms a distinct branch at the periphery of a subclade that encompasses the type strains of S. albiflaviniger, S. iranensis, S. javensis, S. rapamycinicus and S. rhizosphaericus. The S. malaysiensis strain form a distinct lineage between this and a sister subclade composed of the type strains of S. antimycoticus, S. melanosporofaciens and S. violaceusniger. The two remaining members of the S. violaceusniger clade, S. himastatinicus ATCC 58653 T and S. hygroscopicus subspecies hygroscopicus NBRC 16556T form single membered lineages. The close phylogenomic relationships between the type strains of S. milbemycinicus and S. sporogenes and S. violaceusniger clade is in agreement with the earlier study by Nouioui et al. (2018).
Fig. 2.
Maximum-likelihood phylogenomic tree based on 453 single copy core genes showing relationships between isolate PRKS01-29T and closely related type strains which belong to the Streptomyces violaceusniger clade. Numbers at the nodes are bootstrap support values based on 100 replicates. GenBank accession numbers are shown in parentheses. The scale bar indicates 0.03 substitutions per nucleotide position. The tree is rooted using the type strain of Streptomyces albus subsp. albus
The recommended thresholds used to distinguish between closely related prokaryotic species based on ANI and dDDH similarities are 95 to 96% (Richter and Rosselló-Móra 2009; Chun et al. 2018) and 70% (Meier-Kolthoff 2013a; Chun et al. 2018), respectively. Table 2 shows that on this basis the isolate can be separated from the type strains of its closest phylogenomic neighbours, as shown in Fig. 2. It is most closely related to S. albiflaviniger DSM 41598T based on a dDDH similarity of 53.9% and an ANI value of 93.5% though this latter value is shared with S. javensis DSM 41764T and S. iranensis HM 35T.
Table 2.
Average nucleotide identities and digital DNA:DNA hybridisation values between the isolate and Streptomyces species belonging to the S. violaceusniger clade
| Phylogenomic neighbours | ANI (%) | dDDH (%) |
|---|---|---|
| S. albiflaviniger DSM 41598T | 93.5 | 53.9 |
| S. antimycoticus NRRL B-24289T | 91.3 | 44.7 |
| S. himastatinicus ATCC 53653T | 84.9 | 29.0 |
| S. hygroscopicus subsp. hygroscopicus NBRC 16556T | 90.8 | 41.1 |
| S. iranensis HM 35T | 93.5 | 52.0 |
| S. javensis DSM 41764T | 93.5 | 52.8 |
| S. malaysiensis DSM 41697T | 91.6 | 44.4 |
| S. melanosporofaciens DSM 40318T | 91.5 | 44.9 |
| S. rapamycinicus NRRL 5491T | 93.4 | 51.1 |
| S. rhizosphaericus NRRL B-24034T | 93.3 | 52.6 |
| S. violaceusniger NBRC 13459T | 93.7 | 52.7 |
Identical results were obtained for the duplicated cultures in all of the phenotypic tests. It is also encouraging that the results of the biochemical, degradative and tolerance tests are in agreement with those from earlier analyses on the reference strains that were performed under the same experimental procedures (Al-Tai et al. 1999; Sembiring et al. 2000; Saintpierre et al. 2003; Goodfellow et al. 2007; Kumar and Goodfellow 2008; Hamedi et al. 2010; Zhou et al. 2017). Table 3 shows that the isolate can be separated from the type strains of all of its closest phylogenomic neighbours using a combination of phenotypic properties. It can, for instance, be distinguished from S. albiflaviniger DSM 14548T, its closest neighbour, as it is positive for esterase (C4), α-glucosidase and lipase (C14), casein, Tween 20 and uric acid, hydrolyses allantoin and grows in the presence of 7% w/v NaCl. In contrast, the S. albiflaviniger strain, unlike the isolate, hydrolyses arbutin. Additional combinations of phenotypic properties distinguish the isolate from the remaining reference strains and also the latter from one another.
Table 3.
Phenotypic characteristics which distinguish isolate PRKS01-29T from the type strains of closely related species classified in the Streptomyces violaceusniger clade
| Characteristics | Strains | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| API-ZYM tests | |||||||
| Esterase (C4) | + | − | − | + | + | + | + |
| α-Fucosidase | − | − | − | − | + | − | + |
|
α- and β-Galactosidase, α-mannosidase, trypsin |
+ | + | + | + | + | − | + |
| β-Glucuronidase | − | − | − | + | − | − | − |
| α-Glucosidase | + | − | − | − | − | − | − |
| β-Glucosidase | − | − | + | + | − | − | − |
| Lipase (C14) | + | − | + | − | − | + | − |
| Biochemical tests | |||||||
| Nitrate reduction | − | − | − | + | − | + | + |
| Degradation tests | |||||||
| Adenine (0.5%, w/v) | + | + | − | + | + | + | + |
| Aesculin (0.1%, w/v) | − | − | + | + | + | − | + |
| Allantoin (0.5%,w/v) | + | − | + | + | − | + | − |
| Arbutin (0.5%, w/v) | − | + | + | + | − | + | − |
| Casein (1%, w/v) | + | − | + | + | + | − | + |
| Guanine (0.3%, w/v) | − | − | − | + | − | + | − |
| Starch (0.1%, w/v) | + | + | + | + | + | − | − |
| Tween 20 (1%, v/v) | + | − | + | + | + | − | − |
| Uric acid (0.4%, w/v) | + | − | + | + | + | + | − |
| Xylan (0.4%, w/v) | + | + | + | + | − | + | + |
| Tolerance tests | |||||||
| Growth in presence of 7% w/v, NaCl | + | − | − | + | + | + | − |
| Growth at pH 9.0 | − | − | + | – | + | + | − |
| Growth at 45 °C | + | + | − | + | − | − | + |
Strains: 1. Isolate PRKS01-29T., 2. S. albiflaviniger DSM 14598T., 3. S. iranensis DSM 41954T., 4. S. javensis DSM 41764T., 5. S. rapamycinicus NRRL 5491T., 6. S. rhizosphaericus NRRL B-24304T., 7. S. violaceusniger DSM 40563T
All of the strains were positive for acid and alkaline phosphatases, α-chymotrypsin, cystine, leucine and valine arylamidases, esterase (C4), esterase lipase (C8), N-acetyl-β-glucoronidase and naphthol-AS-BI-phosphohydrolase (API-ZYM tests), hydrolysed urea and degraded hypoxanthine (0.4%, w/v), Tweens 40, 60 and 80 (all 1%, v/v) and L-tyrosine (0.4%, w/v), but not chitin (1%, w/v), elastin (0.3%, w/v), tributyrin (0.1%, w/v) or xanthine (0.4%, w/v)
+ positive., − negative, n.d. not determined
The aerial spore mass and substrate mycelial colours produced by the respective reference strains on the ISP media are in agreement with those from earlier analyses (Al-Tai et al. 1999; Goodfellow et al. 2007; Kumar and Goodfellow 2008; Hamedi et al. 2010). Table S2 shows that the isolate and its closest phylogenomic neighbours grew well on nearly all of the ISP media forming a grey-yellowish substrate mycelium bearing a grey aerial spore mass that became moist and black on prolonged incubation on ISP 3 agar, as is the case with the type strains of S. antimycoticus (Kumar and Goodfellow 2008; Komaki and Tamura 2020a), S. griseiniger (Goodfellow et al. 2007), S. hygroscopicus (Labeda and Lyons 1991) and S. yatensis (Saintpierre et al. 2003). The isolate and the S. albiflaviniger can be distinguished by their ability to produce diffusible pigments, for instance, only the reference strain produced diffusible pigments on ISP media 3 and 7.
The isolate showed activity in the primary and secondary screens. Growth of the S. aureus strain was inhibited when the isolate was grown on ISP 2, ISP 3, MMM and 410 agar media. Similarly, it inhibited the B. subtilis, C. albicans and M. luteus strains following cultivation on all of the nutrient formulations, apart from medium 410. In contrast, it did not show any activity against the E. coli strain though it did inhibit the growth of the P. aeruginosa strain when grown on ISP 3 and MMM agar. In the secondary screens, the isolate formed blue halos around inhibition zones against B. subtilis reporter strains YpuAER, YvqIER, YjaxER and DinBCH indicating its ability to inhibit cell envelope, DNA, fatty acid and RNA synthesis, respectively. It also inhibited the growth of the other reporter strains, YvgSER and YheH, without forming blue halos thereby suggesting an ability to produce bioactive compound(s) with unknown modes of action.
Biosynthetic potential of isolate PRKS01-29T and members of the S. violaceusniger clade
The isolate and the type strains of species classified in the S. violaceusniger clade have large genomes (10.1–12.7 Mb) predicted to encode for chemically diverse specialised metabolites. The genome mining studies showed that all of the strains are genetically equipped with bioclusters predicted to encode for ‘core secondary’ metabolites, such as albaflavenone/geosmin, ectoines, hopenes, melanin and spore pigments, results in good agreement with those of Ward and Allenby (2018). In contrast, most of the bioclusters predicted to encode for druggable molecules, notably antibiotics, were discontinuously distributed in the genomes of the strains with many being strain specific, as has been found in recent studies on streptomycetes (Vicente et al. 2018; Martinet et al. 2020).
The genome of all of the strains contained bioclusters predicted to encode for echosides A-E, anti-tumor agents produced by Streptomyces strain LZ35 (Zhu et al. 2014). In contrast, only the genomes of the isolate and the type strains of S. iranensis, S. violaceusniger and S. rapamycinicus contained bioclusters considered to express for meilingmycin, an anti-parasitic macrolide (Sun et al. 2002) and nigericin, which inhibits Gram-positive bacteria (Graven et al.1966). Similarly, the draft genomes of the isolate, S. albiflaviniger DSM 41598T and S. javensis DSM 41764T contained bioclusters predicted to encode for the synthesis of cahuitamycins A-C, which inhibit the formation of bacterial biofilms (Park et al. 2016), pladienolides, anti-tumour antibiotics (Mizui et al. 2004) and funisamine, an aminopolyol polyketide antibiotic which inhibits the growth of wild type strains of Staphylococcus aureus, Escherichia coli and Candida albicans (Covington et al. 2018), respectively. Bioclusters predicted to encode for rapamycin were only detected in the genomes of the S. iranensis and S. rapamycinicus strains.
Conclusion
It can be concluded from the phylogenetic trees and associated colonial and morphological data that isolate PRKS01-29T belongs to the S. violaceusniger clade (Sembiring et al. 2000; Goodfellow et al. 2007; Kumar and Goodfellow 2008, 2010). In addition, the whole genome sequence data show that it belongs to a well-supported monophyletic clade which includes the type strains of S. albiflaviniger, S. iranensis, S. javensis, S. rapamycinicus and S. rhizosphaericus. It can be distinguished from all of these strains by a broad range of phenotypic properties and by low ANI and dDDH values. It is, therefore, proposed that isolate PRKS01-29T represents a novel species within the genus Streptomyces for which the name Streptomyces sabulosicollis sp. nov. is proposed.
Description of Streptomyces sabulosicollis sp. nov.
Streptomyces sabulosicollis (sa.bu.lo.si.col’lis. L. masc. adj. sabulosus sandy; L. masc. n. collis a hill; N.L. gen. n. sabulosicollis of a sandy hill), Gram-stain-positive, catalase positive, aerobic actinobacterium which forms an extensively branched substrate mycelium and aerial hyphae which differentiate into spiral chains of rugose ornamented spores (0.8 × 0.97 µm) on yeast extract-malt extract agar. A yellowish-grey substrate mycelium carries a grey aerial spore mass that becomes moist and black following prolonged incubation on oatmeal agar. Grows from 10 to 45 °C (optimally at 28 °C), from pH 5.5–7.5 (optimally 7.0) and can tolerate up to 7% (w/v) NaCl. Allantoin and urea are hydrolysed but not aesculin or arbutin. Does not reduces nitrate. Degrades adenine, casein, hypoxanthine, starch, L-tyrosine, Tweens 20, 40, 60 and 80, uric acid and xylan, but not chitin, elastin, guanine, tributyrin or xanthine. Positive for acid and alkaline phosphatases, α-chymotrypsin, cystine, leucine and valine arylamidases, esterase lipase, α- and β-galactosidases, α-glucosidase, N-acetyl-β-glucosidase, lipase (C14), α-mannosidase, naphthol-AS.BI-phosphohydrolase and trypsin, but not α-fucosidase, β-glucosidase or β-glucuronidase. Whole organism hydrolysates contain LL-A2pm, galactose, glucose, mannose and ribose., the predominant fatty acids (> 10%) are iso-C15:0 (14.4%), anteiso-C15:0 (13.8%) and iso-C16:0 (27.2%), the major menaquinones MK-9 (H6, H8) with the proportions of 58.4% and 41.6%, respectively, and the polar lipid profile is composed of diphosphatidylglycerol, two phosphatidylglycerols, phosphatidylinositol, two phosphatidylinositol mannosides and two unknown phospholipids. The dDNA G+C content of the strain is 71.7% and its approximate genome size 10.2 Mbp.
The type strain, PRKS01-29T (= CCMM B1303T = ICEBB-02T = NCIMB 15210T) was isolated from a sandy soil sample collected from an arid sand dune system in the Parangkusumo Region of Yogyakarta Province, Java, Indonesia. The GenBank accession number of the assembled draft genome of Streptomyces sabulosicollis is JAEEAP000000000.1.
In the case of the genus Streptomyces genome-based classifications have revealed the presence of well-defined species-groups (Labeda et al. 2012, 2017; Nouioui et al. 2018), the recognition of later heterotypic synonyms of established species (Komaki and Tamura 2020a, b; Madhaiyan et al. 2020) within and outwith the S. violaceusniger phylogenetic clade (Sembiring et al. 2000; Goodfellow et al. 2007; Kumar and Goodfellow 2008, 2010) and the delineation of the genera Embleya and Yinghuangia for species previously included in the genus (Nouioui et al. 2018). Such developments can be expected to continue and in this respect, it is evident from this study that streptomycetes which form rugose-ornamented spores, spiral spore chains and characteristic colonial properties on oatmeal agar belong to a distinct phylogenomic clade the taxonomic status of which merits further investigation.
Data availability statements
The 16S rRNA gene and whole genome sequences of strain PRKS01-29T that support the findings of this study have been deposited in GenBank database with the accession numbers MK503616 and JAEEAP000000000.1, respectively. In turn, corresponding accession numbers for the whole genome sequences of Streptomyces albiflaviniger DSM 42598T and Streptomyces javensis DSM 41764T are JAEEAR000000000.1 and JAEEAQ000000000.1, respectively. All the whole genome sequences described in this paper is version 1.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
ABK is grateful for financial support awarded through the Ph.D Scholarship Scheme of the Indonesian Endowment Fund for Education (LPDP), Ministry of Finance, Indonesia. Thanks are due to Mr. Ja’far Abdurrahman and Mr. Faiz Muhammad for helping to collect the environmental samples. We are indebted to Dr. Arief Budi Wietarto for encouragement and support and to Sausan Nafisah and Sandi Arya Rumintang from the Department of Biotechnology, Sumbawa University of Technology, Indonesia, Dr. Wasu Pathom-aree and Nilita Mukjang from the Department of Biology, Chiang Mai University, Thailand for their valuable assistance in the deposition of the whole genome sequences of the strains in the NCBI genome database. IN is grateful to Newcastle University for a postdoctoral fellowship and MG for an Emeritus Fellowship from the Leverhulme Trust. We also thank Professor Hans-Peter Klenk (Newcastle University, United Kingdom) for his help in initial stages of the study.
Authors contribution
MG and ABK designed the study and prepared the manuscript. ABK helped to collect the soil sample, characterized the strain under the supervision of IN and MG and deposited it in the culture collections. ABK and IN were responsible for the genome sequencing, annotation and the genome analyses. All of the authors approved the final version of the manuscript.
Funding
This research was funded by The Indonesian Endowment Fund for Education (LPDP), Ministry of Finance, Indonesia through PhD scholarship scheme awarded to the first author with the grant number 20160822028928.
Declarations
Conflict of interest
The authors declare that they do not have any conflicts of interest.
Ethical approval
This article does not include any work with human participants and/or animals performed by one of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al-Tai A, Kim B, Kim SB, Manfio GP, Goodfellow M. Streptomyces malaysiensis sp.nov., a new streptomycete species with rugose ornamented spores. Int J Syst Bacteriol. 1999;49:1395–1402. doi: 10.1099/00207713-49-4-1395. [DOI] [PubMed] [Google Scholar]
- Arcamone F, Bertazzoli C, Ghione M, Scotti T. Melanosporin and elaiophylin, new antibiotics from Streptomyces melanosporus (sive melanosporofaciens) n. sp. Giornale di Microbiologia. 1959;7:207–216. [Google Scholar]
- Ayed A, Slama N, Mankai H, Bachkouel S, ElKahoui S, et al. Streptomyces tunisialbus sp. nov., a novel Streptomyces species with antimicrobial activity. Antonie Van Leeuwenhoek. 2018;111:1571–1581. doi: 10.1007/s10482-018-1046-4. [DOI] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, Dejongh M, Disz T, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz RH. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol. 2017;44:573–588. doi: 10.1007/s10295-016-1815-x. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M. SPAdes: a new genome assembly algorithm and its application to single-cell sequencing. J Compu Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranasic D, Gacesa R, Starcevic A, Zucko J, Blazic M, et al. Draft genome sequence of Streptomyces rapamycinicus strain NRRL 5491, the producer of the immunosuppressant rapamycin. Genome Announc. 2013;1:e00581–e613. doi: 10.1128/genomeA.00581-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;2:81–87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Fan Y, Zheng Y, Shen Y. Properties and production of valienamine and its related analogues. Chem Rev. 2003;103:1955–1977. doi: 10.1021/cr0102260. [DOI] [PubMed] [Google Scholar]
- Cheng J, Yang SH, Palaniyandi SA, Han JS, Yoon T-M, et al. Azalomycin F complex is an antifungal substance produced by Streptomyces malaysiensis MJM1968 isolated from agricultural soil. J Korean Soc Appl Biol Chem. 2010;53:545–552. doi: 10.3839/jksabc.2010.084. [DOI] [Google Scholar]
- Chun J, Oren A, Ventosa A, Chirstensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- Clermont N, Legault G, Lerat S, Beaulieu C. Effect of biopolymers on geldanamycin production and biocontrol ability of Streptomyces melanosporofaciens strain EF-76. Can J Plant Path. 2010;32:481–489. doi: 10.1080/07060661.2010.512121. [DOI] [Google Scholar]
- Covington BC, Spraggins JM, Ynigez-Gutierrez AE, Hylton ZB, Bachmann BO. Response of secondary metabolism of hypogean actinobacterial genera to chemical and biological stimuli. Appl Env Microbiol. 2018;84:e01125–e1218. doi: 10.1128/AEM.01125-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. Geldanamycin, a new antibiotic. J Antibiot. 1970;23:442–447. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euzéby J. Validation List No. 119. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2008;58:1–2. doi: 10.1099/ijs.0.65794-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fiedler HP. Screening for bioactivity. In: Bull AT, editor. Microbial diversity and bioprospecting. Washington DC, USA: American Society of Microbiology; 2004. pp. 324–335. [Google Scholar]
- Fitch WM. Toward defining course of evolution - minimum change for a specific tree topology. Syst Zool. 1971;20:406. [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Goodfellow M, Fiedler HA. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek. 2010;98:119–142. doi: 10.1007/s10482-010-9460-2. [DOI] [PubMed] [Google Scholar]
- Goodfellow M, Kumar Y, Labeda DP, Sembiring L. The Streptomyces violaceusniger clade: a home for streptomycetes with rugose ornamented spores. Antonie Van Leeuwenhoek. 2007;92:173–199. doi: 10.1007/s10482-007-9146-6. [DOI] [PubMed] [Google Scholar]
- Graven SN, Estrada-O S, Lardy HA. Alkali metal cation release and respiratory inhibition induced by nigericin in rat liver mitochondria. PNAS. 1966;56:654–658. doi: 10.1073/pnas.56.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamedi J, Mohammadipanah F, Klenk HP, Pötter G, Schumann P, et al. Streptomyces iranensis sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2010;60:1504–1509. doi: 10.1099/ijs.0.015339-0. [DOI] [PubMed] [Google Scholar]
- Hamm PS, Caimi NA, Northup DE, Valdez EW, Buecher DC, et al. Western bats as a reservoir of novel Streptomyces species with antifungal activity. Appl Environ Microbiol. 2017;83:e03057–e3116. doi: 10.1128/AEM.03057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn F, Schroeckh V, Netzker T, Guthke R, Brakhage AA, et al. Draft genome sequence of Streptomyces iranensis. Genome Announc. 2014;17(2):e00616–e714. doi: 10.1128/genomeA.00616-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen HL. Contributions to our knowledge of the Actinomycetales. II. The definition and subdivision of the genus Actinomyces, with a preliminary account of Australian soil actinomycetes. Proc Linn Soc NSW. 1931;56:345–370. [Google Scholar]
- Jukes TH, Cantor CR. Evolution of Protein Molecules. London: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kämpfer P, et al. Genus 1. Streptomyces Waksman and Henrici 1943, 339AL emend. Rainey, Ward-Rainey and Stackebrandt, 1997, 486, emend. Kim, Lonsdale, Seong and Goodfellow 2003b, 113, emend. Zhi, Li and Stackebrandt, 2009 600. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki K-I, et al., editors. The Actinobacteria 5 Part B. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2012. pp. 1455–1767. [Google Scholar]
- Kelly KL. Central notations for the revised ISCC-NBS color name blocks. J Res Nat Bureau Standards USA. 1958;61:427. [Google Scholar]
- Komaki H, Tamura T. Reclassification of Streptomyces castelarensis and Streptomyces sporoclivatus as later heterotypic synonyms of Streptomyces antimycoticus. Int J Sys Evol Microbiol. 2020;70:1099–1105. doi: 10.1099/ijsem.0.003882. [DOI] [PubMed] [Google Scholar]
- Komaki H, Tamura T. Reclassification of Streptomyces diastaticus subsp. ardesiacus, Streptomyces gougerotii and Streptomyces rutgersensis. Int J Syst Evol Microbiol. 2020;70:4291–4297. doi: 10.1099/ijsem.0.004287. [DOI] [PubMed] [Google Scholar]
- Komaki H, Ichikawa N, Oguchi A, Hamada M, Tamura T, et al. Genome analysis-based reclassification of Streptomyces endus and Streptomyces sporocinereus as later heterotypic synonyms of Streptomyces hygroscopicus subsp. hygroscopicus. Int J Syst Evol Microbiol. 2017;67:343–345. doi: 10.1099/ijsem.0.001628. [DOI] [PubMed] [Google Scholar]
- Komaki H, Sakurai K, Hosoyama A, Kimura A, Igarashi Y, et al. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci Rep. 2018;8:6888. doi: 10.1038/s41598-018-24921-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Y, Goodfellow M. Five new members of the Streptomyces violaceusniger 16S rRNA gene clade: Streptomyces castelarensis sp. nov., comb. nov., Streptomyces himastatinicus sp. nov., Streptomyces mordarskii sp. nov., Streptomyces rapamycinicus sp. nov. and Streptomyces ruanii sp. nov. Int J Syst Evol Microbiol. 2008;58:1369–1378. doi: 10.1099/ijs.0.65408-0. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Goodfellow M. Reclassification of Streptomyces hygroscopicus strains as Streptomyces aldersoniae sp. nov., Streptomyces augustmycinicus sp. nov., comb. nov. Streptomyces ascomycinicus sp. nov., Streptomyces decoyicus sp. nov., comb. nov., Streptomyces milbemycinicus sp. nov. and Streptomyces wellingtoniae sp. nov. Int J Syst Ecol Microbiol. 2010;60:769–775. doi: 10.1099/ijs.0.012161-0. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Aiemsum-Ang P, Ward AC, Goodfellow M. Diversity and geographical distribution of members of the Streptomyces violaceusniger 16S rRNA gene clade detected by clade-specific PCR primers. FEMS Microbiol Ecol. 2007;62:54–63. doi: 10.1111/j.1574-6941.2007.00374.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma AB, Nouioui I, Klenk HP, Goodfellow M. Streptomyces harenosi sp. nov., a home for a gifted strain isolated from Indonesian sand dune soil. Int J Syst Evol Microbiol. 2020;70:4874–4882. doi: 10.1099/ijsem.0.004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall LD, Roy MA, O’Neill JJ, Devine TE. Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Evol Microbiol. 1988;38:358–361. [Google Scholar]
- Labeda DP, Lyons AJ. The Streptomyces violaceusniger cluster is heterogeneous in DNA relatedness among strains: Emendation of the descriptions of Streptomyces violaceusniger and Streptomyces hygroscopicus. Int J Syst Bacteriol. 1991;41:398–401. [Google Scholar]
- Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, et al. Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek. 2012;101:73–104. doi: 10.1007/s10482-011-9656-0. [DOI] [PubMed] [Google Scholar]
- Labeda DP, Doroghazi JP, Ju K-S, Metcalf WW. Taxonomic evaluation of Streptomyces albus and related species using multi-locus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int J Syst Evol Microbiol. 2014;64:894–900. doi: 10.1099/ijs.0.058107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, et al. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2017;110:563–583. doi: 10.1007/s10482-016-0824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KS, Hesler GA, Mattei JM, Mamber SW, Forenza S, et al. Himastatin, a new antitumor antibiotic from Streptomyces hygroscopicus. I. Taxonomy of producing organism, fermentation and biological activity. J Antibiot (Tokyo) 1990;43:956–960. doi: 10.7164/antibiotics. [DOI] [PubMed] [Google Scholar]
- Lechevalier MP, Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–443. [Google Scholar]
- Lee I, Ouk Kim Y, Park SC, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66(2):1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- Lee I, Chalita M, Ha SM, Na SI, Yoon SH, Chun J. ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol. 2017;67:2053–2057. doi: 10.1099/ijsem.0.001872. [DOI] [PubMed] [Google Scholar]
- Li K, Guo Y, Wang J, Wang Z, Zhao J, et al. Streptomyces aquilus sp. nov., a novel actinomycete isolated from a Chinese medicinal plant. Int J Syst Evol Microbiol. 2020;70:1912–1917. doi: 10.1099/ijsem.0.003995. [DOI] [PubMed] [Google Scholar]
- Madhaiyan M, Saravanan VS, See-Too WS. Genome-based analyses reveal the presence of 12 heterotypic synonyms in the genus Streptomyces and emended descriptions of Streptomyces bottropensis, Streptomyces celluloflavus, Streptomyces fulvissimus, Streptomyces glaucescens, Streptomyces murinus, and Streptomyces variegatus. Int J Syst Evol Microbiol. 2020;70:3924–3929. doi: 10.1099/ijsem.0.004217. [DOI] [PubMed] [Google Scholar]
- Martinet L, Naômé A, Baiwir D, De Pauw E, Mazzucchelli G, et al. On the risks of phylogeny-based strain prioritization for drug discovery: Streptomyces lunaelactis as a case study. Biomolecules. 2020;10:1027. doi: 10.3390/biom10071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- Miller LT. A single derivatization method for bacterial fatty acid methyl esters including hydroxy acids. J Clin Microbiol. 1982;16:584–586. doi: 10.1128/jcm.16.3.584-586.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin D, O'Donnell A, Goodfellow M, Alderson G, Athalye M, et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241. [Google Scholar]
- Mizui Y, Sakai T, Iwata M, Uenaka T, Okamoto K. Pladienolides, new substances from culture of Streptomyces platensis Mer-11107. III. In vitro and in vivo antitumor activities. J Antibio (Tokyo) 2004;57:188–196. doi: 10.7164/antibiotics.57.188. [DOI] [PubMed] [Google Scholar]
- Murray P, Barron E, Phaller M, Ternover J, Yolkken R. Manual of clinical microbiology. Mycopathologia. 1999;146:107–108. [Google Scholar]
- Nguyen TM, Kim J. Description of Streptomyces fabae sp. nov., a producer of antibiotics against microbial pathogens, isolated from soybean (Glycine max) rhizosphere soil. Int J Syst Evol Microbiol. 2015;65:4151–4156. doi: 10.1099/ijsem.0.000551. [DOI] [PubMed] [Google Scholar]
- Nicault M, Tidjani AR, Gauthier A, Dumarcay S, Gelhaye E, et al. Mining the biosynthetic potential for specialized metabolism of a Streptomyces soil community. Antibiotics (Basel) 2020;23(9):271. doi: 10.3390/antibiotics9050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouioui I, Carro L, Garcia-Lopez M, Meier-Kolthoff JP, Woyke T, et al. Genome-based-taxonomic classification of the phylum Actinobacteria. Front Microbiol. 2018;9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell AG, Falconer C, Goodfellow M, Ward AC, Williams E. Biosystematics and diversity amongst novel carboxydotrophic actinomycetes. Antonie Van Leeuwenhoek. 1993;64:325–340. doi: 10.1007/BF00873091. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. The SEED and the Rapid Annotation of Microbial Genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniyandi SA, Yang SH, Suh JW. Foliar application of extract from an azalomycin-producing Streptomyces malaysiensis strain MJM1968 suppresses yam anthracnose caused by Colletotrichum gloeosporioides. J Microbiol Biotechnol. 2016;28(26):1103–1108. doi: 10.4014/jmb.1601.01018. [DOI] [PubMed] [Google Scholar]
- Park SR, Tripathi A, Wu J, Schultz PJ, Yim I, et al. Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nat Commun. 2016;7:10710. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. How many bootstrap replicates are necessary? J Comp Biol. 2010;17:337–354. doi: 10.1089/cmb.2009.0179. [DOI] [PubMed] [Google Scholar]
- Pridham TG, Hesseltine CW, Benedict RG. A guide for the classification of streptomycetes according to selected groups; placement of strains in morphological sections. Appl Microbiol. 1958;6:52–79. doi: 10.1128/am.6.1.52-79.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe P, Samborskyy M, Leadlay PF, Dickschat JS. Isoafricanol synthase from Streptomyces malaysiensis. Org Biomol Chem. 2017;21(15):2353–2358. doi: 10.1039/c7ob00234c. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. PNAS. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X, Huang Y. Taxonomic evaluation of the Streptomyces hygroscopicus clade using multi-locus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for the systematics of the whole genus. Syst Appl Microbiol. 2012;35:7–18. doi: 10.1016/j.syapm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Rong X, Huang Y. Multilocus sequence analysis: taking prokaryote systematics to the next level. Meth Microbiol. 2014;41:221–251. [Google Scholar]
- Sahin N, Sazak A, Guven K, Dogramaci M. Diversity of members of the Streptomyces violaceusniger 16S rRNA gene clade in the legumes rhizosphere in Turkey. Ann Microbiol. 2010;60:669–684. doi: 10.1007/s13213-010-0112-6. [DOI] [Google Scholar]
- Saintpierre D, Amir H, Pineau R, Sembiring L, Goodfellow M. Streptomyces yatensis sp. nov., a novel bioactive streptomycete isolated from a New-Caledonian ultramafic soil. Antonie Van Leeuwenhoek. 2003;83:21–26. doi: 10.1023/a:1022906325397. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sarwar A, Latif Z, Zhang S, Hao J, Bechthold A. A potential biocontrol agent Streptomyces violaceusniger AC12AB for managing potato common scab. Front Microbiol. 2019;8:202. doi: 10.3389/fmicb.2019.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Inc Technical Notes. 1990;101:1. [Google Scholar]
- Sembiring L, Ward AC, Goodfellow M. Selective isolation and characterisation of members of the Streptomyces violaceusniger clade associated with the roots of Paraserianthes falcataria. Antonie Van Leeuwenhoek. 2000;78:353–366. doi: 10.1023/A:1010226515202. [DOI] [PubMed] [Google Scholar]
- Sembiring L, Ward AC, Goodfellow M. In validation of publication of new names and new combination previously effectively published outline the IJSEM List no. 82. I Syst Evol Microbiol. 2001;51:1619–1620. [Google Scholar]
- Shirling E, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–340. [Google Scholar]
- Society of American Bacteriologists . Manual of microbiological methods. New York: McGraw-Hill; 1957. [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:225–231. doi: 10.1128/am.28.2.226-231.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhou X, Liu J, Bao K, Zhang G, et al. 'Streptomyces nanchangensis', a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiol (Reading) 2002;148:361–371. doi: 10.1099/00221287-148-2-361. [DOI] [PubMed] [Google Scholar]
- Swofford D (2002) PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4.0 b10. Sinauer Associates, Sunderland
- Tindall BJ. On the status of the species name Streptomyces ruanii Kumar and Goodfellow 2008. Int J Syst Evol Microbiol. 2014;64:1068–1070. doi: 10.1099/ijs.0.060640-0. [DOI] [PubMed] [Google Scholar]
- van der Aart LT, Nouioui I, Kloosterman A, Igual J-M, Willemse J, et al. Polyphasic classification of the gifted natural product producer Streptomyces roseifaciens sp. nov. Int J Syst Microbiol. 2019;69:899–908. doi: 10.1099/ijsem.0.003215. [DOI] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal S. Rapamycin (AY-22, 989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycetes and isolation of the active principle. J Antibiot. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Vicente CM, Thibessard A, Lorenzi JN, Benhadj M, Hôtel L. Comparative genomics among closely related Streptomyces strains revealed specialized metabolite biosynthetic gene cluster diversity. Antibiotics (Basel) 2018;2(7):86. doi: 10.3390/antibiotics7040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksman SA, Curtis RE. The Actinomyces of the soil. Soil Sci. 1916;1:99–134. [Google Scholar]
- Waksman SA, Henrici AT. Family III. Streptomycetaceae Waksman and Henrici. In: Breed RS, Murray EGD, Hitchens AP, editors. Bergey’s manual of determinative bacteriology. 6. Baltimore: The Williams & Wilkins Co; 1948. pp. 929–980. [Google Scholar]
- Wang B, Song Y, Luo M, Chun Q, Ma J, et al. Biosynthesis of 9-methylstreptimidone involves a new decarboxylative step for polyketide terminal diene formation. Org Lett. 2013;15:1278–1281. doi: 10.1021/ol400224n. [DOI] [PubMed] [Google Scholar]
- Ward AC, Allenby NE. Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol Let. 2018 doi: 10.1093/femsle/fny240.10.1093/femsle/fny240. [DOI] [PubMed] [Google Scholar]
- Ward AC, Goodfellow M. Phylogeny and functionality: taxonomy as a roadmap to genes. In: Bull AT, editor. Microbial diversity and bioprospecting. Washington DC: American Society for Microbiology; 2004. pp. 288–313. [Google Scholar]
- Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;4(45):535–542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ST, Goodfellow M, Alderson G, Wellington EM, Sneath PH, et al. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- Xie Y, Li Q, Qin X, Ju J, Ma J. Enhancement of himastatin bioproduction via inactivation of atypical repressors in Streptomyces hygroscopicus. Metab Eng Commun. 2019;5(8):e00084. doi: 10.1016/j.mec.2018.e00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamac M, Isik K, Sahin N. Numerical classification of streptomycetes isolated from karstic caves in Turkey. Turk J Biol. 2011;35:473–484. [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, et al. Introducing EzBiocloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yang X, Huang D, Huang X. Streptomyces solisilvae sp. nov., isolated from tropical forest soil. Int J Syst Evol Microbiol. 2017;67:3553–3558. doi: 10.1099/ijsem.0.002166. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen W, Li YY, Deng JJ, Zhu DY, et al. Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene. 2014;546:352–358. doi: 10.1016/j.gene.2014.05.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene and whole genome sequences of strain PRKS01-29T that support the findings of this study have been deposited in GenBank database with the accession numbers MK503616 and JAEEAP000000000.1, respectively. In turn, corresponding accession numbers for the whole genome sequences of Streptomyces albiflaviniger DSM 42598T and Streptomyces javensis DSM 41764T are JAEEAR000000000.1 and JAEEAQ000000000.1, respectively. All the whole genome sequences described in this paper is version 1.