Abstract
Engineering functional human tissues in vitro is currently limited by difficulty replicating the small caliber, complex connectivity, cellularity, and 3D curvature of the native microvasculature. Multiphoton ablation has emerged as a promising technique for fabrication of microvascular structures with high resolution and full 3D control, but cellularization and perfusion of complex capillary‐scale structures has remained challenging. Here, multiphoton ablation combined with guided endothelial cell growth from pre‐formed microvessels is used to successfully create perfusable and cellularized organ‐specific microvascular structures at anatomic scale within collagen hydrogels. Fabrication and perfusion of model 3D pulmonary and renal microvascular beds is demonstrated, as is replication and perfusion of a brain microvascular unit derived from in vivo data. Successful endothelialization and blood perfusion of a kidney‐specific microvascular structure is achieved, using laser‐guided angiogenesis. Finally, proof‐of‐concept hierarchical blood vessels and complex multicellular models are created, using multistep patterning with multiphoton ablation techniques. These successes open new doors for the creation of engineered tissues and organ‐on‐a‐chip devices.
Keywords: biomaterials, microphysiological systems, multiphoton ablation, organ‐on‐a‐chip, tissue engineering
Engineering functional tissues for regenerative medicine and human disease modeling is limited by difficulty re‐creating in vivo‐like microvasculature. This paper reports success combining multiphoton microscopy with directed angiogenesis to precisely generate perfusable 3D organ‐specific capillary beds within natural extracellular matrices. These methods enable full cellularization of complex capillary models and the creation of multicellular, heterogeneous, and hierarchical vasculature.
Introduction
Engineered tissues hold remarkable promise for both disease modeling and regenerative medicine.[ 1 , 2 ] The successful creation of functional tissues in vitro depends on adequate vascularization and perfusion. In vivo, tissues are supported by blood perfusion from interconnected three‐dimensional (3D) capillary networks with diameters down to 5–10 µm.[ 3 ] Adding additional complexity, each human organ relies on structurally distinct microvasculature to support its unique hemodynamic and functional needs. As examples, the brain and heart contain a dense 3D network of thousands of capillaries per cm3 to support their high metabolic activity,[ 4 ] kidneys filter blood via tortuous glomerular capillaries within a spherical space, and in the lung a thin sheet‐like capillary network encircles spherical alveoli to maximize gas exchange.[ 5 ]
Current technology faces challenges in replicating the scale, density, organ‐specific complexity, and 3D curvature of native human capillary networks. Methods to create perfusable, cellularized vascular networks have included the casting of a hydrogel around a supportive material such as a needle,[ 6 ] a lithographically defined mold,[ 7 , 8 ] or a dissolvable material.[ 9 , 10 ] After hydrogel gelation, removal or dissolution of this supportive material results in void spaces within the material into which cells can be introduced to form vascularized lumens. This can generate precise patterning, but does not easily allow full 3D control over vascular geometry. Bioprinting addresses the need for complex 3D patterning,[ 11 , 12 ] but lacks the resolution to create vascular lumens below ≈100 µm in diameter in cell‐remodelable materials.
Multiphoton ablation is an emerging technique capable of patterning biomaterials with both high resolution and precise 3D control. By focusing ultrashort laser pulses within a hydrogel, high‐energy multiphoton events occur principally at the focal point, leading to finely controlled material removal in 3D space. This technique has led to recent advances in the patterning of biomimetic vascular structures in multiple cell‐compatible materials including collagen,[ 13 , 14 ] poly (ethylene glycol)‐based materials,[ 15 , 16 ] silk fibroin,[ 17 ] and specialized photodegradable polymers.[ 18 , 19 , 20 ] Degradation of these materials proceeds through different mechanisms depending on material composition and level of two‐photon absorption.[ 21 ] Prior works have demonstrated creation of void‐spaces with the diameter and geometry of human capillaries and the partial cellularization of larger photoablated lumens.[ 16 ] Despite these achievements, the endothelialization of 3D microvessels at the capillary scale is extremely challenging. Direct cell seeding into capillary‐sized structures generally leads to luminal occlusion and patchy cell coverage. Drawing inspiration from previous reports on the directed migration of fibroblasts, cancer cells, or neural cells into ablated void spaces[ 13 , 20 , 22 ] we recently developed a technique, here termed “laser‐guided angiogenesis,” where multiphoton ablation of linear channels extending from a pre‐cellularized parent vessel guide endothelial cell growth and lead to perfusable endothelialized capillaries.[ 14 ] Here, we use multiphoton ablation approaches to successfully create organ‐specific microvascular structures that are cellularized, perfusable, and exhibit significant geometric complexity at anatomic capillary scale. Highlighting how our techniques enable new approaches to the study of vascular biology, we also demonstrate creation of proof‐of‐concept hierarchical and multicellular microvessels.
Results
Using a two‐photon microscope, we first evaluated the ablation characteristics of three different materials commonly used for tissue engineering: collagen (7.5 mg mL−1), fibrin (10 mg mL−1), and a hydrogel containing 2% (w/v) each of agarose and gelatin. Two channels were formed lithographically within hydrogels and parallel conduits ablated between them using delivered laser powers at a wavelength (λ) of 800 nm ranging from 107 mW up to two exposures at 244 mW. Fluorescent beads were then perfused to evaluate patency (Figure 1a). In collagen and fibrin, laser powers of 157 mW or above led to complete ablation and the formation of a patent lumen with beads adherent only to the vessel wall. Agarose–gelatin gel required a power of 203 mW to generate perfusable structures, however hydrogel material remained incompletely ablated, as perfused beads were seen adherent throughout intraluminal spaces. For successive experiments we chose to use 7.5 mg mL−1 collagen, a laser power of 244 mW and two exposures to ensure full ablation. At these settings we ablated rectangles of decreasing width into collagen, demonstrating precise control down to at least 1 µm in resolution, consistent with previous reports (Figure S1, Supporting Information).[ 13 , 20 ]
Figure 1.
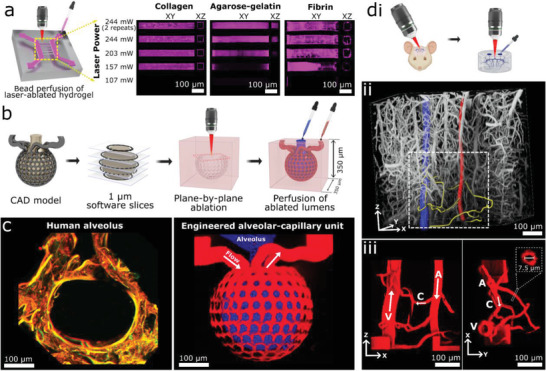
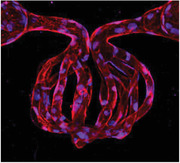
Multiphoton‐guided creation of organ‐specific vascular structures at anatomic scale. a) Hydrogel photoablation. Images shown following perfusion of beads (magenta). b) Schematic of ablation and perfusion process. c) Two‐photon images of a human alveolus (left), and an engineered alveolar‐capillary unit after bead perfusion of capillary (red) and alveolar (blue) compartments (right). d) Recreation of mouse brain microvasculature. i) Schematic of using in vivo imaging data as a mask for ablation. ii) 3D microvascular image traced to isolate an arteriole (red), venule (blue), and capillaries (yellow). iii) Projection images of a replica microvascular unit ablated into collagen and perfused with beads (red). Orthogonal projections and capillary cross‐section shown. Arrows delineate flow from arteriole (A), to capillaries (C), and venule (V).
We next developed a customizable process to use graphical data as a mask to guide the photoablation of complex 3D microvasculature in vitro (Figure 1b). This process allows facile control over laser intensity, pixel dwell time, and line‐repeats for thousands of polygons bounding hundreds of thousands of XYZ points. Using this process, we demonstrated successful ablation of microvascular structures with increasing complexity into collagen hydrogel (Figure 1, Figures S2 and S3, Supporting Information). Structures created in computer‐aided design (CAD) software were ablated either directly between an inlet and outlet or between a lithographically determined arteriole–venule unit.[ 14 ] We chose a helical pattern to demonstrate continuous curvature and smooth walls (Figure S2, Supporting Information), and a meshwork pattern that highlights the achievement of two closely interwoven but separately perfusable channels (Figure S3, Supporting Information). Ablation of each structure was completed in 10–30 min using the above‐listed microscope settings, and faster speeds would likely be possible using a single scan, or a microscope with faster galvonometers and software to allow dwell times < 2 µs (the lower limit of our microscope)—using a laser with higher peak power if necessary to compensate for the reduction in dwell time. When perfused with fluorescent beads, the meshwork structure displayed its intended geometry, including a diameter of 10 µm for individual channels, and a separation of 15 µm between the two interwoven networks. Expanding on this approach, we then created a model of the lung alveolar‐capillary unit at physiologic scale, consisting of a network of capillaries of 10 µm in diameter encircling an alveolus‐like void space with a diameter of 300 µm. When perfused with fluorescent microbeads, the capillary and alveolar compartments remained distinct and separately perfusable (Figure 1c).
In vivo microvascular images have been used previously as masks to guide photoablation of lumens into PEGDA hydrogels with the density of human capillary beds, though to our knowledge perfusion of these structures at this scale has not been shown.[ 16 , 23 ] We extended this technique here into collagen hydrogels, and demonstrate controlled perfusion of a single microvascular unit. Using a multiphoton microscope, we imaged microvasculature in the cerebral cortex of a live mouse down to the capillary scale following perfusion of fluorescent dextran (Figure 1d). Obtained 3D images were used as a mask for spatial control of laser ablation, leading to partial replication of the in vivo microvascular structures in collagen hydrogel (Figure S4, Supporting Information). We then isolated a single microvascular unit from the above imaging data, consisting of an arteriole, capillaries, and a venule defined through filament tracing in Imaris software (Bitplane). Filaments were represented as cylindrical polygons, with a diameter of 7.5 µm used for capillaries. Using this as a mask, a replica microvascular unit was ablated into collagen and perfused with fluorescent beads (Figure 1d), demonstrating patent lumens and faithful recreation of the desired geometry.
We next combined these multiphoton ablation techniques with laser‐guided angiogenesis to successfully vascularize a 3D kidney‐specific microvascular structure—the glomerulus (Figure 2a; Movie S1, Supporting Information). First we formed two parallel microvessels through established lithographic techniques and cellularized these structures with human umbilical vein endothelial cells (HUVECs).[ 7 , 14 ] We then ablated a model glomerulus between these structures, consisting of channels (diameter = 10 µm) spherically encircled by an empty cavity modeling Bowman's space. Afferent and efferent channels were ablated to connect the glomerulus to the pre‐formed parallel vessels. A pressure differential (≈1 mmHg) was applied across the glomerulus and maintained throughout culture. HUVECs were found to migrate into the lumens of the glomerulus within 24 h following ablation, and fully endothelialized the glomerular lumens by day 5–7 (Figure 2b; Figure S5, Supporting Information). These vascular structures displayed an endothelial monolayer with VE‐Cadherin junctions typical of healthy endothelium (Figure 2b). Perfusion of red blood cells demonstrated patent lumens, with single‐cell transit observed in smaller capillaries (Figure 2b; Movie S2, Supporting Information). Perfusion of FITC‐labeled 500 kDa dextran showed that the endothelialized glomerulus exhibits barrier function similar to that of the parent vessel (Figure S6, Supporting Information). Future work is necessary to fully recapitulate the selective barrier function of the human glomerulus and this may require using glomerular endothelial cells, suitable hemodynamic conditions, establishing a glomerular basement membrane, and/or incorporating podocytes.
Figure 2.
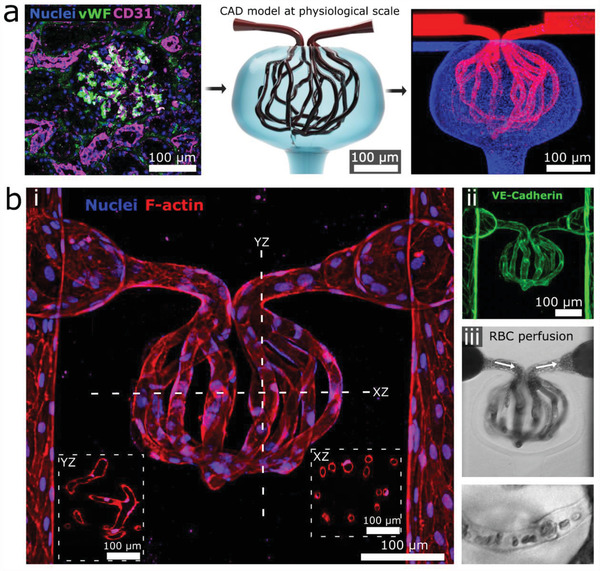
Creation of 3D cellularized microvessels. a) A human glomerulus (left) informed creation of a CAD photomask (middle) for collagen ablation. Image (right) shows ablated structure following bead perfusion of capillaries (red) and Bowman's space (blue). b) Cellularized glomerular structure. i) 3D projection of glomerulus with inset cross‐sectional views through the YZ and XZ plane, respectively; ii) VE‐cadherin staining; iii) RBC perfusion, with magnified view showing single‐cell transit (bottom).
We next demonstrate how these fabrication methods can be used to create increasingly complex vasculature to explore long‐standing vascular biology questions. This includes studies of vascular heterogeneity, the creation of hierarchical vessels, the vascularization of pre‐cellularized tissues, and the patterning of human‐derived matrix materials. Vascular heterogeneity is a key feature of human blood vessels. Even within a single vascular bed, discrete populations of cells exist in close proximity and interact while maintaining unique cellular phenotypes.[ 24 ] To demonstrate the potential of recreating and studying this phenomenon, we fabricated a six‐compartment “color wheel” structure in collagen hydrogel. Compartments were cellularized with HUVECs labeled with one of six combinations of membrane dyes (Figure S7, Supporting Information), grown for 48 h, and then connected via ablation to create one perfusable vascular structure with discrete (separately colored) segments. Cells grew into the area of ablation within 24 h and each segment remained patent without significant cell invasion into adjacent segments over the following 48 h (Figure 3a). Next, to re‐create muscularized hierarchical microvasculature, a parallel arteriole and venule were first fabricated and cultured with primary human coronary artery smooth muscle cells labeled with mCherry (mCherry‐SMCs) for three days, with media manipulated in a manner previously reported to encourage SMC quiescence.[ 25 ] Then a microvascular “grid” was ablated between the arteriole and venule and GFP‐labeled HUVECs seeded throughout all channels, forming a robust endothelium after three further days of culture. This process led to the formation of a hierarchical microvascular unit consisting of a muscularized arteriole and venule, connected by an intervening microvascular network lined only by an endothelial monolayer (Figure 3b). These structures provide a platform for future studies of microvascular tone and hierarchy.
Figure 3.
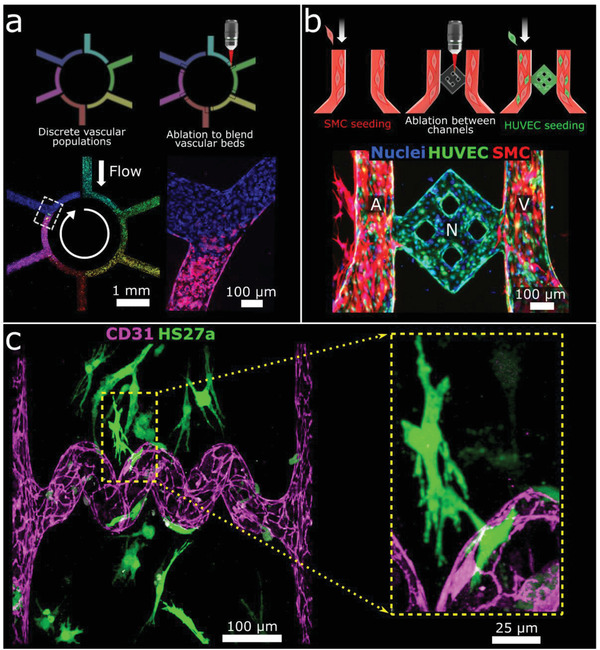
Creation of heterogeneous, hierarchical, and multicellular vasculature. a) Heterogeneous vasculature fabrication. Images 24 h post‐ablation (bottom) show a continuous vessel containing heterogeneous endothelial cell populations. b) Hierarchical vasculature creation. Image 72 h after HUVEC seeding (bottom) show a muscularized arteriole (A) and venule (V) connected by a microvascular network (N) lined by a HUVEC monolayer. c) Vascularization of cellularized tissue. Projection image following ablation of a “spiral” lumen into collagen containing HS27a‐mCherry stromal cells (shown here in green) followed by seeding of HUVECs. Inset shows magnification of stromal cell/HUVEC association.
We then assessed whether ablation is compatible with the survival of cells embedded within matrices, with a focus on the future creation of vascularized parenchymal tissues. We showed that fluorescently labeled stromal cells (HS27a‐mCherry) embedded within collagen matrices remain viable following ablation. In addition, HS27a cells near ablated channels appear to associate with newly created microvessels post‐ablation (Figure 3c). As human tissue is composed of complex heterogeneous extracellular matrices, we sought to show that native human matrices could be ablated and used to create endothelialized microvessels. Our lab has previously derived and characterized human kidney extracellular matrix (kECM) as a material for microvessel fabrication.[ 26 ] Here, we ablated a grid structure extending from cellularized parent vessels within a blended hydrogel consisting of collagen (6 mg mL−1) and kECM (1 mg mL−1). Following ablation, HUVECs migrated into ablated lumens and were observed to remodel the ablated square structures into rounded vessels. The presence of kECM within the bulk matrix was verified via staining for collagen IV (Figure S8, Supporting Information).
Discussion
Overall, we demonstrate the successful use of multiphoton ablation and laser‐guided angiogenesis to create complex 3D cellularized microvessels in biological hydrogels with the scale and geometry of human capillaries. To our knowledge, the presented structures are unique in their combination of caliber, complexity, and cellularity, and could not easily be made via other techniques. All structures took less than 90 min to ablate, with no observed adverse effects on cell viability related to the duration of manufacturing when performed in a temperature‐controlled environment. We demonstrated the ablation of several materials with relevance for the re‐creation of human microvessels. In addition to agarose–gelatin and fibrin, we have shown success using collagen and human decellularized matrices. Collagen is the most abundant protein in the human body, is cell‐remodelable, and has been shown to support cellular growth in complex tissue constructs previously.[ 7 , 27 ] Collagen at 7.5 mg mL−1 has a Young's modulus of ≈500 Pa,[ 28 ] within the range of human soft tissue/organs which have moduli from ≈100 Pa to hundreds of kPa.[ 29 ] We also provide multiple examples showing how multiphoton ablation can be used to replicate key features needed to develop microphysiologic models of increased complexity: cellularization at the capillary scale, perfusability, cellular heterogeneity, vascular hierarchy, and vascularization of pre‐cellularized tissue. Cells within our models show expected behavior including barrier function and interaction with newly created lumens.
The ability to rapidly make perfusable, cellularized microvessels with customizable geometry will have immediate utility for organ‐on‐a‐chip research and tissue engineering for regenerative medicine and disease modeling. Future work will be necessary to reach the eventual goal of patterning complex functional microvessels in dense tissue. Combining photoablation with bioprinting is an especially intriguing possibility for the rapid creation of vascularized organ tissue.
Experimental Section
Microscope System and Settings
A 2.5 W titanium‐sapphire laser (Mai Tai HP, Newport) with a pulse duration of < 100 fs and repetition rate of 80 Mhz attached to a multiphoton microscope (FV1000, Olympus) and focused through a 25×1.05 NA objective (XLPlan N, Olympus) was used for ablation experiments. Dwell time 2 µs per scan and λ = 800 nm were used for ablation, with pixel size set to 497×497 nm for most experiments. Maximum power exiting the objective was 244 mW at 800 nm.
Ablation and Perfusion
CAD models (Fusion 360, Autodesk) or in vivo volumetric imaging data were converted to 3D meshes, sliced into 1‐µm‐thick layers (Netfabb, Autodesk), and exported as vector graphics. Custom R[ 30 ] script (https://git.io/JfXFi) imported vector graphical data and ablation parameters into Fluoview software (Olympus) to drive multi‐area microscope scanning. Lithographically determined microvessels were created as previously described.[ 7 , 14 ] Collagen and kECM matrix were manufactured in house from rat tails or human kidney, respectively.[ 26 , 31 ] Bead perfusion was with 100 nm beads (FluoSpheres, ThermoFisher) and blood for perfusion was obtained from healthy volunteers. Cells were used from passages 4–6. For dextran perfusion, 1 µм) 500 kDa FITC‐dextran (Sigma) was perfused into microphysiological systems under real‐time imaging, using a perfusion pressure of 1 cm H2O. For the “color wheel,” HUVEC aliquots were stained with membrane dyes (Sigma Aldrich): green (PKH67), red (PKH26), claret (“Cellvue Claret”), or a combination.
Tissue Staining and In Vivo Experiments
Human renal cortex (LifeCenter Northwest) was stained for von Willebrand Factor and CD‐31 and imaged. Rejected donor lungs (National Disease Research Interchange) were imaged for tissue autofluorescence. Cerebral cortical microvasculature in a live 4‐month‐old C57Bl6 mouse was imaged following fluorescent dextran perfusion, via chronic cranial window.[ 32 ] Using filament tracing (Imaris, Bitplane), a microvascular unit was traced, with capillary filaments represented as cylinders (diameter = 7.5 µm). Animal studies were approved by Seattle Children's Research Institute IACUC (IACUC00396). Human blood samples were approved by the Institutional Review Board of the University of Washington (#45624‐EA).
Statistics
Laser power determinations were made on two separate occasions and averaged. All cellularized structures were made in at least triplicate, with representative images shown.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
S.G.R., C.C.H., and C.J.M. contributed equally to this work. S.R., C.M., C.H., and Y.Z. conceived the project and designed the experiments. S.R., C.H., and C.M. performed experiments and data analysis. S.S. and A.S. provided mouse brain vessel imaging data. J.H. assisted in designing the glomerulus structure. S.R., C.M., C.H., and Y.Z. wrote the manuscript. All authors edited and approved the manuscript.
Supporting information
Supporting Information
Supporting Movie 1
Supporting Movie 1
Acknowledgements
The authors acknowledge the Lynn and Mike Garvey Imaging Laboratory in the Institute of Stem Cell and Regenerative Medicine and the Washington Nanofabrication Facility. The authors thank Dr. T. Eoin West for providing human lung samples. The authors are thankful for support from the National Institutes of Health R01 HL141570, UH2/UH3 DK107343, and R01 AI141602 (to Y.Z.), UG3 TR002158 (to J.H.), F32 HL143949 (to S.R.), R21 AI130797 (to T. Eoin West for lung procurement), and R01 NS097775 and R21 NS106138 (to A.Y.S.), support from the United Therapeutics Jenesis Innovative Research Award (to S.R.), and support from the National Science Foundation Graduate Student Research Fellowship DGE 1762114 (to C.H.). The authors would like to thank Dr. Brian Hayes for his kind gift of the CMV mCherry plasmid used in this research, Dr. Torok‐Storb for providing HS27a cells, and Daniel Lih who performed the smooth muscle cell transduction with this plasmid.
Rayner S. G., Howard C. C., Mandrycky C. J., Stamenkovic S., Himmelfarb J., Shih A. Y., Zheng Y., Multiphoton‐Guided Creation of Complex Organ‐Specific Microvasculature. Adv. Healthcare Mater. 2021, 10, 2100031. 10.1002/adhm.202100031
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Bhatia S. N., Ingber D. E., Nat. Biotechnol. 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- 2. Marx V., Nature 2015, 522, 373. [DOI] [PubMed] [Google Scholar]
- 3. Lewis M. C., Macarthur B. D., Malda J., Pettet G., Please C. P., Biotechnol. Bioeng. 2005, 91, 607. [DOI] [PubMed] [Google Scholar]
- 4. Rakusan K., Flanagan M. F., Geva T., Southern J., Van Praagh R., Circulation 1992, 86, 38. [DOI] [PubMed] [Google Scholar]
- 5. Neal C. R., Arkill K. P., Bell J. S., Betteridge K. B., Bates D. O., Winlove C. P., Salmon A. H. J., Harper S. J., Am. J. Physiol. 2018, 315, F1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chrobak K. M., Potter D. R., Tien J., Microvasc. Res. 2006, 71, 185. [DOI] [PubMed] [Google Scholar]
- 7. Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz‐Santana A., Kermani P., Hempstead B., Fischbach‐Teschl C., López J. A., Stroock A. D., Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qiu Y., Ahn B., Sakurai Y., Hansen C. E., Tran R., Mimche P. N., Mannino R. G., Ciciliano J. C., Lamb T. J., Joiner C. H., Ofori‐Acquah S. F., Lam W. A., Nat. Biomed. Eng. 2018, 2, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller J. S., Stevens K. R., Yang M. T., Baker B. M., Nguyen D.‐H. T., Cohen D. M., Toro E., a. Chen A., a. Galie P., Yu X., Chaturvedi R., Bhatia S. N., Chen C. S., Nat. Mater. 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolesky D. B., Homan K. A., Skylar‐Scott M. A., Lewis J. A., Science 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee A., Hudson A. R., Shiwarski D. J., Tashman J. W., Hinton T. J., Yerneni S., Bliley J. M., Campbell P. G., Feinberg A. W., Science 2019, 365, 482. [DOI] [PubMed] [Google Scholar]
- 12. Grigoryan B., Paulsen S. J., Corbett D. C., Sazer D. W., Fortin C. L., Zaita A. J., Greenfield P. T., Calafat N. J., Gounley J. P., Ta A. H., Johansson F., Randles A., Rosenkrantz J. E., Louis‐Rosenberg J. D., Galie P. A., Stevens K. R., Miller J. S., Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ilina O., Bakker G.‐J., Vasaturo A., Hofmann R. M., Friedl P., Phys. Biol. 2011, 8, 015010. [DOI] [PubMed] [Google Scholar]
- 14. Arakawa C., Gunnarsson C., Howard C., Bernabeu M., Phong K., Yang E., DeForest C. A., Smith J. D., Zheng Y., Sci. Adv. 2020, 6, eaay7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brandenberg N., Lutolf M. P., Adv. Mater. 2016, 28, 7450. [DOI] [PubMed] [Google Scholar]
- 16. Heintz K. A., Bregenzer M. E., Mantle J. L., Lee K. H., West J. L., Slater J. H., Adv. Healthcare Mater. 2016, 5, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Applegate M. B., Coburn J., Partlow B. P., Moreau J. E., Mondia J. P., Marelli B., Kaplan D. L., Omenetto F. G., Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arakawa C. K., Badeau B. A., Zheng Y., DeForest C. A., Adv. Mater. 2017, 29, 1703156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kloxin A. M., Kasko A. M., Salinas C. N., Anseth K. S., Science 2009, 324, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeForest C. A., Anseth K. S., Nat. Chem. 2011, 3, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pradhan S., Keller K. A., Sperduto J. L., Slater J. H., Adv. Healthcare Mater. 2017, 6, 1700681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarig‐Nadir O., Livnat N., Zajdman R., Shoham S., Seliktar D., Biophys. J. 2009, 96, 4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Culver J. C., Hoffmann J. C., Poché R. A., Slater J. H., West J. L., Dickinson M. E., Adv. Mater. 2012, 24, 2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aird W. C., Circ. Res. 2007, 100, 174. [DOI] [PubMed] [Google Scholar]
- 25. Lavender M. D., Pang Z., Wallace C. S., Niklason L. E., Truskey G. A., Biomaterials 2005, 26, 4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nagao R. J., Xu J., Luo P., Xue J., Wang Y., Kotha S., Zeng W., Fu X., Himmelfarb J., Zheng Y., Tissue Eng., Part A 2016, 22, 1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rayner S. G., Phong K. T., Xue J., Lih D., Shankland S. J., Kelly E. J., Himmelfarb J., Zheng Y., Adv. Healthcare Mater. 2018, 7, 1801120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raub C. B., Putnam A. J., Tromberg B. J., George S. C., Acta Biomater. 2010, 6, 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Zheng H., Poh P. S. P., Machens H. G., Schilling A. F., Int. J. Mol. Sci. 2015, 16, 15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. R Foundation for Statistical Computing, Vienna, Austria, 2018.
- 31. Cross V. L., Zheng Y., Won Choi N., Verbridge S. S., Sutermaster B. A., Bonassar L. J., Fischbach C., Stroock A. D., Biomaterials 2010, 31, 8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holtmaat A., Bonhoeffer T., Chow D. K., Chuckowree J., De Paola V., Hofer S. B., Hübener M., Keck T., Knott G., Lee W.‐C. A., Mostany R., Mrsic‐Flogel T. D., Nedivi E., Portera‐Cailliau C., Svoboda K., Trachtenberg J. T., Wilbrecht L., Nat. Protoc. 2009, 4, 1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Movie 1
Supporting Movie 1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


