Abstract
Valuable diagnostic and prognostic biomarkers are urgently needed for colorectal cancer (CRC), which is one of the leading causes of mortality worldwide. Previous studies have reported altered expression of a mucin-like protein Fc fragment of IgG binding protein (FCGBP) in various types of cancer, but its potential diagnostic, prognostic and immunological roles in CRC remain to be determined. Therefore, the aim of current study was to investigate the potential roles of FCGBP in CRC. The present study investigated FCGBP mutations and changes in its expression levels using a combination of microarray and public dataset analyses, as well as immunohistochemistry. The results demonstrated a 10.5% mutation frequency in the FCGBP coding sequence in CRC tissues, and identified decreased FCGBP mRNA or protein expression levels in colorectal adenoma and CRC (compared with those in normal colorectal tissues from healthy control subjects), including pathologically advanced CRC (stage III+IV vs. I+II). Survival analysis using the GEPIA and Kaplan-Meier Plotter databases revealed that low FCGBP expression levels were associated with short overall, disease-free, relapse-free and event-free survival times in patients with CRC. Notably, analysis using the online Tumor IMmune Estimation Resource database revealed a positive correlation between FCGBP expression levels and the extent of infiltrating immune cells, such as B cells and dendritic cells. Consistently, the expression levels of most markers (51/57) for various types of immune cells were significantly correlated with FCGBP expression levels in CRC tissues. These findings suggested that FCGBP may serve as a diagnostic and prognostic biomarker, and that FCGBP may be associated with immune infiltration in CRC.
Keywords: colorectal cancer, Fc fragment of IgG binding protein, survival, immune cell infiltration, biomarker
Introduction
Globally, colorectal cancer (CRC) ranks third in incidence (10.0%) and second in cancer-related mortality (9.4%) among all types of cancer (1). In China, CRC is the fifth leading cause of death, with continuing increases in incidence (12.2%) and mortality (8.6%) (2). In addition, ~20% of patients with CRC present with distant metastases at the time of initial diagnosis (3). Despite notable advances in treatment strategies for CRC, such as surgery, chemotherapy and radiotherapy, >50% of patients with CRC develop metastasis, leading to a high mortality rate (4,5). Therefore, an improved understanding of the mechanisms underlying CRC pathogenesis is urgently needed to develop effective diagnostic and prognostic biomarkers and to identify potential therapeutic targets.
In our previous study, a gene expression profile microarray (GSE113513) was used to identify a panel of differentially expressed genes between 14 CRC and adjacent noncancerous colorectal tissue samples (6). Among these differentially expressed genes, the expression levels of Fc fragment of IgG binding protein (FCGBP) were lower in CRC tissue specimens compared with those in normal tissues. FCGBP, which was originally isolated from intestinal mucosa, is located on chromosome 19q13 and encodes a large mucin-like protein (>500 kDa) that binds the Fc region of IgG (7,8). FCGBP comprises numerous repeated domains, including 13 von Willebrand factor D, 12 cysteine-rich and 12 trypsin inhibitor-like domains (7–9). FCGBP is broadly expressed in various tissues, including the intestinal epithelium, gall bladder, cystic ducts, bronchi, submandibular glands and uterine cervix, as well as the fluids secreted by cells in these tissues (10).
Mutations and alternative splicing of FCGBP have been reported in hepatocholangiocarcinoma (11) and lung cancer (12). Additionally, differential expression of FCGBP has been demonstrated to occur in various types of malignancy, including gall bladder (13), prostate (14), thyroid (15), lung (12) and ovarian (16) cancer as well as head and neck squamous cell carcinoma (17). FCGBP has been reported to be involved in cancer development and progression and is associated with patient survival in prostate (18), thyroid (19) and head and neck squamous cell (17) carcinoma, gall bladder cancer (13), ovarian adenocarcinoma (16) and osteosarcoma (20).
FCGBP was initially identified as a typical component of the mucus secreted by goblet cells, and low FCGBP levels have been observed in ulcerative colitis (21,22), colorectal adenoma (23), colorectal carcinoma (24–26) and CRC metastatic tissues (27–30). Low levels of FCGBP expression have also been reported to be associated with a short survival time in patients with CRC (27,28), suggesting that FCGBP may serve an important role in the development of CRC and may be used as a diagnostic and prognostic biomarker for CRC. In addition, FCGBP is an important component of immunological mucosal defenses (7). For example, Toxoplasma Gondii infection downregulates the expression levels of a number of secretory genes including FCGBP, which is involved in the transient disruption and reorganization of splenic architecture (31). However, the association between FCGBP expression and tumor immunity in CRC has not been reported to date. A previous study has revealed that goblet cells are not only secretory cells and that they uptake luminal material and deliver it to dendritic cells (DCs) in the lamina propria, suggesting a potential role for FCGBP in the immune response (32). In addition, as components of the tumor microenvironment, immune cells have been demonstrated to act as tumor promoters as well as suppressors (33). Considering their roles during tumor progression and recurrence, including metastasis and therapy resistance, immune cells are crucial determining factors in clinical outcomes and response to immunotherapy (34–36). Therefore, the present study aimed to determine the correlation of FCGBP expression levels with the numbers of immune cells and to assess its potential role as a target for immunotherapy in CRC.
Materials and methods
Gene expression profiling interactive analysis
Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) (37) is a web-based tool that includes 9,736 tumor and 8,587 normal tissue samples from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression projects. The FCGBP expression levels in the colon adenocarcinoma (COAD) dataset were obtained using matching normal tissue data in TCGA. The data were log2(TPM+1)-transformed for differential analysis, and the log2(fold-change) was defined as median (Tumor)-median (Normal). In the COAD dataset, the association between FCGBP mRNA expression levels and overall or disease-free survival was analyzed using the Kaplan-Meier method with the log-rank test. FCGBP expression levels in various CRC stages were compared by one-way analysis of variance to calculate differential FCGBP expression (http://gepia.cancer-pku.cn/detail.php?gene=FCGBP).
cBioPortal database analysis
The online cBioPortal tool (http://www.cbioportal.org), a software for interactive exploration of multiple cancer genomic datasets, was used to analyze FCGBP mutations in CRC samples.
Oncomine database analysis
Oncomine (http://www.oncomine.org) is an online platform that incorporates 264 independent datasets for 35 types of cancer and supports various analysis methods, including molecular concept, interactome and meta-analysis (38). FCGBP mRNA expression levels were compared in multiple datasets (Table I) (39–43) using the following filters: i) Gene, FCGBP; ii) analysis type, differential analysis of cancer vs. normal tissue; and iii) cancer type, CRC. The following thresholds were used: P-value, 0.05; fold-change, 2; and gene ranking, all.
Table I.
Oncomine analysis of Fc fragment of IgG-binding protein expression in colorectal cancer in seven colorectal cancer cohorts.
| Dataset | Samples | Fold-change | P-value | t-value |
|---|---|---|---|---|
| Kaiser Colon | Colon adenocarcinoma (n=41) vs. normal (n=5) | −8.454 | 3.78×10−15 | −11.938 |
| Cecum adenocarcinoma (n=17) vs. normal (n=5) | −11.05 | 7.26×10−9 | −9.516 | |
| Rectosigmoid adenocarcinoma (n=10) vs. normal (n=5) | −6.964 | 1.28×10−4 | −5.559 | |
| Colon mucinous adenocarcinoma (n=13) vs. normal (n=5) | −2.574 | 0.001 | −3.604 | |
| Rectal adenocarcinoma (n=8) vs. normal (n=5) | −5.854 | 0.006 | −3.346 | |
| Rectal mucinous adenocarcinoma (n=4) vs. normal (n=5) | −9.213 | 0.013 | −3.979 | |
| TCGA colorectal | Rectal adenocarcinoma (n=60) vs. normal (n=22) | −4.576 | 2.45×10−27 | −19.081 |
| Colon adenocarcinoma (n=101) vs. normal (n=22) | −4.157 | 9.00×10−25 | −20.029 | |
| Cecum adenocarcinoma (n=22) vs. normal (n=22) | −4.67 | 1.18×10−15 | −13.628 | |
| Colon mucinous adenocarcinoma (n=22) vs. normal (n=22) | −2.849 | 1.63×10−9 | −8.163 | |
| Rectosigmoid adenocarcinoma (n=3) vs. normal (n=22) | −6.682 | 2.92×10−4 | −14.447 | |
| Rectal mucinous adenocarcinoma (n=6) vs. normal (n=22) | −3.279 | 0.008 | −3.520 | |
| Skrzypczak colorectal 2 | Colon adenoma epithelia (5) vs. normal (10) | −11.298 | 2.57×10−9 | −13.767 |
| Colon carcinoma epithelia (n=5) vs. normal (n=10) | −42.143 | 1.43×10−9 | −17.128 | |
| Colon adenoma (n=5) vs. normal (n=10) | −13.483 | 2.05×10−7 | −9.422 | |
| Colon carcinoma (n=5) vs. normal (n=10) | −52.679 | 9.50×10−6 | −9.689 | |
| Notterman colon | Colon adenocarcinoma (n=18) vs. normal (n=18) | −4.521 | 2.68×10−5 | −4.631 |
| Hong colorectal | Colorectal carcinoma (n=70) vs. normal (n=12) | −17.174 | 1.54×10−21 | −12.929 |
| Skrzypczak colorectal | Colorectal carcinoma (n=36) vs. normal (n=24) | −10.911 | 7.31×10−10 | −7.693 |
| Gaedcke colorectal | Rectal adenocarcinoma (n=65) vs. normal (n=65) | −11.899 | 1.00×10−14 | −9.755 |
TCGA, The Cancer Genome Atlas.
R2 database analysis
R2 (http://r2.amc.nl), a web-based genomics analysis and visualization tool that contains gene microarray and RNA-Seq data (44), was used to analyze the association between FCGBP mRNA expression levels and disease-free, relapse-free and event-free survival. The following datasets were used: Tumor Colon - Smith - 232 MAS5.0 - u133p2, Tumor Colon - Sieber - 290 MAS5.0 - u133p2, Tumor Colon - SieberSmith - 355 MAS5.0 - u133p2, Tumor Colon MSI - status (Core-Transcript) - Sveen - 95 rma sketch - huex10t, Tumor Colon CIT (Combat) - Marisa - 566rma - u133p2 and Tumor Colon MVRM - SieberSmith - 345 fRMA(bc) - u133p2.
Tumor IMmune Estimation Resource (TIMER) database analysis
As a comprehensive resource for systematic analysis of immune infiltrates in cancer, TIMER (https://cistrome.shinyapps.io/timer/) (45) includes 10,897 samples from 32 types of cancer in TCGA. Using the COAD dataset, the present study utilized TIMER to analyze the correlation of FCGBP expression levels with the extent of immune infiltrates including B cells, CD4+ and CD8+ T cells, neutrophils, macrophages and DCs, via gene modules. FCGBP expression levels based on tumor purity were also analyzed (46,47). The correlation module generated expression scatter plots between pairs of user-defined genes in CRC, performed Spearman's correlation analysis and estimated the statistical significance. Gene expression levels were displayed as log2[transcripts per million (TPM)] values. Correlations were analyzed between FCGBP expression and markers of tumor-infiltrating immune cells, including those for CD8+ T cells, T cells (general), B cells, monocytes, tumor-associated macrophages (TAMs), M1 and M2 macrophages, neutrophils, natural killer cells, DCs, T-helper 1 (Th1), T-helper 2 (Th2), follicular helper T (Tfh), T-helper 17 (Th17) cells, regulatory T cells (Tregs) and exhausted T cells (48–50).
Immunohistochemistry and image analysis
The tissue microarrays (TMAs) used in the present study (cat no. HColAde080CD01, containing 69 pairs of CRC tissues and adjacent noncancerous colorectal tissues; and HColA180Su15, containing four normal colorectal tissue, seven colorectal adenoma, seven primary and 10 metastatic CRC tissue samples) were purchased from Shanghai Outdo Biotech Co., Ltd. For immunohistochemical staining, the slides were deparaffinized in xylene and rehydrated through graded ethanol series (100, 95, 90, 80 and 70%). Antigen retrieval was performed with 0.1% sodium citrate buffer (pH 6.0) for 20 min at 95°C. Endogenous peroxidase activity was quenched with 3% H2O2 in water, and nonspecific binding was blocked with PBS containing 1% bovine serum albumin (Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at room temperature. Subsequently, the slides were sequentially incubated with an anti-FCGBP antibody (1:200; cat. no. ab217146; Abcam) at 4°C overnight, a ready-to-use biotinylated secondary antibody from the UltraSensitive S-P kit (Fuzhou Maixin Biotech Co., Ltd.) for 10 min at room temperature, and a ready-to-use horseradish peroxidase-conjugated streptavidin antibody from the UltraSensitive S-P kit for 10 min at room temperature. The slides were then incubated with the chromogen diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.) for 2 min at room temperature and counterstained with diluted Harris hematoxylin (Fuzhou Maixin Biotech Co., Ltd.) for 1 min. Images were captured using a Nano Zoomer 2.0 HT slide scanner (Hamamatsu Photonics K.K.) and processed using the Nano Zoomer Digital Pathology View 1.6 software (Hamamatsu Photonics K.K.). Immunohistochemistry scores in various tissue types were independently determined by two experienced pathologists blinded to the clinical and pathological data. Staining intensity and percentage of positively stained cells were determined as previously described (6). Scores of 4–12 and 0–3 were considered to indicate high and low FCGBP protein expression levels, respectively. All experiments were approved by the Ethics Committee of Fujian University of Traditional Chinese Medicine (Fujian, China).
Statistical analysis
Data are presented as the mean ± standard deviation. FCGBP expression is presented as log2(TPM+1)-transformed expression data in the plots. Statistical analyses were performed using SPSS 22.0 software (IBM Corp.). One-way ANOVA was performed to analyze FCGBP expression among pathological stages. Differences between paired CRC and adjacent or distant normal colorectal tissues were assessed using the Wilcoxon paired sample test. Differences between independent samples from two groups were assessed using the Mann-Whitney U test. Differences among three or more groups were assessed using Kruskal-Wallis test with the Mann-Whitney test and the Bonferroni correction for pairwise comparisons. Differences in FCGBP mRNA expression levels between CRC and unpaired normal colorectal tissues in the Oncomine database were analyzed by the unpaired Student's t-test. The associations of FCGBP expression levels between patient groups were evaluated by the Fisher's exact test. Survival rates were determined using the Kaplan-Meier method and compared by the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation and mutations of FCGBP in CRC
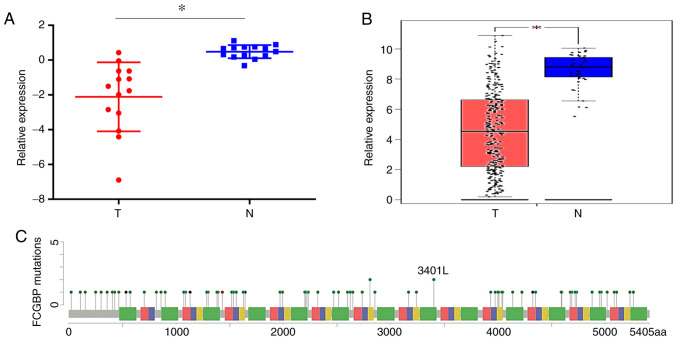
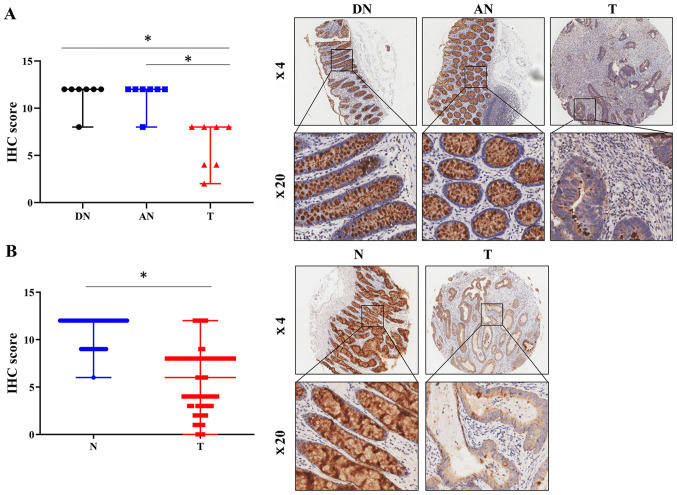
To determine the differences in FCGBP expression between CRC and adjacent noncancerous tissues, FCGBP mRNA expression levels were compared in 14 matched pairs of primary CRC and noncancerous tissues using a cDNA array from our laboratory (GSE113513) (6). The results demonstrated a significant decrease in the FCGBP expression levels in the CRC tissues compared with those in the adjacent noncancerous colorectal tissues (P<0.05; Fig. 1A). The analysis of publicly available tumor expression data in GEPIA (P<0.05; Fig. 1B) and Oncomine (Table I) also revealed that the FCGBP mRNA levels were lower in the CRC tissues compared with those in the noncancerous tissues. The analysis of FCGBP mutations using cBioPortal revealed a somatic mutation frequency of 10.5% in the FCGBP DNA sequence (Fig. 1C). Furthermore, immunohistochemical analysis of a TMA demonstrated a significant decrease in the protein expression levels of FCGBP in the CRC tissues compared with the paired distal and adjacent noncancerous colorectal tissues (n=7 per group; P<0.05; Fig. 2A), which was validated in another TMA comprising 69 pairs of CRC and adjacent noncancerous tissue samples (P<0.05; Fig. 2B). These results demonstrated a significant downregulation of FCGBP mRNA and protein levels in CRC tissues compared with those in noncancerous tissues.
Figure 1.
FCGBP mRNA levels are downregulated in CRC tissues. (A) FCGBP mRNA expression levels in 14 matched CRC and noncancerous colorectal tissue samples from our previously published gene expression microarray profile (GSE113513). (B) FCGBP expression in 275 CRC tissues and 41 noncancerous colorectal tissues from the COAD dataset analyzed by Gene Expression Profiling Interactive Analysis. Each datapoint represents one tissue sample; the error bars represent the mean ± standard deviation. *P<0.05. (C) Schematic representation of gene mutation sites in the coding sequence of FCGBP on the coding strand. Green dots indicate missense mutations, black dots indicate truncation sites, and brown dots indicate in-frame mutations. CRC, colorectal cancer; FCGBP, Fc fragment of IgG-binding protein; T, tumor; N, normal.
Figure 2.
Protein expression levels of FCGBP are downregulated in CRC tissues. (A and B) The protein levels of FCGBP were determined by IHC using two commercially available tissue microarrays in (A) seven pairs of CRC tissues matched with distal and adjacent noncancerous colorectal tissues and (B) 69 pairs of CRC and adjacent noncancerous colorectal tissues. Representative images captured at ×4 and ×20 magnification are presented in the lower right panels. Each datapoint in the plot represents one tissue sample; the error bars represent the median ± range. *P<0.05. IHC, immunohistochemistry; T, colorectal cancer tissue; N, noncancerous colorectal tissue; DN, distal noncancerous colorectal tissue; AN, adjacent noncancerous colorectal tissue; CRC, colorectal cancer; FCGBP, Fc fragment of IgG-binding protein.
Protein expression levels of FCGBP are decreased in colorectal adenoma and carcinoma tissues
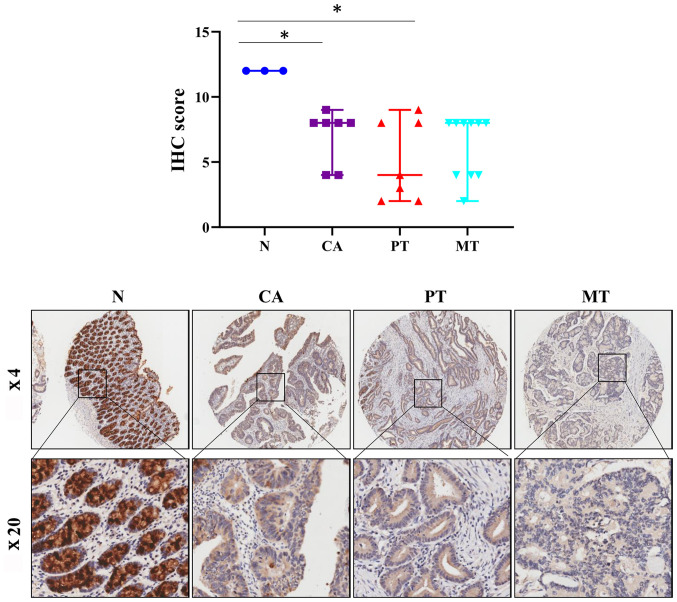
CRC arises in preexisting adenomas, and death frequently occurs due to metastasis in patients with CRC (51). Therefore, the present study next determined the protein levels of FCGBP in colorectal adenoma and primary and metastatic CRC tissues by immunohistochemistry-based TMA analysis. Notably, the protein expression levels of FCGBP were significantly decreased in the colorectal adenoma tissues compared with those in the normal colorectal tissues from healthy controls (P<0.05; Fig. 3) and were comparable to those observed in the primary and metastatic CRC tissues. These results suggested that the decrease in the protein levels of FCGBP may serve a crucial role in malignant transformation during CRC development.
Figure 3.
Protein expression levels of FCGBP are downregulated in colorectal adenoma tissues. FCGBP protein expression levels were determined by IHC using a commercially available tissue microarray. Representative images captured at ×4 and ×20 magnification are presented in the lower panel. Each datapoint in the plot represents one tissue sample; the error bars represent the median ± range. *P<0.05. IHC, immunohistochemistry; N, noncancerous colorectal tissue; CA, colorectal adenoma tissue; PT, primary CRC tissue; MT, metastatic CRC tissue; CRC, colorectal cancer; FCGBP, Fc fragment of IgG-binding protein.
Downregulation of FCGBP expression in advanced-stage CRC tissues
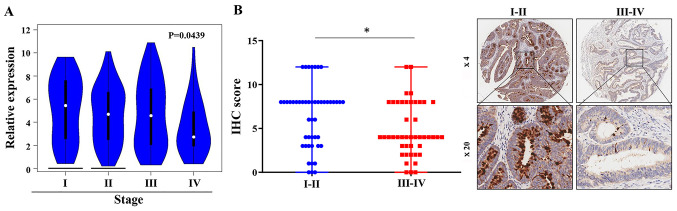
To further assess FCGBP expression in CRC development, FCGBP mRNA expression levels were compared among CRC tissues from various pathological stages. The analysis of the COAD dataset in GEPIA revealed a gradual decrease in FCGBP mRNA expression levels in CRC tissues with advancing pathological stage (P<0.05; Fig. 4;). The analysis of the associations of patient clinicopathological characteristics and the protein levels of FCGBP in CRC TMAs demonstrated that the protein levels of FCGBP were lower in advanced-stage CRC tissues compared with those in early-stage CRC tissues (III+IV vs. I+II, P<0.05; Table II; Fig. 4B); however, a similar association was not observed with other clinicopathological parameters (Table II). Collectively, these results suggested that downregulation of FCGBP expression levels may be associated with advanced-stage CRC.
Figure 4.
Protein expression levels of FCGBP are downregulated in advanced-stage CRC tissues. (A) FCGBP mRNA expression in CRC tissues from different pathological stages in the COAD dataset was analyzed by Gene Expression Profiling Interactive Analysis. (B) The FCGBP protein expression in the different pathological stages of CRC tissues were analyzed in a commercially available TMA containing 93 CRC tissues. The representative images were taken at a magnification ×4 and ×20 and showed in the left panel. Each dot represents 1 tissue; median ± range; *P<0.05, vs. I + II stages. COAD, colon adenocarcinoma; CRC, colorectal cancer; FCGBP, Fc fragment of IgG-binding protein.
Table II.
Associations between FCGBP expression levels and the clinicopathological characteristics of patients with colon cancer.
| FCGBP expression | ||||
|---|---|---|---|---|
| Characteristics | n | Low, n | High, n | Fisher's P-value |
| Total | 93 | 46 | 47 | |
| Sex | ||||
| Male | 51 | 25 | 26 | >0.999 |
| Female | 42 | 21 | 21 | |
| Age, years | ||||
| <65 | 36 | 19 | 17 | 0.673 |
| ≥65 | 57 | 27 | 30 | |
| Clinical stage | ||||
| I+II | 48 | 17 | 31 | 0.007a |
| III+IV | 45 | 29 | 16 | |
| T stage | ||||
| T1+T2 | 7 | 2 | 5 | 0.435 |
| T3+T4 | 86 | 44 | 42 | |
| N stage | ||||
| N0 | 57 | 28 | 29 | >0.999 |
| N1+N2 | 36 | 18 | 18 | |
| M stage | ||||
| M0 | 90 | 44 | 46 | 0.617 |
| M1 | 3 | 2 | 1 | |
| Survival status | ||||
| Alive | 41 | 23 | 18 | 0.300 |
| Dead | 52 | 23 | 29 | |
| Tumor size, cm | ||||
| <5 | 39 | 17 | 22 | 0.403 |
| ≥5 | 54 | 29 | 25 | |
P<0.05. FCGBP, Fc fragment of IgG-binding protein; T, tumor; N, node; M, metastasis.
Downregulation of FCGBP predicts a shorter survival time in patients with CRC
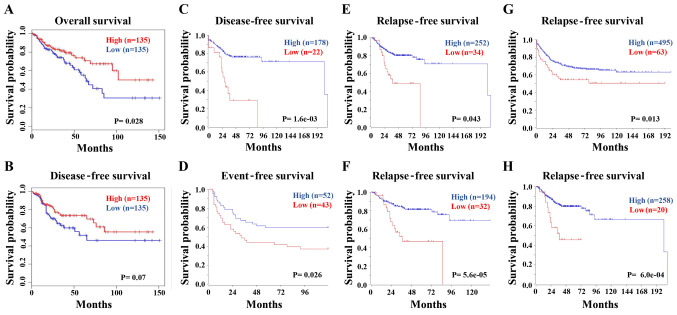
The association between FCGBP expression levels and the survival times of patients with CRC was next analyzed in the COAD dataset using GEPIA. The Kaplan-Meier analysis demonstrated that low FCGBP mRNA levels were associated with a short overall survival (Fig. 5A), but not disease-free survival (P=0.07; Fig. 5B) in patients with CRC. Consistently, the survival analysis from multiple datasets using R2 confirmed the association between low FCGBP levels and short disease-free (Fig. 5C), event-free (Fig. 5D) and relapse-free (Fig. 5E-H) survival times in patients with CRC. These data suggested that low FCGBP mRNA expression levels may serve as an indicator for the prognosis of CRC.
Figure 5.
Low FCGBP expression levels predict a short survival time in patients with CRC. (A and B) The Cancer Genome Atlas colon adenocarcinoma dataset was analyzed by Gene Expression Profiling Interactive Analysis to determine the associations between FCGBP expression levels and (A) overall and (B) disease-free survival using Kaplan-Meier survival curves. (C-E) The R2 database was used to analyze the associations between FCGBP expression levels and (C) disease-free, (D) event-free and (E-H) recurrence-free survival in the (E) Tumor Colon - SieberSmith - 355 MAS5.0 - u133p2, (F) Tumor Colon - Sieber - 290 MAS5.0 - u133p2, (G) Tumor Colon CIT (Combat) - Marisa - 566 rma - u133p2 and (H) Tumor Colon MVRM - SieberSmith - 345 fRMA(bc) - u133p2 datasets of patients with CRC using Kaplan-Meier survival curves. CRC, colorectal cancer; FCGBP, Fc fragment of IgG-binding protein.
Correlation between FCGBP expression levels and markers of immune infiltration in CRC
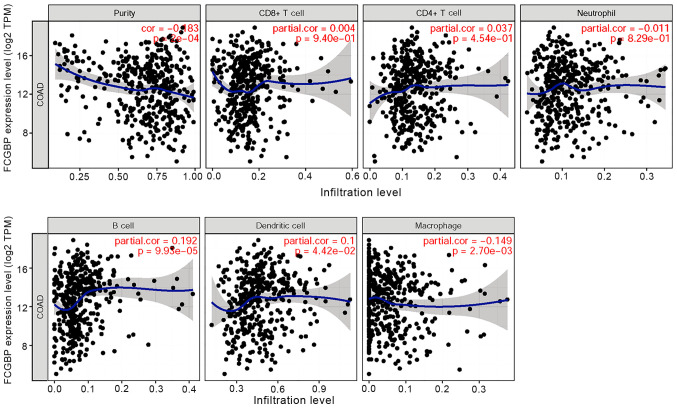
Considering the roles of tumor-infiltrating lymphocytes in CRC development (52), the present study investigated the relationship between FCGBP and immune infiltration in CRC. Correlation analysis using TIMER revealed that FCGBP expression levels were negatively correlated with tumor purity (r=−0.183; P<0.05) and macrophages (r=−0.149; P<0.05) and partially correlated with the levels of infiltrating B cells (r=0.192; P<0.05) and DCs (r=0.100; P<0.05); however, FCGBP expression was not correlated with the levels of infiltrating CD4+ and CD8+ T cells or neutrophils in CRC (all P>0.05; Fig. 6). These results suggested that FCGBP may be involved in the regulation of B cells and DCs, but not T cells, in CRC.
Figure 6.
Correlation between FCGBP expression levels and the extent of immune cell infiltration. The COAD dataset was analyzed by the Tumor IMmune Estimation Resource to determine the correlations between FCGBP expression levels and the extent of immune cell infiltration, including tumor purity, B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells. COAD, colon adenocarcinoma; FCGBP, Fc fragment of IgG-binding protein; cor, correlation; TPM, transcripts per million.
Correlation between FCGBP expression and immune cell markers in CRC
The present study further explored the potential relationship between FCGBP expression and infiltrating immune cells in the COAD dataset. Specific immune cell markers for CD8+ T and all T cells, B cells, TAMs, M1 and M2 macrophages, neutrophils, natural killer cells, DCs, Th1, Th2, Tfh and Th17 cells, Tregs and exhausted T cells in CRC were analyzed using TIMER. As presented in Table III, following adjustment for tumor purity, FCGBP expression levels were significantly correlated with 51 of the 57 immune cell markers in CRC. Notably, FCGBP expression levels were correlated with multiple markers of specific functional T cells, including the CD8+ T cell marker CD8A, all markers of all T cells (CD3D, CD3E and CD2), all markers of Th1 (TBX21, STAT4, STAT1, IFNG and TNF), a number of markers of Th2 (GATA3, STAT6, STAT6 and STAT5A), all markers of Tfh (BCL6 and IL21), Th17 marker STAT3, all markers of Tregs (FOXP3, CCR8, STAT5B and TGFB1) and all markers of exhausted T cells (PDCD1, CTLA4, LAG3, HAVCR2 and GZMB) (all P<0.05). In addition, significant correlation was identified between FCGBP expression levels and a number of cell type-specific markers, including those for TAMs (CD68 and IL10), M1 (NOS2, IRF5 and PTGS2) and M2 (CD163 and VSIG4) macrophages, suggesting that FCGBP may regulate macrophage polarization in CRC.
Table III.
Correlation analysis between FCGBP expression and associated immune cell markers using TIMER.
| No adjustment | Adjusted by tumor purity | |||
|---|---|---|---|---|
| Variables | r-value | P-value | r-value | P-value |
| CD8+ T cell | ||||
| CD8A | −0.021 | 0.492 | −0.114 | 3.25×10−4a |
| CD8B | 0.035 | 0.248 | −0.046 | 0.146 |
| T cell (general) | ||||
| CD3D | 0.008 | 0.786 | −0.098 | 0.002a |
| CD3E | 0.001 | 0.966 | −0.11 | 5.04×10−4a |
| CD2 | −0.015 | 0.615 | −0.122 | 1.10×10−4a |
| B cell | ||||
| CD19 | 0.007 | 0.007 | −0.085 | 0.007a |
| CD79A | −0.02 | −0.02 | −0.132 | 2.98×10−5a |
| CD86 | 0.163 | 0.163 | 0.113 | 3.42×10−4a |
| CSF1R | 0.484 | 8.55×10−66 | 0.462 | 1.06×10−53a |
| Tumor-associated macrophage | ||||
| CCL2 | 0.045 | 0.134 | −0.027 | 0.394 |
| CD68 | 0.117 | 9.67×10−5 | 0.072 | 0.023a |
| IL10 | −0.005 | 0.881 | −0.068 | 0.033a |
| M1 macrophage | ||||
| NOS2 | 0.121 | 5.48×10−5 | 0.113 | 3.40×10−4a |
| IRF5 | 0.11 | 2.45×10−4 | 0.076 | 0.017a |
| PTGS2 | 0.141 | 2.72×10−6 | 0.077 | 0.015a |
| M2 macrophage | ||||
| CD163 | 0.121 | 5.58×10−5 | 0.069 | 0.030a |
| VSIG4 | 0.3 | 2.35×10−24 | 0.266 | 1.48×10−17a |
| MS4A4A | 0.024 | 0.421 | −0.044 | 0.162 |
| Neutrophil | ||||
| CEACAM8 | 0.045 | 0.136 | 0.034 | 0.285 |
| ITGAM | 0.309 | 9.46×10−26 | 0.267 | 1.04×10−17a |
| CCR7 | −0.004 | 0.885 | −0.114 | 3.03×10−4a |
| Natural killer cell | ||||
| KIR2DL1 | −0.019 | 0.552 | −0.078 | 0.014a |
| KIR2DL3 | −0.085 | 0.005 | −0.133 | 2.78×10−5a |
| KIR2DL4 | −0.137 | 5.52×10−6 | −0.199 | 2.71×10−10a |
| KIR3DL1 | −0.017 | 0.563 | −0.068 | 0.031a |
| KIR3DL2 | −0.036 | 0.227 | −0.113 | 3.65×10−4a |
| KIR3DL3 | −0.022 | 0.459 | −0.068 | 0.033a |
| KIR2DS4 | −0.053 | 0.082 | −0.086 | 0.006a |
| Dendritic cell | ||||
| HLA-DPB1 | 0.294 | 1.42×10−23 | 0.247 | 2.82×10−15a |
| HLA-DQB1 | 0.207 | 5.02×10−12 | 0.136 | 1.79×10−5a |
| HLA-DRA | 0.214 | 7.76×10−13 | 0.154 | 1.11×10−6a |
| HLA-DPA1 | 0.243 | 4.18×10−16 | 0.187 | 2.02×10−9a |
| CD1C | 0.328 | 4.84×10−29 | 0.289 | 1.49×10−20a |
| NRP1 | 0.185 | 5.86×10−10 | 0.128 | 5.19×10−5a |
| ITGAX | 0.224 | 4.97×10−14 | 0.176 | 2.49×10−8a |
| T-helper 1 cell | ||||
| TBX21 | −0.001 | 0.965 | −0.102 | 0.001a |
| STAT4 | 0.031 | 0.304 | −0.067 | 0.035a |
| STAT1 | −0.122 | 4.87×10−5 | −0.172 | 5.24×10−8a |
| IFNG | −0.086 | 0.004 | −0.151 | 1.66×10−6a |
| TNF | 0.191 | 1.68×10−10 | 0.158 | 5.63×10−7a |
| T-helper 2 cell | ||||
| GATA3 | 0.024 | 0.423 | 0.069 | 0.030a |
| STAT6 | 0.251 | 3.17×10−17 | 0.218 | 3.33×10−12a |
| STAT5A | 0.303 | 9.40×10−25 | 0.258 | 1.40×10−16a |
| IL-13 | −0.015 | 0.624 | −0.032 | 0.312 |
| Follicular helper T cell | ||||
| BCL6 | 0.294 | 2.45×10−23 | 0.283 | 1.07×10−19a |
| IL21 | −0.054 | 0.073 | −0.117 | 2.20×10−4a |
| T-helper 17 cell | ||||
| STAT3 | 0.289 | 1.42×10−22 | 0.26 | 9.10×10−17a |
| IL17A | −0.002 | 0.937 | −0.044 | 0.168 |
| Regulatory T cell | ||||
| FOXP3 | −0.059 | 0.051 | −0.141 | 7.57×10−6a |
| CCR8 | −0.09 | 0.003 | −0.142 | 7.04×10−6a |
| STAT5B | 0.239 | 1.04×10−14 | 0.215 | 6.91×10−12a |
| TGFB1 | 0.288 | 1.64×10−22 | 0.239 | 2.05×10−14a |
| T cell exhaustion | ||||
| PDCD1 | −0.016 | 0.590 | −0.112 | 3.98×10−4a |
| CTLA4 | −0.077 | 0.011 | −0.161 | 3.17×10−7a |
| LAG3 | −0.103 | 6.32×10−4 | −0.151 | 1.61×10−6a |
| HAVCR2 | 0.189 | 2.61×10−10 | 0.147 | 3.35×10−6a |
| GZMB | −0.122 | 4.87×10−5 | −0.222 | 1.54×10−12a |
P<0.05.
Consistent with the observed correlation between FCGBP expression levels and the extent of DC infiltration in the COAD dataset, all DC markers, including HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, CD1C, NRP1 and ITGAX, were also significantly correlated with FCGBP expression levels. These results further confirmed the association between FCGBP and DC infiltration. Similarly, the correlation between FCGBP levels and B cells in CRC was also observed based on B cell markers, including CD19, CD79A, CD86 and CSF1R. In addition, correlations were identified between FCGBP levels and markers of neutrophils (ITGAM and CCR7) or natural killer cells (KIR2DL1, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2, KIR3DL3 and KIR2DS4). Therefore, these results demonstrated that FCGBP was significantly correlated with infiltrating immune cells in CRC, suggesting a potential role for FCGBP in immune infiltration in the CRC microenvironment.
Discussion
Studies of the underlying mechanisms of CRC pathogenesis are urgently needed to provide potential diagnostic and prognostic biomarkers, as well as therapeutic targets for CRC. The present study identified that CRC tissues exhibited low expression levels of FCGBP, which has been reported in various types of cancer, but its potential diagnostic, prognostic and immunological roles in CRC remain to be further explored. Therefore, the aim of current was to determine the potential roles of FCGBP in CRC. The current study provided evidence supporting the clinical relevance of FCGBP in CRC. Analytical approaches based on microarrays, online databases and TMA immunohistochemistry revealed a significant decrease in FCGBP expression at the mRNA and protein levels in CRC tissues compared with those in noncancerous colorectal tissues, with 10.5% mutation frequency in the FCGBP coding sequence in CRC tissues. Furthermore, the protein expression levels of FCGBP were significantly decreased in colorectal adenoma and primary CRC tissues compared with those in normal colon tissues. In addition, low FCGBP expression levels, which were observed in advanced-stage CRC tissues, were significantly associated with a short survival time in patients with CRC. Notably, the results of the present study revealed that FCGBP expression levels were positively correlated with the levels of infiltrating B cells, macrophages and DCs, and negatively correlated with tumor purity in COAD. These results provided insights into the potential role of FCGBP in tumor immunology and suggested its potential use as a CRC biomarker as well as a therapeutic target.
Due to the relatively high percentage of patients with CRC and distant metastasis at the time of initial diagnosis (3), identification of novel and effective biomarkers for early diagnosis may contribute to the improvement of early screening for CRC. Omics technologies provide powerful methods to identify potential biomarkers for early diagnosis (53). Using microarray and online database analyses, the present study identified mutations and lower expression levels of FCGBP in CRC tissues compared with those in noncancerous colorectal tissues, which was consistent with previous studies on various malignancies, including CRC (12–18,20,24–26). However, other data analysis approaches, including analysis of the original raw data using R or Python software, may provide additional information and will be used in our future studies.
Using immunohistochemistry-based TMA analysis, the present study validated the decreased protein levels of FCGBP in CRC tissues compared with those in proximal or distal adjacent noncancerous colorectal tissues and normal colorectal tissues. Further analysis of the protein levels of FCGBP among normal colorectal, adenomatous and carcinomatous tissues confirmed the findings of a previous study in mice (23) by demonstrating that the levels of FCGBP were decreased in colorectal adenoma and CRC compared with those in normal colon tissues. These results suggested that the reduction in FCGBP levels may be a common event during CRC development, and that FCGBP may be considered as a potential biomarker for the early diagnosis of CRC. However, in the current study, no differences were identified in the protein expression of FCGBP between colorectal adenoma and CRC samples, which may be due to the limitations of IHC or the limited number of samples. Our future study will collect a larger number of samples to detect FCGBP expression levels.
The present study further assessed the associations between the protein levels of FCGBP and the clinicopathological characteristics of patients with CRC. Notably, the protein levels of FCGBP were lower in advanced-stage CRC samples compared with those in early-stage CRC, which was consistent with the results of the GEPIA analysis of FCGBP mRNA levels. Pairwise comparisons among different stages were not available on the GEPIA website, which was a limitation of the current study. However, these data indicated an association between low FCGBP expression levels and an advanced stage in CRC. Consistent with previous analyses of online databases (27,28), the results of the present study demonstrated that low FCGBP expression levels in CRC tissues were significantly associated with a short patient survival time, highlighting the potential role of FCGBP as a prognostic biomarker for CRC. However, due to the lack of survival information of patients with CRC included with the TMAs, the association between the protein expression levels of FCGBP and patient survival could not be validated in the present study. Patient samples and survival information will be collected to verify the results of the database analyses in our future studies. In addition, the aforementioned studies (27,28) and the current study have suggested that the decrease of FCGBP expression levels in tumors function as a promoting factor rather than being a concomitant phenomenon. Therefore, the unexplored biological function and underlying mechanism of the effects of FCGBP in CRC is a limitation of the current study, which should be further addressed in future studies.
FCGBP is secreted by various cell types, including intestinal goblet cells, and is considered to be an important component of mucosal immunological defenses (7). To analyze the roles of FCGBP in colorectal tumor immunology, the present study analyzed the correlation of FCGBP expression levels with the relative abundance of infiltrating immune cells, including B cells, CD4+ and CD8+ T cells, neutrophils, macrophages and DCs, via gene modules. The results of the systematic analysis of immune infiltrates in CRC using the TIMER database demonstrated that FCGBP expression levels were positively correlated with the infiltration levels of B cells, macrophages and DCs and negatively correlated with tumor purity; however, FCGBP expression levels were not correlated with CD4+ and CD8+ T cells or neutrophils in COAD. Collectively, these results suggested that FCGBP may serve an essential role in the regulation of infiltrating immune cells in COAD. Consistently, the results of the present study identified a significant correlation between FCGBP expression levels and multiple immune markers expressed in all tested infiltrating immune cell types in CRC. These results suggested a crucial role of FCGBP in immune cell infiltration into the CRC microenvironment. Although no significant correlations were identified between FCGBP levels and the relative abundance of CD4+ and CD8+ T cells or neutrophils in COAD, the results revealed correlations between FCBGP expression levels and the makers expressed by CD8+ T cells and neutrophils. Therefore, the correlations of FCGBP expression levels with immune cell infiltration levels and the expression of immune markers should be further addressed, and the biological function of FCGBP in the regulation of immune cell infiltration warrants further investigation.
In summary, in the present study, microarray and online database analyses were used to demonstrate that FCGBP expression levels were downregulated in colorectal adenoma and CRC, including advanced-stage CRC, compared with those in normal colorectal tissues from healthy subjects or paired adjacent noncancerous colorectal tissues, and that CRC tissue specimens harbored FCGBP mutations. In addition, low FCGBP expression levels were associated with a short survival time in patients with CRC. FCGBP expression levels were positively correlated with various tumor-infiltrating immune cells and immune markers, suggesting that FCGBP may be involved in the regulation of immune cell infiltration into the CRC microenvironment. Overall, these results suggested that FCGBP may be a potential biomarker for the early diagnosis and prognosis of CRC. The precise role of FCGBP in immune cell infiltration requires further study.
Acknowledgements
Not applicable.
Funding Statement
This study was supported by the National Natural Science Foundation of China (grant nos. 81673721 and 81803882) and the International Cooperative Project of Fujian Department of Science and Technology (grant no. 2017I0007).
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 81673721 and 81803882) and the International Cooperative Project of Fujian Department of Science and Technology (grant no. 2017I0007).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JP and QC conceived and designed the experiments. QZ, AS, MW, YC and LL conducted the bioinformatics analyses. AS, LL, XC, QZ and XL conducted the validation experiments. ZS, XW, JL and WL conducted data analysis. YH, HL and QZ conducted immunohistochemistry analysis and produced the figures. LL and MW confirm the authenticity of all the raw data. AS, QZ, QC and JP wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experiments were approved by the Ethics Committee of Fujian University of Traditional Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Li T. Development of a 15-gene signature for predicting prognosis in advanced colorectal cancer. Bioengineered. 2020;11:165–174. doi: 10.1080/21655979.2020.1718459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poturnajova M, Furielova T, Balintova S, Schmidtova S, Kucerova L, Matuskova M. Molecular features and gene expression signature of metastatic colorectal cancer (Review) Oncol Rep. 2021;45:10. doi: 10.3892/or.2021.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siraj S, Masoodi T, Siraj AK, Azam S, Qadri Z, Ahmed SO, AlBalawy WN, Al-Obaisi KA, Parvathareddy SK, AlManea HM, et al. Clonal evolution and timing of metastatic colorectal cancer. Cancers (Basel) 2020;12:2938. doi: 10.3390/cancers12102938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen A, Chen Y, Liu L, Huang Y, Chen H, Qi F, Lin J, Shen Z, Wu X, Wu M, et al. EBF1-mediated upregulation of ribosome assembly factor PNO1 contributes to cancer progression by negatively regulating the p53 signaling pathway. Cancer Res. 2019;79:2257–2270. doi: 10.1158/0008-5472.CAN-18-3238. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi K, Ogata H, Morikawa M, Iijima S, Harada N, Yoshida T, Brown WR, Inoue N, Hamada Y, Ishii H, et al. Distribution and partial characterisation of IgG Fc binding protein in various mucin producing cells and body fluids. Gut. 2002;51:169–176. doi: 10.1136/gut.51.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamp LA, Braxton DR, Wu J, Akopian V, Hasegawa K, Chandrasoma PT, Hawes SM, McLean C, Petrovic LM, Wang K, et al. The GCTM-5 epitope associated with the mucin-like glycoprotein FCGBP marks progenitor cells in tissues of endodermal origin. Stem Cells. 2012;30:1999–2009. doi: 10.1002/stem.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada N, Iijima S, Kobayashi K, Yoshida T, Brown WR, Hibi T, Oshima A, Morikawa M. Human IgGFc binding protein (FcgammaBP) in colonic epithelial cells exhibits mucin-like structure. J Biol Chem. 1997;272:15232–15241. doi: 10.1074/jbc.272.24.15232. [DOI] [PubMed] [Google Scholar]
- 10.Selbach M, Mann M. Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK) Nat Methods. 2006;3:981–983. doi: 10.1038/nmeth972. [DOI] [PubMed] [Google Scholar]
- 11.Wang A, Wu L, Lin J, Han L, Bian J, Wu Y, Robson SC, Xue L, Ge Y, Sang X, et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat Commun. 2018;9:894. doi: 10.1038/s41467-018-03276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou C, Chen H, Han L, Xue F, Wang A, Liang YJ. Screening of genes related to lung cancer caused by smoking with RNA-Seq. Eur Rev Med Pharmacol Sci. 2014;18:117–125. [PubMed] [Google Scholar]
- 13.Xiong L, Wen Y, Miao X, Yang Z. NT5E and FcGBP as key regulators of TGF-1-induced epithelial-mesenchymal transition (EMT) are associated with tumor progression and survival of patients with gallbladder cancer. Cell Tissue Res. 2014;355:365–374. doi: 10.1007/s00441-013-1752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazi MH, He M, Cheville JC, Young CY. Downregulation of IgG Fc binding protein (Fc gammaBP) in prostate cancer. Cancer Biol Ther. 2008;7:70–75. doi: 10.4161/cbt.7.1.5131. [DOI] [PubMed] [Google Scholar]
- 15.O'Donovan N, Fischer A, Abdo EM, Simon F, Peter HJ, Gerber H, Buergi U, Marti U. Differential expression of IgG Fc binding protein (FcgammaBP) in human normal thyroid tissue, thyroid adenomas and thyroid carcinomas. J Endocrinol. 2002;174:517–524. doi: 10.1677/joe.0.1740517. [DOI] [PubMed] [Google Scholar]
- 16.Choi CH, Choi JJ, Park YA, Lee YY, Song SY, Sung CO, Song T, Kim MK, Kim TJ, Lee JW, et al. Identification of differentially expressed genes according to chemosensitivity in advanced ovarian serous adenocarcinomas: Expression of GRIA2 predicts better survival. Br J Cancer. 2012;107:91–99. doi: 10.1038/bjc.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Liu Y, Liu H, Zhang Q, Song H, Tang J, Fu J, Wang X. FcGBP was upregulated by HPV infection and correlated to longer survival time of HNSCC patients. Oncotarget. 2017;8:86503–86514. doi: 10.18632/oncotarget.21220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witzmann FA, Arnold RJ, Bai F, Hrncirova P, Kimpel MW, Mechref YS, McBride WJ, Novotny MV, Pedrick NM, Ringham HN, et al. A proteomic survey of rat cerebral cortical synaptosomes. Proteomics. 2005;5:2177–2201. doi: 10.1002/pmic.200401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant SG. The synapse proteome and phosphoproteome: a new paradigm for synapse biology. Biochem Soc Trans. 2006;34((Pt 1)):59–63. doi: 10.1042/BST0340059. [DOI] [PubMed] [Google Scholar]
- 20.Dong S, Huo H, Mao Y, Li X, Dong L. A risk score model for the prediction of osteosarcoma metastasis. FEBS Open Bio. 2019;9:519–526. doi: 10.1002/2211-5463.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Lee S, Yang SK, Song K, Lee I. Differential expression in histologically normal crypts of ulcerative colitis suggests primary crypt disorder. Oncol Rep. 2006;16:663–670. [PubMed] [Google Scholar]
- 22.Risques RA, Lai LA, Himmetoglu C, Ebaee A, Li L, Feng Z, Bronner MP, Al-Lahham B, Kowdley KV, Lindor KD, et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res. 2011;71:1669–1679. doi: 10.1158/0008-5472.CAN-10-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Bang S, Song K, Lee I. Differential expression in normal-adenoma-carcinoma sequence suggests complex molecular carcinogenesis in colon. Oncol Rep. 2006;16:747–754. [PubMed] [Google Scholar]
- 24.Zhu H, Wu TC, Chen WQ, Zhou LJ, Wu Y, Zeng L, Pei HP. Screening for differentially expressed genes between left- and right-sided colon carcinoma by microarray analysis. Oncol Lett. 2013;6:353–358. doi: 10.3892/ol.2013.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, Shi J, Zhou Y, Liu T, Zhan F, Zhang K, Liu N. Integrating proteomics and transcriptomics for the identification of potential targets in early colorectal cancer. Int J Oncol. 2019;55:439–450. doi: 10.3892/ijo.2019.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang GL, Pan LL, Huang T, Wang JH. The transcriptome difference between colorectal tumor and normal tissues revealed by single-cell sequencing. J Cancer. 2019;10:5883–5890. doi: 10.7150/jca.32267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma R, Jing C, Zhang Y, Cao H, Liu S, Wang Z, Chen D, Zhang J, Wu Y, Wu J, et al. The somatic mutation landscape of Chinese Colorectal Cancer. J Cancer. 2020;11:1038–1046. doi: 10.7150/jca.37017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi C, Hong L, Cheng Z, Yin Q. Identification of metastasis-associated genes in colorectal cancer using metaDE and survival analysis. Oncol Lett. 2016;11:568–574. doi: 10.3892/ol.2015.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendas G, Borsig L. Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins. Int J Cell Biol. 2012;2012:676731. doi: 10.1155/2012/676731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onstenk W, Sieuwerts AM, Mostert B, Lalmahomed Z, Bolt-de Vries JB, van Galen A, Smid M, Kraan J, Van M, de Weerd V, et al. Molecular characteristics of circulating tumor cells resemble the liver metastasis more closely than the primary tumor in metastatic colorectal cancer. Oncotarget. 2016;7:59058–59069. doi: 10.18632/oncotarget.10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Znalesniak EB, Fu T, Salm F, Händel U, Hoffmann W. Transcriptional responses in the murine spleen after toxoplasma gondii infection: inflammasome and mucus-associated genes. Int J Mol Sci. 2017;18:1245. doi: 10.3390/ijms18061245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EE, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260:8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, Li J, Li F, Tan HB. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol. 2020;11:369. doi: 10.3389/fimmu.2020.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, Hornsteiner L, Punta M, Patil Y, Newey A, et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-egfr treatment in colorectal cancer. Cancer Cell. 2019;36:35–50.e9. doi: 10.1016/j.ccell.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Zeng Z, Huang L, Luo S, Dong J, Zhou FH, Zhou K, Wang L, Kang L. Photothermal therapy technology of metastatic colorectal cancer. Am J Transl Res. 2020;12:3089–3115. [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/annotation/8c585739-a354-4fc9-a7d0-d5ae26fa06ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 42.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 43.Gaedcke J, Grade M, Jung K, Camps J, Jo P, Emons G, Gehoff A, Sax U, Schirmer M, Becker H, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49:1024–1034. doi: 10.1002/gcc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee A, Ahn A, Rodger EJ, Stockwell PA, Eccles MR. A guide for designing and analyzing RNA-Seq data. Methods Mol Biol. 2018;1783:35–80. doi: 10.1007/978-1-4939-7834-2_3. [DOI] [PubMed] [Google Scholar]
- 45.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siemers NO, Holloway JL, Chang H, Chasalow SD, Ross-MacDonald PB, Voliva CF, Szustakowski JD. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One. 2017;12:e0179726. doi: 10.1371/journal.pone.0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, Geller MA, Odunsi K, Beechem J, Fling SP. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sousa S, Määttä J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol. 2016;5:135–138. doi: 10.1016/j.jbo.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siskova A, Cervena K, Kral J, Hucl T, Vodicka P, Vymetalkova V. Colorectal adenomas-genetics and searching for new molecular screening biomarkers. Int J Mol Sci. 2020;21:3260. doi: 10.3390/ijms21093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Idos GE, Kwok J, Bonthala N, Kysh L, Gruber SB, Qu C. The Prognostic implications of tumor infiltrating lymphocytes in colorectal cancer: a systematic review and meta-analysis. Sci Rep. 2020;10:3360. doi: 10.1038/s41598-020-60255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dalal N, Jalandra R, Sharma M, Prakash H, Makharia GK, Solanki PR, Singh R, Kumar A. Omics technologies for improved diagnosis and treatment of colorectal cancer: Technical advancement and major perspectives. Biomed Pharmacother. 2020;131:110648. doi: 10.1016/j.biopha.2020.110648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.