Abstract
Protein folding is crucial for normal physiology including development and healthy aging, and failure of this process is related to the pathology of diseases including neurodegeneration and cancer. Early thermodynamic and kinetic studies based on the unfolding and refolding equilibrium of individual proteins in the test tube have provided insight into the fundamental principles of protein folding, although the problem of predicting how any given protein will fold remains unsolved. Protein folding within cells is a more complex issue than folding of purified protein in isolation, due to the complex interactions within the cellular environment, including post-translational modifications of proteins, the presence of macromolecular crowding in cells, and variations in the cellular environment, for example in cancer versus normal cells. Development of biophysical approaches including fluorescence resonance energy transfer (FRET) and nuclear magnetic resonance (NMR) techniques and cellular manipulations including microinjection and insertion of noncanonical amino acids has allowed the study of protein folding in living cells. Furthermore, biophysical techniques such as single-molecule fluorescence spectroscopy and optical tweezers allows studies of simplified systems at the single molecular level. Combining in-cell techniques with the powerful detail that can be achieved from single-molecule studies allows the effects of different cellular components including molecular chaperones to be monitored, providing us with comprehensive understanding of the protein folding process. The application of biophysical techniques to the study of protein folding is arming us with knowledge that is fundamental to the battle against cancer and other diseases related to protein conformation or protein–protein interactions.
Keywords: biophysical techniques, cancer, neurodegeneration, protein conformation, proteostasis, single molecule detection
Introduction
Newly synthesized polypeptide chains need to fold correctly to form the native protein structure in order to carry out their cellular function [1]. Under normal circumstances, the native protein structure is usually the thermodynamically most stable state, although some proteins maintain an unfolded or partially unfolded state to exert their function in cells [2,3]. The collapse of the polypeptide chain is driven by the hydrophobic effect whereby ordered water molecules are excluded, and new interactions can then form between individual amino acid side chains [1]. In vitro folding studies indicate that most cytosolic proteins are only marginally stable and are in dynamic equilibrium with unfolded or partially folded states, meaning that a protein may unfold and refold many times during its life cycle [4]. Folding and conformational changes of proteins due to interaction with other cellular components, including other proteins, cofactors, metabolites, or nucleic acids, are very common in cells [5]. Anfinsen demonstrated that a small protein can fold spontaneously in the test tube [6]. However complex multi-domain proteins often cannot fold spontaneously to the correct native conformation [7]. There are many factors that can affect the folding process, including solution conditions such as, pH and ionic strength, as well as post-translational modifications and macromolecular crowding in cells [5,8]. To avoid aggregation of folding intermediates and avoid energetic traps during folding, molecular chaperones and folding catalysts assist protein folding through different mechanisms [9,10]. An ATP-independent holdase function or an ATP-dependent remodeling function are the two common types of assistance provided by molecular chaperones [11].
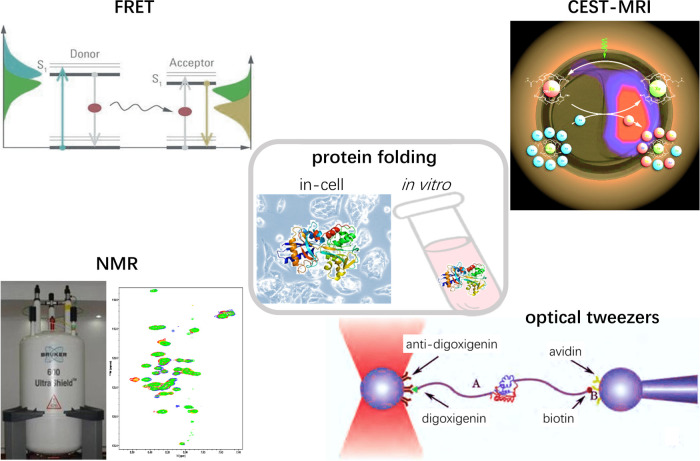
Cancer is an example where disease initiation and progression are often related to the failure of protein folding and quality control [12–14]. Cancer cells are hallmarked by abnormally high proliferation and migration supported by increased metabolism and an enhanced protein quality control system [13,14]. There is often an increase in production of nascent polypeptides, more frequent mutation, and raised levels of reactive oxygen species (ROS), as well as other changes to the cellular environment that challenge the protein folding process in cancer cells [13,15]. Dependence of protein folding in cancer cells on chaperones is indicated by higher expression levels of some chaperones, such as Hsp70 and Hsp90 which have been identified as drug targets for cancer therapy [16,17]. The study of protein folding in cancer is revealing further details of cancer mechanisms and provides new avenues for cancer therapy, and it is valuable to study protein folding in different types of cancer cells and tissues to identify both common characteristics and notable differences. However non-invasive in-cell study is still challenging and so study of the mechanisms by which chaperones and other cellular components affect protein folding carried out using purified protein in simplified environments also provide important clues for the development of anti-cancer drugs [18]. Ligands including drugs often alter the folding, stability and conformation of a protein [19]. Thus changes in protein folding resulting from the introduction of a drug could indicate an interaction between the drug and protein, and identifying drug-binding targets on a proteomic scale based on protein folding status could provide critical information for understanding the mechanism of the efficacy of the drug and possible side effects [20,21]. Protein folding studies provide a very important foundation to understand and overcome cancer. In the recent 20 years biophysical approaches including fluorescence resonance energy transfer (FRET), optical tweezers, nuclear magnetic resonance (NMR) and magnetic resonance imaging (MRI) have significantly extended the scope and depth of protein folding studies based on classical spectroscopic tools such as absorption, circular dichroism (CD) and intrinsic Trp fluorescence of proteins (Figure 1).
Figure 1. In-cell and in vitro study of protein folding has been significantly advanced by using biophysical approaches including FRET, NMR, CEST-MRI and optical tweezers.
In-cell study of protein folding using biophysical probes
The recent trend in studying protein folding is to access the real scene in living cells and tissues. In-cell study of protein folding utilizes the accumulated knowledge regarding living cells as well as the development of new biophysical approaches. Till now most in-cell studies of protein folding have been performed in E. coli or cancer cell lines, which paves the way for further precise study of protein folding in cancer and other diseases.
Clinical magnetic resonance imaging (MRI) is based on imaging of signal contrast of protons from different living components. Chemical exchange saturation transfer (CEST)-MRI has been applied to detect protein states in living tissue. CEST-MRI exploits the spontaneous chemical exchange of protons to indirectly detect the solutes via the abundant water proton signal. CEST-MRI has been extended to a radiofrequency (RF) irradiation scheme at two different frequency offsets (dual CEST-MRI) to sense changes in the state of protein folding in solution and it shows potential application to detect the mobile fraction of the proteome in diverse pathologies [22]. The global status of protein folding in the cancer cell line HepG2 was demonstrated using CEST-MRI of the relayed nuclear Overhauser effect (rNOE) signal which is linked to protein conformation [23]. Signal changes within a 20-min 42°C non-lethal heat-shock were coincident with protein denaturation and aggregation processes induced by the heat shock, and the signal recovery after heat shock was consistent with recovery due to chaperone-induced refolding. This demonstrates the potential of CEST-MRI for monitoring pathological changes to protein folding in cancer and other diseases [23]. While CEST-MRI provides a powerful tool to gauge the global status of protein folding in the cell, it is not suited to the precise study of the folding of individual proteins.
Individual protein studies can provide detailed and precise information about protein folding in cells. The main current biophysical approaches to reveal individual protein folding in situ are in-cell FRET and in-cell NMR. In-cell FRET relies on fluorescent labeling of the target protein to achieve a high signal to noise ratio. There are two basic strategies to deliver exogenous fluorescent probes into living cells: (1) expression of proteins with noncanonical amino acids or fusion expression with fluorescent proteins in cells [24,25]; (2) microinjection of protein labeled with a fluorescent probe (fusion with fluorescence proteins or cysteine labeling with maleimide-conjugated fluorescence dye) into living cells [26]. FRET efficiency is dependent on the distance of two fluorescent labels, enabling FRET methods to explore protein conformational changes, dynamics, folding kinetics and protein interaction in living cells at bulk and single-molecule level while mitigating noise from the complex cellular background [26,27].
Expression of the fluorescent fusion protein GFP-RAF-YFP in HeLa cells for single molecule FRET (smFRET) study of conformational changes in RAF has been carried out [25]. Alternative laser excitation (ALEX) which enables rapid switching between a donor (D)-excitation and an acceptor (A)-excitation laser to identify distinct emission signatures for all diffusing species was used to measure the native state of RAF while undergoing native interactions with other intrinsic proteins and the reaction network of the signal transduction pathway in live cells [25,28]. It was found that cytosolic RAF has at least three conformational states: the inactive closed form, the active open form and the inactive fully-open form. Spontaneous transitions between the conformational states were detected with epidermal growth factor (EGF) stimulation. The S621A mutation shifts the distribution of conformational states to the inactive fully-open form [25]. Thus smFRET is applicable to detect conformational distribution changes resulting from intracellular interactions in live cells for other cytosolic proteins (Figure 2).
Figure 2. SmFRET using alternative laser excitation (ALEX) is applied to detect the structural distribution and changes of cytosolic proteins in live cells.
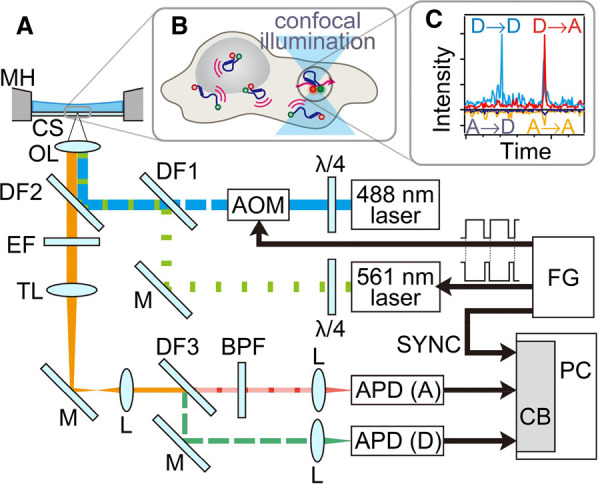
(A) Experimental setup. (B) Single-molecule detection of freely diffusing cytosolic molecules in a live cell. (C) Typical fluorescence burst signals. X → Y denotes the fluorescence signal from dye Y with the excitation of dye X, where X and Y are the donor (D) or the acceptor (A), respectively. FG: function generator; λ/4: quarter wave plate; AOM: acousto-optic modulator; M: mirror; DF: dichroic filter; OL: objective lens; MH: metal holder; CS: coverslip; EF: emission filter; TL: tube lens; L: lens; BPF: band-pass filter; APD: avalanche photodiode; CB: counter board; PC: personal computer. Figure reproduced from ref. [25] with permission.
Membrane proteins undertake important tasks in cells and are major drug targets implicated in many diseases. However, studying the protein folding of membrane proteins has remained very challenging due to the difficulty in applying classical methods including absorption, CD and Trp fluorescence which are more suitable for water-soluble proteins. In recent years, the combination of new technology and development of new membrane-mimicking systems such as nanodiscs and cell unroofing methods has promoted the study of membrane protein folding [29]. Using a noncanonical amino acid insertion strategy together with ACCuRET (Anap Cyclen-Cu2+ resonance energy transfer), maltose binding protein (MBP) was labeled in HEK293T/17 cells and used as a benchmark. Using cell unroofing, it was shown that ACCuRET can accurately monitor rearrangements of proteins in native membranes based on measurement of absolute distances and distance changes [24]. Thus ACCuRET is applicable for measuring conformational dynamics of both soluble proteins and membrane proteins.
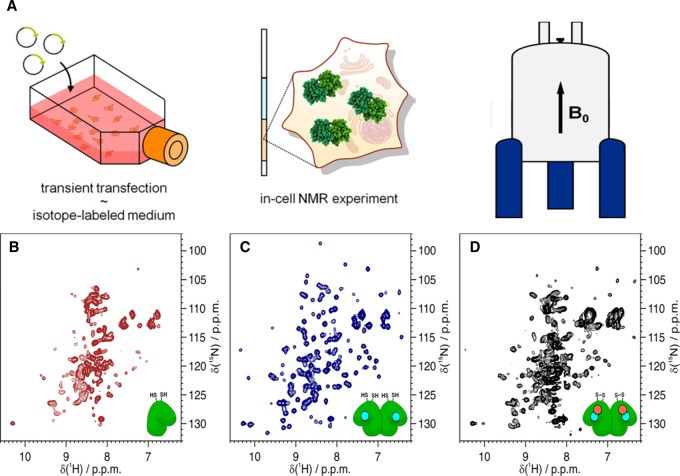
Protein NMR measurements rely on isotope labeling, and classical solution NMR methods are more suitable for small (lower than 30 kDa) proteins, requiring also a high concentration of protein [30]. One challenge for in-cell NMR is to label target proteins in living cells with isotopes, typically 2H, 13C and 15N. Using isotope enriched culture medium to express an isotope-labeled protein has become routine in E. coli, but is still expensive and difficult to achieve in eukaryotic cells. As the endogenous expression level is generally low, an alternative strategy in eukaryotic cells is to microinject or translocate an isotope-labeled protein which is expressed in the E. coli expression system into the cytosol or nuclei of target cells [31,32]. The other labeling strategy is to express 19F labeled protein using noncanonical 19F-amino acids in cells [31]. Another challenge is the lack of sensitivity of in-cell NMR due to the low concentration of target protein in cells, the background noise, and reduced quality of the spectra due to monitoring within the in-cell environment. To improve the signal quality, 19F labeling is a good choice as it is a high-sensitivity isotope, and 19F NMR spectra are virtually background-free as biological molecules do not contain fluorine atoms [33]. Another solution is to optimize NMR detection protocols. Transverse relaxation optimized spectroscopy (TROSY) NMR methods enhance the sensitivity and allow the dynamics and interactions of large (up to 1 MDa) proteins or assemblies to be probed, extending the scope of NMR study [34]. Fast pulsing methods also help maximize the signal-to-noise ratio [32]. The improvement of the sensitivity enables in-cell NMR to measure protein folding in cells at near physiological expression levels (nanomolar to micromolar range) [32]. Relaxation dispersion (RD) and saturation transfer (ST) methods can provide detail about the pathways of biomolecular processes, including transiently populated intermediates of protein folding, enzyme catalysis and binding [35]. Rapid progress in solid-state NMR (ssNMR) and paramagnetic relaxation enhancement (PRE) techniques has also extended the scope of protein folding study in cells [36–38]. A typical in-cell NMR experiment is shown in Figure 3, where NMR spectra of different human superoxide dismutase 1 (SOD1) maturation states in human cells were measured to describe the complete post-translational maturation process of SOD1 [36,39].
Figure 3. In-cell NMR is applied to detect the conformational changes of cytosolic proteins in live cells.
(A) The vector containing a gene of interest (green arrow) is delivered by transient transfection in labeled medium. Cells expressing the protein of interest (green) are collected and placed in a 3 mm Shigemi NMR tube and applied for in-cell NMR analysis. (B–D) 1H−15N NMR spectra of SOD1 in the apo and reduced state (B), the one-zinc-ion (cyan)-per-monomer bound and dimerized state (C) and the fully mature, copper (salmon)-zinc (cyan)-bound and disulfide-oxidized state (D) in human cells. Figure adapted from ref. [36] with permission. (Further permissions related to this figure should be directed to the ACS. Original figures at: https://pubs.acs.org/doi/10.1021/acs.accounts.8b00147).
Intrinsically disordered proteins (IDPs) have strong NMR signals and are suitable for in-cell NMR study. Preliminary in-cell NMR study of protein folding in human cell lines has involved IDPs, such as α-synuclein and Tau, which are implicated in neurodegenerative diseases. After NMR characterization of α-synuclein in E. coli cells [40], 15N labeled α-synuclein was delivered into mammalian cells and an electroporation protocol was used to tightly control the cellular α-synuclein concentration resulting in a uniform distribution of the protein in the cytoplasm with minimal perturbation of cell viability [41]. The conformation of α-synuclein in different cell lines, including neuronal B65 and SK-N-SH cells and RCSN-3 cells was compared by 2D 1H-15N correlation NMR spectra. This study revealed some common features of α-synuclein in different cell lines: (1) The major conformation of the monomeric disordered α-synuclein was similar in different intracellular environments; (2) the N-terminus of α-synuclein undergoes acetylation in cells; (3) α-synuclein does not form stable interactions with cellular membranes; however N- and C-termini of α-synuclein transiently interact with the cytoplasmic components and/or membrane [41]. Further elucidation of how oxidative stress alters the fate of α-synuclein inside the living cells was carried out by delivering 15N-labeled, N-terminally acetylated, methionine-(Met1, Met5, Met116 and Met127)-oxidized α-synuclein into non-neuronal and neuronal cells [42]. Time-resolved in-cell NMR demonstrated that Tyr125 phosphorylation was selectively impaired by the C-terminal methionine oxidation, suggesting that alteration of the cellular environment can selectively affect post-translational modifications of α-synuclein and, thus, potentially control its conformation, conformational landscape, and aggregation propensity [42]. Isotope-enriched Tau was delivered into HEK-293T cells by electroporation and the NMR spectrum of Tau in living cells was acquired [43]. It was found that Tau predominantly binds to microtubules (MT) at its MT-binding repeats in HEK-293T cells. It was also found that disease-associated phosphorylation of Tau was immediately eliminated once phosphorylated Tau was delivered into HEK-293 T cells, implying a potential cellular protection mechanism under stressful conditions [43].
In-cell NMR is also applicable for in situ study of membrane proteins. An ssNMR-based approach has been developed that is supported by dynamic nuclear polarization (DNP) to directly examine the structural and dynamic properties of epidermal growth factor receptor (EGFR) activation by the EGF in native membranes [44]. A431 cells were cultured in the labeled medium to produce [13C, 15N] labeled A431 membrane vesicles. The results show that the ligand-free state of the extracellular domain (ECD) is highly dynamic, while the intracellular kinase domain (KD) is rigid. Ligand binding restricts the overall and local motion of EGFR domains, including the ECD and the C-terminal region. It is suggested that the reduction in conformational entropy of the ECD by ligand binding favors the cooperative binding required for receptor dimerization, causing allosteric activation of the intracellular tyrosine [44]. The accumulation of studies using in-cell NMR highlight the potential of in-cell NMR to study protein folding, structural dynamics and interactions at the residue level in neurodegenerative diseases, cancer and other diseases related to protein conformation.
In vitro study of protein folding using biophysical approaches
It is currently difficult to obtain complete and detailed information about protein folding purely from in-cell study. In vitro study of purified proteins in solution still provides a useful approach to simplify the system of study allowing powerful deductions to be made, and increasingly complex in vitro systems have been developed to imitate the actual cellular environment. There are two attractive trends for in vitro protein folding study: (1) using cell extracts to imitate the actual cellular environment to extrapolate to protein folding in cells; (2) mechanistic study of chaperone-assisted protein folding.
Real-time NMR can provide high-resolution structural information alongside kinetic details of protein folding in a virtually continuous manner, not only for the characterization of folding intermediates but also for the investigation of the molecular mechanisms of assisted protein folding [45,46]. Cell-free protein production is still the best method to achieve high-throughput protein production and selective incorporation of isotope-labeled amino acids into a target protein. When the purified isotope-labeled protein is in the complex environment of native cells, or membrane extracts or tissue homogenates, they are visible by real-time NMR detection, providing residue-level resolution of structural and kinetic changes in conformation during folding, interaction, modification and bioreactions in an environment mimicking that of the cell [45]. The kinase inhibitory domain of the cell cycle regulatory protein p27Kip1 (p27) is intrinsically disordered in isolation. Nuclear spin hyperpolarization using dissolution dynamic nuclear polarization (D-DNP) enabled the real-time observation of 13C NMR signals during p27 folding upon binding to Cdk2/cyclin A on a time scale of several seconds [47]. Time-dependent intensity changes are dependent on the extent of folding and binding, as manifested by differential spin relaxation. This study also followed a partially folded p27 intermediate by analysis of signal decay rates [47].
Mechanisms of chaperone-assisted protein folding has attracted particular attention in recent years as it is possible to manipulate protein folding in cells by adjusting molecular chaperone activity via addition of small molecules and this also opens a new avenue for disease therapy especially for cancer [48–50]. Hsp70s are ATP-dependent foldases with complex and enigmatic mechanisms, and their dynamics and allostery make them challenging to study [51]. Technique development in FRET, NMR and optical tweezers is revealing increasing detail of the precise mechanism by which Hsp70s promote protein folding. Both conformational changes and folding mechanisms of Hsp70s themselves and Hsp70-assisted substrate protein folding have been explored in depth.
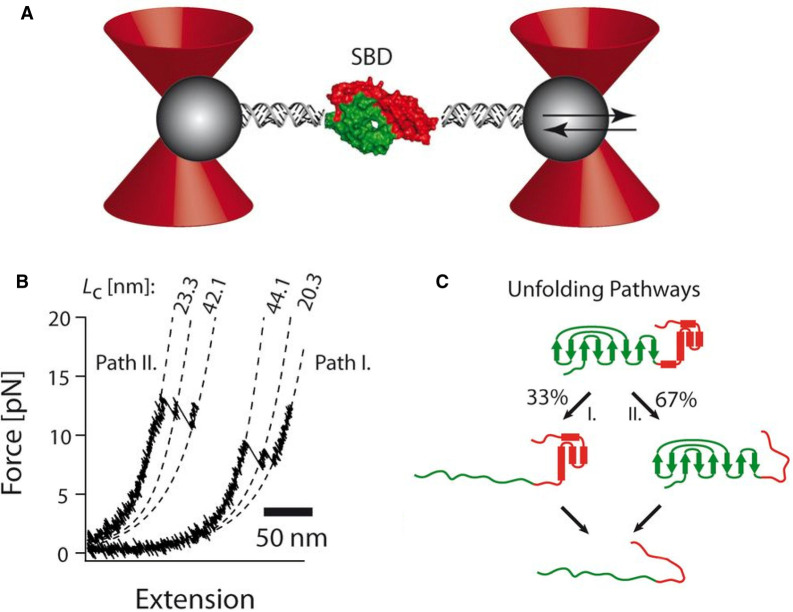
Using smFRET, the interdomain conformational heterogeneity and the kinetics of conformational changes of Hsp70 induced by ATP or the cochaperone Hsp40 was revealed [52]. NMR studies have provided insight into the conformation of Hsp70s with high resolution in different allosteric states [53,54]. Using optical tweezers unfolding and folding of the bacterial Hsp70 homolog (DnaK) was monitored, and a folding nucleus and minimal ATP binding domain of Hsp70 has been identified [55], and bifurcating unfolding pathways for the substrate binding domain (SBD) of Hsp70 and mechanical hinge regions has been revealed (Figure 4) [56].
Figure 4. Single-molecule force experiments of the SBD of the Hsp70 chaperone performed with optical tweezers.
(A) Optical tweezers assay. The SBD (green/red surfaces) is tethered to the beads (gray spheres) by two DNA handles, and beads are trapped in highly focused laser beams (red cones). The connection between the DNA and protein is realized by the modification of the two cysteine residues of the protein by the single-stranded DNA-maleimide oligonucleotide complementary to the DNA-handle overhang. One of the beams is reflected by a steerable mirror, which enables pulling and stretching of a single protein. (B) Force-extension curves of a single SBD domain show the order of the individual unfolding events varies. Pathway I corresponds to unfolding of the larger fragment first, followed by the shorter SBD fragment. For pathway II, the order of unfolding events is reversed. (C) Summary of bifurcating unfolding pathways of the SBD. Figure adapted from ref. [56] with permission.
Combining single-pair FRET and hydrogen/deuterium exchange, the mechanism by which DnaK/DnaJ/GrpE accelerates folding of the multi-domain protein firefly luciferase (FLuc) was explained. Inter-domain misfolding was identified as the cause of slow folding, and DnaK binding causes expansion of the misfolded region and thereby resolves the kinetically trapped intermediates, with folding occurring upon GrpE-mediated release from DnaK, which commits a fraction of FLuc to fast folding, circumventing misfolding [57]. NMR study revealed Hsp70 induced stabilization of the unfolded states of its substrates SRC homology 3 domain (SH3) and hTRF1, and DnaK binding directly affects the unfolded protein ensemble and fine tunes the folding landscape of the substrate proteins [58,59].
In a typical optical tweezers assay, the protein of interest is attached to beads for stretching experiments to monitor the unfolding and folding of an immobilized protein (Figure 4) [56,60]. Monitoring unfolding and folding of MBP using optical tweezers, it was shown that DnaK binds and stabilizes not only extended peptide segments, but also partially folded and near-native protein structures [60]. Integrated optical tweezers monitored unfolding and folding of MBP with fluorescent-particle tracking shows that ClpB translocates both arms of the loop simultaneously and switches to single-arm translocation when encountering obstacles and substrates refold while exiting the pore, analogous to co-translational folding [61]. Using optical tweezers to monitor unfolding and folding of the glucocorticoid receptor (GR), it was observed in detail the steps of folding and hormone binding of GR and identifying a structural element that opens and closes upon hormone binding, forming the basis for understanding GR activation and its regulation by chaperone proteins [62]. A current limitation of optical tweezers is that it is only suited to in vitro study of protein folding.
The above studies demonstrate that biophysical techniques can provide detailed information about the structural and dynamic aspects of protein folding. The study of in-cell protein folding presents challenges, such as short-half lives and degradation of proteins in cells. Each of the techniques described above has strengths and limitations, and a full picture can only be achieved by the combination of multiple approaches. In-cell FRET and NMR require labeling of target proteins and optimizing the signal to noise ratio in a cellular background. To date, only a small fraction of the proteome has been probed by such techniques. In-cell FRET has been more widely applied to membrane proteins than cytosolic proteins. In-cell NMR is still most applicable to small monomeric proteins or intrinsically disordered polypeptide regions, as the relaxation time of the NMR signal is also affected by molecular crowding in the cell, leading to NMR signal attenuation. Careful interpretation of results is required when the techniques employed require overexpression of proteins at levels much higher than their natural cellular protein concentration. The delivery of exogenously labeled proteins can also perturb the natural cell environment, and this must be taken into account. Of the techniques discussed here, smFRET and optical tweezers are particularly powerful for the study of simplified systems at the single molecular level, while NMR studies provide high-resolution residue specific information of the ensemble of protein molecules. In future, the combination of these different biophysical approaches, including further breakthroughs in technique development, will be increasingly required in order to unravel the mysteries of the protein folding process.
Summary
A detailed understanding of protein folding in cancer, neurodegeneration and other diseases will assist early diagnosis and treatment.
A combination of biophysical approaches including FRET, NMR, MRI and optical tweezers, together with cellular manipulations including microinjection and insertion of noncanonical amino acids, is providing a picture of the protein folding process in living cells and tissues.
It is increasingly clear that development of new and better biophysical approaches, and the combination of these techniques, is crucial to obtaining a full picture of protein folding in health and disease.
Abbreviations
- ACCuRET
Anap Cyclen-Cu2+ resonance energy transfer
- ALEX
alternative laser excitation
- CD
circular dichroism
- CEST
chemical exchange saturation transfer
- D-DNP
dissolution dynamic nuclear polarization
- ECD
extracellular domain
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- FLuc
firefly luciferase
- FRET
fluorescence (or Förster) resonance energy transfer
- GR
glucocorticoid receptor
- IDPs
intrinsically disordered proteins
- KD
kinase domain
- MBP
maltose binding protein
- MRI
magnetic resonance imaging
- MT
microtubule
- NMR
nuclear magnetic resonance
- PRE
paramagnetic relaxation enhancement
- RD
relaxation dispersion
- rNOE
relayed nuclear Overhauser effect
- ROS
reactive oxygen species
- SBD
substrate binding domain
- SH3
SRC homology 3 domain
- smFRET
single molecular FRET
- SOD1
superoxide dismutase 1
- ssNMR
solid-state NMR
- ST
saturation transfer
- TROSY
transverse relaxation optimized spectroscopy
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors acknowledge support from the Chinese Ministry of Science and Technology [2017YFA0504000] and the National Natural Science Foundation of China [31920103011, 31770829, 21673278].
Author Contribution
H.Z. and S.P. conceived and wrote the review; all authors revised the review and approved the final version.
References
- 1.Dobson, C.M. (2003) Protein folding and misfolding. Nature 426, 884–890 10.1038/nature02261 [DOI] [PubMed] [Google Scholar]
- 2.Uversky, V.N., Oldfield, C.J. and Dunker, A.K. (2008) Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu. Rev. Biophys. 37, 215–246 10.1146/annurev.biophys.37.032807.125924 [DOI] [PubMed] [Google Scholar]
- 3.Shoemaker, B.A., Portman, J.J. and Wolynes, P.G. (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl Acad. Sci. U.S.A. 97, 8868–8873 10.1073/pnas.160259697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruebele, M. (2005) Downhill protein folding: evolution meets physics. C. R. Biol. 328, 701–712 10.1016/j.crvi.2005.02.007 [DOI] [PubMed] [Google Scholar]
- 5.Gruebele, M., Dave, K. and Sukenik, S. (2016) Globular protein folding in vitro and In vivo. Annu. Rev. Biophys. 45, 233–251 10.1146/annurev-biophys-062215-011236 [DOI] [PubMed] [Google Scholar]
- 6.Anfinsen, C.B. (1973) Principles that govern the folding of protein chains. Science 181, 223–230 10.1126/science.181.4096.223 [DOI] [PubMed] [Google Scholar]
- 7.Han, J.H., Batey, S., Nickson, A.A., Teichmann, S.A. and Clarke, J. (2007) The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 8, 319–330 10.1038/nrm2144 [DOI] [PubMed] [Google Scholar]
- 8.Cheung, M.S. and Gasic, A.G. (2018) Towards developing principles of protein folding and dynamics in the cell. Phys. Biol. 15, 063001 10.1088/1478-3975/aaced2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balchin, D., Hayer-Hartl, M. and Hartl, F.U. (2020) Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 594, 2770–2781 10.1002/1873-3468.13844 [DOI] [PubMed] [Google Scholar]
- 10.Balchin, D., Hayer-Hartl, M. and Hartl, F.U. (2016) In vivo aspects of protein folding and quality control. Science 353, aac4354 10.1126/science.aac4354 [DOI] [PubMed] [Google Scholar]
- 11.Mogk, A., Ruger-Herreros, C. and Bukau, B. (2019) Cellular functions and mechanisms of action of small heat shock proteins. Annu. Rev. Microbiol. 73, 89–110 10.1146/annurev-micro-020518-115515 [DOI] [PubMed] [Google Scholar]
- 12.Bastola, P., Oien, D.B., Cooley, M. and Chien, J. (2018) Emerging cancer therapeutic targets in protein homeostasis. AAPS J. 20, 94 10.1208/s12248-018-0254-1 [DOI] [PubMed] [Google Scholar]
- 13.Hanahan, D. and Weinberg, R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. and Weinberg, R.A. (2000) The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 15.Daher, B., Vučetić, M. and Pouysségur, J. (2020) Cysteine depletion, a Key action to challenge cancer cells to ferroptotic cell death. Front. Oncol. 10, 723 10.3389/fonc.2020.00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee, S. and Burns, T.F. (2017) Targeting heat shock proteins in cancer: a promising therapeutic approach. Int. J. Mol. Sci. 18, 1978 10.3390/ijms18091978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini, J. and Sharma, P.K. (2018) Clinical, prognostic and therapeutic significance of heat shock proteins in cancer. Curr. Drug Targets 19, 1478–1490 10.2174/1389450118666170823121248 [DOI] [PubMed] [Google Scholar]
- 18.Holzbeierlein, J.M., Windsperger, A. and Vielhauer, G. (2010) Hsp90: a drug target? Curr. Oncol. Rep. 12, 95–101 10.1007/s11912-010-0086-3 [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Ruiz, J.M. (2007) Ligand effects on protein thermodynamic stability. Biophys. Chem. 126, 43–49 10.1016/j.bpc.2006.05.021 [DOI] [PubMed] [Google Scholar]
- 20.Liu, Y.K., Chen, H.Y., Chueh, P.J. and Liu, P.F. (2019) A one-pot analysis approach to simplify measurements of protein stability and folding kinetics. Biochim. Biophys. Acta 1867, 184–193 10.1016/j.bbapap.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 21.Martinez Molina, D., Jafari, R., Ignatushchenko, M., Seki, T., Larsson, E.A., Dan, C.et al. (2013) Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 10.1126/science.1233606 [DOI] [PubMed] [Google Scholar]
- 22.Goerke, S., Breitling, J., Zaiss, M., Windschuh, J., Kunz, P., Schuenke, P.et al. (2018) Dual-frequency irradiation CEST-MRI of endogenous bulk mobile proteins. NMR Biomed. 31, e3920 10.1002/nbm.3920 [DOI] [PubMed] [Google Scholar]
- 23.Kleimaier, D., Goerke, S., Nies, C., Zaiss, M., Kunz, P., Bachert, P.et al. (2020) The cellular heat shock response monitored by chemical exchange saturation transfer MRI. Sci. Rep. 10, 11118 10.1038/s41598-020-68022-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, S.E., Munari, M. and Zagotta, W.N. (2018) Visualizing conformational dynamics of proteins in solution and at the cell membrane. eLife 7, e37248 10.7554/eLife.37248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto, K., Hibino, K. and Sako, Y. (2020) In-cell single-molecule FRET measurements reveal three conformational state changes in RAF protein. Biochim. Biophys. Acta Gen. Subj. 1864, 129358 10.1016/j.bbagen.2019.04.022 [DOI] [PubMed] [Google Scholar]
- 26.Sustarsic, M. and Kapanidis, A.N. (2015) Taking the ruler to the jungle: single-molecule FRET for understanding biomolecular structure and dynamics in live cells. Curr. Opin. Struct. Biol. 34, 52–59 10.1016/j.sbi.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 27.Okamoto, K. and Sako, Y. (2017) Recent advances in FRET for the study of protein interactions and dynamics. Curr. Opin. Struct. Biol. 46, 16–23 10.1016/j.sbi.2017.03.010 [DOI] [PubMed] [Google Scholar]
- 28.Kapanidis, A.N., Lee, N.K., Laurence, T.A., Doose, S., Margeat, E. and Weiss, S. (2004) Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl Acad. Sci. U.S.A. 101, 8936–8941 10.1073/pnas.0401690101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krainer, G., Keller, S. and Schlierf, M. (2019) Structural dynamics of membrane-protein folding from single-molecule FRET. Curr. Opin. Struct. Biol. 58, 124–137 10.1016/j.sbi.2019.05.025 [DOI] [PubMed] [Google Scholar]
- 30.Wüthrich, K. (1990) Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 265, 22059–22062 10.1016/S0021-9258(18)45665-7 [DOI] [PubMed] [Google Scholar]
- 31.Luchinat, E. and Banci, L. (2017) In-cell NMR: a topical review. IUCrJ. 4, 108–118 10.1107/S2052252516020625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishida, N., Ito, Y. and Shimada, I. (2020) In situ structural biology using in-cell NMR. Biochim. Biophys. Acta Gen. Subj. 1864, 129364 10.1016/j.bbagen.2019.05.007 [DOI] [PubMed] [Google Scholar]
- 33.Marsh, E.N. and Suzuki, Y. (2014) Using (19)F NMR to probe biological interactions of proteins and peptides. ACS Chem. Biol. 9, 1242–1250 10.1021/cb500111u [DOI] [PubMed] [Google Scholar]
- 34.Xu, Y. and Matthews, S. (2013) TROSY NMR spectroscopy of large soluble proteins. Top. Curr. Chem. 335, 97–119 10.1007/128_2011_228 [DOI] [PubMed] [Google Scholar]
- 35.Zhuravleva, A. and Korzhnev, D.M. (2017) Protein folding by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 100, 52–77 10.1016/j.pnmrs.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 36.Luchinat, E. and Banci, L. (2018) In-cell NMR in human cells: direct protein expression allows structural studies of protein folding and maturation. Acc. Chem. Res. 51, 1550–1557 10.1021/acs.accounts.8b00147 [DOI] [PubMed] [Google Scholar]
- 37.Rahman, S., Byun, Y., Hassan, M.I., Kim, J. and Kumar, V. (2017) Towards understanding cellular structure biology: In-cell NMR. Biochim. Biophys. Acta 1865, 547–557 10.1016/j.bbapap.2017.02.018 [DOI] [PubMed] [Google Scholar]
- 38.Li, C., Zhao, J., Cheng, K., Ge, Y., Wu, Q., Ye, Y.et al. (2017) Magnetic resonance spectroscopy as a tool for assessing macromolecular structure and function in living cells. Annu. Rev. Anal. Chem. 10, 157–182 10.1146/annurev-anchem-061516-045237 [DOI] [PubMed] [Google Scholar]
- 39.Banci, L., Barbieri, L., Bertini, I., Luchinat, E., Secci, E., Zhao, Y.et al. (2013) Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 9, 297–299 10.1038/nchembio.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waudby, C.A., Camilloni, C., Fitzpatrick, A.W., Cabrita, L.D., Dobson, C.M., Vendruscolo, M.et al. (2013) In-cell NMR characterization of the secondary structure populations of a disordered conformation of α-synuclein within E. coli cells. PLoS ONE 8, e72286 10.1371/journal.pone.0072286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theillet, F.X., Binolfi, A., Bekei, B., Martorana, A., Rose, H.M., Stuiver, M.et al. (2016) Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature 530, 45–50 10.1038/nature16531 [DOI] [PubMed] [Google Scholar]
- 42.Binolfi, A., Limatola, A., Verzini, S., Kosten, J., Theillet, F.X., Rose, H.M.et al. (2016) Intracellular repair of oxidation-damaged α-synuclein fails to target C-terminal modification sites. Nat. Commun. 7, 10251 10.1038/ncomms10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, S., Wang, C., Lu, J., Ma, X., Liu, Z., Li, D.et al. (2018) In-cell NMR study of Tau and MARK2 phosphorylated Tau. Int. J. Mol. Sci. 20, 90 10.3390/ijms20010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan, M., Narasimhan, S., de Heus, C., Mance, D., van Doorn, S., Houben, K.et al. (2016) EGFR dynamics change during activation in native membranes as revealed by NMR. Cell 167, 1241–51.e11 10.1016/j.cell.2016.10.038 [DOI] [PubMed] [Google Scholar]
- 45.Smith, M.J., Marshall, C.B., Theillet, F.X., Binolfi, A., Selenko, P. and Ikura, M. (2015) Real-time NMR monitoring of biological activities in complex physiological environments. Curr. Opin. Struct. Biol. 32, 39–47 10.1016/j.sbi.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 46.Kumar, A. and Balbach, J. (2015) Real-time protein NMR spectroscopy and investigation of assisted protein folding. Biochim. Biophys. Acta 1850, 1965–1972 10.1016/j.bbagen.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 47.Ragavan, M., Iconaru, L.I., Park, C.G., Kriwacki, R.W. and Hilty, C. (2017) Real-time analysis of folding upon binding of a disordered protein by using dissolution DNP NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 56, 7070–7073 10.1002/anie.201700464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calderwood, S.K. (2018) Heat shock proteins and cancer: intracellular chaperones or extracellular signalling ligands? Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20160524 10.1098/rstb.2016.0524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi, S., Wang, T., Araujo, T.L.S., Sharma, S., Brodsky, J.L. and Chiosis, G. (2018) Adapting to stress - chaperome networks in cancer. Nat. Rev. Cancer 18, 562–575 10.1038/s41568-018-0020-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun, C.W., Kim, H.J., Lim, J.H. and Lee, S.H. (2019) Heat shock proteins: agents of cancer development and therapeutic targets in anti-Cancer therapy. Cells 9, 60 10.3390/cells9010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenzweig, R., Nillegoda, N.B., Mayer, M.P. and Bukau, B. (2019) The Hsp70 chaperone network. Nat. Rev. Mol Cell Biol. 20, 665–680 10.1038/s41580-019-0133-3 [DOI] [PubMed] [Google Scholar]
- 52.Wu, S., Hong, L., Wang, Y., Yu, J., Yang, J., Yang, J.et al. (2020) Kinetics of the conformational cycle of Hsp70 reveals the importance of the dynamic and heterogeneous nature of Hsp70 for its function. Proc. Natl Acad. Sci. U.S.A. 117, 7814–7823 10.1073/pnas.1914376117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhuravleva, A., Clerico, E.M. and Gierasch, L.M. (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 10.1016/j.cell.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng, W., Clerico, E.M., McArthur, N. and Gierasch, L.M. (2018) Allosteric landscapes of eukaryotic cytoplasmic Hsp70s are shaped by evolutionary tuning of key interfaces. Proc. Natl Acad. Sci. U.S.A. 115, 11970–5 10.1073/pnas.1811105115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauer, D., Meinhold, S., Jakob, R.P., Stigler, J., Merkel, U., Maier, T.et al. (2018) A folding nucleus and minimal ATP binding domain of Hsp70 identified by single-molecule force spectroscopy. Proc. Natl Acad. Sci. U.S.A. 115, 4666–4671 10.1073/pnas.1716899115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandal, S.S., Merz, D.R., Buchsteiner, M., Dima, R.I., Rief, M. and Žoldák, G. (2017) Nanomechanics of the substrate binding domain of Hsp70 determine its allosteric ATP-induced conformational change. Proc. Natl Acad. Sci. U.S.A. 114, 6040–6045 10.1073/pnas.1619843114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imamoglu, R., Balchin, D., Hayer-Hartl, M. and Hartl, F.U. (2020) Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 11, 365 10.1038/s41467-019-14245-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee, J.H., Zhang, D., Hughes, C., Okuno, Y., Sekhar, A. and Cavagnero, S. (2015) Heterogeneous binding of the SH3 client protein to the DnaK molecular chaperone. Proc. Natl Acad. Sci. U.S.A. 112, E4206–E4215 10.1073/pnas.1505173112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sekhar, A., Rosenzweig, R., Bouvignies, G. and Kay, L.E. (2015) Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl Acad. Sci. U.S.A. 112, 10395–10400 10.1073/pnas.1508504112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mashaghi, A., Bezrukavnikov, S., Minde, D.P., Wentink, A.S., Kityk, R., Zachmann-Brand, B.et al. (2016) Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 10.1038/nature20137 [DOI] [PubMed] [Google Scholar]
- 61.Avellaneda, M.J., Franke, K.B., Sunderlikova, V., Bukau, B., Mogk, A. and Tans, S.J. (2020) Processive extrusion of polypeptide loops by a Hsp100 disaggregase. Nature 578, 317–320 10.1038/s41586-020-1964-y [DOI] [PubMed] [Google Scholar]
- 62.Suren, T., Rutz, D., Mößmer, P., Merkel, U., Buchner, J. and Rief, M. (2018) Single-molecule force spectroscopy reveals folding steps associated with hormone binding and activation of the glucocorticoid receptor. Proc. Natl Acad. Sci. U.S.A. 115, 11688–11693 10.1073/pnas.1807618115 [DOI] [PMC free article] [PubMed] [Google Scholar]