Microbiomes form intimate functional associations with their hosts. Much has been learned from correlating changes in microbiome composition to host organismal functions.
KEYWORDS: Caenorhabditis elegans, host-pathogen interactions, innate immunity, microbiome
SUMMARY
Microbiomes form intimate functional associations with their hosts. Much has been learned from correlating changes in microbiome composition to host organismal functions. However, in-depth functional studies require the manipulation of microbiome composition coupled with the precise interrogation of organismal physiology—features available in few host study systems. Caenorhabditis elegans has proven to be an excellent genetic model organism to study innate immunity and, more recently, microbiome interactions. The study of C. elegans-pathogen interactions has provided in depth understanding of innate immune pathways, many of which are conserved in other animals. However, many bacteria were chosen for these studies because of their convenience in the lab setting or their implication in human health rather than their native interactions with C. elegans. In their natural environment, C. elegans feed on a variety of bacteria found in rotting organic matter, such as rotting fruits, flowers, and stems. Recent work has begun to characterize the native microbiome and has identified a common set of bacteria found in the microbiome of C. elegans. While some of these bacteria are beneficial to C. elegans health, others are detrimental, leading to a complex, multifaceted understanding of bacterium-nematode interactions. Current research on nematode-bacterium interactions is focused on these native microbiome components, both their interactions with each other and with C. elegans. We will summarize our knowledge of bacterial pathogen-host interactions in C. elegans, as well as recent work on the native microbiome, and explore the incorporation of these bacterium-nematode interactions into studies of innate immunity and pathogenesis.
INTRODUCTION
Nematodes are the most abundant multicellular organism on Earth and consist of over 25,000 species, both free-living and parasitic (1). Caenorhabditis elegans is a free-living nematode that can be found in the natural environment worldwide (2–6). It feeds on a variety of bacteria found in rotting organic matter and is most commonly isolated from rotting fruits, flowers, and stems (reviewed in reference 7). In the natural environment, C. elegans is in constant contact with many other organisms, including other small invertebrates, bacteria, and fungi. C. elegans travels between locations via vectors, such as isopods and snails (4, 8). Nematodes are also prey to a variety of insects and fungi, in addition to playing host to a number of pathogenic and symbiotic bacteria (3; reviewed in reference 7).
C. elegans is an excellent genetic model organism for studying many biological processes, including development, cell biology, innate immunity, and neurobiology. Some of the features that make C. elegans a tractable model system include its many progeny (up to 300), short generation time (3.5 days), small size (1 mm), and transparency, allowing for visualization of development, bacterial colonization, and protein localization and expression. In addition, there are a variety of tools for determining C. elegans health, including fecundity, life span, and stress response assays. C. elegans is also amenable to genetic manipulation, allowing for an in-depth understanding of the genetics underlying host-microbe interactions. In the laboratory, C. elegans can be grown on plates seeded with Escherichia coli OP50. This artificial lab setting differs significantly from the natural environment and lacks ecologically important biotic components. In addition, many recent studies have determined that bacteria, both in the surrounding environment and in a microbiome of many organisms, influence behavior, aging, and overall health (9–11). These observations led to studies investigating the effects of diverse microorganisms, including bacteria, fungi, and viruses, on C. elegans.
This review will begin with an overview of types of C. elegans-bacterium interactions and then focus more specifically on innate immunity in C. elegans in response to bacteria. The remaining sections will review more recent studies characterizing bacteria found in the natural environment of C. elegans and the role of pathogens in C. elegans’ environment. Although C. elegans encounters numerous pathogenic microbes in its natural environment, including bacteria, fungi, and viruses, extensive identification and characterization of fungi and viruses in C. elegans’ environment has yet to be performed. Therefore, we focus here on C. elegans interactions with bacterial pathogens.
C. ELEGANS-BACTERIUM INTERACTIONS
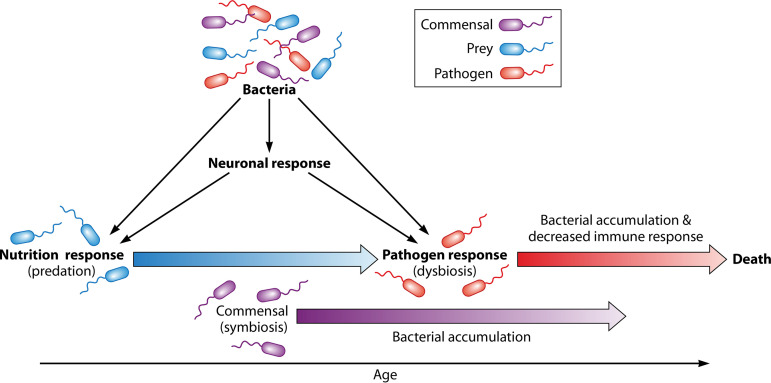
Bacteria interacting with C. elegans can be classified into three different categories: (i) as prey, not influencing the host and having a detrimental relationship with the host; (ii) as mutualists, where both host and bacteria benefit from the association; and (iii) as pathogens, where the bacteria have a detrimental effect on the host. These interactions are dynamic, and a single species can fit into more than one category. This compound relationship causes various responses to bacteria, including a neuronal response, a nutrition response, and a pathogen response (12) (Fig. 1). The neuronal response, or more specifically the olfactory response, is the detection of bacteria as either food or pathogen and leads to further neuronal and endocrine signaling that can affect behavior and longevity (12, 13). For example, C. elegans was shown to preferentially choose food that promotes growth, and this behavior is dependent on the amphid AIY neurons (14). Another study found that tyramine produced by the commensal bacteria Providencia can alter C. elegans neurosensory behavior that is dependent on the octopamine receptor in ASH neurons (15). There is also evidence that neuroendocrine signaling is involved in recognition of pathogens. For example, C. elegans can learn to avoid pathogenic bacteria, such as Pseudomonas aeruginosa and Serratia marcescens, that are dependent on a serotonin signaling pathway (16) and a neurotransmitter NPR-1, in the case of P. aeruginosa (17). Furthermore, signaling between neurons and the intestine plays a role in the immune response to some pathogenic bacteria (18, 19).
FIG 1.
Dynamic interactions between C. elegans and bacteria. The C. elegans response to bacteria involves interactions among three response types: neuronal, nutritional, and pathogenic. The overlapping shifts from predator-prey to pathogen-host as C. elegans age are also shown.
The nutrition response to bacteria is based on both nutrient availability and nutrient quality (12, 20). More specifically, C. elegans requires metabolically active bacteria for optimal fitness (21). C. elegans will arrest development at multiple life cycle points based on the presence of bacteria (20). In addition, some bacteria, including Bacillus anthracis and Bacillus megaterium, are physically harder to ingest, and C. elegans can display lawn avoidance behavior in response to indigestible food (13, 22). C. elegans also respond to different bacterially derived nutrients. For example, C. elegans requires several essential amino acids from its food for survival, and sensing of these amino acids occurs via the DAF-2/16 insulin-like pathway and the TOR pathway (23). More specifically, providing ethanol plus amino acids to starved L1 worms triggered release from developmental arrest (23). Vitamin B12 is produced by some bacteria and is required for many metabolic reactions. Bacterial food sources lacking vitamin B12 affect gene expression in C. elegans, resulting in slowed development and a decrease in fecundity (24, 25).
The pathogen response depends on the activation of several innate immune, defense, and stress pathways that are discussed in detail in the next section. A gradual shift from predator-prey to pathogen-host often occurs as C. elegans ages and correlates with accumulation of bacteria within the intestine (26). This change involves three phases: (i) predation, primarily during development, which involves mastication, or breakdown, of bacteria in the pharyngeal grinder, followed by uptake of nutrients from bacterial cell material in the intestine; (ii) symbiosis, primarily in young adults, in which live bacteria that are able to survive pharyngeal grinding inhabit the intestine and provide nutrients to the nematode through metabolism (21, 23–25); and (iii) dysbiosis, primarily in older adults, where bacteria accumulate in the intestine and cause damage to tissues (27) (Fig. 1). These phases are dynamic and can occur simultaneously, with progression through these phases being largely dependent on the food source. In addition, these stages are dependent on several factors, including the efficiency of the pharynx, the ability of bacteria to proliferate in the intestine, and the ability of the host to reduce bacterial accumulation through digestion and/or defense responses.
The neuronal, nutrition, and pathogen responses in C. elegans are not mutually exclusive, and the interplay of these responses leads to a multifaceted relationship between C. elegans and bacteria. Furthermore, C. elegans encounters a multitude of bacteria in the natural environment, with the dynamic and simultaneous responses becoming even more complex. Thus, to understand the complexity of C. elegans-bacterium interactions in nature, the most tractable approach is to first simplify these interactions to individual bacterial species and responses. For example, one particularly interesting and widely studied relationship is C. elegans’ response to pathogens. Determination of the behavioral and genetic changes C. elegans undergoes upon exposure to virulent bacteria will provide insight into the underlying mechanisms of the pathogen response.
C. ELEGANS PATHOGEN RESPONSE
The study of C. elegans genes and pathways involved in pathogen response is medically relevant, since many of these genes and pathways are conserved. Since approximately 40% of genes found in C. elegans have orthologs in humans, many processes in C. elegans are conserved in mammals (28). For example, conservation of innate immune proteins between nematodes, insects, and mammals has revealed important immune factors in C. elegans, indicating similarities between innate immunity in C. elegans and other metazoa (reviewed in references 29, 30, 30, 31, and 32). However, C. elegans does not have dedicated innate immune cells as are found in vertebrates. Therefore, a primary component of its immune response occurs through physical barriers where pathogenesis begins, such as the cuticle and intestine (28).
In general, the innate immune response involves three steps, each of which is carried out by different classes of proteins (28). The first step is the recognition of the pathogen. This step can be species specific, by proteins that recognize particular toxins or bacterial proteins, or more generally respond to pathogen-induced damage. The second step involves signaling pathways which activate downstream proteins and eventually transcription factors (Fig. 2). The final step involves the expression of effector genes that are regulated downstream of signaling pathways, including antimicrobial peptides (AMPs) (28). Because the scope of this review covers bacterial pathogens and bacteria found in the microbiome of C. elegans (primarily the intestine), our emphasis is on the response to bacterial pathogens in the intestine. However, shared and specific innate immune responses to fungi and viruses occur, as has been studied and reviewed extensively (33–35).
FIG 2.
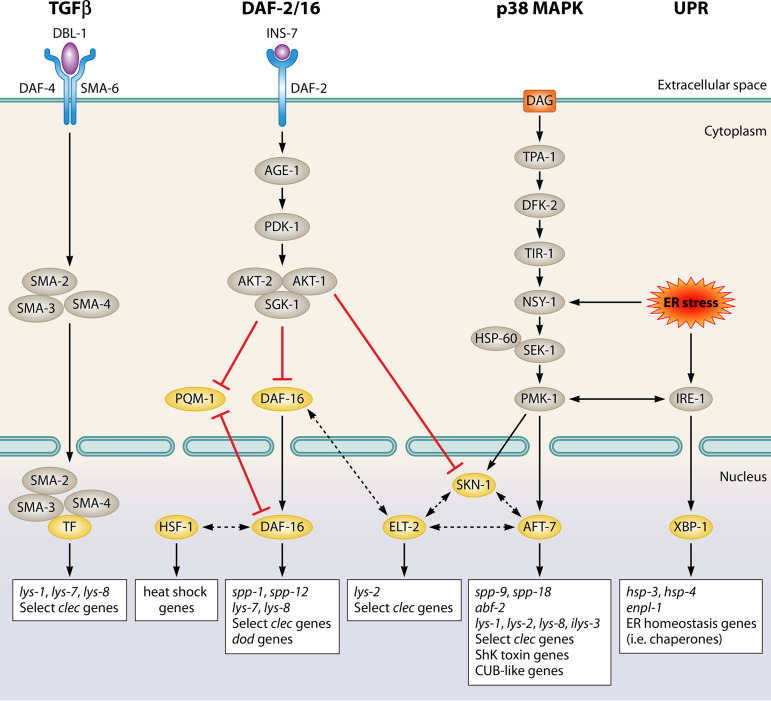
Overview of innate immune and defense pathways: the TGF-β pathway, DAF-2/16 insulin-like pathway, p38 MAPK pathway, and unfolded protein response (UPR) pathway. Purple indicates ligands, blue indicates receptors, gray indicates downstream signaling components, and gold indicates transcription factors. Boxes below pathways identify selected major downstream effectors (65, 80, 82, 83, 86, 88, 93, 96, 99, 104, 109). DAG, diacylglycerol; TF, transcription factor.
Recognition Proteins
Pathogen recognition can involve direct recognition of structural components or secreted proteins of the pathogen, termed microbe-associated molecular patterns (MAMPs), or indirectly via perturbations induced by infection, termed damage-associated molecular patterns (DAMPs). Interestingly, many conserved receptors involved in MAMP recognition, including peptidoglycan recognition proteins, Gram-negative binding proteins, or nucleotide-binding oligomerization domain (NOD)-like receptors, are not found in C. elegans (31). Toll-like pathway receptors play a role in immunity in insects and higher-order metazoa (36, 37). The sole Toll-like receptor (TLR) in C. elegans, TOL-1, is vital for proper development and function of sensory neurons (38). Although the p38 mitogen-activated protein kinase (MAPK) pathway downstream of the TLR is important for response to many pathogens in C. elegans, TOL-1 does not appear to play a role in recognition of or response to Staphylococcus aureus or Pseudomonas aeruginosa (30, 39, 40). The lack of traditional MAMP recognition mechanisms suggests that pathways involved in innate immunity in C. elegans may not be responding directly to the pathogen but instead to cell damage or other stressors that are a consequence of infection.
Exposure to several pathogens, including B. thuringiensis, S. aureus, and P. aeruginosa, leads to intestinal distension (39, 41). Intestinal damage by B. thuringiensis is at least partially dependent on pore-forming cryotoxins (42). Singh and Aballay found that bloating of the intestinal lumen in C. elegans due to bacterial colonization leads to avoidance of pathogens and activates innate immune signaling (43). The mechanisms for how damage induces downstream defense and immune signaling is largely unclear, but Singh and Aballay found that neuroendocrine signaling through NPR-1 and DAF-7 was required for pathogen avoidance behavior (43).
In addition, reactive oxygen species (ROS) are generated during infection due to mitochondrial damage and upon Enterococcus faecalis infection through the NADPH oxidase/dual oxidase BLI-3 (44, 45). ROS then acts as a signal for downstream defense responses, including activation of the DAF-16 and p38 MAPK pathways (reviewed in reference 46).
In other animals, C-type lectin domain (CTLD) containing proteins recognize and bind bacterial cell walls via pathogen recognition receptors (47). In C. elegans, two C-type lectin domain proteins, CLEC-39 and CLEC-49, can directly bind S. marcescens, and mutations that inactivate these genes cause increased susceptibility to S. marcescens infection (48). In another case, mutations in clec gene C54G4.4 results in increased avoidance behavior in response to B. thuringiensis, suggesting that it plays a negative role in B. thuringiensis lawn avoidance (49). However, CTLD containing proteins appear to play a variety of roles in innate immune response in C. elegans, and whether these roles are implicated in pathogen recognition or as downstream effectors is still largely unknown. For example, many clec genes are also differentially expressed in response to several pathogens and are regulated by innate immune pathways (50). Therefore, it is unclear whether these proteins play a role in recognizing pathogens or function as downstream antimicrobial peptides.
The lack of conserved receptors suggests that there may be noncanonical mechanisms involved in pathogen recognition. Recent evidence has shown that the nervous system may be involved in upstream signaling that leads to pathogen responses in the intestine. For example, INS-7, an insulin-like ligand that binds to and activates the insulin-like receptor DAF-2 (discussed below), is expressed mainly in neuronal cells and at low levels in the intestine (51). ins-7 expression is increased upon exposure to P. aeruginosa via excretion from dense core vesicles, leading to activation of the DAF-2 pathway and suppression of the DAF-16 transcription factor (18, 52). Mutations in ins-7 cause resistance to P. aeruginosa, and transgenic expression of INS-7 in neuronal cells alone is able to suppress ins-7 mutant resistance (18, 52).
In addition, several studies have suggested that neuronal G-protein coupled receptors (GPCRs), such as FSHR-1, NPR-1, NPR-9, and OCTR-1, are required for pathogen resistance and downstream innate immune signaling (reviewed in references 53, 54, and 55). The mechanism for how these neuronal receptors activate the immune response and resistance to pathogens remains unclear, but it is possible that neuronal signaling serves as an intermediate between pathogen recognition and innate immune pathway activation in the intestine.
Signaling Pathways
p38 MAPK pathway.
MAPK pathways play significant roles in a variety of cellular responses such as development, differentiation, stress, and apoptosis (56). The p38 MAPK pathway in mammals is activated by cytokines and other stressors in immune cells (57). Analysis of C. elegans mutants that enhanced susceptibility to P. aeruginosa PA14 led to the identification of conserved components of the p38 MAPK signaling pathway, including SEK-1, NSY-1, and PMK-1 (30). NSY-1, a MAPK kinase kinase (MAPKKK), phosphorylates and activates SEK-1, a MAPKK, which signals via the MAPK PMK-1 (30) (Fig. 2). This pathway acts cell autonomously in the intestine in response to bacterial pathogens and in the epidermis in response to fungal pathogens and wounding (58, 59). In mammalian studies, a variety of transcription factors were identified as direct targets of PMK-1, many of which are conserved in C. elegans (60). A forward genetic screen identified AFT-7, ortholog of human AFT2, as being an important p38 MAPK transcription factor regulating transcription of innate immune genes (61). AFT-7 functions as a transcriptional repressor until it is phosphorylated by PMK-1, when it then becomes a transcriptional activator of innate immune genes (61, 62) (Fig. 2).
In Drosophila, TLRs and Toll-Interluekin-1 Receptor (TIR) domain adaptor proteins function upstream of p38 MAPK cascades (36). TIR domain adapter proteins specifically bridge the gap between TLR and MAPK signaling, initiating p38 MAPK pathways. In C. elegans, the TLR pathway does not appear to play a role in intestinal pathogenesis (40, 63), but the sole TIR domain protein in C. elegans, TIR-1, does activate MAPK signaling upstream of PMK-1 (64). The mechanism of TIR-1 activation remains unclear but may be related to diacylglycerol (DAG). TPA-1, a protein kinase C in C. elegans, is activated by DAG and phosphorylates DKF-2, a protein kinase D (63, 65) (Fig. 2). Mutations in dkf-2 result in increased susceptibility to P. aeruginosa and E. faecalis (65). DKF-2 is required for immune response via p38 MAPK signaling, since overexpression of DKF-2 causes increase in phosphorylated PMK-1, but DKF-2 acts independently of PMK-1 as well (65). The mechanism of activation between DKF-2 and MAPK signaling remains unknown, but DKF-2 may directly phosphorylate TIR-1 (28, 63).
HSP-60, a mitochondrial chaperone and heat shock protein, mediates resistance to P. aeruginosa and activates p38 MAPK signaling (66). Further evidence found that HSP-60 binds to and stabilizes SEK-1 in the cytosol, thereby modulating immunity to bacterial pathogens (66).
Pore-forming toxins (PFTs), produced by many human bacterial pathogens, have also been shown to activate MAPK signaling in several organisms, including C. elegans, insects, and mammalian cells (reviewed in references 67 and 68). In C. elegans, mutations in pmk-1 and sek-1 caused increased susceptibility to PFTs produced by B. thuringiensis (69). In addition, a proteomic analysis of the response of C. elegans to a PFT produced by S. aureus revealed the involvement of MAPK pathways, as well as galectins and heat shock proteins (70).
DAF-2/16 insulin-like pathway.
The insulin-like signaling pathway was originally identified in C. elegans for its role in life span, reproduction, and regulating dauer entry, an alternative life stage that occurs under stressful environmental conditions (71, 72). Mutations in the sole insulin/IGF-1-like receptor daf-2 in C. elegans leads to a near doubling of life span when exposed to many bacteria, including pathogens such as P. aeruginosa, E. faecalis, Staphylococcus aureus, and Salmonella Typhimurium (73, 74). This effect is dependent on the Forkhead transcription factor DAF-16 (73). Other components of this pathway include the phosphoinositide 3-kinase, AGE-1, which is phosphorylated by DAF-2 resulting in the recruitment of kinases PDK-1, AKT-1, AKT-2, and SGK-1. PDK-1 phosphorylates AKT-1, AKT-2, and SGK-1, which then form a complex that phosphorylates DAF-16 (75), resulting in its localization to the cytoplasm, preventing it from entering the nucleus and regulating gene expression (reviewed in reference 76) (Fig. 2). When the pathway is inactivated (e.g., in daf-2 mutants), DAF-16 localizes to the nucleus, which results in upregulation of genes involved in longevity and stress resistance. PQM-1, another transcription factor downstream of DAF-2, has an antagonistic relationship with DAF-16, where nuclear localization of one transcription factor excludes nuclear localization of the other (77). Under normal conditions PQM-1 is primarily transcriptionally active and targets genes involved in growth, development, and proteostasis, whereas DAF-16 nuclear localization occurs under stress conditions (77, 78).
Although the DAF-2/16 pathway is involved in both longevity and defense responses, linking these responses, it also plays an important role in innate immunity. In fact, many of the genes targeted by DAF-16 have antimicrobial activities, such as lysozymes and detoxification enzymes (79, 80). Heat shock proteins are also downstream components of DAF-2/16 signaling that may play a role in innate immunity. HSF-1, a transcription factor that regulates a subset of heat shock proteins, is required for life span extension of daf-2 mutants (81) and activation of innate immune genes through DAF-2/16 pathway (82).
In addition, daf-2 mutant worms display a decrease in bacterial packing, suggesting that regulation of genes by DAF-16 defends against accumulation of bacteria in the intestine (74, 80). Interestingly, one study found that P. aeruginosa infection suppresses the activity of DAF-16, which is mediated by several P. aeruginosa virulence factors (52). However, this phenomenon appears to be pathogen specific, since exposure to S. marcescens, E. faecalis, and S. Typhimurium induce the expression of several DAF-16 targets that have putative antimicrobial activity (32, 52). In addition, pqm-1 is required for resistance to P. aeruginosa, suggesting PQM-1 targets may confer resistance in that case, rather than DAF-16 targets (83). Finally, recent evidence suggests that aging plays a role in DAF-2/16 regulated immunity, where DAF-16 becomes more important for immune activation and increased longevity later in adulthood (84). Clearly, there is much yet to be discovered regarding the complexities of the DAF-2/16 pathway involvement in pathogen response.
Unfolded protein response pathway.
The endoplasmic reticulum (ER) is responsible for protein processing, including folding and posttranslational modifications, and transportation. Perturbations resulting in accumulation of unprocessed proteins activates the unfolded protein response (UPR). The IRE-1-XBP-1 branch of the UPR regulates expression of genes involved in ER homeostasis, leading to defense responses and increased longevity (85). The UPRER is conserved in animals, as well as some fungi. This pathway involves activation of IRE-1, which leads to the alternative splicing and activation of xbp-1 mRNA in response to accumulation of unfolded proteins in the ER (Fig. 2). In addition to unfolded proteins, activation of IRE-1-XBP-1 occurs in response to several pathogens likely to protect against ER stress resulting from innate immune responses (86). For example, activation of the UPRER occurs in response to the pore-forming toxins of B. thuringiensis, Cry5, and ire-1 and xbp-1 mutations cause hypersensitivity to Cry5 (87). In addition, intestinal infection of C. elegans with P. aeruginosa induces expression of the heat shock protein HSP-4, a downstream effector of the IRE-1-XBP-1 pathway, and xbp-1 mutants are susceptible to P. aeruginosa and have perturbed ER morphology (86). Both UPRER signaling and p38 MAPK signaling in response to P. aeruginosa are dependent on the oligosaccaryl transferase complex, which mediates protein glycosylation in the ER (88).
In addition to UPR in the ER being involved in innate immunity, another study identified an overlap of upregulated genes in response to mitochondrial stress and infection to P. aeruginosa (89). The activation of several of these genes was dependent on the mitochondrial UPR transcription factor ATFS-1 (89). Therefore, the mitochondrial UPR is also able to protect against pathogens that induce mitochondrial stress by coupling antimicrobial and mitochondrial homeostasis gene expression.
TGF-β pathway.
The transforming growth factor β (TGF-β) pathway is involved in development and embryogenesis (90, 91). In mammals, the TGF-β pathway is required for T cell development and differentiation (reviewed in reference 92). In C. elegans, mutation of the TGF-β ligand dbl-1, causes increased susceptibility to S. marcescens and Salmonella Typhimurium (32, 74). In addition, several families of antimicrobial peptides, including C-type lectin domain proteins and lysosomes, are regulated by this pathway (90, 93). The canonical DBL-1/TGF-β pathway components include the type I and type II receptors SMA-6 and DAF-4, as well as the Smad signal transducers SMA-2, SMA-3, and SMA-4 (91) (Fig. 2). Expression of several AMPs are dependent on SMA-2 (94). Further details on the involvement of other downstream components are largely unknown; however, mutations in dbl-1, sma-6, sma-2, sma-3, and sma-4 in C. elegans cause increase susceptibility to S. maltophilia (95).
Pathway cross talk.
Each of these pathways are complex in nature, involving a variety of coregulators that have been shown to mediate pathway cross talk. For example, the increase in life span of daf-2 mutants is dependent on the p38 MAPK pathway, suggesting that it acts in parallel or downstream of DAF-2/16 (96). In addition, many DAF-16 targets contain a GATA motif, termed the DAF-16 associated element (DAE) (80). Two GATA transcription factors, ELT-3, specific to the epidermis, and ELT-2, specific to the intestine, both regulate expression of DAF-16 targets in a tissue-specific manner (97). Conflicting results on whether ELT-2 or ELT-3 can suppress the longevity of daf-2 mutants suggests that the interplay of these pathways is complex and condition specific (97–99). A meta-analysis of gene expression studies in response to pathogens found that the GATA motif is the most enriched across these studies (100). Mutations in elt-2 cause increased mortality to P. aeruginosa, S. enterica, and E. faecalis and bacterial distention of the intestine (83, 99), and ELT-2 is required for recovery from P. aeruginosa infection (101). ELT-2 activity seems to be specific to pathogen response, since elt-2 mutants are not susceptible to oxidative stress, heat stress, or cadmium exposure (83). Further, elt-2 appears to have a strain-specific role in C. elegans in response to pathogenic B. thuringiensis, since knocking down elt-2 increased survival in response to one strain of B. thuringiensis but decreased survival in response to another (102).
SKN-1, a putative transcription factor involved in stress responses in the intestine, can be phosphorylated by AKT-1, resulting in repression of SKN-1 target gene expression (103) (Fig. 2). Mutations in skn-1 in a daf-2 background suppress the longevity phenotype of daf-2 mutants, suggesting that SKN-1 contributes to increased life span and stress responses (103). To further complicate this response, Block et al. suggest a complex interplay between ELT-2, SKN-1, and ATF-7, where a combination of factors is required for expression of particular immune genes (104) (Fig. 2).
UPRER activation is also dependent on p38 MAPK signaling (86, 87). However, whereas p38 MAPK signaling functions by reducing accumulation of pathogenic bacteria, it appears that the role of the UPRER is to protect from ER damage induced by the immune response (86).
Antimicrobial Peptides
Most known antimicrobial peptides have been identified by homology to other AMPs and by expression profiling. Genes that are commonly differentially expressed in response to bacterial pathogens include caenopores, lysozymes, defensin-like AMPs, and C-type lectin domain proteins (reviewed in reference 29). The function of these proteins in C. elegans has largely been predicted based on sequence structure, and further functional analyses of these AMPs have not been well studied.
Caenopores, or saposin-like proteins in C. elegans, share a structural similarity with saposin-like proteins (SAPLIPS) in protozoa and mammals (105). Although there are 28 saposin-like protein family (SPP) proteins identified in C. elegans, only a few have been identified as immune effectors. spp-9 and spp-18 are regulated by DKF-2 and are upregulated by P. aeruginosa exposure (65). spp-1 and spp-12 are regulated by DAF-16 (106), and mutations in spp-12 result in increased susceptibility to pathogenic B. thuringiensis, whereas mutations in spp-1 result in decreased susceptibility (107). Further, knockdown of spp-1 and spp-5 results in accumulation of bacteria in the intestine (105, 106). Functional analysis of SPP-5 revealed its ability to form pores in and damage bacterial cell walls (105).
Lysozymes are involved in the hydrolysis of peptidoglycan, a major component of bacterial cell walls. Not surprisingly, lysozymes also play a role in digestion (reviewed in reference 29). C. elegans lysozyme genes are classified into 10 protist types (lys-1 to lys-10) and 5 invertebrate types (ilys-1 to ilys-5). This is the largest class of genetically diverse lysozymes found in any organism to date (108). All lysozymes studied to date are expressed mainly in the intestine (reviewed in reference 108). Similar to caenopores, many lysozymes are regulated by defense pathways such as TGF-β (lys-1, lys-7, and lys-8), DAF-2/16 (lys-7 and lys-8), and p38 MAPK (lys-1, lys-2, lys-8, and ilys-3) (80, 93, 96, 109). Many lysozyme proteins are differentially expressed in response to pathogens (29, 110, 111). In fact, ilys-1 and lys-9 are the only lysozymes that are not differentially expressed upon bacterial pathogen exposure (29). In addition, recombinant ILYS-3 exhibits hydrolytic activity and affects cell wall integrity of Gram-positive bacteria, presumably in the pharynx and intestines, where it is expressed (109).
Defensin-like peptides, termed antibacterial factors ABF-1 to ABF-6 in C. elegans, were identified based on sequence homology to proteins of Ascaris suum, an intestinal parasitic nematode (112). ABF-2 displays in vitro antimicrobial activity and knockdown increases pathogen accumulation (106, 112). Regulation of these genes is not well understood, but the M-box motif-class transcription factor HLH-30 appears to be required for abf-2 expression, as well as several other antimicrobial genes, in response to S. aureus (113).
C-type lectin domain (CTLD) proteins were originally characterized for their Ca+-dependent carbohydrate binding ability (reviewed in reference 114). However, this superfamily has now grown to include proteins with structural similarities that do not display these functional characteristics (114). In C. elegans, CTLD proteins are the most diverse group of effector molecules, containing 283 members, 81% of which contain signal peptides, suggesting they are secreted (50). Although CLEC-39 and CLEC-49 have been shown to directly bind to S. marcescens (48), CTLD proteins have not been shown to have antimicrobial activities. Evidence of interaction between several CTLD proteins and LYS-7 could suggest downstream signaling or coregulation of immune partners (115). In addition, CTLD proteins exhibit differential expression in response to a variety of pathogens (29, 39, 93, 96, 111). Mutation or knockdown of many CTLD genes have been analyzed and result in increased and decreased susceptibility to pathogens (39, 48, 49, 116). Although there is a clear role for CTLD proteins in innate immune response, their functional roles in C. elegans are not well known (50).
Analyses of expression data from many experiments in response to a variety of pathogens revealed that expression of effectors is taxon specific, but some members of common classes of AMPs, specifically caenopores and lysozymes, are differentially expressed in response to almost all pathogens. Defensin-like peptides, on the other hand, play a less prominent and more species-specific role in response to bacterial pathogens (29).
In summary, innate immune responses to pathogenic bacteria have been widely studied in C. elegans and have led to the identification of several major, well-defined pathways involved in defense and immune response and their downstream genes. These studies have revealed that many of the pathways are complex, with cross talk between pathways and the genes they regulate. In addition, although many common innate immune pathways and effectors have been identified, there are differences in responses to different bacteria and even strains of bacteria. For example, only 11% overlap in differentially expressed genes was found in a comparison of the responses to the intestinal pathogens S. marcescens, E. faecalis, and Photorhabdus luminescens (117). Other studies found that the C. elegans response to pathogenic strains of bacteria of the same species involve both a common response to both strains and different responses to each of the strains (102, 118). This phenomenon could be due to species- or strain-specific responses to different pathogens or to the ability of bacteria to manipulate different host responses. Therefore, it is essential to study a variety of pathogens, their virulence factors, and responses to these pathogens in order to fully understand the complexity of genetic mechanisms underlying pathogen defense.
BACTERIA FOUND IN THE NATURAL ENVIRONMENT OF C. ELEGANS
Previously, many bacteria used to study C. elegans-bacterium interactions, including S. enterica, P. aeruginosa, S. aureus, and E. faecalis, were chosen because of their convenience in the laboratory setting or their implication in human health rather than their native interactions with C. elegans in the environment. However, in order to understand pathogen response and other host-bacterium interactions, it is necessary to focus efforts on species that have been found to interact with C. elegans in their native environments. C. elegans interact intimately with many bacteria in their environment, and this symbiosis between microbes and host can lead to adaptation and evolution of both species. For example, a study that coevolved C. elegans with a natural pathogen, B. thuringiensis, found that the worms became more resistant to killing by B. thuringiensis when grown together over many generations and this led to an increase in genetic changes in both bacteria and host compared to bacteria and host maintained individually (119). While the previous example refers to antagonistic coevolution, where the organisms evolve as separate entities with opposing goals, coevolution of bacteria and host can also occur in mutualistic relationships, where increased fitness under favorable conditions leads to genetic changes. This later idea supports the hologenome theory, that suggests the genetic information of microbes and their host act as one unit of selection in evolution (120). A key feature of the hologenome theory is transmission of microbes through host generations, and it is important to note that to date, there is no evidence of vertical transmission of microbes in C. elegans across generations. However, because of the essential nature of the C. elegans-microbiome relationship, the effect of microbes on C. elegans fitness, and the ability of these interactions to lead to genetic changes in both host and microbes, it is intriguing to consider the host and microbiome as one unit.
Characterization of the Native Microbiome of C. elegans
Several groups recently determined the bacterial repertoire associated with substrates where C. elegans were found as well as within the worm microbiome (3, 121, 122). Berg et al. created artificial microcosms to simulate natural environments using soil supplemented with various produce, including plants and fruit, and then subjected wild-type worms to these microcosms for several days before characterizing bacterial composition within both worms and soils (121). Samuel et al. identified the bacterial community associated with substrates where C. elegans were collected (122). Dirksen et al. combined the approaches of the previous studies by analyzing the microbial communities in the native habitats of Caenorhabditis species, as well as the microbiome of sampled nematodes from those habitats (3). These researchers also performed “lab enrichment” on some worms, in which the microbial communities of worms found in the natural environment were determined after growing them on E. coli OP50 for several weeks in order to determine bacterial species that persist in the worm and do not simply pass through the intestine. In all three studies, researchers found highly diverse microbiomes within the substrates and within C. elegans, but diversity within the worms was lower than within the substrates (3, 121, 123). In addition, worm microbiomes, regardless of worm origin or experiment, were more similar to each other than to the microbes associated with the substrates (123), suggesting some constraints on microbiome assembly, either by competition of bacteria within the intestine or selection of bacteria by the worm. Furthermore, a recent study found that many of the bacteria found in worm microbiomes are not found in their corresponding substrates, suggesting that these bacteria can persist in the worm microbiome for some time (124).
The similarity of microbial repertoires across experiments resulted in the identification of 14 bacterial families present in all natural worm microbiomes: Xanthomonadaceae, Pseudomonadaceae, Moraxellaceae, Enterobacteriaceae, Oxalobacteraceae, Comamonadaceae, Sphingomonadaceae, Acetobacteraceae, Rhodobacteraceae, Sphingobacteriaceae, Weeksellaceae, Flavobacteriaceae, Microbacteriaceae, and Actinomycetales (123) (Fig. 3). Interestingly, many of these families are also found in higher abundance within the worm microbiome than within the substrate, suggesting they can colonize the intestine (3, 123). Recently, a collaborative consortium has established a 12-species experimental microbiome, termed CeMbio, based on common, representative strains found in C. elegans microbiomes to serve as a tool for future work on microbiome interactions and assembly (125).
FIG 3.
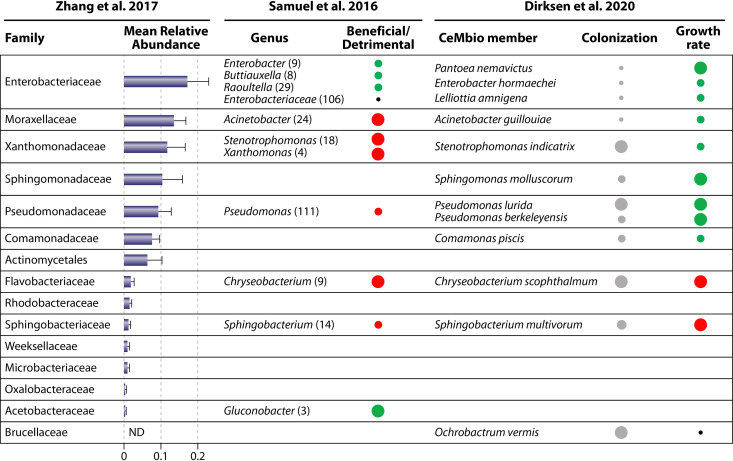
Functional analysis of C. elegans microbiome members. Functional analysis of bacteria representative of common families identified across C. elegans microbiomes (102) and CeMbio members (104). Mean relative abundance indicates the relative abundance of each bacterial family across 23 C. elegans isolates in samples from the natural environment (102). The beneficial/detrimental status was determined for each isolate (the number of isolates for each genus is indicated in parentheses) based on growth rate and induction of stress reporters of C. elegans upon exposure to each isolate (101). Colonization was calculated by analysis of the CFU in individual C. elegans intestines grown on each bacteria at 1 and 3 days of adulthood. Growth rate indicates time to adulthood of C. elegans upon exposure to each bacteria compared to E. coli OP50 (104). The dot size represents the value for each functional assay, green indicates the beneficial/higher growth rate, red indicates detrimental/lower growth rate, and black indicates similarity to OP50. ND, no data. See references 123 (Zhang et al. 2017), 122 (Samuel et al. 2016), and 125 (Dirksen et al. 2020).
Functional Analysis of Microbiota
Previous studies have determined that the microbiome plays a role in life span, aging, and disease. In humans, gut microbiome dysbiosis is linked to several diseases, including colorectal cancer, diarrheal diseases, liver diseases, and diabetes (reviewed in reference 126). In addition, microbiota of elderly people correlated with health markers such as frailty and nutritional status (127).
In C. elegans, microbiota are capable of a variety of metabolic functions, and collectively is able to produce all necessary metabolites needed for C. elegans’ survival (128). Certain metabolites, such as vitamin B12, are only produced by some bacteria, such as Ochrobactrum and Pseudomonas, which could affect nematode fitness (125, 128). Several studies have directly analyzed how bacteria in C. elegans’ natural environment affect their health. Samuel et al. determined the growth rate and induction of stress reporters in response to 565 bacterial isolates found in the natural habitat of C. elegans and used these data to characterize each isolate as either “beneficial,” “detrimental,” or “intermediate” (122) (Fig. 3). Of all 565 isolates, approximately 40% were beneficial, 40% were intermediate, and 20% were detrimental. A range of effects across bacterial genera exists, suggesting differences between strains of closely related bacteria could account for differences in their impact on C. elegans (122). Dirksen et al. also characterized the effects of strains from C. elegans’ microbiome on aspects of its life history and found that a majority (67%) of the 24 strains tested decreased population size of C. elegans (3). Most of these detrimental isolates belonged to Actinobacteridae, Bacilli, Flavobacteriia, and Sphingobacteriia (3). Of the 12 strains chosen for the CeMbio experimental microbiome, all isolates are able to colonize the intestine, but only two slow the growth of C. elegans (125) (Fig. 3).
Utilizing Microbiome Bacteria To Understand Natural Host-Pathogen Interactions
Based on these functional analyses, it is clear that C. elegans encounter bacteria in their microbiome and environment that are pathogenic, but little is known about the molecular mechanisms of these pathogens or their interactions with other species. Samuel et al. found that mixing detrimental strains with beneficial strains found in the environment resulted in protective functions between some species, but not others (122). When mixed in a culture with other core microbiome components, the detrimental effects seen by individual isolates is mitigated, and these mixed cultures are more beneficial for C. elegans (3, 125, 129). This suggests an interplay between species within the microbiome and their interactions with C. elegans (i.e., an innate immune response). For example, Montalvo-Katz et al. found that two species found in compost samples, Pseudomonas mendocina and Bacillus megaterium, both conferred resistance to P. aeruginosa PA14, and the protective function of P. mendocina appears to be an early activation of p38 signaling that primes the immune response to PA14 (130). In addition, several strains of Enterobacter cloacae isolated from C. elegans protected C. elegans upon E. faecalis infection but did not protect C. briggsae (131). There is also evidence for interkingdom interactions within the microbiome, since several strains of Pseudomonas displayed antifungal effects in vitro, and one Pseudomonas strain decreased the susceptibility of C. elegans to fungal infection (3).
These studies have determined that bacteria found in the environment are able to play protective roles in response to clinical pathogens but suggests that they have similar functions in C. elegans natural habitat and within the microbiome. Interestingly, innate immune response does occur in C. elegans upon exposure to compost and associated bacteria (129), suggesting that C. elegans is defending against pathogens in its natural environment. This activated immune response could explain the lack of detrimental effects of a mixed microbiome culture, since C. elegans is able to combat the diluted effects of these pathogens. However, few studies have begun to characterize and molecularly analyze these natural pathogens. In one early study, two strains of Leucobacter were isolated from a Caenorhabditis worm, and each strain displayed distinct, diseased phenotypes in C. elegans (132). Interestingly, worms that were resistant to one strain of Leucobacter were hypersusceptible to the other strain and vice versa, suggesting a trade-off in susceptibility to these strains (132). Page et al. found that two environmental isolates of Chryseobacterium are detrimental to C. elegans (133). These researchers determined that these bacteria are able to break down the pharyngeal wall, and comparative genomics revealed differences in proteases between pathogenic and nonpathogenic strains that could be responsible for the virulence of these strains (133).
Stenotrophomonas is another mildly detrimental bacterium found in high abundance within C. elegans microbiomes (3, 122). A recent comparison of gene expression in C. elegans in response to two pathogenic environmental isolates of S. maltophilia and one nonpathogenic clinical isolate of S. maltophilia showed that C. elegans exhibit both common and strain-specific responses to S. maltophilia strains of various pathogenicities (118). One of these S. maltophilia strains, JCMS, was isolated in association with soil nematodes and evades the DAF-2/16 pathway (95). Another study looking at the transcriptional response to two pathogenic strains of B. thuringiensis also found that both common and strain-specific responses are displayed by C. elegans (102). In that study, the transcription factor ELT-2 was found to have contrasting effects on the two B. thuringiensis strains (102). Further analyses to understand the role of these pathogens within the microbiome will uncover a more representative role of these bacteria in their natural environment and their interaction with C. elegans.
CONCLUSIONS AND FUTURE DIRECTIONS
C. elegans is a major model organism used to uncover gene and protein functions and relate them to homologous functions in other organisms. However, despite decades of research, 40% of gene functions in C. elegans are still unknown (134). One reason for this is that the use of artificial laboratory settings to study C. elegans limits responses to environmental stimuli. C. elegans and bacteria engage in intimate relationships in their natural environment, so utilizing these interactions to understand gene function in C. elegans has already elucidated novel genetic mechanisms (reviewed in reference 134). Before high-throughput characterization of the microbiome, identification of organisms in the natural habitat of C. elegans through simple isolation has encouraged use of these species in further studies (118, 130, 132, 133). Further, recent extensive characterization of the microbial communities within C. elegans, as well as in the surrounding environment, has expanded our understanding of natural C. elegans-bacterium interactions and can be used for further study of microbiome interactions (3, 121, 122). However, because studies of C. elegans ecological interactions are only relatively recent, there is still much to be uncovered.
Of particular interest are interactions between C. elegans and detrimental bacteria, since this could shed light on novel innate immune responses in the host and bacterial virulence mechanisms. As highlighted in this review, much has been learned through studies with medically relevant pathogens. Studying interactions with naturally encountered pathogens can provide further insight into how C. elegans respond to pathogen infection, such as:
To what extent do the major C. elegans innate immune pathways function to combat natural pathogens? Which major pathways play the largest role in natural settings?
Do distinct effectors function in response to different natural pathogens? Recent work has suggested that C. elegans deploys both common and strain-specific responses to different bacterial taxa and strains of the same taxa (102, 118, 135). Does this hold for other natural pathogens?
Do the responses to novel natural pathogens help to elucidate functions of genes whose function has not yet been described?
What virulence mechanisms are employed by natural pathogens in their natural hosts? Do taxa related to natural pathogens employ similar or different virulence mechanisms?
Development of the 12-species experimental microbiome, CeMbio, can be used as a universal tool within the community and allows for direct manipulation of the microbiome (125). This advance opens the door to investigate new questions that are intractable in other study systems. Establishment and initial characterization of CeMbio has laid the groundwork for further studies to explore the role of various microbiome members within the microbial community, as well as on the host (125). Manipulation of the CeMbio microbiome through experiments that omit, add, or substitute bacteria within the community will help elucidate the roles of microbial taxa within the microbiome and the host:
Do microbiome components impact host fitness directly or through interactions with other microbes (i.e., competition, mutualism, pathogenesis, etc. between microbes)? For example, some components might function to limit the growth of other, perhaps more detrimental, microbes (125).
Do the roles fulfilled by microbiome components follow phylogenetic relationships or are they more related by the functions they perform within the community?
What role does the host play on shaping these microbial interactions?
Finally, we find detrimental bacteria in relatively high abundance in natural microbiomes (3, 122). Studying these natural pathogens both individually and as components of experimental microbiomes, we can address the following:
How does the incorporation of microbes of various pathogenicities into the microbiome affect the host? How does this affect the microbiome?
Are microbes that are detrimental to C. elegans also detrimental to other microbes?
What role do natural pathogens play in the microbiome?
As highlighted here, C. elegans has been used for studies involving innate immunity and bacterial virulence for many years, but only recently have we begun to intersect this research with microbiome composition and function. Future research exploring the questions above promises to further our understanding of gene function, host-microbiome interactions, and the evolution of innate immunity in nematodes and other organisms.
ACKNOWLEDGMENTS
This study was supported by the Kansas State University Division of Biology and the University Nebraska—Lincoln School of Biological Sciences. These funding bodies had no role in the design, analysis, interpretation, or writing of the manuscript.
We have no conflicts of interests in presenting this work.
We thank members of the Herman lab for useful comments on the manuscript.
This study was conceptualized by L.J.R. and M.A.H. The manuscript was drafted by L.J.R. L.J.R. and M.A.H. read, revised, and approved the final manuscript.
Biographies

Leah J. Radeke, M.S. received her bachelor’s degree in Biology from the University of Wisconsin-Eau Claire in 2015 and her M.S. degree in Biology from Kansas State University in 2018 under the supervision of Dr. Michael Herman. Her masters research focused on understanding host-pathogen interactions using C. elegans and Stenotrophomonas maltophilia strains of various pathogenicities, exploring both host responses and pathogen virulence mechanisms. Leah is now a Research Technologist II at the University of Nebraska—Lincoln in Dr. Michael Herman’s lab, studying C. elegans interactions with S. maltophilia and Stenotrophomonas strains found in the natural environment of C. elegans.

Michael A. Herman, Ph.D. has a bachelor’s degree from the University of Iowa where he worked with David Soll on dimorphism in Candida albicans. He earned his Ph.D. from the Massachusetts Institute of Technology, where he worked with H. Robert Horvitz on the control of cell polarity in C. elegans. After completing postdoctoral work with Robert Herman at the University of Minnesota, he established his laboratory at Kansas State University in 1997 and was promoted to full professor in 2011. While there, he cofounded the Kansas State Ecological Genomics Institute, and his laboratory shifted to the study of nematode-bacterium interactions. In 2018 he moved to the University of Nebraska—Lincoln to become the Director of the School of Biological Sciences. His laboratory continues work on nematode-bacterium interactions with a focus on C. elegans interactions with Stenotrophomonas.
REFERENCES
- 1.Zhang ZQ. 2013. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (Addenda 2013). Zootaxa 3703:1–82. doi: 10.11646/zootaxa.3703.1.6. [DOI] [PubMed] [Google Scholar]
- 2.Barrière A, Fèlix MA. 2007. Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176:999–1011. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dirksen P, Marsh SA, Braker I, Heitland N, Wagner S, Nakad R, Mader S, Petersen C, Kowallik V, Rosenstiel P, Félix MA, Schulenburg H. 2016. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol 14:38. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Félix MA, Duveau F. 2012. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol 10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frézal L, Félix MA. 2015. Caenorhabditis elegans outside the petri dish. Elife 4:e05849. doi: 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber M, Schüngel M, Putz A, Müller S, Hasert B, Schulenburg H. 2005. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol Biol Evol 22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- 7.Schulenburg H, Félix MA. 2017. The natural biotic environment of Caenorhabditis elegans. Genetics 206:55–86. doi: 10.1534/genetics.116.195511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrière A, Fèlix MA. 2005. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr Biol 15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Pais IS, Valente RS, Sporniak M, Teixeira L. 2018. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol 16:e2005710. doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rae R, Riebesell M, Dinkelacker I, Wang Q, Herrmann M, Weller AM, Dieterich C, Sommer RJ. 2008. Isolation of naturally associated bacteria of necromenic Pristionchus nematodes and fitness consequences. J Exp Biol 211:1927–1936. doi: 10.1242/jeb.014944. [DOI] [PubMed] [Google Scholar]
- 11.Raymann K, Moran NA. 2018. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 26:97–104. doi: 10.1016/j.cois.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DH. 2013. Bacteria and the aging and longevity of Caenorhabditis elegans. Annu Rev Genet 47:233–246. doi: 10.1146/annurev-genet-111212-133352. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y. 2008. Neuronal mechanisms of Caenorhabditis elegans and pathogenic bacteria interactions. Curr Opin Microbiol 11:257–261. doi: 10.1016/j.mib.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Shtonda BB, Avery L. 2006. Dietary choice behavior in Caenorhabditis elegans. J Exp Biol 209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Donnell MP, Fox BW, Chao PH, Schroeder FC, Sengupta P. 2020. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature 583:415–420. doi: 10.1038/s41586-020-2395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Lu H, Bargmann CI. 2005. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 17.Martin N, Singh J, Aballay A. 2017. Natural genetic variation in the Caenorhabditis elegans response to Pseudomonas aeruginosa. G3 (Bethesda) 7:1137–1147. doi: 10.1534/g3.117.039057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawli T, Tan MW. 2008. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat Immunol 9:1415–1424. doi: 10.1038/ni.1672. [DOI] [PubMed] [Google Scholar]
- 19.Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A. 2008. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322:460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid S, Pho KB, Mesbahi H, MacNeil LT. 2020. Nutrient sensing and response drive developmental progression in Caenorhabditis elegans. Bioessays 42:e1900194. doi: 10.1002/bies.201900194. [DOI] [PubMed] [Google Scholar]
- 21.Lenaerts I, Walker GA, Van Hoorebeke L, Gems D, Vanfleteren JR. 2008. Dietary restriction of Caenorhabditis elegans by axenic culture reflects nutritional requirement for constituents provided by metabolically active microbes. J Gerontol A Biol Sci Med Sci 63:242–252. doi: 10.1093/gerona/63.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner MJ, Cox JK, Spellman AC, Stahl C, Bavari S. 2020. Avoidance behavior independent of innate-immune signaling seen in Caenorhabditis elegans challenged with Bacillus anthracis. Dev Comp Immunol 102:103453. doi: 10.1016/j.dci.2019.103453. [DOI] [PubMed] [Google Scholar]
- 23.Fukuyama M, Kontani K, Katada T, Rougvie AE. 2015. The Caenorhabditis elegans hypodermis couples progenitor cell quiescence to the dietary state. Curr Biol 25:1241–1248. doi: 10.1016/j.cub.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 24.Bito T, Matsunaga Y, Yabuta Y, Kawano T, Watanabe F. 2013. Vitamin B12 deficiency in Caenorhabditis elegans results in loss of fertility, extended life cycle, and reduced lifespan. FEBS Open Bio 3:112–117. doi: 10.1016/j.fob.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson E, MacNeil LT, Ritter AD, Yilmaz LS, Rosebrock AP, Caudy AA, Walhout AJM. 2014. Interspecies systems biology uncovers metabolites affecting Caenorhabditis elegans gene expression and life history traits. Cell 156:1336–1337. doi: 10.1016/j.cell.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 26.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161:1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabreiro F, Gems D. 2013. Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med 5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DH, Ewbank JJ. 2018. Signaling in the innate immune response, p 1–35. In Community TCeR WormBook, vol 2018. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dierking K, Yang W, Schulenburg H. 2016. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Philos Trans R Soc B 371:20150299. doi: 10.1098/rstb.2015.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, Ausubel FM. 2002. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 31.Kurz CL, Ewbank JJ. 2003. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet 4:380–390. doi: 10.1038/nrg1067. [DOI] [PubMed] [Google Scholar]
- 32.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. 2002. Inducible antibacterial defense system in Caenorhabditis elegans. Curr Biol 12:1209–1214. doi: 10.1016/S0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, Wang D. 2018. The microbial zoo in the Caenorhabditis elegans intestine: bacteria, fungi and viruses. Viruses 10:85. doi: 10.3390/v10020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ermolaeva MA, Schumacher B. 2014. Insights from the worm: the Caenorhabditis elegans model for innate immunity. Semin Immunol 26:303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taffoni C, Pujol N. 2015. Mechanisms of innate immunity in Caenorhabditis elegans epidermis. Tissue Barriers 3:e1078432. doi: 10.1080/21688370.2015.1078432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsay SA, Wasserman SA. 2014. Conventional and non-conventional Drosophila Toll signaling. Dev Comp Immunol 42:16–24. doi: 10.1016/j.dci.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G, Zhao Y. 2007. Toll-like receptors and immune regulation: their direct and indirect modulation on regulatory CD4+ CD25+ T cells. Immunology 122:149–156. doi: 10.1111/j.1365-2567.2007.02651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandt JP, Ringstad N. 2015. Toll-like receptor signaling promotes development and function of sensory neurons required for a Caenorhabditis elegans pathogen-avoidance behavior. Curr Biol 25:2228–2237. doi: 10.1016/j.cub.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, Ausubel FM. 2010. Distinct pathogenesis and host responses during infection of Caenorhabditis elegans by Pseudomonas aeruginosa and Staphylococcus aureus. PLoS Pathog 6:e1000982. doi: 10.1371/journal.ppat.1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, Ewbank JJ. 2001. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol 11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 41.Borgonie G, Claeys M, Leyns F, Arnaut G, Waele DD, Coomans A. 1996. Effect of nematicidal Bacillus thuringiensis strains on free-living nematodes. 2. Ultrastructural analysis of the intoxication process in Caenorhabditis elegans. Fundam Appl Nematol 19:407–414. [Google Scholar]
- 42.Kho MF, Bellier A, Balasubramani V, Hu Y, Hsu W, Nielsen-LeRoux C, McGillivray SM, Nizet V, Aroian RV. 2011. The pore-forming protein Cry5B elicits the pathogenicity of Bacillus sp. against Caenorhabditis elegans. PLoS One 6:e29122. doi: 10.1371/journal.pone.0029122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh J, Aballay A. 2019. Microbial colonization activates an immune fight-and-flight response via neuroendocrine signaling. Dev Cell 49:89–99. doi: 10.1016/j.devcel.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chávez V, Mohri-Shiomi A, Garsin DA. 2009. Ce-Duox1/BLI-3 generates reactive oxygen species as a protective innate immune mechanism in Caenorhabditis elegans. Infect Immun 77:4983–4989. doi: 10.1128/IAI.00627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 46.Goswamy D, Irazoqui JE. 2020. A unifying hypothesis on the central role of reactive oxygen species in bacterial pathogenesis and host defense in Caenorhabditis elegans. Curr Opin Immunol 68:9–20. doi: 10.1016/j.coi.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg LM, Gringhuis SI, Geijtenbeek TB. 2012. An evolutionary perspective on C-type lectins in infection and immunity. Ann N Y Acad Sci 1253:149–158. doi: 10.1111/j.1749-6632.2011.06392.x. [DOI] [PubMed] [Google Scholar]
- 48.Miltsch SM, Seeberger PH, Lepenies B. 2014. The C-type lectin-like domain containing proteins Clec-39 and Clec-49 are crucial for Caenorhabditis elegans immunity against Serratia marcescens infection. Dev Comp Immunol 45:67–73. doi: 10.1016/j.dci.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Pees B, Kloock A, Nakad R, Barbosa C, Dierking K. 2017. Enhanced behavioral immune defenses in a Caenorhabditis elegans C-type lectin-like domain gene mutant. Dev Comp Immunol 74:237–242. doi: 10.1016/j.dci.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 50.Pees B, Yang W, Zarate-Potes A, Schulenburg H, Dierking K. 2016. High innate immune specificity through diversified C-type lectin-like domain proteins in invertebrates. J Innate Immun 8:129–142. doi: 10.1159/000441475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy CT, Lee SJ, Kenyon C. 2007. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A 104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans EA, Kawli T, Tan MW. 2008. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog 4:e1000175. doi: 10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reboul J, Ewbank JJ. 2016. GPCRs in invertebrate innate immunity. Biochem Pharmacol 114:82–87. doi: 10.1016/j.bcp.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 54.Wani KA, Goswamy D, Irazoqui JE. 2020. Nervous system control of intestinal host defense in Caenorhabditis elegans. Curr Opin Neurobiol 62:1–9. doi: 10.1016/j.conb.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu Y, Zhi L, Wu Q, Jing L, Wang D. 2018. NPR-9 regulates the innate immune response in Caenorhabditis elegans by antagonizing the activity of AIB interneurons. Cell Mol Immunol 15:27–37. doi: 10.1038/cmi.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cargnello M, Roux PP. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson GL, Lapadat R. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 58.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. 2008. Distinct innate immune responses to infection and wounding in the Caenorhabditis elegans epidermis. Curr Biol 18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shivers RP, Kooistra T, Chu SW, Pagano DJ, Kim DH. 2009. Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in Caenorhabditis elegans. Cell Host Microbe 6:321–330. doi: 10.1016/j.chom.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 61.Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, Kamanzi O, Matsumoto K, Hisamoto N, Kim DH. 2010. Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 6:e1000892. doi: 10.1371/journal.pgen.1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fletcher M, Tillman EJ, Butty VL, Levine SS, Kim DH. 2019. Global transcriptional regulation of innate immunity by ATF-7 in Caenorhabditis elegans. PLoS Genet 15:e1007830. doi: 10.1371/journal.pgen.1007830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Irazoqui JE, Urbach JM, Ausubel FM. 2010. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A 101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren M, Feng H, Fu Y, Land M, Rubin CS. 2009. Protein kinase D is an essential regulator of Caenorhabditis elegans innate immunity. Immunity 30:521–532. doi: 10.1016/j.immuni.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jeong DE, Lee D, Hwang SY, Lee Y, Lee JE, Seo M, Hwang W, Seo K, Hwang AB, Artan M, Son HG, Jo JH, Baek H, Oh YM, Ryu Y, Kim HJ, Ha CM, Yoo JY, Lee SV. 2017. Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J 36:1046–1065. doi: 10.15252/embj.201694781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Porta H, Cancino-Rodezno A, Soberon M, Bravo A. 2011. Role of MAPK p38 in the cellular responses to pore-forming toxins. Peptides 32:601–606. doi: 10.1016/j.peptides.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brito C, Cabanes D, Sarmento Mesquita F, Sousa S. 2019. Mechanisms protecting host cells against bacterial pore-forming toxins. Cell Mol Life Sci 76:1319–1339. doi: 10.1007/s00018-018-2992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huffman DL, Abrami L, Sasik R, Corbeil J, van der Goot FG, Aroian RV. 2004. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc Natl Acad Sci U S A 101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mir DA, Balamurugan K. 2019. Global Proteomic Response of Caenorhabditis elegans Against PemKSa Toxin. Front Cell Infect Microbiol 9:172. doi: 10.3389/fcimb.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. 1993. A Caenorhabditis elegans mutant that lives twice as long as wild type. Nature 366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 72.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 73.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. 2003. Long-lived Caenorhabditis elegans daf-2 mutants are resistant to bacterial pathogens. Science 300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 74.Portal-Celhay C, Bradley ER, Blaser MJ. 2012. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol 12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ewbank JJ. 2006. Signaling in the immune response, p 1–12. In Community TCeR WormBook. WormBook. doi: 10.1895/wormbook.1.83.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landis JN, Murphy CT. 2010. Integration of diverse inputs in the regulation of Caenorhabditis elegans DAF-16/FOXO. Dev Dyn 239:1405–1412. doi: 10.1002/dvdy.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Brien D, Jones LM, Good S, Miles J, Vijayabaskar MS, Aston R, Smith CE, Westhead DR, van Oosten-Hawle P. 2018. A PQM-1-mediated response triggers transcellular chaperone signaling and regulates organismal proteostasis. Cell Rep 23:3905–3919. doi: 10.1016/j.celrep.2018.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McElwee J, Bubb K, Thomas JH. 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 80.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 81.Hsu AL, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat shock factor. Science 300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 82.Singh V, Aballay A. 2006. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A 103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, Tan MW. 2006. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A 103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McHugh DR, Koumis E, Jacob P, Goldfarb J, Schlaubitz-Garcia M, Bennani S, Regan P, Patel P, Youngman MJ. 2020. DAF-16 and SMK-1 contribute to innate immunity during adulthood in Caenorhabditis elegans. G3 (Bethesda) 10:1521–1539. doi: 10.1534/g3.120.401166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang K, Kaufman RJ. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem 279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 86.Richardson CE, Kooistra T, Kim DH. 2010. An essential role for XBP-1 in host protection against immune activation in Caenorhabditis elegans. Nature 463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bischof LJ, Kao CY, Los FC, Gonzalez MR, Shen Z, Briggs SP, van der Goot FG, Aroian RV. 2008. Activation of the unfolded protein response is required for defenses against bacterial pore-forming toxin in vivo. PLoS Pathog 4:e1000176. doi: 10.1371/journal.ppat.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeong DE, Lee Y, Ham S, Lee D, Kwon S, Park HH, Hwang SY, Yoo JY, Roh TY, Lee SV. 2020. Inhibition of the oligosaccharyl transferase in Caenorhabditis elegans that compromises ER proteostasis suppresses p38-dependent protection against pathogenic bacteria. PLoS Genet 16:e1008617. doi: 10.1371/journal.pgen.1008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. 2014. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. 2010. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev Biol 10:61. doi: 10.1186/1471-213X-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Savage-Dunn C. 2005. TGF-β signaling, p 1–12. In Community TCeR WormBook. WormBook. doi: 10.1895/wormbook.1.22.1. [DOI] [Google Scholar]
- 92.Letterio JJ. 2005. TGF-beta signaling in T cells: roles in lymphoid and epithelial neoplasia. Oncogene 24:5701–5712. doi: 10.1038/sj.onc.1208922. [DOI] [PubMed] [Google Scholar]
- 93.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. 2007. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol 27:5544–5553. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. 1999. Identification of transforming growth factor-β-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci U S A 96:15020–15025. doi: 10.1073/pnas.96.26.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.White CV, Darby BJ, Breeden RJ, Herman MA. 2016. A Stenotrophomonas maltophilia strain evades a major Caenorhabditis elegans defense pathway. Infect Immun 84:524–536. doi: 10.1128/IAI.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in Caenorhabditis elegans. PLoS Genet 2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang P, Judy M, Lee SJ, Kenyon C. 2013. Direct and indirect gene regulation by a life-extending FOXO protein in Caenorhabditis elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in Caenorhabditis elegans. Cell 134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kerry S, TeKippe M, Gaddis NC, Aballay A. 2006. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS One 1:e77. doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang W, Dierking K, Rosenstiel PC, Schulenburg H. 2016. GATA transcription factor as a likely key regulator of the Caenorhabditis elegans innate immune response against gut pathogens. Zoology (Jena) 119:244–253. doi: 10.1016/j.zool.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 101.Head BP, Olaitan AO, Aballay A. 2017. Role of GATA transcription factor ELT-2 and p38 MAPK PMK-1 in recovery from acute Pseudomonas aeruginosa infection in Caenorhabditis elegans. Virulence 8:261–274. doi: 10.1080/21505594.2016.1222334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zárate-Potes A, Yang W, Pees B, Schalkowski R, Segler P, Andresen B, Haase D, Nakad R, Rosenstiel P, Tetreau G, Colletier JP, Schulenburg H, Dierking K. 2020. The Caenorhabditis elegans GATA transcription factor elt-2 mediates distinct transcriptional responses and opposite infection outcomes towards different Bacillus thuringiensis strains. PLoS Pathog 16:e1008826. doi: 10.1371/journal.ppat.1008826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. 2008. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in Caenorhabditis elegans. Cell 132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Block DH, Twumasi-Boateng K, Kang HS, Carlisle JA, Hanganu A, Lai TY, Shapira M. 2015. The developmental intestinal regulator ELT-2 controls p38-dependent immune responses in adult Caenorhabditis elegans. PLoS Genet 11:e1005265. doi: 10.1371/journal.pgen.1005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roeder T, Stanisak M, Gelhaus C, Bruchhaus I, Grötzinger J, Leippe M. 2010. Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev Comp Immunol 34:203–209. doi: 10.1016/j.dci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 106.Alegado RA, Tan MW. 2008. Resistance to antimicrobial peptides contributes to persistence of Salmonella Typhimurium in the Caenorhabditis elegans intestine. Cell Microbiol 10:1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- 107.Hoeckendorf A, Stanisak M, Leippe M. 2012. The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem J 445:205–212. doi: 10.1042/BJ20112102. [DOI] [PubMed] [Google Scholar]
- 108.Schulenburg H, Boehnisch C. 2008. Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae). BMC Evol Biol 8:114. doi: 10.1186/1471-2148-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gravato-Nobre MJ, Vaz F, Filipe S, Chalmers R, Hodgkin J. 2016. The invertebrate lysozyme effector ILYS-3 is systemically activated in response to danger signals and confers antimicrobial protection in Caenorhabditis elegans. PLoS Pathog 12:e1005826. doi: 10.1371/journal.ppat.1005826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Boehnisch C, Wong D, Habig M, Isermann K, Michiels NK, Roeder T, May RC, Schulenburg H. 2011. Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis. PLoS One 6:e24619. doi: 10.1371/journal.pone.0024619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang W, Dierking K, Schulenburg H. 2016. WormExp: a web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics 32:943–945. doi: 10.1093/bioinformatics/btv667. [DOI] [PubMed] [Google Scholar]
- 112.Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K, Zhang H. 2002. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem J 361:221–230. doi: 10.1042/0264-6021:3610221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. 2014. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J 272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 115.Kesika P, Balamurugan K. 2012. Studies on Shigella boydii infection in Caenorhabditis elegans and bioinformatics analysis of immune regulatory protein interactions. Biochim Biophys Acta 1824:1449–1456. doi: 10.1016/j.bbapap.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 116.O’Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of Caenorhabditis elegans with Mycobacterium nematophilum. Genome Res 16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Engelmann I, Griffon A, Tichit L, Montañana-Sanchis F, Wang G, Reinke V, Waterston RH, Hillier LW, Ewbank JJ. 2011. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in Caenorhabditis elegans. PLoS One 6:e19055. doi: 10.1371/journal.pone.0019055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Radeke LJ, Herman MA. 2020. Identification and characterization of differentially expressed genes in Caenorhabditis elegans in response to pathogenic and nonpathogenic Stenotrophomonas maltophilia. BMC Microbiol 20:170. doi: 10.1186/s12866-020-01771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schulte RD, Makus C, Hasert B, Michiels NK, Schulenburg H. 2010. Multiple reciprocal adaptations and rapid genetic change upon experimental coevolution of an animal host and its microbial parasite. Proc Natl Acad Sci U S A 107:7359–7364. doi: 10.1073/pnas.1003113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 121.Berg M, Stenuit B, Ho J, Wang A, Parke C, Knight M, Alvarez-Cohen L, Shapira M. 2016. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J 10:1998–2009. doi: 10.1038/ismej.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Samuel BS, Rowedder H, Braendle C, Félix MA, Ruvkun G. 2016. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc Natl Acad Sci U S A 113:E3941–E3949. doi: 10.1073/pnas.1607183113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang F, Berg M, Dierking K, Félix MA, Shapira M, Samuel BS, Schulenburg H. 2017. Caenorhabditis elegans as a model for microbiome research. Front Microbiol 8:485. doi: 10.3389/fmicb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnke J, Dirksen P, Schulenburg H. 2020. Community assembly of the native Caenorhabditis elegans microbiome is influenced by time, substrate, and individual bacterial taxa. Environ Microbiol 22:1265–1279. doi: 10.1111/1462-2920.14932. [DOI] [PubMed] [Google Scholar]
- 125.Dirksen P, Assié A, Zimmermann J, Zhang F, Tietje A-M, Marsh SA, Félix M-A, Shapira M, Kaleta C, Schulenburg H, Samuel BS. 2020. CeMbio: the Caenorhabditis elegans microbiome resource. G3 (Bethesda) 10:3025–3039. doi: 10.1534/g3.120.401309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang B, Yao M, Lv L, Ling Z, Li L. 2017. The human microbiota in health and disease. Engineering 3:71–82. doi: 10.1016/J.ENG.2017.01.008. [DOI] [Google Scholar]
- 127.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 128.Zimmermann J, Obeng N, Yang W, Pees B, Petersen C, Waschina S, Kissoyan KA, Aidley J, Hoeppner MP, Bunk B, Spröer C, Leippe M, Dierking K, Kaleta C, Schulenburg H. 2020. The functional repertoire contained within the native microbiota of the model nematode Caenorhabditis elegans. ISME J 14:26–38. doi: 10.1038/s41396-019-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berg M, Monnin D, Cho J, Nelson L, Crits-Christoph A, Shapira M. 2019. TGFβ/BMP immune signaling affects abundance and function of Caenorhabditis elegans gut commensals. Nat Commun 10:604. doi: 10.1038/s41467-019-08379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Montalvo-Katz S, Huang H, Appel MD, Berg M, Shapira M. 2013. Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect Immun 81:514–520. doi: 10.1128/IAI.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Berg M, Zhou XY, Shapira M. 2016. Host-specific functional significance of Caenorhabditis gut commensals. Front Microbiol 7:1622. doi: 10.3389/fmicb.2016.01622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hodgkin J, Félix MA, Clark LC, Stroud D, Gravato-Nobre MJ. 2013. Two Leucobacter strains exert complementary virulence on Caenorhabditis, including death by worm-star formation. Curr Biol 23:2157–2161. doi: 10.1016/j.cub.2013.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Page AP, Roberts M, Félix MA, Pickard D, Page A, Weir W. 2019. The golden death bacillus Chryseobacterium nematophagum is a novel matrix digesting pathogen of nematodes. BMC Biol 17:10. doi: 10.1186/s12915-019-0632-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petersen C, Dirksen P, Schulenburg H. 2015. Why we need more ecology for genetic models such as Caenorhabditis elegans. Trends Genet 31:120–127. doi: 10.1016/j.tig.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 135.White CV, Herman MA. 2018. Transcriptomic, functional, and network analyses reveal novel genes involved in the interaction between Caenorhabditis elegans and Stenotrophomonas maltophilia. Front Cell Infect Microbiol 8:266. doi: 10.3389/fcimb.2018.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]