Abstract
PURPOSE: The magnocellular and parvocellular pathways (M and P pathways) are the major pathways of the visual system, with distinct histologic and physiologic properties that may also have different metabolic characteristics. We hypothesize that the differences of the 2 visual pathways would also manifest as differences in the signal time course of blood oxygen level–dependent functional MR imaging (BOLD fMRI). The differences in BOLD signal time course may provide insight into the metabolic requirements of the 2 pathways.
METHODS: Eleven fMRI sessions on 6 subjects were performed using stimuli that preferentially activated the 2 pathways. Regions commonly activated by both the M and P stimuli in the primary visual cortex (V1) were determined, and the contrast elicited by the stimulus, time-to-peak (TTP), and the full width at half maximum (FWHM) of the BOLD signal time course were measured.
RESULTS: The functional stimuli activated cortical regions described previously in the literature, such as V1, V4, and V5. Within V1, the TTP of the signal time course of the 2 stimuli were statistically different, with the P stimulus generating TTPs that were on average 12% faster than the M stimulus (P = .0037).
CONCLUSION: We have demonstrated the ability to functionally differentiate the M and P stimuli in a commonly activated anatomic region. Because the BOLD response is dependent on the ratio of oxyhemoglobin and deoxyhemoglobin in the blood, the difference in the BOLD time course between the 2 stimuli suggests that the oxygen demand of the 2 pathways may be different.
The magnocellular and parvocellular pathways (M and P pathways) are the major pathways of the visual system, accounting for most of the axons that leave the retina and the perceived vision, as demonstrated by loss of vision when the pathways are destroyed.1,2 Histologic examination reveals that the M and P pathways are distinct. The M ganglion cells in the retina, so called because of their larger cell body,3 project to layers one and 2 in the lateral geniculate nucleus (LGN). The P cells in the retina project to layers 3 through 6 in the LGN. The projections of the M and P cells appear to be distinct in the LGN4,5 and remain so from the LGN to the primary visual cortex (V1), with the M pathway terminating primarily in layer 4Cα of V1 and the P pathway terminating primarily in layers 4A and 4Cβ of V1.6
Cytochrome oxidase (CO) staining shows reactive and nonreactive neurons throughout the layers in LGN, with darkly reactive neurons appearing more frequently in layers 1, 2, and 6.7 Layer 4C of V1 is darkly stained by CO.8 Area V2 of the visual cortex contains CO thick and thin stripes as well as nonstaining interstrips.9 The M pathway thus courses through the CO-rich layers of the LGN, V1, and the thick stripes of V2, whereas the P pathway contains non–CO-rich layers as well as a CO-rich layer of the LGN and the CO-rich layer in V1. The P pathway is further differentiated with a P-B stream, which courses through the CO-rich blobs in V1 and the thin stripes of V2, and a P-I stream, which courses through the non–CO-rich interblobs of V1 and the interstripes of V2.10–14
Physiologically, the M and P pathways are also distinct. The M pathway is considered insensitive to color when the luminance is balanced, has higher contrast sensitivity, is responsive to lower spatial frequencies and higher temporal frequencies, and has transient responses. The P pathway is color sensitive, has lower contrast sensitivity, is responsive to higher spatial frequencies and lower temporal frequencies, and sustained responses.15–18 The P pathway can be further discriminated into the P-B and PI streams, with the P-B stream involved primarily with color discrimination and the P-I stream involved primarily with orientation selectivity. The functional segregation of the P-B and the P-I streams, however, is not complete.19 Another difference between the 2 pathways is that the M pathway has faster impulse conduction.17,20,21 Because of the different physiologic responses of the M and P pathways, a common strategy to preferentially activate the pathways is to use a stimulus that is color-neutral, low in contrast, large in grid size, and reverses contrast at a fast rate for the M pathway. Conversely, the P pathway responds preferentially to stimuli that are different in color, high in contrast, small in grid size, and reverses contrast at a slow rate.
The histologic and physiologic differences of the M and P pathways make them attractive as model systems to study bioenergetic differences in brain pathways. As noted above, the CO-staining patterns of the 2 pathways are different, suggesting that the 2 pathways may have different energetic requirements. The fact that the 2 pathways respond to different stimuli allows for the preferential activation of the individual pathways. Therefore, it may be possible to show metabolic differences in the pathways with the use of particular stimuli.
Functional MR imaging (fMRI) using the blood oxygenation level–dependent (BOLD) contrast mechanism has been used to study many aspects of cognitive function as well as sensory response of the brain. Although the technique has been used primarily to detect regions of activation in the brain, the cortical response to the stimulus has also been studied.22–28
Analysis of the amplitude and time course of the BOLD signal has the potential to elucidate the energetic response of the brain to a particular stimulus. Because the BOLD signal is dependent on the oxyhemoglobin/deoxyhemoglobin ratio ([Hbo]/[Hb]) as well as blood flow, the characteristics of the time course, such as time to peak, contrast, and the width of the response, may be a reflection of the energy demands that a particular stimulus imposes on the brain. The M and P system of the visual cortex are ideal for such a study because of the difference in their physical and functional characteristics. In this study, we hypothesize that the BOLD signal time courses of the responses to brief stimuli that preferentially activate the M and P pathways have different temporal evolutions. This would demonstrate that the functional differences of the 2 pathways can also manifest as a difference in the time course of the signals.
Methods
Eleven fMRI sessions on 6 subjects were performed on a Siemens Trio 3T scanner (Siemens, Munich, Germany). All subjects gave informed consent. Gradient-echo echo-planar imaging was used with the following parameters: repetition time [TR] = 2000 ms; echo time [TE] = 30 ms, field of view [FOV] = 240 mm, matrix size 64 × 64, 5-mm section thickness, no intersection gap, 19 sections.
The subjects viewed a visual stimulus paradigm consisting of a stimulus that preferentially activated the M pathway and a stimulus that preferentially activated the P pathway. The stimulus that preferentially activated the M pathway was a black and white reversing checkerboard with 18% contrast (centered on neutral gray, which is also the background for the rest condition), 2° check size, and reversing rate of 19 Hz. The stimulus that preferentially activated the P pathway was an isoluminant red and green reversing checkerboard with 50% contrast, 0.5° check size, and reversing rate of 2 Hz. The 2 stimuli were designed such that each was more sensitive to the intended pathway, similar to the stimuli used by Kleinschmidt et al,29 and of similar power to produce similar BOLD contrast. The 2 stimuli were presented in a pseudorandom manner, with 20 presentations per session. The stimuli were displayed for the duration of the TR, and 185 measurements were acquired per session. The first 5 measurements were discarded from the functional analysis to ensure that magnetization had reached equilibrium.
Analysis of functional activation was performed with SPM2 software (Wellcome Department of Cognitive Neurology, London, UK). The datasets underwent section timing correction, motion correction, and smoothing with a Gaussian kernel with full width at half maximum (FWHM) of 7.5 × 7.5 × 10 mm. The datasets were also spatially normalized to MNI space and smoothed for group analysis of the response to the stimuli. The normalized datasets were interpolated to 2 × 2 × 2 mm and smoothed with a Gaussian kernel with FWHM of 4 × 4 × 4 mm. For the random effects (group) analysis, a threshold uncorrected for multiple comparison of P = .001 with an extent threshold of voxels set such that P < .05, corrected for multiple comparisons, was used. The resulting activated regions were then analyzed with the use of Talairach Daemon30 to determine the associated Brodmann areas.
The detection of functionally active regions for the time course analysis was performed using an unbiased basis function. Because the goal of this study was to analyze differences in the hemodynamic response function (HRF) of the stimuli, an unbiased design matrix consisting of 8 finite impulse response functions was used for each stimulus31; the first finite impulse response function had the stimulus onset timings, the second basis function was offset by +1 TR from the first basis function, and the third basis function was offset by +2 TRs, and so forth. This ensured that the duration of the HRF (16 seconds) was sampled in an unbiased fashion. The design matrix was then used to determine regions of activation during the M stimulus presentation and regions of activation during the P stimulus presentation. For the group analysis, the SPM2-biased HRF was used to calculate regions of activation during stimulus presentation.
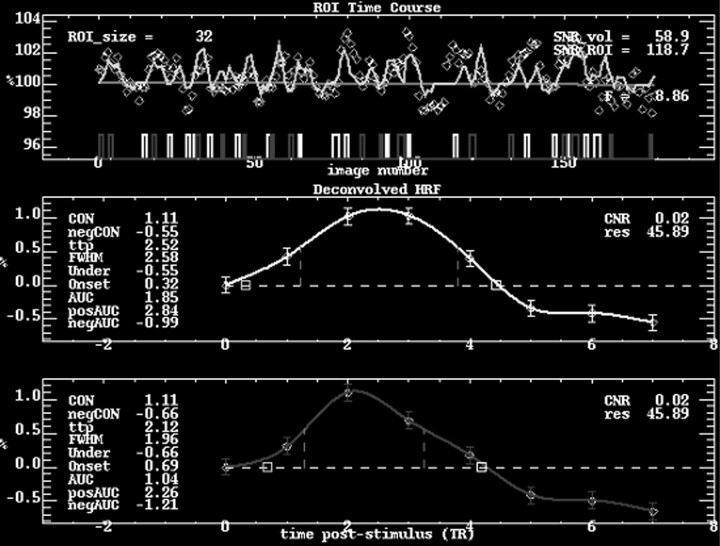
The region responding to both functional stimuli within V1 was used as a region of interest (ROI) for the time course analysis. Using custom software in the IDL programming language (RSI, Boulder, Colo), the M and P activation maps, thresholded at f scores equivalent to P < .05, corrected for multiple comparisons, were obtained. Anatomic regions that were activated by both stimuli were located by subtracting the activated regions and used in the time course analysis. A ROI of V1, manually determined around the calcarine fissure, was further imposed on the voxels commonly activated by both the M and P stimuli. Thus, the time course analyses for the 2 stimuli were limited to the primary visual cortex, ensuring that the differences in HRF were not due to HRF differences caused by anatomy. The HRF of the stimulus was then calculated by deconvolving the signal time course and thus allowing the HRFs to each stimulus presentation to be averaged together to generate a “folded average” HRF for that stimulus. The percentage contrast of the response (percentage change of the maximum BOLD signal from the baseline), the time to peak (TTP) of the HRF, and the FWHM of the HRF were calculated from the spline fit, a method of generating a continuous function from discrete data points using piecewise polynomials,32 of the folded average of the signal time course for each subject and each stimulus (Fig 1). The values of the P stimulus were then normalized to the values of the M stimulus to adjust for individual differences in the magnitude of responses to the stimuli. Therefore, a statistically significant difference between the 2 stimuli would be one in which the ratio was statistically different from 1. Two-tailed, 1-sample t tests were performed on the percentage contrast of the response, TTP, and FWHM for P stimulus compared with that of the M stimulus using InStat 3.06 (GraphPad Software, San Diego, Calif). Regression analysis of TTP versus percentage signal change, FWHM versus percentage signal change, and FWHM versus TTP were also performed, with the metrics of both conditions treated as 1 condition to assess the relationship of the metrics to each other.
Fig 1.
Time course analysis of the response in V1 to the M and P stimuli for a single subject. The average signal time course within V1 for the entire study is seen in the top graph, with the light boxes representing M stimulus onsets and the dark boxes representing P stimulus onsets. The average deconvolved HRFs of the M and P responses are depicted in the middle and bottom graphs, respectively. In this particular case, the P/M contrast ratio is 1.0, the TTP ratio is 0.84, and the FWHM ratio is 0.76.
Results
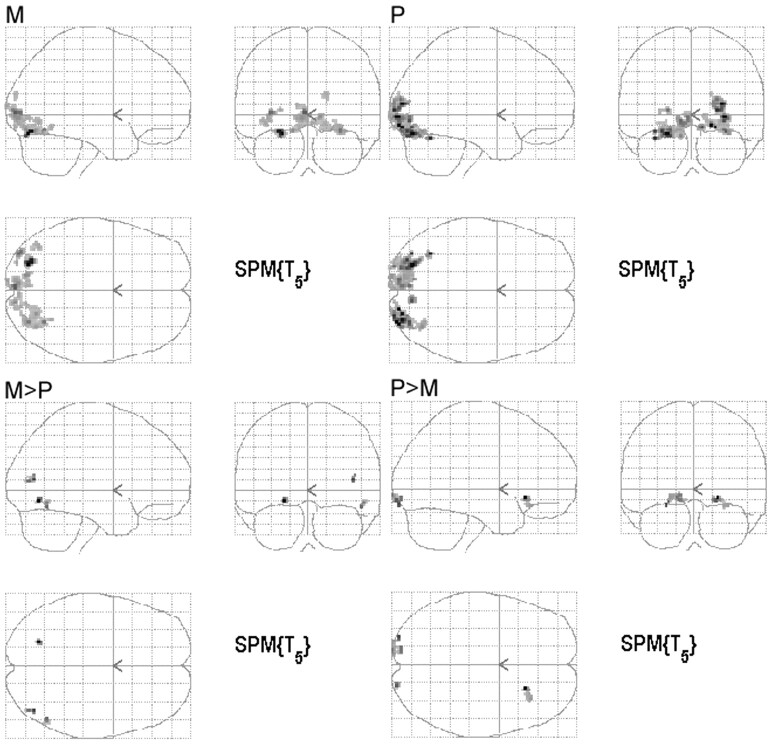
All subjects showed significant activation to the M and P stimuli. Individual statistical parametric maps (SPMs) for the M and P stimuli revealed activations typical for the particular stimulus. The M stimulus activated V5 bilaterally, whereas the P stimulus did not strongly activate V5. For the P stimulus, V4 was more strongly activated compared with the M stimulus (Fig 2). In general, the regions activated by the M stimulus were located superiorly compared with regions activated by the P stimulus, following the dorsal stream of the visual pathway, as described in the literature. Group analysis of the SPMs from the 6 subjects also showed regions known to be activated by the M and P stimuli, though the group response was not as robust as the individual data (Fig 3). Coordinates of the most statistically significant voxels in the activation clusters are listed in Table 1.
Fig 2.
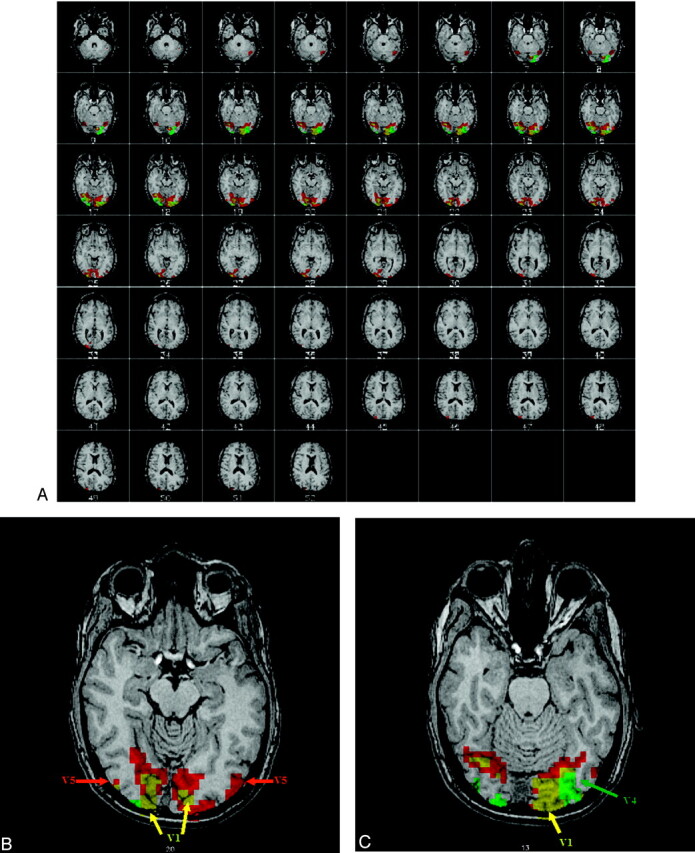
Single subject activation maps to the M (red) and P (green) stimuli. The images are in radiologic convention (ie, left is right). The functional map is overlaid on a high resolution anatomic image. The regions showing overlapping M and P activity are yellow. Note that both stimuli generate robust activation of V1. The sections in the top picture depict active regions in the brain, and the bottom 2 sections show representative regions of activation. For the M stimulus response, V5 is activated. For the P stimulus, V4 is activated.
Fig 3.
Group analysis of the 6 subjects. The images are in neurologic convention (ie, left is left). The SPMs for the response to the M and P stimulus compared with the rest condition as well as the SPMs of regions responding greater for one condition than the other (M>P and P>M) are shown. The maps are created with a threshold uncorrected for multiple comparison of P = .001 with an extent threshold of P < .05, corrected for multiple comparisons.
Table 1:
Coordinates of significantly activated clusters of the group analysis of individual subjects
| Stimulus | Talariach Coordinates |
Cluster Size | Brodmann Area | ||
|---|---|---|---|---|---|
| x | y | z | |||
| M | −24 | −78 | −11 | 59 | 19 |
| −2 | −89 | 6 | 332 | 18 | |
| 30 | −63 | −9 | 19 | ||
| 30 | −78 | −10 | 19 | ||
| −34 | −85 | 6 | 39 | 19 | |
| −26 | −87 | 8 | 19 | ||
| −36 | −82 | −1 | 18 | ||
| −14 | −80 | −8 | 26 | 18 | |
| −40 | −68 | −3 | 20 | 19 | |
| 16 | −86 | 23 | 14 | 18 | |
| P | 30 | −91 | 3 | 301 | 18 |
| 18 | −90 | −4 | 17 | ||
| 28 | −85 | 13 | 19 | ||
| −24 | −76 | −11 | 363 | 19 | |
| −34 | −78 | −10 | 19 | ||
| −20 | −88 | −7 | 18 | ||
| −34 | −61 | −15 | 10 | 37 | |
| 10 | −78 | −6 | 23 | 18 | |
| −22 | −93 | 8 | 20 | 18 | |
| −30 | −91 | 8 | 19 | ||
| M > P | −22 | −68 | −5 | 12 | 19 |
| 42 | −71 | 13 | 13 | 39 | |
| 50 | −61 | −10 | 11 | 37 | |
| P > M | 22 | 23 | −8 | 23 | 47 |
| 32 | 28 | −15 | 47 | ||
| −26 | −92 | −7 | 46 | 18 | |
| −14 | −90 | −4 | 17 | ||
| −14 | −97 | 0 | |||
| 20 | −94 | −5 | 9 | 17 | |
Note:—M indicates magnocellular pathways; P, parvocellular pathways.
For the time course analysis of the BOLD signal, a summary of the comparison of percentage contrast elicited, TTP, and FWHM for the M and P stimuli are shown in Table 2. Both stimuli elicited similar neural responses, as demonstrated by the lack of statistical difference in the percentage signal change of the BOLD signal in response to the 2 stimuli (mean P/M ratio = 0.89, SD = 0.23, P = .16). The HRF of the P stimulus had a statistically significant difference in its TTP response compared with that of the M stimulus, on average being 12% faster than the M stimulus (mean P/M ratio = 0.88, SD = 0.11, P = .0037). The FWHM of the response to the P stimulus was not statistically different from the response to the M stimulus (mean P/M ratio = 0.95, SD = 0.18, P = .36).
Table 2:
Summary of hemodynamic response function metrics of the M and P responses within VI
| Mean P/M Ratio | SD | P Value | 95% Confidence Interval | |
|---|---|---|---|---|
| % Contrast | 0.89 | 0.23 | .16 | 0.74–1.05 |
| TTP | 0.88 | 0.11 | .0037 | 0.81–0.95 |
| FWHM | 0.95 | 0.18 | .36 | 0.82–1.07 |
Note:—V1 indicates primary visual cortex; TTP, time to peak; FWHM, full width at half maximum.
The TTP for the 2 stimuli were not correlated with the magnitude of the BOLD response to the 2 stimuli, as seen by the regression analysis of TTP versus percentage signal change, with r2 = 0.06408 and a slope that was not significantly different from zero (slope = 0.453, P = .2557) (Fig 4A). The FWHM of the HRF was positively correlated with the percentage signal change, with r2 = 0.3114 and a slope that was significantly different from zero (slope = 1.977, P = .0070) (Fig 4B). The FWHM of the HRF was also positively correlated with the TTP, with r2 = 0.2115 and a slope that was significantly different from zero (slope = 0.9105, P = .0313) (Fig 4C).
Fig 4.
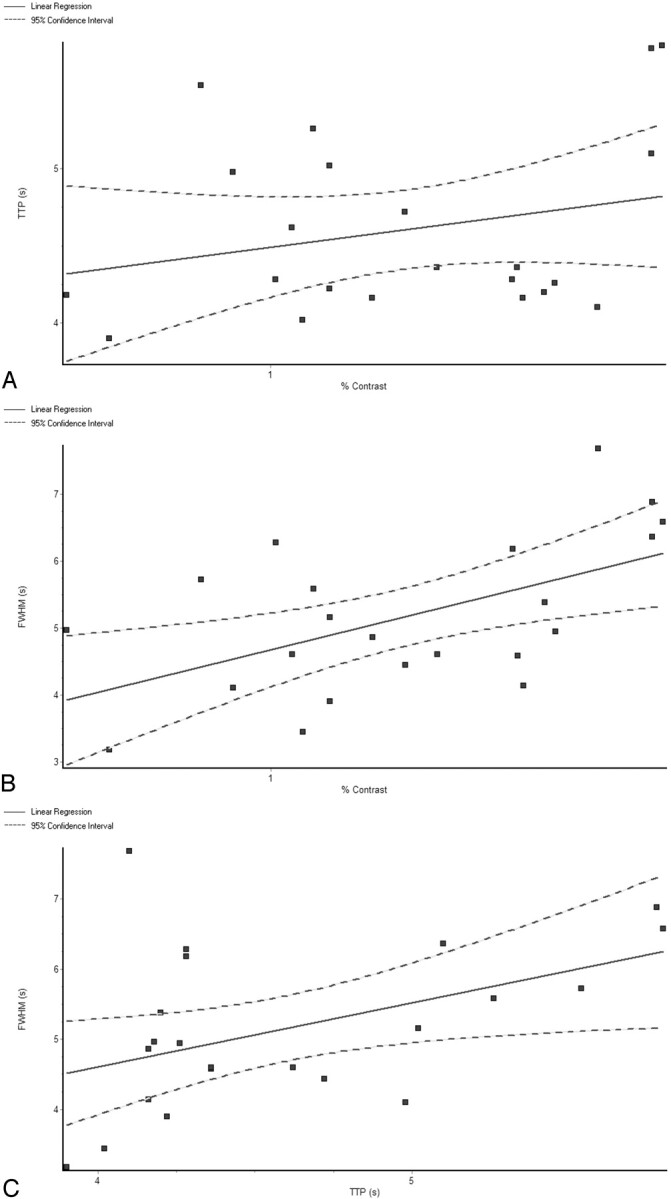
Regression analysis of the HRF metrics, with the M and P responses pooled together for the analysis. TTP versus percentage signal change (A) shows no statistically significant correlation, with r2 = 0.06408 and a slope that is not significantly different from 0 (slope = 0.453, P = .2557). The FWHM of the HRF is positively correlated with the percentage signal change (B), with r2 = 0.3114 and a slope that is significantly different from 0 (slope = 1.977, P = .0070). The FWHM of the HRF is also positively correlated with the TTP (C), with r2 = 0.2115 and a slope that is significantly different from zero (slope = 0.9105, P = .0313).
Discussion
The anatomic and physiologic differences between the M and P pathways offer an opportunity to determine whether BOLD fMRI can detect these differences based solely on signal time course. Our experiments demonstrated that our M and P stimuli, similar to those used by others before to preferentially activate the M and P pathways,29 activate brain regions typically associated with these stimuli.33,34 It is interesting to note that the SPMs of the group analysis for the M and P stimuli, in regions known to be affected by the 2 stimuli, did not show as robust an activation as the individual datasets, demonstrating the anatomic variability of regions activated by the M and P stimuli. It is possible that additional data from more subjects might generate more robust group activation maps.
It has been shown that the time course of the signal is dependent on the ROI used.35 By examining the voxels that were activated in both stimulus conditions and examining activated voxels within an anatomic region, we eliminated the confound of time course differences due to spatial differences in vascular architecture. Our selection of stimulus contrast levels produced no statistically significant differences in the percentage signal change in response to the stimuli, minimizing the confound that the time course differences seen between the 2 stimuli were due to contrast levels.36
Furthermore, regression analysis of TTP versus percentage contrast, with responses from both stimuli pooled together in a single dataset, showed no statistically significant correlation between the 2 metrics. This again suggests that the TTP differences seen from the M and P stimuli are indeed different biologically and are not due simply to a difference in the magnitude of the response to the stimuli. It is not surprising that the FWHM of the HRF, on the other hand, is correlated to percentage contrast, as a greater response to a stimulus resulted in a larger FWHM. The positive correlation of TTP and FWHM is similar to previous findings regarding these 2 parameters of the HRF in response to respiratory CO2 changes.37
In the past, cognitive subtraction has been used to discern the neural activity to different stimuli. For the M and P pathway, cognitive subtraction was used to characterize locations in the brain that differentially respond to 1 of the stimuli. By examining the time course of the neural response to the stimulus, we demonstrate that it is possible to differentiate the 2 stimuli even in regions responding to both stimuli. Not only is it possible to differentiate the 2 stimuli based on the difference in their BOLD time course, the time course itself may also provide information regarding the physiology of the 2 responses. Because the BOLD time course is dependent on [Hbo]/[Hb] as well as the physiologic response of increased blood flow to increase [Hbo]/[Hb], we postulate that the differences in time course between the response to the 2 stimuli also reflect a difference in aerobic metabolism between the 2 stimuli. The fact that the P stimulus had a BOLD time course with faster TTP suggests that the P stimulus produced less [Hb]; thus, the maximum [Hbo]/[Hb] was reached at a faster rate than the time course in response to the M stimulus. The M stimulus thus seems to have a greater aerobic requirement than the P stimulus, consistent with its CO-rich anatomic substrate. Its aerobic requirement generates more [Hb] in response to the stimulus, thus requiring a greater amount of time for the compensatory increase in blood flow to achieve the maximum [Hbo]/[Hb].
The M pathway has been described to have smaller single-unit response latency to visual stimuli compared with the P pathway.38 It is interesting that our results demonstrate that the P pathway, despite being activated at a slower rate, has a faster TTP compared with the M pathway. The difference between the neuronal recordings and the BOLD signal time course further illustrates the fact that the BOLD response is a proxy of the neuronal response and is not necessarily a linear representation of it, a fact that has been demonstrated in the literature.
Because BOLD fMRI is a qualitative measure of neural activity, it is not possible from this experiment to quantitatively describe the metabolic requirements of the 2 visual pathways. Other imaging techniques, such as PET and near-infrared spectroscopy, may further elucidate the metabolic requirements of the 2 stimuli. An imaging sequence that has higher temporal resolution may allow for better characterization of the signal time course and may therefore detect differences in the signal time courses that were not possible with our experimental methods.
Conclusion
It is possible to distinguish the brain responses to 2 distinct stimuli in a commonly activated region by analyzing the HRF to each stimulus. The M and P stimuli have different HRFs, as demonstrated by the difference in TTP. This difference in HRF may be a reflection of the histologic differences in the 2 pathways and suggests that the metabolic requirements of the 2 pathways may be different. The BOLD signal time course could be a qualitative assay of metabolic function.
Acknowledgments
We would like to thank Dr. David Brainard for assistance in designing the stimulus presentation software and for the helpful discussions.
Footnotes
The research was supported by National Heart, Lung, and Blood Institute grant 5T32-HL07614 and National Institutes of Health grants RR02305 and MH64045.
This work was presented in part at the 43rd Annual Meeting of the American Society for Neuroradiology; May 21–27, 2005; Toronto, Ontario, Canada.
References
- 1.Silveria LCL, Perry VH. The topography of magnocellular projecting ganglion cells (M ganglion cells) in the primate retina. Neuroscience 1991;40:217–37 [DOI] [PubMed] [Google Scholar]
- 2.Schiller PH, Logothetis NK, Charles ER. Role of the color-opponent and broadband channels in vision. Vis Neurosci 1990;5:321–46 [DOI] [PubMed] [Google Scholar]
- 3.Perry VH, Oehler R, Cowey A. Retinal ganglion cells that project to the dorsal lateral geniculate nucleus in the macaque monkey. Neuroscience 1984;12:1101–23 [DOI] [PubMed] [Google Scholar]
- 4.Conley M, Fitzpatrick D. Morphology of retinogeniculate axons in the macaque. Vis Neurosci 1989;2:287–96 [DOI] [PubMed] [Google Scholar]
- 5.Michael CR. Retinal afferent arborization patterns, dendritic field orientations, and the segregation of function in the lateral geniculate nucleus of the monkey. Proc Natl Acad Sci U S A 1988;85:4914–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzpatrick D, Lund JS, Blasdel GG. Intrinsic connections of macaque striate cortex: afferent and efferent connections of lamina 4C. J Neurosci 1985;5:3329–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Wong-Riley M. Quantitative light-and electron-microscopic analysis of cytochrome-oxidase distribution in neurons of the lateral geniculate nucleus of the adult monkey. Vis Neurosci 1990;4:269–87 [DOI] [PubMed] [Google Scholar]
- 8.Preuss TM, Qi H, Kaas JH. Distinctive compartmental organization of human primary visual cortex. Proc Natl Acad Sci U S A 1999;96:11601–06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tootell RBH, Silverman MS, De Valois RL, et al. Functional organization of the second cortical visual area in primates. Science 1983;220:737–39 [DOI] [PubMed] [Google Scholar]
- 10.Blasdel, GG, Lund JS, et al. Intrinsic connections of macaque striate cortex: axonal projections of cells outside lamina 4c. J Neurosci 1985;5:3350–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellemen DJ, Van Essen DC. Receptive field properties of neurons in area V3 of macaque monkey extrastriate cortex. J Neurophysiol 1987;57:889–920 [DOI] [PubMed] [Google Scholar]
- 12.DeYoe EA, Van Essen DC. Segregation of efferent connections and receptive field properties in visual area V2 of the macaque. Nature 1985;317:58–61 [DOI] [PubMed] [Google Scholar]
- 13.Livingstone MS, Hubel DH. Connections between layer 4B of area 17 and the thick cytochrome oxidase stripes of area 18 in the squirrel monkey. J Neurosci 1987;7:3371–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipp S, Zeki S. Segregation of pathways leading from area V2 to areas V4 and V5 of macaque monkey visual cortex. Nature 1985;315:322–24 [DOI] [PubMed] [Google Scholar]
- 15.Derrington AM, Lennie P. Spatial and temporal contrast sensitivities of neurons in lateral geniculate nucleus of macaque. J Physiol 1984;357:219–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicks TP, Lee BB, Vidyasagar TR. The responses of cells in the macaque lateral geniculate nucleus to macaque lateral geniculate nucleus to sinusoidal gratings. J Physiol 1983;337:183–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiller PH, Malpeli JG. Functional specificity of lateral geniculate nucleus laminae of the rhesus monkey. J Neurophysiol 1978;41:788–97 [DOI] [PubMed] [Google Scholar]
- 18.Shapley R, Kaplan E, Soodak R. Spatial summation and contrast sensitivity of X and Y cells in the lateral geniculate nucleus of the macaque. Nature 1981;292:543–45 [DOI] [PubMed] [Google Scholar]
- 19.Heywood CA, Cowey A, Newcombe F. On the role of parvocellular (P) and magnocellular (M) pathways in cerebral achromatopsia. Brain 1994;117:245254. [DOI] [PubMed] [Google Scholar]
- 20.Gouras P. Antidromic responses of orthodromically identified ganglion cells in monkey retina. J Physiol 1969;204:407–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan E, Shapley RM. X and Y cells in the lateral geniculate nucleus of macaque monkeys. J Physiol 1982;330:125–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellgowan PS, Saad ZS, Bandettini PA. Understanding neural system dynamics through task modulation and measurement of functional MRI amplitude, latency, and width. Proc Natl Acad Sci U S A 2003;100:1415–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckner R, Koutstaal W, Schacter D, et al. Functional-anatomic study of episodic retrieval. II. Selective averaging of event-related fMRI trials to test the retrieval success hypothesis. NeuroImage 1998;7:163–75 [DOI] [PubMed] [Google Scholar]
- 24.Dilharreguy B, Jones RA, Moonen CTW. Influence of fMRI data sampling on the temporal characterization of the hemodynamic response. NeuroImage 2003;19:1820–28 [DOI] [PubMed] [Google Scholar]
- 25.Hensen RNA, Price CJ, Rugg MD, et al. Detecting latency differences in event-related BOLD responses: application to words versus nonwords and initial versus repeated face presentations. NeuroImage 2002;15:83–97 [DOI] [PubMed] [Google Scholar]
- 26.Miezin F, Maccotta L, Ollinger J, et al. Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. NeuroImage 2000;11:735–59 [DOI] [PubMed] [Google Scholar]
- 27.Neumann J, Lohmann G, Zysset S, et al. Within-subject variability of BOLD response dynamics. NeuroImage 2003;19:784–96 [DOI] [PubMed] [Google Scholar]
- 28.Schacter D, Buckner R, Koutstaal W, et al. Late onset of anterior prefrontal activity during true and false recognition: an event-related fMRI study. NeuroImage 1997;6:259–69 [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt A, Lee BB, Requardt M, et al. Functional mapping of color processing by magnetic resonance imaging of responses to selective P- and M-pathway stimulation. Exp Brain Res 1996;110:279–88 [DOI] [PubMed] [Google Scholar]
- 30.Lancaster JL, Rainey LH, Summerlin JL, et al. Automated labeling of the human brain: a preliminary report on the development and evaluation of a forward-transform method. Hum Brain Mapp 1997;5:238–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burock MA, Dale AM. Estimation and detection of event-related fMRI signals with temporally correlated noise: a statistically efficient and unbiased approach. Hum Brain Mapp 2000;11:249–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unser M. Splines: a perfect fit for signal and image processing. IEEE Signal Process Mag 1999;16:22–38 [Google Scholar]
- 33.Wandell BA, Poirson AB, Newsome WT, et al. Color signals in human motion-selective cortex. Neuron 1999;24:901–09 [DOI] [PubMed] [Google Scholar]
- 34.Bartels A, Zeki S. The architecture of the colour centre in the human visual brain: new results and a review. Eur J Neurosci 2000;12:172–93 [DOI] [PubMed] [Google Scholar]
- 35.Mohamed MA, Yousem DM, Tekes A, et al. Timing of cortical activation: a latency-resolved event-related functional MR imaging study. AJNR Am J Neuroradiol 2003;24:1967–74 [PMC free article] [PubMed] [Google Scholar]
- 36.Stromeyer CF, III, Martini P. Human temporal impulse response speeds up with increased stimulus contrast. Vis Res 2003;43:285–98 [DOI] [PubMed] [Google Scholar]
- 37.Kemna LJ, Posse S. Effect of respiratory CO2 changes on the temporal dynamics of the hemodynamic response in functional MR imaging. NeuroImage 2001;14:642–49 [DOI] [PubMed] [Google Scholar]
- 38.Schmolesky MT, Wang Y, Hanes DP, et al. Signal timing across the macaque visual system. J Neurophysiol 1998;79:3272–78 [DOI] [PubMed] [Google Scholar]