Abstract
Precision medicine has impacted the field of medical oncology by introducing personalized therapies, improving all measurable outcomes. This field, in turn, has expanded to obtaining and analyzing a vast and ever-increasing amount of genomic information. One technique currently applied is the liquid biopsy, which consists of detecting and isolating DNA and exosomes in cancer patients. Newly developed techniques have made it possible to use the liquid biopsy in a wide range of settings. However, challenges regarding the validation of its clinical utility exist because of a lack of standardization across different techniques and tumor types, confounder genomic information, lack of appropriate clinical trial designs, and a non-measured, and therefore not estimated, economic impact on population health. Nowadays, liquid biopsy is not routinely used, but ongoing research is increasing its popularity, and a new era in oncology is developing. Therefore, it is essential to have an in-depth understanding of the liquid biopsy technique. In this review, we summarize the leading techniques and liquid biopsy applications in cancer.
Keywords: Cell-free DNA, Circulating tumor cells, Diagnosis, Liquid biopsy, Screening
Key Summary Points
| Precision medicine has renewed the field of medical oncology, allowing a more personalized treatment. |
| This field in turn has expanded to obtaining and analyzing a vast and every-increasing amount of genomic information. |
| The liquid biopsy technique consists of detecting and isolating circulating tumor DNA and exosomes. |
| Many newly developed techniques and next-generation sequencing analyses allow a broad application of liquid biopsy in a wide range of settings. |
| Some difficulties have been identified in carrying out this procedure, including lack of standardization, presence of confounder genomic information, and lack of appropriate clinical trial designs. |
| This review summarizes the main techniques and applications of the liquid biopsy technique in cancer. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article, go to 10.6084/m9.figshare.13855316.
Introduction
The liquid biopsy (LB) technique has become highly relevant in the clinical field, and the acquisition of cell free DNA (cfDNA), including circulating tumor DNA (ctDNA), has become recognized as an emerging biomarker and a tool for the research, diagnosis, and prognosis of solid tumors [1]. Different studies have confirmed that this type of DNA contains information on specific tumor-related alterations, such as mutations, methylations, and copy number variants/variation (CNVs) [2]. The most striking advantage of LB is its ability to isolate ctDNA through a minimally invasive technique [1, 3].
cfDNA is released into the bloodstream as a consequence of cellular processes, such as apoptosis, pyroptosis, mitotic catastrophe, autophagy, phagocytosis, or a regulated form of neutrophil cell death referred to as NETosis [4]. The levels of cfDNA are higher in patients with cancer and, consequently, structural and epigenetic changes in the DNA sequence can be observed, which in turn reflect the disease process [5, 6]. LB also provides real-time information on the tumor, which is relevant because such information is temporary and spatially heterogeneous. The detection of these variations positively impacts timely treatment modifications that could be beneficial for the patient [5, 7]. Notably, the analysis of nucleic acids in tumor tissue from traditional biopsies provides information on the predominant cells in the tumor while, in contrast, tumor analysis based on cfDNA reveals information on all tumor sites. As such, LB can be used to monitor a patient’s disease burden and progression more accurately and in real-time, thus detecting information on the heterogeneity of different tumors [8].
Another significant advantage of LB is that the cfDNA released by the tumor into the bloodstream has the same variants as those of the tumor cells. Thus, information can be obtained at different times of the disease and, given the ease of sampling, repeated sampling is possible without further inconvenience to the patient [1]. This has led to significant technological advances, including the isolation of cancer cells and the collection of DNA derived from them, that provide accurate and timely information through the use of LB of oncogene-related variants that can be taken into account when designing targeted therapies [9]. In this context, the LB technique is a powerful resource for determining the appearance of additional genetic changes, such as the drug resistance mutations that appear with the use of certain targeted therapies. The possibility of being able to take repeated samples allows for adjustment of the therapy to respond to the evolution of the tumor [6]. It is also possible to obtain ctDNA in several body fluids, such as urine, feces, cerebrospinal fluid, and saliva [10, 11]. Thus, advances in this field will surely change how the treatment of a patient with cancer is selected and monitored.
The development of precision medicine (PM) represents a breakthrough in cancer management [12]. The application of individualized treatments that emphasize tumor biology and predictive responses has significantly improved clinical outcomes. Hence, the clinical applications of PM are broad, encompassing screening, diagnosis, prognosis, prediction of treatment response and resistance, early detection of recurrence/metastasis, and biological cancer stratification [13]. However, challenges related to early diagnosis and real-time treatment monitoring remain. Some of these issues are due to previously described tumor biopsies, including limited tissue availability, the continuously evolving tumor genetic landscape in response to treatment, the emergence of resistant subclones, and the lack of knowledge on spatial and temporal heterogeneity of tumoral cells [14].
The completion of the Human Genome Project (HGP) in 2001 opened the floodgates to a deeper understanding of the diseases and their potential treatment [15]. Therapeutic advances in genome-guided precision oncology rely upon the prospective molecular identification of oncogenic alterations and resistance mechanisms to guide accurate treatments [16]. Technological advances in the genetic sequencing of cfDNA in plasma have increased the advantages of the LB technique so that it now has the potential to identify actionable alterations in tumor-derived DNA present in blood and other fluids and to capture intra-tumoral heterogeneity not addressed by tumor tissue biopsy (that only captures a single site), potentially avoiding the need for invasive measures that consume time and generate unnecessary risks. In this same context, ctDNA is often present in patients without detectable circulating tumor cells, suggesting that these two biomarkers represent distinct entities. At large academic cancer centers, the failure rates from diagnostic tests derived from tissue biopsy-based next-generation sequencing (NGS) are approximately 14%; therefore, ctDNA analysis may be useful in guiding treatment selection in patients for whom tissue-based NGS is not an option [17].
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Basic Concepts of LB
Liquid biopsy is an alternative technique that allows the analysis of different tumor biomarkers, mainly from blood. It is a non-invasive, safe, and effective method with the potential for applications in the clinical diagnosis and treatment management in patients with cancer [18]. The main components released by primary or metastatic tumors in a LB are circulating tumor cells (CTC), cfDNA, ctDNA, microRNA (miRNA), tumor-educated platelets (TEP), and exosomes, among others; the first three components are the most studied [19]. Exosomes are small membrane vesicles released by the cell under both normal and pathological conditions. Their concentrations in the blood correlate with tumor progression, suppression of the immune response, angiogenesis, and metastasis. They are detectable in patients with various types of cancers at concentrations of < 109 vesicles/ml. These biomarkers can be used for diagnosis, evaluation of tumor treatment, and prognosis, even in the setting of drug resistance [20, 21]. TEP are involved in systemic and local responses to tumor growth because tumor cells can alter the mRNA profile of platelets. TEP can splice the circulating mRNA released by the tumor cells and sequester solubilized proteins associated with the tumor. These interactions between platelets and tumor cells may have a significant potential for cancer diagnosis and monitoring of tumor progression. miRNAs are non-coding endogenous RNAs that regulate the expression of approximately 30% of protein-coding genes and function as oncogenes or tumor suppressors under certain conditions; they can also regulate different cellular pathways and they play an essential role in tumor growth and treatment resistance. miRNAs can be transported by exosomes, TEP, apoptotic bodies, or miRNA protein complexes. They have been proposed as possible biomarkers for the diagnosis, prognosis, and prediction of various types of cancers [19].
Circulating tumor cells (CTCs) are cells that have broken away from a primary tumor and/or metastatic lesion and travel through the bloodstream throughout the body. They can be isolated as individual cells or in groups. They are present in the blood at a low concentration (10 cells/ml of blood), even in metastatic environments [19]. CTCs are useful for determining tumor heterogeneity, identifying specific gene alterations on target, and evaluating the presence of treatment-associated markers (such as the programmed cell death ligand 1 [PD-L1]) for immunotherapies. The concentration of cfDNA is higher in advanced stage cancers and also correlates with tumor burden. Although the amount of cfDNA is higher in serum, plasma is preferred for the detection of ctDNA since the tumor DNA fraction is smaller and more variable in serum [19–21].
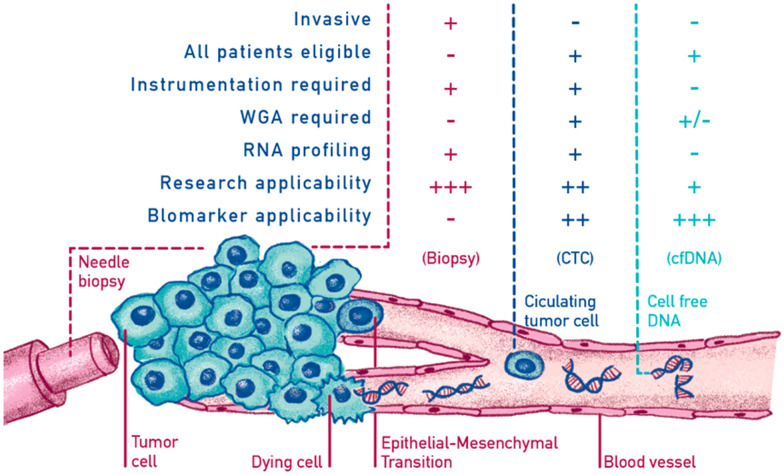
Elevated ctDNA levels were first reported in the serum of patients with cancer in 1977 [22]. More recently, a recent large-scale study reported that ctDNA was detectable in > 75% of patients with advanced pancreatic, ovarian, colorectal, bladder, gastroesophageal, breast, melanoma, hepatocellular, and head and neck cancers, but in < 50% of primary brain, renal, prostate, or thyroid cancers [23]. In the last two decades ctDNA has been explored both as a prognostic or predictive marker, as well as a cancer diagnostic tool [24–26]. The introduction of digital PCR (dPCR) enabled the accurate identification and absolute quantification of diverse mutant fragments. Furthermore, a modification of this technique, using beads in emulsions and flow cytometry, has enabled the quantification of the mutant allele fraction of cancer mutations in the plasma of patients at various stages of colorectal cancer [27–29]. In 2008, Diehl et al. indicated that ctDNA is a highly specific tumor dynamic marker that may reflect the total systemic tumor burden and residual disease [29]. In parallel, allele-specific PCR and other methods were conceived and tested for their ability to identify epidermal growth factor receptor (EGFR) mutations in the serum or plasma of lung cancer patients, following the elucidation of the role of such mutations in predicting response to targeted therapy [30, 31]. An explanation of the differences and potential of LB versus tumor tissue biopsy is provided in Fig. 1 .
Fig. 1.
The molecular characterization of neoplasms allows the correct application of precision medicine, maximizing the results and reducing the treatment’s toxicity. Tissue biopsy is the most widely used method for categorizing tumors and detecting biomarkers. However, it has several limitations: it is an invasive method; it is not always feasible or repeatable; and it provides information limited to a single point in space and time, therefore failing to capture the complexity of cancer. Today it is clear that cancer is an incredibly dynamic disease. During disease, cancers generally become more heterogeneous, with the results that the tumor might include a diverse collection of cells harboring distinct molecular signatures with differential sensitivity levels to treatment. This heterogeneity might result in a non-uniform distribution of genetically distinct tumor-cell subpopulations across and within disease sites (spatial variation) or temporal variations in cancer cells’ molecular makeup (temporal heterogeneity). For this reason, liquid biopsies can help achieve a correct and early detection of various neoplasm diseases, stratify patients to optimize treatment, and monitor the response and resistance mechanisms in the tumor. The non-invasive nature of the liquid biopsy allows systematically repeating of the sampling to monitor changes related to the dynamics of clonal and subclonal variation of the disease.
cfDNA Technologies and Applicability in the Clinical Setting
In early disease stages, specific tumor-derived DNA (i.e., cfDNA) is around 0.01% and identifying such a small quantity is very difficult [32–34]. Two main techniques are used to identify cfDNA. One technique identifies tumoral mutations from the primary tumor, providing the possibility of monitoring minimal residual disease but requiring a priori characterization using traditional strategies on tissue-based analyses. This technique is fast and has high specificity, and it dectects extremely low allele frequencies [35, 36]. The second technique detects aberrations or point mutations without prior tumoral profiling [37–39]. The methodologies used are NGS, dPCR, real-time PCR (qPCR), mass spectrometry, and detection of hypermethylation.
Non-specific methods based on the study of DNA methylation can be classified into site-specific methylation detection and genome-wide methylation detection. After some previous processes, such as bisulfite conversion and methylations of enriched complementary DNA (cDNA), hypermethylation can be detected using different techniques, such as conventional methylation-specific PCR (MSP), quantitative multiplexed specific methylation PCR (QM-PCR), and a modified version of PCR known as bead methylation (BOM), which involves the integration of three different processes in one tube: DNA extraction, bisulfite conversion and conventional PCR using DNA carriers, such as superparamagnetic iron oxide nanoparticles within silica microparticles [40].
Next-generation, deep sequencing has a specificity of around 80% and sensitivity close to 100%, and it detects low allele frequencies. Additionally, it is advantageous in detecting copy number variation with deficient quantities of cfDNA [41–43]. The highest sensitivity for identifying point mutations is offered with tagged amplicon deep sequencing (Tam-Seq), Safe-Sequencing System (Safe-SeqS), Cancer Personalized Profiling by deep Sequencing (CAPP-Seq), Bias Corrected Targeted NGS, and Multiplex PCR NGS, reaching a value close to 98–99%. Safe-SeqS amplifies a tagged region into families and sequences by a prespecified DNA template [43]. Tam-Seq implements an efficient library preparation and algorithms to identify hotspot regions and selected coding regions [33]. To detect low concentrations of cfDNA, CAPP-Seq sequencing involves certain regions previously hybridized by cDNA olygonucleotides and obtains an evaluation of mutations, rearrangements, and alteration in copy number. With further optimization of bioinformatics algorithms, a theoretical detection rate of 0.00025% allele frequency can be achieved [44, 45]. Bias Corrected Targeted NGS results in a high number of zero false positives because it uses markers that bind to small capture probes that are then amplified [46].
Two dPCR techniques can be used to conduct LB, namely, droplet PCR (ddPCR) and beads, emulsion, amplification, magnetics dPCR (BEAMing). The first technique, ddPCR, evaluates specific amplicons with flow cytometry by using specific fluorescent probes in an oil–water emulsion [38, 47]. BEAMing characterizes specific strand termination at divergent positions by flow cytometry. It is highly effective, but the cost and complex workflow make it an option that is complicated to implement [48].
Multiplex PCR NGS is the combination of high-throughput PCR amplification and sequencing and reaches a 99% sensitivity for the detection of mutations in target regions [49]. However, ot is relatively expansive to implement compared to dPCR. Conventional somatic mutation detection methods, such as quantitative PCR, only analyze ctDNA at high concentrations because of their low sensitivity. For that reason, more sensitive and specific methods using a combination of pyrophosphorolysis-activated polymerization and allele-specific amplification during PCR have been used [19]. Another recently developed method is dPCR, which seeks to detect and quantify mutations by analyzing individual molecules; 0.01% of ctDNA can be detected with this method [20, 50].
Finally, qPCR identifies only common mutations, at > 10% allele frequency, by blocking amplification at the oligo 3′-end, and amplifies only the mutant allele or enriches only variant alleles [46]. To overcome the limits of PCR, mass spectrometry is used, which detects up to 40 targets with low DNA input by signal emission from laser excitation or biotin markers that bind to specific regions [51–53].
Clonal Hematopoiesis as a Confounding Factor
When a hematopoietic stem cell expands and produces recurrent, specific, and disruptive genetic variants, a patient without hematological malignancy is known to have clonal hematopoiesis (CH) [54, 55]. When CH is present, somatic mutations in genes recurrently mutate in hematologic malignancies with a variant allele frequency (VAF) ≥ 2%; in the absence of known hematologic malignancy or other clonal disorders, it is defined clonal hematopoiesis of indeterminate potential (CHIP). The most frequent somatic mutations involve the genes DNMT3A, ASXL1, and TET2 [36, 38]. Similar to monoclonal gammopathy of undetermined significance (MGUS), individuals with CHIP have an increased risk of progression to hematologic neoplasia (0.5–1% per year) and all-cause mortality [56]. CH seems more common with aging (approx. 10% of individuals > 65 years, but it is relatively rare in subjects aged < 50 years) [57, 58]. Since cfDNA is a complex mixture of DNA from many sources (including germline, fetal, infectious, and malignant cells), CH may be a potential source of false positives in LB.
CH false positives represent a challenge for minimal residual disease (MRD) profiling and early cancer detection. It can also potentially cause discordances between tumor genotyping and plasma cfDNA genotyping in non-small-cell lung cancer (NSCLC) [59, 60]. Nowadays, the use of highly sensitive plasma NGS increases false positive results due to CH [61]. A comparison of four different NGS platforms to matched tumor–normal tissue pairs revealed that the conventional NGS approach (with both sensitivity and specificity of 95%) could not effectively distinguish between a cfDNA mutation with low VAF (< 0.1%) of hematopoietic origin and a tumor-derived mutation [39, 51]. Recently, a novel approach that incorporates whole blood cell sequencing into cfDNA analysis to filter potential contamination from somatic mutations attributable to CH has been described [62]. This study demonstrated that ultra-deep NGS of plasma cfDNA with CH filtering can accurately detect a wide variety of oncogenic drivers and resistance mechanisms in patients with advanced lung cancers, with a detection sensitivity comparable to that of established dPCR methods [62].
Applicability of LB in Solid Tumors
Over 8.2 million people die of cancer each year due to inaccessibility to appropriate detection procedures and treatments [63]. In this scenario, LBs have been recently gaining attention as an alternative to traditional tissue biopsies, given their minimal invasive nature and the vast amount of information provided [64]. The most commonly used techniques are the quantification of ctDNA and CTCs [65, 66]. Detection of specific genetic abnormalities based on mutation identification and DNA methylation analyses, both genome-wide and site-specific, have also been gaining relevance [67]. Furthermore, ctDNA carries some genomic and epigenomic alterations concordant to the tumor mutational spectrum, such as point mutations, degree of integrity, rearranged genomic sequences, CNV, microsatellite instability (MSI), loss of heterozygosity (LOH), and DNA methylation [68]. These biological characteristics discriminate ctDNA from normal cfDNA and reinforce the latter as a specific biomarker that provides personalized information to detect residual disease or monitor tumor progression during therapy [69]. Under normal circumstances, in healthy individuals, apoptotic and necrotic cells are cleared up by infiltrating phagocytes, and cfDNA levels are relatively low. However, this mechanism does not act effectively with tumoral masses. In solid tumors, ctDNA can also be released through necrosis, autophagy, and other physiological events induced by microenvironmental stress and treatment pressure [7]. The size of most fragments ranges between 180 and 200 bp, suggesting that apoptosis is the predominant source of ctDNA [69].
Advances in LB as a prognostic and predictive tool are mostly associated with lung, colon, and breast cancer (BC). Interestingly, each histology outcome has been preferably associated with a special type of LB. While research in lung or colon cancer has been linked to ctDNA analyses, for BC, the combination of ctDNA and CTCs are more widely used [70]. Advancements have been mainly achieved in the metastatic setting, as the amount of CTCs and ctDNA, which increase with tumoral burden, is the greatest. This, in turn, offers the best disease model for monitoring and assigning new therapies [44].
Specifically in the case of lung cancer, many multidisciplinary efforts have been made to advance the clinical application of LB. In this regard, the standardized isolation methods, such as solid-phase extraction techniques, have been used to isolate ctDNA, and many sequencing or mutation analysis methods have been developed to study and understand the clinical implications [71]. Furthermore, with the current development of high-throughput techniques for NGS, the evaluation of gene panels is expected to be included as a routine analysis, especially for the screening of high-risk individuals. In terms of treatment management, ctDNA has been used in NSCLC for targeted therapy selection, treatment monitoring, and detection of resistance mechanisms through the identification of somatic mutations in the EGFR gene. The Cobas® EGFRMutation Test v2 (Roche Diagnostics, F. Hoffmann-La Roche AG, Basel, Switzerland) was the first LB test approved by the U.S. Food and Drug Administration (FDA) in June 2016 for the detection of EGFR exon 19 deletions or exon 21 (L858R) substitution mutations in patients with NSCLC [72]. This approval represented major progress in establishing the clinical utility of ctDNAs.
In the case of colorectal cancer (CRC), detection of ctDNA in plasma has been associated with worse survival outcomes compared to undetectable levels [73]. Additionally, a strong association between ctDNA positivity and recurrence-free survival (RFS), and overall survival (OS) in patients with CRC irrespective of tumor stage, study size, tumor markers, detection methods, and sample type, is also worth noting [74, 75]. The utility of ctDNA as a prognostic parameter and alternative modality for mutation detection before treatment in metastatic CRC (mCRC) was demonstrated in the randomized CORRECT phase III trial. The progression-free survival (PFS) of patients with KRAS mutations detected in plasma treated with regorafenib was shorter than in those without mutations [76]; this finding established the prognostic value of KRAS mutations in plasma and eliminated the need for tissular confirmation [77]. In turn, it allowed the development of several ctDNA mutation assays and platforms for KRAS mutation screening and treatment selection, such as the Idylla™ (Biocartis, Mechelen, Belgium) and Cobas® platforms, both FDA approved.
BC was one of the first tumor models in which different types of LBs were studied. The prognostic value of ctDNA in BC has been documented in numerous studies, showing the clinical value of detecting of HER2 amplification and the PIK3CA and ESR1 mutations in ctDNA [78].
In the recent years, ctDNA analyses have been reviewed in other different neoplasia, such as liver, pancreas, ovarian, or prostate cancer [79]. In the case of pancreatic cancer (PC), one of the most aggressive and devastating of all malignancies, the absence of a reliable methodology for early identification and the limited therapeutic options for advanced disease set the stage for a possible broad-scale LB implementation [67]. In this context, a study conducted by Shapiro et al. reported that the presence of ctDNA is markedly elevated in patients with PC in comparison to healthy controls, leading these authors to conclude that ctDNA may serve as a useful diagnostic and prognostic biomarker [80]. Similar studies show that patients with PC with notable ctDNA levels have worse survival and greater risk of a diagnosis of an advanced disease stage [72]. Tjensvoll et al. also noted that during chemotherapy, changes in mutant KRAS circulation levels corresponded to radiological response assessments and cancer antigen 19-9 (CA19-9) values, suggesting the utility of ctDNA for monitoring treatment efficacy and tumor progression [81]. These studies demonstrate the potential clinical utility of ctDNA as a prognostic marker in PC.
In the case of ovarian cancer, historically there has been a lack of reliable screening strategies; however, the clinical value of ctDNA-based assays have since been demonstrated [82]. The first study on the application of LB for screening ovarian cancer, conducted by Cohen et al., was based on the detection of CNV in cfDNA and obtained satisfactory results [83]. Similar studies are being developed for other solid tumors, including hepatocellular carcinoma, head and neck cancer, and prostate cancer [84–86]. However, in other tumor types, ctDNA studies are still underexplored. Current standard clinical tools for the diagnosis and monitoring of central nervous system (CNS) tumors have several important limitations [87], of which the most important are the requirement of invasive tissue sampling, with its accompanying higher risk of complications relative to other locations, and the decreased feasibility of repetitive sampling. As a consequence, the use of LB in patients with CNS malignancies is particularly attractive. However, further studies are still necessary [88].
In conclusion, clinical evidence confirms that there are increased levels of ctDNA, particularly in metastatic disease, in patients with any of the solid tumors analyzed to date. In addition, ctDNA levels in cancer patients likely correlate with tumor burden, stage, vascularity, cellular turnover, and response to therapy. However, not all metastatic tumors seem to have the same impact on ctDNA levels, and more research is required since critical clinical standards (including uniform pre-analytic and analytical phase) are scarce and need to be established [17]. The harmonization of studies and comparative studies is crucial to provide clear evidence of the clinical utility of ctDNA in metastatic solid tumors.
Design of Clinical Trials to Assess the Usefulness of LB in Cancer
The design of clinical trials that evaluate the effectiveness of LB are under development. In the context of evaluating the LB technique, multiple objectives can be assessed: evaluating resistance pathways, pharmacodynamics, treatment efficacy, and prognostic and predictive markers. A very comprehensive review of studies assessing the effectiveness of LB in all these settings has been published [11]. Traditionally, LB implementation has been carried out parallel to treatment trials assessing the outcomes mentioned above. However, an interesting approach would be to assess LB as an allocation strategy between the treatment arms of clinical trials. Clinical trials assessing immune checkpoint inhibitors could potentially take advantage of this. PD-L1 expression correlates with responses to pembrolizumab in urothelial carcinoma, NSCLC, head and neck cancer, renal carcinoma, and melanoma [21, 89–91]. The currently running study Targeted Agent and Profiling Utilization Registry (TAPUR) conducted by the American Society of Clinical Oncology (ASCO) and several participating centers in the USA (ClinicalTrials.gov Identifier NCT02693535) recruits patients with advanced cancers with evidence of a potentially targetable genomic alteration who are not candidates for standard treatment. Patients with a LB genomic assessment also candidates for inclusion in the study. A specialized genomic tumoral board defines treatment, and a corresponding medication is administered. Results are expected in the upcoming years. This study is the closest design possible to a master protocol of patient recruitment based on a serum sample test.
Within the traditional trial design, it is difficult to conduct several studies for each possible tumor that carries a specific genomic driver. One strategy by which to address this issue is to implement master protocol trials that run multiple sub-studies in parallel. The three types of master protocol trials are basket, umbrella, and platform trials. Several definitions for each of these trial types are not consistent, but a review published by Hirakawa et al. comprehensively explores each definition and provides caveats [92]. The underlying notion for this avant-garde approach would be to utilize LB assays as a recruitment tool in large prospective clinical trials. Another design is the liquid dynamic medicine model. The basis for this proposal comes from the dynamic variation of molecular profiles in cancer: taking into account that resistance mechanisms arise in virtually all patients receiving targeted therapies, serial evaluation after disease progression could help elucidate the cause of set resistance. Although this concept is not new, the idea would be to administer sequential treatments, i.e., second and further lines, based on LB results, leading to the extreme personalization of care. Difficulties with this approach present in the form of the other lines: as the patient’s disease progresses, there remains fewer possible comparators of efficacy. One possible solution would be to compare the patient against her/himself in a previous treatment line in terms of time to disease progression. This novel strategy is referred to as the ‘N of 1 trial’ [4].
LB has also raised questions regarding the design and conducting of clinical trials. As more evidence on the usage of these assays is gathered, clinical trials should advance in order to provide optimal evidence-based strategies to implement these diagnostic modality.
Usage of LB in Current Clinical Practice
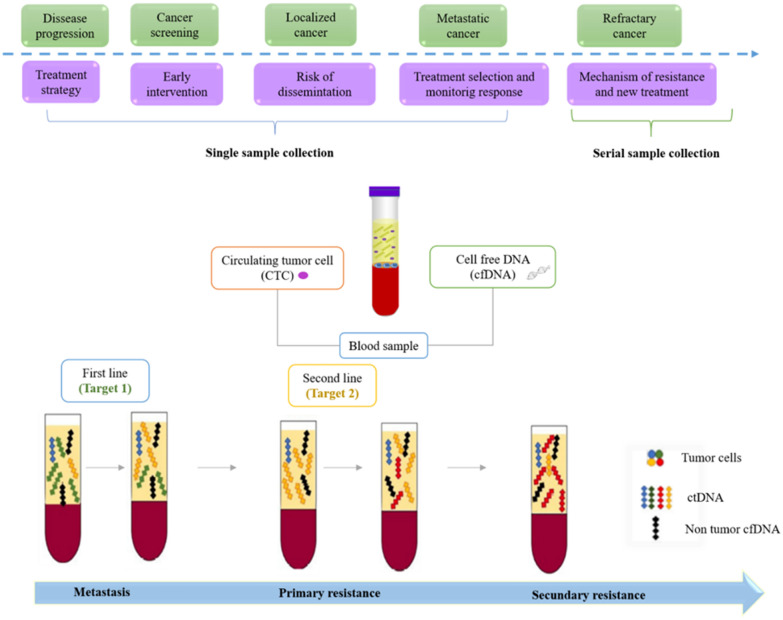
The use of LB in the clinical field has been increasing in recent years, not only due to the ease of obtaining samples from the patient but also because of the possibility of obtaining information at different stages of the disease and of early detection and prognosis [93]. The characteristics of cfDNA are different in cancer patients than in healthy patients [94]. Low levels of cell-free DNA have been reported in healthy patients at levels typically not exceeding 25 ng/ml plasma, while higher levels have been reported in cancer patients, ranging from < 0.1 to approximately 10% of DNA molecules, in association with tumor burden, stage, cell turnover, and accessibility to the bloodstream [95]. Similarly, it has been established that wild-type cfDNA is less fragmented than mutated cfDNA, with a size range of 250–320 and 90–150 bp, respectively [11]. Therefore, LB provides information that is useful for early detection, high-risk determination, choice, response to treatment, and early relapse detection in cancer patients. (Fig. 2). In fact, studies have reported that radiological imaging can only detect tumors when these are about 7–10 mm in size and contain about 1 billion cells [96, 97]. In this context, there is undoubtedly a potential for the use of LB in the early diagnosis of cancer, at a stage in which it can be curable [98].
Fig. 2.
Reported usages of liquid biopsy in cancer patients. The level of cfDNA in cancer patients has been used for the early detection of various types of cancer and patient monitoring at different stages of the disease. This is achieved through the information obtained from the analysis of circulating ctDNA, which provides information on the diagnosis, stage, response to treatment, and mechanisms of resistance that develop after treatment and predict the minimal residual disease and recurrence after surgery or therapy
(Taken and adapted from Bronkhorst et al. [6]; Haber et al. [50], and Vymetalkova et al. [2])
It also has been suggested that the cfDNA found in different body fluid samples, particularly in plasma, could represent a promising biomarker in patients with cancer because the relative levels of cfDNA are related to tumor burden as well as response to therapy. Additionally, more aggressive chemotherapy regimens have also been reported to be more effective in patients whose metastatic cancer is associated with high levels of circulating tumor cells [99, 100]. There is a correlation between cfDNA levels and cancer before treatment begins; as well, persistent and high levels of this type of DNA may indicate relapse and are likely a sign of poor prognosis [101]. Furthermore, for patients with localized cancer, the presence of circulating tumor cells or high levels of ctDNA could contribute towards the identification of patients at a higher risk of recurrence, in which adjuvant therapy could be considered [50]. Also, the high levels of ctDNA favor the determination of specific mutations in the cancer-conducting genes, such as those coding for EGFR, KRAS, BRAF, and PIK3CA, which are predictive of the response to different inhibitors. Another use that can be given is to find personalized biomarkers that are subsequently used to detect residual disease or monitor tumor levels during therapy [102].
Another use of LB in clinical practice is the detection of acquired resistance to targeted therapies. Taking into account that tumors acquire resistance to targeted therapies through the appearance of resistance mutations, which in turn lead to the activation of alternative signaling pathways, increased expression of anti-apoptotic genes, or reduced drug binding, LB is a useful tool in patient relapse, the potential need for continuous follow-upm and the appropriate selection of a second effective line of response for the patient [103].
Usefulness of LB for Screening and Early Detection
Identifying oncogenic driver mutations in asymptomatic or early-stage neoplasms could have a tremendous impact in cancer control and sequels. Breast and lung cancer are among the most prevalent candidates for inclusion in screening programs. An study of a large cohort of patients with BC analyzed promoter methylation in cfDNA in SFN, P16, hMLH1, HOXD13, PCDHGB7, and RASSFI, and a sensitivity and specificity close to 80% was obtained in terms of BC detection. Similar results were obtained with the comparison of benign breast lesions and infiltrating ones [7, 104, 105]. However, larger clinical trials are needed to search for potential markers for BC screening.
Similar conclusions can be drawn from studies on the early detection of lung cancer. Several techniques have provided exciting results. Homologous to the findings for BC, the methylation profiles in cfDNA in APC, CDH13, KLK10, DLEC1, and RASSF1A revealed a higher hypermethylation profile in patients with lung cancer than in healthy controls, achieving a sensitivity close to 84% and specificity of 74% [106]. mRNA analysis, proteomics, transcriptomics, metabolomics, microbiome evaluation, and other techniques have been examined [107]. Of the methodologies previously mentioned, the use of miRNAs has revealed promising results; specifically, the measurements of miR7, mi126, and mi145 in the sputum of 30 healthy controls and patients with lung cancer achieved a high diagnostic performance [108]. Proteomics in a similar setting with 139 cancer patients and 49 controls offered an area under the ROC curve (AUC) of 0.94 with the identification of five proteomic markers [109]. Although encouraging, several disparities exist in the standardization of techniques and studies, yielding a vast amount of results that differ from the reported ones, although similar methodologies were used. Standardization should be addressed in the planning of further studies in the screening and early detection of lung cancer [107]. In conclusion, although LBs are potentially an effective diagnostic tool in this setting, they need further development and characterization of markers.
Biological Fluids, Collection and Management of LB Samples
The feasibility of conducting LB in any fluid with a sufficient ctDNA concentration opens the door to the possibility of characterizing other compartments beyond the circulation. Taking into consideration that pleural effusions, cerebrospinal fluid (CSF), and urine can be populated with malignant cells in much higher concentrations than the circulation, direct analysis of these fluids could be feasible. Detection of EGFR mutations in patients who have lung cancer-related malignant pleural effusions have opened up the possibility to characterize fluids and offer both a diagnostic and prognostic strategy based solely on effusion analysis [110]. Also, detecting ctDNA in CSF for the assessment of primary brain tumors is possible for most patients. Contrary to other tumors, the lack of a widespread validation of targetable molecular targets limits this test’s applicability. On the other hand, metastatic tumors with seeding in the central nervous system could potentially be evaluated for treatment mechanisms as well as molecular disease profiling [111]. Urine has been gaining attention as a reliable source of LB material as it is easy to obtain and does not require any invasive procedure. To an extent, ctDNA is filtered through the kidneys from the blood which also re-collects cellular debris from the urinary tract. Although validated for urological tract neoplasms, the use of LB for non-urological tumors is a matter of controversy. Using both PCR and ddPCR, several studies have published different results across pathologies, using different sample sizes and specific mutation evaluation [112]. Notably, these techniques are not the best for detecting small quantities of DNA, opening the possibility for further studies employing low and extra low allele frequency detection, such as deep sequencing.
Considering that sample processing could also positively influence LB performance, a few points are worth exploring. cfDNA concentration per se is considered to be very low. Therefore, sample extraction and isolation should be efficient. Modern solutions employ columns that have improved extracted quantities compared to the older methods. However, fragments < 100 bp are still lost during the purification process [112]. Blood centrifugation, for example, leads to a release of DNA from cells present in the sample, introducing confounding material into the analysis. Newer techniques, such as dielectrophoresis, nanochip, and nanowire methods, have yielded interesting results that can potentially increase ctDNA detection ocess [113].
Other relevant sources of cfDNA include saliva and pleural fluids, among others. Salivary DNA has been used to detect germline mutations in breast and brain cancers because it provides good quality genomic DNA. Studies have shown that ctDNA isolated from saliva and blood can serve as a biomarker for head and neck squamous cell carcinoma (HNSCC) [114]. Tumor DNA detected in sputum may be useful for the early detection of lung cancer. Although the detection of EGFR mutations in pleural effusion samples is feasible, it is difficult due to the small number of tumor cells. However, studies have shown that the presence of ctDNA in pleural fluid indicate the existence of these mutations in patients with NSCLC [116]. Some studies have also been done to investigate the diagnostic, prognosis, or predictive value of miRNAs in pleural effusion fluid in patients with NSCLC, although more research is needed to validate the usefulness of these biomarkers in this type of sample [114].
Sampling and Handling of Samples
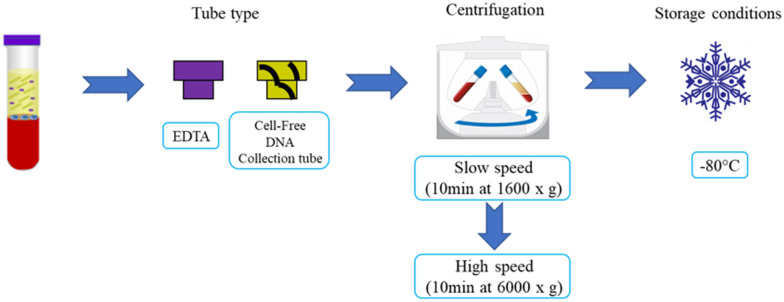
The most common source of ctDNA is plasma. Certain sample processing recommendations should be followed to ensure maximal performance; these are summarized in Fig. 3. A method designed for a selective extraction of small fragments of cfDNA (i.e., 166 bp) or a size selection for short DNA fragments should be performed [115].
Fig. 3.
Recommendations for the processing of plasma samples. Blood should be collected in EDTA-containing tubes (e.g., EDTAK3) as heparin and citrate could inhibit PCR. To prevent lysis of peripheral blood cells and the release of germline DNA, blood samples must be processed immediately after venipuncture by two rounds of refrigerated centrifugation, first at a slow speed to remove cells, followed by high-speed centrifugation of the carefully collected supernatant. Processing can be delayed to a maximum of 4–6 h. Following processing, the samples can be aliquoted and stored at − 80 °C [9]
(Taken and adapted from Bronkhorst et al. [6])
Economic Impact of Liquid Biopsies
An in vitro diagnosis assay requires different levels of investment in each phase of the study. Regarding the LB technique, we are in an initial phase, with the test having been demonstrated to be capable of identifying the presence of certain cancers. Clinical utility, such as clinical decision-making, including treatment regime change, can be taken based on LB results [72]. Positive assessments in areas are essential for a new LB assay to be approved by regulatory agencies, which will also require proof of an overall positive impact on healthcare providers’ systems, as this will enable a high return on investments. One of the main concerns of healthcare providers is the allocation of financial resources; consequently, assessing the impact of a single intervention on the whole system is a crucial task, but also it is not an easy one. To show the full positive economic impact on general health, several impact models have been proposed. One field which can be used as a mirror to develop LB economic impact models is personalized medicine (PM) and within it, economic models developed for genetic testing [116, 117]. As mentioned above, there is no single model to evaluate the economic impact of any clinical intervention; rather, there are many different strategies, of which cost-effective analysis (CEA) and cost–benefit analysis (CBA) are the most widely applied. These two systems differentiate on the normative paradigms which support them, extra-welfarism and welfare, respectively [118]. Extra-welfarism, which supports CEA, focuses on obtaining the maximum overall health status, and welfarism, which supports CBA, looks at individual utilities as its primary outcome. While both have their own individual issues, CEA is the more appropriate in the context of the utility of LBs as it quantifies the impact of the intervention on overall health using parameters such as quality-adjusted life years (QALY) and life-years gained (LYG), both of which measure clinical effectiveness.
While CEA should be a must in developing LB essays, there are not many publications that report such an analysis. One example of a LB assay which has demonstrated its positive economic impact is SelectMDx (MDXHealth, Irvine, CA, USA), an assay based on circulating epigenetic markers for prostate cancer. A CEA in four different countries (Germany, France, Spain, and Italy) reported that, on average, LB intervention results in a savings of €665 and a gain of 0.025 QALY per patient compared to using the PSA alone [119].
Due to this uncertainty and lack of CEA of the different LB assays, it is challenging to calculate the future size of the market for LBs. Also, the full sets of clinical utilities are still to be fully defined. Currently, LB is being used mainly for cancer prognosis and therapy selection in metastatic cancers. However, there are already studies that have proposed that LBs be used for early diagnosis and non-tumoral applications [120–122]. These new LB applications have still to go through the full cycle of regulatory approval and clinical efficacy defined above. The CEA on these applications will determine the full scale of the economic impact of LBs on health systems.
Hence, a broad range of forecasted figures, ranging from $2.4 billion to $100 billion in 2030, can be found. Combining their potential clinical utilities and the broad market that could be created has made investors think it is an opportunity too good to be missed. In the meantime, most LB assays are currently available as laboratory developed tests (LDT) or FDA-approved products and offered as an out-of-pocket test to patients.
Liquid Biopsies for Population Wellbeing
According to the World Health Organization, “a screening program should respond to a recognized need, and its objectives should be defined at the outset. There should be a defined target population and scientific evidence of the screening program’s effectiveness. The program should integrate education, testing, clinical services, and program management, along with quality assurance and mechanisms to minimize potential risks of screening. It should ensure informed choice, confidentiality, and respect for autonomy, and promote equity and access to screening for the entire target population. The evaluation should be planned from the outset, and the overall benefits of screening should outweigh the harm” [123]. In this context, the screening programs for healthy and high-risk individuals for cancer is a crucial application of LB approaches, independently of whether the high-risk population is based, or not, in a developed country [124].
According to the National Cancer Institute (NCI) dictionary of cancer terms, liquid biopsy is “a test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood” [125]. However, from our point of view, this definition is rather imperfect, since this terminology cannot be applied to screening as it does not include: circulating DNAs that are analyzed in the field of prenatal diagnosis, severe/acute inflammation (sepsis), and transplantation, or its application for evaluating pre-tumor or cancer interception (chronic obstructive pulmonary disease [COPD], asthma, or liver disease) [126]. Therefore, all previous concepts should be integrated in the definition, including the concept of cancer interception. This concept could help to develop tools for early cancer detection because it studies the events associated with cancer initiation and progression [127].
Nowadays, many groups are developing cancer-screening tests based on LBs. The majority of these studies are focused on CRC and lung cancer. In CRC, many markers have been identified, such as the methylated Septin9, which tend to be higher in CRC patients than in patients without the disease [128, 129]. These results include the development of a specific test named Epi proColon® 2.0 CE (Epigenomics AG, Berlin, Germany). This test discriminated patients with CRC and healthy controls with high clinical sensitivity and specificity in pivotal case–control studies [130]. Other studies have been conducted with the aim to also evaluate different hypermethylated DNA promoter regions and genes specific to CRC that are present in the plasma [137]. In addition, an age-adjusted panel of four cell-free nucleosomes was developed by Volition (Volition SPRI, Namur, Belgium) for the discrimination between CRC patients and healthy controls. The results of this study showed that the panel had a high sensitivity for early stages (75 and 86% for stages I and II, respectively) at 90% specificity [131]. Regarding lung cancer, the increased levels of cfDNA were also analyzed in a recent study by Esposito et al., where the cfDNA levels from cancer patients were compared to those of healthy individuals [132]; the results of this work demonstrated an increase of cfDNA levels in patients with early LC compared to the cfDNA levels in healthy donors. Similar results were observed for BC with 78% sensitivity and 83% specificity, and ovarian cancer with 70% sensitivity and 90% specificity [133, 134]. Other types of LBs have been used for screening both CCR and lung cancer [129, 135]. However, large studies are still necessary to demonstrate their usefulness as early diagnostic markers.
The patient experience has been recognized as one of the three pillars of quality health care, alongside with clinical quality and patient safety [136]. Patient experience is the most effective measurement of the patient-approach, defined as providing care that is respectful of and responsive to patient preferences, needs, and values. Thus, patients’ groups have become involved in scientific and therapeutic activism to achieve positive feedback for technological development. In the case of LBs and, in particular, in the use of LB as a screening marker, the implication of patients’ associations is especially important to spread knowledge about these new screening methodologies. Note that the potential use of LBs as cancer-screening tests involves the analyses of large populations. Patients’ organizations help promote research and develop processes and methodologies to ensure that their members are fully prepared to get involved in research and clinical trials, and ensure that patients can participate wherever needed. These associations can promote initiatives to bring together patient organizations from different countries and to help others engage as experts in their disease (e.g. EU-funded initiatives) [137]. In addition, patients’ associations could help to involve health centers, encouraging primary care practitioners to engage in early intervention through LB programs, since only a mere blood extraction is needed.
Also, patients’ organizations can play a key role in obtaining funding for LB research, thereby facilitating more research and obtaining money to support early screening programs. However, despite patients’ associations and academic and clinical research in LB programs, implementation of LBs can only become possible if pharmaceutical companies are involved and technology companies provide advice. Based on LB, the screening programs involve the need to work with highly sensitive and specific technologies. In addition, pharmaceutical companies have a double purpose in this context. Their participation promotes research and the dissemination of knowledge on LB, but they are also responsible for the development of “therapeutic responses” that these high-risk populations could need.
Conclusions
Liquid biopsy is an umbrella term that encompasses several techniques for evaluating genomic information that is simple to obtain but complex to perform and analyze. Although several methodologies exist that have been clinically validated, the continuous developmental effort and standardization will lead in the future to further indications for LB that could impact outcomes in cancer patients specifically and population health in general. The implementation of clinical studies with alternate designs will facilitate the evaluation and final measurement of the impact of this novel diagnostic tool.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Andrés F. Cardona discloses financial research support from Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb, and The Foundation for Clinical and Applied Cancer Research (FICMAC). Additionally, he was linked to and received honoraria as advisor, participant in speakers' bureau, and expert from Merck Sharp & Dohme, Boehringer Ingelheim, Roche, Bristol-Myers Squibb, Pfizer, Novartis, Celldex Therapeutics, Foundation Medicine, Eli Lilly, and Foundation for Clinical and Applied Cancer Research (FICMAC). He is also on the Editorial board of Oncology and Therapy. Oscar Arrieta reports personal fees from Pfizer, grants and personal fees from Astra Zeneca, Boehringer-Ingelheim, Lilly, Merck, Bristol Myers Squibb, and Roche, outside the submitted work. Christian Rolfo reports a relation with Mylan, Archer Biosciences, Oncopass, Inivata, Merck Serono Novartis, MSD, Boehringer Ingelheim, Guardant Health, and AstraZeneca as part of Speakers’ Bureau. In addition, he received research funding from Pfizer and had uncompensated associations with OncoDNA, Biomark, and Guardant Health. Gonzalo Recondo discloses travel, accommodations, and expenses from AstraZeneca and Pfizer, and a consulting advisory role with Roche, Amgen, and Pfizer. July Rodríguez, Jenny Avila, Christian Rolfo, Alejandro Ruíz-Patiño, Alessandro Russo, Luisa Ricaurte, Camila Ordóñez-Reyes, and Zyanya Lucia Zatarain-Barrón have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
References
- 1.Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. Role of circulating cell-free DNA in cancers. Mol Diagn Ther. 2015;19(6):339–350. doi: 10.1007/s40291-015-0167-y. [DOI] [PubMed] [Google Scholar]
- 2.Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating cell-free DNA and colorectal cancer: a systematic review. Int J Mol Sci. 2018;19(11):3356. doi: 10.3390/ijms19113356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiley C, de Bruin EC, McGranahan N, Swanton C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol. 2014;15(8):453. doi: 10.1186/s13059-014-0453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silvestris N, Ciliberto G, De Paoli P, et al. Liquid dynamic medicine and N-of-1 clinical trials: a change of perspective in oncology research. J Exp Clin Cancer Res. 2017;36(1):128. doi: 10.1186/s13046-017-0598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif. 2019;17. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6425120/. Accessed 30 Sep 2020. [DOI] [PMC free article] [PubMed]
- 7.Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnedos M, Vicier C, Loi S, et al. Precision medicine for metastatic breast cancer–limitations and solutions. Nat Rev Clin Oncol. 2015;12(12):693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 9.Patel KM, Tsui DWY. The translational potential of circulating tumour DNA in oncology. Clin Biochem. 2015;48(15):957–961. doi: 10.1016/j.clinbiochem.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Otandault A, Anker P, Al Amir Dache Z, et al. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 2019;30(3):374–384. doi: 10.1093/annonc/mdz031. [DOI] [PubMed] [Google Scholar]
- 11.Nandi K, Verma R, Dawar R, Goswami B. Cell free DNA: revolution in molecular diagnostics—the journey so far. Horm Mol Biol Clin Investig. 2020;41(1):/j/hmbci.2020.41.issue-1/hmbci-2019-0012/hmbci-2019-0012.xml. [DOI] [PubMed]
- 12.Moscow JA, Fojo T, Schilsky RL. The evidence framework for precision cancer medicine. Nat Rev Clin Oncol. 2018;15(3):183–192. doi: 10.1038/nrclinonc.2017.186. [DOI] [PubMed] [Google Scholar]
- 13.Di Sanzo M, Cipolloni L, Borro M, et al. Clinical applications of personalized medicine: a new paradigm and challenge. Curr Pharm Biotechnol. 2017;18(3):194–203. doi: 10.2174/1389201018666170224105600. [DOI] [PubMed] [Google Scholar]
- 14.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15(2):81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 15.Green ED, Watson JD, Collins FS. Human genome project: twenty-five years of big biology. Nature. 2015;526(7571):29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional oncogenic driver mutation analysis in lung adenocarcinoma: the lung cancer mutation consortium experience. J Thorac Oncol. 2015;10(5):768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, Zhang Z-H, Wang S, Lang J-H. Circulating cell-free DNA or circulating tumor DNA in the management of ovarian and endometrial cancer. Onco Targets Ther. 2019;27(12):11517–11530. doi: 10.2147/OTT.S227156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulet G, Massias J, Taly V. Liquid biopsy: general concepts. Acta Cytol. 2019;63(6):449–455. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 20.Jia S, Zhang R, Li Z, Li J. Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget. 2017;8(33):55632–55645. doi: 10.18632/oncotarget.17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guibert N, Delaunay M, Lusque A, et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer. 2018;120:108–112. doi: 10.1016/j.lungcan.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- 24.Cappelletti V, Appierto V, Tiberio P, Fina E, Callari M, Daidone MG. Circulating biomarkers for prediction of treatment response. J Natl Cancer Inst Monogrs. 2015;2015(51):60–63. doi: 10.1093/jncimonographs/lgv006. [DOI] [PubMed] [Google Scholar]
- 25.Ou S-HI, Nagasaka M, Zhu VW. Liquid biopsy to identify actionable genomic alterations. Am Soc Clin Oncol Educ Book. 2018;38:978–997. doi: 10.1200/EDBK_199765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. 1999;96(16):9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100(15):8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol. 2018;13(9):1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 31.Sorber L, Zwaenepoel K, De Winne K, et al. A multicenter study to assess egfr mutational status in plasma: focus on an optimized workflow for liquid biopsy in a clinical setting. Cancers. 2018;10(9):290. doi: 10.3390/cancers10090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci. 2008;1137:190–196. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 33.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy SR, Schmitt MW, Fox EJ, et al. Detecting ultralow-frequency mutations by Duplex sequencing. Nat Protoc. 2014;9(11):2586–2606. doi: 10.1038/nprot.2014.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cristofanilli M, Braun S. Circulating tumor cells revisited. JAMA. 2010;303(11):1092–1093. doi: 10.1001/jama.2010.292. [DOI] [PubMed] [Google Scholar]
- 36.Freidin MB, Freydina DV, Leung M, Montero Fernandez A, Nicholson AG, Lim E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem. 2015;61(10):1299–1304. doi: 10.1373/clinchem.2015.242453. [DOI] [PubMed] [Google Scholar]
- 37.Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res. 2014;20(10):2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 40.Marrugo-Ramírez J, Mir M, Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int J Mol Sci. 2018;19(10). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6213360/. Accessed 30 Sep 2020. [DOI] [PMC free article] [PubMed]
- 41.Narayan A, Carriero NJ, Gettinger SN, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72(14):3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res. 2014;20(17):4613–4624. doi: 10.1158/1078-0432.CCR-13-3063. [DOI] [PubMed] [Google Scholar]
- 43.Uchida J, Kato K, Kukita Y, et al. Diagnostic accuracy of noninvasive genotyping of EGFR in lung cancer patients by deep sequencing of plasma cell-free DNA. Clin Chem. 2015;61(9):1191–1196. doi: 10.1373/clinchem.2015.241414. [DOI] [PubMed] [Google Scholar]
- 44.Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy—current status and where we need to progress. Comput Struct Biotechnol J. 2018;16:190–195. doi: 10.1016/j.csbj.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34(5):547–555. doi: 10.1038/nbt.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res. 2016;22(4):915–922. doi: 10.1158/1078-0432.CCR-15-1627-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem. 2009;55(8):1471–1483. doi: 10.1373/clinchem.2008.121962. [DOI] [PubMed] [Google Scholar]
- 48.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4(6):650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wee EJH, Wang Y, Tsao SC-H, Trau M. Simple, sensitive and accurate multiplex detection of clinically important melanoma DNA mutations in circulating tumour DNA with SERS Nanotags. Theranostics. 2016;6(10):1506–1513. doi: 10.7150/thno.15871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harper MM, McKeating KS, Faulds K. Recent developments and future directions in SERS for bioanalysis. Phys Chem Chem Phys. 2013;15(15):5312–5328. doi: 10.1039/c2cp43859c. [DOI] [PubMed] [Google Scholar]
- 53.Mosko MJ, Nakorchevsky AA, Flores E, et al. Ultrasensitive detection of multiplexed somatic mutations using MALDI-TOF mass spectrometry. J Mol Diagn. 2016;18(1):23–31. doi: 10.1016/j.jmoldx.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Gibson CJ, Steensma DP. New insights from studies of clonal hematopoiesis. Clin Cancer Res. 2018;24(19):4633–4642. doi: 10.1158/1078-0432.CCR-17-3044. [DOI] [PubMed] [Google Scholar]
- 55.Shlush LI. Age-related clonal hematopoiesis. Blood. 2018;131(5):496–504. doi: 10.1182/blood-2017-07-746453. [DOI] [PubMed] [Google Scholar]
- 56.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J, Chen X, Wang J, et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann Oncol. 2019;30(3):464–470. doi: 10.1093/annonc/mdy513. [DOI] [PubMed] [Google Scholar]
- 60.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15(9):577–586. doi: 10.1038/s41571-018-0058-3. [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, Ulrich BC, Supplee J, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24(18):4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 62.Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30(4):597–603. doi: 10.1093/annonc/mdz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGuire S. World Cancer Report 2014 Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2015;7(2):418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delgado-Ureña M, Ortega FG, Miguel-Pérez D, et al. Circulating tumor cells criteria (CyCAR) versus standard RECIST criteria for treatment response assessment in metastatic colorectal cancer patients. J Transl Med. 2018;16(1):251. doi: 10.1186/s12967-018-1624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Chang S, Li G, Sun Y. Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med. 2017;11(4):522–527. doi: 10.1007/s11684-017-0526-7. [DOI] [PubMed] [Google Scholar]
- 66.Batth IS, Mitra A, Manier S, et al. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol. 2017;28(3):468–477. doi: 10.1093/annonc/mdw619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yadav DK, Bai X, Yadav RK, et al. Liquid biopsy in pancreatic cancer: the beginning of a new era. Oncotarget. 2018;9(42):26900–26933. doi: 10.18632/oncotarget.24809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chae YK, Davis AA, Jain S, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther. 2017;16(7):1412–1420. doi: 10.1158/1535-7163.MCT-17-0061. [DOI] [PubMed] [Google Scholar]
- 69.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347–376. doi: 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hench IB, Hench J, Tolnay M. Liquid biopsy in clinical management of breast, lung, and colorectal cancer. Front Med. 2018;5:9. doi: 10.3389/fmed.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathai RA, Vidya RVS, Reddy BS, et al. Potential utility of liquid biopsy as a diagnostic and prognostic tool for the assessment of solid tumors: implications in the precision oncology. J Clin Med. 2019;8(3):373. doi: 10.3390/jcm8030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.The ASCO Post. FDA approves cobas EGFR mutation test v2 as companion diagnostic with gefitinib in first-line treatment of NSCLC. 2018. https://www.ascopost.com/News/59214. Accessed 1 June 2019.
- 73.Malapelle U, Sirera R, Jantus-Lewintre E, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn. 2017;17(3):209–215. doi: 10.1080/14737159.2017.1288568. [DOI] [PubMed] [Google Scholar]
- 74.Vietsch EE, Graham GT, McCutcheon JN, et al. Circulating cell-free DNA mutation patterns in early and late stage colon and pancreatic cancer. Cancer Genet. 2017;218–219:39–50. doi: 10.1016/j.cancergen.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basnet S, Zhang Z-Y, Liao W-Q, Li S-H, Li P-S, Ge H-Y. The prognostic value of circulating cell-free DNA in colorectal cancer: a meta-analysis. J Cancer. 2016;7(9):1105–1113. doi: 10.7150/jca.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tabernero J, Lenz H-J, Siena S, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16(8):937–948. doi: 10.1016/S1470-2045(15)00138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Analysis of ESR1 and PIK3CA mutations in plasma cell-free DNA from ER-positive breast cancer patients. Oncotarget. 2017;8(32):52142–52155. doi: 10.18632/oncotarget.18479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Leary B, Hrebien S, Morden JP, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun. 2018;9(1):896. doi: 10.1038/s41467-018-03215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramalingam N, Jeffrey SS. Future of liquid biopsies with growing technological and bioinformatics studies: opportunities and challenges in discovering tumor heterogeneity with single-cell level analysis. Cancer J. 2018;24(2):104–108. doi: 10.1097/PPO.0000000000000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51(11):2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::aid-cncr2820511127>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 81.Tjensvoll K, Lapin M, Buhl T, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016;10(4):635–643. doi: 10.1016/j.molonc.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giannopoulou L, Kasimir-Bauer S, Lianidou ES. Liquid biopsy in ovarian cancer: recent advances on circulating tumor cells and circulating tumor DNA. Clin Chem Lab Med. 2018;56(2):186–197. doi: 10.1515/cclm-2017-0019. [DOI] [PubMed] [Google Scholar]
- 83.Cohen PA, Flowers N, Tong S, Hannan N, Pertile MD, Hui L. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med. 2016;14(1):126. doi: 10.1186/s12916-016-0667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okajima W, Komatsu S, Ichikawa D, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23(31):5650–5668. doi: 10.3748/wjg.v23.i31.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Payne K, Spruce R, Beggs A, et al. Circulating tumor DNA as a biomarker and liquid biopsy in head and neck squamous cell carcinoma. Head Neck. 2018;40(7):1598–1604. doi: 10.1002/hed.25140. [DOI] [PubMed] [Google Scholar]
- 86.Stone L. Prostate cancer: biomarkers from liquid biopsy. Nat Rev Urol. 2016;13(8):434. doi: 10.1038/nrurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 87.Bertero L, Siravegna G, Rudà R, Soffietti R, Bardelli A, Cassoni P. Review: peering through a keyhole: liquid biopsy in primary and metastatic central nervous system tumours. Neuropathol Appl Neurobiol. 2019;45:655–670. doi: 10.1111/nan.12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boire A, Brandsma D, Brastianos PK, et al. Liquid biopsy in central nervous system metastases: a RANO review and proposals for clinical applications. Neuro-oncology. 2019;21(5):571–584. doi: 10.1093/neuonc/noz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer. 2018;17(1):129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yue C, Jiang Y, Li P, et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. Oncoimmunology. 2018;7(7):e1438111. doi: 10.1080/2162402X.2018.1438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y, Kim TH, Fouladdel S, et al. PD-L1 expression in circulating tumor cells increases during radio(chemo)therapy and indicates poor prognosis in non-small cell lung cancer. Sci Rep. 2019;9(1):566. doi: 10.1038/s41598-018-36096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirakawa A, Asano J, Sato H, Teramukai S. Master protocol trials in oncology: review and new trial designs. Contemp Clin Trials Commun. 2018;12:1–8. doi: 10.1016/j.conctc.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Riediger AL, Dietz S, Schirmer U, et al. Mutation analysis of circulating plasma DNA to determine response to EGFR tyrosine kinase inhibitor therapy of lung adenocarcinoma patients. Sci Rep. 2016;6(1):33505. doi: 10.1038/srep33505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kahlert C. Liquid biopsy: is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019;79(10):2462. doi: 10.1158/0008-5472.CAN-19-0019. [DOI] [PubMed] [Google Scholar]
- 95.Francis G, Stein S. Circulating cell-free tumour DNA in the management of cancer. Int J Mol Sci. 2015;16(6):14122–14142. doi: 10.3390/ijms160614122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sorber L, Zwaenepoel K, Deschoolmeester V, et al. A comparison of cell-free DNA isolation kits: isolation and quantification of cell-free DNA in plasma. J Mol Diagn. 2017;19(1):162–168. doi: 10.1016/j.jmoldx.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 97.Wu X, Li J, Gassa A, et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci. 2020;16(9):1551–1562. doi: 10.7150/ijbs.44024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muhanna N, Di Grappa MA, Chan HHL, et al. Cell-free DNA kinetics in a pre-clinical model of head and neck cancer. Sci Rep. 2017;7(1):16723. doi: 10.1038/s41598-017-17079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thijssen MAMA, Swinkels DW, Ruers TJM, de Kok JB. Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res. 2002;22(1A):421–425. [PubMed] [Google Scholar]
- 101.Phallen J, Sausen M, Adleff V, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403):eaan2415. 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed]
- 102.Sausen M, Phallen J, Adleff V, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;7(6):7686. doi: 10.1038/ncomms8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heitzer E, Auer M, Gasch C, et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73(10):2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- 104.Shan M, Yin H, Li J, et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7(14):18485–18494. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Z, Guo X, Tang L, et al. Methylation analysis of plasma cell-free DNA for breast cancer early detection using bisulfite next-generation sequencing. Tumour Biol. 2016;37(10):13111–13119. doi: 10.1007/s13277-016-5190-z. [DOI] [PubMed] [Google Scholar]
- 106.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303(1):21–28. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 107.Calabrese F, Lunardi F, Pezzuto F, et al. Are there new biomarkers in tissue and liquid biopsies for the early detection of non-small cell lung cancer? J Clin Med. 2019;8(3):414. doi: 10.3390/jcm8030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bagheri A, Khorshid HRK, Tavallaie M, et al. A panel of noncoding RNAs in non-small-cell lung cancer. J Cell Biochem. 2018;120:8280–8290. doi: 10.1002/jcb.28111. [DOI] [PubMed] [Google Scholar]
- 109.Uribarri M, Hormaeche I, Zalacain R, Lopez-Vivanco G, Martinez A, Nagore D, et al. A new biomarker panel in bronchoalveolar lavage for an improved lung cancer diagnosis. J Thorac Oncol. 2014;9(10):1504–1512. doi: 10.1097/JTO.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 110.Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer. 2018;18(1):1236. doi: 10.1186/s12885-018-5138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seoane J, De Mattos-Arruda L, Le Rhun E, Bardelli A, Weller M. Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann Oncol. 2019;30(2):211–218. doi: 10.1093/annonc/mdy544. [DOI] [PubMed] [Google Scholar]
- 112.Jain S, Lin SY, Song W, Su Y-H. Urine-based liquid biopsy for nonurological cancers. Genet Test Mol Biomark. 2019;23(4):277–283. doi: 10.1089/gtmb.2018.0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gorgannezhad L, Umer M, Islam MN, Nguyen N-T, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18(8):1174–1196. doi: 10.1039/C8LC00100F. [DOI] [PubMed] [Google Scholar]
- 114.Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol. 2019;141:36–42. doi: 10.1016/j.critrevonc.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 115.Bronkhorst AJ, Wentzel JF, Ungerer V, et al. Sequence analysis of cell-free DNA derived from cultured human bone osteosarcoma (143B) cells. Tumour Biol. 2018;40(9):1010428318801190. doi: 10.1177/1010428318801190. [DOI] [PubMed] [Google Scholar]
- 116.Phillips KA, Ann Sakowski J, Trosman J, Douglas MP, Liang S-Y, Neumann P. The economic value of personalized medicine tests: what we know and what we need to know. Genet Med. 2014;16(3):251–257. doi: 10.1038/gim.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Payne K, Gavan SP, Wright SJ, Thompson AJ. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet. 2018;19(4):235–246. doi: 10.1038/nrg.2017.108. [DOI] [PubMed] [Google Scholar]
- 118.Seixas BV. Welfarism and extra-welfarism: a critical overview. Cad Saude Publica. 2017;33(8):e00014317. doi: 10.1590/0102-311X00014317. [DOI] [PubMed] [Google Scholar]
- 119.Govers TM, Hessels D, Vlaeminck-Guillem V, Schmitz-Dräger BJ, Stief CG, Martinez-Ballesteros C, et al. Cost-effectiveness of SelectMDx for prostate cancer in four European countries: a comparative modeling study. Prostate Cancer Prostatic Dis. 2019;22(1):101–109. doi: 10.1038/s41391-018-0076-3. [DOI] [PubMed] [Google Scholar]
- 120.Oxnard GR, Maddala T, Hubbell E, et al. Genome-wide sequencing for early stage lung cancer detection from plasma cell-free DNA (cfDNA): the Circulating Cancer Genome Atlas (CCGA) study. J Clin Oncol. 2018;36(18_suppl):LBA8501. [Google Scholar]
- 121.Bhan I, Mosesso K, Goyal L, et al. Detection and analysis of circulating epithelial cells in liquid biopsies from patients with liver disease. Gastroenterology. 2018;155(6):2016–2018.e11. doi: 10.1053/j.gastro.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Romero-Palacios PJ, Alcázar-Navarrete B, Díaz Mochón JJ, et al. Liquid biopsy beyond of cancer: circulating pulmonary cells as biomarkers of COPD aggressivity. Crit Rev Oncol Hematol. 2019;136:31–36. doi: 10.1016/j.critrevonc.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 123.Andermann A, Blancquaert I, Beauchamp S, Déry V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ. 2008;86(4):317–9. 10.2471/blt.07.050112. [DOI] [PMC free article] [PubMed]
- 124.Thierry AR. A step closer to cancer screening by blood test. Clin Chem. 2018;64(10):1420–1422. doi: 10.1373/clinchem.2018.287847. [DOI] [PubMed] [Google Scholar]
- 125.National Cancer Institute. Definition: liquid biopsy. https://www.cancer.gov/search/results?swKeyword=liquid+biopsy. Accessed 1 June 2019.
- 126.Ehrich M, Tynan J, Mazloom A, et al. Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet Med. 2017;19(12):1332–1337. doi: 10.1038/gim.2017.56. [DOI] [PubMed] [Google Scholar]
- 127.Beane J, Campbell JD, Lel J, Vick J, Spira A. Genomic approaches to accelerate cancer interception. Lancet Oncol. 2017;18(8):e494–502. doi: 10.1016/S1470-2045(17)30373-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Molnár B, Tóth K, Barták BK, Tulassay Z. Plasma methylated septin 9: a colorectal cancer screening marker. Expert Rev Mol Diagn. 2015;15(2):171–184. doi: 10.1586/14737159.2015.975212. [DOI] [PubMed] [Google Scholar]
- 129.Lamb YN, Dhillon S. Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther. 2017;21(2):225–232. doi: 10.1007/s40291-017-0259-y. [DOI] [PubMed] [Google Scholar]
- 130.Rasmussen SL, Krarup HB, Sunesen KG, et al. Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS ONE. 2017;12(7):e0180809. doi: 10.1371/journal.pone.0180809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rahier J-F, Druez A, Faugeras L, et al. Circulating nucleosomes as new blood-based biomarkers for detection of colorectal cancer. Clin Epigenetics. 2017;9:53. doi: 10.1186/s13148-017-0351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Esposito A, Criscitiello C, Trapani D, Curigliano G. The emerging role of “liquid biopsies”, circulating tumor cells, and circulating cell-free tumor DNA in lung cancer diagnosis and identification of resistance mutations. Curr Oncol Rep. 2017;19(1):1. doi: 10.1007/s11912-017-0564-y. [DOI] [PubMed] [Google Scholar]
- 133.Lin Z, Neiswender J, Fang B, Ma X, Zhang J, Hu X. Value of circulating cell-free DNA analysis as a diagnostic tool for breast cancer: a meta-analysis. Oncotarget. 2017;8(16):26625–26636. doi: 10.18632/oncotarget.15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou Q, Li W, Leng B, et al. Circulating cell free DNA as the diagnostic marker for ovarian cancer: a systematic review and meta-analysis. PLoS ONE. 2016;11(6):e0155495. doi: 10.1371/journal.pone.0155495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang R, Shao F, Wu X, Ying K. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysis. Lung Cancer. 2010;69(2):225–231. doi: 10.1016/j.lungcan.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 136.Doyle C, Lennox L, Bell D. A systematic review of evidence on the links between patient experience and clinical safety and effectiveness. BMJ Open. 2013;3(1):e001570. 10.1136/bmjopen-2012-001570. [DOI] [PMC free article] [PubMed]
- 137.Plueschke K, McGettigan P, Pacurariu A, Kurz X, Cave A. EU-funded initiatives for real world evidence: descriptive analysis of their characteristics and relevance for regulatory decision-making. BMJ Open. 2018;8(6):e021864. doi: 10.1136/bmjopen-2018-021864. [DOI] [PMC free article] [PubMed] [Google Scholar]