Abstract
Late leaf spot (LLS) caused by fungi Passalora personata is generally more destructive and difficult to control than early leaf spot. The aim of this study was to decipher biochemical defense mechanism in groundnut genotypes against P. personata by identifying resistance specific biomarkers and metabolic pathways induced during host–pathogen interaction. Metabolomics of non-infected and infected leaves of moderately resistant (GPBD4 and ICGV86590), resistant (KDG128 and RHRG06083) and susceptible (GG20, JL24 and TMV2) genotypes was carried out at 5 days after infection (65 days after sowing). Non-targeted metabolite analysis using GC–MS revealed total 77 metabolites including carbohydrates, sugar alcohols, amino acids, fatty acids, polyamines, phenolics, terpenes and sterols. Variable importance in projection (VIP) measure of partial least squares-discriminant analysis (PLS-DA) showed that resistant and moderately resistant genotypes possessed higher intensities of ribonic acid, cinnamic acid, malic acid, squalene, xylulose, galactose, fructose, glucose, β-amyrin and hydroquinone while susceptible genotypes had higher amount of gluconic acid 2-methoxime, ribo-hexose-3-ulose and gluconic acid. Heat map analysis showed that resistant genotypes had higher intensities of β-amyrin, hydroquinone in non-infected and malic acid, squalene, putrescine and 2,3,4-trihydroxybutyric acid in infected leaves. Dendrogram analysis further separated resistant genotypes in the same cluster along with infected moderately resistant genotypes. The most significant pathways identified are: linoleic acid metabolism, flavone and flavonol biosynthesis, cutin, suberin and wax biosynthesis, pentose and glucuronate interconversions, starch and sucrose metabolism, stilbenoid biosynthesis and ascorbate and aldarate metabolism. Targeted metabolite analysis further confirmed that resistant genotypes possessed higher content of primary metabolites sucrose, glucose, fructose, malic acid and citric acid. Moreover, resistant genotypes possessed higher content of salicylic, coumaric, ferulic, cinnamic, gallic acid (phenolic acids) and kaempferol, quercetin and catechin (flavonols). Thus metabolites having higher accumulation in resistant genotypes can be used as biomarkers for screening of LSS resistant germplasm. These results unravel that higher amount of primary metabolites leads to stimulate the accumulation of more amounts of secondary metabolites such as phenolic acid, flavanols, stilbenes and terpenoids (squalene and β-amyrin) biosynthesis which are ultimately involved in defense mechanism against LLS pathogen.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-021-00985-5.
Keywords: Groundnut, GC–MS, Late leaf spot, LC-MS-MS, Metabolomics, Organic acids, Phenolics
Introduction
Late leaf spot (LLS) commonly called as tikka disease caused by fungi Passalora personata, is a devastating disease of cultivated groundnut. This disease can cause yield losses up to 70% under favourable conditions (Grichar et al. 1998). Disease produces dark brown to black spots that may or may not have halos. It causes browning and severe defoliation which ultimately cause substantial yield loss. Leaf diseases also affect fodder quality and nutritional value which may reduce milk production in cattle (Mahatma et al. 2019). Disease resistance coupled with desirable agronomic traits are rarely present in cultivated germplasm, thus chemical fungicides are largely used for management of LLS. However, chemical control measures are not economical as they involve huge cost and labour, there are chances of evolution of new races of pathogens. If disease appears, it would be difficult to contain it. These factors have led to the search for new and innovative approaches for plant disease management. An understanding of host–pathogen interactions and the identification of marker metabolites involved in disease resistance and susceptibility can be used to explore disease management strategies.
Plants possess their own defense system involving numerous primary and secondary metabolites which may be pre-formed or induced. Primary metabolites such as amines, amino acids, carbohydrates, organic acid, and lipids are required for energy production and activation of signaling molecules during plant defense responses. However, secondary metabolites mediated plant defense mechanism is exerted by specific functions such as creating toxicity against pathogen or acting as signal molecules to induce defense responses (Gershenzon and Dudareva 2007). Hypersensitive response also triggers the expression of defense responses near the infected area and the onset of systemic acquired resistance (Chitarrini et al. 2017). An integration of all ‘omics’ will allow researchers to identify trait specific genes for crop improvement. Moreover, integrative network analyses reveal molecular interactions between metabolites and genes which improve our understanding about the phenotype-genotype relationship (Vadivel 2015; Kumar et al. 2017). Metabolomics is relatively less studied for trait mapping and plant selection in crop science due to inability to comprehensively profile all of the metabololites having different chemical properties. However, a comprehensive visualization of the metabolome is possible by employing selective extraction and parallel analyses with combination of targeted and non-targeted metabolomics technologies. Non-targeted metabolomics has the ability to detect huge numbers of metabolites from a single extract, thus allowing rapid and accurate analysis of metabolites. Metabolomics facilitates to identify specific metabolic pathways involved in plant responses and reveal function of genes in linked pathway (Wen et al. 2015). This allows to select superior traits for improvement of breeding materials (Zivy et al. 2015) Metabolite-assisted breeding will open a new avenue to select superior traits even when genomic information of crop is not available or when the complexity of the genetic mechanisms underlying the trait is too high. With the availability of whole genome sequence, genome-wide genetic variants and cost-effective genotyping assays metabolomics may effectively integrate in crop breeding programme (Fernie and Schauer 2009).
Various defense-related and signal metabolites such as salicylic acid, linoleic acid, thymol, pentitol, scopolin, hydroquinone and cinnamic acid were observed higher in tolerant/resistant genotypes during various fungal disease in castor and groundnut (Jadhav et al. 2013; Mahatma et al. 2018, 2019). The hypersensitive response signal molecules salicylic acid and jasmonic acid; the antioxidants like glutathione and ascorbic acid are also induced during various host–pathogen interaction to activate plant defense mechanism (Bao et al.2016; Li et al. 2017). Although the defense responses have been well studied in model plants, little information is available on the metabolic changes during infection of groundnut with leaf spot pathogen. Thus, we investigated untargeted and targeted metabolites profiles in LLS susceptible and resistant groundnut genotypes. The aim of this study was to decipher biochemical defense mechanism in groundnut genotypes against P. personata by identifying resistance specific biomarkers and metabolic pathways induced during host–pathogen interaction.
Materials and methods
Seeds of moderately resistant (GPBD4 and ICGV86590), resistant (KDG128 and RHRG06083) and susceptible (GG20, JL24 and TMV2) groundnut genotypes were procured from Plant Breeding Section, ICAR-Directorate of Groundnut Research, Junagadh, Gujarat, India. The cultivars/genotypes having same maturity group (105–110 days) were only included in the study in order to provide same platform for disease reaction. All the genotypes were grown in earthen pots under PII glass house to maintain uniform temperature and humidity. A potting mixture consisting of farm yard manure, sand and Vertisol (1:1:2 ratio) was used. Recommended fertilizers were applied for proper growth of crop. Each genotype was grown in six pots; three pots were kept as non-infected (control) and three pots were inoculated with the pathogen (i.e. two sets). Four plants were kept in each 20 kg soil capacity pot.
The pathogen, Passalora personata was isolated on potato dextrose agar (PDA) as a pure culture from ICAR-DGR GRS field in kharif. The pure culture of P. personata was multiplied by spraying @ 50,000 conidia per ml on 60 days old susceptible cultivar TMV 2 in pots. The conidial spray was done on evening. The macroscopic symptoms were observed on lower leaves, 7–9 days after spraying as minute sesame like brownish-black spots. The spots enlarged and several adjacent spots coalesced and become brownish black spots with concentric black rings representing spores. The typical symptomatic leaves were collected in autoclaved polybags, sealed airtight and placed at 27 ± 2 °C for about 48 h to encourage more conidial production. The conidia were dislodged in sterilized autoclaved water having few drops of tween 20. Conidial suspension was adjusted @ 50,000 spores per ml and spayed on 60 days old test cultivars during evening. Humidity above 80% and temperature 27 ± 2 °C were maintained for next 48 h. Control (non-infected) plants were sprayed with distilled water and placed in another P-II glass house having same growth conditions as in inoculated chamber. Leaf samples were collected 5 days after infection (dai) from non-infected and infected plants. The upper second leaves were taken from each plant in three replications for all the analysis.
Upper second leaves were taken from Passalora personata infected and non-infected plants at 5 dai and immediately immersed into liquid nitrogen for quenching of metabolism. For extraction and derivatization of metabolites the method described by Lisec et al. (2006) was utilized with some modifications. These samples were kept at − 80 °C until further analysis. Metabolites were extracted in methanol, chloroform and deionized water. Ribitol was added as an internal standard during extraction process. Extracted metabolites were derivatized with methoxylamine hydrochloride and N-methyl-N-trimethylsilyl trifluoroacetamide (MSTFA). Single derivatization reaction using an empty reaction tube was prepared as control. One-microliter of derivatized samples were injected into a DB-17 ms capillary column (30 m length, 0.25 mm I.D., 0.25 micron film thickness; Agilent Technologies Inc.) equipped with Shimadzu GC-2010 coupled with MS-QP2010 Plus. The temperature for the ion source was set to 230 °C, for the transfer line to 280 °C, and for the injector to 280 °C. The oven temperature was initially kept at 100 °C for 5 min, followed by increasing 5 °C min−1 to 290 °C for 1 min. Helium as a carrier gas was used at a constant flow rate of 1 ml min−1. Chromatogram acquisition, peak deconvolution, and MS library searches were performed using GCMS Solution version 2.71, Shimadzu Corporation-Japan. Metabolites were putatively identified by matching their mass spectra to spectra in NIST 14 library (National Institute of Standards and Technology, Gaithersburg, MD, USA). Pre-processing of total ion chromatograms (TIC) such as baseline correction, alignment, peak picking, and integration were performed using ACD/Spec Manager v.12.00 (Advanced Chemistry Development, Inc., ACD/Labs, Toronto, Canada). Data were exported as ‘‘.txt’’ files to MS Excel for the creation of data matrices and CSV comma delimited files were created for data analysis (Mahatma et al., 2018).
Targeted metabolite profiling
Identification of phenolic acids and flavonols using LC–MS/MS
Extraction of phenolic acids and flavonols from leaves
Phenolic compounds were extracted following the method described by Jadhav et al. (2013) with minor modifications. One gram of groundnut leaves were immersed in 80% HPLC grade methanol in screw cap glass tubes and kept in refrigerator at 4 °C for 48 h. Leaves were homogenized and extracted with 5 ml of 80% methanol solution (v/v) at room temperature and centrifuged at 10,000 rpm for 10 min. This extraction procedure was repeated four times on the same pellet. The supernatant after every centrifugation was collected in a volumetric flask and volume was made up to 25 ml with 80% methanol. Supernatants of extracted phenolics were evaporated near to dryness in a vacuum concentrator without heating. The obtained residues were re-suspended in 500 μl of absolute methanol.
Separation and identification of phenolic acids and flavonols
Fifteen standard phenolic acid and flavonols (cinnamic acid, caffeic acid, gallic acid, ferulic acid, catechol, chlorogenic acid, coumaric acid, salicylic acid, syringic acid, vanillic acid, catechin, epicatechin, epigallocatechin, kaempferol and quercetin) for identification were purchased from Sigma-Aldrich. ACQUITY Ultra Performance LC system (UPLC; Waters Corporation; Milford, USA) was used for identification of groundnut leaf phenolics and flavonols. Separation of individual phenolics was carried out using a UPLC BEH C18 column at 35º C.
Separation was achieved with a binary mobile phase consisting methanol and acetic acid at 0.4 ml/min (Raval et al. 2018). Sample injection volume was 10 μl. The optimized separation conditions were solvent (A) methanol and (B) 1% (v/v) acetic acid in water as gradient run (Supplementary Table S1). The characterization of each component was carried out by retention time and the accurate molecular mass. Each compound was optimized to its estimated molecular mass in the negative mode before and after fragmentation. ESI ionization conditions include source temperature of 150 °C, negative ionization mode, source voltage of 3.2 kV. High-purity nitrogen (> 99.999%) was used as curtain and auxiliary gas. The quantity of individual phenolics and flavonols was calculated on the basis of area and concentration of standards.
Soluble sugars profile of groundnut leaves using Ion chromatography
Oligosaccharides from groundnut leaves (500 mg) were extracted in 80% ethanol as described by Swami et al. (2015). Extracted sugar separated using CarboPac PA10 analytical column, amino trap column and CarboPac PA10 guard column equipped with ion chromatograph (Dionex, ICS 3000). Separation of sugars was achieved using 150 mM NaOH as an isocratic mobile phase with a flow rate of 1 ml min−1. Different sugars i.e. glucose, fructose, myo-inositol, mannitol, trehalose, sucrose, lactose, raffinose, stachyose, and verbascose (Sigma-Aldrich) were used as standards. Electrochemical detector was used for sugar profiling (Mahatma et al. 2016). During the analysis lactose was used as an internal standard. The Chromeleon software supplied with the equipment was used for data integration.
Organic acid Profile of groundnut leaves using Ion chromatography
Organic acids were extracted from leaves in 10 mM Phosphate buffer (pH 7.0) using chilled pestle and mortar. Extracts were centrifuged at 16,000 g for 30 min at 4 °C. Three aliquots of supernatant thus obtained were made and then filtered through 0.22 µm filters and stored in -20 °C for subsequent analysis (Jain et al. 2014). Citric acid, oxaloacetate, malic acid, oxalic acid and succinic acid from Sigma-Aldrich were used as standard. Organic acids were separated using Ion chromatograph (Dionex, ICS 3000) equipped with an IonPac AS11-HC (4 × 250 mm) anion exchange column and guard column (IonPac AG11-HC). The gradient mobile phase consisted 100 mM NaOH, methanol and water with a flow rate of 1.0 ml min-1 for elution of organic acid (Supplementary Table S2). Conductivity detector was used to analyse organic acids. The Chromeleon software was used for data acquisition, peak analysis and calibration.
Data processing and statistical analysis
Triplicate data related to sugar profile and organic acids were analysed by two-way ANOVA and mean differences were compared by critical differences (genotype x treatment) at P < 0.05 for significance. Statistical analysis of phenolics and untargeted metabolites was performed using web-based MetaboAnalyst 4.0 software (Chong et al. 2019). Concentration of phenolics and peak areas of untargeted metabolites were used for statistical analysis. Data were normalized by pareto scaling. Data were normalized with respect to the internal standard (ribitol), log transformation and pareto scaling to put all variables on equal footing, to minimize variable redundancy and to adjust measurement errors (Tugizimana et al. 2016). Chromatography peaks were considered significant where the signal to noise (S/N) ratio was > 50, the fold change (FC) was > 2.0 and P values were 0.05. Important metabolites were identified using Partial Least Squares—Discriminant Analysis (PLS-DA) Variable Importance in Projection (VIP) value (Chong and Jun 2005). When the VIP value of a metabolite exceeded 1.0, this metabolite was considered as potentially regulated metabolite. To identify resistance and susceptibility specific metabolites, data were arranged in two groups as resistant and susceptible, while for remaining analysis data were grouped as non-infected and infected. To visualize the relative levels of metabolites a heat map was also constructed while a dendrogram was constructed to find out relatedness of metabolites in genotypes. Quantitative enrichment analysis of the identified metabolites was utilized for pathway analysis using MetaboAnalyst 4.0 software based on the pathway library of Arabidopsis thaliana (Xia and Wishart 2011). P value of the hypergeometric test and the impact factor was calculated from global test and out-degree centrality algorithm were used to identify the most relevant metabolic pathways involved in groundnut- P. personata interaction.
Results and discussion
LLS disease rating of groundnut genotypes
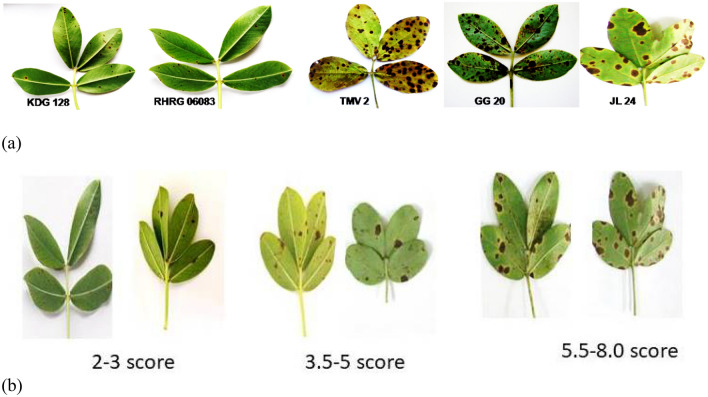
Microscopic disease symptoms appeared at 5 days after inoculation in susceptible cultivars as minute brown flecks. Severity was determined by estimating the percent necrotic area on leaves using 1–9 disease rating scale (Fig. 1 and Table 1) at 90 day after sowing and a day before harvesting. To minimise the impact of varieties/germplasm on disease reaction the observation were recorded till harvest. This method for disease rating was developed by Subrahmanyam et al (1995) for LLS and Rust in groundnut. Based on disease rating for 2 year phenotyping, groundnut genotypes were identified as resistant (KDG128 and RHRG6083 recorded ≤ 3.5 score), moderately resistant (GPBD4 and ICGV86950 recorded 3.5–5 score) and susceptible (GG20, JL24 and TMV2 recorded above 5 score).
Fig. 1.
a Severity of Passalora personata on resistant (KDG128 and RHRG06083) and susceptible (TMV2, GG20 and JL24) cultivars at 90 days after germination. Disease score in groundnut genotypes based on severity. groundnut genotypes were identified as resistant (≤ 3.5 score), moderately resistant (3.5–5 score) and susceptible (5.5–8.0)
Table 1.
Rating scale and reaction of groundnut genotypes for late leaf spots disease caused by Passalora personata
| S. No | Genotypes | Rating scale (LLS) | Reaction |
|---|---|---|---|
| 1 | GG20 | 7 | Susceptible |
| 2 | JL24 | 8 | Susceptible |
| 3 | GPBD4 | 4 | Moderately resistant |
| 4 | ICGV86590 | 4 | Moderately resistant |
| 5 | KDG128 | 3 | Resistant |
| 6 | RHRG06083 | 2 | Resistant |
| 7 | TMV2 | 8 | Susceptible |
Non-targeted metabolomics using Gas Chromatography–Mass Spectrometry (GC–MS)
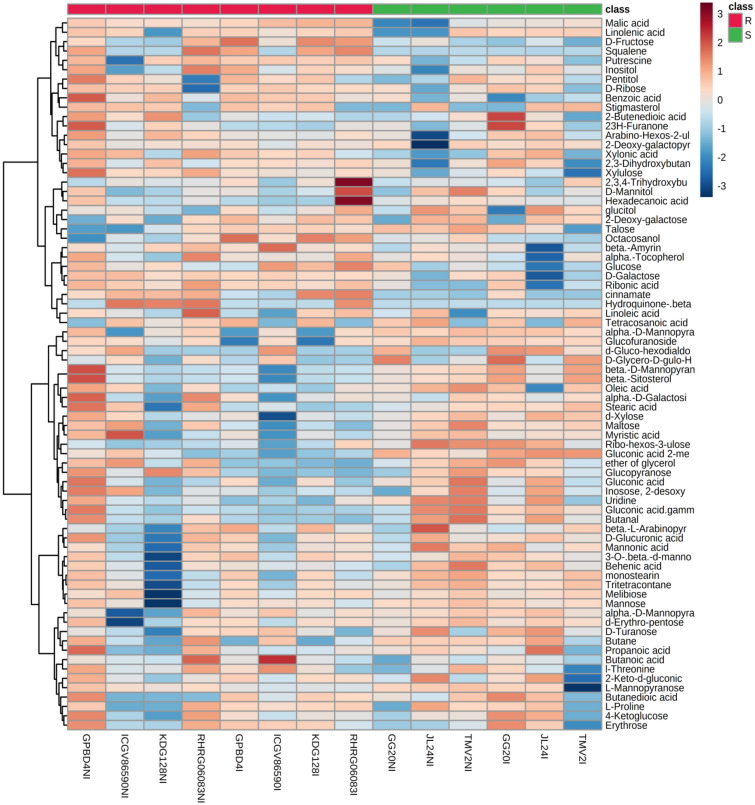
The non-targeted metabolomics approach permits a quantitative assessment of a wide range of metabolites. Total 77 metabolites including amino acids, carbohydrates, sugar alcohols, fatty acids, organic acids, polyamines, phenolics, terpenes and sterols were identified in the leaves of non-infected and infected LLS resistant and susceptible genotypes of groundnut using NIST 14 library. Result of heat map analysis is clearly showing intensity of 77 metabolites in resistant and susceptible genotypes during control and infection stage (Fig. 2). In non-infected leaves, most of metabolites were observed higher in GPBD4 followed by RHRG0608. Moderately resistant (GPBD4 and ICGV86590) and resistant (KDG128 and RHRG06083) genotypes had higher intensity of xylulose, D-galactose, benzoic acid and ribonic acid. Whereas, resistant genotypes showed higher intensity of β-amyrin, hydroquinone in non-infected and malic acid, squalene, putrescine and 2,3,4-trihydroxybutyric acid in infected leaves. Highest intensity of 2,3,4-trihydroxybutyric acid and hexadecanoic acid was observed in infected leaves of RHRG06083. Linoleic acid was observed highest in non-infected and infected leaves of RHRG06083.
Fig. 2.
Heat map analysis showing intensity of metabolite levels in late leaf spot resistant and susceptible genotypes of groundnut during control and infection stage. Colour bar indicates levels of metabolite, brick red colour indicates upregulation and sky blue colour indicates down regulation. Where R: resistant and S: Susceptible, NI: Non-infected and I: Infected (color figure online)
A polyunsaturated fatty acid, linoleic acid oxidizes to oxylipins and its conjugates. These oxylipins act as antimicrobial compounds and also induce the expression of pathogenesis-related proteins (Bollina et al. 2011). Significantly enhanced accumulation of trihydroxybutyrate was also reported in infected leaves of LLS resistant potato cultivar (Hamzehzarghani et al. 2016) and inoculated leaves of Alternaria leaf spot resistant genotypes of groundnut (Mahatma et al. 2019). Thus induction of 2,3,4-trihydroxybutyric acid in resistant genotypes confirms its role as a defense metabolite. The in vitro inhibition of fire blight pathogen (Erwinia amylovora) of apple was reported using gallic acid, phloroglucinol and hydroquinone. Among the tested phenolics only aqueous solution of hydroquinone maintained its protective activity for longer time. It also effectively restricted pathogen spread on apple shoots (Skłodowska et al.2018).
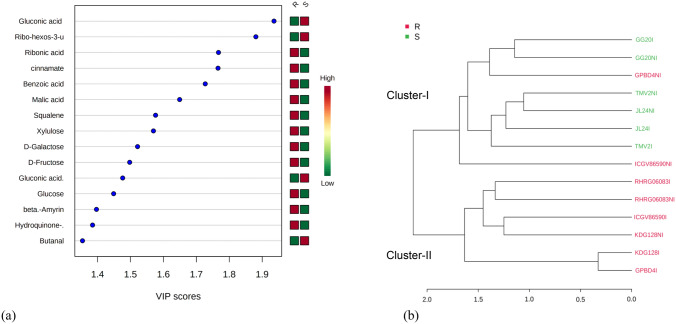
Significantly different metabolites in non-infected and infected plants as well as resistant and susceptible genotypes were determined using VIP measure of PLS-DA. PLS-DA is a chemometric method used to optimize the separation between different groups (Gromski et al. 2015). VIP score (Supplementary Fig S1) of non-infected and infected plants showed that non-infected leaves of groundnut genotypes possessed higher intensity (> 1.5 VIP sore) of glucopyranose, mannopyranose, ether of glycerol, oleic acid, maltose, uridine and myristic acid while malic acid and linolenic acid were observed higher in infected genotypes. Metabolites specific to resistant and susceptible genotypes in potato tubers and mango were also identified using VIP measure of PLS-DA (Steinfath et al. 2010; Augustyn et al. 2014). A total of 20 significantly regulated metabolites (VIP > 1 and p < 0.05) in resistant and susceptible genotypes were identified using the PLS-DA model (Fig. 3a). Resistant and moderately resistant genotypes had higher intensities of ribonic acid, cinnamic acid, malic acid, squalene, xylulose, galactose, fructose (> 1.5 VIP sore), glucose, β-amyrin and hydroquinone (1.4 VIP score) while susceptible genotypes had higher amount of (> 1.5 VIP sore) gluconic acid 2-methoxime, ribo-hexose-3-ulose and gluconic acid. Based on metabolites pattern, a dendrogram was constructed using pearson average method (Fig. 3b). Dendrogram clearly distinguished non-infected and infected resistant genotypes (RHRG06083 and KDG128) and infected moderately resistant genotypes (GPBD4 and ICGV86590) in same cluster (cluster-II) while all non-infected and infected susceptible genotypes were laid on cluster-I along with non-infected moderately resistant genotypes (GPBD4 and ICGV86590). These results imply that resistant and infected moderately resistant genotypes have similar metabolic pattern while susceptible genotypes had different metabolic pattern. Although, non-infected moderately resistant genotype ICGV86590 observed on separate sub-cluster of cluster-I and GPBD4 on separate sub-sub cluster of cluster-I which showed that moderately resistant genotype had somewhat similar metabolic pattern with susceptible genotypes. Intensity of metabolites in different genotypes was also shown by biplot of PCA 1 and PCA2 in non-infected and infected leaves of all genotypes (Supplementary Fig S2). Metabolites (Propanoic acid, butane, 2-keto-d-gluconic acid, myristic acid, β-L-arabinopyranose, monosearin) on PC2 were found higher in susceptible genotypes while 2,3,4-trihydroxybutyric acid, α-tocopherol, hydroquinone and β-amyrin on PC 1 with higher levels in resistant and moderately resistant genotypes. Non-infected and infected leaves of resistant genotype (RHRG06083) had highest intensity of 2,3,4-trihydroxybutyric acid and alpha-tocopherol.
Fig. 3.
a Abundance of metabolites in resistant and susceptible genotypes based on Variable Importance in Projection (VIP) using Partial Least Squares—Discriminant Analysis (PLS-DA).Colour bar indicates high intensity of metabolite levels by brick red colour and low level by green colour. b Dendrogram (Pearson average method) showing clustering of genotypes based on metabolic pattern. Resistant genotypes are denoted by red colour while susceptible genotypes by green colour. Where R: resistant and S: Susceptible, NI: Non-infected and I: Infected (color figure online)
A non-targeted metabolomics of potato genotypes resistant to late blight, revealed that higher production of hydroxycinnamic acid amides such as feruloyltyramine, prevented lesion expansion and thickening of host cell walls (Yogendra et al. 2014). Beside hydroxycinnamic acid amides, other chemical groups, such as flavonoid glycosides benzylisoquinoline and fatty acids also play important role in cell wall thickening (Yogendra et al. 2015). Hamzehzarghani et al. (2008) observed higher accumulation of fructose, inositol and cinnamic acid in fusarium head blight resistant line of wheat. In many plants, triterpenes, triterpenoids and β-amyrin were detected as major components of the intracuticular wax layer. These molecules may constituent plasma membranes and have signaling roles (Buschhaus and Jetter 2011). Triterpene β-amyrin is a cyclization product of 2,3-oxidosqualene and serve as an intermediate in the synthesis of more complex triterpene glycosides associated with plant defense (Kemen et al. 2014). Glycosylated triterpenoid saponins have been observed at higher levels in a healthy plants with potent antifungal activity (Osbourn 1996). Saponin, avenacin A-1 is a β-amyrin derived triterpene, which provides protection against soil-borne diseases caused by Gaeumannomyces graminis var. avenae and Fusarium avenaceum in oat (Papadopoulou et al. 1999). In present study higher levels of squalene and β-amyrin in resistant genotypes confers their role in plant defense mechanism against P. personata.
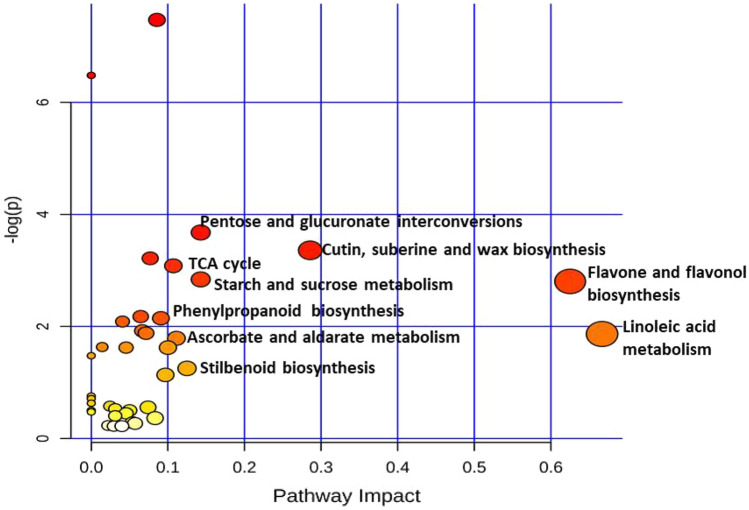
Metabolic pathway analysis
Metabolites from non-targeted and targeted analysis were subjected to pathway analysis. Most significant metabolic pathways activated in groundnut defense mechanism were identified and displayed according to pathway impact in Fig. 4 and Table 2. The most significant pathways (> 0.1 impact) are: Linoleic acid metabolism, Flavone and flavonol biosynthesis, cutin, suberin and wax biosynthesis, pentose and glucuronate interconversions, starch and sucrose metabolism, stilbenoid biosynthesis and ascorbate and aldarate metabolism. While phenylpropanoid (> 0.09 impact), sesquiterpenoid and triterpenoid (> 0.045) and flavonoid (0.040) biosynthetic pathways are also important despite their less pathway impact found in this study. These observations are supported by VIP score of PLS-DA for resistant and susceptible genotypes which showed higher intensity of benzoic acid, cinnamic acid, glucose, fructose and β-amyrin and squalene.
Fig. 4.
Pathway analysis by MetaboAnalyst 4.0 PathwayAnalysis software: Metabolic pathways displayed according to their significance or pathway impact. The graph, presents a view of all the matched pathways arranged by P values on the y-axis, and the pathway impact values on the x-axis. According to graph, high impact pathways are (> 0.1 impact): Linoleic acid metabolism, Flavone and flavonol biosynthesis, cutin suberin and wax biosynthesis, Pentose and glucuronate interconversions, Starch and sucrose metabolism, stilbenoid biosynthesis and ascorbate and aldarate metabolism
Table 2.
Significant metabolic pathways observed in groundnut before and after infection with Passalora personata (Pathway analysis using Metaboanalyst 4.0)
| No | Pathway name | Total | Hits | -log(p) | Impact |
|---|---|---|---|---|---|
| 1 | Linoleic acid metabolism | 4 | 1.000 | 1.866 | 0.667 |
| 2 | Flavone and flavonol biosynthesis | 10 | 2.000 | 2.804 | 0.625 |
| 3 | Cutin, suberine and wax biosynthesis | 18 | 3.000 | 3.359 | 0.286 |
| 4 | Pentose and glucuronate interconversions | 16 | 3.000 | 3.677 | 0.143 |
| 5 | Starch and sucrose metabolism | 22 | 3.000 | 2.840 | 0.143 |
| 6 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | 8 | 1.000 | 1.252 | 0.125 |
| 7 | Ascorbate and aldarate metabolism | 18 | 2.000 | 1.788 | 0.111 |
| 8 | Citrate cycle (TCA cycle) | 20 | 3.000 | 3.083 | 0.107 |
| 9 | Fructose and mannose metabolism | 20 | 2.000 | 1.623 | 0.100 |
| 10 | Inositol phosphate metabolism | 28 | 2.000 | 1.135 | 0.097 |
| 11 | Phenylpropanoid biosynthesis | 46 | 4.000 | 2.147 | 0.091 |
| 12 | Galactose metabolism | 27 | 6.000 | 7.471 | 0.086 |
| 13 | alpha-Linolenic acid metabolism | 28 | 1.000 | 0.364 | 0.083 |
| 14 | Pentose phosphate pathway | 19 | 3.000 | 3.216 | 0.077 |
| 15 | Aminoacyl-tRNA biosynthesis | 46 | 2.000 | 0.556 | 0.074 |
| 16 | Butanoate metabolism | 17 | 2.000 | 1.880 | 0.071 |
| 17 | Amino sugar and nucleotide sugar metabolism | 50 | 4.000 | 1.922 | 0.067 |
| 18 | Glyoxylate and dicarboxylate metabolism | 29 | 3.000 | 2.177 | 0.065 |
| 19 | Arginine and proline metabolism | 34 | 1.000 | 0.269 | 0.057 |
| 20 | Glycine, serine and threonine metabolism | 33 | 1.000 | 0.283 | 0.054 |
| 21 | Valine, leucine and isoleucine biosynthesis | 22 | 1.000 | 0.501 | 0.050 |
| 22 | Propanoate metabolism | 20 | 2.000 | 1.623 | 0.045 |
| 23 | Sesquiterpenoid and triterpenoid biosynthesis | 24 | 1.000 | 0.449 | 0.045 |
| 24 | Flavonoid biosynthesis | 47 | 4.000 | 2.088 | 0.041 |
| 25 | Pyrimidine metabolism | 38 | 1.000 | 0.221 | 0.040 |
| 26 | Carbon fixation in photosynthetic organisms | 21 | 1.000 | 0.530 | 0.031 |
| 27 | Phosphatidylinositol signaling system | 26 | 1.000 | 0.404 | 0.031 |
| 28 | Ubiquinone and other terpenoid-quinone biosynthesis | 38 | 1.000 | 0.221 | 0.029 |
| 29 | Steroid biosynthesis | 45 | 2.000 | 0.578 | 0.024 |
| 30 | Fatty acid degradation | 37 | 1.000 | 0.232 | 0.022 |
| 31 | Fatty acid biosynthesis | 56 | 4.000 | 1.633 | 0.014 |
| 32 | Biosynthesis of unsaturated fatty acids | 22 | 5.000 | 6.481 | 0.000 |
| 33 | Alanine, aspartate and glutamate metabolism | 22 | 2.000 | 1.478 | 0.000 |
| 34 | Sulfur metabolism | 15 | 1.000 | 0.757 | 0.000 |
| 35 | Tyrosine metabolism | 16 | 1.000 | 0.711 | 0.000 |
| 36 | Arginine biosynthesis | 18 | 1.000 | 0.630 | 0.000 |
| 37 | Pyruvate metabolism | 22 | 1.000 | 0.501 | 0.000 |
| 38 | Fatty acid elongation | 23 | 1.000 | 0.474 | 0.000 |
This table shows the detailed results from the pathway analysis. In particular, the Total is the total number of compounds in the pathway; the Hits is the actually matched number from the user uploaded data; the Impact is the pathway impact value calculated from pathway topology analysis
Implication of linoleic acid in defense mechanism is well known which modulate basal, effector-triggered, and systemic immunity in plants. Studies on fatty acid metabolic mutants also reveal an active signaling role for the cuticle in plant defense (Kachroo and Kachroo 2009). Higher accumulation of linoleic acid upregulate lipoxygenase pathway which might contribute to the biosyntheis of oxylipins including jasmonic acid (JA). Phyto-oxylipins act as signal molecules and play an important role in defense mechanism. They are constituents of cutin and acts as a protective compound (Blée 2003). Jasmonic acid is involved in the defense against necrotrophic pathogens, preventing plant cell death, and inducing defense responses to restrict further pathogen infection (Pandey et al. 2016). Cutin embedded in cuticular wax forms a cuticle layer. Cuticle layer is a physical barrier of plant to protect from desiccation as well as from various biotic and abiotic stresses. Plant cuticle also comprises flavonoids and terpenoids, which have antifungal activities (Ziv et al. 2018). Activated plant defense in tomato was observed by induced biosynthesis of terpenoids and flavonoids in response to Colletotrichum gloeosporioides infection (Alkan et al. 2015).
Targeted metabolomics
Phenolic acids and flavonols profile of groundnut leaves
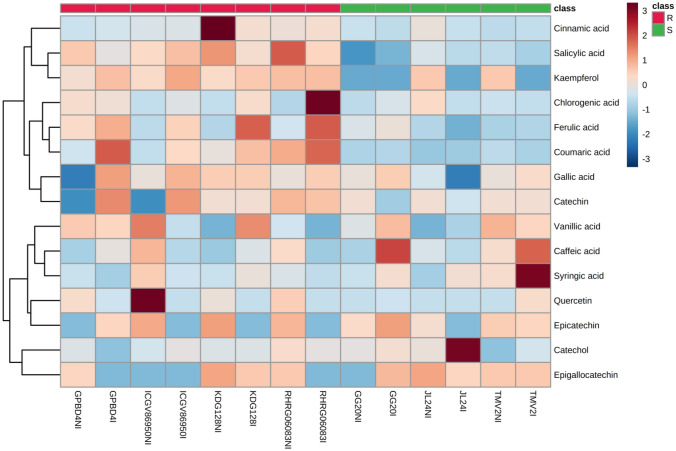
LC–MS/MS was used for better resolution and separation of phenolic acids and flavonols in a single extract. Total fifteen phenolics including phenolic acids (cinnamic acid, caffeic acid, catechol, chlorogenic acid, coumaric acid, gallic acid, ferulic acid, salicylic acid, syringic acid and vanillic acid) and flavonols (catechin, epicatechin, epigallocatechin, kaempferol and quercetin) were identified and quantified in groundnut genotypes (Supplementary Table S3). Heat map analysis (Fig. 5) revealed abundance of phenolics and flavanols in non-infected and infected leaves of resistant and susceptible genotypes. Resistant and moderately resistant genotypes possessed higher amount of cinnamic acid, coumaric acid and salicylic acid at both constitutive and induced level than that of susceptible genotypes. Resistant genotypes also had higher amount of chlorogenic acid, ferulic acid, catechin and Kaempferol at induced level. However, induced level of syringic acid and caffeic acid was observed higher in LLS susceptible genotypes. Resistance and susceptibility specific phenolics were identified by VIP score using PLS-DA (Supplementary Fig. S3) Overall resistant genotypes possessed higher content of salicylic, coumaric, ferulic, cinnamic and gallic acid (phenolic acids) and kaempferol, quercetin and catechin (flavonols), which further confirmed by Principal Component Analysis (PCA) biplot between the selected PC 1 and PC 2 (Supplementary Fig. S4). Phenylpropanoid biosynthesis is an important pathway in plant defense mechanism with different pathosystems which synthesizes different phenolics, including cinnamic acid, hydroxycinnamic acids, coumaric acid, ferulic acid, salicylic acid, and flavonoids (Jadhav et al. 2013; Mahatma et al. 2018, 2019; Islam et al. 2019). Cinnamic acid was reported to reduce the growth rate of Botrytis cinerea. It disrupts the plasma membrane of microorganism and induces the production of intracellular reactive oxygen species (ROS) to restrict the growth of pathogen (Zhang et al. 2015). Pre-treatment with p-coumaric acid Black rot disease symptoms caused by Xanthomonas campestris pv. campestris in Chinese cabbage was alleviated by. The p-coumaric induces jasmonic acid mediated accumulation of phenylpropanoid biosynthesis-related genes and flavonoids to impart resistance in the Brassica napus against X. campestris pv. campestris (Islam et al. 2019). The p-coumaric acid is implicated in cell wall fortification and biosynthesis of flavonoids, stilbenes and lignins to impart disease resistance (Bollina et al. 2011). Catechins are often linked to the antimicrobial activity. It cause membrane damage and oxidative burst by the generation of reactive oxygen species (Fathima and Rao 2016). Catechin was also reported an effective antifungal compound in poplar defense against foliar rust infection (Ullah et al. 2017). Chlorogenic acid inhibited the 87% in vitro mycelial growth of Sclerotium rolfsii followed by ferulic acid (54%) at a concentration of 1 mg ml−1 (Khatediya et al. 2018). An in-vitro studies of phenolic compounds (tyrosol, catechin, and oleuropein) extracted from olive plants also showed antifungal activity and enhance plant resistance against Phytophtora sp. (Del Río et al. 2003). A significant inhibition of spore germination of Pyricularia oryzae fungus was observed using kaempferol. Naringenin and kaempferol improved lipophilicity of medium thereby caused toxicity towards Pyricularia (Padmavati et al.1997). Bollina et al. (2011) identified several conjugates of catechin and kaempferol as resistant related constitutive metabolites in barley against Fusarium graminearum. The role of salicylic acid (SA) accumulation for plant disease resistance was demonstrated by transgenic tobacco and Arabidopsis thaliana where SA biosynthesis was inhibited by expression of bacterial gene salicylate hydroxylase. Transgenic plants were incapable to induce systemic acquired resistance thereby increased susceptibility of plants to bacterial, fungal and viral pathogens (Delaney et al. 1994). The role of SA to confer resistance against a broad-spectrum of pathogens is well documented. Both constitutive and induced defense mechanisms against pathogens are mediated by SA in plants (Chaturvedi and Shah 2007). SA dependent signaling pathway triggers synthesis of antimicrobial phytoalexins and pathogenesis-related proteins which induce hypersensitive responses (HR) and programmed cell death in the infected area, thus restricts colonization of biotrophic pathogens (Kouzai et al. 2018).
Fig. 5.
Heat map analysis showing intensity of phenolic acid s and flavonols levels in late leaf spot resistant and susceptible genotypes of groundnut during control and infection stage. Colour bar indicates levels of metabolite, brick red colour indicates upregulation and sky blue colour indicates down regulation. Where R: resistant; S: susceptible; NI: non-infected and I: infected (color figure online)
Soluble sugars profile of groundnut leaves
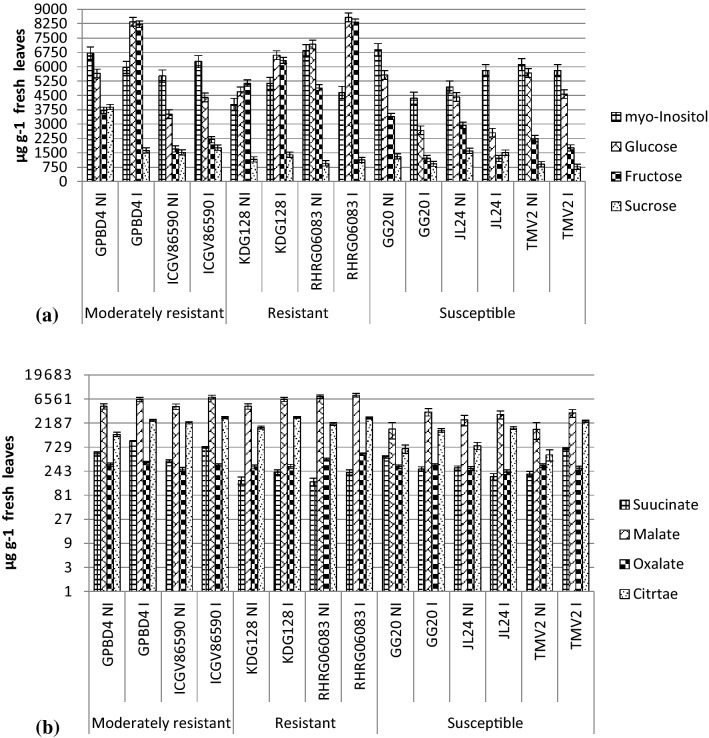
Glucose, fructose, myo-inositol and sucrose were separated from non-infected and infected leaves of groundnut genotypes. Glucose, fructose and sucrose content increased in resistant and moderately resistant genotypes while decreased in susceptible genotypes after infection. Highest glucose and fructose content was observed in the infected leaves of RHRG06083 followed by GPBD4. Specific sugars like glucose, sucrose and galactose are correlated with disease resistance in some plant-pathogen interactions (Daniele et al. 2003). Induced accumulation of sugars plays a role in plant immune system and defense signaling (Morkunas et al. 2013). On the other hand, the reduction of carbohydrates in plants promotes susceptibility towards Colletotrichum higginsianum infection in Arabidopsis (Engelsdorf et al. 2013). An increase in fructose content in tomato plant was associated with lower susceptibility to Botrytis cinerea (Lecompte et al. 2017). Decreased glucose content in susceptible genotypes of groundnut was observed after infection with S. rolfsii (Mahatma et al. 2018). These results suggest that high sugar levels in leaves improve resistance to certain hemibiotrophs and necrotrophs. Carbohydrates are the basic building blocks for the biosynthesis of numerous defense related molecules such as phenolics, phytoalexins and lignin. Thus higher content of sugar may provide more amount of precursor for biosynthesis of secondary metabolites involved in defense mechanism.
Organic acid profile of groundnut leaves
Results of organic acid profile revealed that tricarboxylic acid (TCA) cycle metabolites i.e. citric acid, malic acid, oxalic acid and succinic acid were significantly induced after infection in all the genotypes. Malic and citric acid content were observed higher as compared to oxalic and succinic acid in all groundnut genotypes. Both constitutive and induced levels of malic acid and citric acid observed higher in resistant genotypes than that of susceptible genotypes. Higher content of TCA cycle metabolites suggest that resistant genotypes may generate more energy and substrate to redistribute for the biosynthesis of other metabolites involved in plant defense. Balmer et al. (2018) observed induction in SA and JA pathway in Arabidopsis against the bacterial pathogen Pseudomonas syringae pv. tomato DC3000 using citrate and fumarate priming. Although, any direct antimicrobial effect was not shown by these organic acids. Several diverse functions within and beyond cellular metabolism are governed by TCA cycle intermediates in all plants. (Zhang and Fernie 2018).
The specific role of TCA cycle intermediates in plant defense responses is still not fully understood (Fig. 6). However, these can be associated with the production of various polyphenolic compounds potentially related to defense mechanisms (De Oliveira et al. 2019). During the defense response products of primary metabolism provide carbon fluxes into secondary metabolism. Major flow of carbon from primary metabolism (phosphoenolpyruvate and erthrose-4-phosphate) into secondary metabolism is represented by phenylpropanoid and shikimate pathway (Bolton 2009).
Fig. 6.
a Soluble sugar profiles showing inositol, glucose, fructose and sucrose content of groundnut genotypes b organic acid profiles showing succinic acid, malic acid, oxalic acid and citric acid content of groundnut genotypes. Where R: resistant; S: susceptible; NI: non-infected and I: infected. For statistical significance at P < 0.05, critical differences value for each compound (genotype x treatment) is depicted by error bar
Conclusion
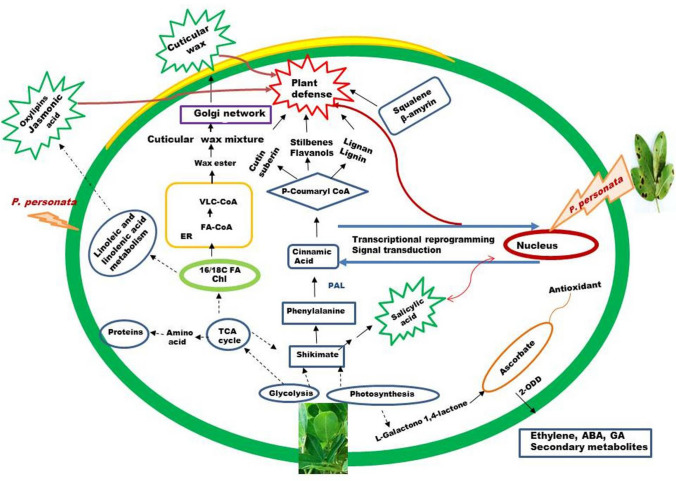
Overall metabolomics of LLS resistant and susceptible genotypes of groundnut revealed that resistant genotypes had higher amount of constitutive sucrose, glucose, fructose, citric acid, malic acid and linoleic acid which further induced after pathogen interaction. Constitutive or induced higher accumulation of salicylic, coumaric, ferulic, cinnamic, gallic acid (phenolic acids) and kaempferol, quercetin and catechin (flavonols), squalene and β-amyrin (terpenoids) in resistant genotypes confirmed that higher pool of primary metabolites are utilized for secondary metabolite synthesis. These identified metabolites can be used as biomarkers. A proposed mechanism for LLS resistance in groundnut is depicted in Fig. 7. Significant pathways identified in the study further demonstrated that primary and secondary metabolites may trigger the defense mechanism by biosynthesis of signaling molecules (oxylipins) and structural barriers like cell wall strengthening and, cutin suberin and wax. Plant cuticle is well known as first layer of plant defense pattern-triggered immunity. Thus, metabolomics can be used to identify metabolic pathways which govern resistance mechanism, and biomarkers for further screening of resistant germplasm for crop breeding.
Fig. 7.
A proposed mechanism of late leaf spot resistance in groundnut based on metabolomic analysis. Where Chl: chloroplast, ER: endoplasmic reticulum, FA: fatty acid FA-CoA: Fatty acyl Co enzyme A, TCA: tricaroxylic acid, VLC-COA: very-long-chain acyl-CoA, dotted arrow indicated (
 ) two or more biosynthesis reactions. The partial model for the cuticular wax biosynthesis was taken from the review of wang et al (2020) and for the phenylpropanoid biosynthesis was taken from the review of Vogt 2010.
) two or more biosynthesis reactions. The partial model for the cuticular wax biosynthesis was taken from the review of wang et al (2020) and for the phenylpropanoid biosynthesis was taken from the review of Vogt 2010.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are thankful to the Director, ICAR-Directorate of Groundnut Research, Junagadh, India for providing all the necessary facilities and support to completing this experiment.
Author contributions
MKM conceived the study, performed and designed the experiments and drafted the manuscript. LKT carried out sugar and organic acid measurements and phenolics and metabolite extraction. KSJ, TPP, and NK carried out the pot and field experiments to screen groundnut genotypes KSJ and TPP also performed inoculation of disease. KJR carried out untargeted metabolites analysis by GC–MS and phenolics profiling by LC–MS/MS and data processing, SKB and AV performed the statistical analysis and interpretation of results of targeted metabolites. BAG contributed to interpretation of the data and editing of the MS.
Funding
This work was carried out under Institute Project. External funning was not received for this work.
Declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alkan N, Friedlander G, Ment D, Prusky D, Fluhr R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015;205:801–881. doi: 10.1111/nph.13087. [DOI] [PubMed] [Google Scholar]
- Augustyn WA, Regnier T, Combrinck S, Botha BM (2014) Metabolic profiling of mango cultivars to identify biomarkers for resistance against Fusarium infection. Phytochem Lett 10:civ–cx
- Balmer A, Pastor V, Glauser G, Mauch-Mani B. Tricarboxylates induce defense priming against bacteria in Arabidopsis thaliana. Front Plant Sci. 2018;9:1221. doi: 10.3389/fpls.2018.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao G, Zhuo C, Qian C, Xiao T, Guo Z, Lu S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol J. 2016;14:206–214. doi: 10.1111/pbi.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2003;7:315–322. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- Bollina V, Kushalappa AC, Choo TM, Dion Y, Rioux S. Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol Biol. 2011;77:355–370. doi: 10.1007/s11103-011-9815-8. [DOI] [PubMed] [Google Scholar]
- Bolton MD. Primary metabolism and plant defense—fuel for the fire. Mol Plant-Microbe Interact. 2009;22:487–497. doi: 10.1094/MPMI-22-5-0487. [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R. Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J Exp Biol. 2011;62:841–853. doi: 10.1093/jxb/erq366. [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Shah J. Salicylic acid in plant disease resistance. In: Hayat S, Ahmad A, editors. Salicylic acid: a plant hormone. Dordrecht: Springer; 2007. pp. 335–370. [Google Scholar]
- Chitarrini G, Soini E, Riccadonna S, Franceschi P, Zulini L, Masuero D, Vecchione A, Stefanini M, Di Gaspero G, Mattivi F, Vrhovsek U. Identification of biomarkers for defense response to Plasmapara viticola in a resistant grape variety. Front Plant Sci. 2017;8:1524. doi: 10.3389/fpls.2017.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong IG, Jun CH. Performance of some variable selection methods when multicollinearity is present. Chemometr Intell Lab. 2005;78:103–112. [Google Scholar]
- Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinform. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Daniele E, Dommes J, Hausman JF. Carbohydrates and resistance to Phytophthora infestans in potato plants. Acta Physiol Plant. 2003;25:171–178. [Google Scholar]
- Del Río JA, Báidez AG, Botía JM, Ortuño A. Enhancement of phenolic compounds in olive plants (Olea europaea L.) and their influence on resistance against Phytophthora sp. Food Chem. 2003;83:75–78. [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- De Oliveira CS, Lião LM, Alcantara GB. Metabolic response of soybean plants to Sclerotinia sclerotiorum infection. Phytochemistry. 2019;167:112099. doi: 10.1016/j.phytochem.2019.112099. [DOI] [PubMed] [Google Scholar]
- Engelsdorf T, Horst RJ, Pröls R, Pröschel M, Dietz F, Hückelhoven R, Voll LM. Reduced carbohydrate availability enhances the susceptibility of arabidopsis toward: Colletotrichum higginsianum. Plant Physiol. 2013;162:225–238. doi: 10.1104/pp.112.209676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathima A, Rao JR. Selective toxicity of Catechin—a natural flavonoid towards bacteria. Appl Microbiol Biotechnol. 2016;100:6395–6402. doi: 10.1007/s00253-016-7492-x. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Schauer N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 2009;25:39–48. doi: 10.1016/j.tig.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N. The function of terpene natural products in the natural world. Nat Chem Biol. 2007;3:408–414. doi: 10.1038/nchembio.2007.5. [DOI] [PubMed] [Google Scholar]
- Grichar WJ, Besler BA, Jaks AJ. Groundnut (Arachis hypogaea L.) cultivar response to leaf spot disease development under four disease management programs. Peanut Sci. 1998;25:35–39. [Google Scholar]
- Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, Goodacre RA. Tutorial review: metabolomics and partial least squares-discriminant analysis–A marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Hamzehzarghani H, Vikram A, Abu-Nada Y, Kushalappa AC. Tuber metabolic profiling of resistant and susceptible potato varieties challenged with Phytophthora infestans. Eur J Plant Pathol. 2016;145:277–287. [Google Scholar]
- Hamzehzarghani H, Paranidhara V, Abu-Nada Y, Kushalappa AC, Dion Y, Rioux S, Comeau A, Yaylayan V, Marshall W. Metabolic profiling coupled with statistical analyses for potential high throughput screening of quantitative resistance to fusarium head blight in wheat cultivars. Can J Plant Pathol. 2008;30:24–36. [Google Scholar]
- Islam MT, Lee BR, Das PR, La VH, Leea H, Jungc WJ, Baed DW, Kim TH. p-Coumaric acid induces jasmonic acid-mediated phenolic accumulation and resistance to black rot disease in Brassica napus. Physiol Mol Plant P. 2019;106:270–275. [Google Scholar]
- Jadhav PR, Mahatma MK, Mahatma L, Jha S, Parekh VB, Khandelwal V. Expression analysis of key genes of phenylpropanoid pathway and phenol profiling during Ricinus communis–Fusarium oxysporum f. sp. ricini interaction. Ind Crop Prod. 2013;50:456–461. [Google Scholar]
- Jain R, Jha S, Adhikary H, Kumar P, Parekh V, Jha A, Mahatma MK, Kumar GN. Isolation and molecular characterization of arsenite-tolerant Alishewanella sp. GIDC-5 originated from industrial effluents. Geomicrobiol J. 2014;31:82–90. [Google Scholar]
- Kachroo A, Kachroo P. Fatty acid–derived signals in plant defense. Ann Rev Phytopathol. 2009;47:153–176. doi: 10.1146/annurev-phyto-080508-081820. [DOI] [PubMed] [Google Scholar]
- Kemen AC, Honkanen S, Melton RE, Findlay KC, Mugford ST, Hayashi K, Haralampidis K, Rosser SJ, Osbourn A. Investigation of triterpene synthesis and regulation in oats reveals a role for β-amyrin in determining root epidermal cell patterning. Proc Natl Acad Sci USA. 2014;111:8679–8684. doi: 10.1073/pnas.1401553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatediya NK, Parmar DV, Mahatma MK, Pareek M. Increased accumulation of phenolic metabolites in groundnut (Arachis hypogaea L.) genotypes contribute to defense against Sclerotium rolfsii infection. Arch Phytopath Plant Protec. 2018;51:530–549. [Google Scholar]
- Kouzai Y, Kimura M, Watanabe M, Kusunoki K, Osaka D, Suzuki T, Matsui H, Yamamoto M, Ichinose Y, Toyoda K, Matsuura T, Mori IC, Hirayama T, Minami E, Nishizawa Y, Inoue K, Onda Y, Mochida K, Noutoshi Y. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018;217:771–783. doi: 10.1111/nph.14849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bohra A, Pandey AK, Pandey MK, Kumar A. Metabolomics for plant improvement: status and prospects. Front Plant Sci. 2017;8:1302. doi: 10.3389/fpls.2017.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte F, Nicot PC, Ripoll J, Abro MA, Raimbault AK, Lopez-Lauri F, Bertin N. Reduced susceptibility of tomato stem to the necrotrophic fungus Botrytis cinerea is associated with a specific adjustment of fructose content in the host sugar pool. Ann Bot. 2017;119:931–943. doi: 10.1093/aob/mcw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li C, Sun J, Peng M. Metabolomic, biochemical, and gene expression analyses reveal 628 the underlying responses of resistant and susceptible banana species during early infection 629 with Fusarium oxysporum f. sp. cubense. Plant Dis. 2017;101:534–543. doi: 10.1094/PDIS-09-16-1245-RE. [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:1–10. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- Mahatma MK, Thawait LK, Jadon KS, Rathod KJ, Sodha KH, Bishi SK, Thirumalaisamy PP, Golakiya BA. Distinguish metabolic profiles and defense enzymes in Alternaria leaf blight resistant and susceptible genotypes of groundnut. Physiol Mol Biol Plants. 2019;25:1395–1405. doi: 10.1007/s12298-019-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahatma MK, Thawait LK, Jadon KS, Thirumalaisamy PP, Bishi SK, Jadav JK, Khatediya N, Golakiya BA. Metabolic profiles of groundnut (Arachis hypogaea L.) genotypes differing in Sclerotium rolfsii reaction. Eur J Plant Pathol. 2018;151:463–474. [Google Scholar]
- Mahatma MK, Thawait LK, Bishi SK, Khatediya N, Rathnakumar AL, Lalwani HB, Misra JB. Nutritional composition and antioxidant activity of Spanish and Virginia groundnuts (Arachis hypogaea L.): a comparative study. J Food Sci Technol. 2016;53:2279–2286. doi: 10.1007/s13197-016-2187-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkunas I, Formela M, Marczak L, Stobiecki M, Bednarski W. Themobilization of defence mechanisms in the early stages of pea seed germination against Ascochyta pisi. Protoplasma. 2013;250:63–75. doi: 10.1007/s00709-012-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell. 1996;8:1821–1831. doi: 10.1105/tpc.8.10.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmavati M, Sakyhivel N, Thara KV, Reddy AR. Differential sensitivity of rice pathogens to growth inhibition by flavonoids. Phytochemistry. 1997;46:499–502. [Google Scholar]
- Pandey D, Rajendran SRCK, Gaur M, Sajeesh PK, Kumar A. Plant defense signaling and responses against necrotrophic fungal pathogens. J Plant Growth Regul. 2016;35:1159–1174. [Google Scholar]
- Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA. 1999;96:12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval SS, Mahatma MK, Chakraborty K, Bishi SK, Singh AL, Rathod KJ, Jadav JK, Sanghani JM, Mandavia MK, Gajera HP, Golakiya BA. Metabolomics of groundnut (Arachis hypogaea L.) genotypes under varying temperature regimes. Plant Growth Regul. 2018;84:493–505. [Google Scholar]
- Skłodowska M, Mikiciński A, Wielanek M, Kuźniak E, Sobiczewski P. Phenolic profiles in apple leaves and the efficacy of selected phenols against fire blight (Erwinia amylovora) Eur J Plant Pathol. 2018;151:213–228. [Google Scholar]
- Steinfath M, Strehmel N, Peters R, Schauer N, Groth D, Hummel J, Steup M, Selbig J, Kopka J, Geigenberger P, van Dongen JT. Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol J. 2010;8:900–911. doi: 10.1111/j.1467-7652.2010.00516.x. [DOI] [PubMed] [Google Scholar]
- Subrahmanyam P, McDonald D, Waliyar F, JReddy L, Nigam SN, Gibbons RW, Ramanatha Rao V, Singh AK, Pande S, Reddy PM, Subba Rao PV (1995) Screening methods and sources of resistance to rust and late leaf spot of groundnut. Information Bulletin no. 47. International Crops Research Institute for the Semi-Arid Tropics Patanchem 502 324, Andhra Pradesh, India.
- Swami RM, Mahatma MK, Parekh MJ, Kalariya KA, Mahatma L. Alteration of metabolites and polyphenol oxidase activity in wilt resistant and susceptible pigeon pea genotypes during Fusarium udum infection. Indian J Agric Biochem. 2015;28:18–23. [Google Scholar]
- Tugizimana F, Steenkamp P, Piater L, Dubery I. A conversation on data mining strategies in LC-MS untargeted metabolomics: pre-processing and pre-treatment steps. Metabolites. 2016;6:1–18. doi: 10.3390/metabo6040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Unsicker SB, Fellenberg C, Constabel CP, Schmidt A, Gershenzon J, Hammerbachera A. Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol. 2017;175:1560–1578. doi: 10.1104/pp.17.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadivel AK. Gel-based proteomics in plants: time to move on from the tradition. Front Plant Sci. 2015;6:369. doi: 10.3389/fpls.2015.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Wang X, Kong L, Zhi P, Chang C. Update on cuticularwax biosynthesis and its roles in plant disease resistance. Int J Mol Sci. 2020;21:5514. doi: 10.3390/ijms21155514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, Florian A, Li W, Pan Q, Nikoloski Z, Yan J, Fernie AR. Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell. 2015;27:1839–1856. doi: 10.1105/tpc.15.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart D. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- Yogendra KN, Kushalappa AC, Sarmiento F, Rodriguez E, Mosquera T. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Funct Plant Biol. 2015;42:284–298. doi: 10.1071/FP14177. [DOI] [PubMed] [Google Scholar]
- Yogendra KN, Pushpa D, Mosa KA, Kushalappa AC, Murphy A, Mosquera T. Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Funct Integr Genomic. 2014;14:285–298. doi: 10.1007/s10142-013-0358-8. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Fernie AR. On the role of the tricarboxylic acid cycle in plant productivity. J Integr Plant Biol. 2018;60:1199–1216. doi: 10.1111/jipb.12690. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Qin G, Li B, Tian S. Effect of cinnamic acid for controlling gray mold on table grape and its possible mechanisms of action. Curr Microbiol. 2015;71:396–402. doi: 10.1007/s00284-015-0863-1. [DOI] [PubMed] [Google Scholar]
- Ziv C, Zhao Z, Gao YG, Xia Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci. 2018;9:1088. doi: 10.3389/fpls.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivy M, Wienkoop S, Renaut J, Pinheiro C, Goulas E, Carpentier S. The quest for tolerant varieties: the importance of integrating “omics” techniques to phenotyping. Front Plant Sci. 2015;6:448. doi: 10.3389/fpls.2015.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.