Abstract
Aims
The aim of this study was to use human genetics to investigate the pathogenesis of sick sinus syndrome (SSS) and the role of risk factors in its development.
Methods and results
We performed a genome-wide association study of 6469 SSS cases and 1 000 187 controls from deCODE genetics, the Copenhagen Hospital Biobank, UK Biobank, and the HUNT study. Variants at six loci associated with SSS, a reported missense variant in MYH6, known atrial fibrillation (AF)/electrocardiogram variants at PITX2, ZFHX3, TTN/CCDC141, and SCN10A and a low-frequency (MAF = 1.1–1.8%) missense variant, p.Gly62Cys in KRT8 encoding the intermediate filament protein keratin 8. A full genotypic model best described the p.Gly62Cys association (P = 1.6 × 10−20), with an odds ratio (OR) of 1.44 for heterozygotes and a disproportionally large OR of 13.99 for homozygotes. All the SSS variants increased the risk of pacemaker implantation. Their association with AF varied and p.Gly62Cys was the only variant not associating with any other arrhythmia or cardiovascular disease. We tested 17 exposure phenotypes in polygenic score (PGS) and Mendelian randomization analyses. Only two associated with the risk of SSS in Mendelian randomization, AF, and lower heart rate, suggesting causality. Powerful PGS analyses provided convincing evidence against causal associations for body mass index, cholesterol, triglycerides, and type 2 diabetes (P > 0.05).
Conclusion
We report the associations of variants at six loci with SSS, including a missense variant in KRT8 that confers high risk in homozygotes and points to a mechanism specific to SSS development. Mendelian randomization supports a causal role for AF in the development of SSS.
Keywords: Sick sinus syndrome, GWAS, KRT8, Mendelian randomization, Atrial fibrillation
Graphical Abstract
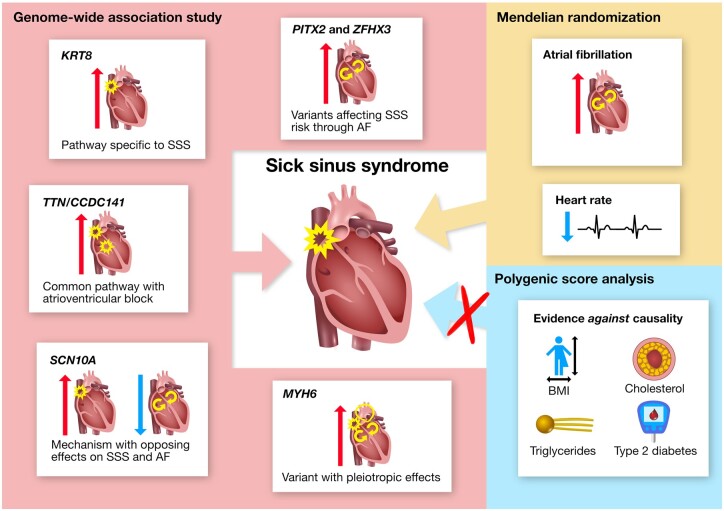
Summary of genetic insight into the pathogenesis of SSS and the role of risk factors in its development. Variants at six loci (named by corresponding gene names) were identified through GWAS and their unique phenotypic associations provide insight into distinct pathways underlying SSS. Investigation of the role of risk factors in SSS development supported a causal role for AF and heart rate and provided convincing evidence against causality for BMI, cholesterol (HDL and non-HDL), triglycerides and T2D. Mendelian randomization did not support causality for CAD, ischaemic stroke, heart failure, PR interval or QRS duration (not shown in figure). Red and blue arrows represent positive and negative associations, respectively.
See page 1972 for the editorial comment on this article (doi: 10.1093/eurheartj/ehab209)
Introduction
Sick sinus syndrome (SSS) is a complex cardiac arrhythmia and the leading indication for permanent pacemaker implantation worldwide.1 It is characterized by pathological sinus bradycardia, sinoatrial block, or alternating atrial brady- and tachyarrhythmias. Symptoms include fatigue, reduced exercise capacity, and syncope.2 , 3 Few studies have been conducted on the basic mechanisms of SSS and therapeutic limitations reflect an incomplete understanding of the pathophysiology. No specific treatment option is aimed at underlying pathways and trying to determine who benefits from cardiac pacing can be challenging.4 A better understanding of mechanisms leading to SSS is crucial for improvements in treatment and prevention and genetic studies provide an opportunity to gain such insight.
Coding variants in several genes, including HCN4, SCN5A, and GNB2, have been implicated in rare familial SSS through linkage analysis and candidate gene methods.5–15 In the only published genome-wide association study (GWAS) of SSS to date, we identified a strong association with a missense variant in MYH6, encoding the alpha-heavy chain subunit of cardiac myosin. This was the first GWAS discovery implicating a cardiac structural or contractile unit as a primary cause of arrhythmia.16
Age is the strongest risk factor for SSS,17 potentially reflecting degenerative fibrosis and electrical remodelling of the SA node and the atria in general.18–20 SSS often coexists with the more common atrial fibrillation (AF)21 and the two arrhythmias are thought to predispose to each other.22 We have previously examined the role of AF in SSS development through Mendelian randomization,23 a method using sequence variants associated with a risk factor (AF) as unbiased proxy indicators to determine whether the risk factor can cause a disease (SSS).24 , 25 The study supported causality,23 likely mediated through atrial remodelling.26 , 27 Several other traits have been associated with SSS while the nature of the associations has not been ascertained. These include body mass index (BMI), height, heart rate, and cardiovascular diseases such as hypertension, myocardial infarction, heart failure, and stroke.17
To gain insight into the pathogenesis of SSS, we performed a large GWAS of over 6000 cases and one million controls of European descent. We used polygenic scores (PGSs) and Mendelian randomization to determine the nature of the associations of risk factors with SSS and in particular whether they contribute to the cause of the disease.
Methods
The study design is two-fold (Supplementary material online, Figure S1). First, we performed a meta-analysis of GWASs, searching for associations of sequence variants with SSS. In total 6469 SSS cases were compared to 1 000 187 controls in a meta-analysis of material from deCODE genetics Iceland, the Copenhagen Hospital Biobank Cardiovascular Study (CHB-CVS)/Danish Blood Donor Study (DBDS), and the UK Biobank,28 with follow-up in the Norwegian Nord-Trøndelag Health Study (HUNT).29 Second, we performed PGS analysis and Mendelian randomization to examine the role of putative risk factors in SSS development (for detailed methods, see Supplementary material online).
Sick sinus syndrome study populations
The deCODE genetics SSS sample consisted of 3577 Icelanders diagnosed with SSS and 347 764 controls. The CHB-CVS included 2209 SSS cases. The control group included blood donors from the DBDS (N > 99 000)30 and SSS cases were also compared to subjects in CHB-CVS with other cardiovascular conditions (N > 89 000). The SSS population from the UK Biobank consisted of 403 cases and 403 181 controls.28 The HUNT cohort consisted of 280 Norwegian SSS cases and 69 141 controls recruited through a population-based health survey conducted in the Nord-Trøndelag County, Norway.29 In all four cohorts, SSS diagnosis was based on International Classification of Diseases 9th (ICD-9: 427.8) or 10th revision (ICD-10: I49.5) from hospitals and/or outpatient clinics. The cohorts are described in more detail in Supplementary material online, Methods and Supplementary material online, Table S1.
Genotyping
The deCODE study was based on whole-genome sequence data from 28 075 Icelanders participating in various disease projects. The 32.9 million variants that passed quality threshold were imputed into 127 175 Icelanders who had been genotyped using Illumina SNP chips. Finally, genotype probabilities for untyped relatives were calculated based on Icelandic genealogy.31 Genotyping for CHB-CVS and DBDS was performed at deCODE using a north European sequencing panel of 15 576 individuals (including 8429 Danes) for imputation into those chip typed (methods manuscript in preparation). Genotyping in the UK Biobank has been described in detail elsewhere.32–35 Illumina HumanCoreExome arrays were used for genotyping in the HUNT cohort.
Genome-wide association study: statistical analysis
We performed a meta-analysis of GWAS of 6189 SSS cases and 931 046 controls from deCODE genetics, CHB-CVS/DBDS, and the UK Biobank. We then tested genome-wide significant and suggestive variants (Supplementary material online, Methods) among 280 cases and 69 141 controls from the Norwegian HUNT study. We used logistic regression to test for association between sequence variants and SSS and other case–control phenotypes, treating phenotype status as the response and allele count as a covariate. Other available individual characteristics that correlate with phenotype status were included in the model as nuisance variables. SSS associations were tested under additive, recessive, and full genotypic models. The full genotypic model includes separate parameters for heterozygotes and homozygotes. We corrected the threshold for genome-wide significance for multiple testing with a weighted Bonferroni adjustment using as weights the enrichment of variant classes with predicted functional impact among association signals estimated from the Icelandic data.36 Significance thresholds were 1.3 × 10−7 for variants with high impact, 2.6 × 10−8 for variants with moderate impact (including missense), 2.4 × 10−9 for low-impact variants, 1.2 × 10−9 for other variants in Dnase I hypersensitivity sites, and 7.5 × 10−10 for all other variants.36 We used linear regression to test variant associations with quantitative phenotypes, treating the quantitative measurement as response and the genotype as covariate. These included endophenotypes of SSS, chronotropic response to exercise (N = 7746), and electrocardiogram (ECG) measurements (N ∼ 73 000) (see Supplementary material online, Methods for detail).
Analysis of genetic risk shared by sick sinus syndrome and putative risk factors
We used PGSs for 17 exposure phenotypes to examine their correlation with SSS (Supplementary material online, Table S2). The phenotypes were chosen because of reported epidemiological associations with SSS or other cardiovascular conditions (see Supplementary material online, Methods). The PGSs were generated using summary statistics from the largest available GWASs (training sample) for each phenotype that do not include deCODE data (Supplementary material online, Table S2 and Supplementary material online, Figure S1). Subsequently, they were tested for association with the risk of SSS among 2556 chip-typed individuals from deCODE (target sample). P-values were corrected using genomic control.37
The use of PGSs to detect association between exposure and outcome is a robust method such that the absence of association in a well-powered PGS analysis provides strong evidence against causality.25 However, finding association in PGS analysis does not confirm causality.38 , 39 For the eight PGSs that associated with SSS, we performed a 2-sample Mendelian randomization analysis, equivalent to a fixed effect MR-Egger40 using published genome-wide significant SNPs from the largest available GWAS on each exposure phenotype as genetic instruments.41–49 To estimate the causal effect, we regressed the published effects of the SNPs on the respective exposure phenotype against their SSS effects in deCODE, CHB-CVS/DBDS, and the UK Biobank, using minor allele frequency (MAF) × (1 − MAF) as weights. For the ECG measurements PR interval and QRS duration, effects in milliseconds were converted to standard deviations (SDs) according to one SD in the Icelandic data so that all causal estimates in a Mendelian randomization forest plot correspond to equal odds ratio (OR) or SD changes for binary and quantitative traits, respectively. For significant associations, we performed further sensitivity analysis to detect outliers (funnel plots) and tested for directional pleiotropy using the MR-Egger intercept test (see Supplementary Material online, Methods).40
Results
Six sick sinus syndrome loci
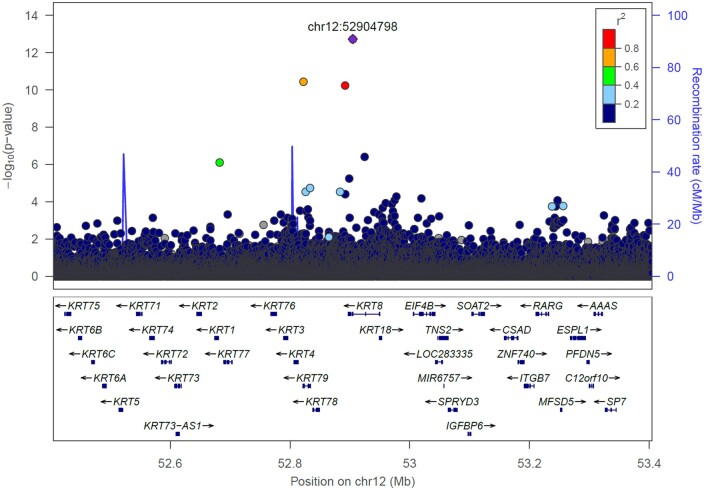
In a GWAS comparing 6469 SSS cases to 1 000 187 controls, variants at six loci satisfied our criteria for genome-wide significance (Table 1, Supplementary material online, Table S3, Figure 1, and Supplementary material online, Figure S2), with ORs ranging from 1.12 to 8.88. One is at a novel SSS locus, a low-frequency missense variant, p.Gly62Cys (MAF = 1.1–1.8% in the four cohorts) in the gene KRT8 on chromosome 12. This variant has not been associated with arrhythmias or ECG traits before. The other variants are the rare Icelandic SSS missense variant p.Arg721Trp in the sarcomere gene MYH6,16 common variants at two AF loci, PITX2 50 and ZFHX3,51 and two ECG loci, TTN/CCDC141 52 and SCN10A.52–54 We have previously reported secondary associations of variants at the four AF and ECG loci with SSS.23 , 52 At the TTN/CCDC141 locus, we report associations of two missense variants, p.Thr811Ile in TTN and p.Glu382Asp in CCDC141, that are weakly correlated (R 2 = 0.20, D’ = 0.64, Supplementary material online, Table S4).
Table 1.
Association results for lead variants at loci reaching genome-wide significance in a meta-analysis of sick sinus syndrome including 6469 cases and 1 000 187 controls
| Locus number | Rs-name/Chr: position (hg38) | Effect allele/other | EAFa (%) | Variant annotation | Coding change | Closest gene | OR (95% CI) | P-value | P-value threshold |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rs387906656b/chr14:23396970 | A/G | 0.34 | Missense | p.Arg721Trp | MYH6 | 8.88 (6.97–11.32) | 7.5 × 10−70 | 2.6 × 10−8 |
| 2 | rs7689774/chr4:110782354 | T/G | 20.07 | Intergenic | – | MIR297, PITX2 | 1.21 (1.15–1.26) | 2.0 × 10−15 | 1.2 × 10−9 |
| 3 | rs11554495/chr12:52904798 | A/C | 1.64 | Missense | p.Gly62Cys | KRT8 | 1.62 (1.43–1.84) | 9.4 × 10−14 | 2.6 × 10−8 |
| 4 | rs12932445/chr16:73035989 | C/T | 20.10 | Intronic | – | ZFHX3 | 1.16 (1.11–1.21) | 8.1 × 10−10 | 1.2 × 10−9 |
| 5 | rs35813871c/chr2:178785681 | A/G | 26.63 | Missense | p.Thr811Ile | TTN | 1.13 (1.09–1.18) | 5.7 × 10−9 | 2.6 × 10−8 |
| rs34883828c/chr2:178905448 | A/C | 15.23 | Missense | p.Glu382Asp | CCDC141 | 1.15 (1.09–1.21) | 1.1 × 10−7 | 2.6 × 10−8 | |
| 6 | rs6795970/chr3:38725184 | A/G | 35.78 | Missense | p.Val1073Ala | SCN10A | 1.12 (1.07–1.16) | 2.5 × 10−8 | 2.6 × 10−8 |
CI, confidence interval; OR, odds ratio.
Effect allele frequency in Iceland (deCODE).
Variant exclusive to Iceland.
p.Thr811Ile in TTN and p.Glu382Asp in CCDC141 are weakly correlated: R2= 0.20, D’ = 0.64.
Figure 1.
Regional plot of the KRT8 locus on chromosome 12q13. The plot depicts association results (P-values) with SSS (N = 6189) in a meta-analysis with data from deCODE, the CHB-CVS/DBDS, and the UK Biobank. The y-axis shows the −log10 P-value and x-axis shows the genomic position (hg38). The lead variant of the signal (p.Gly62Cys) is labelled as a diamond and coloured purple. Other variants are coloured according to correlation (R 2) with the lead variant in the deCODE data. The plot includes variants common to the three datasets as well as variants specific to the Icelandic deCODE data.
High penetrance of sick sinus syndrome among homozygotes for p.Gly62Cys in KRT8
The SSS association of p.Gly62Cys in KRT8 was discovered with the additive model (Table 1 and Supplementary material online, Tables S5 and S6). However, homozygotes of p.Gly62Cys have a higher risk of SSS than assumed under the additive model (Table 2, homozygous genotypic OR > 1.622 = 2.62, P full vs. additive = 2.1 × 10−8). We observed an OR of 1.44 for heterozygotes (95% CI = 1.26–1.65) and 13.99 for homozygotes (95% CI = 8.16–23.98), compared to non-carriers (P-value for the full genotypic model = 1.6 × 10−20, Table 2). None of the other SSS associations deviated from the additive model (P > 0.05). KRT8 encodes the intermediate filament keratin 8 (K8, Supplementary material online, Figure S3) which is widely expressed, including in the heart,55 and we detected its expression (52.1 ± 30.2 transcripts per million) in our cardiac samples from right atria (n = 169, Supplementary material online, Methods). Neither p.Gly62Cys nor two correlated variants (R 2 > 0.6) associated with the expression of KRT8 or nearby genes (1 Mb) in our cardiac samples (Supplementary material online, Table S7) or in GTEx.55
Table 2.
Association of p.Gly62Cys in KRT8 with sick sinus syndrome in the deCODE, CHB-CVS/DBDS, UK Biobank, and HUNT samples under the full genotypic model, calculating independent risk among heterozygotes and homozygotes
| Rs-name/Pos (hg38) | Effect allele/other | Coding change | Gene | Cohort | N | EAF (%) | Risk for heterozygous carriers, OR (95% CI) | Risk for homozygous carriers, OR (95% CI) | P-value for full genotypic modela |
|---|---|---|---|---|---|---|---|---|---|
| rs11554495/chr12:52904798 | A/C | p.Gly62Cys | KRT8 | deCODE | 3577 | 1.64 | 1.42 (1.17–1.72) | 12.95 (4.58–36.63) | 1.5 × 10−9 |
| CHB-CVS | 2209 | 1.77 | 1.34 (1.08–1.66) | 16.13 (6.24–41.67) | 2.6 × 10−10 | ||||
| UK Biobank | 403 | 1.05 | 2.35 (1.42–3.88) | 17.90 (0.74–435.16) | 0.00012 | ||||
| HUNT | 280 | 1.15 | 1.55 (0.77–2.91) | 0.00b | 0.50 | ||||
| Combined | 6469 | – | 1.44 (1.26–1.65) | 13.99 (8.16–23.98) | 1.6 × 10−20 |
CI, confidence interval; EAF, effect allele frequency; OR, odds ratio; Pos, position; SSS, sick sinus syndrome.
The full genotypic model (P full = 1.6 × 10−20) deviates significantly from the additive model (P additive = 9.4 × 10−14). P for deviation from the additive model = 2.1 × 10−8.
Among 14 homozygotes in the HUNT study, none had SSS (P-value for homozygotes = 0.97).
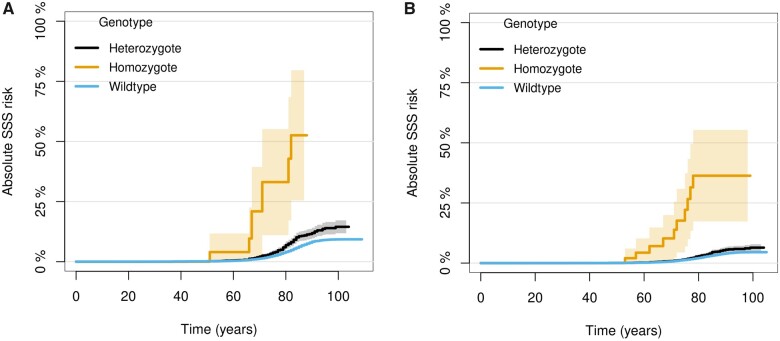
About 1 in 5000 individuals from the four European populations in this study is homozygous for p.Gly62Cys in KRT8, consistent with the Hardy–Weinberg equilibrium (P = 0.071–0.82). To evaluate the penetrance of SSS among homozygous carriers of p.Gly62Cys, we analysed the two largest SSS datasets in the study, deCODE and CHB-CVS/DBDS. Considering that SSS is a late onset disease, the observed penetrance is relatively high although incomplete (Figure 2). Eight out of 36 genotyped Icelandic homozygotes (aged 23–88 years, mean 59.9) had SSS (Supplementary material online, Table S8) and 10 out of 79 homozygotes (aged 27–103 years, mean 62.1) in CHB-CVS/DBDS. Homozygotes were not significantly younger at SSS diagnosis than heterozygotes or non-carriers (P = 0.53 in deCODE and P = 0.26 in CHB-CVS/DBDS, Supplementary material online, Table S9).
Figure 2.
Cumulative incidence curves for SSS age of onset based on KRT8 p.Gly62Cys genotype (A) in deCODE and (B) in CHB-CVS/DBDS. Aalen–Johansen estimator was used, treating death as a competing risk.56 Age of onset was determined by the first registered ICD diagnosis of SSS available to us and thus represents an upper range (Supplementary material online, Methods).
Diverse associations of sick sinus syndrome variants with other phenotypes
We tested the associations of the SSS variants with other phenotypes in deCODE, CHB-CVS/DBDS, and the UK Biobank datasets (Supplementary material online, Tables S10–S14). In addition to SSS, p.Gly62Cys in KRT8 associated with pacemaker implantation (OR heterozygotes = 1.28, 95% CI = 1.13–1.45, OR homozygotes = 9.17, 95% CI = 2.89–29.09, P = 1.9 × 10−8). The variant did not associate with other arrhythmias or cardiovascular diseases, with electrolyte or hormonal disturbances that are linked to the development of SSS (potassium, calcium, thyroid stimulating hormone)57 or with the suggested consequence of p.Gly62Cys in candidate gene studies58 (pancreatitis/lipase, liver disease, Bonferroni-adjusted significance threshold of P = 0.05/23 = 0.0022, Supplementary material online, Tables S10 and S11). Furthermore, p.Gly62Cys did not associate with any of the 122 ECG parameters (N up to 72 825, Supplementary material online, Table S13).
The associations of SSS variants with AF varied. p.Gly62Cys in KRT8 (Supplementary material online, Tables S10 and S11) and the missense variants in TTN/CCDC141 (Supplementary material online, Table S12) did not associate with the risk of AF, applying a Bonferroni-corrected significance threshold of P < 0.05/14 = 0.0036. The TTN/CCDC141 variants are located within 300 kbp from previously reported AF variants41 and are independent of them (R 2 < 0.2, Supplementary material online, Figure S4). The other four SSS loci have been reported to affect AF but their pattern of association with the two phenotypes differ. p.Arg721Trp in MYH6 23 associates stronger with SSS than AF but the reverse applies to variants at PITX2. At ZFHX3, the effects on AF and SSS are comparable.41 , 50 , 51 , 59 , 60 p.Val1073Ala in SCN10A is one of the top AF variants at this locus (Supplementary material online, Figure S5), but the allele that increases the risk of SSS protects against AF (Supplementary material online, Table S12).53 , 61
All the SSS variants increased the risk of pacemaker implantation (Supplementary material online, Tables S10–S12), as expected since SSS is the most common reason for this procedure. We have previously described in detail the associations of p.Arg721Trp in MYH6 with coarctation of the aorta, other congenital malformations of the heart, aortic valve stenosis, and heart failure23 , 62 , 63 (Supplementary material online, Table S12). We note that p.Glu382Asp in CCDC141 associated with both second- and third-degree atrioventricular block (AVB)52 and the association with third-degree AVB was genome-wide significant (P = 1.3 × 10−14, OR = 1.27, 95% CI = 1.20–1.35) and more significant than the SSS association. Another notable association is that of the PITX2 variant with heart failure (Supplementary material online, Table S12) as recently reported in a GWAS of heart failure.47 None of the SSS variants associated with chronotropic response to exercise (Supplementary material online, Tables S15 and S16) in our dataset. However, the SCN10A and CCDC141 loci have been reported to associate with heart rate profile during exercise in larger studies of UK Biobank data.64 , 65
Genetically predicted atrial fibrillation and lower heart rate associate with sick sinus syndrome
PGSs are powerful tools for detecting associations between phenotypes.38 We generated PGSs for 17 exposure phenotypes using summary statistics from the largest available GWASs (Supplementary material online, Table S2) and tested them for association with SSS in deCODE data. Eight PGSs, for AF, heart rate, coronary artery disease (CAD), height, QRS duration, PR interval, ischaemic stroke, and heart failure, associated with SSS while those for BMI, type 2 diabetes (T2D), non-HDL cholesterol, HDL cholesterol, triglycerides, and hypertension/blood pressure traits did not (Bonferroni-adjusted significance threshold of P = 0.05/17 = 0.0029, Supplementary material online, Table S17). The PGS for heart rate was the only one to inversely associate with SSS risk.
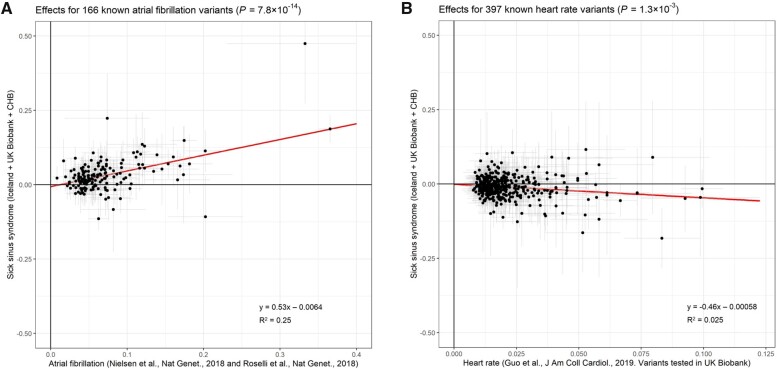
We then performed Mendelian randomization to determine the causality of the associations between the eight exposures and SSS, using genome-wide significant SNPs from GWAS of the exposure phenotypes as instruments (see Methods). Genetic predisposition to AF and genetically determined heart rate and height associated with the risk of SSS but the height association did not remain after accounting for AF in a multivariate analysis (Figures 3 and 4A and B).66–68 With few exceptions, the effects of AF variants on SSS are proportional to their effects on AF (P = 7.8 × 10−14, Figure 4A and Supplementary material online, Figure S6A). The greatest deviation from the expected SSS effect was for the association at SCN10A (tagged by rs6790693, OR for AF = 1.06, OR for SSS = 0.89, Supplementary material online, Figure S6A). Mendelian randomization did not support causality for CAD, ischaemic stroke, heart failure, PR interval, or QRS duration (Figure 3 and Supplementary material online, Figure S7).
Figure 3.
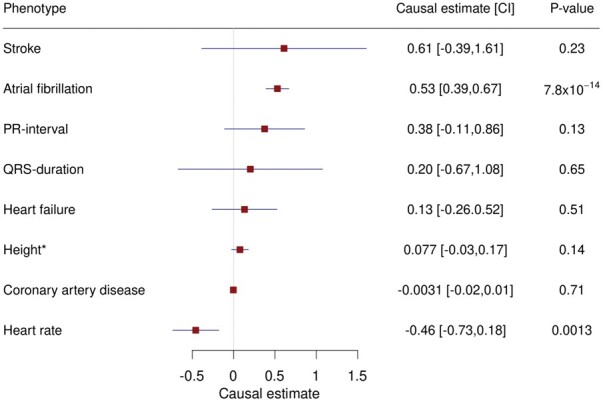
Forest plot showing causal effect estimates from a Mendelian randomization analysis of putative risk factors on SSS. We regressed published effects of SNPs on the respective exposure phenotype against their SSS effects in deCODE, CHB-CVS/DBDS, and the UK Biobank. The causal estimates are the slopes from the regression lines where a one unit change in the exposure phenotypes equals a one standard deviation change for quantitative traits and an odds ratio of 2.7 for binary exposure phenotypes. The forest plot shows the causal estimates, 95% confidence intervals, and corresponding P-values from the Mendelian randomization analysis. *For height, the result from a multivariate analysis accounting for AF effects is shown.
Figure 4.
Visualization of significant associations from Mendelian randomization analysis. Effects of published variants on two phenotypes plotted against their effects on SSS in deCODE, CHB-CVS/DBDS, and the UK Biobank (y-axes, N = 6189). The x-axes depict (A) published AF effects41 , 42 and (B) heart rate effects in UK Biobank.43 The equations for the regression lines are shown and the coefficients of determination (R2).
Discussion
Traditionally, SSS has been considered as a collection of conditions with a variety of causes, intrinsic and extrinsic to the SA node.69 In line with this notion, we identified sequence variants associating with SSS at six loci implicating several distinct pathways in SSS development. One of the SSS associations is novel, with a low-frequency missense variant, p.Gly62Cys in KRT8. One is with the reported p.Arg721Trp in MYH6,16 and four confirm loci previously implicated in SSS secondarily to AF, at PITX2,50 and ZFHX3,51 or ECG traits, at SCN10A 53 and TTN/CCDC141. 52 The leading variants at four of the SSS loci are missense.
KRT8 encodes K8, a cytoskeletal intermediate filament protein widely expressed, including in skeletal muscle and the heart.55 Historically, keratins have been described as epithelial-specific intermediate filaments70 and mutations affecting keratin function have almost exclusively been associated with diseases of the skin and other epithelial tissues.71 , 72 The association of p.Gly62Cys in KRT8 with SSS described here supports the notion of a role for K8 in the human heart, as previously suggested by animal studies.73–76 Experiments have revealed loss-of-function effects of p.Gly62Cys, that is located in the head domain of K8 (Supplementary material online, Figure S3).77 , 78 Glycine at position 62 is a conserved amino acid (GERP79 score 1.73) and the variant introduces cysteine into a protein otherwise devoid of cysteine residues. This interrupts intermediate filament assembly in vitro 77 and prevents K8 from serving as a phosphate ‘sponge’ for stress-activated kinases that mediate apoptosis.78 Thus, the variant could increase the risk of SSS either by affecting the structural or cardioprotective role of K8.77 , 78
In addition to p.Gly62Cys in KRT8, lead variants at two other SSS loci are missense variants in genes encoding structural components of the heart. MYH6 encodes the alpha-myosin heavy chain (αMHC) and TTN encodes the giant protein titin, both of which are key components of the sarcomere.80 , 81 Since the identification of MYH6 in the first GWAS of SSS,16 structural components of cardiomyocytes have increasingly been implicated in atrioventricular conduction and arrhythmias, in particular AF.23 , 41 , 82 , 83 However, the variants at KRT8, MYH6 and TTN do not mediate their risk through AF because they either do not associate with AF (KRT8 and TTN) or have a stronger effect on SSS risk (MYH6).
K8, αMHC, and titin have a common role in cardiac adaptive responses to stress which may be of relevance to their association with SSS. αMHC allows greater economy in force generation than the homologous beta chain and a shift in their relative expression plays a major role in regulating myocardial contractile activity.84 , 85 Likewise, the isoform switch of titin alters ventricular filling81 , 86 and K8 is upregulated under stress to maintain structural integrity.76 The expression of all three genes is modified in human heart failure.76 , 84 , 87 Thus, whether or not the SSS variants in KRT8, MYH6 and TTN directly affect the SA node, it is possible that they could contribute to inadequate cardiac output and symptom development in SSS by interrupting adaptive increase in stroke volume.
The SSS variants at PITX2 and ZFHX3 point to genes encoding transcription factors.88 , 89 They likely mediate their effects on SSS through AF since these have consistently been the strongest AF loci in GWAS.42 , 50 , 51 Conversely, p.Val1073Ala in SCN10A has opposite effects on the two arrhythmias. The allele that increases the risk of SSS protects against AF.53 , 61 It also prolongs the PR interval53 , 54 , 61 and duration of the QRS complex.52 , 54 SCN10A encodes the neuronal sodium channel isoform Nav1.8. A plausible mechanism behind the unique phenotypic effects of p.Val1073Ala involves Nav1.8s role in the way in which the autonomic nervous system affects the heart.90–92 In support of this, animal studies have shown that blockade of Nav1.8 in intracardiac autonomic neurons in the ganglionated plexi (GP) suppresses AF inducibility.90 , 91 The same applies to GP ablation in patients with AF.93–95 However, GP ablation is controversial due to observed adverse events,96–98 including development of SSS and pacemaker implantation.99 The opposite effects of p.Val1073Ala on SSS and AF risk support concerns that GP ablation for AF treatment may cause SSS and links this adverse effect to Nav1.8 function specifically.
Our genetic analyses shed light on pathways contributing to the development of SSS and the role of risk factors. In particular, they provide insight into the intricate and debated relationship between SSS and AF.22 Our Mendelian randomization analysis reveals a strong association between genetic predisposition for AF and risk of SSS. This is consistent with our previous report23 and likely reflects causality. Pacing-induced AF has been shown to impair SA node function in dogs100 and AF causes regional atrial substrate changes around the SA node in humans.27 Pathobiological pathways common to AF and SSS, such as inflammation, interstitial atrial fibrosis, and alterations in intracellular Ca2+ dynamics, may also contribute to this trend.22 , 26 , 101
Importantly, our results also show that SSS does not only occur as a consequence of AF or pathways common to AF. This is evident in the case of SSS variants at KRT8 and TTN/CCDC141 that do not associate with the risk of AF in a large dataset. p.Gly62Cys in KRT8 is the only SSS variant that does not associate with other arrhythmias, ECG traits, or cardiovascular diseases in our dataset. Thus, this variant in particular may point to a mechanism that is specific to SSS development. We observe a genome-wide significant association of p.Glu382Asp in CCDC141 with third-degree AVB, the first one reported for complete heart block. Therefore, this locus points to a mechanism linking the functions of the AV and SA nodes and does not mediate the risk of SSS as a consequence of AF. The complexity of the relationship between SSS and AF is further evident by the opposite effects of the SCN10A locus on the two arrhythmias. Finally, since SSS variants do not consistently associate with AF, our data do not suggest a strong causal role for SSS in the development of AF.
Heart rate variants associate inversely with SSS, consistent with epidemiological observation,17 potentially because bradycardia is an integral part of the SSS diagnosis. However, lower heart rate could also represent a direct marker of biological processes affecting SSS, such as fibrosis or altered autonomic neural input. Finally, lack of associations with powerful PGSs based on large datasets serves as convincing evidence against causality.25 In particular, this applies to BMI, cholesterol, triglycerides, and T2D.
Several of our findings have direct clinical implications. In particular, the causal role of AF in SSS development emphasizes the need for heightened awareness of potential SSS among patients with AF, especially those with unspecific symptoms. Furthermore, the strong association of the SSS variant p.Glu382Asp in CCDC141 with AVB encourages consideration of dual chamber pacemaker implantation in SSS patients carrying this variant. Lastly, the SCN10A locus points to SSS as a potential adverse effect of GP ablation therapy for AF.
The major strength of the study is the large SSS sample set and extensive phenotypic and genotypic data for the same datasets. Different proportions of comorbidities and drug use in SSS cases and controls could constitute confounding; however, the consistent associations of SSS variants with both SSS and pacemaker implantation across cohorts support their true effect on cardiac conduction. Furthermore, if a specific comorbidity would explain one of the SSS associations, this would be evident by a stronger effect on that phenotype. The strength of Mendelian randomization in determining causality includes less concern for confounding and reverse causation than in observational studies since sequence variants associated with the exposure (e.g. AF) are randomized during meiosis and this process is unaffected by the presence of the outcome (SSS) later in life.24 For heart failure, a limitation of the Mendelian randomization analysis is that variants identified in GWAS are not ideal instruments because of their association with potential confounders (e.g. AF, CAD, and BMI).47
Conclusion
In this large genetic study, we found six SSS loci, some of which also associate with other arrhythmias and cardiac electrical function. One of the associations is with a missense variant in the intermediate filament gene KRT8 that confers high risk of SSS among homozygous carriers. Mendelian randomization analysis suggests that AF and lower heart rate are directly involved in SSS development. PGS analysis provides convincing evidence against causality for BMI, cholesterol, triglycerides, and T2D. On the other hand, the data also show that SSS can result from perturbation of pathways unrelated to AF. In particular, p.Gly62Cys at KRT8 does not associate with any other cardiovascular traits and points to a mechanism that is specific to SSS development.
Supplementary material
Supplementary material is available at European Heart Journal online.
Data availability
The Icelandic population WGS data has been deposited at the European Variant Archive under accession code PRJEB15197. We declare that the data supporting the findings of this study are available within the article, its Supplementary material online and upon reasonable request. The genome-wide association scan summary data will be made available at http://www.decode.com/summarydata.
Supplementary Material
Acknowledgements
We thank all the study subjects for their valuable participation as well as our colleagues that contributed to data collection, sample handling, and genotyping.
Contributor Information
Rosa B Thorolfsdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Gardar Sveinbjornsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Hildur M Aegisdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Stefania Benonisdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Lilja Stefansdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Erna V Ivarsdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Gisli H Halldorsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Jon K Sigurdsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Christian Torp-Pedersen, Department of Clinical Research and Cardiology, Nordsjaelland Hospital, Dyrehavevej 29, Hillerød 3400, Denmark.
Peter E Weeke, Department of Cardiology, Copenhagen University Hospital, Blegdamsvej 9, Copenhagen 2100, Denmark.
Søren Brunak, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3A, Copenhagen 2200, Denmark.
David Westergaard, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3A, Copenhagen 2200, Denmark.
Ole B Pedersen, Department of Clinical Immunology, Naestved Hospital, Ringstedgade 77B, Naestved 4700, Denmark.
Erik Sorensen, Department of Clinical Immunology, Copenhagen University Hospital, Blegdamsvej 9, Copenhagen 2100, Denmark.
Kaspar R Nielsen, Department of Clinical Immunology, Aalborg University Hospital North, Urbansgade 36, Aalborg 9000, Denmark.
Kristoffer S Burgdorf, Department of Clinical Immunology, Copenhagen University Hospital, Blegdamsvej 9, Copenhagen 2100, Denmark.
Karina Banasik, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Blegdamsvej 3A, Copenhagen 2200, Denmark.
Ben Brumpton, Department of Thoracic and Occupational Medicine, St. Olavs Hospital, Trondheim University Hospital, Prinsesse Kristinas gate 3, Trondheim 7030, Norway.
Wei Zhou, Department of Computational Medicine and Bioinformatics, University of Michigan, 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218, USA.
Asmundur Oddsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Vinicius Tragante, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Kristjan E Hjorleifsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Department of Computing and Mathematical Sciences, California Institute of Technology, 1200 E California Blvd. MC 305-16, Pasadena, CA 91125, USA.
Olafur B Davidsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Sridharan Rajamani, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Stefan Jonsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Bjarni Torfason, Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland; Department of Cardiothoracic Surgery, Landspitali—The National University Hospital of Iceland, Hringbraut, Reykjavik 101, Iceland.
Atli S Valgardsson, Department of Cardiothoracic Surgery, Landspitali—The National University Hospital of Iceland, Hringbraut, Reykjavik 101, Iceland.
Gudmundur Thorgeirsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland; Department of Medicine, Landspitali—The National University Hospital of Iceland, Hringbraut, Reykjavik 101, Iceland.
Michael L Frigge, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Gudmar Thorleifsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Gudmundur L Norddahl, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Anna Helgadottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Solveig Gretarsdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Patrick Sulem, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Ingileif Jonsdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland; Department of Immunology, Landspitali—The National University Hospital of Iceland, Hringbraut, Reykjavik 101, Iceland.
Cristen J Willer, Department of Computational Medicine and Bioinformatics, University of Michigan, 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218, USA; Department of Internal Medicine: Cardiology, University of Michigan, 1500 East Medical Center Drive, Ann Arbor, MI 48109 -5368, USA; Department of Human Genetics, University of Michigan, 4909 Buhl Building, 1241 E. Catherine St., Ann Arbor, MI 48109 -5618, USA.
Kristian Hveem, K.G. Jebsen Center for Genetic Epidemiology, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Erling Skjalgssons gt. 1, Trondheim 7491, Norway; Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Postboks 8905, Trondheim 7491, Norway; HUNT Research Centre, Department of Public Health and General Practice, Norwegian University of Science and Technology, Forskningsveien 2, Levanger 7600, Norway.
Henning Bundgaard, Department of Cardiology, Copenhagen University Hospital, Blegdamsvej 9, Copenhagen 2100, Denmark.
Henrik Ullum, Department of Clinical Immunology, Copenhagen University Hospital, Blegdamsvej 9, Copenhagen 2100, Denmark; Statens Serum Institut, Artillerivej 5, Copenhagen 2300, Denmark.
David O Arnar, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland; Department of Medicine, Landspitali—The National University Hospital of Iceland, Hringbraut, Reykjavik 101, Iceland.
Unnur Thorsteinsdottir, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland.
Daniel F Gudbjartsson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; School of Engineering and Natural Sciences, University of Iceland, Hjardarhagi 4, Reykjavik 107, Iceland.
Hilma Holm, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland.
Kari Stefansson, deCODE genetics/Amgen, Inc., Sturlugata 8, Reykjavik 101, Iceland; Faculty of Medicine, University of Iceland, Vatnsmyrarvegur 16, Reykjavik 101, Iceland.
DBDS Genomic Consortium:
Steffen Andersen, Christian Erikstrup, Thomas F Hansen, Henrik Hjalgrim, Gregor Jemec, Poul Jennum, Mette Nyegaard, Mie T Bruun, Mikkel Petersen, Thomas Werge, and Per I Johansson
Funding
This work was partly supported by NordForsk through the funding to PM Heart, project number 90580, the Innovation Fund Denmark (IFD) under File No. 8114-00033B and the Technology Development Fund, Iceland, project number 90580.
Conflict of interest: The following authors affiliated with deCODE genetics/Amgen, Inc., are employed by the company: R.B.T., G.S., H.M.A., S.B., L.S., E.V.I., G.H.H., J.K.S., A.O., V.T., K.E.H., O.B.D., S.R., S.J., G.T., M.L.F., G.T., G.L.N., A.H., S.G., P.S., I.J., D.O.A., U.T., D.F.G., H.H., and K.S.. D.W. received grants from Novo Nordisk Foundation during the conduct of the study. S.B. is a board member for Proscion A/S and Intomics A/S.
References
- 1. Mond HG, Proclemer A. The 11th world survey of cardiac pacing and implantable cardioverter-defibrillators: calendar year 2009—a World Society of Arrhythmia's project. Pacing Clin Electrophysiol 2011;34:1013–1027. [DOI] [PubMed] [Google Scholar]
- 2. Ferrer MI. The sick sinus syndrome. Circulation 1973;47:635–641. [DOI] [PubMed] [Google Scholar]
- 3. Fuster V, Harrington RA, Narula J, Eapen ZJ, Hurst’s the Heart. 14th ed. New York: McGraw-Hill Education; 2017. [Google Scholar]
- 4. Brignole M, Auricchio A, Baron-Esquivias G, Bordachar P, Boriani G, Breithardt OA, Cleland J, Deharo JC, Delgado V, Elliott PM, Gorenek B, Israel CW, Leclercq C, Linde C, Mont L, Padeletti L, Sutton R, Vardas PE. 2013 ESC guidelines on cardiac pacing and cardiac resynchronization therapy: the task force on cardiac pacing and resynchronization therapy of the European Society of Cardiology (ESC). Developed in collaboration with the European Heart Rhythm Association (EHRA). Europace 2013;15:1070–1118. [DOI] [PubMed] [Google Scholar]
- 5. Le Scouarnec S, Bhasin N, Vieyres C, Hund TJ, Cunha SR, Koval O, Marionneau C, Chen B, Wu Y, Demolombe S, Song LS, Le Marec H, Probst V, Schott JJ, Anderson ME, Mohler PJ. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci USA 2008;105:15617–15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu YB, Luo JW, Jiang F, Liu G. Genetic analysis of sick sinus syndrome in a family harboring compound CACNA1C and TTN mutations. Mol Med Rep 2018;17:7073–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL. Jr., Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J Clin Investig 2003;112:1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makita N, Sasaki K, Groenewegen WA, Yokota T, Yokoshiki H, Murakami T, Tsutsui H. Congenital atrial standstill associated with coinheritance of a novel SCN5A mutation and connexin 40 polymorphisms. Heart Rhythm 2005;2:1128–1134. [DOI] [PubMed] [Google Scholar]
- 9. Makiyama T, Akao M, Tsuji K, Doi T, Ohno S, Takenaka K, Kobori A, Ninomiya T, Yoshida H, Takano M, Makita N, Yanagisawa F, Higashi Y, Takeyama Y, Kita T, Horie M. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol 2005;46:2100–2106. [DOI] [PubMed] [Google Scholar]
- 10. Ishikawa T, Ohno S, Murakami T, Yoshida K, Mishima H, Fukuoka T, Kimoto H, Sakamoto R, Ohkusa T, Aiba T, Nogami A, Sumitomo N, Shimizu W, Yoshiura KI, Horigome H, Horie M, Makita N. Sick sinus syndrome with HCN4 mutations shows early onset and frequent association with atrial fibrillation and left ventricular noncompaction. Heart Rhythm 2017;14:717–724. [DOI] [PubMed] [Google Scholar]
- 11. Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med 2006;354:151–157. [DOI] [PubMed] [Google Scholar]
- 12. Nof E, Luria D, Brass D, Marek D, Lahat H, Reznik-Wolf H, Pras E, Dascal N, Eldar M, Glikson M. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 2007;116:463–470. [DOI] [PubMed] [Google Scholar]
- 13. Stallmeyer B, Kuß J, Kotthoff S, Zumhagen S, Vowinkel K, Rinné S, Matschke LA, Friedrich C, Schulze-Bahr E, Rust S, Seebohm G, Decher N, Schulze-Bahr E. A mutation in the G-protein gene GNB2 causes familial sinus node and atrioventricular conduction dysfunction. Circ Res 2017;120:e33–e44. [DOI] [PubMed] [Google Scholar]
- 14. Lodder EM, De Nittis P, Koopman CD, Wiszniewski W, Moura de Souza CF, Lahrouchi N, Guex N, Napolioni V, Tessadori F, Beekman L, Nannenberg EA, Boualla L, Blom NA, de Graaff W, Kamermans M, Cocciadiferro D, Malerba N, Mandriani B, Akdemir ZHC, Fish RJ, Eldomery MK, Ratbi I, Wilde AAM, de Boer T, Simonds WF, Neerman-Arbez M, Sutton VR, Kok F, Lupski JR, Reymond A, Bezzina CR, Bakkers J, Merla G. GNB5 mutations cause an autosomal-recessive multisystem syndrome with sinus bradycardia and cognitive disability. Am J Hum Genet 2016;99:704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuß J, Stallmeyer B, Goldstein M, Rinné S, Pees C, Zumhagen S, Seebohm G, Decher N, Pott L, Kienitz MC, Schulze-Bahr E. Familial sinus node disease caused by a gain of GIRK (G-protein activated inwardly rectifying K(+) channel) channel function. Circ Genom Precis Med 2019;12:e002238. [DOI] [PubMed] [Google Scholar]
- 16. Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, Stefansdottir H, Gretarsdottir S, Matthiasson SE, Thorgeirsson GM, Jonasdottir A, Sigurdsson A, Stefansson H, Werge T, Rafnar T, Kiemeney LA, Parvez B, Muhammad R, Roden DM, Darbar D, Thorleifsson G, Walters GB, Kong A, Thorsteinsdottir U, Arnar DO, Stefansson K. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet 2011;43:316–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jensen PN, Gronroos NN, Chen LY, Folsom AR, deFilippi C, Heckbert SR, Alonso A. Incidence of and risk factors for sick sinus syndrome in the general population. J Am Coll Cardiol 2014;64:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demoulin JC, Kulbertus HE. Histopathological correlates of sinoatrial disease. Br Heart J 1978;40:1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lien WP, Lee YS, Chang FZ, Lee SY, Chen CM, Tsai HC. The sick sinus syndrome: natural history of dysfunction of the sinoatrial node. Chest 1977;72:628–634. [DOI] [PubMed] [Google Scholar]
- 20. Morris GM, Kalman JM. Fibrosis, electrics and genetics. Perspectives in sinoatrial node disease. Circ J 2014;78:1272–1282. [DOI] [PubMed] [Google Scholar]
- 21. Gomes JA, Kang PS, Matheson M, Gough WB Jr., El-Sherif N. Coexistence of sick sinus rhythm and atrial flutter-fibrillation. Circulation 1981;63:80–86. [DOI] [PubMed] [Google Scholar]
- 22. Kezerashvili A, Krumerman AK, Fisher JD. Sinus node dysfunction in atrial fibrillation: cause or effect? J Atr Fibrillation 2008;1:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Helgadottir A, Gretarsdottir S, Benonisdottir S, Magnusdottir A, Davidsson OB, Rajamani S, Roden DM, Darbar D, Pedersen TR, Sabatine MS, Jonsdottir I, Arnar DO, Thorsteinsdottir U, Gudbjartsson DF, Holm H, Stefansson K. A missense variant in PLEC increases risk of atrial fibrillation. J Am Coll Cardiol 2017;70:2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, Hartwig FP, Holmes MV, Minelli C, Relton CL, Theodoratou E. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res 2020;4:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akoum N, McGann C, Vergara G, Badger T, Ranjan R, Mahnkopf C, Kholmovski E, Macleod R, Marrouche N. Atrial fibrosis quantified using late gadolinium enhancement MRI is associated with sinus node dysfunction requiring pacemaker implant. J Cardiovasc Electrophysiol 2012;23:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang HY, Lin YJ, Lo LW, Chang SL, Hu YF, Li CH, Chao TF, Yin WH, Chen SA. Sinus node dysfunction in atrial fibrillation patients: the evidence of regional atrial substrate remodelling. Europace 2013;15:205–211. [DOI] [PubMed] [Google Scholar]
- 28. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krokstad S, Langhammer A, Hveem K, Holmen TL, Midthjell K, Stene TR, Bratberg G, Heggland J, Holmen J. Cohort profile: the HUNT study, Norway. Int J Epidemiol 2013;42:968–977. [DOI] [PubMed] [Google Scholar]
- 30. Hansen TF, Banasik K, Erikstrup C, Pedersen OB, Westergaard D, Chmura PJ, Nielsen K, Thorner L, Hjalgrim H, Paarup H, Larsen MAH, Petersen M, Jennum P, Andersen S, Nyegaard M, Jemec GBE, Olesen J, Werge T, Johansson PI, Sorensen E, Brunak S, Ullum H, Burgdorf KS. DBDS genomic cohort, a prospective and comprehensive resource for integrative and temporal analysis of genetic, environmental and lifestyle factors affecting health of blood donors. BMJ Open 2019;9:e028401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E, Sigurdsson GT, Stacey SN, Frigge ML, Holm H, Saemundsdottir J, Helgadottir HT, Johannsdottir H, Sigfusson G, Thorgeirsson G, Sverrisson JT, Gretarsdottir S, Walters GB, Rafnar T, Thjodleifsson B, Bjornsson ES, Olafsson S, Thorarinsdottir H, Steingrimsdottir T, Gudmundsdottir TS, Theodors A, Jonasson JG, Sigurdsson A, Bjornsdottir G, Jonsson JJ, Thorarensen O, Ludvigsson P, Gudbjartsson H, Eyjolfsson GI, Sigurdardottir O, Olafsson I, Arnar DO, Magnusson OT, Kong A, Masson G, Thorsteinsdottir U, Helgason A, Sulem P, Stefansson K. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 2015;47:435–444. [DOI] [PubMed] [Google Scholar]
- 32. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, Ng FL, Evangelou M, Witkowska K,, Tzanis E, Hellwege JN, Giri A, Velez Edwards DR, Sun YV, Cho K, Gaziano JM, Wilson PWF, Tsao PS, Kovesdy CP, Esko T, Magi R, Milani L, Almgren P, Boutin T,, Debette S, Ding J, Giulianini F, Holliday EG, Jackson AU, Li-Gao R, Lin WY, Luan J, Mangino M, Oldmeadow C,, Prins BP, Qian Y, Sargurupremraj M, Shah N, Surendran P, Theriault S, Verweij N, Willems SM, Zhao JH, Amouyel P, Connell J, de Mutsert R, Doney ASF, Farrall M, Menni C, Morris AD, Noordam R, Pare G, Poulter NR, Shields DC, Stanton A, Thom S, Abecasis G, Amin N, Arking DE, Ayers KL, Barbieri CM, Batini C, Bis JC, Blake T,, Bochud M, Boehnke M, Boerwinkle E, Boomsma DI, Bottinger EP, Braund PS, Brumat M, Campbell A, Campbell H, Chakravarti A, Chambers JC, Chauhan G,, Ciullo M, Cocca M, Collins F, Cordell HJ, Davies G, de Borst MH, de Geus EJ, Deary IJ, Deelen J, Del Greco MF, Demirkale CY, Dorr M, Ehret GB, Elosua R, Enroth S,, Erzurumluoglu AM, Ferreira T, Franberg M, Franco OH, Gandin I, Gasparini P, Giedraitis V, Gieger C, Girotto G, Goel A,, Gow AJ, Gudnason V, Guo X, Gyllensten U, Hamsten A, Harris TB, Harris SE, Hartman CA, Havulinna AS,, Hicks AA, Hofer E, Hofman A, Hottenga JJ, Huffman JE, Hwang SJ,, Ingelsson E,, James A, Jansen R, Jarvelin MR, Joehanes R, Johansson A, Johnson AD,, Joshi PK, Jousilahti P,, Jukema JW, Jula A, Kahonen M, Kathiresan S, Keavney BD, Khaw KT, Knekt P, Knight J, Kolcic I, Kooner JS, Koskinen S, Kristiansson K, Kutalik Z, Laan M, Larson M, Launer LJ, Lehne B, Lehtimaki T,, Liewald DCM, Lin L, Lind L, Lindgren CM, Liu Y, Loos RJF,, Lopez LM, Lu Y, Lyytikainen LP, Mahajan A, Mamasoula C, Marrugat J, Marten J, Milaneschi Y, Morgan A, Morris AP, Morrison AC, Munson PJ,, Nalls MA, Nandakumar P, Nelson CP, Niiranen T, Nolte IM, Nutile T, Oldehinkel AJ, Oostra BA, O'Reilly PF, Org E,, Padmanabhan S, Palmas W, Palotie A, Pattie A, Penninx B, Perola M, Peters A, Polasek O, Pramstaller PP, Nguyen QT, Raitakari OT, Ren M, Rettig R, Rice K, Ridker PM, Ried JS, Riese H,, Ripatti S, Robino A, Rose LM, Rotter JI, Rudan I, Ruggiero D, Saba Y, Sala CF, Salomaa V, Samani NJ, Sarin AP, Schmidt R, Schmidt H, Shrine N, Siscovick D, Smith AV, Snieder H, Sober S, Sorice R, Starr JM, Stott DJ, Strachan DP, Strawbridge RJ, Sundstrom J, Swertz MA, Taylor KD, Teumer A, Tobin MD, Tomaszewski M, Toniolo D, Traglia M, Trompet S, Tuomilehto J, Tzourio C, Uitterlinden AG, Vaez A, van der Most PJ, van Duijn CM, Vergnaud AC, Verwoert GC, Vitart V, Volker U, Vollenweider P, Vuckovic D, Watkins H, Wild SH, Willemsen G, Wilson JF, Wright AF, Yao J, Zemunik T, Zhang W, Attia JR, Butterworth AS, Chasman DI, Conen D,, Cucca F, Danesh J, Hayward C, Howson JMM, Laakso M, Lakatta EG, Langenberg C, Melander O, Mook-Kanamori DO, Palmer CNA, Risch L, Scott RA,, Scott RJ, Sever P, Spector TD, van der Harst P, Wareham NJ, Zeggini E, Levy D, Munroe PB, Newton-Cheh C, Brown MJ, Metspalu A, Hung AM, O'Donnell CJ,, Edwards TL,, Psaty BM, Tzoulaki I, Barnes MR, Wain LV, Elliott P, Caulfield MJ. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 2018;50:1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welsh S, Peakman T, Sheard S, Almond R. Comparison of DNA quantification methodology used in the DNA extraction protocol for the UK Biobank cohort. BMC Genomics 2017;18:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham MMcVean G, Leslie S, Donnelly P, Marchini J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, Perry JR, Xu C, Futema M, Lawson D, Iotchkova V, Schiffels S, Hendricks AE, Danecek P, Li R, Floyd J, Wain LV, Barroso I, Humphries SE, Hurles ME, Zeggini E, Barrett JC, Plagnol V, Richards JB, Greenwood CM, Timpson NJ, Durbin R, Soranzo N; UK10K Consortium. The UK10K project identifies rare variants in health and disease. Nature 2015;526:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sveinbjornsson G, Albrechtsen A, Zink F, Gudjonsson SA, Oddson A, Masson G, Holm H, Kong A, Thorsteinsdottir U, Sulem P, Gudbjartsson DF, Stefansson K. Weighting sequence variants based on their annotation increases power of whole-genome association studies. Nat Genet 2016;48:314–317. [DOI] [PubMed] [Google Scholar]
- 37. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richardson TG, Harrison S, Hemani G, Davey Smith G. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. Elife 2019;8:e43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics 2010;186:767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, Mathis MR, Yamazaki M, Crawford RD, Gabrielsen ME, Skogholt AH, Holmen OL, Lin M, Wolford BN, Dey R, Dalen H, Sulem P, Chung JH, Backman JD, Arnar DO, Thorsteinsdottir U, Baras A, O’Dushlaine C, Holst AG, Wen X, Hornsby W, Dewey FE, Boehnke M, Kheterpal S, Mukherjee B, Lee S, Kang HM, Holm H, Kitzman J, Shavit JA, Jalife J, Brummett CM, Teslovich TM, Carey DJ, Gudbjartsson DF, Stefansson K, Abecasis GR, Hveem K, Willer CJ. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 2018;50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, Arking DE, Barnard J, Bartz TM, Benjamin EJ, Bihlmeyer NA, Bis JC, Bloom HL, Boerwinkle E, Bottinger EB, Brody JA, Calkins H, Campbell A, Cappola TP, Carlquist J, Chasman DI, Chen LY, Chen YI, Choi EK, Choi SH, Christophersen IE, Chung MK, Cole JW, Conen D, Cook J, Crijns HJ, Cutler MJ, Damrauer SM, Daniels BR, Darbar D, Delgado G, Denny JC, Dichgans M, Dorr M, Dudink EA, Dudley SC, Esa N, Esko T, Eskola M, Fatkin D, Felix SB, Ford I, Franco OH, Geelhoed B, Grewal RP, Gudnason V, Guo X, Gupta N, Gustafsson S, Gutmann R, Hamsten A, Harris TB, Hayward C, Heckbert SR, Hernesniemi J, Hocking LJ, Hofman A, Horimoto A, Huang J, Huang PL, Huffman J, Ingelsson E, Ipek EG, Ito K, Jimenez-Conde J, Johnson R, Jukema JW, Kaab S, Kahonen M, Kamatani Y, Kane JP, Kastrati A, Kathiresan S, Katschnig-Winter P, Kavousi M, Kessler T, Kietselaer BL, Kirchhof P, Kleber ME, Knight S, Krieger JE, Kubo M, Launer LJ, Laurikka J, Lehtimaki T, Leineweber K, Lemaitre RN, Li M, Lim HE, Lin HJ, Lin H, Lind L, Lindgren CM, Lokki ML, London B, Loos RJF, Low SK, Lu Y, Lyytikainen LP, Macfarlane PW, Magnusson PK, Mahajan A, Malik R, Mansur AJ, Marcus GM, Margolin L, Margulies KB, Marz W, McManus DD, Melander O, Mohanty S, Montgomery JA, Morley MP, Morris AP, Muller-Nurasyid M, Natale A, Nazarian S, Neumann B, Newton-Cheh C, Niemeijer MN, Nikus K, Nilsson P, Noordam R, Oellers H, Olesen MS, Orho-Melander M, Padmanabhan S, Pak HN, Pare G, Pedersen NL, Pera J, Pereira A, Porteous D, Psaty BM, Pulit SL, Pullinger CR, Rader DJ, Refsgaard L, Ribases M, Ridker PM, Rienstra M, Risch L, Roden DM, Rosand J, Rosenberg MA, Rost N, Rotter JI, Saba S, Sandhu RK, Schnabel RB, Schramm K, Schunkert H, Schurman C, Scott SA, Seppala I, Shaffer C, Shah S, Shalaby AA, Shim J, Shoemaker MB, Siland JE, Sinisalo J, Sinner MF, Slowik A, Smith AV, Smith BH, Smith JG, Smith JD, Smith NL, Soliman EZ, Sotoodehnia N, Stricker BH, Sun A, Sun H, Svendsen JH, Tanaka T, Tanriverdi K, Taylor KD, Teder-Laving M, Teumer A, Theriault S, Trompet S, Tucker NR, Tveit A, Uitterlinden AG, Van Der Harst P, Van Gelder IC, Van Wagoner DR, Verweij N, Vlachopoulou E, Volker U, Wang B, Weeke PE, Weijs B, Weiss R, Weiss S, Wells QS, Wiggins KL, Wong JA, Woo D, Worrall BB, Yang PS, Yao J, Yoneda ZT, Zeller T, Zeng L, Lubitz SA, Lunetta KL, Ellinor PT. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo Y, Chung W, Zhu Z, Shan Z, Li J, Liu S, Liang L. Genome-wide assessment for resting heart rate and shared genetics with cardiometabolic traits and type 2 diabetes. J Am Coll Cardiol 2019;74:2162–2174. [DOI] [PubMed] [Google Scholar]
- 44. Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM; the GIANT Consortium. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res 2018;122:433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr., Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen W-M, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu F-C, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee J-M, Lemmens R, Leys D, Lewis CM, Lin W-Y, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O'Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf SConsortium AF, Cohorts for H, Aging Research in Genomic Epidemiology C, International Genomics of Blood Pressure C, Consortium IStarnet Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson ADBioBank Japan Cooperative Hospital G, Consortium C, Consortium E-C, Consortium EP-I, International Stroke Genetics C, Consortium M, Neurology Working Group of the CC, Network NSG, Study UKYLD, Consortium MSanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr., Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50:524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shah S, Henry A, Roselli C, Lin H, Sveinbjörnsson G, Fatemifar G, Hedman ÅK, Wilk JB, Morley MP, Chaffin MD, Helgadottir A, Verweij N, Dehghan A,, Almgren P, Andersson C, Aragam KG, Ärnlöv J, Backman JD, Biggs ML, Bloom HL, Brandimarto J, Brown MR, Buckbinder L, Carey DJ, Chasman DI, Chen X, Chen X, Chung J, Chutkow W, Cook JP, Delgado GE, Denaxas S, Doney AS, Dörr M,, Dudley SC, Dunn ME, Engström G,, Esko T, Felix SB, Finan C, Ford I, Ghanbari M, Ghasemi S, Giedraitis V, Giulianini F, Gottdiener JS, Gross S, Guðbjartsson DF, Gutmann R, Haggerty CM, van der Harst P, Hyde CL, Ingelsson E, Jukema JW, Kavousi M, Khaw K-T, Kleber ME, Køber L, Koekemoer A, Langenberg C, Lind L, Lindgren CM, London B, Lotta LA, Lovering RC, Luan Ja, Magnusson P, Mahajan A, Margulies KB, März W, Melander O, Mordi IR, Morgan T, Morris AD, Morris AP, Morrison AC, Nagle MW, Nelson CP, Niessner A, Niiranen T, O'Donoghue ML, Owens AT,, Palmer CNA,, Parry HM, Perola M, Portilla-Fernandez E, Psaty BMRegeneron Genetics CRice KM, Ridker PM, Romaine SPR, Rotter JI, Salo P, Salomaa V, van Setten J, Shalaby AA, Smelser DT, Smith NL, Stender S, Stott DJ, Svensson P,, Tammesoo M-L, Taylor KD,, Teder-Laving M, Teumer A,, Thorgeirsson G, Thorsteinsdottir U, Torp-Pedersen C, Trompet S, Tyl B, Uitterlinden AG, Veluchamy A, Völker U, Voors AA, Wang X, Wareham NJ,, Waterworth D, Weeke PE, Weiss R, Wiggins KL, Xing H, Yerges-Armstrong LM, Yu B, Zannad F, Zhao JH, Hemingway H, Samani NJ, McMurray JJV, Yang J, Visscher PM, Newton-Cheh C,, Malarstig A, Holm H,, Lubitz SA, Sattar N, Holmes MV, Cappola TP, Asselbergs FW, Hingorani AD, Kuchenbaecker K, Ellinor PT, Lang CC, Stefansson K, Smith JG, Vasan RS, Swerdlow DI, Lumbers RT. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat Commun 2020;11:163–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ntalla I, Weng LC, Cartwright JH, Hall AW, Sveinbjornsson G, Tucker NR, Choi SH, Chaffin MD, Roselli C, Barnes MR, Mifsud B, Warren HR, Hayward C, Marten J, Cranley JJ, Concas MP, Gasparini P, Boutin T, Kolcic I, Polasek O, Rudan I, Araujo NM, Lima-Costa MF, Ribeiro ALP, Souza RP, Tarazona-Santos E, Giedraitis V, Ingelsson E, Mahajan A, Morris AP, Del Greco MF, Foco L, Gögele M, Hicks AA, Cook JP, Lind L, Lindgren CM, Sundström J, Nelson CP, Riaz MB, Samani NJ, Sinagra G, Ulivi S, Kähönen M, Mishra PP, Mononen N, Nikus K, Caulfield MJ, Dominiczak A, Padmanabhan S, Montasser ME, O'Connell JR, Ryan K, Shuldiner AR, Aeschbacher S, Conen D, Risch L, Thériault S, Hutri-Kähönen N, Lehtimäki T, Lyytikäinen LP, Raitakari OT, Barnes CLK, Campbell H, Joshi PK, Wilson JF, Isaacs A, Kors JA, van Duijn CM, Huang PL, Gudnason V, Harris TB, Launer LJ, Smith AV, Bottinger EP, Loos RJF, Nadkarni GN, Preuss MH, Correa A, Mei H, Wilson J, Meitinger T, Müller-Nurasyid M, Peters A, Waldenberger M, Mangino M, Spector TD, Rienstra M, van de Vegte YJ, van der Harst P, Verweij N, Kääb S, Schramm K, Sinner MF, Strauch K, Cutler MJ, Fatkin D, London B, Olesen M, Roden DM, Benjamin Shoemaker M, Gustav Smith J, Biggs ML, Bis JC, Brody JA, Psaty BM, Rice K, Sotoodehnia N, De Grandi A, Fuchsberger C, Pattaro C, Pramstaller PP, Ford I, Wouter Jukema J, Macfarlane PW, Trompet S, Dörr M, Felix SB, Völker U, Weiss S, Havulinna AS, Jula A, Sääksjärvi K, Salomaa V, Guo X, Heckbert SR, Lin HJ, Rotter JI, Taylor KD, Yao J, de Mutsert R, Maan AC, Mook-Kanamori DO, Noordam R, Cucca F, Ding J, Lakatta EG, Qian Y, Tarasov KV, Levy D, Lin H, Newton-Cheh CH, Lunetta KL, Murray AD, Porteous DJ, Smith BH, Stricker BH, Uitterlinden A, van den Berg ME, Haessler J, Jackson RD, Kooperberg C, Peters U, Reiner AP, Whitsel EA, Alonso A, Arking DE, Boerwinkle E, Ehret GB, Soliman EZ, Avery CL, Gogarten SM, Kerr KF, Laurie CC, Seyerle AA, Stilp A, Assa S, Abdullah Said M, Yldau van der Ende M, Lambiase PD, Orini M, Ramirez J, Van Duijvenboden S, Arnar DO, Gudbjartsson DF, Holm H, Sulem P, Thorleifsson G, Thorolfsdottir RB, Thorsteinsdottir U, Benjamin EJ, Tinker A, Stefansson K, Ellinor PT, Jamshidi Y, Lubitz SA, Munroe PB. Multi-ancestry GWAS of the electrocardiographic PR interval identifies 210 loci underlying cardiac conduction. Nat Commun 2020;11:2542. [DOI] [PMC free article] [PubMed]
- 49. van der Harst P, van Setten J,, Verweij N, Vogler G, Franke L, Maurano MT, Wang X, Mateo Leach I, Eijgelsheim M, Sotoodehnia N, Hayward C, Sorice R, Meirelles O, Lyytikainen LP, Polasek O,, Tanaka T, Arking DE, Ulivi S, Trompet S, Muller-Nurasyid M, Smith AV, Dorr M, Kerr KF, Magnani JW, Del Greco MF, Zhang W, Nolte IM, Silva CT, Padmanabhan S, Tragante V, Esko T, Abecasis GR,, Adriaens ME, Andersen K, Barnett P,, Bis JC, Bodmer R,, Buckley BM, Campbell H, Cannon MV, Chakravarti A, Chen LY, Delitala A, Devereux RB, Doevendans PA, Dominiczak AF, Ferrucci L, Ford I, Gieger C, Harris TB, Haugen E, Heinig M, Hernandez DG, Hillege HL, Hirschhorn JN, Hofman A, Hubner N, Hwang SJ,, Iorio A, Kahonen M, Kellis M, Kolcic I,, Kooner IK, Kooner JS, Kors JA, Lakatta EG, Lage K, Launer LJ, Levy D, Lundby A, Macfarlane PW, May D, Meitinger T, Metspalu A, Nappo S, Naitza S, Neph S, Nord AS, Nutile T, Okin PM, Olsen JV, Oostra BA, Penninger JM, Pennacchio LA, Pers TH, Perz S, Peters A, Pinto YM, Pfeufer A, Pilia MG, Pramstaller PP, Prins BP, Raitakari OT, Raychaudhuri S,, Rice KM, Rossin EJ, Rotter JI, Schafer S, Schlessinger D, Schmidt CO, Sehmi J, Sillje HHW, Sinagra G, Sinner MF, Slowikowski K, Soliman EZ, Spector TD, Spiering W, Stamatoyannopoulos JA, Stolk RP, Strauch K, Tan ST, Tarasov KV, Trinh B, Uitterlinden AG, van den Boogaard M, van Duijn CM, van Gilst WH, Viikari JS, Visscher PM, Vitart V, Volker U, Waldenberger M, Weichenberger CX, Westra HJ,, Wijmenga C, Wolffenbuttel BH, Yang J, Bezzina CR, Munroe PB, Snieder H, Wright AF, Rudan I, Boyer LA,, Asselbergs FW, van Veldhuisen DJ, Stricker BH, Psaty BM, Ciullo M, Sanna S, Lehtimaki T, Wilson JF, Bandinelli S, Alonso A,, Gasparini P, Jukema JW, Kaab S, Gudnason V, Felix SB, Heckbert SR, de Boer RA, Newton-Cheh C, Hicks AA, Chambers JC, Jamshidi Y, Visel A, Christoffels VM, Isaacs A, Samani NJ, de Bakker PIW. 52 genetic loci influencing myocardial mass. J Am Coll Cardiol 2016;68(13):1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 51. Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet 2009;41:876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Norland K, Sveinbjornsson G, Thorolfsdottir RB, Davidsson OB, Tragante V, Rajamani S, Helgadottir A, Gretarsdottir S, van Setten J, Asselbergs FW, Sverrisson JT, Stephensen SS, Oskarsson G, Sigurdsson EL, Andersen K, Danielsen R, Thorgeirsson G, Thorsteinsdottir U, Arnar DO, Sulem P, Holm H, Gudbjartsson DF, Stefansson K. Sequence variants with large effects on cardiac electrophysiology and disease. Nat Commun 2019;10:4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Müller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WHL, Köttgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BHC, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Müller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kääb S, Witteman JCM, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of PR interval. Nat Genet 2010;42:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet 2010;42:117–122. [DOI] [PubMed] [Google Scholar]
- 55.Human Genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aalen OO, Johansen S. An empirical transition matrix for nonhomogeneous Markov-chains based on censored observations. Scand Stat Theory Appl 1978;5:141–150. [Google Scholar]
- 57. Semelka M, Gera J, Usman S. Sick sinus syndrome: a review. Am Fam Physician 2013;87:691–696. [PubMed] [Google Scholar]
- 58. Ku NO, Gish R, Wright TL, Omary MB. Keratin 8 mutations in patients with cryptogenic liver disease. N Engl J Med 2001;344:1580–1587. [DOI] [PubMed] [Google Scholar]
- 59. Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D'Agostino RB Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiríksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WHL, Agarwal SK, Stricker BHC, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Köttgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann H-E, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kääb S, Ellinor PT, Witteman JCM. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nielsen JB, Fritsche LG, Zhou W, Teslovich TM, Holmen OL, Gustafsson S, Gabrielsen ME, Schmidt EM, Beaumont R, Wolford BN, Lin M, Brummett CM, Preuss MH, Refsgaard L, Bottinger EP, Graham SE, Surakka I, Chu Y, Skogholt AH, Dalen H, Boyle AP, Oral H, Herron TJ, Kitzman J, Jalife J, Svendsen JH, Olesen MS, Njølstad I, Løchen M-L, Baras A, Gottesman O, Marcketta A, O’Dushlaine C, Ritchie MD, Wilsgaard T, Loos RJF, Frayling TM, Boehnke M, Ingelsson E, Carey DJ, Dewey FE, Kang HM, Abecasis GR, Hveem K, Willer CJ. Genome-wide study of atrial fibrillation identifies seven risk loci and highlights biological pathways and regulatory elements involved in cardiac development. Am J Hum Genet 2018;102:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, Ramirez AH, Mosley JD, Pulley JM, Basford MA, Bradford Y, Rasmussen LV, Pathak J, Chute CG, Kullo IJ, McCarty CA, Chisholm RL, Kho AN, Carlson CS, Larson EB, Jarvik GP, Sotoodehnia N, Manolio TA, Li R, Masys DR, Haines JL, Roden DM. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 2013;127:1377–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bjornsson T, Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Norddahl GL, Helgadottir A, Gretarsdottir S, Magnusdottir A, Danielsen R, Sigurdsson EL, Adalsteinsdottir B, Gunnarsson SI, Jonsdottir I, Arnar DO, Helgason H, Gudbjartsson T, Gudbjartsson DF, Thorsteinsdottir U, Holm H, Stefansson K. A rare missense mutation in MYH6 associates with non-syndromic coarctation of the aorta. Eur Heart J 2018;39:3243–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Helgadottir A, Thorleifsson G, Gretarsdottir S, Stefansson OA, Tragante V, Thorolfsdottir RB, Jonsdottir I, Bjornsson T, Steinthorsdottir V, Verweij N, Nielsen JB, Zhou W, Folkersen L, Martinsson A, Heydarpour M, Prakash S, Oskarsson G, Gudbjartsson T, Geirsson A, Olafsson I, Sigurdsson EL, Almgren P, Melander O, Franco-Cereceda A, Hamsten A, Fritsche L, Lin M, Yang B, Hornsby W, Guo D, Brummett CM, Abecasis G, Mathis M, Milewicz D, Body SC, Eriksson P, Willer CJ, Hveem K, Newton-Cheh C, Smith JG, Danielsen R, Thorgeirsson G, Thorsteinsdottir U, Gudbjartsson DF, Holm H, Stefansson K. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9:987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramŕez J, Duijvenboden SV, Ntalla I, Mifsud B, Warren HR, Tzanis E, Orini M, Tinker A, Lambiase PD, Munroe PB. Thirty loci identified for heart rate response to exercise and recovery implicate autonomic nervous system. Nat Commun 2018;9:1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Verweij N, van de Vegte YJ, van der Harst P. Genetic study links components of the autonomous nervous system to heart-rate profile during exercise. Nat Commun 2018;9:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rosenberg MA, Patton KK, Sotoodehnia N, Karas MG, Kizer JR, Zimetbaum PJ, Chang JD, Siscovick D, Gottdiener JS, Kronmal RA, Heckbert SR, Mukamal KJ. The impact of height on the risk of atrial fibrillation: the Cardiovascular Health Study. Eur Heart J 2012;33:2709–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidt M, Botker HE, Pedersen L, Sorensen HT. Adult height and risk of ischemic heart disease, atrial fibrillation, stroke, venous thromboembolism, and premature death: a population based 36-year follow-up study. Eur J Epidemiol 2014;29:111–118. [DOI] [PubMed] [Google Scholar]
- 68. Levin M, Judy R, Gill D, Vujkovic M, Hyman M, Nazarian S, Rader D, Voight B, Damrauer S. Genetics of height and risk of atrial fibrillation: a Mendelian randomization study. PLoS Medicine 2019;17:e1003288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation 2007;115:1921–1932. [DOI] [PubMed] [Google Scholar]
- 70. Pan X, Hobbs RP, Coulombe PA. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr Opin Cell Biol 2013;25:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol 2002;14:110–122. [DOI] [PubMed] [Google Scholar]
- 72. Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol 2008;129:705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. O'Neill A, Williams MW, Resneck WG, Milner DJ, Capetanaki Y, Bloch RJ. Sarcolemmal organization in skeletal muscle lacking desmin: evidence for cytokeratins associated with the membrane skeleton at costameres. Mol Biol Cell 2002;13:2347–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ursitti JA, Lee PC, Resneck WG, McNally MM, Bowman AL, O'Neill A, Stone MR, Bloch RJ. Cloning and characterization of cytokeratins 8 and 19 in adult rat striated muscle. Interaction with the dystrophin glycoprotein complex. J Biol Chem 2004;279:41830–41838. [DOI] [PubMed] [Google Scholar]
- 75. Stone MR, O'Neill A, Catino D, Bloch RJ. Specific interaction of the actin-binding domain of dystrophin with intermediate filaments containing keratin 19. Mol Biol Cell 2005;16:4280–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Papathanasiou S, Rickelt S, Soriano ME, Schips TG, Maier HJ, Davos CH, Varela A, Kaklamanis L, Mann DL, Capetanaki Y. Tumor necrosis factor-alpha confers cardioprotection through ectopic expression of keratins K8 and K18. Nat Med 2015;21:1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Owens DW, Wilson NJ, Hill AJ, Rugg EL, Porter RM, Hutcheson AM, Quinlan RA, van Heel D, Parkes M, Jewell DP, Campbell SS, Ghosh S, Satsangi J, Lane EB. Human keratin 8 mutations that disturb filament assembly observed in inflammatory bowel disease patients. J Cell Sci 2004;117:1989–1999. [DOI] [PubMed] [Google Scholar]
- 78. Ku NO, Omary MB. A disease- and phosphorylation-related nonmechanical function for keratin 8. J Cell Biol 2006;174:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005;15:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. England J, Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cell Mol Life Sci 2013;70:1221–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Granzier HL, Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res 2004;94:284–295. [DOI] [PubMed] [Google Scholar]
- 82. Gudbjartsson DF, Holm H, Sulem P, Masson G, Oddsson A, Magnusson OT, Saemundsdottir J, Helgadottir HT, Helgason H, Johannsdottir H, Gretarsdottir S, Gudjonsson SA, Njolstad I, Lochen ML, Baum L, Ma RC, Sigfusson G, Kong A, Thorgeirsson G, Sverrisson JT, Thorsteinsdottir U, Stefansson K, Arnar DO. A frameshift deletion in the sarcomere gene MYL4 causes early-onset familial atrial fibrillation. Eur Heart J 2017;38:27–34. [DOI] [PubMed] [Google Scholar]
- 83. Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Nielsen JB, Jonsson S, Halldorsson GH, Melsted P, Ivarsdottir EV, Davidsson OB, Kristjansson RP, Thorleifsson G, Helgadottir A, Gretarsdottir S, Norddahl G, Rajamani S, Torfason B, Valgardsson AS, Sverrisson JT, Tragante V, Holmen OL, Asselbergs FW, Roden DM, Darbar D, Pedersen TR, Sabatine MS, Willer CJ, Lochen ML, Halldorsson BV, Jonsdottir I, Hveem K, Arnar DO, Thorsteinsdottir U, Gudbjartsson DF, Holm H, Stefansson K. Coding variants in RPL3L and MYZAP increase risk of atrial fibrillation. Commun Biol 2018;1:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res 2000;86:386–390. [DOI] [PubMed] [Google Scholar]
- 85. James J, Martin L, Krenz M, Quatman C, Jones F, Klevitsky R, Gulick J, Robbins J. Forced expression of alpha-myosin heavy chain in the rabbit ventricle results in cardioprotection under cardiomyopathic conditions. Circulation 2005;111:2339–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitás K, Labeit S, Granzier H. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res 2000;86:59–67. [DOI] [PubMed] [Google Scholar]
- 87. Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation 2004;110:155–162. [DOI] [PubMed] [Google Scholar]
- 88. Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 1998;94:319–324. [DOI] [PubMed] [Google Scholar]
- 89. Berry FB, Miura Y, Mihara K, Kaspar P, Sakata N, Hashimoto-Tamaoki T, Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J Biol Chem 2001;276:25057–25065. [DOI] [PubMed] [Google Scholar]
- 90. Chen X, Yu L, Shi S, Jiang H, Huang C, Desai M, Li Y, Barajas-Martinez H, Hu D. Neuronal Nav1.8 channels as a novel therapeutic target of acute atrial fibrillation prevention. J Am Heart Assoc 2016;5:e004050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Qi B, Wei Y, Chen S, Zhou G, Li H, Xu J, Ding Y, Lu X, Zhao L, Zhang F, Chen G, Zhao J, Liu S. Nav1.8 channels in ganglionated plexi modulate atrial fibrillation inducibility. Cardiovasc Res 2014;102:480–486. [DOI] [PubMed] [Google Scholar]
- 92. Jabbari J, Olesen MS, Yuan L, Nielsen JB, Liang B, Macri V, Christophersen IE, Nielsen N, Sajadieh A, Ellinor PT, Grunnet M, Haunso S, Holst AG, Svendsen JH, Jespersen T; on behalf of LuCamp. Common and rare variants in SCN10A modulate the risk of atrial fibrillation. Circ Cardiovasc Genet 2015;8:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Katritsis DG, Pokushalov E, Romanov A, Giazitzoglou E, Siontis GC, Po SS, Camm AJ, Ioannidis JP. Autonomic denervation added to pulmonary vein isolation for paroxysmal atrial fibrillation: a randomized clinical trial. J Am Coll Cardiol 2013;62:2318–2325. [DOI] [PubMed] [Google Scholar]
- 94. Nakagawa H, Scherlag BJ, Patterson E, Ikeda A, Lockwood D, Jackman WM. Pathophysiologic basis of autonomic ganglionated plexus ablation in patients with atrial fibrillation. Heart Rhythm 2009;6:S26–34. [DOI] [PubMed] [Google Scholar]
- 95. Pokushalov E, Romanov A, Katritsis DG, Artyomenko S, Shirokova N, Karaskov A, Mittal S, Steinberg JS. Ganglionated plexus ablation vs linear ablation in patients undergoing pulmonary vein isolation for persistent/long-standing persistent atrial fibrillation: a randomized comparison. Heart Rhythm 2013;10:1280–1286. [DOI] [PubMed] [Google Scholar]
- 96. Buckley U, Rajendran PS, Shivkumar K. Ganglionated plexus ablation for atrial fibrillation: just because we can, does that mean we should? Heart Rhythm 2017;14:133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lo LW, Scherlag BJ, Chang HY, Lin YJ, Chen SA, Po SS. Paradoxical long-term proarrhythmic effects after ablating the "head station" ganglionated plexi of the vagal innervation to the heart. Heart Rhythm 2013;10:751–757. [DOI] [PubMed] [Google Scholar]
- 98. Osman F, Kundu S, Tuan J, Jeilan M, Stafford PJ, Ng GA, Ng GA. Ganglionic plexus ablation during pulmonary vein isolation—predisposing to ventricular arrhythmias? Indian Pacing Electrophysiol J 2010;10:104–107. [PMC free article] [PubMed] [Google Scholar]
- 99. Driessen AHG, Berger WR, Krul SPJ, van den Berg NWE, Neefs J, Piersma FR, Chan Pin Yin D, de Jong J, van Boven WP, de Groot JR. Ganglion plexus ablation in advanced atrial fibrillation: the AFACT study. J Am Coll Cardiol 2016;68:1155–1165. [DOI] [PubMed] [Google Scholar]
- 100. Elvan A, Wylie K, Zipes DP. Pacing-induced chronic atrial fibrillation impairs sinus node function in dogs. Electrophysiological remodeling. Circulation 1996;94:2953–2960. [DOI] [PubMed] [Google Scholar]
- 101. Mesubi OO, Anderson ME. Atrial remodelling in atrial fibrillation: CaMKII as a nodal proarrhythmic signal. Cardiovasc Res 2016;109:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Icelandic population WGS data has been deposited at the European Variant Archive under accession code PRJEB15197. We declare that the data supporting the findings of this study are available within the article, its Supplementary material online and upon reasonable request. The genome-wide association scan summary data will be made available at http://www.decode.com/summarydata.