Abstract
Gene therapy for curing congenital human diseases is promising, but the feasibility and safety need to be further evaluated. In this study, based on a pig model that carries the c.740T>C (L247S) mutation in MITF with an inheritance pattern and clinical pathology that mimics Waardenburg syndrome 2A (WS2A), we corrected the point mutation by the CRISPR-Cas9 system in the mutant fibroblast cells using single-stranded oligodeoxynucleotide (ssODN) and long donor plasmid DNA as the repair template. By using long donor DNA, precise correction of this point mutation was achieved. The corrected cells were then used as the donor cell for somatic cell nuclear transfer (SCNT) to produce piglets, which exhibited a successfully rescued phenotype of WS2A, including anophthalmia and hearing loss. Furthermore, engineered base editors (BEs) were exploited to make the correction in mutant porcine fibroblast cells and early embryos. The correction efficiency was greatly improved, whereas substantial off-targeting mutations were detected, raising a safety concern for their potential applications in gene therapy. Thus, we explored the possibility of precise correction of WS2A-causing gene mutation by the CRISPR-Cas9 system in a large-animal model, suggesting great prospects for its future applications in treating human genetic diseases.
Keywords: CRISPR/Cas9, base editor, MITF, gene correction, anophthalmia, hearing loss
Graphical abstract
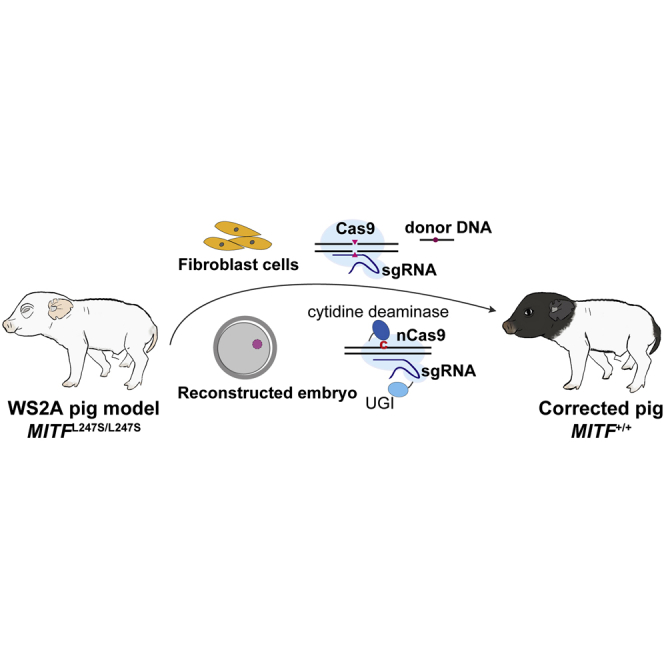
Using a pig model of Waardenburg syndrome 2A, Zhao et al. successfully corrected the causative mutation through genome editing in fibroblast cells and created pigs rescued from the disease, including anophthalmia and hearing loss. Furthermore, precise correction in porcine early embryos provided proof-of-concept evidence for future applications in gene therapy.
Introduction
Gene therapy, a technique to treat or cure diseases by modifying a patient’s genes through replacement or inactivation of disease-causing genes or introduction of modified genes, offers new options to multiple fields of medicine. Several gene therapy products have been approved by the US Food and Drug Administration (FDA), such as LUXTURNATM (voretigene neparvovec-rzyl) from Spark Therapeutics and ZOLGENSMA® (onasemnogene abeparvovec-xioi) from AveXis, highlighting the great advances in treating monogenetic diseases. However, most of the current gene therapy trials in clinics are achieved by using a gene augmentation strategy. Precise gene modifications, which are essential for correcting the pathogenic mutations in most recessive genetic diseases, are still under development, and nuclease-mediated genome editing tools, especially the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) protein 9 (CRISPR-Cas9) system, provide great potential for therapeutic genome editing by mediating accurate gene correction, either ex vivo or in vivo.
Waardenburg syndrome (WS), a dominant inherited auditory-pigmentary syndrome that is characterized by hearing loss and hypopigmentation,1 affects about 1 per 42,000 of the population2 and is classified into four types based on clinical characteristics.3 WS type 2 (WS2) is characterized by sensorineural hearing loss, pigmentary abnormalities in skin, hair, and eyes,1 and no critical vision deficits.4 WS2 cases that contain mutations in the MITF (microphthalmia-associated transcription factor) gene are classified as WS2A.5, 6, 7 Several mouse models of WS2A have been created but only partially present the clinical features of WS2A patients with a recessive or semidominant inheritance pattern.7,8 Due to the high similarities of pigs and humans in physiology, anatomy, and metabolism, pigs serve as an ideal animal for human disease modeling. We previously created a miniature pig model of WS2A using N-ethyl-N-nitrosourea mutagenesis, in which the heterozygous mutants (MITF c.740T>C mutation) resembled the pathology of WS2A and the homozygous mutants exhibited additional anophthalmia,9 which was similarly observed as microphthalmia in patients with biallelic mutations in MITF.10 This dominant MITF c.740T>C mutation leads to WS2A through haploinsufficiency,9 making it impossible to correct the phenotype by disrupting the dominant negative mutant alleles, which was possible to use in the treatment of some types of autosomal dominant hearing loss.11 Considering the complications and immunotoxicity that may arise from the overexpression or ectopic expression of the transgene,12 the repeated therapeutic administration that is often required, and other causes of pathology in addition to haploinsufficiency,13 gene augmentation may not be appropriate for treating diseases that are inherited in an autosomal recessive pattern and dominant pattern due to loss-of-function mutations such as WS2A. Thus, precise correction of the pathogenic mutation is a potential strategy for one-time treatment that would achieve a permanent cure for the disease.
CRISPR-Cas9 has been widely used in genome editing because of its high efficiency and simple design ever since the introduction of CRISPR-Cas9 for multiplexing genome targeting in human and mouse cells.14 Guided by a small guide RNA (sgRNA), the Cas9 nuclease introduces a double-strand break (DSB) at a specific site of the target DNA sequence. This process induces cellular DSB repair pathways, including non-homologous end-joining (NHEJ) that may result in small insertions or deletions (indels) and high-fidelity homology-directed repair (HDR) in the presence of donor DNA fragments.14, 15, 16 CRISPR-Cas9-induced HDR provides an efficient and precise strategy for treating hereditary diseases since most genetic diseases arise from point mutations.17,18 Furthermore, recently developed cytosine base editors and adenine base editors19, 20, 21, 22 provide additional advantages for correction of pathogenic point mutations in genetic diseases without the need for introducing a DSB or a repair template. However, the feasibility and safety for HDR and base editor-mediated gene therapy still need to be further evaluated, especially in large animals, which could be informative for future preclinical applications.
In the current study, based on the WS2A pig model, we performed precise gene correction with CRISPR-Cas9-mediated HDR using single-stranded oligodeoxynucleotide (ssODN) and plasmid DNA with long homology arms as donor DNAs. We successfully rescued the anophthalmia and hearing loss phenotype, which indicates potential applications for gene therapy of autosomal recessive disorders and diseases caused by dominant mutations due to haploinsufficiency. Further attempts were made to achieve precise point mutation correction using an engineered base editor (BE) system in porcine fibroblast cells and early embryos, but results raised safety concerns regarding its application for future gene therapies. Herein, the successful correction of genetic defects in porcine cells and embryos in vitro and rescue of the disease phenotypes in pigs provide proof of concept for the potential of precise gene therapy in treating genetic diseases.
Results
MITF c.740T>C mutation was monoallelically corrected through ssODN strategy
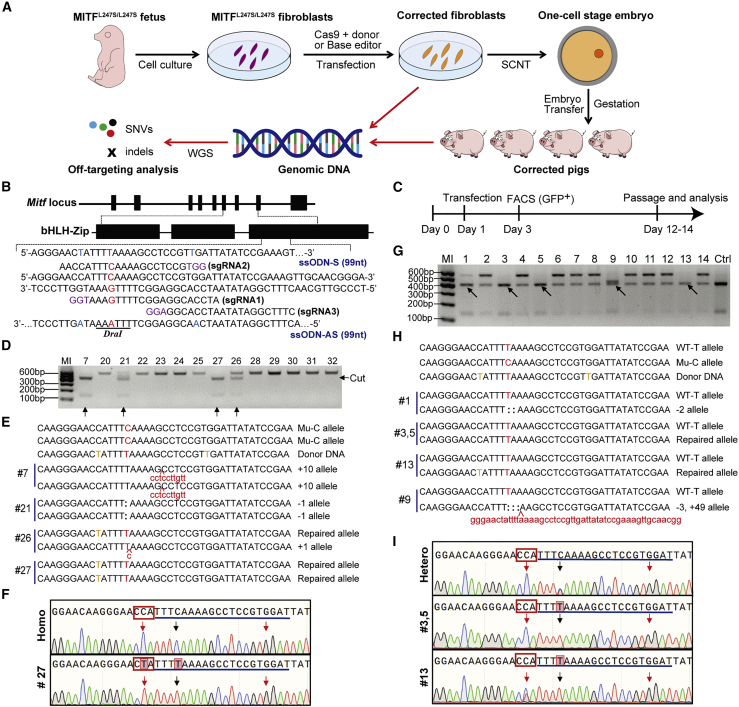
To evaluate the efficiency and safety of nuclease-mediated gene therapy for point mutation-caused diseases, we used the CRISPR-Cas9 system and BEs to precisely correct the causative mutation in a WS2A pig model and then evaluated the off-targeting editing by whole-genome sequencing (WGS) (Figure 1A). We first exploited ssODNs as repair templates to correct the c.740T>C mutation in the porcine MITF gene. We designed three sgRNAs that targeted the mutation site on exon 8 of the MITF gene and two 99-nt ssODN repair templates (sense and antisense) (Figure 1B). The sgRNA-Cas9 expression vector was co-transfected with each of the ssODNs and the pCAG-GFP plasmid (when using PX330) in MITFL247S/L247S porcine fetal fibroblast cells (PFFCs), and GFP-positive cells were collected by fluorescence-activated cell sorting (FACS) for genotyping analysis (Figure 1C). As shown in Figure S1, the correction efficiency with sgRNA1 combined with antisense ssODN (AS) or sgRNA2 combined with sense ssODN (S) was slightly higher than other groups via Hi-TOM analysis, a platform for high-throughput tracking of mutations.23 Therefore, the sgRNA1 and the antisense ssODN (1AS) strategy was exploited in subsequent experiments for the screening of single-cell colonies. A total of 33 colonies were obtained, and genotyping results showed that 1 colony (colony 27) was precisely corrected at the c.740 site (Figures 1D–1F) through genotyping by short-range PCR, which resulted in a 546-bp amplicon. Furthermore, a similar strategy was conducted in MITFL247S/+ PFFCs, and results showed that 3 of the 18 colonies (16.67%) were precisely corrected (colonies 3, 5, and 13, Figures 1G–1I).
Figure 1.
CRISPR-Cas9 mediated MITF c.740C > T repair in porcine fibroblast cells using ssODN as the repair template
(A) Experimental design for the MITF point-mutation repair in the current study. (B) Genome structure of the porcine MITF gene, the sequences of the sgRNAs targeting the c.740 site, and the ssODNs used as HDR templates. The protospacer-adjacent motif (PAM) sequences of the sgRNAs are shown in purple, the intended point mutation is shown in red, and the introduced blocking mutations are shown in blue in the ssODN sequence. (C) Experimental procedure for screening single-cell colonies. (D) Genotyping of single-cell colonies by PCR and DraI digestion that originated from MITFL247S/L247S fibroblasts. The arrows indicate potentially corrected colonies. (E) Sanger sequencing of the colonies cut by DraI. The T base intended to be repaired is shown in red, the blocking mutations in donor DNA are shown in orange, the deleted bases are indicated by colons, and the inserted bases are shown by lowercase red text. (F) Sequencing diagram of the PCR product from colony 27 (#27) and the MITFL247S/L247S fibroblast cells (Homo). The sgRNA sequences are underlined, the PAM sequences of the sgRNA1 are shown in red rectangles, and the repaired base and the introduced blocking mutation are indicated by black and red arrows, respectively. (G) Genotyping of single-cell colonies by PCR and DraI digestion originate from MITFL247S/+ fibroblasts. The arrows indicate the potentially corrected colonies. (H) Sanger sequencing of the colonies cut by DraI. The T base intended to be repaired is shown in red, the blocking mutations in donor DNA are shown in orange, the deleted bases are indicated by colons, and the inserted bases are shown by lowercase red text. (I) Sequencing diagram of the PCR product from colony 3, 5, and 13 and the MITFL247S/+ fibroblast cells (Hetero). The PAM sequences of the sgRNA1 are shown in red rectangle, and the repaired base and the introduced blocking mutation are indicated by black and red arrows, respectively.
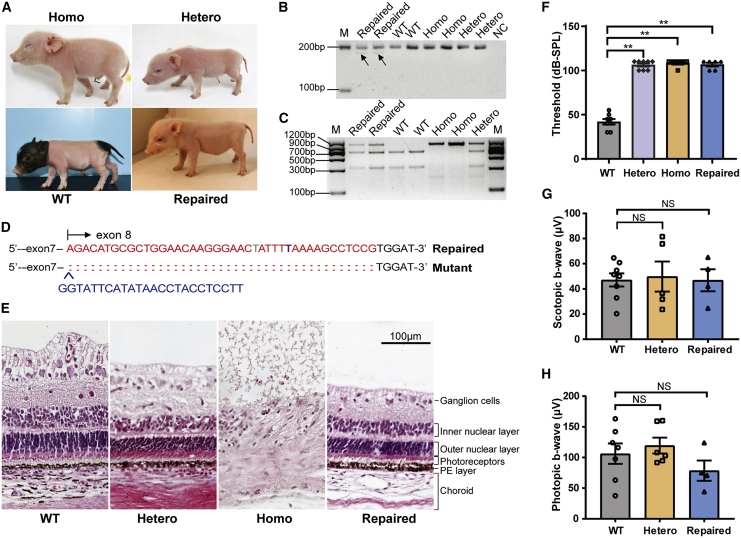
In order to assess the developmental capacity of the corrected cells and phenotype rescue after gene correction, somatic cell nuclear transfer (SCNT) was performed by using colony 27, which was corrected from MITFL247S/L247S PFFCs, as the donor to generate MITF c.740T>C mutation-corrected pigs. A total of 2,380 embryos were transferred to nine surrogates, and three pregnancies were established and developed to term (Table S1). Nine liveborn and three stillborn piglets were obtained in three pregnancies. These piglets were healthy and appeared to have normally developed eyes, which were significantly different from the eyes of MITFL247S/L247S pigs (anophthalmia) (Figure 2A). Unexpectedly, the coat color of all of these piglets was white (Figure 2A), which is similar to the coat color of MITFL247S/+ pigs, but different from that of the desired homozygous corrected pigs, which was expected to be the same as the wild-type (WT) pigs.9 Therefore, we further confirmed the genotypes of these pigs at both the genomic DNA and mRNA level. Unfortunately, a short band was detected in the MITF mRNA (Figure 2B), which could not be cleaved by DraI (Figure 2C). Genomic DNA sequencing of the longer fragment showed a 602-bp deletion at the intron 7 and exon 8 junction of the MITF gene in one allele (41-bp deletion and 24-bp insertion in mRNA) (Figure 2D), resulting in the prematurely truncated MITF open reading frame (ORF) and disruption of the helix-loop-helix domain. This explained why the pigs exhibited a similar phenotype to that of the MITFL247S/+ pigs. Thus, using the ssODN strategy, we successfully corrected the MITF c.740T>C mutation monoallelically while unexpectedly introducing a long fragment deletion in one allele.
Figure 2.
Generation of MITF c.740C>T repaired pigs and phenotype analysis
(A) Photographs of pigs with different genotypes are shown. Homo represents the MITFL247S/L247S pig, Hetero represents the MITFL247S/+ pig, WT represents the wild-type (MITF+/+) pig, and Repaired represents the MITF c.740C>T repaired pig. (B) A short band (indicated by arrows) was detected in mRNA from colony 27-derived pigs using primers MITF-M2F and MITF-M2R. M represents the DNA marker. (C) RT-PCR and DraI digestion were used to genotype mRNAs obtained from the ear tissues of colony 27-derived pigs. (D) Representative sequence of the mutant allele in MITF mRNA. The repaired allele is shown above, which included an introducing mutation shown in green and a repaired base shown in purple bold. The deleted bases are indicated by colons, and the inserted bases are shown by blue uppercase text. (E) H&E staining of the eye sections in different groups of pigs. Scale bar, 100 μm. (F) Comparison of the ABR thresholds in piglets with different genotypes at 2 weeks of age; the numbers of pigs for the WT, hetero, homo, and repaired groups are 4, 4, 3 and 3, respectively. The ABR thresholds were recorded in both ears and are presented as mean ± SEM. ∗∗p < 0.01. (G and H) In different groups of pigs, scotopic and photopic ERGs were performed and are shown as average b-wave amplitudes. Analysis included both eyes of 4 WT, 3 hetero, and 2 MITF-repaired pigs. Data are presented as mean ± SEM. Results of the homo group are not shown in (G) and (H) since their eyes were severely hypoplasia (anophthalmia) and cannot be subjected to the ERG experiment. NS, not significant (p > 0.05).
Eye development, but not the hearing, of colony 27-derived pigs was successfully repaired
A dominant mutation in MITF causes WS2A, resulting in clinical symptoms characterized by sensorineural hearing loss and hypopigmentation. In the repaired pigs, while white coat color was observed (Figure 2A), hearing loss was confirmed by auditory brainstem response (ABR) analysis (Figure 2F). To confirm that the eyes were normal and functional, electroretinography (ERG) was performed on these pigs at 3 and 5 months of age. As Figures 2G and 2H show, the retinal function of the repaired pigs was normal for both the scotopic and photopic response as compared with that of the WT and MITFL247S/+ pigs. Hematoxylin and eosin (H&E) staining of eye sections showed that the retina structure of the repaired pigs was complete and similar to those of WT and MITFL247S/+ pigs (Figure 2E). Taken together, our results demonstrate that the MITFL247S/L247S pigs were successfully rescued from anophthalmia using a strategy combining CRISPR-Cas9 with ssODNs.
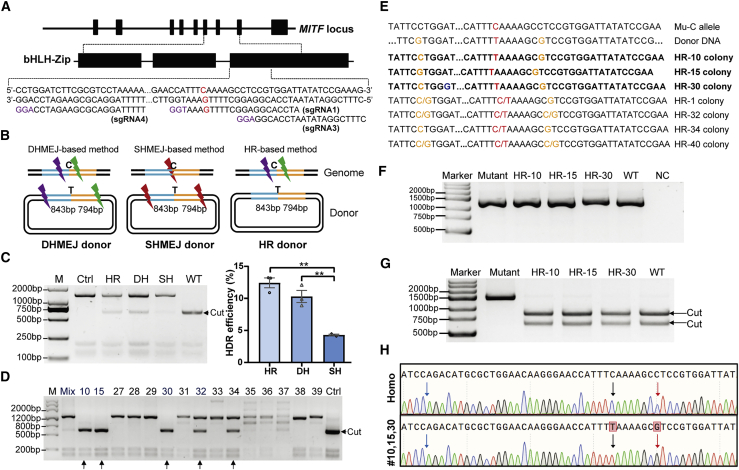
The MITF c.740T>C mutation was precisely corrected in MITFL247S/L247S porcine fibroblast cells through a long donor DNA strategy
Unexpected long-fragment deletion occurred when using ssODNs as the repair template in the correction of the MITF c.740T>C mutation as described above. Furthermore, longer donor DNAs were designed for precise point mutation correction, as previously described for successful gene correction and knockin by TALENs and CRISPR-Cas9 in human cells, rats, and mice.24, 25, 26 Three different schemes were designed, with or without sgRNA sites within the donor DNA to correct the mutation (Figures 3A and 3B): (1) the double homology-mediated end joining (DHMEJ)-based donor contains two different sgRNA target sites flanked by the mutant site and long homology arms, (2) the single homology-mediated end joining (SHMEJ)-based donor contains the same sgRNA target site that harbors the mutant site on each side and long homology arms, and (3) the homologous recombination (HR)-based donor contains only the long homology arms. Different sgRNAs and donor DNA plasmids were co-transfected into MITFL247S/L247S porcine fibroblast cells, and the efficiency of the point mutation correction was evaluated in mixed transfected cells via Hi-TOM analysis. As Figure 3C shows, the correction efficiency in the DHMEJ and HR donor groups was significantly higher than that in the SHMEJ donor group. Considering the potential integration of sgRNA-cleaved donor DNA into the porcine genome for the DHMEJ donor group, we decided to use the HR donor for subsequent screening of corrected single colonies. A total of 75 colonies were obtained, and 3 (HR-10, HR-15, and HR-30) of them were biallelically corrected and 4 (HR-1, HR-32, HR-34, and HR-40) were monoallelically corrected (Figures 3D and 3E). Genotyping of MITF mRNA further confirmed that the three colonies were precisely repaired biallelically, without incorporating unintentional mutations (Figures 3F–3H). These colonies were cryopreserved for the subsequent generation of the MITF c.740T>C mutation-corrected pigs.
Figure 3.
CRISPR-Cas9-mediated MITF point mutation repair in MITFL247S/L247S porcine fibroblast cells using long donor plasmids as the repair templates
(A) Design of different sgRNAs around exon 8 of the porcine MITF gene for the long donor DNA strategy. The PAM sequences of sgRNAs are shown in purple, and the intended point mutation is shown in red. (B) Design of donor DNA and sgRNAs for genome editing. The homology arms are shown in blue and orange lines around the intended T locus, and the different sgRNAs are shown as lightning bolts. The purple lightning bolt represents sgRNA4, the green lightning bolt represents sgRNA3, and the red lightning bolt represents sgRNA1. (C) Analysis of the HDR efficiency for the three different schemes. A representative restriction fragment length polymorphism (RFLP) image of transfected cells is shown on the left, and the Hi-TOM quantitative results of three independent replicates are presented as mean ± SEM (n = 3) on the right. ∗∗p < 0.01. HR, DH, and SH represent HR, DHMEJ, and SHMEJ, respectively. (D) Genotyping of single-cell colonies by PCR and DraI digestion. The arrows indicate the potential repaired colonies. (E) Sanger sequencing of the colonies that were precisely repaired at the intended locus. The biallelically corrected colonies are shown in bold. The T base intended to be repaired is shown in red, the blocking mutations in the donor DNA are shown in orange, and the omitted bases are shown by ellipses. (F and G) Total RNAs were extracted from these colonies and amplified using RT-PCR (F) and then digested with DraI (G). Arrows indicate cleavage. (H) Sequencing diagram of the RT-PCR product of colony HR-10, HR-15, and HR-30 and the MITFL247S/L247S fibroblast cells (Homo). The intended corrected base and the introduced blocking mutation are indicated by arrows in black and red, respectively. The blue arrow indicates the junction of exon 7 and exon 8 of MITF.
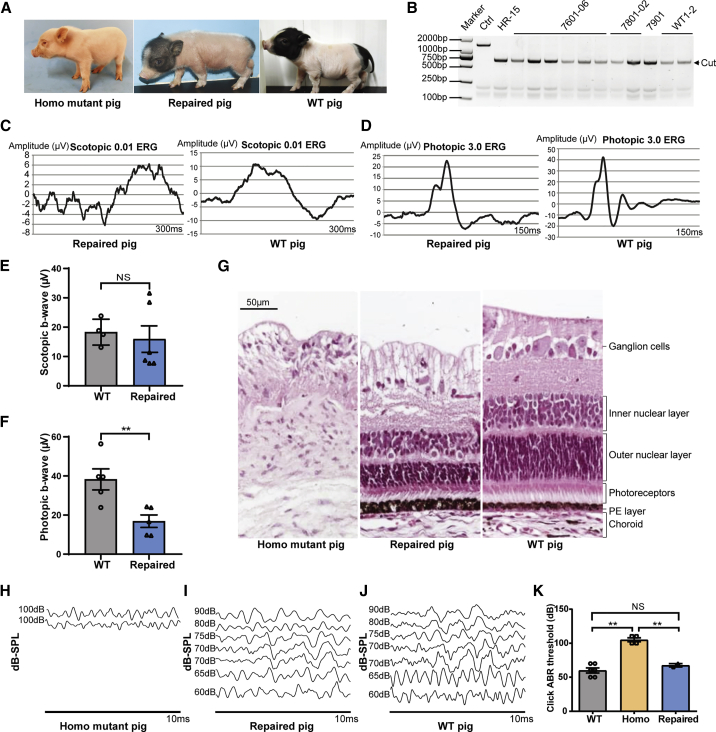
Fully repaired pigs were generated using colony HR-15
In order to assess whether the phenotype could be fully rescued after gene correction by CRISPR-Cas9 combined with long donor DNA, colony HR-15 was used for SCNT to produce pigs. A total of 1,645 embryos were transferred to six surrogates, and three pregnancies were established and developed to term (Table S2). Six liveborn and three stillborn piglets were obtained. All of the piglets had a normal appearance, including the “liang-tou-wu” coat color (black coat on the head and bottom and white coat on the body),27 and the morphology of eyes similar to that of WT pigs (Figure 4A). Genotyping results showed that they were all derived from colony HR-15 (Figure 4B). ERG analysis and H&E staining of eye sections showed that the repaired eyes were functional and intact (Figures 4C–4G). The lower photopic b-wave amplitude in the repaired piglets (Figure 4F) might be due to the cloning process that often led to abnormal development in cloned animals.28,29 Significantly lower birth weight was also observed in the repaired pigs (617.67 ± 27.06 g for the WT piglets versus 194.07 ± 11.78 g for the repaired piglets). The ABR test showed that the repaired pigs had normal hearing, which was comparable to that of WT piglets (Figures 4H–4K). Taken together, colony HR-15-derived pigs were fully rescued from anophthalmia, hearing loss, and an abnormal white coat color caused by the c.740T>C mutation in MITF.
Figure 4.
Generation of fully corrected pigs and analysis of their phenotypes
(A) Representative photographs of piglets with different genotypes. Homo mutant pig represents the MITFL247S/L247S pig. (B) Genotyping of genomic DNA obtained from ear tissue of the HR-15 colony-derived piglets by PCR and DraI digestion. Ctrl represent the MITFL247S/L247S fibroblast cells, and 7601-06, 7801-02, and 7901 represent the repaired pigs. WT1–2 represents two WT pigs. The arrow indicates the cut band with the repaired sequence. (C and D) Retina function analysis in repaired pigs. Scotopic (rod, C) and photopic (cone, D) full-field ERGs were recorded in newborn piglets; representative scotopic and photopic records are shown for the repaired pigs and WT pigs. (E and F) The b-wave amplitudes of scotopic (E) and photopic (F) full-field ERGs were plotted for both eyes of the repaired (n = 3) and WT pigs (n = 3). Data are presented as mean ± SEM. ∗∗p < 0.01. NS, not significant (p > 0.05). Results of the homo group are not shown in (C–F) since their eyes were severely hypoplasia (anophthalmia) and could not be subjected to the ERG experiment. (G) Representative H&E staining image of the retina of newborn piglets with different genotypes. Scale bar, 50 μm. (H–J) Representative images of the ABR test in newborn piglets with different genotypes. (K) Comparison of the ABR threshold in newborn piglets with different genotypes; the numbers of pigs for WT, homozygous mutant, and the repaired group are 3, 2 and 1, respectively. The ABR thresholds were recorded in both ears. Data are presented as mean ± SEM. ∗∗p < 0.01.
Effective MITF c.740C>T repair in MITFL247S/L247S PFFCs by BE
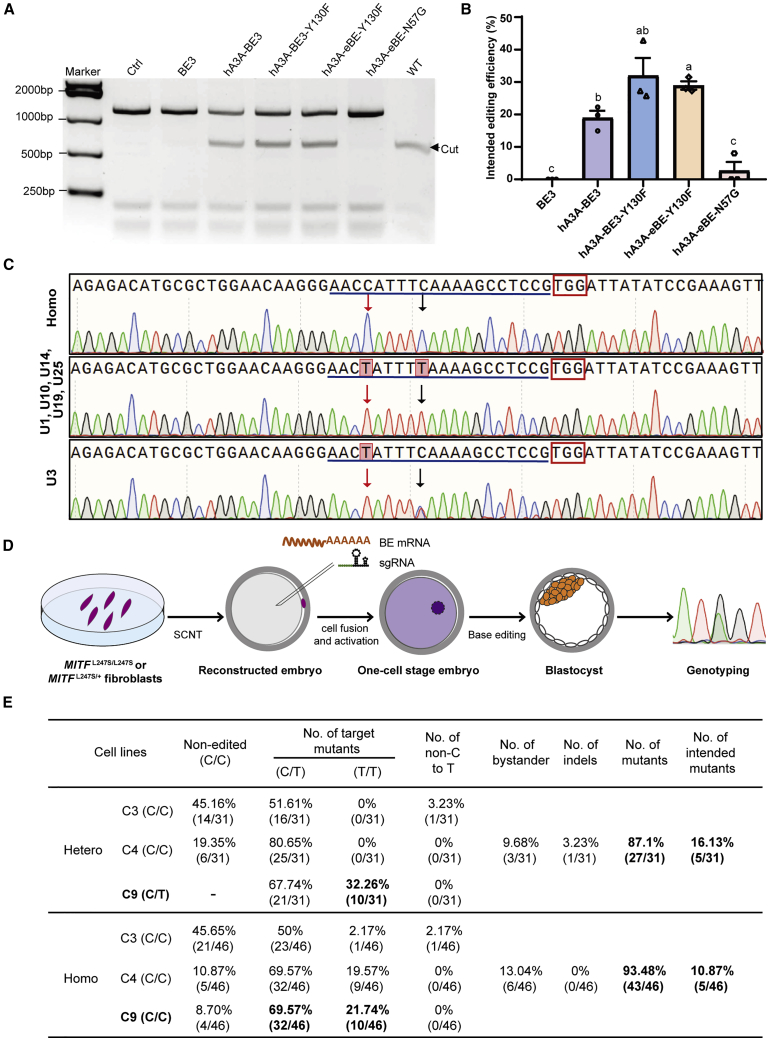
Our results show that our efforts to correct a point mutation by CRISPR-Cas9-mediated HDR were inefficient and need to be further improved to meet the requirements of gene therapy applications. Considering the reported high efficiency and specificity of various engineered BEs in point mutations,30,31 we next turned to evaluate the efficiency of correcting the MITF c.740T>C mutation using several engineered BEs. Restriction fragment length polymorphism (RFLP) results showed that hA3A-BE3, hA3A-BE3-Y130F, and hA3A-eBE-Y130F could efficiently mediate the MITF c.740C>T correction (Figure 5A). The editing efficiency was further analyzed using EditR32 for each C within the sgRNA sequence, and results showed that hA3A-BE3-Y130F and hA3A-eBE-Y130F were superior for editing this site because the third C (C3) cannot be edited, which would otherwise cause a missense mutation (Figures S2 and S3). Further Hi-TOM analysis confirmed that hA3A-BE3-Y130F and hA3A-eBE-Y130F could mediate the intended correction more efficiently (Figure 5B). Thus, hA3A-eBE-Y130F was chosen for further single-cell colony screening. A total of nine colonies were obtained, and five of them (55.56%, U1, U10, U14, U19, and U25) were biallelically corrected and one (11.1%, U3) was monoallelically corrected (Figure 5C). The corrected colonies were cryopreserved for subsequent analysis. In order to evaluate the efficacy of precise correction in early embryonic stage, sgRNA and hA3A-eBE-Y130F mRNA were microinjected into cloned early embryos derived from MITFL247S/L247S and MITFL247S/+ fibroblasts (Figure 5D; Table S3). Genotype analysis showed that up to 16.13% (5/31) of the heterozygous embryos and 10.87% (5/46) of the homozygous embryos were successfully corrected (Figure 5E). These results indicate the potential use of hA3A-eBE-Y130F for gene therapy applications.
Figure 5.
Correction of the MITF c.740T>C mutation using BEs
(A) RFLP analysis in MITFL247S/L247S fibroblast cells that were transfected with different BEs as indicated. (B) Hi-TOM was used to evaluate the editing efficiency of the intended correction with different BEs. The sequences with C9 corrected to T or both C9 and C4 mutated to T were counted as corrected events. Data are presented as mean ± SEM (n = 3 from three independent experiments). a, b, and c values with no letter in common are significantly different (p < 0.05). (C) Representative Sanger sequencing diagrams of the corrected colonies compared with the MITFL247S/L247S cells (Homo). The sgRNA sequences are underlined, and the PAM sequences are shown in red rectangles. The edited bases are indicated by arrows, the black arrows indicate the intended mutations (C9), and the red arrows indicate the C4 mutations in the editing window. (D) Schematic of microinjection in MITFL247S/L247S and MITFL247S/+ fibroblast-derived embryos. (E) Summary of the editing efficiency in heterozygous (hetero) and homozygous (homo) embryos by microinjection of sgRNA and hA3A-eBE-Y130F mRNA. Sequences with C9 corrected to T/T and C3 unedited were counted as intended mutants.
Substantial off-targeting single nucleotide variants were identified in the BE-corrected colony
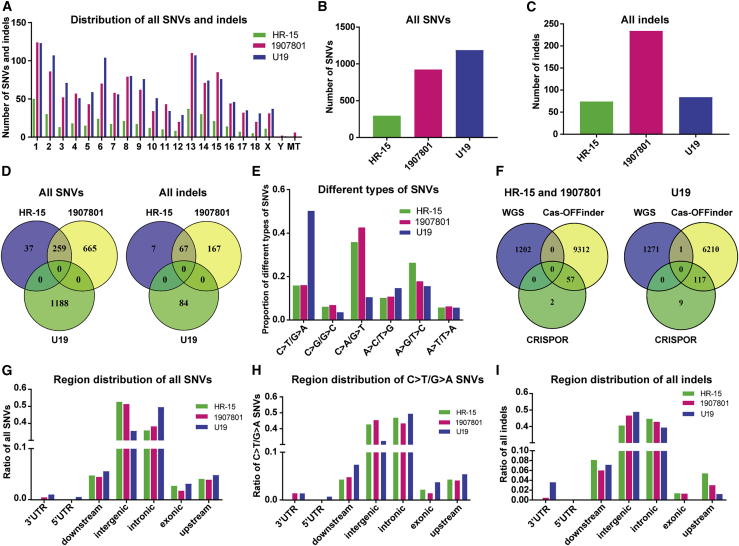
Despite the versatile prospects of the CRISPR-Cas9 and BE system, off-targeting is a safety concern in therapeutic applications. To validate the precision of CRISPR-Cas9 and BE-mediated point mutation correction in the current study, WGS was performed on the CRISPR-Cas9-corrected colony HR-15 and piglet (1907801), and BE-corrected colony (U19), using the Illumina HiSeq X Ten platform (Table S4). Single-nucleotide variants (SNVs) and small indels were identified using three independent algorithms (see Materials and methods). Overall, SNVs and indels were detected in all of the subjected samples, and they were randomly distributed throughout all chromosomes (Figure 6A). A significant number (1,188) of SNVs were detected in the BE-corrected colony (U19), compared with a modest number (296) of SNVs in the Cas9-HDR-corrected colony (HR-15), while the number of indels was comparable in these two colonies (84 and 74, respectively) (Figures 6B and 6C). Interestingly, none of the SNVs and indels found in the Cas9-derived colonies and pigs were shared by those in the U19 colony (Figure 6D), and 87.5% (259/296) of the SNVs and 90.5% (67/74) of the indels detected in the HR-15 colony were also found in the HR-15-derived pig. Moreover, a large amount of novel SNVs (72.0% [665/924] of all SNVs in 1907801) and indels (71.4% [167/234] of all indels in 1907801) occurred during the development process of pig production (HR-15 and 1907801, Figure 6D). Further analysis of the SNVs showed that more than half (50.2%) of the SNVs detected in the U19 colony were a C>T/G>A conversion (Figure 6E). The SNVs and indels identified by WGS were compared with the predicted off-target sites by Cas-OFFinder (http://www.rgenome.net/cas-offinder/)33 and CRISPOR (http://crispor.tefor.net/),34 both depending on sequence similarities of the sgRNA sequences. Interestingly, among all of the SNVs and indels detected by WGS, only one SNV, which was detected in the U19 colony in an intergenic region, was predicted by one of these two software programs (Cas-OFFinder) (Figure 6F). Region distribution analysis showed that most of the SNVs and indels were located on intergenic and intronic regions, and only 0%–3.11% of them were located on exons (Figures 6G–6I), which led to mostly synonymous and missense variations. All of the non-synonymous mutations were confirmed by Sanger sequencing (Tables S5 and S6; Figures S4 and S5) that revealed an overall consistent mutation pattern with WGS results, except that three monoallelic mutations in the WGS data were called as biallelic mutations by Sanger sequencing, and four mutations failed to be validated because of sequencing difficulties.
Figure 6.
Analysis of the off-targeting mutations identified by WGS
(A) The chromosomal distribution of all SNVs and indels throughout the porcine genome. (B and C) Number of all SNVs (B) and indels (C) identified in the three samples. (D) Overlap of all of the SNVs and indels between the three samples. (E) Frequencies of different types of all SNVs detected in the corrected colonies and pig. (F) Overlap of all the SNVs and indels detected by WGS with predicted off-target sites by Cas-OFFinder and CRISPOR. (G–I) The distribution analysis of all SNVs (G), C>T/G>A SNVs (H), and all indels (I) in the given regions.
Discussion
Pigs are considered to be highly useful for human disease modeling and have been widely used in biomedical research. In the current study, we evaluated the efficacy of CRISPR-Cas9- and BE-mediated correction of point mutations in a WS2A pig model carrying a c.740T>C mutation in the MITF gene. CRISPR-Cas9-mediated precise correction combined with SCNT successfully rescued the multi-organ disabilities (anophthalmia, hearing loss, and hypopigmentation), suggesting potential applications in treating monogenetic diseases using current genome-editing tools. Importantly, this strategy of genome editing to mediate gene correction could not only be used for correcting pathogenic mutations that lead to dominant diseases through haploinsufficiency, but it could also be applied to gene therapies for recessive inherited diseases.
Two kinds of donors were designed for CRISPR-Cas9-mediated correction of the c.740T>C mutation, and an unexpected long-fragment deletion was detected in a colony when using a ssODN as the donor, which may occur because the short length of the ssODN may not be able to repair large deletions caused by Cas9 cleavage. Thus, confirmation of repair events at the mRNA level is necessary for gene correction studies that are intolerable of additional indels. Interestingly, the two blocking mutations that were introduced in the donor to avoid recleavage by Cas9 were not simultaneously introduced in the corrected colonies (Figures 1E and 1H). This incomplete or imperfect HDR was previously reported in various species, including pigs,35 mice,36 and human cells.37 In contrast, HDR through long donor DNA seemed to provide a more precise way for correcting this MITF c.740T>C point mutation, which needs to be further investigated. When using the HR donor as the repair template, the efficiency of precise correction reached up to 9.3% (7/75) for single colonies screening and 12.4% for Hi-TOM analysis in MITFL247S/L247S fibroblast cells. However, as we can see from the current study and others, including HDR-mediated gene correction in mouse models,38, 39, 40, 41, 42, 43, 44 the efficiency of CRISPR-Cas9-mediated gene correction still needs further improvement before applying the strategy to treat a variety of diseases. One possibility could be the use of small molecules that inhibit the NHEJ pathway,45, 46, 47, 48 enhance the HDR pathway,47,49 or regulate the cell cycle,50,51 combined with timed delivery of a pre-assembled ribonucleoprotein (RNP) complex of Cas9 protein and sgRNA.51
Recently developed BEs provide an alternative and efficient strategy for precise point mutation modification, and various versions of BEs were engineered to improve the specificity and efficiency of BE-mediated point mutations.30,31 They have already been used for disease modeling and gene therapy studies in human embryos,52 mice,53, 54, 55, 56, 57 rabbits,58 and pigs.59 In the current study, we used several engineered BEs to evaluate the possibility of precise repair of the MITF c.740T>C mutation, and the editing efficiency was greatly improved with use of hA3A-BE3, hA3A-BE3-Y130F, or hA3A-eBE-Y130F, as expected. Further studies were conducted using hA3A-eBE-Y130F, which exhibited a narrowed editing window and increased product purity in a previous study30 and in our preliminary results. The editing efficiency reached up to 66.7% in fibroblast cells as determined by single-colony screening, greatly improving the correction of the point mutation. However, substantial off-targets, primarily SNVs, were detected in the BE-corrected colony, which was four times higher than that in the CRISPR-Cas9 corrected colony. This genome-wide off-targeting was also reported in rice60 and mouse embryos61 using BE3, HF1-BE3, or engineered BE (hA3A-BE3-Y130F),62 probably because of the overexpression of deaminase and/or uracil glycosylase inhibitor (UGI). In contrast, very few off-targeting events were found in the CRISPR-Cas9-HDR-corrected colony, which were even less than the mutations that spontaneously occur during the development process. The rate of spontaneous mutations during the development process is estimated as 3.19 × 10−7 mutations per site, which was calculated as the naturally occurring mutations (the total number of SNVs and indels detected in CRISPR-Cas9 corrected pig 1907801 minus that in the HR-15 colony) divided by the total number of base pairs in porcine genome (2.48 × 109). This estimated rate is higher than that of the average rate of de novo mutations in humans (1.2 × 10−8 mutations per site per generation).63,64 One possible reason for this high occurrence is that the corrected piglets were obtained from SCNT, which is reported to include great genomic instability (DNA damage and chromosome segregation errors) when using fibroblast cells as the donor cell for nuclear transfer in mice.65 Recently, several BE variants, including the introduction of W90Y+R126E in rAPOBEC1 (YE1) and engineered APOBEC3A (eA3A), were reported to significantly reduce DNA and RNA off-targeting editing while maintaining on-targeting efficiency, and several of these variants also had narrowed base-editing windows,62,66 suggesting that rational engineered BEs hold great potential for therapeutic applications. Since there are several Cs in the editing window of the MITF target site, and the editing of C3 would result in missense mutation, BEs with a narrowed editing window are required for precise correction of this causative mutation. Thus, we tried to exploit the A3A (N57G)-BE3 (namely eA3A) that preferably target the TCR (A/G) motif31 to mediate the correction. However, the on-target efficiency of A3A (N57G)-BE3 (namely eA3A) was extremely inefficient for this C>T correction in MITFL247S/L247S porcine fibroblast cells. Thus, they need to be further optimized to correct pathogenic point mutations for future applications.
Similar to WS2A, which displays abnormalities of the retina as early as embryonic day 28 in pigs, there are several diseases that develop before birth and cause irreversible and fatal pathological change. In fetal mouse disease models, prenatal gene therapy has been exploited to prevent disease occurrence at the earliest stage as demonstrated by the treatment of hemophilia B,67 acute neuronopathic Gaucher disease,68 β-thalassaemia,69 spinal muscular atrophy (SMA),70 and hereditary tyrosinemia type 1,55 indicating that prenatal gene editing is a promising approach for treating lethal diseases at the earliest stage. In the current study, we exploited the engineered BE (hA3A-eBE-Y130F) to correct the MITF c.740T>C mutation in SCNT-cloned embryos and demonstrated that efficient correction of this pathogenic mutation could be achieved through one-step microinjection of BE mRNA and sgRNA. Our results provide a proof-of-concept validation of potential applications for in vivo precise correction of pathogenic mutations. Further evaluation of the feasibility of prenatal gene therapy for curing WS2A remains to be investigated, including optimizing the delivery method and verifying safety.
In summary, we show herein that CRISPR-Cas9-mediated gene correction could successfully repair the pathogenic point mutation (MITF c.740T>C) and therefore rescue the vision and hearing loss in a pig model of WS2A, suggesting the potential of nuclease-mediated genome editing in therapeutic applications in the future.
Materials and methods
Animals
Bama miniature pigs were raised at the Beijing Farm Animal Research Center and had ad libitum access to a commercial pig diet (nutrient levels according to the United States National Research Council recommendations) and water throughout the experiment.
Ethics statement
All experiments involving animals were approved by the Animal Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences.
Preparation of sgRNAs and donor DNAs
sgRNAs were designed around the c.740T>C site in exon 8 of the porcine MITF gene using an online tool (http://crispor.tefor.net/).34 Sequences of sgRNAs are shown in corresponding figures. Two complementary oligonucleotide DNAs of sgRNAs for each targeting site were annealed and ligated to the BbsI site of PX330 or PX458 plasmid (Addgene 42230 or 48138) to form the Cas9-sgRNA expression vector. For sgRNA expression vectors used in BE experiments, annealed oligonucleotides were ligated to the BsaI site of pGL3-U6-sgRNA-PGK-puromycin (Addgene 51133, gift from Xingxu Huang from Shanghai Tech University). These sgRNA expression vectors were confirmed by sequencing (Invitrogen).
The 99-nt-long donor DNA was designed to replace the c.740 C to T, and two blocking mutations (one in protospacer adjacent motif (PAM) region, another in the sgRNA sequence) were introduced into the donor DNA sequence. Successful replacement of C>T would generate a DraI restriction site. The donor DNA was synthesized as a single strand (Invitrogen), which was referred to as ssODN-S and ssODN-AS.
To construct long donor DNAs for the c.740C>T correction, about 1,600bp of the genomic sequence around MITF c.740 site (∼800 bp each homology arm) were amplified from the WT porcine fibroblast cells using Phusion high-fidelity PCR master mix (New England Biolabs), with or without sgRNA sequences flanked, and then subcloned into the pLB vector (Tiangen, Beijing, China), which were named as DHMEJ donor, SHMEJ donor, and HR donor. Blocking mutations were introduced into the PAM sequence of sgRNAs to avoid re-cleavage of CRISPR-Cas9 using a Takara MutanBEST kit (Takara, Japan) according to the manufacturer’s instruction. Primers for homology arm amplification and introduction of blocking mutations are listed in Table S7.
Cell transfection and screening of MITF repaired cell colonies
Primary FFCs were isolated from 35-day-old male Bama miniature porcine fetuses (genotyped as MITFL247S/L247S and MITFL247S/+) and cultured as previously described.71 Two micrograms of Cas9-sgRNA expression plasmid and 3 μg of ssODN or 4 μg of long donor DNA plasmid (for CRISPR-Cas9-mediated correction) or 2 μg of pGL3-U6-sgRNA plasmid and 4 μg of BE plasmid (for BE-mediated correction) were co-transfected with 0.2 μg of pCAG-GFP plasmid (when using PX330) into MITFL247S/L247S or MITFL247S/+ FFCs using the 2B Nucleofector device (Lonza, Germany). Forty-eight hours after transfection, cells were subjected to FACS based on GFP fluorescence. For efficiency comparison, all of the GFP-positive cells were collected for genotyping analysis. For single-colony screening, single cells were plated in each well of 96-well plates and cultured for about 10 days in medium supplemented with 2.5 ng/mL basic fibroblast growth factor (Sigma, St. Louis, MO, USA). The cell culture medium was replaced every 4 days. Confluent cell colonies were propagated and genotyped. Cell colonies genotyped with precise correction of the c.740C into T were cryopreserved for subsequent experiments, including SCNT and WGS.
Production of MITF c.740-corrected pigs by SCNT and embryo transfer
Porcine oocyte collection, in vitro maturation, SCNT, and embryo transfer were conducted as previously described.71 Cell colonies with precise correction of the c.740 site (the colony 27 from the ssODN-mediated strategy and the HR-15 colony from the long donor DNA-mediated strategy) were used as the nuclear donor for SCNT. Reconstructed embryos in good condition were transferred into the oviduct of a surrogate the day after observed estrus. Pregnancy was detected by ultrasound 28 days after embryo transfer. All piglets were delivered by natural birth.
Microinjection of BE in MITF c.740T>C mutant early embryos
BE3, hA3A-BE3, hA3A-BE3-Y130F, hA3A-eBE-Y130F, and hA3A-eBE-N57G were kind gifts from Xingxu Huang from Shanghai Tech University30 and Zhanjun Li from Jilin University.58 To obtain the hA3A-eBE-Y130F mRNA for microinjection, the plasmid was linearized by NotI (New England Biolabs, MA, USA), and the capped mRNA was synthesized using mMESSAGE mMACHINE T7 ULTRA transcription kit (Invitrogen, AM1345) and purified using phenol/chloroform extraction and isopropanol precipitation. The sgRNA containing the c.740 site within the editing window (sgRNA2) was in vitro transcribed using the MEGAshortscript T7 transcription kit (Invitrogen, AM1354) after PCR amplification using primers listed in Table S7 and purified using the a MEGAclear kit (Invitrogen, AM1908).
Microinjection of hA3A-eBE-Y130F mRNA (200 ng/μL) and sgRNA (50 ng/μL) was performed as previously described72 after the MITFL247S/L247S and MITFL247S/L247S fibroblast cells were injected into the perivitelline space of enucleated oocytes. Then, the injected oocyte cytoplasm-cell complexes were fused and activated by electric pulse. The resulting reconstructed embryos were cultured in porcine zygote medium (PZM3) in 5% CO2 at 39°C for 6–7 days and each blastocyst was genotyped individually.
Genotyping
Genomic DNA was extracted from mixed GFP-positive cells, cell colonies, and pigs using a TIANamp genomic DNA kit (Tiangen, Beijing, China). Genotyping was conducted by PCR followed by DraI digestion, Sanger sequencing, or Hi-TOM analysis. For genotyping at the mRNA level, total RNA was extracted using TRIzol reagent (Invitrogen) and reverse transcription was performed using the PrimeScript RT reagent kit with gDNA Eraser (Perfect Real time) (Takara, Japan) according to the manufacturer’s instructions. A single blastocyst was lysed by 1× mouse tissue lysis buffer (Vazyme, Nanjing, China) at 55°C for 45 min and 95°C for 15 min, followed by amplification of two rounds of PCR as previously described.72 The PCR products were then sequenced to confirm the sequence around the c.740 site by Sanger sequencing or Hi-TOM.
Off-targeting analysis by WGS
Genomic DNA was extracted from corrected colonies and pigs using a TIANamp genomic DNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions, and DNA integrity was assessed by performing 1% agarose gel electrophoresis. For each sample, about 1 μg of genomic DNA was subjected to WGS, and they were prepared according to the Illumina protocols. For sequencing library preparation, gDNA was first fragmented by a Covaris Focused-ultrasonicator, and adapters were then ligated to both ends of the fragments for each library. WGS was performed using the HiSeq X Ten platform (Novogen, China). The raw sequence reads were split based on index, and the adapters were trimmed out. The Trimmomatic program73 was first used to remove adaptor contamination and trim sequencing reads with low-quality bases. The remaining qualified reads were then mapped to Sscrofa11.1 reference sequence (download from the Ensembl database) using Burrows-Wheeler Aligner (BWA) software74 with default parameters. The SAM files (generated from BWA) containing the read alignments were converted into BAM files, and the processed BAM files (sorted and duplicates removed) were then used to call variants, including SNVs and small indels. To identify the potential off-targets, the somatic mutation detection module of Mutect2 (embedded in GATK v4.1.4.1),75 LoFreq (v2.1.3.1),76 and Varscan2 (v2.4.4)77 software were applied for somatic SNVs calling, and somatic mutation detection module of Mutect2 (embedded in GATK v4.1.4.1), Strelka2 (v2.9.2)78 and VarScan2 (v2.4.4) software were applied for somatic indels calling in corrected colonies and piglet. The somatic SNVs and indels detected by three tools were determined as the potential off-targets. The variants annotation was performed using Variant Effect Predictor (VEP) tools,79 and all of the non-synonymous mutations were validated by Sanger sequencing, using primers listed in Table S5.
H&E staining
Eyes from MITF c.740 corrected piglets and piglets with different genotypes were dissected at birth and fixed in 4% paraformaldehyde, dehydrated in increasing concentrations of ethanol, washed with xylene, and equilibrated in paraffin. Then, the tissues were embedded in paraffin blocks and cut into 5-μm slices. Slices were dewaxed, rehydrated, and stained with H&E by a routine H&E staining protocol. The stained slices were examined under a light microscope for histological analysis.
ERG
Before examination, pigs were placed in a dark room for at least 2 h for dark adaptation. All of the subsequent steps were performed under infrared light. Animals were anesthetized by Zoletil 50 (0.08 mL/kg body weight), and then the corneas of both eyes were anesthetized with a drop of 0.5% proparacaine hydrochloride and both eyes were dilated with compound tropicamide eye drops. Carbomer gel was added to each eye to protect the eye and maintain the electrical connection. ERG was recorded from both eyes simultaneously using a gold ring electrode placed on the cornea of each eye, a reference electrode pierced into the skin of upper eyelid, and a ground electrode placed subcutaneously into the neck. Standard full-field ERGs were performed with a full-field Ganzfeld Q450 stimulator using the commercial RETI-port/scan 21 system (Roland Consult, Germany) according to the standards recommended by International Society for Clinical Electrophysiology of Vision.80 Scotopic ERG was performed with a flash stimuli intensity of 0.0095 cd/m2, a stimulus frequency of 0.2 Hz (eight flashes at an interval of 5 s), and a plot time of 300 ms. After 10 min of light adaptation with a LED background of 25 cd/m2, photopic ERG was recorded using 3 cd/m2 flash stimuli, with a frequency of 0.2 Hz (20 flashes at 5-s intervals) and a plot time of 150 ms.
Measurement of auditory brainstem response
The newborn piglets or pigs at the indicated age were subjected to ABR measurement for hearing evaluation as previously described.81 Pigs were anesthetized with isoflurane, and evoked ABRs with clicks were recorded and analyzed using the Intelligent Hearing System (Intelligent Hearing System, Miami, FL, USA). Sound levels were tested every 10 dB, and the lowest level was confirmed twice at 5-dB intervals. The hearing thresholds and waveforms were recorded and analyzed.
Statistical analysis
Statistical comparison between two groups was conducted by an unpaired, two-tailed t test. Data are presented as mean ± SEM, unless indicated otherwise. A value of ∗p < 0.05 was considered as significantly different and ∗∗p < 0.01 was considered as extremely significantly different.
Acknowledgments
We would like to thank Jingxue Zhang and Shen Wu from Capital Medical University for assistance with ERG analysis, and Lin Jiang from the Chinese Academy of Agricultural Sciences for assistance with the WGS data analysis. This work was supported by the China National Key R&D Program (2020YFC1316600 and 2020YFA0509503); the National Science Fund for Distinguished Young Scholars (31925036); the Strategic Priority Research Programs of CAS (XDA16030304); the National Natural Science Foundation of China (81671274, 31801031, 31601008, 31701073, and 32025034); and the National Transgenic Project of China (2016ZX08009003-006-007) .
Author contributions
Jianguo Z., Yanfang W., J.Y., Yu W., and C.C. conceived the study, designed the experiments, and wrote and edited the manuscript. J.Y., Yu W., C.C., R.S., D.B., H.Z., Y.L., G.Q., N.H., N.Z., Jin Z., W.G., and S.Y. performed the experiments and analyzed the data.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.04.009.
Contributor Information
Yanfang Wang, Email: wangyanfang@caas.cn.
Jianguo Zhao, Email: zhaojg@ioz.ac.cn.
Supplemental information
References
- 1.Kochhar A., Hildebrand M.S., Smith R.J. Clinical aspects of hereditary hearing loss. Genet. Med. 2007;9:393–408. doi: 10.1097/gim.0b013e3180980bd0. [DOI] [PubMed] [Google Scholar]
- 2.Waardenburg P.J. A new syndrome combining developmental anomalies of the eyelids, eyebrows and nose root with pigmentary defects of the iris and head hair and with congenital deafness. Am. J. Hum. Genet. 1951;3:195–253. [PMC free article] [PubMed] [Google Scholar]
- 3.Song J., Feng Y., Acke F.R., Coucke P., Vleminckx K., Dhooge I.J. Hearing loss in Waardenburg syndrome: A systematic review. Clin. Genet. 2016;89:416–425. doi: 10.1111/cge.12631. [DOI] [PubMed] [Google Scholar]
- 4.Ohno N., Kiyosawa M., Mori H., Wang W.F., Takase H., Mochizuki M. Clinical findings in Japanese patients with Waardenburg syndrome type 2. Jpn. J. Ophthalmol. 2003;47:77–84. doi: 10.1016/s0021-5155(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 5.Tassabehji M., Newton V.E., Read A.P. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 6.Nobukuni Y., Watanabe A., Takeda K., Skarka H., Tachibana M. Analyses of loss-of-function mutations of the MITF gene suggest that haploinsufficiency is a cause of Waardenburg syndrome type 2A. Am. J. Hum. Genet. 1996;59:76–83. [PMC free article] [PubMed] [Google Scholar]
- 7.Tassabehji M., Newton V.E., Liu X.Z., Brady A., Donnai D., Krajewska-Walasek M., Murday V., Norman A., Obersztyn E., Reardon W. The mutational spectrum in Waardenburg syndrome. Hum. Mol. Genet. 1995;4:2131–2137. doi: 10.1093/hmg/4.11.2131. [DOI] [PubMed] [Google Scholar]
- 8.Steingrímsson E., Moore K.J., Lamoreux M.L., Ferré-D’Amaré A.R., Burley S.K., Zimring D.C., Skow L.C., Hodgkinson C.A., Arnheiter H., Copeland N.G. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 9.Hai T., Guo W., Yao J., Cao C., Luo A., Qi M., Wang X., Wang X., Huang J., Zhang Y. Creation of miniature pig model of human Waardenburg syndrome type 2A by ENU mutagenesis. Hum. Genet. 2017;136:1463–1475. doi: 10.1007/s00439-017-1851-2. [DOI] [PubMed] [Google Scholar]
- 10.George A., Zand D.J., Hufnagel R.B., Sharma R., Sergeev Y.V., Legare J.M., Rice G.M., Scott Schwoerer J.A., Rius M., Tetri L. Biallelic mutations in MITF cause coloboma, osteopetrosis, microphthalmia, macrocephaly, albinism, and deafness. Am. J. Hum. Genet. 2016;99:1388–1394. doi: 10.1016/j.ajhg.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B., Hu Y.J., Hu J.H., Thompson D.B., Shu Y. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anguela X.M., High K.A. Entering the modern era of gene therapy. Annu. Rev. Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 13.Diakatou M., Manes G., Bocquet B., Meunier I., Kalatzis V. Genome editing as a treatment for the most prevalent causative genes of autosomal dominant retinitis pigmentosa. Int. J. Mol. Sci. 2019;20:2542. doi: 10.3390/ijms20102542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A⋅T to G⋅C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang P., Ding C., Sun H., Xie X., Xu Y., Zhang X., Sun Y., Xiong Y., Ma W., Liu Y. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017;8:811–822. doi: 10.1007/s13238-017-0475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng T.L., Li S., Yuan B., Wang X., Zhou W., Qiu Z. Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat. Commun. 2019;10:3612. doi: 10.1038/s41467-019-11562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q., Wang C., Jiao X., Zhang H., Song L., Li Y., Gao C., Wang K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019;62:1–7. doi: 10.1007/s11427-018-9402-9. [DOI] [PubMed] [Google Scholar]
- 24.Hoban M.D., Lumaquin D., Kuo C.Y., Romero Z., Long J., Ho M., Young C.S., Mojadidi M., Fitz-Gibbon S., Cooper A.R. CRISPR/Cas9-mediated correction of the sickle mutation in human CD34+ cells. Mol. Ther. 2016;24:1561–1569. doi: 10.1038/mt.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponce de León V., Mérillat A.M., Tesson L., Anegón I., Hummler E. Generation of TALEN-mediated GRdim knock-in rats by homologous recombination. PLoS ONE. 2014;9:e88146. doi: 10.1371/journal.pone.0088146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao X., Wang X., Hu X., Liu Z., Liu J., Zhou H., Shen X., Wei Y., Huang Z., Ying W. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–814. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hai T., Cao C., Shang H., Guo W., Mu Y., Yang S., Zhang Y., Zheng Q., Zhang T., Wang X. Pilot study of large-scale production of mutant pigs by ENU mutagenesis. eLife. 2017;6:e26248. doi: 10.7554/eLife.26248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurome M., Geistlinger L., Kessler B., Zakhartchenko V., Klymiuk N., Wuensch A., Richter A., Baehr A., Kraehe K., Burkhardt K. Factors influencing the efficiency of generating genetically engineered pigs by nuclear transfer: Multi-factorial analysis of a large data set. BMC Biotechnol. 2013;13:43. doi: 10.1186/1472-6750-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estrada J., Sommer J., Collins B., Mir B., Martin A., York A., Petters R.M., Piedrahita J.A. Swine generated by somatic cell nuclear transfer have increased incidence of intrauterine growth restriction (IUGR) Cloning Stem Cells. 2007;9:229–236. doi: 10.1089/clo.2006.0079. [DOI] [PubMed] [Google Scholar]
- 30.Wang X., Li J., Wang Y., Yang B., Wei J., Wu J., Wang R., Huang X., Chen J., Yang L. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 2018;36:946–949. doi: 10.1038/nbt.4198. [DOI] [PubMed] [Google Scholar]
- 31.Gehrke J.M., Cervantes O., Clement M.K., Wu Y., Zeng J., Bauer D.E., Pinello L., Joung J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018;36:977–982. doi: 10.1038/nbt.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: A method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae S., Park J., Kim J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Tang X., Liu Y., Xie Z., Zou X., Li M., Yuan H., Ouyang H., Jiao H., Pang D. Efficient generation of orthologous point mutations in pigs via CRISPR-assisted ssODN-mediated homology-directed repair. Mol. Ther. Nucleic Acids. 2016;5:e396. doi: 10.1038/mtna.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inui M., Miyado M., Igarashi M., Tamano M., Kubo A., Yamashita S., Asahara H., Fukami M., Takada S. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci. Rep. 2014;4:5396. doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bialk P., Sansbury B., Rivera-Torres N., Bloh K., Man D., Kmiec E.B. Analyses of point mutation repair and allelic heterogeneity generated by CRISPR/Cas9 and single-stranded DNA oligonucleotides. Sci. Rep. 2016;6:32681. doi: 10.1038/srep32681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H., Li Y., He L., Pu W., Yu W., Li Y., Wu Y.T., Xu C., Wei Y., Ding Q. In vivo AAV-CRISPR/Cas9-mediated gene editing ameliorates atherosclerosis in familial hypercholesterolemia. Circulation. 2020;141:67–79. doi: 10.1161/CIRCULATIONAHA.119.042476. [DOI] [PubMed] [Google Scholar]
- 40.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L., Yang Y., Breton C.A., White J., Zhang J., Che Y., Saveliev A., McMenamin D., He Z., Latshaw C. CRISPR/Cas9-mediated in vivo gene targeting corrects hemostasis in newborn and adult factor IX-knockout mice. Blood. 2019;133:2745–2752. doi: 10.1182/blood.2019000790. [DOI] [PubMed] [Google Scholar]
- 43.Jo D.H., Song D.W., Cho C.S., Kim U.G., Lee K.J., Lee K., Park S.W., Kim D., Kim J.H., Kim J.S. CRISPR-Cas9-mediated therapeutic editing of Rpe65 ameliorates the disease phenotypes in a mouse model of Leber congenital amaurosis. Sci. Adv. 2019;5:eaax1210. doi: 10.1126/sciadv.aax1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y., Chen W., Zhang X., Yu L., Dong W., Pan S., Gao S., Huang X., Zhang L. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13:605–612. doi: 10.1080/15476286.2016.1185591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 47.Pinder J., Salsman J., Dellaire G. Nuclear domain “knock-in” screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song J., Yang D., Xu J., Zhu T., Chen Y.E., Zhang J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016;7:10548. doi: 10.1038/ncomms10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J.P., Li X.L., Li G.H., Chen W., Arakaki C., Botimer G.D., Baylink D., Zhang L., Wen W., Fu Y.W. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng Y., Li J., Li G., Huang S., Yu W., Zhang Y., Chen D., Chen J., Liu J., Huang X. Correction of the Marfan syndrome pathogenic FBN1 mutation by base editing in human cells and heterozygous embryos. Mol. Ther. 2018;26:2631–2637. doi: 10.1016/j.ymthe.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryu S.M., Koo T., Kim K., Lim K., Baek G., Kim S.T., Kim H.S., Kim D.E., Lee H., Chung E., Kim J.S. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 54.Sasaguri H., Nagata K., Sekiguchi M., Fujioka R., Matsuba Y., Hashimoto S., Sato K., Kurup D., Yokota T., Saido T.C. Introduction of pathogenic mutations into the mouse Psen1 gene by Base Editor and Target-AID. Nat. Commun. 2018;9:2892. doi: 10.1038/s41467-018-05262-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossidis A.C., Stratigis J.D., Chadwick A.C., Hartman H.A., Ahn N.J., Li H., Singh K., Coons B.E., Li L., Lv W. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat. Med. 2018;24:1513–1518. doi: 10.1038/s41591-018-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 57.Yeh W.H., Shubina-Oleinik O., Levy J.M., Pan B., Newby G.A., Wornow M., Burt R., Chen J.C., Holt J.R., Liu D.R. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 2020;12:eaay9101. doi: 10.1126/scitranslmed.aay9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z., Chen S., Shan H., Zhang Q., Chen M., Lai L., Li Z. Efficient and precise base editing in rabbits using human APOBEC3A-nCas9 fusions. Cell Discov. 2019;5:31. doi: 10.1038/s41421-019-0099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie J., Ge W., Li N., Liu Q., Chen F., Yang X., Huang X., Ouyang Z., Zhang Q., Zhao Y. Efficient base editing for multiple genes and loci in pigs using base editors. Nat. Commun. 2019;10:2852. doi: 10.1038/s41467-019-10421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 61.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuo E., Sun Y., Yuan T., He B., Zhou C., Ying W., Liu J., Wei W., Zeng R., Li Y., Yang H. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods. 2020;17:600–604. doi: 10.1038/s41592-020-0832-x. [DOI] [PubMed] [Google Scholar]
- 63.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chia G., Agudo J., Treff N., Sauer M.V., Billing D., Brown B.D., Baer R., Egli D. Genomic instability during reprogramming by nuclear transfer is DNA replication dependent. Nat. Cell Biol. 2017;19:282–291. doi: 10.1038/ncb3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waddington S.N., Nivsarkar M.S., Mistry A.R., Buckley S.M., Kemball-Cook G., Mosley K.L., Mitrophanous K., Radcliffe P., Holder M.V., Brittan M. Permanent phenotypic correction of hemophilia B in immunocompetent mice by prenatal gene therapy. Blood. 2004;104:2714–2721. doi: 10.1182/blood-2004-02-0627. [DOI] [PubMed] [Google Scholar]
- 68.Massaro G., Mattar C.N.Z., Wong A.M.S., Sirka E., Buckley S.M.K., Herbert B.R., Karlsson S., Perocheau D.P., Burke D., Heales S. Fetal gene therapy for neurodegenerative disease of infants. Nat. Med. 2018;24:1317–1323. doi: 10.1038/s41591-018-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ricciardi A.S., Bahal R., Farrelly J.S., Quijano E., Bianchi A.H., Luks V.L., Putman R., López-Giráldez F., Coşkun S., Song E. In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 2018;9:2481. doi: 10.1038/s41467-018-04894-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rashnonejad A., Amini Chermahini G., Gündüz C., Onay H., Aykut A., Durmaz B., Baka M., Su Q., Gao G., Özkınay F. Fetal gene therapy using a single injection of recombinant AAV9 rescued SMA phenotype in mice. Mol. Ther. 2019;27:2123–2133. doi: 10.1016/j.ymthe.2019.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao J., Huang J., Hai T., Wang X., Qin G., Zhang H., Wu R., Cao C., Xi J.J., Yuan Z., Zhao J. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci. Rep. 2014;4:6926. doi: 10.1038/srep06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X., Zhou J., Cao C., Huang J., Hai T., Wang Y., Zheng Q., Zhang H., Qin G., Miao X. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci. Rep. 2015;5:13348. doi: 10.1038/srep13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benjamin D., Sato T., Cibulskis K., Getz G., Stewart C., Lichtenstein L. Calling somatic SNVs and indels with Mutect2. bioRxiv. 2019 doi: 10.1101/861054. [DOI] [Google Scholar]
- 76.Wilm A., Aw P.P., Bertrand D., Yeo G.H., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim S., Scheffler K., Halpern A.L., Bekritsky M.A., Noh E., Källberg M., Chen X., Kim Y., Beyter D., Krusche P., Saunders C.T. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 79.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCulloch D.L., Marmor M.F., Brigell M.G., Hamilton R., Holder G.E., Tzekov R., Bach M. ISCEV standard for full-field clinical electroretinography (2015 update) Doc. Ophthalmol. 2015;130:1–12. doi: 10.1007/s10633-014-9473-7. [DOI] [PubMed] [Google Scholar]
- 81.Guo W., Yi H., Ren L., Chen L., Zhao L., Sun W., Yang S.M. The morphology and electrophysiology of the cochlea of the miniature pig. Anat. Rec. (Hoboken) 2015;298:494–500. doi: 10.1002/ar.23095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.