Abstract
Objective
Hepatic miR-34a expression is elevated in diet-induced or genetically obese mice and patients with non-alcoholic steatohepatitis (NASH), yet hepatocyte miR-34a's role in the progression of non-alcoholic fatty liver disease (NAFLD) from non-alcoholic fatty liver (NAFL) to NASH remains to be elucidated.
Methods
Mice overexpressing or deficient in hepatocyte miR-34a and control mice were fed a diet enriched in fats, cholesterol, and fructose (HFCF) to induce NASH. C57BL/6 mice with NASH were treated with an miR-34a inhibitor or a scramble control oligo. The effect of miR-34a on the development, progression, and reversal of NAFLD was determined.
Results
The hepatocyte-specific expression of miR-34a aggravated HFCF diet-induced NAFLD. In contrast, germline or adult-onset deletion of hepatocyte miR-34a attenuated the development and progression of NAFLD. In addition, pharmacological inhibition of miR-34a reversed HFCF diet-induced steatohepatitis. Mechanistically, hepatocyte miR-34a regulated the development and progression of NAFLD by inducing lipid absorption, lipogenesis, inflammation, and apoptosis but inhibiting fatty acid oxidation.
Conclusions
Hepatocyte miR-34a is an important regulator in the development and progression of NAFLD. MiR-34a may be a useful target for treating NAFLD.
Keywords: miR-34a, NAFLD, Lipogenesis, Lipid absorption, Bile acids
Highlights
-
•
Hepatocyte-specific overexpression of miR-34a aggravates high-fat/cholesterol/fructose (HFCF) diet-induced steatohepatitis.
-
•
Germline or adult-onset ablation of hepatocyte miR-34a protects against HFCF diet-induced steatohepatitis.
-
•
Mir-34a inhibitors reverse non-alcoholic steatohepatitis (NASH).
-
•
Mir-34a promotes non-alcoholic fatty liver (NAFL) by inducing lipid absorption and synthesis and inhibiting fatty acid oxidation.
-
•
MiR-34a promotes the progression of NAFL to NASH by inducing inflammation, lipotoxicity, and apoptosis.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is a spectrum of liver diseases spanning non-alcoholic fatty liver (NAFL) to non-alcoholic steatohepatitis (NASH) that may further progress to liver cirrhosis and hepatocellular carcinoma. At present, the pathogenic mechanisms of NAFLD are not fully understood. Several mechanisms have been proposed to underlie the progression of NAFL to NASH, such as insulin resistance, inflammation, apoptosis, mitochondrial dysfunctions, and reactive oxygen species (ROS), among others. Moreover, no drugs have been approved to treat NASH.
MicroRNAs are small non-coding RNA molecules that bind to 3′ UTRs of target mRNAs, usually leading to translational inhibition or mRNA degradation. MiR-34a is expressed in a variety of tissues, such as macrophages, endothelial cells, smooth muscle cells, hepatocytes, and adipocytes, among others. Hepatic miR-34a expression is increased in diet-induced or genetic obese mice and NAFLD patients [1,2]. Adenovirus-mediated miR-34a expression in the liver promotes the development of NAFL whereas inhibition of hepatic miR-34a expression has an opposite effect [1,2]. However, hepatocyte miR-34a's role in the progression of NAFL to NASH or NASH reversal remains to be elucidated.
Bile acids (BAs) are not only critical for intestinal fat and cholesterol absorption, but can also act as signaling molecules to regulate lipid and glucose metabolism and energy homeostasis [3]. Primary BAs are synthesized solely in hepatocytes. Cholesterol 7a-hydroxylase (CYP7A1) and sterol 12a-hydroxylase (CYP8B1) are two rate-limiting enzymes in the classic pathway of BA biosynthesis. MiR-34a has been shown to upregulate hepatic CYP7A1 and CYP8B1 expression [2,4] and intestinal fat and cholesterol absorption [4]. It is unclear whether miR-34a-mediated bile acid metabolism contributes to the development of NAFLD.
In this report, we show that adeno-associated virus serotype 8 (AAV8)-mediated overexpression of miR-34a in hepatocytes aggravates high-fat/cholesterol/fructose (HFCF) diet-induced lipid accumulation, ROS production, apoptosis, inflammation, and fibrosis in the liver. In contrast, germline or adult-onset deletion of hepatocyte miR-34a attenuates the development and progression of HFCF diet-induced NAFLD. In addition, pharmacological inhibition of miR-34a reverses HFCF diet-induced steatohepatitis. Our data indicate that hepatocyte miR-34a is a key regulator of the development and progression of NAFLD by coordinating regulation of lipid metabolism, inflammation, ROS production, and apoptosis.
2. Materials and methods
2.1. Mice, diets, and miR-34a inhibitors
miR-34afl/fl mice, albumin-Cre (Alb-Cre) mice, and C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, Maine, USA) [4]. miR-34afl/fl mice were crossed with Alb-Cre mice to generate germline hepatocyte-specific miR-34a−/− (miR-34agHep−/−) mice and control littermates (miR-34afl/fl mice). AAV8-TBG-Null or AAV8-TBG-Cre was i.v. injected into the miR-34afl/fl mice to generate control mice (miR-34afl/fl mice) or adult-onset hepatocyte-specific miR-34a−/− (miR-34aHep−/−) mice, respectively. Locked nucleic acids (LNA) against scramble sequences (LNA-Scr) or miR-34a (LNA-miR-34a) were synthesized by Qiagen and i.p. injected into the mice at a dosage of 10 mg/kg/week. All of the mice were housed in a temperature- and humidity-controlled room with a 12-h light/12-h dark cycle under pathogen-free conditions. The high-fat/cholesterol/fructose (HFCF) diet contained 40% fat/0.2% cholesterol (AIN-76A Western diet from TestDiet) and 4.2% fructose (in drinking water). Unless otherwise stated, 2-month-old male mice were used and fed an HFCF diet for 16 weeks. The mice were fasted for 5–6 h during the light cycle prior to euthanasia. All the of animal experiments were approved by the Institutional Animal Care and Use Committee at Northeast Ohio Medical University (NEOMED).
2.2. Adeno-associated viruses (AAVs)
Human pri-miR-34a and flanking sequences were cloned into an AAV8 vector under the control of a mouse albumin promoter to generate plasmid AAV-ALB-miR-34a. AAV8-ALB-Null (control), AAV8-ALB-miR-34a, AAV8-ALB-CYP7A1, AAV8-ALB-CYP8B1 [5], AAV8-TBG-Null (control), and AAV8-TBG-Cre were produced and titrated by Vector BioLabs. Each mouse was i.v. injected with 3 × 1011 genome copies (GC) of AAVs.
2.3. Exosome isolation and cell culture
Kupffer cells were isolated from C57BL/6J mice as previously described [6]. In brief, the mice were anesthetized and the liver was perfused through the vena cava cannula with solutions containing collagenase IV (40 mg/mouse, Sigma–Aldrich). Digested liver was gently disrupted using a tissue homogenizer and passed through a 40-μm cell strainer. The cell suspension was centrifuged at 50×g for 3 min to pellet hepatocytes. Kupffer cells in the supernatant fraction were then isolated by gradient centrifugation and cultured in DMEM containing 10% (vol/vol) FBS. Exosomes were isolated from mouse serum using a SmartSEC Single kit from System Biosciences (cat. #SSEC200A-1) and then co-cultured with Kupffer cells.
2.4. Real-time PCR
Total RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific) and mRNA levels were quantified by quantitative real-time PCR (qRT-PCR) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on a 7500 Real-Time PCR machine (Applied Biosystems). mRNA levels were normalized to 36B4. miRNAs were extracted using a mirVana miRNA isolation kit (Thermo Fisher Scientific). MiRNA levels were quantified using a TaqMan MicroRNA assay kit (Thermo Fisher Scientific) and normalized to U6. Primer sequences for qRT-PCR are presented in Supplementary Table 1.
2.5. Western blotting
Total liver lysates and nuclear or microsome extracts of liver samples were used for Western blotting. The antibody against tubulin (cat. #ab4074) was purchased from Abcam. Antibodies against CYP7A1 (cat. #TA351400) or CYP8B1 (cat. #TA313734) were purchased from Origene. Antibodies against acetyl-CoA carboxylase (ACC) (cat. #3662), cleaved caspase 3 (cat. #9661), total caspase 3 (cat. #9662), phospho-Smad2/3 (cat. #8828), or total Smad2/3 (cat. #5678) were purchased from Cell Signaling Technology (Boston, MA, USA). Antibodies against histone (cat. #sc-393358) or SREBP-1 (cat. #SC-8984) were purchased from Santa Cruz Biotechnology. The antibody against calnexin (NB100-1965) was purchased from Novus.
2.6. Analysis of plasma AST, ALT, and β-hydroxybutyrate as well as hepatic lipids, hydroxyproline, malondialdehyde, ROS, and apoptosis
Approximately 100 mg of liver tissue was homogenized in methanol and lipids were extracted using chloroform/methanol (2:1 v/v) as previously described [7]. Triglycerides (TG) and cholesterol in the liver and plasma were determined using Infinity reagents from Thermo Fisher Scientific (Waltham, MA, USA). Plasma ALT and AST levels were measured using Infinity reagents. Hepatic hydroxyproline levels were quantified using a kit from Cell BioLabs (cat. #STA675). Malondialdehyde (MDA) levels in the liver were determined using a TBARS assay kit (cat. #STA-330). Hepatic reactive oxygen species (ROS) levels were measured using an OxiSelect In Vitro ROS/RNS Assay kit (cat. #STA-347) from Cell Biolabs. Liver apoptosis was determined using a TUNEL assay kit from Abcam (cat. #ab206386). Plasma β-hydroxybutyrate levels were measured using a kit from Pointe Scientific (Fisher cat. #23-666-471).
2.7. Bile acid analysis
Bile acids in the liver, gallbladder, and intestine were extracted using ethanol as previously described [4]. Bile acid levels were measured using a bile acid kit from Diazyme (cat. #DZ042A-KY1). Total bile acids in the liver, gallbladder, and intestine were used to calculate the total bile acid pool size. The bile acid composition was measured by HPLC according to the method of Rossi et al. [8,9], and the hydrophobicity index (HI) was calculated.
2.8. Intestinal fat or cholesterol absorption
To determine intestinal fat absorption, the mice were i.v. injected with 100 μl of tyloxapol (500 mg/kg) after a 6 h fast, gavaged with olive oil (15 μl/g body weight), and plasma TG levels at defined time points were determined [5]. Intestinal cholesterol absorption was measured using a dual-isotope plasma ratio method as previously described [4,10]. Briefly, the mice were i.v. injected with 2.5 μCi of 3H-cholesterol dissolved in 160 μl of Intralipids (Sigma–Aldrich, cat. #I141) and then gavaged with 1 μCi of 14C-cholesterol dissolved in 200 μl of medium-chain triglyceride (MCT) oil. After 72 h, plasma was collected to determine the 3H and 14C activity. Cholesterol absorption was calculated using the formula: % cholesterol absorption = (% of gavage dose)/(% of intravenous dose) x 100.
2.9. Fatty acid oxidation (FAO)
Primary hepatocytes were isolated and then cultured in DMEM containing 10% FBS in 12-well dishes. FAO was determined using 3H-palmitic acid as a substrate as previously described [11].
2.10. Analysis of fatty acid levels and fatty acid composition
Hepatic lipids were extracted and total free fatty acids (FFAs) were determined using a kit from Wako Chemicals USA (Richmond, VA, USA). Hepatic fatty acid composition was analyzed by GC–MS as previously described [11].
2.11. Oil Red O (ORO), hematoxylin and eosin (H&E), and Picrosirius red staining and immunostaining
Liver tissues were fixed in 10% formalin and then embedded in OCT or paraffin. Liver sections were stained with ORO, H&E, or Picrosirius red. Liver sections were also immunostained using an F4/80 antibody (Abcam, cat. #ab6640) and an ABC-HRP kit from Vector Laboratories (cat. #PK-4001). Images were acquired using an Olympus microscope.
2.12. Body fat measurement
Body fat was measured using EchoMRI (EchoMRI, Houston, TX, USA) as previously described [12].
2.13. Statistical analysis
All the data were expressed as mean ± SEM. Statistical significance was analyzed using an unpaired Student's t test or analysis of variance (ANOVA, for more than two groups) (GraphPad Prism, San Diego, CA, USA). Differences were considered statistically significant at P < 0.05.
3. Results
3.1. Hepatocyte-specific expression of miR-34a exacerbated HFCF diet-induced NAFL by inducing lipid absorption and synthesis and inhibiting FAO
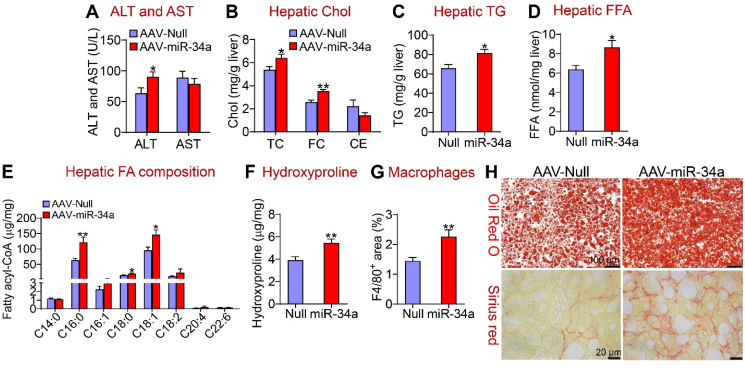
We and others have previously shown that hepatic miR-34a is induced in diabetic or high-fat diet (HFD)-fed mice and NAFLD patients [1,2]. However, hepatocyte miR-34a's role in the development of NAFLD remains to be determined. Therefore, we generated an AAV expressing miR-34a under the control of a mouse albumin promoter (AAV8-ALB-miR-34a). AAV8-ALB-miR-34a or AAV8-ALB-Null (control) was i.v. injected into the C57BL/6J mice, which were then fed a chow diet or an HFCF diet for 16 weeks. On a chow diet, miR-34a over-expression increased hepatic levels of total cholesterol (TC), free cholesterol (FC), triglycerides (TG), free fatty acids (FFAs), and bile acid (BA) pool size (Supplementary Fig. 1A–E). When fed an HFCF diet, overexpression of miR-34a in hepatocytes did not affect body fat content (Supplementary Fig. 2A), but increased plasma ALT levels (Figure 1A) and the ratio of liver weight to body weight (BW) (Supplementary Fig. 2B). MiR-34a overexpression also increased hepatic levels of TC, FC (Figure 1B), TG (Figure 1C), and FFA (Figure 1D). An analysis of the fatty acid composition showed that hepatic C16:0, C18:0, and C18:1 fatty acyl-CoA levels increased by 193%, 139%, and 152%, respectively (Figure 1E). In addition, miR-34a overexpression increased hepatic hydroxyproline levels by 140% (Figure 1F) and hepatic macrophage/Kupffer cell accumulation (Figure 1G). Histological analyses by Oil Red O, Picrosirius red (Figure 1H), or H&E staining and immunostaining using an F4/80 antibody (Supplementary Fig. 2C, top and bottom panels) showed that miR-34a induced neutral lipid accumulation as well as liver inflammation and fibrosis.
Figure 1.
Hepatocyte-specific expression of miR-34a promoted diet-induced steatohepatitis. C57BL/6J mice were i.v. injected with 3 × 1011 GC AAV8-ALB-Null or AAV8-ALB-miR-34a and then fed an HFCH diet for 16 weeks (n = 8 per group). (A) Plasma ALT and AST levels. (B) Hepatic total cholesterol (TC), free cholesterol (FC), and cholesteryl ester (CE) levels. (C) Hepatic triglyceride (TG) levels. (D) Hepatic free fatty acid (FFA) levels. (E) Hepatic individual fatty acyl-CoA levels were determined by GC–MS. (F) Hepatic hydroxyproline levels. (G) Liver sections were immunostained using an F4/80 antibody and the F4/80-positive area was determined. (H) Liver sections were stained by Oil Red O (top panel) or Picrosirius red (lower panel). ∗P < 0.05 and ∗∗P < 0.01.
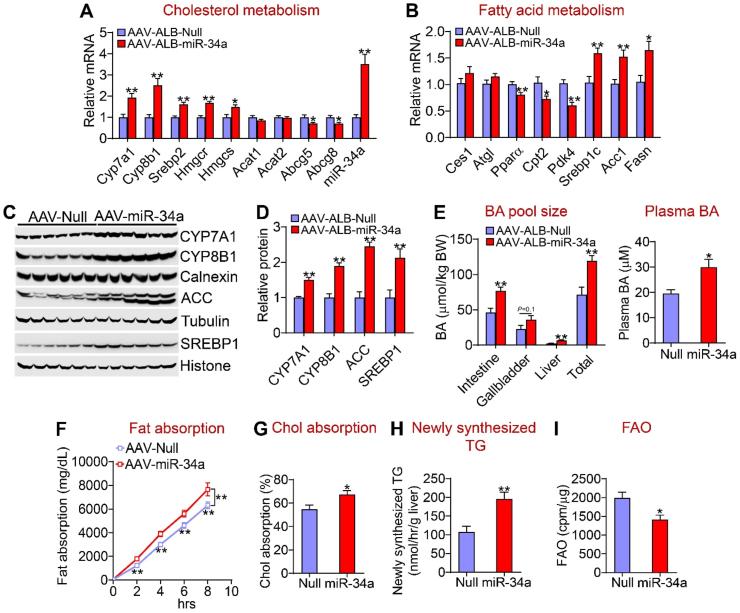
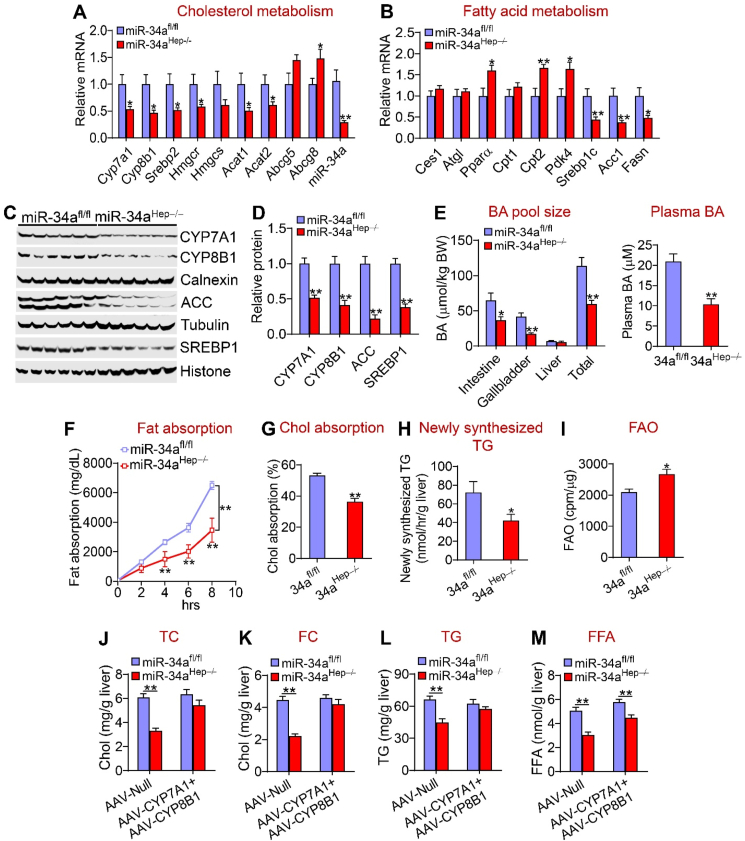
In the liver, hepatocyte-specific expression of miR-34a induced mRNA levels of genes involved in bile acid metabolism (Cyp7a1 and Cyp8b1), cholesterol synthesis [sterol regulatory element-binding protein 2 (Srebp2), HMG-CoA reductase (Hmgcr), and HMG-CoA synthase (Hmgcs)], and fatty acid synthesis [Srebp1c, acetyl-CoA carboxylase 1 (Acc1) and fatty acid synthase (Fasn)] but repressed genes involved in cholesterol excretion into bile [ATP-binding cassette subfamily G member 5 (Abcg5) and Abcg8] and FAO [peroxisome proliferation-activated receptor a (Ppara), carnitine palmitoyltransferase 2 (Cpt2), and pyruvate dehydrogenase kinase 4 (Pdk4)] (Figure 2A,B). Consistent with the changes in mRNA levels, miR-34a significantly increased CYP7A1, CYP8B1, ACC, and SREBP1 protein levels (Figure 2C,D).
Figure 2.
Hepatocyte miR-34a promoted lipid absorption and synthesis but inhibited FAO. The mice are described in the legend in Figure 1 (n = 8 per group). (A) mRNA levels of genes involved in cholesterol metabolism. (B) mRNA levels of genes involved in fatty acid metabolism. (C and D) Western blotting assays (C) were performed and protein levels were quantified (D). (E) Bile acid (BA) pool size (left panel) and plasma BA levels (right panel). (F) Intestinal fat absorption. (G) Intestinal cholesterol absorption. (H) Newly synthesized TGs were quantified by GC–MS. (I) Mouse primary hepatocytes were isolated from mice infected with AAV8-ALB-Null or AAV8-ALB-miR-34a and FAO was determined using 3H-palmitic acid as substrates (n = 5). ∗P < 0.05 and ∗∗P < 0.01.
AAV-mediated expression of miR-34a increased BA levels in the intestine and liver, the total BA pool size by 171%, and plasma BA levels by 153% (Figure 2E). In line with BA's role in promoting lipid absorption, miR-34a overexpression increased intestinal fat (Figure 2F) and cholesterol absorption (Figure 2G). Interestingly, miR-34a overexpression did not affect the BA composition or hydrophobicity index (HI) (Supplementary Fig. 2D and E). In addition, miR-34a overexpression increased the hepatic TG synthesis rate by 182% (Figure 2H), and reduced FAO by 30% in hepatocytes (Figure 2I). The latter data were also supported by the finding that plasma beta-hydroxybutyrate (b-HB) levels were reduced following hepatic miR-34a overexpression (Supplementary Fig. 2F).
Taken together, the data in Figure 2 demonstrate that hepatocyte-specific expression of miR-34a aggravated diet-induced NAFL via a mechanism involving an increase in lipid absorption and synthesis and a reduction in FAO.
3.2. Hepatocyte miR-34a promoted NASH development via inducing inflammation, ROS production, and apoptosis
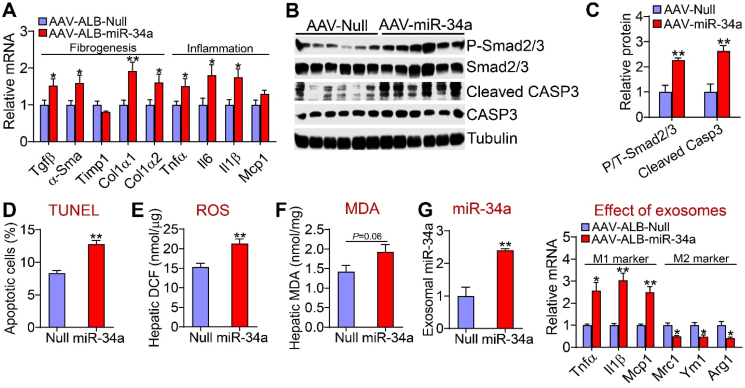
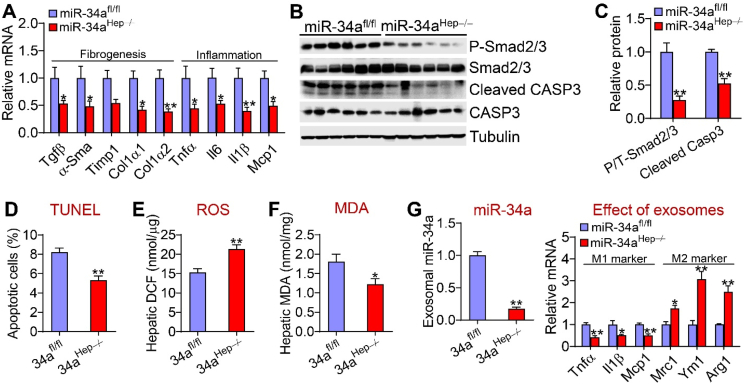
The data in Figure 1 indicate that hepatocyte miR-34a promoted steatohepatitis. Inflammation, ROS, and apoptosis are common mediators of NAFL transition to NASH. Hepatocyte-specific expression of miR-34a induced genes involved in inflammation [tumor necrosis factor a (Tnfa), interleukin 6 (Il6), and Il1b], fibrogenesis (transforming growth factor b (Tgfb), a-smooth muscle actin (a-Sma), collagen type 1 a1(Col1a1), and Col1a2] (Figure 3A). In line with increased Tgfb expression, miR-34a increased hepatic levels of phosphorylated Smad2/3 proteins and cleaved caspase 3 (CAPS3) by >2 fold (Figure 3B,C). TUNEL assays showed that miR-34a increased hepatic apoptosis (Figure 3D). In addition, miR-34a overexpression increased hepatic ROS levels (Figure 3E) and tended to increase hepatic malondialdehyde (MDA) levels (P = 0.06) (Figure 3F).
Figure 3.
Hepatocyte miR-34a induced Kupffer cell activation and hepatic apoptosis. The mice are described in the legend in Figure 1 (n = 8 per group). (A) Hepatic mRNA levels. (B and C) Hepatic proteins were analyzed by Western blotting assays (B) and protein levels were quantified (C). (D) Hepatic apoptotic cells (%) as analyzed by TUNEL assays. (E) Hepatic ROS levels. (F) Hepatic MDA levels. (G) Kupffer cells were treated with exosomes isolated from mice infected with AAV8-ALB-Null or AAV8-ALB-miR-34a. miR-34a levels in exosomes and mRNA levels in Kupffer cells were determined (n = 4). ∗P < 0.05 and ∗∗P < 0.01.
To further understand how hepatocyte miR-34a overexpression induces inflammation, we isolated exosomes from serum of mice infected with AAV8-ALB-miR-34a or AAV8-ALB-Null. The exosomes were then used to treat Kupffer cells. As shown in Figure 3G, exosomes isolated from mice overexpressing miR-34a significantly induced M1 macrophage markers (Tnfa, Il1b, and Mcp1) but repressed M2 macrophage markers [mannose receptor C type 1 (Mrc1), chitinase 3-like 3 (Ym1), and arginase 1 (Arg1)]. These data indicate that hepatocytes may secrete miR-34a to activate Kupffer cells for inflammatory responses.
Together, the data in Figure 3 indicate that miR-34a may promote the progression of NAFL to NASH via induction of inflammation, ROS production, and apoptosis.
3.3. Adult-onset loss of hepatocyte miR-34a was sufficient to prevent HFCF diet-induced development and progression of NAFLD
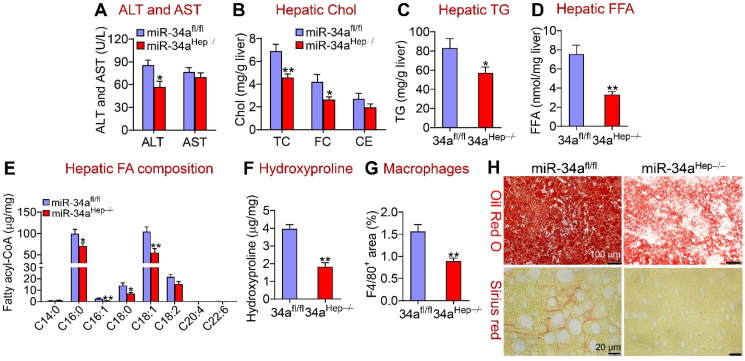
To investigate the effect of loss of hepatocyte miR-34a on diet-induced NAFLD, we used two mouse models, adult-onset miR-34a−/− (miR-34aHep−/−) mice and germline miR-34a−/− (miR-34agHep−/−) mice, which were created by i.v. injection of AAV8-TBG-Cre into miR-34afl/fl mice or crossbreeding miR-34afl/fl mice with albumin-Cre mice, respectively. These mice were then fed an HFCF diet for 16 weeks. Compared to the control mice, the adult-onset miR-34aHep−/− mice had no change in body fat content (Supplementary Fig. 2G) but exhibited a reduction in plasma ALT levels (Figure 4A), the ratio of liver weight to body weight (Supplementary Fig. 2H), and hepatic levels of TC, FC (Figure 4B), TG (Figure 4C) and FFAs (Figure 4D). miR-34aHep−/− mice also had reduced hepatic C16:0, C16:1, C18:0, and C18:1 fatty acyl-CoA levels (Figure 4E). Hepatic hydroxyproline levels were reduced by 54% (Figure 4F), and macrophage infiltration was reduced by 43% (Figure 4G) in the miR-34aHep−/− mice. Histological analyses by Oil Red O, Picrosirius red, or H&E staining and immunostaining further confirmed the results of our biochemical assays (Figure 4H and Supplementary Fig. 2I). Thus, adult-onset deletion of hepatocyte miR-34a prevented the development and progression of diet-induced NAFLD.
Figure 4.
Loss of hepatocyte miR-34a attenuated diet-induced steatohepatitis. The miR-34afl/fl mice and miR-34aHep−/− mice were fed an HFCF diet for 16 weeks (n = 7–8 per group). (A) Plasma ALT and AST levels. (B) Hepatic TC, FC, and CE levels. (C) Hepatic TG levels. (D) Hepatic FFA levels. (E) Hepatic individual fatty acyl-CoA levels. (F) Hepatic hydroxyproline levels. (G) Liver sections were immunostained using an F4/80 antibody and the F4/80-positive area was determined. (H) Representative liver images of Oil Red O staining (upper panel) or Picrosirius red staining (bottom panel). ∗P < 0.05 and ∗∗P < 0.01.
3.4. Loss of hepatocyte miR-34a prevented diet-induced lipid accumulation via repression of CYP7A1 and CYP8B1
In contrast to that observed in mice overexpressing hepatocyte miR-34a, the miR-34aHep−/− mice had reduced hepatic mRNA levels of Cyp7a1, Cyp8b1 (bile acid metabolism), Srebp2, Hmgcr (cholesterol synthesis), Acat1, Acat2 (cholesterol esterification), Srebp1c, Acc1, and Fasn (fatty acid synthesis) but increased hepatic expression of Abcg8 (cholesterol excretion), Ppara, Cpt2, and Pdk4 (FAO) (Figure 5A,B). The miR-34aHep−/− mice also had reduced hepatic CYP7A1, CYP8B1, ACC, and SREBP1 protein levels (Figure 5C,D), BA pool size, and plasma BA levels (Figure 5E). The BA composition did not change (Supplementary Fig. 2J) but there was a small decrease in HI in the miR-34aHep−/− mice (Supplementary Fig. 2K). Consistent with these data, the miR-34aHep−/− mice had reduced intestinal fat absorption (Figure 5F), cholesterol absorption (Figure 5G), and hepatic TG synthesis (Figure 5H) but increased hepatocyte FAO (Figure 5I). The increase in hepatocyte FAO was also consistent with an increase in plasma b-HB levels (Supplementary Figure 2L).
Figure 5.
Loss of hepatocyte miR-34a inhibited lipid absorption and synthesis and induced FAO. (A-I) The mice are described in the legend in Figure 4 (n = 7–8 mice per group). Hepatic mRNA (A and B) and protein levels were determined (C and D). BA pool size (left panel) and plasma BA levels were analyzed (E). Intestinal fat absorption (F), intestinal cholesterol absorption (G), and hepatic newly synthesized TG levels (H) were measured. FAO was determined using hepatocytes isolated from the miR-34afl/fl mice or miR-34aHep−/− mice (I). (J-M) The miR-34afl/fl mice and miR-34aHep−/− mice were i.v. injected with 0.5 × 10ˆ11 GC AAV8-ALB-Null or AAV8-ALB-CYP7A1 plus AAV8-ALB-CYP8B1 (7A1 + 8B1). These mice were then fed an HFCF diet for 16 weeks (n = 8 per group). Hepatic TC (J), FC (K), TG (L), and FFA (M) levels were determined. ∗P < 0.05 and ∗∗P < 0.01.
To investigate the relative contribution of BA signaling to the phenotype observed in the miR-34aHep−/− mice, we normalized hepatic CYP7A1 and CYP8B1 protein levels by i.v. injecting AAV8-ALB-CYP7A1 and AAV8-ALB-CYP8B1 into the miR-34aHep−/− mice, which were then fed an HFCF diet to induce steatohepatitis. Recapitulation of hepatic CYP7A1 and CYP8B1 expression normalized hepatic TC, FC, and TG levels (Figure 5J-L) but had little effect on hepatic FFA levels (Figure 5M). These data suggest that BA signaling played a role in hepatic lipid metabolism in the miR-34aHep−/− mice.
3.5. Hepatocyte miR-34a ablation inhibited inflammation, ROS production, and apoptosis
The data in Figure 5 show a change in lipid and BA metabolism in the miR-34aHep−/− mice. These mice also had reduced hepatic expression of genes involved in inflammation (Tnfa, Il6, Il1b, and Mcp1) or fibrogenesis (Tgfb, a-Sma, Timp1, Col1a1, and Col1a2) (Figure 6A) as well as phosphorylated Smad2/3 and cleaved caspase 3 levels (Figure 6B,C). The miR-34aHep−/− mice also had reduced hepatic apoptosis (Figure 6D), ROS levels (Figure 6E), and MDA levels (Figure 6F). When exosomes isolated from the miR-34aHep−/− mice were incubated with Kupffer cells, M1 macrophage markers (Tnfa, Il1b, and Mcp1) were repressed whereas M2 macrophage markers (Mrc1, Ym1, and Arg1) were induced (Figure 6G). Normalization of hepatic CYP7A1 and CYP8B1 expression in the miR-34aHep−/− mice had no effect on hepatic fibrogenesis or apoptosis (Supplementary Fig. 3A–I). Therefore, hepatocyte miR-34a ablation inhibited intestinal lipid absorption and hepatic TG synthesis, inflammation, ROS production, and apoptosis but induced hepatic FAO, which may account for the mechanism by which hepatocyte miR-34a ablation protects against the development of steatohepatitis. Our data also show that BA signaling plays a role in protecting against NAFL but not NASH in miR-34aHep−/− mice. Interestingly, miR-34a did not regulate Cyp7a1 or Cyp8b1 3′UTR activity (Supplementary Fig. 4A and B), suggesting that miR-34a indirectly regulated Cyp7a1 or Cyp8b1 gene expression.
Figure 6.
Loss of hepatocyte miR-34a inhibited Kupffer cell activation and apoptosis. The mice are described in the legend in Figure 4 (n = 7–8 mice per group). (A) Hepatic mRNA levels. (B and C) Hepatic protein levels. (D) Hepatic apoptotic cells (%) as determined by TUNEL assays. (E) Hepatic ROS levels. (F) Hepatic MDA levels. (G) Kupffer cells were incubated with exosomes isolated from the miR-34afl/fl mice or miR-34aHep−/− mice. miR-34a levels in exosomes and mRNA levels in Kupffer cells were determined (n = 4). ∗P < 0.05 and ∗∗P < 0.01.
3.6. Germline deletion of hepatocyte miR-34a protected against HFCF diet-induced steatohepatitis
To investigate whether germline deletion of hepatocyte miR-34a had similar effects, we also fed the miR-34agHep−/− mice an HFCF diet to induce steatohepatitis or a chow diet to establish the baseline levels. On a chow diet, there was a decrease in hepatic FFA levels in the miR-34agHep−/− mice whereas hepatic TC, FC, or TG levels or BA pool size did not change between the two groups (Supplementary Fig. 5A–D). When fed an HFCF diet, germline deletion of hepatocyte miR-34a did not affect body fat content (Supplementary Fig. 6A) but reduced plasma ALT and AST levels (Supplementary Fig. 6B) and the liver-to-BW ratio (Supplementary Fig. 6C). The miR-34agHep−/− mice also had reduced hepatic levels of TC, FC, TG, FFAs, and hydroxyproline (Supplementary Figs. 6D–G), which were further confirmed by Oil Red O or Picrosirius red staining (Supplementary Fig. 6H).
The miR-34agHep−/− mice also had increased plasma b-HB levels (Supplementary Fig. 7A). At the gene expression levels, the miR-34agHep−/− mice had reduced hepatic expression of genes involved in bile acid synthesis (Cyp7a1 and Cyp8b1), cholesterol biosynthesis (Srebp2 and Hmgcr), fatty acid synthesis (Srebp1c, Acc1, and Fasn), cholesterol esterification (Acat1 and Acat2), inflammation (Tnfa, Il6, Il1b, and Mcp1), and fibrogenesis (Tgfb, a-Sma, Col1a1, and Col1a2) but induced Abcg5, Abcg8, and Pdk4 expression in the liver (Supplementary Figs. 7B–D). Consistent with the changes in gene expression, the miR-34agHep−/− mice had reduced hepatic levels of ROS, MDA, and apoptosis (Supplementary Figs. 7E–H). Thus, germline or adult-onset deletion of miR-34a in hepatocytes had similar effects on preventing diet-induced development and progression of NAFLD.
3.7. Pharmacological inhibition of miR-34a reversed diet-induced steatohepatitis
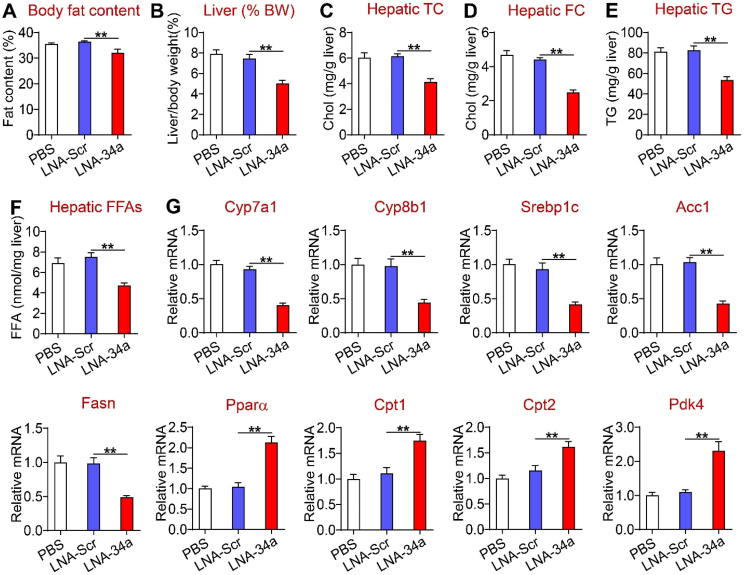
As deficiency in hepatocyte miR-34a prevents diet-induced steatohepatitis, next we determined whether pharmacological inhibition of miR-34a could reverse diet-induced steatohepatitis. The C57BL/6 mice were fed an HFCF diet for 16 weeks. In the last 5 weeks of the feeding study, the mice were i.p. injected with PBS or locked nucleic acid (LNA) against scramble sequences (LNA-Scr) or miR-34a (LNA-miR-34a) once a week. MiR-34a inhibition reduced body weight (Supplementary Fig. 8A), body fat content (Figure 7A), the liver-to-BW ratio (Figure 7B), and hepatic levels of TC, FC, TG, and FFAs (Figure 7C–F). Consistent with these findings, miR-34a inhibitors reduced hepatic expression of genes involved in BA synthesis (Cyp7a1 and Cyp8b1) or fatty acid synthesis (Srebp1c, Acc1, and Fasn) but induced genes involved in FAO (Ppara, Cpt1, Cpt2, and Pdk4). miR-34a inhibitors also reduced BA pool size (Supplementary Fig. 8B).
Figure 7.
Pharmacological inhibition of miR-34a ameliorated liver steatosis. The C57BL/6 mice were fed an HFCF diet for a total of 16 weeks. In the last 5 weeks of the feeding study, the mice were i.p. injected with PBS, locked nucleic acids (LNA) against scramble sequences (LNA-Scr), or miR-34a (LNA-miR-34a) once a week (10 mg/kg). (A) Body fat content. (B) The ratio of liver weight to body weight. (C) Hepatic TC levels. (D) Hepatic FC levels. (E) Hepatic TG levels. (F) Hepatic FFA levels. (G) Hepatic mRNA levels. ∗∗P < 0.01.
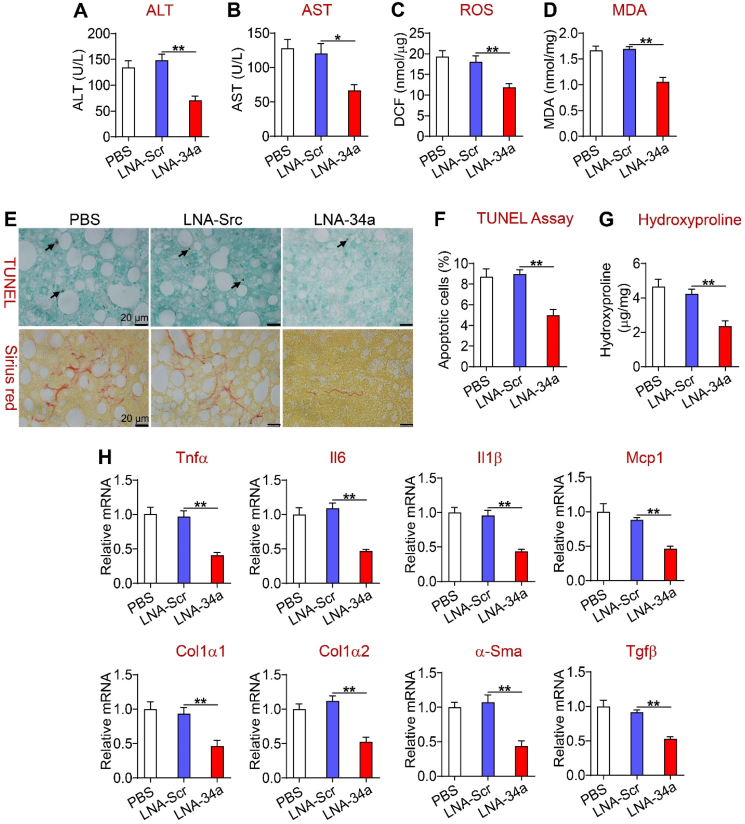
Importantly, miR-34a inhibitors significantly reduced plasma ALT and AST levels (Figure 8A,B) and hepatic levels of ROS (Figure 8C), MDA (Figure 8D), apoptosis, and fibrosis (Figure 8E–G). In support of these findings, miR-34a inhibitors reduced hepatic expression of genes involved in inflammation (Tnfa, Il6, Il1b, and Mcp1) and fibrogenesis (Tgfb, a-Sma, Col1a1, and Col1a2) by > 50% (Figure 8H). Thus, pharmacological inhibition of miR-34a improved steatohepatitis.
Figure 8.
Pharmacological inhibition of miR-34a reversed steatohepatitis. The mice are described in the legend in Figure 7. (A) Plasma ALT levels. (B) Plasma AST levels. (C) Hepatic ROS levels. (D) Hepatic MDA levels. (E) Representative images of liver sections of TUNEL assays or Picrosirius red staining. (F) Hepatic apoptotic cells (%). (G) Hepatic hydroxyproline levels. (H) Hepatic mRNA levels. ∗P < 0.05 and ∗∗P < 0.01.
4. Discussion
Hepatic miR-34a expression is known to be induced under metabolic stress [1,2] but hepatocyte miR-34a's role in the progression of NAFL to NASH has not been previously explored. In this report, we used gain- and loss-of-function approaches to demonstrate that hepatocyte miR-34a is a key regulator of the development and progression of NAFLD. Mechanistically, hepatocyte miR-34a promotes NAFL by inducing lipid absorption and synthesis and inhibiting FAO. MiR-34a also promotes the transition of NAFL to NASH by inducing inflammation, ROS production, and apoptosis. Importantly, pharmacological inhibition of miR-34a reverses diet-induced NAFL and NASH.
Our data show that miR-34a induces the lipogenic program but inhibits FAO. Previous research suggested that the miR-34a-sirtuin 1 (SIRT1)-SREBP1c pathway may contribute to lipogenesis [13,14], in which miR-34a represses SREBP1C activity by inhibiting SIRT1. SIRT1 also induces genes involved in FAO (PPARa, CPT, and PDK4) [15]. However, whether miR-34a induces SREBP-1c and represses genes involved in FAO via SIRT1 remains to be determined. In addition, miR-34a is shown to directly target PPARa to inhibit its activity [16]. In contrast, miR-34a does not regulate lipolysis. Interestingly, our data showed that restoration of hepatic CYP7A1 and CYP8B1 expression did not affect genes involved in FAO but was sufficient to normalize hepatic TG and cholesterol levels in the miR-34aHep−/− mice, suggesting that BA-mediated intestinal lipid absorption was likely responsible for hepatocyte miR-34a to induce steatosis. Fu et al. showed that miR-34a inhibits the expression of b-klotho, the co-receptor of FGF15/19 receptor 4, to attenuate hepatic response to FGF15/19, resulting in an induction of hepatic CYP7A1 and CYP8B1 expression [2]. We showed that miR-34a does not regulate CYP7A1 and CYP8B1 3′UTR activity. Thus, miR-34a regulates CYP7A1 and CYP8B1 likely via inhibition of b-klotho.
The progression of NAFL to NASH needs a second “hit,” such as inflammation, lipotoxicity, and apoptosis, among others. In macrophages, miR-34a is shown to promote M1 macrophage polarization via inhibition of Kruppel-like factor 4 (KLF4) and liver X receptor a (LXRa) [4]. Our data suggest that hepatocytes may regulate Kupffer cell activation via exosomal delivery of miR-34a, which is supported by the finding that exosomes isolated from mice overexpressing or deficient in hepatocyte miR-34a regulate macrophage inflammation. In addition to uptaking exosomes, macrophages may also secrete exosomes that carry endogenous miR-34a. The net miR-34a levels in Kupffer cells may eventually determine the levels of secreted inflammatory cytokines. The role of FC and palmitate in the pathogenesis of NASH has been well established [[17], [18], [19]]. Our data show that miR-34a increases hepatic FC and palmitate levels, which can cause lipotoxicity and the progression of NAFLD. The increased hepatic FC levels may result from decreased cholesterol excretion and increased cholesterol synthesis. LXRs, which are inhibited by miR-34a likely via SIRT1 [20], are known to induce ABCG5 and ABCG8 expression [21]. Therefore, miR-34a inhibits cholesterol excretion likely through suppression of LXR activity. It remains unclear how miR-34a induces genes involved in cholesterol synthesis. One possibly is that miR-34a promotes conversion of FC to bile acids by inducing CYP7A1 and CYP8B1 expression, which leads to a transient reduction in FC levels and subsequent induction of SREBP2 processing and its target genes, including HMGCR.
Previous studies demonstrated that miR-34a promotes apoptosis in various cell types [[22], [23], [24]]. However, the underlying mechanisms are not fully understood. One potential mechanism may involve the miR-34a-SIRT1-p53 pathway, in which miR-34a induces acetylation of p53 by inhibiting SIRT1 [14]. TGFb is shown to mediate hepatocyte apoptosis via Smad3 generation of ROS [25]. Our studies showed that miR-34a activates hepatic TGFb signaling, which may also account for the induction of apoptosis by miR-34a in hepatocytes. Thus, hepatocyte miR-34a promotes the progression of NAFL to NASH likely through, at least in part, induction of Kupffer cell activation/inflammation, lipotoxicity, and apoptosis. Following activation by hepatocyte miR-34a, Kupffer cells can secrete various cytokines, such as TGFb and TNFa, among others. These cytokines may work with ROS and other mediators to activate stellate cells. Whether miR-34a can directly regulate stellate cell activation remains to be determined.
In summary, we used both gain- and loss-of-function approaches to demonstrate that hepatocyte miR-34a is a key player in the development and progression of NAFLD. Since hepatocyte-specific or pharmacological inhibition of miR-34a improves diet-induced steatohepatitis, our data suggest that miR-34a is an attractive target for treating NAFLD.
Author contributions
Y.X., Y.Zhu, L.Y., and Y.Z. designed the studies. Y.X., Y.Zhu, S.H., X.P., and F.C.B. conducted the studies. H.H.W and D.Q-H.W. performed the bile acid composition assays. Y.X. and Y.Z. wrote the manuscript. All of the authors reviewed and approved the manuscript.
Acknowledgments
This study was supported by NIH grants R01DK102619, R01HL142086, R01DK118941, and R01DK118805.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101244.
Conflicts of interest
The authors have no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Xu Y., Zalzala M., Xu J., Li Y., Yin L., Zhang Y. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nature Communications. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu T., Choi S.E., Kim D.H., Seok S., Suino-Powell K.M., Xu H.E. Aberrantly elevated microRNA-34a in obesity attenuates hepatic responses to FGF19 by targeting a membrane coreceptor beta-Klotho. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(40):16137–16142. doi: 10.1073/pnas.1205951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiological Reviews. 2009;89(1):147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y., Xu Y., Zhu Y., Sun H., Juguilon C., Li F. Macrophage miR-34a is a key regulator of cholesterol Efflux and atherosclerosis. Molecular Therapy. 2020;28(1):202–216. doi: 10.1016/j.ymthe.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y., Li Y., Jadhav K., Pan X., Zhu Y., Hu S. Hepatocyte ATF3 protects against atherosclerosis by regulating HDL and bile acid metabolism. Nature Metabolism. 2021;3(1):59–74. doi: 10.1038/s42255-020-00331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li P.Z., Li J.Z., Li M., Gong J.P., He K. An efficient method to isolate and culture mouse Kupffer cells. Immunology Letters. 2014;158(1–2):52–56. doi: 10.1016/j.imlet.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 8.Rossi S.S., Converse J.L., Hofmann A.F. High pressure liquid chromatographic analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. The Journal of Lipid Research. 1987;28(5):589–595. [PubMed] [Google Scholar]
- 9.Wang D.Q., Lammert F., Paigen B., Carey M.C. Phenotypic characterization of lith genes that determine susceptibility to cholesterol cholelithiasis in inbred mice. Pathophysiology of biliary lipid secretion. The Journal of Lipid Research. 1999;40(11):2066–2079. [PubMed] [Google Scholar]
- 10.Turley S.D., Herndon M.W., Dietschy J.M. Reevaluation and application of the dual-isotope plasma ratio method for the measurement of intestinal cholesterol absorption in the hamster. The Journal of Lipid Research. 1994;35(2):328–339. [PubMed] [Google Scholar]
- 11.Li Y., Zalzala M., Jadhav K., Xu Y., Kasumov T., Yin L. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology. 2016;63(6):1860–1874. doi: 10.1002/hep.28472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Ge X., Heemstra L.A., Chen W.D., Xu J., Smith J.L. Loss of FXR protects against diet-induced obesity and accelerates liver carcinogenesis in ob/ob mice. Molecular Endocrinology. 2012;26(2):272–280. doi: 10.1210/me.2011-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponugoti B., Kim D.H., Xiao Z., Smith Z., Miao J., Zang M. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. Journal of Biological Chemistry. 2010;285(44):33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhart-Hines Z., Rodgers J.T., Bare O., Lerin C., Kim S.H., Mostoslavsky R. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. The EMBO Journal. 2007;26(7):1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding J., Li M., Wan X., Jin X., Chen S., Yu C. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Scientific Reports. 2015;5:13729. doi: 10.1038/srep13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannou G.N. The role of cholesterol in the pathogenesis of NASH. Trends in Endocrinology and Metabolism. 2016;27(2):84–95. doi: 10.1016/j.tem.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 18.McGettigan B., McMahan R., Orlicky D., Burchill M., Danhorn T., Francis P. Dietary lipids differentially shape nonalcoholic steatohepatitis progression and the transcriptome of Kupffer cells and infiltrating macrophages. Hepatology. 2019;70(1):67–83. doi: 10.1002/hep.30401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marra F., Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. Journal of Hepatology. 2018;68(2):280–295. doi: 10.1016/j.jhep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Li X., Zhang S., Blander G., Tse J.G., Krieger M., Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Molecules and Cells. 2007;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 21.Repa J.J., Berge K.E., Pomajzl C., Richardson J.A., Hobbs H., Mangelsdorf D.J. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. Journal of Biological Chemistry. 2002;277(21):18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- 22.Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Molecules and Cells. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Molecules and Cells. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Tarasov V., Jung P., Verdoodt B., Lodygin D., Epanchintsev A., Menssen A. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6(13):1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 25.Black D., Lyman S., Qian T., Lemasters J.J., Rippe R.A., Nitta T. Transforming growth factor beta mediates hepatocyte apoptosis through Smad3 generation of reactive oxygen species. Biochimie. 2007;89(12):1464–1473. doi: 10.1016/j.biochi.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.