Abstract
The ongoing COVID-19 pandemic increases the consumption of antimicrobial substances (ABS) due to the unavailability of approved vaccine(s). To assess the effect of imprudent consumption of ABS during the COVID-19 pandemic, we compare the 2020 prevalence of antidrug resistance (ADR) of Escherichia coli (E. coli) with a similar survey carried out in 2018 in Ahmedabad, India using SARS-CoV-2 gene detection as a marker of ABS usage. We found a significant ADR increase in 2020 compared to 2018 in ambient water bodies, harbouring a higher incidence of ADR E.coli towards non-fluoroquinolone drugs. Effective SARS-CoV-2 genome copies were found to be associated with the ADR prevalence. The prevalence of ADR depends on the efficiency of WWTPs (Wastewater Treatment Plants) and the catchment area in its vicinity. In the year 2018 study, prevalence of ADR was discretely distributed, and the maximum ADR prevalence recorded was ~ 60%; against the current homogenous ADR increase, and up to 85% of maximum ADR among the incubated E.coli isolated from the river (Sabarmati) and lake (Chandola and Kankaria) samples. Furthermore, wastewater treatment plants showed less increase in comparison to the ambient waters, which eventually imply that although SARS-CoV-2 genes and faecal pollution may be diluted in the ambient waters, as indicated by low Ct-value and E.coli count, the danger of related aftermath like ADR increase cannot be nullified. Also, Non-fluoroquinolone drugs exhibited overall more resistance than quinolone drugs. Overall, this is probably the first-ever study that traces the COVID-19 pandemic imprints on the prevalence of antidrug resistance (ADR) through wastewater surveillance and hints at monitoring escalation of other environmental health parameters. This study will make the public and policyholders concerned about the optimum use of antibiotics during any kind of treatment.
Keywords: Antidrug resistance, SARS-CoV-2, COVID-19, Wastewater based epidemiology
Graphical Abstract
1. Introduction
The exponential rise in the consumption of antimicrobials in various applications such as medical, veterinary, domestic and agricultural and their leak to aquatic ecosystems has caused the global prevalence of antidrug resistance (ADR), which is being considered a major threat to public health (Rodriguez-Mozaz et al., 2015, Chatterjee et al., 2010, Baker-Austin et al., 2006). The ADR is not only limited to the survival and infection by any particular type of microorganism, but can lead to life threatening diseases for both animals and human (Singer et al., 2008, Ferreira da Silva et al., 2007, Jiang et al., 2013). Due to lack of regulations on the prescription and non-prescription use of antimicrobials, its consumption rate in, for example, India has been increased by 105% from 2000 to 2015 while worldwide it is estimated to increase by 63% during 2010–2030 (Klein et al., 2018, Global Antibiotic Resistance Partnership (GARP)-India Working Group, 2011, Van Boeckel et al., 2015). On top of that, the rate of consumption of certain antimicrobials has escalated during the COVID-19 pandemic in an effort to minimise the risk of severe infections and mortality (Miranda et al., 2020, Liu et al., 2020). Around 70% of COVID-19 patients have received antimicrobial treatment along with overuse of various antibiotics despite only 10% on average show microbial infections (Hsu, 2020, Rawson et al., 2020). As most of the consumed drugs and their metabolites are excreted through urine and faeces, their discharge to aquatic environments depends on the removal efficiency of the WWTPs (Singer et al., 2008, Azuma et al., 2012, Takanami et al., 2010, Auerbach et al., 2007, Kumar et al., 2020a). If the WWTP clearing rate is low, microorganisms exposed to antimicrobials and metabolites develops mutations causing ADR (Aali et al., 2014, Alexander et al., 2020, Guo et al., 2018, Kumar et al., 2020a, Kumar et al., 2021) Thus, the increased use of antimicrobials in the current pandemic will probably pose an increased risk in terms of ADR during post COVID-19 as concerned by a number of recent studies (Kuroda et al., 2021; Lucien et al., 2021; Hsu, 2020; Kumar et al., 2020a; Asaduzzaman et al., 2020).
The high consumption of antimicrobials causes an increase in the prevalence of ADR in several environmental compartments including drinking, waste and groundwater, sludge, sediments and municipal solid waste leachate (Al-Judaibi, 2014, Ferreira da Silva et al., 2007, Kumar et al., 2020d, Kumar et al., 2020e, Ram and Kumar, 2020, Zhang et al., 2015, Storteboom et al., 2010, Threedeach et al., 2012). In the case of for example E.coli isolates from the effluent of WWTPs have shown a higher prevalence of antidrug resistance as compared to the influent, which is probably due to poor treatment conditions, prolonged microbial activities, and chemical properties of the antimicrobial drugs (Reinthaler et al., 2003, Silva et al., 2006, Miranda and Castillo, 1998, Marcinek et al., 1998). Specifically, the conventional treatment processes at WWTPs do not completely mineralise the parent antimicrobial drugs, and generate some residues, metabolites or transformation products that may have the same biological activity as the parent drugs (Zhang et al., 2015, Kumar et al., 2021). Thus, WWTPs are considered hotspots for the spreading ADR due to high microbial density, horizontal gene transfer (HGT), nutritional richness and the availability of antimicrobial metabolites (Zhang et al., 2015, Threedeach et al., 2012, Silva et al., 2006). Previous studies have reported a correlation between the prevalence of ADR and inefficiently treated wastewater discharge, having the abundance of E. coli, extravasating to river and lake waters (Na et al., 2018, Yang et al., 2017, Honda et al., 2016, Honda et al., 2018, Biswas et al., 2015, Akhter et al., 2014, Ram and Kumar, 2020, Kumar et al., 2020d, Kumar et al., 2020e). Thus, a better understanding of the occurrence, distribution and frequency of antidrug resistance in the urban waters is needed to prevent or slower the rate of increase in ADR.
With the same purpose, presumptive actions are needed to study the prevalence of the ADR during wastewater treatment and the water bodies receiving the WWTP effluents. Wastewater based epidemiology (WBE) is an efficient way to trace the prevalence of ADR in highly COVID -19 infected areas, which are potentially major zones of high consumption of drugs, can be identified with the help of the WBE approach for tracing the SARS-CoV-2 genome concentration in wastewaters (Kumar et al., 2020b). Also, with the help of authorised software and apps (for example: Arogya-Setu app in India), the infected population within a certain region can be predicted. Identifying the WWTPs in such infected areas aids in correlating ADR with the elevated cases of COVID-19. Therefore, the impact of such highly contaminated zones on the prevalence of ADR in wastewaters needs to be studied well.
ADR is not included in the water quality standards and guidelines of India mostly due to the lack of proper treatment facilities in many cities where domestic wastewater is directly discharged to aquatic environments (IS10500, 2012). In this study, we select the Ahmedabad City of Gujarat Province in western India with a population of 5.6 million (2011 Census) to assess the prevalence of ADR in WWTP, lake and river locations within various zones of the city. The specific objectives of the present study are: i) to compare and discuss the prevalence of E. coli in the surface water and wastewater in Ahmedabad in order to have a prior knowledge of ADR pervasiveness in different compartments, ii) to analyse a comparative status of the antidrug resistance in the E. coli isolated from the urban waters of the city and iii) to further understand the imprints of COVID-19 situation on the status of SARS-CoV-2 genome concentration and ADR prevalence at various zones of the city.
2. Material and methods
2.1. Sample collection and ADR analyses
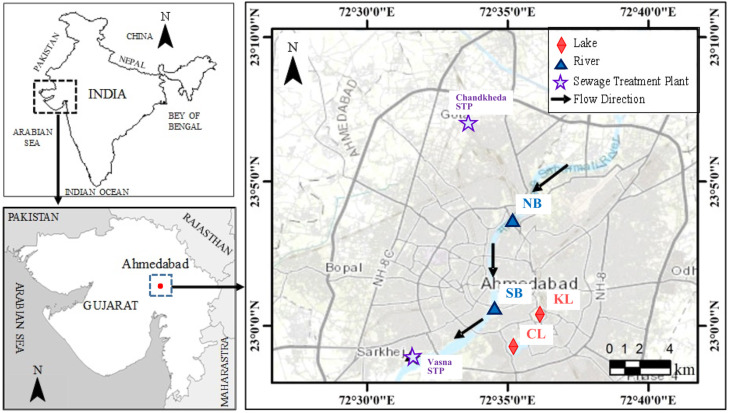
The water samples were collected from 6 different locations of Ahmedabad city on 23rd June 2018, and 16th October 2020 ( Fig. 1). Two locations on the stretch of Sabarmati river: Nehru Bridge (NB) and Sardar Bridge (SB); two lakes: Kankariya Lake (KL) and Chandola Lake (CL), and two WWTP locations: Chandkheda (inlet: CI and outlet: CO) and Vasna, also known as Juhapura (inlet: VI and outlet: VO), selected to assess ADR. For SARS-CoV-2 gene detection, a total of 10 locations were selected to represent various zones of the city that comprises all ADR sampling locations. We kept ADR locations low to match the number of locations tested in 2018 (Ram and Kumar, 2020). The geographical details about the selected locations are well described in our previous study by Ram and Kumar (2020) (See Supplementary Information). Sterile bottles (Tarson-546041) of medical grade were used to collect the samples, which were then kept in iceboxes until arrival at the laboratory. For on-site measurement of pH, EC, ORP, TDS and salinity, a multi-parameter probe, HANNA HI9828 was used. The procedure for testing the isolation of E. coli for ADR is likewise described in Ram and Kumar (2020) (See Supplementary information). Briefly, the water samples were filtered through membranes with 0.45-µm-pore size, and E. coli trapped by the membranes were incubated on Chromocult® Coliform Agar ES (Merck Microbiology, Darmstadt, Germany). Each E. coli isolate was tested for susceptibility to six antibiotics (kanamycin, KM; tetracycline, TC; norfloxacin, NFX; ciprofloxacin, CIP; levofloxacin, LVX; and sulfamethoxazole, ST) by Kirby-Bauer method using PERLCORE® Sensitivity Test (ST) Agar (EIKEN Chemical Co., Ltd, Tokyo).
Fig. 1.
Map showing the sampling locations in Ahmedabad, Gujarat (i) locations at Sabarmati River (Nehru Bridge: NB; Sardar Bridge: SB), (ii) two lakes (Kankaria Lake: KL; Chandola Lake: CL) and (iii) two different Sewage Treatment Plants (STPs) (Chandkheda STP; Vasna STP).
2.2. SARS-CoV-2 RNA detection
The SARS-CoV-2 RNAs were isolated and detected from 30 mL wastewater samples that were centrifuged at 4000g for 40 min, followed by filtration of supernatant using 0.22-micron syringe filter (Mixed cellulose esters syringe filter, Himedia). After filtration, 25 mL of the supernatant was treated with polyethylene glycol and NaCl at 80 g/L and 17.5 g/L respectively, and incubated at 17 °C, 100 rpm overnight. The mixture was centrifuged for 90 min at 14000g and the supernatant was discarded to collect a pellet containing viruses and their fragmented genes. The pellet was re-suspended in 300 µl RNase-free water and kept in 1.5 mL vials at − 40 °C, until further analyses.
RNA isolation from the pellet with the concentrated virus was performed using NucleoSpin® RNA Virus isolation kit (Macherey-Nagel GmbH & Co. KG, Germany). The samples were spiked with MS2 phage as an internal control prior to the RNA extraction provided by TaqPathTM Covid-19 RT-PCR Kit. The nucleic acid was extracted and a Qubit 4 Fluorometer (Invitrogen) was used for RNA concentrations estimation. The molecular process inhibition control was evaluated through the MS2 phage for QA/QC analyses of nucleic acid extraction and PCR inhibition (Haramoto et al., 2018). We have described the methodologies in Kumar et al., 2021, Kumar et al., 2020b.
Briefly, steps were carried out as per the guideline provided with the product manual of Macherey-Nagel GmbH & Co. KG and RNAs were detected using real-time PCR (RT-PCR). An Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (version 2.19 software) was used for SARS-CoV-2 gene detection. A template of 7 µl of extracted RNA was used in each reaction with TaqPath™ 1-Step Multiplex Master Mix (Thermofischer Scientific, USA). Three controls were included: positive control (TaqPath™ COVID-19 Control); negative control (from extraction run spiked with MS2); and a no template control (NTC). Finally, results were interpreted using Applied Biosystems Interpretive Software, and Ct values for three target genes, i.e., ORF1ab, N Protein, and S Protein of SARS-CoV-2, were detected along with MS2 as an internal control.
The samples were considered as positive if at least two genes showed amplification. The average Ct-value of a given sample was then converted to gene copy numbers considering the equivalence of 500 copies of SARS-CoV-2 genes as 26 Ct-value (provided with the kit), and the same was extrapolated to derive approximate copies of each gene. The average effective genome concentration present in a given sample was calculated by multiplying the RNA amount used as a template with the enrichment factor for each sample.
2.3. Quality assurance/quality control (QA/QC) and statistical analysis
To determine the contamination occurred during transport, blanks in the same type of bottle were analysed prior to sampling. Duplicate analysis of samples was conducted to check accuracy and precision. To ensure instrument sensitivity and check cross-contamination, blanks were run for each batch of five samples. Signals were considered significant if the signal-to noise ratio was more than three. The limit of quantification (LOQ) of the overall method was defined as sample concentration equivalent to 1 copy per reaction tube, which was 1.7 × 102 copies/L. We have calculated the gene copy numbers based on the positive control provided with kit i.e., 104 copies/µl and the final concentration of 25 copies per reaction. Based on our experience, the same positive control is providing the same Ct values for all 3 genes analysed in this study. Hence, it is evident that primer efficiency is more or less same. Relative to the Ct values of genes of positive controls, copy numbers have been calculated in test samples of different sources.
ADR analyses were carried out in triplicate for the accuracy and precision of the data generated. Tests were repeated if the standard deviation between the triplicate was higher than 10%. Statistical analysis by Student t-test was done to compare the antidrug resistance caused by all six antibiotics in year 2018 and 2020, and the results were represented by Pearson’s correlation coefficient (p), whose value ranges between zero to unity. The change in percentage resistance of more than 90% (p = 0.10) was considered significant.
3. Results and discussions
3.1. Comparison of prevalence of E. coli
The prevalence of E. coli and environmental parameters is summarised in Table 1. In 2018, the E. coli count was highest in river sampling locations, with maximum count of 76,600 cfu (colonies forming unit) mL-1, which was the highest among the lake and WWTP locations except for the Vasana STP. This critically high prevalence is due to the river-human interactions at the riverfront, wastewater discharge or the stagnant flow conditions near the sampling locations (Pormohammad et al., 2019). This reported prevalence in the Sabarmati River was higher than the reported prevalence in rivers of tropical countries like India and Thailand (Chatterjee et al., 2010; Kumar and Sharma, 2014; Honda et al., 2016, Honda et al., 2018; Hamner et al., 2007; Hu et al., 2008). The higher recreational activities at KL location as compared to the CL location are the main cause of higher E. coli prevalence at KL (15,600 cfu mL-1) than CL (3467 cfu mL-1) (Kumar and Sharma, 2014; Ram and Kumar, 2020). The varying E. coli prevalence at STP locations (inlet and outlet) in 2018 indicates the varying amount of incoming faecal contamination and reduction ratios in the STP.
Table 1.
Sampling locations along with in-situ water quality (pH, EC, TDS, ORP and salinity) and prevalence of E. coli in 2018 and 2020.
| Sampling Location | Year | pH | EC | TDS | ORP | Salinity | E. coli |
|---|---|---|---|---|---|---|---|
| Nehru Bridge (NB) | 2018 | 8.4 | 1320 | 1090 | -16 | 691 | 24,267 |
| 2020 | 7.67 | 554 | 343 | 123.5 | 0.25 | 1400 | |
| Sardar Bridge (SB) | 2018 | 8.00 | 1541 | 1100 | 2 | 691 | 76,600 |
| 2020 | 7.30 | 533 | 352 | 115.7 | 0.27 | 5200 | |
| Kankaria Lake (KL) | 2018 | 8.70 | 3015 | 2050 | 13 | 1350 | 15,333 |
| 2020 | 8.58 | 5934 | 3323 | 30.9 | 2.71 | 13,100 | |
| Chandola Lake (CL) | 2018 | 8.10 | 3240 | 2300 | 29 | 1510 | 3467 |
| 2020 | 7.86 | 1014 | 590 | 43 | 0.44 | ND | |
| Chandkheda Inlet (CI) | 2018 | 6.70 | 2100 | 1480 | -274 | 972 | 4220 |
| 2020 | 6.85 | 3745 | 2324 | -238.6 | 1.87 | 950,000 | |
| Chandkheda Outlet (CO) | 2018 | 7.30 | 1620 | 1400 | -57 | 911 | 2893 |
| 2020 | 7.52 | 3624 | 2249 | 118.6 | 1.81 | 32,500 | |
| Vasna Inlet (VI) | 2018 | 6.60 | 1500 | 1060 | -117 | 674 | 96,393 |
| 2020 | 6.97 | 3254 | 2017 | -231.7 | 1.61 | 4,000,000 | |
| Vasna Outlet (VO) | 2018 | 6.90 | 1506 | 1070 | -193 | 670 | 9467 |
| 2020 | 7.34 | 2767 | 1715 | 90.3 | 1.36 | 19,500 | |
| ND: Not Detected | Unit | – | µS cm-1 | mg L-1 | mV | ppt | cfu mL-1 |
In the year 2020, the E. coli prevalence at STP locations was higher than in 2018 samples ranging from 950,000 to 400,000 cfu mL-1 at inlet locations and 19,500–32,500 cfu mL-1 at outlet locations. This is attributed to the increased domestic wastewater discharge from the Covid-19 lockdown which also increased the burden on municipal WWTPs resulting in less removal of E. coli in WWTPs. It is worth noting that, this critical E. coli prevalence alarms the municipal authorities to advance the disinfection processes in WWTPs, and therefore potential human health effects could be reduced (Pormohammad et al., 2019). Whereas, the reduced E. coli prevalence in the Sabarmati river, can be attributed to the improved water quality and attenuation capacity of the river due to less human and industrial interaction. Another reason can be the dilution level of the samples collected from the river than that of the lake and WWTPs (Pormohammad et al., 2019).
3.2. Mechanism and pathways of antibiotic resistance
Though antimicrobials and antibiotics are among the essential medical interventions, increased antimicrobial resistance threatens the success of patient treatment. Antibiotic resistance has been listed as one of the three major threats to the public health in 21st century by the world health organisation (WHO) (World Health Organization, 2014). Thus, to understand and reduce the consequences of antibiotic resistance, we need to understand its mechanism. Antimicrobial resistance is expected to be the result of the environmental interactions of several organisms. As most antimicrobials consists of naturally produced compounds in nature, many of the bacteria have overcoming molecular mechanism to overcome the drugs thereby being intrinsically resistant to antimicrobials (Blair et al., 2015, Munita and Arias, 2016). However, we are here dealing with the acquired resistance by the bacteria which were originally susceptible to the particular antimicrobial.
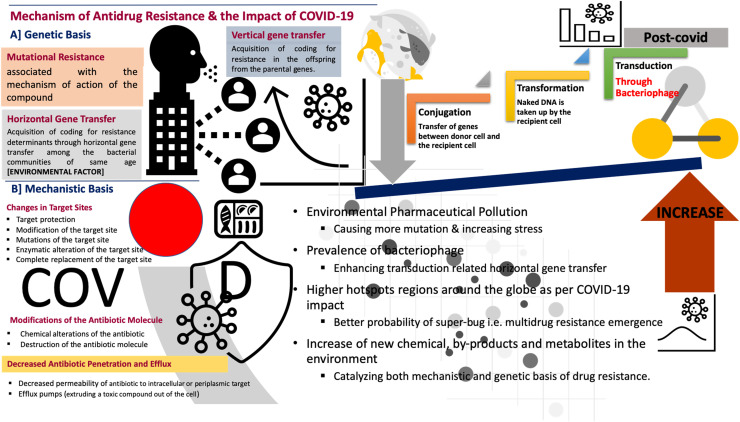
Summarising the molecular and biochemical mechanisms of antibiotic resistance is shown in Fig. 2 (Munita and Arias, 2016). These mechanisms of antidrug are generally categorised based on genetic and mechanistic basis. In a genetic basis, antidrug resistance can be developed due to mutational resistance, horizontal and vertical gene transfer (HGT and VGT). Whereas, in a mechanistic basis, antidrug resistance can be developed due to changes in the target site, modifications of antibiotic molecule, and decreased antibiotic penetration and efflux. Fig. 2 also shows that how COVID-19 spread may impact the development of antidrug resistance. The increased pharmaceutical pollution during COVID-19 spread can increase environmental stress on bacteria or microbes causing more mutation. This catalyses both the genetic and mechanistic basis of drug resistance. Also, the higher prevalence of bacteriophage may enhance the transduction related to HGT. Thus, the highly infected regions or hotspots of COVID-19 spread around the globe have a greater probability of the emergence of super bugs having multidrug resistance. Drugs like Remdesivir, Ivermectin, Azithromycin, Favipiravir, Chloroquine, Umiferovir, Ritonavir, Aspirin, and Hydroxycholoroquinine are going to remain under the scanner.
Fig. 2.
Mechanism of Antidrug Resistance and the impact of COVID-19: Probable changes in molecular and biochemical triggers of an antidrug resistance.
3.3. Comparison of occurrence of ADR
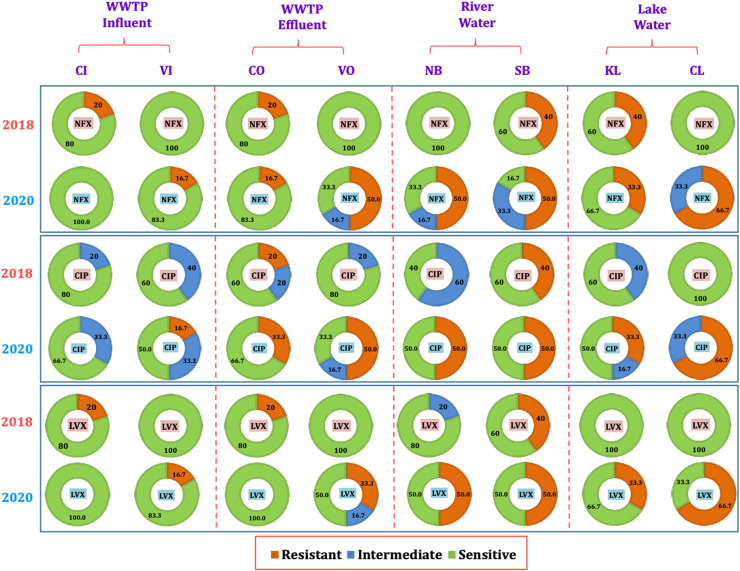
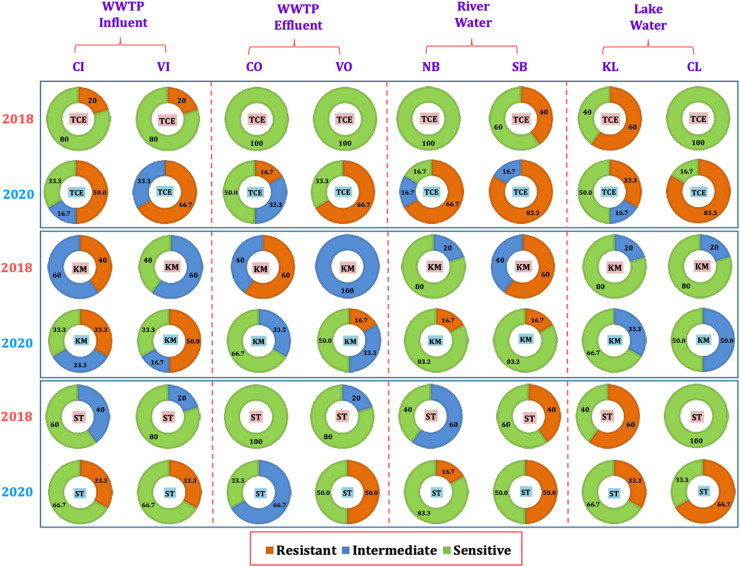
Fig. 3 and Fig. 4 represents the comparative sensitivity of E.coli towards six antibiotics including the fluoroquinolone drugs NFX (norfloxacin), CIP (ciprofloxacin), LVX (levofloxacin) as well as TCE (tetracycline drugs), KM (kanamycin monosulphate), and ST (sulfamethoxazole), at various sampling locations (CI, VI, CO, VO, NB, SB, CL, and KL) in 2018 and 2020. In 2018, the river location NB had 0% resistance for all antibiotics, whereas SB location had 40% resistance towards all antibiotics except 60% resistance for KM. SB is the central urban location. This indicates that the ADR on the urbanisation and the discharge conditions. However, in 2020, this resistance increased at both river locations for all antibiotics, except for KM at SB. For all Quinolone drugs, the antidrug resistance increased to 50% at both river locations in 2020, whereas it was varying for TCE, KM and ST. At location NB, resistance was observed to be increased for TCE, KM and ST. Whereas, at location SB, resistance increased for TCE, ST, but decreased for KM. This indicates inflow or generation of antidrug resistant E.coli in the river water from urbanised sources which reflect increased use of antimicrobials, due to the unavailability of COVID-19 specific drugs (Abelenda-Alonso et al., 2020, Getahun et al., 2020, Hsu, 2020). Though the prevalence of E. coli was highest in 2018, more antidrug resistant E.coli are generated in the year 2020 due to heavy usage of antimicrobials.
Fig. 3.
Percentage of antibiotic resistance in the influents of different water compartments in years 2018 and 2020 against fluroquinolone drugs i.e. NFX (Norfloxacin), CIP (Ciprofloxacin), LVX (Levofloxacin) for locations including WWTPs CI (Chandkheda Inlet), CO (Chandkheda Outlet), VI (Vasna Inlet) and VO (Vasna Outlet); Rivers, NB (Nehru Bridge) and SB (Sardar Bridge), and Lakes, KL (Kankaria Lake) and CL (Chandola Lake).
Fig. 4.
Percentage of antibiotic resistance in the influents of different water compartments in years 2018 and 2020 against tetracycline drugs (TCE), aminoglycosides i.e. KM (kanamycin), and others i.e. ST (sulfamethoxazole) for locations including WWTPs CI (Chandkheda Inlet), CO (Chandkheda Outlet), VI (Vasna Inlet) and VO (Vasna Outlet); Rivers, NB (Nehru Bridge) and SB (Sardar Bridge), and Lakes KL (Kankaria Lake) and CL (Chandola Lake).
In 2018, no ADR was observed for any of the antibiotics at location CL and KL, except for NFX, TCE and ST at location KL. (Fig. 3 and Fig. 4). However, significant resistance was observed for all antibiotics, except KM, at both lake locations with higher values at CL than KL. This indicates more urbanised discharge carrying antidrug resistant E.coli accumulates at the location CL. One of the major reasons for the generated resistance at CL is the occasional discharge to the CL from nearby open Pirana solid waste dumping site (Singh et al. 2008). This call for a monitoring of urban wastewater flows being discharged to the lake ecosystem.
Among the sampled WWTP locations in the year 2018, at locations VI and VO, no resistance was observed for any of the antibiotics except TCE (20% in influent) (Fig. 3 and Fig. 4). Whereas, at CI location resistance for NFX, LVX, TCE, KM, was observed but only found to be increasing towards CIP and KM at location CO. These results show the increase in antidrug resistance after WWTP treatment, which was consistent as reported in the studies from Sweden and Austria (Reinthaler et al., 2003, Flach et al., 2018). Interestingly, ADR increased significantly for all antibiotics in the year 2020 at the VI and VO locations when compared to year 2018. In the year 2020, ADR was observed for all antibiotics at VI and these resistances were observed to be increasing or being constant at VO locations for all antibiotics except KM (decreased by 35%) (Fig. 3, Fig. 4). Such a high increase in the resistance in treated effluent can be attributed to a long residence time of wastewater in WWTP, where E.coli is in contact with the antibiotics or antibiotic residues for a long time (Honda et al., 2018). In the case of CI in the year 2020, no resistance was observed towards the quinolone drugs, whereas the observed ADR for KM, ST, and TCE, was reduced significantly at CO location. However, resistance was observed to be generated for NFX and CIP at CO in year 2020. The high resistance towards quinolone drugs is attributed to the discharge having domestic origin (Threedeach et al., 2012; Auerbach et al., 2007); because these drugs are prescribed for treatments of respiratory and urinary tract infections, their use has increased significantly during the COVID-19 pandemic (Abelenda-Alonso et al., 2020, Getahun et al., 2020, Hsu, 2020).
Overall, domestic municipal wastewater likely possesses higher concentrations of antimicrobials than any other ambient water. Aeration enhances the generation and replication of antidrug resistant E.coli if there is a high density and diversity of the microbial population in a given wastewater (Ram and Kumar, 2020, Kumar et al., 2020f). The advanced or hybrid wastewater treatment processes should be adopted to effectively remove the antimicrobials and their residue in order to reduce the possibility of resistance (Dhangar and Kumar, 2020). Treatment technologies such as MBR-NF/UF, MBR-UV oxidation, AS-gamma radiation was found to be very effective (removal efficiency: 90–100%) for most of the antibiotics and other pharmaceuticals (Dhangar and Kumar, 2020).
The abundance in both antidrug resistance and E. coli count in the STPs was found to be statically related. Previously, in case of the Zenne river of Belgium, the abundance of E. coli and antidrug resistance increased from upstream to downstream after merging the effluent from Brussel’s WWTP (Proia et al., 2018). Thus, proper and timely monitoring should be done to track such load of E. coli and ADR while discharging the treated effluents to the river water. From the current study, it is seen that the antidrug resistance to NFX, CIP, LVX, TCE and ST is found at most sampling locations. Such ADR generated during COVID_19 requires rigorous monitoring at local and international level through wastewater based epidemiology (Kumar et al., 2020b). However, the lack of sanitation and treatment facilities in the undeveloped and developing countries is a big challenge to monitor the spread of ADR in the environmental waters (Pormohammad et al., 2019). Perhaps the current pandemic may accelerate the upgradation of the current status of WWTP processes to tackle the pharmaceuticals and other antimicrobials successfully and to monitor ADR (Kumar et al., 2020a).
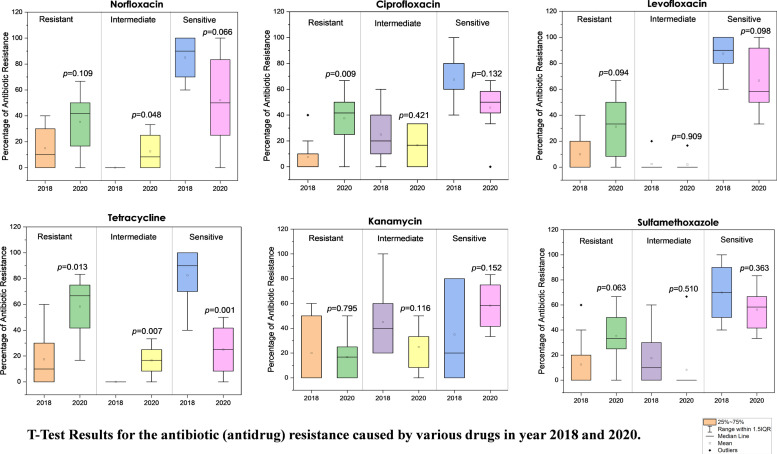
Fig. 5 highlights the statistical comparison of overall ADR in the year 2018 and 2020, whose causes are well described above. It is clearly seen that the mean percentage value of overall ADR was increased for the resistant strains of E. coli in the year 2020 than 2018, except in the case of kanamycin (remains nearly same). Whereas, the mean percentage value of overall ADR observed to be decreasing for the sensitive strains of E. coli in the year 2020 than 2018, except in case of kanamycin (increases). The percentage of ADR (in resistant E.coli strains) for almost all antibiotics: CIP, LVX, TC, KM, ST (except NFX: 89.1% change), was observed to be very significant in the year 2020 than 2018, as p < 0.10. This indicates that the significant change is occurring due to increase in the mean value of percentage of ADR. Overall, the comparison of overall ADR shows a significant increase statistically in the year 2020 than 2018.
Fig. 5.
Comparison of antibiotic (antidrug) resistance against various antibiotics in 2018 and 2020 with the results of a statistical T-test.
3.4. Imprints of COVID-19 spread over ADR distribution
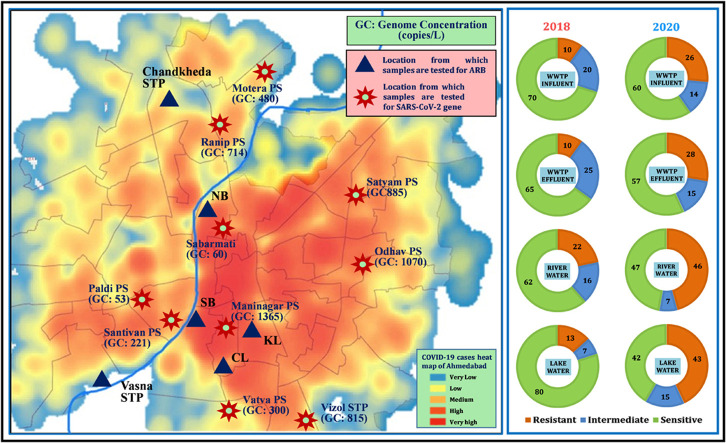
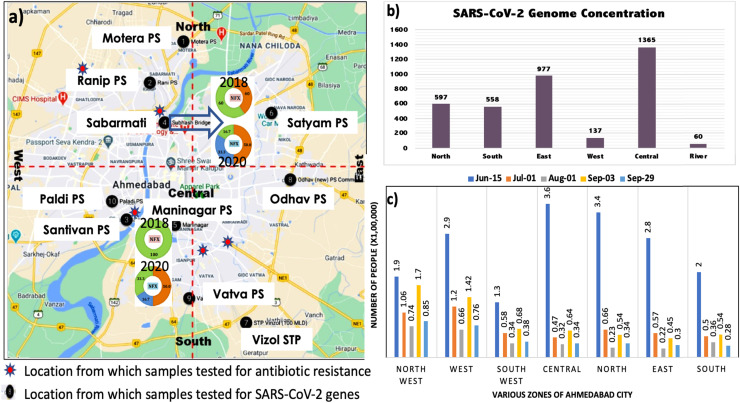
The increased cases of COVID-19 is not surprisingly correlated to SARS-CoV-2 genes in waste and natural waters (Medema et al., 2020, Ahmed et al., 2020, Haramoto et al., 2020, La Rosa et al., 2020, Sherchan et al., 2020; Kumar et al., 2020; Nemudryi et al., 2020; Kocamemi et al., 2020). Fig. 6 represents the population under threat of COVID-19 in Ahmedabad city (as predicted by nationally authorised Arogya-Setu app), Zone-wise scenario of effective SARS-CoV-2 genome concentration (copies/L) in Ahmedabad city, sampling locations for SARS-CoV-2 RNA analyses, and ADR analyses. The population under threat of COVID-19 in various zones of the city has been predicted by Arogya-Setu app based on the confirmed cases and the population of the respective zone. Arogya-Setu is an authorised Indian COVID-19 contact tracing, syndromic mapping and self-assessment digital service provided under the Ministry of Electronics and Information Technology (MeitY), India. The sampling locations were chosen so as to cover various parts of the city. Two locations of the river i.e., SB and NB are shown with their resistance increase for Norfloxacin between 2018 and 2020. These two locations fall in the central zone of the city, which was highly affected by COVID-19, as can be seen from Fig. 6a and b. Table 2 summarises the status of the SARS-CoV-2 gene along copies with their corresponding Ct-values in the water samples collected from various parts of Ahmedabad, Gujarat on 15th October 2020. It also provides the effective genome concentration for the sampled locations. The genome concentrations were observed to be high in central, east, south and north zones of the city, which can be observed at Maninagar (1365 copies/L), Odhav PS (1070 copies/L), Satyam PS (885 copies/L), Vinzole STP (815 copies/L), and Ranip PS (714 copies/L). The sampled river location and the lake locations encompass in the same zones of the city. The high SARS-CoV-2 genome copies in these zones hint at the potential high prescription of antimicrobial drugs as a remedy to the symptoms of COVID-19. This can be the probable reason for a significant increase in ADR towards most of the drugs tested at the sampling locations in these zones. This indicates that the highly infected zones of the city, due to excessive consumption of antimicrobial drugs, have significantly impacted the antidrug resistance generated in the microorganisms. Overall, the spread of COVID-19 in the community has a prodigious correlation with the effective genome concentration of SARS-CoV-2 and with the prevalence of ADR in environmental waters.
Fig. 6.
Illustration depicting: a) zonation of Ahmedabad along with the sampling locations for SARS-CoV-2 RNA analyses, and antibiotic resistance bacteria (ARB) analyses. Two locations of the rivers i.e. SB and NB are shown with their increased resistance against norfloxacin between 2018 and 2020; B) Zone-wise scenario of effective SARS-CoV-2 genome concentrations (copies/L) measured in the samples and C) Number of people under threat of COVID-19 infection as predicted by Aarogya-setu application based on active cases reported and population density of a given area.
Table 2.
SARS-CoV-2 Ct-values along with their corresponding gene copies in the water samples collected from various parts of Ahmedabad, Gujarat on 15th October 2020. Effective genome concentrations have also been provided in the last column.
| Sampling Station | Ct values |
Gene copies/ L |
|||||
|---|---|---|---|---|---|---|---|
| N | ORF | S | N | ORF | S | Effective gene concentration | |
| Motera PS | 35.50 | 32.18 | 33.96 | 123 | 1002 | 317 | 480 |
| Ranip PS | 34.57 | 31.75 | 32.98 | 217 | 1334 | 591 | 714 |
| Paldi PS | 38.36 | 36.47 | 36.53 | 23 | 69 | 66 | 53 |
| Santivan PS | 36.08 | 33.63 | 34.80 | 87 | 390 | 187 | 221 |
| Sanbarmati | 38.46 | 35.67 | 37.14 | 22 | 110 | 47 | 60 |
| Maninagar | 34.17 | 30.77 | 31.89 | 278 | 2605 | 1213 | 1365 |
| Satyam PS | 34.52 | 31.37 | 32.70 | 223 | 1724 | 709 | 885 |
| Vinzole STP | 34.98 | 31.41 | 32.96 | 168 | 1680 | 598 | 815 |
| Odhav PS | 34.54 | 31.06 | 32.41 | 220 | 2131 | 857 | 1070 |
| Vatva PS | 38.51 | 32.58 | 35.69 | 22 | 770 | 109 | 300 |
4. Limitations
The present study compared the anti-drug resistance in E.coli in 2018 and 2020 with the latter prevalence of SARS-CoV-2 genes during the sampling period. Despite the correlations between increased ADR and COVID-19 spread, more future studies with rigorous sampling events are needed to conclude about the cause and effects. In addition, the concentration of pharmaceutical and personal care products (PPCPs) in the ambient environment should be monitored to quantify their increase owing to COVID-19; and then connect back to the corresponding effect on ADR for quantitative evaluation. In this study, we attempted to start a timely discussion about the likely relationships between ADR and COVID-19 spread throughout the globe. Our approach to analyse the ADR prevalence is mostly qualitative and there may be a slight possibility of both false positive and negative results. To obtain the conclusive evidence, the quantification of genetic markers for antimicrobial resistance will be helpful. In addition, one time point data may be argued -inadequate to derive a conclusion especially when samples used for ADR and SARS-CoV-2 genomes studies do not match. Hence we recommend regular monitoring along the consideration of wasetwater flow data for presenting gene flux or E. coli flux.
5. Conclusion
Non-fluoroquinolone drugs showed overall more resistance as compared to fluoroquinolone drugs. Tetracycline followed by norfloxacin has shown more resistance as compared to the other drugs. Despite a decrease in the prevalence of E. coli on the sampled river locations, the percentage resistance had been significantly increased in the year 2020 compared to year 2018. However, the E. coli prevalence in STP samples was increased in the order of 102, but the pattern of antidrug resistance was not consistent. Lake locations also exhibited an increase in the antidrug resistance during the duration of pandemic. The river locations and the lake locations have shown a significant increase in the antidrug resistance, and these locations are from the highly COVID-19 infected zones of the city. The COVID-19 spread in various zones of the city has shown corresponding changes in the SARS-CoV-2 genome concentration and ADR in environmental waters. Overall, due to increased consumption of antimicrobials in the pandemic period, the percentage of antidrug resistance has been increased significantly. Wastewater based epidemiology can be the key tool to monitor the antimicrobials prevalence and antidrug resistance in the pandemic situations.
Notes
The authors declare no competing financial interest.
CRediT authorship contribution statement
Manish Kumar: Conceptualization, Visualization, Project supervision, Writing - review & editing; Kiran Dhangar: Data curation, First draft, Writing - review & editing; Alok Kumar Thakur: Sampling and analyses in 2020, Data curation, First draft, Writing - review & editing; Bhagwana Ram: Sampling and analyses in 2018, Writing - review & editing; Tushara Chaminda: Writing - review & editing; Pradeep Sharma: Writing - review & editing; Abhay Kumar: Writing - review & editing; Nirav Raval: Writing - review & editing; Vaibhav Srivastava: Sampling and analyses, Data curation, First draft, Writing - review & editing; Jörg Rinklebe: Writing - review & editing; Keisuke Kuroda: Writing - review & editing; Christian Sonne: Writing - review & editing; Damia Barcelo: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work is funded by Kiran C Patel Centre for Sustainable Development at IIT Gandhinagar, UNICEF, Gujarat and UK-India Education and Research Initiative (UKIERI). We also acknowledge the help received from Dr. Shyamnarayan Dave, Dr. Madhvi Joshi, Dr. Arbind K Patel, and other GBRC and WET Lab members who contributed towards sample and data analyses.
Editor: Andrew Daugulis
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jhazmat.2021.126125.
Appendix A. Supplementary material
Supplementary material
.
References
- Aali R., Nikaeen M., Khanahmad H., Hassanzadeh A. Monitoring and comparison of antibiotic resistant bacteria and their resistance genes in municipal and hospital wastewaters. Int. J. Prev. Med. 2014;5(7):887–894. [PMC free article] [PubMed] [Google Scholar]
- Abelenda-Alonso G., Padullés A., Rombauts A., Gudiol C., Pujol M., Alvarez-Pouso C., Jodar R., Carratalà J. Antibiotic prescription during the COVID-19 pandemic: a biphasic pattern. Infect. Control Hosp. Epidemiol. 2020;41(11):1371–1372. doi: 10.1017/ice.2020.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter A., Imran M., Akhter F. Antimicrobial resistant coliform bacteria in the Gomti river water and determination of their tolerance level. Bioinformation. 2014;10(4):167–174. doi: 10.6026/97320630010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Judaibi E. Infection and antibiotic resistant bacteria in developing countries: a genetic review. J. Microbiol. Res. 2014;4(6A):10–17. [Google Scholar]
- Alexander J., Hembach N., Schwartz T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-65635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaduzzaman M., Zaman F., Rousham E. Antibiotic consumption may be linked to exaggeration of COVID-19. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach E.A., Seyfried E.E., McMahon K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007;41:1143–1151. doi: 10.1016/j.watres.2006.11.045. [DOI] [PubMed] [Google Scholar]
- Azuma T., Nakada N., Yamashita N., Tanaka H. Synchronous dynamics of observed and predicted values of anti-influenza drugs in environmental waters during a seasonal influenza outbreak. Environ. Sci. Technol. 2012;46(23):12873–12881. doi: 10.1021/es303203c. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Wright M.S., Stepanauskas R., McArthur J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006;14(4):176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Biswas K., Paul D., Sinha S.N. Prevalence of multiple antibiotic-resistant coliform bacteria in the water of river Ganga. Front. Environ. Microbiol. 2015;1(3):44. [Google Scholar]
- Blair J.M., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Chatterjee S.K., Bhattacharjee I., Chandra G. Water quality assessment near an industrial site of Damodar River, India. Environ. Monit. Assess. 2010;161(1–4):177–189. doi: 10.1007/s10661-008-0736-1. [DOI] [PubMed] [Google Scholar]
- Dhangar K., Kumar M. Tricks and tracks in removal of emerging contaminants from the wastewater through hybrid treatment systems: A review. Sci. Total Environ. 2020:140320. doi: 10.1016/j.scitotenv.2020.140320. [DOI] [PubMed] [Google Scholar]
- Ferreira da Silva M., Vaz-Moreira I., Gonzalez-Pajuelo M., Nunes O.C., Manaia C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007;60(1):166–176. doi: 10.1111/j.1574-6941.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Flach C.F., Genheden M., Fick J., Joakim Larsson D.G. A comprehensive screening of Escherichia coli isolates from Scandinavia’s largest sewage treatment plant indicates no selection for antibiotic resistance. Environ. Sci. Technol. 2018;52(19):11419–11428. doi: 10.1021/acs.est.8b03354. [DOI] [PubMed] [Google Scholar]
- Getahun H., Smith I., Trivedi K., Paulin S., Balkhy H.H. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull. World Health Organ. 2020;98(7) doi: 10.2471/BLT.20.268573. 442-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Antibiotic Resistance Partnership (GARP)-India Working Group Rationalizing antibiotic use to limit antibiotic resistance in India+ Indian J. Med. Res. 2011;134(3):281. [PMC free article] [PubMed] [Google Scholar]
- Guo X., Yan Z., Zhang Y., Xu W., Kong D., Shan Z., Wang N. Behavior of antibiotic resistance genes under extremely high-level antibiotic selection pressures in pharmaceutical wastewater treatment plants. Sci. Total Environ. 2018;612:119–128. doi: 10.1016/j.scitotenv.2017.08.229. [DOI] [PubMed] [Google Scholar]
- Hamner S., Broadaway S.C., Mishra V.B., Tripathi A., Mishra R.K., Pulcini E., Pyle B.H., Ford T.E. Isolation of potentially pathogenic Escherichia coli O157: H7 from the Ganges River. Appl. Environ. Microbiol. 2007;73(7):2369–2372. doi: 10.1128/AEM.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Noguchi M., Yamamoto-Ikemoto R., Watanabe T. Effect of sedimentation and aeration on antibiotic resistance induction in the activated sludge process. J. Water Environ. Technol. 2018;16(2):94–105. [Google Scholar]
- Honda R., Watanabe T., Sawaittayotin V., Masago Y., Chulasak R., Tanong K., Chaminda G.T., Wongsila K., Sienglum C., Sunthonwatthanaphong V., Poonnotok A. Impacts of urbanization on the prevalence of antibiotic-resistant Escherichia coli in the Chaophraya River and its tributaries. Water Sci. Technol. 2016;73(2):362–374. doi: 10.2166/wst.2015.502. [DOI] [PubMed] [Google Scholar]
- Hsu J. How covid-19 is accelerating the threat of antimicrobial resistance. BMJ. 2020;369:1983. doi: 10.1136/bmj.m1983. [DOI] [PubMed] [Google Scholar]
- Hu J., Shi J., Chang H., Li D., Yang M., Kamagata Y. Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 2008;42(9):3415–3420. doi: 10.1021/es7026746. [DOI] [PubMed] [Google Scholar]
- IS10500 B.I.S. Bureau of Indian Standards (BIS),; New Delhi: 2012. Indian Standard Drinking Water–Specification (Second Revision) [Google Scholar]
- Jiang L., Hu X., Xu T., Zhang H., Sheng D., Yin D. Prevalence of antibiotic resistance genes and their relationship with antibiotics in the Huangpu River and the drinking water sources, Shanghai, China. Sci. Total Environ. 2013;458:267–272. doi: 10.1016/j.scitotenv.2013.04.038. [DOI] [PubMed] [Google Scholar]
- Klein E.Y., Van Boeckel T.P., Martinez E.M., Pant S., Gandra S., Levin S.A., Goossens H., Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. 2018;115(15):E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 [Google Scholar]
- Kumar A., Sharma M.P. Application of water quality index and diversity index for pollution assessment of Kankaria Lake at Ahmedabad, India. J. Civil & Environ. Eng. 2014;4(3):1. [Google Scholar]
- Kuroda K., Li C., Dhangar K., Kumar M. Predicted occurrence, ecotoxicological risk and environmentally acquired resistance of antiviral drugs associated with COVID-19 in environmental waters. Sci. Total Environ. 2021;776:145740. doi: 10.1016/j.scitotenv.2021.145740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Patel A.K., Patel N., Bhattacharya P., Joshi M., Joshi C.G. Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with Upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 2021;754 doi: 10.1016/j.scitotenv.2020.142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Kuroda K., Dhangar K., Mazumder P., Sonne C., Rinklebe J., Kitajima M. Potential emergence of antiviral-resistant pandemic viruses via environmental drug exposure of animal reservoirs. Environ. Sci. Technol. 2020;54(14):8503–8505. doi: 10.1021/acs.est.0c03105. [DOI] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Mazumder P., Mohapatra S., Thakur A.K., Dhangar K., Taki K., Mukherjee S., Patel A.K., Bhattacharya P., Mohapatra P., Rinklebe J. A chronicle of SARS-CoV-2: seasonality, environmental fate, transport, inactivation, and antiviral drug resistance. J. Hazard. Mater. 2021;405 doi: 10.1016/j.jhazmat.2020.124043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Chaminda T., Patel A.K., Sewwandi H., Mazumder P., Joshi M., Honda R. Prevalence of antibiotic resistance in the tropical rivers of Sri Lanka and India. Environ. Res. 2020;188 doi: 10.1016/j.envres.2020.109765. [DOI] [PubMed] [Google Scholar]
- Kumar M., Chaminda G.T., Honda R. Seasonality impels the antibiotic resistance in Kelani River of the emerging economy of Sri Lanka. npj Clean Water. 2020;3(1):1–8. [Google Scholar]
- Kumar M., Ram B., Sewwandi H., Honda R., Chaminda T. Treatment enhances the prevalence of antibiotic-resistant bacteria and antibiotic resistance genes in the wastewater of Sri Lanka, and India. Environ. Res. 2020;183 doi: 10.1016/j.envres.2020.109179. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Zhou, Q., Li, Y., Garner, L.V., Watkins, S.P., Carter, L.J., Smoot, J., Gregg, A.C., Daniels, A.D., Jervey, S. and Albaiu, D., 2020. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. [DOI] [PMC free article] [PubMed]
- Lucien M.A.B., Canarie M.F., Kilgore P.E., Jean-Denis G., Fénélon N., Pierre M., Cerpa M., Joseph G.A., Maki G., Zervos M.J., Dely P. Antibiotics and antimicrobial resistance in the COVID-19 era: perspective from resource-limited settings. Int. J. Infect. Dis. 2021;104:250–254. doi: 10.1016/j.ijid.2020.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinek H., Wirth R., Muscholl-Silberhorn A., Gauer M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 1998;64(2):626–632. doi: 10.1128/aem.64.2.626-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Miranda C.D., Castillo G. Resistance to antibiotic and heavy metals of motile aeromonads from Chilean freshwater. Sci. Total Environ. 1998;224(1–3):167–176. doi: 10.1016/s0048-9697(98)00354-4. [DOI] [PubMed] [Google Scholar]
- Miranda C., Silva V., Capita R., Alonso-Calleja C., Igrejas G., Poeta P. Implications of antibiotics use during the COVID-19 pandemic: present and future. J. Antimicrob. Chemother. 2020;75(12):3413–3416. doi: 10.1093/jac/dkaa350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munita J.M., Arias C.A. Mechanisms of antibiotic resistance. Virulence Mech. Bact. Pathog. 2016:481–511. [Google Scholar]
- Na G., Lu Z., Gao H., Zhang L., Li Q., Li R., Yang F., Huo C., Yao Z. The effect of environmental factors and migration dynamics on the prevalence of antibiotic-resistant Escherichia coli in estuary environments. Sci. Rep. 2018;8(1):1663. doi: 10.1038/s41598-018-20077-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pormohammad A., Nasiri M.J., Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect. Drug Resist. 2019;12:1181–1197. doi: 10.2147/IDR.S201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia L., Anzil A., Subirats J., Borrego C., Farrè M., Llorca M., Balcázar J.L., Servais P. Antibiotic resistance along an urban river impacted by treated wastewaters. Sci. Total Environ. 2018;628:453–466. doi: 10.1016/j.scitotenv.2018.02.083. [DOI] [PubMed] [Google Scholar]
- Ram B., Kumar M. Correlation appraisal of antibiotic resistance with fecal, metal and microplastic contamination in a tropical Indian river, lakes and sewage. NPJ Clean. Water. 2020;3(1):1–12. [Google Scholar]
- Rawson T.M., Moore L.S., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 2020;71(9):2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinthaler F.F., Posch J., Feierl G., Wüst G., Haas D., Ruckenbauer G., Mascher F., Marth E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003;37(8):1685–1690. doi: 10.1016/S0043-1354(02)00569-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S., Chamorro S., Marti E., Huerta B., Gros M., Sànchez-Melsió A., Borrego C.M., Barceló D., Balcázar J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015;69:234–242. doi: 10.1016/j.watres.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Castillo G., Callejas L., López H., Olmos J. Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electron. J. Biotechnol. 2006;9(5):0. 0-0. [Google Scholar]
- Singer A.C., Howard B.M., Johnson A.C., Knowles C.J., Jackman S., Accinelli C., Caracciolo A.B., Bernard I., Bird S., Boucard T., Boxall A. Meeting report: risk assessment of Tamiflu use under pandemic conditions. Environ. Health Perspect. 2008;116(11):1563–1567. doi: 10.1289/ehp.11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U.K., Kumar M., Chauhan R., Jha P.K., Ramanathan A.L., Subramanian V. Assessment of the impact of landfill on groundwater quality: a case study of the Pirana site in western India. Environ. Monit. Assess. 2008;141(1):309–321. doi: 10.1007/s10661-007-9897-6. [DOI] [PubMed] [Google Scholar]
- Storteboom H., Arabi M., Davis J.G., Crimi B., Pruden A. Tracking antibiotic resistance genes in the South Platte River basin using molecular signatures of urban, agricultural, and pristine sources. Environ. Sci. Technol. 2010;44(19):7397–7404. doi: 10.1021/es101657s. [DOI] [PubMed] [Google Scholar]
- Takanami R., Ozaki H., Giri R.R., Taniguchi S., Hayashi S. Detection of antiviral drugs oseltamivir phosphate and oseltamivir carboxylate in Neya River, Osaka, Japan. J. Water Environ. Technol. 2010;8(4):363–372. [Google Scholar]
- Threedeach S., Chiemchaisri W., Watanabe T., Chiemchaisri C., Honda R., Yamamoto K. Antibiotic resistance of Escherichia coli in leachates from municipal solid waste landfills: comparison between semi-aerobic and anaerobic operations. Bioresour. Technol. 2012;113:253–258. doi: 10.1016/j.biortech.2012.01.086. [DOI] [PubMed] [Google Scholar]
- Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A., Robinson T.P., Teillant A., Laxminarayan R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization,; 2014. Antimicrobial Resistance: Global Report on Surveillance. [Google Scholar]
- Yang Y., Xu C., Cao X., Lin H., Wang J. Antibiotic resistance genes in surface water of eutrophic urban lakes are related to heavy metals, antibiotics, lake morphology and anthropic impact. Ecotoxicology. 2017;26(6):831–840. doi: 10.1007/s10646-017-1814-3. [DOI] [PubMed] [Google Scholar]
- Zhang S., Han B., Gu J., Wang C., Wang P., Ma Y., Cao J., He Z. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere. 2015;135:138–145. doi: 10.1016/j.chemosphere.2015.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material