Several viruses target the human respiratory tract, causing different clinical manifestations spanning from mild upper airway involvement to life-threatening acute respiratory distress syndrome (ARDS). As dramatically evident in the ongoing SARS-CoV-2 pandemic, the clinical picture is not always easily predictable due to the combined effect of direct viral and indirect patient-specific immune-mediated damage.
KEYWORDS: COVID-19, SARS-CoV-2, respiratory tract, viral infections
SUMMARY
Several viruses target the human respiratory tract, causing different clinical manifestations spanning from mild upper airway involvement to life-threatening acute respiratory distress syndrome (ARDS). As dramatically evident in the ongoing SARS-CoV-2 pandemic, the clinical picture is not always easily predictable due to the combined effect of direct viral and indirect patient-specific immune-mediated damage. In this review, we discuss the main RNA (orthomyxoviruses, paramyxoviruses, and coronaviruses) and DNA (adenoviruses, herpesviruses, and bocaviruses) viruses with respiratory tropism and their mechanisms of direct and indirect cell damage. We analyze the thin line existing between a protective immune response, capable of limiting viral replication, and an unbalanced, dysregulated immune activation often leading to the most severe complication. Our comprehension of the molecular mechanisms involved is increasing and this should pave the way for the development and clinical use of new tailored immune-based antiviral strategies.
INTRODUCTION
Acute respiratory tract infections are a major public health problem and a leading cause of morbidity and mortality worldwide, especially in children, elderly, and immunocompromised subjects (1). Annually, almost 29 to 59 million people in the United States become infected with influenza viruses and nearly 500 million people experience two or more non-influenza-related viral respiratory tract infections (2). RNA viruses are the predominant agents, and common pathogens include respiratory syncytial viruses, influenza viruses, parainfluenza viruses, metapneumoviruses, rhinoviruses, enteroviruses, and coronaviruses (2). DNA viruses causing respiratory tract involvement include adenoviruses, bocaviruses (3), and reactivating pathogens in immunosuppressed patients, such as herpes simplex virus, cytomegalovirus, and Epstein-Barr virus (4).
Respiratory tract infections usually exhibit a seasonality, occurring typically during late autumn, winter, and early spring. The hypotheses proposed to explain this phenomenon include increased crowding during the cold seasons, lower temperatures and higher humidity increasing the stability of virions outside the body, and chilling, which may increase host susceptibility and vitamin D deficiency due to lower sun exposure (5). Influenza viruses, coronaviruses, and respiratory syncytial viruses show infection peaks in winter, even if epidemiological differences are observed in tropical and subtropical areas and alteration in seasonality occurs during pandemics (5–8). Adenoviruses, bocaviruses, parainfluenza viruses, metapneumoviruses, and rhinoviruses can be detected throughout the year. Seasonal patterns of parainfluenza viruses can be type specific with types 1 and 3 peaking in the fall and spring-summer, respectively. The prevalence of some enteroviruses increases in summer (5).
CLINICAL AND LABORATORY FEATURES OF RESPIRATORY VIRAL INFECTIONS
Clinical Findings
The diseases caused by these different viruses vary from self-limiting upper respiratory tract infections, such as cold, sore throat, or laryngo-tracheo-bronchitis to life-threatening lower respiratory tract infections, such as bronchiolitis, pneumonia, or acute respiratory distress syndrome (ARDS), as well as occasionally disseminated disease. The involvement of lower respiratory tract can occur through one of the following three mechanisms: (i) direct infection of lung cells without sustained virological replication in the upper respiratory tract (this is the case of several influenza viruses featuring preferential tropism for the sialic acids of the deeper airways), (ii) spread in a contiguous fashion from viral infections involving the upper respiratory tract (e.g., coronaviruses), and (iii) hematogenous spread (e.g., cytomegaloviruses) (9).
The severity of each infection and the possible progression from the upper to the lower respiratory tract depend on the intrinsic virulence of the causing virus, on possible coinfections, and on patient-related factors such as age, underlying illnesses (i.e., concurring respiratory and cardiovascular comorbidities), or degree of immunodeficiency (10).
Considering age, viral respiratory tract infections are a major killer of young children and elderly people. Moreover, in developing countries the incidence of death among children can be up to five times higher than in developed regions (11). Viruses such as respiratory syncytial virus mostly affect young children, who develop rhinitis, laryngitis, or sinusitis, or neonates, who are at risk for bronchiolitis and pneumonia (12). On the other hand, influenza viruses account for an average of 23,607 annual flu-related deaths, 90% of which occur among persons age 65 or older (3).
Comorbidities significantly worsening the prognosis of respiratory infections include chronic heart or lung diseases, diabetes, kidney or liver diseases, blood disorders, malnutrition, and immune deficiencies (2). Immunocompromised patients include transplant recipients, cancer patients on chemotherapy, rheumatic patients treated with immunosuppressant drugs, and subjects with primary immunodeficiencies or infected with human immunodeficiency virus (2). In the latest categories of patients, respiratory viruses are acquired at the same frequency as in immunocompetent individuals but are associated with prolonged infections, increased progression to lower respiratory tract infections, and a higher mortality (2).
Virus-induced ARDS is not common in immunocompetent patients, but its exact prevalence is not known (13). Pandemic viruses, such as severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) in 2002, Middle East respiratory syndrome (MERS) virus in 2012, avian influenza A H5N1 in 1998, influenza A H1N1 in 2009, and recently, the novel coronavirus SARS-CoV-2 in 2019, have a significantly higher frequency of ARDS and higher case fatality rates compared to seasonal viruses (13). In the early phases of SARS-CoV-2 pandemic, up to 30% of hospitalized patients with SARS-CoV-2 disease (COVID-19) progress to ARDS, which is the main cause of mortality (14).
There are no pathognomonic cues for the diagnosis of viral pneumonia as opposed to bacterial pneumonia or other causes of lung disease. Viral pneumonia has a more gradual onset than bacterial pneumonia and more frequent clinical manifestations of concomitant upper respiratory tract infection. Flu-like symptoms, rash or gastrointestinal symptoms, lack of purulent sputum, tachycardia or tachypnea out of proportion to the temperature, paucity of physical findings on pulmonary exam disproportionate to the level of debility, and possible bilateral positive lung findings are also more common in viral pneumonia (9). Immune-mediated interstitial lung diseases (pulmonary fibrosis, sarcoidosis, and hyper-reactive airway disease), lung vascular diseases (pulmonary embolism and pulmonary hypertension), and congestive heart failure should be considered in the differential diagnosis. Laboratory tests in combination with radiography can significantly improve the suspicion of viral pneumonia, while the identification of viruses causing pneumonia is gaining importance considering the new diagnostic tools available.
Radiological Presentation and Laboratory Markers
A thorough description of the radiological findings and the laboratory markers observed in the course of viral pneumonia is well beyond the scope of this review, but nonetheless their role in its clinical diagnosis deserves a general overview.
Chest X-ray is the most widely used imaging technique in the diagnosis of lower respiratory tract infections, followed by computed tomography (CT). A chest radiograph can establish the presence of pneumonia, determine its extent and location, and assess the response to treatment (15). It can also diagnose complications like pleural effusion or pneumothorax. No findings are pathognomonic for different pneumonia etiologies, since there is a substantial overlap in chest X-ray findings between bacterial and viral pneumonia. Features that are suggestive of bacterial pneumonia include alveolar infiltrates with lobar consolidation and coexistent pleural effusion. Features that are more suggestive of viral pneumonia include bilateral interstitial infiltrates with patchy distribution (9). Several chest X-ray-based machine-learning approaches for the detection and differentiation of viral form bacterial pneumonia are under development with encouraging results. A recent meta-analysis calculated 89% sensitivity and 89% specificity of different automated algorithms using chest X-ray images in the differential diagnosis of viral pneumonia but also evidenced several methodological concerns, which still hamper their transfer into the clinic (16).
A CT scan is not used for the initial evaluation of pneumonia but may be used when the clinical suspicion of viral pneumonia is high and chest X-Ray is negative to better characterize the pneumonia pattern, to look for complications, or to detect underlying lung diseases. Pneumonia can appear as lobar pneumonia (confluent areas of consolidation, which are usually confined to one segment or lobe and frequently found in bacterial pneumonia), as bronchopneumonia (centrilobular nodules that coalesce to cause consolidation and involve one or more lobes), and as interstitial pneumonia (involvement of bronchiolar wall and pulmonary interstitium leading to septal thickening, micronodules, and areas of ground-glass opacities). The latter is the most common radiological presentation of viral pneumonia (15). Also, in this case, there are no pathognomonic patterns of different viral pneumonia. Ground-glass opacity seems to be more frequently noticed in patients with adenovirus pneumonia than in patients with other viral infections, while influenza viruses diffusely invade the respiratory epithelium, resulting in necrotizing bronchitis and diffuse alveolar damage, which may manifest as consolidation (17).
The most typical CT features of COVID-19 pneumonia are bilateral multifocal ground-glass opacities; compared to other viruses, the lesions have more frequently a peripheral location, involve the five lobes, and are associated with peribronchovascular thickening (18). In all cases of severe viral pneumonia complicating in ARDS, an evolution from a ground-glass to a consolidation pattern can be observed. Moreover, bacterial coinfections can further complicate the picture concurring to consolidation pattern. Compared to influenza viruses, a lower frequency of bacterial coinfection has been reported in COVID-19, especially in the early phases of hospitalization (19). Of note, pulmonary embolism is highly prevalent in COVID-19 compared to other infectious agents, especially in critically ill patients (20). In case of a clinical-radiological discordance or respiratory worsening, a CT scan with contrast medium injection is useful to diagnose pulmonary embolism (18).
Hematological and biochemical tests may be of help in the correct definition of the clinical framework, contributing to the identification of injury to other organs, to the prognostic stratification of affected patients, and to the monitoring of the disease course (9). Unfortunately, there are no specific peripheral blood diagnostic findings, since viral pneumonia may result in elevated, normal, or decreased white blood cell counts (21). However, viral etiology is less commonly associated with elevated white blood cell counts than bacterial pneumonia. In the case of SARS-CoV-2 infection, for example, lymphopenia (often associated with eosinophylopenia and piastrinopenia) has been correlated with more severe outcomes (22). A correlated marker is the neutrophil/lymphocyte ratio (NLR) which, although not specific, is often increased in severe forms of viral pneumonia and was recently proposed as an early risk stratification marker also for COVID-19 patients (23).
As a reactive phase reactant, the C-reactive protein (CRP) level may be elevated with viral pneumonia, although this is neither a specific nor a sensitive finding (9, 24). In the case of SARS-CoV-2, an early increase of CRP beyond 15 mg/liter correlates with disease severity (25), whereas values higher than 200 mg/liter at admission are a strong predictor of a higher risk of death (26). The detection of procalcitonin (PCT) is less sensitive and even less specific in the case of viral respiratory infection, since its amounts are heavily affected by the parallel secretion of virus-elicited gamma interferon (IFN-γ) (21). Its main role in the course of a viral infection may be to monitor for possible secondary bacterial superinfections (27, 28). Nonetheless, different meta-analyses evidenced the association between higher PCT levels and a significantly increased risk of severe clinical forms of COVID-19 (29, 30).
As an acute-phase protein, ferritin is known to be affected by the inflammatory state (31). Its increasing levels have been proposed as a marker of adverse outcomes in SARS-CoV-2-infected patients (32).
Finally, arterial blood gas parameters (i.e., pO2, pCO2, pH, lactate, and bicarbonate) are obviously of pivotal importance in the continuous monitoring of patients with pneumonia (33).
THE RESPIRATORY VIRUSES AND THEIR DIRECT MECHANISMS OF TISSUE DAMAGE
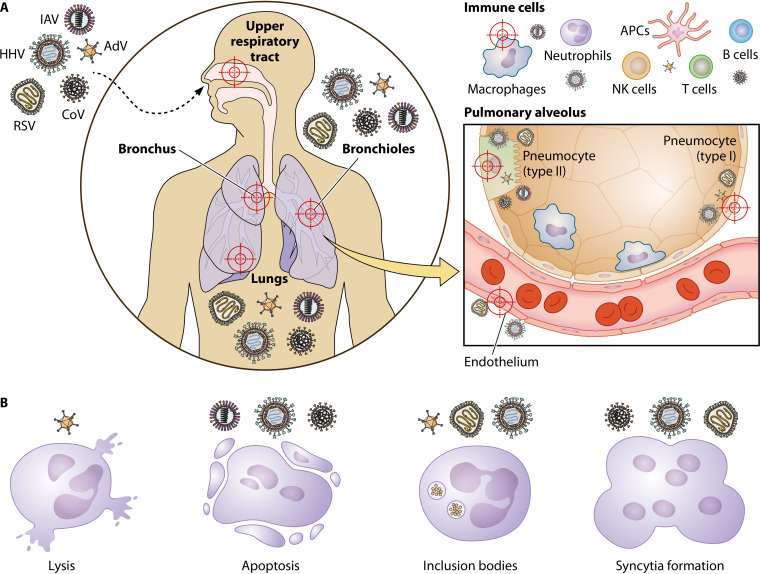
Lung involvement may be the effect of both direct and indirect virus-induced mechanisms of tissue damage. These mechanisms are often dramatically different when considering RNA and DNA viruses (Fig. 1).
FIG 1.
Respiratory viruses and their direct mechanisms of tissue damage. (A) Targets of virus infection. Viral infections by orthomyxoviruses (influenza A virus [IAV], for example) herpesviruses (HHV), adenoviruses (AdV), paramyxoviruses (respiratory syncytial virus [RSV], for example), and coronaviruses (CoV) commonly affect the upper and lower respiratory tract. Thus, all of them might cause of pneumonia, and infection might result in bronchiolitis. RSV is the most common cause of bronchiolitis and pneumonia in children younger than 1 year of age. Focusing on the pulmonary alveolus, CoV and IAV infect directly type II pneumocytes, and RSV, AdV, and HHV infect both type I and II cells. Endothelial cells are targeted by RSV and HHV. On the other hand, immune cells recruited to the site of infection, such as macrophages, neutrophils, T cells, NK cells, B cells, and antigen-presenting cells (APCs), can be infected by AdV, CoV, IAV, and HHV. (B) Direct mechanisms of tissue damage. After infection, viruses alter cellular homeostasis causing different types of damage or alterations. AdV are responsible for the direct lysis of the infected cells, while IAV, CoV, and HHV induce apoptosis of their target cells. Inclusion bodies are a consequence of AdV, RSV, and HHV infection, and syncytium formation has been described for cells infected by RSV, CoV, and HHV.
RNA Viruses
Orthomyxoviruses.
Influenza viruses are responsible for seasonal epidemics and those of type A (influenza A viruses [IAV]) can cause sporadic pandemics. Most influenza cases are caused by self-limiting viral infections typically limited to the upper respiratory tract. However, mainly for IAV, infected patients may evolve possible complications ranging from secondary bacterial infections to primary viral pneumonia and bronchiolitis (34).
Influenza viruses may infect a variety of cells within the respiratory tract, including ciliated epithelial cells, type I and II alveolar cells, and immune cells. The final effect on infected cells is dependent on the specific cell type. Virus tropism is due to the capability of influenza viruses to bind different isoforms of sialic acid present on host cells. As an example, the higher pathogenicity of some influenza subtypes (i.e., the “avian” H5N1) may also be related to their higher affinity for the sialyl-galactosyl residues linked by 2-3-linkage [Neu5Ac(2-3)Gal] present in the deeper respiratory tract, thus leading to a more severe lung involvement (35).
Even if it is evident the cytopathic effect of influenza viruses in vitro, it is important to evidence the role played by the innate immune repose in contributing to massive cell damage during influenza infection. As already evident from the description of cases observed during the 1918 influenza pandemic, the lung damage in the case of primary viral pneumonia is mostly due to the overproduction of inflammatory cytokines that, however, is potently triggered by virus replication in lung cells, type II pneumocytes, alveolar macrophages or, in certain cases, the activation of endothelial cells (36, 37). In this context, both activated macrophages and infected dendritic cells (DCs) further trigger a massive immune response and the cytokine storming (38, 39).
In general, RNA viruses are good inductors of cytokine production; additionally, influenza viruses are peculiar RNA viruses since they replicate their fragmented genomes within the host cell nucleus. Consequently, a description of the main molecular steps involved in virus replication is essential for a better comprehension of influenza pathogenesis at the cellular level. The replicative cycle of influenza viruses starts with the engagement of sialic acid by hemagglutinin (HA), which triggers clathrin-mediated endocytosis into the host cell. The acidification of the endosome triggers both the HA-mediated fusion process and the release of the viral ribonucleocapsids thanks to the M2 ion channel, which allows acidification of viral core responsible for the release of the viral ribonucleoprotein (vRNP) complexes from M1 protein. The vRNP are now free to migrate into the cell nucleus by using the importin-α/importin-β nuclear import pathway (40, 41). After the entry into the nucleus, all genomic segments are then transcribed (two of them are spliced by host cell machinery), capped and polyadenylated by the RNA-dependent RNA polymerase for the cytoplasmic phase of virus protein synthesis. In parallel, the RNA-dependent RNA polymerase complex binds to leader sequence of encapsidated RNA fragments and synthesizes whole complementary antigenomes, which will be further used as a template for the formation of novel RNA genomic segments (42, 43). The viral enzyme passes over the transcription signal of all RNA segments during the genome replication phase. Both structural and nonstructural proteins composing the virus ribonucleocomplexes produced in the cytoplasm reenter the nucleus and assembly. A high level of influenza M1 protein stimulates the nuclear export protein-mediated transfer of neosynthesized virus genome segments back into the cytoplasm (43). The nuclear export of vRNP represents another crucial step for productive infection in lung cells, and it is also dependent on the viral-induced increase of reactive oxygen species (ROS) in the infected cell (44).
The path of neosynthesized envelope virus glycoproteins (HA and neuraminidase [NA]) is different from that of the other influenza proteins. They are synthesized into the cell cytoplasm but do not need to go back into the nucleus. IAV viral envelope proteins are synthesized by endoplasmic reticulum (ER) ribosomes, as well as cellular secretory proteins. Neuraminidase (NA), HA, and M2 are cotranslationally directed to the ER thanks to the interaction of viral hydrophobic targeting sequences with the signal recognition particle. HAs of most IAV infecting humans are endowed with a monobasic cleavage site required for HA activation; this site can be cleaved by different proteases present in human epithelial cells in the airways such as the transmembrane protease serine S-1 member 2 (TMPRSS2) and the human airway trypsin-like protease (HAT). HAT is present at the plasma membrane level, and it can cleave both new synthesized HAs and HA present in cell-associated virions (43, 45–51). TMPRSS2 is also present in the trans-Golgi network; at this level it cleaves HA traveling to the plasma cell membrane. Importantly, the TMPRSS2 expression seems to be higher in the upper and lower respiratory tracts; in contrast, the HAT is mainly represented the upper respiratory tract, suggesting an important role for TMPRSS2 in pulmonary infection (52).
During the assembly phase, the ribonucleocapsids interact with HA and NA and, following M2-mediated membrane depolarization, the viral budding starts, followed by NA-mediated viral release (53). These latest phases directly affect both viral shedding and the clinical evolution of influenza, as demonstrated by NA inhibitors, which are capable of improving flu symptoms only when used in the early phases of infection (54).
During this replicative cycle, the host cell defense machinery is not just a passive bystander, and intracellular innate sensors such as cellular RIG-like receptors (RLR) can recognize the virus and activate the so-called “antiviral state.” In this way, RIG-I recognizes viral RNA and triggers an intracellular signaling cascade, leading to the production of cytokines. Meanwhile, the virus tries to counteract the cellular sensing by blocking RIG-I through its NS1 protein (55). However, these counterstrategies cannot block the production of interferons since Toll-like receptors (TLRs) are also involved in virus recognition (56). Therefore, upon sensing of interferons, host cells are able to transcribe IFN-stimulated genes (ISGs) directly involved in the antiviral state (57).
The activation of these genes is associated with possible viral clearance but may also dramatically contribute to cell damage and clinical severity, especially when the lower airways are involved. Several cellular antiviral defense mechanisms, such as the production of ISGs and mediators altering cholesterol cell homeostasis or cell endocytic pathways, elicited by influenza replication have been described (58–63).
It is evident that influenza-derived cell damage is largely due to a severe deregulation of cell homeostasis even if, in vitro cytopathic effects due to the virus are evident in different cell types. The final effect is highly dependent on the specific cell type infected. In vivo, influenza virus-infected cells are generally cleared via apoptosis and by both innate and adaptive immune response (64, 65). Interestingly, it has been shown that specific populations of epithelial pulmonary cells, named club cells, are able to survive to viral infection thanks to a strong IFN response (66, 67). Moreover, it has been experimentally demonstrated that, after surviving the infection, these cells can acquire an improved antiviral response phenotype possibly required for a nonspecific antiviral immunity after IAV infection, suggesting a tuning of the innate immune response following IAV infection (68).
In fact, even if influenza virus is cytopathic, some cells can also reduce virus replication surviving to the infection. It has been speculated that these “survivor cells” may play an important role in driving the tissue inflammation by altering host response to secondary bacterial infections. Moreover, they could function also as a “long-term” source of viral proteins for the antigen-presenting cells (APCs) (66, 69).
Paramyxoviruses.
The Paramyxoviridae family comprises several viruses involved in human infections, divided into two subfamilies: the Paramyxovirinae and the Pneumovirinae. The former includes parainfluenza viruses (belonging to the Respirovirus and Rubulavirus genera), measles virus, and mumps virus (Morbillivirus) among others; the latter includes respiratory syncytial virus (RSV) and human metapneumovirus (belonging to the genera Pneumovirus and Metapneumovirus, respectively) (70).
Most infections caused by paramyxoviruses are limited to the upper respiratory tract, but the lower respiratory tract may also be involved in specific settings (i.e., RSV bronchiolitis in newborns and pneumonia in children younger than 1 year of age) (71). All paramyxoviruses share several structural analogies: they have a single-stranded nonsegmented RNA genome of negative polarity, they have an envelope, and some of them (Respirovirus and Rubulavirus) have HA and NA on the same spike.
As discussed more fully below, paramyxoviruses bind to different receptors. The docking with cellular receptors is followed by the fusion of virus envelope with host cell plasma membrane that precede ribonucleocapsid release in the cytoplasm. After transcription, viral mRNAs are capped and polyadenylated within the cytoplasm, and replication is supposed to start when an adequate nucleoprotein cytoplasmic amount guarantees the encapsidation of neosynthesized genomes. The virus transcriptase complex, formed by the large protein (L) and phosphoprotein (P), produces new genomes from a double-stranded RNA replicative intermediate. The viral ribonucleocapsid (RNP) is then composed of the association of the transcriptase complex and the nucleocapsid protein (NP) linked to the new genomes. RNP then interacts with the matrix viral protein under the plasma membrane and buds from the infected cell via the ESCRT complex, releasing new virions (72, 73).
Paramyxoviruses have shown to be very efficient in counteracting the mechanisms of innate cell immunity. Many of them encode an immune evasion protein, the V protein, which interferes with RNA recognition in the cytoplasm thus blocking IFN production and induction of the antiviral state (74). V proteins are also known for their ability to interfere with STAT proteins to prevent ISG expression. These proteins also interact with RIG-I-like receptors (RLRs) (75, 76). In particular, inhibition of MDA5 seems to be extremely important to further prevent signaling involved in antiviral effectors production (77, 78).
As anticipated, the tropism of different paramyxoviruses is dependent on their cell receptor usage, and in vitro they can infect a broad variety of cells (79). Respiroviruses and some rubulaviruses can use syaloglycoproteins or glycolipids (80). Interestingly, wild-type clinical isolates of measles virus (WT-MV) cannot use the CD46 receptor as laboratory strains can do. Instead, both lab strains and WT-MV can recognize Signaling Lymphocytic Activation Molecule Family member 1 (SLAMF1) and nectin 4. SLAMF1 can be found on a variety of cells such as activated T, B, and dendritic cells and monocytes (81). Nectin-4 is present on polarized epithelial cells present also in the respiratory tract. The usage of this receptor is important for proper replication of MV also in the upper airways from which the virus can be shed through aerosol. When cultured in vitro, paramyxoviruses cause a lytic infection, although they can also lead to the formation of inclusion bodies and syncytia. The latter represent an important immune-escape strategy allowing the virus to hide from the immune system. Paramyxovirus transmission is mainly airborne. Most infections start and end in the upper respiratory tract, as in the case of most infections caused by human parainfluenza virus 1 (HPIV-1). Sometimes, especially for measles virus (MV), the infection may spread to other sites such as lymphoid or endothelial tissues, while RSV can infect both type I and II pneumocytes (82). The parotid gland, central nervous system (CNS), and endothelial tissues are sites of replication for mumps virus (MuV) (83), whereas Hendra virus (HeV) and Nipah virus (NiV) may target the lungs and CNS (84).
Coronaviruses.
The Coronaviridae is a large family comprising viruses able to infect a broad range of avian and mammal species and includes seven species of human interest: HCoV-229E, HCoV-NL63, HCoV-OC43, and HcoV-HKU1 are endemic and cause seasonal infections; MERS-CoV and SARS-CoV are epidemic viruses; and SARS-CoV-2 is responsible for the current COVID-19 pandemic (85–91). Endemic coronaviruses (CoV) mainly cause self-limiting infections restricted to the upper respiratory tract, although severe manifestations (e.g., pneumonia and bronchiolitis) can happen with higher frequency in young, elderly, and immunocompromised patients. Conversely, epidemic and pandemic viruses frequently replicate in the lower respiratory tracts and are associated with higher lethality rates.
CoVs possess the largest (ca. 27 to 32 kb) monopartite, positive-strand RNA genome of all viruses infecting vertebrates. The genome architecture is conserved and presents the nonstructural proteins (nsp) at the 5′ region and the structural proteins S, E, M, and N at the 3′ region. Furthermore, several species-specific accessory proteins are interspersed among the structural genes and relevantly affect CoV pathogenicity (92). The viral particles are formed by a ribonucleocapsid (viral genome and protein N) with helicoidal symmetry surrounded by an envelope decorated by the S, M, and E proteins and several host proteins, including kinases, cyclophilin A, and APOBEC3G, that can either promote or hamper viral replication (93, 94). Furthermore, HcoV-HKU1 and HcoV-OC43 also expose an HA-esterase that facilitates both viral entry and release (95–97).
All CoVs share the same replicative cycle, that is started by the interaction between protein S and a specific host receptor. Different CoV species, even those belonging to the same lineage, can recognize different receptors or have marked preferentiality to the same receptor of different hosts (98). Also, host receptors can be either proteinaceous or glucidic.
Protein S is a prototypical class I viral fusion protein and thus requires at least one proteolytic cleavage to separate the receptorial (S1) and fusogenic (S2) subunits and mediate fusion (99). Cleavage can happen during biogenesis (usually catalyzed by furin in the Golgi apparatus), upon attachment (catalyzed by membrane proteases such as TMPRSS2), or at the endosomal level (catalyzed by cathepsins). It has been documented that variations in the cleavage sites can alter cellular tropism (100, 101). Once protein S is engaged by host receptors and properly cleaved, it can mediate fusion either at the plasma membrane or in the late endosomes, and the CoV genome is released in the cytoplasm.
CoV genomic RNA (gRNA) is polyadenylated and possesses a 5′-cap synthesized by a viral machinery composed of nsp10, nsp13, nsp14, and nsp16. Therefore, the 5′ portion of the genome, containing a single open reading frame (ORF1ab), can be readily translated. It encodes two polyproteins alternatively translated by ribosomal frameshifting that contain all nsp’s. Mature proteins are released by proteolysis from two viral proteases, the papain-like domains of the multifunctional nsp3 and the chymotrypsin-like protease nsp5 (102, 103).
Once mature, most of the nsp’s participate in the formation of the replication-transcription complex (RTC) that localizes in the double-membrane vesicles (DMV), organelles formed by the virus-induced remodeling of the ER and Golgi apparatus (104, 105). DMVs serve to gather the viral proteins and nucleic acids in order to optimize replication and protect the latter from the host innate immune response. CoV nsp3, nsp4, and nsp6, all possessing transmembrane domains, are implicated in DMV formation and are thought to tether the RTCs to the membranes. Once formed, the RTC begins the synthesis of CoV negative-strand RNAs that can be either continuous or discontinuous, respectively, generating the template for the gRNA replication and subgenomic RNAs (sgRNAs) transcription. Each structural and accessory gene is preceded by a conserved body transcription-regulatory sequence (TRS) and a leader TRS is present in the 5′ untranslated region (UTR). During negative-strand production, every time a TRS complementary is synthesized, the replicative machinery can either proceed or translocate to the leader TRS, producing negative-strand sgRNAs of different lengths, all presenting the 5′-UTR sequence (106). The transcription of negative sgRNAs will produce 5′-capped and polyadenylated mRNAs of different lengths in which the structural or accessory gene at the 5′ will be translated. In the cytoplasm, sgRNAs are translated, and the structural proteins S, M, and E are delivered to the ER, while gRNAs are encapsidated by protein N. The concerted action of proteins N, M, and E leads to virion budding into the ER-Golgi intermediate compartment (ERGIC) and their subsequent release by exocytosis (107).
CoV replication is generally associated with a delayed and dramatically reduced IFN induction in most cell types. The capacity of evading the innate immune response seems to be the highest for SARS-CoV-2, followed by SARS-CoV and MERS-CoV and, generally, human endemic CoVs are worse inhibitors than epidemic and pandemic viruses (108, 109). In the case of SARS-CoV, this leads to the dysregulated activation of inflammatory monocyte-macrophage response, in turn causing vascular leakage and impaired B- and T-cell activation (110). Furthermore, IFN response, particularly of type I, may have either positive or detrimental effect depending on the moment and context in which it is induced (111).
The two main physical factors involved in a dampened infection sensing and IFN induction are the association of viral RNAs with protein N and the protective role of DMVs (112). Recently, SARS-CoV-2 nsp3 has been shown to form pores that allows the passage of molecules from the DMVs lumen to the cytoplasm. Of note, the authors speculate that negative-strand RNAs remain confined in the DMVs and that only mature positive-strand sgRNAs and gRNAs migrate to the cytoplasm. This suggests a selective segregation in the DMVs of single- and double-strand RNA intermediates that are more susceptible to recognition from the host innate response, thus dampening IFN induction (113). Also, CoV RNAs are further protected by complexation with protein N when in the cytoplasm.
Nonetheless, CoV infection is still sensed by the host innate immune response, since viral RNAs activate PKR, OAS, TLR7, MDA5, and RIG-I, whereas proteins E and M activate NF-κB and TLR-like signaling, respectively (114). Therefore, in addition to the importance of protein N and DMV direct shielding of viral nucleic acids, several CoV proteins are further involved in dampening IFN and ISG induction. Of note, most of the factors have been identified in SARS-CoV, but evidence of analogous function in other human CoVs is often documented. Furthermore, functional conservation in murine CoV-MHV-A59 demonstrates common immune escape strategies among phylogenetically distant species.
The CoV proteins whose inhibitory effect on IFN induction has been characterized so far are nsp1, nsp3, nsp16, protein M, and protein N. More specifically, nsp1 is involved in the downregulation of cellular protein expression by selectively mediating host mRNA degradation and directly blocking its translation (115–118).
The papain-like protease domains of nsp3, besides processing the virus polyproteins, also have deubiquitinase activity on innate immune response factors such as IRF3, IRF7, TBK1, and RIG-I (119).
As previously mentioned, mature CoV RNAs are poor IFN inductors since they are single-stranded, 5′ capped, and polyadenylated. MDA5-mediated IFN production is induced upon CoV infection only in specific cell types. The importance of 5′ capping in downregulation is demonstrated by the increased and generalized IFN production triggered by MDA5 when the 2′-O-methylation activity of nsp16 is abrogated (120, 121). Of note, 2′-O-methylation is the last enzymatic step in viral RNA 5′-capping formation, suggesting a similar, although not directly demonstrated, MDA5 sensing suppression by nsp10, nsp13, and nsp14.
Protein M is also involved in preventing IRF3 function by directly binding to TRAF3 (122–124). The abrogation of the TRAF3-TBK1 complex formation in turn blocks phosphorylation and activation of IRF3.
Finally, protein N is also able to suppress type I IFN induction by directly binding to TRIM25, thus blocking RIG-I ubiquitination. Intriguingly, both SARS-CoV and MERS-CoV protein N are able to inhibit RIG-I-induced IFN production, although with partially different pathways (125, 126). Also, EF1α is another ligand of protein N, and its inhibition leads to several effects on cellular physiology, including protein translation block (127).
Altogether, the suppressive role of nsp’s on IFN induction indicates CoV ancestral evolution to counteract the host innate immune response, and it is therefore conserved in the entire family. In addition, several species- and lineage-specific accessory proteins participate in the process, as demonstrated mainly by epidemic and pandemic viruses. In SARS-CoV, ORF3b has an inhibitory effect on RIG-I, MDA5 and, together with ORF6, the phosphorylation of IRF3 (128). Also, two products of ORF8, -8b, and -8ab, directly interact with IRF3, preventing its dimerization and signaling (129). In MERS-CoV, ORF4a dampens MDA5 signaling through the interaction with PRKRA, downregulates the expression of genes under NF-κB control, and, in concert with ORF4b and ORF5, hampers the nuclear trafficking of IRF3 and activation of the IFNB promoter (130). ORF4b activity also affects IRF3 phosphorylation by abrogating the interaction between MAVS and IKKε. Finally, SARS-CoV ORF9b interacts with PCB2 and E3 ubiquitin ligase AIP4, causing TRAF3, TRAF6, and MAVS selective degradation (131, 132).
A growing body of evidence is highlighting the molecular processes at the basis of SARS-CoV-2 strong immune evasion: SARS-CoV-2 nsp1, nsp6, and ORF3b have greater IFN suppressive activity than do SARS-CoV and MERS-CoV orthologs, and a SARS-CoV-2 ORF3b natural variant identified in two patients with severe disease showed increased IFN suppression (133).
CoVs also show a variable ability to inhibit the expression of ISGs in response to IFN signaling through the activity of several nonstructural and accessory proteins. Specifically, nsp1 and nsp6, respectively, inhibit the phosphorylation of STAT1 and STAT2 in a virus species-specific manner, and SARS-CoV-2 proteins are the strongest inhibitors (134, 135). Other inhibitory proteins commonly involved are M, nsp13, and ORF7b in STAT1 phosphorylation; nsp13, ORF7a, and ORF7b in STAT2 phosphorylation; and ORF6 in STAT1 translocation. It has been reported that SARS-CoV-2 can block STAT1 and STAT2 nuclear translocation, altering the proper induction of ISGs through the interaction of Orf6 encoded protein with Nup98-Rae1 and thus resulting in alteration of the docking of cargo-receptor (karyopherin/importin) complex at the nuclear pore complex (NPC) level (128). Other CoV accessory proteins negatively acting on the expression of genes under the ISRE promoter include MERS-CoV ORF4a and ORF4b, SARS-CoV-2 ORF3a, and SARS-CoV ORF3b (133, 136, 137).
Finally, it has been shown that CoVs are able to reduce antigen presentation, thus downregulating the development of an efficient adaptive immune response. MERS-CoV suppresses HLA expression, while SARS-CoV-2 sensing from the host immune response seems to be repressed by ORF8, which has the unique ability to directly bind to MHC-I at the ER membrane and direct it to the lysosomal compartment (138, 139).
All of these immune evasion strategies are at the basis of the SARS-CoV, MERS-CoV, and SARS-CoV-2 clinical course and pathological features that, although distinct, show remarkable similarities. Severe manifestations are indeed directly mediated by an abnormal immune response that progresses into ARDS through the sustained upregulation of proinflammatory cytokines, the delayed/dampened type I IFN response, and the lack of a switch from the innate to the adaptive immune response. Of note, SARS-CoV-2 also often triggers venous thrombosis, both pulmonary and systemic (140, 141).
A relevant source of differentiation in terms of pathogenesis comes from the receptor that SARS-CoV, MERS-CoV, and SARS-CoV-2 exploit to enter the host cells. MERS-CoV binds to the dipeptidyl peptidase-4 (DPP4), a surface protein mainly expressed on alveolar macrophages and, to a lesser extent, on alveolar epithelial cells and T cells (142, 143). This marked tropism for immune rather than lung cells is at the basis of MERS-CoV immunopathogenesis. SARS-CoV and SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2), which is predominantly found on type II pneumocytes in lungs, although it is also present on several other cell types (innate and adaptive immune cells, vascular smooth muscle cells, cardiomyocytes, and enterocytes) (144, 145). The tropism differences may be at the basis of the lower ability of MERS-CoV to abrogate IFN signaling, since it achieves immune evasion by directly infecting immune cells. A remarkable, receptor-mediated pathogenic process comes from the dysregulation of the renin-angiotensin system (RAS) induced by SARS-CoV and SARS-CoV-2. By binding to ACE2, they induce both its internalization and shedding through ADAM17 activation; a reduced ACE2 activity results in increased vascular permeability, enhanced lung edema, and worsening lung damage and proinflammatory response (146–148). Of note, alteration of ACE2 expression seems to be directly related to the stability of the spike-ACE2 complex, since the endemic virus HcoV-NL63 does not trigger the same RAS alterations, and its spike has lower affinity than SARS-CoV for ACE2 (149, 150). In this scenario, SARS-CoV-2, having the highest affinity for ACE2, may also have the highest RAS-disrupting effect.
While the role of inflammation (and coagulopathy for SARS-CoV-2) has been identified as the main pathogenic player in SARS-CoV, MERS-CoV, and SARS-CoV-2, all of these viruses also have a direct cytopathic effect on infected cells. However, the in vitro effect is highly variable depending on the cell line used for the three viruses and some differences are observed between in vitro and autoptic samples. Of note, viral titer and cytopathic effect are independent, since high yield can be achieved in the absence of direct cell damage. The cytopathic effect is directly related to the induction of syncytia and occurs through the induction of apoptosis and autophagy, while necrosis is less associated with CoV-induced cell death, especially in vitro (151, 152). The role of several CoV proteins have been identified in the induction of cell death. Protein N has been shown to interact with several host factors, inducing alterations of the cell cycle. Indeed, by interacting with CDK2, CDK4, and CDK6, protein N can arrest the cell cycle in the G1 or S phase and by binding to EF1α it can block protein translation and cytokinesis, leading to cell proliferation inhibition (127). Intriguingly, the comparison of protein N from SARS-CoV and HcoV-229E has shown a direct relationship between the affinity for EF1α and the rate and frequency in which multinucleate cells are induced, thus suggesting a possible pathogenic mechanism that is more pronounced in SARS-CoV. ORF-8b directly induces the autophagosome formation and causes lysosome malfunctions related to ER stress (153). ORF3a directly inserts into the lysosomal membrane and leads to necrosis by promoting caspase-3 activity (154).
DNA Viruses
Adenoviruses.
Human adenoviruses (HAdVs) are nonenveloped viruses involved in respiratory disease, such as croup, bronchitis, and pneumonia, as well as ARDS in immunocompromised individuals or patients with preexisting comorbidities (155–159). HAdVs may also cause keratoconjunctivitis, gastroenteritis, or cystitis, and they are preferentially transmitted through respiratory droplets or the fecal-oral route (160). As for other respiratory viruses, Adenoviruses may infect respiratory tract also through contaminated hands or fomites (160, 161). HAdVs are included in the genus Mastadenovirus in the Adenoviridae family. Seven species of HAdVs (A to G) are known; these are further divided into more than 50 serotypes and 70 distinct genotypes (162). The HAdV species most involved in upper or lower respiratory tract infections include HAdV B, HAdV C, and HAdV E (163).
The tissue tropism of each HAdV species determines the different clinical manifestations and is driven by the different host cell receptors recognized. However, receptor usage is not the only element driving tissue tropism. As an example, genotypes Ad19 and Ad37, often clinically associated with keratoconjunctivitis, bind to α2,3-sialic acid or CD46 that are broadly expressed in vivo on other, different cell types (164). HAdV C and E recognize the CXADR (coxsackievirus and adenovirus receptor) (165), which is a component of the epithelial apical junction complex also behaving as a cell adhesion molecule important for tight-junction integrity. This receptor is usually expressed in lung, pancreas, brain, heart, small intestine, testis, and prostate and, at a lower level, in liver. Also, it is possibly involved in transepithelial passage of leukocytes (166, 167). CD80, which acts as a cell receptor for HAdV B, is expressed in the tonsil, lymph node, vermiform appendix, gastrocnemius, liver, upper lobe of the lungs, and other tissues, and CD86 is expressed in the tonsil, monocyte, leukocyte, granulocyte, mononuclear cell, lymph node, and vermiform appendix (168).
The adenoviral protein involved in host cell receptor attachment is the fiber glycoprotein mediating the endocytosis and fiber shedding (169). Endosomal acidic pH triggers the release of viral capsid in the cell cytoplasm thanks to the viral protein VI activating membrane repair system, leading to the production of plasma membrane ceramides which promote virus penetration into the endosomal membrane. It has also been suggested that virus entry can be enhanced by elevated levels of autophagy (155, 164). Then, after release, the virus penetrates into the host cell nucleus for replication. Early genes (E) are first transcribed by host RNA polymerase II, and replication of the virus genome by DNA strand displacement occurs. Late genes (L), coding for structural proteins, are then transcribed by the host cell RNA polymerase II, and new virions are assembled into the nucleus. Viral progeny is then released by cell lysis through autophagy and autophagy-mediated caspase activity (170). The lytic infection caused by adenovirus infection probably represents the most typical example of virus direct damage to the host cells. However, as already seen for other viruses, the cell sensing of viral compounds heavily alters the physiology of infected cells starting from the early phases of viral replication. It has been reported that the initial innate immune response to adenoviruses in alveolar macrophages induces pathways that regulate the epithelial polarity of host cell receptors (polarization of CAR and αV integrins involved in virus docking on the apical surfaces of respiratory epithelial cells) by inducing the secretion of interleukin-8 (IL-8) (171). It has also been observed that membrane damage during adenovirus cell entry triggers a cell stress response (172, 173). Further signaling pathways are activated by the presence of virus DNA and the expression of virus-associated small RNAs (174).
Adenoviruses evolved several strategies to counteract cell sensing and cell mechanisms hampering virus intracellular trafficking in different cell types. As for other intracellular pathogens, the adenovirus E3 protein interacts with host immune system elements for immune evasion (175, 176). Moreover, host cell pathways connecting E3 to proinflammatory NF-κB signaling in respiratory epithelial cells were also described. In particular, there are findings suggesting that E3-RIDα regulates NF-κB activity (155, 177). AdV infection affecting the bronchial and bronchiolar mucosa or the pulmonary alveolus (both type I and II pneumocytes are permissible to virus infection) can cause necrotizing infection. Zaki and Paddock described how, in tissue sections, distinct intranuclear basophilic inclusions, surrounded by a small halo or often filling the entire nucleus, may be appreciated (178). The inclusion bodies, composed of adenovirus viral particles within the cell nucleus, were also detected by electron microscopy (179).
Herpesviruses.
The Herpesviridae family comprises human viruses highly prevalent in the global population, the most relevant being herpes simplex virus (HSV), cytomegalovirus (CMV), Epstein-Barr virus (EBV), and varicella-zoster virus (VZV). These viruses all share the ability, upon primary infection, to establish lifelong latency in specific cell types that can lead to reactivations with variable frequency. Primary infections and reactivations are defined as lytic, characterized by the expression of most open reading frame (ORFs) and the production of viral progeny. Conversely, during latency, only a small subset of genes, generally named latency-associated transcripts (LATs), is expressed and necessary to maintain the virus genome as an episome in the nucleus (180). The viral particles carry a large, linear dsDNA genome presenting 100 to 200 ORFs, depending on the species. The genome is surrounded by an icosahedral capsid, a complex tegument, and an envelope decorated by an unusually high number of viral glycoproteins. The general features of the Herpesviridae replicative cycle are conserved, and it takes place as follows. The entry phase is mediated by a cascade of interactions among surface glycoproteins, where a minimal functional unit can be identified in a receptorial protein (gD, following HSV naming), an activator (the gH/gL complex), and a fusion protein (gB). Whether the fusion events happen directly at the plasma membrane or in the endosomes depends on the virus species and the host cell types (181). Once released in the cytoplasm, the nucleocapsid is shuttled to the cell nucleus, where the genome is replicated and transcribed. The ORFs expressed during the lytic phase can be divided according to their temporal expression into three groups: immediate-early, early, and late genes. Immediate-early genes are involved in the protection from the host innate immune response and have a regulatory effect on the transcription of the early genes that encode the replicative machinery. Late genes are translated into the structural proteins necessary to produce the viral progeny (182, 183). The newly formed nucleocapsids are assembled into the nucleus and released by exocytosis after two subsequent budding events from the inner lamella nuclear membrane and the Golgi apparatus (184).
The immune system of healthy individuals can limit the lytic phase; indeed, viral glycoproteins and nucleic acids are recognized as pathogen-associated molecular patterns (PAMPs), thus activating the innate immune response and inducing IFN signaling. Furthermore, viral replication stimulates the host cell stress responses. However, all Herpesviridae members have evolved a plethora of effectors capable of downregulating the sensing and signaling pathways involved in IFN induction, the expression and effect of ISGs, and dysregulating the cell death programs induced by ER stress and DNA damage response (185, 186). Dampening the IFN response allows the herpesviruses to efficiently enter the latency phase following primary infection, whereas modulation of the cell death program is cell type specific and therefore has a multifaceted role. Indeed, herpesviruses are often able to induce apoptosis in immune cells, further promoting evasion, and in cell types where the lytic phase takes place, but can protect from apoptosis those where they enter latency. Two notable examples are HSV and VZV, which are able to induce and block apoptosis in epithelial cells and neurons, respectively (187). Consequently, viral proteins involved in these processes are timely expressed: regulators of the IFN response and promoters of programmed cell death are encoded by immediate-early genes and therefore are readily produced during the lytic phase. Several effectors are also part of the tegument; thus, their role is exerted as soon as the envelope fuses to the cell membranes during entry. Conversely, effectors blocking the apoptotic pathways often are LATs (188, 189).
None of the Herpesviridae family members primarily target the lungs; nonetheless, several of them can cause interstitial pneumonia, bronchopneumonia, and ARDS in specific settings. Specifically, VZV, CMV, EBV, and HSV are associated with nosocomial primary infections and reactivations in ICU patients (190). CMV is also among the most common opportunistic pathogens causing pneumonia in transplanted patients, especially those undergoing hematopoietic cell transplant, HIV-infected patients, and newborns. The latter group is also afflicted by primary perinatal HSV infections, which are characterized by systemic dissemination and have a worse outcome when associated with pneumonia. In general, a fully functional immune response can control the lytic phase of herpesviruses, impede their dissemination, and drive the progression to latency and then maintain it. Conversely, lung involvement is a variably frequent consequence of herpesvirus systemic spread associated with immune deficiencies, particularly those associated with a T-cell response. A notable exception is VZV community-acquired pneumonia, a severe complication associated with primary infection in immunocompetent adults.
Pulmonary involvement can be a consequence of different pathological processes, depending on the Herpesviridae species. HSV pneumonia is mainly caused by HSV type 1 and affects immunocompromised patients and immunocompetent adults subjected to prolonged intubation. Pulmonary manifestations range from necrotizing tracheobronchitis to necrotizing and interstitial pneumonia and are usually caused by HSV orolabial reactivation that progresses into tracheal or lung infection (191). HSV can be found in both type I and type II pneumocytes, epithelial cells, and leukocytes. Infection causes the formation of inclusion bodies and syncytia that results in a marked infiltration of inflammatory cells in the lung parenchyma, necrosis, and alveolar hemorrhage (192). Neonatal infections are mainly due to HSV type 2 infections acquired during delivery predominantly from acutely infected women. Lung involvement is part of a disseminated, multiorgan infection that happens in approximately 25% of neonatal HSV infections and represent the most severe complication (193).
CMV pneumonia can happen both in newborns and immunocompromised adults. Similar to HSV infections, lung involvement in the two categories is radically different. Neonatal CMV pneumonia is preferentially caused by infection acquired during delivery or perinatally, and it is usually caused by direct, descending airway infection, as suggested by a significantly lower lung involvement in congenital infections (194). Conversely, lung involvement in immunocompromised adults is secondary to systemic spread either following primary infection or reactivation. Finally, rare CMV pneumonia has also been reported in immunocompetent individuals (195). Regardless of the immunological state of the patient, CMV pneumonia is often characterized by intermittent symptoms due to recurring immune responses associated with diffuse interstitial infiltrate. The typical CMV cytopathic effect, characterized by intracytoplasmic and intranuclear inclusion bodies, can be identified in type I and type II pneumocytes, alveolar macrophages, and mesenchymal cells, showing CMV broad tropism (196).
VZV is the only Herpesviridae member characterized by a significant prevalence of pneumonia in immunocompetent adults, albeit an impaired cellular immune response favors the pulmonary involvement (197). Pneumonia pathogenesis is caused by a defective control of primary infection that, upon systemic dissemination, reaches the lungs. Rarely, recurrent pulmonary involvement is also associated with VZV reactivation in immunocompromised patients. VZV lung infection rapidly progresses, showing the clinical signs of interstitial pneumonia and ARDS and often leading to death by untreatable hypoxemia (198). Histopathological characterization shows scattered hemorrhagic focal necrosis characterized by the presence of inclusion bodies in infected type I and type II pneumocytes. Of note, VZV shows tropism for monocytes, leukocytes, and dendritic cells, all infiltrating the lung parenchyma, and can deregulate their functionality (199, 200). This feature, although not fully understood, is at the basis of VZV dissemination to the lungs independently from the immune competency of the infected individual.
Human bocaviruses.
Human bocaviruses (HBoVs) are single-stranded DNA (ssDNA) genome naked virus members of the Parvoviridae family. They have been described in respiratory samples from patients with respiratory illness or gastrointestinal clinical symptoms and may be transmitted via the respiratory route, or by ingestion, reaching the gastrointestinal tract (201). HBoV1 was first reported in patients with infections of the lower respiratory tract (202), and it was suggested that it might be associated with acute expiratory wheezing and pneumonia (203–205), whereas other bocaviruses are more related with diarrhea (206). It is estimated that up to 19% children under 2 years old with acute respiratory illness worldwide carry HBoV1 genome, and 10 to 20% of nasopharyngeal aspirates from hospitalized children with lower respiratory illness are also positive for HBoV1 (203, 207–210). Respiratory diseases correlated with the presence of HBoV are usually mild and often characterized by the presence of other respiratory viruses (204, 211–214). Severe respiratory infections, including life-threatening acute bronchiolitis and pneumonia, linked to HBoV1 have also been reported (205, 212, 215–218).
From the few data generated in vitro and the homologies with human parvovirus B19, the proposed replication model for HBoV suggests (216) a first docking with host cell receptors (sialic acid and heparan sulfate) triggering clathrin-mediated endocytosis (219). After permeabilization of the endosome membrane, the virion enters the nucleus via microtubular transport. At the nuclear level, the double-stranded viral DNA is synthesized by host cell enzymes. Viral mRNAs are then transcribed when host cell enters the S phase; alternative splicing of messengers allows expression of eight different mRNAs and then translation occurs. The viral VP2 is translated by “leaky scanning” of the gene encoding VP1, and VP3 is generated via VP2 cleavage (220). Nonstructural virus proteins contribute to genome replication which would occur by a rolling-hairpin mechanism. Then, the NS1 endonuclease, recognizing the 5′ ends of the viral genomes, contributes to the excision of neosynthesized ssDNA genomes thanks to a process known as junction resolution. At this stage of viral replication, the novel ssDNA can both function as a template for transcription and be encapsidated into new virions that can be then released by lysis of the host cell (221).
The pathogenetic mechanisms of HBoV1 have not yet been elucidated due to the lack of animal models and of cell lines allowing a fine and reproducible characterization of its replicative cycle. It has been shown that HBoV1 can replicate in human embryonic kidney HEK293 cells after transfection with an HBoV1 clone and produces progeny virions (222). Even more importantly, it was also shown that HBoV1 can infect polarized differentiated human airway epithelium (HAE) cell preparations on both apical and basolateral surfaces. This evidence suggests that HBoV1 can induce tissue damage at the airway epithelial level by possible alteration of ciliary function, alteration of cell-cell junctions, or through the induction of epithelial cell hypertrophy (223–227).
IMMUNE-MEDIATED DAMAGE INDUCED BY RESPIRATORY VIRUSES
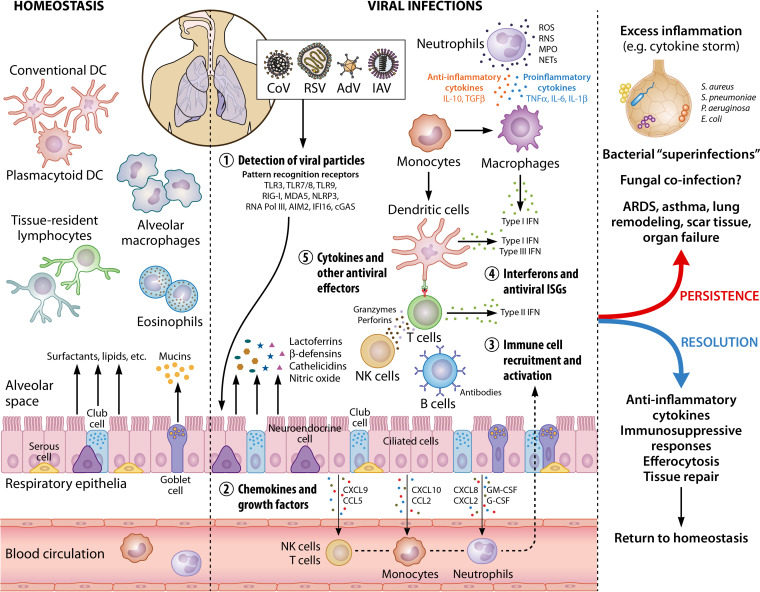
As already anticipated in the previous section, the role of the immune system during a viral infection is balanced on a thin line between protection and exacerbation of virus-induced damage (Fig. 2).
FIG 2.
Immune cell activation in the lungs during viral infections. In healthy lungs, the resident immune cells include primarily alveolar macrophages, conventional and plasmacytoid dendritic cells, and tissue-resident lymphocytes and eosinophils, which patrol the tissue for foreign threats. In addition, respiratory epithelial cells (e.g., club cells and goblet cells) secrete mucins, surfactants, and other molecules that preserve homeostasis, as well as maintain immune cells in their quiescent states. During viral infections, viruses are sensed by innate immune receptors (or pattern recognition receptors) which activate immune responses. Secretion of chemokines and growth factors by respiratory epithelium and resident immune cells leads to the phased recruitment and activation of neutrophils, monocytes, NK cells, and T cells. Type I (IFN-α/β) and type III (IFN-λ) IFNs are the primary cytokines produced upon viral detection, followed by other inflammatory cytokines, as well as ISGs. Other antiviral effectors produced by epithelial, endothelial, and immune cells include lactoferrins, β-defensins, ROS, and RNS. Innate immune cells (neutrophils, macrophages, and DCs) mediate the activation of the adaptive responses (NK cells, B cells, and T cells) that involve IFN-γ secretion, antibody production, and cytotoxic killing of infected cells. Persistence of a viral infection, as well as the accompanying antiviral immune responses, often leads to widespread lung damage and secondary complications such as systemic inflammation (due to dysregulated immune responses) and bacterial coinfections. A prolonged infection can result in oxygen deprivation, ARDS, asthma, remodeled lung structure (e.g., excess collagen deposition, thickening of basal membrane, and scar tissue formation), organ failure, and even death in extreme cases. Resolution of a respiratory viral infection by a kinetically controlled, successful immune activation and a reversal to homeostasis is facilitated by anti-inflammatory cytokines, immunosuppressive molecules, the removal of active, cytotoxic immune cells by efferocytosis, and extensive tissue repair. This image was created using BioRender (BioRender.com). DC, dendritic cells; CoV, coronavirus; RSV, respiratory syncytial virus; AdV, adenovirus; IAV, influenza virus; TLR, Toll-like receptor; RIG-I, retinoic-acid inducible gene I; MDA5, melanoma differentiation-associated protein 5; NLRP3, NOD-like receptor protein 3; RNA Pol III, RNA polymerase III; AIM2, absent in melanoma 2; IFI16, IFN-inducible protein 16; cGAS, cyclic GMP-AMP synthase; CXCL, C-X-C motif chemokine ligand; CCL, C-C motif chemokine ligand; GM-CSF, granulocyte-macrophage colony-stimulating factor; G-CSF, granulocyte colony-stimulating factor; NK cells, natural killer cells; IFN, interferon; ISG, interferon-stimulated genes; IL, interleukin; TNF, tumor necrosis factor; TGFβ, transforming growth factor β; ROS, reactive oxygen species; RNS, reactive nitrogen species; MPO, myeloperoxidase; NET, neutrophil extracellular traps.
Protective Role of Immune System against Respiratory Viruses
The immune system is equipped with pattern recognition receptors (PRRs) that are involved in viral sensing by recognizing PAMPs unique to viruses and other microbes. For viruses, their genetic material acts as PAMPs, which are detected by the host PRRs when viruses enter or replicate in a cell or, as viral replication intermediates from dying cells.
The receptors critical for recognizing viruses are (i) Toll-like receptors (TLRs) such as TLR3 (double-stranded RNA), TLR7/8 (single-stranded RNA), and TLR9 (unmethylated CpG DNA) which signal in a TRIF-dependent manner; (ii) retinoic acid-inducible gene (RIG-I)-like receptors (RLRs), including RIG-I, and melanoma differentiation-associated protein 5 (MDA5) that detect cytosolic, single-stranded, or double-stranded viral RNAs of various lengths to initiate IFN signaling; (iii) nucleotide-binding oligomerization domain (NOD)-like receptors (NLR), such as NLRP3, that recognize RNA viruses and cellular perturbations to activate ASC/Caspase-1-dependent inflammasomes; and (iv) cytosolic DNA virus sensors such as RNA Pol III (converts viral DNA to RNA for detection by RIG-I), AIM2, IFI16, and cGAS/STING that activate inflammasomes or IFNs. These receptors are expressed in the respiratory epithelial cells, as well as in the tissue-resident immune cells. Upon viral sensing, the PRRs activate signaling pathways that trigger the release of type I and III IFNs, as well as proinflammatory mediators, including cytokines, chemokines, and antimicrobial peptides, that assist in the prevention and clearance of respiratory viral infections.
Prevention of respiratory infections.
The respiratory tract is continuously exposed to the external environment that increases the likelihood of encountering pathogens. The lower and upper airways are uniquely fortified for detecting and eliminating external threats to maintain homeostasis and prevent infections. The lung epithelium end endothelium associated with resident immune cells actively survey for foreign threats while after viral encounter, immune cells such as neutrophils, monocytes, dendritic cells (DCs), T cells are recruited to resolve the infection.
(i) Airway epithelium.
The respiratory epithelium consists of ciliated columnar epithelial cells associated with nonciliated airway basal cells and secretory cells (serous, neuroendocrine, goblet, and club cells), which secrete mucus, enzymes, proteases, and antimicrobial peptides. It starts as the nasal epithelium continuing onto the trachea to the bronchus and bronchioles and finally forming the alveolar epithelium (made up of AT-I and AT-II cells).
Cell-cell junctions among epithelial cells, including apically located tight junctions, the underlying adherens junctions, gap junctions, and desmosomes, provide an impermeable structure resistant to infections. In the pseudostratified columnar epithelium, they maintain basal-to-apical directionality by the Na+, K+, and Cl− ion gradient that guides secretion and enables intercellular communication and transport (228, 229). In some cases, despite apoptosis of ciliated cells, the lung barrier functions were retained during H1N1 and H3N2 IAV infections by maintaining transepithelial resistance and expression of tight-junction proteins (230). Others reported that H1N1 and H5N1 IAV infection damaged the epithelial cell tight junctions affecting the epithelial-endothelial barrier and was dependent on the loss of a tight-junction protein, claudin-4 (231). Inflammatory responses during RSV infections destabilizes cell-cell junctions by decreased expression or cleavage of tight-junction proteins such as occludin and claudin-1 and enhanced claudin-2 expression that increases barrier permeability (232, 233).
The mucus layer formed by mucins secreted by the goblet cells protects the epithelium by forming a physical barrier against an incoming virus and limiting access to the lung surface. Several isoforms of mucins are present in the lungs that are either secreted (MUC5B, MUC5AC, and MUC2) or are bound to the epithelium (MUC4, MUC13, MUC16, and MUC21). In addition to its barrier function, in an IAV infection, the secreted MUC5AC isoform acts as decoy for sialic acid receptors (the receptor for IAV binding) and thus limits exposure of the epithelium to IAV (234). Another study showed that the mucin layer senses viruses such as herpes simplex virus (HSV) and facilitates an early release of CXCL10 and recruitment of neutrophils prior to even IFN responses (235).
Lipids and surfactant proteins (released by the epithelial cells) in the alveolar space contribute to the structure and function of pulmonary surfactant and have antiviral functions. AT-II epithelial cells constitutively secrete the surfactant proteins SP-A, SP-B, SP-C, and SP-D. SP-B and SP-C reduce the surface tension in the alveoli to withstand the dynamic changes during ventilation but have limited antiviral properties. SP-A and SP-D, described in detail below, function as “collectins” or soluble PRRs that sense several viruses and microbes and enhance opsonization by alveolar macrophages and neutrophils.
Upon viral encounter, the pulmonary epithelium secretes several cytokines, chemokines, and antimicrobial peptides (AMPs). Cytokines (e.g., TNF-α, IL-6, IL-1β, G-CSF, and GM-CSF) broadly regulate immune responses, as well as cell proliferation and maturation, and control viral spread (described in detail below). Chemokines produced by the lung epithelial cells stimulate the recruitment of neutrophils, T cells, NK cells, DCs, monocytes, and granulocytes to the lungs. In addition, the lung epithelial cells also produce peptides such as β-defensins, lactoferrin, and nitric oxide (NO), which have antiviral activity (described in detail below). Secretory leukocyte protease inhibitor in the lungs has little antiviral abilities but protects the lung tissue integrity from harmful proteolytic enzymes released by the epithelial and inflammatory immune cells.
The airway epithelial cells also maintain homeostasis by regulating the inflammatory state in the lungs. For example, the cells prevent aberrant IL-1β responses by secreting IL-1RA and IL-1RII inhibitors in the steady state. However, during viral infection in the lungs, the anti-inflammatory state can be rapidly reversed by the release of other proinflammatory cytokines.
(ii) Endothelial cells.
Pulmonary endothelium is a dynamic layer of squamous cells on the surface of the lung vasculature and is a critical regulator of vascular homeostasis. At steady state, the endothelial cells have antiplatelet, anticoagulant, and fibrinolytic properties and maintain vascular integrity. Upon microbial infections, the endothelium expresses cytokines, chemokines, adhesion molecules, and growth factors.
While the airway epithelia are the primary target of respiratory viruses, viruses such as RSV that also directly infect the lung vascular endothelium cause respiratory distress and extensive tissue damage due to proximity to the alveoli, as well as increased permeability of inflammatory mediators, leading to pulmonary edema. Endothelium damage affecting vascular permeability and fluid leakage can also be indirectly mediated by leukocyte-driven inflammation and damage to the alveolar epithelium.
During infections, microvascular endothelial cells secrete macrophage inflammatory protein 1β (MIP-1β) and MIP-2 (CXCL2) or IL-8 that recruit neutrophils into the inflamed lungs. The activated endothelium also produces the proinflammatory cytokines, chemokines and interferons (e.g., IL-6, TNF-α, IL-1β, CCL2, CXCL9, CXCL10, and type I and II IFN) (236, 237), and adhesion molecules (e.g., PECAM [CD-31], ICAM–1, VCAM, and E selectin) that recruit leukocytes to the site of infection and mediate leukocyte/endothelial-cell adhesion.
(iii) Alveolar macrophages.
Lung-resident macrophages are present within the alveolar airspaces to protect the lower airways from external pathogens, and they make up >90% of the leukocytes. Alveolar macrophages are highly phagocytic and ingest all types of inhaled, inert particulates in the alveolar spaces such as amorphous silicates, carbon-graphite particles, without triggering inflammation. In healthy lungs, a homeostatic microenvironment is sustained by maintaining the macrophages in an immunosuppressed state by the alveolar epithelium through surfactant release, nitric oxide, IL-10, TGF-β, or GM-CSF signaling (238, 239). The alveolar macrophages also maintain a state of T-cell inactivation by decreased expression of costimulatory molecules (240) or through expression of CD80 that binds the negative T-cell receptor, CTLA-4 (241).
Upon viral infection of the lower respiratory tract, the microenvironment rapidly changes into an inflammatory state that removes the suppressive stimuli to activate the alveolar macrophages that initiate host immune response and viral clearance. Upon resolution of the infection, the acute inflammatory state in the lungs is reversed by efferocytosis, a process by which the inflammatory cells containing cytotoxic mediators are removed. After influenza virus infection, efferocytosis is primarily driven by the alveolar macrophages that remain in the alveolar space after the inflammation is resolved (242). Lipid mediators such as lipoxins, protectins, and resolvins produced by the alveolar macrophages also potentiate efferocytosis (243). Alveolar macrophages return to their original quiescent state by producing TGF-β and IL-10.
Alveolar macrophages are essential in the lungs during viral infections such as RSV infections, where they mediate early antiviral responses by cytokines and IFN secretion. However, during several other respiratory viral infections, namely, IAV and coronavirus infections, the alveolar macrophages are often depleted from the lungs. In fact, in COVID-19 patients, the severity of the disease correlates with the reduction in tissue-resident alveolar macrophages, along with the accumulation of the inflammatory, monocyte-derived macrophages (244).
(iv) Neutrophils.
Neutrophils (245, 246) are the first immune cells to migrate to the lungs during respiratory virus infections and are crucial for removing infected and dying cells. Neutrophils phagocytose viral particles and apoptotic bodies from dying cells that contain viral particles and replication intermediates. Neutrophils release intracellular and extracellular granules containing cytotoxic enzymes, AMP, and ROS (produced by NADPH oxidase) to inactivate or eliminate the viral pathogens. Activated neutrophils also form and release NETs (neutrophil extracellular traps) composed of decondensed chromatin and histones that biophysically incapacitate viruses and avert viral spread. In addition, activated neutrophils also modulate other innate and adaptive immune responses by secreting chemokines that recruit more neutrophils, activate T-cell responses through IFN-γ secretion and antigen presentation, or hone B-cell activity. Efficient neutrophil recruitment and activity check disease pathogenesis in respiratory viral infections. Generally, the activated, inflammatory neutrophils at the sites of infections are short-lived as they undergo apoptosis or NETosis (i.e., programmed cell death accompanied by formation of NETs, which are cleared by tissue-resident macrophages). However, unmitigated neutrophil-mediated activity (e.g., release of ROS, myeloperoxidase, proteolytic enzymes, chemokines, and NETs) in the alveolar space impaired neutrophil cell death or inefficient clearance of dying cells by alveolar macrophages causes hyperinflammation and acute tissue injury.
(v) Dendritic cells.
The lung-resident DCs consist of CD103+ or CD11b+ conventional DCs (cDCs) and plasmacytoid DCs (pDCs). The cDCs are of two types: (i) CD103+, CD8α+, IRF8+ type I (or cDC1) and (ii) CD11b+, CD8α+, IRF4+ type II (or cDC2). In the steady state, CD103+ cDC1 cells remain associated with the lung epithelium and are the primary antigen-presenting cells (APCs) that are responsible for activating both CD8+ and CD4+ naive and memory T cells upon antigen encounter. Upon infection, CD103+ cDCs are the first to migrate to the draining lymph nodes (dLN) to elicit CD8+ T-cell-dependent protective immunity. Deficiency in CD103+ DCs during influenza leads to a reduced cytotoxic-T-cell (CTL) population in the lungs and severe pulmonary inflammation (247). CD11b+ DCs arrive in the dLN later during the infection, and this augments the expansion of effector lymphocytes and the secretion of proinflammatory cytokines. Both cDC populations drive Th1 activation and promote effective memory responses against subsequent infections (248–251).
During homeostasis, cDC1 cells have a greater capacity to present antigens to CD8+ T cells, while cDC2 cells are more efficient in activating CD4+ T cells (252–254). During viral infections such as influenza, some groups showed that cDC1 cells are responsible for inducing CD8+ T-cell responses (255, 256), while others claimed that cDC2 cells are more important (257, 258). Recently, through single-cell analysis, it was demonstrated that during respiratory virus infection, cDC2 cells acquire properties similar to cDC1 cells and monocyte-derived cells (such as the expression of IRF8 and CD64, respectively) to become efficient APCs that induce CD8+ and CD4+ T cells simultaneously. These inflammatory CCR2-expressing cDC2 cells, induced by type I IFN, were distinct from the monocyte-derived cell population, which were unable to function as effectively as APCs (259).
pDCs are also associated with viral sensing, initiating immune responses, and producing large amounts of type I IFN and human β-defensins. Although pDCs can transport viral particles to the dLN, they are poor inducers of naive T-cell responses due to low expression of costimulatory molecules (250, 251). pDCs function differently in respiratory viral infections depending on the viral pathogen. For example, pDCs are dispensable for IAV sensing and clearance. However, during RSV infection, pDCs promote viral clearance and modulate inflammation to reduce lung injury (260).
(vi) Monocyte-derived DCs.
Monocyte-derived DCs (moDCs) and macrophages differentiate from monocytes that migrate into the lungs during infection. The inflammatory monocytes are involved in Th1 differentiation, cytotoxic effector responses, and activation of naive and memory CD8+ T cells and NK cells that expedite efficient viral clearance (250, 261). Type I IFN and chemokines (e.g., CXCL10 and CCL2) produced by moDCs further expand inflammatory responses and increase inflammatory monocyte recruitment to the sites of infection. In fact, increased numbers of CCR2+ inflammatory monocytes or moDCs producing elevated levels of NO and TNF-α during viral infections display greater mortality and morbidity in mice. However, complete depletion of moDCs favored uncontrolled viral dissemination and disease severity. Interestingly, a partial attenuation of inflammatory monocytes in the lungs reduced inflammation-driven lung damage but did not affect viral clearance. These observations allude to the requirement of a fine balance in monocyte numbers and functions in the lungs that benefits the host in controlling respiratory viral infections without pulmonary tissue damage.
(vii) Tissue-resident lymphocytes.
The relatively abundant and diverse population of tissue-resident lymphocytes in the lungs synergizes with the early myeloid responses during respiratory infections to launch an efficient innate immune response before activation of the antigen-specific T-cell responses. Innate lymphoid cells (ILCs), including natural killer (NK) cells, and the “unconventional” T cells, including invariant NKT (iNKT) cells, γδ T cells, and mucosa-associated invariant T (MAIT) cells, constitute the innate tissue-resident lymphocytes (262, 263).
ILCs are a heterogeneous population of cells expressing a variety of genetically coded activating and inhibitory receptors, which are found at epithelial barriers and can initiate rapid immune responses upon infection or injury (262). ILCs can be further differentiated into subgroups based on differential expression of the transcription factors, cell surface receptors, and cytokines produced. Group 1 ILC consists of type 1 ILCs (ILC1s) and mature NK cells, which express the transcription factor Tbx21 (T-bet). However, they differ in their abilities to produce IFN-γ in response to IL-12 and IL-18. Furthermore, ILC1 are primarily tissue-resident cells, whereas NK cells remain in circulation and are recruited in response to chemokines. ILC1s mount an early antiviral response at the local sites of viral infection through IFN-γ production prior to the other tissue-resident lymphocytes. NK cells respond to the viral infection within a few days after neutrophil activation to produce IFN-γ, initiate cytotoxic killing of infected cells, and trigger more specific adaptive immune responses. NK cells eliminate infected cells by releasing cytotoxic granules containing granzymes and perforin, activating death receptors, or through antibody-dependent cell-mediated cytotoxicity (ADCC) to inhibit viral dissemination. NK cells also enhance CTL activity through IFN-γ production. In IAV infection, the virus evades NK cell-mediated killing by inhibitory receptor signaling. Some influenza virus strains infect NK cells, albeit ineffectively, to induce apoptosis, thus limiting NK cell-dependent cytotoxicity. In RSV infection, IFN-γ production by NK cells may cause tissue damage. However, in IAV infection, NK cells have been shown to produce IL-22 and initiate tissue regeneration.
ILC2s are characterized by Th2 cytokines, IL-4, IL-5, and IL-13, which are secreted in response to IL-25, TSLP, and IL-33 produced in the lungs (262) and are primarily implicated in tissue repair after resolution of infection. ILC2s initiate repair by producing amphiregulin (similar to CD4+ T cells) mediated by IL-33 produced by epithelial cells. Furthermore, IL-5 produced by ILC2s, following the clearance of IAV infection, also leads to the accumulation of new eosinophils in the lungs (264). Interestingly, ILC2s are regulated by type I and type II IFNs, which are produced early in a viral infection and can inhibit ILC2 activity in a STAT1-dependent manner (265, 266). ILC3s are distinguished by their expression of retinoid-related orphan receptor γt (RORγt) transcription factor and production of IL-17A and IL-22 in infections (262). ILC3s are implicated in tissue repair responses. Functions of ILC3s have been primarily described in the gut but may also limit airway hyperreactivity during respiratory infections via IL-22 secretion (267). However, similar to ILC2s, the beneficial repair functions of ILC3s can be directly blocked by excess type I IFN signaling during viral infections.
“Unconventional” T cells are defined by their limited T-cell receptor diversity, their ability to recognize antigens without MHC-I or MHC-II presentation, and their swift effector responses that also activate downstream T-cell-mediated adaptive immunity (268). Type 1 (or iNKT) and type 2 NKT cells are differentiated on the basis of their TCRα chain repertoire. iNKT cells are a significant cell population in respiratory viral infections; they produce the inflammatory cytokines IFN-γ, IL-4, IL-17, and TNF-α, they mount antigen-specific cytotoxicity, and they activate innate and adaptive immune responses through APCs to protect against infections (268). Endogenous self-derived lipids (presented by MHC-I like molecule [CD1d]) and cytokines produced during viral infections activate iNKT protective functions. In influenza, iNKT cells were necessary and sufficient for host survival and resolution of infection through the regulation of myeloid-derived suppressor cells and confirmed by correlative data in human influenza (269). iNKT cells were also implicated in CD8+ T-cell activation and expansion in RSV clearance (270).
γδ T cells express the less-diverse γδ TCR (compared to the αβ chains in conventional T cells) that are present in mucosal barrier surfaces and are activated by different multiple microbial pathogens to establish inflammatory responses, as well as regulate tissue homeostasis and repair (271). In patients that survived the 2003 SARS infection, the expansion of memory γδ T-cell population that produced IFN-γ and mediated cytotoxic cell death of SARS-CoV-infected cells may have directly contributed to survival (272).
MAIT cells respond to a diverse range of microbial metabolites from bacterium-infected cells and yeasts via their reactivity to MHC-I like protein, MR1, but do not directly sense viral molecules. However, MAIT cells respond to IL-12 and IL-18 to produce IFN-γ, TNF-α, and IL-17 that augment other antiviral immune responses (273, 274). The observations were corroborated in MR1-deficient mice lacking a MAIT cell population that showed greater weight loss and increased mortality during lethal H1N1 influenza infection (275). However, MAIT cell activation reinforcing an inflammatory response during viral infections has been associated with both protective and pathological functions. It is not yet clear whether MAIT cell activities also correlate with hyperinflammation during severe influenza infections (276).
(viii) B cells and T cells.
The adaptive immune system working though antibodies, B cells and T cells are ultimately responsible for absolute viral clearance and memory formation. B cells produce specific neutralizing antibodies that opsonize and inactivate virions, as well as eliminate virus-infected cells. Neutralizing antibodies encase viral particles to diminish their infectivity and promote phagocytosis by neutrophils and macrophages. The antibodies also bind to viral proteins expressed on the surfaces of infected cells and activate complement-mediated ADCC. Due to its rapid and effective role in viral clearance, antibody-mediated protection is often a coveted outcome of several vaccination strategies. In fact, the presence of neutralizing antibodies against rhinovirus and influenza virus in the blood, prior to infection, correlates strongly with disease protection (277). Neutralizing antibodies can also cross-react and confer extensive protection against a wide variety of viruses. For example, it was recently discovered that neutralizing IgG from plasma of COVID-19 patients recognizes a common epitope that recurs in the spike proteins of SARS-CoV2, SARS, and MERS of the coronavirus family (278). However, as presented for SARS previously (279), while completely protective, an antibody response to COVID-19 may be relatively short-lived (280, 281) compared to a usual cell-mediated immune response.
CD4+ follicular helper T cells (TFH) are important for launching an effective B-cell response in infections through the formation of germinal centers in secondary lymphoid tissues (required for B-cell maturation, proliferation, and memory). Increased TFH cell numbers have been associated with high vaccination-induced, influenza-specific IgM and IgG antibodies (282). Adoptively transferred CD4+ T cells in mice lacking mature T cells promote an effective antibody response to protect from an IAV infection, but not in SCID mice lacking both T cells and B cells. Depletion of CD4+ T cells also correlated with decreased antibody responses during SARS infection (249). Athymic mice that do not express mature, functional T cells are unable to clear virus and succumb to IAV infection. CD8+ T cells are essential in the highly pathogenic SARS infections, and adoptive transfer of SARS-activated T cells into SCID mice promotes survival and reduced viral loads (283). A study analyzing the blood of critical COVID-19 patients identified SARS-CoV2 spike glycoprotein-specific CD4+ T cells in 100% of the patients, whereas 80% of the patients had specific CD8+ T cells, which appeared early in the infections and were enhanced over time (284).
Lungs also harbor resident CD103+ CD8+ T cells, also referred to as tissue-resident memory T cells (TRM), which provide cross-protection against different strains of influenza virus, as well as against coronaviruses (284). The antiviral cytotoxic functions of TRM cells are fortified by infiltrating CD8+ effector T cells (TEFF). The TRM cells undergo rapid proliferation and activation upon reinfection or cross-reaction with a different strain of virus, which are enhanced by an increase in TEFF cells in the lungs. However, with increasing intervals between exposure to cross-reacting viruses, TRM cells are usually replaced by CD103− CD8+ central memory T cells that exhibit a delayed kinetic and more inflammatory phenotype in the lungs (285).
Clearance of viral respiratory infections.
Cytokines, chemokines, and other factors released from epithelial and immune cells in response to respiratory viral infections are central to launching appropriate antiviral responses that limit viral replication and dissemination. However, dysregulation of the protective antiviral pathways by highly pathogenic viruses triggers severe immunopathology that undermines host fitness and causes tissue injury. The initial antiviral cytokines produced in response to viruses are the IFNs and IFN-stimulated genes (ISGs), followed closely by proinflammatory cytokines such as IL-1, IL-6, and TNF-α. The chemokines are responsible for leukocyte migration to the sites of infections and localized release of antiviral cytokines and effectors. Other secreted effectors include β-defensins, NO, and surfactant proteins that function as antimicrobial agents.
(i) Interferons and antiviral ISGs.
Type I and type III IFNs are the primary antivirals that are produced upon viral sensing, which in turn activate numerous ISGs capable of interfering with viral replication and spread and are responsible for critical and effective antiviral host responses. In fact, disease severity in the current COVID-19 pandemic has been linked to genetic deficiencies in IFN or the presence of anti-IFN autoantibodies (286, 287).
Type I IFN (including IFN-α and IFN-β) bind to the ubiquitously expressed IFN-α/β receptor (IFNAR). Meanwhile, type III IFNs (IFN-λ1 or IL-29; IFN-λ2/3 or IL-28A/B) bind to a heterodimer consisting of IL-10Rβ and IFNLR (IL-28R), with the latter being expressed specifically on mucosal epithelium, dendritic cells, and neutrophils. Both IFNs activate JAK/STAT and IRF9 to induce a largely overlapping set of antiviral ISGs. Almost all nucleated cells secrete type I IFNs and IFN-λ are secreted by epithelial cells or cDCs in the lungs, depending on the virus and site of infection. In viral infections, the initial upsurge of type I IFN, followed by IFN-λ, is crucial for removal of virions and infected cells and thus negatively regulates immune cells involved in tissue repair and regeneration to augment apoptosis and inhibit epithelial cell proliferation. Unlike type I and type III IFNs, type II IFN (IFN-γ) does not have direct antiviral effects but promotes adaptive immune responses upon activation by T cells, iNKT cells, NK cells, or ILCs.
During viral infections, human airway epithelial cells produce more IFN-λ than type I IFN. Recent studies have shown that of the two IFNs, type III IFN is more predominant in the airway epithelium (228, 229) and potentially less damaging. In mice, during poly(I·C) stimulation (to mimic RNA virus), IFN-λ is produced primarily by cDC1 cells, while type I IFN is produced by epithelial cells, alveolar macrophages, and moDCs. Furthermore, type I IFN production peaks early upon poly(I·C) stimulation or IAV infection, while IFN-λ production is less potent but sustains longer over the duration of infection. Both type I and type III IFNs have nonredundant and coordinated critical roles in controlling susceptibility and viral clearance in influenza infection, as observed in Ifnar−/− or Ifnlr1−/− mice, and the effects were more pronounced in IFNAR and IFNLR1 doubly deficient mice (288–290). In SARS-CoV-2 infection, disease severity correlated with an early, elevated, and persisting type I IFN signature in the blood and bronchoalveolar fluids, whereas the type III IFN levels increased late and were sustained. Type I IFN responses are associated with effective viral clearance but cause severe immunopathology (288). Comparatively, the initial IFN-λ-induced antiviral state at the mucosal surfaces is less detrimental to the host (291) and is also critical for control of virus dissemination in the upper airways (292).
Autocrine and paracrine IFN responses initiate a cascade of specialized, antiviral ISG expression that is involved in the (i) inhibition of viral entry (e.g., MX1, IFTIM, and TRIM proteins), (ii) prevention of viral release (e.g., Viperin), (iii) apoptosis of infected cells (e.g., PKR), and (iv) regulation of transcriptional and posttranscriptional protein synthesis and posttranslational processes (e.g., Viperin and ISG15). Several ISGs activated downstream of IFN signaling also participate in the innate and adaptive immune systems by triggering recruitment (chemokines), antigen presentation, and the release of other cytokines.
(ii) Chemokines.
Chemokines produced in respiratory infections by the airway epithelium stimulate the migration and recruitment of immune cells responsible for viral clearance. Excess chemokine production leads to excessive leukocyte infiltration and subsequent hyperinflammation and tissue damage. Neutrophil recruitment, survival, and degranulation during viral infection require IL-8 (CXCL8) signaling. Dysregulation of IL-8 causes excess release of neutrophil proteases and ROS, leading to lung tissue damage and ARDS (293). An ISG, IP-10 (CXCL10), mediates the recruitment and activation different types of leukocytes. CXCL10/CCR3 protects against respiratory viruses such as RSV (294), whereas excess signaling during IAV infection caused tissue damage through uncontrolled immune cell-mediated inflammation (295, 296). CCL2 (MCP-1) is a mediator for monocyte recruitment and plays a dual role in respiratory viral infections. A heightened monocyte induction by CCL2/CCR2 signaling in viral infection adversely affects lung pathology (297, 298). However, CCR2-deficient mice also had better outcomes but with delayed clearance of IAV virus (299, 300). In addition, diminished CCL2 expression correlated with the deletion of an immune regulatory factor, A20, in the lung epithelium, which resulted in a better prognosis in late stages of IAV infection (301). CCL5 (RANTES) is an ISG that mediates the chemotaxis and activation of T cells, NK cells, monocytes, DCs, and granulocytes and provides protection against viral infections in the lungs. CCL5 responses favored survival in IAV-infected mice (302) but aggravated inflammation and airway hypersensitivity in response to RSV infection (303).
(iii) Inflammatory cytokines.
In addition to IFN responses, proinflammatory cytokines are produced in the lungs during respiratory viral infections; these cytokines include IL-6, TNF-α, and IL-1, as well as granulocyte colony-stimulating factor (G-CSF), and granulocyte-macrophage colony-stimulating factor (GM-CSF). IL-6 controls the transition to adaptive immune responses by regulating the recruitment, differentiation, and activation of monocytes and T cells, as well as by downregulating neutrophil activity (304). H1N1 IAV infection in Il6r−/− mice underlined the importance of IL-6 in virus-induced neutrophil cell death that negatively affected viral clearance, lung damage, and mortality (305, 306). IL-6 is also critical in viral clearance during subsequent infections by recalling memory CD4+ T cells by inactivating Tregs (307). While high levels in the lungs correlate with disease severity, IL-6 is instrumental in the control of virus replication and spread (308). Similarly, elevated levels of lung TNF-α is associated with severe immunopathology but is required for inhibiting viral replication, enhancing cytotoxicity in infected cells, and cytokine production in leukocytes (308).
Another proinflammatory cytokine, IL-1β, is produced rapidly by the activation of the NLRP3 and/or AIM2 inflammasome upon viral recognition. IL-1β is required for survival during viral infections and augments neutrophil recruitment and CD8+ T-cell activation but also adversely affects lung inflammation (309–311). IL-18 maturation is also regulated by the NLRP3 inflammasome. During influenza infection, IL-18 potentiates cytokine production from activated CD8+ T cells and enhances the cytotoxicity of NK cells (312). However, IL-18 functions can be double-edged, since other studies have shown that IL-18 deficiency can provide protection against viral dissemination (313). Like most other proinflammatory cytokines, a fine balance in IL-18 levels and activity is required which decides whether the cytokine is beneficial or detrimental to the host.
G-CSF and GM-CSF are differentiation factors for myeloid lineage cells. G-CSF regulates the neutrophils by enhancing their differentiation, expansion, and survival (314, 315). GM-CSF promotes the proliferation and activation of lung DCs and macrophages to aid in an effective T-cell response and pathogen clearance. The lungs of GM-CSF-deficient mice are highly vulnerable to viral infections but can be rescued by the exogenous administration of GM-CSF (316, 317).
(iv) Other antiviral factors.
The airway epithelium secretes several antimicrobial peptides, enzymes, protease inhibitors, and oxidants that directly inhibit the respiratory pathogens and/or regulate immune responses. Lactoferrin, β-defensins, cathelicidins, and NO are produced in the alveolar space and interfere with respiratory viral infections.
Lactoferrin is a glycoprotein that inhibits infection by directly binding to both DNA and RNA viruses, as well as by blocking the host receptors required for viral entry (318). β-Defensins are cationic peptides that neutralize viruses by forming a pore-like structure on the pathogen membrane and causing efflux of essential nutrients and ions. β-Defensin 1 (BD1) is constitutively expressed, whereas BD2, -3, and -4 expression is induced upon infection (e.g., IAV) (319). β-Defensins also induce cytokine production by epithelial cells and facilitate the migration of immature DCs and memory T cells (320). Cathelicidins (LL37) are released by epithelial cells and via neutrophil granules (321). The antiviral properties of LL37 include disrupting the viral envelopes and enhancing immune cell recruitment and inflammatory responses (322). The exogenous administration of LL37 lowered the mortality, morbidity, and viral titer in mice infected with IAV (323). Nitric oxide (NO) is constitutively produced by the respiratory epithelia and is further induced by respiratory viruses and proinflammatory cytokines (324). NO and other reactive nitrogen species (RNS) inhibit viral proteins and processes such as replication and transcription. However, RNS activity, like ROS activity, can be indiscriminate and modify host protein functions as well as cause lung injury (324).
As mentioned previously, the highly conserved, hydrophilic surfactant proteins SP-A and SP-D or “collectins” secreted by AT-II epithelial cells have antiviral properties by which the proteins detect viruses in the alveolar space and facilitate their clearance by phagocytes (325). The C-terminal lectin-type domains of SP-A and SP-D act as PRRs that bind with various affinities to complex carbohydrates or associated toxins found on the surfaces of respiratory viruses and other pathogens (326, 327). In IAV infection, SP-A competitively binds viral HA protein through its α-(2,3)-linked sialic acid motifs and thereby blocks viral entry (331, 332). SP-D binds to and promotes the aggregation of HA proteins on IAV to disrupt the optimal functioning of the virus. SP-D-deficient mice with IAV infections had reduced virus clearance and hyperinflammation in the lungs, which is directly dependent on the PRR capabilities of the protein, and independent of its surfactant properties (333). Interestingly, in humans, gene polymorphisms affecting the expression of SP-A, -B, and -D correlated with severe influenza disease (334, 335).
The respiratory epithelium and endothelium, along with lung-resident immune cells, are actively involved in preventing viral infections in the airways. During a viral onslaught, a rapid influx of inflammatory cells occurs that aims to prevent, contain, and clear the infection. The clearance of virions and virus-infected cells, as well as blocking viral dissemination, is greatly reliant on the antiviral cytokines and chemokines secreted by the infected epithelia and activated immune cells at the sites of infection. Cellular infiltration and activation during an infection is a double-edged sword since the protective functions can become damaging to the host unless controlled. Although viruses have evolved with the hosts to evade and manipulate host immune responses, the entry of activated immune cells and the release of cytotoxic, antiviral factors in infected lungs are strictly regulated by the airway epithelium and tissue-resident cells.
Role of Immune System in Lung Damage
Immune responses, including the migration of leukocytes and the release of cytokines and chemokines, are critical for viral clearance and a return to homeostasis but often are also responsible for disease complications and tissue injury that may compromise host fitness during disease, as well as in the long term. Dysregulation of the immune responses directly by viruses or as a result of a persisting infection has far-reaching pathological consequences. Such immunopathological effects arise from (i) increased infiltration of inflammatory cells, (ii) enhanced cellular activity resulting in elevated levels of secreted cytokines and chemokines, and/or (iii) erroneous feedback regulation. Here, we will focus on the roles of cytokines, especially the newly highlighted roles of IFNs, in propagating lung damage. The relevance of neutrophils and T cells in lung immunopathology has been described previously and extensively (328–330).
Uncontrolled inflammation during respiratory viral infections or “cytokine storm” is responsible for extensive lung injury and severe immunopathology. A cytokine storm (also known as the “cytokine release syndrome”) is an excessive immune response caused by interplay between immune cells and cytokines such as TNF-α, IL-6, IL-1, IL-12, IFN-α/β, IFN-γ, MCP-1, and IL-8 and is characterized by systemic elevation in the levels of the cytokines and chemokines (336). Infections with several respiratory viruses—such as IAV, RSV, SARS-CoV, and SARS-CoV-2—cause the dysregulated infiltration of neutrophils and inflammatory monocytes and increased cytokine production that destabilize tissue integrity (337). Neutrophils, inflammatory macrophages, and other hyperactivated immune cells can further propagate structural tissue damage by producing cytotoxic free radicals, reactive nitrogen and oxygen species, proteases, and profibrotic mediators that modify and damage the pulmonary epithelium. Sex may also be an underlying factor dictating cytokine responses and clinical outcomes during respiratory viral infections. In COVID-19 patients, males had higher IL-8 and CCL5 in blood, whereas higher TNFSF10 and IL-15 levels correlated with worse outcomes in females. While both males and females had increased monocyte populations, males had more nonclassical monocytes in circulation. In addition, CD8+ T-cell responses were more robust in females rather than in males (338).
Several respiratory viruses have been associated with complication in the central nervous system (CNS) (339). Cerebrovascular, neurological, and psychiatric symptoms have been observed in COVID-19 patients, which often leads in ischemic stroke, intracerebral hemorrhage, CNS vasculitis, encephalopathy, encephalitis, and psychosis (340). Loss of smell and taste has also been reported in SARS-CoV-2-infected patients (341–343, 390). Patients with neurological complications have been reported in SARS, MERS, and H1N1 IAV cases during acute infection or correlated with disease severity. Severe RSV-induced bronchiolitis in children is also known to affect the CNS (344–346). The neurological complications occur as a direct result of viral infection or indirectly through the cytokine release syndrome and cell- or antibody-mediated immune responses. Interestingly, a recent study by To et al. showed that SARS-CoV-2 could directly infect neurons in human brain organoids without eliciting type I IFN responses (339).
In influenza, TNF-α is the central proinflammatory cytokine contributing to severe immunopathology. TNF receptor 1 (TNFR1)-deficient mice show unaffected viral spread but with lower morbidity during H5N1 IAV infection (391). In addition, in mice lacking both TNF and IL-1 signaling, there was no change in viral clearance but significantly decreased morbidity and mortality (392). IL-1β is beneficial early in IAV infection but leads to disease severity in the later stages of the infection when sustained (311). IL-18 and IL-6 have complicated roles in IAV infection: while they are largely beneficial, increased levels of these cytokines contribute to the “cytokine storm” and are often associated with poor clinical prognosis.
During RSV infections in children, extensive tissue damage is driven by immune cells recruited to sites of infection, which produces the inflammatory cytokines IL-1, IL-6, IL-8, and CCL5 in the upper and lower respiratory tracts. In some patients with severe RSV-induced bronchiolitis, there were several neurological symptoms, such as encephalitis and encephalopathy (346), driven by the induced cytokines. Analysis of cerebrospinal fluid from patients showed augmented IL-6, IL-8, CCL2, and CCL4 levels that correlated with the alterations in the CNS (393).
In COVID-19, the persistent high fever, respiratory distress, and acute lung injury that occur as a result of a hyperactivated immune system are the leading causes of disease complexity. Analyses of the COVID-19 patients from several independent groups unanimously agree upon the detrimental effects of elevated proinflammatory cytokines in driving disease severity and often mortality. An increased level of the inflammatory cytokines IL-1β, IL-18, IL-6, or TNF-α, among others, is a signature of critically ill patients who require hospitalization. Compared to moderate cases, critical COVID-19 patients showed an increased interaction between the pulmonary epithelium and immune cells via receptor-ligand profiling, which was modulated, in part, by heightened chemokine release (e.g., CCL2, CCL3, CCL20, CXCL1, CXCL3, CXCL10, and IL-8) by inflammatory immune cells (394). The SARS-CoV-2 profile shares major similarities with SARS and MERS infections with respect to the proinflammatory cytokines and chemokines found in circulation, which are associated with the coronavirus-mediated “cytokine storm.” However, unlike SARS, SARS-CoV-2 also shows an increased secretion of the anti-inflammatory cytokines IL-4 and IL-10.
The status of the antiviral IFN production in COVID-19 patients is complex; this is compounded by the limited understanding of the newly emerging SARS-CoV-2 virus. Our group observed that type I IFN and IFN-λ2/3 mRNA expressions in the upper respiratory tract of COVID-19 patients were not drastically affected but were greatly enhanced in the bronchoalveolar lavage samples of patients with severe disease (347). Interestingly, tenOever and coworkers reported an attenuated IFN transcriptional profile but high chemokine expression in SARS-Cov-2 infection compared to other respiratory viral infections (348). A larger study by Hadjadj et al. tested 50 patients of different disease severities after 8 to 12 days of initial symptoms and observed a weakened type I IFN signature and ISGs in the blood, which was associated with an increased viral load (349). In contrast, a more recent longitudinal analysis over time led by the Iwasaki group, analyzing a cohort of 113 COVID-19 patients exhibiting either moderate or severe disease, showed a persistent type I and type III signature (350). Elevated levels of both IFN-α and IFN-λ were more sustained in the severely affected patients. However, both studies agreed that IFN-α, and not IFN-β, was the more predominant type I IFN in blood. Moreover, as mentioned earlier, all of these studies observed a similar hyperinflammatory cytokine signature in severely affected COVID-19 patients that eventually drives the pathogenesis, dictates the clinical outcome, and requires therapeutic intervention for disease amelioration.
Unregulated inflammatory cell death provokes excess tissue inflammation and organ failure. Pyroptosis in tissue-resident alveolar macrophages and recruited monocytic macrophages exacerbates lung inflammation by promoting neutrophil infiltration and intense cytokine storm through excess productions of IL-6, TNF-α, and IL-1β (351). IAV infection activates NLRP3 inflammasome in type I IFN- and RIG-I-dependent manner to induce pyroptosis of lung epithelial cells (352–354). Inflammasome and pyroptotic cell death induced by SARS-CoV-2 exacerbates respiratory distress and fever. Acute COVID-19 pathogenesis also affects blood coagulation that often leads to thrombosis in the lungs (e.g., pulmonary embolism) (355, 356). A recent complication of COVID-19 is the occurrence of pathogen-driven Multi-system Inflammatory Syndrome in Children (MIS-C) that presents as Kawasaki disease (357), and the immunological complications may be mediated by autoantibodies (358).
Secondary bacterial infections or “superinfections” is a quite common clinical complication during an acute respiratory viral infection and is a leading cause of mortality in patients. Viral infections such as IAV infections, and now SARS-CoV-2 infections, make the host more vulnerable to Staphylococcus aureus, Streptococcus pneumoniae, Pseudomonas aeruginosa, or Escherichia coli bacterial coinfections (359, 360). The susceptibility of the virally infected lungs is regulated by multiple factors such as increased damage and delayed regeneration of the airway epithelium, inhibition of antibacterial signaling and cytokines, immune cell exhaustion, and altered activation of pulmonary cells that result in an inappropriate or inadequate immune response or reduced defense capabilities.
The antiviral IFN responses initiated during a respiratory viral infection are of immense importance in virus control and clearance. However, a prolonged exposure to IFNs compromises lung health, drastically increasing host vulnerability to secondary coinfections. It has been demonstrated that Ifnar−/− mice, which lack type I IFN signaling, could effectively clear a secondary S. pneumoniae infection after influenza infection. Coinfection with IAV and Mycobacterium tuberculosis also increases the severity of the bacterial disease in a type I IFN-dependent manner (361). In addition, type I IFN can inhibit inflammatory responses by blocking IL-1, inducing IL-10 production (362), and suppressing tissue-resident lymphocytes such as γδ T cells during viral infections (363).
Similar to Ifnar−/− mice, IFNLR1-deficient mice devoid of IFN-λ signaling are completely protected from S. aureus superinfection in a TLR3/TRIF-dependent manner (347). Extended IFN-λ expression in the lung during viral infections induces apoptosis of the infected cells and inhibits the regeneration of the epithelium, leading to a damaged lung barrier and increased permeability. IFN-λ negatively regulates the recovery of the lung epithelium by activating the p53 and/or p21 pathways (347, 364).
Neutrophil-mediated killing is critical for resolving pulmonary bacterial infections. In some cases, such as during nonlethal influenza infection, type I IFN blocks neutrophil recruitment that can favor S. pneumoniae bacterial coinfection (363). IFN-λ is also known to negatively impact neutrophil recruitment (365) and their inflammatory functions (366) and thus may be responsible for restricting antibacterial neutrophil activity in the context of a lung superinfection.
In contrast to bacterial superinfections, the role of IFN-λ in fungal coinfections has not yet been established. IFN-λ is known to prime antifungal responses in neutrophils during Aspergillus fumigatus infection (367). However, in a fungal coinfection model with chronic IFN-λ exposure, the damage to the lung epithelium is likely to be detrimental as well.
Viral infections often perturb the structure and function of the airways, which can be compounded by the host immune responses. Usually, the resolution of an infection is followed by a return to homeostasis, but in many cases it is dependent on the pathogenic nature of the virus, the extent and spread of the infection, and the degree of the inflammatory response.
Common strains of coronavirus, rhinoviruses, and adenoviruses generally manifest as mild to moderate upper respiratory tract infections in patients without preexisting conditions. In severe cases, these infections present increased leakage and edema, increased epithelial cell death, and hypersecretion of mucus, leading to nasal congestion, sneezing, and coughing. More severe viral infections, such as influenza, COVID-19, SARS, MERS, and RSV infections in children, proceed from the upper to the lower respiratory tracts, where they can spread from the bronchi and bronchioles to the alveoli, if uncontrolled. Cellular changes (e.g., desquamation of epithelial cells and loss of cilia in RSV infection, as well as epithelial cell hyperplasia in IAV infection) and increased inflammation in the lower respiratory tract drives severe disease pathogenesis due to compromised respiratory functions. Viral bronchitis and bronchiolitis are characterized by restricted airflow and inefficient gas exchange through increased edema and mucus in the lungs, vascular congestion, and tissue swelling. In serious cases, breathing and respiratory functions may be compromised, and patients may require machine support (368). Pneumonia is caused by the viruses that reach the alveoli and destabilize pulmonary functions. In critical cases, fluid build-up in the alveolar space and necrosis, as well as the loss of surface tension and pulmonary elasticity, can lead to ARDS and respiratory failure. ARDS is a clinical hallmark of critical patients with SARS and MERS infections and is emerging to be the same in SARS-CoV-2 infections. ARDS is driven by “cytokine storms” causing extensive inflammation and pulmonary damage, which leads to multiple organ failure, and often death, in cases of severe, high-risk COVID-19 (369).
Viral infections in lungs can cause extensive inflammation-driven damage, but generally, after effective viral clearance, the lung is able to return to its original pulmonary architecture. However, anomalies in the inflammatory or regeneration pathways can adversely affect the structural and cellular makeup of the lungs, leading to fibrosis, emphysema, bronchiectasis, and pneumatoceles.
Acute lower respiratory tract infections directly affect new and preexisting chronic pulmonary diseases. Preexisting chronic obstructive pulmonary disease or asthma are associated with worsening prognoses in patients with pulmonary infections, which manifest as severely irregular inflammatory responses or defects in repair and regeneration. Exposures to different pathogenic microbes and viruses also promote an immunological imbalance in the lungs that increases a predisposition to asthma. Other lung diseases implicated in respiratory infections include tuberculosis, sarcoidosis, hypersensitive pneumonitis, and pulmonary arterial hypertension (370).
Respiratory viruses change the matrix composition of the lungs via collagen and fibronectin deposition that often leads to scar tissue formation. These respiratory viruses effect remodeling through induction of associated factors, angiogenesis and fibroblast proliferation, which is amplified in an atopic environment. Mice infected RSV, followed by allergen challenge, displayed airway remodeling characterized by increased collagen synthesis and thickening of the bronchial basal membrane, which was mediated by RSV-driven fibroblast growth factor 2 (FGF-2) (371). Lung ILC2s primarily have protective functions during infections but can be dysregulated by viruses or sustained type I IFN signaling, resulting in allergic inflammatory responses and exacerbated asthma. Infection with respiratory viruses such as IAV, RSV, and RV can result in ILC2-dependent airway hypersensitivity mediated by IL-33 and TSLP (372–374). Furthermore, IL-33-primed lung ILC2s acquire “memory” that triggers more robust responses upon rechallenge than do unprimed, primary ILC2s (375). Thus, certain respiratory viral infections present an ILC2-dependent allergic immunopathology that affects lung health in the long term.
INVESTIGATIONAL TOOLS TO IMPACT IMMUNE RESPONSES DURING VIRAL INFECTIONS
Multiple clinical studies and therapeutic interventions target the immune responses to mitigate respiratory virus-driven immunopathology. Complementing and reinforcing vaccine discovery, several studies are currently under way to find an effective cure for patients with influenza, RSV, and the newly emerged, highly contagious SARS-CoV-2 infections.
Interventions in respiratory viral infections, including SARS-CoV-2, such as antibody and convalescent plasma therapy, checkpoint inhibitors, alpha-blockers (e.g., prazosin), steroids (e.g., dexamethasone) and, above all, vaccines would require a much more detailed discussion. Here, we report some additional alternative and complementary approaches involving implicated immune pathways and immunomodulatory molecules that are targeted or commissioned for cure and preventive therapies in respiratory viral infections (Fig. 3).
FIG 3.
Clinical interventions impacting immune responses and viral replication during respiratory viral infections. Several preventive and therapeutic interventions targeting immune responses are approved or currently being tested to combat virus-dependent immunopathology. After initial exposure to a respiratory virus, symptoms begin manifesting within a few days, depending on the virus replication kinetics. The clinical severity of disease can range from mild to moderate, severe, or critical (that define the early, pulmonary, or hyperinflammatory stages) and is often dictated by a persisting infection and/or an aberrant, uncontrolled antiviral immune response. Severe cases of viral infections frequently require hospitalization that can lead to intensive care and ventilation in patients with deteriorating lung functions or other associated systemic complications. In a healthy, uninfected individual, the administration of vaccines is the most effective route to prevent respiratory viral illnesses. Examples include annual vaccinations against the predicted evolving strains of influenza. Currently, several vaccines against COVID-19 have received emergency authorization around the world. Recent authorized SARS-CoV-2 vaccines (with published data from phase 3 clinical trials) include mRNA vaccines from Pfizer-BioNTech and Moderna, as well as an adenovirus-based vaccine from Oxford-AstraZeneca. Prophylaxis with antiviral drugs is another preventive measure for unimmunized children or immunocompromised individuals, or against viruses for which there are no vaccines (e.g., RSV). Prophylactics include oral oseltamivir phosphate for influenza and palivizumab for RSV, and there are considerations for developing nasally administered IFNs for prophylaxis against SARS-CoV-2. In asymptomatic patients or presymptomatic patients with early detection or with mild disease, early complementary type I or type III IFN administration can be beneficial. In severe cases of lung infection, multiple anti-inflammatory immunomodulators such as neutralizing antibodies or inhibitors are used to supplement the antiviral therapies to prevent and mitigate cytokine storm. While mild respiratory illnesses can be relieved with over-the-counter fever-reducing drugs, pathogen-specific antiviral drugs are used in cases of moderate to critical disease, either singularly or in combination with other therapeutic interventions. In COVID-19, patients have also benefited from treatment with antiviral monoclonal antibody cocktails (e.g., bamlanivimab by Eli Lilly or casirivimab and imdevimab by Regeneron) and with convalescent plasma from recovered individuals with high antibody titers. Early administration of the antivirals displays better efficiency in viral clearance. In critical patients with hyperimmune responses and ARDS, treatment with corticosteroids (e.g., dexamethasone) relieves respiratory distress. The effectiveness of steroids in the first phases of a viral infection is controversial, since they may hamper the establishment of a properly effective antiviral response. The image was created using BioRender.
The Possible Therapeutic Role of Interferons
IFNs present an attractive candidate in the treatment of respiratory viral infections due to their well-characterized and pivotal role in virus inhibition and clearance. In fact, many viruses have evolved to bypass or downregulate IFN signaling for a successful infection, which underlines the practicality of administering IFNs exogenously to combat viral infections. Type I and type III IFNs also subdue inflammasome activation and IL-1β expression that contribute to inflammation. Several ongoing clinical trials, worldwide, are assessing the benefits of administering recombinant interferons in patients at various stages of respiratory viral infections. These efforts have also multiplied in the recent, novel COVID-19 pandemic to find an effective treatment for affected people, as well as to formulate IFNs as a preventive measure, while the search for an effective vaccine is underway.
Type I IFNs are the earliest responders to a viral infection that trigger downstream gene expression and cellular activation to launch antiviral effector responses. Type I IFN (IFN-α or IFN-β1) is being tested in clinical trials as an adjuvant to vaccination strategies or as a treatment and/or prophylactic in respiratory viral infections. Multiple clinical trials have been initiated to assess IFN-α as in combination therapy (e.g., with oseltamivir) or as a mucosal adjuvant in vaccines to treat and/or prevent influenza infections.
In the light of the recent COVID-19 pandemic, a clinical trial (NCT04320238) is testing the efficacy of IFN-α1β administered nasally in at-risk frontline medical workers (with thymosin-α, an adjunct used in influenza vaccines, in the high-risk medical staff working directly with COVID-19 patients) as a preventive intervention. Another clinical trial (NCT04293887) will use nebulized IFN-α2β to early-diagnosed COVID-19 patients (within 7 days of onset of symptoms) in combination with other standard therapies (lopinavir-ritonavir, remdesivir, or antibodies against coronavirus) depending on patient requirements. Interestingly, a clinical trial (NCT04379518) is assessing the effects a combinatorial therapy of intravenous IFN-α2b (or Roferon A) and rintatolimod (dsRNA that mimics viral infection) on mild to moderate COVID-19 patients with cancer. Clinical trials with another type I IFN, IFN-β1A/B, used standalone or in combination (NCT04449380, NCT04350671, and NCT04343768) in hospitalized COVID-19 patients are also in progress to treat SARS-CoV-2 infections.
However, type I IFN administration has its limitations since it often presents with severe side effects in healthy participants that manifest clinically as flu-like symptoms, fatigue, weight loss, nausea, vomiting, and diarrhea, among others. Type I IFN failed as an effective antiviral therapy in SARS-CoV 2003 human trials (376) but, based on recent observations, SARS-CoV-2 is likely to be more sensitive to IFN than its ancestor.
Type III IFN, in contrast to type I IFN, has emerged as a more promising antiviral candidate due to its limited proinflammatory effects in the lungs and highly specific signaling repertoire (377). The specific receptor for IFN-λ is restricted to the mucosal epithelium and limited immune cells, such as neutrophils and DCs in humans, but it shares largely overlapping antiviral functions with the type I IFNs. The specificity of IFN-λ signaling drastically minimizes the off-target risks and the accompanying toxicity, and it improves tolerability. Pegylated-IFN-λ is an active form of IFN-λ with a longer half-life, which has been administered successfully for human viral hepatitis treatments in multiple clinical trials and also exhibited strong antiviral effects against influenza virus and coronavirus, among others.
A recent study describing a mouse-adapted SARS-CoV-2 showed that peg-IFN-λ1a can effectively block viral replication in vivo when used early (12 h after infection) or prophylactically (18 h before exposure to virus) Similarly, pretreatment of human airway epithelial cells with peg-IFN-λ1a displayed a dose-dependent decrease in virus production in vitro (378). Following these observations, several clinical trials are under way testing IFN-λ as a prophylactic or as early intervention in COVID-19. Clinical trials testing type III IFN as a treatment (NCT04331899 and NCT04354259) will test mild cases of COVID-19 outpatients with a single subcutaneous (s.c.) administration of peg-IFN-λ1a. Moderate cases of COVID-19 requiring hospitalizations are also being tested with one or two doses of s.c. peg-IFN-λ1a (NCT04354259, NCT04343976, and NCT04388709) in randomized phase 2 clinical trials. Another randomized Phase 2b clinical trial (NCT04344600) will estimate the efficacy of peg-IFN-λ1a as a prophylactic in preventing SARS-CoV-2 in at-risk or asymptomatic participants (e.g., with household exposure to COVID-19). Eiger Biopharmaceuticals is developing the use of peg-IFN-λ as a monotherapy and also with lonafarnib and ritonavir drugs in combination.
However, as we and others have recently demonstrated, timing and duration of type I or type III IFN treatment is of the utmost importance as continued exposure of the inflamed pulmonary tissue to IFNs may cause more harm than good in virus-infected lungs (347, 364). Prolonged exposure to IFN reduces tissue regeneration and escalates host susceptibility to bacterial superinfections, which is directly related to increased morbidity and mortality in patients with respiratory viral infections. Moreover, IFN treatment in mice with SARS-CoV was remedial when applied 6 h postinfection but was unproductive at 12 h postinfection (110), hence proving further that timing is central to the efficacy of IFN treatment.
The Possible Therapeutic Role of Immunomodulators
As discussed above, protective inflammation during infections induced by the immune system can become pathological, leading to severe clinical consequences. In respiratory viral infections, blocking hyperinflammation (often characterized by “cytokine storms”) using immunomodulators often reinforces the necessary antiviral therapy and has favorable clinical outcomes.
IL-6 is an inflammatory cytokine associated with cytokine storm and ARDS in COVID-19 patients and other respiratory viral diseases. IL-1- and IL-18-mediated inflammation via hyperactivated inflammasomes contributes to CRS and ARDS in the late stages of coronavirus and influenza virus infections. IL-1β and IL-18 can induce IL-6 and IFN-γ production by lymphocytes, respectively. A high IL-1/IL-6 profile in blood displays increased neutrophilia and CRP concentrations, whereas a high IL-18/IFN-γ profile is defined by macrophage activation syndrome (MAS), severe ferritinemia, coagulopathy, and cytopenia (379). These disease hallmarks are also observed in other inflammatory conditions, such as hemophagocytosis lymphohistiocytosis (HLH) and Still’s disease. Worsening of the disease conditions transforms from a high IL-1/IL-6 signature to a IL-18/IFN-γ signature in patients, which is similar to what is observed in COVID-19 patients. IL-1 and IL-6 inhibition has been beneficial in Still’s disease, whereas IFN-γ blockade is effective treating HLH.
Anakinra is an IL-1 signaling inhibitor that is an IL-1RA receptor antagonist and also hinders IL-18 production. It has been used clinically for over a decade in autoimmune diseases and Kawasaki disease in infants. Tocilizumab is an IL-6 inhibitor that is effective in blocking cytokine release syndrome (CRS) and is used to treat rheumatoid arthritis (RA) (380). The timing of administration has not been fully evaluated, considering an early IL-6 blockade may favor viral dissemination. Using tocilizumab also runs the risk for contracting secondary infections and may require simultaneous antibiotic treatment. Ruxolitinib and baricitinib are JAK kinase inhibitors that block IFN-γ signaling through receptor inactivation. Baricitinib is approved to treat RA and can mitigate chronic inflammation observed in interferonopathies. Inhibition of signaling through JAK kinase can suppress the detrimental cytokines such as IL-6 and type II IFN, as well as ameliorate the antiproliferative effects of IFN-λ. However, the broad immunosuppressive effects of JAK kinase inhibitors are of paramount consideration when being employed against viral infections. Infliximab is an approved chimeric monoclonal antibody that inhibits TNF-α signaling and is used to treat autoimmune diseases such as RA and inflammatory bowel disease. Preclinical studies in subjects with viral pneumonia and influenza A showed the protective role of GM-CSF (or Sargramostim/Leukine) (381, 382). GM-CSF is beneficial to lung health and maturation of alveolar macrophages; it activates the immune system to help clear virus, reduces the risk of coinfections (383), and diminishes morbidity in patients with ARDS (384).
Ongoing clinical trials aiming to reduce “cytokine storm” and restrain intense immunopathology in COVID-19 include the following:
(i) Use of Anakinra alone or with ruxolitinib, as a treatment in acute COVID-19 patients (NCT04366232).
(ii) Anakinra (phase 3) or Emapalumab (phase 2) intravenous (i.v.) administration with standard care in severely affected COVID-19 inpatients (NCT04324021); Emapalumab is an anti-IFN-γ monoclonal antibody.
(iii) Anakinra or tocilizumab in combination with (in patients with ARDS) or without (in patients with nonhypoxemic pneumonia but no ARDS) ruxolitinib administration in progressed inflammatory COVID-19 disease (NCT04424056).
(iv) A phase 3 clinical trial using one dose of i.v.-administered tocilizumab or siltuximab (anti-IL-6 monoclonal antibody) with or without daily s.c. Anakinra to recover lung homeostasis in ventilated COVID-19 patients (NCT04330638). Currently, a recently completed preliminary clinical trial using Tocilizumab did not show better recovery in COVID-19 patients prompting further studies (NCT04331808) (385).
(v) Comparing IL-6 inhibition in COVID-19 patients with severe pneumonia administered Tocilizumab/RoActemra (i.v. or s.c.) or the anti-IL-6 monoclonal antibody, Sarilumab/Kevzara (s.c.), according to the standard care protocol (NCT04322773).
(vi) Intravenous treatment with Infliximab or Infliximab-abda in phase 2 trials in hospitalized adult COVID-19 patients (NCT04425538).
(vii) XPro1595/Remicade, a second-generation inhibitor that neutralizes soluble inflammatory TNF molecules, will be s.c. administered in COVID-19 patients with breathing complications (NCT04370236).
(viii) Therapy with inhaled (NCT04411680) or vein infusion (NCT04400929) of Sargramostim, or GM-CSF, in acute COVID-19 patients with hypoxic respiratory failure and pneumonia, who need mechanical ventilation.
The Possible Therapeutic Role of Antivirals
Antivirals are drugs that directly target the infecting viruses by interfering with the different stages of a virus life cycle such as viral entry, replication, protein synthesis, and viral particle release. The approved antivirals for influenza include oseltamivir phosphate (Tamiflu), zanamivir (Relenza), peramivir (Rapivab), and baloxavir marboxil (Xofluza). Oseltamivir, zanamivir, and peramivir block the actions of viral neuraminidases (NA proteins) that restrict the movement of virus in the respiratory tract and prevent the release of new viral particles from an infected host cell. Baloxavir, the active hydrolyzed form of Xofluza inhibits influenza virus replication by interfering with the endonuclease activity of the PA subunit of viral polymerase complex. Side effects vary for each medication. The antivirals have some common side effects such as nausea, vomiting, diarrhea, and bronchospasm but are generally considered safe and effective in adults, children, and pregnant women (oseltamivir). Antivirals used to treat severe and high-risk RSV infections include ribavirin (blocks viral polymerase) and palivizumab (monoclonal antibody prevents membrane fusion by binding viral envelope fusion protein). There are no approved antivirals for respiratory adenovirus infections, but the disease can be managed by over-the-counter fever relievers.
During the ongoing COVID-19 pandemic, several other drugs are being repurposed and tested for their efficacies as antivirals.
Remdesivir.
Currently, the most-favored, albeit unapproved, drug in the treatment of COVID-19, remdesivir, is an RNA analogue that blocks viral replication in the coronavirus MERS (386). Remdesivir was not successful in the 2015 Ebola virus infections but is used as the current standard care. In the United States, Gilead Sciences received an emergency use authorization for remdesivir treatment in critical COVID-19 patients.
Lopinavir-ritonavir.
Lopinavir-ritonavir is an antiviral drug combination used in HIV patients and helps control the infection. They form an HIV protease inhibitor complex that leads to the release of noninfectious, immature viral particles. A trial showed no apparent benefits of lopinavir-ritonavir treatment in COVID-19 patients (387), but several physicians recommended continued use of lopinavir-ritonavir as COVID-19 standard care, considering the relative safety of the drug combination. However, another recent trial at the University of Oxford (RECOVERY trial) that was conducted with a much larger cohort (1,596 treated versus 3,376 control patients) ruled out “any meaningful mortality benefit” of lopinavir-ritonavir use in hospitalized COVID-19 patients.
Hydroxychloroquine.
Chloroquine and its less toxic derivative hydroxychloroquine (HCQ) are widely available drugs that have been used to treat malaria and amoebiasis, as well as autoimmune diseases such as RA and systemic lupus erythematosus (SLE). HCQ displayed antiviral properties in vitro, where it inhibited viral entry by altering acidification status of endosome maturation required for membrane fusion and also exhibited anti-inflammatory properties (388). However, multiple, noncontroversial studies found no associated benefits of using HCQ (that outweighs the drastic side effects of HCQ) in hospitalized COVID-19 patients (RECOVERY trial). Recently, U.S. Food and Drug Administration revoked the emergency use authorization for HCQ in the United States.
Colchicine.
Colchicine is a safe, affordable, and orally administered anti-inflammatory drug that is currently used for many conditions (e.g., gout, familial Mediterranean fever, and cardiovascular diseases). Colchicine inhibits microtubule polymerization, as well as IL-1-, IL-6-, GM-CSF-, and NLRP3-dependent inflammasome activation. Mindful of the indirect damage caused by inflammatory responses during SARS-CoV-2 infection on multiple organs, including the lungs and heart, a randomized clinical trial theorized that patients receiving colchicine had significantly better clinical deterioration timing (389). Multiple clinical trials are now recruiting to assess the benefits of colchicine for safeguarding against systemic inflammation in COVID-19 patients.
Atovaquone-azithromycin.
Azithromycin is a macrolide with antimicrobial properties, including against rhinoviruses, while atovaquone is an antifungal and antiparasitic quinone. This combination therapy is under trials for use in cases of confirmed COVID-19 (NCT04339426).
CONCLUDING REMARKS
Viruses can either exert a direct pathogenic effect on the infected cells or can induce immune-mediated damage. Regarding the latter, both innate and adaptive immunity are involved. A translational understanding of the molecular events leading to lung injury during infections by viral respiratory pathogens has undoubtedly theoretical correlates and practical consequences. A balanced interplay of innate and adaptive immunity is of pivotal importance for viral clearance and memory generation. In fact, cytokines, chemokines, and other factors released since the early phases of virus infection can dramatically limit viral replication and diffusion. However, all viruses evolved counterstrategies capable of hampering both branches of the immune system, thus leading to a dysregulated immune response. Many of the most severe clinical manifestations of respiratory viruses are related to this immune interfering mechanisms. Our comprehension of these mechanisms is increasing, and they should be considered in the future development and clinical use of new immune-based antiviral strategies.
ACKNOWLEDGMENT
We declare there are no competing interests.
Biographies

Nicola Clementi, M.S., is Assistant Professor of Medical Microbiology and Virology at University Vita-Salute San Raffaele and Consultant Virologist at the Laboratory of Microbiology and Virology of San Raffaele Hospital, Milan, Italy. His main research interests include the study of hypervariable viruses and their interplay with the antibody response and with antivirals. He is also interested in evaluating the clinical usefulness of next-generation sequencing techniques in the management of viral infections.

Sreya Ghosh completed her Ph.D. in Immunology and Microbiology in 2017 under Dr. Kate Fitzgerald at University of Massachusetts Medical School, Worcester, MA, working on innate immune signaling, inflammasomes, and gene regulation. Thereafter, she joined Dr. Ivan Zanoni’s group at Harvard Medical School and Boston Children’s Hospital as a postdoctoral research fellow, where she works with interferons and immune responses in infectious diseases and cancer. She is interested in understanding how type III interferons affect mucosal immunity and can be harnessed therapeutically in diseases such as respiratory infections and tumorigenesis.

Maria De Santis, M.D., Ph.D., received a degree in Medicine in 2000 and a fellowship in Rheumatology in 2004, both at the Catholic University of Rome, Rome, Italy. She earned the Ph.D. in Proteomics at the University of Verona, Italy, in 2008. She was a medical assistant in the Rheumatology Unit of the Catholic University of Rome between 2004 and 2009. She became a postdoctoral researcher in the BIOMETRA Department at the University of Milan between 2010 and 2015. As of 2016, she is deputy director of the Rheumatology and Clinical Immunology Unit and is responsible for the Scleroderma Unit of the Humanitas Clinical and Research Center in Rozzano, Milan, Italy. Her scientific research focuses on systemic autoimmune and chronic inflammatory diseases with particular interest in systemic sclerosis lung and heart involvement and rheumatoid arthritis pathogenesis.

Matteo Castelli, Ph.D., is a postdoctoral fellow in the Microbiology and Virology Laboratory at the University Vita-Salute San Raffaele, Milan, Italy. He received his Ph.D. in 2016 from the same institution and the Laboratory for Biomolecular Modeling at the École Polytechnique Fédérale de Lausanne, Switzerland, working on the computational modeling of HCV glycoprotein structure. His research interests are focused on the structural and functional characterization of the molecular machineries involved in HCV, HSV, and SARS-CoV-2 entry using experimental and computational techniques.

Elena Criscuolo obtained her Ph.D. in Molecular Medicine at the University Vita-Salute San Raffaele, Milan, Italy, in 2016, working on the antibody response elicited during herpes simplex virus (HSV) infection and the protection conferred in vivo by human monoclonal antibodies. She remained in Massimo Clementi’s Laboratory of Microbiology and Virology as a postdoctoral research fellow, working on synergy evaluation of antiherpetic compounds acting on different steps of the virus life cycle, on the characterization of sera from HSV-infected subjects, and on modeling HSV reactivation dynamics in a patterned human neuron-to-keratinocyte circuit reconstituted in a microfluidic device. She then focused on the characterization of entry mechanisms of SARS-CoV-2, the evaluation of antiviral drugs against the infection, and the dissection of the humoral response of COVID-19 patients.

Ivan Zanoni trained with Paola Ricciardi-Castagnoli, receiving his Ph.D. in Immunology from the University of Roma-Tor Vergata, Rome, Italy. He then performed postdoctoral training with Francesca Granucci at the University of Milano-Bicocca, Milan, Italy. Dr. Zanoni is currently Associate Professor at the Faculty of Medicine at Harvard University and Principal Investigator at Boston Children’s Hospital, Division of Immunology, Boston, MA. The Zanoni lab seeks to understand the events that initiate protective immunity in response to infections and tissue injury or that drive the development of immune-mediated diseases.

Massimo Clementi, M.D., is Full Professor of Microbiology and Virology at the University Vita-Salute San Raffaele and chief of the Laboratory of Microbiology and Virology of San Raffaele Hospital, Milan, Italy. Dr. Clementi is a member of the National Scientific Board of the Italian Society of Microbiology (SIM) and founder and past-President of the Italian Society of Medical Virology (SIVIM). Dr. Clementi has research interests in molecular biology of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) infections, virus-host relationships, microbiology of emerging infectious diseases, infections of immunocompromised patients, and generation and characterization of virus-neutralizing human monoclonal antibodies.

Nicasio Mancini, M.D., is Associate Professor of Medical Microbiology and Virology and Director of Study of the Residency Program in Medical Microbiology and Virology at University Vita-Salute San Raffaele. He is Medical Microbiologist at the Laboratory of Microbiology and Virology of San Raffaele Hospital, Milan, Italy. He graduated in 2000 from the Catholic University of Rome, Rome, Italy, and was Board Certified in Medical Microbiology and Virology in 2004 by the Universitá Politecnica delle Marche, Ancona, Italy. His research interests include the study of the interplay between viral pathogens and the humoral immune response and the development and clinical validation of new diagnostic methods in medical microbiology.
REFERENCES
- 1.Gottlieb J. 2019. Community-acquired respiratory viruses. Curr Opin Organ Transplant 24:311–317. 10.1097/MOT.0000000000000633. [DOI] [PubMed] [Google Scholar]
- 2.Hodinka RL. 2016. Respiratory RNA viruses. Microbiol Spectr 4. 10.1128/microbiolspec.DMIH2-0028-2016. [DOI] [PubMed] [Google Scholar]
- 3.Popescu C-M, Ursache AL, Feketea G, Bocsan C, Jimbu L, Mesaros O, Edwards M, Wang H, Berceanu I, Neaga A, Zdrenghea M. 2019. Are community acquired respiratory viral infections an underestimated burden in hematology patients? Microorganisms 7:521. 10.3390/microorganisms7110521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantan B, Luyt C-E, Martin-Loeches I. 2019. Influenza infections and emergent viral infections in intensive care unit. Semin Respir Crit Care Med 40:488–497. 10.1055/s-0039-1693497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriyama M, Hugentobler WJ, Iwasaki A. 2020. Seasonality of respiratory viral infections. Annu Rev Virol 7:83–101. 10.1146/annurev-virology-012420-022445. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. 2018. Influenza (seasonal) epidemiology. World Health Organization, Geneva, Switzerland. https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal). [Google Scholar]
- 7.Kamigaki T, Chaw L, Tan AG, Tamaki R, Alday PP, Javier JB, Olveda RM, Oshitani H, Tallo VL. 2016. Seasonality of influenza and respiratory syncytial viruses and the effect of climate factors in subtropical-tropical Asia using influenza-like illness surveillance data, 2010 –2012. PLoS One 11:e0167712. 10.1371/journal.pone.0167712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opatowski L, Baguelin M, Eggo RM. 2018. Influenza interaction with cocirculating pathogens and its impact on surveillance, pathogenesis, and epidemic profile: a key role for mathematical modelling. PLoS Pathog 14:e1006770. 10.1371/journal.ppat.1006770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman AM, Leigh TR, Jr.. 2020. Viral pneumonia. In StatPearls. StatPearls Publishing, Treasure Island, FL. [Google Scholar]
- 10.Hirsch HH, Martino R, Ward KN, Boeckh M, Einsele H, Ljungman P. 2013. Fourth European Conference on Infections in Leukaemia (ECIL-4): guidelines for diagnosis and treatment of human respiratory syncytial virus, parainfluenza virus, metapneumovirus, rhinovirus, and coronavirus. Clin Infect Dis 56:258–266. 10.1093/cid/cis844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. 2011. Viral pneumonia. Lancet 377:1264–1275. 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simões EAF, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luyt C-E, Combes A, Trouillet J-L, Nieszkowska A, Chastre J. 2011. Virus-induced acute respiratory distress syndrome: epidemiology, management, and outcome. Presse Med 40:e561–e568. 10.1016/j.lpm.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, McGoogan JM. 2020. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72,314 cases from the Chinese Center for Disease Control and Prevention. JAMA 323:1239–1242. 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 15.Garg M, Prabhakar N, Gulati A, Agarwal R, Dhooria S. 2019. Spectrum of imaging findings in pulmonary infections. 1. Bacterial and viral. Pol J Radiol 84:e205–e213. 10.5114/pjr.2019.85812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Zhang Z, Dai C, Dong Q, Badrigilan S. 2020. Accuracy of deep learning for automated detection of pneumonia using chest X-ray images: a systematic review and meta-analysis. Comput Biol Med 123:103898. 10.1016/j.compbiomed.2020.103898. [DOI] [PubMed] [Google Scholar]
- 17.Koo HJ, Lim S, Choe J, Choi S-H, Sung H, Do K-H. 2018. Radiographic and CT Features of viral pneumonia. Radiographics 38:719–739. 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 18.Jalaber C, Lapotre T, Morcet-Delattre T, Ribet F, Jouneau S, Lederlin M. 2020. Chest CT in COVID-19 pneumonia: a review of current knowledge. Diagn Interv Imaging 101:431–437. 10.1016/j.diii.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. 2020. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect 26:1395–1399. 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson A, Davis S, Murch N. 2020. Pulmonary embolism in acute medicine: a case-based review incorporating latest guidelines in the COVID-19 era. Br J Hosp Med (Lond) 81:1–12. 10.12968/hmed.2020.0300. [DOI] [PubMed] [Google Scholar]
- 21.Ito A, Ishida T. 2020. Diagnostic markers for community-acquired pneumonia. Ann Transl Med 8:609–609. 10.21037/atm.2020.02.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson S, Bohn MK, Mancini N, Loh TP, Wang C-B, Grimmler M, Yuen K-Y, Mueller R, Koch D, Sethi S, Rawlinson WD, Clementi M, Erasmus R, Leportier M, Kwon GC, Menezes ME, Patru M-M, Gramegna M, Singh K, Najjar O, Ferrari M, Lippi G, Adeli K, Horvath A. 2020. IFCC interim guidelines on biochemical/hematological monitoring of COVID-19 patients. Clin Chem Lab Med 0:235. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Liu Y, Xiang P, Pu L, Xiong H, Li C, Zhang M, Tan J, Xu Y, Song R, Song M, Wang L, Zhang W, Han B, Yang L, Wang X, Zhou G, Zhang T, Li B, Wang Y, Chen Z, Wang X. 2020. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med 18:206–212. 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Meer V, Neven AK, van den Broek PJ, Assendelft WJJ. 2005. Diagnostic value of C reactive protein in infections of the lower respiratory tract: systematic review. BMJ 331:26. 10.1136/bmj.38483.478183.EB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. 2020. The role of biomarkers in diagnosis of COVID-19: a systematic review. Life Sci 254:117788. 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell L, Chernyak Y, Tobin KA, Cerfolio RJ, Francois F, Horwitz LI. 2020. Factors associated with hospital admission and critical illness among 5,279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ 369:m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedlund J, Hansson LO. 2000. Procalcitonin and C-reactive protein levels in community-acquired pneumonia: correlation with etiology and prognosis. Infection 28:68–73. 10.1007/s150100050049. [DOI] [PubMed] [Google Scholar]
- 28.Self WH, Balk RA, Grijalva CG, Williams DJ, Zhu Y, Anderson EJ, Waterer GW, Courtney DM, Bramley AM, Trabue C, Fakhran S, Blaschke AJ, Jain S, Edwards KM, Wunderink RG. 2017. Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis 65:183–190. 10.1093/cid/cix317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lippi G, Plebani M. 2020. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chim Acta 505:190–191. 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, Li Q, Jiang C, Zhou Y, Liu S, Ye C, Zhang P, Xing Y, Guo H, Tang W. 2020. Risk factors of critical and mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect 81:e16–e25. 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mei Z, Namaste SM, Serdula M, Suchdev PS, Rohner F, Flores-Ayala R, Addo OY, Raiten DJ. 2017. Adjusting total body iron for inflammation: Biomarkers Reflecting Inflammation and Nutritional Determinants of Anemia (BRINDA) project. Am J Clin Nutr 106:383S–389S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng F, Huang Y, Guo Y, Yin M, Chen X, Xiao L, Deng G. 2020. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis 96:467–474. 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobin MJ. 2020. Basing respiratory management of COVID-19 on physiological principles. Am J Respir Crit Care Med 201:1319–1320. 10.1164/rccm.202004-1076ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boktor SW, Hafner JW. 2020. Influenza. In StatPearls. StatPearls Publishing, Treasure Island, FL. [Google Scholar]
- 35.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 101:4620–4624. 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Short KR, Kuiken T, Van Riel D. 2019. Role of endothelial cells in the pathogenesis of influenza in humans. J Infect Dis 220:1859–1860. 10.1093/infdis/jiz349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinheimer VK, Becher A, Tönnies M, Holland G, Knepper J, Bauer TT, Schneider P, Neudecker J, Rückert JC, Szymanski K, Temmesfeld-Wollbrueck B, Gruber AD, Bannert N, Suttorp N, Hippenstiel S, Wolff T, Hocke AC. 2012. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis 206:1685–1694. 10.1093/infdis/jis455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hussell T, Goulding J. 2010. Structured regulation of inflammation during respiratory viral infection. Lancet Infect Dis 10:360–366. 10.1016/S1473-3099(10)70067-0. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Zhou Y-H, Yang Z-Q. 2016. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol 13:3–10. 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gabriel G, Herwig A, Klenk H-D. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog 4:e11. 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samji T. 2009. Influenza A: understanding the viral life cycle. Yale J Biol Med 82:153–159. [PMC free article] [PubMed] [Google Scholar]
- 42.Ortín J, Martín-Benito J. 2015. The RNA synthesis machinery of negative-stranded RNA viruses. Virology 479–480:532–544. 10.1016/j.virol.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Dou D, Revol R, Östbye H, Wang H, Daniels R. 2018. Influenza a virus cell entry, replication, virion assembly, and movement. Front Immunol 9:1581. 10.3389/fimmu.2018.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amatore D, Sgarbanti R, Aquilano K, Baldelli S, Limongi D, Civitelli L, Nencioni L, Garaci E, Ciriolo MR, Palamara AT. 2015. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell Microbiol 17:131–145. 10.1111/cmi.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniels R, Kurowski B, Johnson AE, Hebert DN. 2003. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell 11:79–90. 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 46.Bos TJ, Davis AR, Nayak DP. 1984. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A 81:2327–2331. 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dou D, da Silva DV, Nordholm J, Wang H, Daniels R. 2014. Type II transmembrane domain hydrophobicity dictates the cotranslational dependence for inversion. Mol Biol Cell 25:3363–3374. 10.1091/mbc.E14-04-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilmore R, Walter P, Blobel G. 1982. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol 95:470–477. 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karamyshev AL, Patrick AE, Karamysheva ZN, Griesemer DS, Hudson H, Tjon-Kon-Sang S, Nilsson I, Otto H, Liu Q, Rospert S, Heijne von G, Johnson AE, Thomas PJ. 2014. Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell 156:146–157. 10.1016/j.cell.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Böttcher E, Matrosovich T, Beyerle M, Klenk H-D, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80:9896–9898. 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Böttcher-Friebertshäuser E, Freuer C, Sielaff F, Schmidt S, Eickmann M, Uhlendorff J, Steinmetzer T, Klenk H-D, Garten W. 2010. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol 84:5605–5614. 10.1128/JVI.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertram S, Heurich A, Lavender H, Gierer S, Danisch S, Perin P, Lucas JM, Nelson PS, Pöhlmann S, Soilleux EJ. 2012. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One 7:e35876. 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gubareva LV, Kaiser L, Hayden FG. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827–835. 10.1016/S0140-6736(99)11433-8. [DOI] [PubMed] [Google Scholar]
- 55.Rückle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. 2012. The NS1 protein of influenza A virus blocks RIG-I-mediated activation of the noncanonical NF-κB pathway and p52/RelB-dependent gene expression in lung epithelial cells. J Virol 86:10211–10217. 10.1128/JVI.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ichinohe T, Iwasaki A, Hasegawa H. 2008. Innate sensors of influenza virus: clues to developing better intranasal vaccines. Expert Rev Vaccines 7:1435–1445. 10.1586/14760584.7.9.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Recum-Knepper von J, Sadewasser A, Weinheimer VK, Wolff T. 2015. Fluorescence-activated cell sorting-based analysis reveals an asymmetric induction of interferon-stimulated genes in response to seasonal influenza A Virus. J Virol 89:6982–6993. 10.1128/JVI.00857-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elias S, Mandelboim O. 2012. Battle of the midgets: innate microRNA networking. RNA Biol 9:792–798. 10.4161/rna.19717. [DOI] [PubMed] [Google Scholar]
- 59.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin T-Y, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Shi P-Y, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature Publishing Group 468:452–456. 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pichlmair A, Lassnig C, Eberle C-A, Górna MW, Baumann CL, Burkard TR, Bürckstümmer T, Stefanovic A, Krieger S, Bennett KL, Rülicke T, Weber F, Colinge J, Müller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol 12:624–630. 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 62.Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang I-C, Farzan M, Jung JU. 2013. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13:452–464. 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Y, Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol 5:65–67. 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 64.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol Rev 227:75–86. 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Julkunen I, Sareneva T, Pirhonen J, Ronni T, Melén K, Matikainen S. 2001. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev 12:171–180. 10.1016/s1359-6101(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 66.Heaton NS, Langlois RA, Sachs D, Lim JK, Palese P, tenOever BR. 2014. Long-term survival of influenza virus infected club cells drives immunopathology. J Exp Med 211:1707–1714. 10.1084/jem.20140488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reuther P, Göpfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346–11317. 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hamilton JR, Sachs D, Lim JK, Langlois RA, Palese P, Heaton NS. 2016. Club cells surviving influenza A virus infection induce temporary nonspecific antiviral immunity. Proc Natl Acad Sci U S A 113:3861–3866. 10.1073/pnas.1522376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heaton NS. 2017. Revisiting the concept of a cytopathic viral infection. PLoS Pathog 13:e1006409. 10.1371/journal.ppat.1006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishio M, Tsurudome M, Ito M, Garcin D, Kolakofsky D, Ito Y. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J Virol 79:8591–8601. 10.1128/JVI.79.13.8591-8601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickles RJ, DeVincenzo JP. 2015. Respiratory syncytial virus (RSV) and its propensity for causing bronchiolitis. J Pathol 235:266–276. 10.1002/path.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wignall-Fleming EB, Hughes DJ, Vattipally S, Modha S, Goodbourn S, Davison AJ, Randall RE. 2019. Analysis of paramyxovirus transcription and replication by high-throughput sequencing. J Virol 93:61. 10.1128/JVI.00571-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noton SL, Fearns R. 2015. Initiation and regulation of paramyxovirus transcription and replication. Virology 479–480:545–554. 10.1016/j.virol.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez KR, Horvath CM. 2014. Paramyxovirus V protein interaction with the antiviral sensor LGP2 disrupts MDA5 signaling enhancement but is not relevant to LGP2-mediated RLR signaling inhibition. J Virol 88:8180–8188. 10.1128/JVI.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramachandran A, Horvath CM. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J Interferon Cytokine Res 29:531–537. 10.1089/jir.2009.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sánchez-Aparicio MT, Feinman LJ, García-Sastre A, Shaw ML. 2018. Paramyxovirus V proteins interact with the RIG-I/TRIM25 regulatory complex and inhibit RIG-I signaling. J Virol 92:e01960-17. 10.1128/JVI.01960-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis ME, Wang MK, Rennick LJ, Full F, Gableske S, Mesman AW, Gringhuis SI, Geijtenbeek TBH, Duprex WP, Gack MU. 2014. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe 16:19–30. 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc Natl Acad Sci U S A 101:17264–17269. 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson SM, McNally BA, Ioannidis I, Flano E, Teng MN, Oomens AG, Walsh EE, Peeples ME. 2015. Respiratory syncytial virus uses CX3CR1 as a receptor on primary human airway epithelial cultures. PLoS Pathog 11:e1005318. 10.1371/journal.ppat.1005318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thibault PA, Watkinson RE, Moreira-Soto A, Drexler JF, Lee B. 2017. Zoonotic potential of emerging paramyxoviruses: knowns and unknowns. Adv Virus Res 98:1–55. 10.1016/bs.aivir.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin L-T, Richardson CD. 2016. The Host cell receptors for measles virus and their interaction with the viral hemagglutinin (H) protein. Viruses 8:250. 10.3390/v8090250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Griffin DE. 2007. Measles virus, p 1042–1069. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 83.Rubin SA, Sauder CJ, Carbone KM. 2013. Mumps virus, p 1024–1041. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 84.Wang LF, Mackenzie JS, Broder CC. 2013. Henipaviruses, p 1070–1085. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 85.Hierholzer JC. 1976. Purification and biophysical properties of human coronavirus 229E. Virology 75:155–165. 10.1016/0042-6822(76)90014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJM, Wolthers KC, Wertheim-van Dillen PME, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brucková M, McIntosh K, Kapikian AZ, Chanock RM. 1970. The adaptation of two human coronavirus strains (OC38 and OC43) to growth in cell monolayers. Proc Soc Exp Biol Med 135:431–435. 10.3181/00379727-135-35068. [DOI] [PubMed] [Google Scholar]
- 88.Woo PCY, Lau SKP, Chu C-M, Chan K-H, Tsoi H-W, Huang Y, Wong BHL, Poon RWS, Cai JJ, Luk W-K, Poon LLM, Wong SSY, Guan Y, Peiris JSM, Yuen K-Y. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79:884–895. 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perlman S, Zhao J. 2013. Human coronavirus EMC is not the same as severe acute respiratory syndrome coronavirus. mBio 4:e00473. 10.1128/mBio.00002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris JSM, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362:1353–1358. 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Woo PCY, Lau SKP, Lam CSF, Tsang AKL, Hui S-W, Fan RYY, Martelli P, Yuen K-Y. 2014. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in gammacoronavirus. J Virol 88:1318–1331. 10.1128/JVI.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Pfefferle S, Schöpf J, Kögl M, Friedel CC, Müller MA, Carbajo-Lozoya J, Stellberger T, von Dall’Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Zust R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel H-J, Schwegmann-Wessels C, Pöhlmann S, Haas J, Drosten C, von Brunn A. 2011. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 7:e1002331. 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang S-M, Wang C-T. 2009. APOBEC3G cytidine deaminase association with coronavirus nucleocapsid protein. Virology 388:112–120. 10.1016/j.virol.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lang Y, Li W, Li Z, Koerhuis D, van den Burg ACS, Rozemuller E, Bosch B-J, van Kuppeveld FJM, Boons G-J, Huizinga EG, van der Schaar HM, de Groot RJ. 2020. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc Natl Acad Sci U S A 117:25759–25770. 10.1073/pnas.2006299117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, Marasco WA, Baric RS, Sims AC, Pyrc K, Li W, Sui J. 2015. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol 89:7202–7213. 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang XM, Kousoulas KG, Storz J. 1992. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology 186:318–323. 10.1016/0042-6822(92)90089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo H, Hu B-J, Yang X-L, Zeng L-P, Li B, Ouyang S, Shi Z-L. 2020. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J Virol 94:3995. 10.1128/JVI.00902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bosch B-J, van der Zee R, de Haan CAM, Rottier PJM. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77:8801–8811. 10.1128/jvi.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, Zhu Y, Wang Q, Lu L, Jiang S. 2020. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther 5:92. 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Le Coupanec A, Desforges M, Meessen-Pinard M, Dubé M, Day R, Seidah NG, Talbot PJ. 2015. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog 11:e1005261. 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao X, Qin B, Chen P, Zhu K, Hou P, Wojdyla JA, Wang M, Cui S. 2021. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm Sin B 11:237–245. 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fang S, Shen H, Wang J, Tay FPL, Liu DX. 2010. Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase. J Virol 84:7325–7336. 10.1128/JVI.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem 279:10136–10141. 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boon den JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 106.Sola I, Almazán F, Zúñiga S, Enjuanes L. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol 2:265–288. 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA, Hogue BG. 2015. Coronavirus envelope (E) protein remains at the site of assembly. Virology 478:75–85. 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shuai H, Chu H, Hou Y, Yang D, Wang Y, Hu B, Huang X, Zhang X, Chai Y, Cai J-P, Chan JF-W, Yuen K-Y. 2020. Differential immune activation profile of SARS-CoV-2 and SARS-CoV infection in human lung and intestinal cells: implications for treatment with IFN-β and IFN inducer. J Infect 81:e1–e10. 10.1016/j.jinf.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, Becker S, Weber F. 2013. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol 87:5300–5304. 10.1128/JVI.03496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Busnadiego I, Fernbach S, Pohl MO, Karakus U, Huber M, Trkola A, Stertz S, Hale BG. 2020. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio 11:e01928-20. 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42:37–45. 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grünewald K, Koster AJ, Snijder EJ, Bárcena M. 2020. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369:1395–1398. 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Liu L. 2016. The membrane protein of severe acute respiratory syndrome coronavirus functions as a novel cytosolic pathogen-associated molecular pattern to promote beta interferon induction via a Toll-like-receptor-related TRAF3-independent mechanism. mBio 7:e01872-15. 10.1128/mBio.01872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng C-TK, Makino S. 2015. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 89:10970–10981. 10.1128/JVI.01352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tanaka T, Kamitani W, DeDiego ML, Enjuanes L, Matsuura Y. 2012. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J Virol 86:11128–11137. 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng C-TK, Makino S. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 82:4471–4479. 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler L-A, Leibundgut M, Thiel V, Mühlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27:959–966. 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 119.Lei J, Kusov Y, Hilgenfeld R. 2018. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res 149:58–74. 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Krafcikova P, Silhan J, Nencka R, Boura E. 2020. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun 11:3717–3717. 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Menachery VD, Yount BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88:4251–4264. 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fang X, Gao J, Zheng H, Li B, Kong L, Zhang Y, Wang W, Zeng Y, Ye L. 2007. The membrane protein of SARS-CoV suppresses NF-κB activation. J Med Virol 79:1431–1439. 10.1002/jmv.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Siu K-L, Kok K-H, Ng M-HJ, Poon VKM, Yuen K-Y, Zheng B-J, Jin D-Y. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKε complex. J Biol Chem 284:16202–16209. 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lui P-Y, Wong L-YR, Fung C-L, Siu K-L, Yeung M-L, Yuen K-S, Chan C-P, Woo PC-Y, Yuen K-Y, Jin D-Y. 2016. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect 5:e39. 10.1038/emi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 91:e02143-17. 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chang C-Y, Liu HM, Chang M-F, Chang SC. 2020. Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J Virol 94:e00099-20. 10.1128/JVI.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou B, Liu J, Wang Q, Liu X, Li X, Li P, Ma Q, Cao C. 2008. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1α. J Virol 82:6962–6971. 10.1128/JVI.00133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, García-Sastre A. 2020. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A 117:28344–28354. 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wong L-YR, Ye Z-W, Lui P-Y, Zheng X, Yuan S, Zhu L, Fung S-Y, Yuen K-S, Siu K-L, Yeung M-L, Cai Z, Woo PC-Y, Yuen K-Y, Chan C-P, Jin D-Y. 2020. Middle East respiratory syndrome coronavirus ORF8b accessory protein suppresses type I IFN expression by impeding HSP70-dependent activation of IRF3 kinase IKKε. J Immunol 205:1564–1579. 10.4049/jimmunol.1901489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. 2019. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio 10:e00319-19. 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jiang H-W, Zhang H-N, Meng Q-F, Xie J, Li Y, Chen H, Zheng Y-X, Wang X-N, Qi H, Zhang J, Wang P-H, Han Z-G, Tao S-C. 2020. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol Immunol 17:998–1000. 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shi C-S, Qi H-Y, Boularan C, Huang N-N, Abu-Asab M, Shelhamer JH, Kehrl JH. 2014. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193:3080–3089. 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ, Nakagawa S, Sato K, USFQ-COVID19 Consortium . 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 32:108185. 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H, Menachery VD, Rajsbaum R, Shi P-Y. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4:951–961. 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. 2009. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One 4:e8342. 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Young BE, Fong S-W, Chan Y-H, Mak TM, Ang LW, Anderson DE, Lee CY-P, Amrun SN, Lee B, Goh YS, Su YCF, Wei WE, Kalimuddin S, Chai LYA, Pada S, Tan SY, Sun L, Parthasarathy P, Chen YYC, Barkham T, Lin RTP, Maurer-Stroh S, Leo Y-S, Wang L-F, Rénia L, Lee VJ, Smith GJD, Lye DC, Ng LFP. 2020. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet 396:603–611. 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Y, Zhang J, Chen Y, Luo B, Yuan Y, Huang F, Yang T, Yu F, Liu J, Liu B, Song Z, Chen J, Pan T, Zhang X, Li Y, Li R, Huang W, Xiao F, Zhang H. 2020. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv 10.1101/2020.05.24.111823. [DOI] [PMC free article] [PubMed]
- 140.Ng JJ, Choong AMTL. 2020. Thromboembolic events in patients with SARS-CoV-2. J Vasc Surg 72:760–761. 10.1016/j.jvs.2020.04.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Criel M, Falter M, Jaeken J, Van Kerrebroeck M, Lefere I, Meylaerts L, Mesotten D, Vander Laenen M, Fivez T, Thomeer M, Ruttens D. 2020. Venous thromboembolism in SARS-CoV-2 patients: only a problem in ventilated ICU patients, or is there more to it? Eur Respir J 56:2001201. 10.1183/13993003.01201-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen Y-H, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Meyerholz DK, Lambertz AM, McCray PB. 2016. Dipeptidyl peptidase 4 distribution in the human respiratory tract: implications for the Middle East respiratory syndrome. Am J Pathol 186:78–86. 10.1016/j.ajpath.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature Publishing Group 426:450–454. 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lambert DW, Yarski M, Warner FJ, Thornhill P, Parkin ET, Smith AI, Hooper NM, Turner AJ. 2005. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J Biol Chem 280:30113–30119. 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. 2014. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 88:1293–1307. 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang S, Guo F, Liu K, Wang H, Rao S, Yang P, Jiang C. 2008. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res 136:8–15. 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dijkman R, Jebbink MF, Deijs M, Milewska A, Pyrc K, Buelow E, van der Bijl A, van der Hoek L. 2012. Replication-dependent downregulation of cellular angiotensin-converting enzyme 2 protein expression by human coronavirus NL63. J Gen Virol 93:1924–1929. 10.1099/vir.0.043919-0. [DOI] [PubMed] [Google Scholar]
- 150.Glowacka I, Bertram S, Herzog P, Pfefferle S, Steffen I, Muench MO, Simmons G, Hofmann H, Kuri T, Weber F, Eichler J, Drosten C, Pöhlmann S. 2010. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J Virol 84:1198–1205. 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, Deng Y, Mao L, Su C, Qiang G, Jiang T, Zhao J, Wu G, Song J, Tan W. 2020. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun 11:3910–3918. 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kaye M. 2006. SARS-associated coronavirus replication in cell lines. Emerg Infect Dis 12:128–133. 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi C-S, Nabar NR, Huang N-N, Kehrl JH. 2019. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov 5:101–112. 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, Han Y, Zhang X-Y, Zhou W, Qiu Y, Zhou X. 2020. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol 17:881–883. 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Carlin CR. 2019. New insights to adenovirus-directed innate immunity in respiratory epithelial cells. Microorganisms 7:216. 10.3390/microorganisms7080216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hayashi S, Hogg JC. 2007. Adenovirus infections and lung disease. Curr Opin Pharmacol 7:237–243. 10.1016/j.coph.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.van Tol MJD, Kroes ACM, Schinkel J, Dinkelaar W, Claas ECJ, Jol-van der Zijde CM, Vossen JM. 2005. Adenovirus infection in paediatric stem cell transplant recipients: increased risk in young children with a delayed immune recovery. Bone Marrow Transplant 36:39–50. 10.1038/sj.bmt.1705003. [DOI] [PubMed] [Google Scholar]
- 158.Majorant D, Qiu F, Kalil AC, Wilson N, Florescu DF. 2018. Adenovirus: a deadly disease in the solid organ transplant population: risk factors and outcomes. Transplant Proc 50:3769–3774. 10.1016/j.transproceed.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 159.Lynch JP, Fishbein M, Echavarria M. 2011. Adenovirus. Semin Respir Crit Care Med 32:494–511. 10.1055/s-0031-1283287. [DOI] [PubMed] [Google Scholar]
- 160.Vasickova P, Pavlik I, Verani M, Carducci A. 2010. Issues concerning survival of viruses on surfaces. Food Environ Virol 2:24–34. 10.1007/s12560-010-9025-6. [DOI] [Google Scholar]
- 161.Goldmann DA. 2000. Transmission of viral respiratory infections in the home. Pediatr Infect Dis J 19:S97–S102. 10.1097/00006454-200010001-00002. [DOI] [PubMed] [Google Scholar]
- 162.Cook J, Radke J. 2017. Mechanisms of pathogenesis of emerging adenoviruses. F1000Res 6:90. 10.12688/f1000research.10152.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ghebremedhin B. 2014. Human adenovirus: viral pathogen with increasing importance. Eur J Microbiol Immunol 4:26–33. 10.1556/EuJMI.4.2014.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wiethoff CM, Nemerow GR. 2015. Adenovirus membrane penetration: tickling the tail of a sleeping dragon. Virology 479–480:591–599. 10.1016/j.virol.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roelvink PW, Lizonova A, Lee JG, Li Y, Bergelson JM, Finberg RW, Brough DE, Kovesdi I, Wickham TJ. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol 72:7909–7915. 10.1128/JVI.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci U S A 98:15191–15196. 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Luissint A-C, Lutz PG, Calderwood DA, Couraud P-O, Bourdoulous S. 2008. JAM-L-mediated leukocyte adhesion to endothelial cells is regulated in cis by α4β1 integrin activation. J Cell Biol 183:1159–1173. 10.1083/jcb.200805061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Short JJ, Vasu C, Holterman MJ, Curiel DT, Pereboev A. 2006. Members of adenovirus species B utilize CD80 and CD86 as cellular attachment receptors. Virus Res 122:144–153. 10.1016/j.virusres.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nakano MY, Boucke K, Suomalainen M, Stidwill RP, Greber UF. 2000. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J Virol 74:7085–7095. 10.1128/jvi.74.15.7085-7095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jiang H, White EJ, Ríos-Vicil CI, Xu J, Gomez-Manzano C, Fueyo J. 2011. Human adenovirus type 5 induces cell lysis through autophagy and autophagy-triggered caspase activity. J Virol 85:4720–4729. 10.1128/JVI.02032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ashbourne Excoffon KJD, Moninger T, Zabner J. 2003. The coxsackie B virus and adenovirus receptor resides in a distinct membrane microdomain. J Virol 77:2559–2567. 10.1128/jvi.77.4.2559-2567.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Smith JS, Xu Z, Tian J, Palmer DJ, Ng P, Byrnes AP. 2011. The role of endosomal escape and mitogen-activated protein kinases in adenoviral activation of the innate immune response. PLoS One 6:e26755. 10.1371/journal.pone.0026755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Di Paolo NC, Miao EA, Iwakura Y, Murali-Krishna K, Aderem A, Flavell RA, Papayannopoulou T, Shayakhmetov DM. 2009. Virus binding to a plasma membrane receptor triggers interleukin-1α-mediated proinflammatory macrophage response in vivo. Immunity 31:110–121. 10.1016/j.immuni.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yamaguchi T, Kawabata K, Kouyama E, Ishii KJ, Katayama K, Suzuki T, Kurachi S, Sakurai F, Akira S, Mizuguchi H. 2010. Induction of type I interferon by adenovirus-encoded small RNAs. Proc Natl Acad Sci U S A 107:17286–17291. 10.1073/pnas.1009823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Horwitz MS. 2004. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J Gene Med 6(Suppl 1):S172–S183. 10.1002/jgm.495. [DOI] [PubMed] [Google Scholar]
- 176.Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A, Greber UF. 2014. Innate immunity to adenovirus. Hum Gene Ther 25:265–284. 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Windheim M, Hilgendorf A, Burgert HG. 2004. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr Top Microbiol Immunol 273:29–85. 10.1007/978-3-662-05599-1_2. [DOI] [PubMed] [Google Scholar]
- 178.Zaki SR, Paddock CD. 2008. Viral infections of the lung, p 426–475. In Dail and Hammar’s pulmonary pathology, 4th ed. Springer, New York, NY. [Google Scholar]
- 179.Martínez-Girón R, Pantanowitz L. 2017. Lower respiratory tract viral infections: diagnostic role of exfoliative cytology. Diagn Cytopathol 45:614–620. 10.1002/dc.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Grinde B. 2013. Herpesviruses: latency and reactivation—viral strategies and host response. J Oral Microbiol 5:22766–22769. 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9:369–381. 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cai W, Schaffer PA. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66:2904–2915. 10.1128/JVI.66.5.2904-2915.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gruffat H, Marchione R, Manet E. 2016. Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front Microbiol 7:869. 10.3389/fmicb.2016.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mettenleiter TC. 2004. Budding events in herpesvirus morphogenesis. Virus Res 106:167–180. 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 185.Zhu H, Zheng C. 2020. The race between host antiviral innate immunity and the immune evasion strategies of herpes simplex virus 1. Microbiol Mol Biol Rev 84:52. 10.1128/MMBR.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gerada C, Campbell TM, Kennedy JJ, McSharry BP, Steain M, Slobedman B, Abendroth A. 2020. Manipulation of the innate immune response by varicella-zoster virus. Front Immunol 11:1. 10.3389/fimmu.2020.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hood C, Cunningham AL, Slobedman B, Boadle RA, Abendroth A. 2003. Varicella-zoster virus-infected human sensory neurons are resistant to apoptosis, yet human foreskin fibroblasts are susceptible: evidence for a cell-type-specific apoptotic response. J Virol 77:12852–12864. 10.1128/jvi.77.23.12852-12864.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ahmed M, Lock M, Miller CG, Fraser NW. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J Virol 76:717–729. 10.1128/jvi.76.2.717-729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang L, Wang M, Cheng A, Yang Q, Wu Y, Jia R, Liu M, Zhu D, Chen S, Zhang S, Zhao X, Huang J, Wang Y, Xu Z, Chen Z, Zhu L, Luo Q, Liu Y, Yu Y, Zhang L, Tian B, Pan L, Rehman MU, Chen X. 2019. Innate immune evasion of alphaherpesvirus tegument proteins. Front Immunol 10:2196. 10.3389/fimmu.2019.02196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Oda Y, Okada Y, Katsuda S, Nakanishi I. 1994. Immunohistochemical study on the infection of herpes simplex virus, human cytomegalovirus, and Epstein-Barr virus in secondary diffuse interstitial pneumonia. Hum Pathol 25:1057–1062. 10.1016/0046-8177(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 191.Luyt C-E, Combes A, Deback C, Aubriot-Lorton M-H, Nieszkowska A, Trouillet J-L, Capron F, Agut H, Gibert C, Chastre J. 2007. Herpes simplex virus lung infection in patients undergoing prolonged mechanical ventilation. Am J Respir Crit Care Med 175:935–942. 10.1164/rccm.200609-1322OC. [DOI] [PubMed] [Google Scholar]
- 192.Chong S, Kim TS, Cho EY. 2010. Herpes simplex virus pneumonia: high-resolution CT findings. Br J Radiol 83:585–589. 10.1259/bjr/51409455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Twagira M, Hadzic N, Smith M, Ramaswamy M, Verma A, Dhawan A, Knisely AS, Mieli-Vergani G, Geretti AM. 2004. Disseminated neonatal herpes simplex virus (HSV) type 2 infection diagnosed by HSV DNA detection in blood and successfully managed by liver transplantation. Eur J Pediatr 163:166–169. 10.1007/s00431-003-1383-8. [DOI] [PubMed] [Google Scholar]
- 194.Coclite E, Di Natale C, Nigro G. 2013. Congenital and perinatal cytomegalovirus lung infection. J Matern Fetal Neonatal Med 26:1671–1675. 10.3109/14767058.2013.794207. [DOI] [PubMed] [Google Scholar]
- 195.Grilli E, Galati V, Bordi L, Taglietti F, Petrosillo N. 2012. Cytomegalovirus pneumonia in immunocompetent host: case report and literature review. J Clin Virol 55:356–359. 10.1016/j.jcv.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 196.Sinzger C, Grefte A, Plachter B, Gouw AS, The TH, Jahn G, 196 . 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol 76:741–750. 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 197.Mirouse A, Vignon P, Piron P, Robert R, Papazian L, Géri G, Blanc P, Guitton C, Guérin C, Bigé N, Rabbat A, Lefebvre A, Razazi K, Fartoukh M, Mariotte E, Bouadma L, Ricard J-D, Seguin A, Souweine B, Moreau A-S, Faguer S, Mari A, Mayaux J, Schneider F, Stoclin A, Perez P, Maizel J, Lafon C, Ganster F, Argaud L, Girault C, Barbier F, Lecuyer L, Lambert J, Canet E. 2017. Severe varicella-zoster virus pneumonia: a multicenter cohort study. Crit Care 21:137–110. 10.1186/s13054-017-1731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, Moffat J, Ku C-C, Zerboni L, Sommer M, et al. 2007. VZV: pathogenesis and the disease consequences of primary infection. In Arvin A (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 199.Arnold N, Girke T, Sureshchandra S, Nguyen C, Rais M, Messaoudi I. 2016. Genomic and functional analysis of the host response to acute simian varicella infection in the lung. Sci Rep 6:34164–34114. 10.1038/srep34164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kennedy JJ, Steain M, Slobedman B, Abendroth A. 2018. Infection and functional modulation of human monocytes and macrophages by varicella-zoster virus. J Virol 93:10980. 10.1128/JVI.01887-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schildgen O. 2013. Human bocavirus: lessons learned to date. Pathogens 2:1–12. 10.3390/pathogens2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102:12891–12896. 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Allander T, Jartti T, Gupta S, Niesters HGM, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung-Lindell A, van den Hoogen BG, Hyypiä T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44:904–910. 10.1086/512196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schildgen O, Müller A, Allander T, Mackay IM, Völz S, Kupfer B, Simon A. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev 21:291–304. 10.1128/CMR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fry AM, Lu X, Chittaganpitch M, Peret T, Fischer J, Dowell SF, Anderson LJ, Erdman D, Olsen SJ. 2007. Human bocavirus: a novel parvovirus epidemiologically associated with pneumonia requiring hospitalization in Thailand. J Infect Dis 195:1038–1045. 10.1086/512163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Vicente D, Cilla G, Montes M, Pérez-Yarza EG, Pérez-Trallero E. 2007. Human bocavirus, a respiratory and enteric virus. Emerg Infect Dis 13:636–637. 10.3201/eid1304.061501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154. 10.1016/j.virol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Foulongne V, Rodière M, Segondy M. 2006. Human bocavirus in children. Emerg Infect Dis 12:862–863. 10.3201/eid1205.051523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lu X, Chittaganpitch M, Olsen SJ, Mackay IM, Sloots TP, Fry AM, Erdman DD. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J Clin Microbiol 44:3231–3235. 10.1128/JCM.00889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nascimento-Carvalho AC, Vilas-Boas A-L, Fontoura M-SH, Xu M, Vuorinen T, Söderlund-Venermo M, Ruuskanen O, Nascimento-Carvalho CM, PNEUMOPAC-Efficacy Study Group . 2018. Serologically diagnosed acute human bocavirus 1 infection in childhood community-acquired pneumonia. Pediatr Pulmonol 53:88–94. 10.1002/ppul.23891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bonzel L, Tenenbaum T, Schroten H, Schildgen O, Schweitzer-Krantz S, Adams O. 2008. Frequent detection of viral coinfection in children hospitalized with acute respiratory tract infection using a real-time polymerase chain reaction. Pediatr Infect Dis J 27:589–594. 10.1097/INF.0b013e3181694fb9. [DOI] [PubMed] [Google Scholar]
- 212.Edner N, Castillo-Rodas P, Falk L, Hedman K, Söderlund-Venermo M, Allander T. 2012. Life-threatening respiratory tract disease with human bocavirus-1 infection in a 4-year-old child. J Clin Microbiol 50:531–532. 10.1128/JCM.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Meriluoto M, Hedman L, Tanner L, Simell V, Mäkinen M, Simell S, Mykkänen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Söderlund-Venermo M. 2012. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg Infect Dis 18:264–271. 10.3201/eid1802.111293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Longtin J, Bastien M, Gilca R, Leblanc E, de Serres G, Bergeron MG, Boivin G. 2008. Human bocavirus infections in hospitalized children and adults. Emerg Infect Dis 14:217–221. 10.3201/eid1402.070851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Moesker FM, van Kampen JJA, van der Eijk AA, van Rossum AMC, de Hoog M, Schutten M, Smits SL, Bodewes R, Osterhaus ADME, Fraaij PLA. 2015. Human bocavirus infection as a cause of severe acute respiratory tract infection in children. Clin Microbiol Infect 21:964.e1–8–964.e8. 10.1016/j.cmi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Christensen A, Nordbø SA, Krokstad S, Rognlien AGW, Døllner H. 2010. Human bocavirus in children: mono-detection, high viral load and viraemia are associated with respiratory tract infection. J Clin Virol 49:158–162. 10.1016/j.jcv.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ziemele I, Xu M, Vilmane A, Rasa-Dzelzkaleja S, Hedman L, Hedman K, Söderlund-Venermo M, Nora-Krukle Z, Murovska M, Gardovska D. 2019. Acute human bocavirus 1 infection in child with life-threatening bilateral bronchiolitis and right-sided pneumonia: a case report. J Med Case Rep 13:290–296. 10.1186/s13256-019-2222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Huang L-Y, Halder S, Agbandje-McKenna M. 2014. Parvovirus glycan interactions. Curr Opin Virol 7:108–118. 10.1016/j.coviro.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Liu X, Hao S, Chen Z, Xu H, Wang H, Huang M, Guan W. 2018. The 5′ untranslated region of human bocavirus capsid transcripts regulates viral mRNA biogenesis and alternative translation. J Virol 92:12891. 10.1128/JVI.00443-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mietzsch M, Pénzes JJ, Agbandje-McKenna M. 2019. Twenty-five years of structural parvovirology. Viruses 11:362. 10.3390/v11040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2017. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol 91:e00710-17. 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Deng X, Zou W, Xiong M, Wang Z, Engelhardt JF, Ye SQ, Yan Z, Qiu J. 2017. Human parvovirus infection of human airway epithelia induces pyroptotic cell death by inhibiting apoptosis. J Virol 91:e01533-17. 10.1128/JVI.01533-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DCM, Chen AY, Li Y, Tang L, Söderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog 8:e1002899. 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Deng X, Yan Z, Luo Y, Xu J, Cheng F, Li Y, Engelhardt JF, Qiu J. 2013. In vitro modeling of human bocavirus 1 infection of polarized primary human airway epithelia. J Virol 87:4097–4102. 10.1128/JVI.03132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J Virol 83:7739–7748. 10.1128/JVI.00614-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Guido M, Tumolo MR, Verri T, Romano A, Serio F, De Giorgi M, De Donno A, Bagordo F, Zizza A. 2016. Human bocavirus: current knowledge and future challenges. World J Gastroenterol 22:8684–8697. 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Davies JA, Garrod DR. 1997. Molecular aspects of the epithelial phenotype. Bioessays 19:699–704. 10.1002/bies.950190810. [DOI] [PubMed] [Google Scholar]
- 229.Roche WR, Montefort S, Baker J, Holgate ST. 1993. Cell adhesion molecules and the bronchial epithelium. Am Rev Respir Dis 148:S79–S82. 10.1164/ajrccm/148.6_Pt_2.S79. [DOI] [PubMed] [Google Scholar]
- 230.Wu N-H, Yang W, Beineke A, Dijkman R, Matrosovich M, Baumgärtner W, Thiel V, Valentin-Weigand P, Meng F, Herrler G. 2016. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci Rep 6:39668. 10.1038/srep39668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Short KR, Kasper J, van der Aa S, Andeweg AC, Zaaraoui-Boutahar F, Goeijenbier M, Richard M, Herold S, Becker C, Scott DP, Limpens RWAL, Koster AJ, Bárcena M, Fouchier RAM, Kirkpatrick CJ, Kuiken T. 2016. Influenza virus damages the alveolar barrier by disrupting epithelial cell tight junctions. Eur Respir J 47:954–966. 10.1183/13993003.01282-2015. [DOI] [PubMed] [Google Scholar]
- 232.Kast JI, McFarlane AJ, Głobińska A, Sokolowska M, Wawrzyniak P, Sanak M, Schwarze J, Akdis CA, Wanke K. 2017. Respiratory syncytial virus infection influences tight junction integrity. Clin Exp Immunol 190:351–359. 10.1111/cei.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Smallcombe CC, Linfield DT, Harford TJ, Bokun V, Ivanov AI, Piedimonte G, Rezaee F. 2019. Disruption of the airway epithelial barrier in a murine model of respiratory syncytial virus infection. Am J Physiol Lung Cell Mol Physiol 316:L358–L368. 10.1152/ajplung.00345.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, O’Neal WK, Sallenave J-M, Pickles RJ, Boucher RC. 2012. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci U S A 109:16528–16533. 10.1073/pnas.1206552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Iversen MB, Reinert LS, Thomsen MK, Bagdonaite I, Nandakumar R, Cheshenko N, Prabakaran T, Vakhrushev SY, Krzyzowska M, Kratholm SK, Ruiz-Perez F, Petersen SV, Goriely S, Bibby BM, Eriksson K, Ruland J, Thomsen AR, Herold BC, Wandall HH, Frische S, Holm CK, Paludan SR. 2016. An innate antiviral pathway acting before interferons at epithelial surfaces. Nat Immunol 17:150–158. 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- 236.Kása A, Csortos C, Verin AD. 2015. Cytoskeletal mechanisms regulating vascular endothelial barrier function in response to acute lung injury. Tissue Barriers 3:e974448. 10.4161/21688370.2014.974448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Short KR, Kroeze EJBV, Fouchier RAM, Kuiken T. 2014. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis 14:57–69. 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- 238.Lipscomb MF, Pollard AM, Yates JL. 1993. A role for TGF-β in the suppression by murine bronchoalveolar cells of lung dendritic cell initiated immune responses. Reg Immunol 5:151–157. [PubMed] [Google Scholar]
- 239.Thepen T, Kraal G, Holt PG. 1994. The role of alveolar macrophages in regulation of lung inflammation. Ann N Y Acad Sci 725:200–206. 10.1111/j.1749-6632.1994.tb39802.x. [DOI] [PubMed] [Google Scholar]
- 240.Chelen CJ, Fang Y, Freeman GJ, Secrist H, Marshall JD, Hwang PT, Frankel LR, DeKruyff RH, Umetsu DT. 1995. Human alveolar macrophages present antigen ineffectively due to defective expression of B7 costimulatory cell surface molecules. J Clin Invest 95:1415–1421. 10.1172/JCI117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Balbo P, Silvestri M, Rossi GA, Crimi E, Burastero SE. 2001. Differential role of CD80 and CD86 on alveolar macrophages in the presentation of allergen to T lymphocytes in asthma. Clin Exp Allergy 31:625–636. 10.1046/j.1365-2222.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- 242.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, Henson PM. 2011. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am J Respir Crit Care Med 184:547–560. 10.1164/rccm.201011-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Schwab JM, Chiang N, Arita M, Serhan CN. 2007. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature Publishing Group 447:869–874. 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z. 2020. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26:842–844. 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 245.Mócsai A. 2013. Diverse novel functions of neutrophils in immunity, inflammation, and beyond. J Exp Med 210:1283–1299. 10.1084/jem.20122220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kruger P, Saffarzadeh M, Weber ANR, Rieber N, Radsak M, Bernuth von H, Benarafa C, Roos D, Skokowa J, Hartl D. 2015. Neutrophils: between host defence, immune modulation, and tissue injury. PLoS Pathog 11:e1004651. 10.1371/journal.ppat.1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ng SL, Teo YJ, Setiagani YA, Karjalainen K, Ruedl C. 2018. Type 1 conventional CD103+ dendritic cells control effector CD8+ T cell migration, survival, and memory responses during influenza infection. Front Immunol 9:3043. 10.3389/fimmu.2018.03043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shi C, Pamer EG. 2011. Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11:762–774. 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, Zaki SR, Subbarao K. 2010. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol 84:1289–1301. 10.1128/JVI.01281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Neyt K, Lambrecht BN. 2013. The role of lung dendritic cell subsets in immunity to respiratory viruses. Immunol Rev 255:57–67. 10.1111/imr.12100. [DOI] [PubMed] [Google Scholar]
- 251.Braciale TJ, Sun J, Kim TS. 2012. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 12:295–305. 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Burgdorf S, Kautz A, Böhnert V, Knolle PA, Kurts C. 2007. Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation. Science 316:612–616. 10.1126/science.1137971. [DOI] [PubMed] [Google Scholar]
- 253.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee H-W, Park CG, Steinman RM, Nussenzweig MC. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science 315:107–111. 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 254.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322:1097–1100. 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.GeurtsvanKessel CH, Willart MAM, van Rijt LS, Muskens F, Kool M, Baas C, Thielemans K, Bennett C, Clausen BE, Hoogsteden HC, Osterhaus ADME, Rimmelzwaan GF, Lambrecht BN. 2008. Clearance of influenza virus from the lung depends on migratory Langerin+ CD11b– but not plasmacytoid dendritic cells. J Exp Med 205:1621–1634. 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, García-Sastre A, Merad M. 2012. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest 122:4037–4047. 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ainsua-Enrich E, Hatipoglu I, Kadel S, Turner S, Paul J, Singh S, Bagavant H, Kovats S. 2019. IRF4-dependent dendritic cells regulate CD8+ T-cell differentiation and memory responses in influenza infection. Mucosal Immunol 12:1025–1037. 10.1038/s41385-019-0173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ballesteros-Tato A, León B, Lund FE, Randall TD. 2010. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat Immunol 11:216–224. 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Bosteels C, Neyt K, Vanheerswynghels M, van Helden MJ, Sichien D, Debeuf N, De Prijck S, Bosteels V, Vandamme N, Martens L, Saeys Y, Louagie E, Lesage M, Williams DL, Tang S-C, Mayer JU, Ronchese F, Scott CL, Hammad H, Guilliams M, Lambrecht BN. 2020. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 52:1039–1056. 10.1016/j.immuni.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Smit JJ, Rudd BD, Lukacs NW. 2006. Plasmacytoid dendritic cells inhibit pulmonary immunopathology and promote clearance of respiratory syncytial virus. J Exp Med 203:1153–1159. 10.1084/jem.20052359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Guilliams M, Lambrecht BN, Hammad H. 2013. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol 6:464–473. 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- 262.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie ANJ, Mebius RE, Powrie F, Spits H. 2018. Innate lymphoid cells: 10 years on. Cell 174:1054–1066. 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 263.Fan X, Rudensky AY. 2016. Hallmarks of tissue-resident lymphocytes. Cell 164:1198–1211. 10.1016/j.cell.2016.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Gorski SA, Hahn YS, Braciale TJ. 2013. Group 2 innate lymphoid cell production of IL-5 is regulated by NKT cells during influenza virus infection. PLoS Pathog 9:e1003615. 10.1371/journal.ppat.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. 2016. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol 17:76–86. 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- 266.Duerr CU, McCarthy CDA, Mindt BC, Rubio M, Meli AP, Pothlichet J, Eva MM, Gauchat J-F, Qureshi ST, Mazer BD, Mossman KL, Malo D, Gamero AM, Vidal SM, King IL, Sarfati M, Fritz JH. 2016. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat Immunol 17:65–75. 10.1038/ni.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Taube C, Tertilt C, Gyülveszi G, Dehzad N, Kreymborg K, Schneeweiss K, Michel E, Reuter S, Renauld J-C, Arnold-Schild D, Schild H, Buhl R, Becher B. 2011. IL-22 is produced by innate lymphoid cells and limits inflammation in allergic airway disease. PLoS One 6:e21799. 10.1371/journal.pone.0021799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. 2015. The burgeoning family of unconventional T cells. Nat Immunol 16:1114–1123. 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- 269.De Santo C, Salio M, Masri SH, Lee LY-H, Dong T, Speak AO, Porubsky S, Booth S, Veerapen N, Besra GS, Gröne H-J, Platt FM, Zambon M, Cerundolo V. 2008. Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest 118:4036–4048. 10.1172/JCI36264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. 2002. NK T cells contribute to expansion of CD8+ T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol 76:4294–4303. 10.1128/jvi.76.9.4294-4303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Latha TS, Reddy MC, Durbaka PVR, Rachamallu A, Pallu R, Lomada D. 2014. γδ T cell-mediated immune responses in disease and therapy. Front Immunol 5:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Poccia F, Agrati C, Castilletti C, Bordi L, Gioia C, Horejsh D, Ippolito G, Chan PKS, Hui DSC, Sung JJY, Capobianchi MR, Malkovsky M. 2006. Anti-severe acute respiratory syndrome coronavirus immune responses: the role played by V gamma 9V delta 2 T cells. J Infect Dis 193:1244–1249. 10.1086/502975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, Mettke E, Kurioka A, Hansen TH, Klenerman P, Willberg CB. 2014. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+ IL-18 in a TCR-independent manner. Eur J Immunol 44:195–203. 10.1002/eji.201343509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bennett MS, Trivedi S, Iyer AS, Hale JS, Leung DT. 2017. Human mucosal-associated invariant T (MAIT) cells possess capacity for B cell help. J Leukoc Biol 102:1261–1269. 10.1189/jlb.4A0317-116R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, Zhao Z, Koutsakos M, Nüssing S, Sant S, Wang Z, D’Souza C, Jia X, Almeida CF, Kostenko L, Eckle SBG, Meehan BS, Kallies A, Godfrey DI, Reading PC, Corbett AJ, McCluskey J, Klenerman P, Kedzierska K, Hinks TSC. 2018. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun 9:4706–4709. 10.1038/s41467-018-07207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wang S, Le TQ, Kurihara N, Chida J, Cisse Y, Yano M, Kido H. 2010. Influenza virus-cytokine-protease cycle in the pathogenesis of vascular hyperpermeability in severe influenza. J Infect Dis 202:991–1001. 10.1086/656044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Alper CM, Doyle WJ, Skoner DP, Buchman CA, Seroky JT, Gwaltney JM, Cohen SA. 1996. Prechallenge antibodies: moderators of infection rate, signs, and symptoms in adults experimentally challenged with rhinovirus type 39. Laryngoscope 106:1298–1305. 10.1097/00005537-199610000-00025. [DOI] [PubMed] [Google Scholar]
- 278.Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, Lorenzi JCC, Finkin S, Hägglöf T, Hurley A, Millard KG, Weisblum Y, Schmidt F, Hatziioannou T, Bieniasz PD, Caskey M, Robbiani DF, Nussenzweig MC, Bjorkman PJ. 2020. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182:828–842.e16. 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Channappanavar R, Zhao J, Perlman S. 2014. T cell-mediated immune response to respiratory coronaviruses. Immunol Res 59:118–128. 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Iyer AS, Jones FK, Nodoushani A, Kelly M, Becker M, Slater D, Mills R, Teng E, Kamruzzaman M, Garcia-Beltran WF, Astudillo M, Yang D, Miller TE, Oliver E, Fischinger S, Atyeo C, Iafrate AJ, Calderwood SB, Lauer SA, Yu J, Li Z, Feldman J, Hauser BM, Caradonna TM, Branda JA, Turbett SE, LaRocque RC, Mellon G, Barouch DH, Schmidt AG, Azman AS, Alter G, Ryan ET, Harris JB, Charles RC. 2020. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol 5:eabe0367. 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, Li Z, Chao G, Rojas OL, Bang YM, Pu A, Christie-Holmes N, Gervais C, Ceccarelli D, Samavarchi-Tehrani P, Guvenc F, Budylowski P, Li A, Paterson A, Yue FY, Marin LM, Caldwell L, Wrana JL, Colwill K, Sicheri F, Mubareka S, Gray-Owen SD, Drews SJ, Siqueira WL, Barrios-Rodiles M, Ostrowski M, Rini JM, Durocher Y, McGeer AJ, Gommerman JL, Gingras A-C. 2020. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 5:eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Herati RS, Reuter MA, Dolfi DV, Mansfield KD, Aung H, Badwan OZ, Kurupati RK, Kannan S, Ertl H, Schmader KE, Betts MR, Canaday DH, Wherry EJ. 2014. Circulating CXCR5+ PD-1+ response predicts influenza vaccine antibody responses in young adults but not elderly adults. J Immunol 193:3528–3537. 10.4049/jimmunol.1302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhao J, Zhao J, Perlman S. 2010. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol 84:9318–9325. 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, van den Akker JPC, Molenkamp R, Koopmans MPG, van Gorp ECM, Haagmans BL, de Swart RL, Sette A, de Vries RD. 2020. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 5:eabd2071. 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Suarez-Ramirez JE, Chandiran K, Brocke S, Cauley LS. 2019. Immunity to respiratory infection is reinforced through early proliferation of lymphoid TRM cells and prompt arrival of effector CD8 T cells in the lungs. Front Immunol 10:1370. 10.3389/fimmu.2019.01370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, et al. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, Dorgham K, Philippot Q, Rosain J, Béziat V, Manry J, Shaw E, Haljasmägi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, et al. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370:eabd4585. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Davidson S, Crotta S, McCabe TM, Wack A. 2014. Pathogenic potential of interferon αβ in acute influenza infection. Nat Commun 5:3864–3815. 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Günther S, Drosten C, Michiels T, Staeheli P. 2010. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol 84:5670–5677. 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mordstein M, Kochs G, Dumoutier L, Renauld J-C, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151. 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wack A, Terczyńska-Dyla E, Hartmann R. 2015. Guarding the frontiers: the biology of type III interferons. Nat Immunol 16:802–809. 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakõiv T, Staeheli P. 2018. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. Elife 7:266. 10.7554/eLife.33354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mukaida N. 2000. Interleukin-8: an expanding universe beyond neutrophil chemotaxis and activation. Int J Hematol 72:391–398. [PubMed] [Google Scholar]
- 294.Lindell DM, Lane TE, Lukacs NW. 2008. CXCL10/CXCR3-mediated responses promote immunity to respiratory syncytial virus infection by augmenting dendritic cell and CD8+ T cell efficacy. Eur J Immunol 38:2168–2179. 10.1002/eji.200838155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang W, Yang P, Zhong Y, Zhao Z, Xing L, Zhao Y, Zou Z, Zhang Y, Li C, Li T, Wang C, Wang Z, Yu X, Cao B, Gao X, Penninger JM, Wang X, Jiang C. 2013. Monoclonal antibody against CXCL-10/IP-10 ameliorates influenza A (H1N1) virus induced acute lung injury. Cell Res 23:577–580. 10.1038/cr.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Ichikawa A, Kuba K, Morita M, Chida S, Tezuka H, Hara H, Sasaki T, Ohteki T, Ranieri VM, Santos dos CC, Kawaoka Y, Akira S, Luster AD, Lu B, Penninger JM, Uhlig S, Slutsky AS, Imai Y. 2013. CXCL10-CXCR3 enhances the development of neutrophil-mediated fulminant lung injury of viral and nonviral origin. Am J Respir Crit Care Med 187:65–77. 10.1164/rccm.201203-0508OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kok WL, Denney L, Benam K, Cole S, Clelland C, McMichael AJ, Ho L-P. 2012. Pivotal Advance: invariant NKT cells reduce accumulation of inflammatory monocytes in the lungs and decrease immune-pathology during severe influenza A virus infection. J Leukoc Biol 91:357–368. 10.1189/jlb.0411184. [DOI] [PubMed] [Google Scholar]
- 298.Cole SL, Dunning J, Kok WL, Benam KH, Benlahrech A, Repapi E, Martinez FO, Drumright L, Powell TJ, Bennett M, Elderfield R, Thomas C, Dong T, McCauley J, Liew FY, Taylor S, Zambon M, Barclay W, Cerundolo V, Openshaw PJ, McMichael AJ, Ho L-P, MOSAIC investigators . 2017. M1-like monocytes are a major immunological determinant of severity in previously healthy adults with life-threatening influenza. JCI Insight 2:e91868. 10.1172/jci.insight.91868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Dessing MC, van der Sluijs KF, Florquin S, van der Poll T. 2007. Monocyte chemoattractant protein 1 contributes to an adequate immune response in influenza pneumonia. Clin Immunol 125:328–336. 10.1016/j.clim.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 300.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol 180:2562–2572. 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 301.Maelfait J, Roose K, Vereecke L, Mc Guire C, Sze M, Schuijs MJ, Willart M, Ibañez LI, Hammad H, Lambrecht BN, Beyaert R, Saelens X, van Loo G. 2016. A20 deficiency in lung epithelial cells protects against influenza A virus infection. PLoS Pathog 12:e1005410. 10.1371/journal.ppat.1005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O’Sullivan MP, Walter MJ, Schwendener RA, Cook DN, Danoff TM, Holtzman MJ. 2005. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nat Med 11:1180–1187. 10.1038/nm1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tekkanat KK, Maassab H, Miller A, Berlin AA, Kunkel SL, Lukacs NW. 2002. RANTES (CCL5) production during primary respiratory syncytial virus infection exacerbates airway disease. Eur J Immunol 32:3276–3284. . [DOI] [PubMed] [Google Scholar]
- 304.Jones SA. 2005. Directing transition from innate to acquired immunity: defining a role for IL-6. J Immunol 175:3463–3468. 10.4049/jimmunol.175.6.3463. [DOI] [PubMed] [Google Scholar]
- 305.Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, Bunn J, Suratt BT, Haynes L, Rincon M. 2012. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol 5:258–266. 10.1038/mi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lauder SN, Jones E, Smart K, Bloom A, Williams AS, Hindley JP, Ondondo B, Taylor PR, Clement M, Fielding C, Godkin AJ, Jones SA, Gallimore AM. 2013. Interleukin-6 limits influenza-induced inflammation and protects against fatal lung pathology. Eur J Immunol 43:2613–2625. 10.1002/eji.201243018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Longhi MP, Wright K, Lauder SN, Nowell MA, Jones GW, Godkin AJ, Jones SA, Gallimore AM. 2008. Interleukin-6 is crucial for recall of influenza-specific memory CD4 T cells. PLoS Pathog 4:e1000006. 10.1371/journal.ppat.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Teijaro JR. 2015. The role of cytokine responses during influenza virus pathogenesis and potential therapeutic options. Curr Top Microbiol Immunol 386:3–22. 10.1007/82_2014_411. [DOI] [PubMed] [Google Scholar]
- 309.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206:79–87. 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Pang IK, Ichinohe T, Iwasaki A. 2013. IL-1R signaling in dendritic cells replaces pattern-recognition receptors in promoting CD8+ T cell responses to influenza A virus. Nat Immunol 14:246–253. 10.1038/ni.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Tate MD, Ong JDH, Dowling JK, McAuley JL, Robertson AB, Latz E, Drummond GR, Cooper MA, Hertzog PJ, Mansell A. 2016. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci Rep 6:27912–27918. 10.1038/srep27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Liu B, Mori I, Hossain MJ, Dong L, Takeda K, Kimura Y. 2004. Interleukin-18 improves the early defence system against influenza virus infection by augmenting natural killer cell-mediated cytotoxicity. J Gen Virol 85:423–428. 10.1099/vir.0.19596-0. [DOI] [PubMed] [Google Scholar]
- 313.van der Sluijs KF, Van Elden LJR, Arens R, Nijhuis M, Schuurman R, Florquin S, Kwakkel J, Akira S, Jansen HM, Lutter R, Van Der Polls T. 2005. Enhanced viral clearance in interleukin-18 gene-deficient mice after pulmonary infection with influenza A virus. Immunology 114:112–120. 10.1111/j.1365-2567.2004.02000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Souza LM, Boone TC, Gabrilove J, Lai PH, Zsebo KM, Murdock DC, Chazin VR, Bruszewski J, Lu H, Chen KK, Barendt J, Platzer E, Moore MAS, Mertelsmann R, Welte K. 1986. Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science 232:61–65. 10.1126/science.2420009. [DOI] [PubMed] [Google Scholar]
- 315.Lieschke GJ, Burgess AW. 1992. Granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor. N Engl J Med 327:28–35. 10.1056/NEJM199207023270106. [DOI] [PubMed] [Google Scholar]
- 316.Huang F-F, Barnes PF, Feng Y, Donis R, Chroneos ZC, Idell S, Allen T, Perez DR, Whitsett JA, Dunussi-Joannopoulos K, Shams H. 2011. GM-CSF in the lung protects against lethal influenza infection. Am J Respir Crit Care Med 184:259–268. 10.1164/rccm.201012-2036OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Shibata Y, Berclaz PY, Chroneos ZC, Yoshida M, Whitsett JA, Trapnell BC. 2001. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity 15:557–567. 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 318.Valenti P, Antonini G. 2005. Lactoferrin: an important host defence against microbial and viral attack. Cell Mol Life Sci 62:2576–2587. 10.1007/s00018-005-5372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Chong KT, Thangavel RR, Tang X. 2008. Enhanced expression of murine beta-defensins (MBD-1, -2,- 3, and -4) in upper and lower airway mucosa of influenza virus infected mice. Virology 380:136–143. 10.1016/j.virol.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 320.Schutte BC, McCray PB. 2002. β-Defensins in lung host defense. Annu Rev Physiol 64:709–748. 10.1146/annurev.physiol.64.081501.134340. [DOI] [PubMed] [Google Scholar]
- 321.Tripathi S, Tecle T, Verma A, Crouch E, White M, Hartshorn KL. 2013. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J Gen Virol 94:40–49. 10.1099/vir.0.045013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Reinholz M, Ruzicka T, Schauber J. 2012. Cathelicidin LL-37: an antimicrobial peptide with a role in inflammatory skin disease. Ann Dermatol 24:126–135. 10.5021/ad.2012.24.2.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Barlow PG, Svoboda P, Mackellar A, Nash AA, York IA, Pohl J, Davidson DJ, Donis RO. 2011. Antiviral activity and increased host defense against influenza infection elicited by the human cathelicidin LL-37. PLoS One 6:e25333. 10.1371/journal.pone.0025333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Xu W, Zheng S, Dweik RA, Erzurum SC. 2006. Role of epithelial nitric oxide in airway viral infection. Free Radic Biol Med 41:19–28. 10.1016/j.freeradbiomed.2006.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Whitsett JA, Wert SE, Weaver TE. 2010. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu Rev Med 61:105–119. 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Hartshorn KL. 2010. Role of surfactant protein A and D (SP-A and SP-D) in human antiviral host defense. Front Biosci 2:527–546. 10.2741/s83. [DOI] [PubMed] [Google Scholar]
- 327.Ariki S, Nishitani C, Kuroki Y. 2012. Diverse functions of pulmonary collectins in host defense of the lung. J Biomed Biotechnol 2012:532071–532077. 10.1155/2012/532071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Pechous RD. 2017. With friends like these: the complex role of neutrophils in the progression of severe pneumonia. Front Cell Infect Microbiol 7:160. 10.3389/fcimb.2017.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Chiu C, Openshaw PJ. 2015. Antiviral B cell and T cell immunity in the lungs. Nat Immunol 16:18–26. 10.1038/ni.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lin S, Wu H, Wang C, Xiao Z, Xu F. 2018. Regulatory T cells and acute lung injury: cytokines, uncontrolled inflammation, and therapeutic implications. Front Immunol 9:1545. 10.3389/fimmu.2018.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Benne CA, Kraaijeveld CA, van Strijp JA, Brouwer E, Harmsen M, Verhoef J, van Golde LM, van Iwaarden JF. 1995. Interactions of surfactant protein A with influenza A viruses: binding and neutralization. J Infect Dis 171:335–341. 10.1093/infdis/171.2.335. [DOI] [PubMed] [Google Scholar]
- 332.Mikerov AN, White M, Hartshorn K, Wang G, Floros J. 2008. Inhibition of hemagglutination activity of influenza A viruses by SP-A1 and SP-A2 variants expressed in CHO cells. Med Microbiol Immunol 197:9–12. 10.1007/s00430-007-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.LeVine AM, Whitsett JA, Hartshorn KL, Crouch EC, Korfhagen TR. 2001. Surfactant protein D enhances clearance of influenza A virus from the lung in vivo. J Immunol 167:5868–5873. 10.4049/jimmunol.167.10.5868. [DOI] [PubMed] [Google Scholar]
- 334.To KKW, Zhou J, Song Y-Q, Hung IFN, Ip WCT, Cheng Z-S, Chan ASF, Kao RYT, Wu AKL, Chau S, Luk W-K, Ip MSM, Chan K-H, Yuen K-Y. 2014. Surfactant protein B gene polymorphism is associated with severe influenza. Chest 145:1237–1243. 10.1378/chest.13-1651. [DOI] [PubMed] [Google Scholar]
- 335.Herrera-Ramos E, López-Rodríguez M, Ruíz-Hernández JJ, Horcajada JP, Borderías L, Lerma E, Blanquer J, Pérez-González MC, García-Laorden MI, Florido Y, Mas-Bosch V, Montero M, Ferrer JM, Sorlí L, Vilaplana C, Rajas O, Briones M, Aspa J, López-Granados E, Solé-Violán J, de Castro FR, Rodríguez-Gallego C. 2014. Surfactant protein A genetic variants associate with severe respiratory insufficiency in pandemic influenza A virus infection. Crit Care 18:R127–R132. 10.1186/cc13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Mangalmurti N, Hunter CA. 2020. Cytokine storms: understanding COVID-19. Immunity 53:19–25. 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ, HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395:1033–1034. 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, Silva J, Mao T, Oh JE, Tokuyama M, Lu P, Venkataraman A, Park A, Liu F, Meir A, Sun J, Wang EY, Casanovas-Massana A, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Yale Shaw A, Fournier JB, Odio CD, Farhadian S, Cruz Dela C, Grubaugh ND, Schulz WL, Ring AM, Ko AI, Omer SB, Iwasaki A, IMPACT Research Team . 2020. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature Publishing Group 395:507–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman O-E, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Jaffar Kazmi SA, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas J-L, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A. 2020. Neuroinvasion of SARS-CoV-2 in human and mouse brain. bioRxiv 16:1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Varatharaj A, Thomas N, Ellul MA, Davies NWS, Pollak TA, Tenorio EL, Sultan M, Easton A, Breen G, Zandi M, Coles JP, Manji H, Al-Shahi Salman R, Menon DK, Nicholson TR, Benjamin LA, Carson A, Smith C, Turner MR, Solomon T, Kneen R, Pett SL, Galea I, Thomas RH, Michael BD, CoroNerve Study Group . 2020. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry 7:875–882. 10.1016/S2215-0366(20)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Mullol J, Alobid I, Mariño-Sánchez F, Izquierdo-Domínguez A, Marin C, Klimek L, Wang D-Y, Liu Z. 2020. The loss of smell and taste in the COVID-19 outbreak: a tale of many countries. Curr Allergy Asthma Rep 20:61–65. 10.1007/s11882-020-00961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Xydakis MS, Dehgani-Mobaraki P, Holbrook EH, Geisthoff UW, Bauer C, Hautefort C, Herman P, Manley GT, Lyon DM, Hopkins C. 2020. Smell and taste dysfunction in patients with COVID-19. Lancet Infect Dis 20:1015–1016. 10.1016/S1473-3099(20)30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Menni C, Valdes AM, Freidin MB, Sudre CH, Nguyen LH, Drew DA, Ganesh S, Varsavsky T, Cardoso MJ, El-Sayed Moustafa JS, Visconti A, Hysi P, Bowyer RCE, Mangino M, Falchi M, Wolf J, Ourselin S, Chan AT, Steves CJ, Spector TD. 2020. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat Med 26:1037–1040. 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Millichap JJ, Wainwright MS. 2009. Neurological complications of respiratory syncytial virus infection: case series and review of literature. J Child Neurol 24:1499–1503. 10.1177/0883073808331362. [DOI] [PubMed] [Google Scholar]
- 345.Antonucci R, Chiappe S, Porcella A, Rosatelli D, Fanos V. 2010. Bronchiolitis-associated encephalopathy in critically-ill infants: an underestimated complication? J Matern Fetal Neonatal Med 23:431–436. 10.1080/14767050903184181. [DOI] [PubMed] [Google Scholar]
- 346.Morichi S, Kawashima H, Ioi H, Yamanaka G, Kashiwagi Y, Hoshika A, Nakayama T, Watanabe Y. 2011. Classification of acute encephalopathy in respiratory syncytial virus infection. J Infect Chemother 17:776–781. 10.1007/s10156-011-0259-5. [DOI] [PubMed] [Google Scholar]
- 347.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Cascio Lo A, Clementi N, De Santis M, Mancini N, Granucci F, Zanoni I. 2020. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 369:706–712. 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045.e9. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, Breillat P, Carlier N, Gauzit R, Morbieu C, Pène F, Marin N, Roche N, Szwebel T-A, Merkling SH, Treluyer J-M, Veyer D, Mouthon L, Blanc C, Tharaux P-L, Rozenberg F, Fischer A, Duffy D, Rieux-Laucat F, Kernéis S, Terrier B. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724. 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, Ellingson MK, Mao T, Oh JE, Israelow B, Takahashi T, Tokuyama M, Lu P, Venkataraman A, Park A, Mohanty S, Wang H, Wyllie AL, Vogels CBF, Earnest R, Lapidus S, Ott IM, Moore AJ, Muenker MC, Fournier JB, Campbell M, Odio CD, Casanovas-Massana A, Herbst R, Shaw AC, Medzhitov R, Schulz WL, Grubaugh ND, Cruz Dela C, Farhadian S, Ko AI, Omer SB, Iwasaki A, Yale IMPACT Team . 2020. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584:463–469. 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.He X, Qian Y, Li Z, Fan EK, Li Y, Wu L, Billiar TR, Wilson MA, Shi X, Fan J. 2016. TLR4-upregulated IL-1β and IL-1RI promote alveolar macrophage pyroptosis and lung inflammation through an autocrine mechanism. Sci Rep 6:31663–31611. 10.1038/srep31663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Pillai PS, Molony RD, Martinod K, Dong H, Pang IK, Tal MC, Solis AG, Bielecki P, Mohanty S, Trentalange M, Homer RJ, Flavell RA, Wagner DD, Montgomery RR, Shaw AC, Staeheli P, Iwasaki A. 2016. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science 352:463–466. 10.1126/science.aaf3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Thomas PG, Dash P, Aldridge JR, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC, Kanneganti T-D. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566–575. 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP-Y. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Klok FA, Kruip MJHA, van der Meer NJM, Arbous MS, Gommers D, Kant KM, Kaptein FHJ, van Paassen J, Stals MAM, Huisman MV, Endeman H. 2020. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res 191:145–147. 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tang N, Li D, Wang X, Sun Z. 2020. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 18:844–847. 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Rowley AH. 2020. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat Rev Immunol 20:453–454. 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Consiglio CR, Cotugno N, Sardh F, Pou C, Amodio D, Rodriguez L, Tan Z, Zicari S, Ruggiero A, Pascucci GR, Santilli V, Campbell T, Bryceson Y, Eriksson D, Wang J, Marchesi A, Lakshmikanth T, Campana A, Villani A, Rossi P, Landegren N, Palma P, Brodin P, CACTUS Study Team . 2020. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell 183:968–981. 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Klugman KP, Chien Y-W, Madhi SA. 2009. Pneumococcal pneumonia and influenza: a deadly combination. Vaccine 27(Suppl 3):C9–C14. 10.1016/j.vaccine.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 360.Chen X, Liao B, Cheng L, Peng X, Xu X, Li Y, Hu T, Li J, Zhou X, Ren B. 2020. The microbial coinfection in COVID-19. Appl Microbiol Biotechnol 104:7777–7785. 10.1007/s00253-020-10814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Redford PS, Mayer-Barber KD, McNab FW, Stavropoulos E, Wack A, Sher A, O’Garra A. 2014. Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J Infect Dis 209:270–274. 10.1093/infdis/jit424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Pasquier Du RA, Romero P, Tschopp J. 2011. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity 34:213–223. 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 363.Li W, Moltedo B, Moran TM. 2012. Type I interferon induction during influenza virus infection increases susceptibility to secondary Streptococcus pneumoniae infection by negative regulation of γδ T cells. J Virol 86:12304–12312. 10.1128/JVI.01269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Major J, Crotta S, Llorian M, McCabe TM, Gad HH, Priestnall SL, Hartmann R, Wack A. 2020. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369:712–717. 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Blazek K, Eames HL, Weiss M, Byrne AJ, Perocheau D, Pease JE, Doyle S, McCann F, Williams RO, Udalova IA. 2015. IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J Exp Med 212:845–853. 10.1084/jem.20140995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Broggi A, Tan Y, Granucci F, Zanoni I. 2017. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat Immunol 18:1084–1093. 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Espinosa V, Dutta O, McElrath C, Du P, Chang Y-J, Cicciarelli B, Pitler A, Whitehead I, Obar JJ, Durbin JE, Kotenko SV, Rivera A. 2017. Type III interferon is a critical regulator of innate antifungal immunity. Sci Immunol 2:eaan5357. 10.1126/sciimmunol.aan5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Grzeœk E, Kołtan S, Dębski R, Wysocki M, Gruszka M, Kubicka M, Kołtan A, Grzeœk G, Manysiak S, Odrowąż-Sypniewska G. 2010. Concentrations of IL-15, IL-18, IFN-γ and activity of CD4+, CD8+, and NK cells at admission in children with viral bronchiolitis. Exp Ther Med 1:873–877. 10.3892/etm.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang F-S. 2020. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8:420–422. 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Mizgerd JP. 2012. Respiratory infection and the impact of pulmonary immunity on lung health and disease. Am J Respir Crit Care Med 186:824–829. 10.1164/rccm.201206-1063PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Tourdot S, Mathie S, Hussell T, Edwards L, Wang H, Openshaw PJM, Schwarze J, Lloyd CM. 2008. Respiratory syncytial virus infection provokes airway remodelling in allergen-exposed mice in absence of prior allergen sensitization. Clin Exp Allergy 38:1016–1024. 10.1111/j.1365-2222.2008.02974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Chang Y-J, Kim HY, Albacker LA, Baumgarth N, McKenzie ANJ, Smith DE, Dekruyff RH, Umetsu DT. 2011. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol 12:631–638. 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Jackson DJ, Makrinioti H, Rana BMJ, Shamji BWH, Trujillo-Torralbo M-B, Footitt J, Jerico D-R, Telcian AG, Nikonova A, Zhu J, Aniscenko J, Gogsadze L, Bakhsoliani E, Traub S, Dhariwal J, Porter J, Hunt D, Hunt T, Hunt T, Stanciu LA, Khaitov M, Bartlett NW, Edwards MR, Kon OM, Mallia P, Papadopoulos NG, Akdis CA, Westwick J, Edwards MJ, Cousins DJ, Walton RP, Johnston SL. 2014. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med 190:1373–1382. 10.1164/rccm.201406-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Stier MT, Bloodworth MH, Toki S, Newcomb DC, Goleniewska K, Boyd KL, Quitalig M, Hotard AL, Moore ML, Hartert TV, Zhou B, McKenzie AN, Peebles RS. 2016. Respiratory syncytial virus infection activates IL-13-producing group 2 innate lymphoid cells through thymic stromal lymphopoietin. J Allergy Clin Immunol 138:814–824. 10.1016/j.jaci.2016.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Martinez-Gonzalez I, Mathä L, Steer CA, Ghaedi M, Poon GFT, Takei F. 2016. Allergen-experienced group 2 innate lymphoid cells acquire memory-like properties and enhance allergic lung inflammation. Immunity 45:198–208. 10.1016/j.immuni.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 376.Sallard E, Lescure F-X, Yazdanpanah Y, Mentre F, Peiffer-Smadja N. 2020. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res 178:104791. 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Prokunina-Olsson L, Alphonse N, Dickenson RE, Durbin JE, Glenn JS, Hartmann R, Kotenko SV, Lazear HM, O’Brien TR, Odendall C, Onabajo OO, Piontkivska H, Santer DM, Reich NC, Wack A, Zanoni I. 2020. COVID-19 and emerging viral infections: the case for interferon lambda. J Exp Med 217:e20200653. 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Dinnon KH, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. 2020. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature Publishing Group 586:560–566. 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Karakike E, Giamarellos-Bourboulis EJ. 2019. Macrophage activation-like syndrome: a distinct entity leading to early death in sepsis. Front Immunol 10:55. 10.3389/fimmu.2019.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Biggioggero M, Crotti C, Becciolini A, Favalli EG. 2019. Tocilizumab in the treatment of rheumatoid arthritis: an evidence-based review and patient selection. Drug Des Devel Ther 13:57–70. 10.2147/DDDT.S150580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Halstead ES, Umstead TM, Davies ML, Kawasawa YI, Silveyra P, Howyrlak J, Yang L, Guo W, Hu S, Hewage EK, Chroneos ZC. 2018. GM-CSF overexpression after influenza a virus infection prevents mortality and moderates M1-like airway monocyte/macrophage polarization. Respir Res 19:3–14. 10.1186/s12931-017-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Unkel B, Hoegner K, Clausen BE, Lewe-Schlosser P, Bodner J, Gattenloehner S, Janßen H, Seeger W, Lohmeyer J, Herold S. 2012. Alveolar epithelial cells orchestrate DC function in murine viral pneumonia. J Clin Invest 122:3652–3664. 10.1172/JCI62139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Hall MW. 2019. Immune modulation in pediatric sepsis. J Pediatr Intensive Care 8:42–50. 10.1055/s-0038-1676607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Herold S, Hoegner K, Vadász I, Gessler T, Wilhelm J, Mayer K, Morty RE, Walmrath H-D, Seeger W, Lohmeyer J. 2014. Inhaled granulocyte/macrophage colony-stimulating factor as treatment of pneumonia-associated acute respiratory distress syndrome. Am J Respir Crit Care Med 189:609–611. 10.1164/rccm.201311-2041LE. [DOI] [PubMed] [Google Scholar]
- 385.Hermine O, Mariette X, Tharaux P-L, Resche-Rigon M, Porcher R, Ravaud P, CORIMUNO-19 Collaborative Group . 2021. Effect of tocilizumab versus usual care in adults hospitalized with COVID-19 and moderate or severe pneumonia: a randomized clinical trial. JAMA Intern Med 181:32–40. 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. 2020. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem 295:6785–6797. 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, Li X, Xia J, Chen N, Xiang J, Yu T, Bai T, Xie X, Zhang L, Li C, Yuan Y, Chen H, Li H, Huang H, Tu S, Gong F, Liu Y, Wei Y, Dong C, Zhou F, Gu X, Xu J, Liu Z, Zhang Y, Li H, Shang L, Wang K, Li K, Zhou X, Dong X, Qu Z, Lu S, Hu X, Ruan S, Luo S, Wu J, Peng L, Cheng F, Pan L, Zou J, Jia C, et al. 2020. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N Engl J Med 382:1787–1799. 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Deftereos SG, Giannopoulos G, Vrachatis DA, Siasos GD, Giotaki SG, Gargalianos P, Metallidis S, Sianos G, Baltagiannis S, Panagopoulos P, Dolianitis K, Randou E, Syrigos K, Kotanidou A, Koulouris NG, Milionis H, Sipsas N, Gogos C, Tsoukalas G, Olympios CD, Tsagalou E, Migdalis I, Gerakari S, Angelidis C, Alexopoulos D, Davlouros P, Hahalis G, Kanonidis I, Katritsis D, Kolettis T, Manolis AS, Michalis L, Naka KK, Pyrgakis VN, Toutouzas KP, Triposkiadis F, Tsioufis K, Vavouranakis E, Martinèz-Dolz L, Reimers B, Stefanini GG, Cleman M, Goudevenos J, Tsiodras S, Tousoulis D, Iliodromitis E, Mehran R, Dangas G, Stefanadis C, GRECCO-19 Investigators . 2020. Effect of colchicine versus standard care on cardiac and inflammatory biomarkers and clinical outcomes in patients hospitalized with coronavirus disease 2019: the GRECCO-19 randomized clinical trial. JAMA Netw Open 3:e2013136. 10.1001/jamanetworkopen.2020.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Xu L, Gao H, Zeng J, Liu J, Lu C, Guan X, Qian S, Xie Z. 2018. A fatal case associated with respiratory syncytial virus infection in a young child. BMC Infect Dis 18:217. 10.1186/s12879-018-3123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Szretter KJ, Gangappa S, Lu X, Smith C, Shieh WJ, Zaki SR, Sambhara S, Tumpey TM, Katz JM. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J Virol 81:2736–2744. 10.1128/JVI.02336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Perrone LA, Szretter KJ, Katz JM, Mizgerd JP, Tumpey TM. 2010. Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J Infect Dis 202:1161–1170. 10.1086/656365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Kawashima H, Kashiwagi Y, Ioi H, Morichi S, Oana S, Yamanaka G, Takekuma K, Hoshika A, Sawai J, Kato Y. 2012. Production of chemokines in respiratory syncytial virus infection with central nervous system manifestations. J Infect Chemother 18:827–831. 10.1007/s10156-012-0418-3. [DOI] [PubMed] [Google Scholar]
- 394.Chua RL, Lukassen S, Trump S, Hennig BP, Wendisch D, Pott F, Debnath O, Thürmann L, Kurth F, Völker MT, Kazmierski J, Timmermann B, Twardziok S, Schneider S, Machleidt F, Müller-Redetzky H, Maier M, Krannich A, Schmidt S, Balzer F, Liebig J, Loske J, Suttorp N, Eils J, Ishaque N, Liebert UG, von Kalle C, Hocke A, Witzenrath M, Goffinet C, Drosten C, Laudi S, Lehmann I, Conrad C, Sander LE, Eils R. 2020. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 38:970–979. 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]