Abstract
Herpesviruses are prevalent throughout the animal kingdom, and they have coexisted and coevolved along with their host species for millions of years. Herpesviruses carry a large (120-230 kb) double-stranded DNA genome surrounded by a protein capsid, a tegument layer consisting of viral and host proteins, and a lipid bilayer envelope with surface glycoproteins. A key characteristic of these viruses is their ability to enter a latent state following primary infection, allowing them to evade the host’s immune system and persist permanently. Herpesviruses can reactivate from their dormant state, usually during times of stress or when the host’s immune responses are impaired. While herpesviruses can cause complications with severe disease in immune-compromised people, most of the population experiences few ill effects from herpesvirus infections. Indeed, herpes simplex virus 1 (HSV-1) in particular has several features that make it an attractive tool for therapeutic gene delivery. Herpes simplex virus 1 targets and infects specific cell types, such as epithelial cells and neurons. The HSV-1 genome can also accommodate large insertions of up to 14 kb. The HSV-1-based vectors have already achieved success for the oncolytic treatment of melanoma. In addition to serving as a vehicle for therapeutic gene delivery and targeted cell lysis, comparative genomics of herpesviruses HSV-1 and 2 has revealed valuable information about the evolutionary history of both viruses and their hosts. This review focuses on the adaptability of HSV-1 as an instrument for gene delivery and an evolutionary marker. Overall, HSV-1 shows great promise as a tool for treating human disease and studying human migration patterns, disease outbreaks, and evolution.
Keywords: Gene therapy, herpesvirus, HSV, neurotrophic vector, oncolytic vector, virus evolution
Introduction
Herpesviruses infect a broad range of animals.1 While these viruses have strict species specificity, they are known to establish lifelong, persistent infections in their hosts. These infections are typically mild or asymptomatic except in immune-compromised individuals. An important characteristic of herpesviruses is their ability to enter latency, a dormant state in which the virus genome is maintained in the host cell without producing infectious virus. Another key feature is their structure. Herpesviruses contain a large genome of double-stranded DNA (dsDNA) enclosed in an icosahedral capsid, a protein tegument layer, and a lipid bilayer envelope. An array of viral glycoproteins are found embedded in the lipid envelope that are used by herpesviruses to dock onto cell surface receptors to initiate entry.
There are currently 9 herpesviruses that infect humans (Table 1). These viruses are divided into 3 subgroups: alpha (α), beta (β), and gamma (γ). These subfamilies differ by the rate of virus replication, range of host cells infected, and site where latency is established.2 This review will focus on herpes simplex virus 1 (HSV-1), an α-herpesvirus that is prevalent in humans.3 This virus is the most characterized and manipulated among the 9 human herpesviruses.4-6
Table 1.
Human herpesviruses.
| Virus | Subgroup | Clinical presentation |
|---|---|---|
| Herpes simplex virus 1 (HSV-1) | α | Fever blisters, cold sores |
| Herpes simplex virus 2 (HSV-2) | α | Genital lesions |
| Varicella-zoster virus (VZV) | α | Chicken pox, shingles |
| Human cytomegalovirus (HCMV) | β | Blueberry muffin rash, birth defects |
| Human herpesvirus 6A (HHV-6A) | β | Roseola infantum |
| Human herpesvirus 6B (HHV-6B) | β | Roseola infantum |
| Human herpesvirus 7 (HHV-7) | β | Roseola infantum, pityriasis rosea |
| Epstein-Barr virus (EBV) | γ | Mononucleosis |
| Kaposi’s sarcoma-associated virus (KSHV) | γ | Kaposi’s sarcoma |
Primary HSV-1 infection typically occurs at the oral mucosal epithelium. Viral gene transcription utilizes host RNA polymerase II and occurs in a temporal fashion, with immediate-early (IE) and late genes expressed in productive infection.7 Then the virus migrates to trigeminal ganglia, where it can establish a latent reservoir.8 During latency, transcription from the viral genome produces only latency-associated transcripts (LATs) that accumulate without additional expression of viral genes or proteins.9 Latency can be punctuated by periods of lytic reactivation, often triggered by stress, hormone-level fluctuations, disease, or aging.10 During these reactivation episodes, HSV-1 is easily transmitted by shedding in bodily fluids. However, many primary infections have few or mild symptoms, contributing to the widespread prevalence of HSV-1 in the general population.8 Indeed, HSV is a valuable marker of human migration patterns that also provides insights into virus and host evolution.
HSV as an Evolutionary Marker
Herpesviruses also provide an informative tool for examining the evolution of humans and viruses. Herpesviruses actually predate Homo sapiens; they infected hominids before their evolutionary split from chimpanzees.11 Because herpesviruses can remain latent in their host, they have a long history of virus-host codivergence. Each herpesvirus has evolved to coexist within its host, such that human HSV-1 is distinct from chimpanzee herpesvirus (ChHV) and other primate herpesviruses.
Evolution of HSV-1 and HSV-2
Herpesviruses are common in primates, including chimpanzees, baboons, macaques, and spider monkeys. However, only humans are infected with more than 1 HSV. Overall, HSV-1 and HSV-2 share 87% nucleotide sequence identity, but the conservation varies between specific strains. Some genes are highly conserved while others are significantly more diverged, especially in the unique short (US) region of the genome.12-14
Wertheim et al11 hypothesized that the 2 distinct human HSVs could have arisen via different mechanisms, either duplication in the viral lineage or introduction of a second virus through cross-species transmission. A variety of methods for phylogenetic and molecular dating support that HSV-2 is the result of a cross-species transmission, whereas HSV-1 arose by virus-host codivergence.11
The HSV-2 sequence appears more closely related to ChHV than HSV-1.15 According to molecular-clock analyses, HSV-1 and ChHV diverged at least 6 million years ago (Ma). Around 1.6 Ma, ChHV transmitted to an ancestor of modern humans, giving rise to HSV-2 (Figure 1). The exact nature of this transmission remains unknown. However, researchers analyzed a probability-based network of proximity to rainforest habitats and duration of species in the fossil records. They found that a common ancestor of the chimpanzee Pan troglodytes, most likely Paranthropus boisei, may have transmitted ChHV to Homo habilis, an extinct species that preceded modern humans.16 These 2 species coexisted near Lake Turkana in Kenya, where they both used stone tools for hunting and scavenging, and they likely shared water sources.16
Figure 1.
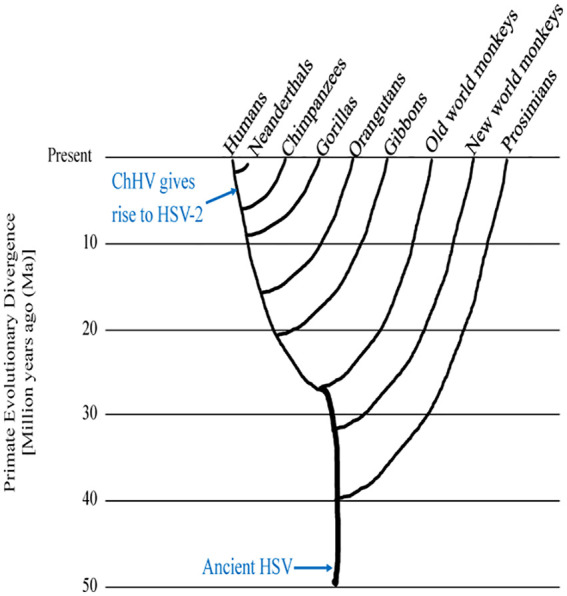
Evolution of herpes and humans. Schematic representation of a phylogenetic tree showing approximate dates known for common ancestors shared between humans and primates. Tree branches represent evolution of distinct species. Evidence suggests HSV infected a common ancestor and then coevolved with each host species, giving rise to HSV-1 in humans. HSV-2 arose due to cross-species transmission that occurred more recently, when early hominids were infected with ChHV. ChHV indicates chimpanzee herpesvirus; HSV, herpes simplex virus.
Humans can be infected with primate simplex viruses, often with dire consequences. For example, in monkeys, macaque simplex virus (MHV or B virus) infection is typically asymptomatic. In humans, however, MHV infection leads to encephalomyelitis, which can cause long-term neurologic impairment or even be fatal.17 Thus, human acquisition of a second herpesvirus was likely due to ChHV establishing less virulent infections in early ancestors of modern humans and adapting to its host, eventually allowing both HSV-1 and HSV-2 to persist in humans.
Recombination between HSV-1 and HSV-2
The potential for 2 simplex viruses to coexist within a single individual has led scientists to study whether recombination occurs between the viruses (Figure 2A). While HSV-1/HSV-2 chimeras have helped identify genes that convey type-specific phenotypes,10,18 only recent studies have revealed that recombination occurs naturally. Evidence for recombination between HSV-1 and HSV-2 came from a study that directly sequenced virus from 150 genital swabs collected from individuals in North America, South America, and Africa.12 In this study, Koelle et al found that 3 HSV-2 genes contained specific segments of the HSV-1 sequence. The UL29 gene encodes a single-stranded DNA-binding protein, the UL30 gene encodes a DNA polymerase catalytic subunit, and the UL39 gene encodes a ribonucleotide reductase subunit.19
Figure 2.
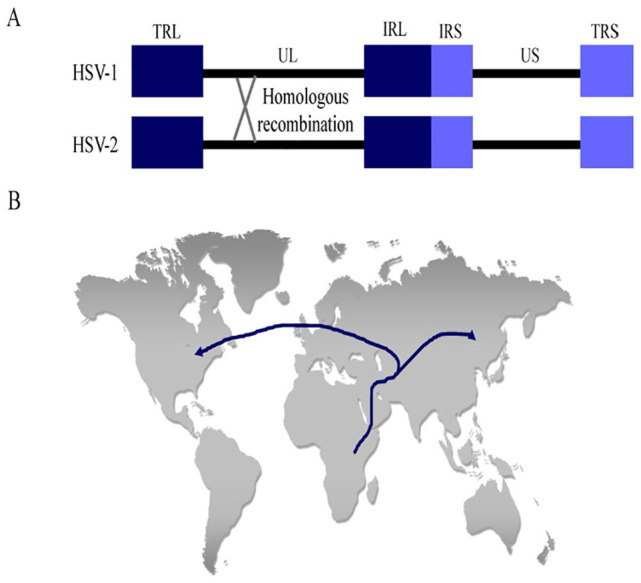
Herpesvirus recombination and human migration patterns. (A) Schematic drawing of the HSV-1 and HSV-2 genomes illustrates how homologous recombination might occur between viral genomes in the same host. (B) Map of human migration patterns out of Africa based on distinct HSV-2 genomes in populations that migrated toward Asia or Europe and North America. HSV indicates herpes simplex virus; IRL, internal repeat long; IRS, internal repeat short; TRL, terminal repeat long; TRS, terminal repeat short; UL, unique long; US, unique short.
With bootscan analysis for recombination sites, it was found that 27% of HSV-2 isolates contained a segment of 457 bp or more of HSV-1 origin, with identical 5′ and 3′ crossover boundaries.12 In addition, most HSV-2 UL30 sequences contained a 539-bp region with sequence identity to HSV-1. This region has more than 70 consecutive single-nucleotide polymorphisms (SNPs) specific to HSV-1,20 whereas surrounding regions of the UL30 gene are distinct and dissimilar to the HSV-1 UL30 sequence.12 Finally, with phylogenetic analysis, Koelle and colleagues identified a 254-bp segment in the UL29 gene. This segment sequence is divergent from the ChHV sequence but is nearly identical to HSV-1. These results suggest that ancestral crossover events incorporated segments of these 3 genes into the HSV-1 gene, which have been conserved in the HSV-2 genome. These findings may have implications for the evolution of virulence or drug resistance. Notably, the current use of Talimogene Laherparepvec (T-VEC) as an oncolytic therapy could enable granulocyte-macrophage colony-stimulating factor (GM-CSF) to be acquired by HSV-2. Likewise, genes deleted from T-VEC to reduce virulence could be regained via recombination with HSV-2. The potential impact of either of these events remains unclear.
Tracking human migration with HSV-1 and HSV-2
Because HSV-1 coevolved with its host for millions of years, the virus represents a useful marker for studying human migration patterns. Phylogenetic analysis of a global sampling of HSV isolates revealed 6 clades and predicted a tree topology that correlated with the geographic origin of the isolate.20 The analyses suggested that HSV originated in Africa and then codiverged in populations that migrated north into Europe and east toward Asia. This theory is consistent with the current “Out of Africa” model of modern human origins (Figure 2B). Interestingly, 4 distinct clades remained prominent within Africa. Kolb et al21 propose that these clades could be associated with the 4 distinct ethnic groups that have been documented in East Africa: the hunter-gatherers, the Cushitic peoples, the Nilotic peoples, and the Bantu groups. While HSV-1 was the primary metric in this study, analysis of HSV-2 sequences also supports the “Out of Africa” model.22 This study analyzed 18 HSV-2 isolates and found 2 main lineages: 1 restricted to sub-Saharan Africa and 1 broadly distributed worldwide.23 Surprisingly, only the worldwide lineage showed evidence of recombination with HSV-1. This finding suggests that HSV-2 first coevolved with its host in Africa before a single lineage containing HSV-1 recombinant fragments was spread via migration of Homo sapiens out of Africa. These results provide compelling evidence that supports current theories of human evolution and migration.
Ancient origins and recent divergence
While this review has focused on primate herpesviruses and human evolution, it is important to note that herpesviruses infect a wide range of species. The viruses that infect birds, reptiles, and mammals are genetically distinct from those that infect fish and amphibians.24 The first herpesvirus identified in invertebrates was reported in the oyster Crassostrea virginica.25 Recently, new variants of oyster herpesviruses have been associated with massive mortality in oyster beds in France.26 This study suggests that recent evolutionary divergence caused increased virulence. Thus, as herpesviruses continue to evolve with their hosts, new strains will likely arise that are more or less well adapted to infecting a broader host range. Meanwhile, in the laboratory, the herpesviruses genome has been modified with the goal of making these viruses effective tools for the treatment of human disease.
HSV as a Therapeutic Tool
Herpesviruses have unique properties that have inspired researchers to manipulate them as therapeutic agents.27 They can target multiple cell types based on their tropism and deliver therapeutics in a controlled fashion through manipulated expression systems.28-32 Herpes simplex virus 1 can cross the blood-brain barrier provided it is disrupted via a drug/agent and infect neurons, making it an attractive vector for treating neurological cancers.33-36 Moreover, HSV-1 can accommodate a significant amount of additional genetic material and does not integrate into the host genome.6,29 These features enable the virus to deliver therapeutic genes while avoiding the potential for insertional mutagenesis, which is inherent to retroviral vectors and also observed with adeno-associated virus.8 Moreover, the HSV genome is maintained as a circular episome.37
Working with virus-manipulated systems comes with risks. However, their potential to treat debilitating diseases may be worth these risks. While more research is needed to better understand the functioning and evolution of these viruses, HSV-1 may contribute to the development of therapies for a number of conditions. The following sections explore the tremendous and versatile potential of HSV-1 as a therapeutic vector for neurological conditions and an oncolytic vector for various malignancies.
HSV as a Neurotrophic Vector for Gene Therapy
Neurological conditions comprise a large part of the global disease burden, and at enormous cost.38 These conditions include disorders and inherited diseases of the brain, spinal cord, and associated nerves.38 Patients with these chronic conditions find only temporary relief with therapeutic interventions, most commonly mechanical devices, surgery, and drugs.39 In recent years, gene therapy has emerged as a promising sustainable treatment for delivery of neurotransmitters, healthy copies of mutant alleles, and other therapeutics.29,40 For many years, adeno-associated viral vectors and lentiviral vectors have been developed as experimental treatments. However, these vector systems are disadvantageous because there are chances of insertional mutagenesis, discontinuous expression of therapeutic genes, and immunogenicity,41 and they can carry only limited genetic material into target cells. The cost of production of these vectors in sufficient quantity is also considerably high. In contrast, HSV-1 overcomes these obstacles and has progressed as a promising neurotrophic vector over the past decade (Figure 3).
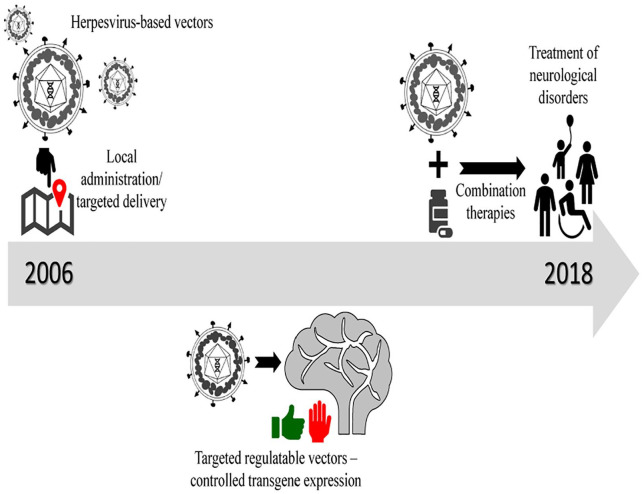
Figure 3.
Timeline of development of HSV as neurotrophic vectors. Vector design started out as localized therapeutic agents that had to be repeatedly administered. Over a decade, these designs progressed to regulatable vectors that could be targeted to specific sites in the brain or specific subsets of cells of the nervous system. These vectors are currently being tested as a part of combination therapies for neurological disorders like glioblastomas in clinical trials. HSV indicates herpes simplex virus.
Early experiments
Research has shown considerable success with using replication defective HSV vectors to treat chronic pain, neuropathies, and neurodegenerative diseases.42,43 One of the earliest studies in the past 15 years used an HSV-1 vector for gene therapy in a model of Parkinson’s disease.44 In this condition, dopamine-producing neurons of the midbrain are progressively depleted, causing motor defects and cognitive impairment. Herpes simplex virus 1 vectors encoding glial-cell-derived factor (GCDF) or erythropoietin (EPO) administered into animal brains were neuroprotective to midbrain dopaminergic neurons.30,44 Success was limited, however, because constant overexpression of EPO led to erythrocytosis or increased production of red blood cells.30 A similar study tested an HSV-1 vector expressing neurturin, a glial cell-line-derived neurotrophic factor (GDNF), that supports survival of sympathetic and sensory neurons.45 In a rat model of induced erectile dysfunction by cavernous nerve injury, HSV-1 expressing neurturin was administered at the site and successfully prevented further damage.46 Thus, HSV vectors can be applied to targeted regions for rejuvenation of damaged nerves.
These studies were encouraging but revealed 2 important limitations. First, the engineered vector needed to be administered locally for effective recovery. Second, transgene expression from the engineered vectors lacked robust on-off regulation, which could lead to adverse effects. However, these early studies opened up the field for seeking experimental solutions to these challenges.
Regulatable HSV vectors
The ability to deliver nerve growth factors to specific neuronal cell populations has great potential for treating neurological disorders. However, prolonged expression of these factors in the central or peripheral nervous system can cause undesirable effects.34 By creating a vector with inducible or repressible expression of the gene of interest, the timing and dosage of the transgene product could be better controlled. Researchers developed such a regulatable vector system using engineered HSV-1 to prevent sensory neuropathy in diabetic mice.47 This vector expressed EPO under control of the human cytomegalovirus (HCMV) IE promoter and contained the tetracycline response element for regulating expression. In primary neurons (of the dorsal root ganglion) infected with the vector, EPO was expressed only when doxycycline was administered. In follow-up studies in Swiss Webster mice, the HSV-1 vector was injected into the plantar surface of both hind feet. Doxycycline (2 mg) was administered by oral gavage 3 days after vector injection. The doxycycline was continued daily for 4 consecutive days, followed by another 3 days of no treatment. This regimen was continued for 4 weeks, after which nerve conduction was assessed. In all 10 mice infected with the HSV-1 vector, the progression of neuropathy was inhibited.47 Numerous studies have shown that on/off regulation can be achieved in various HSV vector systems containing Tet elements or drug-activatable therapeutic genes; however, tight regulation was elusive due to interference of ICP0 (infected-cell polypeptide), an HSV IE gene responsible for activating promoters of HSV-1 genes.48-52
Another proof-of-concept study evaluated a regulatable HSV-1 vector both in vitro and in vivo. Chen et al53 first developed a regulatable HSV-1 vector expressing tetracycline-regulated β-galactosidase (gene encoding β-galactosidase enzyme [LacZ]) that could be tightly controlled by administering tetracycline. When they expressed the vector in cells isolated from the dorsal root ganglion of mice, tetracycline treatment led to robust expression of LacZ in 40% of the cultured cells. This activity tapered off 7 days after the vector was administered. The authors also injected the regulatable vector into the sciatic nerve of CD-1 mice.54 The vector was found upstream in the dorsal root ganglion, and it similarly induced LacZ with tetracycline ingestion.53 This study demonstrated that expression from regulatable HSV-1 vectors was similar in vitro and in vivo, and that this system could potentially be useful for treating sciatic nerve pain.
The ability to control the expression of therapeutics with injected vectors was an important step forward. However, local administration of the vector led to its expression in the entire dorsal root ganglion, which could result in off-target effects. Also, expression of the target gene in different cell types may unintendedly alter signaling pathways. These limitations could be overcome with vectors that targeted specific cell types.
Enhanced HSV vectors
To reduce the possibility of HSV recombinant vectors reactivating and replicating in the host, researchers have stripped away almost all genes responsible for infectivity.27 Removing these genes did not affect cell targeting or transgene delivery, because the HSV vectors were often used along with low amounts of HSV-helper viruses and often propagated in tumor cell lines for efficient production.29,55 Vectors containing the ICP0 locus can be continuously expressed, especially in non-neuronal cells. However, their expression depends on the site of insertion for the gene of interest and the transgene promoter used. For example, Miyagawa et al29 showed that a dystrophin transgene inserted into the ICP0 locus was highly expressed in dystrophin-deficient muscle cells in culture when inserted under the control of a muscle-specific promoter. Other HSV-1 constructs containing transgenes flanked by ICP4 locus sequences were also robustly expressed in non-neuronal cells (osteosarcoma) but not in neuronal cells (primary dorsal root ganglion neurons). As a solution, Verlengia et al32 designed another HSV-1 vector containing a reporter gene construct at the locus for the ICP4 IE gene instead of deleting most of the IE gene cassette. They injected this newer vector into different regions of rat brains and found that the reporter was expressed primarily in neurons and not in other non-neuronal cells like glial cells. They also found that the reporter gene was expressed in the hippocampus for at least 6 months and did not induce neuronal damage or an inflammatory response.32 This study demonstrated that HSV vectors could be designed to deliver large transgenes into neurons, which could be used to treat central nervous system (CNS) diseases. Additional studies published because these outcomes have given nuanced details regarding use of other promoters and gene elements in safe and non-toxic HSV-based vectors.56,57
HSV-1 vectors for controlling latent HSV-1
A major challenge in controlling HSV infections is latency. Upon infection, HSV-1 goes latent in the trigeminal ganglions and resists clearance by the host’s immune system.58 This creates a lifelong infection in which latent HSV-1 can reactivate multiple times throughout life. This reactivation depends on a number of factors, such as stress, aging, and immune compromise due to pregnancy, infection, or disease. Reactivation can have severe consequences, including encephalitis.59
Xu et al60 have published that recombinant HSV vectors can be used to get rid of latent HSV and reduce the risks associated with its reactivation. An HSV-1 vector carrying the gene for interferon gamma (IFN-γ) was able to safely express IFNγ in peripheral neurons both in vitro and in vivo.60 To create this vector, the authors put the IFN-γ gene under control of a strong HCMV promoter, and they deleted the HSV-1 IE and VP16 genes. The VP16 gene encodes a tegument protein of the HSV-1 virion. It is responsible for transcription of IE genes and induces onset of lytic infection that destroys infected cells.61 Replacing VP16 and IE genes inhibited the onset of lytic infection and reduced the cytotoxicity of this vector. In the Xu study, the IFN-γ HSV-1 vector led to robust expression of IFN-γ mRNA and protein, suggesting that the vector could help clear latent HSV-1 by triggering antiviral response in the host. This study supports that HSV-1 vectors may be a promising approach to address reactivated herpesviruses in the CNS and for treating neurotrophic disease.
HSV-1 amplicons for regulating gene expression
Most of the studies evaluating HSV-1 as a vector for gene therapy delivered genes using recombinant HSV-1 with a defective ability to replicate. Vectors based on HSV-1 containing intact viral genes that encoded viral proteins induced cytotoxicity and inflammation.62 To overcome this limitation, researchers designed HSV-1 amplicons. These viral particles completely lacked all protein-encoding genes from the virus, depended on helper viruses for packaging, could accommodate up to 130 kb of transgenes, and were non-toxic to target cells. Thus, such amplicon vectors may be great tools for use in in vitro studies but may prove risky for in vivo experiments due to the chance of reactivation from helper-virus particles.63
Another study evaluated the potential of HSV-1 amplicons to block production of brain-derived neurotrophic factor (BDNF), a protein implicated in epilepsy. Falcicchia et al28 engineered HSV-1-derived amplicons to produce RNA complementary to BDNF. They designed HSV-1 particles with 152 kb of concatemeric repetitions of 2 different expression cassettes. One cassette successfully knocked down the BDNF mRNA in the cytoplasm, and the other cassette successfully repressed its de novo transcription. They tested these amplicons in vitro in various neuronal and non-neuronal cell lines as well as in vivo in the hippocampus of mice with epilepsy.28 They found that there was successful silencing of BDNF in all cases. This study demonstrated that HSV-1 amplicons could be used to treat epilepsy and other CNS disorders. Importantly, these amplicons have the advantage of addressing disease at the transcriptional level, which is beneficial in cases where either the disorder arises from faulty gene expression rather than gene mutations, or the target gene is too large for other vectors. Replication-defective HSV vectors expressing miRNA, siRNA, and transcription factors could also be employed instead of amplicon vectors, provided the gene of interest could be accommodated with robust, regulatable expression.
HSV-1 Vectors for Cancer Therapy
For decades, HSV-1 has been studied as a virus-based therapy for cancer. Initial studies focused on how the virus could be engineered to specifically infect and lyse cancer cells, and new immunotherapy-based techniques have subsequently emerged. There is great potential for HSV-based therapies to treat a range of cancers (Tables 2 and 3).
Table 2.
Recent preclinical studies on HSV-1-based cancer therapy.
| Type of cancer | Cell lines/mice | Modification | Effects | References |
|---|---|---|---|---|
| Medulloblastomas and atypical teratoid/rhabdoid tumors | Orthotopic xenograft models of medulloblastoma and atypical teratoid/rhabdoid tumors | rRp450 HSV-1: lacks ICP6 and expresses rat CYP2B1, enzyme that activates prodrug cyclophosphamide | Prolonged survival, addition of cyclophosphamide enhanced the efficacy of rRp450 | Studebaker et al64 |
| Glioblastoma | High-grade glioma xenograft BALB/c mice | Retargeted to HER2 Expresses murine IL-2 |
Increased overall survival Tumor eradicated in 30% of animals |
Alessandrini et al65 |
| Lung carcinoma | C57BL/6 HER-2-transgenic/tolerant mice | Retargeted to HER2 Armed with IL-12 |
Prevented growth of distant tumors Elicited local and systemic immune response |
Leoni et al66 |
| Liver tumor | C57BL/6 mice Athymic BALB/c mice |
ICP34.5 and ICP47 deleted ICP6 genes replaced by LacZ |
Inhibited tumor growth Enhanced antitumor efficacy via T-cell-mediated immune response |
Nakatake et al67 |
| pHGG, DIPG | Pediatric glioma cell lines, patient-autopsy-derived DIPG cells Mouse orthotopic xenograft model of DIPG |
HSV1716 (ICP34.5 deleted) | Inhibited migration and invasion in cell line Reduced tumor infiltration in a mouse model of orthotropic xenograft DIPG |
Cockle et al68 |
| SCC | Human oral SCC cell line | ICP34.5 deleted | Autophagy-induced cell death | Furukawa et al69 |
Abbreviations: DIPG, diffuse intrinsic pontine glioma; HER2, human epidermal growth factor receptor 2; HSV, herpes simplex virus; pHGG, pediatric high-grade glioma; SCC, squamous cell carcinoma.
Table 3.
Recent clinical trials on HSV-1-based cancer therapy.
| Types of cancer | Therapy | Phase | Study period | Target sample size | Study objectives | Trial No. |
|---|---|---|---|---|---|---|
| Cerebellar brain tumor in children | G207 (ICP34.5 deletion and ICP6 replaced by Escherichia coli LacZ) | I | 2019-2023 | 15 | Evaluate safety | NCT03911388 |
| Glioblastoma | C134 (ICP34.5 deletion and human cytomegalovirus IRS1 expression) | I | 2019-2024 | 24 | Evaluate safety and characterize the activity of C134 | NCT03657576 |
| M032 (ICP34.5 deletion and human IL-12 expression) | I | 2014-2022 | 36 | Evaluate safety and tolerability of the maximum dose | NCT02062827 | |
| Melanoma | T-VEC (now approved by FDA) | III | 2009-2014 | 437 | Compare efficacy and safety of T-VEC combined with granulocyte-macrophage colony-stimulating factor | NCT00769704 |
| T-VEC Ipilimumab |
IB/II | 2013-2019 | 217 | Evaluate safety and compare efficacy of combined therapy versus T-VEC alone | NCT01740297 | |
| Melanoma, Merkel cell carcinoma, or other solid tumors | T-VEC Radiotherapy |
II | 2016-2019 | 34 | Compare efficacy of the combined therapy versus T-VEC alone | NCT02819843 |
| Non-central nervous system solid tumors | HSV1716 | I | 2010-2018 | 18 | Evaluate safety | NCT00931931 |
| Pancreatic cancer | HF10 Gemcitabine Erlotinib |
I | 2013-2020 | 9 | Evaluate safety | UMIN000010150 |
| HF10 Gemcitabine Nab-paclitaxel |
I | 2017-2020 | 76 | Determine recommended dose of HF10 in combination therapy | NCT03252808 | |
| Prostate cancer (castration-resistant) | G47Δ | I | 2013-2016 | 9 | Evaluate safety and efficacy | UMIN000010463 |
Abbreviations: FDA, U.S. Food and Drug Administration; HSV, herpes simplex virus; T-VEC, Talimogene Laherparepvec.
Glioblastoma
Glioblastoma is an aggressive form of brain cancer that is difficult to treat and cannot be completely cured with the currently available treatments.70 Various clinical investigations using oncolytic virus, HSV-1, are underway to develop an effective treatment against glioblastoma. One of the most extensively studied genetically modified HSV-1 for the treatment of glioblastoma is G47Δ, a triple deletion mutant. In this mutant, the ICP34.5 gene is deleted and the ICP6 gene is replaced with Escherichia coli LacZ, which restricts its replication in dividing cells as well as attenuates its virulence.71-75 In addition, deletion of ICP47 enhances viral replication and antigen presentation by major histocompatibility complex (MHC) class I.76-78 Following phase I-IIa clinical trials that demonstrated safety of G47Δ injected into patients with recurrent glioblastoma, a phase II clinical study has been initiated to assess the efficacy and safety of G47Δ in 30 patients with residual or recurrent glioblastoma (UMIN00002661, UMIN000015995). Another type of ICP34.5 deleted HSV-1 investigated for treating glioblastoma is HSV1716, HSV-1 strain 17.75,79 HSV1716 may effectively treat highly invasive cancers, such as pediatric high-grade glioma (pHGG) and diffuse intrinsic pontine glioma (DIPG). The pHGG and DIPG are notoriously difficult to treat and are associated with short survival.80 Cockle et al68 showed that HSV1716 treatment can inhibit invasion of pHGG and DIPG cells by altering cell polarity and stabilizing microtubules. They mixed these tumor cells (taken from human samples) with HSV1716 or phosphate-buffered saline before intracranial injection into a mouse xenograft model of DPIG. There was reduced tumor infiltration into the surrounding cells for the HSV1716 treated DPIG compared with cells treated with saline.68
For the first time, genetically manipulated HSV-1, rQNestin34.5v.2, will be administered to humans in a phase I clinical trial to treat glioma (NCT03152318). Like G47Δ, rQNestin34.5v.2 also lacks ICP6 but contains the ICP34.5 gene under control of a nestin promoter, which is activated only in gliomas. This provides specificity to rQNestin34.5v.2 for replicating only in glioma cells and consequently lysing them.81 This study aims to test the safety and determine the appropriate dose of the treatment in patients with recurrent malignant glioma (NCT03152318). All these studies for the development of an effective treatment against glioblastoma give us hope that one day we will be able to curb this disease, if not cure it.
Melanoma
Melanomas are the most common type of cancer. They comprise malignant tumors of melanocytes that reside in the skin. The lesions are often detected early and treated successfully with radiotherapy, chemotherapy, immunotherapy, and surgery. However, metastatic melanomas are notoriously difficult to treat.82 An oncolytic HSV has been developed that can destroy melanoma cells when injected directly into lesions. In 2016, the U.S. Food and Drug Administration (FDA) approved an HSV-1-based therapy called Talimogene Laherparepvec for treating inoperable metastatic melanoma.83 Talimogene Laherparepvec is an attenuated HSV-1 that specifically infects and lyses tumor cells (Figure 4). Like G47Δ, T-VEC lacks the ICP34.5 gene, required for virulence in non-cancerous cells, and ICP47 which enhances host antitumor response.71-73,75-78 This vector has also been engineered to express GM-CSF, which activates antitumor defense mechanisms by enhancing antigen presentation via dendritic cells.31,71,84
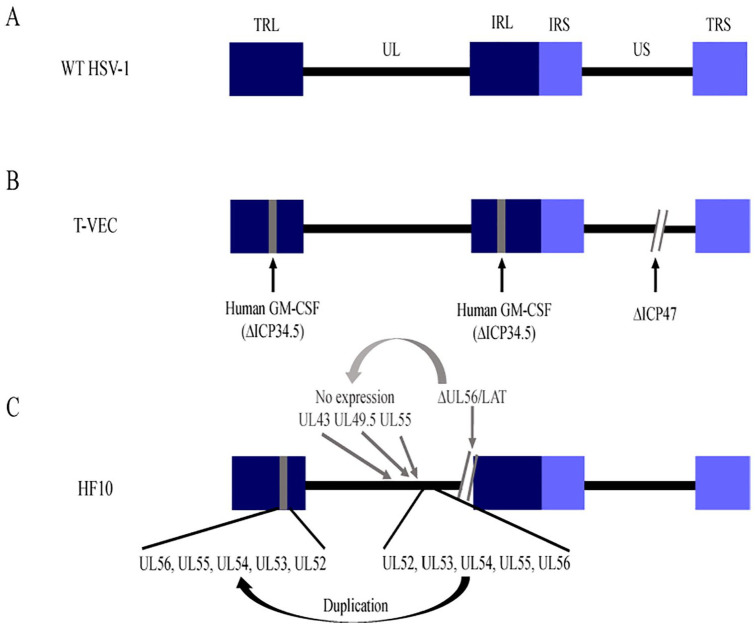
Figure 4.
Schematic diagram of HSV-1 genomes. (A) WT HSV-1 has unique sequences flanked by repeated sequences. (B) T-VEC lacks the gene that encodes ICP34.5 and replaces ICP34.5 with the gene that expresses human GM-CSF. (C) The HF10 genome has partial or completely inverted duplication of UL52-UL56 genes replacing a 2295-bp segment in the TRL region. In addition, UL43, UL49.5, and UL55 genes are not expressed due to frame-shift mutations caused by deletion of UL56 and LATs at the UL56-IRL junction. GM-CSF indicates granulocyte-macrophage colony-stimulating factor; IRL, internal repeat long; IRS, internal repeat short; LATs, latency-associated transcripts; TRL, terminal repeat long; TRS, terminal repeat short; T-VEC, Talimogene Laherparepvec; UL, unique long; US, unique short; WT HSV, wild-type herpes simplex virus.
In clinical trials, T-VEC showed great promise for treating melanomas. A phase III clinical trial was completed on 436 patients with unresected third- or fourth-stage melanomas. In this trial, the efficacy of T-VEC treatment was compared with that of recombinant GM-CSF, a potential adjuvant therapy.85,86 Patients were given either T-VEC by direct injection into lesions or GM-CSF by subcutaneous injection. Compared with GM-CSF treatment, patients treated with T-VEC had significantly reduced tumor size (30%-100%) and increased survival overall. The treatment was well tolerated by the patients; only 2% of patients had severe adverse effects.87 As a result of this study, T-VEC is now approved to treat melanomas that cannot be surgically excised.
Andtbacka et al88 also studied the systemic immune response against tumors after T-VEC treatment, which was attributed to virally expressed GM-CSF. They analyzed 277 unresected melanoma patients with 2116 injected lesions, 177 patients with 981 uninjected non-visceral lesions, and 79 patients with 177 uninjected visceral lesions. Lesion size was reduced by ⩾50% in 64% of injected patients, in 34% of uninjected non-visceral patients, and in 15% of uninjected visceral patients. The observed effect of T-VEC on distal uninjected lesions indicated that the treatment potentially induced a systemic immune response against the tumor.88
Before T-VEC, immune checkpoint inhibitors such as ipilimumab were approved for treating advanced melanoma. Ipilimumab is a monoclonal antibody that suppresses inhibitors of antitumor activity mediated by T cells.89 Talimogene Laherparepvec also enhances this response, albeit through a different mechanism.31 Thus, these 2 cancer therapies may have a synergistic mechanism of action. In a Phase II trial, 198 patients treated with both T-VEC and ipilimumab showed significantly reduced tumor size than patients treated with ipilimumab alone. The combination therapy also showed more efficient treatment of visceral lesions with reduced side effects than treatment with ipilimumab alone.90 While this approach is promising for treating melanomas, further studies are needed that evaluate a larger number of patients and longer follow-up times than 36 months.
As mentioned before, the entire ICP34.5 gene is deleted in the T-VEC version of HSV-1, resulting into tumor-specific viral replication and enhanced antiviral host response. Administration of T-VEC facilitated tumor cell lysis, but these virus-based vectors were replication deficient and multiple administrations were required.91 However, HSV-1 with an N-terminal deletion in ICP34.5 (ΔN146) could efficiently replicate in, and lyse breast carcinoma malignant cells.71 This led to reduced primary breast tumor growth and metastases.71 These findings indicate that ΔN146 could more effectively treat cancers than T-VEC, in which the entire ICP34.5 gene is deleted.
Pancreatic cancer
Pancreatic cancer is responsible for a small percentage of all cancer deaths, but these cancers are difficult to treat because they spread rapidly and are often diagnosed at a late stage.92 A potential treatment option involves oncolytic viruses, which have a different mechanism involving specific cell targeting and lysis, as compared with traditional chemotherapeutics that destroy large sections of tumors and tumor cells, including the surrounding normal cells. The combination of oncolytic viruses and chemotherapy may more efficiently treat pancreatic cancer, especially when chemotherapy cannot be administered at high doses due to its toxicity to normal cells.93
An oncolytic virus that is currently being tested in clinical trials is HF10 that resulted from a spontaneous mutation of an HSV-1 laboratory strain (Figure 4). HF10 lacks expression of UL43, UL49.5, UL55, UL56, and LAT genes. In addition, HF10 has a partial or complete duplication of a few genes, as well as altered amino acid sequences in proteins expressed by several other genes (Figure 4C).94,95 In a phase I clinical trial, 10 patients with inoperable pancreatic cancer were treated with a combination of HF10 and the chemotherapeutics erlotinib (a tyrosine kinase inhibitor targeting epidermal growth factor receptor) and gemcitabine (a nucleoside analogue). This treatment did not cause chronic side effects in patients. It also led to antitumor acquired immunity in the longest surviving patient (3 years as compared with 15.5 months overall survival).96 These results support that HF10 is a potential candidate for virus-based cancer therapy. Further studies are needed to determine the effectiveness of this treatment on pancreatic tumors in a larger population of patients.
Pediatric and young adult cancers
Treatment of cancer in pediatric (0-12 years) and young (13-19 years) patients is challenging because anticancer drugs have long-term toxic effects in these populations than adults.97 Also, there are limitations on drug usage for these groups. An alternative treatment for these patients could be HSV1716, described earlier. To determine the safety of HSV1716, a study was conducted in 9 patients (8-30 years old) with different types of non-CNS solid tumors. Most patients demonstrated efficient virus replication indicated by detection of anti-HSV-1 antibodies in peripheral blood and conversion from seronegative to seropositive status. HSV1716 was determined safe to use in young patients as a cancer therapy because no severe adverse events or dose-limiting toxicities were reported.98 Thus, there is evidence that HSV1716 could be an effective alternative to treating different types of cancer in young patients, including highly invasive cancers.
Limitations of using HSV-1 in cancer therapy
Herpes simplex virus 1 vectors may be a promising therapy for cancer, but this approach does not come without limitations. For example, the genetically modified oncolytic HSV vectors do not replicate efficiently, especially in the tumor microenvironment, as compared with the parental virus they were derived from, which limits their spread among tumor cells. Another limitation is the preexisting immune response which can impede the replication efficiency of oncolytic viruses. One strategy for overcoming this limitation is administering a combination of oncolytic virus and immunotherapy, chemotherapy, or radiotherapy. Several ongoing clinical trials raise hope for more efficient treatment of cancer with HSV-1-based therapies (Table 3). The current opinion in the field is that HSV-based vectors are oncolytic as well as immunotherapeutic because oncolytic viruses have been shown to boost cytotoxic T-cell-mediated immune response by lysis of tumor cells or by neo-antigens generated from virus- or mutation-specific proteins.99-101 However, the studies presented demonstrate that these have great potential for becoming approved, oncolytic therapies.
Herpes simplex virus 1 has been thoroughly characterized and manipulated to produce promising vectors for gene therapy. We have not discussed HSV-based vectors for vaccines as this topic is quite comprehensive and there are some excellent research studies published recently about the advances in vaccine development.102-104
Summary
Herpesviruses are species-specific pathogens that have coevolved with their host species. Over millennia, they have developed tropism for specific cell types and unique characteristics, such as latency, reactivation, and, for HSV, the ability to cross a disrupted blood-brain barrier. Herpes simplex virus vectors have several advantages over the current tools available for treating neurological conditions and cancer. They can evade the host’s immune system, selectively target cells for infection, and can carry larger transgenes than other viral-based vectors. With their huge potential for manipulation, herpesvirus-based vectors are now being tested as potential treatments for various disorders, largely as part of novel combination therapies. These vectors are not only a valuable tool for therapeutics but also for analyzing migration patterns of humans and the epidemiology of herpesviruses. Thus, herpesviruses are highly adaptable for treating human disease and studying virus and human evolution.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health grant AI111232 (to J.V.S.) and Texas Woman’s University (TWU) Biology Department Faculty Development Funds (to L.K.H. and J.V.S.).
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: Concept and design of the review was by JVS and LKH. Writing and editing of the manuscript was done by PHM, SP, and JVS. Critical review of the manuscript was done by JVS. All authors approved the manuscript for publication.
ORCID iDs: Prapti H Mody  https://orcid.org/0000-0002-5530-7785
https://orcid.org/0000-0002-5530-7785
Juliet V Spencer  https://orcid.org/0000-0003-0778-6408
https://orcid.org/0000-0003-0778-6408
References
- 1. Whitley R. Herpesviruses. In: Baron S, ed. Medical Microbiology. 4th ed. Galveston, TX: The University of Texas Medical Branch at Galveston; 1996. [PubMed] [Google Scholar]
- 2. Davison AJ. Overview of Classification. Cambridge, UK: Cambridge University Press; 2007. [PubMed] [Google Scholar]
- 3. McQuillan G, Kruszon-Moran DEW, Flagg E, Paulose-Ram R. Prevalence of Herpes Simplex Virus Type 1 and Type 2 in Persons Aged 14-49: United States, 2015-2016 (NCHS Data Brief No. 304). Hyattsville, MD: National Center for Health Statistics, Centre for Disease Control; 2018. [PubMed] [Google Scholar]
- 4. Agelidis AM, Shukla D. Cell entry mechanisms of HSV: what we have learned in recent years. Future Virol. 2015;10:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun E, Zimmerman T, Hur TB, et al. Neurotropism of herpes simplex virus type 1 in brain organ cultures. J Gen Virol. 2006;87:2827-2837. [DOI] [PubMed] [Google Scholar]
- 6. Rozenberg F, Deback C, Agut H. Herpes simplex encephalitis: from virus to therapy. Infect Disord Drug Targets. 2011;11:235-250. [DOI] [PubMed] [Google Scholar]
- 7. Harkness JM, Kader M, DeLuca NA. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J Virol. 2014;88:6847-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pires de, Mello CP, Bloom DC, Paixao IC. Herpes simplex virus type-1: replication, latency, reactivation and its antiviral targets. Antivir Ther. 2016;21:277-286. [DOI] [PubMed] [Google Scholar]
- 9. Nicoll MP, Proenca JT, Efstathiou S. The molecular basis of herpes simplex virus latency. FEMS Microbiol Rev. 2012;36:684-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertke AS, Patel A, Krause PR. Herpes simplex virus latency-associated transcript sequence downstream of the promoter influences type-specific reactivation and viral neurotropism. J Virol. 2007;81:6605-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wertheim JO, Smith MD, Smith DM, Scheffler K, Kosakovsky Pond SL. Evolutionary origins of human herpes simplex viruses 1 and 2. Mol Biol Evol. 2014;31:2356-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koelle DM, Norberg P, Fitzgibbon MP, et al. Worldwide circulation of HSV-2 × HSV-1 recombinant strains. Sci Rep. 2017;7:44084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baines JD, Pellett PE. Genetic comparison of human alphaherpesvirus genomes. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., eds. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge, UK: Cambridge University Press; 2007:61-69. [PubMed] [Google Scholar]
- 14. Kieff E, Hoyer B, Bachenheimer S, Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972;9:738-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Severini A, Tyler SD, Peters GA, Black D, Eberle R. Genome sequence of a chimpanzee herpesvirus and its relation to other primate alphaherpesviruses. Arch Virol. 2013;158:1825-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Underdown SJ, Kumar K, Houldcroft C. Network analysis of the hominin origin of Herpes Simplex virus 2 from fossil data. Virus Evol. 2017;3:vex026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eberle R, Jones-Engel L. Questioning the extreme neurovirulence of monkey B virus (Macacine alphaherpesvirus 1). Adv Virol. 2018;2018:5248420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morse LS, Buchman TG, Roizman B, Schaffer PA. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977;24:231-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman RM, Lamers SL, Weiner B, et al. Genome sequencing and analysis of geographically diverse clinical isolates of herpes simplex virus 2. J Virol. 2015;89:8219-8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pfaff F, Groth M, Sauerbrei A, Zell R. Genotyping of herpes simplex virus type 1 by whole-genome sequencing. J Gen Virol. 2016;97:2732-2741. [DOI] [PubMed] [Google Scholar]
- 21. Kolb AW, Ané C, Brandt CR. Using HSV-1 genome phylogenetics to track past human migrations. PLoS ONE. 2013;8:e76267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tishkoff SA, Reed FA, Friedlaender FR, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burrel S, Boutolleau D, Ryu D, et al. Ancient recombination events between human herpes simplex viruses. Mol Biol Evol. 2017;34:1713-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanson L, Dishon A, Kotler M. Herpesviruses that infect fish. Viruses. 2011;3:2160-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farley CA, Banfield WG, Kasnic G, Jr, Foster WS. Oyster herpes-type virus. Science. 1972;178:759-760. [DOI] [PubMed] [Google Scholar]
- 26. Burioli EAV, Varello K, Lavazza A, Bozzetta E, Prearo M, Houssin M. A novel divergent group of Ostreid herpesvirus 1 muVar variants associated with a mortality event in Pacific oyster spat in Normandy (France) in 2016. J Fish Dis. 2018;41:1759-1769. [DOI] [PubMed] [Google Scholar]
- 27. Marconi P, Fraefel C, Epstein AL. Herpes simplex virus type 1 (HSV-1)-derived recombinant vectors for gene transfer and gene therapy. Methods Mol Biol. 2015;1254:269-293. [DOI] [PubMed] [Google Scholar]
- 28. Falcicchia C, Trempat P, Binaschi A, et al. Silencing status epilepticus-induced BDNF expression with herpes simplex virus type-1 based amplicon vectors. PLoS ONE. 2016;11:e0150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyagawa Y, Marino P, Verlengia G, et al. Herpes simplex viral-vector design for efficient transduction of nonneuronal cells without cytotoxicity. Proc Natl Acad Sci U S A. 2015;112:E1632-E1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Puskovic V, Wolfe D, Wechuck J, et al. HSV-mediated delivery of erythropoietin restores dopaminergic function in MPTP-treated mice. Mol Ther. 2006;14:710-715. [DOI] [PubMed] [Google Scholar]
- 31. Toda M, Martuza RL, Rabkin SD. Tumor growth inhibition by intratumoral inoculation of defective herpes simplex virus vectors expressing granulocyte-macrophage colony-stimulating factor. Mol Ther. 2000;2:324-329. [DOI] [PubMed] [Google Scholar]
- 32. Verlengia G, Miyagawa Y, Ingusci S, Cohen JB, Simonato M, Glorioso JC. Engineered HSV vector achieves safe long-term transgene expression in the central nervous system. Sci Rep. 2017;7:1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Z, Sun H, Yakisich JS. Overcoming the blood-brain barrier for chemotherapy: limitations, challenges and rising problems. Anticancer Agents Med Chem. 2014;14:1085-1093. [DOI] [PubMed] [Google Scholar]
- 34. Marconi P, Zucchini S, Berto E, et al. Effects of defective herpes simplex vectors expressing neurotrophic factors on the proliferation and differentiation of nervous cells in vivo. Gene Ther. 2005;12:559-569. [DOI] [PubMed] [Google Scholar]
- 35. Nilaver G, Muldoon LL, Kroll RA, et al. Delivery of herpesvirus and adenovirus to nude rat intracerebral tumors after osmotic blood-brain barrier disruption. Proc Natl Acad Sci U S A. 1995;92:9829-9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muldoon LL, Nilaver G, Kroll RA, et al. Comparison of intracerebral inoculation and osmotic blood-brain barrier disruption for delivery of adenovirus, herpesvirus, and iron oxide particles to normal rat brain. Am J Pathol. 1995;147:1840-1851. [PMC free article] [PubMed] [Google Scholar]
- 37. Serquiña AK, Ziegelbauer JM. How herpesviruses pass on their genomes. J Cell Biol. 2017;216:2611-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Feigin VL, Abajobir AA, Abate KH, et al. Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017;16:877-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ling G, Sanchez J, Pankratz K, Clifford D. Editorial of special issue: bio-electronics and prosthetics for neurological diseases. Exp Neurol. 2017;287:435-436. [DOI] [PubMed] [Google Scholar]
- 40. Su W, Kang J, Sopher B, et al. Recombinant adeno-associated viral (rAAV) vectors mediate efficient gene transduction in cultured neonatal and adult microglia. J Neurochem. 2016;136:49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoyng SA, Gnavi S, de Winter F, et al. Developing a potentially immunologically inert tetracycline-regulatable viral vector for gene therapy in the peripheral nerve. Gene Ther. 2014;21:549-557. [DOI] [PubMed] [Google Scholar]
- 42. Mata M, Hao S, Fink DJ. Applications of gene therapy to the treatment of chronic pain. Curr Gene Ther. 2008;8:42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolfe D, Mata M, Fink DJ. Targeted drug delivery to the peripheral nervous system using gene therapy. Neurosci Lett. 2012;527:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Puskovic V, Wolfe D, Goss J, et al. Prolonged biologically active transgene expression driven by HSV LAP2 in brain in vivo. Mol Ther. 2004;10:67-75. [DOI] [PubMed] [Google Scholar]
- 45. Kotzbauer PT, Lampe PA, Heuckeroth RO, et al. Neurturin, a relative of glial-cell-line-derived neurotrophic factor. Nature. 1996;384:467-470. [DOI] [PubMed] [Google Scholar]
- 46. Kato R, Wolfe D, Coyle CH, et al. Herpes simplex virus vector-mediated delivery of neurturin rescues erectile dysfunction of cavernous nerve injury. Gene Ther. 2009;16:26-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Z, Mata M, Fink DJ. Prevention of diabetic neuropathy by regulatable expression of HSV-mediated erythropoietin. Mol Ther. 2011;19:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tannous BA, Christensen AP, Pike L, et al. Mutant sodium channel for tumor therapy. Mol Ther. 2009;17:810-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knopf CW, Zavidij O, Rezuchova I, Rajcani J. Evaluation of the T-REx transcription switch for conditional expression and regulation of HSV-1 vectors. Virus Genes. 2008;36:55-66. [DOI] [PubMed] [Google Scholar]
- 50. Khalique H, Lopez Marco J, Lim F. A haploid HSV-1 genome platform for vector development: testing of the tetracycline-responsive switch shows interference by infected cell protein 0. J Gene Med. 2016;18:302-311. [DOI] [PubMed] [Google Scholar]
- 51. Herrlinger U, Pechan PA, Jacobs AH, et al. HSV-1 infected cell proteins influence tetracycline-regulated transgene expression. J Gene Med. 2000;2:379-389. [DOI] [PubMed] [Google Scholar]
- 52. Smith MC, Boutell C, Davido DJ. HSV-1 ICP0: paving the way for viral replication. Future Virol. 2011;6:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen B, Yao F, Guo L. Regulable transgene expression in dorsal root ganglia of a replication-defective herpes simplex virus type 1 vector by means of sciatic nerve injection. Plast Reconstr Surg. 2016;137:331e-338e. [DOI] [PubMed] [Google Scholar]
- 54. Welser J. CD-1 mice versus C57BL/6 mice. https://sciencellonline.com/blog/cd-1-mice-versus-c57bl6-mice/. Updated 2015. Accessed May 17, 2019.
- 55. Stavropoulos TA, Strathdee CA. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miyagawa Y, Verlengia G, Reinhart B, et al. Deletion of the virion host shut-off gene enhances neuronal-selective transgene expression from an HSV vector lacking functional IE genes. Mol Ther Methods Clin Dev. 2017;6:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han F, Miyagawa Y, Verlengia G, et al. Cellular antisilencing elements support transgene expression from herpes simplex virus vectors in the absence of immediate early gene expression. J Virol. 2018;92:e00536-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Velzen M, van Loenen FB, Meesters RJ, et al. Latent acyclovir-resistant herpes simplex virus type 1 in trigeminal ganglia of immunocompetent individuals. J Infect Dis. 2012;205:1539-1543. [DOI] [PubMed] [Google Scholar]
- 59. Grinde B. Herpesviruses: latency and reactivation—viral strategies and host response [published online ahead of print October 25, 2013]. J Oral Microbiol. doi: 10.1128/JVI.00536-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu K, Pan SY, Song JX, Liu XN, An N, Zheng X. Establishment of a novel therapeutic vector targeting the trigeminal ganglion in rats. Drug Des Devel Ther. 2016;10:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78:9689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sauter MM, Brandt CR. Primate neural retina upregulates IL-6 and IL-10 in response to a herpes simplex vector suggesting the presence of a pro-/anti-inflammatory axis. Exp Eye Res. 2016;148:12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Melendez ME, Fraefel C, Epstein AL. Herpes simplex virus type 1 (HSV-1)-derived amplicon vectors. Methods Mol Biol. 2014;1144:81-98. [DOI] [PubMed] [Google Scholar]
- 64. Studebaker AW, Hutzen BJ, Pierson CR, et al. Oncolytic herpes virus rRp450 shows efficacy in orthotopic xenograft group 3/4 medulloblastomas and atypical teratoid/rhabdoid tumors. Mol Ther Oncolytics. 2017;6:22-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alessandrini F, Menotti L, Avitabile E, et al. Eradication of glioblastoma by immuno-virotherapy with a retargeted oncolytic HSV in a preclinical model. Oncogene. 2019;38:4467-4479. [DOI] [PubMed] [Google Scholar]
- 66. Leoni V, Vannini A, Gatta V, et al. A fully-virulent retargeted oncolytic HSV armed with IL-12 elicits local immunity and vaccine therapy towards distant tumors. PLoS Pathog. 2018;14:e1007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakatake R, Kaibori M, Nakamura Y, et al. Third-generation oncolytic herpes simplex virus inhibits the growth of liver tumors in mice. Cancer Sci. 2018;109:600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cockle JV, Bruning-Richardson A, Scott KJ, et al. Oncolytic herpes simplex virus inhibits pediatric brain tumor migration and invasion. Mol Ther Oncolytics. 2017;5:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Furukawa Y, Takasu A, Yura Y. Role of autophagy in oncolytic herpes simplex virus type 1-induced cell death in squamous cell carcinoma cells. Cancer Gene Ther. 2017;24:393-400. [DOI] [PubMed] [Google Scholar]
- 70. Wirsching HG, Galanis E, Weller M. Glioblastoma. Handb Clin Neurol. 2016;134:381-397. [DOI] [PubMed] [Google Scholar]
- 71. Liu BL, Robinson M, Han ZQ, et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003;10:292-303. [DOI] [PubMed] [Google Scholar]
- 72. Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc Natl Acad Sci U S A. 1992;89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. He B, Chou J, Brandimarti R, Mohr I, Gluzman Y, Roizman B. Suppression of the phenotype of gamma(1)34.5-herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the alpha47 gene. J Virol. 1997;71:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Aghi M, Visted T, Depinho RA, Chiocca EA. Oncolytic herpes virus with defective ICP6 specifically replicates in quiescent cells with homozygous genetic mutations in p16. Oncogene. 2008;27:4249-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 76. Goldsmith K, Chen W, Johnson DC, Hendricks RL. Infected cell protein (ICP)47 enhances herpes simplex virus neurovirulence by blocking the CD8+ T cell response. J Exp Med. 1998;187:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fruh K, Ahn K, Djaballah H, et al. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415-418. [DOI] [PubMed] [Google Scholar]
- 78. York IA, Roop C, Andrews DW, Riddell SR, Graham FL, Johnson DC. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525-535. [DOI] [PubMed] [Google Scholar]
- 79. Ackermann M, Chou J, Sarmiento M, Lerner RA, Roizman B. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986;58:843-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. MacDonald TJ, Aguilera D, Kramm CM. Treatment of high-grade glioma in children and adolescents. Neuro Oncol. 2011;13:1049-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. National Institutes of Health, National Cancer Institute. Oncolytic HSV-1 rQNestin34.5v.2. NCI Drug Dictionary. https://www.cancer.gov/publications/dictionaries/cancer-drug/def/oncolytic-hsv-1-rqnestin345v2 [Google Scholar]
- 82. Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo. 2014;28:1005-1011. [PubMed] [Google Scholar]
- 83. Poh A. First oncolytic viral therapy for melanoma. Cancer Discov. 2016;6:6. [DOI] [PubMed] [Google Scholar]
- 84. Hu JC, Coffin RS, Davis CJ, et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin Cancer Res. 2006;12:6737-6747. [DOI] [PubMed] [Google Scholar]
- 85. Lawson DH, Lee SJ, Tarhini AA, Margolin KA, Ernstoff MS, Kirkwood JM. E4697: phase III cooperative group study of yeast-derived granulocyte macrophage colony-stimulating factor (GM-CSF) versus placebo as adjuvant treatment of patients with completely resected stage III-IV melanoma. J Clin Oncol. 2010;28:8504. [Google Scholar]
- 86. Spitler LE, Weber RW, Allen RE, et al. Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF, sargramostim) administered for 3 years as adjuvant therapy of stages II(T4), III, and IV melanoma. J Immunother. 2009;32:632-637. [DOI] [PubMed] [Google Scholar]
- 87. Andtbacka RH, Kaufman HL, Collichio F, et al. Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33:2780-2788. [DOI] [PubMed] [Google Scholar]
- 88. Andtbacka RH, Ross M, Puzanov I, et al. Patterns of clinical response with Talimogene Laherparepvec (T-VEC) in patients with melanoma treated in the OPTiM phase III clinical trial. Ann Surg Oncol. 2016;23:4169-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. FDA approves YERVOY™ (ipilimumab) for the treatment of patients with newly diagnosed or previously-treated unresectable or metastatic melanoma, the deadliest form of skin cancer (BMS newsroom, press release). Bristol-Myers Squibb. March 25, 2011. https://news.bms.com/press-release/rd-news/fda-approves-yervoy-ipilimumab-treatment-patients-newly-diagnosed-or-previousl
- 90. Chesney J, Puzanov I, Collichio F, et al. Randomized, open-label phase II study evaluating the efficacy and safety of Talimogene Laherparepvec in combination with ipilimumab versus ipilimumab alone in patients with advanced, unresectable melanoma. J Clin Oncol. 2018;36:1658-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu X, He B. Selective editing of herpes simplex virus 1 enables interferon induction and viral replication that destroy malignant cells. J Virol. 2019;93:e01761-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Goral V. Pancreatic cancer: pathogenesis and diagnosis. Asian Pac J Cancer Prev. 2015;16:5619-5624. [DOI] [PubMed] [Google Scholar]
- 93. Cheok CF. Protecting normal cells from the cytotoxicity of chemotherapy. Cell Cycle. 2012;11:2227-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Takakuwa H, Goshima F, Nozawa N, et al. Oncolytic viral therapy using a spontaneously generated herpes simplex virus type 1 variant for disseminated peritoneal tumor in immunocompetent mice. Arch Virol. 2003;148:813-825. [DOI] [PubMed] [Google Scholar]
- 95. Ushijima Y, Luo C, Goshima F, Yamauchi Y, Kimura H, Nishiyama Y. Determination and analysis of the DNA sequence of highly attenuated herpes simplex virus type 1 mutant HF10, a potential oncolytic virus. Microbes Infect. 2007;9:142-149. [DOI] [PubMed] [Google Scholar]
- 96. Hirooka Y, Kasuya H, Ishikawa T, et al. A phase I clinical trial of EUS-guided intratumoral injection of the oncolytic virus, HF10 for unresectable locally advanced pancreatic cancer. BMC Cancer. 2018;18:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ahmad SS, Reinius MA, Hatcher HM, Ajithkumar TV. Anticancer chemotherapy in teenagers and young adults: managing long term side effects. BMJ. 2016;354:i4567. [DOI] [PubMed] [Google Scholar]
- 98. Streby KA, Geller JI, Currier MA, et al. Intratumoral injection of HSV1716, an oncolytic herpes virus, is safe and shows evidence of immune response and viral replication in young cancer patients. Clin Cancer Res. 2017;23:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hunger RE, Brand CU, Streit M, et al. Successful induction of immune responses against mutant ras in melanoma patients using intradermal injection of peptides and GM-CSF as adjuvant. Exp Dermatol. 2001;10:161-167. [DOI] [PubMed] [Google Scholar]
- 100. Andersen MH, Fensterle J, Ugurel S, et al. Immunogenicity of constitutively active V599EBRaf. Cancer Res. 2004;64:5456-5460. [DOI] [PubMed] [Google Scholar]
- 101. Werthmoller N, Frey B, Wunderlich R, Fietkau R, Gaipl US. Modulation of radiochemoimmunotherapy-induced B16 melanoma cell death by the pan-caspase inhibitor zVAD-fmk induces anti-tumor immunity in a HMGB1-, nucleotide- and T-cell-dependent manner. Cell Death Dis. 2015;6:e1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Voellmy R, Bloom DC, Vilaboa N, Feller J. Development of recombinant HSV-based vaccine vectors. Methods Mol Biol. 2017;1581:55-78. [DOI] [PubMed] [Google Scholar]
- 103. Royer DJ, Hendrix JF, Larabee CM, et al. Vaccine-induced antibodies target sequestered viral antigens to prevent ocular HSV-1 pathogenesis, preserve vision, and preempt productive neuronal infection. Mucosal Immunol. 2019;12:827-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bernstein DI, Cardin RD, Pullum DA, Bravo FJ, Kousoulas KG, Dixon DA. Duration of protection from live attenuated vs. sub unit HSV-2 vaccines in the guinea pig model of genital herpes: reassessing efficacy using endpoints from clinical trials. PLoS ONE. 2019;14:e0213401. [DOI] [PMC free article] [PubMed] [Google Scholar]