Abstract
Antibodies against the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein prevent SARS-CoV-2 infection. However, the effects of antibodies against other spike protein domains are largely unknown. Here, we screened a series of anti-spike monoclonal antibodies from coronavirus disease 2019 (COVID-19) patients and found that some of antibodies against the N-terminal domain (NTD) induced the open conformation of RBD and thus enhanced the binding capacity of the spike protein to ACE2 and infectivity of SARS-CoV-2. Mutational analysis revealed that all of the infectivity-enhancing antibodies recognized a specific site on the NTD. Structural analysis demonstrated that all infectivity-enhancing antibodies bound to NTD in a similar manner. The antibodies against this infectivity-enhancing site were detected at high levels in severe patients. Moreover, we identified antibodies against the infectivity-enhancing site in uninfected donors, albeit at a lower frequency. These findings demonstrate that not only neutralizing antibodies but also enhancing antibodies are produced during SARS-CoV-2 infection.
Keywords: ADE, antibody-dependent enhancement, angiotensin converting enzyme 2, cryo-electron microscopy, cryo-EM, docking model
Graphical abstract
A subset of antibodies detected in patients with severe COVID-19 target a specific region of the N-terminal domain of the spike protein and enhance binding of the virus to the ACE2 receptor.
Introduction
SARS-CoV-2 is a novel coronavirus that causes coronavirus disease 2019 (COVID-19) (Zhou et al., 2020b). Although SARS-CoV-2 infection can result in severe symptoms and is associated with high mortality in some patients, most infected individuals do not exhibit such severe symptoms; therefore, additional factors are likely to facilitate the progression to severe COVID-19 (Tabata et al., 2020; Zhou et al., 2020a). SARS-CoV-2, an enveloped positive-strand RNA virus, requires fusion with the host cell membrane for infection (V’kovski et al., 2021). The spike protein is the major envelope protein in SARS-CoV-2 and is composed of S1 and S2 subunits. The S1 subunit is further divided into an N-terminal domain (NTD) and a receptor-binding domain (RBD) (Cai et al., 2020; Wrapp et al., 2020). The interaction between the RBD and the host cell receptor, ACE2, is responsible for SARS-CoV-2 infection of host cells (Hoffmann et al., 2020; Shang et al., 2020).
COVID-19 patients produce antibodies against the RBD of the spike protein, blocking SARS-CoV-2 infection (Brouwer et al., 2020; Robbiani et al., 2020; Zost et al., 2020). Therefore, antibody production against the spike protein plays a pivotal role in host defense against SARS-CoV-2 infection (Brouwer et al., 2020; Robbiani et al., 2020; Zost et al., 2020). However, antibodies against viruses are not always protective (Bournazos et al., 2020). For example, antibodies against dengue virus protein can cause severe diseases mediated by the Fc receptor (Wang et al., 2017). In addition, feline infectious peritonitis coronavirus (FIPV) infection is exacerbated by vaccination of spike protein or adaptive transfer of the serum antibodies from FIPV-challenged animals (Vennema et al., 1990; Weiss and Scott, 1981), suggesting the presence of antibodies that augment coronavirus infection, although the exact mechanism of the enhancement of FIPV infection by such antibodies has remained unclear (Olsen, 1993). Therefore, understanding the function of antibodies produced during SARS-CoV-2 infection is quite important to elucidate the etiology of COVID-19. In this study, we examined various types of anti-spike antibodies, with particular emphasis on those that enhance ACE2 binding and SARS-CoV-2 infection.
Results
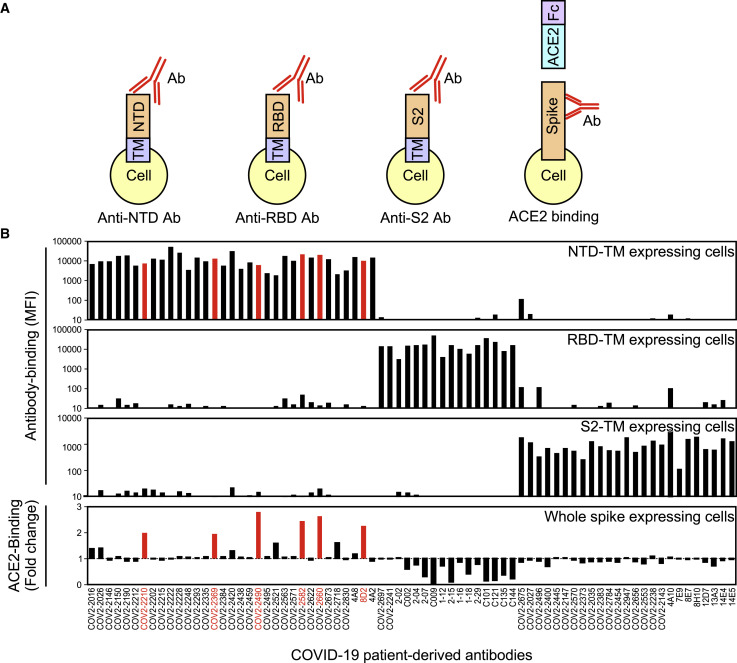
Enhanced ACE2 binding to the spike protein by a subset of anti-NTD antibodies
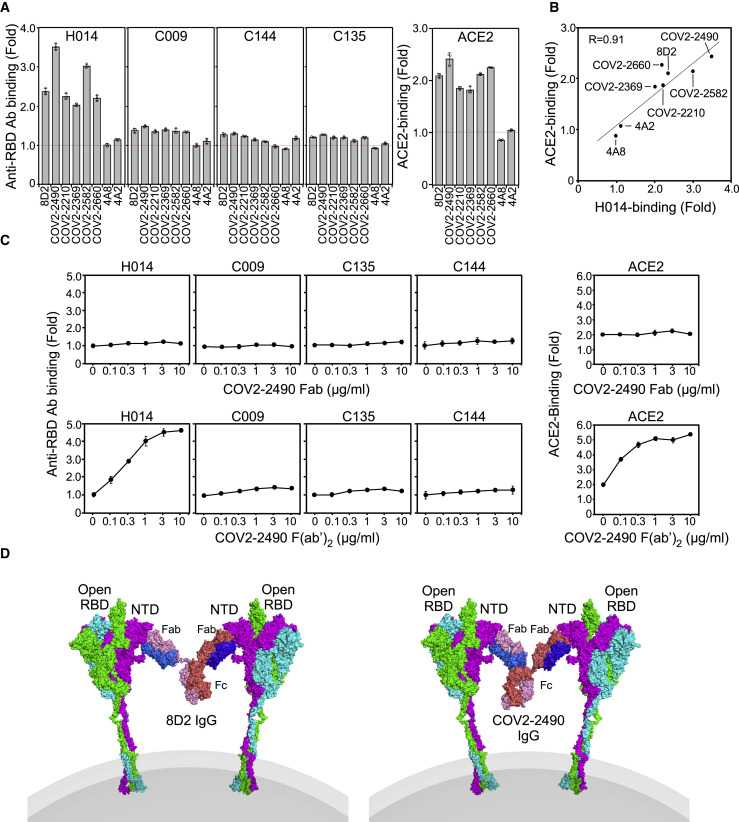
We addressed the function of antibodies against the SARS-CoV-2 spike protein by generating a series of anti-spike monoclonal antibodies from COVID-19 patients (Brouwer et al., 2020; Chi et al., 2020; Robbiani et al., 2020; Zost et al., 2020). We analyzed the effect of antibodies on the binding of recombinant ACE2 to cells expressing the full-length spike protein (Figure 1 A). In order to determine the specificity of antibodies under physiological conditions, we used transfectants expressing FLAG-tagged NTD, RBD, and S2 subunit fused with the transmembrane-cytoplasmic domains of PILRα, a type I membrane protein (Satoh et al., 2008, 2017) (Figure S1 A). Although all of these domains were detected on the cell surface using anti-FLAG antibody, cell-surface expression levels of S2 subunit fused to the PILRα transmembrane region (S2-TM) were lower than those of NTD fused to the PILRα transmembrane region (NTD-TM) or RBD fused to the PILRα transmembrane region (RBD-TM) (Figure S1B). Some anti-spike antibodies that did not to bind to recombinant S1 spike protein in previous reports, such as 8D2 (Chi et al., 2020; Zost et al., 2020), specifically recognized NTD expressed on the cell surface, suggesting that the antigenicity of the NTD expressed on the cell surface is different from that of the recombinant spike protein (Figures 1B and S1B). On the other hand, recombinant ACE2 bound to whole spike and RBD-TM transfectants, but not to NTD-TM or mock transfectants, indicating that recombinant ACE2 specifically binds to the RBD of the spike protein (Figure S2 A). As expected, most of the antibodies against the RBD domain blocked the binding of ACE2 to the spike protein, and antibodies against the S2 domain did not affect ACE2 binding (Figure 1B). However, a specific subset of antibodies targeting the NTD domain (denoted “enhancing antibodies”), including 8D2 (Chi et al., 2020), COV2-2210, COV2-2369, COV2-2490, COV2-2582, and COV2-2660 (Zost et al., 2020), were found to enhance the binding of ACE2 to the spike protein (Figures 1B and S2A). In contrast, other anti-NTD antibodies, such as COV2-2016 (Zost et al., 2020) and 4A8 (Chi et al., 2020), did not affect the binding of ACE2 to the spike protein, although they recognized the full-length spike protein and the NTD domain to a similar degree as the enhancing antibodies (Figures 1B and S1B). The increased ACE2 binding to the spike protein in the presence of enhancing antibodies was not observed in RBD-TM or NTD-TM transfectants (Figure S2A). Furthermore, the increase in ACE2 binding to the spike protein in the presence of enhancing antibodies was dose dependent (Figure 2 A). These data suggested that antibody binding to NTD is involved in enhanced ACE2 binding to RBD. In order to analyze the relationship between antibody binding to NTD and the enhanced ACE2 binding, we compared the half maximal effective concentration (EC50) values of antibody binding and ACE2-binding by each enhancing antibody (Figures S2B and S2C). The EC50 values of antibody binding and ACE2 binding varied, suggesting the affinities of the enhancing antibodies differed. On the other hand, the EC50 of ACE2 binding of each antibody was greater or equal to that of antibody binding.
Figure 1.
Effect of anti-spike antibodies on ACE2 binding
(A) HEK293T cells transfected with vectors expressing NTD-TM, RBD-TM, and S2-TM were stained with anti-spike antibodies. The effect of antibodies on ACE2-binding to spike transfectants was analyzed with ACE2-Fc-fusion protein.
(B) Mean fluorescence intensities (MFIs) of the stained cells are shown (top three columns). The binding of ACE2-Fc-fusion protein to full-length spike transfectants was analyzed in the presence of the indicated antibodies at 1 μg/mL (bottom column). Antibodies that enhanced binding of ACE2-Fc to the spike transfectants by more than 1.9 times are indicated in red.
See also Figures S1 and S2.
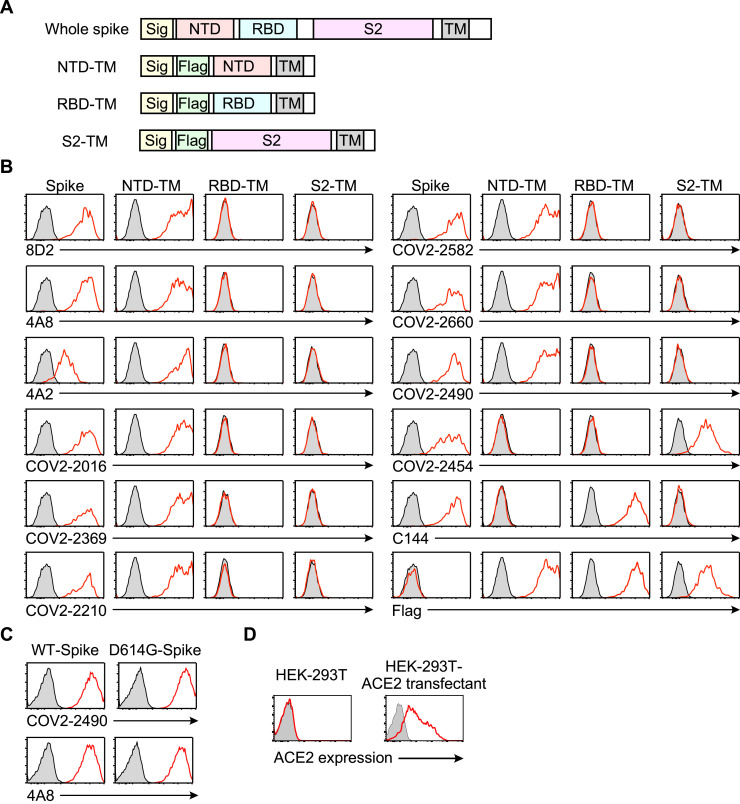
Figure S1.
Specificities of the antibodies used in this study, related to Figures 1 and 3
(A) The expression constructs used to analyze the specificity of the anti-spike antibodies. (B) The plasmids encoding the full-length spike, Flag-NTD-TM, Flag-RBD-TM, and Flag-S2-TM were cotransfected separately with a GFP vector into HEK293T cells. The transfectants were then stained with the indicated antibodies. The antibodies bound to the transfectants were detected with APC-labeled secondary antibodies. The fluoresce intensities of APC on the GFP-expressing cells are shown (red line). Control stainings were shown as shaded histogram. (C) The plasmids expressing the wild-type spike protein and the D614G mutant were cotransfected separately with a GFP vector into HEK293T cells. The transfectants were stained with the anti-NTD infectivity-enhancing antibody COV2-2490 and anti-NTD non-enhancing antibody 4A8. The fluorescence intensities of APC on the GFP-expressing cells are shown (red line). Control stainings were shown as shaded histogram. (D) Parental HEK293T cells and ACE2-transfected HEK293T cells were stained with anti-ACE2 mAb (red line). Control stainings were shown as shaded histogram.
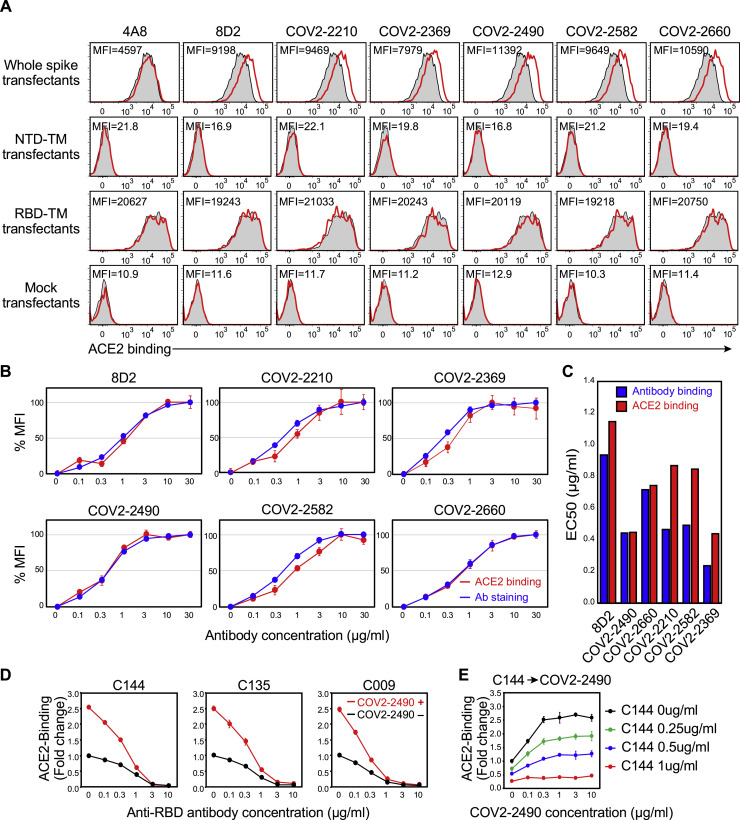
Figure S2.
Enhanced binding of ACE2 to the spike protein by specific anti-NTD antibodies, related to Figures 1 and 2
(A) The plasmids expressing the full-length spike, NTD-TM, RBD-TM, and mock were transfected into HEK293T cells with the GFP vector, and the transfectants were mixed with the indicated anti-NTD antibodies at 10 μg/ml. 4A8 is a non-enhancing antibody and the remaining is enhancing antibodies. Transfectants not mixed with antibodies were used as a control (shaded histogram). Afterward, the cells were stained with biotin-labeled ACE2-Fc fusion protein, followed by APC-labeled streptavidin. The fluorescence intensities of APC on the GFP-expressing cells are shown (red line). Mean fluorescent intensities (MFI) of red lines were shown in the figure.
(B) HEK293T cells transfected with spike protein were mixed with the indicated concentrations of enhancing antibodies. Subsequently, APC-conjugated anti-human IgG Fc antibody (blue) or biotin-ACE2-Fc fusion protein, followed by APC-labeled streptavidin (red), were mixed with the transfectants. The mean fluorescence intensities of cells stained in the absence of the enhancing antibody were set to 0, and the maximum fluorescence intensities in the presence of the enhancing antibody were set to 100. EC50 was calculated from the concentrations of the enhancing antibody that induced half the maximum fluorescence intensities.
(C) EC50s of antibody binding to the spike transfectants and ACE2 binding enhancement by the antibodies were shown.
(D) ACE2-Fc binding to wild-type spike protein in the presence of COV2-2490 antibodies at 3 μg/ml and various concentrations of anti-RBD neutralizing antibodies indicated in the figure (red line). ACE2-Fc binding in the absence of the enhancing antibodies was shown as the control (black line).
(E) Neutralizing C144 Ab was added to the spike transfectants first and then an enhancing antibody COV2-2490 was added. Concentrations of antibodies were indicated in the figure. ACE2-Fc binding to wild-type spike protein was shown. The data are presented as mean ± SD. The representative data from three independent experiments are shown.
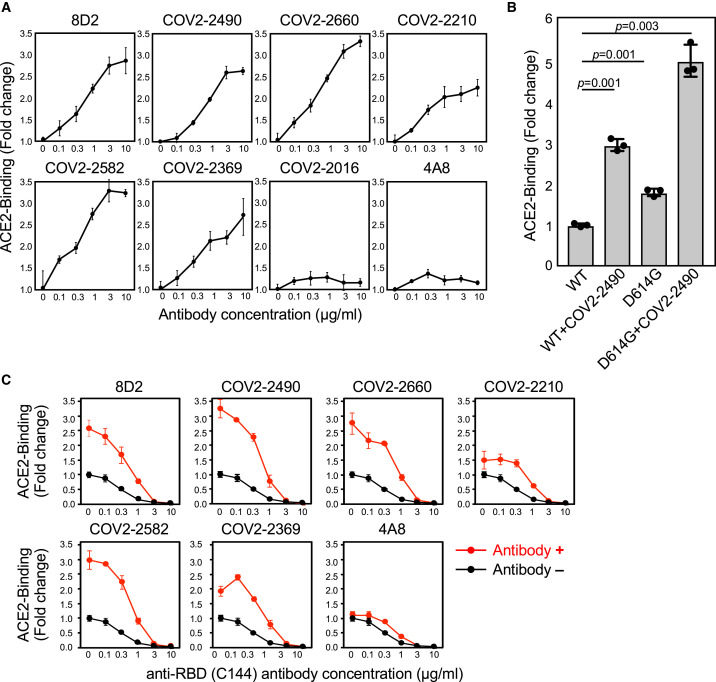
Figure 2.
Enhanced ACE2 binding to spike protein by anti-NTD antibodies
(A) ACE2-Fc binding to the spike transfectants in the various concentrations of antibodies.
(B) ACE2-Fc binding to the wild-type or D614G spike protein in the presence of 3 μg/mL COV2-2490 monoclonal antibody (mAb). The statistical significance derived from an unpaired t test is indicated.
(C) ACE2-Fc binding to wild-type spike protein in the presence of the indicated antibodies at 3 μg/mL and various concentrations of anti-RBD neutralizing antibody C144 (red line). ACE2-Fc binding in the absence of the enhancing antibodies was shown as the control (black line). The data from triplicates are presented as mean ± SD. Representative data from three independent experiments are shown.
See also Figure S2.
Most of recent SARS-CoV-2 viruses harbor the D614G mutation that exhibits higher infectivity than wild-type virus (Grubaugh et al., 2020; Hou et al., 2020; Korber et al., 2020; Li et al., 2020; Plante et al., 2021; Yurkovetskiy et al., 2020). Structural analysis of the mutant has revealed that the RBD is likely to have a more open conformation, with the ACE2 binding interface more exposed than in wild-type spike protein (Li et al., 2020; Plante et al., 2021; Yurkovetskiy et al., 2020). Therefore, we compared the effect of enhancing antibodies with that of the D614G mutation. Compatible with the findings above, the amount of recombinant ACE2 bound to the D614G spike protein was greater than that of the wild-type spike protein (Figure 2B), although the cell surface expression of spike protein was comparable (Figure S1C). Notably, the magnitude of the effect of enhancing antibodies on ACE2 binding to the wild-type spike protein was higher than that of the D614G mutation. Moreover, enhancing antibodies also increased the binding of ACE2 to the D614G spike protein to a similar extent as to wild-type spike protein (Figure 2B).
Since anti-RBD antibodies that block the binding of ACE2 to the spike protein play a central role in antibody-mediated host defense against SARS-CoV-2 infection (Brouwer et al., 2020; Robbiani et al., 2020; Zost et al., 2020), we analyzed how enhancing antibodies affected the ACE2-blocking activity by neutralizing anti-RBD antibodies. We mixed enhancing antibodies with different concentrations of a neutralizing anti-RBD antibody, C144 (Robbiani et al., 2020). We found that the binding of ACE2 to the spike protein increased in the presence of enhancing antibodies, even upon addition of C144 at concentrations up to 1 μg/mL (Figure 2C). Similar results were obtained using other anti-RBD neutralizing antibodies, C009 and C135, that recognize epitopes different from C144 (Robbiani et al., 2020) (Figure S2D). In addition, the enhancing effect was also observed when neutralizing antibodies were added before enhancing antibodies (Figure S2E). These data suggested that the neutralization activity of anti-RBD antibodies is indeed reduced in the presence of enhancing antibodies.
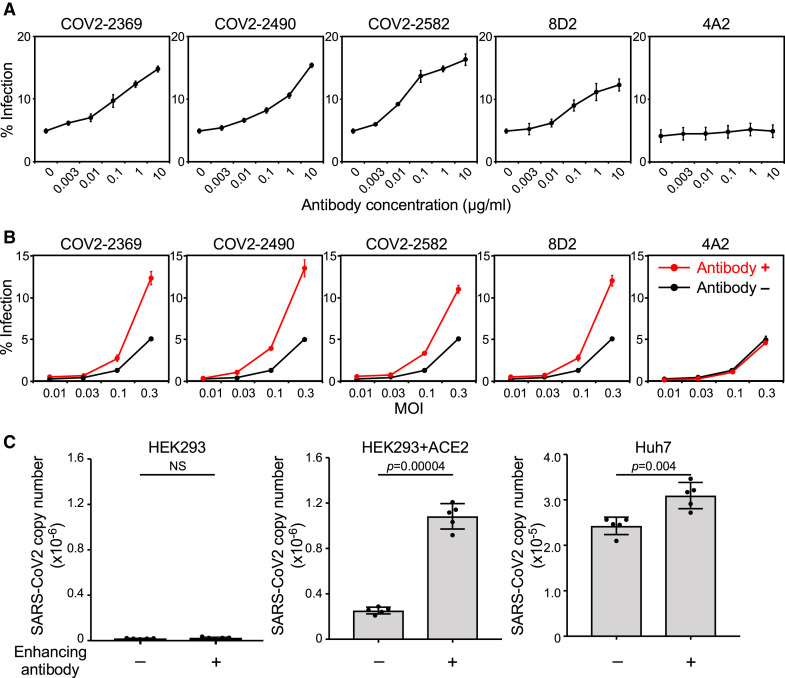
Enhancing antibodies facilitate SARS-CoV-2 infectivity
The effect of enhancing antibodies on ACE2 binding to the spike protein suggested that the infectivity of SARS-CoV-2 would also be increased, similar to the effect of the D614G mutation (Hou et al., 2020; Korber et al., 2020; Li et al., 2020; Plante et al., 2021). To investigate this question, we utilized vesicular stomatitis virus (VSV)/ΔG-GFP SARS-CoV-2 spike pseudovirus (SARS-CoV-2 PV) to quantify the effect of representative enhancing antibodies on SARS-CoV-2 infection. As expected, enhancing antibodies increased the infectivity of SARS-CoV-2 PV to ACE2-transfected HEK293T cells in an enhancing-antibody-dose-dependent manner (Figures 3 A and S1D). In contrast, the anti-NTD antibody (4A2), which did not enhance ACE2 binding to the spike protein (Figure 1B), did not increase infectivity. Moreover, the enhancement of infectivity in the presence of antibodies was observed regardless of the amount of virus (Figure 3B).
Figure 3.
Enhanced SARS-CoV-2 infectivity by specific anti-NTD antibodies.
(A) ACE2-expressing HEK293T cells (MOI: 0.3) were infected by a SARS-CoV-2 spike pseudovirus carrying a GFP reporter gene in the presence of various concentrations of indicated antibodies. The proportion of GFP-positive infected cells is shown.
(B) ACE2-expressing HEK293T cells were infected with a SARS-CoV-2 spike pseudovirus carrying a GFP reporter gene at different MOIs with (red line, 3 μg/mL) or without (black line) the indicated antibodies.
(C) HEK293T cells, ACE2-expressing HEK293T cells, and Huh7 cells were infected with authentic SARS-CoV-2 virus in the presence (+) or absence (−) of enhancing antibody COV2-2490 at 1 μg/mL. The amounts of SARS-CoV-2 virus produced in the cell culture supernatants were analyzed 48 h after infection. The statistical significance derived from an unpaired t test is indicated. NS, not significant. The data are presented as mean ± SD. Representative data from three independent experiments are shown.
See also Figure S1.
Next, we examined the effect of enhancing antibodies using authentic SARS-CoV-2 virus. The replication of the authentic SARS-CoV-2 virus in ACE2-transfected HEK293T cells increased more than four times in the presence of enhancing antibodies (Figure 3C). SARS-CoV-2 infection of Huh7 cells, which express lower levels of ACE2 but are also susceptible to SARS-CoV-2 infection (Chu et al., 2020), was also significantly enhanced by enhancing antibodies (Figure 3C). These data indicate that enhancing antibodies robustly augment infectivity of the SARS-CoV-2 virus to ACE2-expressing cells.
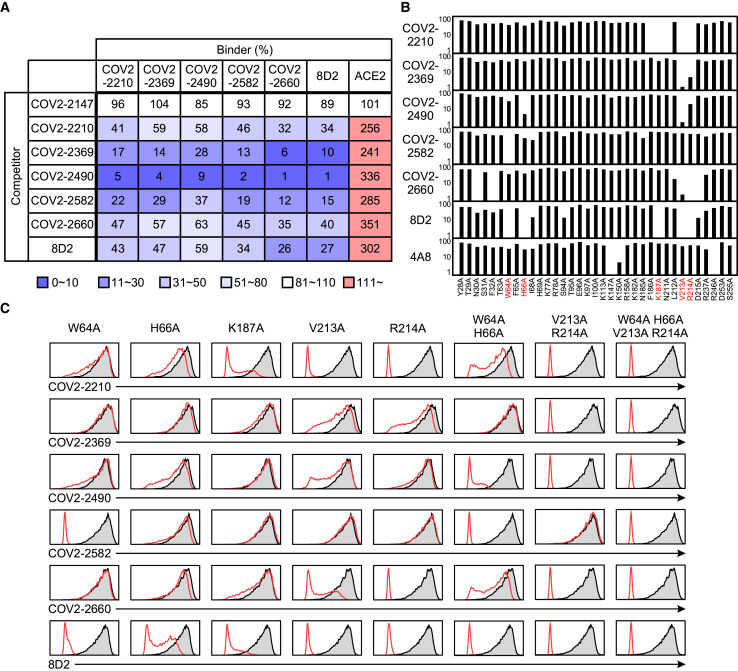
Overlapping epitopes in NTD recognized by enhancing antibodies
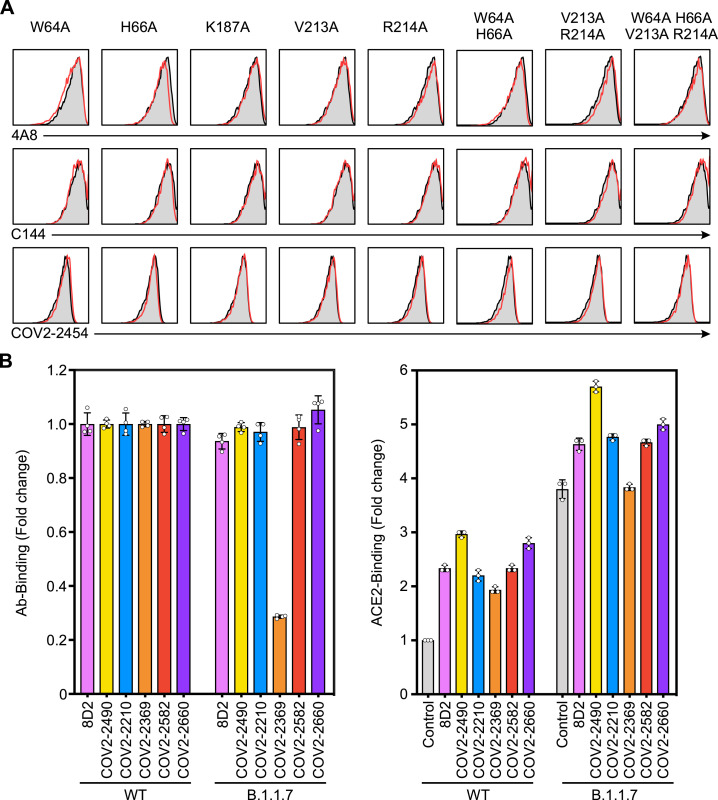
We employed competitive binding assays to identify the epitopes of infectivity-enhancing antibodies. Surprisingly, all enhancing antibodies competed with each other for spike protein binding, indicating that they recognize similar epitopes on the NTD (Figure 4 A). Next, we generated a series of alanine mutants of NTD with the TM domain (Figure S1A) and analyzed their binding to enhancing antibodies. Because the 8D2 antibody contained negatively charged amino acids in the heavy-chain CDR3, we analyzed binding of the 8D2 antibody to lysine- or arginine-mutated NTD. The R214A and K187A mutants were not recognized by the 8D2 antibody (Figure 4B). We then analyzed a series of NTD mutants in which amino acid residues structurally close to R214 or K187 were mutated to alanine and found that binding of enhancing antibodies to the NTD was substantially decreased in W64A, H66A, K187A, V213A, and R214A mutants (Figure 4B). Similar observations were made with the full-length spike protein with mutations in these residues (Figure 4C). Moreover, the W64A-H66A and V213A-R214A double mutants reduced binding of some enhancing antibodies, such as COV2-2490 and COV2-2369, more than single mutants. Furthermore, the quadruple mutant of W64, H66, V213, and R214 was not recognized by any enhancing antibody. Significantly, these mutations did not affect NTD recognition by a non-enhancing anti-NTD antibody (4A8), an anti-RBD antibody (C144), or an anti-S2 antibody (COV2-2454) (Figure S3 A), suggesting that the NTD mutations did not affect the overall structure of the spike protein. These data suggested that the binding of antibodies to a specific site on the NTD is involved in the enhanced binding of ACE2 to spike protein.
Figure 4.
Epitope mapping of SARS-CoV-2 infectivity-enhancing antibodies
(A) Binding of enhancing antibodies (binder) to full-length spike transfectants was analyzed in the presence of the indicated antibodies (competitor). The effect of competitors on ACE2-Fc binding to the spike transfectants was also analyzed. The non-enhancing anti-S2 antibody, COV2-2147, was used as a control. Relative antibody- or ACE2-binding levels observed in the presence of competitor are shown.
(B) Relative antibody-binding levels to a series of NTD mutants compared to wild-type NTD are shown. Non-enhancing anti-NTD antibody 4A8 was used as a control. The most affected residues are shown as red.
(C) Full-length mutant spike proteins were stained with the indicated enhancing antibodies (red line). Staining of wild-type spike is shown as a shaded histogram.
See also Figure S3.
Figure S3.
Binding of anti-spike antibodies against spike mutants with mutated antibody epitopes in the SARS-CoV-2 infectivity site, related to Figure 4
(A) The plasmids expressing the full-length spike proteins with alanine mutations at the indicated amino acid residues were transfected separately with the GFP vector into HEK293T cells, and the binding of 4A8 (anti-NTD non-enhancing antibody), C144 (anti-RBD antibody), and COV2-2454 (anti-S2 antibody) against the GFP-positive cells were analyzed (red line). Control stainings were shown as shaded histogram.
(B) The relative binding of each enhancing antibody to the B.1.1.7 spike transfectants was compared to the wild-type spike transfectants.
(C) The relative bindings of ACE2-Fc to the transfectants of wild-type or B.1.1.7 spike protein were analyzed in the presence or absence of 10 μg/ml of enhancing antibodies. The data are presented as mean ± SD.
The recent B.1.1.7 variant that shows high infectivity possesses a deletion of H69 and V70, very close to the epitopes recognized by most enhancing antibodies. It has been reported that the H69 and V70 deletion enhances the infectivity of SARS-CoV-2 (Kemp et al., 2021). We thus analyzed the effect of enhancing antibodies on B.1.1.7 spike protein. Among six enhancing antibodies, all except COV2-2369 bound to B.1.1.7 spike protein and wild-type spike protein at the same levels (Figure S3B). Although B.1.1.7 spike protein showed a high ACE2-binding capacity, even in the absence of enhancing antibodies, ACE2 binding was further augmented by the five enhancing antibodies that bound to B.1.1.7 (Figure S3C). These data suggest that enhancing antibodies are functional against B.1.1.7 variants, but the effects of enhancing antibodies might vary depending on the SARS-CoV-2 variants.
Structures of infectivity-enhancing antibodies bound to spike protein
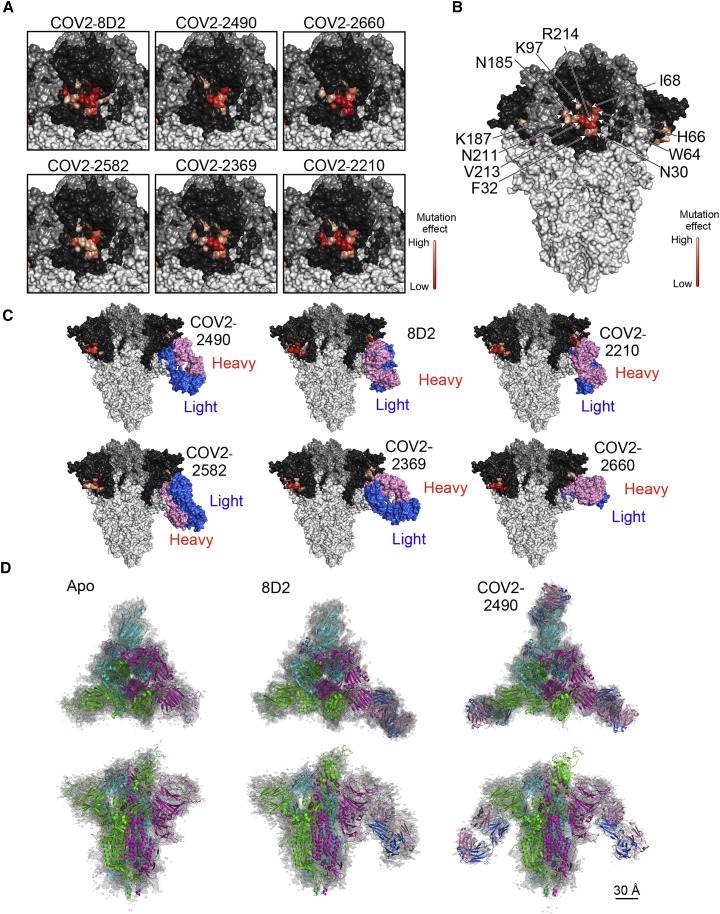
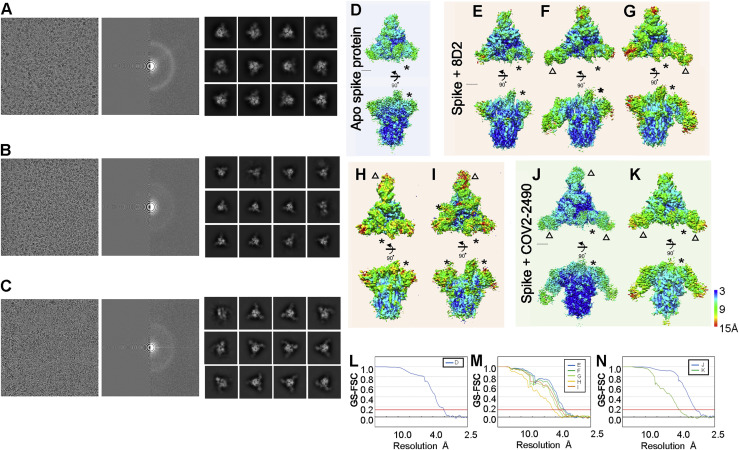
The epitopes for infectivity-enhancing antibodies revealed by alanine mutations spanned a narrow area on the NTD surface (Figures 5A and 5B). Based on the observed epitopes, we generated docking models of the antibody-spike protein complexes and found that each antibody was predicted to bind similarly to the NTD (Figure 5C). To confirm these predictions, single-particle cryoelectron microscopy (cryo-EM) analysis of apo spike protein as well as spike protein complexed with the Fab fragments of enhancing antibodies 8D2 and COV2-2490 was employed. Data were analyzed by heterogenous refinement followed by homogeneous refinement and rigid-body fitting of antibody models and known spike protein trimers (Figures 5D and S4 D). A single apo protein structure with one RBD in the open conformation was observed. For the 8D2 Fab, five structures were obtained (Figures S4E–S4I), one of which was Fab unbound and agreed well with the apo structure (Figure S4E) and four that contained a single 8D2 Fab fragment (Figures 5C and 5D). For the COV2-2490 Fab, two structures were obtained, one containing two and one containing three bound Fab fragments, suggesting higher binding affinity for COV2-2490 compared with 8D2 (Figures S4J and S4K). The binding modes of the COV2-2490 Fab were similar to those of 8D2 (Figure 5D). These data suggested that structures predicted by the docking model agreed well with the cryo-EM densities. Because all of the enhancing antibodies bound to the spike protein in a similar manner, this mode of binding appears to be required for the infectivity-enhancing effect of the antibodies.
Figure 5.
Structures of SARS-CoV-2 infectivity-enhancing antibodies bound to spike protein
(A) Amino acid residues that affected the binding of each enhancing antibody are shown as a heatmap based on their percent reduction of the MFIs in Figure 4B, with higher reduction indicated by darker shades, as shown in bar in the figure. NTD, dark gray; RBD, medium gray; other regions, light gray.
(B) MFI reductions of the affected residues are averaged across the six antibodies and shown as a heatmap, with higher reduction indicated by darker shades, as shown in bar in the figure.
(C) Each SARS-CoV-2 infectivity-enhancing antibody was docked onto the spike protein as described in STAR Methods.
(D) A spike model built from PDB: 7KEB was found to fit the apo density best, while another model built from PDB: 7K8W fit the two other antibody-bound densities best. The spike subunit in the one RBD-“up” form is colored green. Antibodies are colored pink (heavy chain) and blue (light chain). The scale bar represents 30 Å.
Figure S4.
Cryo-EM density map of spike of SARS-CoV-2 with antibody, related to Figure 5
(A-C) A representative micrographs (left), CTF estimation of a micrograph on left panel (middle), and typical 2D class averages. (A) Apo spike protein of SARS-CoV-2. (B) Spike protein with 8D2 antibody. (C) Spike protein with COV2-2490 antibody. The final density map from single particle cryo-EM is colored by local resolution (A-H). (D) Apo spike protein (EMDBID: 30915). (E-I) Spike protein with 8D2 antibody (EMDBID: 30916, 30917, 30918, 30919, and 30920). (J-K) Spike protein with 2490 antibody (EMDBID: 30921 and 30922). Asterisks indicate the rise form of RBD domains. Triangles indicate the binding positions of antibody. (L-N) The GS-FSC curves for each corresponding map are shown. Red line indicates FSC = 0.143 criteria. Scale bars are 30 Å.
Induction of the open conformation of the RBD by enhancing antibodies
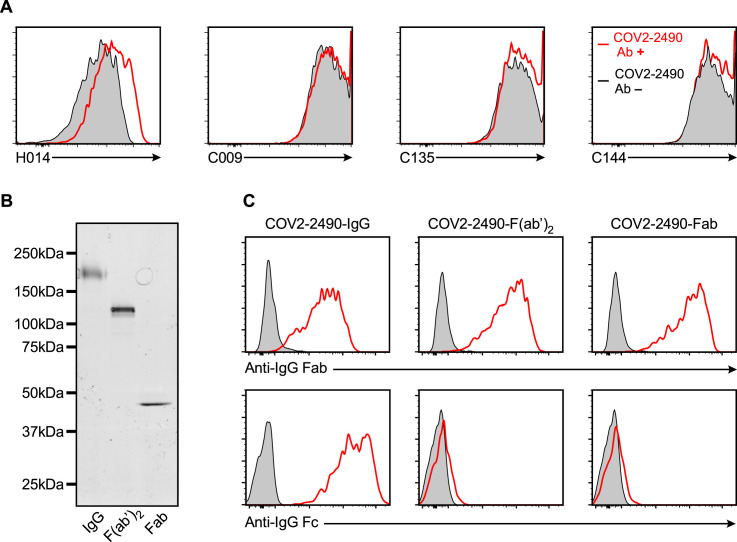
Since other anti-NTD antibodies that recognize other sites on the NTD did not enhance ACE2 binding, it is likely that binding of antibodies to a specific site on the NTD is involved in enhancing ACE2 binding to the spike protein. It has been reported that ACE2 preferentially binds to the open conformation of the RBD (Henderson et al., 2020; Lu et al., 2020). Therefore, it is possible that enhancing antibodies induce the open conformation of the RBD, although the open conformation of the RBD did not increase in the presence of the Fab fragments of enhancing antibodies in cryo-EM analysis (Figure 5). To address this possibility, we analyzed the effect of enhancing antibodies by using a unique antibody, H014, that recognizes a cryptic epitope exposed in the open RBD state (Lv et al., 2020). In addition, we used anti-RBD neutralizing antibodies (C009, C114, and C135) that recognize different epitopes on the RBD but recognize both open and closed RBDs. (Robbiani et al., 2020). The binding of H014 antibody was highly enhanced in the presence of enhancing antibodies (Figures 6 A and S5 A). On the other hand, the binding of H014 antibody to spike protein was not augmented by the non-enhancing anti-NTD antibodies 4A8 and 4A2. Enhancing antibodies did not obviously affect the binding of C009, C114, or C135 anti-RBD antibodies. Furthermore, there was a very strong correlation between the enhancement of ACE2 binding and H014 antibody binding induced by enhancing antibodies (R = 0.91). These data suggest that enhancing antibodies augment the binding of ACE2 by inducing the open RBD state (Figure 6B).
Figure 6.
Induction of the open conformation of the RBD by divalent enhancing antibodies
(A) The binding of antibodies specific to the open RBD (H014) or the open and closed RBD (C009, C144, and C135) against spike transfectants was analyzed in the presence of the indicated enhancing antibodies (3 μg/mL). ACE2 binding to spike protein was also analyzed. The non-enhancing anti-NTD antibodies 4A8 and 4A2 were used as controls. Relative antibody- or ACE2-binding levels observed in the presence of anti-NTD antibodies are shown.
(B) H014 antibody binding and ACE2 binding to spike protein in the presence of indicated antibodies are shown. The correlation coefficient between them is indicated.
(C) H014 antibody binding and ACE2 binding to spike protein in the presence of various concentrations of Fab and F(ab′)2 and fragments of the enhancing COV2-2490 antibody.
(D) A model of spike protein bound with divalent enhancing antibodies (8D2 and COV2-2490). Spike protein and Fab complexes demonstrated in Figure 5D were used as a template for the models.
See also Figure S5.
Figure S5.
Induction of the open conformation of the RBD by divalent enhancing antibodies, related to Figure 6
(A) The binding of antibodies specific to open RBD (H014) or open and closed RBD (C009, C144, C135) against spike transfectants was analyzed in the presence or absence of enhancing (COV2-2490) or non-enhancing (4A8) antibodies (3 ug/ml).
(B) SDS-PAGE analysis of whole IgG antibody, F(ab’)2 and Fab fragments of enhancing COV2-2490 antibody.
(C) Whole spike transfectants were stained with whole IgG antibody, F(ab’)2 or Fab fragment of COV2-2490 antibody. Bound antibodies were detected by APC-labeled anti-human IgG Fc or anti-human IgG Fab specific antibodies.
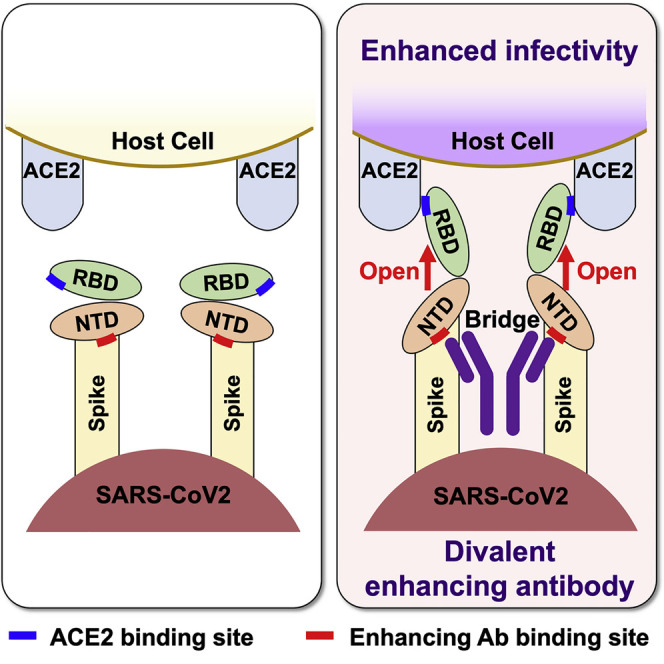
Because antibodies are divalent, we next investigated the role of the valence on the enhancing effect. To address this question, we analyzed the effect of enhancing antibodies on the augmentation of ACE2 binding as well as induction of open RBD using F(ab′)2 or Fab fragments of the enhancing antibody, COV2-2490 (Figure 6C). F(ab′)2 fragments of COV2-2490 antibody induced open RBD and thus enhanced ACE2 binding similar to whole immunoglobulin G (IgG) antibody, whereas the Fab fragments did not. These data suggested that the binding of monomeric enhancing antibodies to NTD alone is not enough to induce an open conformation of the RBD. We next analyzed whether it was possible for both arms to bind to two NTD domains on the same spike. However, we were unable to model such an binding mode using existing full-length IgG antibodies as templates. Subsequently, we investigated the possibility that a single IgG antibody bridges two spike proteins and found that it was possible to generate such a conformation by remodeling the V-C linker regions (Figure 6D). Taken together, it appears that bridging the spike proteins by divalent antibodies is required to induce the open conformation of RBD. These data are compatible with our cryo-EM analysis, in which the induction of the open RBD state was not observed in Fab-bound spike proteins (Figure 5).
Infectivity-enhancing antibodies in COVID-19 patients
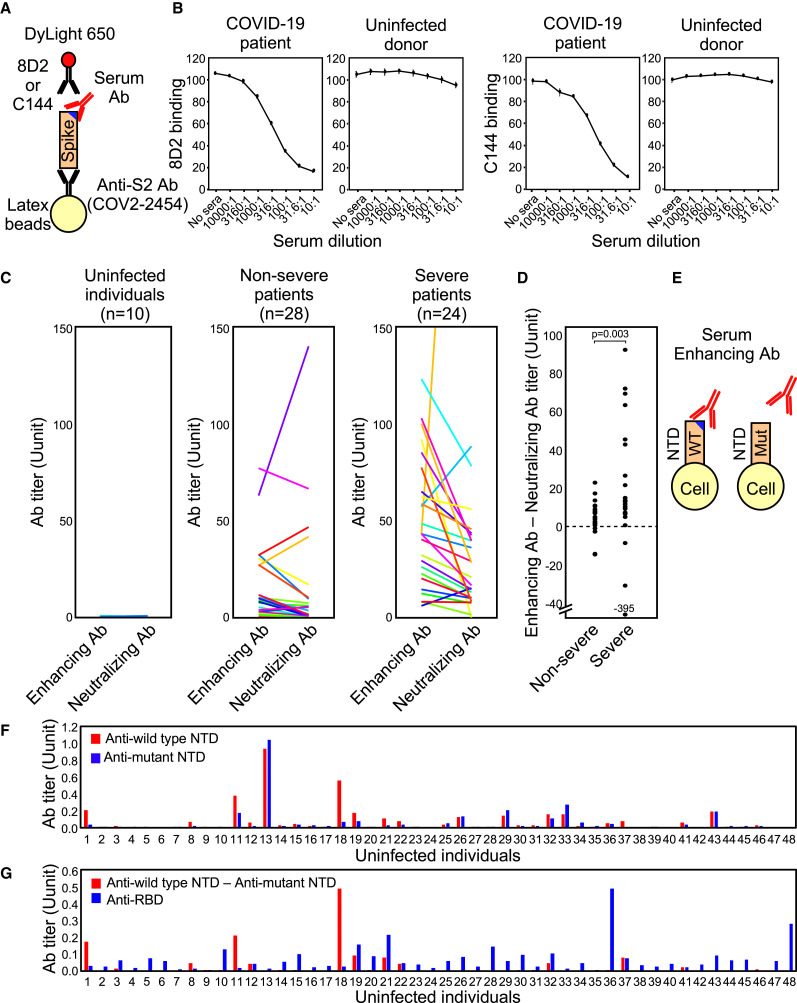
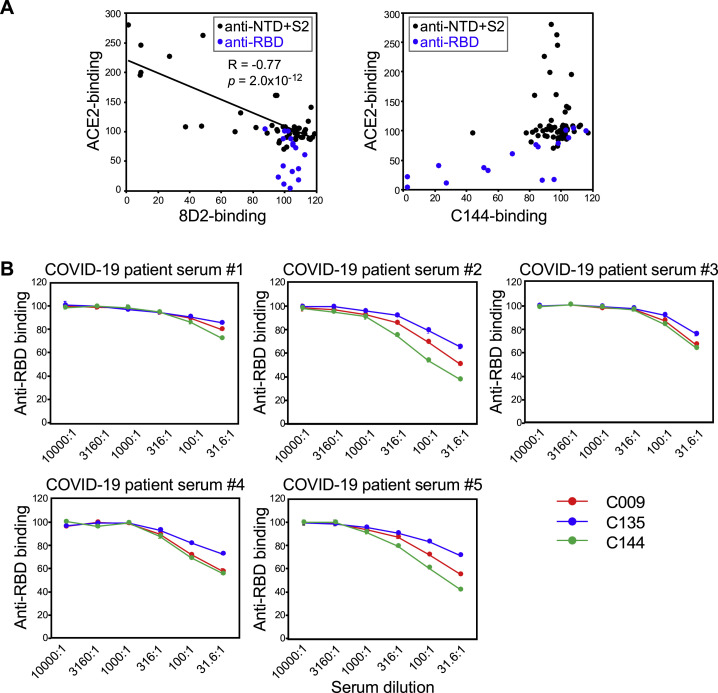
Because enhancing antibodies boost SARS-CoV-2 infectivity, we speculated that their presence might correlate with disease severity. To investigate this possibility, we first compared serum levels of enhancing antibodies in COVID-19 patients and uninfected individuals. We utilized a competitive binding assay using recombinant spike protein and DyLight-fluorescence-labeled 8D2 enhancing antibody to determine serum levels of enhancing antibodies (Figure 7 A). Serum levels of neutralizing antibodies were also determined using fluorescence-labeled C144 anti-RBD neutralizing antibody. Binding of both enhancing antibody 8D2 and neutralizing antibody C144 to spike-protein-coated beads decreased in the presence of serum from a COVID-19 patient, but not from an uninfected donor (Figure 7B). We validated the specificity of the competitive binding assay for enhancing antibodies by analyzing COVID-19-patient-derived anti-NTD and S2 antibodies used in Figure 1A. Although 8D2 binding was partially decreased in the presence of a few antibodies that do not enhance ACE2 binding, enhancing effects on ACE2 binding by antibodies were negatively correlated with 8D2 blocking activity (R = −0.77, p = 2.0 × 10−12) (Figure S6 A). In addition, anti-RBD antibodies did not affect 8D2 binding. Therefore, the levels of enhancing antibodies measured by the 8D2 competition assay appeared to largely reflect serum levels of enhancing antibodies. C144 binding was also not significantly affected by most anti-NTD and S2 antibodies, although some anti-RBD antibodies blocked C144 binding as well as ACE2 binding (Figure S6A). On the other hand, there are several epitopes for anti-RBD neutralizing antibodies, suggesting that the C144 competition assay is not sufficient to detect all neutralizing antibodies. Therefore, we compared C144 antibody with other anti-RBD antibodies, C009 and C135, that recognize different epitopes on the RBD. C144 antibody binding to spike protein was blocked by serum antibodies of COVID-19 patients equally or more than C009 or C135 antibodies, suggesting that the competition assay using C144 antibody is more sensitive than C009 and C135 antibodies to estimate the serum levels of neutralizing antibodies (Figure S6B). These data suggested that the serum levels of enhancing and neutralizing antibodies could be roughly estimated by the competition assay using 8D2 and C144 antibodies, although there remains a possibility that unrelated antibodies affect the competitive binding assay.
Figure 7.
SARS-CoV-2 infectivity-enhancing antibodies in COVID-19 patients and uninfected individuals
(A) A method of detecting the enhancing or neutralizing antibodies using a competitive binding assay. DyLight-650-labeled 8D2 and C144 were used to detect the enhancing and neutralizing antibodies, respectively.
(B) The binding of the enhancing antibody, 8D2, or the neutralizing antibody, C144, to beads coated with the spike protein was analyzed in the presence of the serially diluted serum of a representative COVID-19 patient or an uninfected donor.
(C) The levels of SARS-CoV-2 infectivity-enhancing antibodies and neutralizing antibodies in noninfected individuals and non-severe and severe COVID-19 patients.
(D) Enhancing antibody titers subtracted with neutralizing antibody titers were compared between non-severe and severe COVID-19 patients. Statistical significance derived from Mann-Whitney U test is indicated.
(E) Enhancing antibodies were detected by comparing antibody binding to the wild-type NTD-TM (WT) and the mutant NTD-TM lacking the enhancing antibody epitopes (Mut).
(F) Serum levels of antibodies in uninfected individuals against the wild-type NTD (blue bar) and mutant NTDs whose epitopes for the enhancing antibodies were mutated (red bar).
(G) SARS-CoV-2 infectivity-enhancing antibody titers were calculated by subtracting antibody levels against the mutant NTD from those against the wild-type NTD in uninfected individuals (red bar). Anti-RBD antibody titers were analyzed using RBD-TM transfectants (blue bar).
See also Figure S6.
Figure S6.
Specificity of competitive binding assay to detect enhancing and neutralizing antibodies, related to Figure 7
(A) Specificity of 8D2 and C144 antibody competitive binding assay. Each antibody derived from COVID-19 patients used in Figure 1B was mixed with spike protein transfectants, and DyLight 647-labeled 8D2 or C144 antibody binding as well as ACE2-Fc binding was analyzed. Anti-NTD and S2 antibodies were plotted as black dots and anti-RBD antibodies were plotted as blue dots. Approximate line and correlation coefficient between ACE2 binding and 8D2 binding by anti-NTD and S2 antibodies were shown in the figure.
(B) Comparison of competitive binding assay using C144 antibody with that using C009 and C135 antibody. Spike protein transfectants were mixed with serially diluted COVID-19 patient sera followed by the staining with DyLight 647-labeled C009, C144 and C135 anti-RBD antibodies recognizing different epitopes. Relative fluorescent intensities of the stained cells were shown. The data are presented as mean ± SD.
We then measured the levels of the enhancing and neutralizing antibody titers in COVID-19 patients as well as uninfected individuals (Figure 7C). Enhancing and neutralizing antibodies were not detected in uninfected individuals, whereas both were detected in severe COVID-19 patients. The balance of enhancing and neutralizing antibody titers differed among patients. Because the effect of enhancing antibodies is affected by the level of neutralizing antibodies (Figure 2C), we quantified the overall effect by the difference in titers (enhancing/neutralizing). The observed titer difference was higher in severe patients than in non-severe patients (Figure 7D). Although more detailed and extensive studies on COVID-19 patients are required, our data suggest a correlation between enhancing antibodies and severe COVID-19.
Infectivity-enhancing antibodies in uninfected individuals
It has been reported that a few uninfected individuals possess antibodies against the SARS-CoV-2 spike protein (Anderson et al., 2021; Ng et al., 2020). Therefore, we next investigated whether uninfected individuals carry enhancing antibodies. Because the competitive binding assay used to analyze COVID-19 patients was not sensitive enough to detect low levels of enhancing antibodies in uninfected individuals, we quantified levels of enhancing antibodies by comparing the difference in serum antibody binding to wild-type NTD transfectants with enhancing-epitope-deficient SARS-CoV-2 NTD transfectants (W64A, H66A, K187A, V213A, and R214A) (Figure 7E). Interestingly, some donors (e.g., #13) possessed antibodies that recognized wild-type and mutant NTD to a similar degree, suggesting that they lacked enhancing antibodies (Figure 7F), whereas other donors (e.g., #18) possessed antibodies specific to wild-type NTD, suggesting that they carried enhancing antibodies. Some donors also possessed low levels of anti-RBD antibodies, but there was no correlation between titers of anti-RBD antibodies and enhancing antibodies (Figure 7G). These data suggest that antibody responses against specific sites of the SARS-CoV-2 spike protein vary significantly among individuals.
Discussion
Antibody-dependent enhancement (ADE) of viral infection has been reported for some viruses such as dengue virus (Wan et al., 2020), feline infectious peritonitis virus (FIPV) (Hohdatsu et al., 1998; Vennema et al., 1990), severe acute respiratory syndrome coronavirus (SARS) (Jaume et al., 2011; Kam et al., 2007), and Middle East respiratory syndrome (MERS) (Wan et al., 2020). Binding of the Fc receptor to anti-virus antibodies complexed with virions has been thought to be involved in ADE (Wang et al., 2017). However, Fc-receptor-mediated ADE is restricted to the infection of Fc-receptor-expressing cells such as monocytes or macrophages. In this study, we found a non-canonical, Fc-receptor-independent ADE mechanism. The antibodies against a specific site on the NTD of the SARS-CoV-2 spike protein were found to directly augment the binding of ACE2 to the spike protein, consequently increasing SARS-CoV-2 infectivity. Although the ADE induced by enhancing antibodies is relatively lower than the Fc-receptor-mediated ADE observed in other viruses such as dengue virus, Fc receptors are not involved in this new type of ADE. Therefore, enhancing antibodies could be involved in SARS-CoV-2 infection for a broad range of cells that do not express Fc receptors. So far, the wild-type SARS-CoV-2 has been completely replaced by the D614G mutant SARS-CoV-2. However, the ACE2-binding enhancement effect of the D614G mutation was smaller than that induced by enhancing antibodies. This suggests that an increased ACE2-binding capacity of the spike protein, even if small, plays an important role in SARS-CoV-2 infection.
It is interesting that all of the observed enhancing antibodies recognize a specific site on the NTD. Cryo-EM analysis and docking revealed that all enhancing antibodies bind to the spike protein in a very similar fashion that is distinct from anti-NTD neutralizing antibodies such as 4A8 (Chi et al., 2020). This finding suggests that the specific binding mode is required for this non-canonical ADE. The spike protein RBD is quite flexible, and ACE2 preferentially binds to the open conformation of the RBD (Henderson et al., 2020; Lu et al., 2020). Most structural studies of the wild-type spike protein exhibited one RBD in the open conformation. By contrast, two or three RBDs were observed in the open conformation in the more infective D614G mutant, suggesting that the RBD conformation plays a vital role in the infectivity of SARS-CoV-2 (Henderson et al., 2020; Yurkovetskiy et al., 2020). By using an antibody specific to the open conformation of the RBD, we found that enhancing antibodies induced the open conformation of the RBD upon binding to the NTD. On the other hand, non-enhancing anti-NTD antibodies did not induce an open RBD. This indicated that binding of antibodies to the NTD alone does not induce the open RBD state and that antibodies must bind to a specific site on the NTD to induce this state. Because ACE2 is a dimer, there is a possibility that multimerization of spike protein by antibodies could be involved in enhanced ACE2 binding simply by increasing avidity. However, multimerization of spike protein alone cannot explain why enhanced ACE2 binding is observed with specific antibodies that bind to the specific enhancing site. Furthermore, F(ab′)2 fragments of the enhancing antibody, but not the Fab fragment, induced an open RBD and enhanced ACE2 binding. This could be the reason why the induction of an open RBD was not observed in Fab-bound spike proteins by cryo-EM analysis. Because the affinity of monomeric Fab fragments is lower than that of divalent F(ab′)2 fragments, there is a possibility that Fab fragments did not bind sufficiently to the spike protein to induce the open RBD. However, flow cytometric analysis demonstrated that both Fab and F(ab′)2 fragments bound equally to the spike protein. Furthermore, cryo-EM analysis revealed that COV2-2490 Fab fragments remained attached to all three NTDs on most spike protein, even after the Fab-spike complex was purified by gel filtration column. In addition, we were unable to generate structural models of a full-length IgG molecule with both arms bound to a single spike protein. Together, this suggested that the binding of monomeric enhancing antibodies to the NTD is not sufficient to induce the open conformation of the RBD and that the enhancing site must be bridged by divalent antibodies. In summary, the primary mechanism for enhancing antibodies to augment ACE2 binding seems to be the induction of an open conformation of the RBD by coupling the NTD domains of two spikes.
The RBD packs tightly with the NTD of a neighboring chain (Roy et al., 2020; Wrapp et al., 2020). Recent experimental and computational structural studies have demonstrated that the RBD domains are held in the closed position by their interaction with neighboring NTD domains (Henderson et al., 2020; Mori et al., 2021). Moreover, the spike proteins themselves are highly dynamic, owing to several hinges in their long stalks (Turoňová et al., 2020). It follows that when NTDs located on different spike proteins are bridged by divalent antibodies, intra-spike NTD-RBD interactions will be decoupled, allowing the RBD to assume the open conformation depending on the antibody-binding site on the NTD. These studies are consistent with our mechanistic insights that antibodies that recognize a specific site on the NTD can induce the open RBD state. This implies that the structures around the epitopes recognized by enhancing antibodies play an important role in controlling the conformation of the RBD. It is noteworthy that recent B.1.1.7 and B.1.135 variants contain H69-V70 deletion and D215G mutation, respectively, which are very close to the enhancing antibody-binding site. Recent studies have suggested that H69-V70 deletion increases infectivity of SARS-CoV-2 (Kemp et al., 2021). In fact, ACE2 bound the B.1.1.7 spike protein better than the wild type, even in the absence of enhancing antibodies. Furthermore, a comparison of closely related viruses indicated that the NTD is much more diverse than the rest of the spike protein, implying intense molecular evolution (Saputri et al., 2020). These observations suggest that further mutations around the epitopes of the enhancing site are expected and may affect the infectivity of SARS-CoV-2.
All of the enhancing monoclonal antibodies used in this study were derived from COVID-19 patients, indicating that enhancing antibodies are produced in COVID-19 patients. Using fluorescent-labeled enhancing antibodies, we developed a simple assay to detect enhancing antibodies in serum. With this new assay, we demonstrated that enhancing and neutralizing antibodies are produced in both non-severe and severe patients and that levels of enhancing and neutralizing antibodies are higher in severe patients. These observations are compatible with previous reports showing that anti-spike antibodies are produced at high levels in severe patients (Chen et al., 2020; Lau et al., 2021). Because enhancing antibodies do not work when neutralizing antibodies are at high levels, enhancing antibodies will not function in general. However, before infection or during early infection, when levels of neutralizing antibodies are low, levels of enhancing antibodies may affect the course of COVID-19 disease progression. In addition, a major epitope of neutralizing antibodies has been lost in recent SARS-CoV-2 variants (Supasa et al., 2021; Wang et al., 2021), implying that the effectiveness of enhancing antibodies may vary among SARS-CoV-2 variants. Further large-scale clinical studies are needed to clarify the relationship between emerging variants and enhancing antibodies.
It is noteworthy that uninfected individuals possess antibodies that recognize the infectivity-enhancing site on the NTD, albeit at quite low frequency. Because the epitopes of enhancing antibodies contain charged residues, it is possible that binding to the NTD is mediated by polyreactive antibodies. However, serum antibodies against the infectivity-enhancing site did not bind to the RBD transfectants, suggesting that binding was specific to the infectivity-enhancing site. Recent studies showed that most anti-spike antibodies often detected in uninfected individuals are directed against the S2 subunit, which is relatively similar to that of seasonal human coronaviruses (Anderson et al., 2021; Ng et al., 2020). The sequence homology of the NTD to common human coronaviruses is low compared to the S2 subunit. In particular, antibody epitopes on the enhancing site are not conserved among other coronaviruses. The cause of the production of enhancing antibodies in some uninfected individuals is unknown; certain pathogens with proteins similar to the NTD of SARS-CoV-2 may be involved in the production of enhancing antibodies in some uninfected individuals. There is a possibility that the production of enhancing antibodies might be boosted by SARS-CoV-2 infection or vaccination. It follows that spike proteins that lack the epitopes containing the enhancing site would not stimulate production of enhancing antibodies in individuals with preexisting enhancing antibodies. Transfusion of plasma from recovered COVID-19 patients has been proposed as a possible treatment for COVID-19 (Liu et al., 2020). However, the levels of neutralizing antibodies and enhancing antibodies in serum differ among COVID-19 patients. Therefore, plasma levels of enhancing antibodies in the donor may affect the treatment’s effectiveness. Further studies on the role of enhancing antibodies in both COVID-19 patients and uninfected individuals will be important to understand the complicated pathogenesis of COVID-19.
Limitations of the study
In the present study, we analyzed the levels of the enhancing and neutralizing antibodies in serum by competitive binding assay using 8D2 and C144 antibodies, respectively. Although the competitive binding assay seem to reflect serum levels of the enhancing and neutralizing antibodies as we demonstrated, we cannot exclude the possibility that the levels of enhancing and neutralizing antibodies analyzed by the competitive binding assay may be affected by unrelated antibodies. Indeed, although most of the anti-spike antibodies did not affect the binding of 8D2 and C144 antibodies to spike protein, there were a few antibodies other than the enhancing and neutralizing antibodies that partially inhibited the binding of 8D2 and C144 antibodies. In addition, due to the presence of multiple epitopes for anti-RBD neutralizing antibodies, the levels of neutralizing antibodies detected by the competition assay using C144 antibody may not correctly reflect the total neutralizing antibody levels, although the C144 competitive assay was at least as sensitive as other anti-RBD antibodies. In addition, the effects of enhancing and neutralizing antibodies are expected to vary among SARS-CoV-2 variants, because the mutations on spike protein of the variants would affect the binding of these antibodies. Therefore, there is a possibility that the presence of enhancing antibodies does not always correlate with disease outcome. More extensive studies on enhancing antibodies, neutralizing antibodies, and infected SARS-CoV-2 variant types using a larger number of COVID-19 patient samples are required to clarify the exact function of enhancing antibodies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Anitibodies | ||
| Allophycocyanin (APC) AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, Fcγ fragment specific | Jackson | RRID: AB_2337695 |
| AffiniPure Goat Anti-Mouse IgG, Fcγ fragment specific | Jackson | RRID: AB_2338449 |
| Anti-ACE-2, Mouse-Mono(535919) | R&D | Cat#MAB9332-100 |
| Allophycocyanin (APC) Streptavidin | Jackson | RRID: AB_2337242 |
| APC anti-FLAG Tag Antibody (L5) | Biolegend | Cat#637308 |
| Allophycocyanin (APC) AffiniPure F(ab’)2 Fragment Donkey Anti-Rat IgG (H+L) | Jackson | RRID: AB_2340661 |
| R-Phycoerythrin AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, F(ab’)2 fragment specific | Jackson | RRID: AB_2337677 |
| Chemicals, peptides, and recombinant proteins | ||
| Propidium iodide | Sigma-Aldrich | Cat#25535-16-4 |
| PEI-MAX | Polyscience | Cat#26406 |
| TALON metal affinity resin | Clontech | Cat#635503 |
| Aldehyde/Sulfate Latex Beads, 4% w/v, 4 μm | Thermo Fisher | Cat#A37304 |
| DOTAP | Sigma-Aldrich | Cat#144189-73-1 |
| protein A Sepharose | GE Healthcare | Cat#17127903 |
| Sulfo-NHS-LC-Biotin | Thermo Fisher | Cat#21335 |
| DyLight 650 NHS Ester | Thermo Fisher | Cat#62265 |
| 4% paraformaldehyde | Nacalai Tesque | Cat#09154-85 |
| Wizard Plus SV Minipreps DNA Purification System | promega | Cat#A1465 |
| QIAGEN Plasmid Plus Midi Kit | QIAGEN | Cat#12945 |
| QIAamp viral RNA extraction kit | QIAGEN | Cat# 52904 |
| QuikChange Lightning Multi Site-Directed Mutagenesis Kit | Agilent | Cat#210513-5 |
| Immobilized Pepsin | Thermo Fisher | Prod# 20343 |
| Papain Agarose from papaya latex | Sigma-Aldrich | Prod# P4406 |
| Vivaspin 6, 50,000 MWCO PES | Sartorius | Cat#VS0632 |
| Oriole Fluorescent Gel Stain | Bio-Rad | Cat#1610496 |
| e-PAGEL E-R520L | Atto | Cat#2331730 |
| Zeba Spin Desalting Columns, 7K MWCO, 0.5 mL | Thermo Fisher | Cat#89882 |
| Deposited data | ||
| Cryo-EM map of Apo spike protein | This paper | EMD-30915 |
| Cryo-EM map of spike protein with 8D2 Fab fragment | This paper | EMD-30916 |
| Cryo-EM map of spike protein with (8D2)-1 Fab fragment | This paper | EMD-30917 |
| Cryo-EM map of spike protein with (8D2)-2 Fab fragment | This paper | EMD-30918 |
| Cryo-EM map of spike protein with (8D2)-3 Fab fragment | This paper | EMD-30919 |
| Cryo-EM map of spike protein with (8D2)-4 Fab fragment | This paper | EMD-30920 |
| Cryo-EM map of spike protein with (COV2-2490)-1 Fab fragment | This paper | EMD-30921 |
| Cryo-EM map of spike protein with (COV2-2490)-2 Fab fragment | This paper | EMD-30922 |
| Molecular model fitted to Cryo-EM data of Apo spike protein | This paper | 7DZW |
| Molecular model fitted to Cryo-EM data of spike protein with 8D2 Fab fragment | This paper | 7DZX |
| Molecular model fitted to Cryo-EM data of spike protein with COV2-2490 Fab fragment | This paper | 7DZY |
| Experimental models: Cell lines | ||
| 293T | RIKEN Cell Bank | RCB2202 |
| VERO | Japanese Collection of Research Bioresources | JCRB1819 |
| Huh7 | National Institute of Infectious Diseases | N/A |
| 293T-ACE2 | This paper | N/A |
| PLAT-E | https://doi.org/10.1038/sj.gt.3301206 | N/A |
| Expi293 | Thermo | A14527 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WA-1 | Kanagawa Prefectural Institute of Public Health | KNG19-020 |
| Biological samples | ||
| SARS-CoV-2 patients serum | Kobe City Medical Center General Hospital | N/A |
| Normal humans serum (Taken before June 2019) | George King Bio-Medical | 0070-50 (Lot 1125) |
| Recombinant DNA | ||
| pcDNA3.4 Spike-foldon-his | This paper | N/A |
| pME18S ACE2-Fc | This paper | N/A |
| pME18S Spike | This paper | N/A |
| pME18S Spike D614G | This paper | N/A |
| pME18S Flag-NTD-TM | This paper | N/A |
| pME18S Flag-RBD-TM | This paper | N/A |
| pME18S Flag-S2-TM | This paper | N/A |
| pME18S Flag B.1.1.7 variants Spike (del_70-69, del_144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) | This paper | N/A |
| pMxs-ACE2 | This paper | N/A |
| pME18S-FLAG-COVID-S | This paper | N/A |
| pME18S Flag-NTD-TM | This paper | N/A |
| pME18S-FLAG-COVID-S-V213A | This paper | N/A |
| pME18S-FLAG-COVID-S-H66A | This paper | N/A |
| pME18S-FLAG-COVID-S-W64A | This paper | N/A |
| pME18S-FLAG-COVID-S-K187A | This paper | N/A |
| pME18S-FLAG-COVID-S-R214A | This paper | N/A |
| pME18S-FLAG-COVID-S-W64A H66A | This paper | N/A |
| pME18S-FLAG-COVID-S-V213A R214A | This paper | N/A |
| pME18S-FLAG-COVID-S-W64A H66A V213A R214A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K187A | This paper | N/A |
| pME18S-FLAG-NTD-TM-Y28A | This paper | N/A |
| pME18S-FLAG-NTD-TM-T29A | This paper | N/A |
| pME18S-FLAG-NTD-TM-N30A | This paper | N/A |
| pME18S-FLAG-NTD-TM-S31A | This paper | N/A |
| pME18S-FLAG-NTD-TM-F32A | This paper | N/A |
| pME18S-FLAG-NTD-TM-T63A | This paper | N/A |
| pME18S-FLAG-NTD-TM-W64A | This paper | N/A |
| pME18S-FLAG-NTD-TM-F65A | This paper | N/A |
| pME18S-FLAG-NTD-TM-H66A | This paper | N/A |
| pME18S-FLAG-NTD-TM-I68A | This paper | N/A |
| pME18S-FLAG-NTD-TM-H69A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K77A | This paper | N/A |
| pME18S-FLAG-NTD-TM-R78A | This paper | N/A |
| pME18S-FLAG-NTD-TM-S94A | This paper | N/A |
| pME18S-FLAG-NTD-TM-T95A | This paper | N/A |
| pME18S-FLAG-NTD-TM-E96A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K97A | This paper | N/A |
| pME18S-FLAG-NTD-TM-I100A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K113A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K147A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K150A | This paper | N/A |
| pME18S-FLAG-NTD-TM-R158A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K182A | This paper | N/A |
| pME18S-FLAG-NTD-TM-N185A | This paper | N/A |
| pME18S-FLAG-NTD-TM-F186A | This paper | N/A |
| pME18S-FLAG-NTD-TM-K187A | This paper | N/A |
| pME18S-FLAG-NTD-TM-N211A | This paper | N/A |
| pME18S-FLAG-NTD-TM-L212A | This paper | N/A |
| pME18S-FLAG-NTD-TM-V213A | This paper | N/A |
| pME18S-FLAG-NTD-TM-R214A | This paper | N/A |
| pME18S-FLAG-NTD-TM-D215A | This paper | N/A |
| pME18S-FLAG-NTD-TM-R237A | This paper | N/A |
| pME18S-FLAG-NTD-TM-R246A | This paper | N/A |
| pME18S-FLAG-NTD-TM-D253A | This paper | N/A |
| pME18S-FLAG-NTD-TM-S255A | This paper | N/A |
| pCAGGS H014-human IgG1 | This paper (related to Lv et al., 2020) | N/A |
| pCAGGS H014-human Kappa | This paper (related to Lv et al., 2020) | N/A |
| pCAGGS COV2-2016-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2026-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2146-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2150-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2190-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2212-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2210-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2202-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2215-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2222-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2228-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2248-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2293-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2335-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2369-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2384-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2420-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2438-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2459-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2490-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2495-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2521-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2563-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2571-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2582-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2622-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2660-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2673-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2718-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2830-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 4A8-human IgG1 | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 8D2-human IgG1 | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 4A2-human IgG1 | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS COV2-2697-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2241-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 2-02-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C002-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS 2-04-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-07-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C009-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS 1-12-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-15-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 1-16-human IgG1 | This paper (Brouwer et al., 2020) | N/A |
| pCAGGS 1-18-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-29-human IgG1 | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C101-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C121-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C135-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C144-human IgG1 | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS COV2-2675-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2027-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2496-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2400-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2445-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2147-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2570-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2373-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2035-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2383-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2784-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2454-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2947-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2656-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2553-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2238-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2143-human IgG1 | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 4A10-human IgG1 | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 7E9-human IgG1 | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS COV2-2016-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2026-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2146-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2150-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2190-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2212-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2210-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2202-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2215-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2222-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2228-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2248-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2293-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2335-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2369-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2384-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2420-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2438-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2459-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2490-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2495-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2521-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2563-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2571-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2582-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2622-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2660-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2673-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2718-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2830-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 4A8-human Kappa | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 8D2-human Kappa | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 4A2-human Kappa | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS COV2-2697-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2241-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 2-02-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C002-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS 2-04-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-07-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C009-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS 1-12-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-15-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 1-16-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 1-18-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS 2-29-human Kappa | This paper (related to Brouwer et al., 2020) | N/A |
| pCAGGS C101-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C121-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C135-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS C144-human Kappa | This paper (related to Robbiani et al., 2020) | N/A |
| pCAGGS COV2-2675-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2027-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2496-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2400-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2445-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2147-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2570-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2373-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2035-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2383-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2784-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2454-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2947-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2656-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2553-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2238-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2143-human Kappa | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 4A10-human Kappa | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS 7E9-human Kappa | This paper (related to Chi et al., 2020) | N/A |
| pCAGGS COV2-2210-mouse IgG2a | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2369-mouse IgG2a | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2490-mouse IgG2a | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2582-mouse IgG2a | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS COV2-2660-mouse IgG2a | This paper (related to Zost et al., 2020) | N/A |
| pCAGGS 8D2-mouse IgG2a | This paper (related to Chi et al., 2020) | N/A |
| Software and algorithms | ||
| FlowJo10 | Becton Dicknson | N/A |
| Prism7.0 | GraphPad | N/A |
| cryoSPARC | Structura Biotechnology | N/A |
Requests for further information and reagents should be directed to Hisashi Arase (arase@biken.osaka-u.ac.jp).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hisashi Arase (arase@biken.osaka-u.ac.jp).
Materials availability
All materials generated in this study will be made available on request by the Lead Contact.
Data and code availability
Cryo-EM density maps for SARS-CoV-2 spike protein complexed with the infectivity-enhancing antibodies are deposited at the EMDB under accession code EMD-30915, EMD-30916, EMD-30917, EMD-30918, EMD-30919, EMD-30920, EMD-30921 and EMD-30922, respectively. Molecular models fitted to Cryo-EM data were deposited to PDB under accession code 7DZW, 7DZX and 7DZY. The data that support the findings of this study are available from the Lead Contact on request.
Experimental model and subject details
Cell lines
HEK293T cells (RIKEN Cell Bank), Huh7 cells (the National Institute of Infectious Diseases) and TMPRSS2-expressing VeroE6 cells (Japanese Collection of Research Bioresources Cell Bank, JCRB1819) were cultured in DMEM (Nacalai, Japan) supplemented with 10% FBS (Biological Industries, USA), penicillin (100 U/mL), and streptomycin (100 μg/mL) (Nacalai, Japan) and cultured at 37°C in 5% CO2. The Expi293 cells (Thermo) were cultured with the Expi293 medium. The cells were routinely checked for mycoplasma contamination.
Viruses
SARS-CoV-2 was obtained from the Kanagawa Prefectural Institute of Public Health (KNG19-020). The stock virus was amplified in TMPRSS2-expressing VeroE6 cells. SARS-CoV-2 infection assay was carried out in a Biosafety Level 3 laboratory.
Human samples
The collection and use of human sera and synovial tissues were approved by Osaka University (2020-10 and 19546), Kobe City Medical Center General Hospital (200924), and Osaka South Hospital (2-28). Written informed consent was obtained from the participants according to the relevant guidelines of the institutional review board. The diagnoses of SARS-CoV-2 were PCR-based. The patients required for ventilator were considered as severe patients. The sera from uninfected humans were taken before June 2019 (George King Bio-Medical). All the sera of the SARS-CoV-2 patients were treated with 2% CHAPS for 30 min at room temperature to inactivate the remaining virus.
Method details
Plasmid construction
The SARS-CoV-2 spike gene (NC_045512.2) was prepared by gene synthesis (IDT). The sequences encoding the spike protein lacking C-terminal 19 amino acids (amino acids 1–1254) were cloned into pME18S expression vector. NTD (amino acids 14–333), RBD (amino acids 335–587), and S2 (amino acids 588–1219) were separately cloned into a pME18S expression vector containing a SLAM signal sequence and a PILRα transmembrane domain (Saito et al., 2017). A series of alanine mutants and B.1.1.7 variants (del_70-69, del_144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H) were prepared from SARS-CoV-2 spike using the QuickChange mutagenesis kit or the QuickChange multi-mutagenesis kit (Agilent). The primers for mutagenesis were designed on Agilent’s website (https://www.agilent.com/store/primerDesignProgram.jsp). The cDNA encoding ACE2 (NM_021804.3) was cloned into a pMxs retrovirus vector. The mouse ACE2-IgG2a Fc fusion protein was prepared by cloning the sequence encoding the extracellular domain of Ace2 (amino acid residues 20–740) into the pCAGGS expression vector containing the SLAM signal sequence and the sequence encoding the mouse IgG2a Fc domain as previously described (Hirayasu et al., 2016). The sequence encoding the spike protein’s extracellular domain with a foldon and His-tag at the C terminus (Cai et al., 2020) was cloned into a pcDNA3.4 expression vector containing the SLAM signal sequence. Also, mutations D614G, R686G R687S R689G, K986P, and V987P were introduced using a Quick change multi-mutagenesis kit (Agilent) for stabilization of recombinant spike protein (Yurkovetskiy et al., 2020). The DNA sequences of these constructs were confirmed by sequencing (ABI3130xl).
Transfection
A pME18S expression plasmid containing the full-length or subunit spike protein was transiently transfected into HEK293T cells using PEI max (Polysciences); the pMx-GFP expression plasmids were used as the marker of transfected cells. ACE2 was stably transfected into HEK293T cells using the pMxs retrovirus vector. Briefly, pMxs-ACE2 and amphotropic envelope vectors were co-transfected into PLAT-E packaging cells. The cell culture supernatants containing the retroviruses were harvested 48 hours later and premixed with DOTAP (Roche, Switzerland) before spin transfection at 2400 rpm and 32°C for 2 hours. Afterward, the ACE2-expressing HEK293T cells were purified using a cell sorter (SH800S, Sony).
Anti-spike monoclonal antibodies from COVID-19 patients
The V regions of anti-SARS-CoV-2 spike antibodies from COVID-19 patients were synthesized according to the published sequence (IDT) (Brouwer et al., 2020; Chi et al., 2020; Robbiani et al., 2020; Zost et al., 2020). The cDNA sequences of the variable regions of the heavy chain and light chain were cloned into a pCAGGS vector containing sequences that encode the human IgG1 or kappa constant region. The IgG with the murine IgG2a constant region was used for the competitive binding assay (Figure 4A) or analysis of the open conformation of RBD (Figures 6A, 6B, and S5A). The pCAGGS vectors containing sequences encoding the immunoglobulin heavy chain and light chain were co-transfected into HEK293T cells or Expi293 (Thermo) cells, and the cell culture supernatants were collected according to the manufacturer’s protocols. Recombinant IgG was purified from the culture supernatants using protein A Sepharose (GE healthcare) except for Figure 1B. The concentration of unpurified IgG in the cell culture supernatants used in Figure 1B was measured using the protein A-coupled latex beads (Thermo A37304) and APC-labeled anti-human IgG F(ab’)2 antibodies (Jackson) against IgG standards of known concentration. The concentration of purified IgG was measured at OD280. For competitive binding assay, protein A-purified 8D2 (Chi et al., 2020), C144 (Robbiani et al., 2020), C009 (Robbiani et al., 2020) and C135 (Robbiani et al., 2020) antibodies were labeled using DyLight 650 amine-reactive dye (Thermo) according to the manufacturer’s protocol.
Antibodies and recombinant proteins
Mouse anti-human ACE2 mAb (R&D Systems, USA), rat anti-Flag mAb (L5, Biolegend), allophycocyanin (APC)-conjugated donkey anti-mouse IgG Fc fragment antibody, APC-conjugated anti-human IgG Fc fragment specific antibody, PE-conjugated anti-human IgG Fab fragment specific antibody, APC-conjugated anti-rat IgG specific antibody, and APC-conjugated streptavidin (Jackson ImmunoResearch, USA) were used. The pCAGGS vector containing a sequence that encodes the ACE2-mouse IgG2a Fc fusion protein was transfected into the HEK293T cells. Recombinant ACE2-Fc fusion protein was purified from the cell culture supernatants with the protein A Sepharose (GE Healthcare). The purified ACE2-mouse IgG2a Fc fusion protein was biotinylated with Sulfo-NHS-LC-Biotin (Thermo), followed by buffer exchange with a Zeba spin desalting column. The pcDNA3.4 expression vector containing the sequence that encodes the His-tagged extracellular domain of the spike protein was transfected into Expi293 cells; then, the His-tagged spike protein produced in the culture supernatants was purified with a Talon resin (Clontech). F(ab′)2 fragment was prepared by incubating protein A purified COV2-2490 antibodies with immobilized pepsin (Thermo fisher) in the 20mM sodium acetate pH 4.5 at 37°C 4h. Fab fragment was prepared by incubating protein A purified 8D2 or COV2-2490 antibodies with immobized papain (100 units/ml, Sigma) in the presence of in the presence of 10 mM EDTA and 10 mM cysteine at room temperature for 3h. Immobilized pepsin or papain was precipitated by centrifugation, and the supernatant was loaded onto protein A Sepharose equilibrated with PBS. Flow through fractions were purified with Superdex 200 Increase 10/300 GL gel filtration column using ÄKTA Pure 25 System (GE Healthcare Life Sciences). The purity of the purified F(ab′)2 or Fab fragments were analyzed with SDS/PAGE and the gel was stained with Oriole Fluorescent dye (Bio-Rad) (Figure S5B). For Cryo-EM analysis, the same protein amount of Spike protein and Fab were incubated at 4°C for 1∼2 h, and the complex was separated on a Superose 6 Increase 10/300 GL column equilibrated with 20 mM Tris-HCl (pH 8.0), 0.25 M NaCl. The purified complex was used for Cryo-EM analysis.
The flow cytometric analysis of antibodies
The plasmid expressing the full-length SARS-CoV-2 spike protein, Flag-NTD-PILR-TM, Flag-RBD-PILR-TM, Flag-S2-PILR-TM, mutated spike proteins or B.1.1.7 variant spike protein were co-transfected with the GFP vector into HEK293T cells. The transfectants were incubated with the mAbs, followed by APC-conjugated anti-human IgG Ab. Then, the antibodies bound to the stained cells were analyzed using flow cytometers (Attune, Thermo; FACSCalibur BD bioscience, LSR Fortessa X20 BD bioscience). Antibody binding to the GFP-positive cells were shown in the figures using a FlowJo software (BD bioscience).
ACE2 binding assay
Because SARS-CoV-2 spike protein contains intracellular retention signal at C terminus (Hu et al., 2020; Johnson et al., 2020), we used spike protein lacking C-terminal 19 amino acids for ACE2 binding assay. C-terminal deleted spike protein and GFP were cotransfected into HEK293T cells and the transfectants were mixed with various concentrations of anti-spike antibodies at 4°C for 30 min, followed by incubation with a biotinylated-ACE2-Fc fusion protein at 1 μg / ml and 4°C for 30 min. In some experiments, enhancing antibodies and neutralizing antibodies were added simultaneously or sequentially to the spike transfectants. Thereafter, the ACE2 bound to the spike protein was detected using APC-conjugated streptavidin (2.5 μg/mL). The amount of ACE2 on GFP-positive cells was analyzed by a flow cytometer.
SARS-CoV-2 spike-pseudotyped virus infection assay
The HEK293T cells were transiently transfected with expression plasmids for the SARS-CoV-2 spike protein lacking C-terminal 19 amino acids (Hu et al., 2020; Johnson et al., 2020). Twenty-four hours post transfection, VSV-G-deficient VSV carrying a GFP gene complemented in trans with the VSV-G protein was added for a two-hour incubation. The cells were then carefully washed with DMEM media without FBS and incubated with DMEM with FBS at 37°C in 5% CO2 for 48 hours. The supernatant containing the pseudotyped SARS-CoV-2 virions was harvested and aliquoted before storage at −80°C. The virus titers were determined using TMPRSS2-expressing VeroE6 cells. The pseudotyped SARS-CoV-2 virus was pre-incubated with anti-NTD monoclonal antibodies for 30 minutes and mixed with the ACE2-expressing cells. Twenty-four hours later, dead cells were stained with propidium iodide (Sigma), and the proportions of GFP-expressing cells in the living cells were analyzed by flow cytometry.
The authentic SARS-CoV-2 infection assay
The virus stock (6.25 log TCID50) was prediluted 1: 100 in DMEM supplemented with 2% FBS before being incubated with the cells at 1 × 105 cells/mL at 37°C in 5% CO2 for 3 hours. The virus was then removed by washing the cells with DMEM; the cell culture supernatant was collected for control. The cells were incubated for another 2 days. The cell culture supernatant was collected for viral RNA extraction using the QIAamp viral RNA extraction kit (QIAGEN, Germany) and subsequently quantified using real-time PCR using the N2 primer set (AGCCTCTTCTCGTTCCTCATCAC and CCGCCATTGCCAGCCATTC).
The competitive binding assay among the enhancing and non-enhancing antibodies
The binding of the enhancing antibodies containing a mouse IgG2a constant region (1 μg/ml) to full-length spike transfectants was analyzed in the presence of the human antibodies (10 μg/ml). The effect of competitors on ACE2-Fc binding to the spike transfectants was also analyzed. The non-enhancing anti-S2 antibody, COV2-2147, was used as a control. Relative mean fluorescence intensities observed in the presence of competitor antibodies were calculated.
The docking model of the enhancing antibodies and spike protein
A homology model of the SARS-CoV-2 prefusion spike trimer was built using a template-based method. The prefusion spike trimer structure (PDB ID: 7jji) in the all-RBD-down status was used as the main structural template. To model missing residues within the S2 domain, fragments were sampled from other SARS-CoV-2 spike trimer structures. The best fragment template was selected as the one with the lowest RMSD within flanking regions of the main template and the complete structure was constructed using MODELER (Webb and Sali, 2016). We investigated possible binding modes of the enhancing antibodies through antibody modeling followed by docking. Antibody structures were modeled by Repertoire Builder (Schritt et al., 2019) and docked onto the NTD using the HADDOCK2.4 webserver (Ambrosetti et al., 2020) using observed epitope residues as constraints. The top-scoring docked poses were rendered on the full-length spike model using PyMOL. Residues identified by alanine scanning to affect enhancement were represented as a heatmap onto the surface of the spike model.
Cryo-EM data collection
Spike protein and Fab complex was purified by A 2.5 μl protein solution of the spike complex (4.5mg/ml) or the complex of spike and antibody (2.5 mg/ml) was applied onto the cryo-grid and frozen in liquid ethane using a Vitrobot IV (FEI, 4°C and 100% humidity). In the case of the complex of the spike-COV2-2490 antibody, the graphen corted Quantifoil Au was used. In contrast, Quantifoil Au R0.6/1.0 holey carbon grids were used for the apo and the spike-8D2 antibody complex. Data collection of each sample was carried out on a Titan Krios (FEI, Netherland) equipped with a thermal field emission electron gun operated at 300 kV, an energy filter with a 20 eV slit width and a K3 direct electron detector camera (Gatan, USA) (Figure S5). For automated data acquisition, SerialEM software was used to collect cryo-EM image data. Movie frames were recorded using the K3 camera at a calibrated magnification of × 81,000 corresponding to a pixel size of 0.88 Å with a setting defocus range from −0.8 to −2.0 μm. The data were collected with a total exposure of 3 s fractionated into 50 frames, with a total dose of ∼50 electrons Å2 in counting mode. A total number of movies were collected; 5,075 for the apo spike complex, 2,135 for the spike protein-8D2 antibody complex, and 5,177 for the spike protein-COV-2490 antibody complex, respectively.
Image processing and 3D reconstruction
All of image processes were carried out on cryoSPARC software (Punjani et al., 2017). After motion correction of movies and CTF parameter estimation, the particles were automatically picked by template-based particle picking algorithm. The detailed information is summarized in Table S1. The picked particles were extracted into a box of 360 × 360 pixels for the apo spike complex, 440 × 440 pixels for the spike-8D2 complex, and 380 × 380 pixels for the spike-COV2-2490 complex. For the apo spike complex, after particle extraction, some rounds of heterogenous refinement with C1 symmetry resulted in one class (74,756 particles) that was further used as homogeneous refinement with CTF refinement, and particles polishing. As the result, the density map for the apo spike complex was obtained at 3.36 Å resolution (EMDBID: 30915). For the spike-8D2 antibody complex, 643,616 particles were selected after several rounds of 2D classification. Then, the selected particles were applied to several rounds of heterogenous refinement with C1 symmetry. As the result, five distinguishable classes were obtained. Each of these classified particles (161,974, 196,524, 99,745, 74,399, and 137,974 particles) were applied to homogeneous refinement with CTF refinement. The density maps from the refinement were obtained at 3.46, 3.53, 3.82, 4.16, and 3.74 Å resolution, respectively (EMDBID: 30916, 39017, 30918, 30919, and 30920). For the spike-COV2-2490 antibody complex, using extracted particles, heterogenous classification with C1 symmetry resulted in two distinguishable classes (104,069 and 33,092 particles). As the results of homogeneous refinement with CTF refinement, the two maps of the spike-COV2-2490antibody complex at 3.15 and 4.06 Å resolution (EMDBID: 30921 and 30922) was obtained. Local resolution of the obtained maps were estimated by Local resolution estimation job on cryoSPARC.
To elucidate the relative position between the spike complex and antibody, the homology models of the spike complex and antibody (Fab) were fitted into the obtained maps as rigid bodies by Chimera (Pettersen et al., 2004). A bunch of 36 one-RBD-up trimeric spike complexes were collected from the PDB (dated as of November 5, 2020) and used as templates to construct homology models with alternative conformations as described in methods. Spike homology models were then fitted into the density maps, and the model with the highest correlation was selected. Fab models were fitted independently of the spike. Variable domains of the Fabs were modeled by Repertoire Builder (Schritt et al., 2019) and used as templates to create complete Fab models. The best matched PDB structure 5X8M was selected as the template for Fab constant domains. MODELER (Webb and Sali, 2016) was used to build the complete Fab models from the variable and constant domain templates. After fitting the Fab models into the density maps, binding interface between spike NTD and Fab CDRs was examined, and NTD loops clashing the Fab were remodeled by the MODELER (Webb and Sali, 2016). Finally, refined models were deposited (PDBID: 7DZW for apo, 7DZX for the complex with 8D2, 7DZY for the complex with COV2-2490).
Anti-RBD antibody binding assay
Spike protein and GFP were cotransfected into HEK293T cells and the transfectants were mixed with 3 μg/mL of anti-NTD antibodies of which constant regions were replaced with mouse-IgG2a constant region at 4°C for 30 min, followed by incubation with 1 μg / mL of H014, C009, C135 and C144 anti-RBD antibodies with human IgG1 constant region at 4°C for 30 min. Thereafter, the anti-RBD antibodies bound to the spike protein were detected using APC-conjugated anti-human-IgG Fc specific antibodies that do not cross-react with mouse IgG2a (2.5 μg/mL).
The analysis of the enhancing and neutralizing Ab titers
The 4 μm aldehyde/sulfate latex beads (Thermo A37304) were coated with protein A-purified anti-S2 mAb (COV2-2454) (Zost et al., 2020) and then incubated with recombinant spike protein at 7 μg/ml to capture the recombinant SARS-CoV-2 spike protein on the latex beads. The latex beads containing recombinant SARS-CoV-2 spike were then incubated with 1:30 diluted serum from COVID-19 patients, uninfected individuals or anti-spike antibodies used in Figure 1B (1 μg/ml) and stained with DyLight 650-labeled 8D2 mAb at 3 μg/ml) or C144 mAb at 3 μg/ml. After fixation with 4% paraformaldehyde (Nacalai Tesque), the beads were analyzed using flow cytometry. We defined the high levels of the enhancing and neutralizing antibody titers in a COVID-19 patient’s serum as 1,000 units and used the serum as a standard to calculate antibody titers of other serum samples.
Detection of enhancing antibody in uninfected individuals
The plasmids expressing the wild-type Flag-tagged NTD-TM, which was recognized by the enhancing antibodies, and the Flag tagged NTD-TM mutants (W64A, H66A, K187A,V213A, and R214A) not recognized by all enhancing antibodies were transfected into HEK293T cells with the GFP vector as a transfection marker. Cell surface expression levels of NTD on these transfectants were analyzed using anti-Flag antibody and cells expressing the same levels of NTD were used for comparion between wild-type and mutant NTD. The transfectants were mixed with 1:100 diluted serum from uninfected individuals, and the bound antibodies were detected with the APC-labeled anti-human IgG Fc antibody. The 4A8 anti-NTD antibody (Chi et al., 2020) was used as a standard to calculate the relative concentration of the antibody against wild-type NTD. The stained cells were analyzed by a flow cytometer. The level of serum antibodies specific to the wild-type or mutant NTD was calculated by subtracting the mean fluorescence intensity of the antibodies bound to the GFP-negative cells from those of the GFP-positive cells. The level of the enhancing antibodies was calculated by subtracting antibody levels against the mutant NTD from those against the wild-type NTD. Similarly, the relative level of the anti-RBD antibody was measured using RBD-TM transfectants and C144 as a standard. One μg/ml of 4A8 and C144 was defined as 1 unit.
Data and statistical analysis
FlowJo version 10.7 (BD Biosciences, USA) was used to analyze the flow cytometry data, and Graphpad Prism version 7.0e was used for graph generation and statistical analysis.
Acknowledgments
W.T.S. is supported by a Kishimoto Foundation fellowship. We would like to thank Hiroshi Honda, Kenta Tsutsumi, and Eiki Yamashita for technical assistance and Chikako Kita for administrative assistance. This work was supported by JSPS KAKENHI under grants JP18H05279 and JP19H03478; MEXT KAKENHI under grant JP19H04808; the Japan Agency for Medical Research and Development (AMED) under grants JP19fk0108161 (H.A.), JP20nf0101623 (H.N., H.A., M.Y., and T. Shioda), JP20nk0101602 (H.N., H.A., M.Y., and T. Shioda), 20fk0108403h (Y.N. and H.A.), JP20fk0108263 (T.O.), JP20am0101108 (D.M.S.), and JP20am0101072 (T.K. and Atsushi Nakagawa, BINDS support number 2630); the Japan Science and Technology Agency (JST) (Moonshot R&D grant JPMJMS2025) (Y.M.); and the Panasonic Corporation (H.A. and Y.M.).
Author contributions
Y.L., W.T.S., Y.N., Y.S., T.O., M.Y., H.N., Y.M., Atsushi Nakagawa, T.K., M.O., D.M.S., T. Shioda, and H.A. conceived experiments. Y.L., W.T.S., J.-i.K., M.H., E.E.N., M.S., T. Suzuki, A.T., A.A., S.M., K.A., M. Matsuda, C.O., S.T., K.K., H.J., W.N., M. Matsumoto, and H.A. performed experiments. Atsushi Nakagawa, N.A., N.H., S.O., K.T., and K.O. collected patient sera. J.-i.K., S.L., and D.M.S. constructed a model of antibody binding. Y.L., J.-i.K., M.K., D.M.S, and H.A. wrote the manuscript. All authors read, edited, and approved the manuscript.
Declaration of interests
Osaka University and HuLA immune have filed a patent application on the method to detect the enhancing antibodies and the design of spike protein that does not induce the enhancing antibodies. Y.L., H.A., and Y.S. are listed as inventors. M.Matsumoto, Y.N., and Y.S. are employees of HuLA immune. H.A. and Y.S. are stockholders of HuLA immune.
Published: May 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.05.032.
Supplemental information
References
- Ambrosetti F., Jandova Z., Bonvin A.M. A protocol for information-driven antibody-antigen modelling with the HADDOCK2.4 webserver. arXiv. 2020 doi: 10.1007/978-1-0716-2609-2_14. https://arxiv.org/abs/2005.03283 2005.03283. [DOI] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864 e1810. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhang J., Qin X., Wang W., Xu M., Wang L.F., Xu C., Tang S., Liu P., Zhang L., et al. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomed. Pharmacother. 2020;130:110629. doi: 10.1016/j.biopha.2020.110629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayasu K., Saito F., Suenaga T., Shida K., Arase N., Oikawa K., Yamaoka T., Murota H., Chibana H., Nakagawa I., et al. Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat. Microbiol. 2016;1:16054. doi: 10.1038/nmicrobiol.2016.54. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohdatsu T., Yamada M., Tominaga R., Makino K., Kida K., Koyama H. Antibody-dependent enhancement of feline infectious peritonitis virus infection in feline alveolar macrophages and human monocyte cell line U937 by serum of cats experimentally or naturally infected with feline coronavirus. J. Vet. Med. Sci. 1998;60:49–55. doi: 10.1292/jvms.60.49. [DOI] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. 2020;7:551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R., et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J. Virol. 2020;94:e01062. doi: 10.1128/JVI.01062-20. e01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y.W., Kien F., Roberts A., Cheung Y.C., Lamirande E.W., Vogel L., Chu S.L., Tse J., Guarner J., Zaki S.R., et al. Antibodies against trimeric S glycoprotein protect hamsters against SARS-CoV challenge despite their capacity to mediate FcgammaRII-dependent entry into B cells in vitro. Vaccine. 2007;25:729–740. doi: 10.1016/j.vaccine.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W.H., Chiu S.S., Ko R.L.W., Chan K.H., Cheng S.M.S., Perera R.A.P.M., et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021;12:63. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T.H., Lin H.M., Baine I., Wajnberg A., Gumprecht J.P., Rahman F., Rodriguez D., Tandon P., Bassily-Marcus A., Bander J., et al. Convalescent plasma treatment of severe COVID-19: a propensity score-matched control study. Nat. Med. 2020;26:1708–1713. doi: 10.1038/s41591-020-1088-9. [DOI] [PubMed] [Google Scholar]
- Lu M., Uchil P.D., Li W., Zheng D., Terry D.S., Gorman J., Shi W., Zhang B., Zhou T., Ding S., et al. Real-Time Conformational Dynamics of SARS-CoV-2 Spikes on Virus Particles. Cell Host Microbe. 2020;28:880–891.e8. doi: 10.1016/j.chom.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Jung J., Kobayashi C., Dokainish H.M., Re S., Sugita Y. Elucidation of interactions regulating conformational stability and dynamics of SARS-CoV-2 S-protein. Biophys. J. 2021;120:1060–1071. doi: 10.1016/j.bpj.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W. A review of feline infectious peritonitis virus: molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993;36:1–37. doi: 10.1016/0378-1135(93)90126-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Jaiswar A., Sarkar R. Dynamic Asymmetry Exposes 2019-nCoV Prefusion Spike. J. Phys. Chem. Lett. 2020;11:7021–7027. doi: 10.1021/acs.jpclett.0c01431. [DOI] [PubMed] [Google Scholar]
- Saito F., Hirayasu K., Satoh T., Wang C.W., Lusingu J., Arimori T., Shida K., Palacpac N.M.Q., Itagaki S., Iwanaga S., et al. Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors. Nature. 2017;552:101–105. doi: 10.1038/nature24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saputri D.S., Li S., van Eerden F.J., Rozewicki J., Xu Z., Ismanto H.S., Davila A., Teraguchi S., Katoh K., Standley D.M. Flexible, Functional, and Familiar: Characteristics of SARS-CoV-2 Spike Protein Evolution. Front. Microbiol. 2020;11:2112. doi: 10.3389/fmicb.2020.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Arii J., Suenaga T., Wang J., Kogure A., Uehori J., Arase N., Shiratori I., Tanaka S., Kawaguchi Y., et al. PILRalpha is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schritt D., Li S.L., Rozewicki J., Katoh K., Yamashita K., Volkmuth W., Cavet G., Standley D.M. Repertoire Builder: high-throughput structural modeling of B and T cell receptors. Mol. Syst. Des. Eng. 2019;4:761–768. [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supasa P., Zhou D., Dejnirattisai W., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Nutalai R., Tuekprakhon A., et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211.e7. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Imai K., Kawano S., Ikeda M., Kodama T., Miyoshi K., Obinata H., Mimura S., Kodera T., Kitagaki M., et al. Clinical characteristics of COVID-19 in 104 people with SARS-CoV-2 infection on the Diamond Princess cruise ship: a retrospective analysis. Lancet Infect. Dis. 2020;20:1043–1050. doi: 10.1016/S1473-3099(20)30482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová B., Sikora M., Schürmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bülow S., Gecht M., Bagola K., Hörner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H., de Groot R.J., Harbour D.A., Dalderup M., Gruffydd-Jones T., Horzinek M.C., Spaan W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990;64:1407–1409. doi: 10.1128/jvi.64.3.1407-1409.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020;94:e02015-19. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.T., Sewatanon J., Memoli M.J., Wrammert J., Bournazos S., Bhaumik S.K., Pinsky B.A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K., et al. IgG antibodies to dengue enhanced for FcγRIIIA binding determine disease severity. Science. 2017;355:395–398. doi: 10.1126/science.aai8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.C., Scott F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981;4:175–189. doi: 10.1016/0147-9571(81)90003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps for SARS-CoV-2 spike protein complexed with the infectivity-enhancing antibodies are deposited at the EMDB under accession code EMD-30915, EMD-30916, EMD-30917, EMD-30918, EMD-30919, EMD-30920, EMD-30921 and EMD-30922, respectively. Molecular models fitted to Cryo-EM data were deposited to PDB under accession code 7DZW, 7DZX and 7DZY. The data that support the findings of this study are available from the Lead Contact on request.