Abstract
Leaf trichomes play vital roles in plant resistance and the quality of tea. Basic helix-loop-helix (bHLH) transcription factors (TFs) play an important role in regulating plant development and growth. In this study, a total of 134 CsbHLH proteins were identified in the Camellia sinensis var. sinensis (CSS) genome. They were divided into 17 subgroups according to the Arabidopsis thaliana classification. Phylogenetic tree analysis indicated that members of subgroups IIIc-I and IIIc-II might be associated with trichome formation. The expression patterns of CsbHLH116, CsbHLH133, CsbHLH060, CsbHLH028, CsbHLH024, CsbHLH112 and CsbHLH053 from clusters 1, 3 and 5 were similar to the trichome distribution in tea plants. CsbHLH024 and CsbHLH133 were located in the cell nucleus and possessed transcriptional activation ability. They could interact with CsTTG1, which is a regulator of tea trichome formation. This study provides useful information for further research on the function of CsbHLHs in trichome formation.
Subject terms: Computational biology and bioinformatics, Genetics, Molecular biology
Introduction
Trichomes are developed from epidermal cells and mainly distributed on the undersurface of plant leaves1–4. Plant leaf trichomes are an important basis for botanical classification and play key roles in plant resistance to biotic and abiotic stresses. According to the morphology and function of trichomes, they are classified as nonbranched or branched trichomes and nonglandular or glandular trichomes5–8. Glandular trichomes protect plants from herbivores and insects by accumulating and secreting a series of secondary metabolites, such as alkaloids, nicotine and terpenes9,10. Nonglandular trichomes can enhance plant tolerance in response to extreme temperatures, drought and ultraviolet radiation11–14. Trichome formation has been systematically investigated in Arabidopsis thaliana, Solanum lycopersicum, Cucumis sativus L., Oryza sativa L., Nicotiana tabacum L., Gossypium spp. and Glycine max15–21. Trichome formation is induced by cell differentiation. Arabidopsis thaliana trichome development is an ideal model for the study of cell differentiation22. Many transcription factors (TFs) are associated with trichome development in Arabidopsis thaliana, including R2R3-MYB TFs, bHLH TFs and WD40-repeat (WDR) proteins23–25. The MYB-BHLH-WDR (MBW) complex can positively regulate Arabidopsis thaliana trichome formation26. In addition, most dicots possess a similar regulatory mechanism of trichome formation27.
Tea, one of the three major nonalcoholic beverages, possesses high nutritional and health-benefitting properties28,29. Tender leaves serving as the main raw material are used for tea production. Apical buds and young leaves possess trichomes in most tea plant cultivars; thus, leaf trichomes have become a critical diagnostic characteristic in tea taxonomy. Abundant trichomes are generally indicate high quality in Chinese tea. An abundance of trichomes on tea products indicates that they were processed using tender leaves of tea plants8. Tea trichomes contain abundant metabolites, including theanine, catechins, volatiles and caffeine30,31. These metabolites have different flavors and tastes in tea infusions. Theanine makes the flavor of sweet and umami, and catechins and caffeine make the flavor of bitterness and astringency in tea infusions30. Tea trichomes also possess high contents of benzoic acid derivatives, lipid oxidation derivatives and monoterpene derivatives, which contribute to tea flavor and aroma32. In addition, some signaling genes related to diseases and anti-herbivore and anti-abiotic peptides were specifically transcribed in tea trichomes32.
Basic helix-loop-helix (bHLH) TFs are the second-largest TF family in plants33. Their conserved domains contain two different functional regions, a basic region and a helix-loop-helix (HLH) region, which are composed of 50–60 amino acids34–36. The basic region in the N-terminal domain consists of 13–17 amino acids and binds to the consensus hexanucleotide E-box (CANNTG). The HLH region in the C-terminal domain includes approximately 40 amino acids and contributes to the formation of homodimeric complexes and heterodimeric complexes, as well as the promotion of interactions with other TFs37–40. bHLH TFs play important roles in responses to stresses, secondary metabolism biosynthesis and plant growth and development34,41–44. Numerous studies have demonstrated that bHLH TFs play a critical role in trichome formation. The bHLH proteins GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) are important for the regulation of trichome formation in Arabidopsis thaliana45,46. GL3 and EGL3 interact with the WDR and R2-R3 MYB proteins to induce trichome formation by targeting GLABRA2 (GL2) transcription47–49. GL3 also facilitates trichome branching formation by positively regulating FURCA4 (FRC4) expression50. Trichome formation in tomato plants is independent of SlGL351. Trichome formation is extremely complex in tea plants. Whether bHLH TFs are related to trichome formation in tea plants is less well understood.
In this study, the bHLH family was identified in Camellia sinensis var. sinensis (CSS) genome, and characteristic analyses were systematically performed. The results of phylogenetic tree and expression pattern analyses showed that CsbHLH024 and CsbHLH133 might be associated with tea trichome formation. They were further selected for subcellular localization, transcriptional activation and yeast two-hybrid (Y2H) assays, aiming to preliminarily determine their function. This study provides useful information for further research on the function of CsbHLH TFs in the regulation of trichome formation.
Materials and methods
Identification of the bHLH gene family in tea plants
bHLH TF sequences were acquired from the Tea Plant Information Archive (TPIA) (http://tpia.teaplant.org)31. The SMART database52, National Center for Biotechnology Information (NCBI) conserved domain search service53 and hidden Markov model (HMM) profile of the bHLH domain (PF00010) in the Pfam database54 were used to filter redundant bHLH proteins in tea plants. The full-length amino acid sequences, molecular weights (Mws), theoretical isoelectric points (pIs) and instability index values of these proteins were predicted using the ExPASy server55. The CsbHLHs were renamed CsbHLH001 to CsbHLH134 based on the gene ID order.
Conserved motif and gene structure characterization
The conserved motifs of the bHLH protein were identified using Multiple EM for Motif Elicitation (MEME) with the following parameter settings: site distribution, any number of repetitions; number of motifs, 15; maximum motif width, 100; minimum motif width, 6; maximum number of sites, 100; and minimum number of sites, 556. Both coding DNA sequences (CDSs) and genomic sequences of bHLHs were used for determination of the gene structure with TBtools software57.
Phylogenetic tree analysis
The bHLH protein sequence of Arabidopsis thaliana (128) was downloaded from The Arabidopsis Information Resource (TAIR) database58, and those of Oryza sativa L. (144) and Actinidia chinensis (164) were acquired from the Plant TF Database version 4.059. All sequences were renamed and listed in Table S4. Moreover, the conserved bHLH domains of these proteins were subjected to multiple sequence alignment using ClustalX 2.1 with the default parameters. MEGA X was used to construct neighbor-joining phylogenetic trees with the following parameters: 1000 bootstrap replications; Poisson model; and pairwise deletion60. The phylogenetic trees were optimized using Evolview v361.
Transcriptome data analysis
The transcriptome data of tea plant bHLHs were obtained from TPIA31. The expression patterns of CsbHLHs in different developmental leaf tissues, including apical buds, young leaves, mature leaves and old leaves, were determined with R Language software.
Plant material
‘Fenghuangdancong’ (‘FHDC’), ‘Renhuabaihao’ (‘RHBH’), ‘Yinghongjiuhao’ (‘YH9’) and ‘Baiyedancong’ (‘BYDC’) were cultivated at South China Agricultural University (Guangzhou, China). According to institutional, national and international guidelines, the material used for research purposes does not require specific permissions. The use of rights to these plant materials was obtained by our lab. Apical buds, young leaves, mature leaves and old leaves of the four tea plant cultivars were collected. All the samples were collected in three biological replicates, with two technical replicates for each biological replicate. Some samples were used to conduct trichome observations by stereoscopy (Carl Zeiss, Germany), and the other samples were used for RNA extraction. The samples used for RNA extraction were immediately frozen in liquid nitrogen and stored at -80℃.
qRT-PCR analysis
The total RNA of the tea plant samples was extracted and isolated using the HiPure Total RNA kit (R4111, Magen, China). RNA reverse transcription was carried out with the HiScript III RT SuperMix for qPCR Reagent Kit with gDNA Wiper (R323-01, Vazyme, China). The primers employed for qPCR were designed with the NCBI Primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). All primers are listed in Table S5. SAND1 was used as the reference gene. qRT-PCR analysis was performed with the Bio-Rad CFX384 Touch TM system following routine procedures (Bio-Rad, Hercules, CA, USA)55. Relative expression was calculated using the 2−ΔΔCt method62. All samples were analyzed in three biological replicates, with three technical replicates for each biological replicate.
Subcellular localization
The CDSs of CsbHLH133 and CsbHLH024 without the termination codon were cloned into the pEAQ-EGFP vector. The recombinant plasmids and nuclear localization signal (NLS-DsRed) were transformed into Agrobacterium tumefaciens strain GV3101, which was mixed and injected into tobacco (Nicotiana benthamiana) leaves. After 48 h, the tobacco leaves were collected for fluorescence microscopy observations (Carl Zeiss, Germany).
Dual-luciferase reporter assay
A dual-luciferase reporter assay system (Promega, USA) was used for the determination of transcriptional activation. The full-length CDSs of CsbHLH133 and CsbHLH024 were ligated into the pEAQ-PBD vector and fused with the GAL4 DNA-binding domain under the control of the CaMV 35S promoter. The empty vector, the reporter gene (GAL-LUC) and the generated constructs were transformed into the GV3101 Agrobacterium strain. The Agrobacterium strain containing the empty vector or the constructs and the reporter was coinfiltrated into tobacco leaves. After three days, the tobacco leaves were collected, and the activity of Renilla LUC/firefly was measured according to a previously described protocol44.
Construction of the potential regulatory network
The protein sequences of TFs associated with Arabidopsis thaliana trichome formation, including MYB23, triptychon (TRY), GL3, ENHANCER of TRY and CPC1 (ETC1), GLABRA1 (GL1), GL2, CAPRICE (CPC), TRANSPARENT TESTA GLABRA1 (TTG1), ENHANCER of TRY and CPC2 (ETC2), SENSITIVE TO ABA AND DROUGHT2 (SAD2) and EGL363, were obtained from the TAIR database58. They were submitted to the STRING server for protein–protein interaction network functional enrichment analysis with the default parameters (https://string-db.org).
Yeast two-hybrid assay
The full-length CDSs of CsbHLH024, CsbHLH133 and CsTTG1 were separately cloned into the pGBKT7 and pGADT7 vectors via one-step cloning (C112, Vazyme, China). The resulting positive, negative and recombinant plasmids were subsequently transformed into a Y2HGold yeast strain (YC1002, Weidi Biotechnology, China). A Y2H assay was then performed according to the manufacturer’s instructions (Clontech), and image acquisition was performed via Adobe Illustrator CS2020 (Germany, Zeiss). All primers used are listed in Table S5.
Results and analysis
Identification and conserved domain analysis of CsbHLHs
A total of 134 CsbHLHs were identified in the CSS genome31. The physical and chemical properties of the 134 CsbHLH proteins were predicted. As shown in Table S1, all of the identified proteins encoded 146–1038 amino acids. Their MWs and theoretical pIs ranged from 16.36 kDa to 114.15 kDa and 4.72 to 9.67, respectively. Four CsbHLHs were likely stable (instability index < 40). The spliced sequences in the CCS genome were not clustered on any chromosomes31, and the 134 CsbHLHs were renamed CsbHLH001 to CsbHLH134 based on the gene ID order (Table S1).
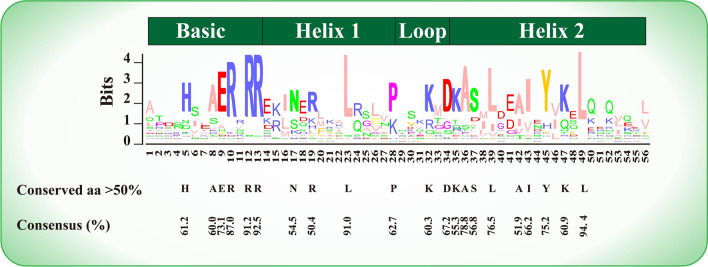
The conserved domains of the CsbHLH proteins were determined via multiple sequence alignment. As shown in Fig. 1, the bHLH domains possessed four conserved regions: basic, first helix, loop and second helix regions. Twenty-one amino acid residues of the bHLH domain were conserved with a consensus ratio greater than 50%: six residues (His-5, Ala-8, Glu-9, Arg-10, Arg-12 and Arg-13) in the basic region; four residues (Asn-17, Arg-19, Leu-23 and Pro-28) in the helix 1 region; two residues (Lys-32 and Asp-34) in the loop region; and nine residues (Lys-35, Ala-36, Ser-37, Leu-39, Ala-42, Ile-43, Tyr-45, Lys-47 and Leu-49) in the helix 2 region. Arg-12, Arg-13, Leu-23 and Leu-49 were highly conserved (Fig. 1). Additionally, the basic region was absent in CsbHLH067, while the loop and helix 2 regions were not present in CsbHLH056 (Fig. S1).
Figure 1.
Conserved residue analysis of bHLH domains. The height of each residue indicates the conservation rate. The black letters represent the residues with a consensus ratio greater than 50%.
Phylogenetic tree analysis of CsbHLH proteins
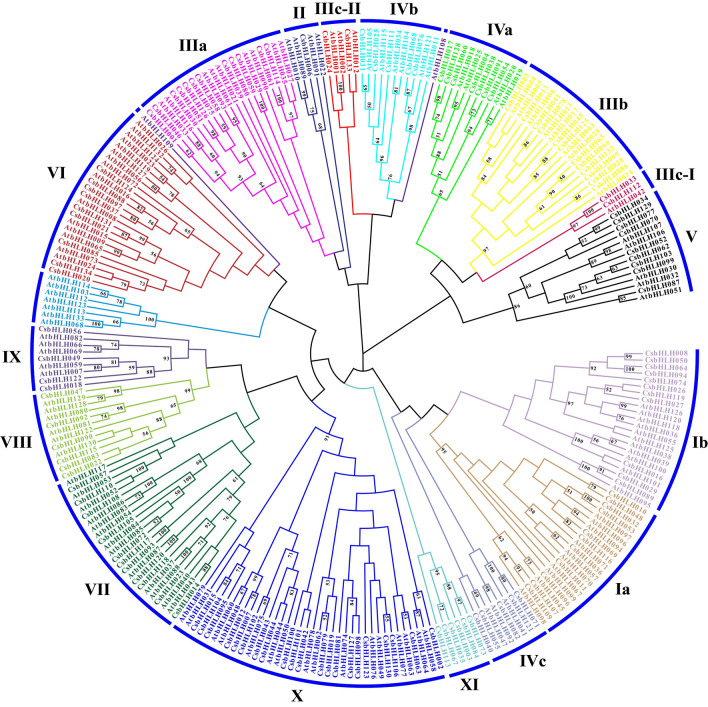
A neighbor-joining phylogenetic tree including all bHLH proteins identified in tea plants and those from Arabidopsis thaliana was constructed for the classification of CsbHLH proteins. The CsbHLH proteins were divided into 17 subgroups according to the classification in Arabidopsis thaliana64 (Fig. 2). Subgroup IIIc was subdivided into subgroups IIIc-I and IIIc-II. The numbers of AtbHLHs and CsbHLHs in each subgroup are listed in Table S2. The members of subgroup II included one CsbHLH and four AtbHLHs. Subgroup X contained the largest numbers of CsbHLHs (21) and AtbHLHs (16). The difference between the members of the CsbHLHs and AtbHLHs within the same group might have resulted from unequal duplication of the bHLH family during species differentiation. To clarify whether the members of 17 subgroups have distinctions in monocots and dicots, all the bHLH proteins of tea plants were used to construct a neighbor-joining phylogenetic tree with those of Arabidopsis thaliana, Oryza sativa L. and Actinidia chinensis (Fig. S2). The results indicated that the members in subgroups IIIc-I and XI were specific to dicots, while the other subgroup included the members of monocots and dicots.
Figure 2.
Phylogenetic tree analysis of tea plant and Arabidopsis thaliana bHLH proteins. Differently colored branches indicate different subgroups. The black Roman numerals indicate the subgroup name of each branch. Two branches did not include CsbHLH members, and they were not included in the subgroups.
TFs classified in the same group in the phylogenetic tree might possess similar functions. Some Arabidopsis thaliana bHLH TFs related to trichome formation were identified, including Transparent Testa8 (TT8)65, EGL366, GL325, and Myelocytomatosis1 (MYC1)67. They were mapped to AtbHLH042, AtbHLH001, AtbHLH002 and AtbHLH012, respectively. All of these TFs were included in subgroups IIIc-I and IIIc-II (Fig. 2). Therefore, subgroups IIIc-I and IIIc-II were defined as ‘trichome-related groups’, and their members might be involved in tea trichome formation.
In addition, the conserved motifs and gene structures of the CsbHLHs were analyzed. Information on 15 identified motifs is listed in Table S3. The results showed that members of the same group might possess similar motifs and gene structures (Fig. S3).
Transcriptome analysis of CsbHLHs in different developmental leaf tissues in tea plants
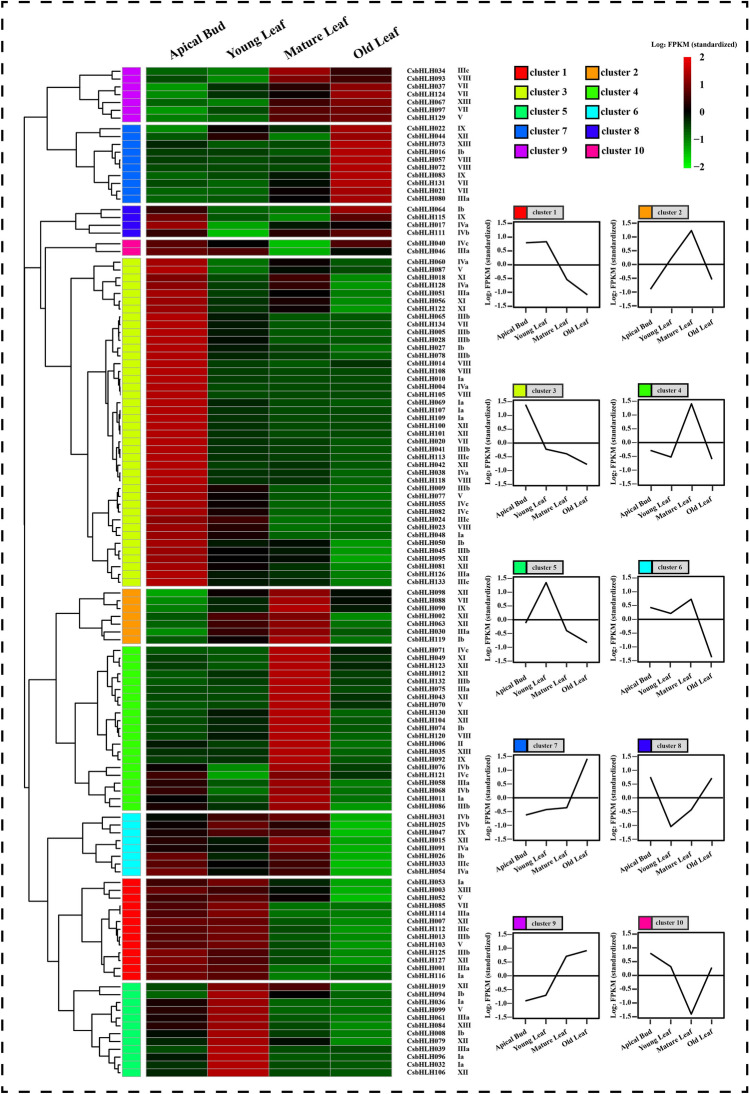
Leaf trichomes are distributed mainly in the apical buds and young leaves. To further understand the potential function of CsbHLH proteins during leaf trichome formation in tea plants, the expression patterns of CsbHLHs in different developmental leaf tissues were determined, including apical buds, young leaves, mature leaves and old leaves. The RNA-seq data of CsbHLHs in different developmental leaf tissues were downloaded from TPIA31. Eight CsbHLHs (CsbHLH029, CsbHLH059, CsbHLH062, CsbHLH066, CsbHLH089, CsbHLH102, CsbHLH110 and CsbHLH117) might be transcribed at low levels in the different developmental leaf tissues, which could not be quantified. According to the similarity of the observed expression patterns, the heatmap was hierarchically clustered into 10 clusters (Fig. 3). The expression patterns of CsbHLHs in clusters 1, 3 and 5 were consistent with the distribution of tea leaf trichomes and primarily associated with apical buds and young leaves. CsbHLHs in clusters 2, 4, 6, 7 and 9 were highly expressed in the mature and old leaves of tea plants, while the expression of CsbHLHs in clusters 8 and 10 was high in the apical buds and old leaves. The members of clusters 1, 3 and 5 might be involved in trichome formation in tea plants.
Figure 3.
Transcriptome analysis of CsbHLHs in different developmental leaf tissues. The name of each gene and the short name of the phylogenetic group are listed on the right of the heatmap. Line charts were generated using the mean value for the whole cluster. Log2 values of fragments per kilobase of exon per million fragments mapped (FPKM) were used to construct the heat map according to the hierarchical clustering analysis.
Additionally, trichome formation is closely related to root hair formation in plants. In Arabidopsis thaliana, an MBW transcriptional activator complex can promote trichome formation and inhibit root hair formation by inducing GL2 expression47. Therefore, the expression profiles of CsbHLHs of cluster 1, cluster 3 and cluster 5 in eight different tissues, including the apical buds, young leaves, mature leaves, old leaves, root, flower, fruit and stem, were investigated. The RNA-seq data of CsbHLHs in eight different tissues were downloaded from TPIA31. The results showed that high expression of CsbHLHs was observed in the tender tissues (apical buds and young leaves), while low expression was observed in the roots of tea plants (Fig. S4).
Expression patterns of CsbHLHs in different developmental leaf tissues in tea plants
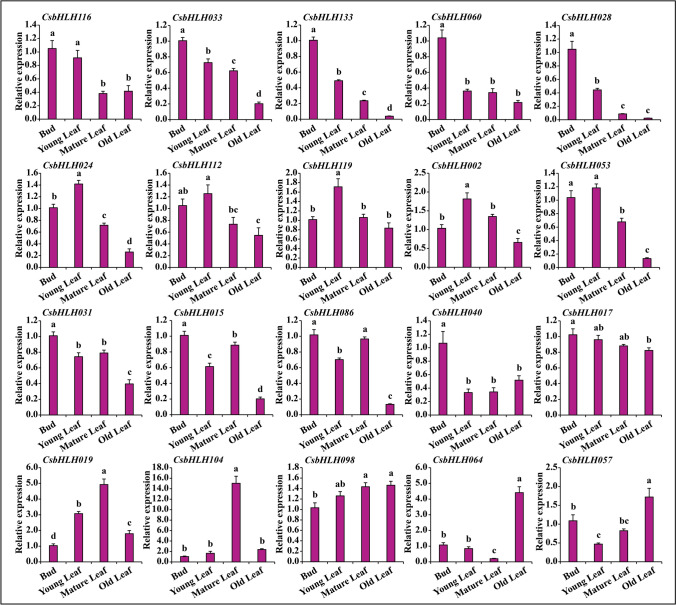
To verify the expression patterns of CsbHLHs in different developmental leaf tissues, twenty CsbHLHs were analyzed using qRT-PCR. The expression of CsbHLH116, CsbHLH033, CsbHLH133, CsbHLH060, CsbHLH028 and CsbHLH040 was upregulated in apical bud tissue, while the expression of CsbHLH024, CsbHLH112, CsbHLH119, CsbHLH002 and CsbHLH053 exhibited a peak in young leaf tissue (Fig. 4). The expression of all of them decreased with leaf maturation, which was similar to the distribution of tea trichomes. Comprehensive and systematic analysis of the topology of the phylogenetic tree and the expression pattern indicated that CsHLH024 and CsbHLH133 were likely candidates for the regulation of trichome formation in tea plants.
Figure 4.
Expression patterns of CsbHLHs in different developmental leaf tissues in the cultivar ‘FHDC’. The different letters in the figures indicate significantly different values (P < 0.05, Tukey’s test). For all developmental leaf tissues, three biological replicates and three technical replicates of each biological replicate were performed.
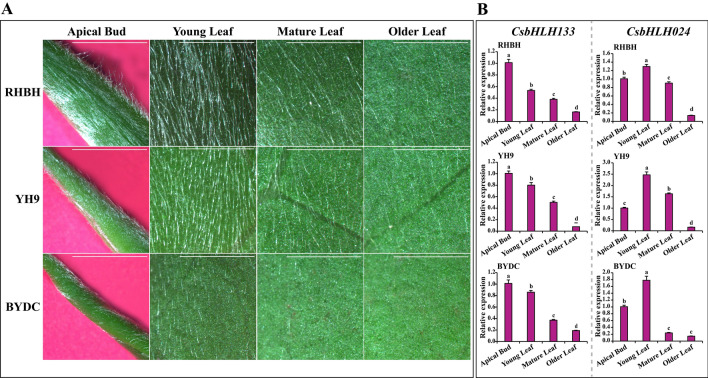
Expression patterns of CsbHLH024 and CsbHLH133 in different tea plant cultivars
To further understand the distribution of leaf trichomes, leaves of different tea plant cultivars were observed using stereoscopy. As shown in Fig. 5A, the tender tissues (apical buds and young leaves) showed more attached trichomes than the mature tissues (mature leaves and old leaves). To understand the expression patterns of CsbHLH024 and CsbHLH133 in different developmental leaf tissues of different tea plant cultivars, their transcriptional levels were determined using qRT-PCR. As shown in Fig. 5B, the expression patterns of CsbHLH133 and CsbHLH024 maintained good agreement with the trichome distribution in different tea plant cultivars. CsbHLH133 was highly expressed in apical bud tissue, while CsbHLH024 expression was upregulated in young leaf tissue. Moreover, the expression of CsbHLH133 and CsbHLH024 decreased with the degree of leaf senescence in different tea plant cultivars, which was similar to the trichome distribution in tea plants.
Figure 5.
The trichome distribution and expression patterns of CsbHLH024 and CsbHLH133 in different tea plant cultivars. A. The trichome distribution in cultivars ‘RHBH’, ‘YH9’ and ‘BYDC’. Scale bars = 5 mm. B. The expression patterns of CsbHLH024 and CsbHLH133 in different developmental leaf tissues of different tea plant cultivars. The different letters in the figures indicate significantly different values (P < 0.05, Tukey’s test). For all samples, three biological replicates and three technical replicates of each biological replicate were performed.
CsbHLH133 and CsbHLH024 act as transcriptional activators
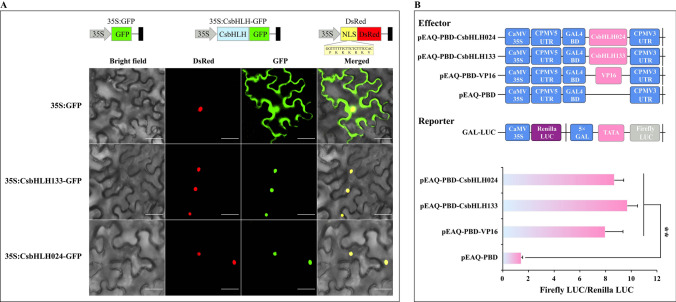
To verify whether CsbHLH133 and CsbHLH024 have transcriptional activation ability, subcellular location and transcriptional activation assays were performed. As shown in Fig. 6A, fluorescence signals from the empty vector were located in the cell nucleus and cytoplasm, while those of the 35S:CsbHLH133-GFP and 35S:CsbHLH024-GFP proteins were found in the cell nucleus. These results indicated that CsbHLH133 and CsbHLH024 were localized in the cell nucleus. A dual-luciferase reporter assay showed that CsbHLH024 and CsbHLH133 could strongly enhance the activity of the reporter. The results confirmed that CsbHLH024 and CsbHLH133 served as activators with transcriptional activity in planta (Fig. 6B).
Figure 6.
Transcriptional activity of CsbHLH133 and CsbHLH024. (A) The subcellular location of CsbHLH133 and CsbHLH024 in Nicotiana benthamiana. Scale bars = 50 μm. (B) The transcriptional activation of CsbHLH133 and CsbHLH024 in planta. The data are presented as the means ± SDs (n = 7). Empty vector (PBD) and pBD-VP16 were used as negative and positive controls, respectively. Significant differences were determined using Student’s t-test by comparison to the negative control (**, P < 0.01).
Potential protein regulatory network of trichome formation
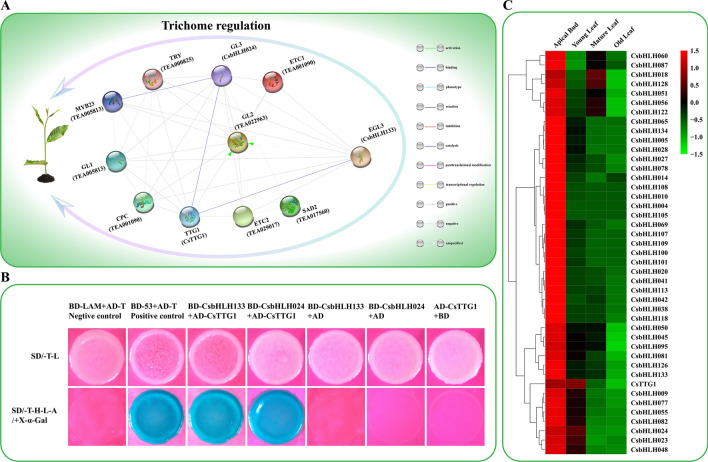
bHLH TFs usually interact with other TFs to regulate plant growth and development. A potential functional protein association network in tea plants was constructed based on the regulation of Arabidopsis thaliana trichome formation by multiple TFs, including MYB23, TRY, GL3, ETC1, GL1, GL2, EGL3, SAD2, ETC2, TTG1 and CPC63 (Fig. 7A). The network showed that CsbHLH024 and CsbHLH133 were likely to bind to multiple TFs. A Y2H assay indicated that CsbHLH024 and CsbHLH133 could interact with CsTTG1, which is a regulator of tea trichome formation (Fig. 7B). In addition, the expression of CsTTG1 maintained good agreement with that of CsbHLH024 and CsbHLH133 (Fig. 7C). These results suggested that CsbHLH024 and CsbHLH133 might regulate trichome formation by interacting with multiple TFs.
Figure 7.
Functional protein association network of trichome formation and a Y2H assay. (A) The potential tea trichome regulatory network. A functional protein association network was constructed based on the TFs associated with Arabidopsis thaliana trichome formation. Homologous genes were found in the CSS genome, including CsbHLH024 and CsbHLH133. The name of homologous CsbHLH is shown in brackets under Arabidopsis thaliana. (B) Yeast two-hybrid assay of protein–protein interactions between CsbHLH024 or CsbHLH133 and CsTTG1. BD and AD represent empty pGBKT7 and pGADT7 vectors, respectively. SD/-T-L: synthetic dextrose medium lacking tryptophan and leucine; SD/-T-H-L-A: synthetic dextrose medium lacking tryptophan, histidine, leucine and adenine. Positive bacteria were stained using X-α-Gal. C. The expression patterns of CsTTG1 and the members of cluster 3 in different developmental leaf tissues. The name of each gene is listed on the right side of the heatmap. Log2 values of FPKM were used to construct the heat map.
Discussion
The identification of gene family members has been widely performed in many plants, and it has contributed to identifying gene functions68–70. Trichomes were conducive not only to plant resistance but also to tea flavor and aroma32. Numerous studies have demonstrated that bHLH TFs contribute to trichome formation45–50. However, whether bHLH TFs are involved in tea trichome formation is still unknown. In this study, genome-wide identification of the tea plant bHLH family was systematically and comprehensively performed. This study provides a further understanding of the relationship between candidate bHLH genes and trichome formation.
A total of 134 CsbHLH genes were identified in the CSS genome. The different plant species possessed different numbers of bHLH members, which ranged from 85 to 31971,72. Members of the bHLH family were identified in Ginkgo biloba (85)71, Solpinganum tuberosum L. (124)73, Solanum lycopersicum (159)74, Oryza sativa L. (167)75, Malus pumila (188)76 and Glycine max (319)72. Twenty-one amino acid residues were conserved in the bHLH domain of tea plants with a consensus rate greater than 50% (Fig. 1), as observed in previous studies38,77. Glu-13 and Arg-16 (according to Glu-9 and Arg-12 in our alignment) could bind to the E-box; His-9, Glu-13 and Arg-17 (according to His-5, Glu-9 and Arg-13 in our alignment) could recognize the G-box39,40,78; Glu-13 and Arg-17 (according to Glu-9 and Arg-13 in our alignment) were important for DNA binding, and Leu-27 (according to Leu-23 in our alignment) played a vital role in dimerization activity in the bHLH domain79,80.
All CsbHLH proteins were divided into 17 subgroups according to the Arabidopsis thaliana classification64. Members of the same group in the phylogenetic tree might possess similar functions. INDUCER OF CBF EXPRESSION1 (ICE1) and INDUCER OF CBF EXPRESSION2 (ICE2) were related to the cold acclimation response and freezing tolerance in Arabidopsis thaliana81,82. They were mapped to AtbHLH116 and AtbHLH033 and located in subgroup IIIa. CsICE1 might be involved in the ICE1-C-repeat binding factor (CBF) cold response pathway in tea plants83, which was mapped to CsbHLH001 and classified into subgroup IIIa. FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) was mapped to AtbHLH029 and included in subgroup IIIa. It was responsive to iron deficiency in Arabidopsis thaliana roots84. The members of subgroup IIIa were likely to be involved in the response to abiotic stress. The bHLHs associated with Arabidopsis thaliana trichome formation were contained in subgroups IIIc-I and IIIc-II (Fig. 2). These two subgroups were defined as ‘trichome-related groups’ in this study. CsbHLH024 and CsbHLH133, the homologs of GL3 and EGL3 in Arabidopsis thaliana, belonged to ‘trichome-related groups’ (Fig. 2). In addition, the members of the same group exhibited similar gene structures and motifs (Fig. S4), which also indicated that the genes within the same group might play similar roles.
Expression pattern analysis facilitated the understanding of gene function. The expression patterns of clusters 1, 3 and 5 were in agreement with the tea trichome distribution (Fig. 3) and focused on tender tissues (apical buds and young leaves). CsbHLH024 and CsbHLH133 were divided into cluster 3. Their expression peaked in the tender tissues of different tea plant cultivars, including apical buds and young leaves (Fig. 5B). Therefore, CsHLH024 and CsbHLH133 might be associated with trichome formation in tea plants. Moreover, CsHLH024 and CsbHLH133 were located in the cell nucleus (Fig. 6A) and possessed transcriptional activity functions (Fig. 6B). The homologs of CsbHLH024 and CsbHLH133 usually regulate trichome formation by interacting with other TFs in Arabidopsis thaliana. CsTTG1 was involved in tea plant trichome formation, and the overexpression of CsTTG1 could enhance the trichome density of Arabidopsis thaliana85; the functions of other Clusters of Orthologous Groups (COGs) (MYB23, TRY, ETC1, GL1, GL2, SAD2, ETC2 and CPC) in trichome formation were less known in tea plants. CsbHLH024 and CsbHLH133 could interact with CsTTG1 in the heterologous system (Fig. 7B). CsbHLH024 and CsbHLH133 might be associated with the regulation of tea plant trichome formation by interacting with CsTTG1.
However, the regeneration rate of tea plant explants in vitro is low because the tea plant is a perennial woody species. Tea plant tissues are rich in the polyphenols. Polyphenols can inhibit the activity of Agrobacterium tumefaciens, which results in low efficiency tea plant genetic transformation86. A stable genetic transformation system for tea plants is still unavailable and needs further exploration. Thus, the functions of candidate CsbHLH TFs in the regulation of trichome formation must be further addressed using multiple methods.
Conclusions
In total, 134 CsbHLH proteins were identified in the CSS genome. Phylogenetic tree, gene structure and protein motif analyses of these proteins were conducted in this study. All CsbHLH proteins were divided into 17 subgroups. Subgroups IIIc-I and IIIc-II were defined as the ‘trichome-related groups’, and their members were likely to be associated with trichome formation. The members of clusters 1, 3 and 5 were candidates for trichome formation in tea plants. Notably, CsbHLH024 and CsbHLH133 classified into the ‘trichome-related group’ and included in cluster 3 were highly expressed in the tender tissues of different tea plant cultivars. The expression of CsbHLH024 and CsbHLH133 was similar to the trichome distribution in tea plants. In addition, CsbHLH024 and CsbHLH133 were located in the cell nucleus. They possessed transcriptional activation ability and might control trichome formation by interacting with CsTTG1. This study provides useful information for the further study of CsbHLH TF function in the regulation of trichome formation in tea plants.
Supplementary Information
Acknowledgements
The authors thank the lab members for assistance.
Abbreviations
- bHLH
Basic helix-loop-helix
- TFs
Transcription factors
- CSS
Camellia sinensis Var. sinensis
- Y2H
Yeast two-hybrid
- WDR
WD40-repeat
- MBW
MYB-BHLH-WDR
- HLH
Helix-loop-helix
- GL3
GLABRA3
- EGL3
ENHANCER OF GLABRA3
- GL2
GLABRA2
- FRC4
FURCA4
- TPIA
Tea plant information archive
- NCBI
National Center for Biotechnology Information
- HMM
Hidden Markov model
- Mws
Molecular weights
- pIs
Isoelectric points
- MEME
Multiple EM for Motif Elicitation
- CDSs
Coding DNA sequences
- TAIR
The Arabidopsis Information Resource
- ‘FHDC’
‘Fenghuangdancong’
- ‘RHBH’
‘Renhuabaihao’
- ‘YH9’
‘Yinghongjiuhao’
- ‘BYDC’
‘Baiyedancong’
- NLS
Nuclear localization signal
- TRY
Triptychon
- ETC1
ENHANCER of TRY and CPC1
- GL1
GLABRA1
- CPC
CAPRICE
- TTG1
Transparent TESTA GLABRA1
- ETC2
ENHANCER of TRY and CPC2
- SAD2
Sensitive to ABA and drought2
- TT8
Transparent Testa8
- MYC1
Myelocytomatosis1
- FPKM
Fragments per kilobase of exon per million fragments mapped
- ICE1
Inducer OF CBF expression1
- ICE2
Inducer OF CBF expression2
- CBF
C-repeat binding factor
- FIT
FER-like iron deficiency-induced transcription factor
- COGs
Clusters of Orthologous Groups
Author contributions
R.J.L. performed most of the experiments and wrote the manuscript. Y.Y.W. analyzed the data. S.T. and J.R.C. provided useful suggestions regarding the study. P.Z., B.M.S. and S.Q.L. supervised the research.
Funding
This work was supported by the National Natural Science Foundation of Guangdong Province (2018A030313089).
Data availability
Most data generated or analyzed during this study are included in this article and its supplemental files. The sequencing data used and analyzed during this study are available in the TPIA database (http://tpia.teaplant.org/index.html).
Materials availability
The tea plant cultivars used in this study were cultivated at South China Agricultural University (Guangzhou, China). The rights of these plant materials were obtained by our lab.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peng Zheng, Email: zhengp@scau.edu.cn.
Binmei Sun, Email: binmei@scau.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90205-7.
References
- 1.Schilmiller AL, Last RL, Pichersky E. Harnessing plant trichome biochemistry for the production of useful compounds. Plant J. 2008;54:702–711. doi: 10.1111/j.1365-313X.2008.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Werker E. Trichome diversity and development. Adv. Bot. Res. 2000;31:1–35. doi: 10.1016/S0065-2296(00)31005-9. [DOI] [Google Scholar]
- 3.Serna L, Martin C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 2006;11:274–280. doi: 10.1016/j.tplants.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- 5.Wagner GJ, Wang E, Shepherd RW. New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 2004;93:3–11. doi: 10.1093/aob/mch011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glas JJ, et al. Plant glandular trichomes as targets for breeding or engineering of resistance to herbivores. Int. J. Mol. Sci. 2012;13:17077–17103. doi: 10.3390/ijms131217077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergau N, Bennewitz S, Syrowatka F, Hause G, Tissier A. The development of type VI glandular trichomes in the cultivated tomato solanum lycopersicum and a related wild species S. habrochaites. BMC Plant Biol. 2015;15:289. doi: 10.1186/s12870-015-0678-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue C, et al. Comparative transcriptome study of hairy and hairless tea plant (Camellia Sinensis) Shoots. J. Plant Physiol. 2018;229:41–52. doi: 10.1016/j.jplph.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Bleeker PM, et al. Improved herbivore resistance in cultivated tomato with the sesquiterpene biosynthetic pathway from a wild relative. Proc. Natl. Acad. Sci. U S A. 2012;109:20124–20129. doi: 10.1073/pnas.1208756109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luu VT, et al. O-acyl sugars protect a wild tobacco from both native fungal pathogens and a specialist herbivore. Plant Physiol. 2017;174:370–386. doi: 10.1104/pp.16.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamasaki S, Murakami Y. Continuous UV-B irradiation induces endoreduplication and trichome formation in cotyledons, and reduces epidermal cell division and expansion in the first leaves of pumpkin seedlings (Cucurbita Maxima Duch. × C. Moschata Duch.) Environ. Control Biol. 2014;52:203–209. doi: 10.2525/ecb.52.203. [DOI] [Google Scholar]
- 12.Agrawal AA, Conner JK, Stinchcombe JR. Evolution of plant resistance and tolerance to frost damage. Ecol. Lett. 2004;7:1199–1208. doi: 10.1111/j.1461-0248.2004.00680.x. [DOI] [Google Scholar]
- 13.Benz BW, Martin CE. Foliar trichomes, boundary layers, and gas exchange in 12 species of epiphytic Tillandsia (Bromeliaceae) J. Plant Physiol. 2006;163:648–656. doi: 10.1016/j.jplph.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Skaltsa H, Verykokidou E, Harvala C, Karabourniotis G, Manetasi Y. UV-B protective potential and flavonoid content of leaf hairs of Quercus Ilex. Phytochemistry. 1994;37:987–990. doi: 10.1016/S0031-9422(00)89514-X. [DOI] [Google Scholar]
- 15.Chang J, et al. Hair, encoding a single C2H2 zinc-finger protein, regulates multicellular trichome formation in tomato. Plant J. 2018;96:90–102. doi: 10.1111/tpj.14018. [DOI] [PubMed] [Google Scholar]
- 16.Sun WQ, et al. Hairy leaf 6, an AP2/ERF transcription factor, interacts with OsWOX3B and regulates trichome formation in rice. Mol. Plant. 2017;10:1417–1433. doi: 10.1016/j.molp.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhang N, et al. Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis. BMC Plant Biol. 2018;18:63. doi: 10.1186/s12870-018-1271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu YH, et al. NbGIS regulates glandular trichome initiation through GA signaling in tobacco. Plant Mol. Biol. 2018;98:153–167. doi: 10.1007/s11103-018-0772-3. [DOI] [PubMed] [Google Scholar]
- 19.Yang S, et al. A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 2018;69:1887–1902. doi: 10.1093/jxb/ery047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, et al. Genetic basis for glandular trichome formation in cotton. Nat. Commun. 2016;7:10456. doi: 10.1038/ncomms10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu SL, et al. A Pd1-Ps-P1 feedback loop controls pubescence density in soybean. Mol. Plant. 2020;13:1768–1783. doi: 10.1016/j.molp.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Marks MD. Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:137–163. doi: 10.1146/annurev.arplant.48.1.137. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirik V, et al. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development. 2005;132:1477–1485. doi: 10.1242/dev.01708. [DOI] [PubMed] [Google Scholar]
- 25.Morohashi K, et al. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 2007;145:736–746. doi: 10.1104/pp.107.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szymanski DB, Jilk RA, Pollock SM, Marks MD. Control of GL2 expression in Arabidopsis leaves and trichomes. Development. 1998;125:1161–1171. doi: 10.1242/dev.125.7.1161. [DOI] [PubMed] [Google Scholar]
- 27.Zheng KJ, et al. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 2016;6:19254. doi: 10.1038/srep19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia EH, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Liu ZH, et al. Leading progress on genomics, health benefits and utilization of tea resources in China. Nature. 2019;566:S15–S19. doi: 10.1038/s41586-019-0908-x. [DOI] [Google Scholar]
- 30.Gilbert N. The science of tea's mood-altering magic. Nature. 2019;566:S8–S9. doi: 10.1038/d41586-019-00398-1. [DOI] [PubMed] [Google Scholar]
- 31.Wei CL, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U S A. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li PH, et al. Metabolite profiling and transcriptome analysis revealed the chemical contributions of tea trichomes to tea flavors and tea plant defenses. J. Agric. Food Chem. 2020;68:11389–11401. doi: 10.1021/acs.jafc.0c04075. [DOI] [PubMed] [Google Scholar]
- 33.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- 34.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin J, et al. A Basic Helix-Loop-Helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2015;2:15059. doi: 10.1038/hortres.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song XM, et al. Genome-Wide analysis of the bHLH transcription factor family in Chinese Cabbage (Brassica rapa ssp. Pekinensis) Mol. Genet. Genom. 2014;289:77–91. doi: 10.1007/s00438-013-0791-3. [DOI] [PubMed] [Google Scholar]
- 37.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, Daughterless, MyoD, and Myc Proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-X. [DOI] [PubMed] [Google Scholar]
- 38.Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 1999;48:501–516. doi: 10.1007/PL00006494. [DOI] [PubMed] [Google Scholar]
- 39.Massari ME, Murre C. Helix-Loop-Helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO. J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ledent V, Vervoort M. The Basic Helix-Loop-Helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun X, Wang Y, Sui N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018;503:397–401. doi: 10.1016/j.bbrc.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 43.Mertens J, et al. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016;170:194–210. doi: 10.1104/pp.15.01645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun BM, et al. Purple foliage coloration in tea (Camellia Sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016;6:32534. doi: 10.1038/srep32534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 47.Rerie WG, Feldmann KA, Marks MD. The GLABRA2 Gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- 48.Zhao MZ, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory Loci. Development. 2008;135:1991–1999. doi: 10.1242/dev.016873. [DOI] [PubMed] [Google Scholar]
- 49.Balkunde R, Pesch M, Hulskamp M. Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 2010;91:299–321. doi: 10.1016/S0070-2153(10)91010-7. [DOI] [PubMed] [Google Scholar]
- 50.Luo D, Oppenheimer DG. Genetic control of trichome branch number in Arabidopsis: The roles of the FURCA Loci. Development. 1999;126:5547–5557. doi: 10.1242/dev.126.24.5547. [DOI] [PubMed] [Google Scholar]
- 51.Tominaga-Wada R, Nukumizu Y, Sato S, Wada T. Control of plant trichome and root-hair development by a tomato (Solanum Lycopersicum) R3 MYB transcription factor. PLoS ONE. 2013;8:e54019. doi: 10.1371/journal.pone.0054019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu SN, et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn RD, et al. The pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gasteiger E, et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen CJ, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian F, Yang DC, Meng YQ, Jin JP, Gao G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48:D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian B, Gao SH, Lercher MJ, Hu SN, Chen WH. Evolview V3: A webserver for visualization, Annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–W275. doi: 10.1093/nar/gkz357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Schellmann S, Hulskamp M. Epidermal differentiation: Trichomes in Arabidopsis as a model system. Int. J. Dev. Biol. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- 64.Heim MA, et al. The Basic Helix-Loop-Helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 65.Nesi N, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis Siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernhardt C, et al. The bHLH genes GLABRA3 (GL3) and ENHANCER of GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis Root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- 67.Pesch M, Schultheiss I, Digiuni S, Uhrig JF, Hulskamp M. Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development. 2013;140:3456–3467. doi: 10.1242/dev.094698. [DOI] [PubMed] [Google Scholar]
- 68.Peng XJ, Liu H, Wang D, Shen SH. Genome-wide identification of the Jatropha Curcas MYB family and functional analysis of the abiotic stress responsive gene JcMYB2. BMC Genom. 2016;17:251. doi: 10.1186/s12864-016-2576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song JL, et al. Systematic analysis of the Capsicum ERF transcription factor family: Identification of regulatory factors involved in the regulation of species-specific metabolites. BMC Genom. 2020;21:573. doi: 10.1186/s12864-020-06983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jue DW, et al. Identification of WRKY gene family from Dimocarpus Longan and its expression analysis during flower induction and abiotic stress responses. Int. J. Mol. Sci. 2018;19:2169. doi: 10.3390/ijms19082169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou X, et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo Biloba. Sci. Rep. 2020;10:13723. doi: 10.1038/s41598-020-69305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hudson KA, Hudson ME. A classification of Basic Helix-Loop-Helix transcription factors of Soybean. Int. J. Genom. 2015;2015:603182. doi: 10.1155/2015/603182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang RQ, et al. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes. 2018;9:51. doi: 10.3390/genes9010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in Tomato. BMC Genom. 2015;16:9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li XX, et al. Genome-wide analysis of Basic/Helix-Loop-Helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mao K, Dong QL, Li C, Liu CH, Ma FW. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 2017;8:480. doi: 10.3389/fpls.2017.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui X, et al. Transcriptome-wide identification and expression profile analysis of the bHLH family genes in Camellia Sinensis. Funct. Integr. Genom. 2018;18:489–503. doi: 10.1007/s10142-018-0608-x. [DOI] [PubMed] [Google Scholar]
- 78.Shimizu T, et al. Crystal structure of PHO4 bHLH domain-DNA complex: Flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Atchley WR, Fitch WM. A natural classification of the Basic Helix-Loop-Helix class of transcription factors. Proc. Natl. Acad. Sci. U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carretero-Paulet L, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chinnusamy V, et al. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fursova OV, Pogorelko GV, Tarasov VA. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene. 2009;429:98–103. doi: 10.1016/j.gene.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Jiang CJ, Li YY, Wei CL, Deng WW. CsICE1 and CsCBF1: Two transcription factors involved in cold responses in Camellia Sinensis. Plant Cell Rep. 2012;31:27–34. doi: 10.1007/s00299-011-1136-5. [DOI] [PubMed] [Google Scholar]
- 84.Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G. Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J. 2011;66:1044–1052. doi: 10.1111/j.1365-313X.2011.04565.x. [DOI] [PubMed] [Google Scholar]
- 85.Sun BM, et al. TRANSPARENT TESTA GLABRA1 (TTG1) regulates leaf trichome density in tea Camellia Sinensis. Nord. J. Bot. 2020 doi: 10.1111/njb.02592. [DOI] [Google Scholar]
- 86.Mondal TK, Bhattacharya A, Laxmikumaran M, Singh Ahuja P. Recent advances of tea (Camellia Sinensis) biotechnology. Plant Cell Tissue Organ Cult. 2004;76:195–254. doi: 10.1023/B:TICU.0000009254.87882.71. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most data generated or analyzed during this study are included in this article and its supplemental files. The sequencing data used and analyzed during this study are available in the TPIA database (http://tpia.teaplant.org/index.html).
The tea plant cultivars used in this study were cultivated at South China Agricultural University (Guangzhou, China). The rights of these plant materials were obtained by our lab.