Abstract
Fibroblast growth factor 21 (FGF21) is a human metabolic hormone whose effects include modification of macronutrient preference and energy homeostasis. In animal models, FGF21 has been shown to have beneficial effects on cardiometabolic outcomes, Alzheimer’s disease risk and lifespan. In this study, the single-nucleotide polymorphism rs838133 in the FGF21 gene region was leveraged to investigate the potential clinical effects of targeting FGF21. The FGF21 G allele was associated with lower intakes of total sugars and alcohol, and higher intakes of protein and fat as well as favourable with lipid levels, blood pressure traits, waist-to-hip ratio, systemic inflammation, cardiovascular outcomes, Alzheimer’s disease risk and lifespan. These findings may be used to anticipate the effects of pharmacologically increasing FGF21 signalling.
Keywords: alcohol, cardiovascular disease, fibroblast growth factor 21, macronutrients, sugar, mendelian randomization
1. Introduction
Fibroblast growth factor 21 (FGF21) is a human metabolic hormone that is expressed in the liver [1]. The effects of FGF21 include altering of macronutrient preference and energy homeostasis [1,2,3]. Increased circulating FGF21 decreases the consumption of sweets, sugar and alcohol [2,4,5], and indirectly increases protein intake by suppressing sugar consumption [6]. In rodents and non-human primates, FGF21 treatment has beneficial effects on cardiometabolic outcomes, such as reduction in fat mass and alleviation of hyperglycaemia, insulin resistance, dyslipidaemia, and cardiovascular diseases [7]. Furthermore, FGF21 has been implicated in protecting against Alzheimer’s disease [8], as well as improving lifespan [9]. However, studies investigating the association between circulating FGF21 and these clinical outcomes in humans have been inconclusive [7,10]. To investigate this further, we aimed to leverage human genetic data within the Mendelian randomization paradigm to investigate the associations of a common allele in the FGF21 gene with cardiometabolic outcomes, Alzheimer’s disease and lifespan [11].
2. Materials and Methods
We used the single-nucleotide polymorphism rs838133 in the FGF21 gene region, previously shown to be associated with intake of macronutrients, alcohol and sweets [10,12], to assess potential effects of increasing FGF21 concentrations. Publicly available summary-level data for the associations of the FGF21 variant with macronutrient and alcohol intake and other outcomes were obtained from the UK Biobank cohort and genome-wide association study consortia [13,14,15,16,17,18,19,20,21,22,23,24]. Details of the outcome datasets used in this study are provided in Table S1. For cardiometabolic diseases, we meta-analysed the estimates from the different data sources using inverse-variance models with fixed effects.
Only summary-level (i.e., aggregated) data were analysed in this study, for which appropriate ethical approval and participant consent had previously been obtained. The present Mendelian randomization analyses were approved by the Swedish Ethical Review Authority. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this work is consistent with those guidelines.
3. Results
The G (major) allele of rs838133 in the FGF21 gene region was associated with lower intakes of total sugars and alcohol, and higher intakes of protein and fat (Figure 1), consistent with the expected effect of an increase in FGF21 concentration. We scaled all results per additional G allele to mimic the effect of elevated FGF21 concentrations.
Figure 1.
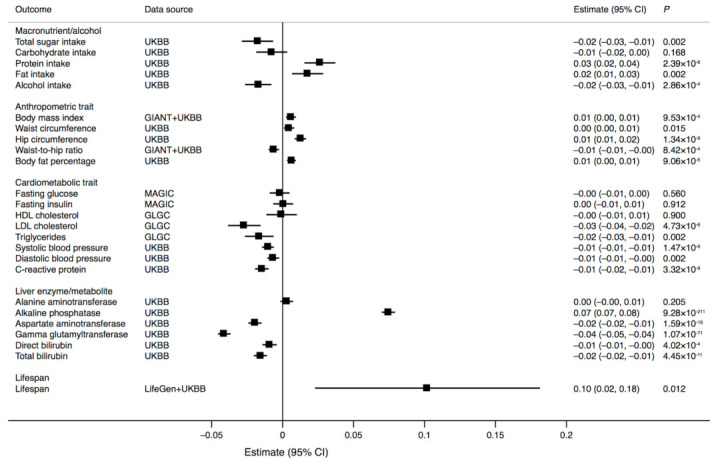
Associations of the FGF21 rs838133 G allele with macronutrient intake, anthropometric and cardiometabolic traits, liver enzymes and metabolites, and lifespan. The outcomes are in standard deviation units except for lifespan (in years). CI: confidence interval.
The FGF21 rs838133 G allele was associated with greater body mass index, body fat percentage and waist and hip circumferences, but with lower waist-to-hip ratio (Figure 1). Additionally, the G allele was associated with lower low-density lipoprotein cholesterol and triglyceride concentrations, lower systolic and diastolic blood pressure, and lower C-reactive protein concentrations, but was not associated with fasting glucose or fasting insulin concentrations (Figure 1). There was a positive association of the FGF21 rs838133 G allele with the liver enzyme alkaline phosphatase, but a negative association with aspartate aminotransferase, gamma glutamyltransferase, and direct and total bilirubin concentrations (Figure 1). There was suggestive evidence of a positive association between the FGF21 rs838133 G allele and lifespan (based on parental lifespan) (Figure 1).
In analyses of cardiovascular diseases, the FGF21 rs838133 G allele was strongly associated with a reduced risk of venous thromboembolism, and had suggestive inverse associations with coronary artery disease, heart failure, and ischemic stroke (Figure 2). There was a suggestive association of the FGF21 rs838133 G allele with reduced risk of Alzheimer’s disease (based on clinically diagnosed Alzheimer’s disease and Alzheimer’s disease by proxy cases and their corresponding controls), but no association with type 2 diabetes (Figure 2).
Figure 2.
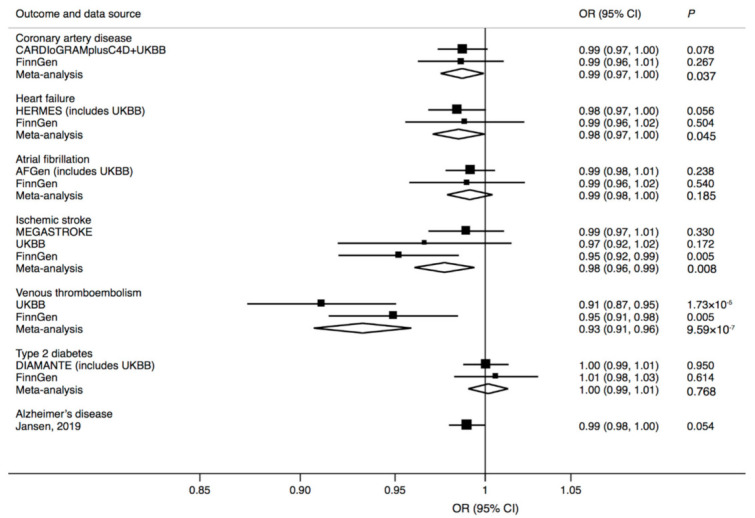
Associations of the FGF21 rs838133 G allele with cardiometabolic diseases and Alzheimer’s disease. CI: confidence interval; OR: odds ratio.
4. Discussion
This work leveraged human genetic data to provide insight into the broad metabolic and clinical effects of the major (G) allele of rs838133 in the FGF21 gene. Our results support previously reported associations between the FGF21 rs838133 variant with macronutrient and alcohol intake, lipid levels, blood pressure, waist-to-hip ratio, and liver enzymes as well as the lack of association with type 2 diabetes in the UK Biobank cohort [10]. It should be noted that the previous study used the minor (A) allele of the rs838133 variant as the effect allele [10] and thus showed associations in the opposite direction to our findings. The present study went further to provide novel evidence that the major allele of rs838133 in the FGF21 gene was associated with decreased systemic inflammation (estimated by C-reactive protein concentration). We also identified a potential beneficial effect of the major allele at FGF21 rs838133 on cardiovascular outcomes, with the strongest association for venous thromboembolism, as well as on Alzheimer’s disease and lifespan.
These findings inform on the potential effects of pharmacologically increasing FGF21 concentrations or signalling. The limitations of this study should also be acknowledged. Critically, it is possible that some of the identified associations may be attributable to genetic confounding, where the FGF21 genetic variant also has pleiotropic associations unrelated to FGF21. As summary genetic association data for circulating FGF21 concentrations were not available, we could not perform colocalization analysis to explore this possibility [25]. Furthermore, the outcomes that we studied were determined by the availability of corresponding large-scale genetic association summary data. As such, it was not possible to perform analyses for other relevant traits, such as non-alcoholic steatohepatitis.
5. Conclusions
In summary, we used a major allele of rs838133 in the FGF21 gene to identify evidence for its effects on macronutrient and alcohol intake as well as favourable effects on a range of cardiometabolic outcomes, Alzheimer’s disease and lifespan. This work anticipates the effects of pharmacologically increasing FGF21 signalling.
Acknowledgments
The authors would like to thank the investigators of the UK Biobank and genetic consortia, including the Coronary ARtery DIsease Genome wide Replication and Meta-analysis plus The Coronary Artery Disease Genetics consortium, DIAMANTE consortium, FinnGen consortium, Genetic Investigation of ANthropometric Traits consortium, Global Lipids Genetics Consortium, Heart Failure Molecular Epidemiology for Therapeutic Targets consortium, International Consortium of Blood Pressure, LifeGen, Meta-Analyses of Glucose and Insulin-related traits Consortium, and MEGASTROKE, for sharing summary statistics genome-wide association study data. The list of investigators of the MEGASTROKE consortium is available at http://megastroke.org/authors.html. Funding of the MEGASTROKE project is specified at megastroke.org/acknowledgements.html.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/nu13051504/s1, Table S1: Data sources for the outcome phenotypes included in the present Mendelian randomization study.
Author Contributions
Conceptualization, S.C.L., D.G.; methodology, S.C.L., D.G.; formal analysis, S.C.L.; writing, S.C.L., D.G.; funding acquisition, S.C.L., D.G. All authors have read and agreed to the published version of the manuscript.
Funding
S.C.L. acknowledges research support from the Swedish Research Council (Vetenskapsrådet, 2016-01042 and 2019-00977), the Swedish Research Council for Health, Working Life and Welfare (Forte, 2018-00123), and the Swedish Heart-Lung Foundation (Hjärt-Lungfonden, 20190247). D.G. is supported by the British Heart Foundation Research Centre of Excellence at Imperial College London (RE/18/4/34215) and by a National Institute for Health Research Clinical Lectureship at St. George’s, University of London (CL-2020-16-001).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Swedish Ethical Review Authority (2019-02793, date of approval 27 May 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data used in this study are publicly available summary statistics data, with relevant data available from cited studies. The summary statistics data analyzed in this study are available on request from the corresponding author.
Conflicts of Interest
D.G. is employed part-time by Novo Nordisk. S.C.L. has no conflict of interest to declare.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Flippo K.H., Potthoff M.J. Metabolic Messengers: FGF21. Nat. Metab. 2021;3:309–317. doi: 10.1038/s42255-021-00354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill C.M., Qualls-Creekmore E., Berthoud H.R., Soto P., Yu S., McDougal D.H., Munzberg H., Morrison C.D. FGF21 and the physiological regulation of macronutrient rreference. Endocrinology. 2020;161:bqaa019. doi: 10.1210/endocr/bqaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tezze C., Romanello V., Sandri M. FGF21 as Modulator of Metabolism in Health and Disease. Front. Physiol. 2019;10:419. doi: 10.3389/fphys.2019.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Holstein-Rathlou S., BonDurant L.D., Peltekian L., Naber M.C., Yin T.C., Claflin K.E., Urizar A.I., Madsen A.N., Ratner C., Holst B., et al. FGF21 Mediates Endocrine Control of Simple Sugar Intake and Sweet Taste Preference by the Liver. Cell Metab. 2016;23:335–343. doi: 10.1016/j.cmet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talukdar S., Owen B.M., Song P., Hernandez G., Zhang Y., Zhou Y., Scott W.T., Paratala B., Turner T., Smith A., et al. FGF21 Regulates Sweet and Alcohol Preference. Cell Metab. 2016;23:344–349. doi: 10.1016/j.cmet.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flippo K.H., Jensen-Cody S.O., Claflin K.E., Potthoff M.J. FGF21 signaling in glutamatergic neurons is required for weight loss associated with dietary protein dilution. Sci. Rep. 2020;10:19521. doi: 10.1038/s41598-020-76593-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng L., Lam K.S.L., Xu A. The therapeutic potential of FGF21 in metabolic diseases: From bench to clinic. Nat. Rev. Endocrinol. 2020;16:654–667. doi: 10.1038/s41574-020-0386-0. [DOI] [PubMed] [Google Scholar]
- 8.Taliyan R., Chandran S.K., Kakoty V. Therapeutic Approaches to Alzheimer’s Type of Dementia: A Focus on FGF21 Mediated Neuroprotection. Curr. Pharm. Des. 2019;25:2555–2568. doi: 10.2174/1381612825666190716101411. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Xie Y., Berglund E.D., Coate K.C., He T.T., Katafuchi T., Xiao G., Potthoff M.J., Wei W., Wan Y., et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. Elife. 2012;1:e00065. doi: 10.7554/eLife.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frayling T.M., Beaumont R.N., Jones S.E., Yaghootkar H., Tuke M.A., Ruth K.S., Casanova F., West B., Locke J., Sharp S., et al. A common allele in FGF21 associated with sugar intake is associated with body shape, lower total body-fat percentage, and higher blood pressure. Cell Rep. 2018;23:327–336. doi: 10.1016/j.celrep.2018.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill D., Burgess S. Use of a Genetic Variant Related to Circulating FXa (Activated Factor X) Levels to Proxy the Effect of FXa Inhibition on Cardiovascular Outcomes. Circ. Genom. Precis. Med. 2020;13:551–553. doi: 10.1161/CIRCGEN.120.003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soberg S., Sandholt C.H., Jespersen N.Z., Toft U., Madsen A.L., von Holstein-Rathlou S., Grevengoed T.J., Christensen K.B., Bredie W.L.P., Potthoff M.J., et al. FGF21 is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metab. 2017;25:1045–1053.e6. doi: 10.1016/j.cmet.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen J.B., Thorolfsdottir R.B., Fritsche L.G., Zhou W., Skov M.W., Graham S.E., Herron T.J., McCarthy S., Schmidt E.M., Sveinbjornsson G., et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., van der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulit S.L., Stoneman C., Morris A.P., Wood A.R., Glastonbury C.A., Tyrrell J., Yengo L., Ferreira T., Marouli E., Ji Y., et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FinnGen Consortium FinnGen Documentation of R4 Release. [(accessed on 10 December 2020)];2020 Available online: https://finngen.gitbook.io/documentation/
- 22.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hagg S., Athanasiu L., et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timmers P.R., Mounier N., Lall K., Fischer K., Ning Z., Feng X., Bretherick A.D., Clark D.W., eQTLGen Consortium. Agbessi M., et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. Elife. 2019;8:e39856. doi: 10.7554/eLife.39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah S., Henry A., Roselli C., Lin H., Sveinbjornsson G., Fatemifar G., Hedman A.K., Wilk J.B., Morley M.P., Chaffin M.D., et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 2020;11:163. doi: 10.1038/s41467-019-13690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are publicly available summary statistics data, with relevant data available from cited studies. The summary statistics data analyzed in this study are available on request from the corresponding author.


