Abstract
The RAS family of oncogenes (HRAS, NRAS, and KRAS) are among the most frequently mutated protein families in cancers. RAS-mutated tumors were originally thought to proliferate independently of upstream signaling inputs, but we now know that non-mutated wild-type (WT) RAS proteins play an important role in modulating downstream effector signaling and driving therapeutic resistance in RAS-mutated cancers. This modulation is complex as different WT RAS family members have opposing functions. The protein product of the WT RAS allele of the same isoform as mutated RAS is often tumor-suppressive and lost during tumor progression. In contrast, RTK-dependent activation of the WT RAS proteins from the two non-mutated WT RAS family members is tumor-promoting. Further, rebound activation of RTK–WT RAS signaling underlies therapeutic resistance to targeted therapeutics in RAS-mutated cancers. The contributions of WT RAS to proliferation and transformation in RAS-mutated cancer cells places renewed interest in upstream signaling molecules, including the phosphatase/adaptor SHP2 and the RasGEFs SOS1 and SOS2, as potential therapeutic targets in RAS-mutated cancers.
Keywords: RAS, KRAS, HRAS, NRAS, SOS1, SOS2, RTK, SHP2, resistance
1. Introduction
The RTK/RAS pathway (Figure 1A) is among the most commonly mutated pathways in cancer [1,2]. The three RAS genes, HRAS, NRAS, and KRAS, encode four highly homologous protein isoforms (HRAS, NRAS, KRAS4A, and KRAS4B), driver mutations in RAS genes occur in ~20% of human tumors (reviewed in [3]). KRAS is the most frequently mutated RAS family member (75% of RAS mutations), including high incidence of mutations in lung [4], colon [5], and pancreatic cancers [6], three of the top four causes of cancer-related death [2,7,8]. HRAS and NRAS mutations are common in other cancer types including head and neck, skin, and hematopoietic cancers [9]. RAS-mutated cancers respond poorly to standard chemotherapy [10,11,12,13,14], so targeted approaches are needed to treat patients with RAS-mutated tumors. While advances in targeting specific mutant RAS proteins have been made [15,16,17,18], the majority of RAS-mutated tumors remain resistant to currently available treatments [4,12,14,19,20]. Novel strategies for targeting the RAS pathway are necessary to provide effective therapeutics to the majority of patients with RAS-mutated cancers. Understanding the signaling context of mutant RAS is key to developing indirect targeting and combination therapy strategies to better manage these cancers.
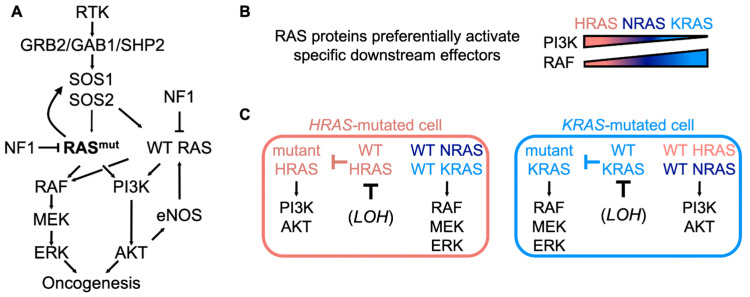
Figure 1.
Mutant RAS and WT RAS cooperate to promote oncogenesis. (A) Schematic showing mutant RAS and WT RAS signaling in RAS-mutated cancer cells. (B) RAS family members show differential activation of downstream RAS effectors. HRAS activates PI3K well but RAF relatively poorly; KRAS activates RAF well but PI3K poorly. (C) Schematic showing proposed activation of RAF/MEK/ERK versus PI3K/AKT signaling in HRAS- and KRAS-mutated cancer cells. In HRAS-mutated cells, mutant HRAS activates PI3K/AKT signaling, whereas RTK-WT N/KRAS activate RAF/MEK/ERK signaling. WT HRAS is tumor suppressive and inhibits mutant HRAS signaling. In KRAS-mutated cells, mutant KRAS activates RAF/MEK/ERK signaling, whereas RTK-WT H/NRAS activate PI3K/AKT signaling. WT KRAS is tumor suppressive and inhibits mutant KRAS signaling.
While typical models of oncogene activation assume that the mutated protein drives oncogenesis separately from the wild-type family members, the evidence that non-mutant wild-type (WT) RAS proteins influence cancer initiation and growth in RAS-mutated cancers is now well-established and several mechanisms for the effects have been proposed. Here, we summarize the current understanding of the effects of WT RAS on RAS-mutated cancers and the proposed mechanisms behind those effects.
2. Contributions of WT RAS to Mutant RAS-Driven Cancers
Non-mutant WT RAS proteins play an important role in modulating downstream effector signaling and oncogenesis in RAS-mutated cancers. While the contributions of WT RAS to RAS-mutated cancers varies based on factors such as the specific RAS isoform that is mutated and the cancer type, WT RAS proteins can be broadly categorized into two groups with opposing biologic functions. The protein product of the WT RAS allele of the same isoform as mutated RAS is tumor-suppressive, whereas the WT RAS proteins from the two non-mutated WT RAS family members are tumor-promoting (Figure 1B and reviewed in [21]).
2.1. The WT RAS Allele of the Same Isoform as Mutated RAS Inhibits Tumorigenesis
Several studies have found that the WT allele corresponding to the specific mutated RAS gene (WT HRAS in HRAS-mutated cancers [22,23]; WT NRAS in NRAS-mutated cancers [24,25]; WT KRAS in KRAS-mutated cancers [23,26,27,28,29,30]) suppresses tumorigenesis. Further, many RAS-mutated cancers have loss of heterozygosity (LOH) at the mutated gene, suggesting that loss of the wild-type allele confers a growth advantage. Evidence for LOH as a frequent event in cancer initiation has been observed in model systems for all three RAS genes (HRAS [23,31,32,33,34], NRAS [21,24,35], KRAS [21,36,37,38,39]). A survey of human tumor samples, cancer cell lines, and xenografts of lung, pancreatic, and colorectal cancers found mutant allele specific imbalance (MASI), where the mutant allele makes up more than half of the gene copies, in 58% of KRAS-mutated samples; over half of these imbalances were due to complete loss of the wild-type allele (uniparental disomy) [40]. Other surveys of both KRAS-mutated [41,42] and HRAS-mutated [43] patient tumor samples have found similar results. KRAS MASI is associated with worse prognosis in colorectal cancer and pancreatic cancer [44] and loss of the WT KRAS allele has been found at a higher rate in metastatic KRAS-mutated lung and pancreatic cancers compared to the primary tumors [36,37]. Several mechanisms have been proposed for inhibition of mutant RAS by the corresponding wild-type RAS. MASI and concomitant loss of the wild-type allele would increase the dosage of the mutant allele, increasing the number of mutant RAS proteins signaling in the cell, potentially increasing the oncogenic growth signal. Increased copy number of KRAS has been correlated with increased cell fitness in AML and CRC cells [41] and more aggressive and undifferentiated states in metastatic murine PDAC cells [45]. In patients, KRAS mutations combined with copy number gains were associated with decreased survival in lung cancer compared to KRAS mutations without copy number gains (LOH due to uniparental disomy or no LOH) [40].
Increased dosage of the mutant allele due to MASI does not, however, fully account for the effects of WT RAS of the same isoform; the level of WT KRAS also plays an important role in determining the extent of mutant KRAS-driven tumorigenesis. To examine the effects of WT KRAS levels on mutant KRAS-driven tumorigenesis, To et al. [27] crossed the Kras2LA2 lung cancer model into different mouse strains that show differing amounts of expression from the endogenous WT Kras allele. They found an inverse correlation between WT KRAS expression and tumorgenesis; mouse strains with lower relative expression of the WT Kras allele showed enhanced tumorigenicity. These data indicated that changes in copy number of the mutant KRAS allele do not fully explain the impact of WT KRAS on inhibiting tumorigenesis. To further probe the effects of WT KRAS on mutant KRAS-driven tumorigenesis, Ambrogio et al. [29] used both mice and RASless MEFs containing floxed WT Kras, and found that removal of WT KRAS enhanced mutant KRAS-driven signaling, proliferation, and tumorigenesis. Mechanistically, they found that the ability of WT KRAS to heterodimerize with mutant KRAS was necessary for its inhibitory function, as a dimerization deficient WT KRAS construct was unable to inhibit tumorigenesis. In contrast, they found that homodimers of mutant KRAS were essential for KRAS oncogenic function. These data corroborated findings that mutant KRAS may act as a dimer, requiring both monomers to be activated to achieve full downstream effector engagement [46,47]. In a cell with both constitutively active mutant KRAS and regulated WT KRAS, only dimers composed of two mutant KRAS proteins would provide a full oncogenic signal. Loss of WT KRAS, either through MASI or decreased expression of the wild-type allele, would increase the fraction of dimers composed of two mutants, increasing the oncogenic signal output. In addition to KRAS, both HRAS and NRAS have all been reported to dimerize [29,46,47,48,49], but the biologic effects of RAS dimerization and whether a similar mechanism underlies the tumor suppressive functions of WT HRAS and NRAS are untested. Of the three RAS family members, inhibition of mutant KRAS by WT KRAS is the most consistently observed across cancer types. KRAS dimerization may be the primary contributor, although changes in dosage of the mutant protein due to loss of the wild-type allele likely also impact the oncogenic signal. In cancers with copies of the wild-type allele remaining, the inhibition of mutant RAS by the corresponding wild-type RAS isoform indicates that therapies that specifically target the oncogenic mutant may perform better than therapies that target the mutant and wild-type proteins [36].
2.2. WT RAS Family Members Distinct from the Mutated RAS Allele Promote Oncogenesis
While WT RAS of the same isoform generally inhibits tumor initiation and growth, the protein products of the two non-mutated, WT RAS genes (for example HRAS and NRAS in a KRAS-mutated cancer; hereafter called WT RAS in all cases) are tumor promoting in RAS-mutated tumors [30,50]. Deletion of WT HRAS lead to decreased proliferation and increased apoptosis in KRAS-mutated endometrial cancer cells [30]. Knockdown of both WT RAS family members decreased proliferation in cell lines with mutated HRAS (T24), NRAS (RD), and KRAS (Mia PaCa 2) [50]. Mechanistically, knockdown of either the mutated RAS isoform or WT RAS differentially altered basal versus RTK-stimulated effector pathway activation [50]. Similar to signaling effects seen with oncogene-targeted inhibitors (see Section 4), these two signals cross-regulated each other: knockdown of either mutated RAS or WT RAS enhanced signaling through the other RAS pathway and simultaneous inhibition of both mutant RAS and WT RAS were required to promote apoptosis [50].
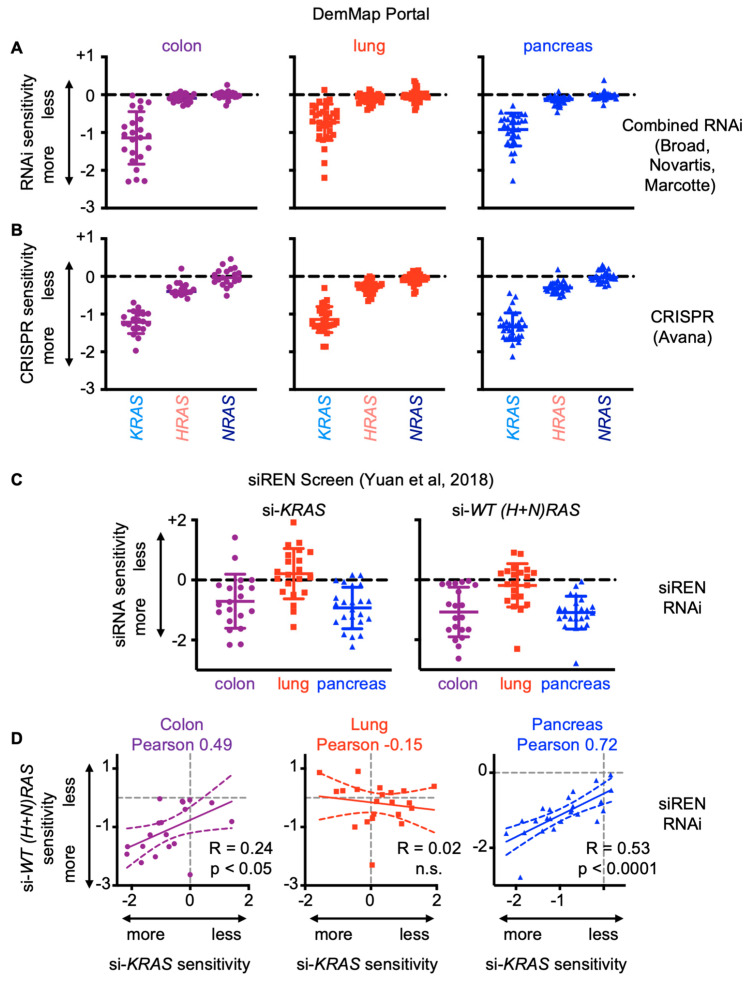
In in vivo models, deletion of individual WT Ras has shown variable effects on mutant RAS-driven tumorigenesis. In skin, WT Kras deletion promoted the progression of Hras-mutated papillomas to invasive squamous carcinomas, whereas WT Nras deletion decreased the formation of Hras-mutated papillomas [23]. Further, WT Nras deletion enhanced whereas WT Hras deletion inhibited Kras-mutated lung carcinogenesis [23]. In KRAS-mutated endometrial cancer cells, individual deletion of WT HRAS or NRAS limited proliferation in cancer cells, but not xenograft tumor growth [30]. These data suggest that the roles of WT RAS isoforms are complex and dependent on specific cellular context. To broadly examine the effects of WT RAS deletion in KRAS-mutated cancer cells, we performed a meta-analysis of mutated KRAS, WT HRAS, and WT NRAS dependencies in KRAS-mutated colorectal cancer, non-small cell lung cancer, and pancreatic cancer cell lines from both the Dependency Map Portal (DepMap) [51,52,53] and a large-scale siRNA knockdown screen that assessed RNAi depletion of RAS pathway ‘nodes’ (siRNEN Screen [54]). Analysis of both combined RNAi (Figure 2A) and AVANA/CRISPR screens (Figure 2B) showed a significant growth disadvantage in cells with mutant KRAS knockdown or deletion, but not in cells where WT HRAS or NRAS were individually removed. In contrast to DepMap data, which analyzed individual gene knockdowns, the siREN screen knocked down all genes of genetic ‘nodes’ simultaneously; thus the WT RAS node knocked down both HRAS and NRAS. In both colorectal cancer and pancreatic cancer cells, simultaneous knockdown of WT HRAS + NRAS limited survival to a similar extent as knockdown of mutated KRAS did (Figure 2C). Further, there was a direct correlation between individual cell line’s sensitivities to knockdown of mutated KRAS and WT (H+N) RAS, suggesting that mutant KRAS and WT (H + N) RAS act together to drive proliferation in these cells [54] (Figure 2D). Interestingly, the lung adenocarcinoma cells did not show dependency to either mutated KRAS or WT RAS in siREN screen data. This observation reflects previous observations that KRAS-mutated lung adenocarcinoma cells are “KRAS-independent” in 2D culture [55,56,57,58,59], but still require KRAS for anchorage-independent growth [60,61,62,63], and some KRASG12C-mutated NSCLC cell lines respond to KRASG12C inhibitors in 3D culture and in vivo but not in 2D adherent culture [16]. Overall, these data highlight the importance of WT RAS signaling to promote tumorigenesis in RAS-mutated cancers.
Figure 2.
WT HRAS+NRAS knockdown correlates with mutant KRAS knockdown in KRAS-mutated colorectal and pancreatic cancer cell lines. (A,B) Gene effect of combined RNAi knockdown (Broad, Novartis, Marcotte, A) or CRISPR-mediated deletion (Avana, B) of KRAS, HRAS, or NRAS from the DepMap Portal data set in KRAS-mutated colorectal, NSCLC, and pancreatic cancer cell lines. (C,D) Effect of siRNA knockdown of mutant KRAS or both non-mutated WT RAS (HRAS and NRAS) genes (C) or linear correlation between mutant KRAS and WT RAS knockdown (D) from the siREN screen [23] in KRAS-mutated colorectal, NSCLC, and pancreatic cancer cell lines. Pearson correlation coefficient is shown. Each symbol indicates an individual cell line.
3. Mechanisms of WT RAS Activation in RAS-Mutated Cancers
Multiple mechanisms have been proposed to describe the activation of WT RAS in the context of mutant RAS. Broadly, WT RAS activation has been described as either mutant RAS-dependent or RTK-dependent, although these two mechanisms are likely interdependent and act in concert to fully activate WT RAS.
3.1. Mutant RAS-Dependent and RTK-Dependent Mechanisms Activate WT RAS in RAS-Mutated Tumor Cells
The Kuriyan and Bar Sagi groups identified an allosteric RAS-GTP binding pocket on SOS1 that is distinct from the catalytic SOS1 domain [64]. RAS-GTP binding to this allosteric pocket relieves SOS1 autoinhibition [65,66], increasing SOS1 catalytic activity 80- to 500-fold [64] and activating a RAS-GTP–SOS1–WT RAS positive feedback that has been proposed to allow for ‘switch-like’ digital RAS activation [67,68,69]. This ‘switch-like’ behavior is important for normal childhood development [70,71], T and B cell development [72,73,74] and activation [75,76], and mutant KRAS-dependent cell proliferation and oncogenesis [77]. While SOS2 contains a homologous allosteric RAS-GTP binding site, whether SOS2 can be allosterically activated remains unconfirmed.
Independent of mutant RAS, RTK-dependent activation of WT RAS promotes activation of downstream effectors in parallel with constitutive mutant RAS signaling [50]. Knockdown studies showed that the non-mutated WT RAS genes are necessary for growth-factor-mediated signaling to RAS effector pathways in HRAS-, NRAS, and KRAS-mutated cancer cells [30,50], indicating that cancer cell response to growth factors may be mediated by WT RAS, not the oncogenic RAS mutant. RTK−WT RAS signaling supplements basal mutant RAS signaling to fully activate RAF/MEK/ERK and PI3K/AKT effector pathways [50,78] to promote proliferation [50] and G2 checkpoint integrity [79] in RAS-mutated cancer cells. Intriguingly, the McCormick lab showed that mutant RAS and WT RAS signals cross-regulate each other; knockdown of mutated RAS enhances RTK−WT RAS signaling to downstream effectors and conversely knockdown of WT RAS enhances basal RAS effector activation [50]. Due to this cross-regulation, which is likely due to rebound signaling (see Section 4), they further showed that combined inhibition of both mutant RAS and WT RAS signaling was necessary to induce apoptosis in RAS-mutated cancer cells.
These two mechanisms of WT RAS activation are not mutually exclusive and likely cooperate in some contexts. For example, positive feedback activation of SOS1 by active RAS-GTP potentiates EGF signaling to downstream effectors in vitro [66] and supports prolonged RAS and ERK activation downstream of T cell and B cell receptors [75,76]. Further, other signaling mechanisms can contribute to WT RAS activation in a context-specific manner. For example, endothelial nitric oxide synthase (eNOS) is phosphorylated and activated by RAS-AKT signaling; eNOS can in turn nitrosylate and activate WT HRAS generating a positive feedback loop that contributes to cellular transformation and tumor maintenance [80].
3.2. The RasGEFs SOS1 and SOS2 May Play Non-Overlapping Roles in Cells Expressing Oncogenic RAS
Data from our lab and others suggests that SOS1 and SOS2 may play non-overlapping roles in RAS-mutated tumors. Mutant KRAS–SOS1–WT RAS allosteric signaling promotes growth of KRAS-mutated pancreatic cancer cell xenografts [77], but has not been assessed for mutant HRAS- or NRAS-dependent transformation. In contrast, we found that RTK-SOS2-WT RAS signaling, but not allosteric SOS2 activation, is a critical mediator of mutant KRAS-driven transformation [81] by protecting KRAS-mutated cancer cells from anoikis [82]. We further showed that there was a hierarchical requirement for SOS2 to drive mutant RAS-dependent transformation, with KRAS > NRAS > HRAS. These data suggest that signaling via SOS1 and SOS2 may promote unique aspects of WT RAS signaling in RAS-mutated tumors.
3.3. WT RAS Cooperates with Mutant RAS to Fully Activate Downstream RAS Effector Pathways
Although the RAS isoforms are highly similar in terms of sequence and structure, the differences in their developmental requirement and mutation rates between cancer types indicate that they are not biologically equivalent. RAS isoforms have differing abilities to activate their downstream effectors [83,84] which are not correlated with a difference in binding affinity [85] or isoform stability [86]. Specifically, HRAS, NRAS, and KRAS show inverse abilities to activate PI3K/AKT signaling and RAF/MEK/ERK signaling: mutant HRAS is a potent activator of PI3K but a relatively poor activator of RAF; conversely, KRAS potently activates RAF but poorly activates PI3K [78,83,84], and NRAS shows intermediate activation of both RAF and PI3K effector pathways (Figure 1C). A recent study has shed light on the mechanism for the differential activation of RAF proteins [87]. Upon RAS activation, RAF proteins form homo- and heterodimers, with BRAF/CRAF heterodimers being the dominant complex responsible for downstream MEK/ERK activation to promote mutant KRAS-driven transformation [88]. BRAF preferentially interacts with KRAS via an interaction between the KRAS (4B) polybasic region and an acidic N-terminal region in BRAF [87]. The ability to directly associate with both BRAF and CRAF makes KRAS a more potent activator of the RAF/MEK/ERK cascade. While the precise mechanism for differential PI3K activation between HRAS and KRAS remains unclear, a major contributor seems to be the polybasic stretch in the hypervariable region (HVR) of KRAS; mutating basic residues in the KRAS (4B) HVR inhibits RAF/MEK/ERK signaling but enhances PI3K/AKT phosphorylation [89].
This differential effector activation by RAS isoforms leads to the proposed model that WT RAS contributes to cancer by signaling to effectors that the mutant RAS isoform cannot activate effectively [90]. HRAS-mutated cancer cells require RTK–WT RAS signaling to activate RAF/MEK/ERK signaling [78,91]. Conversely, in KRAS-mutated colorectal [92] and lung [93] adenocarcinoma cells, PI3K/AKT pathway activation is dependent on RTK signaling. Furthermore, we showed that in KRAS-mutated cancer cells, RTK-SOS2-WT RAS signaling was necessary to provide adequate PI3K/AKT signaling for cells to survive in anchorage-independent growth conditions (protection from anoikis) [81], but HRAS- and NRAS-mutated cancer cells could survive in anchorage-independent conditions without RTK-SOS2 supplemented PI3K signaling [82]. These different requirements for WT RAS isoforms in RAS-mutated cancers are also reflected in mutational activation of RAS and downstream RAS effectors. Analysis of co-mutation frequencies shows that KRAS and BRAF mutations are generally mutually exclusive, while KRAS and PIK3CA (the gene encoding the catalytic p110α subunit of PI3K) mutations co-occur frequently, consistent with the idea that KRAS already highly activates the RAF pathway, but requires supplemental signaling in the PI3K/AKT pathway [94,95]. Thus, WT RAS proteins may contribute to RAS effector activation in RAS-mutated cancer cells through their ability to activate the pathway(s) that the mutant RAS does not activate well (Figure 1).
4. WT RAS Signaling Underlies Resistance to Targeted Therapies in RAS-Mutated Cancers
4.1. Inhibitors of RAS Effector Pathways
Initial efforts to target RAS-mutated cancers focused on inhibiting downstream RAF/MEK/ERK and PI3K/AKT effector signaling, as RAS proteins have been historically difficult to target. Unfortunately, in multiple preclinical models of mutant RAS driven malignancies, single-agent MEK inhibitor treatment is ineffective. In KRAS-mutated cancer cells, single agent MEK inhibitor treatment is ineffective because it both relieves ERK-dependent negative feedback signaling and induces the expression of RTK and ligands [96,97,98,99,100,101,102,103,104,105]. These effects cause rapid RTK–WT RAS-dependent activation of both parallel PI3K/AKT signaling and the inhibited RAF/MEK/ERK cascade to drive therapeutic resistance. Similar rebound signaling occurs after MEK inhibitor treatment in HRAS- and NRAS-mutated cancer cells.
Similar to what is seen after single-agent MEK inhibitor treatment, rebound RTK signaling occurs after PI3K inhibition, with both RAF/MEK/ERK and PI3K/AKT rebound activation [106,107,108]. Combinations of MEK and PI3K/mTOR inhibitors were successful preclinically [98,109], but clinical success has been limited by toxicity [108,110,111]. Unfortunately, toxicity is likely unavoidable with this treatment strategy, since the RAF/MEK/ERK and PI3K/AKT pathways are both key players in normal cell function. To avoid this toxicity, many studies have investigated the efficacy of blocking the PI3K pathway indirectly, or finding other pathways that synergize with MEK or PI3K inhibition [92,112]. In KRAS-mutated colorectal [92] and lung [93] adenocarcinoma cells, PI3K/AKT pathway activation is dependent on RTK–WT RAS signaling (see Section 3.3), and thus inhibition of RTK signaling should indirectly inhibit PI3K activation. For example, Ebi et al. [92] showed that PI3K signaling was most often downstream of IGF-1R in KRAS-mutated colorectal cancer cells and that IGF-1R inhibition did indirectly block PI3K signaling and cooperate with MEK inhibitors to induce cell death.
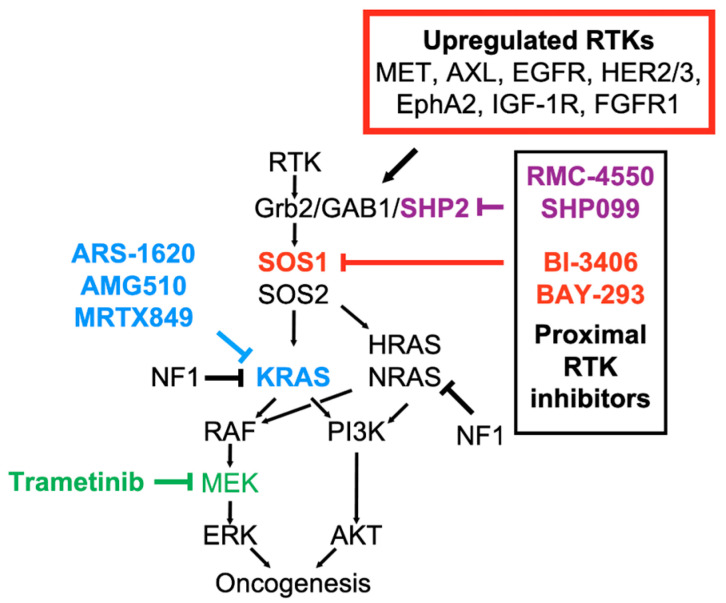
Unfortunately, it is often difficult to identify which RTK must be co-inhibited with either MEK or PI3K inhibitors for a given tumor type. RNA-sequencing studies showed that MEK inhibitor treatment induces simultaneous upregulation of multiple RTKs and ligands [113,114], so co-inhibition of individual RTKs will likely be ineffective in blocking MEK inhibitor resistance [97,104,115]. Further, even specific experiments where a dominant RTK drove MEK inhibitor resistance, the specific RTK involved was either tumor type or more often cell line specific. IGF-IR, MET, ERBB1/2, ERBB3, PDGFRa, AXL and FGFR1 have all been implicated in MEK-inhibitor resistance in KRAS-mutated tumors depending on the anatomical tumor type or specific cell line that was examined [92,97,104,105] (Figure 3). These studies indicate that broad inhibition of proximal RTK signaling will likely be required to block MEK inhibitor resistance.
Figure 3.
In KRAS-mutated cancer cells, treatment with either KRASG12C inhibitors or MEK inhibitors causes rebound RTK signaling leading to therapeutic resistance. Inhibition of proximal RTK signaling using inhibitors of the common proximal RTK signaling intermediates SHP2 or SOS1 can potentially limit resistance to oncogene-targeted therapies, thereby significantly prolonging the initial window of therapeutic efficacy.
4.2. Mutant RAS Inhibition
While most early efforts to target RAS have been unsuccessful, recent breakthroughs in both our understanding of an older ‘unsuccessful’ RAS inhibitor and novel insights into the accessibility of specific RAS mutations have led to renewed hope for successful targeting of mutant RAS in the clinic.
4.2.1. Tipifarnib as an HRAS-Specific Inhibitor
For mutant HRAS, advances in our understanding of the enzymes responsible for post-translational lipid modification of RAS isoforms have showed that farnesyltransferase inhibitors (FTIs), drugs originally designed as pan-RAS inhibitors, specifically inhibit HRAS and have clinical activity for patients with HRAS-mutated tumors. All RAS isoforms are post-translationally modified by the covalent addition of a C15 farnesyl isoprenoid lipid at their C-terminus and this modification is required for their membrane association and biological activity. This process, known as prenylation, is normally catalyzed by the enzyme farnesyltransferase (FTase). Since prenylation is required for RAS biological activity, FTIs were developed and tested as therapeutics in RAS-mutated cancers [12]. Unfortunately, KRAS and NRAS, the major RAS isoforms that are mutated in adult cancers, can be alternatively prenylated by gerangeranyltransferases [116,117,118], and FTIs failed in Phase III clinical trials for KRAS-mutated colorectal [119] and pancreatic [120] cancers. In contrast, HRAS is exclusively farnesylated [121] and its membrane association is inhibited by the FTI tipifarnib. In Phase II trials, patients with HRAS-mutated head and neck squamous cell carcinoma [122], urothelial carcinoma [123], and salivary gland cancer [124] have shown encouraging clinical responses to tipifarnib. Similar to MEK inhibitors (above) and KRASG12C inhibitors (below), tipifarnib treatment of HRAS-mutated cancer cells show adaptive reactivation RTK–WT RAS signaling and enhanced RAF/MEK/ERK pathway activation [91]. Furthermore, rebound RTK signaling after tipifarnib treatment occurred through multiple different RTKs in vivo [91], suggesting the need for novel therapeutic combinations.
4.2.2. Covalent KRASG12C Inhibitors
Unlike other mutant KRAS proteins, the active -SH group on the cysteine of G12C-mutant KRAS allows for covalent modification using therapeutics. The Shokat lab combined this idea with their discovery of a novel binding pocket on KRAS that is only present in the GDP (inactive) state to develop the first KRASG12C inhibitor that binds KRAS in the GDP-bound (inactive) state and covalently modifies the mutant cysteine in KRASG12C [15]. Since this first report, there has been a flurry of novel compounds including the bioavailable tool compound ARS-1620 [16] and several clinical compounds, including AMG 510 [17] and MRTX849 [18], both of which are currently in clinical trials for KRASG12C-mutated solid tumors. Preliminary reports of patient responses to these drugs are encouraging: ~50% of patients with KRASG12C-mutated NSCLC show partial responses to either AMG 510 or MRTX849, and a majority of the remaining patients show disease stabilization [17,18]. Similar to both FTIs and MEK inhibitors, rapid resistance to KRASG12C inhibitors develops. In vitro and in vivo studies revealed upregulated RTK signaling [18,114,125,126] and potential synthesis of new uninhibited KRASG12C [126] as the major drivers of KRASG12C inhibitor resistance. Similar to MEK inhibitors, the specific RTK driving KRASG12C inhibitor resistance is cell type specific, so that while individual RTK inhibitors might be effective in blocking KRASG12C-inhibitor resistance in a specific cancer cell line, broad inhibition of RTK signaling will be required to delay therapeutic resistance and make KRASG12C inhibitors clinically efficacious [18,125,126,127].
4.3. Inhibition of Proximal RTK Signaling Can Overcome MEK- and KRASG12C-Inhibitor Resistance
Unbiased genetic and pharmacologic screens revealed three distinct classes of synthetic lethal targets that synergize with both MEK and KRASG12C inhibitors in KRAS-mutated cancer cells [112,127,128]: (i) individual RTKs or proximal RTK signaling components (including SHP2 and SOS1) whose inhibition can broadly inhibit RTK signaling, (ii) mTOR/PI3K survival signaling components, and (iii) regulators of cell cycle progression. Both mTOR/PI3K pathway inhibitors [128,129] and CDK4/6 inhibitors [127,130] potentiate the effects of MEK inhibitors and KRASG12C inhibitors in xenograft studies, suggesting that targeting these collateral dependencies may be a viable therapeutic strategy. Since PI3K activation is downstream of RTK–WT RAS signaling in KRAS-mutated cancer cells [92,93] and cell cycle progression requires RTK/RAS signaling, these collateral dependent targets may represent a common mechanism for inhibiting MEK- and KRASG12C-inhibitor resistance [127]. Here, the discovery of potent, orally available SHP2 and SOS1 inhibitors has the potential to dramatically augment oncogene-targeted therapies for RAS-mutated cancer.
4.4. SOS1 and SHP2 Are Therapeutic Targets in RAS-Mutated Cancer Cells
SHP2 and SOS1 are common proximal RTK signaling intermediates; the development of potent, specific inhibitors for both SHP2 (SHP099 [131,132]; RMC-4550 [133]) and SOS1 (BAY-293 [134]; BI-3406 [135,136]) has led to new approaches to treating RAS-mutated cancers. Both SHP2 [131,132,133] and SOS1 [134,135,136] inhibitors are effective in inhibiting cell growth in situ as single agents in cells with RTK/RAS pathway mutations that are dependent upon RAS nucleotide cycling, including cells with EGFR mutations, KRAS (G12/13) mutations, LOF NF1 mutations, and BRAF type III mutations, but not in cells with KRAS Q61 mutations, BRAF Type I/II mutations, or concomitant PIK3CA mutations. In xenograft studies using adult lung, pancreas, or colon cancer cell lines, SHP2 inhibitors enhanced the efficacy of covalent KRASG12C inhibitors [18,114] and both SHP2 [113,137] and SOS1 [135] inhibitors enhanced the efficacy of MEK inhibitors. Intriguingly, although neither SHP2 nor SOS1 inhibitors were able to inhibit cancer cells with KRAS Q61 mutations as single agents [133,135], both were able to enhance the efficacy of the MEK inhibitor trametinib in xenograft models harboring KRAS Q61 mutations [113,135], suggesting that inhibiting proximal RTK signaling might be broadly effective in combination therapies for RAS-mutated tumors harboring G12, G13, or Q61 mutations. In HRAS- or NRAS-mutated cells, neither SHP2 or SOS1 inhibitors are effective as single agents [133,135,138], however, a recent study showed that while NRASQ61-mutated neuroblastoma cells were insensitive to SHP2 inhibitors alone, combined SHP2 and MEK inhibition showed synergistic inhibition of cell growth [138], suggesting that proximal RTK (SHP2 or SOS1) inhibitors may be a general therapeutic option to overcome MEK inhibitor resistance in RAS-mutated cancer cells.
4.5. The Spectrum of KRAS Mutations between Different Cancer Types Leads to Cancer-Specific Vulnerabilities to WT RAS Inhibition
KRAS is the most frequently mutated RAS gene; KRAS mutations occur in 32–35% of lung adenocarcinomas (LUAD), 41–50% of colorectal adenocarcinomas (COAD), and 86–88% of pancreatic adenocarcinomas (PAAD) [3,8]. While G12 mutations predominate each of these cancers, there are cancer-specific differences in the KRAS mutational spectrum that have functional consequences for therapeutics targeting WT RAS signaling [8].
In LUAD, 40% of KRAS mutations are G12C, whereas KRASG12C mutations occur less frequently in COAD (7%) and PAAD (1%) [8,11,14]. Due to these mutational differences, covalent G12C inhibitors (see Section 4.2.2) will likely have the greatest impact in LUAD, where ~50% of patients have shown partial responses in Phase I and II trials [17,18]. Similar to MEK inhibition, treatment with covalent KRASG12C inhibitors causes rapid rebound activation of multiple RTKs, making RTK–SOS1/2–WT RAS signaling an important therapeutic target in KRASG12C-mutated LUAD.
In late-stage colorectal adenocarcinoma, the monoclonal antibodies cetuximab and panitumumab, which inhibit the EGFR, improved outcomes for patients with WT KRAS but not with KRAS mutations [139,140,141,142,143]; these anti-EGFR therapies are FDA approved for first-line treatment for patients with WT KRAS colorectal cancers where they used in combination with conventional chemotherapy [144]. Intriguingly, retrospective analysis of the Phase III trial data assessing the efficacy of cetuximab in COAD showed that patients with KRASG13D mutations may benefit from anti-EGFR therapies [145], although subsequent Phase II trials that prospectively assessed anti-EGFR therapies in patients with KRASG13D mutations have shown varying results [146,147,148]. COADs have a high percentage of KRASG13D mutations (17%) compared to either LUAD (3%) or PAAD (<1%) [8]. A recent manuscript by McFall et al. [149] has shed light on why colorectal cancers with KRASG13D mutations might be sensitive to anti-EGFR therapies. In cells with KRASG13D mutations, WT RAS activation is particularly sensitive to EGFR inhibition [149]. KRAS G12 mutant proteins interact strongly with the RasGAP NF1 and this strong interaction competitively inhibits NF1, activating wild-type HRAS and NRAS independent of EGFR. In contrast, mutant KRASG13D proteins have a relatively weak interaction with NF1, allowing NF1 to inactivate wild-type HRAS and NRAS in the absence of EGFR stimulation and making WT RAS signaling EGFR-dependent in these cells [149]. Due to this, downstream signaling in G13D-mutated cells is extremely RTK-dependent, possibly explaining why KRASG13D-mutated colorectal cancers are sensitive to EGFR-TKIs while other KRAS-mutated colorectal tumors are refractory to EGFR-TKI treatment [145]. Rabara et al. [150] confirmed these results and further showed that a subset of KRASG13D-mutated colorectal adenocarcinomas had co-mutation of NF1. Only KRASG13D-mutated cancers with WT NF1 were responsive to EGFR inhibition.
In PAAD, 17% of KRAS mutations are G12R, whereas KRASG12R mutations only occur in ~1% of COAD and LUAD [8]. Hobbs et al. [151] found that KRASG12R-mutated PAAD cells have unique signaling properties that may make them vulnerable to WT RAS inhibition. Pancreatic cancer cells are dependent on RAS-driven macropinocytosis for nutrient uptake and survival [152,153]. Using a panel of KRAS-mutated PAAD cell lines, Hobbs et al. [150] showed that while macropinocytosis was KRAS-dependent in KRASG12D and KRASG12V-mutated cells, macropinocytosis was KRAS-independent in cells with KRASG12R mutations. They found that compared to cells with G12D or G12V mutations, KRASG12R-mutated cells showed defective PI3K-AKT signaling, due to the inability of KRASG12R to interact with the p110α catalytic subunit of PI3K. Macropinocytosis was PI3Kγ-dependent in KRASG12R-mutated cells, suggesting that WT RAS signaling was specifically required for nutrient uptake in these cells. Due to these unique signaling properties, KRASG12R-mutated cells were more sensitive to single-agent PI3K or MEK inhibition compared with KRASG12D and KRASG12V-mutated cells [151]. In addition to its inability to interact with p110α, KRASG12R cannot interact with the catalytic domain of SOS1 [151], and isogenic NCI-H23 cells expressing KRASG12R were insensitive to SOS1 inhibition [135].
To investigate these findings in a controlled model, Zafra et al. [154] recently generated an in vivo Kras allelic series where they directly compared tumorigenesis and drug sensitives of KrasG12C, KrasG12D, KrasG12R, and KrasG13D mutants. In keeping with clinical observations, G12C and G12D mutations showed overall enhanced tumorigenesis in both the colon and pancreas compared to G12R or G13D mutations. Further, when assessing drug sensitivities in pancreatic organoids, KrasG13D-mutated organoids were much more sensitive to EGFR inhibition alone compared with other mutants, and KrasG12C-mutated organoids were sensitive to combining an EGFR inhibitor with covalent KRASG12C inhibition [154], paralleling the findings described above. Taken together, these data indicate that specific KRAS mutations may be more sensitive to inhibitors of WT RAS signaling, leading to organ-specific vulnerabilities based on mutation frequencies.
5. Conclusions
WT RAS signaling is an important modifier of RAS-mutated oncogenesis, and inhibition of WT RAS signaling may be required for effective treatment of RAS-mutated cancers. Understanding the mechanisms by which WT RAS is activated is an important step in determining the best ways to limit WT RAS signaling. The ability to pharmacologically manipulate the common proximal signaling intermediates SHP2 and SOS1/2 may lead to optimized therapeutic combinations that can be used to treat RAS-mutated cancers.
Acknowledgments
We thank Rachel Bagni (NCI) for discussions regarding siREN screen data and Marielle Yohe (NCI) and for thoughtful discussions while preparing this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12050662/s1.
Author Contributions
E.S. and R.L.K. conceptualized the manuscript, analyzed the data, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Congressionally Directed Medical Research Program/Lung Cancer Research Program grant number LC180213.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in supplemental Table S1. Data were downloaded from https://www.depmap.org/depmap, accessed on 27 April 2021 (DepMap Data) and from [54] (siREN Screen Data).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lemmon M.A., Schlessinger J. Cell signaling by receptor-tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papke B., Der C.J. Drugging ras: Know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 3.Prior I.A., Hood F.E., Hartley J.L. The frequency of ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riely G.J., Marks J., Pao W. Kras mutations in non-small cell lung cancer. Proc. Am. Thorac. Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 5.Grady W.M., Markowitz S.D. Genetic and epigenetic alterations in colon cancer. Annu. Rev. Rev. Genom. Hum. Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 6.Jaffee E.M., Hruban R.H., Canto M., Kern S.E. Focus on pancreas cancer. Cancer Cell. 2002;2:25–28. doi: 10.1016/S1535-6108(02)00093-4. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA: A Cancer Journal for Clinicians. Cancer Stat. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 8.Cook J.H., Melloni G.E.M., Gulhan D.C., Park P.J., Haigis K.M. The origins and genetic interactions of kras mutations are allele- and tissue-specific. Nat. Commun. 2021;12:1808. doi: 10.1038/s41467-021-22125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. Ras oncogenes: Weaving a tumorigenic web. Nat. Rev. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knickelbein K., Zhang L. Mutant kras as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015;2:4–12. doi: 10.1016/j.gendis.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis K.M. Kras alleles: The devil is in the detail. Trends Cancer. 2017;3:686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephen A.G., Esposito D., Bagni R.K., McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Gerber D.E., Gandhi L., Costa D.B. Management and future directions in non-small cell lung cancer with known activating mutations. Am. Soc. Clin. Oncol. Educ. Book. 2014:e353-65. doi: 10.14694/EdBook_AM.2014.34.e353. [DOI] [PubMed] [Google Scholar]
- 14.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable ras: Mission possible? Nat. Rev. Rev. Drug. Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-ras(g12c) inhibitors allosterically control gtp affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janes M.R., Zhang J., Li L.S., Hansen R., Peters U., Guo X., Chen Y., Babbar A., Firdaus S.J., Darjania L., et al. Targeting kras mutant cancers with a covalent g12c-specific inhibitor. Cell. 2018;172:578–589.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. The clinical kras(g12c) inhibitor amg 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 18.Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., et al. The kras(g12c) inhibitor mrtx849 provides insight toward therapeutic susceptibility of kras-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick F. Progress in targeting ras with small molecule drugs. Biochem. J. 2019;476:365–374. doi: 10.1042/BCJ20170441. [DOI] [PubMed] [Google Scholar]
- 20.Orgovan Z., Keseru G.M. Small molecule inhibitors of ras proteins with oncogenic mutations. Cancer Metastasis Rev. 2020;39:1107–1126. doi: 10.1007/s10555-020-09911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B., Der C.J., Cox A.D. The role of wild type ras isoforms in cancer. Semin. Cell Dev. Biol. 2016;58:60–69. doi: 10.1016/j.semcdb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spandidos D.A., Frame M., Wilkie N.M. Expression of the normal h-ras1 gene can suppress the transformed and tumorigenic phenotypes induced by mutant ras genes. Anticancer Res. 1990;10:1543–1554. [PubMed] [Google Scholar]
- 23.To M.D., Rosario R.D., Westcott P.M., Banta K.L., Balmain A. Interactions between wild-type and mutant ras genes in lung and skin carcinogenesis. Oncogene. 2013;32:4028–4033. doi: 10.1038/onc.2012.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diaz R., Ahn D., Lopez-Barcons L., Malumbres M., de Castro I.P., Lue J., Ferrer-Miralles N., Mangues R., Tsong J., Garcia R., et al. The n-ras proto-oncogene can suppress the malignant phenotype in the presence or absence of its oncogene. Cancer Res. 2002;62:4514–4518. [PubMed] [Google Scholar]
- 25.Wang J., Liu Y., Li Z., Wang Z., Tan L.X., Ryu M.J., Meline B., Du J., Young K.H., Ranheim E., et al. Endogenous oncogenic nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood. 2011;118:368–379. doi: 10.1182/blood-2010-12-326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz R., Lue J., Mathews J., Yoon A., Ahn D., Garcia-Espana A., Leonardi P., Vargas M.P., Pellicer A. Inhibition of ras oncogenic activity by ras protooncogenes. Int. J. Cancer. 2005;113:241–248. doi: 10.1002/ijc.20563. [DOI] [PubMed] [Google Scholar]
- 27.To M.D., Perez-Losada J., Mao J.H., Hsu J., Jacks T., Balmain A. A functional switch from lung cancer resistance to susceptibility at the pas1 locus in kras2la2 mice. Nat. Genet. 2006;38:926–930. doi: 10.1038/ng1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z., Wang Y., Vikis H.G., Johnson L., Liu G., Li J., Anderson M.W., Sills R.C., Hong H.L., Devereux T.R., et al. Wildtype kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 29.Ambrogio C., Kohler J., Zhou Z.W., Wang H., Paranal R., Li J., Capelletti M., Caffarra C., Li S., Lv Q., et al. Kras dimerization impacts mek inhibitor sensitivity and oncogenic activity of mutant kras. Cell. 2018;172:857–868.e15. doi: 10.1016/j.cell.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Bentley C., Jurinka S.S., Kljavin N.M., Vartanian S., Ramani S.R., Gonzalez L.C., Yu K., Modrusan Z., Du P., Bourgon R., et al. A requirement for wild-type ras isoforms in mutant kras-driven signalling and transformation. Biochem. J. 2013;452:313–320. doi: 10.1042/BJ20121578. [DOI] [PubMed] [Google Scholar]
- 31.Nagase H., Mao J.H., Balmain A. Allele-specific hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res. 2003;63:4849–4853. [PubMed] [Google Scholar]
- 32.Estep A.L., Tidyman W.E., Teitell M.A., Cotter P.D., Rauen K.A. Hras mutations in costello syndrome: Detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 33.Bremner R., Balmain A. Genetic changes in skin tumor progression: Correlation between presence of a mutant ras gene and loss of heterozygosity on mouse chromosome 7. Cell. 1990;61:407–417. doi: 10.1016/0092-8674(90)90523-H. [DOI] [PubMed] [Google Scholar]
- 34.Buchmann A., Bauer-Hofmann R., Mahr J., Drinkwater N.R., Luz A., Schwarz M. Mutational activation of the c-ha-ras gene in liver tumors of different rodent strains: Correlation with susceptibility to hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA. 1991;88:911–915. doi: 10.1073/pnas.88.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helias-Rodzewicz Z., Funck-Brentano E., Terrones N., Beauchet A., Zimmermann U., Marin C., Saiag P., Emile J.F. Variation of mutant allele frequency in nras q61 mutated melanomas. BMC Dermatol. 2017;17:9. doi: 10.1186/s12895-017-0061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu W., Sahin F., Iacobuzio-Donahue C.A., Garcia-Carracedo D., Wang W.M., Kuo C.Y., Chen D., Arking D.E., Lowy A.M., Hruban R.H., et al. Disruption of p16 and activation of kras in pancreas increase ductal adenocarcinoma formation and metastasis in vivo. Oncotarget. 2011;2:862–873. doi: 10.18632/oncotarget.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C.C., Qiu W., Juang C.S., Mansukhani M.M., Halmos B., Su G.H. Mutant allele specific imbalance in oncogenes with copy number alterations: Occurrence, mechanisms, and potential clinical implications. Cancer Lett. 2017;384:86–93. doi: 10.1016/j.canlet.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Zhang Z., Dai Z., Plass C., Morrison C., Wang Y., Wiest J.S., Anderson M.W., You M. Loh of chromosome 12p correlates with kras2 mutation in non-small cell lung cancer. Oncogene. 2003;22:1243–1246. doi: 10.1038/sj.onc.1206192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerrero I., Villasante A., Corces V., Pellicer A. Loss of the normal n-ras allele in a mouse thymic lymphoma induced by a chemical carcinogen. Proc. Natl. Acad. Sci. USA. 1985;82:7810–7814. doi: 10.1073/pnas.82.23.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soh J., Okumura N., Lockwood W.W., Yamamoto H., Shigematsu H., Zhang W., Chari R., Shames D.S., Tang X., MacAulay C., et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (masi) frequently occur together in tumor cells. PLoS ONE. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burgess M.R., Hwang E., Mroue R., Bielski C.M., Wandler A.M., Huang B.J., Firestone A.J., Young A., Lacap J.A., Crocker L., et al. Kras allelic imbalance enhances fitness and modulates map kinase dependence in cancer. Cell. 2017;168:817–829.e15. doi: 10.1016/j.cell.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiosea S.I., Sherer C.K., Jelic T., Dacic S. Kras mutant allele-specific imbalance in lung adenocarcinoma. Mod. Pathol. 2011;24:1571–1577. doi: 10.1038/modpathol.2011.109. [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Makarewicz J.M., Knauf J.A., Johnson L.K., Fagin J.A. Transformation by hras(g12v) is consistently associated with mutant allele copy gains and is reversed by farnesyl transferase inhibition. Oncogene. 2014;33:5442–5449. doi: 10.1038/onc.2013.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bielski C.M., Donoghue M.T.A., Gadiya M., Hanrahan A.J., Won H.H., Chang M.T., Jonsson P., Penson A.V., Gorelick A., Harris C., et al. Widespread selection for oncogenic mutant allele imbalance in cancer. Cancer Cell. 2018;34:852–862.e4. doi: 10.1016/j.ccell.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueller S., Engleitner T., Maresch R., Zukowska M., Lange S., Kaltenbacher T., Konukiewitz B., Ollinger R., Zwiebel M., Strong A., et al. Evolutionary routes and kras dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muratcioglu S., Jang H., Gursoy A., Keskin O., Nussinov R. Pdedelta binding to ras isoforms provides a route to proper membrane localization. J. Phys. Chem. B. 2017;121:5917–5927. doi: 10.1021/acs.jpcb.7b03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer-Smith R., Koide A., Zhou Y., Eguchi R.R., Sha F., Gajwani P., Santana D., Gupta A., Jacobs M., Herrero-Garcia E., et al. Inhibition of ras function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017;13:62–68. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin W.C., Iversen L., Tu H.L., Rhodes C., Christensen S.M., Iwig J.S., Hansen S.D., Huang W.Y., Groves J.T. H-ras forms dimers on membrane surfaces via a protein-protein interface. Proc. Natl. Acad. Sci. USA. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guldenhaupt J., Rudack T., Bachler P., Mann D., Triola G., Waldmann H., Kotting C., Gerwert K. N-ras forms dimers at popc membranes. Biophys. J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young A., Lou D., McCormick F. Oncogenic and wild-type ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- 51.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M., et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dempster J.M., Rossen J., Kazachkova M., Pan J., Kugener G., Root D.E., Tsherniak A. Extracting biological insights from the project achilles genome-scale crispr screens in cancer cell lines. bioRxiv. 2019:720243. [Google Scholar]
- 54.Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., et al. Differential Effector Engagement by Oncogenic KRAS. Cell Rep. 2018;22:1889–1902. doi: 10.1016/j.celrep.2018.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balbin O.A., Prensner J.R., Sahu A., Yocum A.K., Shankar S., Malik R., Fermin D., Dhanasekaran S.M., Chandler B., Thomas D.G., et al. Reconstructing targetable pathways in lung cancer by integrating diverse omics data. Nat. Commun. 2013;4:1–13. doi: 10.1038/ncomms3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. A Gene Expression Signature Associated with “K-Ras Addiction” Reveals Regulators of EMT and Tumor Cell Survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh A., Sweeney M.F., Yu M., Burger A., Greninger P., Benes C., Haber D.A., Settleman J. TAK1 Inhibition Promotes Apoptosis in KRAS-Dependent Colon Cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scholl C., Fröhling S., Dunn I.F., Schinzel A.C., Barbie D.A., Kim S.Y., Silver S.J., Tamayo P., Wadlow R.C., Ramaswamy S., et al. Synthetic Lethal Interaction between Oncogenic KRAS Dependency and STK33 Suppression in Human Cancer Cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 59.Lamba S., Russo M., Sun C., Lazzari L., Cancelliere C., Grernrum W., Lieftink C., Bernards R., Di Nicolantonio F., Bardelli A. RAF Suppression Synergizes with MEK Inhibition in KRAS Mutant Cancer Cells. Cell Rep. 2014;8:1475–1483. doi: 10.1016/j.celrep.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 60.Fujita-Sato S., Galeas J., Truitt M., Pitt C., Urisman A., Bandyopadhyay S., Ruggero D., McCormick F. Enhanced MET Translation and Signaling Sustains K-Ras–Driven Proliferation under Anchorage-Independent Growth Conditions. Cancer Res. 2015;75:2851–2862. doi: 10.1158/0008-5472.CAN-14-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rotem A., Janzer A., Izar B., Ji Z., Doench J.G., Garraway L.A., Struhl K. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. USA. 2015;112:5708–5713. doi: 10.1073/pnas.1505979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z., Jiang G., Yang F., Wang J. Knockdown of mutant K-ras expression by adenovirus-mediated siRNA inhibits the in vitro and in vivo growth of lung cancer cells. Cancer Biol. Ther. 2006;5:1481–1486. doi: 10.4161/cbt.5.11.3297. [DOI] [PubMed] [Google Scholar]
- 63.McCormick F. KRAS as a Therapeutic Target. Clin. Cancer Res. 2015;21:1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Margarit S., Sondermann H., Hall B.E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. Structural Evidence for Feedback Activation by Ras·GTP of the Ras-Specific Nucleotide Exchange Factor SOS. Cell. 2003;112:685–695. doi: 10.1016/S0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 65.Sondermann H., Soisson S.M., Boykevisch S., Yang S.-S., Bar-Sagi D., Kuriyan J. Structural Analysis of Autoinhibition in the Ras Activator Son of Sevenless. Cell. 2004;119:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Boykevisch S., Zhao C., Sondermann H., Philippidou P., Halegoua S., Kuriyan J., Bar-Sagi D. Regulation of Ras Signaling Dynamics by Sos-Mediated Positive Feedback. Curr. Biol. 2006;16:2173–2179. doi: 10.1016/j.cub.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 67.Iversen L.F., Tu H.-L., Lin W.-C., Christensen S.M., Abel S.M., Iwig J.S., Wu H.-J., Gureasko J.M., Rhodes C.P., Petit R.S., et al. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science. 2014;345:50–54. doi: 10.1126/science.1250373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christensen S.M., Tu H.-L., Jun J.E., Alvarez S., Triplet M.G., Iwig J.S., Yadav K.K., Bar-Sagi D., Roose J.P., Groves J.T. One-way membrane trafficking of SOS in receptor-triggered Ras activation. Nat. Struct. Mol. Biol. 2016;23:838–846. doi: 10.1038/nsmb.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang W.Y.C., Alvarez S., Kondo Y., Lee Y.K., Chung J.K., Lam H.Y.M., Biswas K.H., Kuriyan J., Groves J.T. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science. 2019;363:1098–1103. doi: 10.1126/science.aau5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tartaglia M., Pennacchio L.A., Zhao C., Yadav K.K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W., et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 2006;39:75–79. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 71.Umutesi H.G., Hoang H.M., Johnson H.E., Nam K., Heo J. Development of Noonan syndrome by deregulation of allosteric SOS autoactivation. J. Biol. Chem. 2020;295:13651–13663. doi: 10.1074/jbc.RA120.013275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prasad A., Zikherman J., Das J., Roose J.P., Weiss A., Chakraborty A.K. Origin of the sharp boundary that discriminates positive and negative selection of thymocytes. Proc. Natl. Acad. Sci. USA. 2008;106:528–533. doi: 10.1073/pnas.0805981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daniels M.A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G.A., Gascoigne N.R.J., Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nat. Cell Biol. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 74.Kortum R.L., Sommers C.L., Pinski J.M., Alexander C.P., Merrill R.K., Li W., Love P.E., Samelson L.E. Deconstructing Ras Signaling in the Thymus. Mol. Cell. Biol. 2012;32:2748–2759. doi: 10.1128/MCB.00317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roose J.P., Mollenauer M., Ho M., Kurosaki T., Weiss A. Unusual Interplay of Two Types of Ras Activators, RasGRP and SOS, Establishes Sensitive and Robust Ras Activation in Lymphocytes. Mol. Cell. Biol. 2007;27:2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Das J., Ho M., Zikherman J., Govern C., Yang M., Weiss A., Chakraborty A.K., Roose J.P. Digital Signaling and Hysteresis Characterize Ras Activation in Lymphoid Cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeng H.-H., Taylor L.J., Bar-Sagi D. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat. Commun. 2012;3:1–8. doi: 10.1038/ncomms2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hamilton M., Wolfman A. Oncogenic Ha-Ras-dependent Mitogen-activated Protein Kinase Activity Requires Signaling through the Epidermal Growth Factor Receptor. J. Biol. Chem. 1998;273:28155–28162. doi: 10.1074/jbc.273.43.28155. [DOI] [PubMed] [Google Scholar]
- 79.Grabocka E., Pylayeva-Gupta Y., Jones M.J., Lubkov V., Yemanaberhan E., Taylor L., Jeng H.H., Bar-Sagi D. Wild-type H- and N-Ras promote mutant K-Ras-driven tumorigenesis by modulating the DNA damage response. Cancer Cell. 2014;25:243–256. doi: 10.1016/j.ccr.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lim K.-H., Ancrile B.B., Kashatus D.F., Counter C.M. Tumour maintenance is mediated by eNOS. Nat. Cell Biol. 2008;452:646–649. doi: 10.1038/nature06778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheffels E., Sealover N.E., Wang C., Kim D.H., Vazirani I.A., Lee E., Terrell E.M., Morrison D.K., Luo J., Kortum R.L. Oncogenic RAS isoforms show a hierarchical requirement for the guanine nucleotide exchange factor SOS2 to mediate cell transformation. Sci. Signal. 2018;11:eaar8371. doi: 10.1126/scisignal.aar8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sheffels E., Sealover N.E., Theard P.L., Kortum R.L. Anchorage-independent growth conditions reveal a differential SOS2 dependence for transformation and survival in RAS-mutant cancer cells. Small GTPases. 2021;12:67–78. doi: 10.1080/21541248.2019.1611168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan J., Roy S., Apolloni A., Lane A., Hancock J.F. Ras Isoforms Vary in Their Ability to Activate Raf-1 and Phosphoinositide 3-Kinase. J. Biol. Chem. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 84.Voice J.K., Klemke R.L., Le A., Jackson J.H. Four Human Ras Homologs Differ in Their Abilities to Activate Raf-1, Induce Transformation, and Stimulate Cell Motility. J. Biol. Chem. 1999;274:17164–17170. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 85.Herrmann C., Martin G.A., Wittinghofer A. Quantitative Analysis of the Complex between p21 and the Ras-binding Domain of the Human Raf-1 Protein Kinase. J. Biol. Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 86.Maher J., Baker D.A., Manning M., Dibb N.J., Roberts I.A. Evidence for cell-specific differences in transformation by N-, H- and K-ras. Oncogene. 1995;11:1639–1647. [PubMed] [Google Scholar]
- 87.Terrell E.M., Durrant D.E., Ritt D.A., Sealover N.E., Sheffels E., Spencer-Smith R., Esposito D., Zhou Y., Hancock J.F., Kortum R.L., et al. Distinct Binding Preferences between Ras and Raf Family Members and the Impact on Oncogenic Ras Signaling. Mol. Cell. 2019;76:872–884.e5. doi: 10.1016/j.molcel.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freeman A.K., Ritt D.A., Morrison D.K. Effects of raf dimerization and its inhibition on normal and disease-associated raf signaling. Mol. Cell. 2013;49:751–758. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou Y., Prakash P., Liang H., Cho K.-J., Gorfe A.A., Hancock J.F. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell. 2017;168:239–251.e16. doi: 10.1016/j.cell.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Castellano E., Santos E. Functional Specificity of Ras Isoforms: So Similar but So Different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Untch B.R., Dos Anjos V., Garcia-Rendueles M.E.R., Knauf J.A., Krishnamoorthy G.P., Saqcena M., Bhanot U.K., Socci N.D., Ho A.L., Ghossein R., et al. Tipifarnib Inhibits HRAS-Driven Dedifferentiated Thyroid Cancers. Cancer Res. 2018;78:4642–4657. doi: 10.1158/0008-5472.CAN-17-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ebi H., Corcoran R.B., Singh A., Chen Z., Song Y., Lifshits E., Ryan D.P., Meyerhardt J.A., Benes C., Settleman J., et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin. Investig. 2011;121:4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Molina-Arcas M., Hancock D.C., Sheridan C., Kumar M.S., Downward J. Coordinate Direct Input of Both KRAS and IGF1 Receptor to Activation of PI3 kinase in KRAS-Mutant Lung Cancer. Cancer Discov. 2013;3:548–563. doi: 10.1158/2159-8290.CD-12-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turke A.B., Song Y., Costa C., Cook R., Arteaga C.L., Asara J.M., Engelman J.A. MEK Inhibition Leads to PI3K/AKT Activation by Relieving a Negative Feedback on ERBB Receptors. Cancer Res. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pettazzoni P., Viale A., Shah P., Carugo A., Ying H., Wang H., Genovese G., Seth S., Minelli R., Green T., et al. Genetic Events That Limit the Efficacy of MEK and RTK Inhibitor Therapies in a Mouse Model of KRAS-Driven Pancreatic Cancer. Cancer Res. 2015;75:1091–1101. doi: 10.1158/0008-5472.CAN-14-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sos M.L., Fischer S., Ullrich R., Peifer M., Heuckmann J.M., Koker M., Heynck S., Stückrath I., Weiss J., Fischer F., et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc. Natl. Acad. Sci. USA. 2009;106:18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yohe M.E., Gryder B.E., Shern J.F., Song Y.K., Chou H.-C., Sindiri S., Mendoza A., Patidar R., Zhang X., Guha R., et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci. Transl. Med. 2018;10:eaan4470. doi: 10.1126/scitranslmed.aan4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Woodfield S.E., Zhang L., Scorsone K.A., Liu Y., Zage P.E. Binimetinib inhibits MEK and is effective against neuroblastoma tumor cells with low NF1 expression. BMC Cancer. 2016;16:172. doi: 10.1186/s12885-016-2199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jessen W.J., Miller S.J., Jousma E., Wu J., Rizvi T.A., Brundage M.E., Eaves D., Widemann B., Kim M.O., Dombi E., et al. Mek inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J. Clin. Investig. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hymowitz S.G., Malek S. Targeting the mapk pathway in ras mutant cancers. Cold Spring Harb. Perspect. Med. 2018;8 doi: 10.1101/cshperspect.a031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ryan M.B., Corcoran R.B. Therapeutic strategies to target ras-mutant cancers. Nat. Rev. Rev. Clin. Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 104.Sun C., Hobor S., Bertotti A., Zecchin D., Huang S., Galimi F., Cottino F., Prahallad A., Grernrum W., Tzani A., et al. Intrinsic resistance to mek inhibition in kras mutant lung and colon cancer through transcriptional induction of erbb3. Cell Rep. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 105.Manchado E., Weissmueller S., Morris J.P.t., Chen C.C., Wullenkord R., Lujambio A., de Stanchina E., Poirier J.T., Gainor J.F., Corcoran R.B., et al. A combinatorial strategy for treating kras-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012;2:311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chakrabarty A., Sanchez V., Kuba M.G., Rinehart C., Arteaga C.L. Feedback upregulation of her3 (erbb3) expression and activity attenuates antitumor effect of pi3k inhibitors. Proc. Natl. Acad. Sci. USA. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Renshaw J., Taylor K.R., Bishop R., Valenti M., Brandon A.d., Gowan S., Eccles S.A., Ruddle R.R., Johnson L.D., Raynaud F.I., et al. Dual blockade of the pi3k/akt/mtor (azd8055) and ras/mek/erk (azd6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin. Cancer Res. 2013;19:5940–5951. doi: 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Engelman J.A., Chen L., Tan X., Crosby K., Guimaraes A.R., Upadhyay R., Maira M., McNamara K., Perera S.A., Song Y., et al. Effective use of pi3k and mek inhibitors to treat mutant k-ras g12d and pik3ca h1047r murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shimizu T., Tolcher A.W., Papadopoulos K.P., Beeram M., Rasco D.W., Smith L.S., Gunn S., Smetzer L., Mays T.A., Kaiser B., et al. The clinical effect of the dual-targeting strategy involving pi3k/akt/mtor and ras/mek/erk pathways in patients with advanced cancer. Clin. Cancer Res. 2012;18:2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- 111.Jokinen E., Koivunen J.P. Mek and pi3k inhibition in solid tumors: Rationale and evidence to date. Ther. Adv. Med. Oncol. 2015;7:170–180. doi: 10.1177/1758834015571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anderson G.R., Winter P.S., Lin K.H., Nussbaum D.P., Cakir M., Stein E.M., Soderquist R.S., Crawford L., Leeds J.C., Newcomb R., et al. A landscape of therapeutic cooperativity in kras mutant cancers reveals principles for controlling tumor evolution. Cell Rep. 2017;20:999–1015. doi: 10.1016/j.celrep.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fedele C., Ran H., Diskin B., Wei W., Jen J., Geer M.J., Araki K., Ozerdem U., Simeone D.M., Miller G., et al. Shp2 inhibition prevents adaptive resistance to mek inhibitors in multiple cancer models. Cancer Discov. 2018;8:1237–1249. doi: 10.1158/2159-8290.CD-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fedele C., Li S., Teng K.W., Foster C.J.R., Peng D., Ran H., Mita P., Geer M.J., Hattori T., Koide A., et al. Shp2 inhibition diminishes krasg12c cycling and promotes tumor microenvironment remodeling. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vitiello M., Palma G., Monaco M., Bello A.M., Camorani S., Francesca P., Rea D., Barbieri A., Chiappetta G., Vita G., et al. Dual oncogenic/anti-oncogenic role of patz1 in frtl5 rat thyroid cells transformed by the ha-ras(v12) oncogene. Genes. 2019;10:127. doi: 10.3390/genes10020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rowell C.A., Kowalczyk J.J., Lewis M.D., Garcia A.M. Direct demonstration of geranylgeranylation and farnesylation of ki-ras in vivo. J. Biol. Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- 117.James G.L., Goldstein J.L., Brown M.S. Polylysine and cvim sequences of k-rasb dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J. Biol. Chem. 1995;270:6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 118.Whyte D.B., Kirschmeier P., Hockenberry T.N., Nunez-Oliva I., James L., Catino J.J., Bishop W.R., Pai J.K. K- and n-ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 119.Rao S., Cunningham D., de Gramont A., Scheithauer W., Smakal M., Humblet Y., Kourteva G., Iveson T., Andre T., Dostalova J., et al. Phase iii double-blind placebo-controlled study of farnesyl transferase inhibitor r115777 in patients with refractory advanced colorectal cancer. J. Clin. Oncol. 2004;22:3950–3957. doi: 10.1200/JCO.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 120.Van Cutsem E., van de Velde H., Karasek P., Oettle H., Vervenne W.L., Szawlowski A., Schoffski P., Post S., Verslype C., Neumann H., et al. Phase iii trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J. Clin. Oncol. 2004;22:1430–1438. doi: 10.1200/JCO.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 121.Cox A.D., Der C.J., Philips M.R. Targeting ras membrane association: Back to the future for anti-ras drug discovery? Clin. Cancer Res. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ho A., Chau N., Garcia I.B., Ferte C., Even C., Burrows F., Kessler L., Mishra V., Magnuson K., Scholz C., et al. Preliminary results from a phase 2 trial of tipifarnib in hras-mutant head and neck squamous cell carcinomas. Int. J. Radiat. Oncol. Biol. Phys. 2018;100:1367. doi: 10.1016/j.ijrobp.2017.12.156. [DOI] [Google Scholar]
- 123.Lee H.W., Sa J.K., Gualberto A., Scholz C., Sung H.H., Jeong B.C., Choi H.Y., Kwon G.Y., Park S.H. A phase ii trial of tipifarnib for patients with previously treated, metastatic urothelial carcinoma harboring hras mutations. Clin. Cancer Res. 2020;26:5113–5119. doi: 10.1158/1078-0432.CCR-20-1246. [DOI] [PubMed] [Google Scholar]
- 124.Hanna G.J., Guenette J.P., Chau N.G., Sayehli C.M., Wilhelm C., Metcalf R., Wong D.J., Brose M., Razaq M., Perez-Ruiz E., et al. Tipifarnib in recurrent, metastatic hras-mutant salivary gland cancer. Cancer. 2020;126:3972–3981. doi: 10.1002/cncr.33036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ryan M.B., de la Cruz F.F., Phat S., Myers D.T., Wong E., Shahzade H.A., Hong C.B., Corcoran R.B. Vertical pathway inhibition overcomes adaptive feedback resistance to kras(g12c) inhibition. Clin. Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., et al. Rapid non-uniform adaptation to conformation-specific kras(g12c) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lou K., Steri V., Ge A.Y., Hwang Y.C., Yogodzinski C.H., Shkedi A.R., Choi A.L.M., Mitchell D.C., Swaney D.L., Hann B., et al. Kras(g12c) inhibition produces a driver-limited state revealing collateral dependencies. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aaw9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Misale S., Fatherree J.P., Cortez E., Li C., Bilton S., Timonina D., Myers D.T., Lee D., Gomez-Caraballo M., Greenberg M., et al. Kras g12c nsclc models are sensitive to direct targeting of kras in combination with pi3k inhibition. Clin. Cancer Res. 2019;25:796–807. doi: 10.1158/1078-0432.CCR-18-0368. [DOI] [PubMed] [Google Scholar]
- 129.Molina-Arcas M., Moore C., Rana S., van Maldegem F., Mugarza E., Romero-Clavijo P., Herbert E., Horswell S., Li L.S., Janes M.R., et al. Development of combination therapies to maximize the impact of kras-g12c inhibitors in lung cancer. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aaw7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Puyol M., Martin A., Dubus P., Mulero F., Pizcueta P., Khan G., Guerra C., Santamaria D., Barbacid M. A synthetic lethal interaction between k-ras oncogenes and cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 131.Chen Y.N., LaMarche M.J., Chan H.M., Fekkes P., Garcia-Fortanet J., Acker M.G., Antonakos B., Chen C.H., Chen Z., Cooke V.G., et al. Allosteric inhibition of shp2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 132.Garcia Fortanet J., Chen C.H., Chen Y.N., Chen Z., Deng Z., Firestone B., Fekkes P., Fodor M., Fortin P.D., Fridrich C., et al. Allosteric inhibition of shp2: Identification of a potent, selective, and orally efficacious phosphatase inhibitor. J. Med. Chem. 2016;59:7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 133.Nichols R.J., Haderk F., Stahlhut C., Schulze C.J., Hemmati G., Wildes D., Tzitzilonis C., Mordec K., Marquez A., Romero J., et al. Ras nucleotide cycling underlies the shp2 phosphatase dependence of mutant braf-, nf1- and ras-driven cancers. Nat. Cell Biol. 2018;20:1064–1073. doi: 10.1038/s41556-018-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hillig R.C., Sautier B., Schroeder J., Moosmayer D., Hilpmann A., Stegmann C.M., Werbeck N.D., Briem H., Boemer U., Weiske J., et al. Discovery of potent sos1 inhibitors that block ras activation via disruption of the ras-sos1 interaction. Proc. Natl. Acad. Sci. USA. 2019;116:2551–2560. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hofmann M.H., Gmachl M., Ramharter J., Savarese F., Gerlach D., Marszalek J.R., Sanderson M.P., Kessler D., Trapani F., Arnhof H., et al. Bi-3406, a potent and selective sos1::Kras interaction inhibitor, is effective in kras-driven cancers through combined mek inhibition. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramharter J., Kessler D., Ettmayer P., Hofmann M.H., Gerstberger T., Gmachl M., Wunberg T., Kofink C., Sanderson M., Arnhof H., et al. One atom makes all the difference: Getting a foot in the door between sos1 and kras. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c01949. [DOI] [PubMed] [Google Scholar]
- 137.Ruess D.A., Heynen G.J., Ciecielski K.J., Ai J., Berninger A., Kabacaoglu D., Gorgulu K., Dantes Z., Wormann S.M., Diakopoulos K.N., et al. Mutant kras-driven cancers depend on ptpn11/shp2 phosphatase. Nat. Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 138.Valencia-Sama I., Ladumor Y., Kee L., Adderley T., Christopher G., Robinson C.M., Kano Y., Ohh M., Irwin M.S. Nras status determines sensitivity to shp2 inhibitor combination therapies targeting the ras-mapk pathway in neuroblastoma. Cancer Res. 2020;80:3413–3423. doi: 10.1158/0008-5472.CAN-19-3822. [DOI] [PubMed] [Google Scholar]
- 139.Karapetis C.S., Khambata-Ford S., Jonker D.J., O’Callaghan C.J., Tu D., Tebbutt N.C., Simes R.J., Chalchal H., Shapiro J.D., Robitaille S., et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 140.Loupakis F., Ruzzo A., Cremolini C., Vincenzi B., Salvatore L., Santini D., Masi G., Stasi I., Canestrari E., Rulli E., et al. Kras codon 61, 146 and braf mutations predict resistance to cetuximab plus irinotecan in kras codon 12 and 13 wild-type metastatic colorectal cancer. Br. J. Cancer. 2009;101:715–721. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Douillard J.Y., Oliner K.S., Siena S., Tabernero J., Burkes R., Barugel M., Humblet Y., Bodoky G., Cunningham D., Jassem J., et al. Panitumumab-folfox4 treatment and ras mutations in colorectal cancer. N. Engl. J. Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 142.Pinto C., Normanno N., Orlandi A., Fenizia F., Damato A., Maiello E., Tamburini E., di Costanzo F., Tonini G., Bilancia D., et al. Phase iii study with folfiri + cetuximab versus folfiri + cetuximab followed by cetuximab alone in ras and braf wt mcrc. Future Oncol. 2018;14:1339–1346. doi: 10.2217/fon-2017-0592. [DOI] [PubMed] [Google Scholar]
- 143.Peeters M., Douillard J.Y., van Cutsem E., Siena S., Zhang K., Williams R., Wiezorek J. Mutant kras codon 12 and 13 alleles in patients with metastatic colorectal cancer: Assessment as prognostic and predictive biomarkers of response to panitumumab. J. Clin. Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 144.Costas-Chavarri A., Nandakumar G., Temin S., Lopes G., Cervantes A., Correa M.C., Engineer R., Hamashima C., Ho G.F., Huitzil F.D., et al. Treatment of patients with early-stage colorectal cancer: Asco resource-stratified guideline. J. Glob. Oncol. 2019;5:1–19. doi: 10.1200/JGO.18.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Roock W., Jonker D.J., di Nicolantonio F., Sartore-Bianchi A., Tu D., Siena S., Lamba S., Arena S., Frattini M., Piessevaux H., et al. Association of kras p.G13d mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 146.Segelov E., Thavaneswaran S., Waring P.M., Desai J., Robledo K.P., Gebski V.J., Elez E., Nott L.M., Karapetis C.S., Lunke S., et al. Response to cetuximab with or without irinotecan in patients with refractory metastatic colorectal cancer harboring the kras g13d mutation: Australasian gastro-intestinal trials group icecream study. J. Clin. Oncol. 2016;34:2258–2264. doi: 10.1200/JCO.2015.65.6843. [DOI] [PubMed] [Google Scholar]
- 147.Segelov E., Waring P., Desai J., Wilson K., Gebski V., Thavaneswaran S., Elez E., Underhill C., Pavlakis N., Chantrill L., et al. Icecream: Randomised phase ii study of cetuximab alone or in combination with irinotecan in patients with metastatic colorectal cancer with either kras, nras, braf and pi3kca wild type, or g13d mutated tumours. BMC Cancer. 2016;16:339. doi: 10.1186/s12885-016-2389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nakamura M., Aoyama T., Ishibashi K., Tsuji A., Takinishi Y., Shindo Y., Sakamoto J., Oba K., Mishima H. Randomized phase ii study of cetuximab versus irinotecan and cetuximab in patients with chemo-refractory kras codon g13d metastatic colorectal cancer (g13d-study) Cancer Chemother. Pharmacol. 2017;79:29–36. doi: 10.1007/s00280-016-3203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McFall T., Diedrich J.K., Mengistu M., Littlechild S.L., Paskvan K.V., Sisk-Hackworth L., Moresco J.J., Shaw A.S., Stites E.C. A systems mechanism for kras mutant allele-specific responses to targeted therapy. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aaw8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rabara D., Tran H.T., Dharmaiah S., Stephens R.M., McCormick F., Simanshu D.K., Holderfield M. KRAS G13D sensitivity to neurofibromin-mediated GTP hydolysis. Proc. Natl. Acad. Sci. USA. 2019;116:22122–22131. doi: 10.1073/pnas.1908353116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hobbs G.A., Baker N.M., Miermont A.M., Thurman R.D., Pierobon M., Tran T.H., Anderson A.O., Waters A.M., Diehl J.N., Papke B., et al. Atypical kras(g12r) mutant is impaired in pi3k signaling and macropinocytosis in pancreatic cancer. Cancer Discov. 2020;10:104–123. doi: 10.1158/2159-8290.CD-19-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nat. Cell Biol. 2013;497:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kamphorst J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Heiden M.G.V., Miller G., Drebin J.A., Bar-Sagi D., et al. Human Pancreatic Cancer Tumors Are Nutrient Poor and Tumor Cells Actively Scavenge Extracellular Protein. Cancer Res. 2015;75:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zafra M.P., Parsons M.J., Kim J., Alonso-Curbelo D., Goswami S., Schatoff E.M., Han T., Katti A., Fernandez M.T.C., Wilkinson J.E., et al. An In Vivo Kras Allelic Series Reveals Distinct Phenotypes of Common Oncogenic Variants. Cancer Discov. 2020;10:1654–1671. doi: 10.1158/2159-8290.CD-20-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in supplemental Table S1. Data were downloaded from https://www.depmap.org/depmap, accessed on 27 April 2021 (DepMap Data) and from [54] (siREN Screen Data).