Summary
Influenza-associated pulmonary aspergillosis (IAPA) has been reported increasingly since the advent of use of neuraminidase (NA) inhibitors following the 2009 influenza pandemic. We hypothesize that blocking host NA modulates the immune response against Aspergillus fumigatus. We demonstrate that NA influences the host response against A. fumigatus in vitro and that oseltamivir increases the susceptibility of mice to pulmonary aspergillosis. Oseltamivir impairs the mouse splenocyte and human peripheral blood mononuclear cell (PBMC) killing capacity of A. fumigatus, and adding NA restores this defect in PBMCs. Furthermore, the sialic acid-binding receptor SIGLEC15 is upregulated in PBMCs stimulated with A. fumigatus. Silencing of SIGLEC15 decrease PBMC killing of A. fumigatus. We provide evidence that host NA activity and sialic acid recognition are important for anti-Aspergillus defense. NA inhibitors might predispose individuals with severe influenza to invasive aspergillosis. These data shed light on the pathogenesis of invasive fungal infections and may identify potential therapeutic targets.
Keywords: aspergillosis, neuraminidase, oseltamivir, SIGLEC15
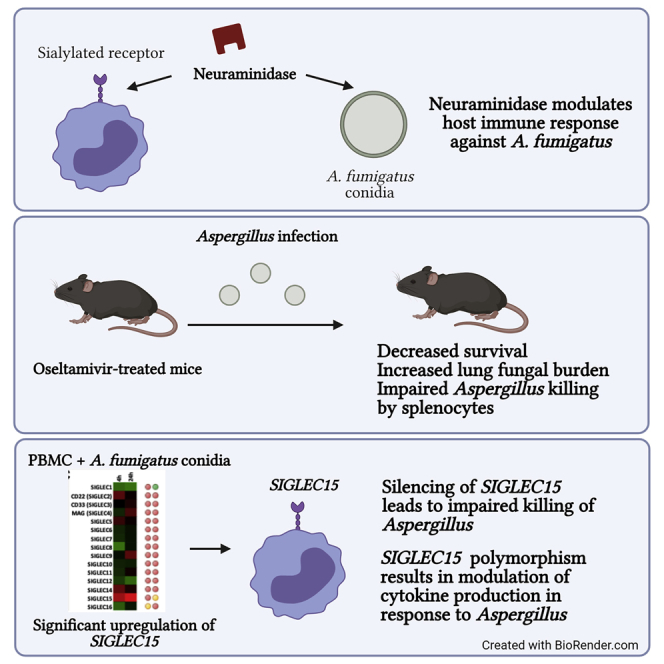
Graphical abstract
Highlights
Neuraminidase modulates the host immune response against A. fumigatus
Oseltamivir increases the susceptibility of animal model to pulmonary aspergillosis
SIGLEC15 is important for host defense against A. fumigatus
Dewi et al. show that neuraminidase plays an important role in host defense against A. fumigatus and that this effect could be mediated by SIGLEC15. Neuraminidase inhibition by oseltamivir might impair antifungal responses. These findings are important for understanding the pathogenesis of influenza-associated pulmonary aspergillosis (IAPA).
Introduction
Influenza-associated pulmonary aspergillosis (IAPA) is an emerging disease caused primarily by the ubiquitous fungus Aspergillus fumigatus. Two recent studies show that the incidence of IAPA among individuals with severe influenza is around 20% and that the mortality rate is more than 50%, whereas the normal background incidence of invasive aspergillosis in the intensive care unit (ICU) is 5%, and the mortality of influenza is around 20%.1,2 A striking feature of IAPA is that the infection occurs in individuals without classic risk factors for invasive aspergillosis, and up to 30% of cases occur in previously healthy individuals.1 The pathogenesis of IAPA is not understood, which limits the clinical possibilities of adequate diagnosis, treatment, and risk stratification. Understanding the underlying mechanisms and interactions between the immune system, the virus, and Aspergillus is essential.
Oseltamivir is a neuraminidase (NA) inhibitor that is administered in virtually all individuals with severe influenza since the H1N1 pandemic in 2009. Oseltamivir inhibits the NA activity of the virus,3 which is needed for successful virus replication. The virus cleaves sialic acid residues on the host cell surface that subsequently allow the mature virion to be released. However, humans and mice also express NAs, NEU1–NEU4, that mediate diverse functions in host immune responses.4, 5, 6, 7 Mammalian cells are covered in dense sialic acid sugars.8 Sialic acid-binding immunoglobulin-like lectin (SIGLEC) receptors are expressed on the surface of immune cells, and these receptors are usually bound and, thus, masked by “self” sialic acids.9 In the presence of NA, the receptors become unmasked, allowing them to bind non-self sialylated ligands. A. fumigatus contains sialic acid sugars on its cell walls that modulate immune responses.10 Interestingly, treatment with oseltamivir has been associated with increased platelet numbers in individuals with idiopathic thrombocytopenic purpura (ITP), supporting the concept that oseltamivir can also block human NA activity.11, 12, 13 Currently, two clinical trials with oseltamivir in individuals with ITP are ongoing (NCT03520049 and NCT01965626), based on the fact that oseltamivir can block human NA activity and, in this way, alter sialic acids on the surface of platelets.
Based on these data, we hypothesized that inhibition of NA for treatment of influenza might predispose individuals to IAPA via the effects of the NA inhibitor on host NA activity. Alteration of host NA activity could potentially affect the host-Aspergillus interaction, leading to increased susceptibility to invasive aspergillosis. The results of this study are important to unravel the underlying mechanisms and pathogenesis of invasive pulmonary aspergillosis following severe influenza, particularly in the setting of clinical use of NA inhibitors.
Results
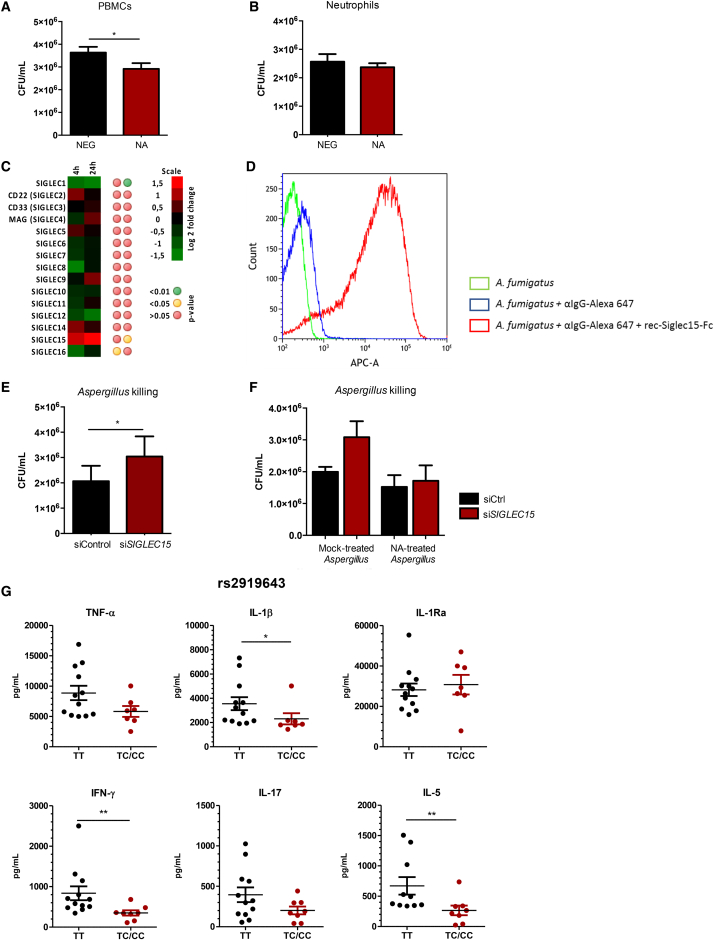
NA activity modulates the host response to A. fumigatus
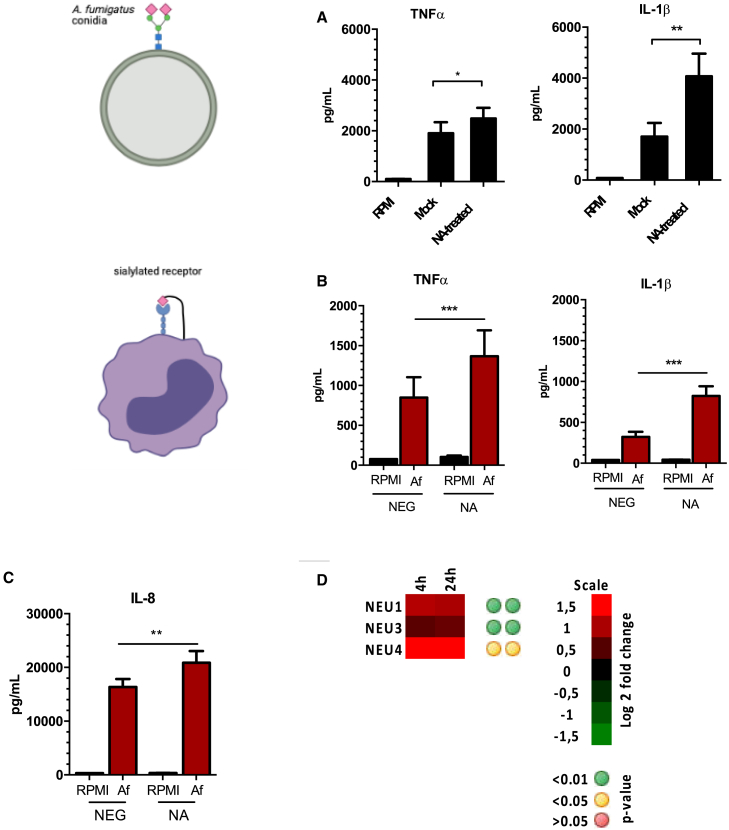
Previous literature has highlighted the role of human NA in mediating the host immune response.14,15 However, the role of these enzymes in modulating the host responses against A. fumigatus has not been described. The cell wall of A. fumigatus contains sialic acid residues that facilitate binding to epithelial cells in the airway.10 To determine the role of NA in mediating host immune responses against A. fumigatus, we first investigated the effect of NA on the capacity of conidia to induce cytokine responses. Peripheral blood mononuclear cells (PBMCs) were stimulated with A. fumigatus conidia pretreated with C. perfringens NA or mock treated. Production of tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) was increased significantly in cells stimulated with NA-treated conidia, suggesting that alteration of sialic acids on the surface of A. fumigatus conidia influences the host response (Figure 1A). To confirm that NA cleaved conidial glycans, A. fumigatus conidia were incubated with Maackia amurensis lectin (MAL) II, Sambucus nigra agglutinin (SNA), or peanut agglutinin (PNA) labeled with a secondary fluorophore and analyzed by flowcytometry. We observed that NA treatment of conidia reduces the amount of (α2,3)-linked and (α2,6)-linked sialic acids (Figure S1).
Figure 1.
NA modulates the host response of PBMCs against A. fumigatus
(A) TNF-α and IL-1β concentrations in culture supernatants of PBMCs (n = 12) stimulated with 1 × 107/mL of mock- or NA-treated A. fumigatus conidia.
(B and C) PBMCs or neutrophils (n = 12) were stimulated with A. fumigatus conidia in the presence or absence of NA, and concentrations of TNF-α, IL-1β, or IL-8 were measured from 24-h-cultured supernatants.
(D) Gene expression profiling of the NEU genes in PBMCs (n = 8) that were stimulated with heat-killed A. fumigatus conidia for 4 and 24 h.16
Data are presented as mean ± SEM. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
Next we determined the effect of NA on host immune cells in response to A. fumigatus conidia. PBMCs and neutrophils were stimulated with A. fumigatus conidia in the presence or absence of NA, and the production of cytokines was measured from 24-h-stimulated culture supernatants. In the presence of NA, PBMCs produced more TNF-α and IL-1β upon stimulation with conidia (Figure 1B). Similarly, IL-8 production against A. fumigatus was increased in neutrophils that were treated with NA (Figure 1C). To confirm that the effect of NA was specific to host immune cells, we also pre-treated cells with NA prior to stimulation with A. fumigatus conidia. Similar to what we observed, exposure of cells to exogenous NA enhanced cytokine production in response to the fungus (Figure S2). To exclude the possibility that the increased cytokine release was due to lipopolysaccharide (LPS) contamination in the enzyme preparation, experiments were repeated in the presence of polymyxin B. Addition of polymyxin B produced similar results: NA-treated cells produced higher cytokine concentrations (Figure S2). We further investigate whether A. fumigatus induces expression of NEU genes in PBMCs. Indeed, NEU1, NEU3, and NEU4 expression was upregulated significantly in PBMCs after 4- and 24-h stimulation with conidia (Figure 1D). These data suggest that NA can modulate host immune responses against A. fumigatus at the level of the pathogen and host immune cells.
NA inhibition increases susceptibility of mice to invasive aspergillosis
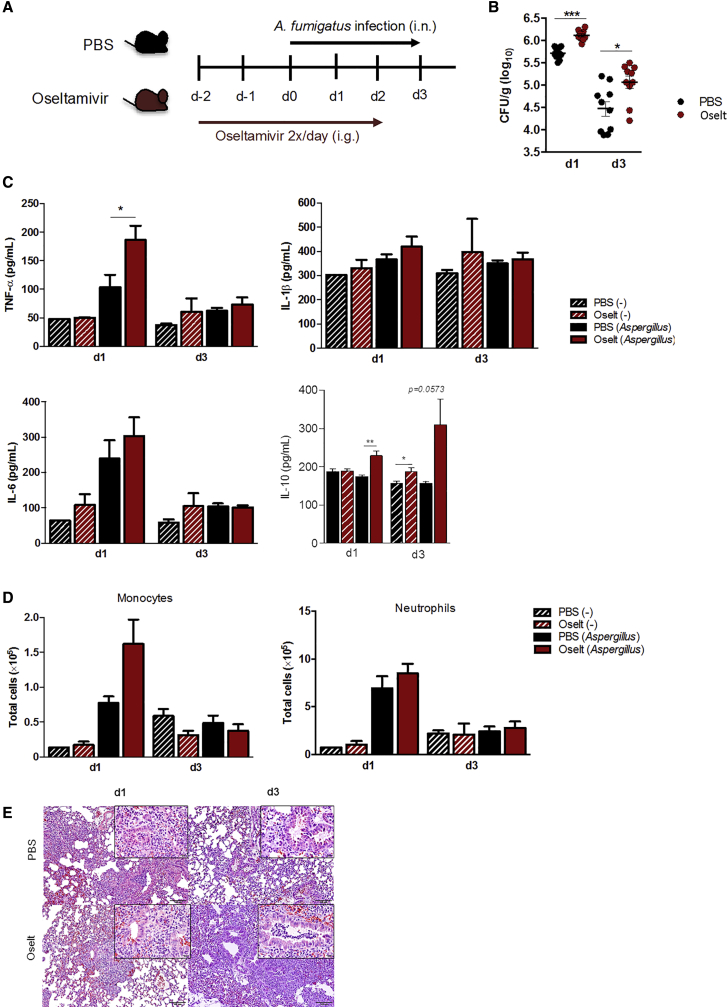
Because NA activity can modulate the host response against A. fumigatus, we investigated whether blocking endogenous NA with oseltamivir would affect cytokine responses against A. fumigatus. Our results demonstrate that oseltamivir carboxylate did not affect production of TNF-α and IL-1β by PBMCs (Figure S3). Therefore, we evaluated the effect of NA inhibition by oseltamivir in vivo and determined whether it would influence the susceptibility of mice to invasive pulmonary aspergillosis. Immunocompetent C57BL/6 mice were treated with oseltamivir twice daily for 5 consecutive days. On day 3 of treatment, mice were infected intranasally with A. fumigatus (Figure 2A). On days 1 and 3 after infection, we measured the concentrations of cytokines and fungal burden in lung homogenates. We observed that mice treated with oseltamivir had a significantly higher fungal burden in the lungs compared with PBS-treated controls at both time points, suggesting that the fungus was not being cleared effectively from the lungs (Figure 2B). Infected immunocompetent mice treated with oseltamivir produced significantly higher TNF-α and IL-10 compared with control mice on day 1 after infection (Figure 2C), but there were no significant differences in IL-1β and IL-6 concentrations. On day 3 after infection, we did not observe any differences between mice treated with oseltamivir and controls in cytokine production except for IL-10. Determination of the total number of immune cells present in the lungs by flow cytometry did not show a significant difference in the total number of monocytes and neutrophils in the lungs between oseltamivir-treated and control mice infected with A. fumigatus (Figure 2D). Histology of the lung tissue showed that, on day 1, there were less inflammatory infiltrates in mice exposed to oseltamivir compared with controls, whereas no differences were observed in the lungs on day 3 between the two groups (Figure 2E). Thus, despite normal cell recruitment and intact cytokine responses, the mice showed a higher fungal burden in the presence of NA inhibition.
Figure 2.
Oseltamivir affects the susceptibility of immunocompetent mice to A. fumigatus infection
(A) Immunocompetent C57BL/6J mice were treated with PBS (n = 20) or oseltamivir (10 mg/kg) (n = 20) twice daily for 5 consecutive days. On day 3 of treatment, mice were infected intranasally with A. fumigatus or remained uninfected (naive). All mice were sacrificed on days 1 and 3 after infection.
(B) Fungal burden in lung homogenates on days 1 and 3 after infection (log10).
(C) Production of TNF-α, IL-1β, IL-6, and IL-10 was measured from lung homogenates of uninfected (striped bars) or infected (empty bars) mice that were treated with oseltamivir (red bars) or remained untreated (black bars).
(D) The total number of immune cells in lung homogenates from infected or uninfected mice treated with oseltamivir or controls.
(E) Representative histology of the lung tissue sections of the mice on day 1 and day 3 after infection, stained with H&E.
Data are presented as mean ± SEM. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
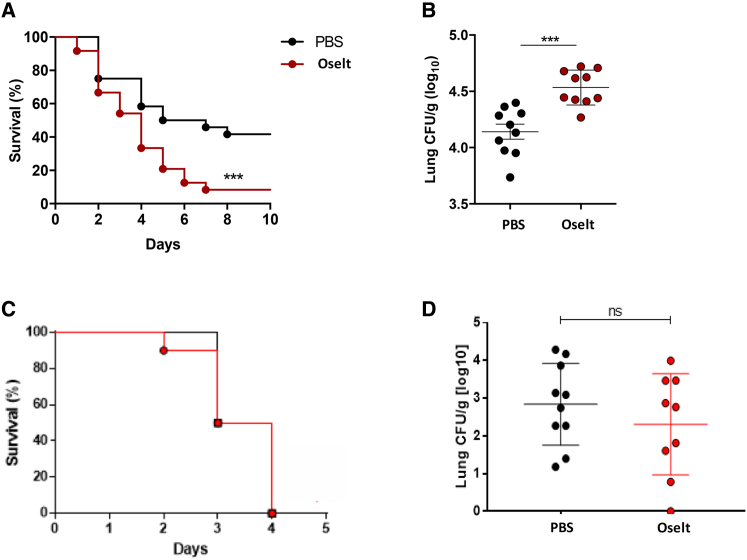
Because invasive pulmonary aspergillosis usually occurs in the setting of an underlying immunosuppressive condition, and in influenza especially in the presence of corticosteroid use, we further determined the effect of blocking NA in vivo in corticosteroid (CS)-treated C57BL/6J mice. Immunosuppressed mice treated with oseltamivir had significant lower survival compared with controls (Figure 3A). Furthermore, the fungal burden in lung homogenates of these mice was significantly higher than in control mice (Figure 3B). We further determined whether oseltamivir could affect the host response against A. fumigatus in the setting of chemotherapy-induced immunosuppression. BALB/c mice were immunosuppressed by intraperitoneal administration of cyclophosphamide. Oseltamivir was given twice daily for 5 days, and A. fumigatus was given intranasally on the third day of treatment. Strikingly, we observed no significant differences in survival and lung fungal burden in oseltamivir-treated mice compared with controls (Figures 3C and 3D). Additionally, bioluminescent signals of regions of interest covering the lungs of A. fumigatus as well as lesion development were not different between the two groups (Figure S4). Because we did not see an effect of oseltamivir on recruitment or cytokine responses, and the effects on survival and fungal burden were lost in the setting of depleted monocytes and neutrophils, we hypothesized that the effect of blocking neuraminidase in vivo on the host response against A. fumigatus might be mediated by altered function of monocytes or neutrophils or both.
Figure 3.
The effect of oseltamivir on immunocompromised mice
(A and B) Corticosteroid-treated C57BL/6J mice were treated with PBS or oseltamivir (10 mg/kg) twice daily for 5 days and infected with A. fumigatus (1 × 107) via the intranasal route.
(A) Kaplan-Meier survival plot of mice infected with A. fumigatus. Results were derived from three independent experiments (total n = 24 mice per group).
(B) Fungal burden in lung homogenates of mice on day 2 after infection. ∗∗∗p < 0.001.
(C and D) BALB/c mice were treated with oseltamivir (10 mg/kg, n = 10) or PBS (n = 10) twice per day for 5 days and infected intranasally with A. fumigatus. Immunosuppression was induced by intraperitoneal administration of cyclophosphamide (150 mg/kg). Shown are Kaplan-Meier survival plot (C) from mice after 3 days of infection and fungal burden (D) measured from lung homogenates.
Blocking NA impairs fungal killing in murine splenocytes and human PBMCs
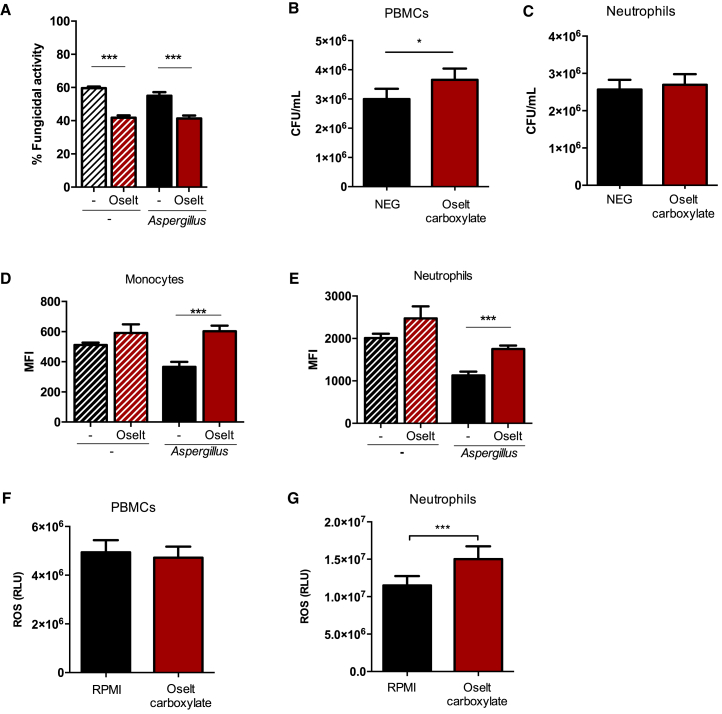
To further investigate whether NA inhibition modulates the antifungal host defense of myeloid cells, we recovered splenocytes from immunocompetent mice treated with oseltamivir or PBS and left uninfected or infected with A. fumigatus. The capacity of these cells to kill A. fumigatus conidia was evaluated ex vivo. We observed that splenocytes from oseltamivir-treated mice had a marked decrease in the capacity to kill conidia compared with splenocytes from untreated mice (Figure 4A). Strikingly, similar findings were observed in murine splenocytes of mice treated with oseltamivir but not infected with A. fumigatus. To validate these findings in the human setting, we analyzed the killing capacity of PBMCs and neutrophils against A. fumigatus in the presence or absence of oseltamivir carboxylate, the active metabolite of oseltamivir. Our data show that the capacity to kill A. fumigatus was decreased in oseltamivir-treated compared with untreated PBMCs (Figure 4B), but killing was unaffected in neutrophils (Figure 4C). NA inhibition did not decrease reactive oxygen species (ROS) production in vivo or in vitro and was even associated with increased ROS production in vitro in neutrophils (Figures 4D–4G).
Figure 4.
Oseltamivir treatment impaired the fungal killing capacity of mouse splenocytes and human PBMCs
(A) Killing capacity of splenocytes from immunocompetent C57BL/6J mice treated with (red bars) or without (black bars) oseltamivir and infected with A. fumigatus (1 × 106/mL) (empty bars) or left uninfected (striped bars).
(B and C) PBMCs and neutrophils were stimulated with A. fumigatus conidia (MOI of 4:1) in the presence or absence of oseltamivir carboxylate (n = 16). A killing assay was performed after 4 h of incubation at 37°C. Cells were lysed with H2O and plated on Sabouraud dextrose agar (SDA) in serial dilutions, and the remaining CFUs were counted after 24-h incubation at 37°C.
(D and E) ROS production from immune cells in lung homogenates of immunocompetent mice treated with or without oseltamivir and infected with A. fumigatus or left uninfected.
(F and G) ROS induction from PBMCs and neutrophils stimulated with A. fumigatus conidia in the presence or absence of oseltamivir carboxylate (n = 11).
Data are shown as mean ± SEM. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
The role of SIGLEC15 in the host immune defense against A. fumigatus
Our data suggest that the fungal capacity of mononuclear cells is altered by NA inhibition. Therefore, we performed killing assays in PBMCs in the presence or absence of NA to determine whether the effect of NA activity acts directly on these cells. In line with these observations, addition of NA enhanced the killing capacity of PBMCs against A. fumigatus (Figure 5A) but did not affect neutrophil killing function (Figure 5B). SIGLECs are a family of receptors that regulate different immune functions,17 and these receptors are densely covered sialic acids from other receptors on the same cell surface. In the presence of NA, SIGLEC receptors become unmasked and can bind other sialylated ligands with higher sialic acid density. To investigate whether A. fumigatus induces expression of SIGLEC genes in PBMCs, we examined RNA sequencing (RNA-seq) from an existing dataset.16 We identified one SIGLEC receptor that was upregulated significantly after exposure to Aspergillus, SIGLEC15 (Figure 5C). Expression of SIGLEC15 was upregulated after 4- and 24-h stimulation with conidia, although, after correction for multiple testing, only 24-h expression remained significant. We further investigated the interaction of this particular receptor with A. fumigatus using flow cytometry. Following incubation of conidia with recombinant SIGLEC15 (recSIGLEC15), we observed an increased fluorescent signal under recSIGLEC15-treated conditions, providing evidence that recSIGLEC15 binds to A. fumigatus (Figure 5D). To further explore the involvement of SIGLEC15 in A. fumigatus killing, SIGLEC15 expression was silenced in PBMCs. Cells treated with SIGLEC15-targeting small interfering RNA (siRNA) exhibited decreased fungicidal activity compared with control siRNA-treated cells (Figure 5E). We also noted that silencing of SIGLEC15 reduced the expression of IL10 and TGFB, both which are known as downstream effectors of the receptor.18 However, we did not observe significant differences in TNF-α production induced by A. fumigatus as well as zymosan-induced ROS in SIGLEC15-silenced cells and controls (Figure S5). The cell wall of Aspergillus spp. conidia contains sialic acid residues.10 Because the SIGLEC15 receptor can bind to sialic acids, we explored whether removal of sialic acids from A. fumigatus would affect killing in the presence or absence of SIGLEC15. Stimulation of SIGLEC15-silenced cells with NA-treated conidia did not affect the killing capacity of these cells compared with mock-treated conidia (Figure 5F). This suggests that SIGLEC15-mediated killing requires the presence of sialic acids on the surface of A. fumigatus conidia. To further determine the role of SIGLEC15 in the host defense against A. fumigatus, we evaluated cytokine production from PBMCs of individuals harboring a polymorphism in SIGLEC15 (rs2919643). In response to A. fumigatus conidia, individuals carrying the disease-associated C allele had significantly lower IL-1β and interferon γ (IFN-γ) concentration, whereas other cytokines were unaffected (Figure 5G). Our results indicate that SIGLEC15 might be important in anti-Aspergillus host defense and further strengthens the theory of a role of NA activity and sialic acid recognition in the host immune response against A. fumigatus.
Figure 5.
The role of SIGLEC15 in the host response against A. fumigatus
(A and B) PBMCs or neutrophils were stimulated with A. fumigatus for 4 h in the presence or absence of exogenous C. perfringens NA (n = 14). Cells were then lysed in H2O, and the remaining conidia were plated in serial dilutions on SDA and incubated for 24 h at 37°C.
(C) Gene expression profiling of SIGLEC genes, using Illumina Human HT-12 Expression BeadChip, in PBMCs stimulated with heat-killed A. fumigatus conidia for 4 and 24 h (n = 8).16
(D) Recombinant SIGLEC15 (recSIGLEC15) binding to A. fumigatus. Green, heat-killed A. fumigatus; blue, heat-killed A. fumigatus + secondary antibody (anti immunoglobulin G [IgG]-Alexa 647); red, heat-killed A. fumigatus + recSIGLEC15 + secondary antibody.
(E) PBMCs were transfected with SIGLEC15 siRNA or scrambled control siRNA for 24 h prior to stimulation with A. fumigatus conidia. After 4 h of stimulation, cells were lysed with H2O, and a killing assay was performed as described previously (n = 12). Data are represented as mean ± SEM (∗p < 0.05).
(F) Killing capacity of PBMCs transfected with SIGLEC15 siRNA or control siRNA, as described previously, and stimulated with NA-treated or mock-treated A. fumigatus conidia (n = 6).
(G) A. fumigatus-induced cytokine production from PBMCs of individuals harboring a polymorphism in the SIGLEC15 gene (rs2919643) (TT = 12, CT/CC = 7). Data are represented as mean ± SEM (∗∗p < 0.01, ∗p < 0.05).
Discussion
IAPA has been reported increasingly since the 2009 flu pandemic, and the pathogenesis of this disease is complex and not well understood. Because sialic acid residues and receptors recognizing sialic acids play a key role in immunity,8,9,17 we hypothesized that the NA inhibitors that are used to treat influenza might impair the host defense against Aspergillus and therefore play a role in the increased susceptibility of individuals with influenza to invasive aspergillosis. We demonstrate that NA influences the host response to A. fumigatus, and inhibition of NA in vivo increased the susceptibility of mice to invasive aspergillosis. We observed that NA activity is important for optimal Aspergillus killing capacity of mononuclear cells in mice and human cells. We further demonstrate that SIGLEC15, a receptor on myeloid cells that recognize sialic acids, plays a role in killing of A. fumigatus. Thus, we provide evidence that NA activity and SIGLEC15 play a role in anti-Aspergillus host defense, opening a field in the pathogenesis of invasive fungal infection that is unexplored.
Our in vitro data suggest that sialic acid recognition and NA activity are vital for host-Aspergillus interaction. Enzymatic removal of sialic acids on fungal and/or host cells modulates host responses against conidia, and this effect was not due to endotoxin-induced effects in the setting of the experiments with the exogenous enzyme. We also demonstrate that A. fumigatus conidia contain (α2,3)-linked and (α2,6)-linked sialic acid residues. This is relevant with regard to previous studies reporting that the cell walls of A. fumigatus conidia contain negatively charged sugars10,19 that are necessary for fungal adhesion to laminin and fibronectin in respiratory epithelial tissues.10,20 Sialic acid residues are also present abundantly on the cell surface of human cells, where they mediate various functions in infection and inflammation.21 Sialic acids comprise an important part of adhesion molecules, which mediate leukocyte migration and adhesion.8 The presence of NA activity, which can cleave off these sialic acids, is thus likely to play a role in modulating host-pathogen interactions. A study by Amith et al.5 shows that binding of pathogen-associated molecular patterns (PAMPs) to Toll-like receptor 2 (TLR2), TLR3, and TLR4 was able to induce NA activity via Neu-1 expression in bone marrow-derived macrophages. Neu-1 is important for TLR4 activation because it removes α-2,3-sialyl residues linked to β-galactosidases on the surface of TLR4. Removal of these sialic acid residues leads to receptor TLR4 dimerization and nuclear factor κB (NF-κB) signaling.6,22 Neutrophils also express NAs that can desialylate several glycoproteins on the cell surface and facilitate binding to other cells.23 NAs present in T-lymphocytes have been shown to remove sialic acids from specific glycoconjugates on the cell surface, leading to enhanced IFN-γ production.7,24 A. fumigatus also produces sialidase (KDNase), which specifically cleaves the sialic acid substrate 2-keto-3-deoxy-D-glycero-D-galacto-nononic acid (KDN), and had lower activity in cleaving Neu5Ac from glycan structures25. Although the fungal sialidase gene could be induced by the presence of serum, it is questionable whether KDNase would affect host glycans because of its substrate specificity and limited abundance in lower vertebrates and bacteria.26,27 Our data add another layer of evidence showing that NA activity can modulate host-pathogen interactions via mononuclear cells.
These observations become especially important in the setting of IAPA. We have seen an increase in invasive aspergillosis in the ICU in individuals with influenza since the pandemic in 2009.1,2,28,29 To date, studies show an incidence of around 20% of IAPA in the ICU,1 with many individuals not having the classic risk factors for IA and even being healthy before admission to the ICU. Virtually all individuals with severe influenza admitted to the hospital or ICU receive NA inhibitors. Notably, the structure of mammalian NAs resembles bacterial and viral NAs in that they all harbor a conserved F(Y)RIP domain followed by a series of “Asp boxes” in the amino-terminal portion of the protein.30 Therefore, we and others have speculated that oseltamivir could also have an inhibitory effect on mammalian NAs.12,31,32 Although previous literature suggests that the drug has limited inhibitory activity against mammalian sialidases, intriguing results were observed in in vitro and in vivo cancer studies.33, 34, 35 Also, the clinical data showing that oseltamivir has effects on platelets in individuals with ITP further support this concept.11,12,32 The effect of oseltamivir on platelets has also been demonstrated in individuals with dengue, where desialylation of platelets occurs, and this effect was reduced by oseltamivir.31 However, we were able to demonstrate that immunocompetent mice treated with oseltamivir were more susceptible to A. fumigatus infection because of the impaired killing capacity of myeloid cells against the fungi. In vivo, NA activity might come from the host cells4,36 or from the fungus.25 However, we also investigated a condition where we did not have the actual Aspergillus infection but just exposed mice to oseltamivir and investigated the killing capacity of splenocytes compared with mice not exposed to oseltamivir. We could already demonstrate impairment of the killing capacity ex vivo of splenocytes exposed to oseltamivir. Therefore, although we do agree that fungus-derived NA could play a role during infection, at least the host NA is largely responsible for our observed killing capacity of monocytes because of oseltamivir. This is in line with previous studies that have elaborated the immunomodulatory effect of oseltamivir in the setting of influenza infection.37,38 We also explored whether NA inhibition affects the susceptibility of mice under immunosuppression conditions. We observed different outcomes in CS- and cyclophosphamide-treated mice. NA inhibition increased the susceptibility to invasive aspergillosis in CS-treated mice but not in mice treated with cyclophosphamide. This suggested that the effects of oseltamivir were dependent on myeloid cells. We observed that oseltamivir impaired the A. fumigatus killing capacity of murine splenocytes and human PBMCs. Additionally, the fact that NA inhibition by oseltamivir carboxylate did not have any effect on cytokine and ROS induction in PBMCs supports our observation that the drug modulates anti-Aspergillus defense by impairing the killing capacity of the cells.
SIGLEC receptors are a family of receptors that play a significant role in regulating host immune responses.9 These receptors bind sialylated structures on the surface of immune cells as well as on pathogens.39 In the absence of a sialylated ligand, SIGLECs are present in a cis transformation state, where they are “masked” by dense sialic acid structures from adjacent receptors. Following cellular activation by exposure to a NA or an encounter with higher-affinity ligands, these receptors are unmasked in a trans form,8,9 allowing binding to other sialylated ligands. Binding of SIGLEC receptors to a sialic acid then signals through downstream proteins, leading to activation or inhibition of different immune functions.17 Based on this concept, it is interesting to speculate that addition of NA can “open” the surface of SIGLEC receptors, allowing them to bind to and recognize sialylated A. fumigatus conidia and activation of downstream signaling pathways. Currently, there are two known SIGLEC families: the highly conserved group 1, which consists of sialoadhesin, CD22, SIGLEC44, and SIGLEC15, and group 2, which consists of CD33-related SIGLECs and varies among species.9 We identified SIGLEC15 as the only SIGLEC receptor that was strongly induced in PBMCs stimulated with A. fumigatus conidia. SIGLEC15 is expressed by myeloid cells40, 41, 42 and has been described recently to play a role in cancer immunology, where it suppresses T cell responses.40 SIGLEC15 binds preferentially to the Neu5Acα2-6GalNAc- (sialyl Tn) structure.41 Moreover, polymorphisms in the SIGLEC15 gene have been identified recently as a risk factor for recurrent vulvovaginal candidiasis (RVVC), and loss of this receptor is associated with altered cytokine responses against the fungal pathogen Candida albicans.43 Our findings showed that PBMCs from individuals carrying the same SNP as the one identified in the RVVC cohort (rs2919643) had altered IL-1β and IFN-γ production in response to A. fumigatus conidia. Most importantly, our results demonstrate that impairment of SIGLEC15 expression altered the killing capacity of mononuclear myeloid cells against A. fumigatus. recSIGLEC15 bound Aspergillus conidia, and knockdown of SIGLEC15 resulted in decreased killing capacity of PBMCs. This effect was lost when Aspergillus conidia were treated with NA, further supporting the concept that SIGLEC15, via recognition of sialic acids on the surface of Aspergillus, regulates fungal killing and that blocking NA alters this killing capacity. Our result was different from those of a previous study by Jaeger et al.43 regarding the fact that SIGLEC15 is important for killing of A. fumigatus but not C. albicans. Upon binding to its sialylated ligand, SIGLEC15 recruits the adaptor protein DNAX adaptor protein 12 (DAP12).18 DAP12 harbors an immunoreceptor tyrosine-based activating motif (ITAM) that, upon activation and phosphorylation, eventually recruits Syk. DAP12-Syk signaling subsequently induces other downstream targets, leading to different effector mechanisms, such as cytokine production and phagocytosis.44 Additionally, activation of DAP12-associated receptors, such as triggering receptors expressed on myeloid cells (TREMs), could further amplify DAP12-mediated signaling. TREM1, for instance, has been reported to mediate immune responses against A. fumigatus conidia in experimental fungal asthma.45 TREM2-DAP12 signaling is also crucial for binding and uptake of bacteria46 but not C. albicans.47 It is interesting to speculate that amplification of DAP12 signaling induced by SIGLEC15 and other DAP12-associated receptors is crucial for phagocytosis and killing of A. fumigatus but not C. albicans.
NA activity is important for protective responses against Aspergillus, and NA inhibitors might lead to decreased fungal killing capacity and increase susceptibility to invasive pulmonary aspergillosis. Furthermore, we identified SIGLEC15 as a receptor with a role in anti-Aspergillus host defense, which underscores the importance of sialic acids in host-pathogen interaction. These findings contribute to improving our understanding of the pathogenesis of IAPA, particularly regarding use of NA inhibition in critically ill individuals with severe influenza. In these people, inhibition of NA may be a crucial risk factor for development of IAPA.
Limitations of study
In this study, we show that NA plays a role in host defense against A. fumigatus. However, we did not report data regarding the efficiency of host NAs in cleaving sialic acids from immune cells and the relative inhibitory effect of these enzymes by oseltamivir. Additional experiments using recombinant mammalian sialidases would provide more detailed information. Furthermore, more work is needed to determine the effect of oseltamivir on other components of the antifungal response, including the effect on different cell types and effector functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BV510 CD45 | BioLegend | RRID:AB_2561392 |
| PerCp-Cy5.5 Ly-6C | BioLegend | RRID:AB_1727558 |
| BV785 Ly-6G | BioLegend | RRID:AB_2566317 |
| BV605 CD11c | BioLegend | RRID:AB_2562415 |
| PE-Cy7 CD11b | BioLegend | RRID:AB_2033994 |
| FITC I-A/I-E | BioLegend | RRID:AB_313321 |
| Alexa Fluor 647 CD103 | BioLegend | 121410 |
| APC F4/80 | BioLegend | RRID:AB_893481 |
| PE Siglec F | BD-Pharmigen | 552126 |
| CellROX™ Green Reagent | Thermo Fisher Scientific | C10444 |
| Goat anti-Human IgG (H+L) Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | RRID:AB_2535862 |
| Fungal strains | ||
| Aspergillus fumigatus V05-27 | Isolated from patient | N/A |
| Aspergillus fumigatus VP145_12 | Isolated from patient | N/A |
| Chemicals, peptides, and recombinant Proteins | ||
| Recombinant human SIGLEC15-Fc | Bio-Techne | 9227-SL-050 |
| Phosphate Buffered Saline (PBS) | Thermo Fisher Scientific | 10010056 |
| Tween-20 | Sigma-Aldrich | P1379 |
| Sabouraud Glucose Agar with Chloramphenicol | Sigma-Aldrich | 89579 |
| Neuraminidase from Clostridium perfringens | Sigma | N2876-6UN |
| RPMI 1640 | Life Technologies | 22409031 |
| Gentamycin | Centraform | N/A |
| Pyruvate | Invitrogen | 11360 |
| Ficoll-Paque Plus | GE Healthcare | 17-1440-03 |
| Bovine Serum Albumin (BSA) | Sigma | A7030 |
| iScript cDNA synthesis kit | Bio-Rad | 1708891 |
| Power SYBR Green | 4368708 | Life Technologies |
| Luminol (3-amino-2,3, dihydro-1,4-phtalazinedione) | Sigma | A8511-5G |
| Zymosan | Sigma | Z4250-1G |
| Oseltamivir carboxylate | Bioconnect Life Sciences | N/A |
| Lipofectamine RNAiMAX Transfection reagent | Fisher Scientific | 10514953 |
| Ketamidor | Mitcher Pharm | N/A |
| Medetomidine (Dormtor®) | Ecuphar | N/A |
| Cortisone-21 acetate | Acros Organics | 448960250 |
| Baytril® | Bayer | N/A |
| Cyclophosphamide | ||
| Oseltamivir (Tamiflu®) | Roche | N/A |
| DMEM medium | Thermo Fisher Scientific | 10938025 |
| Penicillin/Streptomycin | Thermo Fisher Scientific | 15140122 |
| HEPES Buffer Solution | Thermo Fisher Scientific | 15630056 |
| Fetal Bovine Serum | Thermo Fisher Scientific | 10500056 |
| Collagenase D | Sigma-Aldrich | 11088858001 |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | A7906 |
| Formalin | Sigma-Aldrich | HT501128 |
| Percoll | GE Healthcare | 17089102 |
| L-glutamine | Sigma-Aldrich | 25030081 |
| Critical commercial assays | ||
| Human TNF-α ELISA | Bio-Techne/R&D | DY201 |
| Human IL-1β ELISA | Bio-Techne/R&D | DY201 |
| Human IL-8 ELISA | IBL International | M9318 |
| Mouse TNF-α ELISA | BioLegend | 430904 |
| Mouse IL-1β ELISA | BioLegend | 432604 |
| Mouse IL-6 ELISA | BioLegend | 431304 |
| Experimental Models: Organisms/strains | ||
| C57BL/6 mice, 8 weeks old, adult male | Jackson Laboratory | N/A |
| BALB/c mice, 10-weeks old, adult male | Janvier, Le Genest, France | N/A |
| Oligonucleotides | ||
| ON-Target plus human SIGLEC15 siRNA smartpool (Dharmacon) | GE Healthcare | L-023427-02-0005 |
| ON-Target plus non-targeting pool siRNA | GE Healthcare | D-001810-10-05 |
| Forward primer hSIGLEC15: 5′-CGCG GATCGTCAACATCTC-3′ |
Biolegio (Primers designed by BLAST) | N/A |
| Reverse primer hSIGLEC15: 5′-GTTCG GCGGTCACTAGGTG-3′ |
Biolegio (Primers designed by BLAST) | N/A |
| Forward primer hRPL37: 5′-ATTGAA ATCAGCCAGCACG-3′ |
Biolegio (Primers designed by BLAST) | N/A |
| Reverse primer hRPL37: 5′- AGGAAC CACAGTGCCAGAT-3′ |
Biolegio (Primers designed by BLAST) | N/A |
| Deposited data | ||
| RNA seq | NCBI database | GSE162746 |
| Software and algorithms | ||
| GraphPad Prism 5.0 | Graphpad Software | https://www.graphpad.com |
| Kaluza | Beckman Coulter | N/A |
| FlowJo | Tree Star Inc | N/A |
| ImageJ software v1.50i | NHI | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Frank van de Veerdonk at the Radboud University Medical Center, Nijmegen, the Netherlands (Frank.vandeVeerdonk@radboudumc.nl).
Materials availability
This study did not generate new reagents.
Data and code availability
RNA seq data on SIGLEC15 an NEU genes were extracted from an existing RNaseq dataset.16 The datasets were retrieved from Gene Expression Omnibus (GEO) using identifier GEO Series accession number GSE162746.
Experimental model and subject details
Human Subjects
Peripheral Blood Mononuclear Cells (PBMCs) and neutrophils were isolated from venous blood of healthy volunteers after given informed consent. All donors were over 18 years of age. To further validate the findings related to SIGLEC15, ex vivo cytokine production was evaluated in PBMCs from healthy individuals that were recruited in the 200FG cohort study (http://www.humanfunctionalgenomics.org).48 Ethical approval for all the donors in this study was obtained from the ethical board of Arnhem-Nijmegen Medical Ethical Committee.
Animal subjects
For in vivo experiments, eight-week-old gender- and age- matched C57BL/6 mice were bred under specific-pathogen free condition and kept at the Life and Health Sciences Research Institute (ICVS) Animal Facility. Animal experimentation was performed following biosafety level 2 (BSL-2) protocols approved by the Institutional Animal Care and Use Committee (IACUC) of University of Minho and the ethical and regulatory approvals were consented by the Ethical Subcommission for Life and Health Sciences (no. 074/016). To determine the effect of oseltamivir on the susceptibility of cyclophosphamide-induced immunosuppressed mice, we also performed experiments in 10-weeks old, adult male BALB/C mice (Janvier, Le Genest, France), after ethical approval from the animal ethics committee of KU Leuven. All in vivo procedures were carried out according to national and European regulations.
Method details
Preparation of A. fumigatus conidia
Aspergillus fumigatus V05-27 was used for in vitro experiments with PBMCs and neutrophils. To prepare conidia, A. fumigatus was grown on Sabouraud dextrose agar (SDA) for 7 days at 37°C. Colonies were scraped from the agar surface and resuspended with sterile Tween in PBS solution. To remove debris and hyphae fraction, the suspension was filtered over layers of sterile gauze and washed twice with PBS. Conidia were counted and a final concentration of 1x107/mL was used in the experiments, unless otherwise indicated. To acquire NA-treated A. fumigatus, conidia were either treated with 0.5 U/mL NA derived from Clostridium perfringens (Sigma-Aldrich, St. Louis, Missouri, USA) or with a 25 mM KCl solution (mock) for 30 minutes at 37°C in the presence of 2 μg/mL polymyxin B, and washed twice with PBS.19,49 For in vivo experiments, Aspergillus fumigatus VP145_12 was used, with identical procedures for recovery of conidia.
Isolation and stimulation of PBMCs and neutrophils
PBMC fractions were isolated by Ficoll density gradient centrifugation (Ficoll-Paque Plus, GE Healthcare, Zeist, the Netherlands). Cells were washed twice with Phosphate Buffered Saline (PBS) and suspended in RPMI 1640 medium (GIBCO, Invitrogen, Breda, the Netherlands) supplemented with 5 mg/mL gentamicin, 1 mM pyruvate and 2 mM of L-glutamine (GIBCO). Cells were counted in a coulter counter (Beckman Coulter, Pasadena, CA, USA) and adjusted to a final concentration of 2.5x106 cells/mL. After removal of PBMC fractions, neutrophils were isolated from the remaining red blood cells by hypotonic lysis method.50 The remaining red blood cells were lysed twice by adding hypotonic lysis buffer containing 155 mM NH4Cl and 10 mM KHCO3, which was further diluted 10 times. Afterward, cells were washed twice with cold PBS and resuspended in RPMI 1640 medium as mentioned previously.
To determine the role of NA on the host responses against A. fumigatus, isolated PBMCs or neutrophils were plated onto a 96-wells plate and stimulated with RPMI, NA-treated or mock-treated A. fumigatus conidia. In a separate set of experiments, cells were stimulated with A. fumigatus conidia in the presence or absence of 0.2 U/mL C. perfringens NA. For in vitro experiments with NA inhibitors, PBMCs or neutrophils were stimulated with A. fumigatus conidia or medium, and either treated with 0.1 mM oseltamivir carboxylate (Bioconnect Life Sciences, the Netherlands) or remain untreated. The active compound of oseltamivir, which is oseltamivir carboxylate, was used throughout the in vitro experiments. Culture supernatants were collected after 24 hours of incubation at 37°C and 5% CO2, and stored at −20°C for further measurements of cytokines.
Mouse infection
At day 2 before infection and until day 2 after infection of the experimental protocol, 10 mg/kg oseltamivir was administered to the C57BL/6J mice twice daily by oral gavage. The dose was chosen in order to achieve similar oral dose of 75 mg twice daily in humans.51 Control groups consisted of mice to which an equal volume of vehicle (sterile PBS) was administered. At day 0, mice were challenged with 1 × 108 live conidia using a noninvasive intranasal (i.n.) infection procedure upon anesthesia with 75 mg/kg of Ketamine (Ketamidor®, Ritcher Pharma) and 1 mg/Kg of Medetomidine (Domtor®, Ecuphar). At days 1 and 3 post-infection, mice were sacrificed, and the lungs were PBS-perfused and excised, excluding the trachea and major bronchi. For assessment of fungal burden, lung single-cell suspensions were serially diluted and plated on SDA. For histological analysis, the lungs were perfused with PBS, excised and fixed with 10% buffered formalin solution at least 24 hours, processed and paraffin-embedded. Lung paraffin-sections were stained with Hematoxylin and Eosin (H&E) for pathological examination. Images were acquired using a BX61 microscope (Olympus) and a DP70 high-resolution camera (Olympus). Morphometry analysis was carried out on slide images using ImageJ software (v1.50i, NIH, USA).
For survival experiments, animals were immunosuppressed using 112 mg/kg cortisone 21-acetate (Acros Organics), which was administered on days 3 and 1 before infection by subcutaneous (s.c.) injection. At day 0, mice were challenged with 1 × 107 live conidia as mentioned above. To avoid bacterial infections animals were treated with 50μg/mL of chloramphenicol in drinking water ad libitum. Animals were daily weighted and sacrificed in case of 20% loss weight, severe ataxia or hypothermia and other severe complications.
BALB/c mice were rendered immunosuppressed by intraperitoneal injection of cyclophosphamide (150 mg/kg) on days 4 and 1 prior to instillation of A. fumigatus conidia. A broad-spectrum antibiotic (Baytril, 5-8 mg/kg/day) was added to the drinking water to prevent bacterial infection. Oseltamivir (10 mg/kg) was administered twice daily via oral gavage, from day 2 prior to infection until day 2 after infection. Control mice were sham-treated in the same time period. Immunosuppressed mice were infected with 5 × 105 A. fumigatus conidia in 20 μL of PBS (C2/7/1 bioluminescent strain) via intranasal instillation. Directly after infection, the animals were positioned upright until normal breathing resumed. Bioluminescence live-imaging assay and computed tomography (CT) scan took place on day 4 prior to infection (baseline scan) and from day 1 after infection onward on a daily basis. During image acquisition, animals were anesthetised with 1,5%–2% isoflurane (Abbott Laboratories, Queenborough, UK) in 100% oxygen (O2) administered via a nasal cone. Body weight was monitored daily. The animals were killed when humane end points were reached, including a strong loss in body weight (> 25%), lethargy and labored breathing. After euthanasia, the lungs were isolated for fungal load quantification and histologic analysis.
Isolation of mouse splenocytes
Immunocompetent C57BL/6J mice infected with A. fumigatus conidia as above were sacrificed at day 1 and 3 post-infection with posterior spleen excision. The excised spleen was milled into small fragments with a plunger end of a syringe and forced through a 70-μm cell strainer (Corning Inc.) Upon washing the cells with cold and sterile PBS, the resulting cell pellet was resuspended in 2 mL of pre-warmed (at 37°C) ACK lysis buffer (0.15 M NH4Cl, 10 Mm KHCO3 and 0.1 mM EDTA). Subsequently, the cells were centrifuged at 1,600 rpm for 5 min at room temperature. Finally, the cells were counted and adjusted to a final concentration of 5x106 cells/mL of pre-warmed RPMI (GIBCO, Thermo Fisher Scientific) enriched with 10% FBS (GIBCO, Thermo Fisher Scientific) and 200 μL of the cellular suspension were seeded in round bottom 96-well plates (Corning Inc.).
Measurement of cytokines
Concentrations of TNF-α, IL-1β, and IL-8 were measured from the in vitro culture supernatants using enzyme-linked immunosorbent assay (ELISA, R&D Systems), according to protocols supplied by the manufacturer. Cytokine measurements were performed on the supernatants of lung single-cell suspensions on day 1 and 3 post-infection using ELISA MAX Deluxe Set kits (BioLegend) for mouse IL-1β, IL-6, TNF-α, and IL-10 according to the manufacturer’s instructions.
Fungicidal/killing assay of Aspergillus fumigatus
Following isolation, PBMCs (5x105 cells) or neutrophils (2x105 cells) were plated onto a 96-well plate, and were stimulated with A. fumigatus conidia (MOI 4:1), in a final volume of 200 μL. After incubation for 4 hours at 37°C, cells were resuspended in H2O, and lysates were plated on Sabouraud dextrose agar in serial dilutions. Colony-forming units (CFUs) were then counted after 24 hours incubation at 37°C.
To evaluate fungicidal activity of mice splenocytes, cells (1 × 106/well in 96-well plates) were infected with A. fumigatus conidia at a 1:5 ratio for 2 hours at 37°C and 5% CO2. After incubation, culture plates were frozen at −80°C and thawed at 37°C to cause cell lysis and release of ingested conidia. Serial dilutions of cell lysates were plated on solid growth media and, following a 24 hours incubation at 37°C, the number of CFUs was enumerated and the percentage of CFU inhibition was calculated.
Measurement of ROS production in PBMCs and neutrophils
The production of reactive oxygen species (ROS) from PBMCs and neutrophils were measured by oxidation of luminol (3-amino-2,3, dihydro-1,4-phtalazinedione). PBMCs (5x105) or neutrophils (5x105) were stimulated with A. fumigatus conidia (1x107/mL) or zymosan (75 μg/mL) in a dark 96-well plate, together with 20 μL of 1 mM luminol. Chemiluminescence was measured using the BioTek Synergy HT reader at 37°C for 1 hour. The production of ROS within the area under the curve (AUC) was analyzed.
Flow cytometry analysis
The interaction of SIGLEC15 and A. fumigatus was tested with flowcytometry. Recombinant human SIGLEC15-Fc (1μg/mL) (R&D Systems) was incubated with A. fumigatus for 2 hours at 37°C. Subsequently, samples were washed twice with PBS and 1% BSA and stained with secondary antibody (Alexa 647 goat anti-human IgG (H+L) (Thermo Fisher) for 20 minutes. After additional washing steps, samples were measured using CytoFLEX flow cytometer (Beckman Coulter). To determine the sialic acid properties of A. fumigatus, conidia were incubated with either Maackia amurensis lectin (MAL) II, Sambucus nigra agglutinin (SNA), or Peanut agglutinin (PNA), which bind to (α2, 3)-linked, (α2, 6)-linked sialic acids, or galactose residues, respectively. This was followed by labeling with a secondary fluorophore (Streptavidin) and analysis by flow cytometry. Data were analyzed using the Kaluza analyses software version 1.5a.
After infection of the immunocompetent mice, the lungs and spleens were excised and collected in incomplete Dulbecco’s Modified Eagle Medium (iDMEM) culture medium (GIBCO, Thermo Fisher Scientific). The spleens were minced and forced through a 70-μm cell strainer (Corning Inc., New York, USA) and red blood cells were lysed with ACK lysis buffer (0.15 M NH4Cl, 10 Mm KHCO3 and 0.1 mM EDTA). Perfused lungs were sliced in small fragments and digested at 37°C for 30 min in incomplete DMEM culture medium containing 1 mg/mL of Collagenase D (Sigma-Aldrich). Afterward, tissue was forced through a 70-μm cell strainer and remaining red blood cells were lysed. Leukocytes were isolated by Percoll (GE Healthcare Bio-Sciences Ab, Uppsala, Sweden) density gradient and, then resuspended in FACS buffer (PBS containing 2% FBS and 2 mM EDTA). For surface marker staining, cell suspensions were stained for 30 min on ice with the indicated antibodies. Pellets were washed and resuspended in fresh FACS buffer prior to analysis. The composition of lung-infiltrating cells was determined using the combination of the following antibodies: BV510 CD45 (clone 30-F11), PerCp-Cy5.5 Ly-6C (clone HK1.4), BV785 Ly-6G (clone 1A8), BV605 CD11c (clone N418), PE-Cy7 CD11b (clone M1/70), FITC-I-A/I-E (clone M5/114.15.2), APC F4/80 (cloneBM8) and Alexa Fluor 647 CD103 (clone 2E7) from BD Biolegend. PE-Siglec-F (clone E50-2440) was from BD-PharMingen. Data were obtained on a BD FACS LSRII instrument (BD Biosciences) and analyzed using FlowJo (Tree Star Inc, Ashland, OR).
Measurement of ROS production in the lungs of the mice
Upon incubation with the different surface markers mentioned above, lung cells (1 × 105/well) were plated in 96-well plates (Sigma-Aldrich), and 2.5 mM CellROX Green Reagent (Thermo Fisher Scientific) was added to each well and incubated at 4°C for 30 min in the dark. Pellets were washed and resuspended in fresh FACS buffer prior to analysis. ROS production was measured on a BD FACS LSRII instrument (BD Biosciences) according to the manufacturer’s instructions.
Silencing of SIGLEC15
PBMCs were isolated from healthy volunteers as described previously. Cells were transfected with either 25 nM SIGLEC15 siRNA or scrambled control siRNA (SmartPool) using lipofectamine (Thermo Fisher Scientific) for 24 hours at 37°C. Afterward, cells were stimulated with live A. fumigatus conidia (1x107/mL) or medium for 4 hours and 24 hours. Subsequently, RNA was isolated using Trizol (Thermo Fisher Scientific) and synthesized into cDNA. The efficiency of the silencing experiment was tested using qPCR with the following primers: hSIGLEC15 (forward: 5′-CGCGGATCGTCAACATCTC-3′, reverse: 5′-GTTCGGCGGTCACTAGGTG-3′) and hRPL37 (forward: 5′-ATTGAAATCAGCCAGCACG-3′, reverse: 5′- AGGAACCACAGTGCCAGAT-3′).
RNA-seq
RNA seq data on SIGLEC15 and NEU genes were extracted from an existing RNaseq dataset.16 PBMCs (1x106/well) were stimulated with heat-killed A. fumigatus conidia (1x107/mL) or medium for 4 and 24 hours at 37°C in 5% CO2. RNA was isolated using the mirVANA RNA isolation kit (Applied Biosystems) prior to RNA sequencing.16 The datasets were retrieved from Gene Expression Omnibus (GEO) series with the accession number GSE162746.
Quantification and statistical analysis
Statistical comparisons between groups were analyzed using Mann-Whitney-U-test or Wilcoxon signed rank test for paired analysis. Data are presented as mean ± standard error of the mean (SEM). A p value of < 0.05 was considered statistically significant (∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001). Data were analyzed using GraphPad Prism software version 5.0 (La Jolla, CA, USA).
Acknowledgments
F.L.v.d.V. was supported by a VIDI grant from the Netherlands Organization for Scientific Research (NWO). I.M.W.D. was supported by the Indonesian Endowment Fund for Education (LPDP) from the Ministry of Finance of the Republic of Indonesia. A.C. and C.C. were supported by the Northern Portugal Regional Operational Program (NORTE 2020) under the Portugal 2020 Partnership Agreement through the European Regional Development Fund (FEDER) (NORTE-01-0145-FEDER-000013) and the Fundação para a Ciência e a Tecnologia (FCT) (CEECIND/04058/2018 to C.C., SFRH/BD/141127/2018 to C.D.-O., and CEECIND/03628/2017 to A.C.). J.W. was supported by clinical research funding from the Flemish Research Foundation (FWO). L.V. was supported by an FWO fellowship. G.V.V. and K.L. received a research grant from the FWO (1506114N).
Author contributions
Methodology, I.M.W.D., F.L.v.d.V., C.C., C.D.-O., A.C., G.V.V., and L.V.; investigation, I.M.W.D., C.C., C.D.-O., M.J., M.S.G., M.E.G., C.F.C., L.V., A.R.S., and F.M.G.; writing – original draft, I.M.W.D. and F.L.v.d.V.; writing – review & editing, C.C., C.D.-O., A.C., M.J., M.S.G., M.G.N., L.A.B.J., P.E.V., R.J.B., Q.d.M., A.J.A.M.v.d.V., K.L., G.V.V., and J.W.; supervision, F.L.v.d.V., A.C., and J.W.
Declaration of interests
The authors declare no competing interests.
Published: May 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100289.
Supplemental information
References
- 1.van de Veerdonk F.L., Kolwijck E., Lestrade P.P., Hodiamont C.J., Rijnders B.J., van Paassen J., Haas P.J., Oliveira Dos Santos C., Kampinga G.A., Bergmans D.C., Dutch Mycoses Study Group Influenza-Associated Aspergillosis in Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2017;196:524–527. doi: 10.1164/rccm.201612-2540LE. [DOI] [PubMed] [Google Scholar]
- 2.Schauwvlieghe A.F.A.D., Rijnders B.J.A., Philips N., Verwijs R., Vanderbeke L., Van Tienen C., Lagrou K., Verweij P.E., Van de Veerdonk F.L., Gommers D., Dutch-Belgian Mycosis study group Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respir. Med. 2018;6:782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 3.Laborda P., Wang S.Y., Voglmeir J. Influenza Neuraminidase Inhibitors: Synthetic Approaches, Derivatives and Biological Activity. Molecules. 2016;21:E1513. doi: 10.3390/molecules21111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyagi T., Yamaguchi K. Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- 5.Amith S.R., Jayanth P., Franchuk S., Siddiqui S., Seyrantepe V., Gee K., Basta S., Beyaert R., Pshezhetsky A.V., Szewczuk M.R. Dependence of pathogen molecule-induced Toll-like receptor activation and cell function on Neu1 sialidase. Glycoconj. J. 2009;26:1197–1212. doi: 10.1007/s10719-009-9239-8. [DOI] [PubMed] [Google Scholar]
- 6.Abdulkhalek S., Szewczuk M.R. Neu1 sialidase and matrix metalloproteinase-9 cross-talk regulates nucleic acid-induced endosomal TOLL-like receptor-7 and -9 activation, cellular signaling and pro-inflammatory responses. Cell. Signal. 2013;25:2093–2105. doi: 10.1016/j.cellsig.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Nan X., Carubelli I., Stamatos N.M. Sialidase expression in activated human T lymphocytes influences production of IFN-gamma. J. Leukoc. Biol. 2007;81:284–296. doi: 10.1189/jlb.1105692. [DOI] [PubMed] [Google Scholar]
- 8.Varki A., Gagneux P. Multifarious roles of sialic acids in immunity. Ann. N Y Acad. Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 10.Warwas M.L., Watson J.N., Bennet A.J., Moore M.M. Structure and role of sialic acids on the surface of Aspergillus fumigatus conidiospores. Glycobiology. 2007;17:401–410. doi: 10.1093/glycob/cwl085. [DOI] [PubMed] [Google Scholar]
- 11.Revilla N., Corral J., Miñano A., Mingot-Castellano M.E., Campos R.M., Velasco F., Gonzalez N., Galvez E., Berrueco R., Fuentes I. Multirefractory primary immune thrombocytopenia; targeting the decreased sialic acid content. Platelets. 2019;30:743–751. doi: 10.1080/09537104.2018.1513476. [DOI] [PubMed] [Google Scholar]
- 12.Shao L., Wu Y., Zhou H., Qin P., Ni H., Peng J., Hou M. Successful treatment with oseltamivir phosphate in a patient with chronic immune thrombocytopenia positive for anti-GPIb/IX autoantibody. Platelets. 2015;26:495–497. doi: 10.3109/09537104.2014.948838. [DOI] [PubMed] [Google Scholar]
- 13.Bigot P., Auffret M., Gautier S., Weinborn M., Ettahar N.K., Coupé P. Unexpected platelets elevation in a patient with idiopathic thrombocytopenia treated with oseltamivir for influenza infection. Fundam. Clin. Pharmacol. 2016;30:483–485. doi: 10.1111/fcp.12213. [DOI] [PubMed] [Google Scholar]
- 14.Miyagi T., Wada T., Yamaguchi K., Shiozaki K., Sato I., Kakugawa Y., Yamanami H., Fujiya T. Human sialidase as a cancer marker. Proteomics. 2008;8:3303–3311. doi: 10.1002/pmic.200800248. [DOI] [PubMed] [Google Scholar]
- 15.Glanz V.Y., Myasoedova V.A., Grechko A.V., Orekhov A.N. Sialidase activity in human pathologies. Eur. J. Pharmacol. 2019;842:345–350. doi: 10.1016/j.ejphar.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Bruno M., Dewi I.M.W., Matzaraki V., Ter Horst R., Pekmezovic M., Rösler B., Groh L., Röring R.J., Kumar V., Li Y. Comparative host transcriptome in response to pathogenic fungi identifies common and species-specific transcriptional antifungal host response pathways. Comput. Struct. Biotechnol. J. 2020;19:647–663. doi: 10.1016/j.csbj.2020.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takamiya R., Ohtsubo K., Takamatsu S., Taniguchi N., Angata T. The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-β secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology. 2013;23:178–187. doi: 10.1093/glycob/cws139. [DOI] [PubMed] [Google Scholar]
- 19.Wasylnka J.A., Simmer M.I., Moore M.M. Differences in sialic acid density in pathogenic and non-pathogenic Aspergillus species. Microbiology (Reading) 2001;147:869–877. doi: 10.1099/00221287-147-4-869. [DOI] [PubMed] [Google Scholar]
- 20.Bouchara J.P., Sanchez M., Chevailler A., Marot-Leblond A., Lissitzky J.C., Tronchin G., Chabasse D. Sialic acid-dependent recognition of laminin and fibrinogen by Aspergillus fumigatus conidia. Infect. Immun. 1997;65:2717–2724. doi: 10.1128/iai.65.7.2717-2724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amith S.R., Jayanth P., Franchuk S., Finlay T., Seyrantepe V., Beyaert R., Pshezhetsky A.V., Szewczuk M.R. Neu1 desialylation of sialyl α-2,3-linked β-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell. Signal. 2010;22:314–324. doi: 10.1016/j.cellsig.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 23.Feng C., Zhang L., Almulki L., Faez S., Whitford M., Hafezi-Moghadam A., Cross A.S. Endogenous PMN sialidase activity exposes activation epitope on CD11b/CD18 which enhances its binding interaction with ICAM-1. J. Leukoc. Biol. 2011;90:313–321. doi: 10.1189/jlb.1210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatos N.M., Carubelli I., van de Vlekkert D., Bonten E.J., Papini N., Feng C., Venerando B., d’Azzo A., Cross A.S., Wang L.X., Gomatos P.J. LPS-induced cytokine production in human dendritic cells is regulated by sialidase activity. J. Leukoc. Biol. 2010;88:1227–1239. doi: 10.1189/jlb.1209776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telford J.C., Yeung J.H., Xu G., Kiefel M.J., Watts A.G., Hader S., Chan J., Bennet A.J., Moore M.M., Taylor G.L. The Aspergillus fumigatus sialidase is a 3-deoxy-D-glycero-D-galacto-2-nonulosonic acid hydrolase (KDNase): structural and mechanistic insights. J. Biol. Chem. 2011;286:10783–10792. doi: 10.1074/jbc.M110.207043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue S., Kitajima K. KDN (Deaminated neuraminic acid): Dreamful past and exciting future of the newest member of the sialic acid family. Glycoconj. J. 2006;23:277–290. doi: 10.1007/s10719-006-6484-y. [DOI] [PubMed] [Google Scholar]
- 27.Bulai T., Bratosin D., Pons A., Montreuil J., Zanetta J.-P. Diversity of the human erythrocyte membrane sialic acids in relation with blood groups. FEBS Lett. 2003;534:185–189. doi: 10.1016/s0014-5793(02)03838-3. [DOI] [PubMed] [Google Scholar]
- 28.Wauters J., Baar I., Meersseman P., Meersseman W., Dams K., De Paep R., Lagrou K., Wilmer A., Jorens P., Hermans G. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: a retrospective study. Intensive Care Med. 2012;38:1761–1768. doi: 10.1007/s00134-012-2673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crum-Cianflone N.F. Invasive Aspergillosis Associated With Severe Influenza Infections. Open Forum Infect. Dis. 2016;3:ofw171. doi: 10.1093/ofid/ofw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monti E., Preti A., Nesti C., Ballabio A., Borsani G. Expression of a novel human sialidase encoded by the NEU2 gene. Glycobiology. 1999;9:1313–1321. doi: 10.1093/glycob/9.12.1313. [DOI] [PubMed] [Google Scholar]
- 31.Riswari S.F., Tunjungputri R.N., Kullaya V., Garishah F.M., Utari G.S.R., Farhanah N., Overheul G.J., Alisjahbana B., Gasem M.H., Urbanus R.T. Desialylation of platelets induced by Von Willebrand Factor is a novel mechanism of platelet clearance in dengue. PLoS Pathog. 2019;15:e1007500. doi: 10.1371/journal.ppat.1007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jansen A.J.G., Peng J., Zhao H.-G., Hou M., Ni H. Sialidase inhibition to increase platelet counts: A new treatment option for thrombocytopenia. Am. J. Hematol. 2015;90:E94–E95. doi: 10.1002/ajh.23953. [DOI] [PubMed] [Google Scholar]
- 33.O’Shea L.K., Abdulkhalek S., Allison S., Neufeld R.J., Szewczuk M.R. Therapeutic targeting of Neu1 sialidase with oseltamivir phosphate (Tamiflu®) disables cancer cell survival in human pancreatic cancer with acquired chemoresistance. OncoTargets Ther. 2014;7:117–134. doi: 10.2147/OTT.S55344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Oliveira J.T., Santos A.L., Gomes C., Barros R., Ribeiro C., Mendes N., de Matos A.J., Vasconcelos M.H., Oliveira M.J., Reis C.A., Gärtner F. Anti-influenza neuraminidase inhibitor oseltamivir phosphate induces canine mammary cancer cell aggressiveness. PLoS ONE. 2015;10:e0121590. doi: 10.1371/journal.pone.0121590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thulasiraman P., Kerr K., McAlister K., Hardisty S., Wistner A., McCullough I. Neuraminidase 1 regulates proliferation, apoptosis and the expression of Cadherins in mammary carcinoma cells. Mol. Cell. Biochem. 2019;462:207–215. doi: 10.1007/s11010-019-03623-7. [DOI] [PubMed] [Google Scholar]
- 36.Stamatos N.M., Liang F., Nan X., Landry K., Cross A.S., Wang L.X., Pshezhetsky A.V. Differential expression of endogenous sialidases of human monocytes during cellular differentiation into macrophages. FEBS J. 2005;272:2545–2556. doi: 10.1111/j.1742-4658.2005.04679.x. [DOI] [PubMed] [Google Scholar]
- 37.Wong Z.X., Jones J.E., Anderson G.P., Gualano R.C. Oseltamivir treatment of mice before or after mild influenza infection reduced cellular and cytokine inflammation in the lung. Influenza Other Respir. Viruses. 2011;5:343–350. doi: 10.1111/j.1750-2659.2011.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marois I., Cloutier A., Garneau É., Lesur O., Richter M.V. The administration of oseltamivir results in reduced effector and memory CD8+ T cell responses to influenza and affects protective immunity. FASEB J. 2014;29:973–987. doi: 10.1096/fj.14-260687. [DOI] [PubMed] [Google Scholar]
- 39.Chang Y.-C., Nizet V. The interplay between Siglecs and sialylated pathogens. Glycobiology. 2014;24:818–825. doi: 10.1093/glycob/cwu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Sun J., Liu L.N., Flies D.B., Nie X., Toki M., Zhang J., Song C., Zarr M., Zhou X. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019;25:656–666. doi: 10.1038/s41591-019-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angata T., Tabuchi Y., Nakamura K., Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17:838–846. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- 42.Kameda Y., Takahata M., Komatsu M., Mikuni S., Hatakeyama S., Shimizu T., Angata T., Kinjo M., Minami, A, Iwasaki N. Siglec-15 regulates osteoclast differentiation by modulating RANKL-induced phosphatidylinositol 3-kinase/Akt and Erk pathways in association with signaling Adaptor DAP12. J. Bone Miner. Res. 2013;28:2463–2475. doi: 10.1002/jbmr.1989. [DOI] [PubMed] [Google Scholar]
- 43.Jaeger M., Pinelli M., Borghi M., Constantini C., Dindo M., van Emst L., Puccetti M., Pariano M., Ricaño-Ponce I., Büll C. A systems genomics approach identifies SIGLEC15 as a susceptibility factor in recurrent vulvovaginal candidiasis. Sci. Transl. Med. 2019;11:eaar3558. doi: 10.1126/scitranslmed.aar3558. [DOI] [PubMed] [Google Scholar]
- 44.Turnbull I.R., Colonna M. Activating and inhibitory functions of DAP12. Nat. Rev. Immunol. 2007;7:155–161. doi: 10.1038/nri2014. [DOI] [PubMed] [Google Scholar]
- 45.Buckland K.F., Ramaprakash H., Murray L.A., Carpenter K.J., Choi E.S., Kunkel S.L., Lukacs N.W., Xing Z., Aoki N., Hartl D., Hogaboam C.M. Triggering receptor expressed on myeloid cells-1 (TREM-1) modulates immune responses to Aspergillus fumigatus during fungal asthma in mice. Immunol. Invest. 2011;40:692–722. doi: 10.3109/08820139.2011.578270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.N’Diaye E.-N., Branda C.S., Branda S.S., Nevarez L., Colonna M., Lowell C., Hamerman J.A., Seaman W.E. TREM-2 (triggering receptor expressed on myeloid cells 2) is a phagocytic receptor for bacteria. J. Cell Biol. 2009;184:215–223. doi: 10.1083/jcb.200808080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daws M.R., Sullam P.M., Niemi E.C., Chen T.T., Tchao N.K., Seaman W.E. Pattern recognition by TREM-2: binding of anionic ligands. J. Immunol. 2003;171:594–599. doi: 10.4049/jimmunol.171.2.594. [DOI] [PubMed] [Google Scholar]
- 48.Netea M.G., Joosten L.A.B., Li Y., Kumar V., Oosting M., Smeekens S., Jaeger M., Ter Horst R., Schirmer M., Vlamakis H. Understanding human immune function using the resources from the Human Functional Genomics Project. Nat. Med. 2016;22:831–833. doi: 10.1038/nm.4140. [DOI] [PubMed] [Google Scholar]
- 49.Soares R.M., de A Soares R.M., Alviano D.S., Angluster J., Alviano C.S., Travassos L.R. Identification of sialic acids on the cell surface of Candida albicans. Biochim. Biophys. Acta. 2000;1474:262–268. doi: 10.1016/s0304-4165(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 50.Gresnigt M.S., Joosten L.A.B., Verschueren I., van der Meer J.W., Netea M.G., Dinarello C.A., van de Veerdonk F.L. Neutrophil-mediated inhibition of proinflammatory cytokine responses. J. Immunol. 2012;189:4806–4815. doi: 10.4049/jimmunol.1103551. [DOI] [PubMed] [Google Scholar]
- 51.Ward P., Small I., Smith J., Suter P., Dutkowski R. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 2005;55(Suppl 1):i5–i21. doi: 10.1093/jac/dki018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA seq data on SIGLEC15 an NEU genes were extracted from an existing RNaseq dataset.16 The datasets were retrieved from Gene Expression Omnibus (GEO) using identifier GEO Series accession number GSE162746.