Abstract
Swine influenza virus (SIV) belongs to family Orthomyxoviridae and can cause acute respiratory infection in pigs. Several pandemic H1N1 human fatal influenza cases were reported in India. Though pigs are predisposed to both avian and human influenza virus infections with the potential to generate novel reassortants, there are only a few reports of SIV in Indian pigs. We conducted a serological survey to assess the status of H1N1 infection in pigs of various states in India, between 2009 and 2016. Based on Haemagglutination inhibition (HI) assay, seroprevalence rate of H1N1 virus ranged between 5.2% (2009) and 36.3% (2011). Widespread prevalence of antibody was observed in eastern Uttar Pradesh from 6.2 to 37.5% during the study period. Co-circulation of seasonal H1N1 virus along with pandemic H1N1 virus was indicated by the presence of specific antibodies against seasonal H1N1 virus in eastern part of Uttar Pradesh. Seroprevalence rate in pigs and influenza infection trend in human shows the possible spill over transmission of influenza to pigs from human. Hence, besides serological surveillance, continuous and systematic molecular surveillance should be implemented in pig population to reduce/quantify the risk and emergence of pandemic influenza.
Introduction
In early April 2009, there was an outbreak of H1N1 influenza A detected in Mexico [1], spreading to many countries. It was declared as pandemic influenza by World Health Organization (WHO) in June 2009 [2]. Since 2009, pandemic influenza H1N1 has been also reported in various states of India [3]. Pig plays an important role in epidemiology and transmission of influenza virus due the presence of sialic acid (SA) linked to galactose (Gal) by α-2,3 linkage (SA α-2,3-Gal) and α-2,6 receptors (SA α-2,6-Gal) for both avian and mammalian influenza viruses, respectively [4, 5]. The assessment of antibodies for swine influenza in pigs is needed due to its susceptibility to both avian and mammalian influenza viruses and its involvement in interspecies transmission [6].
Swine influenza is an acute respiratory infection with production losses in pig population that is reported worldwide. The disease is caused by type A influenza virus, belonging to family Orthomyxoviridae, genus Influenza virus [7, 8]. Besides morbidity and production losses, maintenance and evolution of influenza viruses in pigs pose obvious public health risk. There are reports of circulation of classical swine H1N1 viruses in pigs for at least 84 years [9]. The emergence of novel influenza viruses with pandemic potential to human population is known to be mediated by antigenic shift in domestic pigs (as a mixing vessel) [10]. All three pandemics of the previous century were believed to originate from reassortment events in either pigs or humans. Co-circulation and mixing of the triple reassortant H3N2 with swine lineage and emergence of pandemic swine origin influenza virus is reported since 2009 [11–14]. Transmission of 2009 pandemic H1N1 virus from humans to pigs (reverse zoonosis) has also been reported in many countries, including India [15–20]. Hence, monitoring of pig population is essential to predict the status and epidemiology of influenza in humans.
There are very few studies of influenza in pigs from India. Serological evidence of swine influenza (H1N1) virus in pigs was reported in 1981 from Andhra Pradesh, West Bengal and Uttar Pradesh [21]. After the emergence of pandemic H1N1 in 2009, there was a report confirming the isolation of influenza H1N1 virus from Indian pigs sharing the close homology with human pandemic H1N1 2009 virus [15]. Another study reported the seroprevalence of 37% of pandemic H1N1 (pdm H1N1 2009) antibodies in abattoir specimen of pigs in Maharashtra and Gujarat [22]. Since 2009, there were continuing outbreaks of swine influenza in humans in India. Pig farming in India is semi-organized, and intermingling of pigs with other species helps in maintenance of virus circulation and emergence of novel influenza viruses. Considering the importance of monitoring swine population, this study was conducted to assess the antibody level against swine influenza A (pdm H1N1 2009) in Indian pigs from 2009 to 2016.
Materials and Methods
Samples
A total of 2632 porcine serum samples, nasal swabs (n = 2001) received / collected from pigs of organized and backyard farms from various states of India during May 2009–February 2016 were tested to assess the status of H1N1 influenza virus infection. Pigs sampled were mostly from backyard holdings, stray animals with different age varying from young to adult, besides pigs of organized sectors. Due to biosafety concerns, permissions for sampling were obtained and samples were collected by concerned veterinarians. Nasal swabs were collected using moistened cotton swabs in sterile PBS, stored at 4 ℃ and transported using storage containers. Whole blood was collected by anterior vena cava or ear vein puncture, followed by serum separation, and stored at − 20 ℃.
As a national referral laboratory, we received samples from various states of India based on outbreaks in the human and pig population. Samples received were mostly from stray/backyard pigs from villages, the remaining samples were collected from organized farms near to urban areas.
Viruses and Sera
Strain A/swine/India/220966/2009 (H1N1) similar to A/Kansas/22/2009 (H1N1) of lineage A (pdm09) influenza virus were used from NIHSAD Repository, and seasonal H1N1 virus similar to A/New Caledonia/20/1999 were obtained from influenza virus repository of National Institute of Virology, Pune, India. Viruses were propagated in Specific Pathogen Free (SPF) embryonated chicken eggs once to ascertain Haemagglutination (HA) titre. Handling of virus, biological samples, HA and Haemagglutination inhibition (HI) assay, were performed in biosafety level 3 containment laboratory at NIHSAD, Bhopal, in conformity with the institute biosafety guidelines. Animal experiments were conducted with Institute Animal Ethics Committee’s approval (IAEC. No. 45/IAEC/HSADL/10).
In the current study, antisera were raised against (H1N1) influenza virus (A/swine/India/220966/2009) in SPF chickens after inactivation. Briefly, virus was formalin-inactivated and checked for inactivation by embryonated chicken egg inoculation for three blind passages followed by HA test to confirm the inactivation. Emulsion prepared by mixing inactivated virus with 28 HA titre with Freund’s complete adjuvant at the ratio of 1:1, 0.5 ml was injected intramuscularly at multiple sites, followed by booster dose with Freund’s incomplete adjuvant after 21 days of primary dose. Serum collected after 7 days of booster was tested by HI assay and used as positive control sera for following HI tests.
Serological Tests
Swine Influenza (H1N1) Antibody ELISA
All serum samples were inactivated at 56 °C for 30 min and were adsorbed with packed chicken red blood cells for 60 min at room temperature prior to testing. Initially, serum samples were tested by using commercial ELISA kit. Once the HI assay was established, it replaced the ELISA due to the specific detection of H1N1 antibodies. Commercial ELISA kit (The HerdChek Swine Influenza H1N1 Antibody test Kit (IDEXX Laboratories, Inc., USA) was used to detect antibodies against H1 SIV HAs according to the manufacturer’s instructions. Briefly, the samples were tested in duplicates with kit positive and negative controls and samples with an S/P ratio (net optical density of test sample/net optical density of positive control) equal or greater to 0.4 were considered positive for antibody against H1N1 virus.
Haemagglutination Inhibition Assay (HI)
All serum samples were inactivated at 56 ℃ for 30 min to remove non-specific serum inhibitors. HI assay was carried out in accordance with Office International des Epizooties guidelines [23]. For serologic surveillance, virus strains A/swine/India/220966/2009 (H1N1) (pdmH1N1 2009 like) and A/New Caledonia/20/1999 (classical H1N1) were used as test antigens, SPF chicken raised antisera was used as positive control for each assay. Briefly, 50 µl of PBS was dispensed to each row. Two-fold serial dilutions of 25 µl of serum samples were carried out. 25 µl of 4 HA units of virus was mixed in microtiter plates and incubated at room temperature for 30 min. 25 µl of 1% chicken RBCs was added to each well and incubated after shaking at room temperature for 30 min. The reciprocal of the highest serum dilution that completely inhibits haemagglutination reaction was determined as HI antibody titre. Antibody titres less than or equal to 20 were considered negative; greater titres were considered positive [24].
Serologic Cross-Reactivity Between Seasonal H1N1 and Pandemic H1N1
Randomly selected serum samples were tested for HI activity against seasonal H1N1 influenza virus similar to A/New Caledonia/20/1999 and A/swine/India/220966/2009 (H1N1) similar to (A/Kansas/22/2009(H1N1)) of lineage A (pdm09) influenza virus. The mean geometric HI titre was calculated to assess the statistical significance.
Virus Isolation
According to the Office International des Epizooties (OIE) recommended protocol [23], isolation of virus was carried out in 9–11 days old embryonated chicken eggs. Nasal swabs were vortexed for 1 min and centrifuged at 2000 g for 15 min at 4 ℃. The samples were pooled so that each pool consisted of five samples originating from the same location/farm of collection. Pooled samples were treated with 2X antibiotic (streptomycin 10,000 units/ml and penicillin 10,000 units/ ml) for one hour at 37 °C prior to inoculation in SPF chicken embryos.
Isolation was attempted by inoculating 200 µl of each pooled sample into amnio-allantoic cavity of three SPF embryonated chicken eggs incubated at 37 ℃. Amnioallantoic fluid (AAF) was harvested after death of embryo or after 5 days of incubation and tested for the presence of influenza A virus by HA test. HA-positive AAF samples were further subjected to characterization of Influenza A virus. Each sample was processed for 3 blind passages before declaring them negative.
Statistical Analysis
HI titres were expressed as reciprocal of highest serum dilution, and geometric mean titre was calculated for each animal from duplicate. Prevalence rate, 95% confidence interval (CI) with upper and lower limits based on Asymptotic (Wald) method for normal approximation, was calculated using epitools software (http://epitools.ausvet.com.au.). Statistical analysis (Independent t test and sample t test between percents) was performed using Statistical Package for Social Sciences (SPSS 16.0).
Results
A total of 1151 out of 2632 sera (43.73%) were tested by HerdChek Swine Influenza H1N1 Antibody test kit for the initial two years of the study (2009 and 2010). State wise prevalence based on ELISA is presented in Table 1. The prevalence rate was 16.1%, (12.9, 19.3) with confidence interval of 95% during the year 2009, whereas in 2010, it increased to 41.7%, CI (38.3, 46.1) based on ELISA.
Table 1.
Serum samples tested for H1N1 swine influenza antibodies by ELISA
| S. No | State | No. of sera tested by ELISA | 2009 | 2010 | ||||
|---|---|---|---|---|---|---|---|---|
| No. of serum tested | No. of serum + ve | % ELISA + ve | No. of serum tested | No. of serum + ve | % ELISA + ve | |||
| 1 | Andhra Pradesh | 61 | 16 | 10 | 62.5 | 45 | 38 | 84.4 |
| 2 | Assam | 60 | 0 | 0 | 0 | 60 | 19 | 32.4 |
| 3 | Himachal Pradesh | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| 4 | Madhya Pradesh | 447 | 246 | 28 | 11.4 | 201 | 37 | 18.4 |
| 5 | Maharashtra | 5 | 3 | 1 | 33.3 | 2 | 0 | 0 |
| 6 | Meghalaya | 65 | 39 | 0 | 0 | 26 | 8 | 30.8 |
| 7 | Nagaland | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| 8 | Odisha | 74 | 64 | 6 | 9.4 | 10 | 0 | 0 |
| 9 | Tamil Nadu | 73 | 9 | 1 | 11.1 | 64 | 43 | 67.2 |
| 10 | Uttar Pradesh | 356 | 110 | 34 | 30.9 | 246 | 128 | 52 |
| Total | 1151 | 497 | 80 | 16.1 | 654 | 273 | 41.74 | |
All 2632 sera received/collected were also tested by HI assay. State wise seroprevalence of H1N1 antibodies from May 2009 to February 2016 is given in Table 2 and depicted in Fig. 1. The trend of prevalence rate was 5.2%, CI (3.3, 7.2) in 2009, and it increased to 18.9%, CI (15.6, 21.7) in 2010, reaching peak in 2011 as 36.3% with CI (31, 41.7). Afterwards, it was reduced to 29.1%, CI (23.9, 34.3) in 2012, 20.8%, CI (17.1, 24.5) in 2013 and again increased to 32.4%, CI (25.6, 39.2) in 2014. From 2015 to February 2016, the prevalence rate reduced drastically to 8.9%, CI (5.3, 12.5).
Table 2.
Serum samples tested by haemagglutination inhibition (HI) assay for H1N1 antibodies (2009–2011) and (2012–2016)
| S. No | State | No. of sera tested by HI assay | 2009 | 2010 | 2011 | |||
|---|---|---|---|---|---|---|---|---|
| No. of Serum tested | % HI Positive | No. of Serum tested | % HI Positive | No. of Serum tested | % HI Positive | |||
| 1 | Andhra Pradesh | 61 | 16 | 37.5 | 45 | 15.6 | 0 | 0 |
| 2 | Assam | 60 | 0 | 0 | 60 | 11.7 | 0 | 0 |
| 3 | Himachal Pradesh | 2 | 2 | 0 | 0 | 0 | 0 | |
| 4 | Madhya Pradesh | 458 | 246 | 4.9 | 201 | 10.0 | 11 | 45.5 |
| 5 | Maharashtra | 19 | 3 | 0 | 2 | 0 | 14 | 14.3 |
| 6 | Meghalaya | 65 | 39 | 0 | 26 | 7.7 | 0 | 0 |
| 7 | Nagaland | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| 8 | Odisha | 74 | 64 | 1.6 | 10 | 0 | 0 | 0 |
| 9 | Tamil Nadu | 73 | 9 | 0 | 64 | 28.1 | 0 | 0 |
| 10 | Uttar Pradesh | 642 | 110 | 6.4 | 246 | 26.4 | 286 | 37.1 |
| Total | 1462 | 497 | 5.2% | 654 | 18.9% | 311 | 36.3% | |
| S. No | State | No. of Sera tested by HI assay | 2012 | 2013 | 2014 | 2015 – up to Feb. 2016 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of Serum tested | % HI Positive | No. of Serum tested | % HI Positive | No. of Serum tested | % HI Positive | No. of Serum tested | % HI Positive | |||
| 1 | Madhya Pradesh | 418 | 62 | 25.8 | 310 | 12.0 | 0 | 0 | 46 | 0 |
| 2 | Punjab | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 66.7 |
| 3 | Uttar Pradesh | 748 | 230 | 30 | 152 | 37.5 | 182 | 32.4 | 184 | 9.24 |
| Total | 1170 | 292 | 29.1 | 452 | 20.8 | 182 | 32.4 | 236 | 8.9 | |
Fig. 1.
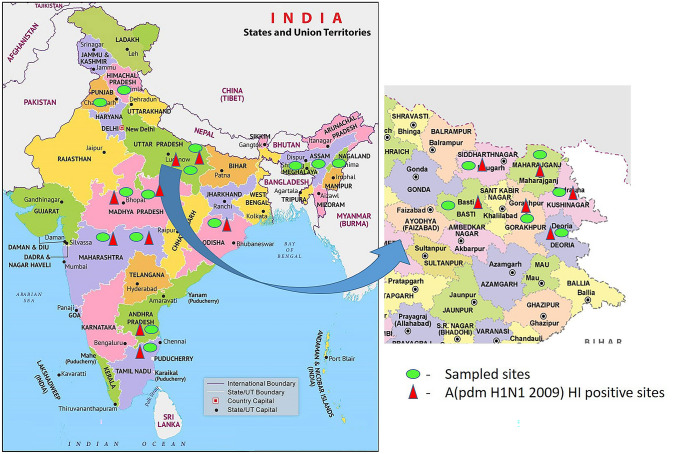
Map showing sampled sites of eastern Uttar Pradesh [(Gorakhpur (n-121), Deoria (n-135), Siddharth Nagar (n-111), Basti (n-126), Kushinagar (n-128), Maharajganj (n-127)] and other influenza A (pdm H1N1 2009) antibody positive sites of India
Based on the receipt of samples, pig population and density, different districts (Gorakhpur, Deoria, Siddharth Nagar, Basti, Kushinagar, Maharajganj, and Kanpur) of Uttar Pradesh were selected for further analysis. Seroprevalence rate of H1N1 antibodies among pigs during the year 2009 to 2016 of selected districts of Uttar Pradesh is shown in Fig. 1 and Table 3. The sero-prevalence was evidenced in all places sampled in eastern Uttar Pradesh, whereas highest prevalence was observed in Kanpur followed by Gorakhpur, Deoria and other districts. The trend of infection through 2009 to 2016 showed a drastic reduction in prevalence rate during January 2015 to February 2016 (Fig. 2).
Table 3.
Prevalence of antibodies against Influenza A (pdmH1N1 2009) in eastern part of Uttar Pradesh from 2009 to 2011 and 2012 to 2016
| S. No | Location | Geometric Coordinates | 2009 | 2010 | 2011 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serum tested | ELISA + ve (%) | HI + ve (%) | Serum tested | ELISA + ve (%) | HI + ve (%) | Serum tested | HI + ve (%) | |||
| 1 | Gorakhpur | 26.7606° N, 83.3732°E | 14 | 42.9 | 7.1 | 33 | 57.6 | 33.3 | 50 | 36.0 |
| 2 | Deoria | 26.4862° N, 83.7838°E | 22 | 36.4 | 9.1 | 32 | 53.1 | 28.1 | 40 | 37.5 |
| 3 | Siddharth Nagar | 27.2716° N, 82.8210°E | 7 | 0.0 | 0.0 | 30 | 73.3 | 23.3 | 39 | 41.0 |
| 4 | Kanpur | 26.4499° N, 80.3319°E | 30 | 46.7 | 13.3 | 46 | 52.2 | 30.4 | 50 | 56.0 |
| 5 | Basti | 26.8177° N, 82.7633°E | 15 | 13.3 | 0.0 | 15 | 86.7 | 26.7 | 54 | 24.1 |
| 6 | Kushinagar | 26.7399° N, 83.8870°E | 20 | 20.0 | 0.0 | 26 | 46.2 | 42.3 | 25 | 12.0 |
| 7 | Maharaiganj | 27.1446° N, 83.5622°E | – | – | – | 64 | 52.2 | 30.4 | 28 | 35.7 |
| Total | 110 | 30.9 | 6.4 | 246 | 52.0 | 26.4 | 286 | 37.1 | ||
| S. No | Location | 2012 | 2013 | 2014 | 2015–March 2016 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Serum tested | HI + ve (%) | Serum tested | HI + ve (%) | Serum tested | HI + ve (%) | Serum tested | HI + ve (%) | ||
| 1 | Gorakhpur | 39 | 30.8 | 25 | 44 | 22 | 40.9 | 35 | 11.4 |
| 2 | Deoria | 39 | 25.6 | 33 | 27.3 | 33 | 27.3 | 30 | 10.0 |
| 3 | Siddharth Nagar | 35 | 22.9 | 19 | 36.8 | 35 | 36.8 | 22 | 9.1 |
| 4 | Basti | 33 | 18.2 | 28 | 35.7 | 30 | 35.7 | 35 | 8.6 |
| 5 | Kushinagar | 43 | 41.9 | 20 | 40 | 30 | 40 | 35 | 8.6 |
| 6 | Maharajganj | 41 | 36.6 | 27 | 44.4 | 32 | 44.4 | 27 | 7.4 |
| Total | 230 | 30 | 152 | 37.5 | 182 | 32.4 | 184 | 9.23 | |
Fig. 2.
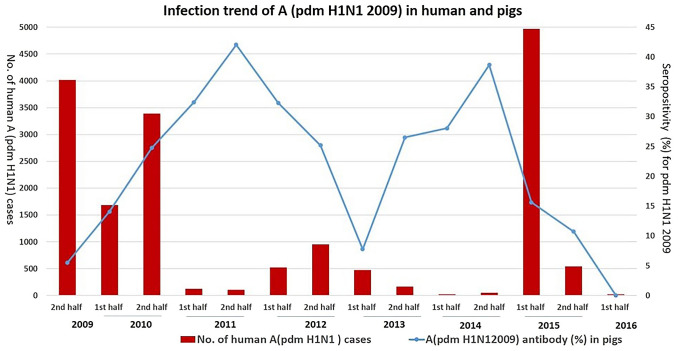
Distribution of percentage of serum samples positive for influenza A (pdm H1N1 2009) antibodies and number of human cases reported. Right Y axis represents the percentage of swine serum samples seropositive for influenza A (pdm H1N1 2009) and left Y axis represents the number of human cases reported. [Source: Global Influenza Surveillance and Response System, World Health Organization (WHO)]
Randomly selected serum samples (192) of three selected districts (Gorakhpur, Deoria and Kushinagar) were also tested for antibodies against seasonal human H1N1 influenza virus. The mean geometric HI titre against seasonal H1N1 virus (13.72), swine H1N1 (pdm) 2009 (152.22) with significant (P < 0.001) difference of 138.5 was observed during 2010–2011. Around 14.3% of tested samples showed HI activity only against seasonal H1N1 with geometric mean HI titre of 24.68 and no HI activity against pdm H1N1 (2009) virus.
To assess the chance of reverse zoonotic transmission from human to pigs, the trend in human influenza (H1N1) infection was compared using WHO database (https://extranet.who.int/sree/Reports?op=vsandpath=/WHO_HQ_Reports/G5/PROD/EXT/Influenza%20Surveillance). The antibody prevalence in pigs and the number of human influenza cases reported is depicted in Fig. 2.
A total of 391 nasal swabs from organized and 1610 from backyard piggeries were attempted for virus isolation in embryonated chicken embryos. In December 2009, two H1N1 viruses were isolated from pigs of an organized farm in Tamilnadu. The isolates were confirmed by haemagglutination assay, followed by HI assay against H1N1 specific reference antisera and by sequencing (data not shown).
Discussion
Swine influenza is a highly contagious respiratory disease in pigs with low mortality but high morbidity with production losses. Pigs possess both avian and human influenza receptors; hence, it poses a major public health threat in evolution of novel reassortant strains and transmission to humans [5, 12, 24]. In 2009, a new triple reassortant virus with pandemic potential spreads rapidly worldwide. In India, 31,974 pandemic H1N1 Influenza cases had been reported with 1895 human death as per March 2015 report of the Health Ministry [25]. Interestingly, transmission of pandemic H1N1 from humans to pigs (reverse zoonosis) has been reported in many countries, including India [15–20].
The prevalence and status of pandemic H1N1 in pigs can be better judged by sero-epidemiology, which provides valuable information on infection and risk posed by pig handlers. In India, pig husbandry is mostly semi-organized or backyard with intermingling of chicken, ducks and other poultry and provides every opportunity to evolve reassortant viruses by crossing the species barrier. Previously, a study on serological prevalence in Indian pigs showed 37.8% positivity for H1N1 Pdm 2009 virus infection based on samples collected from abattoirs of two states during May to August 2011 [22]. The study was limited to short period and region; moreover, sample collected from animals sent to abattoir were mostly adults. The clear picture, trend of prevalence can be evinced by surveillance in farm, backyard and stray pigs for a course of period. Therefore, a seroprevelance study on H1N1 and avian influenza viruses was carried out in pigs to assess the public health risk. To the best of author’s knowledge, there is no report of seroprevalence study against swine origin pdm H1N1 2009 at farm and backyard piggery in India.
Serological evidence of pdm H1N1 infection in pigs has been earlier documented in many countries like Vietnam [26], Thailand [27], Combodia [28], Italy [29], Brazil [18, 30], Srilanka [31], India [22], China [32], Luxemburg [33] and Norway [19]. In the present study, the trend of pdm H1N1 antibody prevalence in pigs (selected randomly) of various states of India was found to be increased constantly from 2009 to 2011 which indicated the constant circulation amongst swine population. Whereas, during 2015 to February 2016, 236 serum samples were tested, the prevalence rate was only 8.9%, indicating lower infection rate of H1N1 pdm 2009 like virus in pigs or which could be overtaken by antigenically different H1N1 strain circulating in human in India as reported by Tharakaraman and Ram (2014) [34]. Around 25% of sera tested during the study period showed high HI titre more than or equal to 256 indicating repeated infection of pdm H1N1 virus in pigs.
During second half of 2011 and 2014, two peaks of A (H1N1) pdm 2009 antibody detection (Fig. 2) were observed in pigs. In late 2015, there was a drastic fall in seroprevalence rate, the trend clearly correlates with human pandemic waves reported in 2009–2011, early 2015 and very few cases in 2016 in India [Global Influenza Surveillance and Response System, World Health Organization (WHO)]. (https://extranet.who.int/sree/Reports?op=vsa&path=/WHO_HQ_Reports/G5/PROD/EXT/Influenza%20Surveillance). Raise in number of human influenza infection was followed by antibody prevalence rate in pigs, indicating the possible reverse zoonosis (transmission from human to pigs), as also reported earlier in other countries.
Some of the districts of Uttar Pradesh were selected based on sample availability for further detailed study. The prevalence rate of eastern Uttar Pradesh also showed similar trend as that of overall prevalence of pdm H1N1 antibodies. In January 2015 to February 2016, the antibody prevalence was reduced drastically between 7 and 11% indicating absence of fresh exposure to pandemic H1N1 virus and waning of antibodies in already exposed animals.
Randomly selected serum samples collected in 2010 and 2011 were tested for antibodies against human seasonal H1N1 influenza A virus as well. Mean geometric HI titre against pdm H1N1 virus was 152.22, indicating repeated and constant circulation of virus in the population during this period. All pdm H1N1 antibody positive serum showed HI activity against seasonal H1N1 virus with significantly (P < 0.001) lesser mean geometric titre (13.72) and could be due to cross-reactivity and antigenic variations as reported earlier [34].
Around 14.3% of tested samples showed HI activity only against seasonal H1N1 virus, indicating the co-circulation of both viruses in the same swine population. Co-circulation could also be due to spill-over infection from human cases, as it was clearly indicated by a report of around 200 human cases of seasonal influenza A (H1N1) in 2009 and 2010 from India [Global Influenza Surveillance and Response System, World Health Organization (WHO)]. There are reports showing the reassortment of endemic swine influenza with pandemic H1N1 virus [35–39]. Hence, co-circulation of seasonal and pandemic H1N1 virus over a period may lead to evolution of reassortant viruses with pandemic potential.
Despite high antibody prevalence, our attempts to isolate virus from nasal swabs resulted in only two positive isolates from an organized farm (representing only 0.5%). Low virus detection/isolation from nasal swabs was also reported earlier [30, 40]. We received many unfrozen samples in our laboratory due to logistics, which might also be the cause for low virus isolation. Apart from this, virus isolation could be successful from the nasal or pharyngeal secretions collected during acute febrile phase which is difficult in influenza, which is mostly mild or asymptomatic in pigs.
This study presents the widespread serological prevalence of pdm H1N1 2009 like virus with chance of co-circulation of seasonal H1N1 in Indian pigs. Widespread H1N1 pdm 2009 infection could be by reverse zoonosis from human, indicating changed epidemiology of influenza in Indian swine population and should be monitored constantly for emergence of novel H1N1 reassortant strains of public health importance. Pigs also can be utilized as sentinels to provide useful information on influenza virus circulating in human population. Besides serological surveillance of influenza virus, an improved and continuous molecular surveillance programme is urgently needed in pig population to study the evolutionary changes in genetic components, reassortment which may pose great threat to public health.
Acknowledgements
Authors thank the Indian Council Agricultural Research, Director, ICAR-IVRI, Director, ICAR—NIHSAD for providing necessary facilities and funds under institute project to conduct this study. We thank Dr. M. S. Chadha, National Institute of Virology, Pune, India for providing seasonal H1N1 virus and all persons involved in sample collection.
Author Contribution
DSK, DDK, GV contributed to conceptualization, methodology & investigation, data analysis and writing-original draft; VG, PP, MD, BS performed some of the experiments and data curing; SB, CT contributed to resources and visualization, writing- review and editing; SCD, VPS contributed to resources and supervision.
Funding
This study was funded by Indian Council of Agricultural research, Govt. of India in the form of institute funded Research project.
Declarations
Conflict of interest
The authors declare that they have no competing interests.
Disclosure
Funding – This study was funded by Indian Council of Agricultural research, Govt. of India in the form of institute funded Research project.
Ethical Approval
The animal experiments were carried out at the Biosafety level 3 containment facility of National Institute of High Security Animal Diseases, Bhopal, India, after due approvals from Institute Animal Ethical Committee (Approval no. 45/IAEC/HSADL/10) and Institute Bio-safety Committee (Approval no. HSADL/IBSC/2010–11). Infected tissues and other bio wastes were safely disposed as per biosafety norms.
Consent for Publication
We also inform that the all authors have contributed significantly, and that all authors are in agreement with the content of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Córdova JA, Hernández M, López-Gatell H, Bojorquez I, Palacios E, Rodríguez G, de la Rosa B, Ocampo R, Alpuche C, Flores R, Hernandez JE. Update: novel influenza A (H1N1) virus infection—Mexico, March–May, 2009. MMWR Morb Mortal Wkly Rep. 2009;58(21):585. [PMC free article] [PubMed] [Google Scholar]
- 2.Fineberg HV. Pandemic preparedness and response—lessons from the H1N1 influenza of 2009. N Engl J Med. 2014;370(14):1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 3.Tandale BV, Pawar SD, Gurav YK, Chadha MS, Koratkar SS, Shelke VN, Mishra AC. Seroepidemiology of pandemic influenza A (H1N1) 2009 virus infections in Pune. India BMC Infect Dis. 2010;10(1):255. doi: 10.1186/1471-2334-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wentworth DE, McGregor MW, Macklin MD, Neumann V, Hinshaw VS. Transmission of swine influenza virus to humans after exposure to experimentally infected pigs. J Infect Dis. 1997;175(1):7–15. doi: 10.1093/infdis/175.1.7. [DOI] [PubMed] [Google Scholar]
- 5.Wright PF. Fields virology. Philadelphia: Lippincott Williams Wilkins; 2007. Orthomyxoviruses; pp. 1691–1740. [Google Scholar]
- 6.Webster RG, Shortridge KF, Kawaoka Y. Influenza: interspecies transmission and emergence of new pandemics. FEMS Immunol Med Microbiol. 1997;18(4):275–279. doi: 10.1111/j.1574-695X.1997.tb01056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easterday B, Van Reeth K (1999). Swine influenza. In: Straw BE, DAllaire S, Mengeling WL, Taylor DJ (Eds). Diseases of Swine 8th edn. Blackwell Sciences, Hoboken, p 277–290
- 8.Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol. 2000;74(1–2):29–46. doi: 10.1016/S0378-1135(00)00164-4. [DOI] [PubMed] [Google Scholar]
- 9.Lewis PA, Shope RE. Swine influenza: II. A hemophilic bacillus from the respiratory tract of infected swine. J Exp Med. 1931;54(3):361–371. doi: 10.1084/jem.54.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977;196(4297):1454–1455. doi: 10.1126/science.867041. [DOI] [PubMed] [Google Scholar]
- 11.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JM. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 12.Kong W, Liu Q, Sun Y, Wang Y, Gao H, Liu L, Qin Z, He Q, Sun H, Pu J, Wang D. Transmission and pathogenicity of novel reassortants derived from Eurasian avian-like and 2009 pandemic H1N1 influenza viruses in mice and guinea pigs. Sci Rep. 2016;6:27067. doi: 10.1038/srep27067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osbjer K, Berg M, Sokerya S, Chheng K, San S, Davun H, Magnusson U, Olsen B, Zohari S. Influenza A virus in backyard pigs and poultry in rural Cambodia. Transbound Emerg Dis. 2017;64(5):1557–1568. doi: 10.1111/tbed.12547. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Sun H, Jiang Z, Liu L, Zhang X, Wei K, Hou D, Pu J, Sun Y, Tong Q, Bi Y, Chang KC, Liu S, Gao GF, Liu J. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci USA. 2020;117(29):17204–17210. doi: 10.1073/pnas.1921186117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereda A, Cappuccio J, Quiroga MA, Baumeister E, Insarralde L, Ibar M, Sanguinetti R, Cannilla ML, Franzese D, Cabrera OEE, Craig MI. Pandemic (H1N1) 2009 outbreak on pig farm. Argentina Emerging infectious diseases. 2010;16(2):304. doi: 10.3201/eid1602.091230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagarajan K, Saikumar G, Singh Arya R, Gupta A, Somvanshi R, Pattnaik B. Influenza A H1N1 virus in Indian pigs & its genetic relatedness with pandemic human influenza A 2009 H1N1. Indian J Med Res. 2010;132(2):160. [PubMed] [Google Scholar]
- 17.Cardinale E, Pascalis H, Temmam S, Hervé S, Saulnier A, Turpin M, Barbier N, Hoarau J, Quéguiner S, Gorin S, Foray C. Influenza a (H1N1) pdm09 virus in pigs, Reunion Island. Emerg Infect Dis. 2012;18(10):1665. doi: 10.3201/eid1810.120398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajão DS, Costa AT, Brasil BS, Del Puerto HL, Oliveira FG, Alves F, Braz GF, Reis JK, Guedes RM, Lobato ZI, Leite RC. Genetic characterization of influenza virus circulating in Brazilian pigs during 2009 and 2010 reveals a high prevalence of the pandemic H1N1 subtype. Influenza Other Respir Viruses. 2013;7(5):783–790. doi: 10.1111/irv.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Er C, Skjerve E, Brun E, Framstad T, Lium B. Occurrence and spread of influenza A (H1N1) pdm09 virus infection in Norwegian pig herds based on active serosurveillance from 2010 to 2014. Epidemiol Infect. 2016;144(15):3148–3165. doi: 10.1017/S0950268816001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arunorat J, Charoenvisal N, Woonwong Y, Kedkovid R, Thanawongnuwech R. Determination of current reference viruses for serological study of swine influenza viruses after the introduction of pandemic 2009 H1N1 (pdmH1N1) in Thailand. J Virol Methods. 2016;236:5–9. doi: 10.1016/j.jviromet.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Prabhudas K, Malick BB. Note on the prevalence of influenza antibodies in swines. Indian J Animal Sci. 1981;51:907–908. [Google Scholar]
- 22.Sabale SS, Pawar SD, More BK, Mishra AC. Seroprevalence of pandemic influenza H1N1 (2009) & seasonal influenza viruses in pigs in Maharashtra & Gujarat States, India, 2011. Indian J Med Res. 2013;138(2):267. [PMC free article] [PubMed] [Google Scholar]
- 23.Influenza A virus of swine, 2018. OIE Terrestrial Manual 2018, chapter 3.8.7
- 24.Kitikoon P, Gauger PC, Vincent AL. Animal influenza virus. New York: Humana Press; 2014. Hemagglutinin inhibition assay with swine sera; pp. 295–301. [DOI] [PubMed] [Google Scholar]
- 25.Swine flu deaths at 1895; number of cases near 32K mark (2015) Press Trust of India (March 21, 2015).
- 26.Trevennec K, Cowling BJ, Peyre M, Baudon E, Martineau GP, Roger F. Swine influenza surveillance in East and Southeast Asia: a systematic review. Anim Health Res Rev. 2011;12(2):213–223. doi: 10.1017/S1466252311000181. [DOI] [PubMed] [Google Scholar]
- 27.Sreta D, Tantawet S, Ayudhya SNN, Thontiravong A, Wongphatcharachai M, Lapkuntod J, Bunpapong N, Tuanudom R, Suradhat S, Vimolket L, Poovorawan Y. Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg Infect Dis. 2010;16(10):1587. doi: 10.3201/eid1610.100665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rith S, Davis CT, Duong V, Sar B, Horm SV, Chin S, Ly S, Laurent D, Richner B, Oboho I, Jang Y. Identification of molecular markers associated with alteration of receptor-binding specificity in a novel genotype of highly pathogenic avian influenza A (H5N1) viruses detected in Cambodia in 2013. J Virol. 2014;88(23):13897–13909. doi: 10.1128/JVI.01887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guercio A, Purpari G, Conaldi PG, Pagano V, Moreno A, Giambruno P, Di Trani L, Vaccari G, Falcone E, Boni A, Cordioli P. Pandemic influenza A/H1N1 virus in a swine farm house in Sicily. Italy J Environ Biol. 2012;33(2):155. [PubMed] [Google Scholar]
- 30.Ciacci-Zanella JR, Schaefer R, Gava D, Haach V, Cantão ME, Coldebella A. Influenza A virus infection in Brazilian swine herds following the introduction of pandemic 2009 H1N1. Vet Microbiol. 2015;180(1–2):118–122. doi: 10.1016/j.vetmic.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 31.Perera HK, Wickramasinghe G, Cheung CL, Nishiura H, Smith DK, Poon LL, Perera AK, Ma SK, Sunil-Chandra NP, Guan Y, Peiris JS. Swine influenza in Sri Lanka. Emerg Infect Dis. 2013;19(3):481. doi: 10.3201/eid1903.120945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Zhang J, Qiao C, Yang H, Zhang Y, Xin X, Chen H. Co-circulation of pandemic 2009 H1N1, classical swine H1N1 and avian-like swine H1N1 influenza viruses in pigs in China. Infect Genet Evol. 2013;13:331–338. doi: 10.1016/j.meegid.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 33.Lutteke N, Sausy A, Black AP, Wildschutz F, Muller CP. Neutralizing antibodies against influenza A in pigs before and after the 2009 pandemic, Luxembourg. Vet Microbiol. 2014;169(1–2):96–101. doi: 10.1016/j.vetmic.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 34.Tharakaraman K, Sasisekharan R. Influenza surveillance: 2014–2015 H1N1 “swine”-derived influenza viruses from India. Cell Host Microbe. 2015;17(3):279–282. doi: 10.1016/j.chom.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Qiao C, Tang X, Chen Y, Xin X, Chen H. Human infection from avian-like influenza A (H1N1) viruses in pigs. China Emerg Infect Dis. 2012;18(7):1144. doi: 10.3201/eid1807.120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray GC, McCarthy T, Capuano AW, Setterquist SF, Olsen CW, Alavanja MC, Lynch CF. Swine workers and swine influenza virus infections. Emerg Infect Dis. 2007;13(12):1871. doi: 10.3201/eid1312.061323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers KP, Olsen CW, Gray GC. Cases of swine influenza in humans: a review of the literature. Clin Infect Dis. 2007;44(8):1084–1088. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, Smith GJD, Peiris JSM, Guan Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328(5985):1529–1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno A, Di Trani L, Faccini S, Vaccari G, Nigrelli D, Boniotti MB, Falcone E, Boni A, Chiapponi C, Sozzi E, Cordioli P. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet Microbiol. 2011;149(3–4):472–477. doi: 10.1016/j.vetmic.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Corzo CA, Culhane M, Juleen K, Stigger-Rosser E, Ducatez MF, Webby RJ, Lowe JF. Active surveillance for influenza A virus among swine, mid-western United States, 2009–2011. Emerg Infect Dis. 2013;19(6):954. doi: 10.3201/eid1906.121637. [DOI] [PMC free article] [PubMed] [Google Scholar]