Abstract
Introduction
Breast cancer (BC) is the second most frequent cancer worldwide. It is known that a subset of BC has amplification, and overexpression of the epidermal growth factor receptor (EGFR) and high expression of the insulin-like growth factor receptor-1 (IGF-1R) are correlated with a favorable prognosis. This study aimed to evaluate the prognostic and predictive values of the EGFR and IGF-1R in tumor samples from patients with BC and their correlation with socio-epidemiological features.
Patients and Methods
We analyzed socio-epidemiological, clinical–pathological data and tumor tissues from 124 patients with BC undergoing treatment, to assess levels of EGFR and IGF-1R mRNA and protein. The predictive performance included the calculation of area-under-the-curve (AUC) to discriminate groups of patients with high and low mRNA expression associated with survival analysis within each molecular group of BC.
Results
We found a significant expression increase (p <0.001) in EGFR associated with body mass index, angiolymphatic invasion, compromised lymph nodes and follow-up in 58.1% of the triple-negative and HER overexpression tumors. The increase in IGF-IR was significant (p <0.001) in 41.9% of luminal tumors A and B. ROC analysis showed that EGFR had a higher predictive performance (AUC = 0.891) than IGF-1R (AUC = 0.60). The Kaplan–Meier analysis indicated that only the high expression of EGFR was associated with a decreased probability of survival for patients, what did not happen with IGF-1R.
Conclusion
Our results suggest that EGFR and IGF-1R expression patterns associated with the clinical characteristics of patients and biological profile influenced the evolution of BC.
Keywords: breast cancer, hormonal receptors, EGFR, IGF-IR, predictive performance
Introduction
Breast cancer (BC) became in 2020 the most frequently diagnosed cancer worldwide according to data from the Global Cancer Observatory (GCO).1 This neoplasm is of uncontrolled growth and rapid proliferation of cells, originating from the breast lobules or ducts and migrating to other regions of the body.2 About ~70–80% of the patients with non-metastatic disease at an early stage are benefit with a superior survival rate. But when in an advanced stage with distant organ metastases, it is considered incurable in currently available therapies.3 Early detection of BC can significantly reduce long-term mortality rates, and the identification of early cancer cells that will eventually define therapeutic success is the most critical point for the best prognosis.4,5
The risk factors for the development of BC are many; however, studies have pointed that the most important factor is related to gender, followed by age, obesity, physical activity, diet, alcohol consumption.6 There is an urgent need to develop a highly sensitive and rapid early-stage breast cancer diagnosis method.4,7 A successful diagnosis in the early stages of BC allows for better treatment, thereby increasing the patient’s likelihood of survival. The cost of treating BC is high, especially in the advanced stages of the disease, due to late diagnosis and biological profiles.6
Various molecular diagnostic tools have been developed to improve the diagnosis of BC in the clinical setting, aiming at the detection of early diagnosis capable of improving survival rates that have increased significantly worldwide in the last decades.8–11 The expansion of the use of different BC biomarkers has been used clinically, and this includes tissue markers, such as hormone receptors, human epidermal growth factor 2, urokinase plasminogen activator, genetic markers, such as breast cancer 1 (BRCA1) and BRCA2, and serum markers, such as cancer-associated antigen (CA 27.29).8,12
Estrogen receptors (ER) and progesterone receptors (PR) are hormone receptors used in clinical studies as predictive and prognostic markers to guide therapy decisions, especially for endocrine and HER2-targeted regimens.13–15 It is known that a subset of BC has amplification and overexpression of the epidermal growth factor receptor (EGFR). EGFR is a receptor tyrosine kinase (RTK) that belongs to the ErbB family, and a transmembrane protein comprising an extracellular ligand-binding domain, transmembrane domain, and cytoplasmic tyrosine kinase domain.16,17 Overexpression of EGFR in cancer is partly due to gene amplification.18 Constitutive activation of EGFR can promote tumor processes including angiogenesis and metastasis and is associated with poor prognosis. EGFR-targeted therapy has finally shown some promise in terms of improving outcomes in breast cancer patients, but molecular prognostic and predictive factors need to be identified to optimize selection of patients for EGFR-targeted therapies.19–21
Several studies show the association of the insulin-like growth factor receptor-1 (IGF-1R) with the establishment and progression of BC.21–23 Recent studies on BC have identified high expression of IGF-1R correlated with positive BC for hormone receptors associated with a favorable prognosis, while low expression was correlated with the triple-negative ER-/PR-/HER2-. IGF-1R has been shown to act as a tumor and metastasis suppressor, probably because IGF-1R acts as a suppressor of tumorigenesis by regulating the tumor microenvironment through protecting tumor epithelial cells from endoplasmic reticulum stress. However, other results support the possibility that the IGF-1R has a dual function as both a tumor suppressor and an oncogene.22
A large body of laboratory, epidemiological and clinical evidence has explored this pathway in cancer, with the active form of IGF-1R being expressed in up to 50% of BC tumors. The Insulin-Like Growth Factor 1 (IGF-1) system, which includes IGF-1, IGF-binding proteins (IGFBPs) and the IGF-1 receptor (IGF-1R), plays a significant role in human physiology, particularly in the development and function of many tissues, including the mammary gland.24
This study aimed to evaluate the prognostic and predictive values of the EGFR and IGF-1R genes in tumor samples from patients with BC who underwent undergoing oncological and pharmacological treatment, crossing the level of gene expression with socio-epidemiological and clinical–pathological variables for understanding the response of patients in this type of treatment.
Patients and Methods
Tumor Samples
Clinical and pathological information and biopsy samples of tumor tissues were collected from 124 patients with breast cancer registered at the Hospital Ophir Loyola – HOL (Pará State, Brazil). The patients signed an Informed Consent Form and underwent neoadjuvant chemotherapy according to the HOL protocol.25 These patients were treated between 2010 and 2017 in the mastology sector at HOL. The study was approved by the HOL ethics committee (Protocol 432008), and all methods were carried out in accordance with Declaration of Helsinki guidelines and regulations.
In this study, exclusion criteria were set to patients who presented heart disease, pregnancy, lactation, or any clinical condition in which the patient could not undergo neoadjuvant chemotherapy, were considered. The guidelines for notification of tumor marker studies were followed according to the characteristics of the patients as recommended.26 Table 1 presents the clinical and pathological characteristics of the 124 studied patients. Fragments of normal breast tissue were also collected from 22 patients who underwent reduction mammoplasty.
Table 1.
Patient Characteristics
| Characteristics | No. of Patients | % |
|---|---|---|
| Age | ||
| <40 years | 26 | 21.0 |
| 41–60 years | 68 | 54.8 |
| >60 years | 30 | 24.2 |
| Parity | ||
| Parous | 110 | 88.7 |
| Nulliparous | 14 | 11.3 |
| Personal background | ||
| Smoking | 27 | 21.8 |
| Ethics | 2 | 1.6 |
| Smoking and alcoholism | 13 | 10.5 |
| Others | 82 | 66.1 |
| Family Background | ||
| Breast cancer | 29 | 23.4 |
| Other cancers | 39 | 31.5 |
| NDN | 56 | 45.2 |
| BMI | ||
| ≤25 kg/m2 | 47 | 37.9 |
| >25 kg/m2 | 68 | 54.8 |
| Not evaluated | 9 | 7.3 |
| Exame access time | ||
| 1–3 months | 35 | 28.0 |
| 4–6 months | 27 | 21.7 |
| >6 months | 29 | 23.3 |
| Not evaluated | 33 | 26.7 |
| Exame used for diagnosis | ||
| Breast ultrasound | 16 | 12.9 |
| Mammography | 33 | 26.6 |
| Mammography and ultrasound | 42 | 33.8 |
| Not evaluated | 33 | 26.6 |
| Biopsy access time | ||
| Up to 1 month | 12 | 9.6 |
| 2–6 months | 49 | 39.5 |
| >6 months | 61 | 49.2 |
| Surgery | 2 | 1.6 |
| Neoadjuvant chemotherapy | ||
| Up to 01 month | 22 | 17.7 |
| 02–06 months | 78 | 62.9 |
| More than 06 months | 24 | 19.3 |
| Surgery | ||
| Up to 1 month | 2 | 1.6 |
| 2–6 months | 28 | 22.6 |
| >6 months | 21 | 16.9 |
| Surgery performed | ||
| Quadrantectomy | 33 | 26.6 |
| Mastectomy | 91 | 73.4 |
| Beginning of radiotherapy after surgery | ||
| Up to 01 month | 1 | 0.8 |
| 02–04 months | 24 | 19.4 |
| More than 04 months | 53 | 42.7 |
| Did not perform | 46 | 37.1 |
| Presence of IAL and/or IPN | ||
| Present | 55 | 44.4 |
| Absent | 67 | 54.0 |
| Without residual neoplasia | 2 | 1.6 |
| Diagnostic stage | ||
| Stage 1 | 3 | 2.4 |
| Stage 2A | 19 | 15.0 |
| Stage 2B | 21 | 17.0 |
| Stage3A | 43 | 34.7 |
| Stage3B | 36 | 29.0 |
| Stage 4 | 0 | 0.0 |
| Tx | 2 | 1.6 |
Abbreviations: IAL, angiolymphatic invasion; IPN, perineural Invasion; Tx, primary tumor cannot be evaluated.
This collection is intended to study the relative quantification, between tumor tissue and normal tissue, of mRNA and proteins of EGFR (Epidermal growth factor receptor) and IGF-1R (insulin-like growth factor receptor 1), for this reason, a pool was built of mRNA and another of proteins with these 22 samples. Tumor samples, obtained by incisional biopsy, embedded in paraffin and fixed in formalin (FFPE) were prepared for histological and immunohistochemical evaluation.
Immunohistochemistry
The expression of estrogen and progesterone receptor, HER-2, Ki-67 and cytokeratin 8/18 for the diagnosis and classification of breast cancer were evaluated in breast tumors using the streptavidin-biotin-peroxidase method with commercial primary antibodies as previously described by our research group.27 Tumors have the following molecular groups of breast cancer: Luminal A (ER or PR positive, HER-negative, Ki-67 <14%), Luminal B (ER or PR positive, HER-negative, Ki-67> 14%), Luminal B HER (ER or PR positive, HER-positive and any Ki-67), Triple-negative (ER and PR negative, HER negative, any Ki-67) and HER Super Express (ER and PR negative and HER positive in +++).28 Immunoreactivity of proteins encoded by the selected genes EGFR (1:200 dilution; MA5-13070; Thermo Fisher Scientific, USA), IGF-1R (dilution 1:200; MA5-13817; Thermo Fisher Scientific, USA), was evaluated by immunohistochemistry on paraffin-embedded tissue sections, as suggested by Calcagno et al.29
mRNA Expression
To quantitate mRNA levels of EGFR and IGF-1R, total RNA was isolated from a pool of 22 normal and 124 tumor tissues using Trizol (Thermo Fisher Scientific, USA). After the total mRNA was isolated, it was reverse-transcribed using the High-Capacity cDNA kit according to the manufacturer’s protocol (Thermo Fisher Scientific, USA). Complementary DNA was then amplified by real-time quantitative PCR (qPCR) using TaqMan probes: EGFR: Hs01076090_m1, IGF-1R: Mm00802831_m1. All qPCRs were performed in triplicate in 7500 Fast Real-Time PCR instrument (Thermo Fisher Scientific, USA). ACTB (4333762F; Thermo Fisher Scientific, USA) gene was selected as an internal control.30 All qPCRs were performed in triplicate in 7500 Fast Real-Time PCR instrument (Thermo Fisher Scientific, USA).
The relative quantification of gene expression was calculated according to the method of Livak and Schmittgen.31 The cDNA pool of 22 samples of normal breast tissue extracted from reductive mammoplasty was designated as a calibrator from each tumor. For the expression changes of mRNAs and protein, we used the sections used in the literature for analysis of tumor tissue.27 When the relative quantification value is equal to 1 it means that the amount of mRNA is equal both in the tumor and in the pool of mRNAs extracted from of pool of 22 samples of normal breast tissue extracted from reductive mammoplasty. When the value is 1.5, it means that the relative quantification of that mRNA is 50% higher in the tumor than in the pool of mRNAs extracted from the same pool. Following this reasoning, when the relative quantification is equal to 0.5 it means that there is 50% less of this mRNA in the tumor than in the cDNA pool of 22 samples of normal breast tissue extracted from reductive mammoplasty. This same correlation was performed at Western Blot using the pool of proteins extracted from 22 samples.
Protein Expression
Western blot analysis was performed as described previously.27 Reduced protein (25 μg) from each sample was applied to SDS–polyacrylamide gel and electrophoresed. Then, the individual proteins in the electrophoresis gel were transferred to a polyvinylidene fluoride membrane and labeled with antibodies specific for the corresponding proteins: anti-EGFR (dilution 1:2000; MA5-13070; Thermo Fisher Scientific, USA), anti-IGF-1R (1:200 dilution; MA5-13817; Thermo Fisher Scientific, USA) and ACTB (dilution 1:250; Ac-15) was used as a loading reference control.
Follow-Up
Patients were followed up with an average of 2 years, where we assessed overall survival (OS), disease-free survival (DFS) and disease-free events. The last follow-up was completed in December 2019.
Statistical Analysis
The validation data are shown as frequency, median and interquartile range (IQR). The association between categorical variables was analyzed using the χ2 test, with p-value <0.05 being considered significant. The data were submitted to the Shapiro–Wilk test to assess the distribution of mRNA and protein expression, then the Kruskal–Wallis test was used to investigate possible comparisons between mRNA and EGFR and IGF-1R protein in the socio-epidemiological and clinical–pathological data. The median and the IQR were used to assess the degree of dispersion of the data around the centrality measure. The difference between the upper and lower quartiles was determined for the interquartile range. Heatmaps with hierarchical groupings were made to show the expression pattern EGFR and IGF-1R in BC samples using the Z-score metric. The predictive performance of the two genes was analyzed using the area-under-the-curve (AUC) calculation on the ROC curve to discriminate groups of patients with high and low gene expression. Sensitivity was assumed as the percentage of true positives and specificity, the percentage of true negatives. The Kaplan–Meier estimator and the Log rank test were used to estimate the probability of survival of the high and low expression groups, within each molecular group of BC, considering a 95% confidence level and p-value <0.0001. All statistics were performed using R (https://www.r-project.org).
Results
In this study, 124 patients with breast cancer between 28 and 89 years of age were included. Most patients are in the age group between 41 and 60 years old (54.8%), with a body mass index (BMI) above 25 (54.8%), and they gestated (88.7%) and breastfeeding (11.3%) (Tables 1 and 2).
Table 2.
Socio-Epidemiological and Clinical–Pathological Variables in Breast Cancer
| Variable | N (%) | p-value a | EGFR mRNA | EGFR Protein | IGF-1R mRNA | IGF-1R Protein | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RQ (Median ± IQR) | p-value b | RQ (Median ± IQR) | p-value b | RQ (Median ± IQR) | p-value b | RQ (Median ± IQR) | p-valueb | ||||
| Age (years) | 18–40 | 26 (21.0) | <0.001* | 1.85 ± 1.06 | 0.286 | 1.71 ± 0.92 | 0.546 | 1.30 ± 0.49 | 0.042* | 1.19 ± 0.48 | 0.056 |
| 41–60 | 68 (54.8) | 1.86 ± 1.31 | 1.77 ± 0.93 | 1.43 ± 0.86 | 1.31 ± 0.86 | ||||||
| >60 | 30 (24.2) | 1.57 ± 1.27 | 1.46 ± 1.24 | 1.81 ± 1.70 | 1.50 ± 1.59 | ||||||
| Parity | Parous | 110 (88.7) | <0.001* | 1.79 ± 1.29 | 0.453 | 1.65 ± 1.08 | 0.434 | 1.45 ± 0.94 | 0.125 | 1.33 ± 0.89 | 0.109 |
| Nulliparous | 14 (11.3) | 1.98 ± 1.27 | 1.81 ± 0.78 | 1.23 ± 0.65 | 1.09 ± 0.60 | ||||||
| Personal background | Smoking | 27 (21.8) | <0.001* | 1.59 ± 1.31 | 0.372 | 1.51 ± 0.74 | 0.439 | 1.47 ± 1.27 | 0.782 | 1.29 ± 1.31 | 0.698 |
| Ethylism | 2 (1.6) | 1.42 ± 0.09 | 1.33 ± 0.08 | 1.92 ± 0.02 | 1.81 ± 0.02 | ||||||
| Smoking+Ethylism | 13 (10.5) | 2.46 ± 1.62 | 2.12 ± 1.45 | 1.31 ± 1.76 | 1.22 ± 1.63 | ||||||
| NDN | 82 (66.1) | 1.87 ± 1.16 | 1.77 ± 0.99 | 1.42 ± 0.86 | 1.32 ± 0.87 | ||||||
| Family background | Breast cancer | 29 (23.4) | 0.011* | 2.00 ± 1.24 | 0.413 | 1.84 ± 0.83 | 0.537 | 1.31 ± 1.99 | 0.519 | 1.20 ± 1.80 | 0.478 |
| Other cancers | 39 (31.5) | 1.86 ± 1.33 | 1.71 ± 1.07 | 1.60 ± 1.14 | 1.49 ± 1.24 | ||||||
| NDN | 56 (45.2) | 1.68 ± 1.21 | 1.58 ± 1.12 | 1.43 ± 0.69 | 1.30 ± 0.63 | ||||||
| BMI (kg/m2) | ≤25 | 47 (37.9) | <0.001* | 1.96 ± 1.13 | 0.041* | 1.78 ± 1.03 | 0.022* | 1.42 ± 0.78 | 0.851 | 1.29 ± 0.82 | 0.789 |
| > 25 | 68 (54.8) | 1.82 ± 1.36 | 1.67 ± 1.09 | 1.46 ± 1.33 | 1.30 ± 1.30 | ||||||
| IMC not rated | 9 (7.3) | 1.35 ± 0.44 | 1.28 ± 0.33 | 1.68 ± 1.56 | 1.47 ± 1.52 | ||||||
| Angiolymphatic invasion | Present | 55 (44.4) | <0.001* | 2.51 ± 1.05 | 0.001* | 2.14 ± 0.92 | 0.001* | 1.42 ± 0.87 | 0.087 | 1.27 ± 0.90 | 0.072 |
| Absent | 67 (54.0) | 1.55 ± 0.61 | 1.41 ± 0.52 | 1.49 ± 0.85 | 1.38 ± 0.81 | ||||||
| Not rated | 2 (1.6) | 1.21 ± 0.17 | 1.18 ± 0.17 | 0.88 ± 0.03 | 0.82 ± 0.01 | ||||||
| Compromised lymph nodes | 0 | 55 (44.4) | 0.009* | 1.57 ± 1.24 | 0.003* | 1.41 ± 0.93 | 0.001* | 1.42 ± 1.36 | 0.570 | 1.26 ± 1.22 | 0.491 |
| 1–3 | 27 (21.8) | 1.85 ± 0.98 | 1.74 ± 0.89 | 1.62 ± 1.38 | 1.52 ± 1.38 | ||||||
| >3 | 42 (33.9) | 2.37 ± 1.27 | 1.91 ± 1.01 | 1.40 ± 0.53 | 1.25 ± 0.35 | ||||||
| Tumor size (cm) | <2 | 14 (11.3) | <0.001* | 2.12 ± 1.53 | 0.067 | 1.78 ± 1.12 | 0.114 | 1.42 ± 1.88 | 0.123 | 1.28 ± 1.80 | 0.282 |
| 2–5 | 69 (55.6) | 1.59 ± 1.24 | 1.51 ± 1.00 | 1.54 ± 1.10 | 1.40 ± 0.97 | ||||||
| >5 | 41 (33.1) | 2.47 ± 1.27 | 1.89 ± 0.97 | 1.33 ± 0.61 | 1.22 ± 0.56 | ||||||
| Immunohistochemistry | ER >10 | 105 (84.7) | 0.002* | 1.80 ± 1.28 | 0.718 | 1.61 ± 1.11 | 0.752 | 1.44 ± 1.37 | 0.584 | 1.29 ± 1.38 | 0.562 |
| ER ≤10 | 19 (15.3) | 1.96 ± 1.09 | 1.77 ± 0.84 | 1.42 ± 0.56 | 1.33 ± 0.56 | ||||||
| Follow-up | DFS | 81 (65.3) | <0.001* | 1.58 ± 1.14 | 0.023* | 1.50 ± 0.89 | 0.012* | 1.60 ± 1.31 | 0.14 | 1.45 ± 1.25 | 0.292 |
| Relapse or metastasis | 16 (12.9) | 1.93 ± 1.10 | 1.79 ± 0.68 | 1.44 ± 1.49 | 1.27 ± 1.59 | ||||||
| Death | 27 (21.8) | 2.57 ± 1.27 | 2.11 ± 1.02 | 1.29 ± 0.30 | 1.19 ± 0.38 | ||||||
|
Access time to treatment (months) |
1 | 27 (21.8) | 0.015* | 1.58 ± 1.23 | 0.502 | 1.45 ± 0.88 | 0.322 | 1.42 ± 0.79 | 0.396 | 1.33 ± 0.70 | 0.414 |
| 02–03 | 53 (42.7) | 1.89 ± 1.34 | 1.74 ± 1.15 | 1.38 ± 0.98 | 1.24 ± 0.92 | ||||||
| >03 | 44 (35.5) | 1.83 ± 1.01 | 1.71 ± 0.92 | 1.47 ± 1.21 | 1.37 ± 1.32 | ||||||
| Biological profile | Luminal A | 25 (20.2) | <0.001* | 1.54 ± 0.65 | 0.001* | 1.39 ± 0.61 | 0.001* | 1.99 ± 1.18 | 0.001* | 1.81 ± 1.17 | 0.001* |
| Luminal B | 33 (26.6) | 1.58 ± 1.17 | 1.51 ± 1.03 | 2.78 ± 1.87 | 2.47 ± 1.74 | ||||||
| Luminal B HER | 8 (6.5) | 1.38 ± 0.39 | 1.33 ± 0.30 | 1.01 ± 0.21 | 0.94 ± 0.24 | ||||||
| Triple-negative | 50 (40.3) | 2.48 ± 1.11 | 1.93 ± 0.82 | 1.25 ± 0.43 | 1.14 ± 0.34 | ||||||
| HER overexpression | 8 (6.45) | 2.74 ± 1.86 | 2.62 ± 1.75 | 1.28 ± 0.36 | 1.19 ± 0.30 | ||||||
Notes: aP-value of the χ2 test; bp-value of the Kruskal–Wallis test. *p-value <0.05, significantly different between groups. n: number of samples; RQ: relative quantification in which the combined non-neoplastic sample was designated as the calibrator for each neoplastic sample; T/N: the protein expression ratio between corresponding neoplastic and non-neoplastic samples.
Abbreviations: BMI, body mass index; ER, estrogen receptor; DFS, disease-free survival; NDN, nothing noteworthy; IQR: interquartile range.
Personal history did not show a predominance of smokers or alcohol users in the evaluated patients. Some comorbidities have been reported, such as asthma, diabetes, systemic arterial hypertension, but without statistical significance. Concerning family history, the predominance was of patients without a history of cancer (45.2%).
The majority of patients after the disease’s perception sought the mastology service in up to 3 months (28%) (Table 1). The exams performed most frequently were mammography and breast ultrasound (33.6%). Most of the biopsy was performed 6 months after the disease was suspected (49.2%). In our sample, three patients did not undergo a biopsy and were directly submitted to the surgical procedure with intraoperative frozen section technique.
Neoadjuvant chemotherapy started only 2–6 months after the biopsy in (62.9%) of the cases. Of the patients, 73.6% underwent a mastectomy and (26.4%) under conservative treatment. Complementary treatment with radiotherapy was delayed in a large number of patients (42.07%), just 4 months after surgery. Stage III was the most found (63.7%).
Expression Profile of EGFR and IGF-1R in Tumor Samples from Breast Cancer Patients
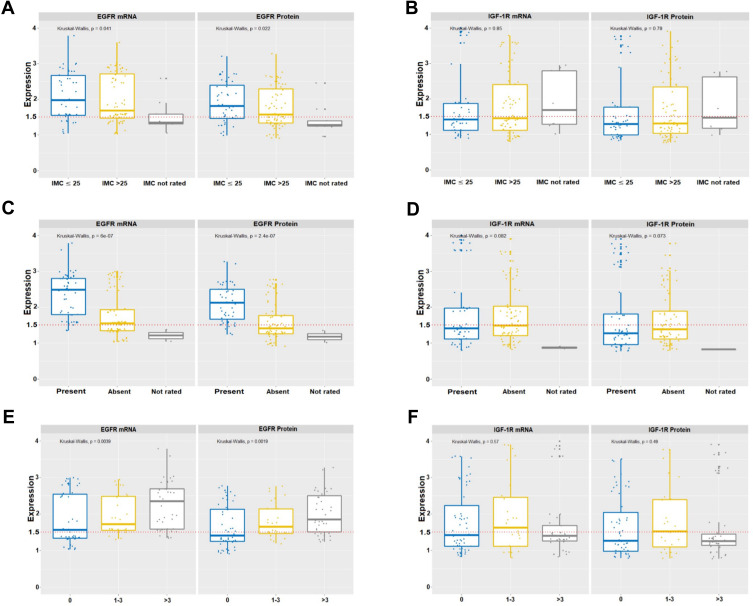
We evaluated the quantitative expression of mRNA and protein of EGFR and IGF-1R in samples of tumor fragments from 124 patients with BC. We identified the increase and decrease in the levels of EGFR gene expression with statistical significance (p <0.001) for BMI, angiolymphatic invasion, compromised lymph nodes, follow-up and biological profiles.
Table 2 shows these results as follows. Patients with BMI ≤ 25, had higher levels of EGFR [median mRNA (interquartile range, IQR): 1.96 (1.13); protein median (IQR): 1.78 (1.03)]. We also observed that the levels of mRNA and EGFR protein increased more than 1.5-fold (at least 50% expression) in 54.8% of the tumor samples of these patients.
We identified patients who were positive for angiolymphatic invasion (Table 2), with higher levels of EGFR [median mRNA (IQR): 2.51 (1.05); protein median (IQR): 2.14 (0.92)]. In this case, the increase in EGFR mRNA and protein levels was more than 1.5-fold in 57% of these tumor samples.
Patients with compromised lymph nodes >3 (Table 2) of the cases, had higher levels of EGFR [median mRNA (IQR): 2.37 (1.27); protein median (IQR): 1.91 (1.01)]. This increase was identified more than 1.5-fold in 40.9% of tumors with a higher number of lymph nodes.
The results of the follow-up of patients who died (Table 2) of the cases showed a significant increase in EGFR [median mRNA (IQR): 2.57 (1.27); protein median (IQR): 2.11 (1.02)]. EGFR mRNA and protein levels increased more than 1.5-fold in only 22.6% of these tumor samples. All of these results are shown in Figure 1.
Figure 1.
Levels of gene expression of EGFR and IGF-1R in socio-epidemiological and pathological–clinical data. Variations in levels of gene expression were significant (p <0.001) for EGFR (A) BMI, (C) angiolymphatic invasion and (E) compromised lymph nodes. IGF-1R did not show statistical significance (B) BMI, (D) angiolymphatic invasion and (F) compromised lymph nodes. In all graphs, the expression of breast tumors was normalized by non-neoplastic breast tissue. The dotted lines represent the 1.5-fold-change.
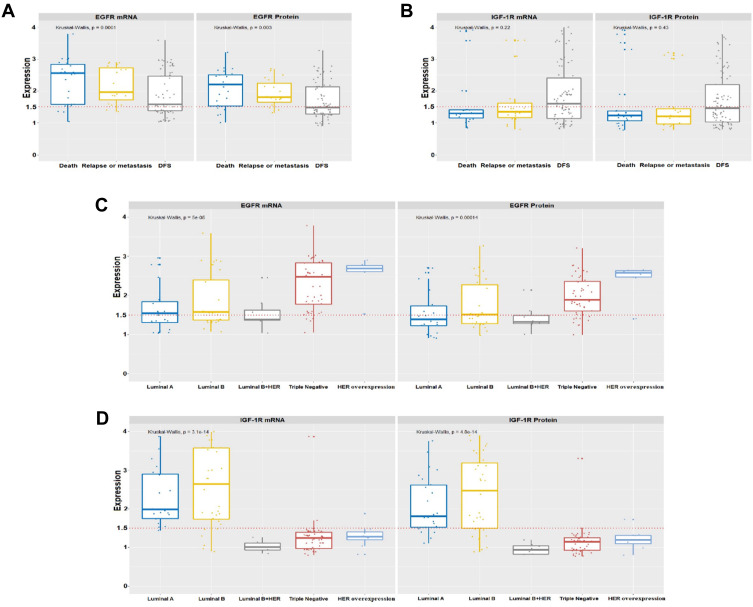
Patients with a negative ER biological profile (40.3%) triple-negative showed a significant increase in EGFR [median mRNA (IQR): 2.48 (1.11); protein median (IQR): 1.93 (0.82)] and HER overexpressed with an increase in [median mRNA (IQR): 2.74 (1.86); protein median (IQR): 2.62 (1.75)]. The levels of mRNA and protein of this gene increased more than 1.5-fold in 58.1% of the tumor samples from patients (Table 2 and Figure 2).
Figure 2.
Levels of gene expression of EGFR and IGF-1R in the follow-up variables and biological profiles. The increase in gene expression levels was significant (p <0.001) for EGFR (A) only at follow-up. IGF-1R (B) did not show statistical significance. The increase in gene expression levels was significant (p <0.001) for EGFR (C) in the triple-negative and Her overexpressed negative receptors, whereas IGF-1R (D) was higher in the positive receptors, Luminal A and Luminal B. In all graphs, the expression of breast tumors was normalized by non-neoplastic breast tissue. The dotted lines represent the 1.5-fold-change.
We identified a significant increase in IGF-1R (p <0.001) only for the ER positive biological profile variables. There were of Luminal A with [median mRNA (IQR): 1.99 (1.18); protein median (IQR): 1.81 (1.17)], of Luminal B with [median mRNA (IQR): 2.78 (1.87); protein median (IQR): 2.47 (1.74)] and of Luminal B HER with [median mRNA (IQR): 1.01 (0.21); protein median (IQR): 0.94 (0.24)]. We found IGF-1R mRNA and protein levels increased more than 1.5-fold in 41.9% of these tumor samples (Table 2 and Figure 2) compared to the pool of 22 samples of normal breast tissue extracted from reductive mammoplasty.
Relationship Between EGFR and IGF-1R Expressions
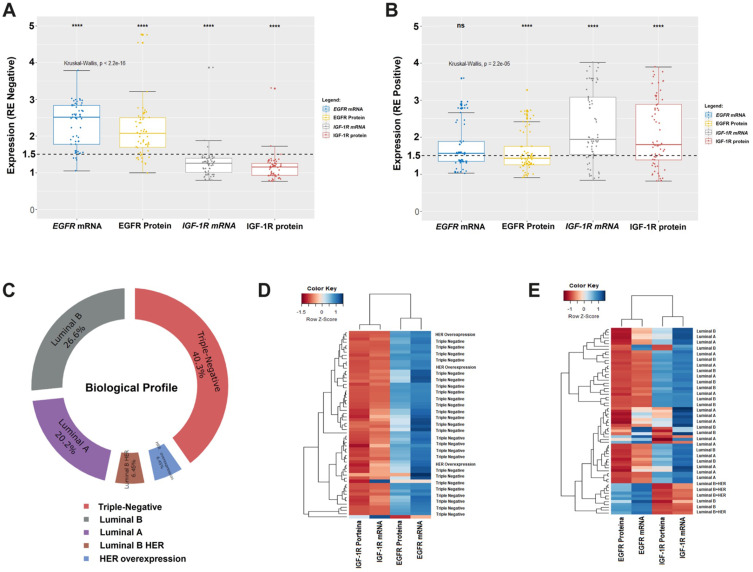
We identified different variations of EGFR and IGF-1R gene expression in the types of biological profiles using the status of estrogen receptors. For example, we identified EGFR receptor mRNA and protein levels elevated more than 1.5-fold in 58.1% of tumors with negative receptor profiles (Figure 3A). On the other hand, levels of mRNA and IGF-1R protein were 1.5-fold higher in only 41.9% in positive receptor tumors (Figure 3B). Our BC tumor samples were classified as 46.75% for negative receptors in triple-negative and HER overexpression and 53.25% for positive receptors in luminal A, B and B/HER (Figure 3C).
Figure 3.
(A and B) Box Plot shows average levels of normalized expression of mRNA and proteins from EGFR and IGF-1R. The boxes are drawn from the 75th to the 25th percentile. The vertical lines above and below the box define the maximum and minimum values and the dots indicate outliers, the horizontal line inside the box representing the median. Kruskal–Wallis test (p <0.0001) was applied to compare the means between the four groups. In all graphs, expression in breast tumors was normalized by the non-malignant tissue removed from reduction mammoplasty. RQ: relative quantification; (T) tumor sample; (N) normal non-malignant tissue; The whiskers indicate the minimum and maximum values. The dotted lines represent the 1.5-fold-change. (C) Donut Chart shows the ranking of the positions of the biological profiles. (D and E) The heat map shows an expression profile defined by the unsupervised cluster to group by the similarity of gene expression the samples of profile RE negative and RE positive, respectively. Z-score was the metric applied to infer the best grouping between profiles. Gradients with a red color trend represent profiles with a lower Z-score and gradients with a blue color tendency with a higher Z-score. (ns, not significant, ****p < 0.0001).
We didactically analyze the relationship of expressions between EGFR and IGF-1R through the unsupervised grouping on the heat map, showing the influence of gene expression associated with negative and positive receptors. The patients that most express EGFR represent 50% in our samples with an expression value of this gene greater than 1.5 (Figure 3D) and who died after 1 year of treatment. While the patients that most express IGF-1R, express positive receptors representing in our samples 50% of the expression of this gene greater than 1.5 (Figure 3E), being those patients with disease-free survival.
Survival Analysis
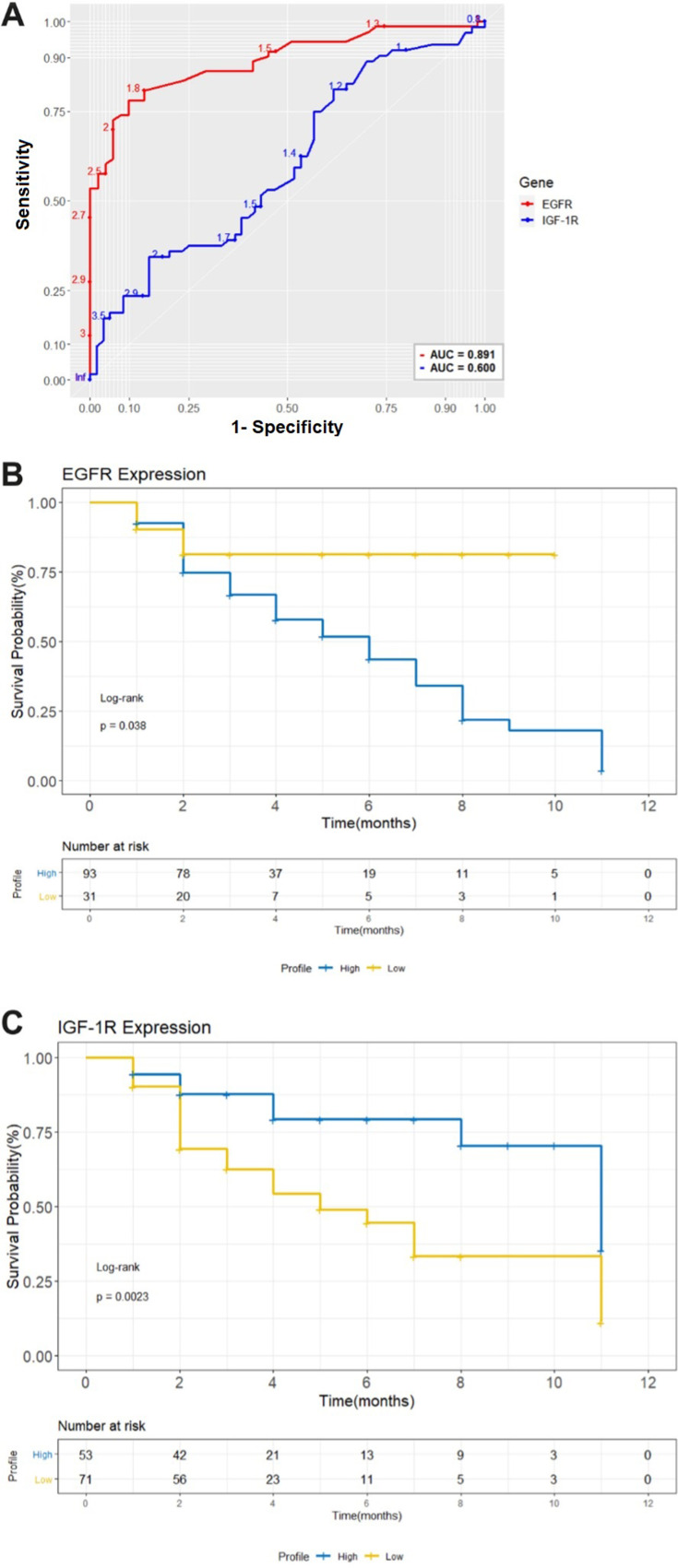
We used survival analysis to assess the contribution of high expression of EGFR and IGF-1R to the overall survival of 124 studied patients. The analysis of the ROC curve classified the patients of high and low expression, associated with follow-up in two groups: Disease-free survival and poor outcome (recurrence/metastasis and death) to understand the prognostic value of these genes after 1 year of treatment disease. The highest point on the AUC curve was chosen as the cutoff point for (EGFR: AUC = 0.891; sensitivity = 94.2%, specificity = 75.4% and for IGF-1R: AUC = 0.60; sensitivity = 80.1%, specificity = 69.5%). Sensitivity is the percentage of true positives and specificity is the percentage of true negatives. (Figure 4A). From these data, the expression values were considered 1.5 for both recipients. The Kaplan–Meier analysis showed that there is an association between high expression of EGFR (Log rank test, p = 0.038) and decreased overall survival in the group of patients who expressed a lot of EGFR within the negative receptor profiles (triple-negative and HER overexpression). This did not happen with the group of patients who expressed IGF-1R (Log rank test, p = 0.0023), revealing an association between low IGF-1R expression and the decrease in overall survival of patients (Figure 4B and C).
Figure 4.
(A) Analysis of the ROC curve to separate patients into groups of high and low expression of EGFR and IGF-1R associated with follow-up. The largest area was for the EGFR receptor, AUC = 0.891 which represents a cut-off point of 1.5 from normalized expression. The IGF-1R receiver had a smaller area AUC = 0.60 for a cut-off point of 1.5. (B) and (C) Kaplan–Meier analysis of the overall survival in months of patients with breast cancer as a function of the expression of the EGFR and IGF-1R receptors. The high gene expression ≥1.5 (blue line) for the two EGFR receptors as opposed to the low expression (yellow line), is strongly associated with a lower probability of survival for patients who do not express hormone receptors in one year after treatment. The low gene expression of IGF-1R was shown to be associated with the lower probability of survival of patients. ROC, receiver operating characteristic. Log-rank p-value <0.0001.
Discussion
World Health Organization (WHO) defines breast cancer (BC) as a heterogeneous disease, with molecular classification criteria that have emerged as an alternative capable of better translating the clinical heterogeneity of this disease, and the treatment strategies differ according to the molecular subtype. The molecular characteristics of BC include expression of human epidermal growth factor receptor 2 (HER2, encoded by ERBB2), hormonal receptors (estrogen receptor and progesterone receptor) and/or BRCA mutations.3
This study analyzed 124 breast cancer patients whose sociodemographic characteristics mentioned in Tables 1 and 2 are in accordance with Brazilian and South American population statistics,32,33 however the average age of development of breast cancer and BMI is lower than the indices of the North American Women’s Health Initiative (WHI) program,34 on the other hand the percentage of individuals who gave birth and breastfed is higher in this present study than the WHI. Most patients of this study are in the age group between 41 and 60 years old (54.8%), with a body mass index (BMI) above 25 kg/m2 (54.8%), parous (88.7%) and breastfeeding (11.3%).
We investigated the influence of EGFR and IGF-1R gene expression on the evolution of BC, as there are promising indications of the activation of different cell signaling pathways due to the deregulation of some growth factors, acting on the progression of malignancy, prevention of apoptosis, drug resistance and metastasis.35,36
Our results of qPCR and Western blot, obtained from tumor samples from patients with breast cancer, showed a significant increase in EGFR expression for BMI, angiolymphatic invasion, compromised lymph nodes, follow-up and biological profiles (p <0.001). The analysis of these variables showed that the levels of EGFR mRNA and protein increased more than 1.5-fold in the patients’ tumors. On the other hand, the increase in IGF-1R expression levels by more than 1.5 in patients’ breast tumors was significant only in biological profiles.
We observed that EGFR was more expressed in patients aged 41–60 years, nulliparous, smokers and alcoholics, who had tumors larger than 5 cm in size, with more than three lymph nodes affected, expressing an estrogen receptor rate <10 and that evolved to death. The analysis of the expression of 58.1% of the tumor samples of these patients, showed high levels of EGFR protein in triple-negative profiles and HER overexpression.
Triple-negative Breast Cancers (TNBC) is a group with the worst prognosis and accounts for ~ 10–20% of human breast cancers.20,37,38 In our data, we found EGFR protein expression in 40.3% (50 cases) of triple-negative and 6.5% (8 cases) of HER overexpression. High gene expression was observed in our clinically negative breast cancer data for the expression of estrogen and progesterone receptors (ER/PR) and HER2 protein, which can occur as a result of underlying genetic changes, such as the number copies of the altered EGFR gene. These EGFR abnormalities exhibit geographic and ethnic variations, their expression varies in different regions of the world.39–43
The IGF-IR expressed more in patients over 60 years of age who had children and alcoholics, who had tumors between two and five centimeters in size, with one and up to three affected lymph nodes, expressing an estrogen receptor rate >10. The cohort of samples with higher expression of IGF-1R had an increase in disease-free survival. The expression analysis showed 41.9% of the tumor samples with high levels of IGF-IR protein with biological luminal profiles A and B.
Luminal A tumors are estrogen receptor-positive, cytokeratin 8 and 18 positives and have a good prognosis, whereas luminal B tumors express the estrogen receptor and cytokeratins 8 and 18 but have a poorer prognosis than luminal A tumors.44–46 As most breast cancers fall under the luminal classification, positive for hormone receptors (ER +, PR +), comprising ~50% luminal A and ~10–20% luminal B, in general, with luminal tumors being the better prognosis.43 In our data, we found the highest IGF-IR expression in luminal B with 26.6% (33 cases), luminal A with 20.2% (25 cases) and luminal B HER with 6.5% (only eight cases), respectively.
Consistently, our analysis of overall survival with Kaplan–Meier estimator, showed that the survival time for the group of patients with high expression of EGFR after 1 year of treatment was shorter to patients with low expression (Log rank test, p = 0.038). These data are confirmed in the study by Zeng et al who demonstrated that high EGFR is significantly associated with distant metastasis and severely reduced survival time.47 Probably because EGFR promotes breast cancer invasion/growth through activation of the MAPK (mitogen-activated protein kinase) pathway.48
Meanwhile, the survival time for the group of patients with high expression of IGF-1R was longer compared to patients with low expression (Log rank test, p = 0.0023). This result confirms the influence of EGFR in patients who do not express hormone receptors, with high sensitivity (94.2%) and specificity (75.4%) in contrast to a decrease in sensitivity (80.1%) and specificity (69.5%) of IGF-1R on the ROC curve, revealing an association between low IGF-1R expression and decreased overall survival for the group of patients.
A hypothesis to explain the reduced survival in the group of patients with reduced IGF-1R expression can be supported by the possibility that these tumors that express hormone receptors and evolved from unfavorable events during hormone therapy, present a downregulated expression of IGF-1R. Pennisi et al, showed that the downregulated expression of IGF-1R in breast cancer cells was associated with decreased E-cadherin expression and increased cellular motility.49 E-cadherin is frequently expressed in epithelial cells of the breast and its presence is related to a non-metastatic phenotype.50,51 Clinical studies have shown that decreased levels of E-cadherin expression in breast cancer are associated with poor prognostic indicators.49
A study by Obr et al analyzed the data set of the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), where they found an association between low expression IGF-1R and decreased overall patient survival. According to this study, the expression of this gene inversely correlated with the patient’s survival indicates that the reduced overall survival is not only due to the low expression of IGF-1R, but to the increase in cellular stress due to the increase in reactive oxygen species (ROS) and the production of cytokines such as interleukin (IL)-6 and C-C motif chemokine ligand 2 (CCL2), to promote the aggressive tumor microenvironment through the infiltration of immune cells, matrix remodeling and collagen deposition observed in rat and lineage tumors breast cancer cells.22
Studies focused on the identification of therapeutic target genes in hormone-responsive BC, show that resistance to hormonal therapy occurs in more than 50% of the patients sustained by new or acquired resistance, the latter emerging after a first treatment or in case of recurrence.52 Such conditions can occur due to the combination of genetic or epigenetic alterations, such as the deregulation of the growth factor pathways that intersect with ER signaling, modification of chromatin remodeling, loss of ER expression, polymorphic variations of ER.53
Our study gathered important information to conclude that patients with molecular subtypes that do not respond to hormonal block and that overexpress EGFR, tend to evolve badly, whether, for recurrence, metastasis, or death, when undergoing oncological and pharmacological treatment.
In conclusion, our results suggest that the analysis of the expression of EGFR and IGF-1R associated with the clinical characteristics of patients, alone or together, influenced the evolution of CM. Patients who have high expression of both growth factor receptors studied have a correlation with disease-free survival and tend to have a worse outcome.
Acknowledgments
The authors thank the financial support of this study, supported by Brazilian funding agencies National Council for Scientific and Technological and Development (CNPq; grants #301350/2019-1 to RMRB) and Coordination for the Improvement of Higher Education Personnel (CAPES; to JHSM.), as well as PROPESP/UFPA for payment of the publication fees.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Day CM, Hickey SM, Song Y, Plush SE, Garg S. Novel tamoxifen nanoformulations for improving breast cancer treatment: old wine in new bottles. Molecules. 2020;25(5):1182. doi: 10.3390/molecules25051182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Prim. 2019;5(1):66. doi: 10.1038/s41572-019-0111-2 [DOI] [PubMed] [Google Scholar]
- 4.Wang L. Early diagnosis of breast cancer. Sensors. 2017;17:1572. doi: 10.3390/s17071572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koriech OM. Breast cancer and early detection. J Family Community Med. 1996;3(1):7–9. [PMC free article] [PubMed] [Google Scholar]
- 6.Santiago-Montero R, Sossa H, Gutiérrez-Hernández DA, Zamudio V, Hernández-Bautista I, Valadez-Godínez S. Novel mathematical model of breast cancer diagnostics using an associative pattern classification. Diagnostics. 2020;10:136. doi: 10.3390/diagnostics10030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Zhang P, Dou L, Wang L, Sun K. Detection of circulating tumor cells in breast cancer patients by nanopore sensing with aptamer-mediated amplification. ACS Sensors. 2020;8:2359–2366. doi: 10.1021/acssensors.9b02537 [DOI] [PubMed] [Google Scholar]
- 8.Fan S, Shahid M, Jin P, Asher A, Kim J. Identification of metabolic alterations in breast cancer using mass spectrometry-based metabolomic analysis. Metabolites. 2020;10(4):170. doi: 10.3390/metabo10040170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- 10.Cossetti RJD, Tyldesley SK, Speers CH, Zheng Y, Gelmon KA. Comparison of breast cancer recurrence and outcome patterns between patients treated from 1986 to 1992 and from 2004 to 2008. J Clin Oncol. 2014;33:65–73. doi: 10.1200/JCO.2014.57.2461 [DOI] [PubMed] [Google Scholar]
- 11.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25, 676, 887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donepudi MS, Kondapalli K. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506–511. doi: 10.4103/0973-1482.137927 [DOI] [PubMed] [Google Scholar]
- 13.Harris LN, Ismaila N. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2016;34:1134–1150. doi: 10.1200/JCO.2015.65.2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryce CJ. Tamoxifen in early breast cancer. Lancet. 1998;352(9125):403. doi: 10.1016/S0140-6736(05)60500-4 [DOI] [PubMed] [Google Scholar]
- 15.Abe O, Abe R, Enomoto K, et al. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- 16.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21(2):177–184. doi: 10.1016/j.ceb.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 17.Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263–274. doi: 10.1080/08977190802312844 [DOI] [PubMed] [Google Scholar]
- 18.Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 2006;118(5):1173–1180. doi: 10.1002/ijc.21454 [DOI] [PubMed] [Google Scholar]
- 19.Levva S, Kotoula V, Kostopoulos I, et al. Prognostic evaluation of epidermal growth factor receptor (EGFR) genotype and phenotype parameters in triple-negative breast cancers. Cancer Genom Proteom. 2017;14:181–195. doi: 10.21873/cgp.20030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashmi AA, Naz S, Hashmi SK. Epidermal growth factor receptor (EGFR) overexpression in triple-negative breast cancer: association with clinicopathologic features and prognostic parameters. Surg Exp Pathol. 2019;2(1):6. doi: 10.1186/s42047-018-0029-0 [DOI] [Google Scholar]
- 21.Masuda H, Zhang D, Bartholomeusz C, Hiroyoshi Doihara GN, Naoto T, Ueno NT. Ueno. 2012. “Role of epidermal growth factor receptor in breast cancer.”. Breast Cancer Res Treat. 2012;136(2):331–345. doi: 10.1007/s10549-012-2289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopoulos PF, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer. 2015;14(1):43. doi: 10.1186/s12943-015-0291-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obr AE, Kumar S, Chang Y-J, et al. Insulin-like growth factor receptor signaling in breast tumor epithelium protects cells from endoplasmic reticulum stress and regulates the tumor microenvironment. Breast Cancer Res. 2018;20:138. doi: 10.1186/s13058-018-1063-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franks SE, Campbell CI, Barnett EF, et al. Transgenic IGF-IR overexpression induces mammary tumors with basal-like characteristics, whereas IGF-IR-independent mammary tumors express a claudin-low gene signature. Oncogene. 2012;31(27):3298–3309. doi: 10.1038/onc.2011.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farabaugh SM, Boone DN, Lee AV. Role of IGF1R in breast cancer subtypes, stemness, and lineage differentiation. Front Endocrinol. 2015;6:59. doi: 10.3389/fendo.2015.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira CBL, Leal MF, Abdelhay ESFW, et al. MYC amplification as a predictive factor of complete pathologic response to docetaxel-based neoadjuvant chemotherapy for breast cancer. Clin Breast Cancer. 2017;17:188–194. doi: 10.1016/j.clbc.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 27.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumour marker prognostic studies (REMARK). Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leal MF, Ribeiro HF, Rey JA. YWHAE silencing induces cell proliferation, invasion and migration through the up regulation of CDC25B and MYC in gastric cancer cells: new insights about YWHAE role in the tumor development and metastasis process. Oncotarget. 2016;7:85393–85410. doi: 10.18632/oncotarget.13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, Koo JS, Kim MS, Park HS, Lee JS, Lee JS. Characteristics and outcomes according to molecular subtypes of breast cancer as classified by a panel of four biomarkers using immunohistochemistry. The Breast. 2012;21:50–57. doi: 10.1016/j.breast.2011.07.008 [DOI] [PubMed] [Google Scholar]
- 30.Calcagno DQ, Leal MF, Seabra AD, et al. Interrelationship between chromosome 8 aneuploidy, C-MYC amplification and increased expression in individuals from northern Brazil with gastric adenocarcinoma. World J Gastroenterol. 2006;12:6207–6211. doi: 10.3748/wjg [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 32.Wallace R. 22nd Brazilian Diabetes Society Congress. Diabetol Metab Syndr. 2019;11(S1). doi: 10.1186/s13098-019-0473-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De-la-cruz-ku G, Luyo M, Morante Z, et al. “Triple-negative breast cancer in Peru: 2000 patients and 15 years of experience.”. PLoS One. 2020;15(8):e0237811. doi: 10.1371/journal.pone.0237811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson GL, Manson J, Wallace R, et al. Prentice. 2003. “Implementation of the women’s health initiative study design.”. Ann Epidemiol. 2003;13(9):S5–17. doi: 10.1016/s1047-2797(03)00043-7 [DOI] [PubMed] [Google Scholar]
- 35.Witsch E, Sela M, Yarden Y. Roles for growth factors in cancer progression. Physiology. 2010;25(2):85–101. doi: 10.1152/physiol.00045.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis NM, Sokolosky M, Stadelman K, et al. Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget. 2014;5:4603–4650. doi: 10.18632/oncotarget.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carotenuto P, Roma C, Rachiglio AM, Botti G, D’Alessio A, Normanno N. Triple negative breast cancer: from molecular portrait to therapeutic intervention. Crit Rev Eukaryot Gene Expr. 2010;20:17–34. doi: 10.1615/CritRevEukarGeneExpr.v20 [DOI] [PubMed] [Google Scholar]
- 38.Aysola K, Akshata D, Welch K, et al. Triple negative breast cancer – an overview. Hered Genet. 2013;1(supl 2). doi: 10.4172/2161-1041.s2-001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakajima H, Ishikawa Y, Furuya M, Sano T. Protein expression, gene amplification, and mutational analysis of EGFR in triple-negative breast cancer. Breast Cancer. 2014;21:66–74. doi: 10.1007/s12282-012-0354-1 [DOI] [PubMed] [Google Scholar]
- 40.Grob TJ, Heilenkötter U, Geist S, et al. Rare oncogenic mutations of predictive markers for targeted therapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012;134(2):561–567. doi: 10.1007/s10549-012-2092-7 [DOI] [PubMed] [Google Scholar]
- 41.Secq V, Villeret J, Fina F, et al. Triple negative breast carcinoma EGFR amplification is not associated with EGFR, Kras or ALK mutations. Br J Cancer. 2014;110:1045–1052. doi: 10.1038/bjc.2013.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacot W, Lopez-Crapez E, Thezenas S, Senai R, Fina F, Bibeau F. Lack of EGFR-activating mutations in European patients with triple-negative breast cancer could emphasise geographic and ethnic variations in breast cancer mutation profiles. Breast Cancer Res. 2011;13:R133. doi: 10.1186/bcr3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tilch E, Seidens T, Cocciardi S, Reide LE, Byrne D, Simpson PT. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast Cancer Res Treat. 2014;143:385–392. doi: 10.1007/s10549-013-2798-1 [DOI] [PubMed] [Google Scholar]
- 44.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 45.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sørlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng Y, Tang C-H, Wang Y, et al. Combined High Resistin and EGFR Expression Predicts a Poor Prognosis in Breast Cancer. Biomed Res Int. 2020;2020(November):8835398. doi: 10.1155/2020/8835398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C-Q, Yang L, Huang B-F, et al. EGFR Conjunct FSCN1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Sci Rep. 2017;7(1):15654. doi: 10.1038/s41598-017-15939-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennisi PA, Barr V, Nunez NP, Stannard B, Le Roith D. Reduced expression of insulin-like growth factor I receptors in MCF-7 breast cancer cells leads to a more metastatic phenotype. Cancer Res. 2002;62(22):6529–6537. [PubMed] [Google Scholar]
- 50.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Wanda A. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 52.Salvati A, Gigantino V, Nassa G, et al. Global view of candidate therapeutic target genes in hormone-responsive breast cancer. Int J Mol Sci. 2020;21:4068. doi: 10.3390/ijms21114068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.AlFakeeh A, Brezden-Masley C. Overcoming endocrine resistance in hormone receptor–positive breast cancer. Curr Oncol. 2018;25:18. doi: 10.3747/co.25.3752 [DOI] [PMC free article] [PubMed] [Google Scholar]