Abstract
Transfer RNAs (tRNAs) mainly function as adapter molecules that decode messenger RNAs (mRNAs) during protein translation by delivering amino acids to the ribosome. Traditionally, tRNAs are considered as housekeepers without additional functions. Nevertheless, it has become apparent from biological research that tRNAs are involved in various physiological and pathological processes. Aging is a form of gradual decline in physiological function that ultimately leads to increased vulnerability to multiple chronic diseases and death. Interestingly, tRNA metabolism is closely associated with aging and lifespan. In this review, we summarize the emerging roles of tRNA-associated metabolism, such as tRNA transcription, tRNA molecules, tRNA modifications, tRNA aminoacylation, and tRNA derivatives, in aging and lifespan, aiming to provide new ideas for developing therapeutics and ultimately extending lifespan in humans.
Subject terms: Ageing, Genetics research
Facts
tRNAs are important participants in protein translation and are involved in various physiological and pathological processes.
tRNA-associated metabolism is closely associated with aging and lifespan.
The enzymes related to tRNA metabolism could be potential targets for future therapeutic interventions in aging and lifespan.
Open questions
Is tRNA metabolism involved in the regulation of aging and lifespan mainly by affecting protein synthesis?
What is the molecular mechanism by which tRNA derivatives regulate aging and lifespan?
Is there potential for practical clinical applications based on findings concerning tRNAs in the context of aging and lifespan?
Introduction
Transfer RNAs (tRNAs) are important participants in protein translation, which transport their cognate amino acids to the ribosome. There are more than 600 putative tRNA genes in the human genome, some of which are transcribed into precursor tRNAs (pre-tRNAs) by RNA polymerase III (Pol III)1. Subsequently, pre-tRNAs are transformed into mature tRNAs after a series of processing and modification processes, which are characterized by a “clover” secondary structure as well as an L-shaped tertiary structure2. After maturation, tRNAs are charged with their cognate amino acids through the aminoacylation reactions mediated by aminoacyl-tRNA synthetases (ARSs), thereby participating in protein translation3. Of note, tRNAs will be cleaved into fragments with regulatory functions under stress conditions4–6. In general, normal tRNA metabolism is essential to maintain the stability and functions of tRNA molecules, while the defects in certain tRNA biogenesis proteins cause various human diseases, including cancer, neurological disorders, immunodeficiency, and diabetes mellitus7–10.
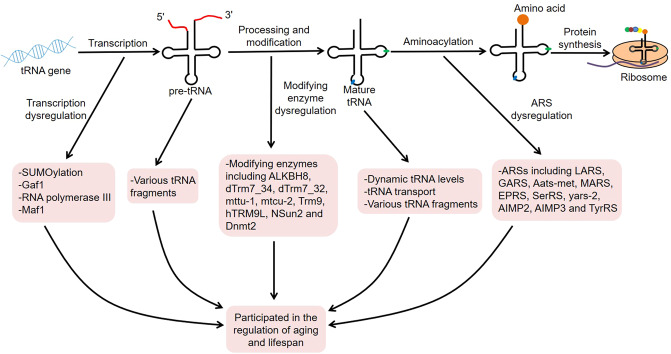
Aging is a complex physiological process, usually manifested by a gradual decline in organ function, as well as an increase in disease incidence and risk of death. It is reported that the global population over 65 will reach 1.6 billion by 205011. In fact, delaying biological aging or extending healthspan is an eternal theme of human health12,13. Strikingly, tRNAs play an important role in aging and lifespan. For example, the serum levels of mitochondrial tRNAs and ribosomal RNAs (rRNAs) will increase with age, which may be related to mitochondrial dysfunction during the aging process14. Another research discovered that Pol III was a downstream molecule of Target of Rapamycin Complex 1 (TORC1), and its inhibition could extend organismal lifespan15. Moreover, the deletion of nucleoporin Nup100 could regulate the life span of Saccharomyces cerevisiae by inhibiting the nuclear export of specific tRNAs16. Therefore, tRNAs are closely related to aging biology and thus participate in the regulation of age-related diseases and lifespan. Here, we focus on the roles of tRNA-associated metabolism, such as tRNA transcription, tRNA molecules, tRNA modifications, tRNA aminoacylation, and tRNA derivatives, in aging and lifespan, which may serve as novel targets for lifespan extension (Fig. 1).
Fig. 1. Roles of tRNA-associated metabolism in aging and lifespan.
The tRNA genes are transcribed into pre-tRNAs, which are then transformed into mature tRNAs after a series of processing and modifications. Subsequently, mature tRNAs are involved in protein translation. During this process, tRNA-associated metabolism, such as tRNA transcription, tRNA molecules, tRNA modifications, tRNA aminoacylation, and tRNA derivatives, plays an important role in aging and lifespan.
Roles of tRNA metabolism in aging and lifespan
tRNA transcription in aging and lifespan
Small ubiquitin-related modifier modification (SUMOylation) is a highly dynamic post-translational modification that has been confirmed to be related to transcriptional repression17,18. Meanwhile, many studies have linked SUMOylation to aging process19,20. In eukaryotes, three essential RNA polymerases are evolutionarily conserved enzymes responsible for the transcription of their nuclear genomes. Of these, Pol I mainly transcribes the 25S rRNA precursor, Pol II transcribes various messenger RNAs (mRNAs), while Pol III transcribes short RNAs such as tRNAs and 5S rRNA21,22. Interestingly, SUMO machinery was widely distributed in the genome, especially at the promoters of histone and protein biogenesis genes, as well as Pol I-transcribed rRNA genes and Pol III-transcribed tRNA genes23. Surprisingly, the SUMO machinery was selectively retained on histone and tRNA genes and released in large quantities from other chromatin in senescent cells, indicating that maintaining the suppression of histone and tRNA loci was beneficial to the stability of the aging state23. These data support that SUMOylation-mediated coordinated repression of a transcriptional program is associated with cell growth and proliferation.
It is well-known that TORC1 is an important longevity determinant among many species24–26. Recent studies observed that the GATA transcription factor Gaf1 deficiency could shorten the normal chronological lifespan and reduce the lifespan extension caused by TORC1 inhibition in yeast27. Specifically, upon TORC1 block, Gaf1 served as a transcription factor downstream of TORC1 that directly bound to Pol III-transcribed tRNA genes and inhibited their transcription, thereby promoting longevity by inhibiting translation. Strikingly, Pol III mediated the longevity-promoting effects of TORC1 inhibition15. In this condition, systemic Pol III deficiency could facilitate the longevity in yeast, flies, and worms, and gut-specific inhibition of Pol III in adult worms or flies was sufficient to prolong the lifespan, which might be related to the reduced protein synthesis and increased resistance to proteotoxic stress. Importantly, the effects of Pol III inhibition and rapamycin treatment on lifespan extension were not additive15. Rapamycin treatment suppressed the phosphorylation of TORC1 substrate in the intestine, while the gut-specific Pol III inhibition did not, indicating that Pol III acted as a downstream molecule of TORC1 to regulate lifespan.
TORC1 directly phosphorylated MAF1 homolog negative regulator of Poll III (Maf1) at multiple sites and thus controlled its localization and Pol III-mediated transcription28–30. Notably, inhibition of TORC1 by rapamycin treatment reduced pre-tRNA levels in whole flies, and overexpression of gut-specific Maf1 reduced pre-tRNA levels and prolonged lifespan, indicating that Maf1-mediated Pol III inhibition might be involved in the regulation of lifespan by mTOR pathway15. Moreover, maf1Δ cells showed a shorter lifespan under lower glucose conditions, and the short lifespan was rescued by introducing the plasmid encoding maf1 gene, which suggested that Maf1 was required for lifespan extension of Schizosaccharomyces pombe31. Maf1 was phosphorylated by TORC1 under high-calorie conditions, while it was dephosphorylated by PP2A and PP4 under calorie-restricted conditions. The phosphorylation status of Maf1 was associated with S. pombe lifespan and tRNA levels. Importantly, Maf1-dependent inhibition of tRNA transcription extended lifespan in fission yeast mainly by preventing genomic instability at tRNA genes, rather than inhibiting protein synthesis31. Further studies discovered that the break of tRNA genes was caused by replication-transcription conflicts, while Maf1 could limit Pol III-mediated transcription to maintain genomic integrity32. These findings indicate that transcription-related genomic instability may play an important role in the aging process. Intriguingly, Maf1 deletion increased the lifespan in worms and mice, which also indicated that Maf1 participated in lifespan regulation through complex mechanisms, not just by regulating Pol III output33,34.
tRNA molecules in aging and lifespan
In addition, tRNA molecules are also involved in the regulation of aging and lifespan. Sagi et al.35 demonstrated that tRNA expression decreased with age in worms, and the higher sup-7 tRNA levels at day 6 were associated with a longer lifespan. The decline in tRNA expression might cause protein misfolding, leading to the development of age-related diseases. Moreover, nuclear tRNA accumulation was related to the increased replicative lifespan in yeast36. In this context, deletion of tRNA exporter Los1 could significantly extend lifespan. Mechanistically, dietary restriction excluded Los1 from the nucleus in a manner dependent on Rad53 and mTOR, thereby promoting nuclear tRNA accumulation and transcription factor Gcn4 activation. Analogously, deletion of Nup100 facilitated the expression of Gcn4 by suppressing the nuclear export of tRNAs and thus contributed to the increased longevity in S. cerevisiae16. nup100Δ cells did not show tRNA splicing and aminoacylation defects, indicating that Nup100 was mainly responsible for the re-export of several mature tRNAs, such as tRNATyr, tRNATrp, and tRNAIle. Of note, the localization of Los1 and Msn5 (another protein involved in tRNA export) was not regulated by Nup100, which supported that Nup100 could regulate tRNA export in a manner distinct from them16. Together, the dysregulation of tRNA levels and transport may affect the lifespan of organisms.
tRNA modifications in aging and lifespan
tRNAs always undergo a variety of post-transcriptional modifications, which affect tRNA stability, codon recognition, and even aminoacylation37. Strikingly, many studies have demonstrated that certain tRNA-modifying enzymes are involved in the regulation of cellular senescence and lifespan (Table 1). Alkylation repair homolog 8 (ALKBH8) is a tRNA methyltransferase involved in the formation of 5-methoxycarbonylmethyluridine (mcm5U), 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), and 5-methoxycarbonylhydroxymethyluridine (mchm5U) at the anticodon wobble position of tRNAs38–40. ALKBH8-deficient mouse embryonic fibroblasts showed selenoprotein loss as well as a senescence phenotype characterized by increased levels of senescence-associated β-galactosidase (SA-β-Gal), heterochromatin foci, p16INK4a, and senescence associated secretory phenotype markers41. Another research found that dTrm7_34 and dTrm7_32, as functional orthologs of yeast TRM7 and human FtsJ RNA 2′-O-methyltransferase 1 (FTSJ1), catalyzed 2′-O-Methylation (Nm) at specific tRNAs in Drosophila42. Interestingly, Nm at position G34 limited the cleavage of tRNAPhe, while the 3′ terminal Cm32 might stabilize the tRNAPhe fragments that were produced in dTrm7_34 mutants. Meanwhile, the mutant animals of dTrm7_34 and dTrm7_32 exhibited small RNA pathway dysfunctions, increased susceptibility to RNA virus infection, and shortened lifespan, suggesting that these two methyltransferases appeared to modulate the small RNA silencing and lifespan in adult flies42.
Table 1.
Roles of tRNA-modifying enzymes in aging and lifespan.
| tRNA-modifying enzymes | Subjects | Function | Effects | Mechanisms | References |
|---|---|---|---|---|---|
| ALKBH8 | MEFs | Anti-aging | ALKBH8-deficient MEFs showed a senescence phenotype | Characterized by increased levels of SA-β-Gal, HCF, p16INK4a, and SASP markers | 41 |
| dTrm7_34 and dTrm7_32 | Drosophila | Anti-aging | dTrm7_34 and dTrm7_32 mutant flies displayed reduced lifespan | – | 42 |
| mttu-1 and mtcu-2 | C. elegans | Pro-aging | The lifespan of mtcu-2 and mttu-1 double mutants was significantly extended | Associated with OXPHOS dysfunction | 44 |
| Trm9 | Yeast | Pro-aging | Trm9 deletion extended lifespan | Enhanced heat-shock resistance of mutants | 45 |
| hTRM9L | Cancer tissues | Pro-aging | Inhibited tumor growth via a senescence-like arrest | Related to SA-β-Gal activity and p21 expression | 46 |
| NSun2 | Human fibroblasts | Anti-aging | Delayed the process of replicative senescence | Inhibited the translation of p27 by methylating p27 mRNA | 48 |
| NSun2 | Human vascular endothelial cells | Pro-aging | Facilitated premature senescence induced by oxidative stress or high glucose | Promoted the translation of Shc adapter proteins by methylating Shc mRNA, thus activating p38MAPK and increasing ROS production | 49 |
| Dnmt2 | Drosophila | Anti-aging | Dnmt2 was indispensable for maintaining the normal lifespan, and its overexpression prolonged lifespan | – | 52 |
| Dnmt2 | Mouse fibroblasts | Anti-aging | Dnmt2 knockdown fibroblasts were susceptible to senescence under control conditions | Manifested by increased levels of p53 and p21, telomere shortening, oxidative stress and DNA damage | 53 |
| Dnmt2 | Human fibroblasts | Anti-aging | Dnmt2 silencing induced cellular senescence | – | 54 |
tRNA transfer RNA, ALKBH8 alkylation repair homolog 8, MEFs mouse embryonic fibroblasts, SA-β-Gal senescence-associated β-galactosidase, HCF heterochromatin foci, SASP senescence-associated secretory phenotype, OXPHOS oxidative phosphorylation, Trm9 tRNA methyltransferase 9, hTRM9L human Trm9-like protein, NSun2 NOP2/Sun domain family member 2, Shc Src homology and collagen, p38MAPK p38 mitogen-activated protein kinase, ROS reactive oxygen species, Dnmt2 DNA methyltransferase-2.
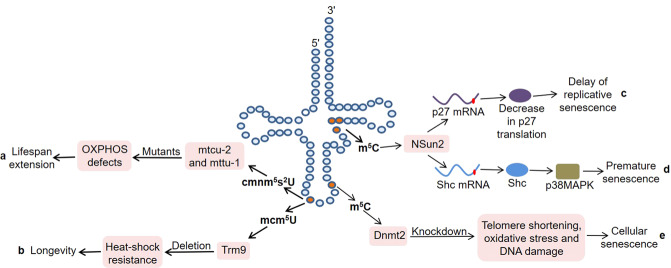
In mammals, mitochondrial translation optimization factor 1 (MTO1) catalyzed the formation of τm5U at anticodon position 34 in certain mitochondrial tRNAs (mt-tRNAs)43. The loss of MTO1 affected translation fidelity through defective tRNA modification in mice, resulting in tissue-specific oxidative phosphorylation (OXPHOS) defects. mttu-1 and mtcu-2 in Caenorhabditis elegans were the homologs of tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (TRMU) and MTO144 (Fig. 2a). Notably, the lifespan of mttu-1 mutants was slightly extended at 20 °C, and that of mtcu-2 and mttu-1 double mutants was significantly extended, which was associated with the OXPHOS dysfunction in C. elegans. These findings indicate that these two modifying enzymes may be synergistic in regulating the lifespan of worms. But the underlying molecular mechanism requires further research. Fabrizio et al.45 discovered that the deletion of acyl-CoA binding protein (Acb1), tRNA methyltransferase 9 (Trm9) and CKA2 could significantly extend lifespan by performing a screen of a yeast homozygous deletion collection. Among them, Trm9 was responsible for the formation of 5-methoxycarbonyl-methyluridine (mcm5U) at position 34 in tRNAGlu and tRNAArg3. Importantly, their deletion enhanced the heat-shock resistance of mutants, thereby supporting the link between longevity and cellular protection (Fig. 2b). Furthermore, human Trm9-like protein (hTRM9L) was down-regulated in a variety of cancer tissues, and its re-expression significantly inhibited tumor growth in vivo46. hTRM9L induced a senescence-like phenotype related to SA-β-Gal activity and p21 expression. Meanwhile, hTRM9L could upregulate LIN9 and inhibit the hypoxic response, thereby exerting antitumor activity.
Fig. 2. The dysregulation of tRNA-modifying enzymes in aging and lifespan.
a The mtcu-2 and mttu-1 mutants show OXPHOS dysfunction, resulting in lifespan extension. b Trm9 deletion enhances heat-shock resistance of mutants, thereby supporting the link between longevity and cellular protection. c NSun2 inhibits the translation of p27 by methylating p27 mRNA, thereby delaying the process of replicative senescence. d NSun2 promotes the translation of Shc adapter proteins by methylating Shc mRNA. Subsequently, the increased Shc proteins activate p38MAPK, thereby facilitating premature senescence. e Dnmt2 knockdown fibroblasts are susceptible to senescence under control conditions, manifested by telomere shortening, oxidative stress, and DNA damage.
Mammalian NOP2/Sun domain family member 2 (NSun2) is responsible for the cytosine-5 methylation (m5C) in specific tRNA molecules, such as tRNAGly, tRNAAsp, and tRNAVal47. Interestingly, NSun2 inhibited the translation of p27 by methylating the 5′-untranslated region (UTR) of p27 mRNA, thereby delaying the process of replicative senescence48 (Fig. 2c). At the same time, overexpression of the p27 5´UTR fragment could rescue the decrease of p27 and the increase of cyclin-dependent kinase 1 caused by NSun2 overexpression in 2BS cells, indicating that NSun2-mediated mRNA methylation played an important role in replicative senescence48. Cai et al.49 demonstrated that NSun2 promoted the translation of Src homology and collagen (Shc) adapter proteins, p66SHC, p52SHC, and p46SHC, by methylating Shc mRNA at multiple sites (Fig. 2d). Subsequently, the increased Shc proteins activated p38 mitogen-activated protein kinase (p38MAPK) and increased cellular reactive oxygen species production, thereby facilitating the premature senescence of human vascular endothelial cells induced by oxidative stress or high glucose.
In addition, DNA methyltransferase-2 (Dnmt2) specifically methylated cytosine 38 in the anticodon loops of tRNAs50,51. It has been confirmed that Dnmt2 is indispensable for maintaining the normal lifespan of Drosophila, and its overexpression can prolong lifespan52. Moreover, Dnmt2 was also related to the condition-dependent apoptosis and senescence in mouse fibroblasts53. On the one hand, Dnmt2 knockdown fibroblasts were more prone to apoptosis under the stimulation of hydrogen peroxide. On the other hand, these cells were more susceptible to senescence under control conditions, manifested by increased levels of p53 and p21, telomere shortening, oxidative stress, and DNA damage (Fig. 2e). Consistently, Dnmt2 silencing inhibited the proliferation of human fibroblasts and induced cellular senescence54. These findings indicate that Dnmt2 may serve as a novel regulator of longevity.
tRNA aminoacylation in aging and lifespan
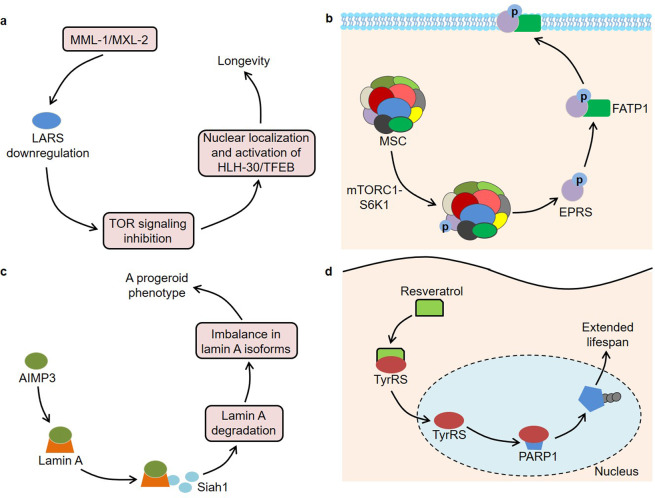
It is well known that tRNAs bind to their homologous amino acids through ARS-mediated aminoacylation, thereby transporting amino acids to the ribosome to participate in protein synthesis. In mammalian cells, one part of ARSs exists in free form, while the other part interacts with three ARS-interacting multi-functional proteins (AIMPs) to form a multi-tRNA synthetase complex (MSC)55. Intriguingly, ARSs and AIMPs are closely associated with aging and lifespan (Table 2). Previous studies found that a null mutation in mitochondrial leucyl-tRNA synthetase 2 (LARS2) was associated with longevity by screening 5690 genes of C. elegans56. The long-lived worms had impaired mitochondrial functions, manifested by lower ATP content and oxygen consumption. Furthermore, Mondo/Max-like complex (MML-1/MXL-2) played an important role in the lifespan extension induced by germline removal57 (Fig. 3a). In this context, MML-1/MXL-2 inhibited TOR activity by downregulating LARS, leading to the nuclear localization and activation of HLH-30/TFEB. Another research observed that prolyl-hydroxylase domain protein 1 (PHD1) levels were reduced in aging muscles, and PHD1 knockout mice had lower muscle mass58. PHD1 increased the stability of LARS by interacting with it, thereby ensuring leucine-mediated mTORC1 activation and maintaining muscle mass. These findings indicate that LARS participates in the biology of aging through different signaling pathways. Niehues et al. built a Drosophila model for Charcot–Marie–Tooth neuropathy by three mutations in glycyl-tRNA synthetase (GARS)59. Of note, the expression of these mutants, including GARS_E71G, GARS_G240R, and GARS_G526R, not only induced defects in neuronal morphology but also shortened the lifespan of flies in a dosage-dependent manner. In-depth research found that the mutant GARS proteins showed normal subcellular localization, but the overall protein synthesis in neurons was significantly reduced59. Interestingly, the heterozygous GARSC201R mice had a normal lifespan, while this mutation significantly rescued the shortened lifespan caused by the SOD1G93A mutation60. Therefore, more studies are still needed to explore the roles of GARS in lifespan regulation.
Table 2.
Roles of ARSs in aging and lifespan.
| ARSs | Subjects | Function | Effects | Mechanisms | References |
|---|---|---|---|---|---|
| LARS2 | C. elegans | Pro-aging | A null mutation in LARS2 was associated with longevity | Associated with impaired mitochondrial functions, manifested by lower ATP content and oxygen consumption | 56 |
| LARS | C. elegans | Pro-aging | MML-1/MXL-2 promoted longevity | MML-1/MXL-2 inhibited TOR activity by down-regulating LARS, leading to the nuclear localization and activation of HLH-30/TFEB | 57 |
| LARS | Mouse | Anti-aging | PHD1 levels were reduced in aging muscles, and PHD1 knockout mice had lower muscle mass | PHD1 increased the stability of LARS, thereby ensuring leucine-mediated mTORC1 activation | 58 |
| GARS | Drosophila | Anti-aging | GARS mutations shortened the lifespan of flies in a dosage-dependent manner | Associated with impaired protein synthesis | 59 |
| Aats-met | Drosophila | Anti-aging | Aats-met mutations caused photoreceptor degeneration and reduced lifespan | Associated with increased ROS, oxidative phosphorylation defects and upregulation of mitochondrial unfolded protein response | 61 |
| MARS | Drosophila | Anti-aging | MARS inhibition shortened the lifespan of flies | Reduced the expression of AMPs genes | 62 |
| EPRS | Mouse | Pro-aging | Homozygous EPRS S999A mice exhibited low body weight, reduced adipose tissue mass and increased lifespan | mTORC1-S6K1 phosphorylated EPRS and induced its release from the MSC. Then, the EPRS bound to FATP1 to promote its translocation to the plasma membrane | 63 |
| SerRS | Cancer cells | Pro-aging | Induced cellular senescence | SerRS bound to telomere DNA repeats and enriched POT1 proteins to telomeres, leading to the shortening of telomeres | 64 |
| yars-2 | C. elegans | Pro-aging | NMD-mediated RNA quality control was crucial for longevity | The down-regulation of yars-2, an NMD target, extended the lifespan of mutants | 65 |
| AIMP2 | Mouse | Pro-aging | Contributed to the development of PD | Overexpression of AIMP2 activated PARP1, thereby resulting in an age-dependent dopaminergic neuronal loss | 67 |
| AIMP2-DX2 | Cancer cells | Anti-aging | Blocked oncogene-induced apoptosis and senescence | Inhibited p14/ARF | 68 |
| AIMP3 | Mouse | Pro-aging | AIMP3 levels were increased in aging human tissues, and AIMP3 transgenic mice had a premature aging phenotype | AIMP3 interacted with lamin A and recruited Siah1, which led to the degradation of lamin A and an imbalance in its isoform levels | 69 |
| AIMP3 | hMSCs | Pro-aging | AIMP3 levels were increased, while the levels of miR-543 and miR-590-3p were decreased during the senescence of hMSCs | The two miRNAs inhibited the expression of AIMP3 by binding to AIMP3 transcripts | 70 |
| TyrRS | Mouse | Anti-aging | Mediated the lifespan extension regulated by resveratrol | Resveratrol bound to TyrRS and facilitated its nuclear translocation. Then, TyrRS interacted with PARP1 and promoted its activation | 94 |
| AIMP3 | Mesenchymal stem cells | Pro-aging | AIMP3 down-regulation improved the age-related senescence of stem cells | HIF1α activated autophagy and inhibited mitochondrial respiration via suppressing the expression of AIMP3 | 95 |
ARSs aminoacyl-tRNA synthetases, LARS2 leucyl-tRNA synthetase 2, MML-1/MXL-2 Mondo/Max-like complex, TOR target of rapamycin, PHD1 prolyl-hydroxylase domain protein 1, GARS glycyl-tRNA synthetase, ROS reactive oxygen species, MARS methionyl-tRNA synthetase, AMPs anti-microbial peptides, EPRS glutamyl-prolyl-tRNA synthetase, S6K1 S6 kinase 1, MSC multi-tRNA synthetase complex, FATP1 fatty acid transport protein 1, SerRS seryl-tRNA synthetase, POT1 Protection of Telomeres 1, NMD nonsense-mediated mRNA decay, AIMP2 ARS-interacting multi-functional protein 2, PD Parkinson’s disease, PARP1 poly(ADP-ribose) polymerase-1, AIMP2-DX2 AIMP2 lacking exon 2, Siah1 seven in absentia homolog 1, hMSCs human mesenchymal stem cells, TyrRS tyrosyl-tRNA synthetase, HIF1α hypoxia-inducible factor 1α.
Fig. 3. The dysregulation of ARSs in aging and lifespan.
a MML-1/MXL-2 inhibits TOR activity by down-regulating LARS, leading to the nuclear localization and activation of HLH-30/TFEB. b mTORC1-S6K1 phosphorylates EPRS and thus induces its release from the MSC. The phosphorylated EPRS binds to FATP1 to promote its translocation to the plasma membrane. c AIMP3 interacts with lamin A and recruits Siah1, which leads to the degradation of lamin A as well as an imbalance in its isoform levels. d Resveratrol binds to TyrRS and facilitates its nuclear translocation. Then, TyrRS interacts with PARP1 and promotes its activation by stimulating NAD+-dependent auto-poly-ADP-ribosylation.
The mutations in Aats-met, a homolog of human methionyl-tRNA synthetase 2 (MARS2), caused photoreceptor degeneration and reduced lifespan of flies, which was associated with the increased ROS, oxidative phosphorylation defects and upregulation of mitochondrial unfolded protein response61. Moreover, inhibition of MARS could shorten the lifespan of flies by reducing the expression of anti-microbial peptides genes62. Arif et al. discovered that glutamyl-prolyl-tRNA synthetase (EPRS) was a downstream effector of the mTORC1 and p70 ribosomal protein S6 kinase 1 (S6K1) axis, which was involved in the biological processes of obesity and aging63 (Fig. 3b). In terms of mechanism, mTORC1-S6K1 phosphorylated EPRS at Ser999 and thus induced its release from the MSC. Subsequently, the phosphorylated EPRS bound to fatty acid transport protein 1 (FATP1) to promote its translocation to the plasma membrane and long-chain fatty acid uptake. Consistently, homozygous phospho-deficient EPRS S999A mice showed reduced adipose tissue, weight loss and longer lifespan, while replacement of the phospho-mimetic S999D allele restored the weight of s6k1-deficient mice to normal63. Furthermore, seryl-tRNA synthetase (SerRS) not only bound to telomere DNA repeats but also enriched Protection of Telomeres 1 (POT1) proteins to telomeres via direct interaction with POT1 in the nucleus64. The enrichment of POT1 led to the shortening of telomeres, thereby inhibiting the growth of HeLa cells by inducing cellular senescence. Intriguingly, the activity of nonsense-mediated mRNA decay (NMD) decreased with the age of C. elegans, and NMD could contribute to the longevity in daf-2 mutant worms65. Further research has shown that downregulation of yars-2/tyrosyl-tRNA synthetase 2 (TyrRS2), an NMD target, effectively extends the lifespan of mutants, indicating that NMD-mediated RNA quality control plays an important role in organismal aging65.
Moreover, AIMPs, which mainly act as scaffolds in the MSC, is also associated with the aging process. It was reported that AIMP2 was a parkin substrate and contributed to the development of Parkinson’s disease (PD)66. Overexpression of AIMP2 could activate poly(ADP-ribose) polymerase-1 (PARP1), thereby resulting in an age-dependent dopaminergic neuronal loss in mice67. The PARP1 inhibitor AG014699 inhibited the degeneration of dopaminergic neurons in AIMP2 transgenic mice, indicating that PARP1 could be used as a target for PD treatment67. Notably, AIMP2-DX2, a splicing variant of AIMP2 lacking exon 2, was induced by oncogenes in human lung cancer cells and could block oncogene-induced apoptosis and senescence by inhibiting p14/ARF68. In addition, endogenous AIMP3 levels increased in aging human tissues, and AIMP3 transgenic mice had an obvious premature aging phenotype, which was manifested as earlier cessation of weight gain, hair loss, reduced bone mineral deposits in female and bone thickness, lordokyphosis, as well as wrinkled skin with reduced adipocytes69 (Fig. 3c). Mechanistically, AIMP3 interacted with lamin A and recruited seven in absentia homolog 1 (Siah1), which led to the degradation of lamin A. The lamin A degradation would result in an imbalance in its isoform levels, thus inducing organismal aging. Analogously, Lee et al.70 demonstrated that AIMP3 levels were increased, while the levels of miR-543 and miR-590-3p were decreased during the senescence of human mesenchymal stem cells. These two microRNAs (miRNAs) could inhibit the expression of AIMP3 by binding to AIMP3 transcripts, thereby delaying cellular aging.
tRNA derivatives in aging and lifespan
Particularly, pre-tRNAs or mature tRNAs are cleaved into diverse subtypes of fragments under stress conditions, which are named tRNA derivatives71. Victoria et al. discovered that the circulating levels of 5′ tRNA halves derived from tRNACys(GCA) and tRNALys(CTT) were decreased, and those derived from tRNAHis(GTG) and tRNAAsp(GTC) were increased with age in normal mice72. Importantly, the alterations in the levels of these 5′ tRNA halves were mitigated in the long-lived Ames dwarf mice. Likewise, another study found that the serum levels of certain specific 5′ tRNA halves changed significantly with age in mice, and their levels could be regulated by calorie restriction73. These results suggest that tRNA fragments may play vital roles in the anti-aging effects of dwarfism or calorie restriction. In some cases, RNA molecules harboring a 2′,3′-cyclic phosphate (cP-RNAs) at the 3′ end are generated from endoribonuclease-mediated RNA cleavage74. It was worth noting that cP-RNAs, mainly from tRNAs, rRNAs, and mRNAs, were abundantly present in mouse tissues, and their levels declined in an age-dependent manner75. Among them, the cP-RNAs derived from tRNAs were produced from the cleavage of anticodon loops and 3′-terminal CCA sequences. However, more studies are needed to explore the roles of tRNA derivatives in aging.
Of note, tRNA derivatives have been confirmed to be related to some age-related pathological processes, especially neurodegenerative diseases. Karaiskos et al.76 observed that the abundance of tRNA fragments in rat brains changed dynamically under the background of age. On the one hand, the levels of tRNA fragments derived from the 3′ end usually increased with age. On the other hand, the levels of tRNA fragments derived from the 5′ end were lower in the brains of middle-aged rats, while their levels were higher in the young and old rats. Interestingly, the potential targets of these fragments appeared to be enriched in neuronal functions and development, indicating that tRNA fragments might be involved in human aging and neurodegeneration76. Similarly, eight tRNA fragments were found to be differentially expressed in the brains of senescence-accelerated mouse prone 8 (SAMP8) mice, and these fragments seemed to regulate the brain function-associated pathways in a miRNA-like pattern77. For example, AS-tDR-011775 could act on myelin-associated oligodendrocyte basic protein or parkin (PARK2), thus contributing to the development of brain aging-associated diseases77. Conspicuously, cleavage and polyadenylation factor I subunit 1 (CLP1) could facilitate the efficient generation of tRNA exons by maintaining the integrity of the tRNA splicing endonuclease complex, and CLP1 kinase-dead mice showed progressive loss of lower motor neurons78. At the mechanistic level, loss of CLP1 activity led to the accumulation of 5′ leader exon tRNA fragments derived from pre-tRNATyr (5′ Tyr-tRF) and p53-mediated cell death. Further research found that the 5′ Tyr-tRF promoted the p53-dependent neuronal cell death by interacting with pyruvate kinase M2 (PKM2)79. In addition, Balaskas et al. discovered that various tRNA and tRNA fragments were differentially expressed between young and old equine chondrocytes80. Importantly, certain 5′ tiRNAs, such as tiRNA His-GTG and tiRNA Glu-TTC, were induced in both old equine chondrocytes and high-grade osteoarthritis cartilage, indicating that tRNA fragments might be involved in the development of age-related cartilage diseases80.
Conclusion and future perspective
Traditionally, tRNAs are considered as housekeeping molecules that mainly transport amino acids to the ribosome to participate in protein translation. After transcription, each tRNA needs to undergo a series of complex maturation processes to become functional81. In their metabolic process, defects in any step may cause various human diseases82–84. For example, the tRNA-modifying enzyme FTSJ1 was down-regulated in non-small cell lung cancer (NSCLC) tissues85. Importantly, FTSJ1 inhibited the growth of NSCLC cells by reducing the expression of DNA damage-regulated autophagy modulator 1. Furthermore, certain ARSs, including asparaginyl-tRNA synthetase, aspartyl-tRNA synthetase 2, and GARS, were associated with the development of neurological disorders86–88. As described above, tRNA-related metabolism, including tRNA transcription, tRNA molecules, tRNA modifications, tRNA aminoacylation, and tRNA derivatives, not only participates in cellular senescence but also plays a vital role in aging and longevity of organisms. In this context, studying tRNAs seems to provide new ideas for lifespan extension. However, the related molecular mechanism research is still in the initial stage, especially in the aspect of tRNA derivatives.
Indeed, some studies have begun to explore clinical transformations based on tRNA metabolism. Mutations in the human mitochondrial DNA (mtDNA) are implicated in age-associated disease phenotypes and aging89,90. Notably, specific mitoTALENs monomers for the tRNAAla m.5024C > T mutation could reduce the mutant mtDNA load and restore the tRNAAla levels in the muscle and heart of a mouse model of heteroplasmic mtDNA mutation91. It was reported that the natural phenol resveratrol contributed to extending the lifespan of organisms92,93. Further research showed that resveratrol could bind to the active site of TyrRS and facilitate its nuclear translocation94 (Fig. 3d). Then, TyrRS interacted with the C-domain of PARP1 and promoted its activation by stimulating NAD+-dependent auto-poly-ADP-ribosylation. Moreover, AIMP3 overexpression inhibited the functions of mesenchymal stem cells under hypoxic conditions, while the down-regulation of AIMP3 significantly improved the age-related senescence of stem cells95. Interestingly, hypoxia-inducible factor 1α (HIF1α) could activate autophagy and inhibit mitochondrial respiration via suppressing the expression of AIMP3, thereby delaying aging95. These findings provided a possible target for the regulation of aging. Another study found that the tRNA-derived fragments from the prefrontal cortex, cerebrospinal fluid and serum were differently expressed between PD patients and control samples, and they could distinguish PD from controls, indicating that tRNA fragments might serve as potential biomarkers for age-associated disease96. In conclusion, tRNA metabolism is closely related to aging and lifespan, and studying their relationship may become a hot topic in the future.
Acknowledgements
The authors wish to acknowledge Chunsheng Zhu from the First Affiliated Hospital of Zhengzhou University, China for editing of English grammar and syntax of the paper.
Author contributions
Z.Z. and B.S. contributed to the drafting and revising of the article; M.B. and D.S.Y. contributed to the conception and design; all authors approved the final version.
Funding
This project was supported by the National Natural Science Foundation of China (81900468).
Ethical approval
Not Applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by B. Zhivotovsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zheng Zhou, Bao Sun
Contributor Information
Dongsheng Yu, Email: dongshyu@163.com.
Meng Bian, Email: bianmeng0208@163.com.
References
- 1.Schaffer AE, Pinkard O, Coller JM. tRNA metabolism and neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 2019;20:359–387. doi: 10.1146/annurev-genom-083118-015334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu C, Sun B, Nie A, Zhou Z. The tRNA-associated dysregulation in immune responses and immune diseases. Acta Physiol. 2020;228:e13391. doi: 10.1111/apha.13391. [DOI] [PubMed] [Google Scholar]
- 3.Kwon NH, Fox PL, Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019;18:629–650. doi: 10.1038/s41573-019-0026-3. [DOI] [PubMed] [Google Scholar]
- 4.Boskovic A, Bing XY, Kaymak E, Rando OJ. Control of noncoding RNA production and histone levels by a 5’ tRNA fragment. Genes Dev. 2020;34:118–131. doi: 10.1101/gad.332783.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mo D, et al. A tRNA fragment, 5’-tiRNAVal, suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019;457:60–73. doi: 10.1016/j.canlet.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Deng J, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosselló-Tortella M, et al. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc. Natl Acad. Sci. USA. 2020;117:20785–20793. doi: 10.1073/pnas.2003358117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blocquel D, et al. CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells. Proc. Natl Acad. Sci. USA. 2019;116:19440–19448. doi: 10.1073/pnas.1908288116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giannelou A, et al. Aberrant tRNA processing causes an autoinflammatory syndrome responsive to TNF inhibitors. Ann. Rheum. Dis. 2018;77:612–619. doi: 10.1136/annrheumdis-2017-212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Sun B, Huang S, Jia W, Yu D. The tRNA-associated dysregulation in diabetes mellitus. Metabolism. 2019;94:9–17. doi: 10.1016/j.metabol.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Ferrucci L, et al. Measuring biological aging in humans: a quest. Aging Cell. 2020;19:e13080. doi: 10.1111/acel.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusu ME, et al. Antioxidant effects of walnut (Juglans regia L.) kernel and walnut septum extract in a D-galactose-induced aging model and in naturally aged rats. Antioxidants. 2020;9:424. doi: 10.3390/antiox9050424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, et al. 1,25-Dihydroxyvitamin D exerts an antiaging role by activation of Nrf2-antioxidant signaling and inactivation of p16/p53-senescence signaling. Aging Cell. 2019;18:e12951. doi: 10.1111/acel.12951. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Dluzen DF, et al. Extracellular RNA profiles with human age. Aging Cell. 2018;17:e12785. doi: 10.1111/acel.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filer D, et al. RNA polymerase III limits longevity downstream of TORC1. Nature. 2017;552:263–267. doi: 10.1038/nature25007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord CL, Ospovat O, Wente SR. Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs. RNA. 2017;23:365–377. doi: 10.1261/rna.057612.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser FJ, Lüdecke HJ, Weger S. SUMOylation modulates transcriptional repression by TRPS1. Biol. Chem. 2007;388:381–390. doi: 10.1515/BC.2007.051. [DOI] [PubMed] [Google Scholar]
- 18.Singh AK, et al. SUMOylation of ROR-γt inhibits IL-17 expression and inflammation via HDAC2. Nat. Commun. 2018;9:4515. doi: 10.1038/s41467-018-06924-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stankova T, Piepkorn L, Bayer TA, Jahn O, Tirard M. SUMO1-conjugation is altered during normal aging but not by increased amyloid burden. Aging Cell. 2018;17:e12760. doi: 10.1111/acel.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rytinki MM, et al. Overexpression of SUMO perturbs the growth and development of Caenorhabditis elegans. Cell Mol. Life Sci. 2011;68:3219–3232. doi: 10.1007/s00018-011-0627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Gerber A, Chen WY, Roeder RG. Functions of paralogous RNA polymerase III subunits POLR3G and POLR3GL in mouse development. Proc. Natl Acad. Sci. USA. 2020;117:15702–15711. doi: 10.1073/pnas.1922821117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Neyret-Kahn H, et al. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013;23:1563–1579. doi: 10.1101/gr.154872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y, Zhang YJ, Cai Y. Growth or longevity: the TOR’s decision on lifespan regulation. Biogerontology. 2013;14:353–363. doi: 10.1007/s10522-013-9435-6. [DOI] [PubMed] [Google Scholar]
- 25.Perić M, et al. TORC1-mediated sensing of chaperone activity alters glucose metabolism and extends lifespan. Aging Cell. 2017;16:994–1005. doi: 10.1111/acel.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carroll B, et al. Persistent mTORC1 signaling in cell senescence results from defects in amino acid and growth factor sensing. J. Cell Biol. 2017;216:1949–1957. doi: 10.1083/jcb.201610113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez-López M, et al. The GATA transcription factor Gaf1 represses tRNAs, inhibits growth, and extends chronological lifespan downstream of fission yeast TORC1. Cell Rep. 2020;30:3240–3249.e4. doi: 10.1016/j.celrep.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Y, Tsang CK, Zheng XF. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl Acad. Sci. USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Li X, Wang HY, Steven Zheng XF. Beyond regulation of pol III: Role of MAF1 in growth, metabolism, aging and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:338–343. doi: 10.1016/j.bbagrm.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Shetty M, et al. Maf1-dependent transcriptional regulation of tRNAs prevents genomic instability and is associated with extended lifespan. Aging Cell. 2020;19:e13068. doi: 10.1111/acel.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguchi C, et al. Maf1 limits RNA polymerase III-directed transcription to preserve genomic integrity and extend lifespan. Cell Cycle. 2021;20:247–255. doi: 10.1080/15384101.2021.1874697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai Y, Wei YH. Stress resistance and lifespan are increased in C. elegans but decreased in S. cerevisiae by mafr-1/maf1 deletion. Oncotarget. 2016;7:10812–10826. doi: 10.18632/oncotarget.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonhoure N, et al. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagi D, et al. Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet. 2016;12:e1006264. doi: 10.1371/journal.pgen.1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCormick MA, et al. A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura S, Srisuknimit V, Waldor MK. Probing the diversity and regulation of tRNA modifications. Curr. Opin. Microbiol. 2020;57:41–48. doi: 10.1016/j.mib.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu D, et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Songe-Møller L, et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leihne V, et al. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 2011;39:7688–7701. doi: 10.1093/nar/gkr406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee MY, Leonardi A, Begley TJ, Melendez JA. Loss of epitranscriptomic control of selenocysteine utilization engages senescence and mitochondrial reprogramming(☆) Redox Biol. 2020;28:101375. doi: 10.1016/j.redox.2019.101375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Angelova MT, et al. tRNA 2’-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res. 2020;48:2050–2072. doi: 10.1093/nar/gkaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tischner C, et al. MTO1 mediates tissue specificity of OXPHOS defects via tRNA modification and translation optimization, which can be bypassed by dietary intervention. Hum. Mol. Genet. 2015;24:2247–2266. doi: 10.1093/hmg/ddu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro-González C, et al. Mutations in the Caenorhabditis elegans orthologs of human genes required for mitochondrial tRNA modification cause similar electron transport chain defects but different nuclear responses. PLoS Genet. 2017;13:e1006921. doi: 10.1371/journal.pgen.1006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabrizio P, et al. Genome-wide screen in Saccharomyces cerevisiae identifies vacuolar protein sorting, autophagy, biosynthetic, and tRNA methylation genes involved in life span regulation. PLoS Genet. 2010;6:e1001024. doi: 10.1371/journal.pgen.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Begley U, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol. Med. 2013;5:366–383. doi: 10.1002/emmm.201201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Haute L, et al. NSUN2 introduces 5-methylcytosines in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8720–8733. doi: 10.1093/nar/gkz735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H, et al. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany NY) 2015;7:1143–1158. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai X, et al. RNA methyltransferase NSUN2 promotes stress-induced HUVEC senescence. Oncotarget. 2016;7:19099–19110. doi: 10.18632/oncotarget.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 51.Schaefer M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J. Biol. Chem. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- 53.Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Wnuk M. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J. Cell Physiol. 2017;232:3714–3726. doi: 10.1002/jcp.25848. [DOI] [PubMed] [Google Scholar]
- 54.Lewinska A, et al. Reduced levels of methyltransferase DNMT2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation-related miRNA expression. Redox Biol. 2018;14:20–34. doi: 10.1016/j.redox.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Z, Sun B, Huang S, Yu D, Zhang X. Roles of aminoacyl-tRNA synthetase-interacting multi-functional proteins in physiology and cancer. Cell Death Dis. 2020;11:579. doi: 10.1038/s41419-020-02794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SS, et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura S, et al. Mondo complexes regulate TFEB via TOR inhibition to promote longevity in response to gonadal signals. Nat. Commun. 2016;7:10944. doi: 10.1038/ncomms10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D’Hulst G, Soro-Arnaiz I, Masschelein E, Veys K. PHD1 controls muscle mTORC1 in a hydroxylation-independent manner by stabilizing leucyl tRNA synthetase. Nat. Commun. 2020;11:174. doi: 10.1038/s41467-019-13889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niehues S, et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat. Commun. 2015;6:7520. doi: 10.1038/ncomms8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banks GT, et al. Mutant glycyl-tRNA synthetase (Gars) ameliorates SOD1(G93A) motor neuron degeneration phenotype but has little affect on Loa dynein heavy chain mutant mice. PloS ONE. 2009;4:e6218. doi: 10.1371/journal.pone.0006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bayat V, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suh YS, et al. Methionyl-tRNA synthetase regulates lifespan in drosophila. Mol. Cells. 2020;43:304–311. doi: 10.14348/molcells.2019.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arif A, et al. EPRS is a critical mTORC1-S6K1 effector that influences adiposity in mice. Nature. 2017;542:357–361. doi: 10.1038/nature21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, et al. Seryl tRNA synthetase cooperates with POT1 to regulate telomere length and cellular senescence. Signal Transduct. Target Ther. 2019;4:50. doi: 10.1038/s41392-019-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Son HG, et al. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat. Commun. 2017;8:14749. doi: 10.1038/ncomms14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corti O, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 67.Lee Y, et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat. Neurosci. 2013;16:1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oh AY, et al. Inhibiting DX2-p14/ARF interaction exerts antitumor effects in lung cancer and delays tumor progression. Cancer Res. 2016;76:4791–4804. doi: 10.1158/0008-5472.CAN-15-1025. [DOI] [PubMed] [Google Scholar]
- 69.Oh YS, et al. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell. 2010;9:810–822. doi: 10.1111/j.1474-9726.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 70.Lee S, et al. miR-543 and miR-590-3p regulate human mesenchymal stem cell aging via direct targeting of AIMP3/p18. Age. 2014;36:9724. doi: 10.1007/s11357-014-9724-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun C, et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16–25. doi: 10.1016/j.canlet.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 72.Victoria B, et al. Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell. 2015;14:1055–1066. doi: 10.1111/acel.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dhahbi JM, et al. 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shigematsu M, Kawamura T, Kirino Y. Generation of 2’,3’-cyclic phosphate-containing RNAs as a hidden layer of the transcriptome. Front. Genet. 2018;9:562. doi: 10.3389/fgene.2018.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shigematsu M, Morichika K, Kawamura T, Honda S, Kirino Y. Genome-wide identification of short 2’,3’-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 2019;15:e1008469. doi: 10.1371/journal.pgen.1008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karaiskos S, Grigoriev A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Res. 2016;5:ISCB Comm J-2758. doi: 10.12688/f1000research.10116.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S, et al. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging (Albany NY) 2019;11:10485–10498. doi: 10.18632/aging.102471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanada T, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Inoue M, et al. Tyrosine pre-transfer RNA fragments are linked to p53-dependent neuronal cell death via PKM2. Biochem. Biophys. Res. Commun. 2020;525:726–732. doi: 10.1016/j.bbrc.2020.02.157. [DOI] [PubMed] [Google Scholar]
- 80.Balaskas P, et al. Small non-coding RNAome of ageing chondrocytes. Int J. Mol. Sci. 2020;21:5675. doi: 10.3390/ijms21165675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ignatova VV, et al. METTL6 is a tRNA m(3)C methyltransferase that regulates pluripotency and tumor cell growth. Sci. Adv. 2020;6:eaaz4551. doi: 10.1126/sciadv.aaz4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Auré K, et al. Homoplasmic mitochondrial tRNA(Pro) mutation causing exercise-induced muscle swelling and fatigue. Neurol. Genet. 2020;6:e480. doi: 10.1212/NXG.0000000000000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu W, et al. Systematic analysis of tRNA-derived small RNAs discloses new therapeutic targets of caloric restriction in myocardial ischemic rats. Front. Cell Dev. Biol. 2020;8:568116. doi: 10.3389/fcell.2020.568116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He Q, et al. FTSJ1 regulates tRNA 2’-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 2020;11:348. doi: 10.1038/s41419-020-2525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang L, et al. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat. Commun. 2020;11:4038. doi: 10.1038/s41467-020-17454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rumyantseva A, Motori E, Trifunovic A. DARS2 is indispensable for Purkinje cell survival and protects against cerebellar ataxia. Hum. Mol. Genet. 2020;29:2845–2854. doi: 10.1093/hmg/ddaa176. [DOI] [PubMed] [Google Scholar]
- 88.Mo Z, et al. Aberrant GlyRS-HDAC6 interaction linked to axonal transport deficits in Charcot-Marie-Tooth neuropathy. Nat. Commun. 2018;9:1007. doi: 10.1038/s41467-018-03461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: an update. Cell Metab. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 90.Ma H, et al. Germline and somatic mtDNA mutations in mouse aging. PLoS ONE. 2018;13:e0201304. doi: 10.1371/journal.pone.0201304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bacman SR, et al. MitoTALEN reduces mutant mtDNA load and restores tRNA(Ala) levels in a mouse model of heteroplasmic mtDNA mutation. Nat. Med. 2018;24:1696–1700. doi: 10.1038/s41591-018-0166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 93.Yoon DS, Cha DS, Choi Y, Lee JW, Lee MH. MPK-1/ERK is required for the full activity of resveratrol in extended lifespan and reproduction. Aging Cell. 2019;18:e12867. doi: 10.1111/acel.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sajish M, Schimmel P. A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature. 2015;519:370–373. doi: 10.1038/nature14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim C, Park JM, Song Y, Kim S, Moon J. HIF1α-mediated AIMP3 suppression delays stem cell aging via the induction of autophagy. Aging Cell. 2019;18:e12909. doi: 10.1111/acel.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Magee R, Londin E, Rigoutsos I. TRNA-derived fragments as sex-dependent circulating candidate biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 2019;65:203–209. doi: 10.1016/j.parkreldis.2019.05.035. [DOI] [PubMed] [Google Scholar]