Abstract
The increasing availability of single-cell data revolutionizes the understanding of biological mechanisms at cellular resolution. For differential expression analysis in multi-subject single-cell data, negative binomial mixed models account for both subject-level and cell-level overdispersions, but are computationally demanding. Here, we propose an efficient NEgative Binomial mixed model Using a Large-sample Approximation (NEBULA). The speed gain is achieved by analytically solving high-dimensional integrals instead of using the Laplace approximation. We demonstrate that NEBULA is orders of magnitude faster than existing tools and controls false-positive errors in marker gene identification and co-expression analysis. Using NEBULA in Alzheimer’s disease cohort data sets, we found that the cell-level expression of APOE correlated with that of other genetic risk factors (including CLU, CST3, TREM2, C1q, and ITM2B) in a cell-type-specific pattern and an isoform-dependent manner in microglia. NEBULA opens up a new avenue for the broad application of mixed models to large-scale multi-subject single-cell data.
Subject terms: Computational models, Transcriptomics, Alzheimer's disease
The application of negative binomial mixed models (NBMMs) to single-cell data is computationally demanding. To address this issue, Liang He et al. have developed NEBULA, an efficient algorithm that can analyze differential gene expression or co-expression networks in multi-subject single-cell data sets, and validate it on snRNA-seq and scRNA-seq data sets comprising ~200k cells from cohorts of Alzheimer’s disease and multiple sclerosis patients.
Introduction
Single-cell genomic profiling technology has revolutionized our understanding of biology to an unprecedented resolution. With the recent advances in high-throughput single-cell profiling techniques1–3, large-scale multi-subject single-cell RNA-seq (scRNA-seq) and ATAC-seq (scATAC-seq) data are becoming increasingly accessible. Notably, through droplet-based technology, tens to hundreds of thousands of cells from multiple subjects can be sequenced in parallel within a single batch2,4. As an example, a recent large-scale population single-nucleus RNA-seq (snRNA-seq) data in the human frontal cortex involving ~80,000 nuclei from a cohort comprising 24 Alzheimer’s disease (AD) patients and 24 healthy controls5 provide biological information in much finer granularity compared to bulk RNA-seq data. Projects profiling more than one million single cells from hundreds of subjects are underway.
The drastically increasing magnitude of sample size, however, poses a serious computational challenge when trying to apply conventional transcriptomics analysis for differential expression, expression quantitive trait loci (eQTLs), and co-expression to large-scale scRNA-seq data. This situation is in contrast to that of statistical models in bulk RNA-seq analysis, in which more emphasis is placed upon building a robust estimate under a small sample size (e.g., regularization of standard errors and robust estimation of overdispersion parameters6–8). Unlike bulk RNA-seq data, multi-subject scRNA-seq data, particularly using the droplet-based technology, often comprise many cells but are characterized by high sparsity and a hierarchical structure. Owing to the hierarchical nature of multi-subject scRNA-seq data, it is appropriate to decompose the overdispersion into the subject and cell levels. One of the standard approaches to handle hierarchical count responses is a negative binomial mixed-effects model (NBMM), which introduces independent random effects to take into account the overdispersion at both levels. NBMMs increase the statistical power by a more accurate specification of the overdispersion structure, and also helps in eliminating spurious associations to detect marker genes of a cell cluster, as we will show in our simulation. Here, we focus on NBMMs rather than a zero-inflated model because multiple recent studies show that a zero-inflated model might be redundant for unique molecular identifiers (UMI)-based single-cell data9–11. This is also consistent with the observations for the vast majority of genes in our analysis of real data.
A plethora of estimation methods have been proposed for NBMMs12–17, and it is the invention of these fast algorithms that popularizes its practical use. However, the computational efficiency of existing tools15,18,19 is still insufficient for its broad application to large-scale multi-subject scRNA-seq data. Owing to the intractable marginal likelihood in NBMMs, current estimation algorithms generally rely on a two-layer iteration procedure, which often converges slowly. To address the computational burden, we propose a NEgative Binomial mixed model Using Large-sample Approximation (NEBULA), a novel fast algorithm for association analysis of scRNA-seq data using an NBMM. As the core idea in NEBULA, we developed an analytical approximation of the high-dimensional integral for the marginal likelihood of the NBMM, by leveraging the common feature of scRNA-seq data of having many cells per subject. The improvement of computational efficiency is achieved by avoiding the two-layer optimization and converging within fewer steps.
We demonstrated the efficiency and accuracy of NEBULA through an extensive simulation study. As NEBULA uses approximation based on large numbers, we investigated the number of cells and subjects required to accurately estimate the subject-level and cell-level overdispersions. We also investigated the performance of NEBULA in dealing with lowly expressed genes and controlling the false-positive errors when testing fixed-effects predictors. It should be noted that a predictor of interest can be classified as a subject-level or a cell-level variable. The first category includes variables that share the same value across all cells from the same subject, for example, disease or treatment groups, genetic variants, and subject-level covariates such as age and sex. Variables with different values across individual cells such as cell cycle stages, subpopulation memberships, and cell-level gene expression belong to the second category. For testing associations with cell-level variables, it is known that ignoring the subject-level random effects would lead to inflated p-values20. In contrast, there is much less literature on the performance of testing subject-level predictors in cell-level data. It is well known that the subject-level random effects need to be included in the model when the predictor of interest is also a subject-level variable. Otherwise, the type I error rate would be inflated due to an underestimated standard error of the effect size. A detailed derivation of such underestimation is given in21,22. In this study, we found that testing a subject-level variable was highly sensitive to the estimation of the subject-level variance component and the assumption of the distribution of the random effects when the number of subjects is small.
To date, few studies have investigated the relative contributions of the subject-level and cell-level overdispersions to cell-type-specific gene expression. Overdispersion often results from failing to include important explanatory variables or account for a hierarchical structure in the data23. To explore these problems in a specific biological context, we used NEBULA to decompose gene-specific overdispersion of large-scale snRNA-seq data in the human frontal cortex from an AD cohort5. We further explored what factors might contribute to both overdispersions. We showed that NEBULA reduced false positives in selecting cell-type marker genes, especially when the numbers of cells across subjects were unbalanced. As a direct application of NEBULA, we performed a cell-level transcriptomic co-expression analysis of APOE, the strongest genetic risk factor of AD, and investigated its cell-type-specific regulatory mechanisms in microglia and astrocytes, both of which are known to abundantly express APOE and to undergo cell activation transitions in the context of AD pathology. This application of NEBULA allowed us to identify both cell-type and isoform-specific coregulatory interactions of APOE that might be relevant for mediating its disease-modifying effects at a molecular level.
Results
Overview of NEBULA
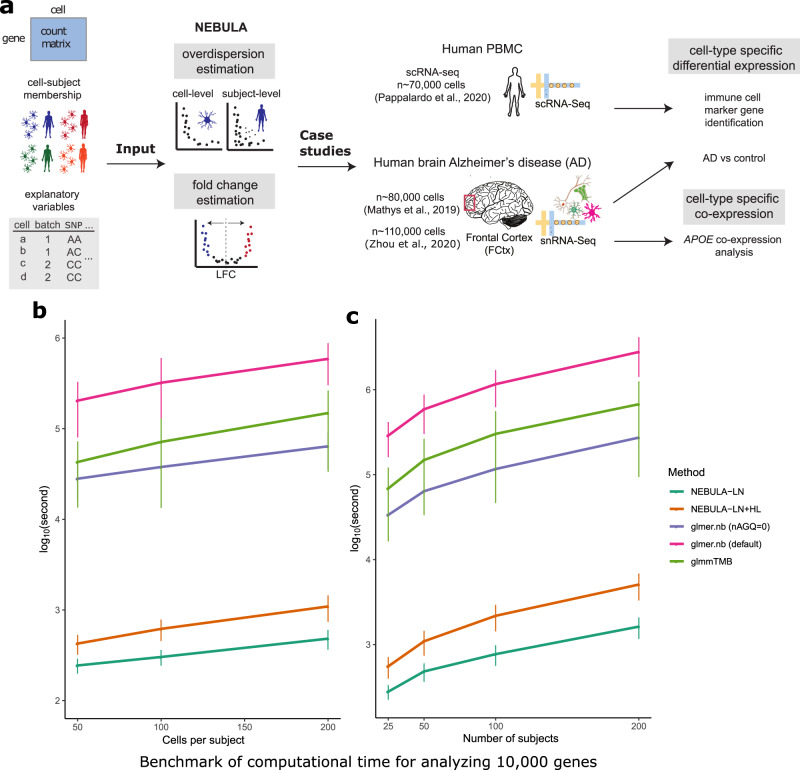
NEBULA takes as an input the raw count matrix of a single-cell data set and a mapping between cells and subjects (Fig. 1a). The variation of raw counts in scRNA-seq data comes from two sources, a Poisson sampling process and overdispersion originating from biological heterogeneity and technical or experimental noise. Cells in large-scale single-cell data are often collected from multiple subjects or batches, leading to an apparent hierarchical structure. To model this structure, NEBULA decomposes the total overdispersion into subject-level (i.e., between-subject) and cell-level (i.e., within-subject) components using a random-effects term parametrized by and the overdispersion parameter in the negative binomial distribution (Fig. 1a). The subject-level overdispersion captures the contributions from, e.g., eQTLs, technical artifacts, batch effects, and other subject-level covariates. The cell-level overdispersion reflects the variation of gene expression due to differences in, e.g., cell cycle, cell-type subpopulation, and other cell-level covariates such as ribosome RNA fraction. Generally, the estimation of an NBMM is computationally intensive because the marginal likelihood is intractable due to high-dimensional integrals of the random effects. A standard estimation method for generalized linear mixed models (GLMMs) using penalized quasi-likelihood (PQL)12 involves a two-step iterative procedure in which the variance of the random effects is first estimated and is subsequently used to estimate the coefficients of the variables of interest. In genome-wide association studies (GWAS), the variance component is often estimated only once under the null model. In contrast, in scRNA-seq or scATAC-seq data, such a two-step procedure has to be carried out for each of tens of thousands of genes or peak regions, giving rise to a considerably more challenging requirement. NEBULA achieves its speed gain by introducing an approximate analytical marginal likelihood derived by leveraging the fact that single-cell data often include a large number of cells for each subject. This approximation, referred to as NEBULA-LN, eliminates the computationally demanding two-layer high-dimensional optimization procedure, and therefore substantially reduces the computational time for estimating the overdispersions. For situations in which NEBULA-LN fails to achieve accurate estimates of the subject-level overdispersion, we use NEBULA-LN to estimate only the cell-level overdispersion and implemented a fast estimation procedure, referred to as NEBULA-HL, based on a hierarchical likelihood (h-likelihood) for estimating the subject-level overdispersion. As only one overdispersion is optimized in the h-likelihood, NEBULA-HL converges in much fewer steps than standard estimation methods.
Fig. 1. Overview of the input, output, and computational efficiency of NEBULA.
a A flowchart of NEBULA including the input data, the major estimation steps in NEBULA, and the analyses that were conducted using NEBULA in the application to the real data5,27,78. b The computational time (measured in log10(seconds)) of fitting an NBMM for 10,000 genes with respect to CPS by NEBULA, glmer.nb18, and glmmTMB19. The number of subjects was set at 50. c The computational time (measured in log10(seconds)) of fitting an NBMM for 10,000 genes with respect to the number of subjects. The error bars represent one standard deviation of n = 10,000 genes. The CPS value was set at 200. Two fixed-effects predictors were included in the NBMM. The average benchmarks were summarized from scenarios of varying subject-level and cell-level overdispersions and the CPC value of a gene ranging from exp(−4) to 1. CPS: cells per subject. CPC: counts per cell.
NEBULA is computationally efficient
We compared NEBULA with existing statistical tools for estimating NBMMs to evaluate its computational efficiency. In addition to NEBULA-LN, we assessed a combination of NEBULA-LN and NEBULA-HL (denoted by NEBULA-LN + HL), in which we first used NEBULA-LN to estimate the cell-level overdispersion, and then fixed this estimate in NEBULA-HL to obtain the subject-level overdispersion. For comparison, we included the glmer.nb function from the lme4 R package18, the glmmTMB function from the glmmTMB R package19, and the inla function from the INLA R package15. The glmmTMB package utilizes the Template Model Builder automatic differentiation engine, and the INLA package uses an efficient Bayesian framework based on integrated nested Laplace approximations (LAs). We did not include methods based on adaptive Gaussian quadrature (AGQ) (glmer.nb with nAGQ>1)24,25 or Bayesian methods using Markov chain Monte Carlo (MCMC)26 because of their computational intensity. The Gaussian variational approximation method14 can be promising but has not been implemented for the NBMM.
Given a simulated data set comprising 50 subjects and cells per subject (CPS) = 200 (a total of ~10,000 cells), it took ~400 s for NEBULA-LN and ~1000 s for NEBULA-LN + HL to analyze 10,000 genes using one CPU thread (Fig. 1b). These benchmarks were on average >100-fold and ~50-fold faster than glmer.nb with nAGQ = 0, respectively. It took much longer for glmer.nb with the default setting and glmmTMB. We failed to obtain the full benchmarks for inla because it took an extremely long time to finish in some scenarios. Generally, the running time of inla was between glmmTMB and the default glmer.nb. The computational time of all methods increased almost linearly with CPS, which was expected because the random effects are assumed to be independent. We also observed that the computational time scaled in a similar trend across these methods with the increasing number of subjects (Fig. 1c). In all settings, NEBULA-LN + HL was ~2–3x slower than NEBULA-LN but was still much faster than glmer.nb. In our following analysis of ~34,000 excitatory neurons from 48 subjects in the snRNA-seq data adopted from ref. 5, NEBULA accomplished an association analysis of ~16,000 genes for identifying marker genes in ~40 min, compared to ~67 hours using glmer.nb with nAGQ = 0. When the number of fixed-effects predictors increased from two to ten, the computational time of both NEBULA and glmer.nb with nAGQ=0 grew modestly, while the default glmer.nb increased by ~10x (Supplementary Fig. S1). This substantial increase of the computational time in the default glmer.nb is expected because it estimates the fixed effects along with the overdispersions in the outer layer of the iterative procedure using a derivative-free method. The number of iterations of such an optimization method rises drastically when the dimension of the parameter space increases. Altogether, these comparisons demonstrate the superior computational efficiency of NEBULA relative to existing alternatives.
NEBULA decomposes cell-level and subject-level overdispersions
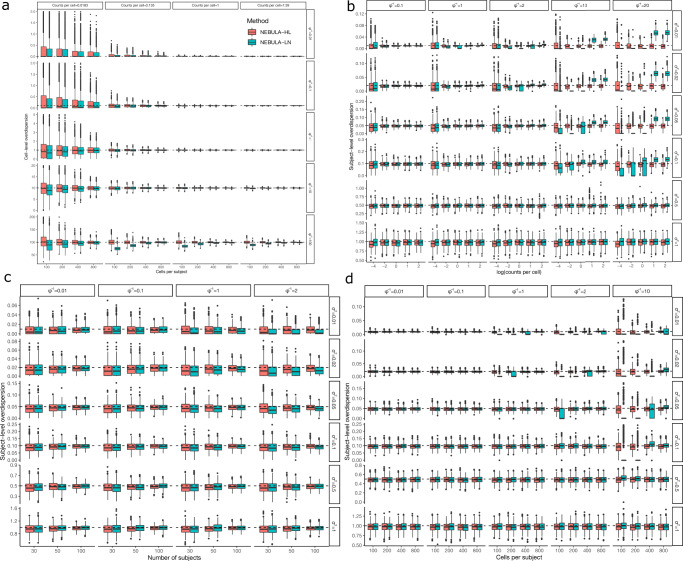
The approximation proposed in NEBULA-LN provides asymptotically consistent estimates (Supplementary Note A.5). Nevertheless, it is crucial to assess its practical performance under a finite sample size. As the above benchmarks indicated that NEBULA-LN had a huge speed advantage over NEBULA-HL, the basic strategy in NEBULA is to first fit the data using NEBULA-LN and apply NEBULA-HL only in situations where NEBULA-LN performs poorly. We, therefore, conducted a comprehensive simulation study to investigate under which situations NEBULA-LN could produce sufficiently accurate estimates. The estimation accuracy of NEBULA-LN is primarily affected by CPS, the magnitude of the cell-level dispersion, and the coefficient of variation (CV) of the scaling factor (see the Methods section). We thus focus on determining a threshold in terms of these parameters. We used NEBULA-HL, which is based on a standard h-likelihood method, as a reference to assess the accuracy of NEBULA-LN in terms of mean squared error (MSE).
We first evaluated the performance in a well-designed study in which the numbers of cells were highly homogeneous across the subjects and were assumed to follow a Poisson distribution. We considered the CPS value varying between 100 and 800, and the number of subjects ranging from 30 to 100, which are common settings consistent with empirical observations in droplet-based scRNA-seq data. We simulated counts based on an NBMM with the subject-level overdispersion ranging from 0.01 to 1 and the cell-level overdispersion ranging from 0.01 to 100 (A larger corresponds to higher overdispersion). These ranges were selected because they were close to our estimates observed in the real snRNA-seq data set in ref. 5 and the scRNA-seq data set in ref. 27. We simulated genes of different mean expression by tuning between −4 and 2 (When the subject-level overdispersion is small, approximately equals the counts per cell (CPC) defined by the total counts of a gene divided by the total number of cells). We considered the following two situations, (i) all cells share the same scaling factor and (ii) the scaling factor across cells varied substantially with the CV equal to 1.
We found that the MSE of NEBULA-LN for estimating the cell-level overdispersion decreased uniformly with increasing CPS (Fig. 2a). The convergence of the estimates to their true values supported our theoretical conclusion that NEBULA-LN is asymptotically consistent. Both methods struggled to accurately estimate for low-expression genes as their MSEs increased with the decreasing mean expression, suggesting that genes with a low CPC value could be filtered during the quality control (QC) unless the number of cells is very large. This is because it is difficult to accurately estimate the overdispersions for these genes due to the high sparsity. In most scenarios, NEBULA-LN showed higher MSEs and was asymptotically less efficient than NEBULA-HL for estimating . Nevertheless, the MSEs between the two methods were comparable when CPS was >400. When was very large (=100), NEBULA-LN produced a more biased estimate if CPS is small. A larger CV of the scaling factor showed little impact on the MSE of NEBULA-LN (Supplementary Fig. S2A). Additionally, increasing the number of subjects alone improved the variance, but not the bias of , suggesting that the bias of depended mainly on CPS (Supplementary Fig. S3).
Fig. 2. Comparison of estimated cell-level and subject-level overdispersion parameters between NEBULA-LN and NEBULA-HL.
a The cell-level overdispersion estimated by NEBULA-LN and NEBULA-HL under different combinations of CPS, (a lower corresponding to a lower CPC value), and . The number of subjects was set at 50. b The subject-level overdispersion estimated by NEBULA-LN and NEBULA-HL under different combinations of , , and . The number of subjects was set at 50. The CPS value was set at 400. c The subject-level overdispersion estimated by NEBULA-LN and NEBULA-HL under different combinations of the number of subjects, , and . The CPS value was set at 400, and was set at 0.05. d The subject-level overdispersion estimated by NEBULA-LN and NEBULA-HL under different combinations of CPS, , and . The number of individuals was set at 50, and was set at 1. The summary statistics were calculated from n = 500 simulated replicates in each of the scenarios. The estimates from NEBULA-HL presented in these panels were based on the first-order Laplace approximation, and the results comparing the first-order and higher-order LA methods in NEBULA-HL were presented in Supplementary Fig. S17. CPC: counts per cell. Center line: median. Box limits: upper and lower quartiles.
On the other hand, NEBULA-LN showed comparable or even lower MSEs than NEBULA-HL for estimating the subject-level overdispersion when it is large () (Fig. 2b). However, when was small, NEBULA-LN began to produce CPC-dependent biased estimates when turned larger (Fig. 2b). Increasing CPS alleviated this bias rapidly (Fig. 2d). When the CV of the scaling factor increased from zero to one, an approximately 2-fold increase of was required to achieve similar MSEs by comparing Supplementary Figs. S2B, S2D with Fig. 2b, d. These observations suggest that the accuracy of NEBULA-LN for estimating approximately depends on ( is the CV of the scaling factor), which is derived in the Methods section. These empirical results suggest that determined the lower bound of for which an accurate estimate could be achieved by NEBULA-LN. Empirically, we found that NEBULA-LN produced good estimates for as low as 0.02 when (corresponding to the column with in Fig. 2b, c), which was sufficient to reliably test cell-level predictors, as we will show in the next section. Both NEBULA-LN and NEBULA-HL underestimated particularly when the number of subjects was small (Fig. 2c). We found that this underestimate was not specific to NEBULA, but also present in other tools (e.g., the glmer.nb function with nAGQ = 0). Increasing the number of subjects alleviated this bias, and the influence of such bias on testing subject-level preditors will be discussed in the next section.
We further evaluated the performance of NEBULA in unbalanced samples, where the numbers of cells varied substantially across the subjects. An unbalanced sample is common when analyzing a cell subpopulation obtained from clustering. For example, in the snRNA-seq data in ref. 5, several individuals had only ~20 inhibitory neurons, although the CPS value among the 48 individuals was ~200. Overall, the results from the unbalanced sample (Supplementary Fig. S4) were comparable to those from the balanced sample (Fig. 2), suggesting the distribution of the number of cells had little impact on estimating and . Overall, NEBULA is able to provide accurate estimates under the parameter regimes relevant for real-data settings and with finite sample sizes.
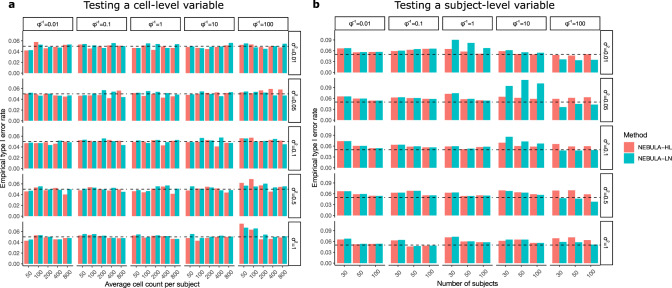
NEBULA controls type I error rate
As shown above, both NEBULA-LN and NEBULA-HL might yield somewhat biased estimates of the overdispersions in specific scenarios. We then assessed the impact of such biases on controlling the type I error rate by testing the fixed-effects predictors under a wide range of settings. As mentioned above, predictors can be classified as either a cell-level or subject-level variable. For a cell-level variable, Fig. 3a shows that both methods controlled the type I error rate well in almost all scenarios. The type I error rate was only slightly inflated when both overdispersions were very large, and the CPS value was as small as 100 (i.e., the bottom-right panels in Fig. 3a). No inflation of the type I error rate was observed for low-expression genes, for which was more underestimated in NEBULA-LN (Fig. 2a and Supplementary Fig. S5). Besides, the biased estimates of the subject-level overdispersion in NEBULA-LN had little effect on testing a cell-level variable. In a simulation study with very small CPS, we found that noticeable inflation of the type I error rate began to manifest in NEBULA-LN when the CPS value dropped to <30.
Fig. 3. Empirical type I error rate of testing cell-level and subject-level variables using NEBULA-LN and NEBULA-HL.
a The empirical type I error rate of testing a cell-level variable using NEBULA-LN (blue) and NEBULA-HL (red) under different combinations of CPS, , and . The number of subjects was set at 50. b The empirical type I error rate of testing a subject-level variable using NEBULA-LN (blue) and NEBULA-HL (red) under different combinations of the number of subjects, , and . The CPS value was set at 400. The type I error rate was calculated from n = 500 simulated replicates in each of the scenarios and was evaluated at the significance level of 0.05 (the dashed lines). We used the higher-order Laplace approximation in NEBULA-HL for scenarios of low CPC. CPS: cells per subject. CPC: counts per cell.
In contrast, the type I error rate of testing a subject-level predictor was highly sensitive to a biased estimate of the subject-level overdispersion . The type I error rate in both methods was inflated in the case of 30 subjects and the inflation diminished with the increasing number of subjects (Fig. 3b). This is because both methods underestimated the subject-level overdispersion in this scenario (Fig. 2c). The comparison between Fig. 2c and Fig. 3b suggests that even a small underestimated subject-level overdispersion would lead to an inflated type I error rate of testing a subject-level predictor when the number of subjects is small. Matching Fig. 2b and Supplementary Fig. S6 suggests that NEBULA-LN had inflated or deflated type I error rate of testing a subject-level predictor whenever was overestimated or underestimated, respectively, even for being as low as 0.01. Besides, we found that misspecification of the random effects could lead to an inflated type I error rate as well, particularly if the sample size is small. These results suggest that NEBULA or other methods based on a mixed model should be used with caution when testing a subject-level predictor, particularly when the number of subjects is small (<100).
We also evaluated the performance of a Poisson mixed model for testing a subject-level predictor. If we ignore the cell-level overdispersion in NEBULA, we end up with a Poisson gamma mixed model (PGMM)28, the likelihood function of which has a simple analytical form. Therefore, it does not suffer from the problem of underestimating as in NEBULA-HL and is computationally much faster than the Poisson mixed model estimated by glmer.nb with nAGQ=10 adopted in ref. 5. As the cell-level overdispersion is ignored in the PGMM, it definitely cannot be used to test a cell-level predictor. However, it is still interesting to check its type I error rate when used for testing a subject-level predictor. Supplementary Fig. S7 shows that overall the PGMM controlled the type I error rate well in scenarios where the number of subjects was 100, except for situations where both the cell-level and subject-level overdispersions were large ( and ). Such situations account for a very small fraction of genes (usually low-expression genes) in real data and such genes are often excluded during quality control. For instance, we observed <0.1% genes with CPC > 0.1% having in the major cell types in the snRNA-seq data in ref. 5, and the percentage became almost zero when we filtered out lowly expressed genes (CPC < 1%) during quality control. Therefore, the PGMM might be considered as a fast tool for testing subject-level predictors followed by a second-round sensitivity analysis for top signals if the computational intensity is a major concern.
Based on these simulation results, the strategy that we recommend for testing the fixed-effects predictors, as summarized in Supplementary Data 1, is to fit the data first using NEBULA-LN to obtain initial estimates and . Both overdispersion parameters are re-estimated using NEBULA-HL if or . Otherwise, we fix and only use NEBULA-HL to re-estimate if and , where is the CV estimated from the data and we chose for testing a cell-level and subject-level predictor, respectively, based on the simulation results. Otherwise, we report the estimates directly from NEBULA-LN because in this case, NEBULA-LN produces a reliable estimate for a sufficiently small . We used this strategy in our following analysis of the real data sets. In the snRNA-seq data set generated by5, ~90% and ~80% of genes with CPC > 0.1% were analyzed using NEBULA-LN alone under this strategy in the excitatory neurons (CPS = ~700) and oligodendrocytes (CPS = ~380). We found that of the cell library size was ~0.8 for the neurons and ~0.25 for all other cell types. The larger CV in the neurons was probably because the neurons consisted of highly heterogeneous subpopulations (Supplementary Fig. S8).
Subpopulation heterogeneity and known covariates contribute to overdispersions
The subject-level and cell-level overdispersion parameters likely reflect some intrinsic biological features of a gene. A gene with a larger subject-level overdispersion might indicate that the gene is strongly regulated by subject-level factors such as age, sex, disease status, exposure, and genetic variation. We used NEBULA to dissect the overdispersions of genes in the snRNA-seq data in ref. 5. We carried out the analysis in each of the six major cell types, including excitatory neurons, inhibitory neurons, oligodendrocytes, astrocytes, oligodendrocyte progenitor cells (OPCs), and microglia.
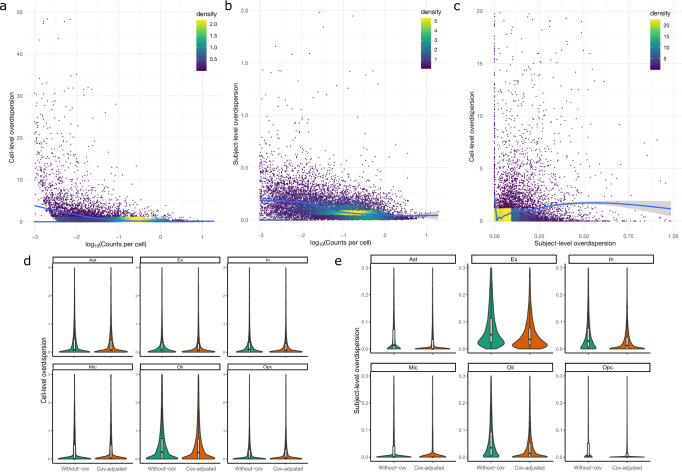
We observed that the mean cell-level overdispersion grew steadily with lower mean gene expression in the excitatory neurons (Fig. 4a) and also in the other cell types (Supplementary Fig. S9). This mean-overdispersion trend is reminiscent of that observed in bulk RNA-seq data, in which low-expression genes often show higher variance6. Less than 0.1% of genes in the excitatory neurons had an estimated cell-level overdispersion larger than 100, and all of them were low-expression genes. In contrast, the subject-level overdispersion exhibited a similar mean-overdispersion trend, except that the overdispersion leveled off for abundantly expressed genes with CPC>1 in the excitatory neurons (Fig. 4b). Similar patterns were observed in the other cell types (Supplementary Fig. S10). We observed a higher variation of both estimated overdispersions in lower-expression genes, which is due to a larger standard error for those genes as shown in the simulation (Fig. 2). There was no clear correlation between the subject-level and the cell-level overdispersions (Fig. 4c).
Fig. 4. Decomposed cell-level and subject-level overdispersions in major neural cells.
a The cell-level overdispersion and b the subject-level overdispersion of 16,207 genes in the excitatory neurons estimated by NEBULA across different CPC values. No covariates other than the intercept were included in the model. The color indicates the two-dimensional kernel density computed using the kde2d R function with n = 100. c The cell-level overdispersion of 16,207 genes in the excitatory neurons estimated by NEBULA versus their subject-level overdispersion. d The estimated cell-level overdispersion and e the estimated subject-level overdispersion in each of the six brain cell types before and after adjusting for age, sex, race, AD status, the total number of features of the cell, and the percentage of ribosomal genes. Only genes with CPC > 0.1% in the relevant cell type were included in the above results. The cells in this analysis are from the 48-subject snRNA-seq data set5 in the frontal cortex.
We then explored which factors might contribute to the gene-specific overdispersions in each of the cell types. First, we found that the median cell-level overdispersion dropped markedly in the different subpopulations within a given cell type (Supplementary Fig. S11A), suggesting that differential expression between subpopulations was one of the major sources for the cell-level overdispersion. In contrast, the median subject-level overdispersion in the subpopulations dropped only mildly (Supplementary Fig. S11B). We then investigated subject-level covariates, including age, sex, AD diagnosis, race, and cell-level covariates, including the number of total features (i.e., detected genes) of a cell and the percentage of counts mapping to ribosomal protein genes. NEBULA enables readily exploring the role of such covariates by including them in the regression model. We found that the overall subject-level overdispersion dropped substantially in most of the cell types after adjustment for these covariates, but the cell-level overdispersion remained almost at the same level (Fig. 4d, e). These results suggest that the subject-level overdispersion was largely attributed to these factors, while the cell-level overdispersion resulted from subpopulation heterogeneity and other unknown factors, e.g., drop-out rates.
In terms of models, the estimated cell-level overdispersions were highly comparable between NEBULA and glmer.nb (Supplementary Fig. S12), except for an outlier in glmer.nb. The estimated subject-level overdispersions were also strongly correlated for those genes with a small to moderate subject-level overdispersion (Supplementary Fig. S13), although it is not straightforward to compare the absolute values because the random effects in these two NBMMs are assumed to follow slightly different distributions (see the Methods section). The larger difference between NEBULA and glmer.nb observed in larger subject-level overdispersions was expected, as previously shown that the two models are highly consistent when the subject-level overdispersion is small28.
NEBULA accurately identifies cell-type marker genes
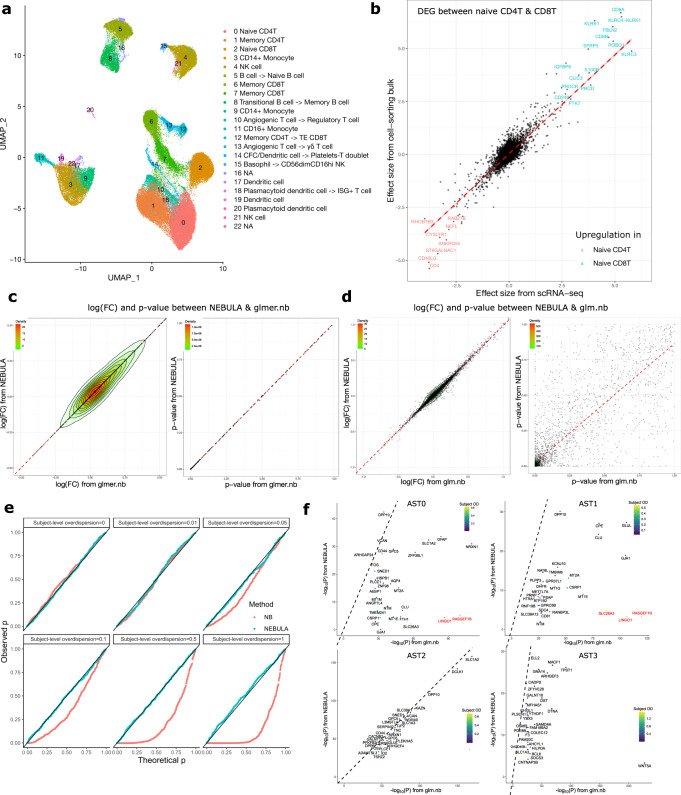
To demonstrate that NEBULA is an efficient tool to find marker genes for annotating cell clusters, we next analyzed a recent multi-subject peripheral blood mononuclear cell (PBMC) scRNA-seq data set27. The PBMC scRNA-seq data contain 69,516 cells from the peripheral blood of 11 subjects. We first classified the cells into 22 clusters using Seurat (Fig. 5a). The cells within each cluster were distributed almost evenly across the subjects. We performed a differential expression analysis using NEBULA for each of the clusters to identify marker genes that showed significantly higher expression in one cluster than the cells of the other clusters. We then used the top marker genes (see the Methods section for the selection of the marker genes) as the input to scCATCH29 to annotate these clusters. We found that this automatic pipeline produced consistent results for the major cell types in PBMC compared to our refined manual annotation (Fig. 5a). Discrepancies appeared mainly in the annotation of subcell types in T cells due in large part to the low resolution of the T-cell subpopulations in the reference databases in scCATCH. To validate the differentially expressed genes (DEGs) identified by NEBULA from the scRNA-seq data, we compared the effect sizes from the DEG analysis between the naive CD4+ T cell (cluster 0) and the naive CD8+ T cell (cluster 2) with those obtained from a reference cell-sorting bulk RNA-seq data from the Database of Immune Cell Expression (DICE)30. We found that the estimated effect sizes were highly concordant between these two independent data sets and different models (Fig. 5b).
Fig. 5. Identification of cell marker genes using NEBULA.
a A UMAP plot of the cells in the PBMC scRNA-seq data27. The automatic cell-type annotation was performed using NEBULA and scCATCH29. The corrected or refined cell type by the manual annotation was shown after the arrow. Cluster 16 and 22 were not annotated because of either too few cells or marker genes. b Comparison between log(FC) estimated by NEBULA in the scRNA-seq data27 and the effect size estimated by glm.nb in the cell-sorting bulk RNA-seq data30 in the analysis of differentially expressed genes between naive CD8+ and CD4+ T cells. Genes having both log(FC) and an effect size >2.5 were highlighted in blue for naive CD8+ and red for naive CD4+ T cells. Comparison between the log(FC) and p-values from c NEBULA and glmer.nb and d NEBULA and glm.nb in the differential expression analysis of detecting marker genes for Cluster 1 (memory CD4+ T cells) in the scRNA-seq data27. e Q–Q plots of p-values for testing the association between simulated counts and cell clusters using NEBULA and a simple negative binomial regression (shown as NB in the plot). The counts were simulated under the null model (i.e., no association between the counts and cell clusters) based on an NBMM with the subject-level overdispersion () ranging from 0 to 1 and a cell-level overdispersion fixed at 1. f Comparison of the p-values of the top 30 genes identified by glm.nb associated with each of the four astrocyte subpopulations (AST0-AST3) in the snRNA-seq data5 with those reported by NEBULA. The labels of the four astrocyte subpopulations are corresponding to those presented in ref. 5. Subject OD: the subject-level overdispersion estimated by NEBULA. Genes highlighted in red showed substantially reduced significance in NEBULA compared to the p-values in the simple negative binomial regression.
To compare different NBMMs, we found that the estimated effect sizes and p-values in the 11-subject PBMC scRNA-seq data were highly consistent between NEBULA and the NBMM using glmer.nb (Fig. 5c) while NEBULA was faster by orders of magnitude. We also compared the performance in the 48-subject snRNA-seq data. Estimated fixed effects of a subject-level variable were also consistent between NEBULA and glmer.nb (Supplementary Fig. S14), although the consistency was lower than that of a cell-level variable (Fig. 5c). In contrast, many genes showed very different p-values between NEBULA and a naive negative binomial model (NBM) using glm.nb (Fig. 5d). The difference can be justified by the fact that using a negative binomial regression without the subject-level random effects could result in substantially inflated false positives20. Disproportionate cell frequency among individuals may exacerbate the inflation of spurious associations, similar to Simpson’s Paradox31. In this case, subjects can be a confounding factor between gene expression and cell clusters. On the other hand, including subjects as fixed effects would lead to too many parameters in the model, especially when the number of subjects is large. Therefore, an NBMM is an ideal approach for the identification of marker genes for subpopulations in multi-subject scRNA-seq data.
To illustrate this problem, we used the 3386 astrocytes from the snRNA-seq data in the frontal cortex, which were classified into four subclusters (AST0-AST3). Some of the subclusters were distributed across the subjects in a highly unbalanced pattern. For example, the vast majority of the cells in the AST3 cluster came from only four subjects (Supplementary Fig. S15). We simulated the counts of gene expression using an NBMM under the null hypothesis (i.e., not associated with any of the subclusters) with both subject-level and cell-level overdispersions. We assessed the false-positive rates (FPR) under four scenarios with the variance component of the subject-level overdispersion ranging from 0 to 0.1. As we can see in Fig. 5e, even with the subject-level overdispersion as small as 0.05, the FPR started to show inflation. The inflation rose rapidly with the increased subject-level overdispersion. In contrast, NEBULA controlled the FPR very well.
To evaluate how this inflation of FPR might affect the selection of marker genes, we carried out an association analysis between the gene expression and the four subclusters in the astrocytes in the snRNA-seq data5 using a negative binomial regression and NEBULA. Among the top 30 genes identified by a negative binomial regression, most of the p-values were consistent between the two methods in the subclusters AST2 and AST3. However, some of the top genes in the subclusters AST0 and AST1, such as RASGEF1B and LINGO1, were much less significant in NEBULA than in the negative binomial regression (Fig. 5f). Both genes showed uniformly strong subject-level overdispersion in the astrocytes (Fig. 5f) and the other five cell types, indicating that the significant p-values in the negative binomial regression could result from failing to account for the subject-level overdispersion. Furthermore, an independent cell-type-specific bulk RNA-seq data32 showed that the expression of RASGEF1B and LINGO1 was much lower in astrocytes than that in microglia and oligodendrocytes, respectively. Given the evidence, care should be taken when classifying these genes as subcell-type marker genes based on the results from the negative binomial regression without the random effects. Besides, GJA1 was identified by NEBULA as a marker gene of AST1 but not AST0, while a negative binomial regression identified GJA1 as significant in both subclusters (Fig. 5f). Thus, NEBULA provides a principled way to identify genes with preferential expression patterns in cellular subpopulations that are consistent across individuals, by effectively accounting for subject-level random effects.
NEBULA robustly estimates cell-level co-expression
Estimating co-expression between genes is crucial for inferring functional associations among genes and defining molecular pathways and coherent gene modules. However, co-expression patterns estimated from bulk RNA-seq data are heavily affected by many hidden confounders such as cell type composition and experimental noise. In contrast, scRNA-seq data opens up new possibilities for investigating co-expression at the cellular level. We interpret the cell-level co-expression as the co-occurrence of two genes in a cell. Nevertheless, we will show that co-expression analysis of scRNA-seq data using common methods poses special challenges. NEBULA has unique advantages to explore cell-level co-expression over common distance metrics such as Pearson or Spearman correlation coefficient because it is more robust and adjusts for overdispersions, sequencing depth, and covariate effects.
To give a demonstration of these advantages and their potential to explore specific biological problems, we compared the performance of estimating cell-level co-expression of TCF7 and BCL3, two of the major transcription factors in adaptive immune cells, with 14,770 genes having CPC > 0.05% in the memory CD4+ T cells in the PBMC scRNA-seq data. We evaluated four methods and assessed their difference including (i) NEBULA without adjusting for any confounders, (ii) NEBULA adjusting for total features, percentage of mitochondrial genes, and percentage of ribosomal genes, (iii) Pearson’s correlation of normalized expression, (iv) Spearman’s rank-order correlation of normalized expression. In bulk RNA-seq data, the Spearman correlation is often preferable to the Pearson correlation because it is more robust against outliers. We found that NEBULA and Pearson’s correlation produced more concordant results with each other than the Spearman correlation in both analyses (Supplementary Data 2). We observed many biological related genes such as LEF1 and CCR7 with TCF7 and NFKBIA and NFKB2 with BCL3 among the top co-expressed genes, respectively. In contrast, the Spearman correlation yielded problematic results. For example, many biologically irrelevant and very low-expression genes such as AC124068.2 showed very significant p-values associated with TCF7. Additionally, in the analysis of BCL3, which was less abundantly expressed than TCF7, 11,821 out of the 14,770 genes had a significant p-value < 1E-7, suggesting strong inflated FPRs using the Spearman correlation. This issue probably resulted from the excessive zeros for the low-expression genes, which led to too many ties in both variables. On the other hand, the Pearson correlation was extremely sensitive to outliers. For example, CX3CR1 and LINC02446 showed very significant p-values (p<2E-7) with BCL3 using the Pearson correlation but were only nominally or even not significant by using NEBULA (Supplementary Data 2). We found that the significant p-value from the Pearson correlation was completely driven by only one outlier cell and it became non-significant after removing this cell (Supplementary Fig. S16). In contrast, NEBULA showed very robust and more accurate co-expression estimates. Furthermore, through the adjustment of these confounders, NEBULA eliminated most ribosomal genes from the top list and thus prioritized more biologically relevant genes (Supplementary Data 2).
Cell-level co-expression analysis of APOE
By utilizing NEBULA, we focused on identifying the genes the expression of which were correlated with the expression of APOE, the strongest genetic risk factor for AD, at the single-cell level. As APOE is abundantly expressed only in astrocytes and microglia cells, we performed the analysis in these two cell types using two large-scale snRNA-seq data sets in the frontal cortex from the ROSMAP, which included 48 and 32 subjects from an elderly non-Hispanic white population, respectively. APOE is known to affect multiple cellular functions and molecular processes in an isoform-specific manner33. The APOE gene is primarily present in the form of one of three main alleles APOE e3, the most common allele; APOE e2, the less frequent and underrepresented in AD-affected subjects, and APOE e4, the relatively frequent and highly overrepresented in AD-affected subjects. Therefore, APOE e2 and APOE e4 are usually considered as, respectively, having a protective or a risk-amplifying role in AD. The molecular and cellular mechanism through which such risk effects are mediated is not understood. One way by which APOE could contribute to such effects is by exerting a differential influence on molecular pathways relevant to the biology of different cell types. NEBULA enables the empirical exploration of this hypothesis by testing (1) whether APOE presents selective functional interaction partners in different cell types, and (2) whether such selection is also influenced by having a given allele. Our results support both possibilities.
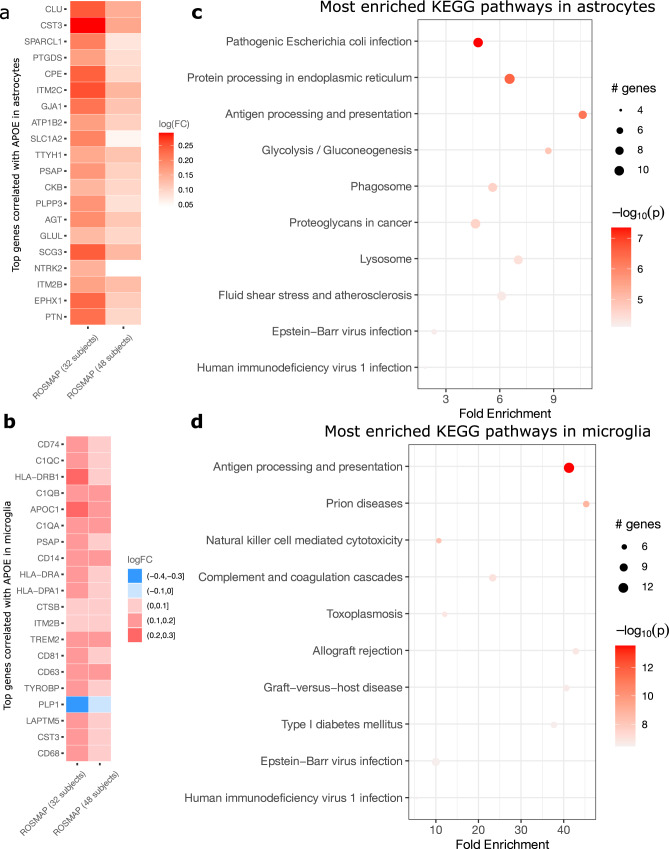
Genes that are highly correlated with the APOE expression are reproducible across the subjects and data sets (Supplementary Data 3). In both data sets, the APOE expression in astrocytes was most strongly correlated with CLU, which is also a genetic risk factor for AD34, on top of other AD-related genes such as CST335 (Fig. 6a). In contrast, APOE was co-expressed in microglia with multiple immune-related genes, particularly TREM2, TYROBP, and members of the complement system including C1QA, C1QB, and C1QC (Fig. 6b), all of which have been implicated in microglia response to amyloid pathology36. We also found examples of recurrent association across cell types. ITM2B, an inhibitor of the amyloid-beta peptide aggregation, was strongly co-expressed with APOE in both cell types, consistent with the role of APOE in both glial cell types in amyloid-beta clearance33. The pathway enrichment analysis revealed that the top co-expressed genes were enriched for pathways involving antigen processing and presentation, prion disease, and the complement system in microglia (Fig. 6d), while APOE-associated pathways in astrocytes included protein processing in the endoplasmic reticulum, and antigen processing and presentation (Fig. 6c), again suggesting both recurrent and cell-type-specific processes mediated by distinct APOE molecular partners. It should be noted that the identified co-expression genes were based on the within-subject expression, thus ruling out effects from subject-level confounders such as AD status. The pathways identified in such a way could be used in downstream analyses to test whether the presence of AD pathology increases or decreases their activity, thus providing a more directed approach as compared, for example, to the conventional GO enrichment analysis, which often leads to insignificant results due to overtesting.
Fig. 6. Analysis of cell-level co-expression of APOE in astrocytes and microglia.
a and b Top 20 genes whose expression in astrocytes (panel a) and microglia (panel b) were correlated with the expression of APOE at the single-cell level. The correlation measured by log(FC) was obtained by NEBULA in two snRNA-seq data sets5,78 in the frontal cortex from the ROSMAP. c and d Top 10 KEGG pathways in which the genes most correlated with APOE in astrocytes (panel c) and microglia (panel d) were enriched. The size of the circles indicates how many genes were found in that pathway. The color indicates the p-value of the enrichment.
Finally, to test the possibility of an allele-specific influence on APOE co-expression, in addition to cell-type specificity, we carried out an isoform-specific co-expression analysis by dividing cells into APOE e3e3, APOE e4+, or APOE e2e3 genotype groups. The co-expression patterns showed substantial differences among the isoform groups in microglia, but not in astrocytes (Supplementary Data 4). Particularly, strong co-expression was observed in the APOE e2e3 cell population with CST3, and the three genes (C1QA, C1QB, C1QC) in the complement system, but not with TREM2 (Supplementary Data 5), which might suggest different biological mechanisms in microglia behind these two APOE isoforms. These preliminary analyses demonstrate how NEBULA can be used to both produce and test hypotheses about gene function in a cell type and isoform-specific fashion, which could complement more in-depth experimental studies.
Discussion
In this work, we propose a fast NBMM for the association analysis of large-scale multi-subject single-cell data. We evaluated and demonstrated the performance of NEBULA using a comprehensive simulation study and multiple droplet-based multi-subject single-cell data sets in the human frontal cortex and PBMC. Overall, our results show that combining methods based on the approximated marginal likelihood and the h-likelihood, NEBULA managed to achieve considerable speed gain and also practically preserve estimation accuracy for analyzing scRNA-seq data. We expect that NEBULA can also be appropriate for other single-cell data such as scATAC-seq data.
The asymptotic efficiency and estimation accuracy of NEBULA-LN are primarily determined by CPS and the cell-level overdispersion. Specifically, our results suggest that was sufficient to produce a sufficiently accurate estimate of the cell-level overdispersion. This means that genes with, e.g., a cell-level overdispersion <10 in a single-cell data set with ~200 cells per subject can benefit from NEBULA-LN. This criterion covers the vast majority of genes in many large-scale real data. On the other hand, a larger CPS value enables a more accurate estimate for a smaller subject-level overdispersion. Accurate estimation requires a larger for the subject-level overdispersion than the cell-level overdispersion and also depends on the CV of the scaling factor and the contribution from the cell-level variables. Compared to a standard h-likelihood method, we see that NEBULA-LN sacrifices some asymptotic efficiency in estimating the cell-level overdispersion, which means that more cells and counts per cell are needed to achieve the same MSEs.
NEBULA is robust in controlling the FPR for testing cell-level variables such as subpopulation marker genes. The simulation study suggests that although the estimated overdispersions for low-expression genes were often less accurate, there was little influence on the FPR. It is possible to treat subjects as batch effects and regress them out in identifying marker genes, but this approach would compromise statistical power if the number of subjects is large. NEBULA is a powerful tool for robustly identifying marker genes. On the other hand, our simulation study suggests that testing a subject-level predictor is sensitive to the estimate of the subject-level overdispersion if the number of subjects is small, which is consistent with a previous simulation study involving only three plates (conceptually equivalent to the subjects)37. We further noticed that when the number of subjects is small, misspecification of the distribution of the random effects may also lead to inflated FPR in testing a subject-level predictor. The downward bias of NEBULA-HL in estimating the subject-level overdispersion is in line with theoretical results for binary data38 and count responses39 for NBLMM. This bias is more severe in the NBGMM and genes having very low expression and a large subject-level overdispersion because the h-likelihood is highly skewed when the response is sparse. The efficient higher-order LA method that we developed substantially alleviated this issue. If the computational efficiency is a major concern, we found that using the PGMM followed by a sensitivity analysis for top findings could be a fast and viable option for testing a subject-level predictor. Another strategy for testing a subject-level variable is to pool all cells of each subject and treat the data as a bulk RNA-seq data set (i.e., the pseudo-bulk method37,40). One potential disadvantage of this strategy is that it cannot adjust for cell-level covariates such as mitochondrial and ribosomal gene concentration to increase the statistical power. The pseudo-bulk method might also be underpowered when the CPS values are imbalanced across the subjects41.
The trend of higher average cell-level overdispersion in low-expression genes is in line with those reported in bulk RNA-seq data6,7. A justification for this observation unique to scRNA-seq data can be that low-expression genes might have a higher drop-out rate or have larger variation across subpopulations. In bulk RNA-seq data, a large proportion of overdispersion results from heterogeneous cell type composition across the individuals. While this heterogeneity is much better controlled in the scRNA-seq data, subpopulation composition still accounts for most of the cell-level overdispersion. The observed smaller cell-level overdispersion in abundant genes might be attributed to a lower drop-out rate and stable expression across subpopulations. In contrast, the subject-level overdispersion is explained by many known covariates. With increased gene expression, the cell-level overdispersion drops while the subject-level overdispersion stabilizes. More work is needed to examine to what extent the subject-level overdispersion is explained by the effects of cis-eQTLs and whether eGenes (i.e., genes having ≥1 eQTL) show a larger subject-level overdispersion.
The co-expression of APOE in astrocytes and microglia is intriguing. Among the top co-expressed genes, multiple SNPs in CLU and TREM2 are AD GWAS loci34,42–44. CLU, CST3, and ITM2B are involved in regulating beta-amyloid production or its fibril formation45–49. CST3 and ITM2B are culprits implicated in hereditary cerebral amyloid angiopathy50. The co-expression results in astrocytes suggest that APOE might be involved in the same biological pathway of these genes regulating beta-amyloid production. On the other hand, the co-expression results in microglia support a recent finding51 that APOE is involved in the immune system through the complement system, particularly C1q. We further found that this co-expression pattern is APOE2+ and APOE4+ dependent in microglia, which might provide more insights into the different roles of APOE isoforms in AD. These results suggest that the effect of APOE on AD is related to regulating both beta-amyloid deposition and the immune system. It should be noted that, because of the data sparsity in the scRNA-seq data, a drawback of this co-expression analysis is that there is little statistical power to identify low-expression genes because the counts of these genes have very small variation. Therefore, no observed correlation does not necessarily mean that the two genes are not co-expressed. Interpretation of the co-expression results for low-expression genes should be cautious. Overall, our analyses of APOE co-expression demonstrate how NEBULA can be used as a tool to explore gene function in a cell type and isoform-specific fashion or to identify modules of co-regulated genes while correcting for subject-level cofounders. Both of these approached can complement more in-depth experimental studies to mechanistically dissect the robust observation detected in human tissue.
Methods
The model specification in NEBULA
Consider a raw count matrix of cells from individuals in a single-cell data set. We choose to use an NBM to take into account the uncertainty originating from the Poisson sampling process, biological effects, and technical noise. Denote by the raw count of a gene and by a row vector of fixed-effects predictors and the intercept () of cell ( and ) in individual (). We model the raw count using the following NBMM
| 1 |
where is a negative binomial distribution parametrized by its mean and cell-level dispersion parameter such that the variance is , is a vector of the coefficients of the predictors, are subject-level random effects capturing the between-subject overdispersion, and is a known cell-specific scaling factor reflecting the sequencing depth, which can be the library size or some global normalizing factor (e.g., ref. 52). Note that can also be gene-specific as recent studies show that a single normalizing factor might be insufficent to normalize scRNA-seq data53. The negative binomial log-linear mixed model (NBLMM) proposed in ref. 54 assumes a log-normal distribution for the random effects , i.e.,
| 2 |
where is the variance component of the between-subject random effects.
The estimation of the NBLMM poses a computational challenge because the marginal likelihood is intractable after integrating out the random effects . Booth et al.54 originally propose an estimation method based on an Expectation-Maximization (EM) algorithm. As the NBLMM falls in the framework of the GLMM, conventional estimation methods for the GLMM can also be applied, including penalized likelihood-based methods12,13, Laplace approximation16, AGQ24,25. These methods, implemented in the lme4 R package18 through the argument nAGQ, differ in how to approximate the high-dimensional integral in the marginal likelihood, among which the penalized likelihood-based method is the fastest but the least accurate while AGQ is more accurate and much slower. We refer to ref. 55 for more extensive reviews of these methods. More recently, Zhang et al.17 propose an estimation procedure for the NBLMM by transforming the problem into repeatedly fitting a linear mixed model using iteratively reweighted least square. All these methods involve at least a two-layer optimization procedure. Although the time complexity is under the assumption of the independent random effects, the two-layer procedure often requires hundreds of iterations to converge, which becomes the major computational bottleneck. On the other hand, Bayesian methods such as MCMC26 offer better accuracy, but are even slower than the above methods.
The rationale behind NEBULA is to skip the time-consuming two-layer optimization algorithm for as many genes as possible and to substantially reduce the number of iterations if such an algorithm has to be applied. The key idea is to derive an approximate marginal likelihood in a closed-form so that the model can be estimated using a simple Newton-Raphson (NR) or quasi-Newton method. Inspired by ref. 28, instead of the log-normal distribution in eq. (2), we assume that the between-subject random effects follow the following gamma distribution
| 3 |
where and are the shape and rate parameters, respectively. One advantage of this parametrization is that it matches the first two moments to the NBLMM, so that even in a situation where eq. (2) is the true distribution of the random effects, Eq. (3) can still provide an accurate estimate of when it is not large28. We call the NBMM with the random effects defined in Eq. (3) as a negative binomial gamma mixed model (NBGMM). Under the NBGMM, the marginal mean and variance of are given by
| 4 |
from which we see that the first overdispersion term in Eq. (4) is controlled by the between-subject variance component alone, and the other includes both and (The derivation can be found in Supplementary Note A.1). Previous studies20,56,57 indicate that it has practically little influence on the estimate of fixed-effects predictors, particularly at the cellular level, to assume a log-normal, gamma, or even misspecified distributions for the random effects. Nevertheless, misspecified random-effects distribution has a larger impact on the estimate of subject-level predictors if the number of subjects is small. Our real data analysis (Fig. 5c and Supplementary Fig. S14) showed concordant results with these studies. In general, the marginal likelihood of the NBGMM after integrating out cannot be obtained analytically. However, we show in the next section how to achieve an approximated closed form of the marginal likelihood when is large. We first focus on the derivation in an asymptotic situation, and then investigate its practical performance in a finite sample.
Approximation of marginal likelihood in NEBULA-LN
We first decompose the NBGMM defined by Eq. (1) and Eq. (3) into a mixture of Poisson distributions as follows
| 5 |
where has the mixing gamma distribution. We then switch the order of the integral in the marginal likelihood to first integrate out . The PGMM proposed in ref. 28, which has a closed-form likelihood, is a special case of the NBGMM if we assume in Eq. (5). As is a conjugate random effect for the Poisson model58, a partial marginal likelihood after integrating out for the cells in individual can be expressed explicitly as
| 6 |
where is the gamma function, , and is the gamma density function defined in the NBGMM. It is not trivial to solve the high-dimensional integral in Eq. (6) because of the entanglement of in, but when , we expect to obtain a simple approximation of the summation in asymptotically by the law of large numbers (LLN). With this notion in mind, we rewrite in Eq. (7) as
| 7 |
where . Now consider a Monte Carlo method to approximate the integral in Eq. (6) by sampling a large number of from and summing up the integrand evaluated at these . We show in Supplementary materials A.2 that under very mild conditions, given any , we have for almost all Monte Carlo samples when , that is, in probability. Given this observation, applying a first-order Taylor expansion to the logarithm in Eq. (7), we can approximate the integral in Eq. (6) (See Supplementary Note A.3 for the derivation) by
| 8 |
where . The integrand in Eq. (8) is now recognized as the kernel of a gamma distribution with respect to . Calculating this integral explicitly and substituting it into Eq. (6) give an approximated marginal log-likelihood
| 9 |
Thus, we may estimate by optimizing , leading to an M-estimator59–61. Compared to the algorithms based on the LA, the optimization procedure of , described in a later section, becomes straightforward because the analytical form is available. As zero counts are prevalent in single-cell data, the evaluation of the log-gamma function in can be saved for most cells. We obtain estimating equations by taking the first derivative of (see Supplementary Note A.4), through which we show in Supplementary Note A.5 that provides an asymptotically consistent estimate of when and . The consistency is also supported by the results in our simulation study described in the Results section.
As is never achieved in real data, we then investigate the performance of using in single-cell data with finite . The major question is under which conditions the method works well and how large is required to produce an accurate estimate of and . We see from Eq. (7) that the accuracy of depends on how close is to zero, and it becomes an exact maximum likelihood estimate (MLE) if this term equals zero. By the Lindeberg central limit theorem, under mild conditions, follows a zero-mean normal distribution with a variance at the rate of
where is the CV of the scaling factor under the null hypothesis that cell-level variables have no fixed effects. Under an alternative hypothesis, the CV should also include the contribution from cell-level variables (see Supplementary Note A.6 for the detailed derivation). Intuitively, the estimation accuracy of NEBULA-LN should depend on the dominant term when is large, and this relationship was supported by the simulation study presented in the Results section by comparing the estimates between NEBULA-LN and NEBULA-HL, which uses an h-likelihood method (described in the next section). Interestingly, we find that a good estimate of in terms of MSE can be achieved even if is as small as 20, where is the CPS value. However, the estimate of requires a larger , and determines the resolution of estimating small . The simulation study in the Results section shows that can empirically provide a good estimate of as small as ~0.02 and ~0.005, respectively. As shown in the analysis of the droplet-based scRNA-seq data5, only ~1% of genes, all of which are very low-expression genes, have very large cell-level overdispersion () in the exitatory neurons. Consequently, NEBULA-LN alone is sufficient for most genes when CPS > 200. The standard error (SE) of can be obtained by the sandwich covariance estimator62. We find that the SE based on the Hessian matrix of alone is practically accurate when is large. Instead, for better accuracy and efficiency, we choose to use the h-likelihood for computing its SE.
Estimation using h-likelihood in NEBULA-HL
We use NEBULA-LN as a first-line solution to estimate and . If or is smaller than a predefined threshold, we then resort to NEBULA-HL to obtain a more accurate estimate. NEBULA-HL is based on an h-likelihood method63 requiring a two-layer iterative procedure. To derive the h-likelihood, we switch the integral order in and first integrate out , which leads to
| 10 |
The h-likelihood method first optimizes the integrand in Eq. (10) with respect to in the inner loop. Note that cannot be optimized with directly because they are not in the canonical scale63. We thus convert the integrand in Eq. (10) to an h-likelihood by re-parametrizing the integral using , which leads to the h-likelihood for individual
| 11 |
We optimize by calculating its first and second derivatives of Eq. (11) (see Supplementary materials A.7 for more detail). As the log-h-likelihood is concave (both the negative binomial term and the penalizing term are concave), an NR algorithm practically converges fast and properly. The submatrix corresponding to in the Hessian matrix is diagonal, so the computational time for solving the inverse Hessian matrix is (m), which is ignorable. We also use this step to compute the SE of in NEBULA-LN by plugging in estimated from .
Given the current estimate , we then optimize using the following marginal log-likelihood
| 12 |
where is the logarithm of the absolute value of the determinant of the second derivative with respect to evaluated at . The second term in Eq. (12) comes from the first-order LA and replacing with gives a restricted maximum likelihood (REML) estimate of . Similar to lme418 and coxmeg64, we use the derivative-free algorithm bobyqa65 implemented in the nloptr R package66 for the optimization of . The number of iterations in this step grows rapidly with the dimension of the parameters. To reduce the iterations, we plug in in as we find that from NEBULA-LN is accurate under mild conditions (e.g., ). Thus, for most genes estimated by NEBULA-HL, we only perform the optimization for , which substantially reduces the number of iterations. In most cases, the algorithm can reach convergence within ~10 iterations.
The first-order LA proposed in Eq. (12) generally produces accurate estimates (as shown in Fig. 2). It, however, underestimates the subject-level overdispersion unignorably when the average count per subject is ≤2 (Supplementary Fig. S17). We find that this problem arises because the h-likelihood of the NBGMM becomes highly skewed if the counts among the cells are very sparse. This issue is unique to the NBGMM because the penalizing term in the h-likelihood of the NBGMM is exponential while it is quadratic in the NBLMM (Supplementary Fig. S18). We, therefore, developed an efficient higher-order LA method67,68 to correct for this skewness for analyzing low-expression genes (See Supplementary Note A.9). This correction greatly improved the accuracy of the estimated subject-level overdispersion for low-expression genes (Supplementary Fig. S17).
Computational implementation of NEBULA-LN
We assessed three algorithms for optimizing in Eq. (8), including (i) the L-BFGS algorithm69 with box constraints implemented in nloptr66, (ii) the NR algorithm implemented in the nlm R function70, and (iii) a trust-region algorithm71 implemented in the trust R package (https://cran.r-project.org/web/packages/trust/index.html). The NR and the trust region algorithms require the Hessian matrix of . Inspired by72, we derive an approximate Hessian matrix (see Supplementary Note A.8) by taking the expectation for some elements to simplify the computation. As are restricted to be positive values, we reparameterize them using logarithm for the unconstrained optimization. In contrast, the L-BFGS algorithm requires only the estimating equations. We find that the NR algorithm using nlm is generally the fastest, and often converges within 10 iterations. However, the NR algorithm may not converge well for low-expression genes with a small CPC value, and occasionally the convergence depends on the initial values. In contrast, the L-BFGS and trust region algorithms are often slower, but much more robust, and both algorithms produce highly consistent estimates. The trust region algorithm takes fewer iterations, but can be slower than the L-BFGS algorithm when the dimension of becomes larger. This is because the computation of the Hessian matrix is more computationally expensive with an increasing number of the parameters. As most genes in single-cell data have low-expression or zero counts, the evaluation of most log-gamma, and digamma functions in the marginal likelihood and the estimating equations can be saved or replaced using a recursive formula.
Processing of the scRNA-seq data in PBMC
The scRNA-seq UMI raw count data in PBMC from the peripheral blood of 11 subjects (six healthy subjects and five patients with multiple sclerosis (MS) from a multiethnic population ranging from age 20 to 40) were obtained from the authors of ref. 27. The original count matrix contained 33,538 genes and 72,600 cells. In the QC step, we excluded outlier cells that satisfied any of the following criteria: total counts >5 median absolute deviations (MADs) away from the median, total features <100, total features >5 MADs, the percentage of counts from mitochondrial genes >5 MADs, and the percentage of counts from ribosomal genes <10%. We included 69,516 cells that passed the QC step in the downstream analysis. Clustering was performed using a routine provided by the Seurat package73. Specifically, the raw count matrix was normalized, highly variable genes were identified, and the normalized data were then scaled. Principal component (PC) analysis was performed on the scaled data and the top 20 PCs were selected based on an elbow plot of the eigenvalues for clustering and visualizing the cells using UMAP74.
Analysis of marker genes and cell-type annotation
For each of the clusters in the PBMC scRNA-seq data, we built a binary variable of whether the cell belongs to this cluster. We performed a differential expression analysis of this binary variable using NEBULA, in which we included the number of features, the percentage of mitochondrial and ribosomal genes as covariates, and subjects as random effects. In each cluster, we defined marker genes as those with CPC > 1, the logarithm of fold change (log(FC)) > 0.2, and p < 1e-50 in the differential expression analysis. These criteria ensured that these marker genes had abundant expression and showed significantly higher expression in that cluster than in the remaining clusters. We then ranked them according to their log(FC) and used the top 40 genes (Supplementary Data 6) as an input to scCATCH29 to annotate these clusters (Some clusters had <40 marker genes based on the above criteria). In the first-round annotation, we used “Blood” as the tissue-specific cell taxonomy reference in scCATCH. The results showed that this reference database did not have refined cell types within T cells (e.g., naive or memory). Therefore we performed a second-round annotation using “Peripheral blood” as the reference for those clusters annotated as T cells in the previous round to further classify naive and memory T cells.
Analysis of cell-sorting bulk RNA-seq data
The cell-sorting bulk RNA-seq data for naive CD4+ and CD8+ T cells were downloaded from the DICE (https://dice-database.org/downloads). The expression data sets for the two cell types included 103 and 104 healthy subjects, respectively, and were given as transcripts per million (TPM). We added a constant of 0.01 and used the log transformation of TPM as the response variable. As no information was available to match the subjects between the two cell types for each gene, we performed a differential expression analysis using simple linear regression with the cell type as the explanatory variable.
Processing of the snRNA-seq data in the frontal cortex
The 48-subject snRNA-seq UMI raw count data from the ROSMAP, including 17,926 genes and 70,634 cells in the human frontal cortex were downloaded from Synapse (https://www.synapse.org/#!Synapse:syn21261143). All cells and genes included in the data passed the QC steps performed in the original study, more details of which are described in ref. 5. To better compare the marker genes identified by NEBULA with those from the original study, we adopted the clustering and cell-type annotation results provided by5, which included 34,976 excitatory neurons, 9196 inhibitory neurons, 18,235 oligodendrocytes, 3392 astrocytes, 1920 microglia, and 2627 OPCs. We did not include pericytes or endothelial cells due to their small sample sizes. To be more stringent, we performed a further QC step using scater75 to remove outlier cells whose total counts or total features were >3 MADs away from the median, which resulted in 69,458 cells. We also removed genes with CPC < 0.1% from the analysis of each cell type (CPC for a gene was calculated as its total UMI counts divided by the number of cells in the cell type). The subcell-type clustering annotation provided in ref. 5 was used in the analysis of marker genes for subclusters. There were 11, 12, 5, 4, 4, and 3 subclusters in the excitatory neurons, inhibitory neurons, oligodendrocytes, astrocytes, microglia, and OPCs, respectively. The percentage of ribosomal genes (RPS and RPL genes) was computed using the PercentageFeatureSet function in the Seurat R package73. Subject-level covariates, including age, sex, AD status, race, and APOE genotype in the ROSMAP project76,77, were downloaded from Synapse (https://www.synapse.org/#!Synapse:syn3157322).
The 32-subject snRNA-seq UMI raw count data from the ROSMAP and its cell type annotation were downloaded from Synapse (https://www.synapse.org/#!Synapse:syn21670836). The original count matrix comprised 33,694 genes and 114,972 cells in the human frontal cortex. We selected the 66,311 cells (14,675 excitatory neurons, 4256 inhibitory neurons, 29,478 oligodendrocytes, 9019 astrocytes, 3986 microglia, 841 endothelial cells, and 3243 OPCs) that passed the original QC step and had cell-type annotation as described in ref. 78. We further excluded 6259 very low-expression genes that showed positive counts in <3 cells.
Co-expression analysis of the scRNA-seq and snRNA-seq data
The cell-level co-expression analysis of BCL3 using 10,476 cells in the memory CD4+ T-cell population (Cluster 1) was performed for 14,770 genes (CPC > 0.05%) using four methods, including NEBULA without adjustment for confounders, NEBULA adjusting for total features, percentage of mitochondrial genes, and percentage of ribosomal genes, the Pearson correlation, and the Spearman correlation. In the two analyses using NEBULA, we first normalized the raw UMI count of BCL3 by the library size of each cell. We then computed the mean of the normalized expression for each subject and subtracted it from the normalized expression. We included this centered expression of BCL3 as the explanatory variable, the raw count of the other gene as the response variable, and 11 subjects as random effects. For the Pearson correlation and the Spearman correlation, we generated the centered expression of both genes and computed the correlation coefficient and its p-value using the cor.test R function.
In the co-expression analysis of APOE using NEBULA, we used the library size as the normalizing factor and included total features and percentage of ribosomal protein genes as the covariates. This is because genes in those cells with larger sequencing depth, more captured genes, or lower ribosomal protein gene expression had a higher co-occurrence rate. We did not include the percentage of mitochondrial genes because their values were very low in the snRNA-seq data and were not associated with the expression of most genes. The subjects were treated as random effects. To build the explanatory variable for the APOE expression, we first normalized APOE expression by dividing the raw count of APOE by its library size of each cell. We then subtracted from it the mean value of the normalized APOE expression across all cells of each subject so that the centered normalized APOE expression did not correlate with subject-level covariates. We only included genes with CPC ≥ 0.1% within each of the cell types. In the isoform-specific analysis, we separated all cells into three categories (e2e3, e3e3, or e4+) based on the APOE genotypes. We then applied the same normalizing and centering procedure as in the co-expression analysis of APOE. The meta-analysis of the summary statistics from the two snRNA-seq data sets was performed using the following fixed-effects model, and , where and are the log(FC) and its standard error of the combined effect, and is the weight for study .
The KEGG pathway enrichment analysis of the top genes correlated with APOE was performed using pathfindR79 with its default setting. In microglia, we used the genes having an FDR p < 0.05 as an input to pathfindR. In astrocytes, we found that no enriched pathway was identified when using all genes with FDR p < 0.05. This is probably because too many significant genes (>700) were included based on this cutoff, which failed to prioritize the top signals. Hence, we instead used the top 200 most significant genes as the input.
Simulation study
We generated the number of CPS using a Poisson distribution for the balanced design and a negative binomial distribution with size = 3 for the unbalanced design. We evaluated scenarios with the number of subjects and the CPS value . The count data were generated based on the following generative model
where we considered ranging from −5 to 2, ranging from 0.01 to 100, and ranging from 0.01 to 1. Under the scenarios of a constant scaling factor, we set , and otherwise we sampled from . We simulated a cell-level variable and a subject-level variable using a standard normal distribution. Under each of the settings, we generated 500 data sets to calculate the summary statistics. The MSE of an estimate (e.g., ) was computed by across the 500 data sets.
Statistics and reproducibility
We compared the computational performance or summary statistics of NEBULA with four commonly used R packages (lme418, glmmTMB19, MASS, and INLA15) for estimating the NBMMs and NBMs. We downloaded the lme4 and glmmTMB R packages via https://cran.r-project.org/ and installed the INLA R package via http://www.r-inla.org/download. In glmer.nb, we assessed the default setting (nAGQ = 1), which is based on the LA16, and a faster but less accurate setting (nAGQ = 0), which is based on a penalized likelihood method13. The difference is that the LA method estimates the fixed effects in the marginal likelihood together with the variance components, while the latter estimates the fixed effects with the random effects in the penalized likelihood. In glmmTMB, we set family=nbinom2. In INLA, we set family = “nbinomial”, and control.predictor = list(compute = FALSE). We set the other arguments as default. We adopted the L-BFGS optimization algorithm in NEBULA in all comparisons because we found that an NR algorithm70 was generally faster but less robust in terms of convergence. All benchmarks were run in R 3.5 on Windows 10 and Linux, separately. In the comparison with an NBM, we used the glm.nb function in the MASS R package.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank the editor and two anonymous reviewers for their constructive comments and suggestions to improve the manuscript. This research was supported by Grants from the National Institute on Aging R01 AG061853, R01 AG065477, and R01 AG070488 to A.M.K., the National Institute on Health R01 AG058002, U01 MH119509, R01 MH109978, R01 AG067151, R01 AG062335, U01 NS110453, UG3 NS115064, RF1 AG054012, RF1 AG062377 to M.K., and the National Institute on Health U19 AI089992, R25 NS079193, P01 AI073748, U24 AI11867, R01 AI22220, UM 1HG009390, P01 AI039671, P50 CA121974, R01 CA227473 to D.A.H. The funders had no role in study design, data collection, and analysis, decision to publish, or manuscript preparation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Study data were provided by the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago. Data collection was supported through funding by NIA grants P30AG10161 (ROS), R01AG15819 (ROSMAP; genomics and RNAseq), R01AG17917 (MAP), R01AG30146, R01AG36042 (5hC methylation, ATACseq), RC2AG036547 (H3K9Ac), R01AG36836 (RNAseq), R01AG48015 (monocyte RNAseq) RF1AG57473 (single-nucleus RNAseq), U01AG32984 (genomic and whole-exome sequencing), U01AG46152 (ROSMAP AMP-AD, targeted proteomics), U01AG46161(TMT proteomics), U01AG61356 (whole-genome sequencing, targeted proteomics, ROSMAP AMP-AD), the Illinois Department of Public Health (ROSMAP), and the Translational Genomics Research Institute (genomic). Additional phenotypic data can be requested at www.radc.rush.edu.
Author contributions
L.H. conceived, developed, and implemented the algorithms. L.H. designed the simulation study and analyzed the simulated and real data. J.D.V. processed the snRNA-seq data in the frontal cortex and participated in the visualization. T.S. performed cell-type annotation in the PBMC data. M.K., A.K., and D.H. contributed to generating or acquiring the real data and discussing the final results. All authors contributed to the writing of the manuscript.
Data availability
This manuscript was prepared using limited access data sets obtained through Synapse (https://www.synapse.org/#!Synapse:syn3219045, https://www.synapse.org/#!Synapse:syn21670836, https://www.synapse.org/#!Synapse:syn18485175) and dbGaP (accession numbers: phs002222.v1.p1 (MS PBMC), phs001703.v3.p1 (DICE)).
Code availability
The code of NEBULA (v1.1.7) can be downloaded and installed via https://github.com/lhe17/nebula, and archived in Zenodo80. The computational tools used for the data analysis include scCATCH v2.1 (https://github.com/ZJUFanLab/scCATCH), lme4 v1.1-26 (https://cran.r-project.org/web/packages/lme4/index.html), glmmTMB v1.0.2.1 (https://cran.r-project.org/web/packages/glmmTMB/index.html), MASS 7.3-53.1 (https://cran.r-project.org/web/packages/MASS/index.html), INLA v20.04.18 (https://www.r-inla.org/download-install).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Liang He, Email: liang.he@duke.edu.
Manolis Kellis, Email: manoli@mit.edu.
Alexander M. Kulminski, Email: alexander.kulminski@duke.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02146-6.
References
- 1.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein AM, et al. Droplet barcoding for single cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 4.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law CW, Chen Y, Shi W, Smyth G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, et al. UMI-count modeling and differential expression analysis for single-cell RNA sequencing. Genome Biol. 2018;19:70. doi: 10.1186/s13059-018-1438-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi K, Chen Y, Skelly DA, Churchill GA. Bayesian model selection reveals biological origins of zero inflation in single-cell transcriptomics. Genome Biol. 2020;21:183. doi: 10.1186/s13059-020-02103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breslow NE, Clayton DG. Approximate inference in generalized linear mixed models. J. Am. Stat. Assoc. 1993;88:9–25. [Google Scholar]
- 13.Lindstrom MJ, Bates DM. Nonlinear mixed effects models for repeated measures data. Biometrics. 1990;46:673–687. doi: 10.2307/2532087. [DOI] [PubMed] [Google Scholar]
- 14.Ormerod JT, Wand MP. Gaussian variational approximate inference for generalized linear mixed models. J. Comput. Graph. Stat. 2012;21:2–17. doi: 10.1198/jcgs.2011.09118. [DOI] [Google Scholar]
- 15.Rue H, Martino S, Chopin N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. Ser. B Stat. Methodol. 2009;71:319–392. doi: 10.1111/j.1467-9868.2008.00700.x. [DOI] [Google Scholar]
- 16.Tierney L, Kadane JB. Accurate approximations for posterior moments and marginal densities. J. Am. Stat. Assoc. 1986;81:82–86. doi: 10.1080/01621459.1986.10478240. [DOI] [Google Scholar]
- 17.Zhang X, et al. Negative binomial mixed models for analyzing microbiome count data. BMC Bioinforma. 2017;18:4. doi: 10.1186/s12859-016-1441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates, D., Mächler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. Preprint at 10.18637/jss.v067.i01 (2014).
- 19.Brooks ME, et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R. J. 2017;9:378–400. doi: 10.32614/RJ-2017-066. [DOI] [Google Scholar]
- 20.Milanzi E, Alonso A, Molenberghs G. Ignoring overdispersion in hierarchical loglinear models: possible problems and solutions. Stat. Med. 2012;31:1475–1482. doi: 10.1002/sim.4482. [DOI] [PubMed] [Google Scholar]
- 21.Landeghem GV, Fraine BD, Damme JV. The consequence of ignoring a level of nesting in multilevel analysis: a comment. Multivar. Behav. Res. 2005;40:423–434. doi: 10.1207/s15327906mbr4004_2. [DOI] [PubMed] [Google Scholar]
- 22.Moerbeek M. The consequence of ignoring a level of nesting in multilevel analysis. Multivar. Behav. Res. 2004;39:129–149. doi: 10.1207/s15327906mbr3901_5. [DOI] [PubMed] [Google Scholar]
- 23.Hilbe, J. M. Negative Binomial Regression (Cambridge University Press, 2011).
- 24.Pinheiro JC, Bates DM. Approximations to the log-likelihood function in the nonlinear mixed-effects model. J. Comput. Graph. Stat. 1995;4:12–35. [Google Scholar]
- 25.Pinheiro JC, Chao EC. Efficient laplacian and adaptive gaussian quadrature algorithms for multilevel generalized linear mixed models. J. Comput. Graph. Stat. 2006;15:58–81. doi: 10.1198/106186006X96962. [DOI] [Google Scholar]
- 26.Vestal BE, et al. MCMSeq: Bayesian hierarchical modeling of clustered and repeated measures RNA sequencing experiments. BMC Bioinforma. 2020;21:375. doi: 10.1186/s12859-020-03715-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pappalardo, J. L. et al. Transcriptomic and clonal characterization of T cells in the human central nervous system. Sci. Immunol. 5, eabb8786 (2020). [DOI] [PMC free article] [PubMed]
- 28.Sutradhar BC, Qu Z. On approximate likelihood inference in a poisson mixed model. Can. J. Stat. 1998;26:169–186. doi: 10.2307/3315682. [DOI] [Google Scholar]
- 29.Shao X, et al. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23:100882. doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmiedel BJ, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson EH. The interpretation of interaction in contingency tables. J. R. Stat. Soc. Ser. B Methodol. 1951;13:238–241. [Google Scholar]
- 32.Zhang Y, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamazaki Y, Zhao N, Caulfield TR, Liu C-C, Bu G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 2019;15:501–518. doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harold D, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng A, Irizarry MC, Nitsch RM, Growdon JH, Rebeck GW. Elevation of cystatin C in susceptible neurons in Alzheimer’s disease. Am. J. Pathol. 2001;159:1061–1068. doi: 10.1016/S0002-9440(10)61781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasemann S, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lun ATL, Marioni JC. Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostat. Oxf. Engl. 2017;18:451–464. doi: 10.1093/biostatistics/kxw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breslow NE, Lin X. Bias correction in generalised linear mixed models with a single component of dispersion. Biometrika. 1995;82:81–91. doi: 10.1093/biomet/82.1.81. [DOI] [Google Scholar]
- 39.Lin X. Estimation using penalized quasilikelihood and quasi-pseudo-likelihood in Poisson mixed models. Lifetime Data Anal. 2007;13:533–544. doi: 10.1007/s10985-007-9071-z. [DOI] [PubMed] [Google Scholar]
- 40.Crowell HL, et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 2020;11:6077. doi: 10.1038/s41467-020-19894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmerman KD, Espeland MA, Langefeld CD. A practical solution to pseudoreplication bias in single-cell studies. Nat. Commun. 2021;12:738. doi: 10.1038/s41467-021-21038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambert J-C, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 45.Bell RD, et al. Transport pathways for clearance of human Alzheimer’s amyloid β-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow. Metab. J. Int. Soc. Cereb. Blood Flow. Metab. 2007;27:909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeser SA, et al. Cystatin C modulates cerebral beta-amyloidosis. Nat. Genet. 2007;39:1437–1439. doi: 10.1038/ng.2007.23. [DOI] [PubMed] [Google Scholar]
- 47.Kim J, et al. BRI2 (ITM2b) inhibits Aβ deposition in vivo. J. Neurosci. 2008;28:6030–6036. doi: 10.1523/JNEUROSCI.0891-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsubara E, Frangione B, Ghiso J. Characterization of apolipoprotein J-Alzheimer’s Aβ interaction. J. Biol. Chem. 1995;270:7563–7567. doi: 10.1074/jbc.270.13.7563. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda S, et al. The familial dementia BRI2 gene binds the Alzheimer gene amyloid-beta precursor protein and inhibits amyloid-beta production. J. Biol. Chem. 2005;280:28912–28916. doi: 10.1074/jbc.C500217200. [DOI] [PubMed] [Google Scholar]
- 50.Revesz T, et al. Genetics and molecular pathogenesis of sporadic and hereditary cerebral amyloid angiopathies. Acta Neuropathol. (Berl.) 2009;118:115–130. doi: 10.1007/s00401-009-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin C, et al. ApoE attenuates unresolvable inflammation by complex formation with activated C1q. Nat. Med. 2019;25:496–506. doi: 10.1038/s41591-018-0336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bacher R, et al. SCnorm: robust normalization of single-cell RNA-seq data. Nat. Methods. 2017;14:584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Booth JG, Casella G, Friedl H, Hobert JP. Negative binomial loglinear mixed models. Stat. Model. 2003;3:179–191. doi: 10.1191/1471082X03st058oa. [DOI] [Google Scholar]
- 55.Tuerlinckx F, Rijmen F, Verbeke G, De Boeck P. Statistical inference in generalized linear mixed models: a review. Br. J. Math. Stat. Psychol. 2006;59:225–255. doi: 10.1348/000711005X79857. [DOI] [PubMed] [Google Scholar]
- 56.Neuhaus JM, McCulloch CE. Estimation of covariate effects in generalized linear mixed models with informative cluster sizes. Biometrika. 2011;98:147–162. doi: 10.1093/biomet/asq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neuhaus JM, McCulloch CE, Boylan R. Estimation of covariate effects in generalized linear mixed models with a misspecified distribution of random intercepts and slopes. Stat. Med. 2013;32:2419–2429. doi: 10.1002/sim.5682. [DOI] [PubMed] [Google Scholar]
- 58.Molenberghs G, Verbeke G, Demétrio CGB, Vieira AMC. A Family of generalized linear models for repeated measures with normal and conjugate random effects. Stat. Sci. 2010;25:325–347. doi: 10.1214/10-STS328. [DOI] [Google Scholar]
- 59.Huber PJ. Robust estimation of a location parameter. Ann. Math. Stat. 1964;35:73–101. doi: 10.1214/aoms/1177703732. [DOI] [Google Scholar]
- 60.Huber, P. J. Robust Statistics. (John Wiley & Sons, 2004).
- 61.Serfling, R. J. Approximation Theorems of Mathematical Statistics (John Wiley & Sons, 2009).
- 62.Huber, P. J. The behavior of maximum likelihood estimates under nonstandard conditions. In Proc. Fifth Berkeley Symposium on Mathematical Statistics and Probability, (ed. Lucien M. Le Cam, Jerzy Neyman) Vol. 1 (University of California Press, 1967).
- 63.Lee, Y., Nelder, J. A. & Pawitan, Y. Generalized Linear Models with Random Effects: Unified Analysis Via H-likelihood. (Chapman and Hall/CRC, 2006).
- 64.He, L. & Kulminski, A. M. Fast algorithms for conducting large-scale GWAS of age-at-onset traits using cox mixed-effects models. Genetics10.1534/genetics.119.302940 (2020). [DOI] [PMC free article] [PubMed]
- 65.Powell, M. J. The BOBYQA algorithm for bound constrained optimization without derivatives. Report DAMTP 2009/NA06. 26–46 (Centre for Mathematical Sciences, University of Cambridge, UK, 2009).
- 66.Ypma, J. Introduction to nloptr: an R interface to NLopthttps://cran.r-project.org/web/packages/nloptr/vignettes/nloptr.pdf (2014).
- 67.Barndorff-Nielsen, O. E., Cox, D. R. & Cox, H. F. D. R. Asymptotic Techniques for Use in Statistics (Springer US, 1989).
- 68.Raudenbush SW, Yang M-L, Yosef M. Maximum likelihood for generalized linear models with nested random effects via high-order, multivariate laplace approximation. J. Comput. Graph. Stat. 2000;9:141–157. [Google Scholar]
- 69.Byrd R, Lu P, Nocedal J, Zhu C. A limited memory algorithm for bound constrained optimization. SIAM J. Sci. Comput. 1995;16:1190–1208. doi: 10.1137/0916069. [DOI] [Google Scholar]
- 70.Dennis, J. E. & Schnabel, R. B. Numerical Methods for Unconstrained Optimization and Nonlinear Equations (Society for Industrial and Applied Mathematics, 1996).
- 71.Fletcher, R. Practical Methods of Optimization (Wiley, 1987).
- 72.Gilmour AR, Thompson R, Cullis BR. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics. 1995;51:1440–1450. doi: 10.2307/2533274. [DOI] [Google Scholar]
- 73.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2020).
- 75.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinforma. Oxf. Engl. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bennett DA, et al. Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Y, et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 2020;26:131–142. doi: 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ulgen, E., Ozisik, O. & Sezerman, O. U. pathfindR: An R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front. Genet. 10, 858 (2019). [DOI] [PMC free article] [PubMed]
- 80.He, L. NEBULA: a fast negative binomial mixed model for differential or co-expression analysis of multi-subject single-cell data. 10.5281/zenodo.4659374 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
This manuscript was prepared using limited access data sets obtained through Synapse (https://www.synapse.org/#!Synapse:syn3219045, https://www.synapse.org/#!Synapse:syn21670836, https://www.synapse.org/#!Synapse:syn18485175) and dbGaP (accession numbers: phs002222.v1.p1 (MS PBMC), phs001703.v3.p1 (DICE)).
The code of NEBULA (v1.1.7) can be downloaded and installed via https://github.com/lhe17/nebula, and archived in Zenodo80. The computational tools used for the data analysis include scCATCH v2.1 (https://github.com/ZJUFanLab/scCATCH), lme4 v1.1-26 (https://cran.r-project.org/web/packages/lme4/index.html), glmmTMB v1.0.2.1 (https://cran.r-project.org/web/packages/glmmTMB/index.html), MASS 7.3-53.1 (https://cran.r-project.org/web/packages/MASS/index.html), INLA v20.04.18 (https://www.r-inla.org/download-install).