Abstract
The human innate immune system responds to both pathogen and commensal bacteria at the molecular level using bacterial peptidoglycan (PG) recognition elements. Traditionally, synthetic and commercially accessible PG monosaccharide units known as muramyl dipeptide (MDP) and N-glycolyl MDP (ng-MDP) have been used to probe the mechanism of innate immune activation of pattern recognition receptors, such as NOD-like receptors. However, bacterial PG is a dynamic and complex structure, with various chemical modifications and trimming mechanisms that result in the production of disaccharide-containing elements. These molecules pose as attractive targets for immunostimulatory screening; however, studies are limited because of their synthetic accessibility. Inspired by disaccharide-containing compounds produced from the gut microbe Lactobacillus acidophilus, a robust and scalable chemical synthesis of PG-based disaccharide ligands was implemented. Together with a monosaccharide PG library, compounds were screened for their ability to stimulate proinflammatory genes in bone-marrow-derived macrophages. The data reveal distinct gene induction patterns for monosaccharide and disaccharide PG units, suggesting that PG innate immune signaling is more complex than a one activator–one pathway program, as biologically relevant fragments induce transcriptional programs to different degrees. These disaccharide molecules will serve as critical immunostimulatory tools to more precisely define specialized innate immune regulatory mechanisms that distinguish between commensal and pathogenic bacteria residing in the microbiome.
Short abstract
Synthesis of biologically inspired peptidoglycan fragments reveals a complex immune pathway activating transcriptional programs that induce genes associated with inflammatory bowel disease.
Introduction
The human body is responsible for maintaining ∼39 trillion bacterial cells that constitute the microbiome.1−3 The gut microflora is one area of the body teeming with hundreds of species of bacterial cells.4−6 The bacteria in the gastrointestinal (GI) tract are benefactors to the human host, performing essential biological chemical transformations and producing key essential vitamins and amino acids.7−10 While many of these organisms serve to maintain a healthy state for the human host, bacterial pathogenesis disrupts this symbiotic relationship. Dysbiosis in the human microbiome can lead to a variety of inflammatory diseases, including ulcerative colitis and Crohn’s disease (CD), rheumatoid arthritis, GI cancer, and asthma.11,12 Therefore, the host–microbiome interface is an attractive target for therapeutic intervention.13 In order to develop novel immunotherapies and antibiotics, it is critical to fully understand the molecular mechanisms by which nature recognizes and responds to bacteria.
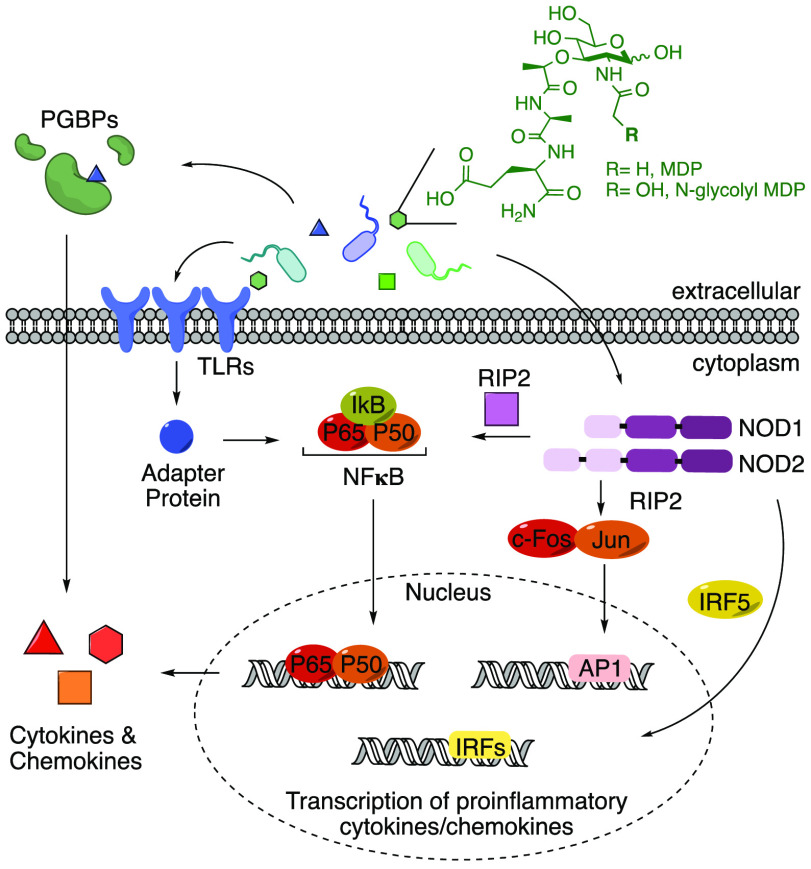
Humans have developed host defense mechanisms to combat infectious diseases, including the innate immune system, the body’s first line of defense against invading pathogens such as bacteria.14 Pattern recognition receptors (PRRs) are programmed in this system to interact with essential components of bacterial cells such as flagella, lipopolysaccharide (LPS), lipoteichoic acids, and bacterial cell wall peptidoglycan (PG) components (Scheme 1).15−18 Since the pioneering PRR discovery, multiple families of PRRs have been classified, such as toll-like receptors (TLRs), C-type lectin receptors (CLRs), NOD-like receptors (NLRs), RIG-I-like receptors (RLRs), AIM2-like receptors (ALRs), peptidoglycan binding proteins (PGBPs), the SLAM family (SLAMF), and OAS-like receptors (OLRs).19−21 How this system provides a tuned innate immune response toward pathogens while ignoring symbiotic microorganisms that constitute the microbiome is not fully understood.
Scheme 1. PRR Signaling.
Common activation pathways of proinflammatory cytokine and chemokine production upon stimulation by molecular signatures known as pathogen-associated molecular patterns (PAMPs) are depicted.15 In the advent of the microbiome, because of similarities between pathogenic and nonpathogenic organisms, these signals are now called microbe-associated molecular patterns (MAMPs).18 Here the model of activation by PG (and synthetic mimics, MDP and ng-MDP) is shown.
The bacterial microbe-associated molecular pattern (MAMP) PG is sensed by a variety of PRRs, including NOD1, NOD2, NLRP3, NLRP1, and PGBPs (Scheme 1).22−33 These ligands are small fragments derived from a large PG polymer that surrounds the bacterial cell, consisting of repetitive units of β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) with short peptide chains containing both l- and d-amino acids present on the muramic acid residue (Figure 1). Although the bacterial cell wall is highly conserved among species, differences arise.34,35 Variations in cross-linking (3–3 vs 3–4 vs 2–4 linkages) as well as substitution of amino acids, primarily at the third position (e.g., meso-diaminopimelic acid (m-DAP), l-Lys, l-Orn, l-Ala, l-Glu, l-homoserine), are observed in both Gram-positive and Gram-negative bacteria (Figure 1, shown in blue).35 Modifications of the carbohydrate backbone of PG have also been identified in a variety of bacterial species, including N-deacetylation in Listeria,36O-acetylation in Helicobacter pylori,37N-glycolylation in Mycobacterium tuberculosis,38 and muramic δ-lactam in Bacillus subtilis,39 all of which block PG lytic enzymatic digestion, leaving the 1,6-anhydro ring40 as the lytic product (Figure 1, shown in red). From this knowledge of PG complexity, one can easily imagine a pool of tunable immunostimulatory fragments that are critical for mediating host–pathogen interactions. However, the small-molecule details in this signaling landscape are incomplete because of the limited amount of biologically significant MAMP PG chemical probes.
Figure 1.
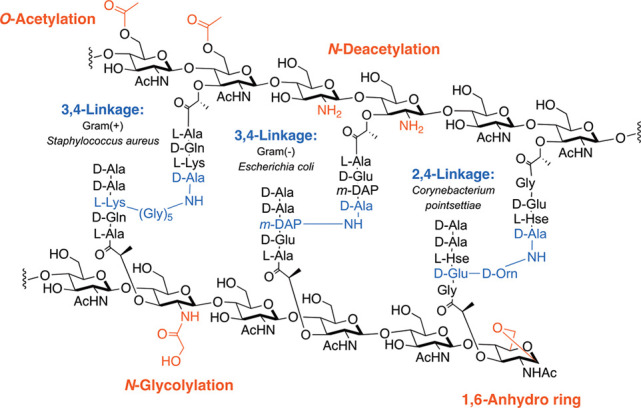
Peptidoglycan structure and modifications. The chemical functionalities of the peptide side chain (blue) and carbohydrate backbone (red) in bacterial PG across species are shown.
Variations in PG structure among bacterial species have led chemists to synthesize a variety of small-molecule probes based on monomeric units of this structure. In particular, many of these building blocks are now commercially available. For example, N-acetyl muramyl dipeptide (MDP) is a representative small-molecule PG mimic for “general” bacteria and has been shown to interact with NLRs, such as NOD2 and NLRP1, which are associated with a variety of diseases including irritable bowel diseases (IBDs) and vitiligo (Scheme 1).24,27,28,31,41,42 In addition, a “modified” bacterial PG fragment, N-glycolyl MDP (ng-MDP), derived from the hydroxylated PG product of Mycobacterium, has been heavily investigated because of the direct link of Mycobacterium paratuberculosis with CD (Scheme 1).43 Both “general” and “modified” PG fragments have been shown to stimulate a potent NOD2-dependent immune response and thus are the primary ligands of choice for immunologists because of the ligand synthetic simplicity and commercial availability (Scheme 1).24,44,45
However, MDP and ng-MDP represent only a defined element of the PG fragment pool and fail to capture the major PG hydrolase degradation products, disaccharide muropeptides.46,47 These disaccharide PG fragments are synthetically complex, limiting their accessibility and restricting their analysis in traditional immunostimulatory assays such as NFκB luciferase-reporter and ELISA screens.26,41,48−50 Questions in the field surrounding the natural ligand of NLRs, like NOD2, as well as the biological influence of other N-acetyl muramic acid-containing PG fragments still remain unanswered without an expanded cell wall PG library.51 In this study, disaccharide PG-based fragments from the gut microbe Lactobacillus acidophilus(52,53) were identified. Inspired by the generation of these PG products, a reliable and scalable synthetic route to several PG disaccharide fragments was implemented, leading to the first fully characterized N-acetylglucosamine N-acetylmuramic acid tripeptide (GMTP). These disaccharides were combined with a library of monosaccharide PG derivatives and screened for gene transcription activation, cytokine production, and phosphorylation profiles using bone-marrow-derived macrophages (BMDMs). Interestingly, GMTP was discovered to be a more potent activator for select gene transcriptional programs, many of which are related to IBD, compared with their monosaccharide counterparts. Measurements of cytokine production and downstream phosphorylation programs complement these genetic studies. Gene expression profiles comparing the disaccharide to the monosaccharide fragments reveal a complex innate immune signaling pattern, validating a highly intricate molecular mechanism for sensing of PG fragments.
Results and Discussion
Identification of Disaccharide PG Fragments from L. acidophilus Cultures
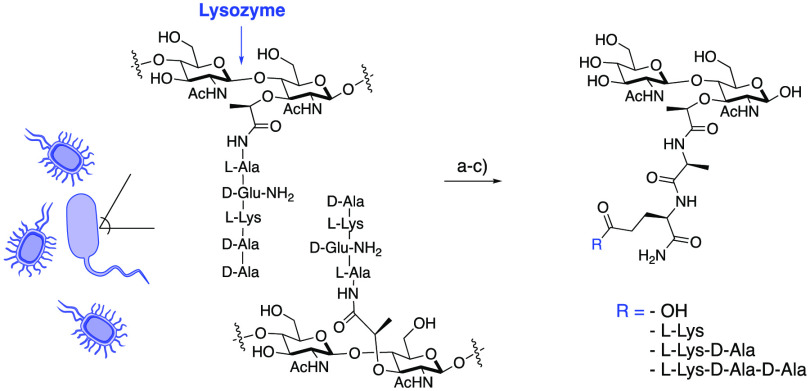
Previous exploration of Gram-negative and Gram-positive bacteria has led to the identification of several disaccharide PG fragments that activate an immune response.24,26,42,54−57 In order to screen for biologically relevant fragments from Gram-positive bacteria residing in the gut microbiome, a lysozyme degradation assay of L. acidophilus was implemented (Scheme 2). L. acidophilus is a major commensal inhabitant of the human microbiota in intestinal, oral, and vaginal tracts.58 Since the 1970s, L. acidophilus has been commercially produced as a probiotic with reported therapeutic effects.53,59,60 To screen this organism for PG small molecule production, L. acidophilus cultures were treated with lysozyme,61 a muramidase found in high concentrations in both the human mouth and gut.62 The lysed PG was then subjected to high-resolution liquid chromatography–mass spectrometry (HR-LCMS) analysis, and four disaccharide PG fragments were identified: N-acetylglucosamine N-acetylmuramic acid dipeptide (GMDP), N-acetylglucosamine N-acetylmuramic acid tripeptide (GMTP), N-acetylglucosamine N-acetylmuramic acid tetrapeptide (GMTTP), and N-acetylglucosamine N-acetylmuramic acid pentapeptide (GMPP) (Scheme 2). GMTP was observed in high abundance compared with other disaccharide fragments (SI-Table 2, entry 2, in Supporting Information 1 (SI1)). This result is in agreement with previously published work in which GMDP, GMTP, GMTTP, and GMPP were identified with variations at the third amino acid residue.42,52,56,61,63−65 However, lack of accessibility to these disaccharides because of the complexity of chemical synthesis has prevented a rigorous investigation of their immunological activity. This motivated the synthetic development of well-characterized disaccharides in larger and more accessible quantities.
Scheme 2. Lysozyme-Induced PG Degradation of L. acidophilus.
L. acidophilus produces several disaccharide products. (a) Whole bacterial cells were treated with lysozyme. (b) Lysed PG was subjected to a 3 kDa spin filter. (c) PG fragmentation was analyzed by high-resolution liquid chromatography–mass spectrometry (HR-LCMS), and four fragments were identified (SI-Table 2). The assay was replicated on three occasions. None of the identified masses were observed in the controls (medium, wash buffer, and enzymatic treatment buffer).
Synthesis and Characterization of Disaccharide PG Fragments
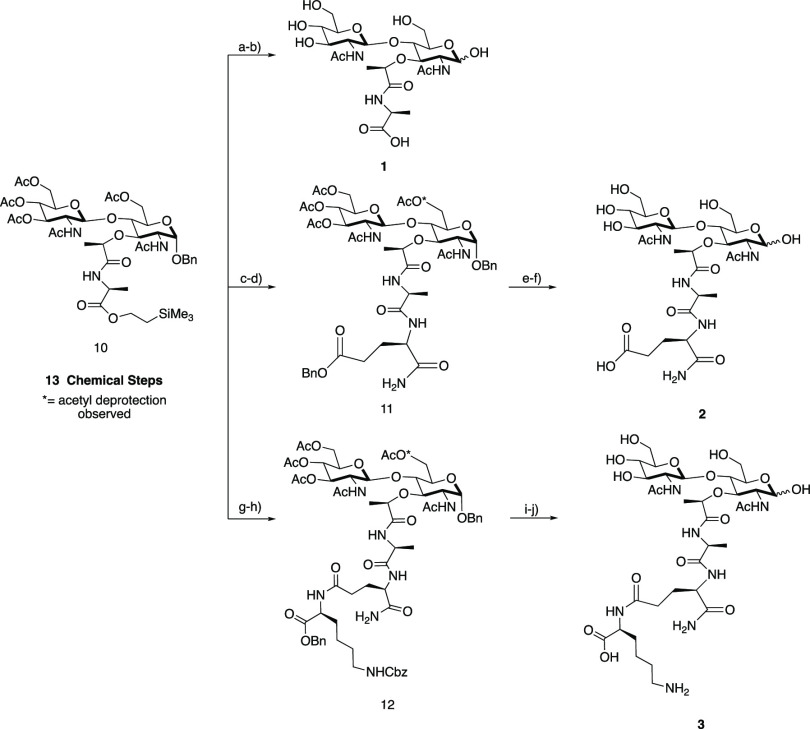
A total synthesis using Schmidt glycosylation was implemented to obtain protected β-1,4-linked intermediate 10 over 13 chemical transformations.42,66,67 From this intermediate, a modular strategy was utilized to access PG fragments 1–3 (Scheme 3). GMMP (1) was first produced through global deprotection of the acetyl protecting groups and the (trimethylsilyl)ethyl ester (TMSE) of compound 10 followed by direct hydrogenation with 20% Pd(OH)2. GMDP (2) was obtained by deprotection of TMSE with 1 N tetra-n-butylammonium fluoride (TBAF) followed by direct coupling of d-isolgn-OBn to yield intermediate 11, deacetylation of 11 using aqueous LiOH, and hydrogenation with 20% Pd(OH)2. Finally, to obtain GMTP (3), by a slightly modified synthesis the protected dipeptide d-isolgn-l-Lys(Z)-OBzl was synthesized over two steps starting from commercially available H-Lys(Z)-OBzl and Boc-d-glutamic acid α-amide.68 Then deprotection of TMSE from 10 followed by direct coupling to d-isolgn-l-Lys(Z)-OBzl yielded 12, which was then deprotected in two subsequent steps to generate the final product 3. To ensure purity for biological testing, all of the final compounds were purified via reversed-phase chromatography using a mass-directed autopurification system. From intermediate 10, compounds 1–3 were obtained in overall yields of 56% (two steps), 9% (four steps), and 21% (four steps), respectively. Full NMR characterization and spectral data are presented for each synthetic compound in the SI1; such data have been unavailable to date for synthetic fragment 3.50
Scheme 3. Synthesis of PG Fragment Disaccharides.
Intermediate 10 was synthesized over 13 chemical steps. GMMP (1): (a) LiOH, ACN/H2O; (b) 20% Pd(OH)2, H2, THF/H2O; 56% yield over two steps. Compound 11: (c) 1 N TBAF in THF; (d) DIPEA, HBTU, HOBt, d-isogln-OBn, DMF; 24% yield over two steps. GMDP (2): (e) LiOH, ACN/H2O; (f) 20% Pd(OH)2, H2, THF/H2O; 37% yield over two steps. Compound 12: (g) 1 N TBAF in THF; (h) DIPEA, HBTU, HOBt, d-isogln-l-Lys(Z)-OBzl, DMF; 58% yield over two steps. GMTP (3): (i) LiOH, ACN/H2O; (j) 20% Pd(OH)2, H2, THF/H2O; 37% yield over two steps.
The assignment of 3 was confirmed utilizing a variety of 2D NMR experiments, including 1H–13C HSQC-TOCSY, 1H–1H COSY, 1H–13C HSQC, and 1H–13C HMBC (Figure 2 and SI1). Upon hydrogenation, the anomeric hydroxyl mutarotates into a mixture of two α/β isomeric species, with the α anomer being favored. Through detailed NMR experiments, the saccharide residues and peptide chains were elucidated, confirming the structure and purity of the synthetic PG fragments.
Figure 2.
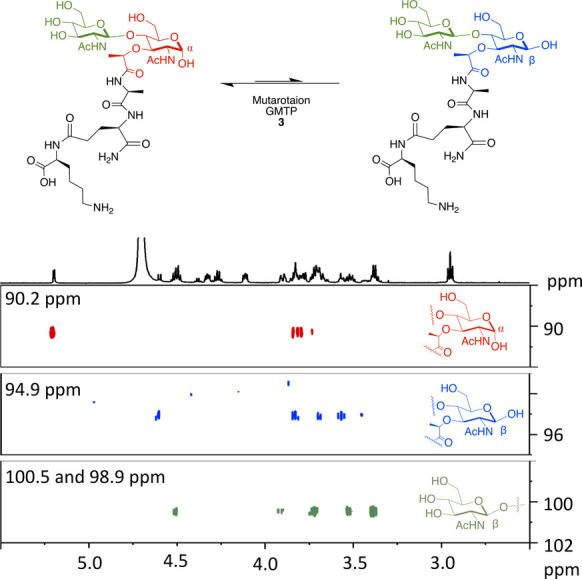
1H–13C HSQC-TOCSY spectra of GMTP (3) with magnification of the anomeric region for the major and minor isomer carbohydrate ring spin systems. Spectra were recorded on a 600 MHz Bruker NMR spectrometer at 298 K in D2O with the following settings: D9 = 0.12; O1P = 3 ppm; O2P = 60 ppm; SW= 6 ppm; 1SW = 100 ppm; D1= 2 s; NS= 24.
Genome-Wide Transcriptional Analysis Reveals PG-Regulated Target Genes
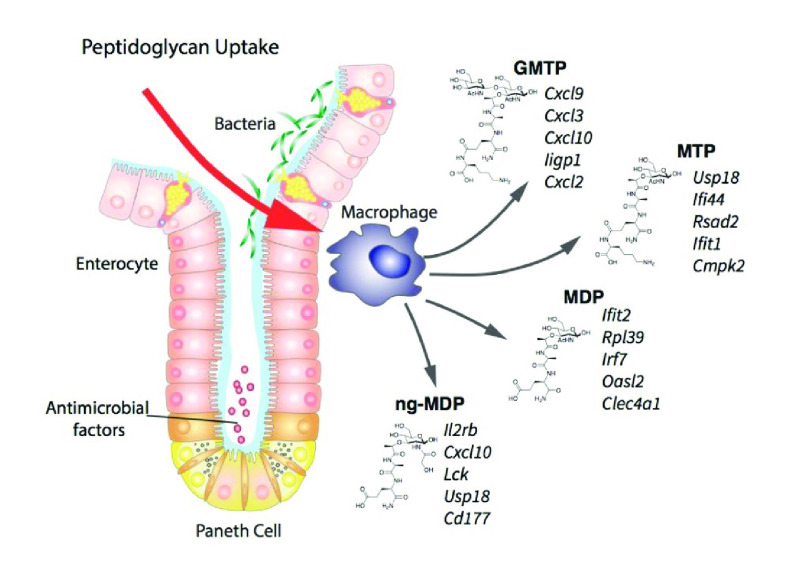
With access to large quantities of compounds 1–3, an investigation of the regulation programs in mouse BMDMs in response to both mono- and disaccharide PG fragments was implemented. To investigate gene regulation mediated by PG units, a qRT-PCR assay was first utilized to analyze the expression of immune response indicator genes. A subset of nine compounds containing both synthesized disaccharides 1–3 and six additional monosaccharides were used to stimulate macrophages (SI-Figure 1). In this initial investigation, target genes were selected on the basis of previous RNA sequencing analysis with ng-MDP.69 From this screen, the mRNA expression levels of the tested genes increased significantly in BMDMs treated with GMTP compared with MDP and ng-MDP (Figure 3A,B and SI-Figure 2). Gene expression regulation was observed after 4 h of stimulation with 20 μM compound treatment (Figure 3A,B and SI-Figure 2). Importantly, this upregulation was not observed when GlcNAc was removed from the GMTP structure to yield the monosaccharide N-acetylmuramic acid tripeptide (MTP), demonstrating that the β-1,4-linked GlcNAc residue plays a critical role in the observed gene activation (SI-Figures 1 and 2). Additionally, the third amino acid, lysine, could play a critical role in generating a significant gene response, as other disaccharide fragments such as GMMP and GMDP did not activate as robustly (Scheme 2 and SI-Figure 2). Upon further investigation, GMTP was determined to activate Tnf-α and Cxcl10 significantly more than MDP and ng-MDP (Figure 3B and SI-Figure 3). Il-1β and Cox2 transcripts were also potently induced by GMTP, whereas MDP had no activation (Figure 3B). These results showcase for the first time a specific disaccharide PG unit that is capable of initiating gene expression programs differently than the previous monosaccharide PG standards, MDP and ng-MDP.
Figure 3.
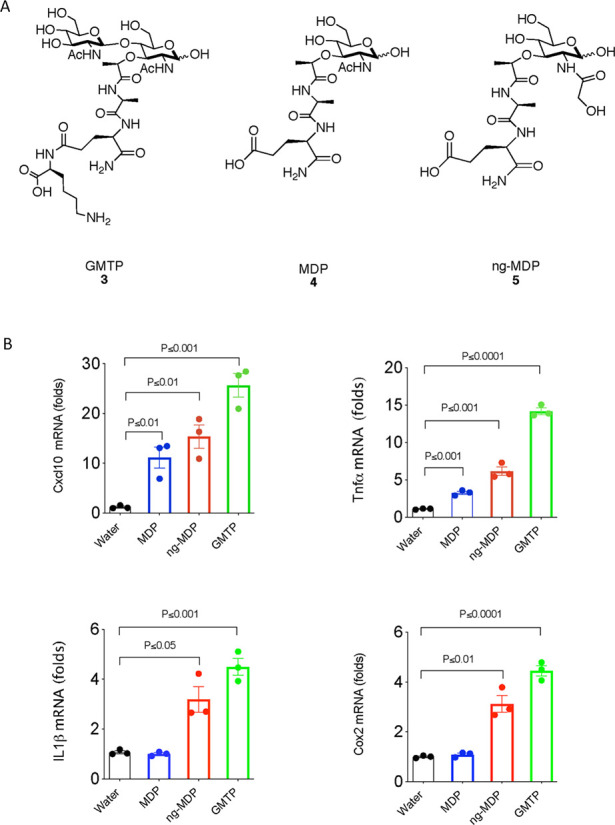
GMTP induces proinflammatory cytokine gene production. (A) Molecular structures of compounds GMTP (3), MDP (4), and ng-MDP (5). (B) Gene expression statistical analysis of qRT-PCR data for Tnfα, Il-1β, Cox2, and Cxcl10 in BMDMs treated with 20 μM 3, 4, or 5 or control (water) for 4 h. Total RNA was harvested, and the expression levels of selected genes were analyzed by qRT-PCR. Individual ΔΔCT values are shown in SI-Table 1. Error bars indicate mean ± SEM. Statistical significance was calculated using the two-tailed Student’s t test (n = 3).
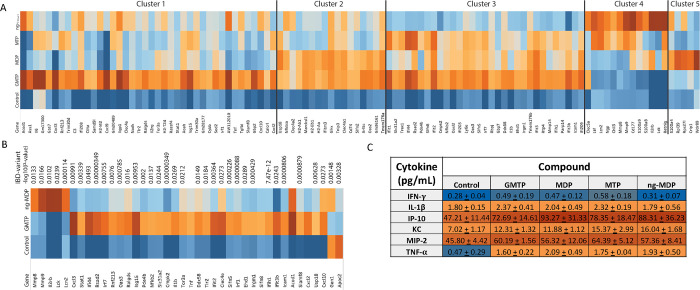
To more precisely understand the transcriptional programs upon PG fragment stimulation and measure the differences observed in the qRT-PCR analysis, a whole-genome RNA sequencing (RNAseq) study was performed on BMDMs treated for 18 h with 20 μM GMTP, MDP, MTP, or ng-MDP (Figure 4A and SI-Figure 1). Gene regulation in unstimulated (water) BMDMs was used as a control. DESeq2 and intensity difference analysis70,71 revealed significantly regulated genes in each stimulation group, which were combined for gene set enrichment and hierarchical clustering in Seqmonk (Supporting Information 2 (SI2), Tables 1–8). Here we show that GMTP has significantly enhanced immune stimulating capacity compared with MDP, MTP, and ng-MDP (Figure 4A and SI2 Table 9). GMTP induced a unique gene expression signature distinct from that of MDP (Figure 4A and SI2 Tables 1, 2, 5, and 6). Hierarchical clustering (HC) analysis identified genes primarily induced by GMTP, such as Acod1, Ass1, Il6, Gm17300, and Cx3cl (Figure 4A, cluster 1). Transcriptional responses to GMTP and MDP similarly induced genes of cluster 2, including Slfn2, Arl5c, Oas2, Gm36161, and Tmem176a (Figure 4A). In contrast, expression of S100a4, Rpl39, Rpns27l, Crip1, and Wdr89 was specifically induced by MDP (and to a lesser extent ng-MDP) but not the other PG fragments (Figure 4A, cluster 5; SI2 Tables 1–9). MTP was less efficient in upregulating genes that were characteristically induced by GMTP in cluster 1, consistent with the finding that the GlcNAc component of GMTP is critical for gene activation (SI2 Tables 5 and 7). ng-MDP, the “modified” PG fragment, induced a strong response of a cluster of genes that included Ltf, Lcn2, Ngp, Chil3, Mmp8, Mmp9, Cd177, S100a9, S100a8, Il2rb, Lck and Retnlg (cluster 4), but GMTP did not (SI2 Tables 4 and 8), indicating a monosaccharide-specific expression pattern. Genes in cluster 3 of the analysis were induced by GMTP, MDP, and MTP (but not the modified PG ng-MDP), representing a core “general” PG response signature that includes Il1b, Irf7, Slfn5, Slfn8, Icam1, Ifit2, Ifit3, Oas3, Rsad2, Ifi209, and Tmem176b (Figure 4A). Overall, macrophages responded to different PGs with identifiable unique gene expression signatures (clusters 1–5) based on their chemical structures.
Figure 4.
RNA sequencing analysis reveals differential gene expression in BMDMs after treatment with PG fragments. BMDMs derived from wild-type mice were treated with PG fragments GMTP, MDP, MTP, or ng-MDP at 20 μM for 18 h (n = 3 biological replicates for each sample). (A, B) Total RNA was subjected to RNA sequencing analysis. (A) Heat map of top genes differentially expressed (p < 0.01, FDR < 0.05, and logFC > 1) in BMDMs treated with water (control) or PG fragments GMTP, MDP, MTP, or ng-MDP. Hierarchical clustering separated the genes into five clusters depending on the PG activation pattern. Color scales represent upregulation (red) or downregulation (blue) of respective genes. (B) Heat map showing all of the top IBD gene sets that are upregulated (red) or downregulated (blue) differentially between GMTP and ng-MDP compared with the control (water). (C) Concentrations of cytokines (in pg/mL) released by BMDM + 20 μM compound treatment for 18 h as determined by Luminex analysis. Data are represented as mean ± SEM (n = 3 biological replicates).
GMTP emerged from these experiments as a new and efficient activator of innate immune responses in macrophages. Remarkably, 31 genes that were identified by DEseq2 analysis70,71 as significantly induced by GMTP have genetic variants associated with either Crohn’s disease or ulcerative colitis (IBD Exomes Browser, ibd.broadinstitute.org) (Figure 4B and SI2 Table 10). The relative expression of these IBD-associated genes and the P values of the highest IBD-associated variant are shown (Figure 4B and SI2 Table 10). While a few of these genes were also part of the ng-MDP-induced pathways, we also identified IBD-associated genes that characterized the response to ng-MDP, including Mmp8, Mmp9, Il2rb, Lck, and Lcn2 (Figure 4B and SI2 Table 10). These findings indicate that the pathway associated with the recognition of GMTP may play an important role in regulating mucosal immune responses. An ELISA–Luminex-based assay was used to screen the expression of a panel of inflammation-associated proteins, and the results showed that IL-1β, CXCL10 (IP10), KC (IL8 homologue), MIP2, IFN-γ, and TNF-α protein expression was induced in the PG-fragment-treated macrophages (Figure 4C), confirming the RNAseq results.
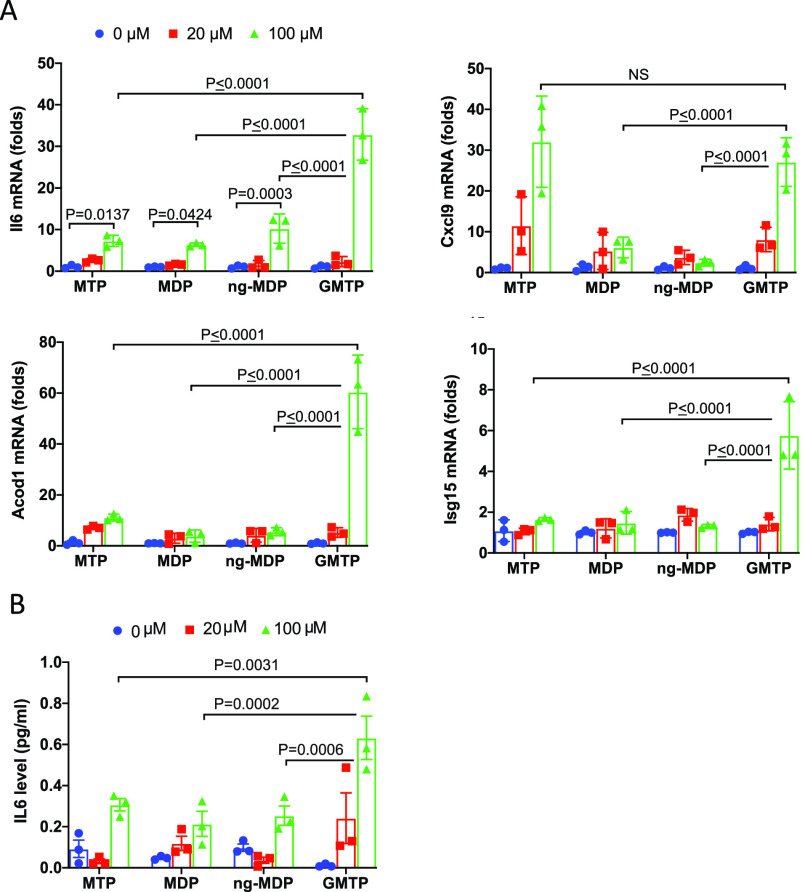
We next carried out qRT-PCR analysis of mRNA expression from genes from cluster 1 (Il6, Isg15, Acod1, and Cxcl9) in three independent experiments in which BMDMs were treated for 18 h with 20 or 100 μM GMTP, MDP, MTP, or ng-MDP (Figure 5A). GMTP induced significantly more Il6, Acod1, and Isg15 mRNA expression compared with MDP, MTP, and ng-MDP (Figure 5A). GMTP induced a 33-fold increase in Il6 mRNA expression, while MDP, MTP, and ng-MDP induced a 7–10-fold increase. Acod1 expression was increased up to 60-fold in the presence of 100 μM GMTP, while the other compounds achieved an up to 10-fold increase. Remarkably, GMTP and MTP had similar ability to induce Cxcl9 expression at 20 or 100 μM compound. Among the studied genes, Isg15 mRNA expression was significantly induced only by GMTP in these experiments (Figure 5A). These data emphasize that even at higher concentrations, different gene programs are activated. Finally, we measured IL6 secretion in the supernatants of BMDMs after 18 h of stimulation. These experiments confirmed that GMTP induced significantly higher levels of IL6 compared with MDP, ng-MDP, or MTP (Figure 5B). Together, these results revealed that the synthesized peptidoglycan fragments were potent innate immune stimuli that were able to induce shared and fragment-specific gene expression profiles that may be able to elicit unique immune responses.
Figure 5.
Gene expression analysis of cluster 1 genes and cytokine analysis confirm GMTP’s reactivity. (A) Gene expression and statistical analysis of qRT-PCR data for Il6, Acod1, Cxcl9, Lcn2, and Isg15 in BMDMs treated with 0, 20, or 100 μM 3, 4, or 5 or control (water) for 18 h. Total RNA was harvested, and the expression levels of selected genes were analyzed by qRT-PCR. Data were analyzed using one-way ANOVA and the ordinary ANOVA test. Error bars represent mean ± SD. (B) IL6 expression in the supernatant of BMDMs from (A) after 18 h of stimulation with the indicated peptidoglycan fragments and concentrations. Data were analyzed using one-way ANOVA and the ordinary ANOVA test. Error bars represent mean ± SD.
Cellular Biochemical Characterization
To further extend this study from the gene to protein level, we next analyzed the activation (i.e., phosphorylation) of common PG signaling pathways. Phosphorylation events are an essential component of the downstream production of cytokines and chemokines upon PG stimulation (Scheme 1).69 Through immunoblot analysis, STAT1, IRF5, cJUN, and p65 NFκB phosphorylation was screened (SI-Figure 4). Phosphorylation was observed for IRF5, cJUN, and P65-NFκB 1 h and 4 h after treatment with 20 μM MDP, ng-MDP, and GMTP, confirming that all the three compounds were able to activate these signaling pathways. The transcriptional activation of immunoregulatory genes observed for GMTP in combination with the qRT-PCR and RNAseq analyses (Figures 3B, 4A,B, and 5) could be a result of combinatorial activation of these pathways (Figure 1) or involve yet uncharacterized transcriptional regulators. The stability, interaction strength, or component recruitment of PRRs (Figure 1) into the subsequently forming signaling complexes may be distinct and responsible for the observed fragment-specific transcriptional responses. The observed gene transcriptional differences could also be the result of different binding affinities of mono- and disaccharide PG fragments for their appropriate PRRs. Analysis of these commonly studied components in peptidoglycan signaling indicates that the transcriptional regulators and intermediaries that induce fragment-specific gene expression will need to be further defined. Future studies probing the potential PRR(s) binding mechanism and complex components are needed to further understand these PG-associated innate immune responses.
Conclusion
Key lysozyme products of the Gram-positive commensal gut bacterium L. acidophilus were identified and confirmed. Corresponding biologically relevant disaccharide PG fragments were subsequently synthesized and fully characterized on scale. Whole-genome RNA sequencing identified GMTP as a key disaccharide PG fragment that serves as a significant activator of multiple gene transcriptional and cytokine programs (Figure 4A,B). By screening beyond traditional PG fragments for gene activation events, we have identified unique induction patterns for the production of a variety of IBD-associated gene transcripts (Figure 4B). qRT-PCR analysis of mRNA expression of genes from cluster 1 and cytokine analysis confirmed that the synthesized peptidoglycan fragments were able to induce shared and fragment-specific gene expression profiles (Figure 5). Finally, the known PG signaling pathways STAT1, IRF5, cJUN, and p65 NFκB were found to be activated by GMTP, MDP, and ng-MDP (SI-Figure 4). These results complement the sophisticated yet limited reports in the field that use adjuvants other than MDP, such as GMTP–N-dipalmitoylpropylamide (DPG)72 and mifamurtide,73 for potent stimulation of PRRs. The knowledge of these specific activation pathways will allow for more tunable adjuvants to be compiled.
This work indicates that distinct PG fragments activate shared pathways with different signaling strengths. Interestingly, we also observed PG-fragment-specific gene signatures that could be due to the activation of distinct recognition pathways or the specific recruitment and activation of additional signaling components. Altogether, the results shown that innate immune stimulation by bacterial peptidoglycan is much more complex than previously observed with the small synthetic PG mimics muramyl dipeptide (MDP) and N-glycolyl MDP (ng-MDP). These unique molecular signatures of pathogen and commensal bacteria could enable PRRs to control microbiome-specific homeostasis and regulate immune responses, as bacteria with modifications (e.g., N-glycolyl) of the carbohydrate backbone of PG are known to evade immune recognition. A next obvious step forward from this work is to determine potential PRR(s) required to detect the various PG fragments, interaction strengths for individual PG fragments therein, and the signaling mechanism that lead to the distinct gene expression signatures that may be linked to yet to be determined innate immune regulation. An important future step will be to catalog all of the biologically relevant PG components that are generated during microbe–host interactions. With the structural details and the newly synthesized molecules now available to the scientific community, harnessing the mechanisms responsible for this differential PG signaling paradigm will be essential in developing novel therapeutics and adjuvants to control inflammation.
Acknowledgments
This work was supported by the Delaware COBRE Program, with a grant from the National Institute of General Medical Sciences (NIGMS P20 GM104316 to C.L.G.) and the National Science Foundation (NSF 1554967). C.L.G. is a Pew Biomedical Scholar and Sloan Fellow and thanks the Pew and Sloan Foundations. H.-C.R. and C.L.G. acknowledge Grant GM138599 from the National Institutes of Health; H.C.R. acknowledges Grants AI11333 and DK068181 from the National Institutes of Health. K.L.B. and K.E.D. thank the University of Delaware for their support through the University Doctoral and Dissertation Fellowship Programs. K.E.D. and K.A.W. thank the NIH for support (5T32GM008550). Instrumentation support was provided by the Delaware COBRE and INBRE Programs, supported by the National Institute of General Medical Sciences (P30 GM110758, P20 GM104316, and P20 GM103446). We thank Lisa Hester and the University of Maryland Cytokine Core Laboratory for assistance with ELISAs, Mark Shaw and Bruce Kinngham for their help with RNA-seq, Dr. PapaNii Asare-Okai for mass spectrometry and liquid chromatography support, and especially Dr. Shi Bai for NMR support in obtaining the spectral data for 1D and 2D experiments.
Glossary
Abbreviations
- MDP
muramyl dipeptide
- ng-MDP
N-glycolyl MDP
- PAMP
pathogen-associated molecular pattern
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- TLR
toll-like receptor
- CLR
C-type lectin receptor
- NLR
NOD-like receptor
- RLR
RIG-I-like receptor
- ALR
AIM2-like receptor
- PGBP
peptidoglycan binding protein
- SLAMF
SLAM family
- OLR
OAS-like receptor
- DC
dendritic cell
- PG
peptidoglycan
- GMDP
N-acetylglucosamine muramyl dipeptide
- GMTP
N-acetylglucosamine muramyl tripeptide
- GMTTP
N-acetylglucosamine muramyl tetrapeptide
- GMPP
N-acetylglucosamine muramyl pentapeptide
- NOD2
nucleotide-binding oligomerization domain-containing protein 2
- MurNAc
N-acetyl muramic acid
- GlcNAc
N-acetylglucosamine
- HSQC
heteronuclear single-quantum coherence spectroscopy
- HMBC
heteronuclear multiple bond correlation
- COSY
correlation spectroscopy
- TOCSY
total correlation spectroscopy
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00200.
Author Contributions
⊥ K.L.B. and K.E.D. contributed equally.
Author Contributions
# H.C.R. and C.L.G. share senior authorship.
The authors declare no competing financial interest.
Supplementary Material
References
- Bäckhed F.; Ley R. E.; Sonnenburg J. L.; Peterson D. A.; Gordon J. I. Host–bacterial mutualism in the human intestine. Science 2005, 307 (5717), 1915. 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Sender R.; Fuchs S.; Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14 (8), e1002533. 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Ley R. E.; Mahowald M. A.; Magrini V.; Mardis E. R.; Gordon J. I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444 (7122), 1027. 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Blaut M.; Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J. Nutr. 2007, 137 (3), 751S. 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J.; Mahurkar A.; Rahnavard G.; Crabtree J.; Orvis J.; Hall A. B.; Brady A.; Creasy H. H.; McCracken C.; Giglio M. G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550 (7674), 61. 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J.; Ley R. E.; Hamady M.; Fraser-Liggett C. M.; Knight R.; Gordon J. I. The Human Microbiome Project. Nature 2007, 449 (7164), 804. 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppel N.; Maini Rekdal V.; Balskus E. P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356 (6344), eaag2770. 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M.; Zimmermann-Kogadeeva M.; Wegmann R.; Goodman A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462. 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiser H. J.; Gootenberg D. B.; Chatman K.; Sirasani G.; Balskus E. P.; Turnbaugh P. J. Predicting and Manipulating Cardiac Drug Inactivation by the Human Gut Bacterium Eggerthella lenta. Science 2013, 341 (6143), 295. 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff J. R.; Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013, 14, 676. 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D.; Liwinski T.; Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30 (6), 492. 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins L. J.; Monga M.; Miller A. W. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci. Rep. 2019, 9 (1), 12918. 10.1038/s41598-019-49452-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora N.; Soffer E.; Elinav E. Transforming medicine with the microbiome. Sci. Transl. Med. 2019, 11 (477), eaaw1815. 10.1126/scitranslmed.aaw1815. [DOI] [PubMed] [Google Scholar]
- Janeway C. A.; Medzhitov R. Innate Immune Recognition. Annu. Rev. Immunol. 2002, 20 (1), 197. 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Roychowdhury A.; Ember B.; Wang Q.; Guan R.; Mariuzza R. A.; Boons G. J. Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins. J. Biol. Chem. 2005, 280 (44), 37005. 10.1074/jbc.M506385200. [DOI] [PubMed] [Google Scholar]
- Palm N. W.; Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009, 227 (1), 221. 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Medzhitov R.; Janeway C. A. Decoding the Patterns of Self and Nonself by the Innate Immune System. Science 2002, 296 (5566), 298. 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- Didierlaurent A.; Simonet M.; Sirard J. C. Innate and acquired plasticity of the intestinal immune system. Cell. Mol. Life Sci. 2005, 62 (12), 1285. 10.1007/s00018-005-5032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss C. A.; Levy M.; Itav S.; Elinav E. Integration of Innate Immune Signaling. Trends Immunol. 2016, 37 (2), 84. 10.1016/j.it.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Bahia D.; Satoskar A. R.; Dussurget O. Editorial: Cell Signaling in Host–Pathogen Interactions: The Host Point of View. Front. Immunol. 2018, 9, 221. 10.3389/fimmu.2018.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnich J. M.; Muruve D. A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019, 670, 4–114. 10.1016/j.abb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- Wolf A. J.; Underhill D. M. Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 2018, 18, 243. 10.1038/nri.2017.136. [DOI] [PubMed] [Google Scholar]
- Inohara N.; Ogura Y.; Fontalba A.; Gutierrez O.; Pons F.; Crespo J.; Fukase K.; Inamura S.; Kusumoto S.; Hashimoto M.; et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J. Biol. Chem. 2003, 278 (8), 5509. 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- Girardin S. E.; Boneca I. G.; Viala J.; Chamaillard M.; Labigne A.; Thomas G.; Philpott D. J.; Sansonetti P. J. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003, 278 (11), 8869. 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- Chamaillard M.; Hashimoto M.; Horie Y.; Masumoto J.; Qiu S.; Saab L.; Ogura Y.; Kawasaki A.; Fukase K.; Kusumoto S.; et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003, 4 (7), 702. 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- Girardin S. E.; Boneca I. G.; Carneiro L. A. M.; Antignac A.; Jéhanno M.; Viala J.; Tedin K.; Taha M.-K.; Labigne A.; Zäthringer U.; et al. Nod1 Detects a Unique Muropeptide from Gram-Negative Bacterial Peptidoglycan. Science 2003, 300 (5625), 1584. 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- Grimes C. L.; Ariyananda L. D. Z.; Melnyk J. E.; O’Shea E. K. The innate immune protein Nod2 binds directly to MDP, a bacterial cell wall fragment. J. Am. Chem. Soc. 2012, 134 (33), 13535. 10.1021/ja303883c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. K.; Melnyk J. E.; Baksh M. M.; Lazor K. M.; Finn M. G.; Grimes C. L. Membrane Association Dictates Ligand Specificity for the Innate Immune Receptor NOD2. ACS Chem. Biol. 2017, 12 (8), 2216. 10.1021/acschembio.7b00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro M. L.; D’Ambrosio E. A.; Bahnson B. J.; Grimes C. L. Molecular Recognition of Muramyl Dipeptide Occurs in the Leucine-rich Repeat Domain of Nod2. ACS Infect. Dis. 2017, 3 (4), 264. 10.1021/acsinfecdis.6b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J.; Boyle J. P.; Howard C. B.; Monie T. P.; Davis B. K.; Duncan J. A. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 2012, 287 (27), 23057. 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio E. A.; Bersch K. L.; Lauro M. L.; Grimes C. L. Differential Peptidoglycan Recognition Assay Using Varied Surface Presentations. J. Am. Chem. Soc. 2020, 142 (25), 10926. 10.1021/jacs.0c03933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-C.; Westcott N. P.; Griffin M. E.; Hang H. C. Peptidoglycan Metabolite Photoaffinity Reporters Reveal Direct Binding to Intracellular Pattern Recognition Receptors and Arf GTPases. ACS Chem. Biol. 2019, 14 (3), 405. 10.1021/acschembio.8b01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R.; Brown P. H.; Swaminathan C. P.; Roychowdhury A.; Boons G.-J.; Mariuzza R. A. Crystal structure of human peptidoglycan recognition protein I alpha bound to a muramyl pentapeptide from Gram-positive bacteria. Protein Sci. 2006, 15 (5), 1199. 10.1110/ps.062077606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008, 32 (2), 287. 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W.; Blanot D.; De Pedro M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32 (2), 149. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Boneca I. G.; Dussurget O.; Cabanes D.; Nahori M.-A.; Sousa S.; Lecuit M.; Psylinakis E.; Bouriotis V.; Hugot J.-P.; Giovannini M.; et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (3), 997. 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Lo L. F.; Forsberg L. S.; Maier R. J. Helicobacter pylori Peptidoglycan Modifications Confer Lysozyme Resistance and Contribute to Survival in the Host. mBio 2012, 3 (6), e00409-12. 10.1128/mBio.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. M.; Golchin S. A.; Veyrier F. J.; Domenech P.; Boneca I. G.; Azad A. K.; Rajaram M. V. S.; Schlesinger L. S.; Divangahi M.; Reed M. B.; et al. N-Glycolylated Peptidoglycan Contributes to the Immunogenicity but Not Pathogenicity of Mycobacterium tuberculosis. J. Infect. Dis. 2014, 209 (7), 1045. 10.1093/infdis/jit622. [DOI] [PubMed] [Google Scholar]
- Warth A. D.; Strominger J. L. Structure of the Peptidoglycan of Bacterial Spores: Occurrence of the Lactam of Muramic Acid. Proc. Natl. Acad. Sci. U. S. A. 1969, 64 (2), 528. 10.1073/pnas.64.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs C. N.; Poggio S.; Charbon G.; Pucheault M.; Vollmer W.; Jacobs-Wagner C. MreB Drives Rod Morphogenesis in Caulobacter crescentus via Remodeling of the Cell Wall. J. Bacteriol. 2010, 192 (6), 1671. 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellouz F.; Adam A.; Ciorbaru R.; Lederer E. Minimal structural requirements for adjuvant activity of bacterial peptidoglycan derivatives. Biochem. Biophys. Res. Commun. 1974, 59 (4), 1317. 10.1016/0006-291X(74)90458-6. [DOI] [PubMed] [Google Scholar]
- Lazor K.; Zhou J.; DeMeester K.; D’Ambrosio E.; Grimes C. L. Synthesis and application of methyl N,O-hydroxylamine muramyl peptides. ChemBioChem 2019, 20 (11), 1369. 10.1002/cbic.201800731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees A. L.; Markesich D.; Zayyani N. R.; Graham D. Y. Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev. Gastroenterol. Hepatol. 2015, 9 (12), 1523. 10.1586/17474124.2015.1093931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F.; Divangahi M.; Veyrier F.; de Léséleuc L.; Gleason J. L.; Yang Y.; Kelliher M. A.; Pandey A. K.; Sassetti C. M.; Reed M. B.; et al. Increased NOD2-mediated recognition of N-glycolyl muramyl dipeptide. J. Exp. Med. 2009, 206 (8), 1709. 10.1084/jem.20081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk J. E.; Mohanan V.; Schaefer A. K.; Hou C.-W.; Grimes C. L. Peptidoglycan Modifications Tune the Stability and Function of the Innate Immune Receptor Nod2. J. Am. Chem. Soc. 2015, 137 (22), 6987. 10.1021/jacs.5b01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W.; Joris B.; Charlier P.; Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008, 32 (2), 259. 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Molinaro R.; Mukherjee T.; Flick R.; Philpott D. J.; Girardin S. E. Trace levels of peptidoglycan in serum underlie the NOD-dependent cytokine response to endoplasmic reticulum stress. J. Biol. Chem. 2019, 294 (22), 9007. 10.1074/jbc.RA119.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.; Huang C. Y.; Hasegawa M.; Inohara N.; Fujimoto Y.; Fukase K. Glycan sequence-dependent Nod2 activation investigated by using a chemically synthesized bacterial peptidoglycan fragment library. ChemBioChem 2013, 14 (4), 482. 10.1002/cbic.201200655. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y.; Konishi Y.; Kubo O.; Hasegawa M.; Inohara N.; Fukase K. Synthesis of crosslinked peptidoglycan fragments for investigation of their immunobiological functions. Tetrahedron Lett. 2009, 50 (26), 3631. 10.1016/j.tetlet.2009.03.081. [DOI] [Google Scholar]
- Inamura S.; Fujimoto Y.; Kawasaki A.; Shiokawa Z.; Woelk E.; Heine H.; Lindner B.; Inohara N.; Kusumoto S.; Fukase K. Synthesis of peptidoglycan fragments and evaluation of their biological activity. Org. Biomol. Chem. 2006, 4 (2), 232. 10.1039/B511866B. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio E. A.; Drake W. R.; Mashayekh S.; Ukaegbu O. I.; Brown A. R.; Grimes C. L. Modulation of the Nod-like receptors NOD1 and NOD2: A Chemist’s Perspective. Bioorg. Med. Chem. Lett. 2019, 29 (10), 1153. 10.1016/j.bmcl.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho Fernandez E.; Pot B.; Grangette C. Beneficial effect of probiotics in IBD. Gut Microbes 2011, 2 (5), 280. 10.4161/gmic.2.5.18255. [DOI] [PubMed] [Google Scholar]
- Altermann E.; Russell W. M.; Azcarate-Peril M. A.; Barrangou R.; Buck B. L.; McAuliffe O.; Souther N.; Dobson A.; Duong T.; Callanan M.; et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (11), 3906. 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti G. J.; Chen J.; Schaefer J.; Gross M. L. Characterization of structural variations in the peptidoglycan of vancomycin-susceptible Enterococcus faecium: understanding glycopeptide-antibiotic binding sites using mass spectrometry. J. Am. Soc. Mass Spectrom. 2008, 19 (10), 1467. 10.1016/j.jasms.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMeester K. E.; Liang H.; Jensen M. R.; Jones Z. S.; D’Ambrosio E. A.; Scinto S. L.; Zhou J.; Grimes C. L. Synthesis of Functionalized N-Acetyl Muramic Acids To Probe Bacterial Cell Wall Recycling and Biosynthesis. J. Am. Chem. Soc. 2018, 140 (30), 9458. 10.1021/jacs.8b03304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M. W.; Gray G. R. Processing of Bacillus subtilis peptidoglycan by a mouse macrophage cell line. Infect. Immun. 1984, 46 (2), 476. 10.1128/IAI.46.2.476-483.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Matsuo Y.; Pradipta A. R.; Inohara N.; Fujimoto Y.; Fukase K. Synthesis of characteristic Mycobacterium peptidoglycan (PGN) fragments utilizing with chemoenzymatic preparation of meso-diaminopimelic acid (DAP), and their modulation of innate immune responses. Org. Biomol. Chem. 2016, 14 (3), 1013. 10.1039/C5OB02145F. [DOI] [PubMed] [Google Scholar]
- Bull M.; Mahenthiralingam E.; Marchesi J.; Plummer S. The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 2013, 349 (2), 77. 10.1111/1574-6968.12293. [DOI] [PubMed] [Google Scholar]
- Gomes A. M. P.; Malcata F. X. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci. Technol. 1999, 10 (4), 139. 10.1016/S0924-2244(99)00033-3. [DOI] [Google Scholar]
- Wasilewski A.; Zielińska M.; Fichna J.; Storr M. Beneficial Effects of Probiotics, Prebiotics, Synbiotics, and Psychobiotics in Inflammatory Bowel Disease. Inflammatory Bowel Dis. 2015, 21 (7), 1674. 10.1097/MIB.0000000000000364. [DOI] [PubMed] [Google Scholar]
- Liang H.; DeMeester K. E.; Hou C.-W.; Parent M. A.; Caplan J. L.; Grimes C. L. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat. Commun. 2017, 8, 15015. 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankiewicz J.; Swierczek E. Lysozyme in human body fluids. Clin. Chim. Acta 1974, 57 (3), 205. 10.1016/0009-8981(74)90398-2. [DOI] [PubMed] [Google Scholar]
- Molinaro R.; Mukherjee T.; Flick R.; Philpott D. J.; Girardin S. E. Trace levels of peptidoglycan in serum underlie the NOD-dependent cytokine response to endoplasmic reticulum stress. J. Biol. Chem. 2019, 294 (22), 9007. 10.1074/jbc.RA119.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M. S.; Whiteside S.; Jewett J. C.; Aditham A.; Cava F.; Bertozzi C. R. d-Amino Acid Chemical Reporters Reveal Peptidoglycan Dynamics of an Intracellular Pathogen. ACS Chem. Biol. 2013, 8 (3), 500. 10.1021/cb3004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S. E.; Travassos L. H.; Herve M.; Blanot D.; Boneca I. G.; Philpott D. J.; Sansonetti P. J.; Mengin-Lecreulx D. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 2003, 278 (43), 41702. 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- VanNieuwenhze M. S.; Mauldin S. C.; Zia-Ebrahimi M.; Winger B. E.; Hornback W. J.; Saha S. L.; Aikins J. A.; Blaszczak L. C. The First Total Synthesis of Lipid II: The Final Monomeric Intermediate in Bacterial Cell Wall Biosynthesis. J. Am. Chem. Soc. 2002, 124 (14), 3656. 10.1021/ja017386d. [DOI] [PubMed] [Google Scholar]
- Schwartz B.; Markwalder J. A.; Wang Y. Lipid II: Total Synthesis of the Bacterial Cell Wall Precursor and Utilization as a Substrate for Glycosyltransfer and Transpeptidation by Penicillin Binding Protein (PBP) 1b of Escherichia coli. J. Am. Chem. Soc. 2001, 123 (47), 11638. 10.1021/ja0166848. [DOI] [PubMed] [Google Scholar]
- Burch J. M.; Mashayekh S.; Wykoff D. D.; Grimes C. L. Bacterial Derived Carbohydrates Bind Cyr1 and Trigger Hyphal Growth in Candida albicans. ACS Infect. Dis. 2018, 4 (1), 53. 10.1021/acsinfecdis.7b00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Zagani R.; Park S.-M.; Yoshida N.; Shah P.; Reinecker H.-C. Microbial recognition by GEF-H1 controls IKKε mediated activation of IRF5. Nat. Commun. 2019, 10 (1), 1349. 10.1038/s41467-019-09283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15 (12), 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorianu I.; Bretman A.; Smith D. T.; Fowler E. K.; Dalmay T.; Chapman T. Comparison of alternative approaches for analysing multi-level RNA-seq data. PLoS One 2017, 12 (8), e0182694. 10.1371/journal.pone.0182694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast D. J.; Vosika G. J. The muramyl dipeptide analog GMTP-N-DPG preferentially induces cellular immunity to soluble antigens. Vaccine 1997, 15 (16), 1748. 10.1016/S0264-410X(97)00110-2. [DOI] [PubMed] [Google Scholar]
- Frampton J. E. Mifamurtide: a review of its use in the treatment of osteosarcoma. Paediatr. Drugs 2010, 12 (3), 141. 10.2165/11204910-000000000-00000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.