Significance
We identify AP-3–mediated targeting of flippase ATP8A1 to lamellar bodies, lysosome-related organelles that store pulmonary surfactant for secretion from alveolar epithelial cells. Genetic AP-3 loss in cultured alveolar epithelial cells and in the pearl mouse model causes ATP8A1 retention in early sorting and/or recycling endosomes and concomitant accumulation of phosphatidylserine on endosomal membranes. This in turn elicits a toxic gain-of-function through Yes-activating protein and initiates lung injury programs that may contribute to progressive lung fibrosis associated with Hermansky Pudlak syndrome type 2.
Keywords: lysosome, endosome, pulmonary fibrosis, lung epithelium
Abstract
Lamellar bodies (LBs) are lysosome-related organelles (LROs) of surfactant-producing alveolar type 2 (AT2) cells of the distal lung epithelium. Trafficking pathways to LBs have been understudied but are likely critical to AT2 cell homeostasis given associations between genetic defects of endosome to LRO trafficking and pulmonary fibrosis in Hermansky Pudlak syndrome (HPS). Our prior studies uncovered a role for AP-3, defective in HPS type 2, in trafficking Peroxiredoxin-6 to LBs. We now show that the P4-type ATPase ATP8A1 is sorted by AP-3 from early endosomes to LBs through recognition of a C-terminal dileucine-based signal. Disruption of the AP-3/ATP8A1 interaction causes ATP8A1 accumulation in early sorting and/or recycling endosomes, enhancing phosphatidylserine exposure on the cytosolic leaflet. This in turn promotes activation of Yes-activating protein, a transcriptional coactivator, augmenting cell migration and AT2 cell numbers. Together, these studies illuminate a mechanism whereby loss of AP-3–mediated trafficking contributes to a toxic gain-of-function that results in enhanced and sustained activation of a repair pathway associated with pulmonary fibrosis.
Pulmonary surfactant is a mixture of proteins, phospholipids, and neutral lipids secreted from alveolar type 2 (AT2) epithelial cells to reduce alveolar surface tension, ensuring that alveoli remain open throughout the respiratory cycle (1). Interruption of surfactant maturation by preterm birth, inactivation of surfactant components, and genetic defects affecting surfactant constituents impair surfactant function and contribute to lung disease. Lamellar bodies (LBs) in AT2 cells are lysosome-related organelles (LROs) that receive surfactant constituents, serve as a site for modification of surfactant components, and store pulmonary surfactant for secretion. Transmembrane proteins, like adenosine triphosphate (ATP)-binding cassette protein A3 (ABCA3) that promotes surfactant phospholipid enrichment, and secreted proteins, like surfactant proteins B and C (SFTPB, SFTPC), are components of LBs (1). However, the critical pathways delivering proteins to LBs and the impact of disrupted LB protein targeting on AT2 cell homeostasis and lung disease are poorly understood.
Trafficking pathways that target membrane proteins to LROs include the biogenesis of LRO complexes (BLOC)-1, -2, and -3 and the Adaptor Protein 3 complex (AP-3), and defects in these pathways are found in genetic diseases known collectively as the Hermansky Pudlak syndromes (HPSs) (2, 3). Investigation of BLOC- and AP-3–mediated protein trafficking to melanosomes has fostered a rich understanding of the role of protein trafficking in loss of pigmentation in HPS patients (3), but few LB proteins are known to utilize BLOCs or AP-3, and mechanisms directly linking loss of BLOC and AP-3 to lung disease in HPS patients are lacking. We previously identified AP-3 as a critical element in trafficking the soluble protein Peroxiredoxin-6 (PRDX6) to nascent LBs through its association with lysosomal integral membrane protein 2 (LIMP2) (4). However, fibrotic lung repair in patients with HPS type 2 (HPS2) or in the pearl mouse, both lacking the essential beta subunit of AP-3 (2, 5–8), could not be explained by mistargeting of PRDX6 or LIMP2. Consequently, evidence that LIMP2 engages AP-3 via a cytoplasmic dileucine-like signal (9, 10) led us to investigate whether dileucine-mediated AP-3 targeting employed by other LB transmembrane proteins might explain how AP-3 loss of function contributes to fibrotic lung repair.
ATP8A1 is a P4 ATPase transmembrane protein identified as a limiting membrane component of LBs in AT2 cells (11). P4 ATPases exploit ATP hydrolysis to flip phospholipids from one side of a membrane to the other (12, 13). ATP8A1, a Class 1 P4 ATPase and previously known as APLT or ATPase II, was first recognized as a flippase with specificity for translocating phosphatidylserine (PtdSer) from the exofacial leaflet of the plasma membrane to the cytofacial leaflet, thus eliminating PtdSer as an “eat me” signal for phagocytes (14, 15). Trafficking of P4 ATPases beyond the endoplasmic reticulum (ER) requires association with a member of the CDC50/LEM3 family of proteins, and ATP8A1 heterodimerizes selectively with CDC50A (16). In addition to the plasma membrane, mammalian ATP8A1 and its yeast ortholog, Drs2, are targeted to Golgi and endosomal membranes at steady state (12, 17), where PtdSer-enrichment of membranes facilitate protein sorting and binding (18, 19). Cytosol-exposed N- or C-terminal di-leucine–based signals mediate Golgi and endosomal targeting of some P4 ATPases (20, 21), whereas yeast Drs2 targeting to endosomes requires interaction with the adaptor AP-1 (22). There is currently no evidence for ATP8A1 trafficking to LROs.
Localization of ATP8A1 to endosome membranes has important effects on Hippo/Yes-activating protein (YAP) signaling, a pathway that mediates a variety of cell behaviors (23, 24). YAP is a transcriptional coactivator, and binding between YAP and TEA domain (TEAD) transcription factors mediates the downstream effects of canonical Hippo/YAP signaling (24). Activated YAP is central to the maintenance of alveolar epithelial cell populations and their transcriptional programs (25). YAP is a critical regulator of AT2 cell proliferation after injury (26), and loss of YAP during recovery after infection can delay regenerative responses of AT2 cells (27). Recently, endosomal targeting of ATP8A1 and cytosolic exposure of PtdSer on endosome membranes were shown to activate YAP in selected cell lines (23). Based on these studies, we hypothesized that AP-3 was involved in ATP8A1 targeting to LBs, such that disruption of AP-3 might lead to retention of ATP8A1 in endosomes, altering the YAP signaling.
Here, we identify an acidic di-leucine–based targeting signal within the C-terminal tail of mammalian ATP8A1 and show that it specifies targeting beyond endosomes to LBs. This di-leucine–based motif shares well-described characteristics of similar motifs that engage AP-3 to direct proteins, such as PI4K2A (28) to endosomes, LIMP2 (9, 10) and HIV-1 negative regulatory factor (NEF) proteins (29) to late endosomes and lysosomes, and Tyrosinase (30, 31) and Oculocutaneous Albinism 2 (OCA2) (32) to LROs. We demonstrate further that disruption of AP-3 in AT2 cells impairs trafficking of ATP8A1 to LBs, resulting in ATP8A1 accumulation in early endosomes. Loss of trafficking of ATP8A1 triggers a toxic gain of function by promoting YAP activation and enhanced expression of YAP gene targets in vitro and in vivo. Together, these data identify a critical target for AP-3–dependent trafficking to LBs in AT2 cells and provide insight into the impact of disrupted trafficking on AT2 cell biology and the etiology of HPS2-associated lung disease.
Results
ATP8A1 Is a P4-ATPase that Is Targeted to LBs in an AP-3–Dependent Manner.
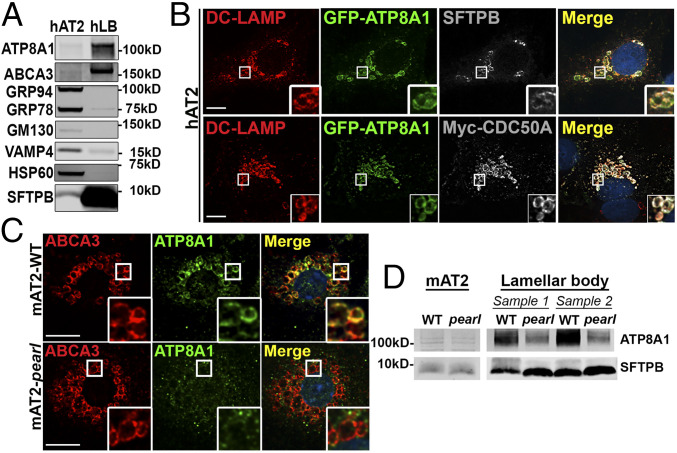
ATP8A1 mRNA is expressed in human AT2 (hAT2) cells [NCBI/Gene Expression Omnibus GDS1404; GSE19699; probes 213196_at and 210192_at (33, 34)] and ATP8A1 has been reported in a LB proteome (11). Immunoblotting of hAT2 cell lysates and subcellular fractions enriched for known LB constituents SFTPB and ABCA3 showed substantial LB enrichment of ATP8A1 but was depleted of markers of ER (GRP94, GRP78), Golgi (GM130), endosomes (VAMP4), and cytosol (HSP60; Fig. 1A). Despite excellent detection of endogenous ATP8A1 by immunoblotting of human samples, the signal to noise ratio of immunofluorescence microscopy (IFM) images of endogenous ATP8a1 in primary hAT2 cells was poor. We therefore constructed a plasmid with an internal ribosome entry site to express both green fluorescent protein (GFP)-tagged ATP8A1 (GFP-ATP8A1) and myc-tagged CDC50A (myc-CDC50A), as ATP8A1 exportation from the ER is dependent on dimerization with CDC50A (16). Transfection of hAT2 cells resulted in colocalization of GFP-ATP8A1 with myc-CDC50A, and with the endogenous LB marker DC-LAMP (Fig. 1B and SI Appendix, Fig. S1) (35). By comparison, IFM demonstrated endogenous ATP8A1 in ABCA3-containing LB of primary AT2 cells from wild-type (WT) C57BL/6J mice (mAT2-WT; Fig. 1C and SI Appendix, Fig. S1). Immunoblotting showed that ATP8A1 was also enriched in LB fractions prepared from adult mouse lung tissue (Fig. 1D). We conclude that ATP8A1, like ABCA3, localizes predominantly to LBs in both human and mouse AT2 cells.
Fig. 1.
Loss of AP-3 disrupts LB localization of ATP8A1. (A) Endogenous ATP8A1 in primary hAT2 cells. Composite representative immunoblot (n = 6) of lysates from hAT2 cells and enriched LB isolates from human lung (hLB). (B) Colocalization of exogenous GFP-ATP8A1 with DC-LAMP/CD208 in primary hAT2 cells. Isolated AT2 cells transfected to express GFP-tagged ATP8A1 and myc-tagged CDC50A were immunostained for endogenous DC-LAMP/CD208 and either endogenous SFTPB or Myc. (Scale bar, 10 μm.) (C) Endogenous ATP8A1 in primary mouse AT2 cells. AT2 cells isolated from C57BL/6 (mAT2-WT) or Ap3b1pe/pe (mAT2-pearl) adult mice were immunostained for endogenous ABCA3 and ATP8A1. (Scale bar, 10 μm.) (D) ATP8A1 expression in primary mouse AT2 cells. Representative immunoblot (n = 4) using lysates from AT2 cells and enriched LB isolates from WT and pearl mice demonstrating ATP8A1 and SFTPB expression.
We wondered whether ATP8A1 trafficking to LBs in AT2 cells, like LIMP2 (4), requires AP-3. IFM analysis of AT2 cells isolated from pearl (Ap3b1pe/pe) mice carrying an inactivating mutation of the Ap3b1 subunit of AP-3 (mAT2-pearl) (36) showed that ATP8A1 colocalization with ABCA3 at LBs was reduced compared with mAT2-WT cells (Fig. 1C and SI Appendix, Fig. S1). Consistently, LB-containing subcellular fractions from pearl mice harbored less ATP8A1 than similar fractions from WT mice, despite similar levels of Atp8a1 mRNA (Fig. 1D and SI Appendix, Fig. S1). We therefore concluded that ATP8A1 localizes to surfactant-containing LBs in an AP-3–dependent fashion.
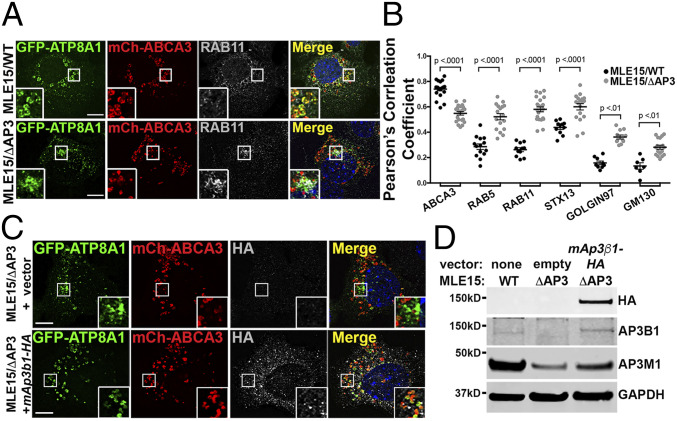
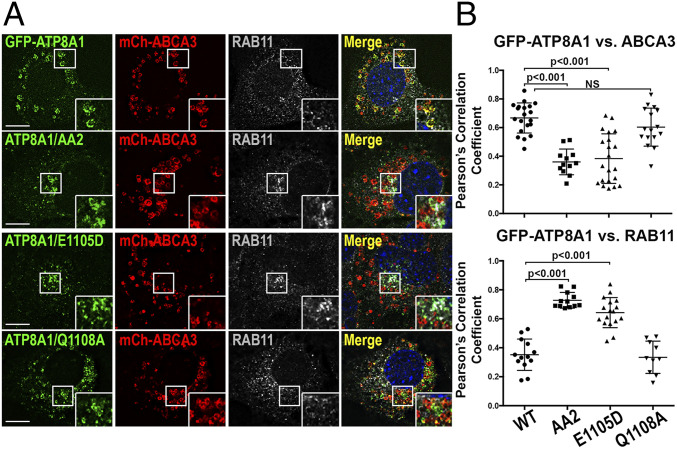
To investigate the role of AP-3 trafficking of ATP8A1 in AT2 cells in greater detail, we used variants of the AT2-like mouse lung epithelial (MLE)-15 cell line (37). We previously generated gene-targeted variants of MLE-15 cells lacking Ap3b1 (MLE15/ΔAP3 cells) that reproducibly model primary AT2 cells from pearl mice (38). We also reported that exogenous expression of ABCA3, in either WT MLE-15 (MLE15/WT) or MLE15/ΔAP3 cells, enlarged LBs for improved identification by fluorescence microscopy (FM) (38). Using FM analyses of transfected MLE15/WT cells, we found that expressed GFP-ATP8A1 colocalized well with mCherry-ABCA3 on LB-like organelles (Fig. 2A), in good agreement with our observations in primary AT2 cells (Fig. 1 B and C), and significantly less well with markers of early endosomes (RAB5, RAB11A, or STX13) and Golgi (GOLGIN97 and GM130) (Fig. 2B and SI Appendix, Fig. S2). By contrast, colocalization between GFP-ATP8A1 and mCherry-ABCA3 was substantially reduced in MLE15/ΔAP3 cells, with GFP-ATP8A1 accumulating in mCherry-ABCA3-negative small cytosolic vesicles and larger vesicles in the juxtanuclear region. Expression of GFP-ATP8A1 exhibited increased colocalization with early sorting and/or recycling endosome markers RAB5, RAB11A, and STX13 and to a lesser extent with Golgi markers (Fig. 2 A and B and SI Appendix, Fig. S2), suggesting that ATP8A1 is retained in proximal compartments of the secretory pathway in the absence of AP-3. To determine whether mislocalization of ATP8A1 in MLE15/ΔAP3 cells reflected only the absence of AP3B1, we coexpressed GFP-ATP8A1, mCherry-ABCA3, and the hemagglutinin (HA)-tagged AP3B1 subunit (AP3B1-HA) to reconstitute AP-3 (Fig. 2 C and D). Expression of exogenous AP3B1-HA restored higher colocalization between GFP-ATP8A1 and mCherry-ABCA3 in MLE15/ΔAP3 cells (Pearson’s correlation coefficient, GFP-ATP8A1 versus mCherry-ABCA3: empty vector 0.55 ± 0.06, Ap3b1-HA vector 0.77 ± 0.6; n = 2 experiments; 15 to 25 cells per group; P < 0.0001; mean ± SD) and stabilized endogenous AP3M1, indicating restoration of intact AP-3. We conclude from these data that AP-3 is required for efficient ATP8A1 trafficking from the early endosomal system to LBs.
Fig. 2.
GFP-ATP8A1 localizes to a RAB11A-positive compartment in AP-3–defective MLE15/ΔAP3 cells. (A) Representative confocal images of MLE15/WT and MLE15/ΔAP3 cells expressing exogenous GFP-ATP8A1 and mCherry-ABCA3 and immunostained for endogenous RAB11A. (Scale bar, 10 μm.) (B) Pearson’s correlation coefficient acquired from images described in A and SI Appendix, Fig. S2, comparing GFP-ATP8A1 to either exogenous mCherry-ABCA3 or endogenous RAB5, RAB11A, STX13, GOLGIN97, and GM130 (n = 2; 10 to 20 cells per group; mean ± SE). (C) Representative confocal images comparing GFP-ATP8A1 and mCherry-ABCA3 localization in MLE15/WT cells and MLE15/ΔAP3 cells after transfection with HA-tagged mouse Ap3β1 or empty vector. (Scale bar, 10 μm.) (D) Representative composite immunoblots (n = 3) using cells described in C comparing the expression of HA, AP3B1, AP3M1, and GAPDH.
AP-3–Dependent LB Targeting of ATP8A1 Requires an Acidic Di-Leucine Motif.
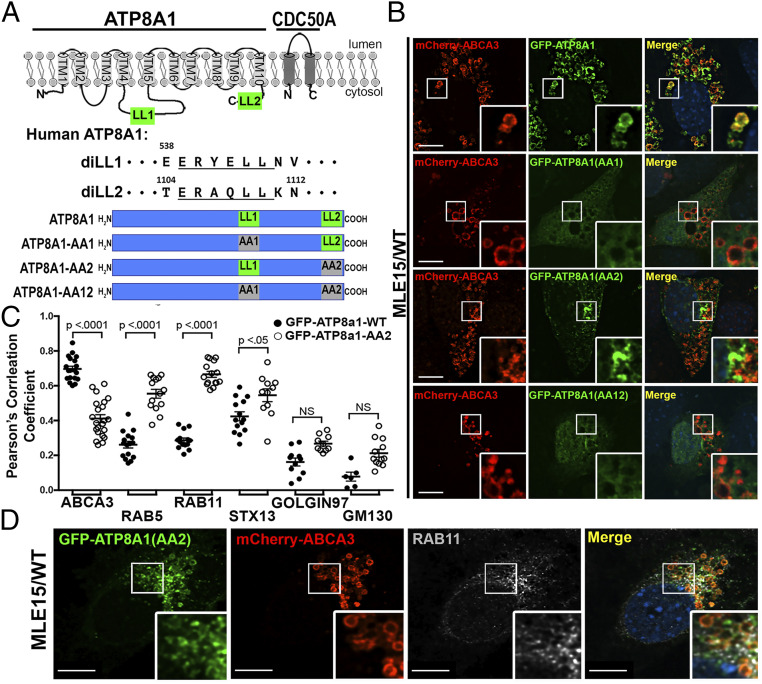
The cytosolic AP complexes, AP-1, -2, and -3, accomplish protein sorting by binding to accessible targeting signals in the cytoplasmic domains of transmembrane protein cargoes. Many of these signals correspond to conserved tyrosine- or acidic di-leucine–based motifs matching the consensus sequences Yxxϕ or [D/E]xxxL[L/I] [where x is any amino acid and ϕ is a bulky hydrophobic residue (3, 39, 40)]. As illustrated in Fig. 3A, we identified two accessible acidic di-leucine consensus sequences in human ATP8A1: 539ERYELL, predicted to fall within a cytosolic loop between transmembrane domains 4 and 5, and 1105ERAQLL, predicted to fall within the cytosolic C-terminal domain (41, 42). Both 539ERYELL and 1105ERAQLL are broadly conserved in ATP8A1 orthologs across mammalian species but not in yeast Drs2 (43–45). Thus, we hypothesized that one or both of these di-leucine motifs facilitates ATP8A1 binding with AP-3 to foster postendosomal targeting. We mutagenized the di-leucines (LL) of human ATP8A1 to di-alanines (AA), separately and in combination (subsequently referred to as GFP-ATP8A1-AA1 inactivating the proximal motif, GFP-ATP8A1-AA2 inactivating the C-terminal tail motif, and GFP-ATP8A1-AA12 inactivating both motifs) and coexpressed them with myc-CDC50A. When coexpressed in human embryonic kidney 293 (HEK293) cells, each of the GFP-ATP8A1 protein variants and myc-CDC50A could be detected in cell lysates by immunoblotting (IB; SI Appendix, Fig. S3), but only GFP-ATP8A1-WT and GFP-ATP8A1-AA2 were successfully co-immunoprecipitated (IP) with myc-CDC50A. Thus, we infer that 539ERYEAA likely destabilizes GFP-ATP8A1-AA1 and GFP-ATP8A1-AA12 proteins.
Fig. 3.
A C-terminal di-leucine motif, 1105ERAQLL, confers LB targeting of ATP8A1. (A) Schematic representation of ATP8A1 and CDC50A protein topology [adapted from ATP8A2 in Coleman et al. (41) and refined by Hiraizumi et al. (42)] and strategy used for mutagenesis (LL = di-leucine; AA = di-alanine). (B) Representative confocal images of MLE15/WT cells expressing mCherry-ABCA3, and WT GFP-ATP8A1-WT or mutagenized GFP-ATP8A1 (AA1 539ERYEAA; AA2 1105ERAQAA; AA12 539ERYEAA + 1105ERAQAA) as described in A. (Scale bar, 10 μm.) (C) Pearson’s correlation coefficients acquired from images of MLE15/WT cells described in B and D, and in SI Appendix, Fig. S3), comparing colocalization of GFP-ATP8A1-WT or GFP-ATP8A1-AA2 with organelle markers (n = 2 experiments; 15 to 20 cells; mean ± SE). (D) Representative confocal images of MLE15/WT cells expressing mCherry-ABCA3 and GFP-ATP8A1-AA2, and immunostained for RAB11A. (Scale bar, 10 μm.)
When coexpressed with CDC50A in MLE15/WT cells and analyzed by FM, GFP-ATP8A1-AA1 and GFP-ATP8A1-AA12 displayed a weakly fluorescent, diffuse pattern (Fig. 3B), consistent with protein destabilization and degradation. By contrast, GFP-ATP8A1-AA2 expressed well but localized to a juxtanuclear region and to small cytosolic vesicles that exhibited reduced colocalization with mCherry-ABCA3 (Fig. 3 B and C). This distribution pattern resembled that of GFP-ATP8A1-WT in MLE15/ΔAP3 cells (Fig. 2 and SI Appendix, Fig. S2). Indeed, by IFM analyses, GFP-ATP8A1-AA2 in MLE15/WT cells demonstrated the strongest colocalization with RAB11A (recycling endosomes), and less robust localization to sorting endosomes (RAB5) and pan-early endosomes (STX13; Fig. 3 C and D and SI Appendix, Fig. S3). By comparison to Fig. 2B, differences in colocalization of GFP-ATP8A1-WT and GFP-ATP8A1-AA2 with Golgi markers were less robust and did not reach statistical significance (Fig. 3C and SI Appendix, Fig. S3). We conclude that AP-3–dependent targeting of ATP8A1 from early endosomes to LBs requires the 1105ERAQLL di-leucine–based motif in the cytosolic tail of ATP8A1.
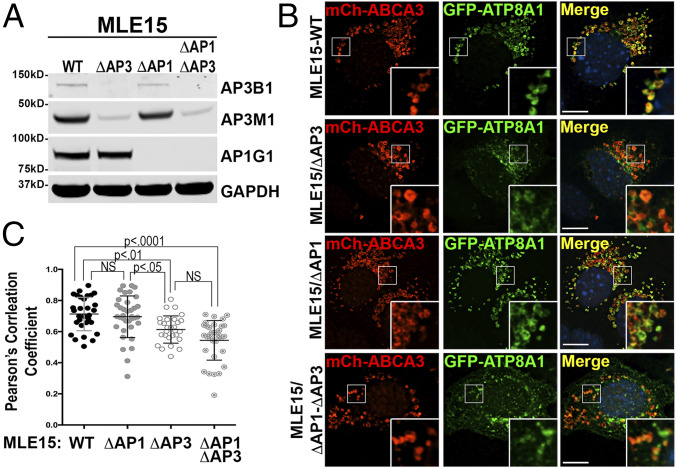
Di-leucine– and tyrosine-based sorting signals can often display promiscuous binding to multiple AP complexes (10, 32, 46–49). In light of prior evidence for AP-1 targeting of Drs2 to the plasma membrane (50), we tested whether AP-1 might also influence 1105ERAQLL-dependent trafficking of ATP8A1 to LBs. We compared the localization of GFP-ATP8A1-WT expressed in MLE15/WT cells in which AP-1 and/or AP-3 were inactivated by using gene editing to delete the first exon of Ap1γ1 to disrupt AP-1 (MLE15/ΔAP1) alone and in combination with Ap3β1 inactivation (MLE15/ΔAP1-ΔAP3; Fig. 4). By FM, the loss of AP-1 expression alone in MLE15/ΔAP1 did not significantly impair ATP8A1 localization to LBs as assessed by colocalization of GFP-ATP8A1 with mCherry-ABCA3 (Fig. 4). Whereas loss of AP-3 expression in MLE15/ΔAP3 significantly reduced colocalization, additional loss of AP-1 in MLE15/ΔAP1-ΔAP3 did not result in further reduction in colocalization of GFP-ATP8A1 with mCherry-ABCA3. We thus conclude that interactions between AP-1 and the 1105ERAQLL di-leucine motif are unlikely to play a prominent role in targeting ATP8A1 to LBs.
Fig. 4.
AP-3 is the dominant AP complex used to target ATP8A1 to LBs. (A) Representative composite immunoblot (n = 3) of lysates from MLE15/WT, MLE15/ΔAP3, MLE15/ΔAP1, and MLE15/ΔAP1-ΔAP3 cells. (B) Representative confocal images of MLE15/WT, MLE15/ΔAP3, MLE15/ΔAP1, and MLE15/ΔAP1-ΔAP3 cells expressing mCherry-ABCA3 and GFP-ATP8A1-WT. (Scale bar, 10 μm.) (C) Pearson’s correlation coefficients acquired from images described in B (n = 2; 10 to 20 cells per group, mean ± SD).
Like the AP-3–binding acidic di-leucine motifs of LIMP2 (9, 10), NEF (10), PI4K2A (28), and OCA2 (32), the ATP8A1 1105ERAQLL sequence features a glutamate in the -4 position relative to the di-leucine motif. The glutamate in the OCA2 dileucine motif is essential for AP-3 binding and for melanosome targeting (32). To test whether 1105E in ATP8A1 was required for AP-3–dependent targeting to LBs, we substituted aspartate for 1105E (GFP-ATP8A1-E1105D). When expressed in MLE15/WT cells, this single amino acid substitution resulted in loss of GFP-ATP8A1-E1105D colocalization with ABCA3-mCherry and enhanced colocalization with RAB11A, much like di-alanine substitution for the di-leucine in GFP-ATP8A1-AA2 (Fig. 5). By contrast, substitution of alanine for the glutamine residue at the -1 position of 1105ERAQLL in GFP-ATP8A1-Q1108A, a position that impacted AP-1 binding in the OCA2 di-leucine–based signal (32), did not interfere with LB localization (Fig. 5). Both GFP-ATP8A1-E1105D and GFP-ATP8A1-Q1108A expressed well and co-IP with CDC50A when expressed in HEK cells (SI Appendix, Fig. S4), indicating that failure of GFP-ATP8A1-E1105D to target to LBs was not due to either misfolding or failure to bind CDC50A. These data suggest that sequence features of the ATP8A1 di-leucine–based motif known to impact AP-3 binding for other proteins are required for its function in ATP8A1 as an LB sorting signal.
Fig. 5.
Preference for Glu at position -4 relative to the di-leucine of 1105ERAQLL contributes to LB targeting by AP-3. (A) Representative confocal images of MLE15/WT cells expressing mCherry-ABCA3, and WT GFP-ATP8A1, GFP-ATP8A1-AA2, GFP-ATP8A1-E1105D, or GFP-ATP8A1-Q1108A, and immunostained for endogenous RAB11A. (Scale bar, 10 μm.) (B) Pearson’s correlation coefficient acquired from images described in A (n = 2; 15 to 20 cells per group, mean ± SD).
AP-3 Trafficking of ATP8A1 Is Facilitated by 1105ERAQLL Interaction with AP-3.
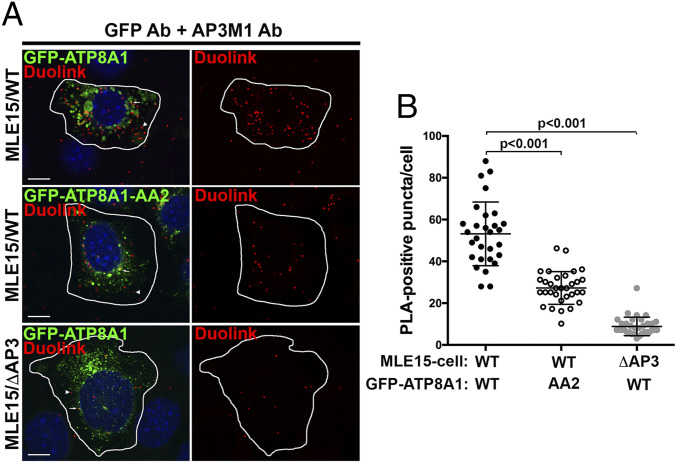
Despite the sequence similarity of 1105ERAQLL to other AP-3–binding motifs and the similar requirements for AP-3 in ATP8A1 trafficking from early endosomes to LBs, we were unable to detect a physical interaction between the ATP8A1 cytoplasmic domain and the di-leucine motif-binding δ/σ3 hemicomplex of AP-3 using a sensitive yeast 3-hybrid (Y3H) assay (SI Appendix, Fig. S5). Therefore, to test whether ATP8A1 and AP-3 are in close proximity within cells, we performed proximity ligation assays (PLAs) in MLE15 cell variants expressing GFP-ATP8A1-WT or GFP-ATP8A1-AA2. The antibodies to the μ subunit of endogenous AP-3 (AP3M1) and to the GFP tag of GFP-ATP8A1 were near enough to enable rolling circle amplification of modified secondary antibodies, incorporating Alexa-594-labeled nucleotides into aggregates that are detected by FM, while GFP-ATP8A1 aided in the identification of transfected cells. In MLE15/WT cells expressing GFP-ATP8A1-WT (Fig. 6), PLA produced numerous cytoplasmic puncta (53 ± 15 puncta/cell, n = 29 cells, mean ± SD). The distribution of PLA products was widespread and was not restrained to the immediate area around GFP-ATP8A1-WT-positive LBs, consistent with localization of PLA products near transport vesicles. By contrast, many fewer fluorescent PLA products (27 ± 8 puncta/cell, n = 32 cells) were observed in MLE15/WT cells expressing GFP-ATP8A1-AA2, implying reduced association between GFP-ATP8A1-AA2 and endogenous AP-3. MLE15/ΔAP3 cells expressing GFP-ATP8A1-WT served as negative controls, as disruption of the AP-3 complex results in loss of expression of all AP-3 subunits (as seen in Fig. 4A), and yielded very few fluorescent PLA products (8.8 ± 4.4 puncta/cell, n = 33 cells). These data suggest that the puncta observed in MLE15/WT cells expressing GFP-ATP8A1 represent sites of AP-3/ATP8A1 interaction en route to LBs, likely in transport carriers emerging from early endosomes.
Fig. 6.
GFP-ATP8A1 and AP-3 are closely associated with LB-like organelles in MLE15/WT cells. (A) Representative confocal images of proximity ligation assays using MLE15/WT or MLE15/ΔAP3 cells expressing exogenous GFP-ATP8A1-WT or GFP-ATP8A1-AA2. (Scale bar, 10 μm.) Arrowheads identify cytosolic PLA reaction products, and arrows indicate PLA products adjacent to organelles expressing GFP-ATP8A1-WT or GFP-ATP8A1-AA2. Cells were also transfected to express untagged ABCA3 thereby enhancing LB volume for identification. (B) Quantification of AlexaFluor594-positive PLA reaction products in cells described in Fig. 5A (n = 2; 15 to 20 cells; mean ± SD).
Retention of ATP8A1 in RAB11A-Positive Endosomes Increases Cytosolic Exposure of PtdSer and Activates YAP in MLE15/ΔAP3 Cells.
ATP8A1 is expressed constitutively on recycling endosomes in many cell lines (19), where it flips PtdSer from the exofacial to the cytofacial leaflet of membranes (19, 51, 52). Recent studies indicate that ATP8A1-dependent accumulation of PtdSer in the cytofacial leaflet of recycling endosome membranes in Cercopithecus aethiops kidney (COS-1) and HEK293 cells promotes signaling by YAP, a transcription coactivator of the Hippo pathway (23). We therefore wondered whether PtdSer is enriched in endosome membranes and whether YAP signaling is enhanced in AP-3–deficient AT2 cells expressing endosomal ATP8A1, compared with WT AT2 cells in which ATP8A1 is largely localized to LBs at steady state.
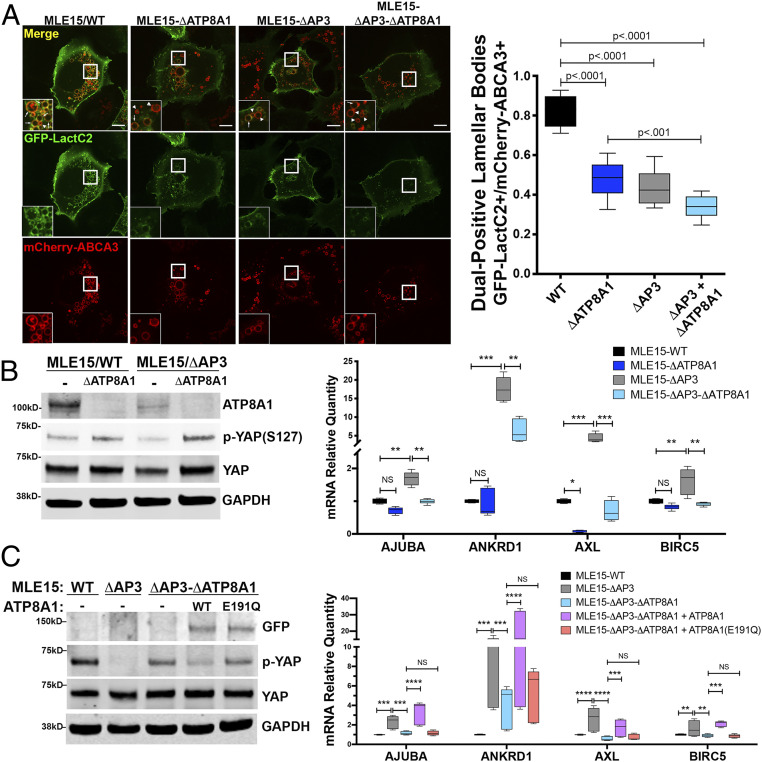
We first tested whether mistargeting of endogenous ATP8A1 in MLE15/ΔAP3 cells alters cytofacial PtdSer distribution. We expressed the cytoplasmic PtdSer biosensor GFP- or mCherry-LactC2 (53) in MLE15/WT and MLE15/ΔAP3 cells and analyzed cells by live cell imaging (Fig. 7A and SI Appendix, Fig. S6 and Movies S1–S8). In MLE15/WT cells, GFP-LactC2 localized preferentially to LB-limiting membranes, overlapping with mCherry-ABCA3, and to the plasma membrane (Fig. 7A). By contrast, in MLE15/ΔATP8A1 cells lacking ATP8A1 (Fig. 7A), the fraction of mCherry-ABCA3-containing LBs labeled by GFP-LactC2 was dramatically reduced, as was binding of GFP-LactC2 to the plasma membrane. This suggests that cytofacial PtdSer accessible to GFP-LactC2 at the LB-limiting membrane and plasma membrane is largely, though not completely, dependent on endogenous ATP8A1 expression and targeting to these subcellular sites. GFP-LactC2 association with LBs was reduced in AP-3–deficient MLE15/ΔAP3 cells, whereas mCherry-LactC2 association with GFP-Rab11A-positive compartments was substantially increased (SI Appendix, Fig. S6). Unlike MLE15/WT cells that demonstrated binding of GFP-LactC2 to both plasma membrane and LBs, loss of AP-3 reduced binding of GFP-LactC2 to only LBs in MLE15/ΔAP3 cells, but not to the plasma membrane, suggesting divergence of postendosomal ATP8A1 trafficking in AT2 cells. Colocalization between mCherry-LactC2 and GFP-RAB11A was greatly diminished in cells lacking both AP-3 and ATP8A1 (MLE15/ΔAP3-ΔATP8A1 cells; SI Appendix, Fig. S6), indicating that PtdSer accumulation on RAB11A-containing structures in AP-3–deficient cells requires ATP8A1. Importantly, inactivation of Ap3b1 and/or Atp8a1 in MLE15 cells had no impact on the expression or localization of mCherry-ABCA3 or GFP-RAB11A. Together, these findings demonstrate that loss of AP-3 results in an ATP8A1-dependent shift in PtdSer enrichment from LB-limiting membranes to endosomes, notably in RAB11A-positive endosomes.
Fig. 7.
Mistargeting of ATP8A1 in the absence of AP-3 activates YAP. (A) Representative images captured from live-cell imaging of MLE15/WT, MLE15/ΔAP3, MLE15/ΔATP8A1, and MLE15/ΔAP3-ΔATP8A1 cells expressing mCherry-ABCA3 and the biosensor GFP-LactC2 (arrow: GFP-LactC2+/mCherry-ABCA3+ dual-positive LBs; arrowhead: mCherry-ABCA3+/GFP-LactC2- LBs). Live-cell imaging was obtained using identical microscope settings, and still images were derived from the first frame to avoid photobleaching. GFP-LactC2/mCherry-ABCA3 dual-positive organelles as a fraction of total mCherry-ABCA3-positive organelles (n = 2; 10 cells; box and whiskers plot showing minimum, 25th percentile, median, 75th percentile, and maximum). (Scale bar, 10 μm.) (B) ATP8A1-dependency of YAP signaling in MLE15 cells described in A. (Left) Composite representative IB (n = 3; 15 μg cell lysate per lane) for endogenous ATP8A1, phospho-YAP-Ser127, total YAP, and GAPDH. (Right) RT-qPCR for Ajuba, Ankrd1, Axl, Birc5 RNA (n = 3; box and whiskers plot as described in A; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (C) Flippase-dependency of YAP signaling in MLE15/ΔAP3 cells. (Left) Composite representative IB of MLE15/WT and MLE15/ΔAP3cells, and MLE15/ΔAP3-ΔATP8A1 cells expressing GFP, GFP-ATP8A1, or GFP-ATP8A1(E191Q) (n = 3; 15 μg cell lysate per lane) for GFP, p-YAP-Ser127, total YAP, and GAPDH. (Right) RT-qPCR for Ajuba, Ankrd1, Axl, Birc5 RNA (n = 3; box and whiskers plot and P values as described in B).
To assess whether increased PtdSer accumulation on RAB11A-containing compartments in AP-3–deficient AT2 cells leads to YAP activation, we exploited the fact that YAP phosphorylation (p-YAP) at Ser127 promotes binding to 14-3-3 proteins and YAP sequestration in the cytosol, thereby reducing levels of nuclear YAP and consequent transcription of YAP target genes (54–56). We found that the expression of p-YAP relative to total YAP in MLE15/ΔAP3 was decreased compared to MLE15/WT cells (Fig. 7B), suggesting that unphosphorylated YAP and YAP activity are increased in MLE15/ΔAP3 cells. Consistent with previous observations (23), YAP activity in both MLE15/WT and MLE15/ΔAP3 cells was ATP8A1-dependent, as indicated by an increase in p-YAP relative to total YAP in lysates of both MLE15/ΔATP8A1 and MLE15/ΔAP3-ΔATP8A1 (Fig. 7B). Importantly, we observed significant changes in YAP signaling despite reduction in endogenous ATP8A1 expression in MLE15/ΔAP3 cells compared with MLE15/WT cells. The mRNA expression of gene targets of YAP signaling, including Ajuba, Ankrd1, Axl, and Birc5, paralleled changes in YAP activation, with increased expression of YAP targets in MLE15/ΔAP3 cells and normalized expression in MLE15/ΔAP3-ΔATP8A1 cells. To determine whether YAP activation and downstream signaling alters MLE15 cell behavior, we examined the response of MLE15 cell variants to scratch wounding (SI Appendix, Fig. S7). MLE15/ΔAP3 cells closed a scratch wound faster than MLE15/WT cells, and loss of ATP8A1 slowed wound closure for both MLE15/WT and MLE15/ΔAP3 cells. Proliferation as assessed by bromodeoxyuridine (BrdU) incorporation was not different in MLE15/WT cells or variants. Together, these data support the conclusion that increased ATP8A1 residence on RAB11A-positive endosomes in MLE15/ΔAP3 cells contributes to enhanced YAP activity, YAP-dependent downstream signaling, and cell migration.
To validate that PtdSer exposure resulting from the flippase activity of ATP8A1 in RAB11A-positive endosomes is necessary and sufficient to stimulate YAP signaling in MLE15/ΔAP3 cells, we assessed whether a flippase-dead form of ATP8A1 would support YAP activation. Using AP-3– and ATP8A1-deficient MLE15/ΔAP3-ΔATP8A1 cells, we expressed either GFP-ATP8A1-WT or GFP-ATP8A1(E191Q), which is unable to hydrolyze ATP and flip PtdSer (19) (Fig. 7C). Expression of GFP-ATP8A1-WT decreased the phosphorylation of YAP and increased expression of YAP gene targets, whereas expression of the ATPase defective GFP-ATP8A1-E191Q had little or no impact, relative to GFP alone, on p-YAP or target transcription in MLE15/ΔAP3-ΔATP8A1 cells. These data indicate that ATP8A1 flippase activity is required for the AP-3–dependent decrease in YAP activation in MLE15/ΔAP3-ΔATP8A1 cells. To determine whether restoration of AP-3 function would also normalize YAP signaling in MLE15/ΔAP3 cells in addition to restoring trafficking of ATP8A1 to LBs (as shown in Fig. 2C), we examined YAP signaling in MLE15/ΔAP3 cells expressing AP3B1-HA. Reconstitution of AP-3 function upon expression of AP3B1-HA resulted in decreased YAP activation in MLE15/ΔAP3 cells, specifically increased p-YAP levels and decreased expression of Ajuba, Ankrd1, Axl, and Birc5 mRNA (SI Appendix, Fig. S8). Taken together, these studies demonstrate that ATP8A1 retention in RAB11A-positive endosomes of MLE15/ΔAP3 cells promotes YAP signaling mediated by PtdSer exposure resulting from ATP8A1 flippase activity.
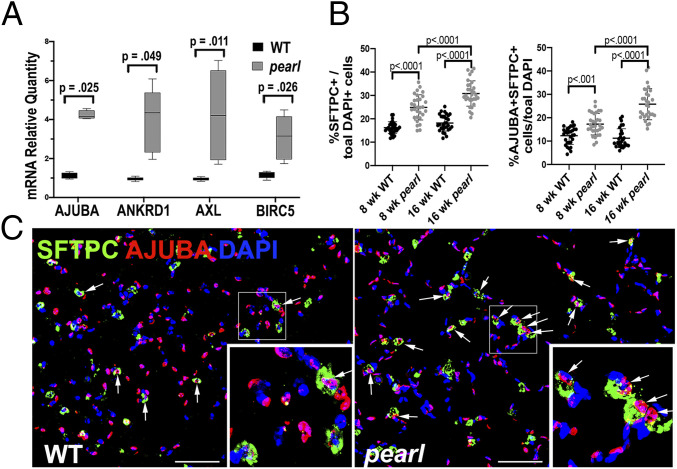
To test whether our observations using MLE15 cells reflected the biology of AP-3–deficient lung tissue, we investigated whether YAP activation was evident in the distal lungs of pearl mice with a mutation in Ap3b1. We found that AT2 cells isolated from the lungs of 8- to 10-wk-old pearl mice expressed increased Ajuba, Ankrd1, Axl, and Birc5 mRNA relative to cells isolated from WT mice (Fig. 8A), suggesting increased activation of YAP in AP-3–deficient AT2 cells. Examination of lung sections from 8- and 16-wk-old mice using IFM showed increased numbers of SFTPC-positive AT2 cells in pearl lungs as previously described (57). Importantly, we found increased numbers of SFTPC-positive AT2 cells expressing nuclear AJUBA in pearl lungs relative to WT lungs, suggesting increased YAP activation in AT2 cells of pearl mouse lungs. We also noted that total SFTPC-positive AT2 cells and dual-positive AJUBA/SFTPC positive cells increased with age in the lungs of pearl mice relative to lungs of WT mice (Fig. 8 B and C). We conclude that our findings of YAP activation and downstream signaling in MLE15/ΔAP-3 cells are recapitulated in vivo in the lung epithelium of HPS2 model mice.
Fig. 8.
YAP activation in AT2 cells of pearl mice. (A) RT-qPCR using mRNA from AT2 cells of WT and pearl mice for Ajuba, Ankrd1, Axl, Birc5 (n = 3; box and whiskers plot showing minimum, 25th percentile, median, 75th percentile, and maximum). (B) Analysis of IFM shown in C of lungs from WT and pearl mice at 8 and 16 wk of age. HALO image analysis software was used to count all nuclei (DAPI-positive) and all AT2 cells (SFTPC-positive). Graphic representation of SFTPC+ AT2 cells (Left) and SFTPC+/nuclear AJUBA+ AT2 cells (Right), both as a percentage of total DAPI-positive nuclei (n = 10 nonoverlapping 40× fields of lung sections from 3 mice per group). (C) Representative images of lungs from WT and pearl mice at 8 wk of age immunostained for SFTPC (green) and AJUBA (red) and costained with DAPI (blue). Arrows point to cells that exhibit cytosolic SFTPC surrounding DAPI-positive nuclei that also exhibit nuclear AJUBA. (Scale bar, 50 μm.)
Discussion
The most lethal complication experienced by patients with HPS is pulmonary fibrosis. Alveolar epithelial cell injury is central to the pathogenesis of pulmonary fibrosis (58), and evidence that injured AT2 cells initiate aberrant fibrotic responses is especially strong in HPS (5–8). Interstitial lung disease with pulmonary fibrosis is highly penetrant in HPS patients lacking AP-3 or BLOC-3, including patients with HPS2 due to mutations of AP3B1 (8, 59), but the mechanisms by which loss of AP-3 contributes to the pathogenesis of pulmonary fibrosis in HPS2 patients have been elusive. Here, we demonstrate that membrane protein ATP8A1 is trafficked to LBs in AT2 cells in an AP-3–dependent manner using an acidic di-leucine motif within the cytoplasmic C-terminal tail. Disruption of ATP8A1 interactions with AP-3, via either loss of the AP-3 complex or mutation of the ATP8A1 acidic di-leucine motif, results in ATP8A1 retention and accumulation in early sorting and recycling endosomes. Because ATP8A1 is a flippase, its retention in endosomes alters the local membrane composition by enriching PtdSer in the cytosolic leaflet. As a result, increased functional ATP8A1 in early endosomes in AP-3–deficient AT2 cells, but not LB ATP8A1 in WT AT2 cells, is able to augment YAP signaling. Together, these findings emphasize the importance of AP-3 as a mediator of LB trafficking in AT2 cells, place the sorting and recycling endosomes as critical upstream organelles in the biogenesis of LBs, and provide a primary mechanism whereby genetic disruption of AP-3 may alter alveolar epithelial cell homeostasis through a toxic gain-of-function of a critical cargo. Further studies will be necessary to assess the impact of YAP signaling due to disrupted ATP8A1 trafficking in HPS2 on the complex process of fibrotic alveolar repair and to address how our findings integrate with other mechanisms implicated in the pathogenesis of HPS2-associated pulmonary fibrosis (7, 8).
When compared with other LROs, trafficking of membrane proteins to LBs has been understudied. We previously reported the AP-3 dependency of LIMP2, a lysosomal protein also targeted to LB in AT2 cells using a well-described dileucine motif (4). Transmembrane proteins ABCA3 and DC-LAMP are targeted to LB at steady state and served as comparators in our present studies. DC-LAMP has a candidate tyrosine-mediated targeting motif—YQRI—that conforms to other tyrosine-mediated AP-3 sorting signals but has not been investigated to date (3, 39, 40). We specifically focused on ABCA3 for comparisons of LB colocalization because ABCA3 utilizes an N-terminal xLxxKN motif for exiting the Golgi (60) but no cytosol-exposed candidate di-leucine–or tyrosine-based sorting signals that could mediate LB targeting (60). Importantly, we previously demonstrated that ABCA3 retains LB targeting in the absence of BLOC-1, -2, -3, or AP-3 in MLE-15 cells, making ABCA3 a reliable LB marker for trafficking membrane proteins to LBs (38). Together, colocalization between ABCA3 and ATP8A1 in mouse AT2 cells, loss of colocalization in AP-3–deficient AT2 cells, and candidate di-leucine motifs with strong homology to other AP-3–binding consensus sequences suggested the potential for AP-3 targeting of ATP8A1 to LBs, as in our prior studies of LIMP2 (4).
The acidic di-leucine motif 1105ERAQLL in the C terminus of ATP8A1 satisfied criteria for an AP-3 binding motif based on evidence from other AP-3 cargo proteins targeted to lysosomes and LROs (9, 10, 28, 31, 32, 49, 61). Importantly, ATP8A1 targeting to LBs—but not binding to its chaperone CDC50A—was disrupted by mutagenesis of key amino acid positions in 1105ERAQLL. Whereas ATP8A1 and CDC50A form extensive intermolecular interactions that span the cytoplasmic and luminal membrane leaflets, the 1105ERAQLL motif resides in a disordered region of the cytoplasmic C terminus, L1065 through K1115, that is free from interaction with CDC50A, likely facilitating access to AP-3 (42). We found that AP-3 and the 1105ERAQLL motif of ATP8A1 were spatially close using a proximity ligation assay, and cytosolic PLA reaction products, likely adjacent to transport vesicles in MLE15/WT cells, were greatly reduced either by mutation of the 1105ERAQLL di-leucine motif or by inactivating Ap3b1 in MLE15 cells. However, we were unable to demonstrate direct interaction between AP-3 and ATP8A1 using a sensitive Y3H assay. There are several potential explanations for this. The intermolecular relationships within the L1065 through K1115 disordered region of ATP8A1 remain elusive, and as a result, it is not possible to determine how changes in the position of the flanking autoregulatory domain in response to nucleotide and/or PtdSer binding might affect the availability of the 1105ERAQLL motif. It is also difficult to predict how expressing the ATP8A1 disordered region alone in the Y3H assay impacts protein conformation, or whether binding with CDC50A may be important for di-leucine motif exposure. Furthermore, we suspect that posttranslational modifications are necessary for the AP-3/ATP8A1 interaction. While our studies demonstrate that 1105ERAQLL is constitutively active like most di-leucine–based sorting signals, the presence of a favorable phosphorylation site in close proximity [PhosphoSitePlus (62); site group ID 4712266] suggests that dynamic phosphorylation may impact AP recognition and/or function (61, 63, 64).
Our studies support the growing evidence that membrane constituents of maturing LROs derive from the early endosomal system (65). Melanosomes mature from precursors derived from multivesicular endosomes (3), and prior studies indicate that this is also true for LBs (66–68). Melanosomes mature by delivery of enzymes and transporters that foster melanin synthesis from tubular and vesicular early endosomal intermediates whose formation, cargo-sorting function, and targeting require AP-3 and BLOCs (3). AP-3 deficiency impairs melanosome targeting and fosters endosomal retention of a subset of cargo proteins, contributing to reduced pigment deposition (31, 32, 69). We now show that AP-3 in AT2 cells captures ATP8A1 into transport carriers from early endosomes for delivery to LBs and that ATP8A1 remains within the early endosomal system in the absence of AP-3. We also noted overlap of GFP-ATP8A1 in Golgi compartments of cells lacking AP-3 and less dramatic overlap between Golgi markers and GFP-ATP8A1-AA2 expressed in MLE15/WT cells. While consistent with prior work demonstrating Golgi/trans Golgi network trafficking for ATP8A1 and other P4 ATPases in other mammalian cells (19, 20, 41, 70, 71), our observations of Golgi overlap may be due to Golgi membranes and early endosomes in close proximity in the perinuclear region that may be confounding our analysis. Another possible explanation is potential recycling of ATP8A1 between endosomes and Golgi in AP-3–deficient cells, as described for TYRP1 in BLOC-2-deficient melanocytes (72). ATP8A1 accumulation in early endosomes in AP-3–deficient cells occurs even though steady-state levels of endogenous ATP8A1 in MLE15/ΔAP3 cells tended to be lower than in MLE15/WT cells by IB (Fig. 7B). This contrasts with the behavior of TYR in AP-3–deficient melanocytes, which largely accumulates on intraluminal vesicles of multivesicular endosomes and is efficiently targeted for degradation (31). Together, our observations suggest a unique adaptation of an AP-3 sorting and trafficking mechanism for ATP8A1 in AT2 cells that defaults to early endosomes, and perhaps Golgi, when AP-3 trafficking is disrupted.
Our studies did not address the role of ATP8A1 at the LB. As PtdSer is a component of exosomes in many cell types (73, 74), it is likely that internal luminal vesicles of multivesicular endosome LB precursors may contribute PtdSer to LB lamellae. However, PtdSer is a membrane phospholipid that contributes <10% to secreted surfactant phospholipids (75–77). Although recent evidence suggests that LB lamellae attach to the LB limiting membrane (78), there is no evidence for membrane continuity that would enable ATP8A1 to flip Ptd-Ser from internal LB lamellae to the cytofacial membrane of LBs. A more likely role of ATP8A1 flippase activity is in preparation of LBs for exocytosis. PtdSer enrichment by the family of P4 ATPases across the secretory pathway is well described (12, 17), and silencing of flippases impairs insulin secretion from pancreatic beta cells (79). Recent studies using AT2 cells derived from induced pluripotent stem cells from an HPS2 patient demonstrated reduced surfactant secretion from LBs (80). Further studies are necessary to determine whether loss of ATP8A1 flippase function from AT2 cells reduces LB secretion.
We and others have implicated AT2 cell injury in initiating and amplifying aberrant fibrotic repair in HPS2 and the pearl mouse model (5–8, 57, 81–83), but to date no plausible mechanism has explained how loss of AP-3 contributes directly to AT2 cell injury. While our data do not exclude an important loss of ATP8A1 function at LBs in AP-3 deficiency, our studies show that ATP8A1 on endosomes results in a toxic gain-of-function model that may contribute to aberrant fibrotic repair. We now find that YAP activation in AP-3–deficient MLE15 cells requires ATP8A1 flippase function in endosomes. Our cell culture observations were corroborated by experiments using primary AT2 cells and distal lung tissue from control and AP-3–deficient pearl mice, averting concerns that aberrant YAP activation in these cell culture models biased our findings (84, 85). Furthermore, expression of four well-known YAP target genes (Ajuba, Ankrd1, Axl, Birc5) was increased in response to AP-3 deficiency in both MLE15 and primary AT2 cells. Of these, Ajuba stood out as a YAP transcriptional target that may have particular significance in the setting of HPS2.
A consistent feature in patients with HPS-associated fibrosis and their corresponding mouse models is AT2 cell hyperplasia (57, 86–89). While critical for surfactant homeostasis, AT2 cells also serve as regional progenitor cells for alveolar type 1 (AT1) cells during lung development or in response to alveolar epithelial cell loss (90, 91). Recent evidence suggests that the Hippo/YAP pathway regulates the restoration of epithelial continuity after alveolar injury (92–95). As expected from prior observations (57), we found more SFTPC-positive AT2 cells in pearl lungs than control lungs, and the difference increased with age. We found increased numbers of AT2 cells exhibiting nuclear AJUBA in pearl lungs, suggesting that YAP activation may be a driver of the AT2 cell hyperplasia associated with HPS2-related pulmonary fibrosis. We also found that YAP activation stimulated migration of MLE15/ΔAP3 cells in an ATP8A1-specific fashion. A reliable reporter of YAP activation, AJUBA mediates aspects of proliferation, migration, and differentiation that are relevant to the pathogenesis of pulmonary fibrosis (25, 96–98). Taken together, our findings add to the growing complexity of YAP signaling in repair of the alveolar epithelium, warranting further investigation of how dysregulated YAP signaling in the setting of AP-3 deficiency contributes to fibrotic alveolar repair.
Materials and Methods
Cell Lines.
MLE-15 cells (RRID:CVCL_D581) were originally obtained from Jeffrey Whitsett, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, and have been described (37). MLE-15 cells were cultured in HITES medium, comprising DMEM/F12 (Thermo Fisher Scientific), supplemented with 10% fetal bovine serum (Corning Life Sciences), 2 mM L-Glutamine (Thermo Fisher Scientific), 10 mM Hepes (Thermo Fisher Scientific), 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 5 ng/mL selenious acid (ITS premix, Corning Life Science), 10 nM hydrocortisone (Sigma-Aldrich), 10 nM β-estradiol (Sigma-Aldrich). Absence of mycoplasma in MLE-15 cell cultures was verified with the LookOut Mycoplasma PCR Detection Kit (Sigma). HEK293 cells (RRID:CVCL_0045) were obtained from American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Corning Life Sciences). All cells were grown in media that included 1% Antibiotic-Antimycotic (Thermo Fisher Scientific) and were incubated at 37 °C and 5% CO2 unless stated otherwise.
Antibodies.
Monoclonal antibodies were procured as follows: ABCA3 (ab23751, 3C9), AP3M1 (ab201227, EPR16385), GFP (ab1218, 9F9.F9) from Abcam; ABCA3 (WMAB-ABCA3-13, 13-H2-57) from Seven Hills Bioreagents; Adaptin γ (610385, Clone88) and GM130 (610822, Clone35) from BD Bioscience; DC-LAMP (IM3448, Clone 104.G4) from Beckman Coulter Life Sciences; GAPDH (39-8600, Clone ZG003) from Thermo Fisher Scientific; Golgin-97 (13192, D8P2K), HA-Tag (2367, 6E2), HA-Tag (3724, C29F4), Myc-Tag (2278, 71D10), Myc-Tag (2276, 9B11), Rab5 (3547, C8B1), Rab11 (5589, D4F5), YAP (12395, 1A12) and p-YAP (Ser127) (13008, D9W21) from Cell Signaling Technology; KDEL (SPA-827, 10C3) from Stressgen Bioreagents. Polyclonal antibodies were procured as follows: Ajuba (4897) and HSP60 (4870) from Cell Signaling Technology; ATP8a1 (21565-1-AP) from Proteintech; GFP (ab290) and Pro and Mature SFPTB (ab40876) from Abcam; HPS2 (GTX113878) from GeneTex; Mature SFPTC (N/A) from Seven Hills Bioreagents; Syntaxin13 (a gift from M. Marks, The Children’s Hospital of Philadelphia, Philadelphia, PA); Vamp4 (a gift from A. Peden, Department of Biomedical Science, University of Sheffield, Sheffield, United Kingdom). Alexa Fluor 488-, 594-, 647-conjugated goat anti-rabbit and anti-mouse or donkey anti-rabbit and anti-mouse were obtained from Jackson ImmunoResearch. IRDye 680LT Donkey anti-rabbit or anti-mouse Immunoglobulin G (IgG) secondary antibody and IRDye 800CW Goat anti-rabbit or anti-mouse IgG secondary antibody were purchased from LI-COR.
Generation of CRISPR Knockout Cell Lines.
Generation of CRISPR knockout MLE-15 cell lines was accomplished as previously described (38). The CRISPR/Cas9 with T2A-puromycin coexpression vector, pSpCas9(BB)-2A-Puro (pX459) vector was purchased from Addgene (plasmid no. 48139). The CHOPCHOP online webtool (https://chopchop.rc.fas.harvard.edu) was used for sgRNA design. Cloning of sgRNA into pX459 was performed following a modified single-step digestion-ligation protocol from the Zhang laboratory, available online at http://genome-engineering.org. Oligos containing guide sequence and BbsI ligation adaptors were synthesized by Eurofins. Annealing of oligos was followed by simultaneous digestion and ligation of the annealed oligo and pX459 vector, followed by transformation into chemically competent DH5α cells. SI Appendix, Table S1 lists the single-guidance RNA sequences used to generate plasmids for each novel MLE-15 knockout cell line developed herein as well as flanking genomic PCR primers used in screening and sequencing of genomic PCR and reverse transcription-polymerase chain reaction (RT-PCR) products. Parental MLE-15 cells were transfected using nucleofection using the Amaxa Cell Line Nucleofactor Kit T (VCA-1002) and the Nucleofector II device with the ×005 cycle (Lonza). Media supplemented with 5 μg/mL puromycin were added 24 h after transfection to select for positive clones over a 3-d period. After a 3-d recovery period, transfected and selected cells were trypsinized and plated as single cells into 96-well plates to expand clones for validation.
DNA Constructs.
The following plasmids were obtained for use in these studies: GFP-LactC2 and mCherry-LactC2 were gifts from Gregory Fairn (18); GFP-Rab11A (99) was a gift from James R. Goldenring, Vanderbilt University Medical Center, Nashville, TN; pEGFP-Atp8a1 and pcDNA-Myc-Cdc50a (19) were gifts from Hiroyuki Arai, Department of Health Chemistry, Graduate School of Pharmaceutical Sciences, University of Tokyo, Tokyo, Japan.
pIRES2-EGFP-Atp8a1-Myc-Cdc50a was generated as follows: Myc-Cdc50a was amplified by PCR with pcDNA-Myc-Cdc50a using the following primers: 5′-GGCCACAACCATGGAGCAGAAGCTGATC-3′ (Myc-Cdc50a; sense primer, BstX1 site is underlined) and 5′-GTCGCGGCCGCTTAAATGGTAATGTCAGC-3′ (Myc-Cdc50a; antisense primer, NotI site is underlined). The product encoding Myc-Cdc50a was introduced into pIRES2-EGFP (Clontech) after EGFP coding region was excised using BstXI and NotI restriction endonucleases to generate pIRES2-Myc-Cdc50a, and EGFP-ATP8A1 was subsequently cloned into pIRES2-Myc-Cdc50a at NheI and SmaI to create pIRES2-EGFP-Atp8a1_Myc-Cdc50a. Point mutations changing di-leucines to di-alanines in conserved ExxxLL motifs were targeted as shown in Fig. 3. Point mutations were constructed by a two-step PCR method using primers indicated in SI Appendix, Table S2 with Platinum Hot Start PCR Master Mix (Invitrogen) using pIRES2-EGFP-Atp8a1_Myc-Cdc50a as the template. The ATP8A1-AA1 and AA2 fragments synthesized by mutagenic two-step amplification of the pIRES2-EGFP-Atp8a1_Myc-Cdc50a template were subcloned into pIRES2-EGFP-ATP8A1_Myc-Cdc50a using EcoRI/SbfI for AA1 and SbfI/SmaI for AA2. pmCherry2-N1-ABCA3 has been previously described (38). HA-tagged Ap3b1 was constructed by amplifying the Ap3b1 C terminus by PCR from pCMV-SPORT6-mAp3b1 (Transomic Technologies, Inc), while also adding BglII and NotI restriction sites at the 5′ (5′-TTCTAGATCTGGACGACTTTA-3′) and 3′ ends (5′-CTCTAGAGCGGCCGCCCTTTTTTTTTTT-3′), respectively. HA tag, an additional StuI restriction site, and stop codon were subsequently added by PCR using a distinct forward primer (5′TTAAGCGTAATCTGGAACATCGTATGGGTAAAGAGGCCTAGTCCCCTGGGACAGGACAGGCTTCAG-3′) and the same reverse primer. The PCR product was directionally subcloned into BglII and NotI restriction sites of pCMV-SPORT6-mAp3b1.
Human Primary Lung Epithelial Cell and LB Isolation.
Second-trimester human lung tissues provided to us by our collaborator Laura Goetzl were deidentified prior to shipping. Human lung epithelial cells were isolated as previously published (4) in keeping with protocols approved by the Vanderbilt University Institutional Review Board. Explanted lung tissue was cultured for 5 d in 10 nM dexamethasone (Sigma-Aldrich) and 0.1 mM each of 8-Bromo-cyclic adenosine monophosphate (8-Br-cAMP) (Sigma-Aldrich) and isobutyl methylxanthine (Sigma-Aldrich) to enhance LB maturation (100). LB fractions were isolated from lung homogenates of explanted tissues via upward flotation through a sucrose step-gradient as previously described (101). Preparation of primary AT2 cells from human lung was carried out as previously described (100).
Animals.
WT C57BL/6J and Ap3b1pe/Ap3b1pe (pearl) were bred and maintained at the Laboratory Animal Facility at Vanderbilt University Medical Center as previously described (4). All animal protocols were reviewed and approved by the Institutional Animal Care and Use Committees of Vanderbilt University Medical Center and adhered to the principles of the National Institutes of Health Guide for the Care and Use of Laboratory Animals (102). Both male and female mice were used between 8 and 10 wk of age. Preparation of paraffin-embedded lung tissue was described previously (83)
Mouse Primary Lung Epithelial Cell and LB Isolation.
Enriched LB fractions were obtained from postlavage, freshly prepared mouse lung homogenates via upward flotation through a sucrose step-gradient as described above. Additional lung homogenates used for biochemical analyses were generated from flash frozen lung tissue after lavage as previously described (83). Preparation of primary AT2 cells from mouse lung was carried out as previously described (4).
Transfection.
Transfection of MLE-15 cells.
MLE-15 cells were transfected using either nucleofector (Lonza) or JetOptimus transfection reagent (Polyplus transfection). Plasmid expression was achieved by electroporating 2 × 106 MLE-15 cells with 4 μg of plasmid using the Amaxa Cell Line Nucleofactor Kit T (VCA-1002) and the Nucleofector II device with the ×005 cycle (all from Lonza). A total of 106 cells were seeded on 60 mm cell culture dish 1 d before transfection and transfected using JetOptimus transfection reagent (Polyplus transfection) for plasmids following the manufacturer’s instructions.
Transfection of human primary lung epithelial cells.
Human primary lung epithelial cells were transfected using the Basic Nucleofector kit for Primary Mammalian Epithelial cells (Lonza, VPI-1005). About 1.0 × 106 cells were resuspended with 2 μg of pIRES2-EGFP-Atp8a1-Myc-Cdc50a plasmid in 100 μL nucleofector reagent, nucleofected with Amaxa Nucleofector II device (M005 cycle; Lonza), and plated on 10% Matrigel-coated round glass coverslip (Electron Microscopy Sciences) in Waymouth’s media with 10% FBS for 24 h. One day after transfection, the culture medium was changed with Waymouth’s media with 10 nM dexamethasone and 0.1 mM each of 8-Br-cAMP and isobutyl methylxanthine.
Quantitative Real-Time PCR Analysis.
Cells (1 × 106) were plated into 60 mm culture dish and grown for 18 h at 37 °C. Total RNA was isolated using the Qiagen RNeasy kit (Qiagen), and cDNA was synthesized from 400 ng of total RNA using SuperScript VILO cDNA synthesis kit (Invitrogen). Quantitative PCR (qPCR) reactions were performed in triplicate using Taqman qPCR Fast Advanced Master Mix (Applied Biosystems, Thermo Fisher Scientific) on a StepOnePlus RT-PCR system (Applied Biosystems) using the following TaqMan probes (Applied Biosystems, Thermo Fisher Scientific): Ajuba Mm00495049_m1, Ankrd1 Mm00496512_m1, Axl Mm00437221_m1, Birc5 Mm00599749_m1, GAPDH Mm99999915_g1, and 18S Hs99999901_s1. Data were normalized to the housekeeping gene GAPDH and 18S.
IB.
Whole cell lysates, lung tissue lysates, and LB-enriched fractions were fractionated using precast 4 to 12% NUPAGE gels (ThermoFisher Scientific) and analyzed by immunoblotting as previously described (38). Proteins were extracted using RIPA lysis buffer (Sigma-Aldrich) with protease inhibitor and phosphatase inhibitor mixtures. Protein concentration was measured by DC Protein Assay Kit (Bio-Rad), and samples were taken as follows: 20 μg total protein from cell lysates, 5 μg from LB lysates, and 80 to 100 μg from cell lysates after gene-editing to demonstrate the loss of protein expression, unless otherwise specified in figure legends. Samples were prepared by boiling after mixing with NuPAGE LDS sample buffer (4×) and NuPAGE Reducing Agent (10×). After transfer to nitrocellulose membranes (Bio-Rad) and blocking in Odyssey blocking solution (LI-COR Biosciences), membranes were probed sequentially primary antibodies and (after rinsing in TBS-T) with IRDye 800CW donkey anti-rabbit (LI-COR Biosciences 926-32213, 1:15,000) or IRDye 680LT donkey anti-mouse secondary antibody (LI-COR Biosciences 926-68022, 1:10,000) diluted in Odyssey blocking solution, and were imaged (after rinsing) with an Odyssey CLx imaging system (LI-COR Biosciences).
IP.
Transfected HEK293 cells were scraped from 100 mm culture plates, collected by centrifugation in phosphate-buffered saline, and resuspended in the IP buffer (IPB) containing 50 mM Tris⋅HCl, 2 mM EDTA, 250 mM NaCl, 10% (vol/vol) glycerol, 0.5% Nonidet P-40, 20 mM NaF, 1 mM sodium orthovanadate, and 10 mM N-ethylmaleimide. Benzamidine and phenylmethylsulfonyl fluoride (to final concentrations of 2 mM and 1 mM, respectively) were added immediately before use. Cells were lysed at 4 °C for 1 h, centrifuged to remove debris, and precleared by incubating with 25 to 30 μL of Protein G Agarose for 1 h at 4 °C. Target proteins were IP by incubating supernatant overnight at 4 °C with appropriate antibodies (1 to 2 μg per 60 mm dish) and 20 to 25 μL of Protein G agarose. Beads were washed three times with IPB, and the proteins were eluted by boiling in Laemmli SDS buffer for 5 min.
Immunostaining.
Cells were plated onto 0.1% Matrigel coated coverslips and fixed with 4% paraformaldehyde for immunostaining. Primary antibodies were used in dilution as recommended by manufacturer, followed by species-specific secondary antibodies. For immunohistochemistry, paraffin-embedded sections provided by the Vanderbilt Translational Pathology Shared Resource were deparaffinized and labeled with primary and secondary antibodies (83), mounted in Prolong Gold (Thermo Fisher Scientific), and analyzed by FM. Where indicated, cells and tissues were stained with DAPI before mounting for imaging.
PLA.
Molecular interaction in vivo between GFP-ATP8A1 and Ap3μ1 was evaluated using the Duolink in situ proximity ligation assay (PLA; Olink Bioscience; Sigma), as previously described (4). Briefly, MLE15/WT and MLE15/ΔAP3 cells transfected with pIRES2-GFPAtp8a1_MycCdc50a and untagged pcDNA3-ABCA3 were cultured onto 0.1% Matrigel coated coverslips (Electron Microscopy Sciences) and fixed with cold ethanol/acetone mixture (1:1 in volume). After fixation, cells were washed with cold PBS, followed by 1% Triton X-100 solution in PBS and blocked in a 3% solution of bovine serum albumin in PBS containing 0.2% Triton X-100. Samples were incubated with primary antibodies against GFP and AP3M1 (1:100 dilution in 0.2% Triton X-100 solution in PBS; GFP MAB3478 mouse monoclonal antibody, Millipore; Ap3μ1 LS-B3225 rabbit polyclonal antibody, LifeSpan BioScience), rinsed in PBS containing 0.2% Triton X-100, and incubated with Duolink PLA probes. Unbound Duolink PLA probes were removed by washing, and the samples were incubated in the ligation solution. Amplification was accomplished by applying diluted polymerase and followed by diluted Duolink detection stock solution for detection of the amplified probe. After washing, the coverslips were mounted with Duolink mounting medium and imaged. Each puncta was considered a positive protein/protein interaction, and the number per AT2 cell was determined by manual counting using NIS Elements software.
Fluorescence Image Acquisition and Analysis.
Confocal images were acquired using Nikon Ti2 Spinning disk confocal microscopy equipped with a Yokogawa CSU-X1 spinning disk head, a motorized stage, a camera (Andor DU-897 EMCCD), 60× or 100× Apo TIRF oil immersion objective (1.49 NA WD 0.12 mm) and NIS-Elements Advanced Research imaging software (Vanderbilt University Medical Center Cell Imaging Shared Resource). Single-cell images shown in the figures were processed from large fields using NIS-Elements Advanced Research imaging software and Photoshop by cropping and converting to RGB file format and adding scale bars.
Fluorescent image acquisition and analysis of immunohistochemistry was performed using a Keyence BZ-X800E with BZ-X Viewer software with a 40× objective (0.95 NA WD 0.25 to 0.17 mm). The images were processed using BZ-X Viewer software by converting to RGB, generating maximum-intensity projections, and adding scale bars. Image analysis was performed with automated HALO image analysis software (INDICA Labs). Total number of DAPI-positive and AJUBA-positive nuclei were quantified. AT2 cell number was determined by counting DAPI-positive nuclei having SFTPC based on HALO cell segmentation. AJUBA-positive AT2 cells were assessed as AT2 cells, as quantified above, having AJUBA-positive nuclei. Both SFTPC-positive AT2 cells and AT2 cells expressing both SFTPC and nuclear AJUBA were represented as a percentage of total DAPI-positive nuclei.
Dual Expression Live Cell Imaging.
MLE-15 cells transfected using JetOptimus transfection reagent were plated on glass-bottom MatTek dishes coated with 0.1% Matrigel Growth Factor Reduced Basement membrane Matrix (Corning Life Sciences) and maintained in HITES media with 10% fetal bovine serum in a 37 °C incubator with 5% CO2 for 24 h after transfection. All live-imaging experiments were performed at 37 °C, 5% CO2 in normal growth medium using Nikon Ti2 Spinning disk confocal microscopy equipped with a Yokogawa CSU-X1 spinning disk head, a motorized stage, a camera (Andor DU-897 EMCCD), 100× Apo TIRF oil immersion objective (1.49 NA WD 0.12 mm) and NIS-Elements software. Time-lapse microscopy was performed by capturing image streams over 1 min at 1 frame/s and collected at 512 × 512 pixels. Still images were derived from the first frame to avoid photobleaching.
Y3H Assay.
The Y3H assay for analysis of ATP8A1 targeting signal binding to AP-1, AP-2, and AP-3 hemicomplexes was performed essentially as described (32). Briefly, cotransformation of Saccharomyces cerevisiae strain HF7C with pBridge and pGADT7 plasmids was performed by a modification of the lithium acetate procedure as describe in the Yeast Protocols Handbook from Clontech. HF7c transformants were selected by spreading on Drop Out Base with Agar (DOBA) plates containing Yeast Nitrogen Base but lacking leucine, tryptophan, and methionine. For colony growth assays, HF7c transformants were pooled and spotted in fivefold serial dilutions on duplicate DOBA plates lacking either leucine, tryptophan, and methionine (+His) or leucine, tryptophan, methionine, and histidine (-His) and allowed to grow at 30 °C for 3 to 5 d before photography. Assays were repeated at least three times with identical results. pBridge plasmids containing the small subunit (σ1A, σ2, or σ3A) of human AP-1, AP-2, or AP-3, respectively, and either the C-terminal cytoplasmic domain of mouse tyrosinase [positive control; (31)] or a mutant form of the OCA2 N-terminal domain lacking the three di-leucine–like motifs [OCA2-AA123N, negative control; (32)] fused to the yeast GAL4 DNA binding domain, and the pGADT7 plasmids containing fusions of the large subunit (mouse γ1, rat αC, or human δ) of AP-1, AP-2, or AP-3, respectively, to the yeast Gal4-Activation Domain (10) have been previously described. pBridge plasmids containing the C-terminal cytoplasmic domain of either WT human ATP8A1 (aa 1071 to 1164) or the AA2 mutant fused to the yeast GAL4 DNA binding domain and the small subunit (σ1A, σ2, or σ3A) of human AP-1, AP-3, or AP-3, respectively, were constructed by PCR amplification of the C-terminal cytoplasmic domain (residues 1071 to 1164) of human ATP8A1 from pIRES2-EGFP-Atp8a1_Myc-Cdc50a and pIRES2-EGFP-Atp8a1-AA2_Myc-Cdc50a, digestion of the resulting fragments with EcoRI and PstI, and subcloning into pBridge/σ1A, σ2, or σ3A. Primers used were: forward, 5′-TCG CCG GAA TTC GTT ATC AAG AGG ACT GCT-3′; reverse, 5′-TGG CTG CAG GTC GAG TCA CCA TTC GTC-3′.
Quantification of Overlap by Markers in AT2 Cells.
For colocalization analysis of whole cells using Pearson’s colocalization coefficient, cells were imaged and processed using NIS Elements Advanced Research imaging software. Region of interest including each single cell was selected, and Pearson’s correlation coefficients (R) were calculated between two channels without threshold using the “colocalization” in NIS Elements software. Two or more independent experiments were repeatedly performed under each condition.
Quantitation of overlap in the cell periphery between mCherry-LactC2 and GFP-RAB11A was performed using ImageJ (NIH) on still images from the first frame of live cell movies to avoid photobleaching and used a method similar to that previously used by Dennis et al. to image melanosomes (72). Single cell images were cropped to exclude the plasma membrane, and binary fluorescence images were generated by subtracting the local background before thresholding using parameters of a rolling ball radius of 2 pixels with sliding paraboloid. The Image Calculator function was used to generate an image representing the area of overlap between channels by multiplying the binary images of each of the two channels. The areas of overlap and of total fluorescence labeling in structure larger than 1 pixel were quantified using the Analyze Particles function to get the ratio of overlap pixels (Rab11A*LactC2) to total pixels in the channel of interest (Rab11A).
Scratch Wound Assay.
The wound‐healing response was observed using a scratch assay. A total of 1 × 106 cells/well were seeded on 12-well plates under normal culture conditions and incubated for 18 h. At 100% confluence, a sterile 200 μL pipette tip was used to scrape a straight line across the cell monolayer to create a “wound.” Images were taken at 10× magnification at 0, 2, 4, 7, 12, and 24 h. The distance between leading edges analyzed (wound width at each time points/wound width at 0 h × 100 = % relative wound width).
BrdU Incorporation.
Cell proliferation was measured using the BrdU Cell Proliferation Colorimetric ELISA Kit (ab126556; Abcam, Cambridge, MA, USA) according to the manufacturer’s instructions. Briefly, cells were plated at 5 × 104 cells/mL in 100 μL/well of 96‐well plates and incubated for 18 h. On the next day, cells were incubated for 8 h with BrdU according to manufacturer’s recommendation. Absorbance was measured at 450 nm using SpectraMax 190 (Molecular Devices, Canada). Assays were completed in technical quadruplicate.
Statistical Methods.
PRISM 8 (version 8.4.2; GraphPad Software, La Jolla, CA) was used for all statistical analysis, and measures of center and variability are provided in the figure legends. For experiments comparing two groups, an unpaired t test was used and values of P < 0.05 were considered as significant. For experiments comparing >2 groups, one-way ANOVA was used with Tukey’s correction for multiple comparisons, and values of P < 0.05 were considered as significant.
Supplementary Material
Acknowledgments
We are indebted to Jim Goldenring, Greg Fairn, and Hiroyuki Arai for gifts of reagents that were used in these studies. The Vanderbilt Translational Pathology Shared Resource is supported by National Cancer Institute/NIH Cancer Center Support Grant 2P30 CA068485-14. The Vanderbilt Cell Imaging Shared Resource and Nikon Center of Excellence are supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126. Finally, we acknowledge the continued encouragement and support of the patients and families in the HPS community who inspire our work. This work was supported by grants from the NIH, National Institute of General Medical Sciences (R01 GM108807, S.H.G., M.S.M., L.G.), the National Eye Institute (R01 EY015625, M.S.M.), and the National Heart, Lung, and Blood Institute K08HL133484 (J.T.B.), and K08HL143051 (J.M.S.S.). J.M.S.S. is a Parker B. Francis Foundation Fellow. J.J.G. is a Pulmonary Fibrosis Foundation Scholar. S.H.G. is the Julia Carell Stadler Professor of Pediatrics at the Vanderbilt University School of Medicine. This work was made possible by the Pulmonary Fibrosis Foundation, by an independent grant from Boehringer Ingelheim Pharmaceuticals, Inc. who provided the financial support. The authors meet criteria for authorship as recommended by the International Committee of Medical Journal Editors (ICMJE) and were fully responsible for all aspects of publication development.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025208118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
Change History
October 7, 2021: The Acknowledgments have been updated.
References
- 1.Whitsett J. A., Wert S. E., Weaver T. E., Diseases of pulmonary surfactant homeostasis. Annu. Rev. Pathol. 10, 371–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vicary G. W., Vergne Y., Santiago-Cornier A., Young L. R., Roman J., Pulmonary fibrosis in hermansky-Pudlak syndrome. Ann. Am. Thorac. Soc. 13, 1839–1846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman S. L., Bi-Karchin J., Le L., Marks M. S., The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic 20, 404–435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kook S., et al., Impaired lysosomal integral membrane protein 2-dependent peroxiredoxin 6 delivery to lamellar bodies accounts for altered alveolar phospholipid content in adaptor protein-3-deficient pearl mice. J. Biol. Chem. 291, 8414–8427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahavadi P., et al., Epithelial stress and apoptosis underlie Hermansky-Pudlak syndrome-associated interstitial pneumonia. Am. J. Respir. Crit. Care Med. 182, 207–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young L. R., et al., The alveolar epithelium determines susceptibility to lung fibrosis in Hermansky-Pudlak syndrome. Am. J. Respir. Crit. Care Med. 186, 1014–1024 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young L. R., et al., Epithelial-macrophage interactions determine pulmonary fibrosis susceptibility in Hermansky-Pudlak syndrome. JCI Insight 1, e88947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hengst M., et al., Hermansky-Pudlak syndrome type 2 manifests with fibrosing lung disease early in childhood. Orphanet J. Rare Dis. 13, 42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Höning S., Sandoval I. V., von Figura K., A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 17, 1304–1314 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janvier K., et al., Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes. J. Cell Biol. 163, 1281–1290 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridsdale R., Na C.-L., Xu Y., Greis K. D., Weaver T., Comparative proteomic analysis of lung lamellar bodies and lysosome-related organelles. PLoS One 6, e16482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Best J. T., Xu P., Graham T. R., Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 59, 8–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmgren M., Østerberg J. T., Nintemann S. J., Poulsen L. R., López-Marqués R. L., Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Biophys. Acta Biomembr. 1861, 1135–1151 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Verhoven B., Schlegel R. A., Williamson P., Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. J. Exp. Med. 182, 1597–1601 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X., Halleck M. S., Schlegel R. A., Williamson P., A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272, 1495–1497 (1996). [DOI] [PubMed] [Google Scholar]
- 16.van der Velden L. M., et al., Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 285, 40088–40096 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hankins H. M., Baldridge R. D., Xu P., Graham T. R., Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairn G. D., et al., High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S., et al., Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34, 669–688 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takatsu H., et al., ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286, 38159–38167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto S., et al., The N- or C-terminal cytoplasmic regions of P4-ATPases determine their cellular localization. Mol. Biol. Cell 31, 2115–2124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K., Surendhran K., Nothwehr S. F., Graham T. R., P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol. Biol. Cell 19, 3526–3535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsudaira T., et al., Endosomal phosphatidylserine is critical for the YAP signalling pathway in proliferating cells. Nat. Commun. 8, 1246 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Soldt B. J., Cardoso W. V., Hippo-Yap/Taz signaling: Complex network interactions and impact in epithelial cell behavior. Wiley Interdiscip. Rev. Dev. Biol. 9, e371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gokey J. J., et al., YAP regulates NFI/KLF5 transcriptional and epigenetic networks directing alveolar epithelial cell differentiation. Ssrn Electron J. [Preprint] (2020). 10.2139/ssrn.3684853. [DOI]
- 26.Hu C., et al., The Hippo-YAP pathway regulates the proliferation of alveolar epithelial progenitors after acute lung injury. Cell Biol. Int. 43, 1174–1183 (2019). [DOI] [PubMed] [Google Scholar]
- 27.LaCanna R., et al., Yap/Taz regulate alveolar regeneration and resolution of lung inflammation. J. Clin. Invest. 129, 2107–2122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craige B., Salazar G., Faundez V., Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol. Biol. Cell 19, 1415–1426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aiken C., Konner J., Landau N. R., Lenburg M. E., Trono D., Nef induces CD4 endocytosis: Requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76, 853–864 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Calvo P. A., Frank D. W., Bieler B. M., Berson J. F., Marks M. S., A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J. Biol. Chem. 274, 12780–12789 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Theos A. C., et al., Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell 16, 5356–5372 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sitaram A., et al., Differential recognition of a dileucine-based sorting signal by AP-1 and AP-3 reveals a requirement for both BLOC-1 and AP-3 in delivery of OCA2 to melanosomes. Mol. Biol. Cell 23, 3178–3192 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade K. C., et al., Gene induction during differentiation of human pulmonary type II cells in vitro. Am. J. Respir. Cell Mol. Biol. 34, 727–737 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballard P. L., et al., Regulated gene expression in cultured type II cells of adult human lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 299, L36–L50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salaun B., et al., CD208/dendritic cell-lysosomal associated membrane protein is a marker of normal and transformed type II pneumocytes. Am. J. Pathol. 164, 861–871 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dell’Angelica E. C., Shotelersuk V., Aguilar R. C., Gahl W. A., Bonifacino J. S., Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the β 3A subunit of the AP-3 adaptor. Mol. Cell 3, 11–21 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Wikenheiser K. A., et al., Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 90, 11029–11033 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kook S., et al., Gene-edited MLE-15 cells as a model for the hermansky-Pudlak syndromes. Am. J. Respir. Cell Mol. Biol. 58, 566–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanger A., Hirst J., Davies A. K., Robinson M. S., Adaptor protein complexes and disease at a glance. J. Cell Sci. 132, jcs222992 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Dell’Angelica E. C., Bonifacino J. S., Coatopathies: Genetic disorders of protein coats. Annu. Rev. Cell Dev. Biol. 35, 131–168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman J. A., Molday R. S., Critical role of the β-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiraizumi M., Yamashita K., Nishizawa T., Nureki O., Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 365, 1149–1155 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Andersen J. P., et al., P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front. Physiol. 7, 275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Mark V. A., Elferink R. P. J. O., Paulusma C. C., P4 ATPases: Flippases in health and disease. Int. J. Mol. Sci. 14, 7897–7922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebastian T. T., Baldridge R. D., Xu P., Graham T. R., Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohno H., et al., The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273, 25915–25921 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Kyttälä A., Yliannala K., Schu P., Jalanko A., Luzio J. P., AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J. Biol. Chem. 280, 10277–10283 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Doray B., Knisely J. M., Wartman L., Bu G., Kornfeld S., Identification of acidic dileucine signals in LRP9 that interact with both GGAs and AP-1/AP-2. Traffic 9, 1551–1562 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattera R., Boehm M., Chaudhuri R., Prabhu Y., Bonifacino J. S., Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J. Biol. Chem. 286, 2022–2030 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu K., Hua Z., Nepute J. A., Graham T. R., Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell 18, 487–500 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paterson J. K., et al., Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II). Biochemistry 45, 5367–5376 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Soupene E., Kuypers F. A., Identification of an erythroid ATP-dependent aminophospholipid transporter. Br. J. Haematol. 133, 436–438 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Kay J. G., Koivusalo M., Ma X., Wohland T., Grinstein S., Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23, 2198–2212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basu S., Totty N. F., Irwin M. S., Sudol M., Downward J., Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 11, 11–23 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Zhao B., et al., Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong J., et al., Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyerla T. A., et al., Aberrant lung structure, composition, and function in a murine model of Hermansky-Pudlak syndrome. Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L643–L653 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Selman M., Pardo A., The leading role of epithelial cells in the pathogenesis of idiopathic pulmonary fibrosis. Cell. Signal. 66, 109482 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Badolato R., Parolini S., Novel insights from adaptor protein 3 complex deficiency. J. Allergy Clin. Immunol. 120, 735–741, quiz 742–743 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Beers M. F., et al., A novel conserved targeting motif found in ABCA transporters mediates trafficking to early post-Golgi compartments. J. Lipid Res. 52, 1471–1482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bonifacino J. S., Traub L. M., Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Hornbeck P. V., et al., PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pallesen L. T., et al., PAK kinases target sortilin and modulate its sorting. Mol. Cell. Biol. 40, e00411-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Awwad H. O., Millman E. E., Alpizar-Foster E., Moore R. H., Knoll B. J., Mutating the dileucine motif of the human β(2)-adrenoceptor reduces the high initial rate of receptor phosphorylation by GRK without affecting postendocytic sorting. Eur. J. Pharmacol. 635, 9–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delevoye C., Marks M. S., Raposo G., Lysosome-related organelles as functional adaptations of the endolysosomal system. Curr. Opin. Cell Biol. 59, 147–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clark J. C., et al., Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. U.S.A. 92, 7794–7798 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Akinbi H. T., et al., Rescue of SP-B knockout mice with a truncated SP-B proprotein. Function of the C-terminal propeptide. J. Biol. Chem. 272, 9640–9647 (1997). [DOI] [PubMed] [Google Scholar]
- 68.deMello D. E., et al., Ultrastructure of lung in surfactant protein B deficiency. Am. J. Respir. Cell Mol. Biol. 11, 230–239 (1994). [DOI] [PubMed] [Google Scholar]
- 69.Huizing M., et al., AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol. Biol. Cell 12, 2075–2085 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu P., Baldridge R. D., Chi R. J., Burd C. G., Graham T. R., Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J. Cell Biol. 202, 875–886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wehman A. M., Poggioli C., Schweinsberg P., Grant B. D., Nance J., The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21, 1951–1959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dennis M. K., et al., BLOC-2 targets recycling endosomal tubules to melanosomes for cargo delivery. J. Cell Biol. 209, 563–577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haraszti R. A., et al., High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles 5, 32570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skotland T., Hessvik N. P., Sandvig K., Llorente A., Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 60, 9–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postle A. D., et al., Lipidomics of cellular and secreted phospholipids from differentiated human fetal type II alveolar epithelial cells. J. Lipid Res. 47, 1322–1331 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Griese M., et al., Quantitative lipidomics in pulmonary alveolar Proteinosis. Am. J. Respir. Crit. Care Med. 200, 881–887 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Agudelo C. W., et al., Decreased surfactant lipids correlate with lung function in chronic obstructive pulmonary disease (COPD). PLoS One 15, e0228279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klein S.et al., Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Comm. Biology 4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ansari I. U., et al., Characterization of P4 ATPase phospholipid translocases (flippases) in human and rat pancreatic beta cells: Their gene silencing inhibits insulin secretion. J. Biol. Chem. 290, 23110–23123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korogi Y., et al., In vitro disease modeling of hermansky-Pudlak syndrome type 2 using human induced pluripotent stem cell-derived alveolar organoids. Stem Cell Reports 12, 431–440 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young L. R., Borchers M. T., Allen H. L., Gibbons R. S., McCormack F. X., Lung-restricted macrophage activation in the pearl mouse model of Hermansky-Pudlak syndrome. J. Immunol. 176, 4361–4368 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young L. R., Pasula R., Gulleman P. M., Deutsch G. H., McCormack F. X., Susceptibility of Hermansky-Pudlak mice to bleomycin-induced type II cell apoptosis and fibrosis. Am. J. Respir. Cell Mol. Biol. 37, 67–74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Atochina-Vasserman E. N., et al., Early alveolar epithelial dysfunction promotes lung inflammation in a mouse model of Hermansky-Pudlak syndrome. Am. J. Respir. Crit. Care Med. 184, 449–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu M., et al., YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell Death Dis. 9, 464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou B., et al., Claudin-18-mediated YAP activity regulates lung stem and progenitor cell homeostasis and tumorigenesis. J. Clin. Invest. 128, 970–984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng L., et al., The Hermansky-Pudlak syndrome 1 (HPS1) and HPS2 genes independently contribute to the production and function of platelet dense granules, melanosomes, and lysosomes. Blood 99, 1651–1658 (2002). [PubMed] [Google Scholar]
- 87.Tang X., Yamanaka S., Miyagi Y., Nagashima Y., Nakatani Y., Lung pathology of pale ear mouse (model of hermansky-Pudlak syndrome 1) and beige mouse (model of chediak-higashi syndrome): Severity of giant lamellar body degeneration of type II pneumocytes correlates with interstitial inflammation. Pathol. Int. 55, 137–143 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Gautam R., et al., Interaction of Hermansky-Pudlak Syndrome genes in the regulation of lysosome-related organelles. Traffic 7, 779–792 (2006). [DOI] [PubMed] [Google Scholar]
- 89.Nakatani Y., et al., Interstitial pneumonia in hermansky-Pudlak syndrome: Significance of florid foamy swelling/degeneration (giant lamellar body degeneration) of type-2 pneumocytes. Virchows Arch. 437, 304–313 (2000). [DOI] [PubMed] [Google Scholar]
- 90.Treutlein B., et al., Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kobayashi Y., et al., Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao R., et al., Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev. Cell 30, 151–165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Z., et al., MAPK-mediated YAP activation controls mechanical-tension-induced pulmonary alveolar regeneration. Cell Rep. 16, 1810–1819 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Otsubo K., et al., MOB1-YAP1/TAZ-NKX2.1 axis controls bronchioalveolar cell differentiation, adhesion and tumour formation. Oncogene 36, 4201–4211 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Volckaert T., et al., Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Dev. Cell 43, 48–59.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kanungo J., Pratt S. J., Marie H., Longmore G. D., Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 11, 3299–3313 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lange A. W., et al., Hippo/Yap signaling controls epithelial progenitor cell proliferation and differentiation in the embryonic and adult lung. J. Mol. Cell Biol. 7, 35–47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gokey J. J., et al., Active epithelial Hippo signaling in idiopathic pulmonary fibrosis. JCI Insight 3, e98738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baetz N. W., Goldenring J. R., Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol. Biol. Cell 24, 643–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sucre J. M. S., et al., Successful establishment of primary type II alveolar epithelium with 3D organotypic coculture. Am. J. Respir. Cell Mol. Biol. 59, 158–166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fisher A. B., Dodia C., Lysosomal-type PLA2 and turnover of alveolar DPPC. Am. J. Physiol. Lung Cell. Mol. Physiol. 280, L748–L754 (2001). [DOI] [PubMed] [Google Scholar]
- 102.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.