Significance
Most sequence variant analyses rely on a linear reference genome that is assumed to lack millions of bases that occur in the genomes of other individuals. To quantify the extent and functional relevance of such missing bases, we integrate six genome assemblies from cattle and related species into a pangenome. This allows us to uncover more than 70 million bases that are not included in the Bos taurus reference genome. Through complementary bioinformatics, genomics, and transcriptomics methods, we discover putative genes from nonreference sequences that are differentially expressed and thousands of polymorphic sites that were unused so far. Our work provides a computational framework, broadly applicable to many species, to make a so-far neglected source of genomic variation amenable to genetic investigations.
Keywords: pangenome, genome graphs, reference genome, genetic diversity
Abstract
Many genomic analyses start by aligning sequencing reads to a linear reference genome. However, linear reference genomes are imperfect, lacking millions of bases of unknown relevance and are unable to reflect the genetic diversity of populations. This makes reference-guided methods susceptible to reference-allele bias. To overcome such limitations, we build a pangenome from six reference-quality assemblies from taurine and indicine cattle as well as yak. The pangenome contains an additional 70,329,827 bases compared to the Bos taurus reference genome. Our multiassembly approach reveals 30 and 10.1 million bases private to yak and indicine cattle, respectively, and between 3.3 and 4.4 million bases unique to each taurine assembly. Utilizing transcriptomes from 56 cattle, we show that these nonreference sequences encode transcripts that hitherto remained undetected from the B. taurus reference genome. We uncover genes, primarily encoding proteins contributing to immune response and pathogen-mediated immunomodulation, differentially expressed between Mycobacterium bovis–infected and noninfected cattle that are also undetectable in the B. taurus reference genome. Using whole-genome sequencing data of cattle from five breeds, we show that reads which were previously misaligned against the Bos taurus reference genome now align accurately to the pangenome sequences. This enables us to discover 83,250 polymorphic sites that segregate within and between breeds of cattle and capture genetic differentiation across breeds. Our work makes a so-far unused source of variation amenable to genetic investigations and provides methods and a framework for establishing and exploiting a more diverse reference genome.
A well-annotated reference genome enables systematic characterization of sequence variation within and between populations, as well as across species. The reference genome of domestic cattle (Bos taurus taurus) was generated from the inbred Hereford cow «L1 Dominette 01449» (1). Long-read sequencing and sophisticated genome assembly methods have enabled spectacular improvements in the contiguity and quality of the Bos taurus reference genome. The contig (contiguous sequence formed by overlapping reads without gaps) N50 size (i.e., 50% of the genome is in contigs of this size or greater) of the B. taurus reference genome has increased from kilo- to megabases over the past 5 y (2). Recent method and sequencing technology developments have facilitated the assembly of multiple reference-quality genomes. The application of trio binning (3) resulted in chromosome-scale haplotype-resolved assemblies for three taurine (Hereford, Angus, and Highland) and one indicine (Brahman) cattle breeds as well as for yak (Bos grunniens), a closely related species to domestic cattle (4, 5).
DNA sequences from taurine and indicine cattle are typically aligned to the Hereford-based reference genome to discover and genotype variable sites. Reference-guided read alignment and variant genotyping has revealed millions of polymorphic variants that segregate within and between taurine and indicine cattle breeds (6–8). However, using the linear reference in this alignment approach is susceptible to reference-allele bias, particularly for DNA samples that are greatly diverged from the reference (9, 10). Moreover, reference-guided methods are blind to variations in sequences that are not present in the reference genome (11). Recent estimates suggest that millions of bases are missing in mammalian reference genomes (12, 13), indicating a high potential for bias.
Efforts to mitigate reference-allele bias and increase the genetic diversity of reference genomes have led to graph-based references (14, 15). We have previously shown that a genome graph, which integrates linear reference coordinates and preselected variants, improves the mapping of reads and enables unbiased variant genotyping in different breeds of cattle (16, 17). However, previous attempts focused on augmenting the B. taurus reference genome with small variations [<50 base pairs (bp)], not the larger class of structural variations. Despite being an important source of genotypic and phenotypic diversity (18, 19), little is known about the prevalence and functional impact of structural variations in the cattle genome. The availability of reference-quality assemblies and long-read sequencing data from different breeds of cattle now provides an opportunity to characterize sequence diversity beyond small variations (20, 21).
In this paper, we integrate reference-quality assemblies from multiple taurine breeds as well as two close relatives into a multiassembly graph with minigraph (21). We detect autosomal sequences that are missing in the B. taurus reference genome and investigate their functional significance using transcriptome data. We show that the nonreference sequences contain transcripts that are differentially expressed as well as polymorphic sites that segregate within and between breeds of cattle.
Results
Construction of a Bovine Multiassembly Graph.
We considered the Hereford-based B. taurus reference genome and five reference-quality assemblies from three breeds of taurine (B. t. taurus) cattle (Angus, Highland, and Original Braunvieh) (2, 4, 5) and their close relatives Brahman (Bos taurus indicus) (4) and yak (B. grunniens) (5). All assemblies, except for the Original Braunvieh breed, were generated prior to this study. The reference-quality assembly for an Original Braunvieh female calf was created with 28-fold PacBio high-fidelity (HiFi) read coverage (SI Appendix, Note S1). The contig and scaffold N50 values of the six assemblies ranged from 21 to 80 Mb and 86.2 to 108 Mb, respectively (Table 1).
Table 1.
Details of six bovine genome assemblies
| Assembly (Species) | Sex* | Primary data used for the assembly† | Type of assembly | Assembler | Contig N50 (Mb) | Scaffold N50 (Mb) | Length of the autosomes |
| Hereford (Bos taurus taurus) | F | PacBio (80-fold CLR) | Primary | Falcon | 21 | 108 | 2,489,385,779 |
| Angus (Bos taurus taurus) | M | PacBio (136-fold CLR) | Haplotype-resolved | TrioCanu | 29.4 | 102.8 | 2,468,157,877 |
| Highland (Bos taurus taurus) | F | PacBio (125-fold CLR) | Haplotype-resolved | TrioCanu | 71.7 | 86.2 | 2,483,452,092 |
| Original Braunvieh (Bos taurus taurus) | F | PacBio (28-fold HiFi) | Primary | Hifiasm | 86.0 | 96.3 | 2,607,746,442 |
| Brahman (Bos taurus indicus) | F | PacBio (136-fold CLR) | Haplotype-resolved | TrioCanu | 23.4 | 104.5 | 2,478,073,158 |
| Yak (Bos grunniens) | F | PacBio (125-fold CLR) | Haplotype-resolved | TrioCanu | 70.9 | 94.7 | 2,478,308,164 |
Female (F) and male (M) assemblies contain either X or Y chromosomal sequences.
Additional data may have been used to polish the assemblies and facilitate scaffolding; CLR: continuous long reads; HiFi: high-fidelity.
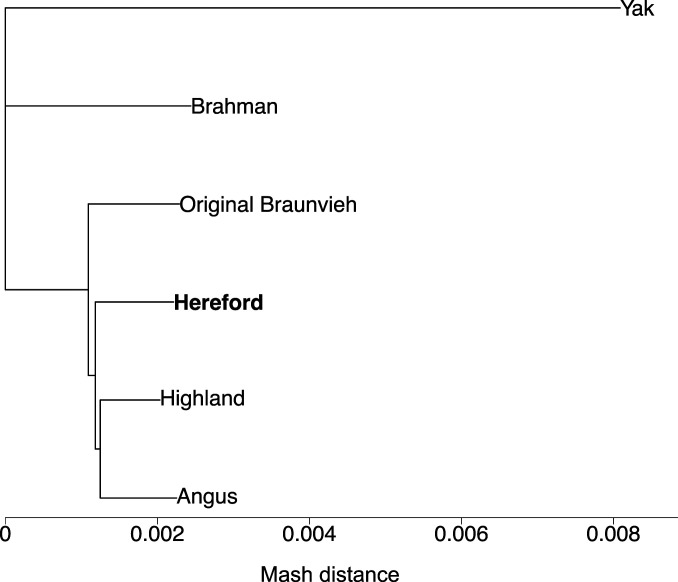
The six assemblies were integrated into a multiassembly graph with minigraph. We only considered autosomal sequences because the haplotype-resolved assemblies represent either paternal or maternal haplotypes, thus lacking either X or Y chromosomal sequences. The Hereford-based linear reference genome (ARS-UCD1.2) formed the backbone of the bovine multiassembly graph. The graph was then augmented with the five additional assemblies, added in order of increasing Mash distance from the ARS-UCD1.2 reference (22) (Fig. 1). Constructing this multiassembly graph took 4.1 CPU (central processing unit) hours and 58 GB of RAM, taking 36 min of wall-clock time when using 10 threads.
Fig. 1.
Phylogenetic distance between six genome assemblies. A Mash-based phylogenetic tree derived from six bovine assemblies, including the current Hereford-based B. taurus reference genome (bold). The yak assembly was used as the outgroup to root the tree during building.
Recovery of Nonreference Sequences from the Multiassembly Graph.
Our bovine multiassembly graph represents 2,558,596,439 nucleotides, spread across 182,940 nodes connected by 258,396 edges. On average, a node spans 13,985 nucleotides and is connected by 1.4 edges. Of the edges, 141,086, 113,332, and 3,978 connect two reference nodes, a reference and nonreference node, or two nonreference nodes, respectively.
The vast majority (2,489,385,779 or 97.29%) of nucleotides in the multiassembly graph originate from the linear reference backbone, covered in 123,483 nodes. These reference nodes span 23,088 bases on average, ranging from 100 to 1,398,882 bases. The incremental integration of the Highland, Angus, Original Braunvieh, Brahman, and yak assemblies added 8,847, 4,613, 3,555, 11,996, and 30,446 nonreference nodes, respectively containing 14,679,286, 5,537,769, 7,013,258, 11,116,220, and 30,864,127 nonreference bases. The resulting multiassembly graph contained 59,457 nonreference nodes spanning 69,210,660 bases.
To determine the support of the nonreference nodes, we aligned individual assemblies back to the multiassembly graph. Nodes were then labeled according to which assembly path traversed them (SI Appendix, Figs. S1 and S2). This approach enabled a straightforward confirmation of minigraph’s mapping accuracy. Only reference nodes should contain a Hereford label, since this assembly was used as the backbone of the graph. Mapping was highly accurate, as indicated by an F1 score of 99.97%.
The nonreference nodes of the multiassembly graph had a cumulative length of 43,341,418, 23,644,772, 18,202,102, 14,453,112, and 15,542,368 bases in the yak, Brahman, Original Braunvieh, Angus, and Highland assemblies, respectively. Yak and Brahman nonreference nodes were shorter on average compared to the taurine assemblies (SI Appendix, Fig. S3). Most nonreference nodes (41,855 or 70.40%) and nonreference sequences (42.52 Mb, 69.52%) were either private to yak (29,854 nodes, 29.9 Mb), Brahman (7,843 nodes, 8.22 Mb), or shared by both assemblies (4,158 nodes, 3.05 Mb) (S1 Appendix, Fig. S4). The Original Braunvieh, Highland, and Angus assemblies contributed 4.51, 2.78, and 2.39 Mb in 2,016, 1,938, and 1,759 nodes, respectively, that were not detected in any other assembly. The three taurine assemblies shared 668 nodes containing 0.77 Mb not detected in ARS-UCD1.2, yak, or Brahman. There were also 1,318 nonreference nodes with a cumulative length of 4.4 Mb supported by all five additional assemblies.
The core genome of the multiassembly graph (i.e., nodes shared by all assemblies) is contained in 67,482 nodes with a cumulative length of 2,402,561,410 bases. About 6.10% of the pangenome (115,458 nodes containing 156,035,029 bases) is flexible (i.e., not shared by all assemblies). Of the flexible part, 69,697 nodes containing 97,106,100 bases are shared by at least two assemblies, and 45,761 nodes with 58,928,929 bases are only found in one assembly. The profile of the multiassembly graph changes markedly when distant assemblies (e.g., Brahman and yak) are added (SI Appendix, Note S2).
The minigraph approach used to construct the multiassembly graph does depend on an initial sequence forming a backbone. The choice of backbone consequently impacts the amount of nonreference sequence detected from each additional assembly (SI Appendix, Note S3). However, the overall effect on the sequence content of the multiassembly graph is relatively minor, with 68.72 ± 3.17 Mb of nonreference sequence identified across all possible backbones.
Structural Variation Discovery from the Multiassembly Graph.
Using the bubble popping algorithm of gfatools (21), we identified 68,328 structural variations present in the multiassembly graph. To reveal true alleles within these structural variations, we traversed all possible paths through the bubbles (i.e., alleles) and retained only those that were supported by at least one assembly (SI Appendix, Fig. S2). Most of the structural variations had two alleles (64,224 or 94%). The remaining 4,104 structural variations were multiallelic, most of which had three alleles (3,324 or 81%). We identified 141,747 alleles at the structural variations, including 73,506 nonreference alleles with a cumulative length of 74,453,929 bases.
We overlapped the breakpoints of the structural variations with the Ensembl annotation (build 101) of ARS-UCD1.2. Almost all structural variations were either intergenic (47,642 or 69.81%) or intronic (20,227 or 29.64%). There were 170 and 202 exons and coding sequences, respectively, of 338 unique genes affected by structural variations. A Panther GO-Slim Biological Process (23) analysis indicated that these genes are enriched for genes related to the adaptive immune response (4.35-fold, P = 0.04), T cell mediated immunity (6.37-fold, P = 0.04), actin filament depolymerization (8.54-fold, P = 6.56 × 10−3), microtubule cytoskeleton organization (10.48-fold, P = 1.85 × 10−4), and iron–sulfur cluster assembly (9.96-fold, P = 0.02).
The nonreference alleles consisted of 40,369 insertions and 33,137 deletions with an average length of 1,181 and 1,210 bases, respectively (SI Appendix, Table S1). The cumulative length (absolute difference between reference and nonreference allele) was longer for insertions (47,691,942 bases) than deletions (40,101,303 bases). This pattern was similar for biallelic variations (35,748 and 28,476 biallelic insertions and deletions, respectively, encompassing 37,388,222 and 28,373,582 bases with an average variant length of 1,045 and 996 bases). The multiassembly graph contained more complete insertions (20,432; i.e., only nonreference sequences present in the bubbles, thus reference length is 0) than alternate insertions (15,316; i.e., both reference and nonreference sequences present but nonreference allele is longer). The pattern was similar for deletions. The multiallelic structural variations had 13,299 alleles including 9,282 nonreference alleles with 4,621 insertions and 4,661 deletions, respectively, affecting 11,727,721 and 10,303,720 bases. Bubbles with multiallelic structural variations contained more mixed mutations (1,941; both deletions and insertions detected within the same bubble) than multiple mutations of the same type (994 and 1,082 for multiple insertions and deletions, respectively).
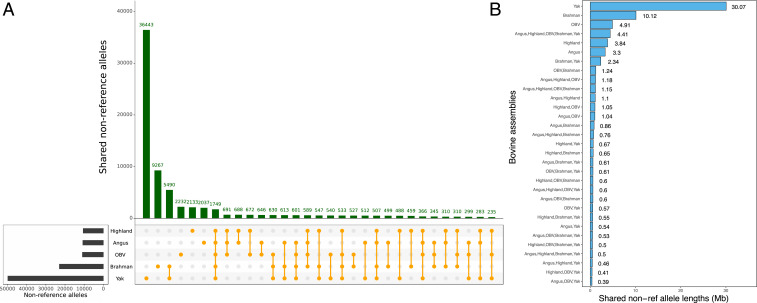
When compared to the ARS-UCD1.2 backbone, the yak, Brahman, Original Braunvieh, Angus, and Highland assemblies contained respectively 49,836, 22,976, 10,965, 10,735, and 10,560 nonreference alleles (Fig. 2). Most nonreference alleles (36,443, total length: 30 Mb) were private to the yak assembly. We detected 9,267, 2,232, 2,133, and 2,037 nonreference alleles, respectively, containing 10.1, 4.9, 3.8, and 3.3 Mb that were private to the Brahman, Original Braunvieh, Highland, and Angus assembly (Fig. 2 and SI Appendix, Fig. S5). We also found 1,749 alleles within the 4.4 Mb of nonreference sequence (2.1 Mb of which is nonrepetitive) shared by all assemblies except ARS-UCD1.2.
Fig. 2.
Nonreference alleles detected across assemblies. Intersection of nonreference alleles (A) and cumulative length of the alleles (B) found in five assemblies when compared to ARS-UCD1.2. OBV: Original Braunvieh.
We mapped PacBio HiFi reads from a Nellore (B. t. indicus) × Brown Swiss (B. t. taurus) crossbred bull to the multiassembly graph to examine support for the nonreference alleles. Nearly one-third of the structural variation breakpoints had support from the hybrid cattle, while this rose to approximately three-quarters after excluding nodes with only yak labels. Since neither parental breed is present in the multiassembly graph, this suggests that the discovered structural variation may be prevalent in different breeds of taurine and indicine cattle.
Sequence Content of the Structural Variations.
In order to investigate the functional relevance of the nonreference sequences, we extracted 45,357 nonreference alleles from the 70,329,827 nonreference bases in the multiassembly graph (SI Appendix, Fig. S6). These sequences originate from 38,906 biallelic and 6,451 multiallelic structural variations, respectively, that have a cumulative length of 43,003,591 and 27,326,236 bases. On average, the alleles of multiallelic structural variations were 4 times longer than that of biallelic bubbles (4,205 versus 1,104 bases).
The nonreference sequences are largely composed of repetitive elements (53,690,260 bases or 76.34%, SI Appendix, Fig. S7). LINE/L1 and LINE/RTE-BovB account for 28.04 (52.22%) and 6.77 (12.61%) Mb repetitive nonreference bases, respectively. Repetitive sequences (both interspersed and simple repeats) are more evenly distributed across the autosomes than nonrepetitive sequences. Both repetitive and nonrepetitive nonreference sequences were detected at two regions on bovine chromosomes 18 and 23 that encompass the leukocyte receptor complex and the major histocompatibility complex (SI Appendix, Fig. S8).
We hypothesized that the 16,639,567 nonrepetitive nonreference bases contain transcribed sequences. A BLASTX search of these sequences against a protein sequence database of Bos and related species revealed hits for 403 structural variations containing 299,337 nonreference bases. As a complementary approach, we predicted genes from the nonrepetitive sequences using the Augustus software tool. The ab initio prediction revealed 857 gene models from 768 distinct structural variations that had a minimum coding sequence length of 150 bp, including 374 complete gene models with transcription start site, start codon, exons, stop codon, and transcription termination site (SI Appendix, Table S2). On average, the transcript, coding sequence, and protein length of the complete gene models is respectively 4,742 bp, 794 bp, and 264 amino acids.
De Novo Transcript Assembly from the Nonreference Sequences.
As the two complementary gene prediction methods indicated that these nonreference sequences contain transcribed features, we sought experimental evidence. We appended the 70 Mb of repeat-masked nonreference sequences contained in 45,357 additional contigs to the ARS-UCD1.2 reference, making an extended reference genome. This renders the nonreference sequences amenable to current methods of linear mapping of transcriptome data. Using HISAT2, we aligned liver transcriptomes from 39 cattle across taurine (Angus, Holstein, and Jersey) and indicine (Brahman) breeds to both the linear reference as well as the extended reference. We also aligned transcriptomes from Dominette, the animal sequenced to assemble the B. taurus reference genome. A greater portion of reads mapped to the extended reference compared to the original reference for all examined samples (SI Appendix, Fig. S9). Across the 40 samples, the overall mapping rate increased by 0.037%, which corresponds to ∼18,000 reads for a paired-end RNA-sequencing (RNA-seq) dataset of 25 million reads. The mapping improvements were larger for samples with greater genetic distance from the reference genome. Brahman had the largest improvement (0.060%), followed by the taurine breeds: Angus (0.032%), Holstein (0.026%), and Jersey (0.030%). As expected, Dominette benefitted the least (0.010%) but still demonstrated an improvement over using the original reference.
Next, we used StringTie2 (24), guided with gene models predicted by Augustus (see above), to assemble reads which aligned to nonreference sequences into 1,431 nonreference genes. Of these, 885 were expressed at transcripts per million (TPM) ≥ 1 in at least one breed, including 405 that were originally predicted by Augustus. We selected these 405 putative genes, supported by both ab initio prediction and de novo transcript assembly for further analyses.
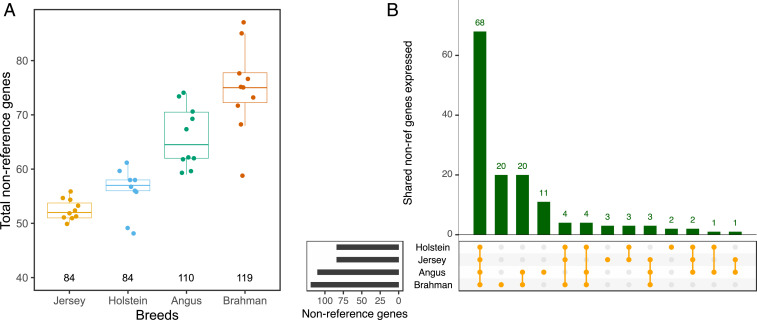
Only 263 of the 405 putative genes were expressed at TPM ≥ 1 in Dominette, with BLASTP queries indicating they may mostly be divergent copies of ribosomal proteins or olfactory receptors. The remaining 142 genes were expressed at TPM ≥ 1 in Angus, Holstein, Jersey, or Brahman cattle. Most were expressed in Brahman cattle (Fig. 3A), including 20 genes specific to this indicine breed. Among the taurine breeds, Angus contributed more genes than either Holstein or Jersey cattle. Approximately half of these genes, 68 of the 142, were common to all four nonreference breeds (Fig. 3B). The average expression was significantly higher (P = 0.004, one-tailed Student’s t test) for genes that were expressed in at least two breeds (N = 106, TPM = 13.48) than genes expressed in only one breed (N = 36, TPM = 1.64). BLASTP queries provided additional support for 57 out of the 142 genes (SI Appendix, Fig. S10). The top hits suggest that these genes encode proteins related to the following: immune response (antigen-presenting glycoprotein, immunoglobulin, Bovine Leukocyte Antigen [BOLA], killer T cell, interferon, Ig-like lectin, CMRF35, MHC [major histocompatibility complex], and cytokine), signaling (G protein–signaling protein and tyrosine phosphatase), cytoskeleton regulations (myosin, actin, twinfilin, and KANTB1), lipid metabolism (apolipoprotein and lipid-binding protein), and protein modifications (heat-shock chaperone, ubiquitin conjugating enzyme, and rhoA ubiquitin).
Fig. 3.
Transcribed genes detected from nonreference sequences. (A) Number of nonreference genes expressed ≥1 TPM in liver tissue from taurine (Jersey, Holstein, and Angus) and indicine (Brahman) cattle breeds. Each point represents the number of nonreference genes detected per animal. The number of distinct nonreference genes detected for each breed is indicated below the boxplots. (B) Expression of 142 nonreference genes in four cattle breeds.
Nonreference Sequences Contain Differentially Expressed Genes.
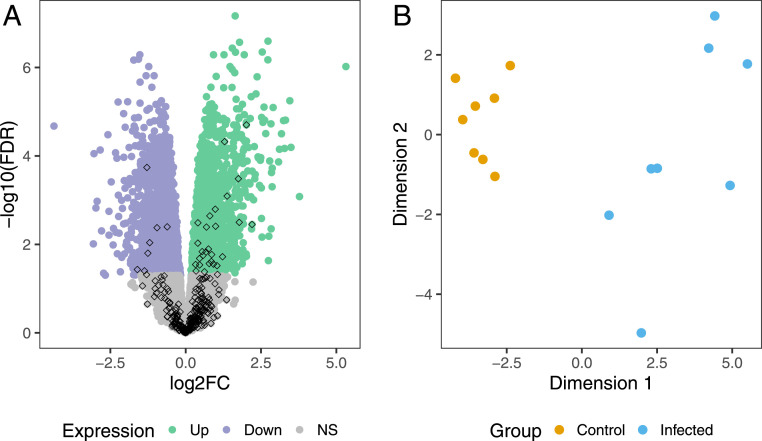
To investigate if the nonrepetitive sequences also encode transcripts that are differentially expressed between individual B. taurus cattle, we obtained publicly available peripheral blood leukocyte transcriptome data for eight M. bovis–infected and eight noninfected Holstein cattle (25). Following the transcriptome analysis introduced earlier, the RNA-seq reads were aligned to both the standard and extended ARS-UCD1.2 reference genome sequence. Between 8,616,414 and 23,940,699 RNA-seq reads aligned to the standard and between 8,631,277 and 23,977,859 RNA-seq reads aligned to the extended reference genome. The subsequent de novo transcript assembly from the nonreference sequences produced 949 transcripts encoded by 661 nonreference genes. We appended them to the Ensembl ARS-UCD1.2 annotation, yielding a total of 28,268 genes. Considering only unique alignments, we detected expression levels ≥1 counts per million (CPM) in at least eight samples for 13,085 genes, including 272 nonreference genes. We subsequently tested these genes for differential expression, finding 3,646 genes, including 36 nonreference genes, which were differentially expressed (false discovery rate (FDR) ≤ 0.05) between M. bovis–infected and noninfected cattle (Fig. 4A). The top differentially expressed genes from our extended Ensembl ARS-UCD1.2 annotation as well as their transcript abundances in cases and controls agreed well with the original findings from McLoughlin et al. (25) that were based on the previous UMD3.1 annotation (Pearson R log2 fold-change: 0.99) as well as with those from the standard ARS-UCD1.2 reference genome annotation (Pearson R log2 fold-change: 0.99, SI Appendix, Note S4).
Fig. 4.
Differentially expressed nonreference genes. (A) A volcano plot representing results from the differential expression analysis. The green and purple color indicates genes that are up- and down-regulated (FDR ≤ 0.05), respectively, in peripheral blood leukocytes of M. bovis–infected cattle. The diamond shapes indicate the 272 genes found in nonreference sequences. (B) Multidimensional scaling plot of 36 differentially expressed nonreference genes in M. bovis–infected (blue) and noninfected (orange) Holstein cattle.
Within the 36 differentially expressed nonreference genes, 28 and 8 are respectively up- and down-regulated in peripheral blood leukocytes of M. bovis–infected cattle, with an average twofold change compared to noninfected controls (SI Appendix, Fig. S11). Multidimensional scaling representations of transcript abundance estimates of the 36 differentially expressed genes separated M. bovis–infected from noninfected cattle (Fig. 4B). BLASTX queries against a protein reference database provided additional support for 13 out of 36 differentially expressed genes (SI Appendix, Table S3). The top up-regulated nonreference gene supported by the BLASTX query (4.04-fold increase, P = 1.98 × 10−5) encodes the Workshop Cluster (WC) 1.1-like protein [i.e., a receptor expressed on gamma delta T cells that modulates the immune response to M. bovis infections (26–28)].
The top down-regulated nonreference gene supported by the BLASTX query encodes a protein with high similarity (79.80%) to leukocyte immunoglobulin–like receptor A5 (LILRA5). LILRA5 triggers the strength of the innate immune response to Mycobacterium infections (29) and might serve as a target for pathogen-mediated immunomodulation. Many genes of the leukocyte receptor complex are missing in the assembled chromosomes of the ARS-UCD1.2 reference (30); instead, LILRA5 (LOC100139766) is annotated on a 236 kb long unplaced scaffold (NW_020190675). A nonreference gene encoding a protein similar to LILRA5 is located within a 20.4 kb insertion of the multiassembly graph at 62,471,732 bp on chromosome 18. Both taurine (Original Braunvieh) and indicine (Brahman) assemblies support this insertion. The gene encoding LILRA5 is expressed at 9.59 ± 2.54 and 23.10 ± 8.30 CPM, respectively, in M. bovis–infected and noninfected cattle, corresponding to a 2.19-fold decrease (P = 1 × 10−4) in infected cattle (SI Appendix, Table S3).
Variant Discovery from the Nonreference Sequences.
Next, we mapped short sequencing reads, with an average of 19-fold sequencing coverage, from 45 cattle representing five taurine breeds against ARS-UCD1.2 and the extended ARS-UCD1.2 reference genome. An average number of 34,342 reads per sample mapped perfectly within 50 bp of the breakpoints of the newly added contigs, indicating that the addition of 100 bp flanking sequence was sufficient to facilitate accurate alignments. Across 45 samples, the average mapping rate increased by 0.0176% over ARS-UCD1.2, corresponding to ∼100,000 sequencing reads for a DNA sample sequenced at 30-fold coverage. The mapping rate increased more noticeably for Brown Swiss (0.024%) and Original Braunvieh (0.021%) than Holstein (0.015%) and Simmental (0.016%) cattle (SI Appendix, Fig. S12). Similarly, to the transcriptome mapping, sequence reads from Dominette benefitted the least from the extended reference genome (0.006%). However, the increase in mapping rate was greater (0.013%) for other Hereford cattle. For all breeds, the extended reference genome also enabled more perfect alignments (alignments without difference from the reference), less partially mapped (i.e., clipped) reads, and less reads with supplementary alignments. However, the proportion of reads with unique alignment was lower for the extended than standard reference genome (SI Appendix, Table S4).
We next investigated the alignments against the 2,115,702 nonrepetitive nonreference bases detected in all assemblies except ARS-UCD1.2. Among these, 919,761 bases were covered by confident alignments (≥10-fold) from Dominette. This suggests that, although absent from the autosomal assembly, these sequences do occur in the animal used to construct the reference. However, 1,195,941 bp were not covered with reads from Dominette but instead from Brown Swiss, Holstein, Original Braunvieh, or Simmental samples. Strikingly, reads from non-Dominette Hereford samples covered 745,392 of the 1,195,941 bases. This directly implies that Dominette has individual-specific deletions, which are either rare or absent in other Hereford cattle.
Mapping against the extended reference resulted in many reads changing alignment location to the nonreference additions. Most (85.55%) of the reads mapping at nonreference sequences already mapped to the original ARS-UCD1.2 reference genome, although 5% of these mapped to unplaced contigs, while 14.45% were previously unmapped. These mappings displayed an increase in the average mapping quality (22 to 44), alignment score (110 to 142), and alignment identity (0.975 to 0.995). The proportion of clipped reads decreased from 39 to 4%. The subset of these reads which were previously unmapped showed even greater improvements (SI Appendix, Fig. S13).
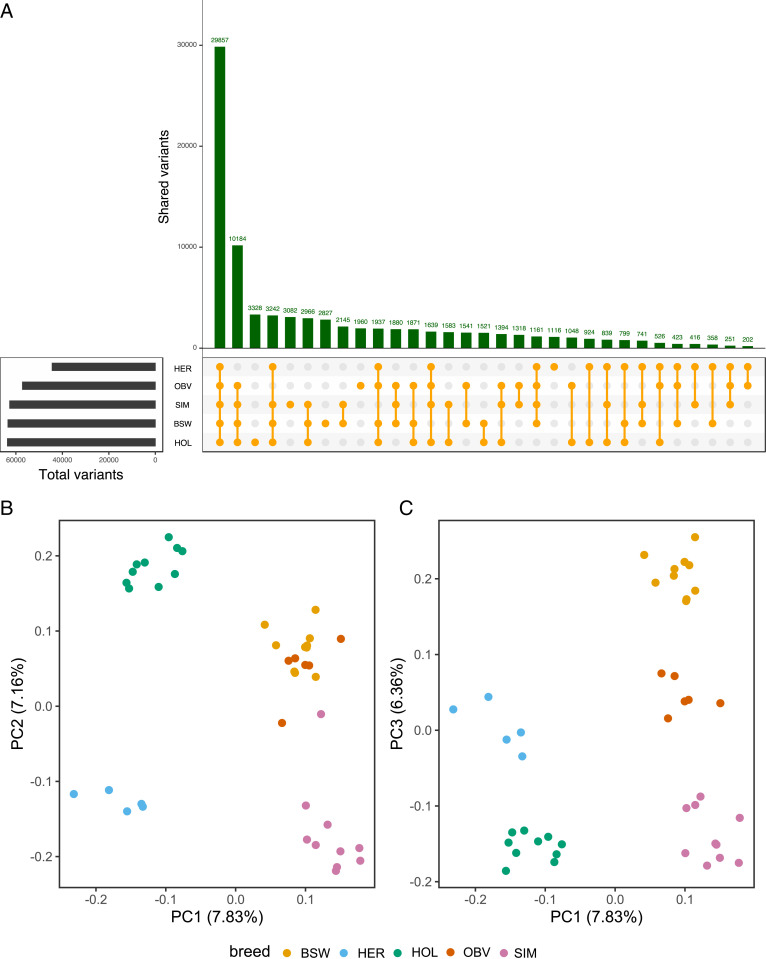
Using reads with mapping quality greater than 10 for reference-guided sequence variant genotyping yielded 83,250 filtered variants (73,709 single-nucleotide polymorphisms [SNPs], 9,541 insertion/deletion polymorphisms [indels]) in nonreference sequences that were identified by both SAMtools and GATK. These variants formed 80,995 biallelic and 2,255 multiallelic sites, with a Ti:Tv (transition:transversion) ratio of 1.91, averaging 1.18 variants per kb. A total of 3,890 small variations (Ti:Tv ratio: 1.79) were detected within 50 bp of the breakpoints of the newly added contigs. On average, each Brown Swiss, Original Braunvieh, Holstein, Simmental, and Hereford animal, respectively, had 31,028, 29,685, 29,851, 30,309, and 15,845 variant sites in nonreference bases (Fig. 5A). A DNA sample from Dominette had considerably fewer polymorphic sites at nonreference bases, only 7,531. Most variants (32.67%) had alternate allele frequency less than 0.1, and 193 were fixed for the alternate allele (SI Appendix, Fig. S14). The top principal components from a genomic relationship matrix that was built from the 83,250 nonreference variants separated the animals by breeds (Fig. 5 B and C). Functional annotation based on the gene models predicted from Augustus indicated that most nonreference variants were either intergenic (83%) or intronic (7.5%). A total of 1,138 variants (Ti:Tv ratio: 1.83) were in putative coding sequences, of which 54 were classified as “HIGH IMPACT” variants (SI Appendix, Table S5).
Fig. 5.
Polymorphic sites detected from nonreference sequences in five breeds. (A) Sharing of 83,250 variants across five taurine cattle breeds (BSW: Brown Swiss, HER: Hereford, HOL: Holstein, OBV: Original Braunvieh, SIM: Simmental). (B and C) The top three principal components (PC) of a genomic relationship matrix constructed from nonreference sequence variants separate the animals by breed.
Discussion
We utilize a bovine multiassembly graph to uncover sequences that are not included in the B. taurus reference genome. Nonreference contigs can also be assembled from unmapped reads, but placing them onto reference coordinates is difficult (12, 31). Our approach provides physical coordinates for the nonreference sequences because the breakpoints anchor them onto the reference genome. Despite including the genetically distant yak, constructing the multiassembly graph using minigraph (21) was computationally efficient and scalable. Our multiassembly graph utilizes a well-annotated backbone assembly to identify nonreference sequences from other assemblies. We show that the choice of the backbone as well as its genetic distance to all other assemblies influences the amount of nonreference bases uncovered through the multiassembly graph. Sophisticated algorithms facilitate the reference-free alignment of thousands of assemblies (32). To determine the origin of the nonreference sequences, we developed an approach to assign labels to all nodes in the multiassembly graph. Our evaluation showed that this strategy is highly accurate.
By systematically characterizing structural variations in multiple assemblies from domestic cattle and their close relatives, we detect 45,357 autosomal segments with a cumulative length of 70,329,827 bases that are not part of the B. taurus reference genome. To obtain continuous nonreference sequences spanning multiple nonreference nodes, we recovered the nonreference alleles from structural variations. The number of bases detected in our study that are not in the B. taurus reference genome is comparable to values reported for pigs [72.5 Mb (33)] and goats [38.3 Mb (34)], based on multiassembly graphs constructed from 11 and 8 animals representing different breeds, respectively. In our study, many nonreference sequences originate from yak. Hybridizing between yak and cattle is widely practiced and results in fertile female descendants. However, multiple generations of backcrossing are required for males to resume fertility (35). A pangenome constructed from domestic cattle and their extant relatives as recently proposed by the Bovine Pangenome Consortium (36) will reveal variants that were lost during domestication and the separation of cattle into specialized breeds (37). For instance, some of the 8 million nonreference bases specific to Brahman might contribute to the adaptation of indicine cattle to harsh environments. Individual taurine assemblies also contain between 14 and 18 million bases that are missing in the Hereford-based reference assembly, many of which are shared between individuals. This value is somewhat higher than the 5 to 10 million nonreference bases detected per human genome (38–40), possibly because cattle breeds have diverged more strongly than human populations due to intense artificial selection. Each of the three taurine assemblies contains ∼3 million autosomal nonreference bases that were not detected in any other assembly. There were also 4.4 million nonreference bases, of which 2.1 million were nonrepetitive, that were present in all assemblies except the reference. This includes 1.2 million bases that are either specifically deleted in the Hereford breed or the animal used to build the reference, inadvertently propagating reference-bias.
A reference graph may integrate linear reference coordinates, nonreference sequences, and shorter variants (20). However, as many genome analysis tools still rely on a linear coordinate system, we append the nonreference sequences linearly to the ARS-UCD1.2 reference genome. Adding 100 bp flanking sequence on either side of the breakpoints facilitated accurate alignment of sequencing reads at the boundaries of the contigs. A graph-based approach might enable the mapping of sequencing reads spanning breakpoints (20). We considered only variations larger than 100 bp because integrating smaller variations increases the complexity of the resulting reference with limited benefit for downstream analyses (21). We show that our extended ARS-UCD1.2 reference genome leads to improved DNA and RNA-seq read mapping in indicine and taurine cattle, even for breeds that did not contribute to the multiassembly graph. However, excessively adding nonreference contigs to the reference genome carries the risk of increasing the number of ambiguous alignments.
The nonreference sequences comprise more repetitive elements than the overall ARS-UCD1.2 reference genome (76% versus 48%) but less than nonreference insertions detected from human pangenomes (88%) (12, 38). Many nonreference sequences with repetitive elements were observed at immune gene complex loci, corroborating that these regions are highly repetitive (41). The immune gene complex loci also contain many nonrepetitive nonreference sequences suggesting great allelic diversity which may cause assembly problems (30), thus resulting in gaps and missing sequences in the primary ARS-UCD1.2 assembly.
We show that the 16.6 million nonrepetitive nonreference bases encompass transcribed features. An ab initio approach predicted 857 gene models from these sequences. The de novo assembly of RNA-seq read alignments from liver samples provided additional support for more than 400 of these gene models. As these analyses were only conducted on liver transcriptomes, it is highly likely that the nonreference sequences contain additional coding sequences that are transcribed in other tissues. The discovery of distinct nonreference genes in an independent RNA-seq dataset from peripheral blood leukocytes of Holstein cattle supports this hypothesis. Some of the nonreference genes, including genes encoding olfactory receptors, were also present in the animal used to build the reference genome. Olfactory receptors have been observed to undergo frequent duplication and rapid evolution in mammalian genomes (42, 43). Segments encompassing duplicated genes may either be collapsed in primary assemblies or result in unplaced contigs that represent variants of the sequence in the assembled chromosomes (44, 45), hence the presence of paralogous copies among nonreference genes is expected. In order to obtain a confident set of nonreference genes, we retained only genes that were not expressed in Dominette. Many of the proteins encoded by these nonreference genes are predicted to play roles in the immune response. Pangenome analyses in species other than cattle have also revealed nonreference genes with immune-related functions (42, 46, 47). Our findings show that more nonreference transcripts can be assembled in breeds that contribute to the multiassembly graph (Brahman and Angus) than those not included (Holstein and Jersey), suggesting that individual assemblies contain breed-specific, functionally relevant bases. We detect the largest number of nonreference genes using RNA samples from Brahman, suggesting that breeds with great genetic distance from the reference benefit the most from a more diverse reference genome. Importantly, some nonreference genes are differentially expressed between M. bovis–infected and noninfected cattle, including genes that encode proteins that either contribute to the immune response against Mycobacterium infections or may serve as targets for immunomodulation by the pathogen. These differentially expressed genes remained undetected when the transcriptomes were aligned against the standard linear reference genome (25). Thus, our multiassembly graph uncovers functionally active and biologically relevant genomic features that are missing in the B. taurus reference genome.
Our extended reference genome also leads to substantial improvements over ARS-UCD1.2 in reference-guided alignment and variant discovery. First, the sequence read mapping rate increases for samples from all breeds investigated. Using the extended reference genome would enable mapping ∼100,000 previously unmapped reads for samples sequenced at 30-fold coverage. Second, the mapping quality increases for reads that were previously aligned to other positions in ARS-UCD1.2, suggesting that the appended nonreference sequences resolve misalignments. These findings agree well with results from species other than cattle, including goats, pigs, and humans (33, 34, 39). In addition, we show that the nonreference sequences contain polymorphic sites that remained hitherto undetected; we discover 83,250 variants that segregate within and between breeds of cattle. A cluster analysis based on these variants separated individuals by breed, suggesting that variable nonreference bases might be associated with breed-specific traits. This hypothesis is further supported by the “HIGH IMPACT” classification of 54 variants affecting nonreference bases. Considering that the Ti/Tv ratio of the nonreference variants in putative coding sequences was only 1.83, they need to be scrutinized for false positives (48). In any case, our multiassembly graph makes a previously neglected source of inherited variation amenable to genetic investigations.
The size of the bovine multiassembly graph will grow as additional reference-quality assemblies from the Bovinae subfamily become available. Assemblies which are more distant will contribute correspondingly to the overall pangenome growth, increasing the flexible part of graph and reducing the size of the core genome (SI Appendix, Note S2). In its current implementation, our multiassembly graph only contains insertions and deletions, as other types of structural variations (e.g., translocations and inversions) that distort the collinearity of the assembly graph cannot be integrated accurately with minigraph. We provide a versatile workflow that facilitates constructing and characterizing multiassembly graphs for a flexible number of assemblies (https://github.com/AnimalGenomicsETH/bovine-graphs, SI Appendix, Note S5). Our workflow provides tools to determine the origin of nonreference bases, derive structural variations from multiassembly graphs, predict nonreference genes, and append the nonreference sequences linearly to a reference genome. We anticipate that the latter will become obsolete as soon as accurate and fast base-level alignment and split-read graph mapping enables the full suite of genome analyses from a reference graph (49).
Methods
Construction of the Multiassembly Graph.
We used minigraph (21) (version 0.12-r389) with option -xggs to integrate six reference-quality genome assemblies into a multiassembly graph. The current bovine reference genome (B. t. taurus, ARS-UCD1.2, GCF_002263795.1) and four assemblies that were generated previously are accessible at the public repository of the National Center for Biotechnology Information (NCBI): Angus (B. t. taurus, UOA_Angus_1, GCA_003369685.2) (4), Brahman (B. t. indicus, UOA_Brahman_1, GCF_003369695.1) (4), Highland (B. t. taurus, ARS_UNL_Btau-highland_paternal_1.0_alt, GCA_009493655.1) (5), yak (B. grunniens, ARS_UNL_BGru_maternal_1.0_p, GCA_009493645.1) (5). Additionally, we constructed an assembly from a female Original Braunvieh calf (B. t. taurus) using PacBio HiFi reads (SI Appendix, Note S1). The sampling of blood from the original Braunvieh animal and its parents was approved by the veterinary office of the Canton of Zurich (animal experimentation permit ZH 200/19).
The genetic distance among the six assemblies was estimated using Mash (version 2.2) (22). We performed genomic sketching separately for each assembly with mash sketch using a sketch and k-mer size of s = 1,000 and k = 21, respectively. Sketches were combined using mash paste, and mash dist was used to estimate the distances between the assemblies. A phylogenetic tree was built from the estimated pairwise distances using the neighbor-joining method (50) as implemented in the R package ape (version 5.4) (51). The tree was visualized with the phylo.plot function, using the yak assembly as the outgroup to root the tree.
Identification of Nonreference Segments from the Multiassembly Graph.
We refer to nodes that are not in the Hereford-based reference genome (ARS-UCD1.2) as nonreference nodes. We separately aligned (with minigraph parameters “--cov -x asm”) each of the six assemblies back to the multiassembly graph to determine the support for nonreference nodes. For each alignment, all nodes with nonzero coverage (i.e., nodes traversed by this specific assembly) were labeled. After iterating through all the alignments, each node then contained labels for every assembly which passed through it. As such, each node necessarily had at least one label, while a node traversed by all six assemblies would have six labels (SI Appendix, Fig. S1).
It was possible to assess minigraph’s alignment accuracy for the path of the Hereford-based reference genome (ARS-UCD1.2) because all reference nodes in the multiassembly graph were from this assembly. Nodes were considered true positive (TP) and true negative (TN) when reference and nonreference nodes were correctly assigned Hereford labels, respectively. Reference nodes aligned as nonreference nodes were assigned false negative (FN), and nonreference nodes aligned as reference nodes were assigned false positive (FP). We characterized alignment recall (TP / (TP + FN)), precision (TP / (TP + FP)), and overall F1 score (2 × (precision × recall) / (precision + recall)).
Identification of Structural Variations from the Multiassembly Graph.
We used the bubble popping algorithm of gfatools (version 0.4) (21) to derive the structural variations from the multiassembly graph. In the reference graph model of minigraph, a bubble is a branching region in the graph for which the start and end node are reference sequences. A path traversing the start and end nodes represents an allele of a structural variant.
The version of gfatools considered in our study reports the shortest and longest path for each bubble. To detect and classify all paths within a bubble, we applied the following stepwise procedure (SI Appendix, Fig. S2):
-
•
Determine the start and stop node for each bubble using the bubble popping algorithm of gfatools.
-
•
Traverse all possible paths in the bubble using a recursive depth-first search.
-
•
Retain only paths with color-consistent labels (see above).
-
•
Classify a path as a reference path when all nodes and edges are part of the Hereford-based reference assembly and as nonreference otherwise.
-
•
Compare reference and nonreference paths to classify the type of the structural variations.
Structural variations were classified as biallelic if two paths were observed in a bubble and multiallelic if a bubble contained more than two paths. The structural variations were further classified into:
-
•
Alternate deletion: when the nonreference path was shorter than the reference path (but the reference path has nonzero length).
-
•
Complete deletion: when the nonreference path has a length of zero.
-
•
Alternate insertion: when the nonreference path was longer than the reference path.
-
•
Complete insertion: when the reference path has a length of zero.
Breakpoints of structural variations were determined according to ARS-UCD1.2 reference coordinates. We overlapped the breakpoints with annotations from Ensembl (build 101) to identify structural variations in coding sequences. Affected genes were subjected to a gene set enrichment analysis using PANTHER (pantherdb.org/) (23) for which the B. taurus reference gene list was supplied as a baseline.
To validate the structural variations, we mapped 6,803,270 (∼46-fold coverage) PacBio HiFi reads to the multiassembly graph using GraphAligner (version 1.0.12) (52) with preset -x vg (variation graph mapping). The HiFi reads were generated from a Nellore × Brown Swiss crossbred bull (SAMEA7765441), representing taurine and indicine breeds that were not used to build the multiassembly graph. The veterinary office of the Canton of Zurich approved the sampling of blood from the crossbred animal and its parents (animal experimentation permit ZH 200/19). The mean read length was 20,612 bases with an average accuracy of 99.76%. We calculated coverage (number of reads aligned) at each node and edge in the graph based on the graphical alignment format output from GraphAligner.
We combined all nonreference alleles (excluding complete deletions, paths without nonreference bases, and paths with length less than 100 bp) to obtain a comprehensive set of nonreference bases from the multiassembly graph. To facilitate the mapping of short reads to the segment edges, we added 100 bp of flanking sequences (derived from sequences at the source and sink nodes) on either side of the structural variations. The flanking sequences were not considered for length calculations or gene predictions (see below).
To investigate the repeat content of the nonreference sequences, we used the RMBlastn search engine (version 2.10.0) to run RepeatMasker version 4.1.1 (option -species cow) (53) using the database of repetitive DNA elements from Repbase (release 20181026) (54).
Bioinformatic Characterization of Nonreference Sequences.
In order to reveal functionally active nonreference sequences, we performed two complementary analyses:
First, we compared the repeat-masked nonreference sequences against a local protein database using DIAMOND BLASTX (version 0.9.30) (55). Using DIAMOND makedb, the local protein database was built from the RefSeq protein sequences of
-
•
Taurine cattle (B. t. taurus, GCF_002263795.1_ARS-UCD1.2_protein.faa)
-
•
Indicine cattle (B. t. indicus, GCF_003369695.1_UOA_Brahman_1_protein.faa)
-
•
Yak (Bos mutus, GCF_000298355.1_BosGru_v2.0_protein.faa)
-
•
Human (Homo sapiens, GCF_000001405.39_GRCh38.p13_protein.faa)
-
•
Mouse (Mus musculus, GCF_000001635.26_GRCm38.p6_protein.faa)
-
•
Bison (Bison bison, GCF_000754665.1_Bison_UMD1.0_protein.faa)
-
•
Water buffalo (Bubalus bubalis, GCF_003121395.1_ASM312139v1_protein.faa)
-
•
Goat (Capra hircus, GCF_001704415.1_ARS1_protein.faa)
-
•
Sheep (Ovis aries, GCF_002742125.1_Oar_rambouillet_v1.0_protein.faa)
-
•
the curated protein databases of SwissProt and PDB (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/)
To query the nonreference sequences against the local protein database, we ran BLASTX with the parameters “--more-sensitive --e-value 1 × 10−10 --outfmt 6.” We considered only the top hit for each queried sequence with minimum coverage and identity of 80%.
Second, we performed an ab initio gene structure prediction from the repeat-masked nonreference sequences using a local instance of Augustus (version 3.3.3) (56) using default parameters trained on the human genome. From the Augustus Gene transfer format (GTF) output file, we extracted the number of gene models, the number of gene models with transcription start and termination site, transcript length, exon count and length per gene, coding sequence count and length per gene, and protein length of the protein-coding sequences. To classify the domain and family of the nonreference proteins, we converted the Augustus GTF output to the fasta format and performed a query against the local protein database (as above) using DIAMOND BLASTP with the same parameters and thresholds as the BLASTX query.
De Novo Transcript Assembly from Nonreference Sequences.
We downloaded between 12,361,440 and 34,421,106 paired-end RNA-seq reads from liver tissue from 10 Angus (57), 10 Brahman (58), 9 Holstein (59), and 10 Jersey (59) cattle as well as from Dominette—the animal used to construct the ARS-UCD1.2 reference genome (2). Adapter sequences and low-quality bases were removed from the raw RNA-seq data using default parameters of fastp (version 0.19.4) (60). The filtered reads were then aligned using HISAT2 (version 2.1.0) (61), with option “--dta” to facilitate the downstream transcriptome assembly, to the original ARS-UCD1.2 reference as well as the extended version of the ARS-UCD1.2 reference. The extended reference was constructed by appending repeat-masked nonreference sequences as unplaced contigs (61).
Nonreference transcripts were assembled de novo using StringTie2 (version 2.1.1) (24) from RNA-seq reads that aligned to the nonreference sequences. To facilitate transcript assembly, we supplied the ARS-UCD1.2 Ensembl annotation (build 101) and the gene models predicted by Augustus (see above). Transcripts were assembled de novo separately for all RNA-seq samples. Subsequently, we used StringTie2 merge to create a unique set of transcripts across all samples and facilitate the assembly of full-length transcripts from partially assembled transcripts. We quantified gene expression for each sample with StringTie2 using a fixed (merged) GTF file that was generated previously (without predicting new transcripts, option -e). Gene abundance was quantified in TPM.
Differential Gene Expression Analysis.
We utilized publicly available peripheral blood leukocyte transcriptomes of eight M. bovis–infected and eight age-matched healthy Holstein cattle (25) to detect differentially expressed genes from nonreference sequences. The RNA-seq data contain between 9,272,629 and 25,358,979 single-end reads of length 78 bp. We performed quality control on the raw sequencing reads using fastp (version 0.19.4) (60) with default parameters. The filtered reads were then mapped to the extended ARS-UCD1.2 reference genome that contained the nonreference sequences using HISAT2 (61). Potential nonreference transcripts were assembled de novo with StringTie2 (see above). Gene-level read counts were estimated based on a custom annotation file that contained the Ensembl (build 101) ARS-UCD1.2 genome annotation and the nonreference annotation as generated by StringTie2 using the featurecounts function of the Rsubread package (option countMultiMappingReads = FALSE to exclude multimapping reads). The read count matrix was used as input for EdgeR version 3.24.3 (62). We normalized transcript abundance by sequencing depth using the trimmed-mean of M-values approach. Genes that were expressed at ≥1 CPM in at least eight samples were tested for differential expression in peripheral blood leukocytes between M. bovis–infected and control animals using a generalized linear model (GLMQfit) with dispersion parameter estimated using the Cox–Reid method. Genes were considered to be differentially expressed at a Benjamini–Hochberg–corrected FDR ≤ 0.05. Multidimensional scaling of the normalized read count matrix of the differentially expressed genes was performed using the cmdscale function in R.
Mapping and Variant Calling from Whole-Genome Short-Read Data.
We considered the original ARS-UCD1.2 reference genome and an extended version of the reference that additionally contained 70,329,827 nonreference bases detected from five assemblies. We used paired-end short-read sequencing data from 45 samples representing five breeds: Original Braunvieh, Brown Swiss, Holstein, Simmental (63), and Hereford (including Dominette, the animal used to construct the ARS-UCD1.2 reference genome) (2, 64) that had average sequencing coverage of 18.94-fold. Quality control of the short-read sequencing reads was performed using fastp (version 0.19.4) (60) with default parameter settings. The filtered reads were subsequently mapped to the original ARS-UCD1.2 reference and the extended ARS-UCD1.2 reference that also contained nonreference sequences using the mem-algorithm of Burrows–Wheeler Aligner (BWA version 0.7.17) (65) with default parameters. Duplicate reads were marked with Samblaster (version 0.1.24) (66).
We performed multisample variant calling (SNP and indels) on the nonreference sequences using SAMtools (version 1.10) (67) and GATK (version v4.1.9.0) (68) as detailed in Crysnanto et al. (17). Base quality scores were recalibrated using known variants from the 1,000 bull genomes project database (www.1000bullgenomes.com/doco/ARS1.2PlusY_BQSR_v3.vcf.gz). We applied the GATK modules HaplotypeCaller, GenomicsDBImport, and GenotypeGVCFs to discover and genotype polymorphic sites. The variants were subsequently hard filtered using recommended parameters (SNP filters: QD < 2 || QUAL < 30 || FS > 60 || MQ < 40 || MQRankSum < −12.5 || ReadPosRankSum < −8 || AN < 10, Indel filters: QD < 2 || QUAL < 30 || FS > 200 || ReadPosRankSum < −20.0 || AN < 10) (17). A second independent variant discovery and genotyping approach was performed using SAMtools mpileup and bcftools call (67). The resulting genotypes were subsequently hard filtered according to parameters recommend by the 1,000 bulls genomes project (QUAL < 20 || MQ < 30 || DP < 10 || AN < 10) (7). To create a consistent variant representation across both datasets, variants were normalized using vt (version 0.5) (69). We retained only filtered variants, which were identified by both SAMtools and GATK. Functional consequences of variants affecting nonreference bases were predicted based on the GTF file from Augustus (see above) using Ensembl’s Variant Effect Predictor (70).
Code Availability.
Workflows to construct multiassembly graphs and custom scripts to characterize nonreference sequences are available via Github (https://github.com/AnimalGenomicsETH/bovine-graphs). All workflows were built using Snakemake (version 5.30.1) (71), and custom scripts were written in R (version 3.5.1) (72) and Python (version 3.7.1).
Supplementary Material
Acknowledgments
We are thankful for the excellent technical support provided by the Functional Genomics Center Zurich (https://fgcz.ch/). Computing was done at the Leonhard High Performance Compute cluster at ETH Zürich. This study was supported by grants from the Swiss NSF (310030_185229) and the Swiss Federal Office for Agriculture, Bern.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101056118/-/DCSupplemental.
Data Availability
Short sequencing reads are available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) with study accessions PRJNA436715 (Transcriptome: Brahman) (58), PRJNA392196 (Transcriptome: Angus) (57), PRJNA357463 (Transcriptome: Holstein and Jersey) (59), PRJNA294306 (Transcriptome: Dominette) (2), PRJNA257841 (Differential expression analysis: Holstein) (25), PRJEB18113 (Whole-genome sequencing (WGS): Brown Swiss, Original Braunvieh, Holstein, and Simmental) (63), PRJNA494431 (WGS: Hereford) (64), and PRJNA391427 (WGS: Dominette) (2). Accession numbers for all samples are provided in Dataset S1. PacBio HiFi reads for an Original Braunvieh animal used to construct a de novo assembly are available at study accession PRJEB42335 (73) under sample accession SAMEA7759028. PacBio HiFi reads for a Nelore × Brown Swiss bull are available at study accession PRJEB42335 (73) under sample accession SAMEA7765441. Data supporting this study, including the multiassembly graph, nonreference sequences, nonreference genes, transcript abundances, and sequence variants detected from nonreference sequences are available via Zenodo (https://zenodo.org/record/4385983#.YHQwER8zbIU) (74).
References
- 1.Bovine Genome Sequencing and Analysis Consortium; Elsik C.G., The genome sequence of Taurine cattle: A window to ruminant biology and evolution. Science 324, 522–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen B. D., et al., De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 9, giaa021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koren S., et al., De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol. 36, 1174–1182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low W. Y., et al., Haplotype-resolved genomes provide insights into structural variation and gene content in Angus and Brahman cattle. Nat. Commun. 11, 2071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice E. S., et al., Continuous chromosome-scale haplotypes assembled from a single interspecies F1 hybrid of yak and cattle. Gigascience 9, giaa02 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K., et al., The mosaic genome of indigenous African cattle as a unique genetic resource for African pastoralism. Nat. Genet. 52, 1099–1110 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Daetwyler H. D., et al., Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46, 858–865 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Koufariotis L., et al., Sequencing the mosaic genome of Brahman cattle identifies historic and recent introgression including polled. Sci. Rep. 8, 17761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballouz S., Dobin A., Gillis J. A., Is it time to change the reference genome? Genome Biol. 20, 159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritt J., Chen N.-C., Langmead B., FORGe: Prioritizing variants for graph genomes. Genome Biol. 19, 220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong K. H. Y., et al., Towards a reference genome that captures global genetic diversity. Nat. Commun. 11, 5482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman R. M., et al., Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 51, 30–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitacre L. K., et al., What’s in your next-generation sequence data? An exploration of unmapped DNA and RNA sequence reads from the bovine reference individual. BMC Genomics 16, 1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrison E., et al., Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol. 36, 875–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggertsson H. P., et al., Graphtyper enables population-scale genotyping using pangenome graphs. Nat. Genet. 49, 1654–1660 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Crysnanto D., Pausch H., Bovine breed-specific augmented reference graphs facilitate accurate sequence read mapping and unbiased variant discovery. Genome Biol. 21, 184 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crysnanto D., Wurmser C., Pausch H., Accurate sequence variant genotyping in cattle using variation-aware genome graphs. Genet. Sel. Evol. 51, 21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song J. M., et al., Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehr B., et al., Diversity in non-repetitive human sequences not found in the reference genome. Nat. Genet. 49, 588–593 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hickey G., et al., Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 21, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Feng X., Chu C., The design and construction of reference pangenome graphs with minigraph. Genome Biol. 21, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ondov B. D., et al., Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovaka S., et al., Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLoughlin K. E., et al., RNA-seq transcriptional profiling of peripheral blood leukocytes from cattle infected with Mycobacterium bovis. Front. Immunol. 5, 396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGill J. L., et al., Specific recognition of mycobacterial protein and peptide antigens by γδ T cell subsets following infection with virulent Mycobacterium bovis. J. Immunol. 192, 2756–2769 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Damani-Yokota P., Telfer J. C., Baldwin C. L., Variegated transcription of the WC1 hybrid PRR/Co-receptor genes by individual γδ T cells and correlation with pathogen responsiveness. Front. Immunol. 9, 717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy H. E., et al., Modulation of immune responses to Mycobacterium bovis in cattle depleted of WC1(+) γ δ T cells. Infect. Immun. 70, 1488–1500 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bah S. Y., Forster T., Dickinson P., Kampmann B., Ghazal P., Meta-analysis identification of highly robust and differential immune-metabolic signatures of systemic host response to acute and latent tuberculosis in children and adults. Front. Genet. 9, 457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakshy K., et al., Development of polymorphic markers in the immune gene complex loci of cattle. J. Dairy Sci., 10.3168/jds.2020-19809 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Golicz A. A., et al., The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7, 13390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong J., et al., Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587, 246–251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian X., et al., Building a sequence map of the pig pan-genome from multiple de novo assemblies and Hi-C data. Sci. China Life Sci. 63, 750–763 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Li R., et al., Towards the complete goat pan-genome by recovering missing genomic segments from the reference genome. Front. Genet. 10, 1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi X. B., Jianlin H., Wang G., Rege J. E. O., Hanotte O., Assessment of cattle genetic introgression into domestic yak populations using mitochondrial and microsatellite DNA markers. Anim. Genet. 41, 242–252 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith T., Individual breed genome assembly to create the cattle pangenome in Online Abstracts in International Plant and Animal Genomes XXVIII Conference, B. D. Rosen, E. Memili, D. Hagen, Eds. (Scherago International, Livingston, NJ, 2020), p. W120.
- 37.Khan A. W., et al., Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 25, 148–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ameur A., et al., De novo assembly of two Swedish genomes reveals missing segments from the human GRCh38 reference and improves variant calling of population-scale sequencing data. Genes (Basel) 9, 486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Audano P. A., et al., Characterizing the major structural variant alleles of the human genome. Cell 176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan Z., et al., HUPAN: A pan-genome analysis pipeline for human genomes. Genome Biol. 20, 149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz J. C., et al., The evolution of the natural killer complex; A comparison between mammals using new high-quality genome assemblies and targeted annotation. Immunogenetics 69, 255–269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., et al., Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 27, 865–874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes G. M., et al., The birth and death of olfactory receptor gene families in Mammalian niche adaptation. Mol. Biol. Evol. 35, 1390–1406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vollger M. R., et al., Long-read sequence and assembly of segmental duplications. Nat. Methods 16, 88–94 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley D. R., Salzberg S. L., Detection and correction of false segmental duplications caused by genome mis-assembly. Genome Biol. 11, R28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon S. P., et al., Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8, 2184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golicz A. A., Bayer P. E., Bhalla P. L., Batley J., Edwards D., Pangenomics comes of age: From bacteria to plant and animal applications. Trends Genet. 36, 132–145 (2020). [DOI] [PubMed] [Google Scholar]
- 48.DePristo M. A., et al., A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siren J., et al., Genotyping common, large structural variations in 5,202 genomes using pangenomes, the Giraffe mapper, and the vg toolkit. Biorxiv [Preprint] (2020). 10.1101/2020.12.04.412486 (Accessed 8 January 2021). [DOI]
- 50.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 51.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Rautiainen M., Marschall T., GraphAligner: Rapid and versatile sequence-to-graph alignment. Genome Biol. 21, 253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smit A., Hubley R., Green P., RepeatMasker Open-4.0 (2015). http://www.repeatmasker.org. Accessed 8 January 2021.
- 54.Bao W., Kojima K. K., Kohany O., Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Stanke M., Waack S., Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, ii215–ii225 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Xiang R., et al., Genome variants associated with RNA splicing variations in bovine are extensively shared between tissues. BMC Genomics 19, 521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen L. T., et al., P1012 Liver transcriptome from pre versus post-pubertal Brahman heifers. J. Anim. Sci. 94, 20–21 (2016). [Google Scholar]
- 59.Salleh S. M., Mazzoni G., Løvendahl P., Kadarmideen H. N., Gene co-expression networks from RNA sequencing of dairy cattle identifies genes and pathways affecting feed efficiency. BMC Bioinformatics 19, 513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Häfliger I. M., et al., An IL17RA frameshift variant in a Holstein cattle family with psoriasis-like skin alterations and immunodeficiency. BMC Genet. 21, 55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young A. E., et al., Genomic and phenotypic analyses of six offspring of a genome-edited hornless bull. Nat. Biotechnol. 38, 225–232 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint] (2013). https:/doi.org/arxiv.org/abs/1303.3997 (Accessed 8 January 2021).
- 66.Faust G. G., Hall I. M., SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H.et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poplin R., et al., Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv [Preprint] (2017). 10.1101/201178 (Accessed 26 March 2018). [DOI]
- 69.Tan A., Abecasis G. R., Kang H. M., Unified representation of genetic variants. Bioinformatics 31, 2202–2204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLaren W., et al., The Ensembl variant effect predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Köster J., Rahmann S., Snakemake–A scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012). [DOI] [PubMed] [Google Scholar]
- 72.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2017). [Google Scholar]
- 73.Pausch H., Long-read sequencing data from cattle for the purpose of de-novo genome assembly. ENA. https://www.ebi.ac.uk/ena/browser/view/PRJEB42335. Deposited 8 January 2021.
- 74.Crysnanto D., Leonard A. S., Fang Z. H., Pausch H., Supporting data for novel functional sequences uncovered through a bovine multi-assembly graph. Zenodo. 10.5281/zenodo.4385983. Deposited 8 January 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Short sequencing reads are available at the European Nucleotide Archive (https://www.ebi.ac.uk/ena) with study accessions PRJNA436715 (Transcriptome: Brahman) (58), PRJNA392196 (Transcriptome: Angus) (57), PRJNA357463 (Transcriptome: Holstein and Jersey) (59), PRJNA294306 (Transcriptome: Dominette) (2), PRJNA257841 (Differential expression analysis: Holstein) (25), PRJEB18113 (Whole-genome sequencing (WGS): Brown Swiss, Original Braunvieh, Holstein, and Simmental) (63), PRJNA494431 (WGS: Hereford) (64), and PRJNA391427 (WGS: Dominette) (2). Accession numbers for all samples are provided in Dataset S1. PacBio HiFi reads for an Original Braunvieh animal used to construct a de novo assembly are available at study accession PRJEB42335 (73) under sample accession SAMEA7759028. PacBio HiFi reads for a Nelore × Brown Swiss bull are available at study accession PRJEB42335 (73) under sample accession SAMEA7765441. Data supporting this study, including the multiassembly graph, nonreference sequences, nonreference genes, transcript abundances, and sequence variants detected from nonreference sequences are available via Zenodo (https://zenodo.org/record/4385983#.YHQwER8zbIU) (74).