Abstract
Tularemia is a highly dangerous zoonotic infection due to the bacteria Francisella tularensis. Low genetic diversity promoted the use of polymorphic tandem repeats (MLVA) as first-line assay for genetic description. Whole genome sequencing (WGS) is becoming increasingly accessible, opening the perspective of a time when WGS might become the universal genotyping assay. The main goal of this study was to describe F. tularensis strains circulating in Kazakhstan based on WGS data and develop a MLVA assay compatible with in vitro and in silico analysis. In vitro MLVA genotyping and WGS were performed for the vaccine strain and for 38 strains isolated in Kazakhstan from natural water bodies, ticks, rodents, carnivores, and from one migratory bird, an Isabellina wheatear captured in a rodent burrow. The two genotyping approaches were congruent and allowed to attribute all strains to two F. tularensis holarctica lineages, B.4 and B.12. The seven tandem repeats polymorphic in the investigated strain collection could be typed in a single multiplex PCR assay. Identical MLVA genotypes were produced by in vitro and in silico analysis, demonstrating full compatibility between the two approaches. The strains from Kazakhstan were compared to all publicly available WGS data of worldwide origin by whole genome SNP (wgSNP) analysis. Genotypes differing at a single SNP position were collected within a time interval of more than fifty years, from locations separated from each other by more than one thousand kilometers, supporting a role for migratory birds in the worldwide spread of the bacteria.
Author summary
Genotyping of Francisella tularensis has become a routine practice in epidemiology. Despite rapidly accumulating knowledge, the phylogeography of the pathogen is still poorly understood and discussions about geographic and temporal origins continue. One important reason is the poor characterization of the pathogen in many tularemia-endemic countries. This article describes the genetic diversity of Francisella tularensis subsp. holarctica in Kazakhstan using tandem repeat polymorphisms as well as whole genome sequencing. Thirty-nine strains were analyzed and two lineages were identified, namely B.4 and B.12. The study demonstrates a wider distribution of genotype B.4 in Asia, and identified a more basal branching point in this subclade. The obtained data support the Asian origin hypothesis for F. tularensis. The finding of identical genotypes in strains separated in time by decades and a thousand-kilometers geographic distance, confirms the ability of the bacteria for long-term preservation and fast long distances spread. The isolation of F. tularensis subsp. holarctica from the bird species Isabellina wheatear allows speculating about a major contribution of birds to the phylogeography of the pathogen. A genotyping protocol was developed utilizing seven polymorphic tandem repeats, two of which were identified within the framework of this work. The in vitro and in silico results are identical when using sequencing reads of 300 base-pairs or more.
Introduction
Precise knowledge of the genetic diversity of microorganisms is useful for multiple purposes, including understanding of the emergence of pathogens, tracking and controlling their spread, as well as certifying and patenting strains of interest for biotechnologies [1–3]. DNA "fingerprinting" of bacteria began to develop with the understanding of the role of this molecule and improved throughout the development of DNA technologies. Genotyping based on DNA fragmentation and resolution by Pulsed-Field Gel Electrophoresis (PFGE), Random Amplified Polymorphic DNA (RAPD), Restriction Fragment Length Polymorphism (RFLP) have been replaced by simpler, more accurate and reproducible (numeric) methods including Multiple-Locus Variable-number of tandem-repeats (VNTR) Analysis (MLVA), MultiLocus Sequence Typing (MLST) and selected single nucleotide polymorphism (canSNP) typing [4–6]. Currently, whole-genome sequencing (WGS) of bacterial strains, the ultimate genotyping method, is becoming increasingly accessible but is still too costly especially in developing countries to allow its use as a first-line assay. In the ongoing transition period, highly discriminatory and cheap DNA based genotyping methods can be used as a first-line method for selecting strains for WGS, especially in highly endemic regions and as a low-cost assay for routine quality control of strain collections [7].
Most laboratories are using one method or more among WGS, MLST, canSNP, MLVA and/or PFGE in parallel for a number of pathogens in order to carry out epidemiological control adapted to local context, capacities, and needs [8]. In this regard, the development of in silico genotyping methodology based on WGS data and allowing backward compatibility is particularly useful for pathogens for which MLVA and/or MLST were considered as the “golden standard” of genotyping such as Brucella spp, Bacillus anthracis, Yersinia pestis, F. tularensis or Neisseria meningitidis [9–12]. Carrying out in silico MLST on WGS data is not difficult owing to the availability of appropriate algorithms and allows comparing the results obtained previously [13]. Obtaining a relevant in silico MLVA profile is a more challenging task due to the occurrence of errors in the assembly of short reads and contig breaks in VNTR locations [14]. Nevertheless, the use of appropriate sequencing reads length allows to conduct in silico MLVA with high reliability. It has been established for Brucella that read sizes of 200 base-pairs or more allow to correctly assemble tandem repeat arrays for most loci used in the MLVA assay and to produce a reliable MLVA profile [15]. Simple scripts have been developed to run in silico MLVA (https://github.com/i2bc/MLVA_finder). FASTA files of assemblies (complete genomes or contigs) are commonly used as input. However, currently available assemblers often use k-mer values shorter than read length, which can be detrimental when assembling tandem repeat arrays and does not allow to take full advantage of reads length [16].
In silico MLVA analysis was not previously performed for F. tularensis, although it is considered to be among the six most dangerous pathogens with potential for use as an agent of bioterrorism [17] and MLVA typing has been shown to be relevant [4,18]. Twenty-five VNTRs have been described so far in the F. tularensis genome and various schemes using a subset of these have been proposed. MLVA correctly classifies F. tularensis into the three subspecies, of which the most pathogenic for humans is F. tularensis subsp. tularensis common in North America with a single strain of unclear origin described in Europe [18,19]. F. tularensis subsp. holarctica distributed in most of the northern hemisphere is less pathogenic for humans [20,21]. F. tularensis subsp. mediasiatica present in Central Asian region and southern Siberia has not been associated with human infections [22]. F. novicida was once proposed as subspecies based on its high genetic similarity (97%) with F. tularensis but this assignment is controversial and confusing in terms of ecology, evolution, and metabolism [23,24]. F. novicida detected in North America and Australia is weakly virulent and can cause disease in people with immunosuppression [23,25]. The evolution of F. tularensis is clonal [26] and the species constitute a monophyletic lineage in contrast to F. novicida which is a clear outgroup with a very different behavior [24]. The geographic origin and the date of emergence of F. tularensis are unknown, and the population structure is reflecting a capacity to travel long distances, as well as a very low mutation rate [26]. The clonal evolution of F. tularensis allows to use selected single nucleotide polymorphisms commonly called canSNPs [27] to robustly designate sublineages. Subspecies holarctica is currently subdivided in four clades identified by canSNPs, B.4, B.6, B.12 and B.16 (alias japonica). B.4 is the dominant holarctica lineage in North America, B.6 is dominant in Western Europe, whereas B.12 is dominant across Asia and Eastern Europe [5]. Currently available data on the B.6 holarctica sublineage in Western Europe is suggesting that Western Europe was contaminated from Eastern Europe [26,28,29]. The most basal B.16 lineage has been reported in Japan, China [30,31] and Turkey [32]. This feature combined with the geographic distribution of mediasiatica which was found only in Central Asia [22] is supporting an Asian or Central Asian origin for F. tularensis [30].
Due to the endemicity of tularemia in Kazakhstan, extensive field work has been done during the past seventy years. This work has allowed to map a number of natural foci accross the country [33]. Natural reservoirs of tularemia are registered in twelve among the 14 administrative regions of Kazakhstan and the total area of the reservoirs is about a fifth of the country’s territory (552 thousand square kilometers). During the past 90 years (1928-2018) about 10.000 human cases of tularemia were registered in Kazakhstan. The majority of cases occured in the 1950s and were caused by labor migration of unvaccinated populations to endemic areas [34]. In the 1928-1970s period, the majority of cases occured as outbreaks involving from 105 to 1791 cases. Improving sanitary conditions and routine vaccination reduced the tularemia incidence to sporadic cases [35,36]. Nevertheless, F. tularensis is present in East Kazakhstan, Akmola, West Kazakhstan, Aktobe, North Kazakhstan and Pavlodar regions, as shown by periodic epizootics in rodents [37]. Strains of F. tularensis subsp. holarctica are routinely isolated from at least 26 vertebrate species including humans, rodents, hares and birds as well as invertebrate vectors (blood-sucking insects and arthropods) from natural habitats across Kazakhstan. Also, strains of F. tularensis subsp. mediasiatica are isolated in the Almaty and Zhambyl regions from rodents, hares and invertebrate vectors.
There is however currently little information about the genetic diversity of circulating strains. The present report is a contribution in order to fill this gap. We applied whole genome sequencing as well as in vitro MLVA genotyping on a relevant subset of 39 strains isolated in Kazakhstan. We compared the in vitro and in silico MLVA data deduced from WGS data, as well as the clustering achieved by MLVA and whole genome SNP (wgSNP) analysis. A panel of seven VNTR loci which can be amplified in a single PCR reaction and is well adapted to the genetic diversity observed in Kazakhstan was defined in order to provide a first-line genotyping assay usable on a large scale. In conclusion, we describe the genetic diversity of F. tularensis subsp. holarctica determined by MLVA, canSNP and wgSNP methods and compare with currently available WGS data of worldwide origins. This is the first report of WGS data for holarctica strains from Kazakhstan.
Materials and methods
Strain and DNA extraction
A total of 39 strains of F. tularensis subsp. holarctica were obtained from the Kazakhstan “Republican collection of microorganisms and the depository of pathogens of especially dangerous infections” of the Republican State Enterprise on the Right of Economic Management “National Scientific Center for Especially Dangerous Infections named by Masgut Aykimbayev” of the Ministry of Healthcare of the Republic of Kazakhstan (NSCEDI). Thirty-eight strains were isolated from rodents (14 strains), ticks (19 strains), carnivores (two strains), one migratory bird and two water samples from eight regions of Kazakhstan in the period from 1951 to 2018. The vaccine strain F. tularensis 15 NIIEG (alias LVS) originating from Kazakhstan was included as a reference strain (Table 1). The strains were isolated during routine monitoring or during periods of epizootological outbreaks, in accordance with applicable laws. All vertebrate hosts but one were trapped, and tick were host-seeking, pasture ticks collected from the vegetation. Work with cultures of tularemia pathogens was carried out in accordance with current regulatory and methodological documents on the organization and conduct of laboratory diagnosis of tularemia in Kazakhstan.
Table 1. Characterization of F. tularensis subsp. holarctica strains from Kazakhstan.
| Key | Region within Kazakhstan | Landscape* | Coordinates | Date of isolation | Host** |
|---|---|---|---|---|---|
| Tul-106_KZ | West-Kazakhstan | S | 49°34’ 49°26’ | 12.12.2014 | Microtus arvalis (corpse found) |
| Tul-11_KZ | East Kazakhstan | B | 47°51’ 84°28’ | 01.06.2001 | Tick |
| Tul-112_KZ | Pavlodar | S | 52°22’ 78°58’ | 15.06.2016 | Tick - Haemaphysalis concinna |
| Tul-128_KZ | West-Kazakhstan | S | 48°50’ 49°52’ | 18.02.2015 | Meriones tamariscinus |
| Tul-13_KZ | West-Kazakhstan | S | 49°26’ 49°48’ | 30.01.2012 | Mus musculus |
| Tul-132_KZ | Aktobe | S | 50°50’ 54°53’ | 29.05.2017 | Mustela nivalis |
| Tul-133_KZ | West-Kazakhstan | S | 49°31’ 50°34’ | 21.12.2012 | Mus musculus |
| Tul-135_KZ | Almaty | B | 45°36’ 80°28’ | 15.07.1953 | (water) |
| Tul-139_KZ | Akmola | S | 51°47’ 68°19’ | 1955 | Tick - Dermacentor |
| Tul-149_KZ | West-Kazakhstan | S | 51°1’ 49°55’ | 23.05.2013 | Tick - D. marginatus |
| Tul-15_KZ | Aktobe | S | 50°50’ 54°53’ | 15.05.2017 | Mustela nivalis |
| Tul-151_KZ | Aktobe | S | 49°42’ 55°3’ | 10.05.2011 | Spermophilus pygmaeus |
| Tul-153_KZ | West-Kazakhstan | S | 49°35’ 53°6’ | 24.05.2011 | Oenanthe isabellina |
| Tul-154_KZ | Almaty | S | 46°13’ 78°54’ | 27.08.1975 | Tick - Dermacentor |
| Tul-155_KZ | East Kazakhstan | S | 49°54’ 82°45’ | 1951 | Arvicola amphibius |
| Tul-161_KZ | (vaccine strain 15 NIIEG) | ||||
| Tul-17_KZ | East Kazakhstan | S | 49°54’ 82°45’ | 1951 | Arvicola amphibius |
| Tul-18_KZ | Almaty | B | 45°37’ 80°26’ | 1953 | (water) |
| Tul-19_KZ | West-Kazakhstan | S | 49°23’ 51°24’ | 25.01.2017 | Apodemus uralensis |
| Tul-2_KZ | Aktobe | S | 49°27’ 54°51’ | 22.05.2016 | Tick |
| Tul-20_KZ | Pavlodar | S | 51°8’ 50°18’ | 07.09.2016 | Tick |
| Tul-30_KZ | West-Kazakhstan | S | 49°15’ 49°28’ | 03.12.2015 | Mus musculus |
| Tul-52_KZ | Aktobe | S | 48°27’ 61°23’ | 22.04.2013 | Allactaga major |
| Tul-6_KZ | West-Kazakhstan | S | 50°25’ 52°33’ | 18.10.2012 | Mus musculus |
| Tul-61_KZ | Aktobe | S | 49°13’ 54°33’ | 27.03.2010 | Tick – D. pictus |
| Tul-66_KZ | Almaty | S | 45°47’ 81°20’ | 29.05.2004 | Tick - Dermacentor |
| Tul-67_KZ | Karaganda | S | 48°40’ 71°39’ | 1989 | Tick |
| Tul-68_KZ | Almaty | S | 45°19’ 82°18’ | 28.04.2002 | Tick - Dermacentor |
| Tul-7_KZ | West-Kazakhstan | S | 49°27’ 49°48’ | 29.09.2004 | Tick |
| Tul-71_KZ | Almaty | T | 45°12’ 74°44’ | 17.05.1963 | Tick - D. daghestanicus |
| Tul-76_KZ | Almaty | B | 45°23’ 80°3’ | 10.05.2018 | Tick - D. marginatus |
| Tul-78_KZ | West-Kazakhstan | S | 49°49’ 53°46’ | 19.04.2007 | Tick |
| Tul-86_KZ | Karaganda | S | 49°12’ 73°3’ | 1986 | Tick - D. marginatus |
| Tul-87_KZ | Zhambyl | T | 44°49’ 71°9’ | 2004 | Tick - D. daghestanicus |
| Tul-92_KZ | West-Kazakhstan | S | 51°1’ 54°1’ | 09.07.1988 | Tick - D. marginatus |
| Tul-93_KZ | West-Kazakhstan | S | 51°37’ 52°18’ | 23.01.2004 | Mus musculus |
| Tul-97_KZ | Pavlodar | S | 52°28’ 78°9’ | 30.05.2013 | Tick - Haemaphysalis conсinna |
| Tul-98_KZ | West-Kazakhstan | S | 49°29’ 50°30’ | 19.12.2011 | Cricetulus migratorius |
| Tul-99_KZ | West-Kazakhstan | S | 49°5’ 50°10’ | 24.05.2011 | Spermophilus major |
* S, Floodplain swamp; B, Foothill brook; T, Turgay.
** All ticks were pasture ticks, collection from vegetation. Vertebrate hosts were trapped except when indicated otherwise (one instance).
The lyophilized strains were plated on transparent agar nutrient medium for cultivation and isolation of tularemia microbe with vitamins and mineral additives (“FT-agar", FBIS SRCAMB, Obolensk, Russia) and cultured for 48 hours at a temperature of 37 ± 1°C. A bacterial suspension was prepared and inactivated by the addition of merthiolate sodium to a final concentration of 0.01%, heated for 30 min at a temperature of 56°C. DNA was isolated from inactivated suspension with QIAamp DNA Mini Kit (Qiagen, Germany).
Genome sequencing, single nucleotide polymorphism (SNP) calling and canSNP assignment
Sequencing libraries were prepared using Nextera XT DNA Library Preparation Kit (Illumina, USA). Sequencing was performed using MiSeq Reagent Kit v3 (600 cycles) on the Illumina MiSeq platform. Sequencing quality was checked using FastQC v0.11.15 [38]. High-quality sequence selection was performed using MultiQC v3 [39]. Raw reads can be accessed from the European Nucleotide Archive [7] under study accession PRJNA639508 (https://www.ebi.ac.uk/ena/browser/view/PRJNA639508; run accessions SRR13617503-SRR13617541).
SNPs were called by mapping raw sequencing reads on a reference genome using BioNumerics version 7.6.3 (Applied-Maths, Laethem-Saint-Martin, Belgium) with default parameters as previously described except when mapping Ion Torrent PGM sequence data for which the “allow gapped alignment” option was activated [22]. Publicly available assemblies and raw reads were downloaded via EBI-ENA (last download on August 25th 2020). The list of public assemblies and sequence reads archives evaluated for this report is provided in S1 Table, as well as the list of accessions used for each figure. Assemblies were split into 50 bp long artificial reads with a 10X cover and no error model. These artificial reads were then mapped on the reference genome for SNP calling as done with raw sequencing reads. BioNumerics was used with default parameters (no resampling) for Maximum Parsimony analysis [40] and dendrogram drawing.
Raw reads datasets were assembled using SPAdes [41] version 3.13.1 and SKESA [42] version 2.3.0. The canSNP assignment of each strain was deduced from genome assemblies using CanSNPer2 (https://github.com/FOI-Bioinformatics/CanSNPer2) [43] and the available F. tularensis canSNPer2 scheme [18,28,43,44].
In vitro MLVA typing
In vitro MLVA genotyping was performed with the previously described 25 VNTR loci [4]. The ten most variable loci (Ft-M02, Ft-M03, Ft-M04, Ft-M05, Ft-M06, Ft-M10, Ft-M20, Ft-M22, Ft-M23 and Ft-M24) were analysed using capillary electrophoresis with fluorescent primers (Applied Biosystems genetic analyzers) [18]. The remaining 15 loci (Ft-M01, Ft-M07, Ft-M08, Ft-M09, Ft-M11, Ft-M12, Ft-M13, Ft-M14, Ft-M15, Ft-M16, Ft-M17, Ft-M18, Ft-M19, Ft-M21 and Ft-M25) were typed using monoplex PCR and agarose gel electrophoresis [22]. The resulting MLVA data were deposited in the public Francisella tularensis MLVA database accessible at https://microbesgenotyping.i2bc.paris-saclay.fr/.
In silico MLVA on genome assemblies and on raw sequence reads
In silico MLVA was run on genome assemblies by applying the MLVA_finder.py script with default settings (https://github.com/i2bc/MLVA_finder) and primers listed in S2 Table.
The size of the polymorphic VNTR arrays being smaller than the 300 bp reads produced here, we also typed raw reads data. A simple script based on bbduk (BBTools, https://sourceforge.net/projects/bbmap/) was developed to identify reads containing VNTRs and both flanking sequences (https://github.com/Vladislav-Shevtsov/search-primers-in-reads/). Primers were selected that were located as close as possible to the VNTR loci (S2 Table). When selecting primers for in silico instead of in vitro detection, attention was paid only to uniqueness and to the formation of only the target fragment. The script combines all reads containing a VNTR locus of interest in a single multifasta file on which the MLVA_finder.py script can subsequently be applied.
Identification of additional VNTR loci
The search for additional VNTR markers was performed using Tandem Repeats Finder, version 4.07b [45] and microsatellite repeats finder (http://insilico.ehu.es/mini_tools/microsatellites/). Tandem repeats were searched in F. tularensis subsp. tularensis SCHU S4 substr. NR-28534 (assembly accession GCA_000628925.1) and F. tularensis subsp. holarctica FTNF002-00 (assembly accession GCF_000017785.1). New VNTR loci showing two alleles or more among the twelve genomes F. tularensis subsp. holarctica (NZ_CP010289, NC_019551, NC_017463, NZ_CP010288 and NC_009749), F. tularensis subsp. mediasiatica (NC_010677), F. novicida (NC_008601), F. tularensis subsp. tularensis (NC_006570, NC_016933, NZ_CP009753, NZ_CP012037 and NC_009257) were further investigated in silico on 276 assemblies of F. tularensis deposited in EBI-ENA. The discriminatory power of each locus was evaluated using Hunter-Gaston Discrimination Index (HGDI) [46].
Results
Whole genome SNP analysis
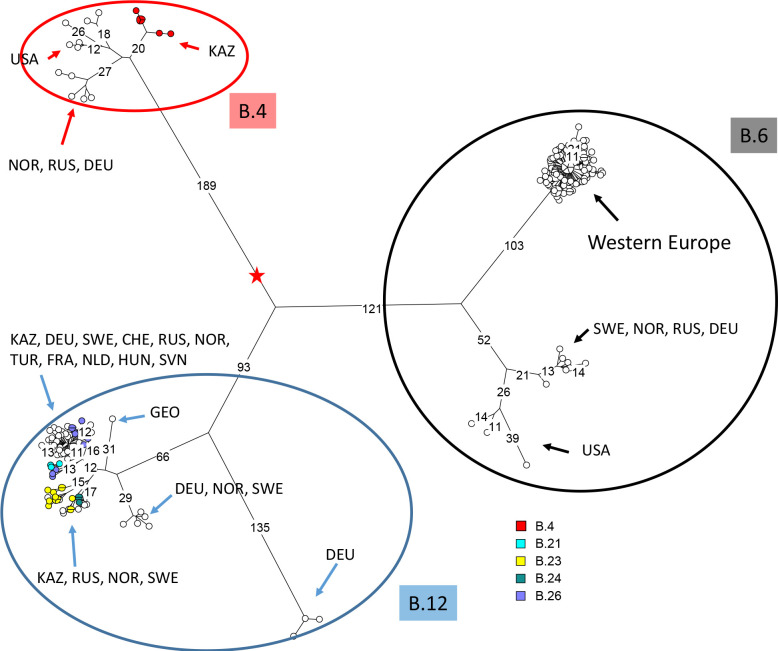
The WGS data obtained from the 39 strains was used for wgSNP sequence phylogenetic analysis together with representative publicly available WGS data (S1 Table). Fig 1 shows the population structure of the holarctica subspecies excluding the most basal holarctica bv. japonica lineage (currently designated B.16). The three sublineages, B.4, B.6 and B.12 are clearly separated. The distances from the root indicated by the red star to the tips are very similar. Each of the three sublineages is itself divided in a limited number of distinct sublineages. These sublineages show a strong geographic association with the exceptions of B.12 sublineages B.23 and particularly B.42. In agreement with current knowledge regarding its geographic distribution, B.6 is not detected in Kazakhstan in the present collection [28].
Fig 1. Positioning the Kazakhstan strains within the holarctica subspecies by wgSNP and Maximum Parsimony analysis.
Two thousand and twenty-two SNPs were called among 219 strains comprising 39 strains from Kazakhstan and 180 public data sets belonging to holarctica B.4, B.6 or B.12 (B.16 alias japonica strains were used to root the tree and are not shown). The red star shows the position of the MRCA. Branch lengths of more than ten SNPs are indicated. The branch scale is linear. The total tree size is 2034 (homoplasia 0.6%). The circles representing strains from Kazakhstan are colored according to canSNP assignment as indicated. The geographic origin of strains constituting secondary sublineages is indicated using three-letters country codes.
Six strains isolated in 1951-1989 in East Kazakhstan, Akmola, Almaty and Karaganda regions define a new basal lineage within holarctica clade B.4 (Figs 1 and S1). The rest of the B.4 group is split in two subgroups, one showing a strong geographic association with North-America, and the other with Russia and Western Europe (Norway and Germany). Branch expansions from the B.4 most recent common ancestor (MRCA) to the tips are similar and vary from a minimum 46 SNPs up to 56 SNPs (S1 Fig).
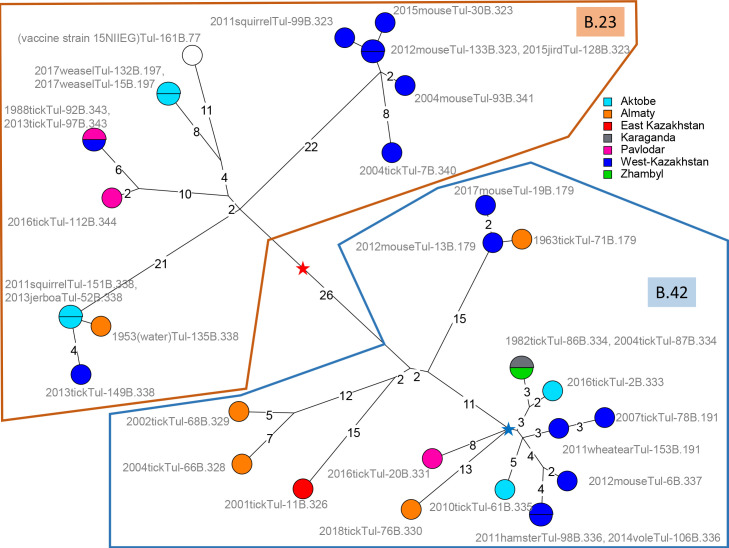
All the other strains belong to holarctica clade B.12. Sixteen belong to subclade B.12_B.23 and seventeen to subclade B.12_B.42 [47]. Fig 2 illustrates that identical whole genome SNP genotypes are observed in strains collected in distant locations at different times. Strain Tul-92_KZ collected in West Kazakhstan in 1988 is identical to Tul-97_KZ collected in Pavlodar in 2013, i.e. twenty-five years later more than one thousand kilometers away. A single SNP separates Tul-13_KZ collected in 2012 in West-Kazakhstan from Tul-71_KZ collected 49 years earlier in the Almaty region, or Tul-52_KZ isolated in 2013 in Aktobe from Tul-135_KZ isolated sixty years earlier in the Almaty region. In both instances, the most recent strain showed the ancestral genotype, and the single SNP in the derived genotype is a G to A transition, in agreement with the previously observed AT mutation bias [26]. This indicates that identical wgSNP profile can be perpetuated for twenty-five years and strengthens previous observation of identical wgSNP genotypes in strains collected in Sweden 15 years apart [48] and in France, Switzerland and Germany up to 31 years apart [28]. Some sub-branches show a strong geographic homogeneity, in particular a group of six B.23 strains isolated in West Kazakhstan in years 2004-2015. They constitute a three-branches polytomy up to eight SNPs in length. The closest strain (seven up to 14 SNPs away) among public WGS data is strain NO-18/2011 isolated in Norway in 2011 (S1 Table).
Fig 2. Focus on the 33 Kazakhstan strains belonging to sublineages B.23 and B.42.
Two hundred and fifty-four SNPs were called by mapping on reference SCHU_S4 (assembly accession GCA_000008985). The Maximum Parsimony tree size is 254 (no homoplasy). The red star indicates the position of the MRCA. The blue star indicates the root of the B.66 polytomy shown in a larger context in S2 Fig. Strains are labelled with year of isolation, host, strain Id and CanSNP assignment. The color code reflect the geographic origin within Kazakhstan (region level). Branch lengths longer than one are indicated. The branch scale is linear.
In contrast, eleven strains coming from six regions within Kazakhstan constitute a polytomy with three branches radiating from the blue star within the B.42 sublineage (Fig 2). S2 Fig includes all publicly available WGS data assigned to this polytomy. In terms of canSNP nomenclature, the polytomy belongs to B.12 B.42 and includes all branches subordinate to B.66. The 64 strains assigned to the B.66 polytomy have been collected between 1928 and 2018 (S2 Fig and S1 Table) and define four branches arbitrarily labelled here L1 to L4. From the MRCA of the polytomy indicated by the red star in S2 Fig to the tips, distances are in the range 5-19 SNPs. Branch L1 in the B.66 polytomy is represented by a single strain, B-8364 isolated in the Far East of Siberia in 1966. Similarly branch L3 is represented by strain Tul-76 from Kazakhstan. L4 with 48 strains is by far the most represented in available WGS data. In addition to strain Tul-20 from Kazakhstan and strain B-7558 presumably from Russia, it comprises strains from Western Europe, particularly Germany (29 strains) and including Scandinavia. Within Europe, Sweden appears to have a special status, as strains from Sweden are present also in L2, and contribute one basal branch within L4. This observation is in agreement with previous reports of a relatively high genetic diversity in Scandinavia, which might suggest that Scandinavia is the primary source for tularemia in Western Europe [47] or that Scandinavia provides more ecological opportunities for long-term maintenance of F. tularensis lineages. Regarding polytomy B.66, Central Asia appears to be a better candidate for being the source since all four branches are represented.
Both lineages L2 and L4 contain secondary polytomies (S2 Fig). The secondary polytomy in L2 contains six branches, with length from root (blue star) to tips varying from five to 14 SNPs. This polytomy is remarquable by its geographic diversity. Two branches are represented by strains isolated in Sweden, and two branches are constituted by strains from West Kazakstan. One of them was isolated from a migratory bird (wheatear Oenanthe isabellina) [49]. The main polytomy in L4 (purple star) contains eleven radiating branches and is shown with a more specific country color code in S3 Fig. Eight branches are represented by strains from Germany and two by strains from Switzerland. The last branch is the most diverse, and was observed in Scandinavia, France, Hungaria and Germany. The most parcimonious interpretation of available data is that Scandinavia (possibly Sweden) was contaminated once by each of the two lineages, L2 and L4, possibly via migratory birds and that secondary contaminations occurred from Scandinavia towards Western Europe, mainly Germany. Much more will need to be known regarding the implication of migratory birds [50] in order to evaluate this preliminary hypothesis.
In vitro genotyping by MLVA
In order to develop a first line genotyping assay, we evaluated the discriminatory power of MLVA in the present collection. Five among the 25 VNTR loci constituting the full MLVA typing scheme are polymorphic, Ft-M3, Ft-M4, Ft-M6, Ft-M20 and Ft-M22 (Tables 2 and S3). The highest discrimination was observed at the FT-M3 locus in agreement with previous reports. Twelve alleles and an HGDI of 0.90 were observed among the 39 strains. Ft-M6 locus was the second most variable, with four alleles. At the Ft-M4, Ft-M20 and Ft-M22 loci, three, two and two alleles were observed respectively.
Table 2. Discrimination of VNTR loci for strains circulating in Kazakhstan.
| Locus name | Number of alleles/genotypes | HGDI (95% confidence interval) |
|---|---|---|
| Ft-M3 | 12 | 0.9042 (0.8730,0.9354) |
| Ft-M4 | 3 | 0.3104 (0.1416,0.4792) |
| Ft-M6 | 4 | 0.6073 (0.4904,0.7242) |
| Ft-M20A | 2 | 0.1457 (0.0013,0.2902) |
| Ft-M22 | 2 | 0.2672 (0.1072,0.4272) |
| MLVA5 Nur-Sultan | 18 | 0.9325 (0.8950,0.9700) |
| insilico-FT-4 | 2 | 0.0513 (0.0000,0.1473) |
| insilico-FT-8 | 2 | 0.2672 (0.1072,0.4272) |
| MLVA7 Nur-Sultan | 19 | 0.9406 (0.9074,0.9738) |
The five polymorphic loci among the classic 25 VNTR resolve 18 genotypes in the present collection (MLVA5 Nur-Sultan, Table 2). Two additional VNTR loci polymorphic in the present collection were identified by in silico analysis, insilico-FT-4_6bp_97bp_2u and insilico-FT-8_4bp_152bp_3u (S2 Table). According to in silico reads analysis, they are polymorphic in the present collection and allow to resolve one additional genotype (Tables 2 and S3). All seven loci can be amplified in a single PCR reaction with different fluorescent dyes and analysed on a genetic analyzer in one run.
In silico MLVA for F. tularensis subsp. holarctica isolates from Kazakhstan
To satisfy future needs in terms of compatibility between in silico and in vitro MLVA genotyping we evaluated two different approaches to deduce the MLVA genotype from sequence data. In the first and more traditional approach, sequencing reads were assembled before analysis. Two popular assemblers were compared, SPAdes and SKESA. In the second approach, raw sequencing reads corresponding to the VNTR loci were extracted and directly analysed to produce the in silico MLVA genotype. The approach takes advantage of the 300 base-pairs reads produced which should be long enough to enclose in a subset of reads the whole tandem repeat and a few flanking nucleotides from both sides, even for the largest Ft-M3 alleles.
The results obtained by direct reads analysis were fully identical to in vitro data (S3 Table). In contrast, discrepancies were observed in the assemblies. SKESA failed to assemble the Ft-M6 allele in two strains. Ft-M3 was not assembled in eleven and five strains with SPAdes and SKESA respectively (S3 Table). Furthermore Ft-M3 SPAdes assemblies showed an incorrect size in five additional strains. SKESA assemblies predicted an incorrect size for insilico-FT-4 in all strains but one.
In summary, direct analysis of 300 bp reads performed significantly better than assemblies analysis. As expected, the number of recovered reads is proportionally dependent on the size of the VNTR allele as illustrated for the Ft-M3 VNTR in S4 Table.
Clustering of F. tularensis subsp. holarctica strains using MLVA and congruence with canSNP
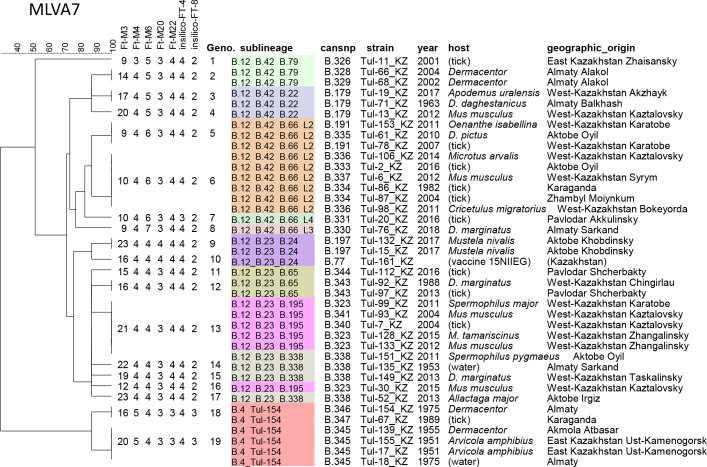
Based on seven variable loci, the 39 strains were clustered into 19 genotypes (Fig 3). Ten genotypes are unique, five genotypes are shared by two strains. The four most frequent genotypes are shared by 3, 4, 6 and 6 strains respectively. The eleven strains belonging to the B.66 polytomy are correctly clustered. More generally, the B.4, B.12_B.23 and B.12_B.42 groups are globally clustered as expected from the wgSNP analysis. Some phylogenetic inconsistencies of terminal branches where MLVA is not congruent with wgSNP phylogeny are visible, due to the homoplasia inherent to tandem repeat polymorphism. Most of these MLVA terminal branches are defined only by the highly variable Ft-M3 locus.
Fig 3. MLVA clustering of the 39 Kazakhstan strains.
The MLVA data for the seven polymorphic loci was clustered using the UPGMA method. The main canSNP sublineages are colored.
Discussion
We present the results of genotyping of thirty-nine F. tularensis subsp. holarctica strains isolated in Kazakhstan including the vaccine strain 15 NIIEG, using MLVA, canSNP and wgSNP. MLVA genotyping was performed by the classical genotyping scheme including 25 loci [4]. In agreement with previous reports five among the subset of ten loci selected by Vogler et al. were variable among the analyzed strains [4,18,51]. For genotyping of isolates circulating in the Central Asian region, we propose a MLVA7 panel combining these five classical VNTR loci and two additional VNTRs identified by in silico analysis of public WGS data. The proposed in silico MLVA genotyping script based on the direct analysis of 300 bp long raw reads made it possible to correctly identify all alleles in the 39 strains. Consequently, MLVA7 constitutes a one PCR assay compatible with in silico WGS analysis which should facilitate the selection of strains for sequencing and the quality control of strain identity.
Based on wgSNP analysis, four genetic groups are distinguished in F. tularensis subsp. holarctica: B.4, B.6, B.12 and B.16 [20,44,52]. In our study, six strains isolated during 1951-89 in Eastern and Central Kazakhstan were assigned to the B.4 genetic group. Strains belonging to the B.4 genetic group were previously found in North America, China, and Northern-Eastern Europe [18,20,31,53,54]. The B.4 genotype might be relatively prevalent in Asia and the sequenced isolates from Kazakhstan allow to define a more basal branching point. This and the presence of B.16 subclade in Japan, Turkey, Tibet and Australia increase the likelihood that F. tularensis subsp. holarctica has an Asian origin [30]. The remaining 33 strains from Kazakhstan were assigned to the B.12 genetic group, which is widely distributed in Central to Eastern Europe [29,52].
Tularemia has been and still is the subject of important monitoring in Kazakhstan because of its zoonotic potential. A number of natural foci have been characterised in Kazakhstan and elsewhere, however the global ecology and phylogeography of this important pathogen remains poorly understood [55]. For instance, although the strict clonality of the evolution of the causal agent predicts that it emerged in one location in space and time, the geographic origin of F. tularensis is unknown. One reason for this situation is that in spite of major progress achieved in the past fifteen years owing to the emergence of powerful genotyping tools, and important efforts in terms of WGS production, data is missing for many countries. Another reason is that the phylogeography of F. tularensis is characterised by the finding of almost identical strains in very distant places and/or at very different dates [26,28,54]. A third reason is the poor knowledge of F. tularensis closest neighbors including F. novicida. It is the knowledge of closest neighbors which provided the strongest argument for an African origin of both Mycobacterium tuberculosis and B. anthracis [56,57].
The work presented here is confirming the remarkable features of the phylogeography of F. tularensis, in particular the growing evidence in favor of the capacity of the bacteria for long-term maintenance combined with a capacity for long-distance spread. This would suggest that F. tularensis has a capacity of «perpetuation», i.e. long-term maintenance without genetic changes, in a state implying a low level of replication [58]. A water-borne viable but non-culturable state has been proposed to explain long-term maintenance [59,60]. However, the available evidence is still fragile and limited. Also this behavior does not seem to fit with the relatively homogeneous branch length from root to tips observed in F. tularensis [22]. Data from many geographic areas is lacking and in addition, most investigations so far were restrospective analyses of historical collection. The analyses required the reculturing of the strains to prepare new DNA extractions. One cannot formally exclude cross-contaminations and additional studies will be required to confirm the present observations. In particular, it would be highly recommended to prepare a DNA batch for later sequencing purposes at the time of the very first isolation and growth of the pathogen. This DNA batch could be validated by MLVA7 in vitro genotyping and the resulting data could be archived in an online F. tularensis MLVA database. This initial genotype could serve as a DNA identification control of the eventual WGS data which could be in silico genotyped at least if sequencing reads are longer than 300 bp.
Interestingly, owing to the extensive field sampling in natural foci regularly done in Kazakhstan, a strain was recovered from an Isabellina wheatear incidentally trapped out of a rodent burrow in West Kazakhstan [49]. Although birds have been suspected as potential spreaders of F. tularensis [61], we found very little direct evidence in the litterature involving birds, either migratory [49,50] or scavenger [62]. The Isabellina wheatear is a migratory insectivorous bird. In spring, it lay eggs in burrows of rodents. Between March and August, birds will visit a number of burrows. After that, winter migrations will start, and rodents will come back to burrows to hibernate. Some fleas are shared between wheatear and rodents [63,64]. The Isabellina wheatear breeding geographic distribution ranges from North-East Greece to North East China. It winters in Africa, the Arabian Peninsula and India where it shares wintering areas with the Northern wheatear (Oenanthe oenanthe), which has an even more widespread geographic range, including Scandinavia, Alaska, North-East Canada. A potential role of the Isabellina wheatear in the propagation of plague has previously been proposed [64,65].
Such an implication would help explain some aspects of the phylogeography of the bacteria, including long-distance transportation and long term preservation (within the soil ecosystem of burrows). This would imply that the birds are healthy carriers. Birds would be infected in their breeding area, and would contaminate other migratory birds, such as the Northern wheatear, in their feeding grounds. The implication of migratory birds might also provide a working hypothesis regarding the dating of the emergence of the clonal F. tularensis. It would have followed the end of the last glaciation peak about 20000 years ago. The Isabelline wheatear is considered to have arrived in the lower Volga region approximately 10000 years ago (reviewed in [64]).
Supporting information
The WGS data from the 17 strains belonging to B.4 were mapped on reference genome OSU18 (assembly accession GCA_000014605) for SNP identification. Two hundred and sixty-six SNPs were called, the Maximum Parsimony tree has a size of 269 (homoplasia 1.12%). For each strain, the country of origin (three letters code), year of isolation, host, strain Id and canSNP assignment are indicated when known. Coloring reflects sublineage assignment. Branches longer than one are labelled with size. The blue star indicates the position of the MRCA.
(TIF)
Three hundred and twenty-five SNPs were identified by mapping on the FSC200 genome sequence (assembly accession GCA_000168775). The Maximum Parsimony tree has a size of 325 (no homoplasy). Branch length is proportional to number of SNPs and ranges from one SNP up to 14. The red star indicates the position of the MRCA of the polytomy. Four branches arbitrarily labelled L1 to L4 radiate from the red star. Distances from the red star to the tips vary from five (L4 strain 4534-12-01 isolated in 2012 in Germany) up to 19 (L4 strain 14T0052 isolated in 2014 in Germany) SNPs. Circles are labelled with year of isolation and strain Id, and colored according to geographic origin as indicated.
(TIF)
The polytomy comprises 20 strains isolated in Europe, predominantly Germany, in the years 2000-2018. Ninety-six SNPs were identified by mapping on the FSC200 genome sequence (assembly accession GCA_000168775). The Maximum Parsimony tree has a size of 96 (no homoplasy). Branch length from the MRCA indicated by the purple star to the tips varies from two up to eleven SNPs. Circles are labelled with the three-letters country code, year of isolation, strain Id and host. They are colored according to geographic origin.
(TIF)
The “map_SCHU_S4_percent” column indicates for each dataset the percentage of the SCHU_S4 genome (assembly accession GCA_000008985) which does not contribute to the SNP search. This percentage corresponds to regions deleted in the strain being mapped, and also to repeated sequences. The “remarks” column provides a list of duplicate entries (usually strains for which both assemblies and raw data have been deposited, and also reference strains which have been independently sequenced). Entries labelled as “redundant” are entries showing an identical wgSNP profile with an other entry. The last four columns indicate which public datasets were used for the making of the indicated figures.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
AShe was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant No. AP05131460). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Savin C, Criscuolo A, Guglielmini J, Le Guern A-S, Carniel E, Pizarro-Cerdá J, et al. Genus-wide Yersinia core-genome multilocus sequence typing for species identification and strain characterization. Microbial genomics. 2019;5(10). 10.1099/mgen.0.000301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunne WM Jr, Pouseele H, Monecke S, Ehricht R, van Belkum A. Epidemiology of transmissible diseases: array hybridization and next generation sequencing as universal nucleic acid-mediated typing tools. Infection, Genetics and Evolution. 2018;63:332–45. 10.1016/j.meegid.2017.09.019 [DOI] [PubMed] [Google Scholar]
- 3.Terletsky VP, Tyshchenko VI, Novikova II, Boikova IV, Tyulebaev SD, Shakhtamirov IY. An efficient method for genetic certification of Bacillus subtilis strains, prospective producers of biopreparations. Microbiology+. 2016;85(1):71–6. 10.1134/S0026261716010136 WOS:000370791300008. [DOI] [PubMed] [Google Scholar]
- 4.Johansson A, Farlow J, Larsson P, Dukerich M, Chambers E, Byström M, et al. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. Journal of bacteriology. 2004;186(17):5808–18. 10.1128/JB.186.17.5808-5818.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogler AJ, Birdsell D, Price LB, Bowers JR, Beckstrom-Sternberg SM, Auerbach RK, et al. Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J Bacteriol. 2009;191(8):2474–84. Epub 2009/03/03. 10.1128/JB.01786-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell CL, Andrianaivoarimanana V, Colman RE, Busch J, Hornstra-O’Neill H, Keim PS, et al. Low cost, low tech SNP genotyping tools for resource-limited areas: Plague in Madagascar as a model. PLoS Negl Trop Dis. 2017;11(12):e0006077. Epub 2017/12/12. 10.1371/journal.pntd.0006077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shevtsova E, Vergnaud G, Shevtsov A, Shustov A, Berdimuratova K, Mukanov K, et al. Genetic diversity of Brucella melitensis in Kazakhstan in relation to World-wide diversity. Front Microbiol. 2019;10:1897. 10.3389/fmicb.2019.01897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen M, Schjørring S, Björkman J, Torpdahl M, Litrup E, Nielsen E, et al. External quality assessment for molecular typing of Salmonella 2013–2015: performance of the European national public health reference laboratories. European Journal of Clinical Microbiology & Infectious Diseases. 2017;36(10):1923–32. 10.1007/s10096-017-3015-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz H, Vergnaud G. Molecular characterisation of Brucella species. Rev Sci Tech. 2013;32(1):149–62. 10.20506/rst.32.1.2189 [DOI] [PubMed] [Google Scholar]
- 10.Levy H, Fisher M, Ariel N, Altboum Z, Kobiler D. Identification of strain specific markers in Bacillus anthracis by random amplification of polymorphic DNA. FEMS microbiology letters. 2005;244(1):199–205. 10.1016/j.femsle.2005.01.039 [DOI] [PubMed] [Google Scholar]
- 11.Antwerpen MH, Prior K, Mellmann A, Höppner S, Splettstoesser WD, Harmsen D. Rapid high resolution genotyping of Francisella tularensis by whole genome sequence comparison of annotated genes (“MLST+”). PLoS One. 2015;10(4):e0123298. 10.1371/journal.pone.0123298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rouphael NG, Stephens DS. Neisseria meningitidis: biology, microbiology, and epidemiology. Neisseria meningitidis: Springer; 2012. p. 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura B. Will the emergence of core genome MLST end the role of in silico MLST? Food microbiology. 2018;75:28–36. 10.1016/j.fm.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 14.Nadon C, Van Walle I, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, et al. PulseNet International: Vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro surveill. 2017;22(23):30544. 10.2807/1560-7917.ES.2017.22.23.30544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergnaud G, Hauck Y, Christiany D, Daoud B, Pourcel C, Jacques I, et al. Genotypic expansion within the population structure of classical Brucella species revealed by MLVA16 typing of 1404 Brucella isolates from different animal and geographic origins, 1974–2006. Frontiers in microbiology. 2018;9:1545. 10.3389/fmicb.2018.01545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambroise J, Irenge LM, Durant J-F, Bearzatto B, Bwire G, Stine OC, et al. Backward compatibility of whole genome sequencing data with MLVA typing using a new MLVAtype shiny application for Vibrio cholerae. PloS one. 2019;14(12):e0225848. 10.1371/journal.pone.0225848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerging infectious diseases. 2002;8(2):225. 10.3201/eid0802.010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogler AJ, Birdsell D, Wagner DM, Keim P. An optimized, multiplexed multi-locus variable-number tandem repeat analysis system for genotyping Francisella tularensis. Lett Appl Microbiol. 2009;48(1):140–4. Epub 2008/11/21. 10.1111/j.1472-765X.2008.02484.x . [DOI] [PubMed] [Google Scholar]
- 19.Guryčová D. First isolation of Francisella tularensis subsp. tularensis in Europe. European journal of epidemiology. 1998;14(8):797–802. 10.1023/a:1007537405242 [DOI] [PubMed] [Google Scholar]
- 20.Appelt S, Köppen K, Radonić A, Drechsel O, Jacob D, Grunow R, et al. Genetic diversity and spatial segregation of Francisella tularensis subspecies holarctica in Germany. Front Cell Infect Microbiol. 2019;9:376. Epub 2019/11/30. 10.3389/fcimb.2019.00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samrakandi M, Zhang C, Zhang M, Nietfeldt J, Kim J, Iwen PC, et al. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol Lett. 2004;237(1):9–17. 10.1016/j.femsle.2004.06.008 [DOI] [PubMed] [Google Scholar]
- 22.Timofeev V, Bakhteeva I, Titareva G, Kopylov P, Christiany D, Mokrievich A, et al. Russian isolates enlarge the known geographic diversity of Francisella tularensis subsp. mediasiatica. PLoS One. 2017;12(9):e0183714. 10.1371/journal.pone.0183714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingry LC, Petersen JM. Comparative review of Francisella tularensis and Francisella novicida. Front Cell Infect Microbiol. 2014;4:35. Epub 2014/03/25. 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson A, Celli J, Conlan W, Elkins KL, Forsman M, Keim PS, et al. Objections to the transfer of Francisella novicida to the subspecies rank of Francisella tularensis. Int J Syst Evol Microbiol. 2010;60(Pt 8):1717–8. Epub 2010/08/07. 10.1099/ijs.0.022830-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whipp MJ, Davis JM, Lum G, de Boer J, Zhou Y, Bearden SW, et al. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. Journal of medical microbiology. 2003;52(9):839–42. 10.1099/jmm.0.05245-0 [DOI] [PubMed] [Google Scholar]
- 26.Dwibedi C, Birdsell D, Larkeryd A, Myrtennas K, Ohrman C, Nilsson E, et al. Long-range dispersal moved Francisella tularensis into Western Europe from the East. Microb Genom. 2016;2(12):e000100. Epub 2017/03/30. 10.1099/mgen.0.000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, Ravel J, et al. Global genetic population structure of Bacillus anthracis. PLoS One. 2007;2(5):e461. Epub 2007/05/24. 10.1371/journal.pone.0000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kevin M, Girault G, Caspar Y, Cherfa MA, Mendy C, Tomaso H, et al. Phylogeography and Genetic Diversity of Francisella tularensis subsp. holarctica in France (1947-2018). Front Microbiol. 2020;11:287. Epub 2020/03/21. 10.3389/fmicb.2020.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koene M, Rijks J, Maas M, Ruuls R, Engelsma M, van Tulden P, et al. Phylogeographic Distribution of Human and Hare Francisella Tularensis Subsp. Holarctica Strains in the Netherlands and Its Pathology in European Brown Hares (Lepus Europaeus). Front Cell Infect Microbiol. 2019;9:11. Epub 2019/02/26. 10.3389/fcimb.2019.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Yu Y, Feng L, Li Y, He J, Zhu H, et al. Phylogeography of Francisella tularensis from Tibet, China: Evidence for an asian origin and radiation of holarctica-type Tularemia. Ticks Tick Borne Dis. 2016;7(5):865–8. Epub 2016/05/07. 10.1016/j.ttbdis.2016.04.001 . [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Peng Y, Hai R, Xia L, Li H, Zhang Z, et al. Diversity of Francisella tularensis subsp. holarctica lineages, China. Emerg Infect Dis. 2014;20(7):1191–4. Epub 2014/06/26. 10.3201/eid2007.130931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilic S, Birdsell DN, Karagoz A, Celebi B, Bakkaloglu Z, Arikan M, et al. Water as Source of Francisella tularensis Infection in Humans, Turkey. Emerg Infect Dis. 2015;21(12):2213–6. Epub 2015/11/20. 10.3201/eid2112.150634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunitsa T, Sadovskaya V, Izbanova U. [Current state of epidemiological monitoring of tularemia in natural foci of Kazakhstan]. In: Aikimbayev A, editor. Collection of works on tularemia dedicated to the 100th anniversary of doctor of medical Sciences, Professor Masgut Aikimbayev. Almaty: Kazakh scientific center for quarantine of zoonotic infections named after M. Aikimbayev; 2016. p. 190–211. Russian. 10.1039/c5cp06476g [DOI] [Google Scholar]
- 34.Meka-Mechenko T, Aikimbayev A, Kunitza T, Ospanov K, Temiralieva G, Dernovaya V, et al. Clinical and epidemiological characteristic of tularemia in Kazakhstan. Przeglad epidemiologiczny. 2003;57(4):587–91. [PubMed] [Google Scholar]
- 35.Kunitsa T, Izbanova U, Meka-Mechenko T, Yakupov V. [The modern clinical and epidemical peculiarities of manifestation of tularemia in Kazakhstan urbanized territories]. Life without danger Health Prevention Longevity. 2013;8(2):41–6. Russian. [Google Scholar]
- 36.Kunitsa T, Meka-Mechenko T, Lukhnova L. [Incidence of tularemia in Kazakhstan]. Probl especially dangerous infections 2001;1:52. Russian. [Google Scholar]
- 37.Izbanova U, Kunitsa T, Meka-Mechenko T, Kazakov S, Kyraubayev K, Ayazbayev T. [Epizootic and epidemic situation on tularemia in Kazakhstan in 2011–2012]. Disinfection Antiseptic 2013;4(2):28–31. Russian. [Google Scholar]
- 38.Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom; 2010. [Google Scholar]
- 39.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–8. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20(4):406–16. [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome biology. 2018;19(1):153. 10.1186/s13059-018-1540-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lärkeryd A, Myrtennäs K, Karlsson E, Dwibedi CK, Forsman M, Larsson P, et al. CanSNPer: a hierarchical genotype classifier of clonal pathogens. Bioinformatics. 2014;30(12):1762–4. 10.1093/bioinformatics/btu113 [DOI] [PubMed] [Google Scholar]
- 44.Wittwer M, Altpeter E, Pilo P, Gygli SM, Beuret C, Foucault F, et al. Population Genomics of Francisella tularensis subsp. holarctica and its Implication on the Eco-Epidemiology of Tularemia in Switzerland. Front Cell Infect Microbiol. 2018;8:89. Epub 2018/04/07. 10.3389/fcimb.2018.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic acids research. 1999;27(2):573–80. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. Journal of clinical microbiology. 1988;26(11):2465–6. 10.1128/JCM.26.11.2465-2466.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson E, Svensson K, Lindgren P, Bystrom M, Sjodin A, Forsman M, et al. The phylogeographic pattern of Francisella tularensis in Sweden indicates a Scandinavian origin of Eurosiberian tularaemia. Environ Microbiol. 2013;15(2):634–45. Epub 2012/12/21. 10.1111/1462-2920.12052 . [DOI] [PubMed] [Google Scholar]
- 48.Johansson A, Larkeryd A, Widerstrom M, Mortberg S, Myrtannas K, Ohrman C, et al. An outbreak of respiratory tularemia caused by diverse clones of Francisella tularensis. Clin Infect Dis. 2014;59(11):1546–53. Epub 2014/08/07. 10.1093/cid/ciu621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kdyrsikh BG, Sundukov RI, Kuspanov AK, Kdyrsikhova GG, Ayazbaev TZ, Mikanov NS. About the isolation of the tularemia agent from the Isabelline wheatear (Oenanthe isabellina) in West Kazakhstan region. Quarantinable and zoonotic infections in Kazakhstan. 2013;28(2):71–2. [Google Scholar]
- 50.de Carvalho IL, Zé-Zé L, Alves AS, Pardal S, Lopes RJ, Mendes L, et al. Borrelia garinii and Francisella tularensis subsp. holarctica detected in migratory shorebirds in Portugal. Eur J Wildl Res. 2012;58:857–61 10.1007/s10344-012-0617-3 [DOI] [Google Scholar]
- 51.Ariza-Miguel J, Johansson A, Fernández-Natal MI, Martínez-Nistal C, Orduña A, Rodríguez-Ferri EF, et al. Molecular investigation of tularemia outbreaks, Spain, 1997-2008. Emerg Infect Dis. 2014;20(5):754–61. Epub 2014/04/23. 10.3201/eid2005.130654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gyuranecz M, Birdsell DN, Splettstoesser W, Seibold E, Beckstrom-Sternberg SM, Makrai L, et al. Phylogeography of Francisella tularensis subsp. holarctica, Europe. Emerg Infect Dis. 2012;18(2):290–3. Epub 2012/02/07. 10.3201/eid1802.111305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sissonen S, Rossow H, Karlsson E, Hemmilä H, Henttonen H, Isomursu M, et al. Phylogeography of Francisella tularensis subspecies holarctica in Finland, 1993-2011. Infect Dis (Lond). 2015;47(10):701–6. Epub 2015/05/26. 10.3109/23744235.2015.1049657 . [DOI] [PubMed] [Google Scholar]
- 54.Myrtennäs K, Marinov K, Johansson A, Niemcewicz M, Karlsson E, Byström M, et al. Introduction and persistence of tularemia in Bulgaria. Infect Ecol Epidemiol. 2016;6:32838. Epub 2016/10/30. 10.3402/iee.v6.32838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telford SR 3rd, Goethert HK. Ecology of Francisella tularensis. Annu Rev Entomol. 2020;65:351–72. Epub 2019/10/11. 10.1146/annurev-ento-011019-025134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vergnaud G. Bacillus anthracis evolutionary history: taking advantage of the topology of the phylogenetic tree and of human history to propose dating points. Erciyes Med J. 2020;42(4):362–9. Epub 21/04/2020. 10.14744/etd.2020.64920 [DOI] [Google Scholar]
- 57.Blouin Y, Hauck Y, Soler C, Fabre M, Vong R, Dehan C, et al. Significance of the identification in the Horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PLoS One. 2012;7(12):e52841. Epub 2013/01/10. 10.1371/journal.pone.0052841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Telford SR 3rd, Goethert HK. Toward an understanding of the perpetuation of the agent of tularemia. Front Microbiol. 2010;1:150. Epub 2010/01/01. 10.3389/fmicb.2010.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hennebique A, Boisset S, Maurin M. Tularemia as a waterborne disease: a review. Emerg Microbes Infect. 2019;8(1):1027–42. Epub 2019/07/10. 10.1080/22221751.2019.1638734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forsman M, Henningson EW, Larsson E, Johansson T, Sandstrom G. Francisella tularensis does not manifest virulence in viable but non-culturable state. FEMS Microbiol Ecol. 2000;31(3):217–24. Epub 2000/03/17. 10.1111/j.1574-6941.2000.tb00686.x . [DOI] [PubMed] [Google Scholar]
- 61.Splettstoesser WD, Mätz-Rensing K, Seibold E, Tomaso H, Al Dahouk S, Grunow R, et al. Re-emergence of Francisella tularensis in Germany: fatal tularaemia in a colony of semi-free-living marmosets (Callithrix jacchus). Epidemiol Infect. 2007;135(8):1256–65. Epub 2007/02/20. 10.1017/S0950268807008035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Padeshki PI, Ivanov IN, Popov B, Kantardjiev TV. The role of birds in dissemination of Francisella tularensis: first direct molecular evidence for bird-to-human transmission. Epidemiol Infect. 2010;138(3):376–9. Epub 2009/08/12. 10.1017/S0950268809990513 . [DOI] [PubMed] [Google Scholar]
- 63.Burdelov AS, Kassenova AK. [Contacts of isabelline wheatear and its specific fleas with the plague pathogen]. Quarantinable and zoonotic infections in Kazakhstan. 2001;(4):17–9. Russian. [Google Scholar]
- 64.Popov NV, Sludsky AA, Zavialov EV, Udovikov AI, Tabachishin VG, Anikin VV, et al. [A probable role of Oenanthe isabellina and other birds in the plague enzootic mechanism]. Povolzhsky journal of ecology. 2007;(3):215–26. Russian. [Google Scholar]
- 65.Balakhonov SV, Korzun VM, Verzhutsky DB, Mikhaylov EP, Rozhdestvensky EN, Denisov AV. The First Case of Yersinia pestis subsp. pestis Isolation in the Territory of the Altai Mountain Natural Plague Focus. Communication 2. Probable Ways and Mechanisms of Plague Agent Main Subspecies Importation into the Territory of the Focus. Problems of Particularly Dangerous Infections. 2013;(2):5–10. 10.21055/0370-1069-2013-2-5-10 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The WGS data from the 17 strains belonging to B.4 were mapped on reference genome OSU18 (assembly accession GCA_000014605) for SNP identification. Two hundred and sixty-six SNPs were called, the Maximum Parsimony tree has a size of 269 (homoplasia 1.12%). For each strain, the country of origin (three letters code), year of isolation, host, strain Id and canSNP assignment are indicated when known. Coloring reflects sublineage assignment. Branches longer than one are labelled with size. The blue star indicates the position of the MRCA.
(TIF)
Three hundred and twenty-five SNPs were identified by mapping on the FSC200 genome sequence (assembly accession GCA_000168775). The Maximum Parsimony tree has a size of 325 (no homoplasy). Branch length is proportional to number of SNPs and ranges from one SNP up to 14. The red star indicates the position of the MRCA of the polytomy. Four branches arbitrarily labelled L1 to L4 radiate from the red star. Distances from the red star to the tips vary from five (L4 strain 4534-12-01 isolated in 2012 in Germany) up to 19 (L4 strain 14T0052 isolated in 2014 in Germany) SNPs. Circles are labelled with year of isolation and strain Id, and colored according to geographic origin as indicated.
(TIF)
The polytomy comprises 20 strains isolated in Europe, predominantly Germany, in the years 2000-2018. Ninety-six SNPs were identified by mapping on the FSC200 genome sequence (assembly accession GCA_000168775). The Maximum Parsimony tree has a size of 96 (no homoplasy). Branch length from the MRCA indicated by the purple star to the tips varies from two up to eleven SNPs. Circles are labelled with the three-letters country code, year of isolation, strain Id and host. They are colored according to geographic origin.
(TIF)
The “map_SCHU_S4_percent” column indicates for each dataset the percentage of the SCHU_S4 genome (assembly accession GCA_000008985) which does not contribute to the SNP search. This percentage corresponds to regions deleted in the strain being mapped, and also to repeated sequences. The “remarks” column provides a list of duplicate entries (usually strains for which both assemblies and raw data have been deposited, and also reference strains which have been independently sequenced). Entries labelled as “redundant” are entries showing an identical wgSNP profile with an other entry. The last four columns indicate which public datasets were used for the making of the indicated figures.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.