Summary
The spine is a defining feature of the vertebrate body plan. However, broad differences in vertebral structures and morphogenetic strategies occur across vertebrate groups, clouding the homology between their developmental programs. Analysis of a zebrafish mutant, spondo, whose spine is dysmorphic, promoted us to reconstruct paleontological evidence, highlighting specific transitions during teleost spine evolution. Interestingly, the spondo mutant recapitulates characteristics present in basal fishes, not found in extant teleosts. Further analysis of the mutation implicated the teleost-specific notochord protein, Calymmin, as a key regulator of spine patterning in zebrafish. The mutation in cmn results in loss of notochord sheath segmentation, altering osteoblast migration to the developing spine, and increasing sensitivity to somitogenesis defects associated with congenital scoliosis in amniotes. These data suggest that signals from the notochord define the evolutionary identity of the spine and demonstrate how simple shifts in development can revert traits canalized for about 250 million years.
Keywords: notochord, vertebrae, evolution, zebrafish, teleosts
Graphical Abstract
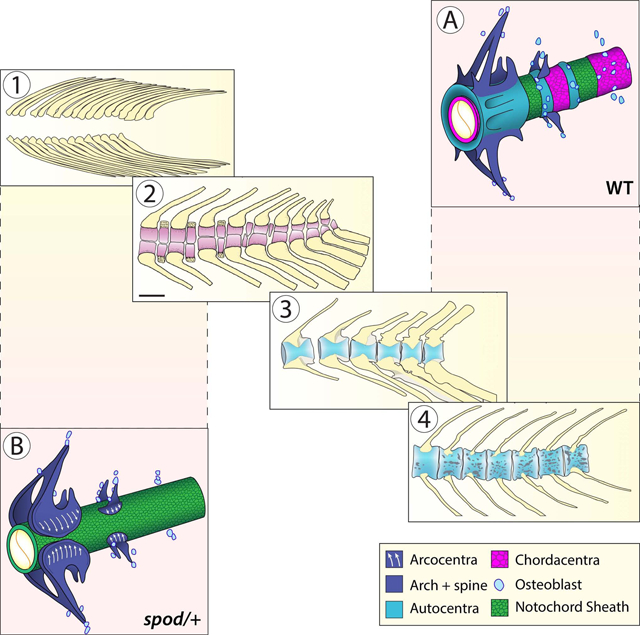
eTOC
Peskin et al. show that a zebrafish mutation alters vertebral morphogenesis to recapitulate features present in the spine of basal fishes, thus unmasking ancestral traits hidden for 250 million years. Using genetics and live imaging, they link this developmental shift to notochord signals that define the evolutionary identity of the teleost spine.
Introduction
The vertebral column or spine has evolved to provide structural support along the body axis as well as to protect the main neural and vascular components of the trunk. The intricate segmentation of the spine is critical for locomotion and regional sub-functionalization of the different vertebrae [1]. While all vertebrates share endoskeletal elements comprising the vertebral axis, the vertebrae themselves are not structurally homologous across broad classes of vertebrates and develop through distinct patterning mechanisms [2, 3]. A fundamental structural difference between vertebrae of teleost fishes and vertebrae of amniotes is the formation of mineralized rings surrounding the notochord sheath, called the chordacentra. This is a defining trait for all teleost fishes [2, 4–6]. In teleosts, vertebrae form through periosteal ossification of these prior mineralized foundations, ultimately forming the autocentra. By contrast, in amniotes, sclerotomal cells give rise to the elements of the vertebrae surrounding the notochord through endochondral ossification, forming an arcocentrum [3].
Recent genetic work in zebrafish revealed an instructional role of the notochord in setting up the initial pattern of mineralization during teleost spine development [7–9]. Prior to chordacentra formation, notochord sheath cells undergo a segmentation event that dictates where the prospective centra and intervertebral discs (IVD) will form at later stages [9]. Initially, the sheath contains a uniform epithelial layer of cells that express markers such as type II and type IX Collagen. During notochord segmentation, alternating domains of chondrocyte-like sheath cells transition into mineralizing cells in a manner dependent on notch signaling [9]. These mineralizing cells, marked by the expression of entpd5a, produce the chordacentra and serve as a template for the mature autocentra that are formed by sclerotomal osteoblasts [9, 10]. The appearance of autocentra are first annotated during the early Jurassic period (circa 180 Ma) and constitute the most advanced step of spine evolution in teleosts [11–15]. While the fossil record and the structures present in extant teleosts suggest a gradual and stepwise emergence of fish vertebral structure and pattern [11–13], the morphogenetic events and transitions underlying their appearance throughout evolution have not been detailed as a transitional series.
Genetic and embryological studies in zebrafish, chicken, and mice have shown that while in all vertebrates the notochord provides an attractive signal nucleating vertebral body formation [3, 9, 16, 17], in amniotes, vertebral patterning is predominately influenced by somite segmentation [16]. Consequently, mutations affecting somitogenesis, such as tbx6, often lead to congenital vertebral malformations and spine patterning defects in amniotes [18, 19]. Conversely, zebrafish centra are largely unaffected by aberrant somite patterning [5, 7, 9]. The apparent diversity in vertebral formation and patterning mechanisms can obscure a clear picture of evolutionary divergence and analysis of the similarities that exist between key vertebrate structures [3]. Moreover, because of this duality of developmental mechanisms among major vertebrate classes, it has been difficult to accurately model human spinal disorders in teleost experimental models such as the zebrafish or medaka. In this study, we have identified core patterning homologies of vertebrates retained in teleosts, normally suppressed by dominant signals stemming from the notochord. This discovery highlights the importance of the notochord during evolution in defining the traits of a major vertebrate group.
Results
In a recent screen for mutations affecting adult skeletal phenotypes [20], we isolated a mutant, spondo (spod), that exhibits a unique vertebral patterning phenotype (Figure 1). In contrast to wild-type siblings, heterozygous spondo adults form hemicentra with different polarity across the spine (Figure 1C,D). Hemicentra form with clefts separating either dorsal/ventral or left/right halves, depending on the presence of ribs or ventral hemal spines, with a distinct bias in lateral clefts occurring caudally (Figure 1D, D’). Additionally, in spondo mutants we detected a low frequency of the formation of two vertebral centra per segment, or diplospondyly, as registered by the position of neural and hemal spines at several points (Figure 1C, F’ arrows).
Figure 1. The spondo mutant impairs axial patterning and leads to the formation of hemicentra.
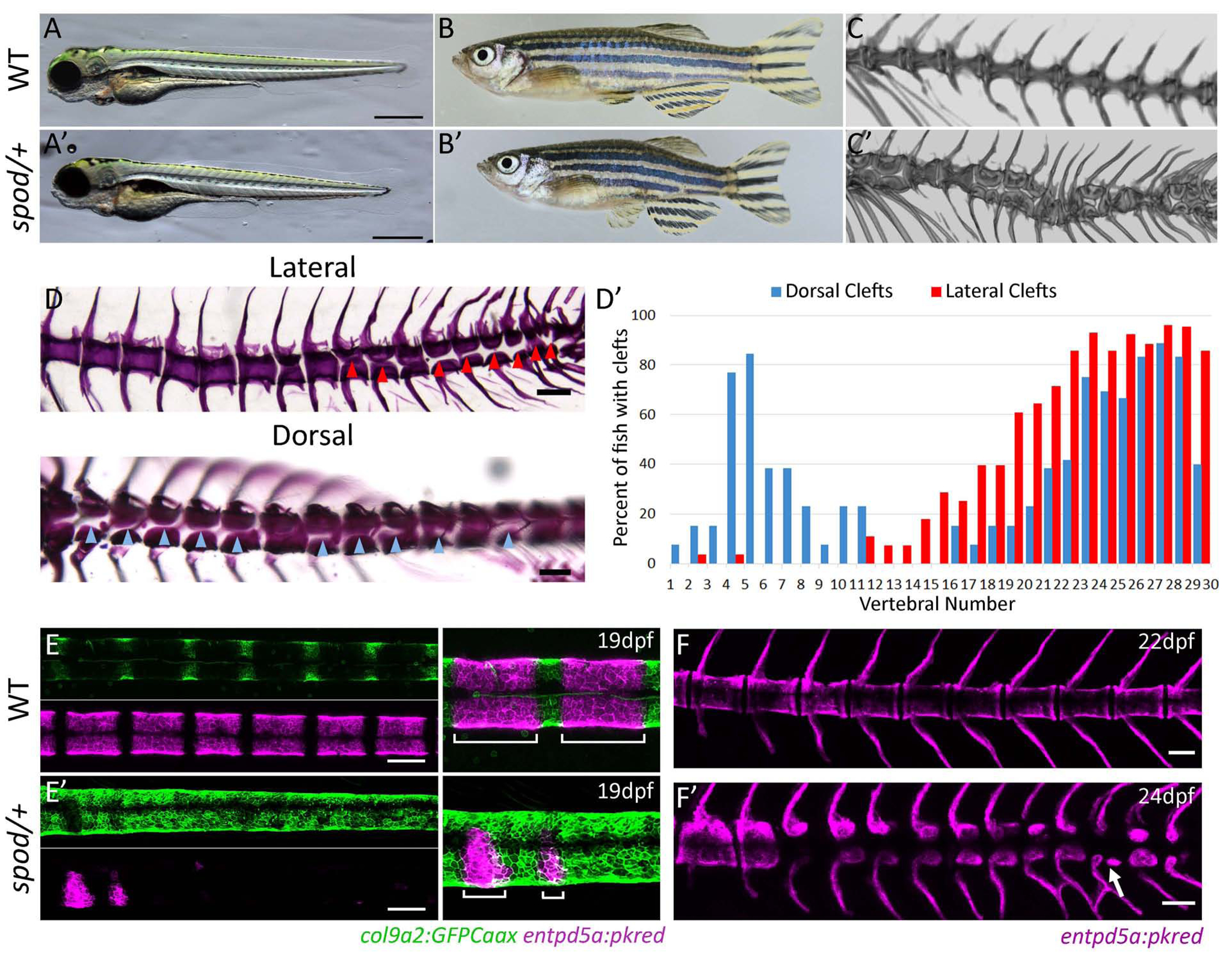
A) spondo mutant zebrafish are viable. 4dpf larval morphology of wild-type and cmnspod heterozygous larvae. Mutants show normal patterning of larval structures, however, are shorter in length. B) cmnspod/+ mutants are viable and exhibit short stature as adults. Tomographic reconstruction of vertebrae of wild-type C) and spondo heterozygous C’) adult fish showing formation of hemicentra and periodic diplospondyly (arrow). Homozygous fish show an increase in phenotype, (Figure S1). D) Lateral and dorsal views of cmnspod/+ alizarin red stained skeletal preps. Hemicentra are present along the body axis. Arrows indicate vertebral clefts. D’) The frequency of vertebral clefts along the body axis was quantified. Dorsal clefts were found bimodally in the rostral and caudal regions of the vertebral column while lateral clefts were most prominent in the caudal region and associated with non-rib-bearing vertebrae. E) Early specification of notochordal patterning is disrupted in spondo mutants. In wild-type fish, col9a2:GFPCaax expression is restricted to the prospective intervertebral discs (IVD) while entpd5a expression is activated in the mineralizing domains (future chordacentra) of the notochord sheath. bottom) E’) Specification of the IVD and mineralizing domains is absent in spondo mutants, while the expression domain of col9a2 is expanded. Overlay of entpd5a and col9a2 transgenes showing lack of meristic patterning of chordacentra in spondo mutants. F) entpd5a expression in juvenile fish showing altered pattering of centra mineralization in the spondo, F’) heterozygous mutant; arrow points to a developing diplospondylous vertebrae.
We traced the origin of these vertebral abnormalities in spondo heterozygous fish back to early patterning defects during notochord segmentation and chordacentra formation (Figure 1E,F). In wild-type zebrafish, the outer layer of the notochord consists of an epithelial-like sheath in which all cells initially express col9a2 uniformly, along with other genes typically associated with cartilage formation. During larval development, notochord sheath cells undergo a segmentation process that establishes a blueprint for the formation of vertebral bodies and IVDs at later stages [9]. During notochord segmentation, alternating segments of sheath cells differentiate into mineralizing cells marked by the expression of entpd5a and the downregulation of col9a2 expression. The entpd5a+ domains then produce mineralized chordacentra, while cells that do not differentiate and retain col9a2 expression become the prospective IVDs [9]. Throughout this study, we have used in vivo imaging of notochord segmentation markers (col9a2:GFPCaax and entpd5a:pKred) [9, 21] to directly visualize the process of notochord segmentation in the developing notochord of larval spondo mutants and wild-type siblings. Compared to wild-type siblings, spondo mutants show a general deficiency of entpd5a activation and an absence of col9a2 downregulation in a segmented manner (Figure 1E). Consequently, this leads to failure in the establishment of alternating centra and intervertebral disc domains early in vertebrae development, precluding chordacentra formation along the body axis (Figure 1F). While homozygous spondo mutant fish retain viability, they display highly dysmorphic vertebrae including fusions and formation of hemicentra (Figure S1). Interestingly, the outward features of the heterozygous spondo mutant vertebrae are not highly dysmorphic, rather closely resemble the morphologies present in the spines of primitive teleost fishes, including the re-emergence of a distinct arcocentral-type centrum, lack of epineural spines, a reversion to the formation of diplospondyly and the presence of lateral clefts [2].
The morphological transitions that took place during teleost spine evolution, specifically the changes associated with crown group vertebral characters have not been systematically integrated and defined. To investigate the evolution of the teleost spine and compare ancestral morphologies to the spondo mutant phenotype, we reconstructed fossil evidence of vertebral anatomy across stem Teleostei or Teleosteomorpha and integrated this data with morphological transitions observed in the phylogeny of early fishes and stem teleosts (Figure 2). The primitive condition for all stem teleosts or teleosteomorphs is the absence of a vertebral centrum. Instead, they retain a functional and persistent notochord that supports the neural arches and spines, or arcualia. The arcualia form from somite-derived mesenchyme over a notochord that persists as a continuous rod into mature stages without mineralization [2]. This condition illustrated by the pachycormiform Orthocormus (Figure 2A, E: state “0”) is also represented in extant basal lineages of vertebrates including early sarcopterygian lineages such as the coelacanth (Actinistia) and the lungfish (Dipnoi) as well as early actinopterygian lineages such as the sturgeon (Chondrostei).
Figure 2. Patterning of the spine and the evolution of the teleost vertebral centrum.
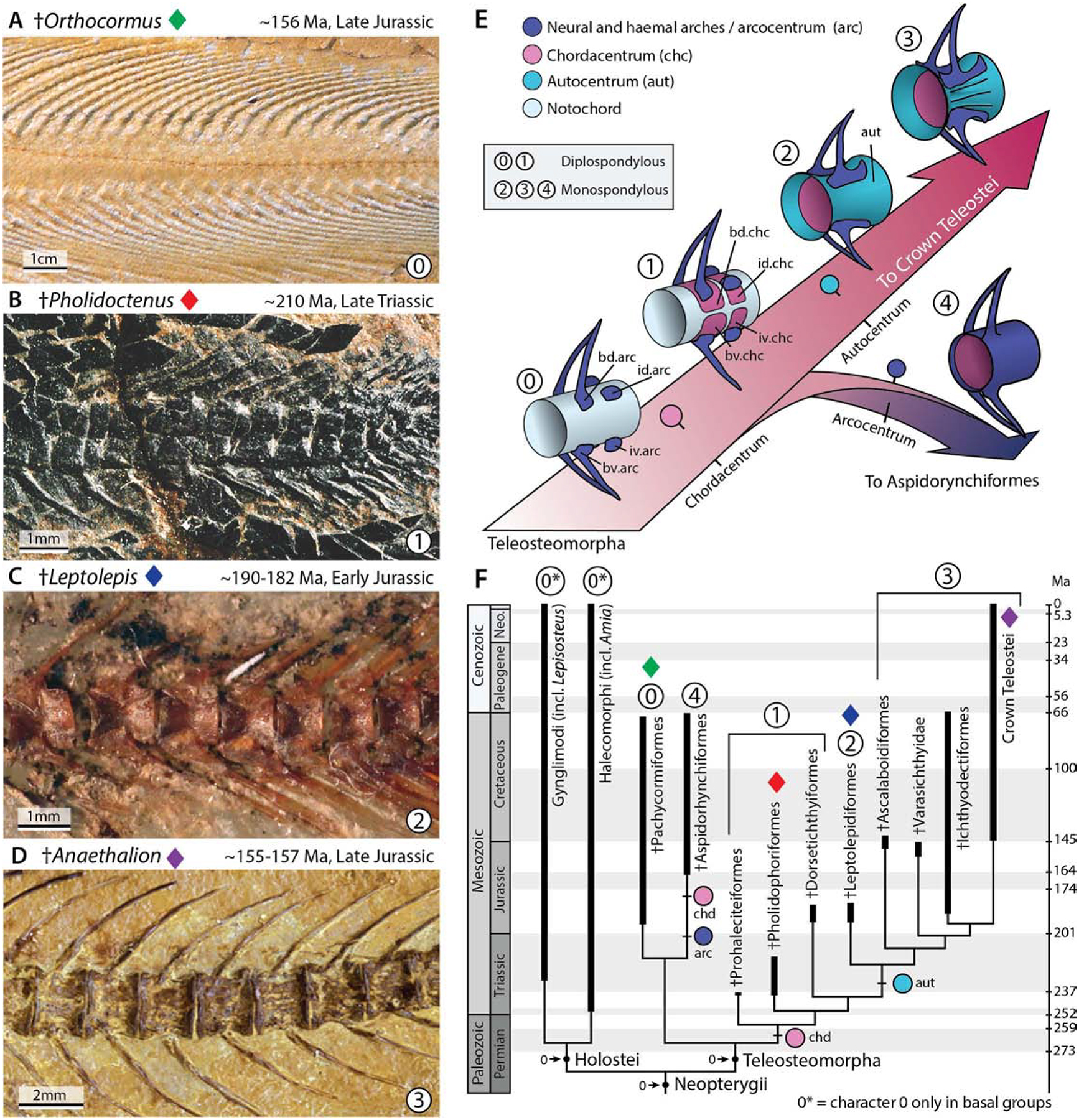
Reconstructing the changes in spine patterning and morphology during the evolution of teleosts. A) The earliest members of stem teleosts lacked a vertebral centrum, and instead retained a functional notochord supporting the arches and spines. This is illustrated by the stem pachycormiform †Orthocormus roeperi; see also E) state “0”. B) †Pholidoctenus serianus illustrates a more derived condition present among some pholidophoriforms, with diplospondylous vertebral centra formed by hemi-chordacentra; E) state “1”. C) †Leptolepis coryphenoides illustrates a major transformation within the spine of stem teleosts, characterized by the appearance of a thin hourglass-shaped autocentrum surrounding a ring-like chordacentrum; E) state “2”. D) Subsequently, the autocentrum incorporated a thickened lateral wall, and a series of ornaments, such as crest, grooves and pits, as illustrated by the oldest fossil member of the teleost crown group †Anaethalion sp., a condition retained in, and defining all living teleosts; E) state “3”. E) Reconstructed scenario of the evolution of the teleosteomorph spine—from a basal functional notochord supporting the arches and spines—leading to a chordacentrum plus autocentrum in recent teleosts, or to a chordacentrum plus arcocentrum among extinct aspidorhynchiforms. F) Consensus phylogenetic hypothesis of the total-group teleosts or teleosteomorphs mapped on the geological time scale. The phylogenetic hypothesis was adapted from Arratia et al. (2019). As outgroup taxa, the analysis included five members of Holostei based on a total of 198 morphological characters.
Early teleosts acquired a vertebral centrum by a remarkable variety of mechanisms; aspidorynchiforms, the sister-group of pachycormiforms, acquired a monospondylous vertebral centrum by extending and fusing the neural and haemal arches over the surface of the notochordal sheath. This kind of centrum is referred to as an arcocentral type (Figure 2E: state “4”), a condition also present in the extant gar (Gynglimodi). By contrast, other early stem teleosts formed a vertebral centrum through the mineralization of robust and diplospondylous hemi-chordacentra that surrounded the notochord, as shown by Pholidoctenus (Figure 2B, E: state “1”) and other pholidophoriforms. A major shift in the mechanism of vertebral spine formation is observed in Leptolepis coryphaenoides and more advanced teleosts, with the appearance of an hourglass shaped autocentrum (Figure 2C, E: state “2”). Among more advanced teleosts, the autocentrum acquired a series of pits, grooves and ornaments considered defining traits for extant teleost fishes, as illustrated by the oldest known crown teleosts Anaethalion (Figure 2D, E: state “3”)[4, 11]. In light of these key morphological transitions defining extant teleosts, the vertebral morphology of spondo mutant zebrafish represents a developmental shift from the formation of an autocentral vertebra, characteristic of modern teleosts, into an arcocentral-type — a characteristic lost during the evolution of teleosts. These vertebral characteristics resemble the vertebral centra of more primitive actinopterygian and sarcopterygian lineages [11, 12], including the absence of the hallmark monospondylous chordacentrum and autocentrum that define the vertebral anatomy of extant teleosts.
Using whole exome sequencing of mutant and wild-type sibling pools, we identified a missense mutation within the calymmin (cmn) gene linked to the spondo mutation (Figure 3A)[20]. Cmn is predicted to be an extracellular matrix (ECM) protein with weak similarity to Elastin (Figures 3E, S2)[22]. We found that, similar to a previous report [22], cmn is developmentally regulated and expressed almost exclusively in the developing notochordal sheath (Figures 3B,C,D and S3). Interestingly, in late larval stages, cmn expression becomes segmented, restricted to the col9a2+ domain as well as the segment boundaries where notochord sheath cells are transitioning into mineralizing cells and express both col9a2 and entpd5a (Figure 3D,D’ arrows, S3). Because of the expression pattern and similarity of cmn to elastins, we analyzed the sheath ECM by transmission electron microscopy (TEM) in wild-type and spondo mutants. In spondo mutants, the collagen layer of the ECM was thinner and more loosely organized, suggesting that the altered gene product causes compositional deficiencies of the notochord sheath (Figure S3).
Figure 3. spondo is a dominant, gain-of-function mutation in cmn.
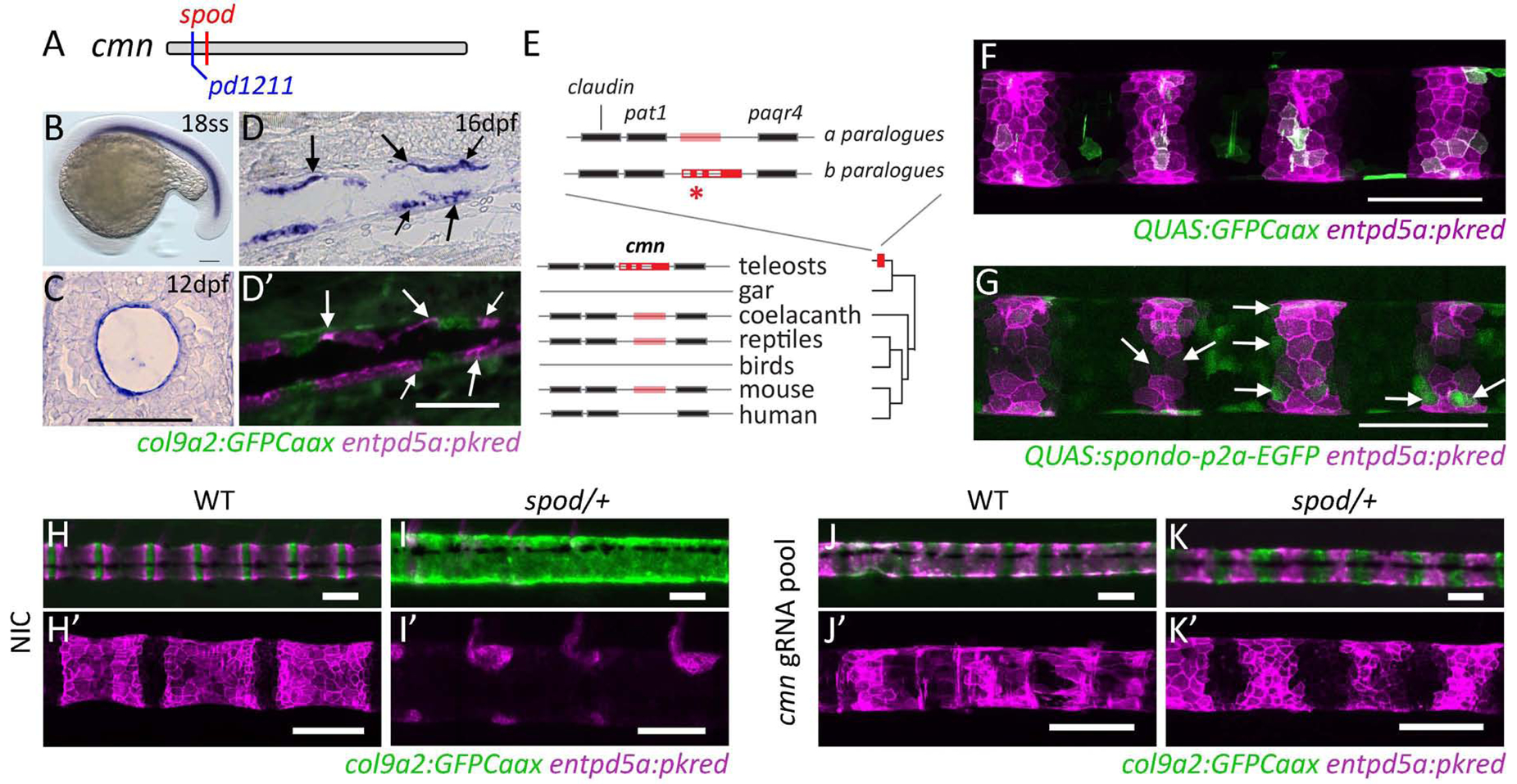
A) The spondo mutant phenotype is due to an early, nonsynonymous mutation in the calymmin gene (M10R). pd1211 mutation is a 2bp deletion upstream of spondo that results in an early stop codon. B) Expression of cmn during early zebrafish development is confined to the notochord (Figure S3). C) Cross section of 12dpf larvae depicting the restriction of cmn expression to the notochord sheath cells. The spondo mutation causes disruption of the notochordal sheath (Figure S3). D,D’) At later developmental stages, cmn expression becomes segmented and downregulated in cells that form the chordacentra. The expression is most highly upregulated in the col9a2 domain and in cells that are actively differentiating into mineralizing cells and express both col9a2 and entpd5a (arrows). E) cmn is a highly derived paralogue of an elastin-like extracellular protein. The locus containing cmn is duplicated in zebrafish, retaining synteny with neighboring genes. The gene altered in spondo (asterisk) is highly differentiated from its other paralogue (‘a’) as well as orthologues found in sarcopterygian lineages and is generally conserved among teleost fishes (Figure S2). F,G) Overexpression of cmn containing the spondo mutation inhibits sheath cell differentiation into mineralizing cells. DNA constructs containing either QUAS:GFPCaax or QUAS:spod-p2A-GFP sequences were mosaically overexpressed in a col9a2:QF2 transgenic line. Gaps in entpd5a:pkred expression within mineralizing domains were quantified (arrows). In control conditions, 14.1% of GFP+ sheath cells did not express entpd5a+. In the experimental group of fish overexpressing the spondo mutation, 42.2% of GFP+ sheath cells were entpd5a negative. Two tailed p-value = 0.0006. Over expression of wild-type cmn does not have any effect (Figure S4). H-K, H’-K’) Reversion of spondo phenotype by creation of disruptive alleles in cis and trans to cmnspondo. Pooled guides targeting cmn injected into wild-type or cmnspod/+ embryos lead to mosaic deletions within the cmn locus. Targeting deletions to cmn reverted the spondo phenotype and partially restored notochord segmentation in 11/11 cmnspod/+ fish analyzed. Phenotype of injected fish closely resembles individuals containing the loss of function allele, pd1211 (Figure S5). See also Table S1.
Previous analysis of the cmn gene suggested that it may be unique to teleosts [22]. However, the taxonomic depth of this work was limited by available genomic information. Although orthologues of cmn are generally not well annotated, we find that cmn likely derived from an ancestral vertebrate elastin-like gene consistently positioned between a set of genes (claudin, pat-1, and paqr4), with high synteny among vertebrates (Figure 3E). Alignment of cmn orthologues revealed retention of two paralogues in teleosts that substantially diverge in sequence identity (Figure S2). The orthologue altered in zebrafish spondo mutants shows unique coding sequence blocks shared among teleosts, not seen in the other teleost paralogue or in ancestral or sarcopterygian orthologues such as coelacanth and lungfish (Figure S2). The locus is missing from the gar genome annotation (Lepisosteus), however, we were able to find evidence for an ancestral elastin-like gene and syntenic paralogues in Bowfin (Amia) and Reedfish (Erpetoichthys) (Braasch pers. Communication) suggesting the presence of an ancestral orthologue in non-teleost basal ray-finned fish (Figure S2). Taken together, the teleost cmn-elastin orthologue appears to have gone through significant modification after whole genome duplication in teleosts with function associated with chordacentral formation and patterning.
The mutation underlying the spondo phenotype lies within the signal peptide domain of cmn and is predicted to impair signal peptide processing. To confirm the identified mutation was causative for the spondo phenotype, we overexpressed spondo mutant cmn in the notochord sheath using the QF2/QUAS system (Figure 3F,G) [23, 24]. Embryos containing col9a2:QF2 and entpd5a:pkred transgenes were injected at the single cell stage with a QUAS:spondo-p2A-EGFP DNA construct, driving the expression of spondo mutant cmn specifically in the notochord sheath. Injection of QUAS:GFPCaax was used as a control (Figure 3A). Overexpression of spondo in the developing zebrafish notochord led to cell-autonomous impaired entpd5a activation in the mineralizing domain of the notochord sheath, similar to that seen in spondo mutants (Figure 3F,G). By contrast, misexpression of wild-type cmn in the col9a2+ domain did not cause ectopic activation of entpd5a:pkred expression (Figure S4). These data suggest that the mutation in cmn is responsible for the spondo phenotype, but that cmn is not sufficient to induce notochord sheath maturation.
Next, we used CRISPR-Cas9 genome editing to generate mutations in cis and trans to the spondo mutation. To this end, we injected single cell stage spondo mutant embryos with a pool of CRISPR guides targeting cmn upstream and downstream of the spondo mutation. Strikingly, notochord sheath segmentation was partially restored in injected spondo mutants compared to non-injected controls (NIC), although segment boundaries remained irregular (Figure 3H–K’). This was particularly apparent in more caudal areas of the notochord. This phenotype is similar to that of wild-type fish injected with the CRISPR pool (Figure 3J,J’) as well as fish carrying a heterozygous inactivating mutation in cmn (Figure S5) and is consistent with the caudally enriched expression of cmn during the later phase of embryonic notochordal patterning (Figure S3, 32hpf). Together, these data indicate that the spondo mutation results in a dominant, gain-of-function cmn variant that disrupts notochord sheath segmentation by impairing the differentiation of col9a2+ sheath cells into entpd5a+ mineralizing cells. Moreover, our findings suggest that cmn normally plays a role in maintaining the boundaries between cartilage-like and mineralizing domains during notochord sheath segmentation.
The spondo mutant uncovers unique properties of the developing spine. The phenotype is not simply an arrest of development nor a manifestation of simple pathology. Carriers of the spondo mutation show consistent formation of meristically patterned arcualia/spines and retain the ability to form hemicentric vertebrae in the adult. These phenotypic states closely resemble early morphologies shared in stem teleosts and extant outgroup and sister species such as the coelacanth, Latimeria, and the holostean Amia, or Bowfin, respectively. For this reason, we investigated the developmental consequences of the spondo mutation on sclerotomal osteoblast migration and interaction with the notochord. Osteoblasts, marked by an osterix promoter driven transgene (osx:mtagbfp-2A-CreER) [25], normally begin to populate the developing zebrafish spine after distinct centra domains have been specified during notochord segmentation (Figure 4A; [9]). They overlay the mineralizing chordacentra (Figure 4A) and are most prominent in the rostral and caudal regions of the centra (Figure 4B), establishing growth sites for the arches or ribs in these regions (Figure 4C). In contrast, in spondo mutants, osteoblasts show delayed and altered migration to the notochord. Instead of migrating directly towards the medial regions of each mineralized notochord segment as is the case in wild-type fish, they situate at puncta on the dorsal and ventral sides where the arches originate (Figure 4E,F). These arches remain patterned presumably through the influence of somite pattering cues. This pattern of somatic mesoderm contribution is reminiscent of sclerotomal cell migration in amniotes during vertebrae formation [26]. Notably, the alteration in cmn does not inhibit entpd5a expression globally, as evidenced by entpd5a expressing osteoblasts during arcualia/spine development (Figure 4F, J). Rather, the absence of normal cmn function leads to an overall shift in patterning and specification of the mineralized components of the spine. Instead of being recruited to centra domains that have been specified by notochord segmentation, our evidence suggests that sclerotomal osteoblasts anchor to defined locations, guided by somite boundaries, and subsequently spill over these points of attachment to form hemicentra (Figure 4K). Since these hemicentra derive from the arches and are not overlaying a chordacentra rudiment, our data suggest an arcocentral origin for these hemicentra structures, most similar to state 4 depicted in Figure 2E.
Figure 4. The spod mutation results in a developmental shift in centra patterning and enhanced osteoblast contribution to arcualia/spines.
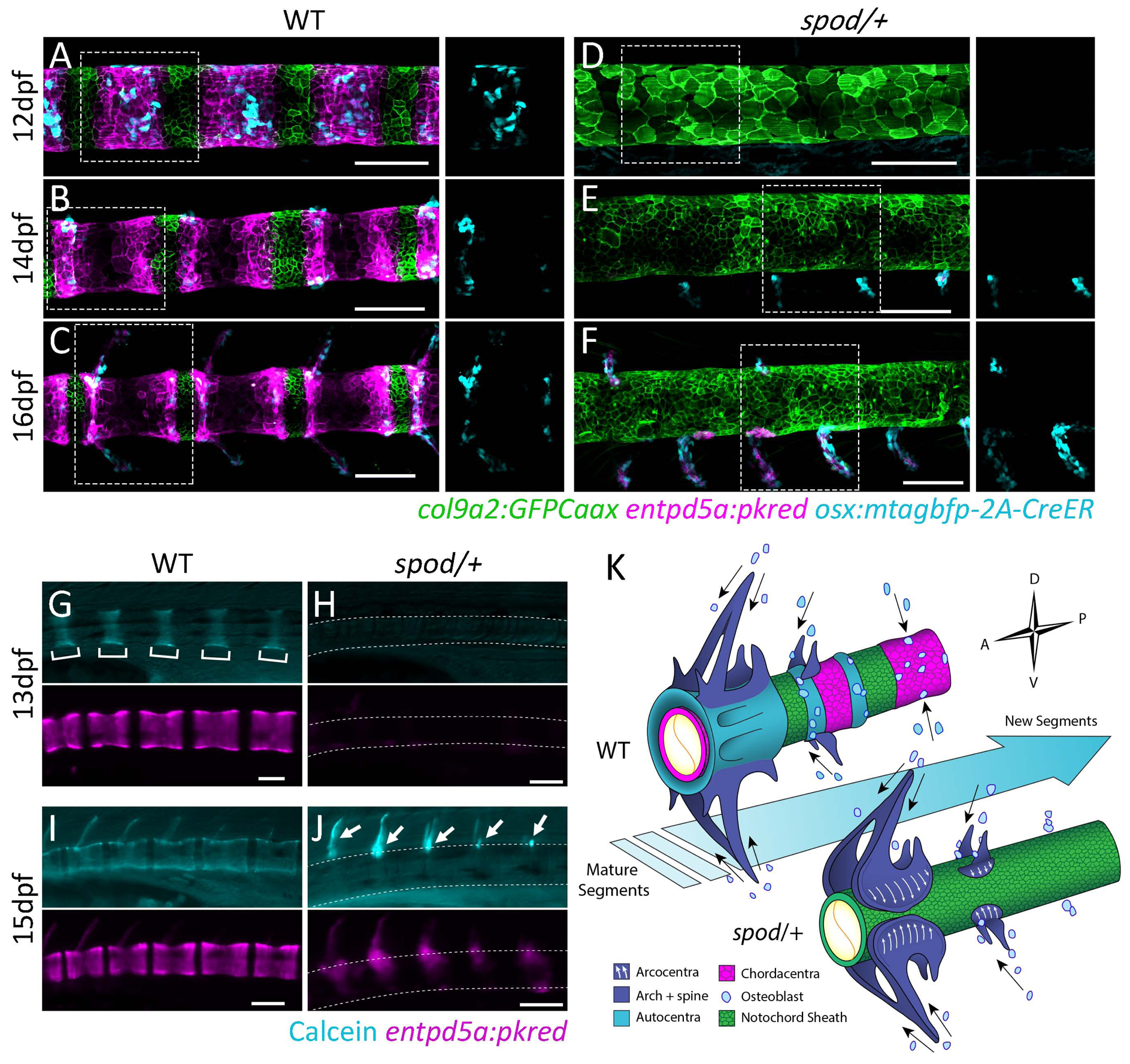
A) Following notochord segmentation in wild type zebrafish, sclerotomal osteoblasts are recruited to the mineralizing domains of the notochord sheath and overlay the chordacentra. B,C) Osteoblasts anchor dorsally and ventrally on each mineralizing domain and establish growth sites for the neural and hemal arches. D) Notochord sheath cells are irregular and do not differentiate into mineralizing cells of the chordacentra in spondo mutants. E) Osteoblasts prematurely migrate to focal points on the notochord sheath that will form the arches. F) Osteoblasts never migrate to centra domains and only contribute to neural and hemal arcualia. Insets represent regions outlined by the dotted lines in which the blue channel (osteoblasts) has been isolated. G-J) Live calcein staining of the same wild type and spondo mutant fish imaged over time. G,I) Chordacentra mineralize from the center of the entpd5a positive domain outwards in wild-type fish. I) Arches are ossified after the centra. H,J) spondo mutants fail to form mineralized chordacentra. Ossification only occurs in the arches to which osteoblasts have been recruited. K) Hypothesis of osteoblast migration patterns and their relationship to notochord segmentation. Arrows represent migratory paths. In wild-type fish, notochord segmentation guides osteoblast migratory patterns to form the final vertebral body structure. Subsequently, osteoblasts rely on cues from somite segmentation and migrate along segment boundaries to form vertebral arches. In spondo mutants, where notochord segmentation does not occur, osteoblasts fail to migrate to form centra structures. Osteoblasts solely migrate along somite segment boundaries to form vertebral arches. Hemicentra form as osteoblasts secrete osteoid matrix in regions of arch attachment and spill over to form partial centra (arcocentra extensions), closely resembling the basal condition of all Osteichthyes including early sarcopterygians lineages that lead to tetrapods.
To investigate if the altered morphogenesis observed in spondo mutants was indeed due to a developmental shift of axial patterning towards dependency on paraxial mesoderm signals, we crossed spondo mutant zebrafish to fused somite/tbx6 mutants (fss)[27, 28]. In contrast to phenotypes observed in humans [19] and mice [18] where loss of Tbx6 leads to the formation of fused ribs and vertebrae, zebrafish deficient for tbx6 do not show comparable vertebral defects [29]. In these zebrafish mutants, the vertebral arches are deformed while the centra still retain a segmented pattern and are largely unaffected, aside from minor alterations in vertebral body length [5, 7, 9] (Figure 5B, F). However, in the background of the spondo mutation, loss of tbx6 function in zebrafish leads to vertebral patterning defects comparable to those seen in mice as well as patients deficient for TBX6 [19](Figure 5D). Analysis of centra formation in these compound mutants shows that osteoblasts migrate towards the notochord along remaining partial somite boundaries (Figure 5H, arrows) but are incapable of reaching the notochord in regions where boundaries are absent (Figure 5H, bracket). This ultimately leads to a loss of the meristic patterning across the forming spine as osteoblasts extend from their initial sites of attachment to the surrounding areas without discernable patterning cues (Figure 5H). Thus, through simple abrogation of a signal within the notochord, zebrafish demonstrate similar dependency on the paraxial mesoderm for spine patterning to that of amniotes.
Figure 5. Sensitization of cmnspod mutants to tetrapod segmentation programs.
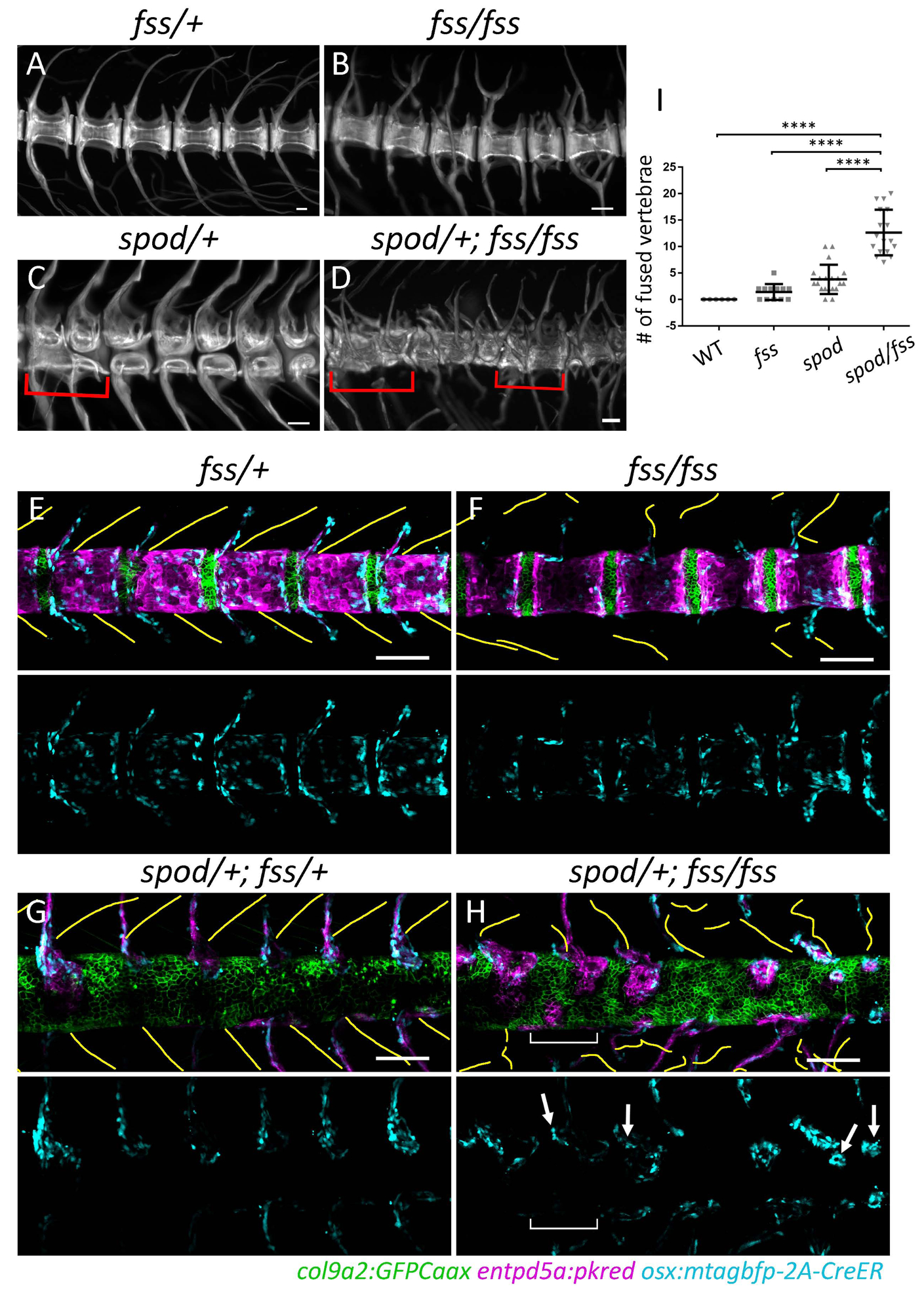
A-D) Alizarin red stained skeletal preparations at 6wpf. A) Wild-type skeletal preps demonstrate meristically patterned autocentra and arches. B) tbx6fss/fss mutants display irregular arch morphology and patterning while centra remain regularly segmented. C) cmnspod/+ mutants have the opposite phenotype wherein arch patterning is normal and the centra contain large vertebral clefts. D) Double mutants in which both notochord and somite segmentation have been disrupted are not simply a combination of the two aforementioned phenotypes. In contrast, double mutants form highly irregular vertebral structures with very little discernable patterning. Red brackets indicate vertebral fusion events. E) The number of fused vertebrae in each condition was quantified along the body axis using alizarin red stained skeletal preparations. The number of fused vertebrae in double mutants was significantly greater than in all other conditions. Wild-type n = 6; tbx6fss/fss n = 12; cmnspod/+ n = 20; cmnspod/+;tbx6fss/fss n = 17 fish. Error bars indicate standard deviation. E-H) Alteration In notochordal patterning demonstrating alteration in early sensitivity of spondo mutants to tbx6 mutations; yellow lines indicate traced somite segment boundaries; blue channel (osteoblasts) has been isolated below each composite image. E,G) In wild-type fish, regions of somite boundaries correlate with articulated pattern of vertebral arches. F) The tbx6fss/fss mutant has impaired somite segmentation where partial segment boundaries form without any metameric pattern. Lack of paraxial mesoderm patterning alters the morphology and arrangement of developing vertebral arches while notochord segmentation is largely unaffected. There is some variability in size of the centra domains (not quantified). H) In spondo;tbx6 double mutants, both notochord and somite segmentation are impaired. Osteoblasts are found nearby partial boundaries. Bracket indicates region devoid of segment boundaries as well as osteoblasts. Arrows mark osteoblasts in close proximity to partial somite boundaries that have begun to spill over to form hemicentra structures.
Discussion
In this study we have shown that novel molecular processes within the notochordal sheath acquired in stem Teleostean lineages led to suppression of underlying core somatic contributions towards spinal patterning. We have demonstrated that cmn, which encodes a highly divergent elastin-like protein, is required for proper notochord segmentation in zebrafish. When this gene product is altered, as in the spondo mutants, notochord patterning signals are lost and sclerotomal osteoblasts do not form autocentra in the adults. Instead, osteoblasts migrate medially along the arches to form structures similar to arcocentra seen in basal vertebrate lineages. Importantly, we find that the spondo mutant zebrafish also revert to a dependence on cues generated during somitogenesis for vertebral patterning, a mechanism that is conserved across vertebrates. Thus, our work demonstrates maintenance of ancestral programs of spine development even in highly derived lineages. This is evident in both the retention of basal conditions, such as the persistent, continuous notochord and simple arcualia seen in sturgeon and even in sarcopterygian lineages such as lungfish, but also through the formation of hemicentra and diplospondyly present in Prohalecites and Triassic pholidophorids [13, 30]. As documented here, these traits resemble axial structures in stem teleosts, lost in all extant teleost lineages. These results are consistent with the idea of broad-scale homology of vertebral patterning [3, 31]. Interestingly, our data points to signaling from the notochord as essential for specifying the evolution of phylogenetically defining traits. While it is still unclear when notochord sheath cells and the process of notochord segmentation emerged during evolutionary history, our data suggest that fixation of this event in teleosts correlates with the appearance of the highly differentiated gene, cmn. As many fish develop externally during embryonic stages and the larva must swim to acquire food and avoid predation, the precocious patterning and robustness imparted by the notochord sheath may provide significant fitness advantages [32].
Our findings provide a working hypothesis that simple changes in the content or structure of the notochordal sheath ECM lead to the formation of broad taxon-specific morphologies. Through uncovering shared ancestral patterning states between teleost fishes and early ancestors of tetrapods, we reconciled mechanisms of spine development occurring over the course of evolution and demonstrated the utility of teleost fish to model human spinal disorders in a manner that was not previously achievable.
STAR METHODS
• LEAD CONTACT AND MATERIALS AVAILABILITY
Lead Contact:
Further information and requests for zebrafish lines and molecular reagents described in this paper should be directed to, and will be fulfilled by the Lead Contact, Matthew Harris (harris@genetic.med.harvard.edu) or Michel Bagnat (michel.bagnat@duke.edu).
Materials Availability
All zebrafish resources generated in this study are available without restriction.
Data and Code Availability Statement
This study did not generate/analyze and datasets no generate code for use in analysis.
• EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fossil specimens and description of ancestral vertebral structure.
The studied fossil specimens were mechanically prepared with fine needles to remove remains of sediment on their surface; except for Leptolepis that was acid-prepared. The specimens of Orthocormus, Pholidoctenus, and Anaethalion were photographed under “normal” light, whereas Leptolepis was photographed under UV light using the technique described in Tischlinger and Arratia [33].
Catalogue numbers: Orthocormus roeperi: BSPG 1993 XVIII-VFKO 816; Prohalecites porroi: MCSNIO P348; Pholidoctenus: MCSNB 3377; Leptolepis: BGHan 1957–5; Ebertichthys ettlingensis: JME-ETT 00063; Anaethalion sp.: BSPG uncatalogued. Museum acronyms: BGHan: Bundesanstalt für Geowissenschafen und Rohstoffe, Niedersächsisches Landesamt für Bodenforschung, Hannover, Germany; BSPG: Bayerische Staatssammlung für Paläontologie und historische Geologie, München, Germany; JME: Jura Museum, Eichstätt, Germany; CMSNIO: Civico Museum Insubrico di Storia Naturale di Induno Olona, Italy.
Zebrafish husbandry and alleles
Zebrafish used in this study were raised and maintained under standard conditions [34] in compliance with internal regulatory review at Boston Children’s Hospital and Duke University School of Medicine. Mutants and transgenic lines used in this study: cmnspondo(dmh16) [20], cmnpd1211, tbx6ti1/fss [29], Tg(col9a2:GFPCaaX)pd1151 [9, 21], Tg(osx:mTagBFP-2A-CreER)pd45 [25], TgBAC(entpd5a:pkRED)hu7478 [7]
• METHOD DETAILS
In situ Hybridization
For the production of in situ hybridization probes, cmn was amplified from cDNA produced from 7dpf zebrafish larvae. The following primers were used for in vitro transcription in conjunction with T7 RNA polymerase (NEB) – cmn_ISHprobe_F: 5’ GATGTTCCAGGAGCAGGAGA 3’; cmn_ISHprobe_R: 5’ GAAATTAATACGACTCACTATAGGGCCTTGCCCCAGGATACTAAAG 3’. The T7 promoter was included in the reverse primer sequence. DIG RNA Labeling Mix (Roche) was used to label probe RNA with digoxygenin. Whole mount in situ hybridization was performed on 15ss-48hpf embryos as described previously (Navis et al., 2013). For later stage larvae (12dpf and 16dpf), larvae were fixed in 4% paraformaldehyde and then cryo-preserved in 30% sucrose. 12μm cryo-sections were generated and then dehydrated in methanol. in situ hybridization was performed using the InSituPro robot (Intavis) as described previously [35]. Whole mount and fluorescence imaging were performed using the AX10 Zoom V116 Zeiss microscope and tissue sections were imaged using a Leica DM6000 compound microscope.
Genome editing
For the pooled guide experiment, 4 sgRNAs were pooled together targeting the 5’ UTR and exons 1 and 2 of cmn. A working solution containing 10ng/μL of each sgRNA and 150ng/μL Cas9 mRNA was injected at the single cell stage into an outcross of spod heterozygous fish and wild type EK. The following target sites were used to generate each sgRNAs: 5’ gttatttagcacatgtggtaagg 3’; 5’ TGTTGAGATGGACAATTCTAAGG 3; 5’ GATGACAAGAGTAACACTTCAGG 3’; 5’ GCAGCACCGATTTCACCAGGAGG 3’.
A heteroduplex mobility shift assay was performed to test guide activity using the following primers: F: 5’ atggcccttaagaggaggtg 3’; R: 5’ GACCAGAAACCCTAACCTCAA 3’ (Figure S5).
The loss of function allele of cmn (pd1211) was generated using CRISPR/Cas9. The following target site in exon 1 was used for the generation of sgRNA: 5’ TGTTGAGATGGACAATTCTAAGG 3’. Zebrafish embryos were injected at the single cell stage with 150ng/μL Cas9 mRNA and 40ng/μL sgRNA.
Genotyping
For genotyping of spondo mutants, the following primers were used to introduce the BseRI restriction site into the mutant PCR product: spod_dCAPS_F: 5’ TGAGATGGACAATTCTAAGGAGGA 3’; spod_dCAPS_R: 5’ CAGAAACCCTAACCTCAAAAGCT 3’. The PCR product was then digested with BseRI (NEB) for 2 hours at 37˚C and run on a 3% TBE gel.
For genotyping of pd1211 mutants, the following primers were used: cmn_pd1211_F: 5’ GAGCACAGTATCTGTTCCCTGT 3’; cmn_pd1211_R: 5’ CCTGTGCCACCCTGAAGTGTT 3’. The PCR product was then digested with DdeI (NEB) for 2 hours at 37˚C and run on a 3% TBE gel.
spod overexpression
The construct used for the cmnspod overexpression experiment was generated using the Tol2kit gateway cloning system and the following vectors: p5E-QUAS [23], pME-MCS, and p3E-p2a-EGFPpA. Full length cmn was amplified from cDNA using the following primers: full_length_cmn_F: 5’ ATGTTGAGATGGACAATTCTAAGG 3’; full_length_cmn_R: 5’ CCTGTCTGGGAGTTGCTGT 3’. The Q5 Site-Directed Mutagenesis Kit (NEB) was used to generate the spod mutation (29T>G) and add the 54 remaining nucleotides to the 3’ end of the cmn sequence up to, but not including the stop codon. To drive expression of the QUAS:spod-p2a-EGFP construct, Tg(col9a2:QF2); TgBAC(entpd5a:pkRED)hu7478 [9] embryos were injected at the single cell stage. Fish expressing the QUAS:spod-p2a-EGFP construct were imaged on a Fluoview FV3000 (Olympus) confocal microscope. Using ImageJ (NIH), sheath cell differentiation was assessed by quantifying gaps in the mineralizing domain. To do this, entpd5a expression was used to delineate the mineralizing domains within the notochord sheath. The total number of GFP positive cells within each mineralizing region was then counted and the percentage of these cells that were also entpd5a negative was calculated. This was compared to control embryos injected with QUAS:GFPCaax [9].
Calcein staining and skeletal preparations
Zebrafish larvae were stained with calcein (Sigma-Aldrich) in egg water for 30 minutes at two developmental time points, 13 and 15dpf. Fish were then anesthetized in 1x tricaine and mounted in 3% methylcellulose for imaging. Imaging was performed using an AX10 Zoom V116 Zeiss microscope. For skeletal preparations, 6-week-old zebrafish were skinned and eviscerated in 80% ethanol and then fixed in 4% PFA. Fish were then stained with alizarin red as previously described [36]. After staining, fish were placed in 1% KOH to clear the remaining tissue. Skeletal preparations were imaged, and vertebral fusions were quantified using the AX10 Zoom V116 Zeiss microscope.
Microscopy
Live confocal microscopy was performed using a Fluoview FV3000 (Olympus) confocal microscope, a 30x/1.05 silicone objective, and Fluoview software (Olympus). Fish were anesthetized in 1X tricaine and then mounted onto glass bottom dishes in 1.3% low-melt agarose dissolved in egg water. Digital stitching of tile scans was performed using Fluoview software (Olympus). Images were false colored and minimally processed for brightness and contrast using ImageJ software (NIH). Additional imaging was performed using the AX10 Zoom V116 Zeiss microscope. Images taken on Zeiss microscope were minimally processed for brightness and contrast using Zen 2.3 lite software (Zeiss) when necessary.
Electron microscopy
7dpf zebrafish larvae were fixed in 0.1M sodium cacodylate, and 2.5% glutaraldehyde (GA). Specimen preparation and staining was performed as previously described [21]. Following fixation, samples were stained with osmium tetroxide and uranyl acetate and were then dehydrated in ethanol solutions of increasing concentration. Samples were then embedded in resin blocks overnight and then cured at 60 degrees for 48 hours. Thin sections were cut and post-stained with lead citrate and uranyl acetate and placed on copper grids for imaging. Thickness quantifications were carried out using ImageJ software (NIH).
Skeletal staining for microcomputed tomography
Fish were euthanized at the indicated stages with 22% MS-222 and fixed in 3.7% formaldehyde over night at room temperature. After fixation fish were washed in PBS. For staining of mineralized matrix, fish were transferred into alizarin red staining solution (100 mg/ml alizarin red (Sigma) in 0.5% KOH) and stained over night at room temperature. Following staining, fish were washed several times in 0.5% KOH before being transferred through a series of glycerol into 80% glycerol/0.5% KOH for clearing.
Prior to mCT imaging, fish were embedded in 1% agarose in 15ml culture tubes to reduce movement during imaging. Fish were imaged using a Skyscan 1173 (Bruker), 240-dregree scan with 0.2 rotational step and 1500 msec exposure time. The X-ray source voltage was set to 70 kV and the current to 80 mA. The scan resolution is 7.14 mm per pixel. Amira software package, version 6.0 was used for image processing.
Genomic Identification and Phylogeny of calymmin orthologues
Calymmin is not well annotated in vertebrate genomes. Additionally, it is both repetitive and evolving rapidly, such that finding orthologs via sequence identity alone was problematic. We used syntenic landmarks to locate cmn orthologs across teleosts and tetrapods. The locus containing parqr4b, pelo, claudins, perforins, and cox6a genes consistently maintained synteny. In thisregion, calymmin was variously annotated as elastin, spidroin, shematrin, glycine-rich cell wall component and others. Occasionally, cmn putative orthologues are split into two consecutive annotations, including pufferfish, python, and killifish, so these coding regions were spliced together in silico for the alignment. All protein sequences were aligned together usingPrank v.170427(http://wasabiapp.org/software/prank/) with default settings. FastTree was used for tree construction with 20 rate categories and pseudocounts enabled.
• QUANTIFICATION AND STATISTICAL ANALYSIS
For the overexpression experiments, p values were calculated through a t-test assuming un-equal variances. For quantification of vertebral fusions, a one-way ANOVA was performed using GraphPad Prism version 7.0c for Mac, GraphPad Software, La Jolla California USA, www.graphpad.com. Additional quantification data can be found within the figure legends.
Supplementary Material
Highlights.
Demonstrate that the notochordal sheath ECM defines phyletic character of the spine.
The emergence of cmn shapes spinal features defining for all teleost fishes.
Disruption of cmn reverts spine development to a mechanism shared by amniotes.
Underlying homologies of spine development across vertebrates are revealed
Acknowledgements:
We would like to thank Jennifer Bagwell for her help with confocal microscopy, Daniel Levic for his help with cryosectioning and molecular biology, Ricardo Vancini for his help with electron microscopy, Kelsey Oonk for her assistance with in situ hybridizations on cryosections, the Duke and Boston Children’s Hospital Aquatics Core for fish care, and members of the Bagnat lab for discussions. This work was supported by Orthopedic Research Foundation at Boston Children’s Hospital (M.P.H.), scholarship grant from ANID, Phd Fellowiship Program, CHILE/2015 - 21150789 (N.C.), NIH grant RO1 AR065439 (M.B.) and in part by a Faculty Scholar grant, HHMI 55108501, from the Howard Hughes Medical Institute (M.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors have nothing to declare
REFERENCES
- 1.Jones KE, Angielczyk KD, Polly PD, Head JJ, Fernandez V, Lungmus JK, Tulga S, and Pierce SE (2018). Fossils reveal the complex evolutionary history of the mammalian regionalized spine. Science 361, 1249–1252. [DOI] [PubMed] [Google Scholar]
- 2.Arratia G, Schultze HP, and Casciotta J (2001). Vertebral column and associated elements in dipnoans and comparison with other fishes: development and homology. J Morphol 250, 101–172. [DOI] [PubMed] [Google Scholar]
- 3.Fleming A, Kishida MG, Kimmel CB, and Keynes RJ (2015). Building the backbone: the development and evolution of vertebral patterning. Development 142, 1733–1744. [DOI] [PubMed] [Google Scholar]
- 4.Arratia G (1991). The caudal skeleton of Jurassic teleosts: A phylogenetic analysis. In Early Vertebrates and Related Problems of Evolutionary Biology, Chang M-M, Hai L and Zhang G-R, eds. (Beijing, China: Science Press; ), pp. 249–340. [Google Scholar]
- 5.Fleming A, Keynes R, and Tannahill D (2004). A central role for the notochord in vertebral patterning. Development 131, 873–880. [DOI] [PubMed] [Google Scholar]
- 6.Grotmol S, Kryvi H, Nordvik K, and Totland GK (2003). Notochord segmentation may lay down the pathway for the development of the vertebral bodies in the Atlantic salmon. Anat Embryol (Berl) 207, 263–272. [DOI] [PubMed] [Google Scholar]
- 7.Forero L, Narayanan R, Huitema LFA, VanBergen M, Apschner A, Peterson-Maduro J, Logister I, Valentin G, Morelli LG, Oates A, et al. (2018). Segmentation of the zebrafish axial skeleton relies on notochord sheath cells and not on the segmentation clock. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pogoda HM, Riedl-Quinkertz I, Lohr H, Waxman JS, Dale RM, Topczewski J, Schulte-Merker S, and Hammerschmidt M (2018). Direct activation of chordoblasts by retinoic acid is required for segmented centra mineralization during zebrafish spine development. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wopat S, Bagwell J, Sumigray KD, Dickson AL, Huitema LFA, Poss KD, Schulte-Merker S, and Bagnat M (2018). Spine Patterning Is Guided by Segmentation of the Notochord Sheath. Cell Rep 22, 2026–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inohaya K, Takano Y, and Kudo A (2007). The teleost intervertebral region acts as a growth center of the centrum: in vivo visualization of osteoblasts and their progenitors in transgenic fish. Dev Dyn 236, 3031–3046. [DOI] [PubMed] [Google Scholar]
- 11.Arratia G (1999). The monophyly of Teleostei and stem group teleosts. Consensus and disagreements. In Mesozoic Fishes 2, Arratia G and Chultze H-P, eds. (Muchen, Germany: Verlag; ), pp. 265–334. [Google Scholar]
- 12.Arratia G (2017). New Triassic teleosts (Actinopterygii, Teleosteomorpha) from northern Italy and their phylogenetic relationships among the most basal teleosts. Journal of Vertebrate Paleontology 37, e1312690. [Google Scholar]
- 13.Arratia G (2013). Morphology, taxonomy, and phylogeny of Triassic pholidophorid fishes (Actinopterygii, Teleostei). Journal of Vertebrate Paleontology 33, 1–138. [Google Scholar]
- 14.Arratia G (2015). Complexities of early Teleostei and the evolution of particular morphological structures through time. Copeia 103, 999–1025. [Google Scholar]
- 15.Arratia G, Schultze H-P, and Tischlinger H (2019). On a remarkable new species of Tharsis, a Late Jurassic teleostean fish from southern Germany: its morphology and phylogenetic relationships. Fossil Record 22, 1–23. [Google Scholar]
- 16.Ward L, Pang ASW, Evans SE, and Stern CD (2018). The role of the notochord in amniote vertebral column segmentation. Dev Biol 439, 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi K-S, and Harfe B (2011). Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. PNAS 108, 9484–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White PH, Farkas DR, McFadden EE, and Chapman DL (2003). Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development 130, 1681–1690. [DOI] [PubMed] [Google Scholar]
- 19.Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, Shen Y, Yu G, Liu J, Xie H, et al. (2015). TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med 372, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henke K, Daane JM, Hawkins MB, Dooley CM, Busch-Nentwich EM, Stemple DL, and Harris MP (2017). Genetic Screen for Postembryonic Development in the Zebrafish (Danio rerio): Dominant Mutations Affecting Adult Form. Genetics 207, 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia J, Bagwell J, Njaine B, Norman J, Levic DS, Wopat S, Miller SE, Liu X, Locasale JW, Stainier DYR, et al. (2017). Sheath Cell Invasion and Trans-differentiation Repair Mechanical Damage Caused by Loss of Caveolae in the Zebrafish Notochord. Curr Biol 27, 1982–1989 e1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerda J, Grund C, Franke WW, and Brand M (2002). Molecular characterization of Calymmin, a novel notochord sheath-associated extracellular matrix protein in the zebrafish embryo. Dev Dyn 224, 200–209. [DOI] [PubMed] [Google Scholar]
- 23.Subedi A, Macurak M, Gee ST, Monge E, Goll MG, Potter CJ, Parsons MJ, and Halpern ME (2014). Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods 66, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potter CJ, Tasic B, Russler EV, Liang L, and Luo L (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh SP, Holdway JE, and Poss KD (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wake D (1979). The endoskeleton: The comparative anatomy of the vertebral column and ribs. In Hyman’s Comparative Vertebrate Anatomy, Wake D, ed. (Chicago: University of Chicago; ), pp. 192–237. [Google Scholar]
- 27.Nikaido M, Kawakami A, Sawada A, Furutani-Seiki M, Takeda H, and Araki K (2002). Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat Genet 31, 195–199. [DOI] [PubMed] [Google Scholar]
- 28.Windner SE, Bird NC, Patterson SE, Doris RA, and Devoto SH (2012). Fss/Tbx6 is required for central dermomyotome cell fate in zebrafish. Biol Open 1, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, et al. (1996). Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development 123, 153–164. [DOI] [PubMed] [Google Scholar]
- 30.Tintori A (1990). The actinopterygian fish Prohalecites from the Triassic of northern Italy. Palaeontology 33, 155–174. [Google Scholar]
- 31.West-Eberhard MJ (2003). Developmental Plasticity and Evolution, (Oxford: Oxford University Press; ). [Google Scholar]
- 32.Harris MP, and Arratia G (2018). Patterning the spine. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tischlinger H, and Arratia G (2013). Ultraviolet light as a tool of investigating Mesozoic fishes with a focus on the ichthyofauna of the Solnhofen Limestone In Mesozoic Fishes 5 – Global Diversity and Evolution, Arratia G, Schultze HP and Wilson WHV, eds. (München: Verlag; ), pp. 549–560. [Google Scholar]
- 34.Nusslein-Volhard C, and Dahm R (2002). Zebrafish: a practical approach, (Oxford: Oxford University Press; ). [Google Scholar]
- 35.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, and Keating MT (2002). Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development 129, 5141–5149. [DOI] [PubMed] [Google Scholar]
- 36.Ellis K, Hoffman BD, and Bagnat M (2013). The vacuole within: how cellular organization dictates notochord function. Bioarchitecture 3, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze and datasets no generate code for use in analysis.


