Abstract
Six strictly anaerobic Gram-negative bacteria representing three novel species were isolated from the female reproductive tract. The proposed type strains for each species were designated UPII 199-6T, KA00182T and BV3C16-1T. Phylogenetic analyses based on 16S rRNA gene sequencing indicated that the bacterial isolates were members of the genus Megasphaera. UPII 199-6T and KA00182T had 16S rRNA gene sequence identities of 99.9 % with 16S rRNA clone sequences previously amplified from the human vagina designated as Megasphaera type 1 and Megasphaera type 2, members of the human vaginal microbiota associated with bacterial vaginosis, preterm birth and HIV acquisition. UPII 199-6T exhibited sequence identities ranging from 92.9 to 93.6 % with validly named Megasphaera isolates and KA00182T had 16S rRNA gene sequence identities ranging from 92.6–94.2 %. BV3C16-1T was most closely related to Megasphaera cerevisiae with a 16S rRNA gene sequence identity of 95.4 %. Cells were coccoid or diplococcoid, non-motile and did not form spores. Genital tract isolates metabolized organic acids but were asaccharolytic. The isolates also metabolized amino acids. The DNA G+C content for the genome sequences of UPII 199-6T, KA00182T and BV3C16-1T were 46.4, 38.9 and 49.8 mol%, respectively. Digital DNA–DNA hybridization and average nucleotide identity between the genital tract isolates and other validly named Megasphaera species suggest that each isolate type represents a new species. The major fatty acid methyl esters include the following: C12 : 0, C16 : 0, C16 : 0 dimethyl acetal (DMA) and summed feature 5 (C15 : 0 DMA and/or C14 : 0 3-OH) in UPII 199-6T; C16 : 0 and C16 : 1 cis 9 in KA00182T; C12 : 0; C14 : 0 3-OH; and summed feature 5 in BV3C16-1T. The isolates produced butyrate, isobutyrate, and isovalerate but there were specific differences including production of formate and propionate. Together, these data indicate that UPII 199-6T, KA00182T and BV3C16-1T represent novel species within the genus Megasphaera. We propose the following names: Megasphaera lornae sp. nov. for UPII 199-6T representing the type strain of this species (=DSM 111201T=ATCC TSD-205T), Megasphaera hutchinsoni sp. nov. for KA00182T representing the type strain of this species (=DSM 111202T=ATCC TSD-206T) and Megasphaera vaginalis sp. nov. for BV3C16-1T representing the type strain of this species (=DSM 111203T=ATCC TSD-207T).
Keywords: bacterial vaginosis, genital tract bacteria, human vagina, Megasphaera, Negativicutes, Veillonellaceae
The genus Megasphaera was created by Rogosa when describing the type species Megasphaera elsdenii, originally classified as Peptostreptococcus elsdenii [1] but revised when the bacterium did not exhibit a key characteristic of the genus Peptostreptococcus. M. elsdenii is Gram-negative by direct staining and by electron microscopy while Peptostreptococcus species are Gram-positive [1]. Eight additional Megasphaera species belonging to the phylum Firmicutes, class Negativicutes, order Veillonellales and family Veillonellaceae have subsequently been validly named [2–7]. Megasphaera species are Gram-negative, with a peculiar diderm cell wall structure typical for the Negativicutes [8], obligate anaerobes that are often detected in intestinal environments [9]. Megasphaera indica and Megasphaera massiliensis were isolated from human faeces [5, 10], and Megasphaera micronuciformis from a human liver abscess [7]. M. elsdenii and Megasphaera hexanoica were cultivated from sheep and cow rumen respectively [1, 3], while Megasphaera stantonii was cultured from a chicken cecum [6]. Three Megasphaera species have been isolated from brewery samples or spoiled beer including Megasphaera cerevisiae, Megasphaera sueciensis and Megasphaera paucivorans [2, 4].
Cultivation-independent molecular investigations have reported that Megasphaera species in the female genital tract are associated with adverse health outcomes in women. Novel Megasphaera sequences were first noted in the human vagina by Zhou et al. in a study of five women using cloning and sequencing methods [11]. Fredricks et al. identified two distinct Megasphaera sequence types in a study evaluating association of vaginal bacteria with the common dysbiotic condition, bacterial vaginosis (BV) [12] and validated these observations using targeted PCR assays in women with and without BV [13, 14]. Subsequently, several studies have reported the association of these two Megasphaera sequence types with BV using molecular approaches [15–26]. Megasphaera type 1 was shown to be useful for the molecular diagnosis of BV [13, 14, 18, 22, 27–31] and has been included as a target in commercially available nucleic acid amplification tests for the diagnosis of BV [28]. Vaginal Megasphaera species have also been shown to be associated with increased risk for HIV acquisition [32, 33] and among a group of mostly South African women, Megasphaera type 2 was associated with increased risk but not Megasphaera type 1 [34]. Furthermore, pregnant women with a prior history of preterm delivery and increasing levels of Megasphaera type 1 through mid-pregnancy were more likely to experience spontaneous preterm delivery [35]. An association between Megasphaera species and spontaneous preterm birth was also noted in a case-control study of mostly African American women [36]. A recent study showed that women with pelvic inflammatory disease were more likely to test positive for Megasphaera species among other anaerobes [37].
Our groups have previously isolated Megasphaera bacterial species [38–40] and have demonstrated that Megasphaera type 1 and type 2 bacterial isolates are susceptible to clindamycin and the nitroimidazoles used to treat BV including metronidazole, tinidazole and secnidazole [38, 39]. Megasphaera species have also been detected in the human oral cavity [17, 19, 41, 42], rectum [17, 19], stool [43–45] and male genitourinary microbiome [46–48]. Here, we systematically characterize three novel Megasphaera species from the human genital tract including isolates previously designated as Megasphaera type 1 and type 2 and compare them to validly published Megasphaera species. We propose the names Megasphaera lornae sp. nov. for UPII 199-6T, Megasphaera hutchinsoni sp. nov. for KA00182T and Megasphaera vaginalis sp. nov. for BV3C16-1T as they represent novel species within the genus Megasphaera. For each of these proposed type strains, we have also characterized a second isolate including DNF00751 for M. lornae, UPII 135-E for M. hutchinsoni and HL562 for M. vaginalis.
The novel Megasphaera species characterized here were isolated from either an endometrial biopsy or a vaginal fluid sample obtained from women participating in two separate research studies. The research study at the University of Pittsburgh evaluated women for pelvic inflammatory disease (PID); endometrial biopsy samples were collected in a protocol approved by the University of Pittsburgh Review Board (IRB approval number: PRO 010010112) [39, 49]. The research study at the Fred Hutchinson Cancer Research Center sought to isolate novel anaerobes from the human vagina [40] and was approved by the institutional review board at the Fred Hutchinson Cancer Research Center (IRB approval number: IR 7363). All participants provided written informed consent. UPII 199-6T was isolated from an endometrial biopsy tissue sample from a woman being evaluated for PID. An endometrial sampling device (Pipelle) was inserted through the cervix into the uterine cavity and a biopsy was aspirated by suction. The biopsy was inoculated onto Brucella agar supplemented with 5 % laked sheep blood, hemin and Vitamin K (Brucella blood agar) (bioMérieux or Hardy Diagnostics) and incubated anaerobically at 37 °C for 4–7 days. An anaerobic atmosphere for all experiments was created using a trimix with 90 % N2, 5 % H2 and 5 % CO2 (Airgas). KA00182T was isolated on Brucella blood agar from a vaginal fluid sample which was diluted, plated and incubated anaerobically at 37 °C for 5–7 days. BV3C16-1T was isolated from a vaginal sample obtained from a woman with BV through enrichment in a liquid basic medium (BM) and incubated at 37 °C for 2 to 4 days, followed by isolation on trypticase yeast extract blood agar plates with anaerobic incubation at 37 °C for at least 2 weeks. BM comprised Solution A and a reducing agent solution which had the following components per litre of media: Solution A contained 1 g yeast extract, 1 g casamino acids, 1.04 g KH2PO4 and 1.11 g K2HPO4; reducing agent solution contained 0.4 g NH4Cl, 0.1 g MgCl2·6H2O, 0.05 g CaCl2·2H2O, 1 ml trace elements SL10, 0.0025 g resazurin, 0.05 g FeCl2·4H2O and 0.5 g l-cysteine HCl. The two solutions were prepared separately to avoid precipitation and chemical interactions during autoclaving. After preliminary identification using 16S rRNA gene sequencing, isolates were frozen at −80 °C in litmus milk (Becton Dickinson) or glycerol stocks (10 % v/v). Prior to use of the bacterial isolates in experiments, scrapings from frozen stocks were plated on Brucella blood agar and incubated anaerobically for 48–72 h at 37 °C. Individual colonies were sub-cultured at least twice and examined by Gram stain or 16S rRNA gene sequencing to ensure purity.
Validly named Megasphaera species were used as reference strains for comparisons with the genital tract strains. M. cerevisiae DSM 20462T [2], M. elsdenii DSM 20460T [1], M. indica DSM 25563T [5], M. massiliensis DSM 26228T [10], M. micronuciformis DSM 17226T [7], M. paucivorans DSM 16981T and M. sueciensis DSM 17042T [4] were obtained from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (DSMZ). Strains were maintained anaerobically on Brucella blood agar or peptone–yeast–glucose modified (PYG-mod) medium [50] supplemented with 1 % yeast extract and 1 % glucose (PYG-mod-YG) at 37 °C. Bacterial characteristics for M. stantonii AJH 120T [6] and M. hexanoica MHT [3] were obtained from the original manuscripts.
Optimal temperature was tested by growing the isolates on Brucella blood agar and incubating them at different temperatures. Optimal pH was tested by growing the isolates in PYG-mod-YG broth adjusted to varying pH with 2 M HCl or 1 N NaOH solutions. OD600 measurements were conducted in an Epoch 2 microplate reader (BioTek). UPII 199-6T, KA00182T and BV3C16-1T were strict anaerobes. UPII 199-6T exhibited growth between 35–37 °C (optimal, 37 °C) and pH 5.0–6.5 (optimal, 6.0–6.5). KA00182T grew between 30–42 °C (optimal, 35–37 °C) and pH 5.5–7.5 (optimal, 5.5). BV3C16-1T had a growth range between 30–37 °C (optimal, 35–37 °C) and pH 5.0–7.5 (optimal, 7.0). Motility and spore formation were assessed in duplicate using previously described methods [51, 52]. For the motility assay, Paeniclostridium sordellii DSM 2141 was used as a positive control and Gardnerella vaginalis ATCC 14018 was used as a negative control. P. sordellii and Escherichia coli DNF00564 were used as positive and negative controls, respectively, for the spore formation assay. All three Megasphaera isolate types from the genital tract were non-motile and non-spore-forming.
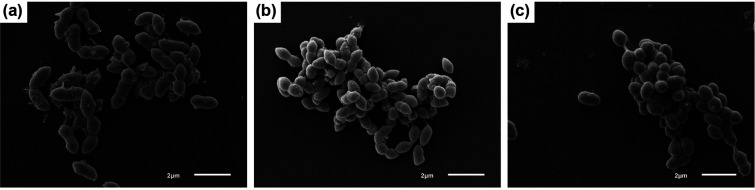
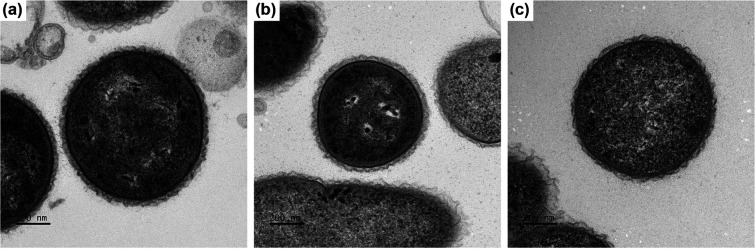
UPII 199-6T, KA00182T and BV3C16-1T were grown anaerobically at 37 °C for 2 days on Brucella blood agar to assess colony and cellular morphologies. Colonies of strain UPII 199-6T were convex, entire, glossy and off-white in colour with a diameter of 0.5–0.7 mm. Colonies of strain KA00182T were convex, entire, glossy and off-white in colour with a diameter of 0.4–0.7 mm. Colonies of strain BV3C16-1T were small, convex, and translucent with a diameter of 0.3–0.4 mm. All isolates stained as Gram-negative cocci or short coccobacilli. Scanning electron microscopy (SEM; Fig. 1) and transmission electron microscopy (TEM; Fig. 2) were performed to further evaluate cellular morphology. Bacterial cultures grown in PYG-mod-YG medium were centrifuged to form a pellet and fixed in 0.5 strength Karnovsky’s fixative (2.5 % glutaraldehyde, 2 % paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.3) overnight at 4 °C for both SEM and TEM. For SEM, duplicates of each sample (50 µl) were applied in a pool on poly-l-lysine coated coverslips for 30 min, rinsed with 0.1 M sodium cacodylate buffer and treated with 1 % osmium tetroxide for 1 h. The coverslips were rinsed with cacodylate buffer, dehydrated through a graded series of alcohols, infiltrated with hexamethyldisilazane and allowed to air dry. Coverslips were mounted on stubs and sputter coated with gold/palladium. Samples were imaged on a jeol 6610 LV SEM at 5kV. For TEM, after fixation, cells were pelleted, rinsed with 0.1 M cacodylate buffer, treated with 1 % osmium tetroxide for 1 h, rinsed with cacodylate buffer and dehydrated through a graded series of alcohols and propylene oxide. Pellets were embedded in Eponate12 resin (Ted Pella). Sections (70 nm) were cut using a Leica EM UC7 ultramicrotome, contrasted with uranyl acetate and lead citrate, and imaged on a jeol JEM 1400 transmission microscope at 120kV. Digital images were acquired with a Gatan Ultrascan 1000XP digital camera system. UPII 199–6T cells had rounded ends, were often arranged in pairs and 0.9–1.5 µm long and 0.9–1.1 µm wide (Table 1, Figs 1 and 2). KA00182T cells were oval with tapered ends and were arranged in singlets, pairs and chains along their tapered ends. KA00182T cells were 0.5–0.7 µm long and 0.5–0.7 µm wide (Table 1, Figs 1 and 2). BV3C16-1T cells were larger with rounded ends, often occurring in pairs with cells being 1.0–1.4 µm long and 0.7–1.1 µm wide (Table 1, Figs 1 and 2).
Fig. 1.
Scanning electron micrograph of cells of (a) UPII 199-6T, (b) KA00182T, and (c) BV3C16-1T. Cells were cultured for 2 days in PYG-mod-YG. Bar, 2 µm.
Fig. 2.
Transmission electron microscope image of cells of (a) UPII 199-6T, (b) KA00182T, and (c) BV3C16-1T. Cells were cultured for 2 days in PYG-mod-YG. Bar, 200 nm.
Table 1.
Comparison of the characteristics of genital tract Megasphaera species with validly named species of the genus Megasphaera
Strains: 1, M. lornae UPII 199-6T; 2, M. lornae DNF00751; 3, M. hutchinsoni KA00182T; 4, M. hutchinsoni UPII 135-E; 5, M. vaginalis BV3C16-1T; 6, M. vaginalis HL562; 7, M. cerevisiae DSM 20462; 8, M. elsdenii DSM 20460; 9, M. hexanoica MH; 10, M. indica DSM 25563; 11, M. massiliensis DSM 26228; 12, M. micronuciformis DSM 17226; 13, M. paucivorans DSM 16981; 14, M. stantonii AJH120; 15, M. sueciensis DSM 17042. Substrate utilization was evaluated using the Biolog anaerobe identification panel. +, Positive; −, negative; w, weak; ng, no growth; nd, not determined; R, resistant; S, susceptible. For tinidazole and secnidazole, the lowest concentrations (μg ml−1) at which we noted a clearance zone are shown. Production of SCFAs was measured after growth in PYG-mod-YG at 37 °C for 24–72 h. ;
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9* |
10 |
11 |
12 |
13 |
14* |
15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Cell size (µm)† |
0.9–1.5 |
− |
0.5–0.7 |
− |
1.0–1.4 |
− |
1.4 |
1.2–1.9 |
0.8–1.2 |
1.2–2.3 |
0.87 |
0.4–0.6 |
1.2–1.9 |
1.1–1.8 |
1.0–1.4 |
|
Growth at 45 °C† |
− |
nd |
− |
nd |
− |
nd |
− |
− |
− |
− |
+ |
nd |
− |
+ |
− |
|
Utilization of: |
|
||||||||||||||
|
d-Glucose‡ |
− |
− |
− |
− |
− |
− |
+/− |
−/+ |
− |
+ |
+ |
− |
− |
− |
− |
|
d-Fructose‡ |
ng |
nd |
− |
ng |
− |
− |
+ |
+ |
+ |
+ |
+ |
− |
− |
+ |
− |
|
Lactose‡ |
− |
nd |
− |
ng |
− |
− |
− |
− |
nd |
− |
− |
− |
− |
nd |
− |
|
Sucrose |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
− |
+ |
− |
− |
nd |
− |
|
Dextrin |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
+ |
+ |
− |
− |
nd |
− |
|
d-Mannitol |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
− |
|
d,l-Lactate |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
w |
+ |
+ |
+ |
+ |
− |
+ |
|
Pyruvate |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Succinate |
w |
w |
+ |
+ |
w |
+ |
− |
− |
nd |
− |
− |
− |
− |
nd |
− |
|
α-Ketovalerate |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
nd |
+ |
+ |
+ |
+ |
nd |
+ |
|
l-Alanine |
− |
− |
− |
− |
+ |
+ |
− |
+ |
nd |
+ |
+ |
+ |
+ |
nd |
− |
|
l-Alanyl-l-Glutamine |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
nd |
+ |
+ |
+ |
+ |
nd |
− |
|
l-Glutamine |
− |
− |
− |
− |
+ |
+ |
+ |
− |
nd |
− |
+ |
+ |
+ |
nd |
− |
|
l-Glutamate |
− |
+ |
+ |
+ |
+ |
+ |
− |
+ |
nd |
− |
+ |
− |
+ |
nd |
− |
|
l-Serine |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
nd |
+ |
+ |
+ |
+ |
nd |
+ |
|
Production of: |
|
||||||||||||||
|
Gas |
nd |
− |
− |
nd |
− |
− |
+ |
+ |
+ |
+ |
+ |
− |
− |
nd |
− |
|
H2S |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
+ |
− |
− |
nd |
− |
|
Indole |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
− |
|
Presence of: |
|
||||||||||||||
|
Oxidase |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
− |
|
Catalase |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
− |
|
Nitrate reduction |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
− |
nd |
− |
|
Susceptibility to: |
|
||||||||||||||
|
Bile (1 mg) |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
nd |
S |
|
Colistin (10 µg) |
R |
R |
S |
R |
R |
R |
S |
S |
S |
S |
S |
S |
R |
R |
R |
|
Vancomycin (5 µg) |
R |
S |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
R |
|
Kanamycin (1000 µg) |
S |
S |
S |
S |
S |
S |
S |
S |
nd |
S |
S |
S |
S |
nd |
S |
|
Clindamycin |
S |
S |
S |
S |
S |
S |
S |
S |
nd |
S |
S |
S |
S |
nd |
nd |
|
Metronidazole |
S |
S |
S |
S |
S |
S |
S |
S |
nd |
S |
S |
S |
S |
nd |
nd |
|
Tinidazole |
nd |
0.5 |
0.125 |
nd |
0.25 |
0.125 |
0.25 |
1 |
nd |
0.5 |
1 |
0.5 |
0.25 |
nd |
nd |
|
Secnidazole |
nd |
0.5 |
0.25 |
nd |
0.25 |
0.125 |
0.25 |
0.5 |
nd |
0.25 |
0.5 |
0.5 |
0.25 |
nd |
nd |
|
Short chain fatty acids: |
|
||||||||||||||
|
2-Aminobutyric acid |
w |
− |
+ |
w |
+ |
+ |
+ |
+ |
nd |
+ |
+ |
+ |
+ |
nd |
+ |
|
2-Methylbutyric acid |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
nd |
w |
w |
+ |
w |
nd |
+ |
|
Acetic acid |
w |
+ |
− |
− |
− |
− |
− |
+ |
+ |
w |
− |
− |
w |
+ |
+ |
|
Butyric acid |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Formic acid |
w |
+ |
w |
w |
+ |
− |
w |
+ |
nd |
+ |
− |
− |
w |
+ |
w |
|
Fumaric acid |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
nd |
+ |
+ |
+ |
+ |
nd |
+ |
|
Isobutyric acid |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
w |
w |
+ |
w |
nd |
+ |
|
Isovaleric acid |
+ |
− |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
|
Lactic acid |
w |
w |
w |
w |
+ |
+ |
− |
− |
nd |
w |
w |
w |
− |
− |
− |
|
n-caproic acid |
− |
− |
− |
− |
− |
− |
− |
+ |
+ |
+ |
+ |
− |
− |
nd |
− |
|
n-valeric acid |
− |
− |
− |
− |
− |
− |
− |
− |
+ |
− |
− |
− |
+ |
nd |
+ |
|
Propionic acid |
+ |
+ |
− |
+ |
+ |
+ |
− |
+ |
+ |
− |
− |
+ |
+ |
+ |
+ |
|
Succinic acid |
− |
− |
− |
− |
− |
− |
+ |
+ |
nd |
+ |
+ |
− |
− |
nd |
− |
*Characteristics for 9, M. hexanoica MH are from Jeon et al. [3], and for 14, M. stantonii AJH120 are from Maki and Looft [6].
†Data for cell size and growth at 45 °C was determined for UPII 199-6T, KA00182T and BV3C16-1T. Cell size and growth at 45 °C for the other species were obtained from the original publications [1–7, 9].
‡d-Glucose, d-fructose and lactose fermentation was also assessed using the Pre-Reduced Anaerobically Sterilized (PRAS) medium. +, Positive by both methods, −, negative by both methods; +/−, positive by Biolog and negative by PRAS; −/+, negative by Biolog and positive by PRAS; ng, no growth; nd, not determined.
The cellular fatty acid composition of UPII 199-6T, KA00182T and BV3C16-1T were compared with the reference Megasphaera strains except M. hexanoica and M. stantonii. All bacterial strains were grown on Brucella blood agar and colonies were harvested at 2 days growth, resuspended in DNAse free water and pelleted by centrifugation at 12 000 r.p.m. for 2 min. Cell pellets were frozen at −80 °C. Fatty acid analysis was conducted using gas chromatography with the Sherlock Fatty Acid Analysis System by Microbial ID. The most abundant fatty acids of the three novel Megasphaera species from the genital tract were: UPII 199-6T, C12 : 0, C16 : 0, C16 : 0 dimethyl acetal (DMA) and summed feature 5 (C15 : 0 DMA and/or C14 : 0 3-OH); KA00182T, C16 : 0 and C16 : 1 cis 9; BV3C16-1T, C12 : 0, C14 : 0 3-OH and summed feature 5 (Table 2). Similar patterns were noted in the second isolate of each proposed type strain (Table 2). While the three novel species shared some similarities with other Megasphaera type strains, proportions were different, and differences were also noted in other cellular fatty acids thereby distinguishing them from other validly named Megasphaera type strains.
Table 2.
Fatty acid methyl ester (FAME) analysis of genital tract Megasphaera species and comparison with validly named species of the genus Megasphaera
Strains: 1, UPII 199-6T; 2, DNF00751; 3, KA00182T; 4, UPII 135-E; 5, BV3C16-1T; 6, HL562; 7, M. cerevisiae DSM 20462; 8, M. elsdenii DSM 20460; 9, M. hexanoica MH; 10, M. indica DSM 25563; 11, M. massiliensis DSM 26228; 12, M. micronuciformis DSM 17226; 13, M. paucivorans DSM 16981; 14, M. stantonii AJH120; 15, M. sueciensis DSM 17042. Values are percentages of total fatty acids. Fatty acids less than 1 % of the total in all of the strains are not listed. –, Not detected. Major components are marked in bold text.
|
Fatty acid |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9* |
10 |
11 |
12 |
13 |
14* |
15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
C9 : 0 |
– |
– |
– |
– |
– |
– |
– |
– |
1.6 |
– |
– |
– |
– |
– |
|
|
C10 : 0 |
3.1 |
2.8 |
2.8 |
3.3 |
0.3 |
0.3 |
7.0 |
– |
1.9 |
0.5 |
0.7 |
0.9 |
– |
0.3 |
– |
|
C11 : 0 |
0.8 |
0.3 |
– |
0.9 |
– |
0.1 |
0.9 |
– |
3.0 |
– |
1.1 |
– |
– |
– |
– |
|
C12 : 0 |
13.5 |
12.7 |
9.0 |
10.1 |
13.2 |
13.1 |
9.2 |
13.7 |
16.6 |
16.1 |
12.8 |
13.5 |
15.8 |
18.9 |
16.5 |
|
C13 : 0 |
– |
– |
– |
– |
– |
– |
– |
– |
3.2 |
– |
0.9 |
– |
– |
– |
0.8 |
|
C14 : 0 |
– |
1.0 |
1.1 |
0.7 |
4.0 |
2.4 |
5.4 |
5.3 |
8.1 |
2.4 |
5.3 |
4.1 |
3.3 |
0.9 |
4 |
|
C16 : 0 aldehyde |
6.1 |
3.0 |
1.6 |
2.1 |
1.8 |
– |
2.4 |
2.2 |
– |
0.8 |
1.1 |
3.7 |
2.0 |
1.1 |
0.8 |
|
C15 : 0 |
– |
– |
– |
– |
1.1 |
– |
0.9 |
0.7 |
6.3 |
– |
3.3 |
– |
1.1 |
– |
0.9 |
|
C16 : 0 |
21 |
20.4 |
13.1 |
10.9 |
9.9 |
8.0 |
7.9 |
12.8 |
12.7 |
3.8 |
7.8 |
7.8 |
8.8 |
8.8 |
3.2 |
|
C17 : 0 |
– |
– |
0.6 |
0.5 |
0.4 |
– |
0.5 |
5.4 |
– |
1.5 |
– |
0.7 |
– |
– |
|
|
C18 : 0 |
3.2 |
1.6 |
2.0 |
3.4 |
1.2 |
1.3 |
– |
1.5 |
9.4 |
– |
1.3 |
2.0 |
1.5 |
2.3 |
– |
|
C11 : 0 DMA |
– |
0.4 |
0.3 |
– |
0.1 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
|
C14 : 0 DMA |
– |
– |
1.0 |
– |
1.4 |
– |
– |
– |
2.2 |
– |
1.4 |
– |
2.6 |
||
|
C16 : 0 DMA |
11.4 |
8.3 |
4.3 |
4.3 |
4.9 |
4.7 |
4.5 |
5.6 |
0.3 |
1.4 |
4.1 |
4.8 |
3.9 |
3.3 |
1.4 |
|
C16 : 1 cis 7 DMA |
– |
– |
0.6 |
– |
7.8 |
11.6 |
– |
– |
– |
2.0 |
0.3 |
– |
– |
– |
– |
|
C16 : 1 cis 9 DMA |
– |
3.0 |
8.8 |
– |
– |
– |
4.8 |
9.8 |
– |
7.1 |
7.6 |
7.5 |
7.3 |
7.1 |
6.2 |
|
C19 : 0 cyc 9,10 DMA |
– |
– |
0.4 |
– |
0.5 |
– |
3.9 |
0.5 |
– |
– |
1.7 |
– |
– |
0.5 |
1.4 |
|
C17 : 0 DMA |
– |
– |
– |
– |
– |
– |
– |
– |
2.3 |
– |
1.0 |
– |
– |
– |
|
|
C18 : 0 DMA |
– |
– |
– |
– |
– |
– |
– |
0.5 |
– |
– |
0.6 |
– |
– |
0.4 |
– |
|
C18 : 1 DMA |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
0.7 |
– |
– |
0.9 |
|
|
C18 : 1 cis 7 DMA |
– |
– |
– |
– |
– |
3.6 |
– |
– |
– |
1.2 |
– |
– |
– |
– |
|
|
C18 : 1 cis 9 DMA |
– |
– |
0.9 |
0.5 |
1.4 |
– |
1.1 |
4.5 |
– |
3.7 |
4.9 |
4.7 |
2.9 |
4.8 |
8.5 |
|
C16 : 1 cis 7 |
– |
– |
2.6 |
– |
– |
10.2 |
– |
1.7 |
4.2 |
10.4 |
1.4 |
– |
1.3 |
– |
|
|
C16 : 1 cis 9 |
3.0 |
6.3 |
13.4 |
9.5 |
10 |
1.1 |
7.6 |
8.9 |
– |
7 |
6.4 |
4.5 |
10.2 |
7.1 |
9.6 |
|
C17 : 1 cis 6 |
– |
12.3 |
1.8 |
2.8 |
5.1 |
– |
7.0 |
1.3 |
– |
9.2 |
1.8 |
0.7 |
4.1 |
0.7 |
2.3 |
|
C17 : 1 cis 11 |
– |
– |
– |
– |
– |
2.2 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
C18 : 2 cis 9,12 |
– |
– |
– |
– |
– |
– |
– |
– |
3.7 |
– |
– |
– |
– |
– |
|
|
C18 : 1 cis 6 |
– |
– |
– |
– |
– |
– |
2.5 |
– |
– |
1.1 |
1 |
– |
– |
– |
– |
|
C18 : 1 cis 7 |
– |
– |
0.8 |
– |
– |
– |
– |
– |
– |
1.1 |
– |
1 |
– |
1.3 |
– |
|
C18 : 1 cis 9 |
8.5 |
1.3 |
7.1 |
3.7 |
4.0 |
9.9 |
2.5 |
6.7 |
8.2 |
3.7 |
4.6 |
13.8 |
6.6 |
15.9 |
10 |
|
C19 cyc 9,10 |
3.7 |
3.6 |
6.9 |
5.1 |
4.4 |
3.4 |
5.5 |
1.5 |
– |
3.6 |
3.2 |
– |
4.3 |
2.0 |
1.3 |
|
C11 : 0 iso |
0.6 |
0.3 |
1.2 |
3.0 |
0.4 |
0.3 |
0.6 |
– |
– |
– |
– |
0.4 |
– |
– |
– |
|
C13 : 0 iso |
– |
– |
– |
0.5 |
2.9 |
1.9 |
– |
– |
– |
– |
0.2 |
1.1 |
– |
– |
0.7 |
|
C13 : 0 iso 3-OH |
0.9 |
0.4 |
– |
3.7 |
1.2 |
0.9 |
– |
– |
– |
– |
– |
0.5 |
0.7 |
– |
– |
|
C15 : 0 iso |
– |
– |
– |
– |
1.1 |
0.5 |
– |
– |
– |
– |
– |
0.5 |
– |
– |
– |
|
C15 : 0 iso 3-OH |
– |
– |
– |
1.0 |
1.1 |
0.7 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
C17 : 0 iso |
0.6 |
0.3 |
0.9 |
1.8 |
1.5 |
1.0 |
– |
– |
– |
– |
– |
0.7 |
– |
– |
– |
|
C12 : 0 3-OH |
– |
6.8 |
4.6 |
– |
– |
1.6 |
3.0 |
2.2 |
– |
3.1 |
1.4 |
1.8 |
2.0 |
1.8 |
2.8 |
|
C13 : 0 3-OH |
– |
1.4 |
– |
1.5 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
|
|
C14 : 0 3-OH |
– |
12.2 |
9.0 |
– |
13.8 |
13.0 |
17.3 |
12.7 |
– |
7.8 |
14.3 |
17.2 |
14.8 |
16.2 |
13.9 |
|
C17 : 2 at 16 760 |
– |
– |
– |
– |
– |
1.0 |
– |
– |
– |
– |
– |
– |
– |
1.5 |
– |
|
Unknown at 14.762 |
– |
1.7 |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
– |
3.8 |
– |
|
Unknown at 17.223 |
– |
– |
– |
– |
0.4 |
– |
– |
– |
1.5 |
– |
1.1 |
– |
– |
– |
0.8 |
|
Summed feature 2† |
7.2 |
6.8 |
4.6 |
5.7 |
1.5 |
1.6 |
3 |
2.2 |
– |
3.2 |
1.4 |
1.8 |
2 |
– |
2.8 |
|
Summed feature 4† |
0.8 |
1.7 |
5.4 |
4.2 |
4.8 |
3.5 |
3.8 |
5.8 |
1.1 |
5.7 |
3.6 |
5.7 |
5.5 |
– |
4.9 |
|
Summed feature 5† |
14.2 |
12.2 |
9 |
13.8 |
13.8 |
13.0 |
17.3 |
12.7 |
6.6 |
7.8 |
14.3 |
17.2 |
14.8 |
16.2 |
13.9 |
|
Summed feature 6† |
0.9 |
3.0 |
8.8 |
5.4 |
7.8 |
11.6 |
4.8 |
9.8 |
2.4 |
7.1 |
7.6 |
7.5 |
7.3 |
– |
6.2 |
|
Summed feature 7† |
– |
– |
– |
– |
0.9 |
1.0 |
0.8 |
1.9 |
1.4 |
2.2 |
1.7 |
2.7 |
2.1 |
– |
5.4 |
|
Summed feature 10† |
– |
– |
0.8 |
0.5 |
– |
0.4 |
– |
– |
– |
1.1 |
– |
1 |
– |
– |
– |
*Characteristics for 9, M. hexanoica MH are from Jeon et al. [3], and for 14, M. stantonii AJH120 are from Maki and Looft [6]. All other data were generated in this study.
†Summed features consist of one or more fatty acids that could not be separated by the MIDI system. Summed feature 2 consists of C13 : 1 cis 12, C14 : 0 ALDE, and/or C11 : 1 2-OH; summed feature 4 consists of an unknown at 14.762 and/or C15 : 2; summed feature 5 consists of C15 : 0 DMA and/or C14 : 0 3-OH; summed feature 6 consists of C15 : 0 anteiso 3-OH and/or C16 : 1 cis 9 DMA; aummed feature 7 consists of C17 : 2 and C17 : 1 cis 8; aummed feature 10 consists of C18 : 1 cis 7 and/or an unknown at 17.834.
Initially, simple biochemical tests were performed for presumptive identification to the genus level [9, 53, 54] and included catalase (Fisher Scientific), spot indole (prepared in-house), oxidase (Becton Dickinson), nitrate reduction (Anaerobe Systems) and H2S production (Hardy Diagnostics). Biochemical characterization of the genital tract isolates and the reference strains were conducted in duplicate using the Biolog Anaerobe Identification Test Panel to evaluate metabolism of sugars and amino acids [54, 55] according to the manufacturer’s instructions. We also used the Pre-Reduced Anaerobically Sterilized (PRAS) medium to assess sugar fermentation [56] and gas production. The genital tract Megasphaera isolates that were tested did not produce gas, H2S, or indole and were negative for catalase, oxidase and nitrate reductase (Table 1). Like M. paucivorans and M. sueciensis, the genital tract Megasphaera species did not metabolize sugars including glucose, fructose, sucrose and lactose. Moreover, none of the Megasphaera species tested metabolized gelatin. They were positive in the Biolog tests for organic acids such as lactate, pyruvate and α-ketovalerate. KA00182T, UPII 135-E and HL562 metabolized succinate but UPII 199-6T, DNF00751 and BV3C16-1T yielded weak results; none of the other Megasphaera type strains with available results metabolized succinate. Specific differences between the genital tract Megasphaera species were noted in amino acid metabolism. Of the amino acids tested, UPII 199-6T only metabolized serine, which was also positive for all other Megasphaera strains tested. In contrast, BV3C16-1T and HL562 metabolized all five amino acids tested including alanine, alanyl-l-glutamine, glutamine, glutamate and serine while KA00182T tested positive for alanyl-l-glutamine, glutamate and serine.
To characterize the metabolic end products, all isolates were grown in PYG-mod-YG at 37 °C in triplicate for 24–48 h. Cell supernatants were used for detection of short chain fatty acids and organic acids using 1H-NMR spectroscopy. NMR analyses of bacterial supernatants were made using a Bruker AVANCE III 800 MHz spectrometer equipped with a cryoprobe or a room temperature probe suitable for 1H inverse detection with Z-gradients at 298 K. 500 µl media was mixed with 100 µl phosphate buffer prepared in deuterated water (0.1 M; pH=7.4) containing 50 µM TSP (3-(trimethylsilyl) propionic-2,2,3,3-d4 acid sodium salt). The solution was placed in a 5 mm NMR tube and one-dimensional 1H NMR spectra were obtained using a one pulse sequence that included residual water signal suppression by pre-saturation during the relaxation delay. For each sample, 32 k data points were acquired using a spectral width of 9615 Hz and a relaxation delay of 10 s. The data were processed using a spectral size of 32 k points and by multiplying with an exponential window function with a line broadening of 0.5 Hz. The resulting spectra were phase and baseline corrected and referenced with respect to the internal TSP signal. Metabolite peaks in the spectra were then assigned using chemical shift databases [57–59] and the peak assignments were confirmed based on the spectra of standard compounds obtained under identical conditions. The peak areas were obtained by integration with reference to the internal reference signal from TSP. Using these peak areas, along with the known concentration of TSP and the number of protons each peak represented in the molecule, short chain fatty acid metabolites concentrations in the supernatants were determined. Bruker Topspin version 3.0 and 3.1 software packages were used for NMR data acquisition and processing, respectively. NMR experiments were performed at the Northwest Metabolomics Research Center at University of Washington. Similar to other validly named Megasphaera type strains, UPII 199-6T, KA00182T and BV3C16-1T produced butyrate, fumarate, isovalerate, 2-methylbutyrate and 2-aminobutyrate (Table 1, Fig. S1, available in the online version of this article). The genital tract isolates did not produce n-caproate, or n-valerate. Moreover, none of the genital tract Megasphaera species produced succinate (Fig. S1), which is notably higher in concentration in vaginal fluid from women with BV [60]. BV3C16-1T produced lactate while the other genital tract isolates were weak producers.
Antimicrobial susceptibility testing was performed using the agar dilution method on Brucella blood agar as per the Clinical and Laboratory Standards Institute (CLSI) guidelines [61]. The US Centers for Disease Control Sexually Transmitted Diseases Treatment guidelines recommend metronidazole, tinidazole or clindamycin for treatment for BV [62], hence these antibiotics (Sigma Aldrich) were selected for testing. We also tested secnidazole (Symbiomix), a single-dose antibiotic that has been recently approved for the treatment of BV [38, 63–65]. Concentrations tested ranged from 0.03 to 128 µg ml−1. The lowest antibiotic concentration that yielded marked reduction or no growth was recorded as the minimum inhibitory concentration (MIC). The CLSI defined breakpoints to clindamycin (≤2 µg ml−1 sensitive, ≥8 µg ml−1 resistant) and metronidazole (≤8 µg ml−1 sensitive, ≥32 µg ml−1 resistant) were used for interpretation of MIC results [61]. There are no CLSI defined breakpoints for tinidazole or secnidazole. Other compounds that we examined for activity against Megasphaera species included bile (1 mg), colistin (10 µg), vancomycin (5 µg) and kanamycin (1000 µg). Testing for bile sensitivity was conducted using bile discs with 20 % bile (Becton Dickinson) [54] that were prepared in-house. Brucella plates were inoculated using the quadrant method and the bile disc was placed in the first quadrant; any zone of inhibition around the bile disc was considered sensitive. The same approach was used for discs containing colistin, vancomycin or kanamycin. If a bacterium exhibited a zone of clearance ≥10 mm, it was marked as sensitive to colistin, vancomycin or kanamycin while if the clearance zone was <10 mm, the bacterium was considered resistant [54]. Genital tract isolates and validly named Megasphaera species were sensitive to bile and kanamycin (Table 1). All genital tract isolates were resistant to colistin while KA00182T was sensitive. All Megasphaera species tested were sensitive to antibiotics typically used to treat BV including clindamycin and metronidazole. The lowest concentrations of secnidazole and tinidazole resulted in clearance zones suggesting sensitivity to these antibiotics. Similar to other Gram-negative bacteria, all Megasphaera isolates tested with the exception of DNF00751 were resistant to vancomycin.
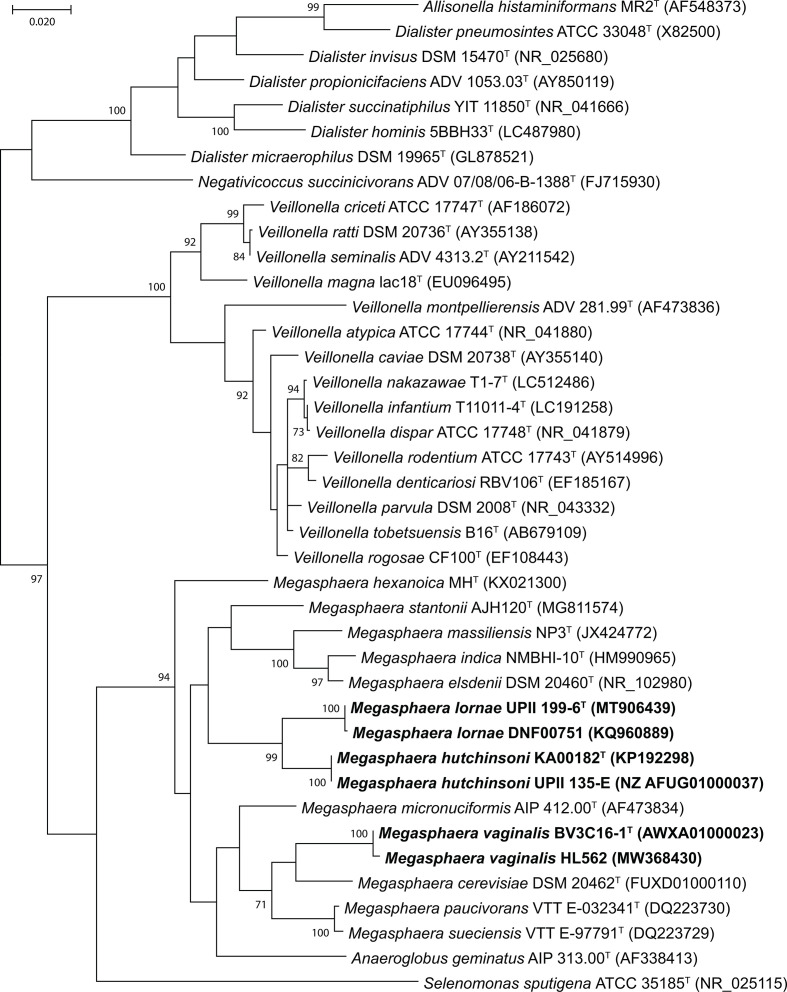
Genomic DNA was extracted using the ZR Fungal/Bacterial DNA MidiPrep Kit (Zymo Research) according to the manufacturer’s instructions for whole genome shotgun sequencing and submitted to one of two Human Microbiome Project (HMP) sequencing centres. UPII 199-6T and BV3C16-1T were sequenced at the J. Craig Venter Institute, Rockville, Maryland while KA00182T was sequenced at the Genome Institute at Washington University School of Medicine, St. Louis, Missouri. The whole genome sequences of the genital tract isolates have been available to the scientific community since they were first deposited by the HMP (Accessions in Table 3). The 16S rRNA gene sequences for each isolate were obtained from the WGS projects or by sequencing the 16S rRNA gene (accessions in Fig. 3). All vaginal isolates characterized in this study each had one copy of the 16S rRNA gene. Validly published Megasphaera species harbour one (M. cerevisiae) to seven gene copies (M. elsdenii) of the 16S rRNA gene. UPII 199-6T had a sequence identity of 96.4 % to KA00182T and 93.7 % to BV3C16-1T, while KA00182T had a sequence identity of 92.8 % with BV3C16-1T. Evaluation of the UPII 199-6T 16S rRNA sequence in the EZ Taxon database of validly named type strains [66] showed that this isolate had a sequence identity of 93.6 % to M. stantonii and ranged in identity from 92.9–93.6 % to validly named Megasphaera species. KA00182T had the closest sequence identity to M. micronuciformis (94.2 %), while BV3C16-1T had the closest identity to M. cerevisiae (95.4 %). The 16S rRNA gene from KA00182T ranged in sequence identity from 92.6–94.2 % among all validly named Megasphaera isolates, while the range for BV3C16-1T was between 93.0 and 95.4 % suggesting that all three isolates are novel species within the Megasphaera genus. NCBI blast searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi) [67] of the 16S rRNA sequences from UPII 199-6T, KA00182T and BV3C16-1T resulted in matches to uncultivated bacterial clones from the human vagina and skin. Importantly, UPII 199-6T had a sequence identity of 99.9 % with the uncultured clone 127-Q 35 (AY738672) designated as Megasphaera type 1. KA00182T (AY738697) had a sequence identity of 99.9 % with the uncultured clone 123-Q 3 designated as Megasphaera type 2. We have previously demonstrated these clones to be associated with BV [12]. A multiple sequence alignment of the 16S rRNA gene from the genital tract isolates along with validly named members of the family Veillonellaceae was created using the ClustalW algorithm and the evolutionary relationships were inferred by using the maximum- likelihood method based on the Tamura–Nei model [68] in mega X [69] (Fig. 3). All three isolates fall within the Megasphaera clade of validly named species. The closest neighbour of UPII 199-6T was KA00182T and both isolates were phylogenetically distinct from M. elsdenii, M. indica and M. massiliensis, the most closely related species. BV3C16-1T was most closely related to M. cerevisiae. Similar results were obtained when evolutionary relationships were inferred using the neighbour-joining method (Fig. S2).
Table 3.
Comparison of the genome characteristics of genital tract Megasphaera isolates with validly named species of the genus Megasphaera.
Strains: 1, UPII 199-6T; 2, DNF00751; 3, KA00182T; 4, UPII 135-E; 5, BV3C16-1T; 6, HL562; 7, M. cerevisiae DSM 20462; 8, M. elsdenii DSM 20460; 9, M. hexanoica MH; 10, M. indica DSM 25563; 11, M. massiliensis DSM 26228; 12, M. micronuciformis DSM 17226; 13, M. paucivorans DSM 16981; 14, M. stantonii AJH120; 15, M. sueciensis DSM 17042. nd, No data.
|
Megasphaera strains |
BioProject accession |
Genome length (Mb) |
No. of protein coding genes |
No. of tRNA genes |
No. of stable RNA genes |
DNA G+C content (mol%)* |
|---|---|---|---|---|---|---|
|
1 |
PRJNA64689 |
1.64 |
1577 |
49 |
52 |
46.4 |
|
2 |
PRJNA257377 |
1.73 |
1819 |
45 |
47 |
45.9 |
|
3 |
PRJNA272074 |
1.57 |
1514 |
45 |
48 |
38.9 |
|
4 |
PRJNA64691 |
1.65 |
1517 |
50 |
53 |
38.9 |
|
5 |
PRJNA89631 |
2.21 |
2262 |
49 |
52 |
49.8 |
|
6 |
nd |
nd |
nd |
nd |
nd |
nd |
|
7 |
PRJEB19539 |
3.15 |
3354 |
52 |
54 |
44.8 |
|
8 |
PRJNA437124 |
2.48 |
2329 |
66 |
80 |
52.8 |
|
9 |
PRJNA287738 |
2.88 |
2858 |
53 |
60 |
49 |
|
10 |
nd |
nd |
nd |
nd |
nd |
54.9 |
|
11 |
PRJEB645 |
2.66 |
2422 |
56 |
61 |
50.2 |
|
12 |
nd |
nd |
nd |
nd |
nd |
46.4 |
|
13 |
PRJEB16204 |
2.91 |
2915 |
51 |
53 |
40.2 |
|
14 |
PRJNA471687 |
2.65 |
2590 |
57 |
67 |
52.6 |
|
15 |
nd |
nd |
nd |
nd |
nd |
43.1 |
*DNA G+C content (mol%) for M. indica, M. micronuciformis, and M. sueciensis were obtained from the original references [4, 5, 7] as genomes were not available. Genome information is also not available for M. vaginalis HL562.
Fig. 3.
Molecular phylogenetic analysis by the maximum-likelihood method based on 16S rRNA gene sequences showing the phylogenetic positions of M. lornae, M. hutchinsoni and M. vaginalis with closely related members of the family Veillonellaceae. The genital tract isolates clustered within the Megasphaera clade. Bootstrap values (based on 1000 replications) greater than or equal to 70 % are shown as percentages at each node. Bar, 0.02 substitutions per nucleotide position. Selenomonas sputigena ATCC 35185 (NR_025115) from the family Selenomonadaceae was added as an outgroup.
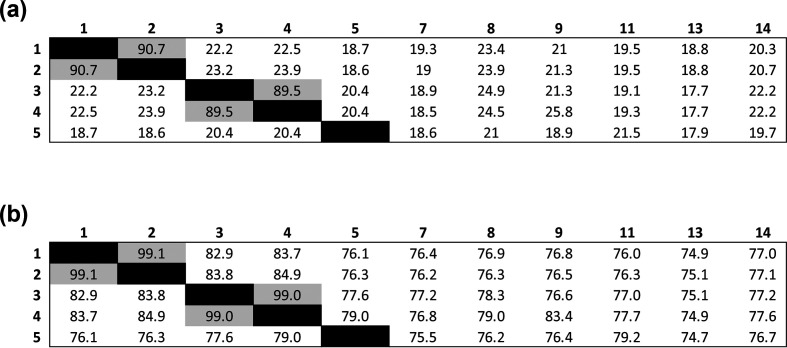
Information regarding genome lengths, predicted protein-coding genes and RNA genes were extracted from the bacterial Bioinformatics Database and Analysis Resource Center, patric (Table 3) [70]. The DNA G+C content was calculated using the online service at http://ggdc.dsmz.de [71] and were different in UPII 199-6T, KA00182T and BV3C16-1T from other validly named Megasphaera species whose genomes were available (Table 3). Genome data is not available for the type strains of M. indica, M. micronuciformis and M. sueciensis and DNA G+C content data was obtained from the original papers [4, 5, 7]. The DNA G+C content for UPII 199-6T was similar to the DNA G+C content reported for M. micronuciformis DSM17226 (46.4 mol%) which was determined using a wet lab approach, HPLC [7], hence the values are not directly comparable. The striking differences between UPII 199-6T and M. micronuciformis DSM17226 using several lines of evidence including phylogenetic analyses, biochemical characterization, FAME analyses and metabolite end product testing show that they are different species. Digital DNA–DNA hybridization (dDDH) was used to evaluate the relatedness of UPII 199-6T, KA00182T and BV3C16-1T to each other and to the reference genomes available for validly named isolates (Fig. 4a). Genome-based species delineation was conducted using the genome blast distance phylogeny approach (GBDP) version 2.1 using the online web tool with the recommended settings for Formula 2 independent of genome length and robust against incomplete draft genomes (http://ggdc.dsmz.de) [71, 72]. DDH values of <70 % of UPII 199-6T, KA00182T and BV3C16-1T, in comparison with M. cerevisiae, M. elsdenii, M. hexanoica, M. massiliensis, M. paucivorans and M. stantonii, warrant designation as novel species [71]. These results were further supported with average nucleotide identity (ANI) analyses using a web-based ANI tool (http://enve-omics.ce.gatech.edu/aai) [73]. None of the genital tract isolates characterized here had an identity >95 % on comparison with validly named Megasphaera species which is the suggested cut-off for a novel species (Fig. 4b).
Fig. 4.
Genome-based species delineation using the (a) genome blast distance phylogeny approach. Values less than 70 % are indicative of a different species. Strains: 1, UPII 199-6T; 2, DNF00751; 3, KA00182T; 4, UPII 135-E; 5, BV3C16-1T; 7, M. cerevisiae DSM 20462; 8, M. elsdenii DSM 20460; 9, M. hexanoica MH; 11, M. massiliensis DSM 26228; 13, M. paucivorans DSM 16981; 14, M. stantonii AJH120; 15, M. sueciensis DSM 17042. M. vaginalis HL562, M. indica, M. micronuciformis and M. sueciensis did not have genomes available for this analysis. UPII 199-6T and DNF00751 belong to the same species with a DDH value of 90.7 % (M. lornae). KA00182T and UPII 135-E belong to the same species with a DDH value of 89.5 % (M. hutchinsoni). (b) Average nucleotide identity analyses. Identities of <95 % indicate a novel species. The genital tract isolates (1–5) are sufficiently different from validly named Megasphaera species (7–15) to warrant designation as novel species. UPII 199-6T and DNF00751 are the same species (99.1 % ANI) while KA00182T and UPII 135-E are the same species (99 % ANI).
In summary, several lines of evidence including 16S rRNA gene-based phylogeny, growth and biochemical features, cellular fatty acid composition, short-chain fatty acids produced, determination of DNA G+C content, dDDH and ANI analyses demonstrate that the six isolates characterized in this study represent three novel species within the genus Megasphaera. The proposed type strains are designated M. lornae sp. nov. UPII 199-6T, M. hutchinsoni sp. nov. KA00182T and M. vaginalis sp. nov. BV3C16-1T.
Description of Megasphaera lornae sp. nov.
Megasphaera lornae (lor′ nae. N.L. fem. n. lornae of Lorna, named in honour of Lorna Rabe, a microbiologist who isolated and contributed to the characterization of Megasphaera species from the female genital tract).
Strictly anaerobic, Gram-negative, non-spore forming, non-motile, coccoid-shaped micro-organism that grows as single cells, in pairs or occasional chains. Cells are 0.9–1.5 µm. Colonies are visible on Brucella blood agar after 2 days of growth and appear as convex, entire, glossy and off-white in colour and a diameter of 0.5–0.7 mm. Growth of the bacterium occurs between 35–37 °C with an optimal temperature of 37 °C in PYG-mod-YG broth. The major SCFAs produced are butyric acid and propionic acid when grown in PYG-mod-YG broth for 48 h. The bacterium is sensitive to metronidazole and clindamycin, antibiotics typically used to treat BV. Bile and kanamycin inhibit growth, while colistin and vancomycin have no impact on growth. The bacterium does not metabolize sugars including glucose, fructose, lactose, sucrose and dextrin, but metabolizes organic acids including lactate, pyruvate, α-ketoglutarate and succinate. This bacterium can metabolize serine, but not alanine, alanyl-l-glutamine, glutamine or glutamate.
The type strain is UPII 199-6T (=DSM 111201T,=ATCC TSD-205T), which was isolated from an endometrial biopsy sample from a woman being evaluated for PID. The DNA G+C content of the type strain is 46.4 mol%. GenBank accession numbers for the 16S rRNA gene sequence and whole genome sequence are MT906439 and PRJNA64689, respectively.
Description of Megasphaera hutchinsoni sp. nov.
Megasphaera hutchinsoni (hutch. in. so′ ni. N.L. fem. adj. hutchinsoni, named in honour of the Fred Hutchinson Cancer Research Center where this cultivation study was conducted).
Strictly anaerobic, Gram-negative, non-spore forming, non-motile, coccoid-shaped micro-organism that grows as singlets, pairs and chains along their tapered ends. Cells are 0.5–0.7 µm. Colonies are visible on Brucella blood agar after 2 days of growth and appear as convex, entire, glossy and off-white in colour with a diameter of 0.4–0.7 mm. Growth of the bacterium occurs between 30–42 °C with an optimal temperature of 35–37 °C in PYG-mod-YG broth. The major SCFAs produced are 2-methylbutyric acid, butyric acid, isobutyric acid, fumaric acid and isovaleric acid when grown in PYG-mod-YG broth for 24 h. The bacterium is sensitive to metronidazole, clindamycin, tinidazole and secnidazole, antibiotics used to treat BV. Bile, colistin and kanamycin inhibit growth, while vancomycin has no impact on growth. The bacterium does not metabolize sugars including glucose, fructose, lactose, sucrose and dextrin, but metabolizes organic acids including lactate, pyruvate, α-ketoglutarate and succinate. The bacterium metabolizes amino acids including alanyl-l-glutamine, glutamate and serine, but not alanine or glutamine.
The type strain is KA00182T (=DSM 111202T=ATCC TSD-206T), which was isolated from human vaginal fluid obtained from a woman with BV. The DNA G+C content of the type strain is 38.9 mol%. GenBank accession numbers for the 16S rRNA gene sequence and whole genome sequence are KP192298 and PRJNA272074, respectively.
Description of Megasphaera vaginalis sp. nov.
Megasphaera vaginalis sp. nov. (va.gi.na′ lis. N.L. fem. adj. vaginalis, pertaining to the vagina).
Strictly anaerobic, Gram-negative, non-spore-forming, non-motile, coccoid-shaped micro-organism that grows as single cells with rounded ends, often occurring in pairs. Cells are 1.0 to 1.4 µm. Colonies are visible on Brucella blood agar after 2 days of growth and appear as small, convex and translucent with a diameter of 0.3–0.4 mm. Growth of the bacterium occurs between 30–37 °C with an optimal temperature of 35–37 °C in PYG-mod-YG broth. The major SCFAs produced are 2-aminobutyric acid, 2-methylbutyric acid, butyric acid, isobutyric acid, isovaleric acid, and propionic acid when grown on PYG-mod-YG broth for 48 h. The bacterium is sensitive to metronidazole, clindamycin, tinidazole and secnidazole, antibiotics used to treat BV. Bile and kanamycin inhibit growth, while colistin and vancomycin have no impact on growth. The bacterium does not metabolize sugars including glucose, fructose, lactose, sucrose and dextrin, but metabolizes organic acids including lactate, pyruvate, α-ketoglutarate and succinate. The bacterium metabolizes several amino acids including alanine, alanyl-l-glutamine, glutamine, glutamate and serine.
The type strain is BV3C16-1T (=DSM 111203T=ATCC TSD-207T), which was isolated from human vaginal fluid obtained from a woman with BV. The DNA G+C content of the type strain is 49.8 mol%. GenBank accession numbers for the 16S rRNA gene sequence and whole genome sequence are JN809775 and PRJNA89631, respectively.
Supplementary Data
Funding information
The work was supported by the National Institutes of Health (grant R01 HG005816 to D. N. F.) as part of the Human Microbiome Project initiative focused on technology development and NIH grant U19AI120249 to S. L. H.
Acknowledgements
We appreciate Sean Proll’s help with figures and tables. Steve MacFarlane and Bobbie Schneider provided support with preparation of samples for electron microscopy.
Conflicts of interest
D. N. F. and T. F., have received a royalty from B. D., around molecular diagnosis of B. V. S. S., has received speaking honoraria from Lupin Inc. S. L. H., has served as a consultant to Hologic related to the development of diagnostic tests for bacterial vaginosis and her institution has received research funding from Cepheid and Becton-Dickinson.
Ethical statement
The research study at the University of Pittsburgh evaluated women for pelvic inflammatory disease; endometrial tissue samples were collected in a protocol approved by the University of Pittsburgh Review Board (IRB approval number: PRO 010010112). The research study at the Fred Hutchinson Cancer Research Center was designed for the isolation of novel anaerobes from the human vagina and was approved by the institutional review board at the Fred Hutch (IRB approval number: IR 7363). All participants provided informed consent.
Footnotes
Abbreviations: ANI, average nucleotide identity; BM, basic medium; BV, bacterial vaginosis; dDDH, digital DNA–DNA hybridization; DMA, dimethyl acetal; FAME, fatty acid methyl ester; PID, pelvic inflammatory disease; PYG-mod, peptone–yeast–glucose modified; PYG-mod-YG, PYG-mod medium supplemented with 1% yeast extract and 1% glucose; SCFA, short chain fatty acid.
Two supplementary figures are available with the online version of this article
References
- 1.Rogosa M. Transfer of Peptostreptococcus elsdenii to a new genus, Megasphaera [M. elsdenii Gutierrez, et al. comb. nov.]. Int J Syst Bact. 1971;21:187–189. doi: 10.1099/00207713-21-2-187. [DOI] [Google Scholar]
- 2.Engelmann U, Weiss N. Megasphaera cerevisiae sp. nov.: a new Gram-negative obligately anaerobic coccus isolated from spoiled beer. Syst Appl Microbiol. 1985;6:287–290. doi: 10.1016/S0723-2020(85)80033-3. [DOI] [Google Scholar]
- 3.Jeon BS, Kim S, Sang BI. Megasphaera hexanoica sp. nov., a medium-chain carboxylic acid-producing bacterium isolated from a cow rumen. Int J Syst Evol Microbiol. 2017;67:2114–2120. doi: 10.1099/ijsem.0.001888. [DOI] [PubMed] [Google Scholar]
- 4.Juvonen R, Suihko ML. Megasphaera paucivorans sp. nov., Megasphaera sueciensis sp. nov. and Pectinatus haikarae sp. nov., isolated from brewery samples, and emended description of the genus Pectinatus . Int J Syst Evol Microbiol. 2006;56:695–702. doi: 10.1099/ijs.0.63699-0. [DOI] [PubMed] [Google Scholar]
- 5.Lanjekar VB, Marathe NP, Ramana VV, Shouche YS, Ranade DR. Megasphaera indica sp. nov., an obligate anaerobic bacteria isolated from human faeces. Int J Syst Evol Microbiol. 2014;64:2250–2256. doi: 10.1099/ijs.0.059816-0. [DOI] [PubMed] [Google Scholar]
- 6.Maki JJ, Looft T. Megasphaera stantonii sp. nov., a butyrate-producing bacterium isolated from the cecum of a healthy chicken. Int J Syst Evol Microbiol. 2018;68:3409–3415. doi: 10.1099/ijsem.0.002991. [DOI] [PubMed] [Google Scholar]
- 7.Marchandin H, Jumas-Bilak E, Gay B, Teyssier C, Jean-Pierre H, et al. Phylogenetic analysis of some Sporomusa sub-branch members isolated from human clinical specimens: description of Megasphaera micronuciformis sp. nov. Int J Syst Evol Microbiol. 2003;53:547–553. doi: 10.1099/ijs.0.02378-0. [DOI] [PubMed] [Google Scholar]
- 8.Antunes LC, Poppleton D, Klingl A, Criscuolo A, Dupuy B, et al. Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. eLife. 2016;5:e14589. doi: 10.7554/eLife.14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchandin H, Juvonen R, Haikara A, et al. Megasphaera. In: Whitman WB, DeVos P, Dedysh S, Hedlund B, Kampfer P, editors. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc., in association with Bergey’s Manual Trust; 2015. [Google Scholar]
- 10.Padmanabhan R, Lagier JC, Dangui NP, Michelle C, Couderc C, et al. Non-contiguous finished genome sequence and description of Megasphaera massiliensis sp. nov. Stand Genomic Sci. 2013;8:525–538. doi: 10.4056/sigs.4077819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, et al. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–2573. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 12.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 13.Fredricks DN, Fiedler TL, Thomas KK, Mitchell CM, Marrazzo JM. Changes in vaginal bacterial concentrations with intravaginal metronidazole therapy for bacterial vaginosis as assessed by quantitative PCR. J Clin Microbiol. 2009;47:721–726. doi: 10.1128/JCM.01384-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol. 2007;45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruciani F, Biagi E, Severgnini M, Consolandi C, Calanni F, et al. Development of a microarray-based tool to characterize vaginal bacterial fluctuations and application to a novel antibiotic treatment for bacterial vaginosis. Antimicrob Agents Chemother. 2015;59:2825–2834. doi: 10.1128/AAC.00225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fethers K, Twin J, Fairley CK, Fowkes FJI, Garland SM, et al. Bacterial vaginosis (bv) candidate bacteria: associations with bv and behavioural practices in sexually-experienced and inexperienced women. PLoS One. 2012;7:e30633. doi: 10.1371/journal.pone.0030633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fredricks DN, Plantinga A, Srinivasan S, Oot A, Wiser A, et al. Vaginal and extra-vaginal bacterial colonization and risk for incident bacterial vaginosis in a population of women who have sex with men. J Infect Dis. 2020;28 doi: 10.1093/infdis/jiaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilbert DW, Smith WL, Chadwick SG, Toner G, Mordechai E, et al. Development and validation of a highly accurate quantitative real-time PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol. 2016;54:1017–1024. doi: 10.1128/JCM.03104-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrazzo JM, Fiedler TL, Srinivasan S, Thomas KK, Liu C, et al. Extravaginal reservoirs of vaginal bacteria as risk factors for incident bacterial vaginosis. J Infect Dis. 2012;205:1580–1588. doi: 10.1093/infdis/jis242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrazzo JM, Thomas KK, Fiedler TL, Ringwood K, Fredricks DN. Relationship of specific vaginal bacteria and bacterial vaginosis treatment failure in women who have sex with women. Ann Intern Med. 2008;149:20–28. doi: 10.7326/0003-4819-149-1-200807010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzny CA, Blanchard E, Taylor CM, Aaron KJ, Talluri R, et al. Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J Infect Dis. 2018;218:966–978. doi: 10.1093/infdis/jiy243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shipitsyna E, Roos A, Datcu R, Hallen A, Fredlund H, et al. Composition of the vaginal microbiota in women of reproductive age--sensitive and specific molecular diagnosis of bacterial vaginosis is possible? PLoS One. 2013;8:e60670. doi: 10.1371/journal.pone.0060670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, et al. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamrakar R, Yamada T, Furuta I, Cho K, Morikawa M, et al. Association between Lactobacillus species and bacterial vaginosis-related bacteria, and bacterial vaginosis scores in pregnant Japanese women. BMC Infect Dis. 2007;7:128. doi: 10.1186/1471-2334-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48:1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zozaya-Hinchliffe M, Martin DH, Ferris MJ. Prevalence and abundance of uncultivated Megasphaera-like bacteria in the human vaginal environment. Appl Environ Microbiol. 2008;74:1656–1659. doi: 10.1128/AEM.02127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartwright CP, Lembke BD, Ramachandran K, Body BA, Nye MB, et al. Development and validation of a semiquantitative, multitarget PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol. 2012;50:2321–2329. doi: 10.1128/JCM.00506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman JS, Gaydos CA. Molecular diagnosis of bacterial vaginosis: an update. J Clin Microbiol. 2018;56:e00342–18. doi: 10.1128/JCM.00342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaydos CA, Beqaj S, Schwebke JR, Lebed J, Smith B, et al. Clinical validation of a test for the diagnosis of vaginitis. Obstet Gynecol. 2017;130:181–189. doi: 10.1097/AOG.0000000000002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusters JG, Reuland EA, Bouter S, Koenig P, Dorigo-Zetsma JW. A multiplex real-time PCR assay for routine diagnosis of bacterial vaginosis. Eur J Clin Microbiol Infect Dis. 2015;34:1779–1785. doi: 10.1007/s10096-015-2412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh R, Ramsuran V, Mitchev N, Niehaus AJ, Han KSS, et al. Assessing a diagnosis tool for bacterial vaginosis. Eur J Clin Microbiol Infect Dis. 2020;39:1481–1485. doi: 10.1007/s10096-020-03862-3. [DOI] [PubMed] [Google Scholar]
- 32.Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, et al. Lactobacillus-deficient cervicovaginal bacterial communities are associated with increased HIV acquisition in young South African women. Immunity. 2017;46:29–37. doi: 10.1016/j.immuni.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClelland RS, Lingappa JR, Srinivasan S, Kinuthia J, John-Stewart GC, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis. 2018;18:554–564. doi: 10.1016/S1473-3099(18)30058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srinivasan S, Richardson BA, Wallis J, Fiedler TL, Dezzutti CS. Conference on Retroviruses and Opportunistic Infections. USA: Boston; 2018. Vaginal microbiota and HIV acquisition risk among African women. [Google Scholar]
- 35.Nelson DB, Hanlon A, Nachamkin I, Haggerty C, Mastrogiannis DS, et al. Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery. Paediatr Perinat Epidemiol. 2014;28:88–96. doi: 10.1111/ppe.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, et al. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun. 2019;10:1305. doi: 10.1038/s41467-019-09285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haggerty CL, Ness RB, Totten PA, Farooq F, Tang G, et al. Presence and concentrations of select bacterial vaginosis-associated bacteria are associated with increased risk of pelvic inflammatory disease. Sex Transm Dis. 2020;47:344–346. doi: 10.1097/OLQ.0000000000001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrina MAB, Cosentino LA, Rabe LK, Hillier SL. Susceptibility of bacterial vaginosis (BV)-associated bacteria to secnidazole compared to metronidazole, tinidazole and clindamycin. Anaerobe. 2017;47:115–119. doi: 10.1016/j.anaerobe.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrina MAB, Cosentino LA, Wiesenfeld HC, Darville T, Hillier SL. Susceptibility of endometrial isolates recovered from women with clinical pelvic inflammatory disease or histological endometritis to antimicrobial agents. Anaerobe. 2019;56:61–65. doi: 10.1016/j.anaerobe.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan S, Munch MM, Sizova MV, Fiedler TL, Kohler CM, et al. More easily cultivated than identified: classical isolation with molecular identification of vaginal bacteria. J Infect Dis. 2016;214 Suppl 1:S21–S28. doi: 10.1093/infdis/jiw192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang AT, Cotton S, Sankaran-Walters S, Li C-S, Lee CY, et al. Evidence of an increased pathogenic footprint in the lingual microbiome of untreated HIV infected patients. BMC Microbiol. 2012;12:153. doi: 10.1186/1471-2180-12-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato N, Kakuta M, Hasegawa T, Yamaguchi R, Uchino E, et al. Metagenomic analysis of bacterial species in tongue microbiome of current and never smokers. NPJ Biofilms Microbiomes. 2020;6:11. doi: 10.1038/s41522-020-0121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velásquez-Mejía EP, Carmona JA, et al. Metformin Is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 44.Gaike AH, Paul D, Bhute S, Dhotre DP, Pande P, et al. The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. mSystems. 2020;5:e00578–19. doi: 10.1128/mSystems.00578-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patrone V, Vajana E, Minuti A, Callegari ML, Federico A, et al. Postoperative changes in fecal bacterial communities and fermentation products in obese patients undergoing bilio-intestinal bypass. Front Microbiol. 2016;7:200. doi: 10.3389/fmicb.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frolund M, Falk L, Ahrens P, Jensen JS. Detection of ureaplasmas and bacterial vaginosis associated bacteria and their association with non-gonococcal urethritis in men. PLoS One. 2019;14:e0214425. doi: 10.1371/journal.pone.0214425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson DE, Dong Q, Van der Pol B, Toh E, Fan B, et al. Bacterial communities of the coronal sulcus and distal urethra of adolescent males. PLoS One. 2012;7:e36298. doi: 10.1371/journal.pone.0036298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan S, Chambers LC, Tapia KA, Hoffman NG, Munch MM, et al. Urethral microbiota in men: association of Haemophilus influenzae and Mycoplasma penetrans with nongonococcal urethritis. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesenfeld HC, Meyn LA, Darville T, Macio IS, Hillier SL. A randomized controlled trial of ceftriaxone and doxycycline, with or without metronidazole, for the treatment of acute pelvic inflammatory disease. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DSMZ https://www.dsmz.de/microorganisms/medium/pdf/DSMZ_Medium104.pdf
- 51.Atlas RM. Handbook of Microbiological Media. Fourth Edition. Washington D. C: ASM Press; 2010. [Google Scholar]
- 52.Koransky JR, Allen SD, Dowell VR. Use of ethanol for selective isolation of sporeforming microorganisms. Appl Environ Microbiol. 1978;35:762–765. doi: 10.1128/AEM.35.4.762-765.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song Y, Finegold S, et al. Peptostreptococcus, Finegoldia, Anaerococcus, Peptoniphilus, Veillonella, and other anaerobic cocci. In: Jorgensen J, Pfaller M, Carroll K, Funke G, Landry M, editors. Manual of Clinical Microbiology. 11th edn. Washington D. C: ASM Press; 2015. pp. 909–919. [Google Scholar]
- 54.Summanen P, Baron EJ, Citron DM, Strong CA, Wexler HM, Belmont Wadsworth Anaerobic Bacteriology Manual. 5th ed. Belmont, CA: Star Publishing; 1993. [Google Scholar]
- 55.Miller JM, Rhoden DL. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J Clin Microbiol. 1991;29:1143–1147. doi: 10.1128/JCM.29.6.1143-1147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaucage CM, Onderdonk AB. Evaluation of a prereduced anaerobically sterilized medium (PRAS II) system for identification anaerobic microorganisms. J Clin Microbiol. 1982;16:570–572. doi: 10.1128/JCM.16.3.570-572.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagana Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal Chem. 2015;87:706–715. doi: 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, et al. HMDB 3.0-the human metabolome database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srinivasan S, Morgan MT, Fiedler TL, Djukovic D, Hoffman NG, et al. Metabolic signatures of bacterial vaginosis. mBio. 2015;6:e00204-15. doi: 10.1128/mBio.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wayne PA, editor; Performance Standards for Antimicrobial Susceptibility Testing. 26th ed. 2016. editor. [Google Scholar]
- 62.Workowski KA. Centers for disease control and prevention sexually transmitted diseases treatment guidelines. Clin Infect Dis. 2015;61 Suppl 8:S759–S762. doi: 10.1093/cid/civ771. [DOI] [PubMed] [Google Scholar]
- 63.Hillier SL, Nyirjesy P, Waldbaum AS, Schwebke JR, Morgan FG, et al. Secnidazole treatment of bacterial vaginosis: a randomized controlled trial. Obstet Gynecol. 2017;130:379–386. doi: 10.1097/AOG.0000000000002135. [DOI] [PubMed] [Google Scholar]
- 64.Pentikis H, Adetoro N, Tipping D, Levy S. An integrated efficacy and safety analysis of single-dose secnidazole 2 g in the treatment of bacterial vaginosis. Reprod Sci. 2020;27:523–528. doi: 10.1007/s43032-019-00048-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwebke JR, Morgan FG, Koltun W, Nyirjesy P. A phase-3, double-blind, placebo-controlled study of the effectiveness and safety of single oral doses of secnidazole 2 G for the treatment of women with bacterial vaginosis. Am J Obstet Gynecol. 2017;217:678.e1–67678. doi: 10.1016/j.ajog.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 66.Yoon SH, Ha S-M, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: a taxonomically United database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 68.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. mega X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meier-Kolthoff JP, Klenk HP, Goker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 72.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-R LM K. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. Peer J Prepr. 2016;4:e1900v1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.