Summary
Most human embryos are aneuploid. Aneuploidy frequently arises during the early mitotic divisions of the embryo, but its origin remains elusive. Human zygotes that cluster their nucleoli at the pronuclear interface are thought to be more likely to develop into healthy euploid embryos. Here, we show that the parental genomes cluster with nucleoli in each pronucleus within human and bovine zygotes, and clustering is required for the reliable unification of the parental genomes after fertilization. During migration of intact pronuclei, the parental genomes polarize toward each other in a process driven by centrosomes, dynein, microtubules, and nuclear pore complexes. The maternal and paternal chromosomes eventually cluster at the pronuclear interface, in direct proximity to each other, yet separated. Parental genome clustering ensures the rapid unification of the parental genomes on nuclear envelope breakdown. However, clustering often fails, leading to chromosome segregation errors and micronuclei, incompatible with healthy embryo development.
Keywords: aneuploidy, human embryo, fertilization, chromosome segregation, genome organization, centrosomes, microtubules, spindle, nuclear pore complex, dynein
Graphical abstract
Highlights
-
•
The parental genomes cluster at the pronuclear interface in human and bovine zygotes
-
•
Clustering is driven by centrosomes, which often reside at the pronuclear interface
-
•
Dynein orients chromosomes toward centrosomes via nuclear pore complexes as adaptors
-
•
Clustering defects lead to aneuploidy and micronuclei, impairing embryo development
In human and bovine zygotes, parental genomes cluster and polarize toward each other in a highly error-prone process driven by centrosomes, dynein, microtubules, and nuclear pore complexes. Failure to cluster the parental genomes leads to chromosome segregation errors and micronuclei, which are incompatible with healthy embryo development.
Introduction
Around 50%–70% of human cleavage embryos are aneuploid—they carry an incorrect number of chromosomes (McCoy et al., 2015; van Echten-Arends et al., 2011; Vanneste et al., 2009; Vera-Rodriguez et al., 2015). Most aneuploid embryos do not develop to term, making aneuploidy in embryos a leading cause of miscarriages and infertility (Benkhalifa et al., 2005; Fritz et al., 2001). The majority of aneuploidy is thought to arise during the mitotic divisions of the embryo (Lee and Kiessling, 2017; McCoy et al., 2015). Mitotic errors have been linked to abnormal division events during early embryo development (Fragouli et al., 2013; Kort et al., 2016; Lee and Kiessling, 2017; McCoy, 2017). However, the cellular origins of mitotic aneuploidy remain unclear.
Interestingly, the zygote stage, when the parental genomes are enclosed in two separate pronuclei in the fertilized egg (zygote), seems to be of particular relevance for healthy embryo development. Zygotes that display clustered nucleoli at the interface of the two pronuclei were proposed to be more likely to develop into blastocysts and less likely to give rise to aneuploid embryos (Coskun et al., 2003; Gámiz et al., 2003; Scott, 2003; Tesarik and Greco, 1999). Moreover, the time interval between nuclear envelope breakdown (NEBD) and cytokinesis in the zygote can predict embryo development and the level of aneuploidy (Vera-Rodriguez et al., 2015). However, why the zygote stage is so important for embryo development and aneuploidy is unclear.
Studies in human zygotes are limited by ethical considerations, the lack of available biological material, and legal restrictions. Bovine embryos closely resemble human embryos in their development: they contain centrosomes (Fishman et al., 2018; Navara et al., 1994), display similar timings of early embryonic divisions (Faramarzi et al., 2018; Lequarre et al., 2003; Wong et al., 2010) and similar rates of aneuploidy (Destouni et al., 2016; Lee and Kiessling, 2017), and their embryonic lineages are specified by related mechanisms (Daigneault et al., 2018; Gerri et al., 2020a, 2020b; Simmet et al., 2018). Thus, we employed high-resolution live cell imaging of human zygotes and bovine embryos to identify potential causes of errors during early mammalian embryogenesis.
Results
The parental genomes cluster with nucleoli at the pronuclear interface in live human zygotes
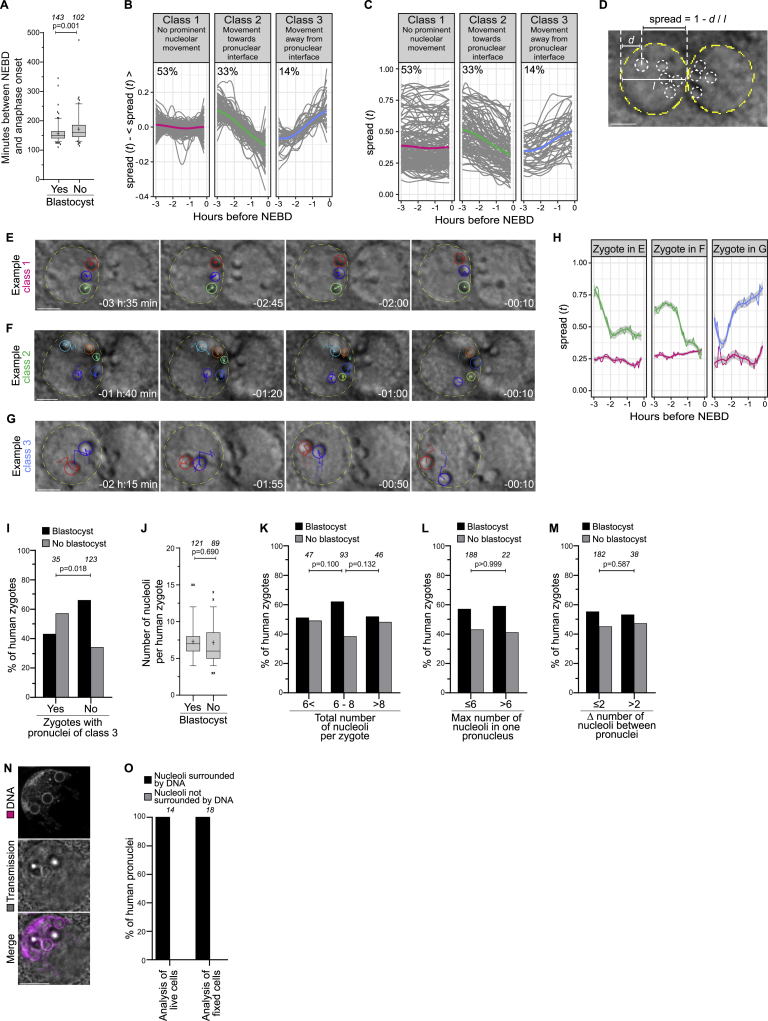
Upon fertilization, the maternal and paternal chromosomes become enclosed in two separate pronuclei in the periphery of the zygote. The pronuclei subsequently migrate to the zygote’s center where they meet, and later undergo NEBD and cell division (Clift and Schuh, 2013). We analyzed time-lapse videos of 245 human embryos and found that the progression from NEBD to cytokinesis was faster in the zygotes that developed into blastocysts compared to those that arrested (Figure S1A), consistent with previous reports (Vera-Rodriguez et al., 2015).
Figure S1.
The parental genomes cluster with nucleoli at the pronuclear interface in live human zygotes, related to Figure 1
(A) Time between NEBD and cytokinesis in human zygotes that develop into blastocysts or developed abnormally.
(B) Classes of nucleolar spread (see (D)) changes during the last 3 hours before NEBD. Each spread trace was smoothed using local regression and mean subtracted. Soft-DTW and k-mean clustering resulted in 3 classes. Class 1 (magenta) contains nuclei with minor changes in nucleolar distribution. Class 2 (green) contains nuclei displaying active nucleolus compaction. Class 3 (blue) contains nuclei displaying active nucleolus decompaction. The percentage numbers indicate the fraction of each class. In total n = 316 nuclei were analyzed (158 zygotes). The colored lines show the barycenter of the classes.
(C) Classes of nucleolar distribution changes in (B) represented without mean subtraction. The colored lines show the barycenter of the classes.
(D) Schematic illustrating calculation of the nucleolar spread.
(E-G) Representative stills from time-lapse movies of zygotes with the indicated pronucleus belonging to class 1 (E), 2 (F), or 3 (G). The tracks of the nucleoli are shown for the indicated pronuclei, see also (H). Time, h:min, 00:00 is NEBD.
(H) Nucleolar spreads during the last 3 hours before NEBD for the zygotes shown in E, F, and G.
(I) Zygotes with or without pronuclei of class 3 that develop into blastocysts or developed abnormally.
(J) Number of nucleoli in zygote that develop into blastocysts or developed abnormally.
(K) Zygotes having less than 6 (< 6), 6, 7, or 8 (6 - 8), or more than 8 (> 8) nucleoli that develop into blastocysts or developed abnormally.
(L) Zygotes having a pronucleus with 6 or less (≤6) or more than 6 (> 6) nucleoli that develop into blastocysts or developed abnormally.
(M) Zygotes having a difference in nucleolar numbers between pronuclei of 2 or less (≤2) or more than 2 (> 2) that develop into blastocysts or developed abnormally.
(N) Representative immunofluorescence images of a zygote with 3 pronuclei. Magenta, DNA (DAPI). Gray, transmission.
(O) Pronuclei in live zygotes (as in Figure 1E) or fixed zygotes (as in (N)) were scored for the presence of DNA around nucleoli.
The number of analyzed zygotes (A, I, J, K, L, M) and pronuclei (F) is specified in italics. p values were calculated using unpaired two-tailed Student’s t test (A, J) and Fisher’s exact test (I, K, L, M). Scale bars, 10 μm.
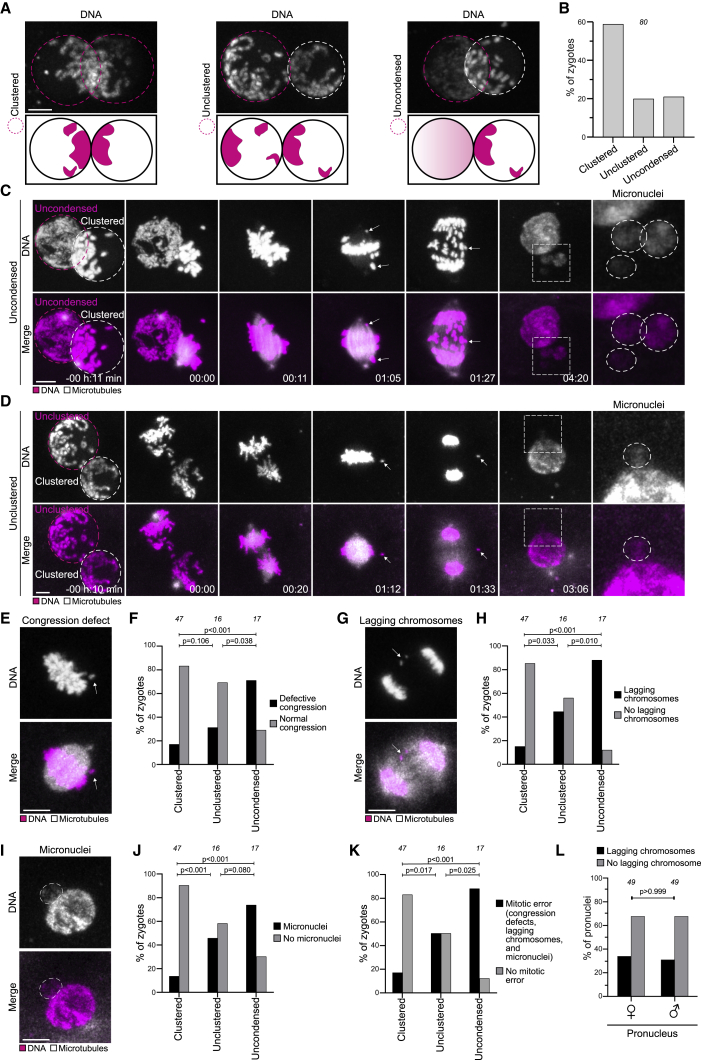
Next, we quantified nucleolus distribution with a semi-automated workflow and found that, at the time of NEBD, 52% of human zygotes had clustered their nucleoli at the interface of the two pronuclei (Figures 1A and 1B; Video S1). Importantly, human zygotes with clustered nucleoli were significantly more likely to develop into blastocysts than zygotes with scattered nucleoli (Figure 1C; Video S1). For 158 zygotes, we were able to measure changes in nucleolus distribution from 3 h before NEBD onward, which corresponds to a time when pronuclear migration is typically completed. In 53% of the pronuclei (class 1), the nucleoli had already reached their final position at this time, often residing in proximity of the pronuclear interface. In 33% of the pronuclei (class 2), the nucleoli moved toward the pronuclear interface, indicating an active compaction process that can last up to shortly before NEBD. In 14% of the pronuclei (class 3), the nucleoli moved away from the pronuclear interface (Figures 1D and S1B–S1G; Video S1). Interestingly, zygotes in this class were less likely to develop into blastocysts than zygotes in the other classes (Figure S1I).
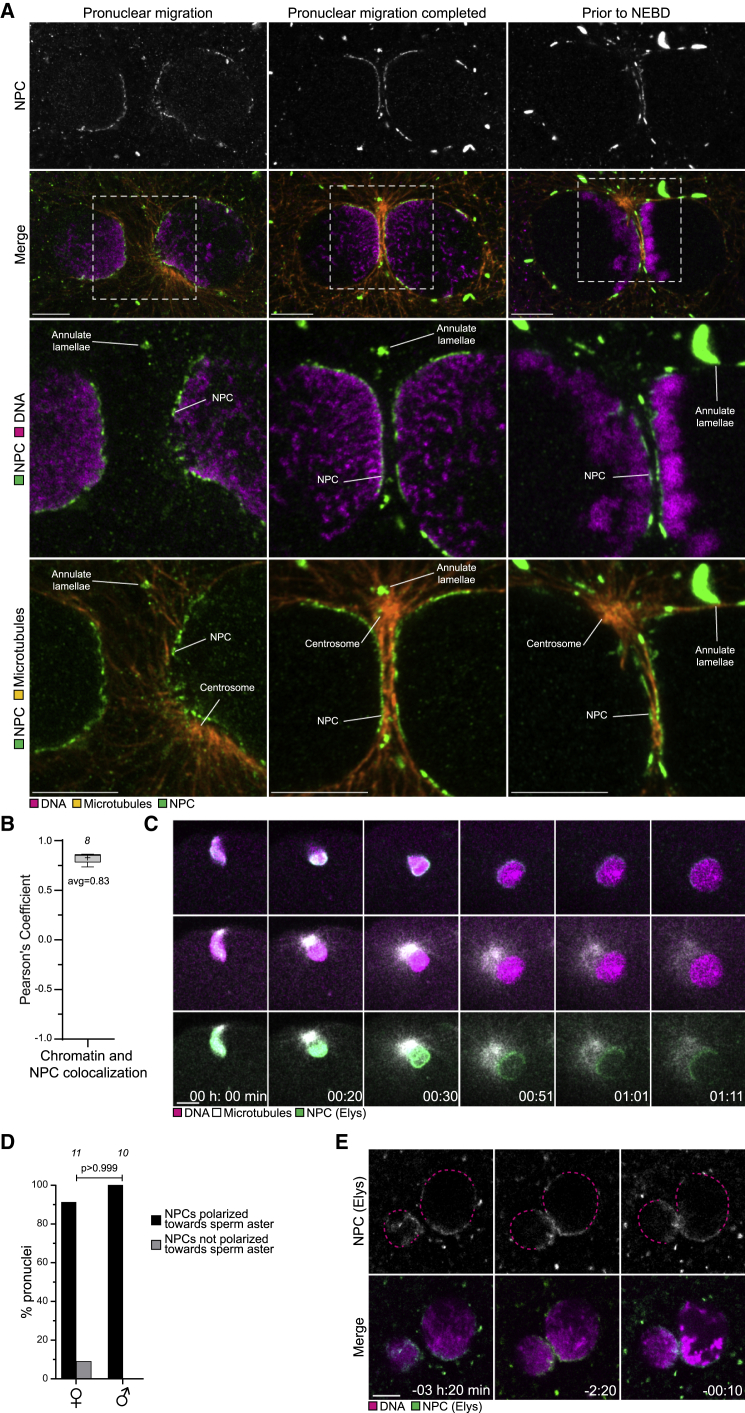
Figure 1.
The parental genomes cluster with nucleoli at the pronuclear interface in live human zygotes
(A) Top: representative stills from time-lapse movies of a human zygote that develops (left) or fails to develop (right) into a blastocyst. Zygotes have clustered (left) or unclustered (right) nucleoli at the pronuclear interface. Middle: magnifications of the regions outlined above. Dashed lines indicate nucleoli and pronuclei. Bottom: schematics of the pronuclei and nucleolar distribution. Time, h:min, 00:00 is NEBD.
(B) Human and bovine zygotes with clustered or unclustered nucleoli (human) or DNA (bovine).
(C) Zygotes with clustered or unclustered nucleoli that develop into blastocyst or abnormally.
(D) Top: representative stills from a time-lapse movie of a zygote. Dashed lines indicate nucleoli and pronuclear outlines. Bottom: schematics of the pronuclei and nucleolar distribution. Arrows indicate a nucleolus that moves toward the pronuclear interface.
(E) Representative stills from time-lapse movies of zygotes. Magenta, DNA (5-580CP-Hoechst). Gray, transmission.
(F) Top: schematics of chromatin organization during pronuclear migration in zygotes. Bottom: representative stills from time-lapse movies of pronuclear migration in zygotes. White, DNA (5-580CP-Hoechst). Outlined regions magnified above. Yellow dashed lines indicate the cell surface. Time, h:min, 00:00 is NEBD. Z projections of 27 (−25:10), 12 (−22:10 and −16:10), 15 (−12:00), and 11 (−06:20 and −00:20) sections every 1.00 μm. Images generated with Imaris 3D view.
(G–I) Schematics (left) and representative stills from time-lapse movies (right) of pronuclei in zygotes before NEBD classified as clustered (G), unclustered (H), and uncondensed (I). White, DNA (5-580CP-Hoechst). Magenta dashed line marks the pronucleus determining the specific category. The arrows in (H) mark chromatin around an unclustered nucleolus. Time, h:min, 00:00 is NEBD. Images generated using Imaris 3D view.
The number of analyzed zygotes (B and C) is specified in italics. p values were calculated using Fisher’s exact test. Scale bars, 10 μm.
Part I: Time-lapse movies of two human embryos. Left embryo at the zygote stage has clustered nucleoli before NEBD and develops into a blastocyst. Right embryo has unclustered nucleoli and develops abnormally. Time, hours:minutes, 00:00 is NEBD in the zygote. Part II: Nucleolar tracking in time-lapse movies of the three human zygotes shown in Figures S1E–S1G. Time, hours:minutes, 00:00 is NEBD in the zygote. Part III: High resolution live cell microscopy of a human zygotes at pronuclear stage. DNA, (5-580CP-Hoechst). Time, hours:minutes, 00:00 is NEBD in the zygote. Part IV: High resolution live cell microscopy of three human zygotes at pronuclear stage. DNA, (5-580CP-Hoechst). Images were generated with Imaris 3D view. Time, hours:minutes, 00:00 is NEBD in the zygote. Part V: Parental genomes remain separated in their respective pronuclei. DNA, (5-580CP-Hoechst). Images were generated with Imaris 3D view.
Together, these data establish that nucleolus clustering is an active process positively associated with human embryo development, as previously suggested (Scott, 2003; Tesarik and Greco, 1999). In contrast, other nucleolar distribution parameters, such as differences in nucleolar numbers between pronuclei, did not correlate strongly with blastocyst formation (Figures S1J–S1M).
By analyzing videos of fluorescently labeled chromatin in live human zygotes, we discovered that chromatin position correlated with the position of nucleoli in each pronucleus (Figures 1E, S1N, and S1O). The chromatin adopted an unusual configuration during pronuclear migration: instead of being homogeneously distributed throughout the pronuclear volume, it became polarized inside the intact pronuclei (Figure 1F; Video S1). In addition, the chromatin became gradually more compact and recruited to the nuclear periphery, localizing within a small volume in each pronucleus multiple hours before NEBD (Figure 1F). Eventually, most of the paternal and maternal chromosomes were highly condensed and had clustered at the interface of the two pronuclei (Figures 1F and 1G; Video S1).
These observations are in sharp contrast to those with intact somatic nuclei and the pronuclei of other systems such as mice, where the chromatin remains uniformly distributed and much less condensed prior to cell division (Courtois et al., 2012; Gönczy et al., 1999; Magidson et al., 2011). Nevertheless, our observation of genome clustering is consistent with previous fixed-cell studies of human zygotes with 2 and 3 pronuclei (Van Blerkom et al., 1995; van de Werken et al., 2014) and is hence not an artifact of live cell microscopy.
Although the parental genomes were in close proximity before NEBD, they remained completely separated in the two apposed pronuclei, as revealed by live super-resolution microscopy (Video S1). This finding suggests that genome editing on one parental genome, using the other as a template, would not be possible at this stage, as previously proposed (Egli et al., 2018; Zuccaro et al., 2020).
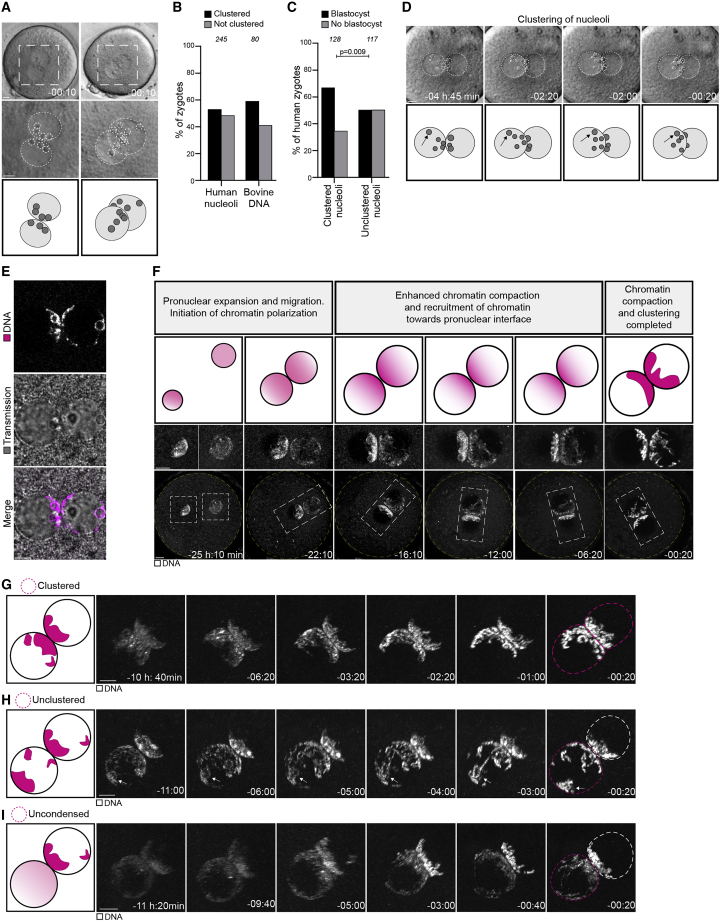
Given that nucleoli do not cluster in all human zygotes, particularly those with compromised development, we asked if chromatin clustering also fails in some cases. Indeed, we observed two types of clustering defects: (1) “unclustered” chromatin that was peripherally localized and condensed but not at the interface of the two pronuclei, including chromatin that previously enclosed a nucleolus (2/8 zygotes) (Figure 1H), and (2) “uncondensed” and unclustered chromatin in one of the two pronuclei (2/8 zygotes) (Figure 1I). Interestingly, differences in chromatin status between the two pronuclei have previously been proposed as a cause for embryo developmental failure (Egli et al., 2011).
Together, our findings reveal that clustering of chromatin and nucleoli at the interface of the pronuclei in zygotes correlates with proper human development.
The parental genomes cluster at the pronuclear interface in bovine zygotes
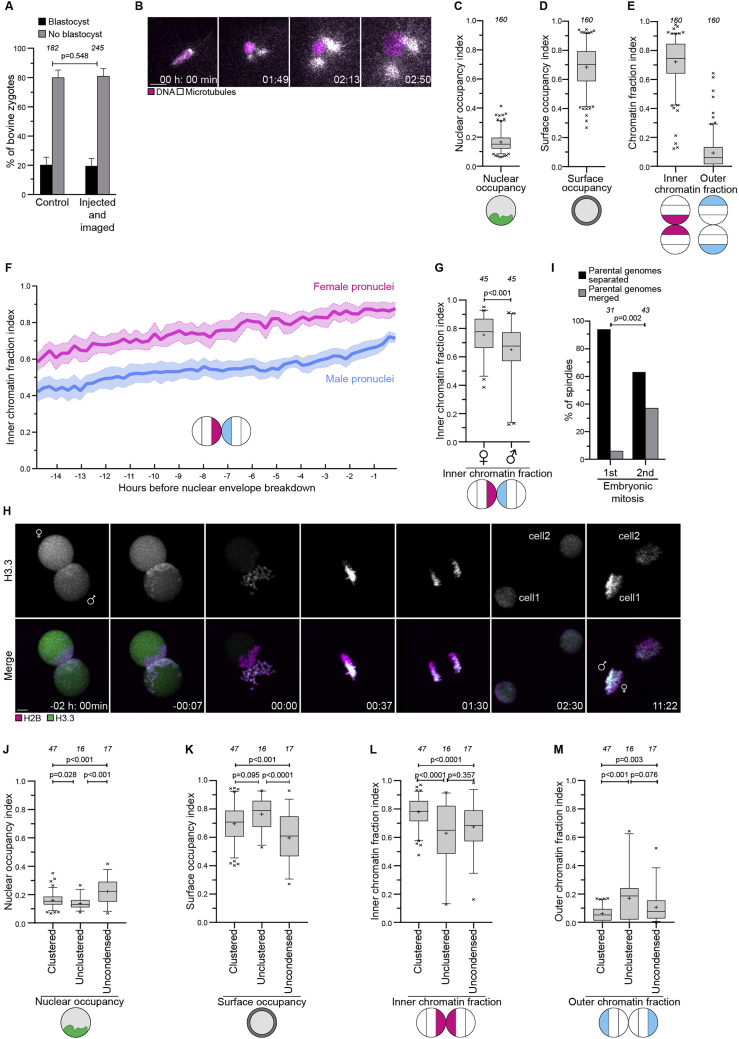
Next, we investigated the function and mechanism of parental genome clustering and its relationship with embryo aneuploidy. Studies in human zygotes are limited by the lack of available biological material and legal restrictions, which in Germany preclude studies of fluorescently labeled human zygotes beyond NEBD. We hence established a high-resolution live cell imaging system for bovine embryos as a model system for human embryos (see STAR Methods; Video S2), confirming that imaged embryos developed into blastocysts with a similar efficiency as non-imaged embryos (Figure S2A).
Figure S2.
The parental genomes cluster at the pronuclear interface in bovine zygotes, related to Figure 2
(A) Bovine zygotes non-injected and non-imaged or injected and imaged that developed into blastocysts or developed abnormally.
(B) Representative stills from time-lapse movies of male pronuclei imaged shortly after egg fertilization. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Time, h:min, 00:00 corresponds to start of image acquisition. First time point shows a single confocal section, while the other time points are Z-projections of 2 (01:49) or 4 (02:13 to 03:14) sections every 2.50 μm.
(C-E) Quantification of chromatin distribution within pronuclei before NEBD using the nuclear occupancy (I), surface occupancy (J), inner and outer chromatin fraction (K), indices.
(F) Inner chromatin fraction indices in female (magenta) and male (cyan) pronuclei. Solid lines represent means of five pronuclei belonging to five zygotes. Shaded areas represent standard error of the mean.
(G) Inner chromatin fraction indices in female (♂) and male (♀) pronuclei before NEBD.
(H) Representative stills from a time-lapse movie of a zygote undergoing cell division. Green, paternal DNA (H3.3-mClover3). Magenta, DNA (H2B-mScarlet). ♀ and ♂ indicate the female and the male pronucleus and the DNA of maternal and paternal origin in the second cell division. Time, h:min, 00:00 is NEBD. Z-projections, 10 sections every 2.50 μm. Scale bar, 10 μm.
(I) Embryos at the indicated stages were scored for spatial separation or merging of the parental genomes on the metaphase spindle.
(J-M) Quantification of chromatin distribution within pronuclei using the nuclear occupancy index (J), surface occupancy index (K), inner chromatin fraction index (L), and outer chromatin fraction index (M) in indicated groups in zygotes.
Data are from five (A, I), eleven (C, D, E, J, K, L, M), three (G), or seven (I) independent experiments. The number of analyzed zygotes (A), pronuclei (C, D, E, G, J, K, L, M), or spindles (I) is specified in italics. p values were calculated using Fisher’s exact test (A, I) and unpaired two-tailed Student’s t test (G, J, K, L, M). Scale bars, 10 μm.
Time-lapse movies of bovine zygotes undergoing the first cell divisions. Time, hours:minutes, 00:00 is NEBD. Part I: A system for high resolution live cell microscopy of early bovine embryo development. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 8 sections every 2.50 μm. Part II: Chromosome segregation errors during the first mitotic division (mild). Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 19 sections every 2.50 μm. Part III: Chromosome segregation errors during the first mitotic division (severe). Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 17 sections every 2.50 μm. Part IV: The maternal and paternal chromosomes occupy partially distinct territories in the metaphase plate of the spindles of bovine zygotes and 2-cell embryos. Time-lapse movie of a zygote expressing mClover3-H3.3B (green, DNA) and H2B-mScarlet (magenta, DNA). ♀ and ♂ indicate the female and the male pronucleus, and the DNA of maternal and paternal origin in the second cell division. Time, hours:minutes, 00:00 is NEBD. Z-projections, 10 sections every 2.50 μm.
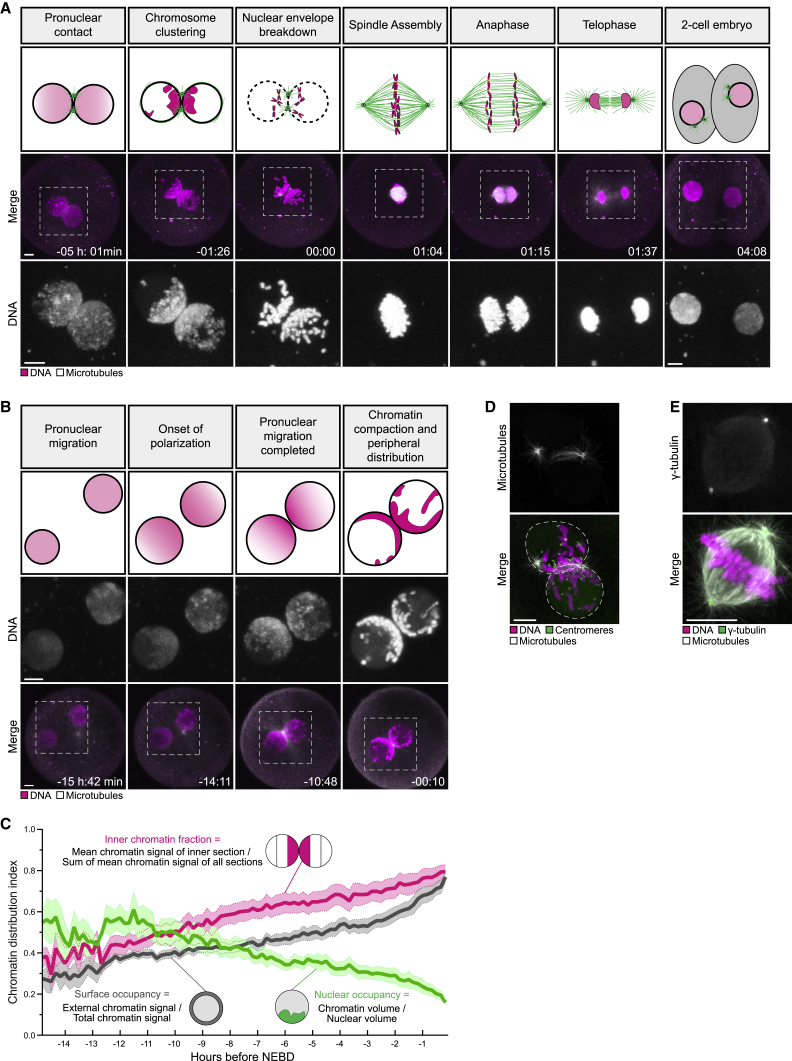
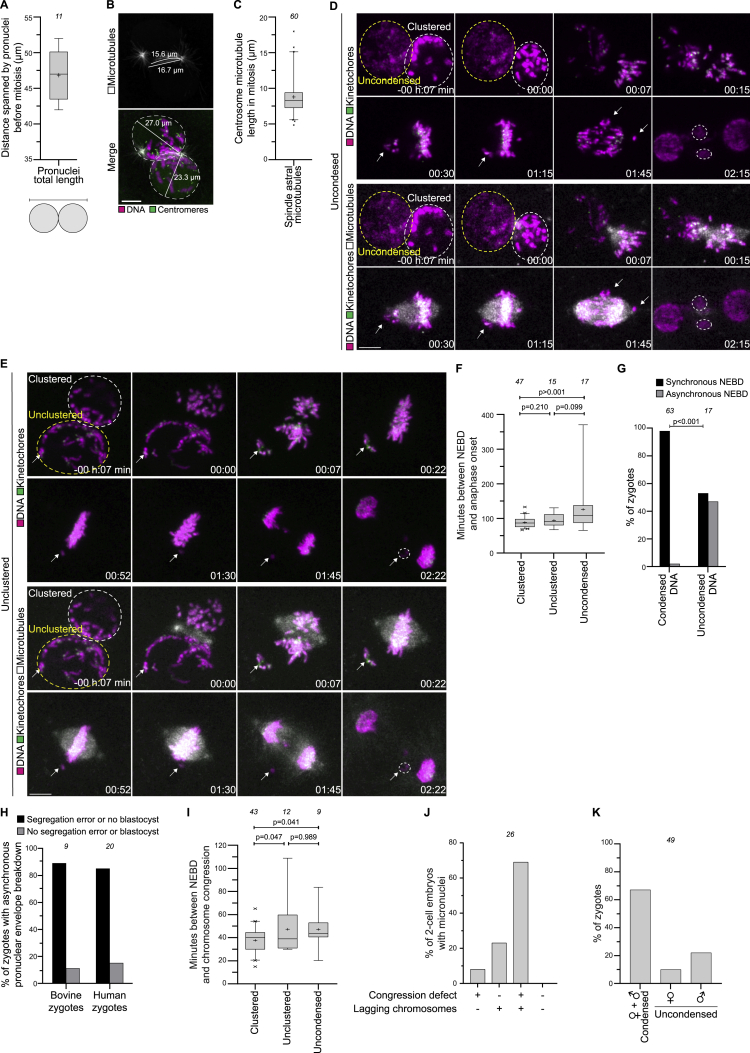
We applied our imaging system to study the first mitotic division in the bovine zygote (Video S2). Upon fertilization, we detected formation of the two pronuclei, pronuclear migration inward, and duplication of the sperm centrosome (Figure S2B). As observed in human zygotes, the chromatin in bovine zygotes became polarized inside intact pronuclei and concentrated in the direction of migration (Figures 2A and 2B). Chromatin polarization initiated early during pronuclear migration and accelerated as the chromosomes condensed during the final 3–4 h before NEBD (Figure 2C). Just before NEBD, the chromatin occupied less than 20% of the nucleus, the chromosomes were at the periphery of the pronuclei, and the parental genomes were clustered at the pronuclear interface (Figures 2A–2C and S2C–S2E). Upon NEBD, the chromosomes were captured by microtubules emanating from the two centrosomes, which served as major microtubule organizing centers and rapidly assembled a spindle with two focused poles (Figures 2D and 2E; Video S2).
Figure 2.
The parental genomes cluster at the pronuclear interface in bovine zygotes
(A) Top: schematics of chromatin organization and spindle assembly in bovine zygotes. Magenta, chromatin. Green, microtubules. Bottom: representative stills from time-lapse movies of bovine zygotes. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Outlined regions are magnified below. Time, h:min, 00:00 is NEBD. Z projections, 11 sections every 2.50 μm.
(B) Top: schematic of chromatin organization during pronuclear migration. Bottom: representative stills from time-lapse movies of pronuclear migration. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Outlined regions magnified above. Time, h:min, 00:00 is NEBD. Z projections, 8 sections every 2.50 μm.
(C) Quantification of chromatin distribution within pronuclei using the inner chromatin fraction index (magenta), nuclear occupancy index (green), and surface occupancy index (gray). Solid lines represent means of ten pronuclei belonging to five zygotes obtained from three independent experiments. Shaded areas represent the standard error of the mean.
(D) Representative immunofluorescence images of a zygote at NEBD. White, microtubules (α-tubulin). Magenta, DNA (DAPI). Green, centromeres (ACA).
(E) Representative immunofluorescence images of a zygotic spindle. White, microtubules (α-tubulin). Magenta, DNA (DAPI). Green, γ-tubulin.
Scale bars, 10 μm.
Interestingly, the chromatin clustered earlier and more strongly in female pronuclei than in male pronuclei (Figures S2F and S2G), similar to nucleolus clustering in human zygotes (Coticchio et al., 2018). To distinguish the behavior of female and male chromosomes, we imaged zygotes expressing a marker for Histone 3.3 (Santenard et al., 2010), which stains only the male chromatin. Using this tool, we found that the maternal and paternal chromosomes continued to occupy partially distinct territories upon NEBD and on the first mitotic spindle, and in some cases also during the following mitotic divisions (Figures S2H and S2I; Video S2), consistent with work in C. elegans, mouse, and humans (Bolková and Lanctôt, 2016; Mayer et al., 2000; van de Werken et al., 2014).
Parental genome clustering promotes accurate chromosome segregation
Similar to human zygotes, some bovine zygotes failed to cluster and to condense the parental genomes. We manually identified zygotes with an uncondensed pronucleus based on their chromatin configuration in the frame before NEBD (Figure 3A) and discerned zygotes with an unclustered pronucleus from zygotes with clustered pronuclei using algorithms that quantify chromatin distribution (see STAR Methods) (Figures 2C, 3A, and S2J–S2M). Overall, the fraction of bovine zygotes with clustered chromosomes was very similar to the fraction of human zygotes with clustered nucleoli (Figures 1B and 3B) and in line with the fraction of human zygotes with clustered chromatin reported above.
Figure 3.
Parental genome clustering promotes accurate chromosome segregation in bovine zygotes
(A) Representative stills from time-lapse movies (top) and schematics (bottom) of pronuclei in bovine zygotes classified as clustered (left); unclustered (middle); uncondensed (right). White, DNA (H2B-mScarlet). Magenta dashed line marks the pronucleus determining the specific category. Z projections, 10 sections every 2.50 μm.
(B) Frequency of clustering defects in zygotes.
(C and D) Representative stills from time-lapse movies of zygotes before and after NEBD. (C) A zygote classified as uncondensed. (D) A zygote classified as unclustered. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Arrows indicate misaligned and lagging chromosomes that form micronuclei, as highlighted by dashed box (magnification on the right). Time, h:min, 00:00 is NEBD. Z projections, 12 sections every 2.50 μm.
(E–K) Representative images and frequencies of zygotes in indicated groups having defective chromosome congression or unattached chromosomes at metaphase (E and F), lagging or unattached chromosomes during anaphase (G, H), micronuclei in 2-cell embryos (I and J), and abnormal mitosis (K). White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Z projections of 5 (E and I) and 4 (G) sections every 2.50 μm.
(L) Frequency of lagging chromosomes originating from female (♀) and male (♂) pronuclei.
Data are from eleven (B, F, H, J, and K) or five (L) independent experiments. The number of analyzed zygotes (B, F, H, J, and K) or pronuclei (L) is specified in italics. p values were calculated using Fisher’s exact tests. Scale bars, 10 μm.
See also Figures S3 and S4 and Videos S3 and S4.
The fact that human zygotes with clustered nucleoli are more efficient in developing into blastocysts than zygotes with unclustered nucleoli suggested an important function for nucleolar and parental genome clustering. Given that the two bovine pronuclei spanned ∼47 μm (Figure S3A), which is much longer than the 5–10 μm diameter of a typical mammalian somatic nucleus (Milo et al., 2010; Milo and Phillips, 2016), and longer than the mitotic microtubules in bovine zygotes (8.7 ± 2.6 μm) (Figures S3B and S3C), we hypothesized that chromosome capture of uniformly distributed (unclustered) chromosomes would be inefficient. On the other hand, clustering the chromosomes at the pronuclear interface reduces their volume and could facilitate chromosome capture.
Figure S3.
Defective clustering leads to a delay in chromosome congression in bovine zygotes, related to Figure 3
(A) Distance spanned by pronuclei before NEBD in bovine zygotes.
(B) Adaptation of Figure 2G with measurements of pronuclear diameter and of microtubule length.
(C) Length of astral microtubules at metaphase in zygotes.
(D-E) Representative stills from time-lapse movies of zygotes classified as uncondensed (D) and unclustered (E). White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-miRFP). Green, kinetochores (mScarlet-hCenpC). Dashed lines indicate pronuclei with uncondensed or unclustered chromatin (yellow) and clustered chromatin (white). Arrows point to uncondensed or distal chromosomes that join the metaphase plate later. Several of these chromosomes subsequently form micronuclei, highlighted by white dashed lines. Time, h:min, 00:00 is NEBD. Z-projections, respectively 4 and 7 sections every 3.08 μm. Scale bars, 10 μm.
(F) Time between NEBD and anaphase onset in indicated zygote groups.
(G) Zygotes in indicated groups that underwent NEBD synchronously or asynchronously.
(H) Bovine and human zygotes with asynchronous NEBD (delay between pronuclei > 10 min) were scored for the presence of chromosome segregation errors (bovine zygotes) or development into blastocysts (human zygotes).
(I) Time between NEBD and the completion of chromosome congression on the metaphase plate in indicated zygote groups. Zygotes that failed to align all chromosomes before anaphase onset were excluded.
(J) 2-cell embryos with micronuclei that displayed chromosome congression defects and/or lagging chromosomes during zygote mitosis.
(K) Zygotes entering mitosis with two condensed pronuclei or with only the female (♀) or male (♂) pronucleus being uncondensed.
Data are from two (A), eleven (F, G, H, I, J), and five (K) independent experiments. Data in (C) are from three zygotes obtained from two independent experiments. The number of analyzed zygotes (A, F, G, H, I, K), microtubules (C), and 2-cell embryos (J) is specified in italics. p values were calculated using unpaired two-tailed Student’s t test. Scale bars, 10 μm.
To test this hypothesis, we followed the clustered, unclustered, and uncondensed groups of zygotes as they progressed through the first mitotic division (Figures 2A, 3C, 3D, S3D, and S3E; Videos S2, S3, and S4) and scored for phenotypes associated with defective chromosome capture, including compromised congression on the metaphase plate or unattached chromosomes, lagging or unattached chromosomes during anaphase, and formation of micronuclei (Figures 3E–3J).
Time-lapse movies of zygotes having one uncondensed pronucleus undergoing mitosis. Arrowheads indicate chromosomes that fail to align at the metaphase plate, lag after anaphase, and eventually form micronuclei, dotted circles. Time, hours:minutes, 00:00 is NEBD. Part I: Zygotes with an uncondensed pronucleus have chromosome segregation errors. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 12 sections every 2.50 μm. Part II: Kinetochores in the uncondensed pronucleus are captured less efficiently than those at the surface of condensed pronuclei. Zygote expressing mScarlet-hCenpC (green, kinetochores), mClover3-MAP4-MTBD (white, microtubules) and H2B-miRFP (magenta, DNA). Z-projections, 4 sections every 3.08 μm. Part III: Chromosome segregation errors originate from uncondensed pronucleus. Zygote expressing mClover3-H2B (green, DNA) and H2B-mScarlet (magenta, DNA). Rectangle indicates region bleached on the condensed pronucleus, using the 561 nm laser line. Z-projections, 10 sections every 1.76 μm.
Time-lapse movies of zygotes having one unclustered pronucleus undergoing mitosis. Arrowheads indicate chromosomes that fail to align at the metaphase plate, lag after anaphase, and eventually form micronuclei, dotted circles. Time, hours:minutes, 00:00 is NEBD. Part I: Zygotes with an unclustered pronucleus and chromosome segregation errors. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 12 sections every 2.50 μm. Part II: Chromosomes that do not cluster at the pronuclear interface congress late and segregate incorrectly. Zygote expressing mScarlet-hCenpC (green, kinetochores), mClover3-MAP4-MTBD (white, microtubules) and H2B-miRFP (magenta, DNA). Z-projections, 4 sections every 3.08 μm. Part III: Chromosomes distal from the pronuclear interface missegregate. Zygote expressing mClover3-H2B (green, DNA) and H2B-mScarlet (magenta, DNA). Rectangle indicates region bleached at pronuclear interface, using the 561 nm laser line. Z-projections, 17 sections every 1.76 μm
The vast majority of zygotes with an uncondensed pronucleus showed serious defects during mitosis (Figure 3K). Chromosomes from the uncondensed pronucleus showed delays in both association with the spindle and congression on the metaphase plate (Figures 3C and S3D; Video S3). Indeed, 71% of uncondensed zygotes failed to congress all their chromosomes on the metaphase plate by the time of anaphase onset (Figure 3F), and anaphase onset was significantly delayed (Figure S3F). Additionally, 88% of these zygotes had multiple lagging chromosomes during anaphase (Figure 3H).
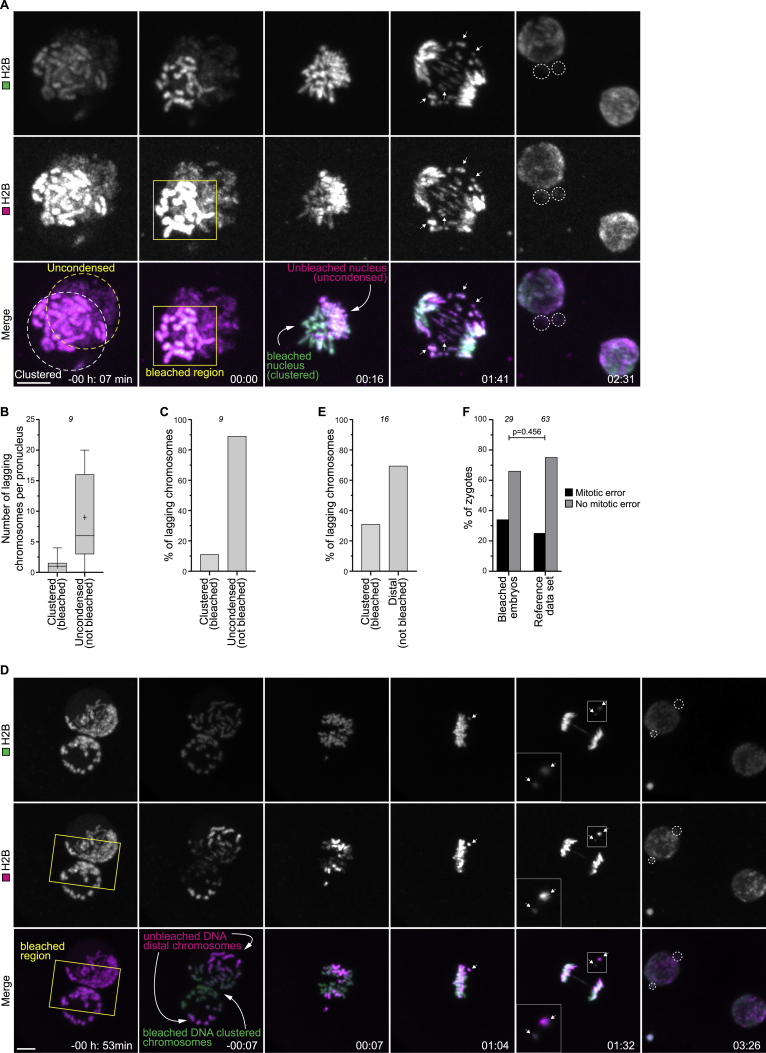
To investigate which pronucleus gives rise to the lagging chromosomes, we selectively photobleached the chromosomes in the clustered pronucleus only (Figure S4A; Video S3). Strikingly, 89% of lagging chromosomes originated from the unbleached, uncondensed pronucleus (Figures S4B and S4C; Video S3). Moreover, NEBD was highly asynchronous in 47% of the zygotes with an uncondensed pronucleus but in only 2% of zygotes with two condensed pronuclei (Figure S3G). Consistently, asynchronous NEBD was observed in 8% of human zygotes and prevented blastocyst formation in over 80% of these cases (Figure S3H), similar to previous observations (Coticchio et al., 2018). Additionally, chromosomes within an uncondensed pronucleus may not yet be fully mature for mitosis, as previously proposed for zygotes undergoing mitosis after somatic nuclear transfer (Chia et al., 2017).
Figure S4.
In bovine zygotes, unclustered chromosomes are more likely to missegregate, related to Figure 3
(A) Representative stills from time-lapse movies of a bovine zygote with one pronucleus having uncondensed chromatin (yellow dashed line). The H2B-mScarlet chromatin signal in the pronucleus with clustered chromatin (left) was bleached upon NEBD in the region indicated by the yellow rectangle. The bleached DNA has a green signal, while the unbleached DNA is visible both in green and magenta. Lagging chromosomes are magenta and green (arrows), indicating that they originated from the uncondensed pronucleus. Green, DNA (H2B-mClover3). Magenta, DNA (H2B-mScarlet). Dashed lines indicate micronuclei. Time, h:min, 00:00 is NEBD. Z-projections, 9 sections every 1.76 μm.
(B) Number of lagging chromosomes that originated from pronuclei with clustered DNA (bleached) or uncondensed DNA (not bleached).
(C) Percentage of total lagging chromosomes that originated from pronuclei with clustered DNA (bleached) or uncondensed DNA (not bleached).
(D) Representative stills from time-lapse movies of a zygote with unclustered chromosomes. The H2B-mScarlet chromatin signal between pronuclei was bleached before NEBD in the region indicated by the yellow rectangle. Lagging chromosomes are magenta and green (arrows), indicating that they were not bleached and were peripheral chromosomes. Green, DNA (H2B-mClover3). Magenta, DNA (H2B-mScarlet). Dashed lines indicate micronuclei. Time, h:min, 00:00 is NEBD. Z-projections, 18 sections every 1.76 μm.
(E) Percentage of total lagging chromosomes that originated from clustered DNA (bleached) or distal DNA (not bleached).
(F) Zygotes displaying mitotic errors after bleaching and in reference dataset (Figure 2J, merge of clustered and unclustered groups).
Data are from four (B, C, E, F-bleached embryos) and eleven (F-reference dataset) independent experiments. The number of analyzed zygotes is specified in italics. p values were calculated using Fisher’s exact test. Scale bars, 10 μm.
Zygotes with an unclustered pronucleus also showed prominent defects in mitosis. Chromosomes in the distal regions of the pronuclei often remained separated from the main chromosome cluster upon NEBD, and the time of chromosome congression on the metaphase plate was delayed (Figures S3E and S3I; Video S4). Over 30% of the zygotes in this class had chromosomes outside the spindle, yet in proximity of the metaphase plate (Figures 3E and 3F). In addition, 50% of zygotes with an unclustered pronucleus had lagging chromosomes during anaphase, indicating incorrect attachment to microtubules (Figure 3H). Interestingly, anaphase onset was not significantly delayed (Figure S3F), revealing that zygotes can progress into anaphase without delay despite incorrect kinetochore-microtubule attachments.
To test if the unclustered chromosomes were more likely to lag in these zygotes, we selectively photobleached the clustered chromosomes at the pronuclear interface leaving the distal chromosomes unbleached (Figure S4D; Video S4). Consistent with our hypothesis, 69% of lagging chromosomes were unbleached and thus originated distal from the pronuclear interface (Figure S4E). Bleaching did not harm the zygotes, given that most bleached zygotes lacked chromosome segregation defects, similar to untreated zygotes (Figure S4F). Together, these data show that distal chromosomes are more likely to lag during anaphase than clustered chromosomes. Consistent with the bleaching experiments, high resolution microscopy showed that the capture of distal chromosomes is more difficult, and in some cases, these chromosomes fail to join the spindle (Figure S3E) and subsequently lag behind during anaphase.
Lagging chromosomes are generally more frequent in zygotic mitosis than in mitosis of somatic cells (Thompson and Compton, 2008), indicating an increased rate of abnormal kinetochore-microtubule attachments. The lagging or unattached chromosomes were often subsequently encapsulated in micronuclei (Figures 3I and 3J). Most embryos with micronuclei had both chromosome congression defects and lagging chromosomes (Figure S3J). Consistently, the frequency of micronuclei in uncondensed zygotes was significantly increased in comparison to clustered zygotes (Figure 3J). Importantly, micronuclei in human embryos correlate with developmental failure (Kort et al., 2016; Meriano et al., 2004).
In total, 90% of zygotes with an uncondensed pronucleus and 50% of zygotes with an unclustered pronucleus had defects during the first mitotic division (Figure 3K). In stark contrast, only 20% of zygotes with two clustered pronuclei had defects (Figure 3K). Thus, parental genome clustering at the pronuclear interface promotes accurate chromosome segregation and prevents the formation of micronuclei.
Selective labeling of the paternal chromosomes with the marker H3.3 revealed that there was no significant difference between the frequencies of paternal or maternal chromosome segregation errors (Figure 3L). The paternal pronucleus was slightly more likely to be uncondensed than the maternal pronucleus, but this difference was not significant (Figure S3K).
Centrosome positions determine the sites of chromosome clustering and accuracy of chromosome segregation
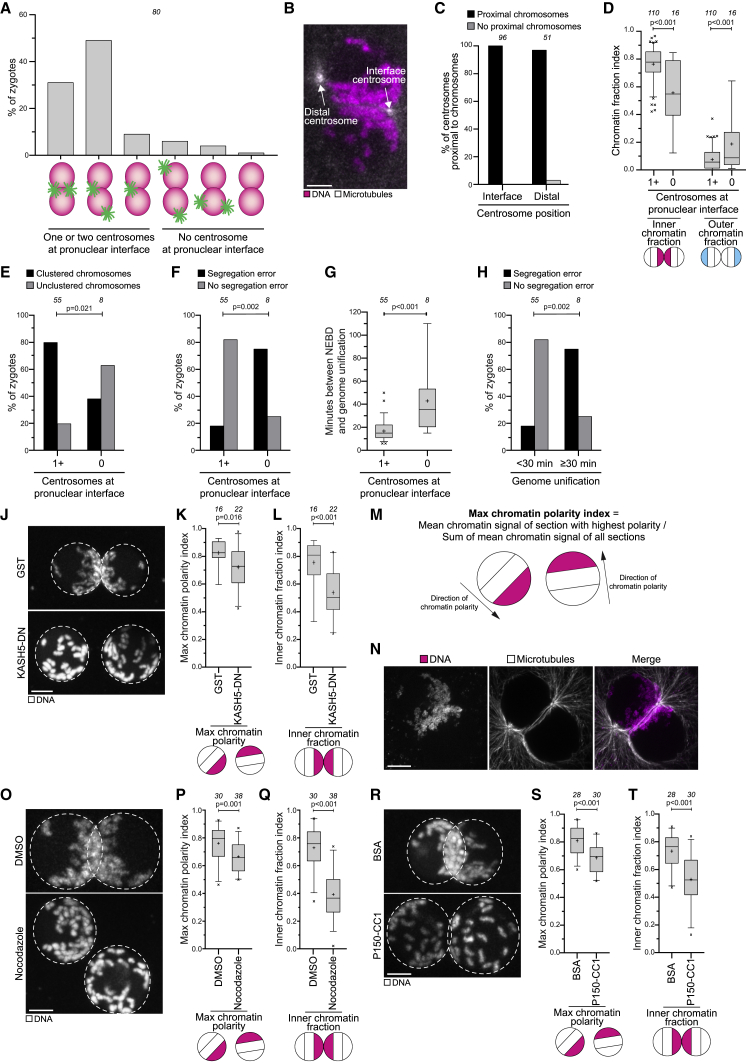
Next, we investigated the mechanism of parental genome clustering. The chromosomes normally clustered at the position of the duplicated centrosomes (Figure 2A; Video S2). In the vast majority of zygotes, one or both centrosomes were located at the pronuclear interface (Figure 4A) and were always associated with chromosomes (Figures 4B and 4C). One of the two centrosomes was sometimes localized distal from the pronuclear interface but was also associated with chromosomes in 98% of all cases (Figures 4B and 4C). Interestingly, zygotes that lacked centrosomes at the pronuclear interface also typically failed to cluster the parental genomes at the pronuclear interface (Figures 4D and 4E), were more likely to have chromosome segregation errors (Figure 4F), and took longer to unify their parental genomes after NEBD (Figure 4G). Zygotes with a delay in genome unification were also generally more likely to have mitotic errors (Figure 4H), emphasizing the importance of rapidly capturing chromosomes and unifying the parental genomes upon NEBD. Together, these data suggest that centrosomes determine the sites of chromosome clustering and their presence at the pronuclear interface promotes rapid and error-free unification of the parental genomes.
Figure 4.
In bovine zygotes, centrosome positions determine the sites of chromosome clustering and accuracy of chromosome segregation
(A) Centrosome localization before NEBD in bovine zygotes.
(B) Representative still from a time-lapse movie of a zygote before NEBD. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Arrows specify distal and the interface centrosomes.
(C) Distal and interface centrosomes as in (B) were scored for proximity to chromosomes.
(D) Inner and outer chromatin fraction indices in zygotes with or without centrosomes at the pronuclear interface.
(E) Chromosome clustering was scored in zygotes with or without centrosomes at the pronuclear interface.
(F) Zygotes with or without centrosomes at the pronuclear interface were scored for the presence of chromosome segregation errors.
(G) Time between NEBD and the unification of the parental genomes in zygotes with or without centrosomes at the pronuclear interface.
(H) Zygotes in which parental genome unification took place within 30 min after NEBD or later were scored for the presence of chromosome segregation errors.
(J) Representative images of zygotes expressing GST or KASH5-DN. White, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Z projections, 9 sections every 3.08 μm.
(K and L) Max chromatin polarity (see M) and inner chromatin fraction indices in zygotes expressing GST or KASH5-DN.
(M) Schematic illustrating calculation of the max chromatin polarity index.
(N) Representative immunofluorescence images of a zygote with microtubules wrapped around the pronuclei. White, microtubules (α-tubulin). Magenta, DNA (DAPI). Z projection, 35 sections every 0.1 μm.
(O–T) Representative images, max chromatin polarity and inner chromatin fraction indices in zygotes treated with DMSO or nocodazole before pronuclear juxtaposition (O–Q) or injected with BSA or P150-CC1 (R–T). The last time point before NEBD is shown. White, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Z projections, 8 (O) and 12 (R) sections every 2.50 μm.
Zygotes having a pronucleus with uncondensed chromatin at NEBD were excluded from the quantifications in (C)–(H), (K), (L), (S), (T), (P), and (Q) to avoid accounting for the role of incomplete chromosome condensation at NEBD. Data are from eleven (A–H), four (K, L, S, and T), or six (P and Q) independent experiments. The number of analyzed zygotes (A and E–H), centrosomes (C), and pronuclei (D, K, L, S, T, P, and Q) is specified in italics. p values were calculated using unpaired two-tailed Student’s t test (D, G, K, L, S, T, P, and Q) and Fisher’s exact test (E, F, and H). Scale bar, 10 μm.
See also Figures S5 and S7 and Video S5.
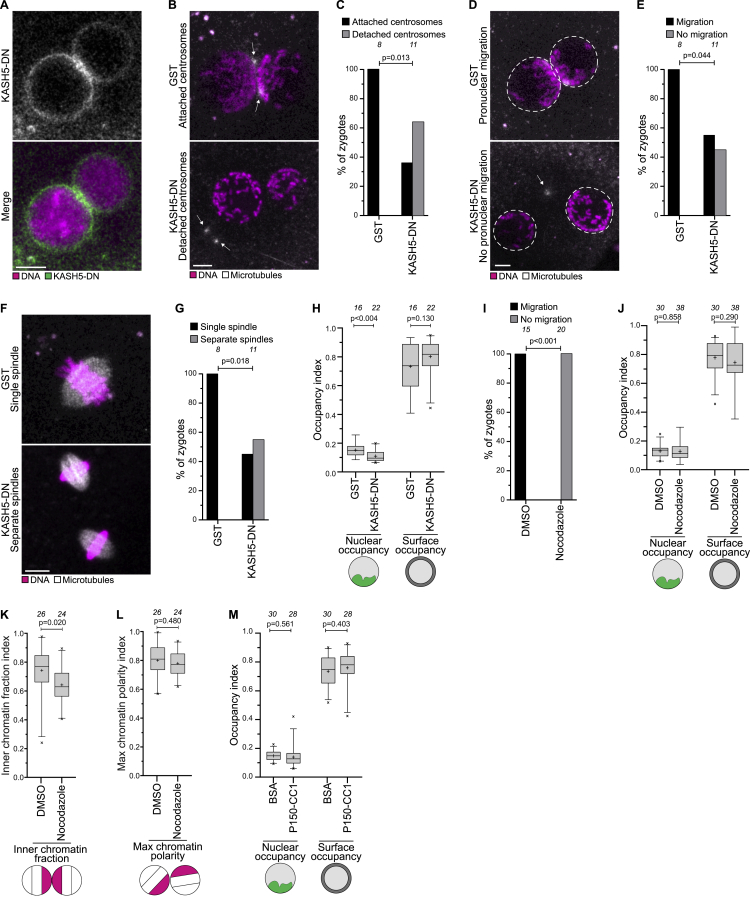
To test directly if centrosomes drive chromosome clustering, we aimed to detach the centrosomes from the pronuclei. According to our hypothesis, this disruption should lead to defects in chromosome clustering. Work in C. elegans and zebrafish zygotes has established that centrosomes are coupled to nuclei through the linker of nucleoskeleton and cytoskeleton (LINC) complex (Bone and Starr, 2016; Lindeman and Pelegri, 2012). This coupling requires the KASH5 subunit of the LINC complex, which can be blocked with a KASH5 dominant-negative (KASH5-DN) fragment (Stewart-Hutchinson et al., 2008). KASH5-DN caused the displacement of centrosomes from the nuclear envelope, and in addition failure in pronuclear migration, and, eventually, assembly of two separate spindles (Figures S5A–S5G; Video S5).
Figure S5.
Effects of KASH5-DN, nocodazole, and P150-CC1 treatments in bovine zygotes, related to Figure 4
(A) Representative image of a bovine zygote expressing mClover3-KASH5-DN (green) and H2B-mScarlet (DNA, magenta).
(B-G) Representative images and quantification of zygotes in indicated groups displaying, upon KASH5-DN treatment, detached centrosomes (B and C), pronuclear migration defects (D and E) or separate spindles at anaphase onset (F and G). White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Arrows indicate detached centrosomes and dashed lines indicate pronuclear envelopes. Z-projection, respectively 4, 7, 5, 8, 4, and 10 sections every 2.50 μm.
(H) Nuclear and surface occupancy indices in zygotes expressing GST or KASH5-DN.
(I) Zygotes treated with DMSO or nocodazole before pronuclear juxtaposition were scored for pronuclear migration defects.
(J) Nuclear and surface occupancy indices in zygotes treated with DMSO or nocodazole before pronuclear juxtaposition.
(K-L) Inner chromatin fraction and max chromatin polarity indices in zygotes treated with DMSO or nocodazole after pronuclear juxtaposition.
(M) Nuclear and surface occupancy indices in zygotes injected with BSA or P150-CC1.
Data are from four (C, E, G, H, M), six (I,J), three (K,L) independent experiments. The number of analyzed zygotes (C, E, G, I) and pronuclei (H, J, K, L, M) are specified in italics. p values were calculated using Fisher’s exact test (C, E, G, I) and unpaired two-tailed Student’s t test (H, J, K, L, M). Scale bars, 10 μm.
Time-lapse movies of zygotes under different perturbation conditions to affect the function of centrosomes, microtubules, or dynein. Time, hours:minutes, 00:00 is NEBD. Part I: Centrosomes are required for chromosome clustering. Three zygotes expressing mClover3-MAP4-MTBD (white, microtubules), H2B-mScarlet (magenta, DNA), and GST (left) or KASH5-DN (middle and right). Arrowheads indicate detached centrosomes before and after NEBD. Z-projections, 9 sections every 3.08 μm. Part II: Microtubules are required for chromosome clustering. Two zygotes expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA) treated with DMSO (left) or nocodazole (right). Z-projections, 7 (left) and 8 (right) sections every 2.50 μm. Part III: Dynein is required for chromosome clustering. Two zygotes expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA) injected with BSA (left) or P150-CC1 (right). Z-projections, 8 (left) and 13 (right) sections every 2.50 μm
We then asked if the detachment of centrosomes affects the polarization of chromatin inside the two pronuclei. KASH5-DN zygotes displayed significantly decreased chromatin polarization, showing that centrosomes contribute to chromatin polarization within the pronuclei (Figures 4J and 4K; Video S5). Importantly, recruitment of chromosomes to the nuclear envelope was not affected (Figure S5H). To score if the centrosomes also function in orienting the polarized chromatin toward each other, we next quantified the fraction of chromatin in the inner regions of the two pronuclei that were closest to each other. KASH5-DN zygotes displayed a significantly lower inner chromatin fraction, showing that centrosomes orient the polarized chromatin toward the pronuclear interface (Figures 4J and 4L; Video S5).
Chromosome clustering at centrosomes is driven by dynein and microtubules
Next, we investigated how centrosomes direct chromosome clustering. Centrosomes are active sites of microtubule nucleation, and microtubules wrapped around the pronuclei (Figure 4N). To test whether microtubules are required for chromosome clustering, we treated zygotes with the microtubule-depolymerizing drug nocodazole. Nocodazole significantly decreased chromatin polarization (Figures 4O and 4P), and blocked pronuclear migration (Figure S5I; Video S5). Moreover, the inner chromatin fraction was strongly reduced, showing that the parental genomes were no longer oriented toward each other (Figures 4O and 4Q). Consistent with the effects of KASH5-DN, chromosomes still relocated to the nuclear envelope, showing that the recruitment of chromosomes into the nuclear periphery is independent of microtubules and centrosomes (Figure S5J). Interestingly, adding nocodazole after pronuclear juxtaposition also reduced the inner chromatin fraction (Figure S5K), but had no significant effect on the polarization of chromatin (Figure S5L). These data show that chromatin polarization is completed by the end of pronuclear migration, but that the orientation of the polarized parental genomes toward each other is further adjusted on pronuclear juxtaposition.
Transport toward centrosomes in interphase is typically mediated by the minus-end directed motor protein dynein (Malone et al., 2003; Quintyne et al., 1999). Thus, we hypothesized that dynein might transport the chromosomes to the centrosome via microtubules. To test for an involvement of dynein, we purified and injected the C terminus of the dynein interaction partner dynactin (P150-CC1), which blocks the activity of the dynein-dynactin complex (Quintyne et al., 1999). P150-CC1 led to a significant reduction of chromatin polarization and of the inner chromatin fraction at the pronuclear interface (Figures 4R–4T and S5M; Video S5). Overall, our data show that dynein mediates the clustering of parental genomes toward the centrosomes via cytoplasmic microtubules.
Nuclear pore complexes cluster with chromatin at the pronuclear interface
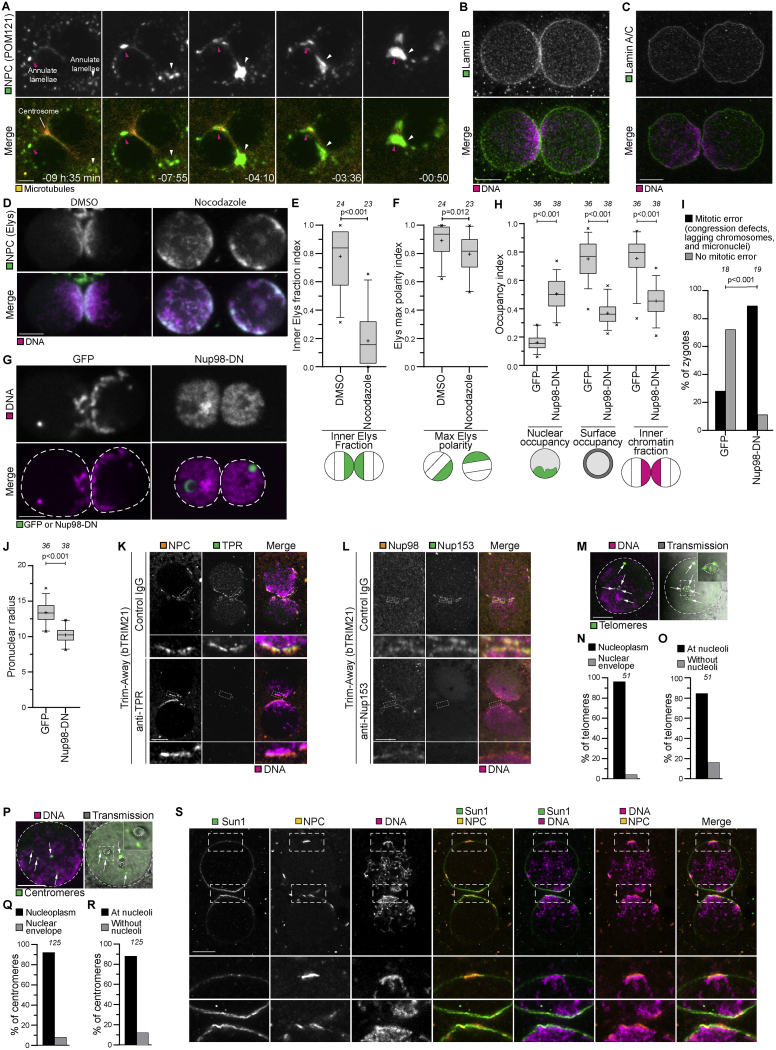
We next set up to identify the adaptor that connects cytoplasmic dynein with chromosomes across the nuclear envelope. The nuclear pore complexes (NPCs) are prominent structures that bridge the nuclear envelope and bind to chromatin as well as cytoplasmic dynein. NPCs associate with chromatin in somatic cells (Capelson, 2018; Liang et al., 2013; Sood and Brickner, 2014), and interactions between chromatin and NPCs have also been reported for later stage bovine embryos (Popken et al., 2015). Moreover, NPC components were reported to co-immunoprecipitate with the dynein complex subunit dynactin in bovine zygotes, and antibodies against components of the NPC interfere with dynein-dependent pronuclear migration (Payne et al., 2003). Consistent with dynein acting on NPCs, we observed transport of NPCs toward centrosomes in live zygotes (Figure S6A; Video S6). Although individual pores could not be resolved on intact nuclei, annulate lamellae, which are membrane stacks enriched in NPCs, were prominently recruited toward centrosomes as the two pronuclei moved inward (Figure S6A; Video S6, arrowheads).
Figure S6.
Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes, related to Figure 5
(A) Representative stills from time-lapse movies of bovine zygotes expressing mClover3-MAP4-MTBD (microtubules, orange) and POM121-mScarlet (NPCs, green). Magenta and white arrowheads indicate two patches of annulate lamellae moving toward centrosomes and pronuclear interface. Time, h:min, 00:00 is NEBD. Single confocal microscopy sections.
(B-C) Representative immunofluorescence images of zygotes stained with Lamin B (B) or Lamin A/C (C) (Green) and DNA (DAPI, Magenta). Single sections Airyscan microscopy (B) and single sections confocal microscopy (C).
(D) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green) and H2B-mScarlet (DNA, magenta) treated with DMSO or nocodazole before pronuclear expansion. The time point at 4 hours before NEBD is shown. Z-projections of 2 (left) and 3 (right) sections every 2.50 μm.
(E-F) Inner Elys polarity and max Elys polarity indices at 4 hours before NEBD in zygotes treated with DMSO or nocodazole before pronuclear expansion. Analysis was not possible later because Elys redistributed to the chromatin before NEBD. Note that nocodazole was added after the formation of the midbody from the meiosis II spindle. Midbody microtubules are known to be very stable and, based on our experiments in parthenotes (Figures S7), might contribute to residual NPC polarization in nocodazole treated zygotes.
(G) Representative stills from time-lapse movies of zygotes before NEBD expressing mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN). Green, GFP or Nup98-DN. Magenta, DNA (H2B-mScarlet). Dashed lines mark pronuclei. Single section confocal microscopy.
(H) Nuclear occupancy, surface occupancy, and inner chromatin fraction indices in zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN).
(I) Zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN) having an abnormal mitosis.
(J) Pronuclear radius in zygotes injected with mClover3 (GFP) or Nup98-DN-mClover3 (Nup98-DN).
(K-L) Representative immunofluorescence images of pronuclei in zygotes expressing bTrim21 (bovine Trim21) and treated with the indicated antibodies. Orange, NPC (NPC-mAb414 (K) or Nup98 (L)). Green, TPR (K) or Nup153 (L). Magenta, DNA (DAPI). Note that antibody injections were performed before fertilization to ensure protein depletion before pronuclear assembly.
(M) Representative immunofluorescence images of telomere distribution within a pronucleus. Gray, transmission. Magenta, DNA (DAPI). Green, telomeres (Trf1). Single sections confocal microscopy. Dashed lines indicate pronucleus and nucleoli. Arrows indicate telomeres. Outlined regions magnified in the top right corner.
(N) Telomeres in the nucleoplasm or at the nuclear envelope.
(O) Telomeres at nucleoli or away from nucleoli.
(P) Representative immunofluorescence images of centromere distribution within a pronucleus. Gray, transmission. Magenta, DNA (DAPI). Green, centromeres (ACA). Single sections confocal microscopy. Dashed lines indicate pronucleus and nucleoli. Arrows indicate centromeres. Outlined regions magnified in the top right corner.
(Q) Centromeres in the nucleoplasm or at the nuclear envelope.
(R) Centromeres at nucleoli or away from nucleoli.
(S) Representative immunofluorescence images of zygotes stained with Sun1 (Green), NPC (NPC-mAb414, Orange) and DNA (DAPI, Magenta). Outlined regions magnified in bottom two rows. Telomere bouquet formation during the early stages of meiosis relies on the LINC inner nuclear membrane protein SUN1. However, in bovine zygotes, SUN1 was distributed along both pronuclear envelopes, without specific enrichment on peripheral chromosomes (magnified region). This localization is in contrast to the clustered appearance of SUN1 in proximity to the centrosome in meiotic cells, but it is consistent with observations in C. elegans zygotes (Minn et al., 2009). Single sections Airyscan microscopy.
Data are from four (E, F) and five (H, I, J) independent experiments. Data in (N, O, Q, R) are from five embryos (two pronuclei each) generated in a single experiment. The number of analyzed pronuclei (E, F, H, J), zygotes (I), telomeres (N,O), and centromeres (Q, R) are specified in italics. p values were calculated using unpaired two-tailed Student’s t test (E, F, H, J) and Fisher’s exact test (I). Scale bars, 10 μm.
Part I: Time-lapse movie of a zygote expressing mClover3-MAP4-MTBD (orange, microtubules) and POM121-mScarlet (green, NPC). Arrowheads indicate annulate lamellae moving toward the centrosomes. Time, hours:minutes, 00:00 is NEBD. Single sections confocal microscopy. Part II: Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes. Airyscan sections of a bovine zygote stained for microtubules (orange, α-tubulin), NPCs (green, NPC-Mab414), and DNA (magenta, DAPI). Outlined regions magnified in bottom row. Centrosomes, NPCs, and annulate lamellae are indicated. One section corresponds to 0.18 μm, as indicated. Part III: Nuclear pore complexes cluster with chromatin at the pronuclear interface in human zygotes. Airyscan sections of a human zygote with three pronuclei stained for microtubules (orange, α-tubulin), γ-tubulin (white, g-tubulin), NPCs (green, NPC-Mab414), and DNA (magenta, DAPI). Outlined regions magnified in bottom row. Centrosomes, NPC, and annulate lamellae are indicated. One section corresponds to 0.16 μm, as indicated.
To gain better resolution, we performed Airyscan super-resolution microscopy of NPCs in fixed zygotes. Strikingly, we found that NPCs were unevenly distributed along the nuclear envelope, clustered at the pronuclear interface, and were in direct proximity to the chromatin located near the nuclear envelope (Figure 5A; Video S6). We quantified this co-localization, obtaining a Pearson’s correlation coefficient of 0.83 (Figure 5B). In contrast, lamin A/C and lamin B1 were distributed along the entire nuclear envelope (Figures S6B and S6C). Intriguingly, the clusters of NPCs were closely associated with microtubules running along the interface between the two pronuclei (Figures 5A; Video S6), consistent with dynein clustering the parental genomes via microtubules and NPCs.
Figure 5.
Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes
(A) Representative immunofluorescence images of bovine zygotes during pronuclear migration (left), in early stages on pronuclear juxtaposition when chromatin condensation is incomplete (middle), and in late stages after increased chromatin condensation (right). Orange, microtubules (respectively, β-tubulin, α-tubulin, and α-tubulin). Magenta, DNA (DAPI). Green, NPC (respectively Nup98, NPC-Mab414, and NPC-Mab414). Outlined regions magnified in the bottom two rows. Centrosomes, NPCs, and annulate lamellae are indicated. Single sections Airyscan microscopy.
(B) Pearson’s coefficient quantifying the co-localization of NPCs and chromatin at the nuclear envelope. +1 indicates perfect co-localization, −1 indicates exclusion.
(C) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green), mScarlet-MAP4-MTBD (microtubules, white), and H2B-miRFP (DNA, magenta). 00:00 is start of image acquisition. First two time points are single confocal microscopy sections, whereas the other time points are Z projections of 3 (00:30) or 5 (00:51 to 01:11) sections every 2.50 μm.
(D) Female (♀) and male (♂) pronuclei were scored for polarization toward the sperm aster.
(E) Representative stills from time-lapse movies of zygotes expressing bElys-mClover3 (NPC, green) and H2B-mScarlet (DNA, magenta). The right pronucleus re-orients, leading to a more clustered organization of chromatin and NPCs. Dashed line indicates regions of the nuclear envelope devoid of NPCs. Time, h:min, 00:00 is NEBD. Single confocal microscopy sections.
Data are from two (B) or three (D) independent experiments. The number of analyzed zygotes (C) or pronuclei (D) is specified in italics. p value in (D) was calculated using unpaired two-tailed Student’s t test. Scale bars, 10 μm.
Live imaging of the NPC component Elys, which binds directly to chromatin and initiates NPC assembly on chromatin in mouse zygotes (Gillespie et al., 2007; Inoue and Zhang, 2014), revealed that NPC polarization was already established during the initial stages of pronuclear migration as the pronuclei expand (Figure 5C). NPC polarization in the female pronucleus was oriented toward the male pronucleus, whereas polarization of the male pronucleus was oriented toward the sperm-associated centrosomes (Figures 5C and 5D). Centrosomal microtubules (sperm aster) are thought to reach all the way from the male pronucleus to the female pronucleus and serve as tracks for dynein-dependent migration of the female pronucleus toward the male pronucleus (Gönczy et al., 1999; Kim et al., 1996; Payne et al., 2003). The centrosomal microtubules could hence similarly serve as tracks for dynein-mediated chromatin and NPC polarization toward the sperm-associated centrosomes. In support of this hypothesis, nocodazole treatment before pronuclear expansion impaired NPC clustering at the pronuclear interface (Figures S6D and S6E) and led to a significant reduction of overall NPC polarization, consistent with the reduction in chromatin polarization reported earlier (Figures 4P and S6F).
In some zygotes, where the NPCs were polarized but not yet clustered at the pronuclear interface, the pronuclei re-oriented to reach NPC clustering even after the pronuclei had reached juxtaposition (Figure 5E). These data show that parental genome clustering initiates with NPC polarization at the start of pronuclear migration and is completed during pronuclear migration when the NPCs and associated chromosomes orient toward each other or in a few cases even after pronuclear juxtaposition, as suggested from the experiments where nocodazole was added after pronuclear juxtaposition (Figures S5K and S5L).
To test directly if NPCs are involved in chromosome clustering, we expressed a fragment of Nup98 that acts as a dominant-negative (Nup98-DN); specifically, unlike Nup98, Nup98-DN binds to chromatin but not to the NPCs (Griffis et al., 2002; Liang et al., 2013). Bovine zygotes expressing Nup98-DN displayed reduced chromosome clustering, reduced chromatin recruitment to the periphery, and more mitotic errors (Figures S6G–S6I). Notably, the pronuclear volume was decreased suggesting that NPC function was partially impaired (Figure S6J). The nuclear basket of the NPC, which protrudes into the nucleoplasm, could mediate chromatin binding (Arlucea et al., 1998; Ibarra and Hetzer, 2015; Pérez-Garrastachu et al., 2017). However, depletion of the nuclear basket proteins TPR and Nup153 by Trim-Away did not affect chromosome clustering, recruitment into the outer nuclear region, or pronuclear size (Figures S6K and S6L). Together, our data show that the nuclear basket is dispensable for chromatin recruitment, and chromatin clustering is prevented by Nup98-DN. Nup98-DN could impair clustering directly be preventing the recruitment of chromatin to NPCs, or indirectly, by decreasing the functionality of NPCs.
NPCs were associated with chromosome arms but not with telomeres or centromeres, which were located away from the nuclear envelope in the central region of zygotic pronuclei, often enriched at nucleoli (Figures S6M–S6R), consistent with reports in mouse and human zygotes (Dozortsev et al., 2000; van de Werken et al., 2014). This localization is distinct from telomere localization in the early stages of meiosis (Zeng et al., 2018), when telomeres cluster in proximity to the centrosome in preparation for meiotic recombination within the telomere bouquet, together with SUN1, which was distributed along both pronuclear envelopes, without specific enrichment on peripheral chromosomes in bovine zygotes (Figure S6S) (Sato et al., 2009; Shibuya et al., 2014). Together, these data indicate that NPCs interact with chromatin via chromosome arms, but not via telomeres or centromeres.
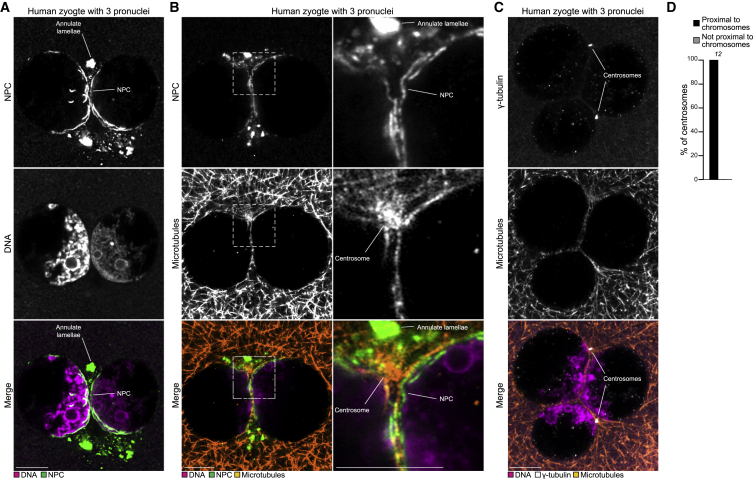
We next investigated if NPCs also cluster with chromatin at the pronuclear interface in human zygotes. Staining of zygotes with three pronuclei (fertilized by two sperm) showed that the NPCs clustered at the interface of the pronuclei, closely associated with the parental genomes, and were depleted from other pronuclear regions (Figures 6A and 6B; Video S6). As in bovine zygotes, the position of the chromosomes and NPCs correlated with the position of the centrosomes and microtubules (Figures 6B–6D; Video S6), strongly indicating that the clustering mechanism is conserved in humans.
Figure 6.
Nuclear pore complexes cluster with chromatin at the pronuclear interface in human zygotes
(A) Representative immunofluorescence images of a human zygote with 3 pronuclei. Magenta, DNA (DAPI). Green, NPC (NPC-Mab414). Single sections Airyscan microscopy.
(B) Representative immunofluorescence images of a zygote with 3 pronuclei. Orange, microtubules (α-tubulin). Magenta, DNA (DAPI). Green, NPC (NPC-Mab414). Outlined regions magnified on the right. Airyscan microscopy, Z projections of 3 sections every 0.16 μm.
(C) Representative immunofluorescence images of a zygote with 3 pronuclei. Orange, microtubules (α-tubulin). Magenta, DNA (DAPI). Green, NPC (NPC-Mab414). White, γ-tubulin (γ-tubulin). Single sections Airyscan microscopy. This is the same zygote shown in (B), restained with additional antibodies.
(D) Zygotes with 3 pronuclei were scored for the presence of chromosomes in proximity of the centrosomes.
The number of analyzed centrosomes is specified in italics. Scale bars, 10 μm.
See also Video S6.
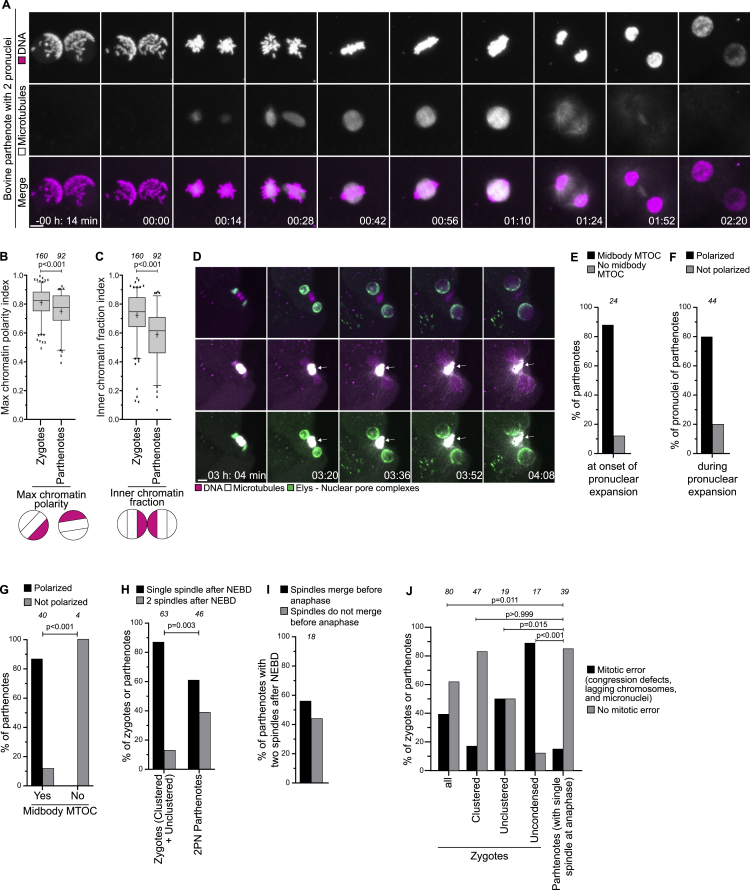
Parthenotes cluster chromosomes toward the midbody of the meiosis II spindle
Our data establish an important role for centrosomes in determining where the parental genomes cluster during the first cell division. Interestingly though, parthenotes progress through the first embryonic divisions in the absence of centrosomes. Parthenotes are eggs that activate and start embryonic development without fertilization, and thus without male chromatin and centrosomes. Activation can be triggered chemically and, when cytokinesis is inhibited by drugs, it generates parthenotes with two pronuclei of maternal origin (Brevini et al., 2012; Navara et al., 1994).
To investigate parental genome clustering in parthenotes, we generated bovine parthenotes with two maternal pronuclei and performed live cell imaging. We discovered that the chromatin still polarized and clustered at the pronuclear interface, although less than in zygotes (Figures S7A–S7C). To understand how polarization is established in parthenotes, we imaged NPCs, chromosomes, and microtubules shortly after activation and found that the pronuclei formed in close proximity of the remnants of the meiosis II spindle after telophase (Figure S7D). During pronuclear expansion, NPCs and chromatin polarized in the direction of the midbody (Figure S7D, arrows). The midbody develops out of the spindle and functions as a microtubule organizing center in early mouse embryos (Zenker et al., 2017). Our data suggest that the midbody can functionally replace the centrosome as microtubule organizing center to direct chromatin polarization in parthenotes (Figures S7D–S7G).
Figure S7.
Bovine parthenotes cluster chromosomes toward the midbody of the meiosis II spindle, related to Figure 4
(A) Representative stills from time-lapse movies of bovine parthenotes with 2 pronuclei. White, microtubules (mClover3-MAP4-MTBD). Magenta, DNA (H2B-mScarlet). Time, h:m, 00:00 is NEBD. Z-projections, 15 sections every 2.50 μm.
(B-C) Max chromatin polarity and inner chromatin fraction indices before NEBD in zygotes (same as Figure S2E) and parthenotes.
(D) Representative stills from time-lapse movies of parthenotes with 2 pronuclei. Green, NPC (bElys-mClover3). White, microtubules (mScarlet-MAP4-MTBD). Magenta, DNA (H2B-miRFP). Arrows point to the meiosis II microtubule spindle that becomes a midbody and acts as microtubule organizing center, persisting for long after the end of meiosis. In the top row, the microtubule signal bleeds into the DNA channel. The midbody is thus also visible in magenta. Time, h:min, 00:00 is egg activation. Z-projections, 3 (first three frames) and 2 (last two frames) non-consecutive sections to display both pronuclei.
(E) Parthenotes with or without a midbody acting as microtubule organizing center (MTOC) at the onset of pronuclear expansion.
(F) Parthenotes with or without a polarized distribution of Elys during pronuclear expansion.
(G) Parthenotes with or without a polarized distribution of Elys during pronuclear expansion in the presence or absence of a midbody MTOC.
(H) Zygotes and parthenotes having one or two spindles after NEBD. Zygotes are those in the clustered and unclustered categories from Figure 3B.
(I) Parthenotes with two spindles after NEBD were scored for spindle merging before anaphase onset.
(J) Zygotes and parthenotes in indicated groups having abnormal mitosis. Zygotes are the same analyzed in Figure 4K.
Data are from four (parthenotes groups in B, C, H, J, and E, F, G, I) and eleven (zygote groups in B, C, H, J) independent experiments. The number of analyzed pronuclei (B, C), parthenotes (E, F, G, H, I, J), and zygotes (H, J) are specified in italics. p values were calculated using unpaired two-tailed Student’s t test (B, C) and Fisher’s exact test (G, H, J). Scale bars, 10 μm.
Chromosome condensation and NEBD occurred synchronously in parthenotes (Figure S7A). Moreover, the chromosomes in both pronuclei simultaneously attached to microtubules, which in 60% of the cases formed as a single array of microtubules across all chromosomes and in 40% of the cases as two separated asters between the two chromosome masses that later merged in almost 60% of the cases (Figures S7H and S7I). As described in a recent study, bovine zygotes sometimes also assembled two separate spindles, although at lower frequency than parthenotes (Figure S7H) (Schneider et al., 2020).
Overall, although parthenotes displayed significantly lower chromosome clustering, they had higher synchrony in chromosome condensation, NEBD, and chromosome capture than zygotes. Consistently, chromosome segregation in parthenotes was more reliable than in unclustered and uncondensed zygotes and, instead, was similar to clustered zygotes (Figures S7A, S7C, and S7J).
Discussion
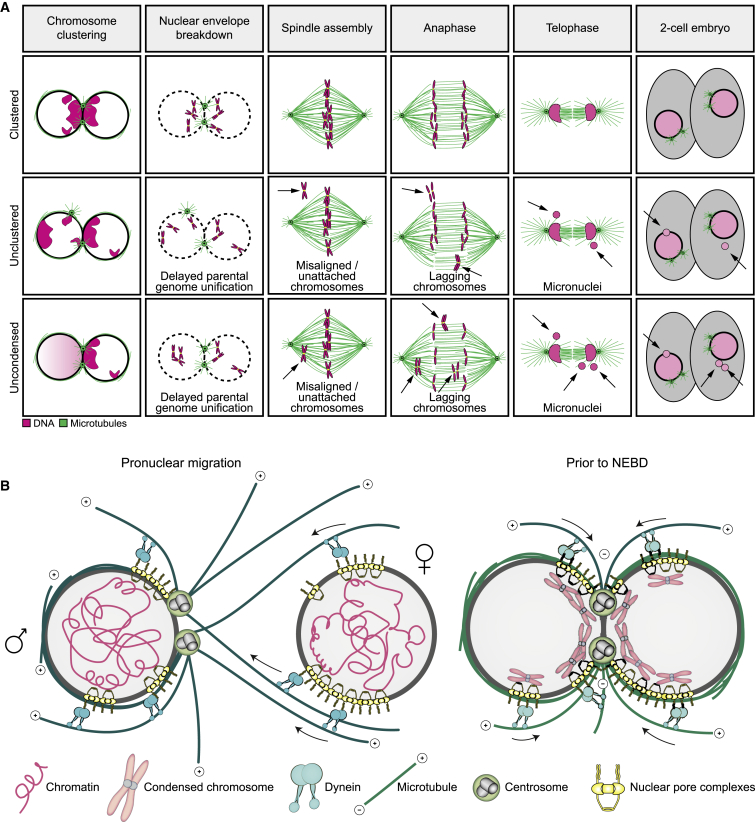
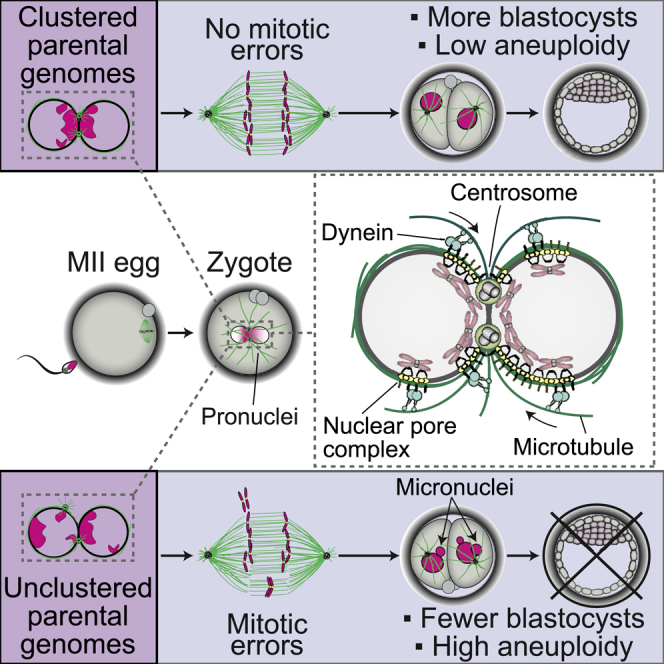
By employing high-resolution live cell microscopy of human zygotes and bovine embryos, we discovered that the unification of maternal and paternal chromosomes in fertilized zygotes is error-prone and depends on an inefficient process that we defined as parental genome clustering. Parental genome clustering increases the speed of the unification of the parental genomes after fertilization, enhances the efficiency of chromosome capture by the newly assembling spindle, and thus prevents chromosome segregation errors and the formation of micronuclei (Figure 7A).
Figure 7.
Models illustrating the function and mechanism of parental genome clustering
(A) Schematics of mitosis in zygotes having clustered (top), unclustered (middle), or uncondensed (bottom) chromosomes before NEBD. Magenta, chromatin. Green, microtubules. Arrows point to defects causing chromosome segregation errors, such as misaligned, unattached, and lagging chromosomes, and to micronuclei.
(B) Model for the mechanism of chromatin clustering at the pronuclear interface before (left) and after (right) completion of pronuclear migration. Gray, nucleoplasm. Magenta, chromatin and chromosomes. Yellow, NPCs. Cyan, dynein. Green, microtubules and centrosomes. Microtubule polarity is indicated by + and −. Arrows indicate dynein directionality. Female and male pronuclei are marked by ♀ and ♂, respectively.
Parental genome clustering initiates early during pronuclear migration. As the two pronuclei expand and start to migrate inward, their NPCs become asymmetrically distributed within the nuclear envelope, polarizing in the direction of the sperm centrosome. The NPCs are linked to chromatin, which also polarizes toward the centrosome. Polarization toward the centrosome is driven by dynein and microtubules and ultimately leads to the clustering of the parental genomes in close proximity of each other once the pronuclei have reached juxtaposition (Figure 7B).
Pronuclear migration and chromosome clustering are not only established together but also the molecular players driving both processes—microtubules, dynein, NPCs, and centrosomes—are intriguingly similar (Hu et al., 2013; Lindeman and Pelegri, 2012; Malone et al., 2003; Payne et al., 2003). Our data are consistent with a unified model whereby pronuclear migration and chromatin clustering are two tightly interwoven processes, established together by the same cellular machinery, with the common aim of uniting the parental genomes. In this model, dynein associates with NPCs and transports the pronuclei toward each other along centrosome-nucleated microtubules. Pulling via NPCs not only brings the two pronuclei into close proximity, but also polarizes the NPCs and parental genomes in the direction of migration, ultimately clustering them at the pronuclear interface to facilitate their rapid capture and union on the first mitotic spindle (Figure 7B).
As previously shown and confirmed by this study, prolonged mitosis and asynchronous NEBD in human zygotes correlate with defective human embryo development (Coticchio et al., 2018; Vera-Rodriguez et al., 2015). Our work in bovine zygotes provides a cellular explanation for this correlation, by demonstrating that prolonged mitosis and asynchronous NEBD are typically linked to a different state of chromosome condensation between the two pronuclei and asynchronous chromosome capture, resulting in chromosome segregation errors and the formation of micronuclei. Altogether, these data establish that the unification of the parental chromosomes is a particularly critical and sensitive step in embryo development. Consistent with our results, a recent preprint demonstrates that chromosome segregation in human zygotes is highly error-prone, and the first mitotic division is more frequently abnormal than the second mitotic division (Ford et al., 2020).
A recent study reported increased aneuploidy on depletion of the spindle assembly checkpoint (SAC) components BUB1B/BUBR1 in bovine zygotes (Brooks et al., 2020), indicating the presence of an active SAC. Our data support the presence of an active SAC in bovine zygotes: we found a delay in anaphase onset in zygotes with an uncondensed pronucleus, which often have unattached chromosomes and eventually progress into anaphase with multiple lagging chromosomes. Our data also show that the SAC is not stringent enough to block anaphase when only a few chromosomes are unattached, as in the case of unclustered zygotes. A low stringency of the SAC has also been reported for mouse and human oocytes and for mouse embryos (Thomas et al., 2021; Vázquez-Diez et al., 2019). The low stringency of the SAC might explain why 17% of the zygotes with clustered genomes display chromosome segregation errors. Overall, our data suggest that accurate chromosome segregation in zygotes requires rapid chromosome capture and is generally challenging, consistent with high aneuploidy levels in human embryos.
Chromosome condensation and recruitment into the nuclear periphery may help to expose kinetochores and bring them into close proximity with the centrosomes to facilitate their capture. Kinetochores buried within an uncondensed chromatin mass, such as those in uncondensed pronuclei, make microtubule contacts later and often stay unattached or incorrectly attached, as evident from a large number of misaligned and lagging chromosomes in this group. The fact that 21% of zygotes have an uncondensed pronucleus before NEBD implies that asynchronous chromosome condensation in the two pronuclei is a frequent phenomenon in bovine zygotes and a major cause of errors during the first mitotic division. This number is strikingly similar to the 25% of human cleavage embryos that are aneuploid for more than 3 chromosomes (McCoy et al., 2015), also referred to as chaotic aneuploidy (McCoy, 2017).
Chromosome clustering might occur specifically in zygotes because their chromosomes are spread over a much larger volume than in somatic mitotic cells, and are hence more difficult to capture and unite by two centrosomes. The position of the centrosomes plays a crucial role in this process, because zygotes with centrosomes at the pronuclear interface unify the parental genomes more rapidly, and are less likely to show chromosome segregation errors.
Interestingly, mouse zygotes have multiple acentriolar microtubule organizing centers distributed on the surface of the two pronuclei, which may facilitate the synchronous capture of chromosomes upon NEBD (Courtois et al., 2012). This alternative and potentially more efficient capture mechanism may explain why mice do not cluster their chromosomes at the pronuclear interface and may underlie the lower aneuploidy rates in mouse embryos compared to human and bovine embryos (Destouni et al., 2016; Lee and Kiessling, 2017; Lightfoot et al., 2006). Consistent with this model, bovine parthenotes, which also employ a centrosome-independent spindle assembly process and undergo NEBD, chromosome condensation, and chromosome capture in a highly synchronous manner, also have less clustered genomes and fewer chromosome segregation errors than fertilized bovine zygotes.
Our results suggest a cellular explanation for why zygotes with clustered nucleoli are more likely to develop into healthy embryos (Scott, 2003; Tesarik and Greco, 1999) and to be euploid (Coskun et al., 2003). The clustering of nucleoli in human zygotes is an indicator of efficient chromosome clustering, and anticipates accurate chromosome segregation and healthy embryo development. Our results thus support the use of scoring nucleolar clustering in zygotes as one of the indicators for embryo quality when selecting the best embryos for transfer in fertility treatments.
Limitations of study
Although our work presents intriguing insights into the causes of aneuploidy in mammalian embryos, at least three main issues remain:
-
(1)
Further studies are required to assess how closely human and bovine zygotes are related. Our data show that chromosome clustering in human and bovine zygotes is achieved by similar mechanisms, and clustering and synchrony between the two pronuclei correlate with healthy embryo development in both systems, indicating that both systems are closely related. However, we were not able to analyze human zygotes after NEBD. Interestingly, human zygotes were reported to undergo multipolar divisions (Ford et al., 2020; Ottolini et al., 2017), which are uncommon in bovine zygotes. There might thus also be differences between human and bovine zygotes.
-
(2)
Further work is required to understand the upstream mechanisms that cause defects in parental genome clustering and asynchronous chromosome condensation.
-
(3)
Our work did not investigate if chromosome and nucleolar clustering has functions beyond promoting the rapid unification of the parental genomes and the accuracy of chromosome segregation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human anti-centromere antibody (ACA) (1:250) | Antibodies Incorporated | Cat# 15-234-0001; RRID:AB_2687472 |
| Rat anti-Nup98 (1:50) | Abcam | Cat# ab50610; RRID:AB_881769 |
| Mouse anti-NPC/MAb414 (1:100) | Covance | Cat# MMS-120P; RRID:AB_291294 |
| Rabbit anti-TPR (1:100) | Abcam | Cat# ab84516; RRID: AB_1861454 |
| Mouse anti-TPR (1:100) | Kuznetsov et al., 2002 | PMID: 12424524 |
| Mouse anti-Nup153 (1:100) | Abcam | Cat# ab96462; RRID: AB_10710699 |
| Rabbit anti-Elys (1:100) | Novus biological | Cat# NBP1-87952; RRID: AB_11006121 |
| Rabbit anti-Elys (1:100) | Sigma-Aldrich | Cat# HPA031658; RRID: AB_10601968 |
| Normal mouse IgG | Millipore | Cat# 12-371; RRID: AB_145840 |
| Rat anti-α-tubulin (1:1000) | AbD Serotec | Cat# MCA78G; RRID: AB_325005 |
| Rabbit anti-β-8-tubulin (1:500) | Sigma-Aldrich | Cat# SAB2700070 |
| Mouse anti-Trf1 (1:250) | Biotrend | Cat# TRF12-S; RRID:AB_2201456 |
| Mouse anti-Histones (1:100) | Millipore | Cat# MAB3422; RRID:AB_2114845 |
| Mouse anti-γ-tubulin (1:250) | Sigma-Aldrich | Cat# T5326; RRID:AB_532292 |
| Rabbit anti-γ-tubulin (1:500) | Sigma-Aldrich | Cat# T3559; RRID:AB_ 477575 |
| Rabbit anti-Sun1 (1:100) | Sigma-Aldrich | Cat# HPA008346; RRID:AB_1080462 |
| Rabbit anti-Lamin B1 (1:100) | Abcam | Cat# ab16048; RRID:AB_443298 |
| Mouse anti-Lamin A/C (1:50) | Sigma-Aldrich | Cat# MABT1340 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID:AB_141607 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A10037; RRID:AB_2534013 |
| Donkey anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571; RRID:AB_162542 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID:AB_2535792 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-10042; RRID:AB_2534017 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21208; RRID:AB_2535794 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat# A-11077; RRID:AB_2534121 |
| Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-21247; RRID:AB_141778 |
| Goat anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11013, RRID:AB_2534080 |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Thermo Fisher Scientific | Cat# D1306; RRID:AB_2629482 |
| 5-580CP-Hoechst | Bucevičius et al., 2018 | PMID: 30881625 |
| Dimethyl sulfoxide Hybri-Max | Sigma-Aldrich | Cat# D2650 |
| Lipase from Candida rugosa | Sigma-Aldrich | Cat# (L8525) |
| Heparin | Merck - Millipore | Cat# 375095-100KU |
| Nocodazole | Sigma-Aldrich | Cat# M1404 |
| Cytochalasin B | Sigma-Aldrich | Cat# C6762 |
| Ionomycin | Sigma-Aldrich | Cat# I0634 |
| Cyclohexamide | Merck - Millipore | Cat# 239763 |
| Critical commercial assays | ||
| HiScribe T7 ARCA mRNA Kit | New England Biolabs | Cat# E2065S |
| Qubit RNA HS Assay Kit | Thermo Fisher Scientific | Cat# Q32852 |
| pENTR/D-TOPO Cloning Kit | Thermo Fisher Scientific | Cat# K240020 |
| SensiFAST cDNA synthesis kit | Bioline | Cat# BIO-65053 |
| Experimental models: Organisms/strains | ||
| Bovine ovaries (Bos taurus) | Local abattoir | N/A |
| Human embryos | IVF clinic | N/A |
| Oligonucleotides | ||
| KASH5-DN cloning sense 5′- CAC CCACCTCCTGATCCCAGCG −3′ |
Sigma-Aldrich | N/A |
| KASH5-DN cloning antisense 5′- ACA TTGCTCTGCAGGGGTAG −3′ |
Sigma-Aldrich | N/A |
| bElys cloning sense 5′- CAC CATGCGAGACCTAACGGCGCA −3′ |
Sigma-Aldrich | N/A |
| bElys cloning antisense 5′- CAGC ATTTTTCTGCGTAGAACT −3′ |
Sigma-Aldrich | N/A |
| mH3.3B cloning sense 5′- CACC ATGGCCCGAACCAAGCAGA −3′ |
Sigma-Aldrich | N/A |
| mH3.3B cloning antisense 5′- AGC TCTCTCCCCCCGTATCC-3′ |
Sigma-Aldrich | N/A |
| bNup98-DN cloning sense 5′- CACCATG TTTAATAAGTCATTTGGAACGC-3′ |
Sigma-Aldrich | N/A |
| bNup98-DN cloning antisense 5′- TTGGAGAACAGCCTGCTGGG −3′ | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| pGEMHE | Liman et al., 1992 | PMID: 1419000 |
| pGEX-6P1 | Panic et al., 2003 | PMID: 12620189 |
| pGEMHE-miRFP-MAP4-MTBD | So et al., 2019 | PMID: 31249032 |
| pGEMHE-H2B-mClover3 | So et al., 2019 | PMID: 31249032 |
| pcDNA5-hCenpC | Klare et al., 2015 | PMID: 26124289 |
| pGEMHE-POM121-EGFP3 | Beaudouin et al., 2002 | PMID: 11792323 |
| pGEMHE-mClover3-MAP4-MTBD | So et al., 2019 | PMID: 31249032 |
| pGEMHE-H2B-miRFP | So et al., 2019 | PMID: 31249032 |
| pGEMHE-H2B-mScarlet | So et al., 2019 | PMID: 31249032 |
| pGEMHE-bTRIM21 | So et al., 2019 | PMID: 31249032 |
| pGEMHE-mRFP-MAP4-MTBD | This paper | N/A |
| pGEMHE-mScarlet-MAP4-MTBD | This paper | N/A |
| pGEMHE-H2B-mClover3 | This paper | N/A |
| pGEMHE-mScarlet-hCenpC | This paper | N/A |
| pGEMHE-Pom121-mScarlet | This paper | N/A |
| pGEMHE-mClover3-KASH5-DN | This paper | N/A |
| pGEMHE-GST-KASH5-DN | This paper | N/A |
| pGEMHE-GST | This paper | N/A |
| pGEMHE-mClover3-bElys | This paper | N/A |
| pGEMHE-bNup98-DN-mClover3 | This paper | N/A |
| pGEMHE-mH3.3B-mClover3 | This paper | N/A |
| pGEMHE-mClover3 | This paper | N/A |
| pENTR/D-TOPO vector | Thermo Fisher Scientific | Cat# K240020 |
| pET28-p150-CC1-His | Courtois et al., 2012 | PMID: 22851319 |
| Software and algorithms | ||
| Prism 8 | GraphPad Software | N/A |
| Fiji (Fiji Is Just ImageJ) | Schindelin et al., 2012 | PMID: 22743772 |
| Imaris version 9.2.1 | Bitplane | N/A |
| QuPath (0.2.3) | (Bankhead et al., 2017) | N/A |
| R | (R Development Core Team, 2019) | N/A |
| Zen 2.3 (Blue edition) | Zeiss | N/A |
| Other | ||
| Zeiss LSM800 microscope | Zeiss | N/A |
| Zeiss LSM880 microscope | Zeiss | N/A |
| Zeiss LSM900 microscope | Zeiss | N/A |
| GERI time lapse system | Genea Biomedx | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Melina Schuh (melina.schuh@mpibpc.mpg.de).
Materials availability
Plasmids generated in this study have been deposited with Addgene, in the Melina Schuh Lab plasmid collection: https://www.addgene.org/Melina_Schuh/
Data and code availability
Scripts and plugins are available at link: https://gitlab.gwdg.de/schuh-meiosis/cavazza_etal_cell_2021_parental_genome_unification
Experimental model and subject details
Generation of bovine zygotes and parthenotes
Bovine ovaries were obtained from cows of different breeds (mostly German and Red Holstein, and less frequently Limousin), all being at least 12 months old, slaughtered by local abattoirs and transported to the laboratory in a thermo-flask within 3 hours of retrieval. Oocyte isolation, culture, and fertilization were performed using the IVF Bioscience media suite and the manufacturer protocol, with small changes. In brief, cumulus-oocyte complexes (COCs) were aspirated from antral follicles using an 18-gauge needle mounted on a 1 mL disposable syringe. The aspirated follicular fluid was transferred to a 50 mL falcon tube containing 140 μl of 5000 IU/ml Heparin (Merck Millipore #375095-100KU). COCs were allowed to sediment and then washed extensively with pre-warmed TCM199 (HEPES-buffered medium 199; Sigma-Aldrich #M2520) supplemented with 0.05 g/ml Gentamycin Sulfate (Roth #0233), 1 mM Na-Pyruvate (GIBCO #11360-039), 0.022 g/ml NaHCO3 (Sigma-Aldrich #S5761-500G), and 5% FBS (GIBCO #16000-044). Only fully-grown oocytes with a homogeneous cytoplasm and at least 3-5 complete layers of compact cumulus cells were selected for the experiments. COCs were washed and transferred to pre-warmed and equilibrated BO-IVM media (IVF Biosciences) and incubated at 38.8°C (5% CO2). After 14 hours, COCs were partially denuded using a transfer pipette with a 175 μm tip (Origio #MXL3-175) in warm TCM199 media. 6 hours later (20 hours after COCs retrieval), COCs were washed into pre-warmed and equilibrated BO-IVF media (IVF Biosciences). Insemination was performed by adding 1x106 spermatozoa to a maximum of 40 COCs in a final volume of 500 μl BO-IVF media. Spermatozoa were purified from frozen bull semen obtained from a bull of proven fertility (Bernal-Ulloa et al., 2016). Frozen semen was thawed, resuspended in 3 mL of pre-warmed BO-Semen media (IVF Bioscience) and centrifuged for 5 min at 300g. The pellet was resuspended in 2 mL of pre-warmed BO-Semen media and ccentrifuged for 5 min at 300g. The supernatant was discarded leaving 300-500 μl and sperm cells were gently resuspended and counted using a Bürker chamber. Insemination was performed at 38.8°C (5% CO2) for 7 to 18 hours. For short inseminations (7-10 hours), the spermatozoa concentration was doubled. Zygotes were retrieved 7 to 18 hours after insemination and gently denuded of cumulus cells and spermatozoa using a transfer pipette with a 135 μm tip (Origio #MXL3-135) in pre-warmed TCM199 media. To improve imaging, zygotes were transferred to a 2 mL tube containing 500 μl of warm TCM199 media and centrifuged for 3 min at 9000g in a pre-warmed rotor. After spinning, zygotes were washed into pre-warmed and equilibrated BO-IVC media (IVF Biosciences) and incubated at 38.8°C (5% CO2 + 6% O2).
For parthenogenesis, oocytes were matured for at least 27 hours. After maturation, eggs were gently denuded of cumulus cells using a transfer pipette with a 135 μm tip and activated by a 5 min incubation in BO-IVM supplemented with 5 μM Ionomycin (Sigma-Aldrich #I0634) pre-warmed and equilibrated at 38.8°C (5% CO2). Subsequently, activated eggs were washed into BO-IVM supplemented with 5 μg/ml Cytochalasin B (Sigma-Aldrich #C6762) and 10 μg/ml Cyclohexamide (Merck #239763) pre-warmed and equilibrated at 38.8°C (5% CO2). Cytochalasin B inhibited polar body extrusion and thus promoted the generation of parthenotes with 2 pronuclei, while Cyclohexamide supported egg activation by inhibiting protein synthesis. 3 to 5 hours after activation, eggs were washed into BO-IVC media pre-warmed and equilibrated at 38.8°C (5% CO2 + 6% O2) and imaged. Parthenotes imaged immediately after activation were imaged in in BO-IVC media supplemented with + 5 μg/ml Cytochalasin B (Sigma-Aldrich #C6762) + 10 μg/ml Cyclohexamide (Merck #239763). The centrifugation for improving imaging was performed either pre-activation or pre-imaging.
Human embryos
Transmitted light videos of human embryos used in this study were obtained from Bourn Hall Clinic (Cambridge, UK) and were generated using embryos from 62 couples who underwent standard infertility treatment. Embryos were imaged as part of the routine fertility treatment after having obtained fully informed patient consent, under the Human Fertilization and Embryology Authority (HFEA) license for Center 0100. Videos were provided to the scientists in an anonymized manner and without any information on the patients. The provision of these videos was covered by Bourn Hall Clinic’s general consent for treatment.
All human zygotes that were fluorescently labeled were obtained by Newcastle Fertility Centre (Newcastle upon Tyne, UK) and experiments conducted under an HFEA- research license (R0152) with Health Research Authority approval from Newcastle and North Tyneside Research Ethics Committee. Informed consent was obtained from all donors by research nurses who were not directly involved in the research, or in the provision of clinical treatment. Immunofluorescence data was obtained from abnormally fertilized zygotes (n = 3) donated for research by couples undergoing standard infertility treatment. Live cell imaging experiments were conducted as part of an ongoing project to map cell cycle transitions during the first zygotic division. For this purpose, vitrified oocytes donated specifically for research (n = 8 oocytes from 4 donors, age range 26-35 years) were fertilized by intracytoplasmic sperm injection (ICSI) using sperm donated for research. Vitrification and warming were performed using RapidVit and RapidWarm oocyte kits (Vitrolife, Sweden). In accordance with HFEA Directions (Human Fertilisation and Embryology Authority, 2018), egg donors received financial compensation of £500 per donation cycle as approved by ethics committee and HFEA.
Method details
Microinjection of bovine oocytes and zygotes
Bovine oocytes were microinjected between 14 and 20 hours after the onset of maturation. Microinjections were performed as previously described (Jaffe and Terasaki, 2004; Schuh and Ellenberg, 2007). Briefly, oocytes and zygotes were loaded on an ‘injection shelf’ generated by assembling two coverslips around a spacer made of two layers of 100 μm thick piece of double stick tape. Oocytes and zygotes were pushed to the end of the shelf using the injection needle, in order to have the adjacent tape keeping the oocytes and zygotes in place during the microinjection procedure. 4 pl of the following mRNAs were microinjected at the indicated needle concentrations: mClover3-MAP4-MTBD at 200 ng/μl, mScarlet-MAP4-MTBD at 248 ng/μl, miRFP-MAP4-MTBD at 300 ng/μl, H2B-miRFP at 219 ng/μl, H2B-mScarlet at 60 ng/μl, mH3.3B-mClover3 at 326 ng/μl, mScarlet-hCenpC at 41 ng/μl, bElys-mClover3 at 864 ng/μl, H2B-mClover3 at 120 ng/μl, and POM121-mScarlet at 50 ng/μl. 10 pl of the following mRNAs were microinjected at the indicated needle concentrations: GST at 1964 ng/μl, mClover3-bKASH5-DN at 1950 ng/μl, GST-bKASH5-DN at 1951 ng/μl, bNup98DN-mClover3-NLS at 612 ng/μl, mClover3 at 618 ng/μl, and bTrim21 at 960ng/μl.
For protein injections, bovine zygotes were microinjected between 8 and 16 hours post insemination with 14 pl of protein solution. BioXtra BSA (Sigma-Aldrich #A3311) and P150-CC1 were injected at a needle concentration of 20 μg/μl, resulting in a protein concentration in zygotes of 4.7 mM and 7.1 mM, respectively. Protein injection buffer was PBS supplemented with 0.03% NP40 and 70 kD Dextran 647 to select for injected cells.
Expression constructs, messenger RNA (mRNA) synthesis, protein expression and purification
All mRNAs were synthesized using HiScribe T7 ARCA mRNA Kit (NEB # E2065S) following the manufacturer’s protocol and quantified using the Qubit RNA HS Assay Kit (Thermo Fisher Scientific # Q32852). mClover3-MAP4-MTBD, H2B-miRFP, H2B-mScarlet, and bTrim21 mRNAs were synthesized from previously published constructs (So et al., 2019). The following plasmids were generated specifically for this study by subcloning the previously published sequences into a pGEMHE plasmid: GST (Panic et al., 2003), mClover3 (So et al., 2019), mRFP-MAP4-MTBD (So et al., 2019), mScarlet-MAP4-MTBD (So et al., 2019), H2B-mClover3 (So et al., 2019); mScarlet-hCenpC (Klare et al., 2015), and Pom121-mScarlet (Beaudouin et al., 2002). mClover3-KASH5-DN, GST-KASH5-DN, mClover3-bElys, mH3.3B-mClover3, bNup98-DN-mClover3-NLS constructs were cloned from bovine fibroblast, bovine oocyte, or mouse oocyte cDNA libraries made using a SensiFAST cDNA synthesis kit (Bioline #BIO-65053). The primers (bKASH5-DN cloning, sense and antisense, bElys cloning sense and antisense, mH3.3B cloning, sense and antisense, bNup98-DN cloning sense and antisense) were used to clone the bovine KASH5-DN, bovine Elys, mouse H3.3B, and bovine Nup98-DN into a pENTR/D-TOPO vector (Invitrogen). bKASH5-DN corresponds to the C-terminal domain of bovine KASH5 (amino acids 483-525), as previously shown in human cells (Stewart-Hutchinson et al., 2008). bNup98-DN corresponds to the N-terminal domain of bovine Nup98 (amino acids 1-501), as shown in Drosophila and human cells (Kalverda et al., 2010; Liang et al., 2013).
His-P150-CC1 (Courtois et al., 2012) was expressed in and purified from BL21(DE3)pLysS competent cells (Promega) as previously described (Courtois et al., 2012) with some modifications. Briefly, recombinant proteins were purified using Ni-NTA Agarose (QIAGEN 30210). Buffer was exchanged by dialysis to PBS. Protein was concentrated using an Amicon 10 kD filter column (Merck #UFC801008D) and purity tested by SDS-PAGE.
Drug addition
Nocodazole (Sigma-Aldrich #M1404) was diluted freshly in hybridoma grade DMSO (Sigma-Aldrich #D2650) to make a 20 mM stock and was added to zygotes at the final concentration of 20 μM, at least 9 hours before NEBD. Ionomycin (Sigma-Aldrich #I0634) was diluted in hybridoma grade DMSO (Sigma-Aldrich D2650) to make a 1.35 mM stock (1 mg/ml). The stock was stored at −20°C for no longer than 3 months. Ionomycin was added to eggs at the final concentration of 5 μM (3.7 μg/ml) for 5 minutes. Cytochalasin B (Sigma-Aldrich #C6762) was diluted in hybridoma grade DMSO (Sigma-Aldrich D2650) to make a 10 mg/ml stock and was added to activated eggs at the final concentation of 5 μg/ml. Cyclohexamide (Merck #239763) was diluted in embryo tested water (Sigma-Aldrich #TMS-006-C) to make a 10 mg/ml stock and was added to activated eggs at a final concentation of 10 μg/ml. Heparin (Merck Millipore #375095-100KU) was diluted in embryo tested water (Sigma-Aldrich #TMS-006-C) to make a 5000 IU/ml stock.
Trim-Away
Only affinity-purified antibodies were used in this study for Trim-Away–mediated protein depletion. Antibodies were transferred into injection buffer (83 mM KCl, 17 mM NaCl, 2 mM MgCl2, 20 mM HEPES pH7.2) and concentrated using Ultra-0,5 mL Amicon 100 kD centrifugal filters (Merck #UFC510024). Before injection, the concentrated antibodies were supplemented with NP40 (Merck #492016) to a final concentration of 0.05% and spun down for 10 min at 14,000g.
Trim-away was performed by injecting 10 pl of the mRNA for bTrim21 into bovine eggs at a needle concentration of 960 ng/μl. After 6 hours, 14 pl of the following antibodies were injected at the indicated needle concentrations: control mouse IgG (Millipore #12-371) 2.6 mg/ml, mouse anti-TPR (Kuznetsov et al., 2002) 1 mg/ml, mouse anti-Nup153 (Abcam #ab96462) 2.4 mg/ml. The eggs were later inseminated and fixed between 20 - 24 hours after antibody injection.
Immunofluorescence
Bovine zygotes and human zygotes with 3 pronuclei were fixed in 100 mM HEPES (pH 7.0, titrated with KOH), 50 mM EGTA (pH 7.0, titrated with KOH), 10 mM MgSO4, 2% methanol-free formaldehyde and 0.5% Triton X-100 at 37°C for 25 min. Fixed zygotes were extracted in PBS with 0.5% Triton X-100 (PBST) overnight at 4°C and blocked in PBST with 5% BSA (Fisher Scientific #BP1605) (PBST-BSA) for 6 hours at room temperature. After blocking, bovine zygotes were process for optical clearing using the Lipase treatment, as previously described (So et al., 2019). Briefly, zygotes were incubated for 1 hour at 37°C into Lipase buffer (50 mM Tris-HCl pH 7.2, 400 mM NaCl, 5 mM CaCl2, 0.2% sodium taurocholate (Sigma-Aldrich #86339), 1:20 mini cOmplete EDTA free protease inhibitor (Roche #11836170001), 5% BSA). After Lipase treatment, the zygotes were washed in PBST-BSA and incubated overnight at 4°C with primary antibodies in PBST-BSA at the concentration listed in the following paragraph. Secondary antibody incubations were performed in PBST-BSA for 1 hour at room temperature at 20 μg/ml.
As primary antibodies we used human anti-centromere antibody (ACA) at 1:250 dilution (Antibodies Incorporated #15-234), rat anti-Nup98 at 1:50 (Abcam #ab50610), mouse anti-NPC/MAb414 at 1:100 (Covance #MMS-120P), rat anti-α-tubulin at 1:1000 (AbD Serotec #MCA78G), rabbit anti-β-8-tubulin at 1:500 (Sigma-Aldrich #SAB2700070), mouse anti-Trf1 at 1:250 (Alpha diagnostic international #TRF12-S), mouse anti-Histone at 1:100 (Merck #MAB3422), mouse anti-γ-tubulin at 1:250 (Sigma-Aldrich #T5326), rabbit anti-Sun1 at 1:100 (CST #8886), rabbit anti-Lamin B1 at 1:100 (Abcam #ab16048) and mouse anti-Lamin A/C at 1:50 (Sigma-Aldrich #MABT1340), rabbit anti-TPR at 1:100 (Abcam #ab84516), rabbit anti-Elys at 1:100 (Novus biological #NBP1-87952), mouse anti-Nup153 at 1:100 (Abcam #ab96462).
As secondary antibodies we used Alexa Fluor 405-, 488-, 568- or 647-conjugated anti-human IgG, anti-mouse IgG, anti-mouse IgM, anti-rabbit IgG, or anti-rat IgG, all raised in donkey or goat (Thermo Fisher Scientific). DNA was stained with DAPI at a final concentration of 20 μg/ml (Thermo Fisher Scientific).
Confocal and super-resolution microscopy of bovine zygotes
For confocal imaging, embryos were imaged in 20 μl of BO-IVC medium at 38.8°C, 5% CO2, 6% O2 (for live zygotes) or PBS with 1% polyvinylpyrrolidone (PVP) at room temperature (for fixed zygotes) under paraffin oil in a 35 mm dish with a #1.0 coverslip. Images were acquired with LSM800, LSM880, or LSM900 confocal laser scanning microscopes (Zeiss) equipped with an environmental incubator box and a 40x C-Apochromat 1.2 NA water-immersion objective. A volume of 65 μm × 65 μm × 60 μm centered on the chromosomes was typically recorded. If full zygotes were imaged we used a volume of 100 μm × 100 μm × 72.5 μm centered on the zygote center. The optical slice thickness was 3.00 μm and we recorded a section every 2.5 μm, except for the bleaching experiments where we imaged every 1.76 μm and for the kinetochore imaging where the optical slice thickness was 3.08 μm and we imaged every 3.08 μm. Each zygote was typically imaged every 5 or 10 minutes, using the lowest possible laser intensity (> 0.2% for the 488nm laser; > 0.2% for the 561 nm laser; > 0.3% for the 633 nm laser). During imaging, we did not immobilize the zygotes as they did not move substantially.
mClover3 was excited with a 488 nm laser line and detected at 493 - 571 nm. mScarlet was excited with a 561 nm laser line and detected at 571 - 638 nm. miRFP was excited with a 633 nm laser line and detected at 638 - 700 nm. Images of the control and experimental groups were acquired under identical imaging conditions on the same microscope. For some images, noise was reduced with a Gaussian filter in ZEN (Zeiss). Airyscan images were acquired using the Airyscan module on LSM800, LSM880, or LSM900 confocal laser scanning microscopes (Zeiss) and processed in ZEN (Zeiss) after acquisition. Care was taken that the imaging conditions (laser power, pixel-dwell time and detector gain) did not cause noticeable phototoxicity (for live imaging), photobleaching or saturation. Selective photobleaching experiments were performed on zygotes expressing H2B-mClover3 and H2B-mScarlet. Selective photobleaching was done once chromosomes had already been recruited into the nuclear periphery or right after NEBD. Bleaching was performed using the bleaching tool of ZEN (Zeiss) with 60% 561 laser power, scan speed 4, and three repeats. Z-projections were generated by maximum intensity projections of the indicated Z-slices.
Live cell confocal microscopy of human zygotes
For live cell fluorescence imaging of human zygotes, chromosomes were visualized using 5-580CP-Hoechst (Bucevičius et al., 2018) or H2B-mScarlet mRNA. For 5-580CP-Hoechst, zygotes were incubated for at least 2 hours in 500 nM or 1000 nM 5-580CP-Hoechst before imaging and imaged with 5-580CP-Hoechst. For H2B-mScarlet mRNA, oocytes were microinjected before ICSI with H2B-mScarlet mRNA at a needle concentration of 60 ng/ul using a pressure injector (Narishige) on a Nikon Diaphot microscope fitted with micromanipulators prior to ICSI. Injection needles were made from borosilicate glass capillaries using a P-97 micropipette puller (Sutter Instruments). Zygotes were imaged in 2 μl of G-TL medium (Vitrolife) under Ovoil (Vitrolife) in a 35 mm dish with a #1.5H coverslip (ibidi) at 36.6°C, 6% CO2, 5% O2. Fast Airyscan images were acquired with the Airyscan module on an LSM 880 confocal laser scanning microscope (Zeiss) equipped with an environmental incubator box and a C-Apochromat 40x/1.2 Korr FCS M27 water-immersion objective. A volume of 115 μm x 115 μm x 73.00 μm centered on the chromosomes was typically recorded. The optical slice thickness was 0.48 μm and we recorded a section every 1.0 μm to reduce phototoxicity. Zygotes were imaged every 4-6 hours from soon after pronucleus formation (6-8 hours post-ICSI) to 17 hours post-ICSI and every 20 minutes thereafter, using the lowest possible laser intensity. Care was taken that the imaging conditions (laser power, pixel-dwell time, and detector gain) did not cause phototoxicity, photobleaching or saturation. During imaging, zygotes were not immobilized as they did not move substantially.
Human embryo imaging
Time-lapse images of human embryos were recorded using a GERI time lapse system (Genea Biomedx) as part of the routine IVF treatment with patient consent at Bourn Hall Clinic, under the HFEA license for Center 0100. Imaging was offered to patients for the purpose of selecting embryos with optimal implantation potential. Archived videos were shared with the scientists after anonymization without any information on the patient.
Normally fertilized oocytes were placed in the GERI time lapse system (Genea Biomedx) immediately after fertilization check (17-19 hours post insemination/ICSI). Zygotes were transferred to an individual well of a GERI Dish (GERI-DSH-20, Genea Biomedx) containing pre-equilibrated Continuous Single Culture Medium (CSCM-C, Irvine Scientific #90165) with a mineral oil overlay (Light mineral oil, Irvine Scientific #9305) at 37°C in an atmosphere of 6% CO2, 5% O2, 89% N2. Time-lapse videography captured images every 5 minutes at 11 sequential focal planes (Z stacks) set at 10 μm apart from 0 to 96 hours of development, 15 μm apart between 96-120 hours and 20 μm apart thereafter.
Quantification and statistical analyses
Statistical analysis
Statistical significance based on paired or unpaired, two-tailed Student’s t test (for absolute values) and two-tailed Fisher’s exact test (for categorical values) were calculated in Prism (GraphPad). All boxplots show median (horizontal black line), mean (small black squares), 25th and 75th percentiles (boxes), 5th and 95th percentiles (whiskers) and 1st and 99th percentiles (crosses). All data are from at least three independent experiments. p values are indicated.
Image analysis
Human nucleolar distribution was quantified with a semi-automated workflow. For each zygote we identified the last frame before NEBD with well detectable nucleoli (∼16 ± 10 min before NEBD) and manually annotated the pronuclei and nucleoli boundaries using QuPath (Bankhead et al., 2017). The pronuclei interface was then defined by a watershed operation applied to the 2 pronuclei. Each image was registered along the pronuclei axis. We computed the shortest distance d of nucleoli centroids to the pronuclei membrane away from the interface (Figure S1D). This quantity was more reliable than the distance to the interface due to the fact that occasionally pronuclei were oriented along the imaging axis. Finally we computed the nucleoli spread before NEBD as
Where is the mean pronuclei size l over all pronuclei (24.8 ± 3.2 μm, n = 490). Pronuclei that have a < 0.5 are classified to have clustered nucleoli. When at least one pronucleus with spread < 0.5 the whole zygote is classified as unclustered.
We used the manual annotation for training two machine learning models (StarDist) (Schmidt et al., 2018) to predict labeling masks for nuclei and nucleoli over time. Due to frequent loss of focus in the videos, nucleoli labeling often required additional manual correction. These corrections were used to further improve the models. Overall we identify 158/245 zygotes that allowed for a more detailed analysis of the nucleoli distribution within 3h before NEBD. For each zygote, we tracked the pronuclei using TrackMate (Tinevez et al., 2017) and registered the images along the pronuclei axis. For each pronuclei we then measured and compute the over time
Where is the time averaged pronuclear size for each pronucleus. This time series were then smoothed using local regression (loess() function in R (R Development Core Team, 2019), with span = 0.5). The smoothed traces minus their time average were used for clustering. We computed the distance between traces with soft dynamic time warping (soft-DTW) and clustering using k-mean clustering (python 3.7.9, Tslearn 0.4.1) (Tavenard et al., 2020). Overall, we were able to identify 3 main classes of movement. The single nucleoli tracks for selected zygotes were computed using trackmate.
DNA-NPC co-localization analysis was performed using Imaris version 9.2.1 (Bitplane). We used the Imaris spot tool to create a sphere for each nucleus. The subsequent analysis was performed within the spheres. Using the surface tool on the NPC channel, we identified the membrane regions containing NPCs. With the Imaris co-localization analysis tool we computed the Pearson correlation coefficient between the DNA and NPC signals.
The quantification of microtubule length was performed using the Imaris spot tool. For microtubules originating from the centrosome we marked the centrosome and the opposite microtubule end and measured the distance between the two points. The microtubule length at metaphase was measured from astral microtubules. For this quantification we used zygotes fixed at metaphase and stained for microtubules (α-tubulin) and DNA (DAPI).
The quantification of bleached and unbleached lagging chromosomes in zygotes with condensed chromosomes (Figure S4E) was performed using Imaris. The Imaris surface detection tool was applied on the unbleached channel (laser 488 – H2B-mClover3) to identify bulk chromatin and individual chromosomes. For each zygote, we computed the average ratio of the H2B-mClover3 and the H2B-mScarlet signals for three surfaces with bleached DNA (B) and for three surfaces with unbleached DNA (U). We also computed the average ratio of the H2B-mClover3 and H2B-mScarlet for each lagging chromosome (LC). B and U were calculated just after the bleaching. Lagging chromosomes with \B - LC\ < \U - LC\ were scored as bleached (clustered). Lagging chromosomes with \B - LC\ > \U - LC\ were scored as unbleached (distal).
The quantification of bleached and unbleached lagging chromosomes in zygotes that contained uncondensed chromosomes in one pronucleus (Figures S4B and S4C) was performed using Imaris. The Imaris surface detection tool was applied on the unbleached channel (laser 488 – H2B-mClover3) to identify bulk chromatin and individual chromosomes. For each zygote, we computed the ratio of the H2B-mClover3 and the H2B-mScarlet signals in the bleached pronucleus (B) and in the unbleached pronucleus (U). We also computed the average ratio of the H2B-mClover3 and the H2B-mScarlet for each lagging chromosome (LC). The quantities B and U were calculated just after the bleaching. Lagging chromosomes with \B - LC\ < \U - LC\ were scored as bleached (clustered). Lagging chromosomes with \B - LC\ > \U - LC\ were scored as unbleached (uncondensed).
The DNA distribution indices (nuclear and surface occupancy indices and the inner, outer and maximal polarity indices) were computed using Fiji (Rueden et al., 2017; Schindelin et al., 2012) (IJ2 v2.0.0-rc-71, IJ1 v1.52r) and MATLAB (v2018a, The MathWorks, Inc.). A sphere is used to approximate each pronucleus, the center coordinates S = (SX, SY, SZ) and radius R were calculated from points placed manually in the image (Fit Sphere, BoneJ2 v6.1.1) (Doube et al., 2010). We developed the ImageJ plugin liveim-drawspheres to compute pronucleus masks using S and R and a maximal polarity direction per nucleus. For this, Gaussian filtered images of the DNA signal using [σX, σY, σZ] equal to [8, 8, 0], [16, 16, 0], [16, 16, 1], [32, 32, 0], [32, 32, 1], and [32, 32, 2] pixels are calculated. The DNA intensity image and the pronucleus mask is used to compute the coordinates, CM, of the center of mass for the intensity distribution and for extended maxima regions (MorphoLibJ v1.4.1) (Legland et al., 2016). For each center of mass, the mean intensity in three equally spaced sphere sections along the vector is computed. This operation is performed for the raw and Gaussian filtered DNA images and the orientation with the highest normalized mean fluorescence intensity in the first region (the region facing the center of mass) approximates the direction of maximal polarity . The ImageJ plugin also computes distance transform for each pronucleus and defines sphere sections oriented along the line connecting the center of the nuclei. These are used to compute the inner and outer polarity indices.
In the last part of the workflow, a MATLAB script is used to extract the relevant quantities per pronucleus. We first smoothed the DNA signal using a 2D median filter (3x3 pixels) and a Gaussian filter (σ = 1 pixel). To minimize the contribution of unbound H2B-fluorescent protein, we derived a mask for the DNA, D, using an Otsu’s threshold thr(pre-NEBD) (multithresh). The pixel values outside D are set to 0. The ratio of number of pixels in D to the whole nucleus gives the:
The surface occupancy is calculated from:
Where and are, respectively, the mean intensities in the region adjacent to the nucleus surface and inside the nucleus. The distance transform is used to define the region boundaries in order to have an equal number of pixels in both regions. For a perfect sphere this is ∼0.8 R. The sphere sections have been chosen to have equal heights h = 2R/3, yielding equal outer surface areas, i.e., the area without the base of the sections. For sphere sections oriented toward we computed the mean fluorescence intensity Ik and normalized it to obtain
Similarly, we computed mean intensities for sections oriented toward the opposite nucleus to obtain , k = 1, 2, 3. Where section 1 is the section in the direction of maximal polarity/toward the other nucleus, 2 the central section, and 3 the section which is oriented in the opposite direction. The maximum polarity is given by , the inner and outer polarities are given by and , respectively.
For time lapse data (Figure 2C) the threshold thr(t) to segment the DNA is adapted per time point. We computed the total intensity in D and adapted the threshold so that the total intensity remains within 10% of the pre-NEBD value. Variation in intensity during migration, is accounted for by computing a correction factor from the total non-segmented DNA intensity at pre-NEBD and the respective time point.
Acknowledgments
We thank L. Pieper and M.W. Kriesten (Westfleisch SCE mbH, Hamm, Germany) for enabling this research by donating bovine ovaries; the MPI-BPC drivers for collecting ovaries; M. Daniel (MPI-BPC) for assistance in coordinating ovary collection; J. Uraji and K. Menelaou (MPI-BPC) for help in providing the human embryo videos; N. Sharma, C. So, A. Webster (MPI-BPC), and A. Zielinska (Imperial College, London, UK) for technical advice; S. Munro (LMB, Cambridge, UK), A. Musacchio (MPI-Molecular Physiology, Dortmund, Germany), and J. Ellenberg (EMBL, Heidelberg, Germany) for plasmids; V. Cordes (MPI-BPC) for antibodies; Z. Holubcová (Department of Histology and Embryology, Masaryk University, Czech Republic) for discussions about human zygotes; M.E. Torres-Padilla (IES, Munich, Germany) for discussion about Nup98; the Bioimaging Unit (Newcastle University, UK), the Live-cell imaging facility (MPI-BPC), and P. Lénárt (MPI-BPC) for discussion and support; C. Thomas (MPI-BPC), L. Wartosch (MPI-BPC), and Life Science Editors for critical comments on the manuscript; members of the Schuh lab for discussions. The work on human zygotes was funded by the Wellcome Human Developmental Biology Initiative (HDBI; 215116/Z/18/Z) and a Wellcome Centre grant (203105/Z/16/Z). The research leading to these results received financial support from the Max Planck Society and Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC 2067/1-390729940) and a DFG Leibniz Prize (SCHU 3047/1-1) to M.S.
Author contributions
T.C and M.S. conceived the study. T.C. and M.S. designed experiments. T.C. performed all bovine experiments. M.H. and Y.T. designed the human zygote experiments. Y.T. performed all human experiments, except for the recording of the human zygote in Figure 1F and in part III of Video S1, done by M.A. T.C., A.Z.P., and M.S. designed methods for data analysis. A.Z.P. wrote all in-house-developed scripts and plugins. T.C. and A.Z.P. analyzed the data. P.A., A.L.-H., and H.N. supervised the establishment of the bovine experimental system. Y.T. and M.H. established the human live imaging system. M.H. and M.C. supervised the work at the Newcastle Fertility Centre at Life. M.B. and K.E. supervised the work at Bourn Hall clinic. C.B. recorded the human embryo GERI videos. J.B. and G.L. produced the 5-580CP-Hoechst. T.C. and M.S. wrote the manuscript and prepared the figures with input from all authors. M.S. supervised the study.
Declaration of interests
The authors declare no competing interests.
Published: May 7, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.04.013.
References
- Arlucea J., Andrade R., Alonso R., Aréchaga J. The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J. Struct. Biol. 1998;124:51–58. doi: 10.1006/jsbi.1998.4054. [DOI] [PubMed] [Google Scholar]
- Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R., Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Benkhalifa M., Kasakyan S., Clement P., Baldi M., Tachdjian G., Demirol A., Gurgan T., Fiorentino F., Mohammed M., Qumsiyeh M.B. Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat. Diagn. 2005;25:894–900. doi: 10.1002/pd.1230. [DOI] [PubMed] [Google Scholar]
- Bernal-Ulloa S.M., Heinzmann J., Herrmann D., Hadeler K.G., Aldag P., Winkler S., Pache D., Baulain U., Lucas-Hahn A., Niemann H. Cyclic AMP Affects Oocyte Maturation and Embryo Development in Prepubertal and Adult Cattle. PLoS ONE. 2016;11:e0150264. doi: 10.1371/journal.pone.0150264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolková J., Lanctôt C. Live imaging reveals spatial separation of parental chromatin until the four-cell stage in Caenorhabditis elegans embryos. Int. J. Dev. Biol. 2016;60:5–12. doi: 10.1387/ijdb.150222cl. [DOI] [PubMed] [Google Scholar]
- Bone C.R., Starr D.A. Nuclear migration events throughout development. J. Cell Sci. 2016;129:1951–1961. doi: 10.1242/jcs.179788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevini T.A.L., Pennarossa G., Vanelli A., Maffei S., Gandolfi F. Parthenogenesis in non-rodent species: developmental competence and differentiation plasticity. Theriogenology. 2012;77:766–772. doi: 10.1016/j.theriogenology.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Brooks K.E., Daughtry B.L., Davis B., Yan M.Y., Fei S.S., Carbone L., Chavez S.L. Molecular Contribution to Embryonic Aneuploidy and Genotypic Complexity During Initial Cleavage Divisions of Mammalian Development Short. bioRxiv. 2020 doi: 10.1242/dev.198341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucevičius J., Keller-Findeisen J., Gilat T., Hell S.W., Lukinavičius G. Rhodamine-Hoechst positional isomers for highly efficient staining of heterochromatin. Chem. Sci. (Camb.) 2018;10:1962–1970. doi: 10.1039/c8sc05082a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M. Nuclear Pores and the Genome. In: Lavelle C., Victor J.-M., editors. Nuclear Architecture and Dynamics, Translational Epigenetics. Volume 2. Academic Press; 2018. pp. 369–385. [Google Scholar]
- Chia G., Agudo J., Treff N., Sauer M.V., Billing D., Brown B.D., Baer R., Egli D. Genomic instability during reprogramming by nuclear transfer is DNA replication dependent. Nat. Cell Biol. 2017;19:282–291. doi: 10.1038/ncb3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D., Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat. Rev. Mol. Cell Biol. 2013;14:549–562. doi: 10.1038/nrm3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun S., Hellani A., Jaroudi K., Al-Mayman H., Al-Kabra M., Qeba M. Nucleolar precursor body distribution in pronuclei is correlated to chromosomal abnormalities in embryos. Reprod. Biomed. Online. 2003;7:86–90. doi: 10.1016/s1472-6483(10)61734-8. [DOI] [PubMed] [Google Scholar]
- Coticchio G., Mignini Renzini M., Novara P.V., Lain M., De Ponti E., Turchi D., Fadini R., Dal Canto M. Focused time-lapse analysis reveals novel aspects of human fertilization and suggests new parameters of embryo viability. Hum. Reprod. 2018;33:23–31. doi: 10.1093/humrep/dex344. [DOI] [PubMed] [Google Scholar]
- Courtois A., Schuh M., Ellenberg J., Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 2012;198:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigneault B.W., Rajput S., Smith G.W., Ross P.J. Embryonic POU5F1 is Required for Expanded Bovine Blastocyst Formation. Sci. Rep. 2018;8:7753. doi: 10.1038/s41598-018-25964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destouni A., Zamani Esteki M., Catteeuw M., Tšuiko O., Dimitriadou E., Smits K., Kurg A., Salumets A., Van Soom A., Voet T., Vermeesch J.R. Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 2016;26:567–578. doi: 10.1101/gr.200527.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doube M., Kłosowski M.M., Arganda-Carreras I., Cordelières F.P., Dougherty R.P., Jackson J.S., Schmid B., Hutchinson J.R., Shefelbine S.J. BoneJ: Free and extensible bone image analysis in ImageJ. Bone. 2010;47:1076–1079. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozortsev D., Coleman A., Nagy P., Diamond M.P., Ermilov A., Weier U., Liyanage M., Reid T. Nucleoli in a pronuclei-stage mouse embryo are represented by major satellite DNA of interconnecting chromosomes. Fertil. Steril. 2000;73:366–371. doi: 10.1016/s0015-0282(99)00491-4. [DOI] [PubMed] [Google Scholar]
- Egli D., Chen A.E., Saphier G., Ichida J., Fitzgerald C., Go K.J., Acevedo N., Patel J., Baetscher M., Kearns W.G. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nat. Commun. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Zuccaro M.V., Kosicki M., Church G.M., Bradley A., Jasin M. Inter-homologue repair in fertilized human eggs? Nature. 2018;560:E5–E7. doi: 10.1038/s41586-018-0379-5. [DOI] [PubMed] [Google Scholar]
- Faramarzi A., Khalili M.A., Omidi M., Agha-Rahimi A., Taheri F. Pronuclear pattern does not predict morphokinetics behavior in human embryos. Gynecol. Endocrinol. 2018;34:248–251. doi: 10.1080/09513590.2017.1388365. [DOI] [PubMed] [Google Scholar]
- Fishman E.L., Jo K., Nguyen Q.P.H., Kong D., Royfman R., Cekic A.R., Khanal S., Miller A.L., Simerly C., Schatten G. A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 2018;9:2210. doi: 10.1038/s41467-018-04678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E., Currie C., Taylor D., Erent M., Marston A., Hartshorne G., McAinsh A. The First Mitotic Division of the Human Embryo is Highly Error-prone. bioRxiv. 2020 doi: 10.1101/2020.07.17.208744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E., Alfarawati S., Spath K., Jaroudi S., Sarasa J., Enciso M., Wells D. The origin and impact of embryonic aneuploidy. Hum. Genet. 2013;132:1001–1013. doi: 10.1007/s00439-013-1309-0. [DOI] [PubMed] [Google Scholar]
- Fritz B., Hallermann C., Olert J., Fuchs B., Bruns M., Aslan M., Schmidt S., Coerdt W., Müntefering H., Rehder H. Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH)-Re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur. J. Hum. Genet. 2001;9:539–547. doi: 10.1038/sj.ejhg.5200669. [DOI] [PubMed] [Google Scholar]
- Gámiz P., Rubio C., de los Santos M.J., Mercader A., Simón C., Remohí J., Pellicer A. The effect of pronuclear morphology on early development and chromosomal abnormalities in cleavage-stage embryos. Hum. Reprod. 2003;18:2413–2419. doi: 10.1093/humrep/deg458. [DOI] [PubMed] [Google Scholar]
- Gerri C., McCarthy A., Alanis-Lobato G., Demtschenko A., Bruneau A., Loubersac S., Fogarty N.M.E., Hampshire D., Elder K., Snell P. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature. 2020;587:443–447. doi: 10.1038/s41586-020-2759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerri C., Menchero S., Mahadevaiah S.K., Turner J.M.A., Niakan K.K. Human Embryogenesis: A Comparative Perspective. Annu. Rev. Cell Dev. Biol. 2020;36:411–440. doi: 10.1146/annurev-cellbio-022020-024900. [DOI] [PubMed] [Google Scholar]
- Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., Blow J.J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A.A. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis E.R., Altan N., Lippincott-Schwartz J., Powers M.A. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Fertilisation and Embryology Authority . HFEA Code of Practice. 9th Edition. Human Fertilisation and Embryology Authority; 2018. HFEA Guidance on Payments for Donors. [Google Scholar]
- Hu D.J.-K., Baffet A.D., Nayak T., Akhmanova A., Doye V., Vallee R.B. Dynein recruitment to nuclear pores activates apical nuclear migration and mitotic entry in brain progenitor cells. Cell. 2013;154:1300–1313. doi: 10.1016/j.cell.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Hetzer M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A., Zhang Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014;21:609–616. doi: 10.1038/nsmb.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe L.A., Terasaki M. Quantitative microinjection of oocytes, eggs, and embryos. Methods Cell Biol. 2004;74:219–242. doi: 10.1016/s0091-679x(04)74010-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kim N.H., Simerly C., Funahashi H., Schatten G., Day B.N. Microtubule organization in porcine oocytes during fertilization and parthenogenesis. Biol. Reprod. 1996;54:1397–1404. doi: 10.1095/biolreprod54.6.1397. [DOI] [PubMed] [Google Scholar]
- Klare K., Weir J.R., Basilico F., Zimniak T., Massimiliano L., Ludwigs N., Herzog F., Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kort D.H., Chia G., Treff N.R., Tanaka A.J., Xing T., Vensand L.B., Micucci S., Prosser R., Lobo R.A., Sauer M.V., Egli D. Human embryos commonly form abnormal nuclei during development: a mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum. Reprod. 2016;31:312–323. doi: 10.1093/humrep/dev281. [DOI] [PubMed] [Google Scholar]
- Kuznetsov N.V., Sandblad L., Hase M.E., Hunziker A., Hergt M., Cordes V.C. The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma. 2002;111:236–255. doi: 10.1007/s00412-002-0208-2. [DOI] [PubMed] [Google Scholar]
- Lee A., Kiessling A.A. Early human embryos are naturally aneuploid-can that be corrected? J. Assist. Reprod. Genet. 2017;34:15–21. doi: 10.1007/s10815-016-0845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legland D., Arganda-Carreras I., Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics. 2016;32:3532–3534. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- Lequarre A.S., Marchandise J., Moreau B., Massip A., Donnay I. Cell cycle duration at the time of maternal zygotic transition for in vitro produced bovine embryos: effect of oxygen tension and transcription inhibition. Biol. Reprod. 2003;69:1707–1713. doi: 10.1095/biolreprod.103.017178. [DOI] [PubMed] [Google Scholar]
- Liang Y., Franks T.M., Marchetto M.C., Gage F.H., Hetzer M.W. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot D.A., Kouznetsova A., Mahdy E., Wilbertz J., Höög C. The fate of mosaic aneuploid embryos during mouse development. Dev. Biol. 2006;289:384–394. doi: 10.1016/j.ydbio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Liman E.R., Tytgat J., Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Lindeman R.E., Pelegri F. Localized products of futile cycle/lrmp promote centrosome-nucleus attachment in the zebrafish zygote. Curr. Biol. 2012;22:843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V., O’Connell C.B., Lončarek J., Paul R., Mogilner A., Khodjakov A. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Mayer W., Smith A., Fundele R., Haaf T. Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 2000;148:629–634. doi: 10.1083/jcb.148.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy R.C. Mosaicism in Preimplantation Human Embryos: When Chromosomal Abnormalities Are the Norm. Trends Genet. 2017;33:448–463. doi: 10.1016/j.tig.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy R.C., Demko Z.P., Ryan A., Banjevic M., Hill M., Sigurjonsson S., Rabinowitz M., Petrov D.A. Evidence of Selection against Complex Mitotic-Origin Aneuploidy during Preimplantation Development. PLoS Genet. 2015;11:e1005601. doi: 10.1371/journal.pgen.1005601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriano J., Clark C., Cadesky K., Laskin C.A. Binucleated and micronucleated blastomeres in embryos derived from human assisted reproduction cycles. Reprod. Biomed. Online. 2004;9:511–520. doi: 10.1016/s1472-6483(10)61635-5. [DOI] [PubMed] [Google Scholar]
- Milo R., Phillips R. Garland Science, Taylor & Francis Group; 2016. Cell Biology by the Numbers. [Google Scholar]
- Milo R., Jorgensen P., Moran U., Weber G., Springer M. BioNumbers--the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn I.L., Rolls M.M., Hanna-Rose W., Malone C.J. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol. Biol. Cell. 2009;20:4586–4595. doi: 10.1091/mbc.E08-10-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navara C.S., First N.L., Schatten G. Microtubule organization in the cow during fertilization, polyspermy, parthenogenesis, and nuclear transfer: the role of the sperm aster. Dev. Biol. 1994;162:29–40. doi: 10.1006/dbio.1994.1064. [DOI] [PubMed] [Google Scholar]
- Ottolini C.S., Kitchen J., Xanthopoulou L., Gordon T., Summers M.C., Handyside A.H. Tripolar mitosis and partitioning of the genome arrests human preimplantation development in vitro. Sci. Rep. 2017;7:9744. doi: 10.1038/s41598-017-09693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B., Whyte J.R., Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 2003;13:405–410. doi: 10.1016/s0960-9822(03)00091-5. [DOI] [PubMed] [Google Scholar]
- Payne C., Rawe V., Ramalho-Santos J., Simerly C., Schatten G. Preferentially localized dynein and perinuclear dynactin associate with nuclear pore complex proteins to mediate genomic union during mammalian fertilization. J. Cell Sci. 2003;116:4727–4738. doi: 10.1242/jcs.00784. [DOI] [PubMed] [Google Scholar]
- Pérez-Garrastachu M., Arluzea J., Andrade R., Díez-Torre A., Urtizberea M., Silió M., Aréchaga J. Nucleoporins redistribute inside the nucleus after cell cycle arrest induced by histone deacetylases inhibition. Nucleus. 2017;8:515–533. doi: 10.1080/19491034.2017.1320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popken J., Graf A., Krebs S., Blum H., Schmid V.J., Strauss A., Guengoer T., Zakhartchenko V., Wolf E., Cremer T. Remodeling of the Nuclear Envelope and Lamina during Bovine Preimplantation Development and Its Functional Implications. PLoS ONE. 2015;10:e0124619. doi: 10.1371/journal.pone.0124619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Gill S.R., Eckley D.M., Crego C.L., Compton D.A., Schroer T.A. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2019. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santenard A., Ziegler-Birling C., Koch M., Tora L., Bannister A.J., Torres-Padilla M.E. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010;12:853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Isaac B., Phillips C.M., Rillo R., Carlton P.M., Wynne D.J., Kasad R.A., Dernburg A.F. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U., Weigert M., Broaddus C., Myers G. Cell Detection with Star-convex Polygons. arXiv. 2018 doi: 10.1007/978-3-030-00934-2_30. [DOI] [Google Scholar]
- Schneider I., de Ruijter-Villani M., Hossain M.J., Stout T.A.E., Ellenberg J. Non-rodent mammalian zygotes assemble dual spindles despite the presence of paternal centrosomes. bioRxiv. 2020 doi: 10.1101/2020.10.16.342154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Scott L. Pronuclear scoring as a predictor of embryo development. Reprod. Biomed. Online. 2003;6:201–214. doi: 10.1016/s1472-6483(10)61711-7. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Morimoto A., Watanabe Y. The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet. 2014;10:e1004821. doi: 10.1371/journal.pgen.1004821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmet K., Zakhartchenko V., Philippou-Massier J., Blum H., Klymiuk N., Wolf E. OCT4/POU5F1 is required for NANOG expression in bovine blastocysts. Proc. Natl. Acad. Sci. USA. 2018;115:2770–2775. doi: 10.1073/pnas.1718833115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C., Seres K.B., Steyer A.M., Mönnich E., Clift D., Pejkovska A., Möbius W., Schuh M. A liquid-like spindle domain promotes acentrosomal spindle assembly in mammalian oocytes. Science. 2019;364:eaat9557. doi: 10.1126/science.aat9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood V., Brickner J.H. Nuclear pore interactions with the genome. Curr. Opin. Genet. Dev. 2014;25:43–49. doi: 10.1016/j.gde.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson P.J., Hale C.M., Wirtz D., Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp. Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavenard R., Faouzi J., Vandewiele G., Divo F., Androz G., Holtz C., Payne M., Yurchak R., Russwurm M., Kolar K. Tslearn, A Machine Learning Toolkit for Time Series Data. J. Mach. Learn. Res. 2020;21:1–6. [Google Scholar]
- Tesarik J., Greco E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum. Reprod. 1999;14:1318–1323. doi: 10.1093/humrep/14.5.1318. [DOI] [PubMed] [Google Scholar]
- Thomas C., Cavazza T., Schuh M. Aneuploidy in human eggs: contributions of the meiotic spindle. Biochem. Soc. Trans. 2021;49:107–118. doi: 10.1042/BST20200043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J., Davis P., Merriam J., Sinclair J. Nuclear and cytoplasmic dynamics of sperm penetration, pronuclear formation and microtubule organization during fertilization and early preimplantation development in the human. Hum. Reprod. Update. 1995;1:429–461. doi: 10.1093/humupd/1.5.429. [DOI] [PubMed] [Google Scholar]
- van de Werken C., van der Heijden G.W., Eleveld C., Teeuwssen M., Albert M., Baarends W.M., Laven J.S., Peters A.H., Baart E.B. Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nat. Commun. 2014;5:5868. doi: 10.1038/ncomms6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Echten-Arends J., Mastenbroek S., Sikkema-Raddatz B., Korevaar J.C., Heineman M.J., van der Veen F., Repping S. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum. Reprod. Update. 2011;17:620–627. doi: 10.1093/humupd/dmr014. [DOI] [PubMed] [Google Scholar]
- Vanneste E., Voet T., Le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Vázquez-Diez C., Paim L.M.G., FitzHarris G. Cell-Size-Independent Spindle Checkpoint Failure Underlies Chromosome Segregation Error in Mouse Embryos. Curr. Biol. 2019;29:865–873.e3. doi: 10.1016/j.cub.2018.12.042. [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M., Chavez S.L., Rubio C., Reijo Pera R.A., Simon C. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat. Commun. 2015;6:7601. doi: 10.1038/ncomms8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., Reijo Pera R.A. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- Zeng X., Li K., Yuan R., Gao H., Luo J., Liu F., Wu Y., Wu G., Yan X. Nuclear Envelope-Associated Chromosome Dynamics during Meiotic Prophase I. Front. Cell Dev. Biol. 2018;5:121. doi: 10.3389/fcell.2017.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker J., White M.D., Templin R.M., Parton R.G., Thorn-Seshold O., Bissiere S., Plachta N. A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science. 2017;357:925–928. doi: 10.1126/science.aam9335. [DOI] [PubMed] [Google Scholar]
- Zuccaro M.V., Xu J., Mitchell C., Marin D., Zimmerman R., Rana B., Weinstein E., King R.T., Palmerola K.L., Smith M.E. Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell. 2020;183:1650–1664.e15. doi: 10.1016/j.cell.2020.10.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Part I: Time-lapse movies of two human embryos. Left embryo at the zygote stage has clustered nucleoli before NEBD and develops into a blastocyst. Right embryo has unclustered nucleoli and develops abnormally. Time, hours:minutes, 00:00 is NEBD in the zygote. Part II: Nucleolar tracking in time-lapse movies of the three human zygotes shown in Figures S1E–S1G. Time, hours:minutes, 00:00 is NEBD in the zygote. Part III: High resolution live cell microscopy of a human zygotes at pronuclear stage. DNA, (5-580CP-Hoechst). Time, hours:minutes, 00:00 is NEBD in the zygote. Part IV: High resolution live cell microscopy of three human zygotes at pronuclear stage. DNA, (5-580CP-Hoechst). Images were generated with Imaris 3D view. Time, hours:minutes, 00:00 is NEBD in the zygote. Part V: Parental genomes remain separated in their respective pronuclei. DNA, (5-580CP-Hoechst). Images were generated with Imaris 3D view.
Time-lapse movies of bovine zygotes undergoing the first cell divisions. Time, hours:minutes, 00:00 is NEBD. Part I: A system for high resolution live cell microscopy of early bovine embryo development. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 8 sections every 2.50 μm. Part II: Chromosome segregation errors during the first mitotic division (mild). Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 19 sections every 2.50 μm. Part III: Chromosome segregation errors during the first mitotic division (severe). Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 17 sections every 2.50 μm. Part IV: The maternal and paternal chromosomes occupy partially distinct territories in the metaphase plate of the spindles of bovine zygotes and 2-cell embryos. Time-lapse movie of a zygote expressing mClover3-H3.3B (green, DNA) and H2B-mScarlet (magenta, DNA). ♀ and ♂ indicate the female and the male pronucleus, and the DNA of maternal and paternal origin in the second cell division. Time, hours:minutes, 00:00 is NEBD. Z-projections, 10 sections every 2.50 μm.
Time-lapse movies of zygotes having one uncondensed pronucleus undergoing mitosis. Arrowheads indicate chromosomes that fail to align at the metaphase plate, lag after anaphase, and eventually form micronuclei, dotted circles. Time, hours:minutes, 00:00 is NEBD. Part I: Zygotes with an uncondensed pronucleus have chromosome segregation errors. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 12 sections every 2.50 μm. Part II: Kinetochores in the uncondensed pronucleus are captured less efficiently than those at the surface of condensed pronuclei. Zygote expressing mScarlet-hCenpC (green, kinetochores), mClover3-MAP4-MTBD (white, microtubules) and H2B-miRFP (magenta, DNA). Z-projections, 4 sections every 3.08 μm. Part III: Chromosome segregation errors originate from uncondensed pronucleus. Zygote expressing mClover3-H2B (green, DNA) and H2B-mScarlet (magenta, DNA). Rectangle indicates region bleached on the condensed pronucleus, using the 561 nm laser line. Z-projections, 10 sections every 1.76 μm.
Time-lapse movies of zygotes having one unclustered pronucleus undergoing mitosis. Arrowheads indicate chromosomes that fail to align at the metaphase plate, lag after anaphase, and eventually form micronuclei, dotted circles. Time, hours:minutes, 00:00 is NEBD. Part I: Zygotes with an unclustered pronucleus and chromosome segregation errors. Zygote expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA). Z-projections, 12 sections every 2.50 μm. Part II: Chromosomes that do not cluster at the pronuclear interface congress late and segregate incorrectly. Zygote expressing mScarlet-hCenpC (green, kinetochores), mClover3-MAP4-MTBD (white, microtubules) and H2B-miRFP (magenta, DNA). Z-projections, 4 sections every 3.08 μm. Part III: Chromosomes distal from the pronuclear interface missegregate. Zygote expressing mClover3-H2B (green, DNA) and H2B-mScarlet (magenta, DNA). Rectangle indicates region bleached at pronuclear interface, using the 561 nm laser line. Z-projections, 17 sections every 1.76 μm
Time-lapse movies of zygotes under different perturbation conditions to affect the function of centrosomes, microtubules, or dynein. Time, hours:minutes, 00:00 is NEBD. Part I: Centrosomes are required for chromosome clustering. Three zygotes expressing mClover3-MAP4-MTBD (white, microtubules), H2B-mScarlet (magenta, DNA), and GST (left) or KASH5-DN (middle and right). Arrowheads indicate detached centrosomes before and after NEBD. Z-projections, 9 sections every 3.08 μm. Part II: Microtubules are required for chromosome clustering. Two zygotes expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA) treated with DMSO (left) or nocodazole (right). Z-projections, 7 (left) and 8 (right) sections every 2.50 μm. Part III: Dynein is required for chromosome clustering. Two zygotes expressing mClover3-MAP4-MTBD (white, microtubules) and H2B-mScarlet (magenta, DNA) injected with BSA (left) or P150-CC1 (right). Z-projections, 8 (left) and 13 (right) sections every 2.50 μm
Part I: Time-lapse movie of a zygote expressing mClover3-MAP4-MTBD (orange, microtubules) and POM121-mScarlet (green, NPC). Arrowheads indicate annulate lamellae moving toward the centrosomes. Time, hours:minutes, 00:00 is NEBD. Single sections confocal microscopy. Part II: Nuclear pore complexes cluster with chromatin at the pronuclear interface in bovine zygotes. Airyscan sections of a bovine zygote stained for microtubules (orange, α-tubulin), NPCs (green, NPC-Mab414), and DNA (magenta, DAPI). Outlined regions magnified in bottom row. Centrosomes, NPCs, and annulate lamellae are indicated. One section corresponds to 0.18 μm, as indicated. Part III: Nuclear pore complexes cluster with chromatin at the pronuclear interface in human zygotes. Airyscan sections of a human zygote with three pronuclei stained for microtubules (orange, α-tubulin), γ-tubulin (white, g-tubulin), NPCs (green, NPC-Mab414), and DNA (magenta, DAPI). Outlined regions magnified in bottom row. Centrosomes, NPC, and annulate lamellae are indicated. One section corresponds to 0.16 μm, as indicated.
Data Availability Statement
Scripts and plugins are available at link: https://gitlab.gwdg.de/schuh-meiosis/cavazza_etal_cell_2021_parental_genome_unification