SUMMARY
Viral genetic tools that target specific brain cell types could transform basic neuroscience and targeted gene therapy. Here, we use comparative open chromatin analysis to identify thousands of human-neocortical-sub-class-specific putative enhancers from across the genome to control gene expression in adeno-associated virus (AAV) vectors. The cellular specificity of reporter expression from enhancer-AAVs is established by molecular profiling after systemic AAV delivery in mouse. Over 30% of enhancer-AAVs produce specific expression in the targeted subclass, including both excitatory and inhibitory subclasses. We present a collection of Parvalbumin (PVALB) enhancer-AAVs that show highly enriched expression not only in cortical PVALB cells but also in some subcortical PVALB populations. Five vectors maintain PVALB-enriched expression in primate neocortex. These results demonstrate how genome-wide open chromatin data mining and cross-species AAV validation can be used to create the next generation of non-species-restricted viral genetic tools.
Graphical Abstract

In brief
Viral genetic tools that target specific brain cell types could transform basic neuroscience and targeted gene therapy. Mich et al. use comparative open chromatin analysis to identify human neocortical enhancers that can drive gene expression from AAV vectors in a subclass-specific fashion in multiple species.
INTRODUCTION
A major goal in neuroscience is to establish the distinct role of each cell population in brain circuitry, how they give rise to complex function, and how their dysfunction can cause disease. Most basic research in neuroscience and neurological diseases occurs in rodents, although it is often not known if the functional roles of cell populations are conserved. Comparison of gene expression between mouse and human shows strong conservation of molecular features across brain cell classes (e.g., inhibitory, excitatory, and glial classes) and subclasses (e.g., Parvalbumin [PVALB], Somatostatin [SST], and Vasoactive intestinal polypeptide [VIP] subclasses). However, direct cross-species correspondences between the most granular divisions in the cell type taxonomy (i.e., cell types) can be challenging due to cross-species variation, with the exception of a handful of highly distinctive cell types (Hodge et al., 2019). New somatic genetic tools to label orthologous neuronal subclasses across species will be highly impactful to directly target and compare conserved and divergent properties of orthologous subclasses.
Viral vectors have recently been shown to allow transgene delivery and genetic marking of neurons from mouse to humans (Dimidschstein et al., 2016; Andersson et al., 2016; Ting et al., 2018; Schwarz et al., 2019; Vormstein-Schneider et al., 2020). Adeno-associated viruses (AAVs) are ubiquitous nonpathogenic viruses that allow transduction of adult post-mitotic neurons and could be leveraged to build tools for genetic access to specific brain cell subclasses. AAV capsids have also been engineered to deliver specific transgenes in many tissues (Tervo et al., 2016; Deverman et al., 2016; Chan et al., 2017; Greig et al., 2018; Song et al., 2019), and specific promoters and enhancers can be used to control transgene expression from recombinant AAVs (Nord et al., 2013; Visel et al., 2013; Silberberg et al., 2016; Dimidschstein et al., 2016; Xiong et al., 2019; Jüttner et al., 2019; Nair et al., 2020; Markenscoff-Papadimitriou et al., 2020; Vormstein-Schneider et al., 2020). However, few suitably compact cell-class- or subclass-specific regulatory elements are known that function across mammalian species and can readily fit into an AAV genome (Dimidschstein et al., 2016; Mehta et al., 2019; Vormstein-Schneider et al., 2020). A complete set of compact enhancers for specific transgene expression in the brains of multiple species, including humans, will help realize the promise of AAVs for manipulating specific brain cell classes, subclasses, and types.
Open chromatin profiling with single-cell resolution techniques matched across multiple organisms allows direct discovery of conserved compact gene regulatory elements. Detailed single-cell assay for transposase accessible chromatin with sequencing (scATAC-seq) datasets profiling mouse brain now exist (Cusanovich et al., 2018; Fang et al., 2021; Lareau et al., 2019; Liu et al., 2020; Li et al., 2020; Preissl et al., 2018), but open chromatin datasets from human brain have been limited (Luo et al., 2017; Fullard et al., 2018; Lake et al., 2018). More high-quality human single-nucleus ATAC-seq (snATAC-seq) data (Bakken et al., 2020) will reveal the regulatory elements that confer human cell type molecular identity and could enable their specific genetic access via viral vectors.
Here, we present a multistep process to generate AAV vectors that drive cell-subclass-specific reporter expression across species. We established a robust snATAC-seq methodology using fresh neurosurgically resected human temporal cortex tissue and used the resulting data to generate a subclass-resolution human neocortex catalog of putative functional enhancers. Comparison to a similar mouse dataset (Graybuck et al., 2021) revealed conserved and divergent subclass-specific putative regulatory elements, which we leveraged to build reporter-AAV vectors. A cross-species enhancer validation process was established to evaluate reporter expression brain-wide in mouse, and in vivo or ex vivo in primate neocortex, followed by molecular confirmation of cell subclass or type with multiplexed fluorescence in situ hybridization (mFISH), immunohistochemistry (IHC), and single-cell RNA sequencing (scRNA-seq). We generated a collection of subclass-specific AAV vectors that drove neocortical transgene expression patterns predicted by enhancer accessibility profiles from both excitatory and inhibitory subclasses. A collection of PVALB-specific vectors was identified that labeled the PVALB subclass in the mouse visual cortex (VISp), and some vectors also labeled distinct subsets of subcortical Pvalb+ cells. We further tested PVALB-specific AAV vectors and show they maintain specificity in non-human primate (NHP). These results provide a generalizable strategy for the identification of enhancers that function in AAV vectors to drive gene expression in cell classes and subclasses across the brain and across mammalian species.
RESULTS
Open chromatin analysis of human neurons
To find distinguishing neocortical-cell-subclass-specific enhancers, we generated high-quality chromatin accessibility profiles from multiple middle temporal gyrus (MTG) neurosurgical specimens that were never frozen (bulk, n = 5; single nucleus, n = 14; Table S1) using ATAC-seq (Buenrostro et al., 2015; Gray et al., 2017; Graybuck et al., 2021) on bulk populations (Figure S1) and sorted single nuclei (Table S2). We prepared 3,660 individual snATAC-seq libraries from single nuclei that were targeted for sorting and analysis according to the presence or absence of neuronal nuclear protein (NeuN) (median of 48,542 uniquely mapped reads per nucleus). Of these, we used 2,858 quality-control-filtered nuclei for clustering and mapping to human snRNA-seq data (Hodge et al., 2019; Figure 1A; Table S2). We excluded nuclei with fewer than 10,000 unique reads, a transcription start site (TSS) enrichment score of <4, or <15% of reads overlapping with known DNase I hypersensitivity peaks isolated from human prefrontal cortex (ENCODE Project Consortium, 2012). We defined 27 robustly detectable snA-TAC-seq clusters (Figure S2) that were mapped by Cicero (Pliner et al., 2018) to the transcriptomic classification at the level of cell subclasses or cell types (Figures 1B and S3). Overall, the cells mapped to all three major classes of brain cells: excitatory, inhibitory, and non-neuronal, which we subdivided into 11 subclasses: excitatory layer 2/3 (L23), L4, L5/6 intra-telencephalic projecting (L56IT), and deep layer non-intratelencephalic projecting neurons (DL); inhibitory LAMP5, VIP, SST, and PVALB neurons; and non-neuronal astrocytes (Astro), microglia (Micro), and oligodendrocytes/oligodendrocyte precursor cells (OPCs) (OligoOPC). In support of the accuracy of mapping to transcriptomic cell subclasses, nuclei microdissected and sorted from superficial neocortical layers usually mapped to superficial cell subclasses (L23, LAMP5, or VIP), nuclei microdissected from deep layers mapped to cells found in infragranular neocortical layers (DL or L56IT), and NeuN-negative cells predominantly mapped to non-neuronal cell subclasses (Figures 1C and S3G). Several snATAC-seq clusters mapped to the same subclass (Figure S3E); in particular, L23 cells contained several clusters showing donor-specific signatures (Figure 1D) that could have arisen from either true inter-individual variation or inexact regional targeting of the surgical specimens. Regardless, all subclasses contained nuclei from multiple specimens (Figure 1D).
Figure 1. A database of human neocortical cell subclass-specific accessible chromatin elements.
(A) Workflow for human neocortical open chromatin characterization. See STAR Methods for details.
(B–D) High-quality nuclei (2,858 from 14 specimens) visualized by t-distributed stochastic neighbor embedding (t-SNE) and colored according to mapped transcriptomic cell types grouped into cell type subclass (B), sort strategy (C), or specimen (D).
(E) Transcriptomic abundances of 11 cell subclass-enriched marker genes (median counts per million [CPM] within subclass) for 11 subclasses of cell types identified in human MTG (Hodge et al., 2019).
(F) Eleven example subclass-specific marker genes demonstrating uniquely accessible chromatin elements in their vicinity (less than 50 kb distance to gene). Pileup heights are scaled proportionally to read number, and yellow bars highlight subclass-specific peaks for visualization. Dashed lines, introns; thick bars, exons; arrows, direction to gene body.
To identify putative regulatory elements within each subclass, we aggregated the data for all nuclei within each subclass and identified peaks (median length of 411 bp across subclasses) using the peak-calling program Homer (Heinz et al., 2010). This analysis revealed peaks proximal to recently identified transcriptomic subclass-enriched marker genes (Hodge et al., 2019), further confirming our clustering and mapping strategy (Figures 1E, 1F, S2, and S3). We then used chromVAR (Schep et al., 2017) to identify differentially enriched transcription factor (TF) family motifs for known neuronal regulators. These TF motifs were strongly correlated with their TF transcript abundances from snRNA-seq data (Figures S4A-S4D; Hodge et al., 2019). Together, these analyses demonstrated strong concordance between snRNA-seq and snATAC-seq data modalities at the cell subclass level.
Concordance of epigenetic marks in human neurons from distinct profiling techniques
We calculated the overlap between subclass snATAC-seq peaks and differentially methylated regions (DMRs) previously identified from human frontal cortex single-nucleus methylcytosine sequencing (snmC-seq; Table S3; Lister et al., 2013; Luo et al., 2017). For every cell subclass, we observed a greater overlap of snATAC-seq peaks with DMRs than expected by chance (Figure S4E), revealing thousands of independently observed neocortical regulatory elements (from 1,253 in microglia to 123,665 in L23 neurons) by the intersection of both DMR and snATAC-seq data. In total, 27% ± 20% (mean ± SD) of all human peaks were also identified as DMRs. Peaks from all subclasses displayed greater than random conservation of primary DNA sequence as measured by phyloP scores (Figure S4F; Pollard et al., 2010). Together, these analyses suggest that snATAC-seq faithfully detects DNA elements that have undergone positive selection through evolution, and likely play a functional role in these diverse cell types.
Conserved and divergent functional genomic elements across species
To identify regions of chromatin accessibility shared with mouse (“conserved”), as well as those present only in human or mouse (“divergent”), we aggregated mouse scATAC-seq peaks (Graybuck et al., 2021) to match our human dataset and then computed Jaccard similarity coefficients between human and mouse subclasses by counting peak overlaps (STAR Methods). All mouse subclasses displayed the highest similarity to the orthologous human subclasses, and all but one human subclass, hL56IT, matched reciprocally (Figure 2A). Non-neuronal classes displayed the strongest cross-species similarity, followed by inhibitory neurons, whereas excitatory neurons displayed the weakest correspondence (Figure 2A). The weak correspondence of excitatory neurons was likely partially due to regional mismatch between the mouse (VISp) and human (MTG) sc/snA-TAC-seq datasets (Graybuck et al., 2021), since excitatory cortical neurons are known to have distinct expression profiles across regions (Tasic et al., 2018). Nevertheless, this analysis yielded many more conserved peaks than expected by chance alone (Figure 2B, **false discovery rate [FDR] <0.01 in each subclass). In sum, 34% ± 10% (mean ± SD) of all human peaks were also detected in matching mouse subclasses. Conserved peaks exhibited significantly greater primary sequence conservation than divergent peaks in both species (heteroscedastic t test; human t = 10.3, p < 0.001; mouse t = 6.6, p < 0.001; Figure 2C), supporting the notion that snATAC-seq reveals genomic elements that perform evolutionarily conserved functions. Consistent with this idea, using linkage disequilibrium score correlation (LDSC; Bulik-Sullivan et al., 2015; Finucane et al., 2015), we found that SNPs linked to educational attainment and schizophrenia were more closely associated with conserved neuronal peaks than with divergent neuronal peaks (Figures 2D-2F; see STAR Methods for details). However, a notable counterpoint is the association between microglia and Alzheimer’s disease (Cusanovich et al., 2018; Girdhar et al., 2018; Skene et al., 2018; Nott et al., 2019), which showed stronger association within divergent human peaks than within conserved peaks (Figure 2E), suggesting that Alzheimer’s-related microglial dysfunction is associated with human regulatory domains not present in mice (Zhou et al., 2020).
Figure 2. High conservation of human neocortical accessible genomic elements and association with disease.
(A) Jaccard similarity coefficient enrichments (ratio of real to randomized peak positions) between human and mouse neocortical cell subclasses. Subclass-specific peaksets almost always best match their orthologous peakset across species.
(B) Visualization of conserved (Cons.) and divergent (Div.) peak counts across cell subclass in human and mouse. Conserved peaks are more frequent than expected by chance (**FDR < 0.01).
(C) Greater primary sequence conservation for conservedly accessible peaks than for divergently accessible peaks in both human and mouse. ***p < 0.001 by heteroscedastic t test (human t = 10.3, df = 18.5; mouse t = 6.6, df = 19.9). Dashed line indicates no difference between real and randomized peak positions.
(D) Associations between GWAS-identified loci and subclass ATAC-seq peaksets (top) and methylation DMRs (bottom; Lister et al., 2013; Luo et al., 2017). Heatmap fill represents ratio of the proportion of heritability contained by that peakset’s linked SNPs, to the proportion of that peakset’s linked SNPs, as calculated by LDSC (Bulik-Sullivan et al., 2015; Finucane et al., 2015). Outline color marks significance; Bonferroni correction for multiple hypothesis testing (180 tests for ATAC-seq peaks and 150 tests for DMRs).
(E) Associations between conserved and divergent human ATAC-seq peaks, and GWAS-identified loci. Outline color marks significance; Bonferroni-corrected p values are employed (345 tests performed).
(F) Total summed heritability of all SNPs associated with conserved peaks versus those associated with divergent peaks, for three studies with multiple significant neuron subclass associations. ***p < 0.01 by heteroscedastic t test, t = 3.8, degrees of freedom (df) = 45.6.
Additionally, we sought to understand how global genetic regulation differs across species and among cell subclasses. We first performed unbiased de novo identification of DNA sequence element motifs using MEME-CHIP (Bailey et al., 2009), which were then filtered for expression of a possible binding site-correlated TFs by RNA sequencing (RNA-seq) (Figure S4G; Tasic et al., 2018; Hodge et al., 2019). This analysis revealed several known cell-subclass-specific TFs (e.g., SPI1/PU.1 in microglia and OLIG2 in oligodendrocytes/OPCs) and many unappreciated subclass-specific TFs (e.g., the TEAD motif is the most significant motif observed in human astrocytes but absent from mouse astrocytes; Figure S4G). We also measured the association of peaks with common genomic repetitive elements. Across cell subclasses and species, divergent peaks more commonly overlap with mobile repetitive genetic elements than conserved peaks do (Figures S4H and S4I), suggesting a means for their dispersal, duplication, and mutagenesis during mammalian evolution (Van’t Hof et al., 2016; Gao et al., 2018). As a whole, these comparative analyses of single-cell open chromatin data furnish a wealth of knowledge about cell-type identity determinants and origins.
Identifying functional enhancers using AAV reporter vectors
To determine whether ATAC-seq peaks might provide useful enhancers for developing novel genetic tools as had been previously shown (Dimidschstein et al., 2016; Nair et al., 2020), we cloned DNA corresponding to several peaks into a super yellow fluorescent protein-2 (SYFP2) reporter-AAV vector backbone and packaged viral particles with the mouse blood-brain-barrier-penetrant capsid PHP.eB (Figure 3A; Chan et al., 2017). We found that 1 × 1011 vector genomes (vgs) of AAV2/PHP.eB vector delivered intravenously (retro-orbital injection) demonstrated wide tropism for many brain neurons, as shown using the pan-neuronal promoter hSyn1 (Figure 3B; McLean et al., 2014). Furthermore, we could also drive reporter expression in specific brain regions and defined neuron classes using enhancers, such as telencephalic interneurons with hDLXI56i (Figure 3C; Zerucha et al., 2000; Dimidschstein et al., 2016).
Figure 3. Accessible chromatin elements furnish cell subclass-specific AAV genetic tools.
(A) AAV2/PHP.eB viral reporter vector design for testing presumptive enhancers cloned upstream of a minimal promoter and SYFP2 reporter expression cassette in mouse retro-orbital assay.
(B) Transgene expression from AAV-hSyn1-H2B-SYFP2 in most neurons throughout mouse brain.
(C) Transgene expression from AAV-hDLXI56i-minBG-SYFP2 in mouse forebrain interneurons, in agreement with previous reports (Zerucha et al., 2000; Dimidschstein et al., 2016).
(D) Several identified enhancers showing ATAC-seq peaks in distinct target cell subclasses. Each selected enhancer is highlighted in yellow on read pileups, and heatmap below demonstrates ATAC-seq read CPM in all cell subclasses.
(E) Distinct expression patterns from these enhancer-AAV vectors in live 300-μm-thick slices of primary visual cortex (VISp) after retro-orbital delivery, consistent with different subclass-specific expression patterns.
(F) Multiplexed FISH in VISp region revealing differing subclass specificities from various enhancer-AAV vectors. Text represents mean ± SD for labeling specificity across three independent mice.
(G)scRNA-seq on sorted individual SYFP2+ cells from VISp region confirming distinct cell subclass transcriptomic identities labeled by the highlighted enhancer-AAV vectors.
We took several strategies to identify enhancers with cell-class- and subclass-specific activity. In one approach, we manually identified peaks in the locus of known subclass marker genes from snRNA-seq (Hodge et al., 2019), as shown for eHGT_078h, 058h, hDLXI56i (previously known), 019h, and 017h (Figure 3D). We selected these peaks from the bulk-layer-specific open chromatin data, based on neuronal (NeuN+), and layer enrichment (Figure S1; Hodge et al., 2019). In a second approach, we identified subclass-specific peaks that were conserved or divergent across human and mouse sn/scATAC-seq and snmC-seq data (e.g., eHGT_128h; Figure 3D; Luo et al., 2017; Graybuck et al., 2021). All enhancer-AAV vectors were systemically administered to mouse and cell subclass- and type-specific reporter expression was validated by both mFISH (Choi et al., 2018) and scRNA-seq from the VISp (Figures 3E-3G and S5; Tasic et al., 2016, 2018).
We discovered several enhancer-AAV vectors that drove distinct reporter expression patterns consistent with their accessibility profiles in neocortical cells (Figures 3D and 3E). These vectors drove reporter expression in excitatory neurons (eHGT_078h), inhibitory neurons (hDLXI56i, Zerucha et al., 2000; Dimidschstein et al., 2016), Rorb+ L4 and L56IT excitatory neurons (eHGT_058h), LAMP5 inhibitory neurons, (eHGT_019h), SST and VIP inhibitory neurons (eHGT_017h), and PVALB (eHGT_128h) inhibitory neurons . As demonstrated by mFISH, some enhancer-AAVs had low specificity (defined as 45%–80% on-target labeled cells); for example, eHGT_019h labeled cells in VISp that were 68% ± 9% Lamp5+ interneurons (Figure 3F). Other enhancer-AAVs showed high specificity (defined as >80% on-target labeled cells), such as eHGT_058h, which labeled cells that are 82% ± 1% Slc17a7+Rorb+ L4 and L56IT neurons (Figure 3F). Finally, scRNA-seq confirmed the transcriptomic identity of the labeled cells with each of these viral vectors at the subclass (Figure 3G) and type levels (Figure S5).
A collection of enhancer-AAVs that label the PVALB subclass
We sought to identify a collection of enhancers to enable access to PVALB interneurons that are important for cortical microcircuit regulation and are implicated as dysfunctional in epilepsy, schizophrenia, and Alzheimer’s disease (Cheah et al., 2012; Verret et al., 2012; Mukherjee et al., 2019). Since we identified many PVALB-specific open chromatin regions, we tested if they could confer PVALB-subclass-specific expression in AAV vectors, similar to recent reports (Mehta et al., 2019; Vormstein-Schneider et al., 2020). We identified, cloned, and tested 20 independent enhancers that showed differing levels of specific accessibility for PVALB interneurons (Figure 4A). The first 10 enhancers were selected using the strategy of identifying neuronal-enriched open chromatin regions near PVALB-subclass marker genes from layer-microdissected bulk population ATAC-seq data (Figure S1). Two of the first 10 enhancer-AAV subset showed low specificity (eHGT_023h and 064h) of reporter expression in PVALB cells, and one (eHGT_079h) demonstrated high specificity (Figures 4A-4D), which agreed with retrospective assessment of the snATAC-seq data showing that only eHGT_079h demonstrated strong and exclusive accessibility in the PVALB subclass (Figure 4A). The remaining 10 enhancer-AAVs in the collection used enhancers selected based on single-cell-resolution open chromatin data that showed strong PVALB-subclass-specific peaks. Four enhancer-AAVs exhibited high specificity for PVALB neocortical neurons as predicted from the human open chromatin data (Figures 4E-4G and S6). These specificity levels were confirmed by both mFISH and scRNA-seq (Figures 4B-4M and S6). Neocortical VISp cells labeled by eHGT_023h were 47% ± 4% Pvalb+ interneurons (Figures 4B and 4H), whereas neocortical VISp cells labeled by eHGT_079h, 082h, 128h, 140h, and 359h were highly specific for Pvalb+ interneurons (92%–99% cells expressed Pvalb mRNA; Figures 4D-4G, 4J-4M, and S6). Intermediate to these is eHGT_064h that labels both Pvalb+ (50% ± 6%) and Sst+ neurons in VISp (54% ± 1%; Figures 4C and 4I), suggesting it enhances the nearby gene CRHBP which is primarily expressed in medial ganglionic eminence (MGE)-derived PVALB and SST interneurons (Tasic et al., 2018; Hodge et al., 2019). In agreement, 99% (183/185) of eHGT_064h-labeled cells expressed the MGE-derived inhibitory neuron marker Lhx6. Overall, 7 out of 20 (35%) of the tested enhancer-AAVs showed some level of specificity for PVALB neocortical cells in mouse. While only 1 out of 10 (10%) enhancers produced highly specific transgene expression after selection based on bulk ATAC-seq data and proximity to a marker gene, 4 out of 10 (40%) enhancer-AAVs showed high specificity for PVALB interneurons after selection based on enhancers enriched for the PVALB subclass in the snA-TAC-seq data.
Figure 4. PVALB neocortical interneuron enhancers display distinct subcortical expression patterns.
(A) Twenty putative PVALB enhancers from snATAC-seq data cloned into AAV vectors. Seven of the 20 (35%) exhibited low or high specificity for PVALB cells in mouse retro-orbital assay (indicated with green boxes).
(B–G) mFISH in L2/L3 of VISp demonstrating positive labeling of Pvalb+ cells (arrows) by each of the indicated enhancer-AAV vectors. eHGT_023h and eHGT_064h also label non-Pvalb+ cells (asterisks). Percentages indicate the mean ± SD of SYFP2 labeling specificity for Pvalb+ cells across three independent mice.
(H–M) scRNA-seq in VISp confirming the PVALB transcriptomic cell subclass identity of enhancer-AAV vector-labeled cells. Bar graph shows the percentage of single cells that map to a transcriptomic cell type within that subclass. In contrast, the percentages given in the text are the percentage of cells recovered that expressed the indicated gene. Note that although only 65% of the eHGT_079h-marked cell types mapped to the PVALB subclass, 94% of the eHGT_079h-marked cells expressed Pvalb mRNA. This is because several SST subclass cell types also express Pvalb mRNA.
(N) Pvalb mRNA expression pattern (Allen Institute public in situ hybridization data) with multiple sites of expression throughout mouse brain.
(O–T) Labeling of both neocortical PVALB cells and various subcortical brain regions by PVALB-specific enhancers. These subcortical brain regions are also seen in the endogenous Pvalb mRNA expression pattern. Two enhancers (eHGT_079h and 140h) show exceptional specificity to neocortical PVALB cells. CTX, cerebral cortex; HPF, hippocampal formation; MOB, main olfactory bulb; MB, midbrain nuclei; MY, medulla nuclei; P, pons; IC, inferior colliculus; CBX, cerebellar cortex; CBN, cerebellar nuclei.
(U and V) Subcortical labeling by eHGT_023h in Purkinje cells (U) and eHGT_082h in CBN (V). eHGT_023h-labeled Purkinje cells are Pvalb+Gad1+, and eHGT_082h-labeled CBN cells are either Pvalb+Gad1+ or Pvalb+Gad1−.
We were surprised to find that the least specific PVALB enhancer eHGT_023h is located within an intron of the PVALB gene itself, while the most specific PVALB enhancer eHGT_140h is not in the proximity of any known PVALB marker gene, instead being in an intron of NRF1, which is expressed by most cell types of the neocortex. This highlights the importance of genome-wide enhancer discovery and demonstrates that restriction of the enhancer search to known marker genes may not support comprehensive development of the most specific or useful viral tools. Most surprisingly, eHGT_079h, eHGT_128h and eHGT_140h produced enhancer-AAVs that were highly specific for mouse PVALB cells despite being accessible in human, but not mouse PVALB cells (Figure S7), showing that the human enhancer sequence is sufficient to confer specificity even in a species that does not use that particular enhancer endogenously.
Pvalb+ neurons are also located outside of the cortex (Figure 4N). We observed that some PVALB enhancer-AAV vectors labeled cells in known regions of Pvalb expression outside of the neocortex (Figures 4O-4T). For example, eHGT_023h-based reporter expression marked Purkinje cells in cerebellar cortex, mid/hindbrain nuclei, hippocampus, and main olfactory bulb neurons (Figures 4O and 4U), similar to Pvalb mRNA expression. eHGT_082h-based reporter expression labeled midbrain structures, deep cerebellar nuclei, and main olfactory bulb, but not Purkinje cells (Figures 4R and 4V). In contrast, eHGT_079h and eHGT_140h-based reporter expression labeled mostly neocortical interneurons (Figures 4Q and 4T). These results show that the identified enhancer elements contain both cortical PVALB cell subclass specificity, as well as differential specificity for Pvalb+ cells in other brain regions, which our enhancer selection strategy did not take into account.
Enhancer-AAV vectors enable genetic access to NHP neocortical PVALB cell types in vivo
To determine if our vectors maintain PVALB-subclass-specific expression in primates, we injected our enhancer-AAV vectors intraparenchymally in three NHP animals into multiple regions of the neocortex. We then evaluated expression specificity 51–113 days after injection (Figure 5A). We tested five PVALB-specific vectors identified from our mouse primary screening (eHGT_079h, 082h, 128h, 140h, and 359h) in the occipital cortex (Figures 5B-5F). By immunohistochemistry, these vectors were highly specific, with most vector-labeled cells expressing PVALB protein (86 to 98% of SYFP2+ cells). Furthermore, nearly all the PVALB+ cells throughout the cortical column within the core injection sites expressed SYFP2 from the enhancer-AAVs containing eHGT_128h and 140h (89% and 92%, respectively). This finding indicates not only that these vectors are highly specific for primate neocortical PVALB cells but also that PVALB cell labeling can be nearly complete (Figures 5B-5F). Next, we also injected eHGT_140h into three additional cortical regions (temporal, somatosensory, and motor cortex) and found that both specificity and completeness were moderate or high in each area (specificity range, 77%–95%; completeness range, 71%–92%), despite differing abundances of PVALB-immunore-active cells in each area. Enhancer-AAVs with eHGT_140h also occasionally labeled large pyramidal L5 neurons in addition to PVALB+ neurons (Figures 5G and 5I), which was also observed infrequently in sagittal mouse brain sections (less than an average of one cell per sagittal section), but never in VISp (data not shown). Finally, we performed brain-slice patch-clamp recordings of NHP motor cortex neurons labeled with eHGT_140h in vivo and demonstrated that all patched SYFP2+ interneurons showed fast-spiking properties (narrow action potentials [APs], large fast afterhyperpolarization [AHP] and the ability to sustain firing rates ≥200 Hz for 1 s), consistent with their identity as PVALB+ inhibitory neurons (Figure 5J). These observations confirm that our PVALB-specific enhancer-AAV vectors can provide prospective marking and experimental access to fast-spiking PVALB neurons in multiple cortical areas in mouse and macaque. We demonstrate cross-species conservation for five of five tested human genomic enhancers for PVALB subclass that were first validated in mouse.
Figure 5. Multiple PVALB enhancer vectors demonstrate cell subclass specificity across the NHP neocortex.
(A) Workflow for in vivo AAV vector testing by multisite intraparenchymal injection in NHP brain.
(B–E) Injection of eHGT_079h, 082h, 128h, and 359h AAV vectors into NHP occipital cortex. These four vectors label PVALB cells throughout the cortical column with high specificity and completeness. Colored dots indicate the positions of immunophenotypic counted cells observed by coimmunostaining with anti-GFP and anti-PVALB antibodies.
(F–I) Injection of eHGT_140h AAV vector into different NHP neocortical areas. This vector labels PVALB cells across multiple cortical areas with moderate or high specificity and completeness. Colored dots represent immunophenotypes of counted cells. Red arrows indicate rare labeled large L5 pyramidal neurons. Quantifications in each panel (B–I) represent >200 cells counted per vector in one experiment.
(J) Electrophysiological characterization of eHGT_140h+ neurons in motor cortex. Compared to unlabeled pyramidal neurons, eHGT_140h+ neurons display more and narrower APs and greater fast AHP amplitude, confirming their fast-spiking neuron identity. Data represent 14 recorded eHGT_140h+ neurons in one experiment and six recorded pyramidal neurons from a second experiment provided for contrast.
Enhancer-AAV testing in NHP and human ex vivo brain slices
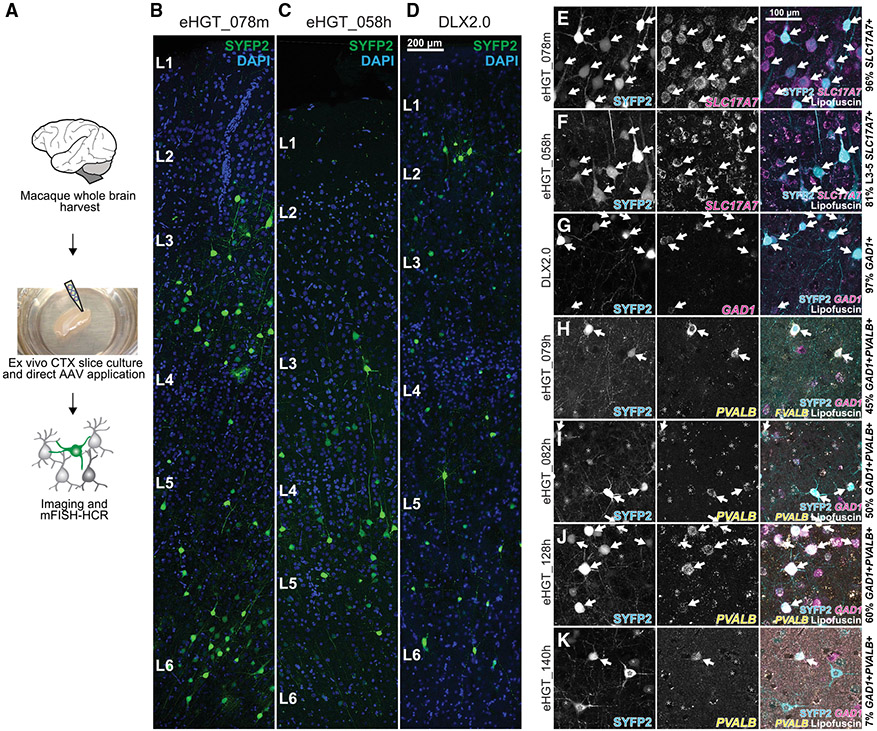
In vitro neocortical cultured slices can be used to characterize the cellular properties of primate neuronal subclasses in their native environment, and are the only viable option to evaluate them in human (Ting et al., 2018). We obtained NHP (Macaca nemestrina) temporal cortex tissue from the Washington National Primate Research Center (Figure 6A), and virally transduced ex vivo slices. After 1–2 weeks, select reporter vectors yielded expression consistent with that seen in mouse, including eHGT_078m (the mouse ortholog of eHGT_078h) in L2–L6 excitatory neurons (Figure 6B) and eHGT_058h in L3–L5 pyramidal neurons (Figure 6C). We could also consistently label GABAergic neurons with hDLXI56i (Dimidschstein et al., 2016), and an optimized version of that enhancer we call DLX2.0. DLX2.0 contains three core elements of the hDLXI56i enhancer in tandem, which resulted in stronger reporter expression in GABAergic neurons (Figure 6D). We confirmed specificity of labeling in these cultures by mFISH (Figures 6E-6G), which demonstrated these three vectors were highly class- or subclass-specific in NHP ex vivo slices just as in mouse (compare Figures 3E-3G with Figures 6E-6G). We further tested these vectors in human neocortical ex vivo slice culture to confirm our findings from NHPs (Figures 7A and 7B). scRNA-seq confirmed that hDLXI56i and DLX2.0 labeled human GAD1+ inhibitory neurons (97%–99%) of all major subclasses (Figures 7C and 7D). However, the PVALB vectors containing enhancers eHGT_079h, 082h, 128h, and 140h displayed low or no PVALB specificity in NHP slice cultures, unlike the high specificity seen in mouse or NHP in vivo (compare Figures 4D-4G, 5B-5D, and 5F with Figures 6H-6K). This loss of specificity was particularly profound in the case of eHGT_140h: 99% of transduced neocortical cells in mouse in vivo were Pvalb+, 77%–95% of transduced cells in NHP neocortex were PVALB+, while only 7% in NHP ex vivo neocortical slice cultures were PVALB+ (Figures 4G, 4M, 5F-5I, and 6K). Therefore, some cell-subclass-specific enhancers do not retain specificity in primate ex vivo slice culture, whereas others faithfully mark the same subclasses as in vivo.
Figure 6. Enhancer-AAV testing in NHP ex vivo neocortical slices.
(A) Workflow for acquiring fresh NHP neocortical tissue for AAV vector testing ex vivo.
(B–D) Transduction of ex vivo NHP neocortical tissue with various AAV2/PHP.eB enhancer-reporter vectors, resulting in diverse expression patterns. eHGT_078m labels excitatory neurons throughout all layers (B), eHGT_058h labels excitatory neurons primarily in L3–L5 (C), and DLX2.0 labels inhibitory neurons (D).
(E–K) NHP neocortical cell subclass specificity of AAV-vector labeling confirmed by mFISH. eHGT_078m, 058h, and DLX2.0 demonstrate high specificity similar to that seen in mouse retro-orbital assay, but eHGT_079h, 082h, 128h, and 140h show reduced specificity compared to that seen in mouse retro-orbital assay and NHP in vivo assay. Arrows highlight specifically labeled on-target cell types, and asterisks mark off-target labeled cells. Text represents mean for labeling specificity across one or two independent transduction experiments (>100 cells counted per vector per experiment).
Figure 7. Enhancer-AAV specificity of hDLXI56i and DLX2.0 vectors in ex vivo human neocortical slices.
(A) Workflow for acquiring fresh human neurosurgical tissue for AAV vector testing.
(B) AAV-DLX2.0-minBG-SYFP2 transduction of human ex vivo brain slice. Reporter fluorescence labels scattered neurons with diverse non-pyramidal cellular morphologies spanning all neocortical layers.
(C and D) Molecular identity of AAV-hDLXI56i-minBG-SYFP2+ (C) or AAV-DLX2.0-minBG-SYFP2+ (D) singly sorted human cells by scRNA-seq. The majority of human cells labeled by these vectors are inhibitory neurons of multiple transcriptomic types. Dendrogram represents human MTG taxonomy (Hodge et al., 2019), leaves represent 75 transcriptomic cell types, and circles represent labeled and sorted cells mapped onto the taxonomy. Circle size represents cell numbers. Circles on intermediate nodes of the dendrogram represent incomplete mapping to cell type. Data from eight independent experiments are shown (four in C and four in D).
DISCUSSION
Here, we present data and methodology to generate and evaluate AAV-based viral tools that drive brain-cell-subclass-specific transgene expression from mouse to primate. First, we report and characterize a subclass-resolution snATAC-seq dataset of human neocortex. Second, we compare this human open chromatin dataset to a comparable mouse cortex dataset (Graybuck et al., 2021) to identify conserved and divergent subclass-specific putative enhancers. Third, we show that many enhancers yield subclass-specific expression in mice once inserted into an AAV vector upstream of a minimal promoter and reporter gene. Fourth, we present a collection of AAV vectors designed to target PVALB interneurons, with an efficient on-target rate of 40% of tested vectors yielding PVALB-subclass-specific expression in the mouse cortex when enhancer selection was based on subclass-specific open chromatin peaks revealed from snATAC-seq data. Fifth, we demonstrate that many enhancers parcellate the expression patterns of marker genes whose expression we are trying to replicate (such as Pvalb), with some labeling multiple subcortical neuron populations and others highly specific for neocortical interneuron populations. Sixth, we show that five vectors (using enhancers eHGT_079h, 082h, 128h, 140h, and 359h) labeled PVALB neocortical inhibitory neurons in NHP after intraparenchymal injection in brain, demonstrating the maintenance of specificity across species. Last, we confirm several class- and subclass-specific AAV-reporters maintained faithful reporter expression in NHP and human ex vivo neocortical slices, whereas the PVALB-specific AAV vectors exhibited substantially reduced specificity for the PVALB subclass. These AAV vectors constitute some of the first genetic tools with validated subclass specificity for neocortical cell types across multiple mammalian species. These results indicate that snATAC-seq-guided enhancer discovery is a generalizable strategy to efficiently identify cell subclass-specific enhancers (Graybuck et al., 2021) for the observation and perturbation of brain cell subclasses and types in a non-species-restricted manner.
Single-cell-resolution open chromatin datasets uncover enhancers
Our subclass-resolution snATAC-seq profiling experiments from the human MTG were critical for the development of the cell-subclass-specific viral tools presented here. Most enhancers identified in our study were not visible in bulk open chromatin datasets (ENCODE Project Consortium, 2012; Fullard et al., 2018). This is partially due to bulk data masking cell-subclass-specific peaks when the target cell population is not abundant. For instance, inhibitory neurons comprise only 20%–30% of all cortical neurons, and each inhibitory subclass is a fraction of the total inhibitory cells (Tasic et al., 2018; Hodge et al., 2019). Single-nuclear-resolution ATAC-seq using freshly isolated nuclei from acutely resected neurosurgical tissue revealed many subclass-specific candidate enhancers.
With high-quality and single-cell-resolution open chromatin data across species, we have made insights into the gene expression regulatory apparatus, and how it varies across subclasses and species. snATAC-seq and snmC-seq studies show high overlap of hypomethylated regions with open chromatin at the single-cell level (27% of snATAC-seq peaks detected as snmC-seq DMRs; Figure S4E; Luo et al., 2017). Human and mouse snATAC-seq data agree strongly between species, with 34% of all human ATAC-seq peaks also detected in the matched mouse subclass (Figures 2A and 2B). We could also predict TFs enriched at cell-subclass- and species-specific peaks (Figure S4G), and through analysis of peak overlap with repetitive genomic elements, we could infer the mechanisms by which enhancers might evolve and expand cell-type diversity (Figures S4H and S4I). Last, we evaluated how different cell subclass-specific peaks associated with genomic intervals highlighted by genome-wide association studies (GWASs) of neurological diseases (Figures 2D-2F), showing that conserved neuron-specific peaks were associated with schizophrenia, while divergent microglial-specific peaks were associated with Alzheimer’s disease. In summary, high-quality open chromatin datasets enabled biological discovery of the evolution, transcriptomic control, and disease association of human brain cell subclasses.
Building the next generation of AAV-based genetic tools
The identification of functional enhancers has been a challenge, and multiple prediction criteria have been used previously. Selection of open chromatin regions proximal to known marker genes is sometimes successful, but this criterion limits the number of putative enhancers to test. The best proximal enhancer to the PVALB gene that we could identify (eHGT_023h) was only weakly specific, while others (eHGT_082h, 128h, and 140h) were highly specific for PVALB-subclass neurons but do not reside in the proximity of any known PVALB cell marker genes. Importantly, removing the restriction of sampling open chromatin regions proximal to known marker genes greatly increases the number of specific elements available to test. We also noted that conservation of sequence or open chromatin from mouse to human is not essential to find functional enhancers that drive subclass-specific expression in mouse and primate. For example, eHGT_079h is not conserved between human and mouse, yet it showed highly specific expression in the PVALB subclass in cortex in both mouse and primate. This demonstrates that identification of conserved enhancers may not be necessary to create vectors for cross-species applications and that a human enhancer sequence can maintain specificity even in a species that does not natively use that particular enhancer. Future studies could reveal insights into the endogenous roles of these nonconserved enhancers.
We have learned several lessons about enhancer discovery through our analysis of the AAV vectors presented in this study. Our technique for screening enhancer-AAV vectors in mouse in vivo enabled us to see whole-brain expression patterns, and we were surprised by the diversity of subcortical cell populations that were labeled by our PVALB vectors. These vectors labeled the PVALB cell subclass within the neocortex as predicted from the open chromatin data, but their subcortical expression patterns vary dramatically. For some vectors, the expression is neocortex-restricted, while other vectors also label various Pvalb+ cell populations in subcortical regions. Since all epigenetic profiling that informed our enhancer discovery is from neocortex (mouse VISp; Graybuck et al., 2021; human MTG; or mouse or human frontal cortex; Luo et al., 2017), we could not have predicted whether these enhancers would drive expression in other brain regions. Clearly, single-cell epigenetic profiling of multiple brain regions will be required to accurately predict the regional specificity of enhancer activity (Li et al., 2020; Liu et al., 2020). Additionally, the distinct patterns of expression for our PVALB enhancer-AAVs hint toward an additive “Lego logic” of enhancers that must act together to yield the complete endogenous expression pattern of a gene of interest. Individual enhancers for a given cell subclass often show a more restricted expression pattern than seen from the best marker genes and could be used to produce highly targeted expression of transgenes in discrete brain regions.
Massively parallel enhancer screens (Shen et al., 2016; Hrvatin et al., 2019) have identified few specific enhancers from libraries of hundreds or thousands of candidate enhancers. In contrast, we show the efficient identification of specific enhancers using a one-by-one enhancer screening strategy informed by single-cell-resolution epigenetic data. Seven of the 20 viral vectors tested for specificity in PVALB cells showed significant on-target reporter expression. Since we identified thousands of putative subclass-specific enhancers from our open chromatin data (range, 1,756 in OligoOPC to 26,688 in L23) and have found vectors with a range of subclass specificities in this study and our companion study (Graybuck et al., 2021), we expect that we will be able to generate many additional enhancer-AAVs specific for each subclass without implementing more complex batched screening strategies. Specific candidate enhancer identification will be further aided by epigenetic datasets with higher resolution (Bakken et al., 2020; Li et al., 2020; Liu et al., 2020).
Enhancer-AAVs function across mammalian species
Testing enhancers in primates is useful to determine if the enhancer-AAVs will be functional across species (Jüttner et al., 2019; Mehta et al., 2019). Two primate preparations were used in this study, in vivo injections into NHP neocortex and ex vivo organotypic slices from NHP or human neocortex. We tested five PVALB enhancer-AAV vectors using intraparenchymal injections into the occipital cortex in macaque monkeys and obtained strong evidence for conservation of cell-subclass-specific expression from mouse to monkey. This confirmed that our selection method and mouse screening strategy can be effective for identifying enhancers that function across species. Since such virus injections in NHP are costly, challenging to execute, cannot be effectively scaled, and cannot be applied in human, we also applied the approach of AAV transduction in ex vivo organotypic slice culture of primate brain tissue (Ting et al., 2018). Using this strategy, we were able to show that DLX2.0 maintained specific expression from mouse to NHP and human and that several other vectors showed matched subclass specificities between mouse in vivo and NHP ex vivo. However, several PVALB enhancers, such as eHGT_140h, produced enhancer-AAVs that exhibited low or no specificity in the ex vivo paradigm. This demonstrated that some, but not all, enhancer-AAVs can be used to mark the same cell subclasses in ex vivo slice cultures as in vivo. Future work is needed to understand why some enhancers behave differently or how to improve in vitro culture methods to more closely mimic the in vivo condition, but these results highlight the importance of in vivo validation in mouse and NHP.
Conclusions
Human brain function and disease are difficult to study because model organisms do not recapitulate human brain circuitry or display clear clinically relevant phenotypes. Here, we describe a process for generating and validating viral genetic tools to allow interrogation of brain circuit components in mouse and primate. We have cataloged human neocortical chromatin accessibility with single-cell resolution, which deepens our knowledge of human brain cell subclass-specific gene regulation. Guided by the epigenetic data, we have built a collection of subclass-specific AAV tools and established an efficient platform to validate enhancer-AAV activity across species. The AAV tools, screening platform, open chromatin data and analyses presented here will accelerate progress toward functional dissection of brain circuits across mammalian species and could improve our understanding and our ability to treat human neurological diseases.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Boaz Levi (boazl@alleninstitute.org).
Materials availability
Plasmids generated in this study have been deposited to Addgene.
Data and code availability
Raw human bulk ATAC-seq data, human snATAC-seq data, human snRNA-seq data, and mouse snRNA-seq data have been deposited to dbGaP.
dbGaP study name: “Development of tools for cell-type specific labeling of neocortical neurons”
The accession number for the data reported in this paper is dbGaP: phs2292.v1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human neurosurgical samples
All human studies are approved by the Western Institutional Review Board, with informed consent obtained from all donors prior to tissue experimentation. Patient demographic information used for collecting open chromatin and transcriptomic data is shown in Table S1. We did not observe any obvious sex- or gender-specific clusters or signatures in snATAC-seq data or in enhancer-AAV vector transduction, but this study was not designed to detect them.
Mouse viral vector testing
All experiments were approved under protocol 1702 by the Institutional Animal Care and Use Committee (IACUC) at the Allen Institute for Brain science. C57BL/6J (stock # 000664) and Gad2-T2a-NLS-mCherry (stock # 023140) mice were purchased from the Jackson Laboratory, and the Gad2-T2a-NLS-mCherry line was maintained by homozygous inbreeding. Male mice between P42-P70 were injected with enhancer-AAV vectors retro-orbitally and sacrificed for expression after 21-28 days. Numbers of mice used per experiment are shown in each figure (n = 2-3 per vector), and no mice were excluded from analysis. No randomization or blinding was performed. Since all experimental mice were male, we did not detect sex differences in enhancer-AAV vector transduction.
Non-human primate viral vector testing
All procedures used with non-human primates conformed to the guidelines provided by the US National Institutes of Health. In vivo injection experiments were approved under University of Washington IACUC protocol number 4167-01. We used three animals housed at the Washington National Primate Research Center (Seattle, WA) for these experiments with multisite injections into occipital, temporal, motor, and somatosensory cortex. Animal 1 was a 10-year-old female 7.2 kg Macaca mulatta and contained the occipital injection sites for eHGT_079h, eHGT_128h, and eHGT_140h, and the temporal injection site for eHGT_140h. Animal 2 was a 6-year-old male 11.6 kg Macaca nemestrina and contained the occipital injection site for eHGT_082h and the somatosensory injection site for eHGT_140h. Animal 3 was a 6-year-old male 12.0 kg Macaca nemestrina and contained the occipital injection site for eHGT_359h and the motor injection site for eHGT_140h. Each animal was healthy prior to, and following, surgery. No randomization or blinding was performed. No recovered injection sites were omitted from analysis. We did not observe any obvious effects of sex on enhancer-AAV vector transduction, but we did not design this study to detect them.
Ex vivo enhancer-AAV vector testing experiments were performed on tissue from healthy Macaca nemestrina animals housed at the Washington National Primate Research Center aged 2-15 years. We obtained these brain samples through the Tissue Distribution Program which is approved by protocol number 4277-01 at the University of Washington IACUC and follows a regular schedule. We did not observe any obvious effects of sex on enhancer-AAV vector transduction, but this study was not designed to detect them.
METHOD DETAILS
Neurosurgical tissue acquisition
We receive regular acute neurosurgical brain tissue donations at the Allen Institute for Brain Science. These samples are excised as a matter of course to access the epileptic focus or tumor. All samples used in this study were derived from temporal cortex, most frequently middle temporal gyrus (MTG). These samples are immersed in pre-carbogenated ACSF.7 (recipe below), transported to the Allen Institute for Brain Science rapidly with carbogenation, and sliced on a compresstome (Precisionary Instruments, Greenville NC USA, catalog #VF-200) into 350 μm slices, and continuously carbogented in ACSF.7 until dissociation.
Bulk tissue ATAC-seq
We harvested MTG tissue slices after carbogen bubbling in ACSF.7 for up to 16 hours, and we treated with NeuroTrace 500/525 (catalog # N21480 from ThermoFisher Scientific, 1/100 in ACSF.7) to highlight layered cortex structure. With fine forceps we trimmed away white matter and meningeal tissues, and then dissected layers 1-6 into six different low-binding Eppendorf 1.5 mL tubes (MilliporeSigma catalog # Z666548) under a fluorescence microscope as in Hodge et al. (2019). We discarded supernatant and replaced with 50-100 μL of Nextera DNA library reaction (#FC-121-1031 from Illumina) containing 0.1% IGEPAL-630 (NP-40 alternative), and then pipetted up and down vigorously 25-50 times using a P200 pipette, and then incubated at 37°C for one hour for transposition. We then added 1 mL of Homogenization Buffer (recipe below) to quench the reaction, pelleted samples at 1000 g for 5 minutes at 4°C, resuspended samples in 1 mL fresh homogenization buffer, released nuclei from samples using ~10-15 strokes of a loose-fitting dounce pestle followed by ~10-15 strokes of a tight-fitting dounce pestle, then filtered nuclei with a 70 μm nylon mesh strainer, and pelleted nuclei at 1,000xg for 10 minutes at 4°C. To stain, we resuspended nuclei in 500 μL of ice-cold Blocking Buffer (recipe below) containing 1/500 PE-NeuN antibody (MilliporeSigma catalog # FCMAB317PE) and 1 μg/mL 4’-diamino-phenylindazole (DAPI, MilliporeSigma catalog # D9542), rocked samples for 30 minutes at 4°C, then pelleted at 1,000xg for 5 minutes at 4°C, and finally resuspended samples in 500 μL fresh ice-cold blocking buffer before sorting cells on a FacsARIA III.
Using scatter profiles to eliminate debris and doublets, we sorted bulk samples as DAPI+NeuN+ from layers 1-6, or as DAPI+NeuN− from layer 1 and layer 5 samples, at 5,000-10,000 cells per sample, into 200 μL of blocking buffer in low-binding Eppendorf 1.5 mL tubes. We pelleted sorted nuclei at 1,000xg for 10 minutes at 4°C, followed by resuspension in 50 μL Proteinase K Cleanup Buffer (recipe below) and 37°C incubation for 30 minutes, and then freezing at −20°C until library prep and sequencing.
For library prep, we purified tagmented DNA with 1.8x vol/vol Ampure XP beads (Beckman-Coulter catalog # A63881), eluted DNA in 11 μL and then PCR-amplified with Nextera Index kit primers (#FC-121-1012 from Illumina) using KAPA HiFi HotStart ReadyMix (KAPA Biosystems #KK2602) in a 30 μL reaction (72° 3:00, 95° 1:00, cycle 17x [98°:20, 65°:15, 72°:15], 72° 1:00). We purified PCR products using 1.8x Ampure XP beads, and quantified libraries using Agilent BioAnalyzer High Sensitivity DNA Chips (catalog #5067-4626). Then sample libraries were pooled evenly and sequenced with paired-end 50bp reads either on Illumina MiSeq (Allen Institute) or NextSeq machines (SeqMatic, Fremont CA USA). We processed fastq files as described below.
Single nuclear ATAC-seq
We modified the single nuclear ATAC-seq workflow from the bulk sample workflow in several ways, most notably performing transposition reactions following sorting rather than prior to sorting, and omitting DAPI except for non-neuronal samples (due to the uncertainty of DAPI possibly interfering with transposition).
We collected and dissected specific MTG tissue layers as for bulk samples, but we immediately dounced the layers to release nuclei, and then stained in blocking buffer containing PE-NeuN antibody but not DAPI. We sorted single NeuN+ nuclei from each layer into wells of a 96-well plate, using scatter profiles to exclude debris and doublets. We confirmed single nucleus-to-event correspondence by test-sorting single NeuN+ events into flat-bottom 96 well plates with 40 μL blocking buffer containing DAPI followed by pelleting 1 min at 3,000xg and microscopic examination. These tests routinely yielded > 95% single nucleus-filled wells and undetectable doublets. In the cases where glial cells were sorted, we first sorted neurons from the sample using PE-NeuN+ staining, and then treated with DAPI (1 μg/μL) for 1-2 minutes prior to sorting glial cells as DAPI+NeuN− events.
We sorted single NeuN+ cells into 1.5 μL of Nextera Tn5 transposition reaction (0.6 μL Tn5 enzyme, 0.75 μL tagmentation buffer, 0.15 μL 1% IGEPAL CA-630) in Eppendorf semi-skirted 96-well plates (MilliporeSigma catalog # EP0030129504). Immediately following sorting we briefly centrifuged plates, vortexed, centrifuged plates again, and then incubated plates at 37°C for 30 minutes for transposition. After transposition we added 0.6 μL Proteinase K Cleanup Buffer (recipe below), vortexed briefly and centrifuged, and incubated at 40°C for an additional 30 minutes, then froze plates at −20°C until library prep. Library prep for single nuclear samples was the same as for bulk samples, except we increased the number of amplification cycles from 17 to 22 cycles due to the lower input DNA content.
Bulk ATAC-seq sample clustering
We called peaks on all 39 bulk samples from five independent specimens using MACS2 (Zhang et al., 2008), and then used DiffBind (Ross-Innes et al., 2012) to identify 73,742 differential peaks for all contrasts among the sample types (sort strategies and specimens). Of these, 1,524 distinguished experimental specimens and were discarded for clustering. With 72,218 remaining peaks found specifically to discriminate any pairwise combinations of sort strategies, we reanalyzed correlation among bulk samples using reads in these peaks. This correlation matrix revealed groupings of non-neuronal samples, upper layer neuronal samples, and lower layer neuronal samples (Figure S1C). One sample was omitted from this analysis (H17.03.009 L1 NeuN+) because this sample appeared intermediate between NeuN+ and NeuN− cells, likely due to a sorting error.
ATAC-seq data preprocessing and quality control
We retrieved sample-specific fastq files using standard built-in Illumina de-indexing protocols. We mapped each fastq file to human genome reference hg38 patch 7 using bowtie2 (Langmead and Salzberg, 2012) and the flags–no-mixed–no-discordant -X 2000 to generate sample-specific bam files, which we then filtered for low-quality mappings, secondary mappings, and unmapped reads using samtools view -q 10 -F 256 -F 4 (Li et al., 2009), and then filtered for duplicate reads using samtools rmdup. We then converted these filtered reads bam files to bed files using bedTools bamToBed (Quinlan and Hall, 2010) for quality control calculations of mean ENCODE overlap and TSS enrichment score. For mean ENCODE overlap we converted bed files to fragment format, and assessed the percentage of unique fragments that overlap with ENCODE project DNaseI hypersensitivity peaks from adult human frontal cortex (studies ENCSR000EIK and ENCSR000EIY; ENCODE Project Consortium, 2012; Sloan et al., 2016) using bedTools intersectBed, and took the mean of these two numbers. For TSS enrichment score we used the published technique of Chen et al. (2016). This technique sums the overlap of reads in 2kb windows surrounding all human TSSs (TSS ± 1kb), then segments this 2kb window into forty 50-bp bins, then normalizes the summed read counts to the outside four bins (first and last two), and finally reports the TSS enrichment score as the maximum height of that normalized read count graph. We noticed that this technique worked well for all bulk samples but gave spurious abnormally high scores for some single nuclei having low read count; as a result we made the modification to set TSS enrichment score to 1 (no enrichment) for single nuclei having fewer than 500 reads or TSSs calculated to be greater than 20 (likely spurious events).
We used these quality control metrics to filter out low quality nuclei (ENCODE overlap < 15% AND TSS score < 4, Table S2). Additionally, we filtered out nuclei having fewer than 10,000 unique read pairs, since we require this many reads for our clustering approach. Of 3,660 initial cells we confined analysis to 2,858 high quality nuclei for clustering.
Clustering single nuclei: bootstrapped clustering
We clustered single nuclei using extended fragment Jaccard distance calculations among cells as implemented by the lowcat package (Graybuck et al., 2021). To accomplish this, we first excluded reads on chromosomes X, Y, and M to prevent differential chromosome-biased clustering. Then we randomly down-sampled to 10,000 unique fragments per nucleus, and then these fragments were extended to a regularized length of 1,000 bp with the same center. With these lists of extended fragments we next calculated the Jaccard similarity score for each nucleus pair, defined as the quotient of the intersecting extended fragment number, by the extended fragment union number. Then we calculated Jaccard distances among all nucleus pairs as 1 minus Jaccard similarity score.
Finally, this 2,858 × 2,858 Jaccard distance matrix was dimensionality reduced to a 2858 × 29 matrix of principal component variates, using axes 2 through 30 calculated by princomp in the R base stats package (R Core Team, 2018). We omitted principal component 1 because it was highly correlated to quality control metrics, suggesting that this axis primarily reflected library quality (Figures S2B-S2D). Principal components beyond 30 contain little cell type information, so excluding them represents a de-noising step (Figure S2A). These resulting 29 PCs are used to call nuclear clusters and to visualize them using tSNE.
To call cell clusters on this 2,858 × 29 principal component matrix, we bootstrapped an iterated PCA then Jaccard-Louvain clustering technique using k = 15 nearest neighbors (after testing k = 5,10,15,20, and finding 15 to give best visual separation of clusters on tSNE coordinates). We repeated each bootstrapping round 200 times, each time including only 80% (2,286) of the nuclei, then performing PCA and using components 2 through 30 for Jaccard-Louvain clustering. Finally, we tabulated the frequency with which each nucleus co-clusters with every other nucleus. This co-clustering frequency matrix was then hierarchically clustered by Euclidean distances, and 27 cell type clusters were called by manually cutting the tree using idendr0 (https://github.com/tsieger/idendr0) to represent visually apparent co-clustered blocks of nuclei (Figure S2E, left). Manual tree-cutting outperformed automatic tree cutting with cutree in the R stats package using either branch height or cluster number specified, likely since clusters have nonuniform separation and tightness.
Next we repeated this process with more stringent bootstrapping criteria: changing the percentage of cell to be re-clustered from 50%–90%, and this analysis resulted in similar cluster structure and nucleus membership (Figure S2E, middle, and Figure S2F). In contrast, randomizing the Jaccard distance matrix prior to bootstrapped clustering yielded no clusters in the dataset (Figure S2E, right). Together these analyses suggest that our identified clusters represent real and reproducible cell groups.
Clustering single nuclei: comparing choice of feature set
We also attempted to cluster nuclei using other feature sets besides Jaccard distances among cells (Figure S2G). These additional feature sets included: 1) the list of all detected peaks from the entire aggregated dataset (236,588 peaks called using Homer findPeaks (Heinz et al., 2010) with -region flag), 2) the list of all RefSeq gene TSS regions, extended ± 10kb (27,021 regions), 3) all 321,184 non-overlapping 10kb windows across the human genome, and 4) the list of “gene bins” defined as the genomic region for each gene between the boundaries of midpoints between each RefSeq gene transcribed region. For each feature set, we initially optimized several parameters including the choice of peak caller, the exact gene list for TSS regions and “gene bins,” the size of the genomic windows, and the size of the TSS regions to consider, so that each featureset could perform best. With parameters chosen, we then computed counts in features for each cell, then identified principal components, and visualized groupings by tSNE of principal components 2:50. For our dataset, Jaccard distances disclosed the qualitatively cleanest separation among nuclei, and among clusters (Figure S2G). Furthermore, a wide range of tSNE perplexity values maintained these separations (Figure S2H). Changing the size of regions around RefSeq TSS sites (from ± 10 kb to ± 500 bp) did not improve the utility of TSS features for clustering our nuclei.
Mapping clusters to transcriptomic cell types: assimilating open chromatin and transcriptomic information
We wished to map our 2,858 high quality ATAC-seq profiled cells to human brain cell types discovered by large-scale RNA-seq studies (Hodge et al., 2019). To do this we first sought the best technique to manufacture gene-level information from the ATAC-seq data, in order to correlate with RNA-seq transcript counts. We tried four techniques: 1) read counts in RefSeq “gene bins” as above, 2) read counts in RefSeq gene bodies, 3) read counts in RefSeg gene TSS regions extended ± 10 kb, and 4) Cicero gene activity scores (Cusanovich et al., 2018; Pliner et al., 2018). With these four sets of gene-level information computed for the 10000 fragment-downsampled library from each nucleus, we then mapped nuclei to RNA-seq cell types as the best correlated (highest Spearman correlation statistic) RNA-seq cluster (using median gene counts per million, CPM) with each nucleus, using each of four gene-level information vectors, resulting in four distinct mappings for each nucleus.
We calculated this correlation using a set of 831 marker genes, which we chose to be both informative marker genes for RNA-seq clustering and to contain abundant epigenetic information. This was accomplished by using the select_markers function with default parameters from the scrattch.hicat R package (Tasic et al., 2018) which yielded 2,791 transcriptomic marker genes, which was further filtered by intersecting with the top ten percent of genes with the highest summed Cicero gene activity scores across all 2,858 cells, to yield 831 combined transcriptomic and epigenetic marker genes for mapping.
The four sets of cellwise mappings yielded four tables of cell type abundances within our dataset. Next, taking the RNA-seq dataset (Hodge et al., 2019) as a true gold standard, we compared the four cell type abundance tables with the ‘expected’ cell type abundances, which was calculated as the sum of numbers of cells sorted in each sort strategy, times the expected cell type frequencies in each sort strategy. Correlating the four cell type abundance tables with the expected abundance table (Pearson correlations of log-transformed abundance values plus one) revealed that, of the four techniques to compute gene-level information from ATAC-seq data, Cicero gene activity scores supply the most dependable gene-level information for the purpose of epigenetic to transcriptomic mapping (Figure S3A).
Mapping clusters to transcriptomic cell types: bootstrapped mapping
Using Cicero gene activity scores, we bootstrapped the cellwise mapping procedure 100 times with retention of a variable 50%–90% of genes each round and applied the most frequently mapped transcriptomic cell type to each single ATAC-seq nucleus. Then we report the percentage of each cluster’s constituent cells mapping to each cell type in Figure S3B, and summed by cell type subclass in Figure S3D.
We also performed clusterwise mapping for each of the 27 ATAC-seq clusters using the same bootstrapped mapping procedure, except that we aggregated Cicero gene activity scores by mean across cells within each cluster prior to mapping. We report the number of 100 times that each cluster is mapped to each cell type in Figure S3C, and summed by transcriptomic subclass in Figure S3E.
We observe that clusterwise mapping largely agrees with, but is cleaner than, cellwise mapping (compare Figures S3B and S3C and also Figures S3D-S3F); hence we elect clusterwise mapping as the final mapping procedure. Each cell is thus assigned a final mapped cell type subclass (shown in Figure S3E) as a result of its ATAC-seq cluster membership. For all downstream analyses of peaks, we use aggregations at the cell type subclass level as in Figure S3E.
Peak calling
We called peaks on both bulk and aggregated single-nucleus data using Homer findPeaks with -region flag (Heinz et al., 2010). We found this program to be superior to Hotspot (v4), MACS2 (Zhang et al., 2008), and SICER (https://home.gwu.edu/~wpeng/Software.htm) to identify small regions corresponding to likely enhancers, while still capturing the peak boundaries. In preliminary experiments we observed that Hotspot returned small regions of a constant size (150bp or 250bp) that did not always align to peak summits, but it was relatively insensitive to read depth. MACS2 performed better than hotspot at picking full peak sizes but peak numbers found were strongly dependent on read depth. SICER returned very large regions (median > 2kb) that did not clearly correspond to visual peaks. Using Homer findPeaks with -region flag, peak sizes are median 300-500 bp across subclasses, and we observed only a shallow dependence of identified peak number on read depth.
Identifying transcription factor motifs using chromVAR
We used chromVAR (Schep et al., 2017) to identify transcription factor motif accessibilities in our single nuclei. Using Homer findPeaks with -region flag, we called peaks on the aggregation of all single nuclear and bulk libraries (236,588 peaks), and then resized them to a standard 150bp size with the same center. We downloaded 452 transcription factor motifs from JASPAR (using JASPAR2018 R package, (Khan et al., 2018), and 1,764 from cisBP (as included in the R package chromVARmotifs, (Schep et al., 2017)), and used chromVAR to aggregate and quantify motif accessibilities in all 2,858 single nuclei. Cell type subclass-distinguishing motifs across were found by ranking subclass-averaged motif accessibilities by standard deviation across subclasses (including DLX1 and NEUROD6, Figures S4A-S4D).
Characterization of peaks by conservation
With peaks called for each subclass, we calculated their phyloP scores as a measure of conservation. For peak phyloP scores, we used bigWigSummary to lookup phyloP values from hg38.phyloP4way.bw (Karolchik et al., 2004). These files quantify the basepair conservation across four mammals: Homo sapiens, Mus musculus, Galeopterus variegatus (Malayan flying lemur), and Tupaia chinensis (Chinese tree shrew). We return ten values evenly spaced across each peak, and calculate the maximum mean of eight three-consecutive-value sets. This is done to find smaller regions on the order of 100 bp highly conserved regions within each peak, and this technique yields greater deviations between real and random phyloP scores than taking a single peak-wise average alone. To compare conservation across groups of peaks, we subtracted the mean phyloP scores of randomized peak positions, from real peak phyloP scores (as in Figure 4F).
Identifying transcriptomic cell type matches for methylation data
Using the dataset of Luo et al. (2015), we correlated the published mCH gene body marker genes (their Table S3 containing 1012 human and 1016 mouse methylation marker genes) with cluster-wise medians for transcriptomic human cell types (Hodge et al., 2019) and for mouse cell types (Tasic et al., 2018). We confined correlation analysis to the top 200 methylation marker genes published by Luo et al. that also have highest variance among transcriptomic cell subclasses. With these genes, we then calculated Pearson correlation coefficients between normalized gene body mCH and RNA-seq clusterwise median CPM, and assigned the best matches as the most anti-correlated mCH and CPM vectors. This analysis was repeated for both human and mouse datasets independently. Importantly, our transcriptomic cell type assignments agree with the previously predicted subclasses by Luo et al.
Quantifying ATAC-seq peak overlaps with DMRs
We first aggregated human DMRs from Luo et al. (2015) and Lister et al. (2013). For neuron types, we downloaded DMRs and merged them using bedtools mergeBed. For non-neuron types, we downloaded raw fastq files from the GEO submission of Lister et al. (2013) corresponding to bulk NeuN-negative cells from two human replicates (GSM1173774 and GSM1173777), and converted these to allc files using the pipeline analysis method of Luo et al. (2017). These allc files were aggregated and used to find DMRs with methylpy DMRfind (minimum differentially methylated sites = 1) against allc files for all human subclasses from Luo et al., and an outgroup of human H1 cells from ENCODE. The same set of bulk non-neuronal DMRs were used for comparison to the ATAC-seq data for Astrocytes, Oligodendrocytes/OPCs, and Microglia subclasses (Figure S4E).
With bed files corresponding to each subclass ATAC-seq peakset and to each subclass DMR set, we used bedtools intersectbed to quantify the overlap between peaks and DMRs. We bootstrapped calculation of real peak overlaps 100x by removing 20 percent of peaks each time and calculating percentage overlap, and the mean of these 100 measurements is reported. Similarly, we randomized peak positions throughout the genome 100x using bedtools shuffleBed, calculated percentage overlap each time, and the mean of these 100 measurements is reported. By definition, disjoint ranges of real versus randomized peak overlap percentages established false discovery rate < 0.01. We also calculated enrichment of DMR overlaps for ATAC-seq peaksets, defined as the ratio of real peak-DMR overlap percentage to the overlap percentage of randomized peak positions.
Mouse to human cross-species comparisons
We used the sets of subclass-specific (uniquely identified in only that subclass) peaks to map between human and mouse subclasses. We first mapped subclass-specific mouse peaks to hg38 using liftOver with minMatch parameter set to 0.6. This setting gave successful mapping for the majority of snATAC-seq peaks (range 58 to 76% across subclasses) while retaining the original peak size distribution (data not shown). Then we bootstrapped calculation of human peak overlap against all mouse peaks 100x with random retention of 80% of human peaks each time, and we took mean of Jaccard similarity coefficients (intersection over union) over 100 runs. In addition, we shuffled genomic peak positions 100x, and calculated mean Jaccard similarity coefficients each time. We report the enrichment of Jaccard similarity coefficients as the ratio of the real over random (Figure 2A). To visualize set intersections in Venn diagram format we display results using all mouse and human peaks (not subclass-specific, Figure 2B).
For characterization of human conserved and divergent peaks, we start with all human peaks and partition to those intersecting (“Conserved”) or not intersecting (“Divergent”) with mouse peaks identified within the same orthologous subclass and mapped to hg38 by liftOver with minMatch parameter set to 0.6. To characterize mouse conserved and divergent peaks, we intersect all mouse peaks with reciprocal mm10-mapped human peaks. Then we calculated phyloP scores as above.
De novo sequence motif identification
We used all mouse peaks and all human peaks to identify enriched sequence motifs using MEME-CHIP (Bailey et al., 2009). These motifs were then matched against known TF motifs in HOCOMOCO database v11 using TomTom. We then filtered the MEME-CHIP output by first excluding all motifs with -log10(E-value) <5; E-value represents the enrichment p value (by Fisher’s exact test) times the number of candidate motifs tested. We further filtered by second excluding all motif matches with TFs not expressed (median CPM = 0) in that cell subclass from RNA-seq studies (Tasic et al., 2018; Hodge et al., 2019). Third we filtered by excluding all low-confidence motif matches with E-value > 0.2 and q-value > 0.2; q-value represents the minimal false discovery rate at which the observed similarity would be deemed significant. Finally, these filtered lists of all detected motifs in all cell subclasses were manually curated to a master list of all high-confidence identified TF motifs.
Quantifying repetitive element overlap
To characterize the repetitive element overlap for peaks, we first partitioned mouse and human subclass-specific peaksets to conserved and divergent peaks. Then we calculated the overlap with repetitive genomic elements using hg38 and mm10 RepeatMasker (Smit et al., 2013) files, using a 100x bootstrapped overlap and 100x bootstrapped randomization strategy as described above for DMR overlap. Human L56IT peaks were omitted from this analysis because very few of these peaks are both subclass-specific and conserved.
Cloning enhancers
Enhancers were chosen for cloning from open chromatin data using one of two strategies. For the first strategy we used the following criteria: 1) visible specific peak manually identified in read pileups adjacent to known subclass-specific marker genes, and 2) containing a region of high primary sequence conservation by phyloP score. For the second strategy we used the following criteria: 1) a subclass-specific ATAC-seq peak identified by Homer (with -region flag) in both human and mouse (conserved) or only human (divergent), 2) a subclass-specific DMR in both human and mouse (conserved) or only human (divergent), 3) ranking by human ATAC-seq read counts within region, and 4) manual confirmation by visualization of read pileup by experimenter.
Chosen enhancers were cloned into either scAAV or rAAV (ssAAV) expression vectors. For scAAV vectors we used a plasmid backbone that is a derivative of pscAAV-MCS (Cell Biolabs catalog # VPK-430, for scAAV vectors), which was used for eHGT_017h, eHGT_019h, eHGT_023h, eHGT_025h, and hDLXI56i (Dimidschstein et al., 2016; Chan et al., 2017). For rAAV (ssAAV) vectors we used a plasmid backbone from Addgene plasmid number 51084 (AAV-hSyn1-GCaMP6s-P2A-nls-dTomato, which was itself originally derived from pAAV-GFP [Cell Biolabs catalog # VPK-410]). We used this rAAV backbone for eHGT_058h, eHGT_064h, eHGT_078h/m, eHGT_079h, eHGT_082h, eHGT_096h, eHGT_098h, eHGT_128h, and eHGT_140h, hDLXI56i, and DLX2.0. Enhancers were inserted by standard Gibson assembly approaches, upstream of a minimal beta-globin promoter and the reporter SYFP2, a brighter EGFP alternative that is well tolerated in neurons (Kremers et al., 2006). NEB Stable cells (New England Biolabs # C3040I) or Stbl3 cells (Thermo Fisher # C7373-03) were used for transformations and cultured at 32°C. scAAV plasmids were monitored by restriction analysis and Sanger sequencing for occasional recombination of the left ITR; this left ITR recombination was not observed for rAAV plasmids. We attempted to boost expression level for some enhancers by engineering a triple tandem array of the enhancer core (“concatemer”), for example for DLX2.0 as in Figures 7B and 7D.
Virus production
Enhancer AAV plasmids were maxi-prepped and transfected with PEI Max 40K (Polysciences Inc., catalog # 24765-1) into one 15 cm plate of AAV-293 cells (Cell Biolabs catalog # AAV-100), along with helper plasmid pHelper (Cell BioLabs) and PHP.eB rep/cap packaging plasmid (Chan et al., 2017), with a total mass of 150 μg PEI Max 40K, 30 μg pHelper, 15 μg rep/cap plasmid, and 15 μg enhancer-AAV vector. The next day medium was changed to 1% FBS, and then after 5 days cells and supernatant were harvested and AAV particles released by three freeze-thaw cycles. Lysate was then treated with benzonase to degrade free DNA (2 μL benzonase, 30 min at 37°C, MilliporeSigma catalog # E8263-25KU), and then cell debris was cleared with low-speed spin (1500 g 10 min). The supernatant containing virus was concentrated over a 100 kDa molecular weight cutoff Centricon column (MilliporeSigma catalog # Z648043) to a final volume of ~150 μL. For highly purified large-scale preps this protocol was altered so that ten plates were transfected and harvested together at 3 days after transfection, and then the crude virus was purified by iodixanol gradient centrifugation.
Mouse virus testing
Mice were retro-orbitally injected at P42-P70 with 10 μL (approximately 2-3 x1011 genome copies) of crude virus prep diluted with 100 μL PBS, then sacrificed at 21-28 days post infection. For live epifluorescence, we perfused mice with ACSF.7 and cut live 350 μm sections with a compresstome from one hemisphere to analyze reporter expression using a 10x objective on a Nikon Ti-Eclipse epifluorescence microscope with built-in real-time deconvolution image processing for thick tissues (Nikon Image Systems Elements software with Advanced Research module). For full sagittal section images of mouse brain, we processed the brain for mFISH and anti-GFP immunostaining (as below), and using a 4x objective on an Olympus FV3000 confocal we took images in a 3x5 grid tiling the brain at two optical slices separated by 4 μm, and in ImageJ we performed z-projections using maximum intensity and stitched images using Grid stitching and linear blending fusion method (Schneider et al., 2012). For antibody staining the other hemisphere was drop-fixed in 4% PFA in PBS for 4-6 hours at 4°C, then cryoprotected in 30% sucrose in PBS 48-72 hours, then embedded in OCT for 3 hours at room temperature, then frozen on dry ice and sectioned at 10 μm thickness, prior to antibody stain using standard practice. We used the following primary antibodies: chicken anti-GFP (Aves # GFP-1020), rabbit anti-Parvalbumin (Swant # PV27), rabbit anti-Somatostatin (Peninsula Biolabs # T-4547), rabbit anti-VIP (BosterBio # RP1108), and mouse anti-RFP (abcam # ab65856) to detect mCherry from Gad2-T2a-NLS-mCherry mice (Peron et al., 2015). Secondary antibodies were 488-, 555-, and 647-conjugated secondary antibodies from ThermoFisher Scientific. We performed single-cell RNA-seq from the mouse visual cortex as described previously (Tasic et al., 2016, 2018).
Multiplexed FISH by hybridization chain reaction (mFISH)
We performed this technique on mouse brain hemispheres fixed by immersion in 4% PFA in PBS for 4-6 hours at 4°C. After fixation, we rinsed hemispheres with PBS and stored them in PBS at 4°C for up to one month. For sectioning, we embedded hemispheres in 1% low-melt agarose in PBS and cut 50 μm sagittal sections on a Leica VT1000S vibratome in cold PBS buffer. We post-fixed sections in 4% PFA in PBS for 2 hours and then rinsed in PBS at room temperature, then dehydrated with 70% ethanol at 4°C. Afterward sections could be stored for up to a month in 4°C. For staining, we cleared sections with 8% SDS in PBS for 2 hours at room temperature then washed three times in 2x SSC for 1 hour each, then with Hybridization Buffer (Molecular Instruments) in a new well before applying Hybridization Buffer containing HCR Probes and hybridized overnight at 37°C. The next day we washed samples with 30% Probe Wash Buffer for 1 hour at 37°C, then rinsed with 2xSSC. During the probe wash, we denatured fluorescently labeled HCR hairpins at 95°C for 90 s and then snap-cooled in a room temperature aluminum block tube holder for 30 minutes. We added the denatured hairpins to Amplification Buffer and applied to tissue sections for 2 hours at room temperature in the dark, then washed with 2x SSC containing DAPI, again with 2x SSC, and finally mounted on SuperFrost Plus slides in Prolong Glass Mounting medium (Thermo Fisher Scientific # P36980). We imaged these HCR stains with an Olympus FV3000 confocal microscope using manufacturer’s software. Molecular Instruments generated HCR probes against the following transcripts: Rorb NM_001043354.2; Lamp5 NM_029530.2; Vip NM_011702.3; Pvalb NM_001330686.1; Sst NM_009215.1; Slc17a7 NM_182993.2; Gad1 NM_008077.5.
Human ex vivo AAV vector testing
We transported neurosurgical temporal cortex samples from the operating suite to the Allen Institute in typically less than 30 minutes, using specialized transportation equipment to maintain sterility and carbogen bubbling throughout processing. We blocked tissue samples and then sliced at 350 μm thickness and then dissected away white matter and pial membranes. Slices then underwent warm recovery (bubbled ACSF.7 at 30 degrees for 15 minutes) followed by reintroduction of sodium (bubbled ACSF.8 at room temperature for 30 minutes, recipe below, (Ting et al., 2018). We then plated slices at the gas interface on Millicell PTFE cell culture inserts (MilliporeSigma # PICM03050) in a 6-well dish on 1 mL of Slice Culture Medium (recipe below). After 30 minutes, we transduced slices by direct application of high-titer AAV2/PHP.eB viral prep to the surface of the slice, 1 μL per slice. Afterward, we replenished slice culture medium every 2 days and monitored reporter expression. For hDLXI56i in human, we performed imaging and single cell RNA-seq in four independent experiments at 8, 13, 28, and 69 days in vitro. A fifth experiment on hDLXI56i (at 11 days in vitro) was excluded from analysis because 36/48 (75%) of sorted cells either failed to map to transcriptomic cell types, or mapped as uncertain non-neuronal types; this is likely due to either a failed sort or poor starting tissue quality given heterogeneity of patient samples. For DLX2.0, we performed imaging and single cell RNA-seq in four independent experiments at 7, 7, 14, and 34 days in vitro
We performed single cell RNA-seq on human virus-infected neurons by 1 hour digestion at 30°C in carbogenated ACSF.1/trehalose + blockers + papain (all recipes below), followed by gentle trituration in Low-BSA Quench buffer, shallow spin gradient centrifugation (100 g 10 minutes at room temperature) into High-BSA Quench buffer, and resuspension into Cell Resuspension Buffer. We also employed Myelin Bead Removal Kit II (Miltenyi catalog # 130-096-733) at 1/20 to remove myelin debris, and PE-anti CD9 clone eBioSN4 (Thermo Fisher catalog # 12-0098-42) at 1/50 to sort away contaminating glial cells. Then we sorted single SYFP2+ labeled human neurons for sequencing using SMARTer V4 as previously described (Tasic et al., 2016, 2018). To map single cells to the transcriptomic taxonomies, we trained a nearest centroid classifier on cell type labels using human and mouse VISp scRNA-seq cluster labels (Tasic et al., 2018), employing informative marker genes chosen by the select.markers function in scrattch.hicat (Tasic et al., 2018). We confined taxonomy mapping analysis to the cells that passed cDNA library generation quality control metrics and showed detectable levels of SYFP2 transcripts. Intermediate-mapping cells are represented as circles on nodes of the cluster dendrograms.
In vivo non-human primate AAV vector testing
All procedures used with macaque monkeys conformed to the guidelines provided by the US National Institutes of Health and were approved by the University of Washington Animal Care and Use Committee. Three animals were used in these experiments: one rhesus macaque (Macaca mulatta) and two pig-tailed macaques (Macaca nemestrina). These animals were injected with a single AAV vector in each of ten injection sites during a single surgery. These sites were left temporal cortex, left and right occipital cortex, left and right motor cortex, and left somatosensory cortex. AAVs were purified by iodixanol gradient ultracentrifugation for this procedure. After craniotomy, using a pneumatic pico pump (World Precision Instruments) a total of 5 μL AAV vector was injected at each site with 500 nL expelled at each of ten depths evenly spaced from 2 mm to 200 μm deep beneath the pial surface. Sites were separated by ~1 cm in each region with multiple injection sites. Eight of the total sites are described in this manuscript (eHGT_079h, 082h, 128h, 140h and 359h in occipital cortex, and 140h in temporal and motor and somatosensory cortex). At 51, 96, or 113 days after injection, the animals were sacrificed. We inspected the brain surface, cut tissue blocks (~2x2x2cm) around each visible fluorescent spot, and fixed each block 4% PFA in PBS for 24 hours at 4°C. After PFA fixation, we embedded blocks in 2% agarose in PBS and cut 350 μm sections and inspect each for fluorescent cells. We then cryopreserved a subset of sections in 30% sucrose in water overnight and subsectioned them on a sliding microtome to 30 μm for immunostaining using the following antibodies: chicken anti-GFP (Aves # GFP-1020), rabbit anti-Parvalbumin (Swant # PV27), and guinea pig anti-GABA (Millipore Sigma # AB175). Images shown are from the region of high labeling close to the needle tract (< 1 mm), but the zone of expression extended for ~3-4 mm orthogonal to the needle tract. Proper recovery of sites was confirmed by PCR on DNA from dissected fixed thick slices (recovered with QIAamp DNA FFPE Tissue Kit, QIAGEN catalog # 56404) using common primers to all vectors: F 5′-ACTCCATCACTAGGGGTTCCTG and R 5′-GGACACGCTGAACTTGTGGC followed by Sanger sequencing with the nested reverse primer 5′-ACGTCGCCGTCCAGCTC.
Ex vivo non-human primate AAV vector testing
Brains from healthy Macaca nemestrina animals housed at the Washington National Primate Research Center aged 2-15 years were obtained through the Tissue Distribution Program. Whole hemispheres or tissue blocks were transported to the Allen Institute and processed for ex vivo culture and AAV vector testing as described above for human neurosurgical samples (Ting et al., 2018). Data are shown for cultures of MTG tissue. Cell subclass specificity was evaluated by mFISH as described above for mouse, except that 350 μm cultured slices were cleared with 67% 2,2′-thiodiethanol in water prior to mounting on slides for microscopy. Molecular Instruments designed probes with the following accession numbers provided to them: Slc17a7 NM_005589901.2; Gad1 NM_005573441.2; Vip NM_005552161.2; Pvalb NM_005567398.2; Sst NM_005545442.2.
Patch clamp physiology and analysis in non-human primate
For patch clamp recordings, we placed slices in a submerged, heated (32-34°C) recording chamber that was continually perfused with Recording aCSF (recipe below) under constant carbogenation. We visualized neurons with an Olympus BX51WI microscope with a 40x water immersion objective and infrared differential interference contrast optics and EYFP filterset. We filled the recording pipette with Recording Pipette Solution (recipe below), and acquired electrical signals using a Multiclamp 700B amplifier and MIES data acquisition software written in Igor Pro (Wavemetrics). We digitized signals at 10-50 kHz and filtered at 2-10 kHz. We compensated pipette capacitance and balanced the bridge balanced throughout whole-cell current clamp recordings. Access resistance was 8-25 MΩ. We analyzed data using custom scripts written in Igor Pro as previously described (Kalmbach et al., 2018).
Inferring GWAS-cell subclass associations
We used linkage disequilibrium score regression (LDSC; Bulik-Sullivan et al., 2015; Finucane et al., 2015) to partition heritability of various brain conditions to regions associated with accessible chromatin in eleven human cortical cell subclasses, whose peaks are grouped into Conserved and Divergent subsets. As outgroup comparators, we also assessed the heritability associated with outgroup populations of human keratinocytes downloaded from ENCODE (ENCODE Project Consortium, 2012). Additionally, we also performed this analysis using DMRs from human cortical neuron subclasses (Luo et al., 2017), human cortical non-neurons (Lister et al., 2013), and H1 human embryonic stem cells (ENCODE Project Consortium, 2012).
Summary statistics from 21 GWAS studies were acquired and evaluated including brain-related (schizophrenia, major depressive disorder, autism spectrum disorder, ADHD, Alzheimer’s disease, Tourette’s syndrome, bipolar disorder, eating disorder, obsessive-compulsive disorder, loneliness, BMI, PTSD) and non-brain-related diseases (Crohn’s disease and asthma) from the PGC and EMBL/EBI GWAS repositories (Anney et al., 2017; Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium, 2015; Demenais et al., 2018; Demontis et al., 2019; Duncan et al., 2017, 2018; Gao et al., 2017; International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS), 2018; Lambert et al., 2013; de Lange et al., 2017; Lee et al., 2018; Liu et al., 2015; Marioni et al., 2018; Okbay et al., 2016; Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011; Ripke et al., 2013; Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium, 2011; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014; Yu et al., 2019; Wray et al., 2018; Yang et al., 2017).
We excluded studies with log10(N * h2) < 3.6, where N is number of patients in the study and h2 represents the sum of heritability across SNPs within the study, which represents the effective power of the study (Finucane et al., 2015). This exclusion removed 6 studies: asthma (Demenais et al., 2018), log10(N * h2) = 3.5), PTSD (Duncan et al., 2018), log10(N * h2) = 2.9), eating disorder (Duncan et al., 2017), log10(N * h2) = 3.5), loneliness (Gao et al., 2017), log10(N * h2) = 3.3), obsessive-compulsive disorder (International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS), 2018), log10(N * h2) = 3.5), and one major depressive disorder study (Ripke et al., 2013), log10(N * h2) = 3.3). The 15 studies with sufficient power for inclusion were all performed on a European descent population. Within these datasets, we confined analysis to 1,389,227 high-confidence SNPs present in the HapMap3 list, and using linkage disequilibrium maps from the 1000 Genomes Project European descent individuals, we analyzed the trait and disease enrichments of cell subclass-associated chromatin along with the LDSC baseline model LDv2.0 with 75 enumerated genomic feature categories. LDSC was performed to associate these 15 studies with both ATAC-seq peaks and methylation DMRs (Figures 2D and 2E; Lister et al., 2013; Luo et al., 2017), and both epigenetic data modalities gave qualitatively similar results although ATAC-seq peaks give stronger enrichments. Generally weak associations were observed between the outgroup disease (Crohn’s disease) with brain cell types, and between the outgroup peak set (Keratinocytes; Figures 2D and 2E; ENCODE Project Consortium, 2012) and brain diseases. For statistical testing of enrichments, we use Bonferroni multiple hypothesis testing correction of LDSC’s block jackknife-estimated p values, as previously suggested (Skene et al., 2018). This correction is 0.05 / 345 disease/subclass combinations = 1.45e-4 significance cutoff in Figure 2E. We similarly use 180 and 150 tests in Figure 2D.
Buffer recipes
Proteinase K cleanup buffer
EDTA 50 mM
Sodium chloride 5 mM
Sodium dodecyl sulfate 1.25% (w/v)
Proteinase K (QIAGEN # 19131) 5 mg/mL
pH 8.0
Nuclei isolation medium
Sucrose 250 mM
Potassium chloride 25 mM
Magnesium chloride 5 mM
Tris-HCl 10 mM
pH 8.0
Homogenization buffer
10 mL Nuclei Isolation Medium
0.1% (w/v) Triton X-100
One pellet Roche Mini cOmplete EDTA-free (Sigma catalog # 4693159001)
Blocking buffer
PBS
0.5% (w/v) BSA (catalog # A2058 from Millipore Sigma)
0.1% (w/v) Triton X-100
ACSF.7
HEPES20 mM
Sodium Pyruvate 3 mM
Taurine 10 μM
Thiourea 2 mM
D-(+)-glucose 25 mM
Myo-inositol 3 mM
Sodium bicarbonate 30 mM
Calcium chloride dihydrate 0.5 mM
Magnesium sulfate 10 mM
Potassium chloride 2.5 mM
Monosodium Phosphate 1.25 mM
HCl 92 mM
N-methyl-D-(+)-glucamine 92 mM
L-ascorbic acid 5.0 mM
N-acetyl-L-cysteine 12 mM
pH adjusted to 7.3-7.4, osmolarity adjusted to 295-305, and carbogenated.
ACSF.8
HEPES 20 mM
Sodium Pyruvate 3 mM
Taurine 10 μM
Thiourea 2 mM
D-(+)-glucose 25 mM
Myo-inositol 3 mM
Sodium bicarbonate 30 mM
Calcium chloride dihydrate 2.0 mM
Magnesium sulfate 2.0 mM
Potassium chloride 2.5 mM
Monosodium Phosphate 1.25 mM
Sodium chloride 92 mM
L-ascorbic acid 5.0 mM
N-acetyl-L-cysteine 12 mM
pH adjusted to 7.3-7.4, osmolarity adjusted to 295-305, and carbogenated.
Slice culture medium
MEM Eagle medium powder 1680 mg (MilliporeSigma catalog # M4642)
L-ascorbic acid powder 36 mg
CaCl2, 2.0 M 100 μL
MgSO4, 2.0 M 200 μL
HEPES, 1.0 M 6.0 mL
Sodium bicarbonate, 893 mM 3.36 mL
D-(+)-glucose, 1.11 M 2.25 mL
Pen/Strep 100x (5k U/mL) 1.0 mL (Thermo catalog # 15070063)
Tris base, 1.0 M 260 μL
GlutaMAX 200 mM 0.5 mL (Thermo catalog # 35050061)
Bovine Pancreas Insulin, 10 mg/mL 20 μL (MilliporeSigma catalog # I0516)
Heat-inactivated horse serum 40 mL (Thermo catalog # 26050088)
Deionized water to 250 mL
pH adjusted to 7.3-7.4, and osmolarity adjusted to 300-305,
ACSF.1/trehalose
HEPES 20 mM
Sodium Pyruvate 3 mM
Taurine 10 μM
Thiourea 2 mM
D-(+)-glucose 25 mM
Myo-inositol 3 mM
Sodium bicarbonate 25 mM
Calcium chloride dihydrate 0.5 mM
Magnesium sulfate 10 mM
Potassium chloride 2.5 mM
Monosodium phosphate 1.25 mM
Trehalose dihydrate 132 mM
HCl 2.9 mM
N-methyl-D-(+)-glucamine 30 mM
L-ascorbic acid 5.0 mM
N-acetyl-L-cysteine 12 mM
pH adjusted to 7.3-7.4, and osmolarity adjusted to 295-305.
ACSF.1/trehalose + blockers
50 mL ACSF.1/trehalose
50 μL 100 μM TTX (final 0.1 μM)
100 μL 25 mM DL-AP5 (final 50 μM)
15 μL 60 mM DNQX (final 20 μM)
5 μL 100 mM (+)-MK801 (final 10 μM)
ACSF.1/trehalose + blockers + papain
15 mL ACSF.1/trehalose + blockers
One vial Worthington PAP2 reagent (150 U, final 10U/mL)
15 μL 10kU/mL DNase I (Roche)
Low-BSA Quench buffer
15 mL ACSF.1/trehalose + blockers
15 μL 10kU/mL DNase I (Roche)
150 μL 20% BSA dissolved in water (final conc 2 mg/mL)
150 μL 10 mg/mL ovomucoid inhibitor (Sigma T9253, final concentration 0.1 mg/mL)
High-BSA Quench buffer
15 mL ACSF.1/trehalose + blockers
15 μL 10kU/mL DNase I (Roche)
750 μL 20% BSA dissolved in water (final concentration 10 mg/mL)
150 μL 10 mg/mL ovomucoid inhibitor (Sigma T9253, final concentration 0.1 mg/mL)
ACSF.1/trehalose + EDTA
HEPES 20 mM
Sodium Pyruvate 3 mM
Taurine 10 μM
Thiourea 2 mM
D-(+)-glucose 25 mM
Myo-inositol 3 mM
Sodium bicarbonate 25 mM
Potassium chloride 2.5 mM
Monosodium phosphate 1.25 mM
Trehalose 132 mM
HCl 2.9 mM
EDTA 0.25 mM
N-methyl-D-(+)-glucamine 30 mM
L-ascorbic acid 5.0 mM
N-acetyl-L-cysteine 12 mM
pH adjusted to 7.3-7.4, and osmolarity adjusted to 295-305.
Cell resuspension buffer
50 mL ACSF.1/trehalose + EDTA
50 μL 100 μM TTX (final concentration 0.1 μM)
100 μL 25 mM DL-AP5 (final concentration 50 μM)
15 μL 60 mM DNQX (final concentration 20 μM)
5 μL 100 mM (+)-MK801 (final concentration 10 μM)
150 μL 20% BSA dissolved in water (final concentration 2 mg/mL)
1 μg/mL 4’-diamino-phenylindazole (DAPI)
Recording aCSF
Sodium chloride 119 mM
Potassium chloride 2.5 mM
Monosodium phosphate 1.25 mM
Sodium bicarbonate 24 mM
Glucose 12.5 mM
Calcium chloride tetrahydrate 2 mM
Magnesium sulfate heptahydrate 2 mM
pH adjusted to 7.3-7.4, and osmolarity adjusted to 295-305. Used with constant carbogenation at 32-34°C.
Recording pipette solution
Potassium gluconate 110 mM
HEPES 10 mM
EGTA 0.2 mM
Potassium chloride 4 mM
Disodium guanosine triphosphate 0.3 mM
Phosphocreatine disodium salt hydrate 10 mM
Magnesium adenosine triphosphate 1 mM
Glycogen 20 μg/ml
RNase Inhibitor 0.5U/μL (Takara catalog # 2313A)
Biocytin 0.5% (Sigma B4261)
pH 7.3
QUANTIFICATION AND STATISTICAL ANALYSIS
Details of the statistical analysis are provided in individual figure legends and in STAR Methods. All heatmaps, dotplots, and barplots were generated using R. In analyses using parametric tests of significance (Figures 2C, 2F, S4H, and S4I), data were confirmed normally distributed by visual inspection and Shapiro-Wilk test as implemented in R.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-NeuN | Millipore Sigma | Cat # FCMAB317PE; RRID:AB_11212465 |
| GFP | Aves | Cat # GFP-1020: RRID:AB_10000240 |
| Parvalbumin | Swant | Cat # PV27; RRID:AB_2631173 |
| Somatostatin | Peninsula Laboratories | Cat # T-4547; RRID:AB_518618 |
| VIP | Boster Bio | Cat # RP1108 |
| RFP | Abcam | Cat # ab65856; RRID:AB_1141717 |
| GABA | Millipore Sigma | Cat # AB175; RRID:AB_91011 |
| PE-CD9 | Thermo Fisher Scientific | Cat # 12-0098-42; RRID:AB_10854122 |
| Bacterial and virus strains | ||
| NEB Stable E. coli | New England Biolabs | Cat # C3040I |
| Stbl3 E. coli | Thermo Fisher Scientific | Cat # C7373-03 |
| Chemicals, peptides, and recombinant proteins | ||
| Nextera Tn5 transposase | Illumina | Cat # FC-121-1031 |
| NeuroTrace 500/525 | Thermo Fisher Scientific | Cat # N21480 |
| KAPA HiFi HotStart ReadyMix | Roche | Cat # KK2602 |
| Myelin Beads Removal Kit II | Miltenyi Biotec | Cat # 130-096-733 |
| Deposited data | ||
| Raw human ATAC-seq and scRNA-seq data | This study | dbGaP:phs2292.v1 |
| Raw mouse snATAC-seq data | Graybuck et al., 2021 | https://assets.nemoarchive.org/dat-7qjdj84 |
| ENCODE frontal human cortex DNaseI-HS data | ENCODE consortium | ENCSR000EIK |
| ENCODE frontal human cortex DNaseI-HS data | ENCODE consortium | ENCSR000EIY |
| ENCODE human keratinocyte ATAC-seq data | ENCODE consortium | ENCSR356KRQ |
| Mouse forebrain processed snmC-seq data | Luo et al., 2017 | Table S5 |
| Human forebrain processed snmC-seq data | Luo et al., 2017 | Table S6 |
| Human and mouse forebrain bulk mC-seq data | Lister et al., 2013 | GEO: GSE47966 |
| Human snRNA-seq data | Hodge et al., 2019 | dbGaP:phs001790.v1.p1 |
| ENCODE human H1 cell bisulfite sequencing | ENCODE consortium | ENCSR617FKV |
| ENCODE human H1 cell bisulfite sequencing | ENCODE consortium | ENCSR000AJJ |
| Experimental models: cell lines | ||
| 293AAV cell lines | Cell Biolabs | Cat # AAV-100 |
| Experimental models: organisms/strains | ||
| C57BL/6J mice | Jackson labs | Stock # 000664 |
| Gad2-T2a-NLS-mCherry mice | Jackson labs | Stock # 023140 |
| Macaca nemestrina animals | Washington National Primate Research Center | N/A |
| Macaca mulatta animal | Washington National Primate Research Center | N/A |
| Oligonucleotides | ||
| Mouse Slc17a7 mFISH-HCR probe | Molecular Instruments | Accession # NM_182993.2 |
| Mouse Rorb mFISH-HCR probe | Molecular Instruments | Accession # NM_001043354.2 |
| Mouse Lamp5 mFISH-HCR probe | Molecular Instruments | Accession # NM_029530.2 |
| Mouse Sst mFISH-HCR probe | Molecular Instruments | Accession # NM_009215.1 |
| Mouse Pvalb mFISH-HCR probe | Molecular Instruments | Accession # NM_001330686.1 |
| Mouse Gad1 mFISH-HCR probe | Molecular Instruments | Accession # NM_008077.5 |
| Mouse Slc17a7 mFISH-HCR probe | Molecular Instruments | Accession # NM_182993.2 |
| Mouse VIP mFISH-HCR probe | Molecular Instruments | Accession # NM_011702.3 |
| Macaque nemestrina SLC17A7 mFISH-HCR probe | Molecular Instruments | Accession # NM_005589901.2 |
| Macaque nemestrina GAD1 mFISH-HCR probe | Molecular Instruments | Accession # NM_005573441.2 |
| Macaque nemestrina VIP mFISH-HCR probe | Molecular Instruments | Accession # NM_005552161.2 |
| Macaque nemestrina PVALB mFISH-HCR probe | Molecular Instruments | Accession # NM_005567398.2 |
| Macaque nemestrina SST mFISH-HCR probe | Molecular Instruments | Accession # NM_005545442.2 |
| Recombinant DNA | ||
| CN1203-scAAV-hDLXI56i-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163492 |
| CN1244-rAAV-hDLXI56i-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163493 |
| CN1390-rAAV-DLX2.0-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163505 |
| CN1402-rAAV-eHGT_058h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163494 |
| CN1457-rAAV-eHGT_078h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163495 |
| CN1466-rAAV-eHGT_078m-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163508 |
| CN1253-scAAV-eHGT_017h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163497 |
| CN1255-scAAV-eHGT_019h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163496 |
| CN1258-scAAV-eHGT_022h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163506 |
| CN1259-scAAV-eHGT_023h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163499 |
| CN1279-scAAV-eHGT_022m-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163507 |
| CN1621-rAAV-hsA2-eHGT_128h-minRho-SYFP2-WPRE3-BGHpA | This study | Addgene # 163498 |
| CN1525-rAAV-hsA2-eHGT_079h-minRho-SYFP2-WPRE3-BGHpA | This study | Addgene # 163501 |
| CN1528-rAAV-hsA2-eHGT_082h-minRho-SYFP2-WPRE3-BGHpA | This study | Addgene # 163502 |
| CN2045-rAAV-3xSP10ins-eHGT_359h-minRho*-SYFP2-WPRE3-BGHpA | This study | Addgene # 163504 |
| CN1408-rAAV-eHGT_064h-minBG-SYFP2-WPRE3-BGHpA | This study | Addgene # 163500 |
| CN1839-rAAV-hSyn 1-SYFP2-10aa-H2B-WPRE3-BGHpA | This study | Addgene # 163509 |
| CN1633-rAAV-hsA2-eHGT_140h-minRho-SYFP2-WPRE3-BGHpA | This study | Addgene # 163503 |
| Software and algorithms | ||
| chromVAR | Schep et al. (2017) | https://github.com/GreenleafLab/chromVAR |
| Cicero | Pliner et al., 2018 | https://cole-trapnell-lab.github.io/cicero-release/ |
| methylpy | Luo et al., 2017 | https://github.com/yupenghe/methylpy |
| HOMER | Heinz et al. (2010) | http://homer.ucsd.edu/homer/ |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg (2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al. (2009) | http://samtools.sourceforge.net/ |
| MACS2 | Zhang et al. (2008) | https://pypi.org/project/MACS2/ |
| DiffBind | Ross-Innes et al., 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Lowcat | Graybuck et al., 2021 | https://github.com/AllenInstitute/lowcat |
| scrattch.hicat | Tasic et al. (2018) | https://github.com/AllenInstitute/scrattch.hicat |
| Bedtools | Quinlan and Hall (2010) | https://bedtools.readthedocs.io/en/latest/# |
| MEME-CHIP | Bailey et al. (2009) | https://meme-suite.org/ |
| repeatMasker | Smit et al. (2013) | http://www.repeatmasker.org |
| R | R Core Team (2018) | https://www.r-project.org/ |
| scrattch.hicat | Tasic et al. (2018) | https://github.com/AllenInstitute/scrattch.hicat |
| LDSC | Bulik-Sullivan et al. (2015) | https://github.com/bulik/ldsc |
Highlights.
Human single-nucleus ATAC-seq dataset reveals neocortical enhancers
The human neocortical open chromatin landscape is compared to mouse
Enhancer-AAV vectors can drive expression in neocortical subclasses
PVALB-specific AAVs function in vivo in mice and primates
ACKNOWLEDGMENTS
We thank Allison Beller, Nathan Hansen, Caryl Tongco, Jae-Guen Yoon, and Gina DeNoble for assistance with obtaining patient consent and human neurosurgical tissue research specimens. We thank Rebecca D. Hodge, Trygve E. Bakken, and Zizhen Yao for assistance with sc/snRNA-seq data. We thank Lisa McConnell for assisting with NHP virus injection surgery and NHP animal care. We thank Allen Institute Tissue Procurement and Facilities teams for institutional support during tissue collections. In addition, we wish to thank the Allen Institute for Brain Science founder, Paul G. Allen, for his vision, encouragement, and support. This work is supported by NIH BRAIN Initiative award 1RF1MH114126-01 from the National Institute of Mental Health to E.S.L., J.T.T., and B.P.L.; National Institute on Drug Abuse award 1R01DA036909-01 to B.T.; the Nancy and Buster Alvord Endowment to C.D.K.; and National Eye Institute award 1R01EY030441-01 to G.D.H. This project was also supported in part by NIH grant P51OD010425 from the Office of Research Infrastructure Programs (ORIP) and grant UL1TR000423 from the National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH, ORIP, NCATS, the Institute of Translational Health Sciences, or the University of Washington National Primate Research Center.
Footnotes
DECLARATION OF INTERESTS
J.K.M., L.T.G., E.E.H., H.Z., B.T., E.L., J.T.T., and B.P.L. are inventors on several U.S. patent applications related to this work. The remaining authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108754.
REFERENCES
- Andersson M, Avaliani N, Svensson A, Wickham J, Pinborg LH, Jespersen B, Christiansen SH, Bengzon J, Woldbye DPD, and Kokaia M (2016). Optogenetic control of human neurons in organotypic brain cultures. Sci. Rep 6, 24818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney RJL, Ripke S, Anttila V, Grove J, Holmans P, Huang H, Klei L, Lee PH, Medland SE, Neale B, et al. ; Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium (2015). Dataset: PGC-ASD summary statistics from a meta-analysis of 5,305 ASD-diagnosed cases and 5,305 pseudocontrols of European descent (based on similarity to CEPH reference genotypes). http://www.med.unc.edu/pgc/results-and-downloads. [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, et al. (2020). Evolution of cellular diversity in primary motor cortex of human, marmoset monkey, and mouse. BioRxiv, 2020.03.31.016972. [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, and Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu W-L, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, and Gradinaru V (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah CS, Yu FH, Westenbroek RE, Kalume FK, Oakley JC, Potter GB, Rubenstein JL, and Catterall WA (2012). Specific deletion of NaV1.1 sodium channels in inhibitory interneurons causes seizures and premature death in a mouse model of Dravet syndrome. Proc. Natl. Acad. Sci. USA 109, 14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, et al. (2016). ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, DeWitt WS, et al. (2018). A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange KM, Moutsianas L, Lee JC, Lamb CA, Luo Y, Kennedy NA, Jostins L, Rice DL, Gutierrez-Achury J, Ji S-G, et al. (2017). Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet 49, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, Barr RG, Beaty TH, Becker AB, Beilby J, et al. ; Australian Asthma Genetics Consortium (AAGC) collaborators (2018). Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet 50, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Baldursson G, Belliveau R, Bybjerg-Grauholm J, Bækvad-Hansen M, et al. ; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu W-L, Yang B, Huber N, Pasca SP, and Gradinaru V (2016). Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol 34, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi G-A, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci 19, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, Bulik-Sullivan B, Ripke S, Thornton L, Hinney A, et al. ; Eating Disorders Working Group of the Psychiatric Genomics Consortium (2017). Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bierut LJ, Bisson J, et al. (2018). Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatry 23, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Preissl S, Li Y, Hou X, Lucero J, Wang X, Motamedi A, Shiau AK, Zhou X, Xie F, et al. (2021). Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nature Communications 12, 1337. 10.1038/s41467-021-21583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru M-D, Reach SM, Motl J, Ehrlich ME, Hurd YL, and Roussos P (2018). An atlas of chromatin accessibility in the adult human brain. Genome Res. 28, 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Davis LK, Hart AB, Sanchez-Roige S, Han L, Cacioppo JT, and Palmer AA (2017). Genome-wide association study of loneliness demonstrates a role for common variation. Neuropsychopharmacology 42, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. (2018). Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248–259.e15. [DOI] [PubMed] [Google Scholar]
- Girdhar K, Hoffman GE, Jiang Y, Brown L, Kundakovic M, Hauberg ME, Francoeur NJ, Wang YC, Shah H, Kavanagh DH, et al. (2018). Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat. Neurosci 21, 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LT, Yao Z, Nguyen TN, Kim TK, Zeng H, and Tasic B (2017). Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. eLife 6, e21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck LT, Daigle TL, Sedeño-Cortés A, Walker M, Kalmbach B, Lenz GH, Morin E, Nguyen TN, Garren E, Bendrick JL, et al. (2021). Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron 109. 10.1016/j.neuron.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig JA, Nordin JML, White JW, Wang Q, Bote E, Goode T, Calcedo R, Wadsworth S, Wang L, and Wilson JM (2018). Optimized adeno-associated viral-mediated human factor VIII gene therapy in cynomolgus macaques. Hum. Gene Ther 29, 1364–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S, Tzeng CP, Nagy MA, Stroud H, Koutsioumpa C, Wilcox OF, Assad EG, Green J, Harvey CD, Griffith EC, and Greenberg ME (2019).A scalable platform for the development of cell-type-specific viral drivers. eLife 8, e48089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS) (2018). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry 23, 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner J, Szabo A, Gross-Scherf B, Morikawa RK, Rompani SB, Hantz P, Szikra T, Esposti F, Cowan CS, Bharioke A, et al. (2019). Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci 22, 1345–1356. [DOI] [PubMed] [Google Scholar]
- Kalmbach BE, Buchin A, Long B, Close J, Nandi A, Miller JA, Bakken TE, Hodge RD, Chong P, de Frates R, et al. (2018). h-Channels contribute to divergent intrinsic membrane properties of supragranular pyramidal neurons in human versus mouse cerebral cortex. Neuron 100, 1194–1208.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Cheneby J, Kulkarni SR, and Tan G (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Research 46 (D1), D260–D266. 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremers G-J, Goedhart J, van Munster EB, and Gadella TWJ Jr. (2006). Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45, 6570–6580. [DOI] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, et al. (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nature Biotechnology 36, 70–80. 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. ; European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Duarte FM, Chew JG, Kartha VK, Burkett ZD, Kohlway AS, Pokholok D, Aryee MJ, Steemers FJ, Lebofsky R, et al. (2019). Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nature Biotechnology 37, 916–924. 10.1038/s41587-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, et al. ; 23andMe Research Team; COGENT (Cognitive Genomics Consortium); Social Science Genetic Association Consortium (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YE, Preissl S, Hou X, Zhang Z, Zhang K, Fang R, Qiu Y, Poirion O, Li B, Liu H, et al. (2020). An atlas of gene regulatory elements in adult mouse cerebrum. BioRxiv, 2020.05.10.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou J, Tian W, Luo C, Bartlett A, Aldridge A, Lucero J, Osteen JK, Nery JR, Chen H, et al. (2020). DNA methylation atlas of the mouse brain at single-cell resolution. bioRxiv, 2020.04.30.069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Coskun V, Liang A, Yu J, Cheng L, Ge W, Shi Z, Zhang K, Li C, Cui Y, et al. (2015). Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, Castanon R, Lucero J, Nery JR, Sandoval JP, et al. (2017). Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, et al. (2018). GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E, Whalen S, Przytycki P, Thomas R, Binyameen F, Nowakowski TJ, Kriegstein AR, Sanders SJ, State MW, Pollard KS, and Rubenstein JL (2020). A chromatin accessibility atlas of the developing human telencephalon. Cell 182, 754–769.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JR, Smith GA, Rocha EM, Hayes MA, Beagan JA, Hallett PJ, and Isacson O (2014). Widespread neuron-specific transgene expression in brain and spinal cord following synapsin promoter-driven AAV9 neonatal intracerebroventricular injection. Neurosci. Lett 576, 73–78. [DOI] [PubMed] [Google Scholar]
- Mehta P, Kreeger L, Wylie DC, Pattadkal JJ, Lusignan T, Davis MJ, Turi GF, Li W-K, Whitmire MP, Chen Y, et al. (2019). Functional access to neuron subclasses in rodent and primate forebrain. Cell Rep. 26, 2818–2832.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Carvalho F, Eliez S, and Caroni P (2019). Long-lasting rescue of network and cognitive dysfunction in a genetic schizophrenia model. Cell 178, 1387–1402.e14. [DOI] [PubMed] [Google Scholar]
- Nair RR, Blankvoort S, Lagartos MJ, and Kentros C (2020). Enhancer-driven gene expression (EDGE) enables the generation of viral vectors specific to neuronal subtypes. iScience 23, 100888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. (2013). Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Holtman IR, Coufal NG, Schlachetzki JCM, Yu M, Hu R, Han CZ, Pena M, Xiao J, Wu Y, et al. (2019). Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen G-B, Emilsson V, Meddens SFW, et al. ; LifeLines Cohort Study (2016). Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peron SP, Freeman J, Iyer V, Guo C, and Svoboda K (2015). A cellular resolution map of barrel cortex activity during tactile behavior. Neuron 86, 783–799. [DOI] [PubMed] [Google Scholar]
- Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, Srivatsan S, Qiu X, Jackson D, Minkina A, et al. (2018). Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, and Siepel A (2010). Detection of non-neutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissl S, Fang R, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang Y, Sos BC, Afzal V, Dickel DE, et al. (2018). Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nature Neuroscience 21, 432–439. 10.1038/s41593-018-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet 43, 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A language and environment for statistical computing (R Foundation for Statistical Computing; ). https://www.R-project.org/. [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, et al. ; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, and Greenleaf WJ (2017). chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium (2011). Genome-wide association study identifies five new schizophrenia loci. Nat. Genet 43, 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz N, Uysal B, Welzer M, Bahr JC, Layer N, Löffler H, Stanaitis K, Pa H, Weber YG, Hedrich UB, et al. (2019). Long-term adult human brain slice cultures as a model system to study human CNS circuitry and disease. eLife 8, e48417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SQ, Myers CA, Hughes AEO, Byrne LC, Flannery JG, and Corbo JC (2016). Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 26, 238–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg SN, Taher L, Lindtner S, Sandberg M, Nord AS, Vogt D, Mckinsey GL, Hoch R, Pattabiraman K, Zhang D, et al. (2016). Subpallial enhancer transgenic lines: a data and tool resource to study transcriptional regulation of GABAergic cell fate. Neuron 92, 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, Giusti-Rodriguez P, Hodge RD, Miller JA, Muñoz-Manchado AB, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genetic identification of brain cell types underlying schizophrenia. Nat. Genet 50, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, Gabdank I, Narayanan AK, Ho M, Lee BT, et al. (2016). ENCODE data at the ENCODE portal. Nucleic Acids Res. 44 (D1), D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, and Green P (2013). RepeatMasker Open-4.0, 2013–2015.. http://www.repeatmasker.org. [Google Scholar]
- Song Y, Morales L, Malik AS, Mead AF, Greer CD, Mitchell MA, Petrov MT, Su LT, Choi ME, Rosenblum ST, et al. (2019). Non-immunogenic utrophin gene therapy for the treatment of muscular dystrophy animal models. Nat. Med 25, 1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Kalmbach B, Chong P, de Frates R, Keene CD, Gwinn RP, Cobbs C, Ko AL, Ojemann JG, Ellenbogen RG, et al. (2018). A robust ex vivo experimental platform for molecular-genetic dissection of adult human neocortical cell types and circuits. Sci. Rep 8, 8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van’t Hof AE, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, and Saccheri IJ (2016). The industrial melanism mutation in British peppered moths is a transposable element. Nature 534, 102–105. [DOI] [PubMed] [Google Scholar]
- Verret L, Mann EO, Hang GB, Barth AMI, Cobos I, Ho K, Devidze N, Masliah E, Kreitzer AC, Mody I, et al. (2012). Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell 149, 708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. (2013). A high-resolution enhancer atlas of the developing telencephalon. Cell 152, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, Allaway K, Sakopoulos S, Schneider G, Stevenson O, et al. (2020). Viral manipulation offunctionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci 23, 1629–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdel-laoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, et al. ; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet 50, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Wu DM, Xue Y, Wang SK, Chung MJ, Ji X, Rana P, Zhao SR, Mai S, and Cepko CL (2019). AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl. Acad. Sci. USA 116, 5785–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Chanda S, Marro S, Ng Y-H, Janas JA, Haag D, Ang CE, Tang Y, Flores Q, Mall M, et al. (2017). Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14, 621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Sul JH, Tsetsos F, Nawaz MS, Huang AY, Zekaya A, Illmann C, Osiecki L, Darrow SM, Hirschtritt ME, et al. ; Tourette Association of America International Consortium for Genetics; the Gilles de la Tourette GWAS Replication Initiative; the Tourette International Collaborative Genetics Study; the Psychiatric Genomics Consortium Tourette Syndrome Working Group (2019). Interrogating the genetic determinants of Tourette syndrome and other tic disorders through genome-wide association studies. Am. J. Psychiatry 176, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerucha T, Stühmer T, Hatch G, Park BK, Long Q, Yu G, Gambarotta A, Schultz JR, Rubenstein JLR, and Ekker M (2000). A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci 20, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med 26, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw human bulk ATAC-seq data, human snATAC-seq data, human snRNA-seq data, and mouse snRNA-seq data have been deposited to dbGaP.
dbGaP study name: “Development of tools for cell-type specific labeling of neocortical neurons”
The accession number for the data reported in this paper is dbGaP: phs2292.v1