Loss of function of NFI transcription factors in hypothalamic tanycytes generates diverse subtypes of functional neurons.
Abstract
Hypothalamic tanycytes, radial glial cells that share many features with neuronal progenitors, can generate small numbers of neurons in the postnatal hypothalamus, but the identity of these neurons and the molecular mechanisms that control tanycyte-derived neurogenesis are unknown. In this study, we show that tanycyte-specific disruption of the NFI family of transcription factors (Nfia/b/x) robustly stimulates tanycyte proliferation and tanycyte-derived neurogenesis. Single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) analysis reveals that NFI (nuclear factor I) factors repress Sonic hedgehog (Shh) and Wnt signaling in tanycytes and modulation of these pathways blocks proliferation and tanycyte-derived neurogenesis in Nfia/b/x-deficient mice. Nfia/b/x-deficient tanycytes give rise to multiple mediobasal hypothalamic neuronal subtypes that can mature, fire action potentials, receive synaptic inputs, and selectively respond to changes in internal states. These findings identify molecular mechanisms that control tanycyte-derived neurogenesis, which can potentially be targeted to selectively remodel the hypothalamic neural circuitry that controls homeostatic physiological processes.
INTRODUCTION
Hypothalamic tanycytes are radial glial cells that line the ventricular walls of the mediobasal third ventricle (1, 2). Tanycytes are subdivided into alpha1, alpha2, beta1, and beta2 subtypes based on dorsoventral position and marker gene expression and closely resemble neural progenitors in morphology and gene expression profile. Tanycytes have been reported to generate small numbers of neurons and glia in the postnatal period, although at much lower levels than in more extensively characterized sites of ongoing neurogenesis, such as the subventricular zone of the lateral ventricles or the subgranular zone of the dentate gyrus (3–6). While tanycyte-derived newborn neurons may play a role in regulating a range of behaviors (3, 7, 8), levels of postnatal tanycyte-derived neurogenesis are low and virtually undetectable in adulthood (9). As a result, little is known about the molecular identity or connectivity of tanycyte-derived neurons (TDNs) (6, 9). A better understanding of the gene regulatory networks that control neurogenic competence in hypothalamic tanycytes would both give insight into the function of TDNs and potentially identify new therapeutic approaches for modulation and repair of hypothalamic neural circuitry.
Studying retinal Müller glia, which closely resemble hypothalamic tanycytes in both morphology and gene expression, provides valuable insight into the neurogenic potential of tanycytes (3, 9, 10). Zebrafish Müller glia function as quiescent neural stem cells and are able to regenerate every major retinal cell type following injury (11). While mammalian Müller glia effectively lack neurogenic competence, in posthatch chick, they retain a limited neurogenic competence that resembles that of mammalian tanycytes (12). Recent studies in the retina have identified the NFI family of transcription factors Nfia/b/x as being essential negative regulators of neurogenesis in both late-stage progenitor cells and in mature mammalian Müller glia (13, 14). Moreover, like in the retina, NFI factors are expressed in late-stage hypothalamic neural progenitors (15), and Nfia is necessary for hypothalamic glia specification (16). These findings raise the possibility that NFI factors may actively repress proliferation and neurogenic competence in tanycytes.
We hypothesize here that suppression of NFI factor activity in tanycytes may enhance their proliferative and neurogenic capacity. To address this possibility, we selectively disrupted Nfia/b/x function in hypothalamic tanycytes of both juvenile and adult mice. We observed that early loss of function of NFI activity in hypothalamic tanycytes led to a robust induction of proliferation and neurogenesis, while Nfia/b/x disruption in adults led to lower levels of tanycyte-derived proliferation that are seen following neonatal loss of function. NFI loss of function activated both Shh and Wnt signaling in tanycytes, and this, in turn, stimulated proliferation and neurogenesis. These TDNs survive, mature, and migrate radially away from the ventricular zone (VZ), express molecular markers of diverse hypothalamic neuronal subtypes, fire action potentials, and receive synaptic inputs. These findings demonstrate that hypothalamic tanycytes have a latent neurogenic competence that is actively suppressed by NFI family transcription factors, which can be modulated to induce generation of multiple hypothalamic neuronal subtypes.
RESULTS
Nfia/b/x loss of function induces proliferation and neurogenesis in neonatal mice
To determine whether hypothalamic tanycytes express Nfia/b/x, we first analyzed bulk RNA sequencing (RNA-seq) data obtained from fluorescence-activated cell sorting (FACS)–isolated green fluorescent protein (GFP)+ cells from the mediobasal hypothalamus of adult Rax-GFP transgenic mice, where GFP is selectively expressed in tanycytes (17). We observe that GFP+ tanycytes, indicated by Rax expression, show highly enriched expression for Nfia, Nfib, and Nfix relative to the GFP−, neuronally enriched fraction of mediobasal hypothalamic cells (Fig. 1A). Immunohistochemical analysis in adult mice reveals that Nfia/b/x proteins are highly expressed in the tanycytic layer and in other hypothalamic glial cells (Fig. 1B) (16). We next generated Rax-CreER;Nfialox/lox;Nfiblox/lox;Nfixlox/lox;CAG-lsl-Sun1-GFP mice (TKO mice hereafter), which allow inducible, tanycyte-specific disruption of Nfia/b/x function while simultaneously tracking the fate of tanycyte-derived cells using Cre-dependent Sun1-GFP expression (Fig. 1C) (18, 19).
Fig. 1. Nfia/b/x genes suppress proliferation and neurogenesis in tanycytes of neonatal mice.
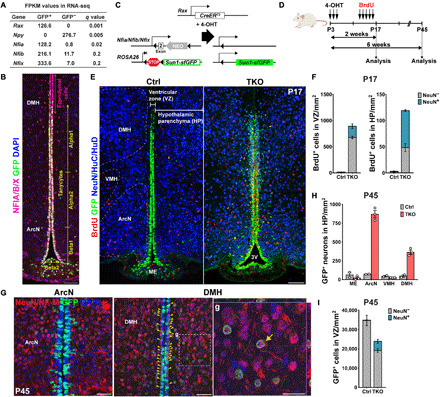
(A) Expression of Nfia/b/x in GFP+ tanycytes isolated from Rax:GFP mice (17) compared to the GFP− cells in adult hypothalamus. The tanycyte-specific marker Rax and the neuronal marker Npy are enriched in GFP+ and GFP− cells, respectively FPKM, fragments per kilobase of transcript per million mapped reads. (B) Distribution of Nfia/b/x protein in Rax-GFP+ tanycytes. Schematic of mouse lines is used in this study. (C and D) Schematic of a genetic approach for simultaneous tanycyte-specific disruption of Nfia/b/x and reporter gene labeling of tanycytes and tanycyte-derived cells using tamoxifen-dependent activation of CreER. (E) Induction of proliferation and neurogenesis in NFI-deficient tanycytes by P17. The Bregma position of the control section is slightly posterior to that of the mutant. 3V, the third ventricle. (F) Quantification of proliferation and neurogenesis in the VZ and hypothalamic parenchyma (HP) at P17 (n = 3 to 5 mice). (G) In NFI TKO mice by P45, mature neuronal marker NeuN and neurofilament M (NF-M) were simultaneously detected in TDNs migrating into the parenchyma of the arcuate nucleus (ArcN) and dorsomedial hypothalamus (DMH), with a small number of neurons remaining in the subventricular zone (yellow arrowheads). Enlarged image of parenchymal TDNs in (g) with the orthogonal view showing costaining within the cell. (H) Substantially increased numbers of NeuN+/GFP+ TDNs are observed in NFI TKO mice in ArcN and DMH relative to wild-type controls, but comparable numbers of neurons are observed in median eminence (ME) and ventromedial hypothalamus (VMH) (n = 2 to 3 mice). (I) The number of GFP+ tanycytes is reduced in NFI-deficient mice at P45, and ectopic neurons are seen in the VZ (n = 2 to 3 mice). Scale bars, 100 μm (B and E), 50 μm (G), and 25 μm (g).
We induced Cre activity using daily intraperitoneal injections of 4-hydroxytamoxifen (4-OHT) between postnatal days 3 and 5 (P3 and P5) (Fig. 1D). At this point, neurogenesis in the mediobasal hypothalamus is low under baseline conditions (3, 9). Following 4-OHT treatment, we observe that NFIA/B/X immunoreactivity is first reduced in the tanycyte layer beginning at P6 following 4-OHT injections between P3 and P5, initially in more ventral regions where Rax expression is strongest (fig. S1A). NFIA/B/X immunoreactivity is largely undetectable by P10, and Cre-dependent GFP expression was correspondingly induced (fig. S1A). 5-Bromo-2′-deoxyuridine (BrdU) incorporation and Ki67 labeling was seen beginning at P6 in dorsally located alpha tanycytes, with labeling spreading to beta1 tanycytes of the arcuate nucleus (ArcN) by P8 and beta2 tanycytes of the median eminence (ME) by P10 (fig. S1, A and B). At P12, Ki67 labeling was observed in the tanycyte layer immediately adjacent to the third ventricle lumen, and a small subset of GFP+ cells in the tanycyte layer closest to the hypothalamic parenchyma (HP) began to express neuronal markers (fig. S1C).
By P17, Nfia/b/x expression was completely lost in the tanycytes, although it was preserved in Rax-negative ependymal cells where GFP expression is not induced (fig. S1D), and tanycyte-derived GFP+ neuronal precursors in the VZ were actively amplified and had begun to migrate outward into the HP in Nfia/b/x-deficient mice (Fig. 1, E and F, and fig. S1D). In contrast, few migrating GFP+ cells were observed in Rax-CreER;CAG-lsl-Sun1-GFP controls (Fig. 1, E and F). At P45, a substantial increase in GFP+ cells expressing mature neuronal markers was observed in the parenchyma of the ArcN and dorsomedial hypothalamic (DMH) nuclei (Fig. 1, G and H), while limited numbers of GFP+ cells expressing neuronal markers remained in the subventricular region in Nfia/b/x-deficient mice [Fig. 1, G (yellow arrowheads) and I].
Loss of function of Nfia/b/x induces tanycyte proliferation in older mice
Our previous studies showed that the loss of function of Nfia/b/x in late-stage retinal progenitors robustly induces proliferation and neurogenesis under baseline conditions, while in adult Müller glia, it induces limited levels of proliferation and neurogenesis but only following neuronal injury (13, 14). This suggests that neurogenic competence in mature murine tanycytes could be lower than that seen in neonates. To test this, we conducted 4-OHT treatment at older ages. While we still observed a robust induction of proliferation and neurogenesis following treatment at P7, this was less effective at P10. Only very low levels were observed at P12, not significantly different from control mice (fig. S2, A to C). However, this low level of proliferation reflected a substantially reduced efficiency of 4-OHT–dependent disruption of Nfia/b/x, as confirmed by the largely intact pattern of immunoreactivity for NFIA/B/X in TKO mice (fig. S2D).
To improve the efficiency of Nfia/b/x deletion and to study the effects of NFI loss of function in adult animals, we applied viral-mediated Cre delivery. Intracerebroventricular injection of adeno-associated virus 1 (AAV1)–Cre-mCherry into both Nfialox/lox;Nfiblox/lox;Nfixlox/lox;CAG-lsl-Sun1-GFP and CAG-lsl-Sun1-GFP control mice at P60 resulted in robust mCherry expression in ventricular hypothalamic cells within 2 weeks, along with Cre-dependent induction of Sun1-GFP expression in both control and Nfia/b/x-floxed conditional mice (fig. S3). We observed efficient loss of NFI expression in GFP+ tanycytes by P74 and coimmunolabeling with BrdU, delivered continuously by osmotic mini pump during the 2 weeks (fig. S3C). To determine the specific induction of proliferation initiated from tanycytes, we administered EdU (5-ethynyl-2′-deoxyuridine) using a once daily intraperitoneal injection between P75 and P77 and analyzed mice at P78 (Fig. 2A). We observed selective EdU incorporation into alpha tanycytes adjacent to the dorsal part of ArcN (Fig. 2, C and D). Much lower levels of EdU incorporation were observed in ventrally located beta tanycytes, while no EdU labeling was observed in controls (Fig. 2, B and D). Although we observed a few GFP+ cells in the mediobasal HP, these cells were not labeled with EdU, and there was no difference in their number between control and TKO animals (Fig. 2E). We confirmed the specific and local induction of tanycyte proliferation with the increased endogenous expression of Ki67, a proliferation marker, only in Nfia/b/x-deficient TKO mice (Fig. 2F and fig. S3C).
Fig. 2. Loss of Nfia/b/x selectively induces tanycyte proliferation in adult mice.
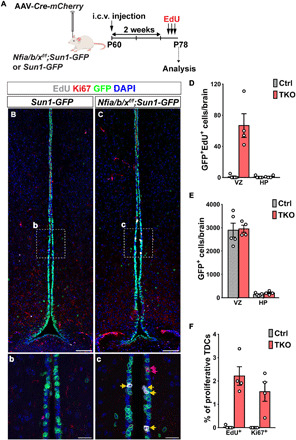
(A) Schematic for intracerebroventricular (i.c.v.) delivery of AAV-Cre and analysis of Nfia/b/x loss of function in P78 mice. (B) AAV-Cre induces Sun1-GFP expression in tanycytes in CAG-lsl-Sun1-GFP control mice at P78 [(b) inset shows alpha tanycytes]. (C) AAV-Cre induces proliferation in alpha tanycytes [shown in inset (c)] of Nfialox/lox;Nfiblox/lox;Nfixlox/lox;CAG-lsl-Sun1-GFP mice. (D) Quantification of GFP+/EdU+ cells in VZ and HP in NFI-deficient mice (n = 4 to 5 mice). (E) Number of GFP+ cells in VZ and HP in control and NFI-deficient mice. (F) Percentage of GFP+ VZ cells in the alpha tanycyte region labeled by Ki67 and EdU in NFI-deficient mice (n = 4 to 5 mice). Scale bars, 100 μm (B and C) and 25 μm (b and c).
Single-cell RNA-seq and assay for transposase-accessible chromatin sequencing analysis identify gene regulatory networks controlling neurogenesis in tanycytes
To obtain a comprehensive picture of the cellular and molecular changes that occur during proliferation and neurogenesis, we conducted single-cell RNA sequencing (scRNA-seq) analysis of FACS-isolated GFP-positive tanycytes and tanycyte-derived cells from both control and Nfia/b/x-deficient mice. To do this, we induced Cre activity in tanycytes between P3 and P5 and harvested the GFP+ cells at P8, P17, and P45 time points in TKO mice. We next profiled a total of >60,000 cells using the Chromium platform (10x Genomics), generated separate uniform manifold approximation and projection (UMAP) plots for control and Nfia/b/x-deficient tanycytes, and then aggregated data obtained from all samples (Fig. 3A and fig. S4). In control mice, we not only could readily distinguish tanycyte subtypes (alpha1, alpha2, beta1, and beta2), based on previously characterized molecular markers (20, 21), but also observed that tanycytes give rise to a range of other hypothalamic cell types (Fig. 3B). In controls, at P8, we observe a small fraction of proliferative tanycytes, from which arise differentiation trajectories that give rise to astrocytes and ependymal cells, as well as small numbers of oligodendrocyte progenitor cells (OPCs) and neurons (Fig. 3, C and D; fig. S4; and table S2). At P17 and P45, however, very few proliferating tanycytes are observed, and evidence for ongoing generation of neurons and glia is lacking (Fig. 3, C and D, and fig. S4). In TKO mice, in contrast, we observe that a remarkably larger fraction of cells are proliferating tanycytes at all ages, along with clear evidence for ongoing neurogenesis (Fig. 3D and fig. S4). Furthermore, a substantial reduction in the relative fraction of nonneuronal cells, including astrocytes, OPCs, and ependymal cells, is observed, in line with previous studies reporting an essential role for NFI family genes in gliogenesis in other central nervous system regions (13, 22, 23). While controls show a higher fraction of tanycyte-derived astrocytes, a much higher fraction of GFP+ cells are neurons in TKO mice (Fig. 3D and table S2). Furthermore, the density and relative fraction of cells expressing alpha2 tanycyte markers is increased in TKO mice, which is consistent with previously published neurosphere and cell lineage analysis (4), demonstrating a higher neurogenic competence for alpha2 tanycytes [Fig. 3, C (bottom) and D, and table S2] (4).
Fig. 3. scRNA-seq analysis of control and NFI-deficient tanycytes.
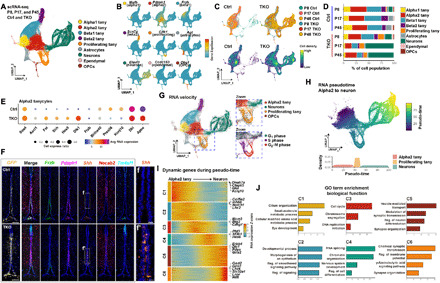
(A) Aggregate UMAP plot of scRNA-seq data from control and NFI-deficient GFP+ tanycytes and tanycyte-derived cells isolated at P8, P17, and P45. Cell types are indicated by color shading. tany, tanycytes. (B) Distribution of cell type–specific marker expression on aggregate UMAP plot. (C) Distribution of cells by age and genotype on aggregate UMAP plot. (D) Percentage of each cell type by age and genotype. (E) Dot plot showing differentially expressed genes (DEGs) in Nfia/b/x-deficient alpha2 tanycytes. Avg, average. (F) HiPlex analysis for tanycyte subtype–specific markers and enhanced Shh expression in a subset of Pdzph1+ alpha2 tanycytes. Necab2+ alpha1 tanycytes and Tm4sf1+ ependymal cells are shown for reference. (G) RNA velocity analysis indicating differentiation trajectories in tanycytes and tanycyte-derived cells. Insets highlight proliferating tanycytes and TDNs. (H) Pseudo-time analysis of differential gene expression in alpha2 tanycytes, proliferating tanycytes and TDNs. (I) Heatmap showing DEGs over the course of tanycyte-derived neurogenesis. (J) Gene ontology (GO) analysis of DEGs in (I), with enrichment shown at −log10 P value. Scale bars, 100 μm (F), and 20 μm (f and f′).
To identify critical regulators of proliferative and neurogenic competence in tanycytes, we performed a differential gene expression analysis between control and TKO mice in each tanycyte subtype. This analysis uncovered differential expression of multiple extrinsic and intrinsic regulators of these processes, particularly in alpha2 tanycytes (Fig. 3E, fig. S5, and table S4). Control tanycytes also selectively expressed many genes expressed by both mature, quiescent tanycytes and retinal Müller glia, whose expression is down-regulated following cell-specific deletion of Nfia/b/x (14). These include genes that are highly and selectively expressed in mature alpha2 tanycytes, such as Apoe and Kcnj10, the Notch pathway target Hes1, the Wnt inhibitor Frzb, and the transcription factors Klf6 and Bhlh40. Nfia/b/x-deficient tanycytes, in contrast, up-regulated Shh, the Notch inhibitor Dlk1, the bone morphogenetic protein inhibitor Fst, the neurogenic factors Ascl1 and Sox4, and the Notch pathway target Hes5 (Fig. 3E). Alpha1 and beta1 tanycytes also showed reduced expression of Tgfb2 and Fgf18 (fig. S5), which were previously shown to be strongly expressed in these cells (21, 24). To validate these results, we conducted multiplexed single-molecule fluorescence in situ hybridization (smfISH) (HiPlex, Advanced Cell Diagnostics Bio-Techne) and observed strong up-regulation of Shh in Pdzph1-positive alpha2 tanycytes at P45 in TKO mice (Fig. 3F).
To infer cell lineage relationships between specific tanycyte subtypes and tanycyte-derived neural progenitors, we conducted RNA velocity analysis (25) on the full aggregated scRNA-seq dataset (Fig. 3G). We observe that alpha2 tanycytes give rise to proliferating tanycytes, which, in turn, give rise to neural precursors following cell cycle exit (Fig. 3G, insets). Note that astrocytes appear to arise directly from alpha1 and alpha2 tanycytes without going through a clear proliferative stage (Fig. 3, G and H).
We then used pseudo-time analysis to identify six major temporally dynamic patterns of gene expression that occurred during the process of alpha2 tanycyte–derived neurogenesis (Fig. 3I and table S5). On the basis of the gene ontology (GO) term enriched in each cluster (Fig. 3J), the transition from a quiescent to an actively proliferating state is associated with the down-regulation of metabolic genes (Glul), ion channels (Kcnj10), transcription factors (Lhx2), and Notch pathway components (Notch1), all of which are expressed at high levels in mature tanycytes (3, 10). In addition, genes regulating ciliogenesis (Dnah7a and Cfap65) are rapidly down-regulated. Following the up-regulation of genes controlling cell cycle progression and DNA replication (Cenpf and Mcm3) and cell cycle exit (Btg2), tanycyte-derived neural precursors up-regulate genes that control chromatin conformation (Phf3), RNA splicing (Sf3b1), and neurogenesis (Hes6). This up-regulation is then followed by the expression of transcription factors that control the specification of specific hypothalamic neuronal subtypes (Dlx1 and Lhx6) and regulators of synaptogenesis (Erbb4), neurotransmitter biogenesis and reuptake (Gad1, Pdyn, and Slc32a1), neurotransmitter receptors (Grin1 and Gria2), and leptin signaling (Lepr).
To investigate changes of chromatin accessibility in TKO mice, we conducted single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) analysis in FACS-isolated GFP+ tanycytes and tanycyte-derived cells in both control and TKO mice at P8. UMAP analysis indicated that cell identity in both control and mutant samples could be readily assigned on the basis of gene expression data obtained from scRNA-seq (Fig. 4A). The overall distribution of cell types was much like that seen for scRNA-seq data (Fig. 3A) with more proliferating tanycytes and TDNs observed in TKO cells compared to controls (Fig. 4B). Accessibility of the consensus NFI motif was assessed in all cell types in Nfia/b/x-deficient mice (Fig. 4C), and reduced levels of bound transcription factors were observed at these sites by footprinting analysis (Fig. 4E), indicating that Nfia/b/x proteins are actively required to maintain accessible chromatin at a subset of their target sites.
Fig. 4. scATAC-seq analysis of P8 wild-type and Nfia/b/x-deficient tanycytes demonstrates the derepression of Shh and increased Wnt signaling.
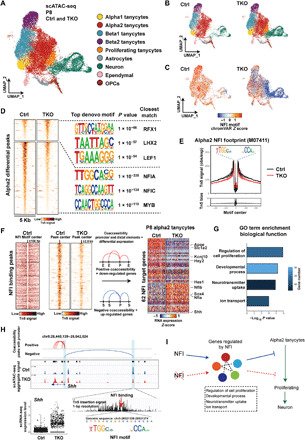
(A) Aggregate UMAP plot of scATAC-seq data from control and NFI-deficient GFP+ tanycytes and tanycyte-derived cells isolated at P8. Cell types are indicated by color shading. (B) Distribution of cell types shown for control and Nfia/b/x-deficient GFP+ cells at P8. (C) Distribution of accessible consensus NFI motif shown for control and Nfia/b/x-deficient GFP+ cells. (D) Transcription factor binding motifs selectively enriched and depleted in control and Nfia/b/x-deficient alpha2 tanycytes. RFX1, regulatory factor X1; LHX1, LIM homeobox 1; LEF1, lymphoid enhancer binding factor 1; MYB, MYB proto-oncogene, transcription factor. (E) Consensus NFI footprint distribution in control and Nfia/b/x-deficient alpha2 tanycytes. obs/exp, observed/expected. (F) Integration of scATAC-seq and scRNA-seq data to identify DEGs in alpha2 tanycytes that are directly regulated by Nfia/b/x. (G) GO analysis of Nfia/b/x-regulated genes expressed in alpha2 tanycytes. (H) Shh is directly repressed by Nfia/b/x in alpha2 tanycytes. (I) Summary of Nfia/b/x action in alpha2 tanycytes.
We observed 1333 chromatin regions that showed increased accessibility and 4564 regions that showed decreased accessibility in Nfia/b/x-deficient alpha2 tanycytes relative to controls (Fig. 4D). As expected, Hypergeometric Optimization of Motif EnRichment (HOMER) analysis indicated that open chromatin regions (OCRs) specific to controls were most highly enriched for consensus sites for NFI family members (Fig. 4D and table S6). In contrast, motifs for the Wnt effector Lef1 were enriched at OCRs specifically detected in Nfia/b/x-deficient alpha2 tanycytes. We observed that a subset of genes with altered expression in scRNA-seq showed altered accessibility in putative associated cis-regulatory sequences, although changes in gene expression and chromatin accessibility often diverged (tables S7 and S8). While putative regulatory elements of the down-regulated genes Aqp4, Hes1, and Fgf18 showed reduced accessibility, elements associated with other down-regulated genes such as Tgfb2 and Sox8 showed increased accessibility. Likewise, while most up-regulated genes showed increased accessibility, such as Shh and Sox4, some showed decreased accessibility. These divergent responses indicate that NFI genes control the expression of a large number of transcription factors in tanycytes, which appear to perform dual roles as both activators and repressors, as has previously been reported in other astroglial cell types (22, 26, 27).
To identify direct targets of NFI factors and to better clarify their function in regulating proliferation and neurogenesis in alpha2 tanycytes, we integrated scRNA-seq and scATAC-seq data from alpha2 tanycytes to identify genes with both altered expression and altered accessibility at sites containing NFI consensus sequences and identified 62 genes in total (Fig. 4F and table S9). These include down-regulated genes such as Kcnj10 and Apoe and Notch pathway effectors such as Hes1 and Hey2, as well as Shh and Sox4, which are up-regulated with direct target genes enriched for genes controlling proliferation and neural development (Fig. 4G). Transcription of Nfia and Nfib are themselves strongly activated by Nfia/b/x, consistent with findings in the retina (13, 14). We found NFI binding sites in peaks that are negatively correlated with the promoter of Shh, suggesting that NFI may directly repress Shh expression (Fig. 4H). Thus, NFI factors may act as both activators and repressors in alpha2 tanycytes that promote quiescence while inhibiting proliferation and neurogenesis (Fig. 4I).
Shh and Wnt signaling regulate tanycyte proliferation and neurogenic competence
The increased expression of Shh and Wnt regulators that are observed in Nfia/b/x-deficient alpha2 tanycytes (Figs. 3E and 5, A and B, and table S4) suggested that Shh and Wnt signaling might promote proliferation and/or neurogenesis in tanycytes. We observe substantially increased expression of Shh in both alpha2 and beta1 tanycytes (Fig. 3F and fig. S5) and more complex regulation of Wnt signaling modulators. We observe increased expression of Sulf1, which, by regulating the synthesis of heparan sulfate proteoglycans, typically enhances Wnt signaling (28). However, the broad-spectrum Wnt inhibitor Notum, which was recently shown to regulate quiescence in adult neural stem cells of the lateral ventricles (29), is up-regulated from P17, potentially acting in a cell-autonomous manner counteracting the effects of increased cellular levels of Wnt signaling (Fig. 5, A and B).
Fig. 5. Shh and Wnt signaling stimulate proliferation and neurogenesis in Nfia/b/x-deficient tanycytes.
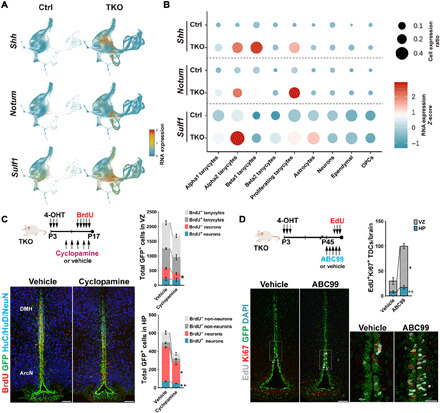
(A) Expression of Shh, Notum, and Sulf1 on aggregate UMAP plot. (B) Dot plot showing expression level of Shh and Wnt pathway genes in each cluster. (C) Shh inhibition by intraperitoneal cyclopamine inhibits neurogenesis in Nfia/b/x-deficient mice. (D) Activation of Wnt signaling by inhibition of Notum via intraperitoneal. ABC99 induces the proliferation of alpha tanycytes and increases numbers of GFP+ proliferating cells in VZ and HP. TDCs, tanycyte-derived cells; n.s., not significant. Scale bars,100 μm (C and D) and 25 μm (D, inset). *P < 0.05, **P < 0.001.
To determine whether this increased Shh inhibition might inhibit regulated tanycyte proliferation and/or tanycyte-derived neurogenesis, we administered the blood-brain barrier–permeable Shh antagonist cyclopamine via intraperitoneal injection to Nfia/b/x-deficient mice every 2 days from P8 to P16 in conjunction with daily intraperitoneal injections of BrdU from P12 to P16 (Fig. 5C). At these ages, Shh is both highly expressed in tanycytes, and levels of proliferation and neurogenesis are high in Nfia/b/x-deficient alpha2 tanycytes. Cyclopamine administration resulted in a significant reduction in both the numbers of total GFP+ cells and GFP/NeuN double-positive neurons in both the tanycytic layers in VZ and HP when compared to vehicle controls, while BrdU incorporation was only significantly altered in parenchymal neurons, indicating a stronger effect on tanycyte-derived neurogenesis than on self-renewing tanycyte proliferation (Fig. 5C).
To determine whether Notum-dependent Wnt signaling played a role in inhibiting the tanycyte proliferation and/or neurogenesis at later ages, we treated P45 Nfia/b/x-deficient mice with the blood-brain barrier–permeable Notum inhibitor ABC99 (30) once daily for 5 days, with EdU coadministered on the last 3 days. At this age, Notum expression is high, and levels of tanycyte proliferation are substantially reduced relative to the early postnatal period. This led to a significant increase in proliferation in alpha2 tanycytes (Fig. 5D), indicating that activation of Wnt signaling stimulates tanycyte proliferation at later ages.
Nfia/b/x-deficient tanycytes give rise to a diverse range of hypothalamic neuronal subtypes
To investigate the identity of TDNs, we analyzed a neuronal subset of scRNA-seq data obtained from both control and Nfia/b/x-deficient mice (Fig. 6A). A total of 582 neurons were obtained from controls, while 15,489 neurons were obtained from Nfia/b/x-deficient mice (table S2). The great majority of control TDNs was obtained at P8, while large numbers of TDNs were seen at all ages in Nfia/b/x-deficient mice. UMAP analysis revealed that both control and Nfia/b/x-deficient TDNs fell into two major clusters each of glutamatergic and γ-aminobutyric acid (GABA)ergic subtypes, with additional clusters corresponding to Ascl1/Hes5-positive postmitotic neural progenitor cells and immature Dlx1/Sox11-positive GABAergic precursors (Fig. 6, A and B). RNA velocity analysis indicated three distinct major differentiation trajectories in both control and Nfia/b/x-deficient TDNs, which give rise to the two major glutamatergic clusters and the GABAergic neurons (Fig. 6C).
Fig. 6. Identification of selective markers of TDNs.
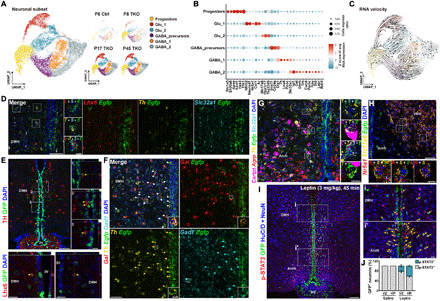
(A) UMAP plot showing major clusters of tanycyte-derived neuronal subset, separated by age and genotype. (B) Dot plot showing major subtype-specific markers of TDNs. (C) RNA velocity analysis indicates differentiation trajectories for TDNs. (D) smfISH analysis and (E) immunohistochemistry demonstrates expression of Th and Lhx6 in Egfp+ TDNs in Nfia/b/x-deficient mice. (F) smfISH analysis of Gal, Gad1, and Th in Egfp+ TDNs in Nfia/b/x-deficient mice. (G) Cart, Agrp, Slc32a1, and Th expression in Egfp+ TDNs in Nfia/b/x-deficient mice. (H) Nr5a1 and Slc17a6 expression in Egfp+ TDNs in Nfia/b/x-deficient mice. All insets are enlarged images of examples of colocalization [white boxes in (D) to (H)]. (I) pStat3 staining 45 min after intraperitoneal administration of leptin (3 mg/kg) in Nfia/b/x-deficient mice (n = 3 mice). Arrows indicate GFP+/pStat3+ TDNs. Insets show higher magnification images in DMH (i) and ArcN (i′). (J) Fraction of pStat3-positive TDNs in VZ and HP after leptin administration. pStat3 was not induced in saline-injected mice (n = 2 mice). Scale bars, 50 μm (D, F, G, and H), 10 μm [insets in (D), (F), (G), and (H)], 100 μm (E and I), and 20 μm [high-magnification images in (E)].
Fifty-three percent of TDNs were GABAergic, as determined by expression of Gad1, Gad2, and/or Slc32a1, while 30% were Slc17a6-positive glutamatergic neurons (Fig. 6B and tables S10 and S11). The glutamatergic cluster Glu_1 was enriched for the transcription factors Nhlh2 and Neurod2, as well as markers of glutamatergic ventromedial hypothalamus (VMH) neurons, such as Nr5a1 and Cnr1, and the androgen receptor Ar, while Glu_2 was enriched for markers of glutamatergic DMH neurons, such as Ppp1r17, and ArcN markers, such as Chgb (Fig. 6B). GABAergic neurons expressed a diverse collection of molecular markers expressed by neurons in the ArcN and DMH as well as the adjacent zona incerta (ZI), which regulates a broad range of internal behaviors including feeding, sleep, and defensive behaviors (31). GABA_1 was enriched for a subset of ZI- and DMH-enriched genes (Lhx6 and Pnoc), while GABA_2 was enriched for genes selectively expressed in ArcN neurons (Isl1), as well as for genes expressed in GABAergic neurons in both the ArcN and DMH (Cartpt, Npy, Sst, Gal, Trh, and Th).
We used both multiplexed smfISH (Fig. 6, D and F) and immunohistochemistry (Fig. 6E and table S12) to confirm the expression of Th, Lhx6, and Gal in GFP+ Nfia/b/x-deficient tanycyte–derived GABAergic neurons in the DMH, as verified by Egfp/Slc32a1 (or Gad1) colabeling. Expression of Cartpt and Th was observed in GABAergic TDNs in both the ArcN and DMH (Fig. 6G and fig. S6A), and coexpression of Lhx6 and Th, as well as Cartpt and Th, was observed in a subset of TDNs and non-TDNs. Small numbers of Nr5a1-expressing glutamatergic (Slc17a6-positive) neurons were detected in the dorsomedial VMH (Fig. 6H). No expression of markers specific to neurons of more anterior or posterior hypothalamic regions (e.g., Avp, Crh, Oxt, Pmch, Hcrt, Vip, etc.) was detected (table S11).
These data demonstrate that TDNs express molecular markers of multiple neuronal subtypes located in the tuberal hypothalamus, including the ArcN and VMH. To determine how closely the gene expression profile of TDNs more broadly resembles the profile of hypothalamic neurons in these regions, we used Linked Inference of Genomic Experimental Relationships (LIGER) analysis (32) to integrate clustered scRNA-seq from a previous study of adult ArcN, in which a small number of VMH neurons was also profiled (fig. S6B and table S13) (21). Integration of these datasets using LIGER (33) and comparison of cell types in each cluster using alluvial plotting (fig. S6C) indicate substantial overlap in cellular gene expression profiles between glutamatergic TDN cluster Glu_1 with both Kiss1/Tac2-positive ArcN neurons and Nr5a1-positive VMH neurons. Glu_2 overlapped with Rgs16/Nmu-positive ArcN neurons. GABAergic tanycyte-derived GABA_1 and GABA_2 clusters overlapped with several different ArcN neuronal clusters, including clusters that contained neurons expressing Th, Ghrh, and/or Trh. In contrast, some subtypes of ArcN neurons were represented only sparsely or not at all among TDNs, while other TDNs appeared to correspond to cell types not found in the published scRNA-seq dataset. For instance, while some TDNs closely resembled Pomc-expressing ArcN neurons, the relative fraction of Pomc-positive TDNs was substantially lower than in ArcN (LIGER cluster 4). Likewise, no TDNs mapped to LIGER cluster 7, which corresponded to Agrp-positive ArcN neurons, and no Agrp-positive TDNs were detected using smfISH (Fig. 6G and fig. S6, B and C). While few immature TDNs mapped to neurons in the mature ArcN scRNA-seq dataset, as expected, two clusters of mature glutamatergic (LIGER clusters 8 and 9) and one cluster of GABAergic (LIGER cluster 11) also showed little correspondence to ArcN neurons and appeared to correspond to DMH-like neurons based on their expression of DMH-enriched genes (e.g., Ppp1r17 and Lhx6).
At P45, 4.4% of TDNs in the GABA_2 cluster expressed the leptin receptor Lepr (Fig. 6B), despite Lepr being essentially undetectable in tanycytes themselves, as previously reported (17). As a result, we tested whether TDNs were capable of responding to leptin signaling. P90 mice that had undergone an overnight fast were injected intraperitoneally with leptin (3 mg/kg) and euthanized after 45 min, providing sufficient time for leptin-responsive neurons throughout the hypothalamus to induce phosphorylation of signal transducer and activator of transcription 3 (pStat3) immunoreactivity (34). We observed robust induction of pStat3 immunoreactivity in GFP+ TDNs under these conditions (Fig. 6I), with low levels of immunoreactivity under unstimulated conditions, as previously reported (34). A total of 42.3 (±2.5)% of parenchymal TDNs in DMH and ArcN show pStat3 induction under these conditions, along with 22.3 (±3.9)% of TDNs in the subventricular region (Fig. 6J), confirming that a subset of TDNs are leptin responsive.
Neurons derived from Nfia/b/x-deficient tanycytes fire action potentials and receive synaptic inputs
Since our scRNA-seq analysis identified the majority of tanycyte-derived cells as neurons in Nfia/b/x-deficient mice, we next investigated whether these cells showed electrophysiological properties of functional neurons. To characterize tanycyte-derived cells, we performed whole-cell patch-clamp recordings from GFP+ parenchymal cells in acute brain slices obtained from Nfia/b/x-deficient mice that had undergone 4-OHT treatment between P3 and P5. Biocytin filling of recorded tanycyte-derived cells revealed neuron-like morphology, typically showing three to five major dendritic processes, similar to GFP− control neurons (Fig. 7A and fig. S7). We found that the majority of GFP+ tanycyte–derived cells in the HP fired action potentials in response to depolarizing current steps (95%, 40 of the 42 cells from P15 to P97; Fig. 7, B and C). In contrast, GFP+ cells located in the tanycytic layer retained nonspiking, glial-like electrophysiological properties in Nfia/b/x-deficient mice (fig. S8).
Fig. 7. Nfia/b/x-deficient tanycytes mostly differentiate into neurons.
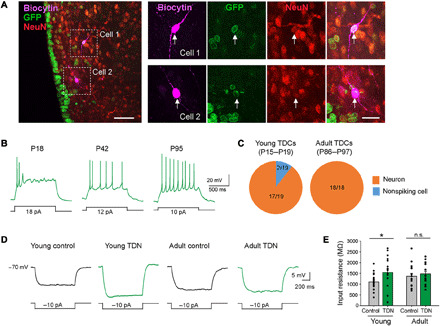
(A) Low- and high-magnification confocal images showing two biocytin-filled GFP+ recorded cells (white arrows) in an NFI TKO brain slice stained with NeuN. (B) Example responses of tanycyte-derived cells to depolarizing current steps. (C) The proportion of TDNs among tested tanycyte-derived cells in young (left) and adult (right) mice. (D) Representative average responses to hyperpolarizing current steps. (E) Summary graphs of input resistance (young control neurons, 18 cells from five mice, 1101 ± 89 MΩ; young TDNs, 17 cells from five mice, 1546 ± 176 MΩ; P = 0.0238, Mann-Whitney U test; adult control neurons, 16 cells from six mice, 1369 ± 132 MΩ; adult TDNs, 18 cells from six mice, 1477 ± 119 MΩ; P = 0.4777, Mann-Whitney U test). Scale bars, 50 μm (A, left) and 20 μm (A, right). *P < 0.05.
Similar to typical hypothalamic neurons (35), many TDNs fired spontaneous action potentials (sAPs) and exhibited relatively depolarized resting membrane potentials (fig. S9A). However, the proportion of TDNs showing sAPs was significantly lower than that of GFP− control neurons in young (P15 to P19) mice (fig. S9B). Nonetheless, TDNs revealed more depolarized resting membrane potentials compared to control neurons in both young (P15 to P19) and adult (P86 to P97) mice (fig. S9B). In addition, we found that the input resistance was significantly higher in TDNs than in GFP− control neurons in young (P15 to P19) mice, although the input resistances were similar for tanycyte-derived and control neurons in adult mice (P86 to P97) (Fig. 7, D and E, and table S14). Together, these data indicate that, although there were some differences with GFP− control neurons, almost all tanycyte-derived cells fired action potentials and shared electrophysiological features of control neurons, indicating that they are neurons.
Since the great majority of tanycyte-derived cells appear to be functional neurons, we next asked whether their evoked action potential firing properties were similar to those of neighboring GFP− hypothalamic neurons. Although TDNs fired action potentials to depolarizing current steps, the average current-frequency curve of TDNs was significantly different from the curve for control neurons in both young and adult mice (Fig. 8, A and B). Although the number of action potentials elicited initially increased with age, the TDNs were unable to reliably generate repetitive actions potentials with larger current steps in contrast to control neurons, leading to saturation of the current-frequency curves (Fig. 8B and fig. S9, C and D).
Fig. 8. TDNs show distinct membrane excitability and synaptic activity and respond to physiological stimuli.
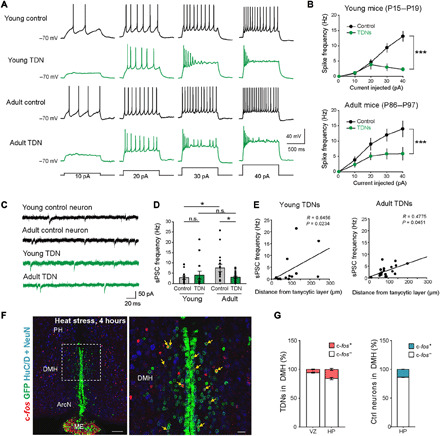
(A) Representative voltage traces recorded from control and TDNs from young and adult mice evoked by depolarizing current steps as indicated. (B) The current-spike frequency relationships measured from control and TDNs from young (top) and adult mice (bottom). The current-frequency relationship was significantly different between TDNs and control neurons at both ages [young control neurons, 14 cells from four mice; young TDNs, 13 cells from four mice; P < 0.0001, two-way analysis of variance (ANOVA); adult control neurons, 16 cells from six mice; adult TDNs, 18 cells from six mice; P = 0.0001, two-way ANOVA]. (C) Representative traces of spontaneous postsynaptic currents (sPSCs). (D) Summary graphs of sPSC frequency (young control neurons, 14 cells from four mice, 2.85 ± 0.74 Hz; young TDNs, 13 cells from four mice, 4.07 ± 1.93 Hz; P = 0.2983, Mann-Whitney U test; adult control neurons, 16 cells from six mice, 7.56 ± 1.82 Hz; adult TDNs, 18 cells from six mice, 2.98 ± 0.54 Hz; P = 0.0324, Mann-Whitney U test). (E) Positive correlation between the distance from the tanycytic layer for each TDN and its sPSC frequency in both young (left: 12 cells from four mice, P = 0.0234, Spearman’s Rho correlation) and adult (right: 18 cells from six mice, P = 0.0451, Spearman’s rho correlation) mice. (F) Four-hour heat stress (38°C) selectively induced the c-fos expression in TDNs in DMH (higher magnification inset shown in right). (G) Fraction of c-fos+ TDNs in VZ and HP of DMH and fraction of all c-fos+ and c-fos− neurons in DMH (n = 3 mice). PH, posterior hypothalamus. Scale bars, 100 μm (F) and 20 μm (F, inset). *P < 0.05 and ***P < 0.001.
These results suggest that TDNs may have a different ion channel composition from control neurons. This was confirmed by a direct comparison of expression levels of voltage-gated ion channels, which was performed using scRNA-seq data obtained from P45 TDNs and arcuate neuronal clusters found to match these TDNs through LIGER analysis (fig. S6). We observed that TDNs expressed lower levels of the pore-forming subunits of multiple voltage-gated potassium (Kcna2, Kcnc1, Kcnq2/3, and Kcnd2), sodium (Scn1a, Scn2b, Scn3a, and Scn8a), calcium (Cacna1b/e, Cacna2d1/2, Cacnb4, and Cacng4), and chloride channels (Clcn3), as well as the calcium-activated potassium channel Kcnma1 (table S15).
We next asked whether TDNs receive synaptic inputs from other neurons. We detected spontaneous postsynaptic currents (sPSCs) in 34 of 35 recorded TDNs (Fig. 8, C and D). However, the frequency of sPSCs in adult TDNs was significantly lower than for control neurons in adult mice (Fig. 8, C and D, and table S14), suggesting that the number of functional synapses received is nonetheless fewer than for wild-type neurons. As our histological data suggest that TDNs undergo progressive radial migration away from the tanycyte layer into the HP as they mature, we asked whether there was any correlation between a TDN’s distance from the tanycyte layer and the number of synaptic inputs it receives. We observed a positive correlation between a TDN’s distance from the tanycyte layer and the sPSC frequency in both young and adult mice, suggesting that the further a TDN migrates from the tanycytic layer, the more functional synapses from local neurons it receives (Fig. 8E).
Having established that TDNs show neuronal electrophysiological properties, fire action potentials, and receive synaptic inputs, we next tested whether TDNs are activated in response to changes in internal states that modulate the activity of nearby hypothalamic neurons. We first investigated the response of TDNs in the DMH to heat stress, which is known to robustly modulate the activity of DMH neurons (36, 37). At P45, we observed that 16.0 ± 1.6% of parenchymal TDNs in the DMH induce c-fos expression following 4 hours of exposure to 38°C ambient temperature in Nfia/b/x-deficient mice, which is essentially equivalent to the portion of GFP− control neurons activated, 13.7 ± 0.3% (Fig. 8, F and G).
DISCUSSION
This study demonstrates that tanycytes retain the ability to generate a broad range of different subtypes of hypothalamic neurons in the postnatal brain and that this latent ability is actively repressed by NFI family transcription factors. Induction of proliferative and neurogenic competence by selective loss of function of Nfia/b/x leads to the robust generation of hypothalamic neuronal precursors that undergo outward radial migration, mature, fire action potentials, and receive synaptic inputs. TDNs respond to dietary signals, such as leptin, and heat stress. This implies that tanycyte-derived neurogenesis can potentially modulate a broad range of hypothalamic-regulated physiological processes.
NFI factors have historically been primarily studied in the context of promoting astrocyte specification and differentiation (38, 39), and loss of function of Nfia/b/x disrupts the generation of tanycyte-derived astrocytes, ependymal cells, and oligodendrocyte progenitors and down-regulates the expression of tanycyte-enriched genes that are also expressed in astrocytes such as Kcnj10 and Aqp4 (Fig. 3). In addition to their role in promoting gliogenesis, however, recent studies have shown that NFI factors confer late-stage temporal identity on retinal progenitors (13), allowing the generation of late-born bipolar neurons and Müller glia and decreasing proliferative and neurogenic competence. Selective loss of function of Nfia/b/x in mature Müller glia likewise induces proliferation and generation of inner retinal neurons (14), although the levels seen are lower than those seen following the loss of function in retinal progenitors (13). As in the retina, Nfia/b/x genes are more strongly expressed in late-stage than early-stage mediobasal hypothalamic progenitors (15), and adult tanycytes show substantially lower levels of proliferation following the loss of function of Nfia/b/x than those seen in neonates (Figs. 1 and 2). However, the levels of proliferation and neurogenesis seen in neonatal tanycytes are much greater than those seen in Müller glia, which likely reflects the fact that mammalian Müller glia proliferate only rarely and essentially lack neurogenic competence (11), while tanycytes retain limited neurogenic competence (9). This implies that NFI factors may be part of a common gene regulatory network that represses proliferation and neurogenic competence in radial glia of the postnatal forebrain and retina.
Using scRNA-seq and scATAC-seq analysis, we identified multiple genes that are strong candidate extrinsic and intrinsic regulators of proliferative and neurogenic competence in tanycytes. We observe that loss of function of Nfia/b/x effectively regresses tanycytes to a progenitor-like state. Both Shh (40, 41) and Wnt (42, 43) signaling are required for progenitor proliferation and neurogenesis in embryonic tuberal hypothalamus, and our study strongly suggests that they play similar roles in Nfia/b/x-deficient tanycytes, although, in the absence of genetic analysis, off-target effects of the small-molecule ligands used for this analysis cannot be formally ruled out (44). Tanycyte-specific loss of function of Nfia/b/x both down-regulates Notch pathway components and up-regulates the Notch inhibitor Dlk1 while also down-regulating Tgfb2. Both Notch signaling and Tgfb2 promote quiescence and inhibit proliferation in retinal Müller glia and cortical astrocytes (45, 46) and likely play a similar role in tanycytes.
Nfia/b/x-deficient tanycytes likewise down-regulate transcription factors that are required for specification of astrocytes and Müller glia—including Sox8/9 (47, 48)—while up-regulating neurogenic basic helix-loop-helix factors such as Ascl1 and Sox4. Ascl1 is both required for differentiation of VMH neurons (49) and also sufficient to confer neurogenic competence on retinal Müller glia (50). NFI factors thus control the expression of a complex network of extrinsic and intrinsic factors that regulate neurogenic competence in tanycytes, and it may be possible to further stimulate tanycyte-derived neurogenesis by modulating select components of this network.
scRNA-seq analysis reveals that TDNs arise from Ascl1+ precursors and are heterogeneous, falling into several molecularly distinct clusters. The gene expression profiles of control and Nfia/b/x-deficient TDNs closely resemble one another, indicating that NFI factors are not obviously required for the differentiation of individual neuronal subtypes in contrast to their role in the retina and cerebellum (13, 51). However, the number of TDNs in control samples drops markedly after P8, with few detected at P45 (Fig. 3). We observe an age-dependent decline in the number of TDNs in controls, potentially in line with the high levels of cell death reported in postnatally generated hippocampal and olfactory bulb neurons (52). However, this decline is insufficient to account for this effect, and it may instead result from the well-established difficulties in obtaining viable, dissociated mature neurons following FACS for whole-cell scRNA-seq analysis (53).
TDNs are predominantly found in the DMH and ArcN, with much smaller numbers detected in the VMH and ME (Fig. 1H). They are mostly GABAergic and express molecular markers of DMH and ArcN neurons (Fig. 6, A and B), and substantial subsets closely match the scRNA-seq profiles of neuronal subtypes obtained from ArcN and VMH (fig. S6) (21). They include neuronal subtypes that regulate feeding and sleep and directly regulate pituitary function and many other subtypes whose function has yet to be characterized. In light of findings that tanycyte-derived neurogenesis can be stimulated by dietary and hormonal signals and potentially modulate body weight and activity levels (3, 7), this raises the possibility that different internal states may trigger generation and/or survival of functionally distinct tanycyte-derived neuronal subtypes, leading to long-term changes in both hypothalamic neural circuitry and physiological function.
We observe that TDNs survive for months and show neuronal electrophysiological properties, with subsets showing c-fos induction in response to heat stress (Figs. 7 and 8). Older TDNs receive more synaptic inputs than younger TDNs, and their input resistance lowers to become equivalent to that of GFP− nearby neurons, demonstrating progressive maturation. However, the frequencies of sPSCs and the evoked action potentials of TDNs remain consistently lower than those of the GFP− control neurons. This may be an intrinsic property of TDNs, distinguishing them with preexisting local neurons. Alternatively, the excess TDNs generated from Nfia/b/x-deficient tanycytes may form synaptic connections less efficiently. Distinguishing these possibilities will require electrophysiological recording of TDNs from control animals, although the far smaller population of these cells in wild-type animals make this experiment very challenging.
While levels of tanycyte-derived neurogenesis are low under baseline conditions, adding small numbers of newly generated neurons to existing hypothalamic neural circuits may have important effects on hypothalamic-regulated behaviors and physiology. Furthermore, selective loss of specific hypothalamic neuronal subtypes, as well as accompanying metabolic and behavior changes, is observed both in neurodegenerative disorders such as Alzheimer’s disease and frontotemporal dementia (54) and under conditions such as obesity and type 2 diabetes (55). Identifying new means of enhancing neurogenic competence in mature tanycytes, while selectively promoting the differentiation of specific tanycyte-derived neuronal subtypes, may thus ultimately prove useful in generating long-term changes in hypothalamic physiology in a variety of therapeutic contexts.
MATERIALS AND METHODS
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, S.B. (sblack@jhmi.edu).
Animals
Rax-CreERT2 mice (the Jackson laboratory, stock no. 025521) generated in the laboratory (18) were crossed with the Cre-inducible Sun1-GFP reporter (Sun1-sfGFP-Myc; the Jackson laboratory, stock no. 021039, provided by J. Nathans) (19). Nfialox/lox (13, 14), Nfiblox/lox (56), and Nfixlox/lox (57) mice were used to generate Nfia/b/x homozygous triple mutant mice previously described (13, 14). To generate tanycyte-specific loss-of-function mutants of Nfia/b/x genes, the triple mutant mice were crossed to Rax-CreERT2;Sun1-GFP mice. To induce the Cre recombination, these mice were intraperitoneally injected with 4-OHT (0.2 mg) dissolved in corn oil for three consecutive days from P3 to P5. Mice were housed on a 14- to 10-hour light/dark cycle (lights on at 07:00 and lights off at 21:00) in a climate-controlled pathogen-free facility. All experimental procedures were preapproved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
BrdU and EdU incorporation assay
To label the proliferating cells, BrdU (Sigma-Aldrich, #B5002) was dissolved in saline solution, and 100 mg/kg of body weight was intraperitoneally injected for five consecutive days for the dates indicated. For the AAV1-Cre-mCherry stereotaxic injection, an osmotic mini pump (ALZET model 1002, #0004317) was filled with BrdU dissolved in artificial cerebrospinal fluid (aCSF) (Tocris Bioscience, #3525) and installed immediately into the hole remaining after the virus injection needle was removed. The 2-cm-long tube connecting the mini pump and cannula was filled with aCSF, so that the actual infusion (30 μg/day) of BrdU was started from the third day following implantation. To avoid potential toxic effects of long-term BrdU exposure, we used EdU (Thermo Fisher Scientific, #A10044) for quantitative studies of cell proliferation. For this purpose, we used a dose of EdU (50 μg/g), which has been previously validated for proliferation studies in the adult brain (58).
Inhibitory drug administration
ABC99 (Sigma-Aldrich, #SML2410) was prepared as previously described (59), except for the fact that a stock solution (16.5 mg/ml) was used. This stock was sequentially mixed with Tween 80 (Sigma-Aldrich, #P1754), polyethylene glycol, molecular weight 400 (Merck, #91893), and 0.9% NaCl in the ratio of 1:1:1:17. P45 Nfia/b/x triple knockout male mice were intraperitoneally injected with ABC99 (10 mg/kg) for five consecutive days. EdU (50 mg/kg of body weight) was injected together from the third day of treatment to profile proliferation induced by Notum inhibition (29). For the cyclopamine stock solution (1 μg/μl), cyclopamine (1 mg; Tocris Bioscience, #1623) powder was dissolved into 2-hydroxylpropyl-β-cyclodextrin (1 ml; Sigma-Aldrich, #C0926) prepared as a 45% solution in phosphate-buffered saline (PBS). Cyclopamine (10 μg/g) was injected from P8 to P16 on alternative days. BrdU (100 μg/g of body weight) was injected together from the third injection for five consecutive days. Control mice were treated with the corresponding vehicle solution.
Tissue processing and immunohistochemistry
Mice were anesthetized with an intraperitoneal injection of tribromoethanol/Avertin, followed by transcardial perfusion with 2% paraformaldehyde (PFA) as previously described (60). Brains were dissected, postfixed in the same fixative, and prepared for the cryopreservation in optimal cutting temperature (OCT)–embedding compound. A series of 25-μm coronal sections were stored in antifreeze solution at −20°C until ready for immunostaining.
After brief washing with 1× PBS to remove the antifreeze solution, sections were mounted on Superfrost Plus slides (Thermo Fisher Scientific) before starting immunohistochemistry and dried at room temperature (RT) for 30 min. To ensure adequate fixation for nuclear staining, sections were immersed into the fixative solution for 10 min at this point. Antigen retrieval was performed by incubating the slides with the prewarmed sodium citrate buffer [10 mM sodium citrate (pH 6.0)] in an 80°C water bath for 30 min. For HuC/HuD (HuC/D) antibody staining, sections were also treated with 0.3% hydrogen peroxide to block endogenous peroxidase activity before the blocking step with 10% horse serum/0.1% Triton X-100 in 1× PBS for an hour. pSTAT3 staining required different pretreatments as described before (17, 60). To detect the overall expression of three different Nfi family members, mouse antibodies recognizing NFIA, NFIA/B, and NFIX were mixed and visualized with the same secondary antibody. After finishing the first round of immunostaining, the fluorescence signal was cross-linked by incubation in 2% PFA for 10 min, followed by either EdU staining using the Click-iT EdU detection kit conjugated with Alexa Fluor 647 (Thermo Fisher Scientific, #C10340) or BrdU antibody staining. For BrdU staining, freshly prepared 2 N HCl was spread on the slides and incubated at 37°C for 30 min on the humidified chamber. Borate buffer (0.1 M) (pH 8.5) was used for acid neutralization by incubating for 10 min at RT. Antibodies used were listed in table S1. After counterstaining with 4′,6-diamidino-2-phenylindole (DAPI), the slides were coverslipped with VECTASHIELD antifade mounting medium (Vector Laboratories, #H-1200) and dried at RT for no more than 30 min. The slides were stored at 4°C and imaged within 2 days to achieve the best quality using a Zeiss LSM 700 confocal microscope at the Microscope Facility at Johns Hopkins University School of Medicine.
RNAscope HiPlex assay
For the RNAscope HiPlex assay, P45 triple knockout and control male mice were euthanized by cervical dislocation, and brains were dissected out. The brains were immediately immersed in 4% PFA in diethyl pyrocarbonate–treated 1× PBS and incubated overnight at 4°C. All other sample preparation procedures were performed as recommended in the manufacturer’s instructions for OCT-embedded fresh-frozen tissue preparation. Fourteen-micrometer sections were cut on a cryostat and briefly washed with 1× PBS before mounting on Superfrost Plus slides (Thermo Fisher Scientific). The slides were dried at −20°C and stored at −80°C before use. The HiPlex assay was performed by following the manufacturer’s instructions using probes listed in table S12. The sections were imaged on a Zeiss LSM 800 confocal microscope at the Multiphoton Imaging Core in the Department of Neuroscience at Johns Hopkins University School of Medicine.
Heat stimulation and leptin injection
P45 male mice were exposed to ambient heat (38°C) for 4 hours (61) by incubating in a prewarmed light-controlled cabinet in the Rodent Metabolic Core Facility at the Center for Metabolism and Obesity Research of the Johns Hopkins University School of Medicine. During the procedure, mice were provided ad libitum access to water and food and carefully monitored. Transcardiac perfusion with 4% PFA in 1× PBS was performed immediately after heat exposure. The dissected brains were processed as described above and used for c-fos immunostaining. Leptin injection was performed on P90 male mice that were fasted for 18 hours before treatment. Leptin (3 mg/kg of body weight; PeproTech, #450-31) dissolved in saline solution was intraperitoneally injected, and, 45 min later, transcardial perfusion was performed using 2% PFA as described above.
Cell counting and statistical analysis
All cell counts were performed blindly and manually by five independent observers using Fiji/ImageJ software. Five sections corresponding to −1.55, −1.67, −1.79, −1.91, and −2.15 mm from bregma were chosen among the serial sections for cell counting. Initially, cell numbers were normalized by the size (in millimeters) of hypothalamic nuclei measured. Because Nfia/b/x-deficient animals did not show any obvious structural differences, in subsequent experiments, we used absolute numbers. All values are expressed as means ± SEM. Comparisons were analyzed by two-tailed Student’s t test using Microsoft Excel unless stated otherwise. A P < 0.05 was considered statistically significant.
Cell preparation for scRNA-seq
P8, P17, and P45 TKO mutant mice and control Rax-CreER;CAG-lsl-Sun1-GFP mice were euthanized by cervical dislocation, and brains were dissected. One biological replicate of each time point and genotype were analyzed, with the exception of P45 TKO mice, where two biological replicates were analyzed. Two-micrometer-thick coronal slices including the hypothalamic protruding ME were collected using adult mouse brain matrix (Kent Scientific). The mediobasal hypothalamic region was microdissected using a surgical scalpel, dampened in Hibernate-A medium supplemented with 0.5 mM GlutaMAX and 2% B-27 (HABG), and chopped with a razor blade. Brain tissues were transferred into preequilibrated papain/deoxyribonuclease I mix (Worthington Papain Dissociation System, #LK003150) and incubated for 30 min at 37°C with frequent agitation using a fire-polished glass pipette. Dissociated cells were filtered through a 40-μm strainer and subjected to density gradient centrifugation to remove the cell debris as suggested in the manufacturer’s protocol. Cells were resuspended in HABG medium, and GFP+ cells were FACS isolated in the Bloomberg Flow Cytometry and Immunology Core at Johns Hopkins University. Cells were resuspended with ice-cold PBS containing 0.04% bovine serum albumin and ribonuclease inhibitor (0.5 U/μl), and 10,000 to 15,000 cells were loaded on a 10x Genomics Chromium Single Cell system (10x Genomics, Redwood City, CA), using the v3 chemistry following the manufacturer’s instructions, and libraries were sequenced on an Illumina NextSeq with ~200 million reads per library. Sequencing results were processed through the Cell Ranger 3.1 pipeline (10x Genomics, Redwood City, CA) with default parameters.
Single-cell ATAC-seq
scATAC-seq was performed using the 10x Genomic scATAC reagent V1 kit following the manufacturer’s instructions. Briefly, FACS-sorted cells (~30,000 cells) were centrifuged at 300g for 5 min at 4°C. The cell pellet was resuspended in 100 μl of lysis buffer, mixed 10× by pipetting, and incubated on ice for 3 min. Wash buffer (1 ml) was added to the lysed cells, and cell nuclei were centrifuged at 500g for 5 min at 4°C. The nuclei pellet was resuspended in 250 μl of 1× nuclei buffer. Cell nuclei were then counted using trypan blue staining. Resuspended cell nuclei (10,000 to 15,000) were used for transposition and loaded into the 10x Genomics Chromium Single Cell system. Libraries were amplified with 10 polymerase chain reaction cycles and were sequenced on an Illumina NextSeq with ~200 million reads per library. Sequencing data were processed through the Cell Ranger ATAC 1.1.0 pipeline (10x Genomics) with default parameters.
scRNA-seq data preprocessing
Raw scRNA-seq data were processed with the Cell Ranger software (62) (version 3.1) for formatting reads, demultiplexing samples, genomic alignment, and generating the cell-by-gene count matrix. First, the “cellranger mkfastq” function was used to generate FASTQ files from BCL files. Second, the “cellranger count” function was used to process FASTQ files for each library using default parameters and the mm10 mouse reference index provided by 10x Genomics. Last, we obtained the cell-by-gene count matrix for each library and used this for all downstream analysis.
Using the Seurat R package (63), we created Seurat objects for each sample with the cell-by-gene count matrix using the function “CreateSeuratObject” (min.cells = 3, min.features = 200). After visual analysis of the violin plot of the total counts for each cell, we filtered out cells with nCount_RNA < 600 or nCount_RNA > 6000. Next, we calculated the fraction of mitochondrial genes for each cell and filtered out the cells with a mitochondrial fraction of >8%. Last, we predicted multiplet artifacts and removed potential doublet cells using Scrublet (64) for each sample. As a result, 6609 (P8 Ctrl), 6494 (P17 Ctrl), 2607 (P45 Ctrl), 12,930 (P8 TKO), 12,413 (P17 TKO), 7531 (P45 KO, replicate 1), and 5886 (P45 KO, replicate 2) cells were used for downstream analysis.
scRNA-seq data analysis
Dimensional reduction, clustering, and visualization
To integrate the cells from different ages and genotypes, we aligned all cells for each sample and obtained an integrated Seurat object using the Seurat function “FindIntegrationAnchors” and “IntegrateData” with the default parameters, respectively. Using the integrated data, we normalized, log-transformed, and scaled the count matrix using the function “NormalizeData” and “ScaleData.” We next selected the variable genes using the function “FindVariableFeatures”(selection.method = “mvp”) and performed dimension reduction analysis with “RunPCA.”
To annotate individual cell types in the integrated dataset, we first clustered all the cells using the function “FindNeighbors” and “FindClusters” with a resolution of 0.3 and 1st to 30th dimensions. Then, we matched the clusters to major cell types using expression of known cell marker genes. As a result, we identified the following nine major cell types: alpha1 tanycytes, alpha2 tanycytes, beta1 tanycytes, beta2 tanycytes, proliferating tanycytes, astrocytes, neurons, ependymal cells, and OPCs. To visualize the integrated data, we used the 1st to 30th dimensions to perform nonlinear dimension reduction and obtained UMAP coordinates with the function “RunUMAP.”
We further characterized molecularly distinct subtypes of TDNs in Fig. 5. First, we restricted our analysis to cells in the neuron cluster and from the following ages and genotypes: P8 Ctrl, P8 TKO, P17 TKO, and P45 TKO. P17 Ctrl and P45 Ctrl were excluded from analysis due to the very small numbers of TDNs present in these datasets. Second, we integrated all the neurons from different conditions using “RunHarmony” in the Harmony R package (65). Next, we used the 1st to 10th harmony dimensions to identify neuronal subclusters with a resolution of 0.5. Last, we aggregated the clusters into individual neuronal subtypes based on known neuron markers and RNA velocity results. To visualize TDNs, we used the 1st to 10th harmony dimensions to obtain UMAP coordinates with the function “RunUMAP.”
Identification of differentially expressed genes
To identify markers for each cell type and differentially expressed genes (DEGs) between Ctrl and TKO samples, we used the Seurat function “FindAllMarkers” and “FindMarkers” with the options: min.pct = 0.2 or 0.1 and logfc.threshold = 0.25. We then retained differential genes with an adjusted P < 0.001.
RNA velocity analysis
To characterize cellular differentiation trajectories associated with tanycyte-derived neurogenesis, we used scVelo software (66) to perform RNA velocity analysis by comparing levels of spliced and unspliced transcripts. Briefly, we converted BAM files for each sample to loom files using a command line tool (25). We then combined these loom files and retained cells that passed filtering in the previous step. Using scVelo, we normalized the spliced and unspliced matrix, filtered the genes, and selected the top 1500 variable genes with the function: “pp.normalize_per_cell,” “pp.filter_genes_dispersion,” and “pp.log1p.” Next, we performed a principal components analysis (PCA) and calculated the velocity vectors and velocity graph using the function: “pp.moments” (n_pcs = 35, n_neighbors = 50), “tl.recover_dynamics,” “tl.velocity” (mode = “dynamical”), and “tl.velocity_graph.” Last, we visualized velocities on the previously calculated UMAP coordinates with the “pl.velocity_embedding_grid” function. We applied the same pipeline to analyze the RNA velocity in differentiating TDNs.
Cell-cycle stage inference
The function “CellCycleScoring” in the Seurat package was used to calculate cell cycle phase scores (S score and G2-M score), with the G2-M and S phase marker genes obtained from Tirosh et al. (67).
Single-cell trajectory inference
Slingshot (68) was applied to infer differentiation trajectories from alpha2 tanycytes to neurons. To construct the trajectory, we included cells in the “alpha2 tanycytes,” “proliferating tanycytes,” and “neuron” clusters. We then then ran the Slingshot using the dimensionality reduction results (UMAP) identified previously. We set the alpha2 tanycytes cluster as the initial cluster to identify lineages with the function “getLineages” and “getCurves” with default parameters. Last, we assigned cells to the lineages and calculated pseudo-time values for each cell using the function “slingPseudotime.”
Monocle 2 (69) was applied to identify developmentally dynamic genes that are significantly altered along the trajectory. First, we converted the expression matrix to Monocle datasets with the function “newCellDataSet,” then processed and normalized the Monocle datasets following the Monocle recommended pipeline, and lastly identified the DEGs using the “differentialGeneTest” function with the following criteria: q < 1 ×10−10 and expressed cell number of >200.
Comparison between TDNs and mature hypothalamic neurons
To further explore the biological similarity between TDNs and the broader population of neurons in mouse hypothalamus, we first used the scRNA-seq datasets for mature neurons in hypothalamic ArcN provided by Campbell et al. (21) and downloaded the cell-by-gene matrix and the annotation file of the mature neuronal cell types from the Gene Expression Omnibus (GEO) database under the accession no. GSE93374. The LIGER (32) package was used to integrate our tanycyte-derived cells identified in the previous rounds of analysis with these mature hypothalamic neurons, using the default pipeline recommended in the LIGER guidelines (https://github.com/welch-lab/liger). After LIGER integration, we reclustered the integrated datasets and calculated new UMAP coordinates using the functions “FindNeighbors,” “FindClusters,” and “RunUMAP” with the following parameters: reduction = “iNMF” and dims = 1:30. Last, we made an alluvial plot to visualize the alignments between the tanycyte-derived neuronal subtypes (six subtypes), LIGER clusters (14 clusters), and the mature arcuate neuronal cell types (34 subtypes).
To compare the gene expression between TDNs and mature neurons, we first reclustered the integrated datasets with a higher resolution (resulting in 22 clusters). To remove the unique cell clusters between TDNs and mature neurons, we only kept the 12 LIGER clusters (C2 to C5, C9, C11, C13, C16 to C18, C20, and C21), which contained both TDNs and mature neurons for downstream comparison. Second, we performed quantile normalization for each cell on the combined and normalized cell-by-gene matrix to reduce the batch effects between TDNs and mature neurons. Last, we used a Wilcoxon rank sum test to identify the differential genes with the function “FindMarkers.”
scATAC-seq data preprocessing
We processed sequencing output data using the Cell Ranger ATAC software (v.1.0) for alignment, deduplication, and identification of transposase cut sites. First, the “cellranger-atac mkfastq” function was used for generating FASTQ files from BCL files. Second, the “cellranger-atac count” function was used to process the FASTQ files for each library using default parameters and the mouse mm10 reference index provided by 10x Genomics (refdata-cellranger-atac-GRCh38-1.2.0). Last, we obtained the barcoded, aligned, and Tn5-corrected fragment file (fragments.tsv.gz) for each library and used these for downstream analysis.
scATAC-seq data analysis
Generating union peaks
We generated the cell-by-peak matrix for each sample using the same method as described in Satpathy et al. (70). First, we constructed 2.5-kb tiled windows across the mm10 genome using the local script. Next, a cell-by-window sparse matrix was computed by counting the Tn5 insertion overlaps for each cell, and this matrix was then binarized and inputted to Signac package (0.2.5) to create a Seurat object using “CreateSeuratObject.” Second, we normalized the cell-by-window matrix by TF-IDF (term frequency–inverse document frequency) methods using “RunTFIDF” and ran a singular value decomposition (SVD) on the TF-IDF normalized matrix with “RunSVD.” We retained the 2nd to 30th dimensions and identified clusters using SNN (shared nearest neighbor) graph clustering with “FindClusters” with a resolution of 0.3. Third, to identify high-quality peaks for each cluster in each sample, we called peaks for each cluster using MACS2 (71) with the command: “-shift -75 --extsize 150 --nomodel --callsummits --nolambda --keep-dup all -q 0.05.” The peak summits were then extended to 250 base pairs (bp) on either side to a final width of 500 bp and then filtered by the mm10 v2 blacklist regions (https://github.com/Boyle-Lab/Blacklist/blob/master/lists/mm10-blacklist.v2.bed.gz). The top 50,000 peaks for each cluster in each sample were kept, converted to Granges, and merged into a union peak set with the function “reduce” in the GenomicRanges package. Last, we obtained 107,377 union peaks and generated a cell-by-peak sparse matrix for all these cells for downstream analysis.
Filter cells by transcription start site enrichment, unique fragments, and nucleosome banding
We calculated the transcription start site (TSS) enrichment, unique fragments, and nucleosome banding for each cell using the Signac package. The cell-by-peak sparse matrices were inputted to the “CreateSeuratObject” function to create a Seurat object with default parameters. Then, we filtered the cells using the following criteria: (i) the number of unique nuclear fragments of >1000, (ii) TSS enrichment score of >2, (iii) nucleosome banding score of <4, and (iv) blacklist_ratio of < 0.05. As a result, 8948 (P8 Ctrl) and 13337 (P8 TKO) cells were identified and used for downstream analysis.
Dimensional reduction, clustering, and visualization
The Harmony package was applied to integrate the scATAC-seq data from control and Nfia/b/x TKO samples. First, we put the Seurat object created in the previous step into the Signac process pipeline. We normalized and obtained a low-dimensional representation of the cell-by-peak matrix using the functions “FindTopFeatures,” “RunTFIDF,” and “RunSVD.” Next, we integrated all the cells from both genotypes (control and TKO) using the “RunHarmony” function with the options: dim.use = 2:50, group.by.vars = “condition,” reduction = “lsi,” and project.dim = FALSE. Third, we used the 2nd to 30th harmony dimensions to identify clusters with a resolution of 0.8 and used the same harmony dimensions to calculate the UMAP coordinates for visualization.
To annotate the cell types for each cluster, we used the integration method in the Seurat package to transfer the previously annotated cell type labels from scRNA-seq data to scATAC-seq data. First, we estimated RNA-seq levels using the function “CreateGeneActivityMatrix” from the scATAC-seq data using the mm10 genome build GTF file. Next, we found anchors between the scATAC-seq datasets (P8 Ctrl and P8 TKO) and the corresponding scRNA-seq datasets (P8 Ctrl and P8 TKO) using the function “transfer.anchors.” Then, with the “TransferData” function, we obtained a matrix of cell type predictions and prediction scores for each cell in the scATAC-seq dataset. We further filtered the cells with max(prediction score) of <0.5. Last, for each cluster, we calculated the number of cells for each predicted cell type and set its final annotation based on the cell type that was most highly represented in the cluster. Using this approach, we identified the following nine major cell types: alpha1 tanycytes, alpha2 tanycytes, beta1 tanycytes, beta2 tanycytes, proliferating tanycytes, astrocytes, neurons, ependymal cells, and OPCs.
Global NFI activity and footprint analysis
We inferred global NFI activity with the chromVAR R package (72). The raw cell-by-peak matrix from the total cells was used as input to chromVAR. The mm10 reference genome was used to correct GC bias. We used the mouse NFI motifs (including Nfia, Nfib, and Nfix) in the TransFac2018 database to generate the transcription factor z-score matrix. The z-score for each cell was used to visualize the global NFI activity using the previously calculated UMAP coordinates.
To analyze the NFI footprint in alpha2 tanycytes, we used the same methods described in Corces et al. (73). First, we used the NFI motifs and all accessible regions to predict the NFI binding sites with the function “match motifs” in the motif matching R package. Second, we calculated the Tn5 insertion bias around every NFI binding site. We generated the aggregated observed 6-bp hexamer table relative to the ±250 bp region from all motif centers and also calculated the aggregated expected 6-bp hexamer table from the mm10 genome. We then obtained the observed/expected (O/E) 6-bp hexamer table by dividing these two hexamer tables. Last, we calculated the observed Tn5 insertion signal at ±250 bp from the motif center and normalized the signal using the O/E 6-bp hexamer table to obtain the final Tn5 bias-corrected signal.
Differential peak analysis
To explore which ATAC regions are changed following Nfia/b/x loss of function, we applied the MAnorm algorithm (74) to perform differential peak analysis between control and Nfia/b/x TKO alpha2 tanycytes. First, we selected cells in the alpha2 tanycytes cluster and then separated these cells by genotype (control and Nfia/b/x TKO). Second, we aggregated the cells of the same genotype by summing the count signals for each peak, then created a new condition-by-peak count matrix, and put it into the MAnorm pipeline. Last, we performed the MAnorm test and identified differential peaks with the cutoff LOG_P > 25 and abs(M_value_rescaled) > 0.5.
De novo motif enrichment analysis
HOMER software (75) was applied to identify motifs enriched in the differential ATAC regions between control and Nfia/b/x TKO alpha2 tanycytes. We analyzed the up-regulated peaks and down-regulated peaks separately using the Homer function “findMotifsGenome.pl” with the default options, except the following: mm10, -size given, -mask.
Identification of genes directly regulated by Nfia/b/x
To further explore the biological function of NFI factors in alpha2 tanycytes, we developed a method to infer potential Nfia/b/x targets with the information in scRNA-seq and scATAC-seq data. Our methods included the following three steps:
-
1)
Identification of Nfia/b/x-binding regions. In the previous motif enrichment analysis, we found that NFI motifs are enriched in the down-regulated peaks (Nfia/b/x TKO/control), so, in the first step, we aimed to identify which down-regulated peaks are bound by NFI factors. Using the NFI motif information in the TransFac2018 database, we first scanned the NFI motifs in the down-regulated peaks with the function “matchMotifs.” Next, with the Tn5 insertion signal in P8 Ctrl alpha2 tanycytes, we calculated the footprint occupancy score (FOS) (76) for each predicted NFI binding region and filtered out the regions with an FOS of <2. Last, we kept only the NFI binding peaks that contain NFI binding sites and used them for downstream analysis.
-
2)
Identification of promoters associated with Nfia/b/x binding regions. To identify genes that are potentially regulated by these NFI binding regions, we used the Cicero algorithm (77) to identify all the distal elements–promoter connections genome wide. First, we converted the cell-by-peak sparse binary matrix into the Cicero pipeline with the functions “make_atac_cds,” “detectedGenes,” and “estimatedSizeFactors.” Next, we created low overlapping cell groups based on the kNN (k-nearest neighbors) in the UMAP dimension and aggregated signals for each cell group with the function “make_cicero_cds.” We then calculated the correlation between each peak-peak pair using the function “run_cicero” with default parameters. Third, we annotated the peak pairs using “annotate_cds_by_site” with mm10 GTF files. We kept the peak pairs with the following criteria: (i) One of the peaks overlapped with ±2 kb of TSS region, and (ii) one of the peaks contained at least one NFI binding motif. Last, we identified NFI-related distal elements–promoter connections from the peak pairs if their coaccessibility score is >0.03 or <−0.03 and their distance is <150 kb.
-
3)
Inference of potential Nfia/b/x targets by integrating with scRNA-seq data. In this step, we aimed to integrate the NFI-related distal elements–promoter connections and differential genes following loss of function of Nfia/b/x to identify NFI target genes. First, we selected enhancer-promoter pairs from the distal elements–promoter connections in step 2 with coaccessibility scores of >0.03. If the gene associated with the promoter in question was down-regulated following the loss of function of Nfia/b/x, then we treated these genes as potential Nfia/b/x targets.
Conversely, we selected silencer-promoter pairs with coaccessibility scores of <−0.03. If the promoter genes were up-regulated following the loss of function of Nfia/b/x, then we also treated these genes as potential Nfia/b/x targets. Using this approach, we identified 62 NFI target genes.
GO term analysis
To understand the biological functions associated with genes dynamically expressed during the process of alpha2 tanycyte–derived neurogenesis, we applied GOrilla algorithm (78) to identify enriched GO terms for each gene cluster using the default parameters (P = 0.001, ontology = “Process”). The output of GO terms from GOrilla was further processed by REVIGO (79) to remove redundant terms. This pipeline was also used to identify the GO term enrichment in NFI-regulated gene sets.
Brain slice preparation and cell type identification
To investigate the electrophysiological characteristics of tanycyte-derived and other hypothalamic neurons, the acute brain slices were generated as previously described (35). TKO mice (P15 to P97, male) were anesthetized with isoflurane and decapitated, and the brains were rapidly removed and chilled in ice-cold sucrose solution containing 76 mM NaCl, 25 mM NaHCO3, 25 mM glucose, 75 mM sucrose, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, and 7 mM MgSO4 (pH 7.3). Acute brain slices (300 μm) including the hypothalamus were prepared using a vibratome (VT 1200 s, Leica) and transferred to warm (32° to 35°C) sucrose solution for 30 min for recovery. The slices were transferred to warm (32° to 34°C) aCSF composed of 125 mM NaCl, 26 mM NaHCO3, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgSO4, 20 mM glucose, 2 mM CaCl2, 0.4 mM ascorbic acid, 2 mM pyruvic acid, and 4 mM l-(+)-lactic acid (pH 7.3, 315 mOsm) and allowed to cool to RT. All solutions were continuously bubbled with 95% O2/5% CO2. For whole-cell patch-clamp recordings, slices were transferred to a submersion chamber on an upright microscope [Zeiss Axio Examiner, objective lens: 5×, 0.16 numerical aperture (NA) and 40×, 1.0 NA] fitted for infrared differential interference contrast and fluorescence microscopy. Slices were continuously superfused (2 to 4 ml/min) with warm, oxygenated aCSF (32° to 34°C). Hypothalamic areas and cells were identified under a digital camera (Sensicam QE, Cooke) using either transmitted light or green fluorescence. Tanycytes were identified as GFP+ cells located in the ependymal cell layer at the third ventricle. Tanycyte-derived cells were identified as GFP+ cells located in the HP but not in the ependymal cell layer. GFP− hypothalamic neurons in the HP, among which were intermingled the sparse tanycyte-derived cells, were targeted as control neurons.
Whole-cell recordings and analysis
Borosilicate glass pipettes (2 to 4 MΩ) were filled with an internal solution containing 2.7 mM KCl, 120 mM KMeSO4, 9 mM Hepes, 0.18 mM EGTA, 4 mM Mg–adenosine 5′-triphosphate, 0.3 mM Na–guanosine 5′-triphosphate, and 20 mM phosphocreatine(Na) (pH 7.3, 295 mOsm). Biocytin (0.25%, w/v) was added to the internal solution for post hoc morphological characterization. Whole-cell patch-clamp recordings were conducted through a MultiClamp 700B amplifier (Molecular Devices) and an ITC-18 (InstruTECH), which were controlled by customized routines written in Igor Pro (WaveMetrics). The series resistance averaged 14.2 ± 5.8 MΩ SD (n = 81 cells, 12 mice, all <36 MΩ, no significant difference between cell types or age groups; P > 0.05, Mann-Whitney U test) and was not compensated. The input resistance was determined by measuring the voltage change in response to a 1-s-long −100-pA hyperpolarizing current step. The current-spike frequency relationship was measured with a series of depolarizing current steps (1-s long, 0 to 50 pA, 10-pA increments, and 5-s interstimulus intervals). Cells were held at −70 mV, and the current steps were applied from −70 mV for the current-spike frequency relationship test. For each current intensity, the total number of action potentials exceeding 0 mV generated during each step was measured and then averaged across the three trials. sPSCs were measured in voltage-clamp mode at −70 mV. sPSCs were recorded for 25 s (250-ms-long current traces, 100 times), and ~110 events, on average, were recorded per cell. High-amplitude, high-frequency depolarizing current steps (10 nA at 100 Hz for 100 ms) were injected into the recorded cells at the end of recording to increase efficiency of biocytin infusion (80). All signals were low-pass filtered at 10 kHz and sampled at 20 kHz for voltage traces and 100 kHz for series resistance and sPSC measurements.
Electrophysiology data analysis and statistical testing
Data analysis was performed in Igor Pro (WaveMetrics), Excel (Microsoft), ImageJ (National Institutes of Health), and Minhee analysis (https://github.com/parkgilbong/Minhee_Analysis_Pack). Data are presented as the means ± SEM unless otherwise noted. A Mann-Whitney U test was used to compare membrane properties and sPSC frequencies between cell types and between age groups. Spearman’s rho test was used to determine the correlation between sPSC frequency and cell location. The location of the cells (distance to tanycytic layer) was measured from low (5×, 0.16 NA) and high (40×, 1.0 NA) magnification images of the recorded cells using ImageJ. The statistical difference in current-spike frequency relationships was tested using a two-way analysis of variance (ANOVA) test with Bonferroni correction. The sPSC events were automatically detected by Minhee analysis software with a 10-pA amplitude threshold. In the figures, the statistical significance is expressed as follows: *P < 0.05, **P < 0.01, or ***P < 0.001.
Visualization of biocytin-filled cells
Following the electrophysiological experiments, slices were fixed in 4% PFA in 0.01 M PBS at least overnight. After rinsing with PBS, slices were incubated in 0.01 M PBS blocking solution containing 2% Triton X-100 (Sigma-Aldrich) and 5% normal donkey serum (NDS) for 1 hour at RT. To visualize biocytin-filled cells, slices were next incubated with a blocking solution containing 1% Triton X-100, 5% NDS, chicken anti-GFP antibody (1:1000; Aves, catalog no. GFP-1020), and Alexa Fluor 647–conjugated streptavidin overnight on shaker at 4°C. The following day, slices were rinsed with 0.01 M PBS and incubated with Alexa Fluor 488–conjugated donkey–anti-chicken (1:500; Jackson ImmunoResearch, catalog no. 705-745-155) for 2 hours at RT. After rinsing, slices were mounted with Aqua-Poly/Mount (Polysciences, 18606-20). A subset of slices was costained with mouse anti-NeuN (1:300; Millipore, MAB377) and Alexa Fluor 568–conjugated donkey anti-mouse (1:300; Thermo Fisher Scientific, A10037) antibodies to confirm neuronal identity of the biocytin-filled cells. Fluorescence images were taken with a confocal microscope (LSM 800, Zeiss; 20× objective lens) as z-stack (2-μm interval) tiled images to cover the extent of the cell’s dendritic and axonal processes.
Acknowledgments
We thank R. Gronostajski for providing Nfia/b/x conditional mutant mice. We thank J. Ling, A. Kolodkin, J. Nathans, D. Lee, J, Bedont, M. Placzek, and W. Yap for helpful comments on the manuscript. We thank the Johns Hopkins Multiphoton Imaging Core for assistance with microscopy and the St. Jude Cloud facility for hosting the scRNA-seq and scATAC-seq data. Funding: This work was supported by grant DK108230 to S.B., a Maryland Stem Cell Postdoctoral Research Fellowship to S.Y., and Imaging Core Grant P30NS050274. Some aspects of this work are covered by a provisional U.S. patent 36406l0015P1. Author contributions: S.Y. and S.B. conceived the study. S.Y., J.K., P.Ly., T.V.H., V.T., L.J., and L.D. generated the data. S.Y., J.K., P.Ly., T.V.H., A.M., V.T., W.D., P.Le., and L.D. analyzed the data. S.B., S.P.B., J.Q., J.-K.W., and S.-H.P. supervised the research. S.Y., J.K., P.Ly., J.Q., S.P.B., and S.B. drafted the manuscript. All authors revised and edited the manuscript. Competing interests: S.Y. and S.B. are inventors on a patent application related to this work, filed by the Johns Hopkins University (no. 36406l0015P1, filed 2 November 2020). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All scRNA-seq and scATAC-seq data presented here are accessible at GEO (GSE160378) and can be queried interactively at https://viz.stjude.cloud/stjude/visualization/tanycyte. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabg3777/DC1
REFERENCES AND NOTES
- 1.Rodríguez E. M., Blázquez J. L., Pastor F. E., Peláez B., Peña P., Peruzzo B., Amat P., Hypothalamic tanycytes: A key component of brain-endocrine interaction. Int. Rev. Cytol. 247, 89–164 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Bolborea M., Dale N., Hypothalamic tanycytes: Potential roles in the control of feeding and energy balance. Trends Neurosci. 36, 91–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee D. A., Bedont J. L., Pak T., Wang H., Song J., Miranda-Angulo A., Takiar V., Charubhumi V., Balordi F., Takebayashi H., Aja S., Ford E., Fishell G., Blackshaw S., Tanycytes of the hypothalamic median eminence form a diet-responsive neurogenic niche. Nat. Neurosci. 15, 700–702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robins S. C., Stewart I., McNay D. E., Taylor V., Giachino C., Goetz M., Ninkovic J., Briancon N., Maratos-Flier E., Flier J. S., Kokoeva M. V., Placzek M., α-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nat. Commun. 4, 2049 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Haan N., Goodman T., Najdi-Samiei A., Stratford C. M., Rice R., El Agha E., Bellusci S., Hajihosseini M. K., Fgf10-expressing tanycytes add new neurons to the appetite/energy-balance regulating centers of the postnatal and adult hypothalamus. J. Neurosci. 33, 6170–6180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman T., Nayar S. G., Clare S., Mikolajczak M., Rice R., Mansour S., Bellusci S., Hajihosseini M. K., Fibroblast growth factor 10 is a negative regulator of postnatal neurogenesis in the mouse hypothalamus. Development 147, dev180950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D. A., Yoo S., Pak T., Salvatierra J., Velarde E., Aja S., Blackshaw S., Dietary and sex-specific factors regulate hypothalamic neurogenesis in young adult mice. Front. Neurosci. 8, 157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Migaud M., Butrille L., Batailler M., Seasonal regulation of structural plasticity and neurogenesis in the adult mammalian brain: Focus on the sheep hypothalamus. Front. Neuroendocrinol. 37, 146–157 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Yoo S., Blackshaw S., Regulation and function of neurogenesis in the adult mammalian hypothalamus. Prog. Neurobiol. 170, 53–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salvatierra J., Lee D. A., Zibetti C., Duran-Moreno M., Yoo S., Newman E. A., Wang H., Bedont J. L., de Melo J., Miranda-Angulo A. L., Gil-Perotin S., Garcia-Verdugo J. M., Blackshaw S., The LIM homeodomain factor Lhx2 is required for hypothalamic tanycyte specification and differentiation. J. Neurosci. 34, 16809–16820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan J., Goldman D., Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 40, 41–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilken M. S., Reh T. A., Retinal regeneration in birds and mice. Curr. Opin. Genet. Dev. 40, 57–64 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Clark B. S., Stein-O’Brien G. L., Shiau F., Cannon G. H., Davis-Marcisak E., Sherman T., Santiago C. P., Hoang T. V., Rajaii F., James-Esposito R. E., Gronostajski R. M., Fertig E. J., Goff L. A., Blackshaw S., Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 102, 1111–1126.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang T., Wang J., Boyd P., Wang F., Santiago C., Jiang L., Yoo S., Lahne M., Todd L. J., Jia M., Saez C., Keuthan C., Palazzo I., Squires N., Campbell W. A., Rajaii F., Parayil T., Trinh V., Kim D. W., Wang G., Campbell L. J., Ash J., Fischer A. J., Hyde D. R., Qian J., Blackshaw S., Gene regulatory networks controlling vertebrate retinal regeneration. Science 370, eabb8598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D. W., Washington P. W., Wang Z. Q., Lin S. H., Sun C., Ismail B. T., Wang H., Jiang L., Blackshaw S., The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat. Commun. 11, 4360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanov R. A., Tretiakov E. O., Kastriti M. E., Zupancic M., Häring M., Korchynska S., Popadin K., Benevento M., Rebernik P., Lallemend F., Nishimori K., Clotman F., Andrews W. D., Parnavelas J. G., Farlik M., Bock C., Adameyko I., Hökfelt T., Keimpema E., Harkany T., Molecular design of hypothalamus development. Nature 582, 246–252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo S., Cha D., Kim D. W., Hoang T. V., Blackshaw S., Tanycyte-independent control of hypothalamic leptin signaling. Front. Neurosci. 13, 240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pak T., Yoo S., Miranda-Angulo A. L., Wang H., Blackshaw S., Rax-CreERT2 knock-in mice: A tool for selective and conditional gene deletion in progenitor cells and radial glia of the retina and hypothalamus. PLOS ONE 9, e90381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo A., Mukamel E. A., Davis F. P., Luo C., Henry G. L., Picard S., Urich M. A., Nery J. R., Sejnowski T. J., Lister R., Eddy S. R., Ecker J. R., Nathans J., Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langlet F., Tanycyte gene expression dynamics in the regulation of energy homeostasis. Front. Endocrinol. 10, 286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell J. N., Macosko E. Z., Fenselau H., Pers T. H., Lyubetskaya A., Tenen D., Goldman M., Verstegen A. M. J., Resch J. M., McCarroll S. A., Rosen E. D., Lowell B. B., Tsai L. T., A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matuzelski E., Bunt J., Harkins D., Lim J. W. C., Gronostajski R. M., Richards L. J., Harris L., Piper M., Transcriptional regulation of Nfix by NFIB drives astrocytic maturation within the developing spinal cord. Dev. Biol. 432, 286–297 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Glasgow S. M., Zhu W., Stolt C. C., Huang T.-W., Chen F., LoTurco J. J., Neul J. L., Wegner M., Mohila C., Deneen B., Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat. Neurosci. 17, 1322–1329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaminskas B., Goodman T., Hagan A., Bellusci S., Ornitz D. M., Hajihosseini M. K., Characterisation of endogenous players in fibroblast growth factor-regulated functions of hypothalamic tanycytes and energy-balance nuclei. J. Neuroendocrinol. 31, e12750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A., Fan J., Borm L. E., Liu Z., van Bruggen D., Guo J., He X., Barker R., Sundström E., Castelo-Branco G., Cramer P., Adameyko I., Linnarsson S., Kharchenko P. V., RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laug D., Huang T.-W., Huerta N. A. B., Huang A. Y.-S., Sardar D., Ortiz-Guzman J., Carlson J. C., Arenkiel B. R., Kuo C. T., Mohila C. A., Glasgow S. M., Lee H. K., Deneen B., Nuclear factor I-A regulates diverse reactive astrocyte responses after CNS injury. J. Clin. Invest. 129, 4408–4418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piper M., Barry G., Hawkins J., Mason S., Lindwall C., Little E., Sarkar A., Smith A. G., Moldrich R. X., Boyle G. M., Tole S., Gronostajski R. M., Bailey T. L., Richards L. J., NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J. Neurosci. 30, 9127–9139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P. Jr., Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293, 1663–1666 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Mizrak D., Bayin N. S., Yuan J., Liu Z., Suciu R. M., Niphakis M. J., Ngo N., Lum K. M., Cravatt B. F., Joyner A. L., Sims P. A., Single-cell profiling and SCOPE-seq reveal lineage dynamics of adult ventricular-subventricular zone neurogenesis and NOTUM as a key regulator. Cell Rep. 31, 107805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suciu R. M., Cognetta A. B. III, Potter Z. E., Cravatt B. F., Selective irreversible inhibitors of the Wnt-deacylating enzyme NOTUM developed by activity-based protein profiling. ACS Med. Chem. Lett. 9, 563–568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Chou X.-l., Zhang L. I., Tao H. W., Zona incerta: An integrative node for global behavioral modulation. Trends Neurosci. 43, 82–87 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch J. D., Kozareva V., Ferreira A., Vanderburg C., Martin C., Macosko E. Z., Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Gao C., Sodicoff J., Kozareva V., Macosko E. Z., Welch J. D., Jointly defining cell types from multiple single-cell datasets using LIGER. Nat. Protoc. 15, 3632–3662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hübschle T., Thom E., Watson A., Roth J., Klaus S., Meyerhof W., Leptin-induced nuclear translocation of STAT3 immunoreactivity in hypothalamic nuclei involved in body weight regulation. J. Neurosci. 21, 2413–2424 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K., Kim J., Kim D. W., Zhang Y. S., Bao H., Denaxa M., Lim S.-A., Kim E., Liu C., Wickersham I. R., Pachnis V., Hattar S., Song J., Brown S. P., Blackshaw S., Lhx6-positive GABA-releasing neurons of the zona incerta promote sleep. Nature 548, 582–587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DiMicco J. A., Zaretsky D. V., The dorsomedial hypothalamus: A new player in thermoregulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R47–R63 (2007). [DOI] [PubMed] [Google Scholar]
- 37.DiMicco J. A., Samuels B. C., Zaretskaia M. V., Zaretsky D. V., The dorsomedial hypothalamus and the response to stress: Part renaissance, part revolution. Pharmacol. Biochem. Behav. 71, 469–480 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Tchieu J., Calder E. L., Guttikonda S. R., Gutzwiller E. M., Aromolaran K. A., Steinbeck J. A., Goldstein P. A., Studer L., NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat. Biotechnol. 37, 267–275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deneen B., Ho R., Lukaszewicz A., Hochstim C. J., Gronostajski R. M., Anderson D. J., The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52, 953–968 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Shimogori T., Lee D. A., Miranda-Angulo A., Yang Y., Wang H., Jiang L., Yoshida A. C., Kataoka A., Mashiko H., Avetisyan M., Qi L., Qian J., Blackshaw S., A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman T. S., Bergendahl S. E., Epstein D. J., Distinct temporal requirements for Sonic hedgehog signaling in development of the tuberal hypothalamus. Development 145, dev167379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman E. A., Wu D., Taketo M. M., Zhang J., Blackshaw S., Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus. Dev. Biol. 442, 236–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. E., Wu S.-F., Goering L. M., Dorsky R. I., Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development 133, 4451–4461 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Meyers-Needham M., Lewis J. A., Gencer S., Sentelle R. D., Saddoughi S. A., Clarke C. J., Hannun Y. A., Norell H., da Palma T. M., Nishimura M., Kraveka J. M., Khavandgar Z., Murshed M., Cevik M. O., Ogretmen B., Off-target function of the Sonic hedgehog inhibitor cyclopamine in mediating apoptosis via nitric oxide–dependent neutral sphingomyelinase 2/ceramide induction. Mol. Cancer Ther. 11, 1092–1102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jadhav A. P., Cho S.-H., Cepko C. L., Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc. Natl. Acad. Sci. U.S.A. 103, 18998–19003 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Todd L., Palazzo I., Squires N., Mendonca N., Fischer A. J., BMP- and TGFβ-signaling regulate the formation of Müller glia-derived progenitor cells in the avian retina. Glia 65, 1640–1655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poché R. A., Furuta Y., Chaboissier M.-C., Schedl A., Behringer R. R., Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J. Comp. Neurol. 510, 237–250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stolt C. C., Lommes P., Sock E., Chaboissier M.-C., Schedl A., Wegner M., The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 17, 1677–1689 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslanpour S., Rosin J. M., Balakrishnan A., Klenin N., Blot F., Gradwohl G., Schuurmans C., Kurrasch D. M., Ascl1 is required to specify a subset of ventromedial hypothalamic neurons. Development 147, dev180067 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Jorstad N. L., Wilken M. S., Grimes W. N., Wohl S. G., VandenBosch L. S., Yoshimatsu T., Wong R. O., Rieke F., Reh T. A., Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature 548, 103–107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piper M., Harris L., Barry G., Heng Y. H. E., Plachez C., Gronostajski R. M., Richards L. J., Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J. Comp. Neurol. 519, 3532–3548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu J. R., Hong C. J., Kim J. Y., Kim E.-K., Sun W., Yu S.-W., Control of adult neurogenesis by programmed cell death in the mammalian brain. Mol. Brain 9, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lafzi A., Moutinho C., Picelli S., Heyn H., Tutorial: Guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 13, 2742–2757 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Vercruysse P., Vieau D., Blum D., Petersén Å., Dupuis L., Hypothalamic alterations in neurodegenerative diseases and their relation to abnormal energy metabolism. Front. Mol. Neurosci. 11, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thaler J. P., Yi C.-X., Schur E. A., Guyenet S. J., Hwang B. H., Dietrich M. O., Zhao X., Sarruf D. A., Izgur V., Maravilla K. R., Nguyen H. T., Fischer J. D., Matsen M. E., Wisse B. E., Morton G. J., Horvath T. L., Baskin D. G., Tschöp M. H., Schwartz M. W., Obesity is associated with hypothalamic injury in rodents and humans. J. Clin. Invest. 122, 153–162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu Y.-C., Osinski J., Campbell C. E., Litwack E. D., Wang D., Liu S., Bachurski C. J., Gronostajski R. M., Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 354, 242–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell C. E., Piper M., Plachez C., Yeh Y.-T., Baizer J. S., Osinski J. M., Litwack E. D., Richards L. J., Gronostajski R. M., The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 8, 52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng C., Pan F., Jones L. A., Lim M. M., Griffin E. A., Sheline Y. I., Mintun M. A., Holtzman D. M., Mach R. H., Evaluation of 5-ethynyl-2′-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res. 1319, 21–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pentinmikko N., Iqbal S., Mana M., Andersson S., Cognetta A. B. III, Suciu R. M., Roper J., Luopajärvi K., Markelin E., Gopalakrishnan S., Smolander O.-P., Naranjo S., Saarinen T., Juuti A., Pietiläinen K., Auvinen P., Ristimäki A., Gupta N., Tammela T., Jacks T., Sabatini D. M., Cravatt B. F., Yilmaz Ö. H., Katajisto P., Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 571, 398–402 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoo S., Cha D., Kim S., Jiang L., Cooke P., Adebesin M., Wolfe A., Riddle R., Aja S., Blackshaw S., Tanycyte ablation in the arcuate nucleus and median eminence increases obesity susceptibility by increasing body fat content in male mice. Glia 68, 1987–2000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneeberger M., Parolari L., Das Banerjee T., Bhave V., Wang P., Patel B., Topilko T., Wu Z., Choi C. H. J., Yu X., Pellegrino K., Engel E. A., Cohen P., Renier N., Friedman J. M., Nectow A. R., Regulation of energy expenditure by brainstem GABA neurons. Cell 178, 672–685.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., McFarland C., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolock S. L., Lopez R., Klein A. M., Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-r., Raychaudhuri S., Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergen V., Lange M., Peidli S., Wolf F. A., Theis F. J., Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Tirosh I., Izar B., Prakadan S. M., Wadsworth M. H. II, Treacy D., Trombetta J. J., Rotem A., Rodman C., Lian C., Murphy G., Fallahi-Sichani M., Dutton-Regester K., Lin J.-R., Cohen O., Shah P., Lu D., Genshaft A. S., Hughes T. K., Ziegler C. G. K., Kazer S. W., Gaillard A., Kolb K. E., Villani A.-C., Johannessen C. M., Andreev A. Y., Van Allen E. M., Bertagnolli M., Sorger P. K., Sullivan R. J., Flaherty K. T., Frederick D. T., Jané-Valbuena J., Yoon C. H., Rozenblatt-Rosen O., Shalek A. K., Regev A., Garraway L. A., Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S., Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu X., Hill A., Packer J., Lin D., Ma Y.-A., Trapnell C., Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satpathy A. T., Granja J. M., Yost K. E., Qi Y., Meschi F., McDermott G. P., Olsen B. N., Mumbach M. R., Pierce S. E., Corces M. R., Shah P., Bell J. C., Jhutty D., Nemec C. M., Wang J., Wang L., Yin Y., Giresi P. G., Chang A. L. S., Zheng G. X. Y., Greenleaf W. J., Chang H. Y., Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schep A. N., Wu B., Buenrostro J. D., Greenleaf W. J., chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corces M. R., Granja J. M., Shams S., Louie B. H., Seoane J. A., Zhou W., Silva T. C., Groeneveld C., Wong C. K., Cho S. W., Satpathy A. T., Mumbach M. R., Hoadley K. A., Robertson A. G., Sheffield N. C., Felau I., Castro M. A. A., Berman B. P., Staudt L. M., Zenklusen J. C., Laird P. W., Curtis C.; Cancer Genome Atlas Analysis Network, Greenleaf W. J., Chang H. Y., The chromatin accessibility landscape of primary human cancers. Science 362, eaav1898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao Z., Zhang Y., Yuan G.-C., Orkin S. H., Waxman D. J., MAnorm: A robust model for quantitative comparison of ChIP-seq data sets. Genome Biol. 13, R16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J., Zibetti C., Shang P., Sripathi S. R., Zhang P., Cano M., Hoang T., Xia S., Ji H., Merbs S. L., Zack D. J., Handa J. T., Sinha D., Blackshaw S., Qian J., ATAC-seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat. Commun. 9, 1364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pliner H. A., Packer J. S., McFaline-Figueroa J. L., Cusanovich D. A., Daza R. M., Aghamirzaie D., Srivatsan S., Qiu X., Jackson D., Minkina A., Adey A. C., Steemers F. J., Shendure J., Trapnell C., Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frandolig J. E., Matney C. J., Lee K., Kim J., Chevée M., Kim S.-J., Bickert A. A., Brown S. P., The synaptic organization of layer 6 circuits reveals inhibition as a major output of a neocortical sublamina. Cell Rep. 28, 3131–3143.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/22/eabg3777/DC1


