Abstract
Melanin is a central polymer in living organisms, yet our understanding of its molecular structure remains unresolved. Here, we apply a biosynthetic approach to explore the composite structures accessible in one type of melanin, eumelanin. Using a combination of solid-state NMR, dynamic nuclear polarization, and electron microscopy, we reveal how a variety of monomers are enzymatically polymerized into their corresponding eumelanin pigments. We demonstrate how this approach can be used to unite structure with an understanding of enzymatic activity, substrate scope, and the regulation of nanostructural features. Overall, this data reveals how intermediate metabolites of the Raper–Mason metabolic pathway contribute to polymerization, allowing us to revisit the original proposal of how eumelanin is biosynthesized.
Melanin is a central polymer in living organisms, yet our understanding of its molecular structure remains unresolved.
Introduction
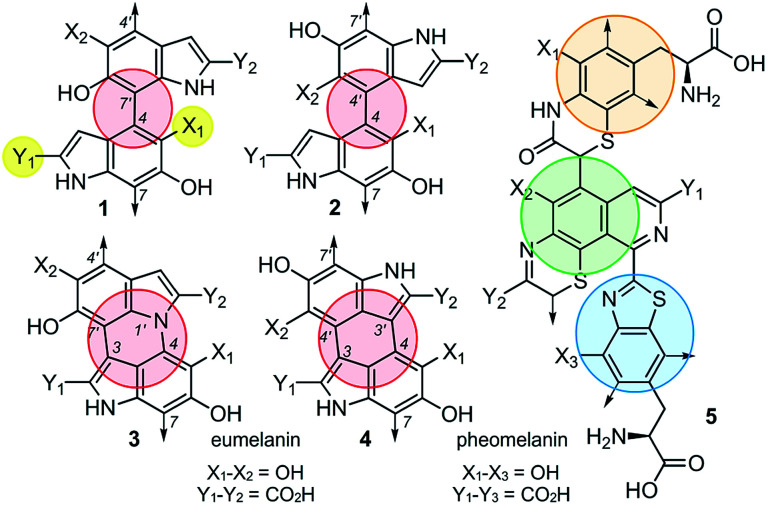
Although the heterogeneous biopolymer melanin1 is ubiquitous in life, there remains a lack in understanding of its intricate molecular structure.2 This structure diverges between three main types of melanin. Eumelanin,3 the most abundant form, is a black/brown pigment derived from tyrosine and is associated with melanoma.4 Pheomelanin,5 the reddish cysteine derivative can be found in hair and skin. Neuromelanin is a dark pigment located in the mammalian brain, which studies have shown to be linked to Parkinson's disease.6 Melanin also has a broad electromagnetic absorption range and plays a role in structural colour.7 These properties are all incredibly useful to harness and emulate in the synthesis of new materials. Although the different melanin types share similar properties,8 they are believed to vary significantly in their molecular composition.9,10 Despite melanin's ubiquity, significant structural uncertainties still remain,11 and a major challenge lies in the unique connectivity and isomeric potential in these polymers. As illustrated by 1–4 (Fig. 1), eumelanin is composed of multiple linkages of discrete monomers. Using a minimal ‘dimeric’ unit, the structural diversity in eumelanin can include disparity in both regioselectivity and connectivity. The complexity becomes even greater in pheomelanin 5 (Fig. 1) which contains multiple structurally discrete units.
Fig. 1. Eumelanin presents multiple levels of structural diversity. The first, 1, arises through different levels of oxidation represented by X1 and Y1 (yellow). The second arises in the monomers connectivity, as noted by the comparison of 1 to 2 (red). The third is the degree of monomer connectivity, such as mono-linked 1/2 to di-linked 3/4. Further complexity can come from different functional linkages, as shown in pheomelanin 5 (orange, green and blue).
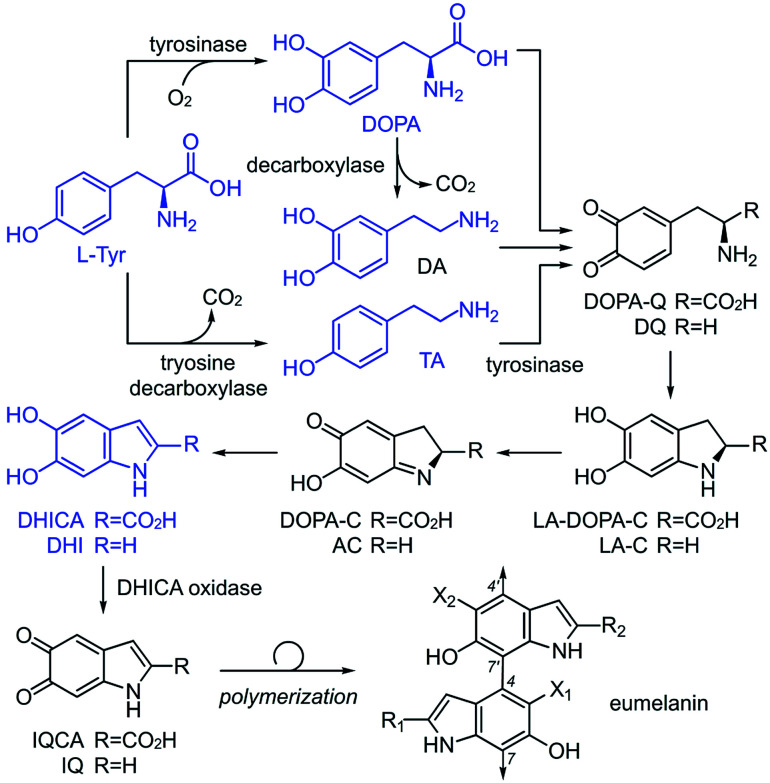
The current understanding of melanin12 is based on the generally accepted biosynthetic pathway for polymerization, the Raper–Mason pathway (RMP),13,14 that was first proposed 70 years ago. The pathway involves a stepwise progression from l-Tyr to dihydroxyindole (DHI) or dihydroxyindole carboxylic acid (DHICA), as shown in Scheme 1. These units then undergo an oxidation to generate indole-5,6-quinone-2-carboxylic acid (IQCA) or indole-5,6-quinone (IQ), the immediate precursors for polymerization. Our findings here show that the general understanding and interpretation of the RMP is incorrect. In addition to the various structural complexities, melanogenesis processes are regulated in vivo through a series of enzymes, including tyrosinases15 and decarboxylases,16 that unite the oxidative and decarboxylative steps shown in Scheme 1. However, our findings suggest that each RMP step need not reach completion prior to polymerization, and heterogeneity may be generated by the relative abundance of pathway intermediates. Here, we explore the link between tyrosinase substrate selectivity by exploring RMP intermediate identity during melanisation.
Scheme 1. The Raper–Mason pathway (RMP) proposes that melanin originates from l-Tyr being oxidized to melanin vial-DOPA, which could be followed by a decarboxylation to dopamine (DA). l-Tyr can also be decarboxylated to tyramine (TA). These substrates are further oxidized to form transient intermediates: dopaquinone (DOPA-Q), dopamine-o-quinone (DQ), leukodopachrome (LA-DOPA-C), leukoaminochrome (LA-C), dopachrome (DOPA-C), and aminochrome (AC), leading to DHI and DHICA units, the proposed building blocks of eumelanin. Prior to polymerization, DHI and DHICA are further oxidized to indole-5,6-quinone (IQ) and indole-5,6-quinone-2-carboxylic acid (IQCA).
To date, our structural understanding of melanin arises from pioneering solid-state NMR (ssNMR) work on fungal melanin by Stark and Casadevall.17,18 These datasets have enabled a hypothesis toward the linkages between DHI and DHICA. Typically, melanin ssNMR spectra are assigned using chemical shift predictions.19 While this method was vital in clarifying the key functionality, the lack of expanded experimental data to support these predictions reduces the accuracy of their assignments.
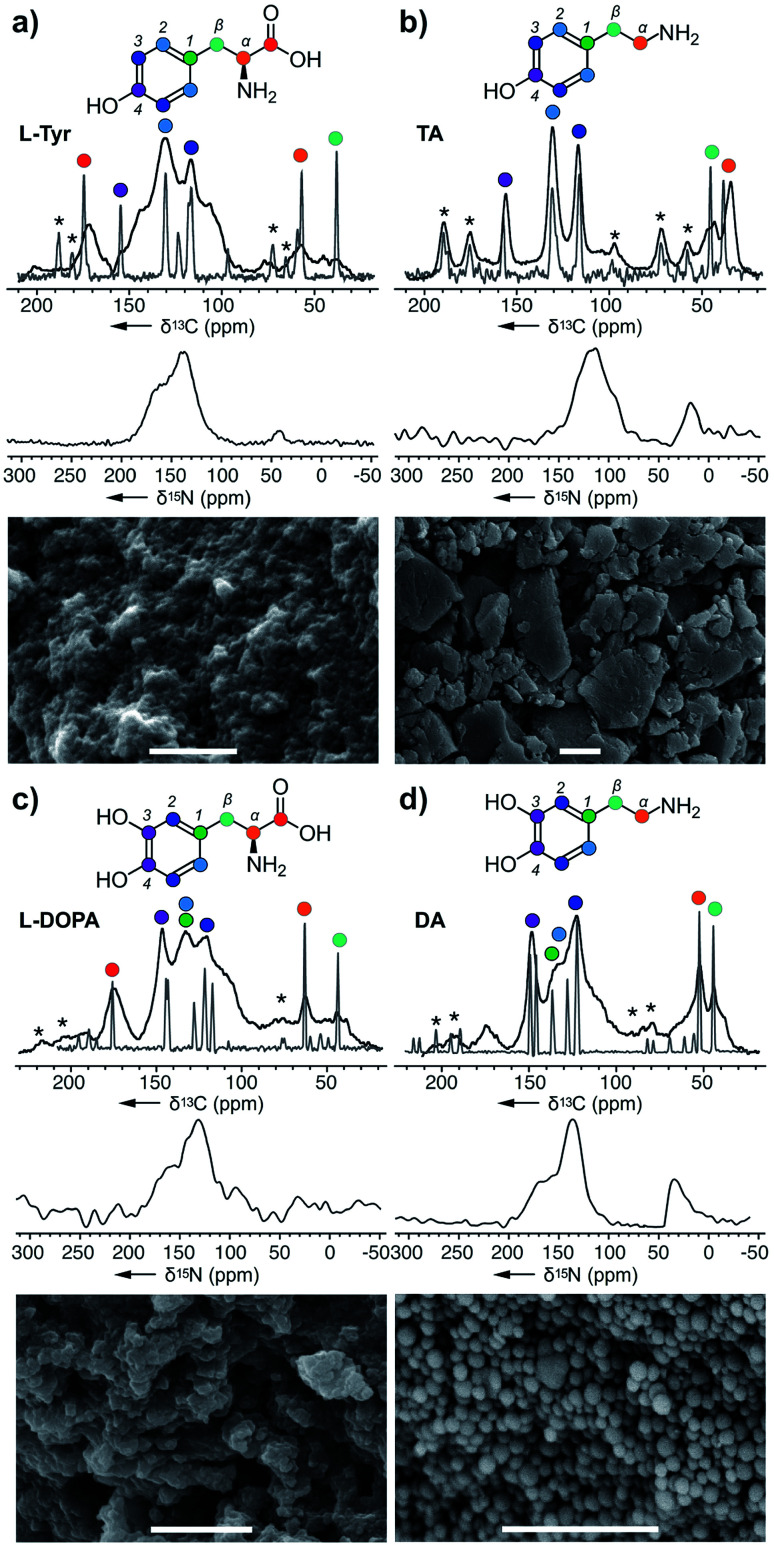
We began by exploring the ability of the well-characterized mushroom tyrosinase from Agaricus bisporus20 for polymerization of different RMP intermediates. Unlike previous studies where the polymerized pigments are treated with 6 M HCl and boiled (SEM images in ESI Fig S1† showing destroyed nanoparticles after acid treatments), which can destroy essential structural features, our pigments were carefully washed with neutral H2O. The tools of ssNMR and dynamic nuclear polarization (DNP)21,22 were applied to evaluate the four critical monomers tyrosine (l-Tyr), tyramine (TA), dopamine (DA),23 and l-DOPA (ESI Fig. S2–S8†) of the RMP (Scheme 1) in their monomeric and polymeric forms.
In this study, we used both 13C chemical shift predictions generated by ChemNMR (ESI Fig. S6†) and ssNMR spectra on the monomers to assign the structures of each polymer (Fig. 2–4). Likewise, corresponding scanning electron microscopy (SEM) images of these pigments provide insight into the different morphologies that can arise from these nanoparticles. From this, one can appreciate the vast differences between melanin pigments at the nanoscale and how it can attribute to different biological functions.24
Fig. 2. Melanin polymers were obtained from the action of A. bisporus tyrosinase on (a) l-Tyr, (b) TA, (c) l-DOPA, and (d) DA. ssNMR spectra of each melanised sample are overlaid with its original monomer, and these peaks were used for assignments. Only the 15N spectra were collected using DNP ssNMR with an enhancement ∼20. Peak assignments are provided in the ESI Fig. S2–S8.† * Denotes the spinning side bands. Bars in the SEM images represent 1 μm.
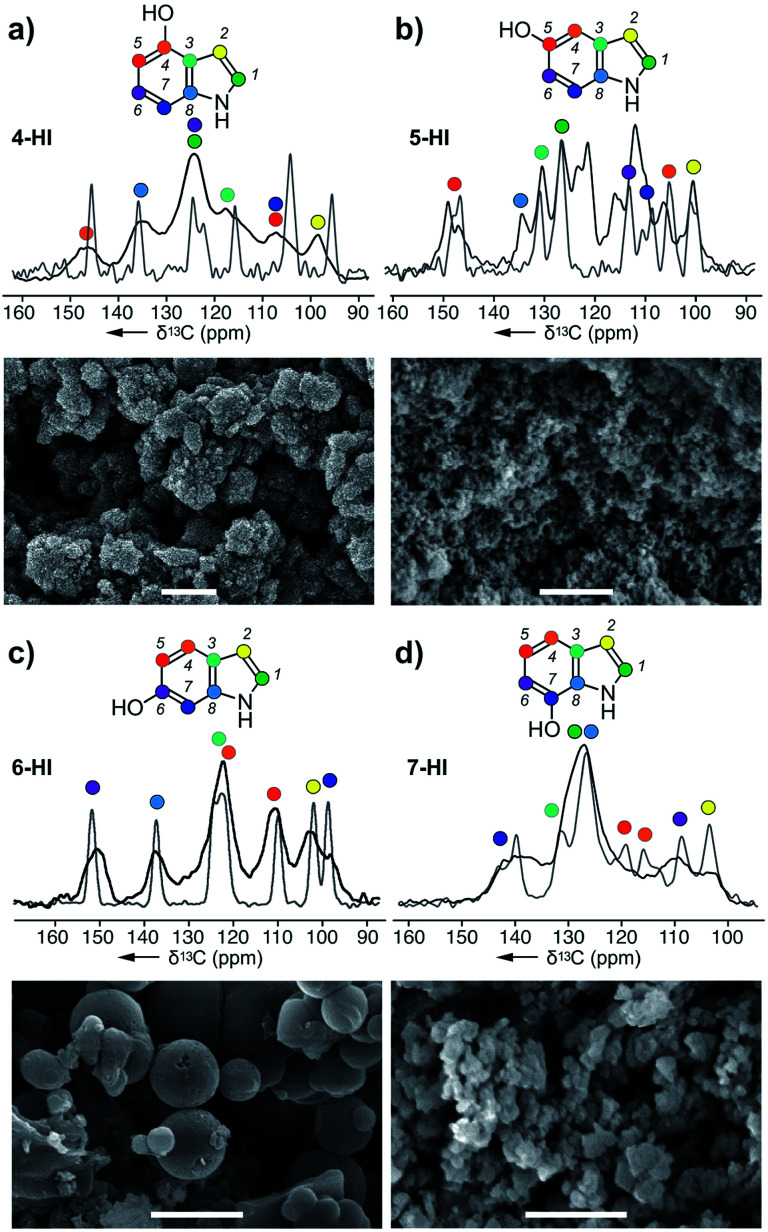
Fig. 3. Melanin particles obtained from the action of A. bisporus tyrosinase on (a) 4-HI, (b) 5-HI, (c) 6-HI, and (d) 7-HI. ssNMR spectra of each melanised sample are overlaid with its original monomer, and these peaks were used for assignments. Bars in the SEM images represent 1 μm. Peak assignments are provided in ESI Fig. S9–S12.†.
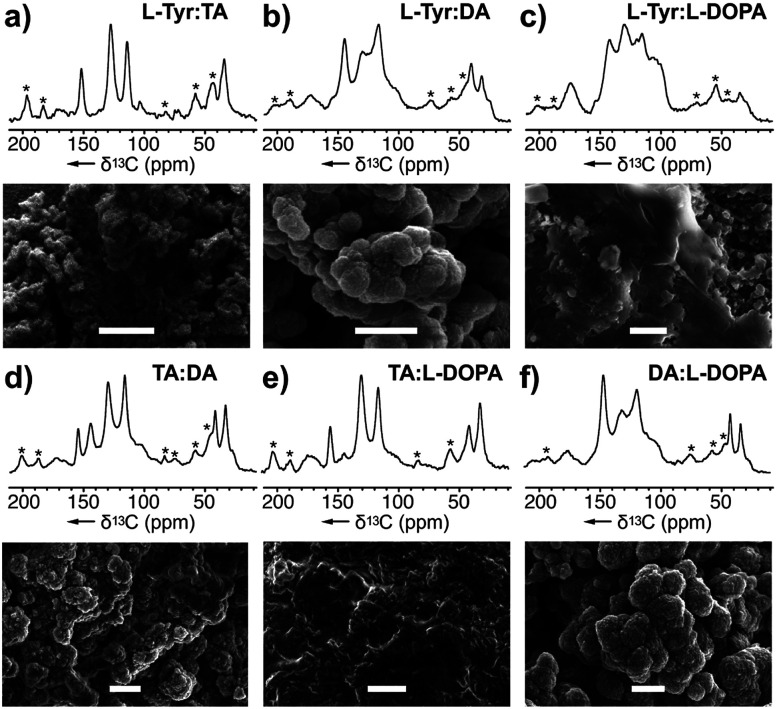
Fig. 4. Melanin produced by copolymerizing two monomers: (a) l-Tyr : TA–melanin generated from a mixture of l-Tyr and TA, (b) l-Tyr : l-DA–melanin, (c) l-Tyr : l-DOPA–melanin, (d) TA : DA–melanin, (e) TA : l-DOPA–melanin, and (f) DA : l-DOPA–melanin using A. bisporus tyrosinase. Comparison of the NMR spectra of these mixtures with that of pure monomers (Fig. 2) was used to explore the monomer selectivity of this enzyme. Peak and monomer assignments are provided in ESI Fig. S13 and S14† as well as the experimental conditions of copolymerization procedure. * Denotes the spinning side bands. Bars in the SEM images represent 1 μm.
l-Tyr–melanin
Starting with l-Tyr (Fig. 2a), a SEM image shows that a black pigment containing rods imbedded within a solid matrix was obtained after treating l-Tyr with tyrosinase. The ssNMR 13C spectrum of the l-Tyr was evaluated against the l-Tyr–melanin. Interestingly, spectral overlays indicate that each of the 13C peaks in the l-Tyr monomer (grey, Fig. 2a) were observed in the l-Tyr–melanin product (black, Fig. 2a). With optimized magic angle spinning NMR cross polarization experimental conditions for both carbonyl and aliphatics, the lower aliphatic α,β peaks at δ 38 and 53 ppm suggest that part of the alkyl carbons didn't convert into an aromatic environment. Meanwhile, a set of peaks with low intensity was detected at δ 105 and 145 ppm, indicative of DHI and DHICA (Scheme 1). The three peaks (δ 160, 135 and 42 ppm) within the 15N spectrum suggested N atoms commonly associated with pyrroles, indoles, and amines, respectively.
TA–melanin
The same method was then applied to tyramine (TA), another RMP intermediate suggested to require processing before polymerization. The SEM image of TA–melanin (Fig. 2b) illustrates a polymer lacking nanostructural features. ssNMR analyses revealed a clear correlation between TA–monomer and TA–melanin (ESI Fig. S3†). As evident in Fig. 2b, the carbon atoms underwent only a modest chemical environment change upon polymerization. The spectral data from TA–melanin was in stark contrast with l-Tyr–melanin, whose broad peaks suggested it contained a nonuniform melanin composition through processes such as incomplete decarboxylation, resulting in mixtures of decarboxylated (R1, R2 = H) and carboxylated (R1, R2 = CO2H) polymers. TA–melanin has an upfield 15N chemical shift of δ 115 ppm resembling peptide linkages. These observations reveal that the action of the mushroom tyrosinase on TA was not restricted to comply with the stepwise nature of the RMP (Scheme 1) but was able to process an expanded monomeric scope.
l-DOPA and DA melanin
To complete the RMP intermediate set, l-DOPA–melanin and DA–melanin spectra were evaluated (Fig. 2c and d, ESI Fig. S4 and S5†). As shown in Fig. 2c, the polymerization of l-DOPA returned a polymer comprised of 100 ± 30 nm sized spheres. Here, the presence of an additional hydroxyl group with respect to l-Tyr (Fig. 2a) resulted in a nanostructural change from a material with imbedded surfaces to more geometrically defined nanometer-sized spheres in l-DOPA–melanin. The ssNMR spectrum from l-DOPA–melanin (Fig. 2c), like l-Tyr–melanin (Fig. 2a), contained both the aromatic and aliphatic peaks of its monomer. Similarly, the presence of the carbonyl at δ 173 ppm, along with α and β carbons at δ 65 and 45 ppm, in l-DOPA–melanin confirmed incomplete decarboxylation and indole ring formation. Meanwhile, the intensity of the aliphatic carbons at δ 53 and 47 ppm from DA–melanin were the strongest of all three polymers, suggesting an increased level of non-indole cyclised materials. This was further supported by the presence of a δ 45 ppm peak in the 15N spectrum. The dopamine-formed particles produced visible 100 ± 10 nm spheres by SEM imaging.
All four spectral sets (Fig. 2) showed that chemical shift values of the melanised particles were similar to their monomeric precursors with the addition of the cyclised DHI or DHICA units. The fact that all four monomers melanised shows the broad selectiveness of the enzyme and the structural diversity arising from the metabolites. This alters the mechanistic model for melanin formation and impacts the resulting nanoparticle structure. SEM images of the four melanin products with different nanoparticle morphologies illustrates each metabolite plays a role in influencing or altering the functional capabilities of melanin. The ssNMR spectra also shows that although the polymer is mostly populated by the monomer itself, it also includes a small amount of cyclised DHI or DHICA. Among them, TA–melanin has the most resolved aromatic region in its 13C spectrum and the most upfield 15N chemical shift at δ 115 ppm which could arise from other types of linkages. All of the melanin polymers in Fig. 2 retained primary amine 15N residues at δ 35 ppm or 25 ppm.
Hydroxyindole–melanins
Next, we explored the ability of the A. bisporus tyrosinase to polymerize indole derivatives of DHI or DHICA (Scheme 1). The goal of this study was to demonstrate the ability of tyrosinase to act on non-natural substrates and further probe the substrate tolerance of this pathway. Using conditions established in the previous study, the tyrosinase was presented with 4 different hydroxyindole (HI) monomers: 4-HI, 5-HI, 6-HI and 7-HI, and the resulting melanins were analysed via ssNMR and SEM (ESI Fig. S9–S12†). Comparable to the acyclic precursors (Fig. 2), the 13C spectra of HI–melanins contained similar chemical shift values to their corresponding monomers. Based on the peak positions, 4-HI–melanin contained either a C5 or C7 downfield shift from δ 105 to 115 ppm, supporting a 5-5 linkage (Fig. 3a). In contrast, the increase in the number of peaks in 5-HI–melanin (Fig. 3b), would result from a 2-4 hetero-linkage between the two 5-HI units, which can be explained by a loss of symmetry and a duplication of its 7 carbons.
Each of the carbons observed in the 13C-NMR spectrum of 6-HI–melanin (Fig. 3c) were identified in the 6-HI monomer, except for C3, which was weakly observed. 6-HI–melanin exhibited the most resolved spectra, indicating a more homogenous environment. Symmetry studies suggest 6-HI–melanin consists of a 2-2 linkage. Unfortunately, the overlap of three residues within the central peak at δ 126 ppm in the spectrum from 7-HI–melanin complicated the elucidation of its linkage (Fig. 3d).
4-HI–melanin (Fig. 3a) and 7-HI–melanin (Fig. 3d) SEM images resembled l-DOPA–melanin with clusters of nanoparticle sizes of 100 ± 25 nm. SEM imaging revealed 5-HI–melanin contained dispersed particles with a high size uniformity of 75 ± 15 nm while 6-HI–melanin formed spherical particles with a large size distribution, 350–900 nm (Fig. 3c). Though the proposed natural intermediate, DHI (Scheme 1), contains oxidation at C5 and C6, the substrate tolerance of the tyrosinase enabled it to polymerize materials with oxidations at non-natural positions.
Copolymerized melanin blends
To investigate its selectivity, the tyrosinase was used to copolymerize various combinations of monomers. The resulting melanin products along with their monomer counterparts were analysed by ssNMR and SEM (Fig. 4 and ESI Fig. 13 and 14†). Among the six copolymers, the ssNMR spectra suggested the resulting melanin was not of 1 : 1 alternating pattern; rather, TA took precedence over the others. Though the overall lineshape follows TA, it is clear that portions of the peaks are derived from the other monomer. Additionally, a set of peaks with distinct DHI or DHICA chemical shifts appeared at low intensity. Furthermore, we observed that DA took precedence over l-DOPA, which took precedence over l-Tyr. Overall, this suggests the polymers generated from the tyrosinase can incorporate monomers at different stages of the RMP and does not require a stepwise conversion as shown in Scheme 1. Instead, the A. bisporus tyrosinase can polymerize a ‘snapshot’ of the monomeric metabolic pool within its local environment. This observation begins to explain the complexity seen within melanin polymers and provides an important advance in understanding the structural diversity accessible in eumelanin.
The SEM images from these blends (Fig. 4) show particles of various morphologies. In particular, the ones containing DA yield products increased sphericity and higher ordered structures. While DA-melanised nanoparticles are around 100 nm, once it is copolymerized with other monomers, the resulting nanoparticles increase in size. We observed 100 nm particles molded into ovoids ranging from 500 nm to 1 μm.
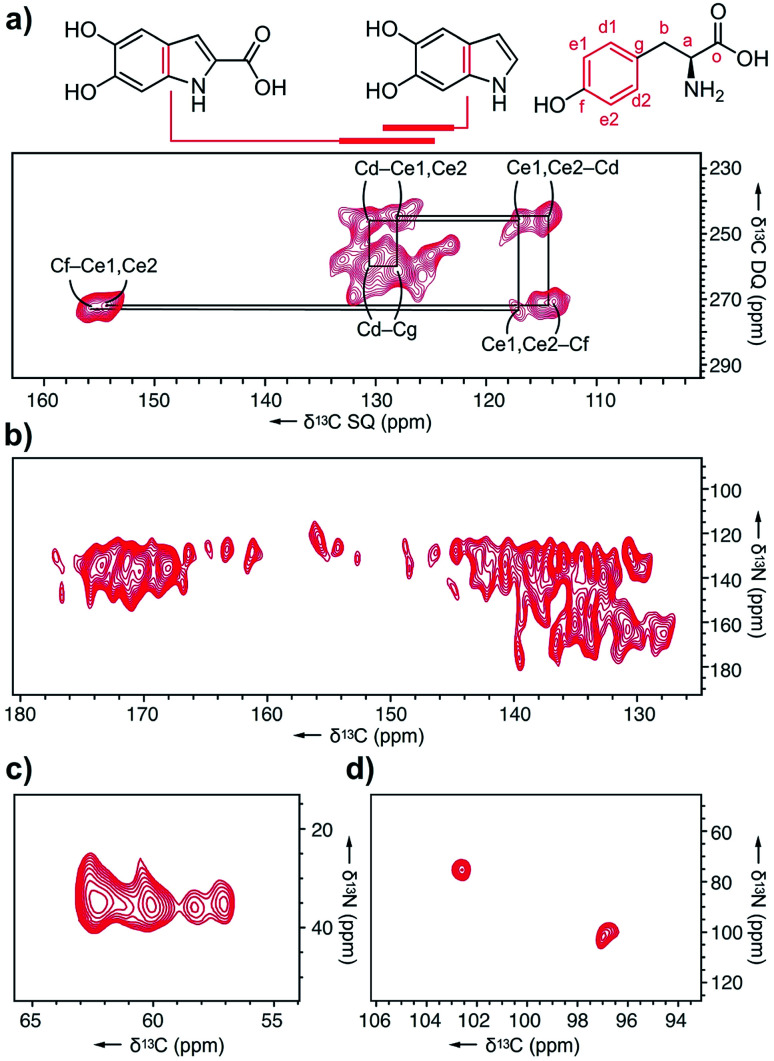
Next, we explored melanin prepared from U-[13C,15N]–l-Tyr using multidimensional DNP NMR (Fig. 5). Correlations observed from the 2D 13C–13C PDSD experiments25 indicated high populations of intramolecular contacts (ESI Fig. S15†). However, this doesn't exclude the possibility of intermolecular interactions between tyrosine units, in the case of pi–pi stacking between monomers.26 The 2D 13C–13C SPC5 experiment27 (Fig. 5a) shows covalent bonds from both the l-Tyr ring and DHI/DHICA units. This further validates the highly populated presence of l-Tyr monomer in this multicomponent melanised polymer. The unique chemical shift values of the tyrosine aromatic ring are clearly assigned, starting with Cf (Fig. 5a) attached to the hydroxyl group at δ 145 ppm to the double resonances of Ce1, Ce2 at δ 115 ppm to another set of double resonances Cd1, Cd2 at δ 130 ppm, and lastly to Cg at δ 128 ppm.
Fig. 5. DNP ssNMR analysis of U-[13C,15N]–l-Tyr–melanin. (a) 2D 13C–13C SPC5 spectrum showing correlations from the tyrosine ring, DHI, and DHICA units. (b) 2D 13C–15N TEDOR spectra acquired with mixing times of 3.2 ms showing pyrrole correlations show only aromatic contacts and no carboxylated motifs. (c) The aliphatic region shows four distinct Ca–N environments. (d) C4–N correlations from DHICA units were detected.
In the 13C–15N TEDOR spectra28 (Fig. 5b–d), nitrogen atoms from hydroxyindole units correlate with both aromatic and carbonyl carbons, suggesting DHI and DHICA units. Pyrrole correlations show only aromatic contacts and no carboxylated motifs, which could result from Baeyer–Villiger type reactions originating from indole metabolites of the RMP. The primary amine 15N is correlated to four distinct 13C atoms, with the two major peaks originating from l-Tyr.
Conclusions
This study suggests the classical Raper–Mason pathway doesn't fully-encompass substrate scope in melanin biosynthesis, as enzymatic polymerization can occur with many of its monomeric intermediates (blue structures, Scheme 1). Here, comparative ssNMR evaluations of monomers (Fig. 2 and 3) provide a useful tool that enables a nuanced understanding of the structural features of each melanin polymer. While copolymerization studies (Fig. 4) indicate some selectivity, the serial nature of the RMP can be misleading. These studies suggest that natural melanin structure likely relies on the local metabolic flux29 to guide the structural composition of the resulting polymers. Overall, tandem SEM and ssNMR analyses enable the development of a structure–function correlation for melanin pigments. Demonstrated here for the A. bisporus tyrosinase, this chemoenzymatic approach offers a general tool to explore the structural selectivity and tolerance of melanin-producing enzymes. Efforts are now underway to apply this union between ssNMR and electron microscopy to further interrogate the diversity of natural melanin.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
The work was supported by a MURI through Air Force Office of Scientific Research (FA9550-18-1-0142). We also thank Stanley Opella, Galia Debelouchina, Sho Konno, Ivan Sergeyev and Melanie Rosay for instrumental assistance and scientific discussions.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc02262d
Notes and references
- D'Alba L. Shawkey M. D. Melanosomes: Biogenesis, Properties, and Evolution of an Ancient Organelle. Physiol. Rev. 2019;99:1–19. doi: 10.1152/physrev.00059.2017. [DOI] [PubMed] [Google Scholar]
- Solano F. Melanin and Melanin-Related Polymers as Materials with Biomedical and Biotechnological Applications-Cuttlefish Ink and Mussel Foot Proteins as Inspired Biomolecules. Int. J. Mol. Sci. 2017;18:E1561. doi: 10.3390/ijms18071561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büngeler A. Hämisch B. Strub O. I. The Supramolecular Buildup of Eumelanin: Structures, Mechanisms, Controllability. Int. J. Mol. Sci. 2017;18:E1901. doi: 10.3390/ijms18091901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski R. Zmijewski M. Slominski A. The role of melanin pigment in melanoma. Exp. Dermatol. 2015;4:258–259. doi: 10.1111/exd.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thody A. J. Higgins E. M. Wakamatsu K. Ito S. Burchill S. A. Marks J. M. Pheomelanin as well as eumelanin is present in human epidermis. J. Invest. Dermatol. 1991;97:340–344. doi: 10.1111/1523-1747.ep12480680. [DOI] [PubMed] [Google Scholar]
- Haining R. L. Achat-Mendes C. Neuromelanin, one of the most overlooked molecules in modern medicine, is not a spectator. Neural Regen. Res. 2017;12:372–375. doi: 10.4103/1673-5374.202928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T. Tamai Y. Yamamoto M. Taniguchi T. Kishikawa K. Kohri M. Melanin Precursor Influence on Structural Colors from Artificial Melanin Particles: PolyDOPA, Polydopamine, and Polynorepinephrine. Langmuir. 2018;34:11814–11821. doi: 10.1021/acs.langmuir.8b02444. [DOI] [PubMed] [Google Scholar]
- Marchetti B. Karsili T. N. Theoretical insights into the photo-protective mechanisms of natural biological sunscreens: building blocks of eumelanin and pheomelanin. Phys. Chem. Chem. Phys. 2016;18:3644–3658. doi: 10.1039/C5CP06767G. [DOI] [PubMed] [Google Scholar]
- Simpson M. J. Wilson J. W. Phipps M. A. Robles F. Selim M. A. Warren W. S. Nonlinear microscopy of eumelanin and pheomelanin with subcellular resolution. J. Invest. Dermatol. 2013;133:1822–1826. doi: 10.1038/jid.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman H. C. Casadevall A. A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012;93:931–940. doi: 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugumaran M. Reactivities of Quinone Methides versus o-Quinones in Catecholamine Metabolism and Eumelanin Biosynthesis. Int. J. Mol. Sci. 2016;17:E1576. doi: 10.3390/ijms17091576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ischia M. Napolitano A. Pezzella A. Meredith P. Sarna T. Chemical and Structural Diversity in Eumelanins: Unexplored Bio-Optoelectronic Materials. Angew. Chem., Int. Ed. 2009;48:3914–3921. doi: 10.1002/anie.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper H. S. The Aerobic Oxidases. Physiol. Rev. 1928;8:245–282. doi: 10.1152/physrev.1928.8.2.245. [DOI] [Google Scholar]
- Mason H. S. The chemistry of melanin; mechanism of the oxidation of dihydroxyphenylalanine by tyrosinase. J. Biol. Chem. 1948;172:83–99. [PubMed] [Google Scholar]
- Casanola-Martin G. M. Le-Thi-Thu H. Marrero-Ponce Y. Castillo-Garit J. A. Torrens F. Rescigno A. Abad C. Khan M. R. Tyrosinase enzyme M. R. 1. An overview on a pharmacological target. Curr. Top. Med. Chem. 2014;14:1494–1501. doi: 10.2174/1568026614666140523121427. [DOI] [PubMed] [Google Scholar]
- Jiang M. Xu G. Ni J. Zhang K. Dong J. Han R. Ni Y. Improving Soluble Expression of Tyrosine Decarboxylase from Lac-tobacillus brevis for Tyramine Synthesis with High Total Turnover Number. Appl. Biochem. Biotechnol. 2019;188:436–449. doi: 10.1007/s12010-018-2925-x. [DOI] [PubMed] [Google Scholar]
- Chatterjee S. Prados-Rosales R. Tan S. Phan V. C. Chrissian C. Itin B. Wang H. Khajo A. Magliozzo R. S. Casadevall A. Stark R. E. The melanization road more traveled by: Precursor substrate effects on melanin synthesis in cell-free and fungal cell systems. J. Biol. Chem. 2018;293:20157–20168. doi: 10.1074/jbc.RA118.005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk J. D. Stark R. E. Casadevall A. Fungal Melanin A. What do We Know About Structure? Front. Microbiol. 2015;6:1463. doi: 10.3389/fmicb.2015.01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiler J. Maier W. Will M. Meusinger R. Using neural networks for 13C NMR chemical shift prediction-comparison with traditional methods. J. Magn. Reson. 2002;157:242–252. doi: 10.1006/jmre.2002.2599. [DOI] [PubMed] [Google Scholar]
- Halaouli S. Asther M. Sigoillot J. C. Hamdi M. Lomascolo A. Fungal tyrosinases: new prospects in molecular characteristics, bioengineering and biotechnological applications. J. Appl. Microbiol. 2006;100:219–232. doi: 10.1111/j.1365-2672.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- Ni Q. Z. Daviso T. Can T. V. Markhasin E. Jawla S. K. Swager T. M. Temkin R. J. Herzfeld J. Griffin R. G. High frequency dynamic nuclear polarization. Acc. Chem. Res. 2013;46:1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly Thankamony S. Wittmann J. J. Kaushik M. Corzilius B. Dynamic nuclear polarization for sensitivity enhancement in modern solid-state NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2017;102–103:120–195. doi: 10.1016/j.pnmrs.2017.06.002. [DOI] [PubMed] [Google Scholar]
- d'Ischia M. Napolitano A. Ball V. Chen C. T. Buehler M. Polydopamine and Eumelanin: From Structure–Property Relationships to a Unified Tailoring Strategy. Acc. Chem. Res. 2014;47:3541–3550. doi: 10.1021/ar500273y. [DOI] [PubMed] [Google Scholar]
- Gatoo M. Naseem S. Arfat M. Dar A. Qasim K. Zubair S. Physicochemical Properties of Nanomaterials: Implication in Associated Toxic Manifestations. Biomed Res. Int. 2014;498420:1–8. doi: 10.1155/2014/498420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegoshi K. Nakamura S. Terao T. 13C-1H dipolar-driven 13C-13C recoupling without 13C rf irradiation in nuclear magnetic resonance of rotating solids. J. Chem. Phys. 2003;118:2325–2341. doi: 10.1063/1.1534105. [DOI] [Google Scholar]
- Li Y. Liu J. Wang Y. Chan H. W. Wang L. Chan W. Mass Spectrometric and Spectrophotometric Analyses Reveal an Alternative Structure and a New Formation Mechanism for Melanin. Anal. Chem. 2015;87:7958–7963. doi: 10.1021/acs.analchem.5b01837. [DOI] [PubMed] [Google Scholar]
- Lee Y. K. Kurur N. D. Helmle M. Johannessen O. G. Neilsen N. C. Levitt M. H. Efficient dipolar recoupling in the NMR of rotating solids. A sevenfold symmetric radiofrequency pulse sequence. Chem. Phys. Lett. 1995;242:304–309. doi: 10.1016/0009-2614(95)00741-L. [DOI] [Google Scholar]
- Hing A. W. Vega S. Schaefer J. Transferred-echo double-resonance NMR. J. Mag. Reson. 1992;96:205–209. [Google Scholar]
- Mekala L. P. Mohammed M. Chintalapati S. Chintalapati V. R. Stable Isotope-Assisted Metabolic Profiling Reveals Growth Mode Dependent Differential Metabolism and Multiple Catabolic Pathways of l-Phenylalanine in Rubrivivax benzoatilyticus JA2. J. Proteome Res. 2018;17:189–202. doi: 10.1021/acs.jproteome.7b00500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.