Significance
The Ycf1p structure provides an atomic model for the TMD0 domain of ABCC transporters and for two segments of the regulatory (R) region that links NBD1 to TMD2. The orientation of TMD0 in Ycf1p differs from that seen in SUR1, the regulatory ABCC protein in KATP channels, demonstrating flexibility in TMD0/ABC core contacts. The structure suggests how posttranslational modifications of the R region modulate ABC protein activity and provides a mechanistic understanding of several diseases that occur due to mutation of human homologs of Ycf1p.
Keywords: ABC transporter, Ycf1p, TMD0, R region, cryo-EM
Abstract
ATP binding cassette (ABC) proteins typically function in active transport of solutes across membranes. The ABC core structure is composed of two transmembrane domains (TMD1 and TMD2) and two cytosolic nucleotide binding domains (NBD1 and NBD2). Some members of the C-subfamily of ABC (ABCC) proteins, including human multidrug resistance proteins (MRPs), also possess an N-terminal transmembrane domain (TMD0) that contains five transmembrane α-helices and is connected to the ABC core by the L0 linker. While TMD0 was resolved in SUR1, the atypical ABCC protein that is part of the hetero-octameric ATP-sensitive K+ channel, little is known about the structure of TMD0 in monomeric ABC transporters. Here, we present the structure of yeast cadmium factor 1 protein (Ycf1p), a homolog of human MRP1, determined by electron cryo-microscopy (cryo-EM). A comparison of Ycf1p, SUR1, and a structure of MRP1 that showed TMD0 at low resolution demonstrates that TMD0 can adopt different orientations relative to the ABC core, including a ∼145° rotation between Ycf1p and SUR1. The cryo-EM map also reveals that segments of the regulatory (R) region, which links NBD1 to TMD2 and was poorly resolved in earlier ABCC structures, interacts with the L0 linker, NBD1, and TMD2. These interactions, combined with fluorescence quenching experiments of isolated NBD1 with and without the R region, suggest how posttranslational modifications of the R region modulate ABC protein activity. Mapping known mutations from MRP2 and MRP6 onto the Ycf1p structure explains how mutations involving TMD0 and the R region of these proteins lead to disease.
ATP binding cassette (ABC) proteins are a large family of membrane proteins found in all kingdoms of life (1, 2). ABC proteins have a core structure composed of two transmembrane (TM) domains (TMD1 and TMD2) and two cytosolic nucleotide binding domains (NBD1 and NBD2) (Fig. 1A and SI Appendix, Fig. S1A) (3–5). Through ATP binding and hydrolysis at the NBDs, ABC proteins actively transport solutes across cell membranes, regulate activities of other proteins, or function as channels (1, 2). Thus, ABC proteins are involved in many biological processes, including lipid homeostasis, cellular metal trafficking, and antigen peptide transport. Mutations in human ABC proteins cause diseases such as Tangier disease, adenoleukodystrophy, cystic fibrosis, Dubin–Johnson syndrome, and pseudoxanthoma elasticum (PXE) (1, 2). Furthermore, the export of a wide range of cancer chemotherapeutics, antibiotics, and anti-fungal drugs by ABC transporters confers multidrug resistance to tumor cells, bacteria, and fungal pathogens, respectively (1, 2, 6, 7).
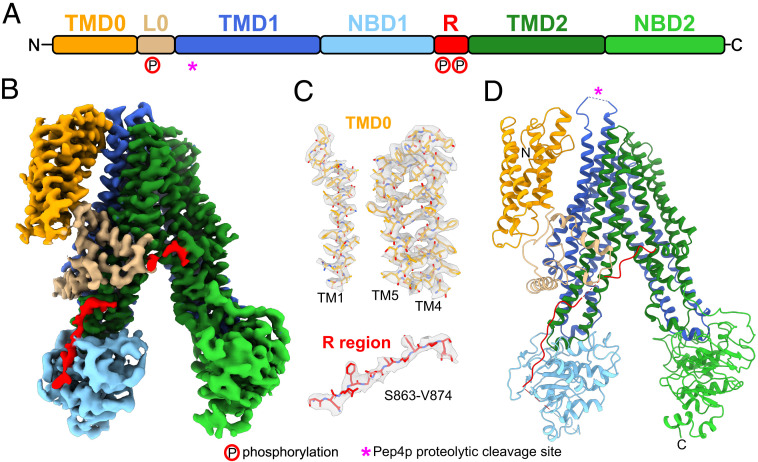
Fig. 1.
Ycf1p structure. (A) Ycf1p domain arrangement. TMD, transmembrane domain; L0, L0 linker; NBD, nucleotide binding domain; and R, regulatory (R) region. The Pep4p proteolytic digestion site within the luminal loop 6 of TMD1 is denoted by a pink “*.” Phosphorylation sites in the L0 linker (S251) and R region (S908 and T911) are depicted with a “P” circled in red. (B) Cryo-EM density of Ycf1p with domains colored as in A. (C) Example of the atomic model for individual TM helices in TMD0 and the R region fit into the corresponding map densities. (D) Schematic ribbon diagram of Ycf1p colored as in A and B and with the proteolytic digestion site denoted by a pink “*.”
Human ABC proteins are divided into seven subfamilies (A to G) based in part on the sequence of their NBDs and TMDs in the core ABC structure (1, 2). The C-subfamily is the most diverse and includes the cystic fibrosis transmembrane conductance regulator (CFTR), the sulphonylurea receptors that form regulatory subunits in ATP-sensitive K+ (KATP) channels, and the multidrug resistance proteins (MRPs). In addition to the ABC core, ABCC proteins contain an N-terminal extension that is either composed of an additional TM domain (TMD0) and L0 linker (Fig. 1A, orange and tan, respectively, and SI Appendix, Fig. S1A) or just an L0 tail (5, 8). A TMD0, but not L0 linker, is also found in some ABCB proteins (3, 5). These N-terminal extensions are involved in trafficking, endosomal recycling, protein interactions, and/or regulation of ABC activity (9–18). The existence of disease-causing mutations in TMD0 and the L0 linker of different ABCC proteins (8, 13, 18) indicates that these regions play important roles in protein function when present.
High-resolution structural information for TMD0 is available only for the atypical ABCC protein SUR1 (19, 20), which is part of the large hetero-octameric KATP channel complex. In contrast, structures of monomeric ABC transporters showed only low-resolution density for TMD0 that was insufficient for building a full atomic model or lacked density for the domain altogether (14, 21–24). The vacuolar ABCC protein yeast cadmium factor 1 (Ycf1p) from Saccharomyces cerevisiae is a close homolog of human MRPs and a model for ABCC proteins that function as monomers. Ycf1p transports glutathione-conjugated heavy metals, such as Cd2+, from the cytosol into the vacuole, detoxifying the cell (25, 26). Human MRP1 can rescue Cd2+ transport activity in a YCF1 deletion strain (27).
Like other ABCC proteins, Ycf1p contains a relatively long and mostly disordered linker that connects NBD1 and TMD2 (25, 28, 29) (Fig. 1A and SI Appendix, Fig. S1A). This linker contains stimulatory phosphorylation sites (25, 28), similar to the phospho-regulatory (R) region in the ABCC protein CFTR (30–32). Ycf1p also contains an inhibitory phosphorylation site in the L0 linker (33). However, how the R region interacts with the ABCC core and how its phosphorylation modulates protein function remain poorly understood for most ABCC proteins. Structural studies of Ycf1p presented here reveal how TMD0 and the R region exert their regulatory functions in MRP-like ABCC proteins.
Results and Discussion
Overall Ycf1p Structure.
To isolate Ycf1p for structural analysis by electron cryo-microscopy (cryo-EM), sequence encoding a 3×FLAG affinity tag was integrated into the yeast chromosomal DNA at the 3′ end of the YCF1 gene. Following growth and harvest of the resulting yeast strain, membranes were solubilized with detergent, and Ycf1p was isolated by affinity chromatography. Although not required for Ycf1p function, proteolytic digestion of Ycf1p at an insertion in loop 6 (Fig. 1 and SI Appendix, Fig. S1A) can affect the substrate specificity of the protein (34). However, the yeast strain used for protein purification lacked the Pep4p protease, and consequently, Ycf1p was isolated without proteolytic digestion, as confirmed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (SI Appendix, Fig. S2A). The Ycf1p preparation showed a specific ATPase activity of 32.2 ± 3.9 nmol Pi/min/mg protein (± SD, n = six replicates, with three replicates from each of two protein batches from independent yeast cultures, SI Appendix, Fig. S2B). Mass spectrometry (MS) indicated that the R region is extensively phosphorylated at multiple sites, while there is limited phosphorylation of the L0 linker (SI Appendix, Fig. S2C).
Cryo-EM of the sample led to a map of the protein with an overall resolution of 3.2 Å (Fig. 1 B and C and SI Appendix, Fig. S3 A–D). Local resolution of the protein ranges from ∼2.7 to ∼6.7 Å. The TMDs (TMD0, TMD1, and TMD2) and L0 linker are at ∼2.7 to ∼3.8 Å resolution, allowing construction of atomic models for these regions (Fig. 1C and SI Appendix, Figs. S3E, S4, and S5). The NBDs are at ∼4.7 to ∼6.7 Å resolution and were interpreted by fitting of homology models of Ycf1p NBD1 and NBD2 (SI Appendix, Figs. S3E and S5). The quality of the map also enabled construction of an atomic model for two segments of the R region from residues S863 to R875 and H928 to K935 (Fig. 1C and SI Appendix, Fig. S5). The atomic model for Ycf1p comprises 94% of its 1,515 residues (L12-N324, H360-L852, S863-R875, and H928-V1512), making it the most complete model of a monomeric ABCC protein to our knowledge.
In the structure, the ABC core of Ycf1p adopts the inward-facing conformation (Fig. 1 B and D and SI Appendix, Fig. S1B) seen for eukaryotic ABCB, ABCC, and ABCD transporters, as well as some bacterial exporters, when bound nucleotides are absent (5). TMD1 and TMD2 each contain six TM segments and are domain-swapped to form two TM bundles that contain residues from both halves of the ABC core (SI Appendix, Fig. S1B). TM bundle 1 is composed of TM helices 6, 7, 8, 11, 15, and 16, while TM bundle 2 is formed by TM helices 9, 10, 12, 13, 14, and 17. The TM domain swapping results in coupling helices 1 and 4 contacting NBD1 and coupling helices 2 and 3 contacting NBD2. The cytosolic NBDs are asymmetrically separated, similar to other ABCC transporters in the inward conformation (8). The inward conformation of Ycf1p also exposes a number of residues likely involved in glutathione binding (SI Appendix, Fig. S6) (22). In addition, several bound lipid molecules are found throughout the TM domains (SI Appendix, Fig. S7A), including one at the interface between TMD0 and TM bundle 1, which is modeled as phosphatidyl ethanolamine (SI Appendix, Fig. S7 B and C), a common lipid in yeast vacuolar membranes (35, 36).
High-Resolution Structure of TMD0 and the L0 Linker.
The hallmark of ABCC transporters is their N-terminal extension, which is either composed of TMD0 and an L0 linker or an L0 tail alone (2, 29). The N-terminal extension has various functions, depending on the ABCC protein (9, 10, 13, 15–17). For example, the N-terminal extensions of MRP2 (9) and CFTR (16) are involved in localization of the proteins to the plasma membrane, and trafficking of these ABCC proteins is regulated through interactions of the N-terminal extension with vesicle fusion machinery (16, 17). TMD0 and L0 in SUR1 (13) and MRP1 (10) mediate protein–protein interactions, and the MRP1 L0 linker can affect transport activity (15). In Ycf1p, TMD0 is required for vacuolar localization and is thought to form interactions with regulatory protein partners (37, 38). The Ycf1p L0 linker is essential for the protein’s role in cadmium resistance (37).
The structure of the Ycf1p TMD0 shows the domain’s five α-helices and N-terminal tail (2, 29) (Figs. 1 and 2, orange). The Ycf1p L0 linker adopts a lasso structure that consists of a membrane-embedded region (R207-S251), an amphipathic helix (S252-Q269) that packs against the elbow helix of TMD1, and an L0 linker/ABC core connecting segment (K270-S274) (Figs. 1 and 2A, tan) (5, 8). Although TMD0 in some ABCC proteins functions as a protein interaction module, no interacting proteins copurified with Ycf1p under the experimental conditions used here (SI Appendix, Fig. S2A).
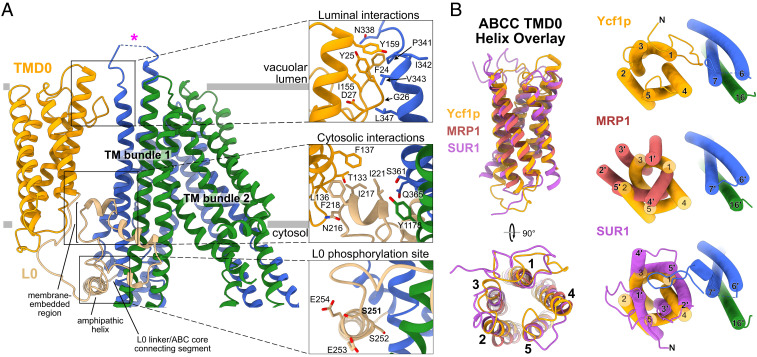
Fig. 2.
TMD0 and L0 interactions in Ycf1p and other ABCC proteins. (A) A Ycf1p ribbon diagram colored as in Fig. 1 (Left). Close-up views of the luminal interaction of TMD0 and TM bundle 1 (Top Right) and cytoplasmic interaction of TMD0, the L0 linker, and TM bundle 1 (Middle Right) are shown with interacting side chains (≤4 Å apart) rendered as sticks. A close-up view of the L0 linker phosphorylation site with side chains rendered as sticks is also shown (Bottom Right). (B) Overlay of TMD0 from Ycf1p, MRP1, and SUR1 (Left) shows that the structure of TMD0 is conserved among these different ABCC proteins. Overlay of the TMD0 (Right) from Ycf1p (shown in orange), MRP1 (shown in red), and SUR1 (shown in purple) after alignment of the ABC core demonstrates that the orientation of TMD0 differs in ABCC proteins. TM helix numbers in MRP1 (Middle Right) and SUR1 (Bottom Right) are denoted with a “´” to distinguish them from Ycf1p TM helices. Loops in TMD0 of Ycf1p are removed in each of the overlays for clarity.
The luminal and cytoplasmic sides of TMD0 contact TM bundle 1 and the L0 linker, respectively, to tether TMD0 to the rest of Ycf1p. On the luminal side of the membrane (Fig. 2 A, Top Right), residues in the N-terminal tail and TM4 in TMD0 contact residues in TM6 and loop 6 of TMD1. Loop 6, which connects TM6 and TM7 in TMD1, contains the Pep4p protease digestion site (34). While most residues in loop 6 are not resolved in the structure, it is possible that this loop interacts transiently with TMD0 and that these interactions change upon proteolytic digestion. In support of this hypothesis, loop 6 in SUR1 contacts TMD0 in some KATP channel structures (18, 39, 40) but is not observed in other KATP channel structures (41–44), likely due to flexibility. On the cytoplasmic side of the membrane, there is a network of interactions formed by TM4 and TM5 in TMD0, the membrane-embedded portion of the L0 lasso structure, the L0 amphipathic helix, and residues in TM6, TM7, TM15, and TM16 in TM bundle 1 (Fig. 2 A, Center and Bottom Right). Notably, TMD0 contacts two regulatory elements in Ycf1p—loop 6 that can be proteolytically cleaved and the L0 linker that contains an inhibitory phosphorylation site—suggesting that the TMD0 interactions may change with different transporter states.
The lasso structure of the L0 linker has implications for Ycf1p activity. MS indicated limited phosphorylation at the S251 inhibitory phosphorylation site, and there is no density in the cryo-EM map corresponding to a phosphate at S251, although the side chain is clearly resolved. Cryo-EM maps show the average structure from the contributing particle images, and together these two observations suggest that the L0 linker is not extensively phosphorylated in our Ycf1p preparation. Notably, residue S251 in the Ycf1p L0 linker is 70% buried within the protein (Fig. 2 A, Bottom Right), making it inaccessible to kinases in this conformation. Furthermore, the side chains of two Glu residues C-terminal to S251 that are thought to be involved in recognition by casein kinase II (CKII) (45), the in vivo kinase for phosphorylation of S251 (33, 46), are also buried. Therefore, accessibility of S251 and the CKII recognition sites requires a conformational change in the L0 linker, which may occur upon proteolytic cleavage or binding of nucleotide. This sort of rearrangement of the L0 linker upon nucleotide binding has been seen in other ABC proteins (41). Because the L0 linker binds the R region (see below), phosphorylation of the R region could also affect the accessibility of S251. Thus, it is possible that the phosphorylation of the L0 linker at S251 may fine tune the Ycf1p activity that results from nucleotide binding and hydrolysis, R region phosphorylation, or proteolytic digestion (28, 37). This putative interplay between different regulatory sites could also occur in other ABCC proteins, such as MRP1, which is similarly phosphorylated at the L0 linker and R region (47, 48).
Variable Association with the ABC Core Suggests How TMD0 Modulates Function in ABC Proteins.
Although TMD0 helices could not be assigned unambiguously and registered in the earlier cryo-EM map of MRP1 (22), fitting of the Ycf1p TMD0 model into the MRP1 density map confirms the identity of the TM helices in MRP1 (SI Appendix, Fig. S8B). A comparison of the TMD0 structures from Ycf1p, MRP1 (22), and SUR1 (39) shows that the domain adopts the same fold in all three proteins (Fig. 2 B, Left). The RMSDs between Cα atoms from Ycf1p and MRP1 TMD0, Ycf1p and SUR1 TMD0, and MRP1 and SUR1 TMD0 are 0.80, 0.89, and 0.96 Å, respectively. Despite this similarity, the orientation of TMD0 relative to the ABC core differs dramatically in the three proteins (Fig. 2 B, Right). The most striking difference is between Ycf1p and SUR1, with a ∼145° rotation of TMD0 between the structures (Fig. 2 B, Bottom Right). In SUR1, unlike Ycf1p, residues in TM2 from TMD0 mediate interactions with TM bundle 1 on the extracellular side of the membrane domains (equivalent to the vacuolar lumen side in Ycf1p), and residues in TM2 and TM3 contact the L0 linker on the cytoplasmic side (19, 20). Another consequence of the different orientation of TMD0 is that residues N-terminal to TM1 in SUR1 contact the adjacent Kir6.2 subunit in the KATP channel complex, while they contact the ABC core in Ycf1p.
The association of TMD0 with the ABC core also differs between Ycf1p and its homolog MRP1 (Fig. 2 B, Middle Right, and SI Appendix, Fig. S8A). While there is no relative rotation of TMD0 between MRP1 and Ycf1p, TMD0 of MRP1 projects away from the ABC core at the extracellular side of the membrane, with distances between backbone atoms in TM4 of TMD0 and TM7 of TM bundle 1 of 16.5 Å in MRP1 versus 9.7 Å in Ycf1p. As a result, there are no observed interactions of helical residues in TMD0 with the ABC core on the extracellular side of MRP1, while these interactions occur in Ycf1p (Fig. 2 A, Top Right). However, direct contacts between TMD0 and TM bundle 1 in MRP1 may be formed by loops in TMD0 or the N-terminal sequence of TMD0. Indeed, in the MRP1 map, there is a previously unidentified region of weak density between TMD0 and TMD1 that, from fitting the Ycf1p model of TMD0, appears to correspond to the N-terminal residues of MRP1 TMD0 (SI Appendix, Fig. S8B). Thus, both the Ycf1p model and the published MRP1 map suggest that N-terminal residues are important for TMD0/ABC core interactions. Furthermore, the similarity in the orientation of TMD0 relative to TM bundle 1 in Ycf1p and MRP1 suggests that this orientation is preserved among MRP and MRP-like ABCC transporters.
The limited protein–protein interactions between TMD0 and the ABC core in Ycf1p suggest that TMD0 is attached flexibly to the ABC core. It is possible that interactions between TMD0 and the ABC core are altered in different functional states of the transporter, which could affect the orientation and/or dynamics of TMD0. Modest changes in TMD0 orientation are seen for SUR1 TMD0 in KATP channel structures under different nucleotide-bound states (41), and differences in TMD0 density in cryo-EM maps of MRP1 obtained at different points in its catalytic cycle imply that TMD0 dynamics can change (22–24). It is also possible that TMD0/ABC core contacts change upon binding of TMD0 to interacting proteins. Another possibility is that TMD0 adopts different orientations in Ycf1p, MRP1, and SUR1 in order to perform various functions in these ABCC proteins. For example, while TMD0 in Ycf1p and a related MRP (MRP2) are required for efficient trafficking of these ABCC proteins (9, 37), SUR1 TMD0 mediates interactions of this ABCC protein with the Kir6.2 pore (19, 20).
Regulatory (R) Region Interactions Explain Its Regulatory Role.
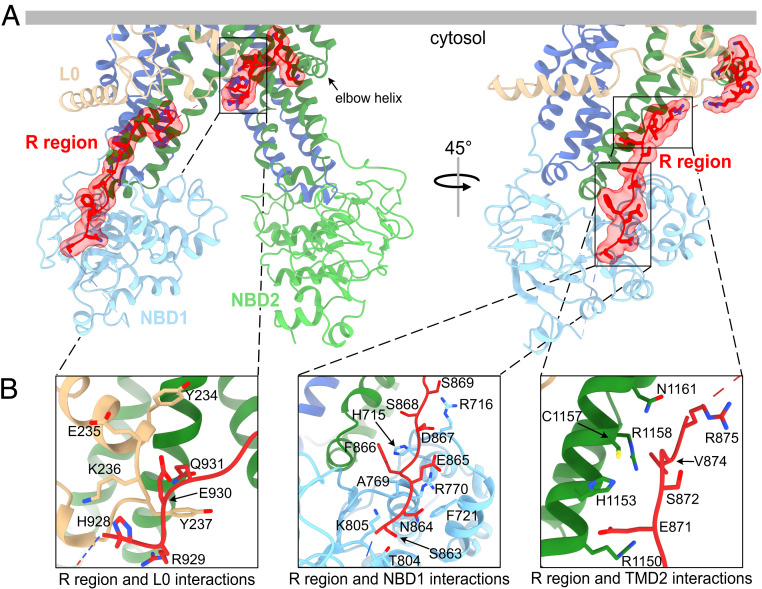
The linker between NBD1 and TMD2 is sometimes referred to as the regulatory (R) region in ABC proteins because its phosphorylation (48, 49) or sumoylation (50) modulates protein activity or cellular protein levels, respectively. In addition, the R region can serve as an interaction hub for associating proteins (30, 51–54). MS showed that the R region of Ycf1p is extensively phosphorylated, particularly at S903, S908, and T911 (SI Appendix, Fig. S2C). As with intrinsically disordered regions of other ABC proteins (3, 4), high-resolution density is absent for most of the R region in Ycf1p. However, the map contains high-resolution density for two segments of the R region (Fig. 3A, red). One of these R region segments consists of residues at the C-terminal end that lead into the elbow helix of TM bundle 2 (Fig. 3 A and B, Left). These Ycf1p residues adopt an extended structure and mediate interactions with the L0 linker and the cytoplasmic extension of TM15. The other well-resolved segment of the R region is located on the peripheral face of NBD1, opposite the ATP binding and NBD dimerization interfaces (Fig. 3 A, Right and Fig. 3 B, Center and Right). This R region segment interacts with residues in NBD1 and in the cytoplasmic extension of TM16. R region interactions in the Ycf1p structure are similar to those in structures of CFTR where the R region is phosphorylated and the NBDs are dimerized (55, 56). In contrast, structures of ABC proteins in which the R region is not phosphorylated and the NBDs are separated show the nonphosphorylated R region binding the NBDs in a manner that prevents dimerization (8, 57–59). As suggested by structural studies of CFTR (56), the Ycf1p structure supports a model in which phosphorylation causes the R region to bind the peripheral side of NBD1 rather than the NBD dimer interface, thus promoting NBD dimerization.
Fig. 3.
Ycf1p regulatory (R) region interacts with the ABC core and L0 linker. (A) The Ycf1p structure in two orientations with the resolved portions of the R region shown in red. (B) Close-up views of the interactions between the R region and the L0 linker (Left), the peripheral side of NBD1 (Center), and the cytoplasmic extensions in TMD2 (Right). Side chains of interacting residues, defined as those within 4 Å of the interacting domain, are shown as sticks.
It is possible that the other portions of the R region are not resolved because of flexibility but still interact transiently with other parts of Ycf1p, such as NBD1, as found for intrinsically disordered regions of other ABC proteins (30, 52, 60–62). To investigate this possibility, we measured tryptophan (Trp) fluorescence quenching with I– and acrylamide for isolated NBD1, NBD1 linked to the R region (NBD1-R), and NBD1-R containing the phosphorylation mimetic mutations S908D and T911E (NBD1-RS908D/T911E) (SI Appendix, Fig. S9A). Phosphorylation of S908 and T911, by protein kinase A, is important for Ycf1p activity, and the mutation of S908 and T911 to acidic residues has been shown to mimic the effect of phosphorylation (25, 28). As these proteins are expressed in Escherichia coli, which lacks protein kinase A, NBD1-R and NBD1-RS908D/T911E have no phosphorylated residues. Because Trp residues are only found in NBD1, these experiments probe changes in the exposure of Trp residues in Ycf1p NBD1 when the R region is present (63) (SI Appendix, Fig. S9B). The different Trp residues are found in different environments, and thus their fluorescence is quenched by different molecules (63). Residue W701 is surrounded primarily by positively charged residues, and thus is efficiently quenched by the negatively charged I–. However, W701 is also near hydrophobic residues, and hence can also be quenched by acrylamide. In contrast, W849 is surface exposed and adjacent to several hydrophobic amino acids on one side and negatively charged residues on the other side and is consequently quenched by acrylamide but not I–.
These fluorescence quenching studies (SI Appendix, Fig. S9 C and D) lead to three main observations. First, Stern–Volmer (KSV) constants for I– and acrylamide are lower for MgATP-bound NBD1, NBD1-R, and NBD1- RS908D/T911E compared to the apo proteins. Binding of MgATP at NBD1 alters the conformation of the γ-phosphate linker that contains W701 as well as the surface exposure of W849, which is located near the nucleotide binding Walker A and Walker B motifs (5). Thus, the decrease in I– and acrylamide quenching values reflects partial burial of W701 and W849 in the MgATP-loaded state. Second, KSV values obtained for I– and acrylamide quenching are smaller for NBD1-R and NBD1-RS908D/T911E than for NBD1 without the R region (SI Appendix, Fig. S9D), in both the absence and presence of MgATP. This observation indicates that the surface accessibilities of W701 and W849 are decreased by the presence of the R region, likely through binding of the R region to NBD1. While interactions of W701 or W849 with the R region are not observed in the Ycf1p map, the fluorescence quenching data suggest that residues outside the two well-resolved R region segments also contact NBD1, albeit transiently. Based on the location of W701, we surmise that additional contacts with NBD1 are mediated by residues in the large unresolved E876-E927 segment of the R region, which also contains the two stimulatory phosphorylation sites of Ycf1p. W849 likely interacts with residues in the N-terminal end of the R region (N853-S863) that are also not resolved in the Ycf1p map. Finally, there is a large difference in acrylamide quenching between the MgATP-loaded states of NBD1-R and NBD1- RS908D/T911E, whereas there is no change between NBD1 and NBD1-R. These results suggest that phosphorylated R region binds NBD1 near W849, whereas nonphosphorylated R region does not. Similarly, structural studies of phosphorylated CFTR show R region density on the peripheral side of NBD1 near the γ-phosphate linker and at the C-terminal end of NBD1 where W849 is located in Ycf1p (56). Notably, changes in interactions of regulatory disordered regions upon phosphorylation have been observed in other ABCC proteins (30, 52, 60–62).
Ycf1p Structure Reveals the Molecular Basis of Human Diseases.
The atomic-level insight into TMD0 and R region interactions in the Ycf1p model shed light on the underlying basis of disease-causing mutations in human MRPs, such as MRP2 and MRP6, for which this structural information is not available. Mutations in MRP2 (ABCC2) that affect transport of bilirubin glucuronides cause jaundice, choleostisis, and Dubin–Johnson syndrome (64). Mutations in MRP6 (ABCC6) manifest as PXE (65). Disease-causing mutations of MRP2 and MRP6 are found throughout the transporters (Fig. 4A; http://www.hgmd.cf.ac.uk/ac/index.php).
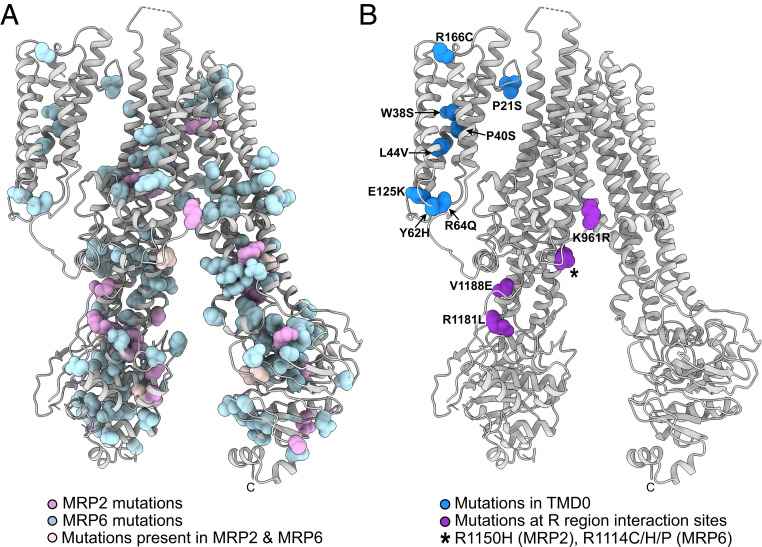
Fig. 4.
MRP2 and MRP6 disease mutations mapped onto the Ycf1p model. Ycf1p residues that are homologous to MRP2 and MRP6 residues mutated in disease are displayed as spheres and colored as follows. (A) MRP2 mutations that cause biliary cirrhosis, choleostasis, Dubin–Johnson syndrome, or drug resistance are shown in pink. MRP6 residues altered in PXE are shown in light blue. Disease-causing mutations of homologous residues in both MRP2 and MRP6 are shown in pale pink. (B) Disease-causing mutations of residues in TMD0 are shown in royal blue, while mutations at R region interaction sites are in purple. The specific mutation is listed next to the homologous Ycf1p residue. A black “*” indicates a conserved Arg residue that is altered in both MRP2 and MRP6 to cause disease, with the specific mutations indicated.
The Ycf1p structure shows the locations of residues that are mutated within TMD0 (Fig. 4B, royal blue). Several MRP6 missense mutations that cause PXE are located within TM helices in TMD0 and likely affect packing of individual helices in TMD0, destabilizing the domain. PXE-causing mutations in the N-terminal tail and in loop 3 of TMD0 may affect association of TMD0 with the luminal side of the ABC core and the L0 linker, respectively. Considering the small number of contacts between TMD0 and other regions of the transporter, even a single mutation could alter how TMD0 interacts with the rest of the ABC transporter. Destabilization of TMD0 and/or altered TMD0/TM helix bundle 1 contacts could have implications for MRP6 trafficking and protein–protein interactions due to this role for TMD0 in ABC proteins (9, 10, 12–14).
Other MRP2 and MRP6 mutations likely compromise R region contacts in the transporters (Fig. 4B, purple). These mutations are found in the second observed R region segment or in the cytoplasmic extensions of TM15 or TM16 that contact the R region. Several of these mutations replace Arg residues with hydrophobic residues or introduce charged residues at hydrophobic positions, which would alter interaction of the R region and TMD2. Other mutations substitute Arg residues with other positively charged amino acids, which subtly alters the charge distribution to affect protein–protein (66) and protein–lipid interactions (67). Cytoplasmic extensions of TM helices in TMD1 and TMD2, and their associated coupling helices (SI Appendix, Fig. S1), link ATP binding and hydrolysis at the NBDs to substrate transport. Thus, changes in how the R region contacts the cytoplasmic extensions of TMD1 and TMD2 could alter substrate transport.
The studies presented here help explain the function of Ycf1p and also give general insights into the ABCC family of proteins. Mapping of MRP mutations onto TMD0 and the R region of the Ycf1p structure suggests how these mutations cause disease. The variable interaction between TMD0 and the ABC core in different ABCC proteins suggests how this domain can serve different roles. The observed binding of the highly phosphorylated R region to the L0 linker and ABC core, in a manner that does not prevent NBD1/NBD2 dimerization, suggests how posttranslational modifications may allow the component parts of ABCC proteins to work together in diverse cellular contexts.
Materials and Methods
Construction of the Ycf1p-3×FLAG Yeast Strain.
The S. cerevisiae strain BJ2168 (MATa, leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407 gal2) was further modified by insertion of sequence encoding a C-terminal 3×FLAG tag, followed by a URA3 marker downstream of the YCF1 gene. The 3×FLAG tag and URA3 gene were amplified from the pJT1 plasmid (68) and integrated into the yeast chromosome by homologous recombination (69). The sequences of the forward and reverse primers used were 5 ′-AATCATTGTTCTATTCACTGTGCATGGAGGC-TGGTTTGGTCAATGAAAATGACTACAAAGACCATGACGG-3′ and 5′-CCATCCTAC-GTACCAGATTGTGCGGGACAGGTTTTTAT TAGTTTCACAGTTTATAATTGGCCAGT-TTTTTTCAAA-3′, respectively, where YCF1 nucleotides are underlined. The resulting PCR products were used to transform BJ2168 cells, with selection of transformants on minimal media lacking uracil. Successful integration of the sequence for the 3×FLAG tag and URA3 marker was confirmed by PCR using a primer corresponding to DNA 193 base pairs upstream from the last coding nucleotide of the YCF1 gene (5′-CGATTCGTACTGCTTTCAAG-3′) and a primer consisting of the reverse and complement of a sequence from within the inserted sequence (5′-GAGCGACCTCATACTATACC-3′). The resulting strain was named ySCB1.
Purification of Ycf1p.
The yeast strain ySCB1 was grown in 11-L yeast extract peptide dextrose (YPD) medium (20 g/L glucose, 20 g/L peptone, and 10 g/L yeast extract) supplemented with 100 µg/mL ampicillin and 0.02% antifoam at 30 °C in a New Brunswick BioFlow fermentor. Yeast was grown for ∼22 h to an optical density at 660 nm (OD660) of 2.0 to 6.0. Cells were harvested by centrifugation at 4,000 × g for 15 min at 4 °C and immediately resuspended in 1 mL/g lysis buffer (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, 80 g/L sucrose, 20 g/L sorbitol, 20 g/L glucose, 5 mM 6-aminocaproic acid, 5 mM benzamidine, 5 mM ethylenediaminetetraacetic acid [EDTA], and 10 mg/L phenylmethylsulfonyl fluoride [PMSF] at pH 7.4). The resuspended cells were lysed with a bead beater (Biospec), cellular debris was removed by centrifugation at 4,000 × g for 15 min at 4 °C, and membranes were collected by ultracentrifugation (Beckman L-90K) at 152,947 × g for 40 min at 4 °C. Membranes were resuspended in 0.5 mL lysis buffer per gram of harvested yeast cells using a Dounce homogenizer and stored at −80 °C prior to protein purification. Frozen membranes were thawed at room temperature and all further purification steps proceeded at 4 °C. Membranes were solubilized with 1% (wt/vol) n-dodecyl-β-D-maltopyranoside (DDM; Anatrace) with mixing for ∼1 h. Insoluble material was removed by ultracentrifugation at 181,078 × g for 70 min (Beckman L-90K). Supernatant containing Ycf1p was filtered with a 0.45-µm syringe filter (Pall) and applied to a 500-µL M2 affinity gel column (Millipore Sigma) preequilibrated with DTBS (50 mM Tris HCl, 150 mM NaCl, 0.04% [wt/vol] DDM, pH 8.0). The column was washed with 12 column volumes of DTBS, followed by 8 column buffers of GTBS (50 mM Tris HCl, 150 mM NaCl, 0.006% [wt/vol] glyco-diosgenin [GDN; Anatrace], pH 8.0). The Ycf1p was eluted with three column volumes of GTBS containing 150 µg/mL 3×FLAG peptide. An additional five column volumes of GTBS without 3×FLAG peptide was applied to wash residual Ycf1p from the column. Fractions containing Ycf1p were concentrated with a 100-kDa molecular weight cutoff (MWCO) Amicon Ultra centrifugal filter (Millipore Sigma) to a volume ∼100 µL and then applied to a Vivaspin 500 100-kDa MWCO concentrator and concentrated to ∼1.5 mg/mL for ATPase assays and ∼5.5 mg/mL for cryo-EM. Protein concentrations were determined by the bicinchoninic acid (BCA) assay.
ATPase activity of freshly purified Ycf1p in GTBS was measured at 30 °C as described previously (70, 71). Briefly, a reaction mixture was prepared containing 50 mM Hepes, 3 mM MgCl2, 0.006% (wt/vol) GDN, pH 8.0, ∼30 µg Ycf1p, 2 mM ATP, 1 mM phosphoenolpyruvate (PEP), 3.2 units of pyruvate kinase, 8 units of lactate dehydrogenase, and 200 µM NADH. The amount of NADH present in the assay was determined spectrophotometrically at 340 nm from a standard curve in the ATPase assay mixture.
MS Analysis of Ycf1p Phosphorylation.
Trypsin digestion of purified Ycf1p followed by liquid chromatography coupled to tandem MS (LC-MS/MS) was performed by SPARC BioCentre (Molecular Analysis), The Hospital for Sick Children. Briefly, purified Ycf1p (1.2 μg/μL) was reduced by incubation with 10 mM dithiothreitol (DTT) at 60 °C for 1 h, alkylated by incubation with 20 mM iodoacetate at room temperature for 45 min in the dark, and subsequently digested with trypsin (1.5 μg) at 30 °C overnight. The digested Ycf1p (0.47 μg) sample was subjected to LC-MS/MS. MS raw files were analyzed using PEAKS Studio software (Bioinformatics Solutions Inc.) and X! Tandem Alanine as part of the Scaffold software suite. Fragment lists were searched against the S. cerevisiae database plus the sequence of Ycf1p-3×FLAG provided. The parent and fragment mass tolerances were set to 10 ppm and 0.02 Da, respectively, and only tryptic peptides with a maximum of two missed cleavage sites were included. The addition of a carbamidomethyl group on Cys was specified as a fixed modification, while deamidation of Asn and Gln, oxidation of Met, acetylation of the N terminus, and phosphorylation of Ser, Thr, or Tyr were set as variable modifications. The ratio of the number of fragments detected from phosphorylated versus nonphosphorylated peptides, when the total number of fragments is high, approximately indicates the extent of phosphorylation, with a large ratio (e.g., 2.5) indicating extensive phosphorylation and a small ratio (e.g., 0.1) indicating limited phosphorylation.
Cryo-EM and Image Processing.
Nanofabricated holey gold grids were prepared as described previously (72). Grids were glow discharged in air (PELCO easiGLOW) for 2 min immediately before use. A 3-µL sample of the purified Ycf1p at ∼4 to 7 mg/mL was applied in a modified Vitrobot grid preparation robot (FEI Company) at ∼100% relative humidity and 4 °C and equilibrated for 20 s before blotting both grid sides for 28 s and plunge freezing in a liquid ethane-propane mixture (73). During optimization of cryo-EM grids, specimens were screened at 200 kV with an FEI Tecnai F20 electron microscope equipped with a Gatan K2 Summit camera. High-resolution data were collected at 300 kV with a Titan Krios G3 electron microscope (Thermo Fisher Scientific), equipped with a prototype Falcon4 camera. Movies were recorded for 9.6 s with a calibrated pixel size of 1.03 Å, a total exposure of ∼45 e−/Å2, and an exposure rate of 5 e−/pixel/s. A dataset of 4,084 movies was collected with EPU software, and data collection was monitored with cryoSPARC Live (74). All image processing was performed with cryoSPARC v.2. Patch motion correction and contrast transfer function (CTF) estimation were performed, followed by particle selection by template matching with a circular template and individual particle motion correction. A total of 2,789,383 particle images were used for two-dimensional classification. Subsequent ab initio structure determination, three-dimensional classification, and nonuniform refinement led to a final map at 3.2 Å resolution from 124,864 particle images.
Model Building and Validation.
An initial model of Ycf1p based on the known structure of bovine MRP1 [5UJ9 (22)] was generated with Phenix (75, 76). Coot (77) was used for manual atomic model building, with real-space refinement in Phenix (75, 76). The TMD0 domain, the observed segments of the R region, and several intracellular and extracellular loops were built de novo. The structure of TMD0 was determined by building TM helices of TM1, TM4, and TM5 first, followed by the N-terminal tail, TM2 and TM3, and the TMD0 loops. Loop 5, which connects TMD0 to the L0 linker, and for which only weak density is observed, was built into the model based on ideal backbone Ramachandran angles. The first observed segment of the R region was built based on the side chains of F866, E871, S872, and V874. Homology models of NBD1 and NBD2 were generated in Modeller (78) based on a structure-based sequence alignment of NBD1 or NBD2, respectively, from ABCC proteins. The homology model of NBD1 was generated using the structures of NBD1 from MRP1 [Protein Data Bank, PDB ID: 2CBZ (79)], ABCC6 (PDB ID: 6NL0), and hamster SUR1 [PDB ID: 6JB1 (40)]. The homology model of Ycf1p NBD2 was generated using the structures of NBD2 from MRP1 [PDB ID: 6BHU (23)] and ABCC6 (PDB: 6BZR). The NBD1 and NBD2 homology models were fit into the low-density NBD regions that possessed defined secondary structure. Phospholipids were added and refined with Coot. Data collection, image processing, and model statistics are in SI Appendix, Table S1.
All figures were generated with UCSF ChimeraX (80) and map-in-model fits with USCF Chimera (81). Structure comparisons were done with the ChimeraX matchmaker tool using the default Needleman-Wunsch BLOSUM-62 matrix. RMSD values were determined comparing Cα positions using the Match > Align tool in UCSF Chimera. Electrostatic surface representation of NBD1-R was generated in MolMol (82).
NBD1, NBD1-R, and NBD1-RS908D/T911E Expression and Purification.
Ycf1p NBD1 (D610-N859), NBD1-R (D610-E930), and NBD1-RS908D/T911E, which contains the phosphorylation mimetic mutations S908D and T911E, were expressed and purified as described previously for other NBDs (62, 63, 83). Briefly, Ycf1p NBD1, NBD1-R, and NBD1-RS908D/T911E were expressed as N-terminal His6-SUMO fusion proteins in E. coli LOBSTR-BL21 (DE3) RIL (Kerafast Inc) using a pET26b-derived expression vector (84). Cell cultures were grown in 1-L of 97.5% M9 minimal media and 2.5% lysogeny broth (LB) media at 37 °C to an OD600 of 0.4 to 0.5, at which point the temperature was gradually lowered so that the OD600 was 0.8 when the temperature reached 18 °C. Cell cultures were incubated at 18 °C for 30 min and gene expression was subsequently induced with 0.75 mM isopropyl β-D-thiogalactoside. Cells were harvested by centrifugation 18 to 20 h postinduction and stored at −20 °C until purification.
All purification steps were conducted at 4 °C. Cell pellets were incubated on ice for 30 min and then resuspended in 15 mL lysis buffer per 1 L of cell culture (20 mM Tris HCl, 10% glycerol, 150 mM NaCl, 5 mM imidazole, 100 mM arginine, 2 mM β-mecraptoethanol, 15 mM MgCl2, 15 mM ATP, 0.2% [vol/vol] Triton X-100, 1 mg/mL lysozyme, 2 mg/mL deoxycholic acid (DCA), 5 mM benzamidine, 5 mM ε-amino n-caproic acid, and 1 mM PMSF, pH 7.6). A trace amount of DNase I was added, and the solution was incubated for 15 min prior to sonication (Misonix Microson Ultrasonic Cell Disruptor). The soluble and insoluble fractions were separated by centrifugation at 17,000 g for 30 min. The lysate was then filtered using a 0.45 µM Acrodisic syringe filter (Pall) and applied to a 5 mL nickel-nitrilotriacetic acid (Ni2+-NTA) affinity column (GE Healthcare) equilibrated with 20 mM Tris HCl, 10% (vol/vol) glycerol, 150 mM NaCl, and 5 mM imidazole, pH 7.6. The column was then washed with five column volumes of the equilibration buffer that also contained 5 mM MgCl2 and 5 mM ATP. The bound His6-SUMO-NBD1, His6-SUMO-NBD1-R, or His6-SUMO-NBD1-RS908D/T911E fusion proteins were eluted with five column volumes of elution buffer (20 mM Tris HCl, 10% [vol/vol] glycerol, 150 mM NaCl, 500 mM imidazole, 15 mM MgCl2, 15 mM ATP, 1 mM benzamidine, and 1 mM ε-amino n-caproic acid). The elution fractions were immediately diluted threefold with elution buffer lacking imidazole and NaCl and dialyzed overnight against 20 mM Tris, 10% (vol/vol) glycerol, 150 mM NaCl, 2 mM β-mercaptoethanol, 2 mM MgCl2, 2 mM ATP, 1 mM benzamidine, and 1 mM ε-amino n-caproic acid, pH 7.6. His6-Ulp1 protease (∼1.8 mg) was added to the dialysis bag in order to remove the His6-SUMO tag from NBD1 (or NBD1-R or NBD1-RS908D/T911E). The resulting solution, containing His6-SUMO and NBD1 (or NBD1-R or NBD1-RS908D/T911E) proteins, was loaded onto a Co2+-agarose affinity column (Thermo Scientific), which was preequilibrated in 20 mM Tris, 10% (vol/vol) glycerol, 150 mM NaCl, 10 mM imidazole, 10 mM MgCl2, and 10 mM ATP, pH 7.6, to isolate NBD1 from the His6-SUMO tag. Fractions containing Ycf1p NBD1 or NBD1-R were applied to a size exclusion column (Superdex 75, GE Healthcare) in 20 mM Na3PO4, 150 mM NaCl, 2% (vol/vol) glycerol, 2 mM DTT, 2 mM MgCl2, 2 mM ATP, 100 µM benzamidine, and 100 µM ε-amino n-caproic acid, pH 7.3, to purify the protein to homogeneity. All protein concentrations were determined by the Bradford protein assay (85).
Trp Fluorescence Quenching Experiments.
Fluorescence quenching experiments were performed on 2 μM Ycf1p NBD1, NBD1-R, and NBD1-RS908D/T911E. Purified Ycf1p NBD1, NBD1-R, and NBD1-RS908D/T911E were exchanged into the fluorescence buffer (20 mM Na3PO4, 2% [vol/vol] glycerol, and 150 mM NaCl, pH 7.3) by size exclusion chromatography (Superdex75, GE Healthcare). MgATP-bound and apo NBD1, NBD1-R, and NBD1-RS908D/T911E were generated by adding 2 mM MgATP or 2 mM EDTA, respectively, to the protein eluents. Fresh reductant (2 mM DTT) was added to quenching solutions. Fluorescence studies were conducted on a Fluoromax-4 fluorimeter (Horiba Scientific) equipped with a Peltier unit for temperature control. Trp emission spectra (310 to 450 nm) were recorded at 15 °C with an excitation wavelength of 298 nm, an excitation slit width of 2 nm, and an emission slit width of 5 nm.
Iodine (I–) quenching experiments were conducted by successive additions of 0.8 M KI dissolved in the fluorescence buffer. The quenching solution was prepared fresh and contained 0.2 mM Na2SO3 to prevent formation of I2 and I3− (86). A parallel titration using 0.8 M KCl was conducted to account for sample dilution and changes in ionic strength during the titration. The initial fluorescence (F○) at each titration point was divided by the fluorescence (F) to give (F○/F)KI or (F○/F)KCl. The change in fluorescence, (F○/F)KI/(F○/F)KCl, was plotted against the concentration of I− to determine the KSV constant (87). Acrylamide quenching experiments were performed by successive addition of a solution containing 1.6 M acrylamide in the fluorescence buffer. The absorbances at 298 (A298) and 352 nm (A352) were measured in order to correct the observed fluorescence for the inner filter effect using the equation: (88, 89). The values of A298 and A352 at the highest acrylamide concentration used were 0.045 and 0.010, respectively. To correct for sample dilution upon addition of acrylamide, parallel titrations with buffer were performed. The normalized fluorescence (F) at each titration point was determined by dividing Fcorrected by the fluorescence of NBD1 proteins obtained from dilution of the sample (Fdilution) so that F = Fcorrected/ Fdilution, and the normalized fluorescence data were analyzed with the Stern–Volmer equation, as described above.
Supplementary Material
Acknowledgments
We thank Stephanie Bueler with assistance in generating the ySCB1 yeast strain and Thamiya Vasanthakumar and Zev Ripstein for helpful discussions regarding ATPase assays and computation, respectively. S.C.B. was supported by a Canada Graduate Scholarship–Master’s Program scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Sarah Cusick Gallop and William George Gallop Memorial Scholarship. J.L.R. was supported by the Canada Research Chairs program. The research was supported by NSERC Grants RGPIN-2015-05372 (to V.K.) and RGPIN-2020-05835 (to V.K.), a Heart and Stroke Foundation of Canada Grant G-18-0022076 (to V.K.), and a Canadian Institutes of Health Research Grant PJT162186 (to J.L.R.). Cryo-EM data were collected at the Toronto High-Resolution High-Throughput cryo-EM facility, supported by the Canada Foundation for Innovation and Ontario Research Fund.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025853118/-/DCSupplemental.
Data Availability
The EM map has been deposited in the Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb/ (EMDB accession code: EMD-23932). The coordinates have been deposited in the Protein Data Bank, http://www.wwpdb.org (PDB ID code: 7MPE).
References
- 1.Vasiliou V., Vasiliou K., Nebert D. W., Human ATP-binding cassette (ABC) transporter family. Hum. Genomics 3, 281–290 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean M., Rzhetsky A., Allikmets R., The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 11, 1156–1166 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Bickers S. C., Sayewich J. S., Kanelis V., Intrinsically disordered regions regulate the activities of ATP binding cassette transporters. Biochim. Biophys. Acta Biomembr. 1862, 183202 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ford R. C., Marshall-Sabey D., Schuetz J., Linker domains: Why ABC transporters ‘live in fragments no longer’. Trends Biochem. Sci. 45, 137–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas C., Tampé R., Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Davidson A. L., Dassa E., Orelle C., Chen J., Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72, 317–364 (2008), table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad R., Banerjee A., Khandelwal N. K., Dhamgaye S., The ABCs of Candida albicans multidrug transporter Cdr1. Eukaryot. Cell 14, 1154–1164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Chen J., Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell 167, 1586–1597.e9 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Bandler P. E., Westlake C. J., Grant C. E., Cole S. P. C., Deeley R. G., Identification of regions required for apical membrane localization of human multidrug resistance protein 2. Mol. Pharmacol. 74, 9–19 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Yang Y., et al., Regulation of function by dimerization through the amino-terminal membrane-spanning domain of human ABCC1/MRP1. J. Biol. Chem. 282, 8821–8830 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Cole S. P. C., Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J. Biol. Chem. 289, 30880–30888 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss K., et al., Role of the N-terminal transmembrane domain in the endo-lysosomal targeting and function of the human ABCB6 protein. Biochem. J. 467, 127–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan F.-F., et al., Congenital hyperinsulinism associated ABCC8 mutations that cause defective trafficking of ATP-sensitive K+ channels: Identification and rescue. Diabetes 56, 2339–2348 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., et al., Cryo-electron microscopy structure of human ABCB6 transporter. Protein Sci. 29, 2363–2374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakos E., et al., Functional multidrug resistance protein (MRP1) lacking the N-terminal transmembrane domain. J. Biol. Chem. 273, 32167–32175 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Naren A. P., Quick M. W., Collawn J. F., Nelson D. J., Kirk K. L., Syntaxin 1A inhibits CFTR chloride channels by means of domain-specific protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 95, 10972–10977 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters K. W., Qi J., Johnson J. P., Watkins S. C., Frizzell R. A., Role of snare proteins in CFTR and ENaC trafficking. Pflugers Arch. 443 (suppl. S1), S65–S69 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Martin G. M., et al., Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. eLife 6, e24149 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin G. M., Sung M. W., Shyng S. L., Pharmacological chaperones of ATP-sensitive potassium channels: Mechanistic insight from cryoEM structures. Mol. Cell. Endocrinol. 502, 110667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J. X., Ding D., Wang M., Chen L., Structural insights into the inhibitory mechanism of insulin secretagogues on the pancreatic ATP-sensitive potassium channel. Biochemistry 59, 18–25 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Oldham M. L., Grigorieff N., Chen J., Structure of the transporter associated with antigen processing trapped by herpes simplex virus. eLife 5, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson Z. L., Chen J., Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085.e9 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Johnson Z. L., Chen J., ATP binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang L., et al., Characterization of the kinetic cycle of an ABC transporter by single-molecule and cryo-EM analyses. eLife 9, 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szczypka M. S., Wemmie J. A., Moye-Rowley W. S., Thiele D. J., A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269, 22853–22857 (1994). [PubMed] [Google Scholar]
- 26.Li Z.-S., Szczypka M., Lu Y.-P., Thiele D. J., Rea P. A., The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 271, 6509–6517 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Tommasini R., et al., The human multidrug resistance-associated protein functionally complements the yeast cadmium resistance factor 1. Proc. Natl. Acad. Sci. U.S.A. 93, 6743–6748 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eraso P., Martínez-Burgos M., Falcón-Pérez J. M., Portillo F., Mazón M. J., Ycf1-dependent cadmium detoxification by yeast requires phosphorylation of residues Ser908 and Thr911. FEBS Lett. 577, 322–326 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Paumi C. M., Chuk M., Snider J., Stagljar I., Michaelis S., ABC transporters in Saccharomyces cerevisiae and their interactors: New technology advances the biology of the ABCC (MRP) subfamily. Microbiol. Mol. Biol. Rev. 73, 577–593 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker J. M. R., et al., CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 14, 738–745 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulhanty A. M., Riordan J. R., Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry 33, 4072–4079 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Ostedgaard L. S., Baldursson O., Vermeer D. W., Welsh M. J., Robertson A. D., A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc. Natl. Acad. Sci. U.S.A. 97, 5657–5662 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paumi C. M., Chuk M., Chevelev I., Stagljar I., Michaelis S., Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J. Biol. Chem. 283, 27079–27088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason D. L., Mallampalli M. P., Huyer G., Michaelis S., A region within a lumenal loop of Saccharomyces cerevisiae Ycf1p directs proteolytic processing and substrate specificity. Eukaryot. Cell 2, 588–598 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hossack J. A., Spencer-Martins I., Lipid composition of the yeast–Lipomyces kononenkoae. Eur. J. Appl. Microbiol. Biotechnol. 5, 273–278 (1978). [Google Scholar]
- 36.Lindberg L., Santos A. X. S., Riezman H., Olsson L., Bettiga M., Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PloS One 8, e73936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason D. L., Michaelis S., Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol. Biol. Cell 13, 4443–4455 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma K. G., et al., Localization, regulation, and substrate transport properties of Bpt1p, a Saccharomyces cerevisiae MRP-type ABC transporter. Eukaryot. Cell 1, 391–400 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin G. M., et al., Mechanism of pharmacochaperoning in a mammalian KATP channel revealed by cryo-EM. eLife 8, e46417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding D., Wang M., Wu J. X., Kang Y., Chen L., The structural basis for the binding of repaglinide to the pancreatic KATP channel. Cell Rep. 27, 1848–1857.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Lee K. P. K., Chen J., MacKinnon R., Molecular structure of human KATP in complex with ATP and ADP. eLife 6, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N., et al., Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Martin G. M., Kandasamy B., DiMaio F., Yoshioka C., Shyng S. L., Anti-diabetic drug binding site in a mammalian KATP channel revealed by cryo-EM. eLife 6, 1–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J. X., et al., Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. Protein Cell 9, 553–567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litchfield D. W., Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 369, 1–15 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pickin K. A., et al., Suppression of Ycf1p function by Cka1p-dependent phosphorylation is attenuated in response to salt stress. FEMS Yeast Res. 10, 839–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stolarczyk E. I., et al., Casein kinase 2α regulates multidrug resistance-associated protein 1 function via phosphorylation of Thr249. Mol. Pharmacol. 82, 488–499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shukalek C. B., et al., Arsenic triglutathione [As(GS)3] transport by multidrug resistance protein 1 (MRP1/ABCC1) is selectively modified by phosphorylation of Tyr920/Ser921 and glycosylation of Asn19/Asn23. Mol. Pharmacol. 90, 127–139 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Cheng S. H., et al., Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell 66, 1027–1036 (1991). [DOI] [PubMed] [Google Scholar]
- 50.Minami S., et al., Posttranslational regulation of Abcc2 expression by SUMOylation system. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G406–G413 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Ambadipudi R., Georges E., Sequences in Linker-1 domain of the multidrug resistance associated protein (MRP1 or ABCC1) bind to tubulin and their binding is modulated by phosphorylation. Biochem. Biophys. Res. Commun. 482, 1001–1006 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Bozoky Z., et al., Regulatory R region of the CFTR chloride channel is a dynamic integrator of phospho-dependent intra- and intermolecular interactions. Proc. Natl. Acad. Sci. U.S.A. 110, E4427–E4436 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bozoky Z., et al., Synergy of cAMP and calcium signaling pathways in CFTR regulation. Proc. Natl. Acad. Sci. U.S.A. 114, E2086–E2095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang X., et al., Phosphorylation-dependent 14-3-3 protein interactions regulate CFTR biogenesis. Mol. Biol. Cell 23, 996–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F., et al., Structural identification of a hotspot on CFTR for potentiation. Science 364, 1184–1188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z., Liu F., Chen J., Molecular structure of the ATP-bound, phosphorylated human CFTR. Proc. Natl. Acad. Sci. U.S.A. 115, 12757–12762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu F., Zhang Z., Csanády L., Gadsby D. C., Chen J., Molecular structure of the human CFTR ion channel. Cell 169, 85–95.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Fay J. F., et al., Cryo-EM visualization of an active high open probability CFTR anion channel. Biochemistry 57, 6234–6246 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Thonghin N., et al., Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 Å resolution. BMC Struct. Biol. 18, 17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Araujo E. D., et al., Phosphorylation-dependent changes in nucleotide binding, conformation, and dynamics of the first nucleotide binding domain (NBD1) of the sulfonylurea receptor 2B (SUR2B). J. Biol. Chem. 290, 22699–22714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sooklal C. R., López-Alonso J. P., Papp N., Kanelis V., Phosphorylation alters the residual structure and interactions of the regulatory L1 linker connecting NBD1 to the membrane-bound domain in SUR2B. Biochemistry 57, 6278–6292 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Kanelis V., Hudson R. P., Thibodeau P. H., Thomas P. J., Forman-Kay J. D., NMR evidence for differential phosphorylation-dependent interactions in WT and DeltaF508 CFTR. EMBO J. 29, 263–277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez C. P., Stagljar M., Muhandiram D. R., Kanelis V., Hyperinsulinism-causing mutations cause multiple molecular defects in SUR1 NBD1. Biochemistry 56, 2400–2416 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Nies A. T., Keppler D., The apical conjugate efflux pump ABCC2 (MRP2). Pflugers Arch. 453, 643–659 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Borst P., Váradi A., van de Wetering K., PXE, a mysterious inborn error clarified. Trends Biochem. Sci. 44, 125–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sokalingam S., Raghunathan G., Soundrarajan N., Lee S. G., A study on the effect of surface lysine to arginine mutagenesis on protein stability and structure using green fluorescent protein. PloS One 7, e40410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L., Vorobyov I., Allen T. W., The different interactions of lysine and arginine side chains with lipid membranes. J. Phys. Chem. B 117, 11906–11920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benlekbir S., Bueler S. A., Rubinstein J. L., Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat. Struct. Mol. Biol. 19, 1356–1362 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Finnigan G. C., Thorner J., Complex in vivo Ligation using homologous recombination and high-efficiency plasmid rescue from Saccharomyces cerevisiae. Bio Protoc. 5, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kornberg A., Pricer W. E. Jr, Enzymatic phosphorylation of adenosine and 2,6-diaminopurine riboside. J. Biol. Chem. 193, 481–495 (1951). [PubMed] [Google Scholar]
- 71.Vasanthakumar T., et al., Structural comparison of the vacuolar and Golgi V-ATPases from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 116, 7272–7277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marr C. R., Benlekbir S., Rubinstein J. L., Fabrication of carbon films with ∼ 500nm holes for cryo-EM with a direct detector device. J. Struct. Biol. 185, 42–47 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Tivol W. F., Briegel A., Jensen G. J., An improved cryogen for plunge freezing. Microsc. Microanal. 14, 375–379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Adams P. D., et al., Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liebschner D., et al., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2 9 1–2 9 37 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Ramaen O., et al., Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J. Mol. Biol. 359, 940–949 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Goddard T. D., et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Koradi R., Billeter M., Wuthrich K., MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29-32 (1996). [DOI] [PubMed] [Google Scholar]
- 83.de Araujo E. D., Kanelis V., Successful development and use of a thermodynamic stability screen for optimizing the yield of nucleotide binding domains. Protein Expr. Purif. 103, 38–47 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Mossessova E., Lima C. D., Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000). [DOI] [PubMed] [Google Scholar]
- 85.Bradford M. M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- 86.Lehrer S. S., Solute perturbation of protein fluorescence. The quenching of the tryptophyl fluorescence of model compounds and of lysozyme by iodide ion. Biochemistry 10, 3254–3263 (1971). [DOI] [PubMed] [Google Scholar]
- 87.Nikolaeva O., Moiseyev G., Rodgers K. K., Ma J. X., Binding to lipid membrane induces conformational changes in RPE65: Implications for its isomerohydrolase activity. Biochem. J. 436, 591–597 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Liu R., Sharom F. J., Fluorescence studies on the nucleotide binding domains of the P-glycoprotein multidrug transporter. Biochemistry 36, 2836–2843 (1997). [DOI] [PubMed] [Google Scholar]
- 89.Lakowicz J. R., Principles of Fluorescence Spectroscopy (Springer, New York, ed. 3, 2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The EM map has been deposited in the Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb/ (EMDB accession code: EMD-23932). The coordinates have been deposited in the Protein Data Bank, http://www.wwpdb.org (PDB ID code: 7MPE).