Abstract
Cancer cell spheroids have been shown to be more physiologically relevant to native tumor tissue than monolayer 2D culture cells. Due to enhanced intercellular communications among cells in spheroids, spheroid secreted exosomes which account for transcellular transportation should exceed those from 2D cell culture and may display a different expression pattern of miRNA or protein. To test this, we employed a widely used pancreatic cancer cell line, PANC-1, to create 3D spheroids and compared exosomes generated by both 2D cell culture and 3D PANC-1 spheroids. We further measured and compared exosomal miRNA and GPC-1 protein expression with qRT-PCR and enzyme-linked immunosorbent assay, respectively. It showed that PANC-1 cells cultured in 3D spheroids can produce significantly more exosomes than PANC-1 2D cells and exosomal miRNA and GPC-1 expression derived from spheroids show more features relevant to the progression of pancreatic cancer. These findings point to the potential importance of using spheroids as in vitro model to study cancer development and progression.
Keywords: Spheroids, Exosome, Pancreatic cancer, PANC-1, miRNA, GPC-1
1. Introduction
Cancer cell lines grown in conventional 2D systems are the most commonly used cancer models and have contributed tremendously to the understanding of the mechanisms governing cancer cell growth and development. However, an increasing body of recent evidence shows that 2D culture cancer cells fairly poorly reflect native tumor tissue [1,2]. Though the possible reasons vary, one of commonly believed rationales is that tumor microenvironment in 2D culture system is not represented in terms of cell–cell and cell–extracellular matrix interactions [3]. In vitro 3D culture technologies, such as spheroids, have opened potential new avenues for developing more physiological human/mouse cancer models and represented an important bridge between traditional 2D cultures and in vivo mouse/human models [1,4,5].
Exosomes, small extracellular vesicles of endocytic origin with a size ranging from 40 to 150 nm [6], have now come to be accepted as ubiquitous mediators in intercellular communication [7,8]. In particular, cancer cells can produce more exosomes than normal cells [9,10], and exosomes derived from cancer cells have a high capacity to mediate cell-to-cell communication [11] and modulate local and distant microenvironments, through horizontal transfer of microRNAs (miRNAs) [9,12,13] as well as other molecules such as proteins [14] or mRNAs [13].
Cancer spheroids, three-dimensional aggregates of cancer cells, are more physiologically relevant to native tumor tissue than monolayer 2D culture models [15]. Therefore, it is reasonable to hypothesize that cancer spheroids should secrete more exosomes than 2D culture cells due to the increased cell-cell contact, and exosomes derived from cancer spheroids may display different properties in terms of miRNA or protein expression. Indeed, in the last few years it has already been shown that tumor cells growing in 3D conditions can recapitulate some properties of exosomes found in native tumors [[16], [17], [18]]. Importantly, enhanced exosome secretion could be observed in 3D spheroid culture using cancer cells compared with 2D cells culture [19,20], and 3D culture derived exosomes showed significantly different RNA expression profiles compared to 2D culture derived exosomes [19]. However, there is little research on the correlation between exosomal miRNA and exosomal protein and comparisons between conventional 2D culture and 3D culture. Considering that 3D cell culture is more recognized for representing an in vivo like tissue microenvironment [21], therefore it is particularly valuable to analyze the differential expression of miRNAs and protein in exosomes and their corresponding parental cells in 3D conditions.
To assess whether cancer spheroids can generate more exosomes than 2D culture cancer cells, we used PANC-1 cells [22], a most widely used pancreatic cancer cell line, to create 3D spheroids, isolated and quantified exosomes from both 2D cell culture and 3D PANC-1 spheroids. To further examine possible changes in exosomal miRNA and protein expression, we measured exosomal miRNA and GPC-1 protein expression with qRT-PCR and enzyme-linked immunosorbent assay (ELISA), respectively. We compared the expression of selected miRNAs and GPC-1 in exosomes generated by PANC-1 2D cells and PANC-1 3D spheroids, as well as in PANC-1 2D cells and PANC-1 3D spheroids. Our results indicated that PANC-1 cells cultured in 3D spheroids show more features relevant to the progression of pancreatic cancer and can produce significantly more exosomes than PANC-1 2D cells. These findings underscore the potential of using spheroids as in vitro model to study cancer development and progression.
2. Materials and methods
2.1. Cell culture
PANC-1 cell line was purchased from American Type Culture Collection (Manassas, VA, USA). After thawing from dry ice, PANC-1 cells were cultured in Nunc™ EasYFlask™ cell culture flasks in 95% humidity and 5% CO2 at 37 °C. The base medium for the cells was Dulbecco's Modified Eagle's Medium (DMEM, Thermofisher) supplemented with 10% exosomes-depleted heat-inactivated fetal bovine serum (FBS, Thermofisher) and 1% Penicillin-Streptomycin (Pen Strep) solution (Sigma). Cells were maintained for at least 2 passages before seeding for exosome experiments and spheroid generation.
For exosome production from 2D cultured PANC-1 cells, 5 × 104 cells were seeded in 1 mL growth medium for a single well of a 24-well plate. After 24 h when cells reached approximately 70% confluency [23], cells were washed with PBS and fresh DMEM supplemented with exosomes-depleted FBS was replaced (Day 0). Medium was changed every other day. At Day 5, culture supernatant (1 mL) was collected and centrifuged at 300×g for 10 min at room temperature to remove cell debris.
2.2. PANC-1 cell line spheroid generation
Spheroids were prepared using a method adapted from previous research [24]. A PANC-1 cell suspension containing 5 × 104 cells/mL of DMEM supplemented with 10% exosomes-depleted FBS and 1% Pen Strep was prepared. One hundred microliters of the PANC-1 cell suspension (5000 cells) were plated in each well of 96-well U-bottom Nunclon™ Sphera™ plates (Thermofisher). These plates were chosen because of their commercial availability and widely validation in the generation of spheroids from multiple cancer cells [[25], [26], [27], [28]]. The plates were centrifugated in a swinging bucket rotor at 1500 rpm for 10 min at room temperature, and then incubated in a humidified atmosphere of 5% CO2 at 37 °C on a rotary shaker under gentle rotation. By using this technique, single spheroids were obtained in each well. After 24 h (Day1), spheroids with loose aggregate morphology were discarded and only spheroids with a compact morphology were cultured and used for exosome collection. Medium was changed every other day.
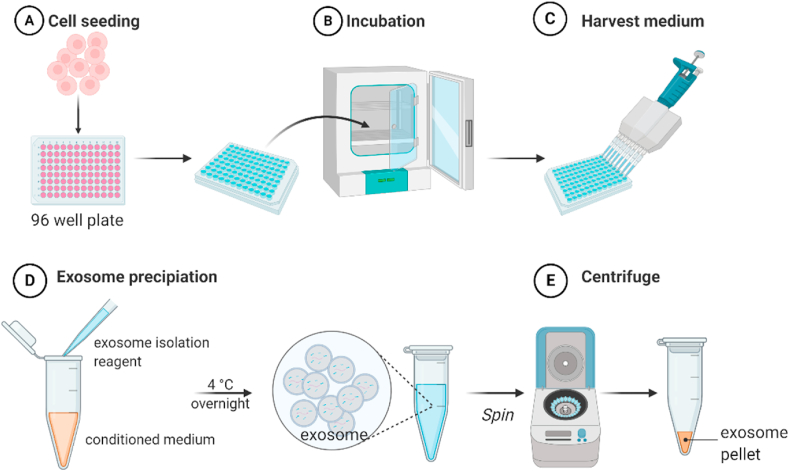
For the exosome production from 3D cultured PANC-1 spheroids (Fig. 1), culture medium was carefully harvested at Day 5 without disturbing the spheroid. The medium from 10 spheroids was mixed together (about 1 mL in total) and centrifuged at 300×g for 10 min at room temperature to remove cell debris.
Fig. 1.
Schematic representation of exosome isolation from PANC-1 3D spheroids. (A) PANC-1 cells (5000 cells per 100 μL) were seeded to 96 well plate. (B) Clusters were incubated in a humidified atmosphere of 5% CO2 at 37 °C, and culture medium was changed every other day. (C) Conditioned medium was harvested at Day 5. (D) Exosome isolation regent was mixed with the conditioned medium and incubated at 4 °C overnight. (E) Samples were centrifuged at 10,000×g for 1 h at 4 °C, and the supernatant was discarded. Exosome pellet was resuspended with PBS. (Created with BioRender.com).
2.3. Exosome isolation, labeling and transmission electron microscope (TEM)
Total exosome isolation reagent from cell culture media (Thermo Fisher Scientific) was used to isolate exosomes following the manufacture's protocol. After that, larger vesicles and cell debris were removed again by passing the samples through the 220 nm filter.
Membrane permeable SYTO™ RNASelect™ green fluorescent cell stain (Thermofisher) was used to fluorescently label the RNA present inside the exosomes, as described by the manufacturer. Briefly, isolated exosomes were incubated at 37 °C for 20min with the stain at a final concentration of 40 μM, and then the remaining free dye was removed by exosome spin columns (MWCO 3 000, Invitrogen). As controls, culture medium without cells was subject to the same isolation and staining procedures. Stained exosomes were visualized by confocal laser scanning microscope equipped with a 20x lens (FV3000, Olympus), irradiated by 488 nm.
To verify the presence of exosomes, TEM imaging was performed with a JEOL 1200EX, 80 kV microscope. A 10 μL drop was incubated on a TEM grid for 1 min, wicked dry, rinsed with water, and stained with uranyl acetate prior to TEM imaging.
2.4. RNA isolation and quantitative PCR
Total RNA from cells and spheroids were isolated with Trizol (Thermofisher) [29] and quantified using a Nanodrop (Thermo Fisher Scientific). cDNA from total RNA (1 μg) was generated using the high-capacity cDNA reverse transcription kit (Thermofisher), and then 100 ng cDNA was used for qRT-PCR to analyze the expression of GPC-1. Relative expression values were calculated by 2−ΔΔCt using GAPDH as an internal control [30]. GPC-1 and GAPDH primers were listed in Appendix A, Appendix A.
miRNA of cells, spheroids and exosomes was isolated with mirVana™️ miRNA Isolation Kit (Thermo Fisher Scientific) according to the protocol provided by the manufacturer, and quantified using NanoDrop (Thermo Fisher Scientific). cDNA was synthesized from miRNA (50 ng) with Mir-X™️ miRNA First Strand Synthesis Kit (Takara) and then qRT-PCR was performed with Mir-X™ miRNA qRT-PCR TB Green® Kit (Takara) with QuantStudio 6 Flex Real-Time PCR System (Thermofisher). Relative expression values were calculated by 2−ΔΔCt using U6 as an internal control [30]. miRNA primers used in this paper were listed in Appendix A, Appendix A.
2.5. Protein isolation and ELISA
Exosomal protein was isolated with total exosome RNA & protein isolation kit (Thermo Fisher Scientific) and GPC-1 protein was measured with human glypican 1 ELISA kit (Thermo Fisher Scientific).
2.6. Statistics
The data were presented as means ± standard error (SE). miRNA expressions were compared in PANC-1 spheroids and cells by Student's t-test. All statistical analyses were carried out using SPSS16.0 software (SPSS, Chicago, IL, USA). P values less than 0.05 were considered statistically significant.
3. Results
3.1. PANC-1 spheroid formation and exosome isolation
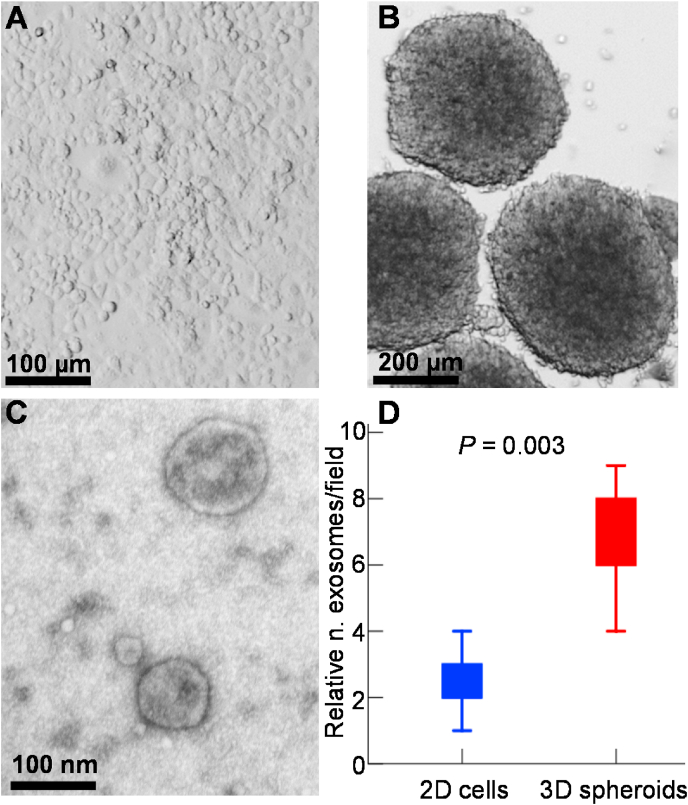
The morphology of 2D cultured PANC-1 cells (Fig. 2A) and 3D cultured PANC-1 spheroids (Fig. 2B) was observed at Day 5. These spheroids were of relatively uniform shape and size (0.5–0.7 mm, Fig. 2B) and showed no significant change in size with time during our experiments. To verify that exosomes were correctly isolated, we utilized TEM to visualize the content of our samples (Fig. 2C). The majority of the detected vesicles range between the size of 40–150 nm, confirming effective enrichment of exosomes from the conditioned media.
Fig. 2.
PANC-1 2D cells, 3D spheroids and exosomes isolated from spheroids culture medium. (A) PANC-1 2D cells. Scale bar: 100 μm. (B) PANC-1 3D spheroids. A total of 5000 cells were seeded for each spheroid. Scale bar: 200 μm. (C) Typical transmission electron microscope (TEM) image of isolated exosomes secreted by PANC-1 3D spheroids. Scale bar: 100 nm. (D) The relative number of exosomes per field (n = 5) analyzed by TEM.
In addition, exosomes labeled with SYTO™ RNASelect™ stain that is selective for RNA could be observed (Appendix A, Appendix A), demonstrating the existence of exosomes in our samples. In addition, from fluorescence images it showed that PANC-1 cells cultured in 3D spheroids secreted much more exosomes than did the 2D-cultured cells, indicating that the PANC-1 spheroids increased exosome production compared with that from 2D culture. In agreement with the fluorescence labeling data, TEM analyses also demonstrated statistically significant enrichment of exosomes derived from PANC-1 3D spheroids compared to those from PANC-1 2D cells (p < 0.01, one-sided unpaired t-test; Fig. 2d).
3.2. Differentially expressed miRNAs in PANC-1 2D cells, 3D spheroids and exosomes derived from 2D cells and 3D spheroids
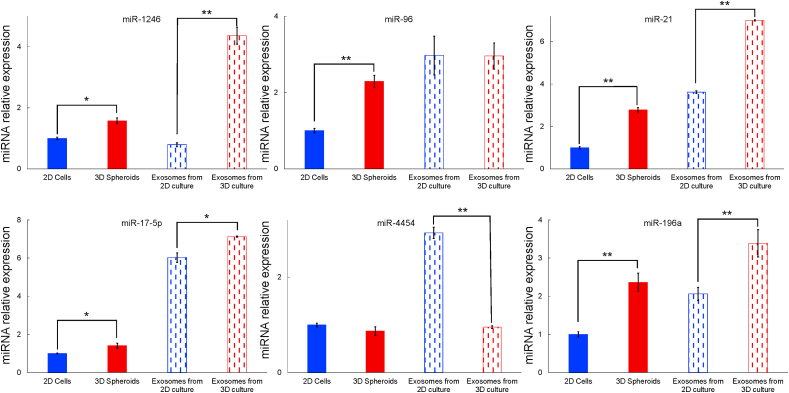
The differentially expressed miRNAs in PANC-1 2D cells and 3D spheroids, and exosomes isolated from PANC-1 2D cells and 3D spheroids were measured using qRT-PCR. A total of 6 differentially expressed miRNAs were examined (Fig. 3), including miR-1246, miR-96, miR-21, miR-17-5p, miR-4454 and miR-196a (Appendix A, Appendix A). It showed that all 6 measured miRNAs in PANC-1 3D spheroids were either significantly higher (miR-1246, miR-96, miR-21, miR-17-5p and miR-196a) than or similar (miR-4454) to those in PANC-1 2D cells. In exosomal miRNAs, there was no significant difference in miR-96 between exosomes derived from PANC-1 2D cells and 3D spheroids. In other exosomal miRNAs, except that miR-4454 was significantly decreased (P < 0.01) in exosomes isolated from 3D spheroids, expression levels of miR-1246, miR-21, miR-17-5p and miR-196a were significantly higher in exosomes from PANC-1 3D spheroids than those from PANC-1 2D cells.
Fig. 3.
Expression levels of selected miRNAs in PANC-1 cells, PANC-1 spheroids and exosomes derived from PANC-1 cells and PANC-1 spheroids. The data were normalized to U6 and expressed as mean ± SEM. Here, miRNAs expression in PANC-1 2D cells was used as control (*P < 0.05, **P < 0.01).
Overall, it could be seen that miRNAs expression in exosomes was higher than that in parental cells. Among the measured miRNAs, expression of miR-1246 in exosomes from PANC-1 2D cells was lower than that in PANC-1 2D cells, all other 5 miRNAs were significantly highly expressed in exosomes from PANC-1 2D cells compared with those from PANC-1 2D cells. In addition, miR-1246, miR-96, miR-21, miR-17-5p and miR-196a expression levels were higher in exosomes isolated from PANC-1 3D spheroids than those in spheroids. There was no significant change between the expression of miR-4454 in PANC-1 3D spheroids or spheroids derived exosomes.
3.3. GPC-1 expression in PANC-1 2D cells, 3D spheroids and exosomes derived from 2D cells 3D spheroids
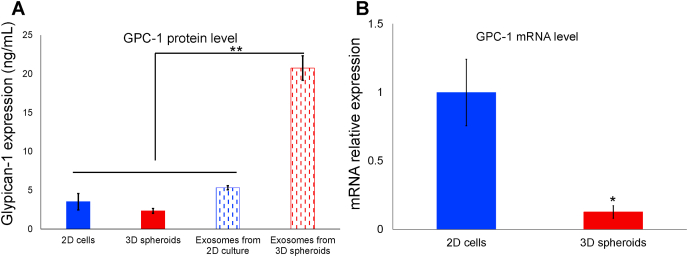
We isolated and quantified protein from exosomes derived from PANC-1 2D cells and 3D spheroids as well as the parental 2D cells and 3D spheroids. The results showed that there was significant difference in GPC-1 levels between exosomes isolated from PANC-1 cells and spheroids (Fig. 4A). There were no significant differences among GPC-1 levels in PANC-1 2D cells, 3D spheroids and exosomes derived from PANC-1 2D cells, however significantly higher concentration of GPC-1 protein in exosomes from PANC-1 3D spheroids could be observed (P < 0.001).
Fig. 4.
GPC-1 expression in PANC-1 2D cells, 3D spheroids and exosomes derived from PANC-1 2D cells and 3D spheroids. (A) GPC-1 protein levels in PANC-1 2D cells, 3D spheroids and exosomes derived from PANC-1 2D cells and 3D spheroids measured with ELISA. The data were mean ± SEM (**P < 0.01). (B) GPC-1 expression in PANC-1 2D cells and 3D spheroids measured with qRT-PCR. The data were normalized to U6 and expressed as mean ± SEM. Here, GPC-1 expression in PANC-1 2D cells was used as control (*P < 0.05).
We also measured GPC-1 mRNA level in PANC-1 2D cells and 3D spheroids with qRT-PCR. It showed that GPC-1 mRNA level significantly decreased (P < 0.05) in the PANC-1 spheroids compared to PANC-1 cells (Fig. 4B).
4. Discussion
In this study, we found PANC-1 3D spheroids secreted much more exosomes than did the 2D-cultured cells. These exosomes were enriched with miRNA and GPC-1 protein compared to their originating PANC-1 cells, consistent with previous reports [[31], [32], [33]]. In particular, significantly increased GPC-1 protein was observed in exosomes derived from PANC-1 3D spheroids, which further indicated that PANC-1 3D spheroids secreted more exosomes than did the 2D-cultured cells.
Exosomes can shuttle various molecular cargo including proteins, mRNA, miRNA, as well as non-coding RNAs, between cells to communicate with each other [34]. Cell cultures in two-dimensional conditions have limited cell-cell communication [4] and might be less efficient in generating exosomes than three-dimensional cell cultures. The 3D culture systems enable increased cell–cell contact communication by allowing spatial cell–cell interactions [35], which provides a favorable environment for exosome synthesis and secretion. In turn, elevated production of exosomes further facilitates communications between cells. The present study showed that miRNAs expression in exosomes increased overall compared with their parental cells or spheroids, which further underscores the role of exosomes in intercellular communication.
Among the miRNAs assayed, expression levels of miR-1246, miR-96, miR-21, miR-17-5p and miR-196a in PANC-1 3D spheroids all significantly increased compared with PANC-1 2D cells. miR-21 and miR-17 are considered to be typical oncomirs which are associated with carcinogenesis, malignant transformation, and metastasis [36], as most of their targets are tumor suppressors [37,38]. miR-1246, along with miR-196a, were found to promote tumor progression, migration and invasion in various malignancies including pancreatic ductal adenocarcinoma [39,40]. The upregulation of miR-21, miR-17, miR-1246 and miR-196a observed in PANC-1 3D spheroids suggests that 3D cultured PANC-1 spheroids show more aggressive features than 2D PANC-1 cells. Interestingly, previous studies found that miR-96 could significantly suppress pancreatic cancer cell proliferation by targeting GPC-1 and downregulation of miR-96 in pancreatic cancer may cause the cancer cells to divide and grow more quickly [41,42]. In the current study, the expression of miR-96 was upregulated in PANC-1 3D spheroids, as verified by decreased GPC-1 mRNA and protein levels (Fig. 4). This indicates that cell proliferation in PANC-1 3D spheroids was influenced probably due to the volume limitation [43], which needs further examination. In the current study miR-4454 was the only one among the assessed miRNAs that showed slight decrease in PANC-1 3D spheroids but significantly downregulation in exosomes derived from PANC-1 3D spheroids (Fig. 3). It is noteworthy that significantly reduced expression of exosomal miR-4454 has been demonstrated previously in pancreatic ductal adenocarcinoma compared to benign pancreatitis [44,45], in line with our speculation that PANC-1 spheroids have enhanced invasive capacities compared with 2D cell culture.
Our finding that GPC-1 protein level was significantly enhanced in exosomes released from 3D culture of PANC-1 cells as compared to 2D culture, underscores the high specificity and sensitivity of the exosomal GPC-1 biomarker in pancreatic cancer diagnosis [32]. Out of our expectation, even though the GPC-1 mRNA level was much higher in 2D culture PANC-1 cells than that in 3D culture, the GPC-1 protein levels were similar in both culture conditions (Fig. 4A). This observation leads to the speculation that very different mechanisms of exosome biogenesis, sorting and releasing exist in 3D culture cells compared with 2D cell culture [19]. Additionally, miRNA expression profile in exosomes secreted by 3D cultured PANC-1 cells showed somewhat different patterns in comparison with 2D derived exosomal miRNA profile. In particular, miR-4454 and miR-96 show distinct expression features between exosomes and their parent cells (Fig. 3), while the expression trends of other miRNAs measured in exosomes coincided with the data obtained from their parent cells. Future more in-depth studies of tumor-derived exosomes in 3D culture are required to elucidate on the precise mechanism of exosome generation and their functioning in 3D culture which has been shown to be more mimicking native tissue and in vivo microenvironment [19,46].
In conclusion, PANC-1 cells cultured in 3D spheroids show more features relevant to progression of pancreatic cancer and can produce significantly more exosomes than PANC-1 2D cells. These results emphasize the potential importance of employing spheroids as in vitro model to study cancer development and progression. More importantly, our findings lead to the suggestion that combining miRNA and protein profiles in 3D culture cells is a preferable and superior approach to determine future exosomal biomarker of disease. Identifying the mechanisms of how these exosomal molecules interact in 3D conditions to contribute to disease progression could potentially reveal new avenues of developing exosomal biomarkers useful in diagnosis and prognosis and of targeted therapy in cancer.
Financial support
This work was supported by Anhui University Natural Science Research Project KJ2019A0697 and KJ2018A0471.
Declaration of competing interest
The authors declare no conflicts of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2021.101026.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Drost J., Clevers H. Organoids in cancer research. Nat. Rev. Canc. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 2.Pan B., Zhao D., Liu Y., Li N., Song C., Li N., Li X., Li M., Zhao Z. Establishment and characterization of breast cancer organoids from a patient with mammary Paget's disease. Canc. Cell Int. 2020;20:1–9. doi: 10.1186/s12935-020-01459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringuette-Goulet C., Bolduc S., Pouliot F. Modeling human bladder cancer. World J. Urol. 2018;36:1759–1766. doi: 10.1007/s00345-018-2369-5. [DOI] [PubMed] [Google Scholar]
- 4.Griffith L.G., Swartz M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006;7:211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 5.Fatehullah A., Tan S.H., Barker N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 6.Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., Fissell W.H., Patton J.G., Rome L.H., Burnette D.T., Coffey R.J. Reassessment of exosome composition. Cell. 2019;177:428–445. doi: 10.1016/j.cell.2019.02.029. e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pluchino S., Smith J.A. Explicating exosomes: reclassifying the rising stars of intercellular communication. Cell. 2019;177:225–227. doi: 10.1016/j.cell.2019.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Masamune A., Yoshida N., Hamada S., Takikawa T., Nabeshima T., Shimosegawa T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem. Biophys. Res. Commun. 2018;495:71–77. doi: 10.1016/j.bbrc.2017.10.141. [DOI] [PubMed] [Google Scholar]
- 9.Melo S.A., Sugimoto H., O'Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., Lucci A., Ivan C., Calin G.A., Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Canc. Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Canc. 2019;1871:455–468. doi: 10.1016/j.bbcan.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diakonov E.E., Selenina A.V., Tomilin A.N., Tsimokha A.S. Evidences against vesicle-dependent trafficking and involvement of extracellular proteasomes into cell-to-cell communications. Biochem. Biophys. Res. Commun. 2019;508:368–373. doi: 10.1016/j.bbrc.2018.11.152. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Choi D.S., Kim D.K., Kim Y.K., Gho Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13:1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 15.Sachs N., Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Villasante A., Marturano-Kruik A., Ambati S.R., Liu Z., Godier-Furnemont A., Parsa H., Lee B.W., Moore M.A., Vunjak-Novakovic G. Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics. 2016;6:1119–1130. doi: 10.7150/thno.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadovska L., Zandberga E., Sagini K., Jekabsons K., Riekstina U., Kalnina Z., Llorente A., Line A. A novel 3D heterotypic spheroid model for studying extracellular vesicle-mediated tumour and immune cell communication. Biochem. Biophys. Res. Commun. 2018;495:1930–1935. doi: 10.1016/j.bbrc.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 18.Eguchi T., Sogawa C., Okusha Y., Uchibe K., Iinuma R., Ono K., Nakano K., Murakami J., Itoh M., Arai K., Fujiwara T., Namba Y., Murata Y., Ohyama K., Shimomura M., Okamura H., Takigawa M., Nakatsura T., Kozaki K.I., Okamoto K., Calderwood S.K. Organoids with cancer stem cell-like properties secrete exosomes and HSP90 in a 3D nanoenvironment. PloS One. 2018;13 doi: 10.1371/journal.pone.0191109. e0191109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thippabhotla S., Zhong C., He M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 2019;9:13012. doi: 10.1038/s41598-019-49671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rocha S., Carvalho J., Oliveira P., Voglstaetter M., Schvartz D., Thomsen A.R., Walter N., Khanduri R., Sanchez J.C., Keller A., Oliveira C., Nazarenko I. 3D cellular architecture affects microRNA and protein cargo of extracellular vesicles. Adv. Sci. 2019;6:1800948. doi: 10.1002/advs.201800948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Q., Hamilton M., Vasquez B., He M. 3D-printing enabled micro-assembly of a microfluidic electroporation system for 3D tissue engineering. Lab Chip. 2019;19:2362–2372. doi: 10.1039/C9LC00046A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieber M., Mazzetta J., Nelson-Rees W., Kaplan M., Todaro G. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int. J. Canc. 1975;15:741–747. doi: 10.1002/ijc.2910150505. [DOI] [PubMed] [Google Scholar]
- 23.Patel G.K., Khan M.A., Zubair H., Srivastava S.K., Khushman M., Singh S., Singh A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-41800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyoshima K.E., Ogawa M., Tsuji T. Regeneration of a bioengineered 3D integumentary organ system from iPS cells. Nat. Protoc. 2019;14:1323–1338. doi: 10.1038/s41596-019-0124-z. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan S., Mehta P., Horst E.N., Ward M.R., Rowley K.R., Mehta G. Comparative analysis of tumor spheroid generation techniques for differential in vitro drug toxicity. Oncotarget. 2016;7:16948–16961. doi: 10.18632/oncotarget.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neeley C., Gaarn L., Marwood T., Scott R., Carter S., Granchelli J. A novel cell culture surface supports effective formation of three dimensional cancer spheroids in suspension. Clin. Canc. Res. 2013;208 doi: 10.1158/1538-7445.AM2013-208. [DOI] [Google Scholar]
- 27.Mansour M.A., Hyodo T., Ito S., Kurita K., Kokuryo T., Uehara K., Nagino M., Takahashi M., Hamaguchi M., Senga T. SATB2 suppresses the progression of colorectal cancer cells via inactivation of MEK5/ERK5 signaling. FEBS J. 2015;282:1394–1405. doi: 10.1111/febs.13227. [DOI] [PubMed] [Google Scholar]
- 28.McClellan S., Slamecka J., Howze P., Thompson L., Finan M., Rocconi R., Owen L. mRNA detection in living cells: a next generation cancer stem cell identification technique. Methods. 2015;82:47–54. doi: 10.1016/j.ymeth.2015.04.022. [DOI] [PubMed] [Google Scholar]
- 29.Rio D.C., Ares M., Hannon G.J., Nilsen T.W. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5439. pdb. prot5439. [DOI] [PubMed] [Google Scholar]
- 30.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 31.Cheng L., Sharples R.A., Scicluna B.J., Hill A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3:23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frampton A.E., Prado M.M., López-Jiménez E., Fajardo-Puerta A.B., Jawad Z.A., Lawton P., Giovannetti E., Habib N.A., Castellano L., Stebbing J., Krell J., Jiao L.R. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006. doi: 10.18632/oncotarget.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L., Liu W., Xiao J., Cao B. The role of exosomes and “exosomal shuttle microRNA” in tumorigenesis and drug resistance. Canc. Lett. 2015;356:339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 35.Edmondson R., Broglie J.J., Adcock A.F., Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Canc. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 37.Hammond S.M. MicroRNAs as tumor suppressors. Nat. Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 38.Jin X.L., Sun Q.S., Liu F., Yang H.W., Liu M., Liu H.X., Xu W., Jiang Y.Y. microRNA 21‐mediated suppression of sprouty1 by Pokemon affects liver cancer cell growth and proliferation. J. Cell. Biochem. 2013;114:1625–1633. doi: 10.1002/jcb.24504. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y.F., Hannafon B.N., Khatri U., Gin A., Ding W.Q. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019;16:770–784. doi: 10.1080/15476286.2019.1585738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y.F., Hannafon B.N., Zhao Y.D., Postier R.G., Ding W.Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget. 2017;8:77028–77040. doi: 10.18632/oncotarget.20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu S., Lu Z., Liu C., Meng Y., Ma Y., Zhao W., Liu J., Yu J., Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Canc. Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 42.Li C., Du X., Tai S., Zhong X., Wang Z., Hu Z., Zhang L., Kang P., Ji D., Jiang X., Zhou Q., Wan M., Jiang G., Cui Y. GPC1 regulated by miR-96-5p, rather than miR-182-5p, in inhibition of pancreatic carcinoma cell proliferation. Int. J. Mol. Sci. 2014;15:6314–6327. doi: 10.3390/ijms15046314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delarue M., Montel F., Vignjevic D., Prost J., Joanny J.F., Cappello G. Compressive stress inhibits proliferation in tumor spheroids through a volume limitation. Biophys. J. 2014;107:1821–1828. doi: 10.1016/j.bpj.2014.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicentini C., Calore F., Nigita G., Fadda P., Simbolo M., Sperandio N., Luchini C., Lawlor R.T., Croce C.M., Corbo V., Fassan M., Scarpa A. Exosomal miRNA signatures of pancreatic lesions. BMC Gastroenterol. 2020;20:137. doi: 10.1186/s12876-020-01287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strobel O., Büchler M.W., Werner J. Surgical therapy of chronic pancreatitis: indications, techniques and results. Int. J. Surg. 2009;7:305–312. doi: 10.1016/j.ijsu.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Hofer M., Lutolf M.P. Engineering organoids. Nat. Rev. Mater. 2021:1–19. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.