Abstract
The outer segment (OS) organelle of vertebrate photoreceptors is a highly specialized cilium evolved to capture light and initiate light response. The plasma membrane which envelopes the OS plays vital and diverse roles in supporting photoreceptor function and health. However, little is known about the identity of its protein constituents, as this membrane cannot be purified to homogeneity. In this study, we used the technique of protein correlation profiling to identify unique OS plasma membrane proteins. To achieve this, we used label-free quantitative MS to compare relative protein abundances in an enriched preparation of the OS plasma membrane with a preparation of total OS membranes. We have found that only five proteins were enriched at the same level as previously validated OS plasma membrane markers. Two of these proteins, TMEM67 and TMEM237, had not been previously assigned to this membrane, and one, embigin, had not been identified in photoreceptors. We further showed that embigin associates with monocarboxylate transporter MCT1 in the OS plasma membrane, facilitating lactate transport through this cellular compartment.
Keywords: retina, photoreceptor, plasma membrane, protein correlation profiling, lactate transport
Abbreviations: MCT, monocarboxylate transporter; OS, outer segment; ROS, rod outer segment; RPE, retinal pigment epithelium
Graphical Abstract
Highlights
-
•
The unique proteome of the photoreceptor outer segment plasma membrane is identified.
-
•
TMEM67, TMEM237, and embigin are novel unique components of this membrane.
-
•
Embigin in this membrane is associated with the monocarboxylate transporter MCT1.
-
•
The photoreceptor outer segment likely facilitates lactate transport.
In Brief
The plasma membrane which envelopes the light-sensitive outer segment organelle of vertebrate photoreceptor cells plays diverse roles in supporting photoreceptor function and health. Protein correlation profiling of this membrane revealed a surprisingly small number of unique protein components. Among them are TMEM67 and TMEM237, whose mutations are associated with various syndromic ciliopathies, and embigin found to be associated with the monocarboxylate transporter MCT1. The MCT1–embigin complex likely facilitates lactate transport through this cellular compartment.
The outer segment (OS) of the vertebrate photoreceptor cell is a ciliary organelle responsible for capturing light and initiating the electrical light signal ultimately transmitted to the brain. The OS consists of a stack of disc-shaped membranes, or “discs”, that are enclosed within the plasma membrane. Whereas proteins responsible for light capture and propagation of the visual signal are located primarily in discs, the plasma membrane harbors proteins responsible for generating electrical responses to light (reviewed in (1)). Despite being contiguous with the plasma membrane surrounding the rest of the photoreceptor cell, the protein composition of the OS plasma membrane is unique because of a combination of targeted protein delivery and protein retention imposed by the diffusional barrier located at the OS base (reviewed in (2, 3, 4, 5)).
Beyond its role in regulating ion currents, the OS plasma membrane serves several additional functions. It is critical for supporting continuous OS renewal, which combines a constant generation of new photoreceptor discs at the OS base with the phagocytosis of old discs at the OS tip by the retinal pigment epithelium (RPE) (6, 7). It is also likely that the OS plasma membrane stabilizes OS architecture through complex interactions with discs, the extracellular matrix and neighboring cells (3, 8). These important processes are thought to be mediated by proteins specific to the OS plasma membrane; however, our knowledge of their identities remains incomplete. Thus, elucidating the protein composition of this membrane would give us a better understanding of its diverse functions.
The goal of this study was to characterize the unique protein components of the OS plasma membrane. The major challenge to achieving this goal is that it is virtually impossible to obtain OS plasma membrane in purity sufficiently high for MS analysis using standard biochemical techniques. This is because this membrane comprises a small fraction of the total OS membrane material and it cannot be completely separated from other membranes. Therefore, we used a strategy known as “protein correlation profiling”, which is a powerful MS approach for analyzing multiprotein complexes or organelles that can be fractionated but not purified to homogeneity (9, 10, 11, 12). In this approach, a membrane preparation is enriched from a crude preparation and the relative abundances of proteins present in both crude and enriched membranes are quantified using label-free quantitative proteomics. The resulting values are compared with the relative abundances of protein markers known to reside uniquely in the membrane of interest. Although the abundances of all unique constituents of the membrane are expected to remain at a constant molar ratio with the markers, the relative abundances of contaminating and/or nonunique proteins are expected to decrease as membrane purity increases.
To conduct protein correlation profiling of the OS plasma membrane, we immunoenriched this membrane from a preparation of total rod OS membranes and compared protein compositions of these preparations. We found that, among the ~800 proteins confidently identified in both membrane preparations, only five were enriched to the same degree as the three well-established OS plasma membrane markers—the Na+/Ca2+/K+ exchanger NCKX1 and two subunits of the cGMP-gated channel, CNGα1 and CNGβ1 (13, 14, 15). Two of these five proteins—prominin-1 and protocadherin-21—have been previously characterized as OS plasma membrane components (16, 17, 18, 19), whereas the other three—TMEM67, TMEM237, and embigin—had not been previously assigned to this membrane. We validated the OS plasma membrane localization of each of these proteins by immunofluorescence. Finally, we showed that embigin, a protein that had not been previously identified in photoreceptor cells, associates with the monocarboxylate transporter MCT1, presumably functioning to control the MCT1 content of the OS plasma membrane and, accordingly, the flux of lactate through the OS.
Experimental Procedures
Antibodies
The PMe 2D9 mAb against an epitope within the extracellular domain on bovine NCKX1 has been previously described and used to obtain enriched OS plasma membranes (20, 21). Rabbit polyclonal antibody against the C-terminal TMEM67 peptide GQKNLATKTLVDERFLI was generated using custom antibody production service from Thermo Scientific Protein Biology. Mouse mAb 3C4 against embigin was prepared using established mAb techniques. Specificity of each newly produced antibody was validated by Western blotting, as shown in supplemental Fig. S1. Rabbit polyclonal antibody against TMEM237 was kindly provided by Dr C.M. Craft, as described (22). Rabbit polyclonal antibody against MCT1 is as described (23). Rabbit polyclonal antibody against R9AP is as described (24). Anti-cMyc mAb 9E10 was from Thermo Fisher Scientific. Mouse mAb PMc1D1against CNGα1 is as described (13).
Antibody Cross-Linking to Protein A/G Magnetic Beads
About 300 to 400 μl of Pierce Protein A/G magnetic beads (#88802, Thermo Scientific) were equilibrated with PBS and incubated with 750 μl of 5 mM bis(sulfosuccinimidyl)suberate (#21580, Thermo Scientific) for 20 min at room temperature. Bis(sulfosuccinimidyl)suberate was removed, and beads were incubated for 40 min at 22 °C with 200 μg of the PMe 2D9 antibody (or the anti-Myc mAb serving as a control) dissolved in 750 μl PBS. The cross-linking reaction was quenched with 100 mM Tris HCl, pH 8.0, followed by equilibrating the beads with PBS. To characterize the ability of cross-linked PMe 2D9 antibodies to precipitate NCKX1, 5 μg of PMe 2D9 beads (or control Myc-beads) was incubated with 100 μg of rod outer segment (ROS) membranes solubilized in PBS containing 0.5% dodecyl maltoside. The beads were washed twice with the same buffer, and bound material was eluted with 100 mM Tris HCl (pH 6.8) containing 2% SDS and 10% glycerol. Eluted proteins were analyzed by Western blotting using the PMe 2D9 antibody.
Isolation of ROS From Bovine Retinas
All procedures were performed under dim red light at 4 °C. First, ROSs were purified following a procedure modified from (25), as described (26). Briefly, 100 frozen bovine retinas (T.A. & W.L. Lawson Co) were thawed and resuspended in 200 ml of buffer A (20 mM Hepes, 100 mM KCl, 2 mM MgCl2, and 0.1 mM EDTA; pH 7.4) containing 25% sucrose. OSs were detached from the retinas by swirling in a 1 l Erlenmeyer flask and separated from retinal debris by centrifugation at 3700g for 6 min. The supernatant was diluted with an equal volume of buffer A, and ROSs were pelleted by centrifugation at 6300g for 8 min. The pellet was resuspended in buffer A containing 20% sucrose, applied on a 27/32% step gradient of sucrose and centrifuged for 1 h in an SW-28 rotor at 83,000g. ROSs were collected from the 27/32% sucrose interphase and immediately used to purify plasma membranes. To obtain a preparation of ROS membranes depleted of most soluble proteins, ROSs were incubated in the hypotonic buffer (10 mM Hepes; pH 7.4) for 1 h on ice and pelleted by centrifugation at 100,000g for 30 min. Membranes were collected and washed with the hypotonic buffer one more time. The resulting preparation was resuspended in buffer A and stored at −20 °C.
OS Plasma Membrane Enrichment
Photoreceptor plasma membrane enrichment was performed as described (27) with the following modifications. Ten milligram of freshly prepared structurally intact ROS was incubated with 200 μg of PMe 2D9 antibody cross-linked to protein A/G magnetic beads (Thermo Fisher) in 4 ml of 20 mM Hepes (pH 7.4), 100 mM KCl, 2 mM MgCl2, and 0.1 mM EDTA for 3 h at 4 °C with gentle rocking. Next, beads were transferred to the hypotonic buffer containing 10 mM Tris HCl (pH 7.5) and 2 mM DTT and incubated for 16 to 20 h to allow OS plasma membranes dissociate from discs and other membranes. Beads were washed three times with the hypotonic buffer, and the bound material was eluted with 200 μl of 100 mM Tris HCl (pH 6.8), 2% SDS, and 10% glycerol and stored at −20 °C until used for MS analysis. To account for nonspecific protein binding to magnetic beads, we repeated these experiments with A/G beads associated with monoclonal anti-Myc antibody 9E10.
Sample Preparation and LC-MS/MS Analysis
Three types of protein samples (hypotonically washed OS membranes, proteins precipitated by PMe 2D9 antibodies and proteins precipitated by 9E10 antibodies) were processed in parallel in three biological repeats. For each sample, 5 to 20 μg total protein was used to prepare peptide mixtures for proteomic profiling. Proteins were cleaved with the trypsin/endoproteinase LysC mixture (V5072, Promega) using the paramagnetic bead–based method (28). Each digest was dissolved in 12 μl of 1/2/97% (by volume) of the trifluoroacetic acid/acetonitrile/water solution, and 3 μl was injected into a 5 μm, 180 μm × 20 mm Symmetry C18 trap column (Waters) in 1% acetonitrile in water for 3 min at 5 μl/min. For protein profiling in serial retinal sections, each section was dissolved in 100 μl of 2% SDS and 0.1 M Tris HCl, pH 8.0 and proteins were cleaved as above. The analytical separation was next performed using 1.8-μm HSS T3 and 75 μm × 250 mm column (Waters) over 90 min at a flow rate of 0.4 μl/min at 55 °C. The 5 to 30% mobile phase B gradient was used, where phase A was 0.1% formic acid in water and phase B 0.1% formic acid in acetonitrile. Peptides separated by LC were introduced into the Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific) using positive electrospray ionization at 1900 V and capillary temperature of 275 °C. Data collection was performed in the data-dependent acquisition mode with 120,000 resolution (at m/z 200) for MS1 precursor measurements. The MS1 analysis utilized a scan from 375 to 1600 m/z with a target automatic gain control value of 2.0e5 ions, the radio frequency lens set at 30%, and a maximum injection time of 50 ms. Advanced peak detection and internal calibration were enabled during data acquisition. Peptides were selected for MS/MS using charge state filtering (2, 3, 4, 5), monoisotopic peak detection, and a dynamic exclusion time of 20 s with a mass tolerance of 10 ppm. MS/MS was performed using higher-energy collisional dissociation with a collision energy of 30 ± 5% with detection in the ion trap using a rapid scanning rate, AGC target value of 3.0e3 ions, maximum injection time of 300 ms, and ion injection for all available parallelizable time enabled.
Protein Identification and Quantification
For label-free relative protein quantification, raw mass spectral data files (.raw) were imported into Progenesis QI for Proteomics 4.2 software (Nonlinear Dynamics) for duplicate run alignment of each preparation and peak area calculations. The peptide peak list was generated using Progenesis 4.2 as well. Peptides were identified using Mascot version 2.5.1 (Matrix Science) for searching a custom IPI bovine database (derived from IPI bovine database v3.73; September 2011) containing 1598 entries of proteins confidently identified in multiple ROS preparations. For mouse samples, we used the UniProt 2019 reviewed mouse database containing 17,008 entrees. Mascot search parameters were as follows: 10 ppm mass tolerance for precursor ions; 0.025 Da for fragment-ion mass tolerance; one missed cleavage by trypsin; fixed modification was carbamidomethylation of cysteine; variable modification was oxidized methionine. Only proteins identified with two or more peptides (Mascot scores >15 for a peptide and >50 for a protein corresponding to protein false discovery rate –1.3% and peptide false discovery rate–0.4% calculated using reversed decoy database) were included in the protein correlation profiling analysis. To account for variations in experimental conditions and amounts of protein material in individual LC-MS/MS runs, the integrated peak area for each identified peptide was corrected using the factors calculated by automatic Progenesis algorithm utilizing the total intensities for all peaks in each run. Values representing protein amounts were calculated based on a sum of ion intensities for all identified constituent nonconflicting peptides (12).
For absolute protein quantification in mouse ROS, samples were spiked with cold isotope-labeled synthetic peptides before trypsin digestion. Peptide mixtures were analyzed by LC-MS/MS using the data-dependent acquisition workflow, as above, for label-free protein quantification. Peptides were identified using Mascot version 2.5.1 by searches against the mouse UniProt database, as described above, with an inclusion of labels: 13C(6)15N(2) for lysine and 13C(6)15N(4) for arginine. The intensities of light and heavy peptides were measured using Progenesis IQ for Proteomics 4.2. Protein amounts were calculated based on the intensities of the corresponding light and heavy peptides and known heavy peptide amounts. The following peptides (all from JPT Peptide Technologies, GmbH) were used: NPLGDDDASATASK and EAAAQQQESATTQK for rhodopsin, VYGSSSPAVDR, IAIISQGR, and ALSLLEENR for ABCA4; LLSSSGSTFEQSIK, SATVPVSIWR, LFVNQDSSSSK, and FVAASYDIR for TMEM67; FTAPTGVSQPVG, AEGSEPPAAELK, and DVALSVHR for TMEM237; and SDDSNGIENNVPR, SLIAYVGDSTVLK, and GTFNIHVPK for embigin.
Experimental Design and Statistical Rationale
For protein correlation profiling, we compared relative amounts of proteins confidently identified in three independent preparations of the OS plasma membrane enriched with the PMe 2D9 antibody and hypotonically washed OS membranes by calculating ratios between their amounts in indicated membrane preparations. Each ratio was normalized by the averaged ratio for three plasma membrane markers—NCKX1, CNGα1, and CNGβ1—contained in the corresponding membrane preparation. In this analysis, the markers and all other unique OS plasma membrane proteins were expected to yield a ratio close to 1. To account for nonspecific protein binding to magnetic beads, we repeated the same analysis for proteins precipitated by control beads coupled to anti-Myc 9E10 antibodies. The resulting values were subtracted from those obtained with experimental samples with PMe 2D9 beads. Only proteins identified and quantified in at least two experiments were used for further protein correlation profiling.
Immunoprecipitation and Western Blotting
For embigin immunoprecipitation, 2.1 mg of bovine ROS were solubilized in 400 μl of 50 mM Hepes (pH 7.4), 150 mM NaCl, and 20 mM CHAPS for 30 min at 4 °C. Insoluble material was removed by centrifugation at 40,000 rpm in a Beckman Optima TLA 110 rotor for 10 min, and the supernatant was added to 100 μl 3C4 conjugated Sepharose beads in a Millipore Ultrafree mC HVPdVDF 0.45-μm cartridge. Beads were incubated at 4 °C with rotation for 1.5 h and washed ten times with 500 μl of the same buffer. Bound material was eluted with two 100 μl aliquots of 2% SDS in the same buffer for 30 min at 20 °C. For MS, samples were prepared as described above. For Western blotting, proteins were subjected to SDS-PAGE, probed with indicated antibodies and the corresponding secondary antibodies, and imaged on an Odyssey infrared scanner (LI-COR Biosciences) in the 700-nm or 800-nm channel.
Immunofluorescence Microscopy
All mouse experiments were conducted under the animal use and care protocol approved by the Duke University Institutional Animal Care and Use Committee. Immunofluorescent protein detection was performed in thick agarose-embedded sections as described (29). Anesthetized mice were transcardially perfused with 15 ml of a fixative solution containing 4% paraformaldehyde in 80 mM Pipes (pH 6.8), 5 mM EGTA, and 2 mM MgCl2 (30). After fixation, the eyes were dissected on ice in Ringer’s solution. Eyecups were embedded in 7% low-melt agarose (A3038; Sigma-Aldrich) and cut by a vibratome (VT1200S; Leica) into 200-μm-thick slices. The sections were blocked in PBS containing 5% donkey serum and 0.5% Triton X-100 for 40 min at 22 °C. Next, slices were incubated for 6 h at 4 °C with the primary antibody in the same blocking buffer. Sections were washed three times in the blocking buffer and then the secondary antibody conjugated with Alexa Fluor 488 was added along with 1 μg/ml wheat germ agglutinin conjugated with Alexa Fluor 647 (W32466; Thermo Fisher Scientific) and 10 μg/ml Hoechst (H3569; Thermo Fisher Scientific) in the same buffer. Sections were incubated for 2 h at 22 °C, washed three times in PBS, and mounted onto slides in glycerol. For MCT1 staining, mice were not perfused or fixed before retina dissection, and fresh vibratome sections were blocked for 25 min, incubated with the primary antibody for 1 h at 4 °C, and fixed for 1 h in the same fixative as above. After fixation, the primary antibody was added for another 6 h and the rest of the procedure was performed as above. Images were taken with a confocal microscope (Eclipse 90i and A1 confocal scanner; Nikon) with a 60× objective (1.4 NA Plan Apochromat VC; Nikon) using Nikon NIS-Elements software. Image analysis and processing was performed with ImageJ.
Isolation of Mouse ROS
The retinas from eight, dark adapted P30 WT C57Bl/6J mice were dissected from eyecups under infrared illumination in mouse Ringer’s solution containing 130 mM NaCl, 3.6 mM KCl, 2.4 mM MgCl2, 1.2 mM CaCl2, and 10 mM Hepes (pH 7.4), adjusted to 314 mOsm. The retinas were pooled into 400 μl ice-cold 16% sucrose in Ringer’s solution and vortexed at maximum speed for 60 s in a 1.5-ml Eppendorf tube. The tube was then centrifuged at 200g for 30 s to sediment large retinal debris. A total of 350 μl of the supernatant was loaded on top of a 1.8-ml step gradient composed of 27% and 32% sucrose in Ringer’s solution and centrifuged for 30 min at 30,000 rpm in a swing-bucket SW-55 rotor (Beckman Coulter) at 4 °C. ROSs were carefully collected from the 27/32% sucrose interface, diluted at least 4-fold in Ringer’s solution and centrifuged for 30 min at 50,000 rpm in a TLA-100.3 rotor (Beckman Coulter). The dark-adapted mouse ROS pellet was washed once with Ringer’s solution before hypotonic shock and centrifugation to remove soluble proteins as described above.
Serial Tangential Sectioning of Frozen Mouse Retinas
Sample preparation was performed essentially as described before (31, 32). A retina punch (2 mm diameter) was cut from a freshly obtained mouse eyecup using a surgical trephine positioned adjacent to the optic disc, transferred onto PVDF membrane with the photoreceptor layer facing up, flat-mounted between two glass slides separated by plastic spacers (ca. 240 μm), and flash-frozen in liquid nitrogen before being stored at −80 °C. Subsequently, the retinal surface was aligned with the cutting plane of a cryostat knife and the edges of the retina were trimmed away. Progressive 10-μm tangential sections were collected into individual tubes on dry ice, solubilized in 100 μl of 2% SDS and 0.1 M Tris HCl (pH 8.0) and prepared for MS analysis.
Results
Protein Correlation Profiling of the OS Plasma Membrane Suggests Eight Unique Components
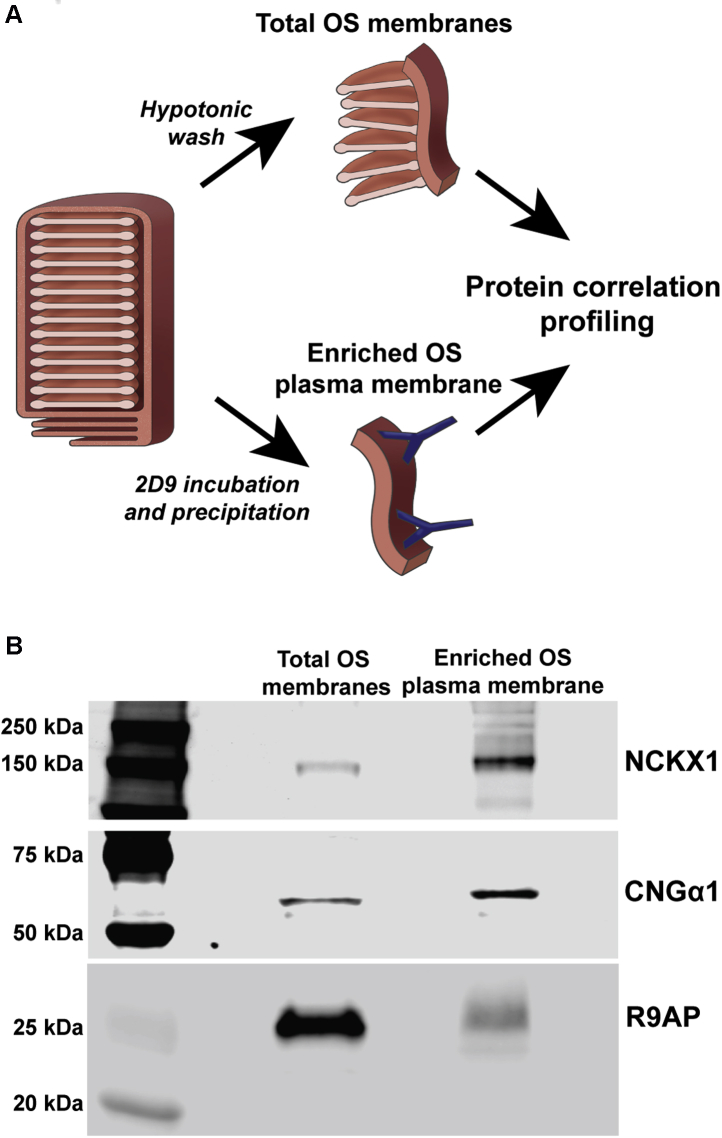
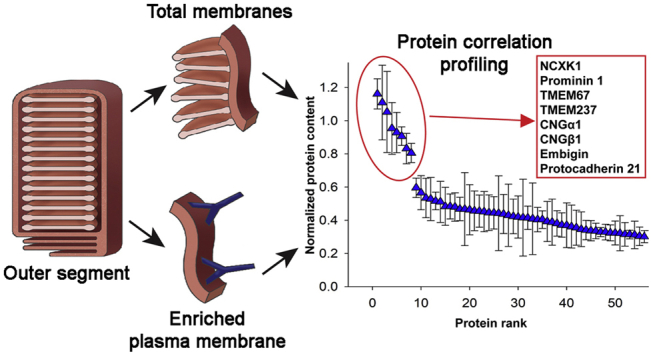
To identify the unique proteome of the OS plasma membrane, we conducted protein correlation profiling of two membrane preparations: total OS membranes and enriched OS plasma membrane. Each preparation was obtained as illustrated in Figure 1A. Total OS membranes were prepared by extensive hypotonic washing of bovine ROSs, which effectively removes soluble and peripheral membrane proteins (33). The OS plasma membrane was enriched from this preparation using a procedure whereby it is dissociated from photoreceptor discs by incubating freshly obtained bovine ROS with magnetic beads conjugated with mAb PMe 2D9 against NCKX1 (27), a protein specifically residing in this membrane (20). This procedure results in substantial separation of plasma membrane from discs, as evidenced by the enrichment in the resulting preparation of both NCKX1 and another OS plasma membrane marker, CNGα1 (Fig. 1B; see also (27)) and the depletion of the disc-specific protein R9AP (Fig. 1B).
Fig. 1.
OS plasma membrane enrichment for protein correlation profiling.A, workflow of obtaining membrane preparations for protein correlation profiling. Total OS membranes were obtained by hypotonic washing of intact bovine ROS, and the OS plasma membrane was prepared by incubating bovine ROS with PMe 2D9 antibody followed by membrane enrichment on protein A/G magnetic beads. B, western blots of total OS membranes and OS plasma membrane preparations (20 μg per lane) probed with anti-NCKX1, anti-CNGα1, and anti-R9AP antibodies. Data are taken from one of three similar experiments. OS, outer segment; ROS, rod outer segment.
To conduct protein correlation profiling, the preparations of total OS membranes and enriched OS plasma membrane were solubilized, digested with trypsin, and subjected to label-free quantitative LC-MS/MS analysis. To account for nonspecific protein binding to the antibody-conjugated beads used for OS plasma membrane enrichment, we used control beads conjugated to an anti-Myc mAb and excluded proteins binding to control beads from the analysis (see Experimental Procedures). Three sets of independently obtained membrane preparations were analyzed, each yielding between 700 and 900 proteins that could be confidently identified and quantified in both crude and enriched preparations.
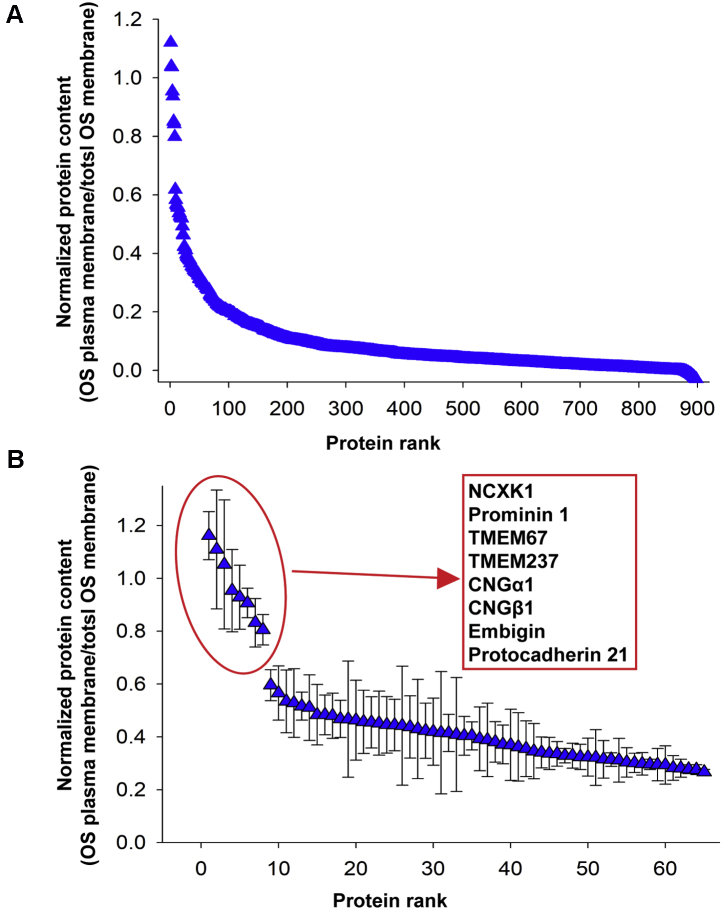
To compare the relative abundance of each identified protein between the two membrane preparations, we first calculated the sum of the ion intensities of its constituent peptides. For each preparation, we then calculated a ratio between protein abundances in the OS plasma membrane sample and the total OS membrane sample, as a measure of protein enrichment. Next, we compared the degree of each protein’s enrichment with that of three well-established rod-specific OS plasma membrane markers, NCKX1, CNGα1, and CNGβ1. This was accomplished by averaging the resulting values for NCKX1, CNGα1, and CNGβ1 and normalizing the ratio values for all other proteins to this average markers’ value. The resulted ratios were plotted in rank order in Figure 2A (see also supplemental Table S1). In this calculation, the average ratio for the markers is equal to 1, so that other proteins yielding ratios close to 1 are likely to be unique OS plasma membrane components. Proteins with lower ratios represent either nonunique constituents of this membrane or, more commonly, contaminations.
Fig. 2.
Protein correlation profiling of the OS plasma membrane.A, a representative protein correlation profiling experiment. Normalized enrichment ratios (OS plasma membrane/total OS membrane preparation) are plotted in the rank order for all proteins identified in both membrane preparations. B, the normalized enrichment ratios for 65 top proteins averaged across three independent profiling experiments plotted in the rank order. Error bars indicate SD. Eight proteins listed in the inset in the rank order profiled with ratios close to 1 across all experiments. See supplemental Table S1 for MS data. OS, outer segment.
A representative protein correlation profiling experiment is shown in Figure 2A, and data for 65 top-ranked proteins averaged among three independently conducted experiments are shown in Figure 2B. Remarkably, only five proteins were clustered with the OS plasma membrane markers. For each of them, the deviation of the normalized ratio value from unity did not exceed the coefficient of variation calculated for the entire dataset (21.2%). These proteins were prominin-1, protocadherin-21, TMEM67, TMEM237, and embigin. Among them, prominin-1 and protocadherin-21 are relatively well characterized. They interact with one another and are thought to participate in the morphogenesis of newly forming photoreceptor discs (3). Their profiling with OS plasma membrane markers suggests either that they sequester into this membrane after disc maturation or that membranes of newly forming discs (whose membranes are still contiguous with the OS plasma membrane) cosegregate with the OS plasma membrane during our fractionation procedure. Two other proteins, TMEM67 and TMEM237, have been previously described as OS proteins (22, 34), although their functions remain unknown, whereas embigin has not been previously reported to be expressed in photoreceptors.
Assessment of OS Plasma Membrane Enrichment
Another utility of protein correlation profiling is that it allowed us to quantitatively evaluate the extent of OS plasma membrane separation from various types of contaminating membranes. For example, we assessed the degree of OS plasma membrane separation from discs. The normalized profiling ratios for two disc-specific proteins ABCA4 and ROM1 were 0.19 and 0.23, suggesting that the degree of OS plasma membrane enrichment over discs was ~4- to 5-fold. On the other hand, rhodopsin, known to be present in both OS plasma membrane and discs (3), yielded a higher profiling ratio of 0.48, consistent with its localization to both membrane types.
The only unexpected observation was that the disc-specific protein peripherin-2 yielded a profiling ratio of 0.53, similar to that of rhodopsin but not in the range of other disc protein markers. The most likely explanation for this anomaly is that peripherin-2 interacts with CNGβ1 across the OS cytoplasm (8), which may result in cosegregation of disc fragments containing peripherin-2 with the OS plasma membrane during its enrichment.
Similarly, we were able to assess the degree of OS plasma membrane enrichment over contaminations representing the inner segment and/or RPE microvilli. The profiling ratios for basigin and GLUT1, two proteins abundantly expressed in each of these membranes but not in the OS, were 0.14 and 0.11, respectively. This indicates an ~8-fold depletion of these membranes from the OS plasma membrane preparation.
TMEM67 and TMEM237 Are Unique OS Plasma Membrane Proteins
In the next set of experiments, we validated the OS plasma membrane localization of TMEM67 (also called meckelin) and TMEM237 suggested by protein correlation profiling. Both of them were previously shown to localize to OSs (22, 34) and to be associated with various syndromic ciliopathies. Mutations in TMEM67 are associated with both COACH (35) and Meckel–Gruber (36) syndromes, whereas mutations in TMEM237 are associated with Joubert syndrome (37). The KO or mutation of Tmem67 in rodents results in defects of OS morphology and severe retinal degeneration (34, 38).
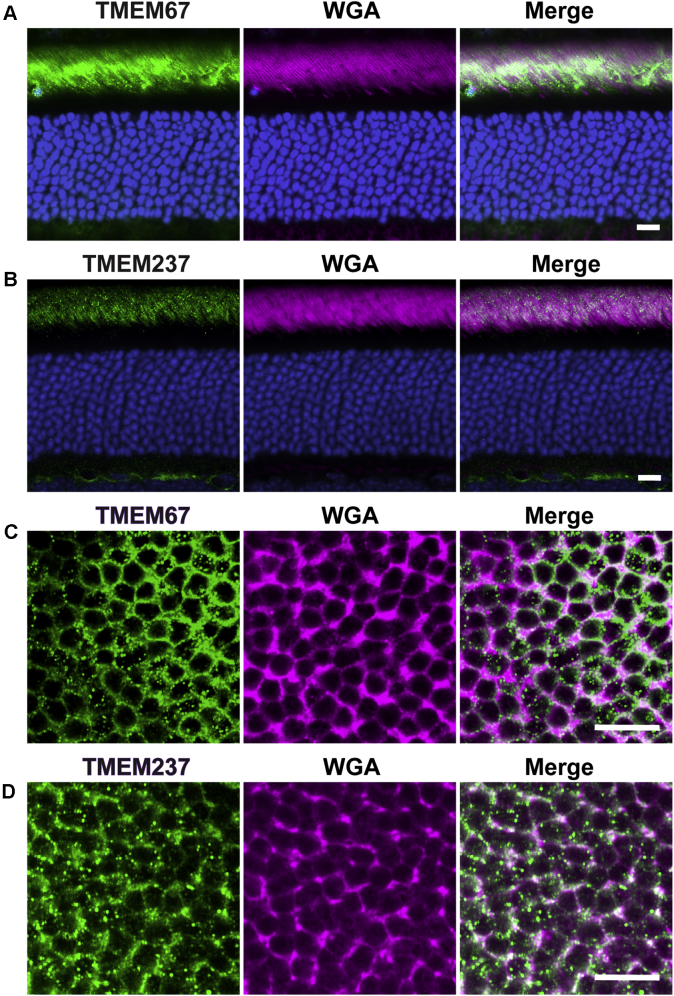
Immunofluorescent staining of TMEM67 and TMEM237 in longitudinal cross-sections of mouse retinas confirmed that each protein is localized exclusively to photoreceptor OS (Fig. 3, A and B), consistent with previous reports. To confirm that these proteins are confined to the OS plasma membrane, we immunostained tangential retinal sections cut through the OS layer (Fig. 3, C and D). The staining pattern of each protein appeared as a ring colocalizing with the OS plasma membrane marker, wheat germ agglutinin. We also noted the presence of punctate staining, which roughly followed the contours of these rings; we suspect that this punctum is an artifact originating from some imperfection in tissue preservation in tangential sections. These results corroborate our findings from protein correlation profiling and establish that TMEM67 and TMEM237 are indeed unique components of the OS plasma membrane.
Fig. 3.
Immunostaining of TMEM67 and TMEM237 inWT mouse retina. TMEM67 and TMEM237 staining in retinal cross-sections (A and B) and tangential sections across the outer segment layer (C and D) is shown in green, as indicated. Staining of the OS plasma membrane marker WGA is shown in magenta. Scale bars are 10 μm for panels A and B and 5 μm for panels C and D. For each panel, data are taken from at least three experiments. OS, outer segment; WGA, wheat germ agglutinin.
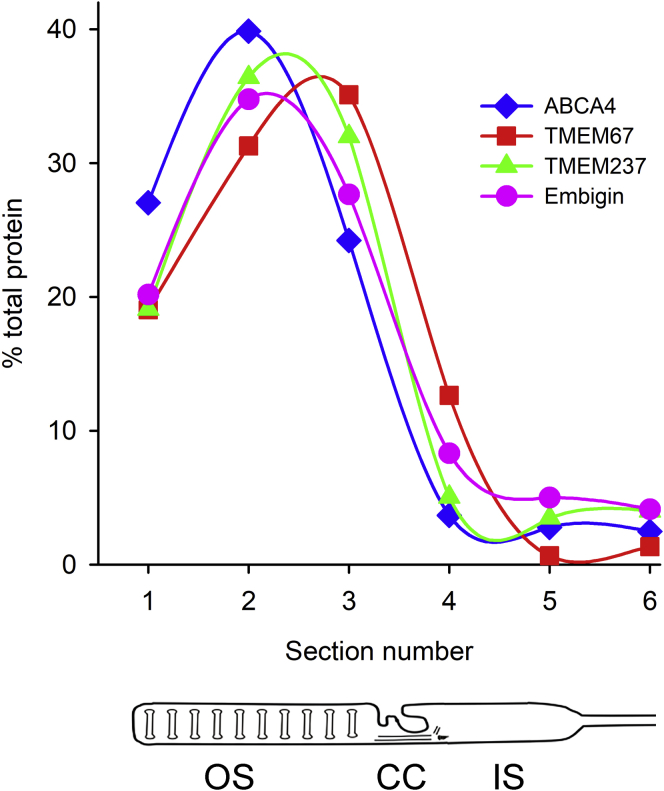
Although previous studies suggested that localization of both TMEM67 and TMEM237 is biased to the OS base (22, 34), their immunostaining in Figure 3, A and B appears to be relatively even throughout the OS. To address this discrepancy, we analyzed the distribution of each protein along the OS length using quantitative MS. We cut serial 10-μm-thick tangential sections through the photoreceptor layer of a frozen flat-mounted mouse retina and determined relative amounts of TMEM67 and TMEM237 in each section using label-free quantitative proteomics (Fig. 4; supplemental Table S2). The distribution of these proteins across individual sections was compared with that of ABCA4, a protein known to be evenly distributed throughout the OS length (39, 40). Whereas the distribution of TMEM237 closely resembled that of ABCA4 with both proteins being most abundant in section #2, TMEM67 was most abundant in section #3, suggesting a shift toward the OS base that could be underappreciated from immunostaining. Nonetheless, both proteins were present in each section originating from the OS layer.
Fig. 4.
Distribution of TMEM67, TMEM237, and embigin in serial tangential sections representing the outer/inner segment layer of the mouse retina. Serial 10-μm-thick tangential sections were cut through the photoreceptor layer of a frozen flat-mounted retina of the WT mouse, and the relative content of TMEM67, TMEM237, embigin and ABCA4 in each section was determined using label-free quantitative proteomics. Cartoon at the bottom illustrates an approximate correspondence between the sections and subcellular compartments of photoreceptor cells. See supplemental Table S2 for MS data. CC, connecting cilium; IS, inner segment; OS, outer segment.
We next determined the absolute amounts of TMEM67 and TMEM237 in purified mouse ROS using MS with cold isotope-labeled peptide standards. Because it is customary to express the OS protein amounts as molar ratios to rhodopsin, we also quantified the content of rhodopsin in the same preparations. As summarized in Table 1 (see also supplemental Table S3), both proteins were found to be expressed at comparable levels corresponding to ~1:3900 and 1:3100 M ratios to rhodopsin for TMEM67 and TMEM237, respectively. Given that a mouse rod contains between 5∙107 and 7∙107 rhodopsin molecules (41, 42), the number of TMEM67 molecules in each rod is ~15,000 and the amount of TMEM237 molecules is ~19,000. For a reference, we similarly quantified the rod content of ABCA4, used as a disc marker in Figure 4, and found that it is expressed in the amount of ~200,000 molecules per rod, somewhat lower than in previous estimates from semiquantitative Western blotting (39).
Table 1.
Absolute quantification of TMEM67, TMEM237, and embigin in mouse rod outer segments
| Protein | Molar ratio with rhodopsin | Molecules per rod |
|---|---|---|
| TMEM237 | 1:3118 ± 412 | ~19,000 |
| TMEM67 | 1:3922 ± 109 | ~15,000 |
| Embigin | 1:6238 ± 1168 | ~10,000 |
| ABCA4 | 1:302 ± 20 | ~200,000 |
Data are averaged from three independent experiments and presented as the mean ± SD. MS data are presented in supplemental Table S3.
Embigin Is a Unique OS Plasma Membrane Protein Associated With Monocarboxylate Transporter MCT1
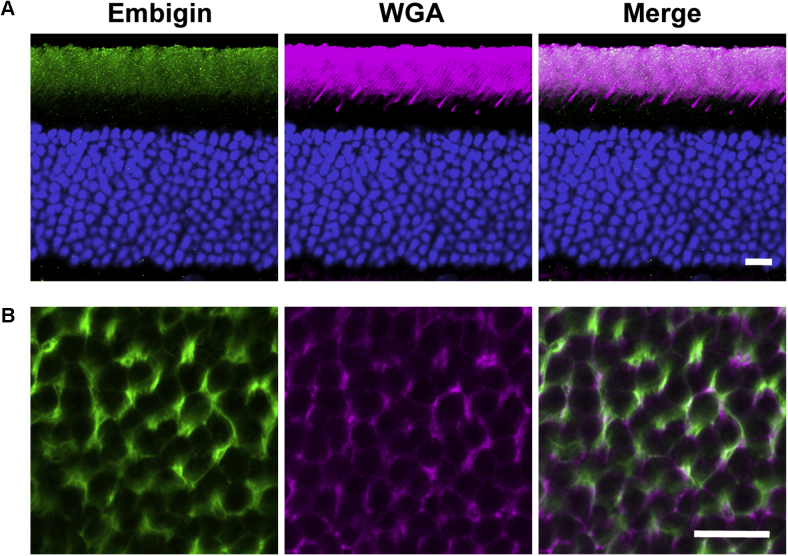
Embigin belongs to a family of integral membrane proteins, alongside basigin and neuroplastin, whose major function is directing monocarboxylate transporters (MCTs) to the plasma membrane of many cell types and regulating MCT function once delivered there (43, 44). However, it had not been previously found in photoreceptors. Immunostaining of retinal cross-sections showed that embigin is localized specifically to photoreceptor OSs (Fig. 5A), and its specific localization to the OS plasma membrane was confirmed by immunostaining of tangential sections cut across the OS layer (Fig. 5B). Embigin appears to be evenly immunostained along the OS length, which was corroborated by quantitative MS of serial tangential sections (Fig. 4; supplemental Table S2). The absolute amount of embigin in mouse rods, estimated as above, is ~10,000 molecules per rod (Table 1; supplemental Table S3).
Fig. 5.
Immunostaining of embigin in WT mouse retina. Embigin staining in retinal cross-sections (A) and tangential sections across the outer segment layer (B) is shown in green. WGA staining is shown in magenta. Scale bars are 10 μm for panel A and 5 μm for panel B. For each panel, data are taken from at least three experiments. WGA, wheat germ agglutinin.
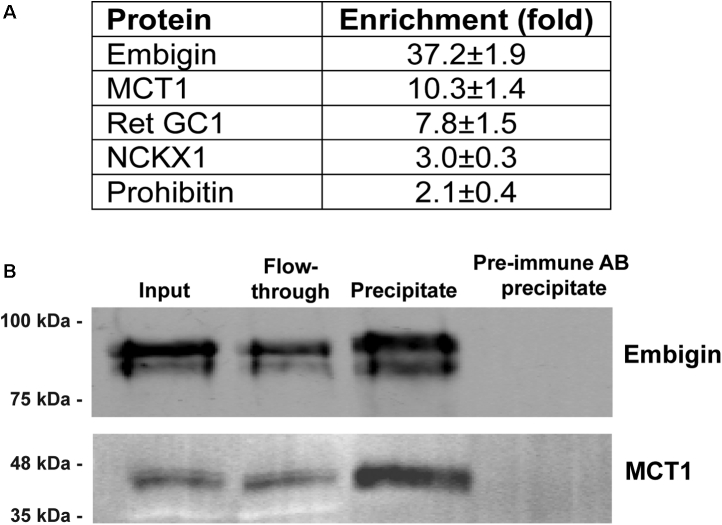
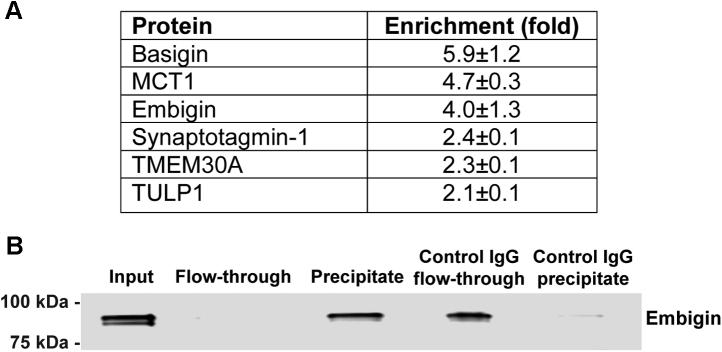
The finding that embigin is a unique component of the OS plasma membrane is the most original result of our profiling analysis. This provoked us to gain further insight into its potential role in photoreceptors. We searched for embigin’s interacting partners by immunoprecipitating proteins from solubilized ROS with the anti-embigin mAb 3C4. To control for interaction specificity, we performed a parallel precipitation with preimmune antibodies. We found that only five proteins—embigin itself, the monocarboxylate transporter MCT1, retinal guanylate cyclase 1, NCKX1, and prohibitin—were enriched by >2-fold in the 3C4 precipitate over the control precipitate (Fig. 6A). The second most enriched protein after embigin was MCT1 whose coprecipitation with embigin was additionally documented by Western blotting (Fig. 6B). Reciprocal coprecipitation using a polyclonal anti-MCT1 antibody confirmed its interaction with embigin, as well as with basigin known to interact with MCT1 in the photoreceptor inner segment (Fig. 7A). The precipitation of embigin by the anti-MCT1 antibody was further confirmed by Western blotting (Fig. 7B).
Fig. 6.
Coprecipitation of monocarboxylate transporter MCT1 with embigin.A, proteins enriched by >2-fold in immunoprecipitates of solubilized bovine ROS proteins by anti-embigin antibody 3C4 over the control precipitate with the preimmune antibody. The relative amounts of proteins in precipitates were determined using label-free MS. Data are shown as the mean ± SD (n = 2). B, MCT1 coprecipitation with embigin using the anti-embigin antibody 3C4, probed by Western blotting with 3C4 and anti-MCT1 antibodies. Preimmune antibody was used in control precipitation experiments. 20 μg of total protein is loaded in each lane. Data are taken from one of two similar experiments. ROS, rod outer segment.
Fig. 7.
Coprecipitation of embigin with MCT1.A, proteins enriched by >2-fold in immunoprecipitates of solubilized bovine ROS proteins by anti-MCT1 antibody over the control precipitate with rabbit immunoglobulin G. The relative amounts of proteins in precipitates were determined using label-free MS. Data are shown as the mean ± SD (n = 2). B, embigin coprecipitation with MCT1 using the anti-MCT1 antibody, probed by Western blotting with anti-embigin antibody 3C4. Rabbit immunoglobulin G was used in control precipitation experiments. The amount of protein loaded to each lane is normalized to 20% of input material. Data are taken from one of three similar experiments. ROS, rod outer segment.
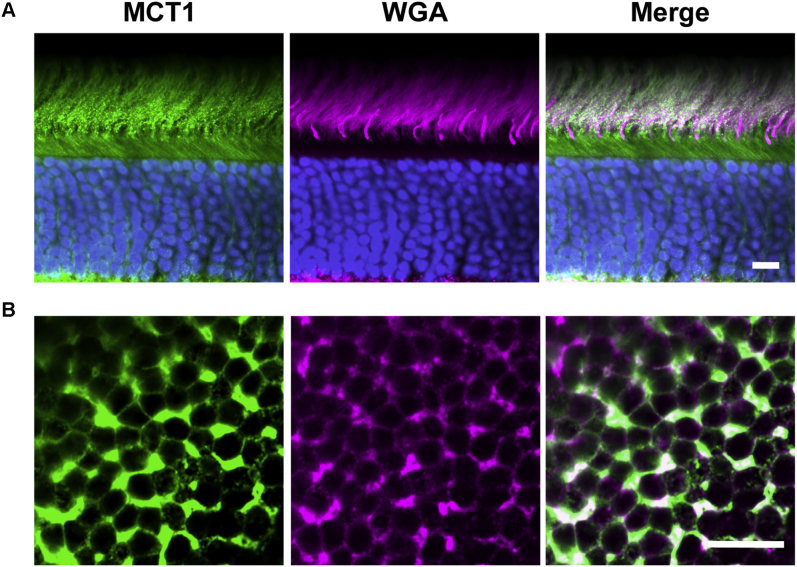
The embigin interaction with MCT1 is consistent with observations in other tissues that embigin is always associated with monocarboxylate transporters (45, 46). What is surprising is that MCT1 has been previously shown to localize to apical membranes of the RPE and the photoreceptor inner segment, in both cases in complex with basigin (47, 48, 49), but not to the OS. Therefore, we investigated whether a portion of photoreceptor MCT1 is in fact localized to the OS. This was technically challenging because the adjacent inner segment and RPE both have very high levels of MCT1 expression. To increase the sensitivity of MCT1 detection, we used an immunostaining procedure whereby the primary antibody is incubated with the retina before tissue fixation. This approach often facilitates antibody staining, presumably because of preservation of epitopes from chemical modification by the fixative (50). As shown in Figure 8A, this procedure yielded strong MCT1 immunostaining in both the inner segment and OS of WT mouse retinas thoroughly detached from the RPE.
Fig. 8.
Immunostaining of MCT1 in WT mouse retina. MCT1 staining in retinal cross-sections (A) and tangential sections across the outer segment layer (B) is shown in green. WGA staining is shown in magenta. Scale bars are 10 μm for panel A and 5 μm for panel B. For each panel, data are taken from at least three experiments. WGA, wheat germ agglutinin.
An independent line of evidence corroborating the presence of MCT1 in the OS plasma membrane follows from protein correlation profiling. The profiling ratio for MCT1 was 0.48; that is, it was depleted by ~2-fold from the starting material. If this MCT1 originated solely from contaminating membranes of the inner segment and/or RPE, then the degree of MCT1 depletion upon OS plasma membrane enrichment should be comparable with that of protein markers representing these contaminating membranes. However, the actual depletion of these membranes combined was ~8-fold, as detailed above. This discrepancy indicates that the majority of MCT1 present in the OS plasma membrane preparation could not have originated from contaminations. Together, these data suggest that both embigin and MCT1 are components of the OS plasma membrane, where they exist in complex with one another.
Discussion
In this study, we used the powerful MS technique of protein correlation profiling to identify unique components of the photoreceptor OS plasma membrane. Remarkably, this analysis revealed that only eight proteins are localized exclusively to this membrane, a surprisingly low number considering the diverse functions that this membrane plays in photoreceptors. Three of these proteins (NCKX1, CNGα1, and CNGβ1) are previously characterized components of this membrane used as marker proteins in our study. Two others (prominin-1 and protocadherin-21) are known to be located within the expanding edges of newly forming discs, which are contiguous with the OS plasma membrane. TMEM67 and TMEM237 had not previously been identified as components of this membrane, although they were shown to localize to OSs. Finally, embigin was not previously known to be expressed in photoreceptors at all.
Beyond these unique proteins, many other profiled as possible nonunique components of the OS plasma membrane. Particularly, a group of ~60 proteins showed profiling ratios between ~0.6 and ~0.25, representing values above profiling ratios for the disc-specific markers. Although some of them are likely to be contaminations from other membrane types, others may represent proteins that are nonunique but present in the OS plasma membrane, such as, for example, rhodopsin. This list of candidate proteins is a valuable resource for future studies, although each protein requires independent validation before any conclusions can be drawn regarding its presence in the OS plasma membrane.
Assessment of Protein Detection Limit
A natural question is whether the OS plasma membrane indeed has only eight unique components or some less-abundant proteins fall below the sensitivity limits of our analysis. Whereas we cannot answer this question directly, we can assess the lower limit of protein detection by our instrumentation. Absolute quantification of TMEM67, TMEM237, and embigin showed that they are expressed in the amount of ~15,000, 20,000 and 10,000 molecules per rod. The total ion intensity for these proteins’ constituent peptides was at least ~1,000,000, which is far above the detection threshold. The corresponding values for the 50 least abundant proteins that were still confidently identified ranged from 12,000 to 227,000 with the average value of 123,000. This suggests that our analysis was sufficiently sensitive to detect proteins expressed at the level of at least 1000 molecules per rod and likely even an order of magnitude less.
We also considered whether any proteins previously reported as unique OS plasma membrane components were not identified as such in our analysis. While the literature describing such proteins is extremely limited, we did expect to find one protein, insulin growth factor 1 receptor, that had been proposed to be predominantly localized to the OS plasma membrane (51). However, despite this protein being confidently identified across all three experiments, it was one of the most depleted in the OS plasma membrane preparation (profiling ratio <0.01). This result strongly suggests that insulin growth factor 1 receptor is not an OS plasma membrane protein.
Profiling of Phototransduction Proteins
Another finding from this study is that the OS plasma membrane is strongly depleted of the three transmembrane proteins involved in phototransduction: retinal guanylate cyclases 1 and 2 and R9AP. The profiling ratios for these proteins varied between 0.17 and 0.25, which is in line with the ratios for disc markers, ABCA4 and ROM1, but not for rhodopsin, a protein shared between discs and the plasma membrane. This suggests that at least the phototransduction events involving these proteins (cascade deactivation and cGMP restoration) occur predominantly on disc membranes. This result is interesting, as there has been limited information on whether the phototransduction cascade is operating primarily on the discs or both discs and plasma membrane. It has even been suggested that guanylate cyclase may be confined to or enriched in the plasma membrane (52), which our current data refute along with our previous proteomic studies demonstrating that both photoreceptor-specific guanylate cyclase isoforms reside in discs (21, 26) and immunolocalization studies observing these isoforms predominantly located at disc edges (53).
Embigin, MCT1, and Lactate Transport in the OS
Our most original observation is that the OS plasma membrane contains embigin associated with the monocarboxylate transporter MCT1. Embigin belongs to a class of cell surface glycosylated proteins which anchor to the plasma membrane through a single transmembrane domain and possess a large glycosylated extracellular portion with two or three immunoglobulin-like domains (two in the case of embigin) (43). Although embigin is typically associated with MCT2, it has been shown to interact with MCT1 in rat erythrocytes, where it directs MCT1 to the plasma membrane and regulates its activity (54). We now demonstrate a similar association of embigin with MCT1 in the photoreceptor OS plasma membrane. This result was unexpected because previous literature had only documented the presence of MCT1 in the photoreceptor inner segment in complex with basigin, where it transports lactate into and out of photoreceptor cells (47, 48, 49).
The export of lactate from photoreceptors is critically important for retinal metabolism. Photoreceptors predominantly rely on aerobic glycolysis for generating ATP, which results in the production of lactate that must be exported out of photoreceptors into both RPE and Müller cells for further energy processing (55). Although it has been previously suggested that this transport occurs solely through the inner segments by MCT1 in complex with basigin (48, 49), our data demonstrate that MCT1 is also expressed in the OS plasma membrane in complex with embigin. Although the precise functional significance of this specialization remains to be determined, one attractive hypothesis is that using two compartment-specific chaperones for MCT1 delivery to the inner segment and OS allows the photoreceptor to achieve an optimal distribution of lactate transport throughout the cell.
In conclusion, our study highlights the power of protein correlation profiling for identifying unique components of cellular membranes or organelles that cannot be obtained in sufficient purity. We showed the plasma membrane enveloping the OS organelle of photoreceptor cells contains only eight unique proteins expressed at levels exceeding several hundred molecules per cell. These findings will undoubtedly facilitate progress toward understanding the functional roles which these proteins play in supporting photoreceptor health and/or the energy metabolism of the retina.
Data Availability
MS data have been deposited to the ProteomeXchange Consortium via the PRIDE[1] partner repository with the dataset identifier PXD022864 and 10.6019/PXD022864.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Unrestricted grant to Duke University by Research to Prevent Blindness.
Author contributions
N. P. S.: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing - original draft & Writing - review and editing, Supervision, Funding acquisition, M. A. C.: Validation, Formal analysis, Investigation, Writing - original draft & Writing - review and editing, L. M.: Formal analysis, Investigation, Resources, J. Y. S. H.: Investigation, T. R. L.: Investigation, Writing - review and editing, W. J. S.: Investigation, Writing - review and editing, W. J. T.: Investigation, S. H.: Investigation, N. J. P.: Conceptualization, Writing - review and editing, Funding acquisition, R. S. M.: Conceptualization, Writing - review and editing, Funding acquisition, V. Y. A.: Conceptualization, Writing - review and editing, Project administration, Funding acquisition.
Funding and additional information
This work was supported by the NIH Grants EY027484 (N. P. S.), EY030451 (V. Y. A.), EY005722 (V. Y. A.), EY002422 (R. S. M.), EY029929 (T. R. L.), and EY012042 (N. J. P.) and Canadian Institutes of Health Research (CIHR) Grant PJT 148649 (R. S. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Nikolai P. Skiba, Email: nikolai.skiba@duke.edu.
Vadim Y. Arshavsky, Email: vadim.arshavsky@duke.edu.
Supplemental Data
References
- 1.Arshavsky V.Y., Burns M.E. Photoreceptor signaling: Supporting vision across a wide range of light intensities. J. Biol. Chem. 2012;287:1620–1626. doi: 10.1074/jbc.R111.305243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearring J.N., Salinas R.Y., Baker S.A., Arshavsky V.Y. Protein sorting, targeting and trafficking in photoreceptor cells. Prog. Retin. Eye Res. 2013;36:24–51. doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg A.F., Moritz O.L., Williams D.S. Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 2016;55:52–81. doi: 10.1016/j.preteyeres.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baehr W., Hanke-Gogokhia C., Sharif A., Reed M., Dahl T., Frederick J.M., Ying G. Insights into photoreceptor ciliogenesis revealed by animal models. Prog. Retin. Eye Res. 2019;71:26–56. doi: 10.1016/j.preteyeres.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wensel T.G., Zhang Z., Anastassov I.A., Gilliam J.C., He F., Schmid M.F., Robichaux M.A. Structural and molecular bases of rod photoreceptor morphogenesis and disease. Prog. Retin. Eye Res. 2016;55:32–51. doi: 10.1016/j.preteyeres.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young R.W. The renewal of photoreceptor cell outer segments. J. Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakkaraju A., Umapathy A., Tan L.X., Daniele L., Philp N.J., Boesze-Battaglia K., Williams D.S. The cell biology of the retinal pigment epithelium. Prog. Retin. Eye Res. 2020:100846. doi: 10.1016/j.preteyeres.2020.100846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poetsch A., Molday L.L., Molday R.S. The cGMP-gated channel and related glutamic acid-rich proteins interact with peripherin-2 at the rim region of rod photoreceptor disc membranes. J. Biol. Chem. 2001;276:48009–48016. doi: 10.1074/jbc.M108941200. [DOI] [PubMed] [Google Scholar]
- 9.Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 10.Foster L.J., de Hoog C.L., Zhang Y., Xie X., Mootha V.K., Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. doi: 10.1016/j.cell.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 11.Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C.P., Almeida J.A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H.R., Schliebs W., Erdmann R., Berger J. Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell. Proteomics. 2007;6:2045–2057. doi: 10.1074/mcp.M700169-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Reidel B., Thompson J.W., Farsiu S., Moseley M.A., Skiba N.P., Arshavsky V.Y. Proteomic profiling of a layered tissue reveals unique glycolytic specializations of photoreceptor cells. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook N.J., Molday L.L., Reid D., Kaupp U.B., Molday R.S. The cGMP-gated channel of bovine rod photoreceptors is localized exclusively in the plasma membrane. J. Biol. Chem. 1989;264:6996–6999. [PubMed] [Google Scholar]
- 14.Reid D.M., Friedel U., Molday R.S., Cook N.J. Identification of the sodium-calcium exchanger as the major ricin-binding glycoprotein of bovine rod outer segments and its localization to the plasma membrane. Biochemistry. 1990;29:1601–1607. doi: 10.1021/bi00458a035. [DOI] [PubMed] [Google Scholar]
- 15.Korschen H.G., Illing M., Seifert R., Sesti F., Williams A., Gotzes S., Colville C., Muller F., Dose A., Godde M., Molday L., Kaupp U.B., Molday R.S. A 240 kDa protein represents the complete beta subunit of the cyclic nucleotide-gated channel from rod photoreceptor. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z., Chen Y., Lillo C., Chien J., Yu Z., Michaelides M., Klein M., Howes K.A., Li Y., Kaminoh Y., Chen H., Zhao C., Chen Y., Al-Sheikh Y.T., Karan G. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J. Clin. Invest. 2008;118:2908–2916. doi: 10.1172/JCI35891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Z., Anderson D.W., Papermaster D.S. Prominin-1 localizes to the open rims of outer segment lamellae in Xenopus laevis rod and cone photoreceptors. Invest. Ophthalmol. Vis. Sci. 2012;53:361–373. doi: 10.1167/iovs.11-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacchigna S., Oh H., Wilsch-Brauninger M., Missol-Kolka E., Jaszai J., Jansen S., Tanimoto N., Tonagel F., Seeliger M., Huttner W.B., Corbeil D., Dewerchin M., Vinckier S., Moons L., Carmeliet P. Loss of the cholesterol-binding protein prominin-1/CD133 causes disk dysmorphogenesis and photoreceptor degeneration. J. Neurosci. 2009;29:2297–2308. doi: 10.1523/JNEUROSCI.2034-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rattner A., Smallwood P.M., Williams J., Cooke C., Savchenko A., Lyubarsky A., Pugh E.N., Nathans J. A photoreceptor-specific cadherin is essential for the structural integrity of the outer segment and for photoreceptor survival. Neuron. 2001;32:775–786. doi: 10.1016/s0896-6273(01)00531-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim T.S., Reid D.M., Molday R.S. Structure-function relationships and localization of the Na/Ca-K exchanger in rod photoreceptors. J. Biol. Chem. 1998;273:16561–16567. doi: 10.1074/jbc.273.26.16561. [DOI] [PubMed] [Google Scholar]
- 21.Kwok M.C., Holopainen J.M., Molday L.L., Foster L.J., Molday R.S. Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol. Cell. Proteomics. 2008;7:1053–1066. doi: 10.1074/mcp.M700571-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuniga F.I., Craft C.M. Deciphering the structure and function of Als2cr4 in the mouse retina. Invest. Ophthalmol. Vis. Sci. 2010;51:4407–4415. doi: 10.1167/iovs.10-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philp N.J., Wang D., Yoon H., Hjelmeland L.M. Polarized expression of monocarboxylate transporters in human retinal pigment epithelium and ARPE-19 cells. Invest. Ophthalmol. Vis. Sci. 2003;44:1716–1721. doi: 10.1167/iovs.02-0287. [DOI] [PubMed] [Google Scholar]
- 24.Keresztes G., Mutai H., Hibino H., Hudspeth A.J., Heller S. Expression patterns of the RGS9-1 anchoring protein R9AP in the chicken and mouse suggest multiple roles in the nervous system. Mol. Cell. Neurosci. 2003;24:687–695. doi: 10.1016/s1044-7431(03)00231-8. [DOI] [PubMed] [Google Scholar]
- 25.McDowell J.H. Preparing rod outer segment membranes, regenerating rhodopsin, and determining rhodopsin concentration. Methods Neurosci. 1993;15:123–130. [Google Scholar]
- 26.Skiba N.P., Spencer W.J., Salinas R.Y., Lieu E.C., Thompson J.W., Arshavsky V.Y. Proteomic identification of unique photoreceptor disc components reveals the presence of PRCD, a protein linked to retinal degeneration. J. Proteome Res. 2013;12:3010–3018. doi: 10.1021/pr4003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molday R.S., Molday L.L. Differences in the protein composition of bovine retinal rod outer segment disk and plasma membranes isolated by a ricin-gold-dextran density perturbation method. J. Cell Biol. 1987;105(6 Pt 1):2589–2601. doi: 10.1083/jcb.105.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., Krijgsveld J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobanova E.S., Herrmann R., Finkelstein S., Reidel B., Skiba N.P., Deng W.T., Jo R., Weiss E.R., Hauswirth W.W., Arshavsky V.Y. Mechanistic basis for the failure of cone transducin to translocate: Why cones are never blinded by light. J. Neurosci. 2010;30:6815–6824. doi: 10.1523/JNEUROSCI.0613-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leyton-Puig D., Kedziora K.M., Isogai T., van den Broek B., Jalink K., Innocenti M. PFA fixation enables artifact-free super-resolution imaging of the actin cytoskeleton and associated proteins. Biol. Open. 2016;5:1001–1009. doi: 10.1242/bio.019570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokolov M., Lyubarsky A.L., Strissel K.J., Savchenko A.B., Govardovskii V.I., Pugh E.N., Jr., Arshavsky V.Y. Massive light-driven translocation of transducin between the two major compartments of rod cells: A novel mechanism of light adaptation. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 32.Lobanova E.S., Finkelstein S., Song H., Tsang S.H., Chen C.-K., Sokolov M., Skiba N.P., Arshavsky V.Y. Transducin translocation in rods is triggered by saturation of the GTPase-activating complex. J. Neurosci. 2007;27:1151–1160. doi: 10.1523/JNEUROSCI.5010-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith H.G., Jr., Stubbs G.W., Litman B.J. The isolation and purification of osmotically intact discs from retinal rod outer segments. Exp. Eye Res. 1975;20:211–217. doi: 10.1016/0014-4835(75)90134-7. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari S., Hudson S., Gattone V.H., 2nd, Miller C., Chernoff E.A., Belecky-Adams T.L. Meckelin 3 is necessary for photoreceptor outer segment development in rat Meckel syndrome. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doherty D., Parisi M.A., Finn L.S., Gunay-Aygun M., Al-Mateen M., Bates D., Clericuzio C., Demir H., Dorschner M., van Essen A.J., Gahl W.A., Gentile M., Gorden N.T., Hikida A., Knutzen D. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis) J. Med. Genet. 2010;47:8–21. doi: 10.1136/jmg.2009.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith U.M., Consugar M., Tee L.J., McKee B.M., Maina E.N., Whelan S., Morgan M.V., Goranson E., Gissen P., Lilliquist S., Aligianis I.A., Ward C.J., Pasha S., Punyashthiti R., Malik Sharif S. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- 37.Huang L., Szymanska K., Jensen V.L., Janecke A.R., Innes A.M., Davis E.E., Frosk P., Li C., Willer J.R., Chodirker B.N., Greenberg C.R., McLeod D.R., Bernier F.P., Chudley A.E., Muller T. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 2011;89:713–730. doi: 10.1016/j.ajhg.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collin G.B., Won J., Hicks W.L., Cook S.A., Nishina P.M., Naggert J.K. Meckelin is necessary for photoreceptor intraciliary transport and outer segment morphogenesis. Invest. Ophthalmol. Vis. Sci. 2012;53:967–974. doi: 10.1167/iovs.11-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun H., Nathans J. Stargardt's ABCR is localized to the disc membrane of retinal rod outer segments. Nat. Genet. 1997;17:15–16. doi: 10.1038/ng0997-15. [DOI] [PubMed] [Google Scholar]
- 40.Molday L.L., Wahl D., Sarunic M.V., Molday R.S. Localization and functional characterization of the p.Asn965Ser (N965S) ABCA4 variant in mice reveal pathogenic mechanisms underlying Stargardt macular degeneration. Hum. Mol. Genet. 2018;27:295–306. doi: 10.1093/hmg/ddx400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyubarsky A.L., Daniele L.L., Pugh E.N., Jr. From candelas to photoisomerizations in the mouse eye by rhodopsin bleaching in situ and the light-rearing dependence of the major components of the mouse ERG. Vis. Res. 2004;44:3235–3251. doi: 10.1016/j.visres.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 42.Nickell S., Park P.S., Baumeister W., Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J. Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iacono K.T., Brown A.L., Greene M.I., Saouaf S.J. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp. Mol. Pathol. 2007;83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Escuredo J., Van Hee V.F., Sboarina M., Falces J., Payen V.L., Pellerin L., Sonveaux P. Monocarboxylate transporters in the brain and in cancer. Biochim. Biophys. Acta. 2016;1863:2481–2497. doi: 10.1016/j.bbamcr.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halestrap A.P. The monocarboxylate transporter family--structure and functional characterization. IUBMB Life. 2012;64:1–9. doi: 10.1002/iub.573. [DOI] [PubMed] [Google Scholar]
- 46.Halestrap A.P., Wilson M.C. The monocarboxylate transporter family--role and regulation. IUBMB Life. 2012;64:109–119. doi: 10.1002/iub.572. [DOI] [PubMed] [Google Scholar]
- 47.Peachey N.S., Yu M., Han J.Y.S., Lengacher S., Magistretti P.J., Pellerin L., Philp N.J. Impact of MCT1 haploinsufficiency on the mouse retina. Adv. Exp. Med. Biol. 2018;1074:375–380. doi: 10.1007/978-3-319-75402-4_46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Philp N.J., Ochrietor J.D., Rudoy C., Muramatsu T., Linser P.J. Loss of MCT1, MCT3, and MCT4 expression in the retinal pigment epithelium and neural retina of the 5A11/basigin-null mouse. Invest. Ophthalmol. Vis. Sci. 2003;44:1305–1311. doi: 10.1167/iovs.02-0552. [DOI] [PubMed] [Google Scholar]
- 49.Han J.Y.S., Kinoshita J., Bisetto S., Bell B.A., Nowak R.A., Peachey N.S., Philp N.J. Role of monocarboxylate transporters in regulating metabolic homeostasis in the outer retina: Insight gained from cell-specific Bsg deletion. FASEB J. 2020;34:5401–5419. doi: 10.1096/fj.201902961R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stradleigh T.W., Ishida A.T. Fixation strategies for retinal immunohistochemistry. Prog. Retin. Eye Res. 2015;48:181–202. doi: 10.1016/j.preteyeres.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dilly A.K., Rajala R.V. Insulin growth factor 1 receptor/PI3K/AKT survival pathway in outer segment membranes of rod photoreceptors. Invest. Ophthalmol. Vis. Sci. 2008;49:4765–4773. doi: 10.1167/iovs.08-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X., Seno K., Nishizawa Y., Hayashi F., Yamazaki A., Matsumoto H., Wakabayashi T., Usukura J. Ultrastructural localization of retinal guanylate cyclase in human and monkey retinas. Exp. Eye Res. 1994;59:761–768. doi: 10.1006/exer.1994.1162. [DOI] [PubMed] [Google Scholar]
- 53.Nemet I., Tian G., Imanishi Y. Organization of cGMP sensing structures on the rod photoreceptor outer segment plasma membrane. Channels (Austin) 2014;8:528–535. doi: 10.4161/19336950.2014.973776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poole R.C., Halestrap A.P. Interaction of the erythrocyte lactate transporter (monocarboxylate transporter 1) with an integral 70-kDa membrane glycoprotein of the immunoglobulin superfamily. J. Biol. Chem. 1997;272:14624–14628. doi: 10.1074/jbc.272.23.14624. [DOI] [PubMed] [Google Scholar]
- 55.Kanow M.A., Giarmarco M.M., Jankowski C.S., Tsantilas K., Engel A.L., Du J., Linton J.D., Farnsworth C.C., Sloat S.R., Rountree A., Sweet I.R., Lindsay K.J., Parker E.D., Brockerhoff S.E., Sadilek M. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife. 2017;6 doi: 10.7554/eLife.28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS data have been deposited to the ProteomeXchange Consortium via the PRIDE[1] partner repository with the dataset identifier PXD022864 and 10.6019/PXD022864.