Abstract
Most proteins assemble into multisubunit complexes1. The persistence of these complexes across evolutionary time is usually explained as the result of natural selection for functional properties that depend upon multimerization, like intersubunit allostery or the capacity to do mechanical work2. In many complexes, however, multimerization does not enable any known function3. An alternative explanation is that multimers could become entrenched if substitutions accumulate that are neutral in multimers but deleterious in monomers; purifying selection would then prevent reversion to the unassembled form, even if assembly per se does not enhance biological function3–7. Here we show that a hydrophobic mutational ratchet systematically entrenches molecular complexes. By applying ancestral protein reconstruction and biochemical assays to the evolution of steroid hormone receptors (SRs), we show that an ancient hydrophobic interface, conserved for hundreds of millions of years, is entrenched because exposing this interface to solvent reduces protein stability and causes aggregation, despite making no detectable contribution to function. Using structural bioinformatics, we show that a universal mutational propensity drives sites that are buried in multimeric interfaces to accumulate hydrophobic substitutions to levels not tolerated in monomers. In a database of hundreds of families of multimers, the majority show signatures of long-term hydrophobic entrenchment. It is therefore likely that many protein complexes persist because a simple ratchet-like mechanism entrenches them across evolutionary time, even when they are functionally gratuitous.
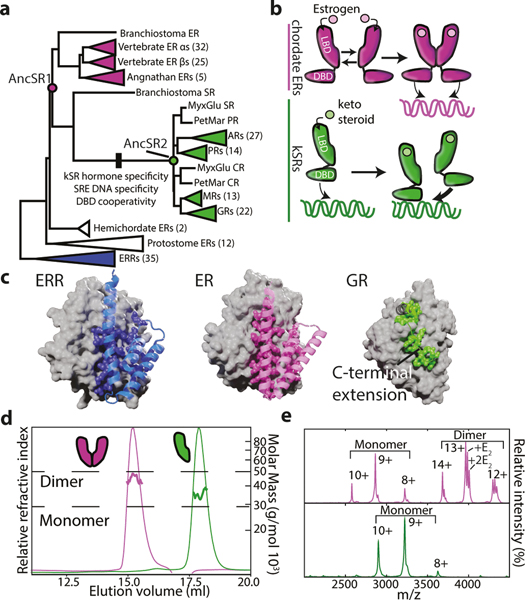
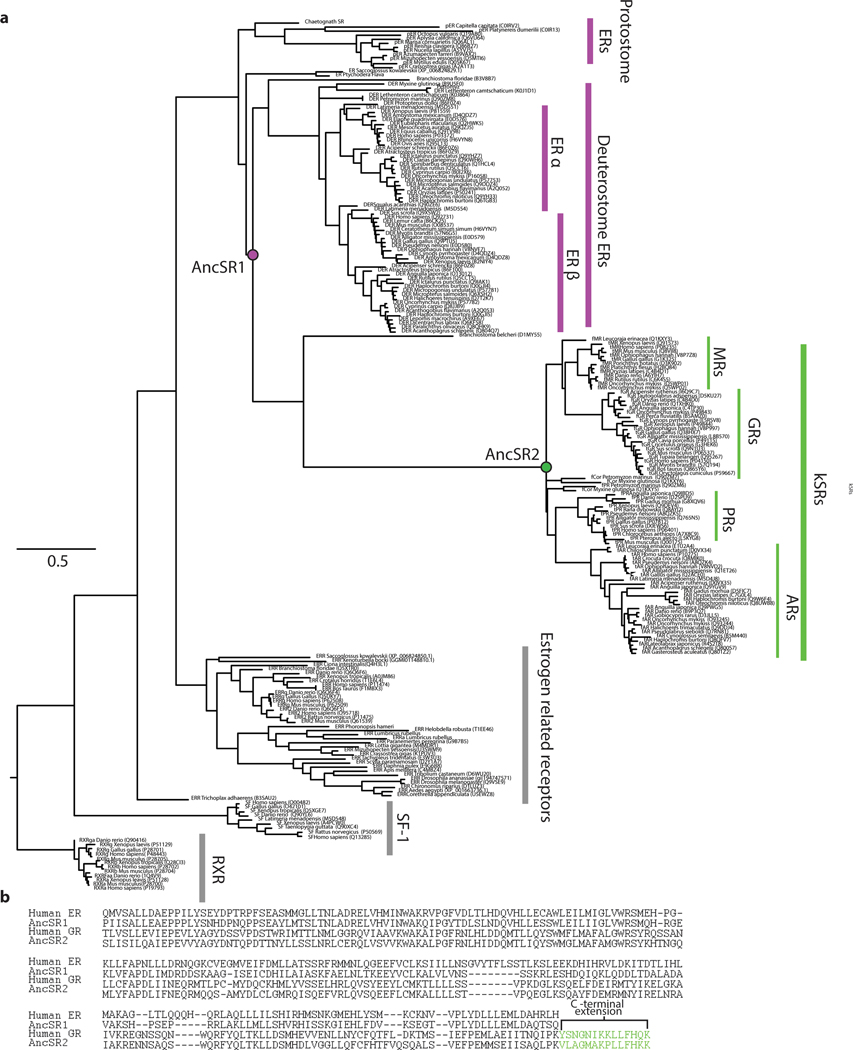
To understand why multimeric interfaces persist and change over evolutionary time, we studied the evolution of SRs, a protein family in which dimerization has been maintained for hundreds of millions of years but the mechanism of dimerization has diversified. SRs are hormone-activated transcription factors that contain structurally distinct DNA-binding and ligand-binding domains (DBD and LBD). There are two major phylogenetic classes of SRs (Fig. 1A,B, Extended Fig. 1A). One class, the estrogen receptors (ERs) homodimerize in solution using a large interface in their LBD8,9 and bind palindromic repeats of a particular six-base-pair DNA response element (ERE).10 The other class, called ketosteroid receptors (kSRs) because of the steroidal ligands that activate them, bind to a different palindromic sequence (SREs) via interactions between DBDs11,12. kSR LBDs are monomeric in solution, and the surface region homologous to ER’s dimerization interface binds instead to a C-terminal extension (CTE) on the same LBD, which is absent on ERs (Fig, 1B, C, Extended Fig. 1B). Previous work showed that the ancestral protein from which the two clades arose by gene duplication (AncSR1, >500 mya) specifically bound estrogens and non-cooperatively bound EREs; specificity for ketosteroids and SREs, as well as DBD-mediated cooperativity, arose on the branch between AncSR1 and AncSR2, the ancient progenitor of kSRs12,13. We reasoned that by identifying the ancestral and derived forms of the LBD interface and characterizing their effects on function and biophysical properties, we could gain insight into factors that caused the interface’s persistence and modification across deep history.
Figure 1: Evolution of self-assembly in SRs.
a) Reduced phylogeny of steroid and related receptors. Vertebrate estrogen receptors (ERs, purple), ketosteroid receptors (kSRs, green), and ancestral proteins are labeled. Black box, functional changes. Complete phylogeny in Extended Data Fig. 1. b) SR dimerization. ERs dimerize via an interface in the LBD, then bind palindromic estrogen response elements. kSR-LBDs are monomeric but cooperatively bind steroid response elements via interactions between DBDs. c) LBD interfaces in SRs and closely related receptors. Left, estrogen-related receptor dimer (2GP7). Gray surface, one LBD subunit. Blue cartoon and spheres, secondary structural elements and residues contributing to the interface on the other subunit. Middle, ER LBD dimer (1ERE). Right, Gluccocorticoid receptor LBD monomer (4P6X) as grey surface; green spheres, CTE on the same subunit. Cartoon, secondary structure elements connecting CTE to the rest of the LBD. d) SEC-MALS of AncSR1 (purple) and AncSR2 (green) at 25 μM. e) nMS at 10μM, with charge series labeled. E2, dimers bound to 1 or 2 estradiol molecules.
Evolutionary history of steroid receptor interfaces.
We first inferred the phylogeny of a large alignment of extant SRs and related proteins (Fig. 1A, Extended Fig. 1A). We then reconstructed the maximum-a-posteriori (MAP) sequences of the LBDs from the last common ancestor of AncSR1 and AncSR2 (Extended Fig. 2A,B).
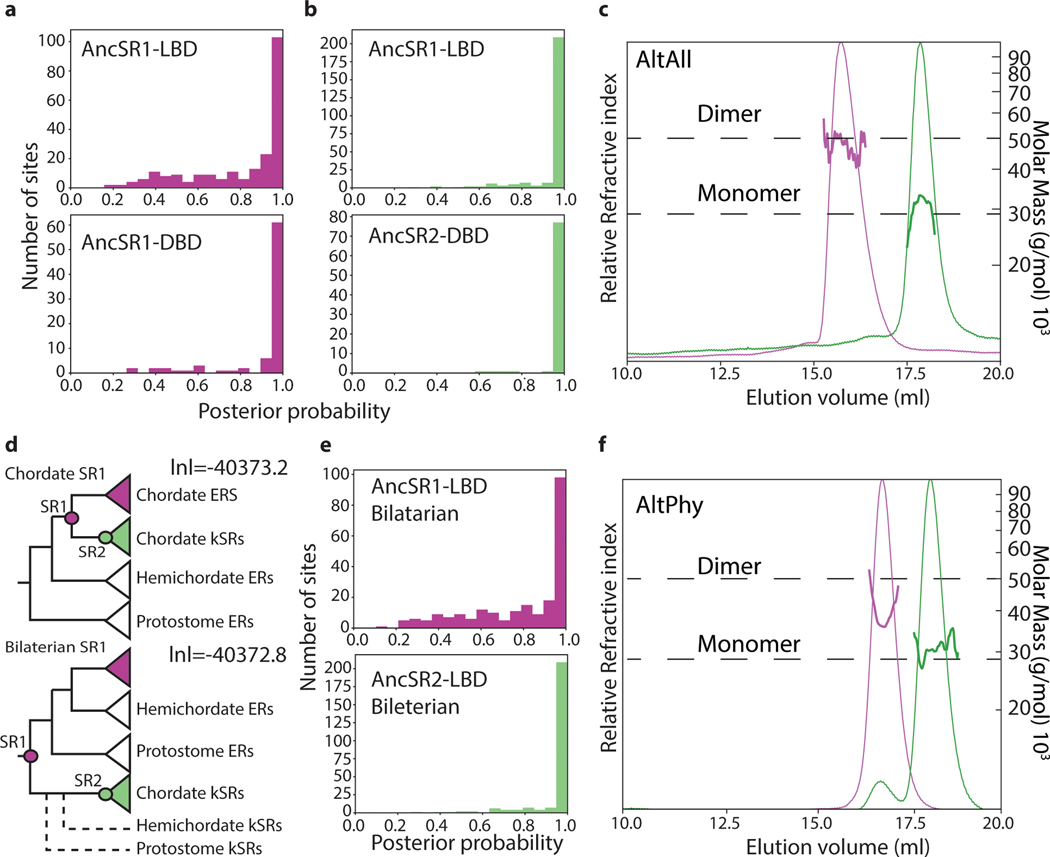
We expressed and purified these LBDs and measured their stoichiometry using size-exclusion chromatography/multi-angle light scattering (SEC-MALS) and native mass spectrometry (nMS) (Fig. 1C,D). AncSR1-LBD was predominantly dimeric at 30 μM and 10μM, indicating a Kd in the high nM, whereas AncSR2-LBD was entirely monomeric at both concentrations. AncSR1 therefore formed LBD-mediated dimers, which were retained in extant vertebrate ERs and lost along the branch leading to AncSR2. Corroborating an ancient origin of LBD dimerization, other nuclear receptor superfamily proteins dimerize through an ER-like interface (Fig. 1C). This inference is robust to statistical uncertainty about the ancestral sequence: alternative versions of AncSR1 and AncSR2 LBDs, which incorporate the second most likely state at all ambiguously reconstructed sites, had the same stoichiometries as the MAP versions (Extended Fig. 2C). Moreover, when LBDs of AncSR1 and AncSR2 were reconstructed using a different plausible phylogeny, AncSR1 remained a dimer and AncSR2 a monomer (Extended Fig. 2D-F).
Entrenchment of dimerization.
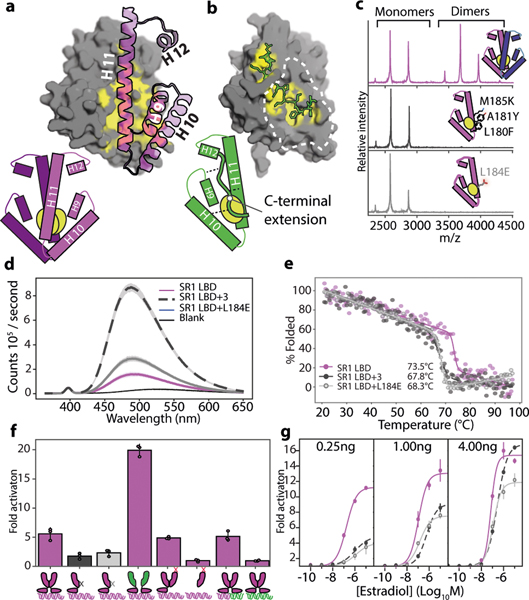
To understand mechanisms underlying the LBD interface’s long-term persistence in ERs and its modification in kSRs, we compared the crystallographic structure of AncSR2-LBD14 to a homology model of AncSR1-LBD. As in modern ERs, AncSR1’s dimer interface comprises a large patch of hydrophobic residues on helices 10 and 11; the patch on each subunit binds to the corresponding patch on the other to form a tight, water-excluding interface (Fig. 2A). AncSR2 and its descendants retain this patch, but it binds the CTE on the same subunit, shielding it from solvent in the monomeric state; this intramolecular interaction is conserved in all descendant kSRs15–18. We reasoned that the patch’s hydrophobicity might have entrenched the ancestral interface in AncSR1, because exposing hydrophobic residues renders many proteins unstable, insoluble, or aggregation-prone19,20; acquisition of the CTE, in turn, would have enabled loss of the multimeric state by replacing the intermolecular hydrophobic interaction with a similar intramolecular interaction.
Figure 2: The ancestral LBD interface was hydrophobically entrenched.
a) Homology model of AncSR1-LBD dimer. Grey surface, one subunit, with carbons shielded by the dimer interface in yellow. Purple, helices on the other subunit involved in dimerization. b) Atomic structure of AncSR2-LBD monomer (4FN9). Grey surface, main body of LBD; yellow, carbons shielded by CTE; green sticks, CTE. Dotted line, surface homologous to AncSR1’s dimer interface. Bottom, schematic of AncSR1 and AncSR2 LBDs. c) nMS of AncSR1-LBD wildtype (purple) or with historical (black) or ahistorical (grey) mutations that disrupt dimerization. Inset, location of mutations. d) bis-ANS fluorescence of AncSR1 and mutant LBDs at 2.5μM. Line, mean.Shaded, 95% CI from 3 technical replicates. e) CD melting curves for AncSR1-LBD mutants. Melting points are shown. f) Activity of AncSR1 and of chimeric and mutant receptors in a dual luciferase assay. Purple bars, receptors with shielded hydrophobic interfaces; black and grey bars, dimerization mutants. Cartoons indicate protein construct (purple, AncSR1; green, AncSR2; black and grey x, dimerization interface mutants as in panel D; red x, activation-function helix mutants) and response element (purple, ERE palindrome; green, SRE palindrome; mixed, hybrid containing one ERE and one SRE half-site). Receptor plasmid concentration was 0.25ng (below saturation for AncSR1, Extended Fig. 3A). Transcription was activated with 10−6μM estradiol or 10−7μM progesterone. Each point shows the average fold change vs. empty receptor control of three technical replicates. Columns and error bars, mean and 95% CI of 3 biological replicates. g) Activation by AncSR1-LBD and mutants on ERE at variable receptor plasmid and estradiol concentrations. Points and error bars, mean and 95% CI of 3 biological replicates.
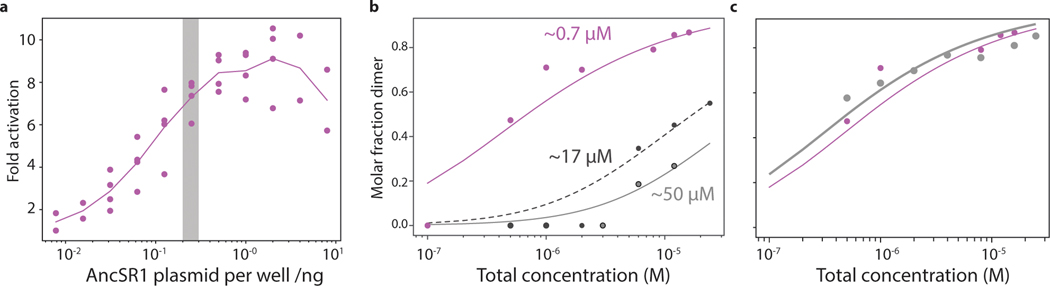
This hypothesis predicts that the LBD interface was already entrenched by the time of AncSR1, and that the CTE interaction that replaced it became quickly entrenched, too. To test the first prediction, we introduced mutations to cause clashes in AncSR1’s dimer interface, preventing dimerization and exposing the interface to solvent (Fig. 2B). We made two mutants, one carrying three historical substitutions from the AncSR1-AncSR2 branch and another with a non-historical mutation that abolishes dimerization in ERs through charge repulsion21. Both mutants were significantly weaker dimers than AncSR1, with Kds >20-fold higher than AncSR1. In both mutants, exposure of hydrophobic surface area is dramatically increased, as shown by binding to bis-ANS, which fluoresces when bound to hydrophobic patches (Fig. 2D). Both mutants had significantly lower Tms than AncSR1 (measured by circular dichroism), although their secondary structures remained largely intact at physiological temperatures (Fig. 2E). Disrupting AncSR1 dimerization without compensating changes would therefore have exposed hydrophobic surface and reduced stability.
Disrupting dimerization also severely impairs function: introducing the LBD dimer-interface mutations into a receptor containing the AncSR1 DBD and LBD dramatically reduced ERE-driven luciferase reporter activation (Fig. 2F). This result could arise for either of two reasons: dimerization might cause the receptor to function better than if it did not have the interface at all by, for example, more effectively occupying DNA response elements or recruiting transcriptional co-activators; alternately, disrupting dimerization could be deleterious if exposing the hydrophobic interface simply impairs the stability and function of each monomer. Four experiments support the latter explanation. First, a chimera containing AncSR1-DBD and AncSR2-LBD – which is fully monomeric – activates from EREs better than AncSR1 (Fig. 2F), demonstrating that dimerization does not enhance function under our assay conditions, as long as the interface is shielded. Second, to test whether having two active DBDs or LBDs in close proximity is necessary for full activation, we coexpressed AncSR1-DBD/AncSR1-LBD with an excess of a disabled AncSR1 that contains no DBD and an LBD in which the activation function is disabled by a point mutation22; the resulting heterodimers, which contain a single DBD and a single active LBD but shield the hydrophobic interface from solvent, activate just as well as wild-type AncSR1-DBD/LBD homodimers (Extended Fig. 3c, Fig. 2f), indicating no direct functional benefit from dimerization. Third, the AncSR1 dimer activates just as well on hybrid response elements containing one ERE half-site and one SRE half-site as it does on ERE palindromes, despite not activating at all from SREs, reinforcing that a single effective receptor/half-site complex can drive full activation under our assay conditions (Fig. 2F). Finally, if exposing the interface explains why interface mutations that reduce AncSR1’s dimerization affinity impair activation, then driving these mutants to re-occupy the dimeric form by increasing their concentration should rescue activation. As predicted, increasing plasmid concentration of the mutant receptor by 4- or 16-fold causes them to progressively recover activation. This effect is far greater than that of increasing concentration of wild-type AncSR1, demonstrating that shielding the interface recovers function (Fig. 2G).
These experiments establish that the ancestral dimeric interaction was entrenched because dissociating it into monomers and exposing the interface to solvent impaired the subunits’ functions, not because dimerization caused each subunit to function better than if it never had the interface. Purifying selection against the deleterious effects of exposing this surface would therefore maintain the dimeric state. It is possible that dimerization could contribute to function under different assay conditions, but our experiments establish that the interface is entrenched even when dimerization does not enhance function per se. This entrenchment has persisted to the present: mutations that interfere with dimerization in human ERs also dramatically impair receptor function21. Because interface-disrupting mutations impair function and reduce stability without unfolding secondary structure at physiological temperatures, the likely mechanism is that exposing the hydrophobic dimerization interface destabilizes the LBD’s active conformation relative to inactive conformations.
Entrenchment of the CTE-LBD interaction.
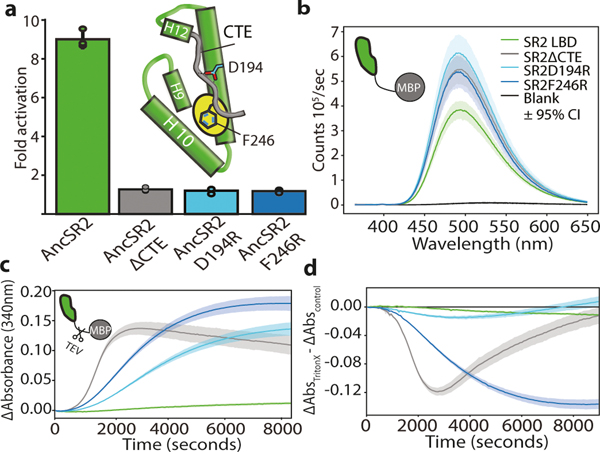
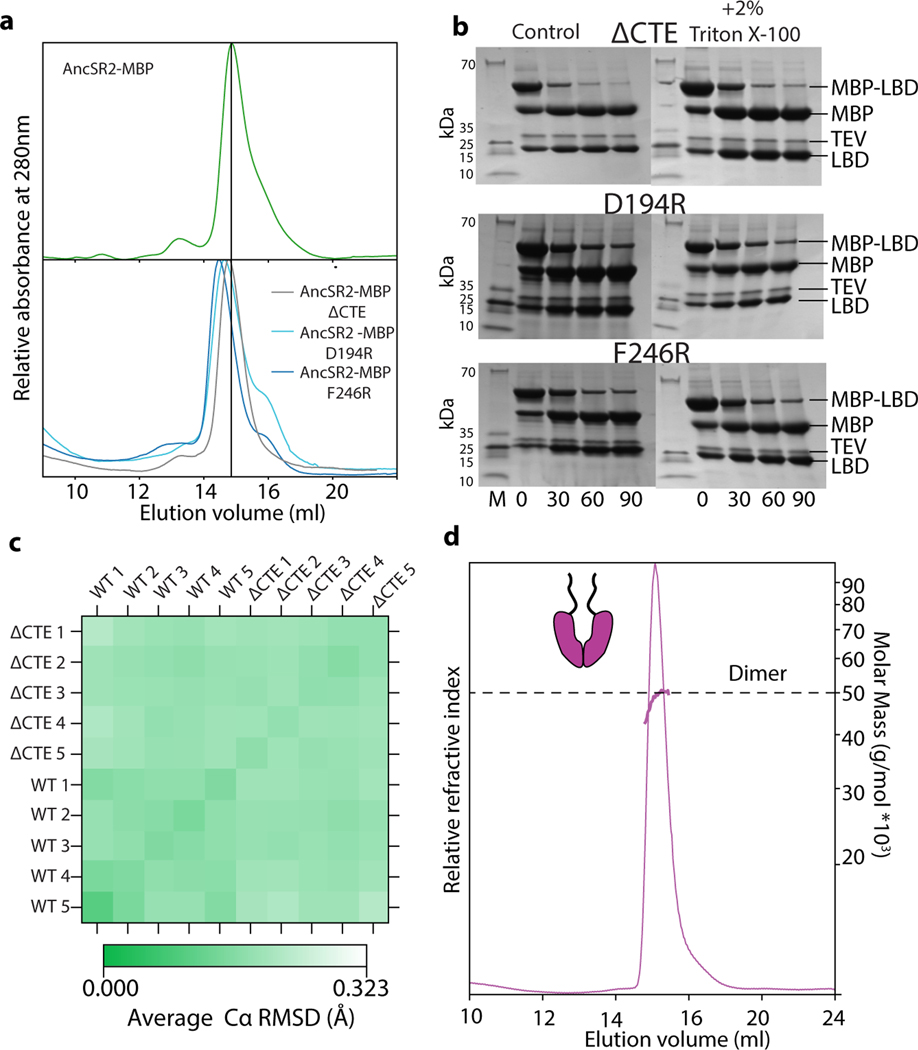
The entrenchment hypothesis implies that AncSR1’s entrenched dimer interaction could be lost in AncSR2 only because the interface became shielded by AncSR2’s new CTE, and that this new intramolecular interaction itself became hydrophobically entrenched. To test this prediction, we made AncSR2 mutants that delete the CTE entirely or disrupt the LBD-CTE interaction through charge repulsion (Fig. 3A). All fail to activate in a reporter assay (Fig. 3A), indicating that the interaction did become indispensable. To test whether the CTE-LBD interaction is entrenched specifically because exposing the hydrophobic patch is deleterious, we purified and characterized the AncSR2-LBD-CTE mutants. As predicted, they are very poorly soluble and produce higher bis-ANS fluorescence than AncSR2 when purified with an MBP tag, confirming that the hydrophobic path is exposed (Extended Fig. 4A) (Fig. 3B). When the tag is removed, all CTE mutants aggregate quickly, whereas intact AncSR2 does not (Fig. 3C). Moreover, shielding hydrophobic surfaces in a micelle by adding a mild non-denaturing detergent slows aggregation of the mutants (Fig. 3D) without affecting TEV cleavage (Extended Fig. 4B).
Figure 3: AncSR2 traded intermolecular for intramolecular entrenchment.
a) Fold activation by AncSR2-LBD and mutants on SREs in HEK293T cells using 4ng receptor plasmid and 10−8μM progesterone. Column and error bars, mean and 95% CI across three biological replicates (points, each of which shows the mean of three technical replicates) Inset: Schematic of AncSR2-LBD. CTE (grey) and sites mutated to disrupt CTE-LBD interaction are indicated. b) bis-ANS fluorescence by AncSR2-LBD and mutants fused to MBP. Line, mean. Shaded area, 95% CI from 3 technical replicates. c) Aggregation of AncSR2-LBD and mutants (colored as in C) when MBP tag is removed by TEV cleavage, measured 340nm absorbance. Line, mean. Shaded area, 95% CI from 10 technical replicates. d) Difference in light scattering between measurements in D and the same experiment with 2% Triton-X100. Line, mean. Shaded area 95% CI from 10 technical replicates.
The interaction between AncSR2’s CTE and hydrophobic patch is therefore entrenched because exposing the patch causes insoluble aggregates, abolishing AncSR2 function. Using 2μsec molecular dynamics simulations, we found no evidence that the protein unfolds completely (Extended Fig. 4C), although denaturation on longer timescales remains possible. Appending AncSR2’s CTE to AncSR1 does not abolish dimerization, indicating that other substitutions during the AncSR1-AncSR2 interval were required to generate a high-affinity interaction with the CTE that outcompetes the dimerization interaction at the same surface (Extended Fig. 4D). AncSR2’s extant descendants inherit the CTE-patch interaction and require the CTE to function23,24, indicating that hydrophobic entrenchment continues to preserve this interaction some 450 million years later.
A universal hydrophobic ratchet.
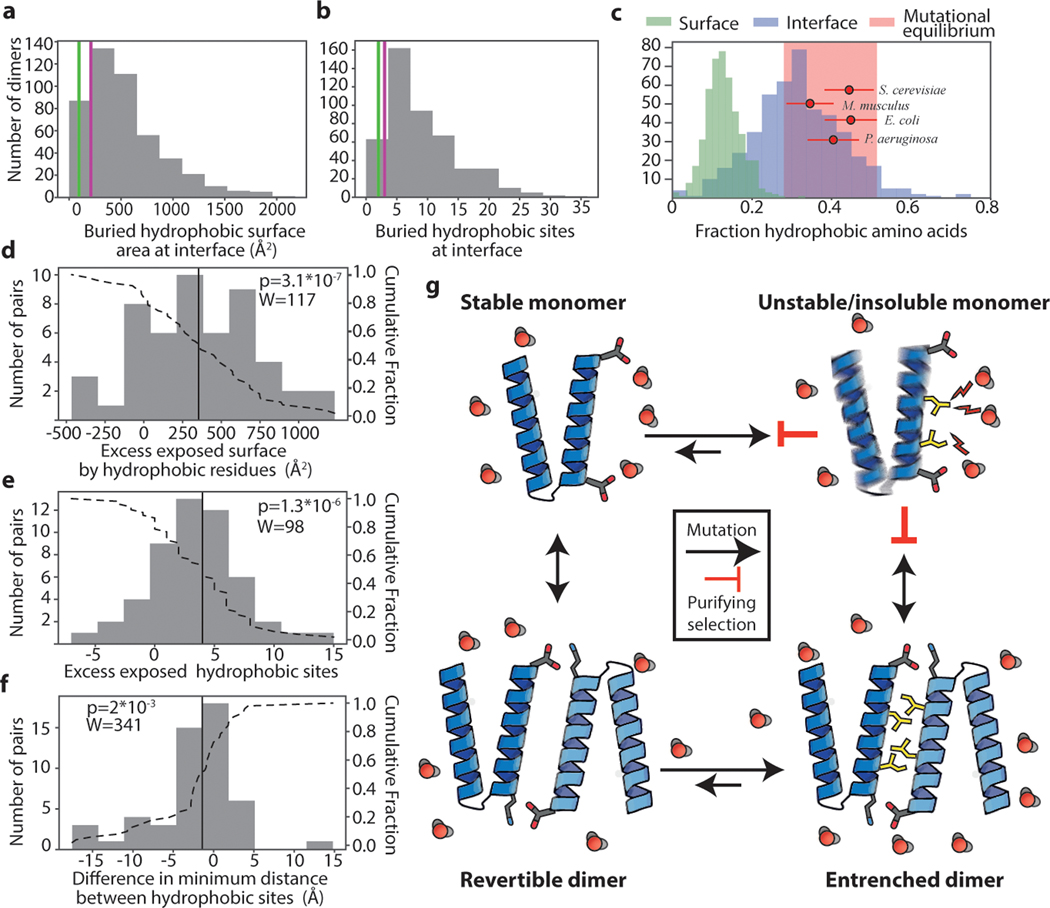
Finally, we investigated whether hydrophobic entrenchment is a general evolutionary phenomenon. We compiled a database containing the atomic structures of 466 homodimers25 and analyzed their solvent-exposed surfaces and interfaces. 83% of dimer interfaces in our dataset are more hydrophobic than AncSR1’s interface, and 94% are more hydrophobic than AncSR2’s CTE-shielded patch (Fig. 4A). Given our experimental finding that the SR interfaces are entrenched, it is likely that most dimers in the database are, too (Fig. 4A,B).
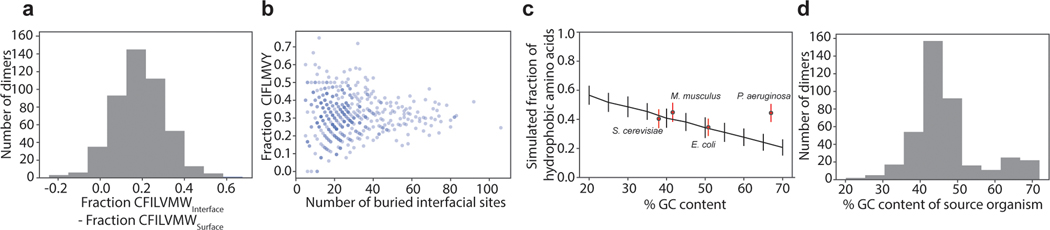
Figure 4: Pervasive hydrophobic entrenchment of molecular complexes.
Surface area (a) and count (b) of hydrophobic residues (amino acids CFILMVY) buried in dimer interfaces in a database of 466 non-redundant dimer structures. Purple and green lines, AncSR1-LBD and AncSR2-LBD interfaces. c) Dimer interfaces are more hydrophobic than is tolerated at surfaces and are close to the hydrophobicity expected by mutation alone. Histograms: Fraction of residues that are hydrophobic on solvent-exposed surfaces (green) or buried in dimer interfaces (blue). Red circles: expected fraction of hydrophobic amino acids from mutation alone, based on spectra from mutation accumulation data in 4 model organisms (see Extended Fig. 5C). Points and error bars, mean and SD from 100 replicates. Pink box, ±1 SD of the mean across all simulations. Red dots are distributed vertically for visual clarity. d-f) Histograms of the difference in surface properties between dimer subunits (when dissociated into monomers) and their monomeric homologs. Dotted line, cumulative fraction of pairs with greater difference. Solid line, median. d) Exposed surface area contributed by hydrophobic residues in dissociated dimer subunits minus that on monomeric homolog. e) Number of hydrophobic residues on surfaces of dimer subunits minus that on monomers. f) Difference in clustering of hydrophobic surface residues between dimer subunits and monomers, calculated as average surface distance from exposed hydrophobic residues to their nearest hydrophobic neighbor. n=51 independent monomer-dimer pairs; P-value and test-statistic W from two-tailed paired Wilcoxon test. g) Mechanism of the hydrophobic ratchet. In monomers, purifying selection counteracts mutational pressure towards increased surface hydrophobicity (yellow sticks), which would be deleterious because of increased propensity to aggregate and/or misfold. Once shielded from solvent (red) in dimers, hydrophobic mutations are free to accumulate in the buried interface. Purifying selection then preserves the complex.
Inspired by prior work on evolutionary entrenchment of complexity 4,26–30, we reasoned that entrenchment will arise if two conditions are met: 1) there is a class of substitutions for which the complex state has a higher tolerance than the simple state, and 2) the mutational process alone generates more of these substitutions than can be tolerated in the simple state. Under these conditions, reversion to the simple state will rapidly become unlikely under purifying selection. Specifically, hydrophobic entrenchment will arise if buried interfaces tolerate higher hydrophobicity than exposed sites do, and if mutation tends to produce a higher fraction of hydrophobic residues than surfaces allow.
To evaluate the first condition, we characterized the hydrophobicity of multimeric interfaces and surface-exposed sites in our structural database. Multimeric interfaces are indeed much more hydrophobic than exposed surface sites are. Across all multimers in our database, the median fraction of hydrophobic residues at interface sites is 31%, whereas the median at exposed surface sites is 12% (Fig. 4C, Extended Fig. 5A,B).
To evaluate the second condition, we examined whether mutation alone is expected to generate more hydrophobic amino acids than surfaces tolerate. Hydrophobic amino acids comprise >40% of all sense codons; moreover, hydrophobic amino acids are AT-rich, and the mutational process universally favours G/C to A/T transitions, irrespective of genomic GC content31. We simulated coding sequence evolution using the universal genetic code and empirical mutation spectra observed in mutation accumulation (MA) experiments in prokaryotes and eukaryotes with a range of GC contents. We found that the fraction of hydrophobic residues expected from mutation alone is 33 to 45%, far greater than is tolerated on exposed surfaces and much closer to the hydrophobicity of buried interfaces (Fig. 4C). This result holds when sequences are simulated using only the universal genetic code and GC contents across a wide empirical range (Extended Fig. 5C). Exposed sites are therefore constrained by purifying selection to maintain lower hydrophobicity than would be generated by mutation, and this constraint is dramatically relaxed once an interface becomes protected in a multimer. The conditions for hydrophobic entrenchment of interfaces are therefore universally satisfied and arise from general properties of surfaces, interfaces, the mutational process, and the genetic code.
For a multimer to escape entrenchment, its surface would have to return to a level of hydrophobicity that can be tolerated in the unassembled state. To understand the extent of entrenchment, we analyzed protein families in our dataset that contain both dimers and monomers. We compared the surface area of all exposed hydrophobic residues on monomers to the total area that would be exposed on the subunits of their homologous dimers if the dimers were dissociated. The degree of apparent entrenchment is large: the hydrophobic surface on dimer subunits is greater than on monomers by a median of 340 Å2, and the exposed hydrophobic residues are more spatially clustered (Fig. 4D-F). Dimers would have to lose a median of 4 hydrophobic residues for their surfaces upon dissociation to become similar to their monomeric relatives; in about 20% of cases, dimers are enriched by ≥7 or more hydrophobic residues. For multimerization to be lost, many or all of these excess hydrophobic residues would have to be mutated or compensated for, but mutational propensity towards high hydrophobicity makes this outcome extremely unlikely (Fig. 4G).
Entrenchment of molecular complexity.
Our findings suggest that many molecular complexes are likely to be entrenched by a biochemical ratchet: mutational propensity drives sites buried in a multimeric interface to accumulate hydrophobic substitutions to a level that renders reversion to the ancestral monomeric state deleterious. Complexes in which multimerization makes no direct contribution to function or fitness will therefore be preserved by purifying selection. Other biochemical mechanisms may also entrench multimers, deepening or broadening the impact of hydrophobic enrichment of buried interfaces in causing molecular complexes to persist5.
Hydrophobic entrenchment explains only the persistence of complexes; it neither explains their origin nor is limited to cases in which the multimeric association was initially acquired by drift. A multimer that originated under selection because it enabled a multimer-dependent function would still become entrenched, preserving the association even if multimerization later becomes functionally dispensable. Entrenchment and functional benefit can coexist: even when multimerization is beneficial because it enables properties such as allostery or cooperativity, hydrophobic entrenchment will further reduce the probability of reversion to the monomeric form, because losing the interaction would impair all functions of the protein, not only those that depend on multimerization. The hydrophobic ratchet could even facilitate the evolution of assembly-associated functions by preserving interfaces that are initially functionally inconsequential, and mutational pressure towards increasing hydrophobicity could quickly strengthen fortuitous interactions. In some cases, multimerization is undoubtedly functionally important; however, given the universal conditions that cause hydrophobic entrenchment, entrenchment should be the null hypothesis to explain persistence of any particular complex in the absence of evidence that multimerization enhances its functions.
Entrenchment does not make multimers impossible to lose. Unlikely trajectories that restore solubility to a hydrophobic interface or otherwise shield it from solvent can in rare cases be followed, as apparently occurred in AncSR2. Selection could increase the probability of overcoming entrenchment if the assembled state became deleterious -- for instance, if inherited interactions after gene duplication produced interference between paralogs32,33. R1cthe other subunit shown as purple helicesCarbons shielded by the CTE are colored yellow. Doted line approximately outlines t In the absence of such pressures, however, even useless interfaces may persist for long periods of time. The cell may therefore be filled with an ever-accumulating stock of entrenched molecular complexes that never performed a useful function, or long ago ceased to do so.
METHODS
Phylogenetics and ancestral reconstruction.
Nuclear receptor LBD and DBD amino acid sequences were aligned using Muscle (version 3.8.31) 34; the alignment was corrected manually, and sites corresponding to lineage-specific insertions were removed. The unalignable hinge region was also removed. We used Protest 335 to identify the best-fit AIC model as JTT with empirical amino acid frequencies and a 4-category gamma distribution of among site rate-variation (JTT+F+G). We used PhyML 3.036 to infer the maximum likelihood phylogeny. We imposed several subsequent rearrangements to reflect prior corroborated phylogenetic information from large-scale studies: 1) We moved the two agnathan ER paralogs to form a monophyletic clade sister to all other vertebrate ERs, rather than successive sister clades to the ERb clade, because this duplication is thought to be a lineage-specific duplication37; 2) we moved the agnathan cortiocosteroid receptors to be sister to the gnathostome GR/MR clade, in accord with prior evidence concerning the timing of vertebrate genome duplications38 and prior work on SR phylogenetics 14; and 3) we moved the ERR sequences of Xenoturbella and hemichordates to form a monophyletic sister clade to chordate ERRs, instead of forming successive outgroups to bilaterian ERRs, in accord with evidence at the time 39, although this grouping has now been revised40. The phylogenies were rooted between RXRs and SF-1s41. Transfer bootstrap values were calculated using the Booster server 42. Approximate likelihood ratio statistics were calculated using PhyML.
This topology (the “Bilaterian” topology, Extended Data Fig. 2d) places the gene duplication that split the chordate ERs from the chordate kSRs deep in the Bilateria, with subsequent losses of kSRs in all protostomes and in all non-chordate deuterostomes. A more parsimonious topology with respect to gene duplications and losses was created by rearranging the two weakly support branches leading to hemichordate and protostome ERs; this topology places the ER/kSR duplication within the chordates and requires no subsequent gene losses (the “Chordate” topology, Fig. 1A, Ext. Data Fig. 1 and Ext. Data Fig. 2d). This topology is only 0.4 lnL uniits less likely than the Bilaterian topology (Ext. Data Fig. 2d). We therefore used Chordate topology for the primary reconstructions of ancestral proteins, but we also produced and tested alternative reconstructions that used the Bilaterian topology as the underlying phylogeny (AltPhy reconstructions, Ext. Data Fig. 2f).
Ancestral sequences were inferred using marginal reconstruction in the codeml module of PAML 4.843 and JTT+F+G. Branch lengths and model parameters were inferred separately for the DBD and LBD and the posterior distribution of states at each site then estimated assuming the alignment, tree, and model parameters. The MAP sequence contains the state with the highest posterior probability at each site. The AltAll sequence contains the MAP state at all sites where only one state has PP>0.2, and the state with the second highest posterior probability at all other sites.
Protein expression and purification.
Codon-optimized sequences coding for SR LBDs were obtained from Integrated DNA technologies and cloned into a pET LIC vector containing an N-terminal, TEV cleavable 6-his MBP tag (Addgene plasmid 27989). Proteins were transformed in BL21(DE3) E. coli, and inoculated into 50mL LB cultures and grown overnight. For expression, starter cultures were used to inoculate 0.5L cultures of TB, which were grown to an optical density of 0.6–0.8. Hormone dissolved in DMSO (estradiol for AncSR1-LBDs and progesterone for AncSR2-LBDs) was then added to a final concentration of 50–100μM. Cultures were induced with 500μM IPTG and incubated with shaking overnight at 22˚C. In the morning cultures were spun down at 5000g, resuspended in PBS, transferred to conical falcon tubes through another 5000g spin and stored at −80˚C until use.
For purification, proteins were re-suspended in buffer A (150mM NaCl, 20 mM Tris, 20mM Imidazole, 10% (w/v) glycerol, pH 8), supplemented with 20mM beta mercaptoethanol and one protease inhibitor tablet (Roche) per 0.5L of culture. Cultures were lysed on ice using a sonicator for 30 one-second-on/one-second-off pulses. The lysate was clarified by first spinning at 20,000g for 20 minutes and passed through a 0.45μm syringe filter. The solution was then loaded at room temperature onto a 5ml HisTrap nickel column (GE) equilibrated with Buffer A. After washing with at least 5 column volumes of Buffer A, the protein was eluted with a linear gradient over 12ml from 0 to 100% buffer B (150mM NaCl, 20mM Tris, 500 mM imidazole, 10% glycerol pH 8). Fractions containing the LBD construct were pooled, approximately 0.5mg of TEV was added, and the solution was then dialyzed overnight at room temperature against 4L of buffer A containing 50–100μM of estradiol or progesterone, depending on the construct. The cut product was passed over a HisTrap column equilibrated in buffer A and the flow-through collected and concentrated. For the last purification step, the sample was injected onto a Superdex 200 10/300 size exclusion column (GE) equilibrated into PBS run at 0.4ml/min. Fractions containing purified LBD were pooled and concentrated. Glyercol was added to a final concentration of 10%, and then flash frozen in liquid nitrogen and stored at −80˚C until use.
Native mass spectrometry.
MS measurements of native protein samples were collected on a Synapt G1 HDMS instrument (Waters Corporation) equipped with a radio frequency generator to isolate higher m/z species (up to 32k) in the quadrupole, and a temperature-controlled source chamber as previously described 44. Instrument parameters were tuned to maximize signal intensity while preserving the solution state of the protein complexes. Data was collected in positive ion mode and typically takes 30 seconds to 60 seconds per sample. Instrument settings are as follows: source temperature of 25 ºC, capillary voltage of 1.7kV, sampling cone voltage of 100V, extractor cone voltage of 5V, trap collision energy of 20V, argon flow rate in the trap was set to 7 ml/min (5.6 × 10−2 mbar), and transfer collision energy set to 10V. The T-wave settings were for trap (300 ms−1/1.0V), IMS (300 ms−1/20V) and transfer (100 ms−1/10V), and trap DC bias (30V). Molar fractions were extracted from spectra analyzed using UniDec 45. Titrations of AncSR1 and its variants were fit to the equation , using a custom Python script where D is the concentration of dimers, [P]0 is the total concentration of monomers, and Kd is the dissociation constant.
SEC-MALS.
SEC-MALS experiments were carried out using a DAWN HELEOS II MALS detector (Wyatt) coupled to an in-line Optilab T-rEX detector for refractive index measurements and a Superdex 200 10/300 size exclusion column (GE). Prior to the experiment, proteins were dialyzed against PBS, and diluted to a final concentration of 0.66mg/ml. 150μL of protein was injected onto the column for each run. The column was run at 0.5ml/min at room temperature. Data analysis was carried out using the ASTRA 6.0 software package.
BIS-ANS incorporation.
For AncSR1 LBD, experiments were carried out at 2.5μM protein concentration and 40μM bis-ANS in PBS. For AncSR2, experiments were carried out with 1μM of protein and 40μM of bis-ANS in PBS. Bis-ANS fluorescence was measured on a HORIBA Flourolog-3 spectrofluorometer, using a 500μL cuvette. The excitation wavelength was set 350nm. Emission was monitored from 350 to 600nm, with gratings set to 120.500.2. Entrance and exit slit widths were set to 2 and 1 nm, respectively.
Protein stability.
CD spectra for thermal melts were recorded on a Jasco J-1500 Circular Dichroism Spectrometer. Proteins were exchanged in 50mM NaPi, 20 mM NaF, pH 7.4 in a concentrator prior to the experiment and diluted to a final concentration of 2.5μM. Spectra were recorded between 260 and 180nm, at a 1nm pitch and a scanning speed of 100nm/min. The temperature was ramped from 20 to 98C in 0.5C steps at a rate of 3 C per minute. The data were analyzed using the calfitter server 46 using a reversible two state model (N=D).
Aggregation assays.
AncSR2 and variant LBDs were purified using and Ni column as previously, but in the presence of 100μM progesterone, and the cleavage and SEC steps were omitted. Their concentrations were determined using a Bradford assay. For the aggregation assay, proteins were diluted to a final concentration of 40μM, in 150mM NaCl, 20mM Tris, pH 7.4, 20 mM Imidazole, 10% glycerol supplemented with 5mM BME, and 0.1mg/ml TEV protease, either with or without 2% TritonX-100. The solution was transferred into a clear 96 well plate, using 100μL of solution per well and followed at 400nm on a Perkin Elmer Victor X5 plate reader at room temperature.
Reporter activation assays.
Ancestral DBD and LBDs were cloned into pcDNA3, separated by the hinge of the human glucocorticoid receptor, which neither confers nor abolishes dimerization10. Response element plasmids contained 4 copies of ERE (AGGTCAGAGTGACCT), SRE (AGAACAGAGTGTTCT), or a hybrid ERE/SRE element (AGGTCAGAGTGTTCT), upstream of a luciferase reporter gene. HEK293T cells were obtained from ATCC. Cells were grown at 37˚C and 5% CO2 in DMEM media (Glibco) supplemented with 5mM sodium pyruvate, 10% fetal bovine serum (Glibco), and Penicillin Streptomycin solution to a final concentration of 1%. For transcriptional activation assays, cells were transfected with a variable amount of receptor plasmid, 40ng of response element plasmid, 1ng of a renilla luciferase plasmid for normalization and pUC19 up to a total amount of 100ng per well. Each well contained 0.05μL of lipofectamine and 0.5μL of plus reagent, to which Optimem (Gilbco) was added to bring the total to 65μL per well. To this, 135μL of cell suspension was added per well. After 18 hours incubation, medium was replaced with 50μL of DMEM with stripped fetal BSA, supplemented with 1% ETOH and variable concentrations of hormones (see figures for details). The cells were incubated for 6h with hormone. 10μL of well solution was then aspirated from each well, and 30μL of luciferase dual-GLO mixture added per well. The mixture was incubated for 2 minutes at room temperature, and 60 μL per well was then transferred into a white 96 well plate, incubated for 8 more minutes, followed by reading of FFL luminescence on a Perkin Elmer Victor X5 plate reader. 30 μL of Stop and Glo (Promega) mixture was then added to each well and the plate incubated at room temperature for 10 minutes before recording Renilla luminescence FFL luminescence was normalized by Renilla luminescence for each well; fold activation is the ratio of the normalized luminescence observed for any treatment divided by normalized luminescence for an empty vector control treated with 1% ETOH.
Homology modeling and molecular dynamics.
The AncSR1 LBD structure was modeled using the SWISS model server using default parameters and specifying the human estrogen receptor (PDB 1ERE) as the template. For MD simulations, the AncSR2-LBD X-ray crystal structure (PDB 4FN9), and a modified version with all CTE residues removed, were used as starting points using Gromacs software47. Each LBD structure was encased in a rhombic-dodecahedral box with a minimal protein–box-edge distance of 1.5 nm. Water and NaCl were added corresponding to a 0.154 M saline solution. Proteins and ions were modelled using the Amber99SB-ildn force field 48 together with the Tip3p water model 49. The hormone was modelled using GAFF/BCC force field parameters 50. Virtual sites 51 for the hormone were constructed using the MkVsites tool 52, all bonds we constrained with LINCS 53), and SETTLE 54 to keep water molecules rigid, enabling a 4-fs time step in all subsequent simulations. Steepest descent energy minimization was carried out for both systems. Each system was replicated fivefold at this stage, each replica subjected to a 100-ps NVT simulation (using different random seeds used for velocity generation for the different replicas) with position restraints applied to all heavy atoms in the protein and the hormone in order to remove internal strain from the structures. Each replica was simulated for 1 ns under NVT conditions and then for 10 ns under NPT conditions for equilibration, using a Berendsen barostat 55. Production simulations were run for 2 μs per replica under NPT conditions using the Parrinello-Rahman barostat 56. The v-rescale thermostat57 was used for all simulations with temperature coupling. The first half of each simulation was excluded from all analysis to allow for structural relaxation. The backbone RMSD with respect to the starting structure of the production runs were calculated for each system to assess overall convergence of the simulations. The RMSD was also calculated between all frames in all trajectories.
Structural bioinformatics.
A curated set of structures was downloaded from the PDB based on the database in ref 25. Structures were downloaded as biological assemblies. We retained monomers and dimers that were annotated as part of a non-redundant set (filtered at <30% sequence identity) and whose quaternary structure was annotated as correct in the database. To find protein families containing both monomers and dimers, we built a BLAST database from the sequences of the monomers and used the dimer sequences as a query using a 20% sequence identity cutoff. We excluded hits that shared more than 70% sequence identity over the aligned portion, to exclude proteins that can populate both stoichoimetries but that were crystallized independently as dimers and monomers in closely related species.
Structures were stripped of atoms labeled as HETEROATOM in the PDB file using a custom Python script to remove ligands. We created a separate PDB file containing only the first subunit of each dimer. We calculated the exposed hydrophobic surface area of dimers, dissociated dimer subunits, and homologous monomers using the Areaimol program 58 with default parameters. Exposed residues were defined as amino acids for which the solvent-accessible surface area (SASA) was greater than 20% of the maximum theoretical surface area obtained from Gly-X-Gly peptides, where X is the residue of interest 59,60. Sites buried at the interface were identified as sites at which the difference between SASA in the dissociated monomer and SASA in the dimer was greater than 10% of value in the dissociated monomer. 59. Hydrophobic residues were defined as amino acids CFILMVW. The number of sites with hydrophobic or non-hydrophobic residues was recorded for both exposed and interfacial sites. The total hydrophobic exposed surface area was calculated as the sum of the solvent-exposed area of all exposed hydrophobic residues. This method is conservative, because hydrophobic portions of amino acids not classified as hydrophobic can contribute to hydrophobic surface area. Buried surface area of hydrophobic sites in dimer interfaces was calculated as Ainterface = Amonomer − ½ Adimer, where Ainterface is the surface area buried by hydrophobic sites at the interface, Amonomer is the SASA of exposed hydrophobic sites in a dissociated single chain of the dimer, and ADimer is the SASA of exposed hydrophobic sites in the dimer. The number of hydrophobic sites buried at the interface was calculated in the same way. lTo compare the exposed hydrophobic surface area of dissociated dimers to their monomeric homologs, we only used dimers for which we found monomeric homologs that differed in the length of the aligned portion of their sequence by no more than 9 residues. Each monomer was only used once, so that dimers that are homologous to only a previously used monomer were excluded from the calculation. Exposed hydrophobic surface area and number of exposed hydrophobic sites was calculated using only the aligned portion of the proteins. The degree of clustering among exposed hydrophobic sites was calculated using the DynamXL program61, which calculates the shortest path along the protein surface between two points on that surface. We calculated all pairwise Cα to Cα distances between all exposed hydrophobic sites, with exposure being defined as above. For each site, we recorded the distance to its closest hydrophobic neighbor. Finally we averaged these distances for all exposed hydrophobic sites within one protein and then calculated the pairwise difference between the averages for dissociated dimers and their monomeric homologs.
Expected hydrophobic content.
GC content of source organisms in our database were obtained from the NCBI genome database62. To produce the expected hydrophobic content of a protein sequence given some specified GC content, we drew nucleotides randomly based on the expected A/T and G/C frequency. The length of the sequence to be drawn was determined by randomly drawing a length from the length distribution in our database of dimers. The sequence was translated using the standard genetic code, and the fraction of hydrophobic amino acids CFILMVW was calculated, with stop codons excluded. This procedure was repeated 200 times to obtain a mean and standard deviation.
To calculate the expected hydrophobic fraction using empirical mutational spectra, we used mutation accumulation experimental data from S. cerevisiae63, M. musculus64, E. coli65 and P. aeruginosa 66. We first constructed an instantaneous DNA mutation rate matrix Q (Supplemental Tables 1–4) for each species by entering the relative frequencies of observed mutations from each wild-type nucleotide to each other possible nucleotide. The matrix has 6 free parameters, because each mutation changes the complementary nucleotide on both DNA strands; for example, every A-to-C mutation is associated with a T-to-G mutation, so the rates of these two kinds of mutations are constrained to be equal. Diagonals were filled so each row adds to zero, and the matrix was scaled so the sum of diagonals = −1. We then calculated the probability matrix P of final nucleotide states given each possible starting state across a branch length of 100 expected substitutions per site as P=e100Q. We simulated a starting DNA sequence given each species’ GC content as previously and assigned a final state at each site given the starting state and P. We translated the resulting DNA sequence using the universal genetic code, excluded stop codons, and calculated the fraction of hydrophobic amino acids. This procedure was repeated 200 times for each species to calculate an average and standard deviation of the expected near-equilibrium fraction of hydrophobic amino acids.
Statistical software.
All statistical tests were carried out using scipy 1.2.1. All plots were produced using matplotlib 2.0.0 in Python 2.7.11.
Data availability.
Data are deposited in the Open Science Framework (DOI 10.17605/OSF.IO/GTJ86),, including alignment, phylogeny, sequences and posterior probability of ancestral reconstructions; list of PDB identifiers for coordinates of dimers and monomers in our structural database; and molecular dynamics trajectories.
Code availability
Scripts and code for structural bioinformatics analysis are deposited at github (https://github.com/JoeThorntonLab).
Extended Data
Extended Data Figure 1: Phylogeny and alignment of steroid and related receptors.
a, Phylogeny of steroid receptors and related nuclear receptor family members. AR, androgen receptors, PR, progestorone receptors, GR, gluccocortociod receptors, MR, mineralocortocoid receptors. Sequence identifiers are in brackets. This topology corresponds to the “Chordate tree” in Extended Data Fig. S2. Scale bar, expected substitutions per site. b, Sequence alignment of the human ER and GR LBDs, with the MAP sequences of AncSR1 and AncSR2. Green, C-terminal extension. Most ERs contain additional sequence on the C-terminus that is unalignable, even among ERs.
Extended Data Figure 2: Robustness of ancestral reconstructions.
a,b, Distribution of posterior probabilities (PP) of the maximum a posteriori (MAP) state at each site in reconstructed LBDs (top) and DBDs (bottom) of AncSR1 (a) and AncSR2 (b). c, Stoichiometry of purified alternative LBD reconstructions (AltAll) of AncSR1 (pink) and AncSR2 (green), as measured by SEC-MALS. AncSR1 is a dimer, AncSR2 a monomer. AltAll reconstructions contain the MAP state at unambiguously reconstructed sites and the state with the next highest PP at all ambiguously reconstructed wites. d, The “chordate” phylogeny (top) was used for primary ancestral reconstructions; it places the gene duplication yielding ERs and kSRs within the chordates. An alternative less parsimonious tree (“Bilaterian,” because it places the duplication deep in the Bilateria, bottom), has very slightly higher likelihood but requires two additional gene losses (dashed lines). The Bilaterian topology was used for alternative reconstructions (AltPhy). Node labels, approximate likelihood ratio test statistic and transfer bootstrap value. lnl, log-likelihood. e, Distribution of per-site posterior probabilities for reconstructed LBDs on the Bilaterian topology for AncSR1 (top) and AncSR2 (bottom). f, Stoichiometry of purified AltPhy versions of AncSR1 (pink) and AncSR2 (green) LBDs, as measured by SEC-MALS. AltPhy-AncSR1-LBD ‘s average molar mass and elution time are between that of a dimer and a monomer, indicating that it is a fast-exchanging, weaker dimer than other AncSR1-LBD versions.
Extended Data Figure 3: Concentration-dependence of activation and dimerization by AncSR1-LBD and mutants.
a, Activation of AncSR1 from 40ng ERE response element plasmid as a function of the AncSR1 plasmid concentration. Grey bar, concentration at which assays in Fig. 2F were performed. b, Molar fraction in the dimeric form measured by nMS as a function of LBD concentration for AncSR1-LBD (purple) and dimerization-interface mutants L180F/A181Y/M185K (black) and 184E (grey). Dissociation constant (Kd) estimated by nonlinear regression is indicated next to each curve. c, Dimeric fraction as a function of LBD concentration for AncSR1-LBD (purple) and activation-helix mutant L126Q (grey), which affects activation but not dimerization.
Extended Data Figure 4: Entrenchment of the CTE in AncSR2.
a, SEC of AncSR2 LBD (top) and mutants that delete the CTE (ΔCTE) or contain point mutations that impair CTE-LBD interactions (bottom), when fused to MBP. The mutants elute in the same fraction as AncSR2, demonstrating that they are monomeric and that re-exposing the patch does not re-establish dimerization. b, TEV cleavage of AncSR2 mutants in the absence (left) and presence (right) of 2% Triton X-100. The positions of bands corresponding to the uncleaved construct, cleaved MBP, cleaved LBD, and TEV protease are indicated. This experiment was replicated once, with similar results. See SI Fig 1 for uncropped gels. c, Average root mean square deviation (RMSD) from replicate 2μsec molecular dynamics simulations of AncSR2-LBD (wt) and ΔCTE mutant. The average Cα RMSD in pairwise comparisons of all simulations is shown as a heatmap. d, SEC-MALS trace of AncSR1-LBD fused to the CTE of AncSR2-LBD. The LBD is still dimeric.
Extended Data Figure 5. Observed hydrophobicity of interfaces compared to expected hydrophobicity from mutation.
a, Difference between the fraction of residues that are hydrophobic in dimer interfaces versus that on solvent-exposed surfaces of the same proteins. The histogram shows the distribution of this difference across every protein in our structural database. b, Fraction of hydrophobic residues in dimer interfaces as a function of the number of interface residues. The variance in the fraction is caused mostly by very small interfaces. c, Expected equilibrium fraction of hydrophobic amino acids from mutation alone. Black: expectation based on GC content and the genetic code. Red dots: Expected hydrophobic fraction based on mutational accumulation experiments (Fig. 5A), plotted against GC content of the organism tested. d, GC content of organisms represented by proteins in our database.
Supplementary Material
Acknowledgments.
We thank Jamie Bridgham for training and extensive advice, Arvind Pillai for assistance with experiments, and members of the Thornton Lab for comments. Molecular dynamics computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Projects SNIC 2019/8-36 and SNIC 2019/3-189. Supported by Chicago fellowship (GKAH), NIH R01GM131128 (JWT) and R01GM121931 (JWT).
Footnotes
Competing interests. The authors declare no competing financial interests.
Works Cited
- 1.Marsh JA & Teichmann SA Structure, dynamics, assembly, and evolution of protein complexes. Annual review of biochemistry 84, 551–575 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Goodsell DS & Olson AJ Structural symmetry and protein function. Annu Rev Biophys Biomol Struct 29, 105–153 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Lynch M. Evolutionary diversification of the multimeric states of proteins. Proc Natl Acad Sci U S A 110, E2821–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lukeš J. et al. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life 63, 528–537 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Manhart M. & Morozov AV Protein folding and binding can emerge as evolutionary spandrels through structural coupling. Proc Natl Acad Sci U S A 112, 1797–1802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schank JC & Wimsatt WC Generative entrenchment and evolution. PSA: Proceedings of the biennial meeting of the philosophy of science association 1986, 33–60 (1986). [Google Scholar]
- 7.Muller HJ Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3, 422 (1918). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moody AD et al. Thermodynamic dissection of estrogen receptor-promoter interactions reveals that steroid receptors differentially partition their self-association and promoter binding energetics. Biochemistry 51, 739–749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamrazi A. et al. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol 16, 2706–2719 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Robblee JP et al. Glucocorticoid receptor–promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry 51, 4463–4472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alroy I. & Freedman LP DNA binding analysis of glucocorticoid receptor specificity mutants. Nucleic acids research 20, 1045–1052 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKeown AN et al. Evolution of DNA specificity in a transcription factor family produced a new gene regulatory module. Cell 159, 58–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harms MJ et al. Biophysical mechanisms for large-effect mutations in the evolution of steroid hormone receptors. Proc Natl Acad Sci U S A 110, 11475–11480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eick GN et al. Evolution of minimal specificity and promiscuity in steroid hormone receptors. PLoS Genet 8, e1003072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagart J. et al. Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension. Nature structural & molecular biology 12, 554 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Kauppi B. et al. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain ru-486 induces a transconformation that leads to active antagonism. Journal of Biological Chemistry 278, 22748–22754 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Sack JS et al. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proceedings of the National Academy of Sciences 98, 4904–4909 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams SP & Sigler PB Atomic structure of progesterone complexed with its receptor. Nature 393, 392–396 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Bowie JU et al. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science 247, 1306–1310 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Pakula AA & Sauer RT Reverse hydrophobic effects relieved by amino-acid substitutions at a protein surface. Nature 344, 363–364 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Valentine JE et al. Mutations in the estrogen receptor ligand binding domain discriminate between hormone-dependent transactivation and transrepression. Journal of Biological Chemistry 275, 25322–25329 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Ince BA et al. Powerful dominant negative mutants of the human estrogen receptor. J Biol Chem 268, 14026–14032 (1993). [PubMed] [Google Scholar]
- 23.Xu J. et al. The extreme C terminus of progesterone receptor contains a transcriptional repressor domain that functions through a putative corepressor. Proc Natl Acad Sci U S A 93, 12195–12199 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S. et al. Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Molecular Endocrinology 10, 24–34 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Ahnert SE et al. Principles of assembly reveal a periodic table of protein complexes. Science 350, aaa2245 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Finnigan GC et al. Evolution of increased complexity in a molecular machine. Nature 481, 360–364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Force A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gray MW et al. Irremediable complexity. Science 330, 920–921 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences 104, 8597–8604 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoltzfus A. On the possibility of constructive neutral evolution. Journal of Molecular Evolution 49, 169–181 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Hershberg R. & Petrov DA Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6, e1001115 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hochberg GKA et al. Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions. Science 359, 930–935 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaltenegger E. & Ober D. Paralogue Interference Affects the Dynamics after Gene Duplication. Trends Plant Sci 20, 814–821 (2015). [DOI] [PubMed] [Google Scholar]
Additional works cited
- 34.Edgar RC MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darriba D. et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Katsu Y. et al. A second estrogen receptor from Japanese lamprey (Lethenteron japonicum) does not have activities for estrogen binding and transcription. General and comparative endocrinology 236, 105–114 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Simakov O. et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nature Ecology & Evolution 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philippe H. et al. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature 470, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cannon JT et al. Xenacoelomorpha is the sister group to Nephrozoa. Nature 530, 89–93 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Bridgham JT et al. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol 8, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemoine F. et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Cong X. et al. Determining Membrane Protein-Lipid Binding Thermodynamics Using Native Mass Spectrometry. J Am Chem Soc 138, 4346–4349 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Marty MT et al. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazurenko S. et al. CalFitter: a web server for analysis of protein thermal denaturation data. Nucleic Acids Res 46, W344–W349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abraham MJ et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015). [Google Scholar]
- 48.Lindorff-Larsen K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jorgensen WL et al. Comparison of simple potential functions for simulating liquid water. The Journal of chemical physics 79, 926–935 (1983). [Google Scholar]
- 50.Wang J. et al. Development and testing of a general amber force field. Journal of computational chemistry 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Feenstra KA et al. Improving efficiency of large time‐scale molecular dynamics simulations of hydrogen‐rich systems. Journal of Computational Chemistry 20, 786–798 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Larsson P. et al. MkVsites: A tool for creating GROMACS virtual sites parameters to increase performance in all‐atom molecular dynamics simulations. Journal of Computational Chemistry (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hess B. et al. LINCS: a linear constraint solver for molecular simulations. Journal of computational chemistry 18, 1463–1472 (1997). [Google Scholar]
- 54.Miyamoto S. & Kollman PA Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of computational chemistry 13, 952–962 (1992). [Google Scholar]
- 55.Berendsen HJC et al. Molecular dynamics with coupling to an external bath. The Journal of chemical physics 81, 3684–3690 (1984). [Google Scholar]
- 56.Parrinello M. & Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied physics 52, 7182–7190 (1981). [Google Scholar]
- 57.Bussi G. et al. Canonical sampling through velocity rescaling. The Journal of chemical physics 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai CJ et al. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci 6, 53–64 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tien MZ et al. Maximum allowed solvent accessibilites of residues in proteins. PLoS One 8, e80635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Degiacomi MT et al. Accommodating Protein Dynamics in the Modeling of Chemical Crosslinks. Structure 25, 1751–1757.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Wang D. GCevobase: an evolution-based database for GC content in eukaryotic genomes. Bioinformatics 34, 2129–2131 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Zhu YO et al. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci U S A 111, E2310–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee H. et al. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A 109, E2774–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dumont BL Significant Strain Variation in the Mutation Spectra of Inbred Laboratory Mice. Mol Biol Evol 36, 865–874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dettman JR et al. The properties of spontaneous mutations in the opportunistic pathogen Pseudomonas aeruginosa. BMC Genomics 17, 27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are deposited in the Open Science Framework (DOI 10.17605/OSF.IO/GTJ86),, including alignment, phylogeny, sequences and posterior probability of ancestral reconstructions; list of PDB identifiers for coordinates of dimers and monomers in our structural database; and molecular dynamics trajectories.