Abstract
Hematological malignancies are characterized by a block in differentiation, which in many cases is caused by recurrent mutations affecting the activity of hematopoietic transcription factors. RUNX1-EVI1 is a fusion protein encoded by the t(3;21) translocation linking two transcription factors required for normal hematopoiesis. RUNX1-EVI1 expression is found in myelodysplastic syndrome, secondary acute my eloid leukemia, and blast crisis of chronic myeloid leukemia; with clinical outcomes being worse than in patients with RUNX1-ETO, RUNX1 or EVI1 mutations alone. RUNX1-EVI1 is usually found as a secondary mutation, therefore the molecular mechanisms underlying how RUNX1-EVI1 alone contributes to poor prognosis are unknown. In order to address this question, we induced expression of RUNX1-EVI1 in hematopoietic cells derived from an embryonic stem cell d i fferentiation model. Induction resulte d in disruption of t he RUNX1-dependent endothelial-hematopoietic transition, blocked the cell cycle and undermined cell fate decisions in multipotent hematopoietic progenitor cells. Integrative analyses of gene expression with chromatin and transcription factor binding data demonstrated that RUNX1- EVI1 binding caused a re-distribution of endogenous RUNX1 within the genome and interfered with both RUNX1 and EVI1 regulated gene expression programs. In summary, RUNX1-EVI1 expression alone leads to extensive epigenetic reprogramming which is incompatible with healthy blood production.
Introduction
The development of acute myeloid leukemia (AML) is a step-wise process wherein cells acquire multiple additional genetic changes following the occurrence of the initial driver mutation which eventually leads to the development of overt disease. A number of driver mutations, such as the t(8;21) translocation which gives rise to the fusion protein RUNX1-ETO are compatible with a pre-leukemic state.1 However, another fusion protein, RUNX1-EVI1 is found most commonly as a secondary mutation2-4 and is associated with a particularly poor prognosis. The RUNX1-EVI1 onco-fusion protein is a product of the t(3;21)(q26;q22) translocation which links sequences from RUNX1 to the entire length of the MDS-EVI1 or EVI1 (also known as MECOM) locus. Elucidating the molecular basis of the phenotypic changes induced by RUNX1-EVI1 alone is complicated by the fact that it is expressed on a background of other mutations and thus unique transcriptional rewiring is seen in each patient.5
Both RUNX1 and EVI1 play important roles in normal hematopoiesis and in various hematological malignancies. RUNX1 (also known as AML1) is a transcription factor essential for initial specification of hematopoietic cells,6 and is frequently found to be mutated in leukemia.7,8 RUNX1 contains a DNA-binding domain – the runt homology domain (RUNT) at the N-terminus, which is preserved in RUNX1- EVI1 and a transactivation domain which is lost.7 MDS-EVI1 and EVI1 arise from alternative transcripts from the MECOM gene which have both overlapping and opposing functions – EVI1 can be a repressor of gene transcription, whereas MDSEVI1 has activating functions.9 MDS-EVI1 is essential for long-term survival of hematopoietic stem cells10 and is also expressed throughout embryonic hematopoiesis.11 EVI1 is able to bind DNA via ten zinc-fingers, but MDS1-EVI1 additionally contains a proline-rich domain with homology to SET domains.12 RUNX1, EVI1 and MDS-EVI1 have all been associated with cell cycle regulation alongside the control of differentiation. 13-15
Mice carrying a RUNX1-EVI1 transgene present with disrupted hematopoiesis and with varying degrees of leukemic transformation that is ultimately embryonic lethal.16-18 In a cell line model of t(3;21) it was shown that RUNX1-EVI1 blocks differentiation by binding to chromatin at both normal RUNX1 binding sites and elsewhere, co-ordinating a transcriptional network that is dependent on GATA2 rather than RUNX1.19 These studies suggest that RUNX1-EVI1 acts in a dominant negative fashion to RUNX1,20 but has additional effects, likely due to interference with EVI1 binding and interactions.
In order to understand the molecular effects of RUNX1- EVI1 expression in the absence of other mutations, we integrated gene expression and chromatin immunoprecipitation followed by sequencing (ChIP-seq) data from blood precursor cells derived from a mouse embryonic stem cell line (mESC) in which we induced RUNX1-EVI1 at the onset of hematopoiesis. We show that RUNX1- EVI1 induction leads to a block in the cell cycle and interferes with both the EVI1 and the RUNX1 driven developmental programs, with cells adopting a multi-lineage gene expression pattern. Moreover, RUNX1-EVI1 orchestrates redistribution and increased binding of endogenous RUNX1, and increases chromatin accessibility at sites enriched in PU.1 motifs. Taken together we show that RUNX1-EVI1 expression is incompatible with normal hematopoietic stem cell function.
Methods
Mouse RUNX1-EVI1 embryonic stem cell line generation
RUNX1-EVI1 from the pME18s-RUNX1-EVI1 plasmid (a gift from Kinuko Mitani, Dokkyo Medical University, Japan) was cloned into the p2lox-targeting vector (a gift from Michael Kyba, University of Minnesota). A2lox mESC (a gift from Michael Kyba) were transduced with 20 g of p2lox-RUNX-EVI1 using the 4D-Nucleofector (Lonza) with the mouse ES program, with the P3 primary cell kit.
Embryonic stem cell line differentiation
ESC were differentiated as previously described. Briefly, cells were plated into bacterial-grade dishes, after 3.25 days the resulting embryoid bodies were dispersed using TrypLE express (Gibco) to single cells and FLK1+ cells were purified by magnetic cells sorting. These FLK1+ cells were then cultured in gelatincoated flasks with mouse vascular endothelial growth factor and mouse interleukin 6. After 1 day 0.5 μg/ml doxycycline was added where appropriate and cells cultured for a further 18 hours.
Fluorescence associated cell sorting
Cell populations were identified and sorted on day 2 of blast culture based on surface markers. For experiments including hemogenic endothelium (HE) the floating and adherent cells were pooled. These cells were stained with KIT-APC (BD pharmingen), Tie2-PE (eBioscience) and CD41-PE-Cy7 (eBioscience) and analyzed on a Cyan ADP flow cytometer (Beckman Coulter) with data analysis using FlowJo, or sorted on a fluorescence associated cell sorting (FACS) Aria cell sorter (BD Biosciences). Progenitors matured in liquid culture were stained with CD11b-PE (eBioscience) and F4/80-APC (eBioscience).
Gene expression analysis
RNA was isolated from sorted cells using the NucleoSpin RNA kit (Macherey-Nagel). RNA sequencing (RNA-seq) libraries were prepared from two biological replicates using the True-Seq stranded total RNA kit (Illumina).
DNaseI-sequencing
DNase I hypersensitive sites sequencing (DNaseI-seq) was performed as previously described.21 3x105 sorted cells were added directly to DNaseI (Worthington Biochemical Corporation) used between 6 and 13 U/mL for 3 minutes at 22°C. The reaction was terminated by addition of sodium dodecyl sulfate to 0.5% and cell lysates treated with 0.5 mg/mL proteinase K. DNA was isolated by phenol/chloroform extraction and used to generate a library using the KAPA hyper prep kit, according to the manufacturer’s instructions.
Chromatin immunoprecipitation sequencing
ChIP-seq was performed essentially as described.22 KIT+ floating progenitor cells were double crosslinked, nuclei prepared22 then sonicated for eight cycles of 30 seconds (s) off using a Picoruptor (Diagenode). Immunoprecipitation was carried out overnight at 4°C, washed and eluted. Extracted DNA was then used to generate a library using the KAPA hyper prep kit.
Data availability
All sequencing data have been deposited at National Center for Biotechnology Information under the number GSE143460. Further methods, including bioinformatic analysis are detailed in the Online Supplementary Appendix.
Results
RUNX1-EVI1 disrupts hematopoietic growth and differentiation
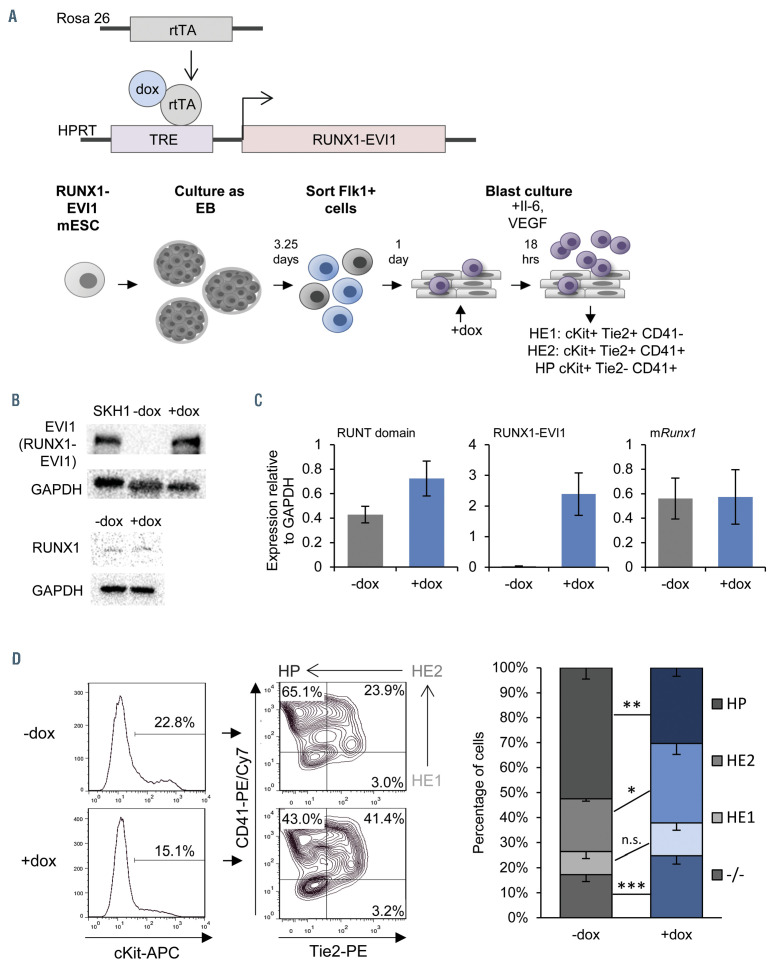
In order to understand the direct effects of RUNX1-EVI1 fusion protein induction on hematopoietic specification we needed to express RUNX1-EVI1 in primary cells in the absence of other mutations. Initial attempts to express the protein in purified human CD34+ cells by retroviral transduction were unsuccessful indicating that expressing uncontrolled levels of this protein may be toxic for the cells (unpublished data). In order to circumvent these problems, we generated a mESC line expressing a human RUNX1-EVI1 cDNA under the control of a doxycycline (dox)-inducible promoter (Figure 1A). The RUNX1-EVI1 transgene was derived from the t(3;21) SKH1 cell line23 and comprised amino acids from the N-terminus of RUNX1 translocated to the MDS1-EVI1 isoform (Online Supplementary Figure S1A). We used a well characterized in vitro differentiation system that recapitulates the different steps of embryonic hematopoietic specification in vitro,11,24 namely mesoderm specification into endothelial cells, followed by endothelial-hematopoietic transition (EHT) which gives rise to multipotent hematopoietic progenitor (HP) cells (Figure 1A, lower panel) during blast culture. The EHT is crucially dependent on the expression of Runx1,6,25 and is also the stage at which Evi1 expression is maximal before being downregulated.11 We therefore induced RUNX1-EVI1 expression in newly forming HP (Online Supplementary Figure S1B), when Runx1 becomes upregulated as shown in the schematic in Figure 1A. These time points were used for all subsequent experiments, with the cells sorted from the day 2 blast culture being used for genome-wide analysis. Since differentiation in this system is transient, we also used cell sorting to obtain cells from earlier differentiation stages which enabled us to study the effect of RUNX1-EVI1 expression at these stages as well. We titrated induction of RUNX1- EVI1 such that the protein expression was at a level similar to that seen in the SKH1 cells, and gene expression was at a similar level to endogenous RUNX1, which itself was stably expressed (Figures 1B and C).
Figure 1.
Induction of RUNX1-EVI1 perturbs Runx1 dependent endothelial to hematopoietic transition. (A) Overview of the generation of dox-inducible RUNX1-EVI1 embryonic stem cells (ESC), in vitro differentiation of ESC to hematopoietic progenitors and timing induction of RUNX1-EVI1. (B) RUNX1-EVI1 was expressed at a comparable level to that in the human t(3;21) cell line SKH-1 shown by western blot. Note that the antibody against EVI-1 does not recognize the endogenous mouse protein. RUNX1 protein levels were unaffected by induction of RUNX1-EVI1. (C) Expression of RUNX1-EVI1 was at a physiological level, equal to that of the endogenous Runx1, shown by quantitative reverse transcriptase polymerase chain reaction, normalized to glyceraldehyde 3-phosphate dehydrogenase (Gapdh). Runt domain primers bind the 5’ end of the gene and so both endogenous Runx1 and RUNX1-EVI1 are detected, mRunx1 primers bind the 3’ end and so only endogenous Runx1 is detected. (D) The composition of the day 2 blast culture, 18 hours following doxycycline (dox) induction, was analyzed by flow cytometry using antibodies against cKit, Tie2 and CD41; representative plots for -dox and +dox samples are shown (left, with the Tie2/CD41 plots pre-gated by cKit+) with the average percentage of each population (right), error bars represent standard error of the mean, n=5, *P<0.05, **P<0.01, ***P<0.005.
Induction of RUNX1-EVI1 led to a partial disruption of the EHT following 18 hours of dox induction (Figure 1D), with a reduction in cKit+ Tie2- CD41+ HP and a reciprocal increase in hematopoietic committed HE cells (HE2, cKit+ Tie2+ CD41+). This result is concordant with the notion that RUNX1-EVI1 acts as a dominant negative to RUNX1, since the earlier uncommitted endothelial cells which had not yet upregulated Runx1 (HE1, cKit+ Tie2+ CD41-) were unaffected. Inducing RUNX1-EVI1 prior to the EHT hampers hematopoietic differentiation leading to a considerably greater proportion of Tie2- CD41- negative cells (Online Supplementary Figure S1C).
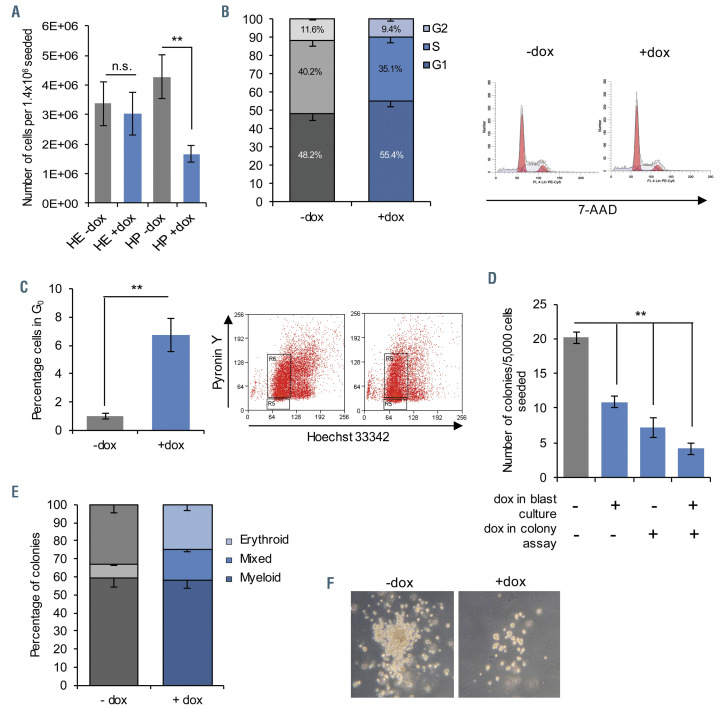
Figure 2.
RUNX1-EVI1 expression causes reduced cell cycling and colony forming capacity in hematopoietic progenitors. (A) Fewer floating hematopoietic progenitor (HC) cells were present in day 2 blast culture, following induction of RUNX1-EVI1, n=5, **P<0.01. Cell cycle stages, n=3. (B) and quiescence, n=5 (C) were assessed in the whole blast culture at the same time-point, showing an increased proportion of cells in G0 and G1. Example flow cytometry plots are shown to the right, *P< 0.05, **P<0.01. (D) Floating progenitor cells with RUNX1-EVI1 induced formed fewer colonies, n=3, **P<0.01 (E) Colonies following doxycycline (dox) induction in blast culture only were comprised of approximately equivalent proportions of granulocyte/macrophage (GM), erythroid (Ery) or mixed colonies, with a slight increase in the mixed-type at the expense of singular lineage, n=3. All error bars (A to E) represent standard error of the mean. (F) Representative brightfield images of colonies with and without dox induction in blast culture only.
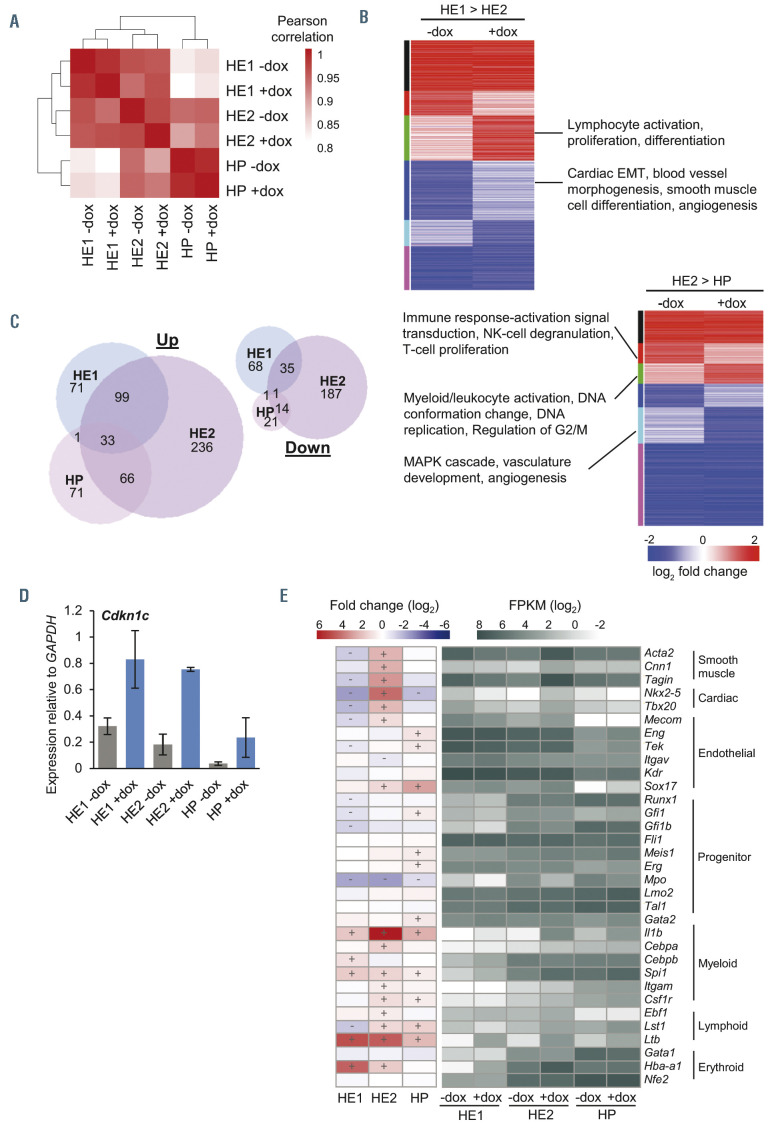
Figure 3.
Gene expression following RUNX1-EVI1 induction is de-regulated in a developmental-stage-specific manner. (A) Heatmap showing the results of hierarchical clustering of hemogenic endothelium 1 (HE1), HE2 and hematopoietic progenitor (HP) cells in both minus doxycycline (-dox) and +dox treated samples. Clustering was performed using sample-wise Pearson correlation coefficients for all genes expressed with fragments per kilobase million (FPKM) >1 in at least one sample. (B) Heatmaps comparing the log2 fold-change of genes that were differentially expressed between differentiation stages (HE1 to HE2, and HE2 to HP) in either -dox or +dox samples. Genes were placed into eight groups based on the difference of the fold-change between -dox and +dox samples, and are indicated as a colored bar to the left of the heatmap. Selected GO terms are shown for those groups that show developmental disruption following RUNX1-EVI1 induction. (C) Venn diagram showing the number of genes that are up- or downregulated in each differentiation stage after induction of RUNX1-EVI1. (D) Quantitative reverse transcriptase polymerase chain reaction expression of Cdkn1c, relative to glyceraldehyde 3-phosphate dehydrogenase (Gapdh), n=3, error bars represent standard error of the mean. (E) Heatmaps showing log2 fold change (left) and log2 FPKM for selected genes involved in various mesodermal lineages, +/- on the heatmap indicates those which show a fold change in expression of at least 1.5. EMT: epithelial-to-mesenchymal transition.
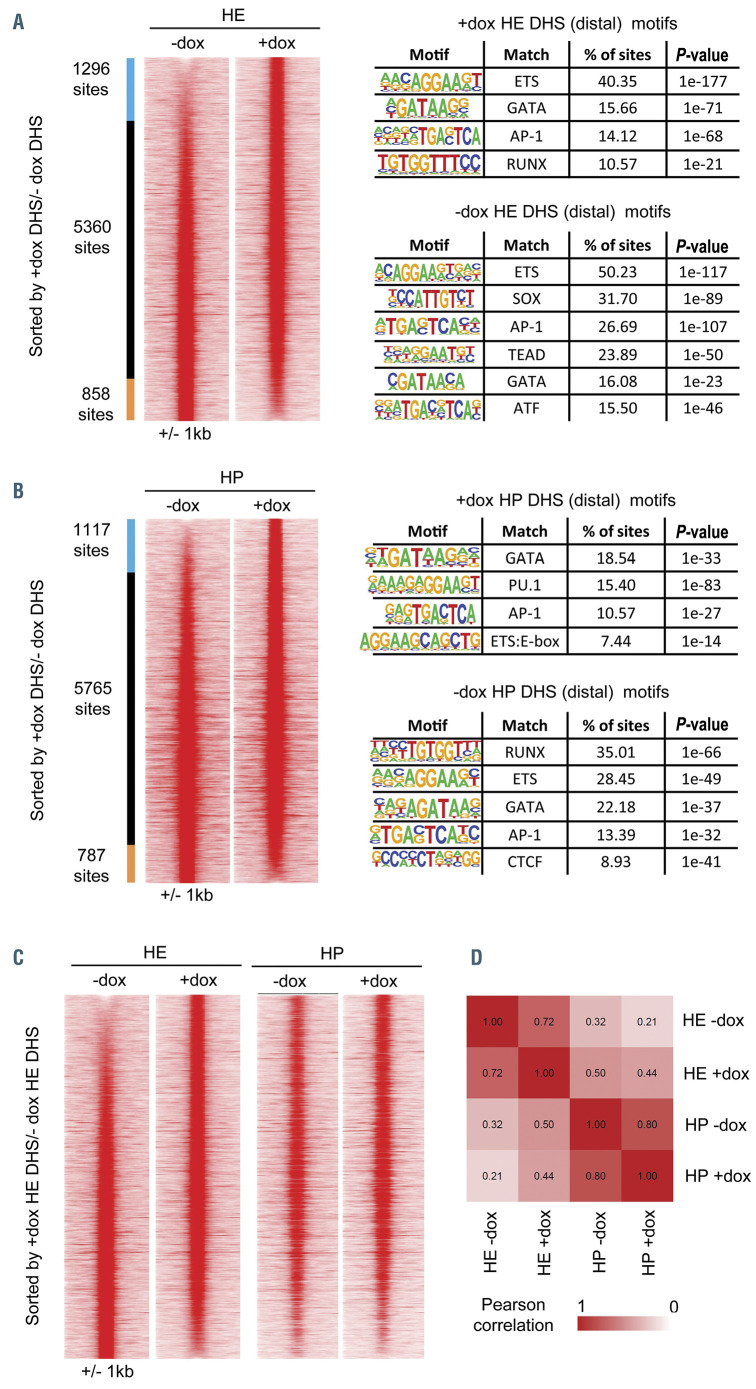
Figure 4.
RUNX1-EVI1 induction causes specific changes to chromatin accessibility. Comparison of distal DNase I hypersensitive sites sequencing (DNaseI-seq) peaks in (A) hemogenic endothelium (HE) cells (cKit+, Tie2+, CD41-/+) and (B) hematopoietic progenitor (HP) cells (cKit+, Tie2-, CD41+). Peaks are ordered according to the fold-difference of the normalized tag-count between -dox and plus doxycycline (+dox) treated cells and are presented as a heatmap of the tag-density for each sample. Peaks that are specific to a sample (fold-difference >2) are indicated as colored bars to the left of the density plots, with the number of peaks in each group shown. The results of a de-novo motif search conducted within the specific sets of peaks are also shown (C) Comparison of the tag-density profiles of the peaks found in HE cells to the same sites measured in HP cells (D) Heatmap showing the results of hierarchical clustering of the pearson correlation values of the distal DNaseI hypersensitive sites (DHS). The actual pearson correlation values are shown on the heatmap.
Figure 5.
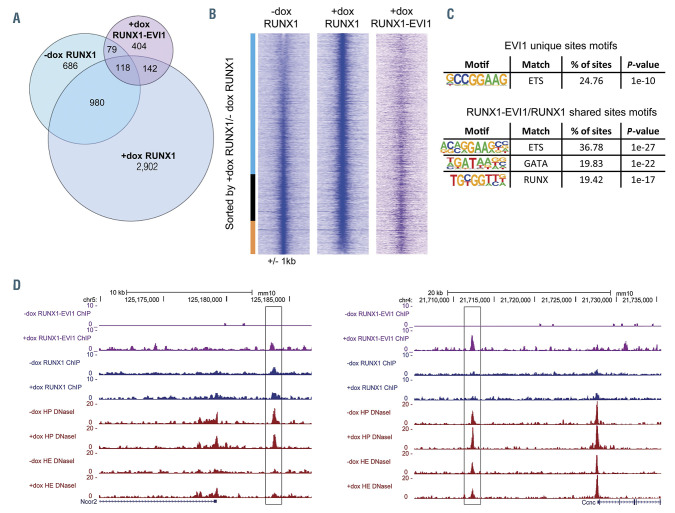
RUNX1-EVI1 disrupts RUNX1 binding but also binds to unique binding sites. (A) Venn diagram showing the number of uniquely called and overlapping RUNX1 and RUNX1-EVI1 chromatin immunoprecipitation sequencing (ChIP-seq) peaks. (B) Comparison of RUNX1 binding in -dox and +dox treated cells, RUNX1 ChIPseq peaks were ranked according to the fold difference of the normalized plus doxycycline (+dox) /-dox tag count across a 2 kilobase (kb) window. The tag-density of the RUNX1-EVI1 ChIP-seq peaks is plotted alongside. The bar alongside indicates the +dox specific sites (blue), shared sites (black) and -dox specific sites (orange). (C) De novo motif enrichment was conducted within the RUNX1-EVI1 ChIP-seq peaks, at both the unique sites and those which were also bound by RUNX1 in either -dox, +dox or both. (D) Genome browser screenshots showing an example site where both RUNX1 and RUNX1-EVI1 bind (left, Ncor2 locus) and where RUNX1-EVI1 binds in the absence of RUNX1 (right, Ccnc locus).
The EHT process is a true cellular transition that is cell cycle independent.26 Whilst we observed a reduction in the proportion of HP related to perturbation of the EHT, we also noted a considerably lower total number of HP than in the control (Figure 2A). This result could be explained by an increase in cells not actively cycling in G0/G1 (48.2% vs. 55.4% without and with RUNX1-EVI1, Figure 2B), specifically by increased numbers of cells in G0 (5.4% with RUNX1-EVI1 compared to <1% without, Figure 2C). We also found a modest increase in apoptotic cells (Online Supplementary Figure S2A).
In order to investigate whether RUNX1-EVI1 expression not only disrupted the EHT, but also affected the ability of progenitor cells to terminally differentiate, we induced RUNX1-EVI1 in newly forming progenitors and placed the progenitors into methylcellulose colony forming unit assays. We first carried out colony-forming unit assays in the absence of dox in the methylcellulose medium. RUNX1-EVI1 protein was quickly lost following the withdrawal of dox (Online Supplementary Figure S2B). Despite RUNX1-EVI1 being absent, we saw an overall reduction in the number of colonies formed (Figure 2D; Online Supplementary Figure S2C) primarily accounted for by a reduction the number of myeloid (-dox 58±13, +dox 42±11 per 5,000 HP seeded) and erythroid colonies (-dox 26±7, +dox 20±4), with a concomitant increase in the proportion of mixed lineage colonies, or those of unclear lineage (-dox 29±9, +dox 26±6, Figure 2E). The colonies which did form were generally smaller with fewer healthy cells (Figure 2F), which may relate to the previously observed increase in apoptosis (Online Supplementary Figure S2A). When RUNX1-EVI1 expression was either induced or maintained in the HP when they were plated into methylcellulose, colony-forming capacity was further reduced (Figure 2D). Importantly, we did not see enhanced myeloid differentiation after RUNX1-EVI1 induction when HP were cultured in liquid or semi-solid medium (Figure 2E; Online Supplementary Figure S2D). These data suggest that whilst RUNX1-EVI1 expression affects the differentiation capacity of HP, there was some reversibility of this phenotype and the continued expression of RUNX1-EVI1 caused ongoing changes.
Figure 6.
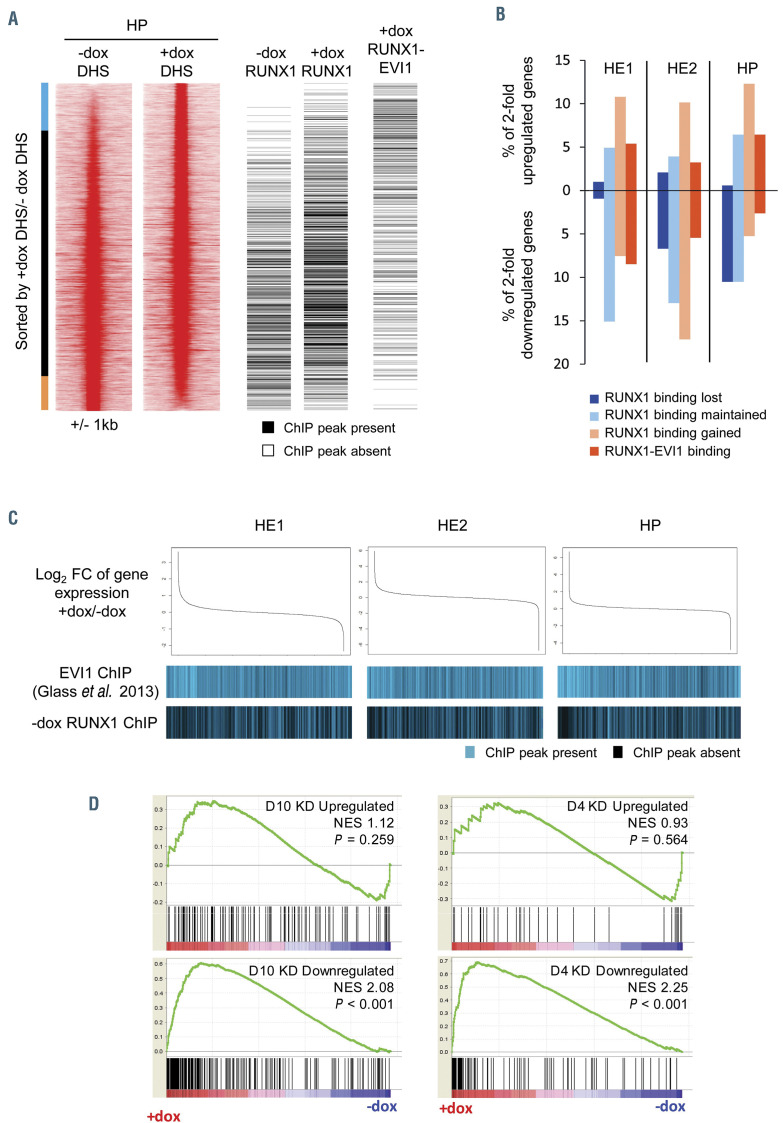
Changes to chromatin organisation and gene expression are modulated by RUNX1 and RUNX1-EVI1 binding. (A) Comparison of RUNX1 and RUNX1-EVI1 binding sites to DNase I hypersensitive sites sequencing (DNaseI-seq) data from hematopoietic progenitor (HE) cells. DNaseI-seq peaks are ranked according to the fold-difference of the plus doxycycline (+dox)/-dox normalized tag-count, with the presence (black) or absence (white) of a chromatin immunoprecipitation sequencing (ChIP-seq) peak indicated alongside. The bar alongside indicates the +dox specific DNaseI sites (blue), shared sites (black) and –dox specific sites (orange). (B) The percentage of 2-fold de-regulated genes at each stage which have an associated RUNX1 or RUNX1-EVI1 binding site. (C) Comparison of changes in gene expression to the binding patterns of wild-type EVI1 and RUNX1. Gene expression was ranked by fold change (top), with the presence or absence of wild-type EVI127 or RUNX1 binding associated with each gene is indicated below in blue. (D) Gene set enrichment analysis comparing changes in gene expression after RUNX1-EVI1 induction in HP to genes that are up- and downregulated following 4 and 10 days of RUNX1-EVI1 knockdown in the SKH1 cell line.19 Genes that are upregulated following RUNX1-EVI1 induction correspond closely to genes that are downregulated after RUNX1-EVI1 knock-down in the SKH1 cell line.
Taken together, our data show that RUNX1-EVI1 expression is incompatible with multipotent precursor development, and when induced in precursors, leads to cell cycle arrest and an increase in apoptosis.
RUNX1-EVI1 induction alters gene expression in a differentiation-stage dependent and independent fashion
We next wanted to understand the molecular basis of the observed phenotypes. To this end, we sorted HE1, HE2 and HP cells on the basis of their surface marker phenotypes, as described in Figure 1A and D and performed RNA-seq on the resulting matched cell populations. Biological duplicates were well correlated (Online Supplementary Figure S3A) and the average was used for further analysis. Hierarchical clustering of these datasets (Figure 3A) showed that the overall gene expression patterns in the different cell types were preserved in the presence of RUNX1-EVI1, but with genes being de-regulated at every stage, particularly in HE2 cells (Online Supplementary Figure S3B, Supplementary Tables S1 to S3). A subset of these genes were validated by quantitative reverse transcriptase polymerase chain reaction (qRTPCR) (Online Supplementary Figure S3C) and the HP gene expression changes, being the target cell for the leukemic transformation, were compared to two previously published t(3;21) patient RNA-seq datasets (Online Supplementary Figure S3D). This analysis showed that those genes which are specific to t(3;21) patients as compared to healthy CD34+ cells were upregulated following induction of RUNX1-EVI1 in HP, with genes such as Cdh5, Hes1, Maff and Arhgef12 being overexpressed in both patients and HP expressing RUNX1-EVI1.
The differentiation of blood cells in the in vitro differentiation system is not entirely synchronous, therefore RUNX1-EVI1 induction occurs in different cell types representing a differentiation trajectory. Many changes normally seen within the differentiation process were maintained after induction. For example, genes which were upor downregulated during the transition from HE1 to HE2 or HE2 to HP continued to be up- or downregulated (Figure 3B; Online Supplementary Tables S4 and S5), including those essential for these transitions such as Tek and Gfi1b. However, a subset of genes failed to be up- or downregulated to the extent it normally should. For example, some regulators of the MAPK pathway including Mapk3 and Dusp6 were downregulated during the transition from HE2 to HP more than they should be following RUNX1-EVI1 induction. Alongside these developmental changes, a core set of genes were upregulated at least 2- fold in two, or all three cell types (Figures 3C and D), including Dusp5, Cdkn1c and Pdgfa. Cdkn1c is a negative regulator of the cell cycle and its universal upregulation may underpin the cell cycle arrest (Figures 2B and C) and the de-regulation of multiple cell cycle associated genes (Online Supplementary Figure S3E).
We also examined how stage-specific gene expression changes related to the differentiation program using known marker genes. In HE cells, the expression of the vascular/smooth muscle program was deregulated (Figure 3E). The smooth muscle genes Acta2, Tagin, Cnn1 and the genes encoding the cardiac regulator TBX20 and homeo - box protein Nkx-2.5 were further downregulated in HE1, but were then upregulated when RUNX1-EVI1 was induced in HE2. When specifically examining hematopoietic lineage gene signatures, we did not see a downregulation of myeloid or erythroid genes as expected from the colony forming assays. Indeed, we found a widespread, albeit modest (>1.5-fold), increase in expression of genes related to a multipotent progenitor identity with the concomitant expression of a multi-lineage gene expression program consisting of myeloid, lymphoid and megakaryocyte/ erythroid genes (Figure 3E; Online Supplementary Figure S3C). Taken together, these results suggest that RUNX1-EVI1 induction causes a cell cycle and differentiation arrest that is associated with a disturbance of the balance between the hematopoietic and vascular/smooth muscle fate.
Disturbed lineage specification is caused by chromatin changes associated with altered RUNX1 binding
In order to understand how RUNX1-EVI1 induction reprograms the chromatin landscape, we performed and integrated ChIP-seq analysis for both RUNX1 and RUNX1-EVI1 in HP, with data from DNaseI-seq experiments performed on sorted cKit+ HP, and HE (cKit+, Tie2+, CD41-/+). Induction of RUNX1-EVI1 led to changes to chromatin accessibility in the HE and HP cells and an increased proportion of distal DNaseI hypersensitive sites (DHS) (Online Supplementary Figure S4A). Few DHS changed at promoter sites (Online Supplementary Figure S4B). We therefore focussed on the analysis of distal DHS and ranked them by the fold change in tag count at each site. 1,296 DHS were gained and 858 lost in the HE when RUNX1-EVI1 was expressed (Figure 4A). The gained sites showed a specific enrichment of RUNX motifs, whilst the sites lost contained SOX, TEAD and AP-1 motifs. In HP cells, RUNX1-EVI1 induction had a completely different effect as here we observed a loss rather than a gain of RUNX motif enrichment (Figure 4B). Taken together, this result suggested a shift in chromatin patterns in HE from those of the vascular/endothelial lineages11,22 towards a HP-like pattern. We confirmed this result by plotting the HP DNaseI-seq peaks alongside those of the HE (Figure 4C) and by performing a correlation analysis (Figure 4D). These analyses demonstrated that the HE chromatin pattern was more similar to that of HP cells following induction of RUNX1-EVI1, despite the cells still displaying surface markers and an overall gene expression signature of the HE (Figures 1C and 3A). Furthermore, in HP we also saw a shift from the ETS motif to a PU.1 specific motif, which was consistent with the upregulation of Spi1 (encoding PU.1) expression, indicating that the chromatin accessibility pattern was being rewired towards myelopoiesis.
In order to test how these results related to the interplay of RUNX1-EVI1 with RUNX1, we compared ChIP-seq for RUNX1 with and without induction of RUNX1-EVI1, to the binding of RUNX1-EVI1 itself in cKit+ HP. The antibody we used against human EVI1 did not recognize the endogenous murine EVI1 and thus exclusively measured binding of the exogenous protein. We analyzed only highconfidence ChIP-seq peaks, which had been filtered for the presence of a DHS at the same site, to minimize noise associated with the technical difficulty of these ChIP experiments.
Around half of RUNX1-EVI1 binding sites overlapped with those of RUNX1 (Figure 5A), including those RUNX1 sites that were either maintained or gained following RUNX1-EVI1 induction. This result suggests that the predominant mechanism of action of RUNX1-EVI1 is not the displacement of RUNX1. This finding was confirmed by examining the proximity of RUNX1 and RUNX1-EVI1 ChIP peaks (Online Supplementary Figure S5A). The RUNX1 and RUNX1-EVI1 peak summits were distributed similarly prior to and following induction of RUNX1-EVI1, both overlapping the same sites and next to each other. More strikingly, induction caused a large movement of RUNX1 within the genome with the loss of over a third of preexisting RUNX1 sites and considerably more gained RUNX1 sites. Most gained sites were not associated with RUNX1-EVI1 binding (Figure 5B) nor did they have any specific motif enrichment not present in the shared sites (Online Supplementary Figure S5B) but were instead found in promoters (Online Supplementary Figure S5C). This movement was a real re-distribution as the overall level of RUNX1 protein was unchanged (Figure 1B).
When RUNX1-EVI1 bound in concert with RUNX1, or displaced RUNX1, its binding sites were enriched for RUNX and GATA motifs (Figure 5C and D; Online Supplementary Figure S5D), suggesting that RUNX1-EVI1 can also bind via the RUNT homology domain. Unique RUNX1-EVI1 ChIP peaks, however, were enriched for ETS-like motifs (Figure 5C), which may be indicative of binding via the EVI1 portion of the protein27 in the absence of RUNX1, an example of which is shown in Figure 5D and the Online Supplementary Figure S5D.
RUNX1-EVI1 disrupts RUNX1 and EVI1 driven gene regulatory networks
We next integrated the RUNX1 and RUNX1-EVI1 ChIPseq data with the DNaseI-seq and RNA-seq data to investigate how the binding of RUNX1-EVI1 and the movement of RUNX1 influenced changes in gene expression. We observed that the DHS that were lost following induction of RUNX1-EVI1 were associated with lost RUNX1 binding as well. However, RUNX1 moved to chromatin which was already accessible and DHS which were gained were associated with RUNX1-EVI1 binding (Figure 6A; Online Supplementary Figure S6A). When considering genes which were at least 2-fold deregulated, those which were downregulated - particularly in HP - were associated with reduced RUNX1 binding (Figure 6B), indicating that RUNX1-EVI1 induction interfered with gene activation by RUNX1. The proportion of changed genes associated with lost RUNX1 binding increased throughout differentiation, as the reliance on RUNX1 increased. Both up- and downregulated genes at all stages were associated with new RUNX1 binding sites, again matching the trend in chromatin accessibility. RUNX1-EVI1 bound de-regulated genes were predominantly upregulated in HP cells, but more were downregulated in HE cells. We therefore compared how the gene expression changes related to endogenous EVI1 and RUNX1 binding sites, by plotting which genes were associated with EVI1 binding from a public EVI1 ChIP dataset,27 and the genes associated with our RUNX1 ChIP in uninduced HP against gene expression changes following RUNX1-EVI1 (Figure 6C; Online Supplementary Figure S6B). Genes which changed expression, were either upregulated or downregulated, were enriched for EVI1 binding and depleted for RUNX1 binding, particularly in the HP. Therefore, neither RUNX1 nor EVI1 is solely associated with up- or downregulation of their target genes but instead we see a complex and stagespecific pattern of interference.
Finally, to examine which changes were a direct response to binding and which were a result of the cells’ changing identity, we employed gene set enrichment analysis comparing the genes upregulated in HP following induction of RUNX1-EVI1 to those downregulated following small interfering RNA knockdown of RUNX1- EVI1 in a human cell line19 (Figure 6D) and observed a good correlation. These genes include hematopoietic genes (Online Supplementary Table S6) such as Gata2 (a RUNX1 target) and Meis1 (a target of both RUNX1 and RUNX1-EVI1). By contrast, the genes downregulated following RUNX1-EVI1 induction in HP did not correlate well with those upregulated following RUNX1-EVI1 knockdown with the exception of a small subset of genes such as Mpo and Rab44 which are neither RUNX1 nor RUNX1-EVI1 targets.
These results suggest that RUNX1-EVI1 is likely to interfere with the repressive activities of both RUNX1 and EVI1 with the balance of lineage decisions depending on the differentiation stage at the time point of induction.
Discussion
RUNX1-EVI1 expression is only found as a secondary event in myeloid malignancies. Our study shows that its expression as sole oncogene in untransformed myeloid progenitor cells is incompatible with blood cell differentiation. We also found that RUNX1-EVI1 induction disrupts the RUNX1 driven endothelial-hematopoietic transition, in a similar fashion to RUNX1-ETO.28 Expression of RUNX1-EVI1 HP cells disrupted their colony forming capacity and led to extensive de-regulation of gene expression. However, the underlying molecular cause was different. As with RUNX1-ETO, genes of the stem cell program were upregulated, but the arrest in differentiation after RUNX1-EVI1 induction was associated with the rapid activation of a multi-lineage gene expression program and a profound disturbance of hematopoietic lineage specification.
Induction of RUNX1-EVI1 in cells committed to the hematopoietic fate is associated with the activation of a pan-lineage hematopoietic gene expression program and a failure in fully downregulating factors associated with a vascular gene expression program. This behavior is reminiscent of mutations in lineage commitment factors, such as PAX5. Knock-out of PAX5 leads to a block in B-cell differentiation which is associated with an inability to activate the B-cell gene expression program, but also an inability to repress the myeloid program,29,30 generating progenitors with a multi-lineage gene expression pattern and the inability to commit to a specific lineage. Alongside the differentiation associated phenotype, we found that RUNX1-EVI1 caused a partial cell cycle arrest and increase in apoptosis which is likely to be associated with increased expression of Cdkn1c, leading to the stage-specific deregulation of cell cycle genes. Cdkn1c encodes the cell cycle inhibitor p57Kip2 which is important in maintenance of the adult hematopoietic stem cell compartment,31 and has been shown to be deregulated by both EVI1 and MDS-EVI1 thus causing to cell cycle mis-regulation.13,32
Our results indicate that the phenotype caused by RUNX1-EVI1 induction is a result of interference with both RUNX1 and EVI1 driven gene regulatory networks. Similar to PAX5, both RUNX1 and EVI1 interact with coactivators and co-repressors to affect gene expression, depending on the genomic context.9,33-35 In accordance with this notion, we did not find RUNX1-EVI1 behaving solely as a repressor or activator of gene expression, with further variation based on the differentiation stage. In early HE, when Mecom expression reaches its peak, we observed a bias towards repression of RUNX1-EVI1 target genes and an activation of the hematopoietic program, indicating that the fusion protein interfered with both the repressive and activating function of EVI1. Conversely, we found a bias towards gene activation in HP for multiple programs where Runx1 expression was upregulated. This result suggests that RUNX1-EVI1 interfered with the repressive activity of RUNX1 which is known to co-operate with other factors to shut down the endothelial gene expression program.36,37 This idea is further supported by our finding that downregulated gene expression in HP can largely be accounted for by lost RUNX1 binding. In contrast, upregulated gene expression is independent of new RUNX1 binding and is therefore likely caused by other transcription factors. These may include PU.1, which is a known mediator of EVI1 function in myeloid malignancy38,39 and which was precociously upregulated following induction of RUNX1-EVI1. PU.1 is a master myeloid regulator which co-operates with RUNX1 in normal hematopoiesis,40,41 and also has roles in cell cycle regulation in stem cells.42 Alongside gene expression being upregulated, open chromatin sites gained in HP were enriched for PU.1 motifs and PU.1 target genes such as Csf1r43 were upregulated.
In conclusion, we found that RUNX1-EVI1 disrupts the function of the endogenous RUNX1 and EVI1 in a developmental program specific fashion leading to loss of cell cycle control and an inability of hematopoietic precursor cells to execute and maintain regulated cell fate commitment decisions. Our results explain why RUNX1-EVI1 is associated with particularly poor prognosis. It adds to a growing number of oncogenes that as sole drivers are incompatible with hematopoietic stem cell function.44 Our results also highlight the fact that whilst targeting transcription factors such as RUNX1 is a therapy currently being developed,45 crosstalk between multiple transcription networks in the presence of several mutated or mis-expressed transcription factors must also be considered.
Supplementary Material
Acknowledgments
The authors would like to thank the Genomics Birmingham Sequencing Facility for their expert sequencing service, and the University of Birmingham Flow Cytometry service and Dr Mary Clarke for expert cell sorting.
Funding Statement
Funding SGK received funding from Kay Kendall Leukemia Fund and PK received funding from Bloodwise, awarded to CB.
References
- 1.Wiemels JL, Xiao Z, Buffler PA, et al. In utero origin of t(8;21) AML1-ETO translocations in childhood acute myeloid leukemia. Blood. 2002;99(10):3801-3805. [DOI] [PubMed] [Google Scholar]
- 2.Rubin CM, Larson RA, Anastasi J, et al. t(3;21)(q26;q22): A recurring chromosomal abnormality in therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 1990;76(12):2594-2598. [PubMed] [Google Scholar]
- 3.Lugthart S, Gröschel S, Beverloo HB, et al. Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3) (q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol. 2010;28(24):3890-3898. [DOI] [PubMed] [Google Scholar]
- 4.Nukina A, Kagoya Y, Watanabe-Okochi N, et al. Single-cell gene expression analysis reveals clonal architecture of blast-phase chronic myeloid leukaemia. Br J Haematol. 2014;165(3):414-416. [DOI] [PubMed] [Google Scholar]
- 5.Assi SA, Imperato MR, Coleman DJL, et al. Subtype-specific regulatory network rewiring in acute myeloid leukemia. Nat Genet. 2019;51(1):151-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457(7231):892-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soderholm J, Kobayashi H, Mathieu C, Rowley JD, Nucifora G. The leukemiaassociated gene MDS1/EVI1 is a new type of GATA-binding transactivator. Leukemia. 1997;11(3):352-358. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka K, Sato T, Yoshimi A, et al. Evi1 is essential for hematopoietic stem cell selfrenewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med. 2011;208(12):2403-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goode Debbie K, Obier N, Vijayabaskar MS, et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell. 2016;36(5): 572-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass C, Wilson M, Gonzalez R, Zhang Y, Perkins AS. The role of EVI1 in myeloid malignancies. Blood Cells Mol Dis. 2014;53(1):67-76. [DOI] [PubMed] [Google Scholar]
- 13.Kustikova OS, Schwarzer A, Stahlhut M, et al. Activation of Evi1 inhibits cell cycle progression and differentiation of hematopoietic progenitor cells. Leukemia. 2012;27(5): 1127-1138. [DOI] [PubMed] [Google Scholar]
- 14.Kilbey A, Stephens V, Bartholomew C. Loss of cell cycle control by deregulation of cyclin-dependent kinase 2 kinase activity in Evi-1 transformed fibroblasts. Cell Growth Differ. 1999;10(9):601-610. [PubMed] [Google Scholar]
- 15.Friedman AD. Cell cycle and developmental control of hematopoiesis by Runx1. J Cell Physiol. 2009;219(3):520-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maki K, Yamagata T, Yamazaki I, Oda H, Mitani K. Development of megakaryoblastic leukaemia in Runx1-Evi1 knock-in chimaeric mouse. Leukemia. 2006;20(8):1458-1460. [DOI] [PubMed] [Google Scholar]
- 17.Cuenco GM, Nucifora G, Ren R. Human AML1/MDS1/EVI1 fusion protein induces an acute myelogenous leukemia (AML) in mice: A model for human AML. Proc Natl Acad Sci U S A. 2000;97(4):1760-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maki K, Yamagata T, Asai T, et al. Dysplastic definitive hematopoiesis in AML1/EVI1 knock-in embryos. Blood. 2005;106(6):2147-2155. [DOI] [PubMed] [Google Scholar]
- 19.Loke J, Assi SA, Imperato MR, et al. RUNX1-ETO and RUNX1-EVI1 differentially reprogram the chromatin landscape in t(8;21) and t(3;21) AML. Cell Rep. 2017;19(8):1654-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Mitani K, Kurokawa M, et al. Dual functions of the AML1/Evi-1 chimeric protein in the mechanism of leukemogenesis in t(3;21) leukemias. Mol Cell Biol. 1995;15(5):2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bert AG, Johnson BV, Baxter EW, Cockerill PN. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol. 2007;27(8):2870-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obier N, Cauchy P, Assi SA, et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development. 2016;143(23):4324-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitani K, Ogawa S, Tanaka T, et al. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 1994;13(3):504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sroczynska P, Lancrin C, Pearson S, Kouskoff V, Lacaud G. In vitro differentiation of embryonic stem cells as a model of early hematopoietic development. ES C.W., ed. Leukemia. Methods in Molecular Biology™ (Methods and Protocols): Humana Press. 2009. [DOI] [PubMed] [Google Scholar]
- 25.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457(7231):887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eilken HM, Nishikawa S-I, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457(7231):896-900. [DOI] [PubMed] [Google Scholar]
- 27.Glass C, Wuertzer C, Cui X, et al. Global identification of EVI1 target genes in acute myeloid leukemia. PLoS One. 2013;8(6): e67134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regha K, Assi SA, Tsoulaki O, Gilmour J, Lacaud G, Bonifer C. Developmental-stagedependent transcriptional response to leukaemic oncogene expression. Nat Commun. 2015;6(1):7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401(6753):556-562. [DOI] [PubMed] [Google Scholar]
- 30.Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 2006;24(3):269-281. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto A, Takeishi S, Kanie T, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9(3):262-271. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Stehling-Sun S, Lezon-Geyda K, et al. PR-domain–containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood. 2011;118(14):3853-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23(24): 4220-4224. [DOI] [PubMed] [Google Scholar]
- 34.Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBP alpha synergize with different regions of AML1. Mol Cell Biol. 1998;18(7):3915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer S, Brouillet J-P, Kilbey A, et al. Evi-1 Transforming and repressor activities are mediated by CtBP co-repressor proteins. J Biol Chem. 2001;276(28):25834-25840. [DOI] [PubMed] [Google Scholar]
- 36.Thambyrajah R, Mazan M, Patel R, et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol. 2016;18(1):21-32. [DOI] [PubMed] [Google Scholar]
- 37.Lancrin C, Mazan M, Stefanska M, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120(2):314-322. [DOI] [PubMed] [Google Scholar]
- 38.Ayoub E, Wilson MP, McGrath KE, et al. EVI1 overexpression reprograms hematopoiesis via upregulation of Spi1 transcription. Nat Commun. 2018;9(1): 4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laricchia-Robbio L, Premanand K, Rinaldi CR, Nucifora G. EVI1 impairs myelopoiesis by deregulation of PU.1 function. Cancer Res. 2009;69(4):1633-1642. [DOI] [PubMed] [Google Scholar]
- 40.Huang G, Zhang P, Hirai H, et al. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40(1):51-60. [DOI] [PubMed] [Google Scholar]
- 41.Hu Z, Gu X, Baraoidan K, et al. RUNX1 regulates corepressor interactions of PU.1. Blood. 2011;117(24):6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staber Philipp B, Zhang P, Ye M, et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol Cell. 2013;49(5):934-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aikawa Y, Katsumoto T, Zhang P, et al. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat Med. 2010;16(5):580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Genua C, Norfo R, Rodriguez-Meira A, et al. Cell-intrinsic depletion of Aml1-ETOexpressing pre-leukemic hematopoietic stem cells by K-Ras activating mutation. Haematologica. 2019;104(11):2215-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushweller JH. Targeting transcription factors in cancer — from undruggable to reality. Nat Rev Cancer. 2019;19(11):611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited at National Center for Biotechnology Information under the number GSE143460. Further methods, including bioinformatic analysis are detailed in the Online Supplementary Appendix.