In this study, Olsen et al. show that alterations in Rb1/Trp53/Myc in the mouse lung induce an ASCL1+ state of small cell lung cancer (SCLC) in multiple cells of origin. Their findings show that in a MYC-driven SCLC model, ASCL1 promotes neuroendocrine fate and represses the emergence of a SOX9+ nonendodermal stem-like fate that resembles neural crest.
Keywords: SCLC, ASCL1, mouse models, lung cancer, cell of origin, plasticity, neuroendocrine, small cell lung cancer
Abstract
ASCL1 is a neuroendocrine lineage-specific oncogenic driver of small cell lung cancer (SCLC), highly expressed in a significant fraction of tumors. However, ∼25% of human SCLC are ASCL1-low and associated with low neuroendocrine fate and high MYC expression. Using genetically engineered mouse models (GEMMs), we show that alterations in Rb1/Trp53/Myc in the mouse lung induce an ASCL1+ state of SCLC in multiple cells of origin. Genetic depletion of ASCL1 in MYC-driven SCLC dramatically inhibits tumor initiation and progression to the NEUROD1+ subtype of SCLC. Surprisingly, ASCL1 loss promotes a SOX9+ mesenchymal/neural crest stem-like state and the emergence of osteosarcoma and chondroid tumors, whose propensity is impacted by cell of origin. ASCL1 is critical for expression of key lineage-related transcription factors NKX2-1, FOXA2, and INSM1 and represses genes involved in the Hippo/Wnt/Notch developmental pathways in vivo. Importantly, ASCL1 represses a SOX9/RUNX1/RUNX2 program in vivo and SOX9 expression in human SCLC cells, suggesting a conserved function for ASCL1. Together, in a MYC-driven SCLC model, ASCL1 promotes neuroendocrine fate and represses the emergence of a SOX9+ nonendodermal stem-like fate that resembles neural crest.
Small cell lung cancer (SCLC) is a highly aggressive neuroendocrine lung tumor with a median survival time of ∼12 mo (Sabari et al. 2017; Rudin et al. 2019). Almost all SCLCs exhibit inactivation of the tumor suppressor genes RB1 and TP53, along with mutually exclusive gain of a MYC family oncogene including MYC, MYCL, or MYCN (George et al. 2015; Brägelmann et al. 2017; Gazdar et al. 2017). While SCLC has historically been treated as a single disease in the clinic, it is increasingly appreciated to be composed of distinct molecular subtypes defined by expression of lineage-related transcription factor genes ASCL1, NEUROD1, POU2F3, or YAP1 (Rudin et al. 2019; Poirier et al. 2020). Approximately 70% of SCLCs are characterized by high expression of ASCL1 and MYCL, along with high expression of neuroendocrine (NE) markers such as synaptophysin (SYP), ubiquitin C-terminal hydrolase L1 (UCHL1) and chromogranin A (CHGA) (George et al. 2015; Gazdar et al. 2017; Mollaoglu et al. 2017; Rudin et al. 2019). NE-high SCLCs typically display classic morphology with small round blue cells and scant cytoplasm. In contrast, ∼25% of SCLCs are associated with high MYC expression, and these tend to have variant morphology with an ASCL1-low/NE-low gene expression profile and high levels of NEUROD1, POU2F3, or YAP1. We reported the first genetically engineered mouse model (GEMM) of MYC-driven SCLC (Mollaoglu et al. 2017) and demonstrated that MYC promotes the variant, ASCL1-low subtype of SCLC that can express NEUROD1, POU2F3, or YAP1, which is impacted by cell of origin and tumor cell plasticity (Ireland et al. 2020).
ASCL1 and NEUROD1 are lineage-specifying basic helix-loop-helix transcription factors that activate neuroendocrine genes and are required for neural differentiation (Borromeo et al. 2016; Rudin et al. 2019). Genetic knockout studies in the mouse demonstrate that ASCL1 is essential for the development of pulmonary neuroendocrine cells (PNECs) (Ito et al. 2000), and ASCL1 is expressed in the adult lung specifically in PNECs (Borges et al. 1997). Importantly, ASCL1 is necessary for the development of classic SCLC as demonstrated in the Rb1fl/fl;Trp53fl/fl;Rbl2fl/fl (RPR2) mouse model where conditional Ascl1 deletion abolishes tumor formation (Borromeo et al. 2016). NEUROD1 is not required for SCLC initiation or progression in the RPR2 model, consistent with the absence of NEUROD1 expression in classic SCLC models (Borromeo et al. 2016; Mollaoglu et al. 2017). NEUROD1 is expressed in the variant MYC-driven SCLC model and has recently been suggested to temporally follow ASCL1 during MYC-driven subtype evolution (Ireland et al. 2020). However, the function of ASCL1 and NEUROD1 in MYC-driven tumors in vivo has not yet been determined.
PNECs have been generally accepted to be the cell of origin for SCLC, although recent studies have challenged whether they are the only relevant initiating cell population (Huang et al. 2018; Yang et al. 2018; Ferone et al. 2020; Ireland et al. 2020). SCLC shares molecular similarities with normal PNECs, including expression of ASCL1 and its neuroendocrine target genes. Studies in mouse models have demonstrated that PNECs are the most permissive for SCLC development in the original classic Rb1fl/fl;Trp53fl/fl (RP) model and in RPR2 mice (Park et al. 2011; Sutherland et al. 2011). Club cells and alveolar type II (AT2) cells in these studies were relatively resistant to SCLC transformation (Park et al. 2011; Sutherland et al. 2011). More recently, a subset of SCLC was revealed to harbor a tuft cell-like signature with dependency on the tuft cell transcription factor POU2F3 (Huang et al. 2018), suggesting that the tuft cell may serve as a cell of origin for SCLC. Basal cells have also been described as cells of origin for SCLC (Park et al. 2018; Ferone et al. 2020). Together, these studies suggest that lung cells may have broad plasticity for SCLC transformation, but the cell(s) of origin for MYC-driven SCLC have not been investigated.
Here, we used new GEM models to determine the function of ASCL1 in MYC-driven SCLC derived from multiple cells of origin.
Results
MYC-driven SCLC can arise in multiple lung cell types
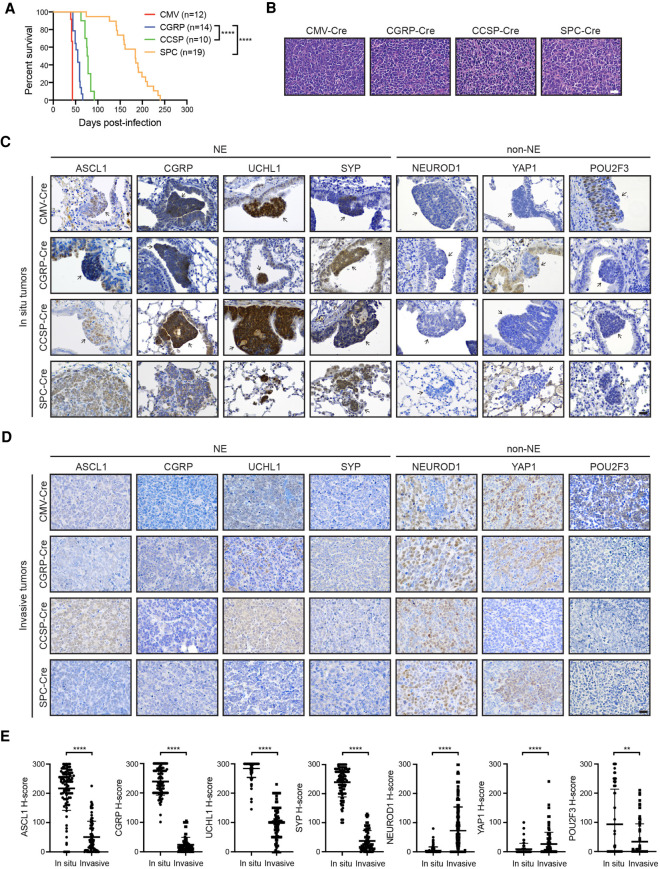
Myc overexpression in the Rb1fl/fl;Trp53fl/fl; MycT58ALSL/LSL (RPM) mouse model promotes the development of a variant, neuroendocrine-low subtype of SCLC that recapitulates features of the human disease (Mollaoglu et al. 2017). To determine which cell types in the normal lung epithelium are capable of initiating tumorigenesis in the RPM model, we intratracheally infected mice with adenoviruses (1 × 108) carrying cell type-specific promoters driving Cre recombinase expression. We used a general Cmv promoter, a neuroendocrine Cgrp promoter, a club cell Ccsp promoter, and an AT2 Spc promoter, whose specificity have been described (Park et al. 2011; Sutherland et al. 2011; Yang et al. 2018; Ferone et al. 2020). The combination of Myc overexpression with Rb1 and Trp53 loss was sufficient to initiate tumorigenesis with all four Cre viruses but with differing tumor latencies and locations in the lung (Fig. 1A,B; Supplemental Fig. S1A; Supplemental Table S1). RPM-CMV tumors had the shortest latency (43 d median survival), followed by RPM-CGRP tumors (55 d median survival). Compared with results from the RP model (Sutherland et al. 2011), targeting of club cells with CCSP-Cre led to tumors with a surprisingly short latency (77 d median survival) and was almost as efficient at tumor initiation as targeting NE cells, suggesting that club cells are highly susceptible to MYC-mediated SCLC transformation. AT2 cells were the most resistant to transformation as RPM tumors initiated with SPC-Cre had the longest latency (184 d median survival) (Fig. 1A). Review by board-certified pathologists (A. Gazdar and B.L. Witt) confirmed that RPM mice infected with different cell type-specific Cre viruses predominantly developed SCLC, with a mixture of classic and variant histopathologies and perilymphatic spread (Fig. 1B; Supplemental Fig. S1A; Supplemental Table S1). CGRP-Cre-initiated RPM tumors were predominantly located in the large airways, while tumors in the CCSP-Cre mice were often located within bronchioles, as well as notable perivascular cuffing; AT2-derived tumors were more commonly found in the alveolar space of the distal lung (Supplemental Fig. S1A). Combined with previous studies in the RP and RPR2 mice using the same viruses (Park et al. 2011; Sutherland et al. 2011; Yang et al. 2018), these results suggest that genetic alterations can dictate the susceptibility of cells to serve as a cell of origin for SCLC.
Figure 1.
MYC-driven SCLC can arise in multiple lung cell types and initially expresses ASCL1. (A) Survival of RPM mice infected with indicated cell type-specific Ad-Cre viruses. Number of mice indicated in the figure. Mantel-Cox log-rank test, (****) P < 0.0001. (B) Representative H&E histology of SCLC from RPM mice initiated with indicated Ad-Cre viruses. Scale bar, 25 µm. (C) Representative immunohistochemistry (IHC) of early in situ tumor lesions for indicated neuroendocrine (NE) or non-NE markers (top) in RPM mice infected with indicated Ad-Cre viruses (left). Arrows indicate in situ tumors. Images shown are from mice collected at approximately the following time points postinfection: CMV 43 d, CGRP 55 d, CCSP 80 d, and SPC 180 d. Scale bar, 25 µm. (D) Representative IHC of large, invasive tumors for indicated neuroendocrine (NE) or non-NE markers (top) in RPM mice infected with indicated Ad-Cre viruses (left). Images shown are from mice collected at approximately the following time points postinfection: CMV 43 d, CGRP 55 d, CCSP 80 d, and SPC 180 d. Scale bar, 25 µm. (E) H-score quantification of IHC in C and D. H-score = percentage positive cells multiplied by intensity score of 0–3 (see the Materials and Methods). Approximately 70–200 tumors from three to 10 mice per condition were quantified. Data are shown as mean ± standard deviation (SD). Mann-Whitney two-tailed t-tests, (**) P < 0.01, (****) P < 0.0001. See also Supplemental Figure S1.
MYC-driven SCLC induces ASCL1 in multiple cells of origin
To further determine whether tumors in this model were neuroendocrine, we examined in situ and invasive tumors for expression of ASCL1 and other NE markers including CGRP, UCHL1, and SYP. Interestingly, in situ tumors from each cohort were dominated by an ASCL1high/NEhigh phenotype even when tumors were initiated in non-NE cells with CCSP-Cre or SPC-Cre and located in regions that should not have ASCL1+ cells (Fig. 1C). NEhigh in situ tumors were proliferative and did not express CCSP or SPC (Supplemental Fig. S1B), suggesting that the original cell fate identities were rapidly lost. Consistent with the classic NEhigh state of these in situ lesions, the majority of in situ RPM tumors expressed low to undetectable levels of NEUROD1, YAP1, and POU2F3 (Fig. 1C), transcription factors associated with NElow SCLC subtypes (Rudin et al. 2019). These results suggest that MYC overexpression in the context of Rb1/Trp53 loss alters the original cellular differentiation state and initially reprograms cells toward a NE-like progenitor.
In contrast to the in situ tumors, invasive RPM tumors were dominated by an ASCL1low/NElow phenotype largely independent of the cell of origin, and tended to express higher levels of NEUROD1 and YAP1 (Fig. 1D,E). Interestingly, tumors initiated in club cells exhibited significantly higher ASCL1 and less YAP1 (Supplemental Fig. S1C), suggesting club cells may not progress as readily as tumors from other cells of origin. Although POU2F3 levels were not significantly elevated in all of the models, we observed an increase in POU2F3+ tumors specifically in the RPM-CMV mice (Supplemental Fig. S1C), consistent with recent studies (Ireland et al. 2020), suggesting that the Cmv promoter may occasionally be targeting a POU2F3+ tuft cell (Huang et al. 2018). Together, this suggests that RPM tumors induce an ASCL1+ NEhigh phenotype in multiple cells of origin that can progress to a NE-low state.
ASCL1 loss delays tumorigenesis and promotes bone and cartilage differentiation
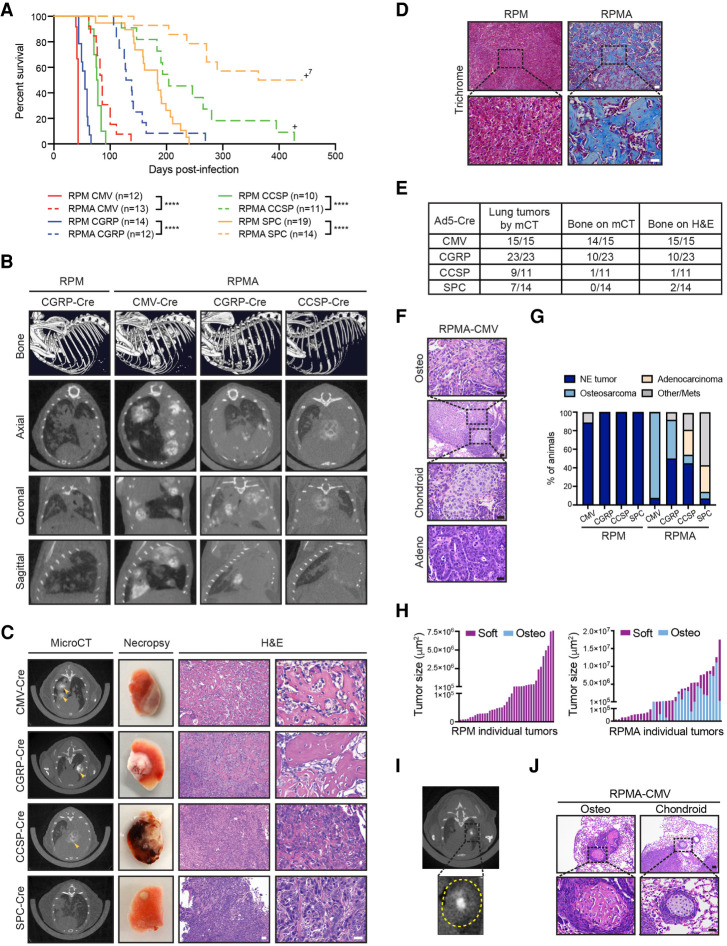
Since ASCL1 has been shown to be required for development of classic SCLC in the RPR2 model (Borromeo et al. 2016), we sought to determine the role of ASCL1 in MYC-driven variant SCLC. We crossed RPM mice to Ascl1-floxed animals to generate Rb1fl/fl;Trp53fl/fl;MycT58ALSL/LSL;Ascl1fl/fl (RPMA) mice and infected RPMA mice with cell type-specific adenoviruses. In contrast to findings in classic SCLC models (Borromeo et al. 2016), RPMA mice developed tumors in the lung, albeit with significantly delayed latencies compared with RPM mice (Fig. 2A). RPMA-CMV mice developed tumors with the shortest median survival of 85 d (2.0-fold longer than RPM-CMV mice), while RPMA mice infected with CGRP-Cre had a median survival of 133 d (2.4-fold longer than RPM-CGRP mice). Median survival in the RPMA-CCSP mice was 204 d (2.6-fold longer than RPM-CCSP mice). RPMA-SPC tumors had the longest latency (median survival 402 d; ∼2.2-fold longer than RPM-SPC mice). Thus, tumor latency differed by cell of origin in both the RPM and RPMA models. Successful Cre-mediated recombination of the Rb1, Trp53, and Ascl1 alleles in these mice was assessed by genomic PCR on individual microdissected RPMA tumors (Supplemental Fig. S2A). We observed recombined Rb1, Trp53, and Ascl1 alleles in ∼94% of tumors. Unrecombined alleles were also detected in the majority of samples, but we cannot rule out that this represents normal tissue contamination in each tumor because it is difficult to cleanly dissect these tumors. We detected tumors frequently in the RPMA-CGRP model (and less frequently in the other models), that expressed ASCL1 protein and did not recombine the Ascl1 alleles and, in these cases, exhibited NE morphology (Supplemental Fig. S2B; Supplemental Table S1).
Figure 2.
ASCL1 loss delays tumorigenesis and promotes bone differentiation in multiple cells of origin. (A) Survival curve comparing RPM (solid lines; data from Fig. 1A) versus RPMA mice (dashed lines) infected with indicated cell type-specific Cre viruses. Number of mice indicated in the figure. (+)Mice censored to end the cohort. Mantel-Cox log-rank test, (****) P < 0.0001. (B, top row) Representative bone analysis of microCT images in RPM versus RPMA mice with advanced lung tumors infected with the indicated Ad-Cre viruses. (Bottom rows) Matched axial, coronal, and sagittal cross-sections of RPM and RPMA lungs used for bone analysis. Images were collected at the following time points postinfection: RPM-CGRP 49 d, RPMA-CMV 111 d, RPMA-CGRP 139 d, and RPMA-CCSP 204 d. (C) Representative microCT axial cross-sections, necropsy images, and H&E staining for tumors from RPMA mice infected with indicated Ad-Cre viruses. Images were from mice collected at the following time points postinfection: CMV 85 d, CGRP 138 d, CCSP 204 d, and SPC 204 d. In microCT panel, yellow arrowheads indicate areas of focal bone formation. Necropsy image shows one whole lung lobe. In H&E panels, scale bar indicates 50 µm in 10× image (left), and 25 µm in the 40× image (right). (D) Representative Trichrome staining in RPM-CMV (43 d postinfection) versus RPMA-CMV (111 d postinfection) tumors. Scale bars: 50 μm for 10× image (top), 25 μm for 40× image (bottom). (E) Table indicating number of RPMA mice per cohort with lung tumors detected by microCT imaging, bone detected by microCT, or bone detected by H&E review. (F) Representative H&E from indicated mouse with adenocarcinoma or osteosarcoma and chondroid differentiation in adjacent tumor areas. Scale bars: 50 μm for 10× image (second image from top), 25 μm for 40× images. (G) Percent of RPM and RPMA mice infected with indicated cell type-specific Cre viruses that succumbed to the indicated tumor histological type. RPMA animals had a mixture of tumor types in each animal but were scored based on the preponderance of tumor type and size. “Other/mets” include animals that were sacrificed without tumors to end the cohort or that succumbed due to lymph node or brain metastases. (H) Size of individual soft tissue tumors and osteosarcomas quantified from PAB-stained slides from a representative RPM-CMV or RPMA-CMV mouse. (I) Representative microCT image from an RPMA-CGRP mouse showing bone density within the center of a soft tissue mass; tumor circled with dashed yellow line in inset. (J) Representative H&E from indicated mouse with osteosarcoma or chondroid differentiation. Scale bar indicates 50 μm for 10× image (top) and 25 μm for 40× image (bottom). See also Supplemental Figure S2, Supplemental Table S1, and Supplemental Movies.
While imaging RPMA mice by microCT, we made the unexpected observation that mice were developing lung tumors with a tissue density consistent with bone (Fig. 2B). Bone analysis by microCT imaging of RPM mice with high lung tumor burden identified only the skeleton (Fig. 2B, top panel; Supplemental Movies). In contrast, bone analysis of RPMA-CMV, RPMA-CGRP, and RPMA-CCSP animals identified both the skeleton as well as multiple bone-like tumors within the lung (Fig. 2B, bottom panels; Supplemental Movies). At necropsy, many RPMA lesions were mineralized and required decalcification prior to sectioning (Fig. 2C). Analysis of H&E-stained tissues by a board-certified pathologist (B.L. Witt) confirmed that RPMA lungs contained high-grade osteosarcoma with well-developed osteoid (Fig. 2C, right panels; Supplemental Fig. S2C). Bone-like lesions in RPMA tumors were confirmed by trichrome (Fig. 2D), Periodic acid–Schiff (PAS) with Alcian blue (PAB) (Supplemental Fig. S2D), and Toluidine blue (Supplemental Fig. S2E) staining.
By microCT imaging, the osteosarcoma-like tumors were frequently observed in RPMA-CMV and RPMA-CGRP mice, less frequent in RPMA-CCSP mice, and not detectable in RPMA-SPC mice (Fig. 2E). However, upon histopathological review, two RPMA-SPC mice did have areas of focal bone formation with osteoid (Fig. 2E). While osteosarcomas were detected in all of the RPMA-CMV mice analyzed, we also frequently observed adenocarcinomas and, more rarely, tumors with chondroid differentiation in these mice (Fig. 2F; Supplemental Table S1). RPMA-CGRP mice more frequently developed SCLC than with other cells of origin, as well as osteosarcomas (Supplemental Table S1). The majority of both in situ and invasive RPMA tumors lacked ASCL1 expression (Supplemental Fig. S2F,G), with the exception of RPMA-CGRP mice, consistent with more SCLC and unrecombined Ascl1 alleles in this model. CCSP- and SPC-RPMA mice developed a mixture of adenocarcinomas, NE tumors (some of which retained ASCL1), and, more rarely, osteosarcomas (Supplemental Table S1). Altogether, in contrast to RPM mice, RPMA mice succumbed to a mixture of osteosarcomas, NE tumors that often retained ASCL1, and adenocarcinomas, whose frequency differed by cell of origin (Fig. 2G). Similar to RPM tumors, in situ RPMA tumors appeared to lack or lose expression of CCSP and SPC early regardless of initial cell of origin (Supplemental Fig. S2F). Consistent with the longer latency and increased frequency of adenocarcinomas and osteosarcomas in RPMA mice (compared with SCLC in RPM mice), RPMA tumors had reduced proliferation as assessed by Ki67 levels compared with RPM tumors (Supplemental Fig. S2H). Some RPMA mice had lymph node metastases, including one animal that appeared to have an osteosarcoma metastasis to the lymph node (Supplemental Fig. S2I).
Interestingly, small RPMA tumors were primarily noncalcified (“soft”), while large invasive RPMA tumors displayed a mix of soft and osteosarcoma phenotypes (Fig. 2H), suggesting that tumors arise in a dedifferentiated state that later adopts a bone phenotype. Consistent with this hypothesis, we often observed bone differentiation arising within the centers of soft tissue tumors by microCT imaging (Fig. 2I). Moreover, osteoid and chondroid differentiation appeared to be surrounded by less differentiated cells (Fig. 2J). Given the differing tumor latencies by cell of origin, we suspect that bone differentiation emerges over time from less differentiated tumors depending on the tumor microenvironment. These data suggest that, in the context of MYC-driven SCLC, ASCL1 represses a latent osteogenic fate and that distinct cells of origin have differing propensities for bone differentiation.
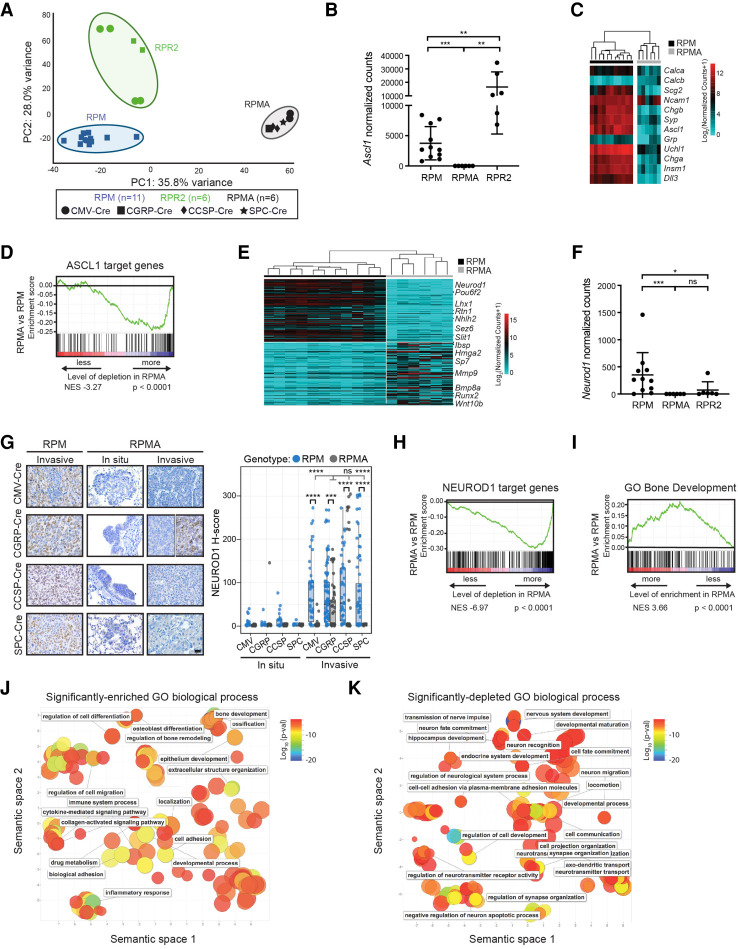
RPMA tumors are transcriptionally distinct with loss of ASCL1 and NEUROD1 target genes
Following the striking observation that loss of ASCL1 in MYC-driven SCLC promotes osteosarcoma formation, we sought to better understand the transcriptional program of RPMA tumors. We performed RNA sequencing (RNA-seq) on variant RPM (n = 11), classic RPR2 (n = 6), and uncalcified RPMA tumors (n = 6). Consistent with their histological differences, principal component analysis of RNA-seq data revealed distinct clustering of all three tumor types (Fig. 3A). We verified that Ascl1 mRNA was significantly depleted in RPMA tumors and established ASCL1 target genes including NE genes were significantly reduced in RPMA compared with RPM tumors (Fig. 3B–D). We performed differential gene expression (DEG) analysis to identify genes and pathways that discriminate the RPMA and RPM tumors. Interestingly, Neurod1 was one of the most significantly down-regulated genes in RPMA versus RPM tumors (Fig. 3E) with complete loss of Neurod1 transcript counts (Fig. 3F). As assessed by IHC, the majority of both in situ and invasive RPMA tumors lacked NEUROD1 protein, in contrast to high levels observed in RPM tumors (Fig. 3G). We did detect NEUROD1 expression particularly in RPMA-CGRP tumors that retained the SCLC phenotype. Gene set enrichment analysis (GSEA) using established human NEUROD1 target genes (Borromeo et al. 2016) combined with NEUROD1 ChIP-seq from mouse RPM tumors revealed a significant depletion of NEUROD1 target genes in RPMA compared with RPM tumors (Fig. 3H). These data reveal that deletion of ASCL1 in RPM tumors during tumor initiation abolishes the NEUROD1+ subtype of SCLC, consistent with recent findings that Ascl1 expression temporally precedes Neurod1 expression during MYC-driven SCLC progression (Ireland et al. 2020).
Figure 3.
RPMA tumors are transcriptionally distinct with loss of ASCL1 and NEUROD1 target genes. (A) Principal component (PC) analysis comparing gene expression by bulk RNA-seq in RPM, RPMA, and RPR2 tumors with Ad-Cre virus indicated in the figure. (B) Ascl1 expression shown as normalized counts by RNA-seq from lung tumors in indicated GEMMs. Mean ± SD. Mann-Whitney two-tailed t-test, (**) P < 0.01, (***) P < 0.0003. (C) Heat map comparing log2 normalized counts for expression of select neuroendocrine genes in RPM versus RPMA tumors. (D) Gene set enrichment analysis (GSEA) in RPMA versus RPM tumors using ASCL1 ChIP-seq target genes from Borromeo et al. (2016). Normalized enrichment score (NES) and P-value indicated in figure. (E) Heat map of top 200 (100 up and 100 down) differentially expressed genes in RPMA versus RPM tumors with select genes indicated. (F) Neurod1 expression as normalized counts by RNA-seq from lung tumors in indicated GEMMs. Mean ± SD. Mann-Whitney two-tailed t-test, (*) P < 0.05, (***) P < 0.001, (ns) not significant. (G) Representative IHC and H-score quantification for NEUROD1 in indicated tumors. Approximately 20–100 tumors were quantified from five to seven mice per condition. Images are from mice collected at approximately the following time points postinfection: RPM-CMV 43 d, RPM-CGRP 45 d, RPM-CCSP 80 d, RPM-SPC 185 d, RPMA-CMV in situ 85 d, RPMA-CMV invasive 95 d, RPMA-CGRP in situ 125 d, RPMA-CGRP invasive 180 d, RPMA-CCSP in situ 265 d, RPMA-CCSP invasive 220 d, RPMA-SPC in situ 290 d, and RPMA-SPC invasive 360 d. Scale bar, 25 µm. RPMA-CGRP IHC panel is split to indicate heterogeneity observed. Data from RPM in situ tumors are also shown in Supplemental Figure S1C. For box plots, the median and interquartile range are shown (top of box is 25th percentile, bottom of box is the 75th percentile). Gray statistics line bars indicate comparisons between RPMA models; black statistics line bars indicate comparisons between RPM and RPMA models initiated with the same virus. Mean ± SD. Mann-Whitney two-tailed t-test, (***) P = 0.0005, (****) P < 0.0001, (ns) not significant. (H) GSEA comparing RPMA versus RPM tumors using NEUROD1 ChIP-seq target genes from RPM tumors (n = 2) and human SCLC cell lines from Borromeo et al. (2016). Normalized enrichment score (NES) and P-value indicated in the figure. (I) GSEA comparing RPMA versus RPM tumor gene expression to a known ossification signature, “GO_Bone_Development.” NES and P-values are indicated in the figure. (J) Scatter plot visualizing semantic similarity of GO biological processes enriched in RPMA versus RPM tumors. (K) Scatter plot visualizing semantic similarity of GO biological processes depleted in RPMA versus RPM tumors.
Consistent with the observed osteoid formation, GSEA demonstrated a significant positive enrichment for bone development genes in RPMA compared with RPM tumors (Fig. 3I). Gene ontology (GO) enrichment analysis revealed that ossification-related processes were significantly up-regulated in RPMA compared with RPM tumors (Fig. 3J). Significantly depleted biological processes in RPMA tumors included those related to neuronal development (Fig. 3K), consistent with the loss of ASCL1 and NEUROD1 in these tumors. Together, these data highlight a critical role for ASCL1 in promoting NE cell fate and repressing an underlying osteosarcoma-like fate in RPM mice.
Network analyses predict transcriptional regulators that drive osteosarcoma cell fate upon ASCL1 loss
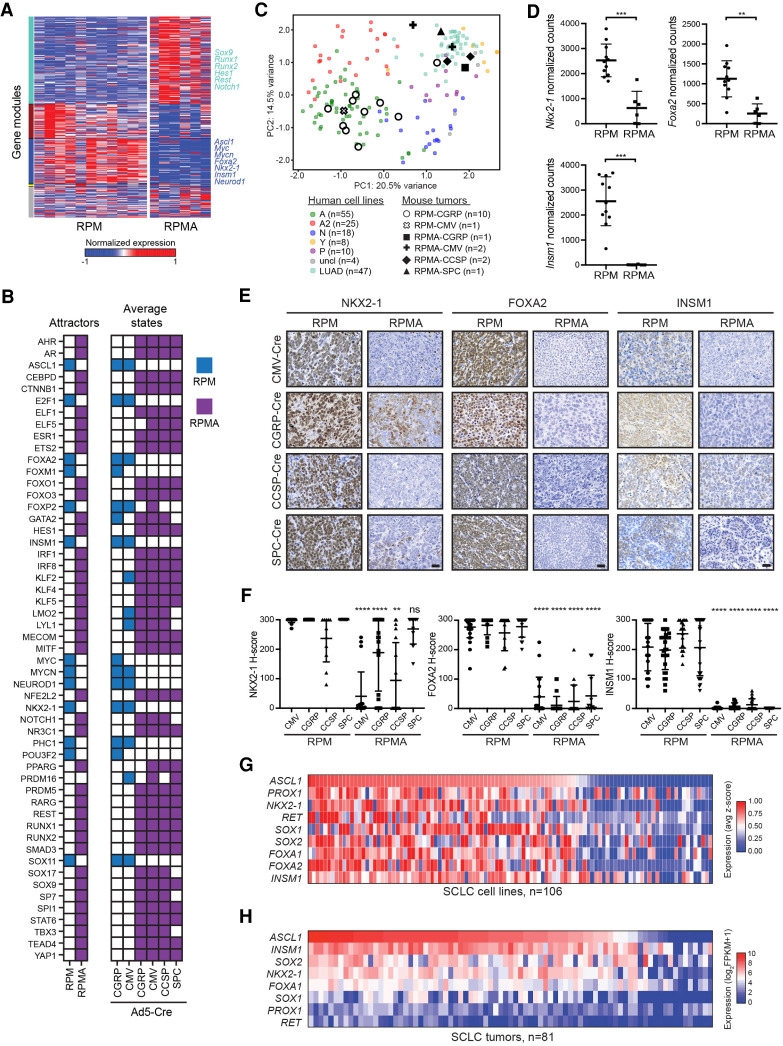
To identify the key transcription factors responsible for this dramatic change in cell fate upon ASCL1 loss, we turned to weighted gene coexpression network analysis (WGCNA). First, we generated a coexpression network of all genes, which allowed the identification of distinct gene modules across all RPM and RPMA samples (Fig. 4A; Supplemental Fig. S3A). Strikingly, approximately one-third of the transcriptome was altered upon ASCL1 loss. To generate a gene regulatory network, we focused on transcription factors predicted by WGCNA to be central to DEG modules, as well as known regulators of lung cancer cell fate. Using BooleaBayes (Wooten et al. 2019), we determined rules of interaction between transcription factors (Supplemental Fig. S3B). For example, ASCL1 is regulated by eight parent nodes (AR, E2F1, HES1, KLF4, MITF, NR3C1, PHC1, and RUNX1) where each ON/OFF combination of these parent nodes determines ASCL1 expression. Likewise, ASCL1 regulates expression of a number of downstream transcription factors. These regulations define how a cell may change its identity or reach a stable phenotype (an “attractor state”). Dynamic simulations identified two attractor states, each corresponding to either RPM or RPMA tumors (Fig. 4B). As described in Wooten et al. (2019), a random walk was used to predict regulators driving these steady states. Satisfyingly, in silico silencing of the ASCL1 node destabilized the RPM attractor, consistent with experimental results (Fig. 4B). Conversely, activation of ASCL1 or NEUROD1 in the RPMA attractor destabilized that steady state, reminiscent of human SCLC, in which ASCL1 is a destabilizer of the nonneuroendocrine subtype (SCLC-Y) (Wooten et al. 2019). By comparing human cell lines (n = 120 total SCLC cell lines from CCLE and cBioPortal, 91 of which are distinct, and n = 47 lung adenocarcinoma cell lines from CCLE) to mouse tumors using PCA, RPMA tumors clustered with the nonneuroendocrine POU2F3 and YAP1 SCLC subtypes and lung adenocarcinoma (Fig. 4C), suggesting a similarity between RPMA tumors and human non-NE tumors.
Figure 4.
Network analyses predict transcriptional regulators that drive osteosarcoma cell fate upon ASCL1 loss. (A) Weighted gene coexpression network analysis (WGCNA) reveals coexpressed gene modules (colored bars on the left Y-axis label) in RPM and RPMA tumors. (B) Binarized average states and state-attractors in RPM (blue) and RPMA (purple) tumors initiated with the indicated Cre viruses. For each gene, colored squares are ON and white squares are OFF. (C) Principal component (PC) analysis comparing bulk RNA-seq expression in human SCLC cell lines (SCLC lines from CCLE and cBioPortal and lung adenocarcinoma [LUAD] cell lines from CCLE) with mouse RPM or RPMA tumors initiated with the indicated viruses. Human SCLC cell lines were classified into subtypes based on high expression of ASCL1 (A and A2 variant), NEUROD1 (N), POU2F3 (P), or YAP1 (Y), or were unclassified (uncl). Mouse tumors were harvested at the following time points postinfection: RPM-CMV 55 d, RPM-CGRP 47–61 d, RPMA-CMV 85–86 d, RPMA-CGRP 111 d, RPMA-CCSP 120–204 d, and RPMA-SPC 204 d. (D) Normalized counts of indicated genes from RNA-seq in indicated GEMMs from C. Data are shown as mean ± SD. Mann-Whitney two-tailed t-test, (***) P < 0.001; (**) P < 0.01. (E) Representative IHC images for indicated antibodies in RPM and RPMA mice infected with cell type-specific Cre viruses. Images were collected from mice at approximately the following time points postinfection: RPM-CMV 43 d, RPM-CGRP 50 d, RPM-CCSP 85 d, RPM-SPC 190 d, RPMA-CMV 95 d, RPMA-CGRP 160 d, RPMA-CCSP 210 d, and RPMA-SPC 310 d. Scale bar, 25 µm. (F) H-Score IHC quantification for indicated proteins in RPM and RPMA tumors. Approximately five to 33 tumors were quantified from n = 3–5 mice per condition. Data are shown as mean ± SD. Mann-Whitney two-tailed t-test, (**) P < 0.01, (****) P < 0.0001, (ns) not significant. (G) Heat map derived from “SCLC-CellMinerCDB” showing relative expression of SCLC transcription factor genes compared with ASCL1. (H) Heat map showing relative expression of SCLC transcription factor genes in human tumors from George et al. (2015) compared with ASCL1. See also Supplemental Figure S3.
Based on these results, we included other known NE-fate specifiers like INSM1 and transcription factors important in endodermal and lung adenocarcinoma fate, such as NKX2-1 and FOXA2, in our network. While all of these genes were significantly reduced in invasive RPMA tumors (Fig. 4D), only FOXA2 was predicted to significantly affect the dynamics of the transcription factor network. We examined protein levels of NKX2-1, FOXA2, and INSM1 in RPM and RPMA tumors. Both RPM and RPMA in situ tumors initially expressed NKX2-1 regardless of cell of origin (Supplemental Fig. S3C), which was lost in invasive osteosarcoma RPMA tumors but less so in the NE tumors and adenocarcinomas (Fig. 4E,F). NKX2-1 is known to be important for adenocarcinoma cell fate, so it is not surprising that the adenocarcinomas maintain NKX2-1. In contrast to RPM in situ tumors, FOXA2 and INSM1 were not highly expressed in RPMA in situ tumors, with the exception of RPMA-CGRP tumors with SCLC morphology that exhibited levels of FOXA2 and INSM1 that were comparable with RPM controls (Supplemental Figs. S2G, S3C). At the invasive stage, NKX2-1, FOXA2, and INSM1 were significantly reduced in RPMA mice compared with RPM (Fig. 4E,F). NFIB, previously implicated as a direct MYC target gene (Mollaoglu et al. 2017), was highly expressed in both in situ and invasive RPMA tumors from all cells of origin, including RPMA osteosarcomas (Supplemental Fig. S3D). Consistently, in human SCLC cell lines from the SCLC-CellMiner database (Tlemsani et al. 2020), ASCL1 expression positively correlated with NKX2-1, FOXA1, FOXA2, and INSM1 (Fig. 4G), with similar results in human SCLC tumors (Fig. 4H). Thus, ASCL1 loss leads to coordinate loss of other key lineage-related transcription factors, which are known to function in the same SCLC superenhancers (Christensen et al. 2014; Borromeo et al. 2016; Pozo et al. 2020).
ASCL1 represses nonendodermal cell fates and Hippo/Notch/Wnt developmental pathway genes in MYC-driven SCLC
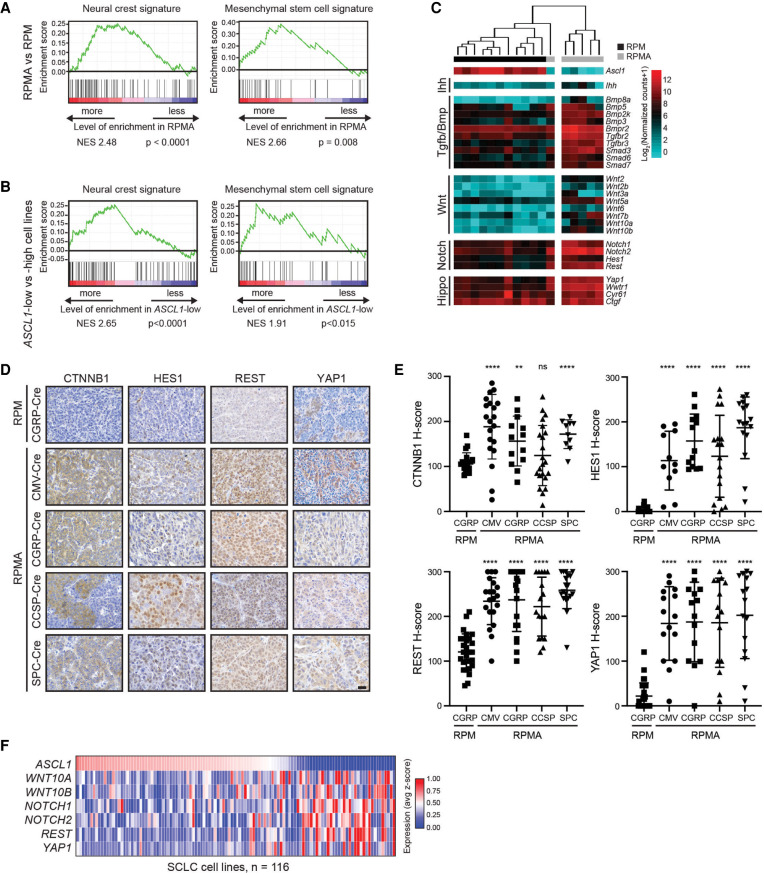
We next focused on understanding the cell fate and predicted regulators gained upon ASCL1 loss. The lung epithelium is believed to derive largely from endoderm, whereas bone and cartilage fates are derived from mesoderm or ectoderm (Serizawa et al. 2019; Zepp and Morrisey 2019). Mesenchymal stem cells (MSCs) from mesodermal tissue and neural crest stem (NCS) cells from ectodermal tissue are known to have the potential to become neurons, bone, or cartilage (Jiang et al. 2002; Achilleos and Trainor 2012). GSEA revealed that both MSC and NC cell signatures were significantly enriched in RPMA versus RPM tumors (Fig. 5A). Consistently, GSEA revealed that both MSC and NC cell signatures were significantly enriched in a panel of 109 human SCLC cell lines when comparing ASCL1-low versus ASCL1-high samples (Fig. 5B). These data suggest that ASCL1 is associated with a more endodermal tumor cell fate and may repress an MSC/NC-stem-like fate.
During development, both MSC and NC-stem-like progenitors have the capacity for bone development given the appropriate developmental signals. Multiple developmental pathways are known to drive bone fate including Indian hedgehog (Ihh), Hippo/Yap1, Transforming growth factor-β (Tgfβ), Bone morphogenic protein (Bmp), Wnt, Notch, and others (Long 2012; Pan et al. 2018; Vanyai et al. 2020). Numerous components of these pathways were transcriptionally up-regulated in RPMA tumors including Ihh, Yap1, Bmp family members (Bmp3, Bmp5, Bmp2k, Bmp8a, and Bmpr2), Tgfβ receptors (Tgfbr2 and Tgfbr3), Smads (Smad3, Smad6, and Smad7), Wnt ligands (Wnt2, Wnt2b, Wnt3a, Wnt5a, Wnt6, Wnt7b, Wnt10a, and Wnt10b), and Notch receptors and target genes (Notch1, Notch2, Hes1, and Rest) (Fig. 5C; Supplemental Fig. S4A). Importantly, network analyses predicted that YAP1, CTNNB1, HES1, NOTCH1, and REST promote the RPMA fate (Fig. 4B). We examined Hippo, Wnt, and Notch pathway proteins for which we could identify good antibodies, including YAP1, CTNNB1 (β-catenin), HES1, and REST (Fig. 5D). Consistent with bioinformatic predictions, YAP1, CTNNB1, HES1, and REST levels were significantly increased in RPMA compared with RPM tumors independent of cell of origin (Fig. 5E). Interestingly, unlike RPM tumors where YAP1 is only detected at the latest stages of progression, a subset of RPMA tumors expressed YAP1 at the in situ stage in all cells of origin, and its expression persisted in invasive tumors and osteosarcomas (Fig. 5E; Supplemental Fig. S4B). Gene expression data from human SCLC cell lines showed an inverse relationship between ASCL1 and select Hippo, Wnt, and Notch pathway genes (Fig. 5F). Together, these data suggest that ASCL1 represses the emergence of an MSC/NC-stem-like state as well as multiple developmental pathway regulators in MYC-driven SCLC.
Figure 5.
ASCL1 represses nonendodermal cell fates and Hippo/Notch/Wnt developmental pathways in MYC-driven SCLC. (A) GSEA in RPMA versus RPM tumors compared with neural crest and mesenchymal stem cell signatures. NES and P-values are indicated in the figure. (B) GSEA in ASCL1-low versus ASCL1-high human cell lines (SCLC-CellMinerCDB, where subtype status is defined by SCLC-CellMinerCDB) compared with neural crest and mesenchymal stem cell signatures. NES and P-values are indicated in the figure. (C) Heat map showing expression of indicated genes in RPM versus RPMA tumors analyzed by RNA-seq for components of the indicated developmental pathways. (D) Representative IHC for indicated antibodies (top) in RPM versus RPMA tumors initiated with the indicated cell type-specific Cre viruses (left). Images were collected from mice at approximately the following time points postinfection: RPM-CGRP, 50 d; RPMA-CMV, 95 d; RPMA-CGRP, 155 d; RPMA-CCSP, 215 d; and RPMA-SPC, 310 d. Scale bar, 25 μm. (E) H-score IHC quantification for indicated antibodies in D. Data are shown as mean ± SD. Approximately 11–28 tumors were used for quantification from three to five animals per condition. Mann-Whitney two-tailed t-test, (**) P < 0.01, (****) P < 0.0001, (ns) not significant. (F) Heat map derived from “SCLC-CellMinerCDB” with expression of selected genes in developmental pathways relative to ASCL1 in human SCLC cell lines. See also Supplemental Figure S4.
ASCL1 represses SOX9 in mouse and human SCLC
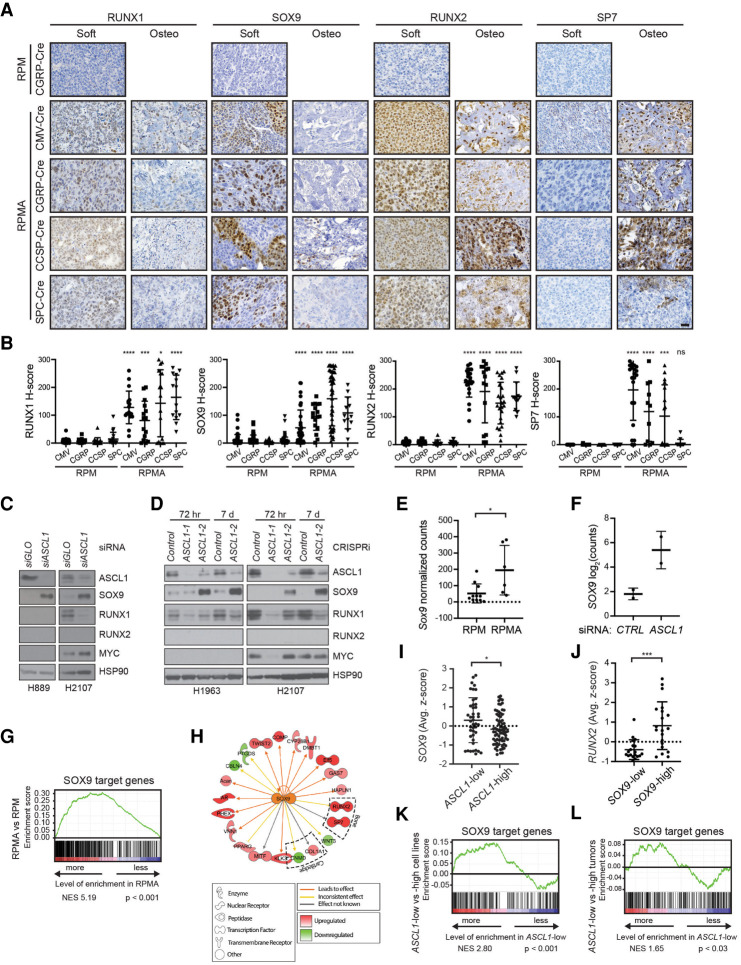
In both the developing limb-bud mesenchyme and neural crest, SOX9 marks osteoblast progenitors and precedes RUNX2 expression and bone differentiation (Long 2012). RUNX1 also precedes RUNX2 expression in developing bone (Lian et al. 2003) and has been suggested to function in chondrogenic lineage commitment of mesenchymal progenitor cells (Smith et al. 2005); RUNX1 can directly regulate multiple genes important for bone development including Sox9 and Runx2 (Soung do et al. 2012; Tang et al. 2020). RUNX2 and SP7 (i.e., Osterix) are required for differentiation of bone-producing osteoblasts, and RUNX2 normally precedes SP7 expression during bone differentiation (Komori et al. 1997; Nakashima et al. 2002; Long 2012). In our network analysis, SOX9, RUNX1, and RUNX2 were predicted drivers of the RPMA attractor state (Fig. 4B). Therefore, we sought to determine whether the MSC/NCS-like tumor cells express these factors upon ASCL1 loss. While RPM tumors rarely expressed SOX9, RUNX1, or RUNX2, all three factors were dramatically increased in RPMA tumors (Fig. 6A,B; Supplemental Fig. S5A). Interestingly, RUNX1 and SOX9 expression were enriched in noncalcified (“soft”) RPMA tumors compared with the more differentiated osteosarcomas. RUNX2 levels were high in soft RPMA tumors and remained high in more differentiated osteosarcomas. In contrast, SP7 was predominantly expressed in the more differentiated osteosarcomas (Fig. 6A). RUNX1 was commonly expressed at the in situ stage in RPMA tumors, while SOX9 and RUNX2 were present in a subset of in situ tumors (Supplemental Fig. S5B). Together, these data are consistent with RUNX1 and SOX9's established roles preceding RUNX2+ and SP7+ differentiation during bone development.
Figure 6.
ASCL1 represses SOX9 in mouse and human SCLC tumor cells. (A) Representative IHC for indicated antibodies (top) in RPM versus RPMA tumors initiated with the indicated cell type-specific Cre viruses (left). RPMA tumors were classified as noncalcified tumors (soft) or osteosarcomas (osteo). Images were collected from mice at approximately the following time points postinfection: RPM-CGRP, 55 d; RPMA-CMV, 85 d; RPMA-CGRP, 150 d; RPMA-CCSP, 215 d; and RPMA-SPC, 310 d. Scale bar, 25 μm. (B) H-score quantification for the indicated proteins in RPM versus RPMA tumors from A. Data are shown as mean ± SD and include both soft and osteo tumors. Approximately 11–85 tumors from three to six mice per condition were quantified. Mann-Whitney two-tailed t-test, (*) P < 0.05, (***) P < 0.001, (****) P < 0.0001, (ns) not significant. (C) Representative immunoblot following 72 h treatment with control (CTRL) or ASCL1 siRNAs in the indicated human SCLC cell lines. HSP90 serves as loading control. (D) Representative immunoblot following stable infection with dCas9-KRAB for CRISPRi-mediated vector control or ASCL1 repression in the indicated human SCLC cell lines at indicated time points. Cells with ASCL1-1 were dying at 72 h and dead by 7 d. HSP90 serves as loading control. (E) Sox9 expression as normalized counts by RNA-seq from lung tumors in indicated GEMMs. Mean ± SD. Two-tailed t-test, (*) P < 0.05. (F) SOX9 expression by RNA-seq from human SCLC cell line NCI-H2107 treated for 72 h with control (CTRL) or ASCL1 siRNAs performed in biological duplicate. Results are reported as log2 normalized counts with mean ± SD. (G) GSEA for SOX9 target genes from Larsimont et al. (2015), in RPMA versus RPM tumors. NES and P-values indicated in the figure. (H) Predicted SOX9 target genes enriched or depleted in RPMA versus RPM tumors by IPA. (I) SOX9 expression in human SCLC cell lines grouped by ASCL1 expression levels in “SCLC-CellMinerCDB.” Data are shown as average z-score ± SD. Mann-Whitney two-tailed t-test, (*) P < 0.05. (J) RUNX2 expression in human SCLC cell lines grouped by SOX9 expression levels in “SCLC-CellMinerCDB.” Data are shown as average z-score ± SD. Mann-Whitney two-tailed t-test, (***) P = 0.0005. (K) GSEA for SOX9 target genes from Larsimont et al. (2015), in ASCL1-low versus ASCL1-high human SCLC cell lines from SCLC-CellMinerCDB, where ASCL1 subtype is defined by SCLC-CellMinerCDB. NES and P-values are indicated in the figure. (L) GSEA for SOX9 target genes from Larsimont et al. (2015), in ASCL1-low versus ASCL1-high human tumors from George et al. (2015). NES and P-values are indicated in the figure. ASCL1 status defined by analysis in Irelend et al. (2020). See also Supplemental Figure S5.
To determine whether ASCL1 repression of SOX9, RUNX1, and/or RUNX2 is conserved in human SCLC cells, we knocked down ASCL1 in human SCLC cell lines using siRNAs or by gene repression using a lentiviral dCas9-KRAB CRISPR interference (“CRISPRi”) approach. SiRNAs were well-tolerated and did not lead to obvious cell death in the 72 h of our assays; CRISPRi led to PARP cleavage during the first 24–72 h following selection but was no longer evident by 3–7 d postselection (Supplemental Fig. S5C). Cells infected with the ASCL1-1 CRISPRi guide died, and we failed to detect most proteins at 72 h (Fig. 6D), but the ASCL1-2 guide led to significant ASCL1 repression that was tolerated for weeks. Importantly, SOX9 was induced upon ASCL1 knockdown in all three cell lines examined regardless of MYC status, while RUNX1 levels decreased in the two cell lines where it was detected (Fig. 6C,D). We did not detect RUNX2 in any of the cell lines examined (Fig. 6C,D). These data suggest that ASCL1 repression of SOX9 is conserved in human cells.
Sox9/SOX9 mRNA is induced in RPMA tumors and human SCLC cell lines transfected with siASCL1 (Fig. 6E,F), respectively, suggesting that ASCL1 represses its expression. Analysis of ASCL1 ChIP-seq data did not identify significant ASCL1 binding sites near SOX9 (Supplemental Fig. S5D,E), suggesting that repression of SOX9 by ASCL1 is likely indirect. Moreover, ASCL1 overexpression was not sufficient to repress SOX9 in three human SCLC cell lines (Supplemental Fig. S5F). GSEA showed that established SOX9 target genes were significantly up-regulated in RPMA compared with RPM tumors (Fig. 6G), suggesting that SOX9 activity is increased in RPMA tumors. Furthermore, Ingenuity Pathway Analysis (IPA) identified SOX9 as a top transcriptional regulator implicated in the transcriptional differences between RPMA and RPM tumors (Fig. 6H; Supplemental Table S3). IPA also predicted other key transcription factor regulators that distinguish RPMA from RPM, such as REST, NKX2-1, CTNNB1, FOXA2, RUNX2, and NOTCH1 (Supplemental Table S3), consistent with changes in their protein levels. In human SCLC cell lines, SOX9 expression was enriched in samples that were ASCL1-low (Fig. 6I), and high SOX9 expression correlated with increased RUNX2 expression (Fig. 6J). GSEA revealed that SOX9 target genes were significantly enriched in ASCL1-low human cell lines (Fig. 6K) and in ASCL1-low human SCLC tumors (Fig. 6L). Together, these data suggest that ASCL1 represses a SOX9+ MSC or NCS-like state that appears to have the capacity for bone or cartilage differentiation, likely dependent on the environmental context.
ASCL1 constrains MYC-driven evolution
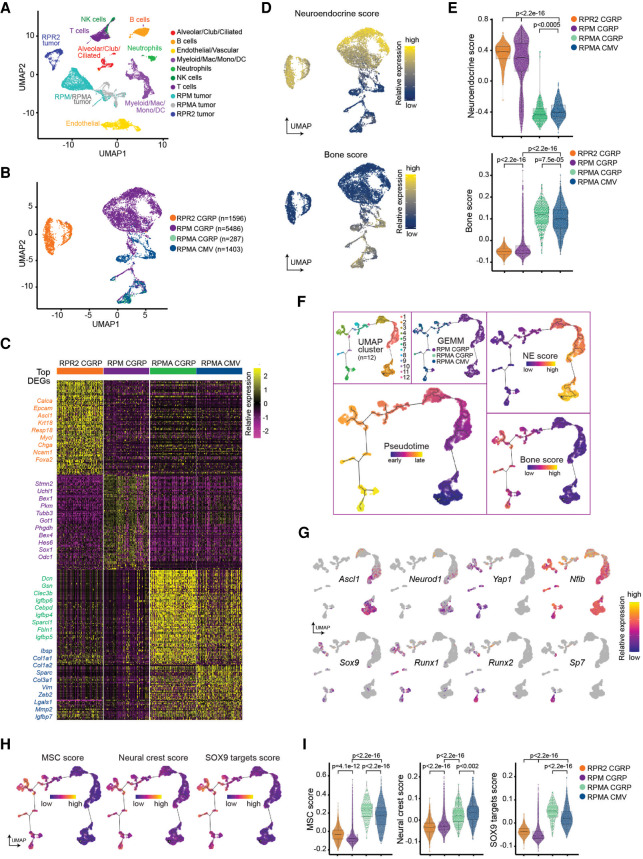
We previously showed using pseudotime-based single-cell RNA sequencing (scRNA-seq) approaches that MYC drives SCLC evolution from a NE-high Ascl1+ to Neurod1+ to Yap1+ non-NE state (Rudin et al. 2019; Ireland et al. 2020). We sought to determine how ASCL1 loss impacts this transcriptional trajectory. To address this, we isolated one Myc-low RPR2-CGRP, five Myc-high RPM-CGRP, one Myc-high, Ascl1 knockout RPMA-CGRP and RPMA-CMV tumor(s) and analyzed samples by scRNA-seq similar to previous methods (Ireland et al. 2020). We performed unbiased clustering and annotated tumor and nontumor cells (immune, endothelial, alveolar, etc.) (Fig. 7A; Supplemental Fig. S6A–C). Then, we reclustered RPR2, RPM, and RPMA tumor cells specifically (Fig. 7B; Supplemental Fig. S6D). Tumor cell clustering was based on the global transcriptome of each cell but independent of cell cycle genes (Supplemental Fig. S6E). RPMA tumor cells clustered distinctly from RPR2 tumors, and more similarly to RPM tumors, but with distinct new populations and signatures (Fig. 7B,C), with both RPMA tumors similar to each other. Consistent with bulk RNA-seq data, RPMA tumor cells have significantly reduced NE scores and significantly increased bone-related transcriptional programs (Fig. 7D,E).
Figure 7.
ASCL1 constrains MYC-driven evolution. (A) Primary RPMA (n = 2), RPR2 (n = 1), and RPM (n = 5) lung tumor and nontumor cell populations captured by scRNA-seq and shown in UMAP space. Predicted cell types were determined based on gene expression in Supplemental Figure S6. (B) Following removal of nontumor and low-quality cells, UMAP of RPM, RPMA, and RPR2 tumor cells only, labeled by genotype and virus used to initiate tumors. Number of cells captured per sample indicated in legend. (C) Heat map of top 100 significant DEGs for each tumor type from B (or fewer if <100 genes were significantly enriched). (Left) Genes of interest are labeled and color-coded by sample. (D) NE score from Zhang et al. (2018) (top) and published bone score (MSigDB “GO_Bone_Development”) (bottom) represented in UMAP space with cells from B. (E) Violin plots of established NE and Bone scores from D applied to scRNA-seq data of tumor samples in B. Student's two-tailed unpaired t test P-values are labeled in the figure. (F) Combined RPM and RPMA tumor cells plotted along a pseudotime trajectory using Monocle 3. (Top left) UMAP clustering along the trajectory. (Top middle) Cells labeled by tumor sample. (Bottom left) Predicted pseudotime ordering where early is dark purple and late is yellow–orange. (Right) Reciprocal expression of NE (top) and Bone scores (bottom) from D over predicted pseudotime. (G) Expression of individual genes in UMAP space as in F. Relative expression levels of each gene per cell are colored based on the panel legend. Cells with no detectable expression are labeled in gray. (H) Single-cell expression scores for a mesenchymal stem cell signature (MSC) (left) and neural crest cell signature (middle) based on MSigDB signatures used for GSEAs in Figure 5. (Right) Single-cell expression score for SOX9 target genes from Larsimont et al. (2015) is also shown. (I) Violin plots of MSC score, Neural crest score, and SOX9 target gene score corresponding with expression maps shown in H. Student's two-tailed unpaired t-test P-values are labeled in the figure. See also Supplemental Figure S6 and Supplemental Table S4.
Next, we constructed pseudotime trajectories to identify predicted transcriptional relationships among the RPM and RPMA tumor cells. Unsupervised pseudotime ordering of the combined tumor cells predicted a trajectory that corresponded with the known MYC-driven evolution from Ascl1+ to Neurod1+ to Yap1+ states—however, RPMA cells progressed farther along this trajectory and occupied multiple new branchpoints (Fig. 7F,G). The most distal branchpoint was occupied only by RPMA cells and was characterized by high Runx2 and Sp7 and a differentiated bone signature (Fig. 7F,G). Consistent with IHC analyses, Yap1, Runx1, and Sox9 appeared to largely precede Runx2 and Sp7 in pseudotime, while Nfib was expressed uniformly throughout the trajectory (Fig. 7G). Finally, MSC, neural crest, and SOX9 target gene signatures were significantly enriched in the latest stages of pseudotime (Fig. 7H,I). Together, these data suggest ASCL1 constrains the transcriptional potential of MYC-driven SCLC evolution to a SOX9+ MSC/NC-stem-like state.
Discussion
The PNEC has been accepted as a major cell of origin for SCLC. This is consistent with multiple similarities between SCLC and PNECs and with the capacity of PNECs for SCLC transformation in mouse models (Park et al. 2011; Sutherland et al. 2011; Song et al. 2012). Our data suggest that specific genetic alterations, namely MYC expression in the context of Rb1 and Trp53 loss, can promote SCLC in other cells of origin, including club and alveolar cells, and potentially other cell types as well. It is possible that Ccsp and Spc are targeting CCSP/SPC-double-positive bronchioalveolar stem cells (BASCs) located at the bronchoalveolar duct junction (BADJ) (Kim et al. 2005), but this warrants further study. While club cells in RP and RPR2 mice were relatively refractory to SCLC development (Park et al. 2011; Sutherland et al. 2011), targeting CCSP+ cells in RPM mice led to SCLC with short latency, comparable with tumor initiation in NE cells. This suggests that club cells may be particularly susceptible to MYC-mediated SCLC transformation. Recent studies reveal that tuft and basal cells may also serve as cells of origin in SCLC (Huang et al. 2018; Park et al. 2018), which could be targeted by CMV in our study; further investigation is needed to determine the spectrum of cells impacted by CMV-Cre. Together with recent findings (Ireland et al. 2020), our data are consistent with the notion that cell of origin, genetic alterations, and tumor cell plasticity can determine SCLC phenotype. Moreover, SCLC diagnosis in the clinic does not necessarily mean that tumors arose in NE cells, and this may need to be considered as we develop predictive models for tumor evolution.
Regardless of the cell of origin, RPM tumors demonstrate NE features at the earliest stages of development, suggesting that NE tumors can arise from non-NE cells. This indicates that multiple differentiated cells in the adult lung have the capacity to dedifferentiate into a NE fate. It is notable, however, that some cell types like AT2 cells appear extremely refractory to transformation even in the context of profound oncogenic changes like Rb1/Trp53/Myc; in contrast, AT2 cells are remarkably sensitive to transformation by MAPK pathway activation, as observed upon expression of mutant EGFR or KRAS in GEMMs. Tumor cell fate plasticity is appreciated in the clinic as it is known that EGFR-driven lung adenocarcinomas (believed to arise in AT2 cells) can convert to SCLC upon resistance to EGFR inhibitors (Oser et al. 2015; Marcoux et al. 2019). A subset of prostate adenocarcinoma that acquires resistance to androgen therapy also converts to a NE fate through transdifferentiation (Terry and Beltran 2014), and these tumors frequently harbor RB1 and TP53 loss. Finally, alteration of RB1, TP53, MYC, BCL2, and AKT in prostate or lung basal epithelial cells in vitro can lead to tumors that resemble NE prostate cancer and SCLC upon xenograft (Park et al. 2018). These observations illustrate the potent capacity of oncogenic changes to alter cell fate, and in particular, how RB1, TP53, and MYC cooperate to promote NE fate.
ASCL1 has been considered as a therapeutic target in SCLC since it is a lineage-specific oncogene that is highly expressed in a significant fraction of tumors (Augustyn et al. 2014). This notion is supported by studies showing that genetic deletion of Ascl1 in classic GEMMs abolishes tumor formation (Borromeo et al. 2016). Here, we find that ASCL1 is not required for MYC-driven tumor development in the RPM model, even though it appears to be required for NE cell fate. Notably, the vast majority of ASCL1+-escaper tumors were from RPMA-CGRP mice, suggesting NE cells are more dependent on ASCL1 than other cells of origin; this is consistent with the fact that NE cells already express ASCL1, while ASCL1 appears to be transiently induced in other cell types upon Rb1/p53/Myc alterations. An important caveat in both studies is that Ascl1 is deleted at the time of tumor initiation. It remains to be tested whether ASCL1 is necessary for the growth of established tumors, with and without MYC expression, which will require the development of more advanced conditional GEMM systems. ASCL1 is lowly expressed in MYC-driven human SCLC (Cardnell et al. 2017; Mollaoglu et al. 2017), suggesting that ASCL1 inhibition may not be sufficient to block the growth of MYC-driven SCLC. Indeed, chemotherapy-relapsed SCLC was found to exhibit significantly reduced ASCL1 expression (Wagner et al. 2018), implying ASCL1 may not be required for SCLC progression. Our data here suggest that loss of ASCL1 could potentially convert SCLC to an alternative cell fate, but fate-tracking approaches will be needed to definitively address this possibility.
We were surprised to find that genetic disruption of Ascl1 led to emergence of an osteosarcoma-like fate. Both RB1 and TP53 alterations are remarkably common in osteosarcomas (RB1, 70%–90%; TP53, 50%–70%) (Berman et al. 2008a; Velletri et al. 2016). In addition, loss of Rb1/Trp53 in mouse osteoblasts or MSCs promotes tumors that highly resemble human osteosarcoma, including high RUNX2 expression (Berman et al. 2008a; Walkley et al. 2008; Calo et al. 2010). During osteogenesis, RUNX2 binds pRB1 and HES1 (Thomas et al. 2001; Lee et al. 2006; Berman et al. 2008b), and we observe up-regulation of HES1 and other indicators of active Notch signaling in RPMA tumors. ASCL1 and NOTCH/REST exhibit a mutually antagonistic relationship in multiple contexts (Masserdotti et al. 2015; Harris and Guillemot 2019). In SCLC, ASCL1 and Notch have an established relationship whereby Notch signaling promotes non-NE fate at least partially through induction of the transcriptional corepressor REST (Lim et al. 2017). We observe induction of NOTCH/REST in RPMA tumors, and we speculate that this event is key to inhibiting NE fate. As Hippo, Notch, Wnt, and other developmental pathways impinge upon RUNX2 to drive bone fate, future studies will be required to elucidate the signals that promote bone differentiation in the context of ASCL1 loss. It is notable that osteosarcomas were most prevalent when an unknown cell type(s) and NE cells were targeted with CMV or CGRP, respectively, whereas adenocarcinomas were found with all cells of origin except from NE cells. Future studies are necessary to decipher how ASCL1 loss impacts these different fates in a cell type-specific fashion.
Our findings suggest that alterations in Rb1, Trp53, and Myc cooperate to dedifferentiate tumor cells to a state that has the potential to be NE in the presence of ASCL1, or bone-like in its absence. One outstanding question prompted by these findings regards the nature of this dedifferentiated cell state; during development, the lung epithelium is believed to arise from endodermal progenitors, whereas bone is derived from mesoderm or ectoderm. Therefore, is there a developmental cell type with NE, bone, and cartilage potential that could be related to SCLC? At least two cell types are known to have the capacity for neural, bone, and cartilage fates during development, MSCs (whose origin is not well understood) and neural crest stem cells, which can give rise to neurons, facial bone, or cartilage (Bruder et al. 1994; Long 2012; Hutchins et al. 2018). Gene expression analyses of RPMA tumors reveal that they harbor MSC and NC stem cell-like gene expression signatures. Other “small, round blue cell” tumors such as Ewing's sarcoma resemble MSCs (Tirode et al. 2007) and neuroblastoma arises from neural crest. The human SCLC H446 cell line, which is MYC-high, has been shown to exhibit multilineage differentiation including neural, bone, and fat cell fates (Zhang et al. 2013). While SCLC was once speculated to arise from neural crest, our data together with previous studies suggest it has the capacity to evolve to a neural crest-like state under specific genetic conditions. While this issue requires further study, it suggests the surprising notion that, in the absence of ASCL1, SCLC can dedifferentiate to a SOX9+ neural crest stem-state that precedes commitment to endodermal lineages.
These findings have led us to question whether SCLC can evolve to a bone-like state in patients. ASCL1 has not been purposely inhibited or knocked out from SCLC tumors in the clinic. In addition, a slow-growing bone phenotype would be under strong negative selective pressure to emerge compared with rapidly growing SCLC, and therefore this should be an extremely uncommon event. There are rare case reports of patients developing carcinoids with ossification (Tsubochi et al. 2013; Özşen et al. 2020) and extraskeletal osteosarcoma (ESOS) in the lung (Qian et al. 2017), including one ESOS following chemotherapy treatment of SCLC (Takamura et al. 2018), but these could represent secondary tumors. Given that ASCL1 repression of SOX9 is conserved, however, it is tempting to speculate that treatments that block ASCL1 potently in the clinic could push tumor evolution to a dedifferentiated NC stem-like state. Further study of chemotherapy-relapsed tissue will be important to further explore this possibility.
Finally, we observed that Neurod1 was one of the most down-regulated genes upon ASCL1 loss in MYC-driven SCLC. This suggests the intriguing possibility that the NEUROD1+ subtype of SCLC requires ASCL1 for development. It has been debated whether the NEUROD1+ SCLC subtype arises in the lung (Borromeo et al. 2016); both our recent data (Ireland et al. 2020) and that shown here indicate that the NEUROD1+ subtype is promoted by MYC but temporally follows ASCL1 expression. There are developmental contexts where ASCL1 expression precedes NEUROD1 such as in the olfactory epithelium and during adult hippocampal neurogenesis (Kim et al. 2011; Schwob et al. 2017). We posit that NEUROD1 expression follows ASCL1 during MYC-driven tumor progression in SCLC. Together, our study suggests that cancer cells demonstrate remarkable plasticity and capacity to evolve to early developmental programs.
Materials and methods
Mice
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Utah and performed in compliance with all relevant ethical regulations. Rb1fl/fl;Trp53fl/fl;MycT58ALSL/LSL (JAX 029971) mice were previously described (Mollaoglu et al. 2017). Rb1fl/fl;Trp53fl/fl; Rbl2fl/fl;Ascl1fl/fl mice were provided by J.E. Johnson (Borromeo et al. 2016). Sperm was collected from mice carrying the conditional Ascl1 allele (Pacary et al. 2011) and used to cross to the RPM strain through in vitro fertilization. Anesthetized mice at 6–8 wk of age were infected by intratracheal instillation (DuPage et al. 2009) with 1 × 108 plaque-forming units of adenovirus carrying Cre recombinase (University of Iowa). Viruses used included Ad-CMV-Cre (VVC-U of Iowa-5), Ad-CGRP-Cre (VVC-Berns-1160), Ad-CCSP/CC10-Cre (VVC-Berns-1166), and Ad-SPC-Cre (VVC-Berns-1168). Viral infections were performed in a biosafety level 2+ room following guidelines from the University of Utah Institutional Biosafety Committee. Male and female mice were divided equally for all experiments. For survival studies, mice were sacrificed due to tumor burden or visibly labored breathing. Mice were euthanized by CO2 asphyxiation followed by necropsy.
MicroCT imaging
Mice were anesthetized with isoflurane and imaged using a small animal Quantum FX or Quantum GX2 microCT (Perkin Elmer). Quantum FX images were acquired with 34-sec scans at 90 kV and 160-µA current, and reconstructed at a 45-µm voxel size. Quantum GX2 images were acquired with 18-sec scans at 90 kV and 88-µA current, and reconstructed at a 90-µm voxel size. For imaging of bone, selected animals were imaged for 2 min on the Quantum GX2 at 90 kV and 88-µA current, and reconstructed at a 90-µm voxel size. Frequency of imaging ranged from twice/week to once/month depending on the tumor growth rate. Resulting images were processed with Analyze 11.0 or 12.0 software (Analyze Direct) as previously described (Mollaoglu et al. 2017). For bone analysis, acquired images were viewed with the Quantum GX2 Simple Viewer.
Immunohistochemistry
At necropsy, lungs were inflated with PBS, fixed overnight in 10% neutral buffered formalin, and then transferred to 70% ethanol. Samples requiring demineralization were incubated in Rapid-Cal Immuno Decal solution (BBC Chemical 6089) prior to paraffin embedding. Tissue was cut in 4-µm sections and stained with H&E to assess tumor pathology or with specific antibodies. Slides were deparaffinized in Citrisolv clearing agent (Fisher Scientific 22-143-975) and rehydrated in an ethanol dilution series. Heat-mediated antigen retrieval was performed in a pressure cooker for 15 min using either 0.01 M citrate buffer (pH 6.0) or 0.01 M TE buffer (pH 9.0), as suggested by the antibody manufacturer. Endogenous peroxidases were blocked by 10-min incubation with 3% H2O2, prior to 30-min blocking in 5% goat serum/PBST (PBS containing 0.2% Tween-20). Primary antibodies were diluted in either 5% goat serum/PBS-T or SignalStain Boost IHC detection reagent (Cell Signaling Technology [CST] 8114S), as recommended by the manufacturer. Slides were incubated in primary antibody overnight at 4°C using the Sequenza coverslip staining system (Thermo Fisher Scientific). The next day, slides were incubated for 30 min at room temperature with VectaStain secondary antibody (Vector Laboratories PK-4001 and BMK-2202), followed by 30-min incubation in ABC reagent and development with DAB (Vector Laboratories SK-4100). Slides were counterstained with hematoxylin and mounted with a Shandon ClearVue coverslipper (Thermo Scientific). Primary antibodies included ASCL1 (1:200; BD Pharmingen 556604), CCSP (1:2000; Millipore Sigma 07623), CGRP (1:250; Sigma-Aldrich C8198), CTNNB1 (1:250; CST 9587S), FOXA2 (1:1000; Abcam ab108422), HES1 (1:600; CST 11988S), KI-67 (1:200; BD Pharmingen 556003), MYC (1:150; Santa Cruz Biotechnology sc-764), NEUROD1 (1:150; Abcam ab109224), NFIB (1:250; Sigma-Aldrich HPA003956), NKX2-1 (1:250; Abcam ab76013), POU2F3 (1:300; Sigma-Aldrich HPA019652-100UL), REST (1:100; Abcam ab202962), RUNX1 (1:250; Proteintech 25315-1-AP), RUNX2 (1:1000; Abcam ab192256), SOX9 (1:1000; Abcam ab185966), SP7 (1:1000; Abcam ab209484), SPC (1:2000; Millipore Sigma AB3786), SYP (1:200; Thermo Fisher Scientific RB1461P1), UCHL1 (1:250; Sigma-Aldrich HPA005993-100UL), INSM1 (1:100; Santa Cruz Biotechnology sc-271408), and YAP1 (1:400; CST 14074S). Images were acquired on a Nikon Ci-L LED microscope with DS-Fi3 camera. Selected slides were digitally scanned with a pannoramic Midi II (3DHISTECH) slidescanner. Staining was manually quantified by H-score on a scale of 0–300 taking into consideration percent positive cells and staining intensity as described (Flowers et al. 1986), where H score = percentage of positive cells multiplied by intensity score of 0–3. For example, a tumor with 80% positive cells with high intensity of 3 = 240 H-score. For SOX9 expression, scanned IHC slides were quantified by H-score using CaseViewer software (3DHISTECH) and an automated nuclear quantification program after manual identification of tumor regions.
PCR for Cre recombination efficiency
Approximately 15 mg of flash-frozen tumor tissue was cut on dry ice and processed with the Qiagen DNeasy kit to isolate tumor genomic DNA. DNA concentrations were measured on a BioTek Synergy HT plate reader. Equal quantities of tumor genomic DNA were amplified by PCR with GoTaq (Promega M7123 and M7832) using the gene-specific primers listed. For Ascl1, Common Fwd (MF1) 5′-CTACTGTCCAAACGCAAAGTGG-3′; WT Rev (MR1) 5′-GCTCCCACAATCCTCGTAAAGA-3′; Flox Rev (VR2) 5′-TAGACGTTGTGGCTGTTGTAGT-3′; and Recombined Fwd (5′ UTR Fwd) 5′-AACTTTCCTCCGGGGCTCGTTTC-3′. PCR conditions were 5 min at 94°C; 30 cycles of 1 min at 94°C, 1.5 min at 64°C, and 1 min at 72°C; and 10 min at 72°C, and then held at 4°C. Expected band sizes were as follows: 5′ UTR Fwd-VR2 (recombined allele) = ∼700–850 bp, MF1-VR2 (floxed allele) = 857 bp, and MF1-MR1 (wild-type allele) = 411 bp.
Primers to detect Rb1 recombination included the following: D1 (5′-GCAGGAGGCAAAAATCCACATAAC-3′), 1lox 5′ (5′-CTCTAGATCCTCTCATTCTTCCC-3′), and 3′ lox (5′-CCTTGACCATAGCCCAGCAC-3′). PCR conditions used were 3 min at 94°C; 30 cycles of 30 sec at 94°C, 1 min at 55°C, and 1.5 min at 72°C; and 5 min at 72°C, and held at 4°C. Expected band sizes were ∼500 bp for the recombined allele and 310 bp for the floxed allele.
Primers to detect Trp53 recombination included the following: A (5′-CACAAAAACAGGTTAAACCCAG-3′), B (5′-AGCACATAGGAGGCAGAGAC-3′), and D (5′-GAAGACAGAAAAGGGGAGGG-3′). PCR conditions were 2 min at 94°C ; 30 cycles of 30 sec at 94°C, 30 sec at 58°C, and 50 sec at 72°C; and 5 min at 72°C, and held at 4°C. Expected band sizes were 612 bp for the recombined allele (A + D) and 370 bp for the floxed allele (A + B). PCR products were run on 1%–2% agarose/TAE gels containing ethidium bromide, and images were acquired using an Azure Biosystem C200 imager.
Mouse tumor RNA-seq
RNA isolation from ∼15 mg of flash-frozen RPM (n = 11), uncalcified RPMA (n = 6), and RPR2 (n = 6) primary tumors was performed using an RNeasy minikit (Qiagen) with the standard protocol. RNA from RPM (n = 11) and RPR2 (n = 2) tumors was subjected to library construction with the Illumina TruSeq stranded mRNA sample preparation kit (RS-122-2101 and RS-122-2102) according to the manufacturer's protocol. RNA from RPR2 (n = 4) and RPMA (n = 6) tumors was subjected to library construction with the Illumina TruSeq stranded total RNA library Ribo-Zero Gold preparation kit (RS-122-2301) according to the manufacturer's protocol. Chemically denatured sequencing libraries (25 pM) from RPM (n = 11) and RPR2 (n = 6) tumors were applied to an Illumina HiSeq v4 single-read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR cluster lit v4-cBot (GD-401-4001). Following transfer of the flow cell to an Illumina HiSeq 2500 instrument (HCSv2.2.38 and RTA v1.18.61), a 50-cycle single-end sequence run was performed using HiSeq SBS kit v4 sequencing reagents (FC-401-4002). Chemically denatured sequencing libraries (1.15 nM) from RPMA tumors (n = 6) were applied to an Illumina NovaSeq S2 flow cell using the standard workflow. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina NovaSeq 6000 S2 reagent kit (20012861). Following transfer of the flow cell to an Illumina NovaSeq 6000 instrument (20012850), a 50-cycle paired-end sequence run was performed.
Bioinformatics
Fastq raw count files were aligned in the R statistical environment (v3.6). The mouse GRCm38 FASTA and GTF files were downloaded from Ensembl release 94, and the reference database was created using STAR version 2.6.1b (Dobin et al. 2013) with splice junctions optimized for 50-bp reads. Optical duplicates were removed using clumpify v38.34, and adapters were trimmed using cutadapt 1.16 (Martin 2011). The trimmed reads were aligned to the reference database using STAR in two pass mode to output a BAM file sorted by coordinates. Mapped reads were assigned to annotated genes in the GTF file using featureCounts version 1.6.3 (Liao et al. 2014). The output files from cutadapt, FastQC, Picard CollectRnaSeqMetrics, STAR, and featureCounts were summarized using MultiQC (Ewels et al. 2016) to check for any sample outliers. To remove sources of unwanted variation from tumor RNA-seq library preparation and sequencing platforms, all noncoding features, histones, and ribosomal RNAs were removed for downstream analyses. The featureCount output files were combined into a single raw count matrix. Differentially expressed genes were identified using a 5% false discovery rate with DESeq2 version 1.24.0 (Love et al. 2014; Supplemental Table S2). These data have been deposited in NCI GEO: GSE155692. PCA was performed on the first two principal components using the regularized log count (rlog) values of the top 500 variable genes. Log2(+1)-transformed, normalized intensity values were obtained to create heat maps with unsupervised hierarchical clustering. A previously described NE marker gene set (Mollaoglu et al. 2017) was used to compare RPMA (n = 6) and RPM (n = 11) tumor expression. Heat maps were generated using Pheatmap version 1.0.12. Semantic similarity-based scatter plot visualizations of enriched and depleted GO biological processes in RPMA versus RPM tumors were created with the REViGO tool using GO biological processes input lists and corresponding P-values (Supek et al. 2011). The scatter plot view visualizes the GO terms in a “semantic space” where the more similar terms are positioned closer together. The color of the bubble and size reflects the log10 P-value obtained in the Panther analysis. Input GO biological processes and log10 P-values were generated using the Panther v14 GO enrichment analysis tool for biological processes (Mi et al. 2019) on enriched (>2.5 log2FC by DESeq2) or depleted (more than −2.5 log2FC by DESeq2) genes in RPMA versus RPM tumors. REViGO and Panther GO enrichment analysis details and curated input gene lists are presented in Supplemental Table S2.
GSEA
Gene set enrichment analysis was performed using GSEA version 3.0 software with default parameters, classic enrichment, inclusion gene set size between 15 and 1000, and the phenotype permutation at 1000 times. GSEA was performed on a preranked gene list representing log2 fold change of RPMA versus RPM tumor bulk RNA-seq expression values (Supplemental Table S2). Normalized enrichment scores and P-values are shown below each respective GSEA plot in the figures. A catalog of functional gene sets from Molecular Signature Database (MSigDB, version 6.2, July 2018, http://www.broad.mit.edu/gsea/msigdb/msigdb_index.html) was used for the “GO bone development,” “mesenchymal stem cell,” and “neural crest signature” gene sets. Gene sets for “ASCL1 targets by ChIP-seq” and “NEUROD1 targets by ChIP-seq” represent conserved transcriptional targets identified by ChIP-seq from mouse SCLC tumor and human cell line SCLC models. The ASCL1 conserved target gene list was previously published (GEO: GSE69398) (Borromeo et al. 2016). The NEUROD1 conserved target gene list was derived from published ChIP-seq data from human SCLC cell lines (GEO: GSE69398) (Borromeo et al. 2016) and NEUROD1 ChIP-seq data from RPM mouse tumors described in this study. The “SOX9 target genes” list was derived from published mouse basal cell carcinoma ChIP-seq data (GEO: GSE68755) (Larsimont et al. 2015). Details of the curated gene sets are presented in Supplemental Table S2.
ASCL1 and NEUROD1 ChIP-seq
Preparation of mouse lung tumors for ChIP-seq was performed as previously described (Chalishazar et al. 2019). Briefly, 20 million cells or ∼250 mg of mouse tumor per ChIP were cross-linked in 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped with 125 mM glycine and nuclei were extracted. Chromatin was sonicated using an Epishear probe sonicator (Active Motif) for 4 min at 40% power. ASCL1 antibody (BD Pharmingen 556604), H3K27Ac antibody (39133, Active Motif), or NEUROD1 antibody (Santa Cruz Biotechnology sc-1084) were used for IP. An input sample of each RPM tumor served as the control. Libraries were sequenced on an Illumina HiSeq 2500 as single-end 50-bp reads to a minimum depth of 35 million reads per sample. Reads were aligned to the mm10 build of the mouse genome with bowtie (Langmead et al. 2009) using the following parameters: -m 1 -t –best -q -S -l 32 -e 80 -n 2. Peaks were called with MACS2 (Zhang et al. 2008) using a P-value cutoff of 10−10 and the mfold parameter bounded between 15 and 100. For visualization, MACS2 produced bedgraphs with the –B and –SPMR options. Binding profiles were visualized using Integrated Genome Viewer (IGV version 2.6.3) aligned to mm10 genome build. ASCL1 and NEUROD1 mouse ChIP-seq data are available on GEO: GSE155692. H3k27Ac mouse ChIP-seq data are available on GEO GSE142496 (Chalishazar et al. 2019).
Human ChIP-seq for ASCL1 was performed as described in Borromeo et al. (2016), and data are published in GEO with accession number GEO GSE69398. Binding profiles of human ASCL1 ChIP-seq were visualized using Integrated Genome Viewer (IGV version 2.6.3) aligned to hg19 human genome build.
Weighted gene coexpression network analysis
WGCNA (Langfelder and Horvath 2008) was performed on RPM (n = 11) and RPMA (n = 6) tumor RNA seq data using the R package “WGCNA.” Low variance genes, with a coefficient of variation <0.2 and median absolute deviation <1, were removed, resulting in 15,286 genes across 17 samples. A signed network of genes, such that only positively correlated genes were grouped into gene modules (negative correlations are given a score of 0), was constructed as follows. WGCNA's pickSoftThreshold function was used to determine the exponent of the correlation coefficients matrix that best produces a scale free network when the matrix is used as weights of network connections. A threshold of 15 was chosen to produce this weights matrix, with the correlation function “Pearson.” A topological overlap matrix was then generated based on the overlap in connections for two genes. This gave a measure of distance between each pair of genes, which was used in average-linkage hierarchical clustering. The “cutTreeDynamic” function was used, with minimum cluster size of 100 to generate clusters, or modules, of coexpressed genes. WGCNA's “sampledBlockwiseModules” was then used to increase robustness of gene modules, which merges or removes genes from modules that are not distinct enough in a majority of the resampled module labels. Differentially expressed gene modules were determined using an ANOVA statistical test between RPM and RPMA tumors. Three gene modules were significant.
Network structure inference
Network structure was inferred as described in Wooten et al. (2019). Briefly, transcription factors were filtered to include those differentially expressed between RPM and RPMA tumors with log fold change >1.5 and adjusted P-value <0.05. Genes were then filtered further to include only those most central to each differentially expressed gene module, calculated as a module membership value (kME) from the WGCNA analysis. Last, expert knowledge was used to add any transcription factors that were deemed central to RPM/RPMA cell identity, including FOXA2, SOX9, and CTNNB1. Network connections were added using the Enrichr tool from the Ma'ayan laboratory (Chen et al. 2013; Kuleshov et al. 2016), which mines various ChIP-seq databases for TF-gene interactions including CHEA, ENCODE, and TRANSFAC. Gene nodes with no downstream nodes (child nodes) were removed, as they do not affect the dynamics of the network, except NKX2-1, INSM1, and SP7, which were kept as biomarkers.
For principal component analysis (PCA) comparing human SCLC and LUAD cell lines with RPM and RPMA GEMM tumors, reduction was based on 35 genes from the human SCLC phenotype network described previously (Wooten et al. 2019). PCA is only fit to human SCLC cell line data. GEMM tumor expression and LUAD cell line expression values were normalized and the human SCLC network-TF expression levels for these samples were binarized to allow inclusion of the samples in the human SCLC PCA projection. Included in the analysis were n = 120 total SCLC cell lines from CCLE and cBioPortal, n = 47 lung adenocarcinoma (LUAD) cell lines from CCLE, n = 11 RPM tumors, and n = 6 RPMA tumors.
Network rule inference and attractors
Network rules of interaction were calculated using BooleaBayes as previously described (Wooten et al. 2019), using the 11 RPM and six RPMA samples. For each TF node in the network, a rule of expression was determined for any ON/OFF combination of parent nodes. BooleaBayes simulations consisted of asynchronous directed random walks, with probabilities of transition between states determined by TF rules. By simulating the network starting at each sampled state, we found pseudoattractors (referred to here as “attractors”) as previously defined (Wooten et al. 2019). Searches for attractor states from each initial condition were cut-off at a TF-basin radius of four steps, and the phenotypes of any attractors were determined by distance to initial condition. Two attractors were found using this method, one for each phenotype. Driving transcription factors for each attractor, or those whose silencing has a destabilizing effect on the attractor, were found using in silico perturbations (Wooten et al. 2019).
ASCL1 knockdown and immunoblot
Human classic SCLC cell lines NCI-H2107, NCI-H889, and NCI-H1963 were obtained from ATCC. Cells were transfected with human ASCL1 stealth siRNA (Thermo Fisher 1299001-HSS100744) using X-tremeGENE siRNA transfection reagent (Sigma 4476115001) according to the manufacturer's protocol. Cells were harvested 72 h after transfection for analysis by immunoblot. Cell pellets were flash-frozen and stored at −80°C until use. For lentiviral CRISPR-mediated repression, the following guideRNAs targeting ASCL1 were cloned into pLV-hU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro (Addgene 71236): ASCL1-1 (5′-TAGGAGAGGAACGCGAGACG-3′) and ASCL1-2 (5′-CGGGAGAAAGGAACGGGAGG-3′). Sequence-verified plasmids were used to generate high-titer lentivirus. Cells (1 × 106/well) in six-well plates were infected with virus in media containing 8 µg/mL polybrene and 12 mM HEPES by spinoculation. Plates were centrifuged at 900g for 75 min at 37°C. Cells were cultured in viral media for 2 d before resuspending in normal growth media and adding puromycin for selection. Cell pellets were collected at the indicated time points and stored at −80°C. Total protein lysates were prepared as previously described, separated via SDS-PAGE, and transferred to a PVDF membrane (Oliver et al. 2011). Membranes were blocked for 1 h in 5% milk followed by overnight incubation with primary antibodies at 4°C. Membranes were washed for four times for 10 min at room temperature in TBS-T. Mouse and rabbit HRP-conjugated secondary antibodies (1:10,000; Jackson ImmunoResearch 711-035-152 and 115-035-205) were incubated for 1 h in 5% milk at room temperature followed by washing four times for 10 min at room temperature in TBS-T. Membranes were exposed to WesternBright HRP quantum substrate (Advansta) and detected on Hyblot CL film (Denville Scientific, Inc.). Primary antibodies included ASCL1 (1:300; BD Pharmingen 556604), SOX9 (1:1000; Abcam ab185966), RUNX1 (1:1000; Proteintech 25315-1-AP), RUNX2 (1:1000; CST 12556), MYC/NMYC (1:1000; CST 13987), and PARP (1:1000; CST 9532S), with HSP90 (1:5000; CST 4877) or ACTIN (1:10,000; Sigma-Aldrich A2066) as a loading control.
ASCL1 overexpression and virus production
Human ASCL1 cDNA was cloned into a lentiviral Tet-On plasmid (Addgene plasmid 110280) with Tet-On-GFP as a control. Tet-On-ASCL1 and GFP were confirmed by direct sequencing. For generation of high-titer virus, HEK-293T cells were transfected with a three-plasmid system including: Tet-On-GFP or Tet-On-ASCL1, pCMV-VSVG, (Addgene 8454), and pCMVδ R8.2 (Addgene 8455). Viruses were harvested at 48 and 72 h posttransfection, concentrated by ultracentrifugation at 24,000g for 1.45 h and stored at −80°C until use. Human cell lines (H524, H526, and GLC1) were infected via spinoculation at 900g for 60 min at 37°C. During spinoculation, 0.5 million to 1 million cells per well of a six-well plate were cultured with 2 mL of RPMI, 25 μL of HEPES buffer (Thermo Fisher 15630080), 8 μg/mL polybrene (Santa Cruz Biotechnology sc-134220), and 25 μL of lentiviral Tet-On-GFP or Tet-On-ASCL1 with titer >106 infectious units per milliliter. Cells were selected with 2.5 μg/mL blasticidin (VWR 71002-676) until all uninfected cells treated in parallel were killed. Following this period, selection was maintained by culturing cells in media plus blasticidin at a concentration of 1 μg/mL. Cells were treated with or without 0.5 μM doxycycline every 48 h and collected at indicated time points for expression analysis by immunoblot.
scRNA-seq sample and library preparataion
One RPMA CMV-Cre infected tumor and one RPMA CGRP-Cre infected tumor at early time points of growth as determined by microCT were digested into a single-cell suspension in preparation for scRNA-seq. Additionally, one RPR2 tumor initiated with CGRP-Cre and one early RPM tumor initiated with CGRP-Cre were collected. Each tumor containing surrounding lung tissue up to ∼200 mg was mechanically dissociated with scissors and digested to a single-cell suspension using 1000 uL of an enzymatic digestion cocktail per sample for 30 min at 37°C. The digestion cocktail consists of 4200 μL of HBSS-free (Thermo Fisher 14175), 600 μL of trypsin-EDTA (0.25%) (Thermo Fisher 25200-072), 600 μL of collagenase type 4 (Worthington Biochemical LS004186) from 10 mg/mL stock prepared in HBSS with calcium and magnesium (Thermo Fisher 14025), and 600 μL of dispase (Worthington Biochemical LS02104). Enzymatic digestion was quenched on ice with 500 μL of quench media containing 7.2 mL of Leibovitz's L15 media (Thermo Fisher 11415-064), 800 μL of FBS (Sigma 12303C), and 30 μL of DNase (Sigma D4527) at 5 mg/mL in HBSS-free media per milliliter of digestion media. Tissue was further dissociated by passing through a 16-gauge syringe to the point of no clogging. The tissue suspension was then passed through a 100-μm cell strainer. Cells were spun at 2000 rpm for 5 min. Supernatant was removed and replaced with 500 μL of ACK (ammonium-chloride-potassium) lysis buffer per sample to remove red blood cell contamination (3-min incubation at 37°C; Thermo Fisher A10492). Reaction was quenched with 10 mL of cold 1× PBS. Cells were spun at 1500 rpm for 5 min and resuspended in 1 mL of cold 0.4% BSA in 1× PBS. Cells were washed twice in cold 0.4% BSA in 1× PBS and submitted at a concentration of 1 × 106 cells/mL for scRNA-seq library preparation.
Single-cell suspensions were further prepared for sequencing according to 10x Chromium platform protocols (https://support.10xgenomics.com/single-cell-gene-expression). In brief, for the RPMA and RPM tumor samples, the Chromium single-cell gene expression solution with 3′ chemistry, version 3 (PN-1000075), was used to barcode individual cells with 16-bp 10x barcodes and to tag cell-specific transcript molecules with 10-bp unique molecular identifiers (UMIs) according to the manufacturer's instructions. The RPR2 tumor was prepared with 2′ chemistry, version 2. The following protocol was performed at the High-Throughput Genomics Shared Resources at Huntsman Cancer Institute, University of Utah. Viability and cell count were assessed and suspensions were equilibrated to targeted cell recovery of 8000 cells (RPMA-CMV, RPM, and RPR2) and 3600 cells (RPMA-CGRP) due to lower total cell number. 10x gel beads and reverse transcription reagents were added and cell suspensions were loaded to the Chromium single-cell controller (PN-120263) to form GEMs—the microdroplets. Within individual GEMs, cDNA generated from captured and barcoded mRNA was synthesized by reverse transcription at the setting of 45 min at 53°C followed by 5 min at 85°C. Subsequent A tailing, end repair, adaptor ligation, and sample indexing were performed in bulk according to the manufacturer's instructions. The resulting barcoding libraries were assessed on Agilent D1000 ScreenTape on an Agilent Technology 2200 TapeStation system and quantified by quantitative PCR using a KAPA Biosystems library quantification kit for Illumina Platforms (KK4842). Multiple libraries were then normalized and sequenced on NovaSeq 6000 with 2x150 paired-end (PE) mode aiming for ∼200 million reads per sample.
10x scRNA-seq data processing
Demultiplexing and data alignment
scRNA-seq data from the RPMA and RPM tumor samples were demultiplexed with 10x cellranger mkfastq version 3.1.0 to create fastq files. The RPR2 sample was demultiplexed with 10x cellranger mkfastq version 3.0.0. For each sample, reads were aligned to the mouse genome (mm10 custom EGFP + Cas9 reference v3.0.0) and count barcodes and UMIs were generated using cellranger count 3.1.0 with expected cells set to 8000 per library. QC reporting, clustering, dimension reduction, and differential gene expression analysis using default parameter inputs (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/reanalyze) were performed for primary data evaluation in 10x Genomics’ Cell Loupe Browser (v5.0). Primary QC reporting on the RPMA-CMV tumor revealed capture of 8363 cells total with 32,584 mean reads per cell and 1206 median genes per cell or ∼273 million reads total. Primary QC reporting on the RPMA-CGRP tumor revealed capture of 2902 cells total with 87,252 mean reads per cell and 1505 median genes per cell or ∼253 million reads total. Primary QC reporting on the early RPM tumor revealed capture of 6578 total cells with 37,955 mean reads per cell and 1474 median genes per cell or ∼250 million reads total. Last, primary QC reporting on the RPR2 tumor revealed capture of 4149 total cells with 60,671 mean reads per cell and 2397 median genes per cell or ∼252 million reads total. For further details of the primary Cell Ranger data processing, see https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/algorithms/overview.
QC, clustering, and cell type identification
All QC, clustering, and cell type identification was performed using common Seurat workflows (Butler et al. 2018; Stuart et al. 2019). All cells sequenced from the early RPM, RPMA-CMV, and RPMA-CGRP samples were read into a sparse matrix in the R statistical computing environment using the Read10X function and converted directly into Seurat objects using the CreateSeuratObject function. RPR2 preidentified tumor cells only (identified based on NE gene expression in the 10X Loupe browser) were likewise called into R and converted into a Seurat object (a list of identified tumor cell barcodes for the RPR2 sample can be found in Supplemental Table S4). Additionally, all cells from previously published scRNA-seq data of four RPM tumors (Ireland et al. 2020) were pulled into R and converted into a Seurat object. Next, all cells from the n = 5 RPM and n = 2 RPMA samples and tumor cells from the n = 1 RPR2 sample were merged into a single Seurat object and subject to QC using a standard Seurat QC workflow. Before QC, there were 18,033 cells from RPM samples, 11,265 cells from the RPMA samples, and 1639 tumor cells from the RPR2 sample. Cells meeting quality standards (nFeature_RNA > 500 and nFeature_RNA < 7500, and percent mitochondrial content <20%) were subsetted for downstream normalization, scaling, and clustering. Cells remaining per sample following QC were as follows: RPM = 13,844 cells, RPMA = 7908 cells, and RPR2 = 1636 cells.
Counts of cells passing QC were then log-normalized and scaled based on all genes with Seurat functions NormalizeData and ScaleData. Linear dimension reduction by PCA was performed and nonlinear dimension reduction and clustering occurred based on the top 25 dimensions using Seurat's RunUMAP function (UMAP shown in Fig. 7A; Supplemental Fig. S6A,C). Each cluster was examined for enrichment of cell type-specific markers to identify cell clusters using the DotPlot function in Seurat (Supplemental Fig. S6B). Additionally, top DEGs per cluster (listed in Supplemental Table S4) were examined to ensure correct cell type identification. Final calling of cell types per cluster for all cells sequenced is shown in Figure 7A and Supplemental Figure S6, A and B, and suspected tumor cell populations are highlighted. Cell barcodes with corresponding cell type can be found in Supplemental Table S4. Next, to perform subsequent analyses on tumor cells only, suspected RPM, RPMA, and RPR2 tumor cells (clusters 22, 20, 18, 16, 7, 2, 4, 8, 10, 17, and 21 from Supplemental Fig. S6A) were subsetted and reclustered in Seurat as demonstrated in Supplemental Figure S6D following cell cycle phase assignment and regression of cell cycle effects using Seurat's CellCycleScoring and ScaleData's command, vars.to.regress, so clustering would not depend on cell cycle phase. Cell cycle genes were derived from Tirosh et al. (2016). Reclustering of suspected tumor cells based on the top 35 dimensions revealed 23 clusters (Supplemental Fig. S6D, middle) and thus allowed higher-resolution analysis of suspected tumor cell populations. Based on the significant DEGs in the 23 suspected tumor cell clusters (log2FC > 0.3), three out of 23 clusters were excluded based on enrichment of cardiac muscle (cluster 22), T-cell (cluster 20), and B-cell/immune (cluster 18) marker expression; these three clusters also occupied distinct transcriptional space from other suspected tumor cells in UMAP (Supplemental Fig. S6D, middle) and comprised cells from multiple genotypes, and therefore are unlikely to be unique RPMA derivatives. Thus, for all subsequent tumor cell-specific analyses, these three clusters were excluded (top DEGS for these excluded clusters are included in Supplemental Table S4). The remaining 20 clusters were subsetted and reclustered based on the top 20 dimensions, revealing n = 19 clusters, independent of cell cycle phase, for downstream analyses (Supplemental Fig. S6D [bottom UMAP],S6E). Cell barcodes identified as tumor cells and used for downstream analyses are listed in Supplemental Table S4.
DEG analyses and score assignments
The heat map of DEGs per tumor type in Figure 7C was created with Seurat's DoHeatmap function and represents, at most, the top 100 most significantly enriched marker genes per sample determined by a log2FC cutoff of 0.25. The complete lists of top significant DEGs per sample in Figure 7C is included in Supplemental Table S4.
NE score for cells of each scRNA-seq sample was determined based on Spearman correlation with an established 50-gene NE versus non-NE expression vector derived from Zhang et al. (2018), where 41 NE and 87 non-NE human cell lines were used to identify a core 50-gene signature comprised of 25 NE genes and 25 non-NE genes that robustly predict NE phenotype. Bone score was assigned per cell of each scRNA-seq sample using Seurat's AddModuleScore function and mouse homologs of the published gene signature “Go_Bone_Development” on MSigDB (both NE and bone signature genes are included in Supplemental Table S4). The Scater package was used to visualize violin plots of NE and Bone scores in the R statistical computing environment.
Pseudotime with Monocle3
Tumor cells identified above from n = 5 RPM and n = 2 RPMA scRNA-seq samples were subsetted into a new Seurat object and converted into a cell data set (CDS) for pseudotime analysis in Monocle3 (Trapnell et al. 2014; Qiu et al. 2017). The CDS was preprocessed with 10 dimensions and reduced with reduction_method set to UMAP. Cells were clustered with resolution set at 1 × 10−5, which revealed n = 12 transcriptionally similar clusters in UMAP space (Fig. 7F). Monocle3's learn_graph was used based on partition assignment. Cells were then ordered in predicted time or pseudotime with the root node selected as the first node with highest NE gene expression (based on published biological knowledge demonstrating SCLC emerges as NE-high and progresses to NE-low in the MYC-driven GEMM) (Ireland et al. 2020) as shown in Figure 7F. NE score and Bone score previously assigned were also visualized along the trajectory. Additional signatures including “MSC score,” “Neural crest score,” and “SOX9 targets score” were assigned via Seurat's AddModuleScore function. Gene lists comprising these scores are the same used in corresponding GSEAs throughout the study. Genes comprising each of these scores can be found in Supplemental Table S4.
Statistics
Statistical analysis was performed with GraphPad Prism 8. Mouse survival studies were analyzed with Mantel-Cox log-rank tests. Immunohistochemistry quantification and normalized RNA-seq counts were compared by nonparametric two-tailed Mann-Whitney t-tests. Measurements were taken from distinct tumors in multiple mice for each experiment, as indicated in the figure legend. P-values < 0.05 were considered statistically significant. Error bars represent mean ± standard deviation unless otherwise specified.
Resource availability
Materials availability
The RPM mice used in this study are deposited at The Jackson Laboratory, JAX 029971. RPMA mice are available upon request with an MTA.
Data and code availability
All software is commercially available or cited in previous publications. Mouse RNA-seq, ChIP-seq, and scRNA-seq data unique to this study are deposited in NCBI GEO: GSE155692. scRNA-seq raw data of n = 4 RPM invasive tumors used in this study were previously deposited in NCBI GEO: GSE149180. All code for scRNA-seq analyses follows standard and publicly available Seurat and Monocle3 workflows. R scripts for this study are available upon reasonable request. Additional raw data for Figures 3–7 are provided in Supplemental Tables S2–S4.
Supplementary Material
Acknowledgments
We appreciate Dr. Anton Berns for permission to use AdCre viruses deposited at University of Iowa. We are grateful to the late Dr. Adi Gazdar, who provided insightful pathological observations for this study. We thank R. Dahlgren and L. Houston for administrative assistance and members of the Oliver lab, especially D. Hansen, P. Ballieu, and A. Micinski for mouse colony management. We acknowledge support from the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award P30CA042014 to Huntsman Cancer Institute for the use of core facilities and shared resources, including Biorepository and Molecular Pathology, High-Throughput Genomics and Bioinformatic Analysis, and DNA Sequencing. This work was supported by the NIH/NCI (R21-CA216504-01A1, U01-CA231844, U01-CA213338, and U24-CA213274).
Author contributions: R.R.O. and T.G.O. conceived the study. R.R.O., D.W.K., A.S.I., K.P., D.P.K., K.B.S., S.J.W., M.R.G., C.P.W., and D.S. acquired the data. R.R.O., D.W.K., A.S.I., K.B.S., S.M.G., B.L.W., V.Q., and T.G.O. analyzed and interpreted the data. R.R.O., A.S.I., and T.G.O. wrote the original draft of the manuscript. R.R.O., A.S.I., S.M.G., V.Q., J.E.J., and T.G.O. reviewed and edited the manuscript. T.G.O. acquired the funding. J.E.J. acquired the resources. V.Q., J.E.J., and T.G.O. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348295.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
T.G.O. has pending patent applications related to subtype stratification of SCLC: US16/335368; JP2019522392, and EP2017865057.
References
- Achilleos A, Trainor PA. 2012. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res 22: 288–304. 10.1038/cr.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, Lee V, Tan C, Sullivan JP, Larsen JE, et al. 2014. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc Natl Acad Sci 111: 14788–14793. 10.1073/pnas.1410419111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, et al. 2008a. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci 105: 11851–11856. 10.1073/pnas.0805462105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. 2008b. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res 6: 1440–1451. 10.1158/1541-7786.MCR-08-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, Baylin SB, Ball DW. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386: 852–855. 10.1038/386852a0 [DOI] [PubMed] [Google Scholar]
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, et al. 2016. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 16: 1259–1272. 10.1016/j.celrep.2016.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brägelmann J, Böhm S, Guthrie MR, Mollaoglu G, Oliver TG, Sos ML. 2017. Family matters: how MYC family oncogenes impact small cell lung cancer. Cell Cycle 16: 1489–1498. 10.1080/15384101.2017.1339849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder SP, Fink DJ, Caplan AI. 1994. Mesenchymal stem cells in bone development, bone repair, and skeletal regeneration therapy. J Cell Biochem 56: 283–294. 10.1002/jcb.240560303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. 2010. Rb regulates fate choice and lineage commitment in vivo. Nature 466: 1110–1114. 10.1038/nature09264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, Ireland AS, Guthrie MR, Bheddah S, Banerjee U, et al. 2017. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 8: 73419–73432. 10.18632/oncotarget.20621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalishazar MD, Wait SJ, Huang F, Ireland AS, Mukhopadhyay A, Lee Y, Schuman SS, Guthrie MR, Berrett KC, Vahrenkamp JM, et al. 2019. MYC-driven small-cell lung cancer is metabolically distinct and vulnerable to arginine depletion. Clin Cancer Res 25: 5107–5121. 10.1158/1078-0432.CCR-18-4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. 2014. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26: 909–922. 10.1016/j.ccell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072. 10.1038/nprot.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Song JY, Krijgsman O, van der Vliet J, Cozijnsen M, Semenova EA, Adams DJ, Peeper D, Berns A. 2020. FGFR1 oncogenic activation reveals an alternative cell of origin of SCLC in Rb1/p53 mice. Cell Rep 30: 3837–3850.e3. 10.1016/j.celrep.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JL, Burton GV, Cox EB, McCarty KS Sr, Dent GA, Geisinger KR, McCarty KS Jr. 1986. Use of monoclonal antiestrogen receptor antibody to evaluate estrogen receptor content in fine needle aspiration breast biopsies. Ann Surg 203: 250–254. 10.1097/00000658-198603000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Bunn PA, Minna JD. 2017. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 17: 725–737. 10.1038/nrc.2017.87 [DOI] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L, Guillemot F. 2019. HES1, two programs: promoting the quiescence and proliferation of adult neural stem cells. Genes Dev 33: 479–481. 10.1101/gad.325761.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, Somerville TDD, Milazzo JP, Wilkinson JE, Demerdash OE, et al. 2018. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 32: 915–928. 10.1101/gad.314815.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins EJ, Kunttas E, Piacentino ML, Howard AGA IV, Bronner ME, Uribe RA. 2018. Migration and diversification of the vagal neural crest. Dev Biol 444 Suppl 1: S98–S109. 10.1016/j.ydbio.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spainhower KB, Conley CC, Chen OS, Guthrie MR, Soltero D, et al. 2020. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell 38: 60–78.e12. 10.1016/j.ccell.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. 2000. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127: 3913–3921. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, et al. 2002. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418: 41–49. 10.1038/nature00870 [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. 2005. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121: 823–835. 10.1016/j.cell.2005.03.032 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. 2011. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One 6: e18472. 10.1371/journal.pone.0018472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764. 10.1016/S0092-8674(00)80258-5 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsimont JC, Youssef KK, Sánchez-Danés A, Sukumaran V, Defrance M, Delatte B, Liagre M, Baatsen P, Marine JC, Lippens S, et al. 2015. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17: 60–73. 10.1016/j.stem.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW. 2006. HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Miner Res 21: 921–933. 10.1359/jbmr.060303 [DOI] [PubMed] [Google Scholar]
- Lian JB, Balint E, Javed A, Drissi H, Vitti R, Quinlan EJ, Zhang L, Van Wijnen AJ, Stein JL, Speck N, et al. 2003. Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. J Cell Physiol 196: 301–311. 10.1002/jcp.10316 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lim JS, Ibaseta A, Fischer MM, Cancilla B, O'Young G, Cristea S, Luca VC, Yang D, Jahchan NS, Hamard C, et al. 2017. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545: 360–364. 10.1038/nature22323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. 2012. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol 13: 27–38. 10.1038/nrm3254 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux N, Gettinger SN, O'Kane G, Arbour KC, Neal JW, Husain H, Evans TL, Brahmer JR, Muzikansky A, Bonomi PD, et al. 2019. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: clinical outcomes. J Clin Oncol 37: 278–285. 10.1200/JCO.18.01585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Masserdotti G, Gillotin S, Sutor B, Drechsel D, Irmler M, Jorgensen HF, Sass S, Theis FJ, Beckers J, Berninger B, et al. 2015. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell 17: 74–88. 10.1016/j.stem.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, Marx A, George J, Heinen C, Chalishazar MD, et al. 2017. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 31: 270–285. 10.1016/j.ccell.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108: 17–29. 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- Oliver TG, Meylan E, Chang GP, Xue W, Burke JR, Humpton TJ, Hubbard D, Bhutkar A, Jacks T. 2011. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol Cell 43: 57–71. 10.1016/j.molcel.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser MG, Niederst MJ, Sequist LV, Engelman JA. 2015. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 16: e165–e172. 10.1016/S1470-2045(14)71180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özşen M, Yalçinkaya U, Akyildiz E, Bayram AS, Gökalp G. 2020. Bronchial carcinoid tumors with massive osseous metaplasia: a case report and review of the literature. Turk Patoloji Derg 36: 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E, Heng J, Azzarelli R, Riou P, Castro D, Lebel-Potter M, Parras C, Bell DM, Ridley AJ, Parsons M, et al. 2011. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron 69: 1069–1084. 10.1016/j.neuron.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JX, Xiong L, Zhao K, Zeng P, Wang B, Tang FL, Sun D, Guo HH, Yang X, Cui S, et al. 2018. YAP promotes osteogenesis and suppresses adipogenic differentiation by regulating β-catenin signaling. Bone Res 6: 18. 10.1038/s41413-018-0018-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Liang MC, Raiser DM, Zamponi R, Roach RR, Curtis SJ, Walton Z, Schaffer BE, Roake CM, Zmoos AF, et al. 2011. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 10: 2806–2815. 10.4161/cc.10.16.17012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, Smith BA, Cheng C, Tsai BL, Cheng D, et al. 2018. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 362: 91–95. 10.1126/science.aat5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier JT, George J, Owonikoko TK, Berns A, Brambilla E, Byers LA, Carbone D, Chen HJ, Christensen CL, Dive C, et al. 2020. New approaches to SCLC therapy: from the laboratory to the clinic. J Thorac Oncol 15: 520–540. 10.1016/j.jtho.2020.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo K, Kollipara RK, Kelenis DP, Rodarte KE, Zhang X, Minna JD, Johnson JE. 2020. Lineage transcription factors co-regulate subtype-specific genes providing a roadmap for systematic identification of small cell lung cancer vulnerabilities. bioRxiv 10.1101/2020.08.13.249029 [DOI] [Google Scholar]
- Qian J, Zhang XY, Gu P, Shao JC, Han BH, Wang HM. 2017. Primary thoracic extraskeletal osteosarcoma: a case report and literature review. J Thorac Dis 9: E1088–E1095. 10.21037/jtd.2017.11.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. 2017. Single-cell mRNA quantification and differential analysis with census. Nat Methods 14: 309–315. 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. 2019. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19: 289–297. 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. 2017. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 14: 549–561. 10.1038/nrclinonc.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Hewitt Coleman J. 2017. Stem and progenitor cells of the mammalian olfactory epithelium: taking poietic license. J Comp Neurol 525: 1034–1054. 10.1002/cne.24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa T, Isotani A, Matsumura T, Nakanishi K, Nonaka S, Shibata S, Ikawa M, Okano H. 2019. Developmental analyses of mouse embryos and adults using a non-overlapping tracing system for all three germ layers. Development 146: dev174938. 10.1242/dev.174938 [DOI] [PubMed] [Google Scholar]
- Smith N, Dong Y, Lian JB, Pratap J, Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O'Keefe RJ, Stein GS, et al. 2005. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J Cell Physiol 203: 133–143. 10.1002/jcp.20210 [DOI] [PubMed] [Google Scholar]
- Song H, Yao E, Lin C, Gacayan R, Chen MH, Chuang PT. 2012. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci 109: 17531–17536. 10.1073/pnas.1207238109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung do Y, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, Speck NA, Drissi H. 2012. Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J Bone Miner Res 27: 1585–1597. 10.1002/jbmr.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. 2011. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19: 754–764. 10.1016/j.ccr.2011.04.019 [DOI] [PubMed] [Google Scholar]
- Takamura K, Ogi T, Yamamoto M, Kikuchi K. 2018. Extraskeletal osteosarcoma of the lung following treatment of primary small-cell lung carcinoma with chemoradiotherapy: a case report. Mol Clin Oncol 8: 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Xie J, Chen W, Tang C, Wu J, Wang Y, Zhou XD, Zhou HD, Li YP. 2020. Runt-related transcription factor 1 is required for murine osteoblast differentiation and bone formation. J Biol Chem 295: 11669–11681. 10.1074/jbc.RA119.007896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry S, Beltran H. 2014. The many faces of neuroendocrine differentiation in prostate cancer progression. Front Oncol 4: 60. 10.3389/fonc.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, Hinds PW. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell 8: 303–316. 10.1016/S1097-2765(01)00327-6 [DOI] [PubMed] [Google Scholar]
- Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. 2007. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 11: 421–429. 10.1016/j.ccr.2007.02.027 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlemsani C, Pongor L, Elloumi F, Girard L, Huffman KE, Roper N, Varma S, Luna A, Rajapakse VN, Sebastian R, et al. 2020. SCLC-CellMiner: a resource for small cell lung cancer cell line genomics and pharmacology based on genomic signatures. Cell Rep 33: 108296. 10.1016/j.celrep.2020.108296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubochi H, Endo S, Oda Y, Dobashi Y. 2013. Carcinoid tumor of the lung with massive ossification: report of a case showing the evidence of osteomimicry and review of the literature. Int J Clin Exp Pathol 6: 957–961. [PMC free article] [PubMed] [Google Scholar]
- Vanyai HK, Prin F, Guillermin O, Marzook B, Boeing S, Howson A, Saunders RE, Snoeks T, Howell M, Mohun TJ, et al. 2020. Control of skeletal morphogenesis by the Hippo-YAP/TAZ pathway. Development 147: dev.187187. 10.1242/dev.187187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velletri T, Xie N, Wang Y, Huang Y, Yang Q, Chen X, Chen Q, Shou P, Gan Y, Cao G, et al. 2016. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis 7: e2015. 10.1038/cddis.2015.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, Kunisaki J, Masood A, Waqar SN, Spies NC, et al. 2018. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 9: 3787. 10.1038/s41467-018-06162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, et al. 2008. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev 22: 1662–1676. 10.1101/gad.1656808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten DJ, Groves SM, Tyson DR, Liu Q, Lim JS, Albert R, Lopez CF, Sage J, Quaranta V. 2019. Systems-level network modeling of small cell lung cancer subtypes identifies master regulators and destabilizers. PLoS Comput Biol 15: e1007343. 10.1371/journal.pcbi.1007343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Denny SK, Greenside PG, Chaikovsky AC, Brady JJ, Ouadah Y, Granja JM, Jahchan NS, Lim JS, Kwok S, et al. 2018. Intertumoral heterogeneity in SCLC is influenced by the cell type of origin. Cancer Discov 8: 1316–1331. 10.1158/2159-8290.CD-17-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp JA, Morrisey EE. 2019. Cellular crosstalk in the development and regeneration of the respiratory system. Nat Rev Mol Cell Biol 20: 551–566. 10.1038/s41580-019-0141-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhou Y, Qian H, Shao G, Lu X, Chen Q, Sun X, Chen D, Yin R, Zhu H, et al. 2013. Stemness and inducing differentiation of small cell lung cancer NCI-H446 cells. Cell Death Dis 4: e633. 10.1038/cddis.2013.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, Ghayee HK, Pacak K, Oliver TG, Minna JD, et al. 2018. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 7: 32–49. 10.21037/tlcr.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software is commercially available or cited in previous publications. Mouse RNA-seq, ChIP-seq, and scRNA-seq data unique to this study are deposited in NCBI GEO: GSE155692. scRNA-seq raw data of n = 4 RPM invasive tumors used in this study were previously deposited in NCBI GEO: GSE149180. All code for scRNA-seq analyses follows standard and publicly available Seurat and Monocle3 workflows. R scripts for this study are available upon reasonable request. Additional raw data for Figures 3–7 are provided in Supplemental Tables S2–S4.